Summary
A population of mesenchymal stem cells, termed CXC chemokine ligand (CXCL)12‐abundant reticular (CAR) cells or leptin receptor‐expressing cells, are the major cellular component of niches for haematopoietic stem cells (HSCs) in murine bone marrow. CAR cells are characterized by several salient features, including much higher expression of CXCL12, stem cell factor (SCF), forkhead box C1 (FOXC1) and early B‐cell factor 3 (EBF3), which are essential for HSC maintenance, than other cells. However, the human counterpart of CAR cells has not been fully described. Here, we show the presence of cells expressing much higher CXCL12 than other cells in human adult bone marrow using a flow cytometry‐based in situ technique that enables high‐throughput detection of mRNA at single‐cell resolution. Most CXCL12hi cells expressed high levels of SCF, FOXC1 and EBF3 and had the potential to differentiate into adipocytes and osteoblasts. Histologically, the nuclei of CXCL12hi cells were identified and quantified by EBF3 expression in fixed marrow sections. CXCL12hi cells sorted from residual bone marrow aspirates of chronic myeloid leukaemia patients expressed reduced levels of CXCL12, SCF, FOXC1 and EBF3 in correlation with increased leukaemic burden. Together, we identified the human counterpart of CAR cells, enabling the evaluation of their alterations in various haematological disorders by flow cytometric and histological analyses.
Keywords: human, haematopoietic stem cells, niche, bone marrow, CXCL12‐abundant reticular cell
Introduction
Most blood cells, including immune cells and leukaemic cells, are generated from haematopoietic stem cells (HSCs) in bone marrow. HSCs are in contact with and maintained by special microenvironments, known as niches, which provide HSCs with critical cytokines, in bone marrow. 1 , 2 , 3 , 4 The identity of HSC niches has been a subject of longstanding debate, but recent studies have demonstrated that a population of bone marrow‐specific mesenchymal stem cells, termed CXC chemokine ligand (CXCL)12‐abundant reticular (CAR) cells, which overlap strongly with leptin receptor‐expressing (Lepr+) cells, are the major component of HSC niches in murine bone marrow. 5 , 6 , 7 , 8 CAR cells have been shown to be specialized mesenchymal stem cells characterized by several salient features, including much higher expression of LEPR and HSC niche factors, such as CXCL12, stem cell factor (SCF), and the transcription factors forkhead box C1 (FOXC1) and early B‐cell factor 3 (EBF3), which are essential for the maintenance of HSCs, than other types of cells. 5 , 6 , 7 , 9 , 10
Alterations of haematopoietic niches might serve as potential diagnostic markers, reliable therapeutic targets and powerful prognostic predictors of haematological disorders. 11 , 12 , 13 , 14 Thus, the specific markers that distinguish mesenchymal stromal cells from other bone marrow cells in humans have been well studied. 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 They are prospectively identified based on the lack of expression of haematopoietic and endothelial markers along with positive expression for CD271 or CD146 in human adult bone marrow. 15 , 16 , 17 , 18 , 19 , 21 However, the human counterpart of CAR cells has not been fully described.
Here, we show the presence of non‐haematopoietic cells which have salient features of CAR cells, including much higher expression of HSC niche factors CXCL12, SCF, FOXC1 and EBF3 than other types of cells, in human adult bone marrow. Histologically, the nuclei of CXCL12hi cells were identified for the first time using anti‐EBF3 antibodies in fixed marrow sections. Furthermore, we showed that CXCL12hi cells from chronic‐phase chronic myeloid leukaemia (CP‐CML) patients expressed reduced levels of HSC niche factors in correlation with increased leukaemic burden.
Materials and methods
Human samples
Bone marrow from femurs or vertebrae and synovial tissues were harvested from patients who underwent orthopaedic surgery. Bone marrow aspirates and biopsies were obtained from patients who underwent bone marrow examination (Table SI). Informed consent was obtained from all participants. This study adhered to the tenets of the Declaration of Helsinki, was approved by the institutional review board of Osaka University Hospital and six related hospitals (HANDAI Clinical Blood Club), and was performed according to the guidelines of the ethical committee of Osaka University Hospital.
Flow cytometry
Crushed bone marrow fragments from femurs or vertebrae, bone marrow aspirates and dissected synovial tissues were digested with collagenase type I (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), and the enzymatically dissociated cells were incubated with red blood cell lysis buffer and then stained with fluorochrome‐conjugated antibodies. The following monoclonal antibodies were purchased from BD Biosciences (Franklin Lakes, NJ, USA), BioLegend (San Diego, CA, USA) or eBioscience (Thermo Fisher Scientific), unless otherwise indicated: anti‐CD31 (WM59), anti‐CD34 (581), anti‐CD45 (HI30), anti‐CD56 (NCAM16.2), anti‐CD71 (CY1G4, OKT9), anti‐CD140a (αR1), anti‐CD146 (P1H12), anti‐CD235a (HIR2), anti‐CD271 (ME20.4), anti‐LEPR (52263; R&D Systems, Bio‐Techne, Minneapolis, MN, USA) and anti‐podoplanin (PDPN; NZ‐1.3). CXCL12hi/LEPR+ cells, osteoblastic cells and bone marrow endothelial cells (ECs) were defined as LEPR+CD45−CD235a−CD71−CD31− cells, LEPR−CD56+CD45−CD235a−CD71−CD31− cells, 23 and CD45−CD235a−CD71−CD31+CD34+ cells, 24 , 25 respectively. Synovial mesenchymal cells were isolated as CD45−CD235a−CD71−CD31−CD146−PDPN+ cells. 26 Dead cells were excluded by staining with propidium iodide. All flow cytometric experiments were performed using a BD FACS Aria IIu (BD Biosciences).
Primeflow RNA assay
In situ hybridization to detect the mRNAs encoding human CXCL12, SCF, FOXC1 and EBF3 with flow cytometry was performed using probes and reagents supplied with the PrimeFlow RNA Assay kit (Thermo Fisher Scientific), according to the manufacturer’s instructions. Briefly, bone marrow cells stained with antibodies targeting the cell surface markers were fixed with the first fixation buffer for 30 min at 4°C, and then washed with RNA permeabilization buffer. Subsequently, the cells were fixed with the second fixation buffer for 60 min at room temperature, and then incubated with target‐specific and control probes at 40°C for 2 h. The probes were amplified and labelled, and cells were subsequently analyzed by flow cytometry. Target probes used were as follows: CXCL12 (VA1‐11035, VA4‐3082401), SCF (VA1‐12982), FOXC1 (VA1‐12673) and EBF3 (VA1‐3015971). Probes for β‐actin (VA1‐10351, VA4‐10293) were used as positive controls to check that cells were permeabilized and probes hybridized and amplified.
qRT‐PCR analysis
Relative mRNA expression was analyzed by a quantitative real‐time (qRT)‐polymerase chain reaction (PCR) analysis performed with the StepOnePlus System (Applied Biosystems, Thermo Fisher Scientific; TOYOBO, Osaka, Japan) and THUNDERBIRD SYBR qPCR Mix (TOYOBO). Total RNA was isolated from sorted or cultured cells using Isogen (Nippon Gene, Tokyo, Japan), and cDNA was synthesized using SuperScript VILO Master Mix (Invitrogen, Thermo Fisher Scientific), according to the manufacturers’ instructions. The expression level of each mRNA was normalized to that of the GAPDH mRNA in each sample. The primers used for PCR are listed in Table SII.
In vitro differentiation assay
Sorted CXCL12hi/LEPR+ cells, LEPR−CD56+ osteoblastic cells and synovial fibroblasts were plated at a density of 1·5 × 102 cells/cm2 and were maintained in mesenchymal stem cell (MSC) expansion medium (130‐104‐182; Miltenyi Biotec, Bergisch Gladbach, Germany) supplemented with 1% penicillin/streptomycin (Nacalai Tesque, Kyoto, Japan) at 37°C in a humidified atmosphere with 5% CO2. For adipogenic differentiation, primary cultured cells were changed to AdipoDiff medium (Miltenyi Biotec) when they reached 80% confluency. After 21 days of differentiation, the cells were washed, fixed in 4% paraformaldehyde and stained with Oil Red O to evaluate lipid droplets. To assess osteogenic differentiation, primary cultured cells at 80% confluency were maintained in OsteoDiff medium (Miltenyi Biotec) for 21 days, fixed with 4% paraformaldehyde and stained with Alizarin Red to detect calcium deposits. To induce chondrogenic differentiation, cell pellets prepared from 2·5 × 105 cells by centrifugation were cultured in ChondroDiff medium (Miltenyi Biotec). After 25 days, the pellets were fixed with 4% paraformaldehyde, embedded in Optimal Cutting Temperature medium (Sakura Finetek, Osaka, Japan) and sectioned. Cryosections were stained with Alcian blue (Fuji Film Wako Pure Chemical, Osaka, Japan).
For differentiation assays of single‐CXCL12hi/LEPR+‐cell‐derived clones, sorted CXCL12hi/LEPR+ cells were seeded into 96‐well plates at a density of three cells per well and were cultured in MSC expansion medium. Colonies were counted at day 7 to exclude the wells with more than one colony from further analysis. When confluent, the cells were differentiated toward the adipogenic or osteogenic lineage as described above.
CFU–F assay
Colony‐forming unit–fibroblast (CFU–F) assays were performed as described previously. 18 Sorted cells were plated at a density of 3–12 cells/cm2 in α‐MEM (Gibco) supplemented with 20% fetal calf serum (FCS) and 1% penicillin/streptomycin. The cultures were incubated at 37°C in a humidified atmosphere with 5% CO2. Colonies were counted after 14 days of culture.
Co‐culture of cord blood CD34+ cells with CXCL12Hi/LEPR+ cells
Sorted human CXCL12hi/LEPR+ cells were seeded at a density of 1·5 × 102 cells/cm2 into 96‐well plates and were cultured in MSC expansion medium for 10 days. Then medium was removed, and 5 000 human cord blood CD34+ cells (RIKEN BRC, Tsukuba, Japan) were co‐cultured with or without the adherent CXCL12hi/LEPR+ cells in StemSpan Serum‐Free Expansion Medium (StemCell Technologies, Vancouver, BC, Canada) supplemented with 25 ng/ml of recombinant human SCF (Fuji Film Wako Pure Chemical), thrombopoietin (TPO; Fuji Film Wako Pure Chemical) and fms‐related receptor tyrosine kinase 3 ligand (FLT3L; Fuji Film Wako Pure Chemical), and 1% penicillin/streptomycin at 37°C with 5% CO2. After one week of co‐culture, cells were harvested and the numbers of CD34+ cells were counted using flow cytometry.
Immunohistochemistry
Human bone marrow samples were fixed in 10% neutral buffered formalin, decalcified with EDTA and embedded in paraffin. Sections (6 µm) were generated via Kawamoto’s film method (Section‐Lab, Hiroshima, Japan) and used for immunohistochemical analysis. In brief, after deparaffinization and hydration, sections were boiled in citrate buffer (pH 6·0), incubated with blocking buffer (X0909, DAKO, Agilent, Santa Clara, CA, USA), and then stained with a primary antibody, followed by incubation with horseradish peroxidase (HRP)‐conjugated secondary antibodies. For detection, tyramide signal amplification (Tyramide SuperBoost Kit, Thermo Fisher Scientific) was used according to the manufacturer’s instructions. For multiplex staining with primary antibodies from the same species, the primary and secondary antibodies bound to the sections were stripped using citrate buffer (pH 6·0) in a microwave before staining with another primary antibody. Primary antibodies used were as follows: mouse anti‐CD31 (JC70A, DAKO), rabbit anti‐CD56 (MRQ‐42, Cell Marque, Rocklin, CA, USA), rabbit anti‐CD271 (HPA004765, Sigma‐Aldrich, St. Louis, MO, USA), rabbit anti‐EBF3 (ab207705, Abcam, Cambridge, UK) and mouse anti‐ runt‐related transcription factor 2 (RUNX2) (8G5, MBL International, Woburn, MA, USA). The nuclei of cells were stained with 4′,6‐diamidino‐2‐phenylindole (DAPI) dye. Confocal tiled Z‐stack images were acquired using a Zeiss LSM 510 META microscope equipped with a Plan‐Apochromat 20×/0·8 objective and a Plan‐Apochromat 63×/1·4 oil objective, and analyzed using Zeiss ZEN (Carl Zeiss, Oberkochen, Germany) and Bitplane Imaris 8.3.1 (Bitplane, Zurich, Switzerland) software.
RNAscope in situ hybridization combined with immunohistochemistry
Sections of formalin‐fixed, paraffin‐embedded decalcified bone marrow biopsies (5 µm) were processed for RNA in situ hybridization and immunohistochemistry using the RNAscope Multiplex Fluorescent Reagent Kit v2 (Advanced Cell Diagnostics, Newark, CA, USA) with the Opal dyes (PerkinElmer, Waltham, MA, USA) according to the manufacturer’s instructions. 27 RNAscope probes used were as follows: CXCL12 (4422991‐C2) and LEPR (410381). Antibodies used were as follows: rabbit anti‐EBF3 (ab207705; Abcam), rabbit anti‐von Willebrand factor (VWF) (D8L8G, Cell Signaling Technology, Danvers, MA, USA) and HRP‐conjugated goat anti‐rabbit IgG (Thermo Fisher Scientific). The nuclei of cells were labelled with DAPI dye. Confocal tiled Z‐stack images were acquired using a Zeiss LSM880 microscope and a Plan‐Apochromat 63×/1·4 oil objective, and analyzed using Zeiss ZEN software.
Results
A population of non‐haematopoietic cells express markedly high levels of CXCL12 as well as SCF, FOXC1 and EBF3 in human adult bone marrow
We evaluated the expression levels of the mRNAs encoding SCF (KITLG), FOXC1 and EBF3 in each bone marrow cell using a flow cytometry‐based in situ technique capable of simultaneous detection of mRNAs and proteins within millions of cells at single‐cell resolution. We identified a population of non‐haematopoietic cells expressing much higher levels of the CXCL12 mRNA than other bone marrow cells in human adult bone marrow (Fig 1A; Figure S1). These CXCL12hi cells expressed high levels of LEPR, and LEPR+ non‐haematopoietic cells expressed markedly high levels of the CXCL12 mRNA (Fig 1A), indicating that CXCL12hi cells overlap strongly with LEPR+ non‐haematopoietic cells in human bone marrow, as reported for murine CAR cells. 7 In addition, CXCL12hi cells uniformly expressed the KITLG, FOXC1 and EBF3 mRNAs (Fig 1B). By contrast, the CXCL12 mRNA was absent or present at very low levels in CD56+ osteoblastic cells 23 and in bone marrow CD31+CD34+ ECs 24 , 25 (Fig 1C; Figure S2A).
Fig 1.
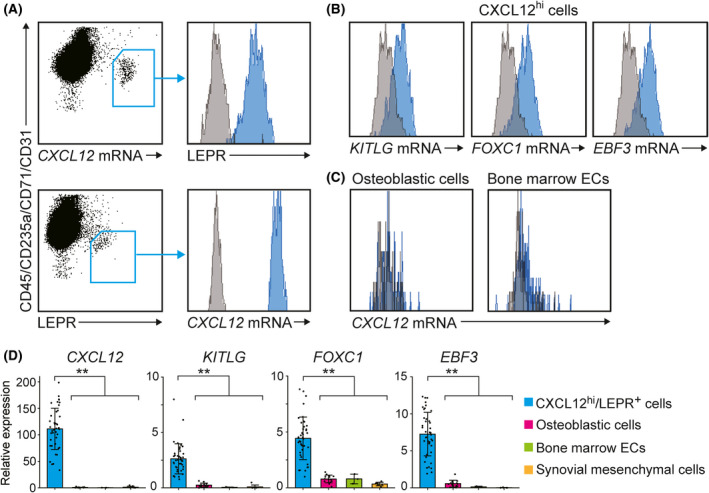
A population of non‐haematopoietic cells express much higher levels of CXCL12, SCF, FOXC1 and EBF3 that other cells in human adult bone marrow. (A–C) Expression of the CXCL12 (A, C), KITLG (B), FOXC1 (B) and EBF3 (B) mRNAs in human adult bone marrow cells as determined by flow cytometry using the PrimeFlow RNA Assay kit. A population of non‐haematopoietic cells expressed markedly high levels of the CXCL12 mRNA, and these CXCL12hi cells expressed high levels of LEPR (A, upper). LEPR+ non‐haematopoietic cells expressed markedly high levels of the CXCL12 mRNA (A, lower). CXCL12hi cells uniformly expressed the KITLG, FOXC1 and EBF3 mRNAs (B). The CXCL12 mRNA was absent or present at very low levels in CD56+ osteoblastic cells and bone marrow CD31+CD34+ ECs (c). The blue histograms represent the target‐specific probes or antibodies, and the gray histograms represent background fluorescence (A–C). (D) Relative expression levels of the CXCL12, KITLG, FOXC1 and EBF3 mRNAs in CXCL12hi/LEPR+ cells (n = 42), osteoblastic cells (n = 14) and endothelial cells (ECs) (n = 3) isolated from human adult bone marrow, as well as human adult synovial mesenchymal cells (n = 8), as determined by quantitative real‐time (qRT)‐polymerase chain reaction (PCR). Data represent mean ± SD (D). Two‐tailed Student’s t tests were used to assess statistical significance (D; **P < 0·01).
qRT‐PCR analysis revealed that CXCL12hi cells that were sorted based on LEPR expression (CXCL12hi/LEPR+ cells) expressed much higher levels of the CXCL12, KITLG, FOXC1 and EBF3 mRNAs than osteoblastic cells, bone marrow ECs and synovial mesenchymal cells (Fig 1D; Figure S2B). The expression levels of the CXCL12, KITLG, FOXC1 and EBF3 mRNAs were similar in CXCL12hi/LEPR+ cells from femurs, vertebrae and iliac crests (Figure S3A). In addition, donor age did not influence the expression levels of the CXCL12, KITLG, FOXC1 and EBF3 mRNAs in CXCL12hi/LEPR+ cells (Figure S3B).
Human adult bone marrow CXCL12Hi/LEPR+ cells are adipo‐osteogenic progenitors with haematopoiesis‐supporting abilities
It was reported previously that the cell surface proteins CD56, CD271, CD140a and CD146 are expressed by a subset of mesenchymal progenitors in human adult bone marrow. 17 , 18 , 22 , 23 A flow cytometric analysis showed that CXCL12hi/LEPR+ cells did not express CD56 (Fig 2A). Both CXCL12hi/LEPR+ cells and LEPR−CD56+ osteoblastic cells had high expression levels of CD271 and low expression levels of CD140a, whereas synovial mesenchymal cells had low expression levels of CD271 and high expression levels of CD140a (Fig 2A). CXCL12hi/LEPR+ cells were heterogenous for CD146 (Fig 2A).
Fig 2.
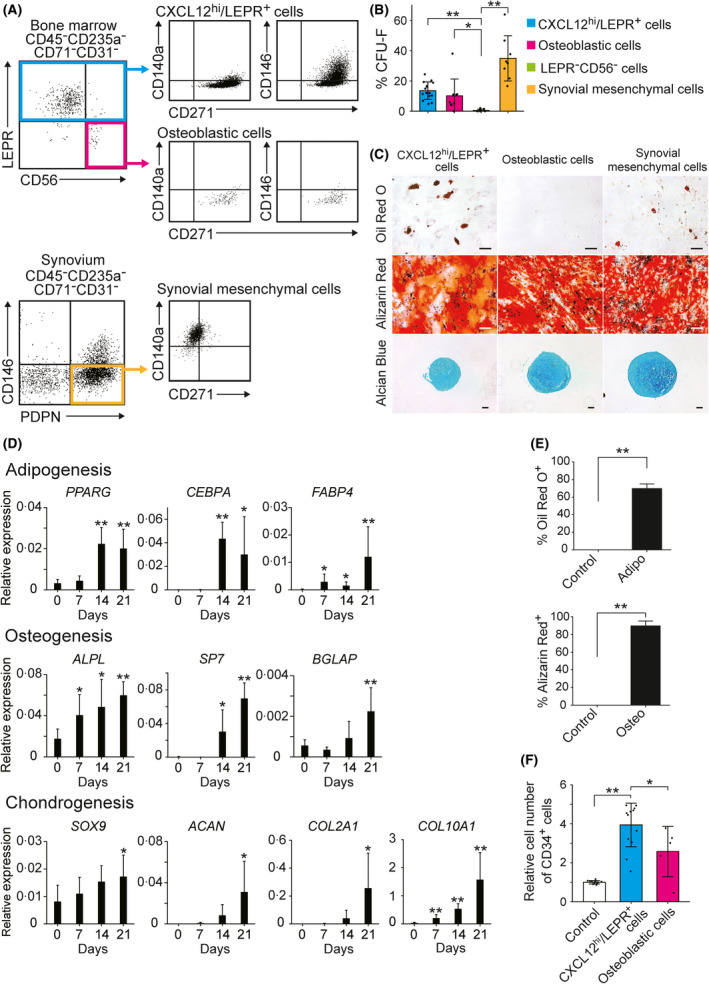
Human adult bone marrow CXCL12hi/LEPR+ cells are adipo‐osteogenic progenitors with haematopoiesis‐supporting abilities. (A) Expression of CD271, CD140a and CD146 in CXCL12hi/LEPR+ cells and osteoblastic cells from human adult bone marrow, as well as synovial mesenchymal cells, as determined by flow cytometry. (B) The colony‐forming unit–fibroblast (CFU–F) frequencies of CXCL12hi/LEPR+ cells (n = 15), LEPR−CD56+ osteoblastic cells (n = 9) and LEPR−CD56− cells (n = 8) within the human adult bone marrow CD45−CD235a−CD71−CD31− population, as well as synovial mesenchymal cells (n = 8). (C) The differentiation potentials of CXCL12hi/LEPR+ cells, LEPR−CD56+ osteoblastic cells and synovial mesenchymal cells towards adipogenic (Oil Red O+), osteogenic (Alizarin Red+) and chondrogenic (Alcian blue+) lineages. Scale bar, 100 µm. (D) Relative expression levels of the mRNAs encoding lineage‐associated markers in CXCL12hi/LEPR+ cells cultured in adipogenic, osteogenic, chondrogenic media, as determined by quantitative real‐time (qRT)‐polymerase chain reaction (PCR; n = 4–6). (E) The percentages of Oil Red O+ (left) or Alizarin Red+ (right) single‐CXCL12hi/LEPR+‐cell‐derived clones cultured in adipogenic (Adipo) or osteogenic (Osteo) medium respectively (n = 3). (F) The numbers of CD34+ haematopoietic cells after one week of co‐culture with CXCL12hi/LEPR+ cells (n = 13) or LEPR−CD56+ osteoblastic cells (n = 5) in serum‐free media containing stem cell factor (SCF), thrombopoietin (TPO) and FMS‐like tyrosine kinase 3 ligand (FLT3L). Data represent mean ± SD (B, D, E and F). Two‐tailed Student’s t tests were used to assess statistical significance (B, D, E and F; *, P < 0·05; **, P < 0·01).
We assessed the CFU–F activities of subpopulations of CD45−CD235a−CD71−CD31− cells in human adult bone marrow; 13% of CXCL12hi/LEPR+ cells and 9% of LEPR−CD56+ osteoblastic cells formed CFU–Fs (Fig 2B). CXCL12hi/LEPR+ cells but not LEPR−CD56+ osteoblastic cells exhibited differentiation potentials toward adipogenic, osteogenic and chondrogenic lineages (Fig 2C). qRT‐PCR analysis showed that the expression levels of lineage‐associated markers gradually increased in CXCL12hi/LEPR+ cells cultured in adipogenic, osteogenic and chondrogenic media (Fig 2D). We analyzed the differentiation potential of individual CXCL12hi/LEPR+ cells, and showed that 70% and 90% of the primary cultured single‐CXCL12hi/LEPR+‐cell‐derived clones differentiated into adipogenic and osteogenic lineages respectively (Fig 2E), suggesting that most CXCL12hi/LEPR+ cells are adipo‐osteogenic bipotential progenitors. In contrast, numbers of cells obtained from single‐CXCL12hi/LEPR+‐cell‐derived clones were not enough to evaluate the chondrogenic differentiation potential.
Subsequently, we assessed the haematopoiesis‐supporting capacity of CXCL12hi/LEPR+ cells and LEPR−CD56+ osteoblastic cells. For this, we cultured cord blood CD34+ cells with CXCL12hi/LEPR+ cells or LEPR−CD56+ osteoblastic cells in serum‐free media containing SCF, TPO and FLT3L. CXCL12hi/LEPR+ cells enhanced the proliferation of CD34+ haematopoietic stem and progenitor cells more effectively than LEPR−CD56+ osteoblastic cells (Fig 2F).
CXCL12Hi cells are identified by EBF3 staining in human adult marrow sections
We tried to identify CXCL12hi cells in fixed human bone marrow sections using an antibody against EBF3, and found that EBF3+ cells were scattered throughout bone marrow (Fig 3A). All EBF3+ cells stained positively with an antibody against CD271 (Fig 3B). Conversely, almost all DAPI+ nuclei of CD271+ cells were positive for EBF3 in the marrow cavity (Fig 3C). EBF3+CD271+ cells had several long processes that formed a reticular network (Fig 3B), as reported for murine CAR cells. 5 CD56+CD271+ cells were bone‐lining osteoblastic cells positive for RUNX2 (Figure S4A). EBF3 was not expressed in bone‐lining osteoblastic cells (Figure S4B) or morphologically identifiable CD31+ ECs (Figure S5). To examine the CXCL12 and LEPR mRNA expression in EBF3+ cells, we performed in situ hybridization of the CXCL12 and LEPR mRNAs combined with immunohistochemistry for EBF3 in human bone marrow sections. EBF3+ cells strongly overlapped with cells expressing the CXCL12 mRNA and cells expressing the LEPR mRNA (Fig 3D). In situ hybridization of the CXCL12 and LEPR mRNAs combined with immunohistochemistry of the pan‐EC marker VWF 28 revealed that CXCL12hi cells were distinct from ECs and approximately 46% of CXCL12hi cells were in contact with ECs of vascular sinuses (Fig 3E).
Fig 3.
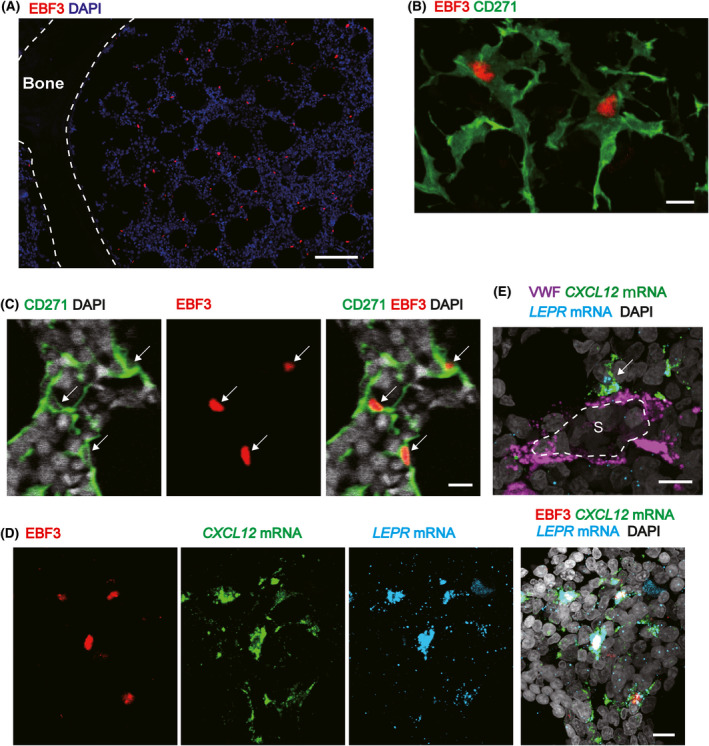
CXCL12hi cells are identified by EBF3 staining in human adult marrow sections. (A) Immunohistochemical analysis for EBF3 in human adult bone marrow. EBF3+ cells were scattered throughout the bone marrow cavity. (B) Immunohistochemical analysis for EBF3 and CD271 in human adult bone marrow. Membranes of EBF3+ cells were positive for CD271, and EBF3+CD271+ cells displayed long processes that formed a reticular network. (C) Immunohistochemical analysis for EBF3 and CD271 in human adult bone marrow. Almost all DAPI+ nuclei of CD271+ cells (white arrows) were positive for EBF3 in the marrow cavity. (D) Combined in situ hybridization of the CXCL12 and LEPR mRNAs and immunohistochemistry for EBF3 in human adult bone marrow. (E) Combined in situ hybridization of the CXCL12 and LEPR mRNAs and immunohistochemistry for von Willebrand factor (VWF) in human adult bone marrow. Cells expressing CXCL12 (white arrow) in contact with endothelial cells (ECs) of vascular sinuses (S) were shown. Scale bar: (A) 100 µm; (B) 10 µm; (C) 10 µm; (D) 10 µm; (E) 10 µm.
CXCL12Hi/LEPR+ cells from CP‐CML patients express reduced levels of CXCL12, SCF, FOXC1 and EBF3 in correlation with increased leukaemic burden
Finally, we focused on CP‐CML because a deeper understanding of niche‐dependent regulation of leukaemic stem cells is required to eradicate the disease. 11 , 14 , 29 We were able to sort thousands of human CXCL12hi/LEPR+ cells from residual clinical bone marrow aspiration samples by flow cytometry. A qRT‐PCR analysis revealed that CXCL12hi/LEPR+ cells from newly diagnosed CP‐CML patients expressed significantly lower levels of the CXCL12, KITLG, FOXC1 and EBF3 mRNAs than those from control patients (Fig 4A). Notably, the expression levels of the CXCL12, KITLG, FOXC1 and EBF3 mRNAs in CXCL12hi/LEPR+ cells from newly diagnosed CP‐CML patients were negatively correlated with the peripheral blood white blood cell (WBC) counts (Fig 4B). The expression levels of the CXCL12, KITLG, FOXC1 and EBF3 mRNAs in CXCL12hi/LEPR+ cells from CP‐CML patients were restored to normal levels when they achieved complete cytogenetic response (CCyR) upon tyrosine kinase inhibitor (TKI) therapy (Fig 4A). These data are consistent with existing murine data, showing that the expression of CXCL12 in whole bone marrow cells was reduced in CML model mice. 30 Flow cytometric analysis and histological analysis revealed that the frequency of CXCL12hi/LEPR+ cells and the density of EBF3+ cells were not significantly different between control patients and patients with myeloproliferative neoplasms (MPNs) (Figure S6).
Fig 4.
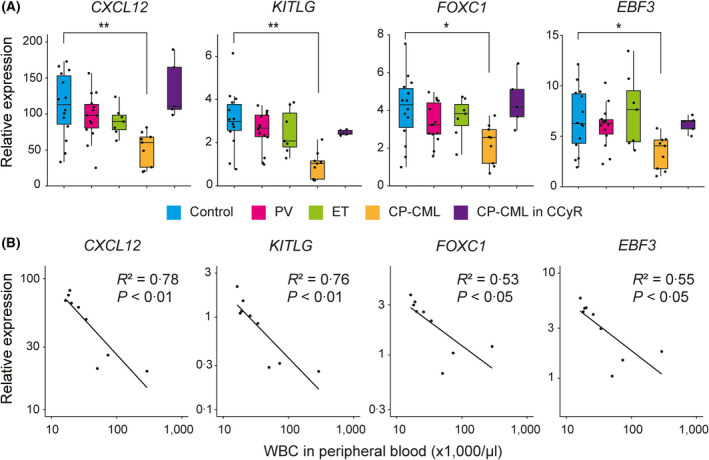
Relative expression levels of CXCL12, KITLG, FOXC1 and EBF3 mRNAs are reduced in human CXCL12hi/LEPR+ cells in correlation with increased CML burden. (A) Relative expression levels of CXCL12, KITLG, FOXC1 and EBF3 mRNAs in CXCL12hi/LEPR+ cells from control patients (n = 14), newly diagnosed polycythaemia vera (PV) patients (n = 14), newly diagnosed essential thrombocythaemia (ET) patients (n = 7), newly diagnosed chronic‐phase chronic myeloid leukaemia (CP‐CML) patients (n = 9) and CP‐CML patients in complete cytogenetic response (CCyR; n = 5), as determined by quantitative real‐time (qRT)‐polymerase chain reaction (PCR). (B) Negative correlations of CXCL12, KITLG, FOXC1 and EBF3 mRNA expression levels in CXCL12hi/LEPR+ cells with peripheral blood white blood cell (WBC) counts in newly diagnosed CP‐CML patients (n = 9). MannºWhitney U tests were used to assess statistical significance (A; *, P < 0·05; **, P < 0·01). Pearson correlation coefficients were used to measure the statistical relationship between the two continuous variables (B).
Discussion
In this study, we identified a population of non‐haematopoietic cells expressing much higher levels of CXCL12, which is involved in HSC behaviour in both mouse and human, 31 , 32 than other bone marrow cells in human adult bone marrow. Most CXCL12hi cells expressed high levels of SCF, FOXC1 and EBF3 and had the potential to differentiate toward adipogenic and osteogenic lineages. These results strongly suggest that CXCL12hi cells are the human counterpart of CAR cells. Prior studies revealed that CD271+ cells expressed the CXCL12 mRNA in human bone marrow; however, it remained unclear whether CD271+ cells expressed much higher levels of key HSC niche factors, including CXCL12, SCF, FOXC1 and EBF3, than other bone marrow cells. 22 , 33 Thus, this study has substantially advanced our understanding of human haematopoietic microenvironments.
Using flow cytometry, we were able to sort thousands of human CXCL12hi cells from residual clinical bone marrow aspiration samples based on LEPR expression. Although CD271 has been used as a marker for bone marrow MSCs in flow cytometric analysis, 18 , 22 CD271+ cells contain osteoblastic cells as well as CXCL12hi cells. Since the MPN development might cause expansion of osteoblastic cells in bone marrow, 34 it is important to discriminate CXCL12hi cells from osteoblastic cells by flow cytometry for the analysis of human CXCL12hi cells in various haematological disorders. These advances will aid the analysis of human CXCL12hi cells in various haematological disorders. 11 , 12 , 13 , 14
In addition, we histologically identified EBF3+ cells in fixed human bone marrow sections. Previously, immunohistochemistry with an anti‐CD271 antibody was the only way to identify mesenchymal stromal cells in fixed human bone marrow sections; 19 however, it was difficult to quantify the number and the density of CD271+ cells in bone marrow because an anti‐CD271 antibody did not stain the nuclei of CD271+ cells. We found that the nuclei of CXCL12hi cells were identified based on EBF3 expression in marrow sections by in situ hybridization of the CXCL12 and LEPR mRNAs combined with immunohistochemistry for EBF3. These findings make it possible for us to easily quantify the number and the density of human CAR cells in fixed bone marrow sections from healthy donors and patients with various haematological disorders.
We found that human CXCL12hi cells showed reduced expression levels of the mRNAs encoding CXCL12 and SCF as well as FOXC1 and EBF3 in correlation with increased leukaemic burden, suggesting that CAR cells from CP‐CML patients have the reduced ability to support normal HSCs. Furthermore, the expression levels of these HSC niche factors in CXCL12hi cells from CP‐CML patients were restored to normal levels when they achieved CCyR upon TKI therapy. These results suggest that TKIs depleted leukaemic cells which affected CXCL12hi cells in CP‐CML patients. However, considering that the platelet‐derived growth factor (PDGF)–PDGFRα axis affects CAR cells, 35 TKIs might have direct effects on PDGFR signalling in CXCL12hi cells in CP‐CML patients.
In summary, we identified the human counterpart of CAR cells and found that CAR cells from CP‐CML patients expressed reduced levels of HSC niche factors in correlation with increased leukaemic burden. This work enables the evaluation of human HSC niches by flow cytometric and histological analyses. Future studies of human CAR cells as well as ECs and osteoblasts in bone marrow aspiration samples and bone marrow sections from patients with various haematological disorders would be important to know how alterations of bone marrow microenvironments serve as potential diagnostic markers, reliable therapeutic targets and powerful prognostic predictors of haematological disorders.
Author contributions
KA, TS, YK and TN conceived the project; KA, MI, TS and TN designed the experiment; KA, KH, MK and TS performed the experiments, analyzed the results and produced the figures; MI, HS, TK, WA, TS and NS provided the study materials; KA, KH, MK, TS and TN wrote the first draft of the manuscript; all the authors approved the final version of the manuscript.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Supporting information
Table SI. Patient characteristics.
Table SII. Primers for qRT–PCR.
Fig S1. Fluorescence signals after hybridization with random probes were not detected in human bone marrow cells.
Fig S2. Osteoblastic cells, bone marrow endothelial cells (ECs), and synovial mesenchymal cells expressed higher levels of the BGLAP, CDH5 and PDPN mRNAs, respectively, than CXCL12hi/LEPR+ cells.
Fig S3. Harvest site and donor age do not influence the expression of the CXCL12, KITLG, FOXC1 or EBF3 mRNAs in human adult bone marrow CXCL12hi/LEPR+ cells.
Fig S4. EBF3 was not expressed in CD56+CD271+ bone‐lining osteoblastic cells.
Fig S5. EBF3 was not expressed in morphologically identifiable CD31+ endothelial cells (ECs) in human adult bone marrow.
Fig S6. The frequency and the density of CXCL12hi cells were similar in bone marrow from control patients and patients with myeloproliferative neoplasms (MPNs).
Acknowledgements
This work was supported by a Grant‐in‐Aid for Scientific Research (KAKENHI) from the Japan Society for the Promotion of Science [19K08814 (MI), 19K16585 (MK), 19K08837 (TS) and 18H03998 (TN)] and Project MEET from Osaka University Graduate School of Medicine and Mitsubishi Tanabe Pharma Corporation (MK). The authors would like to thank patients and families for participating in the study, J. Ishikawa, S. Kosugi, H. Mitsui, M. Kawakami, M. Nakagawa and S. Ueda for providing the study materials, Y. Takashima and H. Shi for technical assistance, and A. Okada, R. Okuyama and M. Ashida for secretarial assistance. Cord blood CD34+ cells were provided by the RIKEN BRC through the National BioResource Project of the MEXT/AMED, Japan.
References
- 1. Mercier FE, Ragu C, Scadden DT. The bone marrow at the crossroads of blood and immunity. Nat Rev Immunol. 2011;12(1):49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505(7483):327–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kumar S, Geiger H. HSC niche biology and HSC expansion ex vivo. Trends Mol Med. 2017;23(9):799–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pinho S, Frenette PS. Haematopoietic stem cell activity and interactions with the niche. Nat Rev Mol Cell Biol. 2019;20(5):303–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12‐CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25(6):977–88. [DOI] [PubMed] [Google Scholar]
- 6. Omatsu Y, Sugiyama T, Kohara H, Kondoh G, Fujii N, Kohno K, et al. The essential functions of adipo‐osteogenic progenitors as the hematopoietic stem and progenitor cell niche. Immunity. 2010;33(3):387–99. [DOI] [PubMed] [Google Scholar]
- 7. Ding L, Saunders TL, Enikolopov G, Morrison SJ. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481(7382):457–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou BO, Yue R, Murphy MM, Peyer JG, Morrison SJ. Leptin‐receptor‐expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 2014;15(2):154–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Omatsu Y, Seike M, Sugiyama T, Kume T, Nagasawa T. Foxc1 is a critical regulator of haematopoietic stem/progenitor cell niche formation. Nature. 2014;508(7497):536–40. [DOI] [PubMed] [Google Scholar]
- 10. Seike M, Omatsu Y, Watanabe H, Kondoh G, Nagasawa T. Stem cell niche‐specific Ebf3 maintains the bone marrow cavity. Genes Dev. 2018;32(5–6):359–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yao JC, Link DC. Concise review: the malignant hematopoietic stem cell niche. Stem Cells. 2017;35(1):3–8. [DOI] [PubMed] [Google Scholar]
- 12. Pronk E, Raaijmakers M. The mesenchymal niche in MDS. Blood. 2019;133(10):1031–8. [DOI] [PubMed] [Google Scholar]
- 13. Ladikou EE, Chevassut T, Pepper CJ, Pepper AG. Dissecting the role of the CXCL12/CXCR4 axis in acute myeloid leukaemia. Br J Haematol. 2020;189(5):815–25. [DOI] [PubMed] [Google Scholar]
- 14. Méndez‐Ferrer S, Bonnet D, Steensma DP, Hasserjian RP, Ghobrial IM, Gribben JG, et al. Bone marrow niches in haematological malignancies. Nat Rev Cancer. 2020;20(5):285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cattoretti G, Schiro R, Orazi A, Soligo D, Colombo MP. Bone marrow stroma in humans: anti‐nerve growth factor receptor antibodies selectively stain reticular cells in vivo and in vitro. Blood. 1993;81(7):1726–38. [PubMed] [Google Scholar]
- 16. Quirici N, Soligo D, Bossolasco P, Servida F, Lumini C, Deliliers GL. Isolation of bone marrow mesenchymal stem cells by anti‐nerve growth factor receptor antibodies. Exp Hematol. 2002;30(7):783–91. [DOI] [PubMed] [Google Scholar]
- 17. Sacchetti B, Funari A, Michienzi S, Di Cesare S, Piersanti S, Saggio I, et al. Self‐renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131(2):324–36. [DOI] [PubMed] [Google Scholar]
- 18. Tormin A, Li O, Brune JC, Walsh S, Schutz B, Ehinger M, et al. CD146 expression on primary nonhematopoietic bone marrow stem cells is correlated with in situ localization. Blood. 2011;117(19):5067–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Flores‐Figueroa E, Varma S, Montgomery K, Greenberg PL, Gratzinger D. Distinctive contact between CD34+ hematopoietic progenitors and CXCL12+ CD271+ mesenchymal stromal cells in benign and myelodysplastic bone marrow. Lab Invest. 2012;92(9):1330–41. [DOI] [PubMed] [Google Scholar]
- 20. Pinho S, Lacombe J, Hanoun M, Mizoguchi T, Bruns I, Kunisaki Y, et al. PDGFRalpha and CD51 mark human nestin+ sphere‐forming mesenchymal stem cells capable of hematopoietic progenitor cell expansion. J Exp Med. 2013;210(7):1351–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johnson RC, Kurzer JH, Greenberg PL, Gratzinger D. Mesenchymal stromal cell density is increased in higher grade myelodysplastic syndromes and independently predicts survival. Am J Clin Pathol. 2014;142(6):795–802. [DOI] [PubMed] [Google Scholar]
- 22. Li H, Ghazanfari R, Zacharaki D, Ditzel N, Isern J, Ekblom M, et al. Low/negative expression of PDGFR‐alpha identifies the candidate primary mesenchymal stromal cells in adult human bone marrow. Stem Cell Rep. 2014;3(6):965–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Battula VL, Treml S, Bareiss PM, Gieseke F, Roelofs H, de Zwart P, et al. Isolation of functionally distinct mesenchymal stem cell subsets using antibodies against CD56, CD271, and mesenchymal stem cell antigen‐1. Haematologica. 2009;94(2):173–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Newman PJ, Berndt MC, Gorski J, White GC 2nd, Lyman S, Paddock C, et al. PECAM‐1 (CD31) cloning and relation to adhesion molecules of the immunoglobulin gene superfamily. Science. 1990;247(4947):1219–22. [DOI] [PubMed] [Google Scholar]
- 25. Soligo D, Delia D, Oriani A, Cattoretti G, Orazi A, Bertolli V, et al. Identification of CD34+ cells in normal and pathological bone marrow biopsies by QBEND10 monoclonal antibody. Leukemia. 1991;5(12):1026–30. [PubMed] [Google Scholar]
- 26. Mizoguchi F, Slowikowski K, Wei K, Marshall JL, Rao DA, Chang SK, et al. Functionally distinct disease‐associated fibroblast subsets in rheumatoid arthritis. Nat Commun. 2018;9(1):789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, et al. RNAscope: a novel in situ RNA analysis platform for formalin‐fixed, paraffin‐embedded tissues. J Mol Diagn. 2012;14(1):22–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pusztaszeri MP, Seelentag W, Bosman FT. Immunohistochemical expression of endothelial markers CD31, CD34, von Willebrand factor, and Fli‐1 in normal human tissues. J Histochem Cytochem. 2006;54(4):385–95. [DOI] [PubMed] [Google Scholar]
- 29. Agarwal P, Isringhausen S, Li H, Paterson AJ, He J, Gomariz A, et al. Mesenchymal niche‐specific expression of Cxcl12 controls quiescence of treatment‐resistant leukemia stem cells. Cell Stem Cell. 2019;24(5):769–84 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang B, Ho YW, Huang Q, Maeda T, Lin A, Lee SU, et al. Altered microenvironmental regulation of leukemic and normal stem cells in chronic myelogenous leukemia. Cancer Cell. 2012;21(4):577–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Broxmeyer HE, Orschell CM, Clapp DW, Hangoc G, Cooper S, Plett PA, et al. Rapid mobilization of murine and human hematopoietic stem and progenitor cells with AMD3100, a CXCR4 antagonist. J Exp Med. 2005;201(8):1307–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McDermott DH, Gao JL, Liu Q, Siwicki M, Martens C, Jacobs P, et al. Chromothriptic cure of WHIM syndrome. Cell. 2015;160(4):686–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ghazanfari R, Li H, Zacharaki D, Lim HC, Scheding S. Human Non‐hematopoietic CD271(pos)/CD140a(low/neg) bone marrow stroma cells fulfill stringent stem cell criteria in serial transplantations. Stem Cells Dev. 2016;25(21):1652–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schepers K, Pietras EM, Reynaud D, Flach J, Binnewies M, Garg T, et al. Myeloproliferative neoplasia remodels the endosteal bone marrow niche into a self‐reinforcing leukemic niche. Cell Stem Cell. 2013;13(3):285–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Decker M, Martinez‐Morentin L, Wang G, Lee Y, Liu Q, Leslie J, et al. Leptin‐receptor‐expressing bone marrow stromal cells are myofibroblasts in primary myelofibrosis. Nat Cell Biol. 2017;19(6):677–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table SI. Patient characteristics.
Table SII. Primers for qRT–PCR.
Fig S1. Fluorescence signals after hybridization with random probes were not detected in human bone marrow cells.
Fig S2. Osteoblastic cells, bone marrow endothelial cells (ECs), and synovial mesenchymal cells expressed higher levels of the BGLAP, CDH5 and PDPN mRNAs, respectively, than CXCL12hi/LEPR+ cells.
Fig S3. Harvest site and donor age do not influence the expression of the CXCL12, KITLG, FOXC1 or EBF3 mRNAs in human adult bone marrow CXCL12hi/LEPR+ cells.
Fig S4. EBF3 was not expressed in CD56+CD271+ bone‐lining osteoblastic cells.
Fig S5. EBF3 was not expressed in morphologically identifiable CD31+ endothelial cells (ECs) in human adult bone marrow.
Fig S6. The frequency and the density of CXCL12hi cells were similar in bone marrow from control patients and patients with myeloproliferative neoplasms (MPNs).


