Summary
Pattern‐triggered immunity (PTI) is activated in plants upon recognition by pattern recognition receptors (PRRs) of damage‐ and microbe‐associated molecular patterns (DAMPs and MAMPs) derived from plants or microorganisms, respectively. To understand better the plant mechanisms involved in the perception of carbohydrate‐based structures recognized as DAMPs/MAMPs, we have studied the ability of mixed‐linked β‐1,3/1,4‐glucans (MLGs), present in some plant and microbial cell walls, to trigger immune responses and disease resistance in plants. A range of MLG structures were tested for their capacity to induce PTI hallmarks, such as cytoplasmic Ca2+ elevations, reactive oxygen species production, phosphorylation of mitogen‐activated protein kinases and gene transcriptional reprogramming. These analyses revealed that MLG oligosaccharides are perceived by Arabidopsis thaliana and identified a trisaccharide, β‐d‐cellobiosyl‐(1,3)‐β‐d‐glucose (MLG43), as the smallest MLG structure triggering strong PTI responses. These MLG43‐mediated PTI responses are partially dependent on LysM PRRs CERK1, LYK4 and LYK5, as they were weaker in cerk1 and lyk4 lyk5 mutants than in wild‐type plants. Cross‐elicitation experiments between MLG43 and the carbohydrate MAMP chitohexaose [β‐1,4‐d‐(GlcNAc)6], which is also perceived by these LysM PRRs, indicated that the mechanism of MLG43 recognition could differ from that of chitohexaose, which is fully impaired in cerk1 and lyk4 lyk5 plants. MLG43 treatment confers enhanced disease resistance in A. thaliana to the oomycete Hyaloperonospora arabidopsidis and in tomato and pepper to different bacterial and fungal pathogens. Our data support the classification of MLGs as a group of carbohydrate‐based molecular patterns that are perceived by plants and trigger immune responses and disease resistance.
Keywords: Arabidopsis thaliana, Capsicum annuum, cell wall, disease resistance, mixed‐linked glucan, Hyaloperonospora arabidopsidis, pattern triggered immunity, plant immunity, Solanum lycopersicum
Significance Statement
A better characterization of plant mechanisms involved in the perception of carbohydrate‐based structures recognized as damage‐/microbe‐associated molecular patterns is needed to understand plant immunity further. We show here that in addition to the β‐1,3‐ and β‐1,4‐glucans damage‐/microbe‐associated molecular patterns, plants are able to perceive mixed‐linked β‐1,3/1,4‐glucans that are present in the cell walls of some plant species and microbes, such as the oomycete Hyaloperonospora arabidopsidis. Mixed‐linked β‐1,3/1,4‐glucans trigger immune responses and disease resistance against different pathogens in several plant species.
INTRODUCTION
As sessile organisms, plants have evolved a complex immune system comprised by several defence layers. One of them is known as pattern‐triggered immunity (PTI), which is based on the recognition of damage‐ and microbe‐associated molecular patterns (DAMPs and MAMPs) derived from plants or microorganisms, respectively, by plasma membrane‐resident pattern recognition receptors (PRRs). Upon DAMP/MAMP recognition, PRRs activate protein kinase signalling cascades that trigger gene reprogramming processes, which ultimately result in plant surveillance to pathogen/pest attack (Bigeard et al., 2015; Boutrot and Zipfel, 2017). PTI relevance is well‐illustrated by the fact that immune responses and disease resistance to pathogens are compromised in plants defective in PRRs perceiving DAMPs or MAMPs of a peptidic nature, such as PEPR1 and FLS2 Receptor Like Kinases that recognize Arabidopsis AtPep1 DAMP and bacterial flg22 MAMP peptides, respectively (Gómez‐Gómez and Boller, 2000; Yamaguchi et al., 2006). Many PRR/peptidic DAMP or MAMP pairs triggering PTI have been elucidated (Boutrot and Zipfel, 2017; Tang et al., 2017). However, the specific mechanisms of plant defence activation by carbohydrate‐based DAMPs and MAMPs, which are highly abundant in plant and microbial extracellular layers, clearly lags behind our knowledge of peptide ligand recognition (Bacete et al., 2018). The first bottleneck in the identification of carbohydrate‐based ligand‐PRR pairs is the relatively low number of carbohydrates known to trigger plant immune responses in contrast to the mammal counterpart (Bacete et al., 2018). Among the carbohydrates recognized by the plant immune system are chitin and β‐1,3‐glucan from fungal/oomycete cell walls, peptidoglycan from bacterial walls, and cellulose (β‐1,4‐glucan), xyloglucan, mannan, xylan and homogalacturonan from plant cell walls (Aziz et al., 2007; Claverie et al., 2018; Galletti et al., 2008; Gust et al., 2007; Kaku et al., 2006; Klarzynski et al., 2000; Mélida et al., 2018, 2020; Wanke et al., 2020; Zang et al., 2019).
In plants, PRR/carbohydrate‐based ligand characterization at the structural level has been mainly limited to PRRs of the LysM family, which are involved in the recognition of several glycoligands such as chitin, peptidoglycans and lipopolysaccharides (Cao et al., 2014; Desaki et al., 2018; Liu et al., 2012; Miya et al., 2007; Willmann et al., 2011). Recent work demonstrated that LysM‐PRRs are also implicated in the perception by plants of β‐1,3‐glucans (Mélida et al., 2018; Wanke et al., 2020). Specifically, a β‐1,3‐glucan hexasaccharide (laminarihexaose or Lam6) is an immune‐active structure whose recognition is dependent in Arabidopsis on the LysM‐PRR CERK1 (Chitin Elicitor Receptor Kinase 1) (Mélida et al., 2018). However, a direct binding of laminarihexaose to CERK1 extracellular ectodomain (ECD) has not been either observed in isothermal titration calorimetry binding assays performed with purified ECD‐CERK1 nor predicted using recently developed in silico structural molecular dynamics simulations (del Hierro et al., 2020). These data suggest that CERK1 function as co‐receptor rather than a receptor in β‐1,3‐glucan hexasaccharide perception. Other plant species, such as rice and tobacco, have been shown to also recognize β‐1,3‐glucans with a higher degree of polymerization (DP) than 6 and this recognition has been demonstrated to be CERK1‐independent, suggesting that β‐glucan recognition may be mediated by multiple receptor/co‐receptor proteins (Wanke et al., 2020). CERK1 does not seem to be involved in the perception of β‐1,4‐glucans (e.g. cellohexaose) in Arabidopsis as immune responses triggered by this DAMP were not impaired in cerk1 mutant, as predicted by in silico structural molecular dynamics simulations (del Hierro et al., 2020).
Glucans represent a group of widely distributed polysaccharides, mainly found in the extracellular layers of numerous phylogenetic groups across the tree of life (Latgé and Calderone, 2006; McIntosh et al., 2005; Mélida et al., 2013; Srivastava et al., 2017). These include a wide variety of structures, mainly with β‐linkages, although α‐linked glucans also occur in many species. Mixed‐linked glucans [MLGs; β‐1,3/1,4‐glucans; (1,3;1,4)‐β‐d‐glucans] consist of unbranched and unsubstituted chains of β‐1,4‐glucosyl residues interspersed by β‐1,3 linkages (Burton and Fincher, 2009). MLGs are widely distributed as matrix polysaccharides in cell walls of the plants from the Poaceae group, but have also been reported in other species such as in the walls of Equisetum spp. and other vascular plants outside the Poaceae (Fincher and Stone, 2004; Fry et al., 2008; Smith and Harris, 1999; Sørensen et al., 2008; Trethewey et al., 2005), bryophytes and algae (Popper and Fry, 2003; Salmeán et al., 2017). MLGs have been also described in lichen‐forming ascomycete symbionts (Perlin and Suzuki, 1962; Gorin et al., 1988; Stone and Clarke, 1992), in fungi and oomycetes (Fontaine et al., 2000; Pettolino et al., 2009; Samar et al., 2015) and bacteria (Lee and Hollingsworth, 1997; Pérez‐Mendoza et al., 2015). β‐Glucans are well‐known modulators of the immune system in mammals, but less is known about their roles in the plant counterpart (Fesel and Zuccaro, 2016; Mélida et al., 2018; Rovenich et al., 2016; Sharp et al., 1984; Wanke et al., 2020). Interestingly, this field of β‐glucan perception by plants has regained momentum due to recent discoveries demonstrating that glucans containing either β‐1,3 or β‐1,4 glycosidic linkages in their main backbones trigger PTI responses in plants (Claverie et al., 2018; del Hierro et al., 2020; Johnson et al., 2018; Mélida et al., 2018; Souza et al., 2017; Wanke et al., 2020; Wawra et al., 2016). However, it remained elusive whether glucans containing both types of linkages (MLGs) can be perceived by plant cells as well (Aziz et al., 2007; Johnson et al., 2018; Klarzynski et al., 2000; Locci et al., 2019; Mélida et al., 2018; Ménard et al., 2004; Souza et al., 2017).
In an effort to characterize additional carbohydrate‐based structures (MAMPs or DAMPs) that are able to activate the plant immune system, we have determined the ability of different MLG structures to trigger early immune responses in Arabidopsis. We tested the capacity to induce PTI hallmarks (Boudsocq et al., 2010; Ranf et al., 2011) of several MLG‐enriched fractions and purified MLGs. Our results demonstrate that MLGs are a group of glycoligands that activate plant immunity and we characterized β‐d‐cellobiosyl‐(1,3)‐β‐d‐glucose (MLG43) as the smallest active structure triggering immune responses in Arabidopsis and boosting Arabidopsis and crops (e.g. tomato and pepper) disease resistance to different pathogens.
RESULTS
MLG oligosaccharides trigger Ca2+ influxes in Arabidopsis
Aequorin‐based Ca2+ Columbia‐0 (Col‐0AEQ) sensor Arabidopsis system (Ranf et al., 2011) was used to monitor whether early cytoplasmic Ca2+ influxes (burst) occurred upon treatment of seedlings with cell wall fractions enriched in MLGs, such as β‐1,3/1,4‐glucans from barley (B‐GLU), an alkali‐extracted fraction isolated from Equisetum arvense cell wall (E‐KOH) and lichenan (a mixture of β‐1,3/1,4‐glucans) from Cetraria islandica (C‐LICH). None of these polymeric fractions induced Ca2+ influxes in comparison with pure MAMPs, such as the chitin hexasaccharide chitohexaose [β‐1,4‐D‐(GlcNAc)6] (Figure 1a). To generate MLG oligosaccharides with lower DP than these polymeric fractions, MLGs were treated with lichenase from Bacillus subtilis (EC 3.2.1.73), a glucan endohydrolase that catalyses the hydrolysis of β‐1,4 bonds immediately following β‐1,3 bonds, but does not catalyse the hydrolysis of purely β‐1,3‐ or β‐1,4‐linked glucans (Henrissat and Bairoch, 1993; Planas, 2000). MLG polysaccharides digested with lichenase released MLGs with a single β‐1,3 linkage placed next to the reducing end. Interestingly, products from lichenase digestions of these MLG polysaccharides activated plant Ca2+ influxes (B‐GLU + L, E‐KOH + L or C‐LICH + L; Figure 1a), indicating that lichenase‐released MLG oligosaccharides can trigger early immune responses in Arabidopsis.
Figure 1.
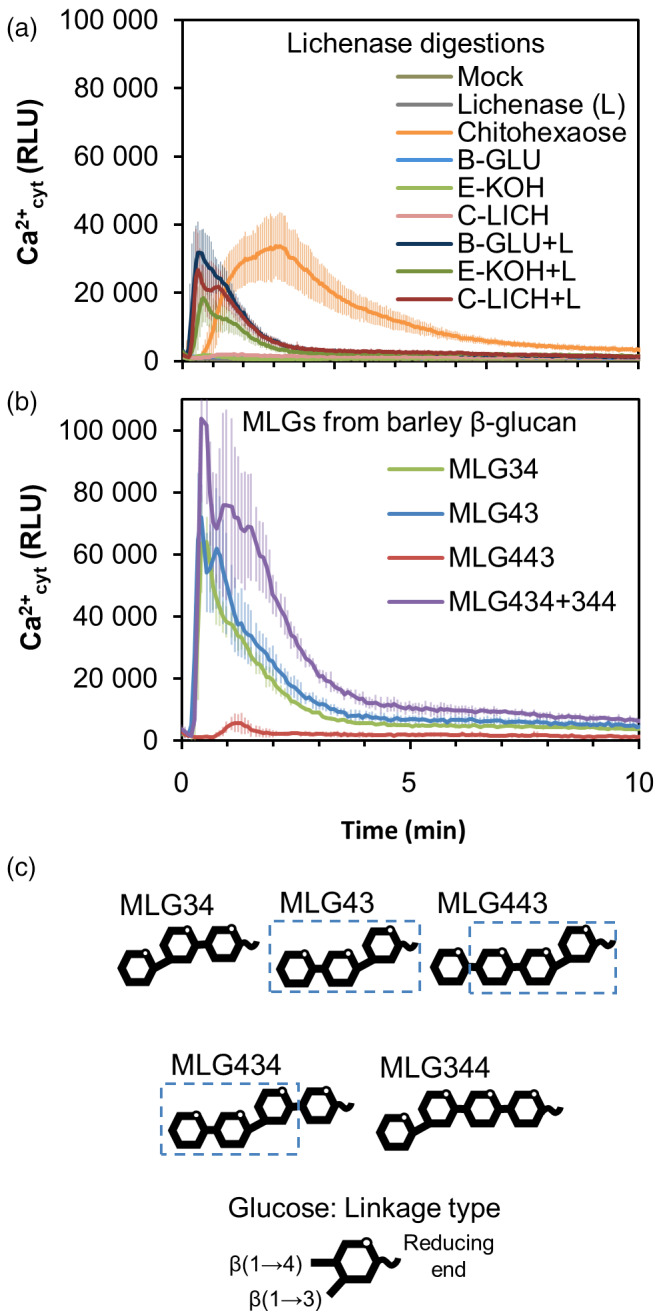
Mixed‐linked glucans (MLG) oligosaccharides trigger cytoplasmic Ca2+ elevations in Arabidopsis.
Ca2+ influxes were measured as relative luminescence units (RLU) over time in 8‐day‐old Arabidopsis Col‐0AEQ seedlings after treatment with (a) β‐1,3/1,4‐glucans of barley (β‐glucan; B‐GLU), 4% KOH fraction of Equisetum arvense cell wall (E‐KOH) and lichenan from Cetraria islandica (C‐LICH) (0.25 mg ml−1 final concentration) untreated or treated with lichenase enzyme (+L). Chitohexaose 50 μm was used as positive control. Undigested materials (0.25 mg ml−1), lichenase suspension (L) and distilled water (mock) were used as negative controls.
(b) Ca2+ influx after treatment with commercial MLGs (50 μm) purified from barley (Megazyme). Data represent mean ± σ (n = 8) in all panels.
(c) Structural scheme of the different MLG oligosaccharides used in the experiments. A β‐d‐cellobiosyl‐(1,3)‐β‐d‐glucose (MLG43) structure is highlighted over the different structures. These data are from one representative experiment of at least three performed that gave similar results.
To confirm, that lichenase‐released oligosaccharides were plant immune‐active structures, we screened the activity of MLG oligosaccharides from commercial sources obtained by the purification of B‐GLU after enzymatic digestions with lichenase and cellulase (Figure 1b). Results from the commercial source clearly demonstrated that, at least, the shortest MLG oligosaccharides with DP 3 (e.g. MLG43 and MLG34) and some with DP 4 (MLG434 + MLG344 mixture and MLG443 in a lesser extent) were able to trigger Ca2+ influxes (Figure 1b). As MLGs consist of unbranched and unsubstituted chains of β‐1,4‐glucosyl residues interspersed by β‐1,3 linkages, the minimal MLGs structures containing both types of linkages would be MLG43 and MLG34 (DP 3; Figure 1c). We decided to compare Ca2+ influxes triggered by MLG43 and MLG34 trisaccharides and by their constituent disaccharides in the Col‐0AEQ sensor lines, and we observed that while cellobiose (β‐1,4‐linked disaccharide) triggered a slight Ca2+ influx, the β‐1,3‐linked glucan disaccharide did not (Lam2; Figure S1). In contrast, cellotriose (Cello3), a well characterized DAMP (Johnson et al., 2018; Locci et al., 2019), triggered Ca2+ influxes that were slightly higher than those activated by MLG43 and MLG34, whereas the β‐1,3‐linked trisaccharide (Lam3) was not active at the concentration tested, as reported previously (Mélida et al. 2018; Figure S1). MLG43 used in these studies can be obtained after MLG (e.g. B‐GLU) digestion with different enzymes, such as lichenases, whereas MLG34 must be obtained through cellulase digestion, which cleave β‐1,4 bonds within the inner cello‐oligosaccharide backbone of the MLG. As illustrated in the thin‐layer chromatography (TLC) at Figure S2, while lichenase digestion products follow a pattern (MLG43, MLG443, MLG4443, etc.), more randomized oligosaccharide structures are released by cellulases, including β‐1,4‐linked (Cello2–Cello6) glucans, which have been described to trigger immune responses (Johnson et al., 2018). Based on these results, we selected the trisaccharide MLG43 as the minimal active MLG oligosaccharide (Figure 1b,c) and lichenase as the best enzyme to release this structure from MLG polymers.
Aiming to support the results obtained with oligosaccharides from commercial sources, we also purified MLG43 from C‐LICH + L, B‐GLU + L and E‐KOH + L by size exclusion chromatography (SEC) (Figure 2; Figure S3). While B‐GLU digestion yielded MLG43 and MLG443, the highest yield of MLG43 was obtained from C. islandica lichenan digestion as revealed by TLC analyses (MLG43‐L in Figure 2a,c). MLG43‐L clearly co‐eluted with the respective commercial compound in a high‐pressure anion exchange (HPAE) chromatogram (Figure 2b,d,e). HPAE chromatography (HPAEC) data also corroborated that, while lichenase hydrolysis of B‐GLU released a clearly noticeable amount of MLG443 tetrasaccharide in addition to the trisaccharide, lichenan structure clearly favoured the release of the trisaccharide (Figure 2c). In Equisetum fractions, according to SEC profiles, oligosaccharides of higher DP than the tetrasaccharide were released (Figure S3). These high DP oligosaccharides are compatible with hexasaccharides and nonasaccharides, previously described for this species (Simmons et al., 2013). Cross‐elicitation experiments in Col‐0AEQ seedlings, by subsequent application of two ligands in 600 sec interval, evidenced that MLG43‐L and commercial MLG43 had a refractory period of Ca2+ influx, indicating that both MLGs have equivalent activity (Figure S4). Dose‐dependence and estimated effective dose (EED; 50% of total signal) of 265 μm were determined for MLG43 on Arabidopsis seedlings using a concentration range between 200 nm and 5 mm (Figure S5). All these concentrations activated a Ca2+ burst to a different extent, and based on Ca2+ kinetics and EED value, 50 μm was selected as an adequate final concentration for further experiments.
Figure 2.
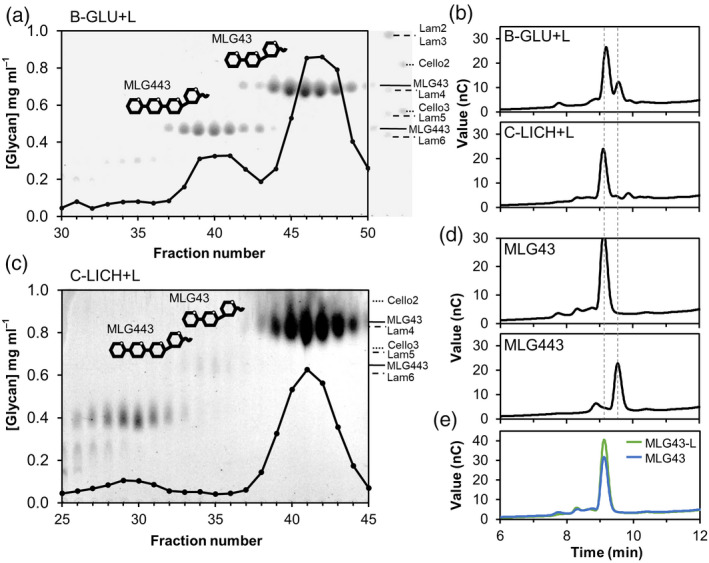
Purification of mixed‐linked glucans (MLG) oligosaccharides from lichenase (L)‐digested polysaccharides.
(a,c) Size exclusion chromatography elution profiles of 5 mg of lichenase digestion products from different β‐1,3/1,4‐glucans sources. (a) Barley (β‐glucan; B‐GLU) and (c) lichenan from Cetraria islandica (C‐LICH). Thin‐layer chromatography profiles of each of the fractions are shown overlaid. Markers indicate the migration of laminarin‐oligosaccharides (Lam), cello‐oligosaccharides (Cell) and MLG oligosaccharides (MLG).
(b,d,e) High pressure anion exchange chromatography profiles of 5 mg of lichenase (L)‐digested polysaccharides and commercial MLG oligosaccharides. (b) B‐GLU + L and C‐LICH + L; (d) MLG43 and MLG443. (e) Overlaid chromatograms comparing commercial MLG43 and purified MLG43 from lichenan (MLG43‐L; pooled fractions 38–44 of the elution profile shown in c). Data shown in all panels are representative chromatograms from one experiment of at least 10 performed that gave similar results.
In addition, we decided to synthesize chemically the pure MLG structures to characterize the PTI activity of additional structures further. The analysis of Ca2+ influxes triggered by these pure, synthetic structures indicated that MLG oligosaccharides with DP3 and DP4 were not active (Figure S6a), probably due to the presence of an aminoalkyl linker at their reducing end, which is required for the synthesis process (Figure S6c; Bartetzko and Pfrengle, 2019; Dallabernardina et al., 2017) and that would affect the tri‐dimensional structures of these synthetic oligosaccharides and may interfere with their perception by PRRs. However, MLG oligosaccharides of higher DP (6 and 8) were active in triggering Ca2+ influxes (Figure S6b; MLG44343′, MLG44434′, MLG43344′, MLG3434443′). Some of the immune active synthetic MLG structures tested cannot be generated through lichenase digestions, that generate MLGs with a single β‐1,3 linkage placed next to the reducing end, further indicating that ‘non‐canonical’ MLG oligosaccharides containing the MLG43/MLG34 ‘signature’ can be also recognized by plant cells.
MLG43 trisaccharide activate PTI hallmarks
Previous reports using luminol‐based assays to quantify reactive oxygen species (ROS) production in leaf discs treated with a β‐1,3‐glucan hexasaccharide (laminarihexaose) or a β‐1,4‐glucan disaccharide (cellobiose) revealed an absence of ROS production, even after applying high concentrations (0.5–1 mm) of these glucans (Mélida et al., 2018; Souza et al., 2017). In contrast, we found that MLG43 triggered an ample ROS burst when applied at 50 μm on Arabidopsis leaf discs, although this ROS burst was weaker than that triggered by chitohexaose (Figure 3a). To confirm PTI activity of MLG43, we next dissected the phosphorylation of protein kinases (MPK3/MPK6/MPK4/MPK11) and the upregulation of PTI‐reporter genes upon treatment of Arabidopsis seedlings with MLG43 (Figure 3b,c). Western blotting showed MPK3 and MPK6 phosphorylation after MLG43 application (50 μm) to Arabidopsis seedlings, with a phosphorylation peak at 10 min post‐treatment (Figure 3b). MPK4/11 phosphorylation was almost undetected in MLG43‐elicited plants, which contrasted with the observed phosphorylation upon chitohexaose treatment (Figure 3b). Expression of five PTI‐marker genes upregulated by chitohexaose (WRKY53, FRK1, NHL10, NUD7 and FH4; Mélida et al., 2018) was assessed by quantitative reverse transcription–polymerase chain reaction (qRT‐PCR), and the results indicated that expression of all these genes was similarly induced after MLG43 or chitohexaose elicitation in comparison with mock‐treated seedlings (Figure 3c), suggesting that these compounds trigger similar transcriptional responses.
Figure 3.
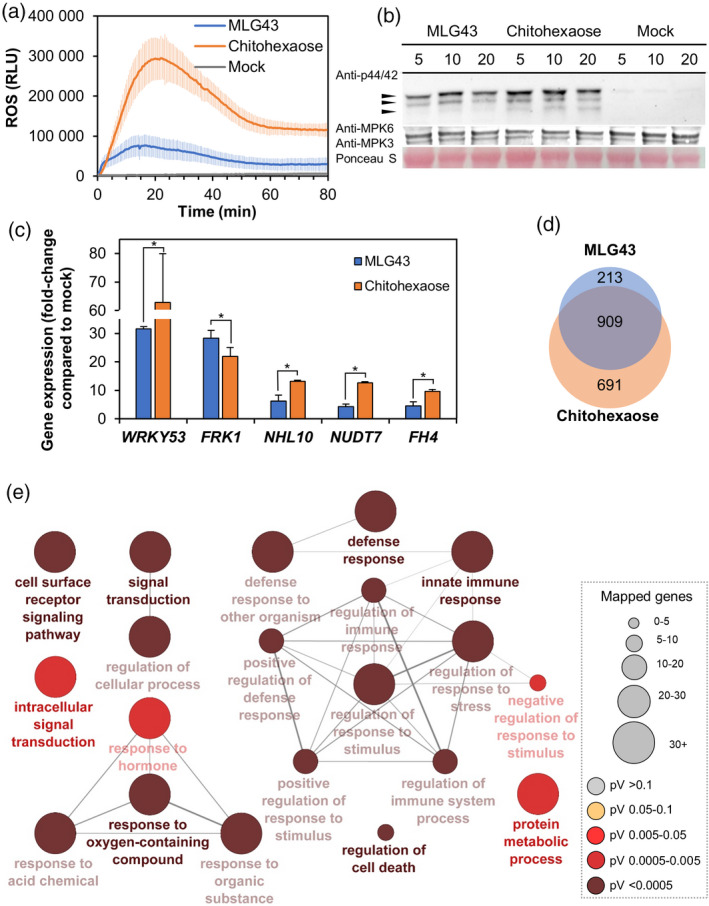
Pattern‐triggered immunity hallmarks activation by mixed‐linked glucans, β‐d‐cellobiosyl‐(1,3)‐β‐d‐glucose (MLG43 50 μm) in Arabidopsis.
Chitohexaose (5 μm (a,b) and 50 μm for gene expression (c,d) and distilled water (mock) were used as controls in all the experiments. (a) Reactive oxygen species (ROS) production in Arabidopsis leaf‐discs of 5‐week‐old Col‐0 plants by Luminol reaction measured as relative luminescence units (RLU) over time. Data represent mean ± σ (n = 8) from one experiment of three performed that produced similar results. (b) Mitogen‐activated protein kinases (MAPK) phosphorylation in 12‐day‐old seedlings determined by western blot using anti‐pTEpY antibody for phosphorylated MAPK moieties at different time points (5, 10 and 20 min). Arrows indicate the position of phosphorylated MPK6 (top), MPK3 (middle) and MPK4/11 (bottom). Ponceau red‐stained membranes show equal loading. Data shown are from one experiment of the five performed that gave similar results. (c) Quantitative reverse transcription–polymerase chain reaction analysis in 12‐day‐old Arabidopsis seedlings. Relative expression levels to UBC21 (At5g25769) gene at 30 min normalized to their expression levels in mock‐treated seedlings were shown. Data represent mean ± σ of three technical replicates from two biological replicates (n = 6) of three independent biological replicates analysed that gave similar results. Statistically significant differences between MLG43 and chitohexaose according to Student's t‐test (*P < 0.05). (d) Venn diagram of shared overexpression between MLG43 and chitohexaose (both at 50 μm). RNA‐sequencing data were obtained from the combination of three biological replicates of 12‐day‐old Arabidopsis Col‐0 plants at 30 min after treatment with MLG43 or chitohexaose.
(e) Biological process Gene Ontology (GO) term enrichment map of the overexpressed genes in 50 μm MLG43. GO term enrichment is expressed by node size. Enrichment P‐value determined by enrichment/depletion (two‐sided hypergeometric) test and corrected by the Bonferroni step down method is represented by colour scale. Only GO terms at P < 0.01 are shown. Links between groups indicate shared genes (κ score level ≥ 0.4).
To characterize the global gene reprogramming triggered by MLG43 further, we performed transcriptomic analyses [RNA‐Sequencing (RNA‐seq)] of Arabidopsis seedlings treated for 30 min with MLG43 or chitohexaose (Figure 3d,e; Tables [Link], [Link], [Link], [Link]). Incubation with MLG43 induced changes in the expression of 1229 genes, most of which (1122) were upregulated (Figure 3d; Table S1). On the other hand, treatments with chitohexaose resulted in 1988 genes whose expression levels were significantly altered, with 691 genes being upregulated exclusively by chitohexaose and 909 after treatment with both structures (Figure 3d; Figure S7, Tables S2 and S3). Gene Ontology (GO) classification of MLG43‐induced genes showed that these mainly grouped in terms related to immune system processes, response to different stimuli, including biotic and abiotic stresses, signal transduction and cell surface receptors signalling pathways, among other GOs (Figure 3e), which are quite similar GOs to those found upon treatment with chitohexaose (Figure S7). These analyses indicate that MLG43‐triggered responses are highly similar to those induced by the well characterized MAMP chitohexaose (Figure 3; Figure S7).
LysM‐PRRs have been described in various species as co‐receptors for glycan‐based molecular patterns such as chitin, peptidoglycan, β‐1,3‐glucans (laminarins) and lipopolysaccharides (Desaki et al., 2018; Mélida et al., 2018; Miya et al., 2007; Willmann et al., 2011). In particular, the LysM‐PRR CERK1 has a crucial role in glycan‐based‐MAMP perception. Given the high similarity in global gene reprograming triggered by MLG43 and the CERK1‐dependent ligand chitohexaose, we wondered whether refractory stages would exist between the application of these two glyco‐ligands to Arabidopsis seedlings. Notably, cross‐elicitation experiments in Col‐0AEQ seedlings demonstrated the absence of such a refractory period, further suggesting that the mechanisms of perception of these glycans and the PRRs involved in their perception are not identical (Figure 4a). In addition to CERK1, the LysM‐PRRs LYK5 and LYK4 have been involved in chitohexaose perception as receptor and co‐receptor, respectively (Cao et al., 2014; del Hierro et al., 2020; Liu et al., 2012). To characterize the molecular mechanisms of MLG43‐mediated immunity further, phosphorylation of mitogen‐activated protein kinase (MAPK) was tested by western blots in wild‐type plants and cerk1 single and lyk4 lyk5 double mutants upon MLG43 treatment and we found that MAPK phosphorylation levels were weaker in these mutants than in wild‐type plants, whereas they were fully impaired upon chitohexaose treatment, as reported previously (Cao et al., 2014; Figure 4b). These data indicate that MLG43 perception partially depends on CERK1, LYK5 and LYK4 PRRs, that might function as co‐receptors, as previously described for the β‐1,3‐glucan elicitor laminarihexaose (Mélida et al., 2018). In addition to LysM‐PRRs, BAK1 and SOBIR1 are frequently involved as PRR co‐receptors in the activation of signal transduction following perception of different DAMP/MAMPs (van der Burgh et al., 2019; Perraki et al., 2018). We tested phosphorylation of MAPK in wild‐type plants and mutant lines impaired in BAK1 and SOBIR1, and we found that MAPK phosphorylation levels were similar in these mutants and wild‐type plants upon MLG43 treatment (Figure S8), suggesting that MLG43 perception does not depend on BAK1 and SOBIR1. These data were in line with those obtained by determining protein–glycan interaction energies applying a molecular dynamics simulation methodology recently described (del Hierro et al., 2020) that confronted optimized CERK1, BAK1 and SOBIR1 ECD structures and MLG43 in solvent boxes (Table S7). ECD‐glycan ΔG determinations resulted in positive or close to zero energy values, which indicate that no direct binding events between the ECDs of these PRRs and MLG43 took place during the molecular dynamics simulations (Table S7). Together, these analyses with the ECDs of main co‐receptors indicated that MLG43 perception and signal transduction is mediated by a not yet characterized immune complex that does not involve BAK1 or SOBIR1 and that probably involves LysM‐PRRs as redundant co‐receptors.
Figure 4.
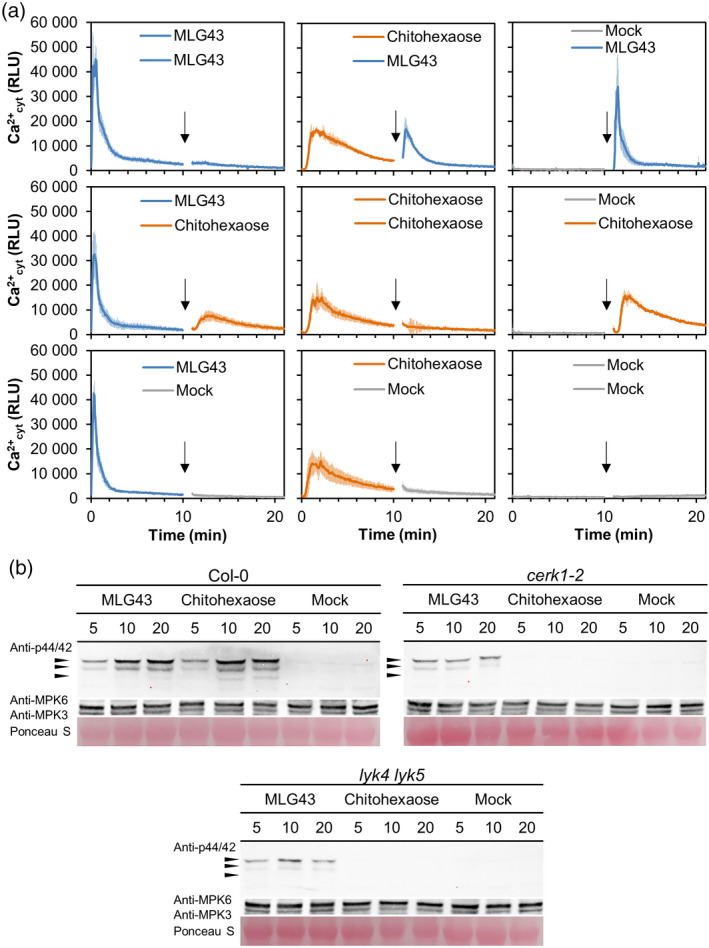
MLG43‐triggered immunity in Arabidopsis is partially dependent on LysM‐PRRs CERK1, LYK4 and LYK5.
(a) Cross‐elicitation during the refractory period of Ca2+ signalling upon application of 50 μm MLG43, 50 μm chitohexaose and distilled water (mock). Data show the elevation of cytoplasmic Ca2+ concentration, measured as relative luminescence units (RLU), over time in 8‐day‐old Arabidopsis Col‐0AEQ seedlings after treatments. Arrow indicates the application time of the second treatment within the refractory period of the first elicitation. Data represent mean ± σ (n = 8) in all panels.
(b) Mitogen‐activated protein kinases (MAPK) phosphorylation in 12‐day‐old Arabidopsis seedlings of Col‐0 plants and cerk1‐2, and lyk4 lyk5 mutants impaired in LysM‐PRR co‐receptors. Western blot using anti‐pTEpY antibody for phosphorylated MAPK moieties at different time points (5, 10 and 20 min). Black arrows indicate the position of phosphorylated MPK6 (top), MPK3 (middle) and MPK4/11 (bottom). Anti‐MPK6 and anti‐MPK3 were used as total protein control. Ponceau red‐stained membranes show equal loading. Chitohexaose (5 µm) and distilled water (mock) were used as controls. These results are from one representative experiment of the three performed that gave similar results.
MLG43 pre‐treatments diminish plant disease symptoms caused by inoculations of pathogens
MLGs are undoubtedly present in the wall of several plants, algae, lichen‐forming ascomycete symbionts, fungi and bacteria, but most likely not in dicot plant species such as Arabidopsis (Burton et al., 2006; Zablackis et al., 1995). Several lines of evidence also point to the presence of MLGs in the cell wall of several plant pathogens harbouring glucan‐rich extracellular envelopes such as oomycetes, fungi and some bacteria, but the presence of MLGs in these organisms has been reported only in a few cases (Fontaine et al., 2000; Lee and Hollingsworth, 1997; Mélida et al., 2013; Pérez‐Mendoza et al., 2015; Pettolino et al., 2009; Samar et al., 2015). We searched for the presence of MLGs in the cell wall of the Arabidopsis oomycete pathogen Hyaloperonospora arabidopsidis, belonging to the Peronosporales order, such as the Phytophthora species, that contain up to 85% of glucans in their cell walls (Mélida et al., 2013). We obtained alcohol insoluble residues (AIR; equivalent to partially purified cell walls) from H. arabidopsidis Noco2 conidiospores, and found that these AIRs contained on average 27.9 µg of MLGs per mg of dry weight (Figure 5a). Next, we purified AIR fractions from non‐inoculated and H. arabidopsidis‐inoculated Arabidopsis plants, which were extensively washed, before mechanical disruption for AIR preparation, to remove H. arabidopsidis sporangiophores and those conidiospores from spray inoculation of the plants that had not germinated from leaves surface. Interestingly, we found that only AIR from H. arabidopsidis‐inoculated plants contained MLGs (29.6 µg per mg of AIR dry weight; Figure 5a), which would necessarily derive from intracellularly grown H. arabidopsidis hyphae inside Arabidopsis leaves, as the conidiospores had been washed out. These data showed that, at least in the Arabidopsis–H. arabidopsidis pathosystem, plant cells are exposed to this oomycete's MLGs. In sight of these data, we tested whether pre‐treatment with MLG43 of Arabidopsis Col‐0 wild‐type plants before infection with the oomycete would improve Arabidopsis resistance to the compatible and virulent Noco2 isolate. Of note, we observed a reduction of up to 60% in conidiospore mg−1 of plant fresh weight at 7 days post‐inoculation (dpi) in plants pre‐treated with MLG43 in comparison with untreated plants (Figure 5b). These data would favour the classification of MLGs as MAMPs triggering PTI in Arabidopsis.
Figure 5.
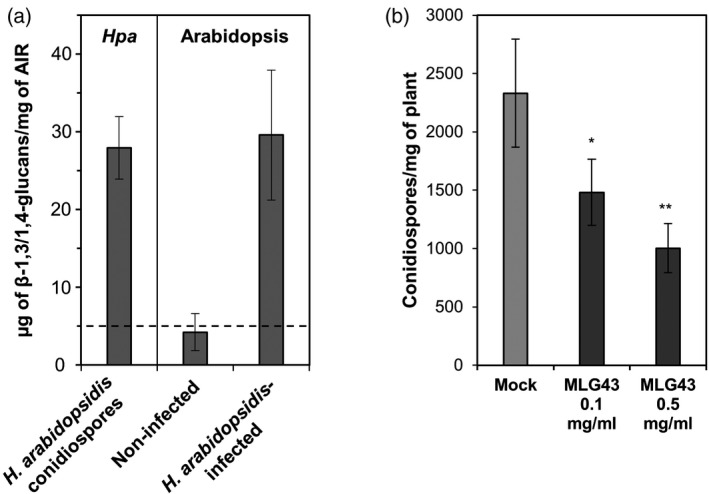
Mixed‐linked glucans are components of the cell walls of Hyaloperonospora arabidopsidis and trigger Arabidopsis disease resistance to this oomycete pathogen.
(a) β‐1,3/1,4‐glucan quantification in alcohol insoluble residues (AIRs) of H. arabidopsidis (Hpa isolate Noco2) conidiospores and of Arabidopsis Col‐0 plants non‐infected (mock‐treated) or infected with H. arabidopsidis (Noco2) 7 days post‐inoculation (dpi) with conidiospores (24‐day‐old Arabidopsis plants). Inoculated plants were extensively washed before AIR preparations in order to release H. arabidopsidis conidiospores from plant tissues. Data represent average ± σ (n = 3). Dashed line indicates the detection limit of the method used.
(b) Arabidopsis plants were foliar pre‐treated with MLG43 at two different concentrations 2 days before inoculation with H. arabidopsidis (Noco2). Presence of H. arabidopsidis in plants was quantified at 7 dpi (24 days old) as the abundance of conidiospores in inoculated per mg plant fresh weight. Data represent mean ± SE (n = 30). Statistically significant differences according to the Student's t‐test (*P < 0.05; **P < 0.01).
Next, we asked whether part of the knowledge gained using the model species Arabidopsis could be translated to crops such as tomato and pepper. In a first instance, we evaluated the MLG43‐protection capacity by pre‐treating tomato plants with the trisaccharide 2 days before challenging them with the bacterium Pseudomonas syringae pv. tomato DC3000. Notably, bacterial growth was significantly reduced in the MLG43‐pre‐treated tomato plants at 11 dpi compared with mock‐treated plants (Figure 6a). Moreover, MLG43 treatment leaded to the upregulation of two tomato PTI‐marker genes, SlWRKY53 and SlPTI5 (Liu et al., 2019) (Figure S9). A similar approach was followed in pepper plants that were pre‐treated with MLG43 2 days before inoculation with the necrotrophic fungi Sclerotinia sclerotiorum or Botrytis cinerea. MLG43‐treated pepper plants showed, in comparison with mock‐treated plants, a reduction in their disease symptoms index (Figure S10) at 9 dpi with S. sclerotiorum (Figure 6b), and at 5 and 9 dpi with B. cinerea (Figure 6c). The activation of MLG43‐triggered immune responses in Arabidopsis, tomato and pepper suggests that these species have the PRRs and co‐PRRs required for MLG perception and PTI activation.
Figure 6.
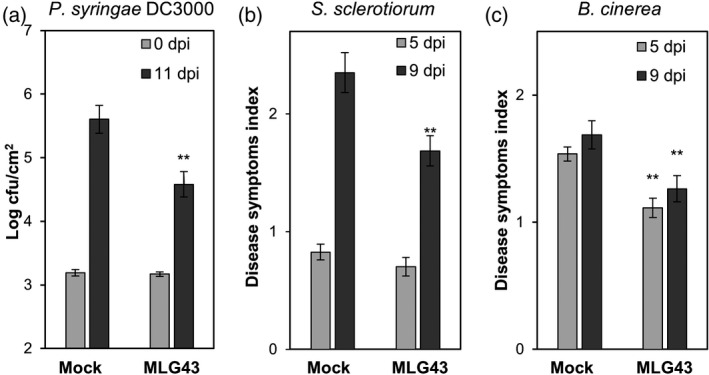
MLG43 pre‐treatment confers enhanced disease resistance against bacterial and fungal pathogens to tomato and pepper plants.
Plants were sprayed with MLG43 (0.25 mg plant−1) 2 days before pathogen challenge.
(a) Colony forming units (cfu) of Pseudomonas syringae pv. tomato DC3000 per leaf area at 0 and 11 days post‐inoculation (dpi) in tomato plants. Data represent mean ± SE (n = 8).
(b) Disease symptoms index produced by Sclerotinia sclerotiorum at 5 and 9 dpi in leaves of pepper plants. Data represent mean ± SE (n = 24).
(c) Disease symptoms index produced by Botrytis cinerea at 5 and 9 dpi in leaves of pepper plants. Data represent mean ± SE (n = 12). Disease indexes (0–5) examples for (b) and (c) are shown in Figure S10. Statistically significant differences according to the Student's t‐test (*P < 0.05; **P < 0.01). All the disease experiments were performed at least four times and one representative experiment is shown.
DISCUSSION
Cell walls are dynamic and highly controlled structures that are the first point of contact during a plant–microbe interaction (Bacete et al., 2018, 2020; Geoghegan et al., 2017; Lampugnani et al., 2018; Rui and Dinneny, 2020). The evolutionary arms race has provided plants and their microbial interactors with a large collection of cell wall‐degrading enzymes to shoot down the opponent's wall (Rovenich et al., 2016). Therefore, cell walls are rich sources of carbohydrate‐based defence signalling molecules (DAMPs and MAMPs), which remain poorly characterized. Here, we demonstrate that some structures contained in MLGs are perceived as molecular patterns by different plant species triggering immune responses. In particular, we identified a minimal structure, the trisaccharide MLG43, which triggers several PTI‐hallmarks in Arabidopsis at low concentrations. Indeed, the transcriptomic study showed that MLG43 and the well‐known MAMP chitohexaose activate transcriptional responses that share almost 81% of the overexpressed genes, further demonstrating a very high gene reprogramming overlap upon treatment with these glycoligands. These data are in line with previous transcriptomic analyses comparing the Arabidopsis‐responsive genes to single‐linked β‐1,3‐glucans (DAMP/MAMP) and β‐1,4‐glucans (DAMPs) and chitin (MAMP) that revealed that a significant percentage of mis‐expressed genes were shared between glucans and chitin treatments (Johnson et al., 2018; Mélida et al., 2018; Souza et al., 2017).
Structurally, the most similar oligosaccharides to MLG43 are glucans with solely β‐1,3 or β‐1,4 linkages (Stone and Clarke, 1992). Cellulose‐derived oligomers (β‐1,4‐glucans) are plant cell wall‐derived DAMPs that trigger signalling cascades sharing some similarities with the responses triggered by chitin and the well‐characterized DAMP oligogalacturonides derived from plant pectic polysaccharides (Aziz et al., 2007; Benedetti et al., 2015; Ferrari et al., 2013; Johnson et al., 2018; Souza et al., 2017). Cello‐oligomers (DP3 or higher) and MLG43, as well as chitin, are active on plants at nanomolar concentrations (Johnson et al., 2018; Kaku et al., 2006), which are significantly lower than those required to trigger PTI responses by β‐1,3‐glucans and xyloglucan (containing a β‐1,4‐linked glucose backbone), which are in the high micromolar range (Aziz et al., 2003; Claverie et al., 2018; Klarzynski et al., 2000; Mélida et al., 2018). Interestingly, in spite of the high β‐1,3‐glucans and xyloglucans doses required to be perceived by plants, they were able to improve protection in several pathosystems. For instance, xyloglucan increased grapevine and Arabidopsis resistance against the fungus B. cinerea or the oomycete H. arabidopsidis, respectively, while β‐1,3‐glucans improved, among others, tobacco and grapevine protection against bacterial (Erwinia carotovora), fungal (B. cinerea) and oomycete (Plasmopara viticola) pathogens (Aziz et al., 2003; Claverie et al., 2018; Klarzynski et al., 2000). Given the high abundance of β‐1,3‐glucans in the cell walls of brown seaweed, laminarin‐based products have been successfully developed to be used in agriculture as activators of plant natural defence against pathogens. Similarly, pre‐treatments with the single‐linked β‐1,4‐glucan cellobiose reduced P. syringae growth on Arabidopsis seedlings, although high doses were required to observe such an effect (Souza et al., 2017).
Notably, we show here that a glucan trisaccharide combining both β‐1,3 and β‐1,4 linkages (MLG43) is also perceived by Arabidopsis and some crops (tomato and pepper) at lower concentrations than the single‐linked counterparts β‐1,3‐ and β‐1,4‐glucans. Indeed, pre‐treatments of Arabidopsis, tomato and pepper with MLG43 before pathogen inoculation confers enhanced disease resistance and significant protection against oomycete, bacterial and fungal pathogens, supporting that different plant species in addition to Arabidopsis harbour the PRRs required for recognition of the MLGs. This perception, at least in Arabidopsis, seems to be independent of the co‐receptors BAK1 and SOBIR1, and only partially dependent on CERK1, LYK4 and LYK5 LysM‐PRRs, contrary to chitin and β‐1,3‐glucans, which were fully impaired in the cerk1‐2 mutant (Cao et al., 2014; Liu et al., 2012; Mélida et al., 2018). Despite these differences in PRR complexes involved in the perception of these carbohydrate‐based MAMPs, they transcriptionally activate a very high similar downstream gene reprograming, as reported in other MAMPs comparisons (Bjornson et al., 2020).
Our data support that MLGs represent a group of molecular patterns perceived by plants, but it raises now several important biological questions about the different source of MLGs that plants are exposed to during plant/pathogen interactions, and the mechanisms of MLG recognition by plants. Notably, some plant phylogenetic groups, such as Poaceae species, harbour MLGs in their cell walls that can release immune‐active MLGs (MLG43) upon the catalytic breakdown during infection by fungal/oomycete cell wall degrading enzymes, such as cellulases, and accordingly MLG43 could be classified as a self‐molecular pattern or DAMP. However, other plant species, including Arabidopsis, tomato and pepper used in this work do not seem to contain MLGs in their cell walls, but we show here that they are able to perceive MLG43 and to trigger immune responses. In these cases, MLG43 will be perceived as non‐self‐molecular pattern or MAMP. We show here that the oomycete H. arabidopsidis harbours MLGs in the cell walls of conidiospores or intracellularly growing hyphae in Arabidopsis‐infected plants. This finding is in line with previous reports indicating the presence of MLGs in the cell wall of several microorganisms, although these glucan structures remain poorly characterized (Fontaine et al., 2000; Lee and Hollingsworth, 1997; Mélida et al., 2013; Pérez‐Mendoza et al., 2015; Pettolino et al., 2009; Samar et al., 2015). There are many groups of microbes whose cell wall contains high proportions of glucans, but as MLG constituent units (1,4‐ and 1,3‐linked glucosyl residues) can be the building blocks of other better microbial characterized polymers (e.g. laminarin, glycogen, cellulose), MLG presence has probably been underestimated to date. For instance, plant pathogens such as oomycetes Phytophthora infestans and Phytophthora parasitica contain over 85% of glucans in their cell walls, including a high proportion of both 1,4‐ and 1,3‐linked glucosyl units (Mélida et al., 2013). However, the presence of polymers combining both types of linkages (MLGs) has not been investigated in detail yet. A good example illustrating this is that the cell wall composition of the closely related oomycete H. arabidopsidis is unknown, but we have demonstrated here that it contains at least a 3% of MLGs in extracted AIR wall fraction. The enhanced resistance to H. arabidopsidis of Arabidopsis plants pre‐treated with MLG43 clearly demonstrate that MLGs are perceived as MAMPs. In this sense, further work unveiling perception mechanisms and the specific immune pathways triggered by MLGs in different species will help to decipher their biological functions further.
In conclusion, our data expand the current knowledge of the diversity of glycan‐based molecular patterns recognized by plant immune systems, support the use of them as products for the modulation of crop immunity and anticipate that their application in agriculture could help towards the transition to a more sustainable agriculture.
Experimental procedures
Plant growth conditions
Col‐0 background lines were used for all the Arabidopsis experiments in the present work. Arabidopsis seedlings used for Ca2+ cyt (Col‐0AEQ), MAPK phosphorylation and gene expression analyses were grown in liquid MS medium and plants in soil–vermiculite (3:1) under 10 h light/14 h dark conditions at 21–20°C (Mélida et al., 2018). Tomato plants (Solanum lycopersicum, Moneymaker) were grown in a greenhouse in soil–vermiculite (3:1) under 14 h of light/10 h of dark at 24–22°C. Pepper plants (Capsicum annuum, Murano) were grown in a greenhouse in soil–vermiculite (3:1) under 14 h of light/10 h of dark at 24–19°C.
Carbohydrates used in the experiments
B‐GLU (#P‐BGBL), MLG43, MLG34, MLG443, MLG434 + 344 and hexaacetyl‐chitohexaose (chitohexaose; β‐1,4‐D‐(GlcNAc)6; #O‐CHI6) were acquired from Megazyme (Wicklow, Ireland). Lichenan from Cetraria islandica (#GLU602) was purchased from Elicityl. Equisetum arvense raw materials were kindly provided by Biosearch Life (Granada, Spain; #COPMCOLO001). MLG34′, MLG434′, MLG44343′, MLG44434′, MLG43344′, MLG34443′ and MLG3434443′ were chemically synthesized using automated glycan assembly as previously described (Dallabernardina et al., 2017). More details can be found in Table S5.
Preparation and digestion of β‐1,3/1,4‐glucan polysaccharides and oligosaccharides purification
Equisetum arvense raw materials were fine‐powdered using a kitchen blender and extracted with MeOH/CHCl3 (1:1) four times during 4 h at 4°C. A vacuum pump filtration was used to separate the soluble fraction after each step. Soluble fractions were discarded, and the insoluble residues were extracted with 70% (v/v) ethanol twice (1 h at 90°C and overnight at room temperature). The residue after filtration was then treated with distilled water twice (1 h at 90°C and overnight at room temperature). The residue after filtration was considered AIR. AIR polysaccharides were chemically extracted using 4% (w/v) KOH (E‐KOH fraction) as previously described (Mélida et al., 2009). B‐GLU, E‐KOH fraction from E. arvense and lichenan (C‐LICH) were suspended in distilled water (5 mg ml−1) containing 1.4 U ml−1 of lichenase (+L; EC 3.2.1.73; Megazyme) and stirred 72 h at 60°C. Digestion products were freeze‐dried and fractionated by SEC (140 cm3 bed volume in a 1.6 cm diameter column; Biogel P2 Extrafine; Bio‐Rad, Hercules, CA, USA) (Mélida et al., 2020). Total carbohydrates in each fraction were quantified by phenol‐sulphuric acid method (Dubois et al., 1956).
β‐1,3/1,4‐glucans content determination
For the determination of MLGs in H. arabidopsidis conidiospores, these were recovered from 24‐day‐old Arabidopsis plants inoculated (4 × 104 conidiospores ml−1) 7 days before tissue harvesting. Plant tissues were extensively washed with distilled water and a conidiospore suspension was collected. The suspension was centrifuged for 10 min at 5000 g to obtain a conidiospore pellet, upon discarding both supernatant and green upper layer of the pellet. Conidiospore pellets were homogenized and then extracted three times with 80% (v/v) ethanol for 1 h, overnight and 1 h. Air‐dried pellets after acetone washings were considered as AIRs (Pettolino et al., 2009). To determine MLGs in Arabidopsis plants inoculated (4 × 104 conidiospores ml−1) or mock‐inoculated with H. arabidopsidis, plants were extensively washed with water to release conidiospores from inoculated plants, and then plant tissues were immediately frozen with liquid nitrogen, and AIRs were prepared as described by Bacete et al. (2017). MLGs were quantified from the different AIR materials using a Mixed Linkage β‐Glucan Assay Kit (Megazyme; #K‐BGLU).
Carbohydrate analyses
Oligosaccharides were analysed by TLC and HPAEC. MLGs (5 µg) were spotted onto TLC plates (Silicagel 60; Merck, Darmstadt, Germany) and run twice using 1‐propanol/ethyl‐acetate/water (9:7:4 by volume). TLC plates were developed by using the thymol‐H2SO4 method (Mélida et al., 2020). HPAEC separations were performed using a CarboPac PA‐200 anion exchange column (4.6 × 250 mm; Dionex, Oakville, ON, Canada) mounted on a Dionex ICS 3000 HPAEC‐PAD system and a pulsed amperometric detector. Oligosaccharides were eluted at 0.5 mL min−1 using a linear saline gradient of 30 mm NaOH to 30 mm NaOH/300 mm sodium acetate over 16 min and equilibration at the initial conditions for 5 min.
Aequorin luminescence measurements
Eight‐day‐old Arabidopsis ‘aequorin‐plants’ (Col‐0AEQ; Ranf et al., 2012) were used for Ca2+ influxes measurements (Bacete et al., 2017). Dose–response curves and EED were calculated using total relative luminescence unit values (areas under kinetic curves) (Mélida et al., 2018).
ROS
H2O2 production upon elicitation was monitored on 4‐mm diameter leaf discs carefully obtained from 5‐week‐old Arabidopsis plants by using the luminol‐peroxidase method (Escudero et al., 2017).
Immunoblot analysis of MAPK activation
Arabidopsis seedlings (12‐day‐old) were treated with different oligosaccharides and distilled water (mock) for 0, 5, 10 and 20 min, and fast‐frozen with liquid nitrogen. Seedlings were homogenized using a FastPrep Bead Beating System (MP Biomedicals, Santa Ana, CA, USA) in extraction buffer (50 mm Tris‐HCl pH 7.5, 200 mm NaCl, 1 mm EDTA, 10 mm NaF, 2 mm sodium orthovanadate, 1 mm sodium molybdate, 10% (v/v) glycerol, 0.1% (v/v) Tween‐20, 1 mm 1,4‐dithiothreitol, 1 mm phenylmethylsulfonyl fluoride and phosphatase inhibitor cocktail #P9599; Sigma‐Aldrich, St. Louis, MO, USA). Total protein extracts were quantified by the Bradford assay (Bio‐Rad). Proteins (40 µg) were separated using 10% Mini‐PROTEAN TGX Precast protein gels and transferred to nitrocellulose membranes using the Invitrogen iBlot Gel Transfer Device. Membranes were blocked with Protein‐Free Blocking Buffer [Tris‐buffered saline (TBS); Thermo Fisher Scientific, Waltham, MA, USA] for 2 h at room temperature. Membranes were incubated overnight at 4°C in TBS containing phospho‐p44/42 MAPK (Erk1/2) (Thr202/Tyr204) antibody (Cell Signaling Technology, Danvers, MA, USA) (1:1000) or anti‐AtMPK3 (1:2500) and anti‐AtMPK6 (1:10 000) antibodies (Sigma‐Aldrich). Membranes were washed with TBS containing 0.1% Tween‐20 and incubated with horseradish peroxidase‐conjugated antirabbit antibody (GE Healthcare, Chicago, IL, USA) (1:5000) in TBS. Blots were finally developed using the ECL western blotting substrate (Thermo Fisher Scientific) and imaged using an iBright FL1000 Image System (Thermo Fisher Scientific). Membranes were also stained with Ponceau‐S Red (Sigma‐Aldrich).
Gene expression analyses
Twelve‐day‐old Arabidopsis seedlings were treated with different oligosaccharides and distilled water (mock) for 0 and 30 min and used for qRT‐PCR and RNA‐seq gene expression analysis. qRT‐PCR analyses were performed as described by Mélida et al. (2020). Oligonucleotides used in the analysis are shown in Table S6.
RNA‐seq analyses were performed by sequencing and analysing three biological replicates for each treatment as previously described (Mélida et al., 2020). RNA‐seq read data can be retrieved from the NCBI Sequence Read Archive (SRA) under BioProject accession ID PRJNA625401 (BioSample accession SAMN15682114). Significant (P < 0.05) enrichments were determined using the hypergeometric test with Bonferroni step down correction. To determine differentially expressed genes, t‐tests were performed for the treatments against mock values. N‐fold ≥ 2 was used to prove upregulation and an n‐fold ≤ 0.5 was applied to select downregulated genes. ClueGO 2.5.6 app for Cytoscape was used to determine which GO categories were statistically overrepresented in the differentially expressed set of genes.
Molecular dynamics simulations
Geometries for MLG43‐PRRs complexes were firstly obtained with docking calculations using the crystal structures of AtCERK1‐ECD (PDB code: 4ebz), AtBAK1‐ECD (PDB code: 4mn8) and AtSOBIR1‐ECD (PDB code: 6rih). MLG43 ligand was built and optimized in vacuum as described (del Hierro et al., 2020). Energy terms contributing to protein–ligand interactions were computed using molecular mechanics following a previously established pipeline (del Hierro et al., 2020).
Crop protection assays
For H. arabidopsidis experiments, Arabidopsis plants were grown in soil as indicated above but at a higher humidity (75%). Two‐week‐old Arabidopsis plants were treated by foliar spray using 0.1 ml of MLG43 solution in water (0.1 mg ml−1 or 0.5 mg ml−1). Two days after treatment, plants were spray‐inoculated with 0.1 ml conidiospore suspension (4 × 104 conidia ml−1) of a H. arabidopsidis isolate Noco2. H. arabidopsidis level of infection in inoculated plants was quantified at 7 dpi (24‐day‐old Arabidopsis plants) as the abundance of conidiospores per mg of plant fresh weight. Conidiospores were recovered from inoculated plants by extensively washing them with distilled water, and released conidiospores in water suspensions were counted using a Neubauer chamber, and relativized to mg of plant fresh weight.
Tomato plants (S. lycopersicum, Moneymaker) were grown in a greenhouse in soil–vermiculite (3:1) under 14 h of light/10 h of dark at 24–22°C. Three‐week‐old plants were sprayed with 2 ml of a MLG43 solution (0.125 mg ml−1) containing 2.5% UEP‐100 (Croda, Snaith, UK) and 2.5% Tween 24 MBAL (Croda) as adjuvants. Adjuvant solutions were used as mocks. Two days after treatments plants were challenged with Pseudomonas syringae pv. tomato DC3000 as described by Santamaría‐Hernando et al., (2019). Tomato leaf discs were collected from four different plants at 0 and 11 dpi and colony forming units (cfu) per foliar area were determined as described (Mélida et al., 2020).
For S. sclerotiorum and B. cinerea experiments, pepper plants (C. annuum, Murano) were grown in a greenhouse in soil–vermiculite (3:1) under 14 h of light/10 h of dark at 24–22°C. Five‐week‐old plants were treated using 5 ml of a MLG43 solution (0.05 mg ml−1; for S. sclerotiorum experiments) or 2 ml of a MLG43 solution (0.125 mg ml−1; for B. cinerea experiments) containing 0.5% UEP‐100 and 0.05% Tween 24 MBAL as adjuvants. Adjuvant solutions were used as mocks. Two days after treatment, plants were spray‐inoculated with 5 ml of 250 colony forming units ml−1 suspension of S. sclerotiorum homogenized mycelia according to Chen and Wang (2005) or with 3 ml of Gamborg's B5 medium containing 106 B. cinerea conidia (Benito et al., 1998) and moved to a greenhouse high‐humidity (75%) chamber. Disease symptoms were determined at 5 and 9 dpi in the first eight leaves of each plant (n = 24 plants for S. sclerotiorum and n = 12 for B. cinerea) using a scale from 0 to 5 where 0 = no symptoms; 1 = little necrotic spots (<10% of leaf area); 2 = two or more notable necrotic spots (10–25% of leaf area); 3 = big necrotic area (25–50% of leaf area); 4 = >50% of leaf area affected and 5 = leaf senescence. Representative images of the disease index scales used to evaluate both pathosystems are shown in Figure S10. All the disease experiments were performed at least four times and one representative experiment is shown.
Conflict of Interest
The authors declare that they have no competing interests.
Author Contributions
HM and AM initiated, conceived and coordinated all the experiments except those related to S. sclerotiorum and B. cinerea which were conceived and initiated by DR, RP and FB. DR performed the experiments described in Figures 1, 2, 3, 4, 5, 6 and Figures [Link], [Link], [Link], [Link], [Link], [Link], [Link], [Link] and S10 with help of GL, LB and HM. IdH performed the RNA‐seq data analysis and the experiments described in Table S7. FV collaborated in HPAEC experiments. PD and FP performed the MLG oligosaccharides synthesis. LJ collaborated in P. syringae experiments and performed those shown in Figure S9. AS‐V collaborated in experiments shown in Figures 5 and 6. DR and HM prepared the tables and figures. HM and AM wrote the paper. FV, FP, LJ and FB edited the paper.
Supporting information
Figure S1. Cytoplasmic calcium elevation triggered by dimer and trimer mixed‐linked glucans. Calcium influx measured as relative luminescence units (RLU) over time in 8‐day‐old Arabidopsis Col‐0AEQ seedlings after treatment with 50 µm of β‐d‐cellobiosyl‐(1,3)‐β‐d‐glucose (MLG43), β‐d‐glucosyl‐(1,3)‐β‐d‐cellobiose (MLG34), cellobiose (Cello2), cellotriose (Cello3) and laminaribiose (Lam2) and laminaritriose (Lam3). Data represent mean ± σ (n = 8). These results are from one representative experiment out of the three performed that gave similar results.
Figure S2. Thin‐layer chromatography of enzymatic hydrolysis of different mixed‐linked glucans sources with either lichenase (EC 3.2.1.73) or cellulase (EC 3.2.1.4): barley β‐glucan (B‐GLU), lichenan from Cetraria islandica (C‐LICH) and 4% KOH fraction of Equisetum arvense cell wall (E‐KOH). Enzyme dilution used and undigested substrates were loaded as controls. Markers at the right side indicate the migration of glucose, laminarin‐oligosaccharides (Lam), cello‐oligosaccharides (Cell) and MLG oligosaccharides (MLG).
Figure S3. Size exclusion chromatography elution profiles of lichenase digestion products (+L) from different β‐1,3/1,4‐glucans sources. (a) Barley β‐glucan (B‐GLU), (b) lichenan from Cetraria islandica (C‐LICH) and (c) 4% KOH fraction of Equisetum arvense cell wall (E‐KOH) were digested with lichenase (+L). Five mg of the digested fractions were loaded in the chromatography column. Data shown in all panels are representative chromatograms from one experiment of at least 10 performed that gave similar results.
Figure S4. Cross‐elicitation during the refractory period of calcium signalling upon application of 50 μm of commercial MLG43 (Megazyme), purified MLG43 (MLG43‐L) or distilled water (mock). Data show the elevation of cytoplasmic calcium concentration, measured as relative luminescence units (RLU), over time in 8‐day‐old Arabidopsis Col‐0AEQ seedlings after treatments. Arrow indicates the application time of the second elicitor within the refractory period of the first elicitation. Data represent mean ± σ (n = 4) of a representative experiment of the three independent experiments performed that gave similar results.
Figure S5. MLG43 dose response analyses. (a) Increase of cytoplasmic calcium concentration in 8‐day‐old Arabidopsis Col‐0AEQ seedlings measured as relative luminescence units (RLU) over time by increasing MLG43 concentrations (from 200 nm to 5 mm). (b) Dose dependence of total cytoplasmic calcium influxes in Arabidopsis Col‐0AEQ seedlings upon treatments with increasing MLG43 concentrations. Calcium saturation curves were adjusted by using Prism 6 Software, and the upper and lower lines represent the 95% confidence intervals. Arrow indicates the MLG43 estimated effective dose (EED = 265 μm).
Figure S6. Calcium influx kinetics triggered by synthetic mixed‐linked glucans. Calcium influx measured as relative luminescence units (RLU) over time in 8‐day‐old Arabidopsis Col‐0AEQ seedlings after treatment with 50 μm of different synthetic MLGs (a, b) or chitohexaose (a). (c) Structural scheme of the different synthetic MLG oligosaccharides used in the experiments. The oligosaccharide linker of the reducing end of the MLGs tested is shown. These data are from one representative experiment of at least three performed that gave similar results.
Figure S7. Biological process Gene Ontology (GO) term enrichment map of the overexpressed genes in 12‐day‐old Arabidopsis Col‐0 plants treated MLG43 or chitohexaose. RNA‐seq data were obtained from the combination of three biological replicates. (a) Common overexpressed genes 30 min after 50 μm MLG43 or chitohexaose treatments. (b) Overexpressed genes 30 min after 50 μm chitohexaose treatment. GO term enrichment is expressed by node size. Enrichment P‐value determined by hypergeometric test and corrected by Benjamini‐Hochberg false discovery rate is represented by colour scale. Links between groups indicates shared genes. Only GO terms with a P‐value <0.01 are shown. Links between groups indicates shared genes (κ score level ≥ 0.4).
Figure S8. Mitogen‐activated protein kinases (MAPK) phosphorylation in 12‐day‐old Arabidopsis seedlings of Col‐0 plants and cerk1‐2, bak1‐5 and sobir1‐12 mutants impaired in PRR co‐receptors treated with 50 µm MLG43. Western blot using anti‐pTEpY antibody for phosphorylated MAPK moieties at different time points (5, 10 and 20 min). Black arrows indicate the position of MPK6 (top), MPK3 (middle) and MPK4/11 (bottom) proteins. Anti‐MPK6 and anti‐MPK3 were used as total protein control. Ponceau red‐stained membranes show equal loading. Chitohexaose (5 µm) and distilled water (mock) were used as controls. These results are from one representative experiment of the three performed that gave similar results.
Figure S9. qRT‐PCR analyses of the expression of PTI‐related genes upregulated in tomato Moneymaker plants treated for 60 min with MLG43 (0.25 mg plant−1). Relative expression levels to the LOC543683 (SlUBC) gene are shown. Values are means ± SD, n = 3 from two independent experiments. Asterisks indicate treatments with significant differences compared with non‐treated control plants (Student's t‐test analysis, *P < 0.05). These results are from one of the two representative experiments performed that gave similar results.
Figure S10. Representative images of disease index scales used to evaluate pepper‐Sclerotinia sclerotiorum (left) or pepper‐Botrytis cinerea (right) pathosystems. Disease index ranges from 0 to 5, where 0 = no symptoms; 1 = little necrotic spots (<10% of leaf area); 2 = two or more notable necrotic spots (10–25% of leaf area); 3 = big necrotic area (25–50% of leaf area); 4 = more than 50% of leaf area affected and 5 = leaf senescence.
Table S1. Differentially expressed genes under treatment with MLG43 in Arabidopsis.
Table S2. Differentially expressed genes under treatment with chitohexaose in Arabidopsis.
Table S3. Classification of Arabidopsis upregulated genes into common and specific after treatment with MLG43 or chitohexaose.
Table S4. Classification of Arabidopsis downregulated genes into common and specific after treatment with MLG43 or chitohexaose.
Table S5. β‐1,3/1,4‐glucan oligosaccharides (MLGs) used in this work.
Table S6. Oligonucleotides used in this work.
Table S7. MLG43‐ectodomains interaction energies determinations.
Acknowledgements
This work was supported by grants IND2017/BIO‐7800 of the Comunidad de Madrid Regional Government, BIO2015‐64077‐R of the Spanish Ministry of Economy and Competitiveness (MINECO), RTI2018‐096975‐B‐I00 of Spanish Ministry of Science, Innovation and Universities, to AM. This work has been also financially supported by the ‘Severo Ochoa Programme for Centres of Excellence in R&D’ from the Agencia Estatal de Investigación of Spain (grant SEV‐2016‐0672 (2017–2021) to the CBGP). Within the framework of this program HM was supported with a postdoctoral fellow. DR was the recipient of an Industrial PhD Fellow (IND2017/BIO‐7800), IdH was the recipient of a PhD FPU fellow from the Spanish Ministry of Education (FPU16/07118) and ASV was the recipient of the RYC2018‐025530‐I grant of Spanish Ministry of Science, Innovation and Universities. FP thanks the Max Planck Society and the German Research Foundation (DFG, Emmy Noether program PF850/1‐1 to FP) for financial support.
These authors should be considered joint senior.
Contributor Information
Antonio Molina, Email: antonio.molina@upm.es.
Hugo Mélida, Email: h.melida@upm.es.
Data Availability Statement
RNA‐seq raw data can be obtained from BioProject PRJNA625401 (BioSample accession SAMN15682114) at the NCBI Sequence Read Archive (SRA) site. Materials and data are available upon request to corresponding authors.
REFERENCES
- Aziz, A. , Gauthier, A. , Bézier, A. , Poinssot, B. , Joubert, J.M. , Pugin, A. et al. (2007) Elicitor and resistance‐inducing activities of beta‐1,4 cellodextrins in grapevine, comparison with beta‐1,3 glucans and alpha‐1,4 oligogalacturonides. Journal of Experimental Botany, 58, 1463–1472. [DOI] [PubMed] [Google Scholar]
- Aziz, A. , Poinssot, B. , Daire, X. , Adrian, M. , Bézier, A. , Lambert, B. et al. (2003) Laminarin elicits defense responses in grapevine and induces protection against Botrytis cinerea and Plasmopara viticola . Molecular Plant‐Microbe Interactions, 16, 1118–1128. [DOI] [PubMed] [Google Scholar]
- Bacete, L. , Mélida, H. , López, G. , Dabos, P. , Tremousaygue, D. , Denancé, N. et al. (2020) Arabidopsis Response Regulator 6 (ARR6) modulates plant cell wall composition and disease resistance. Molecular Plant‐Microbe Interactions, 33, 767–780. [DOI] [PubMed] [Google Scholar]
- Bacete, L. , Mélida, H. , Miedes, E. & Molina, A. (2018) Plant cell wall‐mediated immunity: cell wall changes trigger disease resistance responses. The Plant Journal, 93, 614–636. [DOI] [PubMed] [Google Scholar]
- Bacete, L. , Mélida, H. , Pattathil, S. , Hahn, M.G. , Molina, A. & Miedes, E. (2017) Characterization of plant cell wall damage‐associated molecular patterns regulating immune responses. Methods in Molecular Biology, 1578, 13–23. [DOI] [PubMed] [Google Scholar]
- Bartetzko, M.P. & Pfrengle, F. (2019) Automated glycan assembly of plant oligosaccharides and their application in cell‐wall biology. ChemBioChem, 20, 877–885. [DOI] [PubMed] [Google Scholar]
- Benedetti, M. , Pontiggia, D. , Raggi, S. , Cheng, Z. , Scaloni, F. , Ferrari, S. et al. (2015) Plant immunity triggered by engineered in vivo release of oligogalacturonides, damage‐associated molecular patterns. Proceedings of the National Academy of Sciences of the United States of America, 112, 5533–5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito, E.P. , ten Have, A. , van't Klooster, J.W. and van Kan, J.A.L. (1998) Fungal and plant gene expression during synchronized infection of tomato leaves by Botrytis cinerea . European Journal of Plant Pathology, 104, 207–220. [Google Scholar]
- Bigeard, J. , Colcombet, J. & Hirt, H. (2015) Signaling mechanisms in pattern‐triggered immunity (PTI). Molecular Plant, 8, 521–539. [DOI] [PubMed] [Google Scholar]
- Bjornson, M. , Pimprikar, P. , Nürnberger, T. & Zipfel, C. (2020). The transcriptional landscape of Arabidopsis thaliana pattern‐triggered immunity. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq, M. , Willmann, M.R. , McCormack, M. , Lee, H. , Shan, L. , He, P. et al. (2010) Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature, 464, 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot, F. & Zipfel, C. (2017) Function, discovery, and exploitation of plant pattern recognition receptors for broad‐spectrum disease resistance. Annual review of Phytopathology, 55, 257–286. [DOI] [PubMed] [Google Scholar]
- Burton, R.A. & Fincher, G.B. (2009) (1,3;1,4)‐β‐D‐glucans in cell walls of the poaceae, lower plants, and fungi: a tale of two linkages. Molecular Plant, 2, 873–882. [DOI] [PubMed] [Google Scholar]
- Burton, R.A. , Wilson, S.M. , Hrmova, M. , Harvey, A.J. , Shirley, N.J. , Medhurst, A. et al. (2006) Cellulose synthase‐like CslF genes mediate the synthesis of cell wall (1,3;1,4)‐β‐D‐glucans. Science, 311, 1940–1942. [DOI] [PubMed] [Google Scholar]
- Cao, Y. , Liang, Y. , Tanaka, K. , Nguyen, C.T. , Jedrzejczak, R.P. , Joachimiak, A. et al. (2014). The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin‐induced complex with related kinase CERK1. eLife, 3, e03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. & Wang, D. (2005) Two convenient methods to evaluate soybean for resistance to Sclerotinia sclerotiorum . Plant Disease, 89, 1268–1272. [DOI] [PubMed] [Google Scholar]
- Claverie, J. , Balacey, S. , Lemaître‐Guillier, C. , Brulé, D. , Chiltz, A. , Granet, L. et al. (2018) The cell wall‐derived xyloglucan is a new DAMP triggering plant immunity in Vitis vinifera and Arabidopsis thaliana . Frontiers in Plant Science, 9, 1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallabernardina, P. , Schuhmacher, F. , Seeberger, P.H. & Pfrengle, F. (2017) Mixed‐linkage glucan oligosaccharides produced by automated glycan assembly serve as tools to determine the substrate specificity of lichenase. Chemistry, 23, 3191–3196. [DOI] [PubMed] [Google Scholar]
- del Hierro, I. , Mélida, H. , Broyart, C. , Santiago, J. & Molina, A. (2020) Computational prediction method to decipher receptor‐glycoligand interactions in plant immunity. The Plant Journal, 105. 10.1111/tpj.15133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desaki, Y. , Kouzai, Y. , Ninomiya, Y. , Iwase, R. , Shimizu, Y. , Seko, K. et al. (2018) OsCERK1 plays a crucial role in the lipopolysaccharide‐induced immune response of rice. New Phytologist, 217, 1042–1049. [DOI] [PubMed] [Google Scholar]
- DuBois, M. , Gilles, K.A. , Hamilton, J.K. , Rebers, P.A. & Smith, F. (1956) Colorimetric method for determination of sugars and related substances. Analytical Chemistry, 28, 350–356. [Google Scholar]
- Escudero, V. , Jordá, L. , Sopeña‐Torres, S. , Mélida, H. , Miedes, E. , Muñoz‐Barrios, A. et al. (2017) Alteration of cell wall xylan acetylation triggers defense responses that counterbalance the immune deficiencies of plants impaired in the β‐subunit of the heterotrimeric G‐protein. The Plant Journal, 92, 386–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari, S. , Savatin, D.V. , Sicilia, F. , Gramegna, G. , Cervone, F. & Lorenzo, G.D. (2013) Oligogalacturonides: plant damage‐associated molecular patterns and regulators of growth and development. Frontiers in Plant Science, 4, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fesel, P.H. & Zuccaro, A. (2016) β‐glucan: crucial component of the fungal cell wall and elusive MAMP in plants. Fungal Genetics and Biology, 90, 53–60. [DOI] [PubMed] [Google Scholar]
- Fincher, G.B. & Stone, B.A. (2004) Chemistry of nonstarch polysaccharides. In: Wrigley, C. , Corke, H. and Walker, C.E. (Eds.) Encyclopedia of grain science. Oxford, UK: Elsevier, pp. 206–223. [Google Scholar]
- Fontaine, T. , Simenel, C. , Dubreucq, G. , Adam, O. , Delepierre, M. , Lemoine, J. et al. (2000) Molecular organization of the alkali‐insoluble fraction of Aspergillus fumigatus cell wall. Journal of Biological Chemistry, 275, 27594–27607. [DOI] [PubMed] [Google Scholar]
- Fry, S.C. , Nesselrode, B.H.W.A. , Miller, J.G. & Mewburn, B.R. (2008) Mixed‐linkage (1,3;1,4)‐β‐D‐glucan is a major hemicellulose of Equisetum (horsetail) cell walls. New Phytologist, 179, 104–115. [DOI] [PubMed] [Google Scholar]
- Galletti, R. , Denoux, C. , Gambetta, S. , Dewdney, J. , Ausubel, F.M. , De Lorenzo, G. et al. (2008) The AtrbohD‐mediated oxidative burst elicited by oligogalacturonides in Arabidopsis is dispensable for the activation of defense responses effective against Botrytis cinerea . Plant Physiology, 148, 1695–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan, I. , Steinberg, G. & Gurr, S. (2017) The role of the fungal cell wall in the infection of plants. Trends in Microbiology, 25, 957–967. [DOI] [PubMed] [Google Scholar]
- Gómez‐Gómez, L. & Boller, T. (2000) FLS2: an LRR receptor‐like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Molecular Cell, 5, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Gorin, P. , Baron, M. & Iacomini, M. (1988) Storage products of lichens. In: Galun, M. (Ed.) Handbook of Lichenology. Boca Raton, FL, USA: CRC Press, pp. 9–23. [Google Scholar]
- Gust, A.A. , Biswas, R. , Lenz, H.D. , Rauhut, T. , Ranf, S. , Kemmerling, B. et al. (2007) Bacteria‐derived peptidoglycans constitute pathogen‐associated molecular patterns triggering innate immunity in Arabidopsis. Journal of Biological Chemistry, 282, 32338–32348. [DOI] [PubMed] [Google Scholar]
- Henrissat, B. & Bairoch, A. (1993) New families in the classification of Glycosyl hydrolases based on amino acid sequence similarities. The Biochemical Journal, 293, 781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, J.M. , Thürich, J. , Petutschnig, E.K. , Altschmied, L. , Meichsner, D. , Sherameti, I. et al. (2018) Poly(A) ribonuclease controls the cellotriose‐based interaction between Piriformospora indica and its host Arabidopsis. Plant Physiology, 176, 2496–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaku, H. , Nishizawa, Y. , Ishii‐Minami, N. , Akimoto‐Tomiyama, C. , Dohmae, N. , Takio, K. et al. (2006) Plant cells recognize chitin fragments for defense signalling through a plasma membrane receptor. Proceedings of the National Academy of Sciences of the United States of America, 103, 11086–11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarzynski, O. , Plesse, B. , Joubert, J.M. , Yvin, J.C. , Kopp, M. , Kloareg, B. et al. (2000) Linear β‐1,3 glucans are elicitors of defense responses in tobacco. Plant Physiology, 124, 1027–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampugnani, E.R. , Khan, G.A. , Somssich, M. & Persson, S. (2018) Building a plant cell wall at a glance. Journal of Cell Science, 131(2), jcs207373. [DOI] [PubMed] [Google Scholar]
- Latgé, J.P. & Calderone, R. (2006). The fungal cell wall. In: Kües, U. & Fischer, R. (Eds). The Mycota I. Growth, Differentiation and Sexuality. Heidelberg, Germany: Springer Berlin, pp. 73–104. [Google Scholar]
- Lee, J. & Hollingsworth, R.I. (1997) Oligosaccharide beta‐glucans with unusual linkages from Sarcina ventriculi . Carbohydrate Research, 304, 133–141. [DOI] [PubMed] [Google Scholar]
- Liu, D. , Lu, J. , Li, H. , Wang, J. & Pei, Y. (2019) Characterization of the O‐acetylserine(thiol)lyase gene family in Solanum lycopersicum L. Plant Molecular Biology, 99, 123–134. [DOI] [PubMed] [Google Scholar]
- Liu, T. , Liu, Z. , Song, C. , Hu, Y. , Han, Z. , She, J. et al. (2012) Chitin‐induced dimerization activates a plant immune receptor. Science, 336, 1160–1164. [DOI] [PubMed] [Google Scholar]
- Locci, F. , Benedetti, M. , Pontiggia, D. , Citterico, M. , Caprari, C. , Mattei, B. et al. (2019) An Arabidopsis berberine bridge enzyme‐like protein specifically oxidizes cellulose oligomers and plays a role in immunity. The Plant Journal, 98, 540–554. [DOI] [PubMed] [Google Scholar]
- McIntosh, M. , Stone, B.A. & Stanisich, V.A. (2005) Curdlan and other bacterial (1–>3)‐β‐D‐glucans. Applied Microbiology and Biotechnology, 68, 163–173. [DOI] [PubMed] [Google Scholar]
- Mélida, H. , Bacete, L. , Ruprecht, C. , Rebaque, R. , del Hierro, I. , López, G. et al. (2020) Arabinoxylan‐oligosaccharides act as Damage Associated Molecular Patterns in plants regulating disease resistance. Frontiers in Plant Science, 11, 1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mélida, H. , García‐Angulo, P. , Alonso‐Simón, A. , Encina, A. , Alvarez, J. & Acebes, J.L. (2009) Novel type II cell wall architecture in dichlobenil‐habituated maize calluses. Planta, 229, 617–631. [DOI] [PubMed] [Google Scholar]
- Mélida, H. , Sandoval‐Sierra, J.V. , Diéguez‐Uribeondo, J. & Bulone, V. (2013) Analyses of extracellular carbohydrates in oomycetes unveil the existence of three different cell wall types. Eukaryotic Cell, 12, 194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mélida, H. , Sopeña‐Torres, S. , Bacete, L. , Garrido‐Arandia, M. , Jordá, L. , López, G. et al. (2018) Non‐branched β‐1,3‐glucan oligosaccharides trigger immune responses in Arabidopsis. The Plant Journal, 93, 34–49. [DOI] [PubMed] [Google Scholar]
- Ménard, R. , Alban, S. , de Ruffray, P. , Jamois, F. , Franz, G. , Fritig, B. et al. (2004) β‐1,3 glucan sulfate, but not β‐1,3 glucan, induces the salicylic acid signaling pathway in tobacco and Arabidopsis. The Plant Cell, 16, 3020–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya, A. , Albert, P. , Shinya, T. , Desaki, Y. , Ichimura, K. , Shirasu, K. et al. (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 104, 19613–19618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Mendoza, D. , Rodríguez‐Carvajal, M.Á. , Romero‐Jiménez, L. , Farias Gde, A. , Lloret, J. , Gallegos, M.T. et al. (2015) Novel mixed‐linkage β‐glucan activated by c‐di‐GMP in Sinorhizobium meliloti. Proceedings of the National Academy of Sciences of the United States of America, 112, E757–E765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlin, A.S. & Suzuki, S. (1962) The structure of lichenin: selective enzymolysis studies. Canadian Journal of Botany, 40, 50–56. [Google Scholar]
- Perraki, A. , DeFalco, T.A. , Derbyshire, P. , Avila, J. , Séré, D. , Sklenar, J. et al. (2018) Phosphocode‐dependent functional dichotomy of a common co‐receptor in plant signalling. Nature, 561, 248–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettolino, F. , Sasaki, I. , Turbic, A. , Wilson, S.M. , Bacic, A. , Hrmova, M. et al. (2009) Hyphal cell walls from the plant pathogen Rhynchosporium secalis contain (1,3;1,6)‐β‐D‐glucans, galacto‐ and rhamnomannans, (1,3;1,4)‐β‐D‐glucans and chitin. FEBS Journal, 276, 4122–4133. [DOI] [PubMed] [Google Scholar]
- Planas, A. (2000) Bacterial 1,3–1,4‐β‐glucanases: structure, function and protein engineering. Biochimica et Biophysica Acta, 1543, 361–382. [DOI] [PubMed] [Google Scholar]
- Popper, Z.A. & Fry, S.C. (2003) Primary cell wall composition of bryophytes and charophytes. Annals of Botany, 91, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranf, S. , Eschen‐Lippold, L. , Pecher, P. , Lee, J. & Scheel, D. (2011) Interplay between calcium signalling and early signalling elements during defence responses to microbe‐ or damage‐associated molecular patterns. The Plant Journal, 68, 100–113. [DOI] [PubMed] [Google Scholar]
- Ranf, S. , Grimmer, J. , Pöschl, Y. , Pecher, P. , Chinchilla, D. , Scheel, D. et al. (2012) Defense‐related calcium signaling mutants uncovered via a quantitative high‐throughput screen in Arabidopsis thaliana . Molecular Plant, 5, 115–130. [DOI] [PubMed] [Google Scholar]
- Rovenich, H. , Zuccaro, A. & Thomma, B.P. (2016) Convergent evolution of filamentous microbes towards evasion of glycan‐triggered immunity. New Phytologist, 212, 896–901. [DOI] [PubMed] [Google Scholar]
- Rui, Y. & Dinneny, J.R. (2020) A wall with integrity: surveillance and maintenance of the plant cell wall under stress. New Phytologist, 225, 1428–1439. [DOI] [PubMed] [Google Scholar]
- Salmeán, A.A. , Duffieux, D. , Harholt, J. , Qin, F. , Michel, G. , Czjzek, M. et al. (2017) Insoluble (1 → 3), (1 → 4)‐β‐D‐glucan is a component of cell walls in brown algae (Phaeophyceae) and is masked by alginates in tissues. Scientific Reports, 7, 2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samar, D. , Kieler, J.B. & Klutts, J.S. (2015) Identification and deletion of Tft1, a predicted glycosyltransferase necessary for cell wall β‐1,3;1,4‐glucan synthesis in Aspergillus fumigatus . PLoS ONE, 10, e0117336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaría‐Hernando, S. , Senovilla, M. , González‐Mula, A. , Martínez‐García, P.M. , Nebreda, S. , Rodríguez‐Palenzuela, P. et al. (2019) The Pseudomonas syringae pv. tomato DC3000 PSPTO_0820 multidrug transporter is involved in resistance to plant antimicrobials and bacterial survival during tomato plant infection. PLoS ONE, 14, e0218815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, J.K. , McNeil, M. & Albersheim, P. (1984) The primary structures of one elicitor‐active and seven elicitor‐inactive hexa(β‐D‐glucopyranosyl)‐D‐glucitols isolated from the mycelial walls of Phytophthora megasperma f. sp. glycinea. Journal of Biological Chemistry, 259, 11321–11336. [PubMed] [Google Scholar]
- Simmons, T.J. , Uhrín, D. , Gregson, T. , Murray, L. , Sadler, I.H. & Fry, S.C. (2013) An unexpectedly lichenase‐stable hexasaccharide from cereal, horsetail and lichen mixed‐linkage β‐glucans (MLGs): implications for MLG subunit distribution. Phytochemistry, 95, 322–332. [DOI] [PubMed] [Google Scholar]
- Smith, B.G. & Harris, P.J. (1999) The polysaccharide composition of Poales cell walls: Poaceae cell walls are not unique. Biochemical Systematics and Ecology, 27, 33–53. [Google Scholar]
- Sørensen, I. , Pettolino, F.A. , Wilson, S.M. , Doblin, M.S. , Johansen, B. , Bacic, A. et al. (2008) Mixed‐linkage (1,3;1,4)‐β‐D‐glucan is not unique to the poales and is an abundant component of Equisetum arvense cell walls. The Plant Journal, 54, 510–521. [DOI] [PubMed] [Google Scholar]
- Souza, C. , Li, S. , Lin, A.Z. , Boutrot, F. , Grossmann, G. , Zipfel, C. et al. (2017) Cellulose‐derived oligomers act as damage‐associated molecular patterns and trigger defense‐like responses. Plant Physiology, 173, 2383–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava, V. , McKee, L.S. & Bulone, V. (2017) Plant Cell Walls. eLS. Chichester, UK: John Wiley & Sons Ltd. [Google Scholar]
- Stone, B.A. & Clarke, A.E. (1992) Chemistry and biology of (1,3)‐β‐D‐glucans. Victoria, Australia: La Trobe University Press. [Google Scholar]
- Tang, D. , Wang, G. & Zhou, J.M. (2017) Receptor kinases in plant‐pathogen interactions: more than pattern recognition. The Plant Cell, 29, 618–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trethewey, J.A.K. , Campbell, L.M. & Harris, P.J. (2005) (1,3;1,4)‐β‐D‐Glucans in the cell walls of the poales (sensu lato): an immunogold labeling study using a monoclonal antibody. American Journal of Botany, 92, 1660–1674. [DOI] [PubMed] [Google Scholar]
- van der Burgh, A.M. , Postma, J. , Robatzek, S. & Joosten, M.H.A.J. (2019) Kinase activity of SOBIR1 and BAK1 is required for immune signalling. Molecular Plant Pathology, 20, 410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanke, A. , Rovenich, H. , Schwanke, F. , Velte, S. , Becker, S. , Hehemann, J.H. et al. (2020) Plant species‐specific recognition of long and short β‐1,3‐linked glucans is mediated by different receptor systems. The Plant Journal, 102, 1142–1156. [DOI] [PubMed] [Google Scholar]
- Wawra, S. , Fesel, P. , Widmer, H. , Timm, M. , Seibel, J. , Leson, L. et al. (2016) The fungal‐specific β‐glucan‐binding lectin FGB1 alters cell‐wall composition and suppresses glucan‐triggered immunity in plants. Nature Communications, 27, 13188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann, R. , Lajunen, H.M. , Erbs, G. , Newman, M.‐A. , Kolb, D. , Tsuda, K. et al. (2011) Arabidopsis lysin‐motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proceedings of the National Academy of Sciences of the United States of America, 108, 19824–19829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, Y. , Pearce, G. & Ryan, C.A. (2006) The cell surface leucine‐rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proceedings of the National Academy of Sciences of the United States of America, 103, 10104–10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zablackis, E. , Huang, J. , Müller, B. , Darvill, A.G. & Albersheim, P. (1995) Characterization of the cell‐wall polysaccharides of Arabidopsis thaliana leaves. Plant Physiology, 107, 1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang, H. , Xie, S. , Zhu, B. , Yang, X. , Gu, C. , Hu, B. et al. (2019) Mannan oligosaccharides trigger multiple defence responses in rice and tobacco as a novel danger‐associated molecular pattern. Molecular Plant Pathology, 20, 1067–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Cytoplasmic calcium elevation triggered by dimer and trimer mixed‐linked glucans. Calcium influx measured as relative luminescence units (RLU) over time in 8‐day‐old Arabidopsis Col‐0AEQ seedlings after treatment with 50 µm of β‐d‐cellobiosyl‐(1,3)‐β‐d‐glucose (MLG43), β‐d‐glucosyl‐(1,3)‐β‐d‐cellobiose (MLG34), cellobiose (Cello2), cellotriose (Cello3) and laminaribiose (Lam2) and laminaritriose (Lam3). Data represent mean ± σ (n = 8). These results are from one representative experiment out of the three performed that gave similar results.
Figure S2. Thin‐layer chromatography of enzymatic hydrolysis of different mixed‐linked glucans sources with either lichenase (EC 3.2.1.73) or cellulase (EC 3.2.1.4): barley β‐glucan (B‐GLU), lichenan from Cetraria islandica (C‐LICH) and 4% KOH fraction of Equisetum arvense cell wall (E‐KOH). Enzyme dilution used and undigested substrates were loaded as controls. Markers at the right side indicate the migration of glucose, laminarin‐oligosaccharides (Lam), cello‐oligosaccharides (Cell) and MLG oligosaccharides (MLG).
Figure S3. Size exclusion chromatography elution profiles of lichenase digestion products (+L) from different β‐1,3/1,4‐glucans sources. (a) Barley β‐glucan (B‐GLU), (b) lichenan from Cetraria islandica (C‐LICH) and (c) 4% KOH fraction of Equisetum arvense cell wall (E‐KOH) were digested with lichenase (+L). Five mg of the digested fractions were loaded in the chromatography column. Data shown in all panels are representative chromatograms from one experiment of at least 10 performed that gave similar results.
Figure S4. Cross‐elicitation during the refractory period of calcium signalling upon application of 50 μm of commercial MLG43 (Megazyme), purified MLG43 (MLG43‐L) or distilled water (mock). Data show the elevation of cytoplasmic calcium concentration, measured as relative luminescence units (RLU), over time in 8‐day‐old Arabidopsis Col‐0AEQ seedlings after treatments. Arrow indicates the application time of the second elicitor within the refractory period of the first elicitation. Data represent mean ± σ (n = 4) of a representative experiment of the three independent experiments performed that gave similar results.
Figure S5. MLG43 dose response analyses. (a) Increase of cytoplasmic calcium concentration in 8‐day‐old Arabidopsis Col‐0AEQ seedlings measured as relative luminescence units (RLU) over time by increasing MLG43 concentrations (from 200 nm to 5 mm). (b) Dose dependence of total cytoplasmic calcium influxes in Arabidopsis Col‐0AEQ seedlings upon treatments with increasing MLG43 concentrations. Calcium saturation curves were adjusted by using Prism 6 Software, and the upper and lower lines represent the 95% confidence intervals. Arrow indicates the MLG43 estimated effective dose (EED = 265 μm).
Figure S6. Calcium influx kinetics triggered by synthetic mixed‐linked glucans. Calcium influx measured as relative luminescence units (RLU) over time in 8‐day‐old Arabidopsis Col‐0AEQ seedlings after treatment with 50 μm of different synthetic MLGs (a, b) or chitohexaose (a). (c) Structural scheme of the different synthetic MLG oligosaccharides used in the experiments. The oligosaccharide linker of the reducing end of the MLGs tested is shown. These data are from one representative experiment of at least three performed that gave similar results.
Figure S7. Biological process Gene Ontology (GO) term enrichment map of the overexpressed genes in 12‐day‐old Arabidopsis Col‐0 plants treated MLG43 or chitohexaose. RNA‐seq data were obtained from the combination of three biological replicates. (a) Common overexpressed genes 30 min after 50 μm MLG43 or chitohexaose treatments. (b) Overexpressed genes 30 min after 50 μm chitohexaose treatment. GO term enrichment is expressed by node size. Enrichment P‐value determined by hypergeometric test and corrected by Benjamini‐Hochberg false discovery rate is represented by colour scale. Links between groups indicates shared genes. Only GO terms with a P‐value <0.01 are shown. Links between groups indicates shared genes (κ score level ≥ 0.4).
Figure S8. Mitogen‐activated protein kinases (MAPK) phosphorylation in 12‐day‐old Arabidopsis seedlings of Col‐0 plants and cerk1‐2, bak1‐5 and sobir1‐12 mutants impaired in PRR co‐receptors treated with 50 µm MLG43. Western blot using anti‐pTEpY antibody for phosphorylated MAPK moieties at different time points (5, 10 and 20 min). Black arrows indicate the position of MPK6 (top), MPK3 (middle) and MPK4/11 (bottom) proteins. Anti‐MPK6 and anti‐MPK3 were used as total protein control. Ponceau red‐stained membranes show equal loading. Chitohexaose (5 µm) and distilled water (mock) were used as controls. These results are from one representative experiment of the three performed that gave similar results.
Figure S9. qRT‐PCR analyses of the expression of PTI‐related genes upregulated in tomato Moneymaker plants treated for 60 min with MLG43 (0.25 mg plant−1). Relative expression levels to the LOC543683 (SlUBC) gene are shown. Values are means ± SD, n = 3 from two independent experiments. Asterisks indicate treatments with significant differences compared with non‐treated control plants (Student's t‐test analysis, *P < 0.05). These results are from one of the two representative experiments performed that gave similar results.
Figure S10. Representative images of disease index scales used to evaluate pepper‐Sclerotinia sclerotiorum (left) or pepper‐Botrytis cinerea (right) pathosystems. Disease index ranges from 0 to 5, where 0 = no symptoms; 1 = little necrotic spots (<10% of leaf area); 2 = two or more notable necrotic spots (10–25% of leaf area); 3 = big necrotic area (25–50% of leaf area); 4 = more than 50% of leaf area affected and 5 = leaf senescence.
Table S1. Differentially expressed genes under treatment with MLG43 in Arabidopsis.
Table S2. Differentially expressed genes under treatment with chitohexaose in Arabidopsis.
Table S3. Classification of Arabidopsis upregulated genes into common and specific after treatment with MLG43 or chitohexaose.
Table S4. Classification of Arabidopsis downregulated genes into common and specific after treatment with MLG43 or chitohexaose.
Table S5. β‐1,3/1,4‐glucan oligosaccharides (MLGs) used in this work.
Table S6. Oligonucleotides used in this work.
Table S7. MLG43‐ectodomains interaction energies determinations.
Data Availability Statement
RNA‐seq raw data can be obtained from BioProject PRJNA625401 (BioSample accession SAMN15682114) at the NCBI Sequence Read Archive (SRA) site. Materials and data are available upon request to corresponding authors.


