SUMMARY
The patterns of neuronal connectivity underlying multisensory integration, a fundamental property of many brains, remain poorly characterized. The Drosophila melanogaster mushroom body—an associative center—is an ideal system to investigate how different sensory channels converge in higher order brain centers. The neurons connecting the mushroom body to the olfactory system have been described in great detail, but input from other sensory systems remains poorly defined. Here, we use a range of anatomical and genetic techniques to identify two types of input neuron that connect visual processing centers—the lobula and the posterior lateral protocerebrum—to the dorsal accessory calyx of the mushroom body. Together with previous work that described a pathway conveying visual information from the medulla to the ventral accessory calyx of the mushroom body, our study defines a second, parallel pathway that is anatomically poised to convey information from the visual system to the dorsal accessory calyx.
In Brief
Li et al. find that the dorsal accessory calyx of the Drosophila melanogaster mushroom body integrates visual input from the lobula and the posterior lateral protocerebrum.
Graphical Abstract
INTRODUCTION
Sensory systems use different strategies to detect specific physical features of the outside world. For instance, the olfactory system contains many different types of sensory neuron that are each specialized in detecting a specific class of volatile chemicals. Through only two neuronal layers, olfactory information—the identity of an odor and its concentration—is relayed to higher brain centers (Leinwand and Chalasani, 2011). In contrast, the visual system contains far fewer types of sensory neuron, but through numerous neuronal layers, it relays a range of highly processed information—for instance, color, brightness, motion, and shape—to higher brain centers (Baden et al., 2020). Thus, higher brain centers have to integrate different types of processed information, bind that information into a coherent representation of the outside world, and use such representations to guide behavior (Yau et al., 2015). How higher brain centers achieve this feat remains largely unknown. This gap in our knowledge mainly stems from the fact that higher brain centers are formed by a large number of neurons and that the projection neurons conveying information from different sensory systems to these centers often remain poorly characterized. This makes it difficult to understand whether there are specific patterns of neuronal connectivity that enable multisensory integration and what the nature of these patterns are. Deciphering the fundamental neuronal mechanisms that underlie multisensory integration requires a model system such as the Drosophila melanogaster mushroom body, which consists of a relatively small number of neurons whose connections can be charted reliably.
The Drosophila mushroom body is formed by ∼2,000 neurons—called the Kenyon cells—and has long been studied for its essential role in the formation of olfactory associative memories (Aso et al., 2014; Hige, 2018). The identity of the projection neurons that connect the olfactory system to the mushroom body—and the way Kenyon cells integrate input from these neurons—has been characterized in great detail, highlighting fundamental connectivity patterns that enable this higher brain center to represent olfactory information efficiently (Bates et al., 2020; Caron et al., 2013; Tanaka et al., 2012a, 2012b; Zheng et al., 2018). Evidence in Drosophila melanogaster shows that the mushroom body is more than an olfactory center, as it is also required for the formation of visual and gustatory associative memories (Liu et al., 1999; Masek and Scott, 2010; Vogt et al., 2014). However, the identity of the neurons that connect the mushroom body to other sensory systems remains poorly characterized. Thus, a first step toward understanding how the mushroom body integrates multisensory information is to identify such non-olfactory mushroom body input neurons and the genetic tools necessary to manipulate these neurons.
The mushroom body receives its input through its calyx and sends its output through its lobes. The calyx—a morphologically distinct neuropil containing the synapses formed between projection neurons and Kenyon cells—can be divided into four, non-overlapping regions: one main calyx as well as three accessory calyces named the dorsal, lateral, and ventral accessory calyces (Aso et al., 2014; Yagi et al., 2016). The five output lobes—the α, α’, β, β’, and γ lobes—contain the synapses formed between Kenyon cells, mushroom body output neurons, and dopaminergic neurons (Aso et al., 2014). With respect to these input and output regions, Kenyon cells can be divided into seven distinct types (Aso et al., 2014). Of these seven types, five types—the α/βc, α/βs, α’/β’ap, α’/β’m, and γmain Kenyon cells—extend their dendrites only into the main calyx and their axons along one or two lobes. Most of the neurons that project to the main calyx emerge from the antennal lobe, the primary olfactory center in the Drosophila brain. Thus, α/βc, α/βs, α’/β’ap, α’/β’m, and γmain Kenyon cells receive input primarily from the olfactory system (Caron et al., 2013; Aso et al., 2014; Zheng et al., 2018).
In contrast, the two other classes of Kenyon cells do not extend their dendrites into the main calyx. Instead, the α/βp Kenyon cells extend their dendrites into the dorsal accessory calyx—avoiding completely the main, lateral, and ventral accessory calyces—and their axons along the α and β lobes. Likewise, the γd Kenyon cells extend their dendrites exclusively into the ventral accessory calyx and their axons along the γ lobe (Aso et al., 2014; Vogt et al., 2016). Thus, both the α/βp and γd Kenyon cells are anatomically poised to receive non-olfactory input. There is evidence suggesting that the ventral accessory calyx receives input from the medulla, a region of the optic lobe that specializes in processing brightness and color (Morante and Desplan, 2008; Vogt et al., 2016). Furthermore, a recent study suggests that the dorsal accessory calyx is a multisensory center that integrates input from multiple sensory pathways, including the olfactory, gustatory, and visual systems (Yagi et al., 2016).
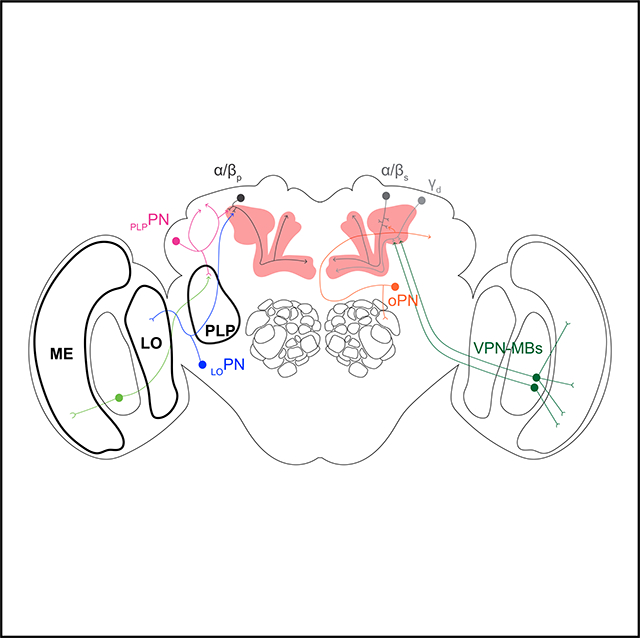
Here, we report a strategy that uses a combination of genetic tools—including transgenic lines that drive expression in few neurons and a photo-labeling technique used to identify individual neurons and their pre-synaptic partners—to characterize the input neurons of the α/βp Kenyon cells. We identify two types of mushroom body input neuron that, together, form about half of the total input the α/βp Kenyon cells receive in the dorsal accessory calyx. The first neuronal type—henceforth referred to as LOPNs—consists of a neuron that projects from the lobula, a region of the optic lobe specialized in detecting visual features, such as shape and motion. The second type of neuron—henceforth referred to as PLPPNs—consists of projection neurons that emerge from the posterior lateral protocerebrum, a brain region that receives input from the optic lobe (Otsuna and Ito, 2006; Keleş and Frye, 2017; Wu et al., 2016). Interestingly, LOPN and PLPPNs do not project to the ventral accessory calyx and do not connect to the γd Kenyon cells. Based on these findings, we conclude that there are two parallel pathways that convey visual information to the mushroom body: a pathway projecting from the medulla to the γd Kenyon cells and another pathway projecting from the lobula and posterior lateral protocerebrum to the α/βp Kenyon cells.
RESULTS
Neurons Projecting to the Dorsal Accessory Calyx Emerge from Different Brain Regions
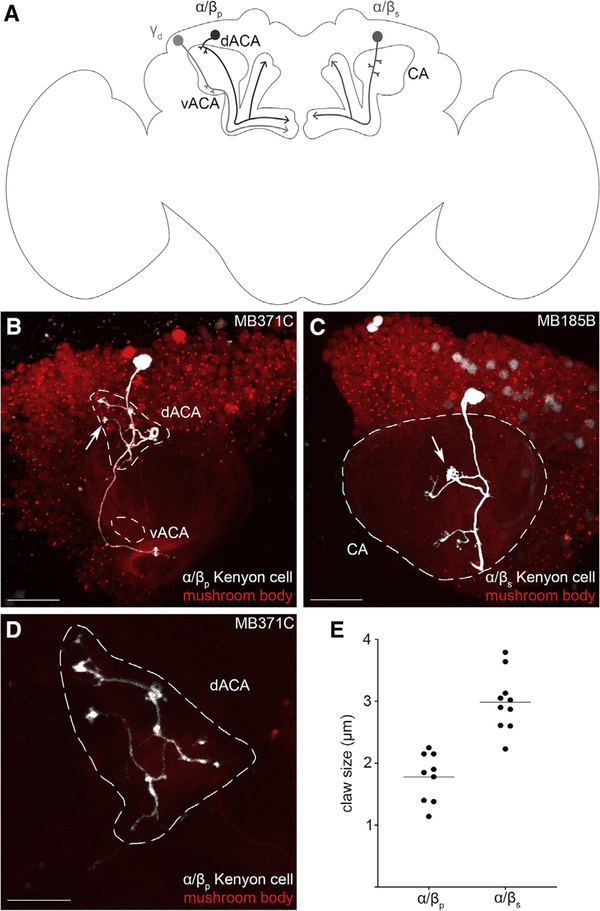
The dorsal accessory calyx is a neuropil formed from the synapses connecting ~90 α/βp Kenyon cells to their input neurons (Figure 1A; Aso et al., 2014). Using transgenic lines that drive expression specifically in the α/βp Kenyon cells (the R85D07-GAL4DBD, R13F02-GAL4AD transgenic line also known as the MB371-GAL4) and transgenic lines that express a photo-activatable form of green fluorescent protein (GFP) or PA-GFP (a combination of the UAS-C3PA-GFP and UAS-SPA-GFP transgenic lines, henceforth referred to as UAS-PA-GFP), we photo-labeled individual α/βp Kenyon cells similarly as described in a previous study (Aso et al., 2014; Datta et al., 2008; Ruta et al., 2010). We found that individual α/βp Kenyon cells extend on average 5 ± 1 claw-shaped dendritic terminals (n = 13) exclusively into the dorsal accessory calyx (Figures 1B and 1D) and project their axons along the α and β lobes (not depicted). The overall morphology of α/βp Kenyon cells is similar to the morphology of other types of α/β Kenyon cell; for instance, individual α/βs Kenyon cells extend on average 6 ± 1 claw-shaped dendritic terminals (n = 12) exclusively in the main calyx (Figure 1C) and project their axons along the α and β lobes (not depicted). It is worth noting that the claws formed by the α/βp Kenyon cells are much smaller in diameter (1.8 ± 0.4 μm; n = 9 claws) than the claws formed by the α/βs Kenyon cells (3.0 ± 0.4 μm; n = 10 claws; Figures 1B, 1C [white arrows], and 1E). It is also worth noting that the border of the dorsal accessory calyx is looser than the compact and well-defined circular border of the main calyx. Individual α/βp Kenyon cells can extend dendrites further away from the core of the dorsal accessory calyx, resulting in an irregularly shaped calyx (Figure 1D). Individual α/βp Kenyon cells are morphologically distinct, much like the other Kenyon cells. Thus, the α/βp Kenyon cells resemble α/βs and α/βc Kenyon cells but also differ from them in one major way: the dendrites of the α/βs and α/βc Kenyon cells exclusively innervate the main calyx—a region known to receive most of its input from the olfactory system—whereas dendrites of the α/βp Kenyon cells exclusively innervate the dorsal accessory calyx—a poorly characterized region of the mushroom body.
Figure 1. The Dorsal Accessory Calyx of the Drosophila melanogaster Mushroom Body as Defined by the Claw-Shaped Dendrites of the α/βp Kenyon Cells.
(A) A schematic of the Drosophila brain shows three types of Kenyon cell associated with the different mushroom body calyces: the α/βp Kenyon cells (dark gray, left) extend their claw-shaped dendrites in the dorsal accessory calyx (dACA); the α/βs (medium gray, right)—as well as the α’/β’ap, α’/β’m, and γmain Kenyon cells (not depicted)—extend their claw-shaped dendrites in the main calyx (CA); and the γd Kenyon cells (light gray, left) extend their claw-shaped dendrites in the ventral accessory calyx (vACA). All Kenyon cells extend their axons along one or two lobes (arrowheads).
(B and C) A single α/βp (B) or α/βs (C) Kenyon cell (white) of the mushroom body (red) was photo-labeled; (B) the photo-labeled α/βp Kenyon cell extends claw-shaped dendritic terminals (arrows) in the dACA (white dashed outline), but not in the vACA (white dashed outline) or CA (not outlined); (C) the photo-labeled α/βs Kenyon cell extends claw-shaped dendritic terminals (arrows) in the CA (white dashed outline) but not in the dACA or the vACA (not outlined).
(D) The dACA is loose and irregularly shaped, but its core can be visualized in the red channel.
(E) α/βp Kenyon cells (n = 13) form smaller claws than the α/βs Kenyon cells (n = 12).
The following genotypes were used in this figure: (B) yw/yw;MB247-DsRedunknown,UAS-C3PA-GFPunknown/UAS-SPA-GFPattP40;UAS-C3PA-GFPattP2,UAS-C3PA-GFPVK00005,UAS-C3PA-GFPVK00027/MB371C-splitGAL4 [R13F02-GAL4ADVK00027,R85D07-GAL4DBDattP2] and (C) yw/yw;MB247-DsRedunknown,UAS-C3PA-GFPunknown/R52H09-GAL4ADattp40;UAS-C3PAattp2,UAS-C3PAVK00005,UAS-C3PAVK00027/MB185B-splitGAL4 [R52H09-GAL4ADattP40, R18F09-GAL4DBDattP2]. Scale bars are 20 μm (B and C) and 10 μm (D).
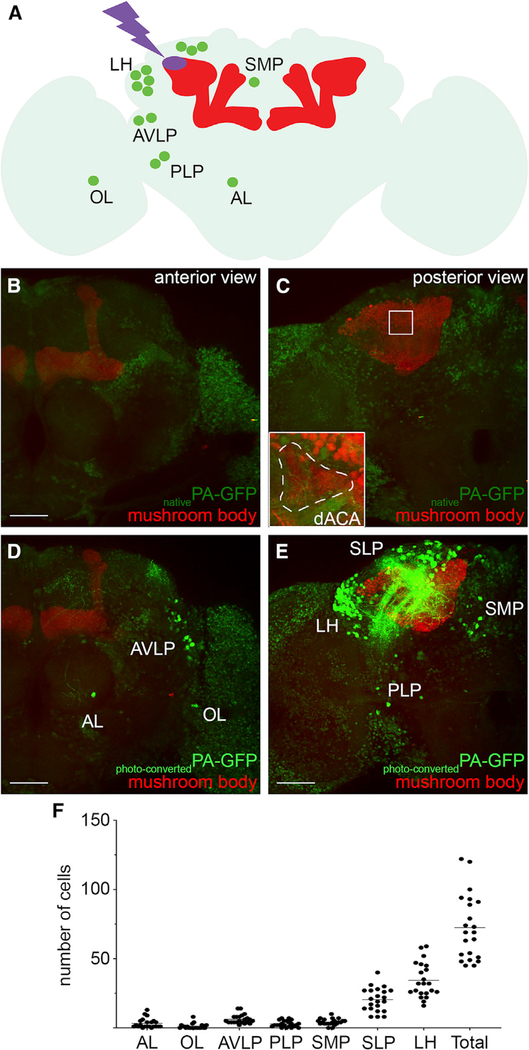
To identify neurons that project to the dorsal accessory calyx and connect to the α/βp Kenyon cells, we used a targeted photo-labeling technique that was adapted from previously published techniques (Aso et al., 2014; Datta et al., 2008; Ruta et al., 2010). In order to only photo-label the neurons projecting to the dorsal accessory calyx, and not the α/βp Kenyon cells, we used a combination of transgenes that, in concert, drive the expression of PA-GFP in all neurons except the Kenyon cells (the N-Synaptobrevin-GAL4, MB247-GAL80, and UAS-PA-GFP transgenes); instead, Kenyon cells were labeled with the red fluorescent protein DsRed using the MB247-DsRed transgene (Figure 2A). The brains of adult flies carrying all the aforementioned transgenes were dissected and imaged using two-photon microscopy (Figures 2B and 2C). Guided by the expression of DsRed, we targeted the dorsal accessory calyx—which is clearly distinct from the main calyx—with high-energy light in order to photo-convert PA-GFP specifically in the neurons projecting to that area (Figure 2C, white dashed outline). Upon photo-labeling, the somata of the neurons that express PA-GFP, and that project either dendrites or axons into the dorsal accessory calyx, were labeled (Figures 2D and 2E). On average, 71 ± 22 neurons (n = 22) were photo-labeled. The somata of these neurons are located in seven distinct clusters that are distributed across the brain (Figures 2A, 2D, and 2E). On the anterior side of the brain, we found three clusters: one located near the antennal lobe (cluster AL containing 3 ± 3 neurons); one located in the optic lobe (cluster OL containing 1 ± 2 neurons); and one located near the anterior ventral lateral protocerebrum (cluster AVLP containing 6 ± 3 neurons; Figures 2D and 2F). On the posterior side of the brain, we found four clusters: one located in the superior medial protocerebrum (cluster SMP containing 4 ± 2 neurons); one located in the superior lateral protocerebrum (cluster SLP containing 20 ± 8 neurons); one located in the lateral horn (cluster LH containing 34 ± 12 neurons); and one located in the posterior lateral protocerebrum (cluster PLP containing 3 ± 2 neurons; Figures 2E and 2F). Altogether, these results suggest that the dorsal accessory calyx receives input from a diverse and distributed collection of projection neurons that can be divided into seven clusters.
Figure 2. Identification of Neurons Projecting to the dACA Using En Masse Photo-labeling.
(A) A schematic of the Drosophila brain shows the seven clusters of neurons identified by photo-labeling the dACA of the mushroom body.
(B and C) All neurons—except for Kenyon cells (red)—express PA-GFP (light green) showing weak fluorescence in structures located on the anterior (B) and posterior (C) sides of the brain.
(C) The dACA is innervated by the α/βp Kenyon cells—and no other Kenyon cells—and is clearly visible in the red channel, forming an irregular shape on the dorsal-anterior side of the CA; (inset) the region outlined by the white dashed line was targeted for photo-labeling.
(D and E) Upon photo-activation of PA-GFP in the dACA, seven clusters of photo-labeled neurons (bright green) are clearly distinguishable in different brain regions located on both the anterior (D) and posterior (E) sides of the brain. The location of these clusters was defined based on the low background fluorescence visible in the unlabeled remaining neurons that were not photo-labeled but that express PA-GFP (light green).
(F) This procedure recovered a total of 71 neuronal cell bodies (71 ± 22; n = 22) in seven different clusters located near or in the antennal lobe (AL), optic lobe (OL), anterior ventral lateral protocerebrum (AVLP), lateral horn (LH), superior lateral protocerebrum (SLP), superior medial protocerebrum (SMP), and posterior lateral protocerebrum (PLP).
The following genotype was used in this figure: yw/yw;MB247-DsRedunkown,UAS-C3PA-GFPunkown/MB247-GAL80unkown,UAS-SPA-GFPattP40;UAS-C3PA-GFPattP2,UAS-C3PA-GFPVK00005,UAS-C3PA-GFPVK00027/N-Synaptobrevin-GAL42–1. Scale bars are 50 μm in all panels.
Identification of Transgenic Lines Driving Expression in the Dorsal Accessory Calyx Input Neurons
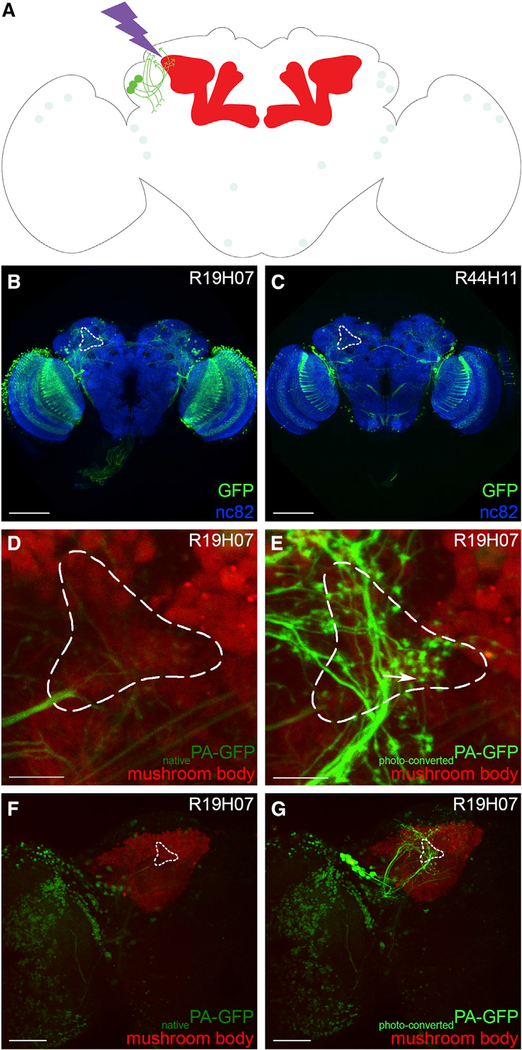
Although en masse photo-labeling of the neurons projecting to the areas within and around the dorsal accessory calyx gives a good approximation of the number of neurons possibly connecting to the α/βp Kenyon cells, this technique cannot confidently identify true pre-synaptic partners. Additionally, the amount of photo-activated PA-GFP within individual neurons varies significantly between trials, and because a large number of neurons project to the dorsal accessory calyx, it is difficult to resolve the morphology of individual neurons using this technique. We, therefore, sought to identify GAL4 transgenic lines that drive expression specifically in these neurons. To identify such lines, we carried out an anatomical screen using the FlyLight collection of GAL4 transgenic lines (Figure 3A; Jenett et al., 2012). As a first step, we screened through the FlyLight database—an online catalog that reports the expression patterns of ∼7,000 GAL4 driver lines—and selected 267 transgenic lines that highlighted neuronal processes in the dorsal accessory calyx (Figures 3B and 3C).
Figure 3. Anatomical Screen to Identify Transgenic Lines Driving Expression in the Neurons Projecting to the dACA.
(A) A schematic of the Drosophila brain shows how transgenic lines driving expression in potential dACA input neurons were identified by photo-labeling; the schematic shows the identification of a transgenic line driving expression in the PLPPNs as an example.
(B and C) First, the expression patterns of each of the transgenic lines available in the FlyLight database were inspected: transgenic lines clearly driving expression strongly (B; R19H07-GAL4) or weakly (C; R44H11-GAL4) in neurons projecting to or from the dACA (white dashed outline) were selected for further investigation. These images were obtained from the FlyLight website.
(D and E) The dACA (white dashed outline) of the mushroom body (red) was visualized in the selected lines, here R19H07-GAL4, and targeted for photo-activation by designing a mask that exposed the outlined region to high-energy light; transgenic lines driving expression in a few neurons extending clear axonal terminals in the dACA (E, arrow) were selected for further investigation.
(F and G) Before photo-labeling, weak fluorescence is visible in many different neurons (F), and upon photo-labeling, the neurons projecting to the dACA (white dashed outline) are visible (G).
The following genotype was used in (D)–(G): yw/yw;MB247-DsRedunknown,UAS-C3PA-GFPunknown/UAS-C3PA-GFPattP40;UAS-C3PAattP2,UAS-C3PAVK00005,UAS-C3PAVK00027/R19H07-GAL4attP2. Scale bars are 100 μm (B and C), 10 μm (D and E), and 50 μm (F and G).
As a second step, we specifically labeled neurons that project to the dorsal accessory calyx using the same technique as described above but with a different combination of transgenes (the R_line-GAL4, UAS-PA-GFP, and MB247-DsRed transgenes). The brains of adult flies that carry this combination of transgenes were dissected and imaged using two-photon microscopy (Figures 3D and 3F). Guided by the expression of DsRed, we targeted the dorsal accessory calyx with high-energy light in order to convert PA-GFP specifically in the neurons that project to that region (Figure 3D, white dashed outline). We screened through the 267 pre-selected transgenic lines and identified 10 lines that we chose to investigate further based on two criteria. First, we selected lines that showed a strong signal after photo-labeling and clear pre-synaptic terminals near or in the dorsal accessory calyx (Figures 3E and 3G; Table S1). Possible pre-synaptic terminals—bouton-shaped (Figure 3E, white arrow)—were distinguished from possible post-synaptic terminals—mesh-shaped—based on the recovered photo-labeling signal. Second, we selected lines based on the strength and specificity of their expression patterns: lines expressing at high level in a few neurons led to clear photo-labeling signals, making it easier to characterize these putative input neurons further (Figure 3G).
Finally, we determined whether the neurons identified with these lines are pre-synaptic partners of the α/βp Kenyon cells. To this end, we used an activity-dependent version of the GFP reconstitution across synaptic partners (GRASP) technique (Figure S1A; Feinberg et al., 2008; Macpherson et al., 2015). In this technique, the GFP is split into two complementary fragments—the GFP1–10 and GFP11 fragments—that do not fluoresce when expressed alone; the GFP1–10 fragment is tagged to an anchor that specifically targets the pre-synaptic membrane (syb::spGFP1–10), whereas the GFP11 fragment is tagged to an anchor that targets the membrane (spGFP11::CD4). When both fragments are in close contact—in this case, when the syb::spGFP1–10 fragment is released at an active synapse—GFP molecules can be reconstituted and recover their fluorescence. The reconstitution of GFP molecules, as indicated by the presence of fluorescent GFP speckles, suggests that the neurons expressing the two complementary fragments form functional synapses (Macpherson et al., 2015). In our study, the syb::spGFP1–10 fragment was expressed in the putative input neurons using the different transgenic lines identified in our screen as well as two transgenic lines that a previous study found to drive expression in putative dorsal accessory calyx input neurons (Figures S1B–S1M; Table S1; Yagi et al., 2016). The expression of the spGFP11::CD4 fragment was driven in most of the Kenyon cells using the MB247-LEXA transgenic line. We look for GFP speckles in the core of the dorsal accessory calyx, which is clearly visible in the red channel as an irregularly shaped structure anterior to the main calyx (Figure S1B, white dashed outline). We detected a large number of GFP speckles in the core of the dorsal accessory calyx for four of the ten lines that were identified in the screen as well as for the two transgenic lines identified by the Yagi et al. (2016) study (Figures S1B–S1E, S1L, and S1M; Table S1). Additionally, we detected a small number of GFP speckles in the same region for three other transgenic lines (Figures S1F–S1H; Table S1). It is worth noting that some of the GFP speckles lay outside of the red signal; most likely, these speckles are formed by Kenyon cells that extend dendrites outside of the core of the dorsal accessory calyx (Figures S1B–S1H, S1L, and S1M). Finally, we did not detect any GFP speckles for the remaining three lines (Figures S1I–S1K; Table S1). Altogether, our screen identified at least nine transgenic lines that drive expression in neurons that form synapses with the α/βp Kenyon cells of the dorsal accessory calyx.
LOPN Connects the Lobula to the α/βp Kenyon Cells
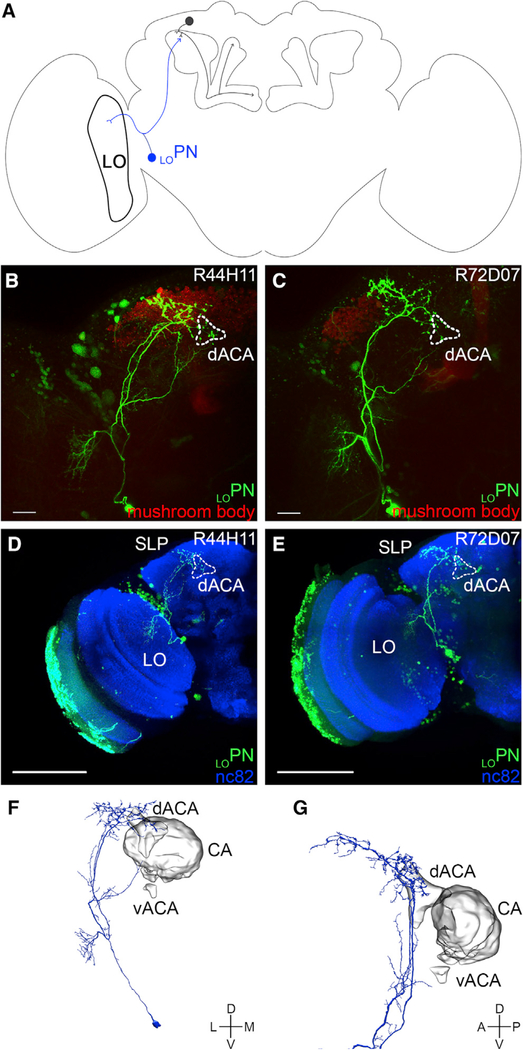
To characterize further the morphology of the projection neurons that connect to the α/βp Kenyon cells, we photo-labeled individual neurons using the transgenic lines recovered from our screen that showed reliable GRASP signal. To this end, neurons were photo-labeled in the dissected brains of flies carrying the R_line-GAL4 and UAS-PA-GFP transgenes; these brains were then immuno-stained and imaged using confocal microscopy. We first focused our attention on the transgenic lines in which we had found the strongest GRASP signal (Figures S1B–S1E; Table S1). Photo-labeling of neurons that project to the dorsal accessory calyx using the R44H11-GAL4 and R72D07-GAL4 transgenic lines revealed a single neuron (Figure 4A). In each line, a neuron projecting from the lobula to the dorsal accessory calyx was clearly photo-labeled (Figures 4B and 4C). Therefore, we named this type of neuron “LOPN.” The soma of LOPN is located in cluster PLP, a region medial to the optic lobe (Figures 2E, 4D, and 4E). LOPN extends its dendrites to the lobula and projects its axon to the superior lateral protocerebrum and the dorsal accessory calyx, completely avoiding the main calyx and the ventral accessory calyx (Figures 4D and 4E). It is worth noting that LOPN is very similar to the OLCT1 neuron that has been previously described using a different transgenic line, R95F09-GAL4; accordingly, we recovered a strong GRASP signal for that transgenic line (Figure S1L; Table S1; Yagi et al., 2016).
Figure 4. LOPN Connecting the Lobula to the dACA.
(A) A schematic of the Drosophila brain shows an α/βp Kenyon cell input neuron—LOPN (blue)—projecting from the lobula (LO) to the dACA.
(B and C) LOPN (bright green) was identified in the screen using two different transgenic lines (B: R44H11-GAL4 and C: R72D07-GAL4); the neurons photo-labeled in each line show an overall similar morphology.
(D and E) LOPN was photo-labeled using either the R44H11-GAL4 (D) or R72D07-GAL4 (E) transgenic lines; the samples were fixed, immuno-stained (nc82 antibody, blue), and imaged. The photo-labeled neurons show an overall similar morphology: their somata are located near the optic lobe; they extend dendritic terminals in a small region of the LO; and they extend axonal terminals in the dACA and the SLP.
(F and G) A LOPN-like neuron was identified in the hemibrain connectome: (F) this neuron projects from the LO to the dACA and the SLP and (G) its axonal terminals innervate the dACA, but not the CA or the vACA. The images in (F) and (G) were taken directly from the Neuprint platform. The neuropil domains are defined by Neuprint platform.
The following genotypes were used in this figure: (B and D) yw/yw;MB247-DsRedunknown,UAS-C3PA-GFPunknown/UAS-C3PA-GFPattP40;UAS-C3PA-GFPattP2,UAS-C3PA-GFPVK00005,UAS-C3PA-GFPVK00027/R44H11-GAL4attP2 and (C and E) yw/yw;MB247-DsRedunknown,UAS-C3PA-GFPattP40/UAS-C3PA-GFPunknown;UAS-C3PA-GFPattP2,UAS-C3PA-GFPVK00005,UAS-C3PA-GFPVK00027/R72D07-GAL4-GFPattP2. Scale bars are 20 μm (B and C) and 100 μm (D and E).
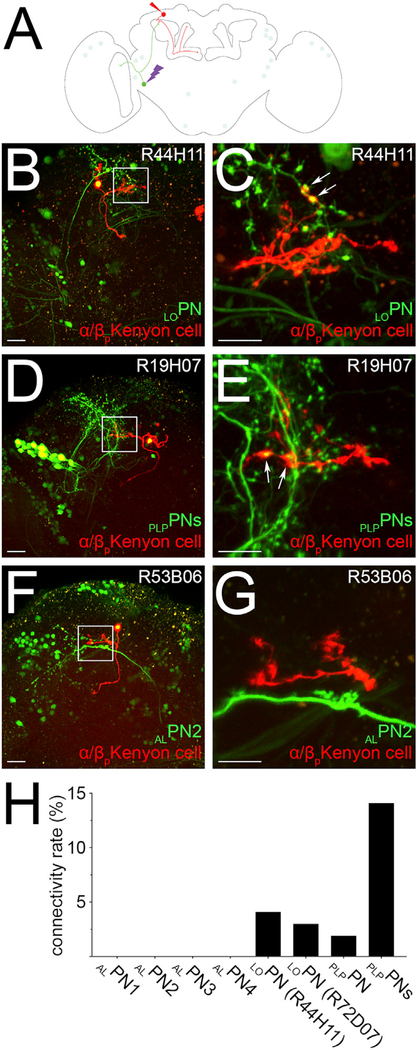
We then set out to confirm whether LOPN does indeed connect to α/βp Kenyon cells, as surmised from the GRASP results. A previous study has demonstrated that a technique, which combines photo-labeling and dye-labeling tools, can be used to identify the complement of input that individual Kenyon cells receive from the antennal lobe and the frequency at which individual projection neurons connect to Kenyon cells (Caron et al., 2013). We have modified this technique in order to directly measure the connectivity rate between a given projection neuron and Kenyon cells. In this modified version of the technique, we used a combination of transgenic lines to photo-label the projection neuron of interest—using the same protocol that we described in the previous section—and to dye label a single Kenyon cell (Figure 5A). We then assessed how many of the dendritic claws formed by the dye-labeled Kenyon cell are connected to a given projection neuron. As a proof of principle, we measured the connectivity rate of the DC3 glomerulus projection neurons to the olfactory Kenyon cells that innervate the main calyx. The connectivity rate of the DC3 projection neurons to the olfactory Kenyon cells has been previously approximated to be 5.1%; this means that there is a 5.1% chance that a given olfactory Kenyon cell claw receives input from the DC3 projection neuron (Caron et al., 2013). Using the modified technique, we found that the DC3 projection neurons connect to Kenyon cells at a connectivity rate of 4.3% (n = 30), a value well within the range measured previously (data not shown). We measured the connectivity rates of LOPN to α/βp Kenyon cells using the two transgenes we identified in our screen. Using the R44H11-GAL4 driver line, we found that 4.1% (n = 27) of the claws formed by the α/βp Kenyon cells connect to LOPN (Figures 5B, 5C, and 5H; Table S1). Likewise, using the R72H07-GAL4 driver line, we found that 3.0% (n = 27) of the claws formed by the α/βp Kenyon cells connect to LOPN (Figure 5H; Table S1). Altogether, based on the results obtained using both the GRASP technique and the dye-/photo-labeling technique, we conclude that LOPN is a true input neuron of the α/βp Kenyon cells and that it conveys information from the lobula, a visual processing center, to the dorsal accessory calyx.
Figure 5. Frequency of Connections between the α/βp Kenyon Cells and Their Input Neurons.
(A) A schematic of the Drosophila brain shows how the frequency of connections between the α/βp Kenyon cells (red) and a given input neuron (green), here LOPN, was measured.
(B–G) A given input neuron was photo labeled (bright green), and a randomly chosen α/βp Kenyon cell was dye filled (red). The total number of claws formed by the dye-filled Kenyon cell was counted, and claws connecting to the axonal terminals of the photo-labeled neuron were detected (arrows). Such connections were found for LOPN (B and C; n = 27) and PLPPNs (D and E; n = 24), but not for the other neurons identified in this study, such as ALPN2 (F and G; n = 30).
(H) The frequency of connections between α/βp Kenyon cells and a given input neuron was calculated by dividing the number of connections detected (for instance, arrows in C and E) by the total number of claws sampled for that particular input neuron.
The following genotype was used in this figure: yw/yw;UAS-C3PA-GFPunknown/UAS-SPA-GFPattP40; R13F02-GAL4ADVK00027,R85D07-GAL4DBDattP2/R_line (as indicated in the panel)-GAL4attP2. Scale bars are 20 μm (B, D, and F) and 10 μm (C, E, and G).
PLPPNs Connect the Posterior Lateral Protocerebrum to the α/βp Kenyon Cells
Photo-labeling of neurons that project to the dorsal accessory calyx using the R19H07-GAL4 and R20G07-GAL4 transgenic lines revealed a group of neurons (Figure 6A). In each line, neurons that project from the posterior lateral protocerebrum to the dorsal accessory calyx were photo-labeled (Figures 6B and 6C). Therefore, we named this type of neuron “PLPPNs.” The somata of the PLPPNs are located in cluster LH, a region near the lateral horn (Figure 6D). We could identify 13 ± 4 PLPPNs (n = 21) and 6 ± 2 PLPPNs (n = 2) per hemisphere using the R19H07-GAL4 and R20G07-GAL4 transgenic lines, respectively (Figures 6B and 6C). All PLPPNs extend projections into many brain centers, including the posterior lateral protocerebrum, the superior clamp, the superior lateral protocerebrum, and the dorsal accessory calyx, avoiding completely the main calyx and the ventral accessory calyx (Figure 6D). Using the dye-/photo-labeling technique described above, we measured the connectivity rate between PLPPNs and α/βp Kenyon cells. We found that the PLPPNs identified using the R19H07-GAL4 transgenic line connect at a high rate: we found that 14.1% (n = 24) of the claws formed by the α/βp Kenyon cells connect to the PLPPNs (Figures 5D, 5E, and 5H). We also measured the connectivity rate of individual PLPPNs using the same transgenic line and found that, on average, an α/βp Kenyon cell claw has 1.90% (n = 32) probability of connecting to a PLPPN (Figure 5H; Table S1).
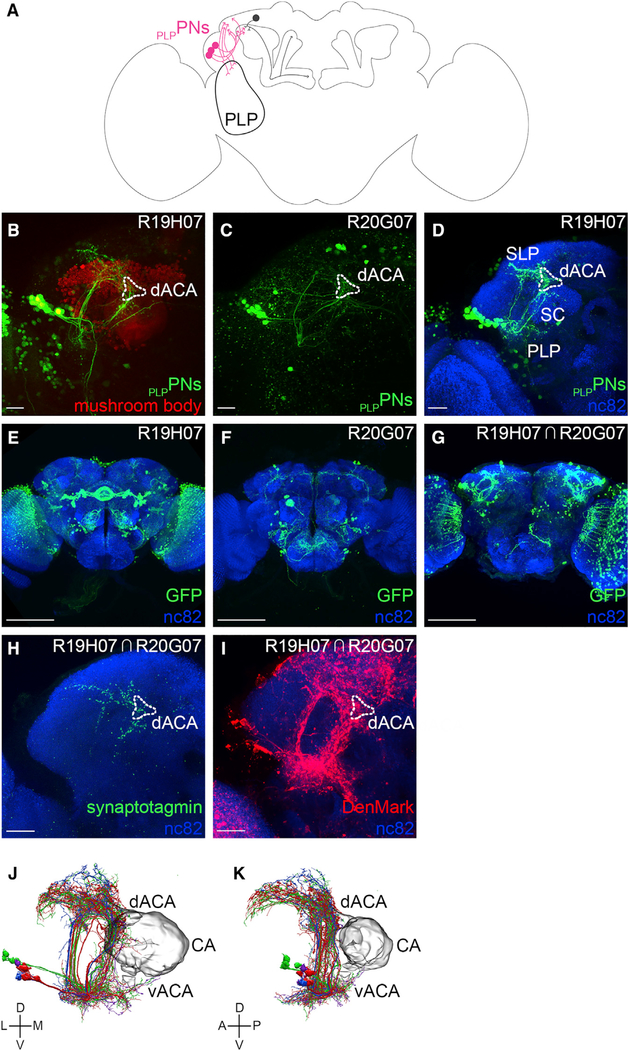
Figure 6. PLPPNs Connecting the Posterior Lateral Protocerebrum to the dACA.
(A) A schematic of the Drosophila brain shows a group of α/βp Kenyon cell input neurons—PLPPNs (pink)—projecting from the posterior lateral protocerebrum (PLP) to the dACA.
(B and C) A group of PLPPNs (bright green) were identified in the screen using two different transgenic lines (B: R19H07-GAL4 and C: R20G07-GAL4).
(D) PLPPNs were photo-labeled using the R19H07-GAL4 transgenic line; the sample was fixed, immuno-stained (nc82 antibody, blue), and imaged. The photo-labeled neurons show an overall similar morphology: their somata are located near the LH; they extend projections in the PLP, the superior clamp (SC), the SLP, and the dACA.
(E and F) The expression patterns observed for each of the GAL4 lines are broad and include many neurons; (E) the R19H07-GAL4 line drives expression strongly in many neurons, including the PLPPNs, (F) whereas the R20G07-GAL4 line drives expression weakly in fewer neurons, including the PLPPNs. These images were obtained from the FlyLight website.
(G) A split-GAL4 transgenic line driving expression strongly in the PLPPNs was engineered using the R19H07 promoter region to drive the GAL4AD domain and the R20G07 promoter region to drive the GAL4DBD.
(H and I) The split-GAL4 line was used to drive the expression of the pre-synaptic marker synaptotagmin-fused GFP (H) or the post-synaptic maker DenMark (I).
(J and K) PLPPN-like neurons were identified in the hemibrain connectome: (J) these neurons project from the PLP to the dACA, the SLP, and the SC; (K) their axonal terminals innervate the dACA, but not the CA or the vACA. The connectome recognizes four different types of PLPPN, but the basis for this categorization is not clear (blue, red, green, and purple). The images in (J) and (K) were taken directly from the Neuprint platform. The neuropil domains are defined by Neuprint platform.
The following genotypes were used in this figure: (B and D) yw/yw; MB247-DsRedunknown, UAS-C3PA-GFPunknown/UAS-C3PA-GFPattP40;UAS-C3PA-GFPattP2, UAS-C3PA-GFPVK00005,UAS-C3PA-GFPVK00027/R19H07-GAL4attP2; (C) yw/yw; UAS-C3PA-GFPunknown/CyO;UAS-C3PA-GFPattP2, R20G07-GAL4attP2/MKRS; (G) yw/yw; MB247-DsRedunknown, UAS-C3PA-GFPunknown/CyO; UAS-C3PA-GFPattP2, UAS-C3PA-GFPVK00005,UAS-C3PA-GFPVK00027/R20G07-GAL4DBDattP2,R19H07-GAL4ADVK00027; (H) yw/yw;Sp/CyO; R20G07-GAL4DBDattP2,R19H07-GAL4ADVK00027/UAS-synaptotagmin::GFPunknown; and (I) yw/yw;UAS-DenMark2/CyO; R20G07-GAL4DBDattP2,R19H07-GAL4ADVK00027/Tm2. Scale bars are 20 μm (A–D), 100 μm (E–G), and 20 μm (H and I).
To determine whether the PLPPNs identified in the R19H07-GAL4 and R20G07-GAL4 transgenic lines are the same or different types of neuron, we used the split-GAL4 technique (Luan et al., 2006). In this technique, the GAL4 transcription factor is split into two complementary fragments—the GAL4AD and GAL4DBD domains—that are both transcriptionally inactive when expressed alone; when both fragments are expressed in the same cell, GAL4 can be reconstituted and recovers its transcriptional activity. We found that 8 ± 3 PLPPNs (n = 4) were visible in flies that carry the R20G07-GAL4DBD and R19H07-GAL4AD transgenes, thus confirming that both lines are expressed in the same group of PLPPNs but that R20G07-GAL4DBD might be expressed in a subset of PLPPNs (Figures 6E–6G). This result revealed, even more clearly, that individual PLPPNs project in many different brain centers (Figure 6G). In order to distinguish the axonal terminals from the dendritic arbors, we used the split-GAL4 combination of transgenic lines (the R20G07-GAL4DBD and R19H07-GAL4AD transgenic lines) to drive the expression of the pre-synaptic marker synaptotagmin in the PLPPNs (Figure 6H). We found that the projections extending into the superior lateral protocerebrum, superior clamp, and dorsal accessory calyx contain pre-synaptic terminals. Similarly, when the expression of the post-synaptic marker DenMark was driven specifically in the PLPPNs, we found that all of the projections made by the PLPPNs contain post-synaptic terminals (Figure 6I). However, the projections extending into the posterior lateral protocerebrum are the only projections formed by PLPPNs that contain only post-synaptic terminals and no pre-synaptic terminals (Figures 6H and 6I).
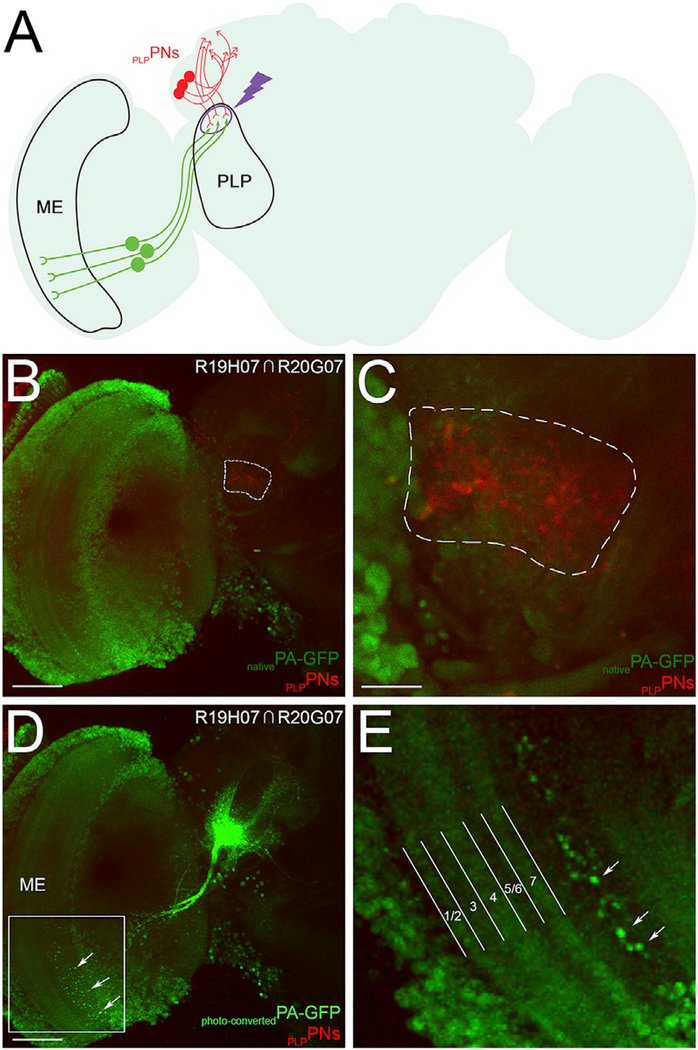
To identify the neurons that project to the post-synaptic terminals formed by the PLPPNs in the posterior lateral protocerebrum, a poorly characterized visual processing center, we used the targeted photo-labeling technique described above (Figure 7A). In short, we used a combination of transgenes that, in concert, drive the expression of PA-GFP in all neurons (the N-synaptobrevin-QF and QUAS-PA-GFP transgenes) and the expression of tdTomato in the PLPPNs (using the R19H07-GAL4AD, R20G07-GAL4DBD, and UAS-tdTomato transgenes). Guided by the expression of tdTomato, we targeted the post-synaptic terminals formed by PLPPNs in the posterior lateral protocerebrum with high-energy light (Figures 7B and 7C). Upon photo-labeling, two types of neuron were clearly photo-labeled: the PLPPNs—showing that the photo-activation was specific to these neurons—and neurons projecting from the ventral medulla (Figure 7D). These photo-labeled neurons project into deeper layers of the medulla (Figure 7E). Altogether, from this set of experiments, we conclude that PLPPNs are one of the major input neurons of the α/βp Kenyon cells and that they convey information from the posterior lateral protocerebrum, and possibly from the ventral medulla, to the dorsal accessory calyx.
Figure 7. Identification of the PLPPNs Input Neurons Using Photo-labeling.
(A) A schematic of the Drosophila brain shows how neurons projecting from the ventral medulla (ME) were identified by photo-labeling the dendritic terminals formed by the PLPPNs in the PLP.
(B and C) All neurons express PA-GFP (light green), and the PLPPNs express tdTomato (red; B). The dendritic terminals formed by the PLPPNs in the PLP are visible in the red channel, and the region outlined by the white dashed line was targeted for photo-labeling (C).
(D and E) Upon photo-activation of PA-GFP in the PLP, neurons projecting from the ventral LO were identified (D); these neurons extend dendritic arbors (white arrows) in a layer of the LO deeper than the well-characterized layers 1–7 (E).
The following genotype is used in this figure: yw/yw; QUAS-C3PA-GFPunknown, QUAS-SPA-GFPunknown/UAS-tdtomatoattP40; R20G07-GAL4DBDattP2,R19H07-GAL4ADVK00027/N-synaptobrevin-QF. Scale bars are 50 μm (B and D) and 10 μm (C).
ALPNs Do Not Contribute Major Input to the α/βp Kenyon Cells
Photo-labeling using the transgenic lines that displayed weak GRASP signal—these are the R30E11-GAL4, R31C03-GAL4, and R53B06-GAL4 lines—or no GRASP signal—these are the R11F07-GAL4, R11F08-GAL4, and R12C04-GAL4 lines—led to the identification of a third type of neuron (Figure S2). The somata of these neurons are all located in the AL cluster, a region near the antennal lobe. Although each of these neurons has a distinct overall morphology, they all project from the antennal lobe to the superior lateral protocerebrum and extend their axons in a region near the dorsal accessory calyx. Therefore, we named this type of neuron “ALPNs.”
The neurons photo-labeled using the R30E11-GAL4 and R31C03-GAL4 driver lines show a nearly identical morphology: in each line, a single neuron that extends its dendrites into the posterior antennal lobe and projects its axons into the superior lateral protocerebrum was visible (Figure S2A). We named this neuron “ALPN1.” It is worth noting that ALPN1 is very similar to the thermosensitive AC neuron that has been previously described (Shih and Chiang, 2011). Using a combination of photo-labeling and dye-tracing techniques, as described above, we measured the connectivity rate between ALPN1 and α/βp Kenyon cells. We could not detect any connections between ALPN1 and α/βp Kenyon cells (n = 26), suggesting that ALPN1 is not a dorsal accessory calyx input neuron (Figure 5H; Table S1). Similarly, the neuron photo-labeled using the R53B06-GAL4 line extends its dendrites into the column region of the posterior antennal lobe, a region known to be activated by high humidity, and the sub-esophageal ganglion, a gustatory processing center (Figure S2B). We named this neuron “ALPN2.” Again, we could not detect any connections between ALPN2 and α/βp Kenyon cells (n = 30), suggesting that ALPN2 is not a dorsal accessory calyx input neuron (Figures 5F–5H; Table S1). Thus, ALPN1 and ALPN2 are most likely not contributing major input to the α/βp Kenyon cells.
We extended our analysis to the transgenic lines that displayed no GRASP signal. The neurons photo-labeled using the R11F08-GAL4 and R12C04-GAL4 driver lines show an overall similar morphology: in each line, a single neuron that extends its dendrites into the arm region of the posterior antennal lobe, a region known to be activated by low humidity, was visible (Figure S2D). We named this neuron “ALPN3.” Not surprisingly, we could not detect any connections between ALPN3 and α/βp Kenyon cells (n = 26), suggesting that ALPN3 does not provide input into the dorsal accessory calyx (Table S1). Similarly, the neuron photo-labeled using the R11F07-GAL4 line extends its dendrites broadly throughout the anterior antennal lobe, an olfactory processing center (Figure S2F). We named this neuron “ALPN4.” Again, we could not detect any connections between ALPN4 and α/βp Kenyon cells (n = 25), suggesting that ALPN4 does not provide input into the dorsal accessory calyx (Table S1). Thus, we could confirm that the transgenic lines that displayed no GRASP signal do not appear to provide major input to the α/βp Kenyon cells. Altogether, these results show that the third type of neuron identified in our screen—ALPNs—project to a region close to the dorsal accessory calyx but are most likely not pre-synaptic to the α/βp Kenyon cells.
LOPN and PLPPNs Are Major Input Neurons of the α/βp Kenyon Cells
Our results suggest that the LOPN and PLPPNs that we identified in our screen connect to the α/βp Kenyon cells and that, together, these two types of projection neuron represent a large fraction of the input neurons of the dorsal accessory calyx. Our results also suggest that the ALPNs do not connect to the α/βp Kenyon cells, as another study suggested (Yagi et al., 2016). Thus, we conclude that the dorsal accessory calyx is anatomically poised to receive information primarily from the lobula and the posterior lateral protocerebrum, two visual processing centers. We verified whether these findings corroborate with the recently released Drosophila hemibrain connectome (Xu et al., 2020). Using the Neuprint 1.0.1 platform, we focused our attention on the 60 α/βp Kenyon cells that have been reconstructed. Altogether, these α/βp Kenyon cells receive input from a large number of neurons, but most of these synapses are most likely axoaxonic synapses, as they are located along the α and β lobes. These input neurons are primarily other Kenyon cells, various dopaminergic neurons, as well as neurons known to connect broadly to all Kenyon cells, such as the APL neuron and the DPM neuron (data not shown). We focused our attention on the 133 input neurons that connect to the α/βp Kenyon cells in the dorsal accessory calyx. Not surprisingly, in accordance with the results obtained from our en masse photo-labeling experiment, the cell bodies of these input neurons can be divided into seven different clusters and the ratio of neurons belonging to a given cluster is largely consistent between both analyses (Figure S3A).
Interestingly, the LH cluster is the most numerous cluster: our study identified 34 ± 12 neurons (n = 22) in this cluster—including the 13 ± 4 PLPPNs (n = 21)—whereas the connectome shows a total of 43 input neurons with cell bodies located in the lateral horn (Figure S3A). Among these neurons, we identified 13 that, based on the location of their pre-synaptic and post-synaptic terminals, are morphologically similar to PLPPNs (Figures 6J and 6K; Table S2). We found that these PLPPN-like neurons connect to 59 of the 60 reconstructed α/βp Kenyon cells, representing 38.49% of the input all α/βp Kenyon cells receive in the dorsal accessory calyx (Figure S3B; Table S2). Interestingly, these neurons are divided into four types in the hemibrain connectome—although the basis for this categorization is not immediately clear—and individual α/βp Kenyon cells do not receive preferential input from neurons within a type. The fact that one of the transgenic lines we identified, R20G07-GAL4, is expressed in a subset of PLPPNs suggests that there is a genetic basis for this categorization. Based on the hemibrain connectome, we found that the 13 PLPPNs receive input from 697 neurons, 207 of these neurons project from known visual centers (either the medulla, the lobula, or the optic glomeruli), and only 8 of these neurons project from the antennal lobe, supporting our assumption that PLPPNs are primarily processing visual information (data not shown). PLPPNs-like neurons connect mostly to α/βp Kenyon cells but also form a few connections to γd and γmain Kenyon cells, reinforcing the observation that the dorsal accessory calyx input neurons and the ventral accessory calyx input neurons form two parallel pathways (Figure S3C). Additionally, our study identified 1 ± 2 neurons (n = 22) in the OL cluster—including LOPN—whereas the connectome shows a total of 4 input neurons with cell bodies located in the optic lobe (Figure S3A). Among these neurons, we identified a neuron very similar to LOPN (Figures 4F and 4G). We found that this LOPN-like neuron connects to 9 of the 60 reconstructed α/βp Kenyon cells, representing 2.14% of the input all α/βp Kenyon cells receive in the dorsal accessory calyx (Figure S3B; Table S1). As we found in our study, this LOPN-like neuron does not connect to any other types of Kenyon cell, forming a pathway parallel to the one conveying visual information to the ventral accessory calyx. Interestingly, PLPPNs and LOPN share 75 common output neurons in the superior lateral protocerebrum, suggesting that other higher brain centers integrate input from this pathway.
Finally, both studies identified a number of neurons with cell bodies located near the antennal lobe: using en masse photo-labeling, we identified a total of 3 ± 3 neurons (n = 22) in the AL cluster, whereas seven such neurons are found in the connectome. Among these neurons, we recognized three of the four ALPNs we characterized: an ALPN2-like neuron and an ALPN4-like neuron that both represent 0.26% of the input that α/βp Kenyon cells receive in the dorsal accessory calyx (Figures S2C, S2G, and S3B; Table S2). We could not recognize an ALPN1-like neuron in the connectome, but we identified an ALPN3-like neuron (Figure S2E; Table S2). The ALPN3-like neuron does not connect to any Kenyon cells (Figures S2E and S3B). Together, the seven antennal lobe input neurons—including the ALPN2-like and ALPN4-like neurons—represent less than 1% of the input that α/βp Kenyon cells receive in the dorsal accessory calyx (Figure S3B). Interestingly, most of the remaining input α/βp Kenyon cells receive in the dorsal accessory calyx (15.14% of the total input α/βp Kenyon cells receive in the dorsal accessory calyx) is from a single neuron projecting from many brain regions, including the main calyx and the pedunculus, two regions of the mushroom body, as well as the superior lateral protocerebrum (Table S2). Altogether, these observations confirm that LOPN-like and PLPPN-like neurons are the major input neurons that connect to the α/βp Kenyon cells in the dorsal accessory calyx. Thus, the dorsal accessory calyx receives most of its input from visual processing centers—the posterior lateral protocerebrum and the lobula—and is thus anatomically poised to process mostly, if not strictly, visual information.
DISCUSSION
In this study, we identified and characterized neurons projecting to the dorsal accessory calyx of the mushroom body and show that these neurons are pre-synaptic to the α/βp Kenyon cells. Using a combination of genetic and anatomical techniques, we could distinguish two different types of projection neuron: LOPN projecting from the lobula—an area of the optic lobe processing visual features, such as shape and motion—and the PLPPNs projecting from the posterior lateral protocerebrum. Although the posterior lateral protocerebrum remains poorly characterized in D. melanogaster, evidence from other insects shows that this brain region receives input from the optic lobe (Paulk et al., 2008, 2009). Interestingly, we found that the dendrites formed by the PLPPNs in the posterior lateral protocerebrum are in close proximity to neurons that project from the ventral medulla. Based on our results—and considering insights from the connectome—we estimate that LOPNs and PLPPNs account for half of total input that α/βp Kenyon cells receive in the dorsal accessory calyx. LOPNs and PLPPNs do not extend axonal terminals into the ventral accessory calyx, the other calyx known to receive visual input, but rather extend axonal terminals into the dorsal accessory calyx and into the superior lateral protocerebrum. Likewise, the α/βp Kenyon cells do not connect to the visual projection neurons that are associated with the ventral accessory calyx (Vogt et al., 2016). These findings suggest that the visual system is connected to the mushroom body via two parallel pathways: the α/βp Kenyon cells receive input from the lobula and the posterior lateral protocerebrum, whereas the γd Kenyon cells receive input directly from the medulla. Further functional studies are necessary to determine what kind of visual information is processed by the α/βp Kenyon cells.
In Drosophila melanogaster, the mushroom body has long been studied as an olfactory processing center. However, evidence from many insects, including the honeybee Apis mellifera, shows that the mushroom body integrates sensory information across different modalities. In honeybees, the input region of the mushroom body, also called the calyx, is divided into different layers, and each layer receives input from either the olfactory or visual system (Gronenberg, 2001). Because the dendrites of Kenyon cells are also restricted to specific layers, it has been suggested that, in the honeybee, multisensory integration does not occur at the level of individual Kenyon cells but rather at the population level (Ehmer and Gronenberg, 2002). Although the honeybee mushroom body differs greatly from the Drosophila mushroom body—it contains about a hundred times as many Kenyon cells and its input region is divided in multiple complex layers—it appears that both mushroom bodies share a common fundamental connectivity principle: the segregation of input based on sensory modality. This connectivity mechanism is immediately apparent in the structural organization of the Drosophila melanogaster mushroom body: the Kenyon cells receiving input from the olfactory system all extend their dendrites into the main calyx, whereas the Kenyon cells receiving input from the visual system extend their dendrites either in the dorsal accessory calyx or the ventral accessory calyx. Many studies have demonstrated that the Kenyon cells that process olfactory information—those associated with the main calyx—integrate input broadly across the different types of olfactory projection neuron (Caron et al., 2013; Zheng et al., 2018). Interestingly, it appears that the Kenyon cells that process visual information are wired differently.
We have a thorough understanding of how olfactory Kenyon cells integrate input from the antennal lobe: most Kenyon cells receive, on average, input from seven projection neurons, and the projection neurons connecting to the same Kenyon cell share no apparent common features (Caron et al., 2013; Zheng et al., 2018). Theoretical studies have shown that this random-like connectivity pattern enables the mushroom body to form sparse and decorrelated odor representations and thus maximizes learning (Litwin-Kumar et al., 2017). Randomization of sensory input is a connectivity pattern that is well suited for representing olfactory information—as an odor is encoded based on the ensemble of olfactory receptors it activates—and might not be suitable for representing visual information. Indeed, our results suggest that specific visual features—the signals processed by the medulla and the ones processed by the lobula and the posterior lateral protocerebrum—need to be represented by two separate subpopulations of Kenyon cells. This observation mirrors anatomical studies of the honeybee brain: the neurons projecting from the lobula terminate in a different layer than the neurons projecting from the medulla (Ehmer and Gronenberg, 2002). This arrangement might be essential to preserve distinct visual features when forming associative memories. Functional and behavioral studies are required to determine whether indeed the mushroom body represents multisensory stimuli in this manner.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Sophie Caron (sophie.caron@utah.edu).
Materials availability
The split-GAL4 Drosophila melanogaster lines generated in this study are available by contacting the Lead Contact directly.
Data and code availability
This study did not generate dataset or codes.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Flies (Drosophila melanogaster) were raised on standard cornmeal agar medium and maintained in an incubator set at 25°C, 60% humidity with a 12 h light/12 h dark cycle. Crosses were set up and reared under the same conditions, but the standard cornmeal agar medium was supplemented with dry yeast. The GAL4-UAS binary transgenic system was used to express transgenes of interest to label neurons in the fly brain. Details of genotypes used in this study and their sources are described in the Key Resources Table. One to 14 days-old flies (depending on the experiment) were used for experiments in this study. Both males and female flies were used in all experiments.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-nc82 | University of Iowa Developmental Studies Hybridoma Bank | RRID: AB_2314866 |
| Mouse monoclonal anti-GFP-20 | Sigma-Aldrich | RRID: AB_259941 |
| Alexa Fluor 488 goat polyclonal anti-mouse IgG | Thermal Fisher | RRID: AB_2576217 |
| Alexa Fluor 633 goat polyclonal anti-mouse IgG | Thermal Fisher | RRID: AB_2535719 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Normal Goat Serum | Jackson ImmunoResearch Laboratories | RRID: AB_2336990 |
| VECTASHIELD mounting medium | Vector Laboratories Inc. | REF#H-1000 |
| 10X phosphate bovine saline | Sigma-Aldrich | Cat#P5493 |
| Triton X-100 | Sigma-Aldrich | Cat#T8787 |
| Paraformaldehyde | Electron Microscopy Sciences | Cat#15710 |
| Collagenase | Sigma-Aldrich | Cat#C5138 |
| TexasRed dye | Thermo-Fisher | REF#D3328 |
| MgCl2 solution (1M in H2O) | Sigma-Aldrich | Cat#63069 |
| CaCl2 solution (1M in H2O) | Sigma-Aldrich | Cat#21115 |
| NaOH solution (10M in H2O) | Sigma-Aldrich | Cat#72068 |
| NaCl | Sigma-Aldrich | Cat#S7653 |
| KCl | Sigma-Aldrich | Cat#P5405 |
| HEPES | Sigma-Aldrich | Cat#H3375 |
| Trehalose | Sigma-Aldrich | Cat#T0167 |
| Sucrose | Sigma-Aldrich | Cat#S1888 |
| NaHCO3 | Sigma-Aldrich | Cat#S5761 |
| NaH2PO4 | Sigma-Aldrich | Cat#S5011 |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: yw;N-Synaptobrevin-GAL42.1;; | Aso et al., 2014 | N/A |
| D. melanogaster: y1w1118;;R11F07-GAL4attP2; | Bloomington Drosophila Stock Center | BDSC: 39414 |
| D. melanogaster: y1w1118;;R11F08-GAL4attP2; | Bloomington Drosophila Stock Center | BDSC: 48467 |
| D. melanogaster: y1w1118;;R12C04-GAL4attP2; | Bloomington Drosophila Stock Center | BDSC: 48494 |
| D. melanogaster: y1w1118;;R19H07-GAL4attP2; | Bloomington Drosophila Stock Center | BDSC: 48867 |
| D. melanogaster: y1w1118;;R20G07-GAL4attP2; | Bloomington Drosophila Stock Center | BDSC: 48613 |
| D. melanogaster: y1w1118;;R30E11-GAL4attP2; | Bloomington Drosophila Stock Center | BDSC: 48100 |
| D. melanogaster: y1w1118;;R31C03-GAL4attP2; | Bloomington Drosophila Stock Center | BDSC: 48103 |
| D. melanogaster: y1w1118;;R53B06-GAL4attP2; | Bloomington Drosophila Stock Center | BDSC: 38863 |
| D. melanogaster: y1w1118;;R44H11-GAL4attP2; | Bloomington Drosophila Stock Center | BDSC: 41268 |
| D. melanogaster: y1w1118;;R72D07-GAL4attP2; | Bloomington Drosophila Stock Center | BDSC: 39770 |
| D. melanogaster: y1w1118;;R91H11-GAL4attP2; | Bloomington Drosophila Stock Center | BDSC: 40596 |
| D. melanogaster: y1w1118;;R95F09-GAL4attP2; | Bloomington Drosophila Stock Center | BDSC: 40712 |
| D. melanogaster: y1w1118;;R13F02-GAL4ADVK00027,R85D07-GAL4DBDattp2; | Aso et al., 2014 | N/A |
| D. melanogaster: y1w1118;R52H09-GAL4ADattP40;R18F09-GAL4DBDattP2; | Bloomington Drosophila Stock Center | BDSC: 68267 |
| D. melanogaster: y1w1118;;R19H07-GAL4ADVK00027;R20G07-GAL4DBDattP2; | Aso et al., 2014 | N/A |
| D. melanogaster: yw;UAS-C3PA-GFPunknown;; | Aso et al., 2014 | N/A |
| D. melanogaster: y1w1118;UAS-C3PA-GFPattP40;; | Aso et al., 2014 | N/A |
| D. melanogaster: y1w1118;;UAS-C3PA-GFPattP2; | Aso et al., 2014 | N/A |
| D. melanogaster: y1w1118;;UAS-C3PA-GFPVK00005; | Aso et al., 2014 | N/A |
| D. melanogaster: y1w1118;UAS-SPA-GFPattP40;; | Aso et al., 2014 | N/A |
| D. melanogaster: yw;UAS-syb::spGFP1-10unknown;; | Aso et al., 2014 | N/A |
| D. melanogaster: yw;UAS-DenMark2;; | Bloomington Drosophila Stock Center | BDSC: 33062 |
| D. melanogaster: yw;; UAS-synaptotagmin::GFP3; | Bloomington Drosophila Stock Center | BDSC: 6926 |
| D. melanogaster: yw;UAS-tdTomatoattP40;; | Aso et al., 2014 | N/A |
| D. melanogaster: yw;MB247-GAL80unknown;; | Bloomington Drosophila Stock Center | BDSC: 64306 |
| D. melanogaster: yw;;MB247-LEXA::VP16unknown; | Pitman et al., 2011 | N/A |
| D. melanogaster: yw;LEXAop-CD4::spGFP11unknown;; | Macpherson et al., 2015 | N/A |
| D. melanogaster: yw;LEXAop-tdTomatoSu(Hw)attP5;; | Bloomington Drosophila Stock Center | BDSC: 56142 |
| D. melanogaster: yw;;N-Synaptobrevin-QF; | Aso et al., 2014 | N/A |
| D. melanogaster: yw;QUAS-C3PAunknown;; | Aso et al., 2014 | N/A |
| D. melanogaster: yw;QUAS-SPAunknown;; | Aso et al., 2014 | N/A |
| D. melanogaster: yw;MB247-DsRedunknown;; | Riemensperger et al., 2005 | N/A |
| Software and Algorithms | ||
| ImageJ | Schindelin et al., 2012 | https://imagej.nih.gov/ij/ |
| Prairie View | Bruker | |
| ZEN microscope software | ZEISS | |
| Other | ||
| FlyLight | Jenett et al., 2012 | http://www.janelia.org/project-team/flylight |
| Virtual Fly Brain | Jenett et al., 2012 | https://v2.virtualflybrain.org |
| NeuPrint web interface for the hemibrain connectome database | Xu et al., 2020 | https://neuprint.janelia.org |
| Borosilicate glass with filament | Sutter Instruments | ITEM#BF100-50-10 |
METHOD DETAILS
Fly stocks
Flies (Drosophila melanogaster) were raised on standard cornmeal agar medium and maintained in an incubator set at 25°C, 60% humidity with a 12 h light / 12 h dark cycle (Percival Scientific, Inc.). Crosses were set up and reared under the same conditions, but the standard cornmeal agar medium was supplemented with dry yeast. The strains used in this study are described in the Key Resources Table.
Photo-labeling neurons using PA-GFP
Two to six-day-old flies were used for all photo-labeling experiments. The protocol was largely based on the one developed in a previous study (Aso et al., 2014). In short, brains were dissected in saline (108 mM NaCl, 5 mM KCl, 5 mM HEPES, 5 mM Trehalose, 10 mM Sucrose, 1 mM NaH2PO4, 4 mM NaHCO3, 2 mM CaCl2, 4 mM MgCl2, pH≈7.3), treated for 1 min with 2 mg/ml collagenase (Sigma-Aldrich) and mounted on a piece of Sylgard placed at the bottom of a Petri dish. Photo-labeling and image acquisition were performed using an Ultima two-photon laser scanning microscope (Bruker) with an ultrafast Chameleon Ti:Saphirre laser (Coherent) modulated by Pockels Cells (Conotopics). For photo-labeling, the laser was tuned to 710 nm with an intensity of 5–30 mW; for image acquisition, the laser was tuned to 925 nm with an intensity of 1–14 mW (both power values were measured behind the objective lens). A 60X water-immersion objective lens (Olympus) was used for both photo-labeling and image acquisition. A 40X water-immersion objective lens (Olympus) was used for image acquisition in some experiments (the ones described in Figures 2 and 3). A GaAsP detector (Hamamatsu Photonics) and PMT detector were used for measuring green and red fluorescence, respectively. Photo-labeling and image acquisition files were visualized on a computer using the Prairie View software version 5.4 (Bruker). Image acquisition was performed at a resolution of 512 by 512 pixels, with a pixel size of 0.39 μm (60X lens) or 0.582 μm (40X) and a pixel dwell time of 4 μs. Each pixel was scanned 8 times.
For photo-labeling of the dorsal accessory calyx (or the posterior lateral protocerebrum), a volume spanning the entire neuropil was divided into eight to 12 planes with a step size of 2 μm. The mask function of the Prairie View software was used to mark the targeted region in every plane, and the boundaries of the mask were determined based on the red fluorescent protein DsRed expressed by the α/βp Kenyon cells. The photo-labeling step was performed using a pixel size of 0.019 μm and a pixel dwell time of 2 μs. Each pixel was scanned four times. Each plane was scanned 30 times with 30 s interval. The entire photo-labeling cycle was repeated two to three times, with a 10-minute resting period between cycles. The entire brain was imaged before and after photo-labeling using the 40X water-immersion objective lens (Olympus, Japan). The number of cell bodies recovered after the photo-labeling was measured using the Multi-Point Tool function of the Fiji software (Schindelin et al., 2012).
For the photo-labeling of single neurons, a slightly modified protocol was used. Instead of a volume, a single square plane of 1.0 μm by 1.0 μm centered on the soma of a neuron cell was scanned 70 to 100 times with a 10 s interval between scans. Photo-labeled brains were fixed in 2% paraformaldehyde diluted in 1X phosphate bovine saline (PFA) for 45 min at room temperature, washed five times in 0.3% Triton X-100 diluted in 1X phosphate bovine saline (PBST) at room temperature, blocked in 5% normal goat serum diluted in PBST (PBST-NGS) for 30 min at room temperature, and incubated with the primary mouse antibody nc82 (1:20 in PBST-NGS; Developmental Studies Hybridoma Bank, University of Iowa) at 4°C overnight. On the following day, brains were washed four times in PBST and incubated with the secondary goat antibody Alexa Fluor 633 anti-mouse (1:500 in PBST-NGS; Life Technologies) at 4°C overnight. On the following day, brains were washed four times in PBST and mounted on a slide (Fisher Scientific) using the mounting media VECTASHIELD (Vector Laboratories Inc.). Immuno-stained brains were imaged using an LSM 880 confocal microscope. (Zeiss). The neuropils innervated by the input neurons were identified by comparing the confocal images with the adult brain template JFRC2 available on Virtual Fly Brain (Jenett et al., 2012).
Immunohistochemistry
For the experiments using the green fluorescent protein reconstitution across synaptic partners (GRASP) technique as well as the experiments using DenMark and synaptotagmin::GFP immunostainings, the brains of five to 14 day-old flies were dissected in saline and fixed in PFA for 45 min at room temperature, washed five times in PBST at room temperature, blocked in PBST-NGS for 30 min at room temperature, and incubated with the primary antibody at 4°C overnight. The mouse anti-GFP-20 antibody was used in GRASP and synaptotagmin experiments (1:100 in PBST-NGS; Sigma-Aldrich); the mouse nc82 antibody was used in synaptotagmin::GFP and DenMark experiments. On the following day, brains were washed four times in PBST and incubated in secondary antibody at 4°C overnight. The AlexaFluor-488 goat-anti-mouse (1:500 in PBST-NGS; Life Technologies) and AlexaFluor-633 goat-anti-mouse (1:500 in PBST-NGS; Life Technologies) were used. On the following day, brains were washed four times in PBST and mounted on a slide (Fisher Scientific) using the mounting media VECTASHIELD (Vector Laboratories Inc.). Immuno-stained brains were imaged using an LSM 880 confocal microscope. (Zeiss). The neuropils innervated by the input neurons were identified by comparing the confocal images with the adult brain template JFRC2 available on Virtual Fly Brain (Jenett et al., 2012).
Measuring the connectivity rate between input neurons and α/βp Kenyon cells
One to three-day-old flies were used when mapping the connectivity rate between input neurons and α/βp Kenyon cells. Brains were dissected in saline, treated for 1 min with 2 mg/ml collagenase (Sigma-Aldrich) and mounted on a piece of Sylgard placed at the bottom of a Petri dish. The imaging protocol is the same as described above but the photo-labeling protocol is different. Each of the input neurons was photo-labeled using a single plane centered on either its soma or its projection and by scanning the plane three to five times. Each pixel was scanned eight times with a pixel size of 0.019 μm and a pixel dwell time of 4 μs. A fire-polished borosilicate glass pipette (0.5 mm I.D., 1.0 mm O.D., 10 cm length; Sutter Instruments) was pulled using the P-2000 micropipette puller (Sutter Instruments) and backfilled with Texas Red dye (lysine-fixable 3000 MW; Life Technologies) dissolved in saline. The tip of the pipette was positioned next to the cell body of a randomly chosen α/βp Kenyon cell under the two-photon microscope. The dye was electroporated into the cell body using three to five 1–5 ms pulses of 20–50 V. The dye was allowed to diffuse within the Kenyon cell for 5 min before the brain was imaged.
Confocal image acquisition and analysis
All confocal images were collected on LSM880 confocal microscope (Zeiss). For imaging whole brain, each sample was imaged twice using a Plan-Apochromat 40X/1.3 Oil M27 objective lens. First, the entire brain was divided into four tiles, each tile was imaged separately (voxel size = 0.46 μm by 0.46 μm by 2 μm, 764 by 764 pixels per image plane) and then stitched together using the Stitch function of the Zen microscope software (Zeiss). For imaging specific neuropil regions, the same objective lens was used, but with higher resolution (voxel size = 0.09 μm by 0.09 μm by 1 μm, 2320 by 2320 pixels per image plane). For imaging brains manipulated using the GRASP technique, a Plan-Apochromat 63X/1.4 Oil M27 objective lens was used in combination with the RGB Airyscan mode. Images were processed using the Airyscan function of the Zen microscope software. All confocal images were analyzed using the Fiji software (Schindelin et al., 2012). All figure panels are maximum intensity projection of confocal stacks or sub-stacks.
QUANTIFICATION AND STATISTICAL ANALYSIS
In the experiment using en masse photo-labeling to identify neurons projecting to the dorsal accessory calyx (Figure 2), the number of cell bodies per brain was manually counted using ImageJ. Sample size (represented by n in the figure legends as well as in the text) indicates the number of fly brains used in the experiment. The average number of recovered cell bodies and the standard deviation across all brain samples were calculated using Microsoft Excel spreadsheet. Statistical graphs (Figures 2F and 5H) were mde using GraphPad Prism 8. No statistical methods were used to determine the size of the sample pool, to compareamong different datasets, nor to determine whether the data met assumptions of the statistical approach.
Supplementary Material
Highlights.
The dorsal accessory calyx, formed by the α/bp Kenyon cells, receives mostly visual input
LOPN projects from the lobula to the dorsal accessory calyx
PLPPNs project from the posterior lateral protocerebrum to the dorsal accessory calyx
ACKNOWLEDGMENTS
We thank Florian Maderspacher, Kaitlyn Ellis, and members of the Caron laboratory for comments on the manuscript; Shannon Torstrom for the initial characterization of the y1w1118;;R19H07-GAL4ADVK00027;R20G07-GAL4DBDattP2; transgenic line; Adam Lin for preparation of the standard cornmeal agar medium; and Shannon Torstrom, Hayley Smihula, and Adam Weinbrom for assistance with general laboratory concerns. This work has been funded in part by grants from the National Institute of Neurological Disorders and Stroke (R01 NS 106018and R01 NS 1079790). Further financial support was provided by the Georges S. and Dolores Eccles Foundation (to S.J.C.C.), the Dale A. String-fellow Fellowship (to J.L.), and the Research Scholar Award (to M.S.J.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108138.
REFERENCES
- Aso Y, Hattori D, Yu Y, Johnston RM, Iyer NA, Ngo TTB, Dionne H, Abbott LF, Axel R, Tanimoto H, and Rubin GM (2014). The neuronal architecture of the mushroom body provides a logic for associative learning. eLife 3, e04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden T, Euler T, and Berens P (2020). Understanding the retinal basis of vision across species. Nat. Rev. Neurosci. 21, 5–20. [DOI] [PubMed] [Google Scholar]
- Bates AS, Schlegel P, Roberts RJV, Drummond N, Tamimi IFM, Turnbull R, Zhao X, Marin EC, Popovici PD, Dhawan S, et al. (2020). Complete connectomic reconstruction of olfactory projection neurons in the fly brain. Curr. Biol. 30, 3183–3199.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron SJC, Ruta V, Abbott LF, and Axel R (2013). Random convergence of olfactory inputs in the Drosophila mushroom body. Nature 497, 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta SR, Vasconcelos ML, Ruta V, Luo S, Wong A, Demir E, Flores J, Balonze K, Dickson BJ, and Axel R (2008). The Drosophila pheromone cVA activates a sexually dimorphic neural circuit. Nature 452, 473–477. [DOI] [PubMed] [Google Scholar]
- Ehmer B, and Gronenberg W (2002). Segregation of visual input to the mushroom bodies in the honeybee (Apis mellifera). J. Comp. Neurol. 451, 362–373. [DOI] [PubMed] [Google Scholar]
- Feinberg EH, VanHoven MK, Bendesky A, Wang G, Fetter RD, Shen K, and Bargmann CI (2008). GFP reconstitution across synaptic partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron 57, 353–363. [DOI] [PubMed] [Google Scholar]
- Gronenberg W (2001). Subdivisions of hymenopteran mushroom body calyces by their afferent supply. J. Comp. Neurol. 435, 474–489. [DOI] [PubMed] [Google Scholar]
- Hige T (2018). What can tiny mushrooms in fruit flies tell us about learning and memory? Neurosci. Res. 129, 8–16. [DOI] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo TTB, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. (2012). A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2, 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleş MF, and Frye MA (2017). Object-detecting neurons in Drosophila . Curr. Biol. 27, 680–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinwand SG, and Chalasani SH (2011). Olfactory networks: from sensation to perception. Curr. Opin. Genet. Dev. 21, 806–811. [DOI] [PubMed] [Google Scholar]
- Litwin-Kumar A, Harris KD, Axel R, Sompolinsky H, and Abbott LF (2017). Optimal degrees of synaptic connectivity. Neuron 93, 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Wolf R, Ernst R, and Heisenberg M (1999). Context generalization in Drosophila visual learning requires the mushroom bodies. Nature 400, 753–756. [DOI] [PubMed] [Google Scholar]
- Luan H, Peabody NC, Vinson CR, and White BH (2006). Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron 52, 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson LJ, Zaharieva EE, Kearney PJ, Alpert MH, Lin TY, Turan Z, Lee CH, and Gallio M (2015). Dynamic labelling of neural connections in multiple colours by trans-synaptic fluorescence complementation. Nat. Commun. 6, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masek P, and Scott K (2010). Limited taste discrimination in Drosophila. Proc. Natl. Acad. Sci. USA 107, 14833–14838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morante J, and Desplan C (2008). The color-vision circuit in the medulla of Drosophila. Curr. Biol. 18, 553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulk AC, Phillips-Portillo J, Dacks AM, Fellous JM, and Gronenberg W (2008). The processing of color, motion, and stimulus timing are anatomically segregated in the bumblebee brain. J. Neurosci. 28, 6319–6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideo O, and Kei I (2006). Systematic analysis of the visual projection neurons of Drosophila melanogaster: Lobula-specific pathways. J. Comp. Neurol. 497, 928–958. [DOI] [PubMed] [Google Scholar]
- Paulk AC, Dacks AM, Phillips-Portillo J, Fellous JM, and Gronenberg W (2009). Visual processing in the central bee brain. J. Neurosci. 29, 9987–9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman JL, Huetteroth W, Burke CJ, Krashes MJ, Lai SL, Lee T, and Waddell S (2011). A pair of inhibitory neurons are required to sustain labile memory in the Drosophila mushroom body. Curr. Biol. 21, 855–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemensperger T, Völler T, Stock P, Buchner E, and Fiala A (2005). Punishment prediction by dopaminergic neurons in Drosophila. Curr. Biol. 15, 1953–1960. [DOI] [PubMed] [Google Scholar]
- Ruta V, Datta SR, Vasconcelos ML, Freeland J, Looger LL, and Axel R (2010). A dimorphic pheromone circuit in Drosophila from sensory input to descending output. Nature 468, 686–690. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih H-W, and Chiang A-S (2011). Anatomical Characterization of Thermo-sensory AC Neurons in the Adult Drosophila Brain. Journal of Neurogenetics 25, 1–6. [DOI] [PubMed] [Google Scholar]
- Tanaka NK, Endo K, and Ito K (2012a). Organization of antennal lobe-associated neurons in adult Drosophila melanogaster brain. J. Comp. Neurol. 520, 4067–4130. [DOI] [PubMed] [Google Scholar]
- Tanaka NK, Suzuki E, Dye L, Ejima A, and Stopfer M (2012b). Dye fills reveal additional olfactory tracts in the protocerebrum of wild-type Drosophila. J. Comp. Neurol. 520, 4131–4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt K, Schnaitmann C, Dylla KV, Knapek S, Aso Y, Rubin GM, and Tanimoto H (2014). Shared mushroom body circuits underlie visual and olfactory memories in Drosophila. eLife 3, e02395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt K, Aso Y, Hige T, Knapek S, Ichinose T, Friedrich AB, Turner GC, Rubin GM, and Tanimoto H (2016). Direct neural pathways convey distinct visual information to Drosophila mushroom bodies. eLife 5, e14009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Nern A, Ryan Williamson W, Morimoto MM, Reiser MB, Card GM, and Rubin GM (2016). Visual projection neurons in the Drosophila lobula link feature detection to distinct behavioral programs. eLife 5, e21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu CS, Januszewski M, Lu Z, Takemura S, Hayworth KJ, Huang G, Shinomiya K, Maitin-Shepard J, Ackerman D, Berg S, et al. (2020). A connectome of the adult Drosophila central brain. bioRxiv. 10.1101/2020.01.21.911859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Mabuchi Y, Mizunami M, and Tanaka NK (2016). Convergence of multimodal sensory pathways to the mushroom body calyx in Drosophila melanogaster. Sci. Rep. 6, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau JM, DeAngelis GC, and Angelaki DE (2015). Dissecting neural circuits for multisensory integration and crossmodal processing. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20140203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Lauritzen JS, Perlman E, Robinson CG, Nichols M, Milkie D, Torrens O, Price J, Fisher CB, Sharifi N, et al. (2018). A complete electron microscopy volume of the brain of adult Drosophila melanogaster. Cell 174, 730–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate dataset or codes.