ABSTRACT
Parabacteroides distasonis is the type strain for the genus Parabacteroides, a group of gram-negative anaerobic bacteria that commonly colonize the gastrointestinal tract of numerous species. First isolated in the 1930s from a clinical specimen as Bacteroides distasonis, the strain was re-classified to form the new genus Parabacteroides in 2006. Currently, the genus consists of 15 species, 10 of which are listed as 'validly named' (P. acidifaciens, P. chartae, P. chinchillae, P. chongii, P. distasonis, P. faecis, P. goldsteinii, P. gordonii, P. johnsonii, and P. merdae) and 5 'not validly named' (P. bouchesdurhonensis, P. massiliensis, P. pacaensis, P. provencensis, and P. timonensis) by the List of Prokaryotic names with Standing in Nomenclature. The Parabacteroides genus has been associated with reports of both beneficial and pathogenic effects in human health. Herein, we review the literature on the history, ecology, diseases, antimicrobial resistance, and genetics of this bacterium, illustrating the effects of P. distasonis on human and animal health.
KEYWORDS: Parabacteroides distasonis, Crohn’s disease, inflammatory bowel disease, gut microbiota, antimicrobial activity
1. Introduction
The intestinal gut is home to the largest collection of microbes, harboring trillions of bacteria and representing hundreds of species. Most of these species fall into two groups – Bacteroidetes and Firmicutes. These were among the first phyla that comprise most of the dominant gut commensal bacteria to be defined. Within the Bacteroidetes phylum, the relatively new genus Parabacteroides (defined as separate from its predecessor, the very broad Bacteroides genus, in 20061), now contains at least ten 'valid species,' which are recognized as being real species in clinical settings, and five 'non-valid species,' which are not recognized as being real species in clinical settings by the List of Prokaryotic names with Standing in Nomenclature. Several of these species were identified in isolates from clinical infections.
Recently, we isolated and fully sequenced multi-drug resistant P. distasonis from deep-gut wall tissue lesions in patients with Crohn’s Disease that underwent surgical removal of the affected bowel further supporting a potential pathogenic role on gut wall health. Recent studies suggest that P. distasonis could exert protective effects against certain diseases, including multiple sclerosis, type II diabetes, colorectal cancer, and inflammatory bowel disease. Furthermore, some reports suggest that this bacterium could even have the potential to serve as a potential probiotic to promote digestive health in humans based on microbiome or animal studies. However, other experimental data show contradictory results, suggesting pathogenic effects in various disease models. This suggests that P. distasonis may serve a dichotomous role depending on the context.
Herein, we hypothesize and illustrate that the pathologic role of these bacteria in human diseases may depend on the context, including the susceptibility of the host to immune suppression, impaired bacterial clearance, and the promotion of hyperinflammatory responses, together with strain-to-strain differences that may account for differences in antimicrobial resistance to human therapies and P. distasonis potential for pathogenicity.
2. The new genus Parabacteroides
As with several other new genera reclassified from the initially broad genus Bacteroides fragilis group, including Alistipes1, Parabacteroides is a relatively new genus with distinctive features shared among other gut commensal bacteria. It is important to highlight that great progress in our understanding of this and other related genera has been facilitated by the taxonomic contributions made by Japanese microbiologists Mitsue Sakamoto and Yoshimi Benno who proposed the genus,2 and by other international groups cited below. According to the NCBI, the full taxonomy lineage for the genus Parabacteroides is: Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides. Of note, the family Tannerellaceae is composed of Parabacteroides and another new genus, Tannerella, with the species T. forsythia and T. pachnodae.
As of May 1, 2021, the Parabacteroides genus (taxonomy ID: 375,288) comprises 15 species (https://lpsn.dsmz.de/search?word=parabacteroides), represented by 493 genome assemblies available in the NCBI. Of these species, 10 are listed as 'validly named', and 5 as 'not validly named' (shown in quotation marks)3: P. acidifaciens (human feces, China, 2019; type strain (TS): 426–9; CGMCC 1.13558; NBRC 113,433),4 “P. bouchesdurhonensis” (healthy human feces, France, 2018; TS: CSUR P3763; Marseille-P3763;),5 P. chartae (wastewater of a paper mill, China, 2011, TS: DSM 24,967; JCM 17,797; NS31-3),6 P. chinchillae (chinchilla feces in a zoo, Japan, 2013; TS: CCUG 62,154; DSM 29,073; JCM 17,104; ST166),7 P. chongii (blood from human with peritonitis, Republic of Korea, 2018; TS: B3181; KACC 19,034; LMG 3006),8 P. distasonis (unclear, human feces, or peritonitis as reported in PATRIC.org, USA, 1933; TS: ATCC 8503; CCUG 4941; CIP 104,284; DSM 20,701; JCM 5825; NCTC 11,152, named in honor to microbiologist A. Distaso),2 P. faecis (human feces; Japan, 2015; TS: 157; CCUG 66,681; JCM 18,682),9 P. goldsteinii (peritoneal fluid, appendix tissue, and intra-abdominal abscess, USA, 2005; TS: ATCC BAA-1180; CCUG 48,944; DSM 19,448; JCM 13,446; WAL 12,034, named in honor to infectious disease clinician Ellie J. C. Goldstein),2,10 P. gordonii (human blood clinical culture, USA, 1930s; TS: CCUG 57,478; DSM 23,371; JCM 15,724; MS-1, named in honor to microbiologist Jeffrey I. Gordon), P. johnsonii (human feces, Japan, 2007; TS: DSM 18,315; JCM 13,406; M-165, named in honor to American molecular taxonomist John L. Johnson, who was the first to describe Bacteroides merdae (P. merdae)),9 “Parabacteroides massiliensis” (healthy human feces, France, 2019; TS:SN4; named after Latin name of Marseille, Massilia),11 P. merdae (human feces, USA, 1978; TS: ATCC 43,184; CCUG 38,734; CIP 104,202; DSM 19,495; JCM 9497; NCTC 13,052; VPI T4-1),2,12 “P. pacaensis” (healthy human feces, France, 2020; TS: Marseille-P4001; named after abbreviation for the region of Provence Alpes Côte d’Azur),13 “P. provencensis” (healthy human feces, France, 2020; TS: Marseille-P3668 T; nomenclature status listed as ‘not validly published’, named after the region of Provence),13 and “P. timonensis” (human feces from healthy pigmy 39-year-old male, Congo, 2019; TS: CCUG 71,183; CSUR P3236; Marseille-P3236; nomenclature status listed as ‘not validly published’, named after French hospital La Timone).14
To provide an example of genetic similarity with other fecal commensals within the Bacteroidetes, based on 16s RNA gene sequence similarity, the three founding species of the Parabacteroides genus-- P. distasonis JCM 5825 T, P. goldsteinii JCM 13,446 T, and P. merdae JCM 9497 T-- are phylogenetically closely related to each other (>92.3% DNA sequence similarity) and related to T. forsythia with about 90% similarity. However, they are distant from their predecessor genus Bacteroides (83.5–88.8%) and other comparable genera including Dysgonomonas (85.9–89.4%), Paludibacter (86.9–88.5%), Porphyromonas (82.2–86.9%), Prevotella (77.2–81.9%) and Proteiniphilum (85.9–87.3%).2
2.1. Parabacteroides distasonis as a referent species for the genus
Of all the species listed previously, P. distasonis is the reference type strain for the genus Parabacteroides. Since the completion of the first genome sequence for the species by investigators at Washington University in St. Louis in 2007, P. distasonis strain ATCC 8503 has become the strain comparator for all Parabacteroides species.15 This type strain is an isolate deposited in 1933 and is a gram-negative, non-spore-forming, rod-shaped, strict anaerobic bacterium present in the gastrointestinal (GI) tract of humans and animals.2,16 A single P. distasonis ATCC 8503 cell is 0.8–1.6 × 1.2–12 μm in size. Its colonies on sheep blood agar plates are 1–2 mm in diameter, appear gray to off-white, and are circular, slightly convex, and smooth in shape.
Recently, studies on P. distasonis have displayed evidence in support of P. distasonis having a potentially beneficial and commensal nature, while others have displayed evidence for a potential pathogenic role. Although P. distasonis is part of the normal gastrointestinal microbiota, it has been isolated from extra-intestinal abdominal infections and abscesses in humans. More recently, P. distasonis has been shown to have ambivalent effects on models of inflammatory bowel disease (IBD) in rodents (“murine models”), with reports describing both pro-inflammatory (pathogenic) and anti-inflammatory (beneficial) effects.17 Thus, P. distasonis currently has unclear mechanistic associations with the main forms of inflammatory bowel disease (IBD), namely Crohn’s Disease (CD) and ulcerative colitis (UC), in both humans and animal models18.
As a result of this and other contradictory findings, P. distasonis has become a highly controversial bacterium in the gastroenterological and microbiological fields of study. In 2020 and early 2021 alone, over twenty studies were published involving P. distasonis, many of which produced controversial findings. Here, we delve into basic concepts on the history, biology, and ecology of the bacterium; its resistance to antibiotics; its genomic features; and its potential clinical relevance in the context of human health. We highlight papers that support both sides of the debate over P. distasonis' pathogenicity and present a summary of potential future perspectives on our analysis of this microbe.
3. History, ecology, and identification
3.1. Historical overview and reclassification
Parabacteroides distasonis is a re-classified bacterium named after A. Distaso, a Romanian bacteriologist who was involved in the description of the Bacteroides phylum species in the 1910s.18,19 Originally, P. distasonis was considered part of the Bacteroides genus, where it bore the name Bacteroides distasonis. Prior to the 1980s, classification of bacteria was based on phenotypic features, which meant that all gram-negative rods with certain phenotypic profiles were designated as being part of Bacteroides.20 For decades, only minor changes in taxonomy were implemented, even though the Bacteroides genus increasingly started to contain vast numbers of strains and species (>50) that greatly differed phenotypically from one another.21
This began to change in the 1980s, when Woese et al. introduced 16S rRNA gene sequencing.20 After using this method in 1989, Shah & Collins formally proposed that the genus Bacteroides be restricted to Bacteroides fragilis and related taxa, resulting in the amendment of the description of the genus.21 This spawned several novel genera over the next decade and into the early 2000s, including Alistipes, Dialister, Dichelobacter, and Tannerella.2
Skepticism about the classification of what was then called 'B. distasonis' and another closely related species, B. merdae, arose in the mid-1990s and early 2000s, when 16S rRNA gene sequence analyses raised the idea that the two species may also have not been members of the Bacteroides genus . Shortly after the discovery of another species closely related to B. distasonis and B. merdae, B. goldsteinii, Sakamoto and Benno analyzed the 16S rRNA gene sequences of all three species, using Tannerella forsynthesis as an outgroup. Phylogenetic analyses showed that the three species were closely related and therefore belonged to the same genus, with Tannerella being the most closely related genus. This was confirmed by analyses of the 16S-23S rRNA gene internal transcribed spacer (ITS) regions, which showed that B. distasonis was phylogenetically distinct from species of the genus Bacteroides, along with other former Bacteroides species including B. goldsteinii and B. merdae.2
In addition to genetic differences, Sakamoto and Benno demonstrated differences in chemo-taxonomic features. Ratios of anteiso-C15:0 to iso- C15:0 ranged from 3.1 to 10.3 in B. distasonis, B. goldsteinii, and B. merdae strains, while those for T. forsythensis ranged from 22.8 to 95.2. Major menaquinones of B. distasonis, B. goldsteinii, and B. merdae were MK-9 and MK-10, while the same for T. forsynthesis and Bacteroides species were MK-10 and MK-11. Taken together, the differences observed prompted Sakamoto and Benno to propose that these species be placed in their own genus, named Parabacteroides, meaning “adjacent to Bacteroides”.2
3.2. Ecology
P. distasonis has been detected in healthy and unhealthy patients, although the loads compared to other members of the Bacteroides group may vary across studies depending on the methods of detection used and factors like diet, which remain largely uncharacterized. Earlier studies utilizing immunoassays and monoclonal antibodies to identify Bacteroides species revealed that more than 30% of bacteria in human feces were Bacteroides species.
As a member of the distal gut microbiome, P. distasonis is present in fecal matter, which allows it to pollute waters and reach other environments passively. Consequentially, this bacterium has been proposed to be used as an indicator of fecal pollution in public environments, including recreational waters. In the waters of the Ohio River in the USA, Kreader et al.22 showed that P. distasonis can survive for up to two weeks if kept under 4°C; however, the bacterium can only survive for two days if kept at 24°C. Kreader et al.22 also showed that in filtered water or in the presence of cycloheximide (a protein synthesis inhibitor) the persistence of P. distasonis in water at 24°C was extended by at least 7 days.22 These are remarkable observations that support the fact that such a strict anaerobe can remain dormant in adverse conditions,22 as also observed in our laboratory. In another study, researchers using PCR-specific primers and Luminex® 100TM based technology (a suspension array that assays multiple analytes in a single well of a microtiter plate) detected P. distasonis in river samples, public water systems, and beach sand.23 Innovative membrane filtration techniques have also shown that P. distasonis and similar Bacteroides species are commonly found in natural waters.24
The sources of fecal pollution in water for this abundant Bacteroidetes species could be traced to homeothermic hosts (including humans) with microbiome-based approaches; however, such methods often lack the resolution to classify sequenced bacteria down to the species level.25 A few examples for the genus may serve to illustrate the ecological range of this species. Recent high-throughput microbiome sequencing has shown that Parabacteroides is one of the main genera present in the GI, ceca, and feces of ducks raised with free access to swimming waters, but not in ducks raised with no access to swimming waters.26 Regarding abundance in this context, at the genus level, Parabacteroides was among the most abundant genera in ducks allowed to swim (Bacteroides, 25%; Escherichia-Shigella, 11%; Peptococcus, 7.7%; and Parabacteroides, 5.86%) compared to the genera in ducks with no swimming access (Bacteroides, 18.1%; Erysipelatoclostridium,10.9%; Ruminococcaceae_unclassified, 10.4%;Lachnospraceae_unclass., 5.2%; Coriobacteriales_unclass., 5.89%; and Faecalibacterium, 4.2%).
The presence of this species in feces and their survival in natural waters supports our proposal that water and migratory birds could serve as an important avenue for Parabacteroides species dissemination between and across avian and mammalian species, possibly increasing the risk of antimicrobial resistant strains appearing in clinical settings. If human-originated isolates of P. distasonis can appear in chicken or other animals, then this bacterium might be able to reach humans via food and livestock, as is common among other aerobic or anaerobic foodborne or food-dwelling microbes.27
3.3. Phenotyping and identification
Numerous methods exist for culturing P. distasonis. As a saccharolytic bacterium, P. distasonis is capable of metabolizing carbohydrates such as mannose and raffinose for energy production and can grow on a medium containing 20% bile.2 Furthermore, the latter enables P. distasonis to selectively grow on Bacteroides bile esculin agar.28
P. distasonis is urease negative, but possesses genes that confer resistance to oxidative stress, such as the KatE gene and the OxyR gene. Deemed advantages of P. distasonis as an aerotolerant commensal stem from the unique properties of its versions of catalase and superoxide dismutase.29 Catalase production enables a detoxifying role against oxidative stress mediated by hydrogen peroxide, which is often produced by inflammatory cells. The decomposition of H2O2 is vitally important for the survival of bacterial cells in conditions where oxygen is present. H2O2 is a major inhibitor of growth for gut bacteria that lack the ability to decompose H2O2 into less reactive molecules. Catalase production is not a universal survival strategy among Bacteroides; P. distasonis and B. fragilis are catalase positive, while B. eggerthii, B. thetaiotaomicron, and B. ovatus have variable catalase production; and B. vulgatus and B. uniformis are catalase-negative.30 Molecular studies have shown that P. distasonis produces a catalase similar to that of B. fragilis, but in contrast, the enzyme is twice the size (250,000).30 Several variables may affect P. distasonis’ catalase production, including the type of medium used, the presence of agar, and the addition of hemin either pre- or post-autoclaving.31 Higher catalase levels occur after hemin is added post-autoclaving and with a high carbohydrate content in the selected medium.31
P. distasonis cannot hydrolyze gelatin in liquefaction tests used to identify bacterial proteolytic enzymes, unlike Bacteroides species such as B. ovatus.2 Gelatin tests primarily seek to assess the presence of bacterial matrix metalloproteinases, which are also abundant in activated (gene expression upregulated) host cells. Although gelatin hydrolysis tests may yield false negatives, tests indicate that P. distasonis’ mechanisms of intestinal modulation are not mediated by gelatinases (gelE gene co-transcribed with sprE, regulated by the fsrA, fsrB, and fsrC gene family) as they are in Enterococcus faecalis, which is important to help the bacteria translocate across polarized T84 human colon cancer cell models.32
In addition to culturing, polymerase chain reaction (PCR) methods enable the identification (with Sanger sequencing of the 16s rRNA gene), quantification of bacterial abundance in tissues when using quantitative reverse transcriptase (qRT-PCR), and differentiation of P. distasonis from other closely related bacteria including P. merdae and Odoribacter splanchnicus. Relative to anaerobic culturing methods, qRT-PCR is a highly accurate and rapid alternative when identifying Bacteroides species and related species, including P. distasonis, as shown in Figure 1.33 Probes designed for P. distasonis ATCC 8503 include the forward primer sequence 5ʹ–TGC CTA TCA GAG GGG GAT AAC- 3ʹ, the reverse primer sequence 5ʹ–GCA AAT ATT CCC ATG CGG GAT-3ʹ, and the probe sequence 5ʹFam–CGA AAG TCG GAC TAA TAC CGC ATG AAGC-3ʹTam.23
Figure 1.
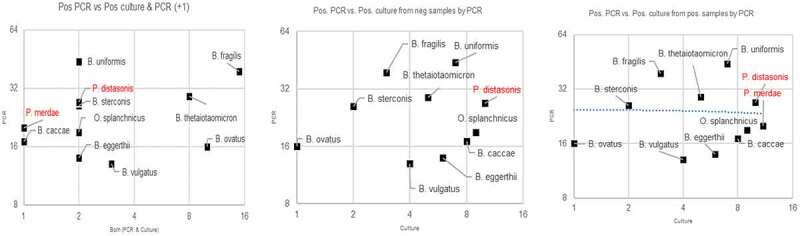
Correlation of PCR identification vs culture isolation from samples that were detected or undetected via qRT-PCR. Eleven Bacteroides and Parabacteroides species detected via qRT-PCR in 400 human surgical wound infection samples or closed abscesses. Target bacteria were detected from 31 samples (8%) via culture vs. 132 samples (33%) via qRT-PCR (p-value < 0.001). For each species, qRT-PCR detected higher counts than culture; this may reflect the detection of DNA of dead organisms by qRT-PCR. Plot created for this manuscript to illustrate the correlation between qRT-PCR and anerobic culture results for Bacteroides species isolated from wound samples using 132 isolates.33 a) y-axis corresponds to number of isolates detected by qRT-PCR; x-axis corresponds to number of isolates detected by both qRT-PCR and culture. b) y-axis corresponds to number of isolates detected by qRT-PCR; x-axis corresponds to number of isolates detected by culture. c) y-axis corresponds to number of isolates detected by qRT-PCR. Adapted from using data from Tong et al.33 with permission. Available from Anaerobe and used with permission from Elsevier
Another identification method for P. distasonis is Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS), which is a fast and cost-effective method for accurately identifying microorganisms based on the profiling of the ionized ribosomal proteins.34 Unlike PCR methods, the clinical value of MALDI-TOF depends on the quality and breadth of coverage of the databases used to identify any given sample.
In a study by Veloo et al.,35 P. distasonis was the most commonly identified Parabacteroides species in human clinical specimens when using MALDI-TOF MS. The role of the MALDI-TOF MS system in correctly identifying emerging species that are either rare or difficult to grow in vitro is influenced by several factors that impair our ability to identify and correctly assign a pathogenic role to clinical specimens for such emergent species1. Using the identification dataset version 5 for MALDI-TOF MS, Veloo et al., on behalf of the numerous laboratories collaborating within the European Network for the Rapid Identification of Anaerobes (ENRIA) project and using 6309 human clinical anaerobic bacterial strains, reported that only four well-established species were represented in the database with most clinical infections linked to P. distasonis (n = 45/54), P. goldsteinii (n = 3/54), P. johnsonii (n = 1/54), and P. merdae (n = 5/54). The MALDI-TOF MS database is regularly updated. Currently, P. faecis and P. gordonii are also represented in the database.
Of these species, P. distasonis was the species with the highest MALDI-TOF MS log-score values indicating a “more reliable identification” profile.35 Current efforts of optimization that exist on MALDI-TOF MS databases show that P. distasonis can be accurately identified by this method, with log-score values of ≥ 2.0 in 44 of 45 isolates.35 In a similar study by Rodíguez-Sánchez et al,34 six P. distasonis isolates were identified by the MALDI-TOF MS system at the species level. This was the same number of isolates detected using 16S rRNA sequencing at the genus level, meaning that all isolates were successfully identfied via the MALDI-TOF MS system. The Rapid ID 32A system, a phenotypic method, was also able to identify all six P. distasonis isolates at the species level.34
In another study, using both the MALDI-TOF MS system and the Rapid ID 32A system, the single P. distasonis isolate (abundance = 0.74%) involved was successfully detected.36 Together, these studies, indicate that P. distasonis is a species of higher abundance, is more readily cultivable, and is identifiable by several detection methods. This is perhaps the reason why it was the first species isolated to be assigned to the Parabacteroides genus. These methods prompt future studies to examine mechanisms by which this species mediates human health; however, emerging strains will need to be considered to help define the genetics behind the modulation of either effect.
4. Evolution, biochemical features, and metabolism
4.1. Evolution and symbiotic adaptation to the gut
According to phylogenetic analyses, P. distasonis diverged from a common ancestor shared with Bacteroides species, as confirmed via nucleotide sequencing of the complete 16S rRNA gene from distinct bacterial species.37 A study by Xu et al.37 explored the driving forces behind the adaptation of Bacteroidetes in the distal gut environment and their importance to the evolution of human gut commensals. To examine how the intestinal environment affects microbial genome evolution, Xu et al.37 sequenced the genomes of two members of the distal human gut microbiota, B. vulgatus and P. distasonis. Through comparison with other sequenced gut and non-gut Bacteroidetes, and analyzing their niches and habitat adaptations, Xu et al. identified three general functions that could illustrate an evolutionary uniqueness for the Bacteroidetes phylum: polysaccharide metabolism, environmental sensing and gene regulation, and membrane transport. These processes are important in aiding with lateral gene transfer, mobile elements, and gene amplification, all of which affect the ability of gut-dwelling Bacteroidetes to vary their cell surface, sense their environment, and harvest nutrients present in the distal colon.37 More recently, genome-based analyses have examined the metabolic features that are typical for a wide array of Bacteroides, which complements the taxonomic classification of the phylumspecies within.38–40
P. distasonis possesses a vast number of laterally transferred genes relative to similar species, which may help it thrive in the distal gut microbiome. Some genes that have been transferred allow hydrogen to be the final electron acceptor in the electron transport chain rather than oxygen. This is an important trait for successful energy utilization in the anaerobic environment of the gut. In addition, P. distasonis contains fewer environmental sensing and gene regulation genes compared to similar species and has a smaller number of carbon source degradation genes such as hemicellulases, pectinases, and other polysaccharides that help break down non-plant-based carbohydrates.37
In a study involving an individual that received ceftriaxone therapy, P. distasonis demonstrated monodominance in the gut microbiota community. Hildebrand et al.41 observed an extreme bloom of P. distasonis in the gut, where it was the second most abundant commensal with a 95% relative abundance. Here, Borkfalki ceftriaxensis was the most common conditionally monodominant taxa (CMT) that was not invasive. The initial colonization by this species was succeeded by P. distasonis.41 Thus, P. distasonis is a key member of the gut microbiota community, rendering it as a highly influential bacterium.
4.2. Lipopolysaccharide and S-layer to blend in with the gut environment
Parabacteroides distasonis’ lack of polysaccharide degradation genes is compensated for by the bacterium’s unique surface layer (S-layer) composed of glycoproteins. Fletcher et al.42 identified at least nine glycoproteins utilized by P. distasonis using the lectin-affinity purification technique. Intriguingly, the researchers found that seven of the nine glycoprotein promoters identified undergo DNA inversion, predominantly in their endogenous human environment.42
In 1996, before P. distasonis was considered a member of the Parabacteroides genus, Corthier et al.43 utilized freeze etch electron microscopy to visualize an S-layer in P. distasonis that was not present in most Bacteroides spp., including B. fragilis and B. thetaiotaomicron.43 This monomolecular layer allows both for the breakdown of complex polysaccharides that cannot be broken down by human enzymes and for the acquisition and synthesis of polysaccharides to “blend in” with the surrounding intestinal tissue. This is possible through the use of an enzyme to coat the S-layer with sugar residues from the surrounding environment. It is thought that this ability to “blend in” with the surrounding gut tissue allows P. distasonis to avoid triggering a strong immune response from the host.42 In fact, in one study, after a lipopolysaccharide (LPS) challenge, the introduction of a membrane fraction of P. distasonis “reduced the release of TNF, IL-6, CCL2 (MCP-1) and CCL12 (MCP-5) by macrophages.”44
This unique S-layer could form the basis of P. distasonis’ effective pathogenic nature. Understanding the potentially pathogenic nature of P. distasonis is crucial for comprehending the interactions between the host or host’s environment and this bacterium.42,45–49
4.3. Hypothetical contribution to local anti-inflammatory methane production
It is thought that fermentation by P. distasonis results in the production of methane. It is unclear if direct production of methane occurs in P. distasonis; however, it is known that P. distasonis produces hydrogen, carbon dioxide, formic acid, acetic acid, carboxylic acid, and succinic acid.2 Other microbes may convert the carbon dioxide and acetic acid to methane. Acetogenic bacteria might then oxidize the acids, obtaining more acetic acid and either hydrogen or formic acid. Finally, in complex gut communities, methanogens may convert acetic acid to methane.
However, there is evidence that methane may also serve a pathogenic role. Methane production has been shown to be involved in the pathogenesis of other intestinal diseases, such as constipation-predominant irritable bowel syndrome (C-IBS), diverticulosis, and colorectal cancer.50 Furthermore, methane production may hinder ileal motility, providing an explanation for its ability to induce constipation.51
Whether P. distasonis contributes(likely indirectly) to local production of methane is at this time speculative, but there is a need to consider that the ultimate beneficial or pathogenic role on any animal model of disease, or clinical disease, might also depend on other species present in the local microbiome.
4.4. Succinic acid as proinflammatory signaling molecule
Comprehensive analysis of the end-products of fermentation have indicated that P. distasonis and other Bacteroides and Prevotella spp. primarily produce acetic acid and succinic acid, in contrast to other microbes that produce other acids in a less specific manner.2 Succinate serves as an inflammatory signal in immune cells to induce IL-1β through HIF-1α (transcription factor induced by hypoxia), a downstream target of succinate.52–54 Succinate can stimulate reactive oxygen species (ROS),55,56 and succinate accumulation in immune cells acts as an inflammatory signal for macrophages via HIF-1α.52 HIF-1α activation attenuates Treg development, induces IL-17 production, and increases RORyt transcription, favoring differentiation of T lymphocytes into pro-inflammatory TH17 cells.57 Succinate is also a ligand for succinate-receptor 1 (SUCNR1; formally GPCR91) expressed by dendritic cells53 and can enhance both the pro-inflammatory cytokine (TNFα and Il-1β) production and the antigen presentation capacity of dendritic cells, thereby inducing adaptive immune responses.53,58 In the colonic mucosa of rats, succinic acid leads to reduced crypt size and inhibition of epithelial cell proliferation rate.59 Notably, leptin is also a well-known HIF-1α-inducible modulator,60 with HIF-1α overexpression observed in obese adipose tissue, and reduction during weight loss.61
5. Antimicrobial resistance
5.1. Antibiotics
Since the isolation of P. distasonis from fecal samples, sites of infection, abscess formation, or the gut wall in IBD patients is potentially linked to clinical outcomes affected by antimicrobial resistance, the sections below provide an overview of studies examining the resistance patterns of P. distasonis to various classes of antibiotics.
5.2. Beta-lactams
Bacteroides spp. and Parabacteroides spp. are becoming increasingly more resistant to certain antibiotics62 because of an increase in the number of antimicrobial resistance-related virulence genes. Within the genus Parabacteroides, P. distasonis has a wide spectrum of resistance to various beta-lactams, being particularly resistant to the penicillin class.63–67 Nakano et al.67 demonstrated that up to 99% of P. distasonis isolates were resistant to penicillin and the first generation cephalosporin, cephalexin, while more than 85% of the P. distasonis isolates could be resistant to amoxicillin and ampicillin. Resistance to the second generation of cephalosporin, cefoxitin, was less frequent; it was found in only 12% of isolates tested.67 Differential resistance to cephalosporins, including cefotetan, cephalothin,63,68 cephalexin,67 cefoxitin,64,68 cefotaxime,64 cefazolin,68 and cefotetan,69 have also been reported, with resistances higher in earlier generations.
The overall resistance to the antibiotic classes penicillin and cephalosporin can be attributed to the presence of various genes encoding beta-lactamases. These include, but are not limited to, cfxA,64,66 cfiA,67 and cepA.66,67 While almost all P. distasonis strains produce beta-lactamase, there are unique strains expressing beta-lactamase genes at significantly elevated rates. Some additional protection from imipenem can be also attributed to the outer membrane of P. distasonis.70,71
P. distasonis is generally susceptible to carbapenems such as meropenem and imipenem.72,73 However, recently it was found that a small percentage of clinical isolates of P. distasonis are resistant to imipenem, ranging from 4%66 to 11%.69 This is clinically relevant considering the apparent current trends of increasing antimicrobial resistance in P. distasonis over time, a pattern that has been reported in the Bacteroides fragilis group in Europe, where high-level resistance to ampicillin (MIC 64 mg/L) increased from 16% to 44.5% over 20 years.69
5.3. Lincosamides
Several studies have reported the various rates of clindamycin resistant isolates of P. distasonis: from 9%74 in the 1990s to about 30% in the 2000s,67,75,76 and even up to 50% in the early 2010s.76 This suggests that a growing proportion of P. distasonis strains have reduced susceptibility to clindamycin.62 It is possible that the indiscriminate use of this antibiotic in clinical settings, especially for members of the Bacteroides and Parabacteroides genera, exerts selective pressure, leading to increased resistance of P. distasonis to clindamycin over time.77
One of the potential underlying mechanisms for clindamycin resistance in P. distasonis is the presence of the resistance gene ermF, known to be frequently found in various anaerobes.78,79 This gene encodes the ribosomal methylase that modifies peptidyl transferase in the ribosome, resulting in resistance to lincosamides, macrolides, and streptogramin drugs.80 However, the data on the frequency of this gene in different P. distasonis isolates remain controversial – it varies from being completely absent in isolates66 to being detected in 37.5% of all samples.67 Further confirmation for the potential role of ermF in clindamycin resistance comes from a study by Kierzkowska et al,77 in which 88.9% (8 out of 9) of clindamycin-resistant isolates were found to harbor the ermF gene.77
5.4. Other antibiotics and antimicrobial agents
P. distasonis demonstrates considerable resistance to tetracycline – up to 87.5% among tested clinical isolates.68 The high frequency of tetracycline resistance among Parabacteroides isolates has been attributed to the presence of the tetQ gene.68,81 The conjugative transposon that tetQ resides on harbors ermF, and exposure to low concentrations of tetracycline can trigger horizontal gene transfer, thereby also triggering the transfer of other transposons present in the genome, like Tn4555 which harbors cfxA. Thus, this has the potential to be horizontally transferred to other susceptible intestinal species of the Bacteroides and Parabacteroides genera.68
An enzyme found in P. distasonis, Pd_dinase (short for P. distasonis protease) has diaminopeptidase activity. This enzyme can hydrolyze some human antimicrobial peptides normally present in the gut, such as keratin-derived antimicrobial peptides (KAMPs), human β-defensin 2, and human neutrophil peptide 3. If secreted into the extracellular milieu of the gut, Pd_dinase may promote intestinal colonization by P. distasonis via the inactivation of the aforementioned host antimicrobial peptides.82
6. Resistance in clinical vs. intestinal isolates
In a study by Sóki et al. conducted in 2020,83 the resistance of intestinal isolates of different bacterial species, including P. distasonis, was evaluated against several antimicrobials. The relevant MIC values were compared between clinical and intestinal isolates. Several notable observations can be made from their findings, summarized in Table 1. For example, in the clinical and intestinal isolates of all species examined, ampicillin resistance is almost 100%; moxifloxacin, cefoxitin, and clindamycin resistance is intermediate (13–44%); and amoxicillin/clavulanate, imipenem, metronidazole, and tigecycline resistance is very low (0–4%).83
Table 1.
Antimicrobial Activity against Parabacteroides distasonis and other species*
 |
*For the intestinal isolates, the study examined 202 Bacteroides and Parabacteroides strains (11.9%, were B. fragilis) collected between 2014 and 2016 in Europe and compared resistance levels between clinical and commensal isolates. Isolates were recovered from feces using Bacteroides Chromogenic Agar (BCA) method and tested via agar dilution for ten antibiotics. For clinical isolates, the study used published data from a 20-year survey of isolates in Europe.69 These results were similar to previous European clinical Bacteroides antibiotic susceptibility survey, all the variations existed across countries and antibiotics.83 Adapted from Table 2 from Sóki et al.83 with permission. Available from Anaerobe and used with permission from Elsevier. Note that that metronidazole, imipenem, cefoxitin, and amoxicillin/clavulanic acid MIC ranges were significantly greater in clinical isolates compared to intestinal isolates. Notably, however, intestinal isolates of P. distasonis are significantly more resistant (p = 0.048 via X2 test) to moxifloxacin than their clinical counterparts.83
However, two findings in particular stand out: first, the significantly greater resistance of P. distasonis to most classes of antibiotics tested compared to other Bacteroides species; and second, the significantly higher resistance of clinical isolates to antibiotics compared to their intestinal counterparts.83 Specifically, for ampicillin, amoxicillin/clavulanic acid, cefoxitin, and in the intestinal isolates, moxifloxacin and tetracycline, resistance was significantly higher in P. distasonis compared to almost all other isolates. In addition, the MIC50 and MIC90 values for P. distasonis in both clinical and intestinal isolates were significantly higher than that of other isolates for cefoxitin and amoxicillin/clavulanic acid. P. distasonis was the most resistant or second-most resistant species in tests with ampicillin, amoxicillin/clavulanic acid, and cefoxitin for both the clinical and intestinal isolates, and was often significantly more resistant across antibiotics compared to the average of all isolates.83
These results highlight the possibility and importance of differential antimicrobial resistance in samples of clinical origin compared to commensal intestinal isolates. In addition, these results also indicate a need for greater study of antimicrobial resistance at the strain level, since multiple genetic factors may confer resistance to antibiotics and/or enable bacteria to induce chronic illness. Culture-based studies and genomic analyses of strains well-defined with respect to the source region (healthy vs diseased) are warranted to understand the drivers that enable some isolates or strains to become pathogenic compared to others that remain as commensals.
7. Diseases
7.1. Dichotomous role in inflammatory bowel disease
Inflammatory bowel disease (IBD) is a spectrum of life-long chronic conditions that affect the digestive tract of humans and animals in a slow, progressive matter. In humans, the prototypic forms of IBD are Crohn’s disease (CD) and ulcerative colitis. Crohn’s disease affects the entirety of the gastrointestinal tract, and patients have chronic, debilitating symptoms. These include abdominal pain, severe diarrhea, stools containing blood, weight loss, and fatigue. In general, IBD has aberrant exaggerated host immune and inflammatory responses to luminal antigens that reveal recurring themes in IBD pathogenesis: first, lesions in IBD predominate in areas of highest bacterial exposure; and second, manipulation of luminal content using selective antibiotics reduces inflammation in IBD patients. Thus, it remains to be determined if IBD is triggered by the presence of an imbalanced microbiota composition and if such composition can be corrected with probiotic-like enhanced anti-inflammatory bacteria like P. distasonis.17
BALB/c mice inoculated with a whole cell lysate of P. distasonis prior to the onset of DSS-induced colitis demonstrated a significant decrease in inflammation compared to the control,17 thus providing some evidence supporting the anti-inflammatory role of P. distasonis in the intestinal microbiome. Several strains of P. distasonis showed anti-inflammatory effects both in vitro and in vivo and were able to restore both the epithelial barrier in a cell culture model and strengthen the gut barrier in a mouse model of 2,4,6-trinitrobenzenesulfonic acid (TNBS)-induced colitis. Intriguingly, P. distasonis stimulated CD4+ T cells to differentiate toward the CD4+ FoxP3+ IL-10+ regulatory phenotype.84 These results correlate with other studies illustrating the potential role of P. distasonis in stimulating regulatory T cell differentiation.85 A study by Koh et al.86 also corroborated similar findings in regard to P. distasonis’ ability to restore the epithelial barrier: the expression of the tight junction proteins Zonula occludens-1 (p < .001) and occludin (p < .001) were significantly increased in mice fed a diet containing 0.04% freeze-dried P. distasonis both at the transcriptional (2-3-fold, p < .01) and post-translational (30–50%, p < .05) levels, regardless of when P. distasonis was introduced into the diet of the mice, but so long as it was introduced.86 In another study, P. distasonis and several Bacteroides species were identified to attenuate E. coli lipopolysaccharide-induced IL-8 release from HT-29 cells and to lack genes to synthesize hexa-acylated, proinflammatory lipid A, exhibiting anti-inflammatory properties. P. distasonis was also found to exert enterocyte monolayer reinforcing action, reinforcing the gut barrier. The study suggested that P. distasonis and the other tested Bacteroides species could be used as “next-generation probiotics.”87
In past literature, it is consistently reported that there is a lack of species diversity in the inflamed regions of the gut overall. Strikingly, however, P. distasonis was identified as a recurring bacterium in the stools of CD patients.88 Notably, there are currently very few studies in humans that provide evidence for the idea that P. distasonis is a bacterium associated with any of the main forms of IBD, and potential evidence of the possible contribution of P. distasonis to IBD pathogenesis remains inconclusive; however, the results from studies in animal models still raise alarm. In one study, researchers demonstrated that in a DSS-induced colitis mouse model, P. distasonis had an anti-inflammatory effect on the gut; however, there is some evidence that, in mice with preexisting abnormal conditions such as SHIP or PGRP gene deficiency, P. distasonis seems to promote intestinal inflammation rather than attenuate it.89 This may be due to P. distasonis ability to protect itself against human immune responses.86 Furthermore, in a murine model of acute and chronic DSS-induced ulcerative colitis, P. distasonis abundance was significantly increased compared to healthy controls.90 More evidence for the potential proinflammatory role of P. distasonis in CD comes from one of our own studies. Here, germ-free senescence-accelerated prone mice (SAMP) were inoculated with P. distasonis. In mice already with Crohn’s disease, myeloperoxidase (MPO) activity was significantly increased, causing further inflammation in the gut.91
Peptidoglycan recognition proteins (PGRPs) are known to control inflammation in the gut, partly by reducing IFN-γ induction and NK cell migration.92 Mice deficient in any of the four types of PGRPs demonstrate markedly increased severity of DSS-induced experimental colitis and a more pro-inflammatory gut microbiome.92 Dziarski et al.93 demonstrated that, in PGRP-deficient mice, gut levels of P. distasonis were consistently elevated, indicating a possible lack of immune control for P. distasonis through PGRPs. Additionally, the gavage of wild type mice with P. distasonis enhanced DSS-induced colitis and possibly predisposed mice to IBD.89
A study by Gonzalez-Paez et al.93 found that C11 proteases could promote host immune responses and bacterial pathogenesis, especially via activation of bacterial pathogenic toxins from the likes of P. distasonis. Here, P. distasonis was found to promote intestinal inflammation in mouse models, degrading mucosal barrier health, and thus potentially contributing to the development of IBD. Additionally, the researchers reported that there was a correlation between elevated proteolytic activity and amino acids with the dysbiosis of the distal gut microbiome in patients. P. distasonis was found to influence gastrointestinal homeostasis and the responding immune activity, especially in the form of enteric cysteine proteases. These proteases were hypothesized to either be tethered to a bacterial cell wall; packaged into outer membrane vesicles (OMVs) that hydrolyze substrates from the parental bacterium surface, other bacteria, or host epithelial cells; or both.93
As previously mentioned, P. distasonis can produce catalase to enable its detoxifying role against oxidative stress mediated by hydrogen peroxide, often produced by inflammatory cells. However, it is suspected that these oxidative agents may be inflammatory triggers for CD.94,95 This is because catalases produced by various species of bacteria, including P. distasonis, catabolize reactive oxygen species (ROS), which may exacerbate inflammation.29
While there are few studies involving humans concerning the potential pathogenicity of P. distasonis in relation to IBD, those published present similarly alarming results. Nagayama et al.96 demonstrated that P. distasonis, along with nine other anaerobic bacteria cultured from the small intestinal mucosa of CD patients, enhanced TH1 and TH17 cell accumulation and intestinal inflammation.96 Furthermore, we have recently identified P. distasonis in the deep-gut wall tissues from patients that underwent surgery for the removal of chronically inflamed bowel segments, specifically patients afflicted with CD,97 supporting a probable pathologic role for this species in CD. This corroberates the observed enrichment of Bacteroidetes in the gut metagenome of the SAMP mouse line, which is naturally prone to CD-like ileitis.98
Taken together, these often-conflicting reports underscore the potential importance of P. distasonis in gut health and the need to elucidate its pathogenic effects so that clinically relevant solutions can be developed to address such effects. An overview of studies reporting an experimental or observational effect on IBD and intestinal health is presented in Table 2.
Table 2.
Examples of studies reporting effects of Parabacteroides distasonis (PD) on intestinal health
| Disease | Study Model | Study Design | Clinical Effect |
|---|---|---|---|
| Familiar Mediterranean Fever99 Inflammation |
Human blood serum and fecal samples. ELISA analysis of varying antibodies that correspond to different bacterial antigens in both FMF patients and healthy controls. DNA extraction was performed with Wizard Genomic DNA Purification kit and sequencing of the 16S rRNA performed with regions V1, V2, V3. | PD, along with other common gut flora, elicited an enhanced nonspecific humoral response to nonspecific antigens present on bacteria in the presence of FMF. | Aggravator |
| Colorectal cancer86 Cancer/Inflammation |
Six-week-old male A/J mouse models treated with different chow diets laced with PD. | Increase in colonic IL-10, TGF-β β and tight junction proteins Zonula occludens and occludin expression in mouse models given the PD long term in comparison to the control diet. Results support a protective role of PD in colonic tumorigenesis. | Protective |
| Colorectal cancer100 Cancer/Inflammation |
6-w-old male A/J mice fed low-fat (LF) diet, high-fat (HF) diet or a HF + whole freeze-dried PD diet (HF + Pd). Mice received 4 weekly injections of azoxymethane after 1 week on diet. PD analyzed with 16s rRNA gene sequencing. | PD membrane fraction (PdMB) largely suppressed production of pro-inflammatory cytokines, lowered MyD88 and pAkt abundance, and induced apoptosis in colon cancer cell lines, suggesting anti-inflammatory and anti-cancer effects. | Protective |
| Colitis84 Inflammation |
Murine Model of 2,4,6-Trinitrobenzenesulfonic Acid (TNBS)-Induced Colitis based on BALB/C ByJ mice. PD sequenced using V3-V4 16s rRNA regions. | PD reinforces the gut barrier and promotes pro-anti-inflammatory profile. This has a positive association with reducing colitis in tested mouse models. In vitro benefits relied heavily reliant on the strain. | Protective |
| Colitis17 Inflammation |
DSS-induced BALB/c mice – oral treatment of PD. PD analyzed via 16s rRNA sequencing. | PD membrane components decreases the severity of gut inflammation in the non-immunocompromised mouse models that had induced acute and chronic colitis. Also, increased PD serum antibodies and decreased pro-inflammatory cytokines. | Protective |
| Colorectal carcinogenesis101 Cancer/Inflammation |
Shotgun metagenomic sequencing analysis between feces of wild-type mice and mice with defects in TGFB signaling. Analysis of microbiota changes prior to colon tumors development. Shotgun metagenomics sequencing was performed using 150 BP, pair-ended sequences through Illumina sequencing. | PD abundance decreased when there were defects in transformation growth factor beta (TGFB) signaling pathway in mouse models. TGFB-deficient mice have more colorectal cancer and lower PD abundance. | Protective |
| Crohn’s Disease and Ulcerative Colitis89 Inflammation |
WT and antibiotic-depleted intestinal microflora mouse models. Pyrosequencing was performed on the variable regions of bacterial 16s rRNA. The sequences were classified with GreenGenes and compared with using QIIME. | Low PD abundance in patients with CD and UC. Higher PD abundance in gut microbiota of the Pglyrp-deficient mouse models promote colitis. | Aggravator |
7.2. Protective role in colorectal cancer
To date, P. distasonis has only been shown to have beneficial effects on colorectal cancer. Multiple researchers have identified that levels of P. distasonis in stool are inversely correlated to the presence of intestinal tumors.101 Koh et al.86 determined that P. distasonis membrane fractions were responsible for suppressing the production of pro-inflammatory cytokines in a colon cancer cell line. Studies by other researchers have suggested that P. distasonis has anti-inflammatory and anti-tumor properties mediated by the reduction of signaling via TLR4, MYD88, and Akt and the stimulation of apoptosis.86 These results are in agreement with observations of reduced microbiome levels of P. distasonis in mouse models of colorectal cancer.44 In a study by Koh et al.86 that was previously noted for revealing P. distasonis effect on the expression of tight junction proteins, the same mice were later treated with the carcinogen azoxymethane (AOM). The investigators stated that TLR4, IL-4, and TNF-α expression were 40% (p < .01 using a one-way ANOVA), 58% (p < .05), and 55% (p < .001) lower in mice fed a chow diet containing P. distasonis throughout the investigation than mice that were never fed a diet containing P. distasonis. In mice fed a diet containing P. distasonis after switching from a chow diet, the IL-10 and TGF-β expression were 217% (p = .05) and 185% (p < .001) higher compared to mice fed a chow diet without P. distasonis.86 Furthermore, Gu et al.101 found that the abundance of P. distasonis, in addition to that of Bacteroides vulgatus, decreased in response to CEACAM proteins disrupting TGF-β signaling. This alteration in the intestinal microbiome in turn promoted colorectal carcinogenesis, thus providing further evidence of P. distasonis potential role in preventing colorectal carcinogenesis.101
The evidence for the potential anti-inflammatory effects of P. distasonis in colorectal cancer is further supported by the inverse correlation between P. distasonis levels and IL-1β production in the gut.44 A comparison of the fecal microbiota composition between patients with spontaneous colorectal adenocarcinomas and patients without any proliferative lesions in the colon revealed a lack of P. distasonis in the patients with tumors.102 Collectively, these studies suggest that P. distasonis has anti-tumorigenic and anti-inflammatory potential in colorectal cancer patients.
7.3. Protective role in obesity
Further potential benefits of P. distasonis have been identified, particularly in relation to obesity. Xu et al.103 found that mice with high-fat diet-induced obesity (DIO) had higher relative abundances of P. distasonis and Akkermansia muciniphila in the gut, resulting in a gut microbiota that was significantly more capable of reducing host adiposity. Here, this change in gut microbiome composition was induced by Panax notoginseng saponins (PNS), often used as a form of traditional Chinese medicine. This suggests a potential ability for PNS and, in turn, P. distasonis, to be used in the treatment of obesity.103 This also raises the potential for P. distasonis to be important in modulating host adiposity. Furthermore, a study by Gallardo-Becerra et al.104 found that the abundance of P. distasonis in the gut microbiota of Mexican children with obesity and metabolic syndrome (MetS), a multicomponent condition associated with obesity, was reduced compared to Mexican children of normal weight. This, the investigators noted, was “correlated with clinical and anthropometric parameters associated with obesity and metabolic syndrome”.104 A study by Haro et al.105 illustrated a link between the gut microbiota, diet, and patients diagnosed with MetS. The study’s assertion implied that P. distasonis was originally diminished in MetS patients who were then presented with a Mediterranean diet for 2 years, partially restoring P. distasonis populations. The data from the MetS patients exhibited a negative relationship between waist circumference and the relative abundance of P. distasonis (R = −0.213). The patients on this Mediterranean diet, which was enriched with antioxidant phenolic-compound-rich foods, saw a statistically significant increase in the abundance of P. distasonis (p = .025 via ANOVA). These results suggest that the Mediterranean diet could potentially be utilized to correct microbial imbalances especially in reference to P. distasonis, providing further evidence that P. distasonis may be involved in mitigating obesity.105
Recently, Wang et al.106 demonstrated that P. distasonis alleviated obesity, hyperglycemia, and hepatic steatosis in ob/ob and high-fat diet mice via the production of secondary bile acids and a previously mentioned byproduct of fermentation: succinate. Here, succinate was found to bind to fructose-1,6-bisphosphatase, a rate-limiting enzyme involved in intestinal gluconeogenesis (IGN), decreasing hyperglycemia in ob/ob mice. Furthermore, treatment with live P. distasonis dramatically altered the bile acid profiles of the mice, increasing the levels of lithocholic acid (LCA) and ursodeoxycholic acid (UDCA), in turn reducing hyperlipidemia by activating the FXR pathway and, as a result, repairing gut barrier integrity, highlighting additional suspected benefits of P. distasonis in relation to obesity and gut barrier integrity.106 Of interest, the abundance of P. goldsteinii in feces has been reported to also have an inverse (protective) correlation with obesity in rats.107
7.4. Dichotomous role in diabetes
As with IBD, P. distasonis has been shown to have both beneficial and detrimental effects on diabetes, complicating its definition as a beneficial commensal or pathogenic bacterium. However, research on P. distasonis’ effects on diabetes is currently very limited. Cai et al.108 found that supplementing extract of propolis (EEP) in high-fat diet mice increased the abundance of P. distasonis, which was identified as an ‘anti-obesity and anti-inflammatory bacterium,’ in line with associated metabolic parameters of insulin resistance.106 Based on these findings, one may conclude that P. distasonis could play a role in decreasing insulin resistance and preventing diabetes.
However, some evidence that P. distasonis may be involved in the pathogenesis of diabetes has begun to emerge. Hasain et al,109 citing several metagenomics studies, concluded that P. distasonis, which has been shown to be enriched in women with gestational diabetes mellitus (GDM), could serve as a gut microbiota signature in women with GDM.109 This suggests that P. distasonis may play a role in the pathogenesis of certain types of diabetes rather than prevent it, as Cai et al.’s106 findings suggest, underscoring the importance of further investigation into P. distasonis’ effects on diabetes.
7.5. Gut microbiome and dichotomous role on autoimmune diseases
Several studies have suggested that P. distasonis may play a role in various forms of autoimmunity. For example, in a recent study addressing the potential functional relationship between gut bacteria and T cell responses in multiple sclerosis (MS), P. distasonis levels were shown to be lower in human MS patients compared to healthy controls.110 P. distasonis was shown to drive T cell differentiation toward an increased percentage of anti-inflammatory CD25+ T cells relative to the total CD3+ CD4+ T cell population. There was also an abundance of CD25+ IL-10+ FoxP3− Tr1 cells, which are strongly associated with the immunoregulatory phenotype.111 Interestingly, these results are corroborated by the elevated proportion of FoxP3+ T regulatory cells in the overall population of CD4+ T cells found in the colonic lamina propria of C57BL/6 J mice monocolonized with P. distasonis.85 Transplantation of the gut microbiome from the MS patients into germ-free mice augmented the severity of experimental autoimmune encephalomyelitis symptoms in comparison to their counterparts that received the microbiome transplant from healthy human donors.110
P. distasonis abundance was significantly elevated in fecal samples from patients with ankylosing spondylitis (AS), a chronic inflammatory disease affecting the spine and the sacroiliac joint. This suggests a possible role for P. distasonis in the development of more diverse autoimmune responses.112,113 In vitro experiments showed that P. distasonis, along with other AS-enriched species including Bacteroides coprophilus, Eubacterium siraeum, Acidaminococcus fermentans, and Prevotella copri, increased the amount of IFN-γ-producing cells through a bacterial peptide of these species, mimicking type II collagen and likely serving as “triggers of autoimmunity by molecular mimicry”.105,110,114
In psoriatic patients, P. distasonis presence was found to be significantly decreased relative to non-psoriatic patients.115 In mouse models of imiquimod-induced skin inflammation (IISI), animals treated with the antibiotic metronidazole showed a significantly higher abundance of P. distasonis in their intestines and demonstrated reduced severity of skin inflammation via downregulation of the TH17 immune response.116 These findings suggest that the intestinal microbiota may play an important role in the regulation of IISI.
Notably, Moreno-Arrones et al.117 found that patients with alopecia areata – an autoimmune disease mediated by T cells – had higher abundances of P. distasonis in their gut microbiota (LDA score > 2). In conjunction with Clostridiales vadin BB60 group, P. distasonis could correctly predict alopecia areata status in 80% of patients, indicating that P. distasonis could be involved in the pathophysiology of alopecia areata.117 However, whether these findings indicate a causative relationship between the abundance of P. distasonis in the gut and alopecia areata and if the bacterium presence serves as a biomarker for the disease remains to be elucidated. At the genus level, elevated systemic antibodies toward commensal gut P. distasonis and P. merdae have been reported to be consistently increased in Familial Mediterranean fever (FMF, which is an autoinflammatory condition characterized by acute, self-limiting episodes of fever and serositis and chronic subclinical inflammation in remission), irrespective of disease activity (remission, 2180 ± 1150 and 2508 ± 1241; vs. flare, 2383 ± 1207 and 2393 ± 1069) compared to controls (658 ± 161 and 995 ± 363, for P. distasonis and P. merdae, respectively).99 The relevance of these findings are not well understood because in IBD, increased IgA antibody concentrations have been found to work against other commensal microbes, including Lactobacillus casei.118
7.6. Dichotomous role in cardiovascular disease
P. distasonis has also been implicated in the pathogenesis of cardiovascular disease (CVD). However, as with its impact on diabetes, research on P. distasonis impact on CVD is limited. A study focused on exploring the relationship between the gut microbiota and CVD in patients with cardiac valve calcifications and coronary artery disease found P. distasonis and other bacterial species to be potential pathogens contributing to CVD.114
Conversely, a study on the role of the intestinal microbial flora in vascular inflammation in rats found a potential anti-inflammatory role for P. distasonis, contributing to potentially beneficial effects on CVD. Here, the relative abundance of P. distasonis was inversely correlated to the neointimal hyperplasia and composite intima+media area after a carotid artery angioplasty.119 Thus, the role of P. distasonis in the pathogenesis of CVD is yet to be determined and remains another point of controversy surrounding this bacterium. It is clear that more research will be needed to determine P. distasonis role in relation to CVD.
7.7. Established pathogen in intestinal and non-intestinal abscess formation
Abscesses are a prime hotspot for numerous infectious bacteria to manifest and thrive. Clinical studies have reported finding culturable P. distasonis isolates in abscesses. Clinical studies and case reports have implied a possible role for P. distasonis in abscess formation in various tissues, including the spleen,120,121 liver,122 and wounds.33 For example, Gunalan et al.120 reported a case of splenic abscess in a 40-year-old man presenting to the hospital with fever, left-side abdominal pain, altered sensorium, and vomiting. After the patient received antimicrobial therapy and underwent a splenectomy, it was discovered that pus aspirated from the splenic abscess grew P. distasonis. Gunalan et al.120 noted that this is one of only a few recorded cases of P. distasonis causing splenic abscess in humans; nonetheless, such a finding is alarming and supportive of a pathogenic role of P. distasonis in human infections.120 Furthermore, CD4+ T-cells were shown to play a key role in the formation of P. distasonis-induced intra-abdominal abscesses in rodent models.123
The mechanism behind P. distasonis pathology regarding abscess formation is still under investigation. One study examined the abscedative and intra-abdominal sepsis role of P. distasonis infections using rats that were intraperitoneally infected with different bacterial pathogens, namely, Staphylococcus aureus, Bacteroides fragilis, and a combination of Enterococcus faecium and P. distasonis.123 The study aimed to define the mechanisms by which i) T-cells mediate abscess induction secondary to intra-abdominal sepsis, ii) the contribution of T-cell activation and iii) the role of co-stimulation of antigen-presenting cells via CD28-B7 pathways. Researchers found that T cells activated with zwitterionic bacterial polysaccharides in vitro required CD28-B7 co-stimulation to induce abscesses when adoptively transferred to the peritoneal cavity of naïve rats, promoting abscess formation. Although not exclusively specific to P. distasonis pathogenesis, the study demonstrated that blockade of T-cell activation via the CD28-B7 with CTLA4Ig prevented abscess formation, while an alarming 82.4% (n = 28) of 34 induced abscess yielded P. distasonis.123
7.8. Cervical cancer, ketogenic diet and glutamate and gamma-aminobutyric acid
It has become apparent that P. distasonis might have a modulatory effect on the predisposition to or protection against numerous other types of diseases. For example, elevated levels of P. distasonis were positively associated with the progression of cervical cancer.124 However, the number of patients in this particular study was rather small, so these results should be interpreted with caution. More possible positive roles for P. distasonis and P. merdae were described in another recent study. Both of these Parabacteroides species were shown to promote the beneficial anti-seizure effects of the ketogenic diet. Presence of these bacterial species strongly correlated with protection against seizures, potentially via increasing levels of glutamate and gamma-aminobutyric acid (GABA) in the hippocampus.125 Reduced levels of GABA are well-known to exacerbate seizures.126 Furthermore, a whole metagenome sequence analysis demonstrated a lower abundance of P. distasonis in children with autism spectrum disorder (ASD) compared to their neurotypical counterparts. Here, metagenomic analysis revealed decreases in the expression of genes linked to the production of melatonin, butyric acid, and GABA.127 A summary of these and additional studies reporting an experimental or observational effect on non-intestinal diseases is presented in Table 3.
Table 3.
Examples of studies reporting effects of Parabacteroides distasonis (PD) on non-intestinal health
| Disease of Interest | Study Model | Study Effect/Type of Association | Effect/PMID |
|---|---|---|---|
| Necrotizing fasciitis (NF)128 Muscular |
Patient of study diagnosed with HIV. Fournier’s Gangrene Polymicrobial mixture isolated from a patient’s tissue culture. Computerized tomographic imaging | Opportunistic/polymicrobial mixture found in perineal and scrotal abscess included PD. | Aggravator |
| Oral Health and Patients with Acrylic partial dentures129 Digestive |
Patients lacking teeth and using prosthetic treatments. Microbial culture using Schaedler K3 solid medium with 5% sheep blood at 37°C after use of active toothpaste containing propolis and tee tree oil-containing hygienic agent versus control group. | PD isolated from some patients before and on day 7 of trial using tested toothpaste. Authors stated that PD was “eliminated” after use of active toothpaste. | N/A |
| Obesity, Hyperlipidemia, hepatic steatosis, Intestinal Gluconeogenesis106 Digestive/Endocrine |
Obese mouse models modulating gut microbiota. In vivo assays that validate beneficial effects of PD. The bacterial 16s rRNA regions V3 and V4 were sequenced using Illumina HiSeq PE250. The primers F341 and R806 were used. | PD associated with reduced weight gain, decrease of hyperglycemia, and hepatic steatosis in obese and high-fat (HDF)-fed mice. PD is lower in patients that are obese. Improved glucose homeostasis and obesity-related abnormalities. | Protective |
| Gestational diabetes mellitus and gestational diabetes109 | Human, fecal samples | GDM is associated with metabolic disorder phenotypes (obesity, low-grade inflammation, insulin resistance). PD has high abundance in women with GDM. Suggested as part of gut microbiota signature for GDM. | PositiveCorrelation (Aggravator?) |
| Amyotrophic lateral sclerosis (ALS)130 Nervous System |
ALS-prone Sod1 transgenic (Sod1-Tg) mouse models. 16s rDNA sequencing was performed on region V4 using a Illumina MiSeq kit with 2 × 250 BP pair-ended sequencing. | PD reportedly exacerbates ALS symptoms whereas other bacteria such as Akkermansia muciniphila (AM) improves ALS symptoms. | Aggravator |
| Multiple Sclerosis110 Inflammation/Neurological |
Germ-free mouse models. | PD was of less abundance in multiple sclerosis patients. However when introduced to mouse model. PD stimulated anti-inflammatory IL-10-expressing human CD4+ CD25 + T cells and IL-10+ FoxP3+ Tregs in mouse models. | Protective |
| Ankylosing spondylitis113 Inflammation/Autoimmune disorder |
Fecal microbial metagenomic analysis of patients. | PD along with other microbiota found in Ankylosing spondylitis (AS) patients. May be a trigger of autoimmunity via molecular mimicry. | Aggravator |
| Alopecia Areata117 Autoimmune disorder |
16S rRNA sequencing of stool samples. | Alopecia areata is T-cell mediated autoimmune disease and gut microbiota has been identified as key modulator of this disease. PD could be used as potential diagnostic tools due to its enriched presence in stool samples. | Aggravator |
| Autism spectrum disorders127 Developmental Disorder |
Metagenomic analysis of fecal specimens of children. | There were decreases in the average abundance of gut microbiota in children with autism spectrum disorder (ASD), including PD. Gut microbiota can be a neurometabolic signature for ASD transcriptional and metabolomic activity. | N/A |
8. Genomics, phages, and genetic engineering
Among the Parabacteroides species associated with the human gut, P. distasonis has the smallest genome (<5Mb, vs >6.5Mb) and the smallest repertoire of genes that are members of the environmental sensing and gene regulation categories. P. distasonis type strain ATCC 8503 possesses a 4,811,369-bp genome, 3,867 protein-coding genes, and shares 1,416 sets of orthologous protein-coding genes with other gut Bacteroidetes. Figure 2 illustrates the genomic relatedness of the Parabacteroides genus with that of other relatively new reclassified genera within the Bacteroidetes phylum. Additionally, P. distasonis has the smallest number of genes associated with carbon source degradation; however, P. distasonis has two classes of carbohydrate-processing enzymes that are more abundant in its proteome than in the proteomes of other gut Bacteroidetes.37 According to the PATRIC database, all P. distasonis sequences strains correspond to single chromosome isolates, with only one phage reported in the system (Parabacteroides phage YZ-2015a, NCBI Taxon ID 1,655,644)131.
Figure 2.
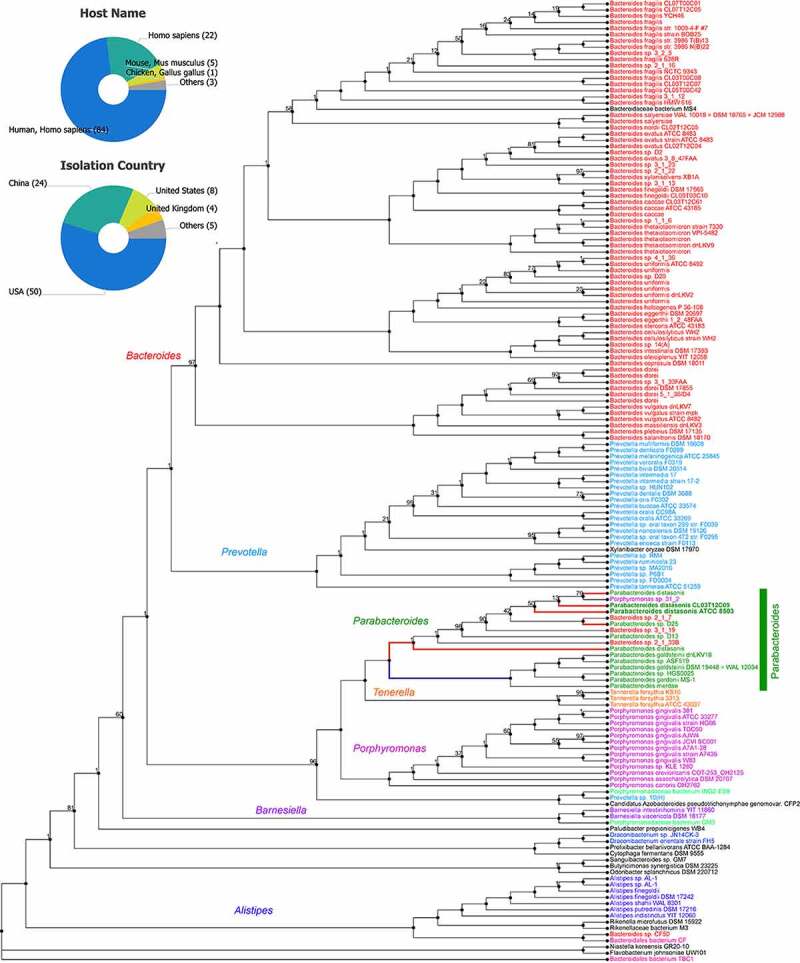
Protein phylogram of 155 complete genomes of the Bacteroidetes phylum to illustrate the potential functional distinction of Parabacteroides distasonis from other species within the genus. The pipeline for genomic phylograms is described in detail based on information from PATRIC, the Pathosystems Resource Integration Center, https://docs.patricbrc.org. In short, the order-level pre-built trees in PATRIC are constructed by an automated pipeline that begins with amino acid sequence files for each genome. For each order-level tree the genomes from that order are used along with a small set of potential outgroup genomes. Branch values are not bootstrap values, which can be overly optimistic for long genomes. Instead, trees are built from random samples of 50% of the homology groups used for the main tree (gene-wise jackknifing). One hundred of these 50% gene-wise jackknife trees are made using FastTree, and the support values shown indicate the number of times a particular branch was observed in the support trees. As of January 12, 2021, there were 133 P. distasonis genomes available, of which 8 are complete (pie charts).131
Recently, a comparative genomic study of Microviridae, a family of bacteria-infecting ssDNA viruses (deemed as a poorly characterized bacteriophage group, even though it includes phage PhiX174, which is one of the main models in virology for genomic and capsid structure studies) across 17 peatlands showed that two new distinct prophages were identified in the genomes of P. merdae and P. distasonis representing a potential new subfamily of Microviridae that matches the protein similarity of the viron capsulatr protein VP1, in viromes of French wetlands.132
Elegantly, Quaiser et al.132 showed through Blast searches that the major capsid protein VP1 sequences from the assembled viral wetland genomes showed similarities to VP1 proteins encoded in the P. merdae and P. distasonis genomes.2 The Parabacteroides VP1 genes show that genes encoding for homolog VP2 and VP4 genes (other capsular phage proteins) were juxtaposed next to the bacterial VP1 genes, supporting the presence of a prophage Microviridae in both Parabacteroides species. Comparing their organization, the identified prophage regions flanking the VP1 gene (5 kbp) were extracted from the genomes and considered circular for analysis, using the VP1 gene as an arbitrary start. Synteny analysis showed, remarkably, that the prophages have the same gene order in such bacteria (P. distasonis: VP1-ORF1-ORF2-ORF3-VP2-VP4, and P. merdae: VP1-ORF1-VP2-VP4) suggesting that both have a common functional prophage ancestor. The gene coding for the arbitrarily assigned protein ORF1 downstream of VP1 in both bacteria were specific to each prophage and did not match to genes from other Microviridae or in NCBI non-redundant databases. This strengthens the hypothesis that these prophages represent a distinct subfamily of Microviridae, suggesting the relevance of this genus and species in water sources previously described.
P. distasonis can make deacetylated products available for itself and other components of the microbiota, devoting a greater proportion of its genome to protein degradation than other Bacteroidetes such as B. thetaiotaomicron.37 This supports the postulate that in vitro tests that quantify the proteolytic activity of this and other potentially pathogenic microbes are not predictive of the genomic potential that exists among gut Bacteroidetes. For example, gelatin hydrolysis and collagenase tests may suggest a lack of proteolytic activity in P. distasonis, but, as previously mentioned, we have recovered this species from micro cavitating (fistulizing) lesions in the gut wall of Crohn’s disease resected bowels, supporting the potential for proinflammatory activity in IBD.97 Additional tests and growth assays will be performed to confirm this observation.
As of January 12, 2021, there are 133 genomes for P. distasonis registered in the bioinformatics resource database PATRIC ver. 3.6.8, which provides integrated data and analysis tools to support biomedical research on infectious diseases, including data on genome sequencing efforts.131 At least seven of these genome sequencing efforts are complete assemblies of strains derived from sources described “feces,” “clinical isolates,” a “peritonitis case,” “sphagnum-peat soil,” and “intramural gut wall lesions from Crohn’s disease patients.”131 Most other registered genomes originate from isolates sourced from feces, while at least seven genomes originate from unspecified regions of the human gut and two additional genomes originate from unspecified parts of the rat gut. At least five genomes originate from sources specified as “tissues.” Numerous isolates originate from mice. Unfortunately, however, most isolates have nonspecific descriptions with respect to the source.131
Other identified P. distasonis strains of interest include P. distasonis ATCC 82G9, P. distasonis NBRC 113,806, P. distasonis ATCC 8503, P. distasonis CavFT-Har46, and P. distasonis FDAARGOS_615. The complete genome sequence of P. distasonis CavFT-Har46 was completed by our team and is of great clinical interest as this strain was isolated from a gut wall-cavitating microlesion in a patient with severe Crohn’s Disease. We have identified that this strain exhibits an 80% match to other P. distasonis strains, including strains P. distasonis ATCC 8503 and 82G9, both isolated from the feces of patients.97
Due to horizontal gene transfer of antimicrobial resistance genes, Bacteroides and Parabacteroides isolates from European and American patients now show tetracycline and erythromycin resistance, complicating genetic selection processes. Furthermore, while genetic manipulation via CRISPR-Cas9 can be done on Bacteroidales, off-target mutations, inefficient transformation, and the need for strain-specific modifications prevent CRISPR-Cas9 from being an effective method for molecular-level genetic manipulation.133 Recently, however, a new avenue for the genetic manipulation of P. merdae and diverse Bacteroides isolates from the human gut microbiota has been identified by García-Bayona & Comstock. This avenue consists of placing a selection cassette based on the dietary fiber inulin (the selection of which P. merdae is thought to be amenable to) into pNBU2, a “pir-dependent suicide vector,” resulting in facilitated transconjugation in P. merdae. This signifies that this combination, dubbed “pLGB28,” could be utilized in the genetic engineering of P. distasonis, thus providing a facilitated method of genetic manipulation for the species.133
9. Conclusions and future directions
P. distasonis is a unique bacterium that is involved in many of the biochemical processes of numerous human diseases. While there are apparent associations between P. distasonis, related IBDs, and numerous other diseases, there is a lack of an established consensus on P. distasonis’ role in modulating the human gut microbiota and, more importantly, the pathogenicity of the bacterium. Going forward, the directive for new studies is to understand and identify the mechanisms of P. distasonis, its pathogenesis, its antimicrobial resistance, and its commensal relationship with the gut mucosal wall. Additionally, it is imperative to understand its impact on our intestinal microbiota.
Ultimately, it may be that P. distasonis does not cleanly fit into either the beneficial commensal bacterium or pathogenic bacterium categories: perhaps P. distasonis, and potentially, other species of bacteria, straddle both definitions. If this were the case, it could have major implications for research on the gut microbiome, including prompting further investigation into other bacteria thought to be beneficial commensals for potential pathogenicity. However, more research is needed to determine whether P. distasonis truly straddles both the pathogenic and beneficial commensal definitions.
Acknowledgments
This project was primarily supported by NIH grant R21 DK118373 to A.R.-P., entitled “Identification of pathogenic bacteria in Crohn’s disease.” We acknowledge the Biorepository Core of the NIH Silvio O. Conte Cleveland Digestive Disease Research Core Center (P30DK097948) for clinical advice and provision of deidentified surgical specimens for the above-mentioned R21 project. Partial support also originated for A.R.-P. from a career development award from the Crohn’s and Colitis Foundation and National Institutes of Health award 2P01DK091222-06 (Germ-Free and Gut Microbiome Core to A.R.-P. and to F.C., Case Western Reserve University). Further support came from the Los Alamos National Laboratory (LANL) internal grants, the Laboratory Directed Research and Development grant 20160340ER. J.C.E. was supported by an internship summer award from The Ohio State University Second-Year Transformational Experience (STEP) program. D.K.S. is supported by a University Scholarship from Case Western Reserve University. A.H. is with the University of Toledo College of Medicine and Life Sciences, H.L.E. is currently affiliated with Cornell University, D.E.C. is with the University of Pennsylvania.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Author’s Contribution
A.R.-P identified the need for and conceived the manuscript. All authors contributed to the review, discussion of the findings, and wrote the manuscript. All authors discussed and edited the manuscript and approved its final version.
Contribution to the field
This is a review of the controversial and intriguing roles of Parabacteroides distasonis (PD) on human and animal health. This is a re-classified bacterium, originally called Bacteroides distasonis, and a part of the Bacteroidetes phylum. This well-established and extensively described bacterium was identified and isolated from patients with clinical infections as well as from microscopic cavitating lesions in the gut wall of a patient with Crohn’s disease, a major prototype of inflammatory bowel disease (IBD). These observations contrast those made by other investigators who have reported potential probiotic effects of P. distasonis for a multitude of diseases, including IBD.
References
- 1.Parker BJ, Wearsch PA, Veloo ACM, Rodriguez-Palacios A. The Genus Alistipes: Gut Bacteria With Emerging Implications to Inflammation, Cancer, and Mental Health. Front Immunol. 2020;11:906. doi: 10.3389/fimmu.2020.00906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sakamoto M, Benno Y. Reclassification of Bacteroides distasonis, Bacteroides goldsteinii and Bacteroides merdae as Parabacteroides distasonis gen. nov., comb. nov., Parabacteroides goldsteinii comb. nov. and Parabacteroides merdae comb. nov. Int J Syst Evol Microbiol. 2006. July;56(Pt7):1599–27. doi: 10.1099/ijs.0.64192-0. [DOI] [PubMed] [Google Scholar]
- 3.Parte AC, Sardà Carbasse J, Meier-Kolthoff JP, Reimer LC, Göker M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol. Nov 2020;70(11):5607–5612. doi: 10.1099/ijsem.0.004332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang YJ, Xu XJ, Zhou N, Sun Y, Liu C, Liu S-J, You X. Parabacteroides acidifaciens sp. nov., isolated from human faeces. Int J Syst Evol Microbiol. 2019. March;69(3):761–766. doi: 10.1099/ijsem.0.003230. [DOI] [PubMed] [Google Scholar]
- 5.Dione N, Ngom II, Valles C, Cadoret F, Fournier PE, Raoult D, Lagier JC. Collinsella provencensis' sp. nov., 'Parabacteroides bouchesdurhonensis' sp. nov. and 'Sutterella seckii,' sp. nov., three new bacterial species identified from human gut microbiota. New Microbes New Infect. 2018. May;23:44–47. doi: 10.1016/j.nmni.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan HQ, Li TT, Zhu C, Zhang XQ, Wu M, Zhu XF. Parabacteroides chartae sp. nov., an obligately anaerobic species from wastewater of a paper mill. Int J Syst Evol Microbiol. 2012. November;62(Pt11):2613–2617. doi: 10.1099/ijs.0.038000-0. [DOI] [PubMed] [Google Scholar]
- 7.Kitahara M, Sakamoto M, Tsuchida S, Kawasumi K, Amao H, Benno Y, Ohkuma M. Parabacteroides chinchillae sp. nov., isolated from chinchilla (Chincilla lanigera) faeces. Int J Syst Evol Microbiol. 2013. September;63(Pt9):3470–3474. doi: 10.1099/ijs.0.050146-0. [DOI] [PubMed] [Google Scholar]
- 8.Kim H, Im WT, Kim M, Kim D, Seo YH, Yong D, Jeong SH, Lee K. Parabacteroides chongii sp. nov., isolated from blood of a patient with peritonitis. J Microbiol. 2018. October;56(10):722–726. doi: 10.1007/s12275-018-8122-3. [DOI] [PubMed] [Google Scholar]
- 9.Sakamoto M, Tanaka Y, Benno Y, Ohkuma M. Parabacteroides faecis sp. nov., isolated from human faeces. Int J Syst Evol Microbiol. 2015. April;65(Pt4):1342–1346. doi: 10.1099/ijs.0.000109. [DOI] [PubMed] [Google Scholar]
- 10.Song Y, Liu C, Lee J, Bolanos M, Vaisanen ML, Finegold SM. “Bacteroides goldsteinii sp. nov.” isolated from clinical specimens of human intestinal origin. J Clin Microbiol. 2005. September;43(9):4522–4527. doi: 10.1128/JCM.43.9.4522-4527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bellali S, Lo CI, Naud S, Fonkou MDM, Armstrong N, Raoult D, Fournier P-E, Fenollar F. Parabacteroides massiliensis sp. nov., a new bacterium isolated from a fresh human stool specimen. New Microbes New Infect. 2019. November;32:100602. doi: 10.1016/j.nmni.2019.100602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson JL, Moore WEC, Moore LVH. Bacteroides caccae sp. nov., Bacteroides merdae sp. nov., and Bacteroides stercoris sp. nov. Isolated from Human Feces. Int J Syst Evol Microbiol. 1986;36(4):499–501. doi: 10.1099/00207713-36-4-499. [DOI] [Google Scholar]
- 13.Benabdelkader S, Naud S, Lo CI, Fadlane A, Traore SI, Aboudharam G, La Scola B. Parabacteroides pacaensis sp. nov. and Parabacteroides provencensis sp. nov., two new species identified from human gut microbiota. New Microbes New Infect. 2020. March;34:100642. doi: 10.1016/j.nmni.2019.100642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bilen M, Mbogning Fonkou MD, Khelaifia S, Tomei E, Cadoret F, Daoud Z, Armstrong N, Bittar F, Fournier P-E, Raoult D, Dubourg G. Taxonogenomics description of Parabacteroides timonensis sp. nov. isolated from a human stool sample. Microbiologyopen. 2019;804(4):e00702. 10.1002/mbo3.702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schoch CL, Ciufo S, Domrachev M, Hotton CL, Kannan S, Khovanskaya R, Leipe D, Mcveigh R, O’Neill K, Robbertse B, Sharma S, Soussov V, Sullivan JP, Sun L, Turner S, Karsch-Mizrachi I. NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database (Oxford). 2020. January 1;2020. doi: 10.1093/database/baaa062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu HY, Hou R, Yang GQ, Zhao F, Dong WG. In vitro effects of inulin and soya bean oligosaccharide on skatole production and the intestinal microbiota in broilers. J Anim Physiol Anim Nutr (Berl). 2018. June;102(3):706–716. doi: 10.1111/jpn.12830. [DOI] [PubMed] [Google Scholar]
- 17.Kverka M, Zakostelska Z, Klimesova K, Sokol D, Hudcovic T, Hrncir T, Rossmann P, Mrazek J, Kopecny J, Verdu EF, Tlaskalova-Hogenova H. Oral administration of Parabacteroides distasonis antigens attenuates experimental murine colitis through modulation of immunity and microbiota composition. Clin Exp Immunol. 2011. February;163(2):250–259. doi: 10.1111/j.1365-2249.2010.04286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Distaso A. Digested and diluted serum as a substitute for broth for bacteriological purposes. Br Med J. 1916;2(2912):555–556. doi: 10.1136/bmj.2.2912.555-a.20768341 [DOI] [Google Scholar]
- 19.Snyder J. Good germs, bad germs: health and survival in a bacterial world. New York City: Hill and Wang; 2007. Vol. 304. [Google Scholar]
- 20.Clarridge JE Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev. 2004. October;17(4):840–862. table of contents. doi: 10.1128/CMR.17.4.840-862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shah HN, Collins MD. Fatty acid and isoprenoid quinone composition in the classification of Bacteroides melaninogenicus and related taxa. J Appl Bacteriol. 1980. February;48(1):75–87. doi: 10.1111/j.1365-2672.1980.tb05209.x. [DOI] [PubMed] [Google Scholar]
- 22.Kreader CA. Persistence of PCR-detectable Bacteroides distasonis from human feces in river water. Appl Environ Microbiol. 1998. October;64(10):4103–4105. doi: 10.1128/AEM.64.10.4103-4105.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baums IB, Goodwin KD, Kiesling T, Wanless D, Diaz MR, Fell JW. Luminex detection of fecal indicators in river samples, marine recreational water, and beach sand. Mar Pollut Bull. 2007. May;54(5):521–536. doi: 10.1016/j.marpolbul.2006.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allsop K, Stickler DJ. An assessment of Bacteroides fragilis group organisms as indicators of human faecal pollution. J Appl Bacteriol. 1985. January;58(1):95–99. doi: 10.1111/j.1365-2672.1985.tb01433.x. [DOI] [PubMed] [Google Scholar]
- 25.Polansky O, Sekelova Z, Faldynova M, Sebkova A, Sisak F, Rychlik I, Dozois CM. Important metabolic pathways and biological processes expressed by chicken cecal microbiota. Appl Environ Microbiol. 2015. December;82(5):1569–1576. doi: 10.1128/AEM.03473-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Y, Li K, Luo H, Duan L, Wei C, Wang M, Jin J, Liu S, Mehmood K, Shahzad M. Comparison of the intestinal microbial community in ducks reared differently through high-throughput sequencing. Biomed Res Int. 2019;2019:9015054. doi: 10.1155/2019/9015054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez-Palacios A, Mo KQ, Shah BU, Msuya J, Bijedic N, Deshpande A, Ilic S. Global and Historical Distribution of Clostridioides difficile in the Human Diet (1981-2019): Systematic Review and Meta-Analysis of 21886 Samples Reveal Sources of Heterogeneity, High-Risk Foods, and Unexpected Higher Prevalence Toward the Tropic. Front Med (Lausanne). 2020;7:9. doi: 10.3389/fmed.2020.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zamani S, Hesam Shariati S, Zali MR, Asadzadeh Aghdaei H, Sarabi Asiabar A, Bokaie S, Nomanpour B, Sechi LA, Feizabadi MM. Detection of enterotoxigenic Bacteroides fragilis in patients with ulcerative colitis. Gut Pathog. 2017;9(53). doi: 10.1186/s13099-017-0202-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gregory EM, Kowalski JB, Holdeman LV. Production and some properties of catalase and superoxide dismutase from the anaerobe Bacteroides distasonis. J Bacteriol. 1977. March;129(3):1298–1302. doi: 10.1128/JB.129.3.1298-1302.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rocha ER, Smith CJ. Biochemical and genetic analyses of a catalase from the anaerobic bacterium Bacteroides fragilis. J Bacteriol. 1995. June;177(11):3111–3119. doi: 10.1128/JB.177.11.3111-3119.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilkins TD, Wagner DL, Veltri BJ, Gregory EM. Factors affecting production of catalase by Bacteroides. J Clin Microbiol. 1978. November;8(5):553–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng J, Teng F, Murray BE. Gelatinase is important for translocation of Enterococcus faecalis across polarized human enterocyte-like T84 cells. Infect Immun. 2005. March;73(3):1606–1612. doi: 10.1128/IAI.73.3.1606-1612.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tong J, Liu C, Summanen P, Xu H, Finegold SM. Application of quantitative real-time PCR for rapid identification of Bacteroides fragilis group and related organisms in human wound samples. Anaerobe. 2011. April;17(2):64–68. doi: 10.1016/j.anaerobe.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 34.Rodríguez-Sánchez B, Alcalá L, Marín M, Ruiz A, Alonso E, Bouza E. Evaluation of MALDI-TOF MS (Matrix-Assisted laser desorption-Ionization time-of-flight mass spectrometry) for routine identification of anaerobic bacteria. Anaerobe. 2016. December;42:101–107. doi: 10.1016/j.anaerobe.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 35.Veloo ACM, Jean-Pierre H, Justesen US, Morris T, Urban E, Wybo I, Kostrzewa M, Friedrich AW, On Behalf of the Enria Workgroup. An overview of the data obtained during the validation of an optimized MALDI-TOF MS biotyper database for the identification of anaerobic bacteria. Data Brief. 2018. June;18:1484–1496. doi: 10.1016/j.dib.2018.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barba MJ, Fernández A, Oviaño M, Fernández B, Velasco D, Bou G. Evaluation of MALDI-TOF mass spectrometry for identification of anaerobic bacteria. Anaerobe. 2014. December;30:126–128. doi: 10.1016/j.anaerobe.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 37.Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, Martens EC, Henrissat B, Coutinho PM, Minx P, Latreille P, Cordum H, Brunt AV, Kim K, Fulton RS, Fulton LA, Clifton SW, Wilson RK, Knight RD, Gordon JI. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007. July;5(7):e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.García-López M, Meier-Kolthoff JP, Tindall BJ, Gronow S, Woyke T, Kyrpides NC, Hahnke RL, Göker M. Analysis of 1,000 type-strain genomes improves Taxonomic classification of Bacteroidetes. Front Microbiol. 2019;10:2083. doi: 10.3389/fmicb.2019.02083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hahnke RL, Meier-Kolthoff JP, García-López M, Mukherjee S, Huntemann M, Ivanova NN, Woyke T, Kyrpides NC, Klenk H-P, Göker M, et al. Corrigendum: genome-based Taxonomic classification of Bacteroidetes. Front Microbiol. 2018;9:304. doi: 10.3389/fmicb.2018.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hahnke RL, Meier-Kolthoff JP, García-López M, Mukherjee S, Huntemann M, Ivanova NN, Woyke T, Kyrpides NC, Klenk H-P, Göker M. Genome-based Taxonomic classification of Bacteroidetes. Front Microbiol. 2016;7:2003. doi: 10.3389/fmicb.2016.02003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hildebrand F, Moitinho-Silva L, Blasche S, Jahn MT, Gossmann TI, Huerta-Cepas J, Hercog R, Luetge M, Bahram M, Pryszlak A, Alves RJ, Waszak SM, Zhu A, Ye L, Costea PI, Aalvink S, Belzer C, Forslund SK, Sunagawa S, Hentschel U, Merten C, Patil KR, Benes V, Bork P. Antibiotics-induced monodominance of a novel gut bacterial order. Gut. 2019. January;68(10):1781–1790. doi: 10.1136/gutjnl-2018-317715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fletcher CM, Coyne MJ, Bentley DL, Villa OF, Comstock LE. Phase-variable expression of a family of glycoproteins imparts a dynamic surface to a symbiont in its human intestinal ecosystem. Proc Natl Acad Sci U S A. 2007. February;104(7):2413–2418. doi: 10.1073/pnas.0608797104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corthier G, Muller MC, L’Haridon R. Selective enumeration of Bacteroides vulgatus and B. distasonis organisms in the predominant human fecal flora by using monoclonal antibodies. Appl Environ Microbiol. 1996. February;62(2):735–738. doi: 10.1128/AEM.62.2.735-738.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pfalzer AC, Nesbeth PD, Parnell LD, Iyer LK, Liu Z, Kane AV, Chen CYO, Tai AK, Bowman TA, Obin MS, Mason JB, Greenberg AS, Choi S-W, Selhub J, Paul L, Crott JW. Diet- and genetically-induced obesity differentially affect the fecal microbiome and metabolome in Apc1638N mice. PLoS One. 2015;10(8):e0135758. doi: 10.1371/journal.pone.0135758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boros M, Ghyczy M, Érces D, Varga G, Tőkés T, Kupai K, Torday C, Kaszaki J. The anti-inflammatory effects of methane. Crit Care Med. 2012. April;40(4):1269–1278. doi: 10.1097/CCM.0b013e31823dae05. [DOI] [PubMed] [Google Scholar]
- 46.Chen O, Ye Z, Cao Z, Manaenko A, Ning K, Zhai X, Zhang R, Zhang T, Chen X, Liu W, Sun X. Methane attenuates myocardial ischemia injury in rats through anti-oxidative, anti-apoptotic and anti-inflammatory actions. Free Radic Biol Med. 2016. January;90:1–11. doi: 10.1016/j.freeradbiomed.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 47.Song K, Zhang M, Hu J, Wang Y, Wang Y, Ma X, Ma X. Methane-rich saline attenuates ischemia/reperfusion injury of abdominal skin flaps in rats via regulating apoptosis level. BMC Surg. 2015. July;15(1):92. doi: 10.1186/s12893-015-0075-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu J, Wang R, Ye Z, Sun X, Chen Z, Xia F, Sun Q, Liu L. Protective effects of methane-rich saline on diabetic retinopathy via anti-inflammation in a streptozotocin-induced diabetic rat model. Biochem Biophys Res Commun. 2015. October;466(2):155–161. doi: 10.1016/j.bbrc.2015.08.121. [DOI] [PubMed] [Google Scholar]
- 49.Zhang X, Li N, Shao H, Meng Y, Wang L, Wu Q, Yao Y, Li J, Bian J, Zhang Y, Deng X. Methane limit LPS-induced NF-κB/MAPKs signal in macrophages and suppress immune response in mice by enhancing PI3K/AKT/GSK-3β-mediated IL-10 expression. Sci Rep. 2016;6(1):29359. doi: 10.1038/srep29359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roccarina D, Lauritano EC, Gabrielli M, Franceschi F, Ojetti V, Gasbarrini A. The role of methane in intestinal diseases. Am J Gastroenterol. 2010. June;105(6):1250–1256. doi: 10.1038/ajg.2009.744. [DOI] [PubMed] [Google Scholar]
- 51.Jahng J, Jung IS, Choi EJ, Conklin JL, Park H.. The effects of methane and hydrogen gases produced by enteric bacteria on ileal motility and colonic transit time. Neurogastroenterol Motil. 2012. February;24(2):185–190. e92. doi: 10.1111/j.1365-2982.2011.01819.x. [DOI] [PubMed] [Google Scholar]
- 52.Tannahill GM, Curtis AM, Adamik J, Palsson-mcdermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, Zheng L, Gardet A, Tong Z, Jany SS, Corr SC, Haneklaus M, Caffrey BE, Pierce K, Walmsley S, Beasley FC, Cummins E, Nizet V, Whyte M, Taylor CT, Lin H, Masters SL, Gottlieb E, Kelly VP, Clish C, Auron PE, Xavier RJ, O'Neill LAJ. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013. April;496(7444):238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mills E, O’Neill LA. Succinate: a metabolic signal in inflammation. Trends Cell Biol. 2014. May;24(5):313–320. doi: 10.1016/j.tcb.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 54.Basson A, Trotter A, Rodriguez-Palacios A, Cominelli F. Mucosal interactions between genetics, diet, and microbiome in inflammatory Bowel disease. Front Immunol. 2016;7:290. doi: 10.3389/fimmu.2016.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guzy RD, Sharma B, Bell E, Chandel NS, Schumacker PT. Loss of the SdhB, but not the SdhA, subunit of complex II triggers reactive oxygen species-dependent hypoxia-inducible factor activation and tumorigenesis. Mol Cell Biol. 2008. January;28(2):718–731. doi: 10.1128/MCB.01338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kietzmann T, Görlach A. Reactive oxygen species in the control of hypoxia-inducible factor-mediated gene expression. Semin Cell Dev Biol. 2005. Aug-Oct;16(4–5):474–486. doi: 10.1016/j.semcdb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 57.Dang EV, Barbi J, Yang HY, Jinasena D, Yu H, Zheng Y, Bordman Z, Fu J, Kim Y, Yen H-R, Luo W, Zeller K, Shimoda L, Topalian SL, Semenza GL, Dang CV, Pardoll DM, Pan F. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011. September;146(5):772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rubic T, Lametschwandtner G, Jost S, Hinteregger S, Kund J, Carballido-Perrig N, Schwärzler C, Junt T, Voshol H, Meingassner JG, Mao X, Werner G, Rot A, Carballido JM. Triggering the succinate receptor GPR91 on dendritic cells enhances immunity. Nat Immunol. 2008. November;9(11):1261–1269. doi: 10.1038/ni.1657. [DOI] [PubMed] [Google Scholar]
- 59.Inagaki A, Ichikawa H, Sakata T. Inhibitory effect of succinic acid on epithelial cell proliferation of colonic mucosa in rats. J Nutr Sci Vitaminol (Tokyo). 2007. August;53(4):377–379. doi: 10.3177/jnsv.53.377. [DOI] [PubMed] [Google Scholar]
- 60.Guerre-Millo M, Grosfeld A, Issad T.. Leptin is a hypoxia-inducible gene. Obes Res. 2002. August;10(8):856. author reply 857-8. doi: 10.1038/oby.2002.116. [DOI] [PubMed] [Google Scholar]
- 61.Clement K, Langin D. Regulation of inflammation-related genes in human adipose tissue. J Intern Med. 2007. October;262(4):422–430. doi: 10.1111/j.1365-2796.2007.01851.x. [DOI] [PubMed] [Google Scholar]
- 62.Kierzkowska MMA, Mlynarczyk G, Mlynarczyk G. Trends and impact in antimicrobial resistance among bacteroides and parabacteroides species in 2007–2012 compared to 2013–2017. Microb Drug Resist. 2020. December;26(12):1452–1457. doi: 10.1089/mdr.2019.0462. [DOI] [PubMed] [Google Scholar]
- 63.Blazevic D. Antibiotic susceptibility of the subspecies of Bacteroides fragilis. Antimicrob Agents Chemother. 1976. March;9(3):481–484. doi: 10.1128/AAC.9.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Avelar KEVJ, Antunes LC, Lobo LA, Antunes EN, Domingues RM, Ferreira MC, Ferreira MCS. Antimicrobial resistance of strains of the Bacteroides fragilis group isolated from the intestinal tract of children and adults in Brazil. Int J Antimicrob Agents. 2001. August;18(2):129–134. doi: 10.1016/S0924-8579(01)00354-5. [DOI] [PubMed] [Google Scholar]
- 65.Kierzkowska MMA, Chmura A, Chmura A, Chmura A, Durlik M, Deborska-Materkowska D, Paczek L, Mlynarczyk G, Mlynarczyk G. Specific character of anaerobic bacterial infections in patients treated in transplantation wards at one of the clinical hospitals in Warsaw. Transplant Proc. 2014. Щсе;46(8):2586–2588. doi: 10.1016/j.transproceed.2014.08.033. [DOI] [PubMed] [Google Scholar]
- 66.Boente RFFL, Falcão LS, Miranda KR, Guimarães PL, Santos-Filho J, Vieira JM, Barroso DE, Emond JP, Ferreira EO, Paula GR, Domingues RMCP. Detection of resistance genes and susceptibility patterns in Bacteroides and Parabacteroides strains. Anaerobe. 2010. June;16(3):190–194. doi: 10.1016/j.anaerobe.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 67.Nakano V, Silva ADNE, Merino VRC, Wexler HM, Avila-Campos MJ, Nascimento e Silva A, Merino VR, Wexler HM, Avila-Campos MJ. Antimicrobial resistance and prevalence of resistance genes in intestinal Bacteroidales strains. Clinics (Sao Paulo). 2011;66(4):543–547. doi: 10.1590/S1807-59322011000400004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Imamura H, Kobayashi T, Murata K, Kohbata S, Watanabe I, Marui T, Watanabe K, Ninomiya K, Ueno K, Suzuki S. [Antibacterial activity of cefoperazone against anaerobic bacteria (author’s transl)]. Jpn J Antibiot. 1980November, 33(11), 1171–1182. [PubMed] [Google Scholar]
- 69.Byun JH, Kim M, Lee Y, Lee K, Chong Y. Antimicrobial susceptibility patterns of anaerobic bacterial clinical isolates from 2014 to 2016, including recently named or renamed species. Ann Lab Med. 2019. March;39(2):190–199. doi: 10.3343/alm.2019.39.2.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Asahi Y, Watanabe K, Ueno K. Biochemical properties of beta-lactamase produced by Bacteroides distasonis. J Antibiot (Tokyo). 1989. June;42(6):960–967. doi: 10.7164/antibiotics.42.960. [DOI] [PubMed] [Google Scholar]
- 71.Hurlbut S, Cuchural GJ, Tally FP. Imipenem resistance in Bacteroides distasonis mediated by a novel beta-lactamase. Antimicrob Agents Chemother. 1990. January;34(1):117–120. doi: 10.1128/AAC.34.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goldstein EJCD, Citron DM. Comparative in-vitro activity of cefoperazone/sulbactam and 11 other agents against multidrug resistant Bacteroides fragilis group species. J Antimicrob Chemother. 1996. October;38(4):733–737. doi: 10.1093/jac/38.4.733. [DOI] [PubMed] [Google Scholar]
- 73.Sheikh WPD, Nadler H, Nadler H. Antibacterial activity of meropenem and selected comparative agents against anaerobic bacteria at seven North American centers. Clin Infect Dis. 1993;16(Suppl4):S361–6. doi: 10.1093/clinids/16.Supplement_4.S361. [DOI] [PubMed] [Google Scholar]
- 74.Bourgault AMLF, Hoban DJ, Dalton MT, Kibsey PC, Smith JA, Smith JA, Low DE, Gilbert H, Gilbert H. Survey of Bacteroides fragilis group susceptibility patterns in Canada. Antimicrob Agents Chemother. 1992. February;36(2):343–347. doi: 10.1128/AAC.36.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Snydman DRJN, McDermott LA, Ruthazer R, Goldstein EJ, Finegold SM, Harrell LJ, Hecht DW, Jenkins SG, Pierson C, Venezia R, Rihs J, Gorbach SL. National survey on the susceptibility of Bacteroides Fragilis group: report and analysis of trends for 1997-2000. Clin Infect Dis. 2002. September 1;35(Suppl 1):S126–34. doi: 10.1086/341934. [DOI] [PubMed] [Google Scholar]
- 76.Aldridge KEAD, Cambre K, Pierson CL, Jenkins SG, Rosenblatt JE, Rosenblatt JE. Multicenter survey of the changing in vitro antimicrobial susceptibilities of clinical isolates of Bacteroides fragilis group, Prevotella, Fusobacterium, Porphyromonas, and Peptostreptococcus species. Antimicrob Agents Chemother. 2001. April;45(4):1238–1243. doi: 10.1128/AAC.45.4.1238-1243.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kierzkowska M, Majewska A, Szymanek-Majchrzak K, Sawicka-Grzelak A, Mlynarczyk A, Mlynarczyk G. In vitro effect of clindamycin against Bacteroides and Parabacteroides isolates in Poland. J Glob Antimicrob Resist. 2018. June;13:49–52. doi: 10.1016/j.jgar.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 78.Arzese ARTL, Botta GA. Detection of tetQ and ermF antibiotic resistance genes in Prevotella and Porphyromonas isolates from clinical specimens and resident microbiota of humans. J Antimicrob Chemother. 2000. May;45(5):577–582. doi: 10.1093/jac/45.5.577. [DOI] [PubMed] [Google Scholar]
- 79.Chung WOWC, Schwarz S, Roberts MC, Roberts MC. Host range of the ermF rRNA methylase gene in bacteria of human and animal origin. J Antimicrob Chemother. 1999. January;43(1):5–14. doi: 10.1093/jac/43.1.5. [DOI] [PubMed] [Google Scholar]
- 80.Arthur MB-NA, Courvalin P, Arthur M, Brisson-Noël A, Courvalin P. Origin and evolution of genes specifying resistance to macrolide, lincosamide and streptogramin antibiotics: data and hypotheses. J Antimicrob Chemother. 1987. December;20(6):783–802. doi: 10.1093/jac/20.6.783. [DOI] [PubMed] [Google Scholar]
- 81.Quesada-Gómez C, Rodríguez-Cavallini E, Rodríguez C. Scarce detection of mobile erm genes associated with tetQ in Bacteroides and Parabacteroides from Costa Rica. Anaerobe. 2013. June;21:18–21. doi: 10.1016/j.anaerobe.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 82.Xu JH, Jiang Z, Solania A, Chatterjee S, Suzuki B, Lietz CB, Hook VYH, O’Donoghue AJ, Wolan DW. A Commensal Dipeptidyl Aminopeptidase with Specificity for N-Terminal Glycine Degrades Human-Produced Antimicrobial Peptides in Vitro. ACS Chem Biol. 2018. October;13(9):2513–2521. doi: 10.1021/acschembio.8b00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sóki J, Wybo I, Hajdú E, Toprak NU, Jeverica S, Stingu C-S, Tierney D, Perry JD, Matuz M, Urbán E, Nagy E. A Europe-wide assessment of antibiotic resistance rates in Bacteroides and Parabacteroides isolates from intestinal microbiota of healthy subjects. Anaerobe. 2020. April;62:102182. doi: 10.1016/j.anaerobe.2020.102182. [DOI] [PubMed] [Google Scholar]
- 84.Cuffaro B, Assohoun ALW, Boutillier D, Súkeníková L, Desramaut J, Boudebbouze S, Salomé-Desnoulez S, Hrdý J, Waligora-Dupriet A-J, Maguin E, Grangette C. In vitro characterization of gut microbiota-derived commensal strains: selection of Parabacteroides distasonis strains alleviating TNBS-induced colitis in mice. Cells. 2020. September 16;9(9):2104. doi: 10.3390/cells9092104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med. 2014. January;6(220):220ra11. doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Koh GY, Kane AV, Wu X, Crott JW. Parabacteroides distasonis attenuates tumorigenesis, modulates inflammatory markers and promotes intestinal barrier integrity in azoxymethane-treated A/J mice. Carcinogenesis. 2020. July 14;41(7):909–917. doi: 10.1093/carcin/bgaa018. [DOI] [PubMed] [Google Scholar]
- 87.Hiippala K, Kainulainen V, Suutarinen M, Heini T, Bowers JR, Jasso-Selles D, Lemmer D, Valentine M, Barnes R, Engelthaler DM, Satokari R. Isolation of Anti-Inflammatory and Epithelium Reinforcing Bacteroides and Parabacteroides Spp. from A Healthy Fecal Donor. Nutrients. 2020. March;12(4):935. doi: 10.3390/nu12040935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lopetuso LR, Petito V, Graziani C, Schiavoni E, Paroni Sterbini F, Poscia A, Gaetani E, Franceschi F, Cammarota G, Sanguinetti M, Luca M, Scaldaferri F, Gasbarrini A. Gut microbiota in health, diverticular disease, irritable bowel syndrome, and inflammatory bowel diseases: time for microbial marker of gastrointestinal disorders. Dig Dis. 2018;36(1):56–65. doi: 10.1159/000477205. [DOI] [PubMed] [Google Scholar]
- 89.Dziarski R, Park SY, Kashyap DR, Dowd SE, Gupta D, Mizoguchi E. Pglyrp-regulated gut Microflora Prevotella falsenii, Parabacteroides distasonis and Bacteroides eggerthii enhance and Alistipes finegoldii Attenuates colitis in mice. PLoS One. 2016;11(1):e0146162. doi: 10.1371/journal.pone.0146162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Okayasu IHS, Yamada M, Ohkusa T, Inagaki Y, Nakaya R, Nakaya R. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology. 1990. March;98(3):694–702. doi: 10.1016/0016-5085(90)90290-H. [DOI] [PubMed] [Google Scholar]
- 91.Rodriguez-Palacios A, Conger M, Hopperton A, Ezeji JC, Erkkila HL, Fiocchi C, Cominelli F. 25 IDENTIFICATION OF PATHOGENIC BACTERIA IN SEVERE CROHN’S DISEASE. Gastroenterology. 2019;156(3):S102. doi: 10.1053/j.gastro.2019.01.238. [DOI] [Google Scholar]
- 92.Saha SJX, Park SY, Wang S, Li X, Gupta D, Dziarski R, Dziarski R. Peptidoglycan recognition proteins protect mice from experimental colitis by promoting normal gut flora and preventing induction of interferon-gamma. Cell Host Microbe. 2010. August 19;8(2):147–162. doi: 10.1016/j.chom.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.González-Páez GE, Roncase EJ, Wolan DW. X-ray structure of an inactive zymogen clostripain-like protease from Parabacteroides distasonis. Acta Crystallographica Section D Structural Biology. 2019. March;75(3):325–332. doi: 10.1107/S2059798319000809. [DOI] [PubMed] [Google Scholar]
- 94.Gregory EM, Fanning DD; Gregory EM, Fanning DD . Effect of heme on Bacteroides distasonis catalase and aerotolerance. J Bacteriol. 1983. December;156(3):1012–1018. doi: 10.1128/JB.156.3.1012-1018.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Iborra M, Moret I, Rausell F, Bastida G, Aguas M, Cerrillo E, Nos P, Beltrán B. Role of oxidative stress and antioxidant enzymes in Crohn’s disease. Biochem Soc Trans. 2011. August;39(4):1102–1106. doi: 10.1042/BST0391102. [DOI] [PubMed] [Google Scholar]
- 96.Nagayama M, Yano T, Atarashi K, Tanoue T, Sekiya M, Kobayashi Y, Sakamoto H, Miura K, Sunada K, Kawaguchi T et al. TH1 cell-inducing Escherichia coli strain identified from the small intestinal mucosa of patients with Crohn’s disease. Gut Microbes. 2020. November 9;12(1):1788898. doi: 10.1080/19490976.2020.1788898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang F, Kumar A, Davenport KW, Kelliher JM, Ezeji JC, Good CE, Jacobs MR, Conger M, West G, Fiocchi C, et al. Complete genome sequence of a Parabacteroides distasonis strain (CavFT hAR46) isolated from a gut wall-cavitating Microlesion in a patient with severe Crohn’s disease. Microbiol Resour Announc. September 2019;8(36). doi: 10.1128/MRA.00585-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rodriguez-Palacios A, Harding A, Menghini P, Himmelman C, Retuerto M, Nickerson KP, Lam M, Croniger CM, McLean MH, Durum SK et al. The artificial sweetener splenda promotes Gut Proteobacteria, Dysbiosis, and Myeloperoxidase reactivity in Crohn’s disease-Like Ileitis. Inflamm Bowel Dis. 2018. April;24(5):1005–1020. doi: 10.1093/ibd/izy060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Manukyan GP, Ghazaryan KA, Ktsoyan ZA, Khachatryan ZA, Arakelova KA, Kelly D, Grant G, Aminov RI. Elevated systemic antibodies towards commensal gut microbiota in autoinflammatory condition. PLoS One. 2008. September;3(9):e3172. doi: 10.1371/journal.pone.0003172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Koh GY, Kane A, Lee K, Xu Q, Wu X, Roper J, Mason JB, Crott JW. Parabacteroides distasonis attenuates toll-like receptor 4 signaling and Akt activation and blocks colon tumor formation in high-fat diet-fed azoxymethane-treated mice. Int J Cancer. 2018. April;143(7):1797–1805. doi: 10.1002/ijc.31559. [DOI] [PubMed] [Google Scholar]
- 101.Gu S, Zaidi S, Hassan MI, Mohammad T, Malta TM, Noushmehr H, Nguyen B, Crandall KA, Srivastav J, Obias V et al. Mutated CEACAMs disrupt transforming growth factor beta signaling and alter the intestinal microbiome to promote colorectal carcinogenesis. Gastroenterology. 2020. January;158(1):238–252. doi: 10.1053/j.gastro.2019.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Polimeno LBM, Mosca A, Viggiani MT, Joukar F, Mansour-Ghanaei F, Mavaddati S, Daniele A, Debellis L, Bilancia M, Santacroce L, et al. Soy metabolism by gut microbiota from patients with precancerous intestinal lesions. Microorganisms. 2020. March 25;8(4):469. doi: 10.3390/microorganisms8040469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xu Y, Wang N, Tan HY, Li S, Zhang C, Zhang Z, Feng Y. Panax notoginseng saponins modulate the gut microbiota to promote thermogenesis and beige adipocyte reconstruction via leptin-mediated AMPKα/STAT3 signaling in diet-induced obesity. Theranostics. 2020;10(24):11302–11323. doi: 10.7150/thno.47746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gallardo-Becerra L, Cornejo-Granados F, García-López R, Valdez-Lara A, Bikel S, Canizales-Quinteros S, López-Contreras BE, Mendoza-Vargas A, Nielsen H, Ochoa-Leyva A et al. Metatranscriptomic analysis to define the Secrebiome, and 16S rRNA profiling of the gut microbiome in obesity and metabolic syndrome of Mexican children. Microb Cell Fact. 2020. March 6;19(1):61. doi: 10.1186/s12934-020-01319-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Haro C, Montes-Borrego M, Rangel-Zúñiga OA, Alcalá-Díaz JF, Gómez-Delgado F, Pérez-Martínez P, Delgado-Lista J, Quintana-Navarro GM, Tinahones FJ, Landa BB et al. Two healthy diets modulate gut microbial community improving insulin sensitivity in a human obese population. J Clin Endocrinol Metab. 2016. January;101(1):233–242. doi: 10.1210/jc.2015-3351. [DOI] [PubMed] [Google Scholar]
- 106.Wang K, Liao M, Zhou N, Bao L, Ma K, Zheng Z, Wang Y, Liu C, Wang W, Wang J, Liu S-J, Liu H. Parabacteroides distasonis alleviates obesity and metabolic dysfunctions via production of succinate and secondary bile acids. Cell Rep. 2019. January;26(1):222–235.e5. doi: 10.1016/j.celrep.2018.12.028. [DOI] [PubMed] [Google Scholar]
- 107.Pan C, Guo Q, Lu N. Role of gut microbiota in the Pharmacological effects of natural products. Evid Based Complement Alternat Med. 2019;2019:2682748. doi: 10.1155/2019/2682748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cai W, Xu J, Li G, Liu T, Guo X, Wang H, Luo L. Ethanol extract of propolis prevents high-fat diet-induced insulin resistance and obesity in association with modulation of gut microbiota in mice. Food Res Int. 2020. April;130:108939. doi: 10.1016/j.foodres.2019.108939. [DOI] [PubMed] [Google Scholar]
- 109.Hasain Z, Mokhtar NM, Kamaruddin NA, Mohamed Ismail NA, Razalli NH, Gnanou JV, Raja Ali RA. Gut microbiota and gestational diabetes mellitus: a review of host-gut microbiota interactions and their therapeutic potential. Front Cell Infect Microbiol. 2020;10:188. doi: 10.3389/fcimb.2020.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cekanaviciute E, Yoo BB, Runia TF, Debelius JW, Singh S, Nelson CA, Kanner R, Bencosme Y, Lee YK, Hauser SL, et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc Natl Acad Sci U S A. 2017;114(40):10713–10718. doi: 10.1073/pnas.1711235114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lee YKMJ, Umesaki Y, Mazmanian SK, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA. 2011;108(Supplement_1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huang RLF, Zhou Y, Zeng Z, He X, Fang L, Pan F, Chen Y, Lin J, Li J, Qiu D, et al. Metagenome-wide association study of the alterations in the intestinal microbiome composition of ankylosing spondylitis patients and the effect of traditional and herbal treatment. J Med Microbiol. 2020. June;69(6):797–805. doi: 10.1099/jmm.0.001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhou C, Zhao H, Xiao XY, Chen B-D, Guo R-J, Wang Q, Chen H, Zhao L-D, Zhang C-C, Jiao Y-H, Ji Y-M, Yang H-X, Fei Y-Y, Wang L, Shen M, Li H, Wang X-H, Lu X, Yang Bo, Liu J-J, Li J, Peng L-Y, Zheng W-J, Zhang C-Y, Zhou J-X, Wu Q-J, Yang Y-J, Su J-M, Shi Q, Wu D, Zhang W, Zhang F-C, Jia H-J, Liu D-P, Jie Z-Y, Zhang X. Metagenomic profiling of the pro-inflammatory gut microbiota in ankylosing spondylitis. J Autoimmun. 2020. February;107:102360. doi: 10.1016/j.jaut.2019.102360. [DOI] [PubMed] [Google Scholar]
- 114.Liu Z, Li J, Liu H, Tang Y, Zhan Q, Lai W, Ao L, Meng X, Ren H, Xu D, et al. The intestinal microbiota associated with cardiac valve calcification differs from that of coronary artery disease. Atherosclerosis. 2019. May;284:121–128. doi: 10.1016/j.atherosclerosis.2018.11.038. [DOI] [PubMed] [Google Scholar]
- 115.Shapiro J, Cohen NA, Shalev V, Uzan A, Koren O, Maharshak N. Psoriatic patients have a distinct structural and functional fecal microbiota compared with controls. J Dermatol. 2019. May;46(7):595–603. doi: 10.1111/1346-8138.14933. [DOI] [PubMed] [Google Scholar]
- 116.Stehlikova Z, Kostovcikova K, Kverka M, Rossmann P, Dvorak J, Novosadova I, Kostovcik M, Coufal S, Srutkova D, Prochazkova P, et al. Crucial role of microbiota in experimental psoriasis revealed by a gnotobiotic mouse model. Front Microbiol. 2019;10:236. doi: 10.3389/fmicb.2019.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Moreno-Arrones OM, Serrano-Villar S, Perez-Brocal V, Saceda‐Corralo D, Morales‐Raya C, Rodrigues‐Barata R, Moya A, Jaen‐Olasolo P, Vano‐Galvan S. Analysis of the gut microbiota in alopecia areata: identification of bacterial biomarkers. J Eur Acad Dermatol Venereol. 2020. February;34(2):400–405. doi: 10.1111/jdv.15885. [DOI] [PubMed] [Google Scholar]
- 118.Hevia A, López P, Suárez A, Jacquot C, Urdaci MC, Margolles A, Sánchez B. Association of levels of antibodies from patients with inflammatory bowel disease with extracellular proteins of food and probiotic bacteria. Biomed Res Int. 2014;2014:351204. doi: 10.1155/2014/351204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cason CAKT, Chen EB, Wun K, Nooromid MJ, Xiong L, Gottel NR, Harris KG, Morton TC, Avram MJ, Chang EB, et al. Microbiota composition modulates inflammation and neointimal hyperplasia after arterial angioplasty. J Vasc Surg. 2020. April;71(4):1378–1389. doi: 10.1016/j.jvs.2019.06.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gunalan ABR, Sridharan B, Elamurugan TP, Elamurugan TP. Pathogenic potential of Parabacteroides distasonis revealed in a splenic abscess case: a truth unfolded. BMJ Case Rep. 2020. December 13;13(12):e236701. doi: 10.1136/bcr-2020-236701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Al-Tawfiq J. Bacteroides (Parabacteroides) distasonis splenic abscess in a sickle cell patient. Intern Med. 2008;47(1):69–72. doi: 10.2169/internalmedicine.47.0428. [DOI] [PubMed] [Google Scholar]
- 122.Higashi TMY, Katsurada K, Katsurada K. A case of gas-containing liver abscess with multiple metastatic lesions. Kansenshogaku Zasshi. 1995. September;69(9):1017–1020. doi: 10.11150/kansenshogakuzasshi1970.69.1017. [DOI] [PubMed] [Google Scholar]
- 123.Tzianabos AO, Chandraker A, Kalka-Moll W, Stingele F, Dong VM, Finberg RW, Peach R, Sayegh MH. Bacterial pathogens induce abscess formation by CD4(+) T-cell activation via the CD28-B7-2 costimulatory pathway. Infect Immun. 2000. December;68(12):6650–6655. doi: 10.1128/IAI.68.12.6650-6655.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mikamo H, Izumi K, Ito K, Watanabe K, Ueno K, Tamaya T. Internal bacterial flora of solid uterine cervical cancer. Kansenshogaku Zasshi. 1993. November;67(11):1057–1061. doi: 10.11150/kansenshogakuzasshi1970.67.1057. [DOI] [PubMed] [Google Scholar]
- 125.Olson CAVH, Yano JM, Liang QY, Nusbaum DJ, Hsiao EY. The gut microbiota mediates the anti-seizure effects of the ketogenic diet. Cell. 1728-1741;2018(173). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Griffiths JA, Mazmanian SK. Emerging evidence linking the gut microbiome to neurologic disorders. Genome Med. 2018;10(1):98. doi: 10.1186/s13073-018-0609-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Averina OV, Kovtun AS, Polyakova SI, Savilova AM, Rebrikov DV, Danilenko VN. The bacterial neurometabolic signature of the gut microbiota of young children with autism spectrum disorders. J Med Microbiol. 2020. April;69(4):558–571. doi: 10.1099/jmm.0.001178. [DOI] [PubMed] [Google Scholar]
- 128.Weimer SB, Matthews MR, Caruso DM, Foster KN. Retroperitoneal necrotizing fasciitis from Fournier’s gangrene in an immunocompromised patient. Case Rep Surg. 2017;2017:5290793. doi: 10.1155/2017/5290793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wiatrak K, Morawiec T, Rój R, Mertas A, Machorowska-Pieniążek, Kownacki P, Tanasiewicz M, Skucha-Nowak M, Baron S, Piekarz T, Wrzol M, Bogacz M. Oral health of patients treated with acrylic partial dentures using a toothpaste containing bee product. Evid Based Complement Alternat Med. 2017;2017:4034179. doi: 10.1155/2017/4034179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Blacher E, Bashiardes S, Shapiro H, Rothschild D, Mor U, Dori-Bachash M, Kleimeyer C, Moresi C, Harnik Y, Zur M et al. Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature. 2019. August;572(7770):474–480. doi: 10.1038/s41586-019-1443-5. [DOI] [PubMed] [Google Scholar]
- 131.Davis JJ, Wattam AR, Aziz RK, Brettin T, Butler R, Butler RM, Chlenski P, Conrad N, Dickerman A, Dietrich EM et al. The PATRIC bioinformatics resource center: expanding data and analysis capabilities. Nucleic Acids Res. 2020. January;48(D1):D606–D612. doi: 10.1093/nar/gkz943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Quaiser A, Dufresne A, Ballaud F, Roux S, Zivanovic Y, Colombet J, Sime-Ngando T, Francez A-J. Diversity and comparative genomics of microviridae in sphagnum- dominated peatlands. Front Microbiol. 2015;6:375. doi: 10.3389/fmicb.2015.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.García-Bayona L, Comstock LE, Lemon KP. Streamlined genetic manipulation of diverse Bacteroides and Parabacteroides isolates from the human gut microbiota. mBio. 2019. August;10(4). doi: 10.1128/mBio.01762-19. [DOI] [PMC free article] [PubMed] [Google Scholar]


