Abstract
Background
Esophageal squamous cell carcinoma (ESCC) is among the most prevalent causes of cancer-related death in adults. Tumor microenvironment (TME) has been associated with therapeutic failure and lethal outcomes for patients. However, published reports on the heterogeneity and TME in ESCC are scanty.
Methods
Five tumor samples and five corresponding non-malignant samples were subjected to scRNA-seq analysis. Bulk RNA sequencing data were retrieved in publicly available databases.
Findings
From the scRNA-seq data, a total of 128,688 cells were enrolled for subsequent analyses. Gene expression and CNV status exhibited high heterogeneity of tumor cells. We further identified a list of tumor-specific genes and four malignant signatures, which are potential new markers for ESCC. Metabolic analysis revealed that energy supply-related pathways are pivotal in cancer metabolic reprogramming. Moreover, significant differences were found in stromal and immune cells between the esophagus normal and tumor tissues, which promoted carcinogenesis at both cellular and molecular levels in ESCC. Immune checkpoints, regarded as potential targets for immunotherapy in ESCC were significantly highly expressed in ESCC, including LAG3 and HAVCR2. Eventually, we constructed a cell-to-cell communication atlas based on cancer cells and immune cells and performed the flow cytometry, qRT-PCR, immunofluorescence, and immunohistochemistry analyses to validate the results.
Interpretation
This study demonstrates a widespread reprogramming across multiple cellular elements within the TME in ESCC, particularly in transcriptional states, cellular functions, and cell-to-cell interactions. The findings offer an insight into the exploration of TME and heterogeneity in the ESCC and provide new therapeutic targets for its clinical management in the future.
Keywords: Esophageal squamous cell carcinoma, Single-cell RNA-seq, Tumor microenvironment, Heterogeneity, Immunotherapy
Research in Context.
Evidence before this study
ESCC is classified among the most malignant and fatal carcinomas in adults. However, its general prognosis is universally poor regardless of the advances in treatments, which is attributed to a high degree of heterogeneity that potentially complicates the treatment and contributes to failure in ESCC patients. The tumor microenvironment (TME) plays an indispensable role in therapeutic failure and lethal outcomes for patients. However, the heterogeneity and TME, particularly in malignant cells are yet to be explored extensively at the single-cell level.
Added-value of this study
In this work, we reveal the great heterogeneity of tumor cells in both gene expression and CNV status at a single cell level. Through metabolic analysis, we suggest that OXPHOS is crucial in cancer metabolic reprogramming. Besides, our exploration of immune cells demonstrates that LAG3 and HAVCR2 are potential checkpoint molecules for immunotherapy in ESCC. This study also constructed a landscape of cellular communications, which revealed the existence of complex intercellular communication networks in ESCC.
Implications of all the available evidence
Our study comprehensively characterized the widespread reprogramming across multiple cellular elements within the TME in ESCC. Also, it established a list of prognostic signatures and potential therapeutic targets for ESCC patients. Therefore, we believe that our findings will serve as a valuable resource, to drive further exploration of ESCC pathogenesis.
Alt-text: Unlabelled box
1. Introduction
As one of the most prevalent causes of cancer-related death in adults, esophageal squamous cell carcinoma is associated with an abysmal overall 5-year survival rate (10%) and a disappointing 5-year post esophagostomy survival rate (15–40%) [1], [2], [3]. With the advancement in research, compelling evidence suggests that ESCC is much more complicated than previously thought since it transpires as a mixture of diverse cells [4]. The tumor microenvironment (TME), which comprises different stromal and tumor cells, is closely related to tumor processes, including proliferation, invasion, metastasis, and resistance to drugs [5,6]. Besides, heterogeneity, a general feature for tumors, is highly associated with therapeutic failure and lethal outcomes for patients [7,8]. Previously, numerous researchers found that heterogeneity could be attributed to the different sensitivity of treatment in malignant tumors [9], [10], [11]. However, heterogeneity and TME in ESCC are yet to be extensively explored.
Over the past decades, most transcriptome analyses of ESCC patients are based on traditional RNA sequencing (RNA-seq) data, raising our awareness of the genesis and development of ESCC. However, with traditional RNA-seq technology, one can primarily explore tumors on a whole sample level, but this is a limited approach to explore the cellular diversity and molecular complexity of tumor cells. In recent years, single-cell sequencing, geared on the cellular and molecular characteristics, has provided an insight into the mechanisms underlying the normal physiological or biochemical processes and diseases at a single cell level [12], [13], [14]. For instance, Lambrechts et al. [15] undertaken a single-cell analysis on lung cancer to explore the stromal cell types of lung tumors. They explored the phenotypes and biological behaviors of the different stromal cell types, which were fundamental in lung cancer diagnosis and treatment. Elsewhere, Zhang et al. [16] applied a single-cell analysis to explore the immune cells in hepatocellular carcinoma. They uncovered a dynamic nature of immune cells and communication across distinct cell types. These findings were valuable in the exploration of the potential therapeutic targets of liver cancer.
Herein, we adopted both traditional RNA-seq and scRNA-seq techniques to investigate cancer cells and the TME of ESCC. Through the analysis of tumor cells and finding their match with non-malignant tissues at a much larger scale, great heterogeneity of tumor cells was revealed in gene expression and CNV status. Moreover, we identified a list of tumor-specific genes and four malignant signatures, which are potential new targets for ESCC. The metabolic analysis demonstrated the pivotal roles of the energy supply-related pathways in cancer metabolic reprogramming. Meanwhile, this work characterized the single-cell expression profiles for immune cell lineages in esophagus normal and cancerous tissues. To better explore the interactions between immune cells and malignant cells in ESCC, a landscape of cellular metabolisms and communications was constructed within the single-cell resolution. We believe these findings will enrich our understanding of cellular and molecular differences between ESCC and non-tumor tissues.
2. Materials and methods
2.1. Ethics statement
Approval for this study was issued by the Ethics Committee of Zhongshan Hospital, Fudan University, China (B2021–137R). Patients gave informed consent at hospitalization.
2.2. Patients
scRNA-seq and clinical data were collected from patients diagnosed with ESCC. We selected 10 samples (five ESCC and five corresponding non-malignant samples) from the Department of Thoracic Surgery, Zhongshan Hospital, Fudan University (FDZSH). Another twenty ESCC and twenty corresponding non-malignant samples were also selected for flow cytometry and qRT-PCR analyses, whereas eight ESCC and eight corresponding non-malignant samples were included for immunohistochemistry and immunofluorescence. Non-malignant samples were collected from match patients, from sites displaced at least several centimeters from the tumor, and were subjected to histopathology review to verify that they lack tumor cells.
Traditional RNA-seq data of ESCC were retrieved from TCGA (https://tcgadata.nci.nih.gov/) database [17] and GEO database (GSE53625) [18].
2.3. Preparation of single-cell suspensions
All fresh samples were processed as follows: Each sample was minced into small pieces and digested in a gentle MACS C Tube containing 200 µL enzyme H, 25 µL enzyme A, 100 µL enzyme R, and 4.7 mL Dulbecco's Modified Essential Medium and incubated for 30 min at 37 °C. To remove the cell aggregates or other residual large particles from the single-cell suspension, the cell suspension was filtered through a 40-um nylon mesh. Red Blood Cell Lysis Solution (10 ×) (Sigma-Aldrich, St. Louis, MO, USA), and Dead Cell Removal Kit (Miltenyi Biotec) were further used to remove erythrocytes, and dead cells, respectively
2.4. Library preparation and sequencing
Library preparation was conducted fusing the 10X Chromium single-cell kit following the manufacturer's protocol. The libraries were sequenced on the Illumina sequencing platform (HiSeq X Ten; Illumina, San Diego, CA, USA).
2.5. Single-cell RNA-seq data preprocessing
The Cell Ranger software pipeline (version 3.0.0) was adopted to process the 10X genomics raw data. The Cell Ranger was applied to demultiplex raw base call files into FASTQ files, and for alignment, filtering, barcode counting, and UMI counting.
2.6. The 10x scRNA-seq data analysis
scRNA-seq data analyses were performed using R version 3.6.1 with the following criteria for cell filtering: (1) The number of expressed genes lower than 200 or larger than 5000; (2) 10% or more of UMIs were mapped to mitochondrial or ribosomal genes cells were eliminated if they met one of the standards. To detect the doublets, first, cells with UMI/gene numbers were eliminated from the limit of mean values ± two-fold of SD, and then the R package “DoubletFinder [19]” was applied with recommendatory parameters. Moreover, to remove the batch effects in single-cell RNA-sequencing data, the mutual nearest neighbors(MNN) presented by Haghverdi et al. was performed with the R package ‘batchelor’ in our study [20].
After quality standardization, the Seurat R package [21] was applied to analyze the scRNA-seq data. First, scRNA-seq data was converted to a Seurat object, and then we used the “FindVariableFeatures” function to detect the top 2000 highly variable genes. After that, principal component analysis (PCA) was applied to reduce the dimension of the scRNA-seq data based on the 2000 genes. The “RunTSNE” function was applied to perform t-distributed stochastic neighbor embedding (TSNE) to analyze the scRNA-seq data. The “FindClusters” (set the “resolution” parameter to 0.5) and “FindAllMarkers” functions were adopted for cell clustering analysis and detect all gene expression markers for identity classes. To identify the marker genes for each cluster, the cutoff threshold values, adj. P value<0.01 and fold change >0.5 were used.
Eventually, according to SingleR package [22], CellMarker dataset [23], and previous reports [15,24], we annotated different cell types (cell clusters were annotated with the dominant expression cell markers).
2.7. Analysis of sub-clusters in epithelial cells, stromal cells, and immune cells
Following primary annotation, epithelial cells, stromal cells, and immune cells were extracted via the “SubsetData” function. Then, we applied the “FindClusters” and “FindAllMarkers functions and reclustered the selected cells by TSNE. The sub-clusters were annotated by the dominant expression cell markers. The following cutoff threshold values were applied to reveal the marker genes for each cluster: adjust P value<0.01 and fold change >0.5. We also inferred the high-risk genes for ESCC using CIPHER as described in previous studies [25,26].
2.8. Estimation of the copy number variations
The R package “scCancer” [27] was used to estimate the initial copy number variations (CNVs) in each region. The expression level of each cell acted as the input file with recommendatory parameters. Immune cells served as the background to calculate the CNVs score. The CNV value of each cell was calculated as the quadratic sum of the CNV region. Also, we applied the R package “inferCNV” to calculate CNVs in tumor cells as described previously [28,29].
2.9. Definition of cell scores and signature
To calculate the M1/M2 polarization and pro-/anti-inflammatory potential of macrophage cells, GSVA analysis was conducted. We retrieved the gene sets associated with the above functions from previous studies [5,30].
To define the resident, cytotoxic, exhausted, and costimulatory score for T cells, we used the average expression of published signature gene lists for resident, cytotoxicity, exhausted, and costimulatory of T cells.
2.10. Identification of gene expression signatures of malignant cells
We selected the identified marker genes in tumor cells compared between malignant and non-malignant epithelial cells to identify the gene expression signatures of malignant cells. Then, we applied unsupervised NMF via the NMF R package [31] to reveal the malignant signatures of tumor cells. Finally, the functional enrichment analyses were performed to define the type of malignant signatures.
2.11. Trajectory analysis
We performed the trajectory analysis via the R package monocle2 to explore the tumor‐reprogramming processes in single cells [32]. First, the function “newCellDataSet” was applied to construct the monocle subject. After that, the differentially expressed genes identified via Seurat were selected for trajectory analysis. The “reduceDimension” function was applied to reduce dimensions, and we placed cells onto a pseudotime trajectory by “orderCells” functions. State, composed of cells primarily from non-malignant tissues, was defined as “root cell”.
2.12. Analyses of metabolic pathways
To assess the pathway activity of each cell type, we applied an algorithm developed by Xiao et al. [9]. Briefly, the analysis of the metabolic program was according to the mean expression level of the metabolic gene across cell types.
Because there is a potential impact by multiple environmental factors on metabolic reprogramming in tumors, such as nutrient supply and location-specific elements, exploration of the intersections between these factors and mitochondrial activity in malignant cells is critical in understanding the metabolic reprogramming of tumors such as ESCC. Therefore, we calculated the average expression level of genes in terms of hypoxia, glycolysis, and OXPHOS which acted as indicators for oxygen supply, glucose supply, and mitochondrial activity, respectively. Data of genes responsive to the three sets of genes (known to respond to hypoxia, glycolysis, and OXPHOS), applied in the calculation were retrieved from the MsigDB database because the nutritional status of cells is not available in the present single-cell samples. Next, with the average expression level of genes in hypoxia, glycolysis, and OXPHOS as indicators for oxygen supply, glucose supply, and mitochondrial activity, we explored the relationship between environmental factors and mitochondrial activity in malignant cells, respectively.
2.13. Survival statistical analysis
CIBERSORTx [33], a new machine-learning method, was applied to evaluate the clinical relevance of genes in bulk RNA sequencing data. Briefly, we first applied our single-cell data for the construction of a signature matrix. Then, based on the signature matrix, gene expression profiles for each cell type were impute in the bulk RNA sequencing data using the ‘High-Resolution mode’. Following the retrieval of the cancer cell-specific gene expression profiles bulk RNA sequencing data, we applied the single-sample gene set enrichment analysis (ssGSEA) [34] to calculate the malignant signatures scores. Then, patients in TCGA and GEO database were categorized into high and low expression groups; this was according to the median value of the ssGSEA score for each malignant signature gene set. Kaplan–Meier and log-rank tests were applied to construct and compare survival curves. Additionally, Cox regression analysis was undertaken to evaluate the prognostic value of each malignant signature. The statistical threshold for significance was adj. P values < 0.05.
2.14. Functional enrichment analyses
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed via Metascape (http://metascape.Org) [35]. Notably, P < 0.01 and the number of enriched genes > 3 acted as the thresholds for a significant difference in GO or KEGG pathways.
2.15. Construction of a cell to cell interaction network
To investigate cell-to-cell interaction between the tumor and non-malignant cells, R package “CellChat” [36] and “CellPhoneDB” Python package [29] were applied. The ligand-receptor pairs in CellChat retrieved from previous studies were divided into four groups including, cytokine/chemokine, immune checkpoint, growth factor, and others [37]. The crosstalk between cell analyses conducted via “CellChat” was as follows: (1) “createCellChat” function was applied to create “CellChat” object; (2) the “computeCommunProb,” “computeCommunProbPathway,” and “aggregateNet” functions were adopted to infer the cellular communication network; (3) “netVisual_aggregate,” “netVisual_signalingRole,” and “netVisual_bubble” were adopted to visualize crosstalk among cells. “CellPhoneDB” was then applied with recommendatory parameters.
2.16. Flow cytometry and qRT-PCR
Phosphate-buffer saline with 20 μg/mL human IgG and 3% fetal bovine serum was applied to block non-specific antibodies for 15 min. Then, cells and allophycocyanin-conjugated mouse anti- human EPCAM (5 μL/106 cells; cat. no.: 566,658, BD Biosciences, RRID: AB_2,869,815) [38], BV510-conjugated mouse anti-human CD45 (5 μL/106 cells; cat. no.: 563,204, BD Biosciences, RRID: AB_2,738,067), or PE-conjugated mouse anti-human KRT5 (10 μL/ 106 cells; cat. no.: AB_224,985, Abcam) were incubated on ice for 30 min. Next, FACSAria III (BD Biosciences) was applied to quantitate and isolate stained cells. Results were analyzed using the FlowJo software (TreeStar, Woodburn, OR, USA). Upon isolation of tumor cells expressing EPCAM and non-malignant cells expressing KRT5, we adopted the kit (Illumina, San Diego, USA) for RNA extraction and reverse transcription on the sorted cells. QuantStudio 5 (Thermo Fisher Scientific) was employed for sequencing.
2.17. Immunohistochemistry and immunofluorescence
Immunohistochemistry was conducted as stipulated by the manufacturer's [39]. Paraffin-embedded slides were incubated with anti-αSMA (Mouse, CY1132, Abways). Primary antibody detection was achieved using avidin-biotin-peroxidase complexes with DAB substrate solution (Gene Tech, China).
Immunofluorescence (IF) specific for IGFBP2 (Rabbit, DF7765, Affinity Biosciences, RRID: AB_2,841,231), ODC1 (Rabbit, DF6712, Affinity Biosciences, RRID: AB_2,838,674), SPINK5 (Rabbit, DF4462, Affinity Biosciences, RRID: AB_2,836,817), and CSTB (Rabbit, AF0256, Affinity Biosciences, RRID: AB_2,833,431) was performed as previously described. Briefly, the paraffin-embedded slides were dewaxed and rehydrated. After antigen retrieval, block of endogenous peroxidase activity and non-specific antigens, and incubation with primary antibodies and horseradish peroxidase-conjugated secondary antibody, the slides were incubated with Opal tyramide signal amplification (TSA) Fuorochromes (Opal 2-Color Manual IHC Kit, G1236, Servicebio Co., Ltd) for 10 min at room temperature. After the second run, the slides were stained with DAPI.
2.18. Statistical analysis
The statistical tools, methods, and threshold for each analysis are explicitly described with the results or detailed in the figure legends or Materials and Methods. Comparison of non-malignant and malignant esophagus was performed by Wilcoxon test in immunohistochemistry and immunofluorescence. Comparison of non-malignant and malignant esophagus was performed by Wilcoxon test in qRT-PCR.
2.19. Role of funding source
The funding sources played no role in the study design, data collection, data analysis, interpretation, writing of the report, and the decision of paper submission.
3. Results
3.1. A single-cell atlas of the non-malignant and malignant esophagus
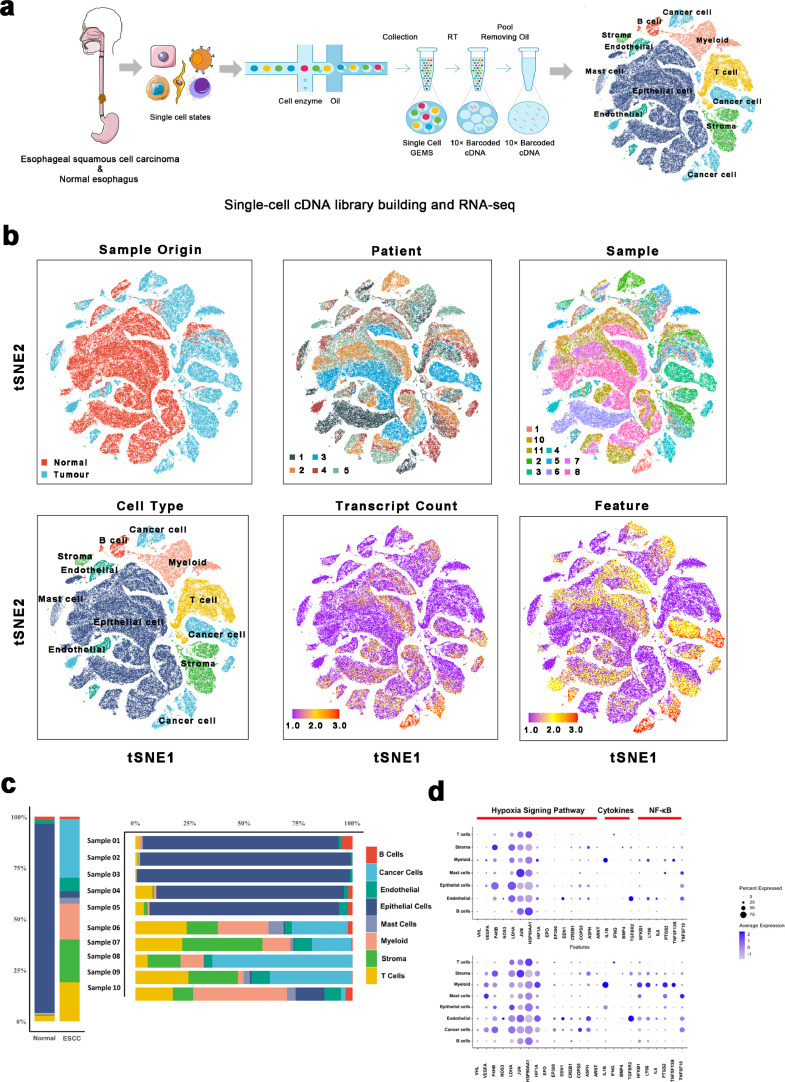
After the selection of five patients, including five tumor samples and five corresponding non-malignant samples in pair, we characterized the single-cell transcriptome atlas of the non-malignant and malignant esophagus (Fig. 1a and Supplementary Table 1). As a result, 6672 doublets were identified, accounting for less than 5% of the total cells, which we eliminated from the analysis (Supplementary Fig. 1a). After quality control check, 128,688 cells were subjected to downstream analyses, of which 53,554 and 75,134 cells were derived from tumor samples and normal samples, respectively (Fig. 1b and c). We identified the cell populations of these cells through dimensionality reduction and unsupervised clustering via the Seurat package. Based on the TSNE plots (Fig. 1b), cells selected for downstream analyses were clustered into eight clusters. According to the cell markers from previous studies (Methods Details), these clusters were revealed as eight cell types: Epithelial cells (marked by KRT13 and SPRR3), mast cells (marked by TPSB2 and CPA3), endothelial cells (marked by VWF and RAMP2), fibroblasts (marked by DCN and COL1A1), T cells (marked by CD3D and CD3E), myeloid cells (marked by LYZ and C1QB), B cells (marked by CD79A and IGKC), and cancer cells (marked by EPCAM and SOX2). Detailed distributions of these marker genes in each cluster are outlined in Supplementary Fig. 1b and c.
Fig. 1.
A Single-cell atlas of esophagus non-malignant tissues and esophageal squamous cell carcinoma.
(a) Workflow depicting collection and processing of specimens of ESCC tumors and non-malignant esophageal tissue for scRNA-seq analysis.
(b) TSNE of the 128,688 cells profiled here, with each cell color coded for: its sample type of origin (ESCC (n = 5) or non-malignant esophageal tissue (n = 5)), the corresponding patient, the corresponding sample, the associated cell type, the transcript counts, and the transcript features.
(c) The proportion of each cell type in non-malignant and ESCC samples.
(d) Dot plot of representative genes in the cytokines, the nuclear factor-kB (NF-kB), and hypoxia signaling pathways mapped onto cell types in non-malignant and ESCC samples, respectively.
Since previous studies had demonstrated that cytokines, nuclear factor-kB (NF-kB), and hypoxia signaling pathways are essential in tumor development [40,41], we plotted these genes in ESCC and non-malignant esophageal cells, respectively. Compared to non-malignant esophageal cells, both VEGFA and HIF1A showed significant elevations in all cell types in ESCC (Fig. 1d). Then, trajectory analysis was applied to explore the dynamic changes in expressions of VEGFA and HIF1A and scores of the hypoxia pathway. Notably, high expressions VEGFA and HIF1A were reported with the larger pseudotime in epithelial cells, especially in tumor cells (Supplementary Fig. 1d). Besides, the hypoxia scores were significantly high in malignant cells (Supplementary Fig. 1e). In summary, these findings demonstrated that hypoxia might be a pivotal role in the promotion of ESCC tumorigenesis. Additionally, myeloid is potentially a more dominant source of cytokines and nuclear factor-kB (NF-kB), especially in ESCC, compared to other cell types. Therefore, myeloid cells have potential regulatory effects on the inflammatory response via the activation of the NF-kB signaling pathway.
3.2. Metabolic reprogramming in ESCC
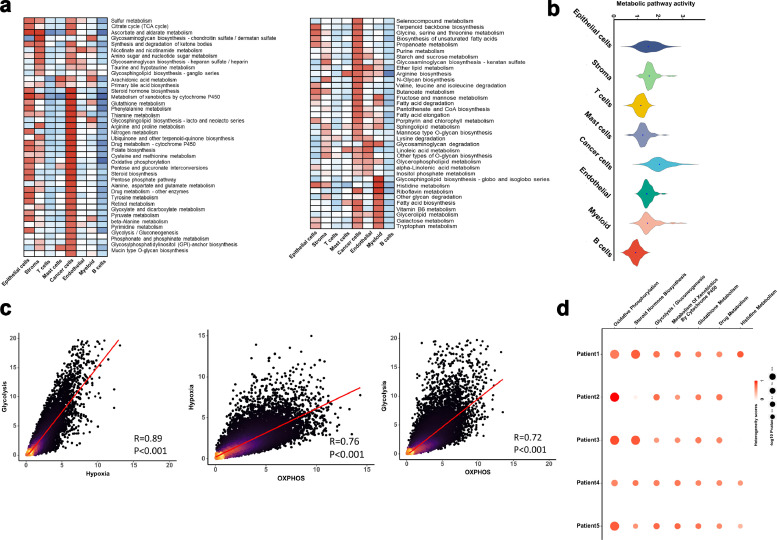
Considering metabolic reprogramming as a hallmark of cancer, we performed a series of analyses over metabolic pathways on different cell types. More significantly up-regulated metabolic pathways were enriched in cancer cells compared to the other cell types (Fig. 2a and b). Notably, the enrichment of the oxidative phosphorylation (OXPHOS), the glycolysis and gluconeogenesis, and the tricarboxylic acid cycle (TCA cycle) depicted a significant mitochondrial activity in the esophageal metabolisms. In Fig. 2c, a significantly close interconnection is depicted between the hypoxia and glycolysis signatures (P < 0.001, Pearson's R = 0.74). OXPHOS activity exhibited a positive correlation with either hypoxia (P < 0.001, Pearson's R = 0.78) or glycolysis (P<0.001, Pearson's R = 0.83).
Fig. 2.
Metabolic landscape for ESCC.
(a) Metabolic pathway activities in each cell type (split metabolic pathways into different spatial positions). Statistically non-significant values (random permutation test p > 0.05) are shown as blank.
(b) Distributions of pathway activities in different cell types.
(c) Comparing activities of glycolysis, OXPHOS, and response to hypoxia in the malignant cells. Spearman's rank correlation was used for analysis.
(d) Metabolic pathways enriched in genes with the highest contribution to the metabolic heterogeneities among malignant cells from different tumors.
Besides, we evaluated the heterogeneity of metabolic pathways in malignant cells for each patient to enhance the reliability of our results. OXPHOS provided the highest contribution to the metabolic heterogeneities among malignant cells from different tumors in the ESCC patients (Fig. 2d). It was inferred that OXPHOS may play an important role in cancer metabolic reprogramming.
3.3. Classification of tumor and normal epithelium in the non-malignant and malignant esophagus
Considering ESCC as a major type of esophageal cancer that transpires from esophageal epithelial cells, we evaluated the characteristics of 86,925 epithelial cells across different esophagus lesions.
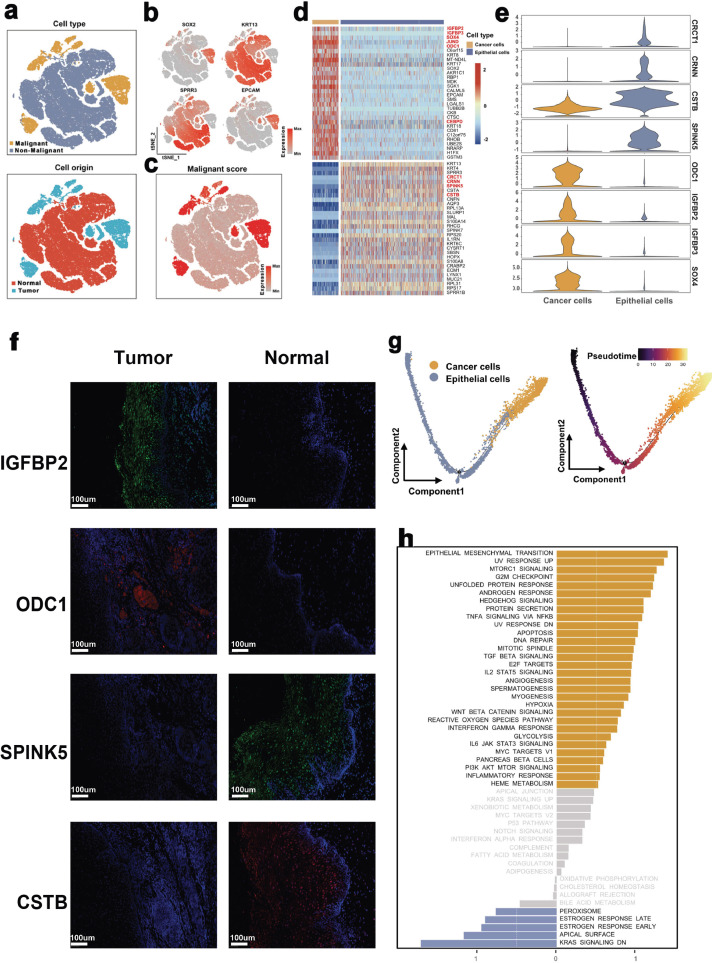
First, these epithelial cells were identified as tumor cells and non-malignant epithelial cells and then followed by a re-clustering process: non-malignant epithelial cells (marked by SPRR3 and KRT13) and cancer cells (marked by EPCAM and SOX2) (Fig. 3a and b). Besides, to verify their cell attributes, we traced back to their origins. Notably, cells expressing high levels of normal epithelial marker genes were mostly derived from normal tissues, whereas all epithelial cells defined in the cancer cluster originated from tumor samples (Fig. 3a).
Fig. 3.
The single-cell transcriptomes of epithelial cells in non-malignant and malignant esophagus.
(a) The TSNE plot and overview of epithelial cells by the cell type and origin of the cells.
(b) Canonical cell markers were used to label epithelial subtypes as represented in the TSNE plot.
(c) The TSNE plot of epithelial cells by the CNV's levels of the cells.
(d) Heatmap showing the expression of marker genes in malignant and non-malignant epithelial cells.
(e) Violin plots showing the marker genes for each epithelial cell type.
(f) Immunofluorescence staining of tumor (n = 8) (up) / normal (n = 8) (down) tissue-specific genes in ESCC and non-malignant tissues. Comparison of non-malignant and malignant esophagus was performed by Wilcoxon test.
(g) Pseudotime analysis for malignant and non-malignant epithelial cells.
(h) Differences in pathway activities scored per cell by GSVA between malignant and non-malignant epithelial cells. The scores of pathways are normalized.
Furthermore, we validated the above clusters by estimating their copy number variations (CNVs) based on the averaged expression patterns across the genome intervals (Supplementary Materials Details). In Fig. 3c, we show that compared to normal epithelial cell types, the cancer cells cluster exhibited remarkably higher CNV levels, which demonstrated the reasonability of our cell annotations.
Upon the identification of malignant epithelium from non-malignant epithelium, we performed differential gene expression analysis between tumor cells and non-malignant epithelial cells. As shown in Fig. 3d and e, a list of tumor-specific genes (such as IGFBP2, IGFBP3, ODC1, and SOX4) were significantly highly expressed in tumor cells (adj. P<0.01). In contrast, genes associated with anti-tumor or pro-inflammatory, including CRCT1, CRNN, SPINK5, and CSTB, were significantly highly expressed in non-malignant epithelial cells (adj. P<0.01). Furthermore, these three genes were validated using flow cytometry and qRT-PCR in twenty patients. The two markers, EPCAM and KRT5, were used to successfully isolate cancer cells from other epithelial cells (Supplementary Fig. 2a). According to the qRT-PCR results, the expression levels of IGFBP2 (P < 0.01), IGFBP3 (P < 0.01), ODC1 (P < 0.01), and SOX4 (P < 0.01) were significantly higher in ESCC cancer cells. In contrast, CRCT1 (P < 0.01), CRNN (P < 0.01), SPINK5 (P < 0.01), and CSTB (P < 0.01) were highly expressed in normal epithelial cells (Supplementary Fig. 2b, comparison of non-malignant and malignant esophagus was performed by Wilcoxon test). To confirm this phenomenon at the protein level, immunofluorescence analyses were applied. We found IGFBP2 and ODC1 were almost expressed in ESCC tissues, while CSTB and SPINK5 were mainly up-regulated in esophagus non-malignant tissues (Fig. 3f). These results implied that the gain of IGFBP2 and ODC1 or the loss of CSTB and SPINK5 can be a new alarm for ESCC.
Following the trajectory analysis to explore the potential transition between malignant and non-malignant epithelium, dynamics changes of marker genes in epithelium revealed the progression of esophagus non-malignant tissues into malignant tissues (Fig. 3g and Supplementary Fig. 2c), which confirmed that the tumor-specific genes promote ESCC.
Subsequently, we performed GSVA analysis to characterize the biological functions between malignant and non-malignant epithelial cells. Compared to non-malignant epithelial cells, pathways closely associated with cell proliferation, cancer progression, and metastasis (such as epithelial-mesenchymal transition, unfolded protein response, and MTORC1 signaling pathways) were highly enriched in tumor cells (Fig. 3h). Pseudotime analysis was then conducted to explore the dynamic changes in expressions of Hallmark pathways in epithelial cells. Notably, we revealed that pathways for tumorigenesis and development were significantly up-regulated with the larger pseudotime (Supplementary Fig. 2d). Collectively, this is the first report on the characterization of the difference between malignant and non-malignant epithelial cells in ESCC at a single-cell level and uncovers a list of potential biomarkers for ESCC.
3.4. Construction of the transcriptional landscape revealing the heterogeneity of ESCC malignant cells
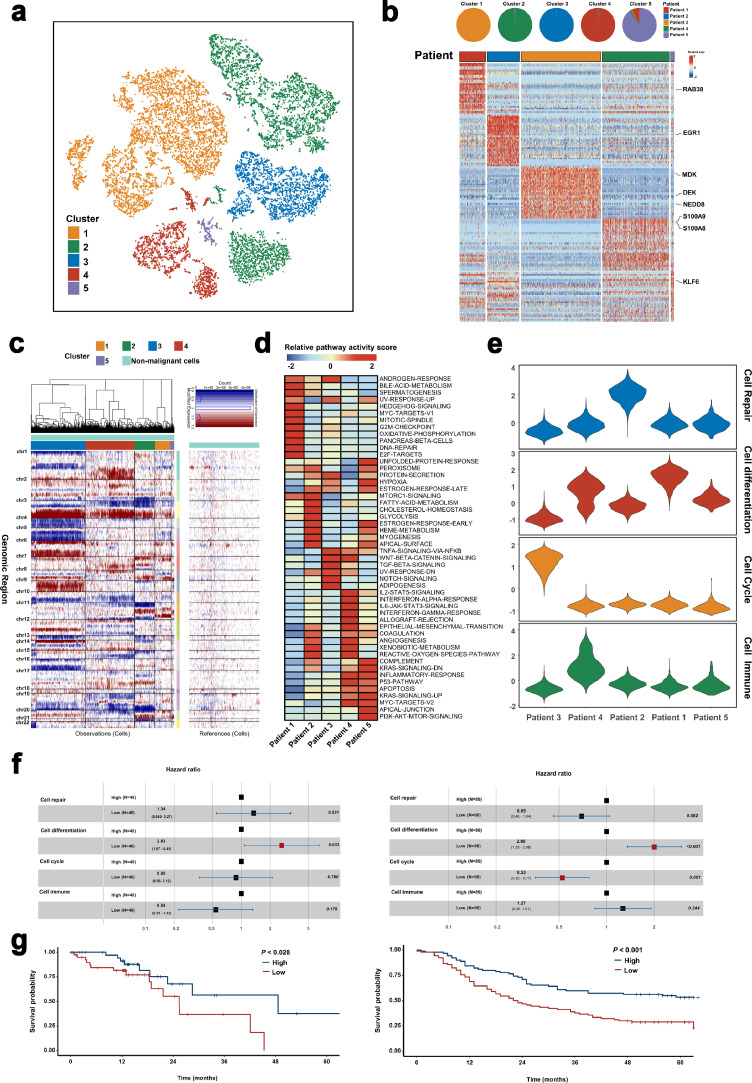
To investigate whether the malignant cells were heterogeneous, we focused on malignant cells and re-clustered 15,644 tumor cells, and identified five sub-clusters (Fig. 4a). Intriguingly, we found that malignant cells formed patient-specific clusters. For instance, cluster 1 was mainly formed by cells from Patient 3, whereas cells in cluster 2 were mostly from Patient 4 (Fig. 4b). Additionally, differential gene expression analysis revealed the high-risk genes for each ESCC patient via the disease-gene prediction algorithm CIPHER [25,26]. Our results demonstrated that different patients exhibited distinct expression patterns, for different high-risk genes (Fig. 4b). For instance, EGR1 was highly expressed in Patient 2, whereas S100A8/9 was found to be a high-risk ESCC gene in Patient 4. In addition, we applied the R package for CNV analysis of malignant cells and found that compared to non-malignant epithelial cells, all tumor cells exhibited a relative variation in CNV profiles; tumor cells in each sub-cluster showed distinct CNV status. Compared to other sub-clusters, cluster 1 demonstrated an apparent CNV loss in chromosome 4 and chromosome 5. However, on cluster 2, an obvious CNV gain in chromosome 1 was detected (Fig. 4c). These findings demonstrated a high heterogeneity of tumor cells in both gene expression and CNV status.
Fig. 4.
Malignant cell clusters and common malignant signatures revealed in ESCC.
(a) The TSNE plot and overview of the malignant cells.
(b) Complex heatmap showing the expression of marker genes in each malignant epithelial sub-cluster. Top: Pie charts showing the proportions of cells originating from each patient detected in each cluster, colored by patient. Bottom: Relative expression map of marker genes associated with each cell sub-cluster. Mean expression values are scaled by mean-centering, and transformed to a scale from −2 to 2.
(c) Heatmap showing large-scale CNVs of each malignant epithelial sub-cluster. The expression values for non-malignant epithelial cells are plotted in the right heatmap, and the cancer cells are plotted in the left heatmap, with genes ordered from top to bottom across the chromosomes.
(d) Differences in pathway activities scored per cell by GSVA in each malignant epithelial sub-cluster. The scores of pathways are normalized.
(e) Violin plots showing the scores for one of the four malignant signatures for malignant cells for each epithelial cell type.
(f) Prognostic values of the malignant signatures in TCGA (Right) and GEO (Left) database. Cox regression survival analyses was used in our study.
(g) Kaplan–Meier curves showing overall survival in the TCGA (Right) and GEO (Left) database according to high vs low expression of the cell differentiation signature. Significant results are indicated with red squares. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Furthermore, the functions of different tumor cell subtypes were explored through a comparison of pathway activities in Hallmark signatures and metabolism. GSVA analyses revealed the distinct pathway activities among these patients. For instance, MYC targets and G2M-checkpoint pathways were mainly enriched in Patient 1. On the contrary, interferon-alpha response and IL2-STAT5 signaling pathways were activated in Patient 4 (Fig. 4d). Through metabolic pathway analyses, we established the different metabolic re-programming patterns in different patients. Patient 2 exhibited significantly higher oxidative phosphorylation and fatty acid elongation in tumor cells compared to other patients. Other energy supply pathways (such as glycolysis and TCA cycle pathways) were enriched in Patient 4 (Supplementary Fig. 3a). These results implied a high degree of intertumour heterogeneity in malignant cells.
Next, after selecting the genes highly expressed in tumor cells, the R package (31) NMF was applied to identify metagenes for malignant cells. Eventually, we found 252 metagenes (Supplementary Table S2), which were enrolled for hierarchical clustering analysis. The results demonstrated high concordance among four malignant signatures, including cell repair, cell differentiation, cell cycle, and immune cells (Supplementary Fig. 3b). Then, the calculated scores of these signatures demonstrated that those of the malignant signatures varied across the malignant cells from different patients (Fig. 4e).
Using CIBERSORTx, we analyzed the correlations between these malignant signatures and survival whereby each cell type was characterized by unique expression profiles in our signature matrix (Supplementary Fig. 3c). This demonstrated the reliability of our signature matrix. Subsequently, based on the signature matrix, we impute gene expression profiles for each cell type in the bulk RNA sequencing data via the ‘High-Resolution mode’ in CIBERSORTx. Notably, the survival analyses demonstrated an obvious association between low cell differentiation scores and poor prognosis of ESCC patients in both TCGA and GEO database cohorts (Fig. 4f and g), which suggested the metagenes for cell differentiation could serve as a prognostic marker for ESCC patients. However, we only applied our own single-cell data for the construction of a cell type specific gene expression profiles of the TCGA data to evaluate the clinical relevance of genes in bulk RNA sequencing data, which might have led to some inaccuracy in our results. Therefore, more research is needed to explore the clinical relevance of these genes in future.
3.5. Stromal cells could contribute to remodeling and angiogenesis
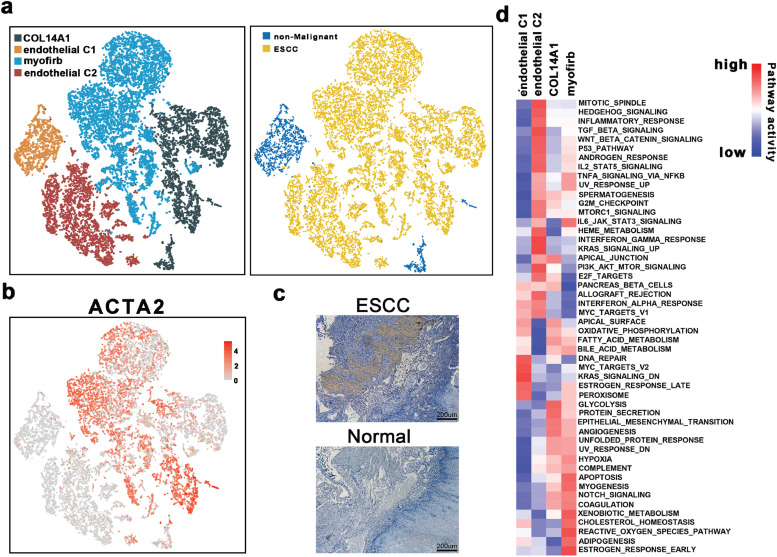
To evaluate the dynamics of stromal cells in the TME, stromal cells (including endothelial cells (ECs) and fibroblasts) were selected and re-clustered. First, four stroma cell clusters were identified after a sub-clustering process, including endothelial C1, endothelial C2, myofibroblasts, and COL14A1-positive fibroblasts (Fig. 5a). Detailed distributions and expression of their marker genes in each cell type are outlined in Supplementary Fig. 4a and Supplementary Fig. 4b. We noted differences between the distribution of each of these four clusters. Endothelial C1 was presented largely in the non-malignant esophageal cell cluster, whereas COL14A1-positive fibroblasts were found in both cell clusters, and endothelial C2 and myofibroblasts were mainly distributed in the ESCC (Fig. 5a). According to previous reports, myofibroblasts exert crucial effects on tumor progression. and were suggested to be associated with cancer- in the stromal environment [42,43]. As such, we used alpha-smooth muscle actin (α-SMA), a product of the ACTA2 gene and the marker protein of myofibroblasts, to verify the infiltration of myofibroblasts. As depicted in Fig. 5b and c, ACTA2 was mainly expressed in the myofibroblasts, similar results were confirmed through IHC staining.
Fig. 5.
The scRNA profiles for stromal cell lineages in non-malignant tissues and esophageal squamous cell carcinoma.
(a) The TSNE plot and overview of the stromal cells.
(b) Feature plot of ACTA2 in stromal cells.
(c) IHC staining of α-SMA on formalin-fixed and paraffin-embedded slides for the independent biospecimens (n = 8). All replicates showed the similar results.
(d) Differences in pathway activities scored per cell by GSVA among the different stromal cell subtypes. The scores of pathways are normalized.
Due to the distinctive distributions among these four clusters of stroma cells, we performed functional enrichment analyses. In particular, GSVA analysis revealed that although myofibroblasts and COL14A1-positive fibroblasts shared several up-regulated pathways, E2F-target, apical-junction-related, and other pathways were exclusively upregulated in myofibroblasts (Fig. 5d). Contrarily, reactive oxygen species pathway, IL-6/Jak/Stat3 signaling, and metabolic pathways (such as Adipogenesis and xenobiotic metabolism pathways) were upregulated in the COL14A1-positive fibroblasts rather than myofibroblasts. Also, endothelial C2 were mainly enriched in the pathways associated with the regulation of the developments and cell fate determiners of stromal cells (e.g., WNT pathways, angiogenesis, and P53 pathway). Besides, pathways related to inflammatory reactions and damage repairs, such as DNA repair and interferon response-related pathways, were up-regulated in endothelial C1. These findings confirmed the dynamics by which stromal cells within the TME promote tumorigenesis and development.
3.6. Diverse B cell subtypes in ESCC
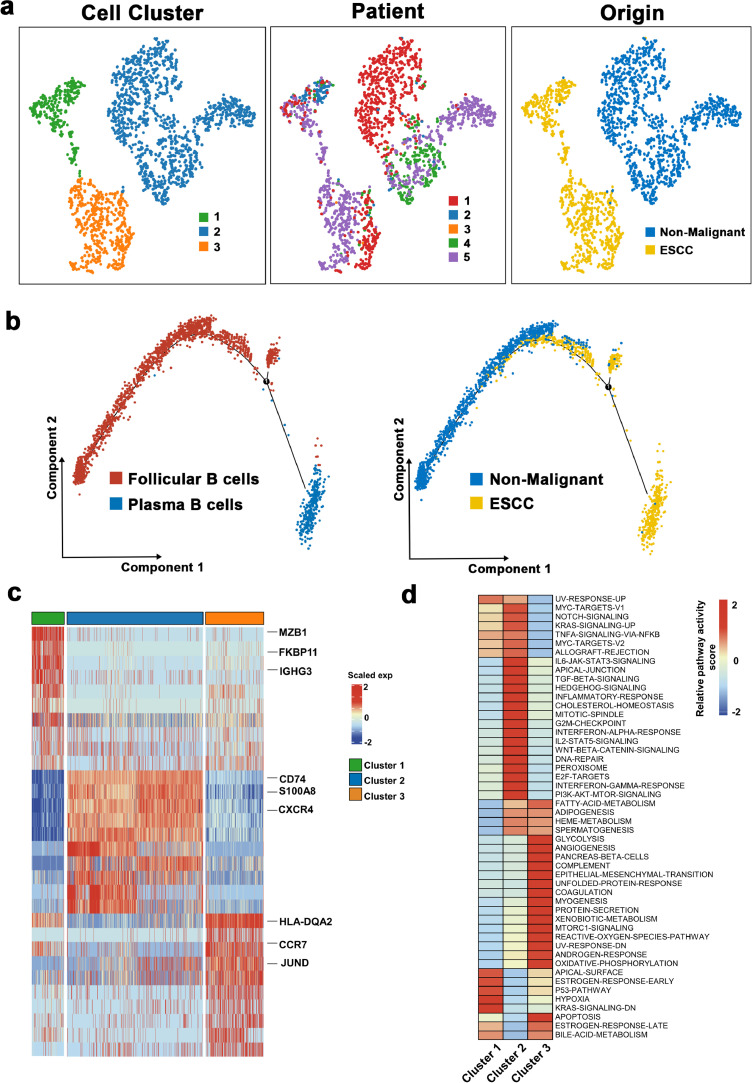
B cells have been revealed to enormously impact molding immune response in the TME and are closely related to the overall survival for patients with tumors [44,45]. Herein, we re-clustered the B cells and identified 3 sub-clusters in our study (Fig. 6a). Then, based on particular cell markers, we defined cluster 1 as plasma B cells (marked by IGHG1 and IGHG4), whereas clusters 2 and 3 were defined as follicular B cells (marked by MS4A1) (Supplementary Fig. 5a). Notably, we also found one of the B cell sub-clusters was mainly seen in normal tissues and the other two B cell sub-clusters were mainly observed in tumor tissues. Additionally, plasma B cells were mainly presented in ESCC tissues, while non-malignant esophageal samples incorporated a substantial number of follicular B cells (Fig. 6a). Analysis of cellular reprogramming at the single-cell level revealed a transitional path of B cells from the non-malignant-derived follicular B cells to tumor-derived plasma B cells and tumor-derived follicular B cells (Fig. 6b); this demonstrated the complex functions of B cells in ESCC.
Fig. 6.
The scRNA profiles for B cells in non-malignant tissues and esophageal squamous cell carcinoma.
(a) The TSNE plot and overview of the B cells.
(b) Trajectory analysis for B Cells.
(c) Heatmap showing the expression of marker genes in each B cell cluster.
(d) Differences in pathway activities scored per cell by GSVA among the different B cell subtypes. The scores of pathways are normalized.
Further, we explored the two types of B cells, along with their features and functions in ESCC. Differential expression analysis revealed high MZB1, FKBP11, and IGHG3 levels in plasma B cells (Fig. 6c). Notably, cluster 2 and cluster 3 were characterized by distinct expression programs. In particular, CD74, S100A8, and CXCR4 were activated in cluster 2, whereas HLA-DQA2, CCR7, and JUND were enriched in cluster 3 (Fig. 6c). Next, we performed GSVA analysis to explore the primary biological functions of these sub-clusters. As shown in Fig. 6d, that cells in cluster 1 exerted essential roles in cell proliferation and differentiation. The primary biological functions of cluster 2 were highly associated with an inflammatory response. Moreover, pathways related to energy supply (such as glycolysis and OXPHO) were activated in cluster 3. Additionally, GSVA analysis of each sub-cluster from each patient was undertaken to explore the heterogeneity of B cells. In Supplementary Fig. 5b, the pattern of the activated pathways varied across different patients and in different sub-clusters in the same patient. However, pathways correlated with tumor progression were more likely to be up-regulated in Patient 5, diagnosed with stage III. Overall, these results provided insights into further exploration of the role of B cells in ESCC.
3.7. Macrophages existed different phenotypes with pro-tumoral functions
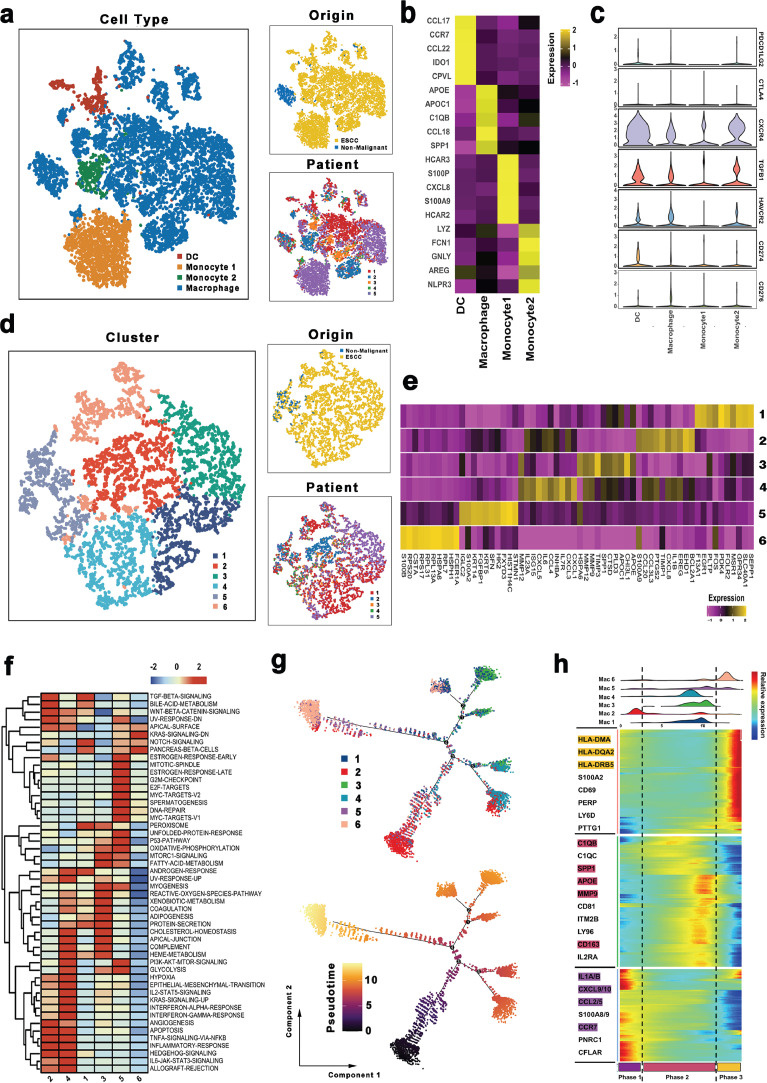
We selected 9801 myeloid cells for downstream analysis. Based on the re-clustering results, macrophages and monocytes were much more presented in the ESCC compared to non-malignant esophageal tissues (Fig. 7a and b). Subsequently, the distribution of immune checkpoints was evaluated in myeloid cells. Of note, some standard immune checkpoints, including PDCD1LG2(PD1-L2) and CTLA4 exhibited relatively low expressions in all myeloid cells, whereas CXCR4, TGFB1, and HAVCR2 were highly expressed in most myeloid cells Fig. 7c and Supplementary Fig. 6a. Moreover, different myeloid clusters had different highly-expressed immune checkpoints, such as CD274 was found to be highly expressed in DCs, and CD276 was up-regulated in most of the macrophages. These results reflected a significant heterogeneity in immunotherapy response in myeloid cells. Therefore, common up-regulated immune checkpoints (e.g., CXCR4, TGFB1, and HAVCR2) may serve as potential immune targets for ESCC.
Fig. 7.
The scRNA profiles for myeloid cells in non-malignant tissues and esophageal squamous cell carcinoma.
(a) The TSNE plot and overview of the myeloid cells.
(b) Heatmap showing the expression of marker genes in each myeloid cell cluster.
(c) Violin plots of immune checkpoints upregulated or downregulated in myeloid cells.
(d) The TSNE plot and overview of the macrophages.
(e) Heatmap showing the expression of marker genes in each macrophage cell cluster.
(f) Differences in pathway activities scored per cell by GSVA among the different macrophage cell subtypes. The scores of pathways are normalized.
(g) Pseudotime-ordered analysis of macrophages. Macrophage cell subtypes are labeled by colors.
(h) Dynamic changes in gene expression of macrophage subtypes during the transition (divided into 3 phases), subtypes are labeled by colors (upper panel).
Because macrophages represent the most prevalent cell type in myeloid cells, they were subjected to further analysis. As shown in Figs. 7d,e and 6 sub-clusters were identified. Based on the known marker genes of M1/M2 and pro/anti-inflammatory macrophages, we estimated the phenotype for each sub-group. A dominant M2/anti-inflammatory phenotype was detected in clusters 1 and 3, whereas cluster 2 displayed an M1/pro-inflammatory phenotype (Supplementary Fig. 6b). Besides, there was no clear distinction of phenotype in clusters 4, 5, and 6. Further comprehensive dissection of pathway analyses demonstrated that distinct biological functions in each sub-cluster in macrophages (Fig. 7f). Collectively, these results revealed the heterogeneity of the sub-clusters.
Furthermore, the transition states of macrophages were explored through trajectory analysis. We found a tendency in macrophages from M1/pro-inflammatory phenotype to M2/anti-inflammatory phenotype to intermediate phenotype (Fig. 7g). According to the pseudotime, three phases were detected in these sub-clusters (Fig. 7h). Also, M1/pro-inflammatory phenotype-related genes (such as CCL2/5 and IL1A/1B) were activated in phase 1, whereas M2/anti-inflammatory phenotype -dominated gene set (such as C1QB, SPP1, and APOE) were mainly up-regulated in phase 2. These observations implied the complex macrophage phenotypes process in ESCC, which was consistent with a previous report [46]. Intriguingly, macrophages in phase 3 expressed more major histocompatibility complex (MHC)-II molecules (such as HLA-DQA2, HLA-DPB1, and HLA-DRB5). Previously, MHC II was revealed to mediate the presentation process of exogenous antigens [47]. Therefore, our results suggest that MHC II that overexpress macrophages could serve as a potential immune therapy target for ESCC.
3.8. T cell sub-cluster show antitumor immune response in ESCC
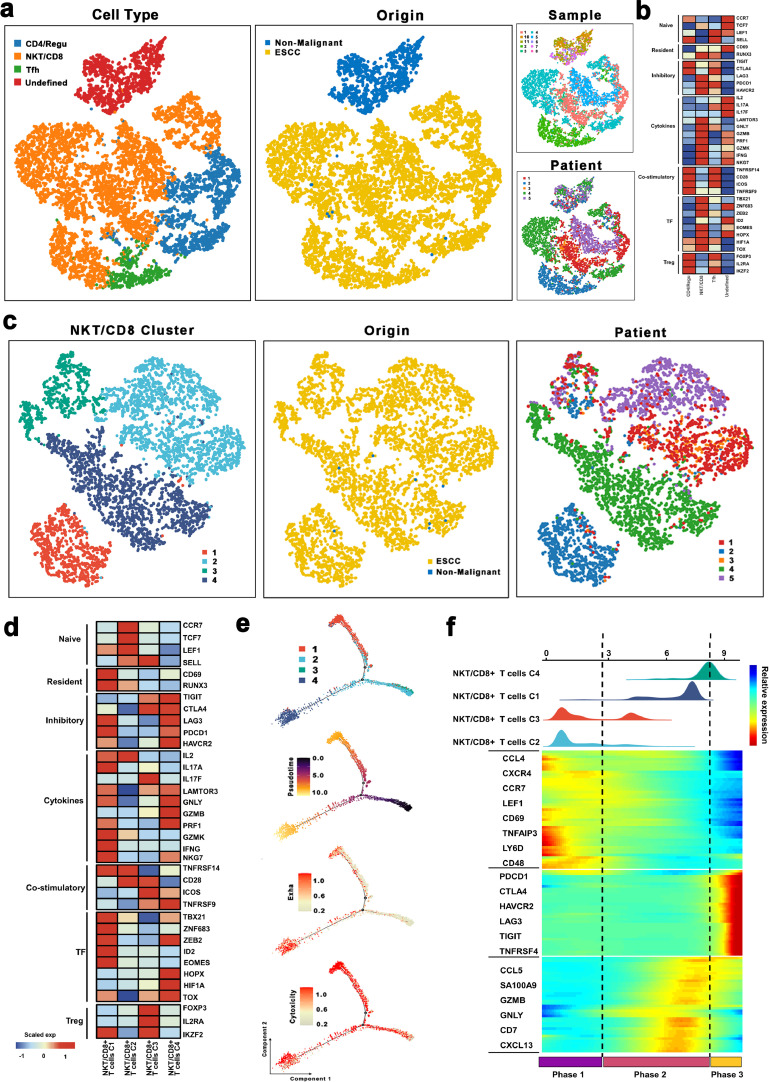
Herein, our focus was on the T cells, the most prevalent immune cell type in ESCC and non-malignant esophageal tissues. The re-clustering of T cells revealed 3 known populations, including NKT/CD8+ T cells (marked by GZMB, NKG7, and GNLY), regulated/CD4+ T cells (CD4, IL2RA, and FOXP3), and T follicular helper cells (MAF, ICOS, and BTAF) (Figs. 8a, S7a and b). Also, NKT/CD8+ T, CD4+/Regu T cells, and Tfh were mostly derived from tumor samples, whereby 6936, 1704, and 906 cells were detected from the malignant samples, respectively.
Fig. 8.
The scRNA profiles for T cells in non-malignant tissues and esophageal squamous cell carcinoma.
(a) The TSNE plot and overview of the T cells.
(b) Average expression of selected T cell function-associated genes of naïve markers, inhibitory receptors, cytokines, and effector molecules, co-stimulatory molecules, and Treg markers in each T cell subtype.
(c) The TSNE plot and overview of the NKT/CD8 + T cells.
(d) Average expression of selected T cell function-associated genes of naïve markers, inhibitory receptors, cytokines, and effector molecules, co-stimulatory molecules, and Treg markers in each NKT/CD8 + T cell subtype.
(e) Pseudotime-ordered analysis of NKT/CD8 + T cells. The dynamics of exhausted (upper panel) or cytotoxic signals (lower panel) in NKT/CD8 + T cells.
(f) Dynamic changes in gene expression of NKT/CD8 + T cell subtypes during the transition (divided into 3 phases), subtypes are labeled by colors (upper panel).
Next, we assessed the genes associated with the function of T cells to explore the functional status of each type of T cells. Of note, cytokine and effector molecules and inhibitory receptors were highly expressed in NKT/CD8+ T cells (Fig. 8b), which demonstrated that NKT/CD8+ T cells in ESCC exert potential cytotoxic and inhibitory effects. Numerous studies had also implicated immunotherapy as a promising therapeutic strategy. The exploration of immune checkpoints has always been essential to the development of immunotherapy [48,49]. Notably, we found that inhibitory receptors, including LAG3, PDCD1, and HAVCR2 were highly expressed in NKT/CD8+ T cells. On the other hand, by exploring the expressions of these immune checkpoints in each patient, LAG3 and HAVCR2 were almost activated in all patients (Fig. 8b). Our flow cytometry and qRT-PCR analysis further demonstrated that NKT/CD8+ T cells accounted for the majority of T cells in ESCC (Supplementary Fig. 7c). Higher expression of LAG3 and HAVCR2 were found in the NKT/CD8+ T cells in ESCC (Supplementary Fig. 7d). In summary, these observations revealed the potential role of LAG3 and HAVCR2 as checkpoint molecules for immunotherapy in ESCC.
3.9. NKT/CD8+ T cells show great heterogeneity in ESCC and tend to be exhausted
NKT/CD8+ T cells accounted for the majority of T cells in ESCC, hence, we re-clustered these NKT/CD8+ T cells, and then categorized them into four sub-groups, including cluster 1 (CCL4hiRGCChi), cluster 2 (CCR7hi RPS4Y1hi), cluster 3 (FOXP3hiSTMN1hi), and cluster 4 (HAVCR2hiCXCL13hi) (Figs. 8c, 8a and b).
Subsequently, we explored the expression of genes associated with the function of T cells in each NKT/CD8+ T cells sub-group. As depicted in Fig. 8d, cluster 2 exhibited higher expressions of naïve T cell markers (such as CCR7, TCF7, and LEF1), whereas genes related to inhibitory status (including TIGIT, CTLA4, and HAVCR2) and cytotoxic status (including GNLY, GZMB, and PRF1) were both highly expressed in cluster 4. Notably, the cytotoxic genes, including IL2, IL17A, GZMK, IFNG, and NKG7 were mainly enriched in cluster 1, whereas T-regulated genes were highly expressed in cluster 3. Since cells in NKT/CD8+ T cluster 1 were mainly originated from Patient 2, cells in NKT/CD8+ T cluster 4 primarily derived from Patient 4, and other clusters were made of several cells from different patients of origin; therefore, GSVA analysis was performed to explore the biological functions of these NKT/CD8+ T cells among different patients. Consequently, pathway analyses revealed that pathways related to tumor development, including IL-6/JAK/STAT3 and Notch pathways, were up-regulated in cluster 4, whereas metabolic pathways (including glycolysis and OXPHO) were enriched in cluster 1. Moreover, cells in cluster 3 were characterized by activated hypoxia, interferon-alpha/gamma response, and inflammatory response pathways. Interestingly, we found that only KRAS-related pathways and wnt/β-catenin signaling were activated in cluster 2 (Supplementary Fig. 8c). Therefore, these findings demonstrated the strong heterogeneity in NKT/CD8+ T cells.
Additionally, we performed pseudotime-ordered analysis to explore the dynamic states and cell transitions of NKT/CD8+ T cells via Monocle (Fig. 8e). To reveal the functional alterations of NKT/CD8+ T cells during the pseudotime, first, the cytotoxic and exhausted scores of each NKT/CD8+ T cell were calculated and then visualized in the monocle plot. Cluster 1 had the highest cytotoxic scores whereas, cluster 2 and cluster 3 had the lowest exhausted scores (Fig. 8e). Intriguingly, cluster 4 both had comparatively higher cytotoxic and exhausted scores. During these transitions, the exhausted score was increased, whereas the cytotoxic score was decreased which might induce tumor progression.
Transcriptional changes associated with transitional states were also explored, and three phases were identified in NKT/CD8+ T cells. Cluster 2 and cluster 3 were predominantly phase 1 cells, expressing higher CCR7 and lower GNLY and GZMB levels. Cells in phase 2 exhibited high expression levels of GNLY, SA100A9, and GZMB (Fig. 8f). Interestingly, phases 2 included most cells from cluster 1 and a few cells from cluster 4, while phases 3 (characterized by exhaustion-related genes) incorporated part of cells from cluster 4 (Fig. 8f). These results confirmed that cluster 4 exerted both inhibitory and cytotoxic effects.
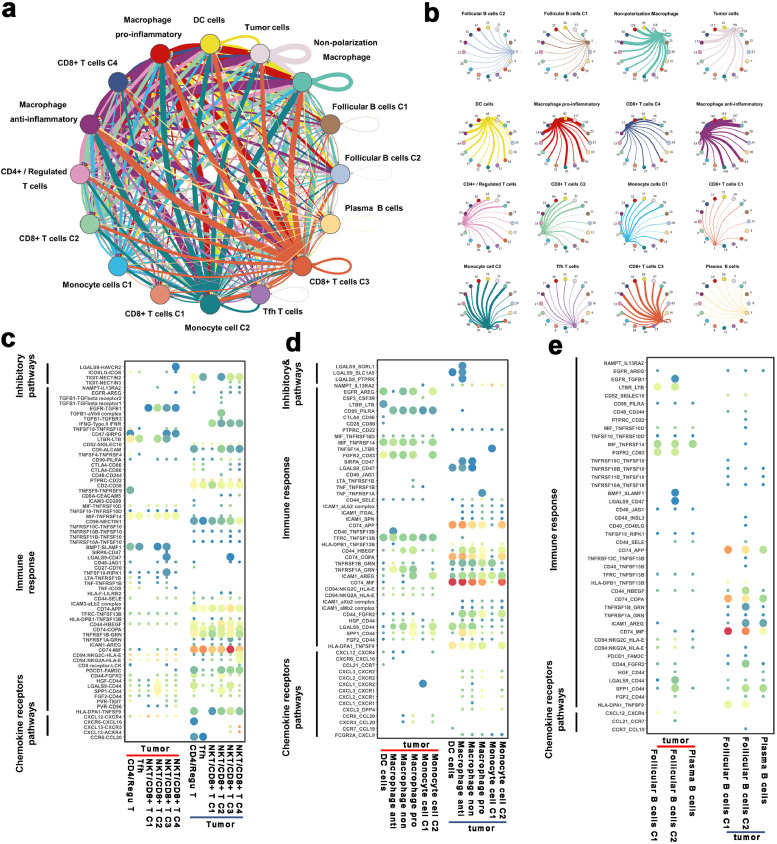
3.10. Crosstalks between cancer and immune cells
Tumorigenesis is an elaborate multistep process that would be affected by numerous factors. Recent studies have highlighted the importance of cell-to-cell communications in the evolution of various tumors [50,51]. The crosstalks between cancer and immune cells were assessed via the R package 'CellChat’ and “cellphonedb” (Materials Details).
The results revealed complex cell-cell interaction networks between cancer and immune cells (Fig. 9a and b). Next, we explored the role of tumor and immune cells in remodeling the TME in ESCC through the exploration of cell-cell communications between tumor and immune cells, respectively. Of note, immune cells exhibited relatively higher expressions of immune response genes (including MIF, ALCAM, COPA), whereas the corresponding genes (especially CD74) were significantly highly expressed in tumor cells (Fig. 9c and e). We inferred that these immune response communication pairs are crucial roles in the promotion of immune cell infiltration into ESCC. Some known inhibitory interactions (such as CD274-PDCD1, PDCD1LG2-PDCD1, and CD86-CTLA4) were not detected between tumor and T cells (Fig. 9c). In contrast, other inhibitory pairs, including LGALS9-HAVCR2 and TIGIT-NECTIN2 were identified between NKT/CD8+ T C4 cluster and tumor cells (Fig. 9c). Also, some critical stimulatory interactions, such as EGFR-TGFB1, CD2-CD58, and MIF-TNFRSF10D pairs were found between tumor and T cells (Fig. 9c). Our qRT-PCR results also revealed that CD74, LGALS9, and TIGIT were highly expressed in tumor cells, while MIF, HAVCR2, and NECTIN2 were highly expressed in T cells (Supplementary Fig. 9, Wilcoxon test was used to compare the gene expression level between T cells and tumor cells).
Fig. 9.
Crosstalk between cancer and immune cells.
(a) Overview of selected ligand-receptor interactions of tumor cells and immune cells. The line thickness is proportional to the number of ligands when cognate receptors are present in the recipient cell type. The loops indicate autocrine circuits.
(b) Detailed view of the ligands expressed by each cell type and the other cell types. Numbers indicate the quantity of ligand-receptor pairs for each intercellular link.
(c–e) Bubble plot showing the selected ligand-receptor interactions between cancer cells and T cell (c), myeloid cells (d), and B cells (e). P values are indicated by circle size, with the scale to the right (permutation test).
Through assessment of the interactions between macrophages and malignant cells, we detected the inhibitory interactions including LGALS9-SORL1, LGALS9-SLC1A5, and LGALS9-PTPRK in malignant cells and M2/anti-inflammatory macrophages (Fig. 9d). Interestingly, immune response-related ligand HLA-DPA1 was expressed in the vast majority of myeloid cells, whereas its receptor, TNFSF9, was identified in tumor cells. Cell-cell interactions between malignant cells and B cells were significantly fewer compared to other immune cells (Fig. 9e). However, there were some common widely expressed interactions between B cells and tumor cells. In summary, our results revealed that TME specific cellular communication has the potential to shape the unique TME of ESCC.
4. Discussion
Reports implicate ESCC as one of the most malignant and fatal carcinomas in adults. However, its general prognosis remains universally poor regardless of the advances in treatments [52,53] and is associated with a high degree of heterogeneity that potentially complicates the treatment and contributes to failure in ESCC patients [54]. Accordingly, extensive exploration of the heterogeneity of ESCC is critical to unraveling the therapeutic targets for the management of ESCC patients. In the present study, we used 128,688 cells retrieved from five patients, including pairs of five tumor samples and five corresponding non-malignant samples to construct a single-cell transcriptome atlas of normal esophageal tissues and ESCC.
Using the single-cell transcriptome atlas, the cellular and molecular characteristics of esophageal epithelial cells were identified in the tumor and normal samples systematically. Although there are published reports on scRNAseq of ESCC [46], our study is far much different because our data including tumor cells. We re-clustered epithelial cells, including tumor cells and non-malignant epithelial cells, and identified a list of tumor-specific genes, which could serve as new targets for ESCC. Besides, the present work also revealed the great heterogeneity of tumor cells in both gene expression and CNV status. Furthermore, the gene expression signatures reflecting the intra-tumoral transcriptional heterogeneity of malignant cells were explored through unsupervised NMF analysis. Consequently, four malignant signatures, including cell repair, cell differentiation, cell cycle, and cell immune were detected, this was followed by the analysis of the correlations between the malignant signatures and survival in both TCGA and GEO databases via CIBERSORTx. As a result, low cell differentiation scores exhibited an obvious association with the poor prognosis of ESCC patients in both TCGA and GEO database cohorts, which implicate the cell differentiation metagenes as a potential prognostic marker for ESCC patients.
Scholars had demonstrated metabolic reprogramming to be an essential player in proliferation and metastasis and the mechanisms of drug resistance in tumors [55,56]. Hence, understanding the characteristics of metabolisms both in malignant and non-malignant cells has potential value as they provide a novel and reliable basis for the treatment of ESCC patients. Herein, a vast metabolic heterogeneity was revealed in ESCC and corroborated the mitochondrial activity, OXPHOS-related signaling. Particularly, as one of the most indispensable metabolic pathways for cellular energy and metabolism which had been demonstrated to be primarily utilized in the ATP synthesis in tumors [57,58]. Furthermore, we found that the OXPHOS-related pathway was up-regulated, which concur with reports from a previous study conducted at a single-cell level [59,9]. Nevertheless, this finding result, to some extent, was discordant with several previous reports, which had reported that the glycolysis pathway was mainly up-regulated in tumor cells, whereas OXPHOS-related pathways were primarily enriched in the non-malignant cells [60,61]. The inconsistency may be mainly attributed to different units used for analyses between the traditional and single-cell RNA sequencing methods. With traditional RNA sequencing, the average expression levels of each gene are evaluated in each tumor sample which comprises multiple cell types other than cancer cells, particularly, T cells that in most cases, account for a large proportion. Of note, we also observed that the hypoxia pathway and the OXPHOS pathway presents obviously positive correlation, which was also discordant with previous studies. To our knowledge, OXPHOS has been reported to regulate the response to hypoxia by working as a sensor of oxygen availability through stabilization of hypoxia-induced factors (HIF) [62,63]. Besides, the interactions between OXPHOS and hypoxia are highly dynamic in living cells and consistent results have been reported in another research using scRNA-seq in tumor [64]. Therefore, we hypothesize that the positive feedback from OXPHOS activity to HIF signaling in response to oxygen availability, which may lead to the interaction between the hypoxia pathway and the OXPHOS pathway, results in a positive correlation between OXPHOS and hypoxia, which is a unique feature of single malignant cells in the tumor environments. However, further studies are warranted to explore the functions of OXPHOS, glycolysis, and hypoxia at a single-cell level in ESCC and other cancer cells. Overall, the present work unraveled the metabolic reprogramming of cell type within the tumor microenvironment, which had been reported in ESCC at the single-cell level but without substantial facts.
With the advanced understanding of tumor immunology, immunotherapy has achieved great success in various neoplasms. Currently, most immunotherapies have been focused on T cells and they exert therapeutic effects by enhancing their aptitude to recognize and kill tumor cells [65]. However, only a minority of patients can benefit from current immunotherapies; thus, it is desirable to explore the immune lineages to uncover novel therapeutic targets for treatments. In our study, we found that two B cell sub-clusters were mainly derived from tumor tissues, which indicated the potential heterogeneity of tumor-associated B cells in our study. Florent Petitprez et al. [66] found that a high infiltration of B cells could form tertiary lymphoid structures to enhance the therapeutic efficacy of immunotherapies, which is associated with a better prognosis. Researchers [67] also reported that B cells could suppress tumors by activating immune effector cells or producing antibodies. Overall, there are numerous suggestions that tumor-associated B cells may play a much more prominent role in immunotherapies than previously appreciated. Howbeit, there is a need to explore the underlying functions of B cells in immunotherapies in the future.
The critical role of myeloid has been confirmed in various tumors using scRNAseq [68], [69], [70]. Herein, explored the distribution of immune checkpoints in myeloid cells and implicated HAVCR2 as potential immune targets for ESCC. We also confirmed the complex macrophage phenotypes process in ESCC, which concurred with other research findings [46]. Besides, in T cell analysis, our main focus was the NKT/CD8+ T cells. Through monocle analysis, the dynamic states and cell transitions of NKT/CD8+ T cells were explored; notably, three phases were identified in NKT/CD8+ cells. Elsewhere, Sun et al. [71] had demonstrated that the exhausted signature of CD8+ cells was upregulated in hepatocellular carcinoma, whereas the cytotoxic signature was decreased. Indeed, we also confirmed the high exhausted score and low cytotoxic score during the cell transitions of NKT/CD8+ T cells. Lastly, the dynamic changes in expressions of transcription factors were explored in macrophages and NKT/CD8+ T cells. Eventually, a single-cell transcriptome atlas was constructed using the scRNA-seq data, which provided a series of in-depth analyses on epithelial cells, stromal cells, and immune cells both in the normal esophagus tissues and ESCC.
Our study had some limitations. First, our limited sample sizes may lead to the bias of our results. Second, the patients included in our study were surgically resected ESCC patients rather than patients with recurrence and metastasis. However, through the above efforts, we believe that our study will contribute to the comprehension of the TME and cellular heterogeneity in ESCC patients and serve as a valuable resource for in-depth exploration of the pathogenesis of ESCC and identification of the potential therapeutic targets in the future.
Declaration of Competing Interest
None
Acknowledgments
Disclosures
The authors declare no conflict of interest relevant to the present manuscript
Data sharing statement
The data used in this study can be obtained by request to C.Z. (czhan10@fudan.edu.cn).
Acknowledgments
This work was supported by the Shanghai Pujiang Program (2020PJD009) and Key R&D and Translational Project in Tibet, China (XZ201901-GB-01) and Research Development Fund of Zhongshan Hospital, Fudan University (2019ZSFZ002 and 2019ZSFZ19). We thank OE Biotech Co., Ltd (Shanghai, China) to help us in sequencing analysis.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103459.
Contributor Information
Cheng Zhan, Email: czhan10@fudan.edu.cn.
Miao Lin, Email: lin.miao@zs-hospital.sh.cn.
Weigang Guo, Email: guo.weigang@zs-hospital.sh.cn.
Appendix. Supplementary materials
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Baba Y., Yoshida N., Kinoshita K., Iwatsuki M., Yamashita Y.I., Chikamoto A. Clinical and prognostic features of patients with esophageal cancer and multiple primary cancers: a retrospective single-institution study. Ann Surg. 2018;267(3):478–483. doi: 10.1097/SLA.0000000000002118. [DOI] [PubMed] [Google Scholar]
- 3.Ma J., Zhan C., Wang L., Jiang W., Zhang Y., Shi Y. The sweet approach is still worthwhile in modern esophagectomy. Ann Thorac Surg. 2014;97(5):1728–1733. doi: 10.1016/j.athoracsur.2014.01.034. [DOI] [PubMed] [Google Scholar]
- 4.Park S., Joung J.G., Min Y.W., Nam J.Y., Ryu D., Oh D. Paired whole exome and transcriptome analyses for the immunogenomic changes during concurrent chemoradiotherapy in esophageal squamous cell carcinoma. J Immunother Cancer. 2020;7(1):128–142. doi: 10.1186/s40425-019-0609-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azizi E., Carr A.J., Plitas G., Cornish A.E., Konopacki C., Prabhakaran S. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174(5):1293–1308. doi: 10.1016/j.cell.2018.05.060. e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao S., Yan L., Wang R., Li J., Yong J., Zhou X. Tracing the temporal-spatial transcriptome landscapes of the human fetal digestive tract using single-cell RNA-sequencing. Nat Cell Biol. 2018;20(6):721–734. doi: 10.1038/s41556-018-0105-4. [DOI] [PubMed] [Google Scholar]
- 7.Peng J., Sun B.F., Chen C.Y., Zhou J.Y., Chen Y.S., Chen H. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019;29(9):725–738. doi: 10.1038/s41422-019-0195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y., Song W., Wang J., Wang T., Xiong X., Qi Z. Single-cell transcriptome analysis reveals differential nutrient absorption functions in human intestine. J Exp Med. 2020;217(2):1–22. doi: 10.1084/jem.20191130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao Z., Dai Z., Locasale J.W. Metabolic landscape of the tumor microenvironment at single cell resolution. Nat Commun. 2019;10(1):3763. doi: 10.1038/s41467-019-11738-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao Z., Locasale J.W., Dai Z. Metabolism in the tumor microenvironment: insights from single-cell analysis. Oncoimmunology. 2020;9(1) doi: 10.1080/2162402X.2020.1726556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou J.X., Taramelli R., Pedrini E., Knijnenburg T., Huang S. Extracting intercellular signaling network of cancer tissues using ligand-receptor expression patterns from whole-tumor and single-cell transcriptomes. Sci Rep. 2017;7(1):8815. doi: 10.1038/s41598-017-09307-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang M.J., Ntranos V., Tse D. Determining sequencing depth in a single-cell RNA-seq experiment. Nat Commun. 2020;11(1):774. doi: 10.1038/s41467-020-14482-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding H., Blair A., Yang Y., Stuart J.M. Biological process activity transformation of single cell gene expression for cross-species alignment. Nat Commun. 2019;10(1):4899. doi: 10.1038/s41467-019-12924-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Z., Zhao M., Li M., Sui Q., Bian Y., Liang J. Identification of differentially expressed genes in lung adenocarcinoma cells using single-cell RNA sequencing not detected using traditional RNA sequencing and microarray. Lab Investig. 2020;100:1318–1329. doi: 10.1038/s41374-020-0428-1. [DOI] [PubMed] [Google Scholar]
- 15.Lambrechts D., Wauters E., Boeckx B., Aibar S., Nittner D., Burton O. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med. 2018;24(8):1277–1289. doi: 10.1038/s41591-018-0096-5. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Q., He Y., Luo N., Patel S.J., Han Y., Gao R. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. 2019;179(4):829–845. doi: 10.1016/j.cell.2019.10.003. e20. [DOI] [PubMed] [Google Scholar]
- 17.Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541(7636):169–175. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W., Zhang L., Guo B., Deng J., Wu S., Li F. Exosomal FMR1-AS1 facilitates maintaining cancer stem-like cell dynamic equilibrium via TLR7/NFkappaB/c-Myc signaling in female esophageal carcinoma. Mol Cancer. 2019;18(1):22. doi: 10.1186/s12943-019-0949-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGinnis C.S., Murrow L.M., Gartner Z.J. Doubletfinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8(4):329–337. doi: 10.1016/j.cels.2019.03.003. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haghverdi L., Lun A.T.L., Morgan M.D., Marioni J.C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol. 2018;36(5):421–427. doi: 10.1038/nbt.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aran D., Looney A.P., Liu L., Wu E., Fong V., Hsu A. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20(2):163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X., Lan Y., Xu J., Quan F., Zhao E., Deng C. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2018;47(D1):D721–D7D8. doi: 10.1093/nar/gky900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madissoon E., Wilbrey-Clark A., Miragaia R.J., Saeb-Parsy K., Mahbubani K.T., Georgakopoulos N. scRNA-seq assessment of the human lung, spleen, and esophagus tissue stability after cold preservation. Genome Biol. 2019;21(1):1. doi: 10.1186/s13059-019-1906-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang P., Yang M., Zhang Y., Xiao S., Lai X., Tan A. Dissecting the single-cell transcriptome network underlying gastric premalignant lesions and early gastric cancer. Cell Rep. 2019;27(6):1934–1947. doi: 10.1016/j.celrep.2019.04.052. e5. [DOI] [PubMed] [Google Scholar]
- 26.Wu X., Jiang R., Zhang M.Q., Li S. Network-based global inference of human disease genes. Mol Syst Biol. 2008;4:189. doi: 10.1038/msb.2008.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo W, Wang D, Wang S, Shan Y, Liu C, Gu J. scCancer: a package for automated processing of single-cell RNA-seq data in cancer. Briefings in bioinformatics. 2021;22(3) doi: 10.1093/bib/bbaa127. [DOI] [PubMed] [Google Scholar]
- 28.Müller S., Liu S.J., Di Lullo E., Malatesta M., Pollen A.A., Nowakowski T.J. Single-cell sequencing maps gene expression to mutational phylogenies in PDGF- and EGF-driven gliomas. Mol Syst Biol. 2016;12(11):889. doi: 10.15252/msb.20166969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel A.P., Tirosh I., Trombetta J.J., Shalek A.K., Gillespie S.M., Wakimoto H. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. doi: 10.1126/science.1254257. (New York, NY) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Y., Wu L., Zhong Y., Zhou K., Hou Y., Wang Z. Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell. 2020;184:404–421. doi: 10.1016/j.cell.2020.11.041. [DOI] [PubMed] [Google Scholar]
- 31.Gaujoux R., Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinform. 2010;11:367. doi: 10.1186/1471-2105-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017;14(10):979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steen C.B., Liu C.L., Alizadeh A.A., Newman A.M. Profiling cell type abundance and expression in bulk tissues with CIBERSORTx. Methods Mol Biol. 2020;2117:135–157. doi: 10.1007/978-1-0716-0301-7_7. (Clifton, NJ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barbie D.A., Tamayo P., Boehm J.S., Kim S.Y., Moody S.E., Dunn I.F. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH. Inference and analysis of cell-cell communication using CellChat. Nature communications. 2021;12(1):1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Wang R, Zhang S, Song S, Jiang C, Han G, et al. iTALK: an R Package to Characterize and Illustrate Intercellular Communication. bioRxiv 507871; doi: 10.1101/507871 [DOI]
- 38.Patriarca C., Macchi R.M., Marschner A.K., Mellstedt H. Epithelial cell adhesion molecule expression (CD326) in cancer: a short review. Cancer Treat Rev. 2012;38(1):68–75. doi: 10.1016/j.ctrv.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Niu H., Huang Y., Yan L., Zhang L., Zhao M., Lu T. Knockdown of SMAD3 inhibits the growth and enhances the radiosensitivity of lung adenocarcinoma via p21 in vitro and in vivo. Int J Biol Sci. 2020;16(6):1010–1022. doi: 10.7150/ijbs.40173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim D., Nam H.J., Lee W., Yim H.Y., Ahn J.Y., Park S.W. PKCα-LSD1-NF-κB-signaling cascade is crucial for epigenetic control of the inflammatory response. Mol Cell. 2018;69(3):398–411. doi: 10.1016/j.molcel.2018.01.002. e6. [DOI] [PubMed] [Google Scholar]
- 41.Xu R., Li Y., Liu Y., Qu J., Cao W., Zhang E. How are MCPIP1 and cytokines mutually regulated in cancer-related immunity? Protein Cell. 2020;11:881–893. doi: 10.1007/s13238-020-00739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Affo S., Yu L.X., Schwabe R.F. The role of cancer-associated fibroblasts and fibrosis in liver cancer. Annu Rev Pathol. 2017;12:153–186. doi: 10.1146/annurev-pathol-052016-100322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim N., Kim H.K., Lee K., Hong Y., Cho J.H., Choi J.W. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat Commun. 2020;11(1):2285. doi: 10.1038/s41467-020-16164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Largeot A., Pagano G., Gonder S., Moussay E., Paggetti J. The B-side of cancer immunity: the underrated tune. Cells. 2019;8(5):449–469. doi: 10.3390/cells8050449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y., Wei G., Cheng W.A., Dong Z., Sun H., Lee V.Y. Targeting myeloid-derived suppressor cells for cancer immunotherapy. Cancer Immunol Immunother. 2018;67(8):1181–1195. doi: 10.1007/s00262-018-2175-3. CII. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng Y., Chen Z., Han Y., Han L., Zou X., Zhou B. Immune suppressive landscape in the human esophageal squamous cell carcinoma microenvironment. Nat Commun. 2020;11(1):6268. doi: 10.1038/s41467-020-20019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marty Pyke R., Thompson W.K., Salem R.M., Font-Burgada J., Zanetti M., Carter H. Evolutionary pressure against MHC class II binding cancer mutations. Cell. 2020;175(2):416–428. doi: 10.1016/j.cell.2018.08.048. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Postow M.A., Sidlow R., Hellmann M.D. Immune-related adverse events associated with immune checkpoint blockade. New Engl J Med. 2018;378(2):158–168. doi: 10.1056/NEJMra1703481. [DOI] [PubMed] [Google Scholar]
- 49.Li B., Chan H.L., Chen P. Immune checkpoint inhibitors: basics and challenges. Curr Med Chem. 2019;26(17):3009–3025. doi: 10.2174/0929867324666170804143706. [DOI] [PubMed] [Google Scholar]
- 50.Raredon M.S.B., Adams T.S., Suhail Y., Schupp J.C., Poli S., Neumark N. Single-cell connectomic analysis of adult mammalian lungs. Sci Adv. 2019;5(12):eaaw3851. doi: 10.1126/sciadv.aaw3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sheikh B.N., Bondareva O., Guhathakurta S., Tsang T.H., Sikora K., Aizarani N. Systematic identification of Cell-Cell communication networks in the developing brain. iScience. 2019;21:273–287. doi: 10.1016/j.isci.2019.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 53.Chen Z, Li M, Ma K, Hu Z, Wang S, Chen H. Clinicopathological features and prognosis of patients with esophageal cancer as the second primary cancer: a large population-based analysis using the SEER program [2000–2015] Translational Cancer Research;2020. 2020;9(2):1113–1124. doi: 10.21037/tcr.2019.12.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan T., Cui H., Zhou Y., Yang B., Kong P., Zhang Y. Multi-region sequencing unveils novel actionable targets and spatial heterogeneity in esophageal squamous cell carcinoma. Nat Commun. 2019;10(1):1670. doi: 10.1038/s41467-019-09255-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nwosu Z.C., Pioronska W., Battello N., Zimmer A.D., Dewidar B., Han M. Severe metabolic alterations in liver cancer lead to ERK pathway activation and drug resistance. EBioMedicine. 2020;54 doi: 10.1016/j.ebiom.2020.102699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wettersten H.I., Aboud O.A., Lara Jr P.N., Weiss R.H. Metabolic reprogramming in clear cell renal cell carcinoma. Nat Rev Nephrol. 2017;13(7):410–419. doi: 10.1038/nrneph.2017.59. [DOI] [PubMed] [Google Scholar]
- 57.Momcilovic M., Jones A., Bailey S.T., Waldmann C.M., Li R., Lee J.T. In vivo imaging of mitochondrial membrane potential in non-small-cell lung cancer. Nature. 2019;575(7782):380–384. doi: 10.1038/s41586-019-1715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lasche M., Emons G., Grundker C. Shedding new light on cancer metabolism: a metabolic tightrope between life and death. Front Oncol. 2020;10:409. doi: 10.3389/fonc.2020.00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deng W., Ma Y., Su Z., Liu Y., Liang P., Huang C. Single-cell RNA-sequencing analyses identify heterogeneity of CD8+ T cell subpopulations and novel therapy targets in melanoma. Mol Ther Oncolytics. 2021;20:105–118. doi: 10.1016/j.omto.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li X., Wenes M., Romero P., Huang S.C.C., Fendt S.M., Ho P.C. Navigating metabolic pathways to enhance antitumour immunity and immunotherapy. Nat Rev Clin Oncol. 2019;16(7):425–441. doi: 10.1038/s41571-019-0203-7. [DOI] [PubMed] [Google Scholar]
- 61.Semenza G.L. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148(3):399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guzy R.D., Hoyos B., Robin E., Chen H., Liu L., Mansfield K.D. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1(6):401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 63.Wallace D.C. Mitochondria and cancer. Nat Rev Cancer. 2012;12(10):685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xiao Z., Dai Z., Locasale J.W. Metabolic landscape of the tumor microenvironment at single cell resolution. Nat Commun. 2019;10(1):3763. doi: 10.1038/s41467-019-11738-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sarkar I., Pati S., Dutta A., Basak U., Sa G. T-memory cells against cancer: remembering the enemy. Cell Immunol. 2019;338:27–31. doi: 10.1016/j.cellimm.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 66.Petitprez F., De Reynies A., Keung E.Z., Chen T.W., Sun C.M., Calderaro J. B cells are associated with survival and immunotherapy response in sarcoma. Nature. 2020;577(7791):556–560. doi: 10.1038/s41586-019-1906-8. [DOI] [PubMed] [Google Scholar]
- 67.Helmink B.A., Reddy S.M., Gao J., Zhang S., Basar R., Thakur R. B cells and tertiary lymphoid structures promote immunotherapy response. Nature. 2020;577(7791):549–555. doi: 10.1038/s41586-019-1922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen Z, Huang Y, Hu Z, Zhao M, Li M, Bi G. Landscape and dynamics of single tumor and immune cells in early and advanced-stage lung adenocarcinoma. Clinical and translational medicine. 2021;11(3):e350. doi: 10.1002/ctm2.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen Y.P., Yin J.H., Li W.F., Li H.J., Chen D.P., Zhang C.J. Single-cell transcriptomics reveals regulators underlying immune cell diversity and immune subtypes associated with prognosis in nasopharyngeal carcinoma. Cell Res. 2020;30(11):1024–1042. doi: 10.1038/s41422-020-0374-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sathe A., Grimes S.M., Lau B.T., Chen J., Suarez C., Huang R.J. Single-cell genomic characterization reveals the cellular reprogramming of the gastric tumor microenvironment. Clin Cancer Res Off J Am Assoc Cancer Res. 2020;26(11):2640–2653. doi: 10.1158/1078-0432.CCR-19-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun Y., Wu L., Zhong Y., Zhou K., Hou Y., Wang Z. Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell. 2021;184(2):404–421. doi: 10.1016/j.cell.2020.11.041. e16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.