Abstract
Several studies have reported association between leukocyte telomere length (LTL) and neuropsychiatric disorders. Although telomere length is affected by environmental factors, genetic variants in certain loci are strongly associated with LTL. Thus, we aimed to identify the genomic relationship between genetic variants of LTL with brain-based regulatory changes and brain volume.
We tested genetic colocalization of seven and nine LTL loci in two ancestry groups, European (EUR) and East-Asian (EAS), respectively, with brain morphology measures for 101 T1-magnetic resonance imaging-based region of interests (n = 21 821). The posterior probability (>90%) was observed for ‘fourth ventricle’, ‘gray matter’ and ‘cerebellar vermal lobules I–IV’ volumes. We then tested causal relationship using LTL loci for gene and methylation expression. We found causal pleiotropy for gene (EAS = four genes; EUR = five genes) and methylation expression (EUR = 17 probes; EAS = 4 probes) of brain tissues (P ≤ 2.47 × 10−6). Integrating chromatin profiles with LTL-single nucleotide polymorphisms identified 45 genes (EUR) and 79 genes (EAS) (P ≤ 9.78×10−7). We found additional 38 LTL-genes using chromatin-based gene mapping for EUR ancestry population. Gene variants in three LTL-genes—GPR37, OBFC1 and RTEL1/RTEL1-TNFRSF6B—show convergent evidence of pleiotropy with brain morphology, gene and methylation expression and chromatin association. Mapping gene functions to drug–gene interactions, we identified process ‘transmission across chemical synapses’ (P < 2.78 × 10−4).
This study provides evidence that genetic variants of LTL have pleiotropic roles with brain-based effects that could explain the phenotypic association of LTL with several neuropsychiatric traits.
Introduction
Telomeres are short sequences of ‘TTAGGG’ at the 3′ end of the chromosomes. With replicating cell cycles, telomere length continues to shorten and represents the mitotic history of the cell. The telomere length shortens (i.e. the number of TTAGGG repeat motifs decreases) over chronological age (i.e. actual age of the person) (1). Additionally, under the effects of influences such as inflammation, oxidative stress, elevated hormone levels under stress or pathological conditions, telomere length can shorten prematurely (2). Perturbations in telomere lengthening enzymes such as telomerase reverse transcriptase (TERT) and telomerase RNA component result in cellular damage and genomic instability, further accelerating damage to physiological processes termed cellular aging (3).
Leukocyte telomere length (LTL) is associated with brain morphology of such structures as hippocampal, temporal and cingulate region volumes among others (4). Teenagers with major depressive disorder (MDD) were reported to have shorter telomeres and smaller right hippocampal volume (5). A longitudinal study measuring the effects of various mental training modules (to improve cognition and behavioral traits) reported a significant association between reduced telomere length and thinning of the cortex structure highlighting neuronal plasticity variation between telomere and certain brain structures (6). The association of telomere length variability with psychiatric disorders has led to the hypothesis of accelerated cellular or biological aging mediating physiological deterioration (7).
Telomerase mediates cell differentiation and is highly expressed in neural stem and progenitor cells and declines with increasing age (8). Dysfunction in telomerase impairs neural circuitry in the hippocampus, which is one of many factors that lead to memory decline (9). The expression of TERT is lower in astrocytes than oligodendrocytes (10), and telomere attrition induces cellular senescence of astrocytes leading to neurological dysregulation (11). Telomere shortening disrupts the neuronal differentiation cell cycle (12). Neurogenesis in both adult and fetal tissue is influenced by functioning telomerase and interaction with sheltrin complex, which protects the telomeric caps (i.e. prevents reduction in telomeric TTAGGG repeat number) (13). Even though telomere length is generally expected to be associated with age-related disorders, telomere length attrition can be influenced by maternal and intrauterine stressors during fetal development exhibiting its effects during an individual’s life course (14).
Twin-based genetic heritability of LTL is estimated at ~30–60% (15). Genome-wide association studies (GWAS) measure relative differences in frequencies of genome-wide single nucleotide polymorphisms (SNPs) for a trait of interest. A relatively high proportion of SNP associations are found in non-coding regions and may have tissue-specific regulatory effects in local or distant regions (16). Tissue- and cell type-specific regulatory effects may be investigated by comparing the association of SNPs with gene expression (termed expression quantitative trait loci, eQTL) and methylation levels of cytosine-phosphate-guanosine (CpG) sites termed methylation QTL (mQTL) (17). Leveraging large studies with genetic, transcriptomic and epigenetic data allow us to identify putative tissue and cell-specific regulatory effects of trait-specific SNPs (18).
We hypothesize that the phenotypic association between telomere length and brain-based profiles is because of pleiotropic effects of genes that are associated with telomere and brain regulatory and volumetric profiles (Fig. 1). To our knowledge, no study has investigated loci that are shared between brain volumetric and expression with telomere length SNP associations.
Figure 1.
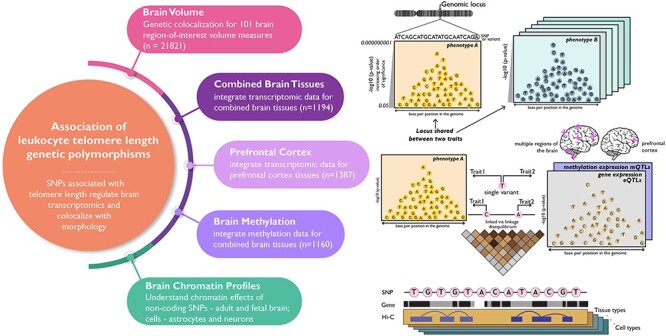
Study overview: schematic presenting the hypothesis and four major domains studied—(i) shared genomic regions between LTL and brain volume measures and gene-based associations for LTL in brain tissue-based (ii) transcriptomics—combined brain tissue (meta-analyzed from different regions of the brains) and prefrontal cortex, (iii) methylomics—combined brain tissues (meta-analyzed) and (iv) Hi-C chromatin profiles—adult and fetal brain, astrocytes and neurons.
Results
Genetic colocalization with brain morphology
We performed colocalization analysis between LTL and 101 brain volume measures to identify causal genomic loci that with respect to telomere length and measures of brain morphology. The ‘coloc’ method measures the posterior probability (PP) for five hypotheses (H0−H4), where H3 and H4 PPs represent sharing two and one genetic variant, respectively. For the LTL-loci in European (EUR) population, we observed gray matter [PPH3 = 99%, PPH4 = 1%; chr10(q24.33)]. The top variants associated with both traits in this locus [chr10(q24.33)] are rs11191849 and rs11191848, both mapping to OBFC1 (alternatively known as STN1).
For the LTL-loci in East-Asian (EAS) population, we found two brain volume measures: cerebellar vermal lobules I–IV [PPH3 = 42%, PPH4 = 50%; chr7 (q31.33)] and fourth ventricle [PPH3 = 4%, PPH4 = 96%; chr20(q13.33)] (Fig. 2).
Figure 2.
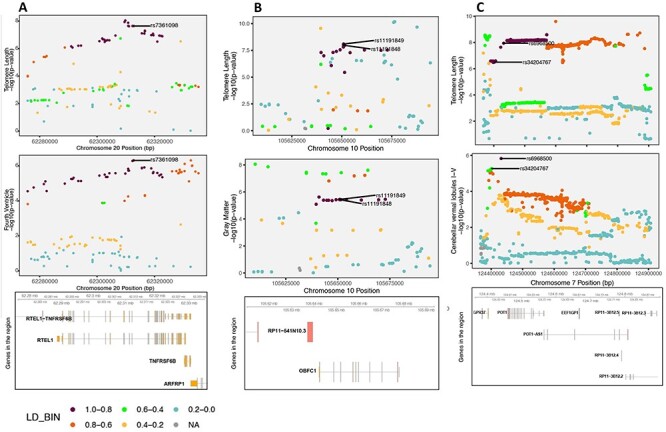
Genetic colocalization. (A) Regional association plot for LTL and fourth ventricle on chr20(q13.33) and the top significant variants for both traits in the locus is shown followed by genes in the region (hg19 build). (B) Regional association plot for LTL and gray matter on chr10(q24.33) and the top significant variants for both traits in the locus is shown followed by genes in the region (hg19 build). (C) Regional association plot for both traits on chr7(q31.33) and the top two most significant variants in the locus are shown followed by genes in the region (hg19 build).
The strongest PP was seen on chr20(q13.33) for sharing risk variants between LTL and fourth ventricle, which is located dorsal to the brain stem and anterior to the cerebellum and drains the cerebrospinal fluid (CSF) into the spinal cord (19). The top variant in this locus rs7361098 is in the intronic region of the RTEL1 (regulator of telomere elongation helicase 1; hg19 and 38) and upstream to the RTEL1-TNFRSF6B. Furthermore, the transethnic meta-analysis of LTL between EAS and EUR (15) reported rs6062498 as genome-wide significant, which is in LD with rs7361098 (EUR—r2 = 0.49; EAS—r2 = 0.53) in the same locus.
On chr7(q31.33), the cerebellar vermal lobules I–IV had high aggregated probability (92%) of shared genomic risk locus between LTL and volume of cerebellar vermal lobule. (Supplementary Material A2). The top variants in this locus were rs34204767 in G-protein coupled receptor 37 (GPR37) and rs6968500 in C7orf77 (upstream). The vermal lobules are located in the cerebellum midline and affect behavioral and cognitive phenotypes (20). Additionally, the top variant rs6968500 was significant in the transethnic meta-analysis of two ancestries for LTL (15).
Brain tissue-based transcriptomic and epigenetic QTLs
We integrated expression cis-QTL data meta-analyzed from different tissues of the brain (brainmeta) with SNPs associated with LTL considering allele frequency and LD structure of the EUR and EAS population separately. For brainmeta eQTL, we found one Bonferroni significant gene for the EAS population (DHRS1; P = 1.55 × 10−6 and PHEIDI = 1.25 × 10−21). None of the genes were Bonferroni significant for the EUR population. Significant P-value for the heterogeneity in dependent instruments (HEIDI) test indicates a linkage model, i.e. more than one variant that are probably in linkage are associated with eQTL and telomere length, whereas non-significant HEIDI P-value indicates pleiotropy under single causal variant model (Fig. 3A and B).
Figure 3.
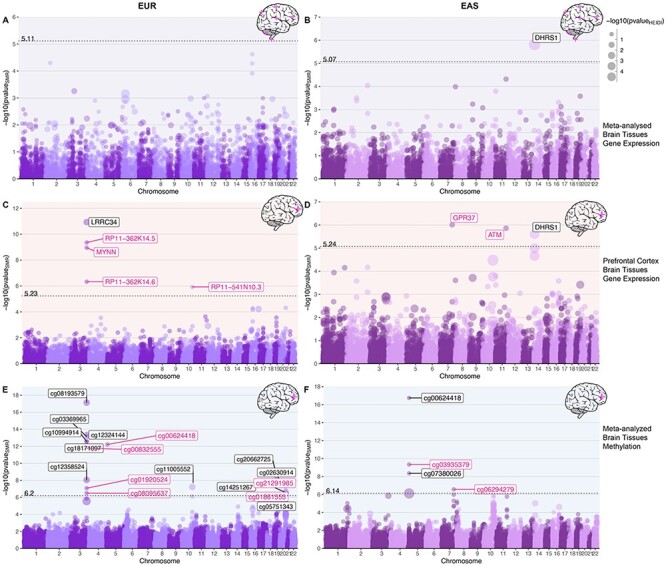
Gene-based associations integrating brain tissue eQTL and mQTL profiles with genetic variants associated with telomere length in EUR and EAS populations. Note: Bokeh plots are intersection between Manhattan and bubble plots. (A, B) Bokeh plots showing LTL-associated genes for gene expression (eQTLs) of combined brain tissues performed using SMR. (C, D) Bokeh plots showing LTL-associated genes for expression in prefrontal cortex. (E, F) Bokeh plots showing LTL-associated genes for methylation in combined brain tissues. The left side panels show association in EUR population and EAS associations are shown on the right. The x-axis shows genomic coordinate for the gene/CpG sites and the y-axis shows the −log10 of P-value of SMR test, so the order of increasing significance moves vertically. The dotted line shows the Bonferroni significance and annotated with respective value on top of the line. The size of the dots are scaled to the −log10(P-value) of HEIDI test, which tests for pleiotropy under single causal variant model defined as PHEIDI > 0.05 or −log10 (P-value) = 1.3 highlighted in pink text label.
Integrating eQTL data from prefrontal cortex (eQTLpfc), we found five genes—leucine-rich repeat containing 34 (LRRC34) (P = 1.15 × 10−11; PHEIDI = 9.64 × 10−4), RP11-362K14.5 (P = 4.38 × 10−10; PHEIDI = 0.13), MYNN (P = 1.11 × 10−9; PHEIDI = 0.17), RP11-362K14.6 (P = 4.67 × 10−7; PHEIDI = 0.17) and RP11-541N10.3 (P = 1.18 × 10−6; PHEIDI = 0.10) in the EUR population. All the significant genes, except LRRC34 had a non-significant HEIDI P-value, suggesting pleiotropic variants in the risk region sharing association with both eQTLpfc and LTL. We found three genes associated in the EAS population; GPR37 (P = 9.75 × 10−7; PHEIDI = 0.22), ataxia telangiectasia mutated (ATM) (P = 1.38 × 10−6; PHEIDI = 0.06) and DHRS1 (P = 2.47 × 10−6; PHEIDI = 1.11 × 10−5). Genes GPR37 and ATM have non-significant HEIDI test statistics, suggesting pleiotropy (single variant) in the region (Fig. 3C and D; Supplementary Material A3).
For the brain tissue mQTL data integrated with LTL association statistics, we observed 17 significant CpG sites (P ≤ 4.54 × 10−7) in EUR ancestry subjects. Six of the 17 CpG sites were associated under the pleiotropic model—cg00624418 (PHEIDI = 0.82), cg00832555 (PHEIDI = 0.06), cg01920524 (PHEIDI = 0.27), cg21291985 (PHEIDI = 0.22), cg08095637 (PHEIDI = 0.14) and cg01861555 (PHEIDI = 0.40). In the EAS population, we found four CpG sites; cg00624418 (P = 1.77 × 10−17; PHEIDI = 0.01), cg03935379 (P = 4.60 × 10−10; PHEIDI = 0.09), cg07380026 (P = 4.37 × 10−9; PHEIDI = 0.01) and cg06294279 (P = 2.65 × 10−7; PHEIDI = 0.35). Of these associations, cg03935379 (TERT) and cg06294279 (GPR37) are associated under the pleiotropic model (Fig. 3E and F; Supplementary Material A3). As methylation studies are mostly performed in blood, and methylation levels vary between tissues, we interpreted the CpG sites in blood and different brain tissues using BECon (21) (Supplementary Material A4; Fig. 4). Using the EWAS Atlas (22), we investigated the eight CpG sites that showed pleiotropy, for their disease-based tissue variability of methylation levels (Supplementary Material A4).
Figure 4.
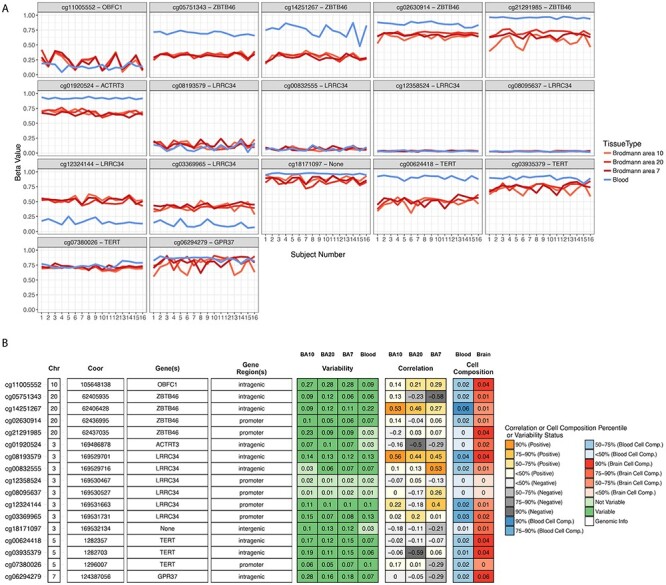
CpG site attributes and blood–brain tissue correlation. To aid in interpretation of the identified CpG sites, we used BECon for visualizing the interindividual and tissue-based variability. (A) The individual variability across three brain and blood tissue methylation levels is shown in each panel for respective CpG site from BECon data. (B) The panel shows genomic location and annotation of the CpG sites and correlation value of blood methylation levels with each of the three brain tissues methylation. Tabular information is listed in Supplementary Material A4.
Brain chromatin association for tissue and cell types
We performed chromatin-based association analysis for LTL, by combining high-resolution chromatin conformation (Hi-C) SNP–gene associations for developing brain—fetal brain, adult brain—dorsolateral prefrontal cortex and cell types—astrocytes and neurons using Hi-C-coupled multimarker analysis of genomic annotation (H-MAGMA) (23). For fetal brain, we observed 20 genes (EUR) and 48 genes (EAS); adult brain (EUR = 23; EAS = 51); astrocytes (EUR = 22; EAS = 50); neurons (EUR = 19; EAS = 49) (Fig. 5). Several genes overlapped across tissue and cell types (unique number of genes—EUR = 45, significant genes lower than P ≤ 9.78−7; EAS = 79; significant genes lower than P ≤ 8.97 × 10−7), and their specific distribution is shown in the Venn diagram (Fig. 6) (Supplementary Material A5). Furthermore, we identified 38 (EUR) and 53 (EAS) novel genes in comparison to with gene-based genome-wide association (Fig. 6). We found that >50% of the genes showed chromatin-based gene associations than gene-based associations.
Figure 5.
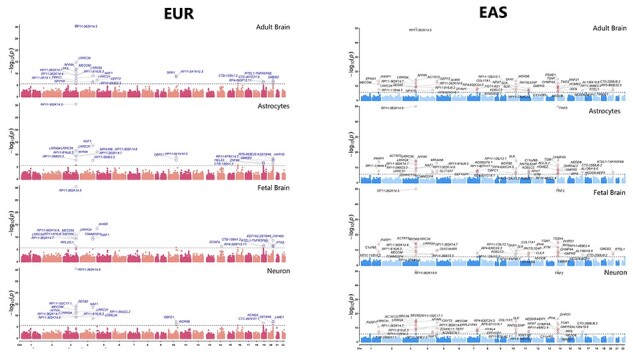
Genes associated with LTL for tissue (fetal and adult brain) and cells (astrocytes and neurons). The genes are shown as circle data points categorized by tissue and cell types (x-axis) and arranged by genomic position (ascending position; y-axis). Details are provided in Supplementary Material A5.
Figure 6.
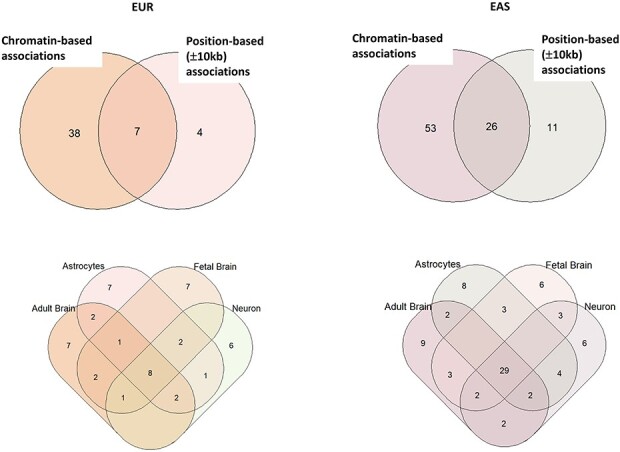
Distribution of top significant genes from the chromatin associations. The Venn diagram on the top shows the distribution of distinct and overlapping genes identified by chromatin-based association versus position-based association. The Venn diagram on bottom panel shows the distribution of overlapping significant genes in tissue and cell types’ chromatin profiles.
Gene–drug interaction and enrichment of gene functions
We analyzed all significant genes identified from analyses for gene–drug interaction. There were 14 genes on whom the drug interaction was available (Supplementary Material, Table S1 in Supplementary Material A6). To connect the influence of these drugs on gene functions, we performed drug set enrichment analysis. Out of 80 drugs, 10 drugs had associated information with gene function sets. Transmission across chemical synapses was false discovery rate (FDR) significant (P = 0.00028; PFDR = 0.019) and 24 nominally significant (P < 0.05) gene functions processes were also observed (Supplementary Material, Fig. S1 and Supplementary Material, Table S2 in Supplementary Material A6).
Discussion
In this study, we investigated the role of genetic variants associated with LTL in the context of brain-based expression, methylation and chromatin profiles of two tissue and cell types. Analyzing different brain morphological traits, we discovered that the volumes of global gray matter, fourth ventricle and cerebellar vermal lobules I–IV have genomic causal loci in common with LTL. The shared locus with global gray matter reported in our results corroborates previous findings of LTL association with cortical gray matter (4). A study of 15 892 individuals including subjects diagnosed with six psychiatric disorders showed gray matter loss across segmental brain regions in comparison to controls (24). The fourth ventricle bordered by the brainstem and the cerebellum is one of the sites for CSF production and is the last ventricle connecting to the subarachnoid space (19). Pathologies involving the fourth ventricle are associated with abnormal CSF volume and flow (25). Different neuropsychiatric disorders such as schizophrenia have demonstrated enlarged fourth ventricle—which reflects a deficit in bordering tissue when compared with brain samples from healthy subjects (26). Schizophrenia also has association with reduced telomere length (27). Furthermore, an extreme pathology (i.e. congenital deformity of the fourth ventricle) causes Dandy–Walker malformation, which exhibits some symptoms similar to those seen in schizophrenia, obsessive compulsive disorder, and type-I and type-II bipolar disorder (28). Furthermore, the cerebellar vermal lobules I–IV are the top four anterior lobes connecting both hemispheres of the cerebellum and are involved in motor, cognitive and behavioral processing in autism spectrum disorders (29). Taken together, our results further corroborate the relationship between central nervous system (CNS) morphology and telomere length.
The brain transcriptomic integrated association studies highlighted genes that are involved with both LTL and neuronal effects. GPR37 is expressed in oligodendrocytes in different tissues of the brain contributing to myelination regulation (30). GPR37 is associated with telomere length and serves as stress response neuroprotective gene. In major depression disorder, GPR37 expression levels in brain tissue were lower compared with healthy participants (31). Furthermore, CpG site—cg06294279—located in the GPR37 intragenic region was also associated under pleiotropic genomic risk (i.e. sharing association with both methylation level of the site and telomere length). The top variants in the colocalization analysis between LTL and cerebellar vermal lobules overlap with GPR37. To understand the role of the CpG site in diseases, we mined the EWAS Datahub (22); CpG site—cg06294279—showed high variability in different brain tissues between cases and controls, for schizophrenia and Alzheimer’s disease (Supplementary Material A4). Similarly, we observe OBFC1 locus associated with LTL in EUR ancestry colocalizing with total gray matter volume and methylation site—cg11005552 (Fig. 4). In the RTEL1 locus, we observe genomic colocalization with volumetric measure of fourth ventricle, and flanking region of ZBTB46, with four methylation CpG sites. Methylation levels of ZBTB46 are associated with cognitive function (32) and schizophrenia in a RNA-sequencing gene expression study (33,34).
LRRC34 gene expression is high in stem cells, which lowers upon cell differentiation (35). Individuals with bipolar disorder who were on lithium showed changes in genetically regulated transcriptome effect of LRRC34 (36). Furthermore, we found six CpG sites in the LRRC34 gene that displayed mQTL associations for SNPs associated with LTL, out of which, site cg08095637 had single-variant pleiotropic association.
Among genes identified from chromatin profile integrated associations, PARP1 (poly(ADP-ribose) polymerase 1) is a regulator of DNA repair and cell death taking part in neurogenesis of neuronal and embryonic stem cells (37). Lowering the expression levels of PARP1 is linked with abnormal ventricle volume in the brain and exhibits psychiatric symptoms including anxiety, depression and behavioral impairment (37,38). ATM is also involved in cellular DNA repair and differentiating pluripotent stem cells (39). ATM regulates GABAergic-based neuronal plasticity and brain development and its dysregulation is associated with onset of different psychopathologies (40). The BECon databank shows that Broadman areas 7, 10 and 20 share little to low negative correlation with blood-based methylation in healthy individuals. The mQTL analysis of brain tissue found CpG sites in addition to LRRC34, in the TERT gene. Methylation-based biological age acceleration is associated with TERT and genetic overlap with schizophrenia, bipolar disorder and dementia (41). TERT is also involved in paternally inherited epigenetic imprint observed in advanced paternal age, which is a risk for several neuropsychiatric disorder (42). Furthermore, pleiotropic gene expression association shows multiple genes from prefrontal cortex, rather than analysis with expression from multiple regions of the brain. This finding may be explained by high correlation between telomere length of whole blood and brain cortex (18).
The drug–gene interaction of significant genes showed enrichment for different transporter systems of chemical synapses, aquaporins and mitochondrial tRNA aminoacylation and transmission in the postsynaptic cell (Supplementary Material, Fig. S1: Supplementary Material A6). Together, these findings indicate the set of LTL-associated genes exhibit plausible pleiotropic causality from regulatory effects of brain tissues to telomere length. Therefore, we believe telomere length genomics is an important avenue for future studies of mental health.
Although our study has provided key evidence of shared genomic causal loci among telomere length, brain morphology and regulatory traits, it has limitations. In our attempt to conduct a dual-ancestry estimation, we capture some population-based differences, we observe most significant genes from one population are nominally significant in the other. However, there were few regions that are significant in one population, but not the other. These differences are because of recombination of the SNPs and allele frequency differences in the locus. Five loci on chromosomes 3, 4, 5, 10 and 20 overlap in both populations; however, the SNPs in these regions show differences in linkage disequilibrium highlighting the importance of studying the population-based genetic association. These differences in SNP peaks are visualized in genome-wide and regional plots (Supplementary Material A1). These differences are also reflected in their regulatory or volumetric genomic relationships. Although LTL is altered during aging and other environmental factors including birth (43), our analyses is limited to genetic determinants of LTL and negates the use of—although informative—longitudinal assessment of telomere length. The results showing genetic colocalizing cannot be interpreted as mediating the direction of causality. Furthermore, colocalization of two traits under a single variant is a conservative model of testing, and there is biological plausibility of more than one variant exhibiting small effects. There are several SNPs in a locus exhibiting different effect directions for LTL and brain volume (Supplementary Material, Fig. S2: Supplementary Material A6). Therefore, it is difficult to assess the phenotypic consequences of each SNP because variants may alter physiological states in tissue- or cell type-specific manner. We assessed regulatory changes using two populations, and therefore, the results may not apply to other major populations and will require separate investigation. Further studies using animal models would narrow targets with specific cellular changes. Our next steps will involve investigating the role of telomere length with respect to neuropsychiatric brain disorders to deepen the understanding of cellular aging in neurological outcomes.
We identified shared genomic causal loci among those affecting telomere length, brain morphology and genetically regulated regulatory traits. The identified genes show involvement in the CNS and provide evidence that shared genetics could explain, in part, the association of LTL with several brain-based outcomes reported previously. These results highlight that telomere length may serve as a marker for certain brain-based diseases such as psychiatric disorders. These findings additionally corroborate that in addition to methylation-based biological aging, telomere-based cellular aging has the potential to provide further resolution on biological mechanisms of brain disorders.
Materials and Methods
Overall, in this study we used large-scale LTL GWAS of 37 505 EUR and 23 096 Chinese individuals (ntotal = 60 601) (15). As ancestry-specific locus for LTL association differs on some loci, we investigated LTL loci reported for each ancestry. We aimed to identify (a) LTL causal loci shared with brain volume measures, (b) transcriptomic and epigenetic regulatory effects and (c) chromatin profiles in brain tissue (Fig. 1). First we analyzed whether LTL risk loci are shared with 101 T1-magnetic resonance imaging (MRI-weighted for longitudinal relaxation time) brain region-of-interest (ROI) phenotypes (n = 21 821) (44). Then, we investigated causality of expression for mQTL (45) (n = 1160) and eQTL associations from different regions of the brain (n = 1194) using summary-based Mendelian randomization (SMR) (46). Similarly, Hi-C-guided H-MAGMA (23) is employed to understand biological mechanisms underlying the effects of non-coding SNPs on LTL association by mapping chromatin profiles from human brain tissue of two developmental stages—fetal and adult, and two cell types—astrocytes and neurons.
Study cohorts
LTL GWAS
The most recent LTL GWAS is available as transethnic meta-analysis of 37 505 EUR [of ENGAGE consortia (47)] and 23 096 Chinese individuals (ntotal = 60 601) (15). The genomic risk regions for LTL were identified from the summary statistics of each population, using FUMA (default values; LD −0.6 and 250 kb locus) resulting in seven genome loci in EUR population, and nine independent loci in EAS population (48). A locus contains several SNPs independent significant SNPs by merging LD blocks within 250 kb. Loci on chromosomes 2 and 19 that are observed in EURs are not present in EAS population. Similarly, there are loci specific to EAS population are on chromosomes 1, 7 and 14. Regions that are positionally similar in both populations are on chromosomes 3, 4, 5, 10 and 20. Length of these genomic regions varies by population because of LD structure and different number of significant variants within these regions. Therefore, in this study we investigated pleiotropic associations of SNPs with LTL and quantitative expression of brain tissues in both populations (Supplementary Material A1).
Brain volume GWAS
To evaluate which loci are shared between LTL and brain volume measures, we used the largest GWAS performed for 101 phenotypes of brain morphology (44). In the reported study, the brain volume measures or ROI were quantified using MRI and mapped to cataloged ROI to identify (i.e. map them to) structural measures (44). The GWAS of 101 brain morphology/ROI measures includes meta-analysis of five cohorts—the Human Connectome Project study, the Pediatric Imaging, Neurocognition, and Genetics study, the Philadelphia Neurodevelopmental Cohort study and the Alzheimer’s Disease Neuroimaging Initiative study (n = 21 821).
Expression-based pleiotropic associations
To identify LTL-gene targets that have pleiotropic associations with brain-based expression, we integrated with eQTL and mQTL information. Data were included from two transcriptomic studies: (a) multiple brain tissue i.e. meta-analysis of gene expression from different regions of the brain (brainmeta) [Genotype-Tissue Expression (49), CommonMind Consortium (50), Religious Orders Study and the Memory and Aging Project]and (b) prefrontal cortex (brainpfc) PsychENCODE consortia (51) and methylation association study using meta-analyzed methylation expression from brain cortical region, fetal brain and frontal cortex region (45). The QTL reference panel weights are provided in the SMR (46).
Cell- and tissue-based chromatin-mapping associations
The H-MAGMA (v1.08) approach aggregates genetic variants to nearest genes derived from Hi-C data of fetal developing cortex, adult dorsolateral prefrontal cortex and cell types—iPSC-derived astrocytes and neurons (52).
All data used in this study were made publicly available by the cited consortia. This study was exempt from institutional review board (IRB) review because of use of deidentified data.
Brain morphology colocalization
The GWAS association statistics for each of the 101 brain morphology measures (44) were evaluated with respect to GWAS of LTL (15) to identify which causal risk loci were shared across both traits using coloc (53). We performed pairwise colocalization analysis for each of the risk loci between LTL and each brain volume phenotypes using the appropriate 1000 Genomes Phase 3 reference panels (i.e. EUR and EAS). The coloc method in R-v3.6 (54) evaluates PP for four alternative hypothesis—(A) H1: association with trait 1, not with trait 2, (B) H2: association with trait 2, not with trait 1, (C) H3: association with trait 1 and trait 2, two independent SNPs and (D) H4: association with trait 1 and trait 2, one shared SNP (53). We used the coloc.abf function using P-values, respective ancestry’s allele frequencies and sample size with default priors. Here we report results for sharing single causal variant with both traits (LTL and brain morphology) having H4 PP >90% in the main text; detailed results can be found in Supplementary Material A2. The coloc approach has also been used by other studies for EAS-based population (15).
Transcriptomic and epigenetic QTLs in brain tissues
The association of LTL was analyzed for gene expression (brainmeta and brainpfc), and methylation association study using meta-analyzed methylation expression from brain tissue using SMR (46). The SMR approach tests for ‘causal estimate’ between genetically expressed expression or methylation and a trait of interest, with the underlying assumption that genetic variant is associated with expression and trait. Bonferroni correction (0.05/number of associations) was applied to identify significant gene associations for each of the six QTL analyses. We performed HEIDI association analysis to test if a single variant is associated with phenotype and QTL (single-variant model of pleiotropy). Significant HEIDI associations (P < 0.05) depart from this assumption of single-variant association, whereas non-significant associations (P > 0.05) do not reject the null, indicating a linkage model (i.e. different SNPs in linkage) (46). The analyses were performed in each ancestry group (i.e. EUR and EAS) separately. Two sets of files were created using LD and SNP allele frequencies of each population from the 1000 Genome Phase 3 reference panel. This resulted in different number of SNPs considered for each population. This approach has been used previously for non-EUR population (55,56). Misaligned alleles between QTL and LTL-SNP dataset were removed. The CpG sites that showed pleiotropic evidence i.e. single causal variant between LTL and brain mQTL were interpreted using BECon for methylation level correlation between brain tissues and blood (21) and extracted from EWAS DataHub for different brain disorders (22).
Brain chromatin profiles using H-MAGMA
The gene-based association study for brain tissue-derived chromatin profiles for fetal and adult brain regions and cells (astrocytes and neurons) was performed using H-MAGMA v.1.08 (23,57). The H-MAGMA approach aids in providing functional/regulatory effects of non-coding SNPs and is complementary to transcriptome-wide association and the coloc approach used in this study (23). Because of telomeres/TERT’s role in the CNS, we tested all four tissue profiles with LTL GWAS association statistics. Each tissue/cell type association was performed for EUR and EAS population reference panels. Bonferroni correction (0.05/number of associations) was applied to identify gene associations for each the four tissue/cell type analyses. The visualizations were created in R and PhenoGram (58).
Gene–drug interaction and overrepresented gene functions based on drugs
All significant genes from aforementioned analyses were investigated for gene–drug interactions using DGidb (59); then identified drugs were analyzed for pathway enrichment using Drug Set Enrichment Analysis (DSEA) (60).
Funding
National Institutes of Health (R21 DC018098, R21 DA047527, F32 MH122058).
Data Availability and URLs
All the data used in this study are publicly available, hosted at the respective consortia portal. Chinese and EUR population telomere length GWAS (https://www.nature.com/articles/s41467-019-10443-2#data-availability); Brain Region Phenotypes GWAS (https://github.com/BIG-S2/GWAS); eQTL and mQTL data are available from SMR (https://cnsgenomics.com/software/smr/#Overview); H-MAGMA (updated 15 September 2020: https://github.com/thewonlab/H-MAGMA). The results generated in this study are provided in the main text and supplementary files.
Author Contributions
G.A.P conceptualized the study design, performed analysis and wrote the first draft. R.P supervised the study, contributed to study design and manuscript revisions and gave final approval on the manuscript. F.R.W, D.F.L, A.P.M, C.H.V.D and J. G contributed to writing and revising the manuscript draft.
Ethics Approval
The study uses publicly shared summary-level data and is therefore exempt from IRB review.
Animal Research
Not applicable.
Consent to Participate
We used publicly shared summary-level data from all studies; therefore, consent to participate is not applicable.
Consent for Publication
All authors have consented for publication.
Supplementary Material
Acknowledgements
We would like to acknowledge the participants and investigators of all consortia whose data helped investigate the research question.
Contributor Information
Gita A Pathak, Department of Psychiatry, Yale School of Medicine, Yale University, New Haven, CT 06551, USA; Veteran Affairs Connecticut Healthcare System, West Haven, CT 06516, USA.
Frank R Wendt, Department of Psychiatry, Yale School of Medicine, Yale University, New Haven, CT 06551, USA; Veteran Affairs Connecticut Healthcare System, West Haven, CT 06516, USA.
Daniel F Levey, Department of Psychiatry, Yale School of Medicine, Yale University, New Haven, CT 06551, USA; Veteran Affairs Connecticut Healthcare System, West Haven, CT 06516, USA.
Adam P Mecca, Department of Psychiatry, Yale School of Medicine, Yale University, New Haven, CT 06551, USA; Alzheimer’s Disease Research Unit, Yale University School of Medicine, New Haven, CT 06511, USA.
Christopher H van Dyck, Department of Psychiatry, Yale School of Medicine, Yale University, New Haven, CT 06551, USA; Alzheimer’s Disease Research Unit, Yale University School of Medicine, New Haven, CT 06511, USA; Department of Neuroscience, Yale University School of Medicine, New Haven, CT 06511, USA; Department of Neurology, Yale University School of Medicine, New Haven, CT 06511, USA.
Joel Gelernter, Department of Psychiatry, Yale School of Medicine, Yale University, New Haven, CT 06551, USA; Veteran Affairs Connecticut Healthcare System, West Haven, CT 06516, USA.
Renato Polimanti, Department of Psychiatry, Yale School of Medicine, Yale University, New Haven, CT 06551, USA; Veteran Affairs Connecticut Healthcare System, West Haven, CT 06516, USA.
Conflict of Interest statement
Drs Polimanti and Gelernter are paid for their editorial work for the journal Complex Psychiatry. The other authors have no conflicts of interest to declare.
References
- 1. Koliada, A.K., Krasnenkov, D.S. and Vaiserman, A.M. (2015) Telomeric aging: mitotic clock or stress indicator? Front. Genet., 6, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vakonaki, E., Tsiminikaki, K., Plaitis, S., Fragkiadaki, P., Tsoukalas, D., Katsikantami, I., Vaki, G., Tzatzarakis, M.N., Spandidos, D.A. and Tsatsakis, A.M. (2018) Common mental disorders and association with telomere length. Biomed. Rep., 8, 111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bär, C. and Blasco, M.A. (2016) Telomeres and telomerase as therapeutic targets to prevent and treat age-related diseases. [version 1; peer review: 4 approved]. F1000Res, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. King, K.S., Kozlitina, J., Rosenberg, R.N., Peshock, R.M., McColl, R.W. and Garcia, C.K. (2014) Effect of leukocyte telomere length on total and regional brain volumes in a large population-based cohort. JAMA Neurol., 71, 1247–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Henje Blom, E., Han, L.K.M., Connolly, C.G., Ho, T.C., Lin, J., LeWinn, K.Z., Simmons, A.N., Sacchet, M.D., Mobayed, N., Luna, M.E.et al. (2015) Peripheral telomere length and hippocampal volume in adolescents with major depressive disorder. Transl. Psychiatry, 5, e676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Puhlmann, L.M.C., Valk, S.L., Engert, V., Bernhardt, B.C., Lin, J., Epel, E.S., Vrticka, P. and Singer, T. (2019) Association of short-term change in leukocyte telomere length with cortical thickness and outcomes of mental training among healthy adults: a randomized clinical trial. JAMA Netw. Open, 2, e199687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aas, M., Elvsåshagen, T., Westlye, L.T., Kaufmann, T., Athanasiu, L., Djurovic, S., Melle, I., van derMeer, D., Martin-Ruiz, C., Steen, N.E.et al. (2019) Telomere length is associated with childhood trauma in patients with severe mental disorders. Transl. Psychiatry, 9, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu, M.-Y., Nemes, A. and Zhou, Q.-G. (2018) The emerging roles for telomerase in the central nervous system. Front. Mol. Neurosci., 11, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou, Q.-G., Liu, M.-Y., Lee, H.-W., Ishikawa, F., Devkota, S., Shen, X.-R., Jin, X., Wu, H.-Y., Liu, Z., Liu, X.et al. (2017) Hippocampal TERT regulates spatial memory formation through modulation of neural development. Stem Cell Rep., 9, 543–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Szebeni, A., Szebeni, K., DiPeri, T., Chandley, M.J., Crawford, J.D., Stockmeier, C.A. and Ordway, G.A. (2014) Shortened telomere length in white matter oligodendrocytes in major depression: potential role of oxidative stress. Int. J. Neuropsychopharmacol., 17, 1579–1589. [DOI] [PubMed] [Google Scholar]
- 11. Cohen, J. and Torres, C. (2019) Astrocyte senescence: evidence and significance. Aging Cell, 18, e12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ferrón, S.R., Marqués-Torrejón, M.A., Mira, H., Flores, I., Taylor, K., Blasco, M.A. and Fariñas, I. (2009) Telomere shortening in neural stem cells disrupts neuronal differentiation and neuritogenesis. J. Neurosci., 29, 14394–14407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lobanova, A., She, R., Pieraut, S., Clapp, C., Maximov, A. and Denchi, E.L. (2017) Different requirements of functional telomeres in neural stem cells and terminally differentiated neurons. Genes Dev., 31, 639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Edlow, A.G., Guedj, F., Sverdlov, D., Pennings, J.L.A. and Bianchi, D.W. (2019) Significant effects of maternal diet during pregnancy on the murine fetal brain transcriptome and offspring behavior. Front. Neurosci., 13, 1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dorajoo, R., Chang, X., Gurung, R.L., Li, Z., Wang, L., Wang, R., Beckman, K.B., Adams-Haduch, J., Yiamunaa, M., Liu, S.et al. (2019) Loci for human leukocyte telomere length in the Singaporean Chinese population and trans-ethnic genetic studies. Nat. Commun., 10, 2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Giral, H., Landmesser, U. and Kratzer, A. (2018) Into the wild: GWAS exploration of non-coding RNAs. Front. Cardiovasc. Med., 5, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhao, T., Hu, Y., Zang, T. and Wang, Y. (2019) Integrate GWAS, eQTL, and mQTL data to identify Alzheimer’s disease-related genes. Front. Genet., 10, 1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Demanelis, K., Jasmine, F., Chen, L.S., Chernoff, M., Tong, L., Delgado, D., Zhang, C., Shinkle, J., Sabarinathan, M., Lin, H.et al. (2020) Determinants of telomere length across human tissues. Science, 369, eaaz6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Roesch, Z.K. and Tadi, P. (2020) Neuroanatomy, fourth ventricle. In Stat Pearls. Stat Pearls Publishing, Treasure Island (FL). [PubMed] [Google Scholar]
- 20. Yucel, K., Nazarov, A., Taylor, V.H., Macdonald, K., Hall, G.B. and Macqueen, G.M. (2013) Cerebellar vermis volume in major depressive disorder. Brain Struct. Funct., 218, 851–858. [DOI] [PubMed] [Google Scholar]
- 21. Edgar, R.D., Jones, M.J., Meaney, M.J., Turecki, G. and Kobor, M.S. (2017) BECon: a tool for interpreting DNA methylation findings from blood in the context of brain. Transl. Psychiatry, 7, 1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li, M., Zou, D., Li, Z., Gao, R., Sang, J., Zhang, Y., Li, R., Xia, L., Zhang, T., Niu, G.et al. (2019) EWAS Atlas: a curated knowledgebase of epigenome-wide association studies. Nucleic Acids Res., 47, D983–D988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sey, N.Y.A., Hu, B., Mah, W., Fauni, H., McAfee, J.C., Rajarajan, P., Brennand, K.J., Akbarian, S. and Won, H. (2020) A computational tool (H-MAGMA) for improved prediction of brain-disorder risk genes by incorporating brain chromatin interaction profiles. Nat. Neurosci., 23, 583–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goodkind, M., Eickhoff, S.B., Oathes, D.J., Jiang, Y., Chang, A., Jones-Hagata, L.B., Ortega, B.N., Zaiko, Y.V., Roach, E.L., Korgaonkar, M.S.et al. (2015) Identification of a common neurobiological substrate for mental illness. JAMA Psychiat., 72, 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Whedon, J.M. and Glassey, D. (2009) Cerebrospinal fluid stasis and its clinical significance. Altern. Ther. Health Med., 15, 54–60. [PMC free article] [PubMed] [Google Scholar]
- 26. Juuhl-Langseth, M., Rimol, L.M., Rasmussen, I.A., Thormodsen, R., Holmén, A., Emblem, K.E., Due-Tønnessen, P., Rund, B.R. and Agartz, I. (2012) Comprehensive segmentation of subcortical brain volumes in early onset schizophrenia reveals limited structural abnormalities. Psychiatry Res., 203, 14–23. [DOI] [PubMed] [Google Scholar]
- 27. Russo, P., Prinzi, G., Proietti, S., Lamonaca, P., Frustaci, A., Boccia, S., Amore, R., Lorenzi, M., Onder, G., Marzetti, E.et al. (2018) Shorter telomere length in schizophrenia: evidence from a real-world population and meta-analysis of most recent literature. Schizophr. Res., 202, 37–45. [DOI] [PubMed] [Google Scholar]
- 28. Lingeswaran, A., Barathi, D. and Sharma, G. (2009) Dandy-Walker variant associated with bipolar affective disorder. J. Pediatr. Neurosci., 4, 131–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. D’Mello, A.M., Crocetti, D., Mostofsky, S.H. and Stoodley, C.J. (2015) Cerebellar gray matter and lobular volumes correlate with core autism symptoms. Neuroimage Clin., 7, 631–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smith, B.M., Giddens, M.M., Neil, J., Owino, S., Nguyen, T.T., Duong, D., Li, F. and Hall, R.A. (2017) Mice lacking Gpr 37 exhibit decreased expression of the myelin-associated glycoprotein MAG and increased susceptibility to demyelination. Neuroscience, 358, 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mamdani, F., Rollins, B., Morgan, L., Myers, R.M., Barchas, J.D., Schatzberg, A.F., Watson, S.J., Akil, H., Potkin, S.G., Bunney, W.E.et al. (2015) Variable telomere length across post-mortem human brain regions and specific reduction in the hippocampus of major depressive disorder. Transl Psychiatry, 5, e636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Starnawska, A., Tan, Q., McGue, M., Mors, O., Børglum, A.D., Christensen, K., Nyegaard, M. and Christiansen, L. (2017) Epigenome-wide association study of cognitive functioning in middle-aged monozygotic twins. Front. Aging Neurosci., 9, 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang, Y., You, X., Li, S., Long, Q., Zhu, Y., Teng, Z. and Zeng, Y. (2020) Peripheral blood leukocyte RNA-Seq identifies a set of genes related to abnormal psychomotor behavior characteristics in patients with schizophrenia. Med. Sci. Monit., 26, e922426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maycox, P.R., Kelly, F., Taylor, A., Bates, S., Reid, J., Logendra, R., Barnes, M.R., Larminie, C., Jones, N., Lennon, M.et al. (2009) Analysis of gene expression in two large schizophrenia cohorts identifies multiple changes associated with nerve terminal function. Mol. Psychiatry, 14, 1083–1094. [DOI] [PubMed] [Google Scholar]
- 35. Lührig, S., Siamishi, I., Tesmer-Wolf, M., Zechner, U., Engel, W. and Nolte, J. (2014) Lrrc 34, a novel nucleolar protein, interacts with npm 1 and ncl and has an impact on pluripotent stem cells. Stem Cells Dev., 23, 2862–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Coutts, F., Palmos, A.B., Duarte, R.R.R., deJong, S., Lewis, C.M., Dima, D. and Powell, T.R. (2019) The polygenic nature of telomere length and the anti-ageing properties of lithium. Neuropsychopharmacology, 44, 757–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hong, S., Yi, J.H., Lee, S., Park, C.-H., Ryu, J.H., Shin, K.S. and Kang, S.J. (2019) Defective neurogenesis and schizophrenia-like behavior in PARP-1-deficient mice. Cell Death Dis., 10, 943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sriram, C.S., Jangra, A., Gurjar, S.S., Mohan, P. and Bezbaruah, B.K. (2016) Edaravone abrogates LPS-induced behavioral anomalies, neuroinflammation and PARP-1. Physiol. Behav., 154, 135–144. [DOI] [PubMed] [Google Scholar]
- 39. Corti, A., Sota, R., Dugo, M., Calogero, R.A., Terragni, B., Mantegazza, M., Franceschetti, S., Restelli, M., Gasparini, P., Lecis, D.et al. (2019) DNA damage and transcriptional regulation in iPSC-derived neurons from ataxia telangiectasia patients. Sci. Rep., 9, 651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pizzamiglio, L., Focchi, E., Murru, L., Tamborini, M., Passafaro, M., Menna, E., Matteoli, M. and Antonucci, F. (2016) New role of ATM in controlling gabaergic tone during development. Cereb. Cortex, 26, 3879–3888. [DOI] [PubMed] [Google Scholar]
- 41. Lu, A.T., Xue, L., Salfati, E.L., Chen, B.H., Ferrucci, L., Levy, D., Joehanes, R., Murabito, J.M., Kiel, D.P., Tsai, P.-C.et al. (2018) GWAS of epigenetic aging rates in blood reveals a critical role for TERT. Nat. Commun., 9, 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hehar, H., Ma, I. and Mychasiuk, R. (2017) Intergenerational transmission of paternal epigenetic marks: mechanisms influencing susceptibility to post-concussion symptomology in a rodent model. Sci. Rep., 7, 7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bijnens, E.M., Zeegers, M.P., Derom, C., Martens, D.S., Gielen, M., Hageman, G.J., Plusquin, M., Thiery, E., Vlietinck, R. and Nawrot, T.S. (2017) Telomere tracking from birth to adulthood and residential traffic exposure. BMC Med., 15, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao, B., Luo, T., Li, T., Li, Y., Zhang, J., Shan, Y., Wang, X., Yang, L., Zhou, F., Zhu, Z.et al. (2019) Genome-wide association analysis of 19, 629 individuals identifies variants influencing regional brain volumes and refines their genetic co-architecture with cognitive and mental health traits. Nat. Genet., 51, 1637–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Qi, T., Wu, Y., Zeng, J., Zhang, F., Xue, A., Jiang, L., Zhu, Z., Kemper, K., Yengo, L., Zheng, Z.et al. (2018) Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat. Commun., 9, 2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhu, Z., Zhang, F., Hu, H., Bakshi, A., Robinson, M.R., Powell, J.E., Montgomery, G.W., Goddard, M.E., Wray, N.R., Visscher, P.M.et al. (2016) Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet., 48, 481–487. [DOI] [PubMed] [Google Scholar]
- 47. Codd, V., Nelson, C.P., Albrecht, E., Mangino, M., Deelen, J., Buxton, J.L., Hottenga, J.J., Fischer, K., Esko, T., Surakka, I.et al. (2013) Identification of seven loci affecting mean telomere length and their association with disease. Nat. Genet., 45, 422–427427e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Watanabe, K., Taskesen, E., vanBochoven, A. and Posthuma, D. (2017) Functional mapping and annotation of genetic associations with FUMA. Nat. Commun., 8, 1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. GTEx Consortium, Laboratory, Data Analysis & Coordinating Center (LDACC)—Analysis Working Group, Statistical Methods groups—Analysis Working Group, Enhancing GTEx (eGTEx) groups, NIH Common Fund, NIH/NCI, NIH/NHGRI, NIH/NIMH, NIH/NIDA, Biospecimen Collection Source Site—NDRI. et al. (2017) Genetic effects on gene expression across human tissues. Nature, 550, 204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fromer, M., Roussos, P., Sieberts, S.K., Johnson, J.S., Kavanagh, D.H., Perumal, T.M., Ruderfer, D.M., Oh, E.C., Topol, A., Shah, H.R.et al. (2016) Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci., 19, 1442–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gandal, M.J., Zhang, P., Hadjimichael, E., Walker, R.L., Chen, C., Liu, S., Won, H., vanBakel, H., Varghese, M., Wang, Y.et al. (2018) Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science, 362, eaat8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rajarajan, P., Borrman, T., Liao, W., Schrode, N., Flaherty, E., Casiño, C., Powell, S., Yashaswini, C., LaMarca, E.A., Kassim, B.et al. (2018) Neuron-specific signatures in the chromosomal connectome associated with schizophrenia risk. Science, 362, eaat4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Giambartolomei, C., Vukcevic, D., Schadt, E.E., Franke, L., Hingorani, A.D., Wallace, C. and Plagnol, V. (2014) Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet., 10, e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Team, R.C. (2017) R: A Language and Environment for Statistical Computing. R Core Team. [Google Scholar]
- 55. Wang, C., Qin, N., Zhu, M., Chen, M., Xie, K., Cheng, Y., Dai, J., Liu, J., Xia, Y., Ma, H.et al. (2017) Metabolome-wide association study identified the association between a circulating polyunsaturated fatty acids variant rs174548 and lung cancer. Carcinogenesis, 38, 1147–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shi, X.-Y., Wang, G., Li, T., Li, Z., Leo, P., Liu, Z., Wu, G., Zhu, H., Zhang, Y., Li, D.et al. (2020) Identification of susceptibility variants to benign childhood epilepsy with centro-temporal spikes (BECTS) in Chinese Han population. EBioMedicine, 57, 102840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yurko, R., Roeder, K., Devlin, B. and G’Sell, M. (2020) H-MAGMA, inheriting a shaky statistical foundation, yields excess false positives. BioRxiv. doi: 10.1101/2020.08.20.260224. [DOI] [PubMed] [Google Scholar]
- 58. Wolfe, D., Dudek, S., Ritchie, M.D. and Pendergrass, S.A. (2013) Visualizing genomic information across chromosomes with PhenoGram. Bio Data Min, 6, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Griffith, M., Griffith, O.L., Coffman, A.C., Weible, J.V., McMichael, J.F., Spies, N.C., Koval, J., Das, I., Callaway, M.B., Eldred, J.M.et al. (2013) DGIdb: mining the druggable genome. Nat. Methods, 10, 1209–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Napolitano, F., Sirci, F., Carrella, D. and diBernardo, D. (2016) Drug-set enrichment analysis: a novel tool to investigate drug mode of action. Bioinformatics, 32, 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data used in this study are publicly available, hosted at the respective consortia portal. Chinese and EUR population telomere length GWAS (https://www.nature.com/articles/s41467-019-10443-2#data-availability); Brain Region Phenotypes GWAS (https://github.com/BIG-S2/GWAS); eQTL and mQTL data are available from SMR (https://cnsgenomics.com/software/smr/#Overview); H-MAGMA (updated 15 September 2020: https://github.com/thewonlab/H-MAGMA). The results generated in this study are provided in the main text and supplementary files.


