Abstract
Purpose
In this paper, we utilized deep learning methods to screen the positive COVID-19 cases in chest CT. Our primary goal is to supply rapid and precise assistance for disease surveillance on the medical imaging aspect.
Materials and methods
Basing on deep learning, we combined semantic segmentation and object detection methods to study the lesion performance of COVID-19. We put forward a novel end-to-end model which takes advantage of the Spatio-temporal features. Furthermore, a segmentation model attached with a fully connected CRF was designed for a more effective ROI input.
Results
Our method showed a better performance across different metrics against the comparison models. Moreover, our strategy highlighted strong robustness for the processed augmented testing samples.
Conclusion
The comprehensive fusion of Spatio-temporal correlations can exploit more valuable features for locating target regions, and this mechanism is friendly to detect tiny lesions. Although it remains in discrete form, the feature extracting in temporal dimension improves the precision of final prediction.
Keywords: COVID-19, Deep Learning, Object Detection, Semantic Segmentation, Chest CT
1. Introduction
Feedbacks from various regions demonstrated that even RT-PCR, the gold standard diagnostic test of COVID-19, has a certain probability of yielding false-negative results[1], [2], [3]. Profiting from deep learning detecting methods, a more productive assessment of COVID-19 cases can be established in time as expected.
One of the intractable challenges of computer-aided diagnosis is to achieve precise segmentation of target lesion after comprehensive analysis. Furthermore, deep learning methods have been widely applied in manifold domains of medical imaging, such as gliomas segmentation [4], lung nodules detection [5], [6], multi-organ segmentation of abdomen [7], [8], pneumonia classification [9], etc. Accordingly, screening the COVID-19 lesion by deep learning algorithms has dramatically blossomed during this outbreak [10]. The study of imaging features contained in COVID-19 lesion has made steady progress profiting from the constant expansion of data sets [11], [12] and the continuous optimization of segmenting methods [13].
In some cases, segmenting methods give out predictions located outside the target regions unreasonably, such as recognizing the artifacts as lesions. Thus, finding the corresponding areas in the whole detecting field has become a preliminary work in lesion segmentation or classification tasks. Some semantic segmentation methods have been exploited in various organ segmenting tasks [14], [15]. To eliminate noise caused by areas outside the body, Jin et al. [16] and li et al. [17], they first extracted the lung regions using UNet at first; After the ROI was acquired, they classified the particular COVID-19 lesions. Their testing showed that their achievements satisfied the requirements of the clinic.
From the constantly updated cases of positive COVID-19 patients on the website of the Italian society of medical and interventional radiology and the Radiopaedia, it is not complex to discover that many tiny lesions are wildly distributed. Suppose the segmenting operations are monotonously completed on the whole image. The positive pixels will be highly unbalanced against the negative pixels in many samples that contain tiny lesions, leading to some calculation mistakes over the tiny lesions. Solutions from small object detection and the corresponding loss functions[18] can subtly optimize the tiny lesion issues. However, in many COVID-19 studies like [19], [20], [21], although they achieved encouraging experimental results, the specific assessment about tiny lesion detection was rarely mentioned explicitly.
According to the spatial continuity of CT images and the temporal performances of COVID-19 lesion [22], analyzing features over 2D slices solely, without any spatial context and temporal hint, may lose essential information over multiple levels [23]. Many studies advocated 3D convolutional operations to handle the vulnerabilities of spatial features extracting from 2D methods. Nevertheless, troubles such as the probable lack of coherence in a specific spatial dimension and the computational complexity should be concerned. Chen et al. [19] extracted lesion regions by adopting a 2D CNN method. They added a logic linking the evaluation of consecutive images to divide predictions into four quadrants. The positive results would only give out when three consecutive images were predicted as positive in the same quadrant. Feedbacks from their internal testing set showed comparable performance to an expert radiologist. Many particular deep learning studies have not taken the temporal features of COVID-19 lesion abundantly, stemming from the absence of data annotation.
In this study, we synthetically considered solutions in multiple stages for the positive COVID-19 screening, such as the ROI (region of interest) selection, the fusion of Spatio-temporal correlations, and tiny lesions detection. For obtaining a more effective detecting area, we first designed a semantic segmentation model featuring CRF to separate lung regions. All data sets [24], [11], [12] then were processed into ROI-only by this model. To evolve the precision of lesion segmenting, we proposed a single-instance segmentation method to predicting lesions in region proposals (intermediate results of Mask R-CNN [25]), which is friendly to tiny lesions. Notably, a Spatio-temporal resolver was designed for processing the spatial correlations and the discrete temporal information (given by our radiologists). Even though our method relied on fully 2D architectures, the crucial 3D contextual information of input images was considered via low computational complexity. We fused temporal and spatial features in a two-stage object detecting pipeline, which reinforced the performance of tiny lesion detecting. The experimental results showed that our method performed a high accuracy and stability for screening the COVID-19 infections.
2. Material and methods
Our study for COVID-19 lesion detection comprises two modules: target region selection and single-instance segmentation. Both temporal and spatial dependences were considered in the proposed method to perform a high-quality prediction. We adopted two kinds of datasets [11], [12], [24], [27] annotated respectively with lung contours only and lesion contours with bounding rectangles to train components from each module mentioned above. The lung region acquiring was explained explanatorily by a semantic segmenting method based on deep learning. To capture and analyze the texture continuity of lung volume, we utilized a reconstructed sagittal view of CT imaging and the corresponding manual mask for the extra 2D segmentation, which supplied the spatial semantic information [26].
By contrast, we rejected the same strategy (reconstructing a sagittal view) on the following lesion detection task due to the presence of abundant isolated lesion slices picked from CT imaging in our dataset. We focused on spatial consecutiveness and discrete temporal features from CT images in our instance segmentation study. Basing on the Mask R-CNN frame, we reconstructed specific generators of the shared feature maps by blending spatiotemporal-wise processing, which provided adequate information from multi-dimensional context. Furthermore, by the advantages of region proposals, the conventional intermedium of Mask R-CNN, the stubborn issues of tiny lesion detection has been eased.
2.1. Segmentation of lung regions
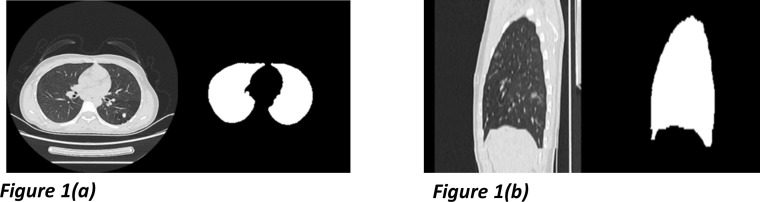
The first step of lesion detection from COVID-19 CT imaging is to segment the lung areas. We labeled lung contours scan-by-scan on CT images of 50 patients with pulmonary lesions and 30 other healthy people [27]. All Chest CT imaging and the corresponding manual masks were additionally reconstructed as sagittal views for the spatial continuity feature learning. The labeled samples and the sagittal reconstructions are shown in Fig. 1 , and each image scan paired with its mask was prepared for the corresponding segmentation model training. According to the comprehensive data distribution analysis, we performed multiple data enhancements in different window-level and window-width (around (−500, 1600), window-level at first) to improve the segmentating stability. In this part, due to the relevant features of imaging data, our presented method optimizes the U-net [28] architecture.
Fig. 1.
Chest CT slices and the corresponding lung region masks from the training samples. The axial imaging showed in Fig. 1 (a) was exploited to train an identically structured model collaborated with the sagittal one shown in Fig. 1(b).
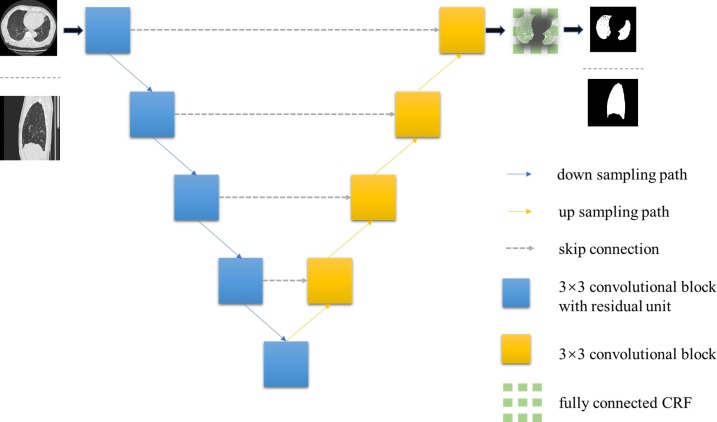
The segmentation architecture is exhibited in Fig. 2 . We trained two parallel homology structures to segment lung regions from both axial and sagittal views. The residual block [29] was added to each downsampling path to reduce the semantic information loss. We replaced the Max-pooling units in the first two downsampling paths with 2-stride convolutional units (filter size as 2 × 2) for feature contracting. We kept the Max-pooling units for the rest downsampling components to maintain feature extraction. The Group Normalization [30] unit was assembled in each down sampling and up sampling path, computing differently to the Batch Normalization. This operation has no concern with the original batch size, and it keeps a stable accuracy in a wide range of batch sizes. The ReLU activator was added following the Group Normalization. Correspondingly, we took a series of upsampling convolutional operations in upsampling paths. To reduce computational complexity, we directly concatenate the features from two paths respectively with skip connection.
Fig. 2.
The architecture of the lung area segmenting model. This end-to-end structure ensures an efficient region of interest (ROI) exporting, which improved subsequential lesion detection efficiency.
The initial segmentation of the lung region was expressed as the probability of each pixel in the original size. However, even though the feature maps can be restored to the original size, it still causes the information loss simultaneously, leading to a fuzzy segmenting boundary. We adopted the fully connected CRF [31] for post-processing to improve final segmentation performance. The method can process the semantic segmentation from the previous block by interfusing relationships between all pixels in original imaging. Furthermore, it corrects the segmented regions by optimizing the rough and uncertain marks. The fully connected CRF conforms to the Gibbs distribution, formulated as equation (1), where x is the observed value, and is the energy function, shown as equation (2). The unary potential is expressed as , which means the category attribution probability of observed pixel. The pairwise potential is displayed as , and is concretely depicted as equation 3. The acts like label compatibility, which restricts the conduction between pixels: only pixels with the same category label can be conducted each other. The represents the weight parameters, and the feature function is shown as equation (4).
| (1) |
| (2) |
| (3) |
| (4) |
In equation (4), the similarity of pixels is expressed as two different feature kernels. Here, we use to represent the weight of the first appearance kernel, and means the weight of the second smoothness kernel. When the energy becomes smaller, the prediction of each pixel will be more precise. We obtained the final post-processing segmentation by iterating to minimize the energy function.
2.2. Lesion detection–single-instance segmentation
After ROI acquiring, we generated different lung volume parts with three sequential axial slices, which subjoined an extra manual feature in discrete digital label form to identify disparate symptom periods. Each lung volume fragment was transported into the detection model as a three-channel input, which contained three continuous spatial slices. Besides, we collected a certain number of isolated lesion slices from different patients. We put them in a three-item sequence to deal with these data, including only one non-zero item located at the middle index.
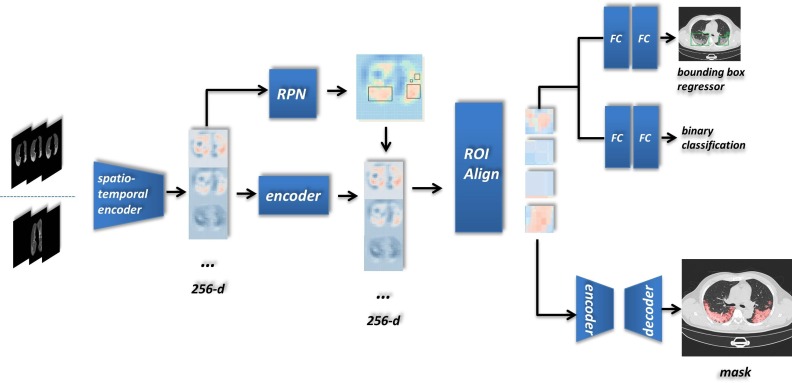
Our detecting architecture is shown in Fig. 3 . We added our proposed components to the Mask R-CNN pattern and achieved a novel end-to-end single-instance segmentation method. Since the class-unicity of the detecting target and the leveraging of the region proposal, we call this case as single-instance segmentation. As shown in Fig. 3, we put one sequence into the model and finally acquired both bounding boxes and instance segmentation of lesion.
Fig. 3.
An example workflow explaining for our detecting architecture: the input array, a three-channel image composed with adjacent CT slices, is transported to the Spatio-temporal encoder in two optional homogenous forms. The key item of the input sequence is immutably located in the middle, and others aim to comprehend the spatial context if they existed.
After the comprehensive feature exploring over multiple dimensions, the output of the Spatio-temporal encoder was formed as a 256-channel feature array. We ulteriorly added an encoder, two serial residual convolutional blocks that keep the input size, for integrating the semantic information based on relationships of the feature. Combined with the RPN region proposals, ROIs in various shapes were delivered into the ROI Align. Each input was rebuilt (along with feature extraction) as the same shape in virtue of the bilinear interpolation. The output then was leveraged by two subsequent branches–the bounding box regression and the mask generator. In the mask branch of network head architecture, we extended it as a tiny U-net, concatenating features from a more miniature encoder and decoder (three times for downsampling and upsampling). All convolutional blocks from tiny U-net contained a four-time convolutional operations (2D Convolution + BatchNormalization + ReLU) which adopted a residual connection.
2.3. Details of spatio-temporal module
Due to the spatial continuity of CT imaging, there is a specific correlation between the spatially adjacent scans. In general, object detection and instance segmentation focus more on 2D feature association. Obtaining feature maps only in a single slice will lose important spatial context, resulting in a loss of prediction accuracy.
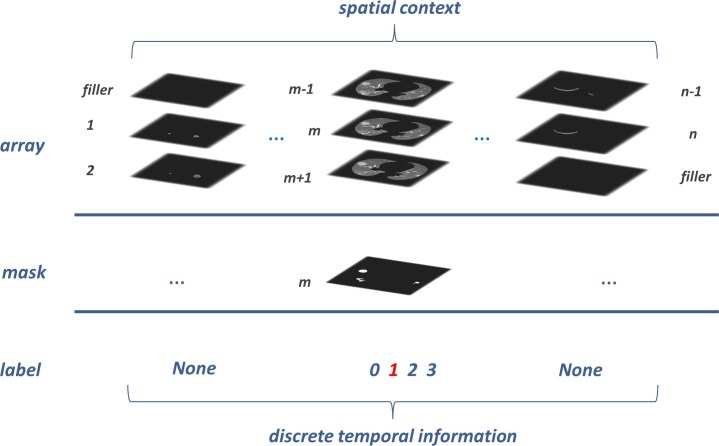
This section first generated the image input array for the Spatio-temporal framework. As shown in Fig. 4 , the whole lung volume has been split into subsequences from top to bottom, with a beginning zero-filler for the first subsequence and an end zero-filler for the last one. We took three adjacent lung slices as an entity, and the middle one was assigned as the key item, which was bound its mask to the corresponding entity uniquely. Each slice had only one opportunity to become the key item for its two neighbors. And for the above reasons, we supplied a zero-filler for the first and last slice when they become the key item.
Fig. 4.
Each prepared input array sequence has two annotations at most: lesion mask (indispensable) and the disparate symptom period (dispensable). From 0 to 3, the discrete labels indicate four lesion statuses: healthy, early stage, advanced stage, and severe stage. Our radiologists gave the label to every isolated lesion slice mentioned in section 2.1. But for data exhibited in this figure, we only gave the label to the lesion entities.
According to the CT imaging diagnosis guidelines of COVID-19 [32], symptom signs are divided into three different periods: early stage, advanced stage, and severe stage. We labeled each lesion entity in {1, 2, 3} form to cover the three steps separately and take 0 for the healthy people. We rejected giving labels for entities that do not possess any lesion in a positive COVID-19 CT imaging to keep a rigorous ground truth. The input array and its annotations were processed as an array tuple for the follow-up Spatio-temporal framework.
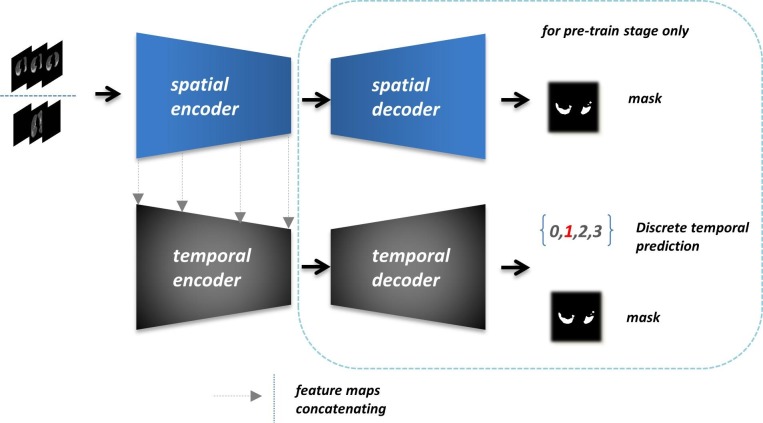
We exploited the temporal information under the supervision of discrete labels mentioned above. Features from different symptom periods showed meaningful diversity, which refined the detail extracting of lesion performances in corresponding stages. Notably, in the early stage of COVID-19, the infection areas sometimes show a highly semblable appearance as the healthy areas, leading to a misdetection or a false positive prediction. Benefitting from the discrete temporal annotations, we optimized the handling mechanism of distinguishing the healthy cases from the early-stage or other inconspicuous infections. The temporal encoder extracted more appropriate features by the multi-class supervision and fused the spatial features adequately from the spatial encoder. The corresponding temporal features of different stages highlight the implicit differences between negative and positive samples.
The Spatio-temporal encoder that appeared in Fig. 3 is a part of our semantic segmenting frame, which is pre-trained for fusing spatial features and discrete temporal features. As illustrated in Fig. 5 , the final output in this module contains temporal classification and lesion segmentation. We first trained the spatial resolver with the mask of key items as ground truth. To intensify the spatial fitting, we only processed all data from [12] (completed CT imaging sequences) as the input array. The 2D convolutional blocks will capture the most weighted spatial correlations in this meaningful truncated context through the three consecutive slices. Then we froze all spatial resolver parameters and closed the rest architecture to train the temporal resolver. The output features from each spatial level were sent to the corresponding temporal encoder block. We took both discrete temporal labels and lesion masks as the supervision of temporal resolver for further integrating the spatial and temporal information. This module leveraged two homogeneous encoder-decoder architectures with a pair of four times down-sampling and up-sampling paths.
Fig. 5.
Overview of the pipeline and essential operations of the Spatio-temporal resolver. All further details like convolutional blocks and feature fusions between encoder and decoder are simplified to display in a simple and straightforward pattern.
When docking with the follow-up network, we cut off all decoder parts and the output layers to provide the comprehensive preconditions of the detecting and segmenting task only for feature acquisition. The output feature maps (256-dimension) were 16 times down-sampled comparing with the original input slices. Moreover, the discrete periods of COVID-19 lesion are just defined for known instances generally. In fact, time granularity acts like a continuous factor, which possesses plenty of implicit characteristics against the current discrete label. We chose to train the spatial resolver separately for the ablation validating between spatial and temporal factors. And on the other aspect, the spatial resolver can be solely deployed for exceptional cases to ensure the primary validity.
2.4. Implementation
We used ResNet50 [29] as the backbone architecture of each encoder, and in the head network, the segmenting branch only adopted three downsampling blocks. Each slice of the input array was processed in 512 × 512 size. In the pre-trained stage, the Spatial-temporal resolver was trained through both dice loss and cross-entropy loss. And then, we only integrated the encoder to the detecting and segmenting pipeline shown in Fig. 3. The loss function of the whole pipeline includes three parts: cross-entropy loss for the class branch, smooth L1 for the bounding box regression, and binary cross-entropy for the mask segmenting brach. Our model was trained for 90 epochs in NVIDIA GeForce RTX 2080Ti with a batch size of 4.
3. Results
In this section, we chiefly reported the ablation experiment results between spatial and temporal factors. Furthermore, we evaluated different metrics between our models and other outstanding methods.
Our original datasets are all from [11], [12], [24], the Italian society of medical and interventional radiology, and the Radiopaedia. We first analyzed all lesion regions in each positive slice:15907 regions from the training set and 7953 from the testing set (both with the corresponding ground truth: bounding boxes, masks, and labels). According to the suggestion of our radiologists, we defined the small regions, which occupy the proportion below 0.059% (the threshold is around 154 pixels) from the whole individual slice (size of 512 × 512), as the tiny lesions to study separately. The detailed statistics are shown in Table 1 .
Table 1.
Statistics about lesion size. Areas are measured in pixels, an image size of 512 × 512.
| Total Regions | Mean | Median | Tiny Lesions (%) | |
|---|---|---|---|---|
| Training set | 15,907 | 954.11 | 230 | 41.28 |
| Testing set | 7953 | 1013.32 | 179 | 43.06 |
In the ablation study, we attended to demonstrate the superiority of the Spatio-temporal resolver. Ulteriorly, the temporal resolver was detached to rebuild a spatial feature encoder for the model in sub-experiment. We took the Mask R-CNN (backbones: ResNet50 + FPN) and U-net (backbones: ResNet50) to compare the testing set.
We evaluated four metrics–the Dice Score [33], accuracy, sensitivity, and sensitivity of tiny lesions–over our models (spatial encoder only and Spatio-temporal encoder) and the comparison models. A valid prediction was confirmed in the experiment if the IOU [34] is above 50%. The accuracy is used for evaluating the segmentation per pixel (without background pixels). Besides, we took the minimum circumscribed rectangle of the U-net segmentation as the predicting bounding box. Table 2 shows the comparison results (Table 3 ).
Table 2.
The ablation analysis on the testing set. The sensitivity of tiny lesions is short for Tiny Sensitivity. The spatial-only model is marked as Ours-SP, the complete one as Ours.
| Mean Dice Score | Mean Accuracy | Sensitivity | Tiny Sensitivity | |
|---|---|---|---|---|
| Ours-SP | 0.9062 | 0.9783 | 0.9689 | 0.9413 |
| Ours | 0.9197 | 0.9839 | 0.9725 | 0.9442 |
| Mask R-CNN | 0.9024 | 0.9717 | 0.9590 | 0.9165 |
| U-net | 0.8801 | 0.9691 | 0.9246 | 0.8408 |
Table 3.
The detailed results of the augmentation experiment.
| Mean Dice Score | Mean Accuracy | Sensitivity | |
|---|---|---|---|
| Coronal view | 0.8907 | 0.9619 | 0.9420 |
| Multi-augmentation | 0.9011 | 0.9703 | 0.9691 |
As shown in Table 2, our two models perform better than Mask R-CNN and U-net across the four metrics. Modestly, the first three models from Table 2 possess a comparable level in instance segmentation tasks. The U-net achieves the segmenting on an integral image, and others take advantage of region proposals, described as the first ablation factor. We demonstrated that segmenting on the reasonable sub-regions is more effective than the integral image. For the second ablation factor–extracting the feature from the Spatio-temporal dimension, our models showed better feedback than Mask R-CNN, especially on the recall of tiny lesions. Finally, as to the third ablation factor–discrete temporal features exploiting, the spatial-only model showed a slightly less performance than the complete one.
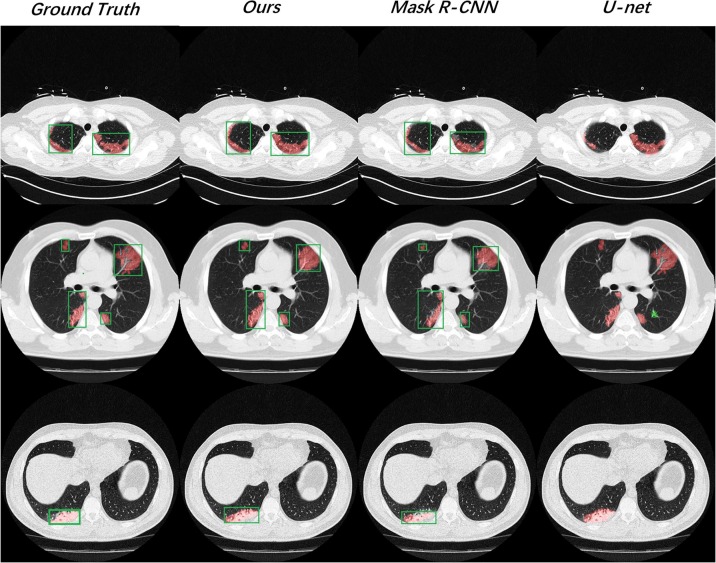
We exhibited three testing results from different patients randomly in Fig. 6 . On the whole, our model gave out better predictions that were closest to the ground truth in both bounding box regression and lesion segmenting. Besides, from the second testing case, Mask R-CNN showed some segmentation deviations. The U-net also worked out a false positive prediction (marked as the green region).
Fig. 6.
We exhibited three positive slices from different testing samples, including solitary lesion, multi-lesion, and tiny lesions.
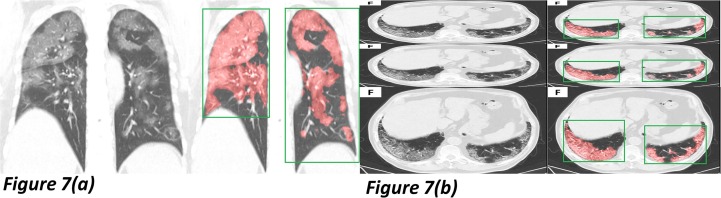
Our models kept strong robustness on the augmentation testing samples for an extension. We transposed the integral CT images of ten patients [12] into the coronal view. Furthermore, 200 positive slices were randomly selected from the testing set for multiple augmentations like squeezing the same or different slices into one integral image, a total of 300 augmented samples. We experiment on our complete model using these augmented samples. Notably, we never trained the model in such data forms, and the morphology of the lung changed a lot in all experiment samples.
As shown in Fig. 7 (a), the testing input is transformed to a coronal view. Both the segmentation and region location showed excellent accuracy. Fig. 7(b) changed the shape ratio and copied three times in one input. We prepare to study the reasons for this robustness and optimize the performance by various mechanisms.
Fig. 7.
Two samples were mixed with different augmentations for the robustness test. Fig. 7(a) exhibits a coronal view testing sample, and Fig. 7(b) showed a multi-augmentation one.
4. Discussion
4.1. Conclusions
In this paper, we proposed a single-instance segmentation method that performed a high accuracy and stability for screening the COVID-19 infections. Due to the exploration of spatial continuity and temporal characters of lesion imaging appearance, the presented Spatio-temporal resolver provides more effective feature maps for reasonable regions generating. And our ablation study showed this mechanism is gracious to detecting tiny lesions. The final estimation is optimized jointly by practical ROI inputs, feature analysis from Spatio-temporal dimension, and sub-regions segmenting.
4.2. Discussion
We achieved both targets locating and semantic segmentation on a two-stage object detection pipeline. Most relevant studies have shown signs of a rise by taking advantage of pure semantic segmentation, which calculates from the integral image. By leveraging different appropriate loss functions, multi-batch deviation sampling, and image augmentations, the matters brought by the unbalanced distribution of positive and negative samples can be alleviated. But taking back to the medical imaging only, COVID-19 lesions possess various performances from size, shape, location, etc., in each imaging slice. Other tissues on the target organ may have extreme similarities that cause certain false-positive estimations.
Furthermore, segmenting on the whole input field sometimes ignores the tiny lesions or recognizes other tiny tissues as a target. The tiny lesions that stay close to each other can be collected in smaller subareas through the two-stage detection pipeline. Our method determines the reasonable subareas for all target lesions, and then the segmenting is completed on each region proposals. These operations maintain the whole spatial context, embodied in the first detection stage, and improve the segmenting performance.
For mitigating the estimation matters caused by tiny lesions further, we processed the samples in various ways of augmentation. Unlike other small object detection tasks, all augmentations from our study need to adhere to medical facts scrupulously. For instance, in the small ball detection task, researchers can copy the target multiple times in proper locations to increase the positive samples and adjust the weights appropriately. However, in our study, if we copy the tiny lesions or other complex targets in the same way, many invalid non-objective samples will be out of thin air. In this case, data becomes untrustworthy, and the model will be misfitting in almost all probability. The fundamental reason is that lesion performances are strongly associated with peripheral tissues and symptom cycles. For COVID-19 CT imaging, the intelligent use of geometrical transformation and other security operations is critical. We implemented augmentations like translation, random scaling, slight rotation, merge individual slices, etc. These procedures improve the robustness and reduce the false positive at the first level.
At the second level, we focused on fully exploiting Spatio-temporal correlations, which contain valuable features. As input sequences, the adjacent CT slices supply the most appropriate context across each spatial dimension. It alleviates the generating of the false-positive via considering the continuous spatial features. The temporal encoder can fuse the Spatio-temporal features by fitting the lesion performances discretized by manual labels. According to the ablation study, this encoder of multi-level feature improved the segmenting precision and recall (especially for tiny lesions) effectively. Moreover, we prepared the healthy samples proportionally as one of the discrete statuses to mitigate the false positive. We aim to focus on self-supervised learning to refine higher fine-grained features in the temporal dimension for further study.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study has received funding by the National Key R&D Program of China (2018YFC1315604, 2018YFC0116901), and the Program for JLU Science and Technology Innovative Research Team (2017TD-27).
References
- 1.H.S. Maghdid, A.T. Asaad, K.Z. Ghafoor, A.S. Sadiq, M.K. Khan, Diagnosing COVID-19 pneumonia from X-ray and CT images using deep learning and transfer learning algorithms, 2020. WHO Director- General's remarks at the media briefing on 2019-nCoV on Feb 11, 2020, (Feb 11, 2020), [Online]. Available: https://www.who.int/dg/speeches/detail/who-director-general-s-remarks-at-the-media-briefing-on-2019-ncov-on-11 -February-2020.
- 2.Huang C., Wang Y., Li X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223) doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Q., Guan X., Wu P., et al. Early transmission dynamics in wuhan, china, of novel coronavirus- infe cte d pneumonia. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001316. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.M. Noori, A. Bahri and K. Mohammadi, Attention-guided version of 2D UNet for Automatic brain tumor segmentation, 2019 9th International Conference on Computer and Knowledge Engineering (ICCKE), Mashhad, Iran, 2019, pp. 269-275, doi: 10.1109/ICCKE48569.2019.8964956.
- 5.Jiang H., Ma H.e., Qian W., Gao M., Li Y. An automatic detection system of lung nodule based on multigroup patch-based deep learning network. IEEE J. Biomed. Health Inf. 2018;22(4):1227–1237. doi: 10.1109/JBHI.2017.2725903. [DOI] [PubMed] [Google Scholar]
- 6.Singh S.P., Wang L., Gupta S., Goli H., Padmanabhan P., Gulyás B. 3D deep learning on medical images: a review. Sensors. 2020;20:5097. doi: 10.3390/s20185097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Y. Liu, J. Zhou, S. Chen, L. Liu, Muscle segmentation of L3 slice in abdomen CT images based on fully convolutional networks, 2019 Ninth International Conference on Image Processing Theory, Tools and Applications (IPTA), Istanbul, Turkey, 2019, pp. 1-5, doi: 10.1109/IPTA.2019.8936106.
- 8.X. Chen, R. Zhang, P. Yan, Feature fusion encoder decoder network for automatic liver lesion segmentation, 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 2019, pp. 430-433, doi: 10.1109/ISBI.2019.8759555.
- 9.J. Garstka, M. Strzelecki, Pneumonia detection in X-ray chest images based on convolutional neural networks and data augmentation methods, 2020 Signal Processing: Algorithms, Architectures, Arrangements, and Applications (SPA), Poznan, Poland, 2020, pp. 18-23, doi: 10.23919/SPA50552.2020.9241305.
- 10.D.-P. Fan et al., Inf-Net: Automatic COVID-19 Lung Infection Segmentation From CT Images, in IEEE Transactions on Medical Imaging, vol. 39, no. 8, pp. 2626-2637, Aug. 2020, doi: 10.1109/TMI.2020.2996645. [DOI] [PubMed]
- 11.Yang X., He X., Zhao J., Zhang Y., Zhang S., Xie P. COVID-CT-dataset: a CT scan dataset about COVID-19. ArXiv e-prints:arXiv. 2020;2003:13865. [Google Scholar]
- 12.J.P. Cohen, P. Morrison, L. Dao, K. Roth, T.Q. Duong, M. Ghassemi (2020) Covid-19 image data collection: Prospective predictions are the future. arXiv preprint arXiv:200611988.
- 13.D. Dong, Z. Tang, S. Wang et al (2020) The role of imaging in the detection and management of COVID-19: a review. IEEE reviews in biomedical engineering. [DOI] [PubMed]
- 14.Nitta J., Nakao M., Imanishi K., Matsuda T. 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada. 2020. Deep learning based lung region segmentation with data preprocessing by generative adversarial nets; pp. 1278–1281. doi: 10.1109/EMBC44109.2020.9176214. [DOI] [PubMed] [Google Scholar]
- 15.N. Goceri, E. Goceri, A neural network based kidney segmentation from MR images, 2015 IEEE 14th International Conference on Machine Learning and Applications (ICMLA), Miami, FL, 2015, pp. 1195-1198, doi: 10.1109/ICMLA.2015.229.
- 16.Boo W., Shuo J., Qingsen Y., et al. AI-assisted CT imaging analysis for COVID-19 screening: building and deploying a medical AI system. Appl. Soft Comput. 2021;98:106897. doi: 10.1016/j.asoc.2020.106897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L., Qin L., Xu Z., et al. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology. 2020;296(2):E65–E71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.K. Doi, A. Iwasaki. The effect of focal loss in semantic segmentation of high resolution aerial image, IGARSS 2018 - 2018 IEEE International Geoscience and Remote Sensing Symposium, Valencia, 2018, pp. 6919-6922, doi: 10.1109/IGARSS.2018.8519409.
- 19.Chen J., Wu L., Zhang J., et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography. Sci. Rep. 2020;10:19196. doi: 10.1038/s41598-020-76282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang M., He B., Li L., et al. CT radiomics can help screen the Coronavirus disease 2019 (COVID-19): a preliminary study. Sci. China Inf. Sci. 2020;63(7):172103. doi: 10.1007/s11432-020-2849-3. [DOI] [Google Scholar]
- 21.Y. Song et al., Deep learning Enables Accurate Diagnosis of Novel Coronavirus (COVID-19) with CT images, in IEEE/ACM Transactions on Computational Biology and Bioinformatics, doi: 10.1109/TCBB.2021.3065361. [DOI] [PMC free article] [PubMed]
- 22.Z. Wu, J.M. McGoogan, Characteristics of and important lessons from the coronavirus disease 2019 (covid-19) outbreak in china: summary of a report of 72 314 cases from the chinese center for disease control and prevention, Jama, 2020. [DOI] [PubMed]
- 23.X. Wang et al., A Weakly-Supervised Framework for COVID-19 Classification and Lesion Localization From Chest CT, in IEEE Transactions on Medical Imaging, vol. 39, no. 8, pp. 2615-2625, Aug. 2020, doi: 10.1109/TMI.2020.2995965. [DOI] [PubMed]
- 24.M. de la Iglesia Vaya, J. Saborit, J. Montell, et al. BIMCV COVID-19: a large annotated dataset of RX and CT images from COVID-19 patients. arXiv(2020):2006.01174.
- 25.K. He, G. Gkioxari, P. Dollar, R. Girshick, “Mask R-CNN” in IEEE Transactions on Pattern Analysis & Machine Intelligence, vol. 42, no. 02, pp. 386-397, 2020. doi: 10.1109/TPAMI.2018.2844175. [DOI] [PubMed]
- 26.Noori, Mehrdad, Bahri, Ali, Mohammadi, Karim. (2019). Attention-Guided Version of 2D UNet for Automatic Brain Tumor Segmentation. 269-275. 10.1109/ICCKE48569.2019.8964956.
- 27.A.Setio, A.Traverso, T.D. Bel, et, al. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: The LUNA16 challenge, Med. Image Anal., 42, 2017,Pages 1-13, https://doi.org/10.1016/j.media.2017.06.015. [DOI] [PubMed]
- 28.Ronneberger O., Fischer P., Brox T. MICCAI 2015. Lecture Notes in Computer Science. Springer; Cham: 2015. U-Net: Convolutional networks for biomedical image segmentation. medical image computing and computer-assisted intervention – MICCAI 2015. https://doi.org/10.1007/978-3-319-24574-4_28. [Google Scholar]
- 29.He, Kaiming, Zhang, Xiangyu, Ren, Shaoqing, Sun, Jian. (2016). Deep Residual Learning for Image Recognition. 770-778. 10.1109/CVPR.2016.90.
- 30.Wu Y., He K. Group normalization. Int. J. Comput. Vis. 2020;128(3):742–755. doi: 10.1007/s11263-019-01198-w. [DOI] [Google Scholar]
- 31.Krähenbühl, Philipp, Koltun, Vladlen. (2012). Efficient Inference in Fully Connected CRFs with Gaussian Edge NIPS 2011; Advances in Neural Information Processing Systems 24 (2011) https://arxiv.org/abs/1210.5644.
- 32.Diagnosis and treatment protocol for COVID-19 (trial version 8). [Online] Available: http://en.nhc.gov.cn/2020-09/07/c_81565.htm.
- 33.A.W. Setiawan, Image Segmentation Metrics in Skin Lesion: Accuracy, Sensitivity, Specificity, Dice Coefficient, Jaccard Index, and Matthews Correlation Coefficient, 2020 International Conference on Computer Engineering, Network, and Intelligent Multimedia (CENIM), Surabaya, 2020, pp. 97-102, doi: 10.1109/CENIM51130.2020.9297970.
- 34.D. Zhou et al., IoU Loss for 2D/3D Object Detection, 2019 International Conference on 3D Vision (3DV), Québec City, QC, Canada, 2019, pp. 85-94, doi: 10.1109/3DV.2019.00019.