Abstract
Higher-order structure governs function for many RNAs. However, discerning this structure for large RNA molecules in solution is an unresolved challenge. Here, we present SHAPE-JuMP (selective 2’-hydroxyl acylation analyzed by primer extension and juxtaposed merged pairs) to interrogate through-space RNA tertiary interactions. A bifunctional small molecule is used to chemically link proximal nucleotides in an RNA structure. The RNA crosslink site is then encoded into complementary DNA (cDNA) in a single, direct step using an engineered reverse transcriptase that “jumps” across crosslinked nucleotides. The resulting cDNAs contain a deletion relative to the native RNA sequence, detectable by sequencing, that indicates the sites of crosslinked nucleotides. SHAPE-JuMP measures RNA tertiary structure proximity concisely across large RNA molecules at nanometer resolution. SHAPE-JuMP is especially effective at measuring interactions at in multi-helix junctions and loop-to-helix packing, enables modeling of the global fold for RNAs up to several hundred nucleotides in length, facilitates ranking of structural models by consistency with through-space restraints, and is poised to enable solution-phase structural interrogation and modeling of complex RNAs.
Introduction
RNA plays key biological roles as ribozymes, translation regulators, scaffolding centers, and ligand binders. These fundamental regulatory functions often occur where an RNA folds into complex secondary and tertiary structures1–3. Discovering and defining these structures provides insights into how RNA molecules function. Chemical probing is a powerful approach for mapping RNA structure at large scales and in a sufficiently unbiased way as to be useful as a function-discovery tool4. Strategies for chemical probing of RNA secondary structure, especially using selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP)5,6, DMS-MaP7,8, and single-molecule correlated chemical probing (RING) methods7,9,10, are transforming understanding of RNA structure-function interrelationships. The broad applicability and accuracy of these methods reflects, in large part, strategies in which the sites of chemical modification in RNA are read out in a complementary DNA (cDNA) in a single direct step. A key insight from prior work emphasizes that methods that detect chemical modifications in RNA directly are superior to those that require multiple intervening biochemical steps11–13.
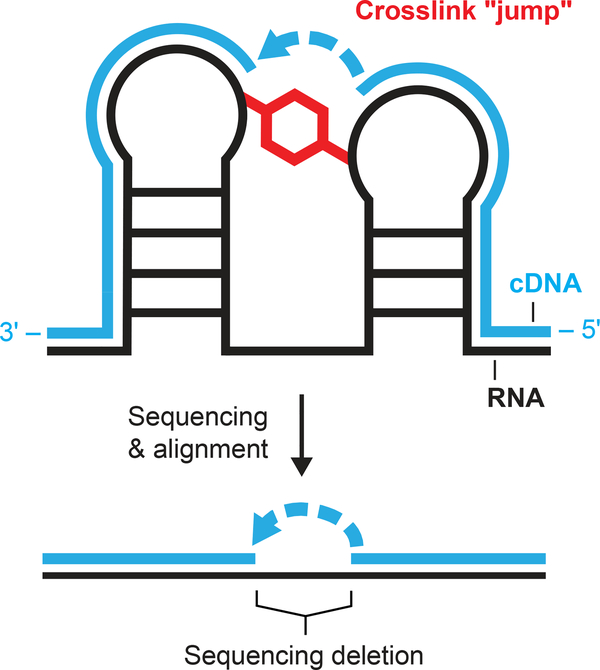
Here, we introduce a concise strategy to map tertiary and higher-order RNA-RNA interactions across large RNA molecules: SHAPE-JuMP, selective 2′-hydroxyl acylation analyzed by primer extension and juxtaposed merged pairs. SHAPE-JuMP monitors tertiary structure via a bi-reactive SHAPE chemical crosslinking reagent that reacts preferentially with the 2′-hydroxyl groups of two nucleotides in close proximity (Fig. 1). An engineered reverse transcriptase (RT) – effectively, a reverse transkiptase – then “jumps” across the crosslink to permanently record the crosslink site as a deletion in the resulting cDNA strand. The ability to traverse chemical crosslinks required an engineered enzyme, RT-C814, which was optimized from a reverse transcriptase previously shown to be permissive toward alternative nucleic acid backbones15. Locations of crosslink-induced deletions are identified by aligning sequencing reads to a reference sequence. We show these internucleotide crosslinks report long-range through-space interactions in complex RNA structures, useful for higher-order structure discovery and modeling.
Figure 1: SHAPE-JuMP overview.
RNA is treated with a bivalent SHAPE crosslinker (red), which covalently links proximal nucleotides. Reverse transcription using an engineered jumping, or crosslink-traversing, polymerase records the crosslinked site as a deletion in the cDNA (blue). The cDNA is sequenced and aligned to the reference RNA sequence to identify deletion sites and, thereby, crosslinked nucleotides.
Results
TBIA crosslinks RNA
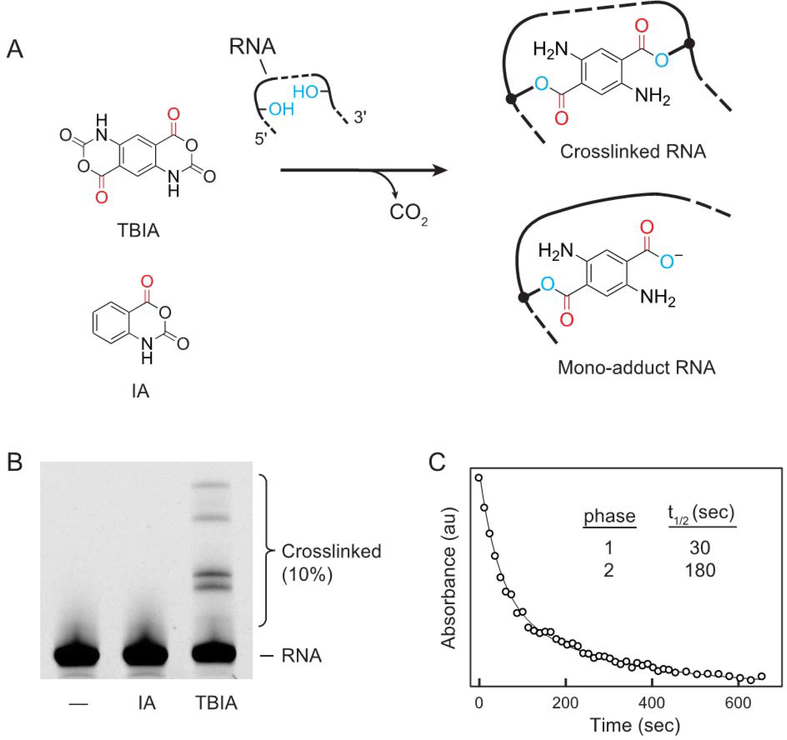
We created a new bifunctional SHAPE reagent, trans bis-isatoic anhydride (TBIA), to crosslink RNA nucleotides (Fig. 2A). TBIA has two electrophilic isatoic anhydride moieties, each of which can react with the nucleotide 2’-OH group. When TBIA reacts with one nucleotide, its second anhydride moiety can react with a second, proximal, nucleotide, forming a covalent crosslink. Crosslinks can occur between nucleotides both adjacent and distant in the primary RNA sequence as long as they are in close three-dimensional physical proximity. SHAPE reagents, like TBIA, react with all four nucleotides5. TBIA will also form mono-adducts when one anhydride reacts with a nucleotide, but the second anhydride is hydrolyzed by water. To distinguish mono-adduct formation from true crosslinks, we compared SHAPE-JuMP signals due to TBIA with mono-adducts created using isatoic anhydride (IA) (Fig. 2A), a molecule with similar structure to TBIA but only one reactive group.
Figure 2: TBIA mechanism and characterization.
(A) Reaction of TBIA with RNA yields both crosslinks and mono-adducts. IA strictly forms mono-adducts. (B) Reaction of the RNase P RNA with no reagent (–), IA (mono-adduct), or TBIA (crosslinking reagent), visualized by denaturing electrophoresis. (C) Hydrolysis of TBIA in buffer. Reaction was monitored by UV absorbance at 296 nm; absorbance as a function of time was fit to a double exponential rate equation.
To examine the ability of TBIA to crosslink RNA, we folded RNA transcripts with the sequence of the Bacillus stearothermophilus RNase P catalytic domain16 or the A756G mutant of the Varkud satellite (VS) ribozyme17 and treated each with TBIA. Product formation was visualized by denaturing gel electrophoresis as a lower mobility band absent in no-reagent and IA-treated controls (Fig. 2B, S1). TBIA induced efficient crosslinking (5–10%) in each RNA and crosslinking was independent of RNA concentration, indicating that crosslinks reflected intramolecular interactions (Fig. S1). To establish optimal reaction times for crosslinking RNA with TBIA, we followed TBIA hydrolysis by monitoring the change in UV absorbance at 296 nm (Fig. 2C). The time decay fit a double exponential function, consistent with two-site reactivity by TBIA, and half-lives (t1/2) of 30 and 180 sec. We implemented crosslinking using a reaction time of 15 min, corresponding to 5 half-lives of the slower reaction.
Identification of an efficient crosslink-traversing RT
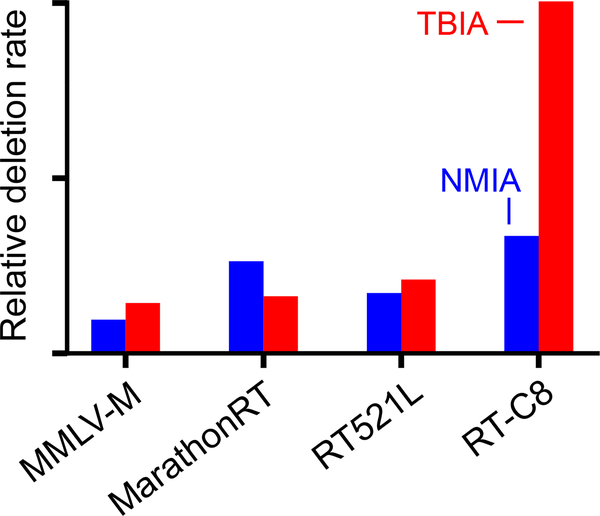
We tested diverse RT enzymes for their ability to use a crosslinked RNA as template. These enzymes included a processive mutant of Moloney murine leukemia virus RT (similar to SuperScript II), Marathon RT18, and multiple enzymes created by directed evolution14,15. We treated the RNase P catalytic domain RNA with either the TBIA crosslinker or N-methyl IA (NMIA, which creates mono-adducts), generated and sequenced cDNA transcripts, aligned these sequences to the parent sequence, and quantified the deletions that formed in the TBIA and IA experiments for each polymerase. We found many enzymes had modest apparent jumping ability, but the most useful enzyme was RT-C814, which consistently showed 3-fold higher absolute rates of deletion detection for TBIA over the NMIA background (Fig. 3). The observed deletion detection rate for TBIA over an IA background is 1–3%, while 5–10% of RNAs form low mobility bands as visualized physically by denaturing gel electrophoresis (Fig. 2B, S1); crosslink detection by RT-C8 is thus roughly 30% efficient. RT-C8 was used for all subsequent experiments.
Figure 3: Identification of reverse transcriptase enzymes with the ability to jump RNA crosslinks.
Comparison of crosslink detection in the RNase P RNA by different RT enzymes. Deletion rates for RNAs treated with TBIA and mono adduct-forming NMIA reagents are red and blue, respectively.
SHAPE-JuMP of RNAs with complex structures
We evaluated the ability of SHAPE-JuMP to detect nucleotides in close proximity by analyzing RNAs with complex structures: the Tetrahymena thermophila group I intron P546 domain (P546, 158 nts)19, the aforementioned VS ribozyme (186 nts)17 and RNase P catalytic domain (268 nts)16 RNAs, and the Oceanobacillus iheyensis group II intron20 (group II, 412 nts). We note that while the VS RNA construct forms a dimer at high concentrations17, it forms crosslinks in a concentration-independent manner over the 0.5–2 μM range, consistent with forming a monomer under our conditions (Fig. S1). These RNAs form diverse structures: P546 adopts a simple tertiary structure involving close packing of two sets of extended helices; the VS ribozyme has a distinct fold but does not form a compact tertiary structure with closely spaced helices, and has served as a target in the RNA-Puzzles structure prediction exercise21; and RNase P and the group II intron form complex structures, and are much larger than can be confidently modeled using current methods. These RNAs allowed us to examine SHAPE-JuMP across diverse structural motifs and degrees of complexity.
Each RNA was transcribed in vitro and refolded under conditions that support native tertiary structure formation. Folded RNAs were then treated with TBIA. The RT-C8 polymerase has a distinctive and valuable crosslinking traversing activity; however, this polymerase does induce mutations and deletions when copying RNA templates that do not contain crosslinks. We controlled for these background deletions by comparing TBIA reactions with parallel experiments performed with IA (Fig. 2A). All TBIA-treated RNAs showed 2- to 5-folder higher per-read deletion rates than samples treated with IA (Fig. S2), suggesting that a majority of deletions detected in TBIA-treated samples correspond to crosslinks resulting from proximal nucleotides rather than background activity of the RT enzyme. We created a specialized alignment algorithm that accounts for the complex observed deletion and sequence mutation patterns (Fig. S2B) to accurately map TBIA-induced deletion sites and rank these by frequency22. RT-C8 does not detect monoadducts as IA-treated and no-reagent experiments showed similar deletion rates (Fig. S2) and positions (Fig. S3). Full experimental replicates showed highly correlated deletion rates, similar deletion sites across replicates, and similar through-space distance distributions (Fig. S4). We observed crosslinks involving all four ribonucleotides, consistent with the expected predominant reaction at the 2’-hydroxyl group5,6.
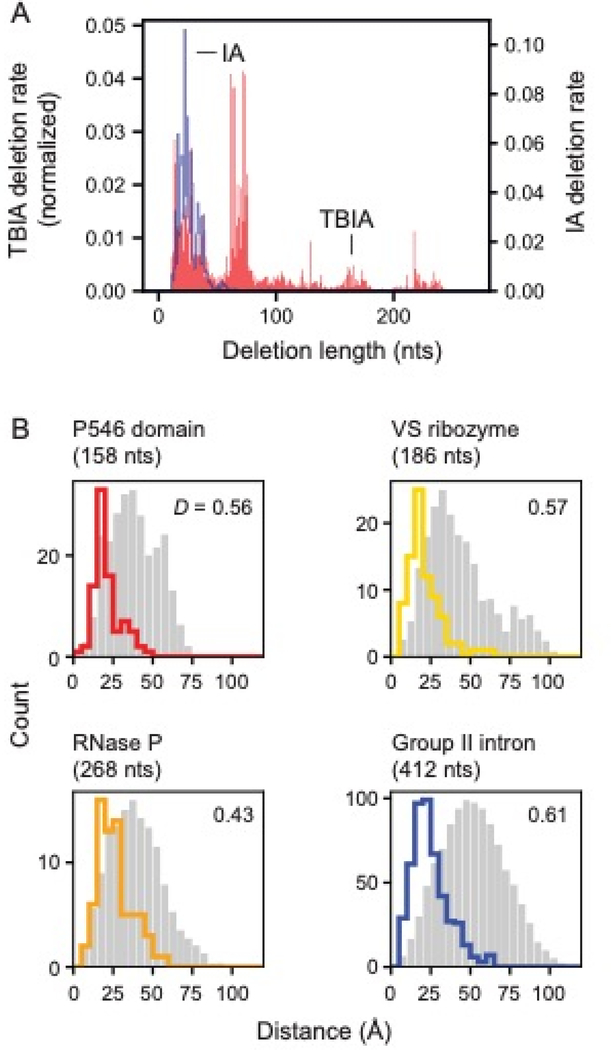
We next characterized deletion lengths as quantified in the cDNAs generated by RT-C8. Across all RNAs, average observed (background) deletion lengths in IA-treated samples were shorter than 50 nucleotides, while deletion lengths in TBIA-treated RNAs varied as a function of RNA structure. For the relatively unstructured VS RNA, deletion lengths for IA and TBIA were similar, whereas, for the highly structured RNase P (Fig. 4A) and group II intron (Fig. S5A), deletion distances in TBIA-treated samples were much longer than those in the IA-treated controls. As measured by RT-C8, TBIA-induced deletion lengths are thus related to the extent of structure in an RNA, but non-crosslink-induced deletions are not.
Figure 4: SHAPE-JuMP deletion detection as a function of intervening sequence length and through-space distance.
(A) Deletion rates of a given length due to treatment with monoadduct-forming IA and crosslinker TBIA for the RNase P RNA. Deletion rates are normalized to sum to 1. (B) Distances between nucleotides that mediate TBIA-induced deletions. The most frequent three percent of deletion rates are shown with colored lines. Random internucleotide distances based on the reference structure, that follow the same sequence length distribution as TBIA-induced contacts, are shown in gray. D, the Kolmogorov-Smirnov metric, quantifies separation between two distributions on a 0 to 1 scale with 0 indicating no separation and 1 indicating complete separation; all D values correspond to p-values ≤ 10–5.
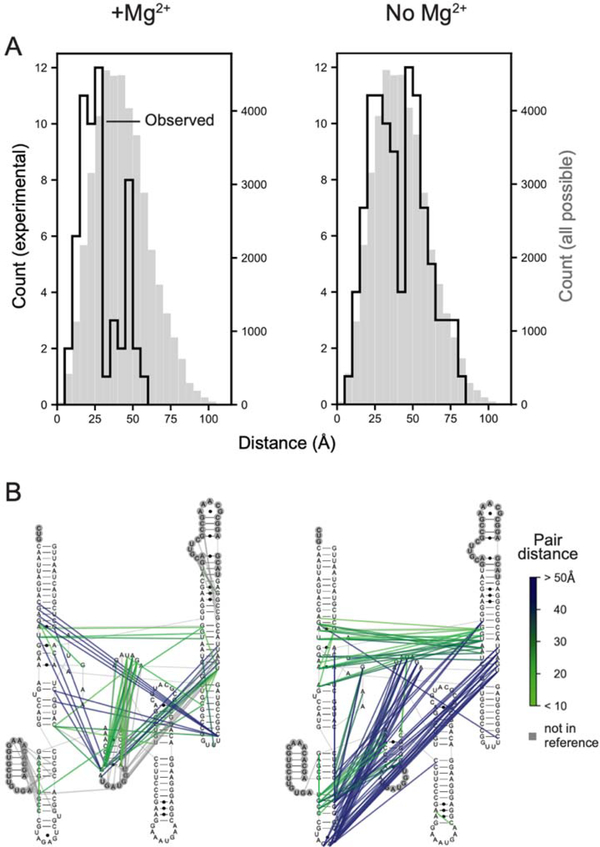
We next examined the physical distances between nucleotides mapped in the JuMP step as a function of crosslink frequency and RNA. For all RNAs, crosslink frequency correlated with shorter through-space distance (Fig. S5D). The most frequent 3% of deletions were between nucleotides separated in space by a mean distance of 21–26 Å, when comparing to the accepted structure for each RNA (Fig. 4B). These distances are significantly shorter than those obtained from random sampling, including after controlling for the sequence length distributions of the TBIA-derived internucleotide pairs (Kolmogorov-Smirnov test, all p-values <10−5). Across all four RNAs, 78% of the distances are less than 30 Å, which we accept as true positive measurements. We also observe a small fraction, 4%, of distances of 45 Å or greater, which likely reflect some combination of false positive measurements, conformational dynamics in these large RNAs, or other features not reflective of internucleotide distances (see Ref. 22). We additionally compared deletions detected for the RNase P RNA in the presence and absence of Mg2+, an ion essential for native tertiary folding16,23. Probing in the absence of Mg2+ yielded a deletion distribution markedly different from the fully folded structure and roughly comparable to the result expected for random sampling of through-space interactions (Fig. 5).
Figure 5: SHAPE-JuMP interactions detected for the RNase P RNA in the presence and absence of Mg2+.
(A) SHAPE-JuMP interactions for the most frequent 3% of deletions for RNase P in the presence (left) and absence (right) of Mg2+. Experimentally measured distances are shown with black lines; gray histograms represent distances of all nucleotide pairs in reference structure. (B) Internucleotide crosslinks, detected as cDNA deletions, superimposed on the secondary structure, colored by through-space distance as calculated from the reference structure16. Nucleotides not visualized in the reference structure are shown within gray circles.
In sum, these data emphasize that SHAPE-JuMP detects proximal through-space interactions at nanometer resolution and, when native interactions are destabilized by omitting Mg2+, the resulting crosslinks capture interactions characteristic of alternative states. Three experimental heuristics facilitate reliable identification of through-space tertiary RNA-RNA internucleotide interactions using the current JuMP strategy: (i) samples should be sequenced to a minimum read depth of 500,000 reads, (ii) the deletion rate for the crosslinked sample should be at least two-fold higher than that of a no-reagent or mono-adduct control, and (iii) the overall crosslink-mediated deletion rate should be greater than 0.01 deletions per read.
SHAPE-JuMP identifies diverse, specific internucleotide interactions
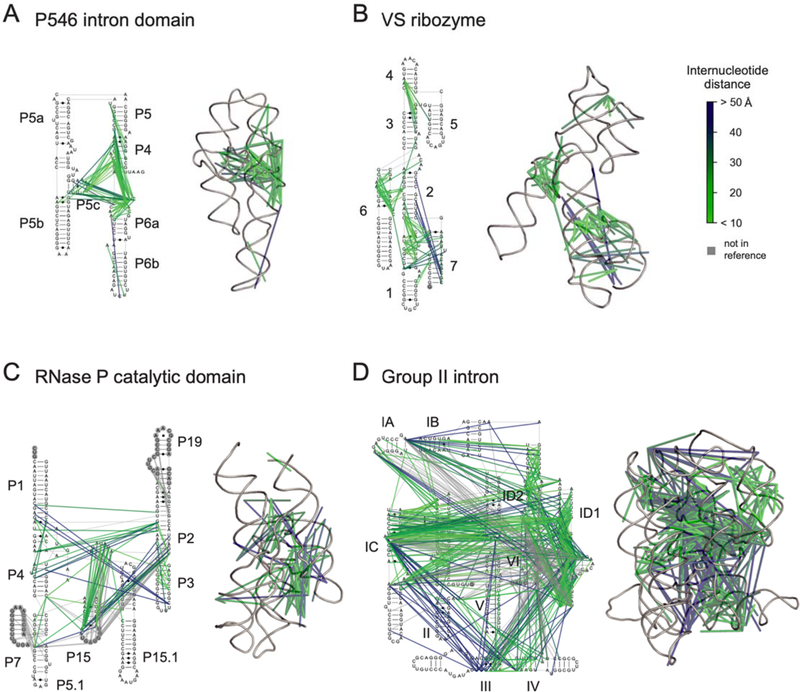
Crosslinked nucleotides identified by SHAPE-JuMP tend to involve at least one unpaired RNA region, consistent with the preferential reactivity of SHAPE reagents for conformationally flexible sites in RNA. Nonetheless, many crosslinks are recorded between an unpaired region and a base-paired region, likely partially because the second reactive group in TBIA can react with structurally diverse nearby nucleotides due to an increased effective local concentration. For example, in P546, interactions extend from single-stranded regions adjacent to helix P6a to the base-paired region in P4 (Fig. 6A). Similar patterns are observed in the group II intron, for example, between the ID2 loop and the IC helix (Fig. 6D).
Figure 6: Visualization of SHAPE-JuMP interactions on complex RNA structures.
Through-space interactions, detected as cDNA deletions, superimposed on secondary and tertiary structure models of (A) P546 intron domain19 (PDB ID 1gid), (B) VS ribozyme17 (4r4p), (C) RNase P catalytic domain16 (3dhs), and (D) group II intron20 (3igi). Internucleotide interactions are shown as lines, colored by through-space distance. Interactions are shown for most frequent 3% of measured deletions. Nucleotides not visualized in three-dimensional structure models are denoted with gray circles.
In the test set of RNAs, SHAPE-JuMP detected internucleotide interactions within every multi-helix junction, as exemplified by the helix 1–2-7 three-helix junction of the VS ribozyme (Fig. 6B) and the helix III-IV-V junction of the group II intron (Fig. 6D). We also identified internucleotide interactions between the single-stranded regions of junctions and their connected helices, as exemplified by the helix 3–4-5 junction of the VS ribozyme (Fig. 6B) and the helix P1-P3-P4 junction of RNase P (Fig. 6C). Internucleotide interactions identified by SHAPE-JuMP are thus not limited to base-paired or single-stranded nucleotides nor to a specific structural motif and are particularly common in multi-helix junctions.
The majority of SHAPE-JuMP internucleotide interactions occur in clusters, as is readily visualized when contacts are mapped onto the known secondary and tertiary structures (Fig. 6). This clustering increases confidence that true proximal through-space interactions are identified by SHAPE-JuMP. Clusters occur throughout the tested RNAs and in both solvent-accessible and inaccessible regions, as supported by comparing the solvent-accessible surface areas of each nucleotide 2′-OH group to the total deletion rate at each nucleotide (Fig. S6). This observation is consistent with prior work that demonstrated that SHAPE reactivity is not governed by solvent accessibility24. Thus, SHAPE-JuMP identifies proximal through-space interactions in both the interior and exterior of RNAs with complex structures.
SHAPE-JuMP-restrained structure modeling
De novo RNA structure modeling is improving rapidly but remains challenging for large RNAs21,25. Experimental restraints can dramatically improve physics-based modeling7,26,27, and SHAPE-JuMP identifies through-space RNA-RNA interactions with mean distances on the sub-25-Å scale (Fig. 4), roughly corresponding to the width of an RNA helix. We therefore explored the usefulness of SHAPE-JuMP measurements for reducing the conformation space sampled during tertiary structure modeling. SHAPE-JuMP restraints can, in principle, be incorporated into any modeling framework. Here, we explored the impact of SHAPE-JuMP restraints using discrete molecular dynamics (DMD)28,29 to model the global folds of the RNAs examined in our study. We represent the RNA structure in a simplified way for DMD simulations. Each nucleotide is modeled as three beads, corresponding to the phosphate, ribose, and nucleobase. SHAPE-JuMP restraints were incorporated by applying an energy bonus to the subset of DMD models during a simulation in which crosslinked nucleotides are within specified distance thresholds7,29.
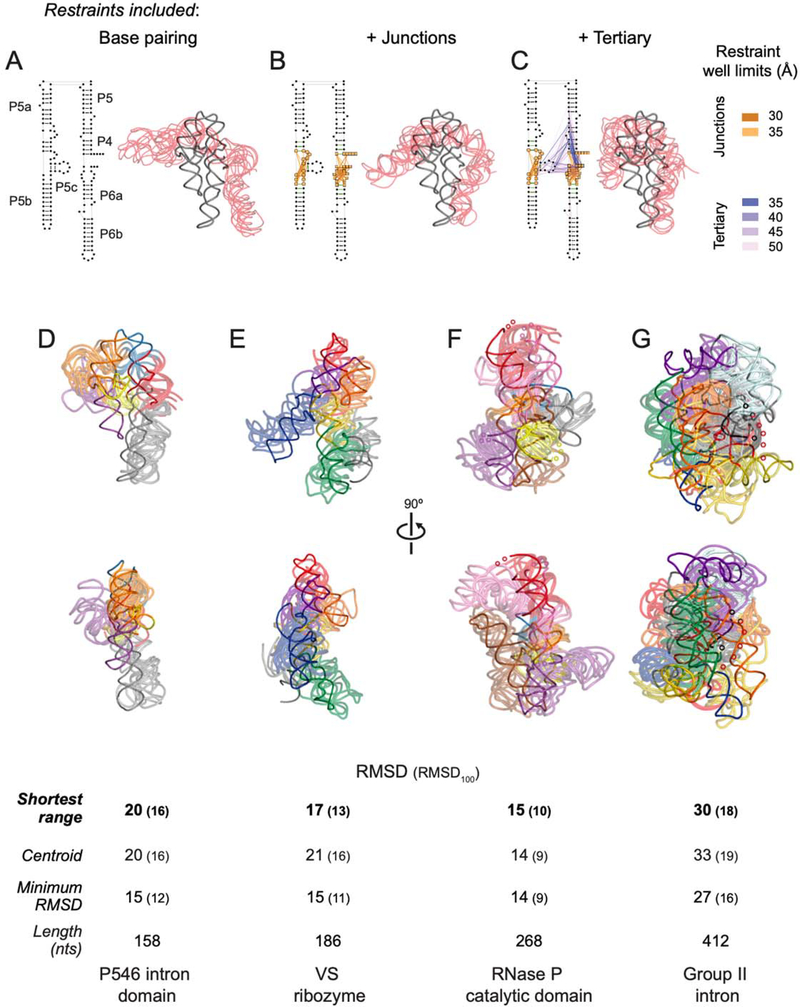
Restraints were applied in three steps during the DMD simulation, motivated by both the classes of internucleotide interactions directly measured by SHAPE-JuMP and the hierarchical nature of RNA folding30. In step 1, secondary structure restraints, corresponding to canonical base pairing, were applied (Fig. 7A). Accurate secondary structure models for most RNAs are readily obtained by SHAPE-MaP and PAIR-MaP chemical probing9,31 and were available for each RNA in the test set6,31,32. This step did not use SHAPE-JuMP data. In step 2, restraints corresponding to SHAPE-JuMP interactions identified within multi-helix junctions were added (Fig. 7B, orange and yellow lines). In step 3, SHAPE-JuMP-informed restraints corresponding to through-space tertiary structure proximity were added (Fig. 7C, blue and purple lines). Restraints corresponding to the most frequently measured deletions were awarded bonuses at shorter distances (Fig. 7C and S5D). A final representative model was obtained from step 3 by selecting the model most consistent with the SHAPE-JuMP data: we calculated distances between SHAPE-JuMP-connected nucleotides for the junction and tertiary restraints for each RNA, and selected the model with the shortest range of restraint distances.
Figure 7: SHAPE-JuMP directed structure refinement.
(A-C) Restraints superimposed on secondary structure and resulting three-dimensional models for the stepwise DMD refinement of the P546 intron domain. Five modeled structures (transparent red), consisting of the centroid and four models with lowest RMSD as compared to this centroid, aligned to the reference structure19 (gray) are shown. Restraints were added stepwise, (A) starting with the base paired secondary structure, (B) adding SHAPE-JuMP restraints at multi-helix junctions (orange lines), and (C) adding high frequency proximity interactions (purple lines). Lengths of restraint wells used during DMD refinement are color-coded. (D-G) Structures obtained using JuMP data-informed DMD aligned to the (D) P546 domain (PDB ID 1gid), (E) VS ribozyme (4r4p), (F) RNase P catalytic domain (3dhs), and (G) group II intron (3igi). JuMP restraints were mapped on to final models. The five models with the shortest restraint distance ranges were taken as representative of the simulation. Structures are colored by major helical elements. Modeled and accepted structures are shown with transparent and solid backbone traces, respectively. RMSD values are shown for: models with shortest restraint distance range, centroid of the largest cluster, and lowest RMSD model obtained. RMSD100 values (in parentheses) report a length normalized RMSD33. Regions not visualized in accepted structures are indicated with small spheres.
SHAPE-JuMP directed refinement yielded final models with RMSD values ranging from 15 to 30 Å, relative to the accepted structure, with longer molecules having larger RMSD values, as expected (Fig. 7D-G). Models show marked improvements in RMSD after modeling steps 2 and 3, emphasizing the value of solution-phase SHAPE-JuMP restraints (Table S1). RMSD values normalized to a length of 100 nucleotides33 (RMSD100) fell into a narrow range of 10 to 18 Å. For P546, the three groups of SHAPE-JuMP restraints yielded a refined model that captured the overall structure of the RNA with the exception of the P5b region, which lacked proximity interactions linking it to the rest of the structure (Fig. 7D). For VS, the refined model clearly captured the global architecture of the reference structure (Fig. 7E). RNase P was an especially favorable case, as the overall fold and helix positions were in good agreement with the accepted structure (Fig. 7F). Similarly, the SHAPE-JuMP directed model of the group II intron has the correct box-like shape of the accepted structure34 and correctly places the helices comprising the catalytic core in the center of the structure (Fig. 7G).
Discussion
SHAPE-JuMP embraces two distinctive learnings informed by our previous work. First, experience with other RNA structure probing technologies has revealed that the sensitivity and accuracy for detection of chemical adducts in RNA is strongly dependent on the directness of the strategy used to record the chemical event in a sequencing readout. In SHAPE-JuMP, sites of crosslinking are read out in a single direct step in which an engineered reverse transcriptase extends through a crosslink site and permanently records this site as a deletion in the synthesized cDNA. Second, the crosslinking reagent, TBIA, reacts preferentially with a generic site in the RNA, the 2’-hydroxyl, such that nearly all high-frequency crosslinks report higher-order internucleotide proximity corresponding to multi-helix junctions, through-space helix packing, and other tertiary interactions.
SHAPE-JuMP thus has unique features relative to the current generation of crosslinking approaches, which are read out by high-throughput sequencing. Several approaches for identifying RNA-RNA interactions use UV-mediated psoralen crosslinking, and primarily detect secondary structure35–40. For psoralen-based methods, crosslink sites are read out by multi-step ligation strategies, and psoralen shows marked sequence and structural preferences11,38,39,41. Psoralen-based methods have the notable advantage of successful use in cells and the ability to detect long-range and intermolecular interactions. There are also several information-rich strategies for mapping through-space interactions that employ extensive mutagenesis of29,42 or introduction of pre-structure-probing chemical modifications into43,44 RNA transcripts. These methods are complex to implement which, to date, has reduced their wide adoption.
The JuMP strategy is thus unique in its experimental directness and concision, can read out multiple classes of crosslinks (including those induced by TBIA, psoralen and UV irradiation)22, and will likely prove a foundational approach for future high-throughput RNA tertiary structure discovery technologies. Current limitations include that SHAPE-JuMP does not detect all tertiary contacts in the benchmark RNAs and the fraction of false positive interactions merits improvement. In addition, SHAPE-JuMP currently works best in an amplicon format in which RNA-specific primers are used to amplify a defined sequence. These limitations are likely addressable by development and use of alternative crosslinking reagents and by further optimization of the jumping activity of the RT-C8 polymerase. SHAPE-JuMP in its current form has the potential to detect very long-range and intermolecular interactions in RNA, which can be examined using alternative benchmarking systems.
Median through-space distances measured by SHAPE-JuMP are about 23 Å. This distance is independent of the size of the RNA in our test set, which range in length from 158 to 412 nucleotides. The 23 Å distance is comparable to the width of an RNA helix but longer than the 7 Å distance between the reactive carbonyl sites in TBIA, indicating that detection of through-space interactions is governed, in part, by features beyond simple physical proximity, likely reflective of RNA flexibility and dynamics. SHAPE-JuMP has a strong propensity toward identifying two classes of through-space interactions. First, SHAPE-JuMP detected multiple interactions across every multi-helix junction present in the RNAs of known structure evaluated here (10 junctions across four RNAs). This property of SHAPE-JuMP will likely prove impactful in future work as RNAs with multi-helix junctions are overrepresented among functional RNAs. Second, SHAPE-JuMP measures through-space interactions between an unpaired loop and either a second loop or a closely packed helix. These interactions are especially valuable in modeling large RNAs with globular folds.
In this study, we focused on large RNAs that are generally difficult to model de novo. There are two core challenges in modeling RNA structure in solution, and SHAPE-JuMP makes a useful contribution to each. SHAPE-JuMP crosslinks can be used, first, as restraints for modeling three-dimensional RNA folds and, second, to rank models output by modeling. The VS ribozyme modeled in our study was a target in the RNA-Puzzles competition, whose entries spanned 26 ± 6 Å RMSD, with the best model yielding a 20 Å RMSD21. Our SHAPE-JuMP directed modeling and selection strategy yielded a structure for the VS ribozyme with a 17 Å RMSD, outperforming the best RNA-Puzzles model. Selecting the best final model by assessing agreement with SHAPE-JuMP restraints was important and improved model accuracy from 21 to 17 Å RMSD, relative to simply selecting the centroid of the lowest energy cluster (Fig. 7E). The RNase P and group II intron RNAs are considerably longer than those either available in the RNA-Puzzles competition or generally attempted by de novo modeling. SHAPE-JuMP restrained DMD modeling outperformed prior data-driven modeling efforts for the RNase P catalytic domain (including prior work from our lab)7,45,46 and for variants of the group II intron47,48.
There are two additional promising features of SHAPE-JuMP directed interrogation of large-scale RNA structure. First, VS lacks long-range tertiary interactions and has an extended topology, and SHAPE-JuMP did not detect tertiary structure where none exists. Second, modeling accuracy, as measured by the length-normalized RMSD100, fell in a compact range, 10–18 Å, despite the variety of architectures modeled and the large sizes of the RNase P (268 nts) and group II intron RNAs (412 nts). These insights suggest that the concise SHAPE-JuMP experiment, coupled with restraint-directed modeling, will specifically prove useful for discovery and modeling of large RNA architectures in viral RNAs, mRNAs, and long non-coding RNAs.
Methods
TBIA synthesis
A sample of 500 mg (2.55 mmol) of 2,5-diaminoterephthalic acid (Sigma-Aldrich c/n 717312) was dissolved in 11 mL fresh anhydrous dioxane (Acros Organics) under nitrogen in a stirred three-neck round-bottom flask with a condenser. Next, 0.5 mL (4.18 mmol) of diphosgene was added dropwise, and the flask was heated to 90 °C for 2 h. A second addition of 0.25 mL (2.09 mmol) diphosgene was added dropwise, the temperature increased to 95 °C, and the reaction continued for a further 3 h. The reaction was then chilled to 4 °C in an ice bath, precipitating the product. The precipitate was filtered in a vacuum flask, washed several times with anhydrous diethyl ether, and dried under vacuum at room temperature. Reactions using diphosgene were performed in a well-functioning hood, all waste was neutralized with a saturated sodium bicarbonate solution. The final product was a green solid, 70% yield [1H NMR: (D6-DMSO) 11.9 (2 NH s), 7.6 (2 ArH s)].
TBIA hydrolysis
Hydrolysis rates were determined by UV (Nanodrop 2000) in a cuvette preheated to 37 °C. Reactions were initiated by adding 900 μL pre-warmed 1× folding buffer (100 mM HEPES, pH 8.0; 100 mM NaCl, 10 mM MgCl2) to 100 μL 40 nM TBIA in DMSO. Spectra were acquired continuously for 11 min. During this time period, a narrow peak at 296 nm was lost, and a broad peak centered at 425 nm shifted to 395 nm. Absorbance at 296 nm was biphasic, consistent with two hydrolysis processes for the reaction; a double-exponential decay model was used to fit (Prism 8) the data:
where Y0 and Yfinal are the initial and final absorbance, ffast is the fraction of signal associated with the fast rate (ideal=0.5), kfast and kslow are the rate constants for each of the sites, and t is time in seconds.
Reverse transcriptase polymerase
The reverse transcriptase RT-C8 was selected using in vitro mutagenesis and optimized using compartmentalized bead labeling14 starting with the previously developed relaxed fidelity, XNA incorporating, reverse transcriptase RT521K15, itself derived from the T. gorgonarius replicative polymerase49,50. Large-scale expression and purification of the RT-C8 was performed as described51. Briefly, RT-C8 was expressed in BL21 CodonPlus-RIL E. coli cells (Agilent Technologies); protein expression was induced at mid-log phase (OD600 0.4–0.6) with anhydrotetracycline (0.4 μg/mL final concentration) for 4 h at 37 °C. A 1 L culture was harvested by centrifugation and resuspended in 10 mL Lysis buffer A [50 mM Tris-HCl (pH 7.4), 1% glucose (w/v)] plus 30 mL Lysis buffer B [10 mM Tris-HCl (pH 7.4), 50 mM KCl, 0.5% NP40, 0.1% Triton X-100], and 10 mL 5 M NaCl, and lysed by incubation at 75 °C for 30 min. Cell debris were pelleted by centrifugation, and the lysate was diluted 20-fold with running buffer [50 mM NaCl, 10% glycerol, 0.02% NaN3, 20 mM Tris-HCl (pH 7.4)] and loaded onto a DEAE Sephadex A-25 column (GE Life Sciences) equilibrated in running buffer. The flow-through was collected and loaded directly onto an equilibrated HiPrep Heparin FF 16/10 column (GE Life Sciences). The heparin column was washed with running buffer, and a linear gradient from 50 mM to 1 M NaCl was used to elute bound protein. RT-C8 typically eluted at 0.5 M NaCl. The protein was exchanged into 2× polymerase storage buffer [1 M KCl, 2 mM EDTA; 20 mM Tris-HCl (pH 7.4)] by filter dialysis (Amicon Ultra-15 Centrifugal Filter Unit, 50 kDa cutoff, MerckMillipore) and stored in 50% glycerol at −20 °C.
In vitro RNA transcript synthesis and folding
DNA templates (IDT) for all RNAs were synthesized with the addition of 5′ and 3′ structure cassettes5. RNAs were transcribed in vitro with T7 polymerase [40 mM Tris (pH 8.0), 25 mM MgCl2, 2.5 mM spermidine, 0.01% (vol/vol) Triton X-100, 10 mM DTT, 2.5 mM each NTP, 15 ng/μl PCR generated template, 0.05 mg/ml T7 RNA Polymerase; 37 °C; 4 h], and the RNA product was treated with TURBO DNase (Invitrogen). Products were purified and exchanged into 0.5× TE buffer [(5 mM Tris (pH 8.0), 0.5 mM EDTA); RNAClean XP beads, Agencourt]. RNAs were refolded by heating for 1 min at 95 °C, placing on ice for 5 min, and incubating at 37 °C for 30 min. Folding buffers were: P546 intron domain [100 mM HEPES (pH 7.5), 100 mM NaCl, 10 mM MgCl2]45, M-Box riboswitch and RNase P catalytic domain [100 mM HEPES (pH 8.0), 100 mM NaCl, 10 mM MgCl2]52, VS ribozyme [200 mM HEPES (pH 8.0), 50 mM KCl, 10 mM MgCl2]53, and group II intron [90 mM HEPES (pH 8.0), 90 mM KCl, 115 mM MgCl2]45.
Visualization of crosslinked RNA by gel electrophoresis
Folded RNA (either RNase P or VS ribozyme) was treated with one-tenth volume 40 mM TBIA dissolved in DMSO, IA dissolved in DMSO, or neat DMSO, and incubated at 37 °C for 15 min. Reactions were performed at 0.5 μM for RNase P and 0.5, 1.0, 1.5 and 2.0 μM for VS RNA. Reactions were purified using a G-50 spin column (GE Healthcare). Aliquots normalized to 200 ng RNA each were denatured in equal volume of loading buffer [95% deionized formamide, 0.025% (w/v) bromophenol blue, 0.025% (w/v) xylene cyanol, 5 mM EDTA] at 95 °C for 2 min. The denatured RNA was loaded into a denaturing acrylamide gel (6% TBE-urea, ThermoFisher) and resolved in 1× TBE running buffer [89 mM Tris-borate (pH 8.3), 2 mM EDTA] at 180 V for 2 or 1 h for the RNase P or VS RNAs, respectively. The gel was stained (30 min with 1× GelStar stain in TBE; Lonza) and imaged using a UV transilluminator, using a non-saturating exposure.
Crosslinking, reverse transcription, and sequencing library preparation
After folding, 5 pmol of RNA in folding buffer were added to 40 mM TBIA, IA, or neat DMSO at 10:1 vol/vol. The samples were incubated at 37 °C for 15 min for TBIA, 30 min for IA, and 30 min for no-reagent controls; the incubation times were chosen to be 5 half-lives of the reagent. Samples were desalted (G-50 spin columns, GE Healthcare). RNAs were incubated with 10 pmol of primer specific to the 3’ structure cassette5 at 95 °C for 1 min and then placed on ice. Samples were then subjected to reverse transcription [20 mM Tris-HCl (pH 8.8), 10 mM (NH4)2SO4, 10 mM KCl, 2 mM MgCl2, 0.1% (vol/vol) Triton X-100, 200 μM dNTPs, 0.04 mg/ml RT-C8] at 65 °C for 4 h. Samples were desalted (G-50 spin column, GE Healthcare). cDNA product was amplified and barcoded via a 2-step PCR approach52 using a high fidelity DNA polymerase (Q5 hot-start, NEB). In step 1, 12 μl of the 50 μl reverse transcription reaction was amplified in a 50-μl PCR reaction for 25 cycles with forward and reverse primers containing Illumina sequencing adapters. Excess primer was removed (DNA beads at a 1:1 sample to bead ratio; Ampure DNA Beads, Agencourt). In step 2, step 1 product was diluted to 0.5 ng/μl, and 1 ng was subjected to 10-cycle PCR using primers complementary to Illumina sequencing adapters and that contained TruSeq (Illumina) barcodes to differentiate samples in the sequencer. Short products were removed prior to sequencing (Ampure DNA beads, Agencourt; 1:1 sample to bead ratio).
To identify an optimal RT enzyme, the above steps were performed for a diverse group of reverse transcriptase enzymes, including MMLV-M (a mutant of MMLV reverse transcriptase with similar activity as SuperScript II), Marathon RT18, RT521K15, and RT-C814, except that NMIA was used as the monoadduct control. Reverse transcriptase conditions were modified to correspond to the reported optimum for each enzyme. For MMLV-M, modified SHAPE-MaP reverse transcription conditions were used52. Treated RNA (7 pmol) was incubated with 2 pmol 3’ structure cassette5 primer and 2 μmols of dNTPs at 65 °C for 5 min and then cooled to 4 °C for 2 min. The RNA-primer mixture was incubated for 2 min at 25 °C in reverse transcription buffer, and then MMLV-M was added [final conditions: 50 mM Tris-HCl (pH 8.0), 75 mM KCl, 10 mM DTT, 1 M betaine, 6 mM MnCl2, 0.05 mg/ml MMLV-M]. Reverse transcription proceeded for 90 min at 42 °C followed by 10 cycles of 2 min at 50 °C and 2 min at 42 °C. The reaction was stopped by heating to 70 °C for 10 min. For Marathon, a modified version of reported reverse transcription conditions was used18; 0.25 pmol treated RNA was incubated with 0.2 pmol 3’ structure cassette5 primer at 95 °C for 30 s and placed on ice. Incubated RNA-primer was then subjected to reverse transcription [50 mM Tris-HCl (pH 8.0), 100 mM KCl, 2 mM MgCl2, 5 mM DTT, 500 μM dNTPs, 0.5 μM MarathonRT] at 42 °C for 1 h. The reaction was stopped by heating in 300 mM NaOH at 95 °C for 5 min, and the reaction was neutralized with an equivalent amount of HCl. For RT521K, the reverse transcription reaction was identical to that for RT-C8 except that 2 mM MnCl2 was substituted for 2 mM MgCl2.
Sequencing and data analysis
Samples were sequenced on an Illumina MiSeq instrument using a 300-nt kit for P546, M-Box, and VS RNAs, a 500-nt kit for RNase P, and a 600-nt kit for the group II intron; kit sizes provided full coverage of the target sequence. A minimum read depth of 500,000 was achieved for all samples. FASTQ files were trimmed, left to right, of nucleotides with an average Q score 10 or less in a window of 5 nucleotides, as described52. Reads were aligned with BWA-MEM54. Custom Python scripts were used to identify deletions longer than 10 nt. Deletion counts were normalized by the square root of the square of median read depth across a 5-nt window at the 5′ and 3′ ends of the deletion; the rate of a given deletion in the control sample was subtracted from the deletion rate in the crosslinked sample. The M-Box RNA was examined in this work (see Fig. S2A) but had a prohibitively low deletion rate and was excluded from further analyses. All computational analysis tools were packaged into a single pipeline; scripts are available at weekslab.com or Github.com/Weeks-UNC. Full independent replicates of all SHAPE-JuMP experiments were performed in parallel. Replicates were reproducible as determined by comparing overall deletion rates (Fig. S2A), correlation between individual deletion rates and locations, and analyzing three-dimensional distance distributions (Fig. S4) between replicate samples.
Analysis of significance of through-space distance measurements
Random sampling was used to access whether observed distributions of three-dimensional crosslinking distances were distinct from the distribution of distances based on random interactions of nucleotides as positioned in the RNA tertiary structure. For each RNA, 1,000,000 random internucleotide distances were sampled from the reference structure. The random internucleotide distances were sampled such that the distribution of sequence lengths followed the same deletion length distribution as observed for the most frequent three percent of TBIA-induced deletions. A Kolmogorov-Smirnov test was performed to test if the sampled and experimentally observed distance distributions were significantly distinct from each other, as quantified by the D statistic. D measures the degree of separation between two distributions on a scale of 0 to 1, where a value of 0 indicates that the two sample distributions are sampled from the same population distribution, and 1 indicates that the two distributions are distinct.
Restraint-directed three-dimensional structure modeling
Three-dimensional RNA structure modeling was performed using the DMD iFoldRNA framework55. We have used this framework for prior (no-restraints) entries in the RNA-Puzzles competitions21,25. Models for all RNAs were generated identically, with the exception that DMD steps were doubled for the group II intron RNA, due to its larger size. Modeling was performed in three steps. In Step 1, a linear, unstructured model was generated, composed of all residues in the RNA, in which each nucleotide was represented by beads for the phosphate, ribose, and nucleobase moieties. A 400,000-step molecular dynamics simulation was performed with canonical base pairs restrained based on the accepted secondary structure. Eight identical sets of ten replicas were run with replica temperature factor values of 0.22, 0.24, 0.26, 0.28, 0.30, 0.32, 0.34, 0.36, 0.38 and 0.40 kT; each set had a unique random seed. Structures extracted from all eight sets were ranked based on free energies, and the 1% lowest free energy structures were clustered using a 15-Å RMSD cutoff. The centroids of the eight largest clusters were then used to seed the eight replica sets of the next step. Step 2 was performed with the same parameters as Step 1, except that modeling iterations were reduced to 200,000 and SHAPE-JuMP internucleotide interactions at multi-helix and external junctions were applied as energy bonuses (Fig. 7B). Internucleotide interactions between single-stranded regions of a junction or between a single-stranded region and a closing junction base pair were ascribed the energy bonus (of −0.12 kcal/mol) when the paired nucleotides were within 30 and 35 Å, respectively (see Fig. 7C). Internucleotide distances used as restraints were filtered by secondary structure proximity7 to prevent artifactual disruption of base pairs by the restraints. Briefly, each SHAPE-JuMP contact was defined as nucleotide positions ni and nj and compared against all base pairs, with each base pair defined as positions mi and mj. If |ni – mi| + |nj – mj| < 4, the contact was not included7. An energy bonus was also applied to each of the first three base pairs for those helices branching from a junction (Fig. 7B, green lines). Bonuses were applied to base pairs within 8.6 to 8.9 Å. Step 3 was executed similarly to Step 2, with the addition that high-frequency tertiary SHAPE-JuMP internucleotide interactions were included as restraints. Nucleotides involved in SHAPE-JuMP interactions within the most frequent 5, 3, 1, and 0.5% of deletion rates were awarded an energy bonus when participating nucleotides were within 50, 45, 40, and 35 Å, respectively (Fig. 7C and S5). These contacts were filtered by secondary structure proximity, as described above, using a distance of 11 instead of 4; this step removed restraints that reflect local secondary structure rather than tertiary structure. At the end of Step 3, the 1% lowest free energy structures were extracted from the eight replica simulations. A representative model was selected by mapping the SHAPE-JuMP (deletion) restraints onto these low energy structures and the five models with the shortest restraint distance range (largest minus shortest distance in model, calculated from those detected by SHAPE-JuMP) were subject to all-atom reconstruction55,56 and aligned to the accepted reference structure (Fig. 7D-G). We also analyzed the final models by a more conventional approach and clustered the low energy structures using an RMSD cutoff equal to the sequence length divided by 10, as described57. The centroid structure of the largest cluster was aligned to the accepted reference structure.
RMSD calculations
RMSD values for RNA structures modeled using SHAPE-JuMP restraints were calculated both using the standard definition of RMSD based on the positions of all atoms:
and also based on normalizing this value to an RNA of 100 nucleotides (RMSD100)33:
where d is the distance between a pair of equivalent atoms and n is the total number of atoms in the structure. The latter metric facilitates comparison of models for RNAs of differing lengths.
Supplementary Material
Acknowledgements
We thank S. Bozal for the initial design of the small RNA constructs. This work was supported by the US National Institutes of Health (R35 GM122532 and R01 AI068462 to K.M.W.; R35 GM134864 to N.V.D.; and R01 GM101237 and R01 HL111527 to A.L.), the Medical Research Council (program grant MC_U105178804 to P.H. and G.H.), and the Passan Foundation (to N.V.D.). T.W.C. and C.A.G. were supported in part by NIH training grants in Bioinformatics (T32 GM067553) and Molecular Biophysics (T32 GM008570), respectively. M.J.S. was an NSF Graduate Research Fellow (DGE-1144081) and was supported in part by an NIH training grant in Molecular Biophysics (T32 GM08570).
Footnotes
Disclosure
K.M.W. is an advisor to and holds equity in Ribometrix. M.J.S. is an employee of Ribometrix.
Supporting Information: One supporting table and six figures (PDF). Supporting datasets contain deletion locations and rates for all RNAs, for replicate experiments. Restraints for DMD simulations with final model centroids. Pymol session files showing SHAPE-JuMP data superimposed on accepted structures and the resulting DMD structure models (from Figures 6 and 7). Supporting datasets are available at https://weekslab.com/. Sequencing data have been deposited in the sequence read archive: PRJNA687281.
References
- (1).Cech TR, and Steitz JA (2014) The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157, 77–94. [DOI] [PubMed] [Google Scholar]
- (2).Mortimer SA, Kidwell MA, and Doudna JA (2014) Insights into RNA structure and function from genome-wide studies. Nat. Rev. Genet. 15, 469–479. [DOI] [PubMed] [Google Scholar]
- (3).Montange RK, and Batey RT (2008) Riboswitches: emerging themes in RNA structure and function. Annu. Rev. Biophys. 37, 117–133. [DOI] [PubMed] [Google Scholar]
- (4).Weeks KM (2021) SHAPE directed discovery of new functions in large RNAs. Acc. Chem. Res. 54, 2502–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Merino EJ, Wilkinson KA, Coughlan JL, and Weeks KM (2005) RNA structure analysis at single nucleotide resolution by selective 2’-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 127, 4223–4231. [DOI] [PubMed] [Google Scholar]
- (6).Siegfried NA, Busan S, Rice GM, Nelson JA, and Weeks KM (2014) RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat. Methods 11, 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Homan PJ, Favorov OV, Lavender CA, Kursun O, Ge X, Busan S, Dokholyan NV, and Weeks KM (2014) Single-molecule correlated chemical probing of RNA. Proc. Natl. Acad. Sci. U. S. A. 111, 13858–13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Zubradt M, Gupta P, Persad S, Lambowitz AM, Weissman JS, and Rouskin S (2017) DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat. Methods 14, 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Mustoe AM, Lama NN, Irving PS, Olson SW, and Weeks KM (2019) RNA base-pairing complexity in living cells visualized by correlated chemical probing. Proc. Natl. Acad. Sci. U. S. A. 116, 24574–24582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Sengupta A, Rice GM, and Weeks KM (2019) Single-molecule correlated chemical probing reveals large-scale structural communication in the ribosome and the mechanism of the antibiotic spectinomycin in living cells. PLoS Biol. 17, e3000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Weeks KM (2015) Review toward all RNA structures, concisely. Biopolymers 103, 438–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Smola M, Calabrese JM, and Weeks KM (2015) Detection of RNA-protein interactions in living cells with SHAPE. Biochemistry 54, 6867–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Busan S, Weidmann CA, Sengupta A, and Weeks KM (2019) Guidelines for SHAPE reagent choice and detection strategy for RNA structure probing studies. Biochemistry 58, 2655–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Houlihan G, Arangundy-Franklin S, Porebski BT, Subramanian N, Taylor AI, and Holliger P (2020) Discovery and evolution of RNA and XNA reverse transcriptase function and fidelity. Nat. Chem. 12, 683–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Pinheiro VB, Taylor AI, Cozens C, Abramov M, Renders M, Zhang S, Chaput JC, Wengel J, Peak-Chew SY, McLaughlin SH, Herdewijn P, and Holliger P (2012) Synthetic genetic polymers capable of heredity and evolution. Science 336, 341–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Kazantsev AV, Krivenko AA, and Pace NR (2009) Mapping metal-binding sites in the catalytic domain of bacterial RNase P RNA. RNA 15, 266–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Suslov NB, DasGupta S, Huang H, Fuller JR, Lilley DMJ, Rice PA, and Piccirilli JA (2015) Crystal structure of the Varkud satellite ribozyme. Nat. Chem. Biol. 11, 840–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Zhao C, Liu F, and Pyle AM (2018) An ultraprocessive, accurate reverse transcriptase encoded by a metazoan group II intron. RNA 24, 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, and Doudna JA (1996) Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science 273, 1678–1685. [DOI] [PubMed] [Google Scholar]
- (20).Toor N, Keating KS, Fedorova O, Rajashankar K, Wang J, and Pyle AM (2010) Tertiary architecture of the Oceanobacillus iheyensis group II intron. RNA 16, 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Miao Z, Adamiak RW, Antczak M, Batey RT, Becka AJ, Biesiada M, Boniecki MJ, Bujnicki JM, Chen S-J, Cheng CY, Chou F-C, Ferre-D’Amare AR, Das R, Dawson WK, Ding F, Dokholyan NV, Dunin-Horkawicz S, Geniesse C, Kappel K, Kladwang W, Krokhotin A, Lach GE, Major F, Mann TH, Magnus M, Pachulska-Wieczorek K, Patel DJ, Piccirilli JA, Popenda M, Purzycka KJ, Ren A, Rice GM, Santalucia JJ, Sarzynska J, Szachniuk M, Tandon A, Trausch JJ, Tian S, Wang J, Weeks KM, Williams B 2nd, Xiao Y, Xu X, Zhang D, Zok T, and Westhof E (2017) RNA-Puzzles Round III: 3D RNA structure prediction of five riboswitches and one ribozyme. RNA 23, 655–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Christy TW, Giannetti CA, Laederach A, and Weeks KM (2021) Identifying proximal RNA interactions from cDNA-encoded crosslinks with ShapeJumper. bioRxiv 2021.06.10.447916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Fang XW, Pan T, and Sosnick TR (1999) Mg2+-dependent folding of a large ribozyme without kinetic traps. Nat. Struct. Biol. 6, 1091–1095. [DOI] [PubMed] [Google Scholar]
- (24).McGinnis JL, Dunkle JA, Cate JHD, and Weeks KM (2012) The mechanisms of RNA SHAPE chemistry. J. Am. Chem. Soc. 134, 6617–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Miao Z, Adamiak RW, Antczak M, Boniecki MJ, Bujnicki J, Chen S-J, Cheng CY, Cheng Y, Chou F-C, Das R, Dokholyan NV, Ding F, Geniesse C, Jiang Y, Joshi A, Krokhotin A, Magnus M, Mailhot O, Major F, Mann TH, Piątkowski P, Pluta R, Popenda M, Sarzynska J, Sun L, Szachniuk M, Tian S, Wang J, Wang J, Watkins AM, Wiedemann J, Xiao Y, Xu X, Yesselman JD, Zhang D, Zhang Y, Zhang Z, Zhao C, Zhao P, Zhou Y, Zok T, Żyła A, Ren A, Batey RT, Golden BL, Huang L, Lilley DM, Liu Y, Patel DJ, and Westhof E (2020) RNA-Puzzles Round IV: 3D structure predictions of four ribozymes and two aptamers. RNA 26, 982–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Ponce-Salvatierra A, Astha Merdas, K., Nithin C, Ghosh P, Mukherjee S, and Bujnicki JM (2019) Computational modeling of RNA 3D structure based on experimental data. Biosci. Rep. 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Li B, Cao Y, Westhof E, and Miao Z (2020) Advances in RNA 3D structure modeling using experimental data. Front. Genet. 11, 574485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Ding F, Sharma S, Chalasani P, Demidov VV, Broude NE, and Dokholyan NV (2008) Ab initio RNA folding by discrete molecular dynamics: from structure prediction to folding mechanisms. RNA 14, 1164–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Gherghe CM, Leonard CW, Ding F, Dokholyan NV, and Weeks KM (2009) Native-like RNA tertiary structures using a sequence-encoded cleavage agent and refinement by discrete molecular dynamics. J. Am. Chem. Soc. 131, 2541–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Brion P, and Westhof E (1997) Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct. 26, 113–137. [DOI] [PubMed] [Google Scholar]
- (31).Hajdin CE, Bellaousov S, Huggins W, Leonard CW, Mathews DH, and Weeks KM (2013) Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc. Natl. Acad. Sci. U. S. A. 110, 5498–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Beattie TL, Olive JE, and Collins RA (1995) A secondary-structure model for the self-cleaving region of Neurospora VS RNA. Proc. Natl. Acad. Sci. U. S. A. 92, 4686–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Carugo O, and Pongor S (2001) A normalized root-mean-square distance for comparing protein three-dimensional structures. Protein Sci. 10, 1470–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Toor N, Keating KS, Taylor SD, and Pyle AM (2008) Crystal structure of a self-spliced group II intron. Science 320, 77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, Davidovich C, Gooding AR, Goodrich KJ, Mattick JS, Mesirov JP, Cech TR, and Chang HY (2016) RNA duplex map in living cells reveals higher-order transcriptome structure. Cell 165, 1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Aw JGA, Shen Y, Wilm A, Sun M, Lim XN, Boon K-L, Tapsin S, Chan Y-S, Tan C-P, Sim AYL, Zhang T, Susanto TT, Fu Z, Nagarajan N, and Wan Y (2016) In vivo mapping of eukaryotic RNA interactomes reveals principles of higher-order organization and regulation. Mol. Cell 62, 603–617. [DOI] [PubMed] [Google Scholar]
- (37).Sharma E, Sterne-Weiler T, O’Hanlon D, and Blencowe BJ (2016) Global mapping of human RNA-RNA interactions. Mol. Cell 62, 618–626. [DOI] [PubMed] [Google Scholar]
- (38).Ziv O, Gabryelska MM, Lun ATL, Gebert LFR, Sheu-Gruttadauria J, Meredith LW, Liu Z-Y, Kwok CK, Qin C-F, MacRae IJ, Goodfellow I, Marioni JC, Kudla G, and Miska EA (2018) COMRADES determines in vivo RNA structures and interactions. Nat. Methods 15, 785–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Weidmann CA, Mustoe AM, and Weeks KM (2016) Direct duplex detection: An emerging tool in the RNA structure analysis toolbox. Trends Biochem. Sci. 41, 734–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Graveley BR (2016) RNA matchmaking: Finding cellular pairing partners. Mol. Cell 63, 186–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Lu Z, and Chang HY (2018) The RNA base-pairing problem and base-pairing solutions. Cold Spring Harb. Perspect. Biol. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Kladwang W, VanLang CC, Cordero P, and Das R (2011) A two-dimensional mutate-and-map strategy for non-coding RNA structure. Nat. Chem. 3, 954–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Homan PJ, Tandon A, Rice GM, Ding F, Dokholyan NV, and Weeks KM (2014) RNA tertiary structure analysis by 2’-hydroxyl molecular interference. Biochemistry 53, 6825–6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Cheng CY, Chou F-C, Kladwang W, Tian S, Cordero P, and Das R (2015) Consistent global structures of complex RNA states through multidimensional chemical mapping. Elife 4, e07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Ding F, Lavender CA, Weeks KM, and Dokholyan NV (2012) Three-dimensional RNA structure refinement by hydroxyl radical probing. Nat. Methods 9, 603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Kerpedjiev P, Höner Zu Siederdissen C, and Hofacker IL (2015) Predicting RNA 3D structure using a coarse-grain helix-centered model. RNA 21, 1110–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Costa M, Michel F, and Westhof E (2000) A three-dimensional perspective on exon binding by a group II self-splicing intron. EMBO J. 19, 5007–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Dai L, Chai D, Gu S-Q, Gabel J, Noskov SY, Blocker FJH, Lambowitz AM, and Zimmerly S (2008) A three-dimensional model of a group II intron RNA and its interaction with the intron-encoded reverse transcriptase. Mol. Cell 30, 472–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Gardner AF, and Jack WE (2002) Acyclic and dideoxy terminator preferences denote divergent sugar recognition by archaeon and Taq DNA polymerases. Nucleic Acids Res. 30, 605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Fogg MJ, Pearl LH, and Connolly BA (2002) Structural basis for uracil recognition by archaeal family B DNA polymerases. Nat. Struct. Biol. 9, 922–927. [DOI] [PubMed] [Google Scholar]
- (51).Ramsay N, Jemth AS, Brown A, Crampton N, Dear P, and Holliger P (2010) CyDNA: synthesis and replication of highly Cy-dye substituted DNA by an evolved polymerase. J. Am. Chem. Soc. 132, 5096–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Smola MJ, Rice GM, Busan S, Siegfried NA, and Weeks KM (2015) Selective 2’-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat. Protoc. 10, 1643–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Hiley SL, and Collins RA (2001) Rapid formation of a solvent-inaccessible core in the Neurospora Varkud satellite ribozyme. EMBO J. 20, 5461–5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Li H, and Durbin R (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Krokhotin A, Houlihan K, and Dokholyan NV (2015) iFoldRNA v2: folding RNA with constraints. Bioinformatics 31, 2891–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Sharma S, Ding F, and Dokholyan NV (2008) iFoldRNA: three-dimensional RNA structure prediction and folding. Bioinformatics 24, 1951–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Boniecki MJ, Lach G, Dawson WK, Tomala K, Lukasz P, Soltysinski T, Rother KM, and Bujnicki JM (2016) SimRNA: a coarse-grained method for RNA folding simulations and 3D structure prediction. Nucleic Acids Res. 44, e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.