Abstract
DNA topology and alternative DNA structures are implicated in regulating diverse biological processes. Although biomechanical properties of these structures have been studied extensively in vitro, characterization in vivo, particularly in multicellular organisms, is limited. We devised new methods to map DNA supercoiling and single-stranded DNA in Caenorhabditis elegans embryos and diapause larvae. To map supercoiling, we quantified the incorporation of biotinylated psoralen into DNA using high-throughput sequencing. To map single-stranded DNA, we combined permanganate treatment with genome-wide sequencing of induced double-stranded breaks. We found high levels of negative supercoiling at transcription start sites (TSSs) in embryos. GC-rich regions flanked by a sharp GC-to-AT transition delineate boundaries of supercoil propagation. In contrast to TSSs in embryos, TSSs in diapause larvae showed dramatic reductions in negative supercoiling without concomitant attenuation of transcription, suggesting developmental-stage-specific regulation. To assess whether alternative DNA structures control chromosome architecture and gene expression, we examined DNA supercoiling in the context of X-Chromosome dosage compensation. We showed that the condensin dosage compensation complex creates negative supercoils locally at its highest-occupancy binding sites but found no evidence for large-scale supercoiling domains along X Chromosomes. In contrast to transcription-coupled negative supercoiling, single-strandedness, which is most pronounced at transcript end sites, is dependent on high AT content and symmetrically positioned nucleosomes. We propose that sharp transitions in sequence composition at functional genomic elements constitute a common regulatory code and that DNA structure and propagation of torsional stress at regulatory elements are critical parameters in shaping important developmental events.
Within the nucleus, the predominant form of DNA is a double-stranded right-handed helix with 10.4 bp per helical turn, termed B-DNA. Essential biological processes such as transcription of genes can either overwind or underwind DNA, creating DNA supercoils (Kouzine and Levens 2007). Supercoils can, in turn, induce formation of single-stranded DNA (ssDNA, hereafter SS DNA) or other non-B DNA structures.
Genome-wide mapping of alternative DNA structures in cell lines and computational predictions in genomes suggested their involvement in diverse biological processes, including gene regulation (Kouzine et al. 2017), genome instability (Paeschke et al. 2013), the formation of centromeres (Kasinathan and Henikoff 2018), and the organization of chromatin into supercoiled domains (Naughton et al. 2013). However, the nature and genomic location of alternative DNA structures and DNA supercoils have not been explored in multicellular organisms, leaving unanswered whether they play significant roles in development and gene expression. To address these questions, we devised and applied new methods to map DNA supercoils and non-B DNA structures genome-wide in embryos and starved L1 larvae of the nematode Caenorhabditis elegans.
Studies before ours showed that actively transcribing RNA polymerase II (Pol II) is the primary source of DNA supercoils in eukaryotes (Teves and Henikoff 2014). To transcribe DNA, Pol II and its nascent RNA must rotate along the DNA's right-handed axis. Either owing to drag force or owing to immobilization inside of transcription factories (Papantonis et al. 2010), Pol II cannot rotate freely, causing the DNA molecule to rotate instead, thereby generating torsional stress (Liu and Wang 1987). In this theory, called the “twin domain model,” positive supercoils would accumulate downstream (overwinding) and negative supercoils would accumulate upstream (underwinding) of transcribing Pol II. However, measurements of supercoils in eukaryotic cells revealed a more complex picture in which the nature of supercoils at sites of active transcription vary considerably (Kouzine et al. 2013a; Naughton et al. 2013; Teves and Henikoff 2014). Measurements from these studies showed supercoils to be a local feature, confined to the immediate vicinity of transcription start sites (TSSs).
Topoisomerases can limit the propagation of supercoils in vivo through their enzymatic activities that relax supercoils (Pommier et al. 2016). Inherent physical properties of DNA such as curvature and bendability, which are determined by DNA sequence, contribute to the regulation of supercoiling in vitro (Kim et al. 2018). Despite these studies, the factors that limit the propagation of supercoils in eukaryotes are not well understood nor are the roles that chromatin might play in forming alternative DNA structures. We address these topics in our genome-wide analysis of DNA structure in nematodes, with a special emphasis on discovering the relationship between transcription and DNA supercoiling during different developmental stages and the role DNA sequence composition might play in restricting supercoil propagation at TSSs.
Supercoils are proposed to regulate higher-order chromosome structure and chromosome-wide gene expression. Although it is well accepted that prokaryotic chromosomes are organized into supercoiled topological domains (Postow et al. 2004), the role of supercoils in eukaryotic chromosome structure remains controversial. Eukaryotic chromosomes are organized into topologically associated domains (TADs) in which loci in one TAD interact predominantly with each other while being insulated from interactions with loci in adjacent TADs (Sikorska and Sexton 2020). Computer simulations suggested that supercoils might create TADs (Benedetti et al. 2014; Yan et al. 2018; Racko et al. 2019), but this hypothesis was not tested in vivo. Human cells have domains of supercoiling (Naughton et al. 2013), but they do not correspond to TAD boundaries. Here we address whether C. elegans chromosomes have domains of supercoiling and investigate the relationship between three-dimensional (3D) chromosome topology and DNA supercoils in the context of the C. elegans X-Chromosome dosage compensation process.
During C. elegans dosage compensation, a specialized condensin complex binds to both hermaphrodite X Chromosomes to reduce X-Chromosome gene expression by half, thereby equalizing expression between XX and XO embryos (Meyer 2018). This dosage compensation complex (DCC) also imposes a 3D topology onto hermaphrodite X Chromosomes by binding to its highest affinity sites on X Chromosomes and creating TAD boundaries (Crane et al. 2015; Anderson et al. 2019). Condensin complexes are renowned for compacting and resolving replicated chromosomes in vivo to promote chromosome segregation (Hirano 2012) and for creating positive supercoils in vitro in the presence of ATP and topoisomerase (Kimura and Hirano 1997; Hagstrom et al. 2002). However, a role for condensin in forming supercoils in vivo had not been shown. Using DCC-driven dosage compensation as a model, we investigated the relationship between 3D chromosome topology, condensin, and supercoiling in vivo.
Lastly, we performed a genome-wide analysis of non-B DNA in C. elegans. Because the formation of non-B DNA structures is linked to molecular events that relieve torsional strain, we investigated roles that nucleosome positioning and DNA sequence might play in shaping the distribution of non-B DNA. The importance of non-B DNA is well illustrated by the fact that duplex DNA in promoter regions must melt and become single-stranded for transcription to initiate. Therefore, we explored the relationship between DNA duplex stability, transcription initiation, transcription termination, and pre-mRNA splicing by measuring the propensity of sequences at TSSs, transcript end sites (TESs), and 5′ trans-splicing acceptor sites (TASs) to melt.
Results
An efficient genome-wide method to map supercoils
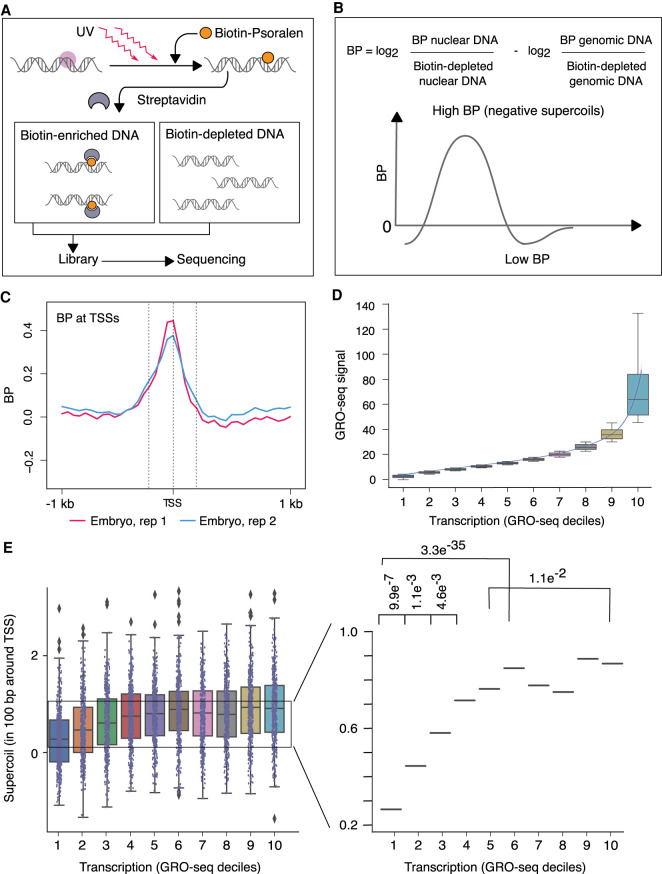
To map supercoils, we developed a method that quantifies genome-wide incorporation of biotinylated psoralen (BP) using high-throughput DNA sequencing (Fig. 1A,B) and overcomes limitations of prior methods that used other psoralen derivatives. Psoralen and its derivatives bind to DNA via intercalation, with a preference for negatively supercoiled DNA over relaxed or positively supercoiled DNA (Sinden et al. 1980). The amount of incorporated psoralen serves as a measure of DNA supercoiling. Upon exposure to UV light, psoralen covalently attaches to DNA through the formation of monoadducts and inter-strand cross-links, thereby permitting enrichment and purification of psoralen-bound DNA fragments.
Figure 1.
Genome-wide mapping strategy for DNA supercoils reveals negative supercoiling at transcription start sites (TSSs) of embryos. (A,B) Strategy for genome-wide mapping of DNA supercoils (BP-seq). (A) When biotinylated psoralen (BP) enters purified nuclei, it preferentially incorporates into negatively supercoiled DNA (purple circle). UV irradiation cross-links the intercalated BP to DNA. After DNA purification and sonication, BP-bound DNA is enriched on streptavidin beads. Libraries are prepared from both BP-enriched and BP-depleted samples and sequenced. As a control, naked genomic DNA is processed through the same steps. (B) The supercoiling profile is represented by the genomic distribution of the log2 ratio of BP-enriched over BP-depleted nuclear DNA adjusted by subtracting the log2 ratio of BP-enriched over BP-depleted naked genomic DNA. High signal corresponds to regions of negative supercoiling. (C) Average profiles of supercoils (BP-seq) are plotted in the 2-kb region surrounding TSSs of 4221 highly expressed genes (Kruesi et al. 2013) at 50-bp resolution for C. elegans embryos. Dashed lines represent locations of TSSs (center) and 200 bp upstream of (left) and downstream from (right) TSSs. (D) Box and whisker plots of GRO-seq values for 4221 genes after partitioning into deciles based on their average GRO-seq signal in the gene body. The lowest decile (decile 1) includes genes with the lowest 10% of GRO-seq values. A trend line through the medians of box plots for each decile shows a graded increase in GRO-seq values up to decile 6, followed by a much sharper ascent from decile 7 to 10. (E) Box and whisker plots of BP-seq signal in 100-bp intervals around TSSs correspond to the GRO-seq deciles shown in D. Magnified view of median values shows that BP signal scales with increases in transcription up to the sixth decile. From the seventh decile, a sharp ascent in GRO-seq values (D) does not correspond to a concomitant increase in BP values. P-values were calculated using the Mann–Whitney U test.
Prior methods to map supercoils (Kouzine et al. 2013a; Naughton et al. 2013; Teves and Henikoff 2014) used the psoralen derivative 4,5′,8-trimethylpsoralen (TMP) in combination with various DNA sequencing schemes. This general approach has the disadvantage that it requires large quantities of starting material and special DNA purification conditions. TMP forms many inter-strand cross-links that prevent DNA from denaturing during exposure to high temperature. This property is used to separate psoralen-enriched DNA using high-temperature gel electrophoresis of denatured samples (Kouzine et al. 2013a; Teves and Henikoff 2014). However, inter-strand cross-links inhibit PCR, an essential step in preparation of sequencing libraries. A cross-link reversal step is essential, and reversal of cross-links is a harsh treatment that can damage the sample. Another method (Naughton et al. 2013) used biotinylated 4,5′,8-trimethylpsoralen (bTMP) to isolate psoralen-enriched DNA fragments on streptavidin beads. The drawback of this method is that bTMP is not commercially available, and in-house synthesis requires expertise and specialized equipment not common to most standard molecular biology laboratories. Also, the reversal of cross-links is required. To measure DNA supercoils in our study, we developed a high-throughput method that permits easy isolation of psoralen-enriched DNA, does not require reversal of cross-links, and uses a commercially available reagent, BP.
Live C. elegans embryos and larvae are generally impenetrable to chemicals. This limitation forced us to use purified nuclei in our experiments. Previous work showed that the nuclei isolation protocol preserves the chromatin structure and transcriptional competence of RNA Pol II (Kruesi et al. 2013; Steiner and Henikoff 2014; Jänes et al. 2018). Because animals were flash-frozen before isolation of nuclei, our results represent DNA supercoiling states arrested at the time of freezing.
BP was used previously to visualize transcription-dependent supercoils in Drosophila polytene chromosomes (Matsumoto and Hirose 2004) via microscopy. BP was also used to develop the Chem-seq method (Anders et al. 2014), but its specificity for detecting supercoils was not shown directly. We therefore tested BP's specificity by incubating negatively supercoiled and relaxed plasmids with BP and quantifying the amount of intercalated psoralen by chemiluminescence using streptavidin-conjugated horseradish peroxidase (Supplemental Fig. S1A–C). We found that negatively supercoiled plasmids incorporated more psoralen than relaxed plasmids, as judged by the intensity of fluorescence, thereby confirming BP as a specific probe for DNA supercoiling. We also confirmed that BP, unlike TMP, does not inhibit PCR, thereby allowing us to amplify sequencing libraries without reversal of psoralen binding.
The PCR compatibility of BP might be explained by its reduced inter-strand cross-linking potential compared with TMP (Kanne et al. 1984). Upon exposure to light, both psoralen and TMP first form monoadducts. Then, upon encountering another photon of light, some monoadducts form cross-links. Monoadducts can be of two types, furan-side or pyrone-side, depending on which site of the psoralen molecule forms the monoadduct. Only furan-side monoadducts can form cross-links. TMP overwhelmingly (98%) forms furan-side monoadducts, whereas BP forms both types (Kanne et al. 1984). TMP would therefore form more inter-strand cross-links than BP and block PCR amplification of DNA more efficiently than BP.
For our mapping strategy (Fig. 1A), we treated nuclei prepared from either C. elegans embryos or starved first-stage larvae (L1) with BP and exposed them to long-wave UV light. DNA was extracted and then fragmented by sonication. Psoralen-bound DNA was enriched on streptavidin beads and further processed to generate a sequencing library. A sequencing library was also prepared from psoralen-depleted DNA that failed to bind streptavidin beads. To correct for sequence-specific psoralen binding rather than supercoil-driven binding, we repeated the procedure with naked genomic DNA. The normalized number of reads from psoralen-enriched DNA over psoralen-depleted DNA from naked genomic DNA was subtracted from the same ratio derived from nuclear DNA (Fig. 1B). The resulting calculation is a measure of supercoils (BP). Regions with high BP signal correspond to negatively supercoiled DNA.
Negative supercoiling is a prominent feature of TSSs in C. elegans embryos
To investigate the magnitude and extent of transcription-induced supercoils in C. elegans, we first visualized supercoiling profiles at TSSs of embryos. C. elegans is unusual in that 70% of its genes undergo a cotranscriptional processing event in which the true 5′ ends of nascent transcripts are replaced by a common 22-nucleotide leader RNA through a trans-splicing mechanism (Allen et al. 2011). The junction of the leader with the 5′ pre-mRNA splice acceptor site is called the trans-splice acceptor site (TAS). The majority of annotated 5′ ends for mature mRNAs correspond to TASs. We initially restricted our analysis to 4221 highly expressed genes whose true TSSs were mapped by recovering nascent RNAs with 5′ caps before their removal by cotranscriptional processing (Kruesi et al. 2013).
We found that strong negative supercoils were confined to a 400-bp region around the TSSs in embryos (Fig. 1C). We assessed whether the magnitude of supercoils correlated with the level of transcription by dividing genes into deciles of expression based on their global run-on sequencing (GRO-seq) level (Fig. 1D) and then plotted total BP signal in 100-bp regions around the TSSs (Fig. 1E). BP signal increased incrementally up to the fifth decile, beyond which the level of supercoils failed to scale with the increase in transcription. This phenomenon may be explained in two ways. A maximum threshold may exist for supercoils induced by transcription-generated torque. Alternatively, supercoil density above certain values may exceed the dynamic range of BP sensitivity.
GC-rich regions flanked by sharp GC/AT boundaries delineate boundaries of supercoil propagation at C. elegans TSSs
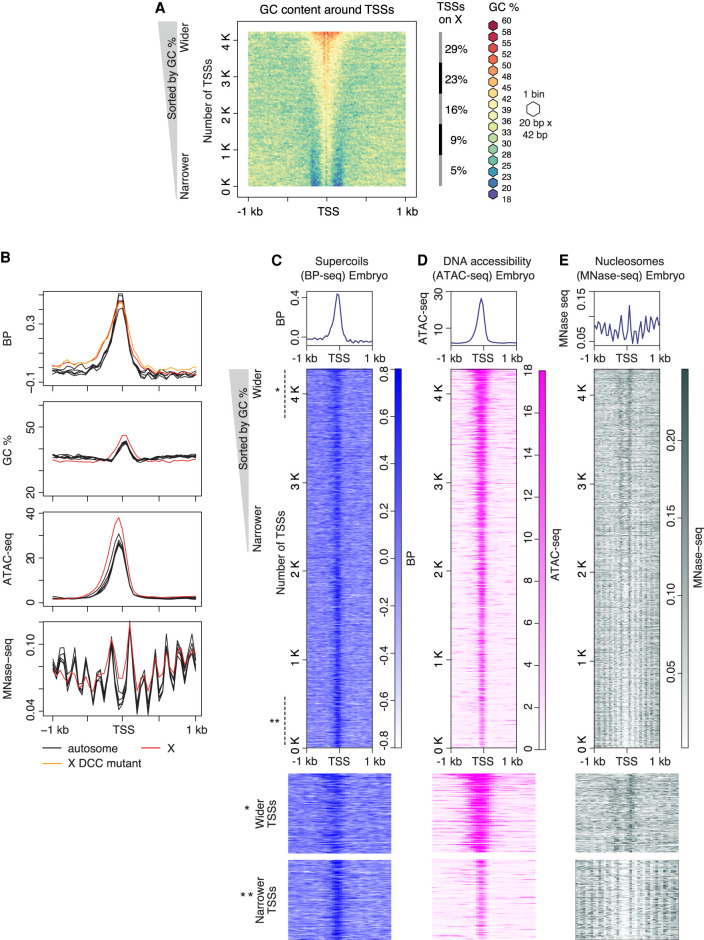
We next investigated parameters that might affect the extent of supercoil propagation. C. elegans TSSs have higher-than-average GC content, with the average GC percentage varying from 39% to 58%. We found that the average span of the GC-rich region around TSSs matches the span of supercoiling. We sorted the 4221 TSSs by their GC content and plotted a GC-content heatmap (Fig. 2A). The TSSs have sharp GC/AT boundaries that flank the GC-rich region, and even TSSs with lower GC content have higher-than-average AT content in sequences surrounding them (Fig. 2A). A graph of supercoils (Fig. 2B) and a heatmap of supercoils plotted by GC content (Fig. 2C) revealed that supercoil propagation follows the extent of the GC-rich region.
Figure 2.
GC-rich regions flanked by sharp GC/AT transition delineate boundaries of supercoil propagation. (A) Heatmap of GC content in the 2-kb region around TSSs of 4221 highly expressed genes measured by GRO-seq (Kruesi et al. 2013). TSSs were sorted by GC content, and adjacent sequences were aligned on top of each other and binned (20 bp × 42 bp). Each bin is colored according to its GC percentage. (B–D) Comparison of profiles for supercoiling, chromatin accessibility, and nucleosome position around TSSs. Heatmaps of supercoils (BP-seq for wild-type embryo replicate 1; B), chromatin accessibility (ATAC-seq; C), and nucleosomes (MNase-seq; D) are plotted in the 2-kb region surrounding TSSs at 50-bp resolution. For B–D, TSSs are sorted by GC content as in A. Lower panels provide a magnified view of the top section (marked with one asterisk) and bottom section (marked with two asterisks) of corresponding heatmaps. (E) Average profiles of supercoils (BP-seq for wild-type embryo replicate 1 and DCC mutant replicate 1), DNA accessibility (ATAC-seq), and nucleosomes (MNase-seq) are plotted separately for the X Chromosome and each autosome in the 2-kb region around TSSs at 50-bp resolution. Averaged GC contents for X Chromosomes and autosomes are plotted separately.
To explore further the correlation between the GC-rich region and propagation of supercoils, we exploited the fact that TSSs on X Chromosomes reside in wider GC-rich regions than do TSSs on autosomes (Fig. 2B). Plots of supercoiling profiles for each autosome and the X Chromosome revealed that supercoils on X propagate over a larger distance than those on autosomes, and the difference corresponds to the difference in extent of the GC-rich region (Fig. 2B). However, because X-linked genes undergo a dosage compensation process in which expression of genes from both hermaphrodite X Chromosomes is reduced by half, we asked whether the dosage compensation process contributes to the difference in supercoil propagation between X Chromosomes versus autosomes. We compared supercoiling profiles between wild-type embryos and embryos carrying a mutation that disrupts dosage compensation by preventing the DCC from binding to X. The span of supercoils on X was not affected by the dosage compensation mutation (Fig. 2B), corroborating the interpretation that GC-rich regions flanked by sharp AT/GC boundaries delineate the span of supercoils. GC percentage and extent of GC stretches around TSSs vary considerably, suggesting that a broad set of combinations for these two parameters facilitates supercoil formation.
We found that the profiles of several other chromatin features at TSSs closely correspond to the span of GC-rich regions, which are delimited by sharp AT/GC transitions. Measurement of DNA accessibility revealed that broader chromatin accessibility correlates with wider stretches of GC-rich regions at TSSs. DNA accessibility was quantified using an approach called assay for transposase-accessible chromatin using sequencing (ATAC-seq) (Jänes et al. 2018). We found that ATAC-seq peaks are wider at TSSs with wider GC-rich regions. Furthermore, nucleosome positioning (Steiner and Henikoff 2014) around TSSs, as monitored by signal for micrococcal nuclease digestion with deep sequencing (MNase-seq), correlates with the width of the GC-rich region. TSSs with narrow GC-rich regions have well-positioned arrays of nucleosomes on both sides of the TSS, but TSSs with wider GC-rich regions have fuzzy +1 and −1 nucleosomes and no other well-positioned nucleosomes (Fig. 2B,E, lower panels).
Analysis of DNA sequence and supercoiling in an expanded set of TSSs from more weakly transcribed coding promoters and enhancers lacking transcription (Jänes et al. 2018) further revealed that TSSs are positioned in GC-rich regions with sharp GC/AT boundaries and that supercoiling at promoters and enhancers scales with the level of transcription (Supplemental Fig. S2A–G). Enhancer heatmaps revealed virtually no BP signal (Supplemental Fig. S2D), but promoter heatmaps showed obvious signal (Supplemental Fig. S2C). BP signal was higher for the highly transcribed TSSs than for the weaker coding promoters, consistent with a robust influence of transcription on BP signal (Supplemental Fig. S2E,G). In contrast, 5862 random GC-rich regions (200–400 bp) lacking enhancers, promoters, or gene bodies had greatly diminished GC/AT boundaries (Supplemental Fig. S3A). Heatmaps revealed no significant supercoiling in random GC-rich or AT-rich sequences (Supplemental Fig. S3B–D). These results support the interpretation that a GC-rich region flanked by a sharp GC/AT boundary is a distinctive feature of a TSS that can delineate the extent of supercoiling. However, the level of supercoiling is influenced by the level of transcription.
Developmental arrest induced by starvation reduces supercoiling
To assess whether developmental state, in addition to DNA sequence, influences supercoiling, we analyzed supercoiling at TSSs in starved L1 larvae, which exhibit growth arrest and increased stress resistance (Baugh and Sternberg 2006), thereby permitting L1s to survive without food for up to a month. Starvation induces Pol II accumulation at TSSs of some genes (Supplemental Fig. S4A; Kruesi et al. 2013), an accumulation similar to transcription pausing in other species. For many genes encoding developmental regulators in flies and humans, elongating Pol II molecules pause after transcribing only a short distance from the TSS. Upon receiving activation signals, paused Pol II molecules resume elongation (Core and Adelman 2019). Pausing is absent in C. elegans embryos, but some promoters in starved L1 larvae show an enhanced peak of Pol II binding and nascent transcription (Kruesi et al. 2013). Alternative DNA structures such as G quadruplexes were proposed to increase pausing of RNA Pol II near TSSs (Szlachta et al. 2018). Hence, we tested whether the transcription program in arrested L1s resulted in differences in supercoiling at TSSs.
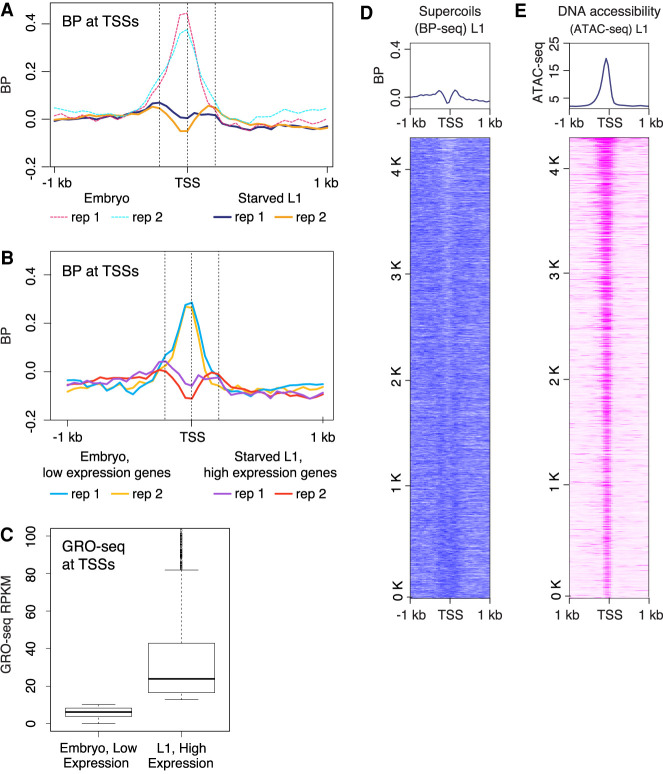
We plotted the supercoiling profile at TSSs of highly transcribed genes in starved L1s, regardless of their paused status, and found that the strong peaks of negative supercoils detected in embryos were absent (Fig. 3A). We asked whether this reduction in supercoiling was owing to a global reduction in transcription. Analysis of gene expression in starved L1s based on GRO-seq deciles (Supplemental Fig. S4B) showed comparable levels of transcription between starved L1s and embryos (Fig. 1E). Comparison of BP-seq signal between the corresponding GRO-seq deciles revealed a clear decoupling of supercoiling and transcription (Supplemental Fig. S4C).
Figure 3.
Developmental arrest through starvation decouples transcription and supercoiling at TSSs. (A) Average profiles of supercoils (BP-seq) are plotted in the 2-kb region surrounding TSSs at 50-bp resolution for starved L1s. For comparison, average profiles of supercoils (BP-seq) for embryos, as shown in Figure 1C, are presented as dashed lines. TSSs are from the 4221 highly expressed genes used in Figure 1C. (B) The reduction of negative supercoils in starved L1s does not correlate with reduction in transcription. Average profiles of supercoils (BP-seq) are plotted for genes with low expression in embryos and for genes with high expression in starved L1s. (C) Average GRO-seq counts in genes shown in B. (D) Heatmap of supercoils from BP-seq signal for starved L1s (replicate 1). (E) Heatmap of ATAC-seq at TSSs of starved L1s. TSS are sorted as shown in Figure 2A.
To further test the degree of decoupling, we compared supercoiling profiles between two groups of genes: the 30% of genes with the highest expression level in arrested L1s and the 30% of genes with the lowest expression level in embryos (Fig. 3B). Although the L1 cohort of highly expressed genes had a 10-fold higher average GRO-seq signal than the lowest-expressed embryo genes (Fig. 3C), they lacked negative supercoils at TSSs, but the embryo genes showed robust negative supercoils (Fig. 3B).
Because GRO-seq signal is normalized (Kruesi et al. 2013), it is possible that starved L1s have uniformly reduced transcription, such that even the highest expressed genes in L1s have reduced transcription compared with the lowest-expressed genes in embryos. However, transcriptomic-based studies showed that starvation in L1s is accompanied by a general up-regulation of mRNA levels (Kruesi et al. 2013; Harvald et al. 2017), further supporting that the global reduction in BP signal in starved L1s is independent of changes in transcription level.
Topoisomerase expression is drastically altered in response to starvation and oxidative stress in eukaryotic models (Ray et al. 2013; Varghese and Ghosh 2020). In prokaryotes, bacterial topoisomerase I is crucial to prevent hypernegative supercoiling at stress response genes during SOS (Liu et al. 2011). Future measurement of topoisomerase activity should reveal whether decoupling of transcription and supercoiling at TSSs in L1 larvae is a result of heightened topoisomerase activity.
We asked whether promoter proximal pausing correlates with the change in supercoiling profile. We identified paused genes in starved L1 larvae using a pause index: the ratio of GRO-seq signal at a promoter to the average GRO-seq signal downstream from the promoter. Genes with a pause index greater than five were considered paused (Supplemental Fig. S4A). In total, we identified 344 paused genes in starved L1s. Pausing was not correlated with any specific level of transcription, because genes with and without pausing had a similar distribution of expression levels (Supplemental Fig. S4E). Even though a general reduction in supercoils occurred at TSSs of starved L1 larvae, the degree of supercoiling at TSSs was not different between the 344 genes with Pol II pausing versus the 3876 genes without Pol II pausing (Supplemental Fig. S4D). Thus, changes in supercoiling at TSSs are not the cause or consequence of Pol II pausing.
Psoralen enrichment at TSSs is not the consequence of high chromatin accessibility
The expanded set of TSSs came from a genome-wide study of open chromatin that was delineated by ATAC-seq (Jänes et al. 2018). The ATAC-seq profile closely matches the profiles of GC-rich regions (Fig. 2B,D,E) that are marked by nucleosomes positioned at their edges. Because psoralen preferentially binds to accessible DNA, it was possible that open chromatin rather than supercoiling was the cause of BP-seq signal.
To assess the degree to which DNA accessibility contributes to psoralen binding, we analyzed 9677 enhancers that have a similar degree of DNA accessibility as the coding promoters and found that these enhancers have significantly lower BP signal than the coding promoters despite having the same level of accessibility (Supplemental Fig. S2F,G). Greatly attenuated supercoiling at enhancers (Supplemental Fig. S2D,G) shows that the presence of open chromatin is not the primary determinant of psoralen enrichment at promoters (Fig. 2C; Supplemental Fig. S2C,G). Furthermore, we found that the level of chromatin accessibility at TSSs in starved L1s (Fig. 3E) was comparable to that of embryos (Fig. 2D), indicating that chromatin accessibility is not the reason underlying the difference in BP signal between embryos (Fig. 2B) and starved L1s (Fig. 3D).
Comparative analysis of supercoiling profiles at nematode, human, and fly TSSs reveals common regulatory themes
Reduced psoralen binding at TSSs of well-transcribed genes has been observed in prior studies. Previous analysis of TSS supercoiling profiles from three human cell lines using both TMP and bTMP revealed different supercoiling profiles in all three studies. TSSs in nontumorigenic human mammary epithelial MCF10A cells (Ghosh et al. 2019) show substantial diminution of negative supercoils with an average propagation of 3 kb surrounding the TSSs (Supplemental Fig. S5A,B). Similar results were obtained for TSSs in Raji human B cells (Kouzine et al. 2013a), where psoralen levels at TSSs were considerably lower than the surroundings, albeit transcriptional inhibition lowered the levels of psoralen intercalation at these TSSs even further. The moderate decrement in psoralen intercalation at TSSs upon transcriptional inhibition was defined by the investigators as “dynamic negative supercoiling.” In contrast to MCF10A and Raji cells, TSSs in the human retinal pigmented epithelial RPE1 cells showed strong peaks of negative supercoiling (Naughton et al. 2013) similar to what we found at C. elegans embryos TSSs, albeit with wider propagation in RPE1 cells (5 kb) than in C. elegans embryos (400 bp). The difference in the spread of supercoils can be explained by the resolution of sequencing methods; however, the shape of the profile is likely owing to the biology of the cells. This observation agrees with other work in human cells, which showed that different supercoiling profiles can be observed in different cell types. Analysis of DNA supercoils in C. elegans supports this interpretation by showing that negative supercoils can be observed at TSSs of embryos and that specific states of development, such as larval arrest, can inhibit supercoiling (Fig. 3A,B). Both studies reinforce the need for further investigation of factors responsible for this phenomenon.
The strong correlation between propagation of supercoils at C. elegans TSSs and the width of corresponding GC-rich regions prompted us to explore sequences surrounding TSSs in human MCF10A cells. By comparing our plots of supercoiling at TSSs of 13,226 highly expressed genes in MCF10A cells (Supplemental Fig. S5B) with plots of GC content around the same set of TSSs (Supplemental Fig. S5C), we found the TSSs to be highly GC-rich. The average width of the GC-rich region surrounding TSSs is ∼3 kb and corresponds to the width of supercoils, suggesting that GC-rich sequences can determine, at least in part, the spread of supercoils around human TSSs.
We also analyzed supercoiling at fly TSSs (Teves and Henikoff 2014). Our comparison of high-resolution supercoiling profiles at 13,883 TSSs of Drosophila melanogaster S2 cells revealed a peak of negative supercoils with an average propagation of 1.2 kb (Supplemental Fig. S5D). Unlike C. elegans and human TSSs, fly TSSs are AT-rich (Supplemental Fig. S5E). The spread of supercoils at fly TSSs corresponds closely to the extent of the AT-rich region. Sharp transitions in sequence composition, either GC-rich to AT-rich or AT-rich to GC-rich, instead of absolute differences in sequence, correlate with the spread of supercoils at TSSs in worms, flies, and humans.
The condensin DCC creates local negative supercoils at its high-occupancy binding sites on X but does not create large domains of supercoiling along X
In C. elegans, a DCC binds to both X Chromosomes of hermaphrodites (XX) to repress transcription by half, thereby equalizing expression with that from the single X of males (XO). DCC binding near an X-linked gene is neither necessary nor sufficient for repression of the gene, implying the DCC acts at a distance to regulate gene expression (Wheeler et al. 2016). Our results thus far indicate that X-Chromosome repression is not achieved by regulating supercoiling at TSSs. However, because the DCC is a condensin complex that alters the 3D topology of hermaphrodite X Chromosomes, it could create domains of supercoiling along the X Chromosome that contribute to gene repression and its unique architecture. Condensin complexes act in vitro to promote DNA supercoiling (Kimura and Hirano 1997; Hagstrom et al. 2002) and formation of DNA loops (Ganji et al. 2018). Moreover, because the DCC creates TADs on X Chromosomes, condensin-driven dosage compensation is an attractive model to explore the relationship between alternative DNA structures and higher-order chromosome organization. To investigate further whether the DCC regulates gene expression by regulating DNA supercoiling, we examined local effects on supercoiling at DCC binding sites and global effects across the entire X Chromosome.
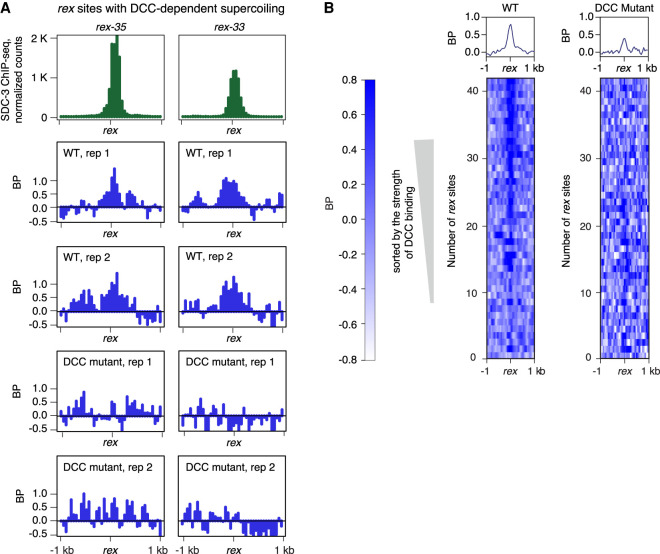
The DCC binds to two classes of sites on the X Chromosomes visualized as ChIP-seq peaks of DCC condensin loader SDC-3, a zinc-finger subunit. One class, called recruitment elements on X (rex sites), has X-enriched DNA motifs that promote high-affinity DCC binding, even when detached from the X Chromosome (Fig. 4A). The highest-affinity rex sites play a prominent role in establishing the DCC-dependent TADs on hermaphrodite X Chromosomes (Anderson et al. 2019). rex sites reside at the eight DCC-dependent TAD boundaries.
Figure 4.
Local DCC-dependent negative supercoils occur at DCC binding sites (rex sites) with the highest DCC occupancy. (A) Examples of rex sites (rex-35 and rex-33) that show DCC-dependent negative supercoils. DCC subunit SDC-3 (green ChIP-seq signal) binds with high affinity to these rex sites. BP-seq signal (blue) is plotted for two replicates at each rex site in wild-type and DCC mutant embryos. Reduction of BP-seq signal in mutants shows that supercoiling is DCC dependent. These rex sites are at TAD boundaries on X. (B) Heatmaps of supercoils (BP-seq) are plotted in 2-kb regions around rex sites at 50-bp resolution in wild-type (replicate 1) and DCC mutant (replicate 1) embryos. rex sites were sorted according to their SDC-3 occupancy in wild-type embryos. Negative supercoiling correlates with strength of DCC occupancy and is eliminated in mutants (sdc-2) that prevent DCC binding.
We found that many high-affinity rex sites have local DCC-dependent negative supercoils: The supercoils are observed in wild-type animals but not in DCC mutants (Fig. 4A,B). The level of supercoiling correlates positively with the strength of DCC binding, and the supercoiling level at rex sites is higher than at TSSs.
The high levels of supercoiling at high-occupancy rex sites could result from condensin translocation, as shown previously for Saccharomyces cerevisiae condensin, which moves processively along DNA over long (≥10 kb) distances (Terakawa et al. 2017). Supercoiling of chromatin fibers driven by friction between chromatin and translocating condensin molecules could potentially extrude loops, as has been suggested for cohesin, another SMC-family member (Rusková and Račko 2021).
The second class of DCC binding sites on X lacks X-enriched motifs, has less DCC occupancy, occurs typically in promoters and enhancers (Supplemental Fig. S6A), and has DCC occupancy only when linked to rex sites on X Chromosomes (Jans et al. 2009). The supercoiling profiles at these secondary sites do not change in DCC mutants (Supplemental Fig. S6B,C) but correspond instead to the transcriptional status of the site, with TSSs having higher levels of supercoiling than enhancers.
To explore the global effects of DCC binding on X Chromosomes, we asked whether the DCC creates supercoiling domains along X Chromosomes and whether supercoils at rex sites influence the formation of TADs. Mechanisms of TAD formation in eukaryotes have been under intense investigation. One proposed mechanism is through DNA supercoiling (Racko et al. 2018, 2019; Yan et al. 2018). Experimental evidence for such a model is lacking. Although supercoiling domains have been observed in human cells (Naughton et al. 2013), they are smaller than TADs. Moreover, only 30% of TAD boundaries in humans are close to boundaries of supercoiling domains.
Even though supercoils occur at the very rex sites that trigger formation of TAD boundaries, the supercoils do not propagate beyond 500 bp (Fig. 4A). This extent of supercoil propagation is less than the size of a TAD or even a TAD boundary itself. Hence, the limited DCC-dependent supercoiling at rex sites cannot directly change the interaction frequencies on a megabase scale to create a TAD. The DCC must create TADs through mechanisms other than DNA supercoiling.
Although the limited spread of supercoils around rex sits points against a direct role of supercoiling in delineating megabase-scale TADs, it is probable that a ∼500-bp supercoil mini-domain could act as barrier to loop extrusion and propagation in C. elegans. In the absence of both CTCF, the zinc-finger CCCTC-binding factor that forms TAD boundaries in mammals, and convergently oriented sequence motifs around loop bases in C. elegans, the ∼500-bp supercoil mini-domains provide a testable alternative for barrier to loop propagation and will be a key direction in future studies.
To assess the possibility that the DCC might create supercoiling domains on X Chromosome that are smaller than TADs, we looked for supercoiling domains of 100 kb to 1 Mb in size by plotting BP signal in embryos along each chromosome using a 1000-bp sliding window (Supplemental Fig. S7). We found the average supercoiling level to follow a uniform random pattern that cannot be attributed to supercoiling domains. It is also not correlated with TADs or TAD boundaries on X Chromosomes or autosomes. Although a higher average BP signal occurs at chromosome ends than centers, the strong enrichment of simple repeats at ends causes more variability in DNA amplification during library preparation and is the likely cause of increased signal. Thus, chromosome-wide supercoiling domains are not evident in C. elegans.
Strategy to map SS DNA
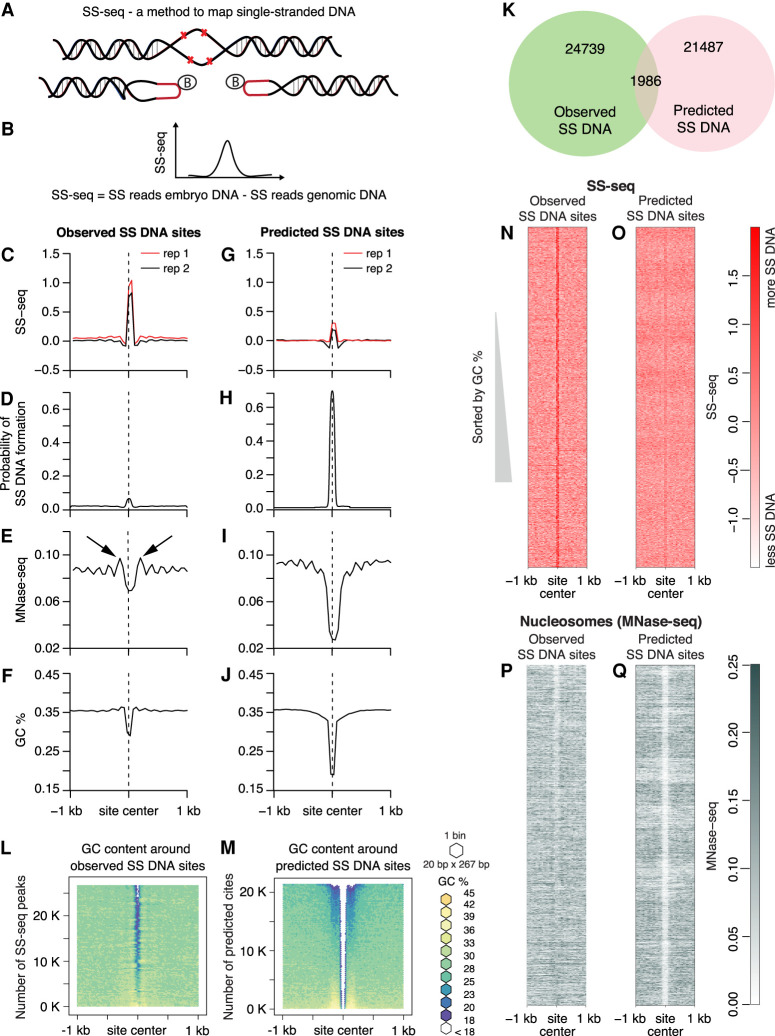
DNA supercoils can facilitate DNA strand separation, creating sites of SS DNA (Kowalski et al. 1988). Such supercoiling-induced strand separation is proposed to have physiological significance, such as activation of gene transcription (Kouzine et al. 2008). To analyze the distribution of SS DNA and non-B DNA structures such as Z-DNA and cruciforms in C. elegans, we mapped genomic sites that are reactive to potassium permanganate (Fig. 5A,B; Supplemental Fig. S8A).
Figure 5.
Nucleosome positioning and AT-rich regions correlate with single-stranded DNA. (A,B) Strategy for genome-wide mapping of single-stranded DNA (SS-seq). (A) When KMnO4 enters purified nuclei, it modifies unpaired pyrimidine bases and prevents reannealing, creating stable single-stranded (SS) DNA. To map non-B DNA, purified KMnO4-treated DNA is digested with the single-strand-specific S1 nuclease, and exposed DNA ends are ligated to biotinylated hairpin adapters. Following DNA sonication to generate 300-bp fragments, biotinylated DNA ends are enriched on streptavidin beads, and a second set of adapters is ligated. After digestion of adapter hairpins, the library is amplified and sequenced. The same procedure is applied to naked genomic DNA as a control. Details of the method are in Supplemental Figure S8A. (B) The distribution of the difference in counts from KMnO4-treated nuclear DNA and genomic control DNA is calculated genome-wide. The SS-seq signal represents the residual signal obtained by subtracting SS reads from naked genomic DNA from the SS reads from embryo DNA. Sites of KMnO4 reactivity are identified by peaks in the SS-seq signal. (C–J) Multiparamteric comparison of observed SS-seq peaks and predicted single-stranded regions. Average profiles of SS-seq in the 2-kb region around observed (C) and predicted (G) SS DNA sites were plotted at 50-bp resolution. Average profiles of SIST-predicted probability of strand separation in the 2-kb region surrounding observed (D) and predicted (H) SS DNA sites. Nucleosome profile (MNase-seq) in the 2-kb region around observed (E) and predicted (I) SS DNA sites. Black arrows indicated well-positioned nucleosomes. Average GC content in the 2-kb region around observed (F) and predicted (J) SS DNA sites is plotted at 50-bp resolution. (K) Venn diagram shows intersection between predicted and observed SS DNA sites. (L,M) Heatmaps of GC content in the 2-kb region around observed SS-seq peaks and predicted SS DNA sites. SS-seq peaks and predicted sites were sorted by GC content; the surrounding sequences were aligned on top of each other and then binned (20 bp × 267 bp for L and 20 bp × 215 bp for M). Each bin was colored according to its GC percentage. (N,O) Heatmaps of SS-seq signal in the 2-kb region surrounding observed SS-seq peaks and predicted SS DNA sites that were sorted as in L and M. (P,Q) Heatmaps of MNase-seq in the 2-kb region around observed SS-seq peaks and predicted SS DNA sites, which were sorted as in L and M.
In prior studies, SS DNA and alternative DNA structures were mapped in mammalian cell lines using potassium permanganate foot printing followed by high-throughput sequencing (ssDNA-seq; Kouzine et al. 2013b). When potassium permanganate enters the nucleus, it modifies unpaired pyrimidine base pairs and prevents them from reannealing, thereby creating stable SS DNA. In these studies, permanganate-treated DNA was purified and then digested with the single-strand-specific nuclease S1, creating double-stranded breaks in regions with unpaired bases. DNA at these breaks was extended with biotinylated nucleotides using terminal transferase (TdT), enriched on streptavidin beads, and converted into a sequencing library. This method has the disadvantage that it requires significantly more starting material than can easily be obtained from worms.
To map SS DNA in worms, we created the method we called SS-seq by combining ssDNA-seq (Kouzine et al. 2013b) with a method to sequence rare double-stranded breaks (END-seq) (Fig. 5A,B; Supplemental Fig. S8A; Canela et al. 2016). Instead of modifying DNA ends using TdT, we ligated biotinylated hairpin adapters to DNA at the breaks. DNA was sonicated, and biotinylated ends were enriched on streptavidin beads. A second adapter was then ligated, hairpins were digested, and the library was amplified and sequenced. This combination of methods, which we refer to as SS-seq, permits mapping of SS DNA using less starting material than the original ssDNA-seq method. To account for any bias arising from sequence-specific binding of potassium permanganate, we performed the same procedure on naked genomic DNA. To calculate SS-seq signal, normalized counts of the genomic DNA sample were subtracted from the nuclear sample. Previous studies showed that SS DNA and other non-B DNA structures, such as Z DNA, correspond to sites of potassium permanganate reactivity (Kouzine et al. 2017). To identify specific types of non-B DNA structure, we complemented our mapping of potassium permanganate reactivity sites with computational predictions of non-B DNA.
Nucleosome positioning correlates with SS DNA at TASs, TESs
We performed SS-seq in wild-type embryos and identified sites of potassium permanganate reactivity genome-wide by calling peaks in the SS-seq signal (Fig. 5C). To identify specific non-B DNA structures at these sites, we used stress-induced structural transitions (SIST) (Zhabinskaya et al. 2015) to computationally analyze the potential of the DNA sequences in SS-seq peaks to form non-B DNA structures. DNA sequence and torsional stress are the main determinants of non-B DNA formation. Although DNA methylation changes mechanical properties of DNA and the energy of strand separation (Severin et al. 2011; Nardo et al. 2015), it is largely absent in C. elegans and therefore cannot be a factor in the formation of non-B DNA. The SIST tool uses a statistical mechanic-based model to predict DNA structural transitions, such as SS DNA, cruciforms, and Z DNA in DNA molecules under supercoiling stress. We found that SS DNA was the only non-B DNA structure predicted at these sites (Fig. 5D). AT-rich sequences have a higher propensity for single-strand formation. Consistent with this prediction, we found DNA sequences corresponding to SS-seq peaks to be highly AT-rich (Fig. 5F). We also asked whether other sites occur in the genome that are predicted to form non-B DNA structures and whether these predicted sites showed potassium permanganate labeling at a level lesser than the threshold used during peak calling. After running SIST on the whole genome, we identified 1519 sites of potential cruciforms and 522 sites of Z DNA. However, we did not see significant SS-seq signal at these sites, suggesting either that cruciform extrusions and Z DNA do not feature in the C. elegans genome or that potassium permanganate reactivity is considerably lower for cruciform and Z DNA than for SS DNA in our experimental setup. We also identified 23,473 sites with high probability to form SS DNA (Fig 5H). Consistently, these sites have very high AT content (Fig. 5J). However, only 1986 of predicted SS DNA sites coincided with the observed SS-seq peaks (Fig. 5K). When we plotted SS-seq signal at these predicted sites, we found it to be much smaller than the SS-seq signal at observed SS-seq peaks (Fig. 5G). Heatmaps of AT content around observed SS peaks (Fig. 5L) and predicted SS DNA sites (Fig. 5M) showed that both observed SS peaks and predicted SS DNA were centered at AT-rich sequences, although predicted SS DNA sites consistently showed higher AT content. Furthermore, when we compared AT-content heatmaps (Fig. 5L,M) with the corresponding SS-seq-signal heatmaps (Fig. 5N,O), the sharpness and the amplitude of the SS-seq signal did not scale with an increase in AT content, suggesting that high AT content is not the sole determinant of single-strandedness.
Two major determinants of DNA strand separation are the AT content of underlying DNA sequence and the magnitude of superhelical stress in DNA. Because predicted SS DNA sites have significantly higher AT content and yet significantly smaller SS-seq signal than observed SS-seq peaks, we speculate that predicted sites are subjected to less supercoiling stress than the observed sites.
Active transcription is known to be the strongest source of torsional stress in vivo. To investigate whether transcription-related supercoiling stress is responsible for the higher SS-seq signal in observed SS DNA sites, we analyzed their positions in the genome. Most of the observed SS DNA sites are located inside genes, most often in introns (Supplemental Fig. S8B). However, the magnitude of SS-seq signal at neither the observed SS peaks nor the predicted SS DNA sites showed any correlation with the strength of transcription (Supplemental Fig. S8C,D).
Because nucleosome assembly, stability, and mobility are strongly dependent on underlying DNA sequence and torsional stress in the DNA, we investigated the relationship between nucleosome positioning and propensity for SS DNA formation by plotting MNase-seq signal (Steiner and Henikoff 2014) around observed and predicted SS-seq sites. We found strong MNase-seq peaks flanking the observed SS DNA sites (Fig. 5E, black arrows), indicating phased nucleosomes, which are arrays of regularly spaced nucleosomes. No such phased nucleosomes were observed around predicted sites (Fig. 5I). Heatmaps of nucleosome positions showed that all SS-seq peaks, including ones with relatively low AT content, have flanking nucleosomes (Fig. 5P,Q). Compared with the predicted sites, more of the observed SS-seq sites have symmetrically positioned nucleosomes on both sides (Supplemental Fig. S8E,F).
Based on this observation, we propose that phased nucleosomes exert torsional stress on to the neighboring DNA, consequently leading to DNA melting. Nucleosomal DNA on average constrains 1.05 supercoils through its interaction with the histone octamer. Torsional strain propagating through a chromatin fiber could be imparted onto linker DNA upon en bloc rotation of a nucleosome (Levens et al. 2016), leading to DNA melting, especially at AT-rich sequences. Additionally, nucleosome motion that can alter inter-nucleosomal spacing can also alter DNA topology. Thus, a change in nucleosome spacing that leads to a reduction in linking number can be expected to release torsional stress through melting of linker DNA (Nikitina et al. 2017).
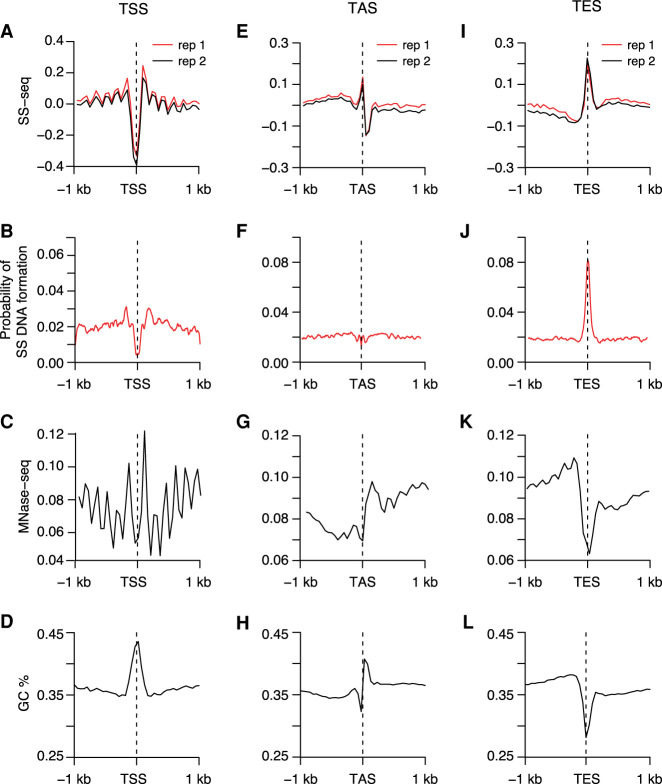
Because DNA melting at TSSs is one of the primary mechanisms of gene regulation, we plotted SS-seq profiles around TSSs of 4221 highly transcribed genes. We observed a strong reduction in potassium permanganate in the immediate vicinity of TSSs (Fig. 6A). SIST modeling predicted that this region would have lower-than-genome-average probability of forming SS DNA (Fig. 6B). Based on the observed SS-seq profiles and computational predictions, we propose that TSSs are resistant to melting under torsional stress, thus mitigating spurious transcriptional up-regulation. Analysis of SS-seq profiles of regions beyond the immediate vicinity of TSSs reveals prominent SS-seq peaks that perfectly coincided with phased nucleosomes (Fig. 6A,C). Formation of SS DNA at these sites can be explained by the partial unwrapping and/or turnover of nucleosomes that is known to accompany strong transcription. Nucleosome unwrapping releases negative supercoils, which in turn can create SS DNA. Such DNA melting accompanying nucleosome disassembly has been observed in vitro (Jordano et al. 1984).
Figure 6.
SS DNA is a universal feature of TASs and TESs but not TSSs. (A,E,I) Average profiles of SS-seq in the 2-kb region around TSSs (A), TASs (E), and TESs (I) are plotted at 50-bp resolution. (B,F,J) Plots show average profiles of the SIST-predicted probability of strand separation in the 2-kb region surrounding TSSs (B), TASs (F), and TESs (J). (C,G,K) Plots of nucleosome profiles (MNase-seq) in the 2-kb region surrounding TSSs (C), TASs (G), and TESs (K) show that nucleosome positioning corresponds with SS DNA at TASs and TESs. (D,H,L) Average GC content is plotted in the 2-kb region around TSSs (D), TASs (H), and TESs (L) at 50-bp resolution.
We analyzed SS-seg signal in two other genomic regions important for gene regulation in C. elegans, TASs and TESs. TASs represent the actual splicing junctions between leader RNAs and 5′ ends of pre-mRNAs. Potentially, Pol II might slow down transcription elongation as it reaches DNA encoding TASs to allow the trans-splicing machinery to bind nascent RNAs (Brodsky et al. 2005; Alexander et al. 2010). Therefore, we investigated formation of SS DNA structures at DNA encoding TASs.
Because the proximity of a TAS to its corresponding TSS might influence the SS-seq signal at the TAS, we chose genes that have TASs positioned at least 50 bp away from known TSSs using the extended data set of TSSs. We identified 9918 such genes and plotted SS-seq signal at their TASs and TESs. Unlike with TSSs (Fig. 6A), we observed strong peaks of SS-seq at TESs (Fig. 6I; Supplemental Fig. 9A,B) and smaller peaks at TASs (Fig. 6E; Supplemental Fig. S9A,B). Both TESs and TASs have AT-rich sequences, with TESs being more AT-rich (Fig. 6H,L; Supplemental Fig. S9C,D), and both different from TSSs with GC-rich regions (Fig. 6D). The percentage and extent of AT-rich sequences at TASs and TESs are highly variable, suggesting that a broad continuum for these two parameters facilitates SS DNA formation.
Predictions of SS DNA formation showed that TES sequences become single-stranded under torsional stress (Fig. 6J). For TASs, which have lower AT content (Fig. 6H) and visibly smaller SS-seq signal (Fig. 6E) compared with that of TESs (Fig. 6I,L), SIST modeling predicted a markedly lower probability of SS DNA formation (Fig. 6F).
Comparable SS-seq peaks were observed for high and low expressed genes (Supplemental Fig. S9A,B), indicating that the formation of SS DNA at TASs and TESs does not depend on transcription. TASs and TESs are flanked by strongly positioned nucleosomes (Fig. 6G,K). Based on these observations, we propose that formation of SS DNA at TASs and TESs is facilitated by torsional stress resulting from nearby nucleosomes rather than transcription.
To summarize, TSSs, which are generally GC-rich, are resistant to DNA melting (Fig. 6A–D). Predicted SS DNA sites, which are AT-rich but do not have phased nucleosomes surrounding them, are also resistant to melting (Fig. 5G–J). TESs, which show a confluence of high AT content and symmetrically positioned phased nucleosomes, show the most pronounced SS-seq peaks (Fig. 6I–L). Thus, single-strandedness of genomic regions is conditional on both high AT content and the presence of symmetrically positioned nucleosomes at their flanks. Either parameter in isolation is not sufficient to cause single-strandedness.
Discussion
The nonrandom distribution of GC/AT content within a genome is common among numerous species (Zhang et al. 2004), but its biological significance has not been well established. We extended the observation that C. elegans promoters are GC-rich (Chen et al. 2013; Kranz et al. 2013) by finding that the hallmark of these promoters is not simply high GC content but rather a GC-rich region bordered by a sharp transition in GC/AT content. The GC-rich regions flanked by sharp GC/AT boundaries are wider at promoters of X Chromosomes than autosomes and correlate strongly with the broader spread of supercoiling. We show that this unique sequence feature, and not simply a stretch of high GC content, demarcates boundaries of supercoiling propagation, chromatin accessibility, and nucleosome positioning in C. elegans.
Enhancers are similar to promoters in having high GC content, sharp GC-to-AT transition, and high chromatin accessibility, but they differ in having weaker transcription and a lack of negative supercoiling. These results suggest that, although supercoil propagation in chromatin can be amplified by specific sequences, a threshold level of torque generated by DNA-based motors is required to initiate supercoiling.
A computational model based on sequence-dependent physical parameters of DNA, such as curvature and bendability, predicted the location of supercoils at TSSs (Kim et al. 2018). The C. elegans supercoiling profiles we measured in vivo provide experimental validation for this prediction. While analyzing the relationship between supercoils and GC content at TSSs in humans and flies, we found that similar to C. elegans, the GC-rich stretch in humans and the AT-rich stretch in flies track with the spread of supercoils. These findings indicate that a direct outcome of nonrandom distribution of GC/AT content at TSSs is a pattern of locally restricted propagation of transcription-coupled supercoiling. Single-molecule studies in vitro showing strong dependence of supercoil propagation on DNA sequence corroborate our conclusions (Kim et al. 2018).
A prominent model of transcription-driven supercoiling (twin-domain) predicts positive supercoils in front of transcribing polymerase and negative in the back (Tsao et al. 1989). This model does not take into account that eukaryotic DNA is organized into nucleosomes that can be destabilized by transcription and can cancel or contribute to polymerase-generated supercoils (Teves and Henikoff 2014). Indeed, the predictions of the twin-domain model have not been validated in human cells. Instead, these studies (Kouzine et al. 2013a; Naughton et al. 2013; Ghosh et al. 2019) revealed wide variations in negative supercoils on both sides of the active TSSs depending on the cell type.
Beyond the nature of supercoiling, our work revealed that the degree of supercoiling also varies considerably across different developmental states. We found a stark reduction in negative supercoiling at promoters in starvation-induced diapause larvae of C. elegans, even though global transcription levels are comparable to that of embryos. Hence, the role of transcription in generating negative supercoils is not absolute, and additional mechanisms must exist that can modulate global levels of transcription-driven supercoiling. Altered supercoiling at promoters might provide specificity for developmental differences that are generally attributed to epigenetic processes, such as nucleosome modification or DNA methylation.
DNA supercoiling has been proposed to regulate higher-order chromatin organization at intermediate genomic-length scales (Naughton et al. 2013). Although our genome-wide map of supercoiling in C. elegans revealed no evidence for such large-scale supercoiling domains, we found that the condensin DCC creates local negative supercoils at its highest occupancy binding sites on the X Chromosome. Condensin-induced supercoiling represents the first demonstration in vivo of changes in local DNA supercoiling induced by a DNA-binding motor protein complex. Whether local changes in supercoiling at DCC binding sites can act as delimiters of topological domains is an important model to test.
Propagation of torsional stress through chromatin has been proposed to alter chromatin topology at various length scales and to introduce distortions in the canonical double helical B-DNA structure. Our computational analysis of the C. elegans genome revealed very few sites with the propensity to form noncanonical DNA structures. However, our potassium permanganate mapping identified thousands of SS DNA sites with high AT content. Computationally predicted single-stranded regions were largely absent from our permanganate-reactivity map, even though these regions had considerably higher AT content than that observed for single-stranded regions. Although AT content alone was a poor predictor of single-strandedness, we found that regions showing high AT content and flanked by phased nucleosomes on both sides were prevalently single-stranded. This combination was particularly prominent at TESs. In eukaryotes, polymerase back-tracking and accumulation occurs as Pol II approaches the 3′ ends of genes (Proudfoot 2016). Accumulation of Pol II at TESs is particularly prominent in C. elegans (Kruesi et al. 2013). Our work suggests that the co-occurrence of phased nucleosomes and sequences prone to duplex-destabilization at TESs might facilitate transcription termination in C. elegans.
Although our work provides a detailed view of the relationship between sequence and nucleosome phasing in governing duplex stability, the ensemble average nature of the data precludes fine-scale analysis of torsional stress propagation and formation of alternative structures at an allelic level in single cells. Essential future steps are to measure the propagation of torsional stress and the rate at which alternative DNA structures form and dissolve at regulatory elements in living cells and multicellular organisms.
Methods
Nematode culture
Wild-type C. elegans nematodes (N2 Bristol) were grown to adulthood on NG agar plates with the Escherichia coli strain HB101. Gravid adults were bleached to obtain embryos, and the embryos were hatched in M9 medium without food to obtain developmentally arrested, synchronized L1 larvae (L1s). To obtain mixed-stage embryos for analysis, approximately 105 starved, synchronized L1s were cultured on NG agar plates with HB101 for 96 h, and the resulting gravid adults were bleached to obtain mixed-stage embryos. These embryos were either frozen for subsequent BP and SS-seq analysis, or they were placed back into culture without food and allowed to hatch overnight, and the resulting synchronized L1s were frozen for BP and SS-seq analysis.
To obtain dosage-compensation-defective embryos for BP and SS-seq analysis, worms carrying the dosage compensation mutation sdc-2(y93) were grown as above, except that after synchronization, the sdc-2 mutant L1s were grown on RNAi plates (NG agar with 1 mM IPTG and 100 μg/mL carbenicillin) seeded with HT115 bacteria carrying a plasmid that produced double-stranded sdc-2 RNA (Ahringer Library). Expression of double-stranded RNA was induced from the plasmid before adding the L1s by incubating the bacteria-containing RNAi plates overnight at 25°C. sdc-2(y93) animals were grown on the RNAi plates for 96 h at 20°C. The resulting gravid adults were bleached and the embryos frozen. The effect of RNAi was confirmed by observing the absence DCC localization on the X Chromosome with immunofluorescence microscopy using a custom antibody (r699) against DCC subunit DPY-27 (Chuang et al. 1994) and by observing lethality after counting viable larvae following incubation of embryos on NG plates.
Preparation of genomic DNA for control samples
Frozen mixed-stage embryos were resuspended 15 mM Tris (pH 7.5), 0.5 M NaCl, 1% SDS and digested with Proteinase K for 1 h at 55°C. DNA was extracted once with phenol-chloroform and precipitated once with ethanol. DNA was resuspended in 1 mL of H2O, treated with 100 μg of RNase A for 1 h at 37°C, extracted with phenol-chloroform, and precipitated with ethanol.
Preparation of BP-seq libraries
The BP-seq protocol was adapted from that of Naughton et al. (2013). Nuclei were isolated from frozen embryos or L1s. L1s were first ground in liquid nitrogen to disrupt them. Approximately 3 × 106 to 4 × 106 embryos or ground L1s were resuspended in 1 mL of buffer A (15 mM Tris at pH 7.5, 2 mM MgCl2, 340 mM sucrose, 0.2 mM spermine, 0.2 mM spermidine, 0.1% Triton X-100, 0.25% NP-40, 0.5 mM PMSF, and a protease inhibitor cocktail set III from Calbiochem 1:1000 dilution), dounced on ice in a 2-mL glass homogenizer with pestle A and pestle B for 15 times each, and spun for 15 min at 4°C at 100g. The nuclei-containing supernatant was saved on ice, the pellet was resuspended in 1 mL of buffer A, and the homogenization procedure above was repeated twice. Nuclei from the three supernatants were pooled together and concentrated to 200 μL by spinning at 1000g for 10 min at 4°C. Nuclei were checked for integrity and quantified using fluorescence microscopy after staining with the DNA-intercalating dye DAPI. Under 10× magnification, intact nuclei appear as small dots with well-resolved edges. Preparations with <95% high-quality nuclei were discarded. We also checked for the absence of debris as an indicator that the nuclei were intact.
EZ-Link psoralen-PEG3-biotin (BP; Thermo Fisher Scientific) was added to the nuclei to a final concentration of 1.38 × 10−2 mg/mL, nuclei were cross-linked for 30 min using a UVL-56 Blak-Ray lamp (long-wave UV 366 nm), and nuclei were pelleted for 10 min at 4°C at 1000g. The supernatant was removed, and the nuclei were resuspended in 200 μL of buffer A and digested with Proteinase K (10 μg/mL final) for 30 min at 55°C. DNA was extracted once with phenol-chloroform and precipitated once with ethanol. DNA was resuspended in 130 μL of H2O, treated with RNase A (10 μg/mL final) for 1 h at 37°C, extracted with phenol-chloroform, and precipitated with ethanol. DNA was resuspended in 130 μL of H2O and sonicated to an average size of 150 bp using a Covaris sonicator.
The BP-containing DNA was then trapped on magnetic Dynabeads MyOne Streptavidin T1 (Invitrogen) for further manipulation. Before incubation with BP-containing DNA, 50 μL of Dynabeads were washed with wash buffer (WB; 10 mM Tris at pH 7.5, 1 M NaCl, 1 mM EDTA), and resuspended in 130 μL of high-salt WB (10 mM Tris at pH 7.5, 2 M NaCl, 1 mM EDTA). The washed Dynabeads were added to 130 μL of DNA (1 M final salt), and incubated for 1 h while rotating. The DNA-containing beads were collected with a magnet, and the supernatant was saved for BP-depleted DNA samples. DNA-containing beads were washed three times with 200 μL of WB and resuspended in 34 μL of EB (10 mM Tris at pH 7.5).
Libraries for BP-enriched samples were prepared on the beads. Libraries for BP-depleted samples were made using ∼1 μg of the unbound, BP-depleted DNA in the supernatant. After each step in library preparation, the DNA-containing beads were washed three times with WB followed by one wash with EB. The BP-depleted DNA samples were cleaned using a Qiagen PCR purification column.
To make libraries, the DNA ends were repaired using the End-It DNA end repair kit from Epicenter (34 μL of BP-depleted DNA or DNA-containing beads, 5 μL 10× end-repair buffer, 5 μL of 2.5 mM dNTP mix, 5 μL of 10 mM ATP, 1 μL end-repair enzyme mix; incubated for 45 min at room temperature). To add 3′-end A overhangs, 34 μL of BP-depleted DNA or DNA-containing beads was mixed with 5 μL NEB buffer 2, 10 μL of 10 mM dATP, 1 U of Klenow fragment (3′–5′-exo) and incubated for 30 min at 37°C. Barcoded adapters were ligated using Quick Ligation from NEB (12 μL of input DNA or beads, 15 μL 2× DNA ligase buffer, 1 μL of 0.125 μM NEXTflex adapters, 2 μL DNA ligase; incubated for 15 min at room temperature). Following adapter ligation, DNA-containing beads were cleaned with WB three times and once with EB and were resuspended in 37 μL of EB. BP-depleted DNA was cleaned using a Qiagen PCR column and eluted in 20 μL of EB. Unligated adapters were removed from BP-depleted samples using 10 μL of AMPure beads and eluted in 50 μL of EB. Libraries were PCR-amplified (37 μL of BP-depleted DNA or DNA-containing beads, 10 μL 5× Phusion HF buffer, 0.4 μL 25 mM dNTP, 2 μL of NEXTflex primer mix at 12 μM, 1 μL Phusion DNA Polymerase) using the following program: 2 min at 98°C; 30 sec at 98°C, 30 sec at 65°C, and 60 sec at 72°C for 16–30 cycles; and 5 min at 72°C. Libraries were cleaned using a Qiagen PCR purification column. Some samples required an additional AMPure bead clean-up to remove unused primers and adapters. Libraries were sequenced using the Illumina HiSeq 4000 platform with 50-bp single end reads.
Control genomic DNA samples were prepared using the same steps outlined above. However, previously purified genomic DNA was used in place of DNA purified from nuclei. BP-seq libraries were prepared from two independent biological replicates of each genotype or developmental stage, and the scatter plots and Pearson's correlations comparing the various independent data sets are presented in Supplemental Figure S10, A through D.
Historically, supercoiling has been detected as a change in psoralen binding following supercoil relaxation upon treatment with topoisomerase inhibitors or ionizing radiation (Sinden et al. 1980; Kouzine et al. 2013a). Instead, our approach is tailored to C. elegans embryos, which are impenetrable to most drugs and have limited information about topoisomerases. Drugs can be fed to adult worms, but their rate of metabolism and nuclear access are not clear, making it unknown whether topoisomerase inhibitors would enter embryo nuclei. Furthermore, the C. elegans supercoiling regions are 400 bp in size on average, and nicks produced by ionizing radiation would not be frequent enough to relax such small regions, precluding use of radiation to study local supercoils. Supercoiling domains in bacteria are much larger and hence compatible with the frequency of nicks from ionizing radiation (Sinden and Pettijohn 1981).
Preparation of SS-seq libraries
The SS-seq protocol was adapted from the method of Kouzine et al. (2013b) and the library preparation from the method of Canela et al. (2016). SS-seq was performed with frozen embryos or L1s. L1s were first ground in liquid nitrogen. Approximately 2 × 106 intact embryos or ground L1s were resuspended in low-salt buffer (15 mM Tris at pH 5, 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 0.5 mM EGTA, 300 mM sucrose). The resuspension buffer used for SS-seq is similar to the one used for BP-seq except for the addition in case of BP-seq of spermine, spermidine, and protease inhibitors, which are added specifically to keep nuclei intact under low-salt conditions. Detergent is also added to the BP-seq buffer to facilitate cell membrane lysis and efficient nuclei isolation. A freshly prepared solution of 100 mM KMnO4 was added to each sample to a final concentration of 40 mM and incubated for 80 sec at room temperature. To stop the reaction, an equal volume of stop solution (700 mM β-mercaptoethanol, 50 mM EDTA, 1% SDS) was added. Samples were digested with Proteinase K (10 μg/mL final) for 1 h at 55°C. DNA was extracted once with phenol-chloroform, precipitated once with ethanol, resuspended in 500 μL of H2O, treated with RNase A (10 μg/mL final) for 1 h at 37°C, extracted with phenol-chloroform, and precipitated with ethanol. As a control, a sample was treated with H2O instead of KMnO4 and processed as described above.
To block free 3′ DNA ends from being cleaved before nuclease treatment by enzymes such as topoisomerase, samples were treated with cordycepin 5′-triphosphate sodium salt. Approximately 25 μg of extracted DNA was incubated with 100 μM cordycepin 5′-triphosphate sodium salt and 40 U of TdT in a final reaction of 150 μL for 1 h at 37°C. DNA was extracted once with phenol-chloroform and precipitated once with ethanol. About 5 μg of precipitated DNA was digested with 100 U of S1 nuclease in a 50 μL reaction at pH 4.6 for 20 min at 37°C. Because the digestion reaction was performed after DNA purification, a step that removes histones and all other proteins, it is unlikely that any potential formation of triplex DNA caused by low pH at this step would alter SS DNA mapping. DNA was extracted again with phenol-chloroform and ethanol-precipitated. Digestion was verified by electrophoresis through an agarose gel. A smear ∼3 kb is expected for a KMnO4 sample, whereas intact genomic-size DNA is expected for an H2O-treated sample.
To prepare libraries, DNA ends were repaired with the DNA end-repair kit from Epicenter (34 μL of DNA, 5 μL 10× end-repair buffer, 5 μL of 2.5 mM dNTP mix, 5 μL of 10 mM ATP, 1 μL end-repair enzyme mix; incubated for 45 min at room temperature). To add 3′-end A overhangs, 34 μL of DNA was mixed with 5 μL NEB buffer 2, 10 μL of 10 mM dATP, 1 U of Klenow fragment (3′–5′-exo) and incubated for 30 min at 37°C. DNA was cleaned using a Qiagen PCR purification column after each step. Hairpin biotinylated adapters (/5Phos/GATCGGAAGAGCGTCGTGTAGGGAAAGAGTGUU/iBiodT/U/iBiodT/UUACACTCTTTCCCTACACGACGCTCTTCCGATC*T) were ligated using 2 μL of Quick Ligation (NEB) in a 30 μL reaction at room temperature with 0.08 μM adapter for 15 min. DNA was purified with a Qiagen PCR purification column and eluted in 20 μL of EB. To remove unligated adapters, 20 μL of sample was mixed with 10 μL of AMPure beads. DNA was eluted with 130 μL of EB and sonicated to an average size of 300 bp using a Covaris sonicator. Before DNA was incubated with 50 μL of magnetic Dynabeads MyOne Streptavidin T1 (Invitrogen), the beads were washed with wash buffer WB and resuspended in 130 μL of high-salt WB. Next, 50 μL of Dynabeads was added to 130 μL of DNA (1 M final salt) and incubated for 1 h while rotating. Beads were washed three times with WB and once with EB, resuspended in 34 μL of EB, and subjected to end repair and 3′-end overhang addition reactions as described above. After the final wash, beads were resuspended in 22 μL of EB. Second hairpin biotinylated adapters (/5Phos/GA TCG GAA GAG CAC ACG TCU UUU UUU UAG ACG TGT GCT CTT CCG ATC* T) were ligated in 50 μL reactions with 0.08 μM adapter concentration and 3 μL of quick DNA ligase. After ligation, beads were washed and resuspended in 45 μL of EB. Hairpins were digested to remove DNA from the beads using 1 U of USER enzyme (NEB) in a 50 μL reaction for 1 h at 37°C. DNA was purified on a Qiagen column and PCR-amplified with barcoded TruSeq primers (37 μL of DNA, 10 μL 5× Phusion HF Bbuffer, 0.4 μL 25 mM dNTP, 1 μL of TruSeq universal primer at 6.5 μM, 1 μL of TruSeq barcoded primer at 6.5 μM, 1 μL Phusion DNA Polymerase) using the following program: 2 min at 98°C; 30 sec at 98°C, 30 sec at 65°C, and 60 sec at 72°C for 16 cycles; and 5 min 72°C. Libraries were cleaned using a Qiagen PCR purification column. Some samples required an additional AMPure bead cleanup step to remove unused primers and adapters. Libraries were sequenced on Illumina HiSeq 4000 platform with 50-bp single end reads.
Control genomic DNA samples were prepared using the same steps listed above. However, previously isolated genomic DNA was used in place of DNA purified from animals.
SS-seq libraries were generally prepared from two independent biological replicates of each genotype or developmental stage, and the scatter plots and Pearson's correlations comparing the various independent data sets are presented in Supplemental Figure S11.
Data processing and visualization
For each Illumina library, DNA sequences were first processed with cutadapt (Martin 2011) to remove read-through adapter sequences and then mapped to the WS230 C. elegans genome sequence edition using Bowtie (Langmead et al. 2009), retaining only uniquely mapped reads. Normalized bigWig ratio files were generated using deepTools2 (Ramírez et al. 2016) with bamCoverage and bigWigCompare commands. For SS-seq samples, only the first base pair of the read was included in the bigWig file using the “offset” bamCoverage command option. For BP samples, the whole read was included. Metaprofiles and heatmaps were generated using deepTools2 computeMatrix and plot heatmap commands. Output of computeMatrix was plotted by a custom R script (R Core Team 2019).
To call SS-seq peaks, reads were separated into forward and reverse strands using SAMtools (Li et al. 2009), and peaks were called using Sicer (Zhang et al. 2008) on each strand separately with the following parameters: species, ce (C. elegans); redundancy threshold, five; window size, 10; fragment size, one; effective genome size, 0.8; gap size, 10; false-discovery rate, 10−3. A custom R script was used to obtain sequences of various genome features (e.g., TASs, TESs) from the WS230 FASTA file, to calculate GC content, and to sort and plot GC-content heatmaps.
To predict sites with high probability of formation of non-B DNA structures, the genome sequence was split into 5-kb sequences. The SIST tool (Zhabinskaya et al. 2015) was run on each sequence with default parameters. Results were aggregated, and sequences with probability greater than 0.2 were identified using a custom R script.
Published data sets used in this study
MNase-seq data are from Steiner and Henikoff (2014): NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) accession number GSE44412. TSS annotations used in Figures 1, 2, 3, and 6 are from Kruesi et al. (2013): GSE43087. ATAC-seq data and annotations for coding promoters and putative enhancers used in Supplemental Figure S2 are from Jänes et al. (2018): GSE114494. SDC-3 ChIP-seq data are from Anderson et al. (2019): GSE128568. Human supercoiling data are from Ghosh et al. (2019): GSE123292. Fly supercoiling data are from Teves and Henikoff (2014): GSE47795.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE153373. Custom R scripts, a readme file, and examples of applications have been uploaded as a Supplemental Code file.
Supplementary Material
Acknowledgments
We thank members of the Meyer laboratory and D. Levens for discussions, N. Fuda and S. Henikoff for critical comments on the manuscript, and D. Stalford for assistance with figures. This work was supported in part by National Institutes of Health grant R35 GM131845 to B.J.M. B.J.M. is an investigator of the Howard Hughes Medical Institute.
Author contributions: K.K. performed all experiments. K.K. and R.P.G. performed data analysis. K.K. and B.J.M. designed the experiments, and K.K., R.P.G., and B.J.M. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.270082.120.
Competing interest statement
The authors declare no competing interests.
References
- Alexander RD, Innocente SA, Barrass JD, Beggs JD. 2010. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell 40: 582–593. 10.1016/j.molcel.2010.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MA, Hillier LW, Waterston RH, Blumenthal T. 2011. A global analysis of C. elegans trans-splicing. Genome Res 21: 255–264. 10.1101/gr.113811.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders L, Guenther MG, Qi J, Fan ZP, Marineau JJ, Rahl PB, Lovén J, Sigova AA, Smith WB, Lee TI, et al. 2014. Genome-wide determination of drug localization. Nat Biotechnol 32: 92–96. 10.1038/nbt.2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson EC, Frankino PA, Higuchi-Sanabria R, Yang Q, Bian Q, Podshivalova K, Shin A, Kenyon C, Dillin A, Meyer BJ. 2019. X chromosome domain architecture regulates Caenorhabditis elegans lifespan but not dosage compensation. Dev Cell 51: 192–207.e6. 10.1016/j.devcel.2019.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, Sternberg PW. 2006. DAF-16/FOXO regulates transcription of cki-1/Cip/Kip and repression of lin-4 during C. elegans L1 arrest. Curr Biol 16: 780–785. 10.1016/j.cub.2006.03.021 [DOI] [PubMed] [Google Scholar]
- Benedetti F, Dorier J, Burnier Y, Stasiak A. 2014. Models that include supercoiling of topological domains reproduce several known features of interphase chromosomes. Nucleic Acids Res 42: 2848–2855. 10.1093/nar/gkt1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky AS, Meyer CA, Swinburne IA, Hall G, Keenan BJ, Liu XS, Fox EA, Silver PA. 2005. Genomic mapping of RNA polymerase II reveals sites of co-transcriptional regulation in human cells. Genome Biol 6: R64. 10.1186/gb-2005-6-8-r64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela A, Sridharan S, Sciascia N, Tubbs A, Meltzer P, Sleckman BP, Nussenzweig A. 2016. DNA breaks and end resection measured genome-wide by end sequencing. Mol Cell 63: 898–911. 10.1016/j.molcel.2016.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RA-J, Down TA, Stempor P, Chen QB, Egelhofer TA, Hillier LW, Jeffers TE, Ahringer J. 2013. The landscape of RNA polymerase II transcription initiation in C. elegans reveals promoter and enhancer architectures. Genome Res 23: 1339–1347. 10.1101/gr.153668.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang P-T, Albertson DG, Meyer BJ. 1994. DPY-27: a chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell 79: 459–474. 10.1016/0092-8674(94)90255-0 [DOI] [PubMed] [Google Scholar]
- Core L, Adelman K. 2019. Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev 33: 960–982. 10.1101/gad.325142.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, Uzawa S, Dekker J, Meyer BJ. 2015. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523: 240–244. 10.1038/nature14450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganji M, Shaltiel IA, Bisht S, Kim E, Kalichava A, Haering CH, Dekker C. 2018. Real-time imaging of DNA loop extrusion by condensin. Science 360: 102–105. 10.1126/science.aar7831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh RP, Shi Q, Yang L, Reddick MP, Nikitina T, Zhurkin VB, Fordyce P, Stasevich TJ, Chang HY, Greenleaf WJ, et al. 2019. Satb1 integrates DNA binding site geometry and torsional stress to differentially target nucleosome-dense regions. Nat Commun 10: 3221. 10.1038/s41467-019-11118-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom KA, Holmes VF, Cozzarelli NR, Meyer BJ. 2002. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev 16: 729–742. 10.1101/gad.968302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvald EB, Sprenger RR, Dall KB, Ejsing CS, Nielsen R, Mandrup S, Murillo AB, Larance M, Gartner A, Lamond AI, et al. 2017. Multi-omics analyses of starvation responses reveal a central role for lipoprotein metabolism in acute starvation survival in C. elegans. Cell Syst 5: 38–52.e4. 10.1016/j.cels.2017.06.004 [DOI] [PubMed] [Google Scholar]
- Hirano T. 2012. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev 26: 1659–1678. 10.1101/gad.194746.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jänes J, Dong Y, Schoof M, Serizay J, Appert A, Cerrato C, Woodbury C, Chen R, Gemma C, Huang N, et al. 2018. Chromatin accessibility dynamics across C. elegans development and ageing. eLife 7: e37344. 10.7554/eLife.37344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans J, Gladden JM, Ralston EJ, Pickle CS, Michel AH, Pferdehirt RR, Eisen MB, Meyer BJ. 2009. A condensin-like dosage compensation complex acts at a distance to control expression throughout the genome. Genes Dev 23: 602–618. 10.1101/gad.1751109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordano J, Montero F, Palacián E. 1984. Dissociation of single-stranded DNA from nucleosomes following modification with acetic anhydride. Biochem Biophys Res Commun 121: 907–914. 10.1016/0006-291X(84)90763-0 [DOI] [PubMed] [Google Scholar]
- Kanne D, Rapoport H, Hearst JE. 1984. 8-Methoxypsoralen-nucleic acid photoreaction: effect of methyl substitution on pyrone vs. furan photoaddition. J Med Chem 27: 531–534. 10.1021/jm00370a017 [DOI] [PubMed] [Google Scholar]
- Kasinathan S, Henikoff S. 2018. Non-B-form DNA is enriched at centromeres. Mol Biol Evol 35: 949–962. 10.1093/molbev/msy010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Ganji M, Kim E, van der Torre J, Abbondanzieri E, Dekker C. 2018. DNA sequence encodes the position of DNA supercoils. eLife 7: e36557. 10.7554/eLife.36557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Hirano T. 1997. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell 90: 625–634. 10.1016/S0092-8674(00)80524-3 [DOI] [PubMed] [Google Scholar]
- Kouzine F, Levens D. 2007. Supercoil-driven DNA structures regulate genetic transactions. Front Biosci J Virtual Libr 12: 4409–4423. 10.2741/2398 [DOI] [PubMed] [Google Scholar]
- Kouzine F, Sanford S, Elisha-Feil Z, Levens D. 2008. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat Struct Mol Biol 15: 146–154. 10.1038/nsmb.1372 [DOI] [PubMed] [Google Scholar]
- Kouzine F, Gupta A, Baranello L, Wojtowicz D, Ben-Aissa K, Liu J, Przytycka TM, Levens D. 2013a. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat Struct Mol Biol 20: 396–403. 10.1038/nsmb.2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzine F, Wojtowicz D, Yamane A, Resch W, Kieffer-Kwon K-R, Bandle R, Nelson S, Nakahashi H, Awasthi P, Feigenbaum L, et al. 2013b. Global regulation of promoter melting in naive lymphocytes. Cell 153: 988–999. 10.1016/j.cell.2013.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzine F, Wojtowicz D, Baranello L, Yamane A, Nelson S, Resch W, Kieffer-Kwon K-R, Benham CJ, Casellas R, Przytycka TM, et al. 2017. Permanganate/S1 nuclease footprinting reveals non-B DNA structures with regulatory potential across a mammalian genome. Cell Syst 4: 344–356.e7. 10.1016/j.cels.2017.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski D, Natale DA, Eddy MJ. 1988. Stable DNA unwinding, not “breathing,” accounts for single-strand-specific nuclease hypersensitivity of specific A+T-rich sequences. Proc Natl Acad Sci 85: 9464–9468. 10.1073/pnas.85.24.9464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz A-L, Jiao C-Y, Winterkorn LH, Albritton SE, Kramer M, Ercan S. 2013. Genome-wide analysis of condensin binding in Caenorhabditis elegans. Genome Biol 14: R112. 10.1186/gb-2013-14-10-r112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruesi WS, Core LJ, Waters CT, Lis JT, Meyer BJ. 2013. Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. eLife 2: e00808. 10.7554/eLife [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levens D, Baranello L, Kouzine F. 2016. Controlling gene expression by DNA mechanics: emerging insights and challenges. Biophys Rev 8: 23–32. 10.1007/s12551-016-0243-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LF, Wang JC. 1987. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci 84: 7024–7027. 10.1073/pnas.84.20.7024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu I-F, Sutherland JH, Cheng B, Tse-Dinh Y-C. 2011. Topoisomerase I function during Escherichia coli response to antibiotics and stress enhances cell killing from stabilization of its cleavage complex. J Antimicrob Chemother 66: 1518–1524. 10.1093/jac/dkr150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Matsumoto K, Hirose S. 2004. Visualization of unconstrained negative supercoils of DNA on polytene chromosomes of Drosophila. J Cell Sci 117: 3797–3805. 10.1242/jcs.01225 [DOI] [PubMed] [Google Scholar]
- Meyer BJ. 2018. Sex and death: from cell fate specification to dynamic control of X-chromosome structure and gene expression. Mol Biol Cell 29: 2616–2621. 10.1091/mbc.E18-06-0397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardo L, Lamperti M, Salerno D, Cassina V, Missana N, Bondani M, Tempestini A, Mantegazza F. 2015. Effects of non-CpG site methylation on DNA thermal stability: a fluorescence study. Nucleic Acids Res 43: 10722–10733. 10.1093/nar/gkv884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton C, Avlonitis N, Corless S, Prendergast JG, Mati IK, Eijk PP, Cockroft SL, Bradley M, Ylstra B, Gilbert N. 2013. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat Struct Mol Biol 20: 387–395. 10.1038/nsmb.2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitina T, Norouzi D, Grigoryev SA, Zhurkin VB. 2017. DNA topology in chromatin is defined by nucleosome spacing. Sci Adv 3: e1700957. 10.1126/sciadv.1700957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K, Bochman ML, Garcia PD, Cejka P, Friedman KL, Kowalczykowski SC, Zakian VA. 2013. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature 497: 458–462. 10.1038/nature12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papantonis A, Larkin JD, Wada Y, Ohta Y, Ihara S, Kodama T, Cook PR. 2010. Active RNA polymerases: mobile or immobile molecular machines? PLoS Biol 8: e1000419. 10.1371/journal.pbio.1000419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Sun Y, Huang SN, Nitiss JL. 2016. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat Rev Mol Cell Biol 17: 703–721. 10.1038/nrm.2016.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L, Hardy CD, Arsuaga J, Cozzarelli NR. 2004. Topological domain structure of the Escherichia coli chromosome. Genes Dev 18: 1766–1779. 10.1101/gad.1207504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ. 2016. Transcriptional termination in mammals: stopping the RNA polymerase II juggernaut. Science 352: aad9926. 10.1126/science.aad9926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racko D, Benedetti F, Dorier J, Stasiak A. 2018. Transcription-induced supercoiling as the driving force of chromatin loop extrusion during formation of TADs in interphase chromosomes. Nucleic Acids Res 46: 1648–1660. 10.1093/nar/gkx1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racko D, Benedetti F, Dorier J, Stasiak A. 2019. Are TADs supercoiled? Nucleic Acids Res 47: 521–532. 10.1093/nar/gky1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Panova T, Miller G, Volkov A, Porter ACG, Russell J, Panov KI, Zomerdijk JCBM. 2013. Topoisomerase IIα promotes activation of RNA polymerase I transcription by facilitating pre-initiation complex formation. Nat Commun 4: 1598. 10.1038/ncomms2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2019. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Rusková R, Račko D. 2021. Entropic competition between supercoiled and torsionally relaxed chromatin fibers drives loop extrusion through pseudo-topologically bound cohesin. Biology (Basel) 10: 130. 10.3390/biology10020130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin PMD, Zou X, Gaub HE, Schulten K. 2011. Cytosine methylation alters DNA mechanical properties. Nucleic Acids Res 39: 8740–8751. 10.1093/nar/gkr578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorska N, Sexton T. 2020. Defining functionally relevant spatial chromatin domains: it is a TAD complicated. J Mol Biol 432: 653–664. 10.1016/j.jmb.2019.12.006 [DOI] [PubMed] [Google Scholar]
- Sinden RR, Pettijohn DE. 1981. Chromosomes in living Escherichia coli cells are segregated into domains of supercoiling. Proc Natl Acad Sci 78: 224–228. 10.1073/pnas.78.1.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden RR, Carlson JO, Pettijohn DE. 1980. Torsional tension in the DNA double helix measured with trimethylpsoralen in living E. coli cells: analogous measurements in insect and human cells. Cell 21: 773–783. 10.1016/0092-8674(80)90440-7 [DOI] [PubMed] [Google Scholar]
- Steiner FA, Henikoff S. 2014. Holocentromeres are dispersed point centromeres localized at transcription factor hotspots. eLife 3: e02025. 10.7554/eLife.02025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szlachta K, Thys RG, Atkin ND, Pierce LCT, Bekiranov S, Wang Y-H. 2018. Alternative DNA secondary structure formation affects RNA polymerase II promoter-proximal pausing in human. Genome Biol 19: 89. 10.1186/s13059-018-1463-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terakawa T, Bisht S, Eeftens JM, Dekker C, Haering CH, Greene EC. 2017. The condensin complex is a mechanochemical motor that translocates along DNA. Science 358: 672–676. 10.1126/science.aan6516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves SS, Henikoff S. 2014. Transcription-generated torsional stress destabilizes nucleosomes. Nat Struct Mol Biol 21: 88–94. 10.1038/nsmb.2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao YP, Wu HY, Liu LF. 1989. Transcription-driven supercoiling of DNA: direct biochemical evidence from in vitro studies. Cell 56: 111–118. 10.1016/0092-8674(89)90989-6 [DOI] [PubMed] [Google Scholar]
- Varghese SS, Ghosh SK. 2020. Stress-responsive Entamoeba topoisomerase II: a potential antiamoebic target. FEBS Lett 594: 1005–1020. 10.1002/1873-3468.13677 [DOI] [PubMed] [Google Scholar]
- Wheeler BS, Anderson E, Frøkjær-Jensen C, Bian Q, Jorgensen E, Meyer BJ. 2016. Chromosome-wide mechanisms to decouple gene expression from gene dose during sex-chromosome evolution. eLife 5: e17365. 10.7554/eLife.17365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Ding Y, Leng F, Dunlap D, Finzi L. 2018. Protein-mediated loops in supercoiled DNA create large topological domains. Nucleic Acids Res 46: 4417–4424. 10.1093/nar/gky153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhabinskaya D, Madden S, Benham CJ. 2015. SIST: stress-induced structural transitions in superhelical DNA. Bioinformatics 31: 421–422. 10.1093/bioinformatics/btu657 [DOI] [PubMed] [Google Scholar]
- Zhang L, Kasif S, Cantor CR, Broude NE. 2004. GC/AT-content spikes as genomic punctuation marks. Proc Natl Acad Sci 101: 16855–16860. 10.1073/pnas.0407821101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9: R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.