Abstract
Cells use mitophagy to remove dysfunctional or excess mitochondria, frequently in response to imposed stresses, such as hypoxia and nutrient deprivation. Mitochondrial cargo receptors (MCR) induced by these stresses target mitochondria to autophagosomes through interaction with members of the LC3/GABARAP family. There are a growing number of these MCRs, including BNIP3, BNIP3L, FUNDC1, Bcl2-L-13, FKBP8, Prohibitin-2, and others, in addition to mitochondrial protein targets of PINK1/Parkin phospho-ubiquitination. There is also an emerging link between mitochondrial lipid signaling and mitophagy where ceramide, sphingosine-1-phosphate, and cardiolipin have all been shown to promote mitophagy. Here, we review the upstream signaling mechanisms that regulate mitophagy, including components of the mitochondrial fission machinery, AMPK, ATF4, FoxOs, Sirtuins, and mtDNA release, and address the significance of these pathways for stress responses in tumorigenesis and metastasis. In particular, we focus on how mitophagy modulators intersect with cell cycle control and survival pathways in cancer, including following ECM detachment and during cell migration and metastasis. Finally, we interrogate how mitophagy affects tissue atrophy during cancer cachexia and therapy responses in the clinic.
Keywords: Autophagy, Mitochondria, Mitophagy, PINK1/Parkin, BNIP3/BNIP3L, FUNDC1, BCL2-L-13, LC3/GABARAP, Metabolism, Respiration, Fission, DRP1, UPRmt, Electron transport chain, AMPK, ATF4, NAD+, PARP, Sirtuins, FoxOs, Metastasis, ROS, Mitohormesis, Cachexia
Introduction
Mitophagy is a selective form of macro-autophagy in which mitochondria are preferentially targeted for degradation at the autophagolysosome [1, 2]. Mitophagy is required to eliminate mitochondria that have become dysfunctional, and thus serves an important housekeeping role that preserves mitochondrial function and limits production of damaging reactive oxygen species (ROS). Mitophagy can also eliminate healthy mitochondria to reduce overall mitochondrial mass as an adaptive response to stresses such as hypoxia and nutrient deprivation, to ensure efficient use of scarce metabolites and oxygen, and again limit production of excess ROS. The turnover of mitochondria by mitophagy relies on the activity of a growing list of mitochondrial cargo receptors (MCRs), that include BNIP3, BNIP3L (NIX), FUNDC1, Bcl2-L-13, FKBP8, FANC-C, Prohibitin-2 (PHB-2), Metaxin-1, Cardiolipin, and others, in addition to mitochondrial proteins that are substrates of PINK1/Parkin-mediated ubiquitination. These MCRs localize to the mitochondria and promote mitophagy by interacting directly with processed LC3/GABARAP proteins through conserved LC3 Interaction Regions (LIR) (Fig. 1). Importantly, the expression and/or processing of these MCRs is modulated by stresses that induce mitophagy, including mitochondrial depolarization, hypoxia, nutrient deprivation, etc. For example, BNIP3, BNIP3L, and FUNDC1-mediated mitophagy is activated by hypoxia, while cardiolipin and PHB-2 are exposed by mitochondrial damage. In addition to these MCRs that bind LC3/GABARAP proteins directly, there are other signaling molecules that contribute to activation of mitophagy including DRP1, FIS1, and MFF that are better known for their role in other aspects of mitochondrial dynamics. Ceramide and sphingolipids modulate mitophagy partly by recruiting LC3 to mitochondria, while other novel mitophagy modulators such as the adenine nucleotide transporter (ANT) affect mitophagy by controlling PINK1 import into mitochondria for degradation. The growing list of mitophagy modulators (Table 1) underlines the extent to which it is important for cells to control mitochondrial turnover and mitochondrial function in response to different stresses and signals. That there are so many different mitophagy modulators also causes one to question the extent to which these mitophagy pathways are redundant and interact with each other, or alternatively are uniquely required for mitophagy in response to distinct stresses. In this review, we will examine some of these questions in the context of tumorigenesis and metastasis to assess whether mitophagy may be a valid and more targeted therapy in cancer treatment than general autophagy.
Fig. 1.
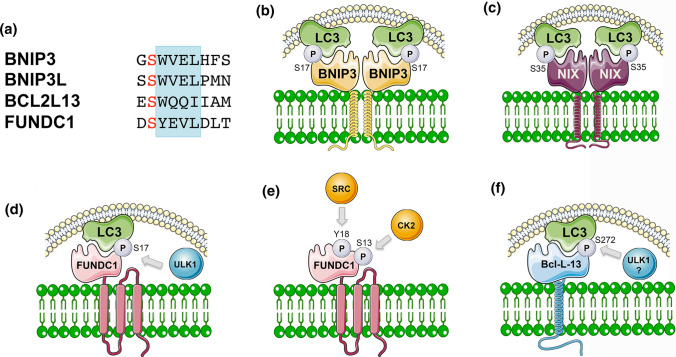
Modulation of mitochondrial cargo receptor interactions with LC3 family members by post-translational modification. a Alignment of the LIR motifs (blue) in BNIP3, BNIP3L, Bcl1L13 and FUNDC1 mitochondrial cargo receptors that are required for them to interact with processed LC3/GABARAP family members, and highlighting the serine residue (red) at − 1 in each LIR motif that is subject to phosphorylation to further promote LC3/GABARAP interaction. b BNIP3 is phosphorylated on S17 (and S24) adjacent to the critical tryptophan reside at W18 which promotes its interaction with LC3B-II. The kinase responsible is currently unknown. c Similarly, BNIP3L is phosphorylated on S35 (and S34) adjacent to W36 of its LIR motif that promotes its interaction with LC3B and GABARAP. d ULK1 phosphorylates FUNDC1 on S17 to promote LC3 interaction, while PGAM5 (not shown) de-phosphorylates inhibitory phospho-S13, also to promote LC3 interaction. e Interaction of FUNDC1 with LC3B is repressed by phosphorylation on Y18 by SRC kinase within its LIR motif and also inhibited by CK2 phosphorylation on S13. f Bcl2-L-13 is phosphorylated on S272 adjacent to its LIR motif and also interacts with ULK1, although whether ULK1 phosphorylates Bcl2-L-13 on S272 has not been shown
Table 1.
Summary of key regulators of mitophagy
| Protein | Mitophagy function | Cancer/disease relevance | References |
|---|---|---|---|
| Autophagy cargo receptors implicated in mitophagy | |||
| NDP52 (Nuclear Dot Protein 52); also known as CALCOCO2 (calcium-binding and coiled-coil domain-containing protein-2) | NDP52 interacts with processed LC3; required for Parkin-dependent mitophagy; recruits LC3, DFCP1 and WIPI1 to focal regions of the OMM | Not known | [3, 238] |
| Optineurin (OPTN) | OPTN is an autophagy cargo receptor that gets recruited to depolarized mitochondria in a Parkin-dependent manner where it interacts with LC3. Phosphorylation by TBK1 required for its ability to bind ubiquitin; OPTN binding to ubiquitin amplifies TBK1 activity | Mutated in ALS. Down-regulated in lung cancer cell lines. Low expression correlates with poor prognosis in human lung cancer | [3, 4, 239] |
| p62/Sqstm1 | Multi-functional protein that acts as a signaling hub integrating numerous stress responses; it promote mitophagy by binding to ubiquitinated proteins at the OMM and to processed LC3 at nascent phagophores. Cargo receptor function is not specific to mitophagy; plays other roles in cancer, including NRF2 activation | Amplified at chr.5 in human kidney cancer, accumulates in pancreatitis and non-alcoholic steatohepatitis, accumulates in autophagy-deficient cancers in mice, required for Ras-mediated transformation in mouse models. Role in cancer is context specific—appears to function distinctly in the tumor from in the tumor microenvironment | [182, 240, 241] |
| TAX1BP1 (Tax1 Binding protein) | Like OPTN1 and NDP52, TAX1BP1 is an autophagy cargo receptor that restores mitophagy in cargo receptor deficient cells exposed to CCCP. However, it does so less well than OPTN or NDP52 | Not known | [3] |
| PINK1-dependent mitophagy | |||
| Parkin | E3 ubiquitin ligase encoded by the PARK2 locus, activated by PINK1 through phosphorylation on S65 in its UBL domain causing it to localize to the OMM; conjugates ubiquitin chains to numerous OMM proteins, including Mfn2, VDAC1, TBK1; phospho-Ub chains are bound by autophagy cargo receptors, like OPTN, NDP52, TAXBP1. Antagonized by USP30 and other mitochondrial de-ubiquitinases. Modulates the mitotic checkpoint via PINK1-dependent and PINK1-independent means | PARK2 inactivating mutations linked to Parkinson’s Disease; deleted in human ovarian, breast, lung and bladder cancers; inactivating mutations found in glioblastoma and other cancers; Parkin null mice develop spontaneous liver tumors, are sensitized to radiation-induced lymphoma, loss of Parkin promotes KRas-driven PDAC | [3, 45, 137, 238] |
| PINK1 | Serine/threonine ubiquitin kinase encoded by the PARK6 locus that undergoes voltage-dependent degradation at the mitochondria; stabilized at the OMM by ΔΨmt; its import is promoted by Atad3a that prevents its aberrant accumulation; import inhibited by ANT to promote mitophagy. PINK1 phosphorylates ubiquitin chains, and Parkin to derepress its auto-inhibitory activity leading to Parkin recruitment to and activity at the OMM. Functions in concert with other E3 Ub ligases at the mitochondria, including ARIH1, to promote mitophagy | PARK6 loss linked to Parkinson’s disease. Reduced PARK6 expression detected in glioblastoma and ovarian cancer; PARK6 mutated in rare cases of neuroblastoma; loss of Pink1 promotes KRas-driven PDAC | [45, 138, 238] |
| LRRK2 | LRRK2 kinase encoded by the PARK8 locus phosphorylates Rab GTPases involved in cellular trafficking (Rab1b, Rab8a, and Rab10); mutant LRRK2 (G2019S) has increased kinase activity for these substrates. LRRK2 promotes the degradation of Miro thereby limiting mitochondrial motility and promoting mitochondrial sequestration and mitophagy | Mutated (e.g., G2019S) in Parkinson’s Disease | [242] |
| ANT | The adenine nucleotide transporter is required for ADP/ATP exchange across the IMM but a screen for genes that modulate mitophagy identified a novel function for ANT in promoting mitophagy that is independent of its ADP/ATP translocase activity. Specifically, ANT promotes PINK1 accumulation and mitophagy by interacting with TIM23 to limit PINK1 uptake and degradation in the mitochondrial matrix | A frameshift mutation in ANT1 in humans associated with mitochondrial abnormalities and cardiomyopathy | [11] |
| Atad3a | Interacts with TOM40 at the OMM and TIM23 at the IMM and promotes PINK1 import into the mitochondrial matrix for degradation | Mutant Atad3a (R528W) associated with increased mitophagy in human fibroblasts. Loss of Atad3a causes aberrant accumulation of PINK1 | [10] |
| ARIH1 | A mitochondrial E3 ubiquitin ligase that can substitute for Parkin, is dependent on PINK1, DRP1, and ZIP1 for mitophagy | Expressed in breast and lung cancer cell lines; protects against cell death induced by chemotherapeutic agents | [16] |
| MARCH5 (also known as MITOL) | A mitochondrial RING-finger E3 ligase that plays a role in mitochondrial dynamics via ubiquitination of DRP1 and Mfn1/Mfn2. Ubiquitinates the FUNDC1 mitophagy receptor resulting in its turnover at the proteasome to attenuate hypoxia-induced mitophagy | Not known | [17, 243] |
| MUL1 | Mitochondrial E3 ubiquitin ligase that is required to eliminate paternal mitochondria during embryogenesis and can compensate for loss of Parkin in mitophagy in flies and other systems | Not known | [14, 15] |
| SIAH1 (Seven in absentia homolog-1) | E3 ubiquitin ligase recruited by synphilin to mitochondria in a PINK1-dependent manner following synphilin-induced mitochondrial depolarization where it ubiquitinates mitochondrial proteins, independent of Parkin | Not known | [244] |
| VPS13D | Required for mitochondrial size and clearance by mitophagy in Drosophila. Binds to K63 UB chains. May function downstream of MFF and DRP1 | Not known | [245] |
| BNIP3/BNIP3L-dependent mitophagy | |||
| BNIP3 | Stress-induced OMM mitochondrial protein that binds directly to LC3; induced by hypoxia (HIF-1 target), nutrient deprivation (PPARα target), FoxO3A, glucocorticoid receptor; repressed by pRB and p53; required for glucagon-induced mitophagy in liver and FoxO3A-induced mitophagy in atrophying muscle | Deleted, silenced or mis-localized in breast, prostate, colon, pancreatic, liver, glioma and other cancers. BNip3 loss accelerates progression to metastasis in mouse models of breast cancer and in human TNBC. A dual role for BNIP3 in promoting metastasis in tumors where its expression is retained is linked to a role in promoting cell migration and preventing anoikis | [22, 23, 35, 156, 161] |
| BNIP3L (NIX) | Functional homolog of BNIP3; induced by hypoxia (HIF-1 target), p53; required for mitophagy during red blood cell differentiation, retinal ganglion cell differentiation and polarization of M1 type macrophages; interacts with Rheb and LC3 | NIX knockdown promoted tumor growth in a mouse mammary tumor xenograft study. Knockout of NIX limited tumorigenesis in KRas-driven PDAC. Prevents cell death following matrix detachment | [21, 38, 40, 49, 50, 54, 153] |
| Lipin-1 | Lipin-1 is a phosphatidic acid phosphatase that regulates triglyceride synthesis. Lipin1 is phosphorylated by mTOR. Lipin-1 mutation causes dysfunctional mitochondria to accumulate in muscle where Lipin-1 is required for BNIP3-dependent mitophagy and autophagosome-lysosome fusion | Heritable mutations found in human myopathies | [80, 81, 246] |
| Rheb | Small GTPase required for mTORC1 activity; interacts with LC3 and BNIP3L to promote mitophagy induced by switch from glycolytic to oxidative metabolism | Rheb point mutations found in genomic analyses of kidney and endometrial cancers | [56, 57] |
| cGAS-STING/Inflammasome activation related mitophagy regulators | |||
| Caspase-1 | Proto-typical caspase involved in cytokine processing, key component of the NLRP3 inflammasome; recruited to the OMM, activated by altered ∆Ψmt ROS and oxidized mtDNA in a positive feedback loop; cleaves Parkin | Caspase-1 is down-regulated in prostate ovarian and colorectal cancers but over-expressed in pancreatic cancer. Caspase-1 null mice are more susceptible to chemically induced colorectal cancer | [125, 129, 247–249] |
| NLRP3 | Core component of NLRP3 inflammasome activated by DAMPs and PAMPs. Binds oxidized mtDNA released by damaged mitochondria, to activate caspase-1 and promote IL-1β secretion | NLRP3-dependent release of IL-1β from dendritic cells is required for anti-tumor immunity. NLRP3 null mice are more susceptible to chemically induced colorectal cancer | [124, 250–252] |
| STING | Activated by cytosolic double-stranded DNA, including mtDNA; promotes autophagosome biogenesis from the ER-Golgi, stimulates TBK1 activity that is required for OPTN and other mitophagy cargo adaptors | Detection of cytosolic mtDNA promotes anti-tumor immunity; some human cancers exhibit reduced cGAS-STING activity. STING agonists being developed for cancer therapy | [121, 132, 252] |
| TBK1 (TANK-binding kinase-1) | Serine/threonine kinase that phosphorylates OPTN, NDP52, TAXBP1 and p62/Sqstm1 promoting their recruitment to depolarized mitochondria in a Parkin/PINK1-dependent manner. Activated by cGAS-STING pathway in response to cytosolic dsDNA. Also, localizes to centrosomes and promotes mitosis until sequestered in PINK1/Parkin-dependent manner at damaged mitochondria | Mutated in ALS; required for K-Ras transformation and tumor cell survival; promotes dormancy of prostate cancer cells | [4, 143, 238] |
| Transcription factors that regulate mitophagy genes | |||
| ATF4 | CREB/ATF family member that is activated in response to amino acid deprivation, ER stress and mitochondrial stress via phosphorylation by PERK, HRI, GCN2, or PKR kinases. HRI plays a unique role in activating ATF4 downstream of mitochondrial stress that activates OMA1 to cleave mitochondrial DELE1 that is released to the cytosol where it binds/activates HRI. Worm homolog Atfs1 required for UPRmt, mitophagy, and OXPHOS gene expression during aging and proteotoxic stress | Target genes involved in amino acid metabolism upregulated in many cancers. Protects against anoikis induced by matrix detachment | [98, 99, 104, 105, 253, 254] |
| FoxOs | FoxO1, FoxO3, FoxO4 and FoxO6 are activated in response to redox stress and by NAD+ dependent Sirtuins; activated by AKT and AMPK in response to nutrient stress. Activity promotes longevity and required for stemness in multiple tissues. Activates target genes involved in cell cycle, anti-oxidant responses, apoptosis and autophagy. Turned over by autophagy. Induces BNIP3 in atrophying muscle and in cachectic muscle | Upregulated in some cancers. FoxO3A induces Puma to promote selective tumor cell killing by genotoxic agents in combination with autophagy inhibition | [35, 255–258] |
| HIF-1 | HIF-1a stabilized by hypoxia to dimerize with HIF-1b. Induces expression of HIF-1 signature that encodes genes in glycolysis, angiogenesis, EMT, stemness etc. Required for induction of BNIP3 and BNIP3L to induce mitophagy that promotes a glycolytic switch, limits ROS, and promotes survival of detached tumor cells | HIF-1 promotes Warburg metabolism, epithelial to mesenchymal transition and cancer stemness. HIF-1 signature is upregulated in many cancers and linked to poor prognosis | [156, 161, 259, 260] |
| MITF/TFE | Related group of transcription factors that induce the CLEAR network of target genes encoding genes involved in autophagy and lysosomal biogenesis. Nuclear localization inhibited by mTOR. Conversely, TFEB and TFE3 are activated by AMPK | Derepressed in various cancers including PDAC, melanoma and renal cell carcinoma | [91, 261] |
| Other | |||
| AMBRA1 (autophagy and beclin1 regulator-1) | AMBRA1 associated with Bcl-2 can localize to the OMM where it binds LC3 via LIR motif to promote mitophagy in a PINK1/Parkin/p62 independent manner; promotes autophagic turnover of oncogenic SRC and cell migration | AMBRA1 expression is upregulated in some cancers and mutated in others; mono-allelic deletion of Ambra1 in mice induces spontaneous liver and lung tumors | [262–266] |
| Bcl2-L13/Bcl-Rambo | Mammalian homologue of yeast Atg32; binds ULK1 and LC3 through conserved LIR motif; over-expression induces mitochondrial fragmentation and mitophagy in a Parkin-independent manner | Not known | [68, 69] |
| Cardiolipin (CL) | IMM phospholipid required for tethering proteins to cristae, including ETC components; mitochondrial stress induces relocalization of CL to OMM; LC3 binds to CL; CL is required for mitophagy involving LC3 | Not known | [74] |
| Ceramide | A bioactive sphingolipid that promotes caspase-independent cell death but also lethal mitophagy by interacting with processed LC3B-II and recruiting autophagolysosomes to the mitochondria | Limits tumor cell growth | [76] |
| CHDH (choline dehydrogenase) | Required for PINK1/Parkin-mediated mitophagy. Located at both the IMM and the OMM but concentrates at the OMM in response to depolarization where it interacts with p62/Sqstm1 and LC3 | Not known | [267] |
| DELE1 | Required for ATF4 activation in response to mitochondrial stress, such as respiratory chain inhibition. Mitochondrial stress activates OMPA1 protease that cleaves mitochondrial DELE1 that is then released to the cytosol where it binds to and activates HRI, an eIF2a kinase, that induces ATF4 levels | Not known | [104, 105] |
| DRP-1 |
DRP1 is a GTPase required for mitochondrial fission and to regulate mitochondrial membrane potential DRP1 is recruited to mitochondria by FIS1 and MFF receptors that both also promote mitophagy. DRP1-dependent fission is required for Parkin-dependent mitophagy. DRP1 plays a key role on identifying regions of the mitochondrial network for turnover through its interaction with ZIP1 |
DRP1 is required for RAS transformation | [180, 181, 188] |
| EndophilinB/Bif-1 | Fatty acyl transferase required for mitochondrial membrane dynamics; interacts with Beclin1 via UVRAG; colocalizes at Atg9 + puncta, involved in membrane lipid trafficking around mitochondria | Haploinsufficiency promotes Myc-driven lymphomagenesis | [268, 269] |
| FANC-C | FANC-C localizes to mitochondria, interacts with Parkin and is required for mitophagy and to prevent inflammasome activation; role in mitophagy is genetically distinct from its role in DNA repair. Dysfunctional mitochondria accumulate in FA cells. Other FA genes also implicated in mitophagy are: FANC-A, FANC-D2, FANC-F, FANC-L, BRCA1, BRCA2 | Inactivated in Fanconi Anemia; other FANC genes include BRCA2 and BRCA1 that are deleted in hereditary breast and ovarian cancer | [174] |
| FIS1 | Formerly considered a mitochondrial receptor for DRP1; phosphorylated by AMPK; recruits TBC1D15 and TBC1D17 Rab-GAPs to mitochondria where they regulate Rab7 GTPase activity to promote proper encapsulation of mitochondria by autophagosomes, required for mitophagy in LSCs | Required for self-renewal and maintenance of leukemia stem cells in AML | [89, 185–187] |
| FKBP8 | A member of the FK506-binding protein family localized to the OMM, possesses a LIR motif and interacts directly with LC3-related protein, LC3A recruiting it to damaged mitochondria in a Parkin-independent manner | Not known | [270] |
| FUNDC1 | OMM protein induced by hypoxia, interacts directly with LC3; LC3 interaction regulated by ULK1, SRC, CK2, and PGAM5; accumulates at ER-mito contact sites through interactions with DRP1 and calnexin; essential for hypoxia-induced mitophagy; interacts with MARCH5 in a ROS-dependent manner to undergo MARCH5-dependent degradation. FUNDC1 has mitophagy-independent functions modulating mitochondrial bioenergetics via interaction with LonP and respiratory chain complexes | Knockout mouse defective for platelet maturation. Fundc1 deletion in the liver promotes DEN-induced HCC as a result of increased release of mtDNA and inflammasome activity. Elevated FUNDC1 expression associated with poor prognosis in breast cancer. Knockdown of FUNDC1 inhibits tumor growth in xenografts but conversely promotes tumor cell migration and metastasis | [17, 59, 62, 169, 170] |
| MFF | Like FIS1, MFF is a mitochondrial receptor for DRP1 in mitochondrial fission. Phosphorylated by AMPK and required for fission induced by AMPK | Not known | [88] |
| Miro | Anchors kinesins to the OMM; phosphorylated by Parkin/PINK1 resulting in its proteolytic cleavage that reduces mitochondrial motility and isolates damaged mitochondria for mitophagy; differential phosphorylation of Miro blocks Parkin recruitment to OMM | Turnover disrupted in PD | [242, 271] |
| MTX1 (Metaxin-1) | Localizes to the OMM, possesses a non-canonical LIR and interacts with LC3C at autophagosomes to promote mitophagy independent of PINK1/Parkin. May be involved in piecemeal mitophagy | Not known | [272] |
| PGAM5 | Serine/threonine phosphatase at the OMM, required for CCCP-induced mitophagy, activates FUNDC1 through S13 de-phosphorylation; interacts with PINK1 at OMM in response to ∆Ψmt; protects PINK1 from degradation at IMM | Pgam5 null mice develop an age-related movement disorder, reminiscent of PD | [63] |
| Prohibitin2 | IMM protein involved in processing OPA1 and cristae remodeling; binds LC3 following Parkin-mediated rupture of OMM, acts as an IMM mitophagy adaptor; required for Parkin-mediated mitophagy. Requires PHB1 for stability. Also has a nuclear function regulating activity of E2F, p53, and the AR | Expression deregulated in breast, prostate and lung cancer; role varies depending on tissue type. PHB1 deletion promotes HCC in mice | [71] |
| PUM-2 | Pumilio-2 is an RNA-binding protein that inhibits protein translation. PUM-2 inhibition of MFF translation inhibited mitochondrial fission and mitophagy | Induced during aging in muscle and other tissues | [273] |
| RIPK1 | Receptor-interacting protein kinase-1 levels and activity are induced by detachment from the extracellular matrix resulting in PINK1-dependent mitophagy causing loss of viability that was dependent on RIPK1 substrate PGAM5 phosphatase, a known modulator of mitophagy | Loss of RIPK1 promotes tumor growth in tumor xenografts, RIPK1 limits spontaneous hepatocellular carcinoma in mice, low expression of RIPK1 in human HCC correlates with poor prognosis | [274, 275] |
| SMURF | Identified in a functional screen for genes required for xenophagy; essential for Parkin-induced mitophagy; recruited to mitochondria by ∆Ψmt; its E3 Ub ligase activity is not required for mitophagy | Not known | [175] |
| STX17 | Syntaxin-17 is a Q-SNARE protein required for autophagosome biogenesis and fusion of the autophagosome with the lysosome. Both activities involve interaction with ATG14L that it recruits to ER–mitochondrial junctions. STX17 recruitment to the phagophore from the Golgi is promoted by phosphorylation by TBK1. STX17 also plays a role via PGAM5 phosphatase in determining the localization of DRP1 during mitochondrial fission and interacts with Fis1 to promote mitophagy | Intron 6 duplication associated with increased melanoma in gray horses | [276–280] |
| TAZ/tafazzin | Phospholipid trans-acylase that remodels cardiolipin; loss of TAZ blocks mitophagy due to failure of LC3 to recognize CL; TAZ deficiency results in reduced acylation of CL and its depletion from mitochondria | Mutated in Barth syndrome | [75] |
| ZIP1 | ZIP1 interacts with DRP1 at the mitochondria to modulate mitochondrial membrane potential by regulating MCU-dependent uptake of Zn2+ ions. ZIP1 promotes ∆Ψmt to control how DRP1 induces mitophagy of depolarized mitochondria | Not known | [188] |
Mitophagy pathways and mitochondrial cargo receptors
Before addressing the role of different mitophagy modulators in cancer, we first review the major molecular pathways that regulate mitophagy with a particular focus on mitochondrial cargo receptors that localize to mitochondria in response to mitochondrial stresses. Table 1 summarizes known mitophagy modulators more comprehensively.
PINK1/Parkin-dependent mitophagy
Mitochondrial membrane depolarization (ΔΨmt) induces mitophagy through activation of PTEN-induced putative kinase-1 (PINK1), a serine/threonine ubiquitin kinase encoded by the PARK6 locus, which accumulates at the outer mitochondrial membrane (OMM) where it recruits E3 ubiquitin ligases, most notably Parkin, encoded by the PARK2 locus [3, 4]. PINK1 phosphorylates Parkin on S65 within its ubiquitin-like (UBL) domain, derepressing Parkin auto-inhibition and amplifying recruitment of active Parkin to the OMM [4]. This leads to the ubiquitination and phosphorylation of proteins at the OMM, such as Mitofusin-2 (Mfn2) and the voltage-dependent anion channel-1 (VDAC-1), that then interact with the Optineurin (OPTN) and NDP52 cargo receptors, but also p62/Sqstm1 in some systems, which contain critical LIR motifs [3, 5–7]. PINK1/Parkin-dependent mitophagy is the major mechanism in the cell for elimination of depolarized mitochondria (Fig. 2).
Fig. 2.
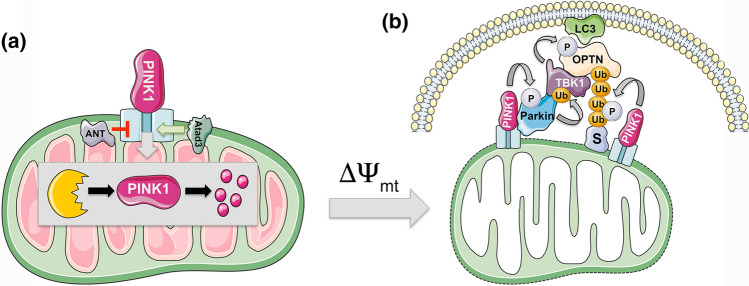
PINK1 degradation controls turnover of depolarized mitochondria. a PINK1 is a ubiquitin kinase that is degraded at functional mitochondria. PINK1 import via the TOM/TIM protein complexes is highly regulated and dependent on mitochondrial membrane polarization. PINK1 uptake into the mitochondria is positively regulated by the Atad3a AAA + ATPase, and negatively regulated by the adenine nucleotide translocase (ANT) that interferes with TOM23 and TIM40 to block PINK1 uptake. This function of ANT is separable from its function in ADP/ATP exchange at the IMM. The MTS of PINK1 is proteolytically cleaved off in the matrix by MPP, while the transmembrane part of PINK1 is cleaved by PARL protease and then retrotranslocated back to the cytosol for proteasomal degradation. b In response to loss of mitochondrial membrane potential (∆Ψmt), PINK1 uptake at the mitochondria is blocked and it accumulates at the OMM, where it recruits the Parkin E3 ubiquitin ligase. PINK1 phosphorylates Parkin within it UBL domain derepressing its activity and also phosphorylates ubiquitin groups on Parkin substrates (S). Parkin ubiquitinates substrates (S) at the OMM such as MFN-2 and Vdac1 and other proteins and these phospho-ubiquitinated substrates are bound by cargo receptors, such as p62/Sqstm1 and OPTN1, which also interact with LC3 to promote mitophagy. Recruitment of OPTN1 and other cargo receptors to the OMM is dependent on TBK1 that is recruited away from centrosomes and its role in mitosis to the OMM in PINK1/Parkin-dependent manner
At healthy mitochondria, the amino-terminal mitochondrial targeting sequence (MTS) of PINK1 is translocated to the mitochondrial matrix where it is cleaved off by mitochondrial processing peptidase (MPP), while the transmembrane portion of PINK1 is cleaved by PINK/PGAM5-associated rhomboid-like (PARL) proteases, and then, the remaining 52 kD fragment of PINK1 is retrotranslocated back out to the cytosol and degraded at the proteasome according to the N-end rule [4, 8, 9]. Mitochondrial import of PINK1 at healthy mitochondria is facilitated by Atad3a, an AAA + ATPase that spans the IMM and OMM [10]. Inactivation of Atad3a increases mitophagy of both depolarized and healthy mitochondria, which is partially rescued by PINK1 deletion, indicating that Atad3a plays an important role in preventing aberrant accumulation of PINK1 [10]. Conversely, the adenine nucleotide transporter-1 and -2 (ANT) plays a role in promoting PINK1 accumulation by suppressing translocase of inner membrane (TIM) proteins, TIM23 and TIM44, that are involved in PINK1 mitochondrial import [11]. This function of the ANT was independent of its role in ADP/ATP exchange at the IMM and loss of ANT decreased levels of both PINK1 and Parkin in mouse brain and heart, causing the accumulation of dysfunctional mitochondria and cardiomyopathy [11]. Interestingly, mutant forms of ANT found in patients with cardiomyopathy and opthalmoplegia were unable to rescue mitophagy defects in ANT null cells, whereas mutant ANT that cannot bind ATP or promote ADP/ATP exchange was able to rescue mitophagy defects. Together with evidence showing mitochondrial abnormalities in these patients, this work indicates that defective mitophagy arising from mutant ANT causes the pathologies in these patients [11].
In addition to their activation by mitochondrial depolarization, PINK1 and Parkin-dependent mitophagy are also activated by accumulation of unfolded proteins in the mitochondrial matrix [12, 13] suggesting a link between the mitochondrial unfolded protein response (UPRmt) and mitophagy, two key mitochondrial quality control processes in the cell.
While most research has focused on the interaction of PINK1 with Parkin, PINK1 interacts functionally with other mitochondrial E3 ubiquitin ligases involved in mitophagy, including ARIH1, MARCH5, and MUL1, which can substitute for Parkin in mitophagy in various contexts [14–17].
BNIP3/BNIP3L-dependent mitophagy
Originally categorized as pro-apoptotic BH3-only proteins, BNIP3 and BNIP3L (NIX) function primarily as stress-induced MCRs. BNIP3 and BNIP3L are tail-anchored proteins that integrate into the OMM via their carboxy terminal transmembrane domains (TMD) with the remaining bulk of both proteins facing into the cytosol where a tightly conserved LIR motif (WVEL) near the amino-terminal end of each protein (W18 to L21 in BNIP3, and W36 to L39 in BNIP3L; Fig. 1a) interacts with processed LC3/GABARAP [18–20]. The putative BH3 domains in BNIP3 and BNIP3L are poorly conserved (2 amino acids out of 11) and are redundant for BNIP3/BNIP3L function in mitophagy [21, 22]. Furthermore, the interaction of BNIP3 and BNIP3L with Bcl-XL (and Bcl-2) is mediated via the amino-terminal region overlapping with the LIR motif, and not dependent on the putative BH3 domains in BNIP3/BNIP3L [23]. This work also suggested that the interaction of BNIP3/BNIP3L with Bcl-XL enhances their interaction with LC3/GABARAP family members to promote mitophagy [23]. The BNIP3 TMD consists of a glycine zipper that mediates homo-dimerization via hydrogen bonding between polar residues on the inside of the glycine zipper, while hydrophobic residues on the outside of the zipper stabilize the dimer in the OMM lipid bilayer [24]. Remarkably, BNIP3 and BNIP3L dimers are resistant to SDS, DDT, and urea, and can be visualized on denaturing gels [25, 26] but to what extent BNIP3 or BNIP3L dimers are stable in vivo has not been examined. While dimerization is not required for integration into the OMM, recent evidence suggests that dimerization is required for efficient LC3/GABARAP interaction [26]. Also, intriguingly, BNIP3 and BNIP3L have been shown to heterodimerize [27], but when and where such heterodimers form in a physiological context has not been determined.
BNIP3 and BNIP3L are rapidly induced at a transcriptional level by stresses, such as hypoxia and nutrient deprivation, but like many stress response proteins, both proteins are also rapidly turned over by mitophagy and at the proteasome, requiring the stressed cell to maintain transcription of BNIP3 and BNIP3L until the stress is removed [2]. Both BNIP3 and BNIP3L are transcriptionally activated by HIF-1 in response to hypoxia and elevated ROS [28, 29], and BNIP3 in particular is a strong hypoxia marker that gets induced rapidly and to high levels as oxygen drops below 2% [30–32]. BNIP3 is also a validated transcriptional target of PPARα explaining its induction in the liver in response to nutrient deprivation and fasting [22, 33, 34], and is induced by FoxO3A in atrophying and cachectic muscle [35–37]. BNIP3 and BNIP3L are both transcriptionally regulated by p53 [38, 39], but while BNIP3L is upregulated transcriptionally by p53 [38], BNIP3 is repressed [39], suggesting that they play differential roles in the response to DNA damage and other signals that activate p53.
In addition to regulation at a transcriptional level, BNIP3 and BNIP3L are modulated post-translationally by phosphorylation [23, 40]. Both proteins are phosphorylated on the serine residue (S17 in BNIP3 and S35 in BNIP3L) adjacent to the critical tryptophan residue in their LIR motifs (W18 in BNIP3 and W36 in BNIP3L) [23, 40] (Fig. 1b, c). These phosphorylation events promote their interaction with LC3/GABARAP [23, 40], and also their interaction with Bcl-XL [23]. BNIP3 and BNIP3L are multiply phosphorylated, but it remains to be determined what kinases are involved in modulating BNIP3/BNIP3L levels and activity.
Beyond phosphorylation, BNIP3L is ubiquitinated by Parkin and promotes PINK1/Parkin-induced mitophagy [41, 42]. However, neither BNIP3 nor BNIP3L requires PINK1 or Parkin to elicit mitophagy [43], and conversely, BNIP3 is not required for PINK1 accumulation, or for Parkin recruitment to the OMM [43]. Nevertheless, Parkin/PINK1 loss causes HIF-1α stabilization [44, 45] but whether BNIP3 or BNIP3L, that are both HIF targets, are upregulated by PINK1/Parkin loss has not been examined, although BNIP3 can compensate in mitophagy for Parkin loss [43].
BNIP3 and BNIP3L play important functions in tissue differentiation and stress responses with each protein exhibiting distinct tissue expression patterns [2]. BNIP3 is induced in a PPAR-α-dependent manner in response to fasting [22, 33, 34] and is required for glucagon-induced mitophagy in the liver [22]. BNIP3 exhibits zonal expression patterning in the liver with high expression around the central vein of the liver lobule where cells are most hypoxic [46], and low expression around the more oxygenated periportal vein [22]. As a result, mitochondrial mass is zonal in the liver with higher mitochondrial mass around the central vein and lower mitochondrial mass around the periportal vein [22]. Metabolic processes are spatially organized across the liver lobule, such that opposing activities, such as urea cycle and glutamine synthesis, fatty acid oxidation and lipogenesis, and gluconeogenesis and glycolysis, are physically separated [47]. This likely improves the efficiency of metabolite re-cycling and avoids competition between metabolic pathways for common substrates [47]. Loss of BNIP3-dependent mitophagy caused mitochondria to accumulate aberrantly and disrupted metabolic zonation in the liver, such that periportal processes such as urea cycle were expanded at the expense of central vein concentrated processes, such as glutamate-glutamine cycling and glycolysis [22].
BNIP3L is markedly upregulated and essential for mitophagy during normal red blood cell differentiation such that loss of BNIP3L resulted in anemia and splenomegaly due to production of defective reticulocytes [48–50]. Sphingosine kinase (SPHK1) contributes to BNIP3L induction during erythroid differentiation [51]. SPHK1 is required for the UPRmt in worms which is a transcriptional response to mitochondrial stress mediated by transcription factors such as ATF4, NRF2, and FoxOs that induce expression of genes that mitigate against mitochondrial stress, including mitochondrial proteases, mitochondrial protein chaperones, autophagy genes, anti-oxidants, etc. [52, 53]. Taken together with the mitochondrial localization of SPHK1 in response to stress, this suggests a mechanism linking activation of the UPRmt to mitophagy through induction of BNIP3L. BNIP3L is also required for differentiation of retinal ganglion cells during embryogenesis, where it is induced in mid-gestation by hypoxia [54]. This results in a BNIP3L-dependent glycolytic switch that is also seen during the polarization of M1 macrophages [54]. BNIP3/BNIP3L-dependent mitophagy was also required for survival of long-lived NK memory cells in response to viral infection [55].
Both BNIP3 and BNIP3L interact with Rheb [56, 57], the small GTPase required to promote lysosomal localization and activity of mTOR, a major growth-promoting kinase in the cell [58]. BNIP3 binding repressed Rheb activity and disrupting the BNIP3–Rheb interaction promoted mTOR activity and cell growth, consistent with a growth suppressive role for BNIP3 via its inhibitory interaction with Rheb [56]. By contrast, the interaction of BNIP3L with Rheb elicited mTOR-independent effects on cell growth [57]. BNIP3L-dependent mitophagy induced by switching cells from growth in glucose-only media to growth in glutamine plus galactose required interaction of BNIP3L with Rheb at the OMM in a tri-molecular complex with processed LC3 [57]. The relative importance of BNIP3/BNIP3L interactions with Rheb has not yet been addressed in a physiological setting.
FUNDC1
Like BNIP3 and BNIP3L, FUNDC1 promotes hypoxia-induced mitophagy, but in contrast to BNIP3 and BNIP3L, the activation of FUNDC1 by hypoxia is mediated primarily at a post-translational level, via regulation of its phosphorylation state [59]. Like BNIP3 and BNIP3L, FUNDC1 integrates into the OMM, although FUNDC1 is a multi-pass protein with three TMDs, and has an amino-terminal LIR motif (YVEL from Y18 to L21), which projects into the cytosol to interact with LC3 family members [59]. Its interaction with LC3B is tightly modulated by phosphorylation on S17 immediately adjacent to its LIR motif that is phosphorylated by ULK1 (Fig. 1d), the catalytic component of the autophagy pre-initiation complex [60, 61], in response to hypoxia to enhance LC3B interaction and mitophagy [62]. FUNDC1 is also phosphorylated by oncogenic SRC on Y18 within the LIR motif (Fig. 1e) to block LC3 interaction and mitophagy [62]. The FUNDC1–LC3B interaction is also inhibited by phosphorylation on S13 by Casein Kinase 2 (CK2). Conversely, PGAM5 phosphatase de-phosphorylates S13 to promote LC3 interaction and mitophagy [63]. Interestingly, PGAM5 also plays a role in mitophagy by promoting PINK1 accumulation [64]. FUNDC1 is also modulated by the MARCH5 E3 Ub ligase that ubiquitinates FUNDC1 on K119 to promote its turnover and limit hypoxia-induced mitophagy [17]. In addition to interacting with LC3B, FUNDC1 interacts with DRP1 and Calnexin at mitochondrial–ER junctions where it appears to be required for recruiting DRP1, since mutant forms of FUNDC1 that cannot bind DRP1 cannot promote mitophagy [65]. FUNDC1 may also play a role in the cellular response to proteostatic stress through its interaction with HSC70 [66]. While the hypoxia-induced microRNA, miR-137 represses both FUNDC1 and BNIP3L expression during hypoxia possibly limiting mitophagy [67], the extent to which FUNDC1 function in hypoxia-induced mitophagy overlaps with that of BNIP3 and BNIP3L has not been examined.
Bcl2-L-13
Bcl2-L-13 is a Bcl-2-related homolog of ATG32 in yeast that integrates into the OMM via a C-terminal TMD and interacts with LC3B via a conserved LIR motif (WQQI) at amino acids 273–276 (Fig. 1f) [68]. Bcl2-L-13 over-expression induces mitochondrial fragmentation and mitophagy, independent of DRP-1 and Parkin, and rescues mitophagy in Atg32-deleted yeast [68]. Bcl2-L-13 expression is induced by mitochondrial depolarization and knockdown of Bcl2-L-13 impeded CCCP-induced mitophagy [68]. Given that Bcl1-L-13 acts independently of Parkin, these results suggest that Parkin is not essential for mitophagy induced by mitochondrial membrane depolarization, but further work is needed to determine whether Bcl2-L-13 depends on PINK1 and other mitochondrial E3 Ub ligases to function. The induction of Bcl2-L-13 by mitochondrial depolarization was associated with its phosphorylation on S272, adjacent to its LIR motif at 273–276 [68]. Recent work has shown that ULK1 binds to Bcl2-L-13 in a manner dependent on the kinase activity of ULK1, in a tri-molecular complex with LC3B, and that ULK1 is required for Bcl2-L-13-dependent mitophagy [69], but whether ULK1 phosphorylates Bcl2-L-13 on S272 was not determined. Physiological studies addressing the importance of Bcl2-L-13 for mitophagy in vivo have not been reported.
Prohibitin-2
Prohibitin-2 (PHB-2) promotes cristae structure and supports electron transport chain (ETC) complex formation at the IMM [70]. PHB-2 interacts directly with LC3B via an LIR motif (YQRL) at amino acids 121–124 that is required for mitophagy induced by mitochondrial membrane depolarization and to eliminate paternal mitochondria in worms [71]. It has been suggested that proteasomal degradation of OMM proteins during mitophagy is required to expose IMM proteins, such as PHB-2 that are turned over by mitophagy [72], and indeed, PHB-2 relies on Parkin-mediated rupture of the OMM to promote mitophagy [71]. Various stresses known to induce mitophagy, including mitochondrial depolarization, happen at the IMM, and thus, it is significant that IMM molecules like PHB-2 and cardiolipin induce mitophagy.
Cardiolipin, ceramide, and lipid-dependent mitophagy
Cardiolipin is a phospholipid required to properly anchor proteins to the IMM, including components of the ETC, and to stabilize mitochondrial cristae [73]. Cardiolipin relocalizes to the OMM in response to stresses, such as mitochondrial membrane depolarization and respiratory chain inhibition, where it binds directly to processed LC3B to promote mitophagy [74]. LC3-dependent mitophagy was attenuated by blocking the LC3B–cardiolipin interaction, knocking down cardiolipin synthase, or preventing transport of cardiolipin to the OMM [74]. LC3 interacts with mature tetralinoleoyl–cardiolipin that is dependent on processing from immature monolyso-cardiolipin by the Tafazzin (TAZ) phospholipid trans-acylase, an enzyme found mutated in patients with Barth syndrome. TAZ deficiency prevents cardiolipin processing and blocks mitophagy leading to mitochondrial dysfunction (reduced respiration and ROS production), cardiomyopathy, and other pathologies found in Barth syndrome patients [75].
Ceramide is a bioactive sphingolipid known to induce caspase-independent cell death that may be attributed to a lethal form of mitophagy [76]. Ceramide C18 induced expression of LC3B-II, co-localized with LC3B-II at mitochondria, and caused recruitment of autophagosomes to mitochondria. The interaction of ceramide with LC3B-II relied on LC3 lipidation, since ceramide did not interact with LC3B-I. Over-expression of ceramide synthase-1 (CS1), but not catalytically dead CS1 (H183A), induced LC3B-II, LC3 puncta and mitophagy [76]. While ceramide-induced mitophagy required DRP1, it did not require p62/Sqstm1 or BNIP3L [76]. Interestingly, serine deprivation of tumor cells caused a marked decrease in ceramide, as well as lower levels of sphingolipids, cardiolipin, and phenylethanolamine, and was associated with mitochondrial fragmentation, mitochondrial dysfunction, and reduced cell proliferation [77]. These defects were rescued by providing ceramide (C16) to cells, although the effects of ceramide on mitophagy were not examined in this study.
Another lipid-dependent modulator of mitophagy is Lipin-1, a phosphatidic acid phosphatase that catalyzes the conversion of phosphatidic acid (PA) to diacylglycerol (DAG), which is required for de novo synthesis of phospholipids and triglycerides [78, 79]. Patients with heritable Lipin-1 mutations develop muscle atrophy that is associated with mitochondrial dysfunction and lipid droplet accumulation due to a failure in autophagy [80]. Muscle-specific over-expression of a Lipin-1 transgene rescued these defects in mice and also ameliorated the myopathy that develops following statin treatment [80]. Ceramide and other phospholipids accumulated in Lipin-1-deficient muscle following statin treatment but the extent to which this contributed to defects in mitophagy was not examined. Lipin-1 is required for activation of Protein Kinase D (PKD) and VPS34 to promote autophagosome fusion with lysosomes [80], but recent work also shows that Lipin-1 is required for the interaction of BNIP3 with LC3 in muscle [81]. How the role of Lipin-1 in mitophagy intersects with its role in control of nuclear transcription factor activity or in lipid signaling pathways is not clear.
Sphingosine kinase 1 (SPHK-1) catalyzes the formation of sphingosine-1-phosphate (S1P) from sphingosine, and while sphingosine (and ceramide) promote cellular senescence or apoptosis, S1P stimulates tumor cell proliferation and migration [82, 83]. SPHK1 is hypoxia-inducible [82, 84] and, as mentioned above, promotes mitophagy during erythropoiesis in part by inducing BNIP3L expression [51]. SPHK1 also activates the UPRmt in response to respiratory chain inhibition, inhibition of mitochondrial protein import or inhibition of mitochondrial fission or fusion [85]. SPHK1 localizes to mitochondria in response to mitochondrial stress and production of S1P regulates a number of mitochondrial proteins involved in the UPRmt to promote mitochondrial functionality [85]. In particular, S1P interacts directly with PHB-2 to promote ETC complex formation, suggesting that PHB-2 (and BNIP3L) may act downstream of lipid signaling to promote mitochondrial function and mitophagy, but much remains to be investigated in this area to understand how signaling lipids regulate mitophagy.
Upstream sensing of mitochondrial stress to induce mitophagy
Mitochondria are subject to many different kinds of stresses, including oxidative damage caused by respiratory chain inhibition, mutation of the mitochondrial genome that can repress respiration, failure to properly uptake mitochondrial proteins that then accumulate in the cytosol and cause an imbalance in mitochondrial to nuclear encoded proteins, defects in mitochondrial translation such as occur in response to certain antibiotics, as well as stresses shown to induce mitophagy, such as low oxygen and membrane depolarization [53]. As discussed above, much is known about how the cell promotes mitophagy in response to membrane depolarization (PINK1 accumulation) or hypoxia (induction of BNIP3/BNIP3L). Here, we examine several signaling pathways that have been linked to induction of mitophagy downstream of mitochondrial stresses (Fig. 3).
Fig. 3.
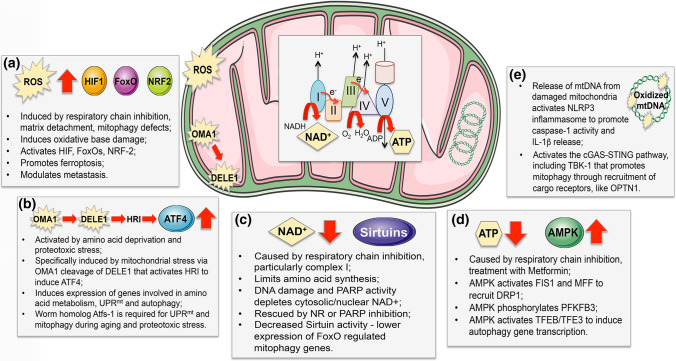
Mitochondrial stress-sensing pathways that modulate mitophagy. Mitochondrial stress and dysfunction is sensed in various ways and feeds forward to activate mitophagy, and other stress response pathways, such as the UPRmt. a Excess ROS produced following inhibition of the electron transport chain (ETC) results in escape of electrons (e−) primarily from complex I and complex III, that react with oxygen and water to form superoxide and peroxides that may be quenched by mitochondrial anti-oxidants or escape to the matrix and cytosol. ROS is also induced in tumor cells by matrix detachment and by mitophagy defects that cause dysfunctional mitochondria to accumulate. ROS activates transcription factors, including HIF-1, FoxOs, NRF2, that induce stress responses, including BNIP3-dependent mitophagy and NRF2 anti-oxidant responses. b While ATF4 is a key component of the cellular integrated stress response, and is activated by amino acid deprivation, it also plays a unique role in sensing mitochondrial stress that is mediated via HRI (heme-regulated eIF2α kinase) that is activated by a cleaved cytosolic form of DELE1, released from damaged mitochondria following OMA1 cleavage. ATF4 induces genes involved in amino acid biosynthesis and transport, but also induces genes that promote autophagy, OXPHOS and the UPRmt. c NAD+ is depleted by mitochondrial dysfunction, particularly in response to defects in respiration and lower complex I activity. NAD+ levels in cells are further decreased by DNA damage although the extent to which the nuclear/cytosolic pools of NAD + affect the mitochondrial pools and vice versa is unclear. Decreased NAD+ limits Sirtuin activity in the nucleus and at the mitochondria, resulting in lower FoxO activity and reduced mitophagy, which can be rescued by replenishing NAD+ with NR or by PARP inhibition. d Decreased ATP levels arising from lower respiratory chain activity and increased AMP activates AMPK, that can promote mitophagy by activitating FIS1 and MFF, or by activating the TFEB/TFE3 transcription factors, amongst other functions. e Mitophagy plays a key role in preventing activation of the cGAS-STING and NRLP3 inflammasome in response to release of mtDNA from damaged mitochondria during innate immune responses and inflammation
Activation of mitophagy by AMPK
AMPK is activated by defects in respiration caused by agents, such as Metformin [86, 87], that can lead to induction of general autophagy via ULK1 activation [60]. AMPK promotes mitophagy specifically via phosphorylation of the FIS1 and MFF receptors for DRP1 [88, 89] and AMPK phosphorylation of glycolysis regulator PFKFB3 promoted mitophagy and survival following extended mitotic arrest [90]. AMPK also activates TFEB and TFE3 in response to glucose deprivation to switch on the CLEAR network of genes that includes many autophagy genes [91]. Inactivation of AMPK caused unprocessed autophagosomes to accumulate in lung tumors driven by Ras and mutant p53 [91]. Loss of LKB1, the regulatory kinase upstream of AMPK [87], also caused mitochondrial dysfunction, including decreased oxygen consumption, increased ROS, and altered mitochondrial metabolism [92–95], but it is unclear if this was due to a failure to activate mitophagy [96, 97]. Nevertheless, the LKB1–AMPK signaling pathway has emerged as a key sensing mechanism of mitochondrial dysfunction that activates mitophagy.
ATF4 and the mitochondrial stress response
To obtain unbiased insight to how mitochondria respond to different kinds of stress, including ΔΨmt, mitochondrial protein translation inhibition, or respiratory chain inhibition, Quiros and colleagues performed transcriptomic, proteomic, and metabolomics profiling of HeLa cells exposed to agents that induce these stresses [98]. Genes involved in amino acid biosynthesis and handling, including ASNS and ASS1, correlated most strongly with mitochondrial stress and many of the induced genes were known ATF4 targets [98]. Consistently, levels of many amino acids were also elevated in response to mitochondrial stress. This ATF4 transcriptional signature was also upregulated in HEK-293T cells depleted for mtDNA or treated with oligomycin to repress respiration, where serine biosynthetic enzymes (PSAT1, PHGDH etc.) in particular were upregulated [99].
The ATF4 signature was also induced in muscle following OPA1 deletion where loss of OPA1 caused small, disrupted mitochondria to accumulate [100]. Interestingly, BNIP3 and LC3 were induced in Opa1 null muscle and deletion of FoxO (known to induce BNIP3) rescued atrophy of Opa1 null muscle fibers. The ATF4 signature has also been seen in other myopathy models [101, 102] and deletion of ATF4 protects muscle from fasting induced atrophy, where again BNIP3 and LC3 were induced [103]. These findings suggest that mitochondrial stress is a major factor in muscle atrophy and myopathies.
Recent work has shed light on how ATF4 is activated by mitochondrial stress (such as treatment with oligomycin and inhibition of respiration) [104, 105]. Mitochondrial stress activates the OMA1 protease to cleave DELE1 at the mitochondria. DELE1 was previously identified in a yeast two-hybrid system as a DAP3-interacting protein involved in modulating rates of apoptosis [106], but until now, little else has been reported about DELE1. Cleavage of DELE1 by OMA1 releases the short form of DELE1 to the cytosol where it interacts directly with HRI (Heme Regulated eIF2a kinase), one of four kinases known to regulate ATF4 activity [104, 105]. Knockdown of DELE1 blocked ATF4 induction by mitochondrial stress, but was not required for ATF4 activation by ER stress [104].
ATF4 has been interrogated as a functional homolog in mammalian systems of Atfs-1, the master regulator of the UPRmt in worms [107, 108]. While ATF4 has not been shown to localize to mitochondria like Atfs-1, both ATF4 and Atfs-1 are induced in response to inhibition of mitochondrial protein translation [109]. Atfs-1 accumulates in the nucleus of cells in response to mitochondrial stresses that block its import to the mitochondrial matrix, where it would be degraded under non-stressed conditions [110]. Atfs-1 target genes include mitochondrial chaperones that are activated by Atfs-1 and components of the ETC that are repressed by Atfs-1 [111]. Atfs-1 was also required to activate the UPRmt in a worm model of Alzheimers Disease (AD) in which amyloid-β was over-expressed [108]. In addition to activating the UPRmt in this model, Atfs-1 was also required for mitophagy via induction of dct-1, the worm homolog of BNIP3/BNIP3L [112]. In a mouse model of AD, mitophagy, UPRmt and OXPHOS gene sets (Mitophagy: Map1lc3b/Lc3b, Bnip3, Sqstm1/p62, Pink1/Park6; UPRmt: Hspa9, Hsp60, Yme1L1, Lonp1; OXPHOS: Cox5a, Cox2, Nd1, Sdhc) were induced [108], although the extent to which their expression was dependent on ATF4 was not examined. The simultaneous induction of the UPRmt and mitophagy in multiple systems in response to mitochondrial stress suggests joint regulation of these survival mechanisms, but there is much more to uncover about how these responses are coordinated.
NAD+ promotes mitophagy
Unresolved DNA damage occurring in cells mutated for enzymes involved in DNA repair, such as the ATM kinase in patients with Ataxia telangiectasia (AT), depletes NAD+ due to constitutive activity of the DNA repair protein Poly-ADP Ribose Polymerase (PARP) that consumes NAD+ [113]. Deficiency for atm1 in worms caused neuronal defects that were rescued by replenishing NAD+ levels with nicotinamide riboside (NR) or via PARP inhibition (Olaprarib/AZD2281) that promoted expression of dct-1, mitophagy, and mitochondrial function [114]. Boosting NAD+ levels also induced mitophagy and the UPRmt in a worm model of AD where amyloid-β was over-expressed, leading to mitochondrial proteostasis that both resolved degenerative disease and extended worm lifespan [108]. Dct-1, sqstm1 and pink1 were among the mitophagy genes induced in response to NR where their induction was dependent on Atfs-1, as described above [108].
Depletion of nuclear NAD+ through elevated PARP activity inhibits NAD+-dependent Sirtuins [113, 115]. Sirtuins modulate many metabolic processes in the cell, including via NAD+-dependent de-acetylation of PGC-1α required for expression of genes involved in mitochondrial biogenesis, fatty acid oxidation, and oxidative phosphorylation [116]. Sirtuins also de-acetylate FoxO3a that is a known inducer of autophagy genes, including BNIP3, LC3B, and Beclin1 [35]. The worm homolog of FoxOs, daf-16, is also activated by sirtuins resulting in dct-1 upregulation that limited aging and extended worm lifespan [112, 114]. Thus, both Atfs-1 and daf-16 induce mitophagy genes in worms in response to nutrient and mitochondrial stress, in a manner modulated by NAD+. Further characterization of these pathways linking NAD+ availability and mitophagy is necessary in mammalian systems [35, 117–119].
Activation of mitophagy in response to cytosolic mtDNA during inflammatory responses
Release of mtDNA to the cytosol following mitochondrial damage or dysfunction activates the cGAS–STING pathway and the NLRP3 inflammasome, and thus by eliminating dysfunctional mitochondria, mitophagy limits activation of cGAS–STING and inflammasomes [120, 121]. During apoptosis, efflux of mtDNA from the mitochondria occurs via IMM balloons that herniate out through BAX/BAK macropores in the OMM [122], but how mtDNA escapes the mitochondria during innate immune responses is not clear, although unlike apoptosis, cytochrome c is not released [123]. Newly synthesized mtDNA not yet packaged into Tfam-positive nucleoids may be most susceptible to oxidation, cleavage and release to activate either cGAS–STING or the NLRP3 inflammasome [124].
Oxidized mtDNA binds directly to the NLRP3 inflammasome, inducing pro-caspase-1 activation at the mitochondria and stimulating IL-1β release [125–127]. Mitochondrial caspase-1 activity causes further mitochondrial damage, including mitochondrial membrane depolarization that induces Parkin-dependent mitophagy [128], although since Parkin is cleaved by caspase-1, mitophagy may be limited depending on the scale of mitochondrial damage and onset of pyroptosis [129].
Activation of cGAS-STING by cytosolic mtDNA induces a Type I interferon response mediated by TANK-binding kinase-1 (TBK1) and IRF3 [121]. Amongst other activities, TBK1 promotes mitophagy by recruiting cargo receptors, such as OPTN to the OMM of damaged mitochondria [130, 131]. STING trafficking also promotes autophagy independent of TBK1 by stimulating autophagosome biogenesis at the ER-Golgi [132]. cGAS–STING and NLRP3 pathways communicate with each other during inflammatory responses, via effects of STING on release of potassium from the lysosome that activates NLRP3 [133]. Thus, by eliminating damaged mitochondria, mitophagy prevents further mtDNA release and attenuates activation of both cGAS–STING and the NLRP3 inflammasome [120, 121, 129, 134].
Mitophagy in cancer
PINK1/Parkin-dependent mitophagy in cancer
PINK1 and Parkin are encoded by genetic loci (PARK6 and PARK2, respectively) that are mutated in human Parkinson’s disease (PD), and also disrupted in various human cancers [135]. The PARK2 locus at 6q25-q26 is frequently deleted in bladder, breast, lung, ovarian, and other cancers [136], while PARK2 mutations have been linked to glioblastoma, colon cancer, and lung cancer [137]. Similarly, PARK6 expression (PINK1) is down-regulated in glioblastoma and ovarian cancer, with rare mutations found in neuroblastoma [138]. Analysis of tumor phenotypes in mouse models of cancer showed Parkin null mice to be susceptible to spontaneous hepatocellular carcinoma (HCC) and sensitized to irradiation-induced lymphomagenesis [139, 140]. Loss of either Parkin or PINK1 increased tumor burden and metastasis in KRas-driven PDAC [45, 141]. Parkin also suppressed the growth and metastatic progression of breast cancer cells [44]. Taken together, these findings indicate that Parkin and PINK1 play tumor suppressor functions in both mouse and human.
Given their canonical function in mitophagy, the tumor suppressor functions of Parkin and PINK1 have largely been attributed to their role in maintaining mitochondrial function and metabolic homeostasis by clearing depolarized mitochondria to prevent Warburg metabolism and excess ROS [45, 140, 142]. However, Parkin and PINK1 also suppress HIF-1α stabilization [44, 45, 138]; Parkin ubiquitinates HIF-1α on K477 to promote its degradation and low Parkin levels correlated with high HIF-1α levels and reduced metastasis-free survival in human breast cancer [44]. Deletion of HIF-1α (or iron chelation) rescued elevated glycolysis and ROS detected in PDAC following deletion of Parkin or PINK1 [45]. These observations indicate that Parkin/PINK1 may suppress tumorigenesis in part by promoting HIF-1α turnover (Fig. 4).
Fig. 4.
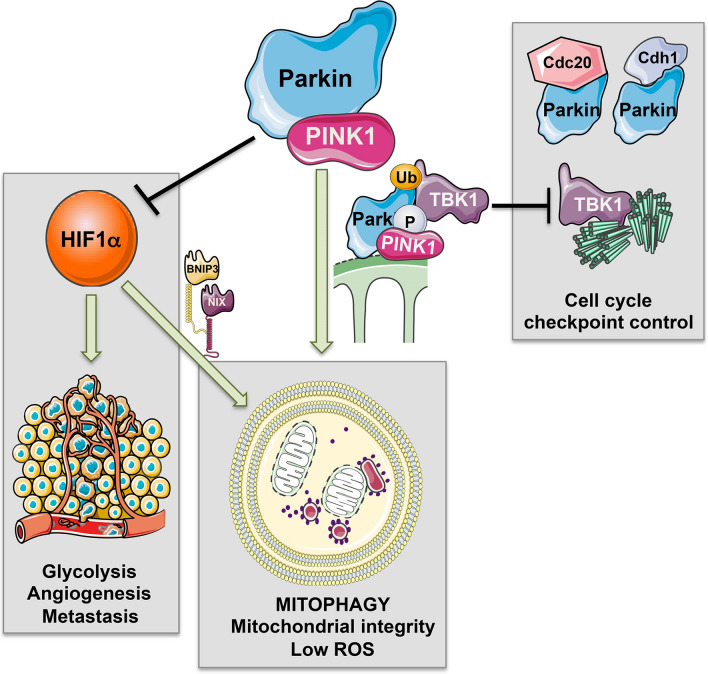
PINK1/Parkin functions relevant to tumor cell growth and tumorigenesis. In addition to their canonical role in promoting mitophagy of depolarized mitochondria, Pink1/Parkin also promote turnover of the HIF-1α transcriptional sub-unit that is known to promote tumorigenesis via induction of HIF target genes that enhance glycolysis, angiogenesis, and metastasis. Loss of Parkin or Pink1 could thus promote tumorigenesis as a result of increased HIF-1α levels. Furthermore, Parkin Pink1/Parkin-induced mitophagy modulates cell cycle checkpoints by sequestering TBK1 away from centrosomes where it promotes spindle assembly and mitosis. Thus increased mitophagy can inhibit cell cycle progression. Parkin also has Pink1-independent roles in regulating cell cycle by promoting Cdc20 and Cdh1 activity
Mitochondrial damage blocked G2-to-M-phase cell cycle transition in a PINK1/Parkin-dependent manner, while loss of Parkin or PINK1 increased rates of cell division [143]. Parkin’s E3 Ub ligase activity modulates cyclin levels and the mitotic checkpoint independently of the Anaphase-Promoting Complex (APC) and its E3 Ub ligase activity, via direct interaction of Parkin with Cdc20 and Cdh1 [144, 145]. This mitotic function for Parkin was independent of PINK1 and required activation of Parkin via phosphorylation on S378 by Polo-like kinase-1 [145]. Thus, Parkin has dual roles in mitophagy and in control of cell cycle. More recently, PINK1-dependent Parkin activity has been reported to modulate mitosis indirectly by sequestering TBK1 at damaged mitochondria [143]. In addition to its role in recruiting OPTN and other cargo receptors to damaged mitochondria [130, 131], TBK1 is a centrosome-associated protein required for microtubule dynamics and mitosis [146]. By sequestering TBK1 at damaged mitochondria and away from centrosomes, PINK1/Parkin activity caused a cell cycle arrest and failure to enter mitosis [143]. Artificially tethering TBK1 to mitochondria blocked cell cycle progression into mitosis, while deletion of Parkin rescued cell cycle progression defects in Drosophila lacking ik2, the fly homoplog of TBK1 [143]. Thus, negative control of cell cycle and de-stabilization of HIF-1α are added aspects to the tumor suppressor functions of PINK1/Parkin, although, arguably, these functions are secondary to their function in eliminating dysfunctional mitochondria (Fig. 4). How the PINK1-independent function of Parkin in promoting anaphase transition via interaction with Cdc20 [145] is coordinated with its PINK1-dependent function in blocking mitosis via TBK1 sequestration [143] may depend on whether mitochondria are functional or not.
There is a growing appreciation that E3 Ub ligases, other than Parkin, play a role in mitophagy and can substitute for Parkin [147]. For example, the MUL1 E3 ubiquitin ligase can compensate for Parkin loss in Drosophila and is required for elimination of paternal mitochondria during embryogenesis [14, 15]. While Parkin is poorly expressed in most cancers and cancer cell lines, ARIH1 is an E3 ubiquitin ligase recruited by PINK1 to mitochondria to promote mitophagy in response to chemotherapeutic agents and in doing so protects cancer cells and causes drug resistance [16]. ARIH1 thus represents a valid therapeutic target. Interestingly, while ARIH1 functions similarly to Parkin in requiring PINK1 and ubiquitinating mitochondrial proteins, its substrates at the mitochondria are different, but remain uncharacterized [147]. The extent to which other mitochondrial E3 Ub ligases perform Parkin-like functions in mitophagy that are relevant to cancer is an area of growing interest.
BNIP3/BNIP3L in cancer
BNIP3 protein levels are induced at early pre-malignant stages of various human solid cancers as tumors become hypoxic, including breast cancer and pancreatic cancer, but frequently down-regulated as these tumors progress to becoming invasive [148–150]. Hyper-methylation of the BNIP3 promoter is a common mechanism of BNIP3 inactivation in human cancers, including in gastric, pancreatic, liver, lung, and hematological malignancies [149, 151]. Epigenetic silencing of BNIP3 in human pancreatic cancer was linked to chemoresistance and poor prognosis [150, 152], consistent with tumor-suppressive functions for BNIP3. In contrast to BNIP3, elevated BNIP3L expression in a cohort of PDAC patients was linked to worse prognosis and loss of BNIP3L in the KPC model of PDAC (LSL-KRasG12D; p53R172H; Pdx1-Cre) delayed tumor progression from the PanIN stage to malignant PDAC [153]. These findings suggest that BNIP3 and BNIP3L may play different roles in PDAC, possibly due to their expression in different cell types or at different stages of disease progression, although this requires further analysis.
The BNIP3 locus at the end of chromosome 10 (chr.10q26.3) is also subject to deletion in a number of cancers including in breast and prostate cancer [154]. BNIP3 is most frequently deleted in triple-negative breast cancer (TNBC) [155, 156]. TNBC is the sub-type of breast cancer that exhibits the strongest HIF signature and Warburg metabolism [157] and loss of BNIP3 combined with high HIF-1 expression predicted poor metastasis-free survival specifically for TNBC patients [156]. TNBC is also the breast cancer sub-type with the highest degree of mitochondrial dysfunction, possibly due to loss of BNIP3 and defective mitophagy, and this could potentially be exploited therapeutically to target mitochondrial function in TNBC and bring a novel treatment option to this deadly form of breast cancer [158, 159]. Supporting data from human studies, BNip3 knockdown promoted tumor growth and metastasis in the 4T1 orthotopic model of mammary tumorigenesis [160]. Similarly, loss of BNip3 in the MMTV-PyMT mouse model of breast cancer increased tumor burden and accelerated progression to invasive carcinoma and lung metastasis [156]. Increased tumor burden and invasiveness of BNip3 null mammary tumors were associated with reduced mitophagy, increased ROS, and elevated HIF-1 levels, contributing to increased glycolysis and angiogenesis [156]. Quenching ROS attenuated HIF-1 levels and reduced metastasis in the BNip3 null mice, indicating that BNIP3 suppressed tumorigenesis in this model by reducing mitochondrial mass and limiting ROS production and explaining why there is selective pressure to inactivate BNIP3 downstream of HIF-1, as tumors progress to becoming invasive [156].
BNIP3 has also been shown in other tumor types to promote tumor cell survival following detachment from extracellular matrix (ECM) and high BNIP3 correlated in these primary human cancers (kidney, sarcoma) with worse patient outcomes [161]. These dichotomies may be explained by differential roles for BNIP3 depending on tumor type or in different cell types within a tumor (cancer stem cell versus non-stem cell, for example). As we have discussed previously, mitophagy plays an important role in maintaining the cancer stem cell phenotype [162]. For example, mitophagy limited p53 levels in HCC stem cells via PINK1-mediated phosphorylation of mitochondrial p53 that gets turned over by mitophagy resulting in de-repression of the stem cell factor, Nanog [163]. Mitophagy also enhances stemness by promoting glycolysis and limiting oxidative metabolism, thus engendering the low energy state required for stemness [162] and is required for segregation of young mitochondria to daughter stem cells that is required to maintain stem cell fitness [164]. Consistently, autophagy reduced replication stress and aging in hematopoietic stem cells [165–167]. Specifically, stimulating mitophagy with nicotinamide riboside (NR) that increases the cellular NAD+/NADH ratio and stimulates Sirtuin activity promoted hematopoietic stem cell self-renewal [168]. The extent to which BNIP3/BNIP3L or other mitophagy pathways promote cancer stemness remains an open area of research.
FUNDC1 in tumor cell proliferation and migration/invasion
The interaction of FUNDC1 with LC3 is inhibited by SRC-mediated phosphorylation on Y18 within the LIR motif of FUNDC1 [62], suggesting that FUNDC1-dependent mitophagy is antagonistic to SRC-driven effects on tumor cell migration and invasion. FUNDC1 also recently emerged from an shRNA screen for genes that modulate tumor cell migration and invasion in response to mitochondrial proteotoxic stress [169]. Silencing FUNDC1 increased rates of focal adhesion assembly and disassembly, and increased cellular motility and invasion in a DRP1-dependent manner through Matrigel of both prostate and glioblastoma cell lines [169]. Conversely, over-expressing FUNDC1 limited cell migration and invasion. The increased migration and invasion caused by decreased FUNDC1 expression was associated with increased liver metastasis in vivo in xenograft models, but conversely also resulted in decreased tumor cell proliferation. RNA-Seq analysis of 33 different tumor types in the TCGA database showed increased FUNDC1 levels to be associated with high levels of expression of genes involved in mitochondrial bioenergetics, while low levels of FUNDC1 were associated with increased ROS signaling and metastasis, particularly in lung cancer. These findings are at odds with previous work in which elevated FUNDC1 in human breast cancer was associated with increased tumor cell proliferation, migration, and invasion and with worse prognosis for breast cancer patients [170], and additional work will be needed to explain these divergent findings.
Nevertheless, it has been suggested that the role of FUNDC1 in tumor cell proliferation and motility is mitophagy-independent and reflects a more significant role for FUNDC1 in mitochondrial bioenergetics [169]. Knockdown of FUNDC1 decreased levels of TCA cycle intermediates, lowered respiration rates, and increased ROS production in tumor cells that was associated with increased degradation of electron transport chain complexes. This was rescued by exogenous expression of the LonP protease that is known to maintain the integrity of electron transport chain complexes and was misfolded as a result of FUNDC1 loss, possibly related to its role in resolving mitochondrial proteostatic stress via interaction with HSC70 [66]. Interestingly, exogenous LonP also rescued the effect of FUNDC1 loss on tumor cell proliferation and invasion, suggesting that FUNDC1 modulates tumor cell behavior independent of its function in mitophagy [169].
Mitophagy defects in XP, AT, and FA cancer predisposition syndromes
Mitophagy defects have emerged as a feature of the cancer predisposition syndromes Xeroderma pigmentosum (XP), Ataxia telangiectasia (AT), and Fanconi Anemia (FA) that arise due to defects in nucleotide excision repair (XP) and double-strand break repair (AT, FA) [114, 115, 171–174].
Work from the lab of Beth Levine identified FANC-C and other Fanconi complex components (FANC-A, FANC-D2, FANC-F, FANC-L, BRCA1, and BRCA2) as required for Parkin-mediated mitophagy as well as virophagy of Sindbis virus [174, 175]. FANC-C co-localized with Parkin at depolarized mitochondria and loss of FANC-C caused dysfunctional mitochondria to accumulate and elevated mitochondrial ROS production, as a result of defective mitophagy. This role of FANC-C in mitophagy was genetically separable from its function in DNA repair [174]. Possibly due to the defect in mitophagy, there was also increased inflammasome activation in FA cells that, along with elevated ROS levels, contributed to bone marrow failure and increased incidence of hematological pathologies in FA individuals, while defective virophagy could explain why FA patients are more prone to infection than others [174]. The FANC-C c.67delG mutation is common in Dutch FA patients and is associated with a milder form of the disease, including older age at diagnosis, lower rate of bone marrow failure, and reduced incidence of malignancy [176]. Intriguingly, this mutant form of FANC-C retained its mitophagy function, but had lost its ability to promote DNA repair, indicating that the loss of mitophagy does contribute significantly to disease pathology [174].
Loss of ATM kinase increased mitochondrial ROS and caused mtDNA depletion [177–179] and neurons lacking ATM exhibited reduced mitophagy [114]. Replenishing NAD+ levels with NR or via PARP inhibition promoted mitophagy, rescued cell death, and stimulated neuronal differentiation of ATM-deficient neurons [114]. Similarly, PARP inhibition rescued mitophagy and neurodegeneration in XPA patients [115]. Boosting NAD+ levels also induced mitophagy in long-term repopulating hematopoietic stem cells and promoted long-term hematopoiesis in mice fed NR, consistent with a role for mitophagy in promoting stemness and longevity [162, 168]. The extent to which NAD+ depletion contributes to the mitophagy defect in FA and whether it also can be rescued by NR or PARP inhibition has not been examined.
From a therapeutic angle, inducing mitophagy with NR or PARP inhibition could be beneficial if it prevents accumulation of DNA mutations arising from mitochondrial dysfunction, but could be detrimental if it promotes tumor cell survival. For example, DNA-damaging agents by depleting NAD+ would be predicted to cause mitochondrial defects and loss of survival if mitophagy gene induction is inhibited. Mitophagy induced via replenishment of NAD+ (via NR or PARP inhibition) may thus limit the efficacy of genotoxic agents, such as cisplatin, or 5-fluorouracil.
Intersection of mitophagy with mitochondrial dynamics
Mitochondrial fission is required for mitophagy in response to certain stresses [180–182] and knocking out DRP1 in the liver resulted in unopposed mitochondrial fusion and caused p62-positive mitophagy intermediates to accumulate [182]. This was rescued by liver-specific knockout of OPA1 [182], consistent with previous work, showing that healthy mitochondria are protected from turnover during nutrient deprivation by OPA1-dependent fusion [180, 181].
Mitophagy may require mitochondrial fission due to the limiting size constraints on autophagosomal cargo that can be engulfed successfully, and since mitochondrial fusion requires membrane polarization, this would also selectively target depolarized mitochondria for turnover [180, 181]. Indeed, DRP1 is activated by loss of MMP (ΔΨmt) and recruited to the OMM via the MFF receptor [183, 184].
While FIS1 is now considered redundant for recruitment of DRP1 to mitochondria [185], FIS1 is required for PINK1 recruitment to mitochondria, acts downstream of Parkin to promote mitophagy and FIS1-dependent mitophagy in leukemia stem cells (LSCs) promoted responses to chemotherapy [89]. FIS1 promotes mitophagy mainly via recruitment of Rab-GAPs TBC1D15 and TBC1D17 that dimerize to modulate Rab7 GTPase activity and also interact with LC3/GABARAP family members ensuring the complete encapsulation of mitochondria by autophagosomes [186]. In this way, FIS1 is less of a fission factor and more a direct mitophagy regulator. Indeed, mutation of FIS1 led to accumulation of large LC3-positive aggregates [187].
At the OMM, DRP1 interacts with ZIP1 to modulate MMP via uptake of Zn2+ through the mitochondrial calcium uniporter (MCU) [188]. DRP1 interacts with ZIP1 at sites of mitochondrial fission, although GTPase-deficient DRP1 retains ability to interact with ZIP1 and to induce ΔΨmt, indicating two separable functions for DRP1 in mitophagy [188]. The DRP1–ZIP1 interaction was required for Parkin recruitment to mitochondria in response to hyperglycemia, and chelating Zn2+ or blocking the DRP1–ZIP1 interaction inhibited Parkin recruitment and mitophagy [188]. As a result, fragmented mitochondria with reduced MMP accumulated as “damaged” mitochondria that could not be turned over by mitophagy. BNIP3 and FUNDC1 also interact with DRP1 and may function as alternative receptors for DRP1, since DRP1 is required for hypoxia-induced mitophagy [65, 189]. These findings highlight how control of mitochondrial dynamics is coordinated to promote mitophagy.
Mitochondrial fission and DRP1 phosphorylation on S616 by ERK2 are required for oncogenic RAS transformation [190, 191]. DRP1-dependent fission was also required for B-RAFV600E transformation in melanoma [192, 193]. Inhibition of DRP1 phosphorylation on S616 induced mitochondrial fusion and blocked tumor cell growth in xenografts, while pancreas-specific deletion of Drp1 inhibited progression from PanIN to pancreatic ductal adenocarcinoma (PDAC) in KPC mice [190, 194]. At this time, it is not understood why mitochondrial fission is required for Ras transformation, but this is clearly of major interest, since ERK inhibitors are used therapeutically against Ras-driven cancers, including human PDAC [195]. Intriguingly, inhibition of Ras or ERK1/2 inhibition induces autophagy that was associated with reduced mitochondrial mass [196], and thus, one could speculate that mitochondrial fission is required for Ras transformation to promote mitophagy and mitochondrial homeostasis. Ras transformation induces expression of BNIP3 and BNIP3L [153, 197] and loss of BNip3L attenuates PDAC formation in KPC mice [153], but studies to demonstrate a requirement for BNIP3/BNIP3L downstream of DRP1 in Ras transformation have not been performed. The combination of ERK1/2 inhibition with autophagy inhibition was shown to be strikingly effective at blocking tumor cell growth in vitro [195, 196], and in patients where the combination of Trametinib with hydroxychloroquine significantly reduced PDAC tumor burden [195]; these studies are now being expanded in clinical trials (NCT03825289).
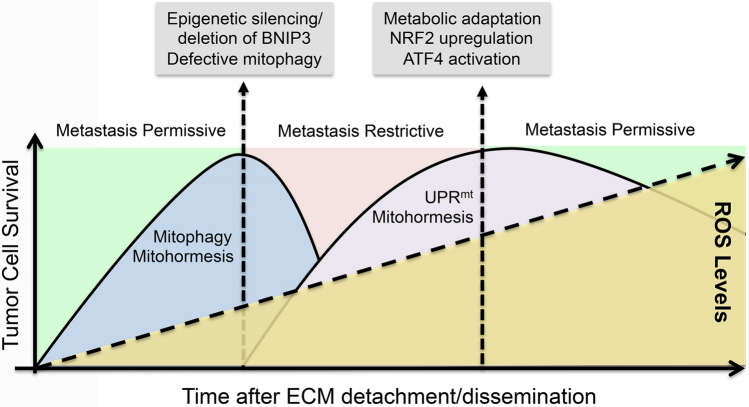
Mitophagy, ROS, and metastasis
Mitochondrial dysfunction has been linked to metastasis most frequently as a result of increased ROS production, for example arising due to mtDNA mutation [198] or ETC inhibition [199]. Indeed, mitochondrial ROS generation following recovery of host mtDNA in mitochondria-depleted tumor cells was required for metastatic progression in both the 4T1 and B16-F10 orthotopic mouse models of metastasis [200, 201]. Increased ROS has been shown to drive the progression to metastasis in a number of other tumor models also, including in PDAC where loss of TIGAR promoted ROS-driven EMT, invasiveness, and metastasis to the lungs, which was mitigated by treatment with N-acetyl cysteine (NAC) [202]. ROS drives metastasis in part by activating HIF-1 and NF-κB to induce EMT and tumor cell invasion, respectively [203, 204]. ROS also inactivates protein phosphatases and activates key signaling kinases, such as ERK, JNK, and p38 that stimulate various pro-metastatic pathways [205, 206]. However, it has also been argued that ROS limits metastasis in melanoma where circulating tumor cells showed higher levels of oxidative stress than primary tumor cells, and treatment of mice with NAC increased metastasis without affecting primary tumor burden, suggesting that many circulating metastatic cells were not surviving in the circulation due to excess ROS [207, 208]. Successfully metastasizing tumor cells had upregulated pathways (such as folate metabolism to produce more NADPH) to manage ROS and survive [207]. Taken together, this suggests that while ROS is required for metastasis, too much ROS is deleterious to the disseminated tumor cell and limits metastasis (Fig. 5).
Fig. 5.
ROS, mitophagy, and metastasis. Extensive evidence shows that ROS is required for tumor cell migration and metastasis. However, other data shows that excess ROS can limit metastasis, which has lead to a model in which levels of ROS need to be maintained within a range, the so-called hormetic zone, to promote metastasis. ROS induces mitohormetic responses, including the UPRmt that promote mitochondrial fitness and limit ROS allowing tumor cells to survive and progress to metastasis. By limiting ROS, mitophagy may inhibit these adaptive responses and work suggests there is selection for BNIP3 loss during progression to invasiveness in breast cancer. However, the ensuing higher levels of ROS that occur once BNIP3 or other mitophagy genes are lost, would not be expected to restrict metastasis due to upregulation of other components of the UPRmt. Selection for tumor cells that survive, possibly by upregulating stress response pathways that manage ROS, such as the NRF2 anti-oxidant response, or ATF4 that promotes amino acid biosynthesis as part of the UPRmt, would promote cell survival and be permissive for metastasis again
This is consistent with data, showing that mitochondrial ROS needs to be within the hormetic zone to promote metastasis [209], as shown in studies reporting how high ROS levels promote breast cancer invasiveness in vitro and in vivo, in part by inducing the UPRmt [210]. ROS-induced UPRmt consisted of a transcriptional response involving CHOP, ATF5, NRF2, FOXO3A, and SIRT3, which activated genes that restored mitochondrial function [209]. Kenny et al. showed that the UPRmt is upregulated in metastatic tumors relative to primary tumors, consistent with the UPRmt conferring a metastatic advantage and high UPRmt in primary human cancer correlated with poor prognosis [210]. Thus, ROS at non-lethal levels may promote mitochondrial fitness, tumor cell viability, and metastasis in part by activating the UPRmt.
Mitophagy genes are induced as part of the UPRmt in response to ROS and nutrient deprivation, including induction of p62/Sqstm1 by NRF2 and BNIP3 by FOXO3A. As discussed above, loss of BNip3 promoted more rapid progression to metastasis in both the autochtonous MMTV-PyMT mammary tumor model and in the 4T1 orthotopic model of breast cancer [156, 160]. Significantly, mammary tumors forming in MMTV-PyMT;BNip3−/− mice had higher mitochondrial mass and higher ROS levels than wild-type control tumors, and expressed elevated nuclear HIF-1 and HIF target genes [156]. Treatment of MMTV-PyMT mice with anti-oxidant attenuated mammary tumor metastasis to the lungs in BNip3 null mice to that seen in wild-type mice, without affecting primary tumor growth [156]. This work indicated that BNIP3 was suppressing metastasis by promoting mitochondrial turnover and limiting ROS that would otherwise cause HIF-1α stabilization [211].
However, BNIP3 and BNIP3L are induced in tumor cells following detachment from the extracellular matrix (ECM) [212] and required for mitophagy and tumor cell survival following detachment [161] suggesting a pro-metastatic function for BNIP3 and BNIP3L. The induction of BNIP3 and BNIP3L in response to detachment was HIF-1 dependent due to hypoxia within tumor cell clusters and associated with a switch to glycolysis and reductive carboxylation of glutamine [161]. Preventing cell clustering, knocking down HIF-1α, or knocking down BNIP3 and BNIP3L inhibited mitophagy and increased mitochondrial mass and ROS levels leading to cell death [161]. BNIP3 has also been shown to promote the migration of B16-F10 melanoma cells through effects on the actin cytoskeleton [213]. Similarly, BNIP3 is induced in the skin by hypoxia where BNIP3-dependent mitophagy is required for keratinocyte migration during wound healing [214]. It is not yet clear how mitophagy modulates the actin cytoskeleton or is otherwise modulating cell migration and this is an avenue for future investigation.
Thus, similar to the impact of ROS on metastasis, these results indicate that BNIP3 (or BNIP3L) may be pro- or anti-metastatic depending on context and whether BNIP3 (or BNIP3L) is lost acutely, or is stably deleted in systems where tumor cells are then able to adapt by upregulating other pathways that may compensate for BNIP3/BNIP3L loss, such as increasing NRF2 target gene expression to mitigate against ROS (Fig. 5). Recent work has identified NRF2 activation as a mechanism by which tumor cells survive autophagy inhibition [215, 216]. NRF2 is upregulated in autophagy-deficient cells due to elevated p62/Sqstm2 levels [215, 217] and may contribute to ATF4 induction in autophagy-deficient cells both due to depletion of amino acids but also by interacting directly with ATF4 [215]. Whether mitophagy inhibition is as effective as inhibition of general autophagy at inducing NRF2, through increased p62/Sqstm1 levels, has not been examined but if not, may indicate that specific mitophagy inhibition would be more effective than autophagy inhibition in cancer therapy.
Mitophagy and cancer cachexia
Another debilitating aspect to terminal cancer is the development of cancer cachexia that is found most commonly in lung, pancreas, and colorectal cancer patients [218, 219]. Cancer cachexia entails rapid body weight loss due to peripheral tissue wasting that negatively affects the long-term survival of cancer patients, partly due to reduced tolerance of and response to therapy [218, 220, 221]. There are many factors playing into cachexia, including release of pro-inflammatory cytokines that affect brain, liver, and immune cell function that feed into breakdown of lipid in adipose tissue and protein mass in skeletal muscle [222]. Previous work identified BNIP3 as a transcriptional target of FoxO3A during skeletal muscle atrophy [35]. It was also shown that FoxO transcriptional activity was required for muscle atrophy in C57/B6 mice implanted with Lewis lung carcinoma (LLC) and that BNIP3 mRNA was elevated in an FoxO-dependent manner in the muscle of these tumor-bearing mice [36, 37]. BNIP3 upregulation has also been detected in cachectic muscle from PDAC patients [223]. Muscle atrophy in cancer cachexia has been linked to reduced oxidative capacity and mitochondrial dysfunction in skeletal muscle [37, 224, 225]. Taken together, this may suggest that BNIP3-dependent mitophagy and lower mitochondrial mass promotes muscle atrophy during cancer cachexia by facilitating the switch away from oxidative metabolism to glycolysis. This awaits further testing in the laboratory, but is clearly an avenue of high significance given its relevance to disease and patient suffering.
Targeting mitophagy as a therapeutic approach
Inhibiting general autophagy is advancing in clinical trials for many different human cancers, primarily in combination with other drug regimens [226]. As mentioned above, autophagy inhibition (with chloroquine) in combination with Trametinib to inhibit ERK signaling showed significant efficacy in reducing tumor burden in KRas-driven PDAC [195]. Autophagy is induced by many therapeutic agents, and by promoting survival elicits drug resistance [227]. This has provided the rationale for combining autophagy inhibitors, such as chloroquine with drugs like temozolomide in the treatment of glioblastoma, where the combined therapy more than doubled patient survival times [228–230]. To what extent these effects are mediated via inhibition of mitophagy versus inhibition of other autophagy functions is not clear.
Functional mitochondria are required for the biosynthesis of certain amino acids, such as aspartate that is required for nucleotide biosynthesis, upon which tumor cells rely for growth, and recent studies showed that inhibiting autophagy with chloroquine synergized with agents that induce replication stress that depletes reserves of nucleotides that rely on amino acids for their synthesis [231]. Furthermore, ferroptosis is a necrosis-like cell death that is activated by oxidized mitochondrial lipids and preceded by ΔΨmt, mitochondrial fragmentation, and disrupted cristae formation, that can be prevented by activating mitophagy through over-expression of Parkin [232]. These studies suggest that inhibiting mitophagy may promote ferroptosis, although it is important to note that mitochondria were only required for ferroptosis induced by cysteine deprivation and not in response to glutathione peroxidase-4 (GPX4) inhibition. Findings such as these provide rationale for developing drugs that specifically inhibit mitophagy to achieve a more targeted effect than with targeting autophagy in general [233] (Fig. 6).
Fig. 6.
Targeting mitophagy in cancer therapy. Mitophagy is induced physiologically in response to hypoxia and nutrient deprivation or can be activated with drugs that induce mitochondrial stress (see Fig. 3), such as Metformin or PARP inhibitors. Alternatively, mitophagy can be inhibited using generic autophagy inhibitors such as chloroquine since as yet specific mitophagy inhibitors remain to be developed. Activating mitophagy promotes mitochondrial homeostasis and cell survival while inhibiting mitophagy, by causing mitochondrial dysfunction, elevated ROS production, metabolite deficiencies, including deficiency for key amino acids, such as cysteine, frequently results in cell death. Based on this rationale, inhibiting mitophagy seems more likely to elicit a clinical benefit in cancer therapy, while activating mitophagy would likely promote cell survival and therefore not be advisable as a cancer therapy, although possibly useful for treatments of myopathies where mitophagy has been shown to promote tissue homeostasis. However, it is likely not a simple as this, since cancer cells may adapt to mitophagy inhibition, for example by upregulating NRF2 signaling to allow cancer cell survival. These studies require much more extensive analysis in the lab and clinic before conclusions can be made
The use of such drugs to inhibit mitophagy would be expected to cause excess dysfunctional mitochondria to accumulate and thus would be expected to generate high levels of ROS as seen when mitophagy is inhibited genetically [156, 161]. This could promote cell killing, but could also induce progression to metastasis depending on the level of ROS generated. As we discussed previously [234], the combination of a drug to inhibit mitophagy with compounds that induce mitochondrial dysfunction and elevated ROS production, such as Metformin or Mitoquinone, may push the tumor cell further in the direction of cell death rather than adaptation [234], although activation of NRF2 in response to some of these agents [235] remains a challenge.
There is also an argument that promoting mitophagy could be beneficial. For example, there was selection for silencing of BNIP3 in drug-resistant PDAC, while re-expression of BNIP3 promoted drug sensitivity [150, 152], although the mechanistic underpinnings of this were not further pursued. The cotton seed derived compound AT-101 induces BNIP3/BNIP3L-dependent mitophagy leading to autophagic cell death in apoptosis-deficient tumor cells [236]. Mitophagy is elicited in response to AT-101 following rapid loss of membrane potential and marked induction of heme oxygenase (HMOX1) [236]. HMOX1 has been shown to induce mitophagy in other systems and the concern here would be the effect of AT-101 on normal tissues with high redox potential, such as the brain and red blood cells. Most of the drugs identified as inducing mitophagy, like AT-101, do so indirectly by depolarizing mitochondria [236] or modulating levels of ROS or NAD+ that activate NRF2 and Sirtuins, respectively [233]. For example, PMI is a Keap1 inhibitor that induces mitophagy through activation of NRF2, and thus, mitophagy is unlikely to be the only effect of such a drug [237]. Compounds that directly activate or modulate MCRs for example are lacking. However, as discussed above by maintaining mitochondrial fitness, stresses that induce mitophagy promote survival [53], which may be a useful approach in the treatment of myopathies and other diseases, but would not be a good outcome in cancer therapy. Overall, acute inhibition of mitophagy in combination with other drugs/stresses that depend on mitophagy for survival remains the approach most likely to yield efficacy in cancer treatment (Fig. 6).
Summary and future directions
Mitophagy is a core component of the mitochondrial quality control system in the cell alongside the UPRmt and other mitochondrial surveillance mechanisms that limit mitochondrial ROS production, ensure metabolic homeostasis, and prevent apoptosis. There are numerous mitophagy pathways in the cell and how each of these different pathways interact with each other, are redundant versus overlapping or are activated by oncogenic signaling specifically remains to be determined in most instances. Furthermore, how mitophagy pathways interact with the UPRmt and other mitochondrial surveillance processes also requires continued investigation. In cancer, various mitophagy pathways appear to be tumor-promoting or tumor-suppressive depending on tumor type or indeed stage of progression, or importantly, in cancer stem cells versus non-stem cells. Teasing these nuanced roles apart will require further study through use of relevant pre-clinical cancer models that target these pathways in appropriate spatial and temporal ways. The role of mitophagy in cancer cachexia is emerging as an area of significant interest given the debilitating effect of tissue wasting on a cancer patient’s long-term survival. Finally, work is on-going to determine how mitophagy might be manipulated therapeutically to better treat cancer, but again, this may depend on tumor type and stage of progression, and this is where the development and use of improved pre-clinical cancer models targeting mitophagy pathways will be essential.
Abbreviations
- AD
Alzheimers disease
- ADP
Adenosine diphosphate
- ALS
Amyotrophic lateral sclerosis
- AMPK
5′ AMP-activated protein kinase
- ANT
Adenine nucleotide transporter
- ARIH1
Ariadne RBR E3 ubiquitin protein ligase 1
- ASNS
Asparagine synthetase
- ASS1
Argininosuccinate synthase-1
- Atad3a
ATPase family AAA-domain-containing protein 3 A
- ATF4
Activating transcription factor 4
- ATM
Ataxia telangiectasia mutated
- ATP
Adenosine triphosphate
- BCL-2
Breakpoint cluster locus-2
- BCL-XL
BCL2-like 1 long
- BNIP3
BCL2 interacting protein 3
- BNIP3L
BCL2 interacting protein 3 like
- BCL2-L-13
BCL2-like-13
- BRCA1
Breast cancer 1
- CCCP
Carbonyl cyanide 3-chlorophenylhydrazone
- CK2
Casein kinase-2
- CL
Cardiolipin
- DAG
Diacyl glycerol
- DELE1
DAP3-binding cell death enhancer 1
- DTT
Dithiothreitol
- DRP1
Dynamin-related protein 1
- ECM
Extracellular matrix
- ERK
Extracellular signal-regulated kinase
- ETC
Electron transport chain
- FA
Fanconi anemia
- FANC-C
FA protein C
- FIS1
Mitochondrial fission 1 protein
- FoxO3
Forkhead box protein O3
- FKBP8
FK506-binding protein 8
- FUNDC1
FUN 14 domain-containing 1
- GABARAP
GABA Type A Receptor-Associated Protein
- GAP
GTPase activating protein
- cGAS
Cyclic GMP–AMP synthase
- GPX4
Glutathione peroxidase-4, GTPase activating protein
- HIF
Hypoxia inducible factor
- HCC
Hepatocellular carcinoma
- HRI
Heme-regulated inhibitor
- IMM
Inner mitochondrial membrane
- IRF3
Interferon response factor 3
- LC3
Microtubule-associated protein 1 light chain 3
- LIR
LC3 interacting region
- LKB1
Liver kinase B1
- LLC
Lewis Lung carcinoma
- MARCH5
Membrane-associated ring finger (C3HC4) 5
- MCR
Mitochondrial cargo receptor
- MITF
Melanocyte inducing transcription factor
- MFF
Mitochondrial fission factor
- MFN
Mitofusin
- MMP
Mitochondrial processing peptidase
- MPP
Mitochondrial processing peptidase
- mtDNA
Mitochondrial DNA
- MUL1
Mitochondrial E3 ubiquitin ligase 1
- NAC
N-acetyl cysteine
- NAD +
Nicotinamde adenine dinucleotide
- NBR1
Near BRCA1
- NDP52
Nuclear dot protein 52
- NLRP3
Nucleotide binding domain and leucine rich repeat pyrin domain-containing 3
- NR
Nicotinamide riboside
- NRF2
Nuclear factor, erythroid 2 like
- OMM
Outer mitochondrial membrane
- OPTN
Optineurin
- OXPHOS
Oxidative phosphorylation, p62/SQSTM1
- PA
Phosphatidic acid
- PARK2
Parkin encoding locus
- PARK6
PINK1 encoding locus
- PARK8
LRRK2 encoding locus
- PARL
PINK/PGAM5-associated rhomboid-like proteases
- PARP
Poly-ADP-ribose polymerase
- PD
Parkinson’s disease
- PDAC
Pancreatic ductal adenocarcinoma
- PFKFB3
6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
- PGAM5
Phosphoglycerate mutase family member 5
- PHB-2
Prohibitin-2
- PHGDH
D-3-phosphoglycerate dehydrogenase
- PINK1
PTEN-induced putative kinase 1
- PMI
P62-mediated mitophagy inducer
- PSAT1
Phosphoserine aminotransferase 1
- RAF
Rapidly accelerated fibrosarcoma oncogene
- RHEB
Ras homolog enriched in brain
- ROS
Reactive oxygen species
- S1P
Sphingosine-1-phosphate
- SDS
Sodium dodecyl sulfide
- SPHK1
Sphingosine kinase 1
- SRC
Rous sarcoma oncogene
- STING
Stimulator of Interferon Genes
- TAX1BP1
Tax1-binding protein 1
- TAZ
Tafazzin
- TBK
TANK-binding kinase 1
- TCA
Tricarboxylic acid cycle
- TIM
Translocase of inner membrane
- TFE
Transcription factor E
- TMD
Transmembrane domain
- TNBC
Triple-negative breast cancer
- TOM
Translocase of outer membrane
- ULK1
Unc-51-like autophagy activating kinase 1
- UPRmt
Mitochondrial unfolded protein response
- VPS34
Vacuolar protein sorting 34
- XP
Xeroderma pigmentosum
- ZIP1
Zinc transporter-1 precursor
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Palikaras K, Lionaki E, Tavernarakis N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat Cell Biol. 2018;20(9):1013–1022. doi: 10.1038/s41556-018-0176-2. [DOI] [PubMed] [Google Scholar]
- 2.Macleod KF. Mitophagy and mitochondrial dysfunction in cancer. Ann Rev Cancer Biol. 2020;4:41–60. [Google Scholar]
- 3.Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harper JW, Ordureau A, Heo JM. Building and decoding ubiquitin chains for mitophagy. Nat Rev Mol Cell Biol. 2018;19(2):93–108. doi: 10.1038/nrm.2017.129. [DOI] [PubMed] [Google Scholar]
- 5.Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12(2):119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Dorn GW. PINK1-phosphorylated Mitofusin-2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarraf SA, Raman M, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP, Harper JW. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;496:372–376. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sekine S, Kanamaru Y, Koike M, Nishihara A, Okada M, Kinoshita H, Kamiyama M, Maruyama J, Uchiyama Y, Ishihara N, Takeda K, Ichijo H. Rhomboid protease PARL mediates the mitochondrial membrane potential loss-induced cleavage of PGAM5. J Biol Chem. 2012;287(41):34635–34645. doi: 10.1074/jbc.M112.357509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sekine S, Wang C, Sideris DP, Bunker E, Zhang Z, Youle RJ. Reciprocal roles of Tom7 and OMA1 during mitochondrial import and activation of PINK1. Mol Cell. 2019;73(5):1028–1043.e1025. doi: 10.1016/j.molcel.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Jin G, Xu C, Zhang X, Long J, Rezaeian AH, Liu C, Furth ME, Kridel S, Pasche B, Bian XW, Lin HK. Atad3a suppresses Pink1-dependent mitophagy to maintain homeostasis of hematopoietic progenitor cells. Nat Immunol. 2018;19(1):29–40. doi: 10.1038/s41590-017-0002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoshino A, Wang WJ, Wada S, McDermott-Roe C, Evans CS, Gosis B, Morley MP, Rathi KS, Li J, Li K, Yang S, McManus MJ, Bowman C, Potluri P, Levin M, Damrauer S, Wallace DC, Holzbaur ELF, Arany Z. The ADP/ATP translocase drives mitophagy independent of nucleotide exchange. Nature. 2019;575(7782):375–379. doi: 10.1038/s41586-019-1667-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin SM, Youle RJ. The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy. 2013;9(11):1750–1757. doi: 10.4161/auto.26122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fiesel FC, James ED, Hudec R, Springer W. Mitochondrial targeted HSP90 inhibitor Gamitrinib-TPP (G-TPP) induces PINK1/Parkin-dependent mitophagy. Oncotarget. 2017;8(63):106233–106248. doi: 10.18632/oncotarget.22287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yun J, Puri R, Yang H, Lizzio MA, Wu C, Sheng ZH, Guo M. MUL1 acts in parallel to the PINK1/parkin pathway in regulating mitofusin and compensates for loss of PINK1/parkin. eLife. 2014;3:e01958. doi: 10.7554/eLife.01958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rojansky R, Cha MY, Chan DC. Elimination of paternal mitochondria in mouse embryos occurs through autophagic degradation dependent on PARKIN and MUL1. eLife. 2016 doi: 10.7554/eLife.17896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villa E, Proics E, Rubio-Patino C, Obba S, Zunino B, Bossowski JP, Rozier RM, Chiche J, Mondragon L, Riley JS, Marchetti S, Verhoeyen E, Tait SWG, Ricci JE. Parkin-independent mitophagy controls chemotherapeutic response in cancer cells. Cell Rep. 2017;20(12):2846–2859. doi: 10.1016/j.celrep.2017.08.087. [DOI] [PubMed] [Google Scholar]
- 17.Chen Z, Liu L, Cheng Q, Li Y, Wu H, Zhang W, Wang Y, Sehgal SA, Siraj S, Wang X, Wang J, Zhu Y, Chen Q. Mitochondrial E3 ligase MARCH5 regulates FUNDC1 to fine-tune hypoxic mitophagy. EMBO Rep. 2017;18(3):495–509. doi: 10.15252/embr.201643309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarten M, Mohrluder J, Ma P, Stoldt M, Thielman Y, Stangler T, Hersch N, Hoffman B, Merkel R, Wilbold D. Nix binds to GABARAP: a possible crosstalk between apoptosis and autophagy. Autophagy. 2009;5(5):690–698. doi: 10.4161/auto.5.5.8494. [DOI] [PubMed] [Google Scholar]
- 19.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Löhr F, Popovic D, Occhipinti A, Reichert AS, Terzic J, Dötsch V, Ney PA, Dikic I. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11(1):45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanna RA, Quinsay MN, Orogo AM, Giang K, Rikka S, Gustafsson AB. Microtubule-associated protein 1 light chain 3 (LC3) interacts with BNip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem. 2012;287:19094–19104. doi: 10.1074/jbc.M111.322933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ney PA. Mitochondrial autophagy: origins, significance, and role of BNIP3 and NIX. Biochim Biophy Acta. 2015;1853:2775–2783. doi: 10.1016/j.bbamcr.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 22.Springer MZ, Poole LP, Drake LE, Bock-Hughes A, Boland ML, Smith AG, Hart J, Chourasia AH, Liu I, Bozek G, Macleod KF (2021) BNIP3-dependent mitophagy promotes cytosolic localization of LC3B and metabolic homeostasis in the liver. Autophagy Jan 17. 10.1080/15548627.2021.1877469 [DOI] [PMC free article] [PubMed]
- 23.Zhu Y, Massen S, Terenzio M, Lang V, Chen-Lindner S, Eils R, Novak I, Dikic I, Hamacher-Brady A, Brady NR. Modulation of serines 17 and 24 in the LC3-interacting region of BNIP3 determines pro-survival mitophagy VS apoptosis. J Biol Chem. 2012;288(2):1099–1113. doi: 10.1074/jbc.M112.399345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sulistijo ES, MacKenzie KR. Structural basis for dimerization of the BNIP3 transmembrane domain. Biochem. 2009;48:5106–5120. doi: 10.1021/bi802245u. [DOI] [PubMed] [Google Scholar]
- 25.Vande Velde C, Cizeau J, Dubik D, Alimonti J, Brown T, Israels S, Hakem R, Greenberg AH. BNIP3 and genetic control of necrosis-like cell death through the mitochondrial permeability transition pore. Mol Cell Biol. 2000;20(15):5454–5468. doi: 10.1128/mcb.20.15.5454-5468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marinković M, Šprung M, Novak I. Dimerization of mitophagy receptor BNIP3L/NIX is essential for recruitment of autophagic machinery. Autophagy. 2020 doi: 10.1080/15548627.2020.1755120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohi N, Tokunaga A, Tsunoda H, Nakano K, Haraguchi K, Oda K, Motoyama N, Nakajima T. A novel adenovirus E1B19K-binding protein B5 inhibits apoptosis induced by Nip3 by forming a heterodimer through the C-terminal hydrophobic region. Cell Death Differ. 1999;6(4):314–325. doi: 10.1038/sj.cdd.4400493. [DOI] [PubMed] [Google Scholar]
- 28.Guo K, Searfoss G, Krolikowski D, Pagnoni M, Franks C, Clark K, Yu KT, Jaye M, Ivashchenko Y. Hypoxia induces the expression of the pro-apoptotic gene BNIP3. Cell Death Diff. 2001;8:367–376. doi: 10.1038/sj.cdd.4400810. [DOI] [PubMed] [Google Scholar]
- 29.Dayan F, Roux D, Brahimi-Horn C, Pouyssegur J, Mazure NM. The oxygen sensing factor-inhibiting hypoxia-inducible factor-1 controls expression of distinct genes through the bi-functional transcriptional character of hypoxia-inducible factor-1a. Cancer Res. 2006;66:3688–3698. doi: 10.1158/0008-5472.CAN-05-4564. [DOI] [PubMed] [Google Scholar]
- 30.Bruick RK. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc Natl Acad Sci USA. 2000;97(6):9082–9087. doi: 10.1073/pnas.97.16.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kasper LH, Boussouar F, Boyd K, Xu W, Biesen M, Rehg J, Baudino T, Cleveland JL, Brindle PK. Two transcriptional mechanisms cooperate for the bulk of HIF-1-responsive gene expression. EMBO J. 2005;24:3846–3858. doi: 10.1038/sj.emboj.7600846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pouyssegur J, Dayan F, Mazure NM. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature. 2006;441:437–443. doi: 10.1038/nature04871. [DOI] [PubMed] [Google Scholar]
- 33.Glick D, Zhang W, Beaton M, Marsboom G, Gruber M, Simon MC, Hart J, Dorn GW, II, Brady MJ, Macleod KF. BNip3 regulates mitochondrial function and lipid metabolism in the liver. Mol Cell Biol. 2012;32(13):2570–2584. doi: 10.1128/MCB.00167-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JM, Wagner M, Xiao R, Kim KH, Lazar MA, Moore DD. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014;516:112–115. doi: 10.1038/nature13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 36.Reed SA, Sandesara PB, Senf SM, Judge AR. Inhibition of FoxO transcriptional activity prevents muscle fiber atrophy during cachexia and induces hypertrophy. FASEB J. 2012;26(3):987–1000. doi: 10.1096/fj.11-189977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown JL, Rosa-Caldwell ME, Lee DE, Blackwell TA, Brown LA, Perry RA, Haynie WS, Hardee JP, Carson JA, Wiggs MP, Washington TA, Greene NP. Mitochondrial degeneration precedes the development of muscle atrophy in progression of cancer cachexia in tumour-bearing mice. J Cachexia Sarcopenia Muscle. 2017;8(6):926–938. doi: 10.1002/jcsm.12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fei P, Wang W, Kim SH, Wang S, Burns TF, Sax JK, Buzzai M, Dicker DT, McKenna WG, Bernhard EJ, El-Deiry WS. Bnip3L is induced by p53 under hypoxia, and its knockdown promotes tumor growth. Cancer Cell. 2005;6:597–609. doi: 10.1016/j.ccr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 39.Feng X, Liu X, Zhang W, Xiao W. p53 directly suppresses BNIP3 expression to protect against hypoxia-induced cell death. EMBO J. 2011;30:3397–3415. doi: 10.1038/emboj.2011.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rogov VV, Suzuki H, Marinkovic M, Lang V, Kato R, Kawasaki M, Buljubasic M, Sprung M, Rogova N, Wakatsuki S, Hamacher-Brady A, Dotsch V, Dikic I, Brady NR, Novak I. Phosphorylation of the mitochondrial autophagy receptor Nix enhances its interaction with LC3 proteins. Sci Rep. 2017;7(1):1131. doi: 10.1038/s41598-017-01258-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding WX, Ni HM, Li M, Liao Y, Chen X, Stolz DB, Dorn GW, 2nd, Yin XM. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem. 2010;285(36):27879–27890. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao F, Chen D, Si J, Hu Q, Qin Z, Fang M, Wang G. The mitochondrial protein BNIP3L is the substrate of PARK2 and mediates mitophagy in PINK1/PARK2 pathway. Hum Mol Genet. 2015;24(9):2528–2538. doi: 10.1093/hmg/ddv017. [DOI] [PubMed] [Google Scholar]
- 43.Zhang T, Xue L, Li L, Tang C, Wan Z, Wang R, Tan J, Tan Y, Han H, Tian R, Billiar TR, Tao WA, Zhang Z. BNIP3 protein suppresses PINK1 kinase proteolytic cleavage to promote mitophagy. J Biol Chem. 2016;291(41):21616–21629. doi: 10.1074/jbc.M116.733410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J, Zhang C, Zhao Y, Yue X, Wu H, Huang S, Chen J, Tomsky K, Xie H, Khella CA, Gatza ML, Xia D, Gao J, White E, Haffty BG, Hu W, Feng Z. Parkin targets HIF-1alpha for ubiquitination and degradation to inhibit breast tumor progression. Nat Commun. 2017;8(1):1823. doi: 10.1038/s41467-017-01947-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li C, Zhang Y, Cheng X, Yuan H, Zhu S, Liu J, Wen Q, Xie Y, Liu J, Kroemer G, Klionsky DJ, Lotze MT, Zeh HJ, Kang R, Tang D. PINK1 and PARK2 suppress pancreatic tumorigenesis through control of mitochondrial iron-mediated immunometabolism. Dev Cell. 2018;46(4):441–455.e448. doi: 10.1016/j.devcel.2018.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kietzmann T. Metabolic zonation of the liver: the oxygen gradient revisited. Redox Biol. 2017;11:622–630. doi: 10.1016/j.redox.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ben-Moshe S, Itzkovitz S. Spatial heterogeneity in the mammalian liver. Nat Rev Gastroenterol Hepatol. 2019;16(7):395–410. doi: 10.1038/s41575-019-0134-x. [DOI] [PubMed] [Google Scholar]
- 48.Diwan A, Koesters AG, Odley AM, Pushkaran S, Baines CP, Spike BT, Daria D, Jegga AG, Geiger H, Aronow BJ, Molkentin JD, Macleod KF, Kalfa TA, Dorn GW. Unrestrained erythroblast development in Nix-/- mice reveals a mechanism for apoptotic modulation of erythropoiesis. Proc Natl Acad Sci USA. 2007;104:6794–6799. doi: 10.1073/pnas.0610666104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, Kundu M, Opferman JT, Cleveland JL, Miller JL, Ney PA. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci USA. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sandoval H, Thiagarajan P, Dasgupta SK, Scumacker A, Prchal JT, Chen M, Wang J. Essential role for Nix in autophagic maturation of red cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang C, Hashimoto M, Lin QXX, Tan DQ, Suda T. Sphingosine-1-phosphate signaling modulates terminal erythroid differentiation through the regulation of mitophagy. Exp Hematol. 2019;72:47–59.e41. doi: 10.1016/j.exphem.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 52.Quiros PM, Langer T, Lopez-Otin C. New roles for mitochondrial proteases in health, ageing and disease. Nat Rev Mol Cell Biol. 2015;16(6):345–359. doi: 10.1038/nrm3984. [DOI] [PubMed] [Google Scholar]
- 53.Mottis A, Herzig S, Auwerx J. Mitocellular communication: shaping health and disease. Science. 2019;366(6467):827–832. doi: 10.1126/science.aax3768. [DOI] [PubMed] [Google Scholar]
- 54.Esteban-Martinez L, Sierra-Filardi E, McGreal RS, Salazar-Roa M, Marino G, Seco E, Durand S, Enot D, Grana O, Malumbres M, Cvekl A, Cuervo AM, Kroemer G, Boya P. Programmed mitophagy is essential for the glycolytic switch during cell differentiation. Embo J. 2017;36(12):1688–1706. doi: 10.15252/embj.201695916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Sullivan TE, Johnson LR, Kang HH, Sun JC. BNIP3 and BNIP3L-mediated mitophagy promotes the generation of natural killer cell memory. Immunity. 2015;43:331–342. doi: 10.1016/j.immuni.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Y, Wang Y, Kim E, Beemiller P, Wang CY, Swanson J, You M, Guan KL. Bnip3 mediates the hypoxia-induced inhibition on mTOR by interacting with Rheb. J Biol Chem. 2007;282:35803–35813. doi: 10.1074/jbc.M705231200. [DOI] [PubMed] [Google Scholar]
- 57.Melser S, Chatelain EH, Lavie J, Mahfouf W, Jose C, Obre E, Goorden S, Priault M, Elgersma Y, Rezvani HR, Rossignol R, Bénard G. Rheb regulates mitophagy induced by mitochondrial energetic status. Cell Metab. 2013;17(5):719–730. doi: 10.1016/j.cmet.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 58.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168(6):960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, Ma Q, Zhu C, Wang R, Qi W, Huang L, Xue P, Li B, Wang X, Jin H, Wang J, Yang F, Liu P, Zhu Y, Sui S, Chen Q. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14(2):177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 60.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor RC, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15(7):741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu WK, Tian W, Hu Z, Chen G, Huang L, Li W, Zhang X, Xue P, Zhou C, Liu L, Zhu Y, Zhang X, Li L, Zhang L, Sui S, Zhao B, Feng D. ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep. 2014;15(5):566–575. doi: 10.1002/embr.201438501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen G, Han Z, Feng D, Chen Y, Chen L, Wu H, Huang L, Zhou C, Cai X, Fu C, Duan L, Wang X, Liu L, Liu X, Shen Y, Zhu Y, Chen Q. A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol Cell. 2014;54(3):362–377. doi: 10.1016/j.molcel.2014.02.034. [DOI] [PubMed] [Google Scholar]
- 64.Lu W, Karuppagounder SS, Springer DA, Allen MD, Zheng L, Chao B, Zhang Y, Dawson VL, Dawson TM, Lenardo M. Genetic deficiency of the mitochondrial protein PGAM5 causes a Parkinson's-like movement disorder. Nat Commun. 2014;5:4930. doi: 10.1038/ncomms5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu W, Lin C, Wu K, Jiang L, Wang X, Li W, Zhuang H, Zhang X, Chen H, Li S, Yang Y, Lu Y, Wang J, Zhu R, Zhang L, Sui S, Tan N, Zhao B, Zhang J, Li L, Feng D. FUNDC1 regulates mitochondrial dynamics at the ER-mitochondrial contact site under hypoxic conditions. Embo J. 2016;35(13):1368–1384. doi: 10.15252/embj.201593102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Y, Xue Y, Xu X, Wang G, Liu Y, Wu H, Li W, Wang Y, Chen Z, Zhang W, Zhu Y, Ji W, Xu T, Liu L, Chen Q. A mitochondrial FUNDC1/HSC70 interaction organizes the proteostatic stress response at the risk of cell morbidity. Embo J. 2019 doi: 10.15252/embj.201798786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li W, Zhang X, Zhuang H, Chen HG, Chen Y, Tian W, Wu WK, Li Y, Wang S, Zhang L, Chen Y, Li L, Zhao B, Sui S, Feng D. MicroRNA-137 is a novel hypoxia-responsive microRNA that inhibits mitophagy via regulation of two mitophagy receptors FUNDC1 and NIX. J Biol Chem. 2014;289:10691–10701. doi: 10.1074/jbc.M113.537050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murakawa T, Yamaguchi O, Hashimoto A, Hikoso S, Takeda T, Oka T, Yasui H, Ueda H, Akazawa Y, Nakayama H, Taneike M, Misaka T, Omiya S, Shah AM, Yamamoto A, Nishida K, Ohsumi Y, Okamoto K, Sakata Y, Otsu K. Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nat Commun. 2015;6:7527. doi: 10.1038/ncomms8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Murakawa T, Okamoto K, Omiya S, Taneike M, Yamaguchi O, Otsu K. A mammalian mitophagy receptor, Bcl2-L-13, recruits the ULK1 complex to induce mitophagy. Cell Rep. 2019;26(2):338–345.e336. doi: 10.1016/j.celrep.2018.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Merkwirth C, Dargazanli S, Tatsuta T, Geimer S, Löwer B, Wunderlich FT, von Kleist-Retzow JC, Waisman A, Westermann B, Langer T. Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev. 2008;22(4):476–488. doi: 10.1101/gad.460708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wei Y, Chiang WC, Sumpter R, Jr, Mishra P, Levine B. Prohibitin 2 is an inner mitochondrial membrane mitophagy receptor. Cell. 2016 doi: 10.1016/j.cell.2016.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoshii SR, Kishi C, Ishihara N, Mizushima N. Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J Biol Chem. 2011;286(22):19630–19640. doi: 10.1074/jbc.M110.209338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dumas JF, Peyta L, Couet C, Servais S. Implication of liver cardiolipins in mitochondrial energy metabolism disorder in cancer cachexia. Biochimie. 2013;95(1):27–32. doi: 10.1016/j.biochi.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 74.Chu CT, Ji J, Dagda RK, Jiang JF, Tyurina YY, Kapralov AA, Tyurin VA, Yanamala N, Shrivastava IH, Mohammadyani D, Qiang Wang KZ, Zhu J, Klein-Seetharaman J, Balasubramanian K, Amoscato AA, Borisenko G, Huang Z, Gusdon AM, Cheikhi A, Steer EK, Wang R, Baty C, Watkins S, Bahar I, Bayir H, Kagan VE. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol. 2013;15(10):1197–1205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hsu P, Liu X, Zhang J, Wang HG, Ye JM, Shi Y. Cardiolipin remodeling by TAZ/tafazzin is selectively required for the initiation of mitophagy. Autophagy. 2015;11(4):643–652. doi: 10.1080/15548627.2015.1023984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sentelle RD, Senkal CE, Jiang W, Ponnusamy S, Gencer S, Selvam SP, Ramshesh VK, Peterson YK, Lemasters JJ, Szulc ZM, Bielawski J, Ogretmen B. Ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Nat Chem Biol. 2012;8(10):831–838. doi: 10.1038/nchembio.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gao X, Lee K, Reid MA, Sanderson SM, Qiu C, Li S, Liu J, Locasale JW. Serine availability influences mitochondrial dynamics and function through lipid metabolism. Cell Rep. 2018;22(13):3507–3520. doi: 10.1016/j.celrep.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Csaki LS, Reue K. Lipins: multifunctional lipid metabolism proteins. Annu Rev Nutr. 2010;30:257–272. doi: 10.1146/annurev.nutr.012809.104729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Harris TE, Finck BN. Dual function lipin proteins and glycerolipid metabolism. Trends Endocrinol Metab TEM. 2011;22(6):226–233. doi: 10.1016/j.tem.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang P, Verity MA, Reue K. Lipin-1 regulates autophagy clearance and intersects with statin drug effects in skeletal muscle. Cell Metab. 2014;20(2):267–279. doi: 10.1016/j.cmet.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alshudukhi AA, Zhu J, Huang D, Jama A, Smith JD, Wang QJ, Esser KA, Ren H. Lipin-1 regulates Bnip3-mediated mitophagy in glycolytic muscle. FASEB J. 2018 doi: 10.1096/fj.201800374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Leong WI, Saba JD. S1P metabolism in cancer and other pathological conditions. Biochimie. 2010;92(6):716–723. doi: 10.1016/j.biochi.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hart PC, Chiyoda T, Liu X, Weigert M, Curtis M, Chiang CY, Loth R, Lastra R, McGregor SM, Locasale JW, Lengyel E, Romero IL. SPHK1 is a novel target of metformin in ovarian cancer. Mol Cancer Res MCR. 2019;17(4):870–881. doi: 10.1158/1541-7786.mcr-18-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ader I, Malavaud B, Cuvillier O. When the sphingosine kinase 1/sphingosine 1-phosphate pathway meets hypoxia signaling: new targets for cancer therapy. Can Res. 2009;69(9):3723–3726. doi: 10.1158/0008-5472.can-09-0389. [DOI] [PubMed] [Google Scholar]
- 85.Kim S, Sieburth D. Sphingosine kinase activates the mitochondrial unfolded protein response and is targeted to mitochondria by stress. Cell Rep. 2018;24(11):2932–2945.e2934. doi: 10.1016/j.celrep.2018.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martinez-Reyes I, Diebold LP, Kong H, Schieber M, Huang H, Hensley CT, Mehta MM, Wang T, Santos JH, Woychik R, Dufour E, Spelbrink JN, Weinberg SE, Zhao Y, DeBerardinis RJ, Chandel NS. TCA cycle and mitochondrial membrane potential are necessary for diverse biological functions. Mol Cell. 2016;61(2):199–209. doi: 10.1016/j.molcel.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Herzig S, Shaw RJ. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol. 2018;19(2):121–135. doi: 10.1038/nrm.2017.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Toyama EQ, Herzig S, Courchet J, Lewis TL, Jr, Loson OC, Hellberg K, Young NP, Chen H, Polleux F, Chan DC, Shaw RJ. Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science. 2016;351(6270):275–281. doi: 10.1126/science.aab4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pei S, Minhajuddin M, Adane B, Khan N, Stevens BM, Mack SC, Lai S, Rich JN, Inguva A, Shannon KM, Kim H, Tan AC, Myers JR, Ashton JM, Neff T, Pollyea DA, Smith CA, Jordan CT. AMPK/FIS1-mediated mitophagy is required for self-renewal of human AML stem cells. Cell Stem Cell. 2018;23(1):86–100.e106. doi: 10.1016/j.stem.2018.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Domenech E, Maestre C, Esteban-Martinez L, Partida D, Pascual R, Fernandez-Miranda G, Seco E, Campos-Olivas R, Perez M, Megias D, Allen K, Lopez M, Saha AK, Velasco G, Rial E, Mendez R, Boya P, Salazar-Roa M, Malumbres M. AMPK and PFKFB3 mediate glycolysis and survival in response to mitophagy during mitotic arrest. Nat Cell Biol. 2015;17(10):1304–1316. doi: 10.1038/ncb3231. [DOI] [PubMed] [Google Scholar]
- 91.Eichner LJ, Brun SN, Herzig S, Young NP, Curtis SD, Shackelford DB, Shokhirev MN, Leblanc M, Vera LI, Hutchins A, Ross DS, Shaw RJ, Svensson RU. Genetic analysis reveals AMPK is required to support tumor growth in murine kras-dependent lung cancer models. Cell Metab. 2019;29(2):285–302.e287. doi: 10.1016/j.cmet.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nakada D, Saunders TL, Morrison SJ. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature. 2010;468(7324):653–658. doi: 10.1038/nature09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gurumurthy S, Xie SZ, Alagesan B, Kim J, Yusuf RZ, Saez B, Tzatsos A, Ozsolak F, Milos P, Ferrari F, Park PJ, Shirihai OS, Scadden DT, Bardeesy N. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature. 2010;468(7324):659–663. doi: 10.1038/nature09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Masand R, Paulo E, Wu D, Wang Y, Swaney DL, Jimenez-Morales D, Krogan NJ, Wang B. Proteome imbalance of mitochondrial electron transport chain in brown adipocytes leads to metabolic benefits. Cell Metab. 2018;27(3):616–629.e614. doi: 10.1016/j.cmet.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kottakis F, Nicolay BN, Roumane A, Karnik R, Gu H, Nagle JM, Boukhali M, Hayward MC, Li YY, Chen T, Liesa M, Hammerman PS, Wong KK, Hayes DN, Shirihai OS, Dyson NJ, Haas W, Meissner A, Bardeesy N. LKB1 loss links serine metabolism to DNA methylation and tumorigenesis. Nature. 2016;539(7629):390–395. doi: 10.1038/nature20132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang K, Blanco DB, Neale G, Vogel P, Avila J, Clish CB, Wu C, Shrestha S, Rankin S, Long L, Kc A, Chi H. Homeostatic control of metabolic and functional fitness of Treg cells by LKB1 signalling. Nature. 2017;548(7669):602–606. doi: 10.1038/nature23665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.He N, Fan W, Henriquez B, Yu RT, Atkins AR, Liddle C, Zheng Y, Downes M, Evans RM. Metabolic control of regulatory T cell (Treg) survival and function by Lkb1. Proc Natl Acad Sci USA. 2017;114(47):12542–12547. doi: 10.1073/pnas.1715363114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Quiros PM, Prado MA, Zamboni N, D'Amico D, Williams RW, Finley D, Gygi SP, Auwerx J. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J Cell Biol. 2017;216(7):2027–2045. doi: 10.1083/jcb.201702058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bao XR, Ong SE, Goldberger O, Peng J, Sharma R, Thompson DA, Vafai SB, Cox AG, Marutani E, Ichinose F, Goessling W, Regev A, Carr SA, Clish CB, Mootha VK. Mitochondrial dysfunction remodels one-carbon metabolism in human cells. eLife. 2016 doi: 10.7554/eLife.10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tezze C, Romanello V, Desbats MA, Fadini GP, Albiero M, Favaro G, Ciciliot S, Soriano ME, Morbidoni V, Cerqua C, Loefler S, Kern H, Franceschi C, Salvioli S, Conte M, Blaauw B, Zampieri S, Salviati L, Scorrano L, Sandri M. Age-associated loss of OPA1 in muscle impacts muscle mass, metabolic homeostasis, systemic inflammation, and epithelial senescence. Cell Metab. 2017;25(6):1374–1389.e1376. doi: 10.1016/j.cmet.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Khan NA, Nikkanen J, Yatsuga S, Jackson C, Wang L, Pradhan S, Kivela R, Pessia A, Velagapudi V, Suomalainen A. mTORC1 regulates mitochondrial integrated stress response and mitochondrial myopathy progression. Cell Metab. 2017;26(2):419–428.e415. doi: 10.1016/j.cmet.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 102.Forsstrom S, Jackson CB, Carroll CJ, Kuronen M, Pirinen E, Pradhan S, Marmyleva A, Auranen M, Kleine IM, Khan NA, Roivainen A, Marjamaki P, Liljenback H, Wang L, Battersby BJ, Richter U, Velagapudi V, Nikkanen J, Euro L, Suomalainen A. Fibroblast growth factor 21 drives dynamics of local and systemic stress responses in mitochondrial myopathy with mtDNA deletions. Cell Metab. 2019;30(6):1040–1054.e1047. doi: 10.1016/j.cmet.2019.08.019. [DOI] [PubMed] [Google Scholar]
- 103.Ebert SM, Dyle MC, Kunkel SD, Bullard SA, Bongers KS, Fox DK, Dierdorff JM, Foster ED, Adams CM. Stress-induced skeletal muscle Gadd45a expression reprograms myonuclei and causes muscle atrophy. J Biol Chem. 2012;287(33):27290–27301. doi: 10.1074/jbc.M112.374777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Guo X, Aviles G, Liu Y, Tian R, Unger BA, Lin YT, Wiita AP, Xu K, Correia MA, Kampmann M. Mitochondrial stress is relayed to the cytosol by an OMA1-DELE1-HRI pathway. Nature. 2020;579(7799):427–432. doi: 10.1038/s41586-020-2078-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fessler E, Eckl EM, Schmitt S, Mancilla IA, Meyer-Bender MF, Hanf M, Philippou-Massier J, Krebs S, Zischka H, Jae LT. A pathway coordinated by DELE1 relays mitochondrial stress to the cytosol. Nature. 2020;579(7799):433–437. doi: 10.1038/s41586-020-2076-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Harada T, Iwai A, Miyazaki T. Identification of DELE, a novel DAP3-binding protein which is crucial for death receptor-mediated apoptosis induction. Apoptosis. 2010;15(10):1247–1255. doi: 10.1007/s10495-010-0519-3. [DOI] [PubMed] [Google Scholar]
- 107.Fiorese CJ, Schulz AM, Lin YF, Rosin N, Pellegrino MW, Haynes CM. The transcription factor ATF5 mediates a mammalian mitochondrial UPR. Curr Biol CB. 2016;26(15):2037–2043. doi: 10.1016/j.cub.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sorrentino V, Romani M, Mouchiroud L, Beck JS, Zhang H, D'Amico D, Moullan N, Potenza F, Schmid AW, Rietsch S, Counts SE, Auwerx J. Enhancing mitochondrial proteostasis reduces amyloid-beta proteotoxicity. Nature. 2017;552(7684):187–193. doi: 10.1038/nature25143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Molenaars M, Janssens GE, Williams EG, Jongejan A, Lan J, Rabot S, Joly F, Moerland PD, Schomakers BV, Lezzerini M, Liu YJ, McCormick MA, Kennedy BK, van Weeghel M, van Kampen AHC, Aebersold R, MacInnes AW, Houtkooper RH. A conserved mito-cytosolic translational balance links two longevity pathways. Cell Metab. 2020;31(3):549–563.e547. doi: 10.1016/j.cmet.2020.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337(6094):587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nargund AM, Fiorese CJ, Pellegrino MW, Deng P, Haynes CM. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPR(mt) Mol Cell. 2015;58(1):123–133. doi: 10.1016/j.molcel.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Palikaras K, Lionaki E, Tavernarakis N. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature. 2015;521:525–528. doi: 10.1038/nature14300. [DOI] [PubMed] [Google Scholar]
- 113.Gupte R, Liu Z, Kraus WL. PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev. 2017;31(2):101–126. doi: 10.1101/gad.291518.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fang EF, Kassahun H, Croteau DL, Scheibye-Knudsen M, Marosi K, Lu H, Shamanna RA, Kalyanasundaram S, Bollineni RC, Wilson MA, Iser WB, Wollman BN, Morevati M, Li J, Kerr JS, Lu Q, Waltz TB, Tian J, Sinclair DA, Mattson MP, Nilsen H, Bohr VA. NAD+ replenishment improves lifespan and healthspan in ataxia telangiectasia models via mitophagy and DNA repair. Cell Metab. 2016;24(4):566–581. doi: 10.1016/j.cmet.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fang EF, Scheibye-Knudsen M, Brace LE, Kassahun H, SenGupta T, Nilsen H, Mitchell JR, Croteau DL, Bohr VA. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell. 2014;157(4):882–896. doi: 10.1016/j.cell.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gerhart-Hines Z, Dominy JE, Jr, Blattler SM, Jedrychowski MP, Banks AS, Lim JH, Chim H, Gygi SP, Puigserver P. The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD(+) Mol Cell. 2011;44(6):851–863. doi: 10.1016/j.molcel.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rzymski T, Milani M, Pike L, Buffa F, Mellor HR, Winchester L, Pires I, Hammond E, Ragoussis I, Harris AL. Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene. 2010;29(31):4424–4435. doi: 10.1038/onc.2010.191. [DOI] [PubMed] [Google Scholar]
- 118.Pike LR, Singleton DC, Buffa F, Abramczyk O, Phadwal K, Li JL, Simon AK, Murray JT, Harris AL. Transcriptional upregulation of ULK1 by ATF4 contributes to cancer cell survival. Biochem J. 2013;449(2):389–400. doi: 10.1042/bj20120972. [DOI] [PubMed] [Google Scholar]
- 119.Anderson CM, Macleod KF. Autophagy and cancer cell metabolism. Int Rev Cell Mol Biol. 2019;347:145–190. doi: 10.1016/bs.ircmb.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469(7329):221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 121.Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol. 2016;17(10):1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- 122.McArthur K, Whitehead LW, Heddleston JM, Li L, Padman BS, Oorschot V, Geoghegan ND, Chappaz S, Davidson S, San Chin H, Lane RM, Dramicanin M, Saunders TL, Sugiana C, Lessene R, Osellame LD, Chew TL, Dewson G, Lazarou M, Ramm G, Lessene G, Ryan MT, Rogers KL, van Delft MF, Kile BT. BAK/BAX macropores facilitate mitochondrial herniation and mtDNA efflux during apoptosis. Science. 2018 doi: 10.1126/science.aao6047. [DOI] [PubMed] [Google Scholar]
- 123.Aarreberg LD, Esser-Nobis K, Driscoll C, Shuvarikov A, Roby JA, Gale M., Jr Interleukin-1β induces mtDNA release to activate innate immune signaling via cGAS-STING. Mol Cell. 2019;74(4):801–815.e806. doi: 10.1016/j.molcel.2019.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhong Z, Liang S, Sanchez-Lopez E, He F, Shalapour S, Lin XJ, Wong J, Ding S, Seki E, Schnabl B, Hevener AL, Greenberg HB, Kisseleva T, Karin M. New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature. 2018;560(7717):198–203. doi: 10.1038/s41586-018-0372-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shimada K, Crother TR, Karlin J, Dagvadorj J, Chiba N, Chen S, Ramanujan VK, Wolf AJ, Vergnes L, Ojcius DM, Rentsendorj A, Vargas M, Guerrero C, Wang Y, Fitzgerald KA, Underhill DM, Town T, Arditi M. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36(3):401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Misawa T, Takahama M, Kozaki T, Lee H, Zou J, Saitoh T, Akira S. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat Immunol. 2013;14(5):454–460. doi: 10.1038/ni.2550. [DOI] [PubMed] [Google Scholar]
- 127.Elliott EI, Sutterwala FS. Initiation and perpetuation of NLRP3 inflammasome activation and assembly. Immunol Rev. 2015;265(1):35–52. doi: 10.1111/imr.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhong Z, Umemura A, Sanchez-Lopez E, Liang S, Shalapour S, Wong J, He F, Boassa D, Perkins G, Ali SR, McGeough MD, Ellisman MH, Seki E, Gustafsson AB, Hoffman HM, Diaz-Meco MT, Moscat J, Karin M. NF-kappaB restricts inflammasome activation via elimination of damaged mitochondria. Cell. 2016;164(5):896–910. doi: 10.1016/j.cell.2015.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yu JJ, Nagasu H, Murakami T, Hoang H, Broderick L, Hoffman HM, Horng T. Inflammasome activation leads to caspase-1 dependent mitochondrial damage and block of mitophagy. Proc Natl Acad Sci U S A. 2014;111(43):15514–15519. doi: 10.1073/pnas.1414859111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Moore AS, Holzbaur EL. Dynamic recruitment and activation of ALS-associated TBK1 with its target optineurin are required for efficient mitophagy. Proc Natl Acad Sci U S A. 2016;113(24):E3349–3358. doi: 10.1073/pnas.1523810113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Richter B, Sliter DA, Herhaus L, Stolz A, Wang C, Beli P, Zaffagnini G, Wild P, Martens S, Wagner SA, Youle RJ, Dikic I. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc Natl Acad Sci U S A. 2016;113(15):4039–4044. doi: 10.1073/pnas.1523926113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gui X, Yang H, Li T, Tan X, Shi P, Li M, Du F, Chen ZJ. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature. 2019;567(7747):262–266. doi: 10.1038/s41586-019-1006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gaidt MM, Ebert TS, Chauhan D, Ramshorn K, Pinci F, Zuber S, O'Duill F, Schmid-Burgk JL, Hoss F, Buhmann R, Wittmann G, Latz E, Subklewe M, Hornung V. The DNA inflammasome in human myeloid cells is initiated by a STING-cell death program upstream of NLRP3. Cell. 2017;171(5):1110–1124.e1118. doi: 10.1016/j.cell.2017.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, Fitzgerald KA, Ryter SW, Choi AM. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12(3):222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Drake LE, Springer MZ, Poole LP, Kim CJ, Macleod KF. Expanding perspectives on the significance of mitophagy in cancer. Semin Cancer Biol. 2017;47:110–124. doi: 10.1016/j.semcancer.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cesari R, Martin ES, Calin GA, Pentimalli F, Bichi R, McAdams H, Trapasso F, Drusco A, Shimizu M, Masciullo V, D'Andrilli G, Scambia G, Picchio MC, Alder H, Godwin AK, Croce CM. Parkin, a gene implicated in autosomal recessive juvenile parkinsonism, is a candidate tumor suppressor gene on chromosome 6q25-q27. Proc Natl Acad Sci U S A. 2003;100(10):5956–5961. doi: 10.1073/pnas.0931262100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Veeriah S, Taylor BS, Meng S, Fang F, Yilmaz E, Vivanco I, Janakiraman M, Schultz N, Hanrahan AJ, Pao W, Ladanyi M, Sander C, Heguy A, Holland EC, Paty PB, Mischel PS, Liau L, Cloughesy TF, Mellinghoff IK, Solit DB, Chan TA. Somatic mutations of the Parkinson's disease-associated gene PARK2 in glioblastoma and other human malignancies. Nat Genet. 2010;42(1):77–82. doi: 10.1038/ng.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Agnihotri S, Golbourn B, Huang X, Remke M, Younger S, Cairns RA, Chalil A, Smith CA, Krumholtz SL, Mackenzie D, Rakopoulos P, Ramaswamy V, Taccone MS, Mischel PS, Fuller GN, Hawkins C, Stanford WL, Taylor MD, Zadeh G, Rutka JT. PINK1 is a negative regulator of growth and the warburg effect in glioblastoma. Can Res. 2016;76(16):4708–4719. doi: 10.1158/0008-5472.can-15-3079. [DOI] [PubMed] [Google Scholar]
- 139.Fujiwara M, Marusawa H, Wang HQ, Iwai A, Ikeuchi K, Imai Y, Kataoka A, Nukina N, Takahashi R, Chiba T. Parkin as a tumor suppressor gene for hepatocellular carcinoma. Oncogene. 2008;27:6002–6011. doi: 10.1038/onc.2008.199. [DOI] [PubMed] [Google Scholar]
- 140.Zhang C, Lin M, Wu R, Wang X, Yang BG, Levine AJ, Hu W, Feng Z. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc Natl Acad Sci U S A. 2011;108(39):16259–16264. doi: 10.1073/pnas.1113884108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bernardini JP, Lazarou M, Dewson G. Parkin and mitophagy in cancer. Oncogene. 2016 doi: 10.1038/onc.2016.302. [DOI] [PubMed] [Google Scholar]
- 142.Kim KY, Stevens MV, Akter MH, Rusk SE, Huang RJ, Cohen A, Noguchi A, Springer D, Bocharov AV, Eggerman TL, Suen DF, Youle RJ, Amar M, Remaley AT, M.N. S, Parkin is a lipid-responsive regulator of fat uptake in mice and mutant human cells. J Clin Invest. 2011;121:3701–3712. doi: 10.1172/JCI44736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sarraf SA, Sideris DP, Giagtzoglou N, Ni L, Kankel MW, Sen A, Bochicchio LE, Huang CH, Nussenzweig SC, Worley SH, Morton PD, Artavanis-Tsakonas S, Youle RJ, Pickrell AM. PINK1/parkin influences cell cycle by sequestering TBK1 at damaged mitochondria. Inhibiting Mitosis Cell Rep. 2019;29(1):225–235.e225. doi: 10.1016/j.celrep.2019.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gong Y, Zack TI, Morris LG, Lin K, Hukkelhoven E, Raheja R, Tan IL, Turcan S, Veeriah S, Meng S, Viale A, Schumacher SE, Palmedo P, Beroukhim R, Chan TA. Pan-cancer genetic analysis identifies PARK2 as a master regulator of G1/S cyclins. Nat Genet. 2014;46(6):588–594. doi: 10.1038/ng.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lee SB, Kim JJ, Nam HJ, Gao B, Yin P, Qin B, Yi SY, Ham H, Evans D, Kim SH, Zhang J, Deng M, Liu T, Zhang H, Billadeau DD, Wang L, Giaime E, Shen J, Pang YP, Jen J, van Deursen JM, Lou Z. Parkin regulates mitosis and genomic stability through Cdc20/Cdh1. Mol Cell. 2015;60(1):21–34. doi: 10.1016/j.molcel.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pillai S, Nguyen J, Johnson J, Haura E, Coppola D, Chellappan S. Tank binding kinase 1 is a centrosome-associated kinase necessary for microtubule dynamics and mitosis. Nat Commun. 2015;6:10072. doi: 10.1038/ncomms10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Villa E, Marchetti S, Ricci JE. No parkin zone: mitophagy without parkin. Trends Cell Biol. 2018;28(11):882–895. doi: 10.1016/j.tcb.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 148.Sowter HM, Ferguson M, Pym C, Watson P, Fox SB, Han C, Harris AL. Expression of the cell death genes BNip3 and Nix in ductal carcinoma in situ of the breast; correlation of BNip3 levels with necrosis and grade. JPath. 2003;201:573–580. doi: 10.1002/path.1486. [DOI] [PubMed] [Google Scholar]
- 149.Okami J, Simeone DM, Logsdon CD. Silencing of the hypoxia-inducible cell death protein BNIP3 in pancreatic cancer. Cancer Res. 2004;64(15):5338–5346. doi: 10.1158/0008-5472.CAN-04-0089. [DOI] [PubMed] [Google Scholar]
- 150.Erkan M, Kleef J, Esposito I, Giese T, Ketterer K, Buchler MW, Giese NA, Friess H. Loss of BNIP3 expression is a late event in pancreatic cancer contributing to chemoresistance and worsened prognosis. Oncogene. 2005;24:4421–4432. doi: 10.1038/sj.onc.1208642. [DOI] [PubMed] [Google Scholar]
- 151.Murai M, Toyota M, Suzuki H, Satoh A, Sasaki Y, Akino K, Ueno M, Takahashi F, Kusano M, Mita H, Yanagihara K, Endo T, Hinoda Y, Tokino T, Imai K. Aberrant methylation and silencing of the BNIP3 gene in colorectal and gastric cancer. Clin Cancer Res. 2005;11(3):1021–1027. [PubMed] [Google Scholar]
- 152.Akada M, Crnogorac-Jurcevic T, Lattimore S, Mahon P, Lopes R, Sunamura M, Matsuno S, Lemoine NR. Intrinsic chemoresistance to gemcitabine is associated with decreased expression of BNIP3 in pancreatic cancer. Clin Cancer Res. 2005;11(8):3094–3101. doi: 10.1158/1078-0432.CCR-04-1785. [DOI] [PubMed] [Google Scholar]
- 153.Humpton TJ, Alagesan B, DeNicola GM, Lu D, Yordanov GN, Leonhardt CS, Yao MA, Alagesan P, Zaatari MN, Park Y, Skepper JN, Macleod KF, Perez-Mancera PA, Murphy MP, Evan GI, Vousden KH, Tuveson DA. Oncogenic KRAS induces NIX-mediated mitophagy to promote pancreatic cancer. Cancer Discov. 2019;9(9):1268–1287. doi: 10.1158/2159-8290.cd-18-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, Mc Henry KT, Pinchback RM, Ligon AH, Cho YJ, Haery L, Greulich H, Reich M, Winckler W, Lawrence MS, Weir BA, Tanaka KE, Chiang DY, Bass AJ, Loo A, Hoffman C, Prensner J, Liefeld T, Gao Q, Yecies D, Signoretti S, Maher E, Kaye FJ, Sasaki H, Tepper JE, Fletcher J, Tabernero J, Baselga J, Tsao MS, Demichelis F, Rubin MA, Janne PA, Daly MJ, Nucera C, Levine RL, Ebert BL, Gabriel S, Rustgi AK, Antonescu CR, Ladanyi M, Letai A, Garraway LA, Loda M, Beer DG, True LD, Okamoto A, Pomeroy SL, Singer S, Golub TR, Lander ES, Getz G, Sellers WR, Meyerson M. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Koop EA, van Laar T, Weger RA, van der Wall E, van Diest PJ. Expression of BNIP3 in invasive breast cancer: correlations with the hypoxic response and clinicopathological features. BMC Cancer. 2009;9:175–182. doi: 10.1186/1471-2407-9-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chourasia AH, Tracy K, Frankenberger C, Boland ML, Sharifi MN, Drake LE, Sachleben JR, Asara JM, Locasale JW, Karczmar GS, Macleod KF. Mitophagy defects arising from BNip3 loss promote mammary tumor progression to metastasis. EMBO Rep. 2015;16(9):1145–1163. doi: 10.15252/embr.201540759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Montagner M, Enzo E, Forcato M, Zanconato F, Parenti A, Rampazzo E, Basso G, Leo G, Rosato A, Bicciato S, Cordenonsi M, Piccolo S. SHARP1 suppresses breast cancer metastasis by promoting degradation of hypoxia-inducible factors. Nature. 2012;487(7407):380–384. doi: 10.1038/nature11207. [DOI] [PubMed] [Google Scholar]
- 158.Guha M, Srinivasan S, Raman P, Jiang Y, Kaufman BA, Taylor D, Dong D, Chakrabarti R, Picard M, Carstens RP, Kijima Y, Feldman M. Avadhani NG (2018) Aggressive triple negative breast cancers have unique molecular signature on the basis of mitochondrial genetic and functional defects. Biochim Biophys Acta Mol Basis Dis. 1864;4:1060–1071. doi: 10.1016/j.bbadis.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lee J, Yesilkanal AE, Wynne JP, Frankenberger C, Liu J, Yan J, Elbaz M, Rabe DC, Rustandy FD, Tiwari P, Grossman EA, Hart PC, Kang C, Sanderson SM, Andrade J, Nomura DK, Bonini MG, Locasale JW, Rosner MR. Effective breast cancer combination therapy targeting BACH1 and mitochondrial metabolism. Nature. 2019;568(7751):254–258. doi: 10.1038/s41586-019-1005-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Manka D, Spicer Z, Millhorn DE. Bcl-2/Adenovirus E1B 19 kDa interacting protein-3 knockdown enables growth of breast cancer metastases in the lung, liver and bone. Cancer Res. 2005;65:11689–11693. doi: 10.1158/0008-5472.CAN-05-3091. [DOI] [PubMed] [Google Scholar]
- 161.Labuschagne CF, Cheung EC, Blagih J, Domart MC, Vousden KH. Cell clustering promotes a metabolic switch that supports metastatic colonization. Cell Metab. 2019;30(4):720–734.e725. doi: 10.1016/j.cmet.2019.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Smith AG, Macleod KF. Autophagy, cancer stem cells and drug resistance. J Pathol. 2019;247(5):708–718. doi: 10.1002/path.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Liu K, Lee J, Kim JY, Wang L, Tian Y, Chan ST, Cho C, Machida K, Chen D, Ou JJ. Mitophagy controls the activities of tumor suppressor p53 to regulate hepatic cancer stem cells. Mol Cell. 2017;68(2):281–292. doi: 10.1016/j.molcel.2017.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Katajisto P, Dohla J, Chaffer CL, Pentinmikko N, Marjanovic N, Iqbal S, Zoncu R, Chen W, Weinberg RA, Sabatini DM. Stem cells. Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science. 2015;348(6232):340–343. doi: 10.1126/science.1260384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ho TT, Warr MR, Adelman ER, Lansinger OM, Flach J, Verovskaya EV, Figueroa ME, Passegue E. Autophagy maintains the metabolism and function of young and old stem cells. Nature. 2017;543(7644):205–210. doi: 10.1038/nature21388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ito K, Turcotte R, Cui J, Zimmerman SE, Pinho S, Mizoguchi T, Arai F, Runnels JM, Alt C, Teruya-Feldstein J, Mar JC, Singh R, Suda T, Lin CP, Frenette PS, Ito K. Self-renewal of a purified Tie2+ hematopoietic stem cell population relies on mitochondrial clearance. Science. 2016;354(6316):1156–1160. doi: 10.1126/science.aaf5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Vannini N, Girotra M, Naveiras O, Nikitin G, Campos V, Giger S, Roch A, Auwerx J, Lutolf MP. Specification of haematopoietic stem cell fate via modulation of mitochondrial activity. Nat Commun. 2016;7:13125. doi: 10.1038/ncomms13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Vannini N, Campos V, Girotra M, Trachsel V, Rojas-Sutterlin S, Tratwal J, Ragusa S, Stefanidis E, Ryu D, Rainer PY, Nikitin G, Giger S, Li TY, Semilietof A, Oggier A, Yersin Y, Tauzin L, Pirinen E, Cheng WC, Ratajczak J, Canto C, Ehrbar M, Sizzano F, Petrova TV, Vanhecke D, Zhang L, Romero P, Nahimana A, Cherix S, Duchosal MA, Ho PC, Deplancke B, Coukos G, Auwerx J, Lutolf MP, Naveiras O. The NAD-booster nicotinamide riboside potently stimulates hematopoiesis through increased mitochondrial clearance. Cell Stem Cell. 2019;24(3):405–418.e407. doi: 10.1016/j.stem.2019.02.012. [DOI] [PubMed] [Google Scholar]
- 169.Li J, Agarwal E, Bertolini I, Seo JH, Caino MC, Ghosh JC, Kossenkov AV, Liu Q, Tang HY, Goldman AR, Languino LR, Speicher DW, Altieri DC. The mitophagy effector FUNDC1 controls mitochondrial reprogramming and cellular plasticity in cancer cells. Sci Signal. 2020 doi: 10.1126/scisignal.aaz8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wu L, Zhang D, Zhou L, Pei Y, Zhuang Y, Cui W, Chen J. FUN14 domain-containing 1 promotes breast cancer proliferation and migration by activating calcium-NFATC1-BMI1 axis. EBioMedicine. 2019;41:384–394. doi: 10.1016/j.ebiom.2019.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cleaver JE. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat Rev Cancer. 2005;5(7):564–573. doi: 10.1038/nrc1652. [DOI] [PubMed] [Google Scholar]
- 172.Kee Y, D'Andrea AD. Molecular pathogenesis and clinical management of Fanconi anemia. J Clin Investig. 2012;122(11):3799–3806. doi: 10.1172/jci58321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14(4):197–210. [PubMed] [Google Scholar]
- 174.Sumpter R, Jr, Sirasanagandla S, Fernandez AF, Wei Y, Dong X, Franco L, Zou Z, Marchal C, Lee MY, Clapp DW, Hanenberg H, Levine B. Fanconi anemia proteins function in mitophagy and immunity. Cell. 2016;165(4):867–881. doi: 10.1016/j.cell.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Orvedahl A, Sumpter R, Jr, Xiao G, Ng A, Zou Z, Tang Y, Narimatsu M, Gilpin C, Sun Q, Roth M, Forst CV, Wrana JL, Zhang YE, Luby-Phelps K, Xavier RJ, Xie Y, Levine B. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480(7375):113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.de Vries Y, Lwiwski N, Levitus M, Kuyt B, Israels SJ, Arwert F, Zwaan M, Greenberg CR, Alter BP, Joenje H, Meijers-Heijboer H. A dutch fanconi anemia FANCC founder mutation in Canadian manitoba mennonites. Anemia. 2012;2012:865170. doi: 10.1155/2012/865170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ambrose M, Goldstine JV, Gatti RA. Intrinsic mitochondrial dysfunction in ATM-deficient lymphoblastoid cells. Hum Mol Genet. 2007;16(18):2154–2164. doi: 10.1093/hmg/ddm166. [DOI] [PubMed] [Google Scholar]
- 178.Eaton JS, Lin ZP, Sartorelli AC, Bonawitz ND, Shadel GS. Ataxia-telangiectasia mutated kinase regulates ribonucleotide reductase and mitochondrial homeostasis. J Clin Investig. 2007;117(9):2723–2734. doi: 10.1172/jci31604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Valentin-Vega YA, Maclean KH, Tait-Mulder J, Milasta S, Steeves M, Dorsey FC, Cleveland JL, Green DR, Kastan MB. Mitochondrial dysfunction in ataxia-telangiectasia. Blood. 2012;119(6):1490–1500. doi: 10.1182/blood-2011-08-373639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13(5):589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rambold AS, Kostelecky B, Elia N, Lippincott-Schwartz J. Tubular network formation protects mitochondrial from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci U S A. 2011;108(25):10190–10195. doi: 10.1073/pnas.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yamada T, Murata D, Adachi Y, Itoh K, Kameoka S, Igarashi A, Kato T, Araki Y, Huganir RL, Dawson TM, Yanagawa T, Okamoto K, Iijima M, Sesaki H. Mitochondrial stasis reveals p62-mediated ubiquitination in parkin-independent mitophagy and mitigates nonalcoholic fatty liver disease. Cell Metab. 2018;28(4):588–604.e585. doi: 10.1016/j.cmet.2018.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mishra P, Chan DC. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat Rev Mol Cell Biol. 2014;15(10):634–646. doi: 10.1038/nrm3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, Mihara K. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol. 2010;191(6):1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wong YC, Ysselstein D, Krainc D. Mitochondria-lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature. 2018;554(7692):382–386. doi: 10.1038/nature25486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yamano K, Fogel AI, Wang C, van der Bliek AM, Youle RJ. Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy. eLife. 2014;3:e01612. doi: 10.7554/eLife.01612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Shen Q, Yamano K, Head BP, Kawajiri S, Cheung JT, Wang C, Cho JH, Hattori N, Youle RJ, van der Bliek AM. Mutations in Fis1 disrupt orderly disposal of defective mitochondria. Mol Biol Cell. 2014;25(1):145–159. doi: 10.1091/mbc.E13-09-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Cho HM, Ryu JR, Jo Y, Seo TW, Choi YN, Kim JH, Chung JM, Cho B, Kang HC, Yu SW, Yoo SJ, Kim H, Sun W. Drp1-Zip1 interaction regulates mitochondrial quality surveillance system. Mol Cell. 2019;73(2):364–376.e368. doi: 10.1016/j.molcel.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 189.Lee YK, Lee HY, Hanna RA, Gustafsson AB. Mitochondrial autophagy by Bnip3 involves Drp1-mediated mitochondrial fission and recruitment of Parkin in cardiac myocytes. Am j Physiol Heart Circ Physiol. 2011;301(5):H1924–1931. doi: 10.1152/ajpheart.00368.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kashatus JA, Nascimento A, Myers LJ, Sher A, Byrne FL, Hoehn KL, Counter CM, Kashatus DF. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol Cell. 2015;57(3):537–551. doi: 10.1016/j.molcel.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Serasinghe MN, Wieder SY, Renault TT, Elkholi R, Asciolla JJ, Yao JL, Jabado O, Hoehn K, Kageyama Y, Sesaki H, Chipuk JE. Mitochondrial division is requisite to RAS-induced transformation and targeted by oncogenic MAPK pathway inhibitors. Mol Cell. 2015;57(3):521–536. doi: 10.1016/j.molcel.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wieder SY, Serasinghe MN, Sung JC, Choi DC, Birge MB, Yao JL, Bernstein E, Celebi JT, Chipuk JE. Activation of the mitochondrial fragmentation protein DRP1 correlates with BRAF(V600E) melanoma. J Invest Dermatol. 2015;135(10):2544–2547. doi: 10.1038/jid.2015.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Trotta AP, Gelles JD, Serasinghe MN, Loi P, Arbiser JL, Chipuk JE. Disruption of mitochondrial electron transport chain function potentiates the pro-apoptotic effects of MAPK inhibition. J Biol Chem. 2017;292(28):11727–11739. doi: 10.1074/jbc.M117.786442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Nagdas S, Kashatus JA, Nascimento A, Hussain SS, Trainor RE, Pollock SR, Adair SJ, Michaels AD, Sesaki H, Stelow EB, Bauer TW, Kashatus DF. Drp1 promotes KRas-driven metabolic changes to drive pancreatic tumor growth. Cell Rep. 2019;28(7):1845–1859.e1845. doi: 10.1016/j.celrep.2019.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kinsey CG, Camolotto SA, Boespflug AM, Guillen KP, Foth M, Truong A, Schuman SS, Shea JE, Seipp MT, Yap JT, Burrell LD, Lum DH, Whisenant JR, Gilcrease GW, 3rd, Cavalieri CC, Rehbein KM, Cutler SL, Affolter KE, Welm AL, Welm BE, Scaife CL, Snyder EL, McMahon M. Protective autophagy elicited by RAF–>MEK–>ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat Med. 2019;25(4):620–627. doi: 10.1038/s41591-019-0367-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Bryant KL, Stalnecker CA, Zeitouni D, Klomp JE, Peng S, Tikunov AP, Gunda V, Pierobon M, Waters AM, George SD, Tomar G, Papke B, Hobbs GA, Yan L, Hayes TK, Diehl JN, Goode GD, Chaika NV, Wang Y, Zhang GF, Witkiewicz AK, Knudsen ES, PetricoinSingh EFPK, Macdonald JM, Tran NL, Lyssiotis CA, Ying H, Kimmelman AC, Cox AD, Der CJ. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat Med. 2019;25(4):628–640. doi: 10.1038/s41591-019-0368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kalas W, Swiderek E, Rapak A, Kopij M, Rak JW, Strzadala L. H-ras up-regulates expression of BNIP3. Anticancer Res. 2011;31(9):2869–2875. [PubMed] [Google Scholar]
- 198.Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yaaguchi A, Imanishi H, Nakada K, Honma Y, Hayashi JI. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 199.Porporato PE, Payen VL, Perez-Escuredo J, De Saedeleer CJ, Danhier P, Copetti T, Dhup S, Tardy M, Vazeille T, Bouzin C, Feron O, Michiels C, Gallez B, Sonveaux P. A mitochondrial switch promotes tumor metastasis. Cell Rep. 2014;8(3):754–766. doi: 10.1016/j.celrep.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 200.Tan A, Baty JW, Dong LF, Bezawork-Geleta A, Endaya B, Goodwin J, Bajzikova M, Kovarova J, Peterka M, Yan B, Pesdar EA, Sobol M, Filimonenko A, Stuart S, Vondrusova M, Kluckova K, Sachaphibulkij K, Rohlena J, Hozak P, Truksa J, Eccles D, Haupt LM, Griffiths LR, Neuzil J, Berridge MV. Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell Metab. 2015;21:81–94. doi: 10.1016/j.cmet.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 201.Dong LF, Kovarova J, Bajzikova M, Bezawork-Geleta A, Svec D, Endaya B, Sachaphibulkij K, Coelho AR, Sebkova N, Ruzickova A, Tan AS, Kluckova K, Judasova K, Zamecnikova K, Rychtarcikova Z, Gopalan V, Andera L, Sobol M, Yan B, Pattnaik B, Bhatraju N, Truksa J, Stopka P, Hozak P, Lam AK, Sedlacek R, Oliveira PJ, Kubista M, Agrawal A, Dvorakova-Hortova K, Rohlena J, Berridge MV, Neuzil J. Horizontal transfer of whole mitochondria restores tumorigenic potential in mitochondrial DNA-deficient cancer cells. eLife. 2017 doi: 10.7554/eLife.22187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cheung EC, DeNicola GM, Nixon C, Blyth K, Labuschagne CF, Tuveson DA, Vousden KH. Dynamic ROS control by TIGAR regulates the initiation and progression of pancreatic cancer. Cancer Cell. 2020;37(2):168–182.e164. doi: 10.1016/j.ccell.2019.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Radisky DC, Levy E, Littlepage LE, Liu H, Nelson CM, Fata JE, Leake D, Godden EL, Albertson DG, Nieto MA, Werb Z, Bissell MJ. Rac1b and reactive oxygen species mediate MMP3-induced EMT and genomic instability. Nature. 2005;436:123–127. doi: 10.1038/nature03688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, Liu CJ, Teng SC, Wu KJ. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol. 2008;10(3):295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 205.Holmstrom KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol. 2014;15:411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 206.Sabharwal SS, Schumacker PT. Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles' heel? Nat Rev Cancer. 2014;14(11):709–721. doi: 10.1038/nrc3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Piskounova E, Agathocleous M, Murphy MM, Hu Z, Huddlestun SE, Zhao Z, Leitch AM, Johnson TM, DeBerardinis RJ, Morrison SJ. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature. 2015;527(7577):186–191. doi: 10.1038/nature15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Le Gal K, Ibrahim MX, Wiel C, Sayin VI, Akula MK, Karlsson C, Dalin MG, Akyurek LM, Lindahl P, Nilsson J, Bergo MO. Antioxidants can increase melanoma metastasis in mice. Sci Transl Med. 2015;7(308):308. doi: 10.1126/scitranslmed.aad3740. [DOI] [PubMed] [Google Scholar]
- 209.Kenny TC, Gomez ML, Germain D. Mitohormesis, UPR(mt), and the complexity of mitochondrial DNA landscapes in cancer. Can Res. 2019 doi: 10.1158/0008-5472.can-19-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Kenny TC, Craig AJ, Villanueva A, Germain D. Mitohormesis primes tumor invasion and metastasis. Cell Rep. 2019;27(8):2292–2303.e2296. doi: 10.1016/j.celrep.2019.04.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Chourasia AH, Macleod KF. Tumor suppressor functions of BNIP3 and mitophagy. Autophagy. 2015;11(10):1937–1938. doi: 10.1080/15548627.2015.1085136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Pedanou VE, Gobeil S, Tabaries S, Simone TM, Zhu LJ, Siegel PM, Green MR. The histone H3K9 demethylase KDM3A promotes anoikis by transcriptionally activating pro-apoptotic genes BNIP3 and BNIP3L. eLife. 2016 doi: 10.7554/eLife.16844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Maes H, Van Eygen S, Krysko DV, Vandenabeele P, Nys K, Rillaerts K, Garg AD, Verfaillie T, Agostinis P. BNIP3 supports melanoma cell migration and vasculogenic mimicry by orchestrating the actin cytoskeleton. Cell Death Dis. 2014;5(3):e1127. doi: 10.1038/cddis.2014.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zhang J, Zhang C, Jiang X, Li L, Zhang D, Tang D, Yan T, Zhang Q, Yuan H, Jia J, Hu J, Zhang J, Huang Y. Involvement of autophagy in hypoxia-BNIP3 signaling to promote epidermal keratinocyte migration. Cell Death Dis. 2019;10(3):234. doi: 10.1038/s41419-019-1473-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 215.Zhang N, Yang X, Yuan F, Zhang L, Wang Y, Wang L, Mao Z, Luo J, Zhang H, Zhu WG, Zhao Y. Increased amino acid uptake supports autophagy-deficient cell survival upon glutamine deprivation. Cell Rep. 2018;23(10):3006–3020. doi: 10.1016/j.celrep.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 216.Towers CG, Fitzwalter BE, Regan D, Goodspeed A, Morgan MJ, Liu CW, Gustafson DL, Thorburn A. Cancer cells upregulate NRF2 signaling to adapt to autophagy inhibition. Dev Cell. 2019;50(6):690–703.e696. doi: 10.1016/j.devcel.2019.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30(13):3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Fearon KC, Glass DJ, Guttridge DC. Cancer cachexia: mediators, signaling, and metabolic pathways. Cell Metab. 2012;16(2):153–166. doi: 10.1016/j.cmet.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 219.Biswas AK, Acharyya S. Understanding cachexia in the context of metastatic progression. Nat Rev Cancer. 2020;20(5):274–284. doi: 10.1038/s41568-020-0251-4. [DOI] [PubMed] [Google Scholar]
- 220.Porporato PE. Understanding cachexia as a cancer metabolism syndrome. Oncogenesis. 2016;5:e200. doi: 10.1038/oncsis.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Petruzzelli M, Wagner EF. Mechanisms of metabolic dysfunction in cancer-associated cachexia. Genes Dev. 2016;30(5):489–501. doi: 10.1101/gad.276733.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Tsoli M, Robertson G. Cancer cachexia: malignant inflammation, tumorkines, and metabolic mayhem. Trends Endocrinol Metab TEM. 2013;24(4):174–183. doi: 10.1016/j.tem.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 223.Stephens NA, Skipworth RJ, Gallagher IJ, Greig CA, Guttridge DC, Ross JA, Fearon KC. Evaluating potential biomarkers of cachexia and survival in skeletal muscle of upper gastrointestinal cancer patients. J Cachexia Sarcopenia Muscle. 2015;6(1):53–61. doi: 10.1002/jcsm.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Julienne CM, Dumas JF, Goupille C, Pinault M, Berri C, Collin A, Tesseraud S, Couet C, Servais S. Cancer cachexia is associated with a decrease in skeletal muscle mitochondrial oxidative capacities without alteration of ATP production efficiency. J Cachexia Sarcopenia Muscle. 2012;3(4):265–275. doi: 10.1007/s13539-012-0071-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Antunes D, Padrao AI, Maciel E, Santinha D, Oliveira P, Vitorino R, Moreira-Goncalves D, Colaco B, Pires MJ, Nunes C, Santos LL, Amado F, Duarte JA, Domingues MR. Molecular insights into mitochondrial dysfunction in cancer-related muscle wasting. Biochem Biophys Acta. 2014;6:896–905. doi: 10.1016/j.bbalip.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 226.Amaravadi R, Kimmelman A, Debnath J (2019) Targeting autophagy in cancer: recent advances, future directions. Cancer Discov Under review [DOI] [PMC free article] [PubMed]
- 227.Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17(9):528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Briceno E, Reyes S, Sotelo J. Therapy of glioblastoma multiforme improved by the antimutagenic chloroquine. Neurosurg Focus. 2003;14(2):e3. doi: 10.3171/foc.2003.14.2.4. [DOI] [PubMed] [Google Scholar]
- 229.Sotelo J, Briceno E, Lopez-Gonzalez MA. Adding chloroquine to conventional treatment for glioblastoma multiforme: a randomized, double-blind, placebo-controlled trial. Ann Intern Med. 2006;144(5):337–343. doi: 10.7326/0003-4819-144-5-200603070-00008. [DOI] [PubMed] [Google Scholar]
- 230.Pascolo S. Time to use a dose of Chloroquine as an adjuvant to anti-cancer chemotherapies. Eur J Pharmacol. 2016;771:139–144. doi: 10.1016/j.ejphar.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 231.Elliott IA, Dann AM, Xu S, Kim SS, Abt ER, Kim W, Poddar S, Moore A, Zhou L, Williams JL, Capri JR, Ghukasyan R, Matsumura C, Tucker DA, Armstrong WR, Cabebe AE, Wu N, Li L, Le TM, Radu CG, Donahue TR. Lysosome inhibition sensitizes pancreatic cancer to replication stress by aspartate depletion. Proc Natl Acad Sci U S A. 2019;116(14):6842–6847. doi: 10.1073/pnas.1812410116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Gao M, Yi J, Zhu J, Minikes AM, Monian P, Thompson CB, Jiang X. Role of mitochondria in ferroptosis. Mol Cell. 2019;73(2):354–363.e353. doi: 10.1016/j.molcel.2018.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Georgakopoulos ND, Wells G, Campanella M. The pharmacological regulation of cellular mitophagy. Nat Chem Biol. 2017;13(2):136–146. doi: 10.1038/nchembio.2287. [DOI] [PubMed] [Google Scholar]
- 234.Chourasia AH, Boland ML, Macleod KF. Mitophagy and cancer. Cancer Metab. 2015;3:4. doi: 10.1186/s40170-015-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Rao VA, Klein SR, Bonar SJ, Zielonka J, Mizuno N, Dickey JS, Keller PW, Joseph J, Kalyanaraman B, Shacter E. The antioxidant transcription factor Nrf2 negatively regulates autophagy and growth arrest induced by the anticancer redox agent mitoquinone. J Biol Chem. 2010;285(45):34447–34459. doi: 10.1074/jbc.M110.133579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Meyer N, Zielke S, Michaelis JB, Linder B, Warnsmann V, Rakel S, Osiewacz HD, Fulda S, Mittelbronn M, Münch C, Behrends C, Kögel D. AT 101 induces early mitochondrial dysfunction and HMOX1 (heme oxygenase 1) to trigger mitophagic cell death in glioma cells. Autophagy. 2018;14(10):1693–1709. doi: 10.1080/15548627.2018.1476812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Georgakopoulos ND, Frison M, Alvarez MS, Bertrand H, Wells G, Campanella M. Reversible Keap1 inhibitors are preferential pharmacological tools to modulate cellular mitophagy. Sci Rep. 2017;7(1):10303. doi: 10.1038/s41598-017-07679-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper J. The PINK1-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol Cell. 2015;60:7–20. doi: 10.1016/j.molcel.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Wong YC, Holzbaur EL. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci USA. 2014;111(42):E4439–4448. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Li L, Shen C, Nakamura E, Ando K, Signoretti S, Beroukhim R, Cowley GS, Lizotte P, Liberzon E, Bair S, Root DE, Tamayo P, Tsherniak A, Cheng SC, Tabak B, Jacobsen A, Hakimi AA, Schultz N, Ciriello G, Sander C, Hsieh JJ, Kaelin WGJ. SQSTM1 is a pathogenic target of 5q copy number gains in kidney cancer. Cancer Cell. 2013;24(6):738–750. doi: 10.1016/j.ccr.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Moscat J, Karin M, Diaz-Meco MT. p62 in cancer: signaling adaptor beyond autophagy. Cell. 2016;167(3):606–609. doi: 10.1016/j.cell.2016.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Hsieh CH, Shaltouki A, Gonzalez AE, Bettencourt da Cruz A, Burbulla LF, St Lawrence E, Schule B, Krainc D, Palmer TD, Wang X. Functional impairment in miro degradation and mitophagy is a shared feature in familial and sporadic Parkinson's disease. Cell Stem Cell. 2016;19(6):709–724. doi: 10.1016/j.stem.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Sugiura A, Nagashima S, Tokuyama T, Amo T, Matsuki Y, Ishido S, Kudo Y, McBride HM, Fukuda T, Matsushita N, Inatome R, Yanagi S. MITOL regulates endoplasmic reticulum-mitochondria contacts via Mitofusin2. Mol Cell. 2013;51(1):20–34. doi: 10.1016/j.molcel.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 244.Szargel R, Shani V, Abd Elghani F, Mekies LN, Liani E, Rott R, Engelender S. The PINK1, synphilin-1 and SIAH-1 complex constitutes a novel mitophagy pathway. Hum Mol Genet. 2016;25(16):3476–3490. doi: 10.1093/hmg/ddw189. [DOI] [PubMed] [Google Scholar]
- 245.Anding AL, Wang C, Chang TK, Sliter DA, Powers CM, Hofmann K, Youle RJ, Baehrecke EH. Vps13D encodes a ubiquitin-binding protein that is required for the regulation of mitochondrial size and clearance. Curr Biol CB. 2018;28(2):287–295.e286. doi: 10.1016/j.cub.2017.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, Sabatini DM. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146(3):408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Winter RN, Kramer A, Borkowski A, Kyprianou N. Loss of caspase-1 and caspase-3 protein expression in human prostate cancer. Can Res. 2001;61(3):1227–1232. [PubMed] [Google Scholar]
- 248.Yang YM, Ramadani M, Huang YT. Overexpression of Caspase-1 in adenocarcinoma of pancreas and chronic pancreatitis. World J Gastroenterol. 2003;9(12):2828–2831. doi: 10.3748/wjg.v9.i12.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Feng Q, Li P, Salamanca C, Huntsman D, Leung PC, Auersperg N. Caspase-1alpha is down-regulated in human ovarian cancer cells and the overexpression of caspase-1alpha induces apoptosis. Can Res. 2005;65(19):8591–8596. doi: 10.1158/0008-5472.can-05-0239. [DOI] [PubMed] [Google Scholar]
- 250.Zaki MH, Vogel P, Body-Malapel M, Lamkanfi M, Kanneganti TD. IL-18 production downstream of the Nlrp3 inflammasome confers protection against colorectal tumor formation. J Immunol. 2010;185(8):4912–4920. doi: 10.4049/jimmunol.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Daley D, Mani VR, Mohan N, Akkad N, Pandian G, Savadkar S, Lee KB, Torres-Hernandez A, Aykut B, Diskin B, Wang W, Farooq MS, Mahmud AI, Werba G, Morales EJ, Lall S, Wadowski BJ, Rubin AG, Berman ME, Narayanan R, Hundeyin M, Miller G. NLRP3 signaling drives macrophage-induced adaptive immune suppression in pancreatic carcinoma. J Exp Med. 2017;214(6):1711–1724. doi: 10.1084/jem.20161707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Vanpouille-Box C, Demaria S, Formenti SC, Galluzzi L. Cytosolic DNA sensing in organismal tumor control. Cancer Cell. 2018;34(3):361–378. doi: 10.1016/j.ccell.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 253.Rzymski T, Milani M, Singleton DC, Harris AL. Role of ATF4 in regulation of autophagy and resistance to drugs and hypoxia. Cell Cycle. 2009;8(23):3838–3847. doi: 10.4161/cc.8.23.10086. [DOI] [PubMed] [Google Scholar]
- 254.Dey S, Sayers CM, Verginadis II, Lehman SL, Cheng Y, Cerniglia GJ, Tuttle SW, Feldman MD, Zhang PJ, Fuchs SY, Diehl JA, Koumenis C. ATF4-dependent induction of heme oxygenase 1 prevents anoikis and promotes metastasis. J Clin Investig. 2015;125(7):2592–2608. doi: 10.1172/jci78031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by tthe autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 256.Zhao Y, Yang J, Liao W, Liu X, Zhang H, Wang S, Wang D, Feng J, Yu L, Zhu WG. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010;12(7):665–675. doi: 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
- 257.Warr MR, Binnewies M, Flach J, Reynaud D, Garg T, Malhotra R, Debnath J, Passegue E. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature. 2013;494(7437):323–327. doi: 10.1038/nature11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Fitzwalter BE, Towers CG, Sullivan KD, Andrysik Z, Hoh M, Ludwig M, O'Prey J, Ryan KM, Espinosa JM, Morgan MJ, Thorburn A. Autophagy inhibition mediates apoptosis sensitization in cancer therapy by relieving FOXO3a turnover. Dev Cell. 2018;44(5):555–565.e553. doi: 10.1016/j.devcel.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Keith B, Simon MC. Hypoxia-inducible factors, stem cells and cancer. Cell. 2007;129:465–472. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Semenza GL. Hypoxia-inducible factors: coupling glucose metabolism and redox regulation with induction of the breast cancer stem cell phenotype. Embo J. 2016 doi: 10.15252/embj.201695204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Perera RM, Stoykova S, Nicolay BN, Ross KN, Fitamant J, Boukhali M, Lengrand J, Deshpande V, Selig MK, Ferrone CR, Settleman J, Stephanopoulos G, Dyson NJ, Zoncu R, Ramaswamy S, Haas W, Bardeesy N. Transcriptional control of the autophagy-lysosome system in pancreatic cancer. Nature. 2015;524:361–365. doi: 10.1038/nature14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Strappazzon F, Nazio F, Corrado M, Cianfanelli V, Romagnoli A, Fimia GM, Campello S, Nardacci R, Piacentini M, Campanella M, Cecconi F. AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell Death Differ. 2015;22(3):419–432. doi: 10.1038/cdd.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Falasca L, Torino F, Marconi M, Costantini M, Pompeo V, Sentinelli S, De Salvo L, Patrizio M, Padula C, Gallucci M, Piacentini M, Malorni W. AMBRA1 and SQSTM1 expression pattern in prostate cancer. Apoptosis. 2015;20(12):1577–1586. doi: 10.1007/s10495-015-1176-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Strappazzon F, Cecconi F. AMBRA1-induced mitophagy: a new mechanism to cope with cancer? Mol Cell Oncol. 2015;2(2):e975647. doi: 10.4161/23723556.2014.975647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Cianfanelli V, Fuoco C, Lorente M, Salazar M, Quondamatteo F, Gherardini PF, De Zio D, Nazio F, Antonioli M, D'Orazio M, Skobo T, Bordi M, Rohde M, Dalla Valle L, Helmer-Citterich M, Gretzmeier C, Dengjel J, Fimia GM, Piacentini M, Di Bartolomeo S, Velasco G, Cecconi F. AMBRA1 links autophagy to cell proliferation and tumorigenesis by promoting c-Myc dephosphorylation and degradation. Nat Cell Biol. 2015;17(1):20–30. doi: 10.1038/ncb3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Schoenherr C, Byron A, Sandilands E, Paliashvili K, Baillie GS, Garcia-Munoz A, Valacca C, Cecconi F, Serrels B, Frame MC. Ambra1 spatially regulates Src activity and Src/FAK-mediated cancer cell invasion via trafficking networks. eLife. 2017 doi: 10.7554/eLife.23172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Park S, Choi SG, Yoo SM, Son JH, Jung YK. Choline dehydrogenase interacts with SQSTM1/p62 to recruit LC3 and stimulate mitophagy. Autophagy. 2014;10(11):1906–1920. doi: 10.4161/auto.32177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Karbowski M, Jeong SY, Youle RJ. Endophilin B1 is required for the maintenance of mitochondrial morphology. J Cell Biol. 2004;166(7):1027–1039. doi: 10.1083/jcb.200407046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Takahashi Y, Hori T, Cooper TK, Liao J, Desai N, Serfass JM, Young MM, Park S, Izu Y, Wang HG. Bif-1 haploinsufficiency promotes chromosomal instability and accelerates Myc-driven lymphomagenesis via suppression of mitophagy. Blood. 2013;121(9):1622–1632. doi: 10.1182/blood-2012-10-459826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Bhujabal Z, Birgisdottir AB, Sjottem E, Brenne HB, Overvatn A, Habisov S, Kirkin V, Lamark T, Johansen T. FKBP8 recruits LC3A to mediate Parkin-independent mitophagy. EMBO Rep. 2017;18(6):947–961. doi: 10.15252/embr.201643147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Shlevkov E, Kramer T, Schapansky J, LaVoie MJ, Schwarz TL. Miro phosphorylation sites regulate Parkin recruitment and mitochondrial motility. Proc Natl Acad Sci U S A. 2016;113(41):E6097–e6106. doi: 10.1073/pnas.1612283113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Le Guerroue F, Eck F, Jung J, Starzetz T, Mittelbronn M, Kaulich M, Behrends C. Autophagosomal content profiling reveals an LC3C-dependent piecemeal mitophagy pathway. Mol Cell. 2017;68(4):786–796.e786. doi: 10.1016/j.molcel.2017.10.029. [DOI] [PubMed] [Google Scholar]
- 273.D'Amico D, Mottis A, Potenza F, Sorrentino V, Li H, Romani M, Lemos V, Schoonjans K, Zamboni N, Knott G, Schneider BL, Auwerx J. The RNA-binding protein PUM2 impairs mitochondrial dynamics and mitophagy during aging. Mol Cell. 2019;73(4):775–787.e710. doi: 10.1016/j.molcel.2018.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Schneider AT, Gautheron J, Feoktistova M, Roderburg C, Loosen SH, Roy S, Benz F, Schemmer P, Büchler MW, Nachbur U, Neumann UP, Tolba R, Luedde M, Zucman-Rossi J, Panayotova-Dimitrova D, Leverkus M, Preisinger C, Tacke F, Trautwein C, Longerich T, Vucur M, Luedde T. RIPK1 suppresses a TRAF2-dependent pathway to liver cancer. Cancer Cell. 2017;31(1):94–109. doi: 10.1016/j.ccell.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 275.Hawk MA, Gorsuch CL, Fagan P, Lee C, Kim SE, Hamann JC, Mason JA, Weigel KJ, Tsegaye MA, Shen L, Shuff S, Zuo J, Hu S, Jiang L, Chapman S, Leevy WM, DeBerardinis RJ, Overholtzer M, Schafer ZT. RIPK1-mediated induction of mitophagy compromises the viability of extracellular-matrix-detached cells. Nat Cell Biol. 2018;20(3):272–284. doi: 10.1038/s41556-018-0034-2. [DOI] [PubMed] [Google Scholar]
- 276.Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, Amano A, Yoshimori T. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495(7441):389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 277.Arasaki K, Nagashima H, Kurosawa Y, Kimura H, Nishida N, Dohmae N, Yamamoto A, Yanagi S, Wakana Y, Inoue H, Tagaya M. MAP1B-LC1 prevents autophagosome formation by linking syntaxin 17 to microtubules. EMBO Rep. 2018 doi: 10.15252/embr.201745584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Sugo M, Kimura H, Arasaki K, Amemiya T, Hirota N, Dohmae N, Imai Y, Inoshita T, Shiba-Fukushima K, Hattori N, Cheng J, Fujimoto T, Wakana Y, Inoue H, Tagaya M. Syntaxin 17 regulates the localization and function of PGAM5 in mitochondrial division and mitophagy. Embo J. 2018 doi: 10.15252/embj.201798899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Xian H, Yang Q, Xiao L, Shen HM, Liou YC. STX17 dynamically regulated by Fis1 induces mitophagy via hierarchical macroautophagic mechanism. Nat Commun. 2019;10(1):2059. doi: 10.1038/s41467-019-10096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Kumar S, Gu Y, Abudu YP, Bruun JA, Jain A, Farzam F, Mudd M, Anonsen JH, Rusten TE, Kasof G, Ktistakis N, Lidke KA, Johansen T, Deretic V. Phosphorylation of syntaxin 17 by TBK1 controls autophagy initiation. Dev Cell. 2019;49(1):130–144. doi: 10.1016/j.devcel.2019.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]