Abstract
Plant hormone abscisic acid (ABA) is essential for regulating plant growth and various stress responses. ABA-mediated signaling depends on local ABA levels rather than the overall cellular ABA concentration. While cellular concentration of ABA can be detected using Förster resonance energy transfer (FRET)-based ABA probes, direct imaging of subcellular ABA levels remains unsolved. Here, we modified the previously reported ABAleon2.1 and generated a new ABA sensor, named ABAleon2.1_Tao3. Via transient expression in tobacco (Nicotiana tabacum) protoplasts, we targeted ABAleon2.1_Tao3s to the endoplasmic reticulum (ER) membrane with the ABA sensing unit facing the cytosol and the ER, respectively, through a nanobody–epitope-mediated protein interaction. Combining FRET with fluorescence lifetime imaging microscopy, ABA-triggered-specific increases in the fluorescence lifetime of the donor mTurquoise in the ABAleon2.1_Tao3 were detected in both transient assays and stably transformed Arabidopsis plants. In tobacco protoplasts, ER membrane-targeted ABAleon2.1_Tao3s showed a generally higher basal level of ABA in the ER than that in the cytosol and ER-specific alterations in the level of ABA upon environmental cues. In ABAleon2.1_Tao3-transformed Arabidopsis roots, mannitol triggered increases in cytosolic ABA in the division zone and increases in ER ABA in the elongation and maturation zone within 1 h after treatment, both of which were abolished in the bg1-2 mutant, suggesting the requirement for BG1 in osmotic stress-triggered early ABA induction in Arabidopsis roots. These data demonstrate that ABAleon2.1_Tao3s can be used to monitor ABA levels in the cytosol and the ER, providing key information on stress-induced changes in the level of ABA in different subcellular compartments.
FRET-based ABA sensors targeted to the ER membrane through a nanobody–epitope-mediated protein assembly reveal specific alterations and dynamics of ABA in the ER upon various environmental stimuli.
Introduction
The phytohormone abscisic acid (ABA) plays pivotal roles in coordinating plant growth, development, and responses to environmental cues like drought, salinity, and pathogen attacks. Regulation of cellular ABA concentration is a dynamic, multistage process involving ABA synthesis, catabolism, and transport. These physiological processes occur in various organelles, into which ABA is partitioned and eventually leads to different signaling outputs (Finkelstein, 2013; Xu et al., 2013). For example, the rate-limiting step for the ABA de novo biosynthesis pathway catalyzed by 9-cis-epoxycarotenoid dioxygenases (NCEDs) occurs in plastids and is essential for various stress responses and plant development (Tan et al., 2003). Two β-glucosidase homologs (AtBG1 and AtBG2) were shown to rapidly activate the ABA pool in the endoplasmic reticulum (ER) and vacuole upon dehydration and osmotic stresses (Lee et al., 2006; Xu et al., 2012). Regulation of cytosolic ABA level either through 8′-hydroxylation by CYP707As or glucose conjugation by uridine diphosphate glucosyltransferases played key roles in plant adaptation to dehydration, osmotic stress, and salinity stresses (Saito et al., 2004; Dong et al., 2014; Liu et al., 2015). Moreover, plasma membrane-localized ABA transporters can modulate drought tolerance through facilitating cytosolic uptake and efflux of ABA (Kang et al., 2010, 2015; Kuromori et al., 2010; Kanno et al., 2012; Zhang et al., 2014). Considering the cellular distribution of ABA and ABA conjugates (Tan et al., 2003; Lee et al., 2006), the initiation of ABA signaling and downstream response outputs upon environmental stimulus are more likely associated with changes in local active ABA levels rather than the overall cellular ABA level. To investigate this, direct monitoring of ABA levels at subcellular resolution is of vital importance.
Imaging approaches using Förster resonance energy transfer (FRET)-based biosensors have been increasingly applied for studying cell signals. FRET sensors contain donor and acceptor fluorophores that are associated with sensory modules, through which analytes are sensed and monitored by changes in FRET signals. A miscellaneous group of FRET sensors has been developed, allowing direct visualization of metabolites, secondary messengers, and plant hormones (Okumoto et al., 2012; Walia et al., 2018). Based on ABA-dependent interactions between its receptors PYR1/PYL1 and a PP2C-type phosphatase ABI1, ABA-specific FRET bioprobes have been engineered in Arabidopsis thaliana and distribution and translocation of ABA were revealed at tissue and cellular levels (Jones et al., 2014; Waadt et al., 2014). However, the spatio-temporal regulation of ABA at subcellular resolution remains elusive.
Targeting of soluble sensors into subcellular organelles can be achieved via the addition of an N-terminal target sequence. Targeting to specific locations in the endomembrane system requires further sorting signals, like the tetrapeptide H/KDEL to/at the C-terminus of the protein of interest for ER targeting (Denecke et al., 1990; Phillipson et al., 2001). In such manners, calcium indicators (Cameleons) have been targeted to the chloroplast stroma, nucleus, mitochondria, and the ER (Iwano et al., 2009; Krebs et al., 2012; Loro et al., 2012, 2016; Bonza et al., 2013). Similarly, organellar-specific pH sensors were targeted to different compartments via fusing specific sorting signals/membrane domains to either C- or N-termini of the sensors (Martiniere et al., 2013; Shen et al., 2013). These targeting strategies, however, tend to render the targeted protein with different conformations in different suborganelles. In the case of pH sensors, they were targeted either as soluble proteins (in nucleus, ER, peroxisome, mitochondria), or were attached to membranes at the N-terminus (in chloroplast and vacuole) and at the C-terminus (in TGN and MVB/PVC; Shen et al., 2013).
A nanobody (Nb)-mediated protein targeting strategy has been recently developed (Kunzl et al., 2016; Fruholz et al., 2018). In this scenario, the coexpression of a soluble Nb fusion protein with an epitope-tagged, compartment-specific membrane protein resulted in Nb–epitope interaction-triggered intramolecular assembly at its corresponding compartment membrane. The established interaction is stable and persists in all compartments of the endomembrane system without any alterations in the topology of sensing domains (Früholz and Pimpl, 2017). Here, we report the generation of ABAleon2.1_Tao3s through modifications of FRET-based ABA sensor ABAleon2.1 and their targeting to the ER membrane, with the central ABA sensing module exposed to the cytosol and the ER lumen, respectively. With the soluble and ER membrane-targeted ABAleon2.1_Tao3s, we show ER-specific increases in the level of ABA under light, cold, and high-sulfate supply conditions compared with cytosolic ABA. Time-resolved analysis of ABAleon2.1_Tao3s revealed distinct kinetics of ABA in the ER and the cytosol upon different osmotic stimuli. ABAleon2.1_Tao3s-transformed Arabidopsis plants showed increases of cytosolic and ER ABA exclusively in root meristem zone and elongation/maturation zone, respectively, upon osmotic stress. Together, these data support fundamental functions of the ER as a major source for regulating subcellular ABA homeostasis in plant growth and stress responses.
Results
Targeting of ABA bioprobes to the ER membrane through a Nb-mediated protein assembly
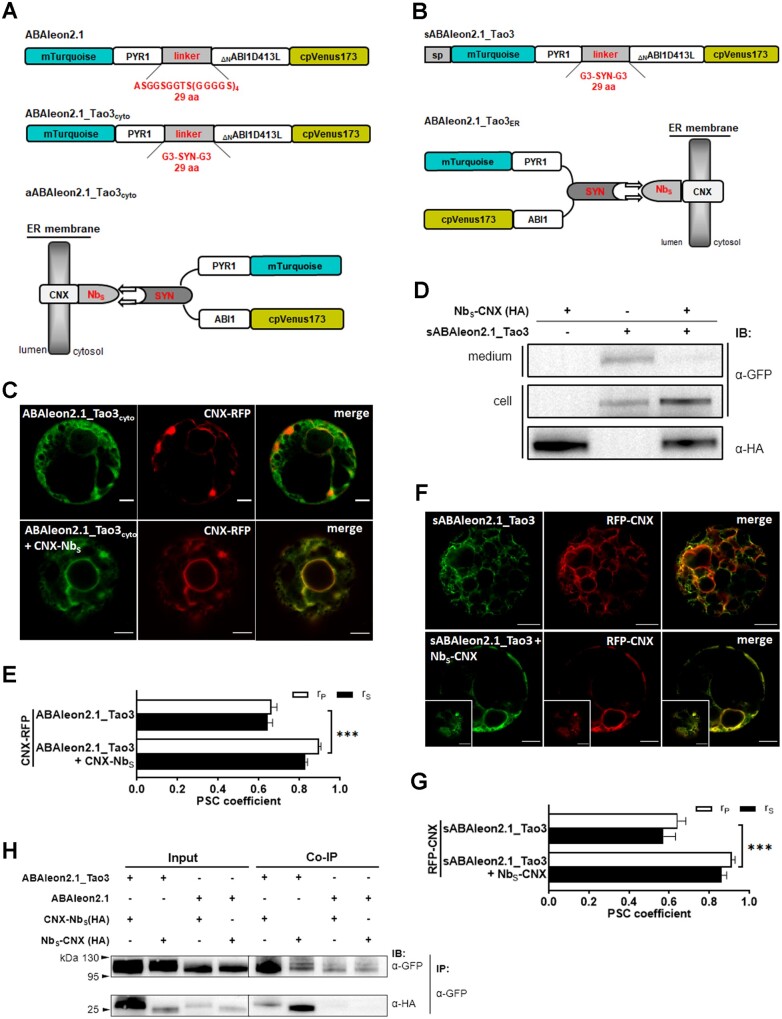
For ER targeting of FRET-based bioprobes, the addition of a C-terminal H/KDEL sequence is generally sufficient, albeit the targeting efficiency varies depending on the nature of fusion proteins (Gomord et al., 1997). However, the extra H/KDEL residuals renders direct comparisons of values obtained from ER-residing sensors with those without sorting signals difficult, especially considering that the emissions or lifetimes of the adjacent fluorophores could be affected through intramolecular quenching (Doose et al., 2009; Chen et al., 2010). To target ABA probes to the ER with similar conformation to their cytosolic counterparts, we first constructed two ER-membrane anchors with a Nb that had been raised against α-synuclein fused to either the C-terminal (CNX-NbS) or the N-terminal of the truncated ER-membrane protein calnexin (NbS-CNX), which has been shown to function in structural integrity of the ER in plants (Niehl et al., 2012; Fruholz et al., 2018). The previously developed ABA sensor, ABAleon2.1 (Waadt et al., 2014), was modified to a version renamed ABAleon2.1_Tao3cyto (Figure 1A) by replacing the 29 amino acid (aa) GS linker with the GGG-SYN-GGG linker, in which SYN was a 23-aa epitope originated from α-synuclein (Iljina et al., 2017). A secretory version of ABAleon2.1_Tao3 (sABAleon2.1_Tao3) was generated by fusing a N-terminal signal peptide originated from a tobacco (Nicotiana tabacum) endochitinase A (Kunzl et al., 2016), allowing the entering of ABAleon2.1_Tao3 into the ER lumen (Figure 1B). The coexpression of ABAleon2.1_Tao3 and sABAleon2.1_Tao3 with the ER-membrane anchor CNX-NbS and NbS-CNX, respectively, would result in ABAleon2.1_Tao3 being anchored at the ER membrane with the ABA sensing module exposed to either the cytosol (aABAleon2.1_Tao3cyto; Figure 1A) or the ER lumen (ABAleon2.1_Tao3ER; Figure 1B). Indeed, the cotransfection of ABAleon2.1_Tao3cyto with CNX-NbS in tobacco protoplasts resulted in the colocalization of ABAleon2.1_Tao3 with its corresponding ER RFP marker (CNX-RFP; Figure 1, C and E), suggesting the assembly of ABAleon2.1_Tao3cyto (aABAleon2.1_Tao3cyto) along the ER membrane. For ER lumen targeting, transfection of sABAleon2.1_Tao3 alone resulted in the sensor either secreted outside the cells (shown by the presence in the medium samples in Figure 1D and no GFP signal in Supplemental Figure S1A) or retained in the cells (Figure 1F;Supplemental Figure S1B). These indicated a normal transport activity of sABAleon2.1_Tao3. When cotransfected with NbS-CNX and the red fluorescent ER marker RFP-CNX (Kunzl et al., 2016), more of sABAleon2.1_Tao3 were retained in the cells compared with those cells that had been transfected with sABAleon2.1_Tao3 alone (Figure 1D). And the sensor proteins colocalized well with the ER marker (Figure 1, F and G;Supplemental Figure S1C), indicating the localization of sABAleon2.1_Tao3 in the ER. Coimmunoprecipitation analysis corroborated the interaction between ABAleon2.1_Tao3s and their corresponding ER-membrane anchors (Figure 1H).
Figure 1.
Targeting and assembly of ABAleon2.1_Tao3s at the ER membrane through Nb-mediated interactions. A, B, Schematic representation of ABAleon2.1_Tao3cyto (A) and sABAleon2.1_Tao3 (B), and their assemblies on the ER membrane through Nb-mediated interaction between ABAleon2.1_Tao3s and the ER-membrane anchor (CNX-NbS (A), NbS-CNX (B)). As in ABAleon2.1, full-length PYR1 and ABI1 that were truncated at S125 were used as an ABA sensor module. C, F, CLSM analysis of tobacco protoplasts cotransfected with ABAleon2.1_Tao3cyto/sABAleon2.1_Tao3 and the RFP markers of ER membrane (CNX-RFP/RFP-CNX), either with or without their corresponding ER-membrane anchors. Insets are cortical sections of their corresponding pictures. Scale bars, 5 μm. D, Immunodetection of sABAleon2.1_Tao3 with and without the ER anchor NbS-CNX using α-GFP. After electrotransfection, protoplasts and the medium buffer were harvested separately and immunoblotted to detect the presence of sensor proteins in the medium (secreted) and in the cell samples (retained in the protoplasts). The HA-tagged anchor NbS-CNX was detected using α-HA. E, G, Pearson’s (rP) and Spearman’s (rS) correlation (PSC) coefficients of ABAleon2.1_Tao3cyto/CNX-RFP and sABAleon2.1_Tao3/RFP-CNX signals from (C) and (F), respectively. Data are presented as means ± SE. Statistical analysis was performed by two-way ANOVA followed by Tukey’s HSD test (***P < 0.001). n = 10–12 cells. H, Immunoblot revealing the Nb-mediated assembly of ABAleon2.1_Tao3s at the ER membrane. ABAleon2.1_Tao3s were assembled by coexpression of ABAleon2.1_Tao3cyto and sABAleon2.1_Tao3 with CNX-NbS and NbS-CNX, respectively, in tobacco protoplasts, and then immunoprecipitated with anti-GFP antibody-coated beads. Immunoblots were probed with antibodies to detect the presence of ABAleon2.1_Tao3cyto and sABAleon2.1_Tao3 (α-GFP) and the Nb–epitope interactions (α-HA). Protoplast samples that had been cotransfected with ABAleon2.1/CNX-NbS or ABAleon2.1/NbS-CNX were harvested and immunoblotted as a negative control
FRET–FLIM responses and characterization of ABAleon2.1_Tao3s in tobacco cells
In ABAleon2.1, ABA-triggered intramolecular interactions between PYR1 and ΔNABI1 resulted in increases in the distance or changes in the orientation of mTurquoise (τmT) and cpVenus173, leading to reductions in FRET efficiency that were measured through emission ratiometric methods (Waadt et al., 2014). Alternatively, FRET signals can be monitored by recording changes of the fluorescence lifetime of the donor protein via fluorescence lifetime imaging microscopy (FLIM). FRET–FLIM is now widely utilized not only for its extremely high sensitivity and being less prone to spectral bleed-through artifacts, which persist in most of the intensity-based FRET signals, but most importantly for being independent of the expression levels of FRET pairs (Wallrabe and Periasamy, 2005).
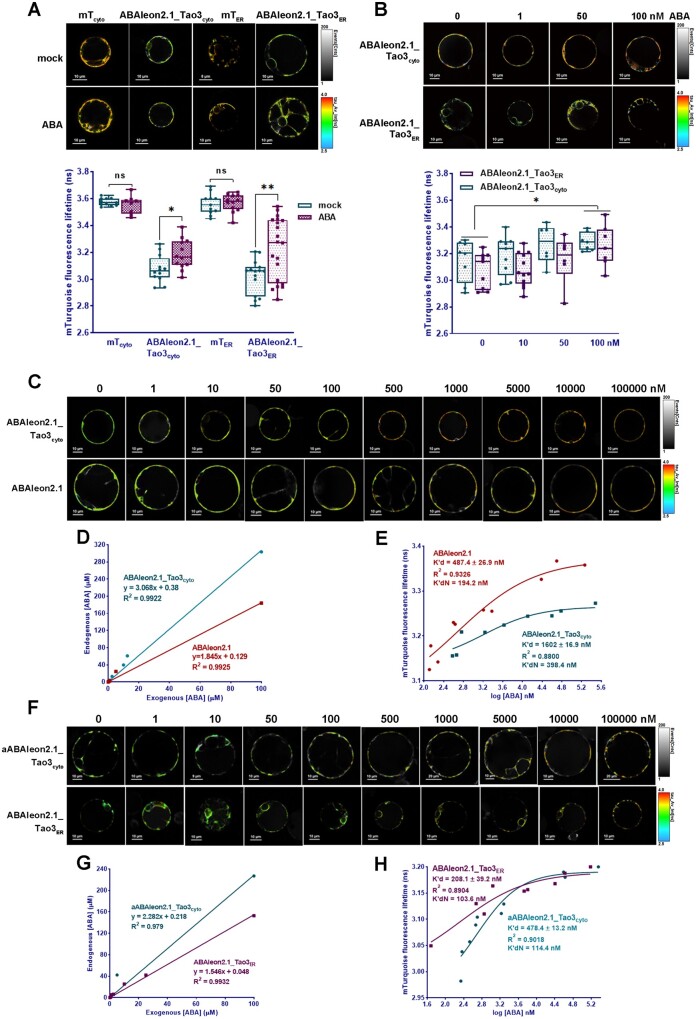
To test whether ABAleon2.1_Tao3 responds to ABA with changes in the fluorescence lifetime of τmT, which acts as the donor fluorophore, we expressed both ABAleon2.1_Tao3cyto and ABAleon2.1_Tao3ER in tobacco protoplasts and tested their responsiveness to exogenous ABA through recording changes in the τmT. Whereas the treatment of exogenous ABA had little effect on the τmT of the respective donor-only controls (mTcyto and mTER), it significantly increased the τmT of both ABAleon2.1_Tao3cyto and ABAleon2.1_Tao3ER (Figure 2A). Moreover, application of ABA had marginal effect on the τmT of a mutated version of ABAleon2.1_Tao3cyto (mABAleon2.1_Tao3cyto; Supplemental Figure S2), in which the ABA receptor PYR1 was mutated at Arg116 to Gly116 (R116G), thus abolishing both ABA binding and ABA-induced PYR1 interaction with ABI1 (Nishimura. et al., 2009). These results together support that ABA triggers specific increases in the τmT in ABAleon2.1_Tao3s. When treated with lower ABA concentrations (1–100 nM), it was noticed that both ABAleon2.1_Tao3cyto and ABAleon2.1_Tao3ER did not show significant increases of the τmT until 100 nM of ABA treatment (Figure 2B).
Figure 2.
Characterization of ABAleon2.1_Tao3s by FRET–FLIM. A, Representative pseudo-color images and data recorded by FLIM show application of ABA (20 μM)-triggered specific increases in the fluorescence lifetime of the donor τmT of cytosolic ABAleon2.1_Tao3 (ABAleon2.1_Tao3cyto) and the ER ABAleon2.1_Tao3 (ABAleon2.1_Tao3ER), but not in their corresponding donor only (mTcyto and mTER) controls. For better visualization and comparisons, the color-scale bar of all the FLIM images in tobacco protoplast assays were normalized to 2.5–4.0 and the intensity bars were from 1 to 100–400 depending on the expression level of proteins in the cells. FLIM data are presented as box plots showing all data points. Significance was calculated using Student’s t test (*P < 0.05; ns, not significant). B, Representative FLIM images and data show increased τmT in ABAleon2.1_Tao3cyto and ABAleon2.1_Tao3ER upon exogenous ABA treatment at 100-nM level. FLIM data are presented as box plots showing all data points. Statistical analysis was performed using two-way ANOVA followed by Tukey’s HSD test (*P < 0.05 for overall difference between 100-nM ABA and mock treatment). C, F, Representative FLIM images of ABAleon2.1_Tao3cyto and ABAleon2.1 (C), aABAleon2.1_Tao3cyto and ABAleon2.1_Tao3ER (F) in response to exogenously applied 0–100-μM ABA treatments. D, G, Shown are the plots of endogenous ABA levels in tobacco protoplasts that had been transfected with ABAleon2.1_Tao3cyto and ABAleon2.1 (D), aABAleon2.1_Tao3cyto, and ABAleon2.1_Tao3ER (G) against exogenous ABA treatments. The linear equations and calculated Rsqr are shown in each plot. E, H, Shown are the τmT of ABAleon2.1_Tao3cyto and ABAleon2.1 (E), aABAleon2.1_Tao3cyto, and ABAleon2.1_Tao3ER (H) plotted against their corresponding endogenous ABA concentrations. The ABA affinities (K′d) were calculated by fitting a four-parameter logistic curve τ = τmin + (τmax-τmin)/1 + ([ABA]/K′d)n using GraphPad Prism 8.0.2. The normalized K′d (K′dN) of each ABA probe was calculated according to its linear relationship with exogenous ABA concentrations
To characterize ABAleon2.1_Tao3s more precisely, we tested dosage-responses of the τmT of both soluble (ABAleon2.1_Tao3cyto) and ER-membrane anchored (aABAleon2.1_Tao3cyto and ABAleon2.1_Tao3ER) sensors in tobacco cells upon exogenous ABA treatments ranging from 1 nM to 100 μM using FRET–FLIM. The total endogenous ABA level was measured upon each exogenous ABA treatment and plotted as linear equations (Figure 2, D and G). The τmT responses of each ABA sensor were fitted against their corresponding endogenous ABA levels in a four-parameter sigmoidal curve (Figure 2, E and H). Considering the basal amount of ABA in the tobacco protoplasts, resulting ABA affinities K′d of each sensor were further calculated according to their respective linear equations (K′dN; Figure 2, D and G). Thus, the K′dN here corresponds to the exogenous ABA concentration. ABAleon2.1 showed an apparent ABA affinity K′dN of ∼200 nM in tobacco cells (Figure 2, C and E), which was two-fold lower than that characterized in vitro (∼100 nM; Waadt et al., 2014). In comparison, the soluble ABAleon2.1_Tao3 exhibited an even lower ABA affinity, which was around 400 nM (Figure 2, C and E). Indeed, ABAleon2.1 exhibited notable increases in τmT at 100-nM ABA treatment, whereas ABAleon2.1_Tao3cyto displayed slight but not significant increases (Supplemental Figure S3), validating a higher sensitivity of ABAleon2.1 compared with that of ABAleon2.1_Tao3cyto in tobacco protoplasts. Nonetheless, when anchored, both aABAleon2.1_Tao3cyto and ABAleon2.1_Tao3ER displayed a higher ABA affinity (∼100 nM) compared to soluble ABA reporters (Figure 2, F and G). Moreover, ABAleon2.1_Tao3cyto showed a dynamic range of ∼15% (Supplemental Table S1), which is relatively larger than that of ABAleon2.1 and anchored ABAleon2.1_Tao3s (∼13%). Together, it appears that the soluble ABAleon2.1_Tao3cyto is more suitable for monitoring cells or tissues with higher ABA levels, whereas the membrane-anchored ABAleon2.1_Tao3s are more competent in situations where more subtle changes in ABA concentration occur.
ABAleon2.1_Tao3s reveal a higher ABA content in the ER in tobacco mesophyll cells and specific increases in the ER ABA upon different environmental stimuli
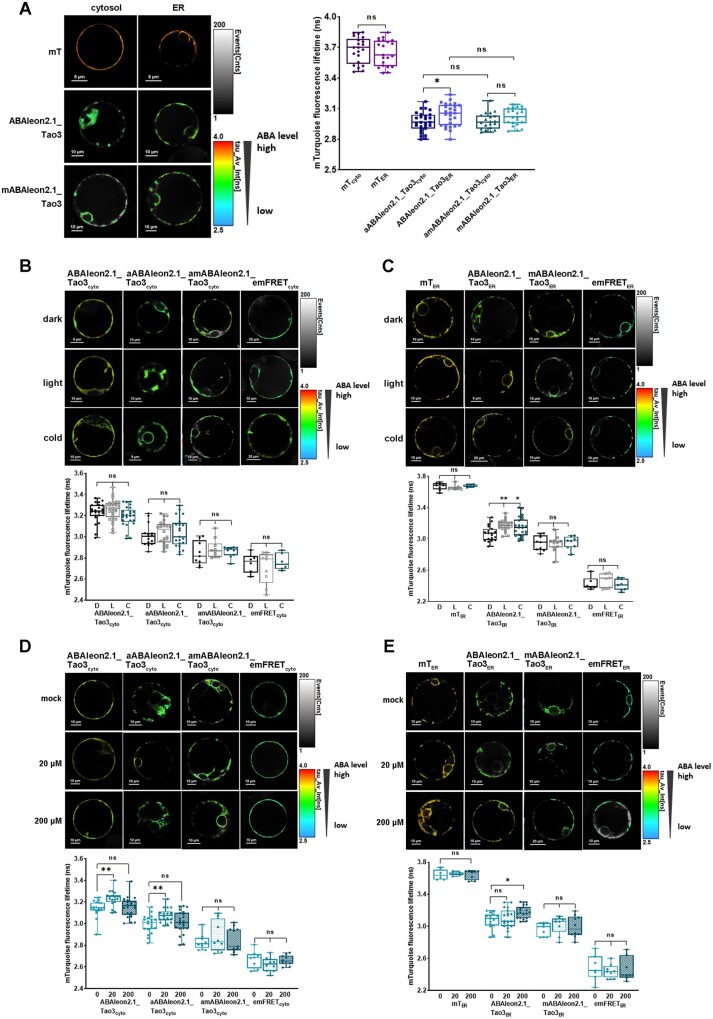
Targeting of ABAleon2.1_Tao3 to the ER membrane with the ABA sensory complex facing the cytosolic side or the ER lumen side allows direct comparison of ABA levels in the cytosol and in the ER. Whereas the ER lumen-residing, donor-only control mTER exhibited a similar level of τmT as the cytosolic donor-only (mTcyto), the ER-located sensor ABAleon2.1_Tao3ER showed notably increased τmT in comparison with the cytosolic sensor aABAleon2.1_Tao3cyto (Figure 3A). Moreover, the ER membrane-anchored, mutated ABAleon2.1_Tao3 in the cytosol (amABAleon2.1_Tao3cyto) displayed comparable levels of τmT compared to its counterpart in the ER (mABAleon2.1_Tao3ER; Figure 3A). These results together suggest a higher amount of ABA in the ER lumen than that in the cytoplasm.
Figure 3.
FRET–FLIM analysis reveals a higher basal amount of ABA in the ER and specific increases in the ER ABA upon different environmental stimuli. A, FLIM images and data showing generally lower τmT in the aABAleon2.1_Tao3cyto compared with that of ABAleon2.1_Tao3ER. The donor-only control mTcyto and ER-membrane anchored non-responsive ABAleon2.1_Tao3 in the cytosol (amABAleon2.1_Tao3cyto) displayed similar levels of τmT compared with their counterparts in the ER (mTER and mABAleon2.1_Tao3ER), respectively. FLIM values are presented as box plot with all data points. Statistical analysis was performed using Student’s t test (*P < 0.05; ns, not significant). B, C, Representative FLIM images and data show little responses of the τmT in the cytosolic ABAleon2.1_Tao3s (B) but evident increases in the τmT in ABAleon2.1_Tao3ER (C) 6 h after dark (D, 25°C), light (L, 25°C), and cold (C, 4°C, dark) treatment. The non-responsive ABAleon2.1_Tao3s (amABAleon2.1_Tao3cyto and mABAleon2.1_Tao3ER), donor-only control mTER, and empty FRET controls (emFRETcyto and emFRETER) showed similar levels of τmT under all three conditions. The fluorescence lifetime values are shown as box bars with all data points. Statistical analysis was performed using one-way ANOVA followed by Dunnett’s multiple comparisons test (**P < 0.01; *P < 0.05 for difference between different stimulus and dark control treatment; ns, not significant). D, E, Representative FLIM images and data show specific responses of the τmT in cytosolic ABAleon2.1_Tao3s (D) and in the ABAleon2.1_Tao3ER (E) at 6 h after 20-μM, 200-μM MgSO4, or mock treatment. The nonresponsive ABAleon2.1_Tao3s (amABAleon2.1_Tao3cyto and mABAleon2.1_Tao3ER), donor-only control mTER, and empty FRET controls (emFRETcyto and emFRETER) showed minor responses to both sulfate treatments. The fluorescence lifetime values are shown as box bars with all data points. The fluorescence lifetime values are shown as bars showing means ± SE. Statistics were performed using one-way ANOVA followed by Dunnett’s multiple comparisons test (**P < 0.01; *P < 0.05 for difference between sulphate and control treatment; ns, not significant)
Cellular ABA levels depend on environmental signals like light and temperature (Seo et al., 2006; Toh et al., 2008; Baron et al., 2012). To test if ABAleon2.1_Tao3s respond to light and cold, we exposed tobacco protoplasts that had been transfected with ABAleon2.1_Tao3s in the dark to light and low temperature for 6 h, respectively. Both soluble (ABAleon2.1_Tao3cyto) and ER membrane-anchored, cytosolic-residing ABAleon2.1_Tao3 (aABAleon2.1_Tao3cyto) displayed little changes in the level of τmT when transferred to light and cold (Figure 3B). In contrast, ER lumen-residing ABAleon2.1_Tao3 (ABAleon2.1_Tao3ER) exhibited notable increases in τmT in response to both light and cold (Figure 3C), suggesting the level of ABA in the ER increased upon both stimuli. The τmT of nonresponsive ABAleon2.1_Tao3 mABAleon2.1_Tao3ER, donor-only mTER, and empty FRET control emFRETER remained similar under all three conditions (Figure 3C), supporting that the tested environmental cues-induced increase in the τmT of the ABAleon2.1_Tao3ER is attributed to increases in ABA level.
It was reported that exogenous supply of sulfate enhanced the steady-state level of ABA (Cao et al., 2014; Batool et al., 2018). To see if both cytosolic and ER ABA were enhanced upon sulfate supply, we recorded the τmT of ABAleon2.1_Tao3s 6 h after exogenous application of sulfate at two concentrations. Whereas the τmT of nonresponsive amABAleon2.1_Tao3cyto and emFRETcyto changed slightly upon both sulphate treatments, both the soluble ABAleon2.1_Tao3cyto and membrane-anchored aABAleon2.1_Tao3cyto displayed notable increases in τmT at low sulphate concentration (20 μM) but exhibited no detectable changes at higher sulfate level (200 μM; Figure 3D). In contrast, the τmT of ABAleon2.1_Tao3ER remained unchanged at low sulphate level but was significantly enhanced at the higher sulfate condition (200 μM; Figure 3E), revealing the enhancement of ABA in the ER at nonlimiting sulfate supply conditions. All three ER controls including the donor-only mTER, the nonresponsive mABAleon2.1_Tao3ER, and emFRETER exhibited similar level of the τmT upon both sulphate treatments, supporting the conclusion that the responses of τmT in the ABAleon2.1_Tao3ER are ABA specific.
ABAleon2.1_Tao3s present ABA inductions in the cytosol and the ER upon osmotic stresses
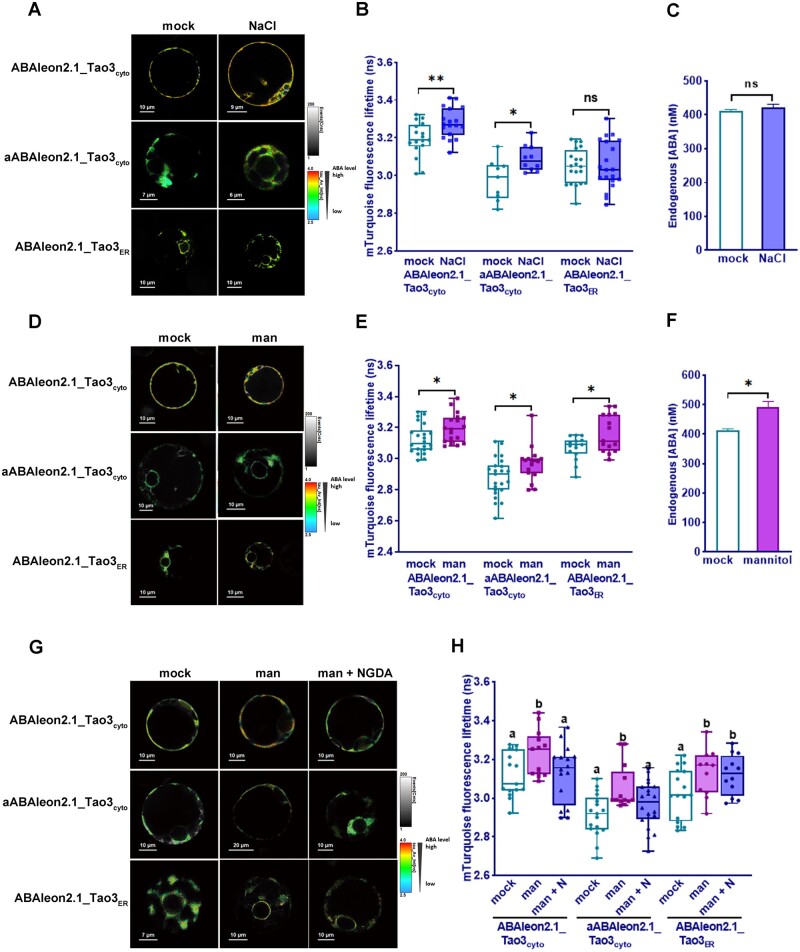
Stresses like high salinity and osmotic stress are also known to induce cellular ABA accumulation in plants through both activation of synthesis and inhibition of degradation (Zhu, 2002). We thus tested the responsiveness of ABAleon2.1_Tao3s to salt and mannitol that are often used as an osmoticum in stress assays (Fujii et al., 2011; Claeys et al., 2014). ABAleon2.1_Tao3cyto exhibited noticeable increases in τmT at 6 h after both 10-mM salt and 50-mM mannitol treatments (Figure 4, B and E), whereas no detectable changes were recorded with mTcyto, mABAleon2.1_Tao3cyto, and emFRETcyto under those conditions (Supplemental Figure S4, A and B), supporting that both osmotic stresses triggered ABA accumulation in the cytosol.
Figure 4.
Osmotic stresses induce accumulation of ABA in the cytosol and the ER in tobacco protoplasts. A–C, FLIM images (A) and data (B) of ABAleon2.1_Tao3cyto, aABAleon2.1_Tao3cyto, and ABAleon2.1_Tao3ER endogenous ABA concentrations (C) at 6 h after NaCl (10 mM) treatment. D–F, FLIM images (D) and data (E) of ABAleon2.1_Tao3cyto, aABAleon2.1_Tao3cyto, and ABAleon2.1_Tao3ER endogenous ABA concentrations (F) at 6 h after 50-mM mannitol (man) treatment. The fluorescence lifetime values are presented as box plots showing all data points. n = 15–24. Endogenous ABA values are indicated as bar plots showing means ± se. n = 9. Statistics was performed using Student’s t test (**P < 0.01; *P < 0.05; ns, not significant). G–H, FLIM images (G) and data (H) showed inhibition of NGDA on osmotic stress induced increases in cytosolic ABA but not ER ABA. Samples were recorded 6 h after treatment with 50-mM mannitol (man) or 100-μM NGDA (N). FLIM data are presented as box plots showing all data points. Statistical analysis was performed using one-way ANOVA, followed by Fisher’s LSD test (different letters indicate statistically significant differences among different treatments, P < 0.05)
More specifically, salt treatment (10 mM) triggered significant increases in the τmT of both ABAleon2.1_Tao3cyto and aABAleon2.1_Tao3cyto, whereas little changes in that of ABAleon2.1_Tao3ER were detected (Figure 4, A and B). With the overall endogenous ABA concentration increased only marginally (Figure 4C), it is likely that salt triggered ABA induction in the cytosol through the translocation of ABA from other compartments/pools. In comparison, all three ABA sensors exhibited notable increases in the τmT upon mannitol treatment (Figure 4, D and E), whereas their corresponding controls showed little changes (Supplemental Figure S4, B and C), suggesting that mannitol induced ABA accumulation in both the cytosol and the ER. Consequently, the overall endogenous ABA concentration evidently increased after mannitol treatment (Figure 4F).
To determine whether mannitol-triggered ABA inductions in the cytosol depended on de novo ABA biosynthesis pathway, we tested one of the ABA biosynthesis inhibitors, nordihydroguaiaretic acid (NDGA), that could inhibit the activity of NCED in vitro (Creelman et al., 1992; Han et al., 2004). Treatment with NDGA fully blocked mannitol-triggered ABA accumulation in the cytosol but not that in the ER (Figure 4, G and H), implying that osmotic stress-induced cytosolic ABA accumulation is mainly attributed to the ABA biosynthesis pathway.
ER-membrane targeted ABAleon2.1_Tao3s show distinct kinetics in the cytosol and the ER during early osmotic stress responses
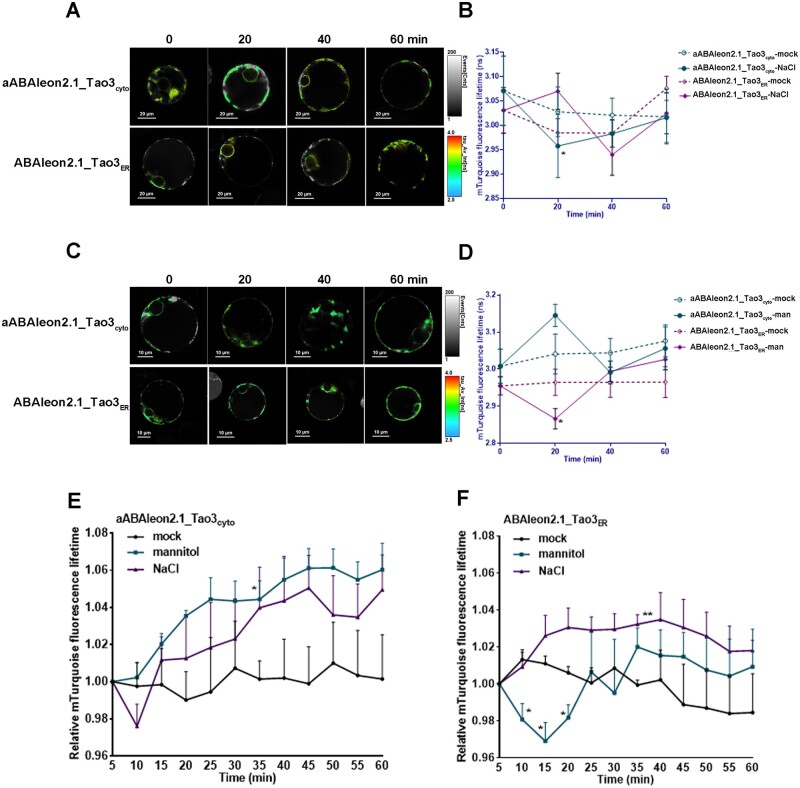
ABAleon2.1_Tao3s also allow visualization of changes in ABA accumulation in a time-resolved manner. To examine how cytosolic and ER ABA respond to osmotic stress at early stage, we tested the responses of ABAleon2.1_Tao3s within 1 h after stress application. When treated with 100-mM salt, a drastic reduction in the τmT of aABAleon2.1_Tao3cyto and a slight increase of the τmT of ABAleon2.1_Tao3ER occurred at 20 min after treatment (Figure 5, A and B), indicating a decrease and rise in the level of cytosolic ABA and ER ABA, respectively. Intriguingly, upon mannitol treatment, the τmT of aABAleon2.1_Tao3cyto rose swiftly within 20 min, followed by a drastic decrease, whereas that of ABAleon2.1_Tao3ER responded firstly with an evident reduction, then regained normal levels, and did not change much thereafter (Figure 5, C and 5D).
Figure 5.
Time-resolved changes of ER-membrane anchored ABAleon2.1_Tao3s in response to salt and osmotic stresses. A, C, Successive FLIM images of aABAleon2.1_Tao3cyto and ABAleon2.1_Tao3ER in protoplasts treated with 100-mM NaCl (A) and 200-mM mannitol (C) at the indicated time after treatments. B, D Time-resolved recording of aABAleon2.1_Tao3cyto and ABAleon2.1_Tao3ER within 1 h upon treatment with 100-mM NaCl (B) or 200-mM mannitol (D) compared with mock treatment. FLIM data are presented as means ± se. n = 6–12. Statistics were performed using Student’s t test (*P < 0.05 compared with mock treated samples). E, F, Time-course responses of aABAleon2.1_Tao3cyto (E) and ABAleon2.1_Tao3ER (F) in a single cell within 1 h upon treatment with 250-mM NaCl, 250-mM mannitol, and mock. FLIM values were recorded from 5 min until 60 min after treatment and at 5-min intervals. Shown are data normalized to FLIM values at starting point (5 min) and representative from at least three independent experiments. Significance analyses were performed using two-way ANOVA followed by Dunnett’s test for multiple comparisons (*P < 0.05, ** P < 0.01 compared with mock treatment)
To see how cytosolic and ER ABA respond to osmoticums in more detail, we monitored the ER membrane-anchored ABAleon2.1_Tao3s in a single cell continuously at 5-min intervals upon treatments. It was evident that 250-mM mannitol treatment stably increased the τmT of aABAleon2.1_Tao3cyto compared with mock treatment, whereas salt treatment first caused a decrease and then an increase of cytosolic τmT (Figure 5E). In comparison, ABAleon2.1_Tao3ER did not respond to mannitol treatment with notable and stable increases in τmT until 20 min later, whereas it showed rapid reduction of τmT within 15 min upon salt treatment and then return to normal level afterwards (Figure 5F). These results together reveal distinct dynamics of ABA in different compartments in response to different osmoticum.
ABAleon2.1_Tao3s responses in Arabidopsis plants and osmotic stress-induced ABA increases are dependent on BG1
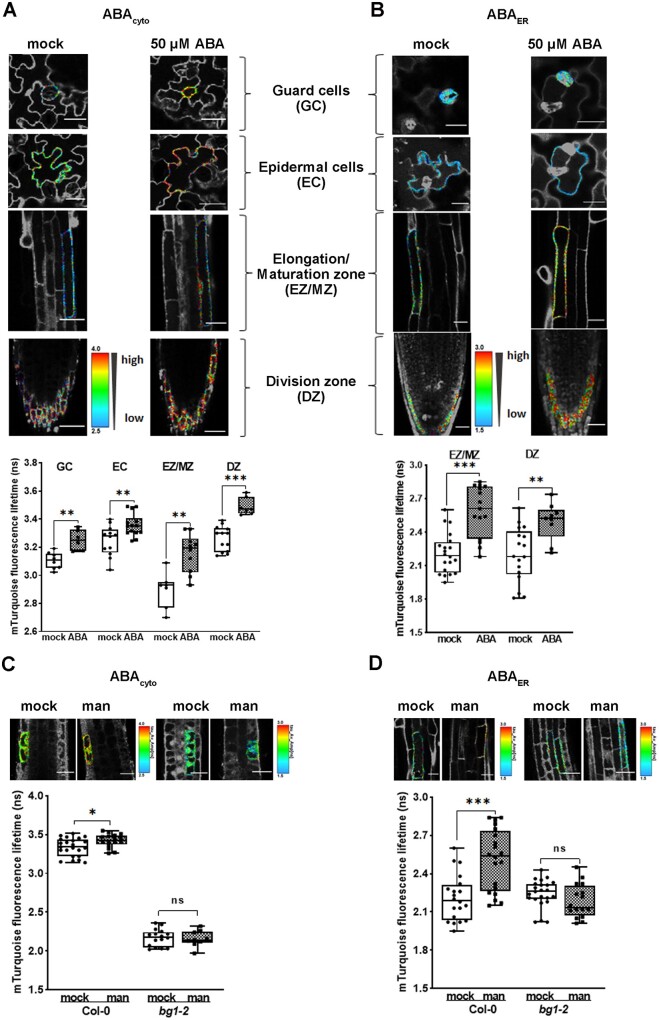
To determine whether ABAleon2.1_Tao3s respond to changes in the ABA level in planta, we transformed ABAleon2.1_Tao3cyto and ABAleon2.1_Tao3ER into the Arabidopsis Col-0 accession to monitor cytosolic (ABAcyto) and ABA in the ER (ABAER), respectively. In ABAleon2.1_Tao3cyto-transformed plants, the τmT increased evidently within 1 h upon ABA treatment (50 μM) in all tissues tested (clear color shift from blue/green to red), including the root division zone (DZ), elongation/maturation zone (EZ/MZ), and guard (GC) and epidermal cells (EC; Figure 6A), indicating substantial increases in ABAcyto. ABA in the ER also increased noticeably in the DZ and EZ/MZ, but not in the aboveground tissues (Figure 6B).
Figure 6.
ABAleon2.1_Tao3s responses in Arabidopsis seedlings upon ABA and mannitol application. A, B, FLIM images and data showing substantial inductions of both cytosolic (A) and ER ABA (B) in different Arabidopsis tissues 1 h after application of 50-μM ABA. The GC/EC data of ABAER was not included as the signals in the leaves of ABAleon2.1_Tao3ER were generally low and did not produce enough recordings for experimental analysis. The colored calibration bars indicate higher ABA concentrations with high τmT (red) and lower ABA concentrations with low τmT (blue). C, D, FLIM images and data reveal increases in ABAcyto (C) and ABAER (D) within 1 h upon 200-mM mannitol treatment in root meristem zone and elongation/maturation zone, respectively, in wild-type Col-0 but not in the root of the mutant bg1-2. FLIM data are presented as means ± se. Statistical analysis performed using Student’s t test (*P < 0.01; ***P < 0.001; ns, not significant)
Osmotic stress enhanced polymerization of BG1, which is required for its hydrolyzing activity and ABA release from the inactive ABA-GE (Lee et al., 2006). It was recently shown that BG1 is involved in ABA accumulation during early stages of dehydration stress (Han et al., 2020). To see if BG1 also plays a role in osmotic stress-triggered ABA inductions, we transformed ABAleon2.1_Tao3s into the bg1-2 mutant. ABAleon2.1_Tao3cyto in the bg1-2 mutant exhibited a generally reduced lifetime compared to that in wild-type Col-0 (Figure 6C). In the roots of wild-type Col-0 plants, mannitol treatment induced notable increases in cytosolic ABA exclusively in root meristematic zone within 1 h (Figure 6C), whereas evident inductions of ABAER occurred in the elongation and maturation zone (Figure 6D), revealing stress-induced regulation of ABA levels at both tissue and organelle levels. No visible changes were recorded in both ABAcyto and ABAER in the mutant bg1-2, suggesting a requirement of BG1 for ABA inductions in Arabidopsis roots during an early stage of osmotic stress.
Discussion
The development of FRET-based bioprobes for direct monitoring of small molecules has exceedingly facilitated our knowledge in cell signaling. Owing to the limited targeting strategies, monitoring of signal molecules at a subcellular level remains challenging. Despite that GFP engineered as a fusion to the N-terminal ER signal peptide and a C-terminal HDEL was adequately accumulated in the ER in diverse plant species (Gnanasambandam et al., 2007), similar efficiency for ER retention of HDEL-fused FRET-based sensors cannot be guaranteed considering their generally large sizes and complex structures. Besides, the overexpression of sensors using the strong CaMV35S or ubiquitin promoter often causes HDEL saturation and possible leakage of sensor proteins to post-Golgi compartments or secretion (Silva-Alvim et al., 2018). To circumvent this, we have devised a Nb-based targeting strategy that involves dual expression of sensor and anchor proteins driven by the CaMV35S promoter, allowing ABAleon2.1_Tao3 that was modified from ABAleon2.1 (Waadt et al., 2014) to be targeted along the ER membrane with fluorescent probes free of sorting signals or attachments to endomembranes. This is particularly favorable for FRET sensors, for which consistent conformation is essential for their downstream applications.
Characterization of ABAleon2.1_Tao3 in vitro (in artificial buffers) cannot reflect the real situation here as when targeted to different organelles, ABAleon2.1_Tao3 is subject to different microenvironments that may affect its innate capacities. For instance, the redox status in the ER lumen is different from that in the cytosol, with a more oxidizing environment within the ER lumen (Braakman et al., 1992; Wan and Jiang, 2016). Thus, the cytosolic- and ER lumen-residing ABAleon2.1_Tao3s were characterized respectively in vivo using tobacco protoplasts. Soluble ABAleon2.1_Tao3 and ABAleon2.1 were also included for comparison. ABAleon2.1_Tao3cyto showed evidently lower sensitivity to ABA compared with ABAleon2.1 (Supplemental Figure S3), suggesting that the swap of the GS linker for a SYN epitope-tagged linker affects ABAleon2.1_Tao3’s flexibility, thus compromising its affinity for ABA. Nonetheless, the ER membrane-anchored ABAleon2.1_Tao3s displayed high sensitivities to ABA, with the apparent ABA affinities of ∼100 nM, corresponding to endogenous ABA concentrations of 17–19 fg per cell (Supplemental Table S1). Cellular ABA concentration was calculated to be around 1–5 fg per cell in unstressed cells (Harris et al., 1988; Supplemental Table S2), and it can be induced up to 6- to 30-fold upon water deficit and salt stresses (Harris et al., 1988; Geng et al., 2013). Inferring from these data, ABAleon2.1_Tao3s are suitable for monitoring ABA levels in mild stress situations.
Anchoring of the soluble ABAleon2.1_Tao3cyto to the ER membrane revealed evidently reduced τmT compared with ABAleon2.1_Tao3ER (Figure 3A). As both the donor-only controls and mutated ABAleon2.1_Tao3s in the cytosol showed similar levels of τmT to those residing in the ER (Figure 3A), it is unlikely that the differing environment, such as the slightly lower environmental pH in the ER (Martiniere et al., 2013; Shen et al., 2013; Schmitt et al., 2014), caused the differences in the τmT of aABAleon2.1_Tao3cyto and ABAleon2.1_Tao3ER. Thus, in contrast to the pH-driven ABA distribution mode, which predicts more accumulation of ABA in a more alkaline microenvironment (Hartung and Slovik, 1991; Boursiac et al., 2013), that is, more ABA in the cytosol, our results instead point to a higher amount of ABA in the ER than in the cytosol. Considering the direct interconnection between the ER and the nucleus (Schwarz and Blower, 2016), it should not be excluded that a certain amount of ER ABA that had come from the nucleus via diffusion through the nucleus pores was sequestered by ABAleon2.1_Tao3ER, resulting in a higher τmT of ABAleon2.1_Tao3 in the ER than in the cytosol.
Environmental signals including light and temperature are essential for seed germination and seedling growth, partly through modulating plant hormonal pathways (Chen. et al., 2008; Lau and Deng, 2010). Light-regulated ABA metabolism in vegetative tissues has been shown by diurnal fluctuations of ABA levels (Adams et al., 2018). In our environmental stimuli assays, 6-h exposure to light triggered minor changes in cytosolic ABA but remarkable inductions of ABA in the ER (Figure 3, B and C). Whereas it is difficult to assess how ABA level changes in the cytosol considering the activation of both biosynthetic and catabolic enzymes upon light exposure (Seung et al., 2012), the notable increases in ER ABA could be attributed to the enhanced hydrolyzing activity of the ER-residing BG1 when plants were shifted from dark to light (Lee et al., 2006). Similarly, cold triggered little changes in cytosolic ABA and substantial increases in ER ABA (Figure 3, B and C).
It has been reported that sulfate also increases cytosolic ABA in guard cells using ABAleon2.1 in a concentration-dependent manner (from 2 mM to 15 mM; Batool et al., 2018). Sulfate response assays with our cytosolic ABAleon2.1_Tao3s, including both soluble and membrane-anchored sensors, revealed also considerable enhancement of ABA in the cytosol upon low (20 µM) sulfate application, though no profound changes were observed at higher sulfate condition (200 µM; Figure 3, D and E). In contrast, ER lumen-residing ABAleon2.1_Tao3ER showed substantially increased τmT only at high sulfate level (Figure 3, D and E). These observations suggest that ABA biosynthesis pathway in the cytosol and ABA hydrolysis pathway in the ER played major roles for cellular ABA accumulation at limit and nonlimit sulfate supply conditions, respectively. It is also possible that cytosolic ABA is transported and stored in the ER at high-sulfate conditions. Moreover, despite that both salt- and mannitol-triggered osmotic stress increased the ABA level in the cytosol, inductions of ABA in the ER occurred only upon mannitol treatment (Figure 4). These findings together support that the ER, like the cytosol, acts as an indispensable source for environmental stimulus-dependent regulation of ABA homeostasis at the subcellular level.
Real-time analysis of ABAleon2.1_Tao3s revealed that cytosolic and ER ABA responded to osmotic stresses with distinct kinetics (Figure 5). The notable reductions in cytosolic ABA observed at 20 min after salt treatment could be explained by the fact that large amounts of ABA were sequestered by its native receptors to activate downstream signaling pathways or secreted to the outside of the cells after initial salt stress. Besides, the rapid and distinct ABA kinetic patterns in the cytosol and the ER could have been a result of a constant exchange of ABA between these two compartments. Other ABA pools like the chloroplast and vacuole (Slovik and Hartung, 1992; Xu et al., 2012) might also play a part. Considering the slightly alkaline microenvironment in the cytosol and the ER (Hartung and Slovik, 1991; Shen et al., 2013), where most ABA molecules exist as nondiffusible anions and require active transporters for transmembrane movement, it is possible that multispecific transporters of ABA and its conjugates exist at the ER membrane and mediate the stimulus-specific ABA translocations between the cytosol and ER.
During transgenic plants screening, it was noticed that ABAleon2.1_Tao3ER-expressing wild-type plants and ABAleon2.1_Tao3cyto- and ABAleon2.1_Tao3ER-expressing bg1-2 mutant plants displayed generally low fluorescent signals, which could be attributed to partial silencing of FRET sensors (Jones et al., 2014). Still, we were able to detect changes in ABA level in those plants with FLIM measurements, which are extremely sensitive and independent on sensor expression levels. In Arabidopsis plants, ABAleon2.1_Tao3s enabled the visualization of inductions of cytosolic ABA in all tissues, whereas ABA induction in the ER was reported most evidently in root tissues (Figure 6, A and B). Previous results have shown that de novo ABA synthesis induced upon dehydration stress occurred in the leaves of Arabidopsis plants, which required root-to-shoot transport of the small peptide CLE25 (Takahashi et al., 2018). It could be that shoot-derived ABA is stored in the ER in the roots, allowing more rapid shoot-independent ABA responses in the roots of Arabidopsis plants once exposed to stresses. It was noticed that the levels of τmT in the cytosol of bg1-2 plants were remarkably lower than those in wild-type Col-0 (Figure 6C), indicating a generally lower level of ABA in the cytosol of bg1-2 plants. Interestingly, within 1 h of osmotic stress, ABAleon2.1_Tao3s revealed substantial ABA accumulation in the cytosol of root meristematic cells and in the ER of root cells in the elongation/maturation zone, both of which were abolished in the bg1-2 mutant (Figure 6, C and D), supporting the indispensable role of BG1 in rapid accumulation of ABA upon water stress (Lee et al., 2006). As the starting point of the secretion pathway, the ER controls protein synthesis and secretion. It is possible that the ER modulates the secretion of small peptides like CLE25 through BG1-dependent ABA in the ER, leading to root-to-shoot transmission of stress signals that ultimately triggers de novo ABA synthesis in the leaves (Takahashi et al., 2018).
Together, our ABAleon2.1_Tao3s not only present a viable toolkit for effective targeting of FRET sensors to subcellular organelles, but also allow for the first time a direct linkage of environmental stimuli with organellar-specific ABA perturbations in tobacco mesophyll cells and in different tissues of Arabidopsis roots. Further studies require the targeting of sensor proteins to other organelles for more comprehensive understanding of environmental stress-triggered ABA redistribution and movement at subcellular levels.
Material and methods
Plant materials and growth conditions
Tobacco (N. tabacum L.) SR1 was grown on Murashige and Skoog (MS) medium supplemented with 2% (w/v) sucrose, 0.5-g L−1 MES, and 0.8% (w/v) agar at pH 5.7 in 16-/8-h light–dark cycles at 22°C.
Plasmid construction
All constructs used in this study were generated according to established standard procedures such as fragment clone, single or double digestion, ligation, and transformation in Escherichia coli DH5α or MC1061. The new fragments/constructs, primers, and clone information are listed in Supplemental Table S3. Unless otherwise indicated, all the constructs start with the CaMV35S promoter and end with a NOS terminator. The ER and cytosolic sensor constructs carrying the point mutation in the ABA receptor PYR1 (R116G) were generated using a seamless cloning kit (TSINGKE, Trelief™ SoSoo Cloning Kit, TSV-S1). Plasmids used for tobacco protoplast transfection were extracted using a plasmid extraction kit (OMEGA bio-tek).
Protoplast isolation and transfection
Tobacco protoplasts were isolated and electrotransfected as previously described (Fruholz et al., 2018). Briefly, protoplasts were prepared from perforated tobacco leaves by overnight incubation in the buffer (3.05-g L−1 Gamborg B5 salt medium, 500-mg L−1 MES, 750-mg L−1 CaCl2·2H2O, 250-mg L−1 NH4NO3 adjusted to pH 5.7 with KOH, supplemented with 0.2% w/v macerozyme and 0.4% w/v cellulase) at 25°C in the dark. They were rebuffered through washing three times in 40-mL electrotransfection buffer (137-g L−1 sucrose, 2.4-g L−1 HEPES, 6-g L−1 KCl, 600-mg L−1 CaCl2·2H2O adjusted to pH 7.2 with KOH). Five hundred microliters protoplasts in a total volume of 600-μL electrotransfection buffer were electrotransfected with 2–6-μg plasmid DNA using the Gene Pulser XcellTM (Bio-Rad) with a pulse at 160 V for 10 ms. After transfection, each sample was supplemented with 2-mL incubation buffer and incubated for 16–24 h at 25°C in the dark.
Harvesting, protein extraction, and immunoblotting
Protoplasts were harvested and proteins were extracted as described (Kunzl et al., 2016). Briefly, protoplasts were collected by floating cells in sealed pre-punctured tubes for 10 min at 80g, and cell-free medium was extracted using syringes. Protein samples in the medium were precipitated with saturated NH4SO4 and 10-mg mL−1 BSA. After sedimentation by centrifugation at 16,000g for 10 min at 4°C, protein pellets were resuspended in 30-μL extraction buffer [100-mM Tris, 200-mM NaCl, 1-mM EDTA, pH 7.8, 2% (v/v) β-mercaptoethanol, 0.2% (v/v) Triton X-100]. After resealing, cells were diluted with 250-mM NaCl and centrifuged as above. The supernatants were removed, and cell samples were extracted 1:1 in extraction buffer by sonication (20 W, 2–3 s for each sample).
For sodium dodecyl sulfate–polyacrylamide gel electrophoresis/western blotting (SDS–PAGE/WB), all processed samples were finally mixed 1:1 with 2× Xtreme loading dye (900 μL of sample buffer (0.1% w/v bromophenol blue, 5-mM EDTA, 200-mM Tris–HCl, pH 8.8, 1-M sucrose) supplemented with 300-μL 10% w/v SDS and 20 μL of 1-M DTT) and denatured for 5 min at 95°C. The antibodies used were rabbit polyclonal anti-GFP (MBL, 1:5,000), rat monoclonal anti-HA–Peroxidase (Roche, 1:2,000).
Confocal laser scanning microscopic analysis
Imaging was performed using Nikon A1plus confocal laser scanning microscope, with a × 40 (1.15 numerical aperture) water immersion objective. The sensor proteins and RFP markers were excited with 488 nm and 561 nm, emission at the range of 500–550 nm and 570–620 nm was detected, respectively. Pinholes were adjusted to 1 Airy unit for each wavelength. Post-acquisition image processing, such as channel merge (Figure 1, C and F;Supplemental Figure S1), addition of scale bars (Figure 1, C and F;Supplemental Figure S1), and signal plotting (Supplemental Figure S1), was performed with ImageJ. The linear Pearson’s correlation coefficient (rP) and the nonlinear Spearman’s rank coefficient (rS) of green and red fluorescent signals were calculated with the PSC colocalization plug-in (http://www.cpib.ac.uk/tools-resources/software/psc-colocalization-plugin/) in ImageJ. The threshold levels were set to 10.
Immunoprecipitation assays
For IP assays, cells that had been cotransfected with ER anchors and ABA sensors were collected as above and mixed 1:1 with 2× binding buffer (40-mM HEPES, 300-mM NaCl, 2-mM CaCl2, 2-mM MgCl2, pH 7.1) and 2% w/v CHAPS. Protease inhibitor cocktail tablets (Roche) were added as instructed. Immunoprecipitations were performed either with anti-HA magnetic beads (Roche) or with anti-GFP mAb-magnetic beads (MBL), overnight at 4°C. Beads were washed five times with binding buffer containing 0.4% w/v CHAPS, and then eluted with 50-μL 1-M glycine (pH 3.0). SDS–PAGE/WB was performed as described above.
Fluorescence lifetime imaging microscopy
FLIM recordings were performed at a Nikon A1R equipped with a PicoHarp time-correlated single-photon counting (TCSPC) module and a PDL800-D multichannel picosecond pulsed diode laser driver. The donor τmT was excited with a 440-nm laser at 20-MHz pulse frequency. Emission was recorded at 482/35 nm by TCSPC until reaching a count of at least 300 photons in the brightest pixel (total count of at least 10,000 photons). At least 10 cells per sample per treatment were recorded. For time-resolved single cell recordings (Figure 6, E and F), cells were mixed gently and mounted to the slides immediately after treatment. FLIM recording started at 5 min after treatment and at 5-min intervals.
FLIM data were analyzed using SymphoTime64 v2.0 (PicoQuant). Values were collected based on the specifically selected fluorescence signals in each cell using the software’s “region of interest” (ROI) tools to avoid background noise. To calculate fluorescence lifetimes of the donor τmT, TCSPC histograms were reconvoluted with an instrumental response function and fitted against a mono- (in the absence of the acceptor) or multiexponential (in the presence of the acceptor, n = 2) decay model. Only fittings giving Chi-squared values between 0.9 and 1.5 were considered.
Stress and chemical assays using tobacco protoplasts
Protoplasts that had been electrotransfected with the same plasmids were collected as described above. The floating cells were transferred to a new tube to remove cell debris and washed twice.
For stress assays, protoplasts were resuspended in a final 2 mL of incubation buffer and aliquoted into 0.5 mL per tube. All the tubes were separated into three groups: one was wrapped in aluminum foil (for dark control treatment), one was exposed to the light (for light treatment), and one was wrapped in aluminum foil and incubated at 4°C (for cold treatment).
For the exogenous chemical assays, protoplasts were aliquoted into 0.9 mL per tube. Stock solutions of ABA (1 mM, 1 μM), MgSO4 (100 mM), mannitol (500 mM), and NaCl (5 M) were prepared in incubation buffer. For ABA tier assay, 0.1 mL of incubation buffer containing 0, 1 μL 1 μM, 10 μL 1 μM, 50 μL 1 μM, 100 μL 1 μM, 0.5 μL 1 mM, 1 μL 1 mM, 5 μL 1 mM, 10 μL 1 mM, or 10 μL 10 mM of ABA was added to each protoplast aliquot, respectively, making the final concentration of ABA at 0, 1, 10, 50, 100, 500, 1,000, 5,000, 10,000, or 100,000 nM. For exogenous sulfate supply, 0.1 mL of incubation buffer containing 0.2-μL and 2-μL 100-mM MgSO4 was added to each protoplast aliquot, making the final concentration of sulfate at 20 μM and 200 μM, respectively. Similarly, for osmotic stress assays, 0.1 mL incubation buffer containing certain amounts of salt or mannitol was added to each protoplast aliquot. For assays with NGDA, a stock solution of 100-mM NGDA was prepared in DMSO. Six hours after treatments, 20 μL and 200 μL from each aliquot were taken for subsequent cell counting (TC20TM, BIO-RAD) and FLIM recording, respectively. The remains of each protoplast aliquot were harvested for ABA quantification by liquid chromatography–tandem mass spectrometry (LC–MS/MS).
ABA quantification by LC–MS/MS
After removal of medium buffer by syringes, protoplasts with stress treatments were sedimented and homogenized in 400 μL of cold extraction buffer (methanol: water: acetic acid= 80: 19: 1) spiked with 20 ng mL−1 of the internal standard ABA-d6 (Toronto Research Chemicals) by whirling at 80 rpm, 4°C (in dark), overnight. For quantifying cellular ABA levels in a single cell, the total live number of protoplasts was counted using the trypan blue staining method (TC20TM, BIO-RAD) before harvest. The homogenates were cleared by centrifugation at 15,000g for 20 min at 4°C. Supernatants were transferred to new tubes and dried under a constant nitrogen flow and a final 200 μL of cold extraction buffer was added to each sample before subjected to LC–MS.
Samples were analyzed in negative full scan mode using liquid chromatography coupled to hybrid quadrupole-Orbitrap mass spectrometer (A Thermo Scientific™ Q Exactive™). Thermo Hypersil GOLD C18 column (2.1 × 100 mm, 1.9 μm) was used to separate 5-μL samples with phase A (0.05% acetic acid in H2O) and phase B (0.05% acetic acid in MeCN) as mobile phases, and the flow rate was 0.3 mL min−1. The gradient was as follows: phase B was increased to 90% in 6 min from 10% at initial, then held for 1 min, it was returned to 10%, and held for 3 min. The capillary temperature of the HESI source was 320°C, and the spray voltages were both 3 kV in negative mode. The sheath gas and aux gas were 30 psi and 10 psi, respectively. The scan range was set to be 200–500 m/z. The resolution was 70,000, the AGC target was 3E6, and the maximum IT was 200 ms. Compounds were detected in the electrospray ionization negative mode.
The ratio of ion intensities of ABA and its internal standard ABA-d6 was used to quantify endogenous ABA. Data were analyzed using Xcalibur 2.2 software (Thermo Scientific™). Endogenous ABA was calculated from the peak area values as Peak areaABA/Peak areaABA-d6 × 40 ng mL−1. A series of external standard solutions (from 3.9 ng mL−1 to 500 ng mL−1) spiked with 20 ng mL−1 ABA-d6 were prepared to yield a standard curve to confirm the concentration of ABA stock solutions used in the study.
Transformation of Arabidopsis plants with ABAleon2.1_Tao3s and stress treatments
ABAleon2.1_Tao3cyto and ABAleon2.1_Tao3ER were transformed into Arabidopsis Col-0 and the mutant bg1-2 (SALK_122533) by the floral dip method (Clough and Bent, 1998). Transformants were selected on 1/2 MS medium supplemented with 50-μg·mL−1 kanamycin. Positive lines were further selected by fluorescence signals under the Nikon A1plus confocal microscope. The mutant bg1-2 was further validated using the left primer 5′-TTACGACAACGGAAAGAAAGC-3′, right primer 5′-CATTCTCGGCAATGATAACTTCT-3′, and LBb1.3 5′-ATTTTGCCGATTTCGGAAC-3′ before subsequent assays.
For ABA and mannitol treatments, T2 seeds were sown on 1/2 MS medium after sterilization with 1.5% (v/v) sodium hypochlorite. Five-day-old seedlings were transferred to a 12-well plate containing 1.6 mL 1/2 MS liquid medium plus kanamycin in each well for 48 h. About 0.4 mL water containing ABA or mannitol was added to each well, making a final concentration of ABA and mannitol at 50 μM and 200 mM, respectively. About 0.4 mL water was added in parallel as mock treatment. Seedlings were subjected to FLIM recording 1 h later. At least three seedlings were recorded in each line for each treatment, and at least three independent lines per transformation were used.
Statistical analysis
In general, data are represented by box plots with all data points. The numbers of biological repeats (n) are provided in each Figure legend. Statistical analysis was performed using GraphPad Prism 8.0.2.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers XXX.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. The secretion and retainment of sABAleon2.1_Tao3 in the ER.
Supplemental Figure S2. Cytosolic ABAleon2.1_Tao3 shows ABA-induced specific increases in τmT.
Supplemental Figure S3. ABAleon2.1_Tao3cyto shows a lower sensitivity of ABA compared with ABAleon2.1.
Supplemental Figure S4. Osmotic stresses triggered specific increases in theτmT of ABAleon2.1_Tao3s but not in that of donor-only controls (mTcyto and mTER), mutated ABAleon2.1_Tao3s (mABAleon2.1_Tao3cyto and mABAleon2.1_Tao3ER), empty FRET controls (emFRETcyto and emFRETER).
Supplemental Table S1. Biochemical properties of ABA probes used in this study.
Supplemental Table S2. Basal level of endogenous ABA in individual cells.
Supplemental Table S3. Oligonucleotides and plasmids used in this study.
Supplementary Material
Acknowledgments
We thank Dr Rainer Waadt (The Centre for Organismal Studies (COS) Heidelberg) for kindly providing the ABAleon constructs. We are deeply grateful for Dr Peter Pimpl for technical support on the targeting part and on manuscript revisions. We thank Dr Yilin Wang for technical help on FRET–FLIM.
Funding
The financial support from the grant from National Natural Science Foundation of China (32070292), Shenzhen government for fundamental researches (JCYJ20170817104523456), Shenzhen Science and Technology Program (Grant No. KQTD20190929173906742), and the grant from the Key Laboratory of Molecular Design for Plant Cell Factory of Guangdong Higher Education Institutes (2019KSYS006) are gratefully acknowledged.
Conflict of interest statement. None declared.
Y. Z. and J. Liang conceived and designed the experiments. Y. Z., Y. W., and J. Li performed the experiments. Y. Z. and J. Liang analyzed the data and wrote the manuscript. All authors reviewed and approved the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructionss) is: Jiansheng Liang (liangjs@sustech.edu.cn).
References
- Adams S, Grundy J, Veflingstad SR, Dyer NP, Hannah MA, Ott S, Carre IA (2018) Circadian control of abscisic acid biosynthesis and signalling pathways revealed by genome-wide analysis of LHY binding targets. New Phytol 220:893–907 [DOI] [PubMed] [Google Scholar]
- Baron KN, Schroeder DF, Stasolla C (2012) Transcriptional response of abscisic acid (ABA) metabolism and transport to cold and heat stress applied at the reproductive stage of development in Arabidopsis thaliana. Plant Sci 188–189:48–59 [DOI] [PubMed] [Google Scholar]
- Batool S, Uslu VV, Rajab H, Ahmad N, Waadt R, Geiger D, Malagoli M, Xiang CB, Hedrich R, Rennenberg H, et al. (2018) Sulfate is incorporated into cysteine to trigger ABA production and stomatal closure. Plant Cell 30:2973–2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonza MC, Loro G, Behera S, Wong A, Kudla J, Costa A (2013) Analyses of Ca2+ accumulation and dynamics in the endoplasmic reticulum of Arabidopsis root cells using a genetically encoded Cameleon sensor. Plant Physiol 163:1230–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursiac Y, Leran S, Corratge-Faillie C, Gojon A, Krouk G, Lacombe B (2013) ABA transport and transporters. Trends Plant Sci 18:325–333 [DOI] [PubMed] [Google Scholar]
- Braakman I, Henlenius J, Henlenius A (1992) Manipulating disulfide bond formation and protein folding in the ER. EMBO J 11:1717–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao MJ, Wang Z, Zhao Q, Mao JL, Speiser A, Wirtz M, Hell R, Zhu JK, Xiang CB (2014) Sulfate availability affects ABA levels and germination response to ABA and salt stress in Arabidopsis thaliana. Plant J 77:604–615 [DOI] [PubMed] [Google Scholar]
- Chen H, Ahsan SS, Santiago-Berrios MB, Abruna HD, Webb WW (2010) Mechanisms of quenching of Alexa fluorophores by natural amino acids. J Am Chem Soc 132:7244–7245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zhang J, Neff MM, Hong S-W, Zhang H, Deng X-W, Xiong L (2008) Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc Natl Acad Sci USA 105:4495–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeys H, Van Landeghem S, Dubois M, Maleux K, Inze D (2014) What is stress? Dose-response effects in commonly used in vitro stress assays. Plant Physiol 165:519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743 [DOI] [PubMed] [Google Scholar]
- Creelman RA, Bell E, MJ E. (1992) Involvement of a lipoxygenase-like enzyme in abscisic acid biosynthesis. Plant Physiol 99:1258–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denecke J, Botterman J, Deblaere R (1990) Protein secretion in plant cells can occur via a default pathway. Plant Cell 2:51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong T, Xu ZY, Park Y, Kim DH, Lee Y, Hwang I (2014) Abscisic acid uridine diphosphate glucosyltransferases play a crucial role in abscisic acid homeostasis in Arabidopsis. Plant Physiol 165:277–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doose S, Neuweiler H, Sauer M (2009) Fluorescence quenching by photoinduced electron transfer: a reporter for conformational dynamics of macromolecules. Chemphyschem 10:1389–1398 [DOI] [PubMed] [Google Scholar]
- Finkelstein R (2013) Abscisic Acid synthesis and response. Arabidopsis Book 11:e0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruholz S, Fassler F, Kolukisaoglu U, Pimpl P (2018) Nanobody-triggered lockdown of VSRs reveals ligand reloading in the Golgi. Nat Commun 9:643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Früholz S, Pimpl P (2017) Analysis of nanobody–epitope interactions in living cells via quantitative protein transport assays. InJiang L, ed, Plant Protein Secretion: Methods and Protocols. Springer New York, New York, pp 171–182 [DOI] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu JK (2011) Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc Natl Acad Sci USA 108:1717–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y, Wu R, Wee CW, Xie F, Wei X, Chan PM, Tham C, Duan L, Dinneny JR (2013) A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell 25:2132–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanasambandam A, Polkinghorne IG, Birch RG (2007) Heterologous signals allow efficient targeting of a nuclear-encoded fusion protein to plastids and endoplasmic reticulum in diverse plant species. Plant Biotechnol J 5:290–296 [DOI] [PubMed] [Google Scholar]
- Gomord V, Denmat L-A, Fitchette-Laine A-C, Satiat-Jeunemaitre B, Hawes C, Loic F (1997) The C-terminal HDEL sequence is sufficient for retenstion of secretory proteins in the endoplasmic reticulum but promotes vacuolar targeting of proteins that escape the ER. Plant J 11:313–325 [DOI] [PubMed] [Google Scholar]
- Han SY, Kitahata N, Sekimata K, Saito T, Kobayashi M, Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K, Yoshida S, Asami T (2004) A novel inhibitor of 9-cis-epoxycarotenoid dioxygenase in abscisic acid biosynthesis in higher plants. Plant Physiol 135:1574–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Watanabe S, Shimada H, Sakamoto A (2020) Dynamics of the leaf endoplasmic reticulum modulate beta-glucosidase-mediated stress-activated ABA production from its glucosyl ester. J Exp Bot 71:2058–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MJ, William H, Outlaw J, Mertens R, Weiler EW (1988) Water-stress-induced changes in the abscisic acid content of guard cells and other cells of Vicia faba L. leaves as determined by enzyme-amplified immunoassay. Proc Natl Acad Sci USA 85:2584–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung W, Slovik S (1991) Physicochemical properties of plant growth regulators and plant tissues determine their distribution and redistribution stomatal regulation by abscisic acid in leaves. New Phytol 119:361–382 [Google Scholar]
- Iljina M, Hong L, Horrocks MH, Ludtmann MH, Choi ML, Hughes CD, Ruggeri FS, Guilliams T, Buell AK, Lee JE, et al. (2017) Nanobodies raised against monomeric a-synuclein inhibit fibril formation and destabilize toxic oligomeric species. BMC Biol 15:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwano M, Entani T, Shiba H, Kakita M, Nagai T, Mizuno H, Miyawaki A, Shoji T, Kubo K, Isogai A, et al. (2009) Fine-tuning of the cytoplasmic Ca2+ concentration is essential for pollen tube growth. Plant Physiol 150:1322–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Danielson JA, Manojkumar SN, Lanquar V, Grossmann G, Frommer WB (2014) Abscisic acid dynamics in roots detected with genetically encoded FRET sensors. Elife 3:e01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Hwang JU, Lee M, Kim YY, Assmann SM, Martinoia E, Lee Y (2010) PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc Natl Acad Sci USA 107:2355–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Yim S, Choi H, Kim A, Lee KP, Lopez-Molina L, Martinoia E, Lee Y (2015) Abscisic acid transporters cooperate to control seed germination. Nature Commun 6:8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Hanada A, Chiba Y, Ichikawa T, Nakazawa M, Matsui M, Koshiba T, Kamiya Y, Seo M (2012) Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc Natl Acad Sci USA 109:9653–9658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs M, Held K, Binder A, Hashimoto K, Den Herder G, Parniske M, Kudla J, Schumacher K (2012) FRET-based genetically encoded sensors allow high-resolution live cell imaging of Ca2+ dynamics. Plant J 69:181–192 [DOI] [PubMed] [Google Scholar]
- Kunzl F, Fruholz S, Fassler F, Li B, Pimpl P (2016) Receptor-mediated sorting of soluble vacuolar proteins ends at the trans-Golgi network/early endosome. Nat Plants 2:16017. [DOI] [PubMed] [Google Scholar]
- Kuromori T, Miyaji T, Yabuuchi H, Shimizu H, Sugimoto E, Kamiya A, Moriyama Y, Shinozaki K (2010) ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc Natl Acad Sci USA 107:2351–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Deng XW (2010) Plant hormone signaling lightens up: integrators of light and hormones. Curr Opin Plant Biol 13:571–577 [DOI] [PubMed] [Google Scholar]
- Lee KH, Piao HL, Kim HY, Choi SM, Jiang F, Hartung W, Hwang I, Kwak JM, Lee IJ, Hwang I (2006) Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126:1109–1120 [DOI] [PubMed] [Google Scholar]
- Liu Z, Yan JP, Li DK, Luo Q, Yan Q, Liu ZB, Ye LM, Wang JM, Li XF, Yang Y (2015) UDP-glucosyltransferase71c5, a major glucosyltransferase, mediates abscisic acid homeostasis in Arabidopsis. Plant Physiol 167:1659–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loro G, Drago I, Pozzan T, Schiavo FL, Zottini M, Costa A (2012) Targeting of Cameleons to various subcellular compartments reveals a strict cytoplasmic/mitochondrial Ca2+ handling relationship in plant cells. Plant J 71:1–13 [DOI] [PubMed] [Google Scholar]
- Loro G, Wagner S, Doccula FG, Behera S, Weinl S, Kudla J, Schwarzlander M, Costa A, Zottini M (2016) Chloroplast-specific in vivo Ca2+ imaging using yellow Cameleon fluorescent protein sensors reveals organelle-autonomous Ca2+ signatures in the stroma. Plant Physiol 171:2317–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiniere A, Bassil E, Jublanc E, Alcon C, Reguera M, Sentenac H, Blumwald E, Paris N (2013) In vivo intracellular pH measurements in tobacco and Arabidopsis reveal an unexpected pH gradient in the endomembrane system. Plant Cell 25:4028–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehl A, Amari K, Gereige D, Brandner K, Mely Y, Heinlein M (2012) Control of Tobacco mosaic virus movement protein fate by CELL-DIVISION-CYCLE protein48. Plant Physiol 160:2093–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED (2009) Structural mechanism of ABA binding and signaling by dimeric PYR1. Science 326:1373–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumoto S, Jones A, Frommer WB (2012) Quantitative imaging with fluorescent biosensors. Annu Rev Plant Biol 63:663–706 [DOI] [PubMed] [Google Scholar]
- Phillipson BA, Pimpl P, daSilva LL, Crofts AJ, Taylor JP, Movafeghi A, Robinson DG, Denecke J (2001) Secretory bulk flow of soluble proteins is COPII dependent. Plant Cell 13:2005–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, Hirai N, Matsumoto C, Ohigashi H, Ohta D, Sakata K, Mizutani M (2004) Arabidopsis CYP707As encode (+)-abscisic acid 8'-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol 134:1439–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt FJ, Thaa B, Junghans C, Vitali M, Veit M, Friedrich T (2014) eGFP-pHsens as a highly sensitive fluorophore for cellular pH determination by fluorescence lifetime imaging microscopy (FLIM). Biochim Biophys Acta 1837:1581–1593 [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Blower MD (2016) The endoplasmic reticulum: structure, function and response to cellular signaling. Cell Mol Life Sci 73:79–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Hanada A, Kuwahara A, Endo A, Okamoto M, Yamauchi Y, North H, Marion-Poll A, Sun TP, Koshiba T, et al. (2006) Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J 48:354–366 [DOI] [PubMed] [Google Scholar]
- Seung D, Risopatron JP, Jones BJ, Marc J (2012) Circadian clock-dependent gating in ABA signalling networks. Protoplasma 249:445–457 [DOI] [PubMed] [Google Scholar]
- Shen J, Zeng Y, Zhuang X, Sun L, Yao X, Pimpl P, Jiang L (2013) Organelle pH in the Arabidopsis endomembrane system. Mol Plant 6:1419–1437 [DOI] [PubMed] [Google Scholar]
- Silva-Alvim FAL, An J, Alvim JC, Foresti O, Grippa A, Pelgrom A, Adams TL, Hawes C, Denecke J (2018) Predominant Golgi residency of the plant K/HDEL receptor is essential for its function in mediating ER retention. Plant Cell 30:2174–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slovik S, Hartung W (1992) Compartmental distribution and redistribution of abscisic acid in intact leaves: III. Analysis of the stress-signal chain. Planta 187:37–47 [DOI] [PubMed] [Google Scholar]
- Takahashi F, Suzuki T, Osakabe Y, Betsuyaku S, Kondo Y, Dohmae N, Fukuda H, Yamaguchi-Shinozaki K, Shinozaki K (2018) A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 556:235–238 [DOI] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng WT, Liu L, Li QB, Cline K, McCarty DR (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35:44–56 [DOI] [PubMed] [Google Scholar]
- Toh S, Imamura A, Watanabe A, Nakabayashi K, Okamoto M, Jikumaru Y, Hanada A, Aso Y, Ishiyama K, Tamura N, et al. (2008) High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol 146:1368–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Hitomi K, Nishimura N, Hitomi C, Adams SR, Getzoff ED, Schroeder JI (2014) FRET-based reporters for the direct visualization of abscisic acid concentration changes and distribution in Arabidopsis. Elife 3:e01739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia A, Waadt R, Jones AM (2018) Genetically encoded biosensors in plants: pathways to discovery. Annu Rev Plant Biol 69:497–524 [DOI] [PubMed] [Google Scholar]
- Wallrabe H, Periasamy A (2005) Imaging protein molecules using FRET and FLIM microscopy. Curr Opin Biotechnol 16:19–27 [DOI] [PubMed] [Google Scholar]
- Wan S, Jiang L (2016) Endoplasmic reticulum (ER) stress and the unfolded protein response (UPR) in plants. Protoplasma 253:753–764 [DOI] [PubMed] [Google Scholar]
- Xu ZY, Kim DH, Hwang I (2013) ABA homeostasis and signaling involving multiple subcellular compartments and multiple receptors. Plant Cell Rep 32:807–813 [DOI] [PubMed] [Google Scholar]
- Xu ZY, Lee KH, Dong T, Jeong JC, Jin JB, Kanno Y, Kim DH, Kim SY, Seo M, Bressan RA, et al. (2012) A vacuolar beta-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis. Plant Cell 24:2184–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhu H, Pan Y, Yu Y, Luan S, Li L (2014) A DTX/MATE-type transporter facilitates abscisic acid efflux and modulates ABA sensitivity and drought tolerance in Arabidopsis. Mol Plant 7:1522–1532 [DOI] [PubMed] [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.