Abstract
Studies have shown bacteria influence the initiation and progression of cancers arising in sites that harbor rich microbial communities, such as the colon. Little is known about the potential for the microbiome to influence tumorigenesis at sites considered sterile, including the upper female genital tract. The recent identification of distinct bacterial signatures associated with ovarian carcinomas suggests microbiota in the gut, vagina, or elsewhere might contribute to ovarian cancer pathogenesis. Here, we tested whether altering the microbiome affects tumorigenesis in a mouse model of high-grade serous carcinoma (HGSC) based on conditional oviduct-specific inactivation of the Brca1, Trp53, Rb1, and Nf1 tumor suppressor genes. Cohorts of control (n=20) and antibiotic-treated (n=23) mice were treated with tamoxifen to induce tumor formation and then monitored for 12 months. The antibiotic cocktail was administered for the first 5 months of the monitoring period in the treatment group. Antibiotic-treated mice had significantly fewer and less advanced tumors than control mice at study endpoint. Antibiotics induced changes in the composition of the intestinal and vaginal microbiota, which were durable in the fecal samples. Clustering analysis showed particular groups of microbiota are associated with the development of HGSC in this model. These findings demonstrate the microbiome influences HGSC pathogenesis in an in vivo model that closely recapitulates the human disease. Because the microbiome can modulate efficacy of cancer chemo- and immunotherapy, our genetically engineered mouse model system may prove useful for testing whether altering the microbiota can improve the heretofore poor response of HGSC to immunotherapies.
INTRODUCTION
High-grade serous carcinoma (HGSC) is the most common and most lethal type of ovarian cancer. Many, if not most HGSCs arise from epithelium in the distal portion of the fallopian tubes, although some HGSCs may arise from the ovarian surface epithelium, and there is a growing consensus to refer to these tumors as tubo-ovarian HGSCs (1–3). Some genetic factors such as germline BRCA1 or BRCA2 mutations clearly increase risk of developing HGSC, but less is known about the contributing roles of lifestyle and/or environmental risk factors in hereditary cases and in apparently sporadic cases. Epidemiological data from population-based studies indicate that high parity, oral contraceptive use, and sterilization via salpingectomy reduce HGSC risk (4). There is a paucity of data about the potential role of the microbiome in HGSC pathogenesis.
The microbiome influences key metabolic, inflammatory, and immunological functions in health and disease. Several studies have shown that gut and oral microbiota contribute to carcinogenesis in mouse models and patients (5–8). Although the association between Helicobacter pylori and gastric cancer is a well-documented direct host-microbial interaction (9), it is broadly accepted that microbial dysbiosis can also directly and indirectly contribute to carcinogenesis at multiple sites. For example, continuous administration of broad-spectrum antibiotics has been shown to reduce the number of tumors in mouse models of colorectal cancer (10). Germ-free mice transplanted with fecal material from patients with colorectal cancer develop colonic epithelial dysplasia, a colorectal cancer precursor lesion (11). Other studies have shown specific bacteria present in the intestinal tract can have systemic effects on the immune system by activating pro-inflammatory Th17 cells or anti-inflammatory Tregs, which could alter the risk of cancer in multiple organs (12).
As in the gut, microbial dysbiosis in the lower genital tract may promote gynecological cancers via ascending spread from the vagina (13). In keeping with this notion, epidemiological studies have shown a modest, but statistically significant increased risk of serous ovarian cancer in women with a history of pelvic inflammatory disease (14). There is emerging evidence that the upper female genital tract, including the uterus, fallopian tubes and ovaries, which was traditionally considered a sterile environment, harbors microbial communities in low abundance (15). Recent studies have shown the microbiome in ovarian cancer tissues is different from that of normal distal fallopian tube tissues or from matched non-neoplastic ovarian tissues (16,17), and that distinct bacterial signatures characterize ovarian carcinoma tissue samples relative to other types of cancer (18). In a recent clinical study, women under 50 years of age with established ovarian carcinoma and those with increased risk of ovarian cancer due to germline BRCA1 or BRCA2 mutations showed dysbiotic vaginal microbiomes, suggesting the vaginal microbiome may not only reflect, but also potentially influence risk of ovarian cancer (19). Other studies have shown vaginal microflora can promote apoptosis of cancer cells (20,21) and induce dendritic cell and Treg differentiation to produce anti-inflammatory cytokines such as IL-10 (22).
The aforementioned studies collectively provide support for the notion that microbial communities in the gut, vagina, and other sites might modulate ovarian cancer risk. However, to our knowledge, there have been no in vivo studies directly testing this premise. We have developed a genetically engineered mouse model of oviductal high-grade serous carcinoma that is based on conditional (tamoxifen [TAM]-inducible) biallelic inactivation of the Brca1, Trp53, Rb1, and Nf1 tumor suppressor genes in the oviductal epithelium, hereafter referred to as BPRN mice (23,24). After transient treatment of BPRN mice with TAM, tumors arise with complete penetrance following a several month latency period and acquire widespread copy number alterations, somatic mutations, and gene expression profiles similar to those observed in human HGSCs, particularly the immunoreactive and mesenchymal subtypes as described by the Cancer Genome Atlas (TCGA) (25). In this study, we sought to test if altering the gut and vaginal microbiome impacts oviductal HGSC development and/or progression in BPRN mice and to demonstrate the utility of the BPRN model as a preclinical experimental platform to advance insights into the role of the microbiome in HGSC pathogenesis.
MATERIALS AND METHODS
Animals and animal care.
Generation of the Ovgp1-iCreERT2;Brca1fl/fl;Trp53fl/fl;Rbfl/fl;Nf1fl/fl (BPRN) mouse model and in vivo induction of oviductal tumors have been described previously in detail (23). Genotypes of individual mice were confirmed by PCR analysis of tail DNA. Mice in both control and antibiotic-treated (Abx) experimental cohorts were cohoused in standard housing in groups of five and fed the same chow diet (Purina 5001) ad libitum. All procedures involving mice for the research described herein have been approved by the University of Michigan’s Institutional Animal Care and Use Committee (PRO00008343).
Antibiotic treatment
Female BPRN mice were randomly assigned to control or Abx treatment groups. Mice in the Abx group (n=23) were treated with a cocktail of metronidazole (0.25g/liter), vancomycin (0.25g/liter), and streptomycin (2g/liter) in drinking water (ad libitum) starting at 6 wk of age, and continuously thereafter for 5 mo. Antibiotics were dissolved in sucrose water (5g/liter) to improve palatability and refreshed every 3 d. Control mice (n=20) were treated with sucrose water without antibiotics for the same period.
Tumor induction and monitoring
At 8 wk of age, BPRN mice in both cohorts were intraperitoneally injected with TAM (0.2g/kg, on d 1 and d 3) to induce Cre-mediated inactivation of the targeted tumor suppressor genes in the oviductal epithelium. Mice were euthanized and necropsied 12 mo after the first TAM injection (or earlier if they reached humane endpoints). The 12 mo time point was selected because based on prior work, BPRN mice are expected to have at least one oviductal serous tubal intraepithelial carcinoma (STIC) by 8 mo post-TAM, and by 12 mo post-TAM, nearly all have more advanced disease, but most have not yet reached humane endpoints.
Sample collection
Fecal and vaginal lavage samples were collected from mice at 6 wk of age (before antibiotic or TAM treatment), 8 wk (just before TAM injection), 10 wk, every 4 wk thereafter during the monitoring period, and at study endpoint (12 mo post-TAM). Oviducts, ovaries, and other major organs were harvested under sterile conditions at the time of necropsy and the presence or absence of grossly metastatic disease or ascites was noted. For grossly visible oviductal tumors, a portion of tumor tissue was collected and snap frozen in liquid nitrogen.
Histopathology and tumor scoring
Histopathological analysis of mouse tissues has been previously described in detail (26). Briefly, formalin-fixed, paraffin-embedded tissues were sectioned and stained with hematoxylin and eosin (H&E) for evaluation by light microscopy. To identify microscopic oviductal lesions, each oviduct and ovary were serially sectioned in their entirety for microscopic examination. Alternate sections were stained with H&E or retained for further analysis. The tissue block from one control mouse oviduct was lost and could not be evaluated. Mice were evaluated for the presence and types of tumors in each oviduct and the extent of disease (no lesion, STIC, early HGSC, HGSC, metastatic disease), and assigned scores of 0, 1, 2, 3 and 4, respectively. Some oviductal tumors were classified as carcinosarcomas, also known as malignant mixed Mullerian tumors, which show a mixture of epithelial and mesenchymal elements and are recognized as variant forms of HGSC (27). For this study, carcinosarcomas and HGSCs were combined into a single HGSC category. Early HGSCs (eHGSCs) were defined as those that are locally invasive (i.e., beyond intraepithelial) but still confined to the oviduct and without invasion into adjacent soft tissue or other organs.
Immunohistochemistry
Advanced oviductal tumors were evaluated for the presence of several immune cell types using standard methods as described previously (26). Antigen retrieval was performed using Tris-EDTA buffer (pH 9.0) for CD3, CD4 and CD8, and citrate buffer pH 6.0 (Biogenex, San Ramon, CA, USA) for CD45. The primary antibodies used were: rabbit anti-CD3, anti-CD4, anti-CD8 (Abcam, Cambridge, UK), anti-FoxP3 (Cell Signaling, Danvers, MA) and rat anti-Macrophage (Abcam) and anti-CD45 (eBioscience, San Diego, CA, USA).
DNA extraction and quantitative PCR analysis of bacterial and mouse DNA
Fecal and vaginal lavage samples collected from mice before antibiotic or TAM treatment (age 6 wk, treatment wk 0), at the end of antibiotic treatment (age 26 wk, treatment wk 20), during the monitoring period off antibiotics (age 50 wk, treatment wk 44), and at study endpoint (age 60 wk, treatment wk 54) were analyzed by 16S rRNA gene sequencing. Microbial genomic DNA was extracted using the PowerSoil-htp 96 Well Soil DNA Isolation kit (Mo Bio Laboratories) with an EpMotion 5075 liquid handling workstation (Eppendorf).
Quantitative real time PCR (qPCR) was performed using the SYBR green PCR master mix and QuantStudio Real-Time PCR (qPCR) system (Applied Biosystems) with gene-specific primers listed in Table S1. Copy numbers of host DNA and bacterial 16S rRNA gene sequences were determined by comparison with known quantities of mouse and E. coli genomic DNA. Relative amounts of individual operational taxonomic units (OTUs) were determined from the ΔCt and normalized by 16S rRNA copies.
16S rRNA gene sequencing and bioinformatic analyses
The V4 region of the 16S rRNA gene from each sample was amplified using primers shown in Table S1, sequenced with the Illumina MiSeq Personal Sequencing platform, and curated with the Mothur software package as described previously (28). Raw 16S rRNA sequence data are available with BioProject number PRJNA713967. The ~250 bp paired-end sequences were assembled to contigs, and unnecessary sequences including non-bacterial sequences and chimeras were eliminated. Contigs were curated to operational taxonomic units (OTUs) at >97 % identity level, then annotated and counted by Mothur as described (28). Samples from two mice in the control group that were euthanized because they reached humane endpoints earlier than the study end point, and 6 additional samples (5 lavage and 1 fecal) that failed to yield >1000 bacterial contigs were excluded from subsequent analyses.
The α-diversity (Shannon diversity and evenness indexes and OTU richness generated by the command summary.single), β-diversity (thetaYC index generated by the command dist.shared), NMDS plot of β-diversity values generated by the command nmds with the options mindim = 2, and iters = 100, and LEfSe Linear discriminant analysis (LDA) values of OTUs were all determined using Mothur (29,30). Positive false discovery rate (FDR) between two groups was calculated with the Marker Selection command of Gene-E (https://www.broadinstitute.org/cancer/software/GENE-E/) with default settings values except 10,000 permutations and maker 0. The samples were hierarchically clustered with Pearson R of log10[abundance] by average linkage with MeV (31). The maximal 1/2 inverse value of total reads per sample was used as a detection threshold for OTUs with zero reads. After hierarchical clustering of samples by MeV, the Newick file of the resulting phylogenetic tree was exported from MeV and visualized by iTol (32). To find OTUs that are associated with tumor scores, Spearman ranking correlation coefficients and p values were calculated by the OTU association command of Mothur with tumor scores and normalized OTU abundances in shared files.
Statistical analyses
Statistical analyses of oviductal lesion presence and type, and microbiome rRNA gene sequence were performed using GraphPad Prism software version 8 (GraphPad Software Inc.). The Mantel-Haenszel Chi-Square test was used to evaluate the effect of antibiotic use on tumor development and/or progression in antibiotic-treated vs. control BPRN mice. Past 4.01 was used for PERMANOVA calculations based on 9999 permutations. The Shapiro-Wilk normality test was used to test the assumption of normal distribution (GraphPad Software Inc.). Differences between two groups were evaluated using two-tailed unpaired t test or Mann-Whitney U test for parametric or non-parametric datasets, respectively. Comparison of more than two groups was performed with one-way ANOVA followed by Dunnett’s multiple comparisons test for normally distributed datasets or by either Kruskal-Wallis or Friedman test followed by Dunn’s multiple comparisons test for non-parametric datasets. For highly repetitive comparisons between two groups, we further performed FDR analysis using Gene-E. PERMANOVA was used for analysis of thetaYC indexes. Differences at p < 0.05 and q < 0.05 were considered significant.
RESULTS
Antibiotic treatment inhibits oviductal HGSC development and progression.
To determine if the microbiome has a contributing effect on tumorigenesis in our BPRN model, littermate mice were randomized to one of two cohorts. The Abx cohort (n=23) received 5 mo of antibiotics with sucrose in the drinking water starting 2 wk before TAM injection, while the control cohort (n=20) received water with sucrose only. Mice were monitored for 12 mo post-TAM and then euthanized. A schematic depicting the experimental design and timing of sample collection is provided in Figure 1A. Two control mice were euthanized at 10.5 and 11.5 mo post-TAM, because their health status reached humane endpoints. Data summarizing the oviductal lesions present in the two cohorts are shown in Figure 1B (photomicrographs of representative lesions are provided in Figure S1A). At study endpoint, each of the two oviducts from the 21 evaluable mice in the control cohort showed STIC or a more advanced lesion and five had metastatic disease. In contrast, two Abx mice had no detectable lesion in either oviduct, and six mice had STIC or eHGSC in one oviduct but no lesion in the other. Only one mouse in the Abx cohort had metastatic disease at study endpoint. Overall, Abx mice had significantly fewer and less advanced oviductal tumors than control mice (Mantel-Haenszel Chi-Square test, P=0.015) (Table 1). Immunohistochemistry was used to assess the advanced tumors seen in the control (n=6) and Abx (n=5) mouse cohorts for the presence of selected immune cells using antibodies for CD3, CD4, CD8, CD45, FoxP3, and macrophage markers. The five HGSCs arising in the Abx cohort were virtually devoid of immune cells, while three of the six HGSCs arising in the control cohort showed detectable infiltration by immune cells expressing one or more of these markers (Figure S1B).
Figure 1. Antibiotic treatment inhibits HGSC development and progression in BPRN mice.
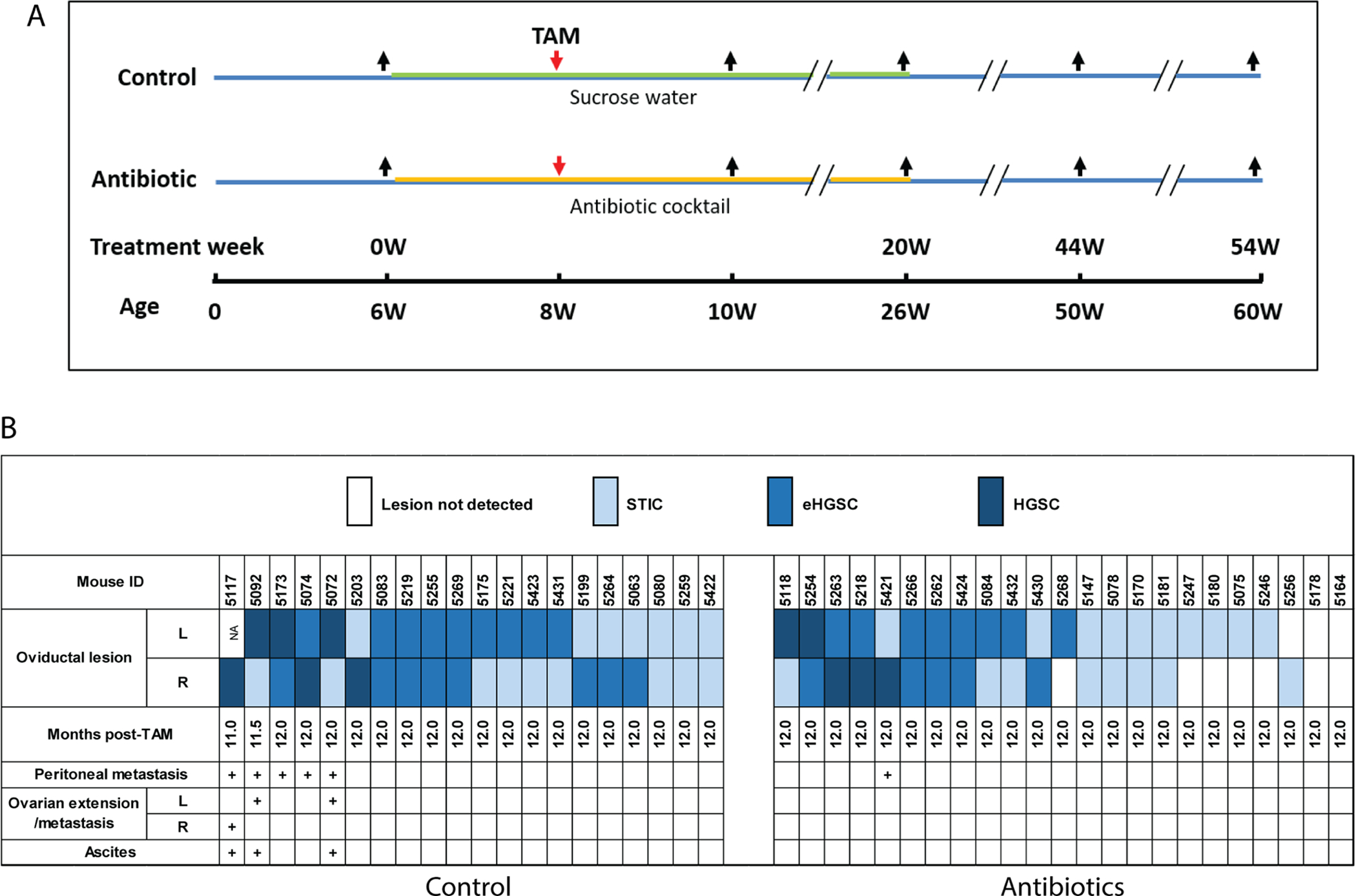
A) Schematic showing experimental design – timing of treatment with antibiotic cocktail or control (sucrose water) is marked with orange and green lines, respectively; red arrows indicate time point of TAM injection; black arrows indicate time points when fecal and vaginal lavage samples were collected. B) Data from all 43 mice (control, n=20; Abx, n=23) are shown. Months post-TAM indicates the time points at which mice were euthanized. STIC: serous tubal intraepithelial carcinoma; eHGSC: early HGSC confined to oviduct; HGSC: invasive HGSC extending beyond oviduct.
Table 1.
Antibiotic treatment inhibits oviductal HGSC development and progression in BPRN mice
| Group | No lesion | STIC | eHGSC | HGSC | Total | P value* |
|---|---|---|---|---|---|---|
| Control | 0 | 16 | 17 | 6 | 39 | 0.015 |
| Antibiotic | 10 | 18 | 13 | 5 | 46 |
Mantel-Haenszel Chi-Square test
Antibiotic treatment induces irreversible transition of the composition of the fecal microbiota.
To further explore the role of microbes in the development of HGSC, we determined the composition of the fecal and vaginal microbiota by analysis of ~25,000 paired reads per mouse sample by amplification of the ~250-bp v4 region of the 16S rRNA gene by Illumina MiSeq sequencing. Using ≥ 97% nucleotide sequence identity level, we detected a total of 3,493 operational taxonomic units (OTUs) with ~350 and ~50 OTUs per mouse in fecal and vaginal samples, respectively in control mice. In contrast, we detected ~50 OTUs per mouse in both fecal and vaginal samples in Abx mice (Figures 2A and S2A). Taxonomic analyses showed high variation of the composition of the microbiota in vaginal samples, compared with fecal samples (Figure 2A). These results were confirmed by non-metric multidimensional scaling (NMDS) plot and medians obtained from thetaYC indexes (a measure of beta diversity) in individual mice (Figures 2B and 2C).
Figure 2. Antibiotic treatment induces irreversible transition of the fecal and vaginal microbiota composition.
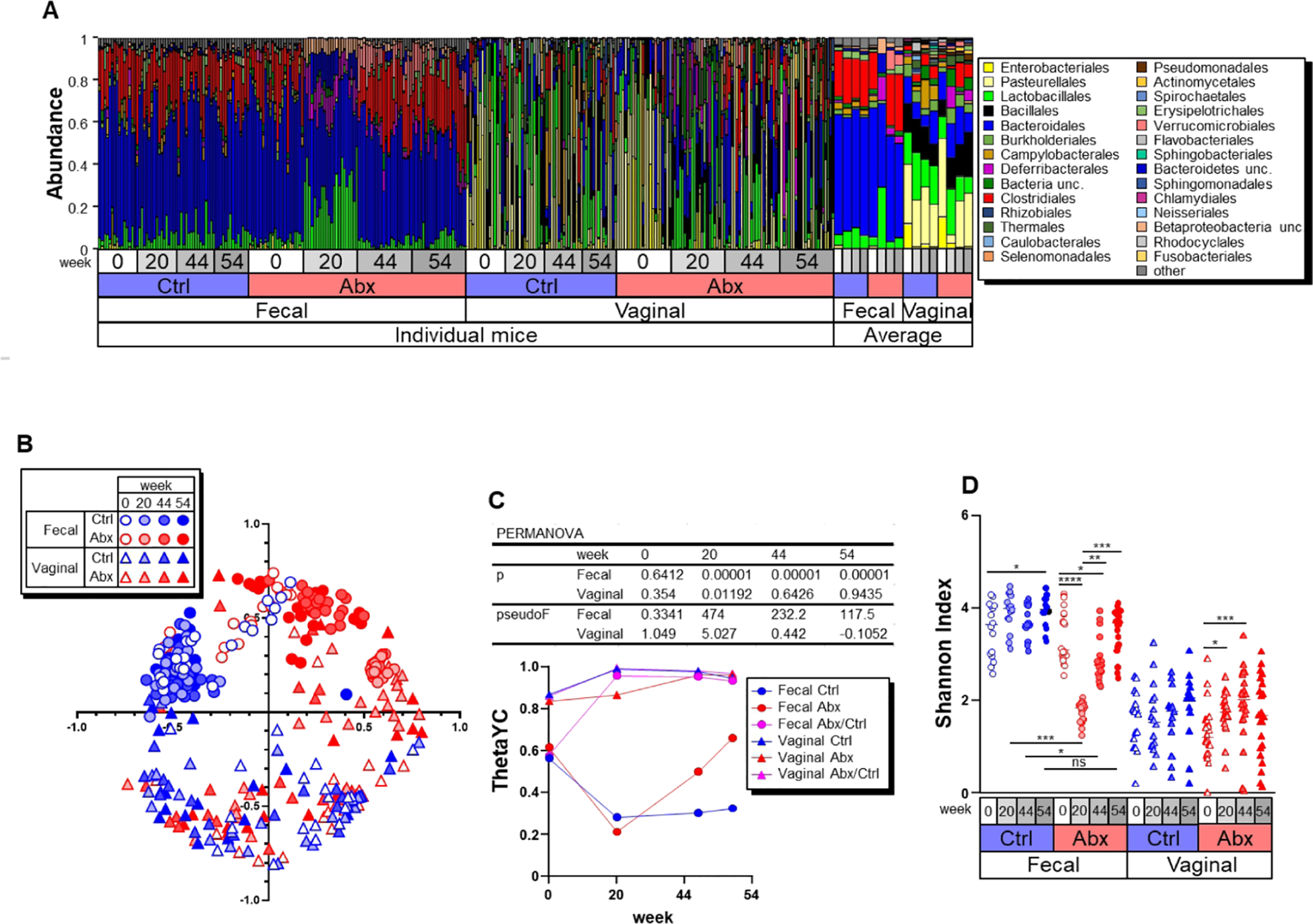
A) Relative abundance of dominant bacterial orders in feces and vaginal lavage from BPRN mice. B) NMDS plot of β-diversity values (θYC indexes) of microbiota in feces (circles) and vaginal lavage (triangles) from BPRN mice is shown with the minimal stress after 100 iterations. C) β diversity at indicated time points in samples from control and Abx BPRN mice. Data are expressed as the median. The p and pseudoF values of PERMANOVA are shown between control and Abx groups in the upper portion of the panel. D) α diversity (Shannon diversity index) of fecal and vaginal microbiotas of Abx and control BPRN mice. Data from individual mice are shown, along with median lines. *, p < 0.05; **, p<0.01; ***, p < 0.005; ****, p<0.001 (one-way ANOVA followed by Friedman and Dunnett’s tests for kinetic and intragroup comparisons, respectively).
Antibiotic treatment significantly changed the microbiota composition during and after antibiotic administration, but quantitative PCR (qPCR) analyses of bacterial 16S rRNA gene copies per host Actb (β-actin) suggests that antibiotic treatment did not significantly reduce total bacterial numbers (Figure S2B). The α-diversity indexes (Shannon diversity and evenness indexes, and OTU richness) were greatly reduced by antibiotic treatment in fecal samples, and the α-diversity returned to high levels once antibiotic treatment was stopped (Figures 2D, S2A and S2C). Comparison of thetaYC beta-diversity indexes between Abx and control samples at wk 20 showed significant differences in both fecal and vaginal samples (p=0.00001 and p=0.01192 by PERMANOVA, respectively) (Fig. 2C). These results indicate that antibiotic treatment induced changes in the composition of both the fecal and vaginal microbiota. Furthermore, the observed changes in the microbiota in fecal samples induced by antibiotic administration were seen up to wk 54 after treatment.
Difference in microbiota composition after antibiotic treatment is associated with oviductal cancer development.
Next, we determined whether particular bacterial species of control or Abx mice were associated with cancer development. To do this, we first classified fecal and vaginal samples based on their microbiota composition by hierarchical clustering. Clustering analysis showed the samples can be classified into seven clusters (Figures 3 and S3) based on microbiota composition associated with site of collection (fecal or vaginal), treatment group (control or Abx), and time point of sampling (“original” = at treatment wk 0 before TAM or antibiotics; “naïve” = no antibiotics; “under Abx” = at treatment wk 20 just at completion of antibiotics; “post Abx” = at treatment wk 44 and 54. A small cluster (cluster 1) labeled “vaginal untreated” was comprised of vaginal samples obtained mostly from mice in the control group obtained at wk 20, 44, or 54). The largest clusters (clusters 3 and 4) were dominated by fecal and vaginal samples from control mice and samples from mice in the Abx group at treatment wk 0, indicating that the microbiota composition is stable over time in untreated mice. The microbiota composition in both fecal and vaginal samples changed as a consequence of antibiotic treatment (cluster 7), and many of the changes were still found several weeks after completion of antibiotic treatment (clusters 2 and 5). Clusters 6 and 7 did not include any samples obtained from control or Abx mice at the time of study completion. We then determined whether tumor score at study end point (wk 54) was differentially associated with individual clusters. Because tumor scores were assigned only at study end point, “projection” tumor scores were assigned to 0, 20 and 44 wk fecal and lavage samples from the same mouse based on its tumor score at study endpoint (wk 54). Assignment of projection tumor scores allowed us to evaluate if earlier changes in the microbiome could help predict extent of tumor development at study endpoint. We found the microbiota associated with cluster 5, which predominantly contains both fecal and vaginal samples at wk 44 and 54 after antibiotic treatment, was significantly associated with lower tumor scores when compared with the microbiota of clusters 2, 3, and 4 (Figure 3). The projection tumor scores of cluster 7, which predominantly contains fecal and vaginal samples under antibiotic treatment (wk 20), were significantly lower than those of cluster 3, composed of fecal samples from control (naïve) mice. This result suggests that antibiotic treatment induces changes in the microbiome that are predictive of cancer outcome in TAM-treated BRPN mice, which is consistent with our finding that antibiotic treatment inhibits HGSC development and/or progression (Figure 1). Collectively, the data suggest antibiotic treatment does not uniformly prevent cancer development, but may delay tumor initiation and/or progression, and antibiotic treatment is associated with effects on selected microbiota.
Figure 3. Differences in microbiota composition after antibiotic treatment are associated with oviductal HGSC development.
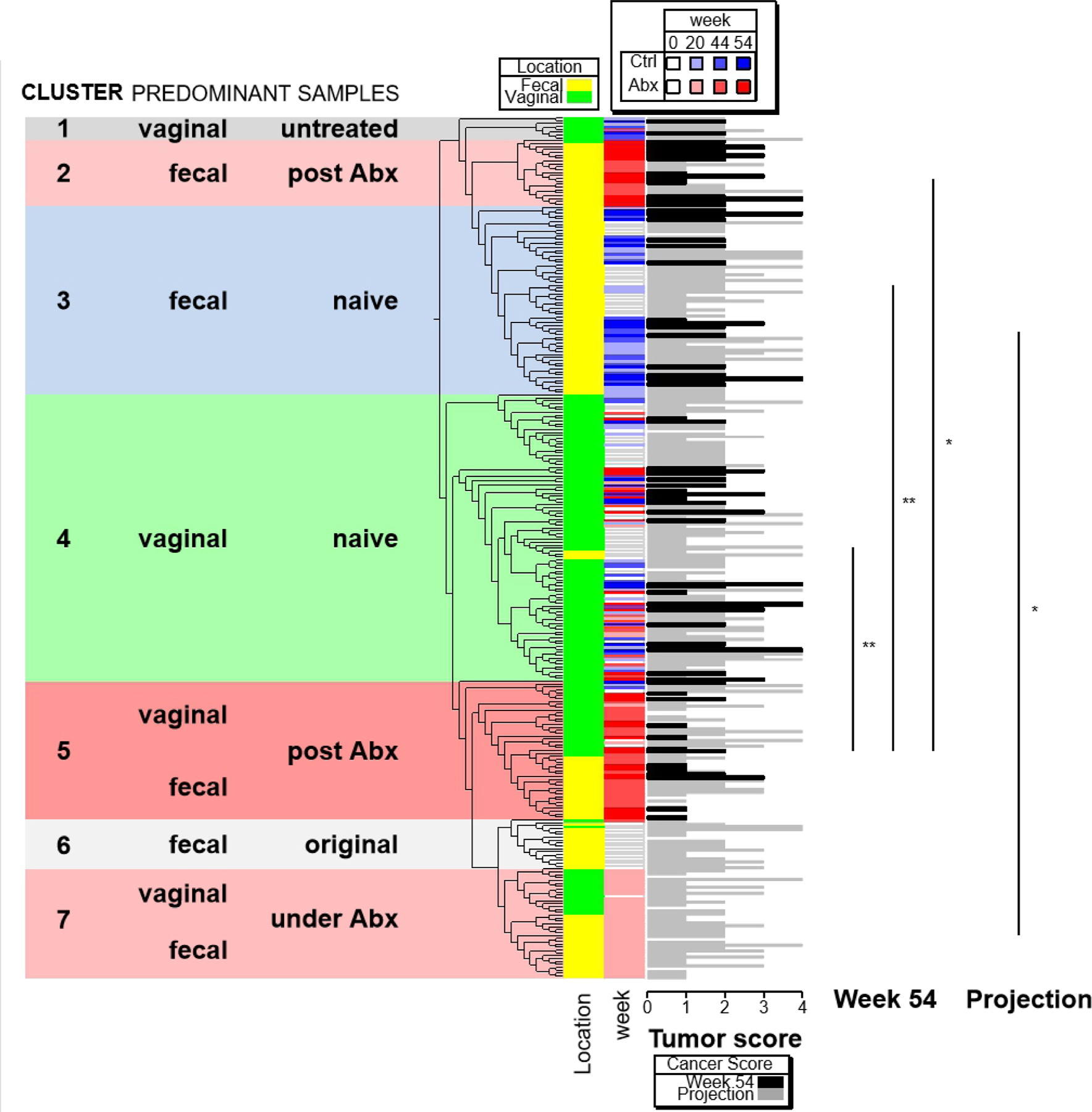
All individual samples from mice that survived until the experimental end point (wk 54) were hierarchically clustered by using logarithmic abundance values of OTUs with maximal abundance cut < 1%. The samples were assigned to seven clusters by type of sample (fecal vs vaginal), type of treatment (control vs Abx) and time point of sampling. Tumor scores of samples at week 54 are shown as black bars, and “projection” tumor scores for samples obtained earlier than 54 wk are shown as grey bars. The difference in tumor scores between specific pairs of clusters are shown at right. *, p < 0.05; **, p<0.01 (one-way ANOVA followed by Dunnett’s tests).
Specific bacteria are associated with HGSC development in BPRN mice
The observations above suggest specific types of bacteria may be associated with oviductal tumor development in the BPRN model. We screened all bacteria taxons associated with tumor development and correlated all tumor-associated bacteria with tumor scores by Spearman ranking coefficient analysis. We found 56 OTUs in fecal samples collected at wk 54 that were positively correlated with overall tumor score, while 9 were negatively correlated (p<0.05). In the vaginal samples, 7 OTUs were positively correlated with overall tumor score, while 6 were negatively correlated (Figures 4A, 4B and S4A). To verify the findings from Illumina MiSeq sequencing, we determined the relative abundance of three representative OTUs that are positively or negatively associated with tumor scores using quantitative PCR with bacteria-specific primers. As expected, we found the abundance of all three OTUs, namely, Otu00011 Clostridium XIVa sp., Otu0004 Bacteroides sp., and Otu00141 Acetatifactor sp., were highly correlated with the abundance of each based on the Illumina MiSeq read counts (Figure S4B).
Figure 4. Specific bacteria are associated with ovarian tumor development.
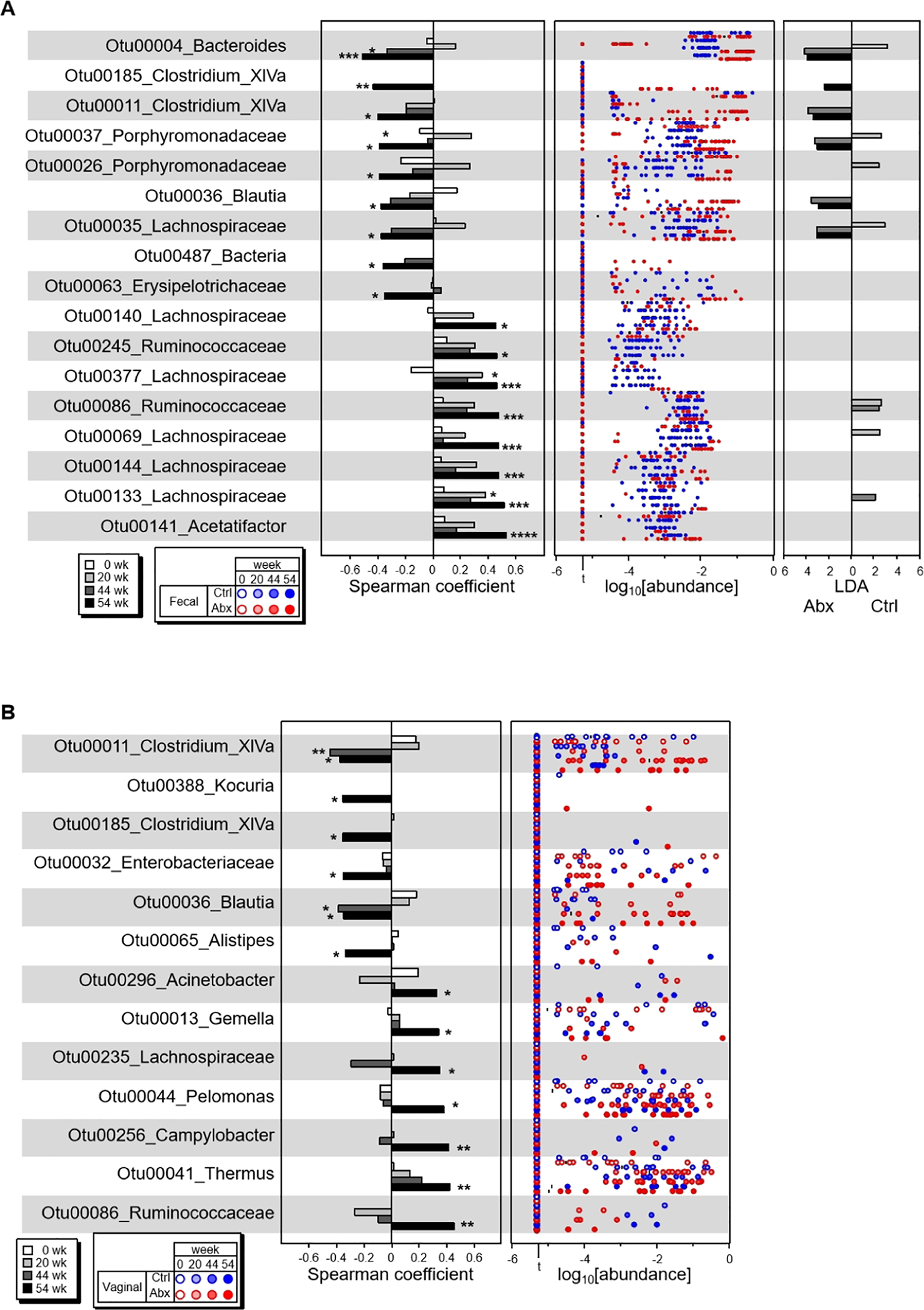
The OTUs whose abundances were highly associated with tumor scores at the end point (wk 54) were screened by Spearman Ranking coefficient. A) All negatively associated and top 8 of 56 positively associated OTUs in fecal microbiota are shown with Spearman Ranking coefficients at wk 54 and early time points with p values below 0.05 (left panel), OTU abundances at indicated weeks (center panel), and LDA values of LEfSe (p<0.05, FDR q<0.05) between Abx-treated and control mice (right panel). B) All negatively associated and positively associated OTUs in vaginal microbiota are shown with Spearman Ranking coefficients at wk 54 and early time points with p value cut 0.05 (left panel) and OTU abundances at indicated weeks (right panel). For vaginal microbiota, no significant difference of indicated OTUs was detected between Abx-treated and control mice with LEfSe p<0.05 and FDR q<0.05. *, p < 0.05; **, p<0.01; ***, p < 0.005; ****, p<0.001. All Spearman coefficient data of fecal bacteria that are positively associated with tumor scores were available in Figure S4A. All bacteria that showed statistically different abundances in Abx-treated and control with LEfSe p<0.05 and FDR q<0.05 are shown in Figures S4E and S4F.
It is possible that the differences in abundance of specific OTUs between the Abx and control BPRN mouse cohorts largely reflects the presence or extent of metastatic disease burden after cancer development rather than a difference in the efficiency of tumor initiation per se. Therefore, we further determined if OTU abundance is associated with the presence of tumors in the right and left oviducts while ignoring the presence or absence of metastases in individual mice. We found that the abundance of fecal and vaginal OTUs that are associated with overall tumor score also correlated with either right or left oviduct tumor scores (p<0.05) (Figures S4C and S4D). These results suggest that OTUs associate with HGSC independently of metastasis development. To further determine the OTUs that are associated with cancer development, we compared OTU abundance in fecal and vaginal samples at wk 44, when most TAM-treated BPRN mice in the absence of antibiotics are expected to have early-stage disease, and at wk 54, when mice are expected to have well-established and often metastatic disease. We found that only Acetatifactor Otu00141, which is the top fecal bacterium associated positively with oviductal cancer scores, increased in abundance from wk 44 to wk 54 (LDA=2.03, p<0.05 in LEfSe, Table S2).
Importantly, 6 of the top 9 OTUs negatively associated with cancer development in wk 54 fecal samples were also found at different abundance in Abx vs. control mice, as assessed by LEfSe followed by FDR (p<0.05 and q<0.05). These results indicate that the majority of the negatively associated fecal OTUs increased after antibiotic treatment. Further inspection of the differences in abundance of these OTUs during antibiotic treatment (represented by wk 20 samples) showed that these OTUs did not increase during antibiotic treatment, suggesting that they are not antibiotic-resistant. Indeed, 4 of 9 of these OTUs were sensitive to antibiotic treatment (Figure 4A). Hence, the findings suggest that antibiotic treatment depletes competitors of these bacteria because they are taxonomically close species that utilize many of same nutrients for survival.
Negatively cancer-associated bacteria include Bacteroidales, Clostridium_group XlVa, Blautia, and Lachnospiraceae species. We investigated close bacterial relatives and found that 19 and 10 of 48 OTUs that are significantly lower in fecal samples at wk 54 from Abx mice compared to control mice, are Bacteroidales and Clostridiales, respectively. Notably, these closely related OTUs are negatively correlated with HGSC-associated OTUs (Figure S4A). However, the abundance of Bacteroides, Bacteroidaceae and Porphymonadaceae are significantly higher in fecal samples of Abx mice, whereas Bacteroidales and Prevotellaceae are lower (Figure S4E). These results suggest that blooming of cancer-associated Bacteroidales species is correlated with the reduction of Prevotellaceae. Indeed, we found that the abundance of Prevotella OTU10011 and Prevotellaceae OTU20008 was positively correlated with tumor score by Spearman coefficients (0.351, p<0.033) and (0.366, p< 0.026), respectively (Table S3). Prevotella OTU00002 is one of 54 OTUs that are positively associated with cancer score (Figure S4A, Table S2). For OTUs that are associated with cancer score, the abundance of none of them was statistically different in Abx and control mice with p<0.05 of LEfSe and q<0.05 of FDR. The abundance of only one bacterium, Mucispirillum, showed an increase in vaginal samples of Abx mice (Figure S4F). Although Mucispirillum was not associated with the tumor score at wk 54, the Spearman coefficient between the abundance of Mucispirillum at wk 44 and tumor score was −0.383 (p<0.020). Two OTUs belonging to Clostridium cluster XlVa Otu00011 and Otu00185, and Blautia Otu00036 that are part of the bacterial group that were negatively associated with cancer score in vaginal samples were also found to be associated with cancer score in fecal samples (Figures 4 and S4A).
Discussion
Some limited and preliminary epidemiological and molecular evidence suggests the microbiome may have a role in human HGSC initiation and/or progression. That said, this is the first study to show that altering the microbiome impacts tumorigenesis in an in vivo model system that closely recapitulates the biology of human HGSC. Here, we performed 16S rRNA gene sequencing to characterize the overall composition of fecal and vaginal microbiota in mice before, during, and after antibiotic treatment in order to test their association with tumor features, and to identify key bacterial phylotypes and potential bacterial biomarkers positively and negatively associated with tubo-ovarian HGSC in this model system.
Antibiotics are often used to eliminate bacterial pathogens associated with infectious diseases, but in this study, antibiotic treatment primarily altered microbiota composition without significantly reducing total bacterial numbers. Mechanisms of dysbiosis, the unfavorable alteration of microbiota composition resulting in increased susceptibility to infectious and/or immune diseases, have been extensively studied by the short-term administration of antibiotics. However, the long-term impact of antibiotic treatment on human health remains unclear. Here, we show that oral administration of antibiotics for a 5 mo period inhibits tumorigenesis in a genetically engineered mouse model of oviductal HGSC, supporting the notion that the microbiota composition can affect development of tubo-ovarian HGSC in humans. Although the antibiotic cocktail was designed to target most or all bacteria, we observed that antibiotic administration induced long-term alteration of the composition of fecal microbiota, and, partially, of the vaginal microbiota in BPRN mice. This limited and selected alteration in the microbiota composition after treatment with a cocktail of antibiotics is consistent with previous studies (33,34). In our study, antibiotic administration significantly reduced the abundance of particular bacterial populations including Prevotella in the fecal microbiota (Fig. S4E and columns I and M in Table S2). The same antibiotic treatment also changed the composition of the vaginal microbiota, but we did not observe a statistically significant reduction in any bacterial groups likely due to the wide mouse-to-mouse variation in the composition of the vaginal microbiota. Clustering analysis of the microbiota composition in fecal and vaginal samples showed two clusters (clusters 2 and 5) as the chief reflections of antibiotic treatment when samples from mice at study endpoint were analyzed. Furthermore, mice with cluster 5 microbiota showed fewer and less advanced tumors than mice harboring cluster 3 or 4 microbiota (naïve/control microbiota composition). Once the mechanisms by which cluster 5 microbiota confer their protective effects are elucidated, the findings could potentially be translated for HGSC risk reduction in cancer-susceptible women.
Although the paucity of immune cells in the tumors of antibiotic-treated mice is a provocative finding, we note that the number of sufficiently advanced tumors were too few to allow us to test statistical significance of this observation. Our previous work showed that advanced oviductal tumors in BPRN mice are enriched in immunosuppressive cell subsets such as MDSCs and regulatory T (Treg) cells (24). It is plausible that antibiotic-induced reduction or elimination of immunosuppressive cells in the tumor microenvironment might enhance “immunoediting” in the early phases of tumor development by mediators such as NK and cytotoxic T cells, to prevent or at least slow tumor initiation and/or progression. In addition, antibiotics could change the composition of the tumor-associated microbiota or reduce certain bacterial molecules/metabolites in the gut that can fuel immune responses including those to tumors (18,35,36). Because we were only able to evaluate tumors at study endpoint, we can only speculate on effects of antibiotics on immune cells in the tumor microenvironment at earlier time points. Larger cohorts of animals can be used in future studies to confirm and further explore this intriguing observation.
Screening of bacteria associated with HGSC development in the BPRN model system showed that Clostridium XIVa species are negatively associated with oviductal cancer development. Notably, Clostridium XIVa species are often strong butyrate producers, and butyrate-producing Clostridium are known to inhibit intestinal tumorigenesis (37). Furthermore, butyrate administration has been shown to inhibit ovarian cancer cell growth in tissue culture models (38). It is possible, however, that bacteria-derived butyrate does not exclusively act locally in the gut to regulate cancer growth. Instead, it is likely that butyrate can also act systemically to regulate cancer development or progression, as butyrate producers have been shown to induce Treg cells (38,39). Because Tregs can contribute to progression of ovarian and other cancers via their immunosuppressive function (38,40), butyrate-producing bacteria might have multiple and opposite roles in HGSC pathogenesis. In our study, we found that high butyrate producers Clostridium XIVa in both fecal and vaginal microbiomes are negatively correlated with tumor scores, suggesting that butyrate’s inhibitory effects are more impactful in this model system. Manipulation of butyrate-producing bacteria at specific sites might clarify the role of these bacteria in the development of HGSC in the BPRN model.
Taxonomic analysis suggests that the blooming of negatively correlated cancer-associated Bacteroidales species after antibiotic treatment is accompanied by reduction or elimination of cancer-associated Prevotella. Some Prevotella species induce a Th17 response (41) and are enriched in the vaginal microbiota of postmenopausal women with gynecological cancer (42). Therefore, administration of treatments that increase cancer-associated Bacteroidales in the fecal microbiota might prevent recolonization of cancer-associated Prevotella and further inhibit HGSC development. This hypothesis could also be directly tested using our mouse model.
We found several bacteria which are positively or negatively associated with HGSC development in our mouse model. Although the precise mechanism(s) by which these bacteria contribute to cancer development are unclear, one possibility is that these bacterial species promote cancer by the regulation of bacteria-sensing innate receptors. This study unexpectedly revealed that two intestinal bacteria, Acetatifactor and Mucispirillum, are associated with HGSC in this model system – both are regulated by the innate receptor Nod2. Mucispirillium and Acetatifactor, are associated with ovarian cancer in a negative and positive manner, respectively. Mucispirillium can induce spontaneous colitis in Nod2/Nox2 double deficient mice whereas Acetatifactor accumulates in Nod2/Cd1d double deficient mice (28,43). Although Crohn’s disease-associated Nod2 mutation 3020InsC alone is not associated with ovarian cancer (44,45), it is possible that Nod2 is involved in ovarian cancer in the presence of particular microbes. Further studies using BPRN and mice deficient in Nod2 would be required to test this hypothesis.
Much remains to be learned about the role of the fecal and vaginal microbes in human diseases, including ovarian cancer pathogenesis, treatment response, and outcomes. Prior studies have shown that human ovarian cancer tissues can harbor particular bacteria, including Roseomonas mucosa, Sphingomonas US_602, and Staphylococcus cohnii are prevalent in ovarian cancers (18), which were not detected in this study. It is unknown how the DNA of bacteria present in human fecal and vaginal samples relate to those found in ovarian cancer tissues, previously thought to be sterile. While microbes that live in microbe-rich environments such as the intestine and vagina can understandably induce strong local immune responses, how such microbes may alter the immune microenvironment at distant sites remains unclear. In vivo model systems such as the one employed here, may help future efforts to reveal the impact of microbes on the initiation, development and progression of tubo-ovarian HGSC.
Although mouse models allow microbiota to be perturbed under controlled experimental conditions not possible in humans, it should be noted that there are substantial differences between the gut and vaginal microbiota in humans and mice. For example, lactobacilli dominance appears to be a unique feature of the human vaginal microbiome while in other mammals, lactobacilli rarely comprise more than one percent of the vaginal microbiota (46). This may be a consequence of vaginal pH, which is lowest in humans (46). Notably, however, lactobacilli dominance is lacking in a significant portion of apparently healthy women (47). Similarly, the gut microbiota in healthy humans and mice are different, as 85% of bacterial genera found in the mouse gut microbiome are not present in humans (48). Some of the observed differences are undoubtedly due to intrinsic factors distinguishing mice and humans, but others are likely due to confounding factors such as housing, diet, and exposure to pathogens. Nonetheless, mechanisms by which various microbes exert their effects on tumor pathogenesis may be shared across species and the data presented here can allow hypotheses regarding mechanism to be proposed and tested.
The use of microbes as medicines for cancer therapy is gaining traction, as recently reviewed by Sawant and colleagues (49). In addition, evidence is accumulating to support a strong association between gut microbiota and clinical outcomes in the context of immunotherapy (50). For example, the gut microbiome has been found to modulate response to anti-PD-1 immunotherapy in patients with melanoma, urothelial carcinoma, renal cell carcinoma, and non-small cell lung carcinoma (51,52). These findings suggest that modulation of the microbiome may prove to be an effective strategy for improving response to cancer therapies. Data showing that probiotics may be efficacious in reducing complications associated with cancer therapy are also accumulating (53). Hence it is not unreasonable to speculate that an in-depth understanding of interactions between vaginal and gut microbiota with the host and tumor phenotypes may ultimately aid in ovarian cancer prevention, optimization of current therapies, and reduction of side effects associated with cancer treatment.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
This study provides strong in vivo evidence for a role of the microbiome in ovarian cancer pathogenesis.
Acknowledgements
The authors thank the Microbiome Explorer Program of the University of Michigan’s Host Microbiome Initiative for supporting this project. This work was supported in part by R01CA226756 (KRC, YZ and ERF) and P30CA046592 (KRC and ERF).
Footnotes
Conflict of Interest Disclosure: The authors have no potential conflicts of interest to disclose.
References
- 1.Crum CP, Drapkin R, Miron A, Ince TA, Muto M, Kindelberger DW, et al. The distal fallopian tube: a new model for pelvic serous carcinogenesis. Curr Opin Obstet Gynecol 2007;19:3–9 [DOI] [PubMed] [Google Scholar]
- 2.Zhang S, Dolgalev I, Zhang T, Ran H, Levine DA, Neel BG. Both fallopian tube and ovarian surface epithelium are cells-of-origin for high-grade serous ovarian carcinoma. Nature communications 2019;10:5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh N, McCluggage WG, Gilks CB. High-grade serous carcinoma of tubo-ovarian origin: recent developments. Histopathology 2017;71:339–56 [DOI] [PubMed] [Google Scholar]
- 4.Reid BM, Permuth JB, Sellers TA. Epidemiology of ovarian cancer: a review. Cancer Biol Med 2017;14:9–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer 2013;13:800–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leystra AA, Clapper ML. Gut Microbiota Influences Experimental Outcomes in Mouse Models of Colorectal Cancer. Genes (Basel) 2019;10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwabe RF, Greten TF. Gut microbiome in HCC - Mechanisms, diagnosis and therapy. J Hepatol 2020;72:230–8 [DOI] [PubMed] [Google Scholar]
- 8.Sun J, Tang Q, Yu S, Xie M, Xie Y, Chen G, et al. Role of the oral microbiota in cancer evolution and progression. Cancer Med 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stewart OA, Wu F, Chen Y. The role of gastric microbiota in gastric cancer. Gut Microbes 2020;11:1220–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen GY, Shaw MH, Redondo G, Nunez G. The innate immune receptor Nod1 protects the intestine from inflammation-induced tumorigenesis. Cancer Res 2008;68:10060–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sobhani I, Bergsten E, Couffin S, Amiot A, Nebbad B, Barau C, et al. Colorectal cancer-associated microbiota contributes to oncogenic epigenetic signatures. Proc Natl Acad Sci U S A 2019;116:24285–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chinen T, Rudensky AY. The effects of commensal microbiota on immune cell subsets and inflammatory responses. Immunol Rev 2012;245:45–55 [DOI] [PubMed] [Google Scholar]
- 13.Laniewski P, Ilhan ZE, Herbst-Kralovetz MM. The microbiome and gynaecological cancer development, prevention and therapy. Nat Rev Urol 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rasmussen CB, Jensen A, Albieri V, Andersen KK, Kjaer SK. Is Pelvic Inflammatory Disease a Risk Factor for Ovarian Cancer? Cancer Epidemiol Biomarkers Prev 2017;26:104–9 [DOI] [PubMed] [Google Scholar]
- 15.Chen C, Song X, Wei W, Zhong H, Dai J, Lan Z, et al. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nature communications 2017;8:875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou B, Sun C, Huang J, Xia M, Guo E, Li N, et al. The biodiversity Composition of Microbiome in Ovarian Carcinoma Patients. Scientific reports 2019;9:1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banerjee S, Tian T, Wei Z, Shih N, Feldman MD, Alwine JC, et al. The ovarian cancer oncobiome. Oncotarget 2017;8:36225–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020;368:973–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nene NR, Reisel D, Leimbach A, Franchi D, Jones A, Evans I, et al. Association between the cervicovaginal microbiome, BRCA1 mutation status, and risk of ovarian cancer: a case-control study. Lancet Oncol 2019;20:1171–82 [DOI] [PubMed] [Google Scholar]
- 20.Nami Y, Abdullah N, Haghshenas B, Radiah D, Rosli R, Yari Khosroushahi A. A newly isolated probiotic Enterococcus faecalis strain from vagina microbiota enhances apoptosis of human cancer cells. J Appl Microbiol 2014;117:498–508 [DOI] [PubMed] [Google Scholar]
- 21.Nami Y, Abdullah N, Haghshenas B, Radiah D, Rosli R, Khosroushahi AY. Assessment of probiotic potential and anticancer activity of newly isolated vaginal bacterium Lactobacillus plantarum 5BL. Microbiol Immunol 2014;58:492–502 [DOI] [PubMed] [Google Scholar]
- 22.Eslami S, Hadjati J, Motevaseli E, Mirzaei R, Farashi Bonab S, Ansaripour B, et al. Lactobacillus crispatus strain SJ-3C-US induces human dendritic cells (DCs) maturation and confers an anti-inflammatory phenotype to DCs. APMIS 2016;124:697–710 [DOI] [PubMed] [Google Scholar]
- 23.Zhai Y, Wu R, Kuick R, Sessine MS, Schulman S, Green M, et al. High-grade serous carcinomas arise in the mouse oviduct via defects linked to the human disease. The Journal of Pathology 2017;243:16–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCool KW, Freeman ZT, Zhai Y, Wu R, Hu K, Liu CJ, et al. Murine Oviductal High-Grade Serous Carcinomas Mirror the Genomic Alterations, Gene Expression Profiles, and Immune Microenvironment of Their Human Counterparts. Cancer Res 2020;80:877–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu R, Zhai Y, Kuick R, Karnezis AN, Garcia P, Naseem A, et al. Impact of oviductal versus ovarian epithelial cell of origin on ovarian endometrioid carcinoma phenotype in the mouse. J Pathol 2016;240:341–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurman RJ, Carcangiu ML, Herrington CS, Young RH, editors. WHO Classification of Tumours of Female Reproductive Organs. 4th ed. Lyon: International Agency for Research on Cancer; 2014. [Google Scholar]
- 28.Caruso R, Ono M, Bunker ME, Nunez G, Inohara N. Dynamic and Asymmetric Changes of the Microbial Communities after Cohousing in Laboratory Mice. Cell Rep 2019;27:3401–12 e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costello EK, Halloy SR, Reed SC, Sowell P, Schmidt SK. Fumarole-supported islands of biodiversity within a hyperarid, high-elevation landscape on Socompa Volcano, Puna de Atacama, Andes. Appl Environ Microbiol 2009;75:735–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 2009;75:7537–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, et al. TM4 microarray software suite. Methods Enzymol 2006;411:134–93 [DOI] [PubMed] [Google Scholar]
- 32.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 2019;47:W256–W9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zackular JP, Baxter NT, Iverson KD, Sadler WD, Petrosino JF, Chen GY, et al. The gut microbiome modulates colon tumorigenesis. mBio 2013;4:e00692–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh JE, Kim BC, Chang DH, Kwon M, Lee SY, Kang D, et al. Dysbiosis-induced IL-33 contributes to impaired antiviral immunity in the genital mucosa. Proc Natl Acad Sci U S A 2016;113:E762–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillere R, Hannani D, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013;342:971–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lo BC, Chen GY, Nunez G, Caruso R. Gut microbiota and systemic immunity in health and disease. Int Immunol 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen D, Jin D, Huang S, Wu J, Xu M, Liu T, et al. Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating Wnt signaling and gut microbiota. Cancer Lett 2020;469:456–67 [DOI] [PubMed] [Google Scholar]
- 38.Mrkvicova A, Chmelarova M, Peterova E, Havelek R, Baranova I, Kazimirova P, et al. The effect of sodium butyrate and cisplatin on expression of EMT markers. PLoS One 2019;14:e0210889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takahashi D, Hoshina N, Kabumoto Y, Maeda Y, Suzuki A, Tanabe H, et al. Microbiota-derived butyrate limits the autoimmune response by promoting the differentiation of follicular regulatory T cells. EBioMedicine 2020;58:102913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang W, Liu JR, Zou W. Immunotherapy in Ovarian Cancer. Surg Oncol Clin N Am 2019;28:447–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Aquino SG, Abdollahi-Roodsaz S, Koenders MI, van de Loo FA, Pruijn GJ, Marijnissen RJ, et al. Periodontal pathogens directly promote autoimmune experimental arthritis by inducing a TLR2- and IL-1-driven Th17 response. J Immunol 2014;192:4103–11 [DOI] [PubMed] [Google Scholar]
- 42.Tsementzi D, Pena-Gonzalez A, Bai J, Hu YJ, Patel P, Shelton J, et al. Comparison of vaginal microbiota in gynecologic cancer patients pre- and post-radiation therapy and healthy women. Cancer Med 2020;9:3714–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee C, Hong SN, Paik NY, Kim TJ, Kim ER, Chang DK, et al. CD1d Modulates Colonic Inflammation in NOD2−/− Mice by Altering the Intestinal Microbial Composition Comprising Acetatifactor muris. J Crohns Colitis 2019;13:1081–91 [DOI] [PubMed] [Google Scholar]
- 44.Lubinski J, Huzarski T, Kurzawski G, Suchy J, Masojc B, Mierzejewski M, et al. The 3020insC Allele of NOD2 Predisposes to Cancers of Multiple Organs. Hered Cancer Clin Pract 2005;3:59–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Magnowski P, Medrek K, Magnowska M, Stawicka M, Kedzia H, Gorski B, et al. The 3020insC NOD2 gene mutation in patients with ovarian cancer. Ginekol Pol 2008;79:544–9 [PubMed] [Google Scholar]
- 46.Miller EA, Beasley DE, Dunn RR, Archie EA. Lactobacilli Dominance and Vaginal pH: Why Is the Human Vaginal Microbiome Unique? Front Microbiol 2016;7:1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang B, Fettweis JM, Brooks JP, Jefferson KK, Buck GA. The changing landscape of the vaginal microbiome. Clin Lab Med 2014;34:747–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A 2005;102:11070–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sawant SS, Patil SM, Gupta V, Kunda NK. Microbes as Medicines: Harnessing the Power of Bacteria in Advancing Cancer Treatment. International journal of molecular sciences 2020;21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chan SL. Microbiome and cancer treatment: Are we ready to apply in clinics? Prog Mol Biol Transl Sci 2020;171:301–8 [DOI] [PubMed] [Google Scholar]
- 51.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018;359:91–7 [DOI] [PubMed] [Google Scholar]
- 52.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018;359:97–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miarons M, Roca M, Salva F. The role of pro-, pre- and symbiotics in cancer: A systematic review. J Clin Pharm Ther 2021;46:50–65 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


