Abstract
Transmembrane protein 229A (TMEM229A) is a member of the TMEM family that plays an important role in tooth differentiation and development. However, the expression level and biological role of TMEM229A in cancer remains unknown. The present study aimed to investigate the expression level of TMEM229A in non-small cell lung cancer (NSCLC), as well as its effect and mechanism on NSCLC progression. Clinical specimens from patients with NSCLC were enrolled from the First People's Hospital of Huzhou (Huzhou, China). TMEM229A expression was detected using reverse transcription-quantitative PCR (RT-qPCR), western blotting and immunohistochemical analysis. The relationship between TMEM229A expression and the survival rate of patients with NSCLC was analyzed using Kaplan-Meier Plotter datasets. The effects of TMEM229A on cell proliferation, migration and invasion were detected using Cell Counting Kit-8, colony formation, soft agar, real-time cellular analysis and Transwell assays. The expression levels of epithelial-mesenchymal transition (EMT)-related proteins, as well as ERK and AKT phosphorylation were determined via RT-qPCR and western blot analysis. The results demonstrated that TMEM229A expression was significantly downregulated in human NSCLC tissues and in several cell lines compared with adjacent normal lung tissues and BEAS-2B cells, respectively. Survival analysis of lung adenocarcinoma and squamous cell lung carcinoma cases identified that low TMEM229A expression was associated with a poor prognosis. The in vitro assays indicated that overexpressing TMEM229A significantly inhibited cell proliferation, migration and invasion, while TMEM229A knockdown had the opposite effect. Mechanistically, TMEM229A overexpression effectively increased E-cadherin expression and reduced N-cadherin, snail family transcriptional repressor 1 and MMP2 expression, indicating that EMT was suppressed. In addition, overexpression of TMEM229A reduced the expression levels of phosphorylated (p)-ERK and p-AKT, and this effect was partially suppressed by the incorporation of specific ERK inhibitor PD98059. Collectively, the results of the present study demonstrated that the effects of TMEM229A on inhibiting cell proliferation, migration and invasion were partially mediated by inactivating the ERK signaling pathway, thereby providing a potential therapeutic target for the treatment of NSCLC.
Keywords: ERK, AKT, non-small cell lung cancer, metastasis, transmembrane protein 229A
Introduction
Lung cancer is one of the most common malignant cancer types and is the leading cause of cancer-associated mortality worldwide (1,2). In China, it is estimated that the number of lung cancer cases reached 2.2 million and 1.8 million individuals succumbed to lung cancer in 2020 (3). Moreover, lung cancer is typically classified as small-cell lung cancer (SCLC) and non-SCLC (NSCLC) (2), with NSCLC being the major type of lung cancer and accounting for 85% of all cases (2). Despite significant improvements in NSCLC treatment, including surgery (4), chemotherapy (5,6), radiotherapy (7), targeted therapy (8–10) and immunotherapy (11), the 5-year survival rate of patients with NSCLC is only 15% (2,12). This is due to the absence of early sensitivity and specificity diagnostic biomarkers (13), drug resistance (14) and distant metastasis (15). Therefore, it is essential to investigate the molecular mechanism underlying NSCLC progression.
The transmembrane protein (TMEM) family includes types of proteins that span the biological membranes, most of which extend through the lipid bilayer of the plasma membrane, while others are located in the cell membranes, such as mitochondrial, endoplasmic reticulum, lysosomal, and Golgi membranes (16,17). Although the functions of most TMEM proteins remain unknown, the associations between TMEM proteins and tumor progression have been a hotspot of research (17,18). Recently, several TMEM proteins have been revealed to act as oncogenes or tumor suppressors in NSCLC, and play a crucial role in the tumorigenesis and progression of NSCLC (17,18). For example, TMEM209, localized to the nuclear envelope and in the cytoplasm of lung cancer cells, is upregulated in lung cancer and promotes cell proliferation and tumor progression by interacting with nucleoporin 205 kDa (19). TMEM88 is a Wnt regulatory protein, and its cytosolic form and the methylation of TMEM88 are highly expressed in lung cancer, which accelerates the tumorigenesis and progression of lung cancer (20,21). Moreover, Liu and Zhu (22) reported that TMEM106A was lowly expressed in NSCLC and suppressed cell proliferation, migration and invasion, as well as induced cell apoptosis via the PI3K/AKT/NF-κB signaling pathway. TMEM17 has also been demonstrated to be aberrantly expressed in various solid tumor types, such as lung (23) and breast cancer (24). Furthermore, TMEM17 has been revealed to be downregulated in NSCLC and to suppress cell invasion and metastasis by inactivating the ERK/ribosomal protein S6 kinase A1/snail family transcriptional repressor 1 (Snail1) pathway (23). It has also been reported that the TMEM39AS41 peptide contributed to cell death and suppressed tumor growth via the suppression of inflammation/autophagy pathways in lung cancer (25). In addition, our previous results revealed that a TMEM229A Q200del mutation was associated with histopathologic type and lymphatic metastasis in lung adenocarcinoma (Zhang et al, unpublished data). However, to the best of our knowledge, the expression level and biological role of TMEM229A in NSCLC has not been previously studied.
The present study aimed to investigate the expression level of TMEM229A in NSCLC tissues and cell lines, and examine the relationship between TMEM229A expression and clinicopathological features. To this end, the biological roles and mechanisms of TMEM229A in the tumorigenesis of NSCLC were determined, providing a potential therapeutic target for the treatment of NSCLC.
Materials and methods
Ethical approval for the study protocol
The present study was approved by the Ethics Committee of The First People's Hospital of Huzhou, Huzhou, China (approval no. 2019012), and informed consent was obtained from all patients.
Patient samples
A total of 240 NSCLC and matched para-carcinoma tissues (>5 cm away from the tumor) were obtained from patients who underwent surgical resections at the First People's Hospital of Huzhou (Huzhou, China) between January 2015 and December 2018. The NSCLC patients consisted of 113 females and 127 males (age range, 27–82 years; mean age, 63). Among them, an NSCLC tissue microarray (TMA), containing 189 pairs of tissues that were formalin-fixed paraffin-embedded (FFPE), was used for immunohistochemical (IHC) analysis, and 51 pairs of fresh tissues were used for reverse transcription-quantitative (RT-q)PCR and western blot analysis. These patients had not received chemotherapy or radiation therapy prior to surgical resection. According to the 8th edition of Union for International Cancer Control (UICC) (26,27), the pathological diagnosis was confirmed and classified by two certified pathologists (Professors Qilin Shi and Hui Xia, Department of Pathology, The First People's Hospital of Huzhou). All patients and clinical information were collected retrospectively and analyzed in this study, including sex, age, smoking history, tumor size, histological type, tumor differentiation, lymph node metastasis, cancer thrombus and TNM stage (Table I). Informed consent was obtained from all patients.
Table I.
Association between TMEM229A expression and clinicopathological characteristics of patients with non-small cell lung cancer.
| TMEM229A expression | |||||
|---|---|---|---|---|---|
| Variables | N=189 | Low | High | χ2 | P-value |
| Sex | 0.058 | 0.810 | |||
| Female | 90 | 53 | 37 | ||
| Male | 99 | 60 | 39 | ||
| Age | 1.883 | 0.170 | |||
| >65 | 88 | 48 | 40 | ||
| ≤65 | 101 | 65 | 36 | ||
| Smoking history | 0.690 | 0.406 | |||
| Ever | 95 | 54 | 41 | ||
| Never | 94 | 59 | 35 | ||
| Tumor size (cm) | 0.009 | 0.925 | |||
| >3.0 | 54 | 32 | 22 | ||
| ≤3.0 | 135 | 81 | 54 | ||
| Histological type | 0.707 | 0.863 | |||
| Squamous cell carcinoma | 47 | 28 | 19 | ||
| Adenocarcinoma | 141 | 94 | 57 | ||
| Large cell carcinoma | 1 | 1 | 0 | ||
| Tumor differentiation | 3.627 | 0.043 | |||
| Well/Moderate | 146 | 82 | 64 | ||
| Poor | 43 | 31 | 12 | ||
| Lymphatic invasion | 7.062 | 0.008 | |||
| Present | 66 | 48 | 18 | ||
| Absent | 123 | 65 | 58 | ||
| Cancer thrombus | 6.354 | 0.012 | |||
| Present | 78 | 55 | 23 | ||
| Absent | 111 | 58 | 53 | ||
| Stage | 4.217 | 0.040 | |||
| I+II | 115 | 62 | 53 | ||
| III+IV | 74 | 51 | 23 | ||
TMEM229A, transmembrane protein 229A.
Kaplan-Meier Plotter
Kaplan-Meier Plotter (http://kmplot.com/analysis/) is a public web server used for the validation of the gene prognostic power using expression data from independent datasets (28). All the related data were downloaded from the Gene Expression Omnibus (GEO) database and The Cancer Genome Atlas (TCGA) (28,29). A total of 1,233 samples of nine independent datasets were integrated, including 1,100 samples in GEO datasets [GSE4573 (30), GSE14814 (31), GSE8894 (32), GSE19188 (33), GSE3141 (34), GSE31210 (35), GSE29013 (36) and GSE37745 (37)] and 133 samples in TCGA datasets (38). TMEM229A expression levels in patients with lung adenocarcinoma (n=601) and patients with squamous cell lung carcinoma (n=632) were collected using the Kaplan-Meier Plotter online bioinformatics datasets. Additionally, patients with lung adenocarcinoma and squamous cell carcinoma were divided into low or high TMEM229A expression groups, according to TMEM229A mRNA expression based on the median value. The overall survival was analyzed using the Kaplan-Meier method.
Cell culture and treatment
The normal human lung epithelial cell (BEAS-2B) and seven lung cancer cell lines (A549, H23, H226, 95D, H1975, PC-9 and H1299) were purchased from the American Type Culture Collection. Cells were cultured in DMEM (cat. no. SH30243.01; HyClone; Cytiva) or RPMI-1640 medium (cat. no. SH30809.01; HyClone; Cytiva) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 IU/ml penicillin and 100 µg/ml streptomycin (Sigma-Aldrich; Merck KGaA) in a humidified incubator under 5% CO2 at 37°C. To inhibit the ERK signaling pathway, cells were treated with 100% dimethylsulfoxide (DMSO) as a negative control or PD98059 (ERK inhibitor; 20 µM; Beyotime Institute of Biotechnology) for 2 h prior to plasmid transfection in a humidified incubator under 5% CO2 at 37°C.
Plasmid construction and cell transfection
To construct the full-length human TMEM229A proteins, the corresponding TMEM229A cDNA was cloned into a pcDNA3.1/myc-his vector (cat. no. V800-20; Invitrogen; Thermo Fisher Scientific, Inc.). In total, two specific small interfering (si)RNAs of TMEM229A (20 µM) and scrambled siRNA (referred to as si-Ctrl; 20 µM) were designed and synthesized by Guangzhou RiboBio Co., Ltd.. Once cells reached 60–70% confluence, cell transfections of different plasmids [pcDNA3.1/myc-his (referred to as vector), pcDNA3.1-TMEM229A/myc-his (referred to as overexpressed (OE)-TMEM229A), si-Ctrl, si-TMEM229A] were carried out using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. The medium was replaced with complete medium and transfection was performed in a humidified incubator under 5% CO2 at 37°C for 6 h. In a 6-well plate, 2 µg vector or OE-TMEM229A and 50 nM si-Ctrl or si-TMEM229A plasmids were added to each well. At 24 h after transfection, cells were harvested for the subsequent experiments. The siRNA sequences used in the experiment were as follows: si-TMEM229A#1 forward, 5′-GGAUGCGCCUCUACUUCUAdTdT-3′ and reverse, 5′-UAGAAGUAGAGGCGCAUCCdTdT-3′; si-TMEM229A#2 forward, 5′-CCUUCGUCUUCAAUUUCCUdTdT-3′ and reverse, 5′-AGGAAAUUGAAGACGAAGGdTdT-3′; and si-Ctrl forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse, 5′-ACGUGACACGUUCGGAGAATT-3′.
Real-time cellular analysis (RTCA)
The RTCA xCELLLigence system (ACEA Biosciences, Inc.; Agilent Technologies, Inc.) was used to uninterruptedly monitor cell morphology, proliferation and migration in vitro in a non-invasive manner, as previously described (39,40). A cell index was used to indicate the cell number and cell adhesion. As cells adhered to the surface of an E-plate and influenced the electrical impedance across the array, the xCELLLigence system provided an electronic record and converted it into the cell index. Cell proliferation and migration assays were performed according to the manufacturer's instructions (ACEA Biosciences, Inc.; Agilent Technologies, Inc.).
Briefly, for cell proliferation assays, 50 µl culture medium was added to measure the background, and then 6×103 A549 cells, 1×104 H23 cells or 1×104 H1299 cells transfected with different plasmids were mixed with 100 µl culture medium and seeded into the E-plate. For cell migration and invasion assays, 165 µl culture medium was added to the lower chamber and 30 µl serum-free culture medium was added to the upper chamber, and then the E-plate was left to stand for 1 h in a humidified incubator under 5% CO2 at 37°C. The cells were mixed with 100 µl serum-free culture medium and seeded into the E-plate. The data were recorded using xCELLLigence software 2.0 (ACEA Biosciences, Inc.; Agilent Technologies, Inc.) and analyzed using GraphPad Prism 5.0 (GraphPad Software, Inc.).
Cell proliferation assay
Cell proliferation was also assessed using a Cell Counting Kit-8 (CCK-8; Beyotime Institute of Biotechnology) assay, according to the manufacturer's instructions. Briefly, 6×103 A549, 1×104 H23 or 1×104 H1299 cells transfected with different plasmids were seeded into 96-well plates and cultured for 24, 48 and 72 h at 37°C, followed by incubation with 10 µl of CCK-8 reagent for 1 h. The absorbance values were measured using a Spectra Max 190 reader (Molecular Devices, LLC) at 450 nm.
Clonogenic assay
The transfected A549, H23 or H1299 cells were seeded into 6-well plates at a density of 1×103 cells/well and cultured with culture medium containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) for 10 days. Cells were fixed in 4% paraformaldehyde for 30 min at room temperature and followed by staining with 1% crystal violet stain solution (Beyotime Institute of Biotechnology) overnight at room temperature. Cell colonies were imaged under an inverted fluorescence microscope with a magnification of ×20 magnification (ZEISS Axio Vert.A1; Carl Zeiss AG) and counted if there were >50 cells in the assay.
Soft agar assay
The cell proliferative ability was also examined using a soft agar assay, as previously described (41,42). At 24 h-post-transfection, A549, H23 or H1299 cells were plated in 24-well plates, with the bottom layer containing 0.7% low-melting point agarose (cat. no. 16520100; Invitrogen; Thermo Fisher Scientific, Inc.). Cells (1,000-2,000 per well) were seeded into the medium with 0.35% agarose. After 14 days of incubation in a humidified incubator under 5% CO2 at 37°C, cell colonies were imaged and counted if there were >50 cells in the assay under an inverted fluorescence microscope with a magnification of ×20 (ZEISS Axio Vert.A1; Carl Zeiss AG).
Transwell assay
Cell migratory and invasive abilities were determined using a Transwell plate chamber (8 µm; Corning; Corning, Inc.), according to the manufacturer's protocols. For the migration assay, the transfected cells (5×104) were mixed in 100 µl serum-free culture medium and seeded into the upper chamber, and 600 µl complete culture medium was added to the lower chamber. For the invasion assay, the upper chamber was firstly coated with Matrigel® Matrix Basement Membrane (cat. no. 356234; BD Biosciences) for 4 h in a humidified incubator under 5% CO2 at 37°C. Then, cells were seeded into the upper chamber, and complete medium was added to the lower chamber. Following 48 h incubation in a humidified incubator under 5% CO2 at 37°C, the cells in the lower chamber were fixed in 4% paraformaldehyde for 30 min at room temperature and stained with 1% crystal violet stain solution (Beyotime Institute of Biotechnology) overnight at room temperature. The images were captured and the cells were decolorized with 30% acetic acid. The absorbance values were measured using a Spectra Max 190 reader (Molecular Devices, LLC) at 570 nm.
RNA extraction and RT-qPCR
Total RNA from tissues and cells was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), as previously described (43). Briefly, samples were treated with 20% chloroform, followed by incubation at room temperature for 5 min. Then, samples were centrifuged and the aqueous phase was transferred to a new tube, followed by the addition of an equal amount of isopropanol and incubation at room temperature for 10 min. The pellets were dissolved in nuclease-free water. cDNA was reverse transcribed using the PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd.), according to the manufacturer's protocol. RT-qPCR was performed using UltraSYBR Green PCR Master mix (cat. no. CW0957H; CWBio) on an ABI 7500 system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions used for the qPCR were as follows: Initial denaturation at 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec and 60°C for 1 min. Gene expression was normalized to 18S ribosomal RNA (18sRNA), and the relative expression level was calculated using the 2−ΔΔCq method (44). The primer sequences used in the experiment were as follows: TMEM229A forward, 5′-CGACCTACCCCGCTTTCTTTT-3′ and reverse, 5′-GCTCCCACCGTAGATGAAGAT-3′; N-cadherin forward, 5′-AGCCAACCTTAACTGAGGAGT-3′ and reverse, 5′-GGCAAGTTGATTGGAGGGATG-3′; E-cadherin forward, 5′-ATTTTTCCCTCGACACCCGAT-3′ and reverse, 5′-TCCCAGGCGTAGACCAAGA-3′; MMP2 forward, 5′-GATACCCCTTTGACGGTAAGGA-3′ and reverse, 5′-CCTTCTCCCAAGGTCCATAGC-3′; Snail1 forward, 5′-ACTGCAACAAGGAATACCTCAG-3′ and reverse, 5′-GCACTGGTACTTCTTGACATCTG-3′; and 18sRNA forward, 5′-GTAACCCGTTGAACCCCATT-3′ and reverse, 5′-CCATCCAATCGGTAGTAGCG-3′.
Western blotting
Tissues or cells were lysed in RIPA buffer (cat. no. P0013B; Beyotime Institute of Biotechnology) containing protease and phosphatase inhibitors (Beyotime Institute of Biotechnology). The total protein concentration was quantified with a BCA assay kit (Beyotime Institute of Biotechnology). An equal amount of total protein (30 µg) was loaded and separated using 8–12% SDS-PAGE. The proteins were transferred onto 0.45-µm PVDF membranes (EMD Millipore). The membranes were blocked with 5% non-fat milk at room temperature for 1 h, followed by incubation with primary antibodies overnight at 4°C. The membranes were subsequently washed with PBS (cat. no. SH30256.01; HyClone; Cytiva) containing 0.1% Tween-20, and incubated with the corresponding HRP-conjugated secondary antibodies [(goat-anti mouse IgG (H+L) antibody; 1:5,000; cat. no. A0216) or (goat-anti rabbit IgG (H+L) antibody; 1:5,000; cat. no. A0208) both from Beyotime Institute of Biotechnology] at room temperature for 1 h. The images were visualized using an ECL reagent (cat. no. P0018S; Beyotime Institute of Biotechnology) and measured using a Tanon 5200 system (Tanon Science & Technology Co., Ltd.). The relative expression level of proteins was normalized to β-actin using ImageJ vl.6.0 software (National Institutes of Health).
The primary antibodies used in the experiment were as follows: Anti-TMEM229A (1:500; cat. no. ab107780; Abcam); anti-E-cadherin (1:1,000; cat. no. 20874-1-AP; ProteinTech Group, Inc.); anti-N-cadherin (1:1,000; cat. no. 22018-1-AP; ProteinTech Group, Inc.); anti-MMP2 (1:1,000; cat. no. 10373-2-AP; ProteinTech Group, Inc.); anti-Snail1 (1:1,000; cat. no. 13099-1-AP; ProteinTech Group, Inc.); anti-ERK1/2 (1:2,000; cat. no. 4695; Cell Signaling Technology, Inc.); anti-phosphorylated (p)-ERK1/2 (1:2,000; cat. no. 4370; Cell Signaling Technology, Inc.); anti-AKT (1:2,000; cat. no. 4691; Cell Signaling Technology, Inc.); anti-p-AKT (1:2,000; Ser473; cat. no. 4060; Cell Signaling Technology, Inc.); anti-p-p38 (1:2,000; Thr180/Tyr182; cat. no. 4631; Cell Signaling Technology, Inc.); anti-p-JNK (1:2,000; Thr183/Tyr185; cat. no. 4668; Cell Signaling Technology, Inc.); anti-p-p65 (1:1,000; Ser536; cat. no. 3033; Cell Signaling Technology, Inc.), and anti-β-actin (1:5,000; cat. no. M1210-2; Hangzhou HuaAn Biotechnology Co., Ltd.).
IHC staining
The tissues were fixed with 10% formalin overnight at room temperature, and the FFPE tissues were sliced into 5-µm thickness sections, followed by dewaxing, hydrating and inactivating of endogenous peroxidase. Sections were then incubated with a polyclonal rabbit anti-TMEM229A antibody (1:500; cat. no. PA5-63294; Invitrogen; Thermo Fisher Scientific, Inc.) overnight at 4°C in a humidified chamber. The sections were subsequently washed with PBS (cat. no. SH30256.01; HyClone; Cytiva), and incubated with the SP Rabbit & Mouse HRP-conjugated secondary antibody kit (cat. no. CW2069S; CWBio) at room temperature for 1 h. Next, the sections were incubated with DAB (included in the aforementioned kit) at room temperature for 20 min. After washing with water, the sections were counterstained with hematoxylin staining solution (cat. no. C0107; Beyotime Institute of Biotechnology) at room temperature for 5 min. The TMA sections were further imaged and digitally scanned at a magnification of ×400 using a KF-PRO-005 platform (Ningbo Jiangfeng Bio-information Technology Co., Ltd.) into whole slide digital images. The scoring of the slides was conducted using HALO Multiplex IHC analysis software version v3.1.1076.308 (Indica Labs). The intensity of TMEM229A was scored as 1 (absent or weak), 2 (moderate) or 3 (high). The percentage of positive cells was assigned as 1 (<25%), 2 (25–50%) and 3 (>50%). The score of each slice was multiplied to acquire a final score of 1–9. A composite score of <3 was defined as TMEM229A low expression, and a score of ≥3 was defined as TMEM229A high expression.
Statistical analysis
All data are presented as the mean ± SEM of three independent experiments. The associations between TMEM229A expression and clinicopathological characteristics of patients were analyzed using the χ2 test, Fisher's exact test and Pearson rank correlation analysis. Kaplan-Meier survival analysis was performed using the log-rank test. Statistical significance between the groups was determined using SPSS 21.0 software (IBM Corp.) with an unpaired Student's t-test or one-way ANOVA followed by a Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
TMEM229A is lowly expressed in NSCLC
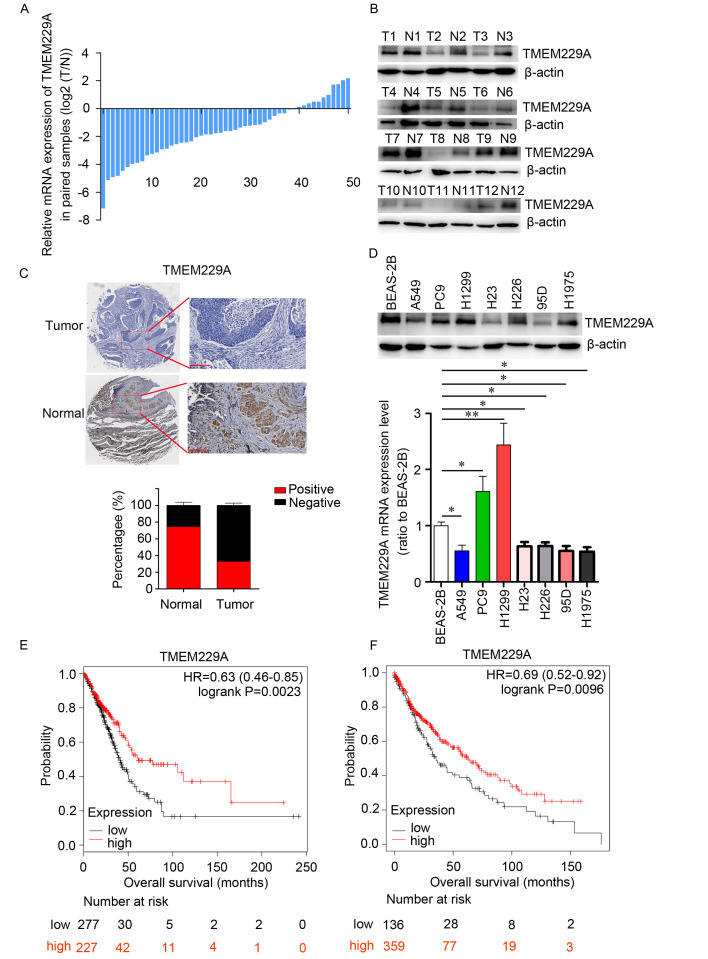
To determine the expression pattern of TMEM229A in NSCLC, the mRNA expression level of TMEM229A was measured in 51 paired NSCLC and corresponding healthy tissues via RT-qPCR. Downregulation of TMEM229A was observed in 40 pairs of tissues, accounting for 78% of the total specimens examined (Fig. 1A). Consistent with the mRNA expression pattern, a reduced protein expression level of TMEM229A was identified in NSCLC tissues compared with the matched adjacent healthy tissues in 10/12 paired samples, as determined using western blotting (Fig. 1B). The low expression of TMEM229A protein in NSCLC tissues was further confirmed via IHC staining analysis (Fig. 1C). Moreover, TMEM229A expression was measured in BEAS-2B cells and seven NSCLC cell lines, including A549, H23, H1299, H226, 95D, PC-9 and H1975. The mRNA and protein expression levels of TMEM229A were decreased in several NSCLC cell lines (A549, H23, 95D, H226 and H1975) compared with BEAS-2B cells (Fig. 1D). Owing to the higher invasiveness of 95D, A549 and H23 were selected for the follow-up overexpressing experiments. Collectively, these data indicated that TMEM229A expression was downregulated in NSCLC.
Figure 1.
TMEM229 is lowly expressed in NSCLC and predicts a poor prognosis. (A) mRNA expression level of TMEM229A was detected via RT-qPCR in 51 paired NSCLC T and N tissues. (B) Western blot analysis of TMEM229A expression in 12 paired NSCLC T and N tissues. β-actin was used as internal control. (C) Immunohistochemical staining of TMEM229A in NSCLC T and N tissues (magnification, ×40 and ×200). (D) RT-qPCR and western blot analysis of TMEM229A expression in BEAS-2B cells and seven NSCLC cell lines. (E and F) Kaplan-Meier Plotter was used to map the overall survival curves for patients with lung adenocarcinoma (n=601, P=0.0023) and patients with squamous cell lung carcinoma (n=632, P=0.0096). *P<0.05 and **P<0.01. TMEM229A, transmembrane protein 229A; NSCLC, non-small cell lung cancer; RT-qPCR, reverse transcription-quantitative PCR; T, tumor; N, normal.
Correlation between TMEM229A expression and the clinicopathological characteristics of NSCLC
To evaluate the clinical significance of TMEM229A in NSCLC, a NSCLC TMA containing 189 specimens was stained with TMEM229A antibody and scored in a standard manner, as previously described (45). The results indicated that 113 patients were classified into the low TMEM229A expression group, while 76 patients were classified into the high TMEM229A expression group (Fig. 1C and Table I). Further analysis of TMEM229A expression and the clinicopathologic characteristics revealed that low TMEM229A expression was associated with tumor differentiation (P=0.043), lymph node metastasis (P=0.008), cancer thrombus (P=0.012) and TNM stage (P=0.04). However, there were no significant associations with sex, age, smoking history, tumor size and histological type (Table I).
Subsequently, Kaplan-Meier Plotter was used to study the association between TMEM229A expression level and overall survival in patients with NSCLC. All the related data were downloaded from the GEO database, the EGA and TCGA. Patients with lung adenocarcinoma and those with squamous cell carcinoma were divided into low- or high-TMEM229A expression groups, according to TMEM229A mRNA expression. The results demonstrated that lung adenocarcinoma cases and squamous cell lung carcinoma cases with low TMEM229A expression had a poor prognosis with a shorter overall survival (P=0.0023 and P=0.0096, respectively; Fig. 1E and F). Thus, these data indicated that low TMEM229A expression was associated with poor prognosis in patients with lung adenocarcinoma and in patients with squamous cell lung carcinoma.
TMEM229A overexpression suppresses NSCLC cell proliferation, migration and invasion in vitro
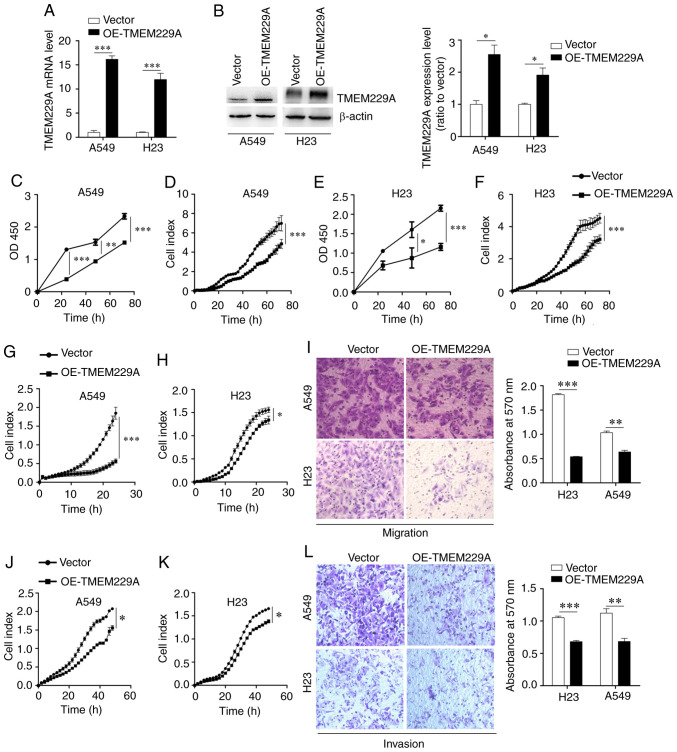
To investigate the effect of TMEM229A on cell proliferation, TMEM229A was overexpressed in A549 and H23 cells (Fig. 2A and B), which had relatively low expression levels of TMEM229A. Subsequently, the proliferative capacity of TMEM229A-overexpressing cells was measured using CCK-8 and RTCA assays. The results demonstrated that overexpression of TMEM229A significantly inhibited A549 and H23 cell proliferation in a time-dependent manner (Fig. 2C-F).
Figure 2.
Overexpression of TMEM229A inhibits NSCLC cell proliferation, migration and invasion in vitro. (A) mRNA expression level of TMEM229A was detected via reverse transcription-quantitative PCR in A549 and H23 cells. ***P<0.001. (B) Western blot analysis of TMEM229A expression in A549 and H23 cells. *P<0.05. (C-F) Cell proliferation was evaluated via Cell Counting Kit-8 and RTCA assays in A549 and H23 cells. (G-L) Migration and invasion were measured using RTCA and Transwell assays in A549 and H23 cells. *P<0.05, **P<0.01 and ***P<0.001. TMEM229A, transmembrane protein 229A; NSCLC, non-small cell lung cancer; OE, overexpressed; RTCA, real-time cellular analysis.
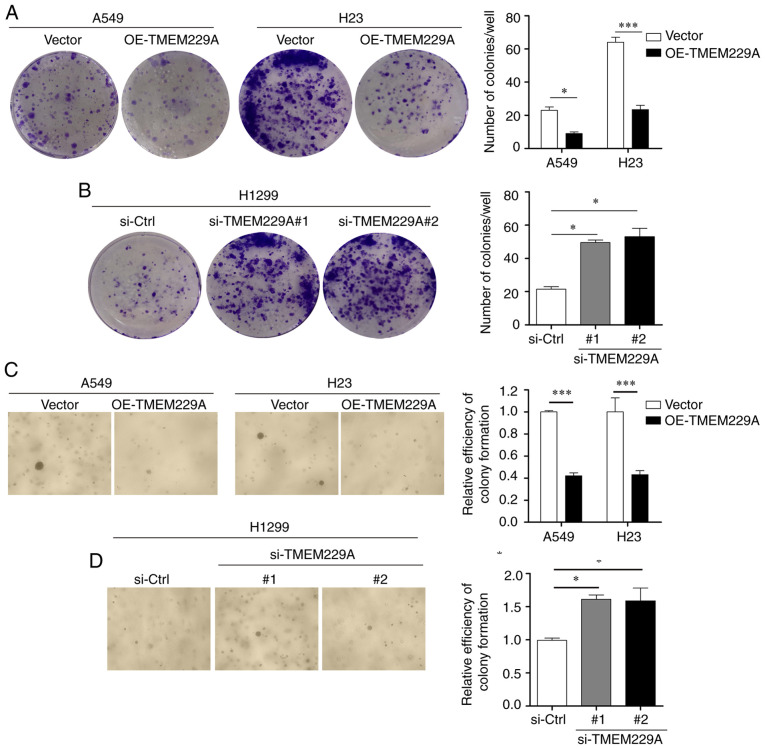
In addition, two TMEM229A siRNAs and si-Ctrl were transfected into H1299 cells, which demonstrated relatively high expression levels of TMEM229A. The RT-qPCR and western blot results revealed that TMEM229A expression was markedly decreased in the si-TMEM229A-transfected H1299 cells compared with si-Ctrl-transfected cells (Fig. S1A and B), and TMEM229A knockdown significantly promoted H1299 cell proliferation (Fig. S1C and D). Furthermore, the effects of TMEM229A overexpression or knockdown on cell proliferation were verified using cell colony and soft agar assays. The data indicated that overexpression of TMEM229A suppressed the number of cell colonies, while TMEM229A knockdown promoted these (Fig. 3A-D). Overall, these findings indicated that TMEM229A plays a crucial role in the proliferation of NSCLC cells.
Figure 3.
TMEM229A influences NSCLC cell colony formation. (A-D) The number of colonies of A549, H23 and H1299 cells was measured using colony formation and soft agar assays. *P<0.05 and ***P<0.001. TMEM229A, transmembrane protein 229A; NSCLC, non-small cell lung cancer; OE, overexpressed; si-Ctrl, scrambled siRNA; si-, small interfering.
The potential influence of TMEM229A overexpression or knockdown on cell migration and invasion was then examined. It was revealed that overexpression of TMEM229A significantly suppressed the migration and invasion of NSCLC cells, as determined using Transwell and RTCA assays (Fig. 2G-L), whereas knockdown of TMEM229A promoted the migratory and invasive abilities of H1299 cells compared with those in the control group (Fig. S1E-G).
TMEM229A regulates the epithelial-mesenchymal transition (EMT) phenotype
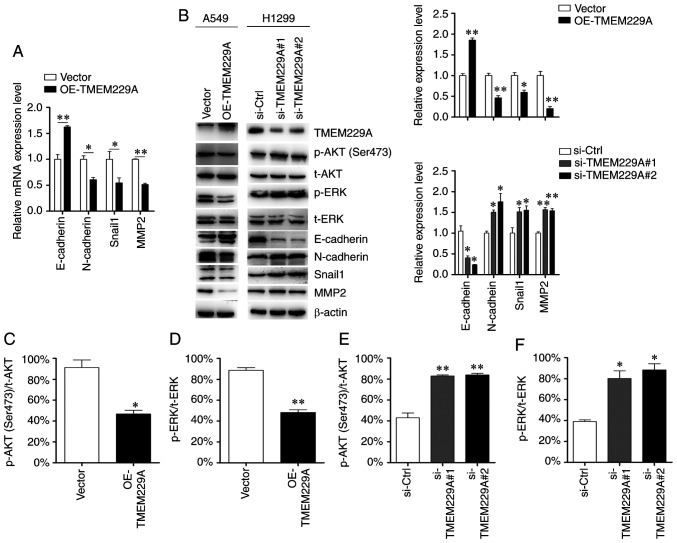
Considering the critical role of EMT in the metastasis of NSCLC (46), the present study examined the role of TMEM229A on EMT, and detected the expression levels of E-cadherin, N-cadherin, MMP2 and Snail1. As demonstrated in Fig. 4A and B, TMEM229A overexpression significantly increased E-cadherin expression and reduced N-cadherin, Snail1 and MMP2 expression, while TMEM229A knockdown exerted the opposite effects. These data indicated that TMEM229A influenced cell migration and invasion by modulating the EMT phenotype.
Figure 4.
TMEM229A regulates epithelial-mesenchymal transition via p-ERK and p-AKT. (A) mRNA expression levels of E-cadherin, N-cadherin, MMP2 and Snail1 were detected using reverse transcription-quantitative PCR. (B-F) Protein expression levels of E-cadherin, N-cadherin, MMP2, Snail1, p-ERK, p-AKT (Ser 473), t-ERK and t-AKT were detected using western blot analysis. The ratios of p-AKT (Ser473)/t-AKT and p-ERK/t-ERK were analyzed. *P<0.05 and **P<0.01. TMEM229A, transmembrane protein 229A; OE, overexpressed; Snail1, snail family transcriptional repressor 1; si-Ctrl, scrambled siRNA; si-, small interfering; p-, phosphorylated; t-, total.
TMEM229A knockdown promotes ERK and AKT phosphorylation
To investigate the mechanisms via which TMEM229A promotes NSCLC progression, key factors of common signaling pathways were screened in NSCLC cells, including p38/MAPK, ERK, AKT, JNK and NF-κB. The expression levels of p-p38, p-ERK, p-AKT (Ser473), p-JNK and p-p65 were primarily analyzed in the present study. It was revealed that knockdown of TMEM229A upregulated p-ERK and p-AKT (Ser473) expression in NSCLC cells, while overexpression of TMEM229A exerted the opposite effect (Fig. 4B-F). Moreover, other signaling pathways showed no obvious changes (Fig. S2).
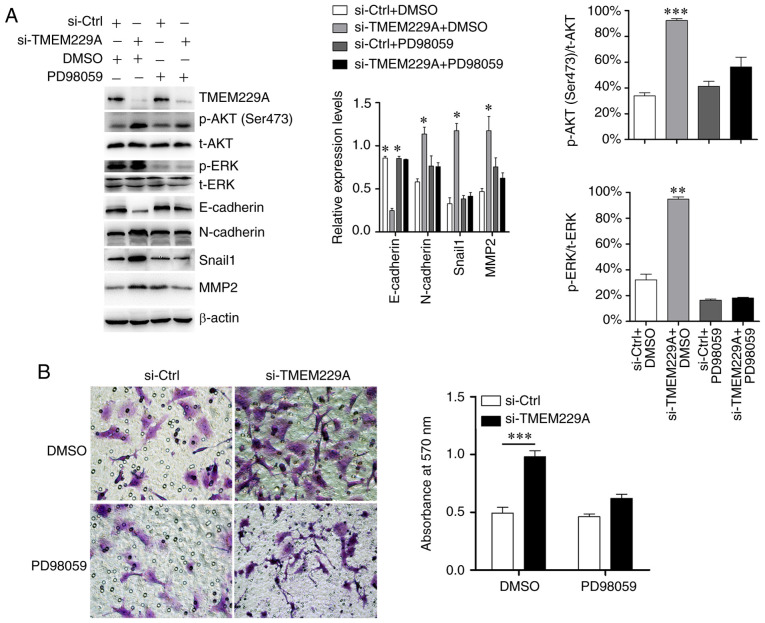
Subsequently, si-TMEM229A-transfected cells were treated with the ERK inhibitor PD98059, and the results demonstrated that PD98059 suppressed the TMEM229A-induced ERK phosphorylation, and partly reversed the expression levels of E-cadherin, N-cadherin, MMP2 and Snail1 (Fig. 5A). Moreover, the Transwell assay results indicated that the promoting effect on cell invasiveness caused by TMEM229A knockdown was also partially reversed by PD98059 (Fig. 5B).
Figure 5.
TMEM229A inhibits NSCLC progression partly via the ERK signaling pathway. (A) Western blotting examination of p-ERK, E-cadherin, N-cadherin, Snail1 and MMP2 expression after TMEM229A knockdown with or without PD98059 treatment (ERK inhibitor; 20 µM). The ratios of p-AKT (Ser473)/t-AKT and p-ERK/t-ERK were analyzed. (B) Cell invasion was measured using a Transwell assay in H1299 cells after knockdown of TMEM229A with or without PD98059 treatment (ERK inhibitor; 20 µM). *P<0.05, **P<0.01 and ***P<0.001. TMEM229A, transmembrane protein 229A; NSCLC, non-small cell lung cancer; si-Ctrl, scrambled siRNA; p-, phosphorylated; t-, total; Snail1, snail family transcriptional repressor 1; si-, small interfering.
Discussion
TMEM229A is a seven-transmembrane protein with poorly defined biology. At present, research has revealed that TMEM229A serves a critical role in the development and differentiation of teeth (47). However, the current understanding of the expression and possible mechanism of TMEM229A is lacking, particularly with regard to that in cancer. Therefore, the present study aimed to investigate the expression level and biological role of TMEM229A in NSCLC. It was identified that TMEM229A was lowly expressed in NSCLC tissues and several cell lines, and low TMEM229A expression was positively associated with poor tumor differentiation, positive lymph node metastasis, positive cancer thrombus and advanced stage of patients with NSCLC, suggesting that TMEM229A may exert a suppressive role in NSCLC development.
Metastasis is the major cause of the poor prognosis of NSCLC (48). Accumulating evidence has revealed that multiple TMEM proteins, such as TMEM209A (19), TMEM88 (20,21), TMEM106A (22), TMEM17 (23), TMEM98 (49) and TMEM48 (50), can serve as prognostic markers for NSCLC. In the present study, survival analysis using the Kaplan-Meier Plotter online bioinformatics datasets revealed that patients with low TMEM229A expression had a poorer prognosis compared with those with high TMEM229A expression, suggesting that TMEM229A could be considered as a prognostic marker for patients with NSCLC.
EMT also plays a critical role in the NSCLC metastasis, and its associated proteins and MMPs are considered as key regulators in NSCLC invasion and metastasis (51–54). The present results demonstrated that overexpression of TMEM229A suppressed cell migration and invasion, and the EMT phenotype, as well as upregulated E-cadherin expression and downregulated N-cadherin, Snail1 and MMP2 expression in vitro. Collectively, these results indicated that TMEM229A promoted the progression of NSCLC by regulating the EMT phenotype.
The ERK signaling pathway exerts an oncogenic role in regulating the proliferation and migration of NSCLC cells (55). Moreover, a previous study reported that TMEM17 suppressed NSCLC progression via the ERK signaling pathway (23). The present study primarily screened for key factors of common signaling pathways (56–58), and revealed that overexpression of TMEM229A reduced the expression levels of p-ERK and p-AKT. Furthermore, other signaling pathways demonstrated no obvious changes. It was also identified that the promoting effect on cell invasiveness caused by TMEM229A knockdown was partly reversed by PD98059, but PD98059 alone did not affect cell invasive capacity. These results were similar to those of a previous study (23), indicating that TMEM229A suppresses NSCLC progression partly by inactivating the ERK pathway.
However, the present study had certain limitations. For example, the mechanism of TMEM229A and the ERK signaling pathway was not fully determined. Thus, future studies will further investigate the underlying mechanisms of TMEM229A on NSCLC progression.
In summary, the present study demonstrated the suppressive role of TMEM229A in NSCLC development and progression. Mechanistically, TMEM229A inhibited NSCLC tumorigenesis, at least partly, via the inhibition of the ERK signaling pathway. These results suggest that TMEM229A may be a potential therapeutic target for NSCLC.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
This study was kindly supported by Zhejiang Provincial Natural Science Foundation of China under grant no. LGF20H160016, the Scientific Technology Projects of Health and Medicine of Zhejiang Province under grant nos. WKJ-ZJ-1830 and 2019KY207, and Huzhou Science and Technology Fund under grant no. 2020GYB01.
Funding
This study was kindly supported by Zhejiang Provincial Natural Science Foundation of China under grant no. LGF20H160016, the Scientific Technology Projects of Health and Medicine of Zhejiang Province under grant nos. WKJ-ZJ-1830 and 2019KY207, and Huzhou Science and Technology Fund under grant no. 2020GYB01.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
XW, HY and XZ designed and conceived the study. XZ, YH and YJ performed the experiments. XZ, YB, DX and QC analyzed the data. XZ wrote and DX revised the manuscript. All authors have read the final manuscript.
Ethics approval and consent to participate
The study was approved (approval no. 2019012) by the Ethics Committee of The First People's Hospital of Huzhou, Huzhou, China. Written informed consent was provided by all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 3.Cao W, Chen HD, Yu YW, Li N, Chen WQ. Changing profiles of cancer burden worldwide and in China: A secondary analysis of the global cancer statistics 2020. Chin Med J (Engl) 2021;134:783–791. doi: 10.1097/CM9.0000000000001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitson BA, Groth SS, Duval SJ, Swanson SJ, Maddaus MA. Surgery for early-stage non-small cell lung cancer: A systematic review of the video-assisted thoracoscopic surgery versus thoracotomy approaches to lobectomy. Ann Thorac Surg. 2008;86:2008–2018. doi: 10.1016/j.athoracsur.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 5.Horinouchi H, Atagi S, Oizumi S, Ohashi K, Kato T, Kozuki T, Seike M, Sone T, Sobue T, Tokito T, et al. Real-world outcomes of chemoradiotherapy for unresectable stage III non-small cell lung cancer: The SOLUTION study. Cancer Med. 2020;9:6597–6608. doi: 10.1002/cam4.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tachihara M, Dokuni R, Okuno K, Tokunaga S, Nakata K, Katsurada N, Yamamoto M, Nagano T, Kobayashi K, Tanaka Y, et al. Phase II study of adjuvant chemotherapy with pemetrexed and cisplatin with a short hydration method for completely resected nonsquamous non-small cell lung cancer. Thorac Cancer. 2020;11:2536–2541. doi: 10.1111/1759-7714.13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delaney GP, Barton MB. Evidence-based estimates of the demand for radiotherapy. Clin Oncol (R Coll Radiol) 2015;27:70–76. doi: 10.1016/j.clon.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Garg A, Batra U, Choudhary P, Jain D, Khurana S, Malik PS, Muthu V, Prasad KT, Singh N, Suri T, Mohan A. Clinical predictors of response to EGFR-tyrosine kinase inhibitors in EGFR-mutated non-small cell lung cancer: A real-world multicentric cohort analysis from India. Curr Probl Cancer. 2020;44:100570. doi: 10.1016/j.currproblcancer.2020.100570. [DOI] [PubMed] [Google Scholar]
- 9.Liu C, Yu H, Chang J, Chen H, Li Y, Zhao W, Zhao K, Zhu Z, Sun S, Fan M, Wang J. Crizotinib in Chinese patients with ROS1-rearranged advanced non-small-cell lung cancer in routine clinical practice. Target Oncol. 2019;14:315–323. doi: 10.1007/s11523-019-00636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Zeng L, Zhou C, Li Y, Wu L, Xia C, Jiang W, Hu Y, Liao D, Xiao L, et al. Detection of nonreciprocal/reciprocal ALK translocation as poor predictive marker in patients with first-line crizotinib-treated ALK-rearranged NSCLC. J Thorac Oncol. 2020;15:1027–1036. doi: 10.1016/j.jtho.2020.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Bylicki O, Paleiron N, Margery J, Guisier F, Vergnenegre A, Robinet G, Auliac JB, Gervais R, Chouaid C. Targeting the PD-1/PD-L1 immune checkpoint in EGFR-mutated or ALK-translocated non-small-cell lung cancer. Target Oncol. 2017;12:563–569. doi: 10.1007/s11523-017-0510-9. [DOI] [PubMed] [Google Scholar]
- 12.Balata H, Fong KM, Hendriks LE, Lam S, Ostroff JS, Peled N, Wu N, Aggarwal C. Prevention and early detection for NSCLC: Advances in thoracic oncology 2018. J Thorac Oncol. 2019;14:1513–1527. doi: 10.1016/j.jtho.2019.06.011. [DOI] [PubMed] [Google Scholar]
- 13.Yang SR, Schultheis AM, Yu H, Mandelker D, Ladanyi M, Büttner R. Precision medicine in non-small cell lung cancer: Current applications and future directions. Semin Cancer Biol. 2020 Jul 27; doi: 10.1016/j.semcancer.2020.07.009. (Epub ahead of print). doi: 10.1016/j.semcancer.2020.07.009. [DOI] [PubMed] [Google Scholar]
- 14.Imakita T, Matsumoto H, Hirano K, Morisawa T, Sakurai A, Kataoka Y. Impact on prognosis of rebiopsy in advanced non-small cell lung cancer patients after epidermal growth factor receptor-tyrosine kinase inhibitor treatment: A systematic review. BMC Cancer. 2019;19:105. doi: 10.1186/s12885-019-5309-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu PC, Tian B, Yang YL, Wang YC, Liu S, Urisman A, Yang CT, Xu Z, Jablons DM, You L. Cucurbitacin E inhibits the Yes-associated protein signaling pathway and suppresses brain metastasis of human non-small cell lung cancer in a murine model. Oncol Rep. 2019;42:697–707. doi: 10.3892/or.2019.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao J, Wu X, Zhou X, Deng R, Ma Y. TMEM205 is an independent prognostic factor and is associated with immune cell infiltrates in hepatocellular carcinoma. Front Genet. 2020;11:575776. doi: 10.3389/fgene.2020.575776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmit K, Michiels C. TMEM proteins in cancer: A review. Front Pharmacol. 2018;9:1345. doi: 10.3389/fphar.2018.01345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marx S, Dal Maso T, Chen JW, Bury M, Wouters J, Michiels C, Le Calvé B. Transmembrane (TMEM) protein family members: Poorly characterized even if essential for the metastatic process. Semin Cancer Biol. 2020;60:96–106. doi: 10.1016/j.semcancer.2019.08.018. [DOI] [PubMed] [Google Scholar]
- 19.Fujitomo T, Daigo Y, Matsuda K, Ueda K, Nakamura Y. Critical function for nuclear envelope protein TMEM209 in human pulmonary carcinogenesis. Cancer Res. 2012;72:4110–4118. doi: 10.1158/0008-5472.CAN-12-0159. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Yu X, Jiang G, Miao Y, Wang L, Zhang Y, Liu Y, Fan C, Lin X, Dong Q, et al. Cytosolic TMEM88 promotes invasion and metastasis in lung cancer cells by binding DVLS. Cancer Res. 2015;75:4527–4537. doi: 10.1158/0008-5472.CAN-14-3828. [DOI] [PubMed] [Google Scholar]
- 21.Ma R, Feng N, Yu X, Lin H, Zhang X, Shi O, Zhang H, Zhang S, Li L, Zheng M, et al. Promoter methylation of Wnt/β-Catenin signal inhibitor TMEM88 is associated with unfavorable prognosis of non-small cell lung cancer. Cancer Biol Med. 2017;14:377–386. doi: 10.20892/j.issn.2095-3941.2017.0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, Zhu H. TMEM106A inhibits cell proliferation, migration, and induces apoptosis of lung cancer cells. J Cell Biochem. 2018 Nov 19; doi: 10.1002/jcb.28057. (Epub ahead of print). doi: 10.1002/jcb.28057. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X, Zhang Y, Miao Y, Zhou H, Jiang G, Wang E. TMEM17 depresses invasion and metastasis in lung cancer cells via ERK signaling pathway. Oncotarget. 2017;8:70685–70694. doi: 10.18632/oncotarget.19977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Y, Song K, Zhang Y, Xu H, Zhang X, Wang L, Fan C, Jiang G, Wang E. TMEM17 promotes malignant progression of breast cancer via AKT/GSK3β signaling. Cancer Manag Res. 2018;10:2419–2428. doi: 10.2147/CMAR.S168723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park S, Kim M, Hong Y, Lee H, Tran Q, Kim C, Kwon SH, Park J, Park J, Kim SH. Myristoylated TMEM39AS41, a cell-permeable peptide, causes lung cancer cell death. Toxicol Res. 2020;36:123–130. doi: 10.1007/s43188-020-00038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asamura H, Chansky K, Crowley J, Goldstraw P, Rusch VW, Vansteenkiste JF, Watanabe H, Wu YL, Zielinski M, Ball D, et al. The international association for the study of lung cancer lung cancer staging project: Proposals for the revision of the N descriptors in the forthcoming 8th edition of the TNM classification for lung cancer. J Thorac Oncol. 2015;10:1675–1684. doi: 10.1097/JTO.0000000000000678. [DOI] [PubMed] [Google Scholar]
- 27.Eberhardt WE, Mitchell A, Crowley J, Kondo H, Kim YT, Turrisi A, III, Goldstraw P, Rami-Porta R, International Association for Study of Lung Cancer Staging. Prognostic Factors Committee, Advisory Board Members, Participating Institutions. The IASLC lung cancer staging project: Proposals for the revision of the M descriptors in the forthcoming eighth edition of the TNM classification of lung cancer. J Thorac Oncol. 2015;10:1515–1522. doi: 10.1097/JTO.0000000000000673. [DOI] [PubMed] [Google Scholar]
- 28.Nagy Á, Munkácsy G, Győrffy B. Pancancer survival analysis of cancer hallmark genes. Sci Rep. 2021;11:6047. doi: 10.1038/s41598-021-84787-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raponi M, Zhang Y, Yu J, Chen G, Lee G, Taylor JM, Macdonald J, Thomas D, Moskaluk C, Wang Y, Beer DG. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 2006;66:7466–7472. doi: 10.1158/0008-5472.CAN-06-1191. [DOI] [PubMed] [Google Scholar]
- 31.Zhu CQ, Ding K, Strumpf D, Weir BA, Meyerson M, Pennell N, Thomas RK, Naoki K, Ladd-Acosta C, Liu N, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol. 2010;28:4417–4424. doi: 10.1200/JCO.2009.26.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee ES, Son DS, Kim SH, Lee J, Jo J, Han J, Kim H, Lee HJ, Choi HY, Jung Y, et al. Prediction of recurrence-free survival in postoperative non-small cell lung cancer patients by using an integrated model of clinical information and gene expression. Clin Cancer Res. 2008;14:7397–7404. doi: 10.1158/1078-0432.CCR-07-4937. [DOI] [PubMed] [Google Scholar]
- 33.Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens JA, Hoogsteden HC, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 35.Yamauchi M, Yamaguchi R, Nakata A, Kohno T, Nagasaki M, Shimamura T, Imoto S, Saito A, Ueno K, Hatanaka Y, et al. Epidermal growth factor receptor tyrosine kinase defines critical prognostic genes of stage I lung adenocarcinoma. PLoS One. 2012;7:e43923. doi: 10.1371/journal.pone.0043923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie Y, Xiao G, Coombes KR, Behrens C, Solis LM, Raso G, Girard L, Erickson HS, Roth J, Heymach JV, et al. Robust gene expression signature from formalin-fixed paraffin-embedded samples predicts prognosis of non-small-cell lung cancer patients. Clin Cancer Res. 2011;17:5705–5714. doi: 10.1158/1078-0432.CCR-11-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Botling J, Edlund K, Lohr M, Hellwig B, Holmberg L, Lambe M, Berglund A, Ekman S, Bergqvist M, Pontén F, et al. Biomarker discovery in non-small cell lung cancer: Integrating gene expression profiling, meta-analysis, and tissue microarray validation. Clin Cancer Res. 2013;19:194–204. doi: 10.1158/1078-0432.CCR-12-1139. [DOI] [PubMed] [Google Scholar]
- 38.Cancer Genome Atlas Research Network, corp-author. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan G, Du Q, Wei X, Miozzi J, Kang C, Wang J, Han X, Pan J, Xie H, Chen J, Zhang W. Application of real-time cell electronic analysis system in modern pharmaceutical evaluation and analysis. Molecules. 2018;23:3280. doi: 10.3390/molecules23123280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X, Guo H, Bao Y, Yu H, Xie D, Wang X. Exosomal long non-coding RNA DLX6-AS1 as a potential diagnostic biomarker for non-small cell lung cancer. Oncol Lett. 2019;18:5197–5204. doi: 10.3892/ol.2019.10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo Z, Zhang X, Zhu H, Zhong N, Luo X, Zhang Y, Tu F, Zhong J, Wang X, He J, Huang L. TELO2 induced progression of colorectal cancer by binding with RICTOR through mTORC2. Oncol Rep. 2021;45:523–534. doi: 10.3892/or.2020.7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J, Li J, Wang K, Liu H, Sun J, Zhao X, Yu Y, Qiao Y, Wu Y, Zhang X, et al. Aberrantly high activation of a FoxM1-STMN1 axis contributes to progression and tumorigenesis in FoxM1-driven cancers. Signal Transduct Target Ther. 2021;6:42. doi: 10.1038/s41392-020-00396-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X, Hu J, Zhong L, Wang N, Yang L, Liu CC, Li H, Wang X, Zhou Y, Zhang Y, et al. Quercetin stabilizes apolipoprotein E and reduces brain Aβ levels in amyloid model mice. Neuropharmacology. 2016;108:179–192. doi: 10.1016/j.neuropharm.2016.04.032. [DOI] [PubMed] [Google Scholar]
- 44.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 45.Stack EC, Wang C, Roman KA, Hoyt CC. Multiplexed immunohistochemistry, imaging, and quantitation: A review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods. 2014;70:46–58. doi: 10.1016/j.ymeth.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 46.Liao H, Liang Y, Kang L, Xiao Y, Yu T, Wan R. miR4543p inhibits nonsmall cell lung cancer cell proliferation and metastasis by targeting TGFB2. Oncol Rep. 2021;45:67. doi: 10.3892/or.2021.8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang J, Bai R, Liu K, Ji XP, Li Y, Han JY. Identification of significantly different modules between permanent and deciduous teeth by network and pathway analyses. Genet Mol Res. 2016;15 doi: 10.4238/gmr15047959. [DOI] [PubMed] [Google Scholar]
- 48.Perlikos F, Harrington KJ, Syrigos KN. Key molecular mechanisms in lung cancer invasion and metastasis: A comprehensive review. Crit Rev Oncol Hematol. 2013;87:1–11. doi: 10.1016/j.critrevonc.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 49.Mao M, Chen J, Li X, Wu Z. siRNA-TMEM98 inhibits the invasion and migration of lung cancer cells. Int J Clin Exp Pathol. 2015;8:15661–15669. [PMC free article] [PubMed] [Google Scholar]
- 50.Akkafa F, Koyuncu I, Temiz E, Dagli H, Dilmec F, Akbas H. miRNA-mediated apoptosis activation through TMEM 48 inhibition in A549 cell line. Biochem Biophys Res Commun. 2018;503:323–329. doi: 10.1016/j.bbrc.2018.06.023. [DOI] [PubMed] [Google Scholar]
- 51.Ombrato L, Malanchi I. The EMT universe: Space between cancer cell dissemination and metastasis initiation. Crit Rev Oncog. 2014;19:349–361. doi: 10.1615/CritRevOncog.2014011802. [DOI] [PubMed] [Google Scholar]
- 52.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 53.Son H, Moon A. Epithelial-mesenchymal transition and cell invasion. Toxicol Res. 2010;26:245–252. doi: 10.5487/TR.2010.26.4.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hida Y, Hamada J. Differential expressions of matrix metalloproteinases, a disintegrin and metalloproteinases, and a disintegrin and metalloproteinases with thrombospondin motifs and their endogenous inhibitors among histologic subtypes of lung cancers. Anticancer Agents Med Chem. 2012;12:744–752. doi: 10.2174/187152012802650156. [DOI] [PubMed] [Google Scholar]
- 55.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M, McNamara K, Perera SA, Song Y, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goodwin JM, Svensson RU, Lou HJ, Winslow MM, Turk BE, Shaw RJ. An AMPK-independent signaling pathway downstream of the LKB1 tumor suppressor controls Snail1 and metastatic potential. Mol Cell. 2014;55:436–450. doi: 10.1016/j.molcel.2014.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qiao M, Sheng S, Pardee AB. Metastasis and AKT activation. Cell Cycle. 2008;7:2991–2996. doi: 10.4161/cc.7.19.6784. [DOI] [PubMed] [Google Scholar]
- 58.Kim SO, Kim MR. [6]-Gingerol prevents disassembly of cell junctions and activities of MMPs in invasive human pancreas cancer cells through ERK/NF-κ B/Snail signal transduction pathway. Evid Based Complement Alternat Med. 2013;2013:761852. doi: 10.1155/2013/761852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.