ABSTRACT
In bacteria, the defense system deployed to counter oxidative stress is orchestrated by three transcriptional factors, SoxS, SoxR, and OxyR. Although the regulon that these factors control is known in many bacteria, similar data are not available for Klebsiella pneumoniae. To address this data gap, oxidative stress was artificially induced in K. pneumoniae MGH78578 using paraquat and the corresponding oxidative stress regulon recorded using transcriptome sequencing (RNA-seq). The soxS gene was significantly induced during oxidative stress, and a knockout mutant was constructed to explore its functionality. The wild type and mutant were grown in the presence of paraquat and subjected to RNA-seq to elucidate the soxS regulon in K. pneumoniae MGH78578. Genes that are commonly regulated both in the oxidative stress and soxS regulons were identified and denoted as the oxidative SoxS regulon; these included a group of genes specifically regulated by SoxS. Efflux pump-encoding genes and global regulators were identified as part of this regulon. Consequently, the isogenic soxS mutant was found to exhibit a reduction in the minimum bactericidal concentration against tetracycline compared to that of the wild type. Impaired efflux activity, allowing tetracycline to be accumulated in the cytoplasm to bactericidal levels, was further evaluated using a tetraphenylphosphonium (TPP+) accumulation assay. The soxS mutant was also susceptible to tetracycline in vivo in a zebrafish embryo model. We conclude that the soxS gene could be considered a genetic target against which an inhibitor could be developed and used in combinatorial therapy to combat infections associated with multidrug-resistant K. pneumoniae.
KEYWORDS: AMR, Klebsiella pneumoniae, mechanisms of resistance, oxidative stress, soxS
INTRODUCTION
Oxygen started accumulating in the biosphere about 2 to 3 billion years ago. Many organisms harvest energy by oxidizing organic compounds, with oxygen acting as the terminal electron acceptor. This molecule, therefore, has become essential for life, at least for aerobic organisms. As a natural consequence of aerobic metabolism, the production of toxic reactive oxygen species (ROS), namely, hydrogen peroxide (H2O2), superoxide radical (O2·−), and the generation of hydroxyl radical (HO·), is inevitable in an oxygen-rich environment. Different ROS will not only oxidize macromolecules (such as DNA, proteins, and lipids) but also extract iron from proteins containing iron-sulfur clusters, creating a highly reactive HO·-rich intracellular environment (1, 2) detrimental for bacteria. Therefore, to survive the effects of ROS, bacteria deploy a variety of adaptive responses. These are well-characterized in bacteria like Escherichia coli (3) but, as yet, not in Klebsiella pneumoniae.
In bacteria, the primary antioxidant defense systems employ superoxide dismutase (SOD) and catalase (CAT) enzymes (1, 4). However, these may prove inadequate to protect bacteria under circumstances of extreme and prolonged oxidative stress. Under these stress conditions, bacteria can activate the OxyR and SoxRS systems in response to hydrogen peroxide (5) and redox-active compounds (6), respectively. Both OxyR and SoxRS work by transcriptionally activating genes whose protein products function either to protect or repair damage caused by intracellular ROS accumulation. In the SoxRS system, the activation of a target gene occurs via a two-step process wherein SoxR acts as a sensory protein recognizing elevated levels of ROS. Under normal conditions (nonstressed), the binuclear iron-sulfur clusters [2Fe-2S] in the SoxR protein remain reduced. In the presence of enhanced levels of superoxides, the [2Fe-2S] clusters are oxidized (7). Oxidization of the SoxR protein enhances an open complex formation with RNA polymerase, thereby activating transcription of soxS (8). The SoxS protein is a transcriptional activator belonging to the XylS/AraC family (7). SoxR-dependent induction of SoxS in turn activates the transcription of many other genes (denoted collectively as the SoxRS regulon) whose primary functions involve antioxidative action, detoxification, efflux of redox-active compounds, changes in membrane permeability, and protecting DNA, thereby rescuing bacteria from the deleterious effects of increased intracellular levels of ROS (9–11). Overall, the biological role of the SoxRS regulon can be summarized as (i) prevention of oxidative damage, (ii) recycling of damaged macromolecules, and (iii) regeneration of NADP.
In E. coli, genes that were regulated by SoxS were identified (12–17). Although the transcriptional organization of soxS is well characterized in some pathogens, the data is lacking for K. pneumoniae. These bacteria are a member of the ESKAPE group, one of six pathogens responsible for most drug-resistant nosocomial infections (18). Since oxidative stress is known to mediate antibiotic resistance in pathogens, we were interested in identifying how K. pneumoniae responds to oxidative stress and what its impact might be on antimicrobial resistance.
In this study, transcriptome sequencing (RNA-seq) was used to describe the transcriptional architecture of K. pneumoniae MGH78578 during exposure to a reactive oxygen species (ROS)-inducing agent, paraquat, revealing that the regulon was controlled by the soxRS two-component system. RNA-seq analysis of the K. pneumoniae MGH78578 ΔsoxS isogenic mutant was carried out and used to describe the oxidative soxS regulon, a stringent set of genes regulated via soxS. K. pneumoniae MGH78578 ΔsoxS was found to be highly susceptible to tetracycline. Susceptibility of the mutant to tetracycline coupled with increased accumulation of tetraphenylphosphonium (TPP+) in the bacterial cytoplasm was supported at least in part by the downregulation of acrAB-tolC and the global regulator marRAB in K. pneumoniae MGH78578 ΔsoxS. Since the mutant was highly avirulent in a zebrafish model, we predict that soxS can be used as a genetic target to inhibit infections associated with multidrug-resistant (MDR) K. pneumoniae.
RESULTS AND DISCUSSION
SoxS is the major transcriptional regulator when K. pneumoniae MGH78578 is exposed to redox compound-based oxidative stress.
Experimentally, oxidative stress can be induced in bacteria by exposing cultures to either redox compounds like PQ (paraquat) or H2O2. PQ is 1,1-dimethyl-4,4-bipyridinium and is a widely used nonselective herbicide, found to induce oxidative stress by enhancing ROS levels, superoxide anion radical (·O2−), in a dose-dependent manner, as exemplified in Vibrio cholerae, E. coli, and others (14, 19). First, we started by assessing the inhibitory concentration of PQ in K. pneumoniae MGH78578 using broth microdilution and determined the MIC to be 15.62 μM. Thereafter, the following transcriptomic experiments were carried out at sub-MICs (half the MIC). Here, we used RNA-seq to investigate the genome-wide transcriptional architecture of multidrug-resistant K. pneumoniae MGH78578 following exposure to a subinhibitory concentration of PQ. K. pneumoniae MGH78578 was exposed to 7.8 μM PQ for 30 min to induce oxidative stress. These PQ-induced cultures (denoted MGHPQ A and B) along with a parallel set of unexposed wild-type (WT) bacterial cells (denoted MGHwt A and B) were subjected to RNA isolation and deep-level sequencing (see Table S1, WS1, in the supplemental material). As discussed in detail later, the soxS gene, an XylS/AraC-type transcriptional regulator of oxidative stress, was one of the most highly induced genes during oxidative stress. Therefore, to understand the soxS-mediated oxidative stress, we constructed the K. pneumoniae MGH78578 ΔsoxS mutant and carried out RNA-seq on the deletion strain grown in the presence of PQ (denoted MGHΔsoxSPQ A and B libraries). Altogether, six RNA-seq libraries were generated in this study.
(WS1) RNA-seq mapping details. This table provides details of the RNA-seq reads mapped against different regions of the K. pneumoniae MGH78578 genome are. A complete list of all primers used in the experiments is also provided. (WS2) The table presents data describing gene expression patterns of Klebsiella pneumoniae MGH75878 (MGHPQ versus MGHWT) and K. pneumoniae MGH75878 ΔsoxS (MGH ΔsoxSPQ versus MGHPQ) during oxidative stress. Column A shows a new gene ID, and column C shows the original name, while column B represents the gene name. Columns X/Y/Z/AA/AB/AC/AD shows raw reads generated from RNA-seq conducted on different libraries, while columns AF/AG/AH/AI/AJ/AK represents the normalized reads. The fold changes were calculated from the normalized reads obtained from different libraries, MGHPQ versus MGHWT (column K) and MGH ΔsoxSPQ versus MGHPQ (column U). The genes with statistically significant data obtained from two biological replicates are indicated by a mark (√) in columns J and T based on the P value indicated in columns M and W. Based on the differential expression indicated in columns K and U (for statistically significant genes), the differential expression of each gene is indicated by a color code in columns E/F/G/H and O/P/Q/R. Genes highlighted in red indicate antimicrobial resistance genes, while those highlighted in blue indicate genes associated with virulence. (WS3) The table shows the oxidative soxS regulon of Klebsiella pneumoniae MGH78578, consisting of a stringent set of genes that are regulated by SoxS. The gene list was obtained from statistically significant genes from oxidative regulon and soxS regulon with a distinct expression pattern. Genes that were upregulated in the oxidative regulon plus downregulated in the soxS regulon and downregulated in oxidative regulon plus upregulated in the soxS regulon are shown. Download Table S1, XLSX file, 2.3 MB (2.3MB, xlsx) .
Copyright © 2021 Anes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Approximately 57 million uniquely mapped reads were generated across all six libraries, accounting for more than 9 million reads/library (Table S1, WS1), data that was sufficient for robust transcriptional analysis (20). The expression levels of 5,185 K. pneumoniae MGH78578 chromosomal genes and the resident plasmid carrying genes (including plasmids pKPN3, pKPN4, pKPN5, pKPN6, and pKPN7) were calculated using the Voom approach (limma package) (21). We confirmed the reproducibility of the RNA-seq data by calculating the Spearman coefficients for the biological replicates of all libraries based on the normalized read counts. In all six libraries, the coefficient was found to be ∼0.96 to 0.99, confirming the statistical significance between replicates (Fig. S1).
Graphs showing the distribution of RNA-seq reads in each dataset. Download FIG S1, PDF file, 0.05 MB (47.2KB, pdf) .
Copyright © 2021 Anes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Here, we describe the oxidative stress regulon of K. pneumoniae MGH78578 by identifying the genes that were differentially regulated in MGHPQ versus MGHwt libraries. The oxidative stress regulon was comprised of 1,366 genes that were differentially regulated (Fig. 1A and Table S1, WS2). Of these, 11.5% (n = 158) were highly upregulated (>4-fold) and 22.5% (n = 309) were upregulated (2- to 4-fold). A total of 49 genes (3.7%) were highly downregulated (>4-fold), while a further 147 (11.12%) were downregulated (2- to 4-fold) (Table S1, WS2). Upon analysis, the most induced K. pneumoniae MGH78578 gene was found to be soxS (145-fold), indicating that the soxRS regulon was highly active in PQ-exposed K. pneumoniae MGH78578. The transcriptomic response of bacteria to oxidative stress is specific to the agent causing oxidative stress; extracellular H2O2 triggers the OxyR regulon, while PQ induces the SoxRS regulon, as exemplified in Escherichia coli (3).
FIG 1.
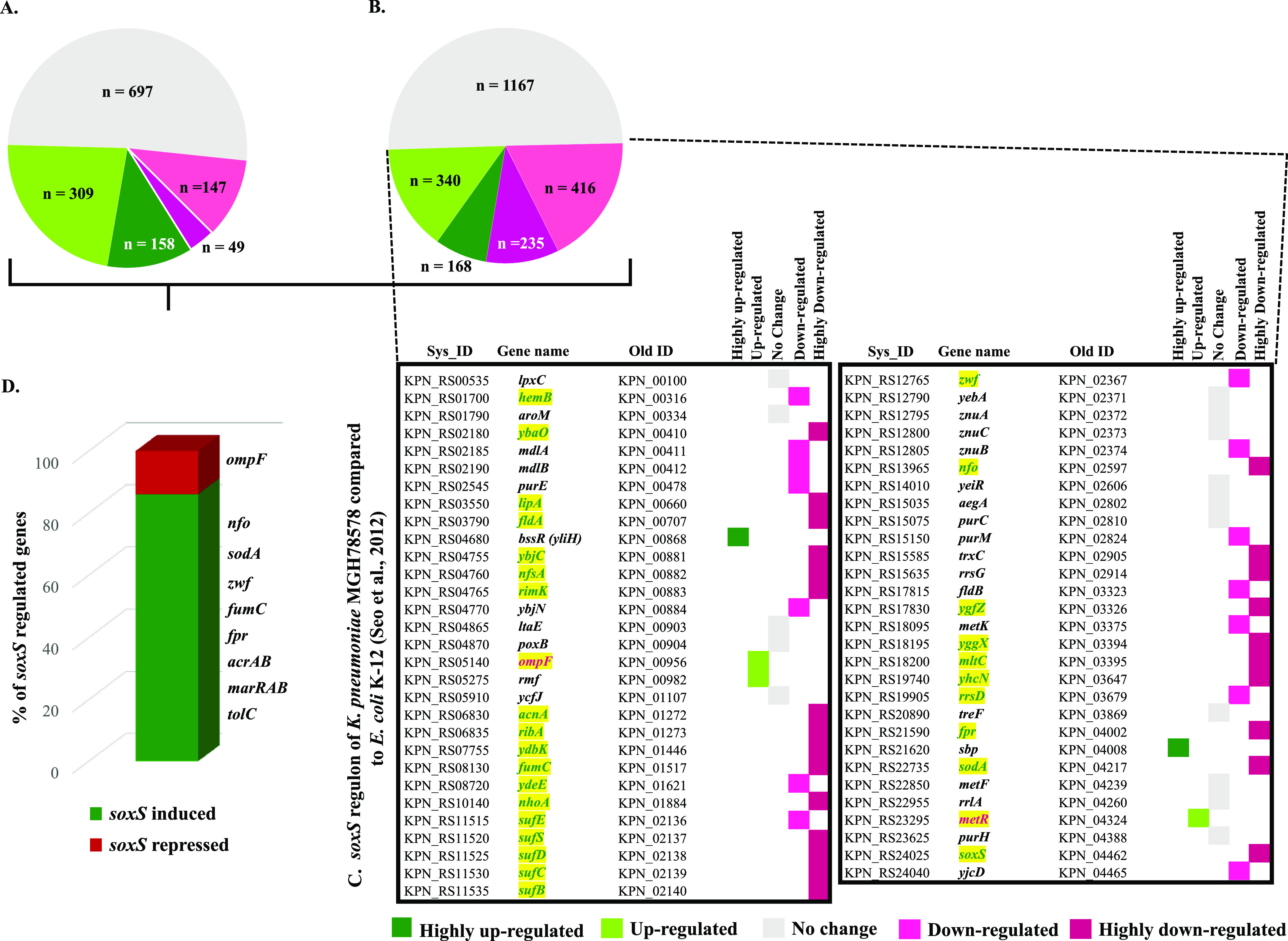
Oxidative, soxS, and oxidative soxS regulon of K. pneumoniae MGH78578. (A) The number of statistically significant genes identified in the oxidative regulon of K. pneumoniae MGH78578. These are categorized according to their expression pattern and depicted in a color code based on the color key given below. (B) The number of statistically significant genes identified in the soxS regulon and categorized according to their expression pattern. (C) Differentially regulated genes common in the soxS regulon of both K. pneumoniae MGH78578 and E. coli K-12 (3). The genes that are in green font and highlighted in yellow are those identified in the oxidative soxS regulon. (D) The number of genes in the oxidative soxS regulon of K. pneumoniae MGH78578 expressed as a percentage. The most significant soxS-induced and -repressed genes are indicated.
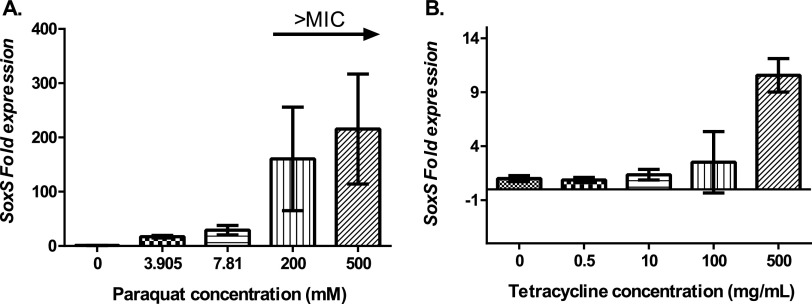
Based on these E. coli data, we hypothesized that exposure to PQ should induce the SoxS regulon in K. pneumoniae MGH78578. Since no data were available in K. pneumoniae, we put our hypothesis to the test using reverse transcription-quantitative PCR (RT-qPCR) targeting the soxS gene. Our RT-qPCR data confirmed that the expression of the soxS transcript improved with increasing concentrations of PQ (Fig. 2A).
FIG 2.
Expression of soxS gene under paraquat stress (A) and tetracycline stress (B). In both cases, the compounds were added to bacteria growing at mid-exponential phase for 30 min. Error bars represent standard deviations (SD) calculated from three biological replicates with three technical replicates each.
Exposure to H2O2, however, generated a different response in other bacteria. Exposure of V. cholerae to oxidative stress increased the activity of SOD and CAT enzymes (19). However, in V. cholerae, the level of CAT did not increase postexposure to PQ but rather increased during exposure to H2O2. Our results describing PQ-exposed K. pneumoniae MGH78578 support this observation: none of the catalases (encoded by genes KPN_RS06170, KPN_RS06615, and KPN_RS09805) were differentially regulated (Table S1, WS2). However, the SOD (encoded by sodA, sodB, and sodC) was highly upregulated; sodA alone was highly upregulated (14-fold), while sodC was upregulated (∼3-fold) in PQ-induced cells. We did not find sodB to be differentially regulated within PQ-treated K. pneumoniae MGH78578. It is tempting to speculate that selective differential regulation of SOD and not CAT in K. pneumoniae MGH78578 is the response to O2·- induced by PQ.
Since SoxS, an XylS/AraC-type transcriptional regulator, was highly induced following exposure to PQ, we were interested in identifying the associated genes that were differentially regulated. For this, we constructed a K. pneumoniae MGH78578 ΔsoxS mutant. We cultured the mutant, exposed the cells to PQ, and again used RNA-seq (MGHΔsoxSPQ library) to identify the differentially regulated genes (MGHΔsoxSPQ library versus MGHPQ library), thereby comprising the soxS regulon. The soxS regulon was made up of 2,326 differentially regulated genes (Table S1, WS2) (Fig. 1B). Of these, 7.2% of the genes (n = 168) were highly upregulated (>4-fold) and 14.6% (n = 340) were upregulated (2- to 4-fold). A total of 235 genes (10%) were highly downregulated (>4-fold), while 416 (17.8%) were downregulated (2- to 4-fold) (Table S1, WS2) (Fig. 1B). To demonstrate the robustness of these data, we compared our K. pneumoniae MGH78578 soxS regulon with the E. coli soxS regulon published earlier (3). Of the 59 soxS genes regulated in E. coli K-12, 44 were also found to be similarly regulated by soxS in K. pneumoniae MGH78578 (Fig. 1C).
To add stringency to our data, we further compared the oxidative stress regulon to the soxS regulon to identify those genes that belonged to the oxidative soxS regulon. The oxidative soxS regulon represented a stringent set of K. pneumoniae MGH78578 genes that were regulated by soxS alone. The genes belonging to this regulon had a characteristic statistically significant expression pattern, upregulated in MGHPQ (w.r.t. [with respect to] MGHwt) and downregulated in MGHΔsoxSPQ (w.r.t. MGHPQ) (i.e., SoxS induced); downregulated in MGHPQ (w.r.t. MGHwt); and upregulated in MGHΔsoxSPQ (w.r.t. MGHPQ) (i.e., SoxS repressed). In total, 256 genes belonged to the oxidative soxS regulon. Of these, 222 genes were found to be SoxS induced, while 34 were SoxS repressed (Table S1, WS3). Examples include soxS, acrAB, tolC, and oqxAB, among others, all of which were soxS induced. Of the 44 genes commonly identified in our soxS regulon and E. coli K-12 (3), 30 were identified to belong to the more stringent oxidative soxS regulon. Our oxidative soxS regulon identified many genes that were previously shown to be regulated by SoxS. A discussion of these genes is included in Text S1.
A text file describing 44 genes commonly identified in our soxS regulon and E. coli K-12 (3), some 30 were identified to belong to the more stringent oxidative soxS regulon. Our oxidative soxS regulon identified many genes that were previously shown to be regulated by SoxS. Download Text S1, DOCX file, 0.03 MB (33KB, docx) .
Copyright © 2021 Anes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
K. pneumoniae MGH78578, isolated from the sputum of a 66-year-old intensive care unit patient in 1994, is a multidrug-resistant isolate, and its antimicrobial resistance profile is well characterized (22). This strain is resistant to ampicillin, oxacillin, ticarcillin, trimethoprim-sulfamethoxazole, nalidixic acid, kanamycin, gentamicin, and tetracycline but is susceptible to amikacin, ciprofloxacin, and imipenem. Our primary interest was in identifying how soxS modulates antimicrobial resistance in K. pneumoniae MGH78578. Thus, we assayed whether any K. pneumoniae MGH78578 genes conferring antimicrobial resistance were captured in our oxidative soxS regulon. We identified 11 antimicrobial resistance-encoding genes (acrAB, acrE, tolC, marRAB, oqxAB, cmr [mdfA], ybhT, KPN_RS15915, and KPN_RS15920) in the oxidative soxS regulon, and all of them were soxS induced (Table S1, WS3). Interestingly, we also found that another member of the XylS/AraC family, tetD, encoding a tetracycline efflux MFS transporter, was identified in the soxS regulon and not in the oxidative regulon. This shows that, at least in K. pneumoniae, tetD is positively regulated by soxS. Although tetD was shown to modulate response against redox compounds and tetracycline (23), we did not find any evidence of differential regulation when K. pneumoniae MGH78578 was exposed to PQ. Since many genes conferring antimicrobial resistance were modulated by soxS, we were interested in examining whether the inactivation of soxS resulted in aberrations in the antimicrobial resistance pattern of K. pneumoniae MGH78578.
Deletion of soxS in multidrug-resistant K. pneumoniae MGH78578 produced a reduction in the minimal bactericidal concentration (MBC) against antimicrobials, particularly tetracycline.
The deletion of a transcriptional regulator like soxS could have a large impact on cell metabolism and stress responses. To globally visualize the metabolic aberrations concerning soxS, we subjected the K. pneumoniae MGH78578 WT and its isogenic ΔsoxS mutant to multiple growing conditions on a phenotypic microarray platform. Of the 1,484 conditions tested, altered phenotypes (WT versus mutant) were observed under 517 conditions, of which only 12 were significantly upregulated, wherein the mutant showed increased respiratory metabolism compared with the WT (Table S2). Significant phenotypic alterations were found to be associated with nitrogen and some nitrogen peptides and with amino acid sources such as l-isoleucine, l-ornithine, and glycine. This analysis also identified 50 conditions determined to be downregulated and in which the mutant showed reduced metabolic respiration compared to the WT. These were found to be associated with high pH (5, 9) sensitivity and antimicrobial drugs such as tetracyclines (doxycycline, demeclocycline, chlortetracycline, and minocycline), aminoglycosides (amikacin), cephalosporins (cephalothin, cefuroxime, and cefotaxime), β-lactams (cloxacillin, oxacillin, and phenethicillin), and others, including polymyxin B (PMB) and colistin (polymyxin E).
Metabolic activities of K. pneumoniae MGH78578 (wild type) and its isogenic ΔsoxS mutant grown under multiple conditions on the OmniLog microarray platform. Significant metabolic changes for delta activities were <−2 and >2. Download Table S2, XLSX file, 0.08 MB (81.2KB, xlsx) .
Copyright © 2021 Anes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Our RNA-seq data showed that the genes encoding antimicrobial resistance, such as acrAB-tolC, marRAB, and others, were differentially regulated in the K. pneumoniae MGH78578 ΔsoxS strain and, thus, classified as soxS induced. This observation, and the phenotypic microarray associated with metabolic profiling, led us to hypothesize that the soxS mutant has a modified antimicrobial resistance profile compared to the wild type. To test our hypothesis, we assayed the MIC/MBC of both K. pneumoniae MGH78578 and K. pneumoniae MGH78578 ΔsoxS strains against a panel of antimicrobial compounds. MIC/MBC assays were carried out on K. pneumoniae MGH78578 and K. pneumoniae MGH78578 ΔsoxS strains against colistin, kanamycin, gentamicin, cefotaxime, and tetracycline (Table 1). Escherichia coli ATCC 25922 was used as a control. Our results showed that there was no significant change in the MIC/MBC values between the mutant and wild type against colistin, kanamycin, and rifampin, even though our phenotypic microarray assay recorded downregulation in the metabolism of the mutant compared to the wild type. It could be that the metabolic downregulation was not sufficient to cause an inhibitory effect. However, there was a significant reduction in the MIC/MBC values for the K. pneumoniae MGH78578 ΔsoxS strain compared to K. pneumoniae MGH78578 when exposed to tetracycline (64- and 8-fold, respectively) and cefotaxime (32- and 64-fold, respectively).
TABLE 1.
Minimum inhibitory and bactericidal concentrations determined for the wild-type K. pneumoniae MGH78578 and the K. pneumoniae MGH78578 ΔsoxS isogenic mutant
| Antimicrobial compound | MICs and MBCsa (μg/ml) |
|||
|---|---|---|---|---|
|
K. pneumoniae MGH78578 |
K. pneumoniae MGH78578 ΔsoxS |
|||
| MIC | MBC | MIC | MBC | |
| Colistin | 0.25 (S) | 0.5 | 0.125 (S) | 0.125 |
| Kanamycin | >512 (R) | >512 | >512 (R) | >512 |
| Gentamicin | 128 (R) | 128 | 64 (R) | 64 |
| Cefotaxime | 32 (R) | 64 | 1 (S) | 1 |
| Tetracycline | 128 (−) | 128 | 2 (−) | 16 |
Results indicate the median value from 3 independent assays. MIC values shown are interpreted according to EUCAST guidelines. (S), susceptible; (R), resistant; (−) not available.
Therefore, using a combination of phenotypic microarray and RNA-seq, we show that tetracycline tolerance was soxS dependent in MDR K. pneumoniae MGH78578. Oxidative stress is a common cause of cell death mediated by antimicrobial agents, irrespective of the class to which the compound belongs (24). Therefore, we were interested to know whether exposure to tetracycline induced any oxidative stress in K. pneumoniae MGH78578. For this, we checked the induction of soxS in tetracycline-exposed K. pneumoniae MGH78578. Proportional induction of soxS expression in response to increasing tetracycline concentration confirmed the exposure to tetracycline-induced soxS-dependent oxidative stress in K. pneumoniae MGH78578 (Fig. 2B).
The soxRS-associated regulation of antibiotic resistance was described earlier in several bacteria (25, 26). Similarly, the induction of ROS was also reported to modulate antibiotic resistance in other pathogenic bacteria. For example, Salmonella enterica serovar Typhimurium was shown to modulate its susceptibility to tetracycline when exposed to an ROS-generating macrolide antibiotic, tylosin (27). In Acinetobacter baumannii, soxR overexpression also led to susceptibility to tetracycline (28). This SoxR-based negative regulation of SoxS could be the reason underpinning the increased susceptibility. Even though the correlation between the expression of soxS and efflux pumps has been shown previously (29), there is no evidence pointing to the cytoplasmic accumulation of antimicrobial compounds due to an inactive soxS-based impaired efflux activity. We therefore proceeded to determine whether an impaired efflux activity led to the accumulation of compounds within the cytoplasm of the K. pneumoniae MGH78578 ΔsoxS mutant, leading to the bactericidal effect.
Reduction in accumulation is due to the impaired efflux pump activity in K. pneumoniae MGH78578 ΔsoxS cells.
Since the K. pneumoniae MGH78578 ΔsoxS mutant was susceptible to tetracycline, we were interested in understanding the mechanism underpinning the observation. Our RNA-seq data revealed that the genes encoding the AcrAB-TolC efflux pump were highly SoxS dependent, because they were >4-fold upregulated in the PQ regulon and >8-fold downregulated in the K. pneumoniae MGH78578 ΔsoxS mutant. Tetracycline is one of several structurally diverse substrates of the efflux pump AcrAB-TolC (30). Hence, we hypothesized that the deletion of the soxS gene could lead to a reduction in the expression of the AcrAB-TolC efflux pump. This feature then could account for the accumulation of tetracycline in the cytoplasm to bactericidal levels.
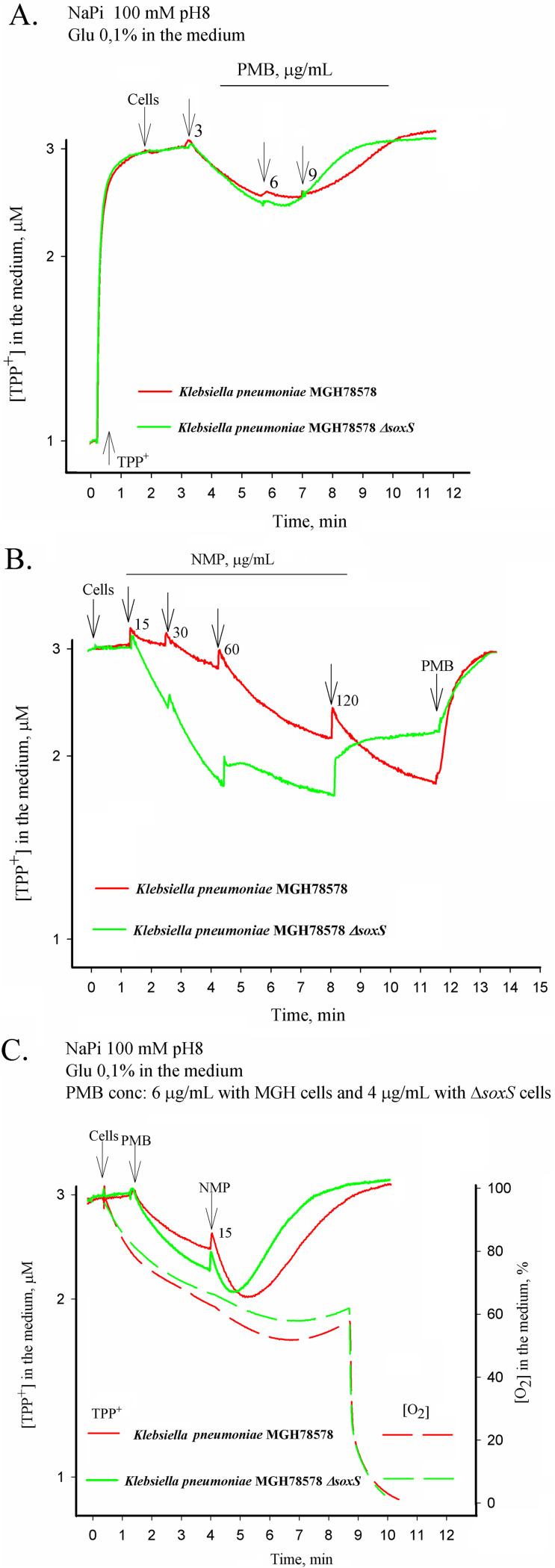
To test our hypothesis, we assayed the efflux activity of wild-type K. pneumoniae MGH78578 and K. pneumoniae MGH78578 ΔsoxS strains by measuring the accumulation of tetraphenylphosphonium (TPP+) ions using previously described protocols (31). We first tested whether the K. pneumoniae MGH78578 ΔsoxS mutant had an intact outer membrane. In this case, both wild-type K. pneumoniae MGH78578 and K. pneumoniae MGH78578 ΔsoxS strains were first exposed to low concentrations of PMB, an antibiotic that causes outer membrane destabilization, and then assayed the accumulation of TPP+. Our results showed that the K. pneumoniae MGH78578 ΔsoxS strain was more susceptible to PMB, and a concentration of 6 μg/ml was sufficient to induce the depolarization of the plasma membrane. In comparison, for the wild-type K. pneumoniae MGH78578, a concentration of PMB of 9 μg/ml was required. Nonetheless, alterations in membrane voltage (maximum amount of TPP+) were similar for both wild-type K. pneumoniae MGH78578 and the isogenic K. pneumoniae MGH78578 ΔsoxS mutant, showing that neither the outer nor the inner plasma membranes were compromised in the K. pneumoniae MGH78578 ΔsoxS strain (Fig. 3A). This finding was supported by our earlier RNA-seq data, which showed that membrane-associated genes that were differentially regulated during 1-(1-naphthyl methyl)-piperazine (NMP) (a chemosensitizer) treatment (32) were not differentially regulated in the soxS regulon.
FIG 3.
TPP+ accumulation in K. pneumoniae MGH78578 and its isogenic mutant MGH78578 ΔsoxS. All measurements were performed in 100 mM NaPi buffer containing 0.1% glucose, pH 8.0. Concentrated cell suspensions were added to obtain OD600 of 1. Final concentrations of NMP (μg/ml) are indicated in panels B and C. The final concentrations of polymixin B (PMB) are indicated in the figure: 3, 6 and 9 μg/ml (A), 50 μg/ml (B), or 6 and 4 μg/ml for wt and ΔsoxS cells, respectively (C).
Next, we investigated whether the efflux pump activity was compromised in the K. pneumoniae MGH78578 ΔsoxS strain compared to that of wild-type K. pneumoniae MGH78578. The aim was to confirm/refute our hypothesis that the impaired pump activity could result in the accumulation of tetracycline within the K. pneumoniae MGH78578 ΔsoxS strain. We previously established that the treatment of K. pneumoniae MGH78578 with NMP destabilized the bacterial outer membrane before efflux pump inhibition and that this phenotype was concentration dependent (32). Hence, we used different concentrations of NMP to test the efflux pump inhibition of K. pneumoniae MGH78578 ΔsoxS cells compared to that of K. pneumoniae MGH78578. Initially, we treated wild-type K. pneumoniae MGH78578 with NMP and assayed the cells for TPP+ accumulation. As expected, in wild-type K. pneumoniae MGH78578, NMP impaired efflux pump activity and induced cytoplasmic TPP+ accumulation at a concentration of 30 μg/ml. However, for the K. pneumoniae MGH78578 ΔsoxS strain, 15 μg/ml NMP was sufficient to inhibit the efflux pump activity and cause TPP+ accumulation (Fig. 3B). The increased sensitivity of the K. pneumoniae MGH78578 ΔsoxS strain to NMP also was observed at 120 μg/ml, where this agent increased the accumulation of TPP+ in K. pneumoniae MGH78578 but induced a partial depolarization of the plasma membrane and leakage of the accumulated cation in the K. pneumoniae MGH78578 ΔsoxS strain. These results show that the efflux pump activity was impaired in the mutant.
To further confirm the effect of the outer membrane destabilization on TPP+ accumulation, we pretreated both K. pneumoniae MGH78578 and K. pneumoniae MGH78578 ΔsoxS strains first with PMB to permeabilize the outer membrane and then retested for NMP-mediated TPP+ accumulation. These data indicated that TPP+ was accumulated at 15 μg/ml for the K. pneumoniae MGH78578 ΔsoxS strain, implying that both efflux pump inhibition and outer membrane destabilization could cause TPP+ accumulation in the bacterial cytoplasm (Fig. 3C). The respiration activity of the K. pneumoniae MGH78578 ΔsoxS strain when measured was very close to that of K. pneumoniae MGH78578. Overall, our experiment describing the accumulation of TPP+ even at very low concentrations of NMP confirmed that the K. pneumoniae MGH78578 ΔsoxS strain exhibited an impaired efflux pump activity, and, as the concentration of NMP increased, membrane stability was affected, resulting in TPP+ accumulation.
Tetracycline is a substrate for the AcrAB-TolC efflux pump. Taking together the downregulation of acrAB-tolC and impaired efflux pump activity in the K. pneumoniae MGH78578 ΔsoxS strain, we conclude that tetracycline accumulates within the cytoplasm of the K. pneumoniae MGH78578 ΔsoxS strain to bactericidal concentrations, making the mutant susceptible to this antibiotic.
K. pneumoniae MGH78578 ΔsoxS strain was avirulent in a zebrafish infection model.
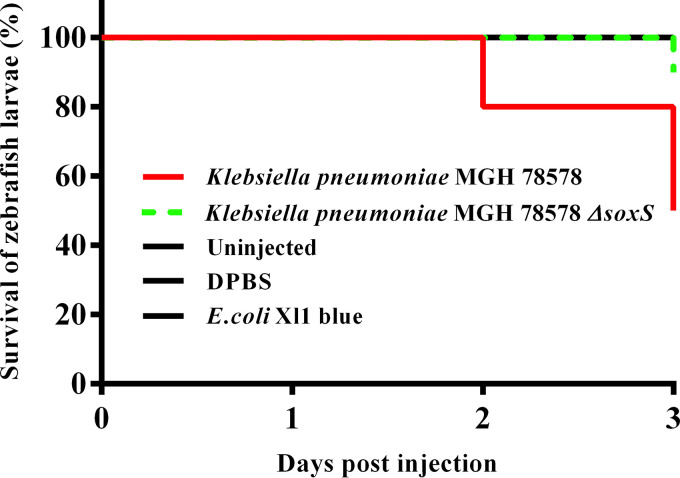
Since SoxS mediates oxidative stress, we sought to characterize the role of soxS to mitigate oxidative stress in an in vivo model. We used a zebrafish (Danio rero) embryo model to investigate the survival of the K. pneumoniae MGH78578 ΔsoxS strain compared with the wild-type K. pneumoniae MGH78578. Zebrafish larvae are generally used as infection models because they are genetically tractable and optically accessible, and they present a fully functional immune system with macrophages and neutrophils that mimic their mammalian counterparts (33). Moreover, a zebrafish larva model was recently used to assay infection associated with K. pneumoniae (34). Wild-type K. pneumoniae MGH78578, K. pneumoniae MGH78578 ΔsoxS, and E. coli Xl1 blue (an avirulent control) bacterial strains and Dulbecco’s phosphate-buffered saline (DPBS; uninoculated control) were directly injected into the caudal vein of 48 hpf (hours postfertilization) zebrafish embryos and survival rates recorded by observing the presence or absence of a heartbeat postinfection. This time point was selected because the innate immune system begins to develop with primitive macrophages at 24 hpf while neutrophils develop later, at 48 hpf (35). Neutrophils are known to use oxidative stress to control bacterial infections in zebrafish larvae. We observed that the K. pneumoniae MGH78578 ΔsoxS strain was inefficient in killing zebrafish larvae compared to the wild type (Fig. 4).
FIG 4.
Survival of larvae in a zebrafish infection model. The survival of larvae infected with the isogenic wild-type K. pneumoniae MGH78578 reduced as the days postinfection increased (red line). The survival of larvae infected with the K. pneumoniae MGH78578 ΔsoxS strain (green line) was maintained at 100% throughout the experiment similar to the negative (avirulent E. coli XI1 blue, uninfected and DPBS) (black line) controls. Log-rank (Mantel-Cox) test, P < 0.0001.
At 1 dpi (days postinfection), a survival rate of 100% was recorded in embryos injected with wild-type K. pneumoniae MGH78578, K. pneumoniae MGH78578 ΔsoxS, and E. coli strains and DPBS. However, at 2 dpi, the survival rate of the embryos injected with wild-type K. pneumoniae MGH78578 decreased to 80%, while the survival rate in those embryos injected with the K. pneumoniae MGH78578 ΔsoxS strain and in the avirulent/uninoculated controls remained unaltered. At 3 dpi, the survival rate dropped further to 50% for the embryos injected with K. pneumoniae MGH78578 while being maintained at 90% for the K. pneumoniae MGH78578 ΔsoxS strain-infected embryos (Fig. 4). The survival rate remained unaltered in the case of E. coli- and DPBS-injected embryos over the time course of infection. Recent studies report that high neutrophil recruitment and zebrafish lethality is observed with K. pneumoniae if directly injected into the blood (34, 36). We anticipate two possibilities for the sensitivity of the K. pneumoniae MGH78578 ΔsoxS strain in zebrafish larvae. First, for vertebrates, extracellular bactericidal action is initiated by neutrophils at a distance by activating the NADPH oxidase-dependent production of superoxide (37). The avirulent phenotype of the K. pneumoniae MGH78578 ΔsoxS strain could be due to the inefficiency in combating the extracellularly produced neutrophil-originated superoxide in the blood. Second, the K. pneumoniae MGH78578 ΔsoxS strain exhibited downregulated expression of acrAB-tolC, which could result in an avirulent phenotype, as seen previously in Salmonella Typhimurium (38).
By impairing soxS, multidrug-resistant K. pneumoniae MGH78578 infections can be treated by tetracycline.
Currently, various strategies are being investigated to mitigate the threat of AMR in bacteria (39). It was thought that restricting the use of a particular antibiotic would restore susceptibility to that compound over time by eliminating the selective advantage, but it has been observed that AMR is persistent over decades (40). Recent research has elaborated on the possibility wherein resistance can be reversed. One strategy here made use of defined drug-adjuvant combinations to reverse resistance so that conventional antibiotics continue to be effective (41). With this broad goal in mind, we endeavored to identify genetic targets that regulate resistance and develop strategies to reverse resistance by inhibiting them. Our transcriptomic and phenotypic data have shown that by inhibiting soxS, susceptibility to tetracycline can be restored in multidrug-resistant K. pneumoniae. We were further interested in investigating whether soxS-mediated tetracycline susceptibility can be demonstrated in an in vivo zebrafish model.
For this, we treated 4-hpf zebrafish embryos with increasing concentrations of tetracycline. At 48 hpf, K. pneumoniae MGH78578, the K. pneumoniae MGH78578 ΔsoxS strain, and DPBS were microinjected into the blood circulation. Postinjection, zebrafish larvae were collected at different time points, and again bacterial counts were enumerated (Fig. 5).
FIG 5.
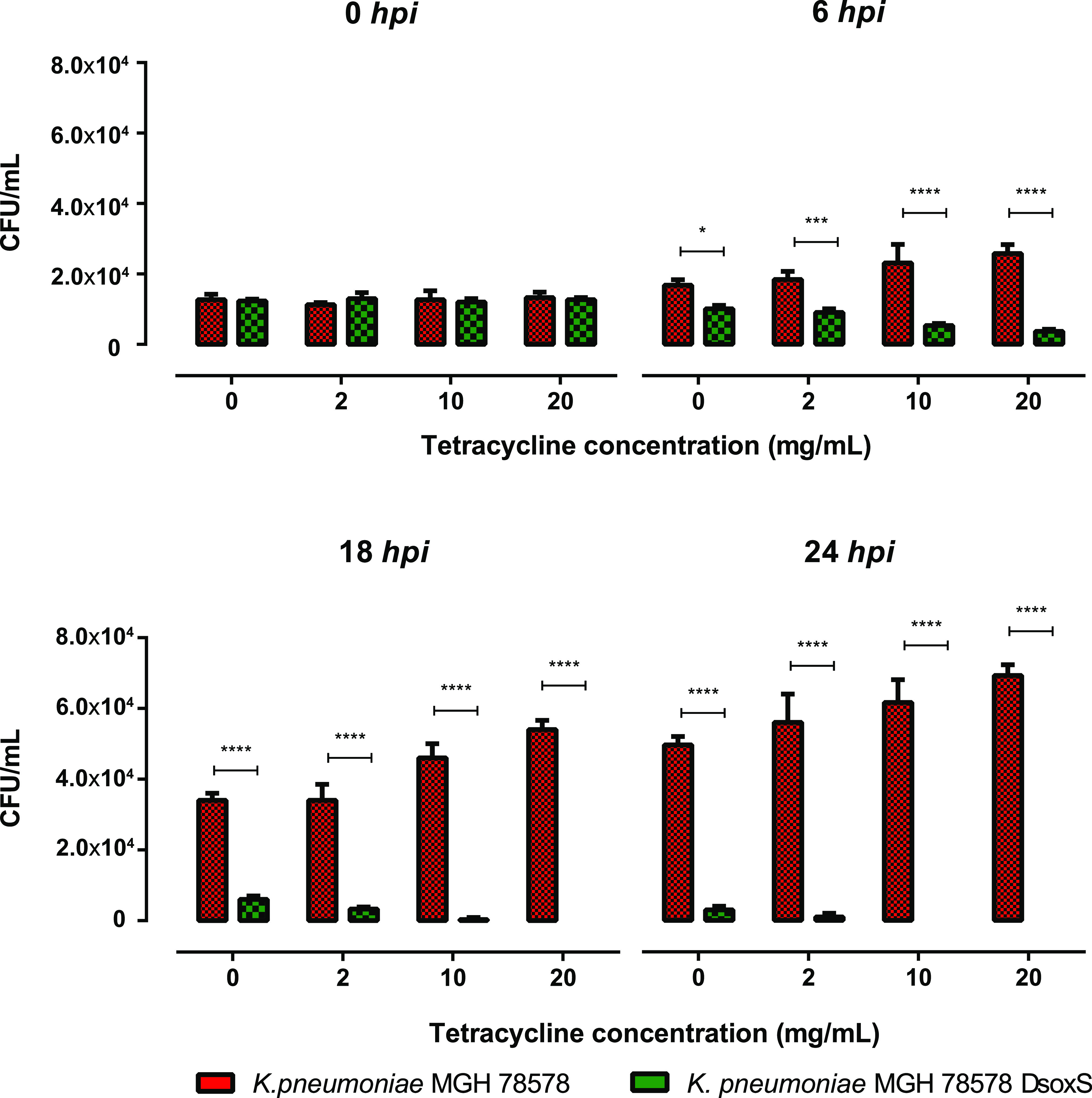
Survival of bacteria in a zebrafish larvae infection model where the larvae are treated with tetracycline. Tetracycline induces ROS generation in the zebrafish larvae. The bar charts show the survival of the different bacterial cultures in the larval blood at 0, 6, 18, and 24 h postinfection and at different concentrations of tetracycline. Error bars represent standard deviations (SD) calculated from 3 independent reads. Significance was determined by two-way ANOVA and Sidak's multiple-comparison test, comparing the different tetracycline concentrations with the control group for each time point.
It was shown recently that exposure to tetracycline induced ROS production in zebrafish larvae (42). Thus, we hypothesized that the K. pneumoniae MGH78578 ΔsoxS strain will be impaired in its ability to survive in tetracycline-treated zebrafish larvae due to increased sensitivity to either ROS production in tetracycline-treated larvae or tetracycline alone. It is also possible that healthy zebrafish larvae could clear the K. pneumoniae MGH78578 ΔsoxS strain from the system due to normal exposure to peroxides synthesized from neutrophils. Confirming our hypothesis, the K. pneumoniae MGH78578 ΔsoxS strain was completely cleared from the tetracycline-treated zebrafish larvae in 24 h. However, the K. pneumoniae MGH78578 ΔsoxS strain was cleared even in untreated zebrafish larvae, confirming that the selective advantage was lost in the bacterial mutant, making it susceptible to the immune system of zebrafish larvae (Fig. 5). It should also be noted that the clearance was much more marked in tetracycline-treated larvae, suggesting that the K. pneumoniae MGH78578 ΔsoxS strain was cleared from the system due to a cumulative effect of both immune system- and tetracycline-induced ROS production. Overall, we show that soxS can be used as a genetic target to treat multidrug-resistant K. pneumoniae infections.
Conclusions.
Apart from elucidating the PQ oxidative stress regulon and the oxidative SoxS regulon, we propose that a combination of tetracycline and an SoxS inhibitor can be used to treat infections associated with MDR K. pneumoniae. A decade ago it was shown that bactericidal antibiotics kill bacteria by unleashing intracellular oxidative stress, a phenomenon independent of the antibacterial action mechanism of antibiotics (24, 43, 44), that harnesses ROS as an effective antibacterial strategy (45). At least in a zebrafish embryo model system, tetracycline is known to induce host oxidative stress (42). Thus, we hypothesized that if tetracycline was used in the presence of an SoxS inhibitor, the overall effect produced was bactericidal, making the tetracycline-SoxS inhibitor combination a potential target for antibiotic drug discovery. Since the target action mechanism is metabolic dysfunction (oxidative stress inhibition), such an approach could work on bacteria regardless of its expressed drug resistance profile.
However, the major limitation of our study is that it is based on a single bacterial strain and its isogenic mutant, and observations recorded in K. pneumoniae MGH78578 may not necessarily apply to other K. pneumoniae strains or other pathogens in a clinical setting. Ours is a proof-of-concept paper, and it will be worthwhile to test if our observations can translate to other isolates of K. pneumoniae and other MDR pathogens, including Salmonella Typhimurium, E. coli, and others. Second, our approach involves oxidative stress inhibition, a mechanism that might not apply if any antibiotic does not employ oxidative stress as a killing mechanism. Nonetheless, since the oxidative stress response is a global bacterial response, the cross-genus application of our predictions, as reported here, may hold some merit. Overall, by providing an interesting target for antibiotic drug discovery, our results address the immediate concern of antimicrobial resistance in pathogens of importance to human health while providing a proof of concept for an approach that requires further experimental investigation to achieve the therapeutic objective.
MATERIALS AND METHODS
Bacterial strain.
MDR Klebsiella pneumoniae MGH78578 (ATCC 700721) was isolated from a sputum sample in 1994 and was purchased from the American Type Culture Collection. This strain was selected mainly because it is a multidrug-resistant type strain (46), and its drug resistance profile was recently published (22). Moreover, the efflux pumps present in this strain are well characterized (47, 48). Further, the whole-genome sequence of this strain is available in NCBI (reference sequence NC_009648.1), which was convenient for mapping RNA-seq data. This bacterium was grown in Müeller-Hinton broth (MHB) and Müeller-Hinton agar (MHA) (Sigma, Dublin, Ireland).
Phenotypic assay (OmniLog).
The comparison of K. pneumoniae MGH78578 WT with the ΔsoxS mutant was evaluated using the OmniLog (Biolog, Inc., Hayward, CA) phenotypic microarray. Microplates PM1 through PM20, except PM5, were used. These plates contain several carbon, nitrogen, sulfur, and phosphorous substrates, ions, osmolytes, and chemicals at different concentrations and pH (49). Klebsiella pneumoniae MGH78578 WT and ΔsoxS strains were grown at 37°C on LB agar plates, and several colonies were picked with a sterile cotton swab and suspended in 15 ml IF-0 until a cell density of 42% transmittance (T) was reached (measured using a Biolog turbidimeter). Each 15-ml suspension was then added to 75 ml of physiological solution IF-0 containing dye A, used to inoculate PM plates 1 and 2. PM plates 3 to 8 were inoculated with IF-0 solution containing sodium pyruvate as a carbon source. PM9 to -20 were inoculated with the physiological solution IF-10. One hundred microliters of each mixture was inoculated into each well of the microplates. All PM microplates were incubated at 37°C in an OmniLog reader and monitored for 72 h. Data were analyzed using DuctApe software v 0.17.4 (50). Each strain was analyzed in duplicate. Results are present in Table S2 in the supplemental material.
Isolation of RNA from oxidatively stressed bacterial cells.
Before RNA isolation, wild-type Klebsiella pneumoniae MGH78578 and the K. pneumoniae MGH78578 ΔsoxS strain were grown until mid-exponential state (MEP) by following an earlier standardized protocol (22). MEP-grown bacterial cells were treated with paraquat (7.81 μM) for 30 min to generate oxidative stress conditions. RNA was then extracted from both oxidative stressed and MEP-grown (control) cells using the Qiagen RNeasy minikit by following the manufacturer’s guidelines. Contaminating DNA was removed from the RNA sample using the Turbo DNase I kit (Thermo Fischer Scientific). RNA was then quantified using both Qubit RNA broad-range assay and the NanoDrop device.
Sequencing RNA isolated from oxidatively stressed bacterial cells.
The library preparation and subsequent sequencing were carried out commercially at the Center For Genomic Research, University of Liverpool. The Ribo-Zero rRNA removal kit for bacteria (Illumina, San Diego, CA) was used to carry out the depletion of ribosomal DNA according to the manufacturer’s instructions. Libraries were created using a NEBNext Ultra directional RNA library prep kit (New England BioLabs, Frankfurt, Germany). Pooled libraries were loaded on the cBot (Illumina, San Diego, CA), and cluster generation was performed according to the manufacturer’s instructions. Single-end sequencing using 125-bp read length was performed on an Illumina HiSeq 2500 platform (Illumina HiSeq control software 2.2.38) using an Illumina HiSeq Flow Cell v4 and TruSeq SBS kit v4 (Illumina). Raw sequencing read data were processed using RTA version 1.18.61 and CASAVA 1.8.4 to generate FASTQ files. Genomic cDNA libraries were prepared using the TruSeq stranded total RNA library prep kit (Illumina, San Diego, CA) with Ribo-Zero to deplete rRNA. An average of 1.48 Gbp of raw sequence data was obtained per sample in 125-bp single-end reads.
Mapping of sequenced reads.
The sequence quality of the RNA-seq reads was analyzed using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Sequence reads were aligned and mapped against the reference genome of K. pneumoniae MGH78578 (reference sequence NC_009648.1) using Segemehl with default mapping parameters (22, 51), and uniquely mapped reads were used and considered for the differential gene expression computational analysis. Read counts (number of reads that aligned to a specific gene) for each gene were quantified using custom Perl scripts.
Computational analysis of RNA-seq data.
Computational analysis of RNA-seq data were performed using R (version 3.5.2; https://www.r-project.org/). To calculate the expression level of genes, the raw read counts were normalized using the VOOM function (21) in the limma package (52). More specifically, counts were converted to log2 counts per million (log2 CPM), quantile normalized, and precision weighted using the VOOM function. A linear model was then fitted to each gene, and empirical Bayes-moderated t-statistics and its corresponding P values were used to assess differences in expression (21, 53). To account for multiple comparisons, Benjamini-Hochberg-corrected P values were computed. As reads for duplicated coding genes (paralogs) or duplicated small RNAs cannot be mapped unequivocally, these genes appear in the analysis as unmapped. The sequence reads can be visualized in the Integrated Genome Browser (version 9.0.0) (54). The read depth was adjusted with the cDNA library with the lowest number of reads (55). RNA sequencing data were analyzed using the following fold change parameters: highly upregulated (>4), upregulated (2- to 4-fold), no change in expression (0.5- to 2-fold), downregulated (0.25- to 0.5-fold), and highly downregulated (less than 0.25-fold).
Construction of the K. pneumoniae MGH78578 ΔsoxS mutant.
A modified λ-Red system was used to construct an in-frame deletion in multidrug-resistant K. pneumoniae MGH78578 (56). Here, three plasmids are employed. The first, plasmid pIJ773, serves as a template to amplify the apramycin resistance gene, aac(3)IV, and flanking FLP recombination target (FRT) sites. The second plasmid, pACBSR-Hyg, contains the λ-Red system comprising beta, gam, and exo genes, which are under the control of an arabinose-inducible promoter and facilitate homologous recombination between the knockout cassette and the target locus in the chromosome. The third plasmid, pFLP-Hyg, contains the FLP recombinase, which was used to excise the apramycin selection marker from the chromosome via the FRT sites. The antibiotic apramycin was used to select plasmid pIJ773, while hygromycin was used to select both plasmids pACBSR-Hyg and pFLP-Hyg. K. pneumoniae MGH78578 was susceptible to both apramycin and hygromycin.
Plasmid pACBSR-Hyg was first introduced into wild-type K. pneumoniae MGH78578 by electroporation. An overnight culture of the bacteria was reinoculated into 100 ml Luria-Bertani (LB) broth at 220-rpm aeration and 30°C temperature until an optical density at 600 nm (OD600) of 0.6 to 0.8 was reached. The cells were washed twice with 50 ml ice-cold 10% (vol/vol) glycerol and resuspended in the residual glycerol solution after the final wash. A 50-μl aliquot of the dense suspension was then mixed with 200 to 400 ng of plasmid DNA and electroporated at 2,500 mV. Bacterial cells were revived in SOC medium, which was added immediately after electroporation, and K. pneumoniae MGH78578[pACBSR-Hyg] was selected by plating on low-salt LB plates containing hygromycin. The knockout cassette consisted of three distinct regions: aac(3)IV, FRT sites that flanked aac(3)IV, and 60-bp regions homologous to the soxS gene were amplified using PCR from the plasmid pIJ773 as the template. Competent K. pneumoniae MGH78578[pACBSR-Hyg] was prepared by growing the cells in low-salt LB with 1 M l-arabinose and 100 μg/ml hygromycin, with washing in ice-cold 10% (vol/vol) glycerol as described above. The PCR-amplified knockout cassette was electroporated into competent K. pneumoniae MGH78578[pACBSR-Hyg], and cells with successful recombination events were selected by plating in LB with apramycin and incubating overnight at 37°C. K. pneumoniae MGH78578 soxS::FRT-aac(3)IV-FRT cells were identified and confirmed using PCR primers targeting regions that flanked the soxS region. Competent K. pneumoniae MGH78578 soxS::FRT-aac(3)IV-FRT cells were prepared and electroporated with plasmid pFLP-Hyg to excise the inserted knockout cassette, and the K. pneumoniae MGH78578 ΔsoxS cells were confirmed using PCR and sequencing. The sequences of all primers used in the experiment are provided in Table S1, worksheet 1 (WS1), in the supplemental material.
Isolation of RNA for qRT-PCR.
Wild-type K. pneumoniae MGH78578 was grown to mid-exponential phase (MEP) at 37°C in Müeller-Hinton broth. Cells were then treated with paraquat at different concentrations (0, 3.905, 7.81, 200, and 500 μM) or similarly with tetracycline (0, 0.5, 10, 100, and 500 μg/ml) for 30 min, and RNA was then extracted. All assays were run in triplicate. Under all conditions, RNA was extracted using an Qiagen RNeasy minikit by following the manufacturer's guidelines. Any contaminating DNA was removed from the RNA sample using the Turbo DNase I kit (Thermo Fischer Scientific). Purified RNA was subsequently quantified using both Qubit RNA broad-range assay and NanoDrop device.
Two-step RT-qPCR.
The reverse transcriptase reaction was carried out on RNA purified earlier from K. pneumoniae MGH78578 under the same conditions mentioned earlier. A high-capacity RNA-to-cDNA preparation kit (Sigma) was used by following the manufacturer’s guidelines. A negative control devoid of RT enzyme was also included. qPCR was then performed by following the prime-time gene expression master mix protocol (IDT, Leuven, Belgium) in an Eppendorf Mastercycler RealPlex ep gradient S (Eppendorf, Arlington, United Kingdom) according to the manufacturer’s instructions. Samples were run for 3 biological replicates, each of which had three technical replicates. Data were analyzed using RealPlex software. The relative fold increases in expression levels (changes in threshold cycle [ΔCT]) were normalized based on the gene expression levels of the housekeeping gene rpoB relative to the soxS gene. Comparative quantification was carried out using the ΔΔCT approach. The sequences of all primers used in the experiment are provided in Table S1, WS1.
Determination of MBC.
Previously, MIC values for paraquat (PQ), colistin (COL), tetracycline (TET), gentamicin (CN), kanamycin (KM), and cefotaxime (CTX) were determined in triplicate using a 96-well microtiter plate 2-fold broth microdilution method (22). The range of concentrations employed was 1 to 512 μg/ml for all antibiotics, excluding colistin, for which a concentration range of 0.03125 to 16 μg/ml and paraquat of 0.97 to 500 μM was utilized. Overnight LB cultures of K. pneumoniae MGH78578 and the K. pneumoniae MGH78578 ΔsoxS strain were diluted in sterile phosphate-buffered saline to 105 CFU/ml. A 96-well plate was used to prepare 2-fold serial dilutions of each antibiotic for MHB and MBC determination of K. pneumoniae MGH78578 and the K. pneumoniae MGH78578 ΔsoxS strain against each compound. A volume of 5 μl of the 105 CFU/ml bacterial culture was then transferred to separate wells containing various concentrations of the compounds to be tested. These plates were then incubated at 37°C for 16 to 18 h according to European Committee on Antimicrobial Susceptibility Testing (EUCAST) 2018 guidelines. Triplicate MBC values for each antibiotic tested were determined using MHB in a 96-well microtiter plate. A steel inoculator was employed to transfer inoculum from the 96-well plate as described above to a fresh 96-well plate containing MHB without any of the antibiotics to be tested. These plates were then incubated at 37°C for 16 to 18 h, following which the MBC values were recorded.
Electrochemical measurement experiments.
The efflux activity of K. pneumoniae MGH78578 and K. pneumoniae MGH78578 ΔsoxS cells was assayed measuring the accumulation of tetraphenylphosphonium (TPP+) ions. Overnight cultures of K. pneumoniae were grown in Luria-Bertani broth containing 0.5% NaCl, diluted 3:100 in fresh medium, and the incubation was continued until the OD600 reached 1.0. The cells were collected by centrifugation at 4°C for 10 min at 3,000 × g. The pelleted cells were resuspended in 100 mM sodium phosphate buffer, pH 8, to obtain 1.4 × 1011 CFU/ml. Concentrated cell suspensions were kept on ice until used but not longer than 3 h.
Changes in TPP+ concentration in the suspensions of thermostated and magnetically stirred cells were monitored using TPP+-selective electrodes as previously described (31, 57). Experiments were performed at 37°C in 100 mM sodium phosphate buffer, pH 8, containing 0.1% glucose. The OD612 of the cell suspension during measurements was 1.
Zebrafish line maintenance, infection, and microinjection experiments.
Zebrafish (Danio rerio) used in this study were wik lines. Adult fish were kept on a 14-h/10-h light/dark cycle at pH 7.5 and 27°C. Eggs were obtained from natural spawning adult fish, which were set up pairwise in individual breeding tanks. Embryos were raised in petri dishes containing E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) supplemented with 0.3 μg/ml methylene blue at 28°C. From 24 hpf, 0.003% 1-phenyl-2-thiourea (PTU) was added to prevent melanin synthesis. The staging of embryos was performed as explained earlier (58).
Microinjections were performed using borosilicate glass microcapillary injection needles (1-mm outer diameter by 0.78-mm inner diameter; 1210332; Science Products) and a PV830 Pneumatic PicoPump (World Precision Instruments). The 48-hpf embryos were manually dechorionated and anesthetized with 200 mg/liter buffered tricine (MS-222; Sigma) before injection. Subsequently, embryos were aligned on an agar plate and injected with 12,000 CFU in a 1- to 4-μl volume of a bacterial suspension in DPBS directly into the blood circulation (caudal vein, n = three sets of 10). Before injection, the volume of the injection suspension was adjusted by injecting a droplet into mineral oil and measuring its approximate diameter over a micrometer scale bar. The number of CFU injected was determined by injection of bacterial suspension into a DPBS droplet on the agar plate. Following injections, injected embryos were allowed to recover in a petri dish with fresh E3 medium for 15 min. To monitor infection kinetics and for survival assays, embryos were transferred into 24-well plates (one embryo per well) containing 1 ml E3 medium per well, incubated at 28°C, and observed for survival under a stereomicroscope twice a day. For survival assays after infection, the number of dead larvae was determined visually based on the absence of a heartbeat. Kaplan-Meier survival analysis and statistics for experiments with zebrafish were done with GraphPad Prism 7 (GraphPad Software). Experiments were performed in triplicate.
Zebrafish tetracycline exposure experiments.
Wild-type (wik strain) zebrafish embryos were used for this study. Four-hpf embryos from three different pairs were examined under stereomicroscope for normal development, and embryos that had reached the blastula stage were selected for the following experiments. Embryos (n = three sets of 10 for each tetracycline concentration used) were randomly transferred into each well of 24-well plates containing 2 ml of E3 medium. A series of tetracycline concentrations (0, 2, 10, and 20 μg/liter) were applied and maintained until 48 hpf at 28°C. The solutions were changed once every 24 h. At 48 hpf, embryos were manually dechorionated, anesthetized, and microinjected directly into the blood circulation as mentioned above. Following injections, injected embryos were allowed to recover in a petri dish with fresh E3 medium for 15 min and subsequently transferred into each well of 24-well plates containing fresh E3 medium and the respective concentration of tetracycline. Embryos or larvae were collected at each time point (0, 6, 18, and 24 hpi) and independently treated for bacterial enumeration. Significance was determined with GraphPad Prism 7 (GraphPad Software) by applying a two-way analysis of variance (ANOVA) and Sidak's multiple-comparison test, comparing the different tetracycline concentrations with the control group for each time point.
Ethics statement.
This study was performed by following the principles and recommendations of the “Ordinance on laboratory animal husbandry, the production of genetically modified animals and the methods of animal experimentation; Animal Experimentation Ordinance” (SR 455.163, 12 April 2010), Swiss Federal Food Safety and Veterinary Office (FSVO/BLV). The maximum age reached by the embryos during experimentation was 5 days postfertilization (dpf), for which no license is required from the cantonal veterinary office in Switzerland, since such embryos will not have reached the free-feeding stage. Husbandry and breeding of the adult zebrafish were performed under the supervision of Stephan Neuhauss, Institute for Molecular Life Sciences, University of Zurich, Zurich, Switzerland (Cantonal Veterinary Office of Zurich, husbandry license no. 150). All animal protocols used were in compliance with internationally recognized standards as well as with Swiss legal ethical guidelines for the use of fish in biomedical research.
Data availability.
All the RNA sequence data generated in the study were deposited in the National Center for Biotechnological Information–Gene Expression Omnibus and are available under the accession number GSE146844. Postanalysis, the differential expression levels of all the genes are given in Table S1 with three worksheets, WS-1, WS-2, and WS-3.
ACKNOWLEDGMENTS
We gratefully acknowledge Enterprise Ireland (IP 2015 0380) for funding J.A. and S.S. J.A. also acknowledges financial support through the research grant 11/F/051 provided by the Department of Agriculture, Food and the Marine (DAFM), Ireland. The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
J.A., S.F., and S.S. conceived and supervised the experiments. J.A., K.D., and Y.C. performed the growth, PQ induction, RNA-seq, KO, phenotypic microarray, and RT-qPCR experiments. S.K.S. and S.N. carried out the bioinformatics analysis of the RNA-seq data sets. S.A.S. and R.D. carried out the TPP+ assays. A.E. and A.L. carried out the zebrafish embryo assays. J.A., S.D., S.F., and S.S. wrote this publication, to which all other authors contributed. All authors read and approved the manuscript.
We have no competing interests to declare.
Dedicated to the memory of Leonard Amaral, who taught us much about the biology and importance of efflux pumps.
Footnotes
This article is a direct contribution from Séamus Fanning, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Christopher Elkins, Centers for Disease Control and Prevention; Alessandra Carattoli, Sapienza University of Rome; and Qijing Zhang, Iowa State University.
Citation Anes J, Dever K, Eshwar A, Nguyen S, Cao Y, Sivasankaran SK, Sakalauskaitė S, Lehner A, Devineau S, Daugelavičius R, Stephan R, Fanning S, Srikumar S. 2021. Analysis of the oxidative stress regulon identifies soxS as a genetic target for resistance reversal in multidrug-resistant Klebsiella pneumoniae. mBio 12:e00867-21. https://doi.org/10.1128/mBio.00867-21.
Contributor Information
Séamus Fanning, Email: sfanning@ucd.ie.
Shabarinath Srikumar, Email: ssrikumar@uaeu.ac.ae.
Eric Brown, McMaster.
Karen Bush, Indiana University Bloomington.
REFERENCES
- 1.Cabiscol E, Tamarit J, Ros J. 2000. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int Microbiol 3:3–8. [PubMed] [Google Scholar]
- 2.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. 2003. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J 17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 3.Seo SW, Kim D, Szubin R, Palsson BO. 2015. Genome-wide reconstruction of OxyR and SoxRS transcriptional regulatory networks under oxidative stress in Escherichia coli K-12 MG1655. Cell Rep 12:1289–1299. doi: 10.1016/j.celrep.2015.07.043. [DOI] [PubMed] [Google Scholar]
- 4.Lushchak VI. 2001. Oxidative stress and mechanisms of protection against it in bacteria. Biochemistry 66:476–489. doi: 10.1023/a:1010294415625. [DOI] [PubMed] [Google Scholar]
- 5.Zheng M, Aslund F, Storz G. 1998. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 6.Nunoshiba T, Hidalgo E, Li Z, Demple B. 1993. Negative autoregulation by the Escherichia coli SoxS protein: a dampening mechanism for the soxRS redox stress response. J Bacteriol 175:7492–7494. doi: 10.1128/JB.175.22.7492-7494.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hidalgo E, Bollinger JM, Bradley TM, Walsh CT, Demple B. 1995. Binuclear [2Fe-2S] clusters in the Escherichia coli soxR protein and role of the metal centers in transcription. J Biol Chem 270:20908–20914. doi: 10.1074/jbc.270.36.20908. [DOI] [PubMed] [Google Scholar]
- 8.Ding H, Hidalgo E, Demple B. 1996. The redox state of the [2Fe-2S] clusters in SoxR protein regulates its activity as a transcription factor. J Biol Chem 271:33173–33175. doi: 10.1074/jbc.271.52.33173. [DOI] [PubMed] [Google Scholar]
- 9.Pomposiello PJ, Demple B. 2000. Identification of SoxS-regulated genes in Salmonella enterica serovar typhimurium. J Bacteriol 182:23–29. doi: 10.1128/JB.182.1.23-29.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z, Demple B. 1994. SoxS, an activator of superoxide stress genes in Escherichia coli. Purification and interaction with DNA. J Biol Chem 269:18371–18377. doi: 10.1016/S0021-9258(17)32317-7. [DOI] [PubMed] [Google Scholar]
- 11.Amabile-Cuevas CF, Demple B. 1991. Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res 19:4479–4484. doi: 10.1093/nar/19.16.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenberg JT, Monach P, Chou JH, Josephy PD, Demple B. 1990. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc Natl Acad Sci U S A 87:6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liochev SI, Benov L, Touati D, Fridovich I. 1999. Induction of the soxRS regulon of Escherichia coli by superoxide. J Biol Chem 274:9479–9481. doi: 10.1074/jbc.274.14.9479. [DOI] [PubMed] [Google Scholar]
- 14.Liochev SI, Fridovich I. 1992. Fumarase C, the stable fumarase of Escherichia coli, is controlled by the soxRS regulon. Proc Natl Acad Sci U S A 89:5892–5896. doi: 10.1073/pnas.89.13.5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liochev SI, Hausladen A, Beyer WF, Fridovich I. 1994. NADPH: ferredoxin oxidoreductase acts as a paraquat diaphorase and is a member of the soxRS regulon. Proc Natl Acad Sci U S A 91:1328–1331. doi: 10.1073/pnas.91.4.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liochev SI, Hausladen A, Fridovich I. 1999. Nitroreductase A is regulated as a member of the soxRS regulon of Escherichia coli. Proc Natl Acad Sci U S A 96:3537–3539. doi: 10.1073/pnas.96.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farr SB, Kogoma T. 1991. Oxidative stress responses in Escherichia coli and Salmonella Typhimurium. Microbiol Rev 55:561–585. doi: 10.1128/MR.55.4.561-585.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pendleton JN, Gorman SP, Gilmore BF. 2013. Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther 11:297–308. doi: 10.1586/eri.13.12. [DOI] [PubMed] [Google Scholar]
- 19.Abrashev R, Krumova E, Dishliska V, Eneva R, Engibarov S, Abrashev I, Maria A. 2011. Differential effect of paraquat and hydrogen peroxide on the oxidative stress response in Vibrio cholerae non O1 26/06. Biotechnol Biotechnological Equip 25:72–76. doi: 10.5504/BBEQ.2011.0118. [DOI] [Google Scholar]
- 20.Haas BJ, Chin M, Nusbaum C, Birren BW, Livny J. 2012. How deep is deep enough for RNA-Seq profiling of bacterial transcriptomes? BMC Genomics 13:734. doi: 10.1186/1471-2164-13-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Law CW, Chen Y, Shi W, Smyth GK. 2014. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anes J, Martins M, Fanning S. 2018. Reversing antimicrobial resistance in multidrug-resistant Klebsiella pneumoniae of clinical origin using 1–(1-naphthylmethyl)-piperazine. Microb Drug Resist doi: 10.1089/mdr.2017.0386. [DOI] [PubMed] [Google Scholar]
- 23.Griffith KL, Becker SM, Wolf RE, Jr.. 2005. Characterization of TetD as a transcriptional activator of a subset of genes of the Escherichia coli SoxS/MarA/Rob regulon. Mol Microbiol 56:1103–1117. doi: 10.1111/j.1365-2958.2005.04599.x. [DOI] [PubMed] [Google Scholar]
- 24.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 25.Koutsolioutsou A, Peña-Llopis S, Demple B. 2005. Constitutive soxR mutations contribute to multiple-antibiotic resistance in clinical Escherichia coli isolates. Antimicrob Agents Chemother 49:2746–2752. doi: 10.1128/AAC.49.7.2746-2752.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Webber MA, Piddock LJ. 2001. Absence of mutations in marRAB or soxRS in acrB-overexpressing fluoroquinolone-resistant clinical and veterinary isolates of Escherichia coli. Antimicrob Agents Chemother 45:1550–1552. doi: 10.1128/AAC.45.5.1550-1552.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mechesso AF, Park SC. 2020. Tylosin exposure reduces the susceptibility of Salmonella Typhimurium to florfenicol and tetracycline. BMC Vet Res 16:22. doi: 10.1186/s12917-020-2246-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Wang Q, Wang R, Zhang Y, Wang X, Wang H. 2017. Global regulator SoxR is a negative regulator of efflux pump gene expression and affects antibiotic resistance and fitness in Acinetobacter baumannii. Medicine 96:e7188. doi: 10.1097/MD.0000000000007188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bratu S, Landman D, George A, Salvani J, Quale J. 2009. Correlation of the expression of acrB and the regulatory genes marA, soxS and ramA with antimicrobial resistance in clinical isolates of Klebsiella pneumoniae endemic to New York City. J Antimicrob Chemother 64:278–283. doi: 10.1093/jac/dkp186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anes J, McCusker MP, Fanning S, Martins M. 2015. The ins and outs of RND efflux pumps in Escherichia coli. Front Microbiol 6:587. doi: 10.3389/fmicb.2015.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mikalayeva V, Sakalauskaitė S, Daugelavičius R. 2017. Interaction of ethidium and tetraphenylphosphonium cations with Salmonella enterica cells. Medicina 53:122–130. doi: 10.1016/j.medici.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 32.Anes J, Sivasankaran SK, Muthappa DM, Fanning S, Srikumar S. 2019. Exposure to sub-inhibitory concentrations of the chemosensitizer 1–(1-naphthylmethyl)-piperazine creates membrane destabilization in multi-drug resistant Klebsiella pneumoniae. Front Microbiol 10:92. doi: 10.3389/fmicb.2019.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torraca V, Mostowy S. 2018. Zebrafish infection: from pathogenesis to cell biology. Trends Cell Biol 28:143–156. doi: 10.1016/j.tcb.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcoleta AE, Varas MA, Ortiz-Severín J, Vásquez L, Berríos-Pastén C, Sabag AV, Chávez FP, Allende ML, Santiviago CA, Monasterio O, Lagos R. 2018. Evaluating different virulence traits of Klebsiella pneumoniae using Dictyostelium discoideum and Zebrafish Larvae as host models. Front Cell Infect Microbiol 8:30. doi: 10.3389/fcimb.2018.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meeker ND, Trede NS. 2008. Immunology and zebrafish: spawning new models of human disease. Dev Comp Immunol 32:745–757. doi: 10.1016/j.dci.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 36.Cheepurupalli L, Raman T, Rathore SS, Ramakrishnan J. 2017. Bioactive molecule from Streptomyces sp. mitigates MDR Klebsiella pneumoniae in zebrafish infection model. Front Microbiol 8:614. doi: 10.3389/fmicb.2017.00614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phan QT, Sipka T, Gonzalez C, Levraud JP, Lutfalla G, Nguyen-Chi M. 2018. Neutrophils use superoxide to control bacterial infection at a distance. PLoS Pathog 14:e1007157. doi: 10.1371/journal.ppat.1007157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang-Kan X, Blair JMA, Chirullo B, Betts J, La Ragione RM, Ivens A, Ricci V, Opperman TJ, Piddock LJV. 2017. Lack of AcrB efflux function confers loss of virulence on. mBio 8:e00968-17. doi: 10.1128/mBio.00968-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghosh C, Sarkar P, Issa R, Haldar J. 2019. Alternatives to conventional antibiotics in the era of antimicrobial resistance. Trends Microbiol 27:323–338. doi: 10.1016/j.tim.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 40.Baym M, Stone LK, Kishony R. 2016. Multidrug evolutionary strategies to reverse antibiotic resistance. Science 351:aad3292. doi: 10.1126/science.aad3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tyers M, Wright GD. 2019. Drug combinations: a strategy to extend the life of antibiotics in the 21st century. Nat Rev Microbiol 17:141–155. doi: 10.1038/s41579-018-0141-x. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Q, Cheng J, Xin Q. 2015. Effects of tetracycline on developmental toxicity and molecular responses in zebrafish (Danio rerio) embryos. Ecotoxicology 24:707–719. doi: 10.1007/s10646-015-1417-9. [DOI] [PubMed] [Google Scholar]
- 43.Kohanski MA, Dwyer DJ, Collins JJ. 2010. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol 8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dwyer DJ, Belenky PA, Yang JH, MacDonald IC, Martell JD, Takahashi N, Chan CT, Lobritz MA, Braff D, Schwarz EG, Ye JD, Pati M, Vercruysse M, Ralifo PS, Allison KR, Khalil AS, Ting AY, Walker GC, Collins JJ. 2014. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc Natl Acad Sci U S A 111:E2100–E2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Memar MY, Ghotaslou R, Samiei M, Adibkia K. 2018. Antimicrobial use of reactive oxygen therapy: current insights. Infect Drug Resist 11:567–576. doi: 10.2147/IDR.S142397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogawa W, Li DW, Yu P, Begum A, Mizushima T, Kuroda T, Tsuchiya T. 2005. Multidrug resistance in Klebsiella pneumoniae MGH78578 and cloning of genes responsible for the resistance. Biol Pharmaceutical Bull 28:1505–1508. doi: 10.1248/bpb.28.1505. [DOI] [PubMed] [Google Scholar]
- 47.Li DW, Onishi M, Kishino T, Matsuo T, Ogawa W, Kuroda T, Tsuchiya T. 2008. Properties and expression of a multidrug efflux pump AcrAB-KocC from Klebsiella pneumoniae. Biol Pharmaceutical Bull 31:577–582. doi: 10.1248/bpb.31.577. [DOI] [PubMed] [Google Scholar]
- 48.Ogawa W, Onishi M, Ni R, Tsuchiya T, Kuroda T. 2012. Functional study of the novel multidrug efflux pump KexD from Klebsiella pneumoniae. Gene 498:177–182. doi: 10.1016/j.gene.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 49.Bochner BR, Gadzinski P, Panomitros E. 2001. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res 11:1246–1255. doi: 10.1101/gr.186501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Galardini M, Mengoni A, Biondi EG, Semeraro R, Florio A, Bazzicalupo M, Benedetti A, Mocali S. 2014. DuctApe: a suite for the analysis and correlation of genomic and OmniLog phenotype microarray data. Genomics 103:1–10. doi: 10.1016/j.ygeno.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 51.Hoffmann S, Otto C, Kurtz S, Sharma CM, Khaitovich P, Vogel J, Stadler PF, Hackermuller J. 2009. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput Biol 5:e1000502. doi: 10.1371/journal.pcbi.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 54.Nicol JW, Helt GA, Blanchard SG, Raja A, Loraine AE. 2009. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skinner ME, Uzilov AV, Stein LD, Mungall CJ, Holmes IH. 2009. JBrowse: a next-generation genome browser. Genome Res 19:1630–1638. doi: 10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang CJ, Wang ZC, Huang HY, Huang HD, Peng HL. 2013. YjcC, a c-di-GMP phosphodiesterase protein, regulates the oxidative stress response and virulence of Klebsiella pneumoniae CG43. PLoS One 8:e66740. doi: 10.1371/journal.pone.0066740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bondarenko OM, Sihtmäe M, Kuzmičiova J, Ragelienė L, Kahru A, Daugelavičius R. 2018. Plasma membrane is the target of rapid antibacterial action of silver nanoparticles in Escherichia coli and Pseudomonas aeruginosa. Int J Nanomed 13:6779–6790. doi: 10.2147/IJN.S177163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. 1995. Stages of embryonic development of the zebrafish. Dev Dyn 203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(WS1) RNA-seq mapping details. This table provides details of the RNA-seq reads mapped against different regions of the K. pneumoniae MGH78578 genome are. A complete list of all primers used in the experiments is also provided. (WS2) The table presents data describing gene expression patterns of Klebsiella pneumoniae MGH75878 (MGHPQ versus MGHWT) and K. pneumoniae MGH75878 ΔsoxS (MGH ΔsoxSPQ versus MGHPQ) during oxidative stress. Column A shows a new gene ID, and column C shows the original name, while column B represents the gene name. Columns X/Y/Z/AA/AB/AC/AD shows raw reads generated from RNA-seq conducted on different libraries, while columns AF/AG/AH/AI/AJ/AK represents the normalized reads. The fold changes were calculated from the normalized reads obtained from different libraries, MGHPQ versus MGHWT (column K) and MGH ΔsoxSPQ versus MGHPQ (column U). The genes with statistically significant data obtained from two biological replicates are indicated by a mark (√) in columns J and T based on the P value indicated in columns M and W. Based on the differential expression indicated in columns K and U (for statistically significant genes), the differential expression of each gene is indicated by a color code in columns E/F/G/H and O/P/Q/R. Genes highlighted in red indicate antimicrobial resistance genes, while those highlighted in blue indicate genes associated with virulence. (WS3) The table shows the oxidative soxS regulon of Klebsiella pneumoniae MGH78578, consisting of a stringent set of genes that are regulated by SoxS. The gene list was obtained from statistically significant genes from oxidative regulon and soxS regulon with a distinct expression pattern. Genes that were upregulated in the oxidative regulon plus downregulated in the soxS regulon and downregulated in oxidative regulon plus upregulated in the soxS regulon are shown. Download Table S1, XLSX file, 2.3 MB (2.3MB, xlsx) .
Copyright © 2021 Anes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Graphs showing the distribution of RNA-seq reads in each dataset. Download FIG S1, PDF file, 0.05 MB (47.2KB, pdf) .
Copyright © 2021 Anes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A text file describing 44 genes commonly identified in our soxS regulon and E. coli K-12 (3), some 30 were identified to belong to the more stringent oxidative soxS regulon. Our oxidative soxS regulon identified many genes that were previously shown to be regulated by SoxS. Download Text S1, DOCX file, 0.03 MB (33KB, docx) .
Copyright © 2021 Anes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metabolic activities of K. pneumoniae MGH78578 (wild type) and its isogenic ΔsoxS mutant grown under multiple conditions on the OmniLog microarray platform. Significant metabolic changes for delta activities were <−2 and >2. Download Table S2, XLSX file, 0.08 MB (81.2KB, xlsx) .
Copyright © 2021 Anes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All the RNA sequence data generated in the study were deposited in the National Center for Biotechnological Information–Gene Expression Omnibus and are available under the accession number GSE146844. Postanalysis, the differential expression levels of all the genes are given in Table S1 with three worksheets, WS-1, WS-2, and WS-3.