ABSTRACT
The mucophilic anaerobic bacterium Akkermansia muciniphila is a prominent member of the gastrointestinal (GI) microbiota and the only known species of the Verrucomicrobia phylum in the mammalian gut. A high prevalence of A. muciniphila in adult humans is associated with leanness and a lower risk for the development of obesity and diabetes. Four distinct A. muciniphila phylogenetic groups have been described, but little is known about their relative abundance in humans or how they impact human metabolic health. In this study, we isolated and characterized 71 new A. muciniphila strains from a cohort of children and adolescents undergoing treatment for obesity. Based on genomic and phenotypic analysis of these strains, we found several phylogroup-specific phenotypes that may impact the colonization of the GI tract or modulate host functions, such as oxygen tolerance, adherence to epithelial cells, iron and sulfur metabolism, and bacterial aggregation. In antibiotic-treated mice, phylogroups AmIV and AmII outcompeted AmI strains. In children and adolescents, AmI strains were most prominent, but we observed high variance in A. muciniphila abundance and single phylogroup dominance, with phylogroup switching occurring in a small subset of patients. Overall, these results highlight that the ecological principles determining which A. muciniphila phylogroup predominates in humans are complex and that A. muciniphila strain genetic and phenotypic diversity may represent an important variable that should be taken into account when making inferences as to this microbe’s impact on its host’s health.
KEYWORDS: Verrucomicrobia, comparative genomics, phylogroups, microbiome, mucin, assimilatory sulfur reduction (ASR), adolescent obesity, phylogenetic analysis
INTRODUCTION
The gastrointestinal (GI) microbiota comprises a complex community of bacteria, fungi, and archaea that significantly influence the metabolic and immunological health of their human and animal hosts (recently reviewed in reference 1). The global obesity epidemic has focused attention on the role that the microbiota plays in regulating energy acquisition and inflammation and how these activities impact the development of metabolic disease and diabetes (2). Western-style diets, in particular, lead to microbiotas of lower taxonomical diversity and metabolic capacity, which in turn, enhance the risk for developing inflammatory disorders such as diabetes, obesity, and cardiovascular disease (3–5).
Akkermansia muciniphila is a Gram-negative anaerobic bacterium of the phylum Verrucomicrobia that can use GI mucins as a sole carbon and nitrogen source (6, 7). A. muciniphila has attracted considerable attention because an increased abundance of Akkermansia in the GI tract correlates with many positive human health outcomes, including protection from obesity, diabetes, and metabolic disease (8–11). Indeed, lean individuals show an increased representation of Verrucomicrobia in their fecal microbiomes as assessed by 16S rRNA gene profiling (9). A. muciniphila is prevalent in the colon and has been reported to comprise between 1 and 4% of the total bacteria in healthy adult fecal samples (12). In mice, repeated administration of A. muciniphila ameliorates the impact of high-fat diets in inducing obesity and strengthens the function of the GI epithelial barrier though the activation of Toll-like receptor 2 (TLR2) (13, 14). In proof-of-concept trials in humans, administration of live or pasteurized A. muciniphila was sufficient to improve insulin sensitivity and reduced insulinemia (15).
Two recent pangenomic studies of Akkermansia, including a comparison of 35 A. muciniphila genomes reconstructed from metagenomic sequences (16) and 39 A. muciniphila isolated strains (17), led to the identification of four distinct phylogroups (clades AmI to IV). An analysis of the genomes of representative members of these phylogroups revealed phylogroup-specific functions such as the ability of AmII strains to synthesize corrin rings (16) and thus potentially outcompete other phylogroups when levels of vitamin B12 precursors in the GI tract are scarce. Given that multiple A. muciniphila phylogroups are found in humans, we asked what is the genomic and phenotypic diversity of A. muciniphila isolated from children, with the long-term goal of determining if there are correlates between strain and phylogroup abundance and specific health outcomes. To begin to address these questions, we used fecal samples collected by the Pediatric Obesity Microbiome and Metabolome Study (POMMS) (18) to isolate 71 unique strains of A. muciniphila. Participants included adolescents with healthy weight at a single time point only and adolescents with obesity at baseline and then at 6 months following a behavioral lifestyle intervention. A subsample of the participants with obesity additionally received pharmacotherapy or bariatric surgery during the study period. We performed a detailed genotypic and phenotypic analysis of these isolates and determined that POMMS A. muciniphila strains belonged to three main phylogroups/clades. Several in vitro traits, such as growth rates in mucin, resistance to ambient oxygen, self-aggregation, and binding to epithelial surfaces were linked to specific phylogroups. Furthermore, we determined that even though specific phylogroups displayed GI tract colonization advantages in antibiotic-treated mice, the parameters influencing Akkermansia colonization in humans are more complex.
RESULTS
The relative abundance of A. muciniphila in fecal samples is highly variable in a cohort of children with obesity.
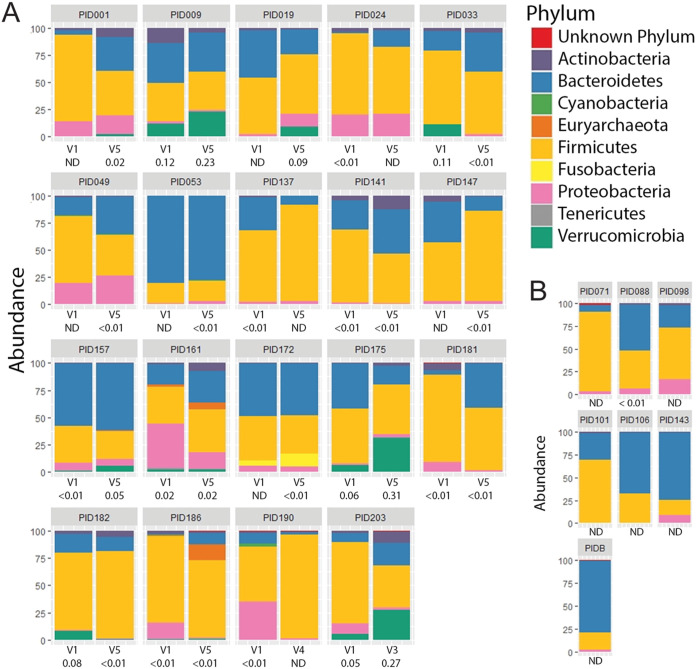
To survey the diversity of A. muciniphila strains present among healthy children and those with obesity, we used fecal samples that had been collected before and after various interventions aimed at decreasing their body mass index (BMI) (18). We selected 5 lean controls (Z-score adjusted BMI, or zBMI, –0.99 to 0.41) and 36 children with extreme obesity (zBMI, –1.63 to 3.18 > 95th percentile) (Table S1). For healthy lean control children, a single stool sample was obtained at the time of enrollment. For the majority of the cohort with obesity (35/36), we used samples collected at baseline and at the end of the study (6 months). We enriched for mucolytic bacteria directly from frozen fecal material by serial passage in liquid medium with gastric porcine mucin as the sole carbon and nitrogen source and then isolated single colonies on mucin agar plates. Bacterial colonies were purified to homogeneity and typed by sequencing the V3-4 region of the 16S rRNA locus. Overall, we cultured 71 strains of A. muciniphila from 35 children and 1 adult. In parallel, we performed a 16S rRNA-based survey of the bacterial communities present in selected fecal samples for which we had baseline and at least one additional visit (Fig. 1 and Table S2). A phylum-level analysis indicated that the relative abundance of Verrucomicrobia ranged from undetectable to 31% of total bacteria (Table S2).
FIG 1.
Relative abundance of Verrucomicrobia in stool samples of children with obesity. (A and B) Phylum-level assessment of the composition of bacterial communities in fecal samples derived from children with obesity before and 6 months after undergoing treatment for weight loss (A) and representative children of healthy weight (B) enrolled in the same study. The identity and relative representation of bacteria within each stool sample were determined by amplification and sequencing of the 16S RNA locus. The normalized fraction (%) of Verrucomicrobia is provided under the visit (V) number. ND, not detected.
Summary of pediatric donors described in this study. Download Table S1, XLSX file, 0.01 MB (14.4KB, xlsx) .
Copyright © 2021 Becken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ASV tables and relative abundance and composition of microbial communities in POMMS samples. Download Table S2, XLSX file, 0.02 MB (20.4KB, xlsx) .
Copyright © 2021 Becken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A. muciniphila clinical isolates are phenotypically diverse.
Previous analysis of microbial metagenomes indicated that there is significant diversity among A. muciniphila strains (19). To begin to address if this genetic diversity correlates with traits of relevance to the colonization and health of the human host, we performed a range of phenotypic tests that have been associated with Akkermansia biology, including variations in (i) growth rates, (ii) the ability to form bacterial aggregates, (iii) adherence to epithelial surfaces, (iv) the generation of short-chain fatty acids (SCFA) during mucin fermentation, (v) sensitivity to oxidative stress, and (vi) activation of Toll-like receptors (TLRs).
Growth rates. The growth rates among individual strains were monitored under anaerobic conditions in a semidefined synthetic medium consisting of glucose, N-acetylglucosamine, soy peptone, and threonine. A. muciniphila strains displayed doubling times ranging from 1.2 to 13 h. These differences were more pronounced when gastric porcine mucin was the sole carbon and nitrogen source, with doubling times ranging from 0.3 to >20 h (Fig. 2A and Table S3).
FIG 2.
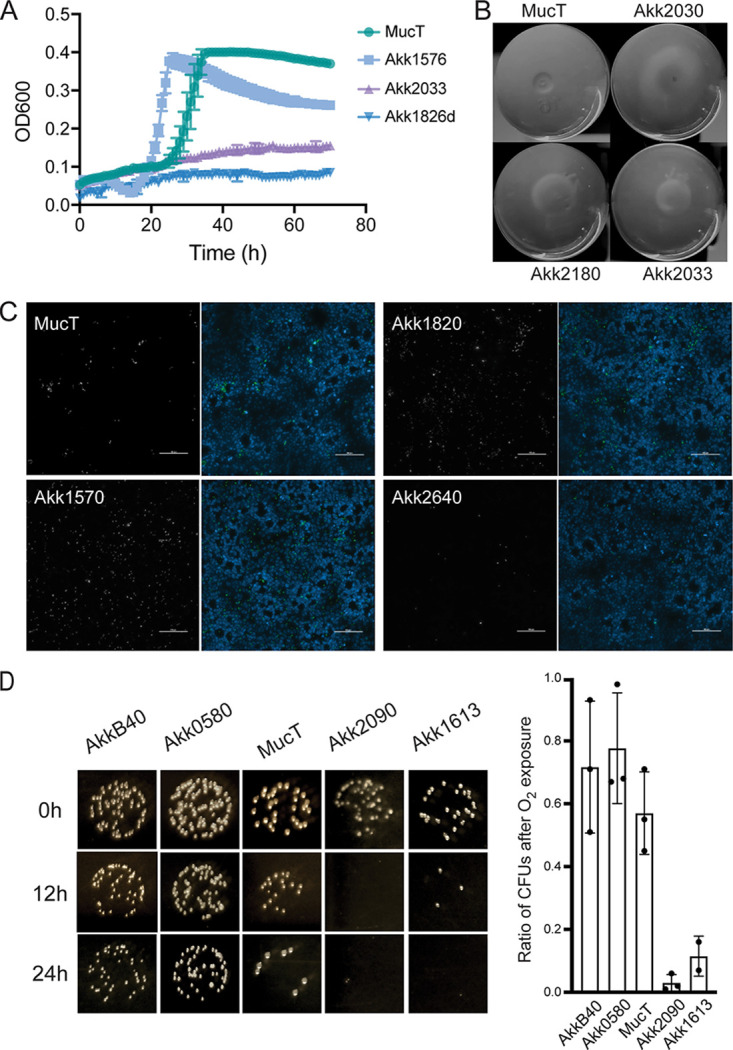
A. muciniphila human isolates display variance in phenotypes relevant to gastrointestinal colonization. (A) A. muciniphila strains display distinct growth rates in porcine gastric mucin. The growth rates of A. muciniphila isolates were monitored in liquid medium over a period of 96 h. Representative examples of fast and slow growers are shown in relation to the typed A. muciniphila strain MucT. (B) A. muciniphila strains agglutinate. Selected strains shown as examples of rapid sedimentation of A. muciniphila grown in mucin medium. (C) A. muciniphila strains vary in their ability to attach to epithelial surfaces. Adhesion of selected A. muciniphila strains to HT29 colonic epithelial cells was assessed by immunofluorescence microscopy. HT29 nuclei were detected with Hoechst (blue) and bacteria with polyclonal anti-Akkermansia antiserum (white, left panels; green, right panels). Scale bar = 100 μm. (D) A. muciniphila strains vary in their tolerance to ambient oxygen. Strains grown in BHI supplemented with mucin were exposed to ambient oxygen for 0, 12, or 24 h on BHI-mucin agar plates followed by outgrowth under anaerobic conditions. Oxygen sensitivity was assessed by enumerating CFU on agar plates.
Summary of A. muciniphila isolates and phylogroups and their phenotypes. Download Table S3, XLSX file, 0.02 MB (24.3KB, xlsx) .
Copyright © 2021 Becken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Aggregation. Some A. muciniphila isolates, unlike the reference strain A. muciniphila MucT, readily sedimented when grown in liquid culture media (Fig. 2B). We measured the extent to which strains aggregated by monitoring the changes in the optical density in samples obtained from the surface of culture tubes that had been grown in mucin medium (Table S3).
Adherence to epithelial cells. We determined if there are strain-level differences in A. muciniphila isolates for their ability to attach to intestinal epithelial cells. Binding was assessed using HT29-MTX colonic epithelial cells grown on microtiter plates for 7 days past confluence, a stage at which they start secreting mucins. As a control for nonspecific binding to biotic surfaces, we used bovine serum albumin (BSA)-coated plates. Adherence was monitored either microscopically or by following the outgrowth of A. muciniphila after addition of synthetic medium to wells. As previously reported, A. muciniphila strain MucT bound to human colonic cells (20), albeit at low levels. Our A. muciniphila isolates displayed a broad range of binding affinities to HT29 monolayers (Fig. 2C, Fig. S1A and B, Table S3) compared to BSA-coated plates. Strains also differed in their binding to BSA-coated plates alone (Fig. S1C).
A. muciniphila binding to HT29 colonic epithelial cell and protein-coated plated. Adherence of A. muciniphila strains isolated from patients enrolled in the POMMS to HT29 colonic epithelial cell- or BSA-coated plates. Bacteria were incubated with HT29-MTX cell- or BSA-coated plates and extensively washed, and binding was indirectly evaluated by adding synthetic medium to the wells and measuring the optical density at 600 nm after 48 to 72 h. (A) Binding is provided as the ratio of the OD600 HT29-MTX to BSA-coated plates. (B and C) The nonnormalized OD600 units for HT29-MT (B) or BSA-coated (C) plates are provided for reference. Bars represent the mean and standard deviation of 3 technical replicates from 3 independent experiments. Download FIG S1, TIF file, 2.7 MB (2.7MB, tif) .
Copyright © 2021 Becken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mucin fermentation. The major end products of mucin metabolism by A. muciniphila are acetate and propionate, with minor amounts of succinate and 1,2-propanediol (16, 21). The ratio of acetate to propionate generated is influenced by how simple sugars are metabolized and the availability of vitamin B12 to activate methylmalonyl-coA and generate propionate (16). We measured SCFAs produced by the various Akkermansia strains after growth in mucin by gas chromatography. All A. muciniphila isolates produced acetate/propionate at a ratio of 1.21 to 1.47 after reaching the stationary phase of growth in 0.5% mucin medium, which is in the range of what has been reported for the A. muciniphila MucT strain (21).
Oxygen sensitivity. Although A. muciniphila is an anaerobe, its growth can be stimulated by low levels of oxygen (22). We predicted that the relative tolerance of A. muciniphila to oxygen may impact its abundance near epithelial surfaces or its resistance to oxidative stress during GI inflammation. We tested the sensitivity of A. muciniphila strains to oxygen by exposing agar plates to ambient air for 12, 18, and 24 h before returning the plates to anaerobic conditions (Fig. 2D). We determined that there is a range of responses, with some strains being fairly tolerant to prolonged exposure to ambient oxygen (∼60% survival at 24 h), while others were extremely sensitive (<0.01% survival at 12 h) (Table S3).
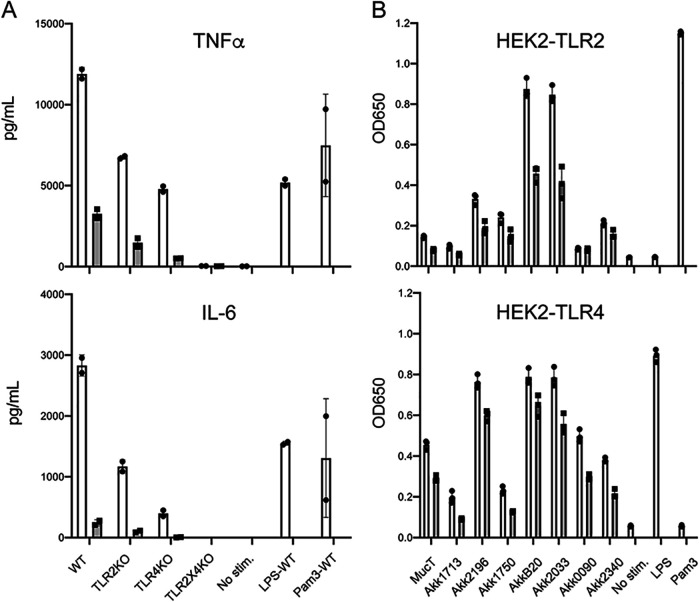
Activation of innate immune sensors. Human monocytes are activated by exposure to A. muciniphila MucT (23), and TLR2-dependent recognition is required for signal transduction events that strengthen barrier function in the GI tract (24). We first tested to be determine the relevant TLRs involved in the recognition of A. muciniphila by stimulating bone marrow-derived macrophages (BMDM) from wild-type C57BL/6J mice and from tlr4, tlr2 tlr4, unc93b1, and tlr2 tlr4 unc93b1 knockout mice (25). Unc93b1−/− mice are defective for the transport of TLRs that sense nucleic acids as well as the expression of TLR5, which recognizes flagellin, at the cell surface (26, 27). BMDM were incubated with A. muciniphila MucT at a ratio of 0.5 or 5 bacteria per cell for 6 h, and the secretion of the cytokine tumor necrosis factor alpha (TNF-α) and interleukin-6 (IL-6) was assessed (Fig. 3A). At the lowest dose of Akkermansia, the response of BMDM was exclusively dependent on TLR4. TLR2-dependent activation was only apparent when bacterial loads were increased by 10-fold. Consistent with previous findings (23), additional TLRs were not required for immune activation of mouse BMDMs.
FIG 3.
Activation of TLR2 and TLR4 by A. muciniphila isolates. (A) TLR2 and TLR4 are the main microbial sensors required for the recognition of A. muciniphila. Bone marrow-derived macrophages from the selected tlr knockout mice were incubated with A. muciniphila MucT at a ratio of 5 (white bars) or 0.5 (gray bars) per BMDM for 6 h, followed by measuring TNF-α or IL-6 as readouts of activation. BMDM activation only occurs in cells expressing either TLR4 or TLR2, with TLR4 displaying a lower threshold for A. muciniphila stimulation. (B) A. muciniphila strains vary in their ability to activate TLR2 and TLR4. Selected strains grown on mucin medium were incubated with HEK293 reporter cell lines expressing either TLR2 or TLR4 at a ratio of 5 or 1 bacteria/cell (white and gray bars, respectively). Stimulation of TLR in these cell lines was determined by assessing the processing of a colorimetric substrate of secreted of alkaline phosphatase.
We next used HEK-TLR reporter cell lines to test the ability of A. muciniphila isolates to specifically activate TLR2 and TLR4. We used cell lines expressing TLR4 or TLR2 and its coreceptors TLR1 and TLR6, which respond to known TLR2 ligands (28). A. muciniphila strains were grown in mucin or synthetic medium and incubated with reporter cell lines expressing TLR2 or TLR4 for 16 h at a ratio of 1 or 5 bacteria per HEK-TLR cell, and levels of TLR-dependent secreted alkaline phosphatase (sAP) were measured 16 h poststimulation (Fig. 3B and Table S3). A subset of strains induced TLR2 or TLR4 activation consistently above or below the mean of all A. muciniphila isolates tested (Table S3 and Fig. S2). The induction of TLR2 and TLR4 reporters was higher for bacteria grown in synthetic medium compared to mucin medium (Fig. S2).
A. muciniphila activation of HEK293-TLR reporter cell lines. (A to D) A. muciniphila strains were grown on mucin or synthetic medium and incubated with HEK293 reporter cell lines expressing either TLR1/2/6 (A and B) or TLR4 (C and D). Activation of the TLR in these cell lines was determined by assessing the secretion of alkaline phosphatase into culture supernatants. Data are reported as the normalized fold induction over the mean OD value of the entire population. Bars represent the mean and standard deviation of 3 technical replicates from 3 independent experiments. Download FIG S2, TIF file, 0.9 MB (923.3KB, tif) .
Copyright © 2021 Becken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparative genomics suggest that phylogroup-specific genetic determinants regulate A. muciniphila replication in mucin and aerotolerance.
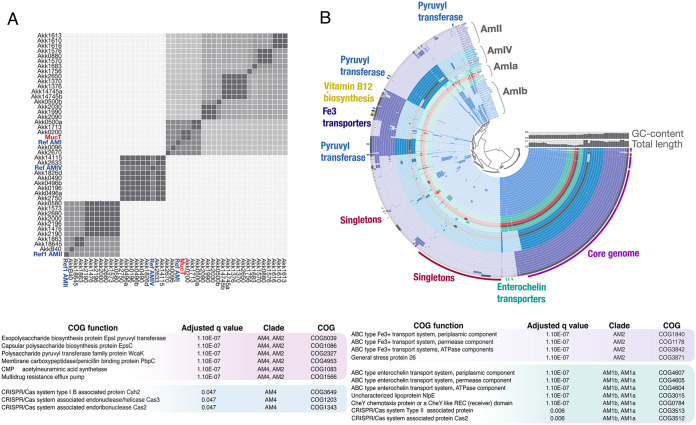
Of the four A. muciniphila phylogroups (clades AmI to IV) the most studied strain (MucT) is a representative of phylogroup AmI (16, 17). To assess the distribution of A. muciniphila phylogroups in the POMMS cohort, we sequenced the genomes of 43 isolates. A comparative analysis of these genomes indicated that we have representatives of phylogroups AmI, AmII, and AmIV, but not AmIII (Fig. 4). We designed phylogroup-specific primers (Table S4) to genotype the isolates whose genomes had not been sequenced and determined that AmI (41/71) was almost twice as prevalent as AmII (22/71) and AmIV (8/71). Based on whole-genome comparisons of AmI members, we propose that AmI can be further subdivided into two related subclades (Ia and Ib) at a threshold of 96% average nucleotide identity (ANI). Genomes ranged in size from 2.6 to 3.3 Mb, and phylogroups AmII and AmIV were consistently larger than the AmI genomes (Fig. 4A and B and Table S3).
FIG 4.
A. muciniphila isolates from children belong to three different phylogroups. (A) Whole-genome comparison of 40 A. muciniphila strains. The average nucleotide identity (ANI) was calculated at 96% identity using Anvi’o and PyANI. Genomes for previously published strains (16, 17) belonging to each Akkermansia phylogroup were included as controls (blue), as well as the type-strain MucT (red). Complete genomes were assembled from PacBio reads. Note that phylogroup AMI can be subdivided into two subtypes at 96% ANI threshold. (B) Circle phylogram of new A. muciniphila strains. The graph displays the pangenome of 40 sequenced isolates, 3 reference genomes (gray), and the type strain MucT (red). The phylogram is clustered based on gene frequency and displays gene cluster presence/absence for each genome. Selected phylogroup-specific gene groups are highlighted to show their distribution, including vitamin B12 biosynthetic gene groups, putative enterochelin transporters, Fe3+ transporters, and capsule genes (labeled as pyruvyl transferases). (C) Gene set enrichment analysis of A. muciniphila phylogroups. Anvi’o was used to identify COG functions associated with specific phylogroups. The selected COGs were detected in all isolates belonging to a given phylogroup(s) and in none of the isolates belong to other phylogroups. The adjusted q value shows the significance of the enrichment between the function and the associated phylogroup corrected for multiple testing.
Primers used for the identification of A. muciniphila phylogroups and quality control for specificity. Download Table S4, XLSX file, 0.01 MB (12KB, xlsx) .
Copyright © 2021 Becken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
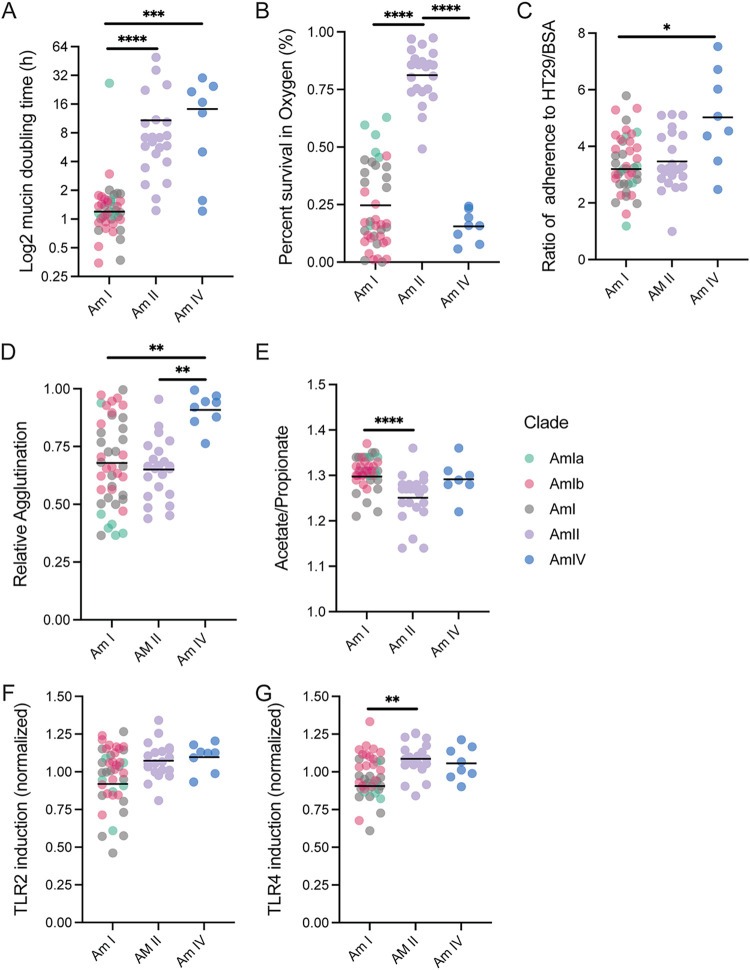
We determined that several phenotypes segregated by phylogroup (Fig. 5). AmI strains displayed rapid doubling times, while most members of AmII and AmIV grew slowly (Fig. 5A). Strains also differed in their sensitivity to ambient oxygen, with strains of AmII being resistant and those of AmIV very sensitive (Fig. 5B). Differential oxygen sensitivity was also observed within phylogroup AmI, with AmIb strains being highly sensitive to exposure to air, while AmIa strains displayed intermediate resistance. AmIV strains had higher adherence to epithelial cells and displayed a greater propensity to aggregate when grown in mucin medium (Fig. 5C and D). We also saw a small drop in the ratio of acetate/propionate fermentation end products of mucin fermentation for AmII strains (Fig. 5E), which we postulate is because they synthesize vitamin B12 (16) and hence generate more propionate as vitamin B12 in the growth medium becomes limiting.
FIG 5.
A. muciniphila strains display phylogroup-dependent and independent phenotypes. (A to G) Distribution of A. muciniphila (A) doubling times in mucin medium, (B) resistance to ambient oxygen on agar plates, (C) adherence, (D) agglutination, (E) mucin fermentation, (F) TLR2 activation, and (G) TLR4 activation. Each symbol represents a strain. AmI strains that were not subtyped into AmIa or AmIb are shown as gray dots. Details are available in Table S3. P values were calculated using Kruskal-Wallis tests followed by Dunn’s multiple-comparison tests. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
For the stimulation of HEK-TLR reporter cell lines, AmII and AmIV strains were more stimulatory for both TLR2 and TLR4 than AmI strains (Fig. 5F and G), but it is unclear if this is simply a reflection of their enhanced binding properties to cell surfaces. AmI strains displayed a broad range of activation of TLR reporter cell lines, and while there was a trend for AmIb strains to be more stimulatory, particularly for TLR4, the differences did not reach statistical significance given the relatively low number of AmIa isolates in our strain collection.
To identify genes that may contribute to these phenotypes, we analyzed the pangenome of our A. muciniphila strains and identified 4,982 total gene clusters, with 1,647 core gene clusters found in all genomes and 506 gene clusters found only in single genomes (Fig. 4A). We found several phylogroup-specific gene groups that may contribute to phenotypic variation (Fig. 4B and C and Table S3). For example, all members of phylogroups AmII and AmIV were predicted to encode distinct capsule and exopolysaccharide genes that were absent in AmI strains. These included putative capsular polysaccharide (CPS) biosynthesis proteins EpsC and EpsI, a CMP-N-acetylneuraminic acid synthetase, and a capsule modifying enzyme, polysaccharide pyruvyl transferase WcaK (29). The AmI isolates have a single Cps locus, while AmII and AmIV genomes contain two additional loci that are largely conserved among these phylogroups and map to similar regions in the chromosome. This suggests common capsule types are present in the phylogroup AmII and IV strains. Phylogroup AmIV also codes for DltB, an enzyme typically involved in modification of lipoteichoic acid in Gram-positive bacteria but that can also modify lipopolysaccharides in some Gram-negative bacteria (30). Conversely, members of phylogroup AmI had additional chemotaxis genes, cytochrome c biosynthetic genes, and code for a quality control sensor protein for outer membrane biogenesis, NlpE (31).
The phylogroups also displayed differences in iron acquisition systems. Anaerobic conditions favor reduced ferrous iron (Fe2+), and aerobic conditions favor the oxidized ferric iron (Fe3+) (32). Although all genomes had a ferrous iron transport system consisting of FeoAB genes (Amuc_1088, Amuc_1089, and Amuc_1090 in MucT), phylogroups AmI and AmII encoded additional mechanisms to acquire ferric iron. Members of AmI had multiple enterochelin transporter gene groups, suggesting that they might use siderophores to scavenge ferric iron. Members of phylogroup AmII also have predicted ferric iron transporters, although these appear to be distinct from the gene groups in phylogroup AmI. In contrast, phylogroup AmIV lacks canonical mechanisms for ferric iron acquisition. Phylogroup AmIV is highly sensitive to ambient oxygen, and it is plausible that defects in iron acquisition or the absence of the oxidative stress protection associated with siderophores (33, 34) may contribute to this phenotype. Additional contributors to differences in oxygen sensitivity include a LexA repressor, indicative of an SOS system, that is present in all phylogroup AmI and AmII strains.
The A. muciniphila strains were predicted to encode approximately 27 glycoside hydrolase (GH) enzyme families, but they varied in the abundance of GH families among the phylogroups, particularly in AmIV genomes (Fig. 3B). GH97 enzymes, which comprise glycoamylases such as Bacteroides thetaiotaomicron SusB (35), were detected in all strains except for AmIV. Similarly, AmIV genomes had fewer GH110 enzymes, a group of galactosidases capable of cleaving blood group B antigens (36). Conversely, AmIV strains were enriched for GH29 and GH95 l-fucosidases, which could potentially cleave the terminal fucose residues that decorate mucin and human milk oligosaccharides (37). In humans, both blood ABH antigens and fucose modifications are more prevalent in ileal mucins, with potential implications for the relative localization of Akkermansia strains in the GI (38).
Another function with phylogroup-specific differences is the CRISPR/cas systems. While only a few of the phylogroup AmII genomes had CRISPR gene clusters, putative CRISPR/cas genes were detected in some AmI and in all AmIV genomes. The class of CRISPR system may be phylogroup-specific since AmIV strains were predicted to encode genes found in type I-B CRISPR/cas systems, while AmI strains were predicted to have genes associated with type II systems (Amuc_2008, Amuc_2009, and Amuc_2010 in MucT) (39, 40), although it is not clear if these represent complete, functional systems. In some instances, the CRISPR genes are located close to predicted phage genes, possibly indicative of horizgontal genes transfer.
Phylogroups AmII and AmIV are deficient for reductive sulfur assimilation.
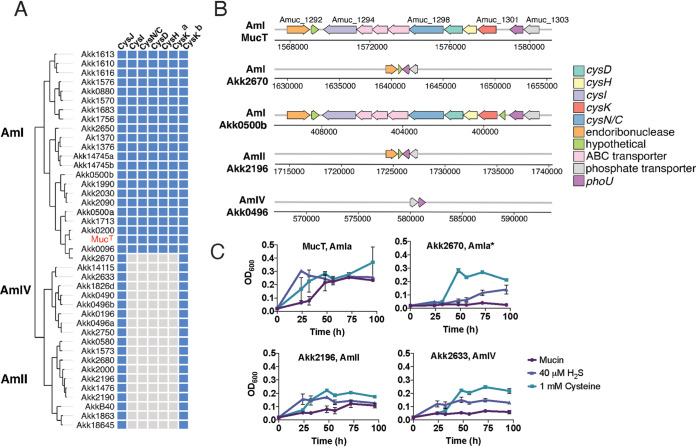
Analysis of the metabolic capabilities of the strains based on genomic sequences revealed additional predicted phylogroup-specific features (Fig. 6 and Fig. S3A). For instance, metabolic enrichment analysis showed that genes required for assimilatory sulfate reduction (ASR) are significantly enriched in phylogroup AmI (adjusted q value, 6.14E-7) (Table S5) but absent in AmII and AmIV isolates (Fig. S3A). Enzymes in the ASR pathway reduce sulfate to hydrogen sulfide for the synthesis of sulfur-containing molecules such as cysteine and methionine. In the canonical ASR pathway, sulfate is first reduced to adenosine phosphosulfate (APS) by ATP sulfurylase (CysN), followed by the formation of phosphoadenosine phosphosulfate (PAPS) by APS kinase (CysD), which is further reduced to sulfite by PAPS reductase (CysH) and, finally, to H2S by sulfite reductase (CysI/J) (41). H2S is a substrate for cysteine synthase (CysK) to generate cysteine. In the MucT strain, ASR genes are clustered in a single locus (Amuc_1294 to Amuc_1301), with the exception of a CysJ homolog (Amuc_0631) and a second cysteine synthase (Amuc_2014). The locus also included a potential inner membrane sulfide permease (Amuc_1295), an ABC transporter-related ATP-binding protein (Amuc_1296), and a substrate-binding protein (Amuc_1297) (Fig. 6A and B).
FIG 6.
AmII and AmIV phylogroups are defective for assimilatory sulfate reduction (ASR). (A) Distribution of ASR genes among A. muciniphila phylogroups. ASR genes (top) from A. muciniphila MucT were used to search for homologs among other sequenced isolates. Blue squares indicate that a gene is present, and gray squares indicate that a gene was not detected. There are two cysK homologs in MucT, Amuc_1301 and Amuc_2014. (B) Genomic context of ASR genes in Akkermansia phylogroups. The majority of the ASR genes are clustered in a single locus in the AmI phylogroup, as represented in strains MucT and Akk0500B. The AmI strain, Akk2670, lacked the entire ASR locus, although the flanking genes remained conserved in other AmI strains. The AmII strain (Akk2196) also lacked the entire ASR locus, while the AmIV strain (Akk0496) missed the locus and flanking genes. Arrows represent genes, polarity of reading frame, and the numbers above each arrow indicate the gene number in the annotated genome. The genome coordinates for each locus are shown below the arrows. (C) Addition of cysteine or NaHS enhances the growth of ASR-deficient A. muciniphila. Representative Am strains from each phylogroup were tested for growth in mucin medium with or without the addition of 1 mM l-cysteine or 40 μM NaHS.
Phylogroup-specific enrichment of metabolic pathways. Download Table S5, XLSX file, 0.02 MB (17.7KB, xlsx) .
Copyright © 2021 Becken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genomic analysis of A. muciniphila strains and predicted functions. (A) Pathway enrichment analysis indicates selective acquisition and loss of metabolic pathways. To compare the metabolic capabilities of isolates, we used Anvi’o to estimate metabolism function to detect KEGG module pathways in the genomes. The lower threshold for detecting a pathway was set to 50%. Strains with 50% or less of the enzymes required for a given pathway are represented as blue squares in the heatmap. (B) Detection of selected glycoside hydrolase families by clade. Bars represent the average number of enzymes per genome. The analysis included genomes from clade AmIa (n = 5), AmIb (n = 20), AmII (n = 10), and AmIV = (n = 8). GH, glycoside hydrolase; NC, nonclassified glycoside hydrolases. Error bars represent the standard error of the mean (SEM). Download FIG S3, GIF file, 1.5 MB (1.5MB, gif) .
Copyright © 2021 Becken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
AmII and AmIV strains lacked the ASR gene cluster found in MucT but retained distally encoded homologs for CysJ and the Amuc_2014 cysteine synthase (CysK-b) (Fig. 6A). All AmI strains were predicted to perform ASR, except for the strain Akk2760 (Fig. 6A and Fig. S3A), which lacked the entire ASR locus (Fig. 6B). Since Akk2670 grew very poorly in mucin medium, with a growth rate comparable to that of AmII and AmIV isolates (Fig. 5A and Table S3), we hypothesized that the inability to generate reduced sulfur may be a limiting factor for their growth on mucin in vitro. To test this, representative strains were grown in mucin medium with or without the addition of cysteine or sodium hydrosulfide as a source of H2S. While cysteine and H2S shortened the lag time for the growth of the MucT strain, it did not affect the maximal biomass achieved (Fig. 6C). In contrast, cysteine significantly enhanced the maximal growth of the predicted ASR-deficient AmIa strain Akk2670, the AmII strain Akk2196, and the AmIV strain Akk0496 (Fig. 6C). These findings suggest that A. muciniphila strains benefit from the addition of reduced sulfur when grown in mucin and that the growth of ASR-deficient phylogroups is significantly enhanced by the addition of cysteine or H2S.
AmIV strains outcompete other phylogroups in a murine colonization model.
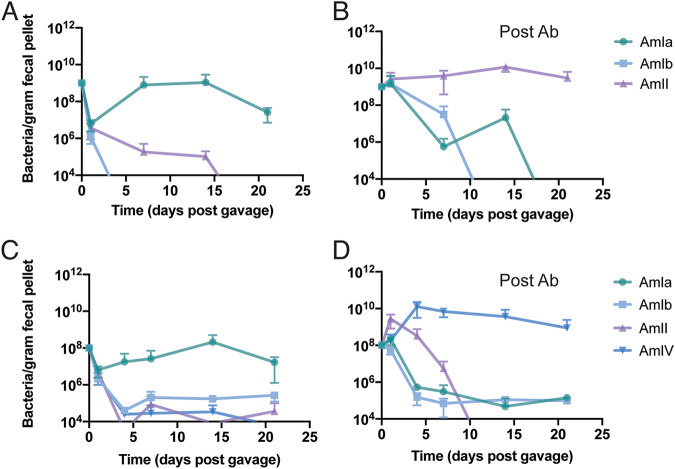
To determine if A. muciniphila phylogroups varied in how they colonize animals, we assessed the ability of representative isolates of each phylogroup to compete in the mouse GI tract. Mice housed in our vivarium are naturally colonized by a mouse Akkermansia strain, which belongs to the AmIa phylogroup (not shown). We first challenged these mice with human strains representing phylogroups AmIa, AmIb, and AmII (Fig. 7A), and their abundance in fecal pellets was monitored over time by quantitative PCR (qPCR). We found that the human AmIa, AmIb, and AmII strains failed to engraft with the endogenous mouse A. muciniphila and associated microbiota, which provided colonization resistance against introduction of additional Akkermansia strains. We cleared mice of their Akkermansia and other microbes with a 14-day regimen of tetracycline and repeated the competition experiments with the same cocktail of human A. muciniphila phylogroup representatives. Under these conditions, the AmII strain (Akk0580) became the dominant phylogroup by 20 days postinoculation (Fig. 7B).
FIG 7.
A. muciniphila phylogroups AmII and AmIV strains outcompete AmI in antibiotic pretreated mice. (A to D) Mice were either untreated (A and C) or treated (B and D) with antibiotics (Ab) and gavaged with a three-phylogroup strain mix containing an equal amount of phylogroups AmIa (MucT), AmIb (Akk1683), and AmII (Akk0580) (A and B) or a four-phylogroup strain mix containing phylogroups AmIa (MucT), AmIb (Akk1570), AmII (Akk0580), and AmIV (Akk0490) (C and D). The AmIa strain identified in mice that had not been pretreated with Ab (A and C) represents the endogenous mouse Akkermansia. Each point represents the average of three cages (n = 4 mice/cage for panels A and B, and n = 2 mice/cage for panels C and D), and error bars represent the standard deviation. The Am1a strain in panels A to C represent the endogenous mouse A. muciniphila strain found at the Duke University vivarium.
Next, we competed strains representing all four phylogroups—AmIa, AmIb, AmII, and AmIV. As with the previous experiment, the human A. muciniphila isolates failed to colonize mice with an intact microbiota (Fig. 7C). However, pretreatment with antibiotics enabled engraftment of the newly introduced strains, but this time the AmIV strain (Akk0490) became the predominant phylogroup (Fig. 7D). These findings were recapitulated with a second set of representative strains, with the AmIV isolate (Akk2750) rapidly becoming the dominant phylogroup (Fig. S4). These findings suggest that AmIV strains, despite their slow growth in mucin medium and high sensitivity to oxygen, overtake other phylogroups in the GI tract when placed in direct competition in mice whose microflora had been depleted.
A. muciniphila phylogroup AmIV outcompetes other phylogroups in colonizing the mouse GI. (A) Fecal samples were collected prior to gavage, and DNA was extracted to assess endogenous Akkermansia levels in mice with and without antibiotic treatment. Akkermansia was detected with primers specific for the Akkermansia 16S rRNA gene (total) and with phylogroup-specific primers. (B) In vivo competition assay using a second set of representative strains for each phylogroup. A representative of each strain from a healthy control was selected as follows: AmIa (Akk2670), AmIb (Akk2650), AmII (2680), and AmIV (2750). Mice were treated with antibiotics until the endogenous Akkermansia was cleared and were subsequently gavaged with a cell suspension containing each of the phylogroups. Colonization was monitored over time by collecting fecal pellets and testing for colonization using qPCR with phylogroup-specific primers. Download FIG S4, TIF file, 2.8 MB (2.8MB, tif) .
Copyright © 2021 Becken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Evidence for phylogroup exclusion and switching in patients colonized with A. muciniphila.
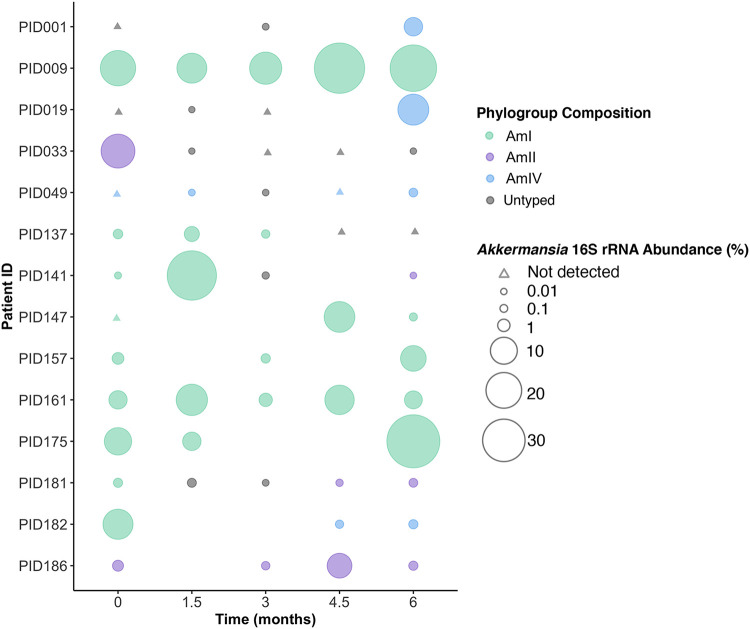
Because AmII and AmIV strains prevented AmI strains from establishing themselves when placed in direct competition in mice, we hypothesized that A. muciniphila phylogroups occupy the same ecological niche and that AmII and AmIV strains have a selective advantage in the GI tract. To assess the natural distribution of major phylogroups in humans, we used phylogroup-specific primers (Table S4) to quantify AmI, II, and IV strains in fecal samples collected from 14 patients at baseline and at 1.5-month intervals after enrolling in POMMS (Fig. 8). In stool samples where a clade-specific signal could be detected above background, we found dominance of a single major phylogroup regardless of the total relative abundance of Akkermansia in that sample. Surprisingly, in three instances, the phylogroup of the A. muciniphila strains cultured (Akk1476, Akk1573, and Akk14115) did not match the dominant phylogroup identified by qPCR in the stool sample, and in one patient we isolated both AmII and AmIV strains (Akk1826b and Akk1826d) from the same fecal material, even though AmIV was the only strain identified by qPCR. This discrepancy between culture-based isolation and molecular quantification may reflect biases in culturing efficiency based on differences in oxygen tolerance that could impact the viability of Akkermansia during the collection, transport, and handling of stool samples or differences in doubling times in porcine mucin medium during the serial enrichment process. Nonetheless, these findings suggest that despite the predominance of any one Akkermansia phylogroup in the GI tract, additional strains can be present and viable despite being below the levels of detection by molecular methods.
FIG 8.
Evidence for single A. muciniphila phylogroup dominance in humans. A subset of stool samples from 14 patients (PID, patient ID) were selected for detection and quantification of specific phylogroups by qPCR. Samples were selected for analysis if patients had provided multiple samples (n > 3) throughout a 6-month period, and 16S rRNA community profiling indicated the presence of Akkermansia at at least one time point. The size of the bubbles represents the relative abundance of Akkermansia as assessed with QIIME analysis. Triangles indicate that no Akkermansia/Verrucomicrobia 16S rRNA sequences were detected. The frequency of phylogroup types was assessed by qPCR with specific primers and is color coded. In all cases, the dominant phylogroup represented >99.9% of total Akkermansia with a threshold for phylogroup identification set at a quantification cycle (Cq) value of 34 or lower (Table S5). Missing data points indicate that no sample was collected at that time point.
In addition to evidence for the coexistence of minor clades, we observed major changes in the overall abundance of Akkermansia phylogroups within the same patient. In 11 of 14 of patients with multiple sampling over a 6-month period, either the relative amount of Akkermansia fluctuated significantly (>100-fold) among samples or the identity of the phylogroups switched (Fig. 8 and Table S6). For instance, three patients (PID141, PID181, and PID182) switched their predominant phylogroup from an AmI strain to either AmII or AmIV strains by 6 months. In two patients that underwent bariatric surgery (PID001, PID019), both converted from almost undetectable levels of Akkermansia at baseline to a phylogroup AmIV-dominant microbiota by the 6-month time point. It is unclear if the emergence of new dominant phylogroups represents blooms of preexisting low-abundance strains or new colonization events.
Phylogroup typing in patients with obesity over a 6-month period. Download Table S6, XLSX file, 0.01 MB (14.2KB, xlsx) .
Copyright © 2021 Becken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Obesity is a multifactorial disease influenced by host genetics, diet, behavior, and the microbial ecosystems that populate the human GI tract. Diet remains a key driver of microbial composition, and the phenotypes associated with these resident microbial communities strongly influence their impact on the immunological and metabolic health of their host (42). However, some bacterial species seem to play an oversized role on the health of their hosts. For instance, A. muciniphila has emerged as a potential probiotic since its abundance in the GI tract positively correlates with decreased incidence of metabolic disease, obesity, and diabetes (9, 10).
There is a growing recognition that there is a great diversity of Akkermansia strains and species. A pangenomic analysis of Akkermansia genomes revealed four A. muciniphila phylogroups (16), and an analysis of >1,000 Akkermansia genomes reconstructed from metagenomic sequences from human samples across the world suggested the existence of up to four new species (43, 44). While metabolic capacities of different Akkermansia strains have been inferred based on genome annotations, experimental validation is largely lacking because isolates of these new phylogroups have either not been collected or not been characterized. A notable exception is a recent analysis of the vitamin B12 biosynthetic properties of a member of the AmII phylogroup (16).
In this work, we leveraged fecal samples collected as part of the POMMS interventional study for childhood obesity (18) to isolate Akkermansia strains that reflect the diversity of phylogroups present in both healthy and diseased states in children. Importantly, the availability of cultured strains enabled the phenotyping of each isolate to identify variances in traits that may impact the health of their hosts. We cultured mucophilic bacteria from 123 fecal samples derived from 49 donors and isolated 71 new strains of A. muciniphila (Table S1 and S3). The relative abundance of Verrucomicrobia amplified sequence variants (ASVs) ranged from nondetectable to >30% of total sequences in children with obesity (Table S2).
Based on the genomes of 43 of new Akkermansia strains, we determined that these isolates belonged to A. muciniphila phylogroups AmI, II, and IV (Fig. 4). AmI strains constituted over half of the isolates, which we propose should be subdivided into two subgroups (AmIa and Ib) based on an ANI cutoff of 96% among complete genome sequences and phylogroup-specific phenotypes, such as increased sensitivity to ambient O2 for AmIb strains (Fig. 5A). In contrast, AmII strains were more resistant to ambient O2 than all the other phylogroups. AmII strains were also more immunostimulatory for TLR2 and TLR4, although this was most apparent when bacteria were grown in mucin, as opposed to synthetic medium (Fig. 5F and G and Fig. S2). AmIV strains also displayed enhanced activation of TLR4 and TLR2 reporter cell lines, which may reflect their increased binding to epithelial cells (Fig. 5C and D). Using BMDM, we confirmed that TLR2 and TLR4 are the relevant pattern recognition receptors for the detection of A. muciniphila as previously reported (23), with TLR2 activation requiring a higher threshold for activation than that of TLR4 (Fig. 4A). TLR2-mediated enhancement of tight junctions in intestinal epithelia has been proposed as a mechanism to explain Akkermansia-dependent enhancement of gut barrier function (23). TLR2 forms heterodimers with TLR1 or TLR6, with TLR2/1 being preferentially activated by triacylated lipopeptides from Gram-negative bacteria, and TLR2/6, by diacylated lipopeptides most commonly expressed on Gram-positive bacteria (45, 46). Amuc_1100, a highly expressed pilus-like protein conserved among all phylogroups, has been proposed to be a major A. muciniphila TLR2 agonist (14, 23). We suspect additional TLR2 agonists exist, given that A. muciniphila can stimulate HEK2923 cells expressing either TLR2/1 or TLR2/6 alone.
Although A. muciniphila is classified as a strict anaerobe, the growth of the reference MucT (AmIa) isolate is enhanced by nanomolar concentrations of O2 (22). The ability to use oxygen as a terminal electron acceptor may provide A. muciniphila an advantage over other colonic bacteria when in proximity to epithelial surfaces. The closely related AmIb strains, however, are very sensitive to ambient oxygen even though the genomes are highly similar and also encode the cytochrome bd complex, which is required to use O2 as a terminal electron acceptor (22). The mechanism underlying this differential sensitivity to oxygen is not apparent from comparative genomics. In contrast, AmII strains’ high resistance to oxygen may be linked to ferrous and ferric iron transport, as genes encoding Fe3+ ABC transporters have expanded in AmII strains and iron transporter genes are highly expressed in A. muciniphila MucT when switched to aerated growth conditions (22). AmIV strains also display high sensitivity to oxygen, which may also be associated with decreased iron acquisition, as this phylogroup lacks components of the enterochelin transport system present in AmI strains.
Given the slow doubling times for both AmII and AmIV strains in vitro, we did not expect these strains to overtake AmI strains in antibiotic-treated mice. One possible explanation is that the overrepresentation of capsular and exopolysaccharide gene clusters in AmII and AmIV phylogroups may provide protection from IgA and host-derived antimicrobial peptides. Unexpectedly, AmIV strains outcompeted AmII strains in mice, even though AmIV was expected to be more sensitive to the oxygen present near colonic epithelium. Given these findings, it is clear that the complexity of the GI ecosystem makes it difficult to predict which in vitro phenotypes are most relevant for in vivo colonization of the GI tract.
A comparison of the predicted metabolic pathways in A. muciniphila strains indicated a clear absence of components of the assimilatory sulfate reduction (ASR) pathway in AmII and AmIV strains and in one AmIa isolate (Akk2670) (Fig. 6). ASR is required to harvest sulfur from imported sulfate for the biosynthesis of amino acids. Akk2670 was unique among AmI strains in that it displayed very slow growth rates in gastric mucin. This led us to postulate that the long doubling times for Akk2670, and AmII and AmIV isolates, reflect a reduced capacity to synthesize sulfur-containing amino acids. Although mucin contains sulfur in the form of cysteine and methionine in the protein backbone and terminally sulfated glycans (47), the cysteine and methionine content may not be sufficient to support rapid growth, and sulfate cannot be used in the absence of a functional ASR pathway. The ASR pathway generates H2S, which is used to synthesize cysteine through condensation with O-acetylserine by cysteine synthase (48). Most AmI strains have two putative cysteine synthases (Amuc_1301 and Amuc_2014 in MucT), with Amuc_2014 being conserved in all Akkermansia (Fig. 6B). Cysteine also plays an essential role as a sulfur source for the biosynthesis of essential cofactors, vitamins, and antioxidants (48). Consistent with this prediction, the growth of Akk2670, AmII, and AmIV strains is significantly enhanced by the addition of exogenous cysteine or H2S to mucin medium (Fig. 6C).
It is clear that the loss of ASR is not essential for GI colonization by A. muciniphila and may even enhance competitiveness given that sulfate transport and reduction is an energy intensive process (49, 50), particularly if other sources of reduced sulfur are available from the host or the microbiota. Potential sources of reduced sulfur in the GI tract include taurine, low levels of cysteine, and H2S (51). For some members of the Bifidobacterium genus, cysteine auxotrophies are prominent, and at least in the case of B. bifidum, cysteine auxotrophy cannot be rescued by supplementation of glutathione or taurine (52). Finally, several bacterial pathogens are cysteine auxotrophs, and even among species with complete ASR systems, clinical isolates have been observed to spontaneously become cysteine auxotrophs (48). Thus, there may be selective pressure for the loss of ASR genes in the presence of alternative reduced sulfur sources.
The loss of ASR in some A. muciniphila strains suggests that AmII and IV strains could be net consumers of any microbiota-derived H2S, especially under conditions where AmII and AmIV strains constitute a significant proportion of the entire microbiota (Fig. 8). If so, their localized detoxification of H2S may contribute to some of the protection that has been ascribed to Akkermansia in the context of inflammatory bowel disease (IBD) and Crohn’s disease (53–55). On the other hand, H2S derived from the breakdown of cysteine by intestinal cystathionine β-synthase has anti-inflammatory properties (56) and may stimulate the production of mucins (57). Under these circumstances, colonization by AmII and AmIV strains may be proinflammatory if they decrease the effective concentration of H2S at epithelial surfaces.
The relative competitive advantage of AmII and AmIV strains in antibiotic-treated mice was unexpected given the relative high prevalence of AmI strains in human populations (16, 44). However, we noted that microbiota of specific-pathogen-free (SPF) mice, which has an endogenous mouse AmI strain, provided colonization resistance, which led us to ask if a similar phylogroup exclusion is observed in humans. In stool samples collected at various time points after various interventions aimed at reducing obesity, we observed dominance by a single phylogroup. In some instances, both 16S rRNA-based community profiling and qPCR indicated that A. muciniphila was not detectable in baseline samples yet appeared by 6 months (PID001 and PID009), and in others, the dominant phylogroup disappeared within 3 months after baseline sample collection (PID033 and PID137), and yet in other patient samples, a phylogroup would disappear and return (PID049) or be replaced by a new phylogroup (PID141, PID181, and PID182). The abrupt disappearance of Akkermansia has been previously documented in densely sampled individuals (58). At this, stage we cannot distinguish between population crashes that are followed by repopulation by a newly acquired Akkermansia phylogroup or blooms of preexisting phylogroups that were present below the limits of detection. Evidence for the latter is supported by our ability to culture A. muciniphila strains that did not belong to the predominant phylogroup within the stool sample. Overall, patients are dominated by a single major phylogroup at any one time, but the abundance and identity of each A. muciniphila phylogroup is subject to fluctuation by environmental factors and ecological pressures that are still unknown.
The phenotypic diversity of the A. muciniphila strains in this cohort of patients suggests that experimental approaches using cultured strains will be critical to understand Akkermansia physiology. A recent survey of more than 10,000 adults in the American Gut Project established a weak inverse correlation between A. muciniphila abundance and BMI, with a protective role against obesity when adjusted for confounders such as sex, age, and diet (11). It is plausible that these correlations may be further strengthened when stratified by what is the most prevalent A. muciniphila phylogroup in an individual. It is certainly possible that while some strains are beneficial, others may be neutral or even potentially harmful (59). Even strains that are considered beneficial, such as MucT, may be harmful in the “wrong” context depending on the host’s inflammatory status, diet, or microbiota (60). The observation that patients can be colonized by different strains at different times suggests that A. muciniphila colonization is a dynamic process, especially considering that its primary food source, host mucins, should not be subject to the same variability as the diet-derived carbohydrates used by other intestinal microbes. Determining which A. muciniphila strains are most beneficial, and what factors influence strain-specific colonization, will be critical for the development of effective A. muciniphila-based probiotics.
MATERIALS AND METHODS
Media, strains, and growth conditions.
Bacteria were isolated and grown in an anaerobic chamber (Coy Laboratory) with the following gaseous characteristics: 5% hydrogen, 5% carbon dioxide, and 90% nitrogen. A. muciniphila was grown in mucin medium based on previous work (7) (3 mM KH2PO4, 3 mM Na2PO4, 5.6 mM NH4Cl, 1 mM MgCl2, 1 mM Na2S · 9H2O, 47 mM NaHCO3, 1 mM CaCl2, and 40 mM HCl, trace elements and vitamins [61], and 0.25% porcine gastric mucin [type III, Sigma-Aldrich]). Additional media used to culture Akkermansia included synthetic media, where porcine gastric mucin was replaced with 0.2% GlcNAc, 0.2% glucose, 16g/liter of soy peptone and 4 g of threonine/liter (14), and BD Bacto brain heart infusion broth (BD; catalog number 237500) with 0.25% porcine gastric mucin. To test growth with cysteine, mucin medium was supplemented with filter-sterilized l-cysteine to a final concentration of 0.5 mM. The A. muciniphila strain MucT (7) was obtained from ATCC (BAA-835). HEK-Blue hTLR2/1/6 and hTLR4 were obtained from InvivoGen (hkb-htlr2, hkb-htlr4) and maintained as described by the manufacturer. HT29-MTX was from Sigma (12040401-1VL) and maintained in Dulbecco’s modified Eagle medium (DMEM) (Gibco 11995-065) supplemented with 10% fetal bovine serum.
Isolation of A. muciniphila from fecal samples.
The recruitment criteria and composition of stool donors in the POMMS Study were previously reported (18). Approximately 75 mg of frozen stool was used to inoculate 1 ml of mucin medium supplemented with vancomycin (6 μg/ml), gentamicin (10 μg/ml), and kanamycin (12 μg/ml) and incubated at 37°C for 48 h. After three sequential passages in mucin medium, a sample of the suspension was streaked on 1% agar mucin medium plates to isolate single colonies and was incubated for 7 days at 37°C. Colonies of unique morphology were restreaked on 1% agar BBL brain heart infusion (BD Biosciences; catalog 211065) plates supplemented with 0.2% mucin and incubated for 4 days. Total DNA was isolated, and the strain was identified by PCR-based amplification of the 16S rRNA gene V3-V4 region. The nomenclature used for new A. muciniphila strains is as follows: AkkXXXYn, with X being the patient number (009-275), Y the month of sample collection, (0,1.5,3,4.5, or 6) and n the clone typed if more that one colony was collected per plate (a to d; no letter indicates that only one colony was picked for analysis). AkkB40 is an isolate from a healthy adult male.
Global analysis of microbial composition by 16S rRNA sequencing.
The composition of total bacteria in stool samples was determined from DNA samples extracted from stool with a Qiagen stool extraction kit (Qiagen; catalog number 51604) by amplification of the 16S rRNA gene by PCR using primers 515 and 806 as described in the Earth Microbiome Project protocols, followed by DNA sequencing of the resulting amplicons on an Illumina MiSeq platform.
Phylogenetic analysis was performed using the Quantitative Insights into Microbial Ecology 2 (QIIME 2) platform version 2019.7 (62). Raw sequence data were demultiplexed using the emp-paired option (63, 64), followed by denoising with DADA2 (65) using the parameters p-trim-left-f 10, p-trim-left-r 10, p-trunc-len-f 233, and p-trunc-len-r 164. Amplicon sequence variants (ASVs) were assigned taxonomy using the feature classifier classify-sklearn (66, 67) and the SILVA 132 99% 515F/806R reference sequences (67).
Phenotypic characterization of A. muciniphila isolates.
Growth rate determination. Growth rates were assessed in both liquid mucin and synthetic medium. Starter cultures were grown to saturation in 3 ml synthetic medium supplemented with 0.25% porcine gastric mucin and diluted 1:5 into fresh medium and grown for an additional 8 h. The resulting cultures were then diluted 1:25 into fresh medium (optical density at 600 nm [OD600], 0.01 to 0.05), and 150-μl aliquots were dispensed into 96-well microplates. Each well was covered with 100 μl of paraffin oil and incubated at 37°C in a BMG SpectroStar Nano plate reader under anaerobic conditions. The optical density (OD600) was measured at 1-h intervals for 72 h. Generation times and growth rates were determined using the R package Growthcurver (version 0.3.0) (68). Results were obtained from three biological replicates per strain.
Growth with cysteine and sodium hydrogen sulfide. Growth was tested in medium supplemented with l-cysteine or the H2S donor sodium hydrogen sulfide (NaSH) (Cayman Chemical; catalog number 10012555). NaSH stock solutions were prepared in phosphate-buffered saline (PBS) under anaerobic conditions. Akkermansia starter cultures were standardized to an OD600 of 0.5 and diluted 1:25 into 3 ml of mucin medium with or without 1 mM l-cysteine or 40 μM NaSH. The cultures were incubated anaerobically at 37°C, and the optical density was measured over 4 days. All assays were run in triplicate.
Agglutination. Actively growing cultures were used to inoculate 1.2 ml of mucin medium in fresh deep 96-well plates and incubated anaerobically at 37°C for 3 days. The degree of bacterial sedimentation was quantified by removing 150 μl of culture from the top of the well. Agglutination was calculated for each strain as:
Three biological replicates of this assay were performed for every strain, and each contained three technical replicates.
Adherence to epithelial cells. HT29-MTX cells were seeded into 96-well plates at a density of 2.5 × 104 cells per well and grown for 7 days past confluence. Wells were washed twice with PBS and incubated with 2.5 × 106 A. muciniphila cells in DMEM for 2 h at 37°C under anaerobic conditions. As a control for nonspecific binding of Akkermansia, UltraCruz high binding enzyme-linked immunosorbent assay (ELISA) (sc-204463) plates were precoated with 100 μl of 1% bovine serum albumin (BSA). Wells with HT29-MTX cells or coated with BSA were washed twice with PBS to remove nonadherent bacteria. Synthetic medium (100 μl) was added to each well, and plates were cultured for either 48 h or 96 h at 37°C under anaerobic conditions. HT29-MTX cell or BSA binding was assessed by measuring the optical density at 600 nm after outgrowth in the assay wells and calculating the ratio of HT29-MTX coated OD:BSA coated OD. Data are reported as the average and standard deviation of 3 technical replicates from 3 to 4 independent biological replicates. For microscopy, HT29-MTX cells were seeded into 24-well plates with 12-mm round glass coverslips at a density of 1 × 105 cells per well and grown for 7 days past confluence. Wells were washed twice with PBS and incubated with 1 × 106 A. muciniphila cells in 500 ml anaerobic-adapted DMEM for 2 h at 37°C under anaerobic conditions. Wells were washed twice, fixed with 3.7% formaldehyde in PBS for 30 min on ice, washed twice with PBS, and blocked overnight at 4°C in blocking buffer (2% [wt/vol] BSA in PBS). Coverslips were incubated with a 1:50 dilution of anti-Akkermansia polyclonal antibody followed by an incubation with goat anti-rabbit-488 (Invitrogen; catalog number A-11008) and Hoechst for 1 h at 25°C. After two washes, coverslips were mounted on slides with Vectashield medium and imaged on a Nikon Eclipse Ti2 inverted microscope with ×20 objective.
Measurement of short-chain fatty acids (SCFA). For each strain, 1 ml of culture supernatants from strains grown in mucin medium was removed for SCFA analysis following the protocol of Holmes et al. (69). In brief, the supernatant was centrifuged at 14,000 relative centrifugal force (rcf) for 5 min at 4°C to pellet debris, and then 750 μl of supernatant was passed through a 0.22-mm spin column filter. The resultant filtrate was then acidified to a pH of <3 with 50 μl of 6N HCL and transferred to a glass autosampler vial for analysis. Filtrates were analyzed on an Agilent 7890 gas chromatograph (GC) equipped with a flame-ionization detector (FID) and an Agilent HP-FFAP free fatty-acid column (69). The concentrations of acetate and propionate in the samples were determined using an 8-point standard curve (0.1 mM to 16 mM).
Sensitivity to ambient oxygen. Strains were grown from frozen stocks in 0.5 ml mucin (0.4%) medium in deep 96-well plates to saturation. After subculturing in mucin medium for 5 h, serial dilutions of each strain were spotted on BBL BHI agar (BD; catalog number 211065) plates supplemented with 0.4% mucin. One plate was left in the anaerobic chamber, while the others were exposed to ambient O2 for 12, 18, or 24 h, before being returned to the chamber. The relative sensitivity to ambient oxygen was determined by monitoring the ratio of CFU with and without exposure to ambient oxygen.
Bone marrow macrophage (BMM) stimulations. BMMs were obtained from 6- to 12-week-old C57BL/6J mice of the following genotypes: wild-type, Tlr2–/– Tlr4–/– Unc93b13d/3d, Tlr2–/– Tlr4–/–, Tlr2–/– Unc93b13d/3d, and Tlr4–/– (25). Bone marrow was dissociated through a 70-μm filter, treated with ACK lysis buffer (Gibco; catalog numberA1049201), and differentiated for 6 days in DMEM complete medium (DMEM supplemented with 10% [vol/vol] fetal bovine serum, l-glutamine, penicillin-streptomycin, sodium pyruvate, HEPES, and 2-mercaptoethanol) supplemented with 10% (vol/vol) of supernatants from 3T3-CSF cells, overproducing macrophage colony-stimulating factor. For stimulations, BMMs were plated in DMEM complete medium supplemented with 10% (vol/vol) M-CSF and incubated with A. muciniphila at the indicated multiplicity of “infection” (MOI), 1 μM CpG-B (InvivoGen; tlr1-1668-1), 500 ng/ml Pam3CSK4 (InvivoGen; tlr1-pms), or 50 ng/ml lipopolysaccharide (LPS) (InvivoGen; tlr1-3pelps). For analysis of secreted cytokines, the supernatant was collected 4 h after stimulation and analyzed with the BD cytometric bead array mouse inflammation kit (BD Biosciences; catalog number 552364) according to the manufacturer’s instructions.
Activation of hTLRs. HEK-Blue hTLR2/1/6- and hTLR4-expressing cells (InvivoGen; hkb-htlr2, hkb-htlr4) were seeded into 96-well plates pretreated with poly-l-lysine. A. muciniphila isolates were added to each well at an MOI of 5 in triplicate. Negative controls included culture medium with 10% heat-inactivated FBS. Positive controls included ultrapure lipopolysaccharide from Escherichia coli 055:B5 at 100 ng/ml and 1 ng/ml. For the experiments using hTLR2/1/6, Pam3CSK4, a synthetic triacylated lipopeptide, was used at concentrations of 100 ng/ml and 5 ng/ml. After a 16-h incubation, levels of sAP were assessed with Quanti-Blue detection medium as detailed by the manufacturer.
Genomic sequencing, annotation, and comparative analysis.
A. muciniphila genomic DNA was extracted using a MagAttract high-molecular-weight (HMW) DNA kit (Qiagen; catalog number 67563) according to the manufacturer’s protocol. The extracted DNA was ethanol precipitated, and the final concentration was determined with a Qubit double-stranded DNA (dsDNA) high-sensitivity (HS) kit (Thermo Scientific). Libraries were generated using a SMRTbell template prep kit version 2.0 (Pacific Biosciences [PacBio]) and sequenced on a PacBio Sequel instrument.
After sequencing, PacBio SMRTLink software (version 8.0.0) was used to demultiplex the samples, and the resulting BAM files were converted to fasta files using SAMtools (70). Genome assembly was performed with Flye version 2.7 with the following parameters: –genome-size 3m, –plasmids, and –meta (71). The assembled, circular genomes were then rotated to set the starting position to the dnaA gene using the fixstart function in Circlator (72). Finally, assembly annotation and quality evaluation were run using the PATRIC RASTtk-enabled genome annotation service (73). Sequences have been deposited in GenBank (NCBI BioProject accession number PRJNA715455.
Comparative analysis of the assembled genomes was run using the pangenomic workflow in Anvi’o version 6.2 (74, 75). First, the 43 assembly fasta files were reformatted into an Anvi’o-compatible contig database by running the script anvi-script-FASTA-to-contigs-db. This command uses Prodigal to identify open reading frames (76). The resulting databases were annotated with the script anvi-run-ncbi-cogs and subsequently combined to make a genome database using anvi-gen-genomes-storage. To compute the pangenome, the command anvi-pan-genome was run with the following parameters: –minbit 0.5, –mcl-inflation 10, and –use-ncbi-blast.
We computed average nucleotide identity (ANI) across the genomes using the command anvi-compute-genome-similarity with the –method pyani parameter (77). We included publicly available Akkermansia genomes as controls in our ANI and pangenome analyses. Representatives of phylogroups AmI, AmII, and AmIII were described in Guo et al. and retrieved from NCBI (17) (GenBank assembly accession numbers GCA_002885425.1, GCA_002885025.1, GCA_002884975.1, GCA_002884915.1, and GCA_002885515.1). The phylogroup AmIV representative genome CDI-150b was obtained from the JGI IMG database (16). Based on the resulting analysis, each genome was assigned to a phylogroup using the function anvi-import-misc-data, and phylogroup-specific gene functions were then obtained using the command anvi-get-enriched-functions-per-pan-group to identify cluster of orthologous groups of proteins (COGs). COGs present in all members of a given phylogroup, and absent in all other phylogroups, were considered to represent phylogroup-specific gene functions. The adjusted q value represents the false-discovery rate adjusted P value corrected for multiple testing as calculated by Anvi-o. Finally, the genomes were analyzed for metabolic pathways in Anvi’o (https://merenlab.org/software/anvio/help/main/programs/anvi-estimate-metabolism/). Each genome was annotated with the KEGG KOfam database using the program anvi-run-kegg-kofams (78, 79). The annotated genomes were then used as input to the program anvi-estimate-metabolism, with the flag –module-completion-threshold set to identify pathways with a minimum of 50% completion in at least one isolate genome. The module output was further analyzed with the command anvi-compute-functional-enrichment to test for phylogroup-specific enrichment. To visualize the data, heatmaps were generated using the pheatmap R package (80), and genes were plotted with the gggenes R package (81) in ggplot2 (82). To search for specific ASR genes among the isolates, custom BLAST databases were generated using the annotated isolate genomes (83). Searches were conducted using the sequences for the ASR genes from A. muciniphila MucT as the query.
To identify glycoside hydrolase families in the sequenced strains, we used dbCAN2 version 2.0.11 to annotate carbohydrate-active enzymes (84). DNA fasta files were used as input, and the annotation was run using the standalone tool run_dbcan. The resulting annotation tables were analyzed to determine the number of each type of glycoside hydrolase family per genome. Annotations detected with Diamond and at least one additional method, Hotpep or Hmmer, were considered positive.
Mouse colonization and phylogroup competitions.
To prepare the inoculum for competition experiments, A. muciniphila cultures were standardized by optical density, combined using equal parts of each phylogroup to be tested, and stored at −80°C in PBS containing 20% glycerol.
All mouse experiments were approved by Duke University’s Institutional Animal Care and Use Committee. In vivo competition experiments were caried out using 6-week-old female C57BL/6J mice obtained from Jackson Laboratories. A. muciniphila colonization was tested in mice both with and without pretreatment with antibiotics (3 g/liter tetracycline suspended in distilled water with 10% sucrose for 2 weeks). Following antibiotic treatment, clearance of residual mouse Akkermansia was determined by PCR using Akkermansia-specific 16S rRNA primers (12).
For in vivo competition assays, mice were inoculated by intragastric gavage with a mixture containing 2.5 × 108 CFU of each phylogroup in a total volume of 140 μl. The three-phylogroup competition experiment used a mixture of the strains MucT (AmIa), Akk1683 (AmIb), and Akk0580 (AmII). Two additional competition experiments were run, each using a combination of four clades. The first four-clade competition used a mixture of strains MucT (AmIa), Akk1570 (AmIb), Akk0580 (AmII), and Akk0490 (AmIV). The second four-clade competition used a mixture of strains Akk2670 (AmIa), Akk2650 (AmIb), Akk2680 (AmII), and Akk2750 (AmIV).
Analysis of A. muciniphila phylogroup distribution in fecal samples from mice and humans.
Phylogroup abundance was assessed by qPCR with phylogroup-specific primers. PCR was run with PowerUp SYBR master mix reagent on a QuantStudio 3 real-time PCR system (Applied Biosystems) using fast cycling mode. The abundance of Akkermansia was calculated as copies per gram fecal material. For human samples, the same DNA used for 16S rDNA sequencing was used as the template for qPCR with phylogroup-specific primers (Table S4).
Unless otherwise noted, statistical analyses and plots were generated with GraphPad Prism version 9.0.0.
ACKNOWLEDGMENTS
We thank the Duke University School of Medicine for the use of the Microbiome Core Facility, which provided fecal samples and 16S rRNA analysis support and the Sequencing and Genomic Technologies Core for sequencing Akkermansia genomes. We also thank Eliud Rivas Hernandez and Ozge Kuddar for assistance with strain isolations, Gabrielle Reiner and Shaina Carroll for assistance with flow analysis, and Gilberto Flores (Cal State) for his useful comments on the manuscript.
This work was supported by NIH awards AI142376 and CA249243 (R.H.V.). POMMS was supported by DK110492. B.B. was supported by the Pediatric Scientist Development Program (K12-HD000850, Eunice Kennedy Shriver National Institute of Child Health and Human Development). L.D. was supported by an AHA grant (18POST34070017). D.R.M. is supported by a fellowship from NIDDK-5T32DK007568-30, and K.D.M. is supported by an NSF institutional predoctoral training grant.
R.H.V. is a founder of Bloom Science, a microbiome therapeutics company. Bloom Science was not involved in the design, funding, or interpretation of the findings in this work.
Footnotes
This article is a direct contribution from Raphael H. Valdivia, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Eric Martens, University of Michigan, and Manuela Raffatellu, University of California San Diego.
Citation Becken B, Davey L, Middleton DR, Mueller KD, Sharma A, Holmes ZC, Dallow E, Remick B, Barton GM, David LA, McCann JR, Armstrong SC, Malkus P, Valdivia RH. 2021. Genotypic and phenotypic diversity among human isolates of Akkermansia muciniphila. mBio 12:e00478-21. https://doi.org/10.1128/mBio.00478-21.
Contributor Information
Raphael H. Valdivia, Email: raphael.valdivia@duke.edu.
Margaret J. McFall-Ngai, University of Hawaii at Manoa
REFERENCES
- 1.Fan Y, Pedersen O. 2021. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol 19:55–71. doi: 10.1038/s41579-020-0433-9. [DOI] [PubMed] [Google Scholar]
- 2.Allaire JM, Crowley SM, Law HT, Chang SY, Ko HJ, Vallance BA. 2018. The intestinal epithelium: central coordinator of mucosal immunity. Trends Immunol 39:677–696. doi: 10.1016/j.it.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, Leonard P, Li J, Burgdorf K, Grarup N, Jørgensen T, Brandslund I, Nielsen HB, Juncker AS, Bertalan M, Levenez F, Pons N, Rasmussen S, Sunagawa S, Tap J, Tims S, Zoetendal EG, Brunak S, Clément K, Doré J, Kleerebezem M, Kristiansen K, Renault P, Sicheritz-Ponten T, De Vos WM, Zucker JD, Raes J, Hansen T, Bork P, Wang J, Ehrlich SD, Pedersen O, Guedon E, Delorme C, Layec S, Khaci G, Van De Guchte M, Vandemeulebrouck G, Jamet A, Dervyn R, Sanchez N, MetaHIT consortium, et al. 2013. Richness of human gut microbiome correlates with metabolic markers. Nature 500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 4.Sonnenburg ED, Sonnenburg JL. 2019. The ancestral and industrialized gut microbiota and implications for human health. Nat Rev Microbiol 17:383–390. doi: 10.1038/s41579-019-0191-8. [DOI] [PubMed] [Google Scholar]
- 5.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derrien M, Collado MC, Ben-Amor K, Salminen S, de Vos WM. 2008. The mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract. Appl Environ Microbiol 74:1646–1648. doi: 10.1128/AEM.01226-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derrien M, Vaughan EE, Plugge CM, de Vos WM. 2004. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol 54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 8.Cani PD, de Vos WM. 2017. Next-generation beneficial microbes: the case of Akkermansia muciniphila. Front Microbiol 8:1–8. doi: 10.3389/fmicb.2017.01765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dao MC, Everard A, Aron-Wisnewsky J, Sokolovska N, Prifti E, Verger EO, Kayser BD, Levenez F, Chilloux J, Hoyles L, Dumas M-EE, Rizkalla SW, Doré J, Cani PD, Clément K, MICRO-Obes Consortium . 2016. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut 65:426–436. doi: 10.1136/gutjnl-2014-308778. [DOI] [PubMed] [Google Scholar]
- 10.Yassour M, Lim MY, Yun HS, Tickle TL, Sung J, Song YM, Lee K, Franzosa EA, Morgan XC, Gevers D, Lander ES, Xavier RJ, Birren BW, Ko GP, Huttenhower C. 2016. Sub-clinical detection of gut microbial biomarkers of obesity and type 2 diabetes. Genome Med 8:1–14. doi: 10.1186/s13073-016-0271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Q, Zhang Y, Wang X, Yang R, Zhu X, Zhang Y, Chen C, Yuan H, Yang Z, Sun L. 2020. Gut bacteria Akkermansia is associated with reduced risk of obesity: evidence from the American Gut Project. Nutr Metab (Lond) 17:90. doi: 10.1186/s12986-020-00516-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collado MC, Derrien M, Isolauri E, De Vos WM, Salminen S. 2007. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl Environ Microbiol 73:7767–7770. doi: 10.1128/AEM.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. 2013. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A 110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, Chilloux J, Ottman N, Duparc T, Lichtenstein L, Myridakis A, Delzenne NM, Klievink J, Bhattacharjee A, van der Ark KCH, Aalvink S, Martinez LO, Dumas M-E, Maiter D, Loumaye A, Hermans MP, Thissen J-P, Belzer C, de Vos WM, Cani PD. 2017. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med 23:107–113. doi: 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- 15.Depommier C, Everard A, Druart C, Plovier H, Van Hul M, Vieira-Silva S, Falony G, Raes J, Maiter D, Delzenne NM, de Barsy M, Loumaye A, Hermans MP, Thissen J-PP, de Vos WM, Cani PD. 2019. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med 25:1096–1103. doi: 10.1038/s41591-019-0495-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirmiz N, Galindo K, Cross KL, Luna E, Rhoades N, Podar M, Flores GE. 2019. Comparative genomics guides elucidation of vitamin B 12 biosynthesis in novel human-associated Akkermansia strains. Appl Environ Microbiol 86:e02117-19. doi: 10.1128/AEM.02117-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo X, Li S, Zhang J, Wu F, Li X, Wu D, Zhang M, Ou Z, Jie Z, Yan Q, Li P, Yi J, Peng Y. 2017. Genome sequencing of 39 Akkermansia muciniphila isolates reveals its population structure, genomic and functional diverisity, and global distribution in mammalian gut microbiotas. BMC Genomics 18:1–12. doi: 10.1186/s12864-017-4195-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCann JR, Bihlmeyer NA, Roche K, Catherine C, Jawahar J, Kwee LC, Younge NE, Silverman J, Ilkayeva O, Sarria C, Zizzi A, Wootton J, Poppe L, Anderson P, Arlotto M, Wei Z, Granek JA, Valdivia RH, David LA, Dressman HK, Newgard CB, Shah SH, Seed PC, Rawls JF, Armstrong SC. 2021. The Pediatric Obesity Microbiome and Metabolism Study (POMMS): methods, baseline data, and early insights. Obesity (Silver Spring) 29:569–578. doi: 10.1002/oby.23081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xing J, Li X, Sun Y, Zhao J, Miao S, Xiong Q, Zhang Y, Zhang G. 2019. Comparative genomic and functional analysis of Akkermansia muciniphila and closely related species. Genes Genomics 41:1253–1264. doi: 10.1007/s13258-019-00855-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reunanen J, Kainulainen V, Huuskonen L, Ottman N, Belzer C, Huhtinen H, de Vos WM, Satokari R. 2015. Akkermansia muciniphila adheres to enterocytes and strengthens the integrity of the epithelial cell layer. Appl Environ Microbiol 81:3655–3662. doi: 10.1128/AEM.04050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belzer C, Chia LW, Aalvink S, Chamlagain B, Piironen V, Knol J, de Vos WM. 2017. Microbial metabolic networks at the mucus layer lead to diet-independent butyrate and vitamin B12 production by intestinal symbionts. mBio 8:1–14. doi: 10.1128/mBio.00770-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ouwerkerk JP, van der Ark KCH, Davids M, Claassens NJ, Finestra TR, de Vos WM, Belzer C. 2016. Adaptation of Akkermansia muciniphila to the oxic-anoxic interface of the mucus layer. Appl Environ Microbiol 82:6983–6993. doi: 10.1128/AEM.01641-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ottman N, Reunanen J, Meijerink M, Pietilä TE, Kainulainen V, Klievink J, Huuskonen L, Aalvink S, Skurnik M, Boeren S, Satokari R, Mercenier A, Palva A, Smidt H, de Vos WM, Belzer C. 2017. Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS One 12:e0173004. doi: 10.1371/journal.pone.0173004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cario E, Gerken G, Podolsky DK. 2007. Toll-like receptor 2 controls mucosal inflammation by regulating epithelial barrier function. Gastroenterology 132:1359–1374. doi: 10.1053/j.gastro.2007.02.056. [DOI] [PubMed] [Google Scholar]
- 25.Sivick KE, Arpaia N, Reiner GL, Lee BL, Russell BR, Barton GM. 2014. Toll-like receptor-deficient mice reveal how innate immune signaling influences Salmonella virulence strategies. Cell Host Microbe 15:203–213. doi: 10.1016/j.chom.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huh JW, Shibata T, Hwang M, Kwon E-H, Jang MS, Fukui R, Kanno A, Jung DJ, Jang MH, Miyake K, Kim YM. 2014. UNC93B1 is essential for the plasma membrane localization and signaling of Toll-like receptor 5. Proc Natl Acad Sci U S A 111:7072–7077. doi: 10.1073/pnas.1322838111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Majer O, Liu B, Woo BJ, Kreuk LSM, Van Dis E, Barton GM. 2019. Release from UNC93B1 reinforces the compartmentalized activation of select TLRs. Nature 575:371–374. doi: 10.1038/s41586-019-1611-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliveira-Nascimento L, Massari P, Wetzler LM. 2012. The role of TLR2 in infection and immunity. Front Immunol 3:1–17. doi: 10.3389/fimmu.2012.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan YJ, Lin TL, Chen CT, Chen YY, Hsieh PF, Hsu CR, Wu MC, Wang JT. 2015. Genetic analysis of capsular polysaccharide synthesis gene clusters in 79 capsular types of Klebsiella spp. Sci Rep 5:15573. doi: 10.1038/srep15573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Percy MG, Gründling A. 2014. Lipoteichoic acid synthesis and function in Gram-positive bacteria. Annu Rev Microbiol 68:81–100. doi: 10.1146/annurev-micro-091213-112949. [DOI] [PubMed] [Google Scholar]
- 31.May KL, Lehman KM, Mitchell AM, Grabowicz M. 2019. A stress response monitoring lipoprotein trafficking to the outer membrane. mBio 10:1–14. doi: 10.1128/mBio.00618-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lau CKY, Krewulak KD, Vogel HJ. 2016. Bacterial ferrous iron transport: the Feo system. FEMS Microbiol Rev 40:273–298. doi: 10.1093/femsre/fuv049. [DOI] [PubMed] [Google Scholar]
- 33.Achard MES, Chen KW, Sweet MJ, Watts RE, Schroder K, Schembri MA, McEwan AG. 2013. An antioxidant role for catecholate siderophores in Salmonella. Biochem J 454:543–549. doi: 10.1042/BJ20121771. [DOI] [PubMed] [Google Scholar]
- 34.Peralta DR, Adler C, Corbalán NS, Paz García EC, Pomares MF, Vincent PA. 2016. Enterobactin as part of the oxidative stress response repertoire. PLoS One 11:e0157799. doi: 10.1371/journal.pone.0157799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitamura M, Okuyama M, Tanzawa F, Mori H, Kitago Y, Watanabe N, Kimura A, Tanaka I, Yao M. 2008. Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides thetaiotaomicron. J Biol Chem 283:36328–36337. doi: 10.1074/jbc.M806115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu QP, Yuan H, Bennett EP, Levery SB, Nudelman E, Spence J, Pietz G, Saunders K, White T, Olsson ML, Henrissat B, Sulzenbacher G, Clausen H. 2008. Identification of a GH110 subfamily of α1,3-galactosidases: novel enzymes for removal of the α3Gal xenotransplantation antigen. J Biol Chem 283:8545–8554. doi: 10.1074/jbc.M709020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu H, Rebello O, Crost EH, Owen CD, Walpole S, Bennati-Granier C, Ndeh D, Monaco S, Hicks T, Colvile A, Urbanowicz PA, Walsh MA, Angulo J, Spencer DIR, Juge N. 2021. Fucosidases from the human gut symbiont Ruminococcus gnavus. Cell Mol Life Sci 78:675–693. doi: 10.1007/s00018-020-03514-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robbe C, Capon C, Maes E, Rousset M, Zweibaum A, Zanetta JP, Michalski JC. 2003. Evidence of regio-specific glycosylation in human intestinal mucins: presence of an acidic gradient along the intestinal tract. J Biol Chem 278:46337–46348. doi: 10.1074/jbc.M302529200. [DOI] [PubMed] [Google Scholar]
- 39.Makarova KS, Koonin EV. 2015. Annotation and classification of CRISPR-Cas systems, p 47–75. In Lundgren M, Charpentier E, Fineran P (ed), CRISPR: methods and protocols. Humana Press, New York, NY. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chylinski K, Le Rhun A, Charpentier E. 2013. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol 10:726–737. doi: 10.4161/rna.24321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sekowska A, Kung HF, Danchin A. 2000. Sulfur metabolism in Escherichia coli and related bacteria: facts and fiction. J Mol Microbiol Biotechnol 2:145–177. [PubMed] [Google Scholar]
- 42.Makki K, Deehan EC, Walter J, Bäckhed F. 2018. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe 23:705–715. doi: 10.1016/j.chom.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 43.Guo X, Zhang J, Wu F, Zhang M, Yi M, Peng Y. 2016. Different subtype strains of Akkermansia muciniphila abundantly colonize in southern China. J Appl Microbiol 120:452–459. doi: 10.1111/jam.13022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lv Q, Li SH, Zhang Y, Wang YC, Peng Y, Zhang XX. 2020. A thousand metagenome-assembled genomes of Akkermansia reveal new phylogroups and geographical and functional variations in human gut. bioRxiv 2020.09.10.292292. [DOI] [PMC free article] [PubMed]
- 45.Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, Paik SG, Lee H, Lee JO. 2007. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell 130:1071–1082. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 46.Kang JY, Nan X, Jin MS, Youn SJ, Ryu YH, Mah S, Han SH, Lee H, Paik SG, Lee JO. 2009. Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity 31:873–884. doi: 10.1016/j.immuni.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 47.Bäckström M, Ambort D, Thomsson E, Johansson MEV, Hansson GC. 2013. Increased understanding of the biochemistry and biosynthesis of MUC2 and other gel-forming mucins through the recombinant expression of their protein domains. Mol Biotechnol 54:250–256. doi: 10.1007/s12033-012-9562-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lensmire JM, Hammer ND. 2019. Nutrient sulfur acquisition strategies employed by bacterial pathogens. Curr Opin Microbiol 47:52–58. doi: 10.1016/j.mib.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 49.Tripp HJ, Kitner JB, Schwalbach MS, Dacey JWH, Wilhelm LJ, Giovannoni SJ. 2008. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature 452:741–744. doi: 10.1038/nature06776. [DOI] [PubMed] [Google Scholar]
- 50.Seif Y, Choudhary KS, Hefner Y, Anand A, Yang L, Palsson BO. 2020. Metabolic and genetic basis for auxotrophies in Gram-negative species. Proc Natl Acad Sci U S A 117:6264–6273. doi: 10.1073/pnas.1910499117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carbonero F, Benefiel AC, Alizadeh-Ghamsari AH, Gaskins HR. 2012. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front Physiol 3:1–11. doi: 10.3389/fphys.2012.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferrario C, Duranti S, Milani C, Mancabelli L, Lugli GA, Turroni F, Mangifesta M, Viappiani A, Ossiprandi MC, van Sinderen D, Ventura M. 2015. Exploring amino acid auxotrophy in Bifidobacterium bifidum PRL2010. Front Microbiol 6:1–11. doi: 10.3389/fmicb.2015.01331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Earley H, Lennon G, Balfe Á, Coffey JC, Winter DC, O’Connell PR. 2019. The abundance of Akkermansia muciniphila and its relationship with sulphated colonic mucins in health and ulcerative colitis. Sci Rep 9:15683. doi: 10.1038/s41598-019-51878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bian X, Wu W, Yang L, Lv L, Wang Q, Li Y, Ye J, Fang D, Wu J, Jiang X, Shi D, Li L. 2019. Administration of Akkermansia muciniphila ameliorates dextran sulfate sodium-induced ulcerative colitis in mice. Front Microbiol 10:1–18. doi: 10.3389/fmicb.2019.02259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Png CW, Lindén SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, Florin THJ. 2010. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol 105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 56.Wallace JL, Vong L, McKnight W, Dicay M, Martin GR. 2009. Endogenous and exogenous hydrogen sulfide promotes resolution of colitis in rats. Gastroenterology 137:569–578.e1. doi: 10.1053/j.gastro.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 57.Motta JP, Flannigan KL, Agbor TA, Beatty JK, Blackler RW, Workentine ML, Da Silva GJ, Wang R, Buret AG, Wallace JL. 2015. Hydrogen sulfide protects from colitis and restores intestinal microbiota biofilm and mucus production. Inflamm Bowel Dis 21:1006–1017. doi: 10.1097/MIB.0000000000000345. [DOI] [PubMed] [Google Scholar]
- 58.David LA, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, Perrotta A, Erdman SE, Alm EJ. 2014. Host lifestyle affects human microbiota on daily timescales. Genome Biol 15:R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cirstea M, Radisavljevic N, Finlay BB. 2018. Good bug, bad bug: breaking through microbial stereotypes. Cell Host Microbe 23:10–13. doi: 10.1016/j.chom.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 60.Seregin SS, Golovchenko N, Schaf B, Chen J, Pudlo NA, Mitchell J, Baxter NT, Zhao L, Schloss PD, Martens EC, Eaton KA, Chen GY. 2017. NLRP6 protects Il10−/− mice from colitis by limiting colonization of Akkermansia muciniphila. Cell Rep 19:733–745. doi: 10.1016/j.celrep.2017.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stams AJM, Van Dijk JB, Dijkema C, Plugge CM. 1993. Growth of syntrophic propionate-oxidizing bacteria with fumarate in the absence of methanogenic bacteria. Appl Environ Microbiol 59:1114–1119. doi: 10.1128/AEM.59.4.1114-1119.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Bin Kang K, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hamady M, Knight R. 2009. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res 19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. 2008. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods 5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Gregory Caporaso J. 2018. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:1–17. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glöckner FO. 2014. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res 42:D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sprouffske K, Wagner A. 2016. Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinformatics 17:17–20. doi: 10.1186/s12859-016-1016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Holmes ZC, Silverman JD, Dressman HK, Wei Z, Dallow EP, Armstrong SC, Seed PC, Rawls JF, David LA. 2020. Short-chain fatty acid production by gut microbiota from children with obesity differs according to prebiotic choice and bacterial community composition. mBio 11:e00914-20. doi: 10.1128/mBio.00914-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 37:540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- 72.Hunt M, De Silva N, Otto TD, Parkhill J, Keane JA, Harris SR. 2015. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol 16:1–10. doi: 10.1186/s13059-015-0849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eren AM, Esen OC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO. 2015. Anvi’o: an advanced analysis and visualization platform for ’omics data. PeerJ 2015:1–29. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Delmont TO, Eren EM. 2018. Linking pangenomes and metagenomes: the prochlorococcus metapangenome. PeerJ 6:e4320. doi: 10.7717/peerj.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. 2016. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods 8:12–24. doi: 10.1039/C5AY02550H. [DOI] [Google Scholar]
- 78.Aramaki T, Blanc-Mathieu R, Endo H, Ohkubo K, Kanehisa M, Goto S, Ogata H. 2020. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 36:2251–2252. doi: 10.1093/bioinformatics/btz859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Muto A, Kotera M, Tokimatsu T, Nakagawa Z, Goto S, Kanehisa M. 2013. Modular architecture of metabolic pathways revealed by conserved sequences of reactions. J Chem Inf Model 53:613–622. doi: 10.1021/ci3005379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kolde R. 2013. pheatmap: pretty heatmaps. R package version 1.0.12. https://cran.r-project.org/web/packages/pheatmap/index.html.
- 81.Wilkins D. 2020. gggenes: draw gene arrow maps in “ggplot2.” R package version 0.4.1. https://cran.r-project.org/web/packages/gggenes/index.html.
- 82.Wickham H. 2009. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
- 83.2008. BLAST command line applications user manual [Internet]. National Center for Biotechnology Information, Bethesda (MD). https://www.ncbi.nlm.nih.gov/books/NBK52640/. [Google Scholar]
- 84.Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, Busk PK, Xu Y, Yin Y. 2018. DbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 46:W95–W101. doi: 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of pediatric donors described in this study. Download Table S1, XLSX file, 0.01 MB (14.4KB, xlsx) .
Copyright © 2021 Becken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ASV tables and relative abundance and composition of microbial communities in POMMS samples. Download Table S2, XLSX file, 0.02 MB (20.4KB, xlsx) .
Copyright © 2021 Becken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary of A. muciniphila isolates and phylogroups and their phenotypes. Download Table S3, XLSX file, 0.02 MB (24.3KB, xlsx) .
Copyright © 2021 Becken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A. muciniphila binding to HT29 colonic epithelial cell and protein-coated plated. Adherence of A. muciniphila strains isolated from patients enrolled in the POMMS to HT29 colonic epithelial cell- or BSA-coated plates. Bacteria were incubated with HT29-MTX cell- or BSA-coated plates and extensively washed, and binding was indirectly evaluated by adding synthetic medium to the wells and measuring the optical density at 600 nm after 48 to 72 h. (A) Binding is provided as the ratio of the OD600 HT29-MTX to BSA-coated plates. (B and C) The nonnormalized OD600 units for HT29-MT (B) or BSA-coated (C) plates are provided for reference. Bars represent the mean and standard deviation of 3 technical replicates from 3 independent experiments. Download FIG S1, TIF file, 2.7 MB (2.7MB, tif) .
Copyright © 2021 Becken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A. muciniphila activation of HEK293-TLR reporter cell lines. (A to D) A. muciniphila strains were grown on mucin or synthetic medium and incubated with HEK293 reporter cell lines expressing either TLR1/2/6 (A and B) or TLR4 (C and D). Activation of the TLR in these cell lines was determined by assessing the secretion of alkaline phosphatase into culture supernatants. Data are reported as the normalized fold induction over the mean OD value of the entire population. Bars represent the mean and standard deviation of 3 technical replicates from 3 independent experiments. Download FIG S2, TIF file, 0.9 MB (923.3KB, tif) .
Copyright © 2021 Becken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used for the identification of A. muciniphila phylogroups and quality control for specificity. Download Table S4, XLSX file, 0.01 MB (12KB, xlsx) .
Copyright © 2021 Becken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogroup-specific enrichment of metabolic pathways. Download Table S5, XLSX file, 0.02 MB (17.7KB, xlsx) .
Copyright © 2021 Becken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genomic analysis of A. muciniphila strains and predicted functions. (A) Pathway enrichment analysis indicates selective acquisition and loss of metabolic pathways. To compare the metabolic capabilities of isolates, we used Anvi’o to estimate metabolism function to detect KEGG module pathways in the genomes. The lower threshold for detecting a pathway was set to 50%. Strains with 50% or less of the enzymes required for a given pathway are represented as blue squares in the heatmap. (B) Detection of selected glycoside hydrolase families by clade. Bars represent the average number of enzymes per genome. The analysis included genomes from clade AmIa (n = 5), AmIb (n = 20), AmII (n = 10), and AmIV = (n = 8). GH, glycoside hydrolase; NC, nonclassified glycoside hydrolases. Error bars represent the standard error of the mean (SEM). Download FIG S3, GIF file, 1.5 MB (1.5MB, gif) .
Copyright © 2021 Becken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A. muciniphila phylogroup AmIV outcompetes other phylogroups in colonizing the mouse GI. (A) Fecal samples were collected prior to gavage, and DNA was extracted to assess endogenous Akkermansia levels in mice with and without antibiotic treatment. Akkermansia was detected with primers specific for the Akkermansia 16S rRNA gene (total) and with phylogroup-specific primers. (B) In vivo competition assay using a second set of representative strains for each phylogroup. A representative of each strain from a healthy control was selected as follows: AmIa (Akk2670), AmIb (Akk2650), AmII (2680), and AmIV (2750). Mice were treated with antibiotics until the endogenous Akkermansia was cleared and were subsequently gavaged with a cell suspension containing each of the phylogroups. Colonization was monitored over time by collecting fecal pellets and testing for colonization using qPCR with phylogroup-specific primers. Download FIG S4, TIF file, 2.8 MB (2.8MB, tif) .
Copyright © 2021 Becken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogroup typing in patients with obesity over a 6-month period. Download Table S6, XLSX file, 0.01 MB (14.2KB, xlsx) .
Copyright © 2021 Becken et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.