ABSTRACT
The depside and depsidone series compounds of polyketide origin accumulate in the cortical or medullary layers of lichen thalli. Despite the taxonomic and ecological significance of lichen chemistry and its pharmaceutical potentials, there has been no single piece of genetic evidence linking biosynthetic genes to lichen substances. Thus, we systematically analyzed lichen polyketide synthases (PKSs) for categorization and identification of the biosynthetic gene cluster (BGC) involved in depside/depsidone production. Our in-depth analysis of the interspecies PKS diversity in the genus Cladonia and a related Antarctic lichen, Stereocaulon alpinum, identified 45 BGC families, linking lichen PKSs to 15 previously characterized PKSs in nonlichenized fungi. Among these, we identified highly syntenic BGCs found exclusively in lichens producing atranorin (a depside). Heterologous expression of the putative atranorin PKS gene (coined atr1) yielded 4-O-demethylbarbatic acid, found in many lichens as a precursor compound, indicating an intermolecular cross-linking activity of Atr1 for depside formation. Subsequent introductions of tailoring enzymes into the heterologous host yielded atranorin, one of the most common cortical substances of macrolichens. Phylogenetic analysis of fungal PKS revealed that the Atr1 is in a novel PKS clade that included two conserved lichen-specific PKS families likely involved in biosynthesis of depsides and depsidones. Here, we provide a comprehensive catalog of PKS families of the genus Cladonia and functionally characterize a biosynthetic gene cluster from lichens, establishing a cornerstone for studying the genetics and chemical evolution of diverse lichen substances.
KEYWORDS: Cladonia, lichen, atranorin, polyketides, secondary metabolism
INTRODUCTION
Lichen-forming fungi (LFF) live in symbiosis with photosynthetic partners, green algae or cyanobacteria—sometimes with both—and LFF are currently estimated to comprise about 20% of all known fungi (1). Lichen symbiosis is one of the most successful mutualisms that enable these organisms to adapt to extremely harsh habitats. A conglutinated, often pigmented, cortical layer made up of dense fungal hyphae provides photosynthetic partners with mechanical stabilization, and algal cells enveloped by hyphae from the medulla provide nutrients to LFF by means of photosynthesis. Secondary metabolites (SMs) of polyketide origin, namely anthraquinones, depsides, depsidones, and dibenzofurans (such as usnic acid), accumulate in cortical or medullary layers of lichen thalli (2). The ecological roles of these lichen SMs are largely unknown, but some studies provide evidence that cortical substances have contributed to habitat expansion (3) and defense from herbivore attacks (4).
Depside and depsidone series compounds are widespread in lichens as cortical and medullary substances, many of which are exclusively found in lichens (2). Lichen depsides are formed by dimerization of either orsellinic acid or 3-methylorsellinic acid (3MOA), and subsequent oxidation of depsides affords tricyclic scaffolds for depsidones. These compounds further undergo different combinations of modification within orsellinic acid and 3MOA moieties, such as alkylation, chlorination, hydroxylation, and O-methylation, yielding hundreds of structurally diverse compounds (5, 6). For this, depside/depsidone biosynthetic pathways in lichens could be a model system for studying the substrate specificity of biosynthetic enzymes resulting in chemodiversity. Thus far, however, ascribing specific biosynthetic genes to cortical or medullary substance has been slow in lichens, due to a paucity of genetic information on LFF and lack of molecular tools for manipulating LFF recalcitrant to genetic transformation.
Nonreducing iterative type I polyketide synthases (NR-PKSs) are multidomain enzymes and have been grouped into seven (7) or eight (8) major groups by protein sequence similarity and PKS domain architecture. Orsellinic acid and 3MOA are basic scaffolds for many different SMs in bacteria, fungi, and plants and are biosynthesized by NR-PKSs in a filamentous fungus, Aspergillus nidulans (9, 10), yet only predictions have been made for two NR-PKSs to be responsible for the biosynthesis of the basic units in lichens (11, 12). Atranorin (a 3MOA-derived depside) and usnic acid (a dibenzofuran) are the most common cortical substances of macrolichens and have attracted great attention because of their taxonomic (13, 14), ecological (15), and pharmaceutical (16–19) importance. Several studies have ascribed an NR-PKS to usnic acid with high likelihood (20–22). However, the effort was unsuccessful for heterologous expression of the putative usnic acid PKS gene in Aspergillus oryzae (23), and thus its precise function remains to be determined.
In recent years, SM research has benefited extensively from genome mining approaches in bacteria, fungi, and plants (24–28). We have sequenced eight lichen genomes, including two Antarctic lichens Cladonia borealis and Stereocaulon alpinum, and a foliose Parmelia sp. strain, KoLRI021559 (Parmelia cf. squarrosa), which are reported in this study. Cladoniaceae is one of the largest families of LFF and closely related to Stereocaulaceae (29). Historically, chemotaxonomy has been used as a polyphasic approach to resolve and delimit species boundaries in Cladoniaceae (30, 31), and the genus Cladonia has been a model for studying PKS genes for the biosynthesis of cortical and medullary substances (11, 12, 32, 33). Here, we investigated biosynthetic gene cluster (BGC) diversity in six Cladonia spp. representing four subsections of the genus and a related lichen, S. alpinum, to provide insight into the metabolic potentials of the genus Cladonia and to identify BGCs involved in cortical or medullary substances by categorizing PKS genes with a homologous relationship. Also, we established a novel heterologous expression system, in that we reconstructed the biosynthetic pathway for atranorin, a major cortical substance found in nearly every family within the Lecanorales (5), the largest and most diverse order of the class Lecanoromycetes.
RESULTS
Metabolic potentials of lichens.
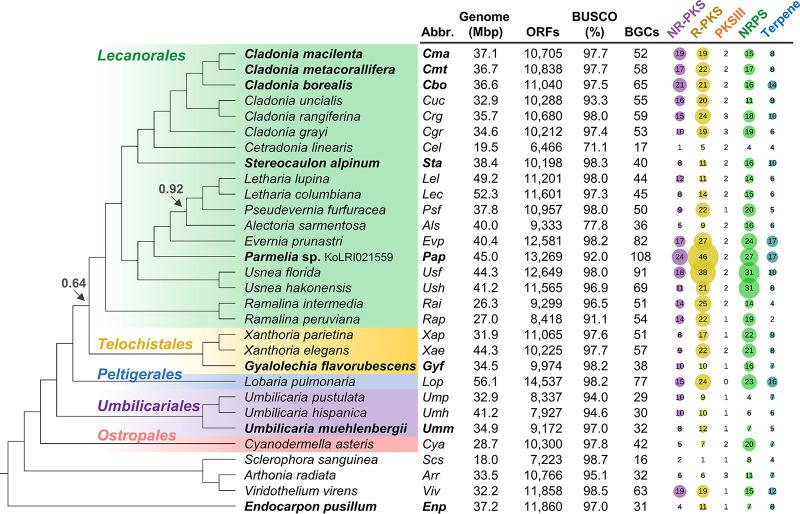
To examine evolutionary relationships of LFF and compare their genome-encoded metabolic potentials, a coalescent-based tree of 393 single-copy orthologous genes was reconstructed, using eight genomes that we have sequenced and 22 genome assemblies available in the NCBI database and JGI website as of September 2020. A majority of the sequenced species (26 out of 30) belong to the Lecanoromycetes, and four LFF were placed outside the Lecanoromycetes: Arthonia radiata (Arthoniomycetes), Endocarpon pusillum (Eurotiomycetes), Sclerophora sanguinea (Coniocybomycetes), and Viridothelium virens (Dothideomycetes). Barring the genomes of Alectoria sarmentosa and Cetradonia linearis, completeness of the genomes assessed by BUSCO analysis was greater than 91% (Fig. 1). An endangered lichen, Ce. linearis (34), and Sc. sanguinea had compact genomes shorter than 20 Mbp (Fig. 1). Except for these two species, all genomes were 26.3 to 56.1 Mbp in size, containing 7,927 to 14,537 open reading frames (ORFs) (Fig. 1). Genome-encoded metabolic potentials of the 30 species were investigated by mining the genomes for BGCs using antiSMASH (35). Metabolic potentials of the sequenced LFF were highly variable, with BGC counts of 16 to 108 (Fig. 1); Parmelia sp. strain KoLRI021559 showed the greatest BGC diversity, whereas the genomes of Ce. linearis and Sc. sanguinea carried the smallest sets of BGCs, which may be attributable to their contracted genome size. Notably, the six Cladonia genomes possess 52 to 65 BGCs and encode similar numbers of PKSs: an average of 16 NR-PKSs and 20 reducing-type PKSs (R-PKSs) (Fig. 1). The six Cladonia spp. produce unique sets of SMs of polyketide origin (see Table S1 and Fig. S1 in the supplemental material), which make them ideal for studying interspecies PKS gene diversity and identifying BGCs responsible for biosynthesis of cortical or medullary substances of lichens through comparative analysis. Full details on genome statistics and BGC information for the 30 sequenced species are provided in Data Set S1 in the supplemental material.
FIG 1.
Genome-encoded metabolic potentials of lichens. A coalescent-based species tree is reconstructed for 29 lichen-forming fungi and Cyanodermella asteris (a plant endophyte). The tree is rooted to Endocarpon pusillum. All local posterior probabilities for nodal supports are higher than 0.98, except those marked by arrows. The five orders of the Lecanoromycetes are highlighted with different colors, and lichen genomes sequenced by the Korean Lichen Research Institute are marked in boldface. BUSCO describes the completeness of the genome assemblies. The numbers of biosynthetic gene clusters (BGCs) are predicted by antiSMASH, and bubble plots indicate the relative abundance of core biosynthetic enzyme categories. NR-PKS, nonreducing type I PKS; R-PKS, reducing type I PKS (including PKS-NRPS hybrid enzymes); PKSIII, type III PKS; NRPS, nonribosomal peptide synthetase (including NRPS-like enzymes); and Terpene, biosynthetic enzymes related to terpenoid production.
Chemical structures of cortical and medullary substances of lichens. Download FIG S1, PDF file, 0.2 MB (238.6KB, pdf) .
Copyright © 2021 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Lichen substances reported in the 30 genome-sequenced species. Download Table S1, DOCX file, 0.02 MB (31.7KB, docx) .
Copyright © 2021 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genome statistics and biosynthetic gene clusters in the sequenced lichen species. Download Data Set S1, XLSX file, 0.02 MB (18.2KB, xlsx) .
Copyright © 2021 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genetic dereplication of Cladonia PKSs.
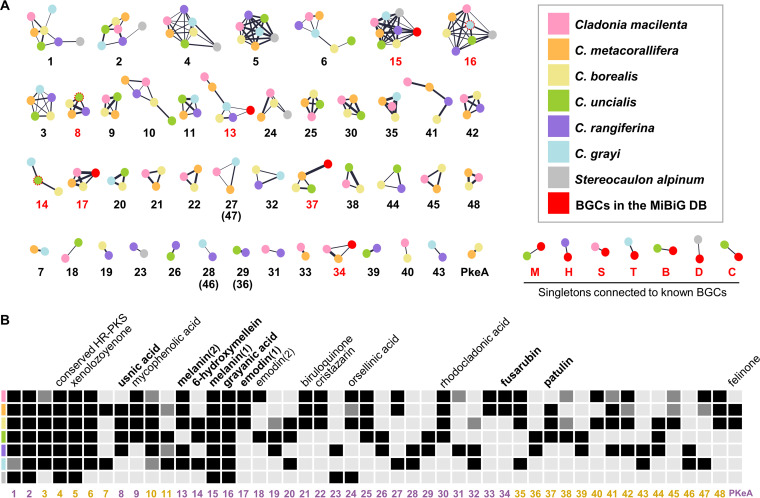
To search for PKS genes involved in biosynthesis of lichen polyketides, namely depside and depsidone series compounds, we focused on 226 BGCs harboring at least one PKS in the six Cladonia spp. plus a related Antarctic lichen, S. alpinum, from which a total of 242 PKSs were identified. We first conducted clustering analysis of the conserved ketosynthase (KS) domains of each PKS and identified a number of clusters that are suggestive of homologous relationships (see Fig. S2 in the supplemental material). Twelve PKS families described in an earlier study on the Cladonia chlorophaea species complex (33) were also detected in our clustering analysis, indicating that a significant proportion of PKS genes are conserved in the genus Cladonia. However, little is known about their products. Therefore, we linked BGCs to known compounds in nonlichenized fungi, using the Big-SCAPE program, which maps BGC diversity onto sequence similarity networks (36). The network analysis was used to graphically summarize three attributes of BGCs: (i) PKS families associated with the network (numbers), (ii) species distribution across the network (nodes), and (iii) degrees of similarity between pairs of BGCs (edges) (Fig. 2A). As depicted by the gene cluster network, the BGCs harboring at least one PKS gene were grouped into 45 gene cluster families (GCFs). Eight GCFs and seven species-specific BGCs (labeled in red) were coupled to previously characterized BGCs deposited in the MIBiG database (37). Figure 2B summarizes the phyletic distribution of Cladonia PKS families, denoting signature SMs linked to Cladonia PKS families. Full annotations of the PKS families linked to known compounds are provided in Text S1 in the supplemental material.
FIG 2.
Polyketide BGC diversity in Cladonia species. (A) Forty-five gene cluster families (GCFs) were identified by gene cluster network analysis in the six Cladonia spp. and Stereocaulon alpinum. GCFs are labeled with their associated PKS families. Numbers in red indicate PKS families connected to characterized BGCs deposited in the MIBiG database. Nodes indicate BGCs and are color coded by species (see the inset). Nodes enclosed by a red dashed circle in the PKS8, PKS14, and PKS16 subnetworks are lichen BGCs in the MIBiG database. Edge width was drawn proportional to squared similarity between a pair of BGCs. Singleton BGCs that connected to known BGCs are labeled as follows: M, monascorubrin BGC; H, hypothemycin BGC; S, sorbicillin BGC; T, terreic acid BGC; B, betaenone BGC; D, depudecin BGC; C, curvupallides BGC. (B) Phyletic distribution of PKS families evidenced by gene cluster network analysis. Numbers in violet and ochre indicate NR-PKS and R-PKS families, respectively. Black squares indicate the presence of PKS families, and gray squares indicate pseudogenes or partial genes due to incomplete gene annotation. Signature secondary metabolites linked to Cladonia PKS families by genetic dereplication are marked in boldface.
Supplemental materials and methods. Download Text S1, DOCX file, 0.2 MB (174.7KB, docx) .
Copyright © 2021 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Overview of the similarity of PKS families. Download FIG S2, PDF file, 0.4 MB (362.2KB, pdf) .
Copyright © 2021 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Identification of a putative atranorin BGC.
The gene cluster network analysis of the six Cladonia spp. and S. alpinum revealed that the PKS23 family among the 45 GCFs may be involved in biosynthesis of atranorin, the major cortical substance of diverse macrolichens, as it was the sole GCF shared by the two atranorin producers, C. rangiferina and S. alpinum (Fig. 2). Atranorin is unique in its structure having a methoxycarbonyl group within the 3MOA moiety, which is rarely found in other depside and depsidone series compounds. A BLAST search with an O-methyltransferase (OMT) in the PKS23 BGC in C. rangiferina showed 37% protein sequence identity to Trt5 (UniProtKB accession no. Q0C8A3), an OMT that mediates the formation of a methoxycarbonyl group within the 3,5-dimethylorsellinic acid moiety in A. terreus during the biosynthesis of terretonin (a meroterpenoid) (38) (Table 1). To examine the presence of conserved PKS23 BGCs in other lichens that produce atranorin, we adopted the CORASON pipeline (36), which is useful for studying conservation and variation of BGCs within and across GCFs. The protein sequence of the OMT in the PKS23 BGC in C. rangiferina was queried against a total of 1,527 BGCs detected in the 30 genomes. We identified 15 BGCs, each harboring an OMT that showed a significant hit (>33% protein sequence identity) (Fig. 3). Among the 15 BGCs, seven BGCs contained four syntenic core genes: the PKS23 gene (coined atr1), a cytochrome P450 monooxygenase gene (atr2), an OMT gene (atr3), and a transporter gene (atr4) (Fig. 3 and Table 1). These syntenic BGCs were exclusively found in the atranorin-producing lichens among the 30 sequenced species (Table S1).
TABLE 1.
The PKS23 biosynthetic gene cluster in Cladonia rangiferina
| ORFa | Size (aa) | BLASTP homologb | Identity (%) | Conserved domain | E value |
|---|---|---|---|---|---|
| 06811 | 744 | MFS monosaccharide transporter | 71 | Sugar transporter (Pfam00083) | 1e−105 |
| 06812 | 306 | Export control protein CHS7-like | 68 | Chitin synthase III catalytic subunit (Pfam12271) | 1e−131 |
| 06813 | 495 | TFIIH complex p47 subunit | 58 | TFIIH complex subunit Ssl1-like (Pfam04056) | 1e−82 |
| 06814 | 549 | MFS multidrug transporter (atr4) | 50 | Major facilitator superfamily (Pfam07690) | 9e−31 |
| 06815 | 346 | Trt5, O-methyltransferase (atr3) | 37 | SAM-dependent methyltransferase (Pfam08241) | 1e−04 |
| 06816 | 508 | Cytochrome P450 monooxygenase (atr2) | 43 | Cytochrome P450 (Pfam00067) | 3e−57 |
| 06817 | 2,513 | Nonreducing polyketide synthase (atr1) | 44 | SAT-KS-AT-PT-ACP-MT-TEc | |
| 06818 | 472 | Hypothetical protein | 33 | Phosphotransferase enzyme family (Pfam01636) | 4e−08 |
| 06819 | 2,377 | Reducing polyketide synthase (R-PKS) | 40 | KS-AT-DH-ER-KR-ACPc | |
| 06820 | 184 | Hypothetical protein | 40 | Domain of unknown function, DUF3237 (Pfam11578) | 5e−22 |
| 06821 | 301 | Hypothetical protein | 34 | Domain of unknown function, DUF2306 (Pfam10067) | 2e−09 |
| 06822 | 236 | Hypothetical protein | 28 | Not detected | |
| 06823 | 356 | Hypothetical protein | 32 | Not detected | |
| 06824 | 565 | Putative dimethylaniline monooxygenase | 41 | Flavin-binding monooxygenase-like (Pfam00743) | 3e−14 |
ORF, C. rangiferina open reading frame. The syntenic core genes are highlighted in boldface.
For BLAST searches, the NCBI nonredundant protein sequence database for four Aspergillus species was used: Aspergillus fumigatus (taxid 746128), A. nidulans (taxid 162425), A. niger (taxid 5061), and A. terreus (taxid 33178).
For PKS domain architecture, see Data Set S2 in the supplemental material.
FIG 3.
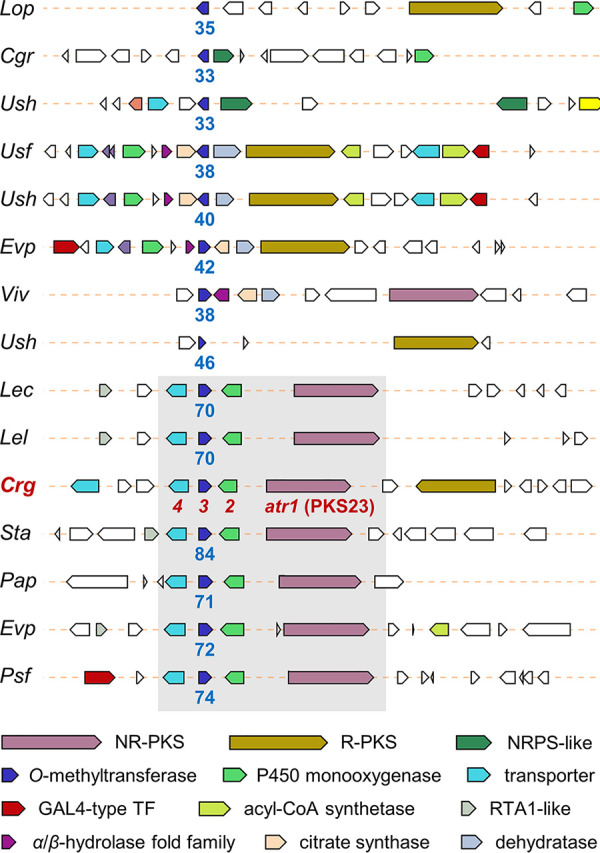
Organization of putative atranorin BGCs. Fifteen BGCs, including an O-methyltransferase that shows significant hits to the one in the PKS23 BGC in C. rangiferina (Crg; labeled in red) are identified from 1,527 BGCs detected in the 30 genomes, using the CORASON analysis pipeline (36). Lichen species that harbor the BGCs are labeled with their abbreviated names, as in Fig. 1. Numbers in blue indicate the percentage of protein sequence identity of homologous OMTs to the one in C. rangiferina. Among the 15 BGCs, seven BGCs harbor four syntenic genes (shaded box): atr1 (PKS23), atr2, atr3, and atr4. Genes related to secondary metabolism are color coded, based on their predicted function. NR-PKS, nonreducing type I PKS; R-PKS, reducing type I PRS; NRPS, nonribosomal peptide synthetase; TF, transcription factor; RTA, resistance to 7-aminocholesterol.
NR-PKSs characterized in nonlichenized fungi and NR-PKSs found in the six Cladonia spp. and Stereocaulon alpinum. Download Data Set S2, XLSX file, 0.5 MB (560.9KB, xlsx) .
Copyright © 2021 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Reconstruction of the atranorin biosynthetic pathway.
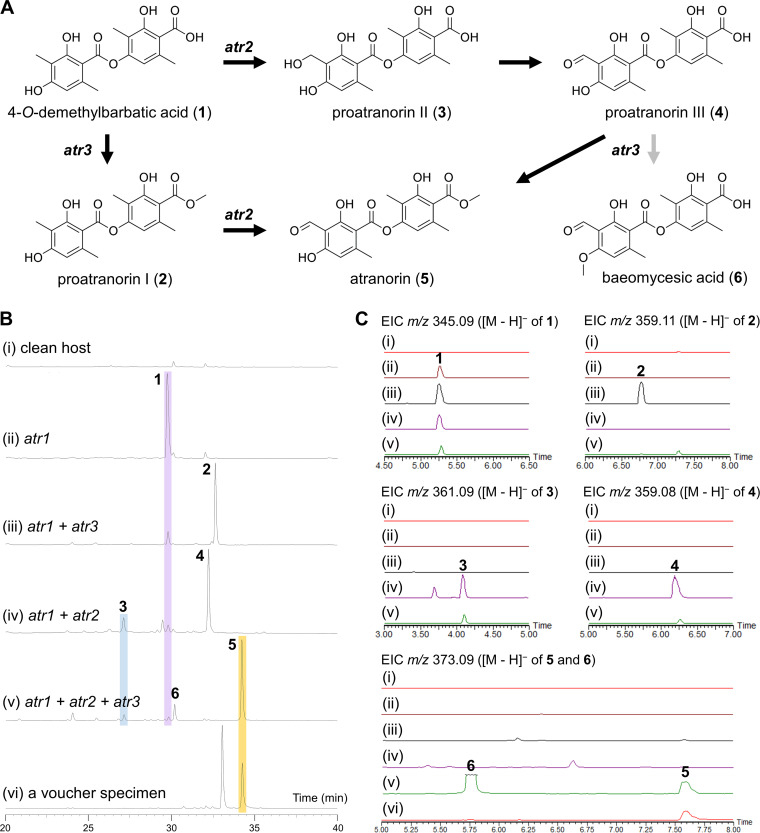
Despite arduous efforts made for heterologous expression of lichen PKS genes in well-established heterologous hosts, such as A. nidulans and A. oryzae, the attempts were unsuccessful for yet unknown reasons (11, 23). Thus, we set out to establish a new heterologous expression system, using a plant-pathogenic fungus, Ascochyta rabiei (Dothideomycetes). On the basis of the chemical structure and the predicted functions of three biosynthetic genes in putative atranorin BGCs, a polyketide pathway for atranorin can be envisioned (Fig. 4A). To investigate the roles of individual genes, we first generated a “clean host” that showed no appreciable metabolite production by removing the BGC for biosynthesis of solanapyrones from the wild-type As. rabiei (39, 40) (Fig. 4B). Then, we introduced the atr1 gene cloned from the S. alpinum genomic DNA (gDNA) into the “clean host.” Expression and correct intron splicing of the atr1 were confirmed by reverse transcription-PCR (RT-PCR) analyses (see Fig. S3 in the supplemental material). The liquid chromatography-tandem mass spectrometry (LC-MS/MS) and nuclear magnetic resonance (NMR) analyses indicated a known lichen depside, 4-O-demethylbarbatic acid (compound 1), was produced by a strain expressing the atr1 gene (Fig. 4B and C; see Fig. S4A in the supplemental material), suggesting that two 3MOA units were joined into a depside by Atr1 itself.
FIG 4.
Functional validation of the atranorin BGC. (A) A proposed biosynthetic pathway for atranorin. (B) HPLC profiles of culture extracts of a “clean host” expressing different sets of biosynthetic genes of the atranorin BGC: with no introduced gene (i), with atr1 only (ii), with atr1 and atr3 (iii), with atr1 and atr2 (iv), and with atr1, atr2, and atr3 (v). The acetone extract of an authentic voucher specimen for the genome-sequenced Stereocaulon alpinum contains atranorin (compound 5) and lobaric acid (tR = 33.1 min) (vi). (C) Extracted-ion chromatograms (EIC) with the indicated m/z values for compounds 1 to 6.
Verification of heterologous expression of PKS23. Download FIG S3, PDF file, 0.1 MB (135.2KB, pdf) .
Copyright © 2021 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
NMR and LC-MS/MS spectroscopic analyses. Download FIG S4, PDF file, 0.7 MB (756KB, pdf) .
Copyright © 2021 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
As the biosynthesis of atranorin from compound 1 requires oxidation of a methyl group and methylation of the carboxyl group within each of the two 3MOA units (Fig. 4A), we individually introduced the atr2 and atr3 genes into the atr1-expressing strain. Coexpression of the atr1 gene with the atr3 gene yielded a 7′-O-methylated analogue of compound 1, proatranorin I (compound 2), indicating that Atr3 is indeed a carboxyl methylase, as is Trt5 in A. terreus (Fig. 4B and C; Fig. S4B). The LC-MS/MS analysis showed that coexpression of atr1 with atr2 yielded compounds 3 and 4, which were annotated as a hydroxylated analogue of compound 1 (proatranorin II) and a further oxidized aldehyde (proatranorin III), respectively (Fig. 4B and C; Fig. S4C). NMR analysis of the isolated proatranorin III (compound 4) confirmed that the oxidation by Atr2 occurs at C-9 of compound 1 (Fig. S4D). Finally, we generated a strain expressing the three genes atr1, atr2, and atr3. The production of atranorin (compound 5) was confirmed by MS/MS spectral matching with the reference spectrum of atranorin in the Lichen Database (41) (Fig. S4E). It was also confirmed by comparison of the chemical profile with an authentic voucher specimen for the sequenced S. alpinum, which produces atranorin (compound 5) and lobaric acid, as a cortical substance and medullary substance, respectively (Fig. 4B and C). Intriguingly, MS/MS spectral matching of compound 6 (Fig. S4E) identified it as another lichen depside, baeomycesic acid, which has not been reported from S. alpinum in nature, suggesting that Atr3 exhibits relaxed substrate specificity in the heterologous host.
A novel phylogenetic clade of PKS responsible for biosynthesis of lichen substances.
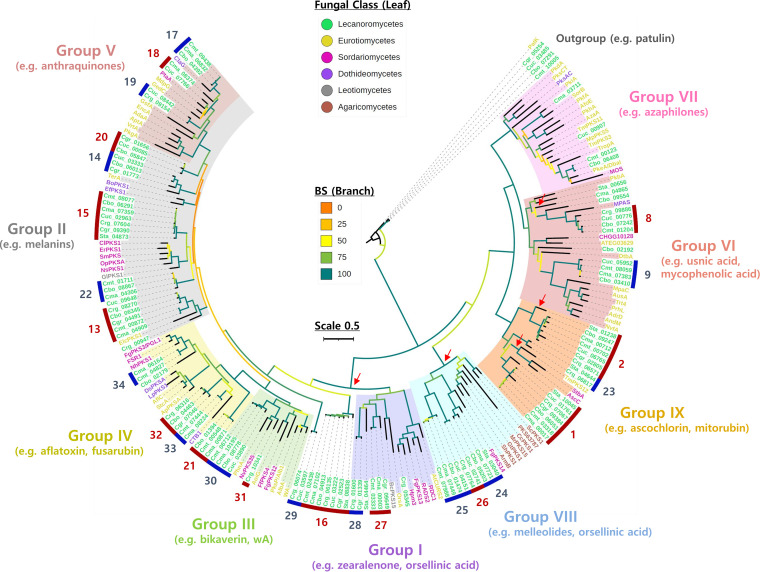
To study the evolutionary relationships of fungal NR-PKS and the Atr1 responsible for atranorin production, we reconstructed a phylogenetic tree of the concatenated sequences of conserved KS and product template (PT) domains of 103 NR-PKSs found in the six Cladonia spp. and S. alpinum and 82 NR-PKSs that have been linked to known compounds in nonlichenized fungi (see Data Set S2 in the supplemental material). Fungal NR-PKSs were hitherto largely classified into eight groups (groups I to VIII) in previous phylogenetic analyses (7, 8, 42–44). Here, we identified a novel NR-PKS group (group IX) supported by a 100% bootstrap, which contained the PKS23 family as well as the PKS1 and PKS2 families (Fig. 5). The PKS1 family was previously proposed to biosynthesize 3MOA-derived lichen substances in C. rangiferina (12). For lichen NR-PKSs in group IX, all but the PKS2 in S. alpinum possessed a C-methyltransferase (cMT) domain involved in the methylation of polyketide intermediates (Data Set S2). Intriguingly, the newly identified group IX included three previously characterized NR-PKSs, AscC (45), StbA (46), and TmPKS12 (47), in nonlichenized fungi (Fig. 5). These NR-PKSs lack the cMT domain and are known to produce orsellinic acids (45–47) (Data Set S2). Moreover, it is noteworthy that the group VIII, basal to group IX, included several NR-PKSs that have been linked to orsellinic acid-derived compounds in mushroom-forming fungi (Agaricomycetes) (48–51) and Fusarium graminearum (52) (Fig. 5; Data Set S2), indicating that the PKS1 and PKS2 families are likely involved in biosynthesis of lichen substances derived from a methylated form of orsellinic acid, as with the PKS23 family.
FIG 5.
The ninth clade of fungal nonreducing PKS genes. A maximum likelihood tree of 103 NR-PKSs in the six Cladonia spp. and Stereocaulon alpinum and 82 NR-PKSs linked to known compounds in nonlichenized fungi was reconstructed using concatenated sequences of ketosynthase and product template domains. The outgroup was set to 6-methylsalicylic acid synthases (6MSAS) found in Cladonia spp. and the 6MSAS for the patulin biosynthesis in Penicillium expansum. Branches are color coded, based on bootstrap (BS) values. The scale represents 0.5 amino acid sequence substitution per site. Blue and red strips with associated numbers at the outermost region indicate PKS families identified by the gene cluster network analysis (Fig. 2). Phylogenetic clades representing nine NR-PKS groups (groups I to IX) are shaded with different colors. The NR-PKS groups are supported by BS values greater than 75, except for group II. Note that the newly identified NR-PKS group (group IX) includes the PKS1, PKS2, and PKS23 families, candidate PKSs for cortical and medullary substances of lichens. Red arrows indicate polyphyletic losses of the C-methyltransferase domain of NR-PKSs.
DISCUSSION
Many lichen SMs are of ecological and pharmaceutical importance; however, lack of genetic tools for studying LFF has hindered identification of BGCs and industrial applications of lichen substances. Our in-depth genomic analysis of the interspecies BGC diversity in the genus Cladonia and heterologous expression of the atranorin BGC identified the complete biosynthetic pathway for atranorin, the major cortical substance of lichens, setting the first precedent for successful genome mining approaches for lichen SMs.
Biosynthesis of cortical and medullary substances in lichens.
Atranorin and usnic acid tend to be found mutually exclusively in lichens, albeit some taxa are known to produce both as cortical substances (e.g., Evernia prunastri) (53). This is true especially for the genus Cladonia; the presence of either usnic acid or atranorin in cortical layers is often used as a key trait for the chemotaxonomy (30). We found that C. rangiferina and Parmelia sp. strain KoLRI021559 carry both atranorin and usnic acid BGCs in the genomes; however, it was not usnic acid but atranorin that was found in their cortex (see Fig. S5A and S5B in the supplemental material), supporting the previous hypothesis that usnic acid has been replaced with atranorin as a major cortical substance independently in different lineages of LFF, especially in parmelioid lichens (13, 21). The PKS1 and PKS23 families are identical in gene structure, with 4 introns at conserved positions. The presence of the PKS1 family in Sc. sanguinea outside the Lecanoromycetes and highly syntenic PKS23 BGCs compared to the PKS1 BGCs (Fig. S5C) suggested that the PKS1 family may be ancestral and that the PKS23 family may have arisen from a duplication event in the common ancestor of atranorin-producing lichens, and then was adopted as a major cortical substance in some lineages or purged from the others, shaping the current phyletic distribution of atranorin in the Lecanoromycetes (5). Although the function of the PKS1 remains to be characterized, we speculate that the PKS1 family is involved in biosynthesis of medullary substances derived from 3MOA units, based on the fact that the expression level of a PKS1 homolog in Usnea rubrotincta (denoted UrPKS5) is highly correlated with in vitro production of medullary depsidones, such as salazinic acid and norsticitic acid (54). The PKS1 and PKS23 families may be an example of neofunctionalization of duplicate genes that have been functionally diverged to produce spatially distinct metabolites in macrolichens.
Lichens with atranorin and usnic acid BGCs. Download FIG S5, PDF file, 0.5 MB (477.5KB, pdf) .
Copyright © 2021 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogenetic relationship of NR-PKSs for depside and depsidone biosynthesis.
The phylogenetic analysis of fungal NR-PKSs revealed two major clades (groups I to V and groups VI to IX). The ancestral NR-PKS was estimated to have a cMT domain, because this domain is present both in fungal NR-PKS and R-PKS, as well as in bacterial R-PKS (55). The groups I to V seem to have diverged after the loss of the cMT domain (55). Also, we observed that there has been polyphyletic loss of the cMT domain in some lichen NR-PKSs within groups VI and IX and presumably during the rise of group VIII diverging from groups I to V. In nonlichenized fungi, several NR-PKSs within groups I and VIII (two basal clades lacking the cMT domain) exhibit intra- or intermolecular cross-linking activities when the enzymes release polyketide intermediates from the active sites, forming depsides, macrolides, and melleolides, all derived from orsellinic acid (48, 50, 56, 57). These two ancestral clades may include NR-PKSs responsible for biosynthesis of depsides and depsidones derived from orsellinic acid, such as the PKS16 family (sister to group I) for grayanic acid production in Cladonia grayi (11). The discovery of the PKS23 family in group IX with both cross-linking and cMT activities is particularly exciting, as this finding directs future research focus on the ninth clade of NR-PKS to resolve long-standing questions of how lichens form structurally diverse depside and depsidone compounds derived from 3MOA.
Lichen BGC diversity treasure chests for novel SMs.
The numbers of NR-PKSs found in the six Cladonia spp. placed their metabolic repertoire on par with those of four representative Aspergillus spp. that have been models for SM studies in nonlichenized fungi (58). More than two-thirds of NR-PKSs were found to be within groups V to VII in each of the four Aspergillus spp. lacking NR-PKS within group IX, whereas Cladonia spp. tended to have diverse NR-PKS groups and possessed 2 to 3 NR-PKSs belonging to group IX (Fig. S5D). Despite huge metabolic potentials encoded in each lichen genome, only a few SMs occur in each lichen thallus as major cortical or medullary substances, implying that most of the BGCs remain silent in nature. Our gene cluster network analysis expanded the knowledge on lichen BGCs connected to known compounds. However, there are still a lot more cryptic BGCs, many of which are species or lineage specific and may be involved in biosynthesis of novel compounds. Given the frequent appearance of unexpected metabolites (not observed in natural lichen thalli) in axenic culture of LFF (59–63), many of these cryptic BGCs appear to be still functional.
Future perspectives.
As lichen genomes become more and more available, we soon will be able to draw a more comprehensive picture of metabolic diversity and evolutionary fate of lichen BGCs. Comparative analyses of BGC contents between lichens producing structurally related compounds will identify biosynthetic enzymes harnessing structural diversity of cortical and medullary substances in lichens. This information can be applied to combinatorial biosynthesis of lichen-derived compounds with improved pharmacological activities. Our newly established heterologous expression system would help us understand the role of biosynthetic enzymes for well-known lichen substances and enable production of novel SMs encoded by cryptic BGCs in lichens.
MATERIALS AND METHODS
Phylogenomic analysis.
To infer the phylogeny of the genome-sequenced 29 LFF and an endophytic fungus Cyanodermella asteris, we pursued a coalescent-based phylogenomics approach. The culture, genome sequencing, and annotation procedures for LFF are described in Text S1. Single-copy ortholog clusters (SCOs) of protein sequences deduced from the 30 annotated genomes were identified using OrthoMCL (v2.0.9) (64) with an inflation factor of 2.5. For each of the 393 SCOs, protein sequences were aligned using MAFFT (v7.310) (65) with the “auto” setting, and the resulting 393 multiple-sequence alignments were trimmed for poorly aligned regions using Gblocks (v0.91b) (66) with the parameter “-b4 = 5.” The RAxML program (v8.2) (67) was used to calculate 100 maximum likelihood trees for each multiple-sequence alignment. To generate coalescent-based trees, we used ASTRAL-III (v5.7.4) (68) with two multilocus bootstrapping options (site-only resampling and gene/site resampling) and with no bootstrapping option. All trees showed the identical topology.
Biosynthetic gene cluster family analysis.
For BGC identification in the 30 genomes, the genome assembly and annotation files were processed by the antiSMASH program (v5.0+) (35), with the parameter setting “–minimal.” For genetic dereplication of BGCs found in the six Cladonia spp. and S. alpinum, we used the BiG-SCAPE program (36), with reference to the MIBiG database (v1.4) (37). To analyze BGCs containing at least one iterative type I PKS, we modified a configuration file, “domain_whitelist.txt,” to include only Pfam domains related to the iterative type I PKS (PF00109 and PF02801) and ran the BiG-SCAPE program with optional arguments “–mix” and “–hybrids-off.” Based on the Jaccard index of domain types, domain sequence similarity, and domain adjacency index, the BiG-SCAPE program calculates a similarity matrix between pairwise combinations of clusters, where smaller values indicate greater BGC similarity (36). We evaluated BGC networks using an edge-length cutoff from 0.3 to 0.8 with a step of 0.1 and considered the network using a cutoff value of 0.5 as a representation of GCFs in the six Cladonia spp. and S. alpinum. Individual networks for PKS families were visualized using a Python package NetworkX (v2.5) (69), in that edge width was weighted by squared similarity between a pair of BGCs calculated by BiG-SCAPE. We used the CORASON program (36), which generates a multilocus, approximately maximum likelihood, phylogenetic tree of BGCs, including PKS families responsible for production of lichen substances. For the PKS23 family, an OMT gene (Crg06815) in C. rangiferina was used as the query gene to search for homologous BGCs from a total of 1,527 BGCs detected in the 30 sequenced species. For the PKS1 family, a GAL4-type transcription factor gene (GeneID Crg07068) in C. rangiferina was used as the query gene.
Phylogenetic analysis of fungal NR-PKS.
The KS domain of iterative type I PKS has been considered evolutionarily conserved (55), and thus it can serve as a proxy for the similarity of the entire PKS. We identified a total of 242 PKSs from the genomes of the six Cladonia spp. and S. alpinum, among which four PKSs (Cgr01615, Cgr03964, Cgr08611, and Cmt10189) (Data Set S2) were missing a KS domain. KS domain sequences were extracted from 238 PKSs using the online tool NaPDoS (70) and aligned using MUSCLE (v3.8.31) (71). For clustering analysis, an all-versus-all similarity matrix for KS domains of 238 PKSs was computed using the AlignBuddy function in the BuddySuite program (72), with an optional argument, “-pi.” A heat map showing the percentage of similarity of KS domains clustered by k-means was generated using the R package Superheat (73). For fungal NR-PKS phylogeny, we used concatenated protein sequences of KS and PT domains of 103 NR-PKSs found in the six Cladonia spp. and S. alpinum and 82 NR-PKSs that have been linked to known compounds in nonlichenized fungi (7, 8, 44) (Data Set S2). We initially identified 106 NR-PKSs in the six Cladonia spp. and S. alpinum, including seven NR-PKSs whose full sequences cannot be reliably defined from the current genome assembly (likely pseudogenes). Among these partial PKSs, three NR-PKSs (Cbo04702, Cma06590, and Cmt06606) (Data Set S2) lacked either the KS or PT domain and were excluded from the phylogenetic analysis. PT domains of lichen NR-PKSs were identified by aligning with those of previously characterized PKSs (8). A 6-methylsalicylic acid synthase (6MSAS) responsible for the biosynthesis of patulin (UniProtKB accession no. A0A075TRC0) in Penicillium expansum was set to be an outgroup to fungal NR-PKS phylogeny. Also, four 6MSASs found in four Cladonia spp. were included in the analysis (Cbo07291, Cgr05254, Cmt10005, and Cuc03485) (Data Set S2). Protein sequences of KS and PT domains were aligned using MAFFT (v7.310) (65) with the “auto” setting, and spurious sequences or poorly aligned regions from each domain were trimmed using the trimAl program (v1.2) (74), with the “gappyout” parameter. The resulting multiple-sequence alignments for KS and PT domains were concatenated with FASconCAT-G (v1.04) (75). From the concatenated sequences, maximum likelihood trees were computed with RAxML (v8.2) (67), using a gamma distribution for substitution rate across sites with the parameter setting “-m PROTGAMMAWAG.” Nodal support was evaluated by 1,000 bootstrap replications. The final tree was rooted to the 6MSAS outgroup and annotated by iTOL (v5.7) (76).
Generation of a “clean host” from Ascochyta rabiei.
A split marker strategy (77) was employed to generate a solanapyrone-negative mutant (“clean host”). A downstream region of the sol1 gene (1,208 bp) and an upstream region of the sol3 gene (1,459 bp) were amplified from gDNA of As. rabiei isolate AR628 (39), using the L5/L3 and R5/R3 primer pairs, respectively. A hygromycin phosphotransferase gene (hph) cassette was amplified from the pCB1004 plasmid (78), using thee primer pair HYG5/HYG3. The L3 and R5 primers have 27-nucleotide (nt)-long overhang sequences complementary to the 3′ and 5′ ends of the hph cassette, respectively, such that the amplified sol1 downstream and sol3 upstream regions can be fused to the hph cassette by overlap extension PCR. Finally, split marker constructs were amplified from the fused construct, using N5/HY-R and YG-F/N3 primer pairs. The split marker constructs were introduced into protoplasts of As. rabiei by polyethylene glycol-mediated genetic transformation as described by Hallen-Adams et al. (79). The primers used in the gene replacement are listed in Table S2 in the supplemental material.
Primers used in this study. Download Table S2, DOCX file, 0.03 MB (32.3KB, docx) .
Copyright © 2021 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Heterologous expression of the atr1 and RT-PCR analysis.
For efficient expression of foreign genes in the “clean host,” we constructed expression vectors that carry either the sol1 gene promoter (pDS35), the sol5 gene promoter (pII95), or the translation elongation factor 1 alpha gene promoter (pII98). Then the atr1, atr2, and atr3 genes were individually cloned to the expression vectors. Details on expression vector construction and cloning procedures are described in Text S1. For heterologous expression of the atr1 gene, the sol1::atr1/pDS35 plasmid (5 to 10 μg) was used to transform the “clean host.” Putative transformants were subcultured on potato dextrose agar (PDA; BD Biosciences) containing 100 μg/ml of nourseothricin sulfate (clonNAT; GoldBio). Two transformants, designated Sta04644-T16 and Sta04644-T25, exhibited resistance to the selective agent and were subcultured on PDA overlaid with a nylon membrane for RNA extraction. After 2 weeks of culture, mycelia growing on PDA were scraped off from the nylon membrane with a single-edge razor blade and were subjected to total RNA extraction. Total RNA was extracted from hyphae ground in liquid nitrogen, using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer's instructions, with the following additional extraction steps: one phenol (pH 4.6)-chloroform-isoamyl alcohol (25:24:1) extraction followed by one chloroform extraction step after the initial TRIzol-chloroform phase separation. RNA pellets were dissolved in 88 μl of nuclease-free water and subjected to genomic DNA digestion by DNase treatment (Qiagen). Then RNA samples were concentrated using the RNA Clean & Concentrator (Zymo Research). We confirmed expression and intron splicing of the atr1 gene with four introns by RT-PCR analysis. Two primer sets were designed: (i) to amplify a flanking region of the first intron located at the 5′ region of the atr1 coding sequence and (ii) to amplify a region harboring the second, third, and fourth (the last) introns at the 3′ region of the atr1 coding sequence. Two hundred nanograms of total RNA was reverse transcribed, and the atr1 gene and actin 1 (as a positive control) were amplified (39) with a Qiagen OneStep RT-PCR kit. The primers used in RT-PCR analysis and plasmid constructions are listed in Table S2.
Generation of a heterologous host producing atranorin.
For introduction of tailoring enzymes into a atr1-expressing strain (Sta04644-T25), the four resulting plasmids (sol5::atr2/pII95, tef1α::atr2/pII98, sol5::atr3/pII95, and tef1α::atr3/pII98) were individually transformed into the Sta04644-T25 strain, and putative transformants were subcultured on PDA containing 200 μg/ml of G418 disulfate (Sigma-Aldrich). Transformants resistant to G418 disulfate were selected for metabolite identification. A strain harboring both sol1:: atr1/pDS35 and sol5:: atr3/pII95 was used for the generation of an atranorin-producing strain, which exhibited the greatest production of compound 2, the immediate precursor of atrarnoin. Since a neomycin phosphotransferase II gene (nptII) cassette that confers resistance to G418 disulfate was already integrated into the strain producing compound 2, we replaced the nptII cassette in the tef1α::atr2/pII98 plasmid with a bleomycin-resistant protein gene (ble), the tef1α::atr2/pII98 plasmid was amplified using a primer pair Inf_BLE_pII99_fwd and Inf_BLE_pII99_rev, and the ble gene was amplified from pAC1750 (80). Then the two PCR products were fused using the In-Fusion HD Cloning kit (TaKaRa). The resulting plasmid was transformed into the strain producing compound 2, and putative transformants were subcultured on PDA containing 200 μg/ml of Zeocin (a member of bleomycin family distributed by ThermoFisher Scientific). To select transformants that produce atranorin, the chemical profiles of transformants resistant to Zeocin were compared to that of an S. alpinum voucher specimen (Korea National Arboretum accession no. KHL0017342), from which the genome-sequenced LFF had been originally isolated. Detailed methodologies for high-performance liquid chromatography (HPLC), LC-MS/MS, and NMR spectroscopic analyses for culture extracts and purified compounds are fully described in Text S1.
Data availability.
The data from this whole-genome shotgun project have been deposited in DDBJ/ENA/GenBank under accession no. JAFEKC000000000, JAFEKC000000000, and JAEUBA000000000 (BioProject no. PRJNA693578, PRJNA693575, and PRJNA693574) for C. borealis, Parmelia cf. squarrosa, and S. alpinum, respectively. Raw LC-MS/MS data have been deposited in the MassIVE database (http://massive.ucsd.edu) under accession no. MSV000087081.
ACKNOWLEDGMENTS
We thank Jung-Ae Ryu and Seol-Hwa Jang for technical support in mycobiont isolation and culture.
This work was supported by the Korea Research Fellowship Program funded by the Ministry of Science, ICT, and Future Planning (2018H1D3A1A01074888) and by The Korean National Research Resource Center Program (2017M3A9B8069471), through the National Research Foundation of Korea (NRF). The genome of Parmelia cf. squarrosa was sequenced with support by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry through the Agricultural Microbiome R&D Program funded by the Ministry of Agriculture, Food and Rural Affairs (918010044SB010). S.W. and K.B.K. were supported by an NRF grant funded by the Ministry of Science, ICT, and Future Planning (2020R1C1C1004046).
Footnotes
Citation Kim W, Liu R, Woo S, Kang KB, Park H, Yu YH, Ha H-H, Oh S-Y, Yang JH, Kim H, Yun S-H, Hur J-S. 2021. Linking a gene cluster to atranorin, a major cortical substance of lichens, through genetic dereplication and heterologous expression. mBio 12:e01111-21. https://doi.org/10.1128/mBio.01111-21.
Contributor Information
Wonyong Kim, Email: wykim@scnu.ac.kr.
Jae-Seoun Hur, Email: jshur1@scnu.ac.kr.
B. Gillian Turgeon, Cornell University
REFERENCES
- 1.Lutzoni F, Miadlikowska J. 2009. Lichens. Curr Biol 19:R502–R503. doi: 10.1016/j.cub.2009.04.034. [DOI] [PubMed] [Google Scholar]
- 2.Calcott MJ, Ackerley DF, Knight A, Keyzers RA, Owen JG. 2018. Secondary metabolism in the lichen symbiosis. Chem Soc Rev 47:1730–1760. doi: 10.1039/c7cs00431a. [DOI] [PubMed] [Google Scholar]
- 3.Gaya E, Fernández-Brime S, Vargas R, Lachlan RF, Gueidan C, Ramírez-Mejía M, Lutzoni F. 2015. The adaptive radiation of lichen-forming Teloschistaceae is associated with sunscreening pigments and a bark-to-rock substrate shift. Proc Natl Acad Sci U S A 112:11600–11605. doi: 10.1073/pnas.1507072112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solhaug KA, Gauslaa Y. 2012. Secondary lichen compounds as protection against excess solar radiation and herbivores, p 283–304. In Lüttge U, Beyschlag W, Büdel B, Francis D (ed), Progress in botany, vol 73. Springer, Berlin, Germany. [Google Scholar]
- 5.Culberson WL, Culberson CF. 1970. A phylogenetic view of chemical evolution in the lichens. Bryologist 73:1–31. doi: 10.2307/3241584. [DOI] [Google Scholar]
- 6.Huovinen K, Ahti T. 1982. Biosequential patterns for the formation of depsides, depsidones and dibenzofurans in the genus Cladonia (lichen-forming ascomycetes). Ann Botanici Fennici 19:225–234. [Google Scholar]
- 7.Ahuja M, Chiang Y-M, Chang S-L, Praseuth MB, Entwistle R, Sanchez JF, Lo H-C, Yeh H-H, Oakley BR, Wang CCC. 2012. Illuminating the diversity of aromatic polyketide synthases in Aspergillus nidulans. J Am Chem Soc 134:8212–8221. doi: 10.1021/ja3016395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu L, Zhang Z, Shao C-L, Wang J-L, Bai H, Wang C-Y. 2015. Bioinformatical analysis of the sequences, structures and functions of fungal polyketide synthase product template domains. Sci Rep 5:10463. doi: 10.1038/srep10463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanchez JF, Chiang Y-M, Szewczyk E, Davidson AD, Ahuja M, Oakley CE, Bok JW, Keller N, Oakley BR, Wang CCC. 2010. Molecular genetic analysis of the orsellinic acid/F9775 gene cluster of Aspergillus nidulans. Mol Biosyst 6:587–593. doi: 10.1039/b904541d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanchez JF, Entwistle R, Corcoran D, Oakley BR, Wang CCC. 2012. Identification and molecular genetic analysis of the cichorine gene cluster in Aspergillus nidulans. Medchemcomm 3:997. doi: 10.1039/c2md20055d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Armaleo D, Sun X, Culberson C. 2011. Insights from the first putative biosynthetic gene cluster for a lichen depside and depsidone. Mycologia 103:741–754. doi: 10.3852/10-335. [DOI] [PubMed] [Google Scholar]
- 12.Elshobary ME, Becker MG, Kalichuk JL, Chan AC, Belmonte MF, Piercey-Normore MD. 2018. Tissue-specific localization of polyketide synthase and other associated genes in the lichen, Cladonia rangiferina, using laser microdissection. Phytochemistry 156:142–150. doi: 10.1016/j.phytochem.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 13.Divakar PK, Kauff F, Crespo A, Leavitt SD, Lumbsch HT. 2013. Understanding phenotypical character evolution in parmelioid lichenized fungi (Parmeliaceae, Ascomycota). PLoS One 8:e83115. doi: 10.1371/journal.pone.0083115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lumbsch HT. 1998. Taxonomic use of metabolic data in lichen-forming fungi, p 345–387. In Frisvad JC, Bridge PD, Arora DK (ed), Chemical fungal taxonomy, CRC Press, New York, NY. [Google Scholar]
- 15.Gadea A, Charrier M, Fanuel M, Clerc P, Daugan C, Sauvager A, Rogniaux H, Boustie J, Le Lamer A-C, Lohézic-Le Devehat F. 2019. Overcoming deterrent metabolites by gaining essential nutrients: a lichen/snail case study. Phytochemistry 164:86–93. doi: 10.1016/j.phytochem.2019.04.019. [DOI] [PubMed] [Google Scholar]
- 16.Jeon Y-J, Kim S, Kim JH, Youn UJ, Suh S-S. 2019. The comprehensive roles of atranorin, a secondary metabolite from the Antarctic lichen Stereocaulon caespitosum, in HCC tumorigenesis. Molecules 24:1414. doi: 10.3390/molecules24071414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y, Nguyen TT, Jeong M-H, Crişan F, Yu YH, Ha H-H, Choi KH, Jeong HG, Jeong TC, Lee KY, Kim KK, Hur J-S, Kim H. 2016. Inhibitory activity of (+)-usnic acid against non-small cell lung cancer cell motility. PLoS One 11:e0146575. doi: 10.1371/journal.pone.0146575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou R, Yang Y, Park S-Y, Nguyen TT, Seo Y-W, Lee KH, Lee JH, Kim KK, Hur J-S, Kim H. 2017. The lichen secondary metabolite atranorin suppresses lung cancer cell motility and tumorigenesis. Sci Rep 7:8136. doi: 10.1038/s41598-017-08225-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Araújo AAS, de Melo MGD, Rabelo TK, Nunes PS, Santos SL, Serafini MR, Santos MRV, Quintans-Júnior LJ, Gelain DP. 2015. Review of the biological properties and toxicity of usnic acid. Nat Prod Res 29:2167–2180. doi: 10.1080/14786419.2015.1007455. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Geng C, Yuan X, Hua M, Tian F, Li C. 2018. Identification of a putative polyketide synthase gene involved in usnic acid biosynthesis in the lichen Nephromopsis pallescens. PLoS One 13:e0199110. doi: 10.1371/journal.pone.0199110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pizarro D, Divakar PK, Grewe F, Crespo A, Dal Grande F, Lumbsch HT. 2020. Genome-wide analysis of biosynthetic gene cluster reveals correlated gene loss with absence of usnic acid in lichen-forming fungi. Genome Biol Evol 12:1858–1868. doi: 10.1093/gbe/evaa189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abdel-Hameed M, Bertrand RL, Piercey-Normore MD, Sorensen JL. 2016. Putative identification of the usnic acid biosynthetic gene cluster by de novo whole-genome sequencing of a lichen-forming fungus. Fungal Biol 120:306–316. doi: 10.1016/j.funbio.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 23.Bertrand RL, Sorensen JL. 2019. Lost in translation: challenges with heterologous expression of lichen polyketide synthases. ChemistrySelect 4:6473–6483. doi: 10.1002/slct.201901762. [DOI] [Google Scholar]
- 24.Nielsen JC, Grijseels S, Prigent S, Ji B, Dainat J, Nielsen KF, Frisvad JC, Workman M, Nielsen J. 2017. Global analysis of biosynthetic gene clusters reveals vast potential of secondary metabolite production in Penicillium species. Nat Microbiol 2:17044. doi: 10.1038/nmicrobiol.2017.44. [DOI] [PubMed] [Google Scholar]
- 25.Tobias NJ, Wolff H, Djahanschiri B, Grundmann F, Kronenwerth M, Shi Y-M, Simonyi S, Grün P, Shapiro-Ilan D, Pidot SJ, Stinear TP, Ebersberger I, Bode HB. 2017. Natural product diversity associated with the nematode symbionts Photorhabdus and Xenorhabdus. Nat Microbiol 2:1676–1685. doi: 10.1038/s41564-017-0039-9. [DOI] [PubMed] [Google Scholar]
- 26.Mullins AJ, Murray JAH, Bull MJ, Jenner M, Jones C, Webster G, Green AE, Neill DR, Connor TR, Parkhill J, Challis GL, Mahenthiralingam E. 2019. Genome mining identifies cepacin as a plant-protective metabolite of the biopesticidal bacterium Burkholderia ambifaria. Nat Microbiol 4:996–1005. doi: 10.1038/s41564-019-0383-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leao T, Castelão G, Korobeynikov A, Monroe EA, Podell S, Glukhov E, Allen EE, Gerwick WH, Gerwick L. 2017. Comparative genomics uncovers the prolific and distinctive metabolic potential of the cyanobacterial genus Moorea. Proc Natl Acad Sci U S A 114:3198–3203. doi: 10.1073/pnas.1618556114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lichman BR, Godden GT, Buell CR. 2020. Gene and genome duplications in the evolution of chemodiversity: perspectives from studies of Lamiaceae. Curr Opin Plant Biol 55:74–83. doi: 10.1016/j.pbi.2020.03.005. [DOI] [PubMed] [Google Scholar]
- 29.Lücking R, Hodkinson BP, Leavitt SD. 2017. The 2016 classification of lichenized fungi in the Ascomycota and Basidiomycota—approaching one thousand genera. Bryologist 119:361–416. doi: 10.1639/0007-2745-119.4.361. [DOI] [Google Scholar]
- 30.Stenroos S, Hyvonen J, Myllys L, Thell A, Ahti T. 2002. Phylogeny of the genus Cladonia s.lat. (Cladoniaceae, Ascomycetes) inferred from molecular, morphological, and chemical data. Cladistics 18:237–278. doi: 10.1111/j.1096-0031.2002.tb00151.x. [DOI] [PubMed] [Google Scholar]
- 31.Stenroos S, Pino-Bodas R, Hyvönen J, Lumbsch HT, Ahti T. 2019. Phylogeny of the family Cladoniaceae (Lecanoromycetes, Ascomycota) based on sequences of multiple loci. Cladistics 35:351–384. doi: 10.1111/cla.12363. [DOI] [PubMed] [Google Scholar]
- 32.Armaleo D, Müller O, Lutzoni F, Andrésson ÓS, Blanc G, Bode HB, Collart FR, Dal Grande F, Dietrich F, Grigoriev IV, Joneson S, Kuo A, Larsen PE, Logsdon JM, Lopez D, Martin F, May SP, McDonald TR, Merchant SS, Miao V, Morin E, Oono R, Pellegrini M, Rubinstein N, Sanchez-Puerta MV, Savelkoul E, Schmitt I, Slot JC, Soanes D, Szövényi P, Talbot NJ, Veneault-Fourrey C, Xavier BB. 2019. The lichen symbiosis re-viewed through the genomes of Cladonia grayi and its algal partner Asterochloris glomerata. BMC Genomics 20:605. doi: 10.1186/s12864-019-5629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Timsina BA, Hausner G, Piercey-Normore MD. 2014. Evolution of ketosynthase domains of polyketide synthase genes in the Cladonia chlorophaea species complex (Cladoniaceae). Fungal Biol 118:896–909. doi: 10.1016/j.funbio.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 34.Allen JL, McKenzie SK, Sleith RS, Alter SE. 2018. First genome-wide analysis of the endangered, endemic lichen Cetradonia linearis reveals isolation by distance and strong population structure. Am J Bot 105:1556–1567. doi: 10.1002/ajb2.1150. [DOI] [PubMed] [Google Scholar]
- 35.Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T. 2019. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Navarro-Muñoz JC, Selem-Mojica N, Mullowney MW, Kautsar SA, Tryon JH, Parkinson EI, De Los Santos ELC, Yeong M, Cruz-Morales P, Abubucker S, Roeters A, Lokhorst W, Fernandez-Guerra A, Cappelini LTD, Goering AW, Thomson RJ, Metcalf WW, Kelleher NL, Barona-Gomez F, Medema MH. 2020. A computational framework to explore large-scale biosynthetic diversity. Nat Chem Biol 16:60–68. doi: 10.1038/s41589-019-0400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Medema MH, Kottmann R, Yilmaz P, Cummings M, Biggins JB, Blin K, de Bruijn I, Chooi YH, Claesen J, Coates RC, Cruz-Morales P, Duddela S, Düsterhus S, Edwards DJ, Fewer DP, Garg N, Geiger C, Gomez-Escribano JP, Greule A, Hadjithomas M, Haines AS, Helfrich EJN, Hillwig ML, Ishida K, Jones AC, Jones CS, Jungmann K, Kegler C, Kim HU, Kötter P, Krug D, Masschelein J, Melnik AV, Mantovani SM, Monroe EA, Moore M, Moss N, Nützmann H-W, Pan G, Pati A, Petras D, Reen FJ, Rosconi F, Rui Z, Tian Z, Tobias NJ, Tsunematsu Y, Wiemann P, Wyckoff E, Yan X, et al. 2015. Minimum information about a biosynthetic gene cluster. Nat Chem Biol 11:625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuda Y, Awakawa T, Itoh T, Wakimoto T, Kushiro T, Fujii I, Ebizuka Y, Abe I. 2012. Terretonin biosynthesis requires methylation as essential step for cyclization. Chembiochem 13:1738–1741. doi: 10.1002/cbic.201200369. [DOI] [PubMed] [Google Scholar]
- 39.Kim W, Park J-J, Gang DR, Peever TL, Chen W. 2015. A novel type pathway-specific regulator and dynamic genome environments of a solanapyrone biosynthesis gene cluster in the fungus Ascochyta rabiei. Eukaryot Cell 14:1102–1113. doi: 10.1128/EC.00084-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim W, Park C-M, Park J-J, Akamatsu HO, Peever TL, Xian M, Gang DR, Vandemark G, Chen W. 2014. Functional analyses of the Diels-Alderase gene sol5 of Ascochyta rabiei and Alternaria solani indicate that the solanapyrone phytotoxins are not required for pathogenicity. Mol Plant Microbe Interact 28:482–496. doi: 10.1094/MPMI-08-14-0234-R. [DOI] [PubMed] [Google Scholar]
- 41.Olivier-Jimenez D, Chollet-Krugler M, Rondeau D, Beniddir MA, Ferron S, Delhaye T, Allard P-M, Wolfender J-L, Sipman HJM, Lücking R, Boustie J, Le Pogam P. 2019. A database of high-resolution MS/MS spectra for lichen metabolites. Sci Data 6:294. doi: 10.1038/s41597-019-0305-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeh H-H, Chang S-L, Chiang Y-M, Bruno KS, Oakley BR, Wu T-K, Wang CCC. 2013. Engineering fungal non-reducing polyketide synthase by heterologous expression and domain swapping. Org Lett 15:756–759. doi: 10.1021/ol303328t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Xu W, Tang Y. 2010. Classification, prediction, and verification of the regioselectivity of fungal polyketide synthase product template domains. J Biol Chem 285:22764–22773. doi: 10.1074/jbc.M110.128504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu L, Zhang Z, Shao C-L, Wang C-Y. 2017. Analysis of the sequences, structures, and functions of product-releasing enzyme domains in fungal polyketide synthases. Front Microbiol 8:1685. doi: 10.3389/fmicb.2017.01685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Araki Y, Awakawa T, Matsuzaki M, Cho R, Matsuda Y, Hoshino S, Shinohara Y, Yamamoto M, Kido Y, Inaoka DK, Nagamune K, Ito K, Abe I, Kita K. 2019. Complete biosynthetic pathways of ascofuranone and ascochlorin in Acremonium egyptiacum. Proc Natl Acad Sci U S A 116:8269–8274. doi: 10.1073/pnas.1819254116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li C, Matsuda Y, Gao H, Hu D, Yao XS, Abe I. 2016. Biosynthesis of LL-Z1272β: discovery of a new member of NRPS-like enzymes for aryl-aldehyde formation. Chembiochem 17:904–907. doi: 10.1002/cbic.201600087. [DOI] [PubMed] [Google Scholar]
- 47.Woo PCY, Lam C-W, Tam EWT, Leung CKF, Wong SSY, Lau SKP, Yuen K-Y. 2012. First discovery of two polyketide synthase genes for mitorubrinic acid and mitorubrinol yellow pigment biosynthesis and implications in virulence of Penicillium marneffei. PLoS Negl Trop Dis 6:e1871. doi: 10.1371/journal.pntd.0001871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lackner G, Bohnert M, Wick J, Hoffmeister D. 2013. Assembly of melleolide antibiotics involves a polyketide synthase with cross-coupling activity. Chem Biol 20:1101–1106. doi: 10.1016/j.chembiol.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 49.Yu P-W, Cho T-Y, Liou R-F, Tzean S-S, Lee T-H. 2017. Identification of the orsellinic acid synthase PKS63787 for the biosynthesis of antroquinonols in Antrodia cinnamomea. Appl Microbiol Biotechnol 101:4701–4711. doi: 10.1007/s00253-017-8196-6. [DOI] [PubMed] [Google Scholar]
- 50.Lackner G, Misiek M, Braesel J, Hoffmeister D. 2012. Genome mining reveals the evolutionary origin and biosynthetic potential of basidiomycete polyketide synthases. Fungal Genet Biol 49:996–1003. doi: 10.1016/j.fgb.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 51.Braesel J, Fricke J, Schwenk D, Hoffmeister D. 2017. Biochemical and genetic basis of orsellinic acid biosynthesis and prenylation in a stereaceous basidiomycete. Fungal Genet Biol 98:12–19. doi: 10.1016/j.fgb.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 52.Jørgensen SH, Frandsen RJN, Nielsen KF, Lysøe E, Sondergaard TE, Wimmer R, Giese H, Sørensen JL. 2014. Fusarium graminearum PKS14 is involved in orsellinic acid and orcinol synthesis. Fungal Genet Biol 70:24–31. doi: 10.1016/j.fgb.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 53.Staples R, LaDuca RL, Roze LV, Laivenieks M, Linz JE, Beaudry R, Fryday A, Schilmiller AL, Koptina AV, Smith B, Trail F. 2020. Structure and chemical analysis of major specialized metabolites produced by the lichen Evernia prunastri. Chem Biodivers 17:e1900465. doi: 10.1002/cbdv.201900465. [DOI] [PubMed] [Google Scholar]
- 54.Kondo H, Hara K, Yamamoto Y, Komine M. 2016. Comprehensive analysis of secondary metabolism-related genes in the mycobiont of Usnea rubrotincta. Lichenology 14:177. (In Japanese.) [Google Scholar]
- 55.Kroken S, Glass NL, Taylor JW, Yoder OC, Turgeon BG. 2003. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc Natl Acad Sci U S A 100:15670–15675. doi: 10.1073/pnas.2532165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gressler M, Hortschansky P, Geib E, Brock M. 2015. A new high-performance heterologous fungal expression system based on regulatory elements from the Aspergillus terreus terrein gene cluster. Front Microbiol 6:184. doi: 10.3389/fmicb.2015.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gaffoor I, Trail F. 2006. Characterization of two polyketide synthase genes involved in zearalenone biosynthesis in Gibberella zeae. Appl Environ Microbiol 72:1793–1799. doi: 10.1128/AEM.72.3.1793-1799.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Romsdahl J, Wang CCC. 2019. Recent advances in the genome mining of Aspergillus secondary metabolites (covering 2012–2018). Medchemcomm 10:840–866. doi: 10.1039/c9md00054b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Molina MC, Crespo A, Vicente C, Elix JA. 2003. Differences in the composition of phenolics and fatty acids of cultured mycobiont and thallus of Physconia distorta. Plant Physiol Biochem 41:175–180. doi: 10.1016/S0981-9428(02)00017-7. [DOI] [Google Scholar]
- 60.Brunauer G, Hager A, Grube M, Türk R, Stocker-Wörgötter E. 2007. Alterations in secondary metabolism of aposymbiotically grown mycobionts of Xanthoria elegans and cultured resynthesis stages. Plant Physiol Biochem 45:146–151. doi: 10.1016/j.plaphy.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 61.Ejiri H, Sankawa U, Shibata S. 1975. Graciliformin and its acetates in Cladonia graciliformis. Phytochemistry 14:277–279. doi: 10.1016/0031-9422(75)85055-2. [DOI] [Google Scholar]
- 62.Yamamoto Y, Matsubara H, Kinoshita Y, Kinoshita K, Koyama K, Takahashi K, Ahmadjiam V, Kurokawa T, Yoshimura I. 1996. Naphthazarin derivatives from cultures of the lichen Cladonia cristatella. Phytochemistry 43:1239–1242. doi: 10.1016/S0031-9422(96)00495-5. [DOI] [Google Scholar]
- 63.Luo H. 2013. Biruloquinone, an acetylcholinesterase inhibitor produced by lichen-forming fungus Cladonia macilenta. J Microbiol Biotechnol 23:161–166. doi: 10.4014/jmb.1207.07016. [DOI] [PubMed] [Google Scholar]
- 64.Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 67.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang C, Rabiee M, Sayyari E, Mirarab S. 2018. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics 19:153. doi: 10.1186/s12859-018-2129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hagberg AA, Schult DA, Swart PJ. 2008. Exploring network structure, dynamics, and function using NetworkX, p 11–15. In Varoquaux G, Vaught T, Millman J (ed), Proceedings of the 7th Python in Science Conference, Pasadena, CA. [Google Scholar]
- 70.Ziemert N, Podell S, Penn K, Badger JH, Allen E, Jensen PR. 2012. The natural product domain seeker NaPDoS: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS One 7:e34064. doi: 10.1371/journal.pone.0034064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bond SR, Keat KE, Barreira SN, Baxevanis AD. 2017. BuddySuite: command-line toolkits for manipulating sequences, alignments, and phylogenetic trees. Mol Biol Evol 34:1543–1546. doi: 10.1093/molbev/msx089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barter RL, Yu B. 2018. Superheat: an R package for creating beautiful and extendable heatmaps for visualizing complex data. J Comput Graph Stat 27:910–922. doi: 10.1080/10618600.2018.1473780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kück P, Longo GC. 2014. FASconCAT-G: extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front Zool 11:81. doi: 10.1186/s12983-014-0081-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Catlett NL, Lee B, Yoder OC, Turgeon BC. 2003. Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genet Newsl 50:9–11. doi: 10.4148/1941-4765.1150. [DOI] [Google Scholar]
- 78.Carroll AM, Sweigard JA, Valent B. 1994. Improved vectors for selecting resistance to hygromycin. Fungal Genet Newsl 41:22. doi: 10.4148/1941-4765.1367. [DOI] [Google Scholar]
- 79.Hallen-Adams HE, Cavinder BL, Trail F. 2011. Fusarium graminearum from expression analysis to functional assays, p 79–101. In Xu J-R, Bluhm BH (ed), Fungal genomics: methods in molecular biology. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 80.Vanegas KG, Jarczynska ZD, Strucko T, Mortensen UH. 2019. Cpf1 enables fast and efficient genome editing in Aspergilli. Fungal Biol Biotechnol 6:6. doi: 10.1186/s40694-019-0069-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Chemical structures of cortical and medullary substances of lichens. Download FIG S1, PDF file, 0.2 MB (238.6KB, pdf) .
Copyright © 2021 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Lichen substances reported in the 30 genome-sequenced species. Download Table S1, DOCX file, 0.02 MB (31.7KB, docx) .
Copyright © 2021 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genome statistics and biosynthetic gene clusters in the sequenced lichen species. Download Data Set S1, XLSX file, 0.02 MB (18.2KB, xlsx) .
Copyright © 2021 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental materials and methods. Download Text S1, DOCX file, 0.2 MB (174.7KB, docx) .
Copyright © 2021 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Overview of the similarity of PKS families. Download FIG S2, PDF file, 0.4 MB (362.2KB, pdf) .
Copyright © 2021 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
NR-PKSs characterized in nonlichenized fungi and NR-PKSs found in the six Cladonia spp. and Stereocaulon alpinum. Download Data Set S2, XLSX file, 0.5 MB (560.9KB, xlsx) .
Copyright © 2021 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Verification of heterologous expression of PKS23. Download FIG S3, PDF file, 0.1 MB (135.2KB, pdf) .
Copyright © 2021 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
NMR and LC-MS/MS spectroscopic analyses. Download FIG S4, PDF file, 0.7 MB (756KB, pdf) .
Copyright © 2021 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Lichens with atranorin and usnic acid BGCs. Download FIG S5, PDF file, 0.5 MB (477.5KB, pdf) .
Copyright © 2021 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download Table S2, DOCX file, 0.03 MB (32.3KB, docx) .
Copyright © 2021 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The data from this whole-genome shotgun project have been deposited in DDBJ/ENA/GenBank under accession no. JAFEKC000000000, JAFEKC000000000, and JAEUBA000000000 (BioProject no. PRJNA693578, PRJNA693575, and PRJNA693574) for C. borealis, Parmelia cf. squarrosa, and S. alpinum, respectively. Raw LC-MS/MS data have been deposited in the MassIVE database (http://massive.ucsd.edu) under accession no. MSV000087081.