Abstract
Radioresistance is one of the main causes of cancer treatment failure, which leads to relapse and inferior survival outcome of cancer patients. Liquid-liquid phase separation (LLPS) of proteins is known to be involved in various biological processes, whereas its role in the regulation of radiosensitivity remains largely unknown. In this study, we characterized NONO, an RNA/DNA binding protein with LLPS capacity, as an essential regulator of tumor radioresistance. In vitro assay showed that NONO involved in DNA repair via non-homologous end joining (NHEJ) manner. NONO knockout significantly reduced DNA damage repair and sensitized tumor cells to irradiation in vitro and in vivo. NONO overexpression was correlated with an inferior survival outcome in cancer patients. Mechanically, NONO was associated with nuclear EGFR (nEGFR). Both irradiation and EGF treatment induced nEGFR accumulation, thereby increased the association between NONO and nEGFR. However, NONO was not a substrate of EGFR kinase. Furthermore, NONO promoted DNA damage-induced DNA-PK phosphorylation at T2609 by enhancing the interaction between EGFR and DNA-PK. Importantly, NONO protein formed high concentration LLPS droplets in vitro, and recruited EGFR and DNA-PK. Disruption of NONO droplets with LLPS inhibitor significantly reduced the interaction between EGFR and DNA-PK, and suppressed DNA damage-induced phosphorylation of T2609-DNA-PK. Taken together, LLPS of NONO recruits nuclear EGFR and DNA-PK and enhances their interaction, further increases DNA damage-activated pT2609-DNA-PK and promotes NHEJ-mediated DNA repair, finally leads to tumor radioresistance. NONO phase separation-mediated radioresistance may serve as a novel molecular target to sensitize tumor cell to radiotherapy.
Keywords: Phase separation, NONO, nuclear EGFR, DNA repair, radioresistance
Introduction
As one of the major cancer treatment strategies, radiotherapy is widely used in most of solid tumor treatment [1,2]. Radioresistance is the major cause of cancer treatment failure, whose underlying mechanism is mainly ascribed to high capacity of tumor cell to repair the irradiation induced DNA damage [3]. In theory, radiation kills cancer cells by producing a large amount of cytotoxic double strand DNA breaks (DSBs), and cells with accelerating DNA repair capacity will survive from radiotherapy [4]. Once DSBs occur, several canonical pathways are activated to arrest the cell proliferation and repair the damaged DNA [5]. DSBs are mainly repaired by non-homologous end joining (NHEJ) and homologous repair (HR) pathways [5-7]. Taking NHEJ for example, radiation induced DSBs triggers KU protein heterodimer (ku70/80), which further recruits DNA protein kinase (DNA-PKcs). Thereafter, XLF-XRCC4-DNA Ligase IV complex is loaded to and ligates the DNA ends [8,9]. Besides to these canonical pathways, some non-canonical mechanism had been confirmed to be an important complement to repair the radiation induced DSBs [10]. For example, we and others all found that membrane EGFR nuclear translocation was one of reasons of tumor cell radioresistance and a cause of Cetuximab resistance [11,12].
Recently, liquid-liquid phase separation (LLPS) or condensation has been reported to be an important mechanism underlying the formation of membraneless bodies, such as nucleoli in cells [13,14]. When undergo LLPS, a part of protein solution condenses into a dense phase which often resembles liquid droplets, and the remaining solution forms a dilute phase [15]. The driving force of LLPS is the weak multivalent interaction of multiple folded domains or intrinsically disordered regions [16]. Additionally, some RNAs have been found to drive LLPS (e.g., long noncoding RNA NEAT1 driving the formation of paraspeckle) [17], which provides a source of multivalency in the protein-RNA interaction. Accumulating evidences show that LLPS participates in various biological process, including DNA damage repair [18,19].
The non-POU domain containing octamer-binding (NONO), an RNA and DNA binding protein, is a member of the Drosophila behavior human splicing (DBHS) family [20,21]. NONO multifunctionally participates in various biological processes, including DNA repair, RNA splicing [22], RNA silencing [23], transcriptional regulation [20] and nuclear mRNA retention [24]. Even NONO has been characterized to participate in DNA repair pathway for decades, the underlying mechanism is still unclear. NONO was firstly reported to increase the infinity of several DNA repair factors, such as ku70, to DNA [25]. Further, SFPQ was identified to form heterodimer with NONO and enhanced NHEJ via in vitro assay [26]. Recently, NONO was found to be recruited to damaged DNA ends by poly (ADP-Ribose) (PAR), a post-translational modification catalyzed by PARP1 at DNA damage sites [27], and complexed with XLF to promote sequence-independent pairing of DNA substrates in NHEJ [28]. Moreover, NONO and other members of DBHS family, including SFPQ and PSPC1, strongly bind to NEAT1 to form paraspeckle, a membraneless bodies driving by LLPS [29]. However, whether NONO phase separation participates in DNA damage repair remains unclear.
Here, our findings show that NONO phase separation contributes to radiation-induced DNA damage repair. Upon irradiation, membrane EGFR translocates to nucleus, where NONO condensates recruit nuclear EGFR (nEGFR) and DNA-PK, following enhance the phosphorylation of DNA-PK at T2609 and accelerate the DNA repair of tumor cells, consequently induce radioresistance.
Materials and methods
Cell lines and tissue specimens
A431 cells were cultured in RPMI 1640 medium (Gibco, ThermoFisher Scientific, Waltham, Massachusetts, USA). HEK293T, MDA-MB-231 and U2OS cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco). All culture medium was supplemented with 10% (vol/vol) fetal bovine serum (FBS, Gibco). NONO knocked-out HeLa (HeLa-KO), MDA-MB-231 (MDA-MB-231-KO) and U2OS (U2OS-KO) cells were generated using CRISPR/cas9 tools, and the sequences of small guide RNA (sgRNA) were 5’-GAGTAATAAAACTTTTAACT-3’.
All of the clinical samples were obtained from the Tissue Bank of the Sixth Affiliated Hospital of Sun Yat-sen University, and approved by Human Medical Ethics Committee of Sun Yat-sen University. Clinicopathological parameters and follow-up information were retrieved from the Follow-up Database of the Sixth Affiliated Hospital of Sun Yat-sen University.
Plasmid constructs
The expression vector pCDH-myc-EGFR and pCDH-Flag-NONO were produced by respectively inserting C-terminal myc-tagged EGFR or Flag-tagged NONO sequence into pCDH-CMV-MCS-EF1-copGFP (pCDH, System Biosciences, Palo Alto, CA, USA), which contains a copGFP expression cassette.
To create different domains of Flag-NONO expression constructs (RRM1, RRM12, 12S, 2NC, NC and CC), pCDH-Flag-NONO was used as a template to perform deletion mutation (SMK-101, TOYOBO, Kita-ku, Osaka, Japan).
Using the pCDH-Flag-NONO expression vector as a template, all 5-tyrosine mutated NONO expression vector pCDH-Flag-NONO-5YF were developed by performing site-directed mutagenesis (SMK-101, TOYOBO) and verified by DNA sequencing.
The plasmids expressing myc-tagged extracellular or intracellular domain of EGFR (myc-ECD, myc-ICD) were described previously [30].
Fractions of cytoplasmic and nuclear proteins
Cells were washed three times with ice-cold PBS, scrapped in 1 mL Cyto-lysis buffer (10 mM Hepes-NaOH, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, and 0.5 mM beta-mercaptoethanol) supplemented with protease inhibitor (0463132001, Roche, Basel, Switzerland) and phosphatase inhibitor (04906837001, Roche), and incubated on ice for 15 minutes, followed by addition of 5 μl 10% NP-40. After kept on ice for 2 minutes, cell lysate was centrifuged at 16000 g, 4°C for 10 minutes and the supernatant (cytoplasmic extract) was collected. The pellet was washed with ice-cold PBS, resuspended in 100 μl Nucl-lysis buffer (10 mM Tris-HCl, pH 7.6, 420 mM NaCl, 0.5% NP-40, and 1 mM DTT, 1 mM PMSF, 2 mM MgCl2 plus protease inhibitor and phosphatase inhibitor), and incubated on ice for 20 minutes with 2-3 vortex. The nuclear extract was collected after centrifugation at 16000 g, 4°C for 15 minutes.
Immunoprecipitation
Cells were lysed in 1 mL ice-cold IP-lysis buffer (25 mM Tris-HCl at pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 5% glycerol) supplemented with protease inhibitor and phosphatase inhibitor at 4°C for 30 minutes. After centrifugation at 16000 g, 4°C for 10 minutes, the supernatant was collected for the following steps. For CoIP assay of Flag-tagged proteins, cell lysates were mixed with Anti-FLAG® M2 Magnetic Beads (M8823, Sigma-Aldrich, St. Louis, Missouri, USA) at 4°C for 2 hours. For CoIP assay, cell lysates were incubated with antibodies or IgG and gently rotated at 4°C for 4 hours, followed by adding of Dynabeads G (Invitrogen, ThermoFisher Scientific) and incubating at 4°C for another 2 hours with gentle rotation. Beads were washed 4-5 times with IP-lysis buffer and IP products were eluted with 50-100 ul 100 mM glycine (pH 3.5).
Real-time quantitative polymerase chain reaction (qPCR)
qPCR assays were performed to evaluate the RNA levels. Total RNA was extracted using TRIzol reagent (Invitrogen) and reverse-transcribed using ReverTra Ace® qPCR RT Master Mix with gDNA Remover (FSQ-301, TOYOBO, Tokoyo, Japan). qPCR was performed on a LightCycler 480 (Roche Diagnostics, Germany) using SYBR® Green Realtime PCR Master Mix (QPK-201, TOYOBO). The cycle threshold (Ct) values differed by less than 0.5 between duplicate wells. The relative expression levels of the target genes were normalized to that of internal control genes, which yielded a 2-ΔCt value. U6 was used as the reference genes. The primer sequences: NONO-F: TATGGAAAGGCAGGCGAAGT; NONO-R: TGGCATATTGTCCAGCTCCA; U6-F: CGGCAGCACATATAC; U6-R: TTCACGAATTTGCGTGTCAT; HuR-F: GCAGAGAGAGCGATCAACAC; HuR-R: GCCCAAACCGAGAGAACATG.
Western blotting and antibodies
Western blotting was used to evaluate protein level as described previously [31]. The primary antibodies used in western blotting: anti-GAPDH (60004-1-Ig, Proteintech, Rosemont, IL, USA), anti-NONO (cat. 611279, BD Bioscience, Franklin Lakes, NJ, USA), anti-EGFR (A11351, ABclonal, Wuhan, China), anti-p-EGFR (3777S, Cell signaling technology, Danvers, MA, USA), anti-Flag (ab49763, abcam, Cambridge, MA, USA), anti-myc (ab1326, abcam), 4G10 (05-321, Merck Millipore), anti-DNA-PK (A1419, ABclonal), anti-p-DNAPK (T2609) (AP0434, ABclonal), anti-LaminA+C (ab108595, abcam), anti-α-Tubulin (66031-1-Ig, Proteintech).
Immunofluorescence
After indicating treatment, cells seeded on glass coverslips were fixed with 4% paraformaldehyde for 15 minutes at room temperature, blocked with blocking buffer (1×PBS containing 5% goat serum and 0.3% Triton X-100) for 1 hour and incubated with primary antibodies for 2 hours at room temperature. After 3 times of washes with 1×PBS, cells were incubated with secondary antibody tagged with Alexa Fluor 488 or 555 (4408S, 4413S, Cell signaling technology) for 1 hour at room temperature. After twice washes with PBS, cells were stained with DAPI (D9542, Sigma-Aldrich) for 5 minutes. Slides were mounted in ProLong™ Diamond Antifade Mountant (P36965, Invitrogen). Primary antibodies used in Immunofluorescence including: anti-NONO (cat. 611279, BD Bioscience), anti-EGFR (A11351, ABclonal).
Duolink proximal ligation assay
Cells were fixed, blocked and incubated with primary antibodies as described in Immunofluorescence. Then, slides were washed gently with 1×PBS for three times and incubated with Duolink® In Situ PLA® Probe Anti-Rabbit MINUS (DUO92005, Sigma-Aldrich) and Duolink® In Situ PLA® Probe Anti-Mouse PLUS (DUO92001, Sigma-Aldrich) at 37°C for 1 hour. Then Duolink® In Situ Detection Reagents was used to perform the following ligation and amplification according to the instructions. After twice washes with PBS, the slides were stained with DAPI and mounted in ProLong™ Diamond Antifade Mountant. Images were captured by confocal microscopy (LSM880, Zeiss, Thornwood, NY, USA).
In vitro NHEJ assay
Restriction enzyme BamHI (R3136S, New England Biolabs, Beverly, MA, USA) was used to linearize pCSCMV-tdTomato plasmid to generate a non-homologous ends DNA fragment, followed by separation with a 0.8% agarose gel and purification using a DNA gel extraction kit (D2500, Omega Bio-tek, Norcross, GA, USA). The linearized pCSCMV-tdTomato plasmid was used as the substrate for the following end-join assay. Nuclear protein of U2OS cells was fractionated as described above. Linearized plasmid was incubated with different amount of nuclear protein in 20 μl NHEJ buffer (20 mM Hepes-KOH at pH 7.5, 80 mM KCl, 10 mM MgCl2, 1 mM ATP, 1 mM DTT and 1 mM dNTP mix) at 30°C for 30 minutes, terminated by the addition of 2 μl 0.5 M EDTA, 2 μl 0.5% sodium dodecylsulfate and 1 μl 10 mg/mL of Proteinase K and incubated at 37°C for 30 minutes. The DNA products were separated by gel electrophoresis.
Phos-tag-SDS-PAGE
Proteins were extracted with a RIPA buffer without EDTA (150 mM NaCl, 50 mM Tris-HCl pH 7.4, 1% NP-40, 0.25% Na-deoxycholate) and treated with/without Alkaline phosphatase (D7027, Beyotime, Shanghai, China) at 37°C for 30 minutes. Then, proteins were separated in 8% SDS-PAGE containing 20 μM Phos-tag-conjugated acrylamide (F4002, Ape×Bio, Houston, USA) and 20 μM MnCl2. The detection of proteins was performed as described in Western blotting and antibodies.
Live-cell imaging
Cells were seeded on glass plates and transfected with NONO-GFP plasmid for 36 hours before imaging. Cells were imaged using LSM880 confocal microscope (Zeiss). ZEN black edition software was used for acquisition and ZEN Lite software were used to process raw images.
Protein expression and purification
pGEX-6P-1-NONO-GFP (N-terminal GST tag) or pGEX-6P-1-GFP were transformed into E.coli strain BL21 (DE3) cells for expression. Cultures were grown to an OD600 of 0.6 and then induced with 0.1 mM IPTG (R0393, Invitrogen), followed by growth at 16°C overnight. The next day, cells were pelleted by centrifugation at 4000 g, 4°C for 10 minutes, resuspended in 1×PBS and lysed with brief sonication. After centrifugation at 10000 g, 4°C for 10 minutes, the supernatant was subjected to the purification of NONO-GFP or GFP proteins using GST-tag protein purification kit (P2262, Beyotime) according to the manufactured protocol.
In vitro droplets formation
GFP or NONO-GFP proteins were diluted to 10 μM in buffers with a final concentration of 150 mM NaCl and 20 mM Tris-HCl (pH=7.4) and then incubated at 25°C for 10 minutes. The protein mix was subjected to glass slide and then images were captured by fluorescence microscope (Zeiss).
Droplets pelleting
The whole cell lysate used below was extracted from U2OS cells with RIPA buffer. NONO-GFP or GFP proteins were co-incubated with U2OS whole cell extract at indicated concentration for 20 minutes at room temperature in buffers with a final concentration of 150 mM NaCl and 20 mM Tris-HCl (pH=7.4). Droplets fractions were obtained after centrifugation at 10,000 g for 10 minutes and then eluted by boiling in SDS sample buffer (2% SDS, 10 mM EDTA, 10% glycerol, 0.1% bromophenol blue, 50 mM Tris-HCl, pH=6.8). The supernatant was kept, followed by western blotting analysis with the droplet fractions.
Colony formation assay
Four thousand HCT116 or U2OS cells were seeded in 6-well plates. Twelve hours later, NU7441 (1 μM) was added for one hour before treated with indicated dose of irradiation. After irradiation, cells were cultured with or without NU7441 for another 24 hours, then cultured with drug-free medium for 8 days. Cells were washed with 1×PBS followed by fixing with methanol for 15 minutes at room temperature. Colonies were stained with 0.1% crystal violet staining solution for 15 minutes at room temperature. After washing, colonies were imaged and analyzed with Image J.
Statistical analysis
Un-paired t test was used to compare the difference between two groups. The data were shown in graphs as the mean ± standard error of the mean (SEM). Experiments were repeated independently for three times. P < 0.05 was considered statistically significant. All statistical tests were two-sided and were performed using GraphPad Prism (GraphPad Software Inc., San Diego, CA, USA).
Results
NONO is essential for DNA damage repair and radio-resistance of tumor cells
To investigate the role of NONO in tumor radioresistance, NONO knocked out (NONO-KO) cell lines (Figure 1A) were generated and exposed to radiation before colony formation assay. In agreement with the previous report [28], compared with wildtype tumor cells, NONO-KO cells formed fewer colony after irradiation (Figure 1B), indicating that NONO knockout sensitized tumor cells to radiation. Moreover, xenograft was conducted to evaluate the impact of NONO on radioresistance in vivo. Similar to in vitro findings, the xenografts derived from NONO-KO MC38 cells were more sensitive to radiotherapy than that from WT cells (Figure 1C). As known, aberrant DNA repair capacity leads to tumor cell radioresistance. We then examined whether NONO enhanced radioresistance by accelerating DNA repair. In vitro non-homologous end joining assay showed that NHEJ capacity was largely reduced in NONO-KO cells (Figure 1D). Furthermore, the analysis with the cancer genome atlas (TCGA) database showed that NONO was highly expressed in breast cancer tissues and NONO expression was correlated with cancer patient survival outcome (Figure 1E, 1F). Consistently, the examination of NONO expression in colorectal cancer (CRC) tissues suggested that NONO was upregulated in CRC tissues and high expression of NONO was correlated with the radioresistance of CRC (Figure 1G, 1H). It has been reported that RNA-binding protein HuR upregulates NONO protein via binding to the 5’ untranslated region of NONO mRNA [32]. We found that both NONO and HuR was upregulated in CRC tissues (Figures 1G, S1A), and their expressions were positively correlated (Figure S1B), suggesting that the increase of NONO expression in CRC may be resulted from the dysregulation of HuR.
Figure 1.
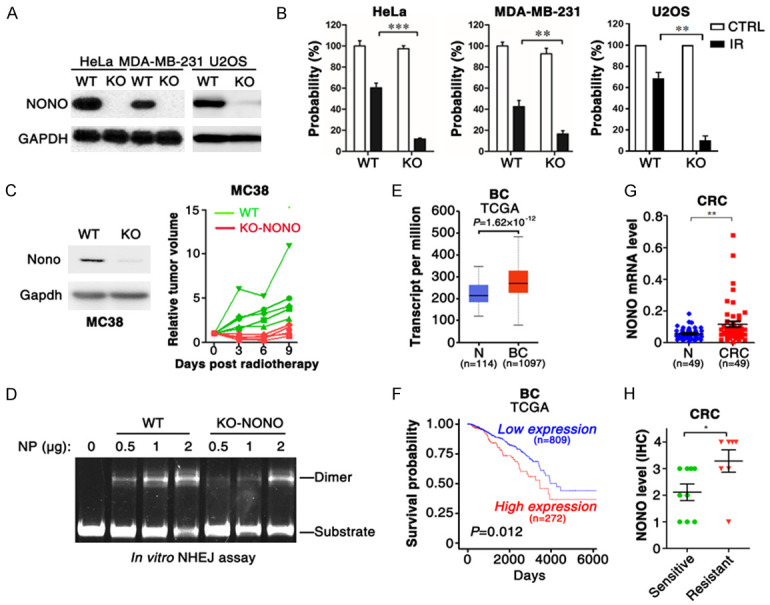
NONO is essential for DNA repair and radioresistance of tumor cells. A. NONO was knocked out in HeLa, MDA-MB-231 and U2OS cells. B. NONO depletion sensitized tumor cells to irradiation. C. NONO-KO cells-derived xenograft was more sensitive to irradiation than that from wildtype cells. Ten days after MC38 cell injection, mice were treated with radiotherapy (RT). D. NONO knockout inhibited NHEJ in in vitro NHEJ assay. NP, nuclear protein extracted from U2OS cells. E, F. NONO was overexpressed in breast cancer (BC) tissues, and high level of NONO was correlated with poor prognosis of breast cancer patients in TCGA data. G. NONO was upregulated in colorectal cancer (CRC). The mRNA level of NONO was analyzed in 49 paired CRC and normal (N) tissues using qPCR. H. NONO was highly expressed in radioresistant CRC tissues. The protein level of NONO was examined in CRC tissues with IHC. **, P < 0.01; ***, P < 0.001.
NONO interacts with EGFR
In our previous study, the interactome of EGFR was investigated by LC-MS and NONO was found to be one of EGFR binding partners [33]. Additionally, nuclear EGFR (nEGFR) have been involved in DNA repair through post-transcriptional regulation of its substrates [30,34] and the analysis of TCGA data showed that NONO-correlated genes were enriched in EGFR pathway (Figure S2A). We then asked if NONO-enhanced DNA repair was mediated by binding to EGFR. Firstly, myc tagged EGFR (myc-EGFR) and Flag tagged NONO (Flag-NONO) were transfected into HEK293T cells before coimmunoprecipitation (coIP) assay using anti-Flag or anti-myc antibodies. Flag-NONO was detected in anti-myc antibody-immunoprecipitates, and vice versa, myc-EGFR was detected in anti-Flag antibody-immunoprecipitates (Figure 2A). Besides, NONO and EGFR was also detected in the immunoprecipitates of endogenous NONO and EGFR protein with anti-NONO or EGFR antibodies (Figures 2B, 2C, S2B), indicating that NONO was associated with EGFR in cells. To visualize the in situ subcellular interaction between nEGFR and NONO, a Duolink proximity ligation assay was performed. As shown, the interaction between NONO and EGFR was detected in nucleus (Figure 2D). Furthermore, a series of truncation mutates of NONO or EGFR was generated and coIP assay was employed to mapping domains responsible for their interaction. Results showed that NONO bound with intracellular domain of EGFR (Figure 2E), and the RRM1 domain of NONO was essential for its interaction with EGFR (Figure 2F).
Figure 2.
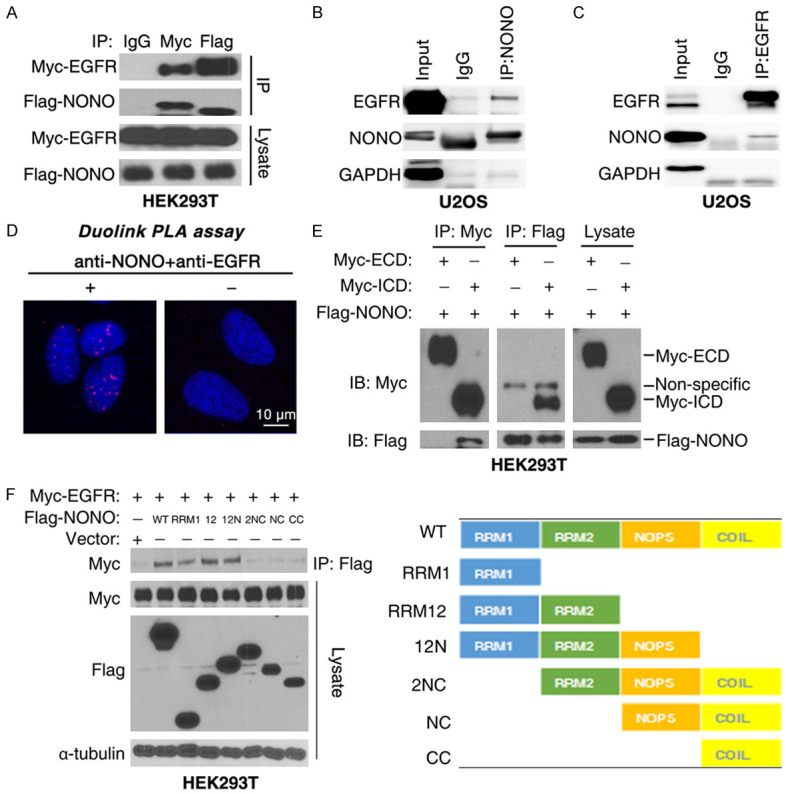
NONO interacts with EGFR. (A) CoIP assay with myc-EGFR and Flag-NONO transfected HEK293T cells. (B, C) CoIP of endogenous NONO or EGFR protein in U2OS cells. (D) The association of NONO and EGFR was verified with Duolink PLA assay. Antibodies, anti-NONO and anti-EGFR antibodies. (E) NONO interacted with the intracellular domain of EGFR. (F) EGFR bound to RRM1 domain of NONO. For (A, E, F), 36 hours after transfection with indicated plasmid, HEK293T cells were collected and subjected to CoIP assay.
IR and EGF induce the association between NONO and EGFR
To further examine whether NONO interacts with EGFR in cytoplasm or nucleus, the nuclear or cytoplasmic proteins were fractionated and subjected to coIP assay. Consistent with previous reports, NONO was only presented in nuclear fraction, and absent in cytoplasmic fraction, whereas EGFR mainly localized in cytoplasmic fraction (Figure 3A, left panel). CoIP assay with nuclear fraction showed that NONO was associated with nEGFR (Figure 3A, right panel; Figure S3A). Significantly, the interaction between NONO and EGFR was remarkably increased upon EGF or irradiation stimulation, but was abrogated by AG1478 (Figures 3B, S3B, S3C), a small compound inhibitor of EGFR. Furthermore, EGF-induced association between NONO and nuclear EGFR was confirmed by Duolink PLA (Figure 3C) and immunofluorescence assay (Figures 3D, S2D). These findings suggest that irradiation-induced internalization of EGFR enhanced the interaction between NONO and nuclear EGFR.
Figure 3.
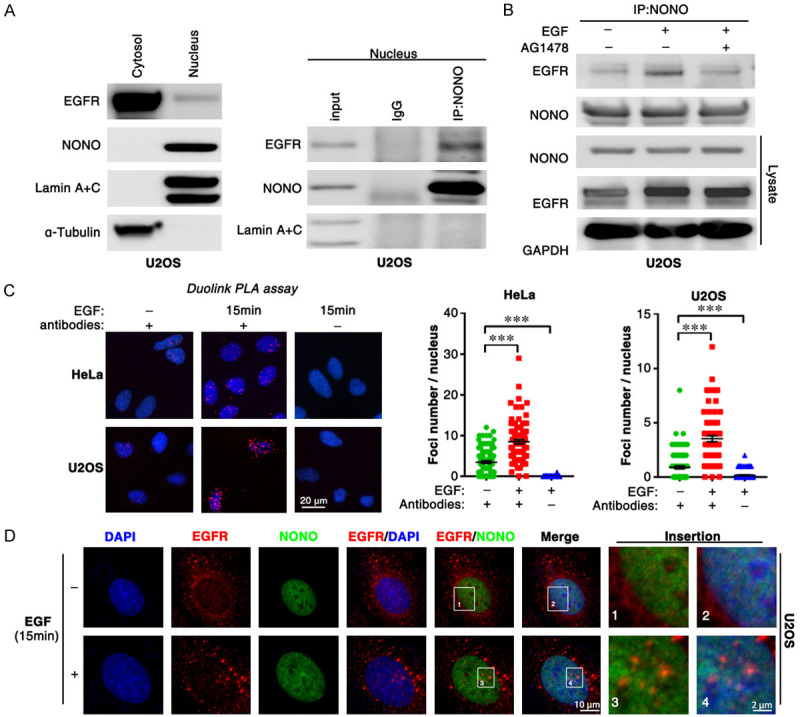
IR and EGF induce the association between NONO and EGFR. (A) The cytoplasmic and nuclear proteins were fractionated from U2OS cells (left panel) and subjected to CoIP assay with anti-NONO antibody (right panel). (B) EGF treatment enhances the interaction of NONO and EGFR in U2OS cell line. After EGF (10 ng/ml) treatment, CoIP assay was conducted with anti-NONO antibody. (C, D) EGF-induced association of NONO and EGFR was confirmed with Duolink PLA (C) and IF assay (D).
NONO is not phosphorylated by EGFR
Nuclear EGFR is a well-known tyrosine kinase that induces the phosphorylation of a various of substrates in nucleus. We next explore whether NONO is one of the nuclear EGFR substrates. Consistent with other reports, we found that anti-phosphotyrosine antibody specially recognized NONO protein, even all 5 tyrosines of NONO (Y97, Y158, Y265, Y267, and Y471) were mutated (Figure 4A), thereby the phosphorylation of NONO tyrosine residues could not be analyzed via this method. Phos-tag is a novel phosphate-binding tag, which could trap phosphorylated proteins and reduce their migration velocity, thereby separates phosphorylated and non-phosphorylated proteins in Phos-tag-SDS-PAGE. We then employed Phos-tag-SDS-PAGE to analyze the phosphorylation of NONO. A shift band of NONO was detected in control cell lysate, but not detected in phosphatase-treated cell lysate (Figure 4B, 4C), indicating that a small fraction of NONO was phosphorylated in cells. However, EGF or irradiation had no impact on the phosphorylation of NONO (p-NONO) (Figures 4B, 4C, S4). Interestingly, p-NONO was also detected in all 5 tyrosine-mutated NONO (Figure 4D), indicating that p-NONO was not phosphorylated at tyrosine residues. Together, these data suggest that NONO is not the substrate of tyrosine kinase EGFR.
Figure 4.
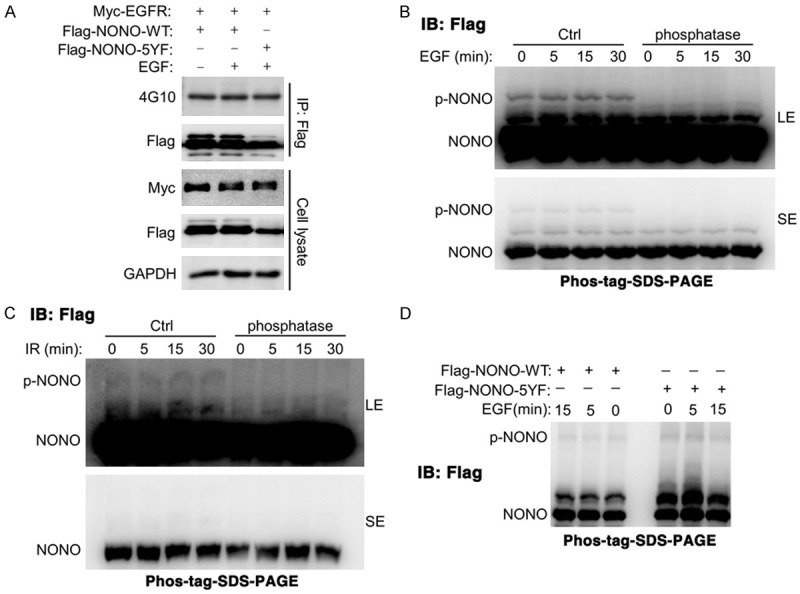
NONO is not phosphorylated by EGFR. (A) anti-phospho-tyrosine antibody (4G10) non-specifically bound to NONO. (B, C) EGF treatment (B) or irradiation (C) had no impact on the phosphorylation of NONO. LE: Long exposure. SE: Short exposure. (D) all of 5-tyrosine sites mutation had no impact on the phosphorylation of NONO. For (A-D), HEK293T cells were transfected with myc-EGFR and Flag-NONO plasmid for 36 hours before treated with IR (10 Gy) or EGF (10 ng/ml) for indicated times. For (B-D), the phosphorylated NONO was analyzed using Phos-tag-SDS-PAGE.
NONO promotes the EGFR-induced DNA-PK activation by enhancing their interaction
nEGFR was reported to interact with and enhance the phosphorylation of DNA-PK [35], a nuclear protein serine/threonine kinase that is essential for NHEJ-mediated DSB repair. Consistently, inhibition of DNA-PK phosphorylation with NU7441 significantly sensitized tumor cells to radiation (Figure S5A-C). We then investigated whether NONO was involved in DNA-PK activation via binding to EGFR. As shown in Figure 5A, 5B, NONO-KO largely reduced the radiation-induced phosphorylation of DNA-PK at T2609, which photocopied the treatment of AG1478. Most importantly, NONO-KO reduced the radiation-induced interaction between nuclear EGFR and DNA-PK (Figure 5C), indicating that NONO may enhance the activation of DNA-PK by increasing the association between nuclear EGFR and DNA-PK.
Figure 5.
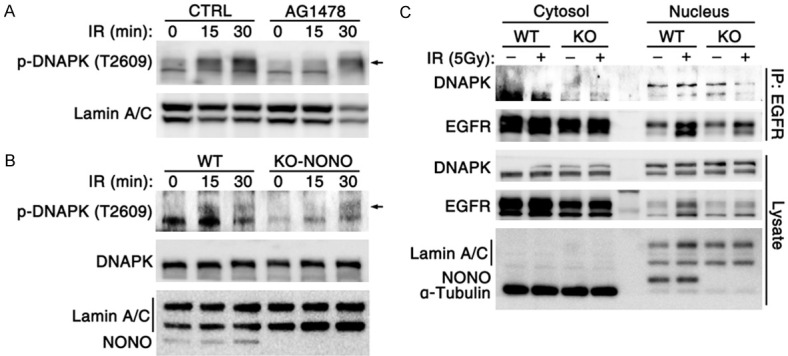
NONO promotes the EGFR-induced DNAPK activation by enhancing their interaction. (A) EGFR inhibitor AG1478 treatment inhibited IR-induced phosphorylation of DNAPK at T2609. (B) NONO depletion inhibited radiation-induced DNA-PK phosphorylation. (C) NONO knockout suppressed the radiation-induced association of EGFR and DNA-PK. For (A-C), the cytoplasmic and nuclear proteins were fractionated from U2OS cells.
LLPS of NONO recruits EGFR and DNA-PK and promotes their interaction
LLPS of NONO is essential for the formation of paraspeckle [23,36], which is involved in DNA damage response. Protein droplets that derived from LLPS could recruit other factors to provide individual place for biological process. We therefore proposed that NONO formed droplets may recruit EGFR and DNA-PK to enhance their interaction. Same to previous report, NONO-GFP formed liquid-like droplets and showed LLPS-like rate of FRAP in nucleus (Figure 6A). Besides, NONO droplets could be disrupted by 1,6-Hexanediol (Figure 6B), a compound known to disrupt liquid-like condensates by disruption of hydrophobic interactions. Furthermore, purified NONO protein could form LLPS in vitro (Figure 6C). After incubating with cell lysate, NONO droplets were separated by centrifugation and subjected to Western blotting analysis. As expected, NONO droplets recruited EGFR and DNA-PK in a dose-dependent manner (Figure 6D). Furthermore, 1,6-Hexanediol, a compound known to disrupt liquid-like condensates, significantly reduced the radiation-induced EGFR/NONO as well as EGFR/DNA-PK complex (Figure 6E, 6F), and inhibited the phosphorylation of DNA-PK at T2609 (Figure 6G).
Figure 6.
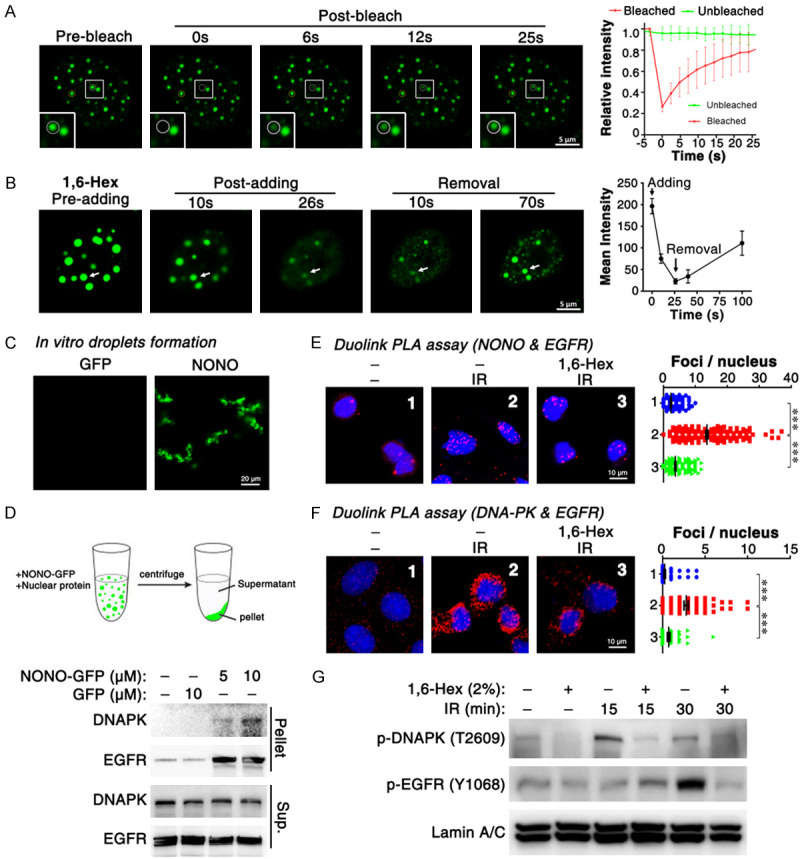
LLPS of NONO promotes the association of EGFR and NONO. (A) FRAP of NONO-GFP droplets in vivo. (B) 1,6-Hexanediol disrupted NONO-GFP droplets. 5% 1,6-Hexanediol in complete medium was added at 0 second and removed at 30 seconds. (C) purified NONO-GFP protein formed LLPS in vitro. RNA extracted from HEK293T cells were added to a concentration of 200 ng/ul. (D) NONO droplets recruit EGFR and DNA-PK. The nuclear protein was fractionated from U2OS cells. (E, F) confirm the impact of 1,6-Hexanediol on EGFR/NONO and EGFR/DNA-PK complex via Duolink PLA assay. (G) 1,6-Hexanediol suppressed IR-induced autophosphorylation of DNA-PK at T2609. For (E-G), 2% 1,6-Hex was added after irradiation and cells were cultured for 15 (E, F) or 30 minutes. ***, P < 0.001.
Taken together, NONO phase separation recruits nuclear EGFR and DNA-PK, enhances their interaction and increases the level of pT2609-DNA-PK, thereby promotes NHEJ-mediated DSB repair.
Discussion
Although NONO is well known to condense with other factors to form paraspeckles [23,36], the functions of NONO condensation in DNA repair and radioresistance remains to be disclosed. In this study, we identified NONO as a key regulator of NHEJ and tumor radioresistance. During the irradiation-induced DDR, NONO condensates recruit DNA-PK and nEGFR, which is activated and translocated into nucleus, to enhance the interaction between EGFR and DNA-PK, thereby increases the autophosphorylation of DNA-PK at T2609, promotes NHEJ-based DNA repair and induces tumor radioresistance (Figure 7).
Figure 7.
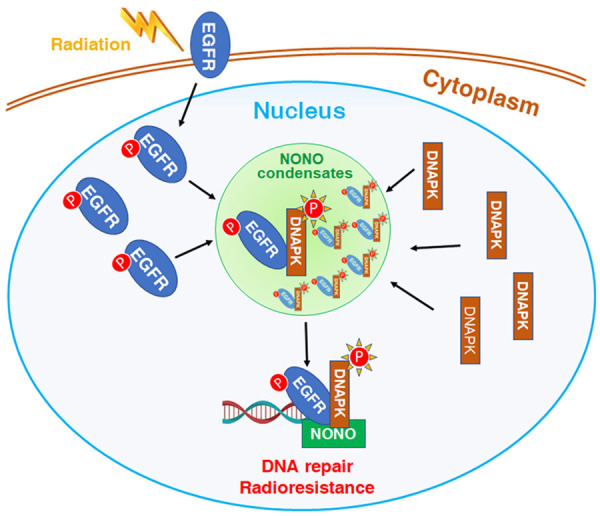
Model of NONO condensates promote DNA repair and radioresistance via acceleration of EGFR-mediated DNA-PK activation. During the irradiation-induced DDR, NONO condensates recruit DNA-PK and nEGFR, which is activated and translocated into nucleus, to enhance the interaction between EGFR and DNA-PK, thereby increases the autophosphorylation of DNA-PK at T2609, promotes NHEJ-mediated DNA repair and induces tumor radioresistance.
In the last decade, LLPS of proteins has been disclosed as an important mechanism underlying the formation and functions of membraneless organelles [16,37], e.g. nucleolus or paraspeckles. LncRNA NEAT1 functions as a scaffold to enrich the proteins of DBSH family, including NONO, SFPQ and PSPC1, to form paraspeckles [23,36]. NONO and NEAT1 are essential for paraspeckle formation, as silencing either of them would disrupt the structure of paraspeckles [28]. Paraspeckles are reported to regulate gene expression by A-I editing, mRNA nuclear retention and microRNA processing [38]. However, its roles in DNA damage repair are still unclear. Our data showed that LLPS of NONO recruited EGFR and DNA-PK and promoted their interaction and the autophosphorylation of DNA-PK. Inhibition of NONO LLPS with 1,6-Hexanediol significantly reduced the interaction between nEGFR and DNA-PK, and suppressed DNA-PK phosphorylation and DNA repair. In lines with our results, it has been shown that depletion of NEAT1, which is required for paraspeckle formation, reduced the genome stability and sensitized tumor cells to chemotherapy [39]. Therefore, LLPS plays a pivotal role in paraspeckles assembly and related DNA repair pathway.
As a tyrosine kinase, nuclear EGFR is well-known to bind and phosphorylate many substrates in nucleus, such as histone H4 [30] and Ago2 [40]. Although NONO was associated with nEGFR in nucleus, we concluded that NONO was not phosphorylated by EGFR according to the following evidences: firstly, EGF stimulation or irradiation had no impact on the phosphorylation level of NONO; secondly, mutation of all 5 tyrosine in NONO to phenylalanine also had no influence on NONO phosphorylation. In agreement with our results, Ahn R. Lee, et al. reported that NONO was not phosphorylated at tyrosine [41]. However, a shifted band was observed in the analysis of NONO using Phos-tag-SDS-PAGE, indicating that a small fraction of NONO was phosphorylated at the residues of other amino acids, rather than tyrosine.
The association between nuclear EGFR and DNA-PK has been reported by several studies, and their interaction increases the phosphorylation of DNA-PK, which is essential for DSB repair pathway of NHEJ [42,43]. In line with these reports, we found that radiation or EGF stimulated EGFR translocation to nucleus followed by interacting with DNA-PK and increasing its phosphorylation. However, the underlying mechanism of recruiting and harboring nuclear EGFR and DNA-PK during NHEJ based DNA repair remained unclear. Here, we uncovered a novel mechanism of NONO promoting DNA repair and enhancing radioresistance: LLPS of NONO formed a liquid droplet, creating a favorable niche to facilitate the accumulation of EGFR and DNA-PK at DSB end. Consistent with the role of EGFR in tumor radiosensitivity, EGFR was significantly down-regulated in radiation-induced enteritis tissues (Figure S6), suggesting that the dysregulation of EGFR may also sensitize normal colon cells to radiotherapy and lead to radiation-induced enteritis.
Targeting DNA repair axis has been reported to be a promising strategy for cancer therapy, e.g., PARP inhibitors are effective in germline BRCA1/2 mutated ovarian and breast cancer [44-47]. Besides PARP inhibitors, inhibitors of several other factors in DNA repair pathways have entered clinical trials [48], including inhibitors of ATM, ATR [49] and DNA-PK [50]. We and others all found that NONO overexpression would promote DNA repair and enhance radioresistance. Besides, radiotherapy-induced DNA damage would upregulate LncRNA NEAT1 and increase the formation of paraspeckle, leading to the enhancement of DNA repair and radioresistance. Indeed, targeting NONO or NEAT1 would sensitize the tumor cells to irradiation in vitro. However, it is difficult to target NONO and paraspeckle with small compound inhibitors. Here, using an alternative way of targeting LLPS by its inhibitor 1.6-hexanediol, we found that NONO enhanced radioresistance, mediated by nuclear EGRF and DNA-PK, was abrogated, providing a novel approach to restore the tumor cell radiosensitivity.
Taken together, NONO phase separation recruits DNA-PK and nuclear EGFR, which facilitates the interaction between EGFR and DNA-PK, thereby increases the phosphorylation of DNA-PK at T2609. Importantly, targeting this molecular pathway by LLPS inhibitor would suppress NHEJ approach-based DNA repair and sensitize tumor cells to radiation.
Acknowledgements
This work was supported by the Guangdong Science and Technology Project (No. 2017B090901065, No. 2019B030316003), Natural Science Foundation of China (No. 81872188, No. 81902867, No. 81903152), Natural Science Foundation of Guangdong Province (No. 2019A1515010901, No. 2020A1515010314), and China Postdoctoral Science Foundation (No. 2019M663301).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Citrin DE. Recent developments in radiotherapy. N Engl J Med. 2017;377:1065–1075. doi: 10.1056/NEJMra1608986. [DOI] [PubMed] [Google Scholar]
- 2.Feng K, Meng X, Liu J, Xing Z, Zhang M, Wang X, Feng Q, Wang X. Update on intraoperative radiotherapy for early-stage breast cancer. Am J Cancer Res. 2020;10:2032–2042. [PMC free article] [PubMed] [Google Scholar]
- 3.Dong Y, Zhang D, Cai M, Luo Z, Zhu Y, Gong L, Lei Y, Tan X, Zhu Q, Han S. SPOP regulates the DNA damage response and lung adenocarcinoma cell response to radiation. Am J Cancer Res. 2019;9:1469–1483. [PMC free article] [PubMed] [Google Scholar]
- 4.Mouw KW, Goldberg MS, Konstantinopoulos PA, D’Andrea AD. DNA damage and repair biomarkers of immunotherapy response. Cancer Discov. 2017;7:675–693. doi: 10.1158/2159-8290.CD-17-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Connor MJ. Targeting the DNA damage response in cancer. Mol Cell. 2015;60:547–560. doi: 10.1016/j.molcel.2015.10.040. [DOI] [PubMed] [Google Scholar]
- 6.Lanz MC, Dibitetto D, Smolka MB. DNA damage kinase signaling: checkpoint and repair at 30 years. EMB0 J. 2019;38:e101801. doi: 10.15252/embj.2019101801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grabarz A, Barascu A, Guirouilh-Barbat J, Lopez BS. Initiation of DNA double strand break repair: signaling and single-stranded resection dictate the choice between homologous recombination, non-homologous end-joining and alternative end-joining. Am J Cancer Res. 2012;2:249–268. [PMC free article] [PubMed] [Google Scholar]
- 8.Blackford AN, Jackson SP. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol Cell. 2017;66:801–817. doi: 10.1016/j.molcel.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 9.Chen MK, Hung MC. Regulation of therapeutic resistance in cancers by receptor tyrosine kinases. Am J Cancer Res. 2016;6:827–842. [PMC free article] [PubMed] [Google Scholar]
- 10.Bregenhorn S, Kallenberger L, Artola-Boran M, Pena-Diaz J, Jiricny J. Non-canonical uracil processing in DNA gives rise to double-strand breaks and deletions: relevance to class switch recombination. Nucleic Acids Res. 2016;44:2691–2705. doi: 10.1093/nar/gkv1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Veggel B, de Langen AJ, Hashemi SMS, Monkhorst K, Heideman DAM, Thunnissen E, Smit EF. Afatinib and cetuximab in four patients with EGFR exon 20 insertion-positive advanced NSCLC. J Thorac Oncol. 2018;13:1222–1226. doi: 10.1016/j.jtho.2018.04.012. [DOI] [PubMed] [Google Scholar]
- 12.Ju H, Hu Z, Lu Y, Wu Y, Zhang L, Wei D, Guo W, Xia W, Liu S, Ren G, Hu J. TLR4 activation leads to anti-EGFR therapy resistance in head and neck squamous cell carcinoma. Am J Cancer Res. 2020;10:454–472. [PMC free article] [PubMed] [Google Scholar]
- 13.Frottin F, Schueder F, Tiwary S, Gupta R, Korner R, Schlichthaerle T, Cox J, Jungmann R, Hartl FU, Hipp MS. The nucleolus functions as a phase-separated protein quality control compartment. Science. 2019;365:342–347. doi: 10.1126/science.aaw9157. [DOI] [PubMed] [Google Scholar]
- 14.Alberti S, Gladfelter A, Mittag T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019;176:419–434. doi: 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, Li CH, Guo YE, Day DS, Schuijers J, Vasile E, Malik S, Hnisz D, Lee TI, Cisse II, Roeder RG, Sharp PA, Chakraborty AK, Young RA. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361:eaar3958. doi: 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science. 2017;357:eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 17.Maharana S, Wang J, Papadopoulos DK, Richter D, Pozniakovsky A, Poser I, Bickle M, Rizk S, Guillen-Boixet J, Franzmann TM, Jahnel M, Marrone L, Chang YT, Sterneckert J, Tomancak P, Hyman AA, Alberti S. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science. 2018;360:918–921. doi: 10.1126/science.aar7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pessina F, Giavazzi F, Yin Y, Gioia U, Vitelli V, Galbiati A, Barozzi S, Garre M, Oldani A, Flaus A, Cerbino R, Parazzoli D, Rothenberg E, d’Adda di Fagagna F. Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nat Cell Biol. 2019;21:1286–1299. doi: 10.1038/s41556-019-0392-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kilic S, Lezaja A, Gatti M, Bianco E, Michelena J, Imhof R, Altmeyer M. Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J. 2019;38:e101379. doi: 10.15252/embj.2018101379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knott GJ, Bond CS, Fox AH. The DBHS proteins SFPQ, NONO and PSPC1: a multipurpose molecular scaffold. Nucleic Acids Res. 2016;44:3989–4004. doi: 10.1093/nar/gkw271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mircsof D, Langouet M, Rio M, Moutton S, Siquier-Pernet K, Bole-Feysot C, Cagnard N, Nitschke P, Gaspar L, Znidaric M, Alibeu O, Fritz AK, Wolfer DP, Schroter A, Bosshard G, Rudin M, Koester C, Crestani F, Seebeck P, Boddaert N, Prescott K, Study DDD, Hines R, Moss SJ, Fritschy JM, Munnich A, Amiel J, Brown SA, Tyagarajan SK, Colleaux L. Mutations in NONO lead to syndromic intellectual disability and inhibitory synaptic defects. Nat Neurosci. 2015;18:1731–1736. doi: 10.1038/nn.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benegiamo G, Mure LS, Erikson G, Le HD, Moriggi E, Brown SA, Panda S. The RNA-binding protein NONO coordinates hepatic adaptation to feeding. Cell Metab. 2018;27:404–418. e407. doi: 10.1016/j.cmet.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang L, Shao C, Wu QJ, Chen G, Zhou J, Yang B, Li H, Gou LT, Zhang Y, Wang Y, Yeo GW, Zhou Y, Fu XD. NEAT1 scaffolds RNA-binding proteins and the Microprocessor to globally enhance pri-miRNA processing. Nat Struct Mol Biol. 2017;24:816–824. doi: 10.1038/nsmb.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alfano L, Costa C, Caporaso A, Altieri A, Indovina P, Macaluso M, Giordano A, Pentimalli F. NONO regulates the intra-S-phase checkpoint in response to UV radiation. Oncogene. 2016;35:567–576. doi: 10.1038/onc.2015.107. [DOI] [PubMed] [Google Scholar]
- 25.Yang YS, Yang MC, Tucker PW, Capra JD. NonO enhances the association of many DNA-binding proteins to their targets. Nucleic Acids Res. 1997;25:2284–2292. doi: 10.1093/nar/25.12.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bladen CL, Udayakumar D, Takeda Y, Dynan WS. Identification of the polypyrimidine tract binding protein-associated splicing factor. p54(nrb) complex as a candidate DNA double-strand break rejoining factor. J Biol Chem. 2005;280:5205–5210. doi: 10.1074/jbc.M412758200. [DOI] [PubMed] [Google Scholar]
- 27.Chu YY, Yam C, Chen MK, Chan LC, Xiao M, Wei YK, Yamaguchi H, Lee PC, Han Y, Nie L, Sun X, Moulder SL, Hess KR, Wang B, Hsu JL, Hortobagyi GN, Litton J, Chang JT, Hung MC. Blocking c-Met and EGFR reverses acquired resistance of PARP inhibitors in triple-negative breast cancer. Am J Cancer Res. 2020;10:648–661. [PMC free article] [PubMed] [Google Scholar]
- 28.Jaafar L, Li Z, Li S, Dynan WS. SFPQ*NONO and XLF function separately and together to promote DNA double-strand break repair via canonical nonhomologous end joining. Nucleic Acids Res. 2017;45:1848–1859. doi: 10.1093/nar/gkw1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirose T, Yamazaki T, Nakagawa S. Molecular anatomy of the architectural NEAT1 noncoding RNA: the domains, interactors, and biogenesis pathway required to build phase-separated nuclear paraspeckles. Wiley Interdiscip Rev RNA. 2019;10:e1545. doi: 10.1002/wrna.1545. [DOI] [PubMed] [Google Scholar]
- 30.Chou RH, Wang YN, Hsieh YH, Li LY, Xia W, Chang WC, Chang LC, Cheng CC, Lai CC, Hsu JL, Chang WJ, Chiang SY, Lee HJ, Liao HW, Chuang PH, Chen HY, Wang HL, Kuo SC, Chen CH, Yu YL, Hung MC. EGFR modulates DNA synthesis and repair through Tyr phosphorylation of histone H4. Dev Cell. 2014;30:224–237. doi: 10.1016/j.devcel.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang YL, Liu JY, Yang JE, Yu XM, Chen ZL, Chen YJ, Kuang M, Zhu Y, Zhuang SM. Lnc-UCID promotes G1/S transition and hepatoma growth by preventing DHX9-mediated CDK6 down-regulation. Hepatology. 2019;70:259–275. doi: 10.1002/hep.30613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alfano L, Costa C, Caporaso A, Antonini D, Giordano A, Pentimalli F. HUR protects NONO from degradation by mir320, which is induced by p53 upon UV irradiation. Oncotarget. 2016;7:78127–78139. doi: 10.18632/oncotarget.13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huo L, Wang YN, Xia W, Hsu SC, Lai CC, Li LY, Chang WC, Wang Y, Hsu MC, Yu YL, Huang TH, Ding Q, Chen CH, Tsai CH, Hung MC. RNA helicase A is a DNA-binding partner for EGFR-mediated transcriptional activation in the nucleus. Proc Natl Acad Sci U S A. 2010;107:16125–16130. doi: 10.1073/pnas.1000743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee HH, Wang YN, Hung MC. Non-canonical signaling mode of the epidermal growth factor receptor family. Am J Cancer Res. 2015;5:2944–2958. [PMC free article] [PubMed] [Google Scholar]
- 35.Dittmann K, Mayer C, Kehlbach R, Rodemann HP. Radiation-induced caveolin-1 associated EGFR internalization is linked with nuclear EGFR transport and activation of DNA-PK. Mol Cancer. 2008;7:69. doi: 10.1186/1476-4598-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamazaki T, Souquere S, Chujo T, Kobelke S, Chong YS, Fox AH, Bond CS, Nakagawa S, Pierron G, Hirose T. Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol Cell. 2018;70:1038–1053. e1037. doi: 10.1016/j.molcel.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 37.Sawyer IA, Sturgill D, Dundr M. Membraneless nuclear organelles and the search for phases within phases. Wiley Interdiscip Rev RNA. 2019;10:e1514. doi: 10.1002/wrna.1514. [DOI] [PubMed] [Google Scholar]
- 38.Pisani G, Baron B. Nuclear paraspeckles function in mediating gene regulatory and apoptotic pathways. Noncoding RNA Res. 2019;4:128–134. doi: 10.1016/j.ncrna.2019.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adriaens C, Standaert L, Barra J, Latil M, Verfaillie A, Kalev P, Boeckx B, Wijnhoven PW, Radaelli E, Vermi W, Leucci E, Lapouge G, Beck B, van den Oord J, Nakagawa S, Hirose T, Sablina AA, Lambrechts D, Aerts S, Blanpain C, Marine JC. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med. 2016;22:861–868. doi: 10.1038/nm.4135. [DOI] [PubMed] [Google Scholar]
- 40.Shen J, Xia W, Khotskaya YB, Huo L, Nakanishi K, Lim SO, Du Y, Wang Y, Chang WC, Chen CH, Hsu JL, Wu Y, Lam YC, James BP, Liu X, Liu CG, Patel DJ, Hung MC. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature. 2013;497:383–387. doi: 10.1038/nature12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee AR, Hung W, Xie N, Liu L, He L, Dong X. Tyrosine residues regulate multiple nuclear functions of P54nrb. J Cell Physiol. 2017;232:852–861. doi: 10.1002/jcp.25493. [DOI] [PubMed] [Google Scholar]
- 42.Liccardi G, Hartley JA, Hochhauser D. EGFR nuclear translocation modulates DNA repair following cisplatin and ionizing radiation treatment. Cancer Res. 2011;71:1103–1114. doi: 10.1158/0008-5472.CAN-10-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Javvadi P, Makino H, Das AK, Lin YF, Chen DJ, Chen BP, Nirodi CS. Threonine 2609 phosphorylation of the DNA-dependent protein kinase is a critical prerequisite for epidermal growth factor receptor-mediated radiation resistance. Mol Cancer Res. 2012;10:1359–1368. doi: 10.1158/1541-7786.MCR-12-0482-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slade D. PARP and PARG inhibitors in cancer treatment. Genes Dev. 2020;34:360–394. doi: 10.1101/gad.334516.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barker HE, Paget JT, Khan AA, Harrington KJ. The tumour microenvironment after radiotherapy: mechanisms of resistance and recurrence. Nat Rev Cancer. 2015;15:409–425. doi: 10.1038/nrc3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chu YY, Yam C, Chen MK, Chan LC, Xiao M, Wei YK, Yamaguchi H, Lee PC, Han Y, Nie L, Sun X, Moulder SL, Hess KR, Wang B, Hsu JL, Hortobagyi GN, Litton J, Chang JT, Hung MC. Blocking c-Met and EGFR reverses acquired resistance of PARP inhibitors in triple-negative breast cancer. Am J Cancer Res. 2020;10:648–661. [PMC free article] [PubMed] [Google Scholar]
- 47.Li JJ, Zhi XL, Chen SY, Shen XQ, Chen C, Yuan L, Guo JY, Meng D, Chen M, Yao LQ. CDK9 inhibitor CDKI-73 is synergetic lethal with PARP inhibitor olaparib in BRCA1 wide-type ovarian cancer. Am J Cancer Res. 2020;10:1140–1155. [PMC free article] [PubMed] [Google Scholar]
- 48.Stover EH, Konstantinopoulos PA, Matulonis UA, Swisher EM. Biomarkers of response and resistance to DNA repair targeted therapies. Clin Cancer Res. 2016;22:5651–5660. doi: 10.1158/1078-0432.CCR-16-0247. [DOI] [PubMed] [Google Scholar]
- 49.Sun LL, Yang RY, Li CW, Chen MK, Shao B, Hsu JM, Chan LC, Yang Y, Hsu JL, Lai YJ, Hung MC. Inhibition of ATR downregulates PD-L1 and sensitizes tumor cells to T cell-mediated killing. Am J Cancer Res. 2018;8:1307–1316. [PMC free article] [PubMed] [Google Scholar]
- 50.Fok JHL, Ramos-Montoya A, Vazquez-Chantada M, Wijnhoven PWG, Follia V, James N, Farrington PM, Karmokar A, Willis SE, Cairns J, Nikkila J, Beattie D, Lamont GM, Finlay MRV, Wilson J, Smith A, O’Connor LO, Ling S, Fawell SE, O’Connor MJ, Hollingsworth SJ, Dean E, Goldberg FW, Davies BR, Cadogan EB. AZD7648 is a potent and selective DNA-PK inhibitor that enhances radiation, chemotherapy and olaparib activity. Nat Commun. 2019;10:5065. doi: 10.1038/s41467-019-12836-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


