Abstract
BET bromodomain inhibitors (BETi) are promising therapeutic regimens for epithelial ovarian cancer (EOC). However, early-stage clinical trials indicate that drug tolerance may limit their anti-tumor efficacy. Here, we show that JQ1-refractory EOC cells acquire reversible resistance to BET inhibition and remain dependent on BRD4 function. The insensitivity is driven by a unique non-genetic mechanism that involves clonal selection for a pre-existing cell subpopulation with ample acetylated histones and sufficient nuclear phase-separated BRD4 droplets to counteract BETi antagonism. A vertical combination approach by co-blocking BET proteins and downstream Aurora kinases proves to achieve more complete responses than single inhibitors. Collectively, our study implicates epigenetic heterogeneity in therapeutic resistance to chromatin-targeted agents and proposes a rational strategy to address this anticipated clinical dilemma.
Keywords: Ovarian cancer, BET inhibitor, drug resistance, epigenetic heterogeneity, vertical combination therapy
Introduction
A growing emphasis of the current drug discovery program has focused on modulating the chromatin-modifying factors, including histone acetyltransferases (HAT), histone deacetylases (HDAC) and the bromodomain and extra-terminal (BET) family members [1-6]. Notably, as epithelial ovarian cancer lacks conventional druggable genetic alterations [7], specific inhibition of epigenetic regulators is increasingly recognized as an attractive therapeutic strategy [8-12]. For example, we and other groups recently reported that in preclinical studies, small molecule inhibitors of BET bromodomain proteins exhibited robust efficacy against ovarian cancer via inducing cell cycle arrest and apoptosis [8,13,14]. The selectivity of BET-targeting compounds arose from disproportionate localization of BET proteins to super-enhancer elements that transcriptionally regulated key oncogenes such as FOXM1, ALDH1A1 and MYC [6,8,15,16]. However, early clinical trials of BET inhibitors (BETi) showed disappointing results in ovarian cancer cases, necessitating further investigations to overcome the limitations of these epigenomic agents [17].
Lessons from myriad molecular targeted therapies indicate that drug resistance represents a major obstacle to successful cancer treatment. Substantial efforts have revealed diverse mechanisms underlying BETi tolerance in a spectrum of advanced malignancies [18-23]. In acute myeloid leukemia, adaptive WNT signaling serves as an alternative pathway to circumvent BET inhibition [24,25]. Likewise, receptor tyrosine kinase reprogramming leads to BETi resistance in multiple solid tumors [26-30]. On the other hand, BETi-tolerant triple-negative breast cancer cells preserve dependency on BRD4 function but develop bromodomain-independent chromatin recruitment machinery [31,32]. Therefore, acquired resistance to BET inhibitors can occur through either bypass tracks or target reactivation.
In this study, using JQ1-responsive ovarian cancer as a prototype, we identified an under-appreciated modality of BETi resistance involving epigenetic heterogeneity and clonal evolution. Our data demonstrated that JQ1-resistant ovarian cancer cells emerged from the therapeutic selection of pre-existing subpopulations with desirable basal level of acetylated histone H3 lysine 27 (H3K27ac), and remained addicted to excessive chromatin-bound BRD4. Simultaneous suppression of BRD4 and its downstream Aurora kinases provided a rational combination approach yielding vertical pathway inhibition and synergistic anti-tumor effects. Together, these findings proposed a novel paradigm for epigenetic drug evasion and unveiled a unique opportunity to optimize the clinical efficacy of numerous BET inhibitors under development.
Materials and methods
Cell culture and reagents
Cell lines COV 413B (RRID: CVCL_2423), OVCA420 (RRID: CVCL_3935) and SKOV3 (RRID: CVCL_0532) were obtained from American Type Culture Collection (ATCC) and were cultured in RPMI1640 (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) and 1% penicillin and streptomycin (Gibco). JQ1 was purchased from Millipore. C646, AZD1152, VX680, and MLN8237 were purchased from MedChemExpress. 1,6-hexanediol was purchased from Sigma. For visualization, cells were fixed with formalin and stained with crystal violet.
Cell viability assays and combination matrices
Cell viability assay was performed using Cell Counting Kit-8 (Dojindo Laboratories). Cells were seeded in triplicates in 96-well plates and treated with indicated inhibitors for 96 h before measuring the absorbance at 450 nm according to the manufacturer’s instructions. For evaluating combination efficacy, cells were seeded in 96-well plates at 3000-5000 cells per well and treated with JQ1 and Aurora kinase inhibitors in 6×6 matrices. After 96 h, cell viability was determined using Cell Counting Kit-8. The Bliss synergy score was calculated by the equation (A+B) - A×B. A and B were the fractional growth inhibitions induced by agents A and B at a given dose.
Western blot
Cells were lysed in RIPA buffer (Tris pH 7.4 50 mM, NaCl 150 mM, NP-40 1%, SDS 0.1%, EDTA 2 μM) containing proteinase inhibitors (Roche) and phosphatase inhibitors (Roche), and subjected to SDS-PAGE and Western blot. Antibodies against the following proteins were used: BRD2, BRD3, BRD4 (Abcam); H3K27ac, FoxM1, AURKA, AURKB, H3, Actin, GAPDH (Cell Signaling Technology).
Quantitative PCR assays
Genomic DNA from tumor cells was extracted using QIAampDNA mini kit (Qiagen) according to the manufacturer’s protocol. The genomic DNA was used to perform the quantitative PCR on the Applied Biosystems ViiA7 machine. Three biological replicates were included for each condition. Data were normalized relative to MTHFR. The primer sequences used for PCR were as follows: MTHFR-F: CCATCTTCCTGCTGCTGTAACTG; MTHFR-R: GCCTTCTCTGCCAACTGTCC; BRD4-exon1-F: GACCTCCAACCCTAACAA; BRD4-exon1-R: TTCCATAGTGTCTTGAGCA; BRD4-exon5-F: GAGATGTTTGCCAAGAAG; BRD4-exon5-R: TTGATGATGTCACAGTAGT.
Immunofluorescent staining
Cells were plated in an 8-well culture dish for confocal laser scanning microscopy (Ibidi). For immunofluorescent staining, cells were washed with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100 and blocked with 4% BSA. The primary antibodies (1:200) were incubated overnight, followed by secondary antibodies (1:400) for 30 min and DAPI counterstaining for 5 min in the dark. Alexa Fluor 488 (Goat anti-rabbit IgG), Alexa Fluor 594 (Goat anti-mouse IgG), and DAPI were purchased from Invitrogen.
Flow cytometry analysis
Cells were fixed and permeabilized using BD Cytofix/Cytoperm kit (BD Biosciences) according to the manufacturer’s protocol. The primary antibodies (1:200) were incubated for 1 hour, followed by secondary antibodies (1:200) for 30 min in the dark. Flow cytometric analysis was performed on a FACS AriaII cytometer (BD Biosciences). Flow cytometry data were analyzed using FlowJo software.
Chromatin immunoprecipitation and sequencing
Chromatin precipitation was performed as previously described [33]. Cells were cross-linked with serum-free medium plus 1% formaldehyde for 10 min and quenched with 2.5 M glycine. Cell pellets were lysed and sonicated using Sonics Vibra-Cell 505 ultrasonicator (Sonics and Materials). 50 μl of sonicated DNA was taken as control, and the remaining DNA fragments were cleared and incubated overnight with magnetic beads coated with the H3K27ac antibody (Abcam). Precipitated complexes were rinsed and cross-links were reversed overnight. Samples were digested with RNase A and Proteinase K, and DNA was extracted with QIAquick PCR Purification Kit according to the manufacturer’s instructions (Qiagen). DNA libraries were generated and sequenced on an Illumina Hiseq 2000 platform. All the downstream analyses were based on high-quality clean data. Index of the reference genome was built using BWA v0.7.12 and clean reads were aligned to the reference genome using BWA mem v 0.7.12. After mapping reads to the reference genome, we used the MACS2 version 2.1.0 (model-based analysis of ChIP-seq) peak finding algorithm to identify regions of ChIP enrichment over the background. A q-value threshold of enrichment of 0.05 was used for all data sets. Super-enhancers were identified using the ROSE package.
RNA sequencing and analysis
Cells were treated with DMSO or JQ1 (500 nM) for 6 hours. ERCC Spike-In RNA Mix (Life Technologies) was added to the cell lysates in proportion to cell number as previously described. Total RNA (three biological replicates per condition) was extracted using RNeasy plus mini kit (Qiagen) according to the manufacturer’s protocol, and RNA qualification was evaluated by RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system. Sequencing libraries were prepared using the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs). The clustering of the index-coded libraries was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina), and the library preparations were sequenced on an Illumina Hiseq 2500 platform to generate 30 million 125 bp paired-end reads (Novogene). The following analyses were based on clean data which were obtained by removing low-quality reads and reads containing adapters or ploy-N sequences. The index of the reference genome was built using Bowtie v2.2.3 and clean reads were aligned to the reference genome using TopHat v2.0.12. We used HTSeq v0.6.1 to count the reads mapped to each gene and further normalized read counts to the control ERCC reads. Differential expression analysis was performed using the DESeq R package (1.18.0). P-values were adjusted using the Benjamini-Hochberg procedure for controlling the false discovery rate. Genes with an adjusted P-value of < 0.05 were considered differentially expressed.
Plasmids and sgRNA
The BRD4 open reading frame was amplified from the genomic DNA of SKOV3 cells. Plasmids expressing GFP-tagged BRD4 or GFP were constructed using the Gibson Assembly Cloning Kit (New England Biolabs) according to the manufacturer’s protocol. Stable cell lines were established by infecting COV 413B, OVCA420, and SKOV3 with a lentiviral construct carrying pLVX-BRD4-GFP or pLVX-GFP. The CRISPR-Cas9 technology was employed to knock out indicated genes. Virally infected cells were selected with 2-5 μg/mL puromycin.
The primer sequences used for constructing plasmids were as follows: BRD4-F: GCATGGACGAGCTGTACAAGTCTAGAATGTCTGCGGAGAGCGGCCC; BRD4-R: GGAGGGAGAGGGGCGGGATCCTCAGAAAAGATTTTCTTCAAATATTGACAAT; GFP-F: TAGAGGATCTATTTCCGGTGATGGTGAGCAAGGGCGAG; GFP-R: CGTCATGGTCTTTGTAGTCTTTACTTGTACAGCTCGTCCATG.
The sgRNA sequences used for gene knockout were as follows: EGFP-sgRNA-F-CACCGGAAGTTCGAGGGCGACACCC; EGFP-sgRNA-R-AAACGGGTGTCGCCCTCGAACTTCC; BRD4-sgRNA-1F: CACCGCCAGACCCCTGTCATGACAG; BRD4-sgRNA-1R: AAACCTGTCATGACAGGGGTCTGGC; BRD4-sgRNA-2F: CACCGGTCGATGCTTGAGTTGTGTT; BRD4-sgRNA-2R: AAACAACACAACTCAAGCATCGACC; EP300-sgRNA-1F: GTTCAATTGGAGCAGGCCGA; EP300-sgRNA-1R: TCGGCCTGCTCCAATTGAAC; EP300-sgRNA-2F: ATTCTTCATTGTGCGACAGT; EP300-sgRNA-2R: ACTGTCGCACAATGAAGAAT; EP300-sgRNA-3F: GTGGCACGAAGATATTACTC; EP300-sgRNA-3R: GAGTAATATCTTCGTGCCAC; CREBBP-sgRNA-1F: CGCGTGACCAGTCATTTGCG; CREBBP-sgRNA-1R: CGCAAATGACTGGTCACGCG; CREBBP-sgRNA-2F: TCGACAATGCGGGAGCGAGC; CREBBP-sgRNA-2R: GCTCGCTCCCGCATTGTCGA; CREBBP-sgRNA-3F: AGCTCTAAAGGATCGCCGCA; CREBBP-sgRNA-3R: TGCGGCGATCCTTTAGAGCT.
Statistical analysis
Statistical analysis was performed with the R system version 3.6.0 or GraphPad Prism version 6.00. Cell counting and fluorescence intensity assessment were performed using Image-Pro Plus 6.0. In all experiments, comparisons between two groups were based on a two-sided Student’s t-test. Pearson’s correlation coefficient was used to measure the linear correlation between two variables. P-values of < 0.05 were considered statistically significant.
Results
JQ1-tolerant ovarian cancer cells exhibit reversible resistance to BET inhibition
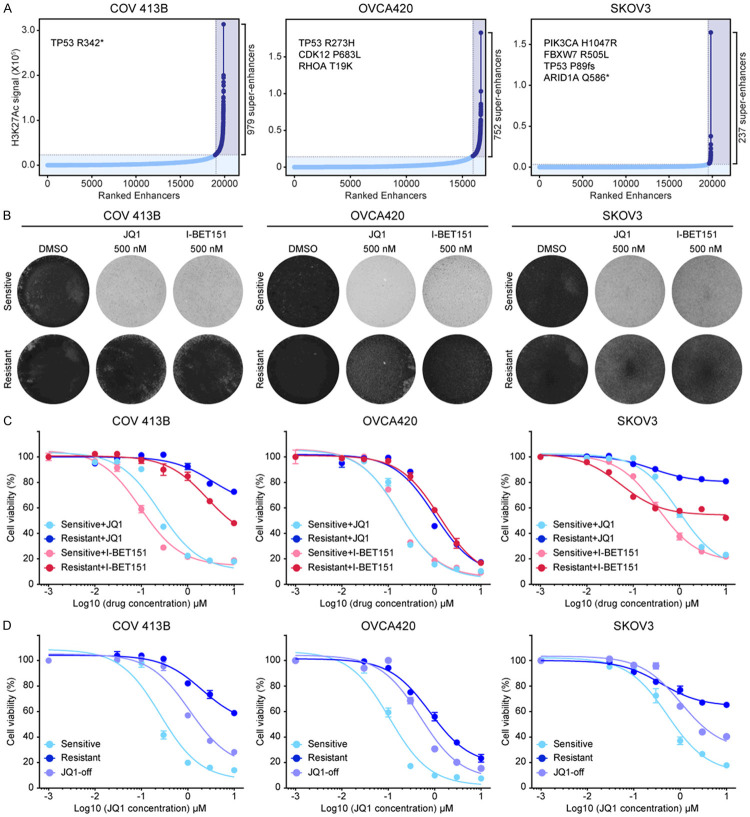
To determine the molecular mechanisms of acquired resistance to BET inhibitors in ovarian cancer, we used multiple models to represent different genetic and epigenetic scenarios. Specifically, three ovarian cancer cell lines were selected, namely COV 413B, OVCA420 and SKOV3 which displayed diverse somatic profiles and super-enhancer landscapes as identified with chromatin immunoprecipitation followed by sequencing (Figure 1A; Supplementary Tables 1, 2, 3). We generated BETi-resistant cells by the long-term culture of these three models in escalating concentrations of JQ1 [3]. Consistent with our previous work [13], JQ1 treatment severely reduced parental cell viability (Figure 1B). In contrast, resistant clones maintained stable growth in the presence of 1 μM JQ1 and showed a greater than ten-fold increase in IC50 (Figure 1C). Cross-tolerance was observed between JQ1 and chemically distinct I-BET151 [6], indicating that both compounds might share common resistance mechanisms (Figure 1B and 1C). More importantly, the resistant phenotype was reversible upon culturing cells in JQ1-free media for over four weeks. Indeed, we found that all three JQ1-resistant cell lines gradually restored drug sensitivity (Figure 1D; Supplementary Figure 1A), and of interest, regained similar morphology of parental cells (Supplementary Figure 1B). Taken together, these findings suggested that JQ1-tolerant ovarian cancer cells exhibited reversible resistance to BET inhibition and the underlying mechanisms were most likely non-mutational.
Figure 1.
JQ1-tolerant ovarian cancer cells exhibit reversible resistance to BET inhibition. A. Enhancers in COV 413B, OVCA420, and SKOV3 cell lines ranked by increasing normalized H3K27ac signal (length × density). Dashed gray lines marked cutoffs distinguishing typical from super-enhancers. These three cell lines harbored different somatic mutations as labeled on the top left of each graph. B. JQ1-sensitive and JQ1-resistant cells (COV 413B, OVCA420, and SKOV3) were treated with DMSO, JQ1 (500 nM), or I-BET151 (500 nM) for 10 days. The remaining cells were stained with crystal violet. C. Cell viability of indicated cells treated with various concentrations of JQ1 or I-BET151 for 5 days. Data were presented as mean ± standard deviation of four biological replicates. D. Cell viability of indicated cells treated with various concentrations of JQ1 for 5 days. Data were presented as mean ± standard deviation of four biological replicates.
JQ1-resistant tumor cells retain oncogene addiction to BRD4
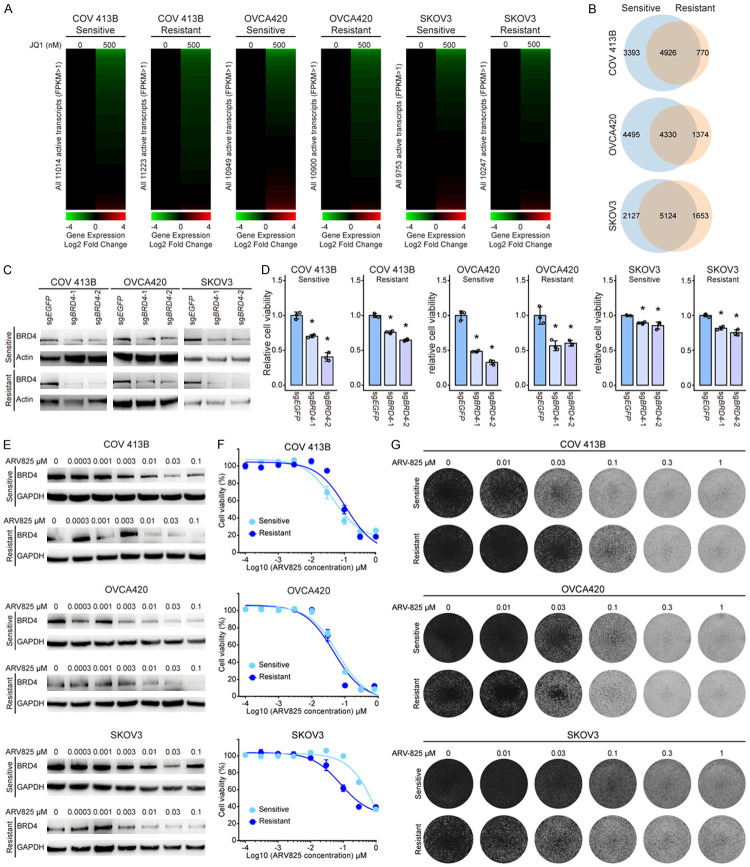
To gain insights into potential molecular underpinnings of the reversible resistance to BET inhibition, we initially probed JQ1-induced differential gene expression by performing RNA sequencing (RNA-seq) of ovarian cancer models. To our surprise, the drastic response of gene transcripts to JQ1 exposure in parental cells was also observed in resistant lines (Figure 2A; Supplementary Tables 4, 5, 6, 7, 8, 9), indicating that BRD4 and its bromodomain remained functionally indispensable for global transcriptional programs. The majority of differentially expressed genes exhibited downregulation upon JQ1 treatment, with considerable overlap between sensitive and resistant clones. Nevertheless, the diversity of significantly altered genes was noticeably reduced in all three resistant cell lines relative to their respective parental cell lines (Figure 2B). These results suggested that albeit to a less extent, BRD4 might be still required in JQ1-resistant cells. Indeed, we observed a significant decrease of cell viability upon BRD4 knockout using the clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 system (Figure 2C, 2D). Consistently, BRD4 depletion with ARV825 (Figure 2E), a BET protein proteolysis-targeting chimera (PROTAC) [34], effectively inhibited resistant cell growth (Figure 2F) in a dose-dependent manner (Figure 2G). Taken together, JQ1-resistant tumor cells retained oncogene addiction to BRD4.
Figure 2.
JQ1-resistant cells retain oncogene addiction to BRD4. A. Heatmaps of global gene expression values in COV 413B, OVCA420, and SKOV3 cells that were treated with JQ1 (500 nM for 6 hours) versus DMSO control. B. Venn diagram showing the overlap of JQ1-induced differentially expressed transcripts between sensitive and resistant cells. C. Immunoblotting of BRD4 in BRD4-depleted cells using the CRISPR-Cas9 system. D. Relative cell viability of BRD4 knockout cells as compared to control cells (*P < 0.05, unpaired Student’s t test). E. Immunoblotting of BRD4 in cells treated with various concentrations of ARV825. F. Cell viability of indicated cells treated with ARV825 for 5 days. Data were presented as mean ± standard deviation of four biological replicates. G. Indicated cells were treated with ARV825 for 5 days and the remaining cells were stained with crystal violet.
JQ1-resistant ovarian cancer cells display excessive chromatin-bound BRD4
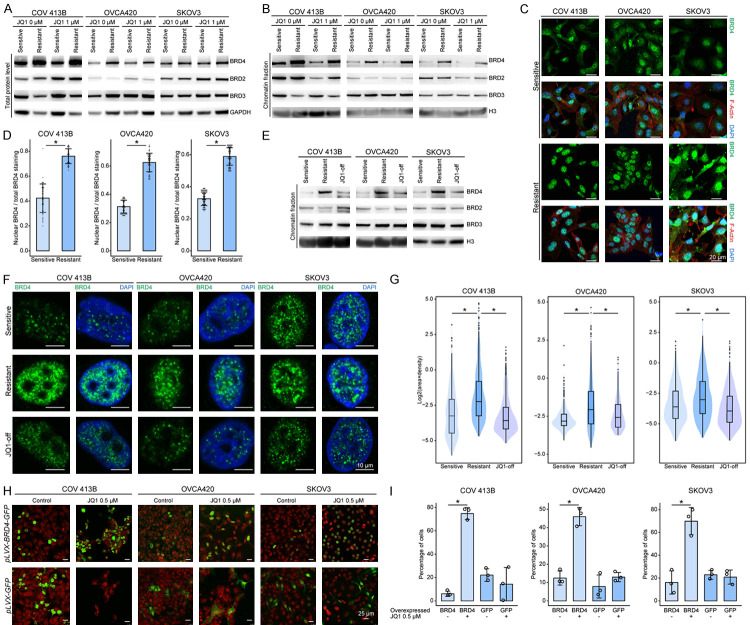
The coexisting JQ1 resistance and BRD4 addiction in ovarian cancer seemed counterintuitive and prompted us to hypothesize that BRD4 might be upregulated in resistant cells to counteract the effects of BET inhibitors. As expected, we found that BRD4 levels (Figure 3A), most obviously the chromatin-associated protein (Figure 3B), were consistently elevated in all three resistant cell lines regardless of JQ1 treatment. Immunofluorescence assay also demonstrated a relative enrichment of BRD4 signals within the nuclei of resistant cells (Figure 3C), whereas in sensitive cells, a substantial proportion of BRD4 molecules resided in the cytoplasm (Figure 3D). In line with these results, restoration of drug sensitivity upon taking off JQ1 was accompanied by decreased chromatin-bound BRD4 protein (Figure 3E). More importantly, reminiscent of recent data showing that BET proteins and other coactivators formed condensates at super-enhancers via liquid-liquid phase separation [35], BRD4 was visualized as discrete puncta within the nuclei of ovarian cancer cells (Figure 3F), and brighter and larger spots accumulated in resistant cells relative to sensitive controls (Figure 3G), indicating that JQ1-tolerant cells were likely dependent on more condensed phase-separated BRD4 to overcome drug inhibition. To support this notion, we treated JQ1-resistant cells with 1,6-hexanediol, a compound known to diminish liquid-like clusters [35]. Following the disruption of BRD4 puncta by pre-exposure to 1,6-hexanediol, JQ1 sensitivity was restored in drug-tolerant cells (Supplementary Figure 2A, 2B). To validate the role of BRD4, we tested whether exogenous BRD4 expression was sufficient to induce JQ1 resistance using a GFP-tagging assay. Specifically, ovarian cancer cells overexpressing BRD4-GFP chimera or GFP alone (Supplementary Figure 2C, 2D) were mixed with parental cells at a starting fraction of approximately 10%. We observed that the proportion of GFP-positive cells remained roughly stable in DMSO (Figure 3H). However, upon JQ1 treatment, the percentage of BRD4-GFP, but not GFP, cell population increased significantly (Figure 3I). Hence, we concluded that high amount of chromatin-bound BRD4 formed phase-separated clusters and conferred BETi resistance in ovarian cancer.
Figure 3.
JQ1-resistant ovarian cancer cells display excessive chromatin-bound BRD4. A. Western blot analysis of total BRD4, BRD2 and BRD3 in sensitive and resistant cells treated with DMSO or JQ1 (1 μM). B. Western blot analysis of chromatin-bound BRD4, BRD2 and BRD3 in sensitive and resistant cells treated with DMSO or JQ1 (1 μM). C. Immunofluorescence imaging (40×) of BRD4 in sensitive and resistant cells. The fluorescence signal of BRD4 was shown alone or merged with fibrous actin and DAPI staining. Scale bar = 20 μm. D. Bar charts with dot plots represented the ratio of nuclear BRD4 to total BRD4 calculated by fluorescence intensity in 50 sensitive and resistant cells. E. Western blot analysis showed the reduced levels of chromatin-bound BRD4 upon JQ1 withdrawal. F. Representative images (63×) of BRD4 puncta in JQ1-sensitive, JQ1-resistant, and JQ1-withdrawn ovarian cancer cells. Scale bar = 10 μm. G. Violin plots represented the quantification of BRD4 puncta in JQ1-sensitive, JQ1-resistant, and JQ1-withdrawn cells (~20 cells per group). H. Representative images (20×) were shown at 7 days after initial cell seeding of approximately 10% BRD4-GFP- or GFP-expressing tumor cells with DMSO or JQ1 treatment. Nuclear DNA was labeled with TO-PRO-3. Scale bar = 25 μm. I. Bar graphs showing the percentage of GFP-positive cells (3 images per group, *P < 0.05, unpaired Student’s t test).
Epigenetic heterogeneity underlies drug resistance to BET inhibition
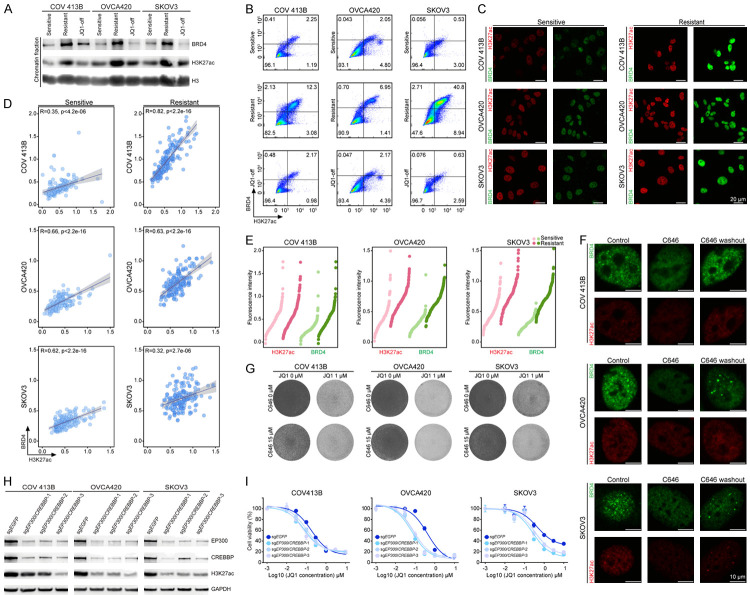
Since BRD4 gene copy number (Supplementary Figure 3A) and mRNA level (Supplementary Figure 3B) were not necessarily elevated, we sought to explore the alternative mechanisms underlying increased BRD4 accumulation and recruitment to chromatin in the nuclei of JQ1-resistant cells. BET family proteins are epigenetic readers that can bind to distal enhancers via acetylated histone H3 lysine 27 (H3K27ac) [16,36]. Intriguingly, resistant cells exhibited a consistent increase in nuclear H3K27ac abundance along with BRD4 protein enrichment (Figure 4A). Flow cytometry (FACS) analysis confirmed this finding and showed that higher expression of H3K27ac and BRD4 were detected in all three resistant models (Figure 4B; Supplementary Figure 4). We performed immunofluorescence staining of ovarian cancer cells (Figure 4C), and found that H3K27ac and BRD4 fluorescent signals (normalized to DAPI) significantly correlated with each other at the single-cell level (Figure 4D). Notably, ovarian tumor cells displayed considerable diversity of these two proteins even at baseline, and their relative distribution in dot plots shifted upwards in resistant cells (Figure 4E). Hence, pre-existing epigenetic heterogeneity might lead to differential JQ1 responses and positive selection for functionally robust subclones under the therapeutic pressure. To corroborate this point, we used histone acetyltransferase inhibitor C646 to eliminate H3K27ac heterogeneity. Following C646 treatment, we observed an obvious reduction of H3K27ac and the disappearance of BRD4 puncta (Figure 4F; Supplementary Figure 5A). When we washed out the compounds, BRD4 phase separation was largely recovered, reinforcing the specific effects of this epigenetic drug (Figure 4F). More importantly, H3K27ac disequilibrium upon inhibitor administration substantially narrowed down BRD4 distribution (Supplementary Figure 5B) and consequently sensitized ovarian cancer cells to JQ1 treatment (Figure 4G). Consistently, we observed that knocking out EP300 and CREBBP using the CRISPR-Cas9 system (Figure 4H) could disperse phase-separated puncta and render ovarian cancer cells more sensitive to JQ1 (Figure 4I; Supplementary Figure 5C). These data highlighted the intrinsic heterogeneity of epigenetic machinery in mediating resistance to chromatin-targeted therapies.
Figure 4.
Epigenetic heterogeneity underlies drug resistance to BET inhibition. A. Western blot analysis showed the increased levels of H3K27ac and chromatin-bound BRD4 in JQ1-resistant cells, and the reduced levels of H3K27ac and chromatin-bound BRD4 upon JQ1 withdrawal. B. Sensitive cells and resistant cells were labeled with BRD4 and H3K27ac antibodies and analyzed by flow cytometry (FACS) analysis. FACS showed a correlation between H3K27ac and BRD4 signals and an enrichment of resistant cells with higher expression of H3K27ac and BRD4. C. Immunofluorescence imaging (40×) of H3K27ac and BRD4 in sensitive and resistant cells. Scale bar = 20 μm. D. Fluorescent signals of H3K27ac and BRD4 (50 cells per group) were quantified and normalized to DAPI. The data were presented in scatter plots and the corresponding regression lines reflected the positive associations between H3K27ac and BRD4. R indicated the Pearson’s correlation coefficient. E. Fluorescent signals of H3K27ac and BRD4 were quantified and presented in dot charts ranked by increasing levels of H3K27ac or BRD4. F. Immunofluorescence imaging (63×) of H3K27ac and BRD4 in sensitive and resistant cells upon treatment of histone acetyltransferase inhibitor C646 (15 μM). Scale bar = 20 μm. G. Cells were treated with JQ1 (0.5 μM) and C646 (15 μM) as indicated. The remaining cells were stained with crystal violet. H. EP300 and CREBBP were depleted using the CRISPR-Cas9 system. Western blot analysis showed knockout efficiency and reduced H3K27ac. I. Cell viability of indicated cells treated with various concentrations of JQ1 for 5 days. Data were presented as mean ± standard deviation of four biological replicates.
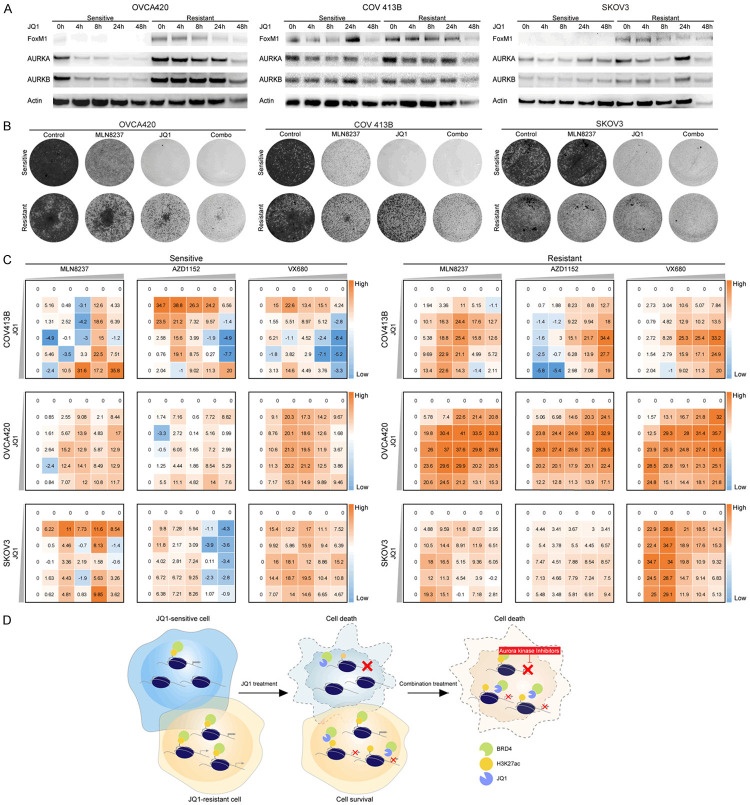
Combined BRD4 and Aurora kinases inhibition eradicates JQ1-resistant ovarian cancer cells
Previous work by our laboratory identified Forkhead box protein M1 (FoxM1) as the functional target of BET inhibitors in ovarian cancer [13]. Based on the aforementioned nongenetic mechanism of acquired resistance, we reasoned that FoxM1 might display distinct kinetics during JQ1 exposure. Indeed, the immunoblots revealed that all three resistant lines exhibited higher expression levels of FoxM1 and its key transcriptional targets Aurora kinases at baseline and upon treatment (Figure 5A). Remarkably, the combination of JQ1 and MLN8237 [37,38], the most clinically advanced Aurora kinase inhibitor, demonstrated a synergistic reduction in resistant cell growth (Figure 5B). The synergy was confirmed by crystal violet staining and Bliss independence assay using two other Aurora kinase agents, AZD1152 and VX680 (Figure 5C; Supplementary Figure 6). Therefore, combined BRD4 and Aurora kinases inhibitors acted synergistically in eradicating JQ1-resistant ovarian cancer cells.
Figure 5.
Combined BRD4 and Aurora kinases inhibition eradicates JQ1-resistant ovarian cancer cells. A. Western blot analysis of FoxM1, AURKA and AURKB in sensitive and resistant cells treated with JQ1 (1 μM). B. Cells were treated with JQ1 (1 μM) and Aurora kinase inhibitor MLN8237 (30 nM) as indicated. The remaining cells were stained with crystal violet. C. Heatmaps of bliss synergy scores demonstrated synergistic activities of JQ1 and Aurora kinase inhibitors in JQ1-sensitive and JQ1-resistant cells. A higher score represented a more potent synergistic efficacy. D. A schematic summary of the study, showing that BETi resistance might emerge from epigenetic heterogeneity and vertical inhibition of BRD4 and its downstream Aurora kinases could overcome drug tolerance in ovarian cancer.
Discussion
Recent investigations have unequivocally revealed transcriptional addiction of ovarian cancer [33,39,40], and nominated BET inhibitors as new transcription-targeted therapeutic candidates. To realize their clinical promise, the molecular determinants of drug sensitivity need to be rigorously elucidated. Here, we documented that BETi resistant clones might emerge from pre-existing cell subsets that transiently acquired increased H3K27ac and chromatin-bound BRD4. As a result, vertical inhibition of BRD4 and its downstream Aurora kinases could deliver synergistic benefit, enabling effective treatment of ovarian cancer (Figure 5D). These findings have important implications for interpreting epigenetic heterogeneity and clonal selection as an unprecedented etiology of BETi resistance and providing rational combination approaches to improve therapeutic efficacy in future studies.
Triggered by early promising clinical trials of multiple BET inhibitors, recent work has started to evaluate the possible mechanisms of anticipated drug resistance using preclinical models. Different trajectories of acquired BETi resistance have been proposed in various cancer types [22,32,41-45]. For example, it has been demonstrated that WNT pathway activation compensates for BRD4 inhibition to drive active transcriptional programs in leukemia cells [24,25]. On the other hand, triple-negative breast cancer (TNBC) is reported to gain hyper-phosphorylated BRD4, which associates with MED1 and supports gene transcription in a bromodomain-independent manner [31]. Remarkably, we identified a new molecular underpinning for reversible BETi resistance in ovarian cancer, which simply involved more abundant chromatin-bound BRD4. In contrast to the dispensable roles of bromodomain in leukemia and TNBC, BRD4 and its bromodomain remained functionally important in JQ1-tolerant ovarian cancer cells. Additionally, it was worth noting that BRD4 molecules in the nuclei of resistant cells formed larger phase-separated condensates relative to sensitive controls. Presumably, the enlarged clusters contained higher levels of BRD4 protein, ensuring the robust expression of pivotal genes to survive the therapeutic attack of BET inhibitors [46,47]. Alternatively, it was lately reported that antineoplastic drugs including JQ1 could partition into transcriptional condensates, raising the possibility that accumulated BRD4 expanded the volumes of liquid-like compartments in which available JQ1 compound might be more diluted and consequently less efficient at displacing BRD4 from super-enhancers [48]. These two potential mechanisms are not mutually exclusive and future studies are required to unravel the regulatory apparatus of BRD4 droplet size and the behavior of BET inhibitors within BRD4 condensates.
We further uncovered that nuclear BRD4 enrichment correlated with elevated H3K27ac marks which were also reversible and attributable to stochastic cellular levels. Hence, we proposed a new pattern for BETi resistance that dynamic regulation of epigenetic heterogeneity provided a reservoir of tumor cells surviving potentially lethal treatment. This model was supported by a serial of evidence. First, H3K27ac along with nuclear BRD4 varied across individual cells at baseline. Second, the diversity distribution of these two proteins allowed for a therapeutic selection of fitness-enhanced cells, leading to the establishment of a new homeostatic state. Third, reducing the variation in basal H3K27ac expression by histone acetyltransferase inhibitors or gene editing could dissipate phase-separated BRD4 droplets and preclude the emergence of the resistant phenotype. Taken together, our findings went beyond the well-defined role of resistance-conferring genetic heterogeneity and implicated an analogous mechanism of non-mutational epigenetic heterogeneity in acquired drug tolerance to chromatin-targeted therapeutics [49-51].
In light of the persistent BRD4 dependence of JQ1-tolerant cells, this research offered mechanism-based combination strategies to overcome BETi resistance in ovarian cancer. Specifically, we identified that pharmacological perturbation of key downstream targets via Aurora kinase inhibitors, which have reached phase 3 trials [52-54], sensitized resistant cells to BRD4-targeted treatment. Although early clinical experience suggests that single BETi agents can be given safely [17], toxicity liabilities must be carefully considered when they are concurrently administrated with other therapies. The proposed approach here substantiates the recently described concept of vertical pathway inhibition, which might elicit powerful anti-tumor synergy even at suboptimal doses with minimal side effects [55,56]. Furthermore, in addition to reactively treating resistant disease, we envision that preventive upfront polytherapy might be more effective to forestall the onset of drug tolerance in patients. Our results call for delicately designed clinical studies testing the combination of BET protein and Aurora kinase inhibitors in advanced ovarian cancer and potentially other human malignancies.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81672714 and 81922047 to G Zhuang; 81974454 to W Di; 81802585 to X Yin; 81802809 to MC Cai; 81802734 to P Ma), Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (20161313 to G Zhuang), Shanghai Sailing Program (18YF1413200 to P Ma), the Incubating Program for Clinical Research and Innovation of Renji Hospital (PY2018-IIC-04 to P Ma), the grants from Shanghai Key Laboratory of Gynecologic Oncology (FKZL-2018-02 to P Ma), School of Medicine, Shanghai Jiao Tong University (YG2017MS54 to Z Gu), Shanghai Natural Science Foundation (20ZR1433100 to X Yin), Shanghai Shenkang Hospital Development Center (SHDC2020CR3057B to X Yin), Shanghai Collaborative Innovation Center for Translational Medicine (TM202004 to X Yin), State Key Laboratory of Oncogenes and Related Genes (SB201802 to Z Zhang), Shanghai Health Committee (201940251 to Z Zhang).
Disclosure of conflict of interest
None.
Supplementary Figures 1-6
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3
Supplementary Table 4
Supplementary Table 5
Supplementary Table 6
Supplementary Table 7
Supplementary Table 8
Supplementary Table 9
References
- 1.Lasko LM, Jakob CG, Edalji RP, Qiu W, Montgomery D, Digiammarino EL, Hansen TM, Risi RM, Frey R, Manaves V, Shaw B, Algire M, Hessler P, Lam LT, Uziel T, Faivre E, Ferguson D, Buchanan FG, Martin RL, Torrent M, Chiang GG, Karukurichi K, Langston JW, Weinert BT, Choudhary C, de Vries P, Van Drie JH, McElligott D, Kesicki E, Marmorstein R, Sun C, Cole PA, Rosenberg SH, Michaelides MR, Lai A, Bromberg KD. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature. 2017;550:128–132. doi: 10.1038/nature24028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogiwara H, Sasaki M, Mitachi T, Oike T, Higuchi S, Tominaga Y, Kohno T. Targeting p300 addiction in CBP-deficient cancers causes synthetic lethality by apoptotic cell death due to abrogation of MYC expression. Cancer Discov. 2016;6:430–445. doi: 10.1158/2159-8290.CD-15-0754. [DOI] [PubMed] [Google Scholar]
- 3.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohammad HP, Barbash O, Creasy CL. Targeting epigenetic modifications in cancer therapy: erasing the roadmap to cancer. Nat Med. 2019;25:403–418. doi: 10.1038/s41591-019-0376-8. [DOI] [PubMed] [Google Scholar]
- 5.Kelly AD, Issa JJ. The promise of epigenetic therapy: reprogramming the cancer epigenome. Curr Opin Genet Dev. 2017;42:68–77. doi: 10.1016/j.gde.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 6.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, Huthmacher C, Gudgin E, Lugo D, Beinke S, Chapman TD, Roberts EJ, Soden PE, Auger KR, Mirguet O, Doehner K, Delwel R, Burnett AK, Jeffrey P, Drewes G, Lee K, Huntly BJ, Kouzarides T. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yokoyama Y, Zhu H, Lee JH, Kossenkov AV, Wu SY, Wickramasinghe JM, Yin X, Palozola KC, Gardini A, Showe LC, Zaret KS, Liu Q, Speicher D, Conejo-Garcia JR, Bradner JE, Zhang Z, Sood AK, Ordog T, Bitler BG, Zhang R. BET inhibitors suppress ALDH activity by targeting ALDH1A1 super-enhancer in ovarian cancer. Cancer Res. 2016;76:6320–6330. doi: 10.1158/0008-5472.CAN-16-0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karakashev S, Zhu H, Wu S, Yokoyama Y, Bitler BG, Park PH, Lee JH, Kossenkov AV, Gaonkar KS, Yan H, Drapkin R. CARM1-expressing ovarian cancer depends on the histone methyltransferase EZH2 activity. Nat Commun. 2018;9:631. doi: 10.1038/s41467-018-03031-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bitler BG, Aird KM, Garipov A, Li H, Amatangelo M, Kossenkov AV, Schultz DC, Liu Q, Shih Ie M, Conejo-Garcia JR, Speicher DW, Zhang R. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med. 2015;21:231–238. doi: 10.1038/nm.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dizon DS, Blessing JA, Penson RT, Drake RD, Walker JL, Johnston CM, Disilvestro PA, Fader AN. A phase II evaluation of belinostat and carboplatin in the treatment of recurrent or persistent platinum-resistant ovarian, fallopian tube, or primary peritoneal carcinoma: a gynecologic oncology group study. Gynecol Oncol. 2012;125:367–371. doi: 10.1016/j.ygyno.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bazzaro M, Lin Z, Santillan A, Lee MK, Wang MC, Chan KC, Bristow RE, Mazitschek R, Bradner J, Roden RB. Ubiquitin proteasome system stress underlies synergistic killing of ovarian cancer cells by bortezomib and a novel HDAC6 inhibitor. Clin Cancer Res. 2008;14:7340–7347. doi: 10.1158/1078-0432.CCR-08-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z, Ma P, Jing Y, Yan Y, Cai MC, Zhang M, Zhang S, Peng H, Ji ZL, Di W, Gu Z, Gao WQ, Zhuang G. BET bromodomain inhibition as a therapeutic strategy in ovarian cancer by downregulating FoxM1. Theranostics. 2016;6:219–230. doi: 10.7150/thno.13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baratta MG, Schinzel AC, Zwang Y, Bandopadhayay P, Bowman-Colin C, Kutt J, Curtis J, Piao H, Wong LC, Kung AL, Beroukhim R, Bradner JE, Drapkin R, Hahn WC, Liu JF, Livingston DM. An in-tumor genetic screen reveals that the BET bromodomain protein, BRD4, is a potential therapeutic target in ovarian carcinoma. Proc Natl Acad Sci U S A. 2015;112:232–237. doi: 10.1073/pnas.1422165112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mazur PK, Herner A, Mello SS, Wirth M, Hausmann S, Sanchez-Rivera FJ, Lofgren SM, Kuschma T, Hahn SA, Vangala D, Trajkovic-Arsic M, Gupta A, Heid I, Noel PB, Braren R, Erkan M, Kleeff J, Sipos B, Sayles LC, Heikenwalder M, Hessmann E, Ellenrieder V, Esposito I, Jacks T, Bradner JE, Khatri P, Sweet-Cordero EA, Attardi LD, Schmid RM, Schneider G, Sage J, Siveke JT. Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nat Med. 2015;21:1163–1171. doi: 10.1038/nm.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falchook G, Rosen S, LoRusso P, Watts J, Gupta S, Coombs CC, Talpaz M, Kurzrock R, Mita M, Cassaday R, Harb W, Peguero J, Smith DC, Piha-Paul SA, Szmulewitz R, Noel MS, Yeleswaram S, Liu P, Switzky J, Zhou G, Zheng F, Mehta A. Development of 2 bromodomain and extraterminal inhibitors with distinct pharmacokinetic and pharmacodynamic profiles for the treatment of advanced malignancies. Clin Cancer Res. 2020;26:1247–1257. doi: 10.1158/1078-0432.CCR-18-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang P, Wang D, Zhao Y, Ren S, Gao K, Ye Z, Wang S, Pan CW, Zhu Y, Yan Y, Yang Y, Wu D, He Y, Zhang J, Lu D, Liu X, Yu L, Zhao S, Li Y, Lin D, Wang Y, Wang L, Chen Y, Sun Y, Wang C, Huang H. Intrinsic BET inhibitor resistance in SPOP-mutated prostate cancer is mediated by BET protein stabilization and AKT-mTORC1 activation. Nat Med. 2017;23:1055–1062. doi: 10.1038/nm.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janouskova H, El Tekle G, Bellini E, Udeshi ND, Rinaldi A, Ulbricht A, Bernasocchi T, Civenni G, Losa M, Svinkina T, Bielski CM, Kryukov GV, Cascione L, Napoli S, Enchev RI, Mutch DG, Carney ME, Berchuck A, Winterhoff BJN, Broaddus RR, Schraml P, Moch H, Bertoni F, Catapano CV, Peter M, Carr SA, Garraway LA, Wild PJ, Theurillat JP. Opposing effects of cancer-type-specific SPOP mutants on BET protein degradation and sensitivity to BET inhibitors. Nat Med. 2017;23:1046–1054. doi: 10.1038/nm.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ambrosini G, Do C, Tycko B, Realubit RB, Karan C, Musi E, Carvajal RD, Chua V, Aplin AE, Schwartz GK. Inhibition of NF-kappaB-dependent signaling enhances sensitivity and overcomes resistance to BET inhibition in uveal melanoma. Cancer Res. 2019;79:2415–2425. doi: 10.1158/0008-5472.CAN-18-3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bandopadhayay P, Piccioni F, O’Rourke R, Ho P, Gonzalez EM, Buchan G, Qian K, Gionet G, Girard E, Coxon M, Rees MG, Brenan L, Dubois F, Shapira O, Greenwald NF, Pages M, Balboni Iniguez A, Paolella BR, Meng A, Sinai C, Roti G, Dharia NV, Creech A, Tanenbaum B, Khadka P, Tracy A, Tiv HL, Hong AL, Coy S, Rashid R, Lin JR, Cowley GS, Lam FC, Goodale A, Lee Y, Schoolcraft K, Vazquez F, Hahn WC, Tsherniak A, Bradner JE, Yaffe MB, Milde T, Pfister SM, Qi J, Schenone M, Carr SA, Ligon KL, Kieran MW, Santagata S, Olson JM, Gokhale PC, Jaffe JD, Root DE, Stegmaier K, Johannessen CM, Beroukhim R. Neuronal differentiation and cell-cycle programs mediate response to BET-bromodomain inhibition in MYC-driven medulloblastoma. Nat Commun. 2019;10:2400. doi: 10.1038/s41467-019-10307-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo L, Li J, Zeng H, Guzman AG, Li T, Lee M, Zhou Y, Goodell MA, Stephan C, Davies PJA, Dawson MA, Sun D, Huang Y. A combination strategy targeting enhancer plasticity exerts synergistic lethality against BETi-resistant leukemia cells. Nat Commun. 2020;11:740. doi: 10.1038/s41467-020-14604-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai X, Gan W, Li X, Wang S, Zhang W, Huang L, Liu S, Zhong Q. Prostate cancer-associated SPOP mutations confer resistance to BET inhibitors through stabilization of BRD4. Nat Med. 2017;23:1063–1071. doi: 10.1038/nm.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fong CY, Gilan O, Lam EY, Rubin AF, Ftouni S, Tyler D, Stanley K, Sinha D, Yeh P, Morison J, Giotopoulos G, Lugo D, Jeffrey P, Lee SC, Carpenter C, Gregory R, Ramsay RG, Lane SW, Abdel-Wahab O, Kouzarides T, Johnstone RW, Dawson SJ, Huntly BJ, Prinjha RK, Papenfuss AT, Dawson MA. BET inhibitor resistance emerges from leukaemia stem cells. Nature. 2015;525:538–542. doi: 10.1038/nature14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rathert P, Roth M, Neumann T, Muerdter F, Roe JS, Muhar M, Deswal S, Cerny-Reiterer S, Peter B, Jude J, Hoffmann T, Boryn LM, Axelsson E, Schweifer N, Tontsch-Grunt U, Dow LE, Gianni D, Pearson M, Valent P, Stark A, Kraut N, Vakoc CR, Zuber J. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature. 2015;525:543–547. doi: 10.1038/nature14898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurimchak AM, Shelton C, Duncan KE, Johnson KJ, Brown J, O’Brien S, Gabbasov R, Fink LS, Li Y, Lounsbury N, Abou-Gharbia M, Childers WE, Connolly DC, Chernoff J, Peterson JR, Duncan JS. Resistance to BET bromodomain inhibitors is mediated by kinome reprogramming in ovarian cancer. Cell Rep. 2016;16:1273–1286. doi: 10.1016/j.celrep.2016.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao S, Maertens O, Cichowski K, Elledge SJ. Genetic modifiers of the BRD4-NUT dependency of NUT midline carcinoma uncovers a synergism between BETis and CDK4/6is. Genes Dev. 2018;32:1188–1200. doi: 10.1101/gad.315648.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chua V, Orloff M, Teh JL, Sugase T, Liao C, Purwin TJ, Lam BQ, Terai M, Ambrosini G, Carvajal RD, Schwartz G, Sato T, Aplin AE. Stromal fibroblast growth factor 2 reduces the efficacy of bromodomain inhibitors in uveal melanoma. EMBO Mol Med. 2019;11:e9081. doi: 10.15252/emmm.201809081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tai F, Gong K, Song K, He Y, Shi J. Enhanced JunD/RSK3 signalling due to loss of BRD4/FOXD3/miR-548d-3p axis determines BET inhibition resistance. Nat Commun. 2020;11:258. doi: 10.1038/s41467-019-14083-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iniguez AB, Alexe G, Wang EJ, Roti G, Patel S, Chen L, Kitara S, Conway A, Robichaud AL, Stolte B, Bandopadhayay P, Goodale A, Pantel S, Lee Y, Cheff DM, Hall MD, Guha R, Davis MI, Menard M, Nasholm N, Weiss WA, Qi J, Beroukhim R, Piccioni F, Johannessen C, Stegmaier K. Resistance to epigenetic-targeted therapy engenders tumor cell vulnerabilities associated with enhancer remodeling. Cancer Cell. 2018;34:922–938. e927. doi: 10.1016/j.ccell.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shu S, Lin CY, He HH, Witwicki RM, Tabassum DP, Roberts JM, Janiszewska M, Huh SJ, Liang Y, Ryan J, Doherty E, Mohammed H, Guo H, Stover DG, Ekram MB, Brown J, D’Santos C, Krop IE, Dillon D, McKeown M, Ott C, Qi J, Ni M, Rao PK, Duarte M, Wu SY, Chiang CM, Anders L, Young RA, Winer E, Letai A, Barry WT, Carroll JS, Long H, Brown M, Liu XS, Meyer CA, Bradner JE, Polyak K. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature. 2016;529:413–417. doi: 10.1038/nature16508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shu S, Wu HJ, Ge JY, Zeid R, Harris IS, Jovanovic B, Murphy K, Wang B, Qiu X, Endress JE, Reyes J, Lim K, Font-Tello A, Syamala S, Xiao T, Reddy Chilamakuri CS, Papachristou EK, D’Santos C, Anand J, Hinohara K, Li W, McDonald TO, Luoma A, Modiste RJ, Nguyen QD, Michel B, Cejas P, Kadoch C, Jaffe JD, Wucherpfennig KW, Qi J, Liu XS, Long H, Brown M, Carroll JS, Brugge JS, Bradner J, Michor F, Polyak K. Synthetic lethal and resistance interactions with BET bromodomain inhibitors in triple-negative breast cancer. Mol Cell. 2020;78:1096–1113. e1098. doi: 10.1016/j.molcel.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z, Peng H, Wang X, Yin X, Ma P, Jing Y, Cai MC, Liu J, Zhang M, Zhang S, Shi K, Gao WQ, Di W, Zhuang G. Preclinical efficacy and molecular mechanism of targeting CDK7-dependent transcriptional addiction in ovarian cancer. Mol Cancer Ther. 2017;16:1739–1750. doi: 10.1158/1535-7163.MCT-17-0078. [DOI] [PubMed] [Google Scholar]
- 34.Saenz DT, Fiskus W, Qian Y, Manshouri T, Rajapakshe K, Raina K, Coleman KG, Crew AP, Shen A, Mill CP, Sun B, Qiu P, Kadia TM, Pemmaraju N, DiNardo C, Kim MS, Nowak AJ, Coarfa C, Crews CM, Verstovsek S, Bhalla KN. Novel BET protein proteolysis-targeting chimera exerts superior lethal activity than bromodomain inhibitor (BETi) against post-myeloproliferative neoplasm secondary (s) AML cells. Leukemia. 2017;31:1951–1961. doi: 10.1038/leu.2016.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, Li CH, Guo YE, Day DS, Schuijers J, Vasile E, Malik S, Hnisz D, Lee TI, Cisse II, Roeder RG, Sharp PA, Chakraborty AK, Young RA. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361:eaar3958. doi: 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Filippakopoulos P. What is the BET on solid tumors? J. Clin. Oncol. 2018;36:3040–3042. doi: 10.1200/JCO.2018.78.8695. [DOI] [PubMed] [Google Scholar]
- 37.Liewer S, Huddleston A. Alisertib: a review of pharmacokinetics, efficacy and toxicity in patients with hematologic malignancies and solid tumors. Expert Opin Investig Drugs. 2018;27:105–112. doi: 10.1080/13543784.2018.1417382. [DOI] [PubMed] [Google Scholar]
- 38.Falchook G, Coleman RL, Roszak A, Behbakht K, Matulonis U, Ray-Coquard I, Sawrycki P, Duska LR, Tew W, Ghamande S, Lesoin A, Schwartz PE, Buscema J, Fabbro M, Lortholary A, Goff B, Kurzrock R, Martin LP, Gray HJ, Fu S, Sheldon-Waniga E, Lin HM, Venkatakrishnan K, Zhou X, Leonard EJ, Schilder RJ. Alisertib in combination with weekly paclitaxel in patients with advanced breast cancer or recurrent ovarian cancer: a randomized clinical trial. JAMA Oncol. 2019;5:e183773. doi: 10.1001/jamaoncol.2018.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bradner JE, Hnisz D, Young RA. Transcriptional addiction in cancer. Cell. 2017;168:629–643. doi: 10.1016/j.cell.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeng M, Kwiatkowski NP, Zhang T, Nabet B. Targeting MYC dependency in ovarian cancer through inhibition of CDK7 and CDK12/13. Elife. 2018;7:e39030. doi: 10.7554/eLife.39030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ge JY, Shu S, Kwon M, Jovanovic B, Murphy K, Gulvady A, Fassl A, Trinh A, Kuang Y, Heavey GA, Luoma A, Paweletz C, Thorner AR, Wucherpfennig KW, Qi J, Brown M, Sicinski P, McDonald TO, Pellman D, Michor F, Polyak K. Acquired resistance to combined BET and CDK4/6 inhibition in triple-negative breast cancer. Nat Commun. 2020;11:2350. doi: 10.1038/s41467-020-16170-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pawar A, Gollavilli PN, Wang S, Asangani IA. Resistance to BET inhibitor leads to alternative therapeutic vulnerabilities in castration-resistant prostate cancer. Cell Rep. 2018;22:2236–2245. doi: 10.1016/j.celrep.2018.02.011. [DOI] [PubMed] [Google Scholar]
- 43.Gobbi G, Donati B, Do Valle IF, Reggiani F, Torricelli F, Remondini D, Castellani G, Ambrosetti DC, Ciarrocchi A, Sancisi V. The Hippo pathway modulates resistance to BET proteins inhibitors in lung cancer cells. Oncogene. 2019;38:6801–6817. doi: 10.1038/s41388-019-0924-1. [DOI] [PubMed] [Google Scholar]
- 44.Cooper JM, Patel AJ, Chen Z, Liao CP, Chen K, Mo J, Wang Y, Le LQ. Overcoming BET inhibitor resistance in malignant peripheral nerve sheath tumors. Clin Cancer Res. 2019;25:3404–3416. doi: 10.1158/1078-0432.CCR-18-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bell CC, Fennell KA, Chan YC, Rambow F, Yeung MM, Vassiliadis D, Lara L, Yeh P, Martelotto LG, Rogiers A, Kremer BE, Barbash O, Mohammad HP, Johanson TM, Burr ML, Dhar A, Karpinich N, Tian L, Tyler DS, MacPherson L, Shi J, Pinnawala N, Yew Fong C, Papenfuss AT, Grimmond SM, Dawson SJ, Allan RS, Kruger RG, Vakoc CR, Goode DL, Naik SH, Gilan O, Lam EYN, Marine JC, Prinjha RK, Dawson MA. Targeting enhancer switching overcomes non-genetic drug resistance in acute myeloid leukaemia. Nat Commun. 2019;10:2723. doi: 10.1038/s41467-019-10652-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A phase separation model for transcriptional control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cho WK, Spille JH. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 2018;361:412–415. doi: 10.1126/science.aar4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klein IA, Boija A. Partitioning of cancer therapeutics in nuclear condensates. Science. 2020;368:1386–1392. doi: 10.1126/science.aaz4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bai X, Fisher DE, Flaherty KT. Cell-state dynamics and therapeutic resistance in melanoma from the perspective of MITF and IFNgamma pathways. Nat Rev Clin Oncol. 2019;16:549–562. doi: 10.1038/s41571-019-0204-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hinohara K, Wu HJ, Vigneau S, McDonald TO, Igarashi KJ, Yamamoto KN, Madsen T, Fassl A, Egri SB, Papanastasiou M, Ding L, Peluffo G, Cohen O, Kales SC, Lal-Nag M, Rai G, Maloney DJ, Jadhav A, Simeonov A, Wagle N, Brown M, Meissner A, Sicinski P, Jaffe JD, Jeselsohn R, Gimelbrant AA, Michor F, Polyak K. KDM5 histone demethylase activity links cellular transcriptomic heterogeneity to therapeutic resistance. Cancer Cell. 2018;34:939–953. e939. doi: 10.1016/j.ccell.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seth S, Li CY, Ho IL, Corti D, Loponte S, Sapio L, Del Poggetto E, Yen EY, Robinson FS, Peoples M, Karpinets T, Deem AK, Kumar T, Song X, Jiang S, Kang Y, Fleming J, Kim M, Zhang J, Maitra A, Heffernan TP, Giuliani V, Genovese G, Futreal A, Draetta GF, Carugo A, Viale A. Pre-existing functional heterogeneity of tumorigenic compartment as the origin of chemoresistance in pancreatic tumors. Cell Rep. 2019;26:1518–1532. e1519. doi: 10.1016/j.celrep.2019.01.048. [DOI] [PubMed] [Google Scholar]
- 52.Shah KN, Bhatt R, Rotow J, Rohrberg J, Olivas V, Wang VE, Hemmati G, Martins MM, Maynard A, Kuhn J, Galeas J, Donnella HJ, Kaushik S, Ku A, Dumont S, Krings G, Haringsma HJ, Robillard L, Simmons AD, Harding TC, McCormick F, Goga A, Blakely CM, Bivona TG, Bandyopadhyay S. Aurora kinase a drives the evolution of resistance to third-generation EGFR inhibitors in lung cancer. Nat Med. 2019;25:111–118. doi: 10.1038/s41591-018-0264-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bertran-Alamillo J, Cattan V, Schoumacher M, Codony-Servat J, Gimenez-Capitan A, Cantero F, Burbridge M, Rodriguez S, Teixido C, Roman R, Castellvi J, Garcia-Roman S, Codony-Servat C, Viteri S, Cardona AF, Karachaliou N, Rosell R, Molina-Vila MA. AURKB as a target in non-small cell lung cancer with acquired resistance to anti-EGFR therapy. Nat Commun. 2019;10:1812. doi: 10.1038/s41467-019-09734-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tischer J, Gergely F. Anti-mitotic therapies in cancer. J Cell Biol. 2019;218:10–11. doi: 10.1083/jcb.201808077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ozkan-Dagliyan I, Diehl JN, George SD, Schaefer A, Papke B, Klotz-Noack K, Waters AM, Goodwin CM, Gautam P, Pierobon M, Peng S, Gilbert TSK, Lin KH, Dagliyan O, Wennerberg K, Petricoin EF 3rd, Tran NL, Bhagwat SV, Tiu RV, Peng SB, Herring LE, Graves LM, Sers C, Wood KC, Cox AD, Der CJ. Low-dose vertical inhibition of the RAF-MEK-ERK cascade causes apoptotic death of KRAS mutant cancers. Cell Rep. 2020;31:107764. doi: 10.1016/j.celrep.2020.107764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fernandes Neto JM, Nadal E, Bosdriesz E, Ooft SN, Farre L, McLean C, Klarenbeek S, Jurgens A, Hagen H, Wang L, Felip E, Martinez-Marti A, Vidal A, Voest E, Wessels LFA, van Tellingen O, Villanueva A, Bernards R. Multiple low dose therapy as an effective strategy to treat EGFR inhibitor-resistant NSCLC tumours. Nat Commun. 2020;11:3157. doi: 10.1038/s41467-020-16952-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.