Summary
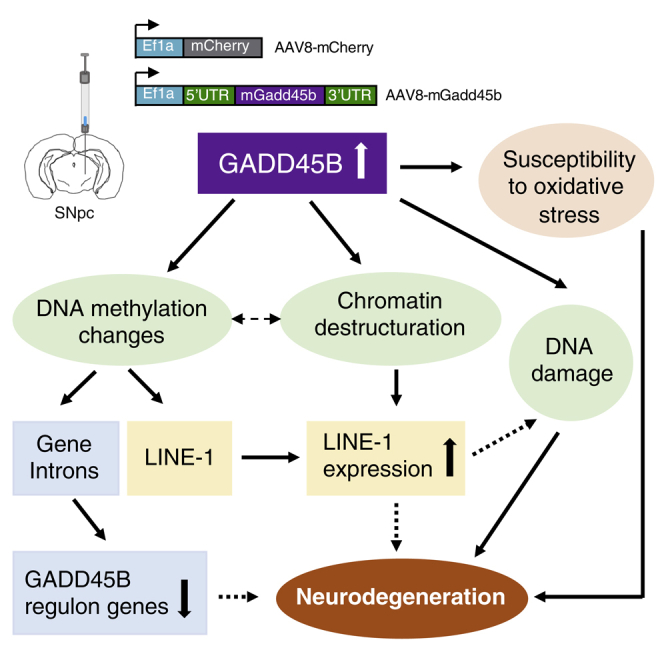
Age is a major risk factor for neurodegenerative diseases like Parkinson’s disease, but few studies have explored the contribution of key hallmarks of aging, namely DNA methylation changes and heterochromatin destructuration, in the neurodegenerative process. Here, we investigated the consequences of viral overexpression of Gadd45b, a multifactorial protein involved in DNA demethylation, in the mouse midbrain. Gadd45b overexpression induced global and stable changes in DNA methylation, particularly in introns of genes related to neuronal functions, as well as on LINE-1 transposable elements. This was paralleled by disorganized heterochromatin, increased DNA damage, and vulnerability to oxidative stress. LINE-1 de-repression, a potential source of DNA damage, preceded Gadd45b-induced neurodegeneration, whereas prolonged Gadd45b expression deregulated expression of genes related to heterochromatin maintenance, DNA methylation, or Parkinson’s disease. Our data indicates that aging-related alterations contribute to dopaminergic neuron degeneration with potential implications for Parkinson’s disease.
Subject areas: molecular biology, epigenetics, molecular neuroscience
Graphical abstract
Highlights
-
•
Altered DNA methylation and chromatin in dopaminergic neurons overexpressing Gadd45b
-
•
GADD45B-induced epigenetic changes affect gene expression and up-regulate LINE-1
-
•
Gadd45b overexpression leads to DNA strand breaks and dopaminergic neuron death
-
•
Dopaminergic neurons overexpressing Gadd45b are more susceptible to oxidative stress
Molecular biology; Epigenetics; Molecular neuroscience
Introduction
Epigenetic marks separate chromatin into actively transcribed euchromatin and repressive heterochromatin domains, and participate in the spatial organization of the genome into highly structured 3D domains (Solovei et al., 2016). These epigenetic signatures include DNA methylation, various post-translational modifications of histones, and attraction forces between different types of genomic repeat elements, including transposable elements (TEs) (Solovei et al., 2016). Changes in chromatin structure regulate DNA accessibility to transcription factors and the physical proximity of enhancers to promoters, thereby regulating gene expression.
It is well established that DNA methylation regulates various important cellular processes during development and cell differentiation. In recent years, however, perturbations of chromatin organization have been linked to the aging process and global changes in DNA methylation are currently the main molecular predictor of chronological age (reviewed in [Benayoun et al., 2015]). Aging-induced epigenetic remodeling of chromatin can impact on genomic stability (Lodato et al., 2018; Pal and Tyler, 2016), and vice versa (Benayoun et al., 2015). Thus, chromatin states and genomic stability are important, interdependent factors associated with the aging process (Burgess et al., 2012). One emerging culprit related to both processes is the un-silencing of transposable elements (TEs) with age. Around half of the human genome is comprised of TEs. The evolutionary most successful TEs in mammals are long interspersed nuclear element-1 (LINE-1 or L1). Mostly fossilized and a few remaining active LINE-1 sequences (around 100 in humans and 3000 in mice) represent around 17% of the human (Lander et al., 2001) and 21% of the mouse genome (Mouse Genome Sequencing Consortium et al., 2002). Young and full-length LINE-1 elements are autonomous retrotransposons, expanding in the genome through a “copy and paste” retrotransposition mechanism and encoding the two necessary proteins, namely ORF1p and ORF2p, required for their mobilization. ORF1p is an RNA binding protein with strong “cis” preference (Martin and Bushman, 2001; Trelogan and Martin, 1995; Wei et al., 2001) and ORF2p encodes an endonuclease and a reverse transcriptase (Feng et al., 1996; Hattori et al., 1986). Several repressive cellular mechanisms, including DNA methylation and heterochromatinization, limit their expression (Pizarro and Cristofari, 2016). When these fail with age, TEs can become derepressed (Van Meter et al., 2014). An increased activity of LINE-1 is associated with genomic instability through the induction of DNA damage (Blaudin de Thé et al., 2018; Gasior et al., 2006; Kazazian and Goodier, 2002; Simon et al., 2019).
How aging and neurodegeneration are linked at the molecular level remains widely unknown. This question is, however, of high relevance as age is the primary risk factor for the most common neurodegenerative diseases (NDs) including Parkinson’s disease (PD) and Alzheimer’s disease (AD) (Collier et al., 2011). Some of the cellular processes defined as the hallmarks of aging (López-Otín et al., 2013) overlap with pathways shown to be dysfunctional in NDs. This is the case for impaired proteostasis, mitochondrial dysfunction, deregulated nutrient sensing, increased oxidative stress, and neuroinflammation (Gan et al., 2018; Hou et al., 2019). Whether two other important nominators of aging, namely perturbations in chromatin organization and genomic instability, are associated with neuronal aging and neurodegeneration has not been unequivocally proven yet.
A cardinal feature of PD is the degeneration of midbrain dopaminergic (mDA) neurons in the substantia nigra pars compacta (SNpc). These neurons project to the striatum and their loss leads to a striatal deficiency in the neurotransmitter dopamine inducing the typical motor symptoms of PD. Decreased global DNA methylation with age in the SNpc has been observed (Fasolino et al., 2017) and DNA methylation changes, mostly on specific genetic risk loci, have been linked to several NDs (Sanchez-Mut et al., 2016) including PD (Labbé et al., 2016). Alterations in histone modifications have also been observed in PD (Park et al., 2016). However, the possible contributions of age-related epigenetic alterations to the pathogenesis of PD and the onset of neurodegeneration have not been demonstrated yet.
Here, we investigated how SNpc mDA neurons react to perturbations of chromatin organization. For this purpose, we overexpressed Gadd45b in the SNpc of wild-type mice using an adeno-associated virus (AAV) vector. GADD45B is a multifunctional protein which coordinates in the nucleus an active DNA demethylation pathway involving cytidine deaminases and DNA glycosylases (Gavin et al., 2015) in association with the base excision repair (BER) pathway (Niehrs and Schäfer, 2012). We chose GADD45B because it has known functions related to DNA demethylation in post-mitotic neurons (Gavin et al., 2015; Ma et al., 2009) and it is highly inducible in conditions of oxidative stress in the SNpc (Rekaik et al., 2015). We show that overexpression of Gadd45b in the SNpc of wild-type mice leads to widespread perturbations of DNA methylation, heterochromatin disorganization, increased vulnerability of mDA neurons to oxidative stress, activation of LINE-1 elements, DNA strand breaks, and neuronal death. Our data reinforces the hypothesis that aging-induced global chromatin disorganization initiates neurodegeneration, possibly via the derepression of LINE-1 elements.
Results
Gadd45b overexpression in the SNpc of wild-type mice leads to early and stable DNA methylation perturbations in introns of genes related to neuronal functions
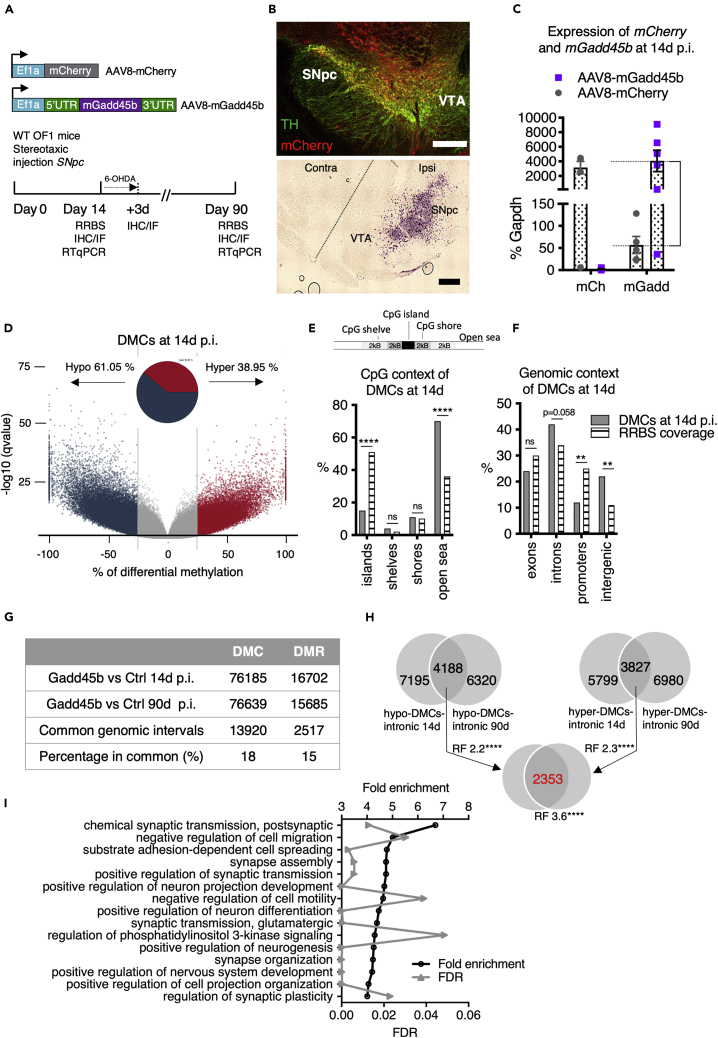
Wild-type mice littermates (6 weeks old) were injected unilaterally in the SNpc using AAV8-mCherry or AAV8-mGadd45b. The animals were sacrificed after 14 or 90 days post-injection (p.i.) and either perfused or dissected as schematized in Figure 1A. The AAV8-mCherry control virus efficiently infected tyrosine hydroxylase positive (TH+) neurons of the SNpc as shown by mCherry expression at 14d p.i (Figure 1B, upper panel). Due to the lack of a good antibody against GADD45B, expression of Gadd45b was verified at 14d p.i. by in situ hybridization (Figure 1B, lower panel) and RT-qPCR after manual micro-dissection of the SNpc at 14d and 90d p.i (Figure 1C). Gadd45b transcripts on the injected ipsilateral side were increased 72-fold on average (4069.46 ± 1463.75; 56.88 ± 19.27; n = 5/6 mice per condition) at 14d p.i. compared to the injected ipsilateral mCherry control and 143-fold (1127 ± 596; 7.89 ± 1.52; n = 5 mice per condition) at 90d p.i. compared to the endogenous transcript levels of the non-injected contralateral side (Figure S1H). Importantly, values of Gadd45b transcript expression in mice injected with AAV8-mGadd45b were comparable to the levels of mCherry control transcripts at 14d p.i. in mice injected with AAV8-mCherry (3145.72 ± 861.74; 2.12 ± 0.87; n = 5/6 mice per condition).
Figure 1.
Gadd45b overexpression in the SNpc of wild-type mice leads to perturbed DNA methylation
(A) Overview of the viral injection protocol into the SNpc of wild-type mice. Wild-type OF1 mice were injected unilaterally in the SNpc with AAV8 virus expressing either mCherry or mouse Gadd45b with its regulatory sequences under the control of the Ef1a promoter. Animals were then sacrificed 14 d or 90 d later to perform RRBS, immunostainings or RT-qPCR.
(B) The AAV8-mCherry virus diffuses within the SNpc and infects mDA neurons. Upper panel: TH and mCherry immunostaining of the SNpc of AAV8-mCherry injected mice showing the diffusion of the virus in the SNpc area while sparing the VTA. Scale bar represents 300 μm. Lower panel: ISH of Gadd45b expression. Gadd45b expression in the SNpc of AAV8-mGadd45b injected mice at 14d p.i. shows that Gadd45b is overexpressed only in the ipsilateral SNpc and not in the ipsilateral VTA, nor on the contralateral side. Scale bar represents 300 μm.
(C) Exogenous Gadd45b RNA is expressed 14d after the injection of AAV8-mGadd45b. Transcript expression analysis by RT-qPCR following manual microdissection of the SNpc of the injected side (upper panel) show a 79-fold increase in mGadd45b transcript level (mCherry = 11.02 ± 4.83; mGadd45b = 869.10 ± 282.80; mean ± SEM) at 14d p.i. (left); n = 6 mice per condition; and a 178 fold increase (7.89 ± 1.52; 1402 ± 683) at 90d p.i. (right); n = 4 mice per condition; ∗p < 0,05; error bars represent SEM. Note that the overexpression of Gadd45b 90d p.i. is shown in Figure S1H.
(D–I) RRBS analysis of differentially methylated CpGs (DMCs) in the SNpc region after injection of AAV8-mGadd45b or AAV8-control.
(D) Volcano plot of differentially methylated CpGs shows widespread perturbations in DNA methylation upon Gadd45b overexpression 14d p.i. 76,185 significantly differentially methylated CpGs were detected with a q-value smaller than or equal to 0.01 and at least 25% difference. The Volcano plot shows the number of regions with changed patterns of methylation between Gadd45b and control samples significantly higher or lower than the 25% difference cut-off and considering a q-value threshold of 0.01. The difference in methylation is reflected in the x-axis while the y axis represents the significance of the difference. Regions that are highly differentially methylated are further to the left and right sides of the plot, while highly significant changes appear higher on the plot. Values on the x- and y-axes are percent methylation differences and negative log10 of the corrected p values, respectively. The pie chart shows the percentage of hyper- and hypomethylated regions.
(E andF) DMCs are located majorly in open sea regions and in introns of genes. DMCs were annotated in relation to the distance to a CpG island (E), as well as based on the genomic regions they are associated with (F). The distributions of DMCs are plotted in gray bars while striped bars represent expected values of RRBS coverage. The enrichment between DMCs (“observed”, light gray) compared to RRBS coverage (=striped bars) for a specific CpG context (left) or genomic location (right) was calculated (two-tailed binominal test, ∗∗p < 0.01).
(G) Comparison of total number, common genetic intervals (minimum overlap of 1 bp) and percentage in common for DMCs and DMRs 14d and 90d p.i..
(H) The “GADD45B-DMC-regulon”. Venn diagrams showing the overlap of genes containing at least one intronic hypomethylated DMC at 14 and 90d (4188 genes, left upper Venn diagram), the overlap of genes containing at least one hypermethylated DMC at 14d and 90d (3827 genes, right upper Venn diagram) and the overlap of both groups (2353 genes, lower Venn diagram) representing the “GADD45B-DMC-regulon”. To test for the statistical significance of these overlaps, we calculated the representation factor (RF) and the corresponding p value. An RF > 1 indicates more overlap than expected of two independent groups. Note that the “GADD45B-DMR-regulon” comprising 447 genes is summarized in the Figure S1E.
(I) GO-analysis of the “GADD45B-DMC-regulon” reveals neuron-related gene categories enriched after Gadd45b overexpression 14d p.i.. Gene ontology analysis (PANTHER version 15.0) and the PANTHER overrepresentation test with the GO-Slim annotation dataset ‘biological process’ identified significantly overrepresented GO categories. The first 15 significantly overrepresented GO categories with the highest fold enrichment are displayed with the fold enrichment on the left y-axis (black points) and the FDR value on the right y-axis (gray points). Note that the RRBS analysis of differentially methylated regions (DMRs) in the SNpc region after injection of AAV8-mGadd45b or AAV8-control is shown in Figures S1A–S1F.
Having verified the efficient overexpression of Gadd45b in the SNpc, we asked whether this perturbs DNA methylation patterns in the SNpc. To this end, we injected wild-type mice with AAV8-mGadd45b (n = 6) or AAV8-mCherry (n = 6) and simultaneously extracted DNA and RNA from manually micro-dissected SNpc biopsies at 14d p.i (Scheme Figure 1A). The DNA of two mice per condition, selected for high expression by RT-qPCR of Gadd45b, was then subjected to reduced representation bisulfite sequencing (RRBS) for DNA methylation analysis. Bioinformatic analysis of RRBS data detected 809117 CpGs that were common between both conditions. Of those, 76,185 individual CpGs were differentially methylated (DMCs) with a q-value smaller than, or equal to, 0.01 and at least 25% difference. Using a window and step size of 1000 bp, differentially methylated regions (DMRs) were defined and 16,702 regions passed the defined significance threshold. A majority of DMCs and DMRs were hypomethylated throughout time, located in open sea regions (defined as regions outside CpG islands, CpG shores, or CpG shelves) and localized in gene bodies, particularly introns. The Volcano plot in Figure 1D illustrates the DMC methylation pattern at 14d p.i. Upon Gadd45b overexpression, 46,508 DMCs (61.05%) were hypomethylated and 29,677 DMCs (38.95%) were hypermethylated. The percentage of hypo- and hypermethylated regions per chromosome was similar (Figure S1G). We then interrogated DMCs in relation to their distance to a CpG island and found 69.73% to be overlapping with open sea regions (Figure 1E), defined as regions outside of a known CpG island, CpG shore (2000 bp flanking the CpG island) or CpG shelves (2000 bp flanking the CpG shores). Analysis of the genomic context revealed that 41.36% of DMCs were located in introns (Figure 1F), followed by 22.3% in the intergenic space, 23.6% in exons, and 12.5% in promoter regions. To understand if this distribution reflects the typical CpG distribution expected in an RRBS experiment, we used the annotation of common CpGs obtained from a set of 16 murine wild-type samples processed by RRBS and displayed the expected compared to the observed localization of CpGs detected by RRBS with respect to the CpG context (Figure 1E) and genomic context (Figure 1F). Comparing observed values with expected, the most prominent enrichment of DMCs upon Gadd45b overexpression were observed in open sea regions (two-tailed binominal test, p < 0.0001) and in introns (two-tailed binominal test, p = 0.058) and intergenic regions (two-tailed binominal test, p < 0.0017). This analysis indicates early and widespread methylation changes primarily on open sea CpGs in introns and intergenic regions upon Gadd45b overexpression.
In order to understand the long-term changes in methylation patterns induced by Gadd45b overexpression, we extracted DNA from mice 90 days after injection of AAV8-mGadd45b or AAV8-mCherry control. Analysis revealed a similar distribution of hypo- and hypermethylated DMRs at 14d p.i (Figure S1A). and at 90d p.i (Figure S1C). The genomic context of DMCs and DMRs, both at 14d p.i (Figure S1B).and at 90d p.i (Figure S1D). was very similar as well, the majority of differential methylation concerning open sea regions and gene bodies, particularly introns (summarized in Table 1). The overall numbers of DMRs and DMCs with AAV8-mGadd45b also resembled that at 14d p.i., but common DMRs or DMCs examination revealed an overlap of only 15% of DMRs (2517) and 18% of DMCs (13,920) (Figure 1G). While this indicates that the specific regions with methylation changes induced by Gadd45b overexpression are not stable over time, the general location in open sea regions and in introns of genes of DMRs and DMCs is maintained as a specific and stable footprint of Gadd45b overexpression, which we termed “GADD45B-regulon”. Furthermore, there was an overlap of more than half of the genes containing intronic hypomethylated or hypermethylated DMCs at 14d and at 90d (Figure 1H). In order to quantify the significance of the overlaps between genes containing hypo- or hypermethylated DMCs at 14d and 90d, we used the calculation of the representation factor, which is the number of overlapping genes divided by the expected number of overlapping genes drawn from two independent groups. We used the MGI international database resource for the laboratory mouse estimate of 24,241 coding genes as the total number of mouse genes. Using this statistical tool, we found that the overlaps of the groups of genes is statistically significant (RF > 1 indicates more overlap than expected, Figure 1H). The extent of overlap was similar for intronic DMRs (hypoDMRs at 14d: 3937 genes; hypoDMRs at 90d: 3467 genes, overlap (1712 common genes); Figure S1E). Of those common genes, 2353 genes contained at least one intronic hypo- and one intronic hypermethylated DMC (Figures 1H) and 447 genes at least one intronic hypo- and one intronic hypermethylated DMR at both 14d and 90d p.i (Figure S1E). Thus, Gadd45b overexpression induces stable changes in methylation patterns in introns of genes.
Table 1.
Summary of the differential methylation analysis comparing Gadd45b overexpression to mCherry control in the SNpc of wild-type mice
| 14d p.i. |
90d p.i. |
|||
|---|---|---|---|---|
| DMCs | DMRs | DMCs | DMRs | |
| Gadd45b/mCherry | ||||
| Total number | 76,185 | 16,702 | 76,639 | 15,685 |
| Hypomethylated (in %) | 61,05 | 64,57 | 54,50 | 56,35 |
| Distance to CpG island (in %) | ||||
| Islands | 15,34 | 1,50 | 16,03 | 1,30 |
| Shelves | 4,23 | 6,30 | 4,18 | 6,60 |
| Shores | 10,71 | 7,60 | 10,73 | 7,60 |
| Open sea | 69,73 | 84,60 | 69,06 | 84,60 |
| Association with genomic regions (in %) | ||||
| Exons | 23,60 | 19,22 | 24,00 | 19,06 |
| Introns | 41,60 | 45,39 | 41,40 | 45,03 |
| Promoters | 12,50 | 8,57 | 12,80 | 9,03 |
| Intergenic | 22,30 | 26,83 | 21,90 | 26,88 |
The results of the RRBS analysis prompted us to identify functional categories associated with this GADD45B-induced methylation footprint corresponding to the 2353 genes stably containing DMCs over time (Figure 1H). We used Gene ontology analysis (PANTHER version 15.0; http://pantherdb.org) and the PANTHER overrepresentation test with the GO-Slim annotation dataset ‘biological process’. 145 GO categories were significantly overrepresented with a fold change >2 and an FDR <0.05. The first 15 significantly overrepresented GO categories are displayed in Figure 1I. Of those, 10 GO categories are explicitly related to neuronal functions with two prevailing categories, namely synaptic function and organization, and neurodevelopment and neurogenesis. This is very similar to what we found for DMRs (Figure S1F) and suggests that Gadd45b is involved in the specific regulation of gene body methylation of neuron-related genes.
Gadd45b overexpression leads to heterochromatin disorganization in mDA neurons
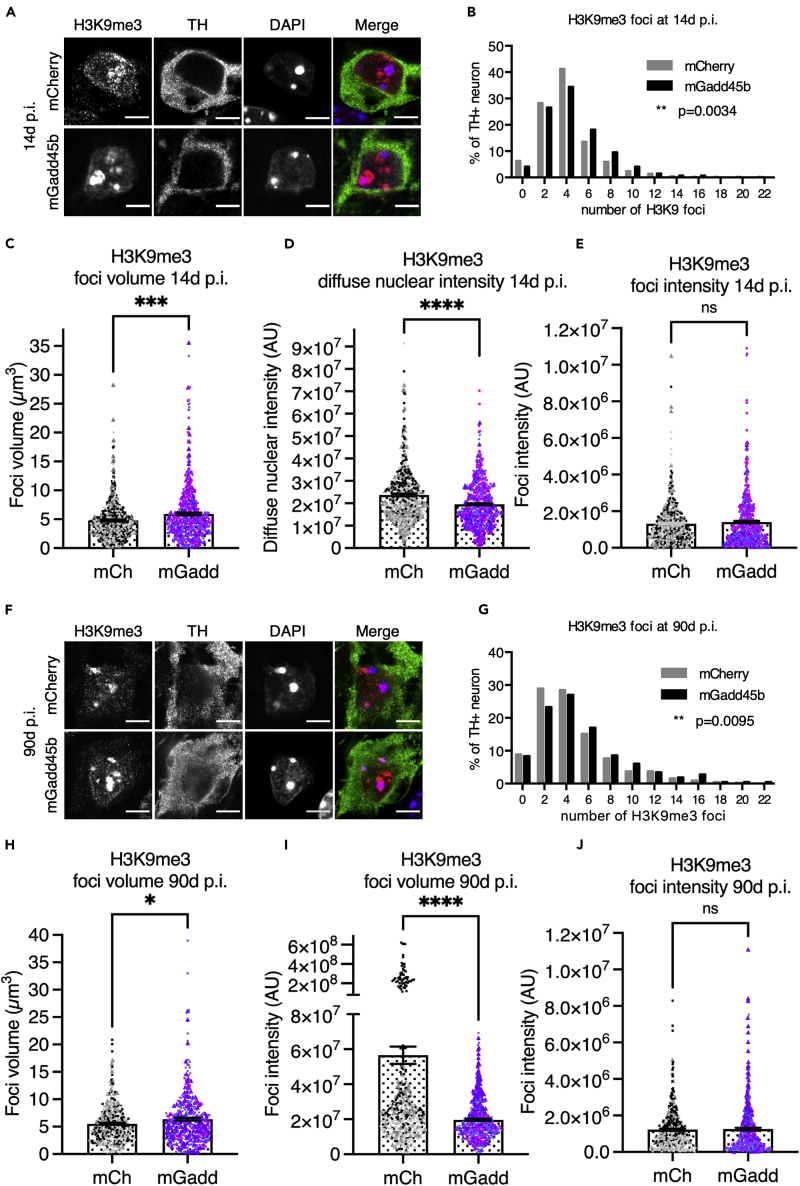
Members of GADD45 protein family have been described to promote heterochromatin relaxation (Chen et al., 2016). We therefore examined whether Gadd45b overexpression, in addition to global methylation changes, would also alter chromatin organization, in particular the organization of heterochromatin. To do so, we stained mDA neurons for histone H3 lysine 9 trimethylation (H3K9me3), a repressive heterochromatin mark. Immunostaining for H3K9me3 shows a perinucleolar pattern composed, on average, of 3 or 4 foci (3.64 ± 0.12) in TH + neurons in the SNpc of AAV8-mCherry injected mice (Figure 2A). This pattern becomes disorganized in AAV8-mGadd45b injected mice at 14d p.i. Semi-automated quantification of H3K9me3 staining specifically in TH + neurons identified a 1.13-fold increase in the number of H3K9me3 foci (4.10 ± 0.12) scattered across the nucleus, an increase by 1.24-fold of the average H3K9me3 foci volume (4.78 ± 0.15; 5.91 ± 0.22 μm3), a reduction by 1.21 fold in the intensity of the diffuse nucleoplasmic H3K9me3 staining (2.37 × 107 ± 557961; 1.96 × 107 ± 441292), but no difference in foci intensity (1.32 × 106 ± 54,236; 1.41 × 106 ± 64,942) (Figure 2A, quantification in Figures 2B–2E). This shift in heterochromatin organization is still detectable at 90d p.i (Figures 2F–2J). These results show that a de-structuration of heterochromatin is already detectable 14d and stable up to 90d after the injection of AAV8-mGadd45b indicating an early and stable perturbation of global heterochromatin organization upon Gadd45b overexpression.
Figure 2.
Gadd45b overexpression leads to heterochromatin destructuration in mDA neurons
(A–E) H3K9me3 heterochromatin staining is perturbed as early as 14d after the injection of AAV8-mGadd45b. TH + neurons in the SNpc of AAV8-mGadd45b injected mice (A) display at 14d p.i. a 1.13-fold increase in the number (3.64 ± 0.12; 4.10 ± 0.12, data represented as a frequency distribution histogramm in (B) and a 1.24-fold increase in the volume (4.78 ± 0.15; 5.91 ± 0.22 μm3, (C) of H3K9me3 foci. The diffuse nucleoplasmic staining intensity (2.37 × 107 ± 557961; 1.96 × 107 ± 441292) is decreased by 1.21-fold (D) while the foci intensity (1.32 × 106 ± 54,236; 1.41 × 106 ± 64,942) remains unchanged (E).
(F–J) H3K9me3 heterochromatin staining remains perturbed up until 90d after the injection of AAV8-mGadd45b. A similar pattern as in A-E was observed at 90d p.i. with a 1.18-fold increase in the number (4.07 ± 0.17; 4.82 ± 0.20, data represented as a frequency distribution histogramm in (G) and a 1.15-fold increase in the volume (5.51 ± 0.18; 6.35 ± 0.24 μm3, (H) of H3K9me3 foci shown in (F). The diffuse nucleoplasmic staining intensity (5.66 × 107±5 × 106; 1.97 × 107 ± 573406) is decreased by 2.87-fold while the foci intensity (1.22 × 106 ± 56,943; 1.26 × 106 ± 74,064) remains unchanged (I; J). Scale bar in A and F represents 5 μm. ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; 3 mice per condition; between 510 and 534 neurons from at least nine different images were quantified per condition at 14d p.i. and 428 neurons from at least nine different images were quantified per condition at 90d p.i.. Error bars represent SEM.
We next examined whether Gadd45b overexpression leads to perturbations in the pattern of the DNA methylation marker MeCP2. Immunostaining of sections from mice injected with AAV8-mGadd45b or AAV8-mCherry did not show any difference in the number of MeCP2 foci in TH + neurons (Figures S2A–S2D), the intensity of the diffuse nucleoplasmic MeCP2 staining nor the volume and intensity of MeCP2 foci, neither at 14d nor at 90d p.i (Figures S2E and S2G–S2J). There was a slight increase in foci intensity at 14d p.i. with AAV8-mGadd45b (Figure S2F). This analysis indicates that DNA methylation perturbation as detected by RRBS does not imply a global change in the organization of MeCP2 distribution.
Gadd45b overexpression leads to the loss of TH + neurons
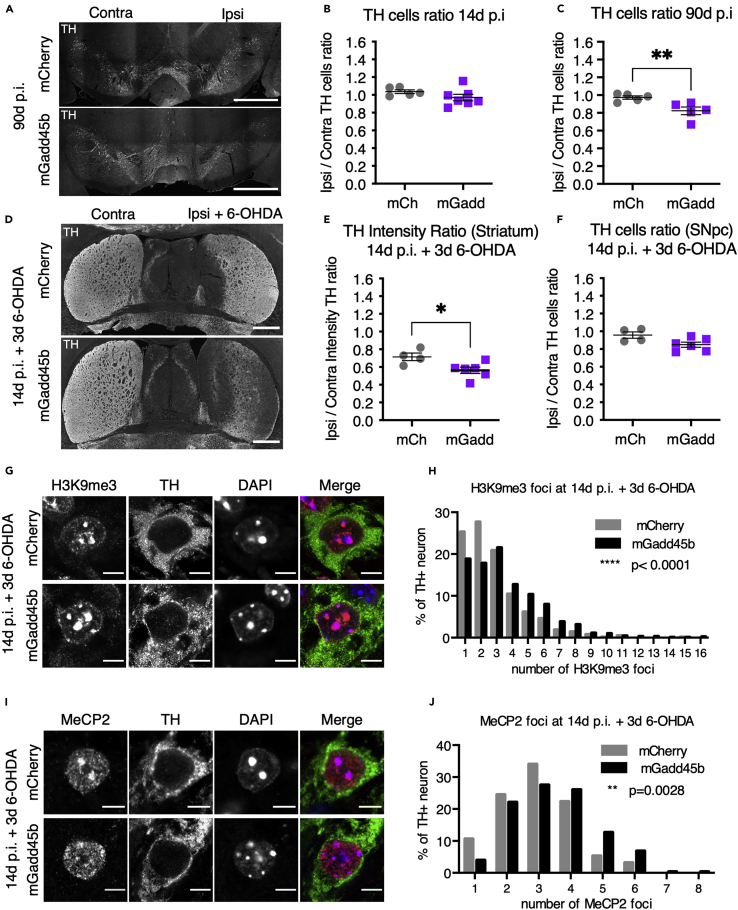
To examine the effect of Gadd45b overexpression on the survival of mDA neurons in the SNpc, we quantified the number of TH + neurons in mice injected with AAV8-mCherry or AAV8-mGadd45b at 14d and 90d p.i (Figure 3A). There was no difference between TH + cell numbers comparing AAV8-mCherry- and AAV8-mGadd45b-injected (ipsilateral) sides to the non-injected side (contralateral) at 14d p.i (Figure 3B: AAV8-mCherry ipsi/contra: 1.04 ± 0.02; AAV8-mGadd45b ipsi/contra: 0.97 ± 0.04). However, 90d after injection of AAV8-mGadd45b, this ratio shifted to an average of 0.82 ± 0.04, indicating a specific loss of 18% of TH + neurons on the AAV8-mGadd45b injected side compared to the contralateral side (Figure 3C). The TH + neurons ratio between the ipsi- and the contralateral side remained unchanged in AAV8-mCherry injected mice (Figure 3C: ipsi/contra: 0.97 ± 0.02). These results show that Gadd45b overexpression in the SNpc can trigger degeneration of mDA neurons in the long-term. In addition, the absence of a significant TH + cell loss at 14d p.i. suggests that alterations in the distribution of methylation at CpGs and in the organization of chromatin precede neuronal death and cannot be due to changes in the cellular composition of the SNpc due to the loss of certain cell types.
Figure 3.
Loss of TH + neurons and their increased vulnerability upon Gadd45boverexpression
(A–C) Dopaminergic cell loss in the SNpc 90d following the injection of AAV8-mGadd45b. The ratio of the number of TH + neurons of the injected side on the non-injected side is similar at 14d p.i. between AAV8-mCherry and AAV8-mGadd4b injected mice and close to 1 (1.04 ± 0.02; 0.97 ± 0.04) (B). At 90d p.i., however, if the ratio of AAV8-mCherry mice is still close to 1 (0.97 ± 0.02), the ratio of AAV8-mGadd45b mice (0.82 ± 0.04) reflect a 18% loss of TH + neurons compared to the non-injected side, shown in (A) and quantified in (C); scale bar represents 1000 μm; n = 5 mice per condition.
(D–F) Dopaminergic neurons are sensitized to 6-OHDA upon Gadd45b overexpression. At 14d p.i., AAV8-mCherry and AAV8-mGadd45b mice were injected with 6-OHDA in the striatum, on the same side as the viral injection and analyzed 3 days after (Figure 1A). TH staining of dopaminergic axons shows that axonal degeneration in the striatum induced by 6-OHDA occurs in AAV8-mCherry mice (0.71 ± 0.04) but is 1.5-fold more pronounced in AAV8-mGadd45b mice (0.56 ± 0.04) shown in (D), quantified in (E). The number of TH + neurons is unaffected (0.96 ± 0.04; 0.85 ± 0.03) after the 3 day 6-OHDA treatment (F); n = 4–6 mice per condition; Scale bar represents 1000 μm.
(G–J)Gadd45b expression accentuates heterochromatin loss and MeCP2 foci dispersion upon striatal 6-OHDA injection. TH + neurons in the SNpc of AAV8-mGadd45b mice injected with 6-OHDA at 14d p.i. display 3 days after the 6-OHDA injection an increase by 1.47-fold in the number of H3K9me3 foci (3.28 ± 0.15; 4.82 ± 0.16), shown in (G), quantified and represented as a frequency distribution histogramm in (H) and by 1.16-fold in the number of MeCP2 foci (2.97 ± 0.12; 3.45 ± 0.08), shown in (I), quantified and represented as a frequency distribution histogramm in (J). Scale bar represents 5 μm; ∗p < 0,05; ∗∗p < 0,01; ∗∗∗∗p < 0,0001; 3 mice per condition; Between 632 and 887 neurons were quantified per condition from at least nine different images for H3K9me3 foci and between 94 and 276 neurons per condition from at least nine different images for MeCP2 foci. Error bars represent SEM.
Enhanced vulnerability of mDA neurons overexpressing Gadd45b to oxidative stress
To examine whether the heterochromatin de-structuration observed in TH + neurons at 14d p.i. upon Gadd45b overexpression could render these neurons more vulnerable to oxidative stress, mice were first injected with AAV8-mCherry or AAV8-mGadd45b and 14d later with 6-hydroxy-dopamine (6-OHDA, 2 μL; 0.5 μg/μL) in the ipsilateral striatum. When injected into the striatum, 6-OHDA induces a specific and retrograde death of mDA neurons in the SNpc and is frequently used to model PD in rodents (Blandini et al., 2008) including mice (Bagga et al., 2015). Three days after the unilateral, striatal injection of 6-OHDA, immunostainings of striatal sections show a 29% loss (mCherry ipsi/contra: 0.71 ± 0.04) of TH intensity in the ipsilateral striatum compared to the contralateral side in mCherry expressing mice (illustrated in Figure 3D, quantified in Figure 3E). This decrease in TH staining intensity reaches 44% (mGadd45b ipsi/contra: 0.56 ± 0.04) in AAV8-mGadd45b injected mice (illustrated in Figure 3D, quantified in Figure 3E), suggesting that Gadd45b overexpression increases the axonal degeneration of mDA neurons induced by 6-OHDA. However, this experimental paradigm did not lead to any significant loss of TH cell bodies in the SNpc (Figure 3F), potentially due to the time of analysis. We analyzed mice 3 days rather than 6 days after 6-OHDA injection, the time point normally used to induce mDA cell death in the SNpc (Blandini et al., 2008), to identify early events in TH + cell bodies. An increased vulnerability to oxidative stress of mDA neurons overexpressing Gadd45b was also reflected at the heterochromatin level. The injection of 6-OHDA changed the nuclear localization and increased the number of H3K9me3 positive foci (Figures 3G and3H) compared to AAV8-mCherry. Under these conditions, the number of MeCP2 positive foci in mDA neurons expressing AAV8-mGadd45b was also increased as compared to AAV8-mCherry (Figures 3I and 3J). This presumably reflects more significant DNA methylation changes upon Gadd45b overexpression under oxidative stress.
Chromatin de-structuration is accompanied by increased DNA damage
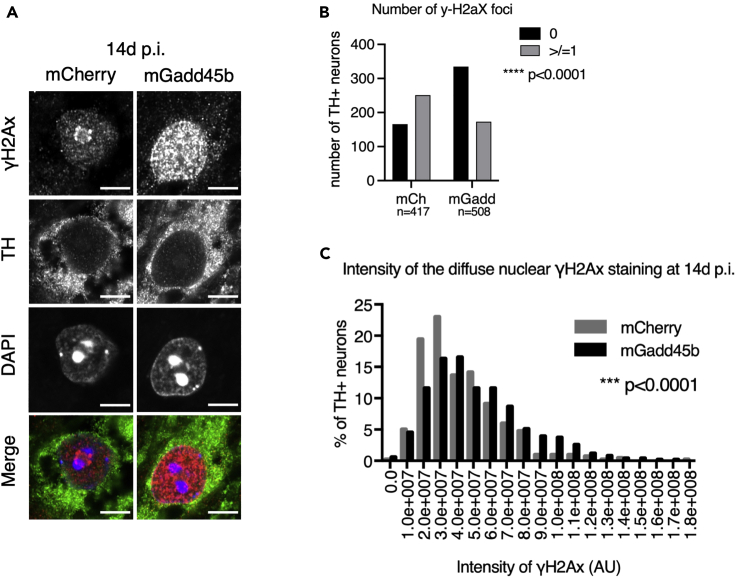
The heterochromatin loss model of aging stipulates that heterochromatin de-condensation is a driving force of cellular aging (Villeponteau, 1997). Loss of proteins involved in heterochromatin maintenance has been shown to lead to increased DNA damage and accelerated aging (Pegoraro et al., 2009; Peters et al., 2001). In the context of NDs, recent studies have reported that chromatin relaxation in the brain could also result in increased DNA damage and genome instability (Blaudin de Thé et al., 2018; Frost et al., 2014; Hajjar et al., 2019; Sun et al., 2018). Since Gadd45b overexpression led to chromatin changes, we investigated whether it might also induce DNA damage. We therefore performed immunostainings for phosphorylated histone H2AX (γ-H2AX), a marker for DNA strand breaks (Figure 4A). TH + neurons contained either a single perinucleolar focus or a diffuse nuclear staining without any foci. After injection of AAV8-mGadd45b (14d p.i.), the majority of TH + neurons (65.94%) displayed a diffuse, intense nuclear staining compared to TH + neurons after AAV8-mCherry injection 39.81% of which the majority showed one or more prominent perinucleolar γ-H2AX-positive foci (Fisher’s exact test, p < 0.0001; Figure 4B). The intensity of the diffuse nuclear γ-H2AX staining was significantly higher after AAV-mGadd45b injection (Kolmogorov-Smironov test, p <0.0001, Figure 4C). The quantification of the diffuse γ-H2AX staining in the nucleus revealed a more intense staining after Gadd45b overexpression, indicating widespread DNA damage.
Figure 4.
DNA methylation changes and heterochromatin de-structuration upon Gadd45b overexpression are associated with DNA damage
(A–C)Gadd45b overexpression induced DNA damage in TH + neurons of the SNpc. The majority (65.94 ± 2.10%) of TH + neurons of the SNpc in AAV8-mGadd45b display no single γ-H2AX foci but instead a diffuse nuclear staining as shown in (A), and quantified in (B) as TH + neurons having 0 or ≥1 foci (two-sided Fisher's Exact test, p < 0.0001). The diffuse nuclear intensity is quantified (Kolmogorov-Smirnov test, p < 0.0001) and represented as a frequency distribution histogram in (C). Scale bar represents 5 μm; 3 mice per condition, between 417 and 518 neurons from at least nine different images were quantified per condition.
LINE-1 methylation is affected by Gadd45b overexpression
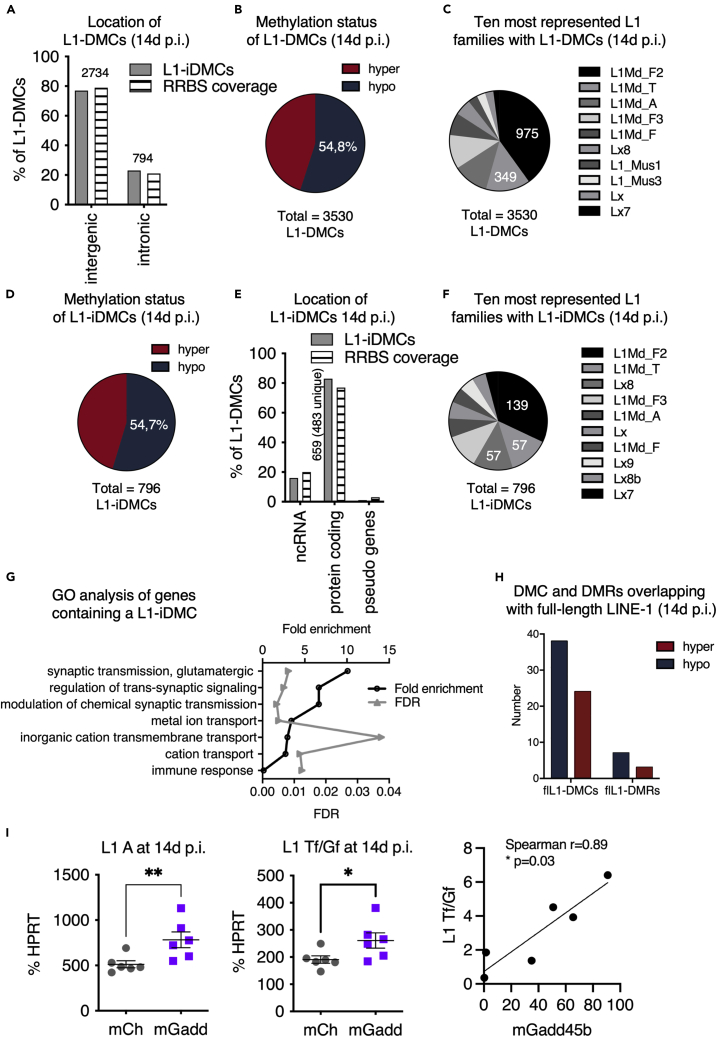
Heterochromatin alterations can unsilence normally repressed TEs, including LINE-1 elements (Van Meter et al., 2014). LINE-1 are a potential source of DNA damage (Belgnaoui et al., 2006; Blaudin de Thé et al., 2018; Gasior et al., 2006; Simon et al., 2019). Repeat elements are detected by RRBS and the Bismark software used for mapping of the RRBS reads only considers uniquely mapped reads to avoid any bias during the methylation calling. Thus, reads that are mapping with the same mapping score to multiple locations on the reference genome, which will be the case for most reads derived from repetitive elements, will not be considered. We exploited these facts to explore the methylation status of LINE-1 elements in our experimental conditions. Using the RRBS data, we used the LINE-1 annotation included in the software HOMER, which contains its own database of LINE-1 elements for the mouse genome to interrogate the overlap of DMRs and DMCs with LINE-1 elements 14 days after injection of AAV8 viruses. This analysis showed that 3530 DMCs and 1030 DMRs overlapped with an annotated LINE-1 element, which we termed L1-DMCs and L1-DMRs respectively. 794 (22.5%) L1-DMCs (Figure 5A) and 264 (25.6%) L1-DMRs (Figure S3A) were located in intronic regions, and 2734 (77.5%) L1-DMCs (Figures 5A) and 766 L1-DMRs (Figure S3A) in intergenic regions. Of all L1-DMCs the majority was hypomethylated (1933, 54,8%, Figure 5B), similar to L1-DMRs (595, 57,8%). We took the same set of common CpGs defined by the analysis of 16 murine samples and annotated them with HOMER to identify the common CpGs falling into the LINE annotation. We then determined the annotated features these LINE annotations were falling into to get an estimation of the proportion of CpGs associated with “LINE”. We then performed a chi-squared test for goodness of fit for the location of L1-DMCs (intronic/intergenic) and L1-intronicDMCs (ncRNA, protein coding and pseudo genes) which did not show any significant differences in the proportion of observed versus expected location of differentially methylated DMCs (Figures 5A and 5E). This suggests that Gadd45b overexpression is accompanied by a differential methylation of CpGs (DMCs and DMRs) in LINE sequences but these DMCs are not enriched in a particular genomic location.
Figure 5.
LINE-1 methylation changes and increased LINE-1 expression following Gadd45b overexpression
(A–C) Analysis of DMCs in LINE-1 sequences (L1-DMCs) 14d p.i. of AAV8-mGadd45b. (A) Location of L1-DMCs. 22.5% of all L1-DMCs are located in introns (L1-iDMCs, light gray), which is similar to the proportion of coverage by the RRBS (=”RRBS coverage, striped bars). (B) Of all the L1-DMCs at 14d p.i., most (54.8%) are hypomethylated (blue). (C) The ten most represented L1 families with at least one L1-DMC.
(D–G) Analysis of DMCs in intronic LINE-1 sequences (L1-iDMCs) 14d p.i. of AAV8-mGadd45b. Most L1-iDMCs (54.7%) are hypomethylated (D), located in protein-coding genes (in 483 genes, E, light gray) and related to neuronal functions (G). The frequency of the location of L1-iDMCs in protein-coding genes is similar to the RRBS coverage (= striped bars). (F) The ten most represented L1 families with L1-iDMCs.
(H) Analysis of the methylation status of full-length LINE-1 (flL1) elements containing a DMC or a DMR. Most DMCs and DMRs in full-length L1 elements are hypomethylated.Note that the analysis of differentially methylated regions (DMRs) in LINE-1 sequences (L1-DMCs) at 14d p.i. of AAV8-mGadd45b is shown in Figures S3A–S3C.
(I) LINE-1 transcripts of the young L1Md-T and L1Md-A families are increased after injection of AAV8-mGadd45b compared to AAV8-mCherry 14d p.i.. RT-qPCR of RNA extracted from manually micro-dissected SNpc of the injected sides shows a 1.5-fold increase in L1 A transcripts level (left) (512.40 ± 38.48; 782.60 ± 87.44) and 1.4-fold increase in L1 Tf/Gf transcripts level (right) (190.80 ± 13.63; 261.20 ± 28.28) in AAV8-mGadd45b mice at 14d p.i. compared to AAV8-mCherry mice. Expression levels of L1Tf/Gf are significantly correlated with expression levels of mGadd45b (Spearman correlation r = 0.89, p = 0.03); ∗p < 0,05; ∗∗p < 0,01; ∗∗∗∗p < 0,0001; n = 6 mice per condition. Error bars represent SEM. Note that the expression analysis of another repetitive element (murine Intracisternal A-particle (IAPs)) is shown in Figure S3D and the analysis of the L1-encoded ORF1 protein expression by Western blot 14d after the injection of AAV8-mGadd45b is shown in Figure S3E.
To determine which LINE-1 families containing at least one L1-DMC upon Gadd45b overexpression were overrepresented, we counted the number of families and ordered them by frequency. Figure 5C shows the ten most represented LINE-1 families with an L1-DMC. Interestingly, of the 94 different families present, three LINE-1 family members, namely L1-MdF2 (975 L1-DMCs), L1Md-T (349 L1-DMCs), and L1Md-A (280 L1-DMCs), were the three most frequently represented. L1Md-T, L1Md-A, and L1-Md_F2 elements are young LINE-1 elements (Sookdeo et al., 2013), which contain full-length, retrotransposition-competent LINE-1 copies. Of LINE-1 associated intronic DMCs (L1-iDMCs, 794) at 14d p.i., the majority (54.7%) was hypomethylated (434 hypomethylated vs 360 hypermethylated, Figure 5D). L1-iDMCs were mostly located in protein-coding genes (82.8%, Figure 5E) and the GO analysis of biological processes of 483 genes, containing at least one L1-iDMC, identified seven enriched categories. Among those categories, three were neuron-related (Figure 5G). The most frequent LINE-1 families in intronic L1-iDMCs were, again, the young L1Md-F2 family (139 of 794) and L1Md-T (57 L1-iDMCs) followed by the old Lx8 family (57 L1-iDMCs) (Figure 5F). Notably, other young LINE-1 families were also overlapping with iDMCs (L1Md-A: 27 L1-iDMCs, L1Md-Gf: 6 L1-iDMCs, not shown) (Figure 5F). Similar to L1-iDMCs, the majority of LINE-1 associated DMRs in introns (L1-iDMRs) was hypomethylated (63,3%, 167 L1-iDMRs; Figure S3A) and belonged to the L1Md-F2 family (39 of 264) (Figure S3B). Hypo- and hypermethylated iDMRs were also found in LINE-1 elements of the active L1Md-T (3 and 2, respectively) and L1Md-A families (3 and 3 elements, respectively). This prompted to examine a database annotating full-length LINE-1 elements (L1Basev2 (Penzkofer et al., 2017)) to see if intronic and intergenic DMRs and DMCs could coincide with possibly active LINE-1. Ten DMRs and 62 DMCs overlapped with a full-length LINE-1. More than half of them (7 out of 10 DMRs and 38 out of 62 DMCs, Figures 5H andS3C), were hypomethylated. This data indicates a widespread change in the methylation status of LINE-1 elements upon Gadd45b overexpression (14d p.i.). The most differentially methylated LINE-1 elements are hypomethylated and belong to young L1 families (L1Md-F2, L1Md-T, L1Md-A) suggesting a possible expression of these individual LINE-1 loci. Interestingly, L1-iDMCs are frequently located in introns of genes related to neuronal functions (Figure 5G).
LINE-1 transcripts are increased upon overexpression of Gadd45b
Having established a change in the methylation pattern of LINE-1 elements after injection of AAV8-mGadd45b, some of which were full-length, we evaluated the expression of the youngest L1 families in mice, namely L1Md-A and L1Md-Tf/Gf. The analysis by RT-qPCR with specific primers located in the 5′UTR of the L1Md-A (Figure 5I, left panel) and L1Md-Gf/Tf families (Figure 5I, middle panel) showed a 1.5-fold increase in LINE-1 transcripts in the SNpc after 14d of Gadd45b overexpression. We plotted L1Tf/Gf expression as a function of Gadd45b expression and found a significant correlation between the expression levels of Gadd45b and L1Tf/Gf (Spearman r = 0.89, p = 0.03, Figure 5I, right panel). The increase in L1 expression was confirmed by Western blot with an in-house rabbit ORF1p antibody (Blaudin de Thé et al., 2018) and actin as a loading control on a membrane loaded with proteins extracted simultaneously with DNA and RNA from the ventral midbrain. Upon Gadd45b overexpression, ORF1p levels were significantly increased at 14d p.i. (two-tailed Mann-Whitney test, p = 0.041, n = 6 mice per condition; Figure S3E). There was no increase in the expression of another repetitive element family, namely murine Intracisternal A-particle (IAP) elements, at 14d p.i. after injection of mGadd45b compared to mCherry control (two-tailed Mann-Whitney test, p = 0.81, n = 6 mice per condition; Figure S3D).
Gadd45b overexpression is accompanied by expression changes in genes with DMCs
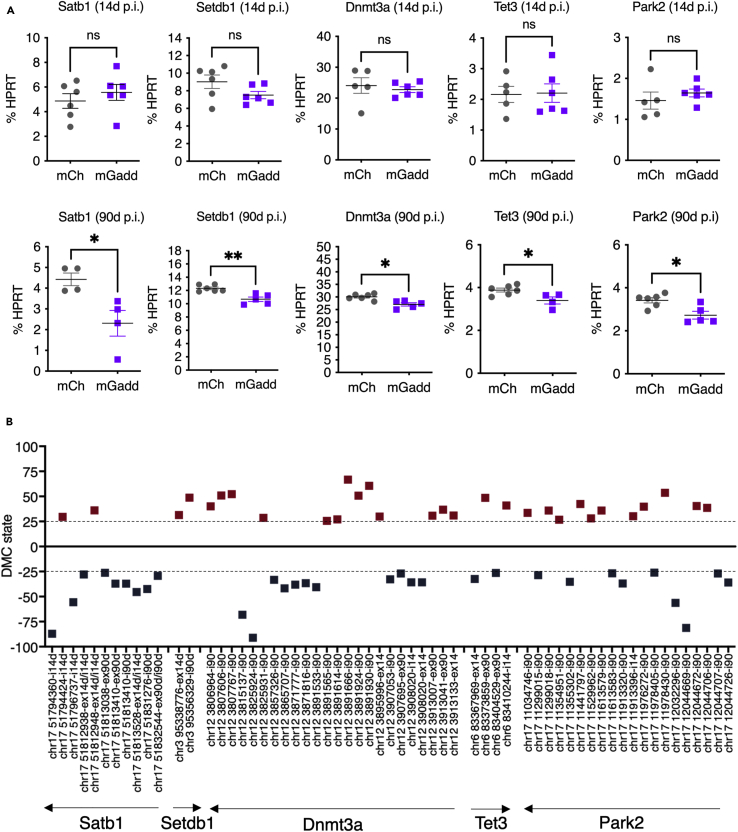
Expression levels of candidate genes with intronic DMCs at either 14d or 90d p.i. were then analyzed by RT-qPCR. We selected 16 genes based on their known function either in chromatin remodeling (Satb1, Setdb1, Wapl), DNA methylation (Tet1, Tet2, Tet3, Dnmt3a, Dnmt3b, Gadd45a), PD relevance (Lrrk2, Park2), synaptic remodeling (Sorcs2), DNA damage (Xpa), or in aging and senescence (Cdkn2a-p19, Cdkn2d, Sirt1). None of the candidate genes showed a change in expression at 14d p.i. (not tested for Dnmt3a, Tet1 and Gadd45a). However, at 90d p.i., the expression of Satb1, Setdb1, Dnmt3a, Tet3, and Park2 decreased significantly in the ipsilateral SNpc injected with AAV8-mGadd45b compared to AAV8-mCherry (Figure 6A). Gadd45a was expressed at very low levels and did not show a change in expression upon Gadd45b overexpression (Figure S4A). Of note, 4 out of these 5 genes belong to the GADD45B-DMC-regulon (Figure 1H). Using RNA-seq data of laser-microdissected SNpc from wild-type mice (GEO GSE72321 (Rekaik et al., 2015); and unpublished), we compared the expression levels of the selected candidate genes to those of dopaminergic neuron-specific markers such as Th, dopamine transporter Slc6a3 or the homeogene, Engrailed 1 (En1) (Blaudin de Thé et al., 2016). We found high levels of expression of Satb1 compared to the other selected genes, indicating that Satb1 is strongly expressed in the SNpc (Figure S4B). Exonic and intronic DMCs within the gene region of downregulated genes comprised hypo-as well as hypermethylated CpGs. Disruption of the normal gene body methylation state of several genes of the GADD45B-DMC-regulon might thus provoke their dysregulation over time but is not associated with a loss of a particular methylation pattern.
Figure 6.
Candidate genes with DMCs upon Gadd45b overexpression show altered expression in the SNpc at 90d p.i
(A) The expression of several gene candidates containing DMCs is dysregulated. Setdb1, Park2, Dnmt3a, Tet3, and Satb1 transcripts are downregulated 90d p.i. of AAV8-mGadd45b. The mRNA analysis in SNpc of the injected side by RT-qPCR shows no significant difference in Satb1 (4.87 ± 0.59; 5.57 ± 0.65), Setdb1 (9.03 ± 0.76; 7.51 ± 0.42), Dnmt3a (24.07 ± 2.52; 22.76 ± 0.92), Tet3 (2.16 ± 0.26; 2.20 ± 0.65) and Park2 (1.46 ± 0.21; 1.65 ± 0.09) transcripts at 14d p.i. (4.87 ± 0.59; 5.57 ± 0.65); n = 5–6 mice per condition. At 90d p.i., there is a decrease in transcript levels of 1.9-fold for Satb1 (4.43 ± 0.30; 2.31 ± 0.62), of 1.15-fold for Setdb1 (12.32 ± 0.20; 10.68 ± 0.34) of 1.11-fold for Dnmt3a (30.06 ± 0.47; 27.05 ± 0.67), of 1.14 fold for Tet3 (3.89 ± 0.09; 3.40 ± 0.17) of 1.25-fold for Park2 (3.42 ± 0.12; 2.73 ± 0.18), in AAV8-mGadd45b mice compared to AAV8-mCherry mice; ∗p < 0,05; n = 4 mice per condition. Error bars represent SEM.
(B) Genes significantly deregulated upon Gadd45b overexpression were analyzed with regard to methylation differences throughout their gene bodies (exonic and intronic DMCs) at 14d and 90d p.i. Results are displayed for Satb1, Setdb1, Dnmt3a, Tet3, and Park2. Exact chromosomal positions of DMCs are given on the x-axis and the orientation of the genes is indicated by an arrow. Hypermethylated are depicted as red squares while hypomethylated DMCs are depicted in blue squares. DMCs were defined as having a methylation difference > or equal to 25 (dashed line) and a q-value <0.01.
Gadd45b expression with age
To evaluate Gadd45b expression levels in mice with increasing age, we measured Gadd45b levels by RT-qPCR at different ages from 30 to 140 days. Expression levels increased significantly about 3-fold between 30 and 140 days (One-way ANOVA, Holm-Sidak’s multiple comparison test, adjusted p-value = 0.022; Figure S4C).
Discussion
Epigenetic alterations and chromatin relaxation are hallmarks of aging (Benayoun et al., 2015; Pal and Tyler, 2016), but evidence of age- and disease-related global changes in the epigenetic landscape of different neuronal populations is still scarce. This information, however, holds important promises for understanding the implication of aging in the neuronal degeneration characterizing age-related NDs and might thus foster the understanding of the pathogenesis of NDs.
Here, we show that Gadd45b overexpression induces early changes in DNA methylation, particularly in introns of genes related to neuronal functions and on young and potentially active LINE-1 elements. This, accompanied by perturbations of heterochromatin organization and increased DNA damage culminate in neuronal cell death after several weeks. At an early time point before the onset of neurodegeneration, Gadd45b overexpression induces a vulnerable state in mDA neurons, increasing their sensitivity to oxidative stress induced by the striatal injection of 6-OHDA. This vulnerability is characterized by an amplified striatal dopaminergic axon terminal loss and an accentuation of perturbations in the organization of heterochromatin and DNA methylation in mDA cell bodies of the SNpc. We also demonstrate that LINE-1 transcripts are increased early on after Gadd45b overexpression. This increase in LINE-1, which are potent inducers of DNA damage in mDA neurons (Blaudin de Thé et al., 2018), could explain the cell death of mDA neurons we observe when overexpressing Gadd45b long-term.
Expression levels of Gadd45b are approximately 70-fold higher than endogenous levels in our experimental condition but, importantly, comparable to expression levels of the mCherry control. This increase in expression must be considered as supra-physiological when compared to the extent of expression changes of Gadd45b upon oxidative stress (our own data and that of others) or in postmortem human SNpc of PD patients. Six hours after a 6-OHDA injection into the SNpc of wild-type mice, Gadd45b levels increase 2-fold compared to sham. This is in a similar range as reported by previous in vitro studies using 6-OHDA (Ryu et al., 2005). In the context of an exposure of cells to zinc or a combination of zinc and dopamine in which PC12 cells undergo degeneration, the mRNA levels of Gadd45b were increased ≈12-fold. Gadd45b overexpression led to an increased susceptibility to cell death and knock-down of Gadd45b by shRNA was protective. Microarray data from human postmortem SNpc of 13 male PD patients showed that Gadd45b transcripts were significantly increased (an estimated 4-fold) compared to neurologically healthy individuals. This was not the case in female PD patients, possibly due to the lower numbers of individuals analyzed (Yang et al., 2016). This data strongly supports our conclusion as to a detrimental effect of an increase in Gadd45b expression on dopaminergic neuron survival. Interestingly in this context, we observed that endogenous Gadd45b levels in wild-type mice increase with age. Whether mDA neurons are particularly susceptible to an increase in Gadd45b expression remains to be determined, but a course analysis of TH-negative cell numbers did not reveal any major loss of TH-negative cells.
A number of recent studies indicate that GADD45 proteins play a key role in active DNA demethylation in post-mitotic neurons in the brain (Gavin et al., 2015; Niehrs and Schäfer, 2012) by serving as scaffolding proteins to recruit DNA repair enzymes such as the thymidine DNA glycosylase (TDG) to the site of DNA demethylation (Li et al., 2015). So far, GADD45B-regulated DNA demethylation has been described in the context of adult neurogenesis (Ma et al., 2009), depressive-like behavior in mice (Labonté et al., 2019), major psychosis in humans (Gavin et al., 2012), and cerebral cortex plasticity (Apulei et al., 2019) on specific promoters, mostly on promoters of the Bdnf gene. Unexpectedly, when overexpressing Gadd45b in the SNpc of wild-type mice, we observed very limited changes in methylation on gene promoters, but rather widespread methylation changes on gene bodies with both hypermethylated and hypomethylated CpGs. This apparent discrepancy with previous studies in terms of extent, localization and pattern of the DNA methylation changes induced by GADD45B might be due to methodology. Indeed, most studies in the brain did not use techniques allowing for unbiased global DNA methylation surveillance.
GADD45B has been shown to associate with Ten-eleven translocation (TET) proteins (Arab et al., 2018; Kienhöfer et al., 2015) which transform 5-methylcytosines (5mC) through a series of sequential oxidations. Those modified cytosines are excised by TDG and then replaced by non-methylated cytosines through BER-dependent mechanisms (Bayraktar and Kreutz, 2018). A recent study has shown that Tet2 expression is increased in PD patients leading to altered 5mC patterns in enhancers of neuronal genes. Conversely, TET2 loss in mDA neurons was neuroprotective (Marshall et al., 2020). Tet2 did not show any changes in expression at either time point after AAV8-mGadd45b injection. This might be explained by the fact that Tet2 has many splice variants, which were not all covered in our RT-qPCR assay. However, Tet3 is down-regulated 90d after AAV8-mGadd45b injection. TET3 has been shown to interact with transcriptional regulators and histone writers such as H3K36 methyltransferases, and to allow the active transcription of certain neuronal genes (Perera et al., 2015). TET3 has also been reported to bind DNA and prevent aberrant methylation at the transcription start site of genes involved in lysosomal functions, mRNA processing and the BER pathway, pointing to a possible relevance of TET3 in the pathogenesis of NDs (Jin et al., 2016). While our knowledge of how genome-wide DNA methylation patterns or epigenetic changes are correlated with PD pathogenesis is still scarce, there is evidence supporting that methylation changes in the SNpc correlate with aging (Fasolino et al., 2017) and with cognitive impairment in PD (Chuang et al., 2019). Most of the existing data concerns methylation patterns of specific genes associated with NDs. Expression of the SNCA gene, encoding α-synuclein and mutated in familial PD, for example, is under the control of DNA methylation (Pihlstrøm et al., 2015; Wüllner et al., 2016). Chromatin modifications on specific genes have also been reported, and the expression of several genes mutated in PD is modulated by histone modifications, including SNCA and MAPT encoding Tau (Labbé et al., 2016). Differentially methylated enhancers have been reported in AD patients, and a dysregulation of histone acetylation in both, AD and PD patients (Berson et al., 2018; Cedar and Bergman, 2009; Li et al., 2019; Nativio et al., 2018).
By serving as an adaptor between repair factors and chromatin, GADD45B can be seen as a communicating platform between DNA repair and epigenetics (Niehrs and Schäfer, 2012; Schäfer, 2013). This could provide an explanation to the fact that we observe global changes in heterochromatin organization in addition to changes in DNA methylation. In line with this, GADD45 proteins have been reported to induce heterochromatin relaxation during cellular reprogramming (Chen et al., 2016). GADD45B might thus be part of protein coordinators which link DNA methylation and histone modifications (Cedar and Bergman, 2009). Perturbations of DNA methylation and global changes in heterochromatin organization, as we show here, induce neurodegeneration of mDA neurons in the SNpc with time and might thus be primary drivers of neurodegeneration.
Another primary hallmark of aging is genomic instability and some data suggest that this is also true in neuronal aging (Hou et al., 2019). In this context, it is important to note that DNA strand breaks are physiologically occurring in post-mitotic neurons. This physiological process, however, needs to be tightly regulated, since the induction of DNA strand breaks is pathologically exacerbated in AD (Suberbielle et al., 2013). We have shown earlier that DNA damage in mDA neurons can be induced by acute and chronic oxidative stress (Blaudin de Thé et al., 2018; Rekaik et al., 2015) and that LINE-1 activity participates in DNA damage (Blaudin de Thé et al., 2018). In the latter study, DNA damage was prevented either by siRNA against LINE-1 ORF2, the LINE-1 repressive protein Piwil1 or a nucleoside analogue reverse transcriptase inhibitor in vitro (Blaudin de Thé et al., 2018). This LINE-1 induced DNA damage is dependent on young and active LINE-1 copies. These young LINE-1 elements belong mainly to the L1Md-Tf/Gf, A and F families. In this context it is interesting to note that we observe the most pronounced changes in DNA methylation upon Gadd45b overexpression on these young LINE-1 families (Figure 5). Intronic DMCs and DMRs, intronic L1-associated DMCs (L1-iDMCs), and DMCs or DMRs overlapping with full-length LINE-1 (flL1-iDMCs/flL1-iDMRs), are majorly hypomethylated and iDMCs are preferentially located in genes related to neuronal functions. It is therefore conceivable that the changes in methylation patterns we observe upon Gadd45b overexpression, particularly on young LINE-1 elements, are functionally linked to the increase in LINE-1 transcripts we observe 14 days after injection of AAV8-mGadd45b. Furthermore, based on previous evidence summarized above, this increase in expression of young L1 elements of the L1Md-Tf/Gf, A and F families might be at the origin of the DNA damage in mDA neurons upon Gadd45b overexpression.
Several lines of evidence suggest that the activation of TEs might be associated with aging and NDs. The expression of LINE-1 in wild-type mice increases in neurons (Simon et al., 2019), liver and muscle during aging (De Cecco et al., 2013). The activation of TEs with aging leads to neuronal decline and shorter lifespan in drosophila (Li et al., 2013; Wood et al., 2016) and increased TE expression has been reported in brain tissue from PD, AD, and ALS patients (Liu et al., 2019; Sun et al., 2018). Recently, elevated transcripts of repetitive sequences have been found in the blood of PD patients (Billingsley et al., 2019). Heterochromatin de-structuration and increased TE activity lead to an AD-like phenotype in a mouse model with targeted disruption of Bmi1, a gene involved in heterochromatin maintenance and altered expression in AD patients (Hajjar et al., 2019). In AD and in ALS, the Tau protein as well as TDP-43 can induce heterochromatin relaxation, especially at the level of LINE-1 elements in the case of TDP-43, leading to increased TE activity and neurotoxicity (Li et al., 2012; Liu et al., 2019; Sun et al., 2018). An aging-induced activation of TEs, which are intrinsic components of the genomes of virtually all eukaryotes, might link genomic instability and epigenetic changes to the aging process.
Changes in chromatin states lead to changes in gene expression as exemplified during the transition from neuronal progenitors to adult neurons (Södersten et al., 2018) and during aging (Zahn et al., 2007), and recent data suggests that this could also be the case in NDs (Berson et al., 2018; Urdinguio et al., 2009). In several NDs (AD and HD (Hodges et al., 2006; Narayanan et al., 2014); ALS (Prudencio et al., 2015)), disease-specific gene expression changes are increasingly recognized, but whether they overlap with expression profiles characteristic to aging is not known yet. In AD patients, widespread loss of heterochromatin was accompanied by a transcriptomic profile resembling the one of a fetal brain (Frost et al., 2014), suggesting that the relaxation of chromatin allows the expression of normally repressed genes which alters several biological processes and leads to neurodegeneration (Soukup and Verstreken, 2014). This is also in line with our data showing the preferential location of iDMCs and iDMRs in genes involved in neurogenesis but it remains to be seen whether their expression is altered. Interestingly, it has been shown that Gadd45b activity promotes adult neurogenesis (Ma et al., 2009). The majority of these genes with iDMCs and iDMRs upon overexpression of Gadd45b belonged to neuronal categories, particularly synapse-related and neurodevelopmental categories. In this context it is interesting to note that synaptic homeostasis is an emerging key player in the pathogenesis of PD (Soukup et al., 2018) and synaptic dysfunction is an early event in neurodegeneration (Burke and O'Malley, 2013). Gadd45b overexpression induced DMCs and DMRs preferentially in introns of genes. The role of gene body or intronic methylation on gene expression is not completely understood (Jones, 2012). In recent years, several studies have reported that gene body methylation influences gene expression levels and/or alternative splicing (Kinde et al., 2015; Neri et al., 2017; Yang et al., 2014). Among the list of genes underlying a direct or indirect regulation through methylation changes triggered by Gadd45b overexpression, the decrease in expression did not correlate with a specific direction of methylation changes toward hypo- or hypermethylated CpGs but rather with a change in the methylation state throughout the gene body. Satb1 has been described as a dopaminergic-specific regulator of senescence (Cancio-Bello and Saez-Atienzar, 2020), a dopaminergic neuron cell survival factor (Riessland et al., 2019) and a regulator of global chromatin structuration (Cai et al., 2003; Yasui et al., 2002). The decline in Satb1 expression at 90d p.i. might help to explain, at least partly, the neurodegeneration of mDA neurons upon Gadd45b overexpression. Setdb1 is a histone-lysine-methyltransferase that specifically trimethylates lysine-9 of histone H3. It is tempting to speculate that the decrease in expression might relate to the change in the organization of H3K9me3 we observe upon Gadd45b overexpression. Dnmt3a, a genome-wide de novo DNA methyltrasferase, and Tet3, are involved in DNA methylation or demethylation, respectively. Interestingly, inactivation of TET and/or DNMT proteins causes gains and losses of DNA methylation, suggesting that the loss of one regulator can lead to the redistribution of other regulators and of DNA modifications (Zhang et al., 2016). Park2, encoding the ubiquitin protein ligase Parkin, is a PD-related gene and loss-of-function mutations in this gene are responsible for familial forms of PD. It is thus possible that a decrease in the expression of Park2 in the context of Gadd45b overexpression might contribute to the degeneration of mDA neurons. Overall, the expression changes might be due to the Gadd45b-induced epigenetic dysregulation of the neuronal genome and participation in the Gadd45b overexpression phenotype and the challenge will be to correlate one with the other.
Altogether, our data are in line with an emerging concept of a so far understudied pathogenic pathway initiating age-related neurodegeneration. Recent evidence, including from our group, suggests that aging-induced chromatin reorganization triggers the activation of LINE-1 retrotransposons and subsequent LINE-1 induced DNA damage cumulating in neuronal cell death. Our group has shown that acute and chronic oxidative stress leads to heterochromatin relaxation and LINE-1 activation in mDA neurons in vivo (Blaudin de Thé et al., 2018; Rekaik et al., 2015). Aging-induced epigenetic alterations might produce a vulnerable pre-ND state. Combining this pre-ND state with a particular genetic susceptibility, a familial gene mutation or an accelerating environmental trigger, could initiate a cascade of secondary events including protein aggregation, metabolic dysregulation, and mitochondrial dysfunctions. NDs share several common pathological features and despite extensive investigation, no disease-modifying treatment is available. Acknowledging aging as a vulnerability factor for neurodegeneration is important not only for understanding the pathogenesis of NDs, but also for modeling, testing, and developing therapeutics for crucially lacking disease-modifying treatments. Our study suggests two therapeutic targets for neuroprotection. Drugs restoring chromatin structure and/or repressing LINE-1 transcription or activity might hold promise for the prevention of age-related neurodegeneration.
Limitations of the study
Although RRBS identifies differential methylation changes, the method is limited by the partial genome coverage that may not reflect on global methylation changes as detected by an enzyme-linked immunosorbent assay (ELISA) or by high-performance liquid chromatography (HPLC).
STAR★methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Orf1p rabbit antibody, clone SY6858 | Eurogentec | N/A |
| Orf1p guinea pig antibody, clone SYC809 | Eurogentec | N/A |
| Anti-phospho-histone H2A.X (Ser139) mouse antibody, clone JBW301 | Millipore | #05-636; RRID:AB_309864 |
| Anti-tyrosine hydroxylase chicken antibody | Abcam | #ab76442; RRID:AB_1524535 |
| Living Colors® mCherry Monoclonal mouse antibody | Clontech | #632543; RRID:AB_2307319 |
| Anti-histone H3 (tri methyl K9) rabbit antibody - ChIP grade | Abcam | #ab8898; RRID:AB_306848 |
| Anti-MeCP2 rabbit antibody, clone 4H7 | Millipore | #MAB328; RRID:AB_2144004 |
| Goat anti-mouse IgG (H + L) highly cross-adsorbed secondary antibody, alexa fluor 546 | ThermoFischer | Cat# A-11030; RRID:AB_2534089 |
| Goat anti-chicken IgY (H + L) secondary antibody, alexa fluor 488 | ThermoFischer | Cat # A-11039; RRID:AB_2534096 |
| Goat anti-Guinea pig IgG (H + L) highly cross-adsorbed secondary antibody, alexa fluor 647 | ThermoFischer | Cat # A-21450; RRID:AB_2735091 |
| Donkey anti-rabbit IgG (H + L) highly cross-adsorbed secondary antibody, alexa fluor 647 | ThermoFischer | Cat# A-31573; RRID:AB_2536183 |
| Goat anti-mouse IgG (H + L) cross-adsorbed secondary antibody, alexa fluor 647 | ThermoFischer | Cat # A-21235; RRID:AB_2535804 |
| Monoclonal anti-β-Actin−Peroxidase antibody produced in mouse, clone AC-15 | Sigma | Cat # A-3854; RRID:AB_262011 |
| Anti-digoxigenin-POD (poly), fab fragments from sheep | Roche | #11633716001; RRID:AB_514499 |
| Anti-rabbit IgG, HRP-linked antibody | Cell signaling | Cat #7074; RRID:AB_2099233 |
| Bacterial and virus strains | ||
| AAV8-Ef1a-mGadd45b, including ORF and UTRs (RefSeq: BC033815) | VectorBiolabs | NA |
| AAV8-Ef1a-mCherry | VectorBiolabs | #VB2098 |
| Chemicals, peptides, and recombinant proteins | ||
| 6-Hydroxydopamine hydrobromide | Sigma | #H116 |
| RNase H | ThermoFischer | #18021071 |
| Proteinase K, recombinant, PCR grade | Roche | #3115836001 |
| Phosphate buffered saline (PBS) | ThermoFischer Scientific | Cat# 10010023 |
| LightCycler® 480 SYBR Green I Master | Roche | Cat # 04707516001 |
| Critical commercial assays | ||
| AllPrep DNA/RNA Micro kit | Qiagen | Cat # 80,284 |
| RNase-free DNase set | Qiagen | Cat # 79,256 |
| QuantiTect reverse transcription kit | Qiagen | Cat # 205313 |
| DNA clean & Concentrator-5 kit | Zymo | Cat # D-4013 |
| Qubit™ 1X dsDNA HS assay kit | Thermo Fisher Scientific | Cat # Q33230 |
| Premium reduced representation bisulfite sequencing (RRBS) kit | Diagenode | Cat. No. C02030032 |
| High sensitivity genomic DNA analysis kit | Agilent | # DNF-488-0500 |
| DIG RNA labeling kit | Roche | Cat # 11277073910 |
| BCIP/NBT substrate kit, alkaline phosphatase (AP), (5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium) | Vector Labs | # SK-5400 |
| AllPrep DNA/RNA/Protein Mini extraction kit | Qiagen | Cat # 80,004 |
| Pierce™ fast western kit, SuperSignal™West pico, mouse | ThermoFischer Scientific | Cat # 35,060 |
| Deposited data | ||
| Reduced representation bisulfite sequencing data (Gadd45b vs. mCherry at 14d and 90d) | Gene Expression Omnibus | GSE176544 |
| Experimental models: Organisms/Strains | ||
| Mouse: WT: RjOrl:SWISS | Janvier Labs | # SN-SWISS-M |
| Oligonucleotides | ||
| Primers | See Table S2 | NA |
| Software and algorithms | ||
| Microsoft excel v16 | Microsoft Office | https://www.microsoft.com/en-ca/microsoft-365/excel |
| GraphPad prism v9.1.2 | GraphPad | https://www.graphpad.com |
| FIJI v2.1.0/1.53c | ImageJ | https://imagej.net/software/fiji/downloads |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Julia Fuchs (julia.fuchs@college-de-france.fr).
Materials availability
This study did not generate new unique reagents or material.
Data and code availability
The RRBS data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE176544 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE176544). The original western blot image and microscopy data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Animals
All animal treatments followed the guidelines for the care and use of laboratory animals (US National Institutes of Health), the European Directive number 2010/68/UE (EEC Council for Animal Protection in Experimental Research and Other Scientific Utilization). This project was validated by the competent ethical committee (CEA 59) and authorized by the Minister of Higher Education, Research and Innovation (n° 00703.01 and n° APAFIS#6605-2016090209434306 v3).
For surgical procedures, animals were anesthetized with Xylazine (Rompun 2%, 5 mg/kg) and Ketamine (Imalgene 1000, 80 mg/kg) by intraperitoneal injection and a local subcutaneous injection of lidocaine (0.5%, 3mg/kg) on the incision site. Post-chirurgical analgesia was insured by an injection of the analgesic Meloxicam (Metacam, 0,5mg/kg) s.c..
Swiss OF1 wild-type mice (Janvier) were maintained under a 12 h day/night cycle with ad libitum access to food and water. A maximum of 5 mice were housed in one cage, and cotton material was provided for mice to build a nest. Experimental groups consisted of five to seven randomly assigned male mice of 6 weeks of age. Sample size calculations were based on previous experiments.
Method details
AAV8 vectors to overexpress Gadd45b
Forced expression of Gadd45b in mDA neurons was achieved using an AAV8 viral vector. The constructs contained cDNAs for either mouse Gadd45b (AAV8-mGadd45b) or mCherry (AAV8-mCherry) under the control of the ubiquitous EF1a promotor. Gadd45b cDNA was flanked by the cognate 5’ and 3’ UTRs (Figure 1A). AAV8 was chosen because it has been previously shown to efficiently infect mDA neurons in the midbrain.
Brain injections
For injections, mice were placed in a stereotaxic instrument, and a burr hole was drilled into the skull 3.3 mm caudal and 1.3 mm lateral to the Bregma. The needle was lowered 3.8 mm from the surface of the skull, and AAV8-Ef1a-mCherry or AAV8-Ef1a-mGadd45b (Vector Biolabs; 2 μL; 4.8 × 1013 GC/ml suspended in NaCl 0,9% with 5% glycerol) injections were performed over 4 min. Where indicated 6-OHDA (2 μL; 0.5 μg/μL; Sigma) injections were performed in the same manner 0.4 mm rostral, 1.8 mm lateral and 3.8 mm ventral to the bregma, over 4 min.
Tissue dissection
For RNA and DNA analyses, biopsies of the SNpc were performed. Brains were put into a custom-made brain slicer for adult mice brain. A coronal slice of ≈2mm encompassing the SNpc was excised (bregma −3.26 mm to −5.2mm) and placed on a cold cover slide with the rostral side facing the experimenter. Dissection of the SNpc was then done following anatomical landmarks: a sagittal cut to separate the two hemisphere, a second parasagittal cut through the fasciculus retroflexus and the mammillothalamic tract (about 2/3 starting from the midline of the distance between the midline and the rostral end of cerebral peduncle) to remove the VTA, a transversal section from the ventral part of the lateral geniculate complex to the midline, a second transversal cut from the ventral end of the cerebral peduncle to the midline. The cerebral cortex was then removed to only keep the midbrain part containing the SNpc which was immediately frozen on dry ice and kept at −80°C until extraction.
RT-qPCR4
Total RNA from dissected SNpc was extracted using the AllPrep DNA/RNA Micro Kit (Qiagen 80,284) adding an on-column DNase I treatment (Qiagen 79,256), followed by RT-qPCR. RNA (200 ng) was reverse-transcribed using the QuantiTect Reverse Transcription kit (Qiagen, 205313). Quantitative PCR reactions were carried out in duplicates with SYBR Green I Master Mix (Roche S-7563) on a LightCycler 480 system (Roche Applied Science). The following primers were used: Hprt (forward: AGCAGGTGTTCTAGTCCTGTGG, reverse: ACGCAGCAACTGACATTTCTAA); LINE-1 A (forward: TTCTGCCAGGAGTCTGGTTC, reverse: TGAGCAGACCTGGAGGGTAG); LINE-1 Tf/Gf (forward: CTGGGAACTGCCAAAGCAAC, reverse: CCTCCGTTTACCTTTCGCCA); Gadd45b (forward: ACTGATGAATGTGGACCCCG, reverse: CCTCTGCATGCCTGATACCC); Satb1 (forward: TCTTTTACCCCCTCCTCCCA, reverse: TCACCTGCCAGAACACTTCA); Tet3 (forward: CTCGGCGGGGATAATGGGAG, reverse: AGCCTGTCTTGACAGTCGCC); Dnmt3a (forward: GCCGAATTGTGTCTTGGTGGATGACA, reverse: CCTGGTGGAATGCACTGCAGAAGGA), Setdb1 (forward: GTTTGCCTGGGTTTGGCAAG, reverse: CTTTGGCCCTCAGTCCGTC); Park2 (forward: GCTCAAGGAAGTGGTTGCTAAG, reverse: CAATACTCTGTTGTTCCAGGTCA); IAP (forward: AAGCAGCAATCACCCACTTTGG, reverse: CAATCATTAGATGTGGCTGCCAAG); Gadd45a (forward: TGAGCTGCTGCTACTGGAGAAC, reverse: GATCCTTCCATTGTGATGAATG); Gapdh (forward: TGACGTGCCGCCTGGAGAAAC, reverse: CCGGCATCGAAGGTGGAAGAG); Eno (forward: TCCGGAACTATCCTGTGGTCT, reverse: CCCACTATCTGGATGCCGAC) summed up in Table S2. Primer efficiencies were tested using 10-fold dilution series of cDNA spanning at least three orders of magnitude. Data were analyzed using the ddCt method and values normalized to hypoxanthine-guanine phosphoribosyl transferase (Hprt) or to glyceraldehyde-3-phosphate dehydrogenase (Gapdh).
DNA extraction and quantification
DNA was also extracted during the same process of RNA extraction using the AllPrep DNA/RNA Micro Kit (Qiagen 80,284). The DNA was then purified, treated with RNase H (ThermoFischer, 18021071) and Proteinase K (PCR grade, Roche, 3115836001) and concentrated with the DNA Clean & Concentrator-5 kit (Zymo, D4013). DNA concentration of each sample was measured using the Qubit fluorometer with dsDNA BR Assay Kit (Thermo Fisher Scientific).
RRBS
RRBS was performed by Diagenode. DNA quality of the samples was assessed with the Fragment AnalyzerTM and the DNF-488 High Sensitivity genomic DNA Analysis Kit (Agilent). DNA was slightly more fragmented than defined by the quality control standards but this fragmentation was minor. RRBS libraries were prepared using the Premium Reduced Representation Bisulfite Sequencing (RRBS) Kit (Diagenode) which uses the Mspl restriction enzyme and size selection to enrich for CpG-rich regions (coverage of about 4 million CpGs). 100ng of genomic DNA were used to start library preparation for each sample. Following library preparation, samples were pooled together by 8. PCR clean-up after the final library amplification was performed using a 1.45x beads:sample ratio of Agencourt AMPure XP (Beckman Coulter). DNA concentration of the pools was measured using the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific). The profile of the pools was checked using the High Sensitivity DNA chip for 2100 Bioanalyzer (Agilent). RRBS library pools were sequenced on a HiSeq3000 (Illumina) using 50 bp single-read sequencing (SR50). Bisulfite conversion and amplification were performed using Diagenode's Premium RRBS Kit. After conversion, the pooled samples were analyzed by qPCR. Sequencing was performed in single-end mode, generating 50 bases reads (SE50) on an Illumina HiSeq 3000/4000. Quality control of sequencing reads was performed using FastQC version 0.11.8 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Adapter removal was performed using Trim Galore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) version 0.4.1. Reads were then aligned to the murine reference genome mm10/GRCm38 using bismark v0.20.0 (Krueger and Andrews, 2011). Bismark is a specialized tool for mapping bisulfite-treated reads such as the ones generated in RRBS-seq experiments. Bismark requires that the referenced genome first undergoes an in-silico bisulfite conversion while transforming the genome into forward (C → T) and reverse strand (G → A) versions. The reads producing a unique best hit to one of the bisulfite genomes were then compared to the unconverted genome to identify cytosine contexts (CpG, CHG or CHH - where H is A, C or T). The cytosine2coverage and bismark_methylation_extractor modules of Bismark were used to infer the methylation state of all cytosines (for every single uniquely mappable read) and their context, and to compute the percentage methylation. The reported cytosines were filtered to get only the CpGs covered in each sample. The spike-in control sequences were used at this step to check the bisulfite conversion rates and to validate the efficiency of the bisulfite treatment. Methylkit v1.7.0 (Akalin et al., 2012), an R/Bioconductor package, was used to perform the differential methylation analysis between the two groups of samples. The CpG dataset was filtered for low coverage (CpGs with coverage less than 10X in all samples per comparative group were discarded) and for extremely high coverage to discard reads with possible PCR bias (CpGs with coverage higher than the 99.ninth percentile were discarded). The data was then normalized for read coverage distribution between samples. A pairwise comparison was performed for first versus second group of samples to identify differentially methylated CpGs (DMCs) and differentially methylated regions (DMRs), the latter with a window and step size of 1000bp. Methylkit uses logistic regression to compare the methylation percentages between groups at a given CpG/region. All DMCs and DMRs were annotated with the R/Bioconductor package annotation (Cavalcante and Sartor, 2017), with the refGene and CpG island annotations from UCSC. The annotation comprised two categories: (i) distance to a CpG island and (ii) regional annotation. The distance related annotation classified DMCs and DMRs whether they overlapped a known CpG island, 2000 bp of the flanking regions of the CpG islands (shores), 2000 bp of the flanking regions of the shores (shelves) or outside these regions (open sea). The regional analysis classified DMCs and DMRs in four groups, namely, exons, introns, promoters and intergenic regions. Details on the number of total reads obtained, number of uniquely aligned reads, mapping efficiency, number of CpGs detected are available in Table S1.
RRBS coverage
To determine the coverage of a typical RRBS experiment done on the same platform as our RRBS experiment, we used the annotation of common CpGs obtained from a set of 16 murine wildtype samples processed by RRBS (provided by Diagenode). This set of detected CpGs therefore constitutes a consensus and displays the expected localization of CpGs detected by RRBS in mice which we then calculated as percent values for a given genomic or CpG context. To test for statistical enrichment compared to the distribution of DMCs identified in our RRBS experiment, a two-tailed binominal test was chosen.
Orf1p antibody production
Orf1p polyclonal antibodies (rabbit and guinea pig) were produced using the speed 28-day protocol (Eurogentec) after injection of the recombinant full-length Orf1 protein (Eurogenix). The final bleeds were then subjected to a protein-A purification step. The rabbit antibody was used for the detection of the Orf1p protein in Western blots, and the guinea pig was used in immunostainings.
Western blot analysis
Cell lysates were obtained according to the protocol contained in the AllPrep DNA/RNA/Protein Mini Extraction kit (Qiagen), allowing for the simultaneous extraction of DNA, RNA and proteins from tissues. Protein lysates were diluted in Laemmli SDS gel loading buffer and boiled at 95°C for 10 min. Equal amount of protein (10 μg) were separated on a 4–12% SDS-polyacrylamide gel and transferred to an Immobilon-P PVDF membrane (Millipore). Membranes were blocked in 5% non-fat milk/TBS/0.1% Tween 20, immunostained in 5% non-fat milk with the primary antibody, followed by incubation with an appropriate secondary antibody (anti rabbit HRP, cell signaling #7074; 1:2000). ECL detection was carried out with Super Signal Pico West (Pierce) and the chemiluminescence of membranes were imaged with the LAS 4000 image quant, images taken by increment of 30s during 4 min. The following primary antibodies were used: anti- ORF1p (rabbit, in house 1:500) and anti-actin-HRP (Sigma A3854; 1:10,000).
Immunostaining
For immunostaining, animals received a lethal intraperitoneal injection of 1 μL/g body weight dose of Euthasol (150mg/kg) and were then perfused with 8 mL of Phosphate Buffer Saline (PBS) then 8 mL of 4% Paraformaldehyde (PFA) at a rate of 300 mL/hr using a syringe pump. Brains were then post-fixated 1 hr at room temperature (RT) in 4% PFA, washed in PBS three times for 30 min at RT and placed in PBS with 20% sucrose overnight at 4°C. After cryoprotection, the brains were embedded in Tissue Freezing Medium (TFM, Microm Microtech), frozen on dry ice and 30 μm sections of mouse striatum and ventral midbrains encompassing the SNpc were prepared using an HM 560 Microm cryostat (Thermo Scientific).
Slides with 30 μm striatum or midbrain sections were washed in PBS and permeabilized with 1% Triton X-100. After 30 min at RT in 100 μM glycine buffer (for TH/mCherry and TH/ORF1p) or 30 min at 100°C in demasking citrate buffer (10 mM, pH 6, 0.05% Tween) (for TH/MeCP2, H3K9 or γ-H2AX), sections were first blocked in 10% Fetal Bovine Serum (FBS, Gibco) in the presence of 0.5% Triton X-100 for 1 hr at RT and incubated with primary antibodies overnight at 4°C, washed and further incubated with secondary antibodies for 1 hr at RT. The following primary antibodies used: anti-γ-H2AX (mouse, 1/200, Millipore, clone JBW301), anti-TH (chicken, 1/500, Abcam, ab76442), anti-ORF1p (guinea pig, 1/200, in-house, clone 09), anti-mCherry (mouse, 1/200, Clontech 632543), rabbit anti-H3K9me3 (rabbit, 1/200, Abcam, ab8898) and anti-MeCP2 (rabbit, 1/200, Millipore MABE328). Sections were incubated with appropriate secondary antibodies (488 anti-chicken, 546 anti-mouse, 647 anti-guinea pig, 647 anti-rabbit, 647 anti-mouse, Alexa Fluor, Life Technologies)
In situ hybridization
Mice were perfused with PBS in RNase-free conditions, and frozen in isopentane (embedded in TissueTek O.C.T). Brain slices (20 μm) were fixed in 4% PFA in PBS for 10 min at RT and then permeabilized twice for 10 min in RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% Na deoxycholate, 0.1% SDS, 1 mM EDTA, 50 mM Tris– HCl pH 8). Brain sections were fixed again for 5 min in 4% PFA, acetylated for 10 min in 0.25% acetic anhydride (in 0. 1 M triethanolamine, pH 8). Sections were then permeabilized for 30 min in PBS with 1% Triton X-100, 10 min in 10mM citrate buffer pH6 at a 100°C and then pre-incubated for 1 hr at 70°C in hybridization buffer (50% formamide, 5× SSC, 5× Denhardt (1% Ficoll, 1% SSC, 1% Tween 20), 500 μg/mL Salmon sperm DNA, 250 μg/mL yeast tRNA). Sections were then hybridized overnight at 70°C with a digoxigenin (DIG)-labeled RNA probes (DIG RNA labeling kit, Roche 11277073910) for Gadd45b mRNA. Sections were washed with FAM/SSC (50% formamide, 2× SSC, 0.1% Tween 20) twice 30 min at 37°C, then twice in 0.2× SCC at 42°C, blocked in B1 buffer (100 mM maleic acid pH 7.5, 150 mM NaCl) with 10% fetal bovine serum (FBS) for 1 hr, and incubated overnight at 4°C in B1 buffer with alkaline phosphatase-conjugated anti-DIG (1/2000; Roche 11633716001). After three washes in B1 buffer and one wash in B3 buffer (100 mM Tris–HCl pH 9, 50 mM MgCl2, 100 mM NaCl, 0.1% Tween 20) for 15 min, slides were stained using the NBT/BCIP kit (Vector labs, SK5400), stopped with ddH20 and slides were mounted with DAKO-mounting medium.
Imaging/microscopy
All large field images used for TH + neuron quantification, level of viral infection quantification and striatal intensity quantification were made on widefield microscope (Axio zoom V16 – Zeiss – Apotome.2) at 2,3 magnification with a zoom factor of 100. ISH image was taken by upright widefield microscope equipped with a color CCD camera (Nikon 90i microscope) at 20× magnification in brightfield. H3K9me3, MeCP2 and γ-H2AX foci quantification as well as ORF1p intensity quantification were made on images taken by spinning disk microscopy (Yokogawa W1 Spinnning-disk head mounted on an inverted Zeiss AxioObserver Z1) at 63× magnification.
The images in the figures are for illustration purposes and were taken by confocal microscopy (LSM 980 with Airyscan 2, Zeiss) at 63× magnification with a zoom factor of 1.4, except for the MeCP2 and ORF1p images (performed on the spinning-disk microscope).
Cell counting and image quantification
TH cell counting in conditions comparing ipsi- (treated) and contralateral (non-treated) sides were done as follows: For every brain, a minimum of four serial sections were stained, and the number of TH cells was counted in the SNpc of both ipsi- and contralateral sides. An ipsi/contra ratio was calculated for each section, and the resulting mean of four sections was used to quantify the difference between the TH cell number of the ipsi- and contralateral side of the same animal. The counting was done blindly.
The quantification of axonal degeneration in the striatum comparing ipsi- and contralateral sides was done as follows: For every brain, a minimum of seven serial sections were stained, and the integrated density of TH staining intensity was measured in ImageJ by determining the entire contralateral striatum as region of interest (ROI) and conserving area for the measurement of ipsilateral sides. An ipsi/contra ratio was calculated for each section, and the mean ratio of sections containing the striatum was used to quantify the difference between TH striatal intensity of the ipsi- and contralateral side of the same animal. The quantification was done blindly.
Quantifications of foci were performed using a 63× magnification and 0.3 μm -thick successive focal planes except for γ-H2AX foci quantification, which was made using 0.2 μm-thick successive focal planes. Immunostainings of one parameter (H3K9, MeCP2, Etc.) were all done in one experiment for all the conditions. For each immunostaining, three images of the SNpc were taken on three different sections from three mice per condition. The same parameters were set-up on the spinning disk microscope to allow for comparison between experimental conditions for the same staining. Images were analyzed by the same experimenter using ImageJ software. For foci quantifications the foci counting Fiji Plug-in was used. In addition, an image analysis plug-in was developed for the ImageJ/Fiji software, using Bio-Format (openmicroscopy.org), mcib3D and GDSC (Alex Herbert from Sussex University) libraries. First, nuclei that belonged to TH + neurons were manually marked with the plug-in Cell Counter,an xml file for each image containing 3D nuclei coordinates was saved. Then, nucleus channel was filtered with a median filter (radius = 4) and a Difference of Gaussian (DOG) (sigma1 = 30, sigma2 = 15), a binary mask was done with an Otsu threshold. Only nuclei that are associated to the nuclei defined in the xml file were kept. MeCP2 and H3K9me3 foci detections were performed using a median filter (radius = 2), DOG (sigma1 = 10, sigma2 = 2), binary mask was done with a MaxEntropy threshold, then 3D objects (foci) that had a volume comprised between 1.5 and 40 μm3 and were inside TH + nuclei or at a 2 μm distance to nucleus was associated to nuclei. γ-H2AX foci detection was performed using a median filter (radius = 2), DOG (sigma1 = 7, sigma2 = 3), binary mask was done with a Moments threshold, then 3D objects (foci) that had a volume comprised between 0.5 and 100 μm3 were inside TH + nuclei or at a 2 μm distance to nucleus was associated to nuclei. For each nucleus, foci number, average foci volume, average foci integrated intensity and average nucleus integrated intensity was computed.
Gene ontology analysis
Gene ontology analysis (PANTHER version 15.0) was done using the PANTHER overrepresentation test with the GO-Slim annotation datasets ‘biological process’, ‘molecular function’ and cellular component’ and the ‘mus musculus’ gene set as the reference list. Fisher's exact test was used to compute statistical significance of overrepresentation with the false discovery rate (FDR) set at p < 0.05. The first 15 categories, ordered by fold enrichment, are displayed along with the corresponding FDR value.
Quantification and statistical analysis
Unless otherwise stated, the graphs represent the mean of replicates and the error bar the standard error of the mean (SEM). Error bars, values of n and mean ± SEM are as stated in the figure legends. Results were considered as statistically significant for p value <0.05; in some cases, the exact p value is given (∗p < 0.5, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 in all experiments). Normality test were performed prior to the statistical test and unless stated otherwise, the non-parametric two-tailed Mann-Whitney test was applied. All statistical analysis was done with the software Prism.
For the bioinformatic analysis of the RRBS, we formulated the null hypothesis that there are no differences in methylation between the two groups. After the p values have been computed, Methylkit, an R package for DNA methylation analysis and annotation, uses the sliding window model (SLIM) to correct p values to q-values for multiple comparison tests. Statistically significant DMCs and DMRs were identified with a q-value cutoff <0.01 and a methylation difference higher than 25%.
In order to quantify the significance of the overlaps between genes containing hypo-or hypermethylated CpGs at 14d and 90d, we used the calculation of the representation factor using the tool provided on http://nemates.org/MA/progs/overlap_stats.html. We used the MGI international database resource for the laboratory mouse estimate of 24,241 coding genes (http://www.informatics.jax.org/mgihome/homepages/stats/all_stats.shtml) to estimate the total number of mouse genes. The representation factor is the number of overlapping genes divided by the expected number of overlapping genes drawn from two independent groups. A representation factor (RF) > 1 indicates more overlap than expected of two independent groups, a representation factor <1 indicates less overlap than expected, and a representation factor of 1 indicates that the two groups share the number of genes expected for independent groups of genes. To test for significant enrichment between the observed distribution of DMCs upon Gadd45b overexpression and the “consensus”’ CpG coverage expected in an RRBS experiment, we used a two-sided binominal test as the chi-squared test was not applicable due to too small numbers for certain categories.
Acknowledgments
This work was supported by grants from the “Fondation de France” (00086320 to J.F.), the“Fondation du Collège de France” (to J.F.) and the French Agence Nationale de la Recherche (ANR) under reference ANR-20-CE16-0022 NEURAGE (to J.F.). We are grateful to all primary donors for their financial contributions to this work. We thank the "Fondation BettencourtSchueller". We thank the animal facility members for their essential contributions. We gratefully acknowledge Julien Dumont and the Collège de France Orion imaging facility (IMACHEM-IBiSA), member of the French National Research Infrastructure France-BioImaging (ANR-10-INBS-04), which received support from the program «Investissements d’Avenir»ANR-10-LABX-54 MEMOLIFE. We thank Yves Dupraz for the manufacturing of a customized mouse brain slicer.
Author contributions
CRG performed most of the experiments and participated in the writing of the manuscript, OMB contributed experimentally, PM designed the semi-automated image analysis workflow, AP co-supervised the beginning of the study, RLJ co-supervised the study and contributed to the writing of the manuscript, JF co-supervised the study, analyzed the RRBS data, wrote the manuscript and received the funding.
Declaration of interests
The authors declare no competing interests.
Published: July 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102756.
Contributor Information
Rajiv L. Joshi, Email: julia.fuchs@college-de-france.fr.
Julia Fuchs, Email: rajiv.joshi@college-de-france.fr.
Supplemental information
References
- Akalin A., Kormaksson M., Li S., Garrett-Bakelman F.E., Figueroa M.E., Melnick A., Mason C.E. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apulei J., Kim N., Testa D., Ribot J., Morizet D., Bernard C., Jourdren L., Blugeon C., Di Nardo A.A., Prochiantz A. Non-cell autonomous OTX2 homeoprotein regulates Visual cortex plasticity through gadd45b/g. Cereb. Cortex. 2019;29:2384–2395. doi: 10.1093/cercor/bhy108. [DOI] [PubMed] [Google Scholar]
- Arab K., Karaulanov E., Musheev M., Trnka P., fer A.S.X., Grummt I., Niehrs C. GADD45A binds R-loops and recruits TET1 to CpG island promoters. Nat. Genet. 2018:1–13. doi: 10.1038/s41588-018-0306-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga V., Dunnett S.B., Fricker R.A. The 6-OHDA mouse model of Parkinson’s disease - terminal striatal lesions provide a superior measure of neuronal loss and replacement than median forebrain bundle lesions. Behav. Brain Res. 2015;288:107–117. doi: 10.1016/j.bbr.2015.03.058. [DOI] [PubMed] [Google Scholar]
- Bayraktar G., Kreutz M.R. The role of activity-dependent DNA demethylation in the adult brain and in neurological disorders. Front. Mol. Neurosci. 2018;11:169. doi: 10.3389/fnmol.2018.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgnaoui S.M., Gosden R.G., Semmes O.J., Haoudi A. Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer CellInt. 2006;6:13. doi: 10.1186/1475-2867-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benayoun B.A., Pollina E.A., Brunet A. Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nat. Rev. Mol. Cell Biol. 2015;16:593–610. doi: 10.1038/nrm4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson A., Nativio R., Berger S.L., Bonini N.M. Epigenetic regulation in neurodegenerative diseases. Trends Neurosci. 2018;41:587–598. doi: 10.1016/j.tins.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billingsley K.J., Lättekivi F., Planken A., Reimann E., Kurvits L., Kadastik-Eerme L., Kasterpalu K.M., Bubb V.J., Quinn J.P., Kõks S. Analysis of repetitive element expression in the blood and skin of patients with Parkinson’s disease identifies differential expression of satellite elements. Sci. Rep. 2019;9:4369. doi: 10.1038/s41598-019-40869-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blandini F., Armentero M.-T., Martignoni E. The 6-hydroxydopamine model: News from the past. Parkinsonism Relat.Disord. 2008;14:S124–S129. doi: 10.1016/j.parkreldis.2008.04.015. [DOI] [PubMed] [Google Scholar]
- Blaudin de Thé F.-X., Rekaik H., Peze-Heidsieck E., Massiani-Beaudoin O., Joshi R.L., Fuchs J., Prochiantz A. Engrailed homeoprotein blocks degeneration in adult dopaminergic neurons through LINE-1 repression. EMBO J. 2018;37:e97374. doi: 10.15252/embj.201797374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaudin de Thé F.-X., Rekaik H., Prochiantz A., Fuchs J., Joshi R.L. Neuroprotective transcription factors in animal models of Parkinson disease. Neural Plast. 2016;2016:6097107. doi: 10.1155/2016/6097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess R.C., Misteli T., Oberdoerffer P. DNA damage, chromatin, and transcription: the trinity of aging. Curr.Opin.Cell Biol. 2012;24:724–730. doi: 10.1016/j.ceb.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke R.E., O'Malley K. Axon degeneration in Parkinson’s disease. Exp. Neurol. 2013;246:72–83. doi: 10.1016/j.expneurol.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S., Han H.-J., Kohwi-Shigematsu T. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat. Genet. 2003;34:42–51. doi: 10.1038/ng1146. [DOI] [PubMed] [Google Scholar]
- Cancio-Bello A., Saez-Atienzar S. SATB1 is a dopaminergic neuron-specific regulator of cellular senescence. Mov. Disord. 2020;35:235. doi: 10.1002/mds.27978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalcante R.G., Sartor M.A. annotatr: genomic regions in context. Bioinformatics. 2017;33:2381–2383. doi: 10.1093/bioinformatics/btx183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H., Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Chen K., Long Q., Wang T., Zhao D., Zhou Y., Qi J., Wu Y., Li S., Chen C., Zeng X. Gadd45a is a heterochromatin relaxer that enhances iPS cell generation. EMBO Rep. 2016;17:1641–1656. doi: 10.15252/embr.201642402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang Y.-H., Lu A.T., Paul K.C., Folle A.D., Bronstein J.M., Bordelon Y., Horvath S., Ritz B. Longitudinal epigenome-wide methylation study of cognitive decline and motor progression in Parkinson’s disease. J. Parkinsons Dis. 2019;9:389–400. doi: 10.3233/JPD-181549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier T.J., Kanaan N.M., Kordower J.H. Ageing as a primary risk factor for Parkinson’s disease: evidence from studies of non-human primates. Nat. Rev. Neurosci. 2011;12:359–366. doi: 10.1038/nrn3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cecco M., Criscione S.W., Peterson A.L., Neretti N., Sedivy J.M., Kreiling J.A. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging (Albany NY) 2013;5:867–883. doi: 10.18632/aging.100621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasolino M., Liu S., Wang Y., Zhou Z. Distinct cellular and molecular environments support aging-related DNA methylation changes in the substantia nigra. Epigenomics. 2017;9:21–31. doi: 10.2217/epi-2016-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q., Moran J.V., Kazazian H.H., Boeke J.D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Frost B., Hemberg M., Lewis J., Feany M.B. Tau promotes neurodegeneration through global chromatin relaxation. Nat. Neurosci. 2014;17:357–366. doi: 10.1038/nn.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L., Cookson M.R., Petrucelli L., La Spada A.R. Converging pathways in neurodegeneration, from genetics to mechanisms. Nat. Neurosci. 2018;21:1300–1309. doi: 10.1038/s41593-018-0237-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior S.L., Wakeman T.P., Xu B., Deininger P.L. The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 2006;357:1383–1393. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin D.P., Kusumo H., Sharma R.P., Guizzetti M., Guidotti A., Pandey S.C. Gadd45b and N-methyl-D-aspartate induced DNA demethylation in postmitotic neurons. Epigenomics. 2015;7:567–579. doi: 10.2217/epi.15.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin D.P., Sharma R.P., Chase K.A., Matrisciano F., Dong E., Guidotti A. Growth arrest and DNA-damage-inducible, beta (GADD45b)-mediated DNA demethylation in major psychosis. Neuropsychopharmacology. 2012;37:531–542. doi: 10.1038/npp.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjar E.J., Chatoo W., Hanna R., Nkanza P., treault N.T.X., Tse Y.C., Wong T.P., Abdouh M., Bernier G. Heterochromatic genome instability and neurodegeneration sharing similarities with Alzheimer’s disease in old Bmi1+/- mice. Sci. Rep. 2019:1–16. doi: 10.1038/s41598-018-37444-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M., Kuhara S., Takenaka O., Sakaki Y. L1 family of repetitive DNA sequences in primates may be derived from a sequence encoding a reverse transcriptase-related protein. Nature. 1986;321:625–628. doi: 10.1038/321625a0. [DOI] [PubMed] [Google Scholar]
- Hodges A., Strand A.D., Aragaki A.K., Kuhn A., Sengstag T., Hughes G., Elliston L.A., Hartog C., Goldstein D.R., Thu D. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum. Mol. Genet. 2006;15:965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- Hou Y., Dan X., Babbar M., Wei Y., Hasselbalch S.G., Croteau D.L., Bohr V.A. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 2019;15:565–581. doi: 10.1038/s41582-019-0244-7. [DOI] [PubMed] [Google Scholar]
- Jin S.-G., Zhang Z.-M., Dunwell T.L., Harter M.R., Wu X., Johnson J., Li Z., Liu J., Szabó P.E., Lu Q. Tet3 reads 5-carboxylcytosine through its CXXC domain and is a potential guardian against neurodegeneration. Cell Rep. 2016;14:493–505. doi: 10.1016/j.celrep.2015.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Kazazian H.H., Goodier J.L. LINE drive retrotransposition and genome instability. Cell. 2002;110:277–280. doi: 10.1016/s0092-8674(02)00868-1. [DOI] [PubMed] [Google Scholar]
- Kienhöfer S., Musheev M.U., Stapf U., Helm M., Schomacher L., Niehrs C., Schäfer A. GADD45a physically and functionally interacts with TET1. Differentiation. 2015;90:59–68. doi: 10.1016/j.diff.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinde B., Gabel H.W., Gilbert C.S., Griffith E.C., Greenberg M.E. Reading the unique DNA methylation landscape of the brain: non-CpG methylation, hydroxymethylation, and MeCP2. Proc. Natl. Acad. Sci. U S A. 2015;112:6800–6806. doi: 10.1073/pnas.1411269112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F., Andrews S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé C., Lorenzo-Betancor O., Ross O.A. Epigenetic regulation in Parkinson's disease. Acta Neuropathol. 2016;132:515–530. doi: 10.1007/s00401-016-1590-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonté B., Jeong Y.H., Parise E., Issler O., Fatma M., Engmann O., Cho K.-A., Neve R., Nestler E.J., Koo J.W. Gadd45b mediates depressive-like role through DNA demethylation. Sci. Rep. 2019;9:4615. doi: 10.1038/s41598-019-40844-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Li P., Marshall L., Oh G., Jakubowski J.L., Groot D., He Y., Wang T., Petronis A., Labrie V. Epigenetic dysregulation of enhancers in neurons is associated with Alzheimer’s disease pathology and cognitive symptoms. Nat. Commun. 2019;10:2246. doi: 10.1038/s41467-019-10101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Jin Y., Prazak L., Hammell M., Dubnau J. Transposable elements in TDP-43-mediated neurodegenerative disorders. PLoS One. 2012;7:e44099. doi: 10.1371/journal.pone.0044099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Prazak L., Chatterjee N., Grüninger S., Krug L., Theodorou D., Dubnau J. Activation of transposable elements during aging and neuronal decline in Drosophila. Nat. Neurosci. 2013;16:529–531. doi: 10.1038/nn.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Gu T.-P., Weber A.R., Shen J.-Z., Li B.-Z., Xie Z.-G., Yin R., Guo F., Liu X., Tang F. Gadd45a promotes DNA demethylation through TDG. Nucleic Acids Res. 2015;43:3986–3997. doi: 10.1093/nar/gkv283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu E.Y., Russ J., Cali C.P., Phan J.M., Amlie-Wolf A., Lee E.B. Loss of nuclear TDP-43 is associated with decondensation of LINE retrotransposons. Cell Rep. 2019;27:1409–1421.e6. doi: 10.1016/j.celrep.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato M.A., Rodin R.E., Bohrson C.L., Coulter M.E., Barton A.R., Kwon M., Sherman M.A., Vitzthum C.M., Luquette L.J., Yandava C.N. Aging and neurodegeneration are associated with increased mutations in single human neurons. Science. 2018;359:555–559. doi: 10.1126/science.aao4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D.K., Jang M.H., Guo J.U., Kitabatake Y., Chang M.L., Pow-anpongkul N., Flavell R.A., Lu B., Ming G.L., Song H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall L.L., Killinger B.A., Ensink E., Li P., Li K.X., Cui W., Lubben N., Weiland M., Wang X., Gordevicius J. Epigenomic analysis of Parkinson’s disease neurons identifies Tet2 loss as neuroprotective. Nat. Neurosci. 2020;23:1203–1214. doi: 10.1038/s41593-020-0690-y. [DOI] [PubMed] [Google Scholar]
- Martin S.L., Bushman F.D. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell. Biol. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium, Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Narayanan M., Huynh J.L., Wang K., Yang X., Yoo S., McElwee J., Zhang B., Zhang C., Lamb J.R., Xie T. Common dysregulation network in the human prefrontal cortex underlies two neurodegenerative diseases. Mol. Syst. Biol. 2014;10:743. doi: 10.15252/msb.20145304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nativio R., Donahue G., Berson A., Lan Y., Amlie-Wolf A., Tuzer F., Toledo J.B., Gosai S.J., Gregory B.D., Torres C. Dysregulation of the epigenetic landscape of normal aging in Alzheimer’s disease. Nat. Neurosci. 2018;21:497–505. doi: 10.1038/s41593-018-0101-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri F., Rapelli S., Krepelova A., Incarnato D., Parlato C., Basile G., Maldotti M., Anselmi F., Oliviero S. Intragenic DNA methylation prevents spurious transcription initiation. Nature. 2017:1–30. doi: 10.1038/nature21373. [DOI] [PubMed] [Google Scholar]
- Niehrs C., Schäfer A. Active DNA demethylation by Gadd45 and DNA repair. Trends CellBiol. 2012;22:220–227. doi: 10.1016/j.tcb.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Pal S., Tyler J.K. Epigenetics and aging. Sci. Adv. 2016;2 doi: 10.1126/sciadv.1600584. e1600584-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G., Tan J., Garcia G., Kang Y., Salvesen G., Zhang Z. Regulation of histone acetylation by autophagy in Parkinson disease. J. Biol. Chem. 2016;291:3531–3540. doi: 10.1074/jbc.M115.675488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegoraro G., Kubben N., Wickert U., Göhler H., Hoffmann K., Misteli T. Ageing-related chromatin defects through loss of the NURD complex. Nat. Cell Biol. 2009;11:1261–1267. doi: 10.1038/ncb1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzkofer T., Jäger M., Figlerowicz M., Badge R., Mundlos S., Robinson P.N., Zemojtel T. L1Base 2: more retrotransposition-active LINE-1s, more mammalian genomes. Nucleic Acids Res. 2017;45:D68–D73. doi: 10.1093/nar/gkw925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera A., Eisen D., Wagner M., Laube S.K., Künzel A.F., Koch S., Steinbacher J., Schulze E., Splith V., Mittermeier N. TET3 is recruited by REST for context-specific hydroxymethylation and induction of gene expression. Cell Rep. 2015;11:283–294. doi: 10.1016/j.celrep.2015.03.020. [DOI] [PubMed] [Google Scholar]
- Peters A.H., O'Carroll D., Scherthan H., Mechtler K., Sauer S., Schöfer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Pihlstrøm L., Berge V., Rengmark A., Toft M. Parkinson’s disease correlates with promoter methylation in the α-synuclein gene. Mov. Disord. 2015;30:577–580. doi: 10.1002/mds.26073. [DOI] [PubMed] [Google Scholar]
- Pizarro J.G., Cristofari G. Post-transcriptional control of LINE-1 retrotransposition by cellular host factors in somatic cells. Front.Cell.Dev. Biol. 2016;4:14. doi: 10.3389/fcell.2016.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudencio M., Belzil V.V., Batra R., Ross C.A., Gendron T.F., Pregent L.J., Murray M.E., Overstreet K.K., Piazza-Johnston A.E., Desaro P. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat. Neurosci. 2015;18:1175–1182. doi: 10.1038/nn.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekaik H., Blaudin de Thé F.-X., Fuchs J., Massiani-Beaudoin O., Prochiantz A., Joshi R.L. Engrailed homeoprotein protects mesencephalic dopaminergic neurons from oxidative stress. Cell Rep. 2015;13:242–250. doi: 10.1016/j.celrep.2015.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riessland M., Kolisnyk B., Kim T.W., Cheng J., Ni J., Pearson J.A., Park E.J., Dam K., Acehan D., Ramos-Espiritu L.S. Loss of SATB1 induces p21-dependent cellular senescence in post-mitotic dopaminergic neurons. Cell Stem Cell. 2019;25:514–530.e518. doi: 10.1016/j.stem.2019.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu E.J., Angelastro J.M., Greene L.A. Analysis of gene expression changes in a cellular model of Parkinson disease. Neurobiol. Dis. 2005;18:54–74. doi: 10.1016/j.nbd.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Sanchez-Mut J.V., Heyn H., Vidal E., Moran S., Sayols S., Delgado-Morales R., Schultz M.D., Ansoleaga B., Garcia-Esparcia P., Pons-Espinal M. Human DNA methylomes of neurodegenerative diseases show common epigenomic patterns. Transl. Psychiatry. 2016;6:e718. doi: 10.1038/tp.2015.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer A. Gadd45 proteins: key players of repair-mediated DNA demethylation. Adv. Exp. Med. Biol. 2013;793:35–50. doi: 10.1007/978-1-4614-8289-5_3. [DOI] [PubMed] [Google Scholar]
- Simon M., Van Meter M., Ablaeva J., Ke Z., Gonzalez R.S., Taguchi T., De Cecco M., Leonova K.I., Kogan V., Helfand S.L. LINE1 derepression in aged wild-type and SIRT6- deficient mice drives inflammation. Cell Metab. 2019;29:1–21. doi: 10.1016/j.cmet.2019.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I., Thanisch K., Feodorova Y. How to rule the nucleus: Divide et impera. Curr.Opin.Cell Biol. 2016;40:47–59. doi: 10.1016/j.ceb.2016.02.014. [DOI] [PubMed] [Google Scholar]
- Sookdeo A., Hepp C.M., McClure M.A., Boissinot S. Revisiting the evolution of mouse LINE-1 in the genomic era. Mob DNA. 2013;4:1. doi: 10.1186/1759-8753-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup S.-F., Verstreken P. PIWIL1 protein power targets tau therapy. Nat. Neurosci. 2014;17:334–335. doi: 10.1038/nn.3659. [DOI] [PubMed] [Google Scholar]
- Soukup S.-F., Vanhauwaert R., Verstreken P. Parkinson’s disease: convergence on synaptic homeostasis. Embo J. 2018;37:5365. doi: 10.15252/embj.201898960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Södersten E., Toskas K., Rraklli V., Tiklova K., Björklund Å.K., Ringnér M., Perlmann T., Holmberg J. A comprehensive map coupling histone modifications with gene regulation in adult dopaminergic and serotonergic neurons. Nat. Commun. 2018;9:1226. doi: 10.1038/s41467-018-03538-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suberbielle E., Sanchez P.E., Kravitz A.V., Wang X., Ho K., Eilertson K., Devidze N., Kreitzer A.C., Mucke L. Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-β. Nat. Neurosci. 2013;16:613–621. doi: 10.1038/nn.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Samimi H., Gamez M., Zare H., Frost B. Pathogenic tau-induced piRNA depletion promotes neuronal death through transposable element dysregulation in neurodegenerative tauopathies. Nat. Neurosci. 2018;21:1038–1048. doi: 10.1038/s41593-018-0194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trelogan S.A., Martin S.L. Tightly regulated, developmentally specific expression of the first open reading frame from LINE-1 during mouse embryogenesis. Proc. Natl. Acad. Sci. U S A. 1995;92:1520–1524. doi: 10.1073/pnas.92.5.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urdinguio R.G., Sanchez-Mut J.V., Esteller M. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol. 2009;8:1056–1072. doi: 10.1016/S1474-4422(09)70262-5. [DOI] [PubMed] [Google Scholar]
- Van Meter M., Kashyap M., Rezazadeh S., Geneva A.J., Morello T.D., Seluanov A., Gorbunova V. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat. Commun. 2014;5:5011. doi: 10.1038/ncomms6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeponteau B. The heterochromatin loss model of aging. Exp. Gerontol. 1997;32:383–394. doi: 10.1016/s0531-5565(96)00155-6. [DOI] [PubMed] [Google Scholar]
- Wei W., Gilbert N., Ooi S.L., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J.G., Jones B.C., Jiang N., Chang C., Hosier S., Wickremesinghe P., Garcia M., Hartnett D.A., Burhenn L., Neretti N. Chromatin-modifying genetic interventions suppress age-associated transposable element activation and extend life span in Drosophila. Proc. Natl. Acad. Sci. U S A. 2016;113:11277–11282. doi: 10.1073/pnas.1604621113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüllner U., Kaut O., deBoni L., Piston D., Schmitt I. DNA methylation in Parkinson’s disease. J. Neurochem. 2016;139:108–120. doi: 10.1111/jnc.13646. [DOI] [PubMed] [Google Scholar]
- Yang T.-C., Wu P.-C., Chung I.-F., Jiang J.-H., Fann M.-J., Kao L.-S. Cell death caused by the synergistic effects of zinc and dopamine is mediated by a stress sensor gene Gadd45b - implication in the pathogenesis of Parkinson's disease. J. Neurochem. 2016;139:120–133. doi: 10.1111/jnc.13728. [DOI] [PubMed] [Google Scholar]
- Yang X., Han H., De Carvalho D.D., Lay F.D., Jones P.A., Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui D., Miyano M., Cai S., Varga-Weisz P., Kohwi-Shigematsu T. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature. 2002;419:641–645. doi: 10.1038/nature01084. [DOI] [PubMed] [Google Scholar]
- Zahn J.M., Poosala S., Owen A.B., Ingram D.K., Lustig A., Carter A., Weeraratna A.T., Taub D.D., Gorospe M., Mazan-Mamczarz K. AGEMAP: a gene expression database for aging in mice. PLoS Genet. 2007;3:e201. doi: 10.1371/journal.pgen.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Su J., Jeong M., Ko M., Huang Y., Park H.J., Guzman A., Lei Y., Huang Y.-H., Rao A. DNMT3A and TET2 compete and cooperate to repress lineage-specific transcription factors in hematopoietic stem cells. Nat. Genet. 2016;48:1014–1023. doi: 10.1038/ng.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RRBS data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE176544 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE176544). The original western blot image and microscopy data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.