Abstract
This study describes the synthesis and vacuole-inducing activity of 5-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)-1H-indole-2-carbohydrazide derivatives, including five potent derivatives 12c, 12 g, 12i, 12n, and 12A that exhibit excellent vacuole-inducing activity. Remarkably, 12A effectively induces methuosis in tested cancer cells but not human normal cells. In addition, 12A exhibits high pan-cytotoxicity against different cancer cell lines but is hardly toxic to normal cells. It is found that the 12A-induced vacuoles are derived from macropinosomes but not autophagosomes. The 12A-induced cytoplasmic vacuoles may originate from the endoplasmic reticulum (ER) and be accompanied by ER stress. The MAPK/JNK signalling pathway is involved in the 12A-induced methuotic cell death. Moreover, 12A exhibits significant inhibition of tumour growth in the MDA-MB-231 xenograft mouse model. The excellent potency and selectivity of 12A prompt us to select it as a good lead compound for further development of methuosis inducers and investigation of the molecular and cellular mechanisms underlying methuosis.
Keywords: 1H-indole-2-carbohydrazide derivatives, Methuosis, Anticancer agent, MDA-MB-231 cells
Graphical Abstract
HIGHLIGHTS
Novel methuosis-inducing anticancer agents are designed and synthesised.
Vacuole inducers 12i and 12A display high anticancer activity and little-toxic to normal cells.
Compound 12A is a selective methuosis inducer involving the activation of MAPK/JNK signalling pathway.
Compound 12A effectively inhibits tumour growth in the MDA-MB-231 xenografted mouse model.
1. Introduction
To date, numerous anticancer agents have been developed and some have been widely used in clinical therapy, but cancer remains one of the leading causes of death worldwide1–3. In general, the existing anticancer drug treatments are mainly based on inducing programmed cell death (PCD). However, tumour suppressor genes controlling PCD in cancer cells are often mutated, which can render cancer cells relatively insensitive to apoptosis and eventually confer them a multidrug resistance phenotype to these conventional chemotherapeutic drugs typically targeting apoptosis. Therefore, small molecules killing cancer cells via non-apoptotic cell death mechanisms are considered as a promising therapeutic strategy for those cancers that are insensitive to apoptosis-inducing drugs. It has been demonstrated that methuosis plays a key role in various physiological processes, including neural development and the death of retinal pigment epithelial cells1,2. In addition, methuosis has been reported as an important non-apoptotic cell death pathway3–5, and thus, methuosis inducers have emerged as a novel class of potential therapeutic agents for cancer treatment, especially the treatment of therapy-resistant cancers. Several types of methuosis inducers with different scaffolds are shown in Figure 16–12. Among them, the (E)-3-(1H-indol-3-yl)-1-(pyridin-4-yl)prop-2-en-1-one (IPP) derivatives have emerged as a promising type of methuosis inducer7,11. Several IPP molecules, including 3–(2-methyl)-IPP (MIPP) and 3–(5-methoxy-2-methyl)-IPP (MOMIPP), have been developed as potent methuosis inducers13. MIPP and MOMIPP could induce vacuolization and lead to non-apoptotic cell death in various cancer cell lines, including non-resistant (e.g. LN229, MCF7, U251, MDA-MB-468, A549) and resistant (adriamycin-resistant MCF7 and temozolomide-resistant U251) cells. Initially, it was found that the indolyl and pyridinyl moieties of IPP molecules exhibited a high degree of structural specificity for the induction of methuosis. Since then, extensive research has been conducted on the analysis of the quantitative structure-activity relationship (QSAR) of IPP derivatives, focussing on the substitutions at the 6- and 2-indolyl position8,13. Notably, in 2018, one azaindole derivative (compound 13) was identified as a highly selective and potent methuosis inducer, which exhibited a remarkable tumour-suppressive effect in the MDA-MB-231 xenograft mouse model10. In recent years, more and more studies have shown that the therapeutic use of methuosis-inducing agents can be considered as a potential anticancer strategy14.
Figure 1.
The chemical structures of several representative methuosis inducers.
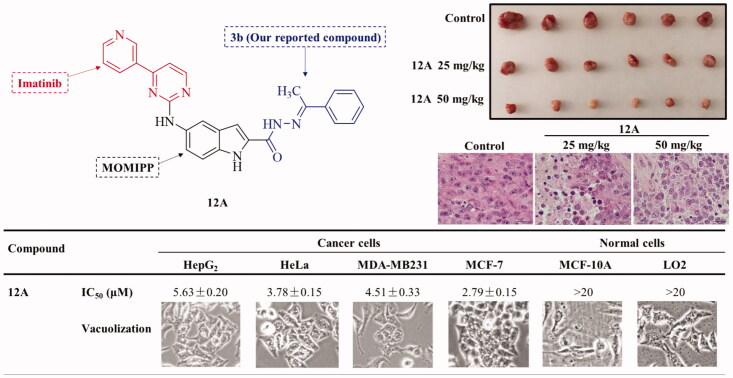
Many compounds containing the 4-(pyridin-3-yl)pyrimidine moiety show good anticancer activity15–19. For example, imatinib, a representative anticancer agent with the 4-(pyridin-3-yl)pyrimidine moiety, is the first thymidine kinase inhibitor to be introduced into the market (in 2002 under the name “Glivec”) and is still being used against certain types of cancer, such as chronic and acute leukaemia. In addition, we previously found that the 1H-indole-2-carbohydrazide derivative 3 b was a good anticancer agent20. Thus, we developed an idea to find new methuotic anticancer agents that involved combining key pharmacophores of imatinib, MOMIPP, and 3 b to produce three classes of target compounds (series A, B, and C) (Figure 2). Firstly, we synthesised the three classes of target compounds (series A, B, and C) and then evaluated them for their vacuolization effect and antitumor activity. Notably, several synthesised compounds (e.g. 12 g, 12i, 12n, 12A) were identified as potent vacuolization-inducing agents with excellent antitumor activity and low toxicity to normal cells in vitro. Furthermore, in vivo antitumor efficacy of compound 12A was evaluated in an MDA-MB-231 xenograft mouse model, and its mechanisms of antitumor activity were probed, by assessing its methuosis, autophagy, and apoptosis inducing effects. In summary, 12A, which showed potent cytotoxic activity against a broad panel of cancer cell lines but low toxicity to human normal cells, was selected as a promising lead compound for further development as a potent methuosis inducer for use in cancer therapy.
Figure 2.
Design and modification strategies of target compounds.
2. Results and discussion
2.1. Chemistry
The synthetic routes of target compounds are outlined in Scheme 1 and Scheme 2. First, we synthesised (E)-N′-methylene-4–(3–(4-methyl-3-(4-(pyridin-3yl)pyrimidin-2- yl)amino)phenyl)ureido)-1H-indole-2-carbohydrazide derivatives (8a–8f). As shown in Scheme 1, the general chemistry for the synthesis of 8a–8f was adapted from previously reported methods20–22. Briefly, 1–(4-nitrophenyl)hydrazine (1) was refluxed with ethyl 2-oxopropanoate in ethanol to give compound 2, which then underwent ring closure with polyphosphoric acid to form intermediate 3. Catalytic hydrogenation of the nitro group of compound 3 with palladium on carbon (Pd/C) afforded 4, which was followed by reaction with triphosgene in tetrahydrofuran (THF) to produce compound 5. Subsequently, the intermediate 5 reacted with 6-methyl-N1-(4-(pyridin-3-yl)pyrimidin-2-yl)benzene-1,3-diamine to give ethyl 4–(3-(4-methyl-3-((4-(pyridin-3-yl)-pyrimidin-2-yl)amino)phenyl)ureido)-1H-indole-2-carboxylate (6). Compound 6 was treated with hydrazine hydrate in refluxing ethanol to afford 7, which was subjected to reactions with various aldehydes to produce the corresponding compounds 8a-8f.
Scheme 1.
Synthetic route of target compounds 8a–8f (Series A). Reagents and conditions: (a) ethyl 2-oxopropanoate, EtOH, reflux, for 2 h; (b) polyphosphoric acid, at 100 °C, for 1 h; (c) 10% Pd/C, H2; (d) triphosgene, THF, triethylamine at 0 °C, for 4 h; (e) 6-methyl-N1-(4-( pyridin-3-yl) pyrimidin-2-yl)-benzene-1,3-diamine, toluene, at 60 °C, 4 h; (f) hydrazine hydrate, EtOH, reflux, for 6 h; (g) RCHO, EtOH, reflux, for 8 h.
Scheme 2.
Synthetic route of target compounds 12a–12r (Series B) and 12A–12F (Series C) Reagents and conditions: (h) ① H2NCN, concentrated HCl, EtOH, reflux, for 24 h, ② NH4NO3 (aq); (i) 3-(dimethyl amino)-1–(3-pyridinyl)prop-2-en-1-one, NaOH, EtOH, reflux, for 72 h; (j) hydrazine hydrate, EtOH, reflux, for 6 h; (k) RCHO/RCOCH3, EtOH, reflux, for 8 h.
The synthesis of compounds 12a–12r (Series B) and compounds 12A–12F (Series C) is shown in Scheme 2. The intermediate 10 was obtained from compound 423. Compound 4 was refluxed with cyanamide in ethanol with concentrated HCl for 24 h, then treated with NH4NO3 to produce ethyl 5-guanidino-1H-indole-2-carboxylate (9). The intermediate 9 was reacted with 3-(dimethylamino)-1–(3-pyridinyl)prop-2-en-1-one in the presence of NaOH under reflux condition in ethanol for 72 h to give 10 which was then converted to 11 upon reaction with hydrazine hydrate in refluxing ethanol. Ultimately, compound 11 was separately reacted with different aldehydes or methyl ketones in refluxing ethanol to produce the corresponding target compounds of series B (12a–12r) and series C (12A–12F), respectively.
2.2. Biological activity
2.2.1. Identification of effective vacuolization inducers with anticancer activity and analysis of the structure-activity relationships
To identify effective methuosis inducing agents for use as anticancer drugs, both the vacuolization effect and antiproliferative activities of target compounds 8a–8f, 12a–12r, and 12A–12F were evaluated in HeLa cells. HeLa cells were treated with different compounds with a low concentration (1.0 µM) for 8 h to evaluate the vacuolization-inducing effect of target compounds. Similarly, the antiproliferative effect of all synthesised compounds at 1.0 and 20.0 µM was evaluated in HeLa cells after 48 h treatment. MOMIPP was used as a positive control. The vacuolization effect and cell growth inhibition results for all target compounds are summarised in Tables 1–3.
Table 1.
Summary of the vacuolization effect and cell growth inhibition evaluation results for compounds 8a–8f (Series A) in HeLa Cells.
 |
Table 2.
Summary of the vacuolization effect and cell growth inhibition evaluation results for compounds 12a–12r (Series B) in HeLa Cells.
 |
Table 3.
Summary of the vacuolization effect and cell growth inhibition evaluation results for compounds 12A–12F (Series C) in HeLa Cells.
 |
First, we investigated the functionality of the key pharmacophores from imatinib at the 4-position of the indole ring. Compounds 8a–8f (Table 1) had no growth inhibitory (GI) activity against Hela cells at the tested concentration range and also did not induce vacuolization at 1.0 µM. However, when the 1-(p-tolyl)urea moiety was deleted and the residue (4-(pyridin-3-yl)pyrimidin-2-yl)amino was transferred to the 5-position of the indole ring, there was a significant increase in the vacuolization-inducing effect. For example, treatment with 8a, 8 b, and 8c had no vacuolization-inducing effect (Table 1), whereas compounds 12c, 12d, and 12p showed a vacuolization-inducing effect to various degrees at 1.0 µM (Table 2), with a vacuolization ratio of 89 ± 0.55%, 74 ± 0.56%, and 73 ± 1.56%, respectively. These data suggested that the functional group should be substituted at the 5-position of the indole ring and the 1-(p-tolyl)urea moiety might be unhelpful for vacuole-inducing effect.
Next, we turned our attention to Series B compounds of the acyl hydrazones (R3=H). More than half of the Series B compounds exhibited a vacuolization-inducing effect on HeLa cells at 1.0 µM (Table 2). Among the alkyl-substituted compounds, 12c (n-butyl) induced extensive vacuolization, while 12a (ethyl) and 12 b (propyl) completely lost the ability to induce vacuolization. These results suggested that the chain space volume of n- alkyl group at R2 plays an important role in determining the vacuolization-inducing effect. Compound 12c had an excellent vacuolization-inducing effect, but a weak antiproliferative effect. Accordingly, we replaced the n-butyl group with a phenyl group to afford compound 12d, which induced moderate vacuolization and showed a modest antiproliferative effect. When the R2 moiety was the ortho-substituted phenyl group, compounds with 2-chlorophenyl (12e), 2-hydroxyphenyl (12f), and 2-methoxylphenyl (12 g) substitutions, showed different vacuolization-inducing effects with vacuolization rate values at 1.0 µM of 41 ± 0.91%, 3 ± 0.81%, and 98 ± 0.45%, respectively. These results indicated that the introduction of a strong electron-donating group (–OCH3) on the 2-positon of the benzene ring was beneficial for the induction of vacuolization, which might be mainly due to the formation of electronic interactions between the R2 group and the target(s). However, para-substituted phenyl acyl hydrazones, such as 12i (4-methylphenyl) exhibited potent vacuolization-inducing effect with vacuolization rate values at 1.0 µM of 94 ± 3.21%, while 12 h (4-methoxylphenyl) showed almost no vacuolization-inducing effect. This result further confirmed that the size of R2 is crucial for maintaining the inhibitory effect. In compounds 12k–12o which contain di- or tri- substituted phenyl ring (R2), the substituted positions of the substitutes showed an important relationship with the vacuolization-inducing effect. Compounds 12k (3-hydroxy-4-methoxylphenyl) and 12 l (2,3-dimethoxylphenyl) showed a similar level of effect. The 3,5-dimethoxy derivative, compound 12n, showed a dramatically increased vacuolization effect at 1.0 µM and exhibited potent inhibition of cell growth at 20.0 µM. However, treatment with compound 12 m (2,4-dimethoxylphenyl) had a weak vacuolization-inducing effect on HeLa cells. Compound 12o with the 3,4,5-trimethoxylphenyl substitution also lost the ability to induce vacuolization at 1.0 µM. Taken together, the shape of the R2 group plays a crucial role in determining the vacuolization-inducing effect. In addition, we investigated the impact of other aromatic groups at R2 on the vacuolization-inducing effect. A comparison of the effects of compounds 12p and 12q with 12d, as shown in Table 2, revealed that the five-membered aromatic heterocycle substitutions, such as thiophen-2-yl and furan-2-yl groups, had a similar effect as a phenyl group, but the replacement of the phenyl group with a 4-methoxynaphthalen-1-yl moiety resulted in the complete loss of the effect. The results also indicated that the introduction of the electron-rich aromatic ring has a beneficial effect on vacuolization and the size and shape of the R2 group play a key role in determining the vacuolization-inducing effect.
To further investigate the influence of the acyl hydrazone substituents on the biological effect of the pharmacophore, we replaced the two hydrogens of the acyl hydrazone methylene with methyl and (un)substituted phenyl groups, obtaining compounds 12A–12F. The vacuolization effect and cell growth inhibition of compounds 12A–12F in HeLa cells are summarised in Table 3. Surprisingly, 12A (R2=phenyl, R3=CH3) showed a significantly stronger effect than 12d (R2=phenyl, R3=H) in both the growth inhibition and vacuolization assays, but 12 D (R2=4-methylphenyl, R3=CH3) dramatically decreased vacuolization and the inhibitory effect on cell growth compared with 12i (R2=4-methylphenyl, R3=H). These results indicated that the global shape and size of the substituents R2 and R3 played an important role in determining the vacuolization-inducing effect, which might be determined by the joint formation of the key interactions of the two substituents of methylene with their targets(s). In addition, we found that the replacement of the phenyl group with 4-chlorphenyl (12B) and 4-nitrobenzene (12 C) moieties also significantly reduced the vacuolization-inducing effect at 1.0 µM. Additionally, when the R3 moiety was the di-substituted phenyl, compounds with 2-hydroxy-5-bromophenyl (12E) and 2-hydroxy-5-methoxyphenyl (12 F) substitutions completely lost the ability to induce vacuolization, but 12E caused a large reduction in the cell growth rate at the concentration of 20 µM. These results further demonstrated that the global shape and size of the substituents R2 and R3 were crucial for maintaining the vacuolization-inducing effect. Finally, the preliminary structure and activity relationships are summarised in an illustration in Figure S1 in the Supporting Information.
Overall, five compounds (12c, 12 g, 12i, 12n, and 12A) at 1.0 µM showed similar vacuolization-inducing effects as the reference compound MOMIPP, thus these five compounds were selected for further evaluation of their antiproliferative activities against human cancer and normal cells using the MTT assay based on the degradation of 3–(4, 5-dimethylthiazol- 2-yl) −2,5-diphenyltetrazolium bromide (MTT). As shown in Table 4, compound 12c did effectively induce vacuolization at 1.0 µM but did not affect the cell growth rate of cancer and normal cells, with half-maximal inhibitory concentration (IC50) values above 50 µM. However, the other four compounds showed both significant vacuolization-inducing effect and a broad spectrum of cytotoxicity against cancer cells with IC50 values in the low micromolar range, but low cytotoxicity to normal cells. This finding implies that one aryl substitution on acyl hydrazone methylene plays a key role in the antitumor activity.
Table 4.
In vitro antiproliferative effects of selected compounds against cancer and normal cells
| ID | Cancer cells (IC50, μM) |
Normal cells (IC50, μM) |
||||
|---|---|---|---|---|---|---|
| HepG2 | HeLa | MDA-MB-231 | MCF-7 | MCF-10A | LO2 | |
| 12c | >20 | >20 | >20 | >20 | >50 | >50 |
| 12g | 10.86 ± 0.20 | 9.75 ± 0.21 | 8.88 ± 0.41 | 3.36 ± 0.17 | >50 | >50 |
| 12i | 8.25 ± 0.15 | 6.43 ± 0.18 | 3.81 ± 0.08 | 2.70 ± 0.13 | >50 | >50 |
| 12n | 7.66 ± 0.24 | 7.46 ± 0.07 | 13.57 ± 0.06 | 2.84 ± 0.08 | >50 | >50 |
| 12A | 5.63 ± 0.20 | 3.78 ± 0.15 | 4.51 ± 0.33 | 2.79 ± 0.15 | 185.7 ± 0.51 | 50.19 ± 0.35 |
| Imatinib | 28.08 ± 0.25 | 37.30 ± 0.22 | 28.19 ± 0.24 | 40.08 ± 0.35 | 39.37 ± 0.42 | 87.67 ± 0.31 |
| MOMIPP | 3.6 ± 0.045 | 5.2 ± 0.05 | 4.98 ± 0.11 | 4.5 ± 0.18 | >50 | >50 |
Cells were treated with indicated compounds for 48 h. Data are means ± SD of triplicate experiments.
2.2.2. Compound 12A effectively induces cytoplasmic vacuolization in cancer cells
After considering both the vacuolization and anticancer effects (Table 4), we selected 12A for further biological assays. First, the ability of 12A to induce vacuolization in different types of cells was evaluated and the morphological changes were examined under a microscope. As shown in Figure 3(A), the number and the size of vacuoles increased markedly in 12A-treated HeLa cells with increasing treatment time from 1 to 24 h. After treatment with 1 µM 12A for 1 h, small vesicles appeared in HeLa cells. The vesicles accumulated, fused, and formed larger vacuoles after treatment with 1 µM 12A for 16 h, becoming larger and more numerous after treatment for 24 h. Therefore, compound 12A induced vacuolization in a time-dependent manner. Similarly, we also evaluated the effect of the treatment with 12A at different concentrations on vacuolization in HeLa cells. As shown in Figure 3(B), vacuoles formation could be observed after treatment with 0.1 µM 12A. Vacuoles accumulated in HeLa cells after treatment with 12A at concentrations ranging from 0.1 to 2 µM, implying that 12A dose-dependently induced vacuole formation in HeLa cells. These results suggest that 12A is an effective vacuolization inducer in Hela cells, which can induce vacuole formation in a time- and dose-dependent manner. We also found that compound 12A effectively induced vacuole formation in Hela cells with a half-maximal effective concentration (EC50) value of 0.26 ± 0.02 µM, which was comparable to MOMIPP (EC50 = 0.18 ± 0.05 µM). Then, to verify whether the 12A-induced vacuolization also occurs in other cell lines, seven other different cancer cell lines (MDA-MB-231, HepG2, MCF-7, NT2, A549, A875, and A375) and three human normal cell lines (HaCat, MCF-10A, and LO2 cells) were evaluated after treatment with 1 µM 12A for 8 h. As shown in Figure 3(C), 12A induced vacuolization, to different extents, in the seven tested cancer cell lines, but not in the three normal cell lines. These data indicate that 12A is an effective vacuolization inducer in cancer cells, but has almost no effect on normal human cells. Therefore, 12A is a potent and selective vacuolization-inducer, which is consistent with the cell cytotoxicity analysis.
Figure 3.
Compound 12A effectively induces cytoplasmic vacuolization in cancer cells. (A) HeLa cells were treated with 1 µM 12A, and imaged by microscopy at the indicated time. (B) HeLa cells were treated with 12A for 8 h at the indicated concentrations, then imaged by microscopy. (C) Different cancer cells were treated with 1 µM 12A for 8 h and normal human HaCat, MCF-10A, and LO2 cells were used as negative controls.
2.2.3. Compound 12A induces the caspase-independent formation of macropinosome-derived cytoplasmic vacuoles
Since compound 12A effectively inhibited cancer cell proliferation, it would be interesting to know whether 12A induces PCD, such as cell apoptosis and cell cycle arrest. We found that the 12A-induced vacuolization was not inhibited by Z-VAD-FMK (a pan-caspase inhibitor) in HeLa cells (Figure 4(A)). In addition, it was found that the cleaved PARP was hardly detected after treatment with 12A at low concentrations (<5 µM) (Supporting Information Figure S2). Consistently, only slight apoptosis was detected after the treatment with 12A at concentrations below 5 µM (Supporting Information Figure S2). These findings indicate that the 12A-induced vacuolization and cell growth inhibition might not be PARP cleavage-dependent. Additionally, very slight effects of 12A on the cell cycle were observed in MDA-MB-231 cells (Supporting Information Figure S3). These data suggest that cell cycle arrest is not significantly involved in the inhibition of cancer cell proliferation by treatment with 12A.
Figure 4.
The effect of different compounds on 12A-induced vacuolation. (A) A pan-caspase inhibitor (Z-VAD-FMK). (B) Two autophagy inhibitors (3-methyladenine (3-MA) and LY294002) and an autophagy activator (rapamycin). (C) Two macropinosome formation inhibitors bafilomycin A1 (Baf-A1) and cycloheximide (CHX) were used to treat HeLa cells alone or co-treated HeLa cells treated with 12A, as indicated for 8 h, cells were imaged by microscopy.
However, considering that in autophagic cells there are some autophagosome-like vacuoles, which sequester damaged cytoplasmic proteins and organelles, we also evaluated the effect of autophagy inhibitors and activators on 12A-induced vacuoles. The results revealed that the 12A-induced vacuolization did not decrease when cells were pre-treated with two autophagy inhibitors, namely 3-methyladenine (3-MA) and LY294002 (Figure 4(B)). In addition, the autophagy activator rapamycin also failed to promote the 12A-induced vacuolization (Figure 4(B)). These results suggest that the 12A-induced vacuoles are not the autophagic vacuoles (AVs).
Methuosis is characterised by the accumulation of cytoplasmic vacuoles derived from macropinosomes. It has been reported that protein synthesis is needed during macropinosome formation. Bafilomycin A1 (Baf-A1), a selective inhibitor of vacuolar-type H + ATPase (V-ATPase), can inhibit nascent macropinosome formation and restore the vacuoles to normal morphology by inhibiting acid influx in cells24. Cycloheximide (CHX) is a protein synthesis inhibitor that can disrupt the process of macropinosome formation24. Accordingly, HeLa cells were treated with 12A or in combination with Baf-A1 or CHX for 8 h, and the morphological changes were then imaged. As shown in Figure 4(C), both CHX and Baf-A1 dramatically blocked the formation of vacuoles, indicating that 12A was a methuosis inducer.
2.2.4. The MAPK/JNK pathway is involved in 12A-induced methuosis
We also examined the methuotic features of 12A-induced vacuolization-mediated cell death. First, examination of the cells by transmission electron microscopy (TEM) revealed ER swelling after treatment with 12A (Figure 5(A)), implying that the compound 12A-induced cytoplasmic vacuoles may originate from the ER and be accompanied by ER stress. To evaluate whether ER stress is involved in 12A-induced methuosis, the mRNA expression level of the endoplasmic reticulum (ER) stress marker CHOP was measured. As shown in Figure 5(B), treatment with 12A increased the CHOP mRNA level in a dose-dependent manner. Together, these data support that 12A induces macropinosome-derived vacuoles. Methuosis can be induced by activation of the MAPK/JNK signalling pathway and the insulin-like growth factor I receptor signalling pathway25–27. Therefore, we evaluated the levels of JNK, p-JNK, ERK1/2, and p-ERK1/2 in HeLa cells and MDA-MB-231 cells with or without 12A treatment. The results shown in Figure 5(C) revealed that p-JNK and p-ERK1/2 were dose-dependently increased in 12A-treated HeLa and MDA-MB-231 cells. The increase of p-JNK and p-ERK1/2 after 12A treatment suggested that the activation of the MAPK/JNK signalling pathway was involved in 12A-induced vacuolization. To further confirm whether the 12A-induced vacuole formation required the activation of the MAPK/JNK signalling pathway, the effects of the MAPK/JNK inhibitors PD98059 and SP600125 were assessed. HeLa cells were pre-treated with these two inhibitors for 2 h, then treated with 12A for 8 h. As shown in Figure 5(D), vacuolization in HeLa cells significantly decreased in the presence of MAPK/JNK inhibitors. These findings imply that the MAPK/JNK signalling pathway is involved in 12A-induced methuosis, demonstrating that 12A-induced vacuoles are the hallmark of methuosis.
Figure 5.
Compound 12A activates the MAPK/JNK signalling pathway and induces methuosis-like vacuoles originated from the ER and accompanied by ER stress. (A) Transmission electron microscopy (TEM) image of HeLa cells after treatment with 12A for 8 h. Red arrows: the ER (red arrows) in HeLa cells. Scale bar: 0.5 µm for 3,000x and 0.1 µm for 10,000x. (B) The CHOP mRNA levels in HeLa cells after treatment with 12A for 8 h. (C) The levels of the indicated proteins in the MAPK/JNK signalling pathway after treatment with 12A for 24 h. (D) The effect of the ERK1/2 inhibitor PD98059 and JNK inhibitor SP600125 on 12A-induced methuosis after treatment for 8 h.
2.2.5. Compound 12A inhibits tumour growth in vivo
The above experimental results imply that compound 12A is a novel potent vacuolization-inducing agent that promotes methuosis-like cell death in different cancer cells (HeLa, HepG2, MDA-MB-231, and MCF-7), but not in normal human cells (HaCat, MCF-10A, and LO2). Remarkably, 12A also shows a broad spectrum of cytotoxicity against cancer cells with IC50 values below 6.0 µM but does not affect non-tumorigenic cell lines, like MCF-10A and LO2 (IC50>50 µM). In particular, 12A exhibits more than 40-fold selectivity between MDA-MB-231 (IC50 = 4.51 ± 0.33 µM) and MCF-10A cells (IC50 = 185.7 ± 0.51 µM). Triple-negative breast cancer (TNBC) is currently one of the trickiest tumours for clinical treatment28,29. In this study, we established a TNBC xenograft model in mice based on the implantation of MDA-MB-231 cells to evaluate the in vivo antitumor effects of 12A. The effects of 12A on MDA-MB-231 cell-mediated tumour progression are shown in Figure 6(A–C). The results showed that the treatment with 12A substantially suppressed tumour growth, achieving 72.6% (50 mg/kg, i.p.) and 57.3% (25 mg/kg, i.p.) tumour growth inhibition (TGI). Also, 12A-treatment did not cause a significant loss of body weight (Figure 6(D)). Additionally, we investigated the effects of 12A on tumour cell morphology using haematoxylin and eosin (H&E) staining (Figure 6(E)). The control sample contains denser packed tumour cells with normal morphological features, while in the 12A-treated samples the tumour cells are loosely and irregularly arranged and many necrotic cells are present showing aberrant nuclear morphology, such as karyolysis and pyknosis. Furthermore, in 12A-treated tumour tissues (especially high-dose treated samples), there is extensive severe necrosis that appears in light pink cells without colourable nuclei, and results in more interspace. Together, these findings suggest that compound 12A has a strong anticancer effect and good tolerability in vivo, indicating that 12A can be used as a potential methuosis inducer for the treatment of cancers, such as TNBC.
Figure 6.
Compound 12A suppresses MDA-MB-231 tumour growth in vivo. (A–B) Nude mice with MDA-MB-231 xenografts were intraperitoneally (i.p.) injected daily with vehicle, 12A (25 mg/kg or 50 mg/kg) for 13 days. Tumours were separated and measured. Tumour size and weight in control and 12A-treated mice were compared. (C–D) Tumour volume and body-weight of mice during treatment. Tumour volume and body weight were measured every day. Error bars indicate mean tumour volume ± SEM and mean body weight ± SEM (n = 6 animals in each group). (E) H&E staining in untreated tumours or after 13 days of the treatment with 12A. Red arrows indicate vacuoles.
3. Conclusion
In summary, three series of pyridine-pyrimidine-indole-carbohydrazide derivatives were designed, synthesised, and evaluated for biological effects in vitro and in vivo. In HeLa cells, the majority of the synthetic analogs exhibited potential vacuolization-inducing effect at 1 µM, and/or had significant antiproliferative activity at 20 µM. We investigated the preliminary structure-activity relationship (SAR) and found four potent compounds (12 g, 12i, 12n, and 12A) that had both similar anti-proliferative and vacuole-inducing effects as MOMIPP. Besides, in HeLa cells, the representative compound 12A induced vacuolization in the other seven cancer cell lines tested, including MDA-MB-231, but not in the three human normal cell lines tested, including MCF-10A, which was consistent with its antiproliferative activity (cancer cell lines: IC50<6 µM; normal cell lines: IC50>50 µM). A subsequent mechanism study demonstrated that compound 12A induced caspase-independent cytoplasmic vacuolization and the vacuoles were derived from macropinosomes, instead of autophagosomes. Compound 12A was also found to induce methuosis through the MAPK/JNK signalling pathway. Furthermore, the results obtained in the MDA-MB-231 xenograft mouse model suggested that 12A could significantly suppress the tumour growth and showed obvious accumulation of vacuoles in tumour tissues, but had almost no influence on the body weight. Taken together, our results demonstrate that 12A acts as a methuosis inducer and has obvious anticancer effects, and lower toxicity to normal cells/tissues. Thus, 12A is a potential candidate for the development of novel anticancer treatment strategies, especially for TNBC treatment. Moreover, 12A can serve as a lead compound to make further structural optimisation. Our main future goals are to develop highly selective and low toxic methuosis-based anticancer agents and identify the targets of these methuosis-inducing agents.
4. Experimental section
4.1. Chemistry
All chemicals and solvents were of analytical grade, and were purchased commercially and used without further purification. All melting points were measured with a SGW X-4 micro-melting point apparatus (INESA Co., Ltd., Shanghai, China) and were uncorrected. High-resolution mass spectrometry (HRMS) was performed on a Q-Exactive mass spectrometer (Thermo Fisher Scientific Inc., Waltham, MA, USA) equipped with an electrospray ionisation source (ESI). Proton nuclear magnetic resonance (1H-NMR) and 13 C-NMR spectra were performed on a Bruker Avance 600 MHz spectrometer (Bruker Biospin GmbH, Rheinstetten, Germany). The purity of synthesised compounds was determined by high-performance liquid chromatography (HPLC) performed on an Agilent 1100 Series HPLC system (Agilent Technologies, Santa Clara, CA, USA) using a COSMOSIL5C18-MS-II Column (4.6ID × 250 mm). The mobile phase was water (A) and acetonitrile (B) in a linear gradient model as follows: (A) from 95 to 0% and (B) from 5 to 100% during 0–30 min. The flow rate was 1 ml·min−1, and the detection wavelengths were 254 and 365 nm.
4.1.1. Synthesis of intermediates 2–5
Compounds 2–5 were prepared by previously described methods29.
4.1.2. Synthesis of intermediate 6
A mixture of ethyl 5-isocyanato-1H-indole-2-carboxylate 5 (0.4 mmol, 0.092 g) in toluene (5 ml) and 6-methyl-N1-(4-(pyridin-3-yl)pyrimidin-2-yl)benzene-1,3-diamine (0.4 mmol, 0.11 g) was heated at 65 °C for 6 h. Then, the mixture was concentrated, and the resulting crude product was purified by flash chromatography on silica gel, eluting with the appropriate mixtures of CH2Cl2 and MeOH to obtain intermediate 6 as a white solid with an 80% yield. 1H NMR (600 MHz, DMSO-d6): δ 11.76 (s, 1H), 9.30 (s, 1H), 8.91 (s, 1H), 8.70 (d, J = 5.1 Hz, 1H), 8.56 (s, 1H), 8.53–8.48 (m, 3H), 7.85 (s, 1H), 7.80 (s, 1H), 7.53 (dd, J = 5.1, 8.1 Hz, 1H), 7.44 (d, J = 5.1 Hz, 1H), 7.37 (d, J = 8.1 Hz, 1H), 7.24 (dd, J = 2.0, 8.1 Hz, 1H), 7.18–7.15 (m, 1H), 7.15–7.11 (m, 1H), 7.06 (s, 1H), 4.33 (q, J = 7.2 Hz, 2H), 2.20 (s, 3H), 1.34 (t, J = 7.2 Hz, 3H); 13 C NMR (150 MHz, DMSO-d6):δ 162.1 (C=O), 161.7 (C=O), 60.8 (CH2), 18.0 (CH3), 14.8 (CH3), aromatic carbon atoms (23 C) gave signals at 161.6, 159.9, 153.4, 151.8, 148.7, 138.4, 134.9, 134.2, 133.2, 132.7, 130.7, 128.1, 127.3, 125.6, 124.3, 119.3, 115.1, 114.9, 113.1, 110.9, 108.0, 107.8; ESI-HRMS (+): m/z calcd for C28H26N7O3+ [M + H]+ 508.2092, found 508.2090; calcd for C28H25N7O3Na+ [M + Na]+ 530.1911, found 530.1910.
4.1.3. Synthesis of intermediate 7
A 12-ml volume of an 80% hydrazine hydrate solution was added to a solution of intermediate 6 (5 mmol, 2.54 g) in 12 ml of ethanol. Then, the mixture was refluxed for 10 h. Afterwards, the reaction mixture was filtered to obtain the intermediate 7 as white solid (85% yield). 1H NMR (600 MHz, DMSO-d6): δ 11.46 (s, 1H), 9.73 (s, 1H), 9.31 (s, 1H), 8.87 (s, 1H), 8.71 (d, J = 5.1 Hz, 1H), 8.55–8.49 (m, 3H), 8.43 (s, 1H), 7.86 (s, 1H), 7.80 (s, 1H), 7.53 (dd, J = 5.1, 8.1 Hz, 1H), 7.43 (d, J = 5.1 Hz, 1H), 7.34 (d, J = 8.1 Hz, 1H), 7.16–7.09 (m, 3H), 7.01 (s, 1H), 4.49 (s, 2H), 2.21 (s, 3H);13C NMR (150 MHz, DMSO-d6): 162.0 (C=O), 161.7 (C=O), 18.0 (CH3), aromatic carbon atoms (23 C) gave signals at 161.6, 159.9, 153.4, 151.9, 148.7, 138.5, 138.4, 135.0, 133.2, 132.8, 132.7, 131.4, 130.7, 127.7, 125.4, 124.2, 117.6, 115.0, 114.8, 112.8, 110.7, 108.0, 102.1; ESI-HRMS (+): m/z calcd for C24H24N7O3+ [M + H]+ 494.2047, found 494.2047; calcd for C24H23N7O3Na+ [M + Na]+ 516.1867, found 516.1864.
4.1.4. General procedure for the synthesis of compounds 8a–8f (series A)
Different aromatic aldehydes (0.4 mmol) were separately added to a solution of 5-isocyanato-1H-indole-adamantine 6 (0.12 g, 0.4 mmol) in 5 ml of toluene. The mixtures were heated at 60 °C for 4 h. Subsequently, the reaction mixtures were concentrated and the resulting crudes were purified by flash chromatography on silica gel, eluting with the appropriate mixtures of CH2Cl2 and MeOH to afford the corresponding target compounds 8a–8f. Their structures were characterised by NMR and ESI-HRMS. The structures and IUPAC names of these target compounds (Series A) are listed in Table S1 (Supporting Information).
Compound 8a: Yellow solid; Yield: 81%; HPLC purity: 95.37%; 1H NMR (600 MHz, DMSO-d6): δ 11.62 (s, 1H), 11.43 (s, 1H), 9.30 (s, 1H), 8.88 (s, 1H), 8.71 (d, J = 3.9 Hz, 1H), 8.54 (s, 1H), 8.53–8.50 (m, 2H), 8.46 (s, 1H), 7.89–7.83 (m, 2H), 7.74 (s, 1H), 7.53 (dd, J = 5.1, 8.1 Hz, 1H), 7.43 (d, J = 5.1 Hz, 1H), 7.36 (d, J = 8.1 Hz, 1H), 7.20–7.06 (m, 4H), 2.33–2.23 (m, 2H), 2.21 (s, 3H), 1.54 (d, J = 8.1 Hz, 2H), 0.95 (t, J = 7.3 Hz, 3H); 13 C NMR (150 MHz, DMSO-d6): δ 162.1 (C=O), 161.6 (C=O), 148.7 (–CH=N), 56.0 (CH2), 55.9 (CH2), 40.4 (CH3), 18.0 (CH3), aromatic carbon atoms (23 C) gave signals at 159.9, 153.4, 151.9, 151.2, 149.5, 147.9, 138.4, 135.0, 133.1, 132.7, 130.7, 127.6, 127.5, 125.5, 124.3, 122.4, 115.1, 114.8, 113.0, 111.9, 108.6, 108.0, 103.6; ESI-HRMS (+): m/z calcd for C30H30N9O2+ [M + H]+ 548.2517, found 548.2519.
Compound 8b: Yellow solid; Yield: 84%; HPLC purity: 94.57%; 1H NMR (600 MHz, DMSO-d6): δ 11.87 (s, 1H), 11.71 (s, 1H), 9.31 (s, 1H), 8.89 (s, 1H), 8.71 (s, 1H), 8.55 (s, 1H), 8.54–8.50 (m, 2H), 8.49 (s, 2H), 7.92 (s, 1H), 7.86 (s, 1H), 7.77 (d, J = 8.1 Hz, 2H), 7.53 (dd, J = 5.1, 8.1 Hz, 1H), 7.47 (dd, J = 5.1, 8.1 Hz, 3H), 7.44 (d, J = 5.1 Hz, 1H), 7.40 (d, J = 8.1 Hz, 1H), 7.25 (s, 1H), 7.20–7.13 (m, 3H), 2.21 (s, 3H);13C NMR (150 MHz, DMSO-d6); δ 162.1 (C=O), 161.6 (C=O), 148.7 (–CH=N), 18.0 (CH3), aromatic carbon atoms (27 C) gave signals at 159.9, 154.9, 153.5, 151.8, 151.3, 138.4, 137.6, 136.7, 134.9, 134.3, 132.7, 130.9, 130.8, 130.7, 130.7, 130.5, 127.5, 125.5, 124.2, 120.3, 115.1, 114.9, 113.0, 108.8, 108.5, 108.0, 94.7; ESI-HRMS (+): m/z calcd for C33H28N9O2+ [M + H]+ 582.2360, found 582.2363.
Compound 8c: Yellow solid; Yield: 83%; HPLC purity: 97.32%; 1H NMR (600 MHz, DMSO-d6): δ 11.83 (s, 1H), 11.69 (s, 1H), 9.31 (s, 1H), 8.89 (s, 1H), 8.71 (s, 1H), 8.68 (s, 1H), 8.56 (s, 1H), 8.54–8.50 (m, 2H), 8.49 (s, 1H), 7.91 (s, 1H), 7.86 (s, 1H), 7.68 (s, 1H), 7.53 (dd, J = 5.1, 8.1 Hz, 1H), 7.49 (brs, 1H), 7.43 (d, J = 5.1 Hz, 1H), 7.40 (d, J = 8.1 Hz, 1H), 7.21 (s, 1H), 7.19–7.12 (m, 4H), 2.21 (s, 3H); 13 C NMR (150 MHz, DMSO-d6): δ 162.1 (C=O), 161.6 (C=O), 148.7(–CH=N), 18.0 (CH3), aromatic carbon atoms (27 C) gave signals at 159.9, 158.0, 153.4, 151.8, 142.7, 139.6, 138.4, 135.0, 133.7, 133.1, 132.7, 131.4, 130.9, 130.7, 129.4, 128.4, 127.6, 125.5, 124.3, 120.6, 118.4, 115.1, 114.9, 113.0, 110.8, 108.0, 103.8; ESI-HRMS (+): m/z calcd for C31H26N9O2S+ [M + H]+ 588.1925, found 588.1926.
Compound 8d: Yellow solid; Yield: 80%; HPLC purity: 97.02%; 1H NMR (600 MHz, DMSO-d6): δ12.04 (s, 1H), 11.72 (s, 1H), 9.31 (s, 1H), 8.89 (s, 2H), 8.71 (s, 1H), 8.64 (s, 1H), 8.57 (s, 1H), 8.53 (d, J = 5.1 Hz, 3H), 8.50 (s, 1H), 8.19 (d, J = 5.1 Hz, 1H), 7.92 (s, 1H), 7.85 (s, 1H), 7.53 (d, J = 5.1 Hz, 2H), 7.44 (d, J = 5.1 Hz, 1H), 7.40 (d, J = 8.1 Hz, 1H), 7.26 (s, 1H), 7.18 (d, J = 8.1 Hz, 1H), 7.15 (s, 2H), 2.21 (s, 3H);13C NMR (150 MHz, DMSO-d6): δ 164.5 (C=O), 162.0 (C=O), 148.6 (–CH=N), 18.0 (CH3), aromatic carbon atoms (27 C) gave signals at 162.0, 161.6, 161.1, 159.9, 153.5, 151.7, 150.9, 149.0, 144.7, 140.0, 135.1, 134.1, 133.8, 133.1, 132.8, 130.7, 128.9, 127.6, 125.5, 124.5, 124.3, 115.1, 118.7, 114.9, 113.0, 110.9, 108.0; ESI-HRMS (–): m/z calcd for C32H25N10O2– [M–H]– 581.2167, found 581.2181.
Compound 8e: Yellow solid; Yield: 86%; HPLC purity: 96.39%; 1H NMR (600 MHz, DMSO-d6): δ 11.73 (s, 1H), 11.67 (s, 1H), 9.30 (s, 1H), 8.89 (s, 1H), 8.71 (s, 1H), 8.55 (s, 1H), 8.54–8.50 (m, 2H), 8.47 (s, 1H), 8.41 (s, 1H), 7.90 (s, 1H), 7.85 (s, 1H), 7.71 (d, J = 8.1 Hz, 2H), 7.53 (dd, J = 5.1, 8.1 Hz, 1H), 7.44 (d, J = 5.1 Hz, 1H), 7.39 (d, J = 8.1 Hz, 1H), 7.21 (brs, 1H), 7.18–7.11 (m, 3H), 7.04 (d, J = 8.1 Hz, 2H), 3.82 (s, 3H), 2.21 (s, 3H); 13 C NMR (150 MHz, DMSO-d6): δ 162.1 (C=O), 161.6 (C=O), 148.7 (–CH=N), 55.8 (OCH3), 18.0 (CH3), aromatic carbon atoms (25 C) gave signals at 161.3, 159.9, 157.9, 153.4, 151.8, 147.5, 138.4, 135.0, 133.6, 133.0, 132.7, 131.1, 130.7, 129.2, 127.6, 127.4, 125.5, 124.3, 118.3, 115.1, 114.9, 114.8, 113.0, 110.8, 108.0, 103.6; ESI-HRMS (+): m/z calcd for C34H30N9O3+ [M + H]+ 612.2466, found 612.2467.
Compound 8f: Yellow solid; Yield: 89%; HPLC purity: 93.88%; 1H NMR (600 MHz, DMSO-d6): δ 11.74 (s, 1H), 11.65 (s, 1H), 9.30 (s, 1H), 8.89 (s, 1H), 8.71 (s, 1H), 8.54 (s, 1H), 8.53–8.50 (m, 2H), 8.47 (s, 1H), 8.39 (s, 1H), 7.89 (s, 1H), 7.84 (s, 1H), 7.53 (dd, J = 5.1, 8.1 Hz, 1H), 7.44 (d, J = 5.1 Hz, 1H), 7.41–7.35 (m, 2H), 7.28–7.20 (m, 2H), 7.18–7.12 (m, 3H), 7.05 (d, J = 8.1 Hz, 1H), 3.85 (s, 3H), 3.82 (s, 3H), 2.21 (s, 3H); 13 C NMR (150 MHz, DMSO-d6): δ 162.1 (C=O), 161.6 (C=O), 148.7 (–CH=N), 56.1 (OCH3), 56.0 (OCH3), 18.0 (CH3), aromatic carbon atoms (29 C) gave signals at 159.9, 157.9, 153.4, 151.8, 151.2, 149.6, 147.9, 138.4, 134.9, 133.6, 133.0, 132.7, 131.0, 130.7, 127.6, 127.6, 127.5, 125.5, 124.3, 122.4, 118.3, 115.1, 114.9, 112.9, 112.0, 110.8, 108.7, 108.0, 103.6; ESI-HRMS (+): m/z calcd for C35H32N9O4+ [M + H]+ 642.2572, found 642.2573.
4.1.5. Synthesis of intermediates 10 and 11
The intermediates 10 and 11 were synthesised as previously described20.
4.1.6. General procedure for the synthesis of compounds 12a-12r (series B)
The mixtures of different aldehydes (1.2 mmol) and 11 (0.345 g, 1 mmol) in ethyl 5 ml of ethanol were heated to reflux for 8 h. The solvent was removed from the reaction mixtures under a vacuum. The resulting crude products were purified by flash chromatography on silica gel, eluting with the appropriate mixtures of CH2Cl2 and MeOH to obtain the corresponding compounds 12a–12r. Their structures were characterised by NMR and ESI-HRMS. Structures and IUPAC names of these target compounds (Series B) are listed in Table S2 (Supporting Information).
Compound 12a: Yellow solid; Yield: 91%; HPLC purity: 95.78%; 1H-NMR (600 MHz, DMSO-d6): δ 11.68 (s, 1H), 11.48 (s, 1H), 9.60 (s, 1H), 9.37 (s, 1H), 8.74 (s, 1H), 8.58 (s, 1H), 8.50 (d, J = 5.1 Hz, 1H), 8.20 (s, 1H), 7.77 (s, 1H), 7.59 (s, 1H), 7.51 (s, 1H), 7.43 (s, 2H), 7.20 (s, 1H), 1.97 (s, 3H); 13 C-NMR (150 MHz, DMSO-d6): δ 161.9 (C=O), 148.6 (–CH=N), 19.0(CH3), aromatic carbon atoms (17 C) gave signals at 161.1, 159.8, 157.8, 151.9, 148.0, 134.8, 133.6, 133.6, 132.9, 131.0, 127.4, 124.4, 119.4, 112.6, 111.6, 107.9, 103.4;ESI-HRMS (+): m/z calcd for C20H18N7O+ [M + H]+ 372.1567, found 372.1565, C20H17N7ONa+ [M + Na]+ 394.1387, found 394.1385.
Compound 12b: Yellow solid; Yield: 90%; HPLC purity: 93.70%; 1H-NMR (600 MHz, DMSO-d6): δ 11.66 (s,1H), 11.50–11.36(m, 1H), 9.59 (s, 1H), 9.35 (s, 1H), 8.72 (s, 1H), 8.56 (d, J = 2.1 Hz, 1H), 8.48 (d, J = 5.1 Hz, 1H), 8.19 (s, 1H), 7.77 (s, 1H), 7.57 (s, 1H), 7.51 (d, J = 5.1 Hz, 1H), 7.46–7.34 (m, 1H), 7.19 (s,1H), 2.31 (s, 2H), 1.05 (t, J = 7.1 Hz,3H); 13 C-NMR (150 MHz, DMSO-d6): δ 161.9 (C=O), 148.6 (–CH=N), 25.9 (CH2), 11.2 (CH3), aromatic carbon atoms (17 C) gave signals at 161.1, 159.8, 157.9, 152.8, 151.9, 134.8, 133.6, 133.6, 132.9, 131.0, 127.4, 124.3, 119.4, 112.6, 111.6, 107.9, 103.6; ESI-HRMS (+): m/z calcd for C21H20N7O+ [M + H]+ 386.1724, found 386.1719, C21H19N7ONa+ [M + Na]+ 408.1543, found 408.1540.
Compound 12c: Yellow solid; Yield: 89%; HPLC purity: 96.70%; 1H-NMR (600 MHz, DMSO-d6): δ 11.67 (s, 1H), 11.46 (s, 1H), 9.60 (s, 1H), 9.37 (s, 1H), 8.74 (s, 1H), 8.58 (s, 1H), 8.50 (d, J = 5.1 Hz, 1H), 8.20 (s, 1H), 7.75 (s, 1H), 7.59 (s, 1H), 7.51 (d, J = 5.1 Hz, 1H), 7.47–7.37 (m, 2H), 7.20 (s, 1H), 2.28 (s, 2H), 1.56 (s, 2H), 0.96 (s, 3H); 13 C-NMR (150 MHz, DMSO-d6): δ 161.9 (C=O), 148.6(–CH=N), 33.6 (CH2), 20.6 (CH2), 14.1 (CH3), aromatic carbon atoms (17 C) gave signals at 161.1, 159.8, 157.9, 151.9, 134.8, 133.6, 133.5, 132.9, 131.0, 127.4, 124.3, 119.4, 112.6, 111.6, 107.9, 103.5; ESI-HRMS (+): m/z calcd for C22H22N7O+ [M + H]+ 400.1880, found 400.1877, C22H21N7ONa+ [M + Na]+ 422.1700, found 420.1696.
Compound 12d: Yellow solid; Yield: 86%; HPLC purity: 95.45%; 1H-NMR (600 MHz, DMSO-d6): δ 11.91 (s,1H), 11.77 (s, 1H), 9.64 (s, 1H), 9.38 (s, 1H), 8.75 (s, 1H), 8.59 (d, J = 3.1 Hz, 1H), 8.54–8.44 (m, 2H), 8.25 (s, 1H), 7.77 (s, 2H), 7.60 (s, 1H),7.53 (s, 1H), 7.51–7.39 (m, 5H), 7.31 (s, 1H); 13 C-NMR (150 MHz, DMSO-d6): δ 163.3 (C=O), 148.6 (–CH=N), aromatic carbon atoms (23 C) gave signals at 161.9, 161.1, 159.9, 158.1, 151.9, 147.4, 145.4, 136.9, 134.9, 134.8, 133.7, 132.9, 132.3, 130.8, 130.5, 129.3, 127.5, 124.4, 119.6, 112.7, 111.6, 107.9, 104.0; ESI-HRMS (+): m/z calcd for C25H20N7O+ [M + H]+ 434.1724, found 434.1718; calcd for C25H19N7ONa+ [M + Na]+ 456.1543, found 456.1537.
Compound 12e: This compound has been reported in our previous work16.
Compound 12f: Yellow solid; Yield: 89%; HPLC purity: 94.45%; 1H-NMR (600 MHz, DMSO-d6): δ 12.15 (s, 1H), 11.78 (s, 1H), 11.24 (s, 1H), 9.71 (s, 1H), 9.43 (s, 1H), 8.84 (s, 1H), 8.73–8.66 (m, 2H), 8.62 (d, J = 5.1 Hz, 1H), 8.20 (s, 1H), 7.77 (s, 1H), 7.60 (d, J = 5.1 Hz, 1H), 7.56 (d, J = 8.1 Hz, 1H), 7.49 (d, J = 5.1 Hz, 1H), 7.46 (d, J = 8.1 Hz, 1H), 7.32 (s, 2H), 6.97 (d, J = 8.1 Hz, 2H); 13 C-NMR (150 MHz, DMSO-d6): δ 161.2 (C=O), 147.7 (–CH=N), aromatic carbon atoms (23 C) gave signals at 160.9, 159.9, 159.0, 157.9, 157.8, 149.9, 146.8, 137.1, 133.9, 133.7, 133.6, 131.8, 130.5, 129.7, 127.5, 125.3, 119.9, 119.8, 119.4, 116.9, 112.8, 112.0, 108.0, 104.2; ESI-HRMS (+): m/z calcd for C25H20N7O2+ [M + H]+ 450.1673, found 450.1672, C25H19N7O2Na+ [M + Na]+ 472.1492, found 472.1492.
Compound 12g: Yellow solid; Yield: 88%; HPLC purity: 96.34%; 1H-NMR (600 MHz, DMSO-d6): δ 11.95 (s, 1H), 11.74 (s, 1H), 9.63 (s, 1H), 9.37 (d, J = 2.0 Hz, 1H), 8.84 (s, 1H), 8.74 (d, J = 5.1 Hz, 1H), 8.58 (d, J = 5.1 Hz, 1H), 8.50 (td, J = 2.0, 8.1 Hz, 1H), 8.23 (s, 1H), 7.91 (d, J = 8.1 Hz, 1H), 7.59 (dd, J = 5.1, 8.1 Hz, 1H), 7.54 (d, J = 8.1 Hz, 1H), 7.47–7.40 (m, 3H), 7.34 (s, 1H), 7.12 (d, J = 8.1 Hz, 1H), 7.04 (t, J = 8.1 Hz, 1H), 3.89 (s, 3H); 13 C-NMR (150 MHz, DMSO-d6): δ 161.9 (C=O), 161.1, 159.8, 158.2, 158.0, 151.9, 148.6 (–CH=N), 56.2 (CH3O), aromatic carbon atoms (23 C) gave signals at 142.8, 134.8, 133.7, 132.9, 131.9, 130.9, 127.5, 126.0, 124.4, 122.9, 121.2, 119.6, 116.6, 112.7, 112.3, 111.6, 107.9, 104.0; ESI-HRMS (+): m/z calcd for C26H22N7O2+ [M + H]+ 464.1829, found 464.1828; calcd for C26H21N7O2Na+ [M + Na]+ 486.1649, found 486.1649.
Compound 12 h: Yellow solid; Yield: 88%; HPLC purity: 94.11%; 1H-NMR (600 MHz, DMSO-d6): δ 11.77 (s, 1H), 11.73 (s, 1H), 9.62 (s, 1H), 9.38 (s, 1H), 8.74 (d, J = 5.1 Hz, 1H), 8.59 (d, J = 5.1 Hz, 1H), 8.51 (d, J = 8.1 Hz, 1H), 8.43 (s, 1H), 8.24 (s, 1H), 7.72 (d, J = 8.1 Hz, 2H), 7.63–7.49 (m, 3H), 7.44 (d, J = 5.1 Hz, 2H), 7.17–6.96(m, 2H), 3.83 (s, 3H); 13 C-NMR (150 MHz, DMSO-d6): δ 161.9 (C=O), 148.6 (–CH=N), 55.8 (CH3O), aromatic carbon atoms (21 C) gave signals at 161.3, 161.1, 159.8, 151.9, 134.8, 133.7, 133.2, 132.9, 131.3, 129.1, 127.4, 119.5, 114.8, 112.7, 112.5, 111.6, 107.8, 103.7, 102.3; ESI-HRMS (+): m/z calcd for C26H22N7O2+ [M + H]+ 464.1829, found 464.1833, C26H21N7O2Na+ [M + Na]+ 486.1649, found 486.1653.
Compound 12i: Yellow solid; Yield:88%; HPLC purity: 96.10%; 1H NMR (600 MHz, DMSO-d6): δ 11.82 (s, 1H), 11.73 (s, 1H), 9.62 (s, 1H), 9.38 (s, 1H), 8.75 (d, J = 5.1 Hz, 1H), 8.59 (d, J = 5.1 Hz, 1H), 8.51 (d, J = 8.1 Hz, 1H), 8.44 (s, 1H), 8.24 (s, 1H), 7.66 (d, J = 8.1 Hz, 2H), 7.59 (dd, J = 5.1, 8.1 Hz, 1H), 7.54 (d, J = 8.1 Hz, 1H), 7.45 (d, J = 5.1 Hz, 2H), 7.30 (s, 3H), 2.37 (s, 3H); 13 C NMR (150 MHz, DMSO-d6): δ 161.9 (C=O), 148.6 (–CH=N), 21.0 (CH3), aromatic carbon atoms (21 C) gave signals at 161.1, 159.8, 158.0, 152.0, 147.5, 140.1, 134.8, 133.7, 133.0, 132.0, 131.0, 129.9, 127.4, 124.3, 119.7, 112.7, 111.6, 107.8, 102.9; ESI-HRMS (+): m/z calcd for C26H22N7O+ [M + H]+ 448.1880, found 448.1882, calcd for C26H21N7ONa+ [M + Na]+ 470.1700, found 470.1705.
Compound 12j: Yellow solid; Yield: 86%; HPLC purity: 93.10%; 1H-NMR (600 MHz, DMSO-d6): δ 11.93 (s,1H), 11.74 (s, 1H), 9.68 (s, 1H), 9.41 (s, 1H), 8.80 (d, J = 5.1 Hz, 1H), 8.67–8.58(m, 2H), 8.47 (s, 1H), 8.21 (brs, 1H), 7.70 (dd, J = 5.1, 8.1 Hz, 1H), 7.55 (d, J = 8.1 Hz, 1H), 7.51 (s, 2H), 7.47 (d, J = 5.1 Hz, 1H), 7.46–7.42(m, 3H), 7.40 (s, 1H), 7.31 (s, 1H), 7.20 (t, J = 7.3 Hz, 1H), 7.10 (d, J = 8.1 Hz, 3H); 13 C-NMR (150 MHz, DMSO-d6): δ 161.5(C=O), 148.4 (–CH=N), 69.8(CH2), aromatic carbon atoms (29 C) gave signals at 161.0, 159.9, 157.7, 156.8, 150.6, 147.4, 146.7, 136.9, 136.2, 133.8, 133.6, 133.4, 131.1, 130.7, 127.4, 124.9, 124.3, 123.2, 120.7, 119.8, 119.4, 116.3, 112.7, 111.8, 108.0, 104.1; ESI-HRMS (+): m/z calcd for C32H26N7O2+ [M + H]+ 540.2142, found 540.2139; calcd for C32H25N7O2Na+ [M + Na]+ 562.1962, found 562.1959.
Compound 12k: Yellow solid; Yield: 85%; HPLC purity: 93.15%; 1H-NMR (600 MHz, DMSO-d6): δ 11.74 (s, 2H), 9.71 (s, 1H), 9.42 (d, J = 2.0 Hz, 1H), 8.84 (d, J = 5.1 Hz, 1H), 8.70 (d, J = 8.1 Hz, 1H), 8.62 (d, J = 5.1 Hz, 1H), 8.33 (s, 1H), 8.18 (s, 1H), 7.80–7.69 (m, 1H), 7.54 (d, J = 8.1 Hz, 1H), 7.49 (d, J = 5.1 Hz, 1H), 7.44 (d, J = 8.1 Hz, 1H), 7.32 (s, 1H), 7.28 (s, 1H), 7.10 (d, J = 8.1 Hz, 1H), 7.00 (d, J = 8.1 Hz, 1H), 3.83 (s, 3H); 13 C-NMR (150 MHz, DMSO-d6): δ 160.9 (C=O), 148.3 (–CH=N), 56.0 (CH3O), aromatic carbon atoms (23 C) gave signals at 159.9, 159.0, 158.7, 157.9, 150.2, 149.9, 147.4, 146.7, 137.1, 133.8, 133.7, 133.5, 131.1, 127.7, 127.5, 125.3, 120.7, 112.7, 112.7, 112.3, 111.9, 108.0, 103.7;ESI-HRMS (+): m/z calcd for C26H22N7O3+ [M + H]+ 480.1779, found 480.1778; calcd for C26H21N7O3Na+ [M + Na]+ 502.1598, found 502.1596.
Compound 12l: Yellow solid; Yield: 91%; HPLC purity: 97.09%; 1H-NMR (600 MHz, DMSO-d6): δ 11.93 (s, 1H), 11.76 (s, 1H), 9.72 (s, 1H), 9.44 (d, J = 5.1 Hz, 1H), 8.87 (d, J = 5.1 Hz, 1H), 8.76 (s, 2H), 8.63 (d, J = 5.1 Hz, 1H), 8.18 (s, 1H), 7.83 (dd, J = 5.1, 8.1 Hz, 1H), 7.55 (dd, J = 5.1, 8.1 Hz, 1H), 7.53–7.49 (m, 2H), 7.45 (d, J = 8.1 Hz, 1H), 7.33 (s, 1H), 7.19–7.11 (m, 2H), 3.86 (s, 3H), 3.84 (s, 3H); 13 C-NMR (150 MHz, DMSO-d6): δ 161.0 (C=O), 148.4 (–CH=N), 61.7 (CH3O), 56.2 (CH3O), aromatic carbon atoms (23 C) gave signals at 160.9, 159.9, 159.0, 158.7, 153.2, 149.2, 146.1, 143.0, 137.8, 134.0, 133.4, 128.3, 127.5, 125.6, 124.8, 119.8, 117.5, 116.9, 115.0, 114.6, 112.7, 108.1, 104.0; ESI-HRMS (+): m/z calcd for C27H24N7O3+ [M + H]+ 494.1935, found 494.1938; calcd for C27H23N7O3Na[M + Na]+ 516.1755, found 516.1758.
Compound 12 m: Yellow solid; Yield: 91%; HPLC purity: 95.14%; 1H-NMR (600 MHz, DMSO-d6): δ 11.75 (s, 1H), 11.70 (s, 1H), 9.68 (s, 1H), 9.42 (s, 1H), 8.83 (dd, J = 2.0, 5.1 Hz, 1H), 8.73 (s, 1H), 8.69 (d, J = 8.1 Hz, 1H), 8.62 (d, J = 5.1 Hz, 1H), 8.16 (s, 1H), 7.84 (d, J = 8.1 Hz, 1H), 7.77 (dd, J = 5.1, 8.1 Hz, 1H), 7.54 (d, J = 8.1 Hz, 1H), 7.49 (d, J = 5.1 Hz, 1H), 7.44 (d, J = 8.1 Hz, 1H), 7.30 (s, 1H), 6.83–6.56 (m, 2H), 3.90 (s, 3H), 3.84 (s, 3H); 13 C-NMR (150 MHz, DMSO-d6): δ 162.9 (C=O), 146.8 (–CH=N), 56.3 (CH3O), 55.9(CH3O), aromatic carbon atoms (23 C) gave signals at 161.2, 161.0, 159.9, 159.6, 158.9, 158.7, 157.8, 149.9, 137.0, 133.8, 133.7, 133.4, 131.1, 127.5, 127.1, 125.2, 119.6, 115.7, 112.7, 112.0, 108.0, 106.9, 103.7; ESI-HRMS (+): m/z calcd for C27H24N7O3+ [M + H]+ 494.1935, found 494.1935; calcd for C27H23N7O3Na+ [M + Na]+ 516.1755, found 516.1754.
Compound 12n: Yellow solid; Yield: 94%; HPLC purity: 94.55%; 1H-NMR (600 MHz, DMSO-d6): δ 11.93 (s, 1H), 11.73 (s, 1H), 9.64 (s, 1H), 9.38 (s, 1H), 8.75 (d, J = 5.1 Hz, 1H), 8.59 (d, J = 5.1 Hz, 1H), 8.55–8.51 (m, 1H), 8.42 (s, 1H), 8.24 (brs, 1H), 7.61 (dd, J = 5.1, 8.1 Hz, 1H), 7.54 (d, J = 8.1 Hz, 1H), 7.49–7.39 (m, 2H), 7.32 (s, 1H), 6.92 (s, 2H), 6.63–6.53 (m, 1H), 3.82 (s, 6H); 13 C-NMR (150 MHz, DMSO-d6): δ 161.9 (C=O), 148.5 (–CH=N), 55.8 (CH3O), aromatic carbon atoms (23 C) gave signals at 161.2, 161.0, 159.9, 158.3, 158.1, 151.7, 148.5, 147.4, 136.9, 134.9, 133.7, 132.9, 130.8, 127.4, 124.4, 119.7, 112.7, 111.6, 107.9, 105.3, 104.1, 102.7; ESI-HRMS (+): m/z calcd for C27H24N7O3+ [M + H]+ 494.1935, found 494.1937; calcd for C27H23N7O3Na+ [M + Na]+ 516.1755, found 516.1758.
Compound 12o: Yellow solid; Yield: 89%; HPLC purity: 92.54%; 1H-NMR (600 MHz, DMSO-d6): δ 11.89 (s, 1H), 11.73 (s, 1H), 9.63 (s, 1H), 9.37 (s, 1H), 8.74 (d, J = 5.1 Hz, 1H), 8.58 (d, J = 5.1 Hz, 1H), 8.50 (d, J = 8.1 Hz, 1H), 8.42 (s, 1H), 8.24 (s, 1H), 7.58 (dd, J = 5.1, 8.1 Hz, 1H), 7.55 (d, J = 8.1 Hz, 1H), 7.47–7.41(m, 2H), 7.32 (s, 1H), 7.06 (s, 2H), 3.86 (s, 6H), 3.73 (s, 3H); 13 C-NMR (150 MHz, DMSO-d6): δ 161.9 (C=O), 148.6 (–CH=N), 60.6 (CH3O), 56.4(CH3O), aromatic carbon atoms (23 C) gave signals at 161.1, 159.8, 158.1, 153.7, 151.9, 148.1, 147.6, 139.6, 134.8, 133.7, 132.9, 130.8, 130.4, 127.4, 124.4, 119.6, 112.7, 111.6, 107.9, 104.7, 104.0; ESI-HRMS (+): m/z calcd for C28H26N7O4+ [M + H]+ 524.2041, found 524.2040; calcd for C28H25N7O4Na+ [M + Na]+ 546.186, found 546.1860.
Compound 12p: Yellow solid; Yield: 84%; HPLC purity: 94.34%;1H NMR (600 MHz, DMSO-d6): δ 11.86 (s, 1H), 11.75 (s, 1H), 9.64 (s, 1H), 9.39 (s, 1H), 8.76 (s, 1H), 8.71 (brs, 1H), 8.59 (d, J = 5.1 Hz, 1H), 8.51 (d, J = 8.1 Hz, 1H), 8.25 (s, 1H), 7.70 (d, J = 5.1 Hz, 1H), 7.60 (dd, J = 5.1, 8.1 Hz, 1H), 7.56 (d, J = 8.1 Hz, 1H), 7.51 (d, J = 5.1 Hz, 1H), 7.48–7.42 (m, 2H), 7.29 (s, 1H), 7.18 (t, J = 5.1 Hz, 1H); 13 C NMR (150 MHz, DMSO-d6): δ 161.9 (C=O), 148.6 (–CH=N), aromatic carbon atoms (21 C) gave signals at 161.1, 159.8, 158.0, 151.8, 148.1, 142.6, 139.7, 134.8, 133.7, 132.9, 131.3, 130.8, 129.3, 128.4, 127.4, 124.4, 119.7, 112.7, 111.6, 107.9, 103.9;ESI-HRMS (+): m/z calcd for C23H18N7OS+ [M + H]+ 440.1288, found 440.1286; calcd for C23H17N7OSNa+ [M + Na]+ 462.1107, found 462.1103.
Compound 12q: Yellow solid; Yield: 83%; HPLC purity: 97.10%; 1H NMR (600 MHz, DMSO-d6): δ 11.86 (s, 1H), 11.78 (s, 1H), 9.70 (s, 1H), 9.42 (s, 1H), 8.84 (d, J = 5.1 Hz, 1H), 8.71 (d, J = 8.1 Hz, 1H), 8.62 (d, J = 5.1 Hz, 1H), 8.37 (s, 1H), 8.18 (s, 1H), 7.88 (s, 1H), 7.81–7.75(m, 1H), 7.55 (d, J = 8.1 Hz, 1H), 7.49 (d, J = 5.1 Hz, 1H), 7.44 (d, J = 8.1 Hz, 1H), 7.27 (s, 1H), 6.97 (s, 1H), 6.67 (s, 1H); 13 C NMR (150 MHz, DMSO-d6): δ 161.2 (C=O), 149.1 (–CH=N), aromatic carbon atoms (21 C) gave signals at 160.9, 159.9, 159.0, 158.7, 150.0, 149.8, 146.7, 145.6, 137.2, 133.9, 133.7, 133.5, 130.8, 127.4, 125.3, 119.8, 117.0, 113.9, 112.7, 112.0, 108.0, 103.9;ESI-HRMS (+): m/z calcd for C23H18N7O+ [M + H]+ 424.1516, found 424.1510; calcd for C23H17N7O2Na+ [M + Na]+ 446.1336, found 446.1331.
Compound 12r: Yellow solid; Yield: 86%; HPLC purity: 94.56%; 1H NMR (600 MHz, DMSO-d6): δ 11.98 (s, 1H), 11.71 (s, 1H), 9.67 (s, 1H), 9.44 (d, J = 8.1 Hz, 1H), 9.41 (s, 1H), 9.20 (s, 1H), 8.78 (d, J = 5.1 Hz, 1H), 8.61 (d, J = 5.1 Hz, 1H), 8.57 (d, J = 8.1 Hz, 1H), 8.26 (s, 1H), 8.07 (d, J = 8.1 Hz, 1H), 7.93 (d, J = 8.1 Hz, 1H), 7.64 (dd, J = 5.1, 8.1 Hz, 2H), 7.59–7.52 (m, 2H), 7.48 (d, J = 8.1 Hz, 1H), 7.46 (d, J = 5.1 Hz, 1H), 7.41 (s, 1H), 4.05 (s,3H); 13 C-NMR (150 MHz, DMSO-d6): δ 161.1 (C=O), 148.4 (–CH=N), 56.4 (CH3O), aromatic carbon atoms (25 C) gave signals at 160.9, 159.9, 159.1, 157.9, 157.1, 147.9, 146.6, 137.3, 133.8, 133.5, 131.6, 131.1, 129.9, 128.3, 127.5, 126.2, 125.5, 125.4, 124.9, 119.7, 112.7, 112.0, 108.0, 104.9, 103.8; ESI-HRMS (+): m/z calcd for C30H24N7O2+ [M + H]+ 514.1986, found 514.1981; calcd for C30H23N7O2Na+ [M + Na]+ 536.1805, found 536.1800.
4.1.7. General procedure for the synthesis of compounds 12A–12F (series C)
Different methyl ketones (1.1 mmol) were separately added to a solution of 11 (1 mmol, 0.345 g) in ethyl 6 ml of alcohol. These mixtures were heated at 78 °C for 10 h, and the solvent was removed from the reaction mixtures under a vacuum. The resulting crude products were purified by flash chromatography on silica gel, eluting with the appropriate mixtures of CH2Cl2 and MeOH to obtain the corresponding compounds 12A–12F (Series C). Their structures were characterised by NMR and ESI-HRMS. Structures and IUPAC names of these target compounds (Series B) are listed in Table S3 (Supporting Information).
Compound 12A: Yellow solid; Yield: 91%; HPLC purity: 93.25%; 1H-NMR (600 MHz, DMSO-d6): δ 11.71 (s, 1H), 9.65 (s, 1H), 9.39 (brs, 1H), 8.78 (s, 1H), 8.59–8.58 (m, 2H), 8.20 (s, 1H), 7.88 (d, J = 8.1 Hz, 2H), 7.70–7.63 (m, 1H), 7.56 (d, J = 8.1 Hz, 1H), 7.51–7.43 (m, 6H), 7.42–7.35 (m, 1H), 2.43 (s, 3H); 13 C-NMR (150 MHz, DMSO-d6): δ 161.7 (C=O), 147.9 (–CCH3=N), 19.0(CH3), aromatic carbon atoms (23 C) gave signals at 161.0, 159.8, 151.0, 147.9 135.7, 133.9, 133.9, 133.6, 133.2, 129.9, 128.9, 127.5, 127.3, 126.9, 124.7, 120.3, 112.7, 111.8, 108.0, 105.2; ESI-HRMS (+): m/z calcd for C26H22N7O+ [M + H]+ 448.1880, found 448.1878, C26H21N7O Na+ [M + Na]+ 470.17, found 470.1698.
Compound 12B: Yellow solid; Yield: 88%; HPLC purity: 96.25%; 1H-NMR (600 MHz, DMSO-d6): δ 11.71 (s, 1H), 10.71 (s, 1H), 9.63 (s, 1H), 9.38 (s, 1H), 8.75 (s, 1H), 8.59 (d, J = 5.1 Hz, 1H), 8.51 (d, J = 8.1 Hz, 1H), 8.25 (s, 1H), 7.91 (d, J = 8.1 Hz, 2H), 7.59 (s, 2H), 7.55 (s, 2H), 7.44 (d, J = 5.1 Hz, 2H), 7.40 (s, 1H), 2.42 (s, 3H); 13 C-NMR (150 MHz, DMSO-d6): δ 164.9 (C=O), 148.6 (–CCH3=N), 19.0 (CH3), aromatic carbon atoms (23 C) gave signals at 161.9, 161.1, 159.8, 153.3, 151.8, 137.5, 134.8, 134.5, 133.7, 132.9, 129.7, 129.0, 128.6, 127.5, 124.4, 121.1, 119.7, 112.7, 111.7, 109.5, 107.9;ESI-HRMS (+): m/z calcd for C26H21ClN7O+ [M + H]+ 482.1491, found 482.1491, C26H20ClN7ONa+ [M + Na]+ 504.1310, found 504.1311.
Compound 12C: Yellow solid; Yield: 91%; HPLC purity: 94.05%; 1H-NMR (600 MHz, DMSO-d6): δ 11.77 (s, 1H), 9.70 (s, 1H), 9.43 (d, J = 2.0 Hz, 1H), 8.83 (dd, J = 2.0, 5.1 Hz, 1H), 8.69 (td, J = 2.0, 8.1 Hz, 1H), 8.62 (d, J = 5.1 Hz, 1H), 8.34 (d, J = 8.1 Hz, 2H), 8.23 (s, 1H), 8.13 (d, J = 8.1 Hz, 2H), 7.76 (dd, J = 5.1, 8.1 Hz, 1H), 7.57 (d, J = 8.1 Hz, 1H), 7.49 (d, J = 5.1 Hz, 1H), 7.46 (d, J = 8.1 Hz, 1H), 7.44 (d, J = 2.0 Hz, 1H), 2.49 (s, 3H); 13 C-NMR (150 MHz, DMSO-d6): δ 161.1 (C=O), 148.0 (–CCH3=N), 27.5 (CH3), aromatic carbon atoms (23 C) gave signals at 160.8, 159.9, 149.5, 146.4, 144.8, 141.7, 137.4, 133.7, 133.4, 130.0, 128.2, 127.9, 127.5, 125.4, 124.2, 124.1, 120.1, 112.9, 112.8, 112.1, 108.0; ESI-HRMS (+): m/z calcd for C26H21N8O3+ [M + H]+ 493.1731, found 493.1732, C26H20N8O3Na+ [M + Na]+ 515.1551, found 515.1552.
Compound 12D: Yellow solid; Yield: 87%; HPLC purity: 94.55%; 1H NMR (DMSO-d6): δ 11.69 (s,1H), 9.62 (s, 1H), 9.37 (s,1H), 8.75 (d, J = 5.1 Hz, 1H), 8.58 (d, J = 5.1 Hz, 1H) , 8.51 (d, J = 8.1 Hz, 1H), 8.22 (s, 1H), 7.78 (d, J = 8.1 Hz, 2H,), 7.62–7.52 (m, 2H,), 7.48–7.42 (m, 2H), 7.39 (s, 1H), 7.29 (s, 1H), 2.40 (s, 3H), 2.37 (s, 3H); 13 C NMR (150 MHz, DMSO-d6): δ 161.9 (C=O), 148.5 (–CCH3=N), 40.3 (CH3), 21.3 (CH3), aromatic carbon atoms (23 C) gave signals at 161.0, 159.8, 156.3, 151.8, 139.6, 135.8, 134.9, 133.6, 132.9, 129.7, 129.5, 128.7, 127.5, 126.8, 124.5, 119.6, 112.7, 111.8, 108.0, 104.7, 102.6; ESI-HRMS (+): m/z calcd for C27H24N7O+ [M + H]+ 462.2037, found 462.2035 C27H23N7O+ [M + Na]+ 484.1856, found 484.1856.
Compound 12E: Yellow solid; Yield: 90%; HPLC purity: 91.25%; 1H-NMR (600 MHz, DMSO-d6): δ 13.45 (s,1H), 11.81 (s, 1H), 11.33 (s, 1H), 9.65 (s, 1H), 9.38 (s, 1H), 8.76 (d, J = 5.0 Hz, 1H), 8.59 (d, J = 5.0 Hz, 1H), 8.54 (td, J = 2.0, 8.1 Hz, 1H), 8.24 (s, 1H), 7.78 (d, J = 2.0 Hz, 1H), 7.63 (dd, J = 5.1, 8.1 Hz, 1H), 7.58 (dd, J = 2.0, 8.1 Hz,1H), 7.49–7.44 (m, 4H), 6.92 (d, J = 8.1 Hz, 1H), 2.54 (s, 3H); 13 C-NMR (150 MHz, DMSO-d6): δ 161.8 (C=O), 148.3 (–CCH3=N), 14.7 (CH3), aromatic carbon atoms (23 C) gave signals at 161.0, 159.8, 159.0, 158.3, 158.0, 151.6, 135.2, 134.0, 133.9, 133.8, 133.0, 131.0, 129.8, 127.4, 124.5, 122.1, 120.0, 120.0, 112.7, 111.7, 110.1, 108.0, 105.8; ESI-HRMS (+): m/z calcd for C26H21BrN7O2+ [M + H]+ 542.0935, found 542.0934, C26H20BrN7O2Na+ [M + Na]+ 564.0754, found 564.0752.
Compound 12F: Yellow solid; Yield: 91%; HPLC purity: 94.67%; 1H-NMR (600 MHz, DMSO-d6): δ 11.83 (s, 1H), 11.24 (s, 1H), 9.78–9.63 (m, 1H), 9.47–9.23 (m, 1H), 8.82 (s, 1H), 8.67 (d, J = 8.1 Hz, 1H), 8.62 (d, J = 5.1 Hz, 1H), 8.21 (s, 1H), 7.75 (s, 1H), 7.59 (d, J = 8.1 Hz, 1H), 7.51–7.40 (m, J = 8.1 Hz, 3H), 7.16 (s, 1H), 6.96 (d, J = 8.1 Hz, 1H), 6.88 (d, J = 8.1 Hz, 1H), 2.55 (s, 3H);13C-NMR (150 MHz, DMSO-d6): δ 161.3 (C=O), 147.0 (–CCH3=N), 56.1 (CH3O), 14.7 (CH3), aromatic carbon atoms (23 C) gave signals at 161.0, 159.9, 159.0, 158.7, 153.0, 151.9, 150.2, 136.7, 134.0, 133.6, 133.6, 130.1, 127.4, 125.1, 120.0, 118.3, 118.0, 117.3, 113.3, 112.7, 111.9, 108.1, 105.6; ESI-HRMS (+): m/z calcd for C27H24N7O3+ [M + H]+ 494.1935, found 494.1938, C27H23N7O3Na+ [M + Na]+ 516.1755, found 516.1758.
4.2. Biologicals
4.2.1. Cell lines and cell culture
The HeLa, MDA-MB-231, HepG2, MCF-7, NT2, A549, A375, A875, HeCat, and LO2 cell lines were cultured in complete Dulbecco’s modified Eagle’s medium (DMEM; Basal Media, D120716) supplemented with 10% foetal bovine serum (900–108; Gemini Bio = Products Inc., Calabasas, CA, USA). The MCF-10A cell line was cultured in Lonza MEGM™ Bullet Kit (CC-3151 & CC-4136; Lonza Group AG, Basel, Switzerland) containing 2 mg/mL insulin, 1 mg/mL hydrocortisone, 100 U/mL penicillin, 100 µg/mL EGF, and 100 µg/mL streptomycin and 5% horse serum. All cells were incubated in a humidified atmosphere (5% CO2, 95% air) at 37 °C.
4.2.2. Cell viability assay
Cancer cells were cultured in DMEM in a 96-well plate and treated with each compound at 0.1, 1, 5, 10, 20, 50 µM, with eight replicates for each concentration. After treatment for 48 h, the cells were subjected to MTT assay with some modifications. Briefly, 5 × 103 cells were seeded in each well of a 96-well plate with 100 µL of DMEM. Before treatment, cells were allowed to adhere for 24 h. All the compounds were dissolved in DMSO with a stock solution of 20 µM and subsequent dilutions were made with DMEM medium. Treatment with 0.5% DMSO was used as control. After 48 h treatment, 20 µL of MTT solution (5 mg/mL) was added into each well, followed by incubation for 4 h. Then, the supernatant medium was removed and 100 µL DMSO was added to each well. The absorbance value of each well was recorded at 490 nm on a Multiskan™ FC microplate reader (Thermo Fisher Scientific Inc.) and the DMSO treatment group was considered as 100% viability. The IC50 and error values were calculated using the GraphPad Prism 8 software (GraphPad software Inc., San Diego, CA, USA) using the absorbance values recorded at 490 nm.
4.2.3. Western blot analysis
After treatment with the compounds, the cells were boiled with 1 × SDS loading buffer at 100 °C, for 10 min. The boiled samples were subjected to 8%–12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), then transferred to polyvinylidene fluoride (PVDF) membrane (03010040001; Hoffmann-La Roche AG, Basel, Switzerland). The proteins were probed with indicated primary antibodies and then secondary antibodies. After incubating with enhanced chemiluminescence (ECL) reagents, the membranes were exposed to films and then films were developed to visualised the immunoreactive protein bands.
4.2.4. Quantitative real-time PCR (qRT-PCR) analysis
The qRT-PCR analysis was performed as previously described with some modifications[30]. Briefly, cells were harvested and total RNA was extracted. Then, the cDNAs were obtained by reverse transcription using the RevertAid RT Reverse Transcription Kit (K1691; Thermo Fisher Scientific Inc.) following the manufacturer’s protocol. Then, qRT-PCR was conducted for the measurement of mRNA expression levels. The following primers were used:
CHOP sense: 5′-TCTTGACCCTGCTTCTCTG-G-3′.
Antisense: 5′-GCTGTGCCACTTTCCTTTCA-3′.
GAPDH sense: 5′-CCAAGGAGTAAGACCCCTGG-3′.
Antisense: 5′-TGGTTGAGCACAGGGTACTT-3′.
4.2.5. Cell apoptosis assay
An Annexin V–fluorescein isothiocyanate (FITC) Apoptosis Detection Kit (BD Biosciences, San Jose, CA, USA) was used to measure cell apoptosis. Cells (about 1 × 105/well) cultured in a 6-well plate were treated with 12A or DMSO for 24 h, and then subjected to apoptosis assay following the kit manufacturer’s protocol. After double-staining with FITC annexin V/propidium iodide (PI), the apoptosis status of the cells was detected using the Attune NxT flow cytometer (Thermo Fisher Scientific Inc.) and the data were analysed with the FlowJo 7.6 software (BD Biosciences).
4.2.6. In vivo tumor model
We established a mice hepatoma homograft model following a previously reported method15. When each tumour grew to about 100 mm3, mice were randomly assigned into groups and treated by i.p. injection with the vehicle [1% DMSO + 10% PEG400 (w/v) (Sigma-Aldrich, Saint Louis, MO, USA)] in normal saline or compound 12A formulated in the vehicle at doses of 20 and 50 mg/kg every other day, respectively. At 6 h after the final dose, mice were euthanized, and tumours were separated, weighed, and subjected to further analysis. All procedures were performed in compliance with the guidelines from the Institutional Animal Care and Use Committee at the Experimental Animal Centre of Xiamen University.
4.2.7. Haematoxylin and eosin staining
The staining procedure for H&E followed a previously reported method15.
Supplementary Material
Acknowledgement
We are grateful to Lijuan Wang, Cuiling Sun, Rong Ding, and Junjie Chen for assistance in the melting point assay, the NMR experiment, the MS experiment, and the surface plasmon resonance assay, respectively.
Funding Statement
The research was supported in part by grants from the Fundamental Research Funds for the Central Universities of China [No. 20720180051], the National Key R&D Program of China [2018YFA0107303], the National Natural Science Foundation of China [No. 81672955], the Fujian Provincial Natural Science Foundation [No. 2018J01132], the Jinhua Science and Technology Plan Project of China [2020–4-088], and the Health Commission of Zhejiang Province of China [2020KY628].
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Wang Y, Xu K, Zhang H, et al. Retinal ganglion cell death is triggered by paraptosis via reactive oxygen species production: a brief literature review presenting a novel hypothesis in glaucoma pathology. Mol Med Rep 2014;10:1179–83. [DOI] [PubMed] [Google Scholar]
- 2.Maltese WA, Overmeyer JH.. Methuosis: nonapoptotic cell death associated with vacuolization of macropinosome and endosome compartments. Am J Pathol 2014;184:1630–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grace NJ, Lopus M.. Cell death mechanisms in eukaryotes. Cell Biol Toxicol 2020;36:145–64. [DOI] [PubMed] [Google Scholar]
- 4.Colin M, Delporte C, Janky R, etal. Dysregulation of macropinocytosis processes in glioblastomas may be exploited to increase intracellular anti-cancer drug levels: the example of temozolomide. Cancers 2019;11:411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maltese WA, Overmeyer JH.. Non-apoptotic cell death associated with perturbations of macropinocytosis. Front Physiol 2015;6:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cingolani F, Simbari F, Abad JL, et al. Jaspine B induces nonapoptotic cell death in gastric cancer cells independently of its inhibition of ceramide synthase. J Lipid Res 2017;58:1500–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson MW, Overmeyer JH, Young AM, et al. Synthesis and evaluation of indole-based chalcones as inducers of methuosis, a novel type of nonapoptotic cell death. J Med Chem 2012;55:1940–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trabbic CJ, Dietsch HM, Alexander EM, et al. Differential induction of cytoplasmic vacuolization and methuosis by novel 2-indolyl-substituted pyridinylpropenones. ACS Med Chem Lett 2014;5:73–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong X, Sun R, Gao Z, et al. Tubeimoside 1 acts as a chemotherapeutic synergist via stimulating macropinocytosis. Front Pharmacol 2018;9:1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang W, Sun X, Li Y, et al. Discovery and identification of small molecules as methuosis inducers with in vivo antitumor activities. J Med Chem 2018;61:5424–34. [DOI] [PubMed] [Google Scholar]
- 11.Overmeyer JH, Young AM, Bhanot H, Maltese WA.. A chalcone-related small molecule that induces methuosis, a novel form of non-apoptotic cell death, in glioblastoma cells. Mol Cancer 2011;10:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pang L, Liu CY, Gong GH, Quan ZS.. Synthesis, in vitro and in vivo biological evaluation of novel lappaconitine derivatives as potential anti-inflammatory agents. Acta Pharm Sin B 2020;10:628–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trabbic CJ, George SM, Alexander EM, et al. Synthesis and biological evaluation of isomeric methoxy substitutions on anti-cancer indolyl-pyridinyl-propenones: effects on potency and mode of activity. Eur J Med Chem 2016;122:79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lertsuwan J, Lertsuwan K, Sawasdichai A, et al. CX-4945 induces methuosis in cholangiocarcinoma cell lines by a CK2-independent mechanism. Cancers 2018;10:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin J, Liu J, Wu C, et al. Synthesis and biological evaluation of (3/4-(pyrimidin-2-ylamino)benzoyl)-based hydrazine-1-carboxamide/carbothioamide derivatives as novel RXRalpha antagonists. J Enzyme Inhib Med Chem 2020;35:880–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu H, Wu J, Ao M, et al. Design, synthesis and biological evaluation of methylenehydrazine-1-carboxamide derivatives with (5-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)-1H-indole scaffold: Novel potential CDK9 inhibitors. Bioorg Chem 2020;102:104064. [DOI] [PubMed] [Google Scholar]
- 17.Cortes-García CJ, Islas-Jácome A, Rentería-Gómez A, et al. Synthesis of 1,5-disubstituted tetrazoles containing a fragment of the anticancer drug imatinib via a microwave-assisted Ugi-azide reaction. Monatshefte für Chemie – Chem Monthly 2016;147:1277–90. [Google Scholar]
- 18.Li YT, Wang JH, Pan CW, et al. Syntheses and biological evaluation of 1,2,3-triazole and 1,3,4-oxadiazole derivatives of imatinib. Bioorg Med Chem Lett 2016;26:1419–27. [DOI] [PubMed] [Google Scholar]
- 19.Buclin T, Thoma Y, Widmer N, et al. The steps to therapeutic drug monitoring: a structured approach illustrated with imatinib. Front Pharmacol 2020;11:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu H, Wu J, Ao M, et al. Synthesis, structure-activity relationship studies and biological evaluation of novel 2,5-disubstituted indole derivatives as anticancer agents. Chem Biol Drug Des 2016;88:766–78. [DOI] [PubMed] [Google Scholar]
- 21.Busschaert N, Kirby IL, Young S, et al. Squaramides as potent transmembrane anion transporters. Angew Chem Int Ed Engl 2012;51:4426–30. [DOI] [PubMed] [Google Scholar]
- 22.Morales S, Aceña JL, García Ruano JL, Cid MB.. Sustainable synthesis of oximes, hydrazones, and thiosemicarbazones under mild organocatalyzed reaction conditions. J Org Chem 2016;81:10016–22. [DOI] [PubMed] [Google Scholar]
- 23.Pan X, Wang F, Zhang Y, et al. Design, synthesis and biological activities of Nilotinib derivates as antitumor agents. Bioorganic Med Chem 2013;21:2527–34. [DOI] [PubMed] [Google Scholar]
- 24.Sperandio S, de Belle I, Bredesen DE.. An alternative, nonapoptotic form of programmed cell death. Proc Natl Acad Sci USA 2000;97:14376–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim SH, Kang JG, Kim CS, et al. The hsp70 inhibitor VER155008 induces paraptosis requiring de novo protein synthesis in anaplastic thyroid carcinoma cells. Biochem Biophys Res Commun 2014;454:36–41. [DOI] [PubMed] [Google Scholar]
- 26.Sperandio S, Poksay K, de Belle I, et al. Paraptosis: mediation by MAP kinases and inhibition by AIP-1/Alix. Cell Death Different 2004;11:1066–75. [DOI] [PubMed] [Google Scholar]
- 27.Li Z, Mbah NE, Overmeyer JH, et al. The JNK signaling pathway plays a key role in methuosis (non-apoptotic cell death) induced by MOMIPP in glioblastoma. BMC Cancer 2019;19:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X, Yang J, Peng L, et al. Triple-negative breast cancer has worse overall survival and cause-specific survival than non-triple-negative breast cancer. Breast Cancer Res Treat 2017;161:279–87. [DOI] [PubMed] [Google Scholar]
- 29.Hwang SY, Park S, Kwon Y.. Recent therapeutic trends and promising targets in triple negative breast cancer. Pharmacol Ther 2019;199:30–57. [DOI] [PubMed] [Google Scholar]
- 30.Wu J, Xue YH, Gao X, Zhou Q. Host cell factors stimulate HIV-1 transcription by antagonizing substrate-binding function of Siah1 ubiquitin ligase to stabilize transcription elongation factor ELL2. er, Nucleic Acids Research 2020;48:7321–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.