Abstract
Mosquito-borne flaviviruses (MBFVs) including dengue, West Nile, yellow fever, and Zika viruses have an RNA genome encoding one open reading frame flanked by 5′ and 3′ untranslated regions (UTRs). The 3′ UTRs of MBFVs contain regions of high sequence conservation in structured RNA elements known as dumbbells (DBs). DBs regulate translation and replication of the viral RNA genome, functions proposed to depend on the formation of an RNA pseudoknot. To understand how DB structure provides this function, we solved the x-ray crystal structure of the Donggang virus DB to 2.1Å resolution and used structural modeling to reveal the details of its three-dimensional fold. The structure confirmed the predicted pseudoknot and molecular modeling revealed how conserved sequences form a four-way junction that appears to stabilize the pseudoknot. Single-molecule FRET suggests that the DB pseudoknot is a stable element that can regulate the switch between translation and replication during the viral lifecycle by modulating long-range RNA conformational changes.
INTRODUCTION
Mosquito-borne flaviviruses (MBFVs) are a widespread group of medically and economically important single-stranded positive-sense RNA viruses. Examples include dengue virus (DENV), the world′s most prevalent arbovirus that is endemic to over 100 countries and infects ∼390 million people annually (1), yellow fever virus (YFV), West Nile virus (WNV) and Zika virus (ZIKV) whose recent spread has demonstrated how rapidly MBFV outbreaks can emerge (2).
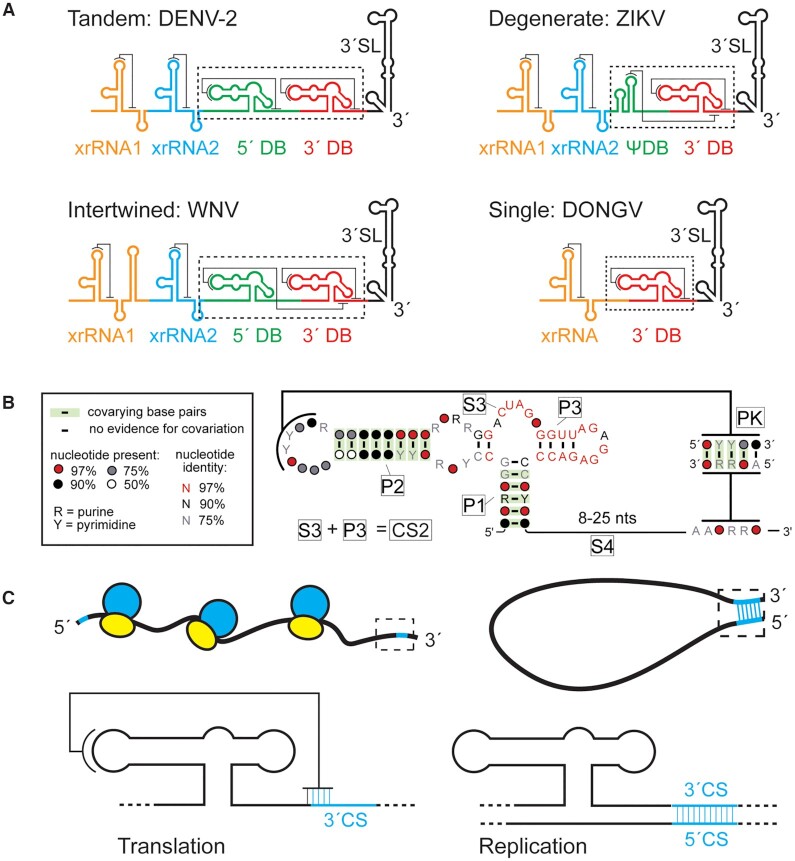
MBFV RNA genomes comprise a single open reading frame flanked by 5′ and 3′ untranslated regions (UTRs), important for regulating viral processes. The genomic RNA is translated to yield viral proteins and is the template for genome replication. These processes cannot occur simultaneously on a single RNA as ribosomes moving in the 5′ to 3′ direction would collide with the replicase machinery moving in the opposite direction. Coordinating these processes requires signals within the viral UTRs, identified by sequence and structure conservation and through mutagenesis studies demonstrating a replication defect when they are disrupted. Two such sequences in MBFV 3′ UTRs are conserved sequence 2 (CS2) and repeat conserved sequence 2 (RCS2) (3). CS2 and RCS2 are contained within secondary structural elements called dumbbell (DB) RNAs that are proposed to contain tertiary structures called pseudoknots (4–7) (Supplementary Figure S1). In dengue virus (DENV), two tandem DBs contain the RCS2 and CS2 motifs, respectively; however, some flaviviruses have only a single DB (8) (Figure 1A).
Figure 1.
Conservation and role of flaviviral DB RNAs. (A) Predicted organization and predicted secondary structures of 3′ UTRs from DENV-2, West Nile virus, ZIKV, and DONGV. The 3′ dumbbell element is conserved but can exist in several configurations. (B) Co-variation analysis of the 3′ DB element from 46 different viruses analyzed using R-scape (40). Predicted helices (P1, P2, P3 and PK) are labeled as are predicted unpaired linker regions (S3, S4). The S3 and P3 regions collectively make up a previously identified conserved sequence (CS2) (3). (C) Model of the 3′ DB element during translation and replication. The pseudoknot overlaps with the 3′ CS, an element responsible for base pairing with the 5′ CS during viral minus-strand synthesis. During viral protein translation (left) the DB pseudoknot is formed. The pseudoknot is incompatible with 5′-3′ CS formation (cyan) during replication (right).
The importance of the DB elements, both virologically and clinically, is well established. Functional investigations using mutagenized DENV replicons showed that DBs have a role in both viral protein production and replication, with particularly strong effects on regulation of replication (9). Mutations that specifically disrupt pseudoknot base pairing showed this pairing was important for both translation and replication (10). In DENV, the two DBs have distinct roles in viral replication in human and mosquito cells, suggesting DBs participate in a complex regulatory network of RNA-RNA interactions (11). A 30-nucleotide deletion from the Dengue virus 4 DB (DENVΔ30) is highly attenuating (12,13), and has been developed into a vaccine candidate against all four DENV strains (14). Given that DB mutations are attenuating in all four DENV strains, mechanistically understanding how a short truncation of a noncoding portion of the DB affects pathogenicity could guide vaccine development (15,16).
Many RNA viruses regulate the switch between translation and replication states through changes in RNA structure. In MBFVs, evidence suggests this switch involves a global conformational change in the genomic RNA, with translation occurring on a ‘linear’ genome conformation and replication on a ‘cyclized’ form with base pairing between the 5′ and 3′ ends (17). Evidence for this includes conserved regions of co-varying base pairs between the 5′ and 3′ ends of the genome, known as the 5′ and 3′ conserved sequences (5′ and 3′ CSs) (3,18–20). Disrupting the base pairing between the 5′ and 3′ CS severely affects viral replication, and atomic force microscopy showed cyclized RNAs with the viral replicase localized to the junction between the 5′ and 3′ ends (17). Other interactions between the 5′ and 3′ ends supplement this base pairing, including the 5′ and 3′ ‘upstream of AUG regions’ and ‘downstream of AUG regions’ (Supplementary Figure S2) (17,21–24). In addition, the location of a replication promoter at the genome′s 5′ end suggests that genome cyclization delivers the viral polymerase from its binding site at the 5′ end to signals at the 3′ end to begin replication (25–27).
The location of the DBs and their relationship to other elements in the 3′ UTR suggests these structures play important regulatory roles. Specifically, the DBs are directly adjacent to the 3′ CS and in the majority of MBFVs the pseudoknot directly overlaps with the 3′ CS. This led to the hypothesis that the DB pseudoknot could act as a sensor of 5′-3′ CS base pairing, or act as a competitor to cyclization, temporally repressing viral replication to maximize viral protein translation (11). In support of this assertion, a simplified DENV minigenome containing only the 5′ and 3′ UTRs demonstrated that the most downstream DB (3′ DB) RNA pseudoknot does not form when the RNA is cyclized (28). This idea is further supported by an analysis of sequence variants in MBFVs and in revertant strains of CS mutants, showing that the 3′ DB pseudoknot regulates genome cyclization during replication (11). Finally, it was proposed that the most upstream DB (5′ DB) from DENV competes with an alternate conformation that base pairs with a region of the capsid protein coding region during viral replication (Supplementary Figure S2) (24). Collectively, this suggests a model in which the DB pseudoknots compete with the cyclized form of the genome. DBs have also been implicated in the formation of non-coding subgenomic flaviviral RNAs (sfRNAs), which are formed through resistance to degradation by the host 5′→3′ exonuclease XRN1, using RNA structures in the 3′ UTR known as XRN1-resistant RNAs (xrRNAs) (29). It has been theorized that DBs, by virtue of their complex secondary structures and tertiary interactions, may act as xrRNAs.
The presence of conserved secondary and tertiary structural features in MBFV DB elements suggests that their 3D fold dictates function (8), but until now this structure was unknown. We therefore used x-ray crystallography to determine the three-dimensional structure of a DB from Donggang virus (DONGV), an insect-specific flavivirus with homology to many medically important MBFVs. The RNA crystallized as a dimer by a domain exchange between CS2 elements, but both copies had folded RNA pseudoknots and the structure provided sufficient information to model the monomeric form. This model contains a four-way junction in which the CS2 sequence folds into two helices, imparting stability to the overall fold and forming an available surface for potential protein interactions. Chemical probing of the DB structure supports this model and shows the same structure is formed by DENV DBs. Using smFRET experiments to explore conformational dynamics, we discovered that the DB could act as a switch and that the pseudoknotted form is heavily favored under physiologic conditions, with implications for its function as a regulator of viral replication.
MATERIALS AND METHODS
In vitro transcription
DNA templates for transcription were ordered as gBlocks from Integrated DNA technologies (IDT) and cloned into pUC19 and sequenced. DNA was amplified for transcription by PCR using custom DNA primers and Phusion Hot Start polymerase (New England BioLabs). Transcription reactions were conducted using 500 μl PCR reactions as template into 5 ml transcriptions. Transcription reactions contained: ∼0.1 μM template DNA, 8 mM each NTP, 60 mM MgCl2, 30 mM Tris pH 8.0, 10 mM DTT, 0.1% spermidine, 0.1% Triton X-100 and T7 RNA polymerase as well as 5 μl RNasin RNase inhibitor (Promega). Tris was buffered at room temperature. Transcription reactions were left overnight, inorganic pyrophosphates were removed by centrifugation, and the reactions were ethanol precipitated and purified by denaturing PAGE purification. RNAs were eluted overnight at 4°C into ∼40 ml of diethylpyrocarbonate (DEPC)-treated milli-Q filtered water (Millipore) and concentrated using Amicon spin concentrators (Millipore).
RNA crystallization and X-ray diffraction data collection
RNAs for crystallization were prepared as described above. The sequence used for in vitro transcription was 5′-GGGGGCCAGATGTCATGTCTCTCAAGCCTAGGAGACACTAGACACTCTGGACTATCGGTTAGAGGAAACCCCCCCAAAAATGTATAGGCTGAGGTGCTTGTATATAACCTCCACGATGGTGCACCTTGGGCAACACTTCGGTGGCAAATCATCTACA-3′ where the underlined sequence represents a slightly modified sequence derived from the Chilo iridescent virus ribozyme (30) which was used to generate homogenous 3′ ends. Ribozyme cleavage was facilitated at the end of the transcription reaction by adding MgCl2 (final conc. 120 mM) and incubating for 15 min at 65°C. The ribozyme-cleaved RNA was purified on a 10% dPAGE gel and re-folded at 65°C for 3 min at 5 mg/ml in a buffer containing 2.5 mM MgCl2 and 10 mM HEPES–KOH pH 7.5 and then allowed to equilibrate to room temperature before a final concentration of 0.5 mM spermidine was added. Crystal Screens I and II, Natrix I and II (Hampton Research) as well as the HELIX and MIDASplus screens (Molecular Dimensions) were used to perform initial screens at 20°C using single-drop vapor diffusion, and initial hits were optimized using custom screens. Crystals were grown by sitting drop vapor diffusion at 20°C in a solution containing 50 mM Bis–Tris pH 7.0, 25 mM NaCl, 25 mM LiCl, 4.5 mM CaCl2, 50 mM guanidine HCl, 23.75% polyethylene glycol 2000. Tris was buffered at room temperature. Crystals appeared over several weeks. For data collection, crystals were gradually equilibrated into a freezing solution (crystallization solution supplemented to 27.5% polyethylene glycol 2000 + 4 mM iridium(III) hexamine) and were flash-frozen using liquid nitrogen. Diffraction data were collected at Advanced Light Source Beamline 4.2.2 using ′shutterless′ collection at the Iridium L-III edge (1.10208 Å) at 100°K. 180° datasets were collected with 0.2° oscillation images. Data were indexed, integrated, and scaled using XDS (31). SHELX (32) was used to obtain phasing information by single-wavelength anomalous dispersion (CFOM = 53.4).
Model building and refinement
Iterative rounds of model building and refinement (simulated annealing, rigid-body, B-factor refinement, occupancy) using COOT (33) and Phenix (34) were used to generate a complete model of the RNA. Crystal diffraction data, phasing, and refinement statistics are contained in Supplementary Table S1.
RNA chemical probing
DNA templates for T7 RNA transcription follow. In all, underlined = T7 promoter, bold = normalization hairpins, italics = handle for FAM-labeled primer annealing
WT DONGV DB
TAATACGACTCACTATA GGAAACGACTCGAGTAGAGTCGAAAATTAGGGGGCCAGATGTCATGTCTCTCAAGCCTAGGAGACACTAGACACTCTGGACTATCGGTTAGAGGAAACCCCCCCAAAAATGTATAGGCTATTATATCGACAGTTGGAGTCGAGTAGACTCCAACAAAAGAAACAACAACAACAAC
PKmut1 DONGV DB
TAATACGACTCACTATA GGAAACGACTCGAGTAGAGTCGAAAATTAGGGGGCCAGATGTCATGTCTCTCAACGGATGGAGACACTAGACACTCTGGACTATCGGTTAGAGGAAACCCCCCCAAAAATGTATAGGCTATTATATCGACAGTTGGAGTCGAGTAGACTCCAACAAAAGAAACAACAACAACAAC
Pkmut2 DONGV DB
TAATACGACTCACTATA GGAAACGACTCGAGTAGAGTCGAAAATTAGGGGGCCAGATGTCATGTCTCTCAAGCCTAGGAGACACTAGACACTCTGGACTATCGGTTAGAGGAAACCCCCCCAAAAATGTAATCCGTATTATATCGACAGTTGGAGTCGAGTAGACTCCAACAAAAGAAACAACAACAACAAC
PKcomp DONGV DB
TAATACGACTCACTATA GGAAACGACTCGAGTAGAGTCGAAAATTAGGGGGCCAGATGTCATGTCTCTCAACGGATGGAGACACTAGACACTCTGGACTATCGGTTAGAGGAAACCCCCCCAAAAATGTAATCCGTATTATATCGACAGTTGGAGTCGAGTAGACTCCAACAAAAGAAACAACAACAACAAC
DENV-2 DB
TAATACGACTCACTATA GGAAACGACTCGAGTAGAGTCGAAAACAACGGGGGCCAAGGTGAGATGAAGCTGTAATCTCACTGGAAGGACTAGAGGTTAGAGGAGACCCCCCGAAAACAAAAACAGCATATTGACGGTTGGAGTCGAGTAGACTCCAACAAAAGAAACAACAACAACAAC
DNA templates were used for T7 RNA polymerase transcription as described above. Purified RNAs were subjected to chemical mapping procedures as previously described (35). Briefly, 1.2 pmoles of RNA was re-folded in 50 mM HEPES pH 8.0, 100 mM NaCl, 10 mM MgCl2 and equilibrated to room temperature before adding chemical agents. In separate reactions RNA was probed using 5 μl of dimethyl sulfoxide (DMSO), 3 mg/mL N-methylisatoic anhydride (NMIA) or 1% dimethyl sulfate (DMS) and incubated at room temperature for 15–30 min. Reactions were quenched using either a final concentration of 2.3 M 2-mercaptoethanol (for DMS reactions) or 83 mM 2-(N-morpholino) ethanesulfonic acid sodium salt (MES) pH 6.0 (for DMSO or NMIA reactions). Chemically modified RNAs were isolated using a Poly(A)Purist™ MAG Kit (Thermo) and reverse transcribed using SuperScript III reverse transcriptase (Thermo) at 42°C for 60 min using a fluorescently labeled primer (IDT): 5′- /5–6FAM/ AAAAAAAAAAAAAAAAAAAAGTTGTTGTTGTTGTTTCTTT-3′. Labeled DNA products were eluted in HiDi formamide spiked with Gene Scan ROX 500 size standard (Thermo). Samples were run on an Applied Biosystems 3500 XL capillary electrophoresis system and the data were analyzed using HiTRACE (https://ribokit.github.io/HiTRACE/) (36) with MatLab (MathWorks). HiTRACE measures the chemical reactivity at each nucleotide by reverse transcriptase stops. The reactivity due to treatment with DMSO was subtracted from the reactivity due to NMIA or DMS to subtract background signal. Secondary structure diagram coloring was generated using MatLab and HiTRACE RiboKit: RiboPaint (https://ribokit.github.io/RiboPaint/tutorial/). Each experiment was performed four times to generate the mean reactivity and standard deviation at each nucleotide position, except for the DENV DMS probing which was performed three times.
Single-molecule FRET construct generation
Fluorescently labeled RNAs for smFRET were prepared as described (37). In brief, a Cy3-labeled RNA fragment was ligated to unlabeled 5′ and 3′ RNA fragments. The modified Cy3 labeled fragment was generated by labeling an amine-modified RNA containing a 5′ monophosphate ordered from Horizon Discovery with monoreactive Cy3 (GE Life Sciences) and the labeled RNA was purified by reverse-phase HPLC. The unlabeled 5′ fragment was ordered from Horizon Discovery. The unlabeled 3′ fragment was transcribed using T7 RNA polymerase and purified by denaturing polyacrylamide gel electrophoresis as described above. The 3′ fragment was then treated with recombinant BdRppH for 2 h at 37°C in a buffer containing 50 mM Tris–HCl pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1 mM DTT followed by phenol:chloroform:isoamyl alcohol extraction and ethanol precipitation to convert the 5′-triphosphorylated RNA to 5′-monophosphorylated RNA (to facilitate ligation by T4 DNA ligase). Tris was buffered at room temperature. The fluorescently labeled fragment was combined with the 5′ and 3′ unmodified RNAs by DNA-splinted RNA ligation using T4 DNA ligase (New England Biolabs) and purified by denaturing polyacrylamide gel electrophoresis. Fragments for splinted ligation are as follows:
5′ fragment
P – GGGGGCCAGAUGUC
Middle fragment
P – AUGUC[5-LC-N-U]*CUCAA (* 5-aminohexylacrylamino-uridine)
3′ fragment Wild-type
GCCUAGGAGACACUAGACACUCUGGACUAUCGGUUAGAGGAAACCCCCCCAAAAAUGUAUAGGCUAUUAUAUCGACACCUAACCACCAAGCCGACCG
3′ fragment PK mutant
GCCUAGGAGACACUAGACACUCUGGACUAUCGGUUAGAGGAAACCCCCCCAAAAAUGUAAUCCGUAUUAUAUCGACACCUAACCACCAAGCCGACCG
3′ fragment S3 mutant
GCCUAGGAGACACUAGACACUCUGGCCUAUCGGUUAGAGCAAACCCCCCCAAAAAUGUAUAGGCUAUUAUAUCGACACCUAACCACCAAGCCGACCG
3′ fragment 4U insert
GCCUAGGAGACACUAGACACUCUGGUUUUCCCCCAAAAAUGUAUAGGCUAUUAUAUCGACACCUAACCACCAAGCCGACCG
3′ fragment 8U insert
GCCUAGGAGACACUAGACACUCUGGUUUUUUUUCCCCCAAAAAUGUAUAGGCUAUUAUAUCGACACCUAACCACCAAGCCGACCG
DNA Splint
AGTGTCTAGTGTCTCCTAGGCTTGAGAGACATGACATCTGGCCCCC
A biotin-labeled DNA handle (Integrated DNA Technologies) was ordered containing a 5′ biotin: 5′ - biotin - TTCGGTCGGCTTGGTGGTTAGGTGTCGA(iAmMC6T)**AT - 3′ (** Internal amine modified C6 dT) and was designed to hybridize with the RNA for surface immobilization and was labeled with Cy5 for FRET measurements. Cy5 labeling was accomplished by incubation with monoreactive Cy5 (GE Lifesciences) and DNA was purified by reverse-phase HPLC.
smFRET data collection and analysis
Cy3-labeled purified RNAs were annealed to biotinylated Cy5-labeled DNA oligos and immobilized on PEGylated cover slides (Ted Pella, Inc.). PEGylated cover slides were prepared as described (38). Slides were imaged on a total internal reflection fluorescence microscope (Nikon Eclipse Ti-E) using an Andor iXon+ DU897 CCD camera with a 100 ms integration time. The image was spectrally separated using the DV2 Dualview (Photometrics) and custom Semrock filters. FRET studies were performed in 100 mM NaCl, 50 mM Tris pH 7.5, 10% glycerol, 0.1 mg/mL BSA, 2 mM Trolox, 1% glucose, 1 mg/ml catalase, 1 mg/ml glucose oxidase and the indicated MgCl2 concentrations at room temperature. Tris was buffered at room temperature. Analysis was performed using custom MATLAB (MathWorks) scripts.
Structure-based sequence alignments
An initial list of 29 flaviviral DB sequences was manually compiled and a structural alignment was performed automatically using Locarna (https://rna.informatik.uni-freiburg.de/LocARNA/Input.jsp) and exported in Stockholm format. A search for structurally similar RNA sequences was performed using Infernal v. 1.1.2 (39) searching a database of Flaviviridae genomes from the NCBI viral genome browser (https://www.ncbi.nlm.nih.gov/genomes/GenomesGroup.cgi?taxid=11050). The consensus sequence and secondary structure model as well as the statistical analysis of covariation were calculated using the RNA Significant Covariation Above Phylogenetic Expectation (R-scape) program (40) visualized using R2R (41) and labeled in Adobe Illustrator.
Accession numbers for sequences used are as follows: NC_015843.2 Tembusu virus, NC_034151.1 T′Ho virus, NC_001477.1 Dengue virus 1, NC_006551.1 Usutu virus, NC_001563.2 West Nile virus lineage 2, NC_001474.2 Dengue virus 2, NC_018705.3 Ntaya virus, NC_000943.1 Murray Valley encephalitis virus, NC_001437.1 Japanese encephalitis virus, NC_012533.1 Kedougou virus, NC_009942.1 West Nile virus lineage 1, NC_002640.1 Dengue virus 4, NC_009028.2 Ilheus virus, NC_032088.1 New Mapoon virus, NC_009029.2 Kokobera virus, NC_007580.2 St. Louis encephalitis virus, NC_012532.1 Zika virus African isolate, NC_001475.2 Dengue virus 3,NC_012534.1 Bagaza virus, NC_035889.1 Zika virus Brazilian isolate, NC_009026.2 Bussuquara virus, NC_024017.1 Nhumirim virus, NC_002640.1 Dengue virus 4, NC_018705.3 Ntaya virus, NC_003635.1 Modoc virus, NC_034151.1 T′Ho virus, NC_001563.2 West Nile virus lineage 2, NC_009942.1 West Nile virus lineage 1, NC_006551.1 Usutu virus, NC_001475.2 Dengue virus 3, NC_007580.2 St. Louis encephalitis virus, NC_001477.1 Dengue virus 1, NC_015843.2 Tembusu virus flavivirus, NC_008718.1 Entebbe bat virus, NC_004119.1 Montana myotis leukoencephalitis virus, NC_009028.2 Ilheus virus, NC_009026.2 Bussuquara virus, NC_005039.1 Yokose virus, NC_033715.1 Nounane virus, NC_000943.1 Murray Valley encephalitis virus, NC_012735.1 Wesselsbron virus, NC_008719.1 Sepik virus, NC_017086.1 Chaoyang virus, NC_016997.1 Donggang virus, NC_002031.1 Yellow fever virus, NC_032088.1 New Mapoon virus.
Protein expression
6x-histidine tagged Kleuveromyces lactis Xrn1 and Bdellovibrio bacteriovorous RNA pyrophosphate hydrolase (RppH) were expressed in BL21 E. coli cells and purified using nickel affinity and size exclusion chromatography by previously described methods (42).
Xrn1 digests
1 μg pure RNA was resuspended in a 20 μl solution containing 50 mM Tris pH 8.0, 100 mM NaCl, 1 mM DTT, 10% glycerol and either 2 mM or 10 mM MgCl2. Tris was buffered at room temperature. The RNA was re-folded at 65°C for 5 min. 1 μl Bdellovibrio bacteriovorus RppH at a concentration of 0.5 μg/μl was added to all reactions and 1 μl Kleuveromyces lactis Xrn1 was added to every other reaction. Reactions were incubated for 1 h at 37°C and separated on a 10% denaturing PAGE gel and visualized by staining with ethidium bromide. DNA constructs for RNA transcription for Xrn1 digestion were as follows:
Zika virus Xrn1-resistant RNA ± leader (leader italicized)
GAAGCACCAATCTTAATGTTGTCAGGCCTGCTAGTCAGCCACAGCTTGGGGAAAGCTGTGCAGCCTGTGACCCCCCCAGGAGAAGCTGGGAAACCAAGCCTAT
Zika virus DB RNA ± leader (leader italicized)
GAAGCACCAATCTTAATGTTCAATCTGGGGCCTGAACTGGAGATCAGCTGTGGATCTCCAGAAGAGGGACTAGTGGTTAGAGGAGACCCCCCGGAAAACGCAAAACAGCATA
Northern blots
Viral infection
A549 cells were plated into six-well plates so that at the time of harvest each well would have approximately 1 million cells. For infection, 2 ml of culture media was aspirated from each well, followed by a wash with 1 mL of 1× PBS. Each well was infected with 500 μl of ZIKV viral inoculum at an MOI of 3. Plates were incubated with viral inoculum at 37°C for 1 h. After incubation viral inoculum was removed and 2 ml of complete Ham′s F-12 media was added to each well. After 48 h, cells were harvested. For harvest, media was aspirated from the plate. Each well was washed once with 1 ml 1× PBS and RNA was extracted using the EZNA Total RNA kit (Omega Biotek). 375 μl of TRK lysis buffer with β-Mercaptoethanol was added to each well and incubated for 3 min at room temperature. Two biological replicates were combined into one sample for RNA extraction (each RNA extraction sample was from an approximate total of 2 million cells) and the RNA was column purified via manufacturer′s instructions.
Preparation of probes
A 35-mer DNA oligo complementary to nucleotides 263–298 of the PRVABC59 strain 3′ UTR (sequence: 5′- TCCTCTAACCACTAGTCCCTCTTCTGGAGATCCAC-3′) as well as a DNA oligo complementary to nucleotides 323–358 (sequence: 5′-CTCATGGAGTCTCTGGTCTTTCCCAGCGTCAATAT-3′) and complementary to the U6 snRNA (sequence: 5″- TATGGAACGCTTCACGAATTTGCGTGTCATCC-3′) was ordered from IDT. 100 pmol of DNA was incubated with 2 μl 5mCi [γ32-P]ATP (Perkin Elmer) and 4 μl T4 polynucleotide kinase (New England BioLabs) in a 100 μl reaction for 2 h and purified using P-30 spin columns (BioRad). The radiolabeled probe was heated to 100°C for 2 min and resuspended in 10 ml of ULTRAhyb Oligo hybridization buffer (Ambion). Blots were incubated with 10 ml of the resuspended probe overnight at 42°C.
Blotting procedure
1 μg of total RNA from mock- and ZIKV-infected cells was resuspended in 2× formamide RNA loading buffer and run on a 6% denaturing PAGE gel (Invitrogen) with an RNA ladder (Thermo Scientific). Gels were stained with ethidium bromide and imaged to obtain the position of the RNA ladder. The RNA from the gels was subsequently transferred to a HyBond-N+ nylon membrane (GE Life Sciences) using an electrophoretic transfer apparatus (Idea Scientific). The membrane was crosslinked using a UV stratalinker and blocked at 42°C using ULTRAhyb Oligo hybridization and blocking buffer (Ambion) for 2 h while rotating. Blots were probed rotating at 42°C with a 35-mer DNA oligo prepared as described above overnight and washed in 2× saline-sodium citrate (SSC) buffer with 0.5% SDS for 10 min at 42°C four times. The blots were imaged with a phosphor screen and a Typhoon scanner (GE Life Sciences) and were aligned with the ethidium-stained image of the gels to obtain the position of size standards from the RNA ladder.
RESULTS
Base pairing covariation identifies a conserved dumbbell RNA in the viral 3′ UTR
We first bioinformatically analyzed MBFV 3′ UTRs to identify conserved secondary structures and features of DBs from diverse sources. Overall, the 5′ DB is more variable between species and can be absent or altered compared to the 3′ DB (Figure 1A). We therefore decided that structural and mechanistic studies of the more conserved 3′ DB would be more representative of multiple MBFV species, an idea supported by the presence of CS2 within it (3). To assemble a list of homologous DB structures, we started with a small sequence alignment of 3′ DB RNAs, identified 46 putative DB sequences by RNA base pairing covariance using the Infernal algorithm (39), then aligned them using R-scape (40). This verified helices (P1 and P2) by base pair covariance (Figure 1B). A third helix (P3) was predicted to fold by the mFold and Vienna RNA secondary structure prediction algorithms (43,44), however its sequence is so conserved that there is no base pairing covariation. Manual alignment of the sequences identified co-varying base pairs consistent with an RNA pseudoknot in all the DB sequences (Figure 1B). This pseudoknot often overlaps with the 3′ CS; in such cases the pseudoknot cannot form when the 5′ and 3′ CSs base pair during viral replication (Figure 1C) (11).
The dumbbell crystal structure reveals a complex three-dimensional fold and RNA pseudoknot
To understand how DB structure may participate in global RNA conformational changes, we used X-ray crystallography to reveal its 3D structure. After screening DBs from multiple MBFVs, we obtained diffracting crystals using a sequence from DONGV, an insect-specific flavivirus (45,46). The DONGV DB sequence is conserved with other MBFVs including DENV, WNV, YFV, and ZIKV, thus structural information from DONGV DB is broadly applicable (Supplementary Figure S3). DONGV DB RNA crystals were soaked with iridium (III) hexammine, which was used to determine phases and solve the 3D structure (47). The final structural model was refined to a resolution of 2.1 Å, providing high-resolution information (Table 1).
Table 1.
Crystallographic data collection and refinement statistics
| Data collection | |
|---|---|
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 65.93, 49.67, 73.46 |
| α, β, γ (°) | 90, 99.73, 90 |
| Wavelength (Å) | 1.102 |
| Resolution (Å) | 44.74–2.1 (2.175–2.1) |
| R merge | 0.1303 (0.8781) |
| R-meas | 0.1367 (0.9265) |
| R-pim | 0.04105 (0.2917) |
| I/σI | 17.95 (3.17) |
| Completeness (%) | 99.81 (99.89) |
| Multiplicity | 10.9 (10.0) |
| CC1/2 (%) | 0.998 (0.834) |
| CC* | 0.999 (0.954) |
| Wilson B-factor | 34.69 |
| Refinement | |
| Resolution (Å) | 44.74–2.1 (2.175–2.1) |
| No. of unique reflections | 27604 (2741) |
| R work / R free | 0.23/0.26 |
| CCwork/CCfree | 0.95 (0.74)/0.94 (0.75) |
| No. atoms | |
| RNA | 3822 |
| Ligand/ion | 357 |
| Water | 127 |
| B-factors | |
| RNA | 37.22 |
| Ligand/ion | 66.96 |
| Water | 33.03 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.039 |
| Bond angles (°) | 0.56 |
| Clashscore | 3.92 |
| Coordinate error | 0.33 |
| Phase error | 29.78 |
*Highest resolution shell is shown in parenthesis.
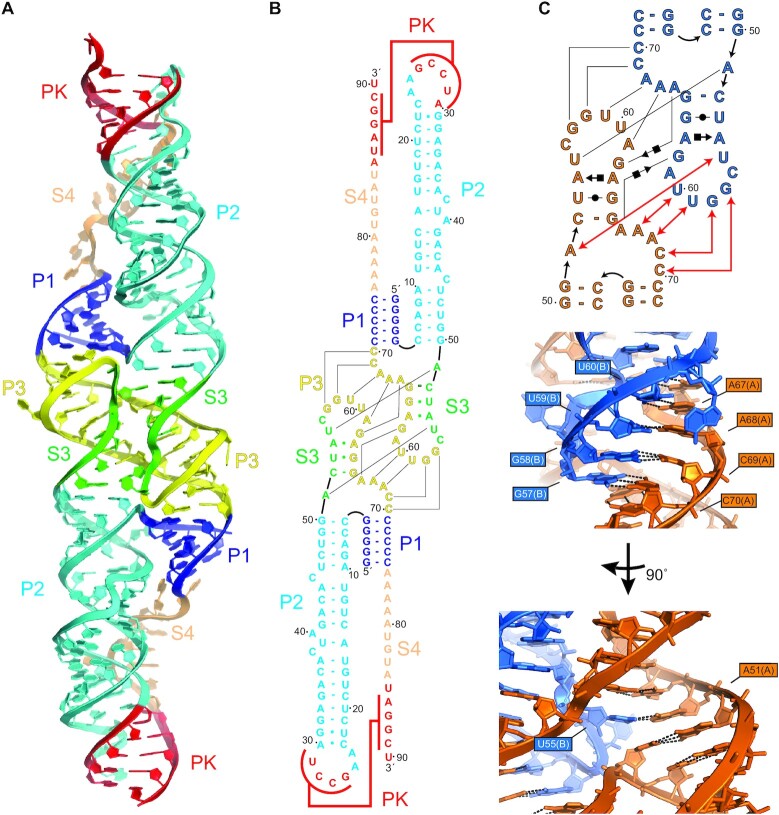
The DONGV DB crystallized as a dimer in the asymmetric unit (Figure 2A, Supplementary Figures S4, S5A), with both copies adopting a similar fold (Supplementary Figure S5B). As predicted, helices P1, P2 and the pseudoknot formed in both copies. However, the predicted P3 stem did not form, instead its sequence made base pairs with a linker predicted to be single stranded (S3) (Figure 2A and B). Additional intermolecular base pairs also formed in this region, forming an extensive interface of crystal contacts (Figure 2B and C). The largest difference between the two molecules in the crystallographic dimer was in the single-stranded RNA linker S4 and there was a slight difference in the orientation of the P2 helix (Supplementary Figure S5B).
Figure 2.
Crystal structure of the DONGV DB RNA. (A) Overall structure of the dimer form of the wild-type DONGV DB observed in the crystal. Bound iridium (III) hexammine ions used for SAD phasing are omitted for clarity. Predicted stems from Figure 1B are labeled (P1 = blue, P2 = cyan, P3 = yellow, pseudoknot = red) as are the predicted unpaired regions (S3 = green, S4 = orange). (B) Base pairing interactions observed in the RNA crystal structure colored as in (A). Lines indicate base pairing observed between copy 1 and copy 2. (C) Secondary structure in Leontis-Westhof notation (top) and detailed 3D view (bottom) of the dimer interface highlighting in trans base pairing interactions between molecule A (orange) and molecule B (blue). Red lines in the secondary structure show base pairing interactions labeled in the 3D view.
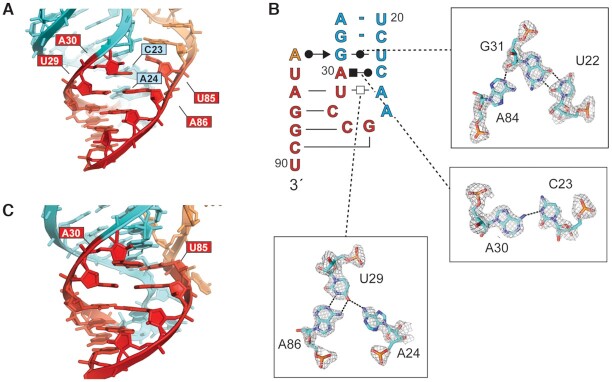
Both copies of the DONGV DB observed in the asymmetric unit formed the predicted pseudoknot with at least four base pairs, but the two copies differed somewhat in this region (Figure 3A and B). Molecule A (chain A in the deposited coordinate file) formed four Watson–Crick (WC) base pairs in the pseudoknot, however the base pair predicted to form between A30 and U85 was not observed (Figure 3A). Instead, A30 participates in an A•C interaction with C23, which is positioned for this interaction by adjacent residues in the P2 loop. A24 stacks underneath C23, participating in an A-U-A base triple interaction with the U29-A86 base pair. U85 stacks on A84, which hydrogen bonds to the 2′OH of G31 (Figure 3B). In contrast, molecule B (chain B in the deposited coordinate file) has five base pairs in its pseudoknot with the A30–U85 base pair formed as predicted, and molecule B has neither the A•C base pair nor the A–U–A base triple observed in molecule A (Figure 3C). The distinct pseudoknot organizations in molecules A and B may represent two alternate conformations of the RNA captured in the crystal.
Figure 3.
Molecular details of the RNA pseudoknot. (A) Detailed view of the DB RNA pseudoknot in molecule A of the asymmetric unit colored as in Figure 2A. Nucleotides A30 and U85 are labeled, this predicted base pair is not formed. The position of A30 is stabilized by non-canonical base pairing interactions in the P2 stem–loop (Figure 3B). The position of bound iridium (III) hexammine ions are not shown. (B) Secondary structure in Leontis-Westhof notation of molecule A pseudoknot loop, insets show details of non-canonical base pairing stabilizing the loop in the absence of the A30–U85 base pair with electron density from a composite omit map of the crystal shown at the 1σ contour level. (C) Detailed view of the pseudoknot of molecule B. A30 and U85 are labeled and are base paired in this copy.
Molecular modeling of a monomeric structure
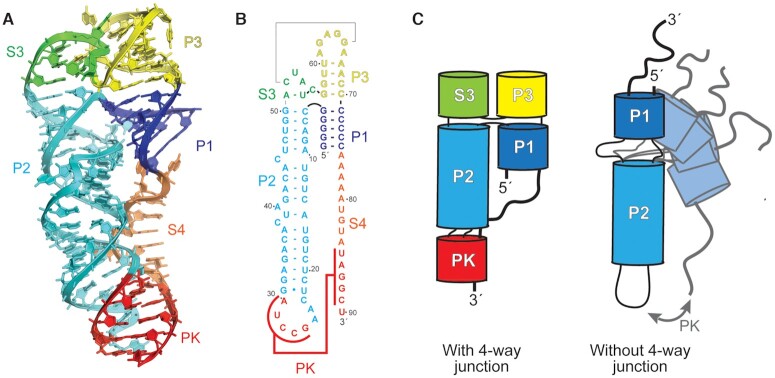
Examination of each copy of the DB individually, outside of the context of the crystal, revealed distorted base pairing interactions in the S3 and P3 regions with far from ideal A-form RNA geometries and large internal solvent channels (Supplementary Figure S5B). This suggests that crystal packing in this region distorts the RNA fold compared to its solution conformation. Close examination of the intermolecular crystal contacts revealed that the predicted base pairs in the P3 stem formed, but in trans, as a domain-swap between the two copies of the RNA (Figure 2C). Domain-swapping is frequently observed in crystallography of both proteins and RNA, due to the high concentration of molecules; interactions that occur in cis at physiologic concentrations form in trans in the crystal (29,42,48,49). We therefore hypothesized that the P3 stem is formed in solution in cis, but forms in trans in the crystal. We used the fact that the base pairing arrangement of the consensus model was observed in the crystal, albeit in a domain-swapped interaction, to model the monomer structure as it would occur in cis. We excised nucleotides 55–63 from copy 2 and transferred them to copy 1, and vice versa, to model the P3 stem formed in cis (Supplementary Figure S6). The remainder of the RNA structure is consistent with this arrangement and few other perturbations were required to create the model, other than repositioning the connecting nucleotides 54 and 64. These two were deleted and positioned without using electron density to connect the two chains, producing a monomeric model of the RNA guided by the crystal structure (Figure 4).
Figure 4.
Model of the monomeric DB RNA. (A) Three-dimensional model of the monomeric solution structure of the DONGV DB guided by the crystal structure. The structure forms a four-way junction with co-axial stacking of the P1 (blue) and P3 (yellow) stems and with the P2 (cyan) and S3 (green) stems. (B) Secondary structure map of the DONGV DB solution structure model. (C) Cartoon model demonstrating a four-way junction with limited conformational space (left) compared against an RNA without a four-way junction (right) that can sample many different conformations.
Chemical probing supports the monomeric structure model of the dumbbell RNA
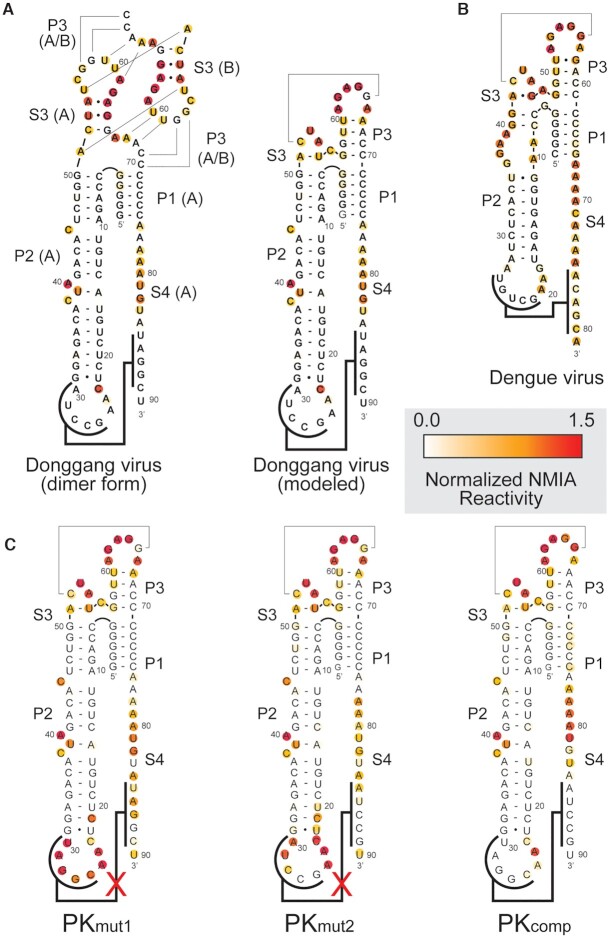
We used chemical probing to test if the resultant modification patterns are consistent with our monomeric structural model. Wild-type DONGV and DENV-2 DBs were used as well as a DONGV DB with a pseudoknot disrupted by mutation. We used N-methylisatoic anhydride (NMIA), which detects flexible regions of the RNA as a proxy for unpaired nucleotides (50), and dimethyl sulfate (DMS), which detects N1 on adenines and N3 on cytosines that are not hydrogen bonded (51). The reactivity pattern was consistent with the monomeric structural model in the P3 and S3 regions using NMIA (Figure 5A and Supplementary Figure S7) and using DMS (Supplementary Figure S8 and S9), with high reactivity in loop regions and low reactivity in Watson-Crick base-paired regions. In contrast, the reactivity pattern did not agree with the dimeric form observed in the crystal in the P3 and S3 regions, providing strong evidence that the monomeric structural model more accurately represents the folded solution state. Probing of the DONGV DB pseudoknot mutants confirmed the formation of the pseudoknot (Figure 5C). Specifically, residues 26–30 were mutated to their complementary sequence (PKmut1) as well as residues 85–89 (PKmut2), and a third RNA contained both mutations to restore the pseudoknot by forming compensatory base pairs (PKcomp). As predicted PKmut1 and PKmut2 showed higher reactivity in the regions forming the pseudoknot, while the PKcomp mutant′s reactivity pattern resembled wild-type RNA.
Figure 5.
NMIA chemical probing of the DB RNA. (A) Secondary structure of the DONGV DB RNA in the form observed in the crystal structure (dimer form) compared with the modeled form. NMIA reactivity of the wild-type DB sequence is color-coded (red is high reactivity, white is low reactivity). (B) NMIA reactivity of the DENV-2 DB plotted on the putative secondary structure based on homology to DONGV. (C) NMIA reactivity of pseudoknot mutant RNAs (PKmut1 and PKmut2) plotted on the modeled RNA secondary structure. The mutations in PKmut1 and PKmut2 were combined to generate compensatory base pairing (PKcomp).
We next probed a DB from DENV-2 to determine if the RNA fold is conserved. The similar reactivity profiles between DONGV and DENV-2 indicated that the overall folding of the RNA is maintained (Figure 5A and B and Supplementary Figure S7). In particular, both had reactivity patterns consistent with the formation of the P1, P2 and P3 stems and were protected in their pseudoknots. This, combined with the conservation in the consensus model (Figure 1B), strongly supports that the DONGV DB structure solved here is representative of DB elements across the flaviviruses.
The modeled monomeric RNA structure contains a four-way junction
A defining feature of the DONGV DB RNA structure is an acute angle between the P1 and P2 stems, which appears to be important for favoring pseudoknot formation. This angle is associated with a sharp kink in the RNA backbone that places the 3′ single-stranded region (S4) proximal to the P2 stem–loop (Figure 4A and B). In the crystal structure, several bases in the S3 and P3 regions stack on the P1 and P2 helices. Base stacking is central to stabilizing folded RNA structures so these interactions likely stabilize the global architecture of the DB, including the acute angle between P1 and P2. In the monomeric structure model, the stacking pattern of P1 and P2 with S3 and P3 create a structural feature that could be called a four-way or a three-way junction (Figure 2C). Specifically, in S3 the A51-U55 pair stacks on the P2 stem, and the C52–G65 pair stacks on A51–U55. The C52–G65 base pair may help lock the angle between the S3 and P3 stems (Figure 6A and B). The P3 stem in the model stacks on the P1 stem to complete the junction, but only contains one A-U base pair and one long-range G–C base pair (Figure 4B and 6A). Comparing the model solution structure with existing RNA structures reveals that the DONG DB junction is very similar to a four-way junction in RNase P that stabilizes the specificity domain, classified as a member of family H four-way junctions (Supplementary Figure S10) (52,53). Based on this, we refer to the DONG DB junction as a four-way junction. Overall, the model suggests a hypothesis that the four-way junction structure locally favors the acute angle between P1 and P2, which globally brings the two complementary sequences of the pseudoknot in proximity (Figure 4C).
Figure 6.
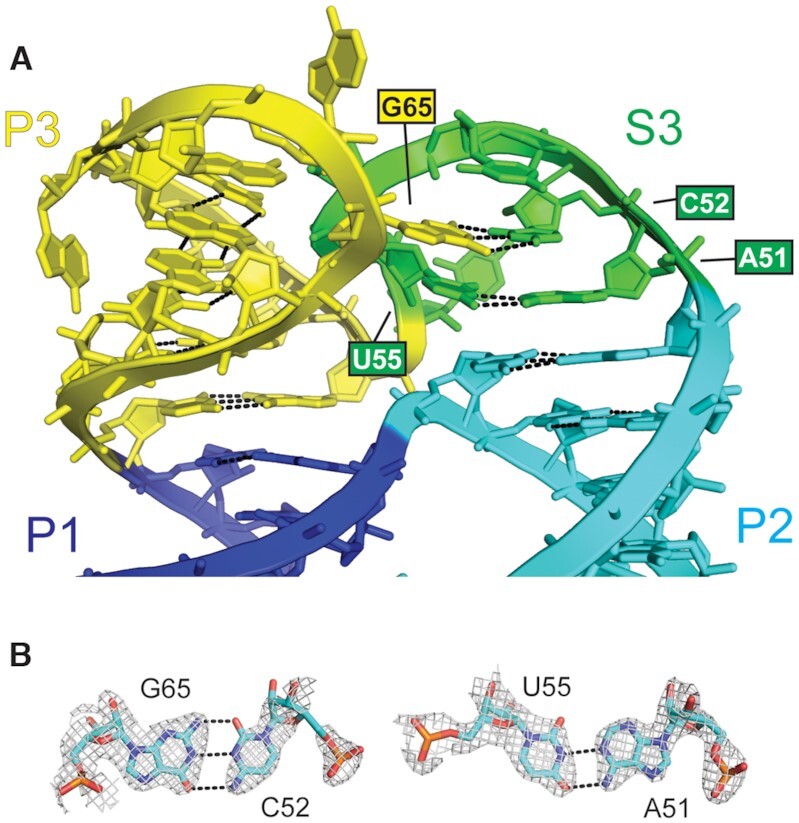
Molecular details of the modeled four-way junction. (A) Detailed view of the four-way junction from the modeled monomeric RNA. The four-way junction is formed by base stacking between the P1 (blue) and P3 (yellow) stems and between the P2 stem (cyan) and S3 (green) stems. Base pairs between A51 and U55 and between C52 and G65 are labeled. (B) Molecular details of the base pairs highlighted in (A). Electron density is from a composite omit map of the crystal structure at 1σ contour level.
The dumbbell pseudoknot is the heavily favored conformation
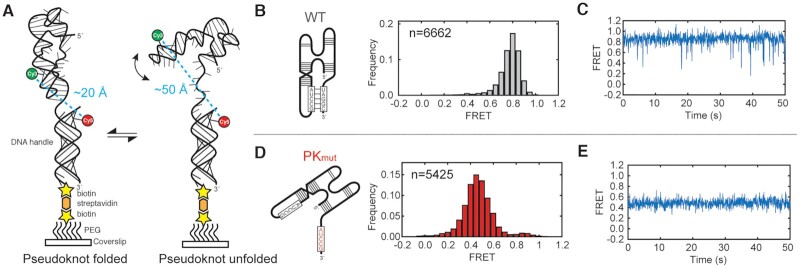
The crystal structure and solution chemical probing suggest that the pseudoknot tertiary interaction is heavy favored. To directly query the stability and dynamics of the pseudoknot, we used single-molecule Förster resonance energy transfer (smFRET) to observe pseudoknot dynamics with temporal resolution on individual molecules. A Cy3 fluorophore was attached to the DONGV DB in the P2 stem (Figure 7A and Supplementary Figure S11A), and a 3′ extension was added. This extension is complementary to a DNA oligonucleotide used to immobilize the complex on a microscope coverslip through a biotin-streptavidin linkage (Figure 7A and Supplementary Figure S11A). This DNA oligo was modified with Cy5 at a position designed to display higher FRET when the pseudoknot is formed relative to when it is not formed. The Cy3-modified RNA was annealed to the Cy5-modified DNA oligo and immobilized on a microscope coverslip, followed by smFRET observation of real-time RNA dynamics (Figure 7A). Most individual spots that photobleached had single photobleaching events, suggesting that each spot on the microscope slide had only a single Cy3 and Cy5 molecule and thus the DB is monomeric (Supplementary Table S1).
Figure 7.
smFRET measurements of pseudoknot stability. (A) Labeling strategy for smFRET experiments. DONGV DB RNA was site-specifically labeled with Cy3. A 3′ extension on the RNA was used to immobilize the RNA construct on a microscope slide based on hybridization to a biotinylated and Cy5-labeled DNA handle. A folded pseudoknot is predicted to have a high FRET value and an unfolded pseudoknot is predicted to have a lower FRET value. (B) smFRET histogram of wild-type DB RNA. (left) Model of the RNA construct. (right) Histogram of the distribution of FRET values, n = number of molecules observed. (C) Representative smFRET trace of a wild-type DB RNA. (D) smFRET histogram of PKmut DB RNA. (left) Model of the RNA construct used. (right) smFRET histogram. (E) Representative smFRET trace of a PKmut DB RNA.
smFRET histograms compiled from wild-type DONGV DBs showed a single Gaussian distribution centered at 0.8 FRET (Figure 7B). To test whether this FRET state corresponded to the pseudoknotted form, we tested pseudoknot mutant (PKmut) RNA in this assay. The PKmut RNA showed a single predominant FRET distribution at ∼0.5 FRET, verifying that the high FRET state observed in the wild-type RNA is the pseudoknotted form (Figure 7D). Thus, smFRET distributions from the wild-type RNA showed that the folded pseudoknot is the heavily favored conformation.
We next examined the conformational dynamics of individual DB molecules, showed in the smFRET traces as a function of time. As expected, PKmut single-molecule traces displayed a stable 0.5 FRET value without any detectable transitions to the higher FRET value (Figure 7E and Supplementary Figure S11C), consistent with the pseudoknotted state being destabilized by the mutation. In contrast, wild-type traces showed molecules predominantly in the folded 0.8 FRET state, with only occasional transient excursions to a 0.5 FRET state. These transient states are so short-lived as to be near the limit of detection of the camera (∼0.1 s) (Figure 7C and Supplementary Figure S11B). The short duration of the unfolded 0.5 FRET state shows that breaking the pseudoknot interaction is followed by rapid re-formation, consistent with a low kinetic barrier in the transition from the unfolded to folded state (Supplementary Figure S12). However, the distribution of states revealed that the pseudoknotted state predominates and therefore is thermodynamically much more stable than the unfolded state. The transient excursions to a lower FRET state were apparent in conditions containing 2 mM MgCl2 but not at 10 mM MgCl2, demonstrating Mg2+′s role in stabilizing the pseudoknot fold (Supplementary Figure S13). This is expected, as Mg2+ dependence is a common feature of RNA tertiary structure (54).
The DB pseudoknot appears to be more stable than many other pseudoknots measured in biophysical assays including the human telomerase RNA pseudoknot (55), the PreQ1 riboswitch pseudoknot (56), and the dianthovirus XRN-resistant RNA pseudoknot (42). Since the DB pseudoknot is mutually exclusive with long-range 5′-3′ base pairing associated with viral replication (Figure 1C), the stability of the DB pseudoknot suggests that the DB folding landscape favors the translation-competent and not replication-competent form of the viral genome.
Coupling between the four-way junction and the pseudoknot
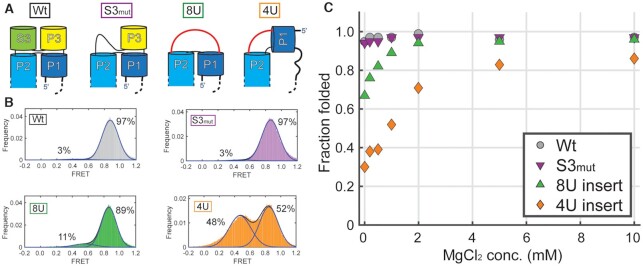
To determine if the stability of the DB pseudoknot is enhanced by the presence and structure of the four-way junction, we designed several mutants to interrogate using smFRET. These (1) disrupted specific base pairs in the four-way junction, (2) reduced base stacking or (3) restricted the flexibility between the P1 and P2 helices (Figure 8A).
Figure 8.
Effect of alterations to the four-way junction on pseudoknot folding. (A) Diagram of the expected tertiary structures of the four-way junction of wild-type, S3mut (A51C/G65C), 8U insert, and 4U insert mutations. Red lines indicate the position of the 8- and 4-nucleotide poly-uridine insertions replacing residues 51–70 of the DB RNA. (B) smFRET histograms of the four constructs in conditions containing 1 mM MgCl2. FRET values of 3289, 7293, 4092 and 5661 molecules respectively were collected over 2 seconds at a 0.1 s integration time to generate histograms of all FRET values observed. Histograms were fit to the sum of two Gaussian distributions, with fits displayed on graph as well as the relative percentage of the ∼0.5 and ∼0.8 FRET states. (C) Effect of MgCl2 on pseudoknot folding of Wt, S3mut, 8U insert, and 4U insert constructs. smFRET histograms (Supplementary Figure S14) were collected as in (B). The fraction folded in the pseudoknot (∼0.8 FRET state) were plotted against the MgCl2 concentration in the experiment.
To interrogate the contribution of specific base pairs and base stacking in the four-way junction, we eliminated the A51-U55 and C52–G65 base pairs that organize the junction (A51C/G65C, labeled S3mut) (Figure 6). smFRET showed that S3mut formed the pseudoknot virtually indistinguishably from wild-type, suggesting that pseudoknot stability tolerates alterations to the junction structure, or that the structure can compensate with non-Watson-Crick base pairs (Figure 8B).
We created additional mutations to the four-way junction by removing the S3 and P3 and replacing them with poly-uridine linkers of either eight uridines (8U) or four uridines (4U). The 8U mutant removes the specific four-way junction's structure and stacking but preserves flexibility between the P1 and P2 helices. Thus, this mutant RNA can still form an acute angle between P1 and P2 (Figure 8A). The 4U mutant not only reduces base stacking, but also potentially restricts the P1 and P2 helical angle (Figure 8A). smFRET experiments at 1 mM MgCl2 showed that the 8U mutant occupies the pseudoknot state 89% of the time and the 4U mutant 52%, compared with 97% for wild-type (Figure 8B). These trends are more pronounced at lower magnesium. For instance, at 0 mM MgCl2 the 8U mutant occupies the pseudoknot state 67% of the time and the 4U mutant 30% of the time, compared with 95% for wild-type. At higher MgCl2 concentrations the 8U and 4U pseudoknot forms were stabilized, with the 8U construct closely resembling wild-type pseudoknot stability (Figure 8C and Supplementary Figure S14).
Taken together these data illustrate that the acute angle between the P1 and P2 helices is important for pseudoknot stability and base stacking in the four-way junction stabilizes pseudoknot folding on the other side of the molecule. However, the data also suggests that the specific structure of the four-way junction is not strictly required, rather the general ability for the P1 and P2 helices to bend into a close conformation is most important. Elevated [MgCl2] heavily favors pseudoknot formation, with high magnesium concentrations compensating for the energetic contributions of interactions lost in mutants. Magnesium may generally reduce electrostatic repulsion between regions of the RNA that must come into proximity or could form specific interactions. We could not definitively identify coordinated magnesium ions within our crystal structure; the unusually high number of bound iridium (III) hexammines used for SAD phasing may have displaced magnesium.
The state of the pseudoknot during infection could be affected by several factors including the amount of free magnesium in the cell as well as temperature, bound protein co-factors, and mechanical stresses, therefore we cannot definitively predict the state of the pseudoknot at any point during infection based on chemical parameters alone. Nevertheless, when we consider the trends of our quantitative magnesium titrations, they suggest that the base stacking and steric interactions in the P3/S3 region can exert subtle effects on the RNA pseudoknot located on the other side of the molecule, approximately 40Å away. Because the S3 and P3 regions are highly conserved in a way that cannot be explained by just RNA structure stability, they likely are a platform for binding of protein factors. Binding of these factors could influence the distal pseudoknot (either stabilizing or destabilizing it) to influence genome circularization and replication.
Dumbbell RNAs do not efficiently resist the 5′→3′ exonuclease XRN1
xrRNA elements in the flaviviral 3′ UTR form 3D folds that resist degradation by host 5′→3′ exonucleases, predominantly the host protein XRN1 (57,58). These xrRNAs form a structure where the 5′ end is surrounded by a distinctive ring-like fold, forming a ‘molecular brace’ preventing the 5′ end from being pulled into the XRN1 active site (29,59,60), creating sfRNAs (57). It has been proposed that DBs may also be XRN1 resistant, as lower molecular weight sfRNAs that could correspond to XRN1 stalled at DB RNA elements have been observed (58). However, in vitro XRN1 resistance assays and northern blots of YFV infections suggest that these low molecular weight sfRNAs may be due to trimming of the 3′ end of the sfRNA, not DB exonuclease resistance. (61,62).
The DONGV DB 5′ end is surrounded by a complex RNA fold (Figure 4B) but lacks the distinctive xrRNA topology in which a continuous strand forms a ring of defined size through which the 5′ end is threaded (59). Therefore, we tested DB XRN1 resistance using a ZIKV model system with an established assay. Briefly in vitro transcribed RNA with a 29 nucleotide 5′ leader was modified to contain a 5′ monophosphate, which allowed recombinant purified K. lactis Xrn1 to load on the 5′ end and begin degradation (62). In 2 mM MgCl2, the ZIKV xrRNA was highly resistant to K. lactis Xrn1, with a significant majority of the input RNA shifted to a lower molecular weight band corresponding to the partially degraded RNA. In contrast, at 2 mM MgCl2 the ZIKV DB was efficiently degraded as no Xrn1-resistant band was observed (Figure 9A). Interestingly, at 10 mM MgCl2 an Xrn1-resistant fragment was observed in the DB, albeit at a much lower intensity (∼20% of input). This is consistent with high magnesium concentrations stabilizing the DB fold (Supplementary Figure S13), although even at these higher magnesium concentrations only a small amount of Xrn1 resistance was observed. Because the context of xrRNAs within the 3′ could influence their ability to resist XRN1 (63), we also tested Xrn1 resistance of a ZIKV 3′ UTR fragment containing other elements upstream and downstream of the 3′ DB. This 3′ UTR exhibited no significant Xrn1 resistance, demonstrating that the local context of the ZIKV DB RNA had no effect (Supplementary Figure S15).
Figure 9.
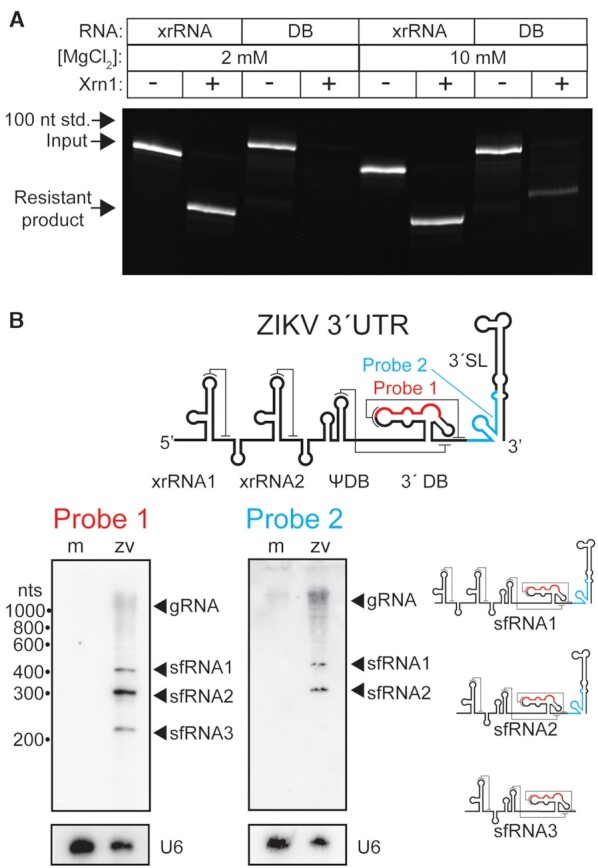
The DB lacks resistance to the 5′→3′ exonuclease Xrn1. (A) Xrn1 digest of ZIKV Xrn1-resistant RNA (xrRNA) or ZIKV DB RNA in the presence of 2 mM or 10 mM MgCl2. The upper bands correspond to RNAs with an intact 5′ single-stranded leader that have not been treated with Xrn1. The lower bands correspond to RNA fragments resistant to Xrn1 digestion, in which Xrn1 has removed the 5′ single stranded linker leaving the resistant fragment behind. The position on the gel where a 100 nucleotide RNA standard migrated to is marked as indicated. (B) Upper panel: the 3′ UTR of ZIKV highlighting the position that northern blot DNA probes hybridize to for probe 1 (red) and probe 2 (cyan). Lower panel: northern blots from identical mock (m) and ZIKV infected (ZV) cellular RNA probed with either probe 1 or probe 2. Blots were also probed with a DNA oligo hybridizing with the U6 snRNA as a loading control. Positions of the genomic RNA (gRNA) and subgenomic flaviviral RNAs (sfRNAs) are as labeled. Cartoon diagrams of sfRNA1, sfRNA2, and sfRNA3 are shown.
Given their limited Xrn1 resistance in in vitro assays, we next determined if MBFV DBs demonstrate resistance during infection. Infection with DENV, WNV, and ZIKV results in two sfRNAs that are clearly due to two xrRNAs, but other lower molecular weight bands as well (29,58,59). These shorter sfRNAs could be formed by two possible mechanisms: either XRN1 resistance by the DBs or trimming of the 3′ end by another nuclease. To distinguish between these mechanisms, we infected A549 cells with ZIKV and detected sfRNAs using northern analysis with two different oligonucleotide probes: one hybridized within the DB and a second hybridized within the terminal 3′ stem-loop. If the DBs are XRN1 resistant during infection, the same sfRNA fragments should be observed with both probes. Conversely, if the additional sfRNA species are formed by trimming of the 3′ end, the northern blot with a probe directed against the 3′ stem-loop should have fewer bands than the northern blot probed against the DB. With the probe directed against the DB, three predominant sfRNAs were observed—sfRNA1, sfRNA2, and sfRNA3—with sfRNA1 and sfRNA2 known to be formed by XRN1 resistance of xrRNA1 and xrRNA2 (Figure 9B). With the probe against the 3′ stem-loop, only two sfRNAs were observed, sfRNA1 and sfRNA2 (Figure 9B). This clearly demonstrates that in ZIKV infection, sfRNA3 is formed due to 3′ trimming of the viral sfRNA and not due to XRN1 resistance of the DB.
Overall, while the DB elements are stable and complex structures containing a pseudoknot, we find no evidence that DB RNAs efficiently resist Xrn1 in vitro or during ZIKV infection in A549 cells, consistent with the lack of a tight ring-like conformation through which the 5′ end is threaded, and prior reports with YFV DBs (61). However, a recent study mapped sequencing reads that terminated at the DBs in sfRNAs of mosquito-adapted DENV (64). Thus, we cannot exclude that XRN1 resistance by DBs is possible with different viruses, in different cell types, or under specific conditions.
DISCUSSION
Here, we describe the 3D structure of the DONGV DB, an important functional RNA element in the flaviviral 3′ UTR implicated in regulating viral replication and whose structural disruption has been linked to attenuated viruses. Using our crystal structure, structure-guided molecular modeling, solution-based chemical probing, and single-molecule experiments, we determined that the DB contains a four-way junction and a global architecture that favors the formation of a stable pseudoknot associated with regulating replication.
Within the structure, numerous interactions in addition to pseudoknot base pairing appear to favor a global conformation that promotes pseudoknot formation. That is, the overall DB conformation is such that it brings the sequences that pair to form the pseudoknot into proximity. The features that favor this fold likely include a four-way junction, whose characteristics may help to constrain two of the emergent helices into an acute angle. Indeed, helical junctions are critical determinants of global RNA structure as they dictate the angle between emerging helices through the formation of a specific pattern of base stacking (52,65). By favoring specific angles, junction-induced global organization brings distal regions near each other in space such that long-range tertiary interactions can form. However, the disruption of the junction affects pseudoknot formation only subtly in vitro, hence other features may also stabilize the pseudoknot that could allow precise regulation of structure-dependent effects.
Our discovery that the DB 3D fold favors pseudoknot formation has implications for this element′s role in viral infection. In MBFVs, the DB is directly adjacent to the 3′ CS, a sequence responsible for the hybridization of 5′ and 3′ RNA elements and the subsequent genome cyclization required for viral replication. In the majority of MBFVs, the DB pseudoknot physically overlaps with the 3′ CS itself (11). This suggests that DB pseudoknot formation is mutually exclusive with 5′-3′ CS base pairing, as has been demonstrated in SHAPE probing of a construct containing the DENV 5′ and 3′ UTRs (28). The stability of the DB pseudoknot suggests that under the right conditions, the DB could act as an efficient competitor to 5′-3′ CS interactions, delaying replication timing until sufficient protein has been translated. Alternatively, a stable DB pseudoknot could also be a sensor of 5′-3′ cyclization, with cyclization inducing conformational changes within the DB and exposing DB sequences for potential RNA-protein interactions associated with replication.
Under the models presented here, the two DB conformations are connected to translation (pseudoknot formed) or genome replication (pseudoknot unformed). Given the high RNA sequence conservation within the DBs, it is tempting to speculate that protein co-factors could help regulate this equilibrium. Indeed, if the DB acts as a switch, RNA binding proteins could selectively stabilize or destabilize the pseudoknot to modulate this switch, with RNA binding helicases being an intriguing class of such proteins. In fact, such helicases have been previously implicated in DB binding and viral replication. Specifically, host factor DDX6 in mammalian cells (66) and its insect homolog Me31B in mosquito cells (67) are candidate RNA helicases that could promote viral replication by capturing the unfolded form or helping to unwind the DB pseudoknot. However, currently there is no direct evidence linking these helicases to changes in DB structure, and DDX6 and Me31B appear to perform distinct roles to either promote or restrict replication in mosquito versus mammalian cells (66,67). Another candidate helicase is NS3, a virally produced protein involved in replication (68). Overall, more research is needed to determine which factors may affect the structure of the DB during infection and how they relate to DB folding dynamics. Such insight will be useful to find ways to trap the DB in a single conformation, inhibiting a critical part of the viral life cycle.
In addition to the pseudoknot, the DBs contain a hyper-conserved region known as CS2. In our model, CS2 forms two helices (S3 and P3), located adjacent to one another on a single side of the 3D fold. These may create platform for interactions with proteins in the viral replication machinery that could explain the role of CS2 in promoting viral replication (10). This RNA region was generally more reactive to the chemical probes, as both DENV-2 and DONGV DBs exhibited partial reactivity in their S3 regions, and this part of the RNA was involved in the intermolecular domain-swap in the crystal. Together, these observations suggest that CS2 may be inherently conformationally flexible. It is tempting to hypothesize that this flexibility may be conducive to pseudoknot formation, protein binding and subsequent larger structural rearrangements. Given the connection of CS2 to the four-way junction and its potential to affect pseudoknot stability, binding of protein co-factors to CS2 that substantially alter its function could alter the angle between P1 and P2 or otherwise alter the overall architecture, linking protein binding to pseudoknot unfolding and viral genome cyclization.
The 3′ UTRs of flaviviruses accumulate during infection as separate fragments known as sfRNAs, linked to cytopathic effects in infected cells and formed due to resistance of the 3′ UTR to 5′→3′ degradation by the host exonuclease XRN1 (57). As part of the 3′ UTR, DBs are contained both within the genomic RNA and in sfRNAs although we found no evidence that they block exonucleases to form sfRNAs. However, given the extraordinary diversity of MBFV infections in multiple hosts we cannot extend this conclusion to all MBFVs. Currently it is unclear if the DBs perform a function as part of sfRNAs, however the fact that they are within the protected sfRNAs suggests they may have some other function in addition to their role in the viral genomic RNA.
DATA AVAILABILITY
Coordinates and structure factors have been deposited in the Protein Data Bank with the accession code 7KGA.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jay Nix (ALS Beamline 4.2.2) and John Hardin (UC Anschutz Medical Campus) for their assistance with crystal data collection as well as Anna-Lena Steckelberg, David Costantino, and Steve Bonilla Rosales for their critical reading of the manuscript.
Contributor Information
Benjamin M Akiyama, Department of Biochemistry and Molecular Genetics, Aurora, CO 80045, USA.
Monica E Graham, Department of Immunology and Microbiology, Aurora, CO 80045, USA.
Zoe O′Donoghue, Department of Immunology and Microbiology, Aurora, CO 80045, USA.
J David Beckham, Department of Immunology and Microbiology, Aurora, CO 80045, USA; Department of Medicine Division of Infectious Diseases, Aurora, CO 80045, USA.
Jeffrey S Kieft, Department of Biochemistry and Molecular Genetics, Aurora, CO 80045, USA; RNA BioScience Initiative, University of Colorado Denver School of Medicine, Aurora, CO 80045, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Health [R35GM118070 to J.S.K.]; Department of Defense [MIIRA PRMRP PR160117 to J.D.B.]; Veterans Affairs Merit Award [I01BX003863 to J.D.B.]; The University of Colorado, Denver Crystallography core is supported by NIH [P30CA046934, S10OD012033]; Beamline 4.2.2 at the Advanced Light Source is partially funded by the NIH [P30GM124169-01]; operated under contract with the U.S. DOE [DE-AC02-05CH11231]. Funding for open access charge: NIGMS.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bhatt S., Gething P.W., Brady O.J., Messina J.P., Farlow A.W., Moyes C.L., Drake J.M., Brownstein J.S., Hoen A.G., Sankoh O.et al.. The global distribution and burden of dengue. Nature. 2013; 496:504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mackenzie J.S., Gubler D.J., Petersen L.R.. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat. Med. 2004; 10:S98–S109. [DOI] [PubMed] [Google Scholar]
- 3. Hahn C.S., Hahn Y.S., Rice C.M., Lee E., Dalgarno L., Strauss E.G., Strauss J.H.. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 1987; 198:33–41. [DOI] [PubMed] [Google Scholar]
- 4. Proutski V., Gould E.A., Holmes E.C.. Secondary structure of the 3′ untranslated region of flaviviruses: similarities and differences. Nucleic Acids Res. 1997; 25:1194–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Proutski V., Gritsun T.S., Gould E.A., Holmes E.C.. Biological consequences of deletions within the 3′-untranslated region of flaviviruses may be due to rearrangements of RNA secondary structure. Virus Res. 1999; 64:107–123. [DOI] [PubMed] [Google Scholar]
- 6. Shurtleff A.C., Beasley D.W., Chen J.J., Ni H., Suderman M.T., Wang H., Xu R., Wang E., Weaver S.C., Watts D.M.et al.. Genetic variation in the 3′ non-coding region of dengue viruses. Virology. 2001; 281:75–87. [DOI] [PubMed] [Google Scholar]
- 7. Olsthoorn R.C., Bol J.F.. Sequence comparison and secondary structure analysis of the 3′ noncoding region of flavivirus genomes reveals multiple pseudoknots. RNA. 2001; 7:1370–1377. [PMC free article] [PubMed] [Google Scholar]
- 8. Villordo S.M., Carballeda J.M., Filomatori C.V., Gamarnik A.V.. RNA structure duplications and flavivirus host adaptation. Trends Microbiol. 2016; 24:270–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alvarez D.E., De Lella Ezcurra A.L., Fucito S., Gamarnik A.V.. Role of RNA structures present at the 3′UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology. 2005; 339:200–212. [DOI] [PubMed] [Google Scholar]
- 10. Manzano M., Reichert E.D., Polo S., Falgout B., Kasprzak W., Shapiro B.A., Padmanabhan R.. Identification of cis-acting elements in the 3′-untranslated region of the dengue virus type 2 RNA that modulate translation and replication. J. Biol. Chem. 2011; 286:22521–22534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Borba L., Villordo S.M., Marsico F.L., Carballeda J.M., Filomatori C.V., Gebhard L.G., Pallares H.M., Lequime S., Lambrechts L., Sanchez Vargas I.et al.. RNA structure duplication in the dengue virus 3′ UTR: redundancy or host specificity?. mBio. 2019; 10:e02506-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Durbin A.P., Karron R.A., Sun W., Vaughn D.W., Reynolds M.J., Perreault J.R., Thumar B., Men R., Lai C.J., Elkins W.R.et al.. Attenuation and immunogenicity in humans of a live dengue virus type-4 vaccine candidate with a 30 nucleotide deletion in its 3′-untranslated region. Am. J. Trop. Med. Hyg. 2001; 65:405–413. [DOI] [PubMed] [Google Scholar]
- 13. Men R., Bray M., Clark D., Chanock R.M., Lai C.J.. Dengue type 4 virus mutants containing deletions in the 3′ noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J. Virol. 1996; 70:3930–3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Whitehead S.S. Development of TV003/TV005, a single dose, highly immunogenic live attenuated dengue vaccine; what makes this vaccine different from the Sanofi-Pasteur CYD vaccine. Expert Rev. Vaccines. 2016; 15:509–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guzman M.G., Alvarez M., Halstead S.B.. Secondary infection as a risk factor for dengue hemorrhagic fever/dengue shock syndrome: an historical perspective and role of antibody-dependent enhancement of infection. Arch. Virol. 2013; 158:1445–1459. [DOI] [PubMed] [Google Scholar]
- 16. Shan C., Muruato A.E., Nunes B.T.D., Luo H., Xie X., Medeiros D.B.A., Wakamiya M., Tesh R.B., Barrett A.D., Wang T.et al.. A live-attenuated Zika virus vaccine candidate induces sterilizing immunity in mouse models. Nat. Med. 2017; 23:763–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alvarez D.E., Lodeiro M.F., Luduena S.J., Pietrasanta L.I., Gamarnik A.V.. Long-range RNA-RNA interactions circularize the dengue virus genome. J. Virol. 2005; 79:6631–6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Khromykh A.A., Meka H., Guyatt K.J., Westaway E.G.. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 2001; 75:6719–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lo M.K., Tilgner M., Bernard K.A., Shi P.Y.. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J. Virol. 2003; 77:10004–10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Corver J., Lenches E., Smith K., Robison R.A., Sando T., Strauss E.G., Strauss J.H.. Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J. Virol. 2003; 77:2265–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alvarez D.E., Filomatori C.V., Gamarnik A.V.. Functional analysis of dengue virus cyclization sequences located at the 5′ and 3′UTRs. Virology. 2008; 375:223–235. [DOI] [PubMed] [Google Scholar]
- 22. Friebe P., Shi P.Y., Harris E.. The 5′ and 3′ downstream AUG region elements are required for mosquito-borne flavivirus RNA replication. J. Virol. 2011; 85:1900–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu Z.Y., Li X.F., Jiang T., Deng Y.Q., Zhao H., Wang H.J., Ye Q., Zhu S.Y., Qiu Y., Zhou X.et al.. Novel cis-acting element within the capsid-coding region enhances flavivirus viral-RNA replication by regulating genome cyclization. J. Virol. 2013; 87:6804–6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de Borba L., Villordo S.M., Iglesias N.G., Filomatori C.V., Gebhard L.G., Gamarnik A.V.. Overlapping local and long-range RNA-RNA interactions modulate dengue virus genome cyclization and replication. J. Virol. 2015; 89:3430–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Filomatori C.V., Lodeiro M.F., Alvarez D.E., Samsa M.M., Pietrasanta L., Gamarnik A.V.. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006; 20:2238–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. You S., Falgout B., Markoff L., Padmanabhan R.. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 2001; 276:15581–15591. [DOI] [PubMed] [Google Scholar]
- 27. Davis W.G., Blackwell J.L., Shi P.Y., Brinton M.A.. Interaction between the cellular protein eEF1A and the 3′-terminal stem-loop of West Nile virus genomic RNA facilitates viral minus-strand RNA synthesis. J. Virol. 2007; 81:10172–10187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sztuba-Solinska J., Teramoto T., Rausch J.W., Shapiro B.A., Padmanabhan R., Le Grice S.F.. Structural complexity of Dengue virus untranslated regions: cis-acting RNA motifs and pseudoknot interactions modulating functionality of the viral genome. Nucleic Acids Res. 2013; 41:5075–5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chapman E.G., Costantino D.A., Rabe J.L., Moon S.L., Wilusz J., Nix J.C., Kieft J.S.. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science. 2014; 344:307–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Webb C.H., Luptak A.. HDV-like self-cleaving ribozymes. RNA Biol. 2011; 8:719–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schneider T.R., Sheldrick G.M.. Substructure solution with SHELXD. Acta Crystallogr. D. Biol. Crystallogr. 2002; 58:1772–1779. [DOI] [PubMed] [Google Scholar]
- 33. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 34. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W.et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cordero P., Kladwang W., VanLang C.C., Das R.. The mutate-and-map protocol for inferring base pairs in structured RNA. Methods Mol. Biol. 2014; 1086:53–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kladwang W., VanLang C.C., Cordero P., Das R.. A two-dimensional mutate-and-map strategy for non-coding RNA structure. Nat. Chem. 2011; 3:954–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Akiyama B.M., Stone M.D.. Assembly of complex RNAs by splinted ligation. Methods Enzymol. 2009; 469:27–46. [DOI] [PubMed] [Google Scholar]
- 38. Selvin P.R., Ha T.. Single-Molecule Techniques: A Laboratory Manual. 2008; NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 39. Nawrocki E.P., Eddy S.R.. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013; 29:2933–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rivas E., Clements J., Eddy S.R.. A statistical test for conserved RNA structure shows lack of evidence for structure in lncRNAs. Nat. Methods. 2017; 14:45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Weinberg Z., Breaker R.R.. R2R–software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics. 2011; 12:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Steckelberg A.L., Akiyama B.M., Costantino D.A., Sit T.L., Nix J.C., Kieft J.S.. A folded viral noncoding RNA blocks host cell exoribonucleases through a conformationally dynamic RNA structure. Proc. Natl. Acad. Sci. USA. 2018; 115:6404–6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gruber A.R., Lorenz R., Bernhart S.H., Neubock R., Hofacker I.L.. The Vienna RNA websuite. Nucleic Acids Res. 2008; 36:W70–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Blitvich B.J., Firth A.E.. Insect-specific flaviviruses: a systematic review of their discovery, host range, mode of transmission, superinfection exclusion potential and genomic organization. Viruses. 2015; 7:1927–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Takhampunya R., Kim H.C., Tippayachai B., Lee D.K., Lee W.J., Chong S.T., Kim M.S., Lee J.S., Klein T.A.. Distribution and mosquito hosts of Chaoyang virus, a newly reported flavivirus from the Republic of Korea, 2008–2011. J. Med. Entomol. 2014; 51:464–474. [DOI] [PubMed] [Google Scholar]
- 47. Keel A.Y., Rambo R.P., Batey R.T., Kieft J.S.. A general strategy to solve the phase problem in RNA crystallography. Structure. 2007; 15:761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu Y., Eisenberg D.. 3D domain swapping: as domains continue to swap. Protein Sci. 2002; 11:1285–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pfingsten J.S., Costantino D.A., Kieft J.S.. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science. 2006; 314:1450–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Merino E.J., Wilkinson K.A., Coughlan J.L., Weeks K.M.. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 2005; 127:4223–4231. [DOI] [PubMed] [Google Scholar]
- 51. Inoue T., Cech T.R.. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc. Natl. Acad. Sci. U.S.A. 1985; 82:648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Laing C., Schlick T.. Analysis of four-way junctions in RNA structures. J. Mol. Biol. 2009; 390:547–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Krasilnikov A.S., Yang X., Pan T., Mondragon A.. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003; 421:760–764. [DOI] [PubMed] [Google Scholar]
- 54. Grilley D., Soto A.M., Draper D.E.. Mg2+-RNA interaction free energies and their relationship to the folding of RNA tertiary structures. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:14003–14008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hengesbach M., Kim N.K., Feigon J., Stone M.D.. Single-molecule FRET reveals the folding dynamics of the human telomerase RNA pseudoknot domain. Angew. Chem. Int. Ed. Engl. 2012; 51:5876–5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Warnasooriya C., Ling C., Belashov I.A., Salim M., Wedekind J.E., Ermolenko D.N.. Observation of preQ1-II riboswitch dynamics using single-molecule FRET. RNA Biol. 2019; 16:1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pijlman G.P., Funk A., Kondratieva N., Leung J., Torres S., van der Aa L., Liu W.J., Palmenberg A.C., Shi P.Y., Hall R.A.et al.. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008; 4:579–591. [DOI] [PubMed] [Google Scholar]
- 58. Funk A., Truong K., Nagasaki T., Torres S., Floden N., Balmori Melian E., Edmonds J., Dong H., Shi P.Y., Khromykh A.A.. RNA structures required for production of subgenomic flavivirus RNA. J. Virol. 2010; 84:11407–11417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Akiyama B.M., Laurence H.M., Massey A.R., Costantino D.A., Xie X., Yang Y., Shi P.Y., Nix J.C., Beckham J.D., Kieft J.S.. Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease. Science. 2016; 354:1148–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Akiyama B.M., Eiler D., Kieft J.S.. Structured RNAs that evade or confound exonucleases: function follows form. Curr. Opin. Struct. Biol. 2016; 36:40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Silva P.A., Pereira C.F., Dalebout T.J., Spaan W.J., Bredenbeek P.J.. An RNA pseudoknot is required for production of yellow fever virus subgenomic RNA by the host nuclease XRN1. J. Virol. 2010; 84:11395–11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chapman E.G., Moon S.L., Wilusz J., Kieft J.S.. RNA structures that resist degradation by Xrn1 produce a pathogenic Dengue virus RNA. Elife. 2014; 3:e01892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pallares H.M., Costa Navarro G.S., Villordo S.M., Merwaiss F., de Borba L., Gonzalez Lopez Ledesma M.M., Ojeda D.S., Henrion-Lacritick A., Morales M.A., Fabri C.et al.. Zika virus subgenomic flavivirus RNA generation requires cooperativity between duplicated RNA structures that are essential for productive infection in human cells. J. Virol. 2020; 94:e00343-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Filomatori C.V., Carballeda J.M., Villordo S.M., Aguirre S., Pallares H.M., Maestre A.M., Sanchez-Vargas I., Blair C.D., Fabri C., Morales M.A.et al.. Dengue virus genomic variation associated with mosquito adaptation defines the pattern of viral non-coding RNAs and fitness in human cells. PLoS Pathog. 2017; 13:e1006265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lescoute A., Westhof E.. Topology of three-way junctions in folded RNAs. RNA. 2006; 12:83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ward A.M., Bidet K., Yinglin A., Ler S.G., Hogue K., Blackstock W., Gunaratne J., Garcia-Blanco M.A.. Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3′ UTR structures. RNA Biol. 2011; 8:1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Goertz G.P., van Bree J.W.M., Hiralal A., Fernhout B.M., Steffens C., Boeren S., Visser T.M., Vogels C.B.F., Abbo S.R., Fros J.J.et al.. Subgenomic flavivirus RNA binds the mosquito DEAD/H-box helicase ME31B and determines Zika virus transmission by Aedes aegypti. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:19136–19144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mazeaud C., Freppel W., Chatel-Chaix L.. The multiples fates of the flavivirus RNA genome during pathogenesis. Front Genet. 2018; 9:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors have been deposited in the Protein Data Bank with the accession code 7KGA.