Abstract
Background:
Surgical tumor resection is the primary treatment option for diffuse glioma, the most common malignant brain cancer. The intraoperative diagnosis of gliomas from tumor core samples can be improved by use of molecular diagnostics. Further, residual tumor at surgical margins is a primary cause of tumor recurrence and malignant progression. This study evaluates a desorption electrospray ionization mass spectrometry (DESI-MS) system for intraoperative isocitrate dehydrogenase (IDH) mutation assessment, estimation of tumor cell infiltration as tumor cell percentage (TCP), and disease status. This information could be used to enhance the extent of safe resection and so potentially improve patient outcomes.
Methods:
A mobile DESI-MS instrument was modified and used in neurosurgical operating rooms (ORs) on a cohort of 49 human subjects undergoing craniotomy with tumor resection for suspected diffuse glioma. Small tissue biopsies (ntotal = 203) from the tumor core and surgical margins were analyzed by DESI-MS in the OR and classified using univariate and multivariate statistical methods.
Results:
Assessment of IDH mutation status using DESI-MS/MS to measure 2-hydroxyglutarate (2-HG) ion intensities from tumor cores yielded a sensitivity, specificity, and overall diagnostic accuracy of 89, 100, and 94%, respectively (ncore = 71). Assessment of TCP (categorized as low or high) in tumor margin and core biopsies using N-acetyl-aspartic acid (NAA) intensity provided a sensitivity, specificity, and accuracy of 91, 76, and 83%, respectively (ntotal = 203). TCP assessment using lipid profile deconvolution provided sensitivity, specificity, and accuracy of 76, 85, and 81%, respectively (ntotal = 203). Combining the experimental data and using PCA-LDA predictions of disease status, the sensitivity, specificity, and accuracy in predicting disease status are 63%, 83%, and 74%, respectively (ntotal = 203).
Conclusions:
The DESI-MS system allowed for identification of IDH mutation status, glioma diagnosis, and estimation of tumor cell infiltration intraoperatively in a large human glioma cohort. This methodology should be further refined for clinical diagnostic applications.
INTRODUCTION
Diffuse gliomas are high morbidity primary brain tumors. The 5-year survival rate of patients with glioblastoma is less than 5% (1). The primary treatment option for gliomas is gross total surgical resection, accompanied by adjuvant chemoradio-therapy (2–4). A central dilemma of the neurosurgeon is preservation of vital brain functions while maximizing extent of resection. Unfortunately, glioma cells are diffusely infiltrative and the high risk of neurological deficits often results in residual tumor at surgical margins, leading to progression and recurrence (5, 6).
Accurate glioma diagnosis and prognosis increasingly relies on molecular and genetic information assessed from tumor core biopsies (2). Currently, brain tumor resections are performed without the aid of a molecular diagnosis, as these slow assays must be performed postoperatively (7). The development of rapid intraoperative molecular diagnostics could help improve glioma patient management and the quality of surgical resection (7–10). Notably, recent studies show that the effect of extent of resection on overall survival and malignant-free progression is significantly different between isocitrate dehydrogenase-wildtype (IDH-wt) and IDH-mutant (IDH-mut) gliomas, suggesting that surgical strategies may be impacted favorably by knowledge of the IDH mutation status at the time of surgery (9, 10). It is important to note that even if tumor cells are found in eloquent brain areas, surgical resection of that tissue may not be performed due to the significant harm it may cause to the patient. However, it may serve as an area to target for postoperative radiotherapy or application of local chemotherapeutics, the choice of which can be guided by information provided by rapid molecular diagnostics.
Mass spectrometry-based methods of tissue analysis may be able to provide clinical diagnostic information on brain tissue at the time of glioma resection. In particular, ambient ionization mass spectrometry (MS) has emerged as a family of rapid methods for intraoperative tissue analysis (11). Ambient ionization MS methods are being evaluated for their ability to assess molecular features of various cancers, as well as in assessing surgical margins for residual tumor (12, 13). Probe-based ambient methods, such as probe electrospray ionization (14) and touch spray ionization (15), have yielded pathologically relevant results on fresh surgical tissue. Rapid evaporative ionization MS has been used intraoperatively in a variety of cancers (16, 17). Several laser ablation systems, based on picosecond-infrared and other optical regimes, have byielded diagnostic MS signals with low/no tissue damage (18, 19). The MassSpec Pen has been used in vivo to assess residual tumor in a minimally invasive, nondestructive approach, recently in human ovarian cancer (20, 21).
Desorption electrospray ionization-mass spectrometry (DESI-MS), the method utilized in this work, is an ambient ionization method in which charged microdroplets of a solvent are sprayed onto a sample surface, desorbing and ionizing molecules present in the sample and transporting them into a mass spectrometer for analysis (11, 22). DESI is used in many laboratories for biological applications (23, 24), including the distinction between cancerous and normal tissues in a variety of human organs (12, 25, 26). In some studies, such as the one described herein, DESI is not used as an imaging modality but as a diagnostic method. Using banked, fresh-frozen brain tissue, we have demonstrated the capability to differentiate gliomas, meningiomas, and pituitary tumors with high accuracy (27), along with the capability to provide glioma subtyping (28–30). Importantly, these brain cancers could be distinguished readily from normal brain tissue.
This study represents the completion of a project which demonstrated intraoperative application of DESI-MS during glioma resection using a customized, stand-alone mass spectrometer. Initial data from a set of 10 human subjects was published early in the project to highlight the potential clinical utility of TCP estimation by intraoperative MS (31) (Supplemental Table 1). Using the same customized mass spectrometer, approximately half way through the study we developed and applied an intraoperative DESI-MS assay for the assessment of IDH-mutation status by detecting the oncometabolite 2-hydroxyglutarate (2-HG), a key glioma prognostic marker, in a set of 51 glioma core biopsies obtained from 25 human subjects and analyzed intrasurgically (32). IDH mutations, specifically R132H mutations, disrupt the conversion of isocitrate to α-ketoglutarate. Additionally, IDH1 mutations result in the ability of IDH1 to catalyze the reduction of α-ketoglutarate to R(−)-2-HG. Consequently, 2-HG levels have been found to be significantly higher in IDH1 mutated human gliomas (33).
In this report, we have combined and improved our previously developed methods of tissue smear classification and analyzed tissue biopsy-smears during tumor resection in a cohort of 49 glioma patients, integrating data on new subjects, and re-examining data for subjects whose data appear in the aforementioned publications. (Supplemental Table 2 indicates the numbers of subjects in each category.) As the development of an online methodology for the determination of IDH mutation status was completed midway through the study, measurement of 2-HG as a predictor of IDH mutation status was possible for only 30 of the 49 recruited patients. While previous publications have analyzed the data with respect to individual tissues smears, in this work, we have elected to analyze the data with respect to biopsy in order to facilitate the understanding of the results in the context of a clinical setting. Three categories of information were acquired using DESI-MS: 1) IDH mutation status, 2) tumor cell percentage (TCP), and 3) disease status. When the DESI-MS results are considered in combination with other available data (e.g., MRI and in vivo brain mapping), they should allow the neurosurgeon to make better informed resection decisions.
MATERIALS AND METHODS
Human Subjects
Human subjects research was performed in accordance with an Institutional Review Board approved study at the Indiana University School of Medicine (IRB #1410342262). Glioma patients undergoing craniotomy with tumor resection were prospectively enrolled after providing written informed consent and Health Insurance Portability and Accountability Act authorization. No DESI-MS results were shared with neurosurgeons during the surgical resection, so as not to affect the standard of care.
Intraoperative DESI-MS
All experiments were performed using a modified linear ion trap mass spectrometer (Thermo LTQ) as previously described (31, 32). For each surgery, the instrument was rolled into the operating room (OR) and turned on. During tumor resection, small stereotactic tissue biopsies (approximately 5–10 mg each) were provided by the neurosurgeon to the mass spectrometer operators for DESI-MS analysis. The number and location of the biopsies were determined according to the surgeon’s best medical judgement. Samples from the tumor core (for assessing diagnostic information) as well as surgical margins (for assessing residual tumor) were provided for each case. The tissue biopsies were smeared on glass slides and then analyzed by DESI-MS using a zigzag raster pattern acquire representative data (Fig. 1, A). Two different negative ion mode DESI-MS methods were used. Using method 1, full-scan lipid (m/z 700—1000) and metabolite (m/z 80—200) mass spectra and a targeted MS2 scan for N-acetyl-aspartic acid (NAA, 174 -> O), were acquired over 3.3 minutes. Using method 2, MS2 (MS/MS) and MS3 data were acquired specific to 2-HG (147 -> O and 147 -> 129 -> O, respectively), along with a full-scan metabolite profile (m/z 50 – 200), all over a period of 3.3 minutes (Fig. 1, B and C). Method 1 utilized 1:1 dimethylformamide (DMF)-acetonitrile (ACN) and method 2 utilized 25:37:38 DMF-ACN-ethanol (EtOH) as solvent. Additional details of the MS methods and conditions are given in the Supplemental Information.
Fig. 1.
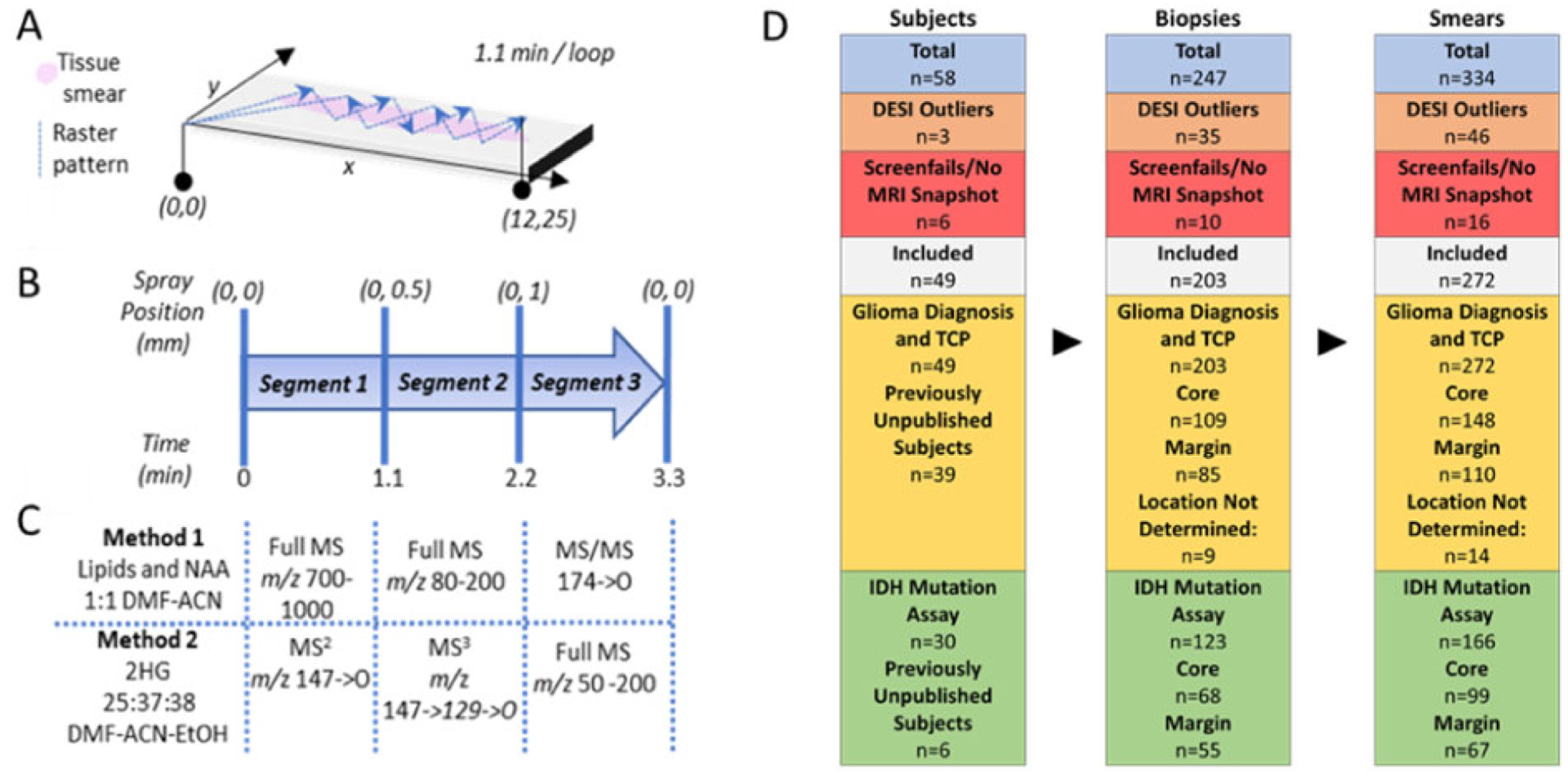
Summary of the DESI-MS method. (A) Smeared tissue (pink) is analyzed with DESI spray using a zigzag raster pattern (dotted lines, with direction noted by arrow heads), spanning 12 mm in x-dimension and 25 mm in y-dimension over 1.1 min; the pattern is repeated 3 times. The DESI spot is offset 0.5 mm in the y-dimension after each raster loop. (B) DESI spray position and the timeline for the DESI-MS method. The (x, y) coordinates denote the starting position of the DESI spot for each raster loop. (C) Description of the 2 DESI-MS methods, shown synchronized with the position of the moving stage. A different set of MS data is collected during each method segment. Note the use of full MS, MS/MS (MS2), and MS3 experiments. (D) Summary of number of patients, biopsies, and smears, noting how many were excluded, and which subjects are new to the study. See Supplemental Tables 2–5 for additional patient cohort data. Data from earlier subjects recruited in the study were published in Reference 27 for glioma diagnosis and TCP, and in Reference 32 for IDH-mutation assessment. DESI data was considered an outlier if no lipid or metabolite profile scans were retained after data filtering due to low signal or high similarity to data collected from a blank glass slide (see Supplemental Methods for data filtering methods).
Data Analysis
De-identified clinical data were obtained for each subject for correlation with the DESI-MS results (Supplemental Tables 3–6). The DESI-MS data were analyzed in MATLAB using custom algorithms to remove background scans (e.g., signal collected from regions of the glass slide containing no tissue) and to perform statistical classifications. MS scans from areas of the glass slide containing no smeared tissue and from smears giving insufficient intensity were excluded by applying a cut-off to the absolute signal intensity. Only scans with mass spectra having the summed ion counts greater than the cutoff value were used for chemical predictions. Additionally, for each selected mass spectrum, the full width at half maximum (FWHM) was calculated for the base peak; all spectra with resolution < 1000 were excluded. Additional information on the filtering and statistical methods used is included in the Supplemental Information (Data Analysis).
Histopathological Analysis
After DESI-MS analysis, the tissue smears were moved from the surgical core to the Indiana University Health Pathology Laboratory and were stained with H&E. The stained smears were then blindly evaluated by an expert neuropathologist (E.M.H.) and interpretations of smear diagnosis, tumor grade, TCP, and smear quality were provided (Supplemental Table 6). The entire smear was evaluated and the interpretations made reflect the average state of the entire smear. Smears with significant heterogeneity (e.g., half the slide diseased and half normal) were rare (2 smears out of 272 in this study).
RESULTS AND DISCUSSION
Summary of Patient Cohort and Tissue Samples
Data were collected from 49 human subjects; 203 biopsies were obtained and 272 smears were analyzed (Fig. 1, D). For some biopsies, multiple smears were created and analyzed. The subject cohort and DESI-MS results are described in detail in the Supplemental Information and in Supplemental Tables 2–6. Supplemental Table 2 summarizes the number of patients and the biopsies obtained, indicates whether they have been included in a previous, preliminary publication, and whether subjects were excluded after recruitment; Supplemental Table 3 provides the demographics, diagnosis, and treatment information; Supplemental Table 4 provides additional histopathology data. The statistical predictions for disease status, TCP, and IDH mutation status for all analyzed smears are tabulated in Supplemental Table 5. The histopathology assessments of all these DESI-MS analyzed smears are tabulated in Supplemental Table 6. The overall subject classifications are described in detail for IDH mutation status, TCP, and disease status in the following sections.
IDH Mutation Status Prediction from 2-HG Abundance
The methodology for online determination of IDH mutation status was developed midway through the study, after off-line method development. From this point onwards, 169 smears from 30 subjects were collected for the measurement of 2-HG as a predictor of IDH mutation status. Three smears were excluded due to lack of location information available and/or determined, resulting in 166 smears from 30 subjects being included. Of these 166 smears, 67 smears were from tumor margins and 99 smears were from tumor cores. In instances where multiple smears were made for the same tumor core biopsy, the average TIC normalized and summed 2-HG product ion intensity of all smears was calculated and used to predict the IDH mutation status of that biopsy. This produced 55 margin biopsies (23 IDH-mut biopsies from 12 subjects, 32 IDH-wt biopsies from 14 subjects), and 68 core biopsies (36 IDH-mut biopsies from 12 subjects, 32 IDH-wt biopsies from 16 subjects). The measurement of 2-HG is qualitative and predictions of IDH mutation status are made solely on the basis of TIC normalized and summed MS3 intensities. Two MS3 product ions were detected at m/z 85 and 101 from the sequential fragmentation of the 2-HG [M-H]− precursor ion (147 -> 129 -> O). The intensities of the two product ions were then summed and used to predict the IDH mutation status of the tumor. While the margin biopsies from IDH-mut subjects contained significantly higher levels of 2-HG compared to margins from IDH-wt subjects (ROC AUC = 0.96, Wilcoxon rank-sum test P < 0.0001), tumor core biopsies were preferred for IDH mutation status assessment, as the concentration of 2-HG is highest at the tumor core (Supplemental Fig. 1). The IDH mutation status of the subjects was obtained from pathology reports and was typically the result of postoperative IDH1 R132H immuno-histochemistry and, when inconclusive, PCR-based IDH1/2 sequencing analysis.
The chemical structures of α-ketoglutartic acid and 2-HG, its mutated form which accumulates in the presence of an IDH mutation, are shown in Fig. 2, A. A statistically significant (Wilcoxon rank-sum test P < 0.0001) increase in the 2-HG signal of tumor core biopsies was observed in the IDH-mut subjects compared to the IDH-wt subjects. A receiver operating characteristic (ROC) curve model provided an area under the curve (AUC) of 0.98 using the tumor cores, as shown in Fig. 2, B. This difference is apparent in the box-plots shown in Fig. 2, C. The summed and normalized 2-HG product ion intensity cutoff of 62.3 (in the MS3 spectrum) resulted in the highest overall accuracy. This normalized cutoff is instrument- and method-dependent and requires further interlaboratory investigation to evaluate whether a universal cut-off can be determined. Using the ROC determined cutoff, the sensitivity (correctly identifying core biopsies from IDH-mut subjects when the IDH mutation was present) and specificity (correctly identifying core biopsies from IDH-wildtype when IDH mutation was absent) were 89% and 100%, respectively, with an overall accuracy of 94%.
Fig. 2.
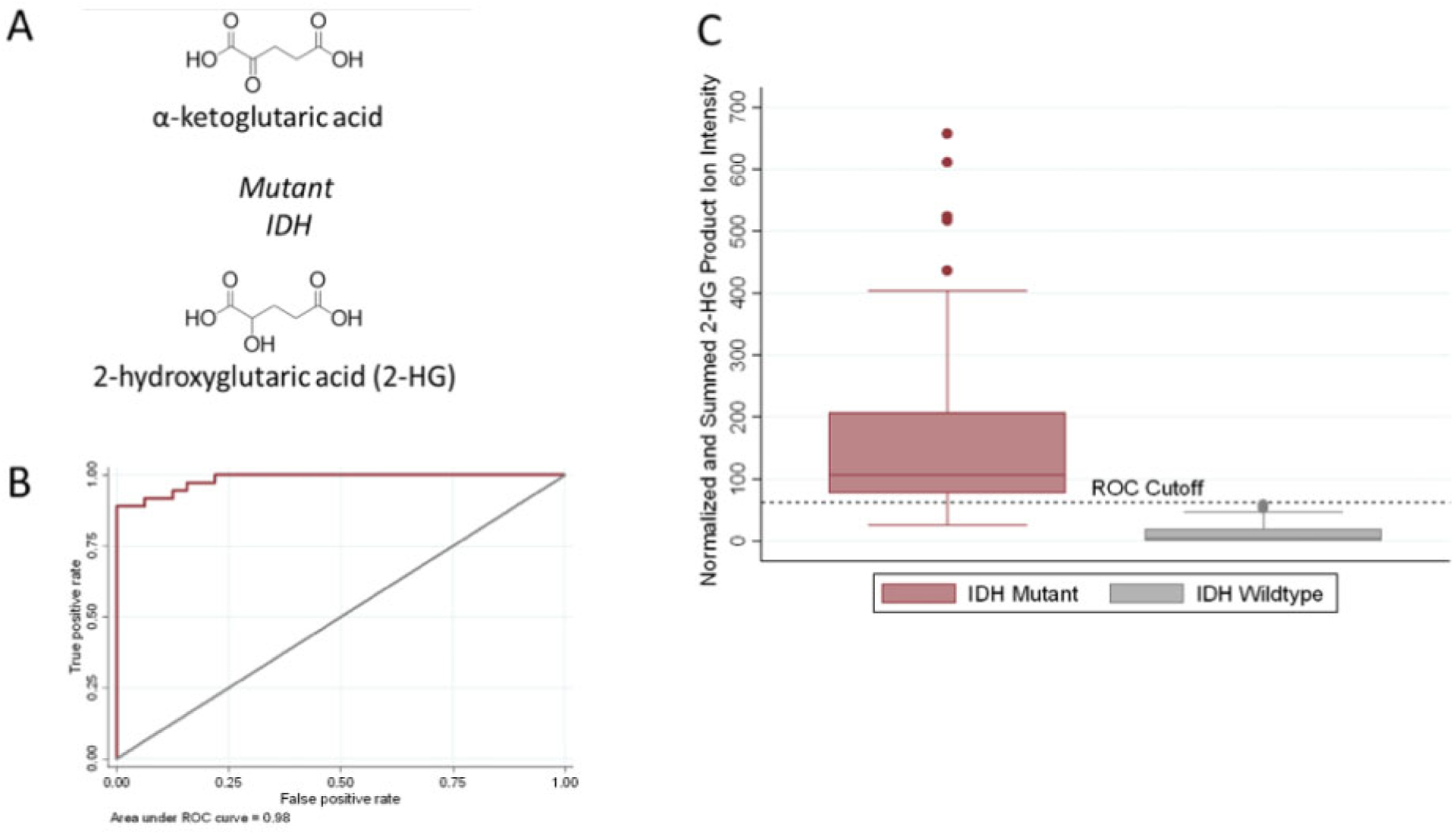
Intraoperative assessment of IDH-mutation status from tumor cores. (A) Chemical structure of α-ketoglutaric acid and 2-hydroxyglutarate (2-HG), the oncometabolite associated with IDH mutations. (B) ROC curve for the IDH-mutation assay using the tumor core biopsies. The area under the curve is 0.98, indicating the high accuracy of the method. (C) Box-plot showing the average summed and TIC normalized MS3 fragment ion intensities (m/z 85 + m/z 101) produced by sequential dissociation (MS3) of 2-HG for all tumor core biopsies (n = 68 biopsies from 28 subjects) by IDH mutation status. The fragment ion intensities from duplicate smears of the same biopsy were averaged to generate one value per biopsy. Error bars represent ± 1.5 times the calculated standard deviations. The black dashed horizontal decision line was calculated from ROC curve analysis and differentiates tumor core biopsies from IDH-mut subjects and IDH-wt subjects with the highest sensitivity and specificity.
For several biopsies included in this study (ndiscordant = 4), discordant mutation status predictions exist between core biopsies from the same subject. Due to the high degree of heterogeneity of glioma, it is likely that 2-HG concentration varies within the tumor cavity, even over small distances. Additionally, cauterized or hemorrhagic tissue results in ion suppression and provides poor diagnostic information. These problems are shared with traditional histopathology. The solution for both methods is to take additional biopsies that are of sufficient diagnostic quality. While this is impractical for histopathology, DESI-MS analysis takes only 3 min, so resampling from the same patient during surgery is a feasible solution. To further limit the impact of signal variation, the average of the normalized 2-HG intensity for all biopsies from a subject was generated and used to predict IDH mutation status. When doing so, only 1 subject (Subject 26) was misclassified with respect to its IDH mutation status. This subject, and subjects close to the ROC cutoff value, had low grade (i.e., WHO grade II and III) astrocytomas (i.e., anaplastic astrocytomas and diffuse astrocytomas). In our experience, these lower grade tumors have lower concentrations of 2-HG than higher grade gliomas, resulting in lower summed 2-HG product ion intensity values. Therefore, for suspected low-grade gliomas, resampling may be necessary to increase the degree of confidence placed in measured summed 2-HG product ion intensities. Biological variability within the tumor cavity is an area of active research; MRI instruments are being used to probe distributions of metabolites within the tumor cavity and surrounding tissue to further understand tumor heterogeneity. As knowledge of tumor heterogeneity and its impacts on the spatial distribution of metabolites increases, so too will our understanding of discordant predictions in IDH mutation status.
Estimating Tumor Cell Percentage from N-Acetyl-Aspartic Acid (NAA) Abundance
Two DESI-MS-based methods for the rapid estimation of TCP, based on NAA and lipid abundances, respectively, have been developed and tested previously on an initial set of 10 human subjects (31). Data for this original set of subjects have been integrated into the complete 49 patient cohort and reexamined, with the results now discussed. In the first method, the standard normal variate (SNV) normalized intensity of m/z 174, corresponding to NAA, measured in the full-scan MS metabolite profile was used to estimate TCP, using previously published data as a training set (27, 34). For biopsies with multiple smears, the average TCP from the corresponding smears was calculated and used to estimate TCP for the specific biopsy. This produced a collection of 203 biopsies (109 core, 85 margin, 9 undermined) from 49 subjects. Biopsies that provided 50% or less glioma contribution were categorized as low TCP and samples providing 51% or more glioma were categorized as high TCP. The histopathology estimates of TCP were categorized using the same decision boundary. Using these categories, an overall accuracy of correctly classifying the biopsies as low or high TCP was 83%, with sensitivity and specificity of 91% and 76%, respectively (Table 1). The full-scan MS abundance of NAA (detected as m/z 174 in the metabolite profile scan) provided slightly higher overall accuracy than did the MS2 data (sensitivity, specificity, and overall accuracy were 79%, 85%, and 82%, respectively, for the MS2 data (nNAA MS2 = 198).) The targeted MS2 scan for NAA (174 -> O) gave product ions at m/z 88, 114, 130, and 156. See the Supplemental Information Data Analysis section for details. Box plots and ROC curves of the NAA full-scan and MS2 data are shown in Supplemental Fig. 2. Principal component analysis (PCA) was performed on the metabolite profiles and the contribution of m/z 174 (NAA) in separating low and high TCP smears was consistent with previous reports (27).
Table 1.
Confusion matrix for assessing correlation between histopathology assessments and the DESI-MS estimates of TCP and disease status of tissue biopsies
| Histopathologya
High TCP/Gliomab |
Low TCP/Infiltrative Marginb | ||
|---|---|---|---|
| Full-scan NAA Intensity TCP Estimate | High TCP/Gliomab | 91 | 25 |
| Lipid Deconvolution TCP Estimate | 76 | 15 | |
| PCA-LDA Diagnosis | 60 | 18 | |
| Full-scan NAA Intensity TCP Estimate | Low TCP/Infiltrative Marginb | 9 | 78 |
| Lipid Deconvolution TCP Estimate | 24 | 88 | |
| PCA-LDA Diagnosis | 35 | 91 |
Histopathology assessments of TCP were used for correlation with lipid deconvolution and NAA intensity classifications of TCP. Histopathology assessments of disease status were used for correlation with PCA-LDA diagnosis of the tissue smears.
For lipid deconvolution and NAA intensity classification, histopathology TCP categories were High = 51% and Low = 50%. Histopathology categories of glioma and infiltrative margin were used for correlation with PCA-LDA diagnosis of glioma and infiltrative margin.
Estimating Glioma Content (Relative Tumor Burden) Using Lipid Profile Deconvolution
The second method for estimating TCP employed a lipid profile deconvolution approach: PCA was used to calculate the percentage of white matter (WM), gray matter (GM), and glioma (G) contributing to the lipid profiles of the subject samples using a linear regression model built from data collected in a previous study (27, 31). The model was based on the presumption that the observed lipid profile of each brain tissue biopsy is composed of a ternary mixture of WM, GM, and G. The summed percentages of these 3 categories are constrained to 100%. The regression model was built from data collected from DESI-MS analysis of banked glioma and normal human brain specimens. Histologically correlated mass spectra were compiled based on histopathological assessment, compressed with PCA, and the average PC1 and PC2 scores for samples of pure WM, GM, and G were calculated. The three extremes (PC1 and PC2 scores corresponding to 100% WM, 100% GM, 100% G) were used to calculate the predicted PC1 and PC2 scores for each possible mixture (Supplemental Fig. 3). To predict the composition of new samples (ntotal = 203), the PCA lipid profile loading matrix from the training set data (27) was used to calculate the PC scores of the new samples. The calculated PC1 and PC2 scores of the new samples were matched to predicted PC1 and PC2 scores using the original training set data; the predicted scores were each associated with a specific percentage combination of GM: WM: G (Supplemental Fig. 3).
The classifications of GM and WM were recategorized as infiltrative margin (IM) to match the histopathology categories, as the nature of the underlying normal brain tissue (specifically white or gray matter) could not always be reliably determined by histopathology. The percentage of G calculated for each unknown sample was used as an estimate of the TCP. See Supplemental Information for additional details. For biopsies with multiple smears, the average percentage of G from corresponding smears was calculated and used to estimate TCP for the specific biopsy. This produced data for a collection of 203 biopsies (109 core, 85 margin, 9 undermined) from 49 subjects. Using 50% TCP delineation value as for the NAA abundance method, an overall accuracy of correctly classifying the biopsies as low or high TCP was 81%, with sensitivity and specificity of 76% and 85%, respectively (Table 1). The low TCP and high TCP smears analyzed in this study are fairly well separated in PCA space, due primarily to the ions of m/z 834, 794, and 888.
Predicting Disease Status from Fused Metabolite and Lipid Profiles
The final category of diagnostic information provided by DESI-MS was disease status. Using PCA-LDA on fused SNV normalized lipid and metabolite profiles obtained from DESI-MS, tissue smears were classified as either G, GM, or WM, with the latter two being recategorized as infiltrative margin (IM) to match the histopathology categories (see Table 1 footnote). Similar overall accuracy was observed when performing PCA-LDA cross-validation with the lipid and metabolite profiles of all 272 tissue smears (Supplemental Tables 7 and 8), however, the sensitivities and specificities were quite different, supporting the use of fused lipid and metabolite profiles for a more robust method that utilizes diagnostic features contained in both parts of a mass spectrum. For biopsies with multiple smears, the average SNV normalized lipid and metabolite profiles from corresponding smears were calculated prior to fusing to generate a representative lipid and metabolite profiles for the biopsy used to predict disease status. This produced a collection of 203 biopsies (109 core, 85 margin, 9 undermined) from 49 subjects. The agreement between PCA-LDA predictions and histopathological assessment of disease status of tissue biopsies is shown in Table 1. The sensitivity, specificity, and accuracy in predicting disease status are 63%, 83%, and 74%, respectively. While the sensitivity using the fused data method was lower than either the lipid or metabolite profiles independently, the use of the fused profiles to predict TCP is likely more robust than using only one segment of the mass spectrum, as diagnostics features appear in both regions of the mass spectrum. A significant fraction of the smears classified as glioma based on histopathology (nmissclasified smears = 43%) were misclassified as either WM or GM using PCA-LDA classification. These cases likely reflect mixed tissue smears which are challenging for histopathology assessment as well for our current DESI-MS system. These types of samples were encountered both at surgical margins and at tumor cores.
Evaluation of Methods for Estimating TCP and Disease Status
Ultimately, we found that both methods (i.e., full-scan NAA abundance and lipid deconvolution) were capable of predicting TCP with similar accuracies (83% and 80%, respectively), with the full-scan NAA abundance having greater sensitivity (87% vs. 76%) and the lipid deconvolution method having greater specificity (79% vs. 84%). In instances of misclassification, it would be beneficial to compare the disease status predictions made using the fused metabolite and lipid profiles with TCP estimations using NAA and lipid deconvolution (i.e., many of the misclassified biopsies have TCP estimates indicating mixed diseased and normal tissue). Additionally, the PCA-LDA method itself may need to be adjusted in the future to improve classification of biopsies with mixed morphologies by using additional categories (i.e., rather than categorizing tissues into 3 rigid categories (G, WM, GM), perhaps 6 categories (G, G/WM, WM, WM/GM, GM, GM/G)).
While it is likely that there are many contributing factors, a possible cause of variability in TCP estimates may be that the full-scan NAA abundance method predicts TCP based on the intensity of a single analyte that exists in very low concentrations within the tumor, while the lipid deconvolution method predicts TCP based on a collection of analytes in a characteristic pattern of intensities. Matrix effects are inherent in the measurements of complex samples, especially biological samples; they are an intrinsic component of the material being analyzed and, consequently, cannot be eliminated. Excepting cases where tissue is substantially different (e.g., cauterized or hemorrhagic tissues), observed changes with matrix should involve total signal but not relative signals, provided the same type of tissue is being analyzed. That said, the measurement of a single analyte (i.e., 2-HG or NAA) is a situation where the matrix effects on signal intensity are of most concern. To minimize variations in signal, the recorded mass spectra are normalized (specifically TIC and/or SNV normalized) before additional comparison or statistical analysis is performed.
There are likely additional experimental factors that may have affected classification in the present study. Improvements are needed to increase the agreement between the DESI-MS predictions of disease status and TCP with histopathology, the current gold-standard method in morphology. The diagnostic sensitivity and specificity for the TCP predictions could be improved by better correlating the positions at which TCP pathology data are taken relative to DESI-MS data, while the biological variation can be addressed with additional cases. To explore the impact of these factors, a new 50 patient study for the optimization of the DESI-MS platform has commenced in which the above factors will be rigorously controlled.
Assessment of Margins of Tumors of Different WHO Grades
The assessment of glioma margins is not routinely performed intraoperatively with pathologic or molecular methods. That said, glioma infiltration is often significant beyond contrast enhancement in MRI, especially in glioblastoma (6). In this section, using our DESI-MS methods, we investigate whether margins from high-grade glioma patients contain higher TCP than margins from low-grade glioma patients. In our study, 85 of the 203 biopsies (110 smears with some replicates) were obtained from margins (based on MRI) and provided acceptable quality for histopathology and DESI-MS analysis. WHO grade information was not available for eight margin biopsies from two subjects (Subjects 41 and 46) and, thus, these biopsies were excluded from this portion of the study.
For low-grade gliomas (WHO grades I–III), 78% of the margin biopsies had low TCP (nlow TCP, low grade margins = 35) and 22% had high TCP based on histopathology (nhigh TCP, low grade margins = 10), using 50% TCP for delineating between low and high TCP. Our accuracy with respect to agreement with histopathology assessment for the grade I–III margin samples (ntotal low grade margins = 45) was 80% and 76% overall using the normalized NAA intensity and lipid deconvolution approaches, respectively (Supplemental Table 9). For the grade IV gliomas, histopathology determined that 59% of the margin biopsies had low TCP (nlow TCP, high grade margins = 19) and 41% had high TCP (nhigh TCP, high grade margins = 13), also using the 50% TCP cutoff. Our agreement with the histopathology assessment for the margin biopsies from grade IV gliomas (ntotal high grade margins = 32) was 97% and 84% overall using the normalized NAA intensities and lipid deconvolution approaches, respectively (Supplemental Table 10).
Literature reports suggest that high TCP may extend beyond MRI contrast enhancement, especially in grade IV glioblastomas (6, 8, 35). We found that the margins of grade IV gliomas contained high TCP more often than the margins of grade I–III gliomas, although the difference in number of high TCP smears obtained from the margins of grade IV compared to grade I–III gliomas using nominal logistic regression and a 50% TCP cutoff to delineate between the low and high TCP categories is not statistically significant (P = 0.0833). In this context, a DESI-MS tool for assessment of local tumor burden must be used in combination with an independent diagnostic method such as MRI neuronavigation and/or brain mapping.
Simplified DESI-MS Outputs That Enable Surgical Decision Making
In this study, we have demonstrated two major applications of the DESI-MS system: providing diagnostic information of two types for tumor cores (IDH mutation status and disease status) and providing surgical margin assessments of residual tumor (i.e., TCP) at discrete locations. With appropriate method validations and permissions, these predictions could be provided to the neurosurgeon in order to improve treatment strategy, including the extent of resection. Here we describe a typical example of a case in which DESI-MS would be useful. Figure 3, A shows the biopsy locations for two stereotactic biopsies from Subject 58. Figure 3, B–D shows the raw lipid, metabolite, and 2-HG MS3 data for biopsy #328, respectively; Fig. 3, E–G shows the same types of data but for biopsy #333. This biopsy was predicted with PCA-LDA to be glioma, by lipid deconvolution to have high TCP, and by 2-HG intensity to be IDH-mut. In contrast, biopsy #333 (blue point in Fig. 3, A) was taken several millimeters outside the MRI contrast enhancing area and was predicted from PCA-LDA to be normal white matter, by lipid deconvolution to have low TCP, and by 2-HG intensity to also be IDH-mut. Each of these predictions matched the histopathology assessments of the tissue smears. Thus, using the DESI-MS system, the neurosurgeon could analyze biopsies from discrete regions of the resection cavity to assess residual tumor when clinical acumen is insufficient.
Fig. 3.
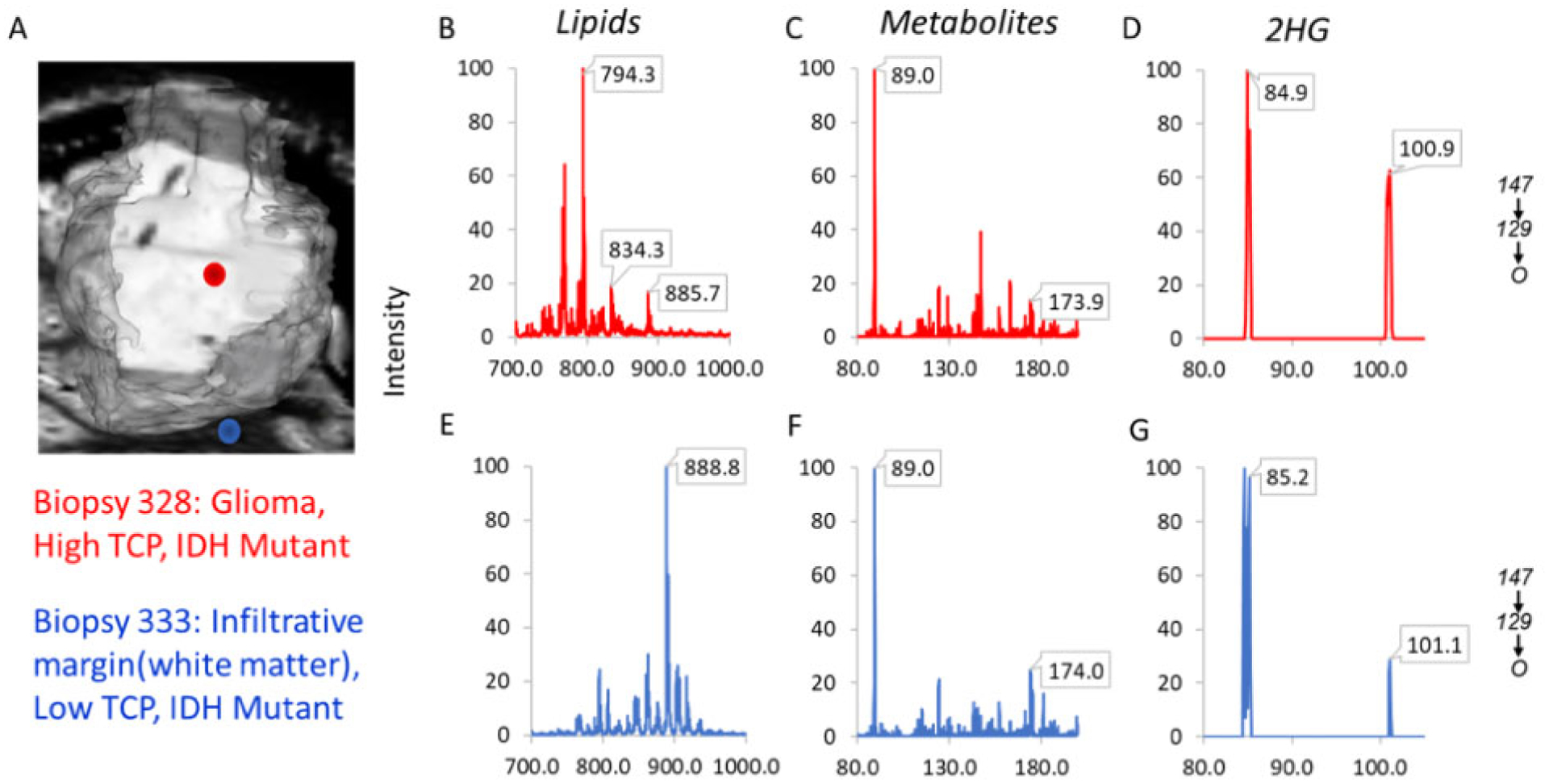
DESI-MS predictions of disease status, TCP, and IDH-mutation status for a core and margin biopsy from Subject 58. (A) Reconstruction of MRI tumor volume showing location of the two biopsies. Biopsy 328 (red dot) is within the contrast enhancing region of the tumor; biopsy 333 (blue dot) is a few mm outside of the contrast enhancing region. (B–D) The lipid, metabolite, and MS3 product ion scans for biopsy 328 (red dot in Fig. 2, A). (E–G) The lipid, metabolite, and 2-HG MS3 product ion scan for biopsy 333 (blue dot in Fig. 2, A). Biopsy 328 was classified as glioma, high TCP, and IDH-mut; biopsy 333 was infiltrative margin (white matter), low TCP, and IDH-mut.
While the initial results presented in this article are promising, it is important to remember that the DESI-MS system is proposed as an ancillary method to support the existing standard of care. Thus, when making the decision to resect, the DESI-MS assessments cannot be interpreted in isolation. In future studies, it will be worthwhile to evaluate whether DESI-MS assessment of areas at the MRI tumor borders can improve extent of resection and whether this in turn can reduce tumor recurrence and increase overall and progression-free survival. It will be worthwhile also to assess whether prediction of IDH mutation status via DESI-MS analysis can increase overall and progression-free survival by allowing for more personalized resection approaches and use of chemotherapeutic drugs targeting IDH mutations at the time of surgery.
CONCLUSIONS
DESI-MS monitoring of 2-HG, along with lipid and metabolite profiling, can improve the differential diagnosis of glioma intraoperatively through the assessment of IDH mutation status and disease status. The system also provides a means of assessing surgical margins for residual tumor through lipid profile deconvolution and measurement of NAA. The clinical accuracy of the IDH mutation assay was high (94%). Implementation of a new standard of care for surgical glioma resection based on IDH mutation status would be enabled by intraoperative DESI-MS assessments in combination with preoperative MRI detection of 2-HG. Additionally, the presence of the IDH mutation in this context is 100% specific for diffuse glioma and can assist in differential diagnosis to confirm the presence of glioma (36). The clinical accuracy of correctly predicting TCP (based on correlations with morphologic pathology evaluations) was moderate (83% and 81% for NAA and lipid profile deconvolution, respectively). Emerging evidence which suggests differences in patient outcome from maximal tumor resection based on IDH mutation status demonstrates a need for accurate and rapid intraoperative IDH mutation status assessment. Overwhelming clinical evidence showing the need to maximize extent of resection in gliomas highlights the need for methods to assess residual tumor at surgical margins. The intraoperative DESI-MS system is capable of these two applications. Advanced development of this DESI-MS system to enable clinical diagnostic applications and rapid assessments of residual tumor could significantly improve glioma resection.
Supplementary Material
IMPACT STATEMENT.
The intraoperative diagnosis of gliomas from tumor core samples can be improved using molecular diagnostics. Residual tumor at surgical margins is a primary contributor to tumor recurrence and malignant progression. Additionally, knowledge of prognostic genetic mutations at the time of surgery can better inform patient management strategies. DESI-MS can be used as a tool to aid intraoperative decision-making during glioma resection, specifically by allowing for the intraoperative assessment of IDH mutations, estimation of tumor cell infiltration, and disease status. Consequently, intraoperative DESI-MS has the potential to increase survival of glioma patients if available at the time of surgical resection.
Acknowledgments:
We thank Professor Tim Ratliff for his support; Heather Cero and Lauren Snyder for patient consent, providing clinical data, and IRB monitoring. Fan Pu, Rong Chen, Alan K. Jarmusch, and Zane Baird are thanked for their assistance with the study.
Role of Sponsor: The funding organizations played a direct role in the design of study, review, and interpretation of data, preparation of manuscript, and final approval of manuscript. The funding organizations played no role in the choice of enrolled patients.
Nonstandard Abbreviations:
- ACN
acetonitrile
- AUC
area under curve
- DESI-MS
desorption electrospray ionization-mass spectrometry
- DMF
dimethylformamide
- FWHM
full width at half maximum
- G
glioma
- GM
gray matter
- 2-HG
2-hydroxyglutarate
- IDH
isocitrate dehydrogenase
- IM
infiltrative margin
- NAA
N-acetyl-aspartic acid
- PCA
principal component analysis
- ROC
receiver operating curve
- SNV
standard normal variate
- TCP
tumor cell percentage
- WM
white matter
Footnotes
Authors’ Disclosures or Potential Conflicts of Interest: Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest. Employment or Leadership: None declared. Consultant or Advisory Role: None declared. Stock Ownership: None declared. Honoraria: None declared. Research Funding: This research was supported by the National Institute of Biomedical Imaging and Bioengineering (NIH grant no. R21EB015722). Support of the Purdue Center for Cancer Research, NIH grant P30 CA023168, is gratefully acknowledged. This publication was made possible with partial support for Clint Alfaro from grant no. UL1TR001108 (to A. Shekhar, principal investigator) from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award. Expert Testimony: None declared. Patents: R.G. Cooks, 7,335,897.
SUPPLEMENTAL MATERIAL
Supplemental material is available at The Journal of Applied Laboratory Medicine online.
Previous presentations: Oral presentation at MSACL 2019 US conference.
REFERENCES
- 1.D’Amico RS, Englander ZK, Canoll P, Bruce JN. Extent of resection in glioma-a review of the cutting edge. World Neurosurg 2017;103:538–49. [DOI] [PubMed] [Google Scholar]
- 2.Brat DJ, Verhaak RGW, Aldape KD, Yung WKA, Salama SR, Cooper LAD; Cancer Genome Atlas Research Network, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 2015;372: 2481–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGirt MJ, Chaichana KL, Gathinji M, Attenello FJ, Than K, Olivi A, et al. Independent association of extent of resection with survival in patients with malignant brain astrocytoma. J Neurosurg 2009;110:156–62. [DOI] [PubMed] [Google Scholar]
- 4.Hervey-Jumper SL, Berger MS. Maximizing safe resection of low- and high-grade glioma. J Neurooncol 2016;130: 269–82. [DOI] [PubMed] [Google Scholar]
- 5.Mampre D, Ehresman J, Pinilla-Monsalve G, Osorio MAG, Olivi A, Quinones-Hinojosa A, et al. Extending the resection beyond the contrast-enhancement for glioblastoma: feasibility, efficacy, and outcomes. Br J Neurosurg 2018;32:528–35. [DOI] [PubMed] [Google Scholar]
- 6.Eidel O, Burth S, Neumann JO, Kieslich PJ, Sahm F, Jungk C, et al. Tumor infiltration in enhancing and non-enhancing parts of glioblastoma: a correlation with histopathology. PLoS One 2017;12:e0169292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen R, Ravindra VM, Cohen AL, Jensen RL, Salzman KL, Prescot AP, et al. Molecular features assisting in diagnosis, surgery, and treatment decision making in low-grade gliomas. Neurosurg Focus 2015;38:E2. [DOI] [PubMed] [Google Scholar]
- 8.Claes A, Idema AJ, Wesseling P. Diffuse glioma growth: a guerilla war. Acta Neuropathol 2007;114: 443–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beiko J, Suki D, Hess KR, Fox BD, Cheung V, Cabral M, et al. IDH1 mutant malignant astrocytomas are more amenable to surgical resection and have a survival benefit associated with maximal surgical resection. Neuro Oncol 2014;16:81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel T, Bander ED, Venn RA, Powell T, Cederquist GY, Schaefer PM, et al. The role of extent of resection in IDH1 wild-type or mutant low-grade gliomas. Neurosurgery 2018;82:808–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takats Z, Wiseman JM, Gologan B, Cooks RG. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science 2004;306: 471–3. [DOI] [PubMed] [Google Scholar]
- 12.Ifa DR, Eberlin LS. Ambient ionization mass spectrometry for cancer diagnosis and surgical margin evaluation. Clin Chem 2016;62:111–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanel L, Kwiatkowski M, Heikaus L, Schluter H. Mass spectrometry-based intraoperative tumor diagnostics. Future Sci OA 2019;5:FSO373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshimura K, Mandal MK, Hara M, Fujii H, Chen LC, Tanabe K, et al. Real-time diagnosis of chemically induced hepatocellular carcinoma using a novel mass spectrometry-based technique. Anal Biochem 2013;441: 32–7. [DOI] [PubMed] [Google Scholar]
- 15.Alfaro CM, Jarmusch AK, Pirro V, Kerian KS, Masterson TA, Cheng L, et al. Ambient ionization mass spectrometric analysis of human surgical specimens to distinguish renal cell carcinoma from healthy renal tissue. Anal Bioanal Chem 2016;408:5407–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balog J, Sasi-Szabo L, Kinross J, Lewis MR, Muirhead LJ, Veselkov K, et al. Intraoperative tissue identification using rapid evaporative ionization mass spectrometry. Sci Transl Med 2013;5:194ra93. [DOI] [PubMed] [Google Scholar]
- 17.Phelps DL, Balog J, Gildea LF, Bodai Z, Savage A, El-Bahrawy MA, et al. The surgical intelligent knife distinguishes normal, borderline and malignant gynaecological tissues using rapid evaporative ionisation mass spectrometry (REIMS). Br J Cancer 2018;118: 1349–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woolman M, Ferry I, Kuzan-Fischer CM, Wu M, Zou J, Kiyota T, et al. Rapid determination of medulloblastoma subgroup affiliation with mass spectrometry using a handheld picosecond infrared laser desorption probe. Chem Sci 2017;8:6508–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fatou B, Saudemont P, Leblanc E, Vinatier D, Mesdag V, Wisztorski M, et al. In vivo real-time mass spectrometry for guided surgery application. Sci Rep 2016;6:25919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, Rector J, Lin JQ, Young JH, Sans M, Katta N, et al. Nondestructive tissue analysis for ex vivo and in vivo cancer diagnosis using a handheld mass spectrometry system. Sci Transl Med 2017;9:eaan3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sans M, Zhang J, Lin JQ, Feider CL, Giese N, Breen MT, et al. Performance of the MasSpec pen for rapid diagnosis of ovarian cancer. Clin Chem 2019;65:674–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swales JG, Hamm G, Clench MR, Goodwin RJA. Mass spectrometry imaging and its application in pharmaceutical research and development: A concise review. Int J Mass Spectrometry 2019;437:99–112. [Google Scholar]
- 23.Kooijman PC, Nagornov KO, Kozhinov AN, Kilgour DPA, Tsybin YO, Heeren RMA, et al. Increased throughput and ultra-high mass resolution in DESI FT-ICR MS imaging through new-generation external data acquisition system and advanced data processing approaches. Sci Rep 2019;9:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, Manicke NE, Yang Q, Zheng L, Shi R, Cooks RG, et al. Direct analysis of biological tissue by paper spray mass spectrometry. Anal Chem 2011;83:1197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woolman M, Zarrine-Afsar A. Platforms for rapid cancer characterization by ambient mass spectrometry: advancements, challenges and opportunities for improvement towards intrasurgical use. Analyst 2018; 143:2717–22. [DOI] [PubMed] [Google Scholar]
- 26.Eberlin LS, Margulis K, Planell-Mendez I, Zare RN, Tibshirani R, Longacre TA, et al. Pancreatic cancer surgical resection margins: molecular assessment by mass spectrometry imaging. PLoS Med 2016;13: e1002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarmusch AK, Pirro V, Baird Z, Hattab EM, Cohen-Gadol AA, Cooks RG. Lipid and metabolite profiles of human brain tumors by desorption electrospray ionization-MS. Proc Natl Acad Sci Usa USA 2016;113:1486–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eberlin LS, Norton I, Orringer D, Dunn IF, Liu X, Ide JL, et al. Ambient mass spectrometry for the intraoperative molecular diagnosis of human brain tumors. Proc Natl Acad Sci USA 2013;110:1611–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eberlin LS, Norton I, Dill AL, Golby AJ, Ligon KL, Santagata S, et al. Classifying human brain tumors by lipid imaging with mass spectrometry. Cancer Res 2012;72:645–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eberlin LS, Dill AL, Golby AJ, Ligon KL, Wiseman JM, Cooks RG, et al. Discrimination of human astrocytoma subtypes by lipid analysis using desorption electrospray ionization imaging mass spectrometry. Angew Chem Int Ed Engl 2010;49:5953–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pirro V, Alfaro CM, Jarmusch AK, Hattab EM, Cohen-Gadol AA, Cooks RG. Intraoperative assessment of tumor margins during glioma resection by desorption electrospray ionization-mass spectrometry. Proc Natl Acad Sci USA 2017;114:6700–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alfaro CM, Pirro V, Keating MF, Hattab EM, Cooks RG, Cohen-Gadol AA. Intraoperative assessment of isocitrate dehydrogenase mutation status in human gliomas using desorption electrospray ionization-mass spectrometry. J Neurosurg 2019;132:1–8. [DOI] [PubMed] [Google Scholar]
- 33.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009;462:739–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moffett JR, Ross B, Arun P, Madhavarao CN, Namboodiri AM. N-Acetylaspartate in the CNS: from neurodiagnostics to neurobiology. Prog Neurobiol 2007; 81:89–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price SJ, Jena R, Burnet NG, Hutchinson PJ, Dean AF, Peña A, et al. Improved delineation of glioma margins and regions of infiltration with the use of diffusion tensor imaging: an image-guided biopsy study. AJNR Am J Neuroradiol 2006;27:1969–74. [PMC free article] [PubMed] [Google Scholar]
- 36.Kim BYS, Jiang W, Beiko J, Prabhu SS, DeMonte F, Gilbert MR, et al. Diagnostic discrepancies in malignant astrocytoma due to limited small pathological tumor sample can be overcome by IDH1 testing. J Neurooncol 2014;118:405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


