Abstract
Background
Human skin color is highly heritable and one of the most variable phenotypic traits. However, the genetic causes and environmental selective pressures underlying this phenotypic variation have remained largely unknown. To investigate whether the pigmentation related-genes polymorphisms are associated with the geographic environmental variables. We selected randomly 795 healthy individuals from eight ethnic groups in nine provinces in China. Six single nucleotide polymorphisms (SNPs) of SLC45A2 and TYR were genotyped using Agena MassARRAY. The Chi-square test and Spearman correlation analysis were used to compare the frequency distribution of genotypes among different ethnic groups and evaluate the relationship between SNP genetic diversity and environmental variables, respectively.
Results
The results indicated that rs28777 and rs183671 (SLC45A2) and rs1042602 (TYR) genotype frequency distributions were significantly different between the Xinjiang-Uighur and other ethnic groups (P < 0.05). Spearman correlation analysis found that rs28777-A (r = − 0.090, P = 0.011), rs183671-G (r = − 0.105, P = 0.003), rs1042602-A (r = − 0.108, P = 0.002), rs1126809-A (r = − 0.151, P < 0.001) allele frequencies were negatively correlated with the longitude; rs183671-G (r = 0.151), rs1042602-A (r = 0.157) and rs1126809-A (r = 0.138) allele frequencies were positively associated with the latitude (P < 0.001); rs183671-G (r = 0.116, P = 0.001), rs1042602-A (r = 0.105, P = 0.003) and rs1126809-A (r = 0.070, P = 0.048) allele frequencies were positively correlated with the sunshine hours; rs183671-G (r = − 0.076, P = 0.033), rs1042602-A (r = − 0.079, P = 0.027) and rs1126809-A (r = − 0.076, P = 0.031) were negatively correlated with the annual average temperature.
Conclusions
Our results confirmed the idea that environmental factors have been an important selective pressure upon pigmentation related gene polymorphisms.
Supplementary Information
The online version contains supplementary material available at 10.1186/s41065-021-00189-7.
Keywords: Association study, Polymorphisms, Environmental factors, Selection pressure
Introduction
Human skin color is highly heritable and one of the most variable phenotypic traits that can vary dramatically within and across ethnic populations [1]. It is known that the human skin color is predominantly determined by pigments include melanin, hemoglobin (red), hemosiderin (brown), carotene (yellow), and bilirubin (yellow) [2]. Among those, the amount, type, and distribution of melanin play key roles in determining human skin pigmentation. Studies indicate that the human skin pigmentation in global populations is highly associated with latitude, and fundamentally, the distribution of ultraviolet (UV) radiation [3, 4]. Moreover, the researchers believe that geographic variation in skin pigmentation was influenced by the concerted action of different types of natural selection, including climate, lifestyle, diet, metabolism [1]. However, the genetic causes and environmental selective pressures underlying this range of skin color variation have remained largely unknown.
With the rapid development of genetics and genomics, researchers have gradually realized that the human skin color diversity is due to the natural positive selection of those genes that impact on human pigmentation, especially in the melanosome biogenesis or the melanin biosynthetic pathways [5, 6]. Recently, a large number of genome-wide association studies (GWAS) for pigmentation have been established and identified that some single nucleotide polymorphisms (SNPs) on TYR, IRF4, TYRP1, OCA2, SLC45A2, MC1R and KITLG genes are significantly associated with human skin color [7–10]. The solute carrier family 45, member 2 (SLC45A2) gene encodes the membrane associated transporter protein (MATP). The SLC45A2 protein expresses in melanocyte cell lines and mediates melanin synthesis by tyrosinase trafficking and proton transportation to melanosomes [11]. SLC45A2 mutations cause oculocutaneous albinism type IV (OCA4) and polymorphisms of SLC45A2 gene are associated with dark skin, hair, and eye pigmentation [12, 13]. In addition, the TYR gene encodes tyrosinase, a multifunctional enzyme that plays a major role in melanin biosynthesis in melanocytes [14]. TYR is commonly known as the albino locus since the homozygous or compound heterozygous mutations of this gene result in oculocutaneous albinism type 1 (OCA1), an autosomal recessive genetic disorder characterized by hypopigmented hair, skin and eyes [15].
However, the genetic causes and environmental selective pressures underlying this range of phenotypic variation have remained largely unknown. Therefore, to investigate whether the six polymorphisms in the two pigmentation related-genes SLC45A2 (rs11568737, rs28777 and rs183671) and TYR gene (rs1042602, rs1393350 and rs1126809) are associated with the geographic environmental variables, we selected randomly a total of 795 healthy individuals from eight ethnic groups in nine provinces in China, while collected the geographic environmental variables (altitude, longitude, latitude, air pressure, sunshine hours, and annual average temperature). The results of this study will improve our understanding of the impact of environmental variables in genetic differentiation and maintenance of genetic variation.
Results
A total of 795 samples including eight ethnic groups from nine provinces in China (Tibet-Tibetan accounted for 13.2%, Inner Mongolia-Ewenki 12.6%, Hainan-Han 6.2%, Ningxia-Hui 12.6%, Hainan-Li 12.5%, Inner Mongolia-Mongolian 12.6%, Guizhou-Miao 11.2%, Xinjiang-Uighur 13.3%, and Shaanxi-Han 5.9%) were collected to study the relationship between skin pigmentation-related gene variants and environmental variables. We also collected the detailed geographical environment information of different ethnic regions (Fig. 1), including altitude (m), longitude (°), latitude (°), atmosphere pressure (kPa), sunshine duration (hours), and year-round average temperature (°C), as shown in Table 1.
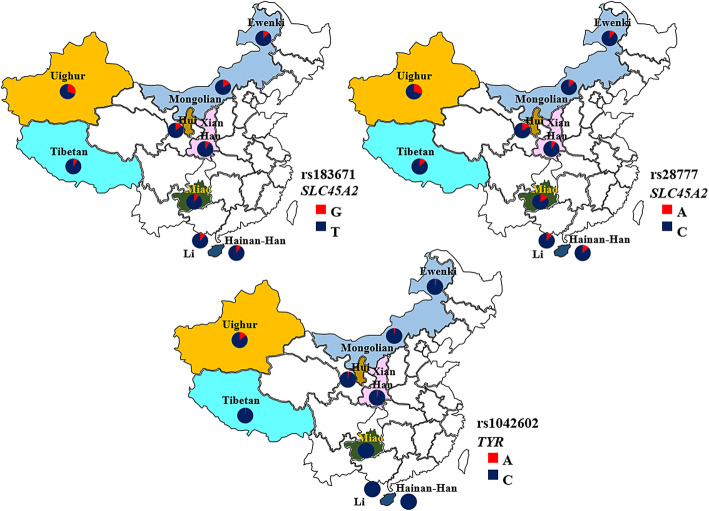
Fig. 1.
Distribution of allele frequencies of rs1042602 (A/C), rs28777 (A/C) and rs183671 (G/T) among different ethnic groups in China
Table 1.
Detailed geographical environment information of different ethnic regions
| Ethnic | Residence | N | Altitude (m) | Longitude (°) | Latitude (°) | Atmosphere pressure (kPa) | Sunshine duration (hours) | Year-round average temperature (°C) |
|---|---|---|---|---|---|---|---|---|
| Tibetan | Naqu | 4 | 4505 | 92.058 | 31.482 | 58 | 2879 | 11 |
| Linzhi | 9 | 2994 | 94.368 | 29.655 | 70 | 2005 | 11 | |
| Shannan | 13 | 3572 | 91.78 | 29.243 | 65 | 2800 | 10 | |
| Shigatse | 20 | 3844 | 88.887 | 29.273 | 63 | 3248 | 8 | |
| Lhasa | 59 | 3651 | 91.129 | 29.659 | 64 | 3055 | 10 | |
| Total | 105 | |||||||
| Ewenki | Ewenki Autonomous Banner Huisumuhakemugacha | 10 | 690 | 119.172 | 48.379 | 93 | 2900 | 3 |
| Yiminhe Town, Ewenki Autonomous Banner | 19 | 673 | 119.791 | 48.583 | 93 | 2900 | 3 | |
| Bayantuohai Town, Ewenki Autonomous Banner | 21 | 617 | 119.762 | 49.143 | 94 | 2900 | 3 | |
| Dayan Town, Ewenki Autonomous Banner | 23 | 682 | 120.558 | 49.237 | 93 | 2900 | 3 | |
| Ewenki Autonomous Banner, Xinihe East Sumu | 27 | 788 | 120.3 | 48.867 | 92 | 2900 | 3 | |
| Total | 100 | |||||||
| Hainan-Han | Haikou City, Hainan | 49 | 9 | 110.339 | 20.035 | 101 | 2041 | 24.4 |
| Hui | Haiyuan County, Zhongwei City, Ningxia | 5 | 1841 | 105.65 | 36.571 | 81 | 1609 | 11 |
| Guyuan City, Ningxia | 10 | 1778 | 106.249 | 36.022 | 82 | 1602 | 9 | |
| Tongxin County, Wuzhong City, Ningxia | 85 | 1316 | 105.816 | 36.986 | 86 | 1690 | 12.5 | |
| Total | 100 | |||||||
| Li | Wangxia Town, Changjiang, Hainan | 5 | 357 | 109.157 | 19.009 | 97 | 2300 | 26 |
| Baoting Li and Miao Autonomous County | 8 | 54 | 109.707 | 18.647 | 101 | 2300 | 26 | |
| Changjiang Li Autonomous County, Hainan | 38 | 140 | 109.062 | 19.304 | 100 | 2300 | 26 | |
| Qicha Town, Changjiang, Hainan | 48 | 107 | 109.062 | 19.118 | 100 | 2300 | 26 | |
| Total | 99 | |||||||
| Mongolian | Chenqiba Town, Inner Mongolia | 19 | 597 | 119.446 | 49.334 | 94 | 3205 | 7.7 |
| Hohhot | 81 | 1056 | 111.668 | 40.819 | 89 | 2588 | 7.3 | |
| Total | 100 | |||||||
| Miao | Gaopo Township, Huaxi District, Guiyang City | 39 | 1459 | 106.819 | 26.302 | 85 | 1060 | 14.8 |
| Mengguan Township, Huaxi District, Guiyang City | 50 | 1196 | 106.755 | 26.415 | 88 | 1060 | 14.8 | |
| Total | 89 | |||||||
| Uighur | Bazhou | 23 | 944 | 86.152 | 41.77 | 90 | 2990 | 11.5 |
| Ili | 23 | 646 | 81.331 | 43.923 | 94 | 2977 | 5.8 | |
| Aqsu | 30 | 1109 | 80.314 | 41.15 | 89 | 2911 | 12.5 | |
| Kashgar | 30 | 1298 | 75.996 | 39.476 | 87 | 2760 | 13 | |
| Total | 106 | |||||||
| Shaanxi-Han | Yan’an | 1 | 1070 | 109.496 | 36.591 | 89 | 2056 | 15.5 |
| Hancheng | 1 | 457 | 110.449 | 35.483 | 96 | 2056 | 15.5 | |
| Fuping | 2 | 520 | 109.364 | 34.95 | 95 | 2056 | 15.5 | |
| Weinan | 3 | 355 | 109.516 | 34.506 | 93 | 2056 | 15.5 | |
| Xi’an | 40 | 381 | 108.947 | 34.27 | 96 | 2056 | 15.5 | |
| Total | 47 |
The six SNPs on the two skin pigmentation-related gene SLC45A2 (rs11568737, rs28777 and rs183671) and TYR (rs1042602, rs1393350 and rs1126809) were successfully genotyped from 795 samples (call rate > 95%). The basic information (SNP-ID, chromosome number, position, alleles and gene name) and polymerase chain reaction (PCR) primer sequence (1st-PCRP, 2nd-PCRP and unique base extension primer sequence (UEP-SEQ) of the six SNPs was showed in Table 2. The minor allele frequency (MAF), genotype frequency and Hardy-Weinberg equilibrium (HWE)-P value of each SNPs are shown in Supplementary Table 1, Tables 2, and 3, respectively. The results showed that except for rs1393350 in TYR was not in accordance with the HWE in Uighur (P < 0.01), other five SNPs were in accordance with the HWE in the nine groups (P > 0.01).
Table 2.
The basic information and primer sequence of SNPs
| SNP-ID | Chromosome | Position | Alleles | Genes | 1st-PCRP | 2nd-PCRP | UEP_SEQ |
|---|---|---|---|---|---|---|---|
| rs11568737 | 5 | 33,944,743 | T > C | SLC45A2 | ACGTTGGATGGTGATCACCACGACGACAAC | ACGTTGGATGATGGTGCAGCTGGCTCAGAT | gGGGCTTTCTGGTCAAC |
| rs28777 | 5 | 33,958,854 | C > A | SLC45A2 | ACGTTGGATGAAAAGGCTTCCACTCAGTTG | ACGTTGGATGCAAGAGTCGCATAGGACAGG | cctcCGTCCCATCCACTCAGAG |
| rs183671 | 5 | 33,964,105 | T > G | SLC45A2 | ACGTTGGATGTCCTCATGCATAGACACTCC | ACGTTGGATGATATCCAGGTTGCCTCTGCT | ggcaTCTGCTGTCTTCAGGG |
| rs1042602 | 11 | 89,178,528 | C > A | TYR | ACGTTGGATGTGACCTCTTTGTCTGGATGC | ACGTTGGATGGGTGCTTCATGGGCAAAATC | TCAATGTCTCTCCAGATTTCA |
| rs1393350 | 11 | 89,277,878 | G > A | TYR | ACGTTGGATGGCATATCCACCAACTCCTAC | ACGTTGGATGGGAAGGTGAATGATAACACG | TTTGTAAAAGACCACACAGATTT |
| rs1126809 | 11 | 89,284,793 | G > A | TYR | ACGTTGGATGAATGGGTGCATTGGCTTCTG | ACGTTGGATGCCTCTGCAGTATTTTTGAGC | catcTTGAGCAGTGGCTCC |
SNP single nucleotide polymorphism, Chr chromosome, PCRP polymerase chain reaction primer, UEP_SEQ unique base extension primer sequence
Table 3.
Differences in genotype distributions of SNPs among different ethnic groups
| SNP-ID | Ethnic | Ewenki | Hainan-Han | Hui | Li | Miao | Mongolian | Tibetan | Uighur | Shaanxi-Han |
|---|---|---|---|---|---|---|---|---|---|---|
| rs28777 | Ewenki | |||||||||
| Hainan-Han | 0.811 | |||||||||
| Hui | 0.279 | 0.561 | ||||||||
| Li | 0.977 | 0.897 | 0.323 | |||||||
| Miao | 0.248 | 0.763 | 0.271 | 0.346 | ||||||
| Mongolian | 0.778 | 0.998 | 0.353 | 0.887 | 0.614 | |||||
| Tibetan | 0.783 | 0.995 | 0.318 | 0.891 | 0.591 | 0.999 | ||||
| Uighur | 1.12E-05 | 5.32E-03 | 9.27E-04 | 2.87E-05 | 6.78E-03 | 1.65E-04 | 1.09E-04 | |||
| Shaanxi-Han | 0.617 | 0.557 | 0.209 | 0.617 | 0.278 | 0.560 | 0.576 | 5.02E-04 | ||
| rs183671 | Ewenki | |||||||||
| Hainan-Han | 0.416 | |||||||||
| Hui | 0.366 | 0.545 | ||||||||
| Li | 0.608 | 0.837 | 0.662 | |||||||
| Miao | 0.503 | 0.322 | 0.155 | 0.401 | ||||||
| Mongolian | 0.506 | 0.142 | 0.265 | 0.197 | 0.102 | |||||
| Tibetan | 0.575 | 0.784 | 0.301 | 0.797 | 0.620 | 0.098 | ||||
| Uighur | 3.20E-05 | 5.18E-05 | 4.80E-05 | 3.29E-06 | 1.43E-06 | 2.46E-03 | 2.59E-07 | |||
| Shaanxi-Han | 0.675 | 0.516 | 0.370 | 0.616 | NA | 0.252 | 0.769 | 2.04E-04 | ||
| rs1042602 | Uighur | 3.53E-04 | 3.71E-04 | 4.56E-05 | 3.05E-07 | 1.09E-06 | 4.56E-05 | 2.19E-06 | 1.58E-02 |
SNP single nucleotide polymorphism
P < 0.05 was considered to be significant
In addition, we used the Chi-square test to evaluate the difference of genotype frequency distribution of the five SNPs among eight ethnic groups, as shown in Table 3. The results indicated that the genotype frequency distribution of rs28777 and rs183671 (SLC45A2) and rs1042602 (TYR) were significantly different between the Xinjiang-Uighur and other ethnic groups (P < 0.05). The allele frequency distribution of these three significantly different SNPs was shown in Fig. 1.
Simultaneously, we analyzed the relationship between SNP genetic diversity and environmental variables using Spearman correlation analysis (Table 4). It was found that the allele frequencies of rs28777-A (r = − 0.090, P = 0.011), rs183671-G (r = − 0.105, P = 0.003), rs1042602-A (r = − 0.108, P = 0.002), rs1126809-A (r = − 0.151, P < 0.001) were negatively correlated with the longitude. However, the positive correlation between the alleles frequencies of rs183671-G (r = 0.151), rs1042602-A (r = 0.157) and rs1126809-A (r = 0.138) and the latitude were extremely significant (P < 0.001). The alleles frequencies of rs183671-G (r = 0.116, P = 0.001), rs1042602-A (r = 0.105, P = 0.003) and rs1126809-A (r = 0.070, P = 0.048) were found to be significantly positively correlated with the sunshine hours. However, the alleles frequencies of rs183671-G (r = − 0.076, P = 0.033), rs1042602-A (r = − 0.079, P = 0.027) and rs1126809-A (r = − 0.076, P = 0.031) were significantly negatively correlated with the annual average temperature. The correlations between the allele frequencies of other SNPs and environmental variables were not significant. These findings indicate that environmental factors have selective pressure on these SNPs.
Table 4.
The association between polymorphisms and geographic environmental variables
| SNP-ID | Altitude | Longitude | Latitude | Air pressure | Sunshine hours | Annual average temperature | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| γ | p | γ | p | γ | p | γ | p | γ | p | γ | p | |
| rs11568737 | 0.038 | 0.286 | −0.031 | 0.383 | −0.007 | 0.851 | −0.037 | 0.302 | 0.010 | 0.783 | 0.011 | 0.748 |
| rs28777 | 0.002 | 0.958 | −0.090 | 0.011 | 0.036 | 0.313 | 0.003 | 0.942 | 0.036 | 0.305 | −0.011 | 0.761 |
| rs183671 | −0.002 | 0.950 | −0.105 | 0.003 | 0.151 | 1.86E-05 | 0.006 | 0.872 | 0.116 | 0.001 | −0.076 | 0.033 |
| rs1042602 | 0.004 | 0.903 | −0.108 | 0.002 | 0.157 | 8.54E-06 | 0.005 | 0.893 | 0.105 | 0.003 | −0.079 | 0.027 |
| rs1126809 | 0.022 | 0.539 | −0.151 | 1.90E-05 | 0.138 | 9.38E-05 | −0.015 | 0.666 | 0.070 | 0.048 | −0.076 | 0.031 |
P < 0.05 was considered to be significant
Discussion
To investigate whether the two pigmentation related genes (SLC45A2 and TYR) polymorphisms are associated with the geographic environmental variables (altitude, longitude, latitude, and air pressure, sunshine hours, and annual average temperature), we selected randomly selected 795 healthy individuals from eight ethnic groups in nine provinces in China. The results of this study found that the genotype frequency distribution of rs28777 and rs183671 in SLC45A2 and rs1042602 in TYR were significantly different between the Xinjiang-Uighur and other ethnic groups (P < 0.05). Simultaneously, the rs28777, rs183671, rs1042602, rs1126809 polymorphisms were found to be correlated with the geographic environmental variables (longitude, latitude, sunshine hours or annual average temperature).
SLC45A2 (as also AIM1 or MATP) encodes a transporter protein that mediates melanin synthesis, which is expressed in a high percentage of melanoma cell lines. It has been reported that some SLC45A2 mutations cause OCA4 and polymorphisms of this gene were found to be significantly associated with human skin, hair, and eye pigmentation, and its mutation frequency varies significantly among the global population. Yuko Abe et al. found that rs11568737 in SLC45A2 (T500P) was significantly associated with melanin index [16]. A multi-stage GWAS of natural hair color in European ancestry found that rs28777 (SLC45A2) was associated with skin color and tanning ability [17]. A large Australian population-based case control study reveal that rs28777 exhibited the strongest crude association with risk of cutaneous malignant melanoma [18]. The study found that rs183671 (SLC45A2) was in strong linkage disequilibrium (LD) with rs16891982 (F374L) in CEU. A previous GWAS declared that the frequency of the rs183671 derived allele increased from Southern to Northern Europe, and this SNP was associated with skin pigmentation, and that each copy of the derived allele lightens the skin by 1.2 M index units [19]. Moreover, a previous GWAS demonstrated that the SNP rs183671 can explain skin color variation in three European studies RS, BTNS, and TwinsUK [20].
TYR is located at human chromosome 11q14.3, and encodes tyrosinase, which regulates the biosynthesis of melanin. Previous study demonstrated that mutations in TYR can cause OCA1 [15]. The non-synonymous polymorphism rs1042602 (Ser192Tyr) in TYR derived allele has specifically high frequency in Europe, and rs1042602 was significantly associated with eye color, freckles and lighter skin pigmentation [21–24]. It has been reported that rs1393350 was also associated with human hair, eye and skin color and tanning ability [23, 25–27]. A GWAS of melanoma conducted by the GenoMEL consortium identifies the locus rs1393350 associated with melanoma risk [28]. The rs1126809 variant is located in exon 4 of TYR gene and encodes a tyrosinase enzyme with an arginine-to-glutamine substitution at codon 402 (R402Q), and is a strong linkage with rs1393350 [29, 30]. The mutation of rs1126809 (A-G) causes the TYR enzyme to be thermosensitive, thus less active [31]. The rs1126809 has previously been used as a marker for skin pigmentation and also influence brown eye color formation [23, 30]. Previous GWAS indicated that the allele A of rs1042602 (TYR) was highly associated with lighter skin color in a South Asian descent population [32]. It has reported that the allele A of rs1042602 was over-represented in the IndoEuropeans population [33]. The two polymorphisms (rs1042602 and rs1126809) in TYR appear at high frequency in Europeans and are largely absent in African populations [34].
This study indicated that the genotype distribution of rs28777 and rs18367 in Xinjiang-Uighur was significantly different from other ethnic groups. Moreover, the allele frequencies of rs28777-A, rs183671-G, rs1042602-A, rs1126809-A were negatively correlated with the longitude; rs183671, rs1042602 and rs1126809 allele frequencies were positively associated with the latitude and the sunshine hours, while were negatively correlated with the annual average temperature in Chinese population. At present, there are few research reports on the association between genetic polymorphism and environmental factors. In 2010, Ji et al. [35] found that the disease-predisposition polymorphisms of the melatonin receptors were associated with sunshine duration in the global human populations. These results indicated that environmental factors had selective pressure on these loci, and their changes were related to environmental variables, that is, differences in selection caused by differences in environmental factors play an important role in genetic differentiation.
However, this study has some limitations that cannot be ignored. First, the sample size is small and the statistical power is relatively low. Second, this study is the first to explore the correlation between the allele frequencies of these six SNPs and geographical environmental factors. Third, we only selected six SNP loci on two genes to explore their correlation with geographical environmental factors. Finally, the effect of these genetic variations on human skin color diversity is not involved in this study. Therefore, we will further collect a larger sample and choose more SNPs and design functional experiments to explore the impact of environmental factors on genetic mutations.
In summary, this study results indicate that rs28777, rs183671 (SLC45A2) and 1,042,602 (TYR) polymorphisms were different among different populations. More importantly, our results confirm the idea that environmental factors have been an important selective pressure upon pigmentation related gene polymorphisms (rs28777, rs183671, rs1042602 and rs1126809). Further association and functional studies need to confirm our results in a large sample and explore the influence of geographical environment factors on the skin pigmentation-related genes polymorphisms and the mechanism of action.
Materials and methods
Study design
This study randomly selected a total of 795 healthy individuals from eight ethnic groups in nine provinces in China, including 105 Tibetan individuals, 100 Ewenki individuals, 49 Hainan Han individuals, 100 Hui individuals, 99 Li individuals, 100 Mongolian individuals, 89 Miao individuals, 106 Uighur individuals, and 47 Shaanxi-Han individuals. The basic situation of each population was shown in Table 1. The climate data (sunshine hours and annual average temperature) are quoted from China’s surface climate data in 2019. The information of altitude, longitude, latitude, and air pressure was collected through online query. Individuals who have a history of skin pigmentation-related diseases (albinism or melanoma), history of serious illness, mental illness, pregnancy were excluded from the study.
DNA extraction
The peripheral venous blood sample (5 mL) from each subjects were taken from fasting in the morning using the Ethylene diamine tetraacetic acid (EDTA) tube, and stored at − 20 °C refrigerator for further experiment. The GoldMag-Mini Whole Blood Genomic DNA Purification Kit (GoldMag. Co. Ltd., Xi’an, China) was used to extract genomic DNA, including blood lysis, adding GoldMag® gold magnetic particles to bind DNA, magnetic separation, washing, elution, magnetic separation to obtain DNA. In order to determine the concentration and purity of the extracted DNA, we use a spectrophotometer (Nanodrop 2000, Thermo Fisher Scientific, Waltham, MA, USA). If the ratio of OD260/OD280 ratios is about 1.8, the extracted DNA is qualified.
SNP selection and genotyping
We randomly selected the six SNPs (rs11568737, rs28777 and rs183671 in the SLC45A2 gene and rs1042602, rs1393350 and rs1126809 in the TYR gene) based on previously published genes related to pigmentation [18, 21, 28, 36–40]. The online software Agena Bioscience Assay Design Suite Version 2.0 (https://agenacx.com/online-tools/) was used to design the primers sequence (Table 2). The Agena MassARRAY platform (Agena Bioscience, San Diego, CA, USA) was used to genotype the six SNPs from 795 samples, according to the manufacturer’s instructions, including DNA sample preparation; polymerase chain reaction (PCR) amplification (95 °C pre-denaturation 2 min; 45 cycles (95 °C denaturation 30s, 56 °C annealing 30s, 72 °C extension 60s); 72 °C extension 5 min; 4 °C storage); shrimp alkaline phosphatase purification; Unique base extension primer (UEP) reaction; resin purification; spotting and mass spectrometry. Genotyping results data management and analysis using the Agena Bioscience TYPER software (version 4.0).
Statistical analysis
We used the Microsoft Excel (Microsoft Corp., Redmond, WA, USA) and Statistical Package for the Social Sciences (SPSS) version 25 (SPSS, Chicago, IL) to perform statistical analysis. The Chi-square test was used to evaluate whether each SNP was consistent with Hardy-Weinberg Equilibrium (HWE), and compare whether there are significant differences in the frequency distribution of genotypes among different ethnic groups. The relationship between SNP genetic diversity and environmental variables was analyzed using Spearman correlation analysis. All statistical analyses were two sided and the P < 0.05 was considered as statistically significant.
Supplementary Information
Additional file 1: Supplementary Table 1. The minor allele frequency of each SNP in different ethnic groups.
Additional file 2: Supplementary Table 2. The genotype frequency of each SNP in different ethnic groups.
Additional file 3: Supplementary Table 3. The HWE-P value of each SNP in different ethnic groups.
Acknowledgements
We are grateful to the individuals for their participation in the study. We also thank the staff of the Medical College of Nanchang University who contributed to sample collection for this study.
Abbreviations
- EDTA
Ethylene diamine tetraacetic acid
- GWAS
Genome-wide association studies
- HWE
Hardy-Weinberg Equilibrium
- LD
Linkage disequilibrium
- MATP
Membrane associated transporter protein
- OCA
Oculocutaneous albinism type
- PCR
Polymerase chain reaction
- SLC45A2
Solute carrier family 45, member 2
- SNPs
Single nucleotide polymorphisms
- SPSS
Statistical Package for the Social Sciences
- UV
Ultraviolet
Author’s contributions
Yuxin Wang contributed to the study conception and design and data analysis and manuscript writing and provided final approval of the manuscript.
Funding
Not applicable.
Availability of data and materials
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
The study design and protocol were approved by the Ethics Committee of the Nanchang University (NCU201908). And the present study was conducted in accordance with the ethical principles of the Declaration of Helsinki. Written informed consent was obtained from all individual participants included in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rocha J. The evolutionary history of human skin pigmentation. J Mol Evol. 2020;88(1):77–87. doi: 10.1007/s00239-019-09902-7. [DOI] [PubMed] [Google Scholar]
- 2.Yamaguchi Y, Hearing VJ. Melanocytes and their diseases. Cold Spring Harb Perspect Med. 2014;4(5):a017046. doi: 10.1101/cshperspect.a017046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deng L, Xu S. Adaptation of human skin color in various populations. Hereditas. 2018;155(1):1. doi: 10.1186/s41065-017-0036-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barsh GS. What controls variation in human skin color? PLoS Biol. 2003;1(1):E27. doi: 10.1371/journal.pbio.0000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sturm RA. Molecular genetics of human pigmentation diversity. Hum Mol Genet. 2009;18(R1):R9–17. doi: 10.1093/hmg/ddp003. [DOI] [PubMed] [Google Scholar]
- 6.Pavan WJ, Sturm RA. The genetics of human skin and hair pigmentation. Annu Rev Genomics Hum Genet. 2019;20(1):41–72. doi: 10.1146/annurev-genom-083118-015230. [DOI] [PubMed] [Google Scholar]
- 7.Crawford NG, Kelly DE, Hansen MEB, Beltrame MH, Fan S, Bowman SL, Jewett E, Ranciaro A, Thompson S, Lo Y, et al. Loci associated with skin pigmentation identified in African populations. Science. 2017;358(6365):eaan8433. doi: 10.1126/science.aan8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobs LC, Hamer MA, Gunn DA, Deelen J, Lall JS, van Heemst D, Uh HW, Hofman A, Uitterlinden AG, Griffiths CEM, Beekman M, Slagboom PE, Kayser M, Liu F, Nijsten T. A genome-wide association study identifies the skin color genes IRF4, MC1R, ASIP, and BNC2 influencing facial pigmented spots. J Invest Dermatol. 2015;135(7):1735–1742. doi: 10.1038/jid.2015.62. [DOI] [PubMed] [Google Scholar]
- 9.Hernandez-Pacheco N, Flores C, Alonso S, Eng C, Mak AC, Hunstman S, Hu D, White MJ, Oh SS, Meade K, et al. Identification of a novel locus associated with skin colour in African-admixed populations. Sci Rep. 2017;7(1):44548. doi: 10.1038/srep44548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adhikari K, Mendoza-Revilla J, Sohail A, Fuentes-Guajardo M, Lampert J, Chacon-Duque JC, Hurtado M, Villegas V, Granja V, Acuna-Alonzo V, et al. A GWAS in Latin Americans highlights the convergent evolution of lighter skin pigmentation in Eurasia. Nat Commun. 2019;10(1):358. doi: 10.1038/s41467-018-08147-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fracasso NCA, de Andrade ES, Wiezel CEV, Andrade CCF, Zanao LR, da Silva MS, Marano LA, Donadi EA, Castelli EC, Simoes AL, et al. Haplotypes from the SLC45A2 gene are associated with the presence of freckles and eye, hair and skin pigmentation in Brazil. Leg Med. 2017;25:43–51. doi: 10.1016/j.legalmed.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 12.Mauri L, Barone L, Al Oum M, Del Longo A, Piozzi E, Manfredini E, Stanzial F, Benedicenti F, Penco S, Patrosso MC. SLC45A2 mutation frequency in Oculocutaneous albinism Italian patients doesn't differ from other European studies. Gene. 2014;533(1):398–402. doi: 10.1016/j.gene.2013.09.053. [DOI] [PubMed] [Google Scholar]
- 13.Le L, Escobar IE, Ho T, Lefkovith AJ, Latteri E, Haltaufderhyde KD, Dennis MK, Plowright L, Sviderskaya EV, Bennett DC, et al. SLC45A2 protein stability and regulation of melanosome pH determine melanocyte pigmentation. Mol Biol Cell. 2020;31:2687. doi: 10.1091/mbc.E20-03-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anello M, Fernandez E, Daverio MS, Vidal-Rioja L, Di Rocco F. TYR gene in llamas: polymorphisms and expression study in different color phenotypes. Front Genet. 2019;10:568. doi: 10.3389/fgene.2019.00568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun W, Shen Y, Shan S, Han L, Li Y, Zhou Z, Zhong Z, Chen J. Identification of TYR mutations in patients with oculocutaneous albinism. Mol Med Rep. 2018;17(6):8409–8413. doi: 10.3892/mmr.2018.8881. [DOI] [PubMed] [Google Scholar]
- 16.Abe Y, Tamiya G, Nakamura T, Hozumi Y, Suzuki T. Association of melanogenesis genes with skin color variation among Japanese females. J Dermatol Sci. 2013;69(2):167–172. doi: 10.1016/j.jdermsci.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 17.Han J, Kraft P, Nan H, Guo Q, Chen C, Qureshi A, Hankinson SE, Hu FB, Duffy DL, Zhao ZZ, Martin NG, Montgomery GW, Hayward NK, Thomas G, Hoover RN, Chanock S, Hunter DJ. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 2008;4(5):e1000074. doi: 10.1371/journal.pgen.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duffy DL, Zhao ZZ, Sturm RA, Hayward NK, Martin NG, Montgomery GW. Multiple pigmentation gene polymorphisms account for a substantial proportion of risk of cutaneous malignant melanoma. J Invest Dermatol. 2010;130(2):520–528. doi: 10.1038/jid.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Candille SI, Absher DM, Beleza S, Bauchet M, McEvoy B, Garrison NA, Li JZ, Myers RM, Barsh GS, Tang H, et al. Genome-wide association studies of quantitatively measured skin, hair, and eye pigmentation in four European populations. PLoS One. 2012;7(10):e48294. doi: 10.1371/journal.pone.0048294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu F, Visser M, Duffy DL, Hysi PG, Jacobs LC, Lao O, Zhong K, Walsh S, Chaitanya L, Wollstein A, Zhu G, Montgomery GW, Henders AK, Mangino M, Glass D, Bataille V, Sturm RA, Rivadeneira F, Hofman A, van IJcken WFJ, Uitterlinden AG, Palstra RJTS, Spector TD, Martin NG, Nijsten TEC, Kayser M. Genetics of skin color variation in Europeans: genome-wide association studies with functional follow-up. Hum Genet. 2015;134(8):823–835. doi: 10.1007/s00439-015-1559-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jonnalagadda M, Norton H, Ozarkar S, Kulkarni S, Ashma R. Association of genetic variants with skin pigmentation phenotype among populations of West Maharashtra, India. Am J Hum Biol. 2016;28(5):610–618. doi: 10.1002/ajhb.22836. [DOI] [PubMed] [Google Scholar]
- 22.Norton HL, Kittles RA, Parra E, McKeigue P, Mao X, Cheng K, Canfield VA, Bradley DG, McEvoy B, Shriver MD. Genetic evidence for the convergent evolution of light skin in Europeans and east Asians. Mol Biol Evol. 2007;24(3):710–722. doi: 10.1093/molbev/msl203. [DOI] [PubMed] [Google Scholar]
- 23.Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Magnusson KP, Manolescu A, Karason A, Palsson A, Thorleifsson G, Jakobsdottir M, Steinberg S, Pálsson S, Jonasson F, Sigurgeirsson B, Thorisdottir K, Ragnarsson R, Benediktsdottir KR, Aben KK, Kiemeney LA, Olafsson JH, Gulcher J, Kong A, Thorsteinsdottir U, Stefansson K. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet. 2007;39(12):1443–1452. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 24.Beleza S, Johnson NA, Candille SI, Absher DM, Coram MA, Lopes J, Campos J, Araujo II, Anderson TM, Vilhjalmsson BJ, et al. Genetic architecture of skin and eye color in an African-European admixed population. PLoS Genet. 2013;9(3):e1003372. doi: 10.1371/journal.pgen.1003372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kastelic V, Drobnic K. A single-nucleotide polymorphism (SNP) multiplex system: the association of five SNPs with human eye and hair color in the Slovenian population and comparison using a Bayesian network and logistic regression model. Croatian Med J. 2012;53(5):401–408. doi: 10.3325/cmj.2012.53.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersen JD, Johansen P, Harder S, Christoffersen SR, Delgado MC, Henriksen ST, Nielsen MM, Sorensen E, Ullum H, Hansen T, et al. Genetic analyses of the human eye colours using a novel objective method for eye colour classification. Forensic Sci Int Genet. 2013;7(5):508–515. doi: 10.1016/j.fsigen.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 27.Nan H, Kraft P, Qureshi AA, Guo Q, Chen C, Hankinson SE, Hu FB, Thomas G, Hoover RN, Chanock S, Hunter DJ, Han J. Genome-wide association study of tanning phenotype in a population of European ancestry. J Invest Dermatol. 2009;129(9):2250–2257. doi: 10.1038/jid.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bishop DT, Demenais F, Iles MM, Harland M, Taylor JC, Corda E, Randerson-Moor J, Aitken JF, Avril MF, Azizi E, Bakker B, Bianchi-Scarrà G, Bressac-de Paillerets B, Calista D, Cannon-Albright LA, Chin-A-Woeng T, Dębniak T, Galore-Haskel G, Ghiorzo P, Gut I, Hansson J, Hočevar M, Höiom V, Hopper JL, Ingvar C, Kanetsky PA, Kefford RF, Landi MT, Lang J, Lubiński J, Mackie R, Malvehy J, Mann GJ, Martin NG, Montgomery GW, van Nieuwpoort FA, Novakovic S, Olsson H, Puig S, Weiss M, van Workum W, Zelenika D, Brown KM, Goldstein AM, Gillanders EM, Boland A, Galan P, Elder DE, Gruis NA, Hayward NK, Lathrop GM, Barrett JH, Newton Bishop JA. Genome-wide association study identifies three loci associated with melanoma risk. Nat Genet. 2009;41(8):920–925. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutton SM, Spritz RA. A comprehensive genetic study of autosomal recessive ocular albinism in Caucasian patients. Invest Ophthalmol Vis Sci. 2008;49(3):868–872. doi: 10.1167/iovs.07-0791. [DOI] [PubMed] [Google Scholar]
- 30.Meyer OS, Lunn MMB, Garcia SL, Kjaerbye AB, Morling N, Borsting C, Andersen JD. Association between brown eye colour in rs12913832:GG individuals and SNPs in TYR, TYRP1, and SLC24A4. PLoS One. 2020;15(9):e0239131. doi: 10.1371/journal.pone.0239131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jagirdar K, Smit DJ, Ainger SA, Lee KJ, Brown DL, Chapman B, Zhen Zhao Z, Montgomery GW, Martin NG, Stow JL, Duffy DL, Sturm RA. Molecular analysis of common polymorphisms within the human Tyrosinase locus and genetic association with pigmentation traits. Pigment Cell Melanoma Res. 2014;27(4):552–564. doi: 10.1111/pcmr.12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stokowski RP, Pant PV, Dadd T, Fereday A, Hinds DA, Jarman C, Filsell W, Ginger RS, Green MR, van der Ouderaa FJ, et al. A genomewide association study of skin pigmentation in a south Asian population. Am J Hum Genet. 2007;81(6):1119–1132. doi: 10.1086/522235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaki M, Sengupta M, Mondal M, Bhattacharya A, Mallick S, Bhadra R. Indian genome variation C, ray K: molecular and functional studies of tyrosinase variants among Indian oculocutaneous albinism type 1 patients. J Invest Dermatol. 2011;131(1):260–262. doi: 10.1038/jid.2010.274. [DOI] [PubMed] [Google Scholar]
- 34.Sturm RA, Duffy DL. Human pigmentation genes under environmental selection. Genome Biol. 2012;13(9):248. doi: 10.1186/gb-2012-13-9-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji LD, Xu J, Wu DD, Xie SD, Tang NL, Zhang YP. Association of disease-predisposition polymorphisms of the melatonin receptors and sunshine duration in the global human populations. J Pineal Res. 2010;48(2):133–141. doi: 10.1111/j.1600-079X.2009.00736.x. [DOI] [PubMed] [Google Scholar]
- 36.Durso DF, Bydlowski SP, Hutz MH, Suarez-Kurtz G, Magalhaes TR, Pena SD. Association of genetic variants with self-assessed color categories in Brazilians. PLoS One. 2014;9(1):e83926. doi: 10.1371/journal.pone.0083926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacobs LC, Liu F, Pardo LM, Hofman A, Uitterlinden AG, Kayser M, Nijsten T. IRF4, MC1R and TYR genes are risk factors for actinic keratosis independent of skin color. Hum Mol Genet. 2015;24(11):3296–3303. doi: 10.1093/hmg/ddv076. [DOI] [PubMed] [Google Scholar]
- 38.Lona-Durazo F, Hernandez-Pacheco N, Fan S, Zhang T, Choi J, Kovacs MA, Loftus SK, Le P, Edwards M, Fortes-Lima CA, et al. Meta-analysis of GWA studies provides new insights on the genetic architecture of skin pigmentation in recently admixed populations. BMC Genet. 2019;20(1):59. doi: 10.1186/s12863-019-0765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rayner JE, McMeniman EK, Duffy DL, De'Ambrosis B, Smithers BM, Jagirdar K, Lee KJ, Soyer HP, Sturm RA. Phenotypic and genotypic analysis of amelanotic and hypomelanotic melanoma patients. J Eur Acad Dermatol Venereol. 2019;33(6):1076–1083. doi: 10.1111/jdv.15446. [DOI] [PubMed] [Google Scholar]
- 40.Reis LB, Bakos RM, Vianna FSL, Macedo GS, Jacovas VC, Ribeiro-Dos-Santos AM, Santos S, Bakos L, Ashton-Prolla P. Skin pigmentation polymorphisms associated with increased risk of melanoma in a case-control sample from southern Brazil. BMC Cancer. 2020;20(1):1069. doi: 10.1186/s12885-020-07485-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Table 1. The minor allele frequency of each SNP in different ethnic groups.
Additional file 2: Supplementary Table 2. The genotype frequency of each SNP in different ethnic groups.
Additional file 3: Supplementary Table 3. The HWE-P value of each SNP in different ethnic groups.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.