ABSTRACT
Antimicrobial resistance (AMR) is an important global health threat that impacts millions of people worldwide each year. Developing methods that can detect and predict AMR phenotypes can help to mitigate the spread of AMR by informing clinical decision making and appropriate mitigation strategies. Many bioinformatic methods have been developed for predicting AMR phenotypes from whole-genome sequences and AMR genes, but recent studies have indicated that predictions can be made from incomplete genome sequence data. In order to more systematically understand this, we built random forest-based machine learning classifiers for predicting susceptible and resistant phenotypes for Klebsiella pneumoniae (1,640 strains), Mycobacterium tuberculosis (2,497 strains), and Salmonella enterica (1,981 strains). We started by building models from alignments that were based on a reference chromosome for each species. We then subsampled each chromosomal alignment and built models for the resulting subalignments, finding that very small regions, representing approximately 0.1 to 0.2% of the chromosome, are predictive. In K. pneumoniae, M. tuberculosis, and S. enterica, the subalignments are able to predict multiple AMR phenotypes with at least 70% accuracy, even though most do not encode an AMR-related function. We used these models to identify regions of the chromosome with high and low predictive signals. Finally, subalignments that retain high accuracy across larger phylogenetic distances were examined in greater detail, revealing genes and intergenic regions with potential links to AMR, virulence, transport, and survival under stress conditions.
IMPORTANCE Antimicrobial resistance causes thousands of deaths annually worldwide. Understanding the regions of the genome that are involved in antimicrobial resistance is important for developing mitigation strategies and preventing transmission. Machine learning models are capable of predicting antimicrobial resistance phenotypes from bacterial genome sequence data by identifying resistance genes, mutations, and other correlated features. They are also capable of implicating regions of the genome that have not been previously characterized as being involved in resistance. In this study, we generated global chromosomal alignments for Klebsiella pneumoniae, Mycobacterium tuberculosis, and Salmonella enterica and systematically searched them for small conserved regions of the genome that enable the prediction of antimicrobial resistance phenotypes. In addition to known antimicrobial resistance genes, this analysis identified genes involved in virulence and transport functions, as well as many genes with no previous implication in antimicrobial resistance.
KEYWORDS: antimicrobial resistance, AMR, machine learning, ML, random forest
INTRODUCTION
Antimicrobial resistance (AMR) threatens global health by preventing effective treatments against bacteria, parasites, viruses, and eukaryotic pathogens. AMR usually arises as a natural consequence of genetic alterations; however, misuse or overuse of antimicrobials accelerates the selection of resistant variants. In 2019, the United States Centers for Disease Control and Prevention reported that antimicrobial-resistant infections occur annually in over 2.8 million people in the United States, with at least 35,000 deaths (1), and the European Centre for Disease Prevention and Control (ECDC) reported 33,000 deaths due to antimicrobial-resistant infections in the European Union and the European Economic Area each year (2). In addition to causing disease and mortality, AMR also causes major economic burdens to health care systems because of longer hospital stays, additional tests, and use of more expensive drugs (3).
In bacteria, AMR mechanisms can be grouped in three general categories, i.e., intrinsic, acquired, and adaptive. Intrinsic resistance involves all of the inherent features of a microorganism that prevent antibiotic effects, such as outer membranes with low permeability in some of the Gram-negative bacteria (4). Acquired resistance refers to the acquisition of mutations or genes on mobilizable elements such as plasmids, transposons, integrons, or to the transformation of naked DNA (4, 5). Adaptive resistance is triggered by environmental factors, such as antibiotic or nutrient stress, and can be revertible. Alterations in gene expressions and duplications of existing genes are known consequences of adaptive resistance (4, 5). These acquired or genetically encoded mutations and cell responses can cause resistance through enzymatic deactivation of antibiotics, reduction in the amount of antibiotic in the cells by efflux mechanisms, and modifications of the cell surface that make the cell less permeable (4).
To provide effective treatments and prevent rapid AMR spread, it is essential to know the resistance phenotypes of the pathogen (6). In the clinic, this is usually done using traditional antimicrobial susceptibility testing (AST), in which a bacterial culture is subjected to various antibiotics (7). As increasing numbers of genome sequences paired with AST phenotypes have become available, several studies have published machine learning (ML) models for predicting AMR phenotypes based on sequence data (8). Different ML algorithms have been used with good effect for making predictions; these include adaptive boosting (9), random forest (10–12), extreme gradient boosting (13, 14), set-covering machines (15), support vector machines (16, 17), and neural networks (10, 18). The choice of features also differs, and studies have been published where the authors used the AMR genes (10, 16, 19), k-mers based on the whole-genome sequence (9, 13, 14), whole-genome alignments (11), and alignments of the entire pangenome (17, 20). Two previous studies have also shown that the phylogeny of the isolates can provide a predictive signal (18, 21).
Although many studies have shown that ML methods can yield accurate models, the robustness of these models is highly dependent on the quality of the training set. For instance, training set size, phylogenetic diversity, and the various drug susceptibility testing methods can impact model accuracies (22–24). Ideally, ML models should be built from data sets that are balanced by class (e.g., resistant versus susceptible), and the number of samples should be greater than the number of features (24, 25). In practice, large and well-balanced training sets of genomes paired with antimicrobial susceptibility test data can be difficult to obtain.
In previous work, Nguyen et al. demonstrated that AMR phenotypes could be predicted using sets of conserved genes that are held in common among the members of a species, even if they are not known to be involved in AMR (26). The reasons relatively high accuracies are observed in the non-AMR genes are not entirely clear. Based on previous studies, it is likely that phylogeny plays a role in providing a predictive signal in these core gene models (18, 21). However, the authors also showed several cocorrelating virulence factor genes with high feature importance values, and other ML studies have been used to identify previously unrecognized epistatic mutations in the genome (17). It is also possible that some core genes could have a previously unrecognized function relating to AMR.
Knowing which conserved parts of the genome provide signal for AMR prediction models could potentially help to enhance our understanding of AMR mechanisms and the compensatory changes that result from the acquisition of resistance. In this study, we generate AMR prediction models from short chromosomal subalignments and use them to systematically identify regions of the genome with predictive power and potential links to AMR.
RESULTS
Chromosomal subalignments can be used to predict AMR phenotypes.
In this study, we wanted to gain a more systematic understanding of the conserved chromosomal regions that are useful for predicting AMR phenotypes. To do this, we selected three species, Klebsiella pneumoniae, Salmonella enterica, and Mycobacterium tuberculosis, which at the time of writing, all had rather large collections of AST data paired with whole-genome sequences (see Fig. S1 to S3 in the supplemental material).
Nucleotide k-mer distance-based tree of Klebsiella pneumoniae genomes used in this study. Surrounding rings around the distance tree are colored according to the antibiotic resistance phenotypes of strains; susceptible strains are colored green and resistant strains are colored red. The antibiotic phenotypes from inner to outer ring are as follows: amikacin, aztreonam, cefepime, cefoxitin, ceftazidime, ciprofloxacin, gentamicin, imipenem, levofloxacin, meropenem, piperacillin-tazobactam, tetracycline, tobramycin, and trimethoprim-sulfamethoxazole. Download FIG S1, PDF file, 1.8 MB (1.8MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The collection of genomes and the AST data were downloaded from the Pathosystems Resource Integration Center (PATRIC) (Table 1; see also Tables S1A to S1C in the supplemental material) (27), and the contigs of each genome were aligned against the chromosomal sequence of a high-quality reference genome. This resulted in a single global nucleotide alignment for the chromosomes of each species. The global chromosomal alignment was then used to build a matrix for machine learning. Susceptibility or resistance to each antibiotic was predicted by building a random forest classifier (28) for each species and antibiotic. For K. pneumoniae, the average area under the receiver operating characteristic curve (AUC) of the test set genomes in the 5-fold cross-validation is 0.878 ± 0.023 (Table 1; see also Fig. S4 in the supplemental material). The AUC values of each antibiotic range from 0.733 ± 0.044 for cefepime to 0.974 ± 0.006 for levofloxacin. For M. tuberculosis, the average AUC for all antibiotics is 0.778 ± 0.028, and the values for individual antibiotics range from 0.676 ± 0.026 for streptomycin to 0.833 ± 0.013 for rifampin. For S. enterica, the AUC for all antibiotics is 0.804 ± 0.029, with individual values ranging from 0.740 ± 0.027 for gentamicin to 0.841 ± 0.033 for sulfisoxazole. When the Salmonella models are recomputed using the chromosome of Salmonella enterica serovar Typhi CT18 as the reference, we also observe nearly identical results (see Table S1D in the supplemental material), indicating that the choice of the reference chromosomes has little impact. Overall, these AUCs and corresponding model statistics, including F1 scores and error rates (see Tables S1E to S1G in the supplemental material), are consistent with previous studies that have used k-mers and AMR genes as input (14, 15, 26).
TABLE 1.
Data set sizes and model performances reported as AUC for models built from the whole chromosomal alignment for each species and from randomly selected subalignments
| Species or antibiotic | No. of susceptible genomes | No. of resistant genomes | Chromosomal alignment AUCa | Subalignment AUCb |
|---|---|---|---|---|
| Klebsiella pneumoniae | ||||
| Amikacin | 1,296 | 100 | 0.897 ± 0.063 | 0.868 ± 0.080 |
| Aztreonam | 208 | 1,388 | 0.797 ± 0.026 | 0.675 ± 0.065 |
| Cefepime | 407 | 950 | 0.733 ± 0.044 | 0.653 ± 0.048 |
| Cefoxitin | 650 | 819 | 0.880 ± 0.043 | 0.749 ± 0.059 |
| Ceftazidime | 128 | 1,470 | 0.930 ± 0.027 | 0.761 ± 0.095 |
| Ciprofloxacin | 189 | 1,413 | 0.963 ± 0.012 | 0.865 ± 0.089 |
| Gentamicin | 909 | 676 | 0.897 ± 0.015 | 0.762 ± 0.065 |
| Imipenem | 1,138 | 473 | 0.915 ± 0.016 | 0.757 ± 0.066 |
| Levofloxacin | 332 | 1,277 | 0.974 ± 0.006 | 0.906 ± 0.091 |
| Meropenem | 1,112 | 478 | 0.915 ± 0.009 | 0.763 ± 0.072 |
| Piperacillin-tazobactam | 417 | 1,040 | 0.850 ± 0.014 | 0.746 ± 0.064 |
| Tetracycline | 725 | 766 | 0.805 ± 0.015 | 0.708 ± 0.049 |
| Tobramycin | 578 | 715 | 0.891 ± 0.015 | 0.821 ± 0.068 |
| Trimethoprim-sulfamethoxazole | 405 | 1,235 | 0.844 ± 0.023 | 0.641 ± 0.058 |
| Avg | 0.878 ± 0.023 | 0.763 ± 0.069 | ||
| Mycobacterium tuberculosis | ||||
| Ethambutol | 2,182 | 246 | 0.807 ± 0.030 | 0.712 ± 0.058 |
| Isoniazid | 1,868 | 570 | 0.760 ± 0.038 | 0.656 ± 0.045 |
| Pyrazinamide | 1,803 | 224 | 0.815 ± 0.033 | 0.723 ± 0.063 |
| Rifampin | 2,061 | 416 | 0.833 ± 0.013 | 0.698 ± 0.053 |
| Streptomycin | 364 | 132 | 0.676 ± 0.026 | 0.659 ± 0.077 |
| Avg | 0.778 ± 0.028 | 0.690 ± 0.059 | ||
| Salmonella enterica | ||||
| Amoxicillin-clavulanate | 1,473 | 398 | 0.792 ± 0.053 | 0.762 ± 0.046 |
| Ampicillin | 1,277 | 702 | 0.779 ± 0.018 | 0.735 ± 0.045 |
| Cefoxitin | 1,584 | 346 | 0.765 ± 0.036 | 0.751 ± 0.050 |
| Ceftiofur | 1,586 | 391 | 0.815 ± 0.015 | 0.762 ± 0.052 |
| Ceftriaxone | 1,585 | 395 | 0.813 ± 0.037 | 0.762 ± 0.055 |
| Chloramphenicol | 1,848 | 88 | 0.820 ± 0.039 | 0.737 ± 0.083 |
| Gentamicin | 1,622 | 328 | 0.740 ± 0.027 | 0.695 ± 0.050 |
| Streptomycin | 378 | 767 | 0.833 ± 0.014 | 0.759 ± 0.064 |
| Sulfisoxazole | 1,093 | 770 | 0.841 ± 0.033 | 0.793 ± 0.052 |
| Tetracycline | 859 | 1,114 | 0.839 ± 0.021 | 0.753 ± 0.051 |
| Avg | 0.804 ± 0.029 | 0.751 ± 0.055 | ||
Data are the results of one model per antibiotic, with the standard deviation of a 5-fold cross-validation.
Data are for 1,066 and 971 5-kb subalignment models for each antibiotic for K. pneumoniae and S. enterica, respectively, and for 441 10-kb subalignment models for M. tuberculosis. Data are the averages of all 5-fold cross-validations with standard deviations.
Nucleotide k-mer distance-based tree of Salmonella enterica genomes used in this study. Surrounding rings around the distance tree are colored according to the antibiotic resistance phenotypes of strains; susceptible strains are colored green and resistant strains are colored red. The antibiotic phenotypes from inner to outer ring are as follows: amoxicillin-clavulanate, ampicillin, cefoxitin, ceftiofur, ceftriaxone, chloramphenicol, gentamicin, streptomycin, sulfisoxazole, and tetracycline. Download FIG S3, PDF file, 1.7 MB (1.7MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Receiver operating characteristic (ROC) curves corresponding to antimicrobial resistance (AMR) prediction models for the corresponding samples in the test sets of the 5-fold cross validation. (Top) Areas under the receiver operating characteristic curve (AUCs) from whole-chromosome alignment-based models. (Bottom) AUCs from subalignment-based models. A subalignment size of 5 kb was used for K. pneumoniae and S. enterica, and 10 kb was used for M. tuberculosis. The ROC curves represent a single 5-fold cross-validated model for each chromosomal alignment-based model, and 1,066, 441, and 971 5-fold cross-validated models for randomly sampled subalignments for K. pneumoniae, M. tuberculosis, and S. enterica, respectively. Results are shown for each antibiotic, and the bright green line indicates the average test performance for all antibiotics. Download FIG S4, PDF file, 0.4 MB (405.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
(Tab A) Klebsiella pneumoniae genomes used in this study, with their laboratory-derived AMR phenotypes (R, resistant; S, susceptible). Blank cells correspond to intermediate or unknown phenotypes. (Tab B) Mycobacterium tuberculosis genomes used in this study, with their laboratory-derived AMR phenotypes (R, resistant; S, susceptible). Blank cells correspond to intermediate or unknown phenotypes. (Tab C) Salmonella enterica genomes used in this study, with their laboratory-derived AMR phenotypes (R, resistant; S, susceptible). Blank cells correspond to intermediate or unknown phenotypes. (Tab D) A comparison of areas under the receiver operating characteristic curve (AUCs) for S. enterica models built using the chromosome of two different reference strains, S. enterica subsp. enterica serovar Typhimurium LT2 (Pathosystems Resource Integration Center [PATRIC] genome identifier [ID] 99287.12) and S. enterica subsp. enterica serovar Typhi CT18 (PATRIC genome ID 220341.7). (Tab E) K. pneumoniae model performances for subalignments ranging in size from 1 kb to 1 Mb, and the model based on the whole chromosomal alignment. Subalignments were drawn randomly from the whole chromosomal alignment. (Tab F) M. tuberculosis model performances for subalignments ranging in size from 1 kb to 1 Mb, and the model based on the whole chromosomal alignment. Subalignments were drawn randomly from the whole chromosomal alignment. (Tab G) S. enterica model performances for subalignments ranging in size from 1 kb to 1 Mb, and the model based on the whole chromosomal alignment. Subalignments were drawn randomly from the whole chromosomal alignment. (Tab H) Some examples of highly performing subalignment models in M. tuberculosis, defined as an AUC of >0.85 for at least three of the tested antibiotics. (Tab I) Some examples of poorly performing subalignment models in K. pneumoniae and S. enterica, defined as an AUC of <0.6 for at least six of the tested antibiotics. (Tab J) K. pneumoniae subalignments with model AUCs greater than 0.80 following clustering at various k-mer similarity thresholds. The table displays all subalignments containing an AMR-, virulence-, or transport-related gene, or that have one within 5 kb upstream or downstream of the subalignment. The subalignments containing an important feature column that falls within an AMR, virulence, or transport gene are marked with an annotation source. PATRIC annotations are shown for the protein-encoding genes containing important feature columns in the subalignment and are separated by two # symbols. An asterisk (*) denotes a match 5 kb upstream or downstream of the subalignment. Two asterisks (**) denote an important subalignment column in an intergenic region. (Tab K) M. tuberculosis subalignments with model AUCs greater than 0.80 following clustering at various k-mer similarity thresholds. The table displays all subalignments containing an AMR-, virulence-, or transport-related gene, or that have one within 5 kb upstream or downstream of the subalignment. The subalignments containing an important feature column that falls within an AMR, virulence, or transport gene are marked with an annotation source. PATRIC annotations are shown for the protein-encoding genes containing important feature columns in the subalignment and are separated by two # symbols. An asterisk (*) denotes a match 5 kb upstream or downstream of the subalignment. Two asterisks (**) denote an important subalignment column in an intergenic region. (Tab L) S. enterica subalignments with model AUCs greater than 0.80 following clustering at various k-mer similarity thresholds. The table displays all subalignments containing an AMR-, virulence-, or transport-related gene, or that have one within 5 kb upstream or downstream of the subalignment. The subalignments containing an important feature column that falls within an AMR, virulence, or transport gene are marked with an annotation source. PATRIC annotations are shown for the protein-encoding genes containing important feature columns in the subalignment and are separated by two # symbols. An asterisk (*) denotes a match 5 kb upstream or downstream of the subalignment. Two asterisks (**) denote an important subalignment column in an intergenic region. (Tab M) K. pneumoniae subalignments with model AUCs greater than 0.80 following clustering at various k-mer similarity thresholds. The table displays subalignments lacking a match to AMR-, virulence-, or transport-related genes. PATRIC annotations are shown for the protein-encoding genes containing important feature columns in the subalignment and are separated by two # symbols. Two asterisks (**) denote an important subalignment column in an intergenic region. (Tab N) M. tuberculosis subalignments with model AUCs greater than 0.80 following clustering at various k-mer similarity thresholds. The table displays subalignments lacking a match to AMR-, virulence-, or transport-related genes. PATRIC annotations are shown for the protein-encoding genes containing important feature columns in the subalignment and are separated by two # symbols. Two asterisks (**) denote an important subalignment column in an intergenic region. (Tab O) S. enterica subalignments with model AUCs greater than 0.80 following clustering at various k-mer similarity thresholds. The table displays subalignments lacking a match to AMR-, virulence-, or transport-related genes. PATRIC annotations are shown for the protein-encoding genes containing important feature columns in the subalignment and are separated by two # symbols. Two asterisks (**) denote an important subalignment column in an intergenic region. (Tab P) Number of randomly selected subalignments used per species. The total length of all subalignments was limited to the length of the reference chromosome. Download Table S1, XLSX file, 0.9 MB (943.4KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
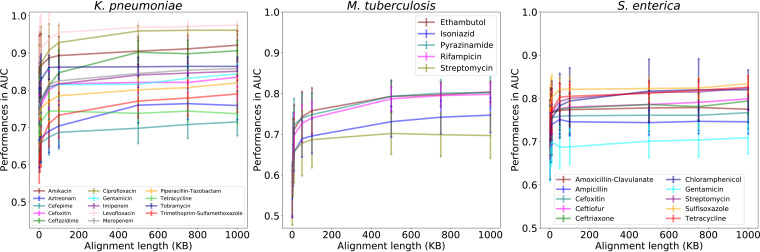
In order to assess how much sequence data are required to make an accurate AMR phenotype prediction, we randomly sampled smaller regions of each chromosomal alignment to generate subalignments of various sizes and built models for each subalignment using the same algorithm and parameters. For all three species, the AUC increases as the subalignment length increases (Fig. 1). This effect is most pronounced in M. tuberculosis, perhaps because of the high similarity between strains of this species (Fig. S2). The accuracies, F1 scores, and Matthews correlation coefficients (MCCs) follow the same trend as the AUCs, and error rates trend downward as alignment length increases (Tables S1E to S1G). These results indicate that small conserved chromosomal regions of only a few thousand bases in length contain a predictive signal that can be identified by the machine learning algorithm.
FIG 1.
The effect of subalignment length on model performance. The y axis depicts model performance for subalignment-based models as area under the receiver operating characteristic curve (AUC) values, and the x axis depicts subalignment nucleotide length (in kilobases). Error bars represent the standard deviation of multiple random samples for each length. The number of random samples for each subalignment is shown in Table S1P in the supplemental material. A separate set of 5-fold cross-validated models was computed for each subalignment and antibiotic.
Nucleotide k-mer distance-based tree of Mycobacterium tuberculosis genomes used in this study. Surrounding rings around the distance tree are colored according to the antibiotic resistance phenotypes of strains; susceptible strains are colored green and resistant strains are colored red. The antibiotic phenotypes from inner to outer ring are as follows: ethambutol, isoniazid, pyrazinamide, rifampicin, and streptomycin. Download FIG S2, PDF file, 1.1 MB (1.1MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Chromosomal regions yielding high- and low-accuracy models.
In order to understand which regions of the chromosome provide high- and low-accuracy AMR predictions, we plotted the AUCs of the subalignment-based models based on their alignment positions. To do this, we chose a subalignment size of 5 kb for K. pneumoniae and S. enterica, and an alignment size of 10 kb for M. tuberculosis. The longer subalignment size was chosen for M. tuberculosis since the increase in accuracy starts to become less dramatic at 10 kb (Fig. 1).
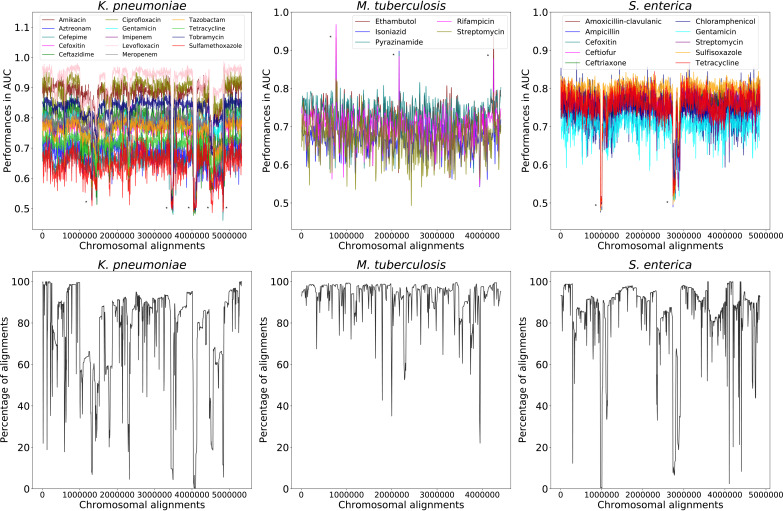
When the AUCs for each small subalignment-based model are plotted based on their chromosomal positions, we observe a consistent pattern of relatively high AUCs across the reference chromosome (Fig. 2), although the average AUCs are generally 5 to 12% lower than those predicted by the corresponding model based on the whole chromosomal alignment (Table 1 and Fig. S4). For each species, there are several peaks that are higher than the background. For example, three dramatic peaks at the approximate positions 770,000, 2,160,000, and 4,250,000 in M. tuberculosis correspond with subalignments containing the rpoB, katG, ermA, and ermB AMR genes (see Fig. S5 and Table S1H in the supplemental material). The rpoB, katG, ermA, and ermB genes encode proteins that can confer resistance to rifamycin, isoniazid, and macrolide antibiotics, respectively (29–31).
FIG 2.
Subalignment model accuracies by chromosomal location with alignment conservation. (Top) AUCs of every subalignment-based model are plotted based on their position in the whole chromosomal alignment. Peaks corresponding with antimicrobial resistance (AMR) genes and valleys corresponding with low alignment conservation are denoted with asterisks and are described in greater detail in Fig. S5 and Tables S1H and S1I in the supplemental material. (Bottom) The alignment conservation for each whole chromosomal alignment. The y axis depicts the percentage of sequences with a nucleotide in each column, and the x axis depicts chromosomal alignment position.
Subalignment model accuracies corresponding to Fig. 2 in the main text, with notable protein annotations corresponding to high- and low-performing subalignments overlaid. Download FIG S5, PDF file, 0.8 MB (798.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
On the other hand, we also observe several valleys where subalignment models fail to predict AMR phenotypes. For example, these include the approximate positions 1,310,000, 3,440,000, 4,035,000, and 4,535,000 for K. pneumoniae. These valleys often correspond with the locations of mobile elements (Fig. S5 and Table S1I in the supplemental material). Indeed, when we plot the alignment conservation for each column, many of the regions with poor alignment conservation corresponded with poor accuracies in the subalignment models (Fig. 2). Overall, these results indicate fairly stable AUCs over the reference chromosome, with higher-than-average predictive power for regions containing chromosomally encoded AMR genes and lower than average AUCs in regions of poor conservation.
Sequence similarity influences subalignment model performance.
The subalignment-based machine learning models can detect signal in most conserved regions of the chromosome, even though many of these small regions do not contain annotated AMR genes (Fig. 2). Since the machine learning models learn from nucleotide similarity that has been observed across samples in the training set, some of the high accuracies are likely due to similar sequences occurring in both the training and testing sets, which may obscure the more broadly conserved nucleotide signatures relating to, or correlating with, resistance. This has been observed previously, and several studies have tried to balance strains based on phylogeny to reduce this effect in the ML models (10, 11, 21, 26). In essence, having related genomes in the training and testing sets can improve accuracies, but may also potentially reduce the generalizability of the models if the overall phylogenetic distribution of the training set is biased.
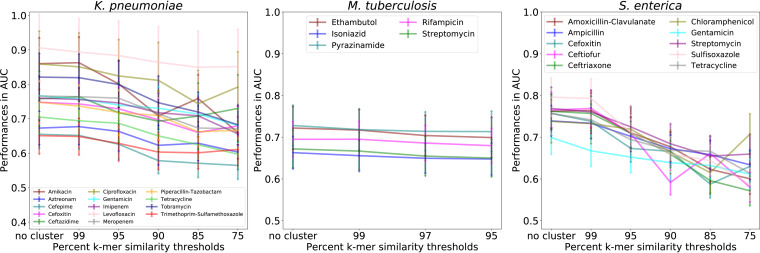
To observe how strain similarity affects the predictions, we clustered each strain using the nucleotide k-mer similarity from the whole chromosomal alignment and evaluated the subalignment-based model performances by changing the clustering threshold. All strains were used in the models; however, strains that were members of the same cluster were restricted to either the testing or the training set in each fold of the 5-fold cross-validation. The similarity thresholds that were used differed depending on the diversity of the sequences for each species. For K. pneumoniae and S. enterica, clustering was evaluated from 99% to 75% k-mer similarity, and from 99% to 95% for M. tuberculosis (see Fig. S6 in the supplemental material).
Number of k-mer-based genome clusters versus clustering threshold. The genomes of each species were clustered based on k-mer similarity, using their aligned sequences in the whole chromosomal alignment. As the clustering threshold decreases, clusters become larger and more inclusive, and there are fewer total clusters. Download FIG S6, PDF file, 0.04 MB (42KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
As expected, when the clustering thresholds decrease in k-mer similarity, the model performances also begin to decrease (Fig. 3). This happens because as the clusters become larger and more inclusive, the genetic distance between strains in the training and testing sets are increasing. Going from no clustering to clustering with the lowest similarity threshold, the average AUCs drop approximately 10% for K. pneumoniae and S. enterica and approximately 2% for M. tuberculosis. This trend is also observed when the analysis is repeated using a k-mer similarity that is based on the individual sequences of each subalignment, rather than the entire chromosomal alignment (data not shown). These results indicate that the random forest models can learn the underlying phylogeny and use this information to aid the phenotype prediction.
FIG 3.
Results of clustering similar genomes on subalignment model performance. All samples were clustered based on their k-mer similarity in the whole chromosomal alignment at various k-mer identity thresholds. Samples belonging to the same cluster were restricted to either the test or the training sets of each 5-fold cross-validation. Data are the average accuracies with standard deviations of 5-fold cross-validations for 971 Klebsiella pneumoniae (5 kb), 1,066 Salmonella enterica (5 kb), and 441 Mycobacterium tuberculosis (10 kb) subalignments, respectively.
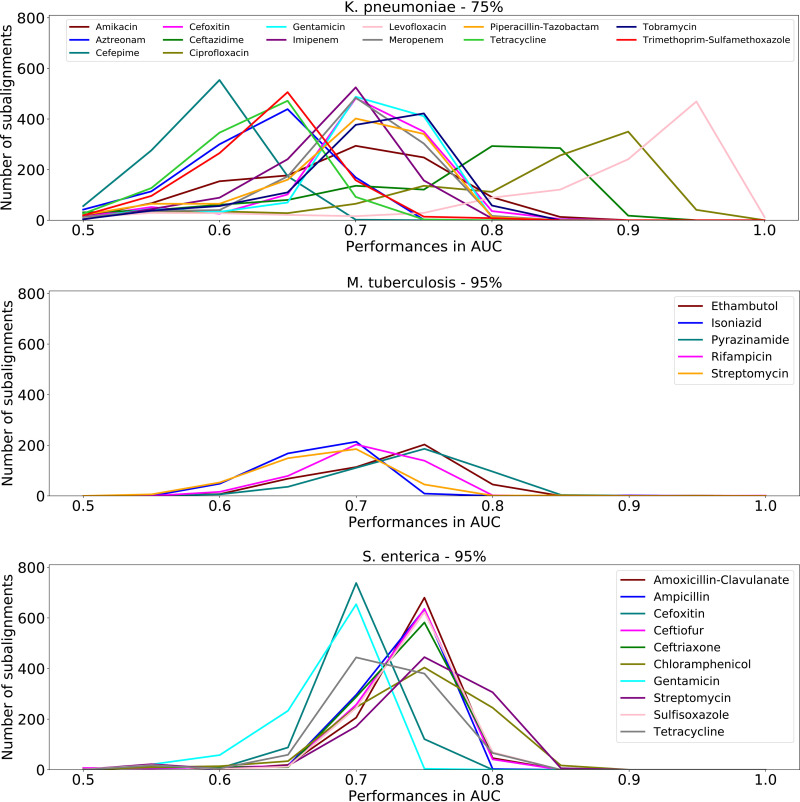
As the clustering becomes more inclusive, the decrease in performance is not uniform across species and antibiotics (see Fig. S7 in the supplemental material). For instance, in K. pneumoniae, ciprofloxacin and levofloxacin have the largest number of subalignments with AUCs of >0.80 at 75% clustering, for M. tuberculosis pyrazinamide retains the largest number of informative subalignments at 95% clustering, and for S. enterica, chloramphenicol and streptomycin have the largest number of informative subalignments at 95% clustering (Fig. 4). The higher accuracies for certain antibiotics, such as ciprofloxacin and levofloxacin in K. pneumoniae, may be due to chromosomally encoded AMR genes, epigenetic effects, or the time at which the AMR gene or mutation became fixed in the population. For instance, previous work in Neisseria has demonstrated larger epistatic effects relating to ciprofloxacin relative to those related to other antibiotics (32).
FIG 4.
The number of subalignments and corresponding to a given model AUC for each antibiotic. Genomes were clustered based on similarity, and samples belonging to the same cluster were restricted to either the testing or the training sets in each fold of the 5-fold cross-validation. A clustering threshold of 75% k-mer similarity is shown for K. pneumoniae, and a clustering threshold of 95% is shown M. tuberculosis and S. enterica. Results for additional thresholds are shown in Fig. S7 in the supplemental material.
Number of subalignments and corresponding model AUCs at various k-mer similarity thresholds. Each panel shows a distribution of AMR prediction performances in AUC for the clustered strains at a given similarity threshold. All samples were clustered based on their k-mer similarity in the whole chromosomal alignment. Samples belonging to the same cluster were restricted to either the testing or training sets of each fold of the 5-fold cross-validation. Download FIG S7, PDF file, 0.3 MB (286.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Some subalignments contain broadly conserved AMR signals.
When similar strains are prevented from occurring in both the training and testing sets, we observe an expected decrease in subalignment model performances. However, we also observe large standard deviations in the average AUCs for the subalignment models, even at the lower similarity thresholds (Fig. 3). This implies that some of the subalignments that retain high AUCs contain sequence signatures that are conserved and are being learned even when the strains are less closely related. We reasoned that these subalignments are likely to contain sequence signatures that are more phylogenetically widespread within the species and could be regions that cooccur with AMR or that are involved with AMR-related functions. We chose to examine these highly performing subalignments in greater detail.
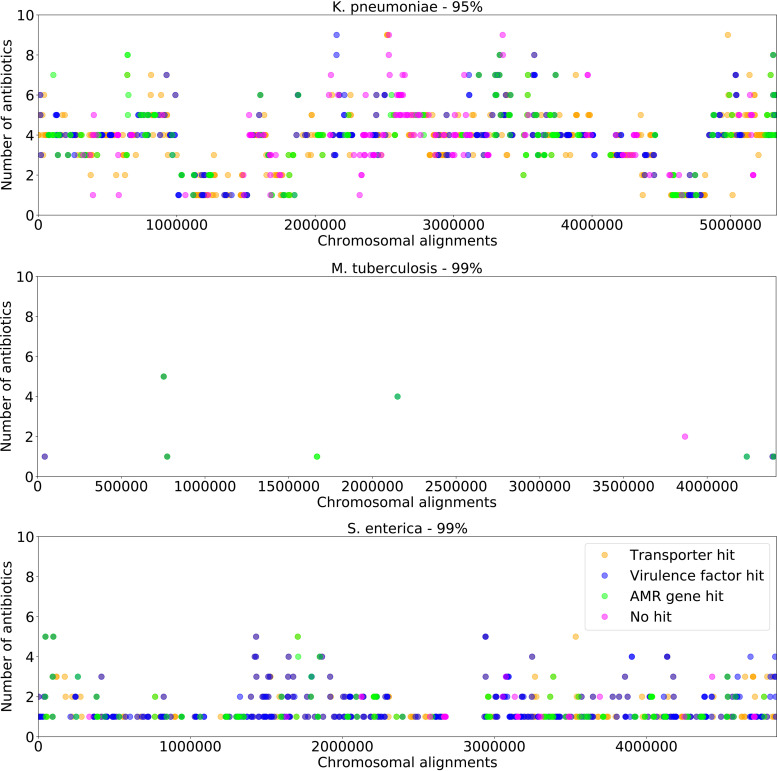
To attempt to distinguish the subalignments containing genes encoding proteins with well-characterized functions from those that are poorly characterized, we plotted the subalignments with AUCs of >0.80 at each clustering threshold based on their chromosomal alignment positions (see Fig. S8 in the supplemental material). We searched within each subalignment for genes that are known to encode functions that are involved in AMR, virulence, or membrane transport. This was done by comparing the genes to the PATRIC (33), Comprehensive Antibiotic Resistance Database (CARD) (34), Virulence Factor Database (VFDB) (35), National Database of Antibiotic Resistant Organisms (NDARO) (36), and Transporter Classification Database (TCDB) (37) resources. We chose to look for virulence factors and transporter genes because these functions often correlate or cooccur with AMR (38–40). The subalignments were then colored in the plot based on whether they contain one of these genes (Fig. 5 and Fig. S8). Overall, there is an enrichment in AMR, virulence factor, and transporter genes in this set of subalignments (see Tables S1J to S1L in the supplemental material). There are also many subalignments that do not have an annotated AMR, virulence, or transporter gene (see Tables S1M to S1O in the supplemental material). M. tuberculosis has the fewest highly predictive subalignments, and most of these have AMR or virulence factor matches. In all three species, there are cases where subalignments are predictive for several antibiotics, possibly indicating a protein function relating to a class of antibiotics. The informative subalignments do not tend to cluster based on their coordinates in the reference chromosome and instead appear to be spread out over the reference chromosome.
FIG 5.
Protein-encoding gene functions for subalignments with high AMR prediction accuracies. Each panel includes predictive subalignments with AUCs of >0.8 after clustering the strains at a given k-mer similarity threshold. Points represent each subalignment, and they are plotted based on the corresponding position on the reference chromosome. Subalignments are colored according to hits for AMR (green), virulence (blue), and transporter genes (orange), respectively. If a subalignment contains multiple gene categories, the color will appear as a mixture. Subalignments that do not produce hits in any of the known AMR-related genes are colored in pink. Results for high-scoring subalignments at other clustering thresholds are shown in Fig. S8 in the supplemental material.
Reference chromosome position corresponding to subalignments with high AMR prediction accuracies. Each panel includes predictive subalignments with AUCs of >0.8 after clustering the strains at a given k-mer similarity threshold. Subalignments were colored according to hits to AMR (green), virulence (blue), and transporter genes (orange), respectively. If a subalignment contains multiple gene categories, the color will appear as a mixture. Subalignments that do not hit any of the known AMR-related genes are colored in pink. Download FIG S8, PDF file, 1.5 MB (1.5MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Many of the subalignments that retain high AUCs in spite of the clustering do not contain annotated AMR, virulence factor, or transporter genes (Tables S1M to S1O). These regions could have a previously unrecognized role in AMR or may be important cocorrelates with AMR in the evolution of resistant strains. For example, in both K. pneumoniae and S. enterica, we observed high-AUC subalignment models that contained the gene encoding the chaperone protein HscA. HscA is a member of the Hsp70 family and is known for its role in maturation of iron-sulfur-cluster-containing proteins (41, 42). Although hscA is not known as an AMR gene, a study tracking the acquisition of resistance mutations in Salmonella has documented single-nucleotide polymorphisms (SNPs) in hscA (43), so a role in AMR could be plausible. In comparison, DnaK, which is another Hsp70, has a role in survival under unfavorable conditions such as exposure to oxidative stress, heavy metals, and antibiotics (44). Additionally, we detected many AMR-related mutations in metabolic genes, including in genes encoding sulfur acceptor protein, cysteine desulfurase, and 3-mercaptopyruvate sulfur transferase. Cysteine desulfurase is known for its role in protection from oxidative stress, which might also be important for protecting the bacterium from antimicrobial stress (45). Recently, Collins and colleagues detected metabolic mutations that confer AMR for streptomycin, ciprofloxacin, and carbenicillin in pathogenic bacteria using in vitro analyses. Overall, our analysis identifies 10 of the 17 metabolic genes originally found by Collins et al., including cycA, gltB, mltA, rsxC, sucA, dppA, yqiK, bcsC, mdfA, and rpoB (46). Taken together, these detected genes point to a relationship between stress conditions and AMR and provide potential targets for further phenotypic characterization at the bench.
DISCUSSION
In this study, we built models for predicting AMR phenotypes for K. pneumoniae, M. tuberculosis, and S. enterica using short chromosomal subalignments that were 5 to 10 kb in length (approximately 0.1 to 0.2% of the length of the reference chromosomes). Overall, these models have average AUCs that are 5 to 12% lower than the AUCs for models that are based on the whole chromosomal alignments. As the subalignment length increases, the performance of the models also tends to increase. These results are consistent with previous work by Nguyen et al. that showed that AMR phenotypes can be predicted using small sets of conserved genes (26). Using the short subalignments, we plotted the model performances, based on their coordinates on the reference chromosome, with a fairly high degree of resolution. In all three organisms, the data show relatively consistent AUCs across the chromosome, with specific regions yielding high- and low-performing models. For instance, in K. pneumoniae and S. enterica, we observed that the poorly performing subalignments included insertions, such as phage and mobile elements. These also tended to correspond with areas of low alignment conservation. In M. tuberculosis, most of the regions with dramatically high prediction performances contain known AMR genes, such as those encoding DNA-directed RNA polymerase beta subunit (rpoB), catalase-peroxidase (katG), and integral membrane indolylacetylinositol arabinosyltransferase (embA, embB, and embC), which confer resistance to rifampin, isoniazid, and macrolides, respectively (47–49). Although it is unsurprising that the subalignments containing the AMR genes have high model performances, we were surprised by the large number of subalignments with residually high AUCs, many of which contained no annotated functions relating to AMR.
In order to understand how genome similarity influences the accuracy of these models, we clustered the strains based on the k-mer similarity using the whole chromosomal alignments. We then prevented similar sequences from existing in both the training and testing sets for each fold of the 5-fold cross-validation. The AUCs dropped gradually as the clustering became more inclusive, indicating that the models rely on sequence similarity. This is consistent with previous studies that have performed similar experiments to control for the effects of related strains in ML models (10, 11, 21, 26). However, we also found that there were many subalignments that retained high AUCs in spite of the clustering, which indicated the presence of sequence signatures that were more strongly conserved relative to their background similarity. These regions were enriched in AMR, virulence, and transport-related functions. Although the cooccurrence between AMR, virulence factors, and mutations in transporters is well known (38–40), we also observed several regions that did not have an obvious role in AMR, virulence, or transport, which merit further analysis.
The ability to predict AMR phenotypes over the genome clearly differs by antibiotic, with the subalignment models for some antibiotics having better overall performances than those of others. This can be seen in the color banding pattern of the plots in Fig. 2. For example, in K. pneumoniae, the AUCs for the 5-kb amikacin models were only 3% lower than those for the whole chromosomal alignment-based model, whereas for trimethoprim-sulfamethoxazole, there was a 20% drop. This difference can also be seen in the subalignments that remained predictive after k-mer similarity was used to prevent similar sequences from occurring in the training and testing sets. For example, for K. pneumoniae, a large number of the subalignments that were accurate after this filtering step had high AUCs for predicting ciprofloxacin and levofloxacin phenotypes, followed by those for amikacin and tobramycin. On the other hand, only two predictive subalignments were obtained for trimethoprim-sulfamethoxazole at 99% k-mer similarity. Previous studies have shown that certain antibiotics may have a larger epistatic impact on the genome than others (50), which might result in the signal that is being detected by the models. However, we note that filtering the subalignments based on sequence similarity may not have entirely eliminated other potentially nonrandom effects, such as biased strain sampling or linkage disequilibrium, which could also cause this difference (51, 52).
Using short chromosomal subalignments to predict AMR phenotypes presented a few drawbacks that are worth noting. First, we used a clustering approach that was based on the k-mer similarity of the whole chromosomal alignment rather than on the k-mer similarities of each individual subalignment. When we clustered based on subalignment similarity, we obtained similar results, but balancing the sets became more difficult due to a lack of diversity in many subalignments. Another drawback of using an alignment-based modeling approach is that it has the potential to result in very large matrix files as the diversity of the training sequences increases. This could eventually limit the number of strains or species that could be used in an alignment-based model, although we did not encounter this problem in this study. In comparison, the size of a k-mer-based matrix is usually scoped to the number of possible k-mers of a given length. Overall, the benefit of using alignment-based models is that they clearly preserve feature importance down to the nucleotide position in the alignment.
The results of this study suggest that a complete genome sequence may not be necessary for predicting AMR phenotypes. However, the antibiotic, subalignment size, and the region of the genome sequence used in the model have clear impacts on the accuracies that can be obtained. This may eventually inform the development of bioinformatic workflows that can make predictions using incomplete genome sequence data. For instance, using read mapping against a known region of a reference genome sequence could be a potentially fast and appealing way to predict AMR in the incomplete genomes that are often found in metagenomic samples, although there would be potential drawbacks, including the reduced accuracy of partial genome models and the need to ensure that similar strains can be adequately differentiated. Nevertheless, the approach outlined in this study provides a potential way forward, and AMR may be only one of many phenotypes that might be predicted in this way.
In conclusion, this study offers a unique approach for identifying AMR-predictive regions in bacterial chromosomes and may eventually provide a means for understanding and predicting the chromosomal changes that accompany the evolution of resistance.
MATERIALS AND METHODS
Data sets.
Three bacterial species were selected for this study, K. pneumoniae, M. tuberculosis, and S. enterica (nontyphoidal serovars). Genomes for 1,664 K. pneumoniae, 16,906 M. tuberculosis, and 5,268 S. enterica isolates were downloaded from the Pathosystems Resource Integration Center (PATRIC) in October 2019 (27, 33). Metadata information, including susceptibility and resistance determinations (48) and MICs, was downloaded from the PATRIC FTP site (ftp://ftp.patricbrc.org/RELEASE_NOTES/PATRIC_genomes_AMR.txt). MIC data were converted into sensitive (S) and resistant (R) phenotypes for K. pneumoniae, using Clinical and Laboratory Standards Institute (53) guidelines (53). For M. tuberculosis, interpretations were based on World Health Organization (WHO)-defined critical concentrations from the original studies (54, 55). For S. enterica, CLSI and United States Food and Drug Administration (56) guidelines were used to interpret MIC values (56).
The large number of M. tuberculosis and S. enterica isolates were downsampled to ∼2,500 and ∼2,000 diverse genomes, respectively, to keep the number of genomes per antibiotic relatively consistent and to reduce computational overhead. Downsampling was performed using whole-genome k-mer distances using overlapping 8-mer oligonucleotides, as in Nguyen et al. (14). KMC (version 2.3.0) (57) was used to compute k-mers, and clustering was performed using the cluster.AgglomerativeClustering function from the Python Scikit-learn library (version 0.19.2) (58) by setting affinity to “l1” and linkage to “complete.” The list of isolates and AMR phenotypes is provided in Tables S1A to S1C in the supplemental material. Due to low counts, the intermediate phenotypes were not modeled in this study. Poor-quality genomes were removed from the analysis based on low chromosomal alignment coverage (<50%), defined as having extreme genome lengths (two times longer than the reference genome or shorter than the half of the reference genome) or poor genome quality scores based on the PATRIC genome quality tool (59).
Alignment generation.
The assembled genomes for each isolate were globally aligned to the chromosome of a relevant high-quality reference genome. In this case, we chose Klebsiella pneumoniae subsp. pneumoniae HS11286 (PATRIC identified [ID] 1125630.4; GenBank accession number CP003200.1), Mycobacterium tuberculosis H37Rv (PATRIC ID 83332.12; GenBank accession no. AL123456.3), Salmonella enterica subsp. enterica serovar Typhimurium strain LT2 (PATRIC ID 99287.12; GenBank accession number AE006468.2). Reference genomes were based on the NCBI reference genome collection (60). Global alignments were generated using KMA (version 1.2.0) (61) with the following parameters: “-dense -ref_fsa -ca -mem_mode -mrs 0 -Mt1 1 -e 1.0.” The KMA tool produces output files that include the mapping information, alignment statistics, and the consensus alignment against the template. The “dense” option prevents the insertion of gaps into the consensus sequence. This preserves contiguity of the reference chromosome and prevents the subsequent matrix files from becoming sparse.
Subalignments were generated by randomly sampling continuous regions of the global chromosomal alignment by choosing an arbitrary starting point. When experiments used many subalignments, the total length of the sampled subalignments was not allowed to exceed the length of the reference chromosome, to help prevent oversampling of the same regions. In practice, this means that a larger number of short subalignments than long subalignments could be generated (see Table S1P in the supplemental material).
Model generation and cross-validation.
For each species, a matrix was generated from either the global chromosomal alignment or the subaligned regions where the rows represent each isolate, the columns represent the positions of the alignment, and the entries are DNA nucleotides for each position. To decrease the matrix size, alignment columns having no variation were removed from consideration, since they provide no discriminative information. We used one-hot encoding to convert nucleotide sequence data into combinations of one(s) and zero(s) as described by Aytan-Aktug et al. (10). The utils.to_categorical function in the Python Keras library (version 2.2.4) (62) was used to perform the one-hot encoding.
Unless otherwise stated, the models generated in this study are binary classifiers that predict susceptible or resistant phenotypes for a single antibiotic. If genomes had intermediate or unknown phenotypes for a given antibiotic, they were excluded from the training, testing, and validation sets for the corresponding model. Random forest (63) was chosen for the machine learning algorithm in this study because it is a robust tree-based method that is relatively easy to interpret and has been used with good effect in in many studies for regression and classification purposes (10–12, 28). The Python Scikit-learn package (version 0.19.2) ensemble.RandomForestClassifier was used for generating the models (58). Unless otherwise stated, we chose to use the random forest parameters defined previously (10), which set the number of trees to 200, and used default settings for the remaining parameters. Additionally, the class weight was set to “balanced” in this study. This adjusted the class weights according to the class frequencies in the input data and was intended to help prevent biased predictions caused by class imbalances. The chosen parameters are intended to be optimal for the majority of the models, although it may be possible to find a more ideal set of parameters for any given subalignment.
Standard 5-fold cross-validations were performed for each model using a training, a testing, and a held-out validation set to monitor models for overfitting. Unless otherwise stated, the average test performances are reported as area under the receiver operating characteristic curve (AUC) values with a standard deviation. In experiments where a subalignment was sampled multiple times, we report the average of all tests with a standard deviation. Model performances were assessed using the AUC, macro F1 scores, and Matthews correlation coefficient (MCC) using the Scikit-learn package (version 0.19.2) (58). Major error (ME) and very major error (VME) rates were also used as metrics. Major errors are defined as susceptible isolates that are classified as being resistant, and very major errors are resistant isolates that are classified as being susceptible.
Clustering subalignments.
Machine learning models can learn AMR phenotypes based on the genome similarity or correlations between training, test, and validation subsets (21, 26). To explore how genome similarity effects the machine learning predictions, we clustered the aligned sequences using the KMA index (version 1.3.7) (61). KMA uses the 16-mer oligonucleotide k-mers and the Hobohm-1 algorithm to generate clusters (63). Whole alignments that are similar to each other within a certain similarity threshold (between 75 and 99% k-mer similarity) were clustered, and the corresponding subalignments were kept in the same partition of the 5-fold cross-validation. Thus, the isolates sharing similarity greater than the given threshold were only used in either the training or testing sets. To help prevent biased predictions due to imbalances between cluster sizes, each subalignment was weighted in inverse proportion to the cluster size. No hold-out set was used in this analysis in order to maximize the number of clusters that could be tested.
The genes encoded within subalignments that had high model performances were evaluated for a potential role in AMR. These subalignments were aligned to the PATRIC (33), CARD (34), and NDARO (36) databases to identify AMR genes, VFDB (35) and Victors (64) to identify virulence factors, and TCDB (37) to identify transporter genes. Furthermore, to explore the precise AMR-related subalignment positions, we calculated feature contributions to the AMR predictions using random forest’s feature importance implementation. The most informative 10 features were considered per fold.
Tree generation.
Distance matrices were computed for the whole chromosomal alignments using KMA (version 1.3.7) using the k-mer distance option. KMA calculates input distances using 16-mers and accepts inputs in single indexed file. Input sequences were indexed using “-NI,” “-Sparse TG,” and “-nbp” parameters. Distance trees were constructed using CCPhylo (version 0.0.15) (65), which generates a neighbor-joining tree, and visualized using iTOL (version 4) (66).
Data availability.
The Python 2.7.15 scripts used for this project are available on Bitbucket (https://bitbucket.org/deaytan/aligned-fragments/). All genomes and metadata can be accessed through the PATRIC resource (https://www.patricbrc.org) using the genome identifiers given in Tables S1A to S1C.
ACKNOWLEDGMENTS
We thank our collaborators at the FDA NARMS program and at Houston Methodist for the generation of the Salmonella and Klebsiella data sets. We also thank Emily Dietrich for her careful editing.
This work was funded by the United States Defense Advanced Research Projects Agency iSENTRY Friend or Foe program award (contract no. HR0011150042) to J.J.D., by the United States National Institute of Allergy and Infectious Diseases Bacterial and Viral Bioinformatics resource center award (contract no. 75n93019c00076) to R.L.S., and by the Novo Nordisk Foundation (grant NNF16OC0021856, Global Surveillance of Antimicrobial Resistance, awarded to F.M.A. and O.L.).
The funding agencies did not play any role in the design of the study or the writing of the manuscript, nor did they have any influence on the data collection, analysis or interpretation of the data, or the results.
We declare no conflicts of interest.
Contributor Information
J. J. Davis, Email: jjdavis@anl.gov.
Tamia A. Harris-Tryon, UT Southwestern Medical Center at Dallas
REFERENCES
- 1.CDC. 2019. Antibiotic resistance threats in the United States. CDC, U.S. Department of Health and Human Services, Atlanta, GA. [Google Scholar]
- 2.European Centre for Disease Prevention and Control. 2020. Antimicrobial resistance in the EU/EEA (EARS-Net)—annual epidemiological report 2019. Stockholm, Sweden. [Google Scholar]
- 3.Llor C, Bjerrum L. 2014. Antimicrobial resistance: risk associated with antibiotic overuse and initiatives to reduce the problem. Ther Adv Drug Saf 5:229–241. doi: 10.1177/2042098614554919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fernández L, Hancock REW. 2012. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev 25:661–681. doi: 10.1128/CMR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boolchandani M, D’Souza AW, Dantas G. 2019. Sequencing-based methods and resources to study antimicrobial resistance. Nat Rev Genet 20:356–370. doi: 10.1038/s41576-019-0108-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katiyar A, Sharma P, Dahiya S, Singh H, Kapil A, Kaur P. 2020. Genomic profiling of antimicrobial resistance genes in clinical isolates of Salmonella Typhi from patients infected with typhoid fever in India. Sci Rep 10:8299. doi: 10.1038/s41598-020-64934-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim SJ. 2005. Drug-susceptibility testing in tuberculosis: methods and reliability of results. Eur Respir J 25:564–569. doi: 10.1183/09031936.05.00111304. [DOI] [PubMed] [Google Scholar]
- 8.McDermott PF, Davis JJ. 2020. Predicting antimicrobial susceptibility from the bacterial genome: a new paradigm for one health resistance monitoring. J Vet Pharmacol Ther 44:223–237. doi: 10.1111/jvp.12913. [DOI] [PubMed] [Google Scholar]
- 9.Davis JJ, Boisvert S, Brettin T, Kenyon RW, Mao C, Olson R, Overbeek R, Santerre J, Shukla M, Wattam AR, Will R, Xia F, Stevens R. 2016. Antimicrobial resistance prediction in PATRIC and RAST. Sci Rep 6:27930. doi: 10.1038/srep27930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aytan-Aktug D, Clausen PTLC, Bortolaia V, Aarestrup FM, Lund O. 2020. Prediction of acquired antimicrobial resistance for multiple bacterial species using neural networks mSystems 5:e00774-19. doi: 10.1128/mSystems.00774-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pataki BÁ, Matamoros S, van der Putten BCL, Remondini D, Giampieri E, Aytan-Aktug D, Hendriksen RS, Lund O, Csabai I, Schultsz C, Matamoros S, Janes V, Hendriksen RS, Lund O, Clausen P, Aarestrup FM, Koopmans M, Pataki B, Visontai D, Stéger J, Szalai-Gindl JM, Csabai I, Pakseresht N, Rossello M, Silvester N, Amid C, Cochrane G, Schultsz C, Pradel F, Westeel E, Fuchs S, Kumar SM, Xavier BB, Ngoc MN, Remondini D, Giampieri E, Pasquali F, Petrovska L, Ajayi D, Nielsen EM, Trung NV, Hoa NT, Ishii Y, Aoki K, McDermott P, SPS COMPARE ML-AMR group. 2020. Understanding and predicting ciprofloxacin minimum inhibitory concentration in Escherichia coli with machine learning. Sci Rep 10:15026. doi: 10.1038/s41598-020-71693-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santerre JW, Davis JJ, Xia F, Stevens R. 2016. Machine learning for antimicrobial resistance. arXiv 1607.01224 [stat.ML]. https://arxiv.org/abs/1607.01224.
- 13.Nguyen M, Brettin T, Long SW, Musser JM, Olsen RJ, Olson R, Shukla M, Stevens RL, Xia F, Yoo H, Davis JJ. 2018. Developing an in silico minimum inhibitory concentration panel test for Klebsiella pneumoniae. Sci Rep 8:421–421. doi: 10.1038/s41598-017-18972-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen M, Long SW, McDermott PF, Olsen RJ, Olson R, Stevens RL, Tyson GH, Zhao S, Davis JJ. 2018. Using machine learning to predict antimicrobial MICs and associated genomic features for nontyphoidal Salmonella. J Clin Microbiol 57:e01260-18. doi: 10.1128/JCM.01260-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drouin A, Letarte G, Raymond F, Marchand M, Corbeil J, Laviolette F. 2019. Interpretable genotype-to-phenotype classifiers with performance guarantees. Sci Rep 9:4071. doi: 10.1038/s41598-019-40561-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyun JC, Kavvas ES, Monk JM, Palsson BO. 2020. Machine learning with random subspace ensembles identifies antimicrobial resistance determinants from pan-genomes of three pathogens. PLoS Comput Biol 16:e1007608. doi: 10.1371/journal.pcbi.1007608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kavvas ES, Catoiu E, Mih N, Yurkovich JT, Seif Y, Dillon N, Heckmann D, Anand A, Yang L, Nizet V, Monk JM, Palsson BO. 2018. Machine learning and structural analysis of Mycobacterium tuberculosis pan-genome identifies genetic signatures of antibiotic resistance. Nat Commun 9:4306. doi: 10.1038/s41467-018-06634-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moradigaravand D, Palm M, Farewell A, Mustonen V, Warringer J, Parts L. 2018. Prediction of antibiotic resistance in Escherichia coli from large-scale pan-genome data. PLoS Comput Biol 14:e1006258. doi: 10.1371/journal.pcbi.1006258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J, Greenberg DE, Pifer R, Jiang S, Xiao G, Shelburne SA, Koh A, Xie Y, Zhan X. 2020. VAMPr: VAriant Mapping and Prediction of antibiotic resistance via explainable features and machine learning. PLoS Comput Biol 16:e1007511. doi: 10.1371/journal.pcbi.1007511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Her H-L, Wu Y-W. 2018. A pan-genome-based machine learning approach for predicting antimicrobial resistance activities of the Escherichia coli strains. Bioinformatics 34:i89–i95. doi: 10.1093/bioinformatics/bty276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Břinda K, Callendrello A, Ma KC, MacFadden DR, Charalampous T, Lee RS, Cowley L, Wadsworth CB, Grad YH, Kucherov G, O’Grady J, Baym M, Hanage WP. 2020. Rapid inference of antibiotic resistance and susceptibility by genomic neighbour typing. Nat Microbiol 5:455–464. doi: 10.1038/s41564-019-0656-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sordo M, Zeng Q. 2005. On sample size and classification accuracy: a performance comparison, p 193–201. In Oliveira JL, Maojo V, Martín-Sánchez F, Pereira AS (ed), Biological and medical data analysis. Springer, Berlin, Germany. [Google Scholar]
- 23.Horne DJ, Pinto LM, Arentz M, Lin SYG, Desmond E, Flores LL, Steingart KR, Minion J. 2013. Diagnostic accuracy and reproducibility of WHO-endorsed phenotypic drug susceptibility testing methods for first-line and second-line antituberculosis drugs. J Clin Microbiol 51:393–401. doi: 10.1128/JCM.02724-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krawczyk B. 2016. Learning from imbalanced data: open challenges and future directions. Prog Artif Intell 5:221–232. doi: 10.1007/s13748-016-0094-0. [DOI] [Google Scholar]
- 25.Raudys SJ, Jain AK. 1991. Small sample size effects in statistical pattern recognition: recommendations for practitioners. IEEE Trans Pattern Anal Machine Intell 13:252–264. doi: 10.1109/34.75512. [DOI] [Google Scholar]
- 26.Nguyen M, Olson R, Shukla M, VanOeffelen M, Davis JJ. 2020. Predicting antimicrobial resistance using conserved genes. bioRxiv doi: 10.1101/2020.04.29.068254. [DOI] [PMC free article] [PubMed]
- 27.Davis JJ, Wattam AR, Aziz RK, Brettin T, Butler R, Butler RM, Chlenski P, Conrad N, Dickerman A, Dietrich EM, Gabbard JL, Gerdes S, Guard A, Kenyon RW, Machi D, Mao C, Murphy-Olson D, Nguyen M, Nordberg EK, Olsen GJ, Olson RD, Overbeek JC, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomas C, VanOeffelen M, Vonstein V, Warren AS, Xia F, Xie D, Yoo H, Stevens R. 2020. The PATRIC Bioinformatics Resource Center: expanding data and analysis capabilities. Nucleic Acids Res 48:D606–D612. doi: 10.1093/nar/gkz943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breiman L. 2001. Random forests. Mach Learn 45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 29.Adékambi T, Drancourt M, Raoult D. 2009. The rpoB gene as a tool for clinical microbiologists. Trends Microbiol 17:37–45. doi: 10.1016/j.tim.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Jaber M, Rattan A, Kumar R. 1996. Presence of katG gene in resistant Mycobacterium tuberculosis. J Clin Pathol 49:945–947. doi: 10.1136/jcp.49.11.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts MC, Sutcliffe J, Courvalin P, Jensen LB, Rood J, Seppala H. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob Agents Chemother 43:2823–2830. doi: 10.1128/AAC.43.12.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schubert B, Maddamsetti R, Nyman J, Farhat MR, Marks DS. 2019. Genome-wide discovery of epistatic loci affecting antibiotic resistance in Neisseria gonorrhoeae using evolutionary couplings. Nat Microbiol 4:328–338. doi: 10.1038/s41564-018-0309-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Antonopoulos DA, Assaf R, Aziz RK, Brettin T, Bun C, Conrad N, Davis JJ, Dietrich EM, Disz T, Gerdes S, Kenyon RW, Machi D, Mao C, Murphy-Olson DE, Nordberg EK, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Santerre J, Shukla M, Stevens RL, VanOeffelen M, Vonstein V, Warren AS, Wattam AR, Xia F, Yoo H. 2019. PATRIC as a unique resource for studying antimicrobial resistance. Brief Bioinform 20:1094–1102. doi: 10.1093/bib/bbx083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen A-LV, Cheng AA, Liu S, Min SY, Miroshnichenko A, Tran H-K, Werfalli RE, Nasir JA, Oloni M, Speicher DJ, Florescu A, Singh B, Faltyn M, Hernandez-Koutoucheva A, Sharma AN, Bordeleau E, Pawlowski AC, Zubyk HL, Dooley D, Griffiths E, Maguire F, Winsor GL, Beiko RG, Brinkman FSL, Hsiao WWL, Domselaar GV, McArthur AG. 2020. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 48:D517–D525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu B, Zheng D, Jin Q, Chen L, Yang J. 2019. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res 47:D687–D692. doi: 10.1093/nar/gky1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feldgarden M, Brover V, Haft DH, Prasad AB, Slotta DJ, Tolstoy I, Tyson GH, Zhao S, Hsu C-H, McDermott PF, Tadesse DA, Morales C, Simmons M, Tillman G, Wasilenko J, Folster JP, Klimke W. 2019. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother 63:e00483-19. doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saier MH, Jr, Reddy VS, Tsu BV, Ahmed MS, Li C, Moreno-Hagelsieb G. 2016. The Transporter Classification Database (TCDB): recent advances. Nucleic Acids Res 44:D372–9. doi: 10.1093/nar/gkv1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beceiro A, Tomás M, Bou G. 2013. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin Microbiol Rev 26:185–230. doi: 10.1128/CMR.00059-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rahman T, Yarnall B, Doyle DA. 2017. Efflux drug transporters at the forefront of antimicrobial resistance. Eur Biophys J 46:647–653. doi: 10.1007/s00249-017-1238-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Severi E, Thomas GH. 2019. Antibiotic export: transporters involved in the final step of natural product production. Microbiology (Reading) 165:805–818. doi: 10.1099/mic.0.000794. [DOI] [PubMed] [Google Scholar]
- 41.Hesterkamp T, Bukau B. 1998. Role of the DnaK and HscA homologs of Hsp70 chaperones in protein folding in E.coli. EMBO J 17:4818–4828. doi: 10.1093/emboj/17.16.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roche B, Aussel L, Ezraty B, Mandin P, Py B, Barras F. 2013. Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochim Biophys Acta 1827:455–469. doi: 10.1016/j.bbabio.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 43.Oladeinde A, Cook K, Lakin SM, Woyda R, Abdo Z, Looft T, Herrington K, Zock G, Lawrence JP, Thomas JC, Beaudry MS, Glenn T. 2019. Horizontal gene transfer and acquired antibiotic resistance in Salmonella enterica serovar Heidelberg following in vitro incubation in broiler ceca. Appl Environ Microbiol 85:e01903-19. doi: 10.1128/AEM.01903-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiappori F, Fumian M, Milanesi L, Merelli I. 2015. DnaK as antibiotic target: hot spot residues analysis for differential inhibition of the bacterial protein in comparison with the human HSP70. PLoS One 10:e0124563. doi: 10.1371/journal.pone.0124563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giordano N, Hastie JL, Smith AD, Foss ED, Gutierrez-Munoz DF, Carlson PE. 2018. Cysteine desulfurase IscS2 plays a role in oxygen resistance in Clostridium difficile. Infect Immun 86:e00326-18. doi: 10.1128/IAI.00326-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lopatkin AJ, Bening SC, Manson AL, Stokes JM, Kohanski MA, Badran AH, Earl AM, Cheney NJ, Yang JH, Collins JJ. 2021. Clinically relevant mutations in core metabolic genes confer antibiotic resistance. Science 371:eaba0862. doi: 10.1126/science.aba0862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mariam DH, Mengistu Y, Hoffner SE, Andersson DI. 2004. Effect of rpoB mutations conferring rifampin resistance on fitness of Mycobacterium tuberculosis. Antimicrob Agents Chemother 48:1289–1294. doi: 10.1128/aac.48.4.1289-1294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramaswamy SV, Amin AG, Göksel S, Stager CE, Dou S-J, El Sahly H, Moghazeh SL, Kreiswirth BN, Musser JM. 2000. Molecular genetic analysis of nucleotide polymorphisms associated with ethambutol resistance in human isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother 44:326–336. doi: 10.1128/aac.44.2.326-336.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rouse DA, Li Z, Bai GH, Morris SL. 1995. Characterization of the katG and inhA genes of isoniazid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother 39:2472–2477. doi: 10.1128/aac.39.11.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong A. 2017. Epistasis and the evolution of antimicrobial resistance. Front Microbiol 8:246. doi: 10.3389/fmicb.2017.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Slatkin M. 2008. Linkage disequilibrium—understanding the evolutionary past and mapping the medical future. Nat Rev Genet 9:477–485. doi: 10.1038/nrg2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mathisen BM, Aamodt A, Bach K, Langseth H. 2020. Learning similarity measures from data. Prog Artif Intell 9:129–143. doi: 10.1007/s13748-019-00201-2. [DOI] [Google Scholar]
- 53.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing, 28th ed. CLSI supplement M100. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 54.Starks AM, Aviles E, Cirillo DM, Denkinger CM, Dolinger DL, Emerson C, Gallarda J, Hanna D, Kim PS, Liwski R, Miotto P, Schito M, Zignol M. 2015. Collaborative effort for a centralized worldwide tuberculosis relational sequencing data platform. Clin Infect Dis 61:S141–S146. doi: 10.1093/cid/civ610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ezewudo M, Borens A, Chiner-Oms Á, Miotto P, Chindelevitch L, Starks AM, Hanna D, Liwski R, Zignol M, Gilpin C, Niemann S, Kohl TA, Warren RM, Crook D, Gagneux S, Hoffner S, Rodrigues C, Comas I, Engelthaler DM, Alland D, Rigouts L, Lange C, Dheda K, Hasan R, McNerney R, Cirillo DM, Schito M, Rodwell TC, Posey J. 2018. Integrating standardized whole genome sequence analysis with a global Mycobacterium tuberculosis antibiotic resistance knowledgebase. Sci Rep 8:15382. doi: 10.1038/s41598-018-33731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Food and Drug Administration. 2013. National Antimicrobial Resistance Monitoring System—enteric bacteria (NARMS): 2011 executive report. Food and Drug Administration, Department of Health and Human Services, Rockville, MD. [Google Scholar]
- 57.Deorowicz S, Kokot M, Grabowski S, Debudaj-Grabysz A. 2015. KMC 2: fast and resource-frugal k-mer counting. Bioinformatics 31:1569–1576. doi: 10.1093/bioinformatics/btv022. [DOI] [PubMed] [Google Scholar]
- 58.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Louppe G, Prettenhofer P, Weiss R. 2012. Scikit-learn: machine learning in Python. arXiv 1201.0490 [cs.LG]. https://arxiv.org/abs/1201.0490.
- 59.Parrello B, Butler R, Chlenski P, Olson R, Overbeek J, Pusch GD, Vonstein V, Overbeek R. 2019. A machine learning-based service for estimating quality of genomes using PATRIC. BMC Bioinformatics 20:486. doi: 10.1186/s12859-019-3068-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O’Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clausen PTLC, Aarestrup FM, Lund O. 2018. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinformatics 19:307. doi: 10.1186/s12859-018-2336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chollet F. 2015. Keras. https://github.com/fchollet/keras.
- 63.Hobohm U, Scharf M, Schneider R, Sander C. 1992. Selection of representative protein data sets. Protein Sci 1:409–417. doi: 10.1002/pro.5560010313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sayers S, Li L, Ong E, Deng S, Fu G, Lin Y, Yang B, Zhang S, Fa Z, Zhao B, Xiang Z, Li Y, Zhao X-M, Olszewski MA, Chen L, He Y. 2019. Victors: a web-based knowledge base of virulence factors in human and animal pathogens. Nucleic Acids Res 47:D693–D700. doi: 10.1093/nar/gky999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hallgren MB, Overballe-Petersen S, Lund O, Hasman H, Clausen PTLC. 2020. MINTyper: an outbreak-detection method for accurate and rapid SNP typing of clonal clusters with noisy long reads. bioRxiv doi: 10.1101/2020.05.28.121251. [DOI] [PMC free article] [PubMed]
- 66.Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nucleotide k-mer distance-based tree of Klebsiella pneumoniae genomes used in this study. Surrounding rings around the distance tree are colored according to the antibiotic resistance phenotypes of strains; susceptible strains are colored green and resistant strains are colored red. The antibiotic phenotypes from inner to outer ring are as follows: amikacin, aztreonam, cefepime, cefoxitin, ceftazidime, ciprofloxacin, gentamicin, imipenem, levofloxacin, meropenem, piperacillin-tazobactam, tetracycline, tobramycin, and trimethoprim-sulfamethoxazole. Download FIG S1, PDF file, 1.8 MB (1.8MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Nucleotide k-mer distance-based tree of Salmonella enterica genomes used in this study. Surrounding rings around the distance tree are colored according to the antibiotic resistance phenotypes of strains; susceptible strains are colored green and resistant strains are colored red. The antibiotic phenotypes from inner to outer ring are as follows: amoxicillin-clavulanate, ampicillin, cefoxitin, ceftiofur, ceftriaxone, chloramphenicol, gentamicin, streptomycin, sulfisoxazole, and tetracycline. Download FIG S3, PDF file, 1.7 MB (1.7MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Receiver operating characteristic (ROC) curves corresponding to antimicrobial resistance (AMR) prediction models for the corresponding samples in the test sets of the 5-fold cross validation. (Top) Areas under the receiver operating characteristic curve (AUCs) from whole-chromosome alignment-based models. (Bottom) AUCs from subalignment-based models. A subalignment size of 5 kb was used for K. pneumoniae and S. enterica, and 10 kb was used for M. tuberculosis. The ROC curves represent a single 5-fold cross-validated model for each chromosomal alignment-based model, and 1,066, 441, and 971 5-fold cross-validated models for randomly sampled subalignments for K. pneumoniae, M. tuberculosis, and S. enterica, respectively. Results are shown for each antibiotic, and the bright green line indicates the average test performance for all antibiotics. Download FIG S4, PDF file, 0.4 MB (405.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
(Tab A) Klebsiella pneumoniae genomes used in this study, with their laboratory-derived AMR phenotypes (R, resistant; S, susceptible). Blank cells correspond to intermediate or unknown phenotypes. (Tab B) Mycobacterium tuberculosis genomes used in this study, with their laboratory-derived AMR phenotypes (R, resistant; S, susceptible). Blank cells correspond to intermediate or unknown phenotypes. (Tab C) Salmonella enterica genomes used in this study, with their laboratory-derived AMR phenotypes (R, resistant; S, susceptible). Blank cells correspond to intermediate or unknown phenotypes. (Tab D) A comparison of areas under the receiver operating characteristic curve (AUCs) for S. enterica models built using the chromosome of two different reference strains, S. enterica subsp. enterica serovar Typhimurium LT2 (Pathosystems Resource Integration Center [PATRIC] genome identifier [ID] 99287.12) and S. enterica subsp. enterica serovar Typhi CT18 (PATRIC genome ID 220341.7). (Tab E) K. pneumoniae model performances for subalignments ranging in size from 1 kb to 1 Mb, and the model based on the whole chromosomal alignment. Subalignments were drawn randomly from the whole chromosomal alignment. (Tab F) M. tuberculosis model performances for subalignments ranging in size from 1 kb to 1 Mb, and the model based on the whole chromosomal alignment. Subalignments were drawn randomly from the whole chromosomal alignment. (Tab G) S. enterica model performances for subalignments ranging in size from 1 kb to 1 Mb, and the model based on the whole chromosomal alignment. Subalignments were drawn randomly from the whole chromosomal alignment. (Tab H) Some examples of highly performing subalignment models in M. tuberculosis, defined as an AUC of >0.85 for at least three of the tested antibiotics. (Tab I) Some examples of poorly performing subalignment models in K. pneumoniae and S. enterica, defined as an AUC of <0.6 for at least six of the tested antibiotics. (Tab J) K. pneumoniae subalignments with model AUCs greater than 0.80 following clustering at various k-mer similarity thresholds. The table displays all subalignments containing an AMR-, virulence-, or transport-related gene, or that have one within 5 kb upstream or downstream of the subalignment. The subalignments containing an important feature column that falls within an AMR, virulence, or transport gene are marked with an annotation source. PATRIC annotations are shown for the protein-encoding genes containing important feature columns in the subalignment and are separated by two # symbols. An asterisk (*) denotes a match 5 kb upstream or downstream of the subalignment. Two asterisks (**) denote an important subalignment column in an intergenic region. (Tab K) M. tuberculosis subalignments with model AUCs greater than 0.80 following clustering at various k-mer similarity thresholds. The table displays all subalignments containing an AMR-, virulence-, or transport-related gene, or that have one within 5 kb upstream or downstream of the subalignment. The subalignments containing an important feature column that falls within an AMR, virulence, or transport gene are marked with an annotation source. PATRIC annotations are shown for the protein-encoding genes containing important feature columns in the subalignment and are separated by two # symbols. An asterisk (*) denotes a match 5 kb upstream or downstream of the subalignment. Two asterisks (**) denote an important subalignment column in an intergenic region. (Tab L) S. enterica subalignments with model AUCs greater than 0.80 following clustering at various k-mer similarity thresholds. The table displays all subalignments containing an AMR-, virulence-, or transport-related gene, or that have one within 5 kb upstream or downstream of the subalignment. The subalignments containing an important feature column that falls within an AMR, virulence, or transport gene are marked with an annotation source. PATRIC annotations are shown for the protein-encoding genes containing important feature columns in the subalignment and are separated by two # symbols. An asterisk (*) denotes a match 5 kb upstream or downstream of the subalignment. Two asterisks (**) denote an important subalignment column in an intergenic region. (Tab M) K. pneumoniae subalignments with model AUCs greater than 0.80 following clustering at various k-mer similarity thresholds. The table displays subalignments lacking a match to AMR-, virulence-, or transport-related genes. PATRIC annotations are shown for the protein-encoding genes containing important feature columns in the subalignment and are separated by two # symbols. Two asterisks (**) denote an important subalignment column in an intergenic region. (Tab N) M. tuberculosis subalignments with model AUCs greater than 0.80 following clustering at various k-mer similarity thresholds. The table displays subalignments lacking a match to AMR-, virulence-, or transport-related genes. PATRIC annotations are shown for the protein-encoding genes containing important feature columns in the subalignment and are separated by two # symbols. Two asterisks (**) denote an important subalignment column in an intergenic region. (Tab O) S. enterica subalignments with model AUCs greater than 0.80 following clustering at various k-mer similarity thresholds. The table displays subalignments lacking a match to AMR-, virulence-, or transport-related genes. PATRIC annotations are shown for the protein-encoding genes containing important feature columns in the subalignment and are separated by two # symbols. Two asterisks (**) denote an important subalignment column in an intergenic region. (Tab P) Number of randomly selected subalignments used per species. The total length of all subalignments was limited to the length of the reference chromosome. Download Table S1, XLSX file, 0.9 MB (943.4KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Nucleotide k-mer distance-based tree of Mycobacterium tuberculosis genomes used in this study. Surrounding rings around the distance tree are colored according to the antibiotic resistance phenotypes of strains; susceptible strains are colored green and resistant strains are colored red. The antibiotic phenotypes from inner to outer ring are as follows: ethambutol, isoniazid, pyrazinamide, rifampicin, and streptomycin. Download FIG S2, PDF file, 1.1 MB (1.1MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Subalignment model accuracies corresponding to Fig. 2 in the main text, with notable protein annotations corresponding to high- and low-performing subalignments overlaid. Download FIG S5, PDF file, 0.8 MB (798.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Number of k-mer-based genome clusters versus clustering threshold. The genomes of each species were clustered based on k-mer similarity, using their aligned sequences in the whole chromosomal alignment. As the clustering threshold decreases, clusters become larger and more inclusive, and there are fewer total clusters. Download FIG S6, PDF file, 0.04 MB (42KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Number of subalignments and corresponding model AUCs at various k-mer similarity thresholds. Each panel shows a distribution of AMR prediction performances in AUC for the clustered strains at a given similarity threshold. All samples were clustered based on their k-mer similarity in the whole chromosomal alignment. Samples belonging to the same cluster were restricted to either the testing or training sets of each fold of the 5-fold cross-validation. Download FIG S7, PDF file, 0.3 MB (286.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Reference chromosome position corresponding to subalignments with high AMR prediction accuracies. Each panel includes predictive subalignments with AUCs of >0.8 after clustering the strains at a given k-mer similarity threshold. Subalignments were colored according to hits to AMR (green), virulence (blue), and transporter genes (orange), respectively. If a subalignment contains multiple gene categories, the color will appear as a mixture. Subalignments that do not hit any of the known AMR-related genes are colored in pink. Download FIG S8, PDF file, 1.5 MB (1.5MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
The Python 2.7.15 scripts used for this project are available on Bitbucket (https://bitbucket.org/deaytan/aligned-fragments/). All genomes and metadata can be accessed through the PATRIC resource (https://www.patricbrc.org) using the genome identifiers given in Tables S1A to S1C.