ABSTRACT
Metabolites have essential roles in microbial communities, including as mediators of nutrient and energy exchange, cell-to-cell communication, and antibiosis. However, detecting and quantifying metabolites and other chemicals in samples having extremes in salt or mineral content using liquid chromatography-mass spectrometry (LC-MS)-based methods remains a significant challenge. Here, we report a facile method based on in situ chemical derivatization followed by extraction for analysis of metabolites and other chemicals in hypersaline samples, enabling for the first time direct LC-MS-based exometabolomics analysis in sample matrices containing up to 2 M total dissolved salts. The method, MetFish, is applicable to molecules containing amine, carboxylic acid, carbonyl, or hydroxyl functional groups, and it can be integrated into either targeted or untargeted analysis pipelines. In targeted analyses, MetFish provided limits of quantification as low as 1 nM, broad linear dynamic ranges (up to 5 to 6 orders of magnitude) with excellent linearity, and low median interday reproducibility (e.g., 2.6%). MetFish was successfully applied in targeted and untargeted exometabolomics analyses of microbial consortia, quantifying amino acid dynamics in the exometabolome during community succession; in situ in a native prairie soil, whose exometabolome was isolated using a hypersaline extraction; and in input and produced fluids from a hydraulically fractured well, identifying dramatic changes in the exometabolome over time in the well.
IMPORTANCE The identification and accurate quantification of metabolites using electrospray ionization-mass spectrometry (ESI-MS) in hypersaline samples is a challenge due to matrix effects. Clean-up and desalting strategies that typically work well for samples with lower salt concentrations are often ineffective in hypersaline samples. To address this gap, we developed and demonstrated a simple yet sensitive and accurate method—MetFish—using chemical derivatization to enable mass spectrometry-based metabolomics in a variety of hypersaline samples from varied ecosystems and containing up to 2 M dissolved salts.
KEYWORDS: exometabolomics, extreme environments, hypersaline, mass spectrometry, microbial communities
INTRODUCTION
Microbial communities are ubiquitous and colonize a wide range of habitats and organisms, often thriving even in extreme environments with physicochemical conditions unsuitable for most other life forms. There is increasing evidence that microbial communities are responsible for a wide range of processes critical to the health of the ecosystems they inhabit and that they impact it in ways of which we currently have limited knowledge. Thriving in complex or extreme environments requires specific adaptations; therefore, studying these organisms lends evolutionary insight into microbial stress responses (1, 2). The balance between cooperation and competition in harsh conditions contributes to the resistance and resilience of these communities (3–7), and elucidating the role of chemical exchange and communication among members will provide an improved understanding of the underlying molecular mechanisms that might be exploited, as well as aid in the identification of beneficial natural products (8–14). While metagenomics studies have been conducted to identify genes encoding novel biosynthetic pathways (15–17), the measurement of primary and secondary metabolites in chemically extreme environments has been hampered by the complexities of the associated sample matrices.
Mass spectrometry is an indispensable analytical tool for identifying, quantifying, and structurally characterizing chemical and biological molecules with high sensitivity and accuracy (18–21). As the central workhorse for proteomics and metabolomics, liquid chromatography coupled with mass spectrometry (LC-MS) has played a critical role in the development of omics technologies that have enabled high-throughput systems biology investigations of organisms (22–24). However, performing exometabolomics analyses in environmental samples can be challenging, due to the complexity of the associated sample matrices. A particular challenge is the presence of high (e.g., mM to M) concentrations of salts and minerals, which can compromise the extraction of metabolites from the samples and suppress the ionization of metabolites during LC-MS analysis, resulting in diminished or skewed quantitative performance (25–27). The majority of the Earth’s water bodies are saline. Hypersaline environments such as soda lakes, acidic hypersaline lakes, solar salterns, and deep-sea brine pools contain salt concentrations that typically far exceed ocean salt levels, the latter of which average 35 g/liter total dissolved salts (28). Studying the metabolisms of and chemical communication among the halophilic microorganisms that inhabit these unique ecosystems could provide important insights into specialized functional adaptations and ecosystem interactions to survive such extreme conditions and are also of astrobiological interest as analogues to lifeforms that might have existed on Mars (29). Researchers have been able to characterize the microbial diversity in these extreme and evolutionally important environments using sequencing-based approaches but have not had nearly the same success using metabolomics (30–34). Until now, samples consisting of or derived from such matrices have precluded the application of LC-MS-based measurements of metabolites and other small molecules. Conventional metabolomics approaches and strategies that have worked well for sample types that contain relatively lower salt concentration, such as seawater (35–38) or human urine (39–41), have not been applied successfully to hypersaline samples.
To address this, we present MetFish, a method based on chemical tagging and extraction for comprehensive and quantitative measurement of metabolites and other small molecules in LC-MS-prohibitive matrices. Named for its ability to selectively “fish” metabolites of interest from sample matrices based upon common functional groups, MetFish is composed of four simple and inexpensive chemical tags targeting amine, carboxyl, carbonyl, and hydroxyl functional groups and allows for sensitive quantification of low-abundance metabolites in both targeted and untargeted approaches. The four functional groups targeted by MetFish represent over 97%, 89%, and 83% of the metabolites contained in the Natural Products Atlas, E. coli Metabolome, and PlantCyc databases, respectively (42–44). The chemical tags can be either used in tandem for untargeted global analysis of the metabolome or individually to profile the submetabolome by targeting the molecules containing a specific functional group. MetFish uses low-cost, commercially available reagents that (i) could be used to study diverse sample types based on the functional groups of interest; (ii) facilitate physical separation of metabolites from salt, mineral, and other matrix components that interfere with quantitative LC-MS-based analysis; and (iii) can be deployed in situ to minimize sample manipulation.
Here, we demonstrate the utility and simplicity of MetFish in LC-MS-based exometabolomics analyses of three types of samples containing or derived from microbial communities from diverse ecosystems, namely, a hypersaline aquatic microbial community, a prairie soil, and fluids injected into and produced from a hydraulically fractured well, each consisting of or derived from hypersaline (i.e., from 400 mM to 2 M) sample matrices. MetFish demonstrated excellent sensitivity, reproducibility, and linear dynamic range, and is a simple, rapid, and effective approach for addressing the needs of the broader research community.
RESULTS AND DISCUSSION
Background and overview of MetFish.
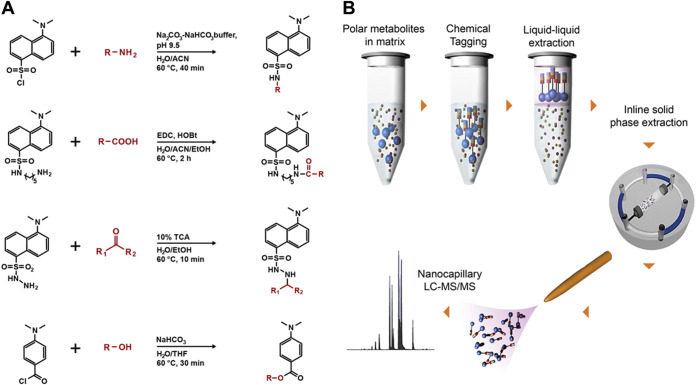
In our search for an effective and simple approach to separate metabolites from interfering matrix constituents such as high concentrations of salts, we evaluated several commercially available solid-phase extraction (SPE) chemistries to capture metabolites from a hypersaline matrix (e.g., 2 M total dissolved salts) but all were unsuccessful (see Table S1 in the supplemental material). We determined that separation methods based on molecular weight (e.g., dialysis or size exclusion) were not suitable, since the masses of low-molecular-weight metabolites (e.g., glycine, 75.07 g/mol) overlap those of salt components (e.g., sulfate, 96.06 g/mol), resulting in loss of metabolites in the lower mass range. We also tested the feasibility of using gas chromatography coupled with mass spectrometry (GC-MS) to detect the presence of amino acid standards in high-salt matrices and found that the presence of salt (both 400 mM and 2 M total dissolved salts) severely affected the measurement, and no analyte peaks were observed in the chromatograms (see Fig. S2 and Table S4 in the supplemental material). Subsequently, we explored chemical tagging and capture techniques, including metabolite enrichment by tagging and proteolytic release (METPR) (45) and a derivatization approach developed by Mattingly et al. (45, 46). Both approaches were time-consuming and required significant solid-/liquid-phase chemical synthesis (e.g., up to 1 week for a single METPR probe for a researcher with basic organic synthesis skills), followed by structural characterization for preparing the capture or derivatization reagents. We were able to obtain a small amount of the QDA (N-[2-(aminooxy)ethyl]-N,N-dimethyl-1-dodecylammonium iodide) (46) derivatizing agent from the authors of the original paper to evaluate its efficacy in hypersaline samples. We found that QDA derivatization was effective for detecting sodium pyruvate, the test analyte, in hypersaline sample matrices containing up to 2 M MgSO4 (see Fig. S3 in the supplemental material). However, it is important to note that the use of QDA as a derivatizing agent is limited to metabolites that contain reactive carbonyl groups. Recognizing the need for a more efficient method that could be readily adopted by researchers from a broad range of disciplines, we adopted a suite of dansylated and related reagents coupled with downstream enrichment. The reagents were selected for their low cost, commercial availability, ease of use to increase accessibility of the method in the research community, and optimal coverage of various major functional groups represented in the metabolome. Dansylation has been used for decades as a derivatization method for quantification of amino acids based on fluorescence detection (47). More recently, Li and colleagues have used dansylated and related reagents for targeted profiling of various submetabolomes using LC-MS (48–51). We postulated that the derivatization chemistries associated with these reagents would be successful when applied in hypersaline matrices, and that we could then efficiently extract derivatized molecules from the samples and away from interfering salts. For MetFish, we selected dansylchloride, dansylhydrazine, dansylcadaverine, and 4-(dimethylamino)benzoyl chloride to specifically tag metabolites containing amine, carbonyl, carboxyl, and hydroxyl functional groups, respectively (Fig. 1a). The one-step derivatization reactions require as few as 10 min to a maximum of 120 min to couple the target metabolite (the “fish”) and the chemical tag (the “hook”), thus increasing its hydrophobicity and facilitating its extraction with organic solvent (the “line”) and concomitant enrichment from interfering components of the sample matrix (Fig. 1b). The tagged and extracted metabolites are subsequently analyzed using reversed-phase liquid chromatography (LC) coupled with MS (48–51). The reversed-phase LC includes inline solid-phase extraction, which focuses the tagged metabolites prior to the analytical separation and separates them from any residual matrix components. Tandem MS (MS/MS) is used to fragment the tagged metabolites, resulting in fragment ions that are uniform for a given reagent and unique for a given metabolite (50), providing identification confidence and metabolite specificity, respectively. Exceptions to the latter are some isomeric metabolites, such as leucine and isoleucine, which do not produce unique fragment ions during collision-induced dissociation.
FIG 1.
Overview of the MetFish method. (a) MetFish reagents and associated derivatization reactions. (b) General workflow of the MetFish method.
(A) GC-MS chromatogram of amino acid mix in water followed by a methanol extraction. (B) GC-MS chromatogram of amino acid mix in water followed by an MPLEx (chloroform-methanol) extraction. (C) GC-MS chromatogram of amino acid mix in 400 μM MgSO4 followed by a methanol extraction. (D) GC-MS chromatogram of amino acid mix in 400 μM MgSO4 followed by a MPLEx extraction. (E) GC-MS chromatogram of amino acid mix in 2 M MgSO4 followed by a methanol extraction. (F) GC-MS chromatogram of amino acid mix in 2 M MgSO4 followed by a MPLEx extraction. Insets show the chromatogram zoomed in. Download FIG S2, DOCX file, 0.3 MB (267.9KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
FTICR-MS spectra of sodium pyruvate in (A) Nanopure H2O, (B) 400 μM MgSO4, and (C) 2 M MgSO4, with the QDA (N-[2-(aminooxy)ethyl]-N,N-dimethyl-1-dodecylammonium iodide)-pyruvate adduct detected at 343.2955 m/z. The top 3 panes in each figure show three independent replicates of QDA derivatization of pyruvate, followed by FTICR-MS analysis. The bottom pane shows an instrument blank. Download FIG S3, DOCX file, 0.04 MB (37.8KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Performance of commercial SPE columns for separation of metabolites from high-salt matrices, as determined by nuclear magnetic resonance (NMR) spectroscopy. Generally, SPE columns were not effective in recovering metabolites from either water or hypersaline media, although some column chemistries were effective to some extent for certain metabolites. Download Table S1, DOCX file, 0.01 MB (15.6KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Metabolite identifications for peaks/features in the chromatogram shown in Fig. S2A. Download Table S4, DOCX file, 0.01 MB (14.1KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
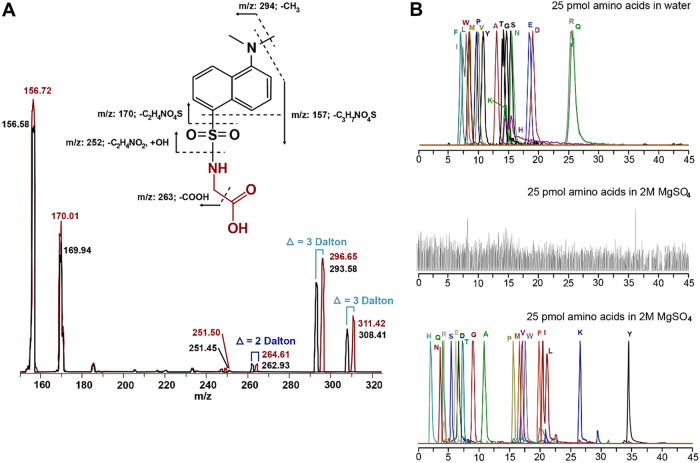
To illustrate metabolite identification using unique fragment ions, the fragmentation spectrum for dansylated glycine is shown in Fig. 2a. Fragment ions due only to the dansyl moiety are, e.g., m/z 157, 170, and 252, whereas fragment ions due to dansyl-glycine are m/z 263 and 294. Some amount of the molecular ion (m/z 308) also appears in the MS/MS spectrum. All metabolites that have been tagged using the dansyl chloride reagent will generate the same fragment ions (e.g., m/z 157, 170, and 252), providing confidence in detection of an appropriately tagged amine-containing metabolite. In contrast, each dansylated metabolite will also generate fragment ions that are specific to the dansyl-metabolite complex and proportional in m/z to the mass of the tagged metabolite. The other MetFish reagents also produce uniform and specific fragment ions upon dissociation (see Table S2 in the supplemental material). These chemical characteristics enable MetFish reagents to be effective for both targeted and untargeted metabolomics applications. An added benefit is that differentially isotopically labeled reagents can be used, allowing for the multiplexing of labeled samples in untargeted metabolomics analysis, analogous to the isobaric tags for relative and absolute quantitation (iTRAQ) and tandem mass tag (TMT) peptide labeling approaches commonly used for multiplexing proteomics sample analyses using LC-MS/MS (52). Differences in abundances of “reporter ions” from MS/MS fragmentation of differentially labeled reagent-metabolite complexes would be used to provide accurate relative or absolute metabolite quantification. Alternatively, labeled metabolites could be incorporated as internal standards in targeted metabolite analysis (48–51). As shown in Fig. 2a, dansylated, uniformly labeled 13C- and 15N-glycine produces fragment ions specific to the dansyl-glycine complex and with mass shifts proportional to the degree and type of isotope labeling.
FIG 2.
Validation of the MetFish method using amino acids. (a) Tandem mass spectra from analysis of a mixture of unlabeled (black spectrum) and 13C and 15N uniformly labeled glycine (red spectrum), both derivatized with dansyl chloride. The m/z of each fragment peak is listed, and the mass shifts due to the isotopic labels are indicated. (b) Overlayed extracted ion chromatograms (EICs) of amino acids in neat solution analyzed by nanocapillary liquid chromatography-tandem mass spectrometry (LC-MS/MS) without chemical tagging (upper chromatogram); total ion chromatogram from analysis of amino acids in 2 M MgSO4 analyzed by nanocapillary LC-MS/MS without chemical tagging (middle chromatogram); EICs of amino acids in 2 M MgSO4, derivatized using dansyl chloride, followed by extraction with organic solvent, and analyzed by nanocapillary LC-MS/MS with dansylation chemical tagging (lower chromatogram). The y axes for all plots have been normalized to the highest intensity peaks in each.
Common fragment ions generated during tandem mass spectrometry (MS/MS) for MetFish reagents. Download Table S2, DOCX file, 0.01 MB (13.1KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Validation of MetFish.
To assess the effectiveness of MetFish for targeted metabolite analysis in MS-prohibitive samples, we analyzed a mixture of 19 proteinogenic amino acids in water containing 2 M MgSO4, with and without the MetFish method and using LC-MS/MS with the mass spectrometer operating in selected reaction monitoring (SRM) mode. MgSO4 was chosen because it is a major salt component of Hot Lake, located in Oroville, WA, where a photoautotrophic microbial mat community resides (53, 54). In typical MS-based metabolomics analyses, amino acids would be enriched from samples using extraction with organic solvents or a solid phase. As described above and shown in Table S1, SPE is not effective for extracting small polar molecules from matrices containing high salt concentrations. Liquid/liquid extraction of amino acids from high-salt matrices either carries over sufficient salt in the extract to cause ionization suppression during analysis or does not effectively extract amino acids due to formation of amino acid-salt complexes that are insoluble in the organic solvent. As shown in Fig. 2b (top), analysis of a 25 pmol mixed amino acid standard dissolved in deionized water was straightforward using hydrophilic interaction liquid chromatography (HILIC)-MS/MS; however, no signal was observed above background for the same 25 pmol mixed amino acid standard dissolved in 2 M MgSO4 (Fig. 2b, middle). Applying the MetFish method using the amine tagging reagent resulted in quantitative measurement of all amino acids using reversed-phase LC-MS/MS (Fig. 2b, lower). In the top and bottom panels of Fig. 2b, the extracted ion chromatograms of each amino acid are normalized to a relative abundance of 100% and overlaid. Because of the increased hydrophobicity of the tagged amino acids, their SRM signals were also more intense (due to enhanced electrospray ionization [55]), and they were better resolved chromatographically using reversed-phase LC compared to their native forms, which were measured using HILIC. In the MetFish analyses, the unique fragment ion from each singly charged, tagged amino acid was used for quantification purposes, and a fragment ion common to all tagged amino acids (e.g., m/z 157, 170, or 252) provided confident identification.
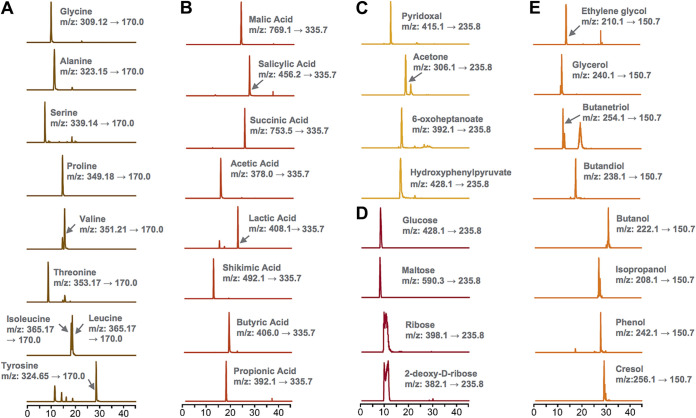
To demonstrate the broad applicability of the MetFish approach for detecting metabolites containing other functional groups, we analyzed metabolites containing carbonyl, carboxyl, and hydroxyl functional groups. As with amino acids (Fig. 2a and 3a), the MetFish method enabled quantification of metabolites with carboxylic acids (Fig. 3b), carbonyl (Fig. 3c), and hydroxyl groups, including sugars (Fig. 3d) and alcohols (Fig. 3e), all in water containing 2 M MgSO4.
FIG 3.
MetFish is applicable to measuring metabolites with a broad range of functional groups in challenging sample matrices. Shown are extracted ion chromatograms with the transitions obtained in selected reaction monitoring (SRM) mode for metabolite quantification from application of MetFish in measurement of (a) amine metabolites, (b) carboxyl metabolites, (c) carbonyl metabolites, (d) hydroxyl metabolites as sugars, and (e) hydroxyl metabolites as alcohols. In all cases, MetFish was deployed in situ in metabolite-salt mixtures containing 2 M MgSO4.
To further validate MetFish, we determined limits of quantification (LOQ), linear dynamic ranges, and relative standard deviations (RSDs) for all four MetFish reagents and in measurements of 45 metabolites containing amine, carboxyl, carbonyl, or hydroxyl functional groups (see Table S3 in the supplemental material) dissolved in water containing 2 M total salt. The amine tagging method provided the lowest LOQ (median of 5 nM), the broadest linear dynamic range (5 to 6 orders of magnitude), and the lowest median interday reproducibility (median of 2.6%) of the four methods, based on data for 19 proteinogenic amino acids (Table S3A). The other tags showed median LOQs ranging from 40 nM (carboxyl; 10 metabolites) to 3.5 μM (hydroxyl; 8 metabolites), linear dynamic ranges of 3 to 5 orders of magnitude, and median interday RSDs of 3.3% (carboxyl; 10 metabolites) to 9.3% (hydroxyl; 8 metabolites) (Table S3B to D). The hydroxyl tagging approach gave the highest LOQ, ranging from sub- to low micromolar. All four MetFish tags showed excellent linearity over the dynamic range of quantification, with an R2 of 0.99.
Limits of quantification (LOQ), linearity, and interday reproducibility of (A) amine, (B) carboxylic acid (C) carbonyl, and (D) hydroxyl tagging approaches. Download Table S3, DOCX file, 0.03 MB (28.9KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Application of MetFish in targeted analyses of proteinogenic amino acids in hypersaline matrices.
After validating that the MetFish method can be used to enrich polar metabolites from a model hypersaline solution, we then applied the amine capture reagent in quantification of amino acids in exometabolomics analyses of two microbial communities, (i) a unicyanobacterial phototrophic microbial community and (ii) a prairie soil.
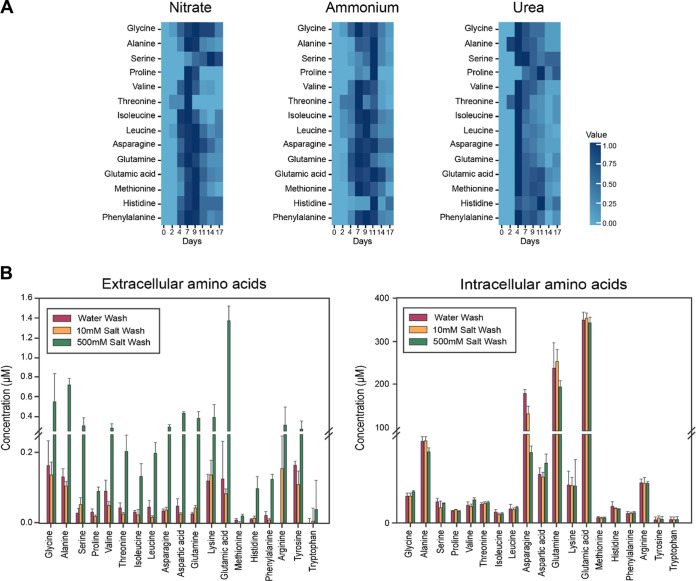
MetFish was used to examine nitrogen metabolism over a 28-day succession in a unicyanobacterial consortial biofilm isolated from a benthic phototrophic microbial mat from a highly saline alkaline lake in northern Washington state (53, 54, 56). During the seasonal cycle, the salt concentration in the lake fluctuates from low hundreds of mM to well over 2 M total dissolved salts (primarily MgSO4); the consortium in this experiment was therefore cultured in a defined medium containing 400 mM MgSO4 (54). As organisms in the consortium are divergent for their ability to incorporate nitrate (57), this experiment aimed to determine how differences in the organismal access to nitrogen for amino acid biosynthesis influenced community dynamics and metabolite exchange. To test the hypothesis that availability of reduced nitrogen would increase the rate of amino acid sharing, the nitrate-containing growth medium was amended with either ammonium or urea. The samples were spiked with 13C and 15N uniformly labeled amino acid standards, and endogenous amino acids in the medium were quantified using isotope dilution MS. The MetFish analysis quantified 14 extracellular proteinogenic amino acids over a 17-day cultivation period (Fig. 4a). The remaining 5 amino acids were below the limit of detection. In general, amino acid concentrations increased to detectable levels early in cultivation until they reached a maximum at ∼7 to 9 days for nitrite and ammonium or 4 d for urea, and decreased thereafter. Surprisingly, this trend did not hold true for all amino acids. For example, serine reached a maximum concentration at 14 days in medium amended with nitrate and at 11 days for ammonium. For proline, the maximum extracellular concentration occurred at 11 days for both ammonium and urea. The exometabolomics analysis of amino acid profiles during the phototrophic consortia succession revealed that availability of extracellular amino acids as community “public goods” differed among nitrogen sources at the level of individual amino acids. MetFish therefore enabled us to conclude that the nitrogen source for amino acid biosynthesis rewires overall community amino acid exchange.
FIG 4.
Application of MetFish in quantification of proteinogenic amino acids in representative microbial communities. (a) Quantification of amino acids during phototrophic microbial community succession with various nitrogen amendments (data shown are normalized average amino acid concentrations from analysis of 3 biological replicate succession experiments). (b) Exo- and endometabolomics analysis of amino acids in soil, using a high-salt wash to increase recovery due to possible disruption of nonspecific binding to soil particles (data shown are mean ± standard deviation from analysis of 3 replicate soil samples).
We next used MetFish in exometabolomics analyses to quantify free proteinogenic amino acids in soil, followed by analysis of biomass-associated molecules. To do so, we modified the classic fumigation-extraction method (58) for measuring microbial biomass-associated carbon content. In the traditional format, soil samples are fumigated with chloroform to lyse microbial cells, followed by immediate extraction with 500 mM K2SO4, which extracts the total of free and biomass-associated molecules but cannot be used to distinguish between the two (59–61). Makarov and colleagues reported that microbial biomass-associated carbon is increasingly extractable with increasing concentration of the K2SO4 extraction solution, with solubility increases of 1.5- to 3.9-fold in 500 mM K2SO4 compared with 50 mM K2SO4.(62) We therefore hypothesized that performing a 500 mM K2SO4 extraction of soil prior to microbial cell lysis would allow us to obtain higher recovery of molecules located in the extracellular milieu and also enable us to follow up with a subsequent measurement of microbial biomass-associated molecules. Because the hypersaline environment of the salt extract would otherwise prohibit a LC-MS-based exometabolomics analysis, MetFish was employed. We used three different extractants—deionized water, 10 mM K2SO4, and 500 mM K2SO4—to extract amino acids from a native prairie soil at the Konza Prairie Biological Station, a long-term ecological research site located in eastern Kansas, USA. Accordingly, we extracted equivalently size aliquots of soil in replicate (see Materials and Methods for details), and subsequently spiked the extracts with 13C- and 15N-labeled amino acid standards and applied the amine tagging MetFish reagent. The extracted soil remaining was then subjected to bead beating to lyse microbial cells, followed by spiking with labeled standards and derivatization of amino acids directly in the soil samples, demonstrating the in situ applicability of MetFish. Nineteen proteinogenic amino acids in both the free and biomass-associated extracts were quantified using isotope dilution MS (Fig. 4b and c). As expected, preextraction of the soil with 500 mM K2SO4 resulted in 2- to 10-fold higher recovery of amino acids from the extracellular milieu compared to preextraction with water and 10 mM K2SO4. Asparagine, glutamine, and glutamic acid were the three most abundant biomass-associated amino acids with concentrations of 70.9 μM/mg, 191.7 μM/mg, and 337.7 μM/mg soil, respectively (Fig. 4c). Intracellular levels of amino acids were similar between the three different preextractants.
Application of MetFish in untargeted metabolomics analysis of fluids injected into and produced from a hydraulically fractured well.
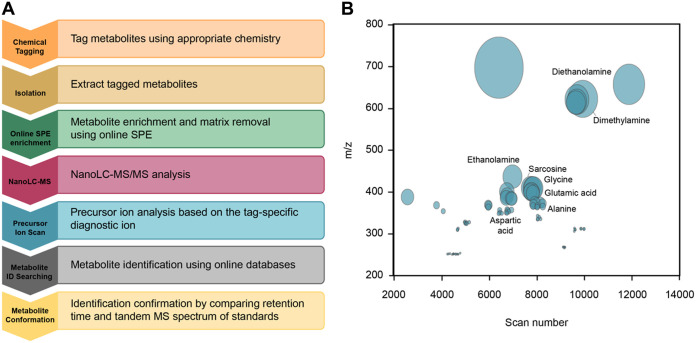
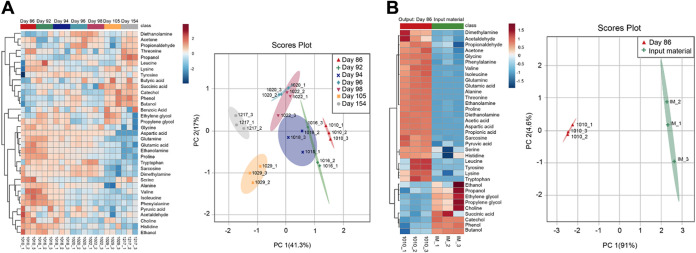
As described above, each MetFish reagent generates one or more tag-specific fragment ions during collision-induced dissociation during MS analysis. These “reporter ions” can be exploited in untargeted exometabolomics analyses to broadly query the metabolome in otherwise MS-prohibitive sample matrices. To demonstrate this, we applied each of the 4 MetFish reagents, separately, in parallel analyses of fluids injected into and produced from a hydraulically fractured well from the Utica-Point Pleasant shale (Ohio, USA), and operated the mass spectrometer in data-dependent MS/MS mode to obtain comprehensive untargeted data (Fig. 5a). Although the complete composition of fracture fluid is typically proprietary, the fracking fluid used in our analyses was known to be complex, with up to 125 g/liter total dissolved solids, including salts, various corrosion inhibitors, and gelling agents. We initially applied each of the 4 MetFish reagents in untargeted exometabolomics analysis of a representative produced fluid sample, in order to identify as many putative molecules as possible (see Materials and Methods for details). It is important to note that the reagents were not used in combination in the same sample to avoid cross-reactivity in derivatization chemistry and confounding of data processing. Figure 5b shows both the putatively identified and unknown features profiled using the MetFish reagent targeting the amine functional group. A total of 17,714 precursor ions were initially detected in the raw data, which was then postprocessed using MASIC (63) to remove low-abundance ions and reduce false identifications and duplicate features, resulting in 100 confidently detected features (see Table S5 in the supplemental material). Low-abundance ions can arise due to chemical noise or to other factors, such as poor ionization of tagged analytes, incomplete derivatization, and matrix effects. To increase the confidence in detected features, three reporter ion matches were used as the criteria for selection instead of one, based on the most abundant fragments in the SRM product ion scan. Precursor ions that generated all three reporter ions were considered potential features and duplicate features having the same precursor ions were removed. We then purchased isotopically labeled standards for putatively identified metabolites and applied MetFish in a targeted exometabolomics analysis to confirm molecular identities in a time series of produced fluid samples collected between 86 and 154 days postinjection (Fig. 6a). Using this approach, we confirmed the identities of 37 metabolites. As shown in Fig. 6a, fluids initially produced from the well at 86 to 98 days showed larger amounts of amino acids than those at 105 and 154 days, while the concentrations of most alcohols and organic acids detected were evenly distributed over the time course. Compared to the input fluids, the metabolite concentrations in produced fluid samples show significant differences (Fig. 6b). Metabolites such as amino acids and organic acids have significantly higher concentrations in produced fluid samples than in the input fluids, indicating the presence of metabolically active microbial communities. The input fluids also contained extremely high levels of diols, such as propylene glycol, which are typical additives in hydraulic fracture fluids. For untargeted discovery of metabolites in the samples, the data-dependent MS/MS spectra were also searched against reference GNPS (64) spectral libraries. This resulted in 99 unique metabolites being identified across all of the fracking fluid data sets (see Table S6 in the supplemental material).
FIG 5.
Untargeted metabolomics using MetFish. (a) Workflow for untargeted metabolomics analysis using MetFish. (b) Global amine tag-based metabolite profile of a produced fluid sample. The size of the circle is proportional to the ion intensity, and putatively identified metabolites are labeled.
FIG 6.
Targeted metabolomics analysis using MetFish in injected and produced fluids from hydraulic fracturing. (a) Quantification of 37 metabolites identified by targeted MetFish analysis. (b) Comparison of metabolite levels in the input material and spent fracking fluid. Data shown are from replicate analysis (n = 3) of each fluid sample. Heatmaps were generated using MetaboAnalyst 3.0 (67), using Pearson’s distance measure and average linkage for the clustering algorithm.
Features detected in the untargeted metabolomics analysis by use of each of the four MetFish tags. Data have been processed to remove low-abundance ions (noise), reduce false identifications, and remove duplicate features. The precursor ions that generated all 3 reporter ions (170, 172, and 252) were considered potential features, and the tag mass was deducted from the precursor mass to obtain the metabolite mass. Download Table S5, XLSX file, 0.01 MB (15.6KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
GNPS spectral library search matches with a cosine score greater than 0.7, for untargeted discovery of metabolites in the fracking fluid datasets collected using data-dependent MS/MS. Download Table S6, XLSX file, 0.03 MB (35.9KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Conclusion.
As with any chemical derivatization-based analysis (65), artifact formation is an inherent limitation of this approach. As with the chemical derivatization used for GC-MS, some compounds may form additional products of derivatization apart from the desired derivative or incompletely derivatized products, known as artifacts. Artifacts cause unexpected or multiple peaks in the LC-MS analysis for the same compound, confounding data interpretation. Related to this, we note that the expected product m/z of cysteine was not observed when using the amine tagging reagent. We suspect that this was due to the formation of oxidative side products.
In summary, the MetFish method enables highly sensitive targeted and untargeted exometabolomics measurements in chemically extreme environments that are otherwise prohibitive to MS-based analyses. We demonstrated use of MetFish in quantification of exometabolites in hypersaline matrices, including spent medium from a phototrophic microbial consortium, salt-extracted soil, and injected/produced fluids from hydraulic fracturing. The combination of a high-salt wash and MetFish was particularly useful for extracting metabolites from the extracellular soil milieu, prior to subsequent in situ application of MetFish for analysis of intracellular metabolites in the same samples after microbial cell lysis. The use of MetFish offers control over the subclass of metabolites being captured, which greatly constrains the chemical search space when attempting to identify unknowns during untargeted exometabolomics analysis. This is particularly useful for samples containing a diversity of high concentration organic constituents, such as soils or those produced from hydrocarbon-bearing, hydraulically fractured wells. We believe that such an approach will aid in the investigation of metabolite exchange in microbial communities and provide a more effective way to understand the microbial metabolism in extreme ecosystems that remain understudied.
MATERIALS AND METHODS
MetFish chemical tagging methods.
(i) Amine reagent. Aliquots (200 μl) of amine-containing metabolites in water, medium, or matrix were combined with 500 μl of dansyl chloride in acetonitrile (40 mM) and diluted to a final volume of 1,200 μl with 0.5 M Na2CO3-NaHCO3 buffer (pH 9.5). The reaction solution was mixed using a ThermoMixer (Eppendorf, Hauppauge, NY) at 60°C for 40 min at 1,500 rpm. The organic portion of the resulting solution was removed using a SpeedVac (Eppendorf) for 30 min, and the pH of the aqueous portion was adjusted to 3 to 4 by using 20% formic acid (vol/vol). Then a liquid-liquid extraction was performed using dichloromethane and water. The organic layer was collected and reconstituted in 1 ml of a mixture of water and methanol (95:5, vol/vol) containing 0.1% formic acid, followed by online SPE-nanocapillary LC-MS/MS analysis.
(ii) Carbonyl reagent. Aliquots (200 μl) of carbonyl-containing metabolites in water, medium, or matrix were combined with 50 μl of 10% trichloroacetic acid in water (wt/vol) and 250 μl dansyl hydrazine in ethanol (10 mM), and diluted to a final volume of 900 μl with 400 μl of water. The reaction solution was mixed using a ThermoMixer (Eppendorf, Hauppauge, NY) at 60°C for 15 min at 1,500 rpm. The organic portion of the resulting solution was removed using a SpeedVac (Eppendorf) for 30 min, and the pH of the aqueous portion was adjusted to 3 to 4 by using 20% formic acid (vol/vol). A liquid-liquid extraction was then performed by using dichloromethane and water. The organic layer was collected and reconstituted in 1 ml of a mixture of water and methanol (95:5, vol/vol) containing 0.1% formic acid, followed by online SPE-nanocapillary LC-MS/MS analysis. Dansylhydrazine (DNSH) is a derivatization agent used for carbonyl bonds, such as ketones and aldehydes (66). It is less reactive with esters because the carbonyl carbon of this functional group has decreased electrophilicity due to resonance stabilization.
(iii) Carboxylic acid reagent. Aliquots (150 μl) of carboxylic acid-containing metabolites in water, medium, or matrix were combined with 300 μl of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide in water (10 mM), 300 μl dansylcadaverine in methanol (10 mM), and 300 μl hydroxybenzotriazole in ethanol, and diluted to a final volume of 1350 μl with 300 μl of water. The reaction solution was mixed using a ThermoMixer (Eppendorf) at 60°C for 120 min at 1,500 rpm. The organic portion of the resulting solution was removed using a SpeedVac (Eppendorf) for 30 min, and the pH of the aqueous portion was adjusted to 3 to 4 by using 20% formic acid (vol/vol). Then, a liquid-liquid extraction was performed using dichloromethane and water. The organic layer was collected and reconstituted in 1 ml of a mixture of water and methanol (95:5, vol/vol) containing 0.1% formic acid, followed by online SPE-nanocapillary LC-MS/MS analysis.
(iv) Hydroxyl reagent. Aliquots (300 μl) of hydroxyl-containing metabolites in water, medium, or matrix were combined with 300 μl 4-dimethylaminobenzoic chloride in tetrahydrofuran (10 mM) and diluted to a final volume of 900 μl with 300 μl of 100 mM sodium carbonate in water. The reaction solution was mixed using a ThermoMixer (Eppendorf) at 30°C for 50 min at 1,500 rpm. The organic portion of the resulting solution was removed using a SpeedVac (Eppendorf) for 30 min, and the pH of the aqueous portion was adjusted to 3 to 4 using 20% formic acid (vol/vol). Then a liquid-liquid extraction was performed using dichloromethane and water. The organic layer was collected and reconstituted in 1 ml of a mixture of water and methanol (95:5, vol/vol) containing 0.1% formic acid, followed by online SPE-nanocapillary LC-MS/MS analysis.
Sample preparation for metabolomics analyses.
(i) Photoautotrophic microbial consortium. The photoautotrophic microbial consortium was routinely cultured as described previously (54). Briefly, biofilms were grown and maintained in T75 Corning cell culture flasks (catalog no. 10-126-37; Fisher) on Hot Lake autotroph medium (HLA; see Table S7 in the supplemental material for details) at room temperature and atmosphere under ∼35 μE/m2/s light (PL/AQ; General Electric) over 28-day succession experiments. Sterile water was replaced weekly to replace volume lost to evaporation. For exometabolomics analysis, 20 ml of cell culture was centrifuged at 6,000 × g for 15 min at 4°C. The supernatant was then transferred into a separate vessel. A portion of the supernatant was spiked with isotopically labeled amino acids as internal standards and analyzed using the amine tagging protocol.
(ii) Fracture fluid and produced fluids. Due to the high acidity of fracking fluid samples, the samples were pretreated by adding 250 μl of 0.5 M NaOH solution. Then, the MetFish methodology was applied to the fracture fluid and produced fluid samples by using each chemical tagging approach followed by LC-MS/MS analysis. A Q Exactive hybrid quadrupole-Orbitrap high-resolution mass spectrometer (Thermo Fisher Scientific, San Jose, CA) was used for metabolite profiling, and data-dependent MS/MS spectra were obtained. Data analysis was performed by using in-house developed software, MASIC, to extract tag-derived metabolite masses with the use of three diagnostic fragments for each tagging approach. The masses of unknown metabolites were matched against the METLIN database and the Human Metabolome Database (HMDB) using a mass accuracy of <10 ppm. Additionally, the data were searched against the GNPS spectral libraries using the online workflow (https://ccms-ucsd.github.io/GNPSDocumentation/) on the GNPS website (http://gnps.ucsd.edu). Data were filtered by removing all MS/MS fragment ions within ±17 Da of the precursor m/z. MS/MS spectra were window filtered by choosing only the top 6 fragment ions in the ±50 Da window throughout the spectrum. The precursor ion mass tolerance was set to 2.0 Da and a MS/MS fragment ion tolerance of 0.5 Da. All matches were required to have a cosine score above 0.7 and more than 6 matched peaks.
(iii) Extracellular soil metabolite extraction. Silty loam soil was collected from the upper 15 cm of a watershed (39°06′11″ N, 96°36′48″ W) located at the Konza Prairie Biological Station. Upon collection, soil samples were shipped on ice to the Pacific Northwest Laboratory, where soils were immediately sieved (<2 mm) and stored at −80°C until further use. Soil pH (in water) was 6.5, and the sulfate concentration was 5.45 ppm. Soil gravimetric water and clay content were determined to be 37% and 2%, respectively. A salt wash of soil samples was performed in order to completely extract extracellular metabolites from soil particulates. K2SO4 (500 mM and 10 mM) solutions were used for salt washes; a water extraction was also performed as a control. Each extraction was performed in triplicate. Sieved soil sample (1 g) and 2 ml K2SO4 salt wash solution or water were added into a centrifuge tube. The mixture was vortexed for 30 min. The resulting mixture was centrifuged at 5,000 rpm for 5 min, and the supernatant was depleted. Then, another salt or water wash was performed on the residual soil sample. Another 2 ml K2SO4 salt wash solution or water was added to the residual soil sample, and the mixture was vortexed for 30 min. The two supernatants were combined and filtered through a 0.22-μm filter. A 20-μl aliquot of 300 μg/ml 13C- and 15N-labeled amino acid mixture standard was added to the filtered supernatant. Then, the amine-specific tagging approach was conducted to analyze extracellular metabolites.
(iv) Intracellular soil metabolite extraction. The residual soil from the extracellular metabolite extraction was used for subsequent intracellular metabolite extraction. A scoop of stainless-steel beads and garnet beads was added into the centrifuge tube. A 1-ml aliquot of water was then added, and bead beating was performed at speed 7 in a Bullet Blender Tissue Homogenizer 24 (Next Advance, NY, USA) for 4 min at 4°C to lyse microbial cells. A 20-μl aliquot of 300 μg/ml 13C- and 15N-labeled amine acid mixture standard was added to the sample. Then, the amine-specific tagging approach was directly conducted in soil to quantify released intracellular amino acids.
Instrumentation.
(i) Online SPE-nanocapillary liquid chromatography system. An online SPE-nanocapillary liquid chromatography system was used for analysis of MetFish-tagged metabolites. The system consists of two parallel subsystems, each of which consists of two Agilent 1200 series nanopumps (Agilent Technologies, Santa Clara, CA), a six-port injection valve (VICI Valco, Houston, TX) with a sample loop, and a six-port injection valve (VICI Valco, Houston, TX) with a micro-solid-phase-extraction (SPE) column coupled in line (see Fig. S1 in the supplemental material). The valves were switched by program and directed the LC flow to carry the analyte from the sample loop into the SPE. After the analyte was enriched by SPE, it was backflushed to a nine-port valve (VICI Valco, Houston, TX), which delivered the analyte into the nanocapillary LC column. The nanocapillary LC column was packed with porous C18 particles (3-μm particle size, 300-Å pore size; Phenomenex, Terrence, CA) in fused silica capillaries (35 cm × 75-μm inside diameter [i.d.] × 360-μm outside diameter [o.d.]; Polymicro Technologies, Phoenix, AZ). The outlet of the nanocapillary LC column was connected to an approximately 3-cm-long nanoESI emitter, which was chemically etched from a 20-μm i.d. × 150-μm o.d. fused silica capillary. An autosampler (LEAP Technologies, Carrboro, NC) with a cooled-drawer sample holder was used for both parallel subsystems. A 5-μl sample loop was used for all experiments except when otherwise noted. Chromatography solvents consisted of 0.1% formic acid in water (mobile phase A) and 0.1% formic acid in acetonitrile (mobile phase B). Chromatographic separation was performed by using gradient elution of 2% to 30% B over 5 min, 30% to 95% B over 35 min, and 95% B for 20 min. The LC system was operated at constant flow rate at 300 nl/min. Data acquisition begins after 10 min of gradient elution to avoid recording of data-poor regions.
(A) Instrumental setup of MetFish methodology. (B) Operation timeline of the two parallel liquid chromatography (LC) columns. Download FIG S1, PDF file, 0.1 MB (149.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
(ii) Mass spectrometry. A TSQ Quantum Ultra mass spectrometer (Thermo Fisher Scientific, San Jose, CA) was used for targeted metabolite quantification. The mass spectrometer was operated with an electrospray voltage of +2,400 V, a capillary offset voltage of 35 V, an ion transfer capillary temperature of 310°C, and a skimmer offset voltage of 0 V. Tube lens voltages were obtained from automatic tuning without further optimization. In selected reaction monitoring (SRM) mode, both Q1 and Q3 were set at a resolution of 0.7 at the full width at half maximum of the LC-MS peak, and Q2 gas pressure was set at 1.5 mTorr. Scan width was set at 0.002 m/z, and dwell time was set at 25 ms. Scan time was typically set at 20 ms for each SRM transition and adjusted as necessary based on transition numbers to provide enough data points across the chromatographic peaks. The collision energy was optimized for each SRM transition by using automatic tuning. Peak identification and quantification were performed using Quan Browser, provided by Xcalibur 2.0 software.
A Q Exactive hybrid quadrupole-Orbitrap high-resolution mass spectrometer (Thermo Fisher Scientific) was used for untargeted metabolite analysis. The mass spectrometer conditions were as follows: electrospray voltage, +3,900 V; ion transfer capillary temperature, 320°C; source DC offset, 21 V; and S-lens radiofrequency (RF) level, 50. The instrument was operated in data-dependent mode acquiring high-resolution full-scan (resolution = 70 000, automatic gain control [AGC] = 3 × 106) spectra followed by MS/MS scans (resolution = 70 000, AGC = 1 × 105) of the top five most abundant ions within the mass range of 200 to 2,000 m/z. An isolation window of 2.0 Da was used. The dynamic exclusion function was not used. Stepped normalized collision energy (NCE) during collision-induced dissociation (CID) was set as 30, 33, and 36. Data were processed using MASIC (63) with a precursor ion tolerance of ±10 ppm and a product ion tolerance of ±0.01 Da. The reporter ions for each chemical tagging approach were identified and the corresponding precursor ion mass spectra were generated by using MASIC. Reporter ions searched were as follows: carboxylic acid tag, 171.104, 234.058, and 336.174 m/z; carbonyl tag, 171.104, 236.074, and 157.088 m/z; hydroxyl tag, 151.063 and 166.087 m/z; amine tag, 157.089, 170.097, and 234.059 m/z.
Analytical method validation.
Fragment ions for each analyte were generated from product ion scans using the TSQ Quantum Ultra mass spectrometer (Thermo Fisher Scientific). The most abundant ion generated during collision-induced fragmentation was selected as the quantification ion, and two other abundant and characteristic fragment ions were used as confirmation ions. The quantification of metabolites was accomplished in the SRM mode by using internal calibration (R2 > 0.99). The calibration curve was produced by plotting the ratio of peak area of analyte to peak area of the internal standard versus the concentration of metabolite standard solution before derivatization. The method detection limits were determined by dilution series of metabolite standard solutions followed by chemical tagging until a signal-to-noise ratio of 10 was noted in the LC-MS chromatogram. The signal-to-noise ratio was calculated based on a Genesis peak detection algorithm provided by the Xcalibur 2.0 software. Reproducibility was examined by evaluating the relative standard deviation (RSD) for 3 days (interday reproducibility) using sample solutions spiked with 5 μM metabolite standards.
Data availability.
The data that support the findings of this study are openly available in MassIVE (accession number MSV000085713, doi:10.25345/C53Q93).
Supplemental text with details on sold-phase extraction (SPE), gas chromatography-mass spectrometry (GC-MS), and Fourier-transform ion cyclotron resonance (FTICR)-MS analysis of metabolites from hypersaline matrices. Download Text S1, DOCX file, 0.03 MB (32.3KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Genomic Science Program (GSP), Office of Biological and Environmental Research (OBER), U.S. Department of Energy (DOE), and is a contribution of the Metabolic and Spatial Interactions in Communities Scientific Focus Area. Additional support was provided by the Pacific Northwest National Laboratory (PNNL), Laboratory Directed Research and Development program, via the Microbiomes in Transition Initiative. P.J.M. was partially supported by funding from the National Sciences Foundation Dimensions of Biodiversity (award no. 1342701).
We thank Susan Welch with the Ohio State University Subsurface Energy Materials Characterization and Analysis Laboratory (SEMCAL) for field sampling assistance, as well as Kelly Wrighton, Mike Wilkins, and Kayla Borton for providing samples and their preparation, Aaron Wright of PNNL for useful discussions and assistance with the MetFish derivatization reactions, and Charles Rice and Ari Jumpponen at Kansas State University for providing the soil samples. We also thank Teresa Fan at the University of Kentucky for kindly providing the QDA reagent. Targeted and untargeted metabolomics analyses using MetFish were performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the U.S. OBER and located at PNNL in Richland, Washington. PNNL is a multiprogram national laboratory operated by Battelle for the Department of Energy (DOE) under contract DE-AC05-76RLO 1830.
C.X. and T.O.M. conceived and designed the method and studies. S.R.L., T.R.C., R.Z., R.J.M., J.K.J., V.L.B., P.J.M., and M.F.R. contributed materials and assisted with experimental design. C.X., R.L.S., N.G.I., Y.M., and B.R.M. performed experiments and data analysis. M.F.R. and J.K.F. provided funding and critical review of the manuscript. C.X., S.P.C., and T.O.M. performed data analysis, interpreted results, and wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Thomas O. Metz, Email: thomas.metz@pnnl.gov.
Marcelino Gutierrez, INDICASAT.
Laura M. Sanchez, University of California, Santa Cruz.
Mauricio Caraballo Rodriguez, University of California, San Diego.
REFERENCES
- 1.Denef VJ, Mueller RS, Banfield JF. 2010. AMD biofilms: using model communities to study microbial evolution and ecological complexity in nature. ISME J 4:599–610. doi: 10.1038/ismej.2009.158. [DOI] [PubMed] [Google Scholar]
- 2.Hemme CL, Deng Y, Gentry TJ, Fields MW, Wu L, Barua S, Barry K, Tringe SG, Watson DB, He Z, Hazen TC, Tiedje JM, Rubin EM, Zhou J. 2010. Metagenomic insights into evolution of a heavy metal-contaminated groundwater microbial community. ISME J 4:660–672. doi: 10.1038/ismej.2009.154. [DOI] [PubMed] [Google Scholar]
- 3.Song HS, Renslow RS, Fredrickson JK, Lindemann SR. 2015. Integrating ecological and engineering concepts of resilience in microbial communities. Front Microbiol 6:1298. doi: 10.3389/fmicb.2015.01298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Konopka A, Lindemann S, Fredrickson J. 2015. Dynamics in microbial communities: unraveling mechanisms to identify principles. ISME J 9:1488–1495. doi: 10.1038/ismej.2014.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson CE, Lopatkin AJ, Craddock TJ, Driscoll WW, Eldakar OT, Lopez JV, Smith RP. 2017. Cooperation and competition shape ecological resistance during periodic spatial disturbance of engineered bacteria. Sci Rep 7:440. doi: 10.1038/s41598-017-00588-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Embree M, Liu JK, Al-Bassam MM, Zengler K. 2015. Networks of energetic and metabolic interactions define dynamics in microbial communities. Proc Natl Acad Sci U S A 112:15450–15455. doi: 10.1073/pnas.1506034112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng K, Zhang Z, Cai W, Liu W, Xu M, Yin H, Wang A, He Z, Deng Y. 2017. Biodiversity and species competition regulate the resilience of microbial biofilm community. Mol Ecol 26:6170–6182. doi: 10.1111/mec.14356. [DOI] [PubMed] [Google Scholar]
- 8.Fredrickson JK. 2015. Ecological communities by design. Science 348:1425–1427. doi: 10.1126/science.aab0946. [DOI] [PubMed] [Google Scholar]
- 9.Mee MT, Collins JJ, Church GM, Wang HH. 2014. Syntrophic exchange in synthetic microbial communities. Proc Natl Acad Sci U S A 111:E2149–E2156. doi: 10.1073/pnas.1405641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phelan VV, Liu W-T, Pogliano K, Dorrestein PC. 2012. Microbial metabolic exchange—the chemotype-to-phenotype link. Nat Chem Biol 8:26–35. doi: 10.1038/nchembio.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson JC, Wan Y, Kim Y-M, Pasa-Tolic L, Metz TO, Peck SC. 2014. Decreased abundance of type III secretion system-inducing signals in Arabidopsis mkp1 enhances resistance against Pseudomonas syringae. Proc Natl Acad Sci U S A 111:6846–6851. doi: 10.1073/pnas.1403248111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bednarek P. 2012. Chemical warfare or modulators of defence responses—the function of secondary metabolites in plant immunity. Curr Opin Plant Biol 15:407–414. doi: 10.1016/j.pbi.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Berdy J. 2005. Bioactive microbial metabolites. The J Antibiotics 58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 14.Harvey AL, Edrada-Ebel R, Quinn RJ. 2015. The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discov 14:111–129. doi: 10.1038/nrd4510. [DOI] [PubMed] [Google Scholar]
- 15.Keller M, Zengler K. 2004. Tapping into microbial diversity. Nat Rev Microbiol 2:141–150. doi: 10.1038/nrmicro819. [DOI] [PubMed] [Google Scholar]
- 16.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov 6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 17.Pan G, Xu Z, Guo Z, Ma M, Yang D, Zhou H, Gansemans Y, Zhu X, Huang Y, Zhao L-X. 2017. Discovery of the leinamycin family of natural products by mining actinobacterial genomes. Proc Natl Acad Sci U S A:201716245 doi: 10.1073/pnas.1716245115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aebersold R, Mann M. 2016. Mass-spectrometric exploration of proteome structure and function. Nature 537:347–355. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 19.Want EJ, Cravatt BF, Siuzdak G. 2005. The expanding role of mass spectrometry in metabolite profiling and characterization. Chembiochem 6:1941–1951. doi: 10.1002/cbic.200500151. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung Y-M, Wu Y, Schauer P, Smith JD, Allayee H, Tang WHW, DiDonato JA, Lusis AJ, Hazen SL. 2011. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mehmood S, Marcoux J, Gault J, Quigley A, Michaelis S, Young SG, Carpenter EP, Robinson CV. 2016. Mass spectrometry captures off-target drug binding and provides mechanistic insights into the human metalloprotease ZMPSTE24. Nature Chem 8:1152–1158. doi: 10.1038/nchem.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilhelm M, Schlegl J, Hahne H, Gholami AM, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, Mathieson T, Lemeer S, Schnatbaum K, Reimer U, Wenschuh H, Mollenhauer M, Slotta-Huspenina J, Boese J-H, Bantscheff M, Gerstmair A, Faerber F, Kuster B. 2014. Mass-spectrometry-based draft of the human proteome. Nature 509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 23.Girolamo FD, Lante I, Muraca M, Putignani L. 2013. The role of mass spectrometry in the “omics” era. Curr Org Chem 17:2891–2905. doi: 10.2174/1385272817888131118162725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patti GJ, Yanes O, Siuzdak G. 2012. Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol 13:263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.King R, Bonfiglio R, Fernandez-Metzler C, Miller-Stein C, Olah T. 2000. Mechanistic investigation of ionization suppression in electrospray ionization. J Am Soc Mass Spectrom 11:942–950. doi: 10.1016/S1044-0305(00)00163-X. [DOI] [PubMed] [Google Scholar]
- 26.Sterner J, Johnston M, Nicol G, Ridge D. 2000. Signal suppression in electrospray ionization Fourier transform mass spectrometry of multi‐component samples. J Mass Spectrom 35:385–391. doi:. [DOI] [PubMed] [Google Scholar]
- 27.Bonfiglio R, King RC, Olah TV, Merkle K. 1999. The effects of sample preparation methods on the variability of the electrospray ionization response for model drug compounds. Rapid Commun Mass Spectrom 13:1175–1185. doi:. [DOI] [PubMed] [Google Scholar]
- 28.Oren A. 2016. Life in hypersaline environments, p 301–339. In Hurst CJ (ed), Their world: a diversity of microbial environments. Springer International Publishing, Cham, Switzerland. doi: 10.1007/978-3-319-28071-4_8. [DOI] [Google Scholar]
- 29.Oren A, Elevi Bardavid R, Mana L. 2014. Perchlorate and halophilic prokaryotes: implications for possible halophilic life on Mars. Extremophiles 18:75–80. doi: 10.1007/s00792-013-0594-9. [DOI] [PubMed] [Google Scholar]
- 30.Jacob JH, Hussein EI, Shakhatreh MAK, Cornelison CT. 2017. Microbial community analysis of the hypersaline water of the Dead Sea using high-throughput amplicon sequencing. Microbiologyopen 6:e00500. doi: 10.1002/mbo3.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vera-Gargallo B, Navarro-Sampedro L, Carballo M, Ventosa A. 2018. Metagenome sequencing of prokaryotic microbiota from two hypersaline soils of the Odiel salt marshes in Huelva, Southwestern Spain. Genome Announc 6:e00140-18. doi: 10.1128/genomeA.00140-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pontefract A, Zhu TF, Walker VK, Hepburn H, Lui C, Zuber MT, Ruvkun G, Carr CE. 2017. Microbial diversity in a hypersaline sulfate lake: a terrestrial analog of ancient Mars. Front Microbiol 8:1819. doi: 10.3389/fmicb.2017.01819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghai R, Pašić L, Fernández AB, Martin-Cuadrado A-B, Mizuno CM, McMahon KD, Papke RT, Stepanauskas R, Rodriguez-Brito B, Rohwer F, Sánchez-Porro C, Ventosa A, Rodríguez-Valera F. 2011. New abundant microbial groups in aquatic hypersaline environments. Sci Rep 1:135. doi: 10.1038/srep00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vera-Gargallo B, Chowdhury TR, Brown J, Fansler SJ, Durán-Viseras A, Sánchez-Porro C, Bailey VL, Jansson JK, Ventosa A. 2019. Spatial distribution of prokaryotic communities in hypersaline soils. Sci Rep 9:1769. doi: 10.1038/s41598-018-38339-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petras D, Minich JJ, Cancelada LB, Torres RR, Kunselman E, Wang M, White ME, Allen EE, Prather KA, Aluwihare LI, Dorrenstein PD. 2019. Non-target tandem mass spectrometry enables the prioritization of anthropogenic pollutants in seawater along the northern San Diego coast. ChemRxiv doi: 10.26434/chemrxiv.9817133.v2. [DOI] [Google Scholar]
- 36.Johnson WM, Kido Soule MC, Kujawinski EB. 2017. Extraction efficiency and quantification of dissolved metabolites in targeted marine metabolomics. Limnol Oceanogr Methods 15:417–428. doi: 10.1002/lom3.10181. [DOI] [Google Scholar]
- 37.Sogin EM, Puskás E, Dubilier N, Liebeke M. 2019. Marine metabolomics: a method for nontargeted measurement of metabolites in seawater by gas chromatography-mass spectrometry. mSystems 4:e00638-19. doi: 10.1128/mSystems.00638-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zabalegui N, Manzi M, Depoorter A, Hayeck N, Roveretto M, Li C, van Pinxteren M, Herrmann H, George C, Monge ME. 2020. Seawater analysis by ambient mass-spectrometry-based seaomics. Atmos Chem Phys 20:6243–6257. doi: 10.5194/acp-20-6243-2020. [DOI] [Google Scholar]
- 39.Bouatra S, Aziat F, Mandal R, Guo AC, Wilson MR, Knox C, Bjorndahl TC, Krishnamurthy R, Saleem F, Liu P, Dame ZT, Poelzer J, Huynh J, Yallou FS, Psychogios N, Dong E, Bogumil R, Roehring C, Wishart DS. 2013. The human urine metabolome. PLoS One 8:e73076. doi: 10.1371/journal.pone.0073076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jain A, Li XH, Chen WN. 2019. An untargeted fecal and urine metabolomics analysis of the interplay between the gut microbiome, diet and human metabolism in Indian and Chinese adults. Sci Rep 9:9191. doi: 10.1038/s41598-019-45640-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller IJ, Peters SR, Overmyer KA, Paulson BR, Westphall MS, Coon JJ. 2019. Real-time health monitoring through urine metabolomics. NPJ Digit Med 2:109. doi: 10.1038/s41746-019-0185-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Santen JA, Jacob G, Singh AL, Aniebok V, Balunas MJ, Bunsko D, Neto FC, Castaño-Espriu L, Chang C, Clark TN, Cleary Little JL, Delgadillo DA, Dorrestein PC, Duncan KR, Egan JM, Galey MM, Haeckl FPJ, Hua A, Hughes AH, Iskakova D, Khadilkar A, Lee J-H, Lee S, LeGrow N, Liu DY, Macho JM, McCaughey CS, Medema MH, Neupane RP, O’Donnell TJ, Paula JS, Sanchez LM, Shaikh AF, Soldatou S, Terlouw BR, Tran TA, Valentine M, van der Hooft JJJ, Vo DA, Wang M, Wilson D, Zink KE, Linington RG. 2019. The natural products atlas: an open access knowledge base for microbial natural products discovery. ACS Cent Sci 5:1824–1833. doi: 10.1021/acscentsci.9b00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sajed T, Marcu A, Ramirez M, Pon A, Guo AC, Knox C, Wilson M, Grant JR, Djoumbou Y, Wishart DS. 2016. ECMDB 2.0: a richer resource for understanding the biochemistry of E. coli. Nucleic Acids Res 44:D495–D501. doi: 10.1093/nar/gkv1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dreher K. 2014. Putting the plant metabolic network pathway databases to work: going offline to gain new capabilities, p 151–171. In Sriram G (ed), Plant metabolism: methods and protocols. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 45.Carlson EE, Cravatt BF. 2007. Chemoselective probes for metabolite enrichment and profiling. Nat Methods 4:429–435. doi: 10.1038/nmeth1038. [DOI] [PubMed] [Google Scholar]
- 46.Mattingly SJ, Xu T, Nantz MH, Higashi RM, Fan TW-M. 2012. A carbonyl capture approach for profiling oxidized metabolites in cell extracts. Metabolomics 8:989–996. doi: 10.1007/s11306-011-0395-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gros C, Labouesse B. 1969. Study of the dansylation reaction of amino acids, peptides and proteins. Eur J Biochem 7:463–470. doi: 10.1111/j.1432-1033.1969.tb19632.x. [DOI] [PubMed] [Google Scholar]
- 48.Guo K, Li L. 2009. Differential 12C-/13C-isotope dansylation labeling and fast liquid chromatography/mass spectrometry for absolute and relative quantification of the metabolome. Anal Chem 81:3919–3932. doi: 10.1021/ac900166a. [DOI] [PubMed] [Google Scholar]
- 49.Zhao S, Dawe M, Guo K, Li L. 2017. Development of high-performance chemical isotope labeling LC-MS for profiling the carbonyl submetabolome. Anal Chem 89:6758–6765. doi: 10.1021/acs.analchem.7b01098. [DOI] [PubMed] [Google Scholar]
- 50.Zhao S, Luo X, Li L. 2016. Chemical isotope labeling LC-MS for high coverage and quantitative profiling of the hydroxyl submetabolome in metabolomics. Anal Chem 88:10617–10623. doi: 10.1021/acs.analchem.6b02967. [DOI] [PubMed] [Google Scholar]
- 51.Guo K, Li L. 2010. High-performance isotope labeling for profiling carboxylic acid-containing metabolites in biofluids by mass spectrometry. Anal Chem 82:8789–8793. doi: 10.1021/ac102146g. [DOI] [PubMed] [Google Scholar]
- 52.Thompson A, Schäfer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AKA, Hamon C. 2003. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem 75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 53.Lindemann SR, Moran JJ, Stegen JC, Renslow RS, Hutchison JR, Cole JK, Dohnalkova AC, Tremblay J, Singh K, Malfatti SA, Chen F, Tringe SG, Beyenal H, Fredrickson JK. 2013. The epsomitic phototrophic microbial mat of Hot Lake, Washington: community structural responses to seasonal cycling. Front Microbiol 4:323. doi: 10.3389/fmicb.2013.00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cole JK, Hutchison JR, Renslow RS, Kim Y-M, Chrisler WB, Engelmann HE, Dohnalkova AC, Hu D, Metz TO, Fredrickson JK, Lindemann SR. 2014. Phototrophic biofilm assembly in microbial-mat-derived unicyanobacterial consortia: model systems for the study of autotroph-heterotroph interactions. Front Microbiol 5:109. doi: 10.3389/fmicb.2014.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. 1989. Electrospray ionization for mass spectrometry of large biomolecules. Science 246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 56.Zachara JM, Moran JJ, Resch CT, Lindemann SR, Felmy AR, Bowden ME, Cory AB, Fredrickson JK. 2016. Geo-and biogeochemical processes in a heliothermal hypersaline lake. Geochim Cosmochim Acta 181:144–163. doi: 10.1016/j.gca.2016.02.001. [DOI] [Google Scholar]
- 57.Lindemann SR, Mobberley JM, Cole JK, Markillie LM, Taylor RC, Huang E, Chrisler WB, Wiley HS, Lipton MS, Nelson WC, Fredrickson JK, Romine MF. 2017. Predicting species-resolved macronutrient acquisition during succession in a model phototrophic biofilm using an integrated ‘omics. Approach Front Microbiol 8:1020. doi: 10.3389/fmicb.2017.01020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vance ED, Brookes PC, Jenkinson DS. 1987. Microbial biomass measurements in forest soils—the use of the chloroform fumigation incubation method in strongly acid soils. Soil Biol Biochem 19:697–702. doi: 10.1016/0038-0717(87)90051-4. [DOI] [Google Scholar]
- 59.Brookes PC, Landman A, Pruden G, Jenkinson DS. 1985. Chloroform fumigation and the release of soil-nitrogen—a rapid direct extraction method to measure microbial biomass nitrogen in soil. Soil Biol Biochem 17:837–842. doi: 10.1016/0038-0717(85)90144-0. [DOI] [Google Scholar]
- 60.Vance ED, Brookes PC, Jenkinson DS. 1987. An extraction method for measuring soil microbial biomass C. Soil Biol Biochem 19:703–707. doi: 10.1016/0038-0717(87)90052-6. [DOI] [Google Scholar]
- 61.Tate KR, Ross DJ, Feltham CW. 1988. A direct extraction method to estimate soil microbial-C—effects of experimental-variables and some different calibration procedures. Soil Biol Biochem 20:329–335. doi: 10.1016/0038-0717(88)90013-2. [DOI] [Google Scholar]
- 62.Makarov MI, Shuleva MS, Malysheva TI, Menyailo OV. 2013. Solubility of the labile forms of soil carbon and nitrogen in K2SO4 of different concentrations. Eurasian Soil Sci 46:369–374. doi: 10.1134/S1064229313040091. [DOI] [Google Scholar]
- 63.Monroe ME, Shaw JL, Daly DS, Adkins JN, Smith RD. 2008. MASIC: a software program for fast quantitation and flexible visualization of chromatographic profiles from detected LC–MS(/MS) features. Comput Biol Chem 32:215–217. doi: 10.1016/j.compbiolchem.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, Nguyen DD, Watrous J, Kapono CA, Luzzatto-Knaan T, Porto C, Bouslimani A, Melnik AV, Meehan MJ, Liu W-T, Crüsemann M, Boudreau PD, Esquenazi E, Sandoval-Calderón M, Kersten RD, Pace LA, Quinn RA, Duncan KR, Hsu C-C, Floros DJ, Gavilan RG, Kleigrewe K, Northen T, Dutton RJ, Parrot D, Carlson EE, Aigle B, Michelsen CF, Jelsbak L, Sohlenkamp C, Pevzner P, Edlund A, McLean J, Piel J, Murphy BT, Gerwick L, Liaw C-C, Yang Y-L, Humpf H-U, Maansson M, Keyzers RA, Sims AC, Johnson AR, Sidebottom AM, Sedio BE, et al. 2016. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol 34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Little JL. 1999. Artifacts in trimethylsilyl derivatization reactions and ways to avoid them. J Chromatogr A 844:1–22. doi: 10.1016/S0021-9673(99)00267-8. [DOI] [PubMed] [Google Scholar]
- 66.Herrington J, Zhang L, Whitaker D, Sheldon L, Zhang J. 2005. Optimizing a dansylhydrazine (DNSH) based method for measuring airborne acrolein and other unsaturated carbonyls. J Environ Monit 7:969–976. doi: 10.1039/b502063h. [DOI] [PubMed] [Google Scholar]
- 67.Xia J, Sinelnikov IV, Han B, Wishart DS. 2015. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res 43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) GC-MS chromatogram of amino acid mix in water followed by a methanol extraction. (B) GC-MS chromatogram of amino acid mix in water followed by an MPLEx (chloroform-methanol) extraction. (C) GC-MS chromatogram of amino acid mix in 400 μM MgSO4 followed by a methanol extraction. (D) GC-MS chromatogram of amino acid mix in 400 μM MgSO4 followed by a MPLEx extraction. (E) GC-MS chromatogram of amino acid mix in 2 M MgSO4 followed by a methanol extraction. (F) GC-MS chromatogram of amino acid mix in 2 M MgSO4 followed by a MPLEx extraction. Insets show the chromatogram zoomed in. Download FIG S2, DOCX file, 0.3 MB (267.9KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
FTICR-MS spectra of sodium pyruvate in (A) Nanopure H2O, (B) 400 μM MgSO4, and (C) 2 M MgSO4, with the QDA (N-[2-(aminooxy)ethyl]-N,N-dimethyl-1-dodecylammonium iodide)-pyruvate adduct detected at 343.2955 m/z. The top 3 panes in each figure show three independent replicates of QDA derivatization of pyruvate, followed by FTICR-MS analysis. The bottom pane shows an instrument blank. Download FIG S3, DOCX file, 0.04 MB (37.8KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Performance of commercial SPE columns for separation of metabolites from high-salt matrices, as determined by nuclear magnetic resonance (NMR) spectroscopy. Generally, SPE columns were not effective in recovering metabolites from either water or hypersaline media, although some column chemistries were effective to some extent for certain metabolites. Download Table S1, DOCX file, 0.01 MB (15.6KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Metabolite identifications for peaks/features in the chromatogram shown in Fig. S2A. Download Table S4, DOCX file, 0.01 MB (14.1KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Common fragment ions generated during tandem mass spectrometry (MS/MS) for MetFish reagents. Download Table S2, DOCX file, 0.01 MB (13.1KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Limits of quantification (LOQ), linearity, and interday reproducibility of (A) amine, (B) carboxylic acid (C) carbonyl, and (D) hydroxyl tagging approaches. Download Table S3, DOCX file, 0.03 MB (28.9KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Features detected in the untargeted metabolomics analysis by use of each of the four MetFish tags. Data have been processed to remove low-abundance ions (noise), reduce false identifications, and remove duplicate features. The precursor ions that generated all 3 reporter ions (170, 172, and 252) were considered potential features, and the tag mass was deducted from the precursor mass to obtain the metabolite mass. Download Table S5, XLSX file, 0.01 MB (15.6KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
GNPS spectral library search matches with a cosine score greater than 0.7, for untargeted discovery of metabolites in the fracking fluid datasets collected using data-dependent MS/MS. Download Table S6, XLSX file, 0.03 MB (35.9KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
(A) Instrumental setup of MetFish methodology. (B) Operation timeline of the two parallel liquid chromatography (LC) columns. Download FIG S1, PDF file, 0.1 MB (149.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Supplemental text with details on sold-phase extraction (SPE), gas chromatography-mass spectrometry (GC-MS), and Fourier-transform ion cyclotron resonance (FTICR)-MS analysis of metabolites from hypersaline matrices. Download Text S1, DOCX file, 0.03 MB (32.3KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
The data that support the findings of this study are openly available in MassIVE (accession number MSV000085713, doi:10.25345/C53Q93).
Supplemental text with details on sold-phase extraction (SPE), gas chromatography-mass spectrometry (GC-MS), and Fourier-transform ion cyclotron resonance (FTICR)-MS analysis of metabolites from hypersaline matrices. Download Text S1, DOCX file, 0.03 MB (32.3KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.