Significance
Innate lymphoid cells hold great promise for the treatment of metastases. Development of effective therapies based on these versatile immune cells, however, is hampered by our limited knowledge of their behavior in the metastatic niche. Here, we describe that defense against liver metastasis requires collaboration between two innate lymphocyte subsets, conventional NK cells (cNKs) and tissue-resident type I innate lymphoid cells (trILC1s). We show that different cancers generate their own particular metastatic niche–inducing specific changes in cNKs and trILC1s. Furthermore, we uncover specific cNK subsets that can be manipulated to improve their antimetastatic potential. Our work contributes to understanding how cancer-specific factors and hepatic innate lymphocytes exert mutual influence and how this can be exploited for therapeutic purposes.
Keywords: innate lymphocytes, metastatic surveillance, tissue-resident ILC1s, conventional NK cells
Abstract
The liver is a major metastatic target organ, and little is known about the role of immunity in controlling hepatic metastases. Here, we discovered that the concerted and nonredundant action of two innate lymphocyte subpopulations, conventional natural killer cells (cNKs) and tissue-resident type I innate lymphoid cells (trILC1s), is essential for antimetastatic defense. Using different preclinical models for liver metastasis, we found that trILC1 controls metastatic seeding, whereas cNKs restrain outgrowth. Whereas the killing capacity of trILC1s was not affected by the metastatic microenvironment, the phenotype and function of cNK cells were affected in a cancer type–specific fashion. Thus, individual cancer cell lines orchestrate the emergence of unique cNK subsets, which respond differently to tumor-derived factors. Our findings will contribute to the development of therapies for liver metastasis involving hepatic innate cells.
Hepatic metastases are a major cause of cancer-related death in disparate cancer types (1). The healthy liver contains a large population of innate lymphocytes including conventional natural killer cells (cNKs) and tissue-resident type I innate lymphoid cells (trILC1s). Although hepatic cNKs and trILC1s share some features, they differ in phenotype, ontogeny, and function (2–4). In the steady state, both cNKs and trILC1s are defined as Lin− CD45+NK1.1+NKp46+ lymphocytes, with cNKs being CD49a−CD49b+ and trILC1s CD49a+CD49b−. Whereas cNKs rely on the transcription factors Eomes (5) and Nfil3 (6) for their development, trILC1s rather depend on T-bet (7), Hobit (8), and the aryl hydrocarbon receptor (AhR) (9). Furthermore, cNKs express higher amounts of markers associated with maturation (CD11b, CD43, KLRG-1) compared to trILC1s, which rather feature the cytotoxic molecule TNF-related apoptosis-inducing ligand and markers for tissue residency (e.g., CD69) (10).
Upon activation, both cNKs and trILC1s produce IFN-γ and are cytotoxic (4, 11). However, trILC1s express more inhibitory receptors (2, 3), suggesting they perform immune regulatory functions. Nevertheless, the main roles ascribed to trILC1s so far include immune responses against haptens and early protection against viral antigens and acute liver injury (3, 12, 13).
The protective role of NK cells against spontaneous and experimental metastasis to the lungs is well established (14–18). At this particular site, cNKs progressively lose their protective capacities, which limits their antimetastatic effect (19–21). Whether innate lymphocytes control hepatic metastases and the individual contributions of cNKs and trILC1s is largely unknown.
The understanding of innate immune defense against liver metastasis is pivotal to exploiting innate lymphocytes for novel cancer therapies. Here, we sought to unravel the mechanism that governs metastatic control in this immunologically unique organ and discovered that both cNKs and trILC1s controlled the metastatic burden in the liver but seemed to have nonredundant roles. In summary, we found that cNKs and trILC1 collaborate in a nonredundant fashion to control liver metastases and that the metastatic niche influences each innate subset differently and in a cancer type–specific way.
Results
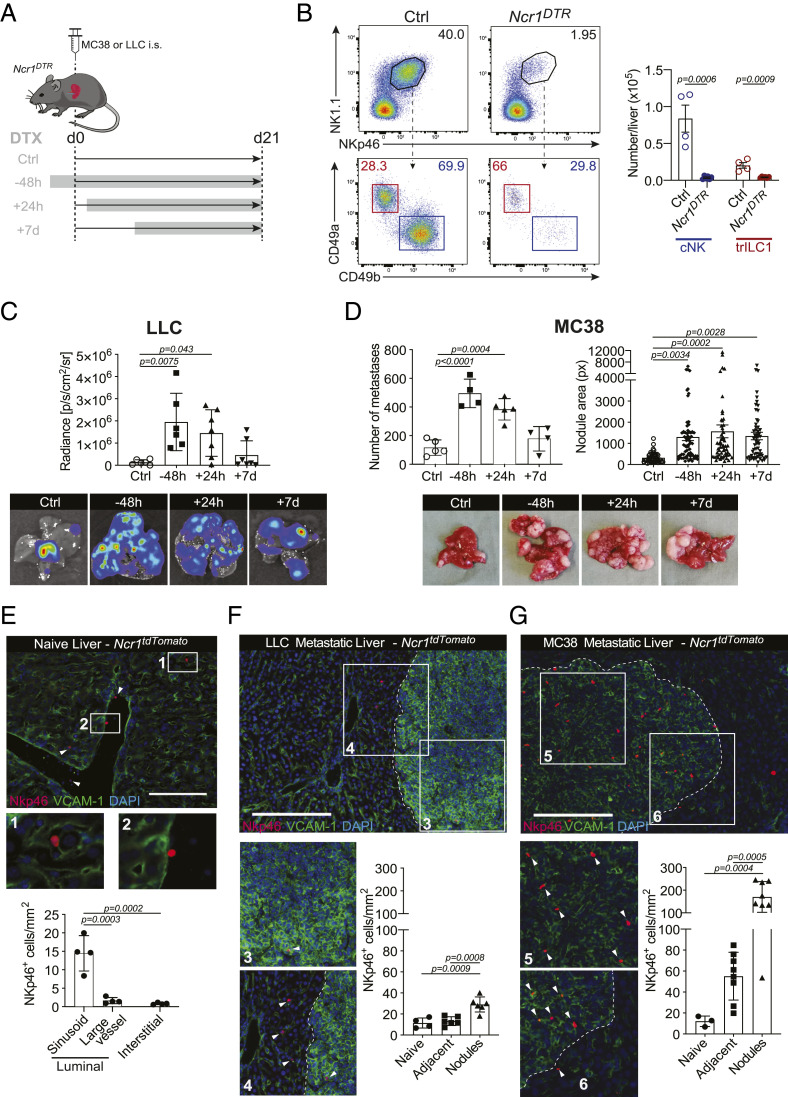
Control of Hepatic Metastases Depends on NKp46+ Innate Cells.
Despite the well-accepted role of NK cells in the control of tumor progression, their influence on hepatic metastases is still poorly understood. We used two different models for liver metastasis to address this question: MC38 colon carcinoma cells and Lewis lung carcinoma cells (LLC). We selected these two models based on their intrinsic difference in the activation of innate immune pathways. Whereas MC38 cells produce cyclic GMP-AMP (cGAMP), resulting in the local production of type I interferon (IFN) and consequent activation of NK and CD8+ T cells, LLC do not produce cGAMP and, consequently, do not activate downstream pathways (22, 23). We induced liver metastasis from MC38 or LLC carcinoma cells and depleted NKp46+ (encoded by Ncr1) subsets at different time points by administering diphtheria toxin (DTX) to Ncr1iCreR26RiDTR mice (Fig. 1 A and B and SI Appendix, Fig. S1A). The metastatic load increased dramatically in both models when NKp46+ cells were depleted 48 h before or 24 h after tumor cell injection (Fig. 1 C and D and SI Appendix, Fig. S1B). However, their depletion 7 d after tumor injection resulted in larger liver metastases from MC38 but not LLC cells (Fig. 1 C and D and SI Appendix, Fig. S1B). Thus, NKp46+ cells control the seeding of MC38 and LLC cells but only the outgrowth of MC38 cells, suggesting that they become ineffective against progressing LLC metastases. In this experimental setup, CD8+ T cells are dispensable for restricting liver metastases (SI Appendix, Fig. S1C).
Fig. 1.
Control of hepatic metastases depends on NKp46+ cells. (A) Experimental schedule for NKp46+ depletion. NKp46+ cells were depleted by an injection of 250 ng DTX i.p. 48 h before, 24 h after, or 7 d after tumor injection. Depletion was maintained until the endpoint. Ncr1DTR = Ncr1iCre/wt.R26RiDTR/wt; Ctrl = undepleted control mice (Ncr1iCre/wt.R26Rwt/wt, group injected at time point −48 h); i.s. = intrasplenic. (B) Representative dot plots (Left) and quantification (Right) of cNKs and trILC1s in Ctrl and Ncr1DTR livers. Samples were pregated on single live CD45+lineage− cells and subsequently gated on NK1.1+NKp46+ cells. cNK = conventional NK cells, CD49a−CD49b+; trILC1 = tissue-resident ILC1s, CD49a+CD49b−. (C, Upper) Quantification of LLC liver nodules by in vivo imaging system (IVIS) at the endpoint. Bioluminescence measurements on the whole liver are shown. (Lower) Representative IVIS images of metastatic livers from each group at the endpoint. The bar represents the mean ± SD, and the symbols represent livers from individual mice. One-way ANOVA with Tukey’s multiple comparisons test. The experiment was performed twice with similar results. (D, Upper) Quantification of MC38 macroscopic liver nodules and their area at the endpoint. The area of each hepatic nodule is measured manually using the drawing tool from ImageJ. Each point represented corresponds to the area of one nodule displayed in pixels (px). (Lower) Representative images of metastatic livers from each group at the endpoint. The bar represents the mean ± SD, and the symbols represent livers from individual mice. One-way ANOVA with Tukey’s multiple comparisons test. The experiment was performed twice with similar results. (E–G) Representative immunofluorescence image and quantification of NKp46+ cell vascular localization. Ncr1tdTomato = Ncr1Cre/wt.R26RAi14/wt. Three livers per experimental condition were analyzed, and each symbol represents an individual section. The distribution of NKp46+ cells was determined based on their positioning relative to the liver vasculature (luminal side of the sinusoids or large vessels, or interstitial). (E) Naïve liver. (Scale bar, 50 µm.) (F) LLC metastatic liver. (G) MC38 metastatic liver. (Scale bar, 100 µm.)
Because NKp46+ cells lose control over LLC metastases after some time, we investigated the location of these cells within the hepatic MC38 and LLC metastatic niche. For this purpose, we used Ncr1iCreTdtomatofl/wt mice, in which NKp46+ cells are irreversibly marked with TdTomato (SI Appendix, Fig. S1D), and visualized tumor cells and the sinusoidal endothelium with an antibody against VCAM-1. In healthy livers, NKp46+ cells were mostly located along the luminal aspect of the sinusoids (Fig. 1E). NKp46+ cells were abundant in MC38 nodules, whereas they were sparse in LLC nodules (Fig. 1 F and G). Thus, failure to infiltrate LLC nodules may explain why NKp46+ cells are relatively ineffective in controlling disseminated LLC cells in the liver.
Together, these findings highlight the crucial contribution of cNKs and trILC1s to control hepatic metastasis and reveal important differences linked to cancer cell–intrinsic traits.
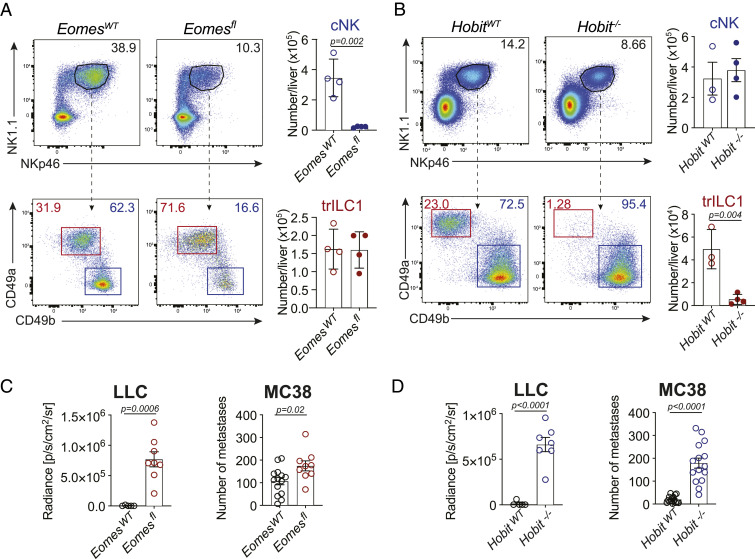
Division of Labor between Hepatic cNKs and trILC1s in the Control of Metastasis.
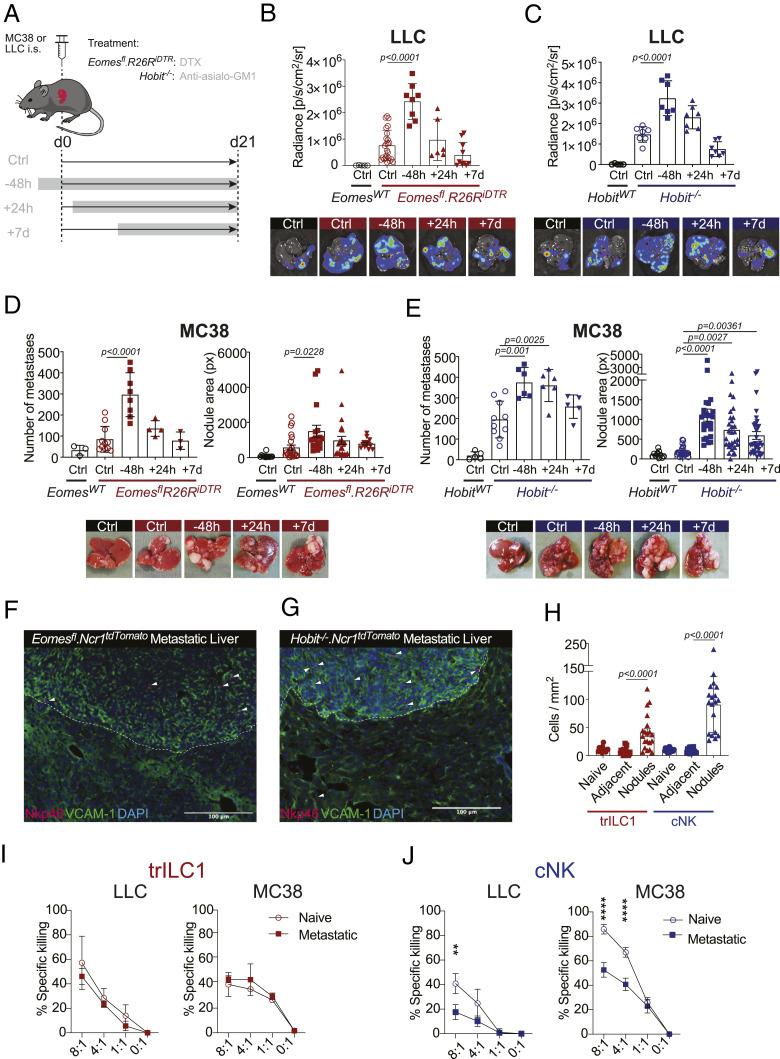
The liver contains a high number of cNKs and trILC1s. Generally, CD49a−CD49b+ cNKs are phenotypically more mature and differentiated, while CD49a+CD49b− trILC1s have a combined signature of activation and immaturity (3). To determine the individual contribution of cNKs and trILC1s to the control of hepatic metastases, we used two genetically modified mouse lines that lack one of the two innate subsets: Ncr1iCreEomesfl/fl mice (5, 12) severely reduced in cNKs but not trILC1s (Fig. 2A) and Hobit-deficient (Hobit−/−) mice (8) selectively lacking trILC1s but not cNKs (Fig. 2B). We found that both cNK and trILC1s restricted the progression of MC38- and LLC-derived liver metastasis as shown by the higher metastatic load in Ncr1iCreEomesfl/fl and Hobit−/− livers in comparison to their respective littermate controls (Fig. 2 C and D). We corroborated the antimetastatic properties of cNKs using Nfil3−/− mice, which are also devoid of cNKs (24) (SI Appendix, Fig. S1 E and F). The difference between LLC and MC38 metastases concerning the time window of NKp46+ cell-dependent control led us to investigate the temporal requirements for cNKs and trILC1s to exert their antimetastatic activity. Therefore, we first crossed mice missing cNKs (Ncr1iCreEomesfl/fl) to R26RiDTR mice and additionally eliminated trILC1s via DTX administration at different time points relative to tumor cell injection as shown in Fig. 3A. The depletion of trILC1s 48 h prior to tumor cell injection—but not thereafter—resulted in an increased load of MC38- or LLC-derived metastases (Fig. 3 B and C and SI Appendix, Fig. S1G), indicating that trILC1s mainly control metastatic seeding. To study the antimetastatic potential of cNKs, we performed a similar time course experiment in trILC1-deficient Hobit−/− mice and depleted cNKs using anti-asialo–GM1 antibodies (as shown in Fig. 3A). cNKs controlled the early stages of both LLC- and MC38-derived metastasis (Fig. 3 D and E and SI Appendix, Fig. S1H). In contrast, however, cNKs were ineffective against late-stage LLC nodules but still restricted late-stage MC38 nodules (Fig. 3 D and E and SI Appendix, Fig. S1H). Using Ncr1iCreEomesfl/fl TdTomatofl/wt and Ncr1iCreHobit−/− TdTomatofl/wt mice, we found that MC38-derived nodules were better infiltrated by cNK cells in comparison to trILC1s (Fig. 3 F–H). In accordance, the immunostaining of MC38 metastases using Eomes revealed that the frequency of cNK cells (Eomes+) in the metastatic nodules was higher than that of trILC1s (Eomes−) in the adjacent tissue or the healthy liver (SI Appendix, Fig. S1I). Furthermore, both the cNK cell and the trILC1 subset remained associated to the sinusoids in the adjacent tissue from metastatic livers (SI Appendix, Fig. S1J).
Fig. 2.
Division of labor between hepatic cNKs and trILC1s in the control of metastasis. (A and B) Representative dot plots and quantification of cNKs and trILC1s. Samples were pregated on single live CD45+lineage− cells and subsequently gated on NK1.1+NKp46+ cells. cNK = conventional NK cells, CD49a−CD49b+; trILC1 = tissue-resident ILC1s, CD49a+CD49b−. (A) EomesWT and Eomesfl livers. EomesWT = Ncr1iCre/wt.Eomeswt/wt; Eomesfl = Ncr1iCre/wt.Eomesfl/fl. (B) HobitWT and Hobit−/− livers. (C and D) Quantification of the metastatic burden from LLC and MC38 cells in (C) EomesWT and Eomesfl mice and (D) HobitWT and Hobit−/− mice. Quantification of LLC liver nodules was performed by in vivo imaging system at the endpoint. Bioluminescence measurements on the whole liver are shown. Quantification of MC38 macroscopic liver nodules and their area at the endpoint. The area of each hepatic nodule is measured manually using the drawing tool from ImageJ. Each point represented corresponds to the area of one nodule displayed in pixels (px). The bar represents the mean ± SD, and the symbols represent livers from individual mice. Student’s unpaired t test. LLC: The experiment was performed twice with similar results. MC38: Pooled data from four experiments.
Fig. 3.
cNKs but not trILC1s control advanced metastatic disease. (A) Kinetics of NKp46+ cell depletion. trILC1s in Eomesfl.R26RiDTR mice were depleted by i.p. injection of DTX. cNKs in HobitWT and Hobit−/− were depleted by i.p. injection of anti-Asialo-GM1. Depletion was performed 48 h before, 24 h after, or 7 d after tumor injection and maintained until the endpoint (day 18). i.s. = intrasplenic; EomesWT Ctrl = Ncr1iCre/wt.Eomeswt/wt.R26Rwt/wt; Eomesfl.R26RiDTR Ctrl = Ncr1iCre/wt.Eomesfl/fl.R26Rwt/wt; Eomesfl.R26RiDTR NKp46+ depleted = Ncr1iCre/wt.Eomesfl/fl.R26RiDTR/wt. (B and C, Upper) Quantification of LLC liver nodules was performed by ex vivo in vivo imaging system imaging at the endpoint. Bioluminescence measurements on the whole liver are shown. (Lower) Representative measurements of metastatic livers at endpoint. The bar represents the mean ± SD, and the symbols represent livers from individual mice. One-way ANOVA with Tukey’s multiple comparisons test. The experiment was performed twice with similar results. (D and E, Upper) Quantification of MC38 macroscopic liver nodules and area at the endpoint. The area of each hepatic nodule is measured manually using the drawing tool from ImageJ. Each point represented corresponds to the area of one nodule displayed in pixels (px). (Lower) Representative images of metastatic livers at endpoint. The bar represents the mean ± SD, and the symbols represent livers from individual mice. One-way ANOVA with Tukey’s multiple comparisons test. The experiment was performed twice with similar results. (F and G) Representative immunofluorescence images of MC38 metastatic livers from (F) Ncr1iCre/wt.Eomesfl/fl.R26RAi14/wt mice and (G) Ncr1iCre/wt.Hobit−/−.R26RAi14/wt mice. (H) Quantification of the localization of cNKs and trILC1s. Groups contained three to four MC38 metastatic livers from Ncr1iCre/wt.Eomesfl/fl.R26RAi14/wt mice (trILC1s) and Ncr1iCre/wt.Hobit−/−.R26RAi14/wt mice (cNKs), each symbol represents a liver section. (I) trILC1s and (J) cNKs were sorted from naïve, LLC metastatic, or MC38 metastatic livers and incubated with the respective cell lines at different effector:target ratios for 12 h. The percentage of specific killing was calculated as follows: [(% cell death in the presence of NKp46+ cells) − (% spontaneous cell death)] ÷ (100 − % spontaneous cell death). The results are displayed as mean of 3 replicates ± SD. Student’s unpaired t test: **P = 0.0076, ****P < 0.0001. The experiment was performed twice with similar results.
The apparent correlation between the capacity of cNKs to control established metastasis and to infiltrate the latter led us to study the functionality of cNKs in advanced metastatic disease (around 3 wk after tumor injection). We found impaired cytotoxicity of cNKs from MC38 and LLC metastatic livers when compared to cNKs isolated from naïve livers (Fig. 3 I and J). In contrast, trILC1s from both tumor models preserved the ability to kill tumor cells even in an environment of advanced metastasis (Fig. 3 I and J).
Collectively, these results point toward an important role of both trILC1s and cNK cells in limiting metastatic seeding. cNKs, however, are more prone to infiltrate metastatic nodules and provide an enduring antimetastatic activity until they become dysfunctional at late stages of the disease.
Metastatic Livers Drive the Emergence of Unique cNK Cell Populations.
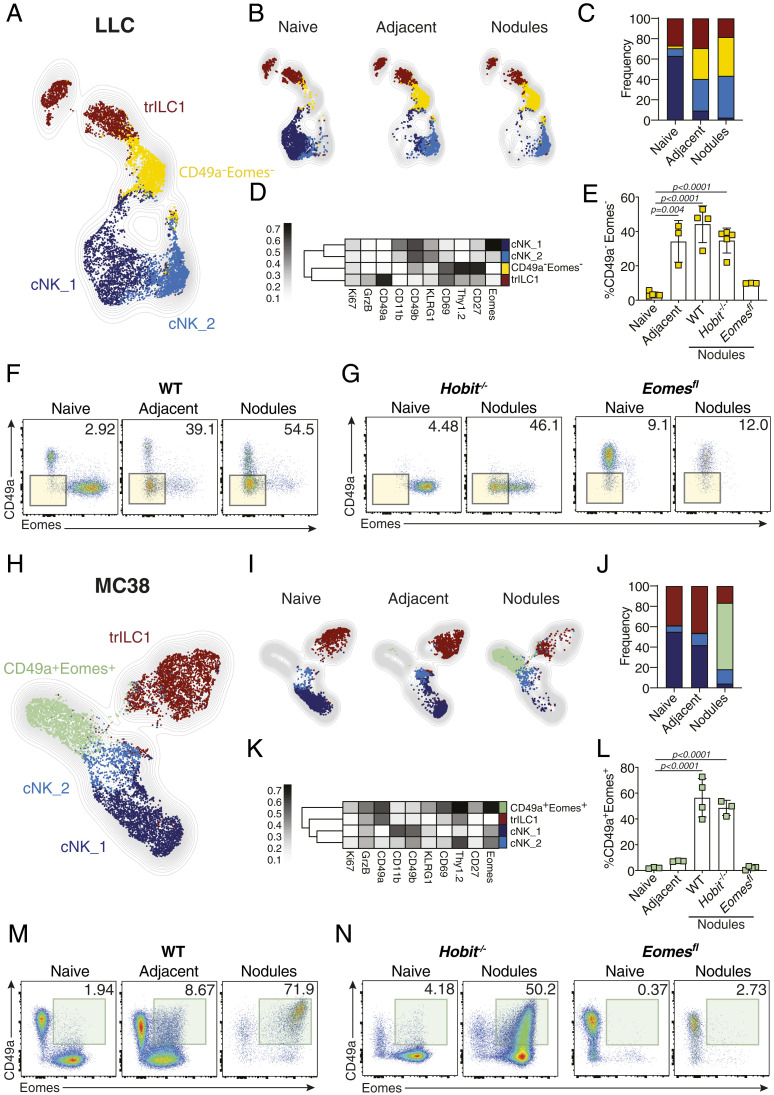
To better understand the influence of the MC38- and LLC-associated hepatic metastatic niche on cNKs and trILC1s, we characterized both NKp46+ cell subsets using high-parametric single-cell cytometry. We used cell suspensions of naïve liver as well as the adjacent tissue and nodules from metastatic sites and focused on the expression of NK cell surface receptors, transcription factors, and effector molecules. To better visualize phenotypical variations, we reduced the high-dimensional dataset into two dimensions using the uniform manifold approximation and projection (UMAP) algorithm. Typical UMAP plots show the normalized expression of each individual marker (SI Appendix, Fig. S2 A and B). We then combined this analysis to the self-organizing map (FlowSOM) metaclustering algorithm (25) to identify distinct populations (Fig. 4 A and H). This approach revealed two clusters of cNKs, termed cNK_1 and cNK_2, corresponding to more-differentiated (CD49bhigh, CD11bhigh, Eomeshigh, Tbethigh) and less-differentiated (CD49bint, CD11blow, Eomesint, Tbetint/low) cNKs, respectively (Fig. 4 A–D and H–K and SI Appendix, Fig. S2C). We found an expansion of cNK_2 cells in the metastatic livers of both MC38 and LLC tumor-bearing mice in agreement with the increased numbers of less-differentiated NK cells reported in the pulmonary metastatic niche (19) (Fig. 4 B, C, I, and J). Besides these expected phenotypes, we found unique NKp46+ cell subsets in metastatic livers that, however, differed in MC38 and LLC metastases, suggesting tumor-driven education. In LLC metastatic livers, a cNK subset emerged that is characterized by a very low expression of CD49b, CD49a, KLRG-1, CD11b, Eomes, T-bet, GrzB, and Ki67 but a high expression of CD69, Thy1.2, and CD27. We refer to this subset as CD49a−Eomes− cNKs, which are reminiscent of very immature NK cells (Fig. 4 B–F and SI Appendix, Fig. S2C). CD49a−Eomes− cNKs accumulated in metastatic livers in Hobit−/− but not in Ncr1iCreEomesfl/fl mice, which confirms their cNK cell origin (Fig. 4 E–G). Also, the blood of mice with LLC liver metastasis contained increased numbers of CD49a−Eomes− cNKs, suggesting that LLC metastases induce systemic changes in the cNK compartment (SI Appendix, Fig. S2D). In MC38-derived metastatic nodules, we detected another unique subset of cNKs. Despite the expression of immature markers CD69, Thy1.2, and CD27, this subset expressed characteristic markers of differentiation (T-bet, Eomes, and GrzB) as well as the integrin CD49a (Fig. 4 H–K). We refer to these cells as CD49a+Eomes+ NKp46+ cells and show that also this subset arose from cNKs since it was only found in the metastatic livers of Hobit−/− but not in those of Ncr1iCreEomesfl/fl mice (Fig. 4 L–N). In contrast to mice with LLC-derived liver metastasis, we did not observe high frequencies of CD49a+Eomes+ cNKs in the blood of mice with MC38-derived metastases, although the systemic population characteristic of this tumor type resembles the less-differentiated cNK_2 cluster (SI Appendix, Fig. S2E). The emergence of CD49a−Eomes− cNKs was not a unique feature of the LLC tumor type or C57BL/6 mice since we found similar CD49a−Eomes− cNKs in livers of BALB/c mice with 4T1-derived metastases (SI Appendix, Fig. S3 A–F). Along the same lines, CD49a+Eomes+ cNKs were not unique to MC38 liver metastases or C57BL/6 mice, as we also found them in BALB/c mice with CT26-derived liver metastases (SI Appendix, Fig. S3 G–L). These findings suggest a dichotomy of cancers that impair cNKs (LLC, 4T1) and those that do not (MC38, CT26). It was recently described that cancer cell–intrinsic cGAS supports NK-mediated control of cancer (22). Because MC38 and CT26 express cGAS in contrast with LLC and 4T1 (23), we analyzed whether cancer cell–intrinsic cGAS may be one of the factors driving the emergence of specific cNK subsets. Therefore, we overexpressed cGAS in LLC and found that this resulted in a reduced metastatic load in the liver (SI Appendix, Fig. S3M) and a reduction of the proportion of Eomes−CD49a− cNKs (SI Appendix, Fig. S3N). We also found important differences in the myeloid compartment when comparing nodules from MC38 and LLC tumors. The proportion of proinflammatory CD11b+Ly6Chigh monocytes as well as dendritic cell subsets 1 and 2 (cDC1 and cDC2) was higher in MC38-derived nodules, whereas the proportion of CD11b+Ly6G+ granulocytes and immunoregulatory CD206+F480+CD64+ macrophages was higher in LLC-derived nodules (SI Appendix, Fig. S4 A and B).
Fig. 4.
Metastatic livers drive the differentiation of unique cNK populations. NKp46+ cells from naïve and day 21 metastatic (nodules and adjacent tissue) livers were analyzed by multiparameter single-cell mapping using flow cytometry. (A–G) LLC metastatic and control livers. (H–N) MC38 metastatic and control livers. Samples were pregated on single live CD45+lineage− cells and subsequently gated on NK1.1+NKp46+ cells. cNK = conventional NK cells, CD49a−CD49b+; trILC1 = tissue-resident ILC1s, CD49a+CD49b−. (A and H) UMAP projections overlaid with FlowSOM-guided manual metaclusters displaying cNKs and trILC1s from all samples. (B and I) UMAP projections overlaid with FlowSOM-guided manual metaclusters separated by sample category (naïve, adjacent, nodules). (C and J) Relative frequency of each cluster in the different sample categories (naïve, adjacent, nodules). (D and K) Heat map summary of median marker expression values of the different markers analyzed for each cluster. (E and L) The frequency of unique metastasis-induced subsets. (E) CD49a−Eomes− NKp46+ cells in LLC metastasis. (L) CD49a+Eomes+ NKp46+ cells in MC38 metastasis. The bar represents the mean ± SD, and the symbols represent livers from individual mice. One-way ANOVA with Tukey’s multiple comparisons test. Experiments were performed at least twice with similar results. (F and M) Representative dot plots of cNKs and trLC1s cells for each sample category based on their expression of CD49a and Eomes. (F) LLC metastasis. Highlighted in yellow is the CD49a−Eomes− population observed in LLC-adjacent tissue and nodules. (M) MC38 metastasis. Highlighted in green is the CD49a+Eomes+ population observed in MC38 nodules. (G and N) Representative dot plots of cNKs and trILC1s isolated from naïve liver or metastatic nodules from Hobit−/− and Eomesfl mice based on their expression of CD49a and Eomes. (G) LLC metastasis. Highlighted in yellow is the CD49a−Eomes− population observed in LLC-adjacent tissue and nodules. (N) MC38 metastasis. Highlighted in green is the CD49a+Eomes+ population observed in MC38 nodules. Groups contained three to five mice. Experiments were performed at least twice with similar results.
Our high-dimensional analyses also confirmed that trILC1s were mostly located outside the metastatic nodules (Fig. 4 B, C, I, and J) as previously revealed by immunofluorescence. In line with their preserved cytotoxicity, trILC1s only underwent slight phenotypic changes in the nodules and their adjacent tissue of metastatic livers from both LLC and MC38 origin (Fig. 4 B, C, I, and J).
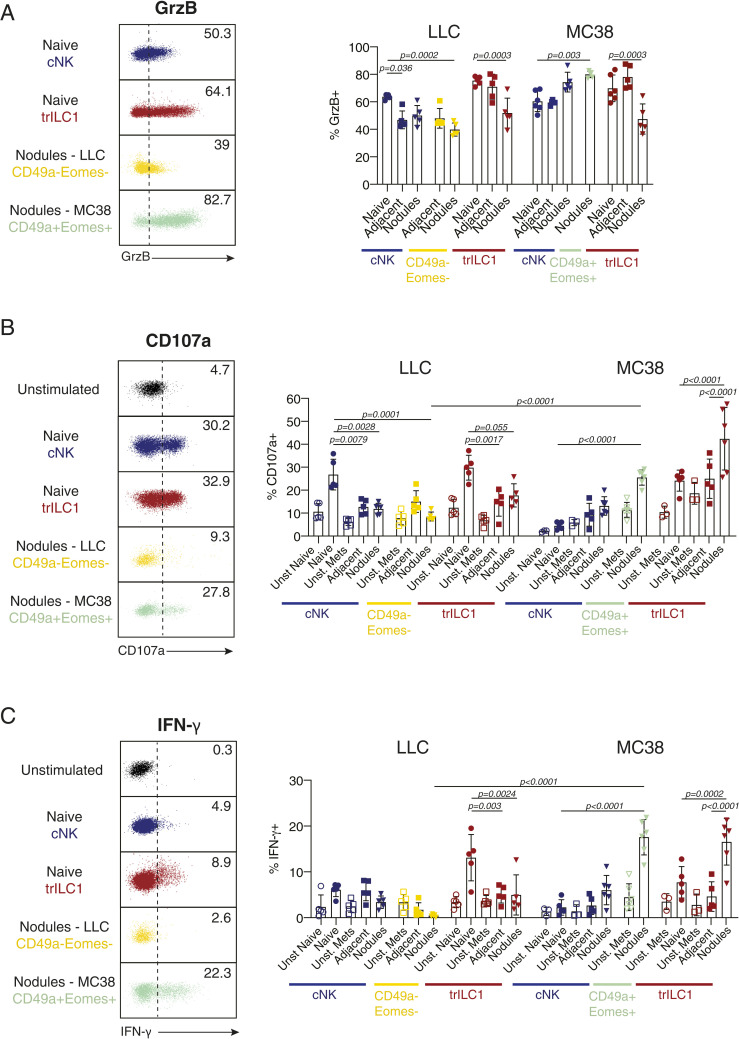
Next, we investigated additional features of LLC- and MC38-driven cNK and trILC1 subsets including the production of granzyme B (GrzB), degranulation (indicated by surface expression of CD107a (LAMP-1) (26), and IFN-γ production in response to different stimuli. cNKs (including the differentiated Eomeshigh and the less-differentiated Eomesint subsets) and especially the intranodular Eomes+CD49a+ subset from MC38 metastatic livers produced high amounts of GrzB (Fig. 5A) in comparison to the lower amount observed for cNKs and Eomes−CD49a− cells from LLC metastatic livers (Fig. 5A). We found that cNKs, Eomes+CD49a+ cells, and trILC1s from MC38 metastatic livers degranulated and produced high amounts of IFN-γ in response to anti-NK1.1 stimulation, in particular those localized in the metastatic nodules (Fig. 5 B and C). This was not the case for cNK cells, trILC1s, and Eomes−CD49a− cells from LLC metastatic livers, which were poorly stimulated in response to anti-NK1.1 (Fig. 5 B and C). We observed similar results when using phorbol 12-myristate 13-acetate (PMA) plus ionomycin as a stimulus (SI Appendix, Fig. S4 C and D). However, the impaired IFN-γ production of LLC-derived cNKs, Eomes+CD49a+ cells, and trILC1s from LLC metastatic livers were rescued by incubation with IL-15 or IL-12 plus IL-18 (SI Appendix, Fig. S5 A–D).
Fig. 5.
The MC38- but not the LLC-derived metastatic microenvironment preserves functional features of cNKs and trILC1s. Flow cytometry analysis of (A) Granzyme B (GrzB), (B) CD107a and (C), IFN-γ expression by cNKs and trILC1s cells from naïve and metastatic livers. Metastatic livers were manually dissected to separate the nodules from the adjacent tissue, and tissues were enzymatically processed into a single-cell suspension. Granzyme B was stained intracellularly directly ex vivo; CD107a and IFN-γ were stained after 4 h of stimulation by plate-bound anti-NK1.1. Samples were pregated on single live CD45+lineage− cells and subsequently gated on NK1.1+NKp46+ cells. cNK cells (CD49a− cells) were divided into CD49b+Eomeshigh/int cells (named here cNKs and present in naïve and both LLC and MC38 metastatic livers), CD49b−Eomes− cells (present in LLC-derived metastatic livers), and CD49b+Eomes+ cells (present in MC38-derived nodules). trILC1s were gated as CD49a+CD49b− cells. (Left) Representative dot plots showing the expression and gating strategy for GrzB+ (A), CD107a+ (B), and IFN-γ+ (C) cells in cNKs (blue), trILC1s (red), CD49a−Eomes− cells (yellow), and CD49a+Eomes+ cells (green). (Right) Percentages of GrzB+ (A), CD107a+ (B), and IFN-γ+ (C) cells. The bar represents the mean ± SD, and the symbols represent livers from individual mice. One-way ANOVA with Tukey’s multiple comparisons test. Groups contained five and six mice. Experiments were performed at least twice with similar results.
Altogether, we uncovered unique populations of cNKs in metastatic livers and found that cancer-specific features drive the differentiation of these cells. Thus, whereas certain tumor types (e.g., LLC and 4T1) promote the emergence of very immature subsets of cNKs, others such as MC38 or CT26 contain a unique intranodular cNK cell subset that retains key features of differentiation and functionality.
Single-Cell RNA Sequencing Reveals Distinctive Transcriptional Signatures of cNKs Populating the Hepatic Metastatic Niche.
To gain further insights into the nature of these unique and previously undiscovered cNK cell subsets populating the hepatic metastatic niche, we performed 10× single-cell RNA sequencing (RNA-seq) on CD45+ lin−NK1.1+NKp46+ cells sorted from naïve as well as from MC38 and LLC metastatic livers. The clusters were subsequently projected on a UMAP plot with each dot representing a single cell. The identity of cNK and trILC1 cells was confirmed by their uniformly high expression of the lineage marker gene Ncr1 (NKp46) as well as the expression of Eomes (Eomes, for cNKs) and Itga1 (CD49a, for trILC1s) (SI Appendix, Fig. S6A).
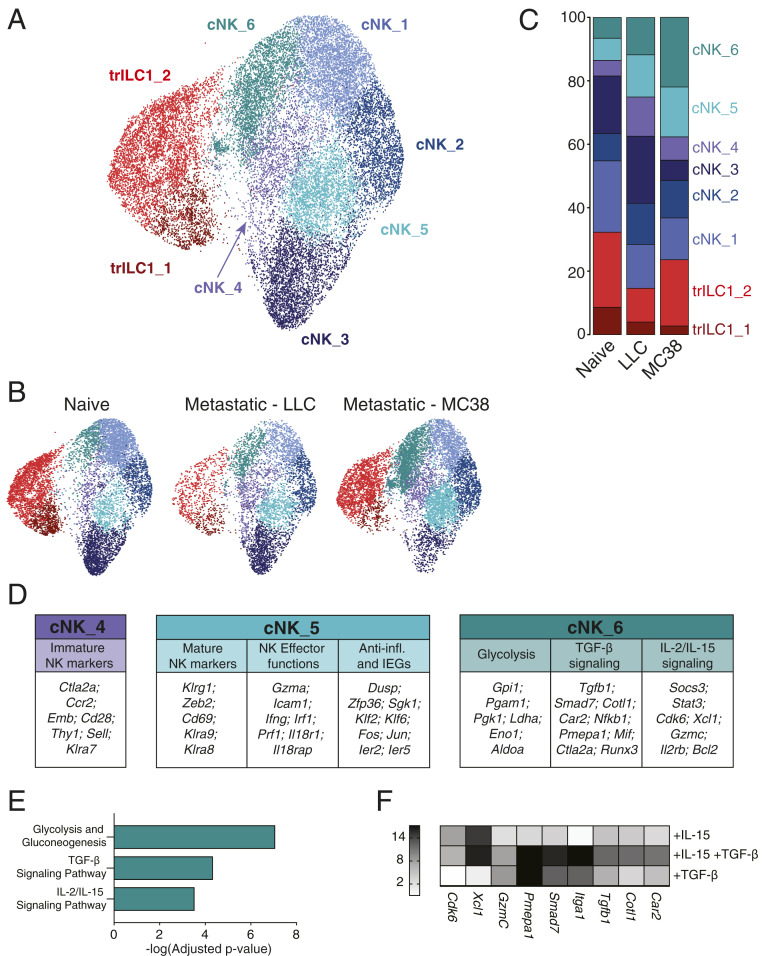
We identified two clusters for trILC1s and six clusters corresponding to cNKs (Fig. 6A). The top genes identifying each cluster are displayed in SI Appendix, Table 1 and in a heat map (SI Appendix, Fig. S6B). We found that cNK_4 and cNK_6 were preferentially enriched in LLC-derived and MC38-derived metastases, respectively (Fig. 6 B and C), while the cNK_5 cluster expanded in either environment (Fig. 6 B and C).
Fig. 6.
Single-cell RNA-seq reveals unique transcriptional signatures of cNKs in the hepatic metastatic niche. Single-cell RNA-seq was performed on NKp46+ cells sorted from naïve, LLC metastatic, and MC38 metastatic livers (six mice per condition pooled into one sample for droplet encapsulation and library preparation). (A–C) UMAP projections identifying the different cNK/trILC1 clusters in (A) all samples or (B) individual samples with (C) bar plots showing the relative frequency of the different clusters in each sample. (D) Top up-regulated genes in cluster cNK_4, cNK_5, and cNK_6 arranged in functional categories. (E) Pathway analysis of significantly up-regulated genes in cluster cNK_6. (F) cNKs and trILC1s were sorted from naïve livers and cultured in vitro for 48 h with 25 ng/mL mouse IL-15/IL-15R complex and/or 10 ng/mL human TGF-β1. The transcripts of cNK_6 cluster-specific genes were quantified by qPCR. The heat map shows the average fold change relative to the untreated condition. The data represent three biological and three technical replicates, and the experiment was performed three times with similar results.
The majority of differentially expressed genes (DEGs) from the cNK_4 cluster suggested an immature phenotype as illustrated by the high expression of Ctla2a, Ccr2, Emb, Cd28, Thy1, Sell (CD62L), or Klra7 (Ly49G2) and the strong down-regulation of Gzmb (SI Appendix, Table 1 and Fig. 6 C and D). Also on the protein level, we found an up-regulation of Thy1.2, CD62L, and Ly49G2 in cNKs from MC38- and LLC-derived metastatic nodules (SI Appendix, Fig. S6 C and D). Furthermore, the transcriptional signature of this cluster showed a striking resemblance with that of an immature NK population previously described in the steady-state spleen (27). In contrast, NKp46+ cells from the cNK_5 cluster, common to both LLC and MC38 metastatic livers, appeared to be more differentiated expressing Klrg1 and Cd69 as well as Klra9 (Ly49I) and Klra8 (Ly49H) (Fig. 6D and SI Appendix, Table 1). This cluster is characterized by the high expression of effector genes (Icam1, Ifng, Gzma, and Prf1), a few genes involved in anti-inflammatory responses (Dusp1, Zfp36, or Sgk1), and several immediate early genes (Fos, Jun, Klf2, Klf6, Ier2, and Ier5) (Fig. 6D and SI Appendix, Table 1). The latter may indicate an “alertness” of cells in cluster cNK_5, allowing them to rapidly respond to environmental cues. Additionally, the cNK_5 cluster strongly resembled the cNK_3 cluster enriched in naïve livers and underrepresented in metastases (SI Appendix, Fig. S6E). Cells from the cNK_5 cluster, however, expressed higher amounts of cytotoxic genes (Prf1 or Gzma) and genes from the IL-18 pathway (Il18r1, Il18rap), suggesting its skewing toward a more effector phenotype.
cNK_6 was the only cNK cell cluster expressing Itga1 (CD49a) and increased amounts of Thy1 (SI Appendix, Fig. S6F) transcripts compared to other clusters and thus showed the highest similarity to the CD49a+Eomes+ cNK subset identified by flow cytometry. About half of the genes that define the immature cNK_4 cluster were highly expressed by cells of the cNK_6 cluster (Fig. 6D and SI Appendix, Table 1 and Fig. S6G). We found an increased expression of several TGF-β–induced genes such as Smad7, Runx3, Pmepa1, Plac8, Car2, Cotl1, and Tgfb1 itself (Fig. 6D and SI Appendix, Table 1). In agreement, EnrichR pathway analysis identified up-regulation of the “TGF-β signaling pathway” in the cNK_6 cluster (Fig. 6E) (28). This analysis also revealed an enrichment of a gene set related to the “IL-2/IL-15 signaling pathway” (Gzmc, Socs3, Stat3, Il2rb, Bcl2, Cdk6, Xcl1) (Fig. 6 D and E) and another gene set related to “glycolysis and gluconeogenesis” (Ldha, Eno1, Pgam1, Gpi1, Pgk1) (Fig. 6 D and E). The latter pathway can be induced by the synergy of hypoxia and IL-15 (29). We validated the importance of the TGF-β and IL-15 signatures in cNK_6 by assessing the expression of selected DEGs upon ex vivo stimulation of isolated cNKs with TGF-β and/or IL-15 (Fig. 6F).
Taken together, whereas the transcriptome of cNK_4 cells expanded in LLC-derived metastases is reminiscent of an immature status, that of cNK_6 cells from MC38-derived metastases appears to be the result of dual TGF-β and IL-15 signaling. This illustrates that tumors can employ differential mechanisms to drive NKp46+ cell phenotype and function.
IL-15 and TGF-β Modulate the Fate of CD49a+Eomes+ cNKs in Metastatic Nodules.
Our discovery that cNKs populating liver metastases have a unique signature suggests that the metastatic microenvironment regulates the differentiation of cNKs. Furthermore, it appears that each tumor influences cNKs differently, presumably dependent on specific features associated with individual tumors. Because the single-cell RNA-seq data pointed toward the TGF-β and IL-15 pathways, we quantified the transcripts for these cytokines in the hepatic metastatic niche. We found high amounts of Tgfb1 in the adjacent tissue and nodules of both MC38 and LLC metastatic livers (Fig. 7A). Furthermore, we detected high amounts of transcripts of Il15 and Il15ra in MC38 but not LLC nodules (Fig. 7 B and C), which may explain that MC38 nodules contained significantly more cNKs than MC38 nodule–adjacent tissue, LLC nodules, or naïve liver (Fig. 1 E–G).
Fig. 7.
IL-15 and TGF-β modulate the fate of CD49a+Eomes+ cNKs in metastatic nodules. (A and B) Quantification of transcripts of (A) Tgfb1, (B) Il15, and (C) Il15ra in lysates of naïve, LLC and MC38 metastatic livers. Bars show the mean ± SD. Each symbol represents an individual mouse. One-way ANOVA with Tukey’s multiple comparisons test. The experiment was performed twice with similar results. (D, Upper) Quantification of LLC liver nodules from Tgfbr2WT and Tgfbr2fl mice by in vivo imaging system (IVIS) 21 d after tumor cell injection. Bars show the mean ± SD. Each symbol represents an individual mouse. One-way ANOVA with Tukey’s multiple comparisons test. The experiment was performed twice with similar results. (Lower) Representative IVIS images of metastatic livers from each group at the endpoint. Tgfbr2WT = Ncr1iCre/wt.Tgfbr2wt/wt ; Tgfbr2fl = Ncr1iCre/wt.Tgfbr2fl/fl. (E, Upper) Quantification of macroscopic MC38 liver nodules (number and area) from Tgfbr2WT and Tgfbr2fl mice 21 d after tumor cell injection. Bars show the mean ± SD. Each symbol represents an individual mouse. One-way ANOVA with Tukey’s multiple comparisons test. The experiment was performed twice with similar results. (Lower) Representative images of metastatic livers from each group at the endpoint. Tgfbr2WT = Ncr1iCre/wt.Tgfbr2wt/wt; Tgfbr2fl = Ncr1iCre/wt.Tgfbr2fl/fl. (F, Upper) Percentage of CD49a+Eomes+ NKp46+ cells in MC38 nodules from Tgfbr2WT and Tgfbr2fl mice. Bars show the mean ± SD. Each symbol represents an individual mouse. One-way ANOVA with Tukey’s multiple comparisons test. The experiment was performed twice with similar results. (Lower) Representative dot plots of cNKs and trILC1s in MC38 nodules from Tgfbr2WT and Tgfbr2fl mice with the CD49a+Eomes+ population highlighted in green.
We then crossed Ncr1iCre to Tgfbr2fl/fl mice to specifically delete TGF-β signaling in NKp46+ cells. Despite the previously reported TGF-β–mediated inhibition of NK maturation during their development (30), we did not observe phenotypical differences in cNKs or trILC1s of Ncr1iCreTgfbr2fl/fl mice compared to control mice in the steady state (SI Appendix, Fig. S7 A–C). The LLC-derived metastatic load as well as the phenotype of cNKs and trILC1s in Ncr1iCreTgfbr2fl/fl mice were similar to those in control mice (Fig. 7D and SI Appendix, Fig. S7 D and E). However, hepatic MC38-derived metastases were significantly reduced in mice lacking the TGF-β receptor on NKp46+ cells (Fig. 7E). Although the NKp46+ cell population did not undergo major phenotypic changes (SI Appendix, Fig. S7F), the CD49a+Eomes+ cNK cell subset infiltrating the metastatic nodules in control mice failed to develop in Ncr1iCreTgfbr2fl/fl mice (Fig. 7F).
Taken together, cytokines produced in the metastatic niche influence the phenotype and function of tumor-associated NKp46+ cells in an individual fashion as exemplified here by IL-15 and TGF-β in case of LLC and MC38 liver metastasis. Unraveling such pathways may lead to a discovery of druggable targets and improved NKp46+ cell-based therapies for metastatic disease.
Discussion
The liver is a preferred metastatic site for many different cancer types (1), suggesting an exceptional immune environment. Indeed, the liver is equipped with mechanisms that promote immune tolerance of CD8+ T cells (31, 32) to prevent deleterious immune responses against innocuous antigens. In addition, the liver is surveilled by passer-by cNKs and by resident trILC1s (33). These abundant innate lymphocytes may be the major effectors in hepatic immune defense.
In the context of viral infection, trILC1s suppressed T cell–mediated viral control (34) and were protective against acute liver injury (13), suggesting a dampening rather than effector function. Using genetically modified mice that lack cNKs or trILC1s, we discovered that both innate subsets collaborated in a nonredundant fashion to control hepatic metastasis. Specifically, we found that trILC1s mainly interfered with the seeding of cancer cells, while cNKs controlled metastatic outgrowth and progression. The doorkeeper function of trILC1s may be explained by their unique location in the hepatic sinusoids (3). A significant proportion of trILC1s expresses CXCR6, which may mediate retention to CXCL16-producing sinusoidal cells (35). Our observation that trILC1s hardly infiltrated metastatic nodules is in line with their inability to control cancer cells beyond seeding. At the same time, keeping their distance from disseminated cancer cells may explain why trILC1s sustain their cytotoxic function even at a late stage of metastatic disease.
The limited capacity to infiltrate metastatic nodules was also a hallmark of cNKs emerging in the context of LLC-derived metastases. The cNK cell subset instructed by this tumor type expressed low amounts of CD49b, Eomes, and T-bet, suggesting stalled maturation (5) or exhaustion (36). Along the same lines, we found that cNKs from LLC metastatic livers were unresponsive to a variety of stimuli. In contrast, CD49a+CD49b+Eomes+ cNKs readily infiltrated MC38-derived metastatic nodules and maintained several of their functions. The expression of CD49a and additional markers of immaturity (Thy1.2, CD69, CD27) associated with this population is reminiscent of an intratumoral ILC1-like subset (CD49a+CD49b−Eomesint/low) arising from NK cells reported in methylcholanthrene-induced sarcomas (37) and of a population of TCR−NK1.1+CD49a+ ILC1-like cells found described in PyMT tumors (38). Both studies described that these particular ILC1-like cells expressed a low amount of the transcription factor Eomes, which we did not observe in the MC38-associated CD49a+Eomes+ cells. Since not only the expression of Eomes but also of T-bet and CD49b in the CD49a+Eomes+ subset remained high, we propose that CD49a+Eomes+ cells in MC38 metastatic nodules are transiently activated immature cNK cells rather than cNK cells converted into the ILC1 subset.
The differences between cNKs associated with LLC- or MC38-derived metastasis suggested that disseminated cancer cells orchestrate the metastatic niche in a cancer-specific fashion, therefore influencing the phenotype and function of cNKs. Indeed, MC38-derived metastatic nodules feature a more proinflammatory myeloid milieu in comparison to the immunoregulatory one observed in metastasis from LLC tumor cells. Furthermore, we detected a high expression of the survival factor IL-15 (39) in MC38- but not LLC-derived metastasis, which may explain the paucity of cNKs in the latter tumor type. Even though metastatic livers of both tumor types expressed TGF-β, only the subset of CD49a+Eomes+ cNKs in MC38-derived metastases depended on TGF-β signaling. Thus, TGF-β directly regulates the expression of CD49a, which is in agreement with previous reports (37, 40), and moreover, directly compromises the antimetastatic efficacy of cNK cells. The CD49a−Eomes− subset associated with LLC-derived metastases was not affected by the lack of TGF-β signaling, nor was the extent of hepatic metastases derived from this tumor cell line. This was unexpected because TGF-β impairs NK cell cytotoxicity and maturation (30, 41, 42), and two previous studies have reported decreased metastasis in mice with ablated TGF-β signaling in NKp46+ cells (37, 43). We thus propose that the TGF-β blockade may only be efficient when cNK cells face a metastatic microenvironment that favors their survival, for example, in the presence of IL-15.
We found that the cNK cell subsets present in the metastatic niche of LLC or MC38 also emerged in hepatic metastases derived from other cell lines. Indeed, CT26 influenced cNKs in a similar way as MC38 did, whereas the impact of 4T1 resembled that of LLC. These findings point toward a dichotomy, which is reminiscent of the concept in which cancers are described as hot or cold, a classification mainly based on the CD8+ T cell landscape within a tumor (44). Recently, a four-category classification was proposed—hot, altered excluded, altered suppressed, and cold—which considers other parameters as well (45). The relevance of such classifications for the quality of innate immune defense is not well studied, but it is likely that at least some factors promoting T cell immunity also support innate lymphoid cells. As discussed above, one such factor may be IL-15 (39, 46, 47). IL-15 is induced by the cancer cell–intrinsic factor cGAS through the activation of the stimulator of interferon genes (STING) pathway (48, 49). STING activation in myeloid cells induces the production of type I IFN, which supports NK and CD8+ T cells responses directly or indirectly through the production of IL-15 (22, 23, 48). Interestingly, hot cancers display a type I IFN signature (50), and both MC38 and CT26 cells contain high amounts of active cGAS and produce spontaneous type I IFN themselves (MC38) or induce its production in adjacent myeloid cells (CT26), whereas LLC and 4T1 do not (23). We did not find an up-regulation of IL-15 in cGAS-overexpressing LLC nodules, and the mechanisms leading to reduced metastases in this setting require further investigation.
Taken together, our study demonstrates collaborative roles of circulating and tissue-resident innate lymphocytes combatting the formation and growth of metastasis. The unique properties of the cNK subsets described here and their differentiation are instructed by the metastatic microenvironment. Thus, understanding the regulatory hierarchy of metastatic-derived factors for each individual tumor type will be crucial to exploit the antimetastatic potential of innate lymphoid cells in metastasis.
Materials and Methods
Mice.
Female 6- to 10-wk-old C57BL/6 mice were obtained from Janvier Labs. Ncr1Cre/wt mice (B6.Ncr1tm1.1(icre)Viv) were provided by Eric Vivier, Rosa26iDTR mice by Ari Waisman, and Eomesfl/fl, Ai14fl/fl mice were obtained from the Jackson Laboratory. Ncr1iCre mice were crossed to Rosa26iDTR mice to obtain Ncr1iCre/wt Rosa26iDTR/wt (Ncr1iCreR26RiDTR) or Ncr1iCre/wtRosa26wt/wt controls (Ncr1iCre). Mice homozygous for the Hobit-targeted mutation (Hobit−/− mice) were provided by Klaas van Gisbergen (8). B6.129-Nfil3(loxP)tm1Rbrc (Nfil3fl/fl) mice were generated by Motomura Y (51) and provided by Henrique Veiga-Fernandes, Champalimaud Foundation, Lisbon, Portugal. Mice were housed in the Laboratory Animal Services Center of the University of Zurich according to institutional guidelines under specific pathogen-free conditions. All animal experiments were approved by the Swiss Cantonal Veterinary office (license numbers 65/2015 and 156/2018).
Model of Experimental Liver Metastasis.
To generate murine liver metastases, 6- to 10-wk-old mice were anesthetized using vaporized isoflurane, and the upper lateral abdominal wall was incised followed by the injection of 5 × 105 MC38 or 2 × 105 LLC-LUC, CT26, or 4T1 cells in 40 µl phosphate-buffered saline (PBS) into the splenic parenchyma. Immediately after injection, the spleen was excised to prevent the growth of splenic tumors. The wound was closed using sutures (70 cm Coated Vicryl, ETHICON) and autoclips (Autoclip 9 mm, Becton Dickinson). Control mice underwent mock surgery. Livers were analyzed 21 to 28 d after tumor cell injection. MC38-derived metastatic nodules were counted using a stereotactic microscope, and the nodule area of each hepatic nodule was quantified using the drawing tool from the ImageJ software. Each point represented in the final graph corresponds to the area of one nodule displayed in pixels. LLC-derived metastases were visualized and quantified using an IVIS 200 imaging system (PerkinElmer) 20 min after intraperitoneal (i.p.) injection of 150 mg/kg D-Luciferin (Promega). The data are presented as total luminescence per liver for each mouse.
Depletion of Cells by Antibodies or DTX.
Mice were treated i.p. with 50 µl anti-asialo GM1 antibodies (Wako Pure Chemical Industries) or 0.2 mg rabbit IgG (Sigma) twice a week starting 48 h before intrasplenic injection of tumor cells. DTX from Corynebacterium diphtheria (Calbiochem, 250 ng/mouse for initial depletion and 125 ng/mouse for the following injections) diluted in PBS was injected i.p. at days −2, −1, 2, 5, 8, 11, 15, and 18 relative to tumor injection. Anti-CD8α (100 µg/mouse, clone YTS169.4, BioXCell) was injected i.p. 2 d before tumor injection. Depletion efficiency was monitored in the blood by flow cytometry.
Flow Cytometry.
Livers were harvested in Roswell Park Memorial Institute medium (RPMI) supplemented with 10% fetal calf serum (FCS) and digested with Collagenase IV (1 mg/mL, Bioconcept) and deoxyribonuclease I (2.6 μg/mL, Thermo Fisher Scientific) for 45 min at 37 °C. Cells were washed with PBS and filtered through a 100-μm filter to remove debris. Lymphocytes were purified using a density gradient centrifugation step (Percoll, GE Healthcare). Blood was collected in PBS containing 2 mM ethylenediaminetetraacetic acid (EDTA). Red blood cells were lysed in all samples using red blood cell lysis buffer (4.15 g NH4Cl, 0.55 g KHCO3, and 0.185 g EDTA in 500 mL ddH2O) for 1.5 min.
Single cells were incubated for 15 min in 250 µg/mL anti-mouse CD16/32 (clone 2.4G2) in PBS to block Fc receptors, washed with PBS, and surface stained in 50 μL antibody mix in PBS. For intracellular cytokine staining, cells were stimulated with 100 ng/mL PMA plus 1 µg/mL ionomycin, 100 ng/mL recombinant mouse IL-18 (MBL International) plus 10 ng/mL recombinant mouse IL-12 p70 (Peprotech), 10 ng/mL recombinant mouse IL-15 (Peprotech) or anti-NK1.1 (clone PK136, BioXCell) for 4 h at 37 °C in the presence of GolgiPlug/GolgiStop (BD Pharmigen). Cells were stained for surface molecules as described above, washed with PBS, and fixed for 60 min on ice using IC Fixation Buffer from the Foxp3/Transcription Factor Staining Buffer Set (eBioscience). Subsequently, cells were stained for intracellular cytokines in 50 μL permeabilization buffer from the Foxp3/Transcription Factor Staining Buffer Set overnight at 4 °C. After washing with permeabilization buffer, samples were suspended in fluorescence-activated cell sorting (FACS) buffer (PBS, 20 mM EDTA pH 8.0, 2% FCS) and acquired with a FACS LSRII Fortessa (BD Bioscience) or FACSymphony (BD Biosciences).
To determine cell numbers, CountBright Absolute Counting Beads were used (Thermo Fisher Scientific). Dead cells were excluded using Live/Dead fixable staining reagents (Invitrogen), and doublets were excluded by FSC-A/FSC-H gating.
Analysis of Flow Cytometry Data.
Flow cytometry data were preprocessed, down-sampled, and exported using FlowJo v10 software (Tree Star). The exported FCS files were uploaded in Rstudio (R software environment, version 3.4.0) and were transformed with an inverse hyperbolic sine (Arsinh) function. UMAP projections were calculated and FlowSOM-based clustering was performed using the guided manual metaclustering (25, 52). All plots were drawn with ggplot2.
Degranulation Assay and IFN-γ Production.
Cell suspensions were cultured in RPMI (GIBCO) supplemented with 10% FCS (Thermo Fisher Scientific) and 2 mM L-Glutamine (Thermo Fisher Scientific) with either 100 ng/mL PMA plus 1 µg/mL ionomycin, 100 ng/mL IL-18 (MBL) plus 10 ng/mL IL-12 p70 (Peprotech), 10 ng/mL IL-15 (Peprotech), or plate-bound anti-NK1.1 (clone PK136, BioXCell) for 1 h in 96-well plates at 37 °C. CD107a antibody (1:200 final dilution, 1D4B BioLegend) was added to the mix. Subsequently, GolgiPlug (10 µg/mL, BD Pharmigen) and GolgiStop (10 µg/mL, BD Pharmigen) were added, and cells were incubated for an additional 3 h. For anti-NK1.1 activation, 96-well plates were coated overnight at 4 °C with 10 µg/mL anti-NK1.1 antibody (PK136) or isotype control (C1.18.4), washed with PBS, and blocked with 20% FCS for 20 min. After incubation, cells were collected and analyzed by flow cytometry.
Killing Assay.
Liver NK cells (live, CD45+CD3−CD5−Ly6G−CD19−CD115−NK1.1+NKp46+CD49a+CD49b− and CD45+CD3−CD5−Ly6G−CD19−CD115−NK1.1+NKp46+CD49a−CD49b+) were sorted with a BD FACSAria III sorter. Target cells (MC38, LLC) were stained with PKH26 Red Fluorescent Cell Linker Mini Kit (Sigma) following manufacturer’s instructions and seeded in 96-well round-bottom plates with 104 cells/well. NK cells were incubated with target cells at effector:target ratios of 1:1, 4:1, and 8:1 for 24 h at 37 °C in 5% CO2. After the removal of medium, 0.8 µM TO-PRO-3 (Thermo Fisher Scientific) was added, and cells were acquired on a LSRII Fortessa flow cytometer (BD). Data were analyzed using FlowJo v10 software (Tree Star). The percentage of specific killing was calculated as follows: [(% cell death in the presence of NK cells) − (% spontaneous cell death)] ÷ (100 − % spontaneous cell death).
Quantitative Real-Time PCR.
Metastatic livers were divided in adjacent tissue and nodules. RNA was isolated from lysates of naïve or metastatic livers from C57BL/6 mice using the Qiagen Micro Kit according to the manufacturer`s protocol. Random primers (Invitrogen) were used for the synthesis of complementary DNA. The quantitative real-time PCR was performed with a CFX384 Cycler (Bio-Rad Laboratories) using the SYBR Green Supermix (Bio-Rad Laboratories). Individual samples were run in technical triplicates. Gene expression relative to the housekeeping gene PolII was calculated with the ∆Ct values as previously described (53).
In Vitro Stimulation of NKp46+ Cells.
Liver NKp46+ cells (live, CD45+CD3-CD5-Ly6G-CD19-CD115-NK1.1+NKp46+) were sorted with a BD FACSAria III sorter and seeded in a 96-well round-bottom plate with 2 × 105 cells per well. Cells were supplemented with 0.1 ng/mL IL-15/IL-15R complex recombinant protein (Thermo Fisher) in all conditions. For IL-15 stimulation, cells were incubated with 25 ng/mL IL-15/IL-15R complex and 50 µg/mL anti–TGF-β blocking antibody (clone 1D11.16.8; BioXCell). For TGF-β stimulation, cells were incubated with 10 ng/mL hTGF-β1 (Peprotech). For the double stimulation, cells were incubated with 25 ng/mL IL-15/IL-15R complex plus 10 ng/mL hTGF-β1. After 48 h, cells were lysed, and the RNA was collected as described in Quantitative Real-Time PCR.
Immunofluorescence.
Livers were fixed in 4% paraformaldehyde (Roti-Histofix 4%, Roth) for 4 h and dehydrated in 30% sucrose for 48 h at 4 °C. Livers were frozen in Tissue-Tek O.C.T. (Optimal Cutting Temperature; Sakura), and 10-µm cryosections were cut and dried for 30 min at 37 °C. Slides were washed with PBS and blocked/permeabilized with PBS/4% bovine serum albumin (BSA; Roth)/0.1% Triton X-100 room temperature (Sigma) blocking buffer. Subsequently, slides were washed with PBS/0.05% Tween 20 (Sigma) and stained with goat-anti-mouse-VCAM-1 (R&D, 1:100 in PBS/1% BSA) at 4 °C overnight. After incubation, slides were washed and stained with donkey-anti-goat IgG-Alexa Fluor 488 (Jackson ImmunoResearch, 1:500 in PBS/1% BSA). Finally, slides were washed and incubated with 0.5 µg/mL DAPI (Invitrogen) for 5 min and mounted with ProlongDiamond medium (Invitrogen).
Stained slides were scanned using the automated multispectral microscopy system Vectra 3.0 (PerkinElmer). Eight to 10 representative areas were imaged at 200-fold magnification. Spectral unmixing, cell segmentation, and quantification was performed using inForm software (PerkinElmer) as described (54).
Statistics.
P values were calculated using GraphPad Prism software (GraphPad Software, Inc.). For the comparison of two experimental groups, the unpaired two-tailed Student’s t test was used, and for the comparison of more than two groups, the one-way ANOVA multiple comparison correction was used. P values smaller than 0.05 were considered significant. *P < 0.05; **P < 0.01; ***P < 0.001. Data are shown as mean ± SD.
Supplementary Material
Acknowledgments
We thank Karina Silina, Mirjam Lutz, Virginia Cecconi (Institute of Experimental Immunology, University of Zurich), the Flow Cytometry Facility (University of Zurich), and Emilio Yangüez and Ge Tan from the Functional Genomics Center Zurich for assistance. This work was supported by grants from the Swiss National Science Foundation (CRSII5_177208 and 310030_175565 to M.v.d.B., PR00P3_179775 to S.T., and 316030_150768 and 310030_146130 to B.B.), the Swiss Cancer League (KLS-4098-02-2017 to M.v.d.B. and KFS-4431-02-2018 to S.T. and B.B.), the Promedica Stiftung (S.T.), the University of Zurich (University Research Priority Project “Translational Cancer Research” to M.v.d.B. and B.B.), and the Monique-Dornonville-de-la-Cour Foundation Zurich (M.v.d.B.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2026271118/-/DCSupplemental.
Data Availability
Single-cell sequencing data have been deposited in the NCBI Bio Sample database (PRJNA656253).
References
- 1.Williamson T., Sultanpuram N., Sendi H., The role of liver microenvironment in hepatic metastasis. Clin. Transl. Med. 8, 21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sojka D. K., et al., Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife 3, e01659 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peng H., et al., Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J. Clin. Invest. 123, 1444–1456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeda K., et al., TRAIL identifies immature natural killer cells in newborn mice and adult mouse liver. Blood 105, 2082–2089 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Gordon S. M., et al., The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity 36, 55–67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamizono S., et al., Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J. Exp. Med. 206, 2977–2986 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daussy C., et al., T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J. Exp. Med. 211, 563–577 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mackay L. K., et al., Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352, 459–463 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Zhang L. H., Shin J. H., Haggadone M. D., Sunwoo J. B., The aryl hydrocarbon receptor is required for the maintenance of liver-resident natural killer cells. J. Exp. Med. 213, 2249–2257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuchs A., ILC1s in tissue inflammation and infection. Front. Immunol. 7, 104 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishiyama K., et al., Difference in cytotoxicity against hepatocellular carcinoma between liver and periphery natural killer cells in humans. Hepatology 43, 362–372 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Weizman O. E., et al., ILC1 confer early host protection at initial sites of viral infection. Cell 171, 795–808.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nabekura T., Riggan L., Hildreth A. D., O’Sullivan T. E., Shibuya A., Type 1 innate lymphoid cells protect mice from acute liver injury via interferon-γ secretion for upregulating Bcl-xL expression in hepatocytes. Immunity 52, 96–108.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.López-Soto A., Gonzalez S., Smyth M. J., Galluzzi L., Control of metastasis by NK cells. Cancer Cell 32, 135–154 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Chiossone L., Dumas P. Y., Vienne M., Vivier E., Natural killer cells and other innate lymphoid cells in cancer. Nat. Rev. Immunol. 18, 671–688 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Ohs I., et al., Interleukin-12 bypasses common gamma-chain signalling in emergency natural killer cell lymphopoiesis. Nat. Commun. 7, 13708 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beffinger M., et al., CSF1R-dependent myeloid cells are required for NK-mediated control of metastasis. JCI Insight 3, e97792 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guillerey C., Smyth M. J., NK cells and cancer immunoediting. Curr. Top. Microbiol. Immunol. 395, 115–145 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Ohs I., et al., Restoration of natural killer cell antimetastatic activity by IL12 and checkpoint blockade. Cancer Res. 77, 7059–7071 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Platonova S., et al., Profound coordinated alterations of intratumoral NK cell phenotype and function in lung carcinoma. Cancer Res. 71, 5412–5422 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Carrega P., et al., Natural killer cells infiltrating human nonsmall-cell lung cancer are enriched in CD56 bright CD16(−) cells and display an impaired capability to kill tumor cells. Cancer 112, 863–875 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Marcus A., et al., Tumor-derived cGAMP triggers a STING-mediated interferon response in non-tumor cells to activate the NK cell response. Immunity 49, 754–763.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schadt L., et al., Cancer-cell-intrinsic cGAS expression mediates tumor immunogenicity. Cell Rep. 29, 1236–1248.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Gascoyne D. M., et al., The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat. Immunol. 10, 1118–1124 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Brummelman J., et al., Development, application and computational analysis of high-dimensional fluorescent antibody panels for single-cell flow cytometry. Nat. Protoc. 14, 1946–1969 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Betts M. R., et al., Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J. Immunol. Methods 281, 65–78 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Crinier A., et al., High-dimensional single-cell analysis identifies organ-specific signatures and conserved NK cell subsets in humans and mice. Immunity 49, 971–986.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen E. Y., et al., Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Velásquez S. Y., et al., Short term hypoxia synergizes with interleukin 15 priming in driving glycolytic gene transcription and supports human natural killer cell activities. J. Biol. Chem. 291, 12960–12977 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marcoe J. P., et al., TGF-β is responsible for NK cell immaturity during ontogeny and increased susceptibility to infection during mouse infancy. Nat. Immunol. 13, 843–850 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Limmer A., et al., Efficient presentation of exogenous antigen by liver endothelial cells to CD8+ T cells results in antigen-specific T-cell tolerance. Nat. Med. 6, 1348–1354 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Zheng M., Tian Z., Liver-mediated adaptive immune tolerance. Front. Immunol. 10, 2525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng H., Tian Z., Diversity of tissue-resident NK cells. Semin. Immunol. 31, 3–10 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Zhou J., et al., Liver-resident NK cells control antiviral activity of hepatic T cells via the PD-1-PD-L1 axis. Immunity 50, 403–417.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Paust S., et al., Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat. Immunol. 11, 1127–1135 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gill S., et al., Rapid development of exhaustion and down-regulation of eomesodermin limit the antitumor activity of adoptively transferred murine natural killer cells. Blood 119, 5758–5768 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao Y., et al., Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat. Immunol. 18, 1004–1015 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Dadi S., et al., Cancer immunosurveillance by tissue-resident innate lymphoid cells and innate-like T cells. Cell 164, 365–377 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ranson T., et al., IL-15 is an essential mediator of peripheral NK-cell homeostasis. Blood 101, 4887–4893 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Rautela J., et al., Therapeutic blockade of activin-A improves NK cell function and antitumor immunity. Sci. Signal. 12, aat7527 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Viel S., et al., TGF-β inhibits the activation and functions of NK cells by repressing the mTOR pathway. Sci. Signal. 9, ra19 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Lee J.-C., Lee K.-M., Kim D.-W., Heo D. S., Elevated TGF-β1 secretion and down-modulation of NKG2D underlies impaired NK cytotoxicity in cancer patients. J. Immunol. 172, 7335–7340 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Best S., et al., Harnessing natural killer immunity in metastatic SCLC. J. Thorac. Oncol. 15, 1507–1521 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Galon J., et al., Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313, 1960–1964 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Galon J., Bruni D., Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 18, 197–218 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Burkett P. R., et al., Coordinate expression and trans presentation of interleukin (IL)-15Ralpha and IL-15 supports natural killer cell and memory CD8+ T cell homeostasis. J. Exp. Med. 200, 825–834 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato N., Patel H. J., Waldmann T. A., Tagaya Y., The IL-15/IL-15Ralpha on cell surfaces enables sustained IL-15 activity and contributes to the long survival of CD8 memory T cells. Proc. Natl. Acad. Sci. U.S.A. 104, 588–593 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicolai C. J., et al., NK cells mediate clearance of CD8+ T cell-resistant tumors in response to STING agonist. Sci. Immunol. 5, 1–14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santana Carrero R. M., et al., IL-15 is a component of the inflammatory milieu in the tumor microenvironment promoting antitumor responses. Proc. Natl. Acad. Sci. U.S.A. 116, 599–608 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gajewski T. F., Schreiber H., Fu Y. X., Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 14, 1014–1022 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Motomura Y., et al., The transcription factor E4BP4 regulates the production of IL-10 and IL-13 in CD4+ T cells. Nat. Immunol. 12, 450–459 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson M. D., et al., CyTOF workflow: Differential discovery in high-throughput high-dimensional cytometry datasets. F1000Res. 6, 1–69 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pfaffl M. W., A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Siliņa K., et al., Germinal centers determine the prognostic relevance of tertiary lymphoid structures and are impaired by corticosteroids in lung squamous cell carcinoma. Cancer Res. 78, 1308–1320 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell sequencing data have been deposited in the NCBI Bio Sample database (PRJNA656253).