Abstract
Soft materials that facilitate the three-dimensional (3D) encapsulation, proliferation, and facile local delivery of cells to targeted tissues will aid cell-based therapies, especially those that depend on the local engraftment of implanted cells. Herein, we develop a negatively charged fibrillar hydrogel based on the de novo-designed self-assembling peptide AcVES3-RGDV. Cells are easily encapsulated during the triggered self-assembly of the peptide leading to gel formation. Self-assembly is induced by adjusting the ionic strength and/or temperature of the solution, while avoiding large changes in pH. The AcVES3-RGDV gel allows cell–material attachment enabling both two-dimensional and 3D cell culture of adherent cells. Gel–cell constructs display shear-thin/recovery rheological properties enabling their syringe-based delivery. In vivo cellular fluorescence as well as tissue resection experiments show that the gel supports the long-term engraftment of cells delivered subcutaneously into mice.
Keywords: Hydrogel, peptide, 3D cell culture, cell delivery, self-assembly
Graphical Abstract
INTRODUCTION
The first FDA-approved Car-T cell therapy, tisagenlecleucel, for B-cell precursor acute lymphoblastic leukemia exemplifies the potential of cell-based therapies, which as a therapeutic class, encompasses an enormous spectrum of clinical indications with cancer being only one.1,2 For example, ex vivo gene modification of hematopoietic stem cells holds promise to cure sickle cell disease,3 and somatic and other cell-based approaches are continuously under development for tissue regenerative therapies.4 Delivery of cells and their maintenance in vivo is of paramount importance to the success of any cell-based therapy. For some clinical indications, delivery leading to systemic distribution is important, and for others, local delivery to a particular tissue. For the later, soft materials such as hydrogels can act as scaffolds to facilitate localized delivery, inform phenotype, and aid in establishing long-term engraftment. Self-assembled peptide-based gels are particularly promising in that biochemical cues can be directly incorporated into the peptide’s sequence, they are chemically defined, cells can be directly incorporated during material formation, and some peptide gels exhibit rheological properties that enable their syringe-based delivery.5-15
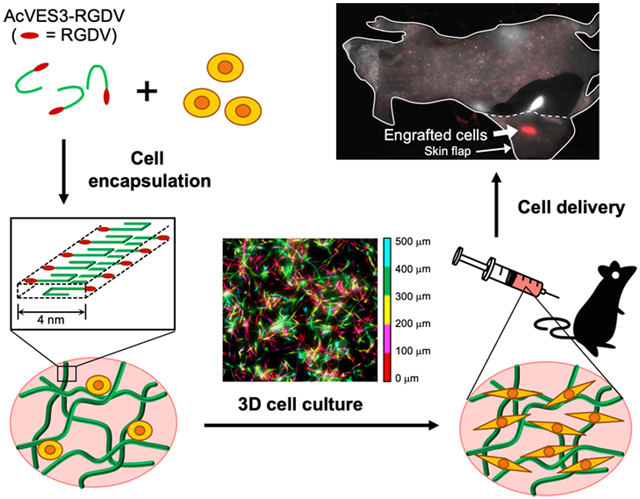
We have been developing a class of β-hairpin peptide gels for use in localized drug delivery,12,16-22 with many of the gels having the potential to deliver cells.23-33 In our system, peptides are designed to be fully soluble when initially dissolved in water but undergo triggered self-assembly leading to the formation of a fibrillar gel network in response to environmental change.33-41 If cells are present during self-assembly, they can be directly encapsulated during gel formation. The resultant peptide gels exhibit shear-thin/recovery behavior following shear stress, allowing their injection by syringe locally to the tissue. Capillary flow studies reported by Pochan et al. indicate that cells experience little or no shear rate during delivery using these gels as a vehicle.42,43 Our early work centered on the development of gels formed by cationic peptides due to their ease of synthesis and purification. Although these peptide gels can be used for cell delivery, the highly positively charged fibrillar networks they form can impact cell viability and phenotype.29 More recently, we turned our attention to developing self-assembling anionic peptides whose negatively charged gel networks might display improved cytocompatibility.44 Our first-generation design comprised carboxylate-rich sequences that undergo triggered self-assembly leading to gelation in response to changes in pH and temperature.32 Although a few of the gels in this family could be used for cell encapsulation, large swings in pH (from pH 11 to 7.4) were needed to trigger material formation. Changes in pH of this magnitude are certainly less than optimal for most cells. Given all the other beneficial characteristics of peptide-based gels, we set out to design a new sequence capable of encapsulating and delivering cells whose gelation did not require large changes in pH. Herein, we report the design, biophysical and mechanical characterization, and utility of a hydrogel formed from the peptide AcVES3-RGDV. The peptide assembles under physiological pH, temperature, and ionic strength conditions affording cytocompatible, semirigid gels capable of encapsulating and delivering cells in vivo for long-term engraftment, Figure 1.
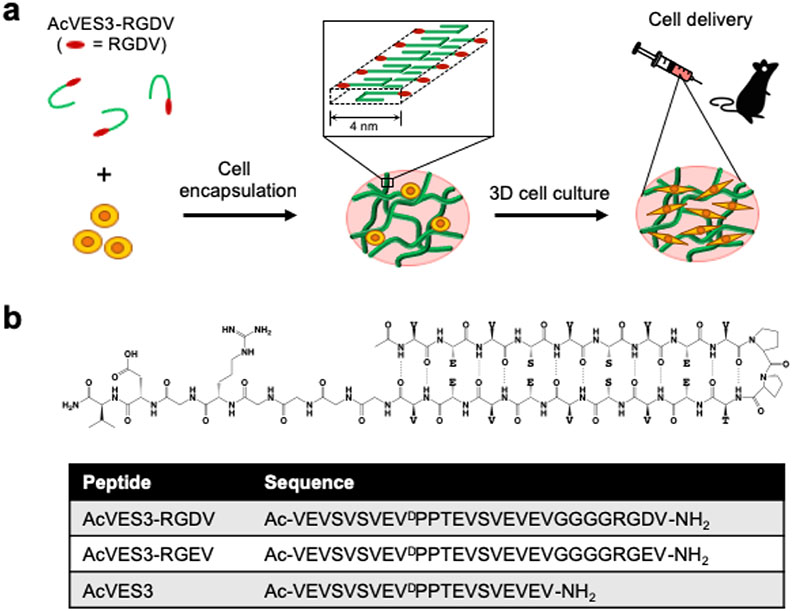
Figure 1.
(a) Cell encapsulation during peptide self-assembly affords fibrillar hydrogel network laden with cells that can be syringe-delivered to target tissues. (b) Schematic of unsymmetrical AcVES3-RGDV hairpin peptide along with the linear sequences of peptides used in this study.
MATERIALS AND METHODS
Peptide Synthesis and Purification.
Peptides were synthesized on PL-rink resin (Agilent Technologies) using an automated ABI 433A peptide synthesizer (Applied Biosystems). Syntheses were performed via solid-phase Fmoc-based chemistry with 1H-benzo-triazolium-1-[bis(dimethylamino)methylene]-5-chloro-hexafluoro-phosphate-(1-),3-oxide (Peptide International) activation. Fmoc-protected amino acids were purchased from Novabiochem. Dried resin-bound peptides were cleaved from the resin and side chain deprotected with a trifluoroacetic acid/thioanisole/ethanedithiol/anisole (90:5:3:2) cocktail for 2 h under an argon atmosphere. Crude peptides were precipitated with cold ethyl ether and lyophilized. Peptides were purified by reversed phase-high-performance liquid chromatography (RP-HPLC) with a Phenomenex PolymerX column employing a linear gradient from 0 to 80% acetonitrile with 20 mM ammonium bicarbonate, peptide peaks fractionated, and finally lyophilized. Peptides were then dissolved in 20 mM ammonium bicarbonate at 1 mg mL−1 and converted to their sodium salts by the addition of a NaOH solution followed by lyophilization. Analytical HPLC and electron spray ionization (negative mode) mass spectrometry (MS) confirmed the purity and composition of the peptides.
Circular Dichroism Spectroscopy.
CD spectroscopy of the peptides was performed on an Aviv 410 spectropolarimeter (Aviv Biomedical). Samples in buffer were prepared by first making a 300 μM stock solution of peptide in MilliQ water. To this, an equal volume of ice-chilled 2× buffer [50 mM N-(2-hydroxyethyl)-piperazine-N′-ethanesulfonic acid (HEPES) containing 300 mM NaCl, pH 7.4] was added resulting in a final 150 μM solution of peptide in 25 mM HEPES, 150 mM NaCl, pH 7.4 buffer. Samples of peptide in pure water were also prepared. Wavelength spectra were collected using a 1 mm path length cell pre-equilibrated at 5 °C, which was then heated to 37 °C. Temperature-dependent data were collected following ellipticity at 216 nm from 2 to 52 °C in 5 °C increments and a 10 min equilibration time for each temperature point. Mean residue ellipticity [θ] was calculated from [θ] = (θobs/10lcr), where θobs is the measured ellipticity in millidegree, l is the cell path length in centimeters, c is the concentration in molarity, and r is the number of residues in the peptide.
Transmission Electron Microscopy.
Fibril morphology was assessed using a Hitachi H-7650 transmission electron microscope at a voltage of 80 kV. Gels were first prepared by mixing 1 wt % solutions of desired peptide and an equal volume of HEPES buffer (50 mM HEPES containing 300 mM NaCl, pH 7.4) to initiate gelation. Gels were incubated overnight at 37 °C. Afterward, samples for transmission electron microscopy (TEM) analysis were prepared by suspending 5 μL of the resulting peptide gel in 195 μL of water to yield a 40-fold dilution. Small amounts (5 μL) of each diluted gel solution were applied to separate carbon-coated copper grids. After 1 min, the grid was washed with water. Then, a 1 w/v % uranyl acetate aqueous solution was placed on each grid for negative staining. Excess staining solution was blotted away, and the grids were then air-dried and imaged.
Oscillatory Rheology.
Rheology experiments were performed on an AR-G2 rheometer (TA Instruments) equipped with a 25 mm stainless steel parallel plate geometry with a 0.5 mm gap height. Gel formation and subsequent shear-thin/recovery was analyzed using the following procedure. For gels formed in HEPES buffer, a 1 wt % peptide solution in water was first prepared on ice, and then an equal volume of buffer (50 mM HEPES containing 300 mM NaCl, pH 7.4) was added to initiate gelation. For the gelation by Dulbecco’s modified Eagle’s medium (DMEM), a 1 wt % peptide solution containing 285 mM sucrose was prepared and then an equal volume of HEPES-supplemented DMEM was added to initiate gelation. After gelation was initiated, 300 μL of the solution was immediately transferred to the rheometer plate equilibrated at 5 °C. Oil was placed around the sample and on the plate to prevent evaporation. The temperature was then ramped to 37 °C (0.5 °C s−1), and a 1 h dynamic time sweep was performed monitoring the storage (G′) and loss (G″) modulus using an angular frequency of 6 rad s−1 and 0.2% strain. After this, 1000% strain was applied for 30 s to dilute the material. Subsequently, the ability of the hydrogel to recover was monitored by measuring G′ at 6 rad s−1 and 0.2% strain for an additional 1 h. Bovine type I collagen gels (0.3 wt %, Thermo Fisher Scientific) were prepared following manufacturer’s instructions.
Rheology measurements of cell-encapsulating gels after 4 days of three-dimensional (3D) cell culturing were performed using an 8 mm stainless steel parallel plate geometry with a 0.5 mm gap height. A cell-encapsulating gel (100 μL) in a transwell insert (24-well plates, Corning) was transferred to the rheometer plate previously equilibrated at 37 °C and oil was placed around the sample. A dynamic time sweep was performed with 6 rad s−1 angular frequency and 0.2% strain, the gel was shear-thinned at 1000% strain for 30 s every 10 min, and allowed to recover by reducing the strain to 0.2%.
Optical Transmittance Measurements.
Optical transmittance measurements were performed using an Agilent 8453 UV–visible Spectroscopy System. A 1 wt % aqueous solution of peptide was prepared, and an equal volume of buffer (50 mM HEPES containing 300 mM NaCl, pH 7.4) was added to initiate gelation. The solution (150 μL) was immediately transferred to a 10 mm path length quartz cell and incubated for 1 h at 37 °C to complete gelation. A 0.3 wt % collagen gel was also prepared following manufacturer’s instructions. The transmittance of the gels was measured in the wavelength range of 380–780 nm.
Cell Maintenance.
Human neonatal dermal fibroblasts (HDFs) and red-fluorescent protein (RFP)-expressing human neonatal dermal fibroblasts (RFP-HDFs) were purchased from Cell Applications and ANGIO-PROTEOMIE, respectively. Cells were maintained in DMEM (low glucose, Thermo Fisher Scientific) containing 10% fetal bovine serum (FBS), 100 U/mL of penicillin, and 100 μg/mL of streptomycin.
Two-Dimensional Cell Culture.
Hydrogels used for two-dimensional (2D) culture (Figures 5 and S3) were prepared as follows. Peptides were initially dissolved in water at 1 wt % and chilled on ice and then mixed with an equal volume of ice-cold HEPES buffer (50 mM HEPES, 300 mM NaCl, pH 7.4). Aliquots of the resulting solution (50 μL per well) were transferred to a nontissue culture-treated 96-well plate and incubated at 37 °C to initiate gelation. After 1 h, HDFs (2 × 104 cells per 200 μL per well) were seeded onto the gels and culture using Essential 8 Medium (data shown in Figure 5) or serum-free DMEM (data in Figure S3a) or 10% FBS-supplemented DMEM (Figure S3b). Cells were cultured for 4 days. Bovine type I collagen gels (0.3 wt %, 50 μL per well) were prepared in 96-well plates following manufacturer’s instructions.
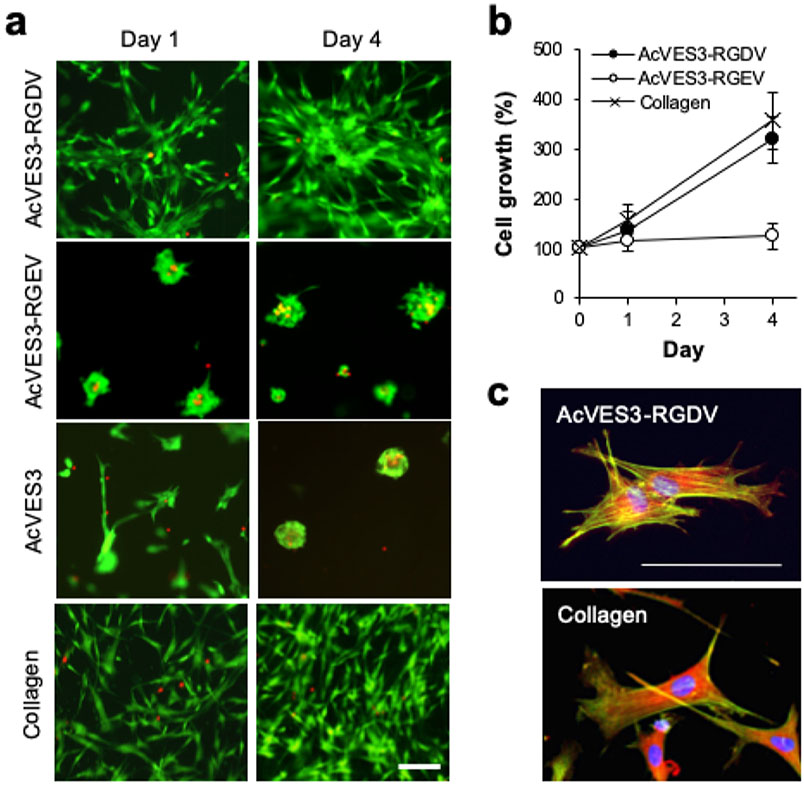
Figure 5.
HDF cells were cultured on the surfaces of 0.5 wt % peptide gels or 0.3 wt % collagen gels in Essential 8 Medium for 4 days. (a) Live and dead cells were visualized using calcein AM and ethidium homodimer-1 staining (scale bar = 100 μm), respectively. (b) Cell proliferation was monitored by quantifying cell viability using a WST-8 assay on days 0, 1, and 4. Data are represented as mean ± standard deviation (SD) of three independent replicates. (c) Cytoskeleton organization and focal adhesions were stained on day 1 against actin (green), vinculin (red), and nuclei (blue, scale bar = 100 μm).
3D Cell Encapsulation and Culture.
For cell encapsulation, peptides are initially dissolved in 285 mM sucrose at 1 wt % and chilled on ice. This solution is mixed with an equal volume of cells suspended in serum-free, HEPES-supplemented DMEM (2 × 106 cells mL−1). A portion of the resulting suspension (5 × 104 cells per 50 μL per insert) was transferred to a Falcon cell culture insert (for 24-well plates, 8 μm pore size, corning) and incubated at 37 °C for 30 min for gelation. These inserts were previously coated with the empty hydrogel (30 μL per insert) to avoid cell adhesion to the bottom surface of the inserts. After gelation of cell-peptide solution, 500 μL of Essential 8 Medium was added to the top and bottom of the gels. Type I collagen gels were prepared following manufacturer’s instructions. Briefly, a 0.4 wt % collagen solution was prepared on ice and mixed with cell suspension in serum-free DMEM (4 × 106 cells mL−1) at a ratio of 3:1 (0.3 wt % collagen, 1 × 106 cells mL−1).
Cell Viability Assay.
Cell viability was evaluated by live and dead cell staining after 2D and 3D cell culture. Serum-free DMEM containing 1 μM calcein-AM and 2 μM ethidium homodimer-1 was added to the cells (200 μL per well for 2D culture and 500 μL each to the top and bottom of the gels for 3D culture). After incubation at 37 °C for 30 min, live and dead cells were visualized using an EVOS FL Auto Cell Imaging System (Thermo Fisher Scientific). For the pseudo-colored Z-stack composite, images of the live-stained cells were captured every 5 μm and then Z-stack images for each 100 μm were flattened using the EVOS software, and the flattened images were pseudo-colored and merged using ImageJ.
Cell Proliferation Assay.
Cell proliferation was assessed using a WST-8 assay, Cell Counting Kit-8 (CCK-8, Dojindo Molecular Technologies) for 2D and 3D cell culture. Serum-free DMEM containing 10% CCK-8 was added to the gels (200 μL per well for 2D culture; 500 μL to the top of the gels for 3D culture) and incubated for 2 h at 37 °C. Subsequently, media were transferred to 96-well plates (150 μL per well) and their absorbance measured at 450 nm relative to control viabilities from day 0 for 2D cell culture and day 1 for 3D cell culture. Experiments were performed in triplicate.
Cytoskeleton Staining.
For 2D cell culture, the peptide and collagen gels (70 μL) were prepared as described above on the glass bottom of 35 mm dishes (7 mm glass diameter, MatTek). HDFs in Essential 8 Medium (1.5 × 105 cells per 3 mL) were added to the dishes and incubated at 37 °C for 24 h. Cells were fixed and permeabilized with 4% paraformaldehyde and 0.1% Triton X-100 in phosphate-buffered saline (PBS) for 15 min and then blocked with 1% BSA in PBS. Cells were incubated with anti-vinculin antibody (hVIN-1, Sigma) for 1 h at room temperature. Then, a goat anti-mouse IgG (H + L) cross-adsorbed secondary antibody, Rhodamine Red-X (2 μg mL−1, Thermo Fisher Scientific), Alexa Fluor 488 phalloidin (5 units mL−1, Thermo Fisher Scientific), and Hoechst 33342 (5 μg mL−1, Invitrogen) in PBS were added to the dishes and incubated for 1 h at room temperature. After washing, the cells were visualized using an EVOS FL Auto Cell Imaging System.
For 3D cell culture, HDF-encapsulating gels were prepared in the Falcon cell culture inserts (5 × 104 cells per 50 μL gel per insert) as described above. Next, the cells were cultured in Essential 8 Medium for 4 days. The cells were then fixed, permeabilized, and stained simultaneously with 4% paraformaldehyde, 0.1% Triton X-100, and Alexa Fluor 488 phalloidin (5 units mL−1) in PBS (500 μL to the top and bottom of the gels) for 1 h at room temperature. After washing with PBS (15 min, three times), Hoechst 33342 (5 μg mL−1, 500 μL to the top and bottom of the gels) was added for 30 min at room temperature. The cells were washed with PBS and visualized using an EVOS FL Auto Cell Imaging System.
In Vivo Implantation.
Six to eight week-old nontumor-bearing female athymic nude mice were used (n = 3). Animal care was in accordance with the procedures outlined in the Guide for Care and Use of Laboratory Animals (National Research Council, 1996; National Academy Press, Washington, D.C.), and animal protocols were approved by the Frederick National Laboratory for Cancer Research (FNLCR) Institutional Animal Care and Use Committee. RFP-HDFs were encapsulated in 0.5 wt % AcVES3-RGDV at 107 cells mL−1 as described in the 3D cell culture section. After gelation, the gel–cell construct (106 cells per 100 μL) was syringe-injected into the left dorsal region of the mouse, and as a control, an RFP-HDF suspension in serum-free DMEM (106 cells per 100 μL) was injected into the right dorsal region of the mouse. Multispectral fluorescence imaging was performed daily (Monday-Friday) for 42 days using a Maestro Flex imager (PerkinElmer). Briefly, mice were anesthetized for 3–4 min in an induction chamber with 3% isoflurane with filtered air (0.2 μm filter) used as a carrier at a flow rate of 1 L per minute. After isoflurane induction, the mice were transferred to an imaging chamber, where isoflurane was reduced to 2% and O2 was used as a carrier. The imaging platform was kept at 37 °C to maintain the mouse’s internal temperature. Image acquisition (Maestro software, ver. 2.10.0) of the RFP fluorescence signal utilized a 533 ± 26 nm excitation filter (placed in front of the 300 W xenon excitation source), a 580 nm long-pass filter (placed in front of the charge-coupled device), and implementing a multispectral imaging acquisition of 550–720 nm range, 10 nm increment, autoexposure (typically 0.5–5 s per emission wavelength), with a 2 × 2 pixel binning, resulting in a stack of images (image cube). The RFP signal was distinguished from the tissue autofluorescence by employing a vendor-specified unmixing protocol. Quantitative analysis was performed by drawing a standard circular region of interest (ROI) over the injection site of the unmixed RFP component image. The total RFP signal (scaled counts second−1) within each ROI was measured and plotted as a function of time.
After in vivo imaging on day 42, mice were euthanized using CO2 and ex vivo images (Maestro Software) were acquired by exposing the gel–cell construct injection site. Image acquisition and processing were performed with the same parameters as performed for in vivo imaging as stated above. The RFP-positive tissue was isolated utilizing the unmixed RFP image to guide its resection. The isolated tissue was homogenized and cells were isolated via a cell strainer. The collected cells were then cultured on a tissue culture plate with FBS-supplemented DMEM for 10 days and observed using an EVOS FL Auto Cell Imaging System.
RESULTS AND DISCUSSION
Peptide Gel Design.
We have expended considerable effort in defining the design rules for the self-assembling cationic β-hairpin peptides that have been previously developed. Careful biophysical,23,34,35,37,40,45-48 mechanical,41-43,45,49,50 and structural41,51-53 studies have provided insight into their mechanism of self-assembly, hydrogel material properties, and their structure in the self-assembled state to molecular detail. When these cationic peptides are initially dissolved in water, they adopt an ensemble of random coil conformations because of the electrostatic repulsion between their protonated side chains affording a stable solution of monomeric peptide. This is an important design feature of these peptides in that it ensures a well-defined starting phase state from which self-assembly and resultant hydrogelation can be triggered. Many other reported peptide systems rely on rapid dissolution of a peptide’s lyophilized solid state, which often takes place concurrently with self-assembly. Dissolution kinetics are highly dependent on particle size and environmental conditions and thus difficult to control. Thus, the self-assembly mechanism, fibril morphology, and mechanical properties of resultant gels can vary dramatically batch-to-batch.
From the unordered state of our peptides, self-assembly is triggered by a number of means but usually entails alleviating the charge repulsion between amino acid side chains by adjusting the pH or ionic strength. Increasing the temperature also facilitates assembly by driving the hydrophobic effect and the burial of hydrophobic side chains into the dry interior of the fibrils. Solid-state NMR indicates that this class of peptide assembles into fibrils where sequences are folded, adopting an amphiphilic β-hairpin structure.53 Peptides assemble with high fidelity into fibrils of reproducible morphology composed of a bilayered cross-β structure. Folded hairpins assemble with their β-strands in-register along a given fibril monolayer. The resulting network formed by these fibrils constitutes a mechanically rigid hydrogel. Having established the core design principles allows one to efficiently design new peptides with targeted characteristics.
AcVES3-RGDV contains 28 residues and is designed to adopt an unsymmetrical amphiphilic β-hairpin in its self-assembled fibril state, Figure 1B. Here, the C-terminal strand is eight residues longer than the N-terminal strand because of the inclusion of a linker and integrin binding motif. Designing unsymmetric β-sheets is not trivial and requires exact turn reversal in the sequence of the peptide main chain. Linear peptides designed to fold into β-sheet structure can easily shift their β-strand registry, even when they contain turn sequences intended to exactly define the location of main chain reversal.54 AcVES3-RGDV incorporates one of the strongest turn-propensity motifs known at sequence positions 9–12, namely—VDPPT. This four-residue turn sequence contains a -DPP- at the i + 1, i + 2 positions of the reverse turn. Balaram’s structural work on DPro-Xxx dipeptide motifs indicate that they form ideal type II′ turn geometries that strictly define chain reversal and facilitate the formation of β-hairpin structure.55,56 Valine was placed at the i-position for its ability to enforce a trans-prolyl bond at the d-proline, a requirement for ensuring chain reversal at this position within the sequence.57 Lastly, threonine was included at the i + 3 position based on its inherent propensity to occupy this position in type II′ turns of naturally occurring proteins.58 The peptide contains two eight-residue segments proximal to the turn region, each containing an alternating sequence of hydrophobic and hydrophilic residues, which gives AcVES3-RGDV facial amphiphilicity when folded in the self-assembled state. Valines, occupying the hairpin’s hydrophobic face, are a β-branched residue with a high propensity for sheet structure.59 Hydrophobic collapse of the valine-rich faces of hairpins drives self-assembly and the formation of the bilayered cross-β structure of the fibrils.53 The design of the hydrophilic face of the AcVES3-RGDV hairpin is more involved. We designed toward a hairpin having a formal charge state of −5 (excluding residues in the integrin binding motif). Previous work suggested that this degree of charge displayed from a facial amphiphile could be screened by solutions of physiological ionic strength.32 Glutamic acid was used to imbibe charge, whose positional placement within the hairpin is important for defining the initial solution phase state of the peptide and actuating self-assembly. Four of the Glu residues were placed in pairs directly opposing each other across the hairpin at positions 2/19 and 8/13. Pairwise charge repulsion keeps the peptide unfolded and monomeric until solution conditions permit self-assembly. The fifth Glu residue was placed at position 17, which can interact diagonally with Glu2 to also inhibit self-assembly. Neutral hydrophilic serine residues were placed at the remaining positions on the hairpin’s hydrophilic face. As will be shown later, screening the Glu-based charged can trigger hydrogelation under physiological conditions. Last, -RGDV-, a known integrin binder was incorporated at the C-terminus through a four-residue Gly linker to facilitate cell adhesion.60 The length of the linker was chosen based on polymer studies showing that four glycines were sufficient to allow cellular adhesion.61,62 Two additional peptides were prepared as controls. The first, AcVES3-RGEV, is a negative control that replaces the aspartic acid residue in the integrin binding domain with glutamic acid. This sequence change should limit the ability of the gel to foster cell adhesion. The second control, AcVES3, lacks the linker and integrin binding domain altogether to determine if placing sequence extensions at the C-terminus influences peptide self-assembly, gelation, and gel mechanical properties. Peptides were prepared by Fmoc-based solid-phase peptide synthesis, purified by HPLC, and their primary structure characterized by electrospray ionization (ESI)–MS, Figure S1.
Triggered Gelation under Physiological Buffer Conditions.
AcVES3-RGDV undergoes triggered self-assembly leading to the formation of a fibrillar gel under various physiologically relevant buffer conditions, including biophysical buffers and cell culture media with or without calf serum. For example, we first studied the self-assembly of AcVES3-RGDV using biophysical buffer. CD spectroscopy shows that when AcVES3-RGDV is initially dissolved in water, the peptide remains unfolded at 37 °C because of the charge repulsion between glutamate side chains, Figure 2a (solid symbols). Adding saline buffer resulting in a solution composed of 25 mM HEPES and 150 mM NaCl (pH 7.4) increases the ionic strength of the solution, which screens the glutamate charge allowing self-assembly and the formation of β-sheet-rich fibrils as evidenced by the transformation of the wavelength spectra from a random coil to the β-sheet (open symbols).34 A similar behavior is observed for the two control peptides, Figure 2b,c. Increasing the temperature also drives self-assembly via the hydrophobic effect. Each of the peptides are also able to undergo temperature-dependent assembly at pH 7.4 in buffered saline as shown in Figure 2d-f (open symbols). Peptides dissolved in pure water are largely temperature-insensitive (closed symbols). Thus, thermal energy can trigger assembly only if the solution conditions (ionic strength and pH) are permissive. Importantly, AcVES3-RGDV shows a capacity to assemble at 37 °C when dissolved in physiologically relevant buffer.
Figure 2.
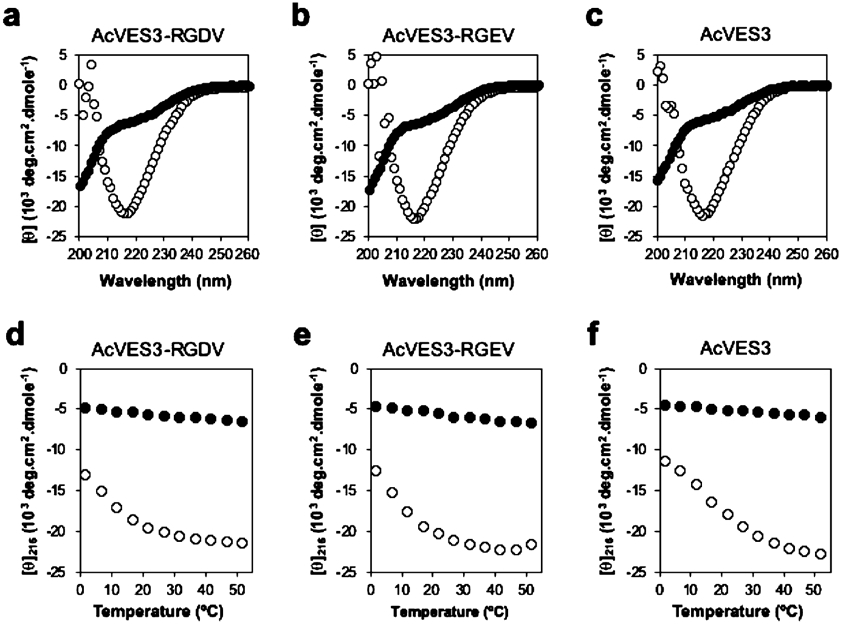
(a–c) CD wavelength spectra of 150 μM peptides in water (●) or in HEPES buffer (○) (25 mM HEPES, 150 mM NaCl, pH 7.4) at 37 °C. (d–f) Temperature-dependent CD following [θ]216 in HEPES buffer. Increased signal at 216 nm is indicative of β-sheet structure formation.
Peptides assemble into morphologically well-defined fibrils as shown in Figure 3. Here, fibrils are isolated from 0.5 wt % gels and interrogated by TEM. The average width of fibrils formed by AcVES3-RGDV is 4.0 ± 0.4 nm, which is consistent with the peptide adopting the desired unsymmetrical hairpin fold in its self-assembled fibrillar state. The width of the AcVES3-RGEV fibrils is nearly identical as expected, and the width of fibrils formed by AcVES3, the control peptide lacking the integrin binding domain, is ~3 nm, consistent with a shorter symmetrical hairpin comprising these fibrils.
Figure 3.
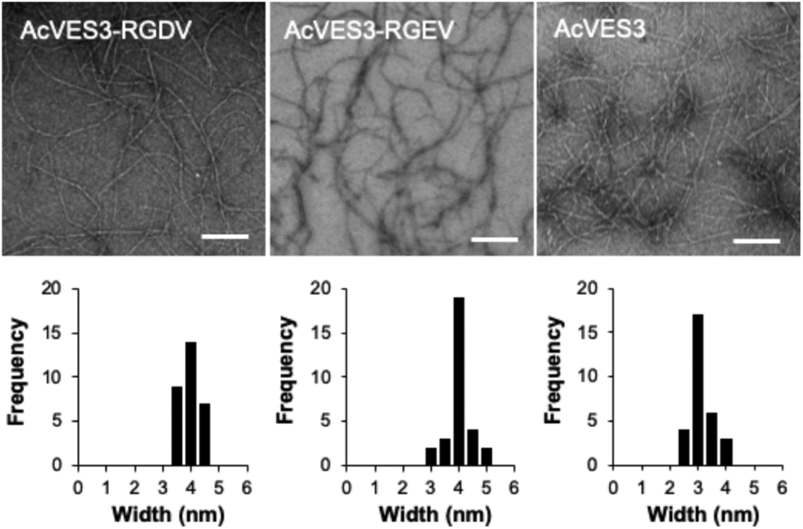
Transmission electron micrographs showing local morphology of fibrils formed by AcVES3-RGDV, AcVES3-RGEV, and AcVES3 peptides. The peptide gels (0.5 wt %) in HEPES buffer (25 mM HEPES containing 150 mM NaCl, pH 7.4) were formed by incubating overnight at 37 °C. Then, the TEM samples were prepared by suspending 5 μL of the resulting peptide gel in 195 μL of water to yield a 40-fold dilution. Fibril widths were determined from 30 independent measurements (scale bar = 100 nm).
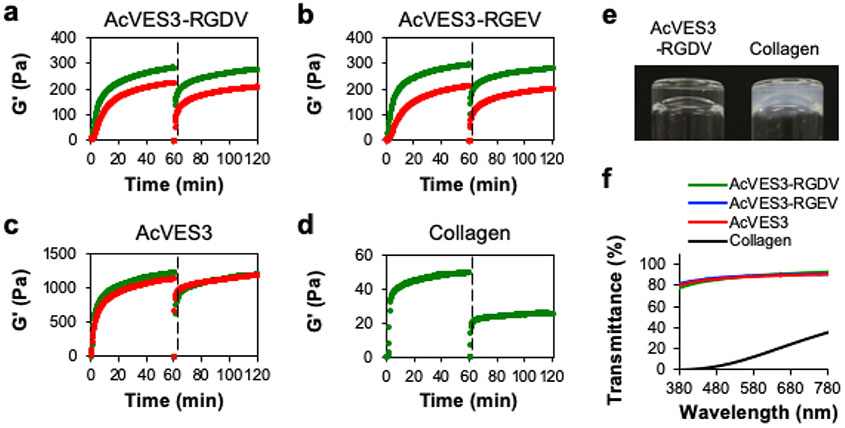
The rheological properties of 0.5 wt % gels formed by the three peptides were characterized by time sweep shear-thin/recovery experiments, Figure 4. Here, self-assembly is triggered and hydrogel formation is followed over 60 min; at that time, the resultant gels are thinned and allowed to recover. Gelation was triggered either by the addition of HEPES saline buffer (green data, final buffer: 25 mM HEPES, 150 mM NaCl, pH 7.4) or cell culture media (red data, final buffer: 0.5× HEPES-supplemented DMEM containing 143 mM sucrose). After 60 min, both the AcVES3-RGDV and control AcVES3-RGEV form moderately rigid gels (G′ ≈ 300 Pa) in HEPES buffer. Although the ionic strength of the DMEM solution is sufficient to screen the glutamate side chain charge to initiate self-assembly and resultant gelation, each gel exhibits a decrease in rigidity in cell culture media (G′ ≈ 200 Pa). This difference is most likely due to the lower ionic strength of cell culture media compared to HEPES buffer. Gels formed by the second control peptide, AcVES3, are significantly more rigid (G′ ≈ 1200 Pa) and show no dependence on the triggering buffers used here. The observed differences in G′ between the gels formed by the AcVES3 control and the other two longer peptides are interesting. Although CD and TEM indicated that the C-terminal integrin binding domain contained in the longer peptides does not influence self-assembly and fibril formation, rheology suggests that this sequence extension does influence network rigidity. The molecular basis for this is currently unknown but could be due to changes in fibril bending moduli or fibril network mesh size.63-65 At any rate, all three peptide gels are of sufficient rigidity to encapsulate cells and are capable of shear-thin/recovery, indicating that they can be delivered in vivo by syringe injection. For comparison, we performed similar experiments on a bovine type I collagen gel, which is FDA approved and widely used for 2D and 3D cell culture,66,67 Figure 4d. The collagen gel is substantially weaker and does not recover well after being shear-thinned, suggesting that its ability to deliver cells by syringe injection may be limited. Dynamic frequency and strain sweep experiments were also performed on the gels and provided in Figure S2.
Figure 4.
(a–c) Oscillatory rheological dynamic time sweeps and shear-thin/recovery of 0.5 wt % peptide gels monitoring the storage modulus (G′). Gelation was triggered with either HEPES buffer (green data, final buffer in gel: 25 mM HEPES, 150 mM NaCl, pH 7.4) or cell culture media (red data, final buffer: 0.5× HEPES-supplemented DMEM containing 143 mM sucrose). (d) 0.3 wt % Type I collagen gel in PBS buffer. The first 60 min displays the onset of gelation under a strain of 0.2% and a frequency of 6 rad s−1. Subsequently, the gels were shear-thinned at 1000% strain for 30 s (indicated by the dashed line) and allowed to recover by reducing the strain to 0.2%. (e) Image of an optically clear 0.5 wt % AcVES3-RGDV gel and an opaque 0.3 wt % collagen gel. (f) Transmittance measurements of 0.5 wt % peptide gels and a 0.3 wt % collagen gel.
Last, we assessed the optical transparency of the peptide gels, Figure 4e,f. Transparency is a useful gel characteristic that allows microscopy to be performed on encapsulated cells. Panel (e) shows a visual comparison of an AcVES3-RGDV gel, which is optically clear, with the collagen gel, which appears opaque. Light transmittance was also measured over a range of wavelengths (380–780) commonly employed in cell microscopy. Panel (f) shows that the peptide gels can transmit significantly more light than the collagen gel. Taken together, the CD, TEM, and rheological analysis indicate that AcVES3-RGDV and its control peptides undergo triggered self-assembly under physiological buffer or cell culture conditions to form gels composed of morphologically well-defined fibrils. Gels exhibit shear-thin/recovery behavior allowing their delivery by syringe injection.
2D Cell Culture on Peptide Gel Surfaces.
Initial 2D experiments were performed to assess the ability of the gels to permit cell adhesion, viability, and proliferation. The data in Figure 5 were collected using gels prepared in HEPES saline buffer. After gel formation, HDF (human dermal fibroblast) cells were introduced and cultured in chemically defined Essential 8 Medium. These serum-free conditions were used to eliminate any possible effects from adsorbed serum proteins. Figure 5a shows a time-dependent live–dead assay that reports viability (green cells are alive, red cells are dead) and also allows qualitative assessment of cell adhesion and morphology. Cells seeded onto the AcVES3-RGDV gel robustly attach and form spread-out morphologies within a day and are viable over the course of 4 days, the last time point tested. The behavior of cells cultured on the AcVES3-RGDV gel was similar to that of cells cultured on the collagen gel. In contrast, cells introduced onto the control AcVES3-RGEV gel were initially attached, but a significant number had lifted off the gel surface over time. Those cells that did remain attached had rounded clumped morphologies and some cells stained red indicating death. Cells introduced onto the AcVES3 control gel behaved similarly. Thus, the control gels, which lack proper integrin adhesion signals, are unable to support long-term cell attachment nor foster adhesion events leading to spread out morphologies. More importantly, the data suggest that cells are able to access the -RGDV- motif presented from the surface of fibrils comprising the AcVES3-RGDV gel, which facilitates adhesion. We also performed similar live–dead assays using serum-free DMEM to culture the cells and similar trends were observed underscoring the importance of the integrin binding domain, Figure S3a. In fact, when cells are cultured in the presence of 10% FBS, cells are unable to properly adhere and adopt spread-out morphologies presumably because serum protein adsorption limits access to the -RGDV- motif displayed from the gel’s surface, Figure S3b. WST-8 cell proliferation assays show that the AcVES3-RGDV gel also supports cell proliferation to the same degree as cells cultured on the collagen gel, Figure 5b. In contrast, although some of the cells cultured on the control AcVES3-RGEV gel were viable, they did not proliferate. Lastly, confocal microscopy was used to further examine cell adhesion. Figure 5c shows that cells seeded onto the AcVES3-RGDV gel contain actin stress fibers and focal adhesion points, the formation of which involves integrin binding.68 Taken together, these 2D studies show that the AcVES3-RGDV gel is cytocompatible, and we proceeded to investigate its potential utility in 3D cell encapsulation, culture, and in vivo delivery.
3D Cell Encapsulation, Culture, and in Vivo Delivery.
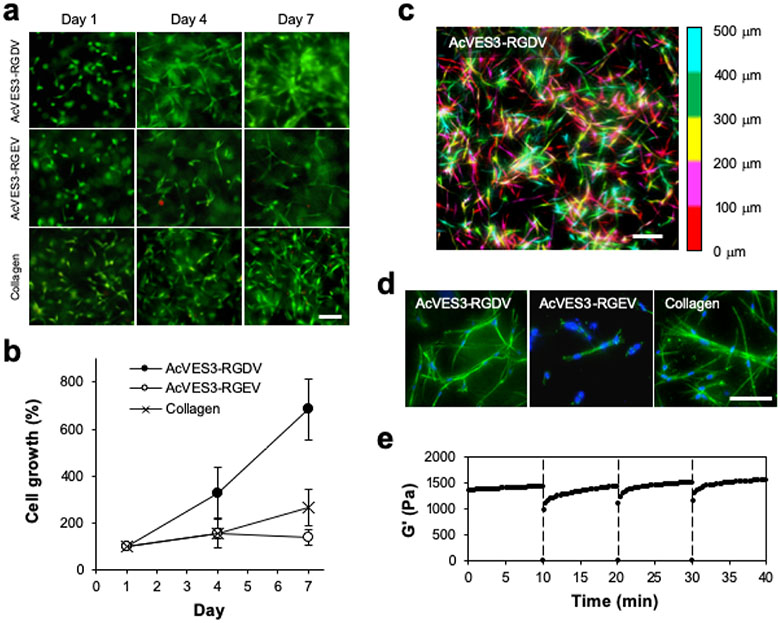
Encapsulation of cells within the AcVES3-RGDV gel is accomplished during peptide self-assembly in a simple procedure. Here, the peptide is initially dissolved in water containing 285 mM sucrose and an equal volume of cells suspended in serum-free, HEPES-supplemented DMEM is added. The resulting solution is incubated at 37 °C to allow gelation. This procedure produces uniform distribution of cells directly encapsulated throughout the hydrogel. Live–dead assays performed over the time course of a week show that cells are viable and able to form spread-out morphologies within the gel’s fibrillar network, Figure 6a. A similar behavior was observed for the collagen gel. In contrast, although cells encapsulated within the AcVES3-RGEV control gel remained viable, only a small population was capable of forming spread-out morphologies. Cell proliferation was quantified over the same time period, which showed that the cells encapsulated within the AcVES3-RGDV gel proliferated nearly 3 times more quickly than cells contained within collagen gels, Figure 6b. Cells within the control AcVES3-RGEV gel did not proliferate over the same 7 day period. Confocal microscopy was then used to study cellular distribution within the gel matrix Figure 6c is a Z-stack image that shows cells embedded throughout 500 microns of gel where cells are false-colored according to their depth within the fibrillar network. Cells are uniformly distributed and appear to have adopted healthy spread-out morphologies, which was later verified via additional confocal studies that monitored actin stress fiber development, Figure 6d. Every observed cell within the AcVES3-RGDV gel formed actin stress fibers, in contrast to those embedded within the control AcVES3-RGEV gel. Importantly, gels containing cells that had been cultured for extended periods of time still maintained their shear-thin/recovery rheological property. Figure 6e shows a series of time sweep experiments on a gel containing cells that had been cultured for 4 days. The gel cell culture was repeatedly shear-thinned every 10 min and showed the potential to recover nearly instantaneously after each thinning cycle. Together, the data in Figure 6 demonstrate that the AcVES3-RGDV gel can be used to easily encapsulate and culture adherent cells in 3D and further demonstrate rheological properties consistent with facilitating in vivo implantation of cell cultures by syringe-based injection.
Figure 6.
HDF cells were encapsulated and 3D-cultured within 0.5 wt % peptide gels or 0.3 wt % collagen gels. (a) Live and dead cells were visualized on days 1, 4, and 7, using calcein AM and ethidium homodimer-1 staining, respectively (scale bar = 100 μm). (b) Cell proliferation was monitored by quantifying cell viability using a WST-8 assay on days 0, 1, and 4. Data are represented as mean ± SD of three independent replicates. (c) A pseudo-colored composite of a Z-stacked image (500 μm) of live cells stained with calcein AM encapsulated within a 0.5 wt % AcVES3-RGDV gel at day 4 of culture. The depths are colored red (0–100 μm), magenta (100–200 μm), yellow (200–300 μm), green (300–400 μm), and cyan (400–500 μm); scale bar = 100 μm. (d) Z-stack image of cytoskeleton organization in the gels on day 4 of culture with actin-stained green and nuclei stained blue, scale bar = 100 μm. (e) Shear-thin/recovery of a 0.5 wt % AcVES3-RGDV gel containing cells that had been 3D-cultured for 4 days. Gels were shear-thinned at 1000% strain for 30 s (indicated by dashed line) every 10 min and allowed to recover by reducing the strain to 0.2%.
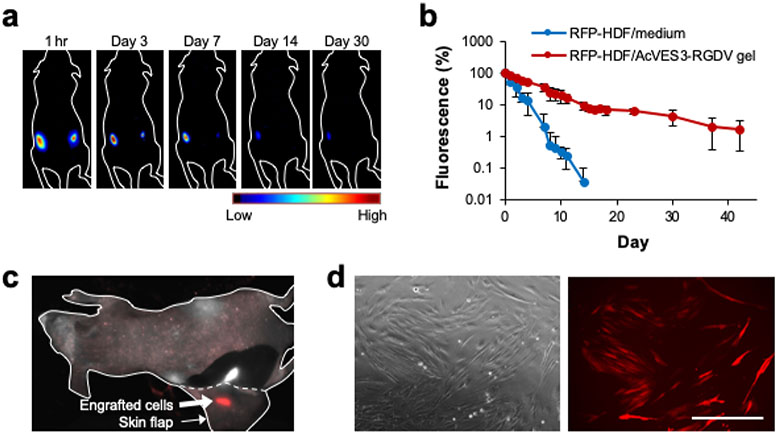
We next assessed the ability of the AcVES3-RGDV gel to deliver cells in vivo and facilitate their long-term engraftment into implanted tissue. RFP-expressing human dermal fibroblasts were encapsulated into the gel and delivered subcutaneously to the dorsal flank of athymic mice via syringe injection. Subsequent fluorescence imaging allowed in situ monitoring of cell residency as a function of time. Figure 7a shows a representative mouse that received an injection of gel/cell construct into its left dorsal flank. Once injected, the cells may experience several different fates including death, migration out of the gel into distal regions, or local engraftment into the tissue at the implant site. The qualitative data in panel (a) and the quantitative data in panel (b) show that a population of cells remains resident at the implant site for at least 42 days (the last time point assayed). In contrast, control injections of cells suspended in media into the right flank maintain residency only about 7 days.
Figure 7.
(a) RFP-HDFs were encapsulated in AcVES3-RGDV gels (106 cells per 100 μL) and injected into the left dorsal region. In the control, the cells were suspended in medium (106 cells per 100 μL) and injected into the right dorsal region of the same mouse. Fluorescent images were recorded over time. (b) RFP fluorescence from an ROI at the implantation site plotted as a function of time. Data are represented as mean ± SD (n = 3). (c) RFP fluorescence from the dissected subcutaneous tissue on day 42. Red and white signals represent RFP fluorescence and tissue autofluorescence, respectively. (d) Cells resected from an RFP-positive tissue were cultured in vitro for 10 days (scale bar = 400 μm).
Cell engraftment was verified at day 42 by resecting the subcutaneous tissue at the gel/cell construct implantation site. Figure 7c shows an isolated skin flap before fully resecting the tissue, engrafted cells are clearly observed. The isolated and surrounding tissue had no visible signs of inflammation nor the presence of intact gel, suggesting that it is biocompatible and fully biodegraded over the time course of the experiment. To further confirm successful engraftment, the cells were harvested from the isolated tissue at day 42 and expanded in 2D culture. Figure 7d shows a typical culture of cells that clearly demonstrate a fluorescent phenotype, indicating that a population of delivered cells locally engrafted into the tissue at the implantation site.
CONCLUSIONS
Gels capable of encapsulating, culturing, and delivering cells could facilitate cell-based therapies, especially treatments that require local engraftment. We report a new hydrogel, based on the de novo-designed, self-assembling peptide AcVES3-RGDV that can be used to easily encapsulate cells during peptide assembly, leading to gel formation. 2D and 3D culture studies show that the gel facilitates cell attachment, the adoption of spread-out cellular morphologies, and allows proliferation. Although we demonstrate the utility of employing adherent cells in this study, the gel should be conducive to delivering nonadherent cells as well. The viscoelastic gel is shear thinning but self-heals, allowing its delivery by syringe injection. Cells can be encapsulated and cultured directly in the gels and then the entire culture directly implanted into mice by syringe. In vivo fluorescent studies as well as tissue resection cell isolation studies show that the biodegradable AcVES3-RGDV gel facilitates long-term engraftment of implanted cells.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsami.9b12152.
Analytical HPLC and ESI–mass spectra of pure peptides; rheology measurements of peptide gels; and 2D culture of HDFs on gels with DMEM or FBS-supplemented DMEM (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Maude SL; Laetsch TW; Buechner J; Rives S; Boyer M; Bittencourt H; Bader P; Verneris MR; Stefanski HE; Myers GD; De Moerloose B; Hiramatsu H; Schlis K; Davis KL; Martin PL; Nemecek ER; Yanik GA; Peters C; Baruchel A; Boissel N; Mechinaud F; Balduzzi A; Krueger J; June CH; Levine BL; Wood P; Taran T; Leung M; Mueller KT; Zhang Y; Sen K; Lebwohl D; Pulsipher MA; Grupp SA Tisagenlecleucel in Children and Young Adults with B-cell Lymphoblastic Leukemia. N. Engl. J. Med 2018, 378, 439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Prasad V Tisagenlecleucel - the first approved CAR-T-cell therapy: implications for payers and policy makers. Nat. Rev. Clin. Oncol 2018, 15, 11. [DOI] [PubMed] [Google Scholar]
- (3).Mapara MY; Tisdale JF; Kanter J; Kwiatkowski JL; Krishnamurti L; Schmidt M; Miller AL; Pierciey FJ Jr.; Shi W; Ribeil J-A; Asmal AA; Walters MC Lentiglobin Gene Therapy in Patients with Sickle Cell Disease: Updated Interim Results from Hgb-206. Biol. Blood Marrow Transplant 2019, 25, S64–S65. [Google Scholar]
- (4).Mount NM; Ward SJ; Kefalas P; Hyllner J Cell-based Therapy Technology Classifications and Translational Challenges. Philos. Trans. R. Soc., B 2015, 370, 20150017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Zhou M; Smith AM; Das AK; Hodson NW; Collins RF; Ulijn RV; Gough JE Self-Assembled Peptide-based Hydrogels as Scaffolds for Anchorage-dependent Cells. Biomaterials 2009, 30, 2523–2530. [DOI] [PubMed] [Google Scholar]
- (6).Webber MJ; Tongers J; Renault M-A; Roncalli JG; Losordo DW; Stupp SI Development of Bioactive Peptide Amphiphiles for Therapeutic Cell Delivery. Acta Biomater. 2010, 6, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Maude S; Miles DE; Felton SH; Ingram J; Carrick LM; Wilcox RK; Ingham E; Aggeli A De Novo Designed Positively Charged Tape-forming Peptides: Self-Assembly and Gelation in Physiological Solutions and their Evaluation as 3D Matrices for Cell Growth. Soft Matter 2011, 7, 8085–8099. [Google Scholar]
- (8).Pérez CMR; Panitch A; Chmielewski J A Collagen Peptide-Based Physical Hydrogel for Cell Encapsulation. Macromol. Biosci 2011, 11, 1426–1431. [DOI] [PubMed] [Google Scholar]
- (9).Kang MK; Colombo JS; D’Souza RN; Hartgerink JD Sequence Effects of Self-Assembling Multidomain Peptide Hydrogels on Encapsulated SHED Cells. Biomacromolecules 2014, 15, 2004–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Szkolar L; Guilbaud J-B; Miller AF; Gough JE; Saiani A Enzymatically Triggered Peptide Hydrogels for 3D cell Encapsulation and Culture. J. Pept. Sci 2014, 20, 578–584. [DOI] [PubMed] [Google Scholar]
- (11).Motamed S; Del Borgo MP; Kulkarni K; Habila N; Zhou K; Perlmutter P; Forsythe JS; Aguilar MI A Self-Assembling β-peptide Hydrogel for Neural Tissue Engineering. Soft Matter 2016, 12, 2243–2246. [DOI] [PubMed] [Google Scholar]
- (12).Sun Y; Li W; Wu X; Zhang N; Zhang Y; Ouyang S; Song X; Fang X; Seeram R; Xue W; He W Functional Self-Assembling Peptide Nanofiber Hydrogels Designed for Nerve Degeneration. ACS Appl. Mater. Interfaces 2016, 8, 2348–2359. [DOI] [PubMed] [Google Scholar]
- (13).Green H; Ochbaum G; Gitelman-Povimonsky A; Bitton R; Rapaport H RGD-Presenting Peptides in Amphiphilic and Anionic β-sheet Hydrogels for Improved Interactions with Cells. RSC Adv 2018, 8, 10072–10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Hainline KM; Gu F; Handley JF; Tian YF; Wu Y; de Wet L; Vander Griend DJ; Collier JH Self-Assembling Peptide Gels for 3D Prostate Cancer Spheroid Culture. Macromol. Biosci 2019, 19, 1800249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Zhou J; Du X; Chen X; Xu B Adaptive Multifunctional Supramolecular Assemblies of Glycopeptides Rapidly Enable Morphogenesis. Biochemistry 2018, 57, 4867–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Branco MC; Pochan DJ; Wagner NJ; Schneider JP Macromolecular Diffusion and Release from Self-Assembled beta-hairpin Peptide Hydrogels. Biomaterials 2009, 30, 1339–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Altunbas A; Lee SJ; Rajasekaran SA; Schneider JP; Pochan DJ Encapsulation of Curcumin in Self-Assembling Peptide Hydrogels as Injectable Drug Delivery Vehicles. Biomaterials 2011, 32, 5906–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Lindsey S; Piatt JH; Worthington P; Sönmez C; Satheye S; Schneider JP; Pochan DJ; Langhans SA Beta hairpin Peptide Hydrogels as an Injectable Solid Vehicle for Neurotrophic Growth Factor Delivery. Biomacromolecules 2015, 16, 2672–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Medina SH; Li S; Howard OMZ; Dunlap M; Trivett A; Schneider JP; Oppenheim JJ Enhanced Immunostimulatory Effects of DNA-Encapsulated Peptide Hydrogels. Biomaterials 2015, 53, 545–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Nagy-Smith K; Yamada Y; Schneider JP Protein Release from Highly Charged Peptide Hydrogel Networks. J. Mater. Chem. B 2016, 4, 1999–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Majumder P; Baxa U; Walsh STR; Schneider JP Design of a Multicompartment Hydrogel that Facilitates Time-Resolved Delivery of Combination Therapy and Synergized Killing of Glioblastoma. Angew. Chem 2018, 130, 15260–15264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Yamada Y; Chowdhury A; Schneider JP; Stetler-Stevenson WG Macromolecule-Network Electrostatics Controlling Delivery of the Biotherapeutic Cell Modulator TIMP-2. Biomacromolecules 2018, 19, 1285–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Kretsinger JK; Haines LA; Ozbas B; Pochan DJ; Schneider JP Cytocompatibility of Self-Assembled Beta-hairpin Peptide Hydrogel Surfaces. Biomaterials 2005, 26, 5177–5186. [DOI] [PubMed] [Google Scholar]
- (24).Haines-Butterick L; Rajagopal K; Branco M; Salick D; Rughani R; Pilarz M; Lamm MS; Pochan DJ; Schneider JP Controlling Hydrogelation Kinetics by Peptide Design for Three-Dimensional Encapsulation and Injectable Delivery of Cells. Proc. Natl. Acad. Sci. U.S.A 2007, 104, 7791–7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Salick DA; Kretsinger JK; Pochan DJ; Schneider JP Inherent Antibacterial Activity of a Peptide-based beta-Hairpin Hydrogel. J. Am. Chem. Soc 2007, 129, 14793–14799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Haines-Butterick LA; Salick DA; Pochan DJ; Schneider JP In vitro Assessment of the Pro-inflammatory Potential of beta-Hairpin Peptide Hydrogels. Biomaterials 2008, 29, 4164–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Rughani RV; Salick DA; Lamm MS; Yucel T; Pochan DJ; Schneider JP Folding, Self-Assembly, and Bulk Material Properties of a De Novo Designed Three-Stranded beta-Sheet Hydrogel. Biomacromolecules 2009, 10, 1295–1304. [DOI] [PubMed] [Google Scholar]
- (28).Giano MC; Pochan DJ; Schneider JP Controlled Biodegradation of Self-Assembling beta-Hairpin Peptide Hydrogels by Proteolysis with Matrix Metalloproteinase-13. Biomaterials 2011, 32, 6471–6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Sinthuvanich C; Haines-Butterick LA; Nagy KJ; Schneider JP Iterative Design of Peptide-based Hydrogels and the Effect of Network Electrostatics on Primary Chondrocyte Behavior. Biomaterials 2012, 33, 7478–7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Veiga AS; Sinthuvanich C; Gaspar D; Franquelim HG; Castanho MARB; Schneider JP Arginine-rich Self-Assembling Peptides as Potent Antibacterial Gels. Biomaterials 2012, 33, 8907–8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Smith DJ; Brat GA; Medina SH; Tong D; Huang Y; Grahammer J; Furtmüller GJ; Oh BC; Nagy-Smith KJ; Walczak P; Brandacher JP A Multiphase Transitioning Peptide Hydrogel for Suturing Ultrasmall Vessels. Nat. Nanotechnol 2016, 11, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Sinthuvanich C; Nagy-Smith KJ; Walsh STR; Schneider JP Triggered Formation of Anionic Hydrogels from Self-Assembling Acidic Peptide Amphiphiles. Macromolecules 2017, 50, 5643–5651. [Google Scholar]
- (33).Shi J; Fichman G; Schneider JP Enzymatic Control of the Conformational Landscape of Self-Assembling Peptides. Angew. Chem 2018, 130, 11358–11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Schneider JP; Pochan DJ; Ozbas B; Rajagopal K; Pakstis L; Kretsinger J Responsive Hydrogels from the Intramolecular Folding and Self-Assembly of a Designed Peptide. J. Am. Chem. Soc 2002, 124, 15030–15037. [DOI] [PubMed] [Google Scholar]
- (35).Pochan DJ; Schneider JP; Kretsinger J; Ozbas B; Rajagopal K; Haines L Thermally Reversible Hydrogels via Intramolecular Folding and Consequent Self-Assembly of a De Novo Designed Peptide. J. Am. Chem. Soc 2003, 125, 11802–11803. [DOI] [PubMed] [Google Scholar]
- (36).Ozbas B; Kretsinger J; Rajagopal K; Schneider JP; Pochan DJ Salt-triggered Peptide Folding and Consequent Self-Assembly into Hydrogels with Tunable Modulus. Macromolecules 2004, 37, 7331–7337. [Google Scholar]
- (37).Rajagopal K; Lamm MS; Haines-Butterick LA; Pochan DJ; Schneider JP Tuning the pH Responsiveness of beta-Hairpin Peptide Folding, Self-Assembly, and Hydrogel Material Formation. Biomacromolecules 2009, 10, 2619–2625. [DOI] [PubMed] [Google Scholar]
- (38).Micklitsch CM; Knerr PJ; Branco MC; Nagarkar R; Pochan DJ; Schneider JP Zinc-Triggered Hydrogelation of a Self-Assembling beta-Hairpin Peptide. Angew. Chem. Int. Ed 2011, 50, 1577–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Knerr PJ; Branco MC; Nagarkar R; Pochan DJ; Schneider JP Heavy Metal Ion Hydrogelation of a Self-Assembling Peptide via Cysteinyl Chelation. J. Mater. Chem 2012, 22, 1352–1357. [Google Scholar]
- (40).Micklitsch CM; Medina SH; Yucel T; Nagy-Smith KJ; Pochan DJ; Schneider JP Influence of Hydrophobic Face Amino Acids on the Hydrogelation of β-hairpin Peptide Amphiphiles. Macromolecules 2015, 48, 1281–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Nagy-Smith K; Beltramo PJ; Moore E; Tycko R; Furst EM; Schneider JP Molecular, Local, and Network-Level Basis for the Enhanced Stiffness of Hydrogel Networks Formed from Coassembled Racemic Peptides: Predictions from Pauling and Corey. ACS Cent. Sci 2017, 3, 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Yan C; Altunbas A; Yucel T; Nagarkar RP; Schneider JP; Pochan DJ Injectable Solid Hydrogel: Mechanism of Shear-Thinning and Immediate Recovery of Injectable beta-Hairpin Peptide Hydrogels. Soft Matter 2010, 6, 5143–5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Yan C; Mackay ME; Czymmek K; Nagarkar RP; Schneider JP; Pochan DJ Injectable Solid Peptide Hydrogel as a Cell Carrier: Effects of Shear Flow on Hydrogels and Cell Payload. Langmuir 2012, 28, 6076–6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Hyland LL; Twomey JD; Vogel S; Hsieh AH; Yu YB Enhancing Biocompatibility of D-oligopeptide Hydrogels by Negative Charges. Biomacromolecules 2013, 14, 406–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Ozbas B; Rajagopal K; Schneider JP; Pochan DJ Semiflexible Chain Networks Formed via Self-Assembly of beta-Hairpin Molecules. Phys. Rev. Lett 2004, 93, 268106. [DOI] [PubMed] [Google Scholar]
- (46).Veerman C; Rajagopal K; Palla CS; Pochan DJ; Schneider JP; Furst EM Gelation Kinetics of beta-Hairpin Peptide Hydrogel Networks. Macromolecules 2006, 39, 6608–6614. [Google Scholar]
- (47).Larsen TH; Branco MC; Rajagopal K; Schneider JP; Furst EM Sequence-Dependent Gelation Kinetics of beta-Hairpin Peptide Hydrogels. Macromolecules 2009, 42, 8443–8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Nagy KJ; Giano MC; Jin A; Pochan DJ; Schneider JP Enhanced Mechanical Rigidity of Hydrogels Formed from Enantiomeric Peptide Assemblies. J. Am. Chem. Soc 2011, 133, 14975–14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Ozbas B; Rajagopal K; Haines-Butterick L; Schneider JP; Pochan DJ Reversible Stiffening Transition in beta-Hairpin Hydrogels Induced by Ion Complexation. J. Phys. Chem. B 2007, 111, 13901–13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Branco MC; Nettesheim F; Pochan DJ; Schneider JP; Wagner NJ Fast Dynamics of Semiflexible Chain Networks of Self-Assembled Peptides. Biomacromolecules 2009, 10, 1374–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Yucel T; Micklitsch CM; Schneider JP; Pochan DJ Direct Observation of Early-Time Hydrogelation in beta-Hairpin Peptide Self-Assembly. Macromolecules 2008, 41, 5763–5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Hule RA; Nagarkar RP; Hammouda B; Schneider JP; Pochan DJ Dependence of Self-Assembled Peptide Hydrogel Network Structure on Local Fibril Nanostructure. Macromolecules 2009, 42, 7137–7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Nagy-Smith K; Moore E; Schneider J; Tycko R Molecular Structure of Monomorphic Peptide Fibrils within a Kinetically Trapped Hydrogel Network. Proc. Natl. Acad. Sci U.S.A 2015, 112, 9816–9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Searle MS; Williams DH; Packman LC A Short Linear Peptide Derived from the N-Terminal Sequence of Ubiquitin Folds into a Water-Stable non-Native β-Hairpin. Nat. Struct. Biol 1995, 2, 999. [DOI] [PubMed] [Google Scholar]
- (55).Nair CM; Vijayan M; Venkatachalapathi YV; Balaram P X-Ray Crystal Structure of pivaloyl-D-Pro-L-Pro-L-Ala-N-methylamide; Observation of a Consecutive β-turn Conformation. J. Chem. Soc., Chem. Commun 1979, 1183–1184. [Google Scholar]
- (56).Chatterjee B; Saha I; Raghothama S; Aravinda S; Rai R; Shamala N; Balaram P Designed Peptides with Homochiral and Heterochiral Diproline Templates as Conformational Constraints. Chem.—Eur. J 2008, 14, 6192–6204. [DOI] [PubMed] [Google Scholar]
- (57).Grathwohl C; Wüthrich K The X-Pro Peptide Bond as an NMR Probe for Conformational Studies of Flexible Linear Peptides. Biopolymers 1976, 15, 2025–2041. [DOI] [PubMed] [Google Scholar]
- (58).Hutchinson EG; Thornton JM A revised set of potentials for β-turn Formation in Proteins. Protein Sci 1994, 3, 2207–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Kim CA; Berg JM Thermodynamic β-sheet Propensities Measured using a Zinc-Finger Host Peptide. Nature 1993, 362, 267. [DOI] [PubMed] [Google Scholar]
- (60).Ruoslahti E RGD and other Recognition Sequences for Integrins. Annu. Rev. Cell Dev. Biol 1996, 12, 697–715. [DOI] [PubMed] [Google Scholar]
- (61).Hersel U; Dahmen C; Kessler H RGD Modified Polymers: Biomaterials for Stimulated Cell Adhesion and Beyond. Biomaterials 2003, 24, 4385–4415. [DOI] [PubMed] [Google Scholar]
- (62).Lee JW; Park YJ; Lee SJ; Lee SK; Lee KY The Effect of Spacer Arm Length of an Adhesion Ligand Coupled to an Alginate Gel on the Control of Fibroblast Phenotype. Biomaterials 2010, 31, 5545–5551. [DOI] [PubMed] [Google Scholar]
- (63).Mackintosh FC; Käs J; Janmey PA Elasticity of Semiflexible Biopolymer Networks. Phys. Rev. Lett 1995, 75, 4425–4428. [DOI] [PubMed] [Google Scholar]
- (64).Morse DC Viscoelasticity of Concentrated Isotropic Solutions of Semiflexible Polymers. 1. Model and Stress Tensor. Macromolecules 1998, 31, 7030–7043. [Google Scholar]
- (65).Morse DC Viscoelasticity of Concentrated Isotropic Solutions of Semiflexible Polymers. 2. Linear Response. Macromolecules 1998, 31, 7044–7067. [Google Scholar]
- (66).Hunt NC; Grover LM Cell Encapsulation Using Biopolymer Gels for Regenerative Medicine. Biotechnol. Lett 2010, 32, 733–742. [DOI] [PubMed] [Google Scholar]
- (67).Kanta J Collagen Matrix as a Tool in Studying Fibroblastic Cell Behavior. Cell Adhes. Migr 2015, 9, 308–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Tojkander S; Gateva G; Lappalainen P Actin Stress Fibers–Assembly, Dynamics and Biological Roles. J. Cell Sci 2012, 125, 1855–1864. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.