Abstract
OBJECTIVE
To report the mutational profile and clinical outcomes of a cohort of patients with KIT-mutant seminomas and nonseminomatous germ-cell tumors (SGCT/NSGCTs).
PATIENTS AND METHODS
Retrospective cohort study of all patients with KIT-mutant GCTs sequenced at Memorial Sloan Kettering between March 2014 and March 2020. Tumors were assessed with MSK-IMPACT, a DNA next-generation sequencing assay for targeted sequencing of up to 468 key cancer genes.
RESULTS
Among 568 patients with GCTs, 8.1% had somatic KIT mutations, including 28 seminomas and 18 mixed/NSGCTs. Exons 17 (67.3%), 11 (22.4%), and 13 (6.1%) were most commonly affected. KIT-mutant cases were enriched for oncogenic RAS/MAPK pathway alterations compared to KIT-wildtype cases (34.8% vs 19.2%, P = .02). Among KIT-mutant cases, concurrent mutations were noted in KRAS (21.7%), RRAS2 (11.8%), CBL (6.5%), NRAS (4.3%), MAP2Kl (2.2%), and RACI (2.2%). Mutations in KRAS, RRAS2, and NRAS were mutually exclusive. In all, 73.9% of patients developed metastases and 95.7% received chemotherapy. No patients received KIT-directed tyrosine kinase inhibitors (TKIs). Classification as a NSGCT rather than a SGCT was associated with an increased risk of death (hazard ratio 9.1, 95% confidence interval 1.1–78.4, P = .04) while the presence of a concurrent RAS/MAPK pathway alteration was not (hazard ratio 0.8, 95% confidence interval 0.1–4.3, P = .76).
CONCLUSION
Mitogenic driver alterations can co-occur with activating KIT mutations, which may explain the lack of efficacy of KIT-directed TKIS in 0070rior trials. Novel KIT-directed TKIS that target exon 17 mutations may benefit chemotherapy-refractory patients with KIT-mutant GCTs without RAS/MAPK alterations. Dual MEK/KIT inhibitor therapy in KIT-mutant GCTs with concurrent RAS/MAPK alterations could also be a plausible therapeutic strategy.
A subset of seminomas and nonseminomatous germ-cell tumors (SGCT/NSGCTs) is characterized by activating mutations in KIT, which encodes the 21-exon cell surface receptor tyrosine kinase protein KIT (CD117) that drives downstream signaling through the RAS/MAPK pathway.1,2 Such mutations have potential therapeutic relevance, as patients with KIT-mutant gastrointestinal stromal tumors experience durable clinical benefit from tyrosine kinase inhibitors (TKIs).3,4 However, a phase 2 clinical trial of imatinib treatment in KIT-expressing, chemotherapy-refractory GCTs showed no evidence of significant antitumor activity.5 That trial enrolled patients based on KIT expression by immunohistochemistry (IHC) rather than the presence of KIT mutations by sequencing. This lack of efficacy has therefore not been rigorously explored by considering underlying mutations in KIT or concurrent downstream genomic alterations in the RAS/MAPK pathway that could potentially explain the inactivity of TKIS in GCTs. Herein, we report the mutational profiles and clinical outcomes in a cohort of patients with KIT-mutant GCTs.
PATIENTS AND METHODS
Design, Setting, and Participants
We performed a retrospective cohort study of all patients with KIT-mutant GCTs sequenced between March 2014 and March 2020 at Memorial Sloan Kettering Cancer Center (MSK) in New York, NY. The study was approved by the MSK Institutional Review Board and reported in accordance with the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) guidelines.
Case Selection and Diagnostic Criteria
All patients with KIT-mutant GCTs were included in the study. KIT-wildtype cases sequenced during the same period were selected for comparison. Cases were diagnosed by experienced subspecialized genitourinary pathologists according to the criteria specified in the 2016 World Health Organization Classification of Tumors of the Urinary System and Male Genital Organs.
DNA-based Molecular Analyses
To assess for the presence of KIT mutations and other concurrent molecular alterations, tumors and matched normal blood samples were analyzed with the MSK-IMPACT DNA next-generation sequencing platform that targets up to 468 genes and select introns to produce data on single nucleotide variants, small insertions and deletions, copy number changes, and structural variants.6 Variants were classified as oncogenic based on their curation in the MSK Precision Oncology Knowledge Base (OncoKB).7 Tumors that did not meet minimum requirements for tumor content or sequencing coverage were excluded.
Clinicopathologic and Survival Data
Clinicopathologic data were extracted from the electronic medical record, including data on age, sex, date of initial diagnosis, pathologic diagnosis including results of KIT IHC, survival time, and treatment history with chemotherapy or targeted therapy. Overall survival was defined as the time from initial pathologic diagnosis until the time of death due to any cause.
Statistical Analyses
Differences among categorical variables were assessed using Fisher’s exact test. Those among continuous variables were assessed using the nonparametric Mann-Whitney-Wilcoxon test. Clinicopathologic variables were examined in Cox proportional hazards models for associations with overall survival. Genomic data were accessed using the internal MSK cBioPortal for Cancer Genomics,8 PathwayMapper,9 and ProteinPaint.10 Statistical analyses were performed using R version 3.6.2 (R Foundation for Statistical Computing). Statistical tests were 2-sided and used a significance threshold of P < .05. Reported P values were not adjusted for multiple testing.
RESULTS
Samples Included in the Study
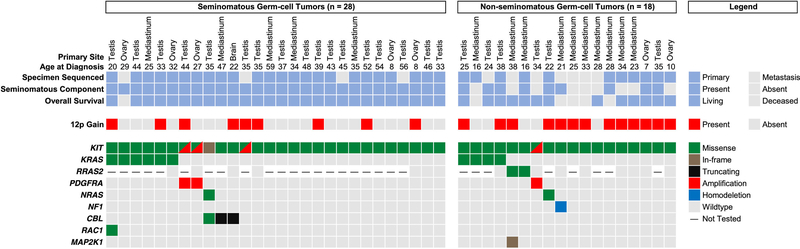
Among 568 patients with GCTs, 8.1% (46/568) had somatic KIT mutations curated in OncoKB as likely oncogenic alterations that may respond to TKIs in other tumor types, including 23 men (of 381 men) with testicular GCTs, 15 men (of 71 individuals) with mediastinal GCTs, 7 women (of 66 women) with ovarian GCTs, and 1 man (of 4 men) with a pineal germinoma. Mediastinal GCTs were more likely to have KIT mutations than were GCTs from other sites (21.1% [15/71] vs 6.2% [31/497], P < .001). The median age at initial pathologic diagnosis of patients with KIT-mutant GCTs was 33.4 years (range, 7.2–58.9 years) and 15.2% (7/46) were female. Patients with KIT-mutant GCTs did not differ from those with KIT-wildtype GCTs with respect to age (P = .62) or sex (P = .99). The histologic classification (eg, SGCT vs NSGCT), presence or absence of a seminomatous component (for NSGCTs), specimen sequenced (eg, primary vs metastatic lesion), clinical details, and associated genetic alterations for each KIT-mutant ease are shown in Figure 1. Specific pathologic diagnoses, as well as additional data on the molecular alterations presented below, are provided in the Supplementary Data.
Figure 1.
Clinical, histologic, and genetic features of patients with KIT-mutant germ-cell tumors. The specimens are stratified according to histologic classification (ie, seminomatous vs nonseminomatous germ-cell tumor). Of the 11 metastatic samples sequenced, 3 were obtained prior to chemotherapy, 1 was obtained prior to chemotherapy but after radiotherapy, and 7 were obtained after chemotherapy.
Spectrum of KIT Mutations
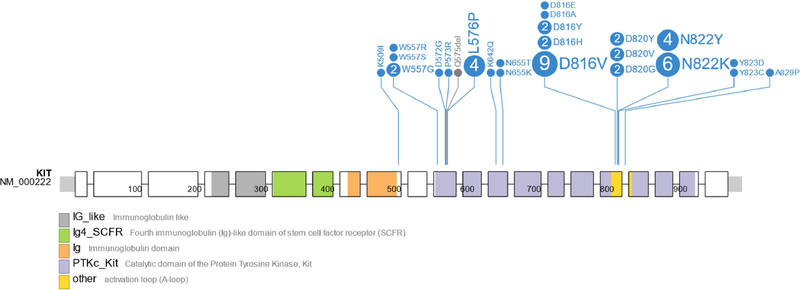
Among the 46 KIT-mutant cases, 95.6% (44/46) had 1, 2.2% (1/46) had 2, and 2.2% (1/46) had 3 KIT mutations. The spectrum of KIT mutations identified among the samples is shown in Figure 2. Exons 17 (67.3% [33/49]), 11 (22.4% [11/49]), and 13 (6.1% [3/49]) were most commonly affected. The exonic distribution did not differ by sex (P = 1.0), primary site (P = .91), or presence of a seminomatous component (P = .56). The most frequently mutated codons overall were D816 and N822, both part of the exon 17 tyrosine kinase II activation loop, in 30.6% (15/49) and 20.4% (10/49) of cases, respectively. The most frequently mutated codons in the exon 11 juxtamembrane domain were L576 (8.2% [4/49]) and W557 (8.2% [4/49]). The most frequently mutated codon in the exon 13 tyrosine kinase I domain was N655 (4.1% [2/49]). Individual cases with mutations in exons 9 and 18 were also identified (K5091 and A829P, respectively). Among the 32.6% (15/46) of eases for which KIT IHC was reported, 93.3% (14/15) exhibited positive staining.
Figure 2.
Spectrum of somatic mutations in KIT. Exons 1–21 are represented from left to right by the enclosed boxes with superimposed amino acid positions. Missense mutations are shown in in the lollipop plot in blue; deletions are in gray. The number within each circle corresponds to the number of times that mutation was identified. Selected protein domains are indicated by the colors shown below the figure.
Prevalence of Concurrent RAS/MAPK Pathway Alterations
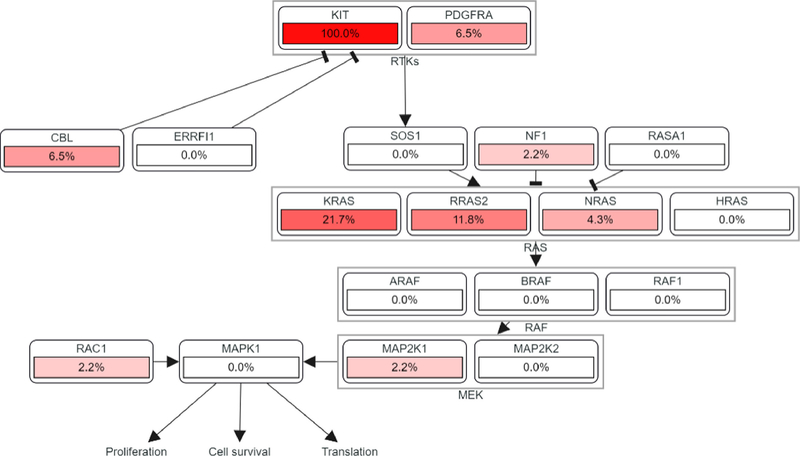
Cases with KIT mutations were significantly enriched for oncogenic RAS/MAPK pathway mutations compared to KIT-wildtype cases (34.8% [16/46] vs 19.2% [100/522], P = .02) (Supplemental Table 1). Among KIT-mutant cases, concurrent mutations were noted in KRAS (21.7% [10/46]), RRAS2 (11.8% [2/17]), CBL (6.5% [3/46]), NRAS (4.3% [2/46]), MAP2K1 (2.2% [1/46]), and RACI (2.2% [1/46]). PDGFRA amplification (6.5% [3/46]) and NF1 homodeletion (2.2% [1/46]) were also rarely noted. Of note, PDGFRA and KIT are adjacent to one another on 4q12, and all 3 PDGFRA-amplified cases exhibited co-amplification of KIT. Alterations in KRAS, RRAS2, NRAS, PDGFRA, and NF1 were mutually exclusive. Copy-number gains in 12p (including KRAS, among other genes) were seen in 47.8% (22/46) of cases, suggesting isochromosome 12p. The spectrum of KRAS, RRAS2, and NRAS mutations identified is shown in Figure 3. A pathway diagram summarizing the frequency of RTK/RAS/MAPK alterations among is eases is shown in Figure 4.
Figure 3.
Spectrum of somatic mutations in KRAS, NRAS, and RRAS2. Missense mutations are shown in the lollipop plots in blue. The number within each circle corresponds to the number of times that mutation was identified. Selected protein domains are indicated by the colors shown below the diagrams. The amino acid sequences and positions are superimposed.
Figure 4.
RTK/RAS/MAPK pathway alterations in KIT-mutant germ-cell tumors. The values shown represent the percentage of KIT-mutant samples (n = 46) with oncogenic alterations in the listed genes.
Clinical Outcomes
The median follow-up time among the 46 patients with KIT-mutant GCTs was 6.0 years (interquartile range, 3.3–9.4). In all, 73.9% (34/46) had metastasis at any point and 95.7% (44/46) received chemotherapy. One patient, a 16-year-old man with a multiply refractory (ie, high-dose chemotherapy resistant) primary mediastinal yolk sac tumor with widespread metastases, received the PD-L1 inhibitor atezolizumab and then the MEK inhibitor cobimetinib after chemotherapy failed, with minimal response. No patients received KIT-directed TKIs, although the majority (n = 36) were cured by other treatments and thus not considered for TKI therapy. At follow-up, 73.9% (34/46) of patients were free of disease, 13.0% (6/46) had disease, and 13.0% (6/46) had died due to disease, including the patient who received cobimetinib. On multivariable Cox proportional hazards regression, classification as a NSGCT rather than a SGCT was associated with an increased risk of death (hazard ratio 9.1, 95% confidence interval 1.1–78.4, P = .04) while the presence of a concurrent RAS/MAPK pathway alteration was not (hazard ratio 0.8, 95% confidence interval 0.1–4.3, P = .76), although the number of events was too small to draw firm conclusions.
COMMENTS
This retrospective cohort study of 568 patients with GCTs from multiple sites demonstrated that 8.1% (46/568) had potentially clinically actionable somatic KIT mutations, of which 34.8% (16/46) exhibited concurrent oncogenic mutations in RAS/MAPK pathway genes, an observation that could potentially explain the lack of efficacy of KIT-directed TKIs in prior case reports and clinical trials.5 In addition, it is known that exon 17 KIT mutations, the most frequent site of mutations in GCT, are not sensitive to the majority of available KIT-directed TKIs including imatinib and sunitinib.11,12 No patients received KIT-directed TKIs in this cohort, so potential reasons for the previously demonstrated resistance to KIT-directed TKIs could not be directly explored.
The 8.1% prevalence of KIT mutations among GCTs identified in this report and their relative distribution between the kinase and juxtamembrane domains are broadly concordant with the results of previously published studies. A 2015 whole-exome sequencing study of 42 testicular GCTs identified KIT mutations in 14.3% (6/42) of cases, 83.3% (5/6) in the exon 17 tyrosine kinase domain II activation loop and 16.7% (1/6) in the exon 11 juxtamembrane domain.1 However, in contrast to the present study, only a single KIT-wildtype case harbored a KRAS mutation. A larger 2018 analysis of 137 primary testicular GCTs demonstrated somatic mutations in KIT, KRAS, and NRAS exclusively in samples with seminoma components.2 KIT mutations were noted in 18.2% (25/137) of cases, including in the exon 17 kinase activation loop (74.1% [20/27]), the exon 11 juxtamembrane domain (22.2% [6/27]), and the exon 13 protein tyrosine kinase I domain (3.7% [1/27]). Concurrent KRAS and NRAS mutations were noted in 16.0% (4/25) and 8.0% (2/25) of the KIT-mutant cases, respectively, and were mutually exclusive of one another.
In a 2018 study of 24 ovarian GCTs, 16.7% (4/24) had oncogenic KIT mutations in exons 11, 13, or 17, including a case of pure dysgerminoma (synonymous to seminoma) with a concurrent NF1 mutation.13 All the KIT-mutant cases were dysgerminomas or mixed forms with a dysgerminomatous component. In all, 8.3% (2/24) had KRAS mutations, although again contrasting with the present study, none had alterations in KIT. Last, in a 2014 study of intracranial GCTs, 25.8% (16/62) had mutations in KIT, including 2 cases with concurrent mutations in the negative RAS pathway regulator CBL,14 similar to the present study. All the KIT-mutant cases had seminomatous components. Cases with KRAS (14.5% [9/62]), NRAS (4.8% [3/62]), and INF1 (3.2% [2/62]) mutations were also identified; however, none had concurrent mutations in KIT.
In contrast to prior reports, our study revealed that KIT and RAS/MAPK pathway alterations also co-occurred in NSGCTs lacking seminomatous components and that RAS/MAPK mutations were more common in KIT-mutant compared to KIT-wildtype tumors. It also confirmed the increased frequency of exon 17 compared to exon 11 mutations in GCTs, contrary to the pattern seen in GISTs, where exon 11 alterations predominate.15 Further, concurrent RTK/RAS/MAPK pathway alterations were noted not only in KRAS, NRAS, CBL, and NF1, but also in RRAS2, MAP2K1, RAC1, and PDGFRA. Mutual exclusivity among concurrent RAS/MAPK alterations was noted not just for KRAS and NRAS, but also for RRAS2, PDGFRA, and NF1.
The identification of activating RRAS2 mutations that were mutually exclusive with KRAS and NRAS in the present study merits special consideration. RRAS2, also known as TC21, is a RAS superfamily oncogene16,17 recently recognized as a rare cause of the RASopathy Noonan syndrome.18 The literature on its role in cancer is relatively sparse, although reports have linked RRAS2 alterations to ovarian and breast carcinomas and radiotherapy-associated gliomas.19–21 RRAS2 encodes a 6-exon protein with significant homology to KRAS and NRAS.10 Specifically, RRAS2 amino-acid positions 20–25 (VGGGGV) have a sequence similar to KRAS and NRAS positions 9–14 (VGAGGV), including a recurrent G23 hotspot variant homologous to the G12 hotspot variant in KRAS and NRAS. No specific RRAS2 inhibitor is available for clinical use, although research into multivalent small molecule pan-RAS inhibitors is ongoing.22
Identification of oncogenic KIT mutations is of potential clinical relevance given that it has been successfully targeted in other solid tumors, GIST being the most notable example.3,4 However, KIT has yet to be successfully targeted in GCTs. A phase 2 clinical trial published in 2006 of imatinib treatment in KIT-expressing, chemotherapy-refractory GCTs showed no evidence of significant antitumor activity. That trial enrolled patients based on KIT expression by IHC rather than the presence of KIT mutations by sequencing. Since IHC has not been demonstrated to be a predictive biomarker for response to KIT inhibitors, the lack of efficacy could thus be explained by possible absence of KIT mutations in this cohort. In addition, most available KIT inhibitors primarily target exon 11 or 13 mutations with limited efficacy against exon 17 mutations, which are more prevalent in GCT. Finally, co-mutations in downstream pathways such as RAS/MAPK could serve to limit TKI activity.
The potential of RAS/MAPK alterations to induce resistance to TKIs like imatinib is relevant given ongoing trials of MEK inhibitors such as trametinib, cobimetinib, and binimetinib.23–25 Interestingly, in melanomas and GISTs where KIT is recurrently mutated, activating KIT mutations represent mitogenic drivers that are generally mutually exclusive with other activating mutations in the RAS/MAPK pathway.26,27 In contrast, our study demonstrates that activating mutations in KIT and RAS/MAPK genes commonly co-occur in GCTs. Although these alterations may seem redundant, these co-mutations may reflect the unique tissue-specific dependencies of germ-cell differentiation and development. Importantly, these data suggest that dual inhibition of KIT and the RAS/MAPK pathway may provide a therapeutic strategy for patients with these co-mutations. Such a strategy could potentially yield successes similar to those noted for dual RAF/MEK inhibition in colorectal adenocarcinoma and malignant melanoma, where synergistic inhibition of both kinase pathways overcomes resistance to therapy with BRAF kinase inhibitors alone.28,29
Since the advent of cisplatin-based chemotherapy for treating patients with testicular cancer, the combination of surgery and chemotherapy can be expected to cure >90% of patients with GCTs.30 Therefore, at present, the possibility of utilizing targeted therapy in patients with GCTs is relevant only to those with multiply relapsed or refractory disease. The clinical outcomes of the KIT-mutant cases in the present study mirrored the generally favorable prognosis of GCTs overall, with 73.9% of patients cured of disease and 13.0% of patients whose treatment was ongoing at the time of this writing. As expected, patients with NSGCTs had worse outcomes than those with SGCTs. The presence of a concurrent RAS/MAPK pathway alteration was not associated with a better or worse outcome in this patient population, although the number of adverse outcomes (ie, persistent disease or death) was low, and no patients received KIT-directed therapy.
This study has limitations. Not all patients at MSK with clinical stage I GCTs are subjected to sequencing, thus patients with metastatic disease and with extragonadal GCTs are overrepresented in this study. Although 46 patients with GCTs with somatic KIT mutations were identified, the presence of RRAS2 alterations was only interrogated in 32.6% (15/46) of patients since it was only recently added to the MSK-IMPACT assay design. Further, the presence of isochromosome 12p was inferred indirectly by copy-number analysis and the sensitivity of the assessment could be adversely affected by low tumor content and the level of genomic instability, thus its prevalence could be underestimated. Last, no patients received KIT-directed TKIS in this cohort, thus future studies will be required to test the hypothesis that ascertaining the mutational status of both KIT and RAS/MAPK pathway genes could successfully inform therapy in patients with chemotherapy-refractory disease. Studies of KIT inhibitors targeting tumors with exon 17 mutations are ongoing (eg, NCT02508532 and NCT03673501).
CONCLUSION
Next-generation sequencing analysis of GCTs can uncover potentially TKI-responsive KIT mutations and associated genomic alterations. KIT mutations occur most commonly in exon 17 and are associated with RAS/MAPK alterations, which may explain the lack of efficacy of TKIS in prior trials. Novel KIT-directed TKIS that target exon 17 mutations may benefit patients with KIT-mutant GCTs without RAS/MAPK alterations. Alternately, dual MEK/KIT inhibitor therapy in KIT-mutant GCTs with concurrent RAS/MAPK alterations may be a plausible therapeutic strategy.
DATA SHARING STATEMENT
Clinical and genetic data are available for download in the provided Supplementary Data file.
Supplementary Material
Acknowledgment.
The authors thank the members of the Molecular Diagnostics Service in the Department of Pathology and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology for the establishment of the MSK-IMPACT dataset.
Funding Sources: This research was supported in part by the National Cancer Institute of the National Institutes of Health (P30 CA008748).
Footnotes
SUPPLEMENTARY MATERIALS
Supplementary material associated with this article can be found in the online version at https://doi.org/10.1016/j.urology.2020.07.027.
References
- 1.Litchfield K, Summersgill B, Yost S, et al. Whole-exome sequencing reveals the mutational spectrum of testicular germ cell tumours. Nat Commun. 2015;6:5973. 10.1038/ncomms6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen H, Shih J, Hollern DP, et al. Integrated molecular characterization of testicular germ cell tumors. Cell Rep. 2018;23:3392–3406. 10.1016/j.celrep.2018.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Demetri GD, von Mehren M, Blanke CD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347:472–480. 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 4.Verweij J, van Oosterom A, Blay J-Y, et al. Imatinib mesylate (STI-571 Glivec, Gleevec) is an active agent for gastrointestinal stromal tumours, but does not yield responses in other soft-tissue sarcomas that are unselected for a molecular target. Results from an EORTC soft tissue and bone sarcoma group phase II study. Eur J Cancer Oxf Engl 1990. 2003;39:2006–2011. [PubMed] [Google Scholar]
- 5.Einhorn LH, Brames MJ, Heinrich MC, Corless CL, Madani A. Phase II study of imatinib mesylate in chemotherapy refractory germ cell tumors expressing KIT. Am J Clin Oncol. 2006;29:12–13. 10.1097/01.coc.0000195086.47548.ef. [DOI] [PubMed] [Google Scholar]
- 6.Cheng DT, Mitchell TN, Zehir A, et al. Memorial sloan kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn JMD. 2015;17:251–264. 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakravarty D, Gao J, Phillips SM, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017;2017. 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bahceci I, Dogrusoz U, La KC, Ö Babur, Gao J, Schultz N. Pathway-Mapper: a collaborative visual web editor for cancer pathways and genomic data. Bioinforma Oxf Engl. 2017;33:2238–2240. 10.1093/bioinformatics/btx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou X, Edmonson MN, Wilkinson MR, et al. Exploring genomic alteration in pediatric cancer using protein paint. Nat Genet. 2016;48:4–6. 10.1038/ng.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao J, Tian Y, Li J, Sun N, Yuan J, Shen L. Secondary mutations of c-KIT contribute to acquired resistance to imatinib and decrease efficacy of sunitinib in Chinese patients with gastrointestinal stromal tumors. Med Oncol Northwood Lond Engl. 2013;30:522. 10.1007/s12032-013-0522-y. [DOI] [PubMed] [Google Scholar]
- 12.Napolitano A, Vincenzi B. Secondary KIT mutations: the GIST of drug resistance and sensitivity. Br J Cancer. 2019;120:577–578. 10.1038/s41416-019-0388-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Nieuwenhuysen E, Busschaert P, Neven P, et al. The genetic landscape of 87 ovarian germ cell tumors. Gynecol Oncol. 2018;151:61–68. 10.1016/j.ygyno.2018.08.013. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Yamaguchi S, Burstein MD, et al. Novel somatic and germline mutations in intracranial germ cell tumours. Nature. 2014;511:241–245. 10.1038/nature13296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corless CL, Barnett CM, Heinrich MC. Gastrointestinal stromal tumours: origin and molecular oncology. Nat Rev Cancer. 2011;11:865–878. 10.1038/nrc3143. [DOI] [PubMed] [Google Scholar]
- 16.Chan AM, Miki T, Meyers KA, Aaronson SA. A human oncogene of the RAS superfamily unmasked by expression cDNA cloning. Proc Natl Acad Sci U S A. 1994;91:7558–7562. 10.1073/pnas.91.16.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drivas GT, Shih A, Coutavas E, Rush MG, D’Eustachio P. Characterization of four novel ras-like genes expressed in a human teratocarcinoma cell line. Mol Cell Biol. 1990;10:1793–1798. 10.1128/mcb.10.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niihori T, Nagai K, Fujita A, et al. Germline-activating RRAS2 mutations cause noonan syndrome. Am J Hum Genet. 2019;104:1233–1240. 10.1016/j.ajhg.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larive RM, Moriggi G, Menacho-Márquez M, et al. Contribution of the R-Ras2 GTP-binding protein to primary breast tumorigenesis and late-stage metastatic disease. Nat Commun. 2014;5:3881. 10.1038/ncomms4881. [DOI] [PubMed] [Google Scholar]
- 20.Li N, Zhan X. Signaling pathway network alterations in human ovarian cancers identified with quantitative mitochondrial proteomics. EPMA J. 2019;10:153–172. 10.1007/s13167-019-00170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.López GY, Van Ziffle J, Onodera C, et al. The genetic landscape of gliomas arising after therapeutic radiation. Acta Neuropathol (Berl). 2019;137:139–150. 10.1007/s00401-018-1906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welsch ME, Kaplan A, Chambers JM, et al. Multivalent small molecule pan-RAS inhibitors. Cell. 2017;168:878–889.e29. 10.1016/j.cell.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robert C, Flaherty KT, Hersey P, et al. METRIC phase III study: efficacy of trametinib (T), a potent and selective MEK inhibitor (MEKi), in progression-free survival (PFS) and overall survival (OS), compared with chemotherapy (C) in patients (pts) with BRAFV600E/K mutant advanced or metastatic melanoma (MM). J Clin Oncol. 2012;30(18_suppl). LBA8509–LBA8509. 10.1200/jco.2012.30.18_suppl.1ba8509. [DOI] [Google Scholar]
- 24.Takahashi RH, Choo EF, Ma S, et al. Absorption, metabolism, excretion, and the contribution of intestinal metabolism to the oral disposition of [14C]Cobimetinib, a MEK inhibitor, in humans. Drug Metab Dispos Biol Fate Chem. 2016;44:28–39. 10.1124/dmd.115.066282. [DOI] [PubMed] [Google Scholar]
- 25.Ascierto PA, Schadendorf D, Berking C, et al. MEK162 for patients with advanced melanoma harbouring NRAS or Val600 BRAF mutations: a non-randomised, open-label phase 2 study. Lancet Oncol. 2013;14:249–256. 10.1016/S1470-2045(13)70024-X. [DOI] [PubMed] [Google Scholar]
- 26.Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–1696. 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rubin BP, Singer S, Tsao C, et al. KIT activation is a ubiquitous fature of gastrointestinal stromal tumors. Cancer Res. 2001;61:8118–8121. [PubMed] [Google Scholar]
- 28.Flaherty KT, Infante JR, Daud A, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–1703. 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kopetz S, Grothey A, Yaeger R, et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated colorectal cancer. N Engl J Med. 2019;381:1632–1643. 10.1056/NEJMoa1908075. [DOI] [PubMed] [Google Scholar]
- 30.Cheng L, Albers P, Berney DM, et al. Testicular cancer. Nat Rev Dis Primer. 2018;4:1–24. 10.1038/s41572-018-0029-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.