Abstract
Cytokines, growth factors or hormones take action through the JAK/STAT5 signaling pathway, which plays a critical role in regulating the intestinal response to infection and inflammation. However, the way in which STAT5 regulates intestinal epithelial compartment is largely ignored due to the lack of genetic tools for proper exploration and because the two STAT5 transcription factors (STAT5A and STAT5B) have some redundant but also distinct functions. In this review article, by focusing on STAT5 functions in the intestinal undifferentiated and differentiated epithelia, we discuss major advances of the growth factor/cytokine-JAK/STAT5 research in view of intestinal mucosal inflammation and immunity. We highlight the gap in the research of the intestinal STAT5 signaling to anticipate the gastrointestinal explorative insights. Furthermore, we address the critical questions to illuminate how STAT5 signaling influences intestinal epithelial cell differentiation and stem cell regeneration during homeostasis and injury. Overall, our article provides a centric view of the relevance of the relationship between chronic inflammatory diseases and JAK/STAT5 pathway and it also gives an example of how chronic infection and inflammation pirate STAT5 signaling to worsen intestinal injuries. Importantly, our review suggests how to protect a wound healing from gastrointestinal diseases by modulating intestinal STAT5.
Keywords: STAT5A; STAT5B, JAK; Intestinal epithelial stem cells; Intestinal homeostasis; Inflammation; Infection
1. An overview of the JAK/STAT5 pathway
The JAK/STAT pathway is one of the major cellular pathways that integrates signals from cytokines, growth factors, and hormones to regulate three main cellular processes: cell fate, cell survival, and genome maintenance. The JAK/STAT signaling cascade regulates gene expression in all eukaryotic cells, promoting cell cycle progression, survival and steering of metabolism. Generally, the JAK/STAT signaling pathway is initiated upon binding of a ligand to a growth factor or cytokine receptor, upon which the receptors are being autophosphorylated in the case of growth factor receptors, which are usually JAK-independent, or efficiently tyrosine phosphorylated by JAK on many target tyrosine residues, that serve as docking sites for SH2 domain-containing molecules, such as STAT5A/B (Fig. 1). Upon tyrosine phosphorylation, STATs change the conformation drastically from an anti-parallel dimer to a parallel dimer, where both complexes were proven in crystallized forms [1,2]. It is thought that parallel dimer formation is most efficient for translocation into the nucleus, even though shuttling in and out of the nuclear pore occurs constantly and differently, most likely dependent on the cell types. Nuclear shuttling of STAT3/5 is facilitated by the Nucleophosmin protein (NPM), frequently translocated in cancer and associated with the DNA damage machinery that associates with BRCA1/2 proteins, serving as scaffold proteins for DNA damage repair, once RAD51 as classic STAT5 target gene products are decorated on BRCA proteins.
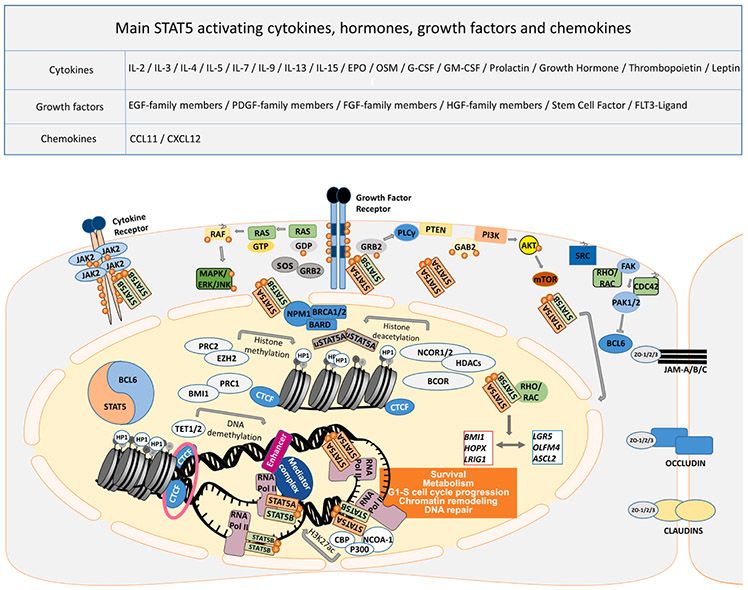
Fig. 1.
Cell signaling pathways involved in cytokine, hormone, growth factor and chemokine signaling in association with STAT5 functions. Signaling pathways, triggered by a plethora of cytokines, hormones, growth factors, and chemokines regulate survival, metabolism, and G1-S cell cycle progression, as well as chromatin remodeling through STAT5 involvement with and without pYSTAT5 function. Overexpression of growth factor/cytokine receptor and/or mutated downstream effectors are often found in the EGFR-RAS-RAF axis and PI3K-AKT-mTOR pathway. In the cytoplasm, STAT5A/B can be activated through phosphorylation of tyrosine residue at position 694/699 by JAK or tyrosine kinase receptor. STAT5A is translocated into the nucleus with the assistance of RHO/RAC, which phosphorylates a serine residue of STAT5A at position 779. There are complex homo- and heterodimers with STAT5B, and RHO/RAC can bind to serine phosphorylated STAT5A homo- or STAT5A/B heterodimers, forming a trimer. Besides, higher-order oligomer formation is facilitated by higher pYSTAT5 levels in a concentration-dependent manner. RHO and RAC can both trigger PAK1/2 kinase activation, which phosphorylates STAT5A to promote nuclear shuttling, or PAK kinases block the BCL6 transcription factor by serine phosphorylation. Interestingly, STAT5 binds to DNA as a transcription factor, competing with BCL6 for identical DNA binding sites, to promote transcription of the target genes. As discussed above, BCL6 acts as a locus repressor of STAT5A/B, autoregulating itself by two unique and distinct promoters. STAT5 can also recruit epigenetic modifiers, such as NCOR1/2, BCOR, HDAC3, EZH2, TET1/2 and CBP-P300. BCL6 and STAT5 also compete for corepressor binding. Even though only NCOR1/2 mapping was reported, BCOR attraction might be relevant as well. Furthermore, STAT5 acts on chromatin topology to consequently regulate the expression of genes, keeping the balance between renewal- (LGR5, OLFM4, ASCL4) and quiescence-associated genes (BMI1, HOPX, LRIG1). Surprisingly, unphosphorylated STAT5A (uSTAT5A) binds to heterochromatin protein 1 (HP1) to promote chromatin compaction. In comparison, the more oncogenic STAT5B gene product binds DNA more abundantly, which is antagonized by STAT5A, being itself under transcriptional regulation by TP53 action. Moreover, uSTAT5 was shown to be colocalized with CTCF in myeloid cells. CTCF transcription factor binding organizes chromatin into topology associated domains, where open and closed chromatin is folded by simplified schematic loop structures in association with Cohesin subunits (pink ring), separating them into different transcriptionally active or closed regions. Cohesin subunits, STAG1/2 are often mutated in colorectal cancer.
As mentioned above, JAK/STAT pathway regulates gene expression in all eukaryotic cells, and a remarkable example of the conservation of the JAK/STAT pathway is the STAT3 species conservation. This is supported by the insights we got by analyzing the Tasmanian devil facial tumor disease, in which STAT3 shares >99% amino acid sequence homology with human STAT3 orthologue. More precisely, human and Tasmanian devil STAT3 differ in 6 amino acids, and additionally Tasmanian devil STAT3 has one extra threonine at position 748 [3]. Interestingly, in comparison to STAT family members, JAK kinases appeared quite late during evolution. The first bona fide JAK-like protein is represented in the basal metazoan porifera lineage, found in Ephydatia fluviatilis. However, this JAK-like protein lacks pseudo-kinase domain. JAK proteins inclusive of the pseudo-kinase domain appear in bilateria [4]. In comparison, proteins resembling metazoan STATs appeared by the time of amoebozoan divergence, although they lack N-terminal and transactivation domain (TAD). Members of anemone and nematode contain proteins similar to STATs, which include N-terminal and TAD, respectively [5]. This suggests that before JAK tyrosine kinases came about, the conserved STAT domains must have evolved with different functions independent of tyrosine phosphorylation. This suggests further that many fundamental STAT functions for genome maintenance, as well as the proliferation and steering of metabolism, during the early stages of evolution, could have evolved first, maintaining STAT function independent of tyrosine phosphorylation. There are seven different mammalian STAT gene products, and the orthologues of all seven of them exist in teleostean fish. This proves, that STAT domain structures were already complete before the teleost-tetrapod split 450 million years ago [6]. In Drosophila melanogaster, a canonical JAK/STAT pathway consists of three UPD ligands, a transmembrane receptor Domeless, a JAK kinase called Hopscotch, and a transcription factor STAT92E [7]. The genetic mapping of mammalian STATs suggests the colocalization of STAT1-STAT4, STAT2-STAT6 and STAT3-STAT5A/5B linked to different chromosomes respectively, which suggests that a primordial gene had duplicated and that the whole duplication was further divided. The Drosophila gene, encoding STAT92E, seems to be most similar to STAT5, which leads to the conclusion that STAT3-STAT5A/B are the most ancestral among all STAT genes [8]. Regarding gastrointestinal relevance, JAK/STAT signaling is in Drosophila melanogaster essential for mid- and hindgut homeostasis. A similar situation occurs in mammals when enterocytes are subjected to apoptosis or stress, in Drosophila, UPD ligands are produced to trigger STAT92E to promote division of enterocytes and epithelial regeneration [9]. Even though there is a lot of literature on STAT3 signaling in intestinal cell types, which is reported in [10,11], there seems to be no comprehensive STAT5 intestinal function overview article to this point. Thus, our primary focus in this contribution will be on STAT5. First, we introduce activation by multiple pathways, then we describe the structure and function of intestinal epithelial monolayer, subsequently, we move to STAT5 function in the intestinal epithelia.
1.1. Different activity of STAT5A and STAT5B gene products
The STAT5 locus encodes two distinct gene products, STAT5A and STAT5B (Fig. 2), both located on chromosome 17q21.2, encoded in two different genomic orientations with two distinct promoters driving in different directions. The locus possibly also has a bi-cistronic element(s) influencing transcription in both directions via chromatin looping events. This is autoregulated by STAT5 and counteracted by the locus repressor BCL6 or other repressor complexes such as YY1/BCOR/NCOR [12,13,14]. STAT5A and STAT5B display approximately 92% identity (731 amino acids out of 794). However, they additionally share 12 similar positions, which makes them approximately 94% similar. Considering that STAT5A consists of 794 amino acids and STAT5B of 787 amino acids, with STAT5B having the C-terminal 12 amino acids shorter, aligning the two proteins excluding the last amino acids of C-terminus, where most differences are located, revealed that the two proteins are approximately 94% identical and 95% similar based on the first 774 and 779 amino acids of STAT5A and STAT5B, respectively (Fig. 3). Most differences are located in both extreme N- and C-termini, but also the DNA binding properties between STAT5A and STAT5B differ, namely, STAT5B binds more promiscuously and strongly to DNA with very different and versatile spacing to palindromic invert repeat sequences, affecting gene regulation differently. Further, some gene product-specific differences exist in secondary post-translational modifications, with functional consequences for nuclear shuttling, chromatin function, activity duration, or protein half-life. STAT5A and STAT5B also have distinct non-redundant functions that are particularly noticeable in distinct cell types. The differences are also related to distinct protein–protein interaction, chromatin assembly, or difference in protein turnover and expression. Moreover, the kinetics of down-regulation by tyrosine phosphatase and docking of phosphatases is unique due to the sequence diversity in the phosphatase binding region following the critical tyrosine residue in a stretch of approximately 30–60 amino acids. STAT5B is more dominantly expressed in natural killer, muscle cells, as well as liver hepatocytes, liver endothelium and cholangiocytes, whereas STAT5A is more abundantly expressed in different epithelial cell types from the mammary gland [15-18]. Therefore, mice with the deletion of Stat5a or Stat5b display distinct phenotypes [19,20]. Stat5a and Stat5b double knockout mice survive on a mixed genetic background of C57BL/6xSv129j, even though this results in perinatal lethality in approximately 92–96% [21]. Only 1–2% of Stat5a/b null phenotype survive, and these survivors have a severe growth retardation and very high pYStat3 activity as a compensatory mechanism. Examination of Stat5 double knockout embryos revealed that these embryos were anemic, leukopenic, had smaller spleens and thymi, and disordered thymic architecture, which results in severe combined immunodeficiency phenotype [22]. The intestinal phenotype of Stat5 knockout mice so far hasn’t been vigorously investigated. However, the analysis of GI tract phenotype of Stat5a/b null mice during embryonic development might not reveal strong difference, since villi differentiation is dependent on microbiota and nutrient intake, which is only present after birth. Therefore, such studies of GI tract architecture should be carried out on the mixed genetic background survivor mice, as well as Stat5a/b single knockout mice. Stat5a or Stat5b knockout strains are viable, albeit both have developmental defects and many subtle abnormalities in blood lineages. Stat5a knockout mice have undeveloped mammary glands and defects during pregnancy due to progesterone signaling. In contrast, Stat5b knockout mice show dwarfism, lower hepatic RNA biosynthesis capacity, lower glucose and lipid metabolism, sexual conversion with marked gender differences affecting, for example, drug metabolism due to sex-specific differences in p450 cytochrome enzyme upregulation downstream of growth hormone action [19,15,23,20,24]. Our previous reports showed that Stat5b-deficient mice have an increased gut mucosal barrier dysfunction [25,26]. Specific deletion of total Stat5a/b in intestinal epithelial cells impairs intestinal tight junction barriers, particularly, ZO-1 expression reduction and disruption in the intestinal and colonic epithelia, resulting in susceptibility to intestinal inflammation [27,28]. This indicates the requirement of Stat5 function in intestinal epithelial physiology [25-28].
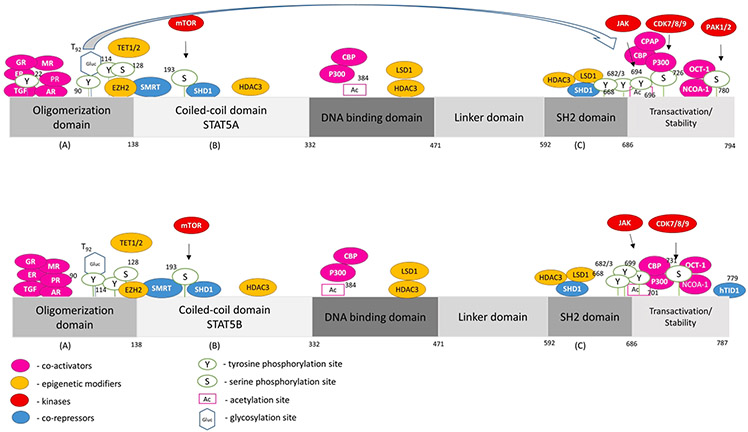
Fig. 2. STAT5A and STAT5B interactome and mutational landscape.
Human STAT5A and STAT5B contain 794 and 787 amino acids, respectively. The proteins consist of an oligomerization domain, a coiled-coil domain, a DNA binding domain, a linker domain, an SH2 domain and a transactivation domain. Three different dimerization domains (A, B, C) allow versatile complex formations of STAT5A/B proteins. We depict proteins physically interacting with STAT5A/B (co-activators, epigenetic modifiers, kinases and co-repressors, shown in colors as indicated), as well as post-translational modifications, with black arrows linking the proteins with their respective post-translational modification above each STAT5 schematic protein structure. The data of post-translational modifications were collected from PhosphoSitePlus® and selected due to biological importance in current literature insight. More precisely, we show acetylation sites reported in HTP (high-throughput papers), phosphorylation reported in more than 5 HTP or 10 LTP (low-throughput papers), and glycosylation [29]. The term “high-throughput papers” refers to records in which a certain modification was assigned using only proteomic discovery mass spectrometry, while the term “low-throughput papers” refers to records which assigned the certain modification using methods other than proteomic mass spectrometry.
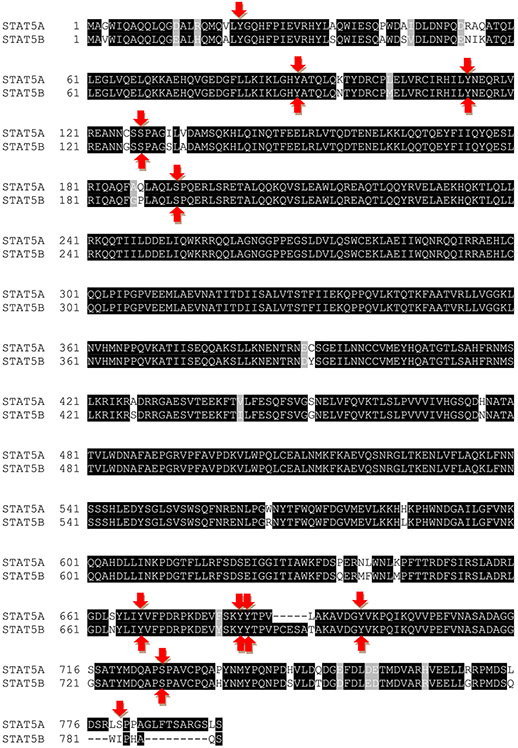
Fig. 3. Human STAT5A and STAT5B share 92% identity.
STAT5A and STAT5B display approximately 92% identity (731 amino acids out of 794). However, they additionally share 12 similar amino acid positions, which makes them approximately 94% similar. STAT5A consists of 794 amino acids and STAT5B of 787 amino acids, with STAT5B lacking 12 C-terminal amino acids present in STAT5A. Based on the first 774 and 779 amino acids of STAT5A and STAT5B, respectively, both proteins are ~94% identical and ~95% similar. Phosphorylation sites are highlighted by red arrows and the data were collected from PhosphoSitePlus® (see also Fig. 2).
1.2. Interactions of STAT5A and STAT5B
Simply put, STAT5A and STAT5B function as homo- or heterodimers or homo- or heterotetramers, assembling with several STAT family members on higher order chromatin. STAT5 is not only a transcription factor, but it also regulates gene expression as an important effector molecule of chromatin topology [30]. A STAT5 protein has three different dimer interfaces for complex formation: (A) the N-domain essential for oligomerization, (B) the coiled-coil domain essential for DNA binding, which occurs even in absence of tyrosine phosphorylation, (C) the SH2 domain essential for dimerization via SH2 domain-pY residue contact.
Both STAT5A/B proteins are tyrosine phosphorylated and efficiently activated by broad spectra of cytokines and hormones, including Prolactin, Growth Hormone, Erythropoietin, Thrombopoietin, Oncostatin M, Granulocyte Colony Stimulating Factor, Granulocyte Macrophage Colony Stimulating Factor [31], many Interleukins, e.g. all those that use the common gamma chain, except for IL-21, to induce gene transcription in concert with co-activator protein interaction. STAT5 can also be activated by growth factors such as Stem Cell Factor, FLT3 Ligand, Hepatocyte Growth Factor-, Epidermal Growth Factor-, Fibroblast Growth Factor-, Platelet-Derived Growth Factor- family members and other cytokines/growth factors/chemokines [16] (Fig. 1). In rare cases, they can also repress gene transcription through co-repressor protein interaction (Fig. 2). STAT5 activation is, however, not only regulated by tyrosine phosphorylation, but also by serine/threonine phosphorylation and other post-translational modifications, such as acetylation/sumoylation at lysine residues and glycosylation at T92, which is known to enhance tyrosine phosphorylation to trigger oncogenic transcription in STAT5A, but different and unexplored in STAT5B, in which the glycosylation at a similar conserved threonine residue has been shown [29,32-34]. The addition of O-GlcNAc to proteins is dependent on UDP-GlcNAc, which serves as substrate for O-GlcNAc transferase, and is synthesized in the hexosamine biosynthetic pathway upon glucose uptake. This post-translational modification regulates the activity of approximately 3000 different proteins, including transcription factors, such as STAT5, P53, c-MYC, FOXO1 and CREB. This essential enzyme is found in different organisms, from humans to Drosophila. In human cells, it is enriched at sites of active transcription and is a part of Polycomb repressor complex in Drosophila [35,36]. Polycomb repressor complex in Drosophila maintains Hox gene expression to make cell fate decisions during organogenesis, and Hox initiates genetic cascade in concert with the JAK/STAT pathway [37]. Furthermore, OGT has been shown to stabilize TET1 in the nuclei of embryonic stem cells at transcription start sites to promote transcription and, as shown in Fig. 1, STAT5 recruits TET1/2 to promote DNA demethylation and transcription. As we previously showed, this post-translational modification in STAT5A, but not STAT5B, controls tyrosine phosphorylation and oncogenic transcription [29]. Taken together, OGT and O-GlcNAc regulate gene expression by directly influencing transcription factors and RNA Pol II, but also by interacting with epigenetic modifiers. Moreover, STAT5A activation by this enzyme might further enhance the effects of OGT, as they both act on similar key proteins controlling the epigenome or gene regulation per se. Furthermore, there are several studies providing proof for cross-talk between STAT5 proteins and members of the steroid receptor family, such as Glucocorticoid Receptor (GR), Mineralocorticoid Receptor (MR), Progesterone Receptor (PR), Estrogen Receptor (ER) alpha and beta, and Androgen Receptor (AR; Fig. 2), as these share high homology in their amino acid sequence and have a conserved domain structure. GR, MR and PR have been shown to functionally synergize with STAT5, while ER antagonizes STAT5 transcription induction from the β-CASEIN gene promoter. The AR was shown to have no effect on STAT5 transcriptional function from the β-CASEIN gene promoter but highly synergizes with STAT5A and STAT5B to promote survival and cell cycle progression in prostate cancer cells [38-40].
2. Intestinal epithelial cells (IEC), intestinal stem cells (ISC) and niche cells
The intestinal epithelial stem cells (IESC) are one of the most rapidly proliferating cells in adult mammals. They function throughout a lifetime to maintain homeostasis by differentiating into intestinal epithelial cells (IEC). The intestine contains a semipermeable physiological barrier, which requires homeostasis [41]. Many commensals important for metabolic functions providing, e.g. essential amino acids and other energy supplies to the body, need to be balanced. It is therefore not surprising, but largely underappreciated, that the highest number of immune cells in the body reside in the GI tract. The intestinal epithelia comprise 6 differentiated lineages: Enterocytes, Goblet, Enteroendocrine, Microfold (M cells), Paneth and Tuft cells, which are renewed every 4 to 5 days in mammals, except for Paneth cells [42]. IESC reside at the crypt bottoms, and they proliferate and differentiate to replenish cells that form a homeostatic intestinal monolayer [43]. IESC differentiate into two distinct progenitor cells; enterocytes are differentiated from absorptive progenitor cells, while goblet and enteroendrocrine cells are differentiated from secretory progenitor cells [44,41]. Differentiating IEC migrate upwards the crypt-villus axis during their life and development, while some fully differentiated cells, like Paneth cells, migrate from the transient amplifying (TA) differentiation zone downwards and reside at the crypt base to live for 6-8 weeks (Fig. 4), where they serve as niche cells to protect IESC from insults and providing complex signals for IESC self-renewal [44-46]. However, the underlying mechanism of upwards Paneth cells during infection and inflammation is still a puzzle. IESC are roughly categorized as either active or quiescent IESC. Active IESC, the majority of which are LGR5+ crypt base columnar cells (CBC), maintain intestinal lineage development and self-renewal with rapid cycling [47,48]. These cells are highly sensitive to intestinal injury, surrounded by Paneth niche cells at a constant number of around 15 LGR5hi IESC and 10 Paneth cells [49]. In contrast, quiescent IESC (also called label-retaining cells, LRC) are present at the “+4” crypt position, contributing to homeostatic regenerative capacity, particularly during recovery from injury [50]. Quiescent IESC express markers such as BMI1, mTERT, and/or LRIG1 [48], and resemble the DLL1 secretory precursors that can convert to active IESC or secretory progenitors in response to injury [51]. Interestingly, quiescent IESC are also labelled as an LGR5lowKi67+ population that can be reactivated to differentiate into hormone-producing enteroendocrine cells by increasing Notch ligands or into Paneth-like cells via activating WNT, STAT5 or MAPK signaling [51-55]. In spite of a long-term debate how Paneth-like cells become niche cells, current studies indicate that IESC/progenitor as well as their niche cells retain a highly hierarchical, heterogeneous, and plastic phenotype during lineage commitment and repair. WNT, Notch, BMP, Hedgehog, Hippo–YAP, or JAK/STAT signaling (Fig. 4) regulate IESC homeostasis and regeneration either IESC-autonomously or through niche factors, such as WNT3a or 5a, R-Spondin, Notch ligands (JAG1/2), TGFβ, inflammatory cytokines, and bacterial metabolites (Butyrate) [56-61]. Furthermore, these niche factors can induce ISC progenitors to differentiate into absorptive enterocytes and secretory cells. However, we still need to further explore the components of niche factors during gut injury and repair (see Fig. 5).
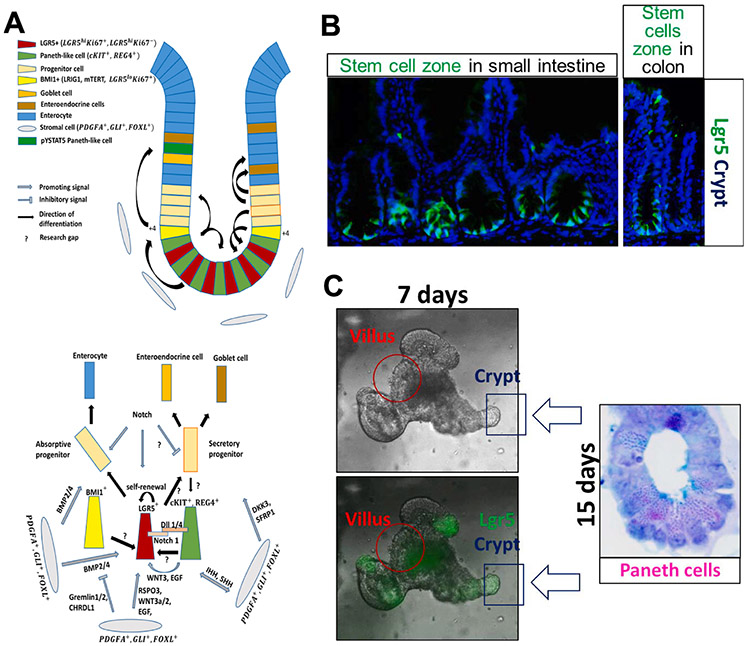
Fig. 4. Homeostasis in the intestinal crypt is maintained by IESC self-renewal and differentiation.
A) LGR5+ cells in the intestinal crypt proliferate to give rise to all other cell types of the intestinal epithelial monolayer, either via absorptive or secretory progenitor. There are 4–6 LGR5+ cells (red cells) in each crypt residing between cKIT+, REG4+ cells (green cells). All cells from the microenvironment contribute to the homeostasis by producing signals that promote either differentiation or self-renewal of LGR5+ stem cells. RSPO3, WNT3, EGF, and Notch support LGR5+ self-renewal, while BMP2/4, DKK3, and SFRP1 support differentiation into progenitor cells. The differentiation is tightly regulated by Gremlin1/2 and CHRDL1, as their increased expression at the base of the crypt inhibits BMP2/4, maintaining LGR5+ lineage at the base, but allowing BMP2/4 to promote enterocyte differentiation in the upper crypt area. Notch signaling is required for absorptive lineage differentiation, while “Notch off/WNT on” determines secretory lineages. Differentiating cells migrate upwards the crypt (black arrows showing the direction of differentiation), facing apoptosis at the top of the villus (not shown). However, fully differentiated, post-mitotic cells, cKIT+ REG4+, migrate towards the base of the crypt. cKIT+ REG4+ double-positive cells together with BMI1+ cells, residing at the +4 position, proliferate to maintain the LGR5+ stem cell pool. Research gaps in the key mechanism, which, in our opinion, could be the focus of future research and should investigate the possible cross talk or interference with STAT5 signaling, are marked by “?”. B and C) Representative images showing mouse intestinal epithelial stem cells and their epithelial niche cells in the in vivo crypt-villus (B) and in vitro organoids (C).
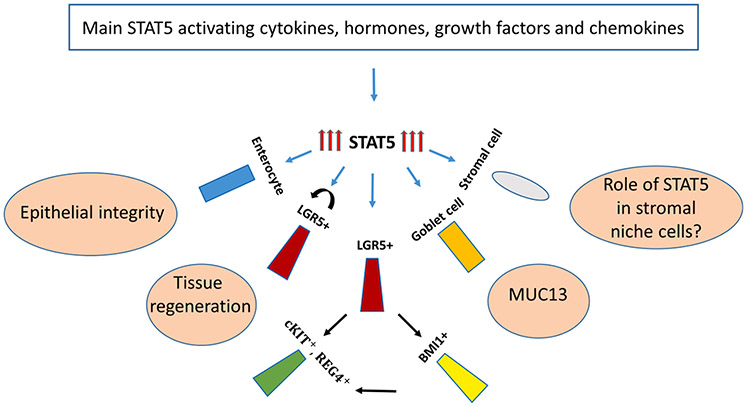
Fig. 5.
STAT5-activating cytokines, hormones, growth factors, and chemokines maintain homeostasis in intestinal cells through STAT5 action. Cytokines, hormones, growth factors, and chemokines activate STAT5 in colonic cells as shown in Fig. 1. STAT5 protects intestinal tissue from C. difficile colitis by regulating tight junction permeability in enterocytes. Additionally, STAT5 protects the intestinal tissue from injury by promoting self-renewal of LGR5+ stem cells and differentiation of LGR5+ towards pYSTAT5+ LGR5− CD24+ Lyso+ or CD44+ cKIT+ cells. Moreover, STAT5 has been found to upregulate MUC13 production in Goblet cells, which modulates intestinal inflammation by playing a protective role in the colonic epithelium. However, the role of STAT5 in mesenchymal niche cells of the colonic tissue is unknown and is yet to be explored (marked by “?”).
As depicted in Fig. 4, the signaling network controlling IESC self-renewal and lineage differentiation is extremely complex, not accounting for immune cell interplay involving, e.g. communication with commensals or the AXL/MER/TERO tyrosine kinase receptor family and their ligands [33]. AXL tyrosine kinase is a direct target of STAT5 in hematopoietic cell types, facilitating immune cell control or prolonged pYSTAT5 action when activated by its ligands [62]. Therefore, the signaling network must be tightly regulated to maintain intestinal homeostasis. Many essential stromal cell interactions or microbiota consequences in the context of cytokine-STAT5 signaling are poorly studied. Overall, the underlying mechanisms that regulate lineage differentiation of secretory cells such as goblet, Paneth, or enteroendocrine cells of the colon are incompletely understood.
The IESC are interspersed at the crypt bottom between fully differentiated CD24+ CD44+ Paneth-like cells in a such way that results in the maximization of the contact area (CD24+ CD44+ -stem cell) [45]. These secretory cells are also residents at the colonic crypt base to serve as an IESC niche, which can be labeled by specific antibodies against cKIT, REG4, MUC2, or by Alcian Blue (AB) staining [44]. These fully differentiated, post-mitotic cells were found to produce WNT3 and WNT11, EGF, TGF-α, and to express the Notch ligand DLL4 to support IESC maintenance [45]. In addition to supporting IESC renewal, these cells are capable to de-differentiate upon injury to acquire stem cell features, to proliferate and to re-differentiate to maintain intestinal homeostasis [63,64]. WNT activation was also shown to be crucial for reverting mTERT+ cells from a quiescent state to fast-proliferating cells. In contrast, increased Notch signaling was indicated upon irradiation injury and Paneth cell metaplasia, whereas acute Notch inhibition induces the niche repair, upon which colonic homeostasis is restored [64,65,66]. Moreover, SCF/c-KIT signaling and downstream amplification via kinase cascades, such as the PI3K/AKT-mTOR activation and GSK3β inhibition, are of functional relevance for Paneth-like cell dedifferentiation during chronic inflammation [63]. Here, it should be noted that mTOR can directly phosphorylate STAT5A/B at the serine residue in the coiled-coil domain (Fig. 2) and that STAT5 can transcriptionally drive AKT isoforms, as well as interact with PI3K-AKT binding directly in a cytoplasmic complex to the p85 subunit of PI3K via GAB1/2 scaffold proteins [15,30]. The exact mechanism and signals required for these fully differentiated, post-mitotic epithelial cells, and whether or not they require STAT5A/B protein function to adopt stem cell features, remains unclear. Besides, other IESC niche cells might have important roles. In particular, mesenchymal niche cells will need further investigation before we can illuminate or review them within a STAT5 centric vision. The exact number of niche cells contributing to intestinal homeostasis maintenance is unknown. However, there is some strong evidence supporting the role of mesenchymal niche cells. Subepithelial myofibroblasts marked by PDGFRα were shown to be an essential source of WNTs and RSPO3 [67]. Importantly, the PDGFRβ chain is in neoplastic T-cells under direct STAT5 transcriptional control, and PDGFR signaling or hyperactivation through translocations with its kinase part captures pYSTAT5 action [68,69]. Of note, intestinal telocytes, marked by FOXL1, PDGFRα, and CD34 play an important role in the regulation of proliferation and differentiation, by expressing high levels of both, activators and repressors [70]. As shown in Fig. 4, these cells secrete WNT2b, WNT5a, RSPO3, and BMP4-7, but also their antagonists, such as DKK3 and sFRP1, CHORDIN-LIKE1 and GREMLIN1, highlighting the capability of these cells to tightly regulate proliferation at the crypt bases, as well as differentiation in the villi [70]. GLI1+ cells are WNT-producing cells localized near the crypt bases [71]. Studies found that these cells could compensate for the absence of WNT-secreting Paneth cells in the small intestine in vivo, accounting for the absence of exogenous WNT3ain the development of colonic organoids. Moreover, the morphology of co-cultured colonic organoids was very similar to those cultured with exogenous WNT3a [71]. ScRNA-seq data on single-sorted colonic GLI1+ cells revealed eight different clusters with PDGFRα transcripts present among all clusters and WNT2, WNT2b, WNT4 transcripts overlapping with CD34 transcripts in two of the clusters. However, only one GLI1+ cluster showed high levels of RSPO3 transcripts, indicating different subsets of PDGFRα expressing cells [71]. The number of different cell types of the epithelial or mesenchymal origin, that orchestrate IESC renewal and maintenance, remains to be explored.
3. The JAK/STAT5 pathway in the intestinal homeostasis and diseases
The gastrointestinal tract is not only a digestive organ, but it is also an immune and endocrine unit [72]. Homeostatic tissue renewal is regulated by epithelial niche, secretory lineages or mesenchymal niche populations. Behavior and fate of LGR5+ stem cells highly depend not only on classical morphogens, which are described above and extensively reviewed in [73], but also on immune-derived factors. The regulation of intestinal mucosal immunity by JAK/STAT5 has been well reported in the past decades [74,75,26]. These findings demonstrate that intestinal JAK/STAT5 is the key signaling in the lamina propria T lymphocytes [76], macrophages/dendritic cells [31,77], M cells [78], and the survival factor of intraepithelial αβ or γδ T-cells during homeostasis [79,80]. The intestinal JAK/STAT5 plays a nodal role in various gut cytokines, hormones, and growth factors-mediated mucosal destruction or protection [81,82]. Immune responses in infection and inflammation of the intestinal epithelium play a key role in regeneration, which is directly dependent on ISC proliferation. Some of the transducers from immune cells to epithelial signaling are not known, e.g. group 3 innate lymphoid cells from stroma regulate epithelial Hippo-YAP pathway, but so far it has been clarified whether this is mediated by IL-6 or group 3 innate lymphoid cells act directly on the epithelium [73]. As reviewed extensively in [83], cytokines produced by immune cells, such as IL-6 and TNF-α, promote regeneration of the injured mucosa, acting directly on epithelial cells. IL-6-related proteins in Drosophila, which are known as UPD ligands, engage STAT5 to stimulate intestinal repair. Immune cells-derived factors, such as cytokines, have been investigated on intestinal organoid 3D cultures. These investigations revealed that organoid treatment with IL-2, which activates STAT5 as a downstream effector, promotes the growth of organoids and expression of markers of mature intestinal epithelial cells. Cytokines such as, IL-4 and IL-13 have been shown to promote Tuft cell maturation, which react to parasite antigens [84]. Furthermore, IL-17 regulates the integrity of intestinal epithelial barrier and induces transcription of antimicrobial peptides, protecting the tissue from microbiota translocation and subsequent inflammation. Interestingly, STAT5A and STAT5B have in particular very important neonatal functions for γδ T-cell generation. γδ T-cells are innate immune cells involved in intestinal epithelial homeostasis by playing a protective role upon epithelial barrier dysfunction and microorganisms translocation, exerting pro- and anti-inflammatory responses. IL-17-producing RORγt± γδ T-cells (γδT17 cells) are lymphocytes that regulate immune responses during infection and inflammation. However, recent studies show that STAT5 is required for the expansion of these cells, as mice with conditional lack of STAT5 in RORγt± γδ T-cells showed loss of γδT17 cells. Furthermore, it was shown that STAT5A promotes IL-17-producing γδT17 cells and down-regulates T-bet, while STAT5B promotes IFNγ-producing γδ T-cells and increases T-bet intestinal expression [79]. In addition, IL-9 induces IL-8 to increase neutrophil infiltration into gut mucosa, leading to the overactivation of STAT5 in the intestinal epithelial compartment that interferes with mucosal healing [85], while an impaired IL-15 delivery results in diminished colonic STAT5 phosphorylation in the Treg cells, which propagates intestinal inflammation in inflammatory bowel diseases (IBD) [86]. Interestingly, the reports regarding the role of JAK/STAT5 in the intraepithelial T-cells or innate lymphoid cells [87] are controversial. It has been reported that IL-15-induced STAT5 activation in intraepithelial T-cells increases the inflammatory severity of human Celiac disease [88], whereas STAT5-deficient Treg cells lose suppression of murine colitis [89]. Furthermore, IL-23- and IL-2-activated STAT5 increases innate lymphoid cell [87] to release IL-22 and maintains the gut epithelial integrity to prevent from C. rodentium-induced colitis [80]. Recently, an inflammatory disease-associated enhancer at 11q13.5 has been reported to suppress colitis by recruiting STAT5 on Lrrc32 in Treg cells [90]. Therefore, given these effects of JAK/STAT5 are dependent on mucosal stimuli, cell types, and even genetic variations, it is plausible to determine the pathophysiological functions of JAK/STAT5 in the context of diseases.
The cytokine-JAK/STAT pathway is critically involved in the regulation of IESC regeneration and IEC lineage differentiation, particularly during intestinal infection and inflammation [9,60]. Importantly, STAT5 activity is predominantly associated with the maintenance of the hematopoietic [91], mammary [92], embryonic stem cell, and IESC self-renewal [27,93], and mammary secretory lineage specification [94]. Loss of STAT5 in specific somatic stem cells results in a defective immune system, impaired myelopoiesis, disturbed liver metabolism function, mammary gland atrophy associated with lactation failure or reproductive tract deficiencies, and intestinal barrier disruption [95,27,28,22]. We and others reported that STAT5 activation increased IESC regeneration, thereby contributing to functional tissue formation upon chronic inflammatory injury [27,92]. Moreover, the absence of STAT5 in the mammary epithelia impairs epithelial adhesion, which was also shown in IEC monolayers, most likely due to increased NF-κB DNA binding promoting the high expression of the long chain of the Myosin Light Chain Kinase that upon enzymatic activity bursts tight junction formation (MLCK promoter), leading to tissue permeability for commensals upon lost epithelial integrity [28], bacterial infection, chronic injury and tissue repair processes, predisposing to colon cancer. However, specific IEC cell types regulated by STAT5 have not been explored yet, which is highly relevant to GI tract diseases and the exploration of new therapeutic targets. Intriguingly, the Hans Clevers group has recently reported that the LGR5lowKi67+ population above Paneth cells contains high STAT5A gene abundance associated with significantly increased signatures of secretory lineages after induced activation. These data and our own investigations strongly suggest that pYSTAT5 activation is required for specifying LGR5lowKi67+ IESCs to pYSTAT5+LGR5−CD24+ or CD44+ DCSs. Importantly, TcdA and TcdB, primary virulence factors in C. difficile-associated colitis, pirate STAT5 signaling. TcdA can inhibit JAK2 [96], and TcdB together with TNFα activates IEC pYSTAT5 [28,97]. Leptin, an adipocytokine that can activate STAT5 (and also STAT1/3) in IECs through JAK2 [98], was reported to protect against C. difficile colitis [99]. Butyrate, a major gut bacterial metabolite, can strongly activate STAT5 to promote cellular proliferation [100]. These findings suggest that the JAK/STAT5 axis could be a critical pathway for protecting against dysbiosis-induced IEC barrier dysfunction.
4. Concluding remarks and future directions
In conclusion, STAT5 plays a significant role in regulating the interaction of microbiota and IEC, mediating chronic inflammatory and promoting mucosal healing. However, which STAT5, STAT5A or B, promotes healing during disease course is unclear, or whether STAT5A and STAT5B act in a YIN-YANG scenario similar to STAT1-STAT3 deserves exploration [101]. Further investigation would be needed to prove its potential as a possible prognostic marker of inflammation and infection, maybe even target for future therapeutic interventions in IEC lineage repair. However, the crucial role of STAT5 in intestinal homeostasis is undeniable. Changing components of the JAK/STAT pathway is not a simple prediction in chronic inflammatory or infectious disease. The way in which targeting the JAK/STAT pathway might affect intestinal homeostasis has to be experimentally explored in relevant model systems to illuminate its involvement in colorectal cancer formation, one of the big five cancer killers. Pharmacologic intervention studies using STAT5 inhibitors are also available for in vivo testing in genetic mouse model systems, where the impact on IESC cycling or tissue regeneration upon injury are important readout systems to be investigated. There is work ahead of multiple groups to illuminate the role of the two STAT5 transcription factors in GI tract diseases, and we hope that this review could be a cornerstone to build on.
Acknowledgements
MS and XH are supported by the National Institutes of Health (1R01DK123299-01 to XH and P30 DK078392). RM is supported by the Austrian Science Fund (FWF; SFB-F04707/SFB-F06105 to RM) and under the frame of ERA-NET (I 4157-B to RM). This project was supported in part by NIH P30 DK078392 of the Digestive Diseases Research Core Center in Cincinnati.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Becker S, Groner B, Muller CW, Three-dimensional structure of the Stat3beta homodimer bound to DNA, Nature 394 (6689) (1998) 145–151. [DOI] [PubMed] [Google Scholar]
- [2].Neculai D, et al. , Structure of the unphosphorylated STAT5a dimer, J. Biol. Chem 280 (49) (2005) 40782–40787. [DOI] [PubMed] [Google Scholar]
- [3].Kosack L, et al. , The ERBB-STAT3 Axis Drives Tasmanian Devil Facial Tumor Disease, Cancer Cell 35 (1) (2019) 125–139 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Liu BA, et al. , The human and mouse complement of SH2 domain proteins-establishing the boundaries of phosphotyrosine signaling, Mol. Cell 22 (6) (2006) 851–868. [DOI] [PubMed] [Google Scholar]
- [5].Liongue C, Ward AC, Evolution of the JAK-STAT pathway, JAKSTAT 2 (1) (2013), e22756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gorissen M, et al. , STAT genes display differential evolutionary rates that correlate with their roles in the endocrine and immune system, J. Endocrinol 209 (2) (2011) 175–184. [DOI] [PubMed] [Google Scholar]
- [7].Arbouzova NI, Zeidler MP, JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions, Development 133 (14) (2006) 2605–2616. [DOI] [PubMed] [Google Scholar]
- [8].Ihle JN, STATs: Signal Transducers and Activators of Transcription, Cell 84 (3) (1996) 331–334. [DOI] [PubMed] [Google Scholar]
- [9].Jiang H, et al. , Cytokine/Jak/Stat Signaling Mediates Regeneration and Homeostasis in the Drosophila Midgut, Cell 137 (7) (2009) 1343–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ernst M, et al. , Epithelial gp130/Stat3 functions: an intestinal signaling node in health and disease, Semin. Immunol 26 (1) (2014) 29–37. [DOI] [PubMed] [Google Scholar]
- [11].Furtek SL, et al. , Strategies and Approaches of Targeting STAT3 for Cancer Treatment, ACS Chem. Biol 11 (2) (2016) 308–318. [DOI] [PubMed] [Google Scholar]
- [12].Cui Y, et al. , The Stat3/5 locus encodes novel endoplasmic reticulum and helicase-like proteins that are preferentially expressed in normal and neoplastic mammary tissue, Genomics 78 (3) (2001) 129–134. [DOI] [PubMed] [Google Scholar]
- [13].Miyoshi K, et al. , Structure of the mouse Stat 3/5 locus: evolution from Drosophila to zebrafish to mouse, Genomics 71 (2) (2001) 150–155. [DOI] [PubMed] [Google Scholar]
- [14].Crispi S, et al. , Characterization of the human STAT5A and STAT5B promoters: evidence of a positive and negative mechanism of transcriptional regulation, FEBS Lett. 562 (1–3) (2004) 27–34. [DOI] [PubMed] [Google Scholar]
- [15].Ferbeyre G, Moriggl R, The role of Stat5 transcription factors as tumor suppressors or oncogenes, Biochim. Biophys. Acta 1815 (1) (2011) 104–114. [DOI] [PubMed] [Google Scholar]
- [16].Hennighausen L, Robinson GW, Interpretation of cytokine signaling through the transcription factors STAT5A and STAT5B, Genes. Dev 22 (6) (2008) 711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Imada K, et al. , Stat5b is essential for natural killer cell-mediated proliferation and cytolytic activity, J. Exp. Med 188 (11) (1998) 2067–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Liu X, et al. , Stat5a is mandatory for adult mammary gland development and lactogenesis, Genes. Dev 11 (2) (1997) 179–186. [DOI] [PubMed] [Google Scholar]
- [19].Feldman GM, et al. , STAT5A-deficient mice demonstrate a defect in granulocyte-macrophage colony-stimulating factor-induced proliferation and gene expression, Blood 90 (5) (1997) 1768–1776. [PubMed] [Google Scholar]
- [20].Udy GB, et al. , Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression, Proc. Natl. Acad. Sci. USA 94 (14) (1997) 7239–7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kerenyi MA, et al. , Stat5 regulates cellular iron uptake of erythroid cells via IRP-2 and TfR-1, Blood 112 (9) (2008) 3878–3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yao Z, et al. , Stat5a/b are essential for normal lymphoid development and differentiation, Proc. Natl. Acad. Sci. USA 103 (4) (2006) 1000–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Nakajima H, et al. , An indirect effect of Stat5a in IL-2-induced proliferation: a critical role for Stat5a in IL-2-mediated IL-2 receptor alpha chain induction, Immunity 7 (5) (1997) 691–701. [DOI] [PubMed] [Google Scholar]
- [24].Engblom D, et al. , Direct glucocorticoid receptor-Stat5 interaction in hepatocytes controls body size and maturation-related gene expression, Genes. Dev 21 (10) (2007) 1157–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Han X, et al. , Signal transducer and activator of transcription 5b promotes mucosal tolerance in pediatric Crohn’s disease and murine colitis, Am. J. Pathol 169 (6) (2006) 1999–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Han X, et al. , Regulation of intestinal barrier function by signal transducer and activator of transcription 5b, Gut 58 (1) (2009) 49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gilbert S, et al. , Activated STAT5 confers resistance to intestinal injury by increasing intestinal stem cell proliferation and regeneration, Stem. Cell Reports 4 (2) (2015) 209–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gilbert S, et al. , Enterocyte STAT5 promotes mucosal wound healing via suppression of myosin light chain kinase-mediated loss of barrier function and inflammation, EMBO Mol. Med 4 (2) (2012) 109–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Freund P, et al. , O-GlcNAcylation of STAT5 controls tyrosine phosphorylation and oncogenic transcription in STAT5-dependent malignancies, Leukemia 31 (10) (2017) 2132–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wingelhofer B, et al. , Implications of STAT3 and STAT5 signaling on gene regulation and chromatin remodeling in hematopoietic cancer, Leukemia 32 (8) (2018)1713–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Han X, et al. , Loss of GM-CSF signalling in non-haematopoietic cells increases NSAID ileal injury, Gut 59 (8) (2010) 1066–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jatiani SS, et al. , Jak/STAT pathways in cytokine signaling and myeloproliferative disorders: approaches for targeted therapies, Genes. Cancer 1 (10) (2010) 979–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Orlova A, et al. , Direct Targeting Options for STAT3 and STAT5 in Cancer, Cancers (Basel) 11 (12) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kramer OH, Moriggl R, Acetylation and sumoylation control STAT5 activation antagonistically, JAKSTAT 1 (3) (2012) 203–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gambetta MC, Oktaba K, Muller J, Essential role of the glycosyltransferase sxc/Ogt in polycomb repression, Science 325 (5936) (2009) 93–96. [DOI] [PubMed] [Google Scholar]
- [36].Liu TW, et al. , Genome-wide chemical mapping of O-GlcNAcylated proteins in Drosophila melanogaster, Nat. Chem. Biol 13 (2) (2017) 161–167. [DOI] [PubMed] [Google Scholar]
- [37].Pinto PB, et al. , JAK/STAT and Hox Dynamic Interactions in an Organogenetic Gene Cascade, PLoS Genet. 11 (7) (2015), e1005412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Stoecklin E, et al. , Interactions in the transcriptional regulation exerted by Stat5 and by members of the steroid hormone receptor family, J. Steroid Biochem. Mol. Biol 69 (1–6) (1999) 195–204. [DOI] [PubMed] [Google Scholar]
- [39].Tan SH, et al. , Transcription factor Stat5 synergizes with androgen receptor in prostate cancer cells, Cancer Res 68 (1) (2008) 236–248. [DOI] [PubMed] [Google Scholar]
- [40].Wyszomierski SL, Yeh J, Rosen JM, Glucocorticoid receptor/signal transducer and activator of transcription 5 (STAT5) interactions enhance STAT5 activation by prolonging STAT5 DNA binding and tyrosine phosphorylation, Mol. Endocrinol 13 (2) (1999) 330–343. [DOI] [PubMed] [Google Scholar]
- [41].Koo BK, Clevers H, Stem cells marked by the R-spondin receptor LGR5, Gastroenterology 147 (2) (2014) 289–302. [DOI] [PubMed] [Google Scholar]
- [42].van der Flier LG, Clevers H, Stem cells, self-renewal, and differentiation in the intestinal epithelium, Annu. Rev. Physiol 71 (2009) 241–260. [DOI] [PubMed] [Google Scholar]
- [43].Sato T, et al. , Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche, Nature 459 (7244) (2009) 262–265. [DOI] [PubMed] [Google Scholar]
- [44].Barker N, Clevers H, Leucine-rich repeat-containing G-protein-coupled receptors as markers of adult stem cells, Gastroenterology 138 (5) (2010) 1681–1696. [DOI] [PubMed] [Google Scholar]
- [45].Sato T, et al. , Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts, Nature 469 (7330) (2011) 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Snippert HJ, et al. , Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells, Cell 143 (1) (2010) 134–144. [DOI] [PubMed] [Google Scholar]
- [47].Barker N, et al. , Identification of stem cells in small intestine and colon by marker gene Lgr5, Nature 449 (7165) (2007) 1003–1007. [DOI] [PubMed] [Google Scholar]
- [48].Sangiorgi E, Capecchi MR, Bmi1 is expressed in vivo in intestinal stem cells, Nat. Genet 40 (7) (2008) 915–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Tian H, et al. , A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable, Nature 478 (7368) (2011) 255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Takeda N, et al. , Interconversion between intestinal stem cell populations in distinct niches, Science 334 (6061) (2011) 1420–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].van Es JH, et al. , Dll1+ secretory progenitor cells revert to stem cells upon crypt damage, Nat. Cell Biol 14 (10) (2012) 1099–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Basak O, et al. , Induced Quiescence of Lgr5+ Stem Cells in Intestinal Organoids Enables Differentiation of Hormone-Producing Enteroendocrine Cells, Cell Stem Cell 20 (2) (2017) 177–190 e4. [DOI] [PubMed] [Google Scholar]
- [53].Basak O, et al. , Mapping early fate determination in Lgr5+ crypt stem cells using a novel Ki67-RFP allele, EMBO J. 33 (18) (2014) 2057–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Yan KS, et al. , Intestinal Enteroendocrine Lineage Cells Possess Homeostatic and Injury-Inducible Stem Cell Activity, Cell Stem Cell 21 (1) (2017) 78–90 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Liu R, et al. , Constitutive STAT5 activation regulates Paneth and Paneth-like cells to control Clostridium difficile colitis, Life Sci. Alliance 2 (2) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Crosnier C, Stamataki D, Lewis J, Organizing cell renewal in the intestine: stem cells, signals and combinatorial control, Nat. Rev. Genet 7 (5) (2006) 349–359. [DOI] [PubMed] [Google Scholar]
- [57].Willert K, et al. , Wnt proteins are lipid-modified and can act as stem cell growth factors, Nature 423 (6938) (2003) 448–452. [DOI] [PubMed] [Google Scholar]
- [58].Carmon KS, et al. , R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling, Proc. Natl. Acad. Sci. USA 108 (28) (2011) 11452–11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Pellegrinet L, et al. , Dll1- and dll4-mediated notch signaling are required for homeostasis of intestinal stem cells, Gastroenterology 140 (4) (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lindemans CA, et al. , Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration, Nature 528 (7583) (2015) 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kaiko GE, et al. , The Colonic Crypt Protects Stem Cells from Microbiota-Derived Metabolites, Cell 165 (7) (2016) 1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Orlova A, Neubauer HA, Moriggl R, The stromal microenvironment provides an escape route from FLT3 inhibitors through the GAS6-AXL-STAT5 axis, Haematologica 104 (10) (2019) 1907–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Schmitt M, et al. , Paneth Cells Respond to Inflammation and Contribute to Tissue Regeneration by Acquiring Stem-like Features through SCF/c-Kit Signaling, Cell Rep. 24 (9) (2018) 2312–2328 e7. [DOI] [PubMed] [Google Scholar]
- [64].Yu S, et al. , Paneth Cell Multipotency Induced by Notch Activation following Injury, Cell Stem Cell 23 (1) (2018) 46–59 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Durand A, et al. , Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1), Proc. Natl. Acad. Sci. USA 109 (23) (2012) 8965–8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Bohin N, et al. , Rapid Crypt Cell Remodeling Regenerates the Intestinal Stem Cell Niche after Notch Inhibition, Stem Cell Reports 15 (1) (2020) 156–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Greicius G, et al. , PDGFRalpha(+) pericryptal stromal cells are the critical source of Wnts and RSPO3 for murine intestinal stem cells in vivo, Proc. Natl. Acad. Sci. USA 115 (14) (2018) E3173–E3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Sternberg DW, et al. , The TEL/PDGFbetaR fusion in chronic myelomonocytic leukemia signals through STAT5-dependent and STAT5-independent pathways, Blood 98 (12) (2001) 3390–3397. [DOI] [PubMed] [Google Scholar]
- [69].Triplett TA, et al. , Endogenous dendritic cells from the tumor microenvironment support T-ALL growth via IGF1R activation, Proc. Natl. Acad. Sci. USA 113 (8) (2016) E1016–E1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Shoshkes-Carmel M, et al. , Subepithelial telocytes are an important source of Wnts that supports intestinal crypts, Nature 557 (7704) (2018) 242–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Degirmenci B, et al. , GLI1-expressing mesenchymal cells form the essential Wnt-secreting niche for colon stem cells, Nature 558 (7710) (2018) 449–453. [DOI] [PubMed] [Google Scholar]
- [72].Worthington JJ, Reimann F, Gribble FM, Enteroendocrine cells-sensory sentinels of the intestinal environment and orchestrators of mucosal immunity, Mucosal. Immunol 11 (1) (2018)3–20. [DOI] [PubMed] [Google Scholar]
- [73].Beumer J, Clevers H, Cell fate specification and differentiation in the adult mammalian intestine, Nat. Rev. Mol. Cell Biol (2020). [DOI] [PubMed] [Google Scholar]
- [74].Snow JW, et al. , Loss of tolerance and autoimmunity affecting multiple organs in STAT5A/5B-deficient mice, J. Immunol 171 (10) (2003) 5042–5050. [DOI] [PubMed] [Google Scholar]
- [75].Zhao HM, et al. , Astragalus polysaccharide attenuates rat experimental colitis by inducing regulatory T cells in intestinal Peyer’s patches, World J. Gastroenterol 22 (11) (2016) 3175–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Ebert EC, Interleukin 21 up-regulates perforin-mediated cytotoxic activity of human intra epithelial lymphocytes, Immunology 127 (2) (2009) 206 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Jaensson E, et al. , Small intestinal CD103+ dendritic cells display unique functional properties that are conserved between mice and humans, J. Exp. Med 205 (9) (2008) 2139–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Tagaya Y, et al. , Identification of a novel receptor/signal transduction pathway for IL-15/T in mast cells, EMBO J. 15 (18) (1996) 4928–4939. [PMC free article] [PubMed] [Google Scholar]
- [79].Kadekar D, et al. , The neonatal microenvironment programs innate gammadelta T cells through the transcription factor STAT5, J. Clin. Invest 130 (5) (2020) 2496–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Bauche D, et al. , IL-23 and IL-2 activation of STAT5 is required for optimal IL-22 production in ILC3s during colitis, Sci. Immunol 5 (46) (2020). [DOI] [PubMed] [Google Scholar]
- [81].Herrera SC, Bach EA, JAK/STAT signaling in stem cells and regeneration: from Drosophila to vertebrates, Development 146 (2) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Salas A, et al. , JAK-STAT pathway targeting for the treatment of inflammatory bowel disease, Nat. Rev. Gastroenterol. Hepatol 17 (6) (2020) 323–337. [DOI] [PubMed] [Google Scholar]
- [83].Karin M, Clevers H, Reparative inflammation takes charge of tissue regeneration, Nature 529 (7586) (2016) 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Bar-Ephraim YE, Kretzschmar K, Clevers H, Organoids in immunological research, Nat. Rev. Immunol 20 (5) (2020) 279–293. [DOI] [PubMed] [Google Scholar]
- [85].Nalleweg N, et al. , IL-9 and its receptor are predominantly involved in the pathogenesis of UC, Gut 64 (5) (2015) 743–755. [DOI] [PubMed] [Google Scholar]
- [86].Tosiek MJ, et al. , IL-15-dependent balance between Foxp3 and RORgammat expression impacts inflammatory bowel disease, Nat. Commun 7 (2016) 10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Amersi FF, et al. , Long-term survival after radiofrequency ablation of complex unresectable liver tumors, Arch. Surg 141 (6) (2006). [DOI] [PubMed] [Google Scholar]
- [88].Malamut G, et al. , IL-15 triggers an antiapoptotic pathway in human intraepithelial lymphocytes that is a potential new target in celiac disease-associated inflammation and lymphomagenesis, J. Clin. Invest 120 (6) (2010) 2131–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Lin JX, et al. , Critical Role of STAT5 transcription factor tetramerization for cytokine responses and normal immune function, Immunity 36 (4) (2012) 586–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Nasrallah R, et al. , A distal enhancer at risk locus 11q13.5 promotes suppression of colitis by Treg cells, Nature 583 (7816) (2020) 447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Kato Y, et al. , Selective activation of STAT5 unveils its role in stem cell self-renewal in normal and leukemic hematopoiesis, J. Exp. Med 202 (1) (2005) 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Vafaizadeh V, et al. , Mammary epithelial reconstitution with gene-modified stem cells assigns roles to Stat5 in luminal alveolar cell fate decisions, differentiation, involution, and mammary tumor formation, Stem Cells 28 (5) (2010) 928–938. [DOI] [PubMed] [Google Scholar]
- [93].Kyba M, et al. , Enhanced hematopoietic differentiation of embryonic stem cells conditionally expressing Stat5, Proc. Natl. Acad. Sci. USA 100 (Suppl 1) (2003) 11904–11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Miyoshi K, et al. , Signal transducer and activator of transcription (Stat) 5 controls the proliferation and differentiation of mammary alveolar epithelium, J. Cell Biol 155 (4) (2001) 531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Cui Y, et al. , Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation, Mol. Cell Biol 24 (18) (2004) 8037–8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Nam ST, et al. , Clostridium difficile toxin A inhibits erythropoietin receptor-mediated colonocyte focal adhesion through inactivation of Janus Kinase-2, J. Microbiol. Biotechnol 22 (12) (2012) 1629–1635. [DOI] [PubMed] [Google Scholar]
- [97].Stankiewicz TR, et al. , Signal transducer and activator of transcription-5 mediates neuronal apoptosis induced by inhibition of Rac GTPase activity, J. Biol. Chem 287 (20) (2012) 16835–16848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Morton NM, et al. , Leptin action in intestinal cells, J. Biol. Chem 273 (40) (1998) 26194–26201. [DOI] [PubMed] [Google Scholar]
- [99].Madan R, et al. , Role of leptin-mediated colonic inflammation in defense against Clostridium difficile colitis, Infect. Immun 82 (1) (2014) 341–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Boosalis MS, et al. , Short-chain fatty acid derivatives stimulate cell proliferation and induce STAT-5 activation, Blood 97 (10) (2001) 3259–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Friedrich K, et al. , Steering of carcinoma progression by the YIN/YANG interaction of STAT1/STAT3, Biosci. Trends 11 (1) (2017) 1–8. [DOI] [PubMed] [Google Scholar]