SUMMARY
BACKGROUND:
Up to fifty percent of the adult human sinoatrial node (SAN), is composed of dense connective tissue. Cardiac diseases including heart failure (HF) may further increase fibrosis within the SAN pacemaker complex, leading to impaired automaticity and conduction of electrical activity to the atria. However, unlike the role of cardiac fibroblasts in pathological fibrotic remodeling and tissue repair, nothing is known about fibroblasts that maintain the inherently fibrotic SAN environment.
METHODS:
Intact SAN pacemaker complex was dissected from cardioplegically arrested explanted non-failing (non-HF, n=22; 48.7±3.1y.o,) and HF human hearts (n=16; 54.9±2.6y.o.). Connective tissue content was quantified from Masson’s trichrome stained head-center and center-tail SAN sections. Expression of extracellular matrix (ECM) proteins, including Collagens 1, 3A1, cartilage intermediate layer protein 1 (CILP1) and periostin, fibroblast and myofibroblast numbers were quantified by in situ and in vitro immunolabeling. Fibroblasts from the central intramural SAN pacemaker compartment (~10×5×2 mm3) and right atria (RA) were isolated, cultured, passaged once, and treated ±transforming growth factor beta-1 (TGFβ1) and subjected to comprehensive high-throughput next-generation sequencing of whole transcriptome, microRNA and proteomic analyses.
RESULTS:
Intranodal fibrotic content was significantly higher in SAN pacemaker complex from HF vs non-HF hearts (57.7±2.6% vs 44.0±1.2% p<0.0001). Proliferating phosphorylated histone3+/vimentin+/CD31- fibroblasts were higher in HF SAN. Vimentin+/alpha smooth muscle actin+/CD31- myofibroblasts along with increased interstitial periostin expression were found only in HF SAN. RNA sequencing and proteomic analyses identified unique differences in mRNA, long non-coding RNA, microRNA and proteomic profiles between non-HF and HF SAN and RA fibroblasts, and TGFβ1-induced myofibroblasts. Specifically, proteins and signaling pathways associated with ECM flexibility, stiffness, focal adhesion and metabolism were altered in HF SAN fibroblasts compared to non-HF SAN.
CONCLUSIONS:
This study revealed increased SAN-specific fibrosis with presence of myofibroblasts, CILP1 and periostin-positive interstitial fibrosis only in HF vs non-HF human hearts. Comprehensive proteo-transcriptomic profiles of SAN fibroblasts identified upregulation of genes and proteins promoting stiffer SAN ECM in HF hearts. Fibroblast-specific profiles generated by our proteo-transcriptomic analyses of the human SAN, provide a comprehensive framework for future studies to investigate the role of SAN-specific fibrosis in cardiac rhythm regulation and arrhythmias.
Keywords: Sinoatrial node, fibrosis, fibroblasts, myofibroblasts, whole transcriptome, proteomics, heart failure, arrhythmias, extracellular matrix
INTRODUCTION
The sinoatrial node (SAN), the primary pacemaker of the human heart, is a heterogeneous composition of specialized pacemaker cardiomyocytes and non-excitable cells (e.g. fibroblasts)1, 2. Histological postmortem evaluations3 and recent structural in vivo and ex vivo 3D electro-structural imaging of human hearts have demonstrated that the distinctive 3D intramural structure of the human SAN pacemaker complex is primarily composed of dense connective tissue1, 4, 5. Even healthy human SANs are composed of 35–55% dense connective tissue, from ~24% in infants with age-induced increases up to ~60% in elderly hearts6 which is unique for human vs animals4, 7. Such high levels of extracellular matrix (ECM) would be considered pathological in any other cardiac region, which could lead to arrhythmias and conduction impairments. In contrast, the distinctive high level of ECM in the SAN is required to electrically insulate pacemaker cardiomyocytes in order to maintain source-sink balance leading to activation of the large right atrial (RA) myocardium by the relatively small SAN pacemaker complex4. However, the cellular basis and molecular mechanisms driving the naturally high fibrotic levels in human SAN is a veritable “terra incognita”. Specifically, no study has examined SAN fibroblasts, the primary cellular source of connective tissue, to determine if there are quantitative and qualitative characteristics that uniquely enable them to secrete and maintain the high levels of connective tissue found in the SAN.
Moreover, SAN ECM is known to increase in several human diseases, including heart failure (HF), which is associated with cardiac arrhythmias, impairment of SAN automaticity and eventually leading to SAN dysfunction (SND)3, 4, 8, 9. Recent ex vivo near-infrared optical mapping studies of diseased human hearts show that SAN fibrosis could specifically interrupt automaticity and introduce conduction blocks, by disrupting electrical connectivity between SAN pacemaker clusters and conduction pathways5, 10, 11 (Figure 1A). However, while clinical and experimental studies have clearly emphasized the critical role of SAN fibrosis as a causal factor of SND, the cellular and molecular basis for the intriguing levels of dense connective tissue in the healthy SAN and pathophysiological HF-induced SAN fibrosis upregulation has never been studied.
Figure 1: Project description and overview of workflow.
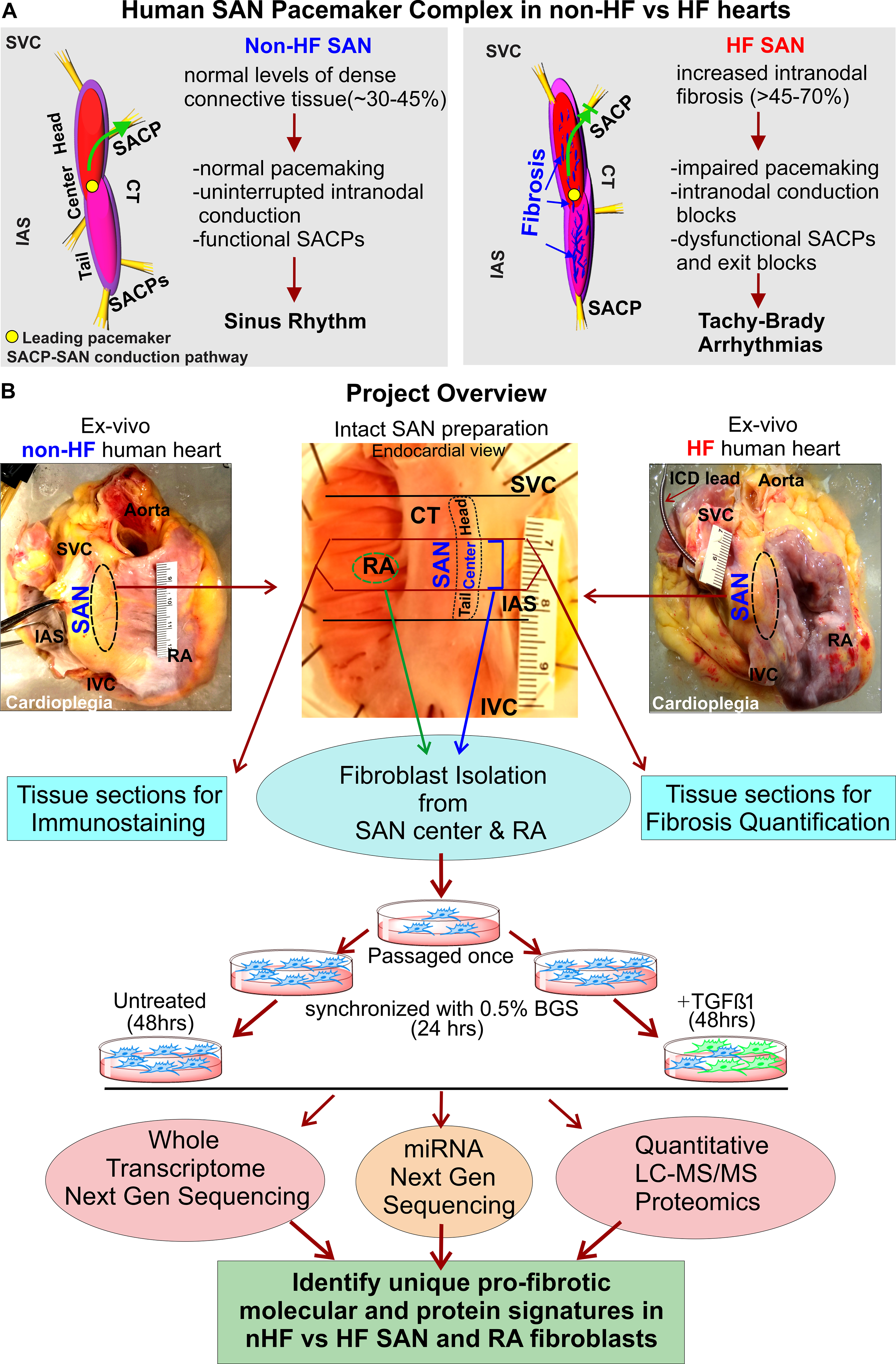
A, 3D models of non-failing (non-HF) human Sinoatrial node (SAN) pacemaker complex (left) and heart failure (HF) (right) depicting their functional/dysfunctional and structural characteristics. Green arrow shows preferential conduction from the leading pacemaker in the SAN to atria through the SAN conduction pathway (SACP) in non-HF hearts, and blocked green arrow indicates exit block in HF hearts. B, Description of the project work flow. Abbreviations: BGS-Bovine growth serum; CT- crista terminalis; IAS- interatrial septum; IVC- inferior vena cava; RA-right atrium; SVC-superior vena cava; TGFβ1-Transforming growth factor beta 1.
Importantly, almost all of our understanding regarding cardiac fibrosis, fibroblasts, and their differentiated states including myofibroblasts, come from studies of acute injury responses including myocardial infarction12, 13 and chronic disease states. Hence, we aimed to develop an in-depth understanding of the phenotypic and molecular landscape of SAN fibroblasts in vitro and in situ together with intranodal ECM composition in non-failing (non-HF) and HF human hearts, which could ultimately be utilized to target pathologic SAN fibrosis. Specifically, we isolated and cultured human SAN and surrounding RA fibroblasts from human non-HF and HF hearts, and subjected the same samples to quantitative proteomics and next generation sequencing (NGS) of the whole cellular transcriptome in order to generate for the first time, a comprehensive atlas of protein expression, coding mRNA, and non-coding RNAs including long non-coding RNA (lncRNA) and microRNA (miRNA) (Figure 1B).
METHODS
Detailed methods provided in Supplemental Materials. Proteomics data are publicly available at MASSIVE (MSV000086692) [https://doi.org/doi:10.25345/C5DN5P;]. RNA sequencing data have been deposited in NCBI’s Gene Expression Omnibus14 and are accessible with GSE164794 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE164794).
Human cardiac tissue collection
Explanted failing hearts from transplant patients (n=16; 54.9±2.6y.o;) non-ischemic or dilated cardiomyopathy with/without implantable cardiac defibrillator and left ventricular assist device), and donor human hearts (non-HF, n=22; 48.7±3.1y.o;) without history of cardiac dysfunction and with intact SAN pacemaker complexes (Table I in the Data Supplement) were obtained from The OSU Cardiac Transplant Team or LifeLine of Ohio Organ Procurement Organization respectively, in accordance with the approved OSU IRB with informed consent.
SAN and RA Fibroblast Isolation
Explanted human hearts were arrested and dissected in ice-cold cardioplegic solution (Figure 1B). Fibroblasts were isolated from the intramural SAN pacemaker compartment (<10×5×1.5 mm3) and neighboring RA tissues by enzymatic digestion (Table II in the Data Supplement), cultured, passaged once and synchronized in DMEM with 0.5% BGS for 24 hrs. Subsequently, cells were treated with ±5 ng transforming growth factor-1 (TGFβ1) for 48 hrs after which the resulting pellet was used for RNA sequencing and proteomics.
Quantitative Proteomics and NGS
Mass Spectrometry data were acquired from 3μg peptides per sample. Database Search of mass spectra from all samples (Excel File I in the Data Supplement) were searched with the OpenMS platform and X!Tandem search engine against a reviewed UniProt human proteome (downloaded 09/04/2019). RNA for NGS was isolated from fibroblast cell pellets using the miRNeasy Kit (QIAGEN) according to the manufacturer’s instructions (Excel File II in the Data Supplement). All data analyses were performed with R and various packages (full analysis pipelines and quality control in Supplemental Material).
Gene Ontology and KEGG Network Analyses
KEGG analysis was performed with kegga and topKEGG functions in R for the top 20 terms. Data were filtered for significant enrichment only (p-value < 0.05).
Statistical Analyses
Results are presented as mean±SEM, in scatter plots with bar. Shapiro–Wilk method was used to test normality of the group. Paired or unpaired Student T-test (two tailed) and one/two way ANOVA (with Tukey’s test) were used to compare two or multiple groups where p < 0.05 was considered statistically significant.
RESULTS
Fibrotic content increases in the HF SAN
Fibrotic content in the SAN pacemaker compartments and adjacent RA tissue were determined by quantifying fibrotic areas identified by Masson’s Trichrome staining (non-HF=15, HF=13) (Figure 2A). Analyses showed that percentage of dense fibrotic content was significantly higher across the SAN compartments vs RA in both non-HF hearts (44.0±1.2 vs 24.7±1.7%, p<0.0001) and in HF hearts (57.6±2.6 vs 34.4±1.3%, p<0.0001). Importantly, fibrotic content in SAN and RA in HF hearts were significantly higher than non-HF.
Figure 2: Quantification of dense connective tissue and composition of extra cellular matrix (ECM) in human sinoatrial node (SAN).
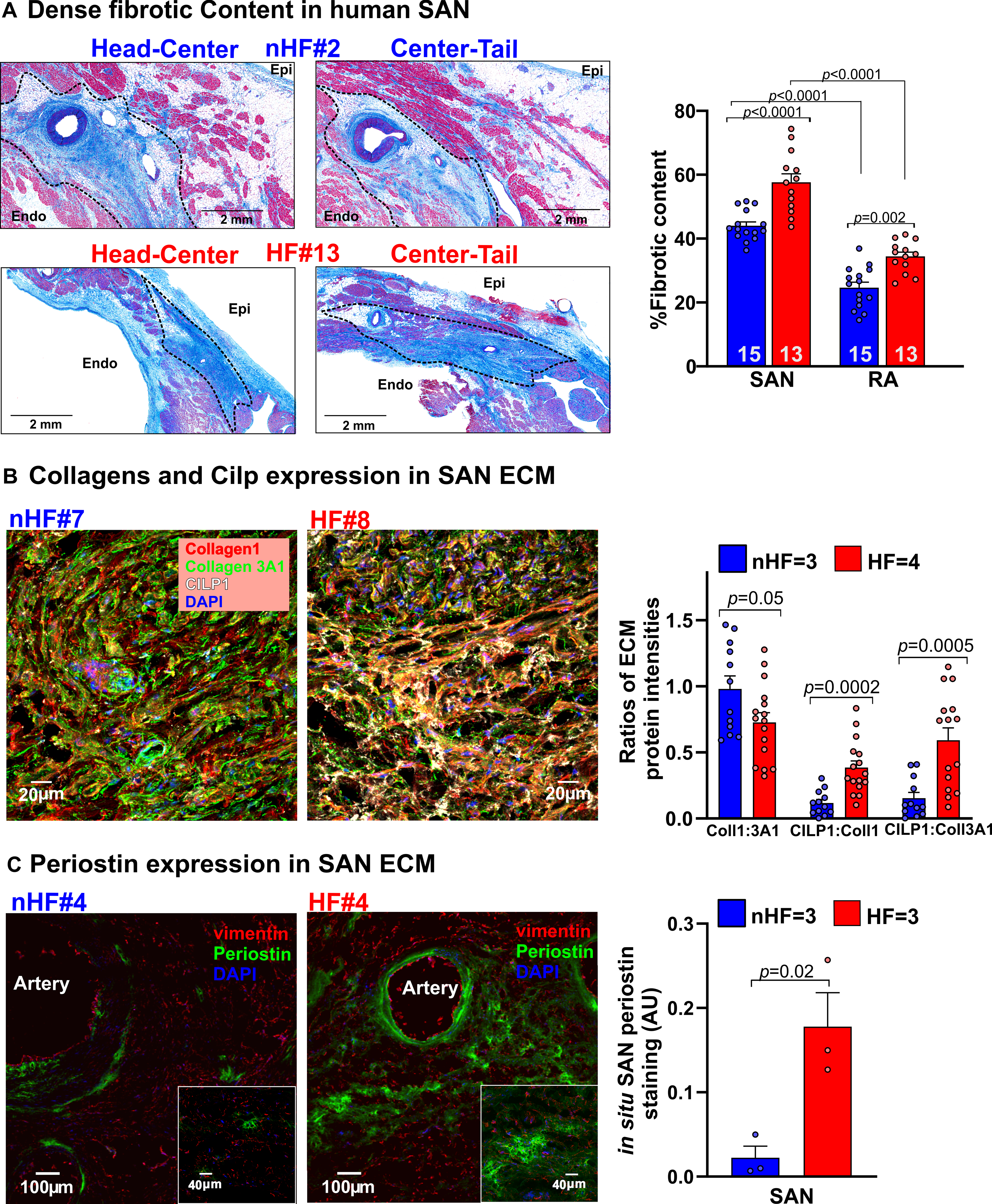
A, Left, representative images showing dense connective tissue and fibrosis (blue) identified in Masson’s Trichrome stained paraffin embedded human non-failing (nHF) and failing (HF) SAN sections respectively. Right, quantification of dense connective tissue from sections of SAN head, center and tail compartments. (Hearts: nHF (n=15) and HF (n=13); each dot is an average of at least 3 measurements per SAN and RA). B, Left, representative images of ECM proteins collagens 1 (red), 3A1 (green) and cartilage intermediate layer protein 1 (CILP1) (white) staining in nHF and HF SAN cryosections (also see Figure IA in the Data Supplement). Right, summary data of their ratios (Hearts: nHF (n=3) and HF (n=4); each dot is an average of measurements from 4 fields-of-view per SAN) C, Left, nHF and HF SAN cryosections stained with vimentin (red) and periostin (green). Cell nuclei were stained with Dapi (blue). Right, summary data of periostin staining density ((nHF (n=3) and HF (n=3); each dot represents measurement from each SAN). (All data analyzed with ANOVA (two or multiple group comparison as applicable) and expressed as mean±SEM, (p<0.05)). CT- crista terminalis; IAS- interatrial septum; Epi/Endo-epi/endocardial; SAN-sinoatrial node; RA-right atria.
Interstitial ECM and fibroblast remodeling in the HF SAN
The first goal of our study was to determine ECM protein composition and characteristics of non-HF vs HF SAN fibroblasts in situ. Sections of non-HF and HF SAN were stained for Collagen1 and Collagen 3A1, cartilage intermediate layer protein 1 (CILP1) and Periostin (POSTN), components of the ECM in pathological remodeling and scars (Figure I, Table III in the Data Supplement). Collagen 1 to 3A1 ratio was lower in SAN HF vs non-HF. CILP1 was higher (non-HF=1.0±0.2%; HF=3.7%±0.4% (p<0.0001)) in HF SAN; ratios of CILP1 to Collagen 1 and 3A1 were higher in HF compared to non-HF SAN (Figure 2B). Interstitial POSTN expression in the SAN pacemaker compartments was found only in HF SAN while it was negligible in non-HF (Figure 2C). Cryosections from non-HF and HF SAN were immunostained with α-actinin, vimentin and CD31 antibodies to identify cardiomyocytes, fibroblasts, and endothelial cells respectively (Figure 3A). Percentage of vimentin+/CD31- cells were quantified as fibroblasts, which was comparable between non-HF (66±6%) and HF (61±5%) (p=0.18) SAN sections. However, vimentin+/CD31-cells positive for the proliferation marker phosphorylated histone H3 (PHH3) were higher in HF compared to non-HF SAN (0.46± 0.05 vs 0.12±0.02) (p=0.001) (Figure 3B).
Figure 3: Composition of fibroblasts in nHF and HF SAN.
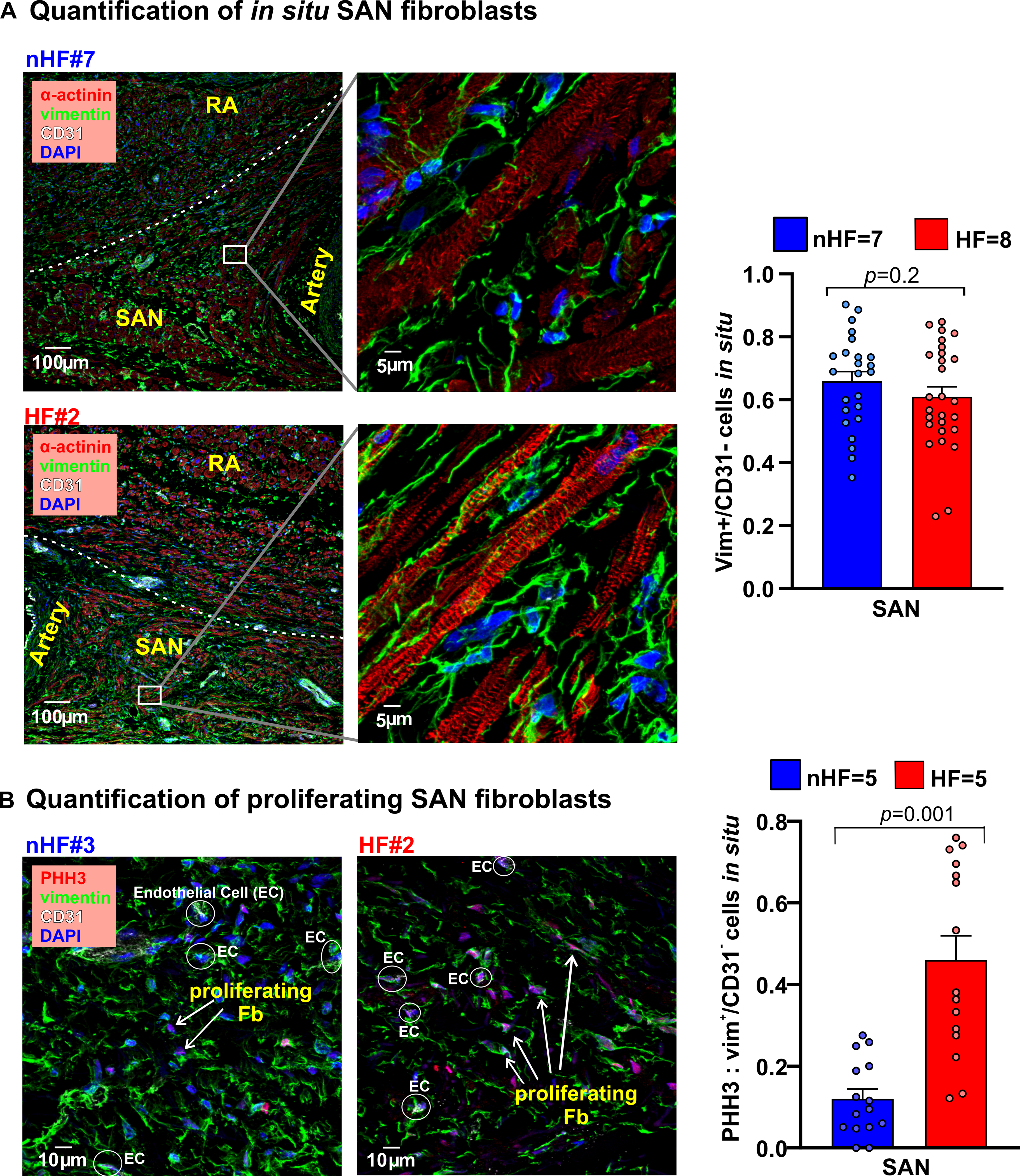
A, Left, Fibroblasts in the non-failing (nHF) and failing (HF) SAN stained in cryosections with vimentin (green), cardiomyocytes with α-actinin (red), and Dapi (blue); right, summary data of vimentin positive cells relative to all cells (Hearts: nHF (n=7) and HF (n=8); each dot is an average of measurements from 3–4 fields-of-view per SAN). B, Left, sections from nHF and HF SANs stained for vimentin (green), phosphorylated histone3 (PHH3; red), CD31(white) and blue (Dapi); right, summary of proliferating fibroblasts quantified as a ratio of PHH3 positive cells to vimentin+/CD31- cells (Hearts: nHF (n=5) and HF (n=5); each dot is an average of measurements from 3 fields-of-view per SAN). (All data analyzed with ANOVA and expressed as mean±SEM, (p<0.05)).SAN-sinoatrial node; RA-right atria.
In situ, myofibroblasts are present in the HF SAN but not in non-HF SAN
Next, we examined the composition of fibroblasts and myofibroblasts in situ in order to determine inherent qualitative differences between the SAN and RA, which could correlate with their fibrotic content. Cryosections were stained for vimentin, CD31 and α-smooth muscle actin (αSMA) (n=3; 3 fields-of-view/heart) to identify differentiated myofibroblasts in the SAN (Figure 4A, Figure II in the Data Supplement). Thirty-nine percent αSMA+/vimentin+/CD31- myofibroblasts were found in HF SANs relative to αSMA-/vimentin+/CD31- fibroblasts, but no definitive myofibroblasts were found in non-HF SAN sections.
Figure 4: Composition of in situ and in vitro nHF and HF SAN fibroblasts.
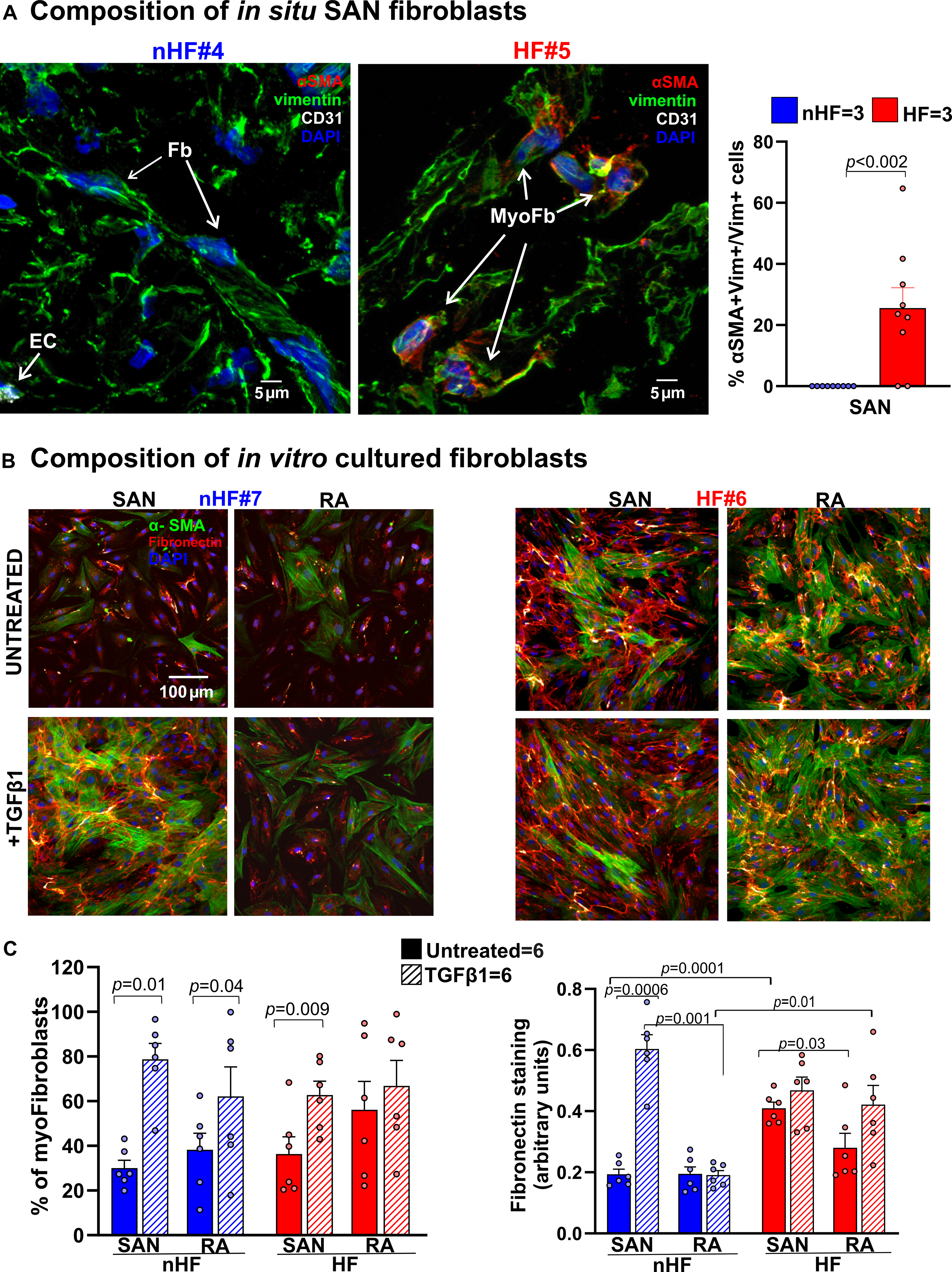
A, Representative images of non-failing (nHF) and failing (HF) human Sinoatrial node (SAN) cryo-sections stained for α smooth muscle actin (αSMA; red), vimentin (vim; green) and CD31 (white) identifying vimentin+/ αSMA- fibroblasts (Fb), vimentin+/αSMA+ myofibroblasts (myoFb) and CD31+/ αSMA+ endothelial cells (EC) (Hearts: nHF (n=3) and HF (n=3); each dot is an average of measurements from 3 fields-of-view per SAN). B, Representative images of isolated SAN and RA fibroblasts from nHF or HF hearts, cultured for 48hrs ± Transforming Growth Factor β1 (TGFβ1). Cells were stained with αSMA (green) and fibronectin (red). C, summary data of myofibroblast percentages in the cultures and quantification of fibronectin staining (Hearts: nHF (n=6) and HF (n=6); each dot is a measurement from SAN/RA±TGFβ1 cultures). Cell nuclei were stained with Dapi (blue). (Data analyzed with ANOVA (two or multiple group comparison as applicable) and expressed as mean±SEM, (p<0.05)).SAN-sinoatrial node; RA-right atria.
In vitro, HF fibroblasts show robust myofibroblast differentiation and fibronectin secretion
We studied the composition of non-HF and HF SAN and RA cultured fibroblasts ±TGFβ1. TGFβ1 was used to activate myofibroblast transdifferentiation in these populations and determine if it would be increased in HF derived cells, since we predicted that SAN and RA fibroblasts from HF hearts would already be activated in situ, compared to non-HF hearts. We also hypothesized that SAN fibroblasts from non-HF hearts would have an inherently higher propensity to transdifferentiate than corresponding RA cells. (Figure 4B). In line with this rationale, αSMA positive myofibroblasts were found in untreated non-HF SAN and HF SAN and RA fibroblast cultures. TGFβ1 treatment significantly increased the number of myofibroblasts in non-HF and HF SAN and non-HF RA (Figure 4C). Fibronectin staining was higher in untreated HF vs non-HF SAN fibroblast cultures; furthermore, it increased after TGFβ1 treatment only in non-HF SAN fibroblast cultures. Untreated HF SAN cultures also showed higher fibronectin staining compared to non-HF SAN cells.
Whole transcriptomic profiles in SAN and RA cultured fibroblasts
Our next goal was to evaluate and quantify the global expression of all fibroblast-specific transcripts in the SAN and RA cultured fibroblast groups. Enrichment of fibroblasts excluding other cell types (endothelial cells and macrophages) was confirmed by immunostaining for all samples. Furthermore, immunostaining with Cyclin A2, D1 and PHH3 antibodies confirmed that fibroblasts from both non-failing and failing cultures are not actively cycling or proliferating (Figure III in the Data Supplement). Whole transcriptome of SAN and RA samples was sequenced for 40 samples from 5 non-HF and 5 HF hearts; (samples from 3 hearts (2 non-HF and 1 HF hearts) were removed from the sequencing and proteomics analyses based on low protein spectral counts from complementary proteomics analyses (Excel File I in the Data Supplement). Genes expressed lower than the minimum count per million (cpm) of 0.59 (equivalent to 10 counts) in at least 3 samples in one group were filtered, and the resulting sequenced reads were normalized with trimmed mean of M-values (TMM). Across the 8 test groups, averages of total counts revealed that majority of the genes detected were protein coding mRNAs (14415 genes, 63%), then in decreasing prevalence long non-coding RNA (lncRNA; 5876, 26%), miscellaneous RNA (1254, 5%), miRNA (677, 3%) and other types of RNA (577, 3%) (Figure 5A) (Table IV in the Data Supplement).
Figure 5: Whole transcriptomic profiles of human SAN fibroblasts.
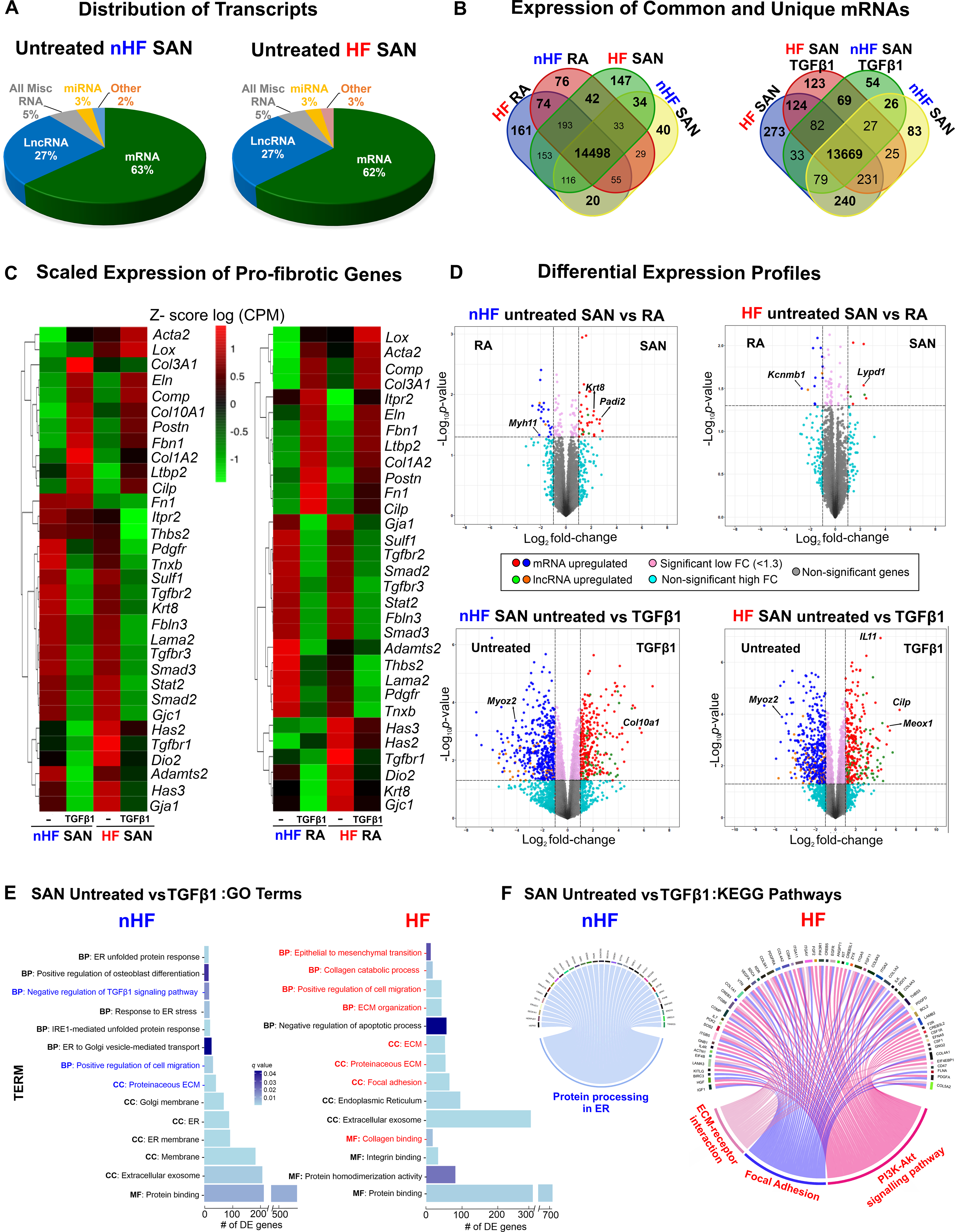
A, Distribution of major coding and non-coding transcript groups detected across untreated non-failing (nHF) and failing (HF) SAN fibroblasts. B, Venn diagrams showing unique and shared mRNAs between SAN shown comparison groups. C, Heatmaps showing scaled expression levels of selected pro-fibrotic genes in nHF and HF SAN vs RA ±TGFβ1 groups. D, Volcano plots of mRNAs and long non-coding RNAs (lncRNA) differentially expressed between groups shown at p <0.05 and log2fold change >1.3. E, Differentially expressed genes (q<0.05) enriched within three gene ontology analyses terms (MF-molecular function, BP-biological process, CC-cellular component) in nHF and HF untreated vs TGFβ1 fibroblasts (selected terms are presented) and F, Circos plots showing differentially expressed genes (p<0.05) associated with KEGG pathways in nHF and HF untreated vs TGFβ1 SAN fibroblasts (nHF=3; HF=4). Magnified images of GO and KEGG plots are presented in Figure IX in the Data Supplement (ER- endoplasmic reticulum; ECM-Extracellular matrix; SAN-Sinoatrial node; RA-right atria; TGFβ1-Transforming Growth Factor beta 1; mRNA- coding messenger RNA; Misc-miscellaneous; miRNA- microRNA).
Protein coding mRNA and lncRNA were further analyzed to determine their distinctive patterns in SAN vs RA fibroblasts; venn diagrams in Figure 5B represent analyses of mRNA counts in SAN and RA fibroblasts from non-HF and HF hearts and reveal different patterns of unique vs commonly expressed mRNAs between non-HF SAN vs RA and HF SAN vs RA. Similarly, unique and common mRNAs were detected after TGFβ1 treatment.
Fibroblast-specific SAN mRNA profiles
In addition to differences in mRNA profiles across the groups (Figures IV–VIII in the Data Supplement), we first examined the expression of several mRNAs associated with fibrosis and with the TGFβ pathway in SAN and RA fibroblasts (Figure 5C). Scaled expression levels (z-scores of CPM) of these selected pro-fibrotic genes across the datasets showed differing patterns between SAN and RA samples from non-HF hearts, indicating specific differences in these cultured fibroblasts and TGFβ1-induced myofibroblast samples (Figure 5C). Notably, mRNAs including Smad2/3, Tgfbr3, and other pro-fibrotic proteins, including Fibulin3 (Fbln3), Sulfatase1 (Sulf1), Keratin8 (Krt8), Tenascin X (Tnxb), Laminin2 (Lama2) and Cx45 (Gjc1), decreased in both SAN non-HF and HF myofibroblasts after TGFβ1 treatment. Similarly, mRNA counts of Collagens (Col3A1, Col1A2), Elastin (Eln), Latent TGF binding protein (Ltbp2), Postn, Cartilage Oligomeric Matrix Protein (Comp), Cilp1, Fibrillin 1 (Fbn1) were increased in TGFβ1 treated non-HF and HF SAN fibroblasts. These trends indicate similar transcriptional activation patterns in non-HF and HF SAN fibroblasts. In contrast, smooth muscle actin (Acta2) and Lysl oxidase (Lox) were increased only in TGFβ1 HF SAN fibroblasts, supporting increased myofibroblastic differentiation in the setting of HF. Similarly, Hyaluronan synthase 3 (Has3) and Cx43 (Gja1) transcripts were detected in untreated non-HF and HF SAN fibroblasts, but decreased in TGFβ1 treated fibroblasts.
Comparing non-HF SAN and RA gene expressions, 20 mRNAs were found upregulated (p-value <0.05 and log FC >1.3) in SAN fibroblasts, including Keratin8 (Krt8), and 14 different mRNAs were upregulated in RA fibroblasts including Myosin heavy chain 11 (Myh11) (Figure 5D, top left; Excel File III in the Data Supplement). SAN fibroblasts from HF hearts showed 13 mRNA upregulated in SAN fibroblasts including Ly6/Plaur domain-containing 1 (Lypd1) and Potassium Channel Tetramerization Domain Containing 8 (Kctd8), and 36 upregulated in the RA fibroblasts including the large-conductance potassium channel b subunit (Kcnmb1) (Figure 5D, top right; Excel File IV in the Data Supplement). Differential expression analyses comparing the activated response of untreated non-HF SAN fibroblasts to TGFβ1, showed 241 mRNAs to be different between the groups (p-value <0.05, logFC >1.3); (Figure 5D, bottom left; Excel File V in the Data Supplement). Between untreated HF SAN vs TGFβ1 treated SAN fibroblasts, 124 were upregulated in untreated including Myozenin 2 (Myoz2) (Figure 5D, bottom right, Excel File VI in the Data Supplement) and 56 in TGFβ1 treated HF SAN fibroblasts including Cilp1 and Mesenchyme homeobox 1 (Meox1).
Gene Ontology (GO) analyses of differentially expressed mRNAs in treated vs untreated non-HF SAN revealed 16 significantly enriched categories (q-value<0.05) including endoplasmic reticulum related functions, proteinaceous ECM, and positive regulation of cell migration. On the other hand, differentially expressed mRNA in treated vs untreated HF SAN were statistically enriched in 21 categories including epithelial to mesenchymal transition, focal adhesion, collagen, and ECM matrix organization (Figure 5E; Excel File VII in the Data Supplement). Analyses with the KEGG database showed differentially expressed mRNAs in TGFβ1 treated non-HF SAN fibroblasts in one signaling pathway related to protein processing in ER (Figure IX in the Data Supplement). Furthermore, ECM-receptor interaction, focal adhesion, and PI3K-Akt signaling pathways were enriched in HF groups compared to non-HF SAN (Figure 5F).
Fibroblast-specific SAN LncRNA profiles
Similar to mRNA profiles, several lncRNAs were also found to be differentially expressed between the SAN and RA groups (Figure 5D). We found several annotated but uncharacterized lncRNAs to be differentially expressed in these groups. Moreover, multiple common and unique lncRNAs were detected in all groups (Figure 6A). We also compared scaled expression of lncRNAs previously associated with fibrosis and/or cardiac diseases12, 15–17 (Figure 6B). Although we did not find significant differences, scaled expression of z-scores showed that Malat1 was higher in untreated non-HF SAN vs TGFβ1 while it increased only after TGFβ1 in RA relative to untreated; TGFβ1 increased Neat1 only in non-HF RA but not in the SAN.
Figure 6: Differential expression of long non-coding RNAs (lncRNA) and microRNAs (miRNA).
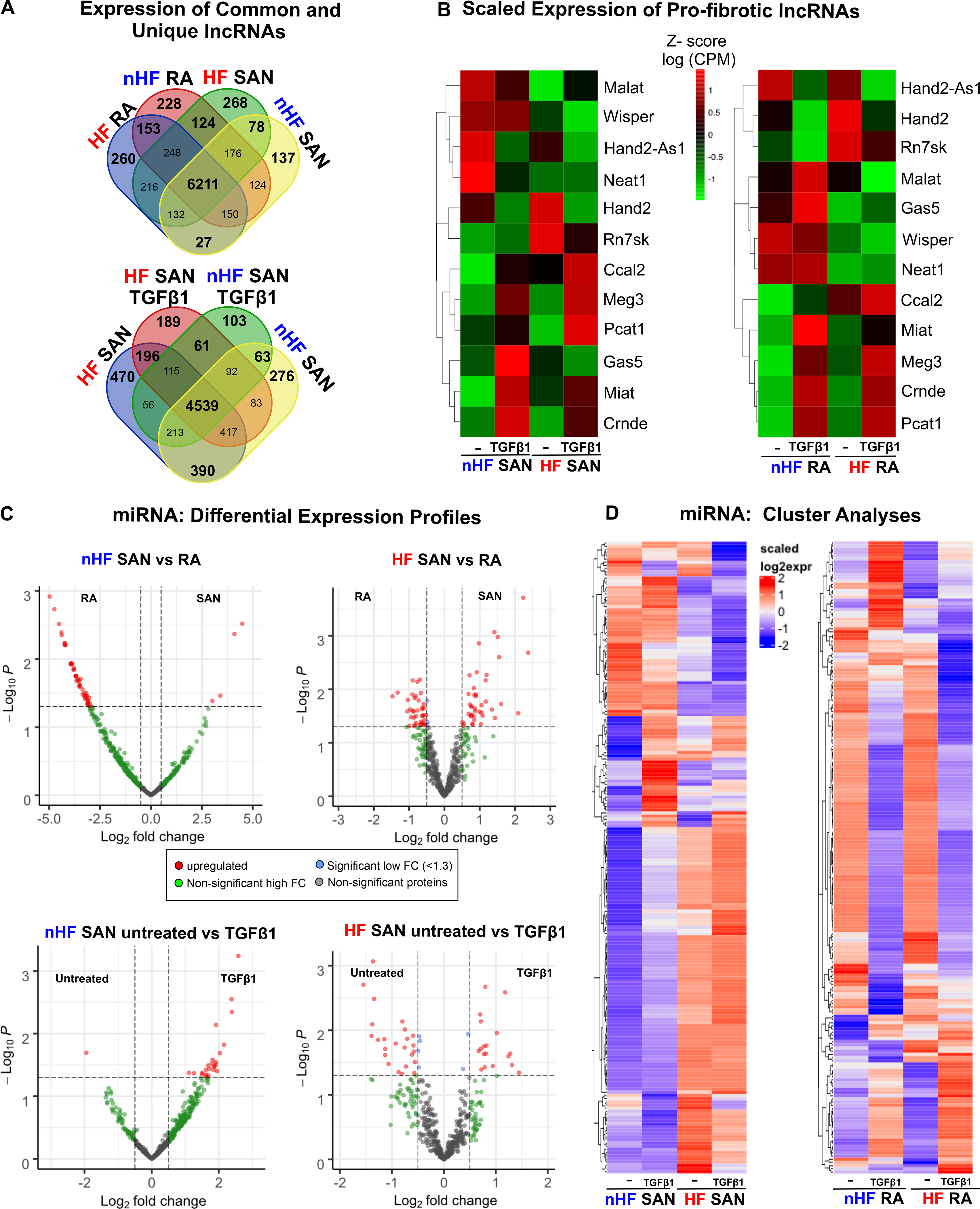
A, Distribution of lncRNAs across untreated non-failing (nHF) and failing (HF) untreated and +TGFβ1 SAN fibroblast comparison groups. B, Heatmaps showing scaled expression levels of selected pro-fibrotic lncRNAs in nHF and HF SAN vs RA ±TGFβ1 groups. C, Volcano plots of miRNAs differentially expressed between shown groups at p-value <0.05 and log2fold change >1.3. D, Heatmaps showing scaled expression levels of miRNAs differentially expressed (p<0.05) in nHF and HF SAN vs RA ±TGFβ1 groups (nHF=3; HF=4). (SAN-sinoatrial node; RA-right atria; TGFβ1-Transforming Growth Factor beta 1).
Fibroblast-specific SAN miRNA profiles
MiRNA quantitation identified over 12 million reads/sample. In non-HF and HF hearts, both common and unique miRNAs were detected in RA and SAN fibroblasts (Figures X, XI in the Data Supplement). Furthermore, differential expression analyses revealed that 4 miRNAs (miRNA-615–3p, miRNA-10b-3p, miRNA-10b-5p and miRNA-1292–5p) were upregulated in the non-HF SAN and 51 miRNAs upregulated in the non-HF RA (Figure 6C, top left). However, in HF hearts, 41 miRNAs were highly expressed in the SAN and 41 miRNAs were highly expressed in the RA (Figure 6C, top right). When comparing differential expression of miRNAs between SAN fibroblasts in non-HF vs HF hearts, a very different pattern was found, with 19 differentially expressed miRNAs: 13 miRNAs were upregulated in the HF SAN fibroblasts and 6 in non-HF. In contrast, 13 miRNAs were upregulated in RA HF fibroblasts while only 3 miRNAs were upregulated in non-HF (Figure XI in the Data Supplement).
MiRNA profiles showed very different patterns between non-HF and HF SAN fibroblasts in response to TGFβ1. While only miRNA-1306–5p, was upregulated in untreated non-HF SAN fibroblasts, 25 miRNAs were found upregulated in TGFβ1 treated non-HF SAN fibroblasts (Figure 6C, bottom left). On the other hand, in HF SAN, 31 miRNAs were upregulated after TGFβ1 compared to 20 miRNAs upregulated in untreated fibroblasts (Figure 6C, bottom right). Hierarchical cluster analyses (p<0.05; Figure 6D and Excel File VIII in the Data Supplement) also revealed that both HF and TGFβ1 affected expression patterns of miRNAs very differently in the SAN and RA fibroblasts.
Predicted interactions and networks of miRNAs and their target mRNAs
Ingenuity pathway analyses of interactions between significantly different miRNAs and mRNAs among non-HF SAN vs HF SAN untreated fibroblasts revealed 33 interactions, of which 9 miRNAs are inverse, suggesting putative miRNA mediated regulation of mRNA targets (Figure 7A; Table V in the Data Supplement). Among the differentially expressed miRNAs between non-HF SAN untreated vs TGFβ1, 47 showed interactions with multiple differentially expressed mRNAs, 22 interactions were inversely paired in expression. Specifically, 12 differentially expressed miRNAs were predicted to interact selected ECM genes, of which 10 were inversely paired (Figure 7B; Table VI in the Data Supplement). Among miRNAs differentially expressed between HF SAN untreated vs TGFβ1, 51 miRNAs were predicted to interact with selected ECM related mRNAs, of which 26 were inversely paired (Figure 7C; Table VII in the Data Supplement).
Figure 7: Predicted interactions and networks between microRNAs (miRNA) and pro-fibrotic mRNAs in fibroblasts.
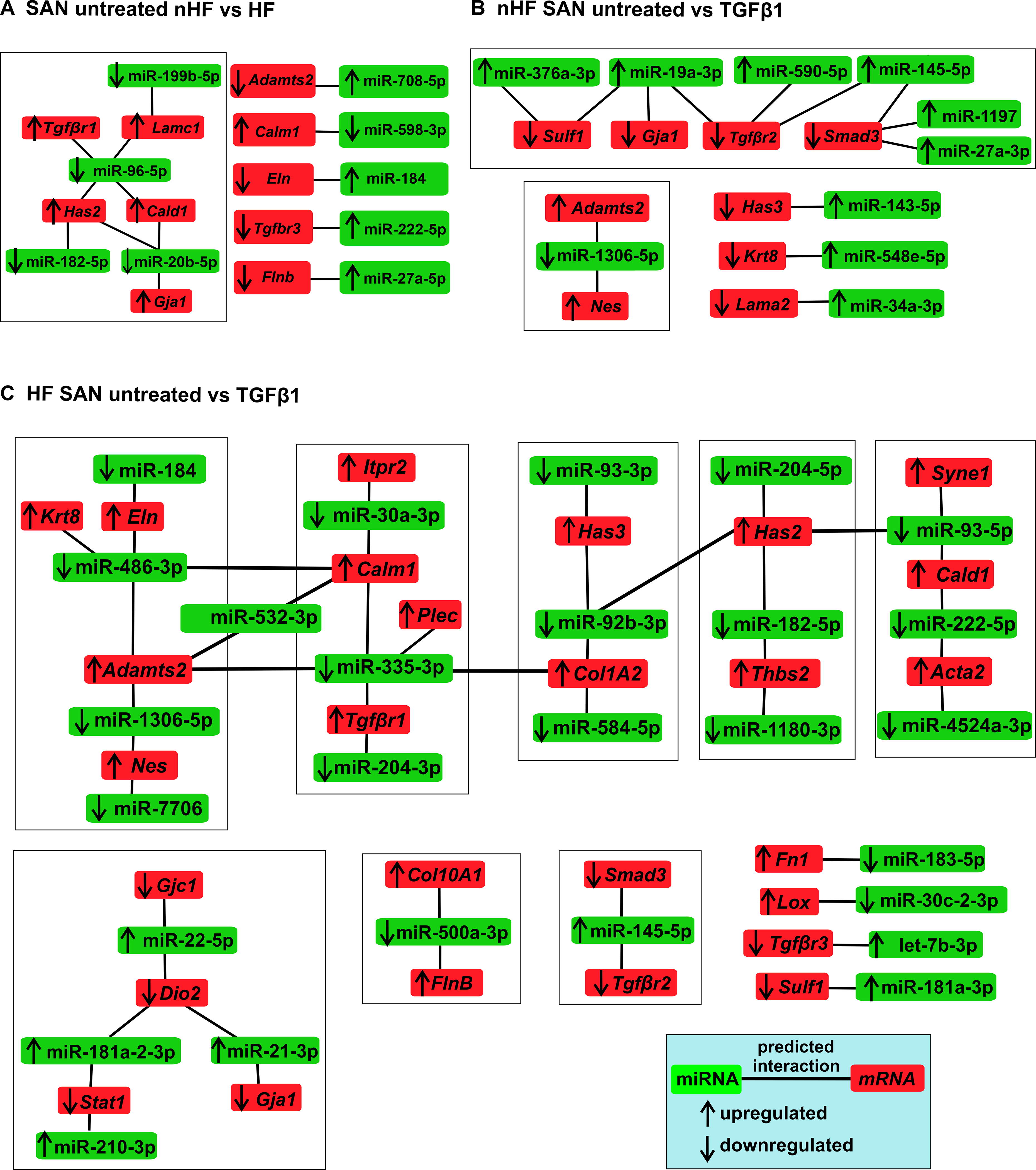
Ingenuity Pathway Analyses predicted interactions and networks between miRNAs (p<0.05) (green) and selected pro-fibrotic mRNAs (red) in A, SAN untreated nHF vs HF B, SAN nHF untreated vs TGFβ1 fibroblasts C, SAN HF untreated vs TGFβ1 fibroblasts (nHF=3; HF=4). Direction of arrows indicate upregulation/downregulation of the respective transcripts. Lines indicate predicted interactions between networks.
Proteomic profiles in SAN and RA fibroblasts
Next, we examined the proteomic profiles in the same cultured fibroblast populations. Data from LC-MS/MS studies (Table VIII in the Data Supplement, Figures XII–XV) were analyzed for unique and common proteins among the groups (Figure 8A). Comparing protein expression in SAN fibroblasts, 176 proteins were unique to untreated non-HF and 89 to untreated HF group, indicating a distinctive protein composition between untreated non-HF and HF SAN fibroblasts, with 51 proteins unique to both groups (Figure 8A, middle venn diagram). Similarly, following TGFβ1 treatment, non-HF and HF SAN groups showed 36 and 192 proteins respectively that were unique to each group, while 42 were shared.
Figure 8: Differential proteomic profiles of human SAN fibroblasts.
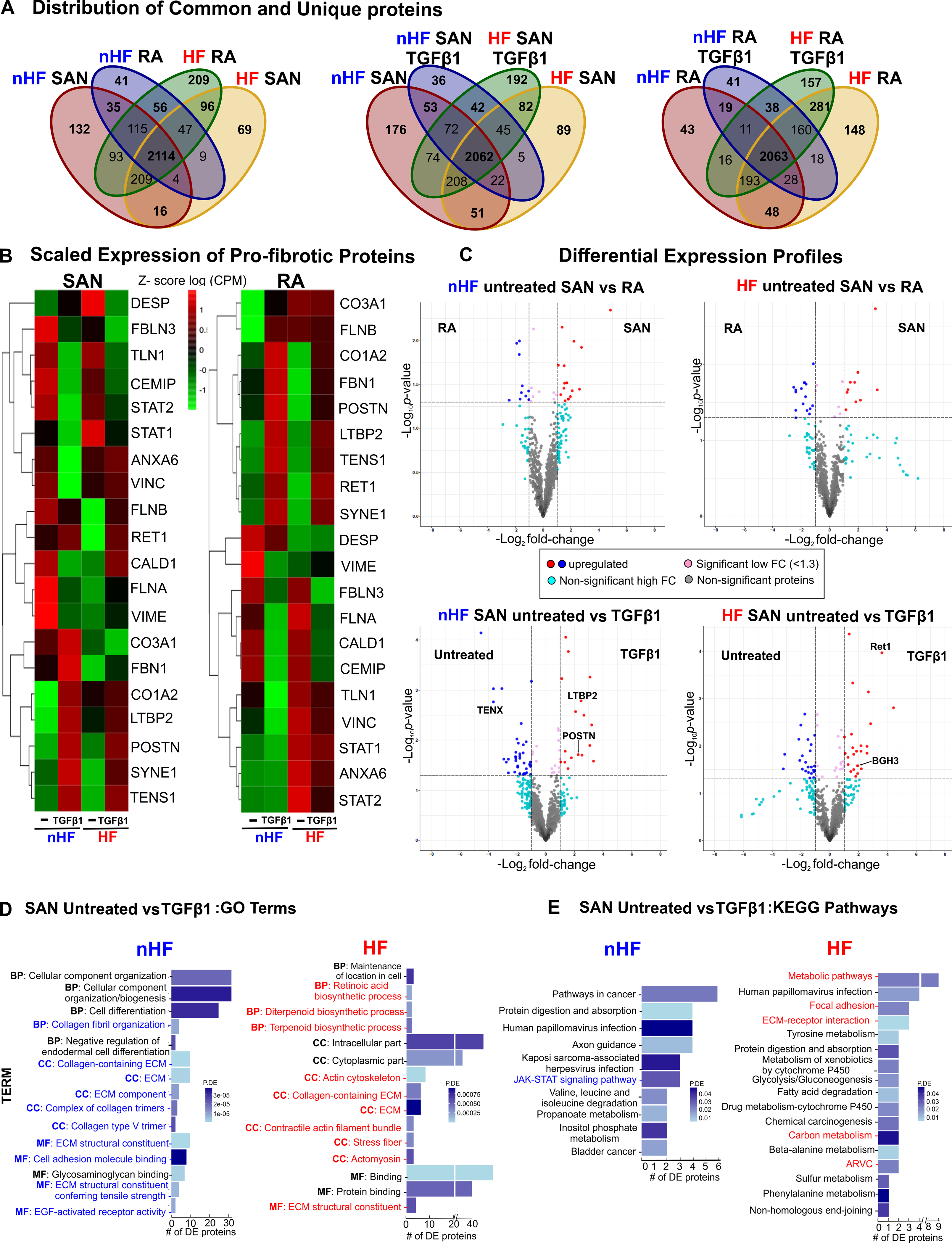
A, Venn diagrams showing unique and shared proteins between nHF, HF SAN vs RA, SAN nHF vs HF and RA nHF vs HF ±TGFβ1 fibroblast populations. B, Heatmaps showing scaled expression levels (counts per million (CPM)) of selected pro-fibrotic proteins in non-failing (nHF) and failing (HF) SAN vs RA ±TGFβ1 groups. C, Volcano plots of proteins differentially expressed at p-value <0.05 and log2fold change (FC) >1.3 are in red or blue. D, Differentially expressed proteins (p<0.05) enriched within three gene ontology analyses terms (MF-molecular function, BP-biological process, CC-cellular component) in nHF and HF untreated vs TGFβ1 fibroblasts (selected terms are presented) and E, KEGG pathways associated with differentially expressed proteins (p<0.05) in nHF and HF untreated vs TGFβ1 SAN fibroblasts (nHF=3; HF=4). Magnified images of GO and KEGG plots are presented in Figure XVI in the Data Supplement. (ARVC-Arrhythmogenic right ventricular cardiomyopathy; ECM- extracellular matrix; SAN-sinoatrial node; RA-right atria; TGFβ1-Transforming Growth Factor beta 1).
Comparing scaled expression patterns in the SAN vs RA non-HF untreated fibroblast of the selected proteins associated with fibrosis and ECM secretion, SAN fibroblasts demonstrated higher levels of Collagen3A1, FilaminA, Reticulin1, STAT2 and focal adhesion proteins Annexin6, Talin1 (TLN1) and Vinculin (Figure 8B). After TGFβ1 treatment, SAN and RA non-HF fibroblasts showed very similar changes in expression of several proteins while Collagen3A1 remained higher in SAN non-HF TGFβ1 fibroblasts. While, SAN vs RA HF untreated cells showed lower levels of Collagen3A1, Caldesmon1, Fibulin3 and Filamin A/B. TGFβ1 treatment similarly increased Annexin6, Collagen1A2, FilaminB, LTBP2, POSTN, Retinol binding protein (RET1), Nesprin (SYNE1), Tensin1 and Vinculin, in both SAN and RA HF fibroblasts. Notably, Collagen3A1, Caldesmon1, Fibrillin1, STAT1, STAT2 and Talin1 were lower in HF SAN after TGFβ1 compared to RA.
We next examined the differential expression of proteins in these groups. An average of 2867 proteins were detected across the 8 comparison groups (Table VIII in the Data Supplement) and differentially expressed proteins were determined significant (p-value<0.05 and logFC >1.3) (Figure 8C). Proteins including Laminin and smooth muscle myosin heavy chain 11 were upregulated in untreated non-HF RA fibroblasts while STAT2, VAV2, Inositol 1,4,5-Trisphosphate Receptor (ITPR2) were upregulated in untreated non-HF SAN fibroblasts. In contrast, a different set of proteins were upregulated in HF SAN and RA fibroblasts. TGFβ1 treatment increased POSTN, LTBP2, Collagen5, A1, 2 in non-HF SAN cells while SYNE1, RET1, Cadherin2 (CADH2), Integrin-α4 (ITA4) were increased in HF SAN cells.
Gene Ontology (GO) analyses of differentially expressed proteins revealed 20 significantly enriched categories in non-HF SAN untreated vs TGFβ1 related to ECM, collagen fibril organization, collagen trimers and epidermal growth factor receptor activation while 20 categories including cytoplasmic functions, actin cytoskeleton, stress fiber, ECM constituents, and biosynthetic pathways were enriched in HF (Figure 8D, Figure XVI in the Data Supplement). Furthermore, analysis with KEGG database identified 10 highly enriched pathways between untreated and TGFβ1 non-HF SAN fibroblasts including JAK-STAT signaling, and 17 signaling pathways including ECM-receptor interaction, focal adhesion, arrhythmogenic right ventricular cardiomyopathy and metabolic pathways among differentially expressed proteins from untreated vs TGFβ1 HF SAN fibroblasts (Figure 8E and Excel File IX in the Data Supplement). Overall, proteomic signatures in SAN and RA fibroblasts were uniquely affected by HF or TGFβ1 treatment.
DISCUSSION
In recent years, extensive research has demonstrated cardiac fibroblasts to be equally important as cardiomyocytes in their impact on structure and function of the healthy and diseased heart18, 19. The human SAN is designed to serve as the leading pacemaker, which persistently generates and conducts electrical activity to the atria. For these functions, the specialized pacemaker cardiomyocytes require the dense fibrotic fibers that form a structural lattice and define conduction pathways into the atria1. Pathological remodeling of ECM composition and fibrosis preferentially increases in the SAN during HF (Figure 2A), which can contribute to automaticity and conduction abnormalities found in diseased hearts9–11. Here, we found that while CILP120 increases, Collagen 3A1 expression decreases, leading to higher ratios of CILP1 to Collagen1 and Collagen3A1 in HF vs non-HF SAN (Figure 2B). This remodeling of ECM protein composition may promote less flexible and stiffer SAN ECM. Furthermore, POSTN, associated with pathological fibrosis and myocardial scars, cell-to-matrix signaling, and epithelial-mesenchymal transition21, 22, was negligible in the human non-HF SAN but detected in all HF SANs studied, and appeared as concentrated islands within the ECM (Figure 2C). Such fibrotic islands/strands can create electrical barriers interrupting mutual entrainment between pacemaker cardiomyocyte clusters and conduction pathways between SAN and RA leading to tachy-bradycardias, chronotropic incompetence and SAN conduction blocks often associated with HF8, 11.
Very little is known about SAN fibroblasts mainly due to the primary focus on SAN pacemaker cardiomyocytes as drivers of SAN function, with no emphasis on the fibrotic framework’s role in efficient automaticity and conduction23. A single report previously showed that SAN fibroblasts form a network in a rabbit model24. We found similar numbers of fibroblasts in non-HF and HF human SAN studied, indicating that disease may not affect overall SAN fibroblast numbers (Figure 3A). However the number of proliferative PHH3+/vimentin+/CD31- fibroblasts were higher in HF SAN (Figure 3B) and in contrast to previous findings that reported higher myofibroblasts in human HF ventricle25, vimentin+/αSMA+/CD31- myofibroblasts were present only in the HF SAN (Figure 4A).
In contrast to in situ findings, in vitro cultured SAN and RA fibroblasts revealed robust myofibroblastic differentiation and fibronectin secretion when treated with TGFβ1 (Figures 4B, C). Our quantitative analyses of proteo-transcriptomes of the same samples of cultured fibroblast populations showed similarities in the overall expression patterns of selected pro-fibrotic mRNAs after TGFβ1 treatment between non-HF and HF SAN fibroblasts, indicating common signaling pathways and mechanisms between them (Figure 5). It is possible that these common pro-fibrotic mRNAs are not additionally affected by HF. Exceptions were Lox, associated with collagen crosslinking and αSMA that were upregulated in HF SAN TGFβ1-induced myofibroblasts, which could promote denser and stiffer ECM properties and an enhanced contractile phenotype, respectively. We also found higher scaled expression of Comp and Cilp1, markers of matrifibrocytes associated with mature scars26, in TGFβ1 treated non-HF and HF SAN fibroblasts implicating a role for similar specialized myofibroblasts, in addition to the classical αSMA+ myofibroblasts, in the accumulation of dense fibrotic SAN tissue.
We found a potential role for inflammation in modulating SAN fibroblasts implicated with upregulation of pro-fibrotic cytokine IL11 mRNA after TGFβ1 treatment in both non-HF and HF SAN and RA fibroblasts (Figure 5D). In human RA fibroblasts, cytokine IL11 was recently shown to play an important role in cardiovascular fibrosis27, suggesting similar pro-fibro-inflammatory roles in the HF SAN. Myoz2 encoding Calsarcin1, previously identified in slow twitch skeletal muscle and enriched in cardiomyocytes28, 29, increased in untreated non-HF and HF SAN and RA fibroblasts suggesting that it is also expressed in fibroblasts and not restricted only to myocytes. Since Myoz2 mutations have been shown to cause human cardiomyopathies, it would be important to determine if these mutations affect non-myocyte cells as well30. Similarly transcription factor Meox1, recently shown to play a central regulatory role in stress-induced myofibroblast activation31 was higher in non-HF and HF SAN TGFβ1 fibroblasts, relative to untreated fibroblasts.
Furthermore, GO and KEGG analyses in SAN HF groups also identified enrichment of genes in pathways of ECM-receptor interaction, focal adhesion, and PI3K-Akt signaling pathway, which is associated with switching TGFβ1 signaling from apoptosis to epithelial-mesenchymal transition with increasing ECM stiffness32 (Figures 5E, F). These gene expression patterns and enriched pathways suggest mature myofibroblasts and increased myofibroblastic differentiation in HF, which could lead to higher levels of fibrosis in HF SAN.
While studies have identified several lncRNAs in the heart and fibroblasts from cardiac tissues, very few data exist to document their relevance in the human heart16, 33,34–36. Notably, this study is the first to document lncRNAs specific to human SAN fibroblasts. Our study identified several unique and common lncRNAs in all groups of fibroblasts studied (Figures 5D, 6A). LncRNAs Malat1 17 and Gas5 37, previously shown to activate fibroblasts, could play a role in modulating non-HF SAN fibrotic content relative to RA. Neat138 may be more specific to TGFβ1 activation in RA fibroblasts but not to SAN cells (Figure 6B). Future studies should systematically investigate if any uncharacterized lncRNAs in our dataset contribute to SAN fibrotic content and serve as novel therapeutic targets to treat cardiac remodeling in HF.
Among miRNAs, miRNA-615–3p, miRNA-10b-3p, miRNA-10b-5p, and miRNA-1292–5p, previously associated with cancer and tumor progression39–41, were specifically higher in the non-HF SAN compared to RA fibroblasts (Figure 6C). MiRNA-1306–5p is known to be released by the myocardium, and its circulating levels are considered a biomarker of HF42. Notably we found that miRNA-1306–5p is expressed in human SAN fibroblasts as well, with higher levels in untreated non-HF SAN fibroblasts. Network analyses also showed that many of the significantly different SAN miRNAs paired inversely with selected ECM and pro-fibrotic genes, which predicted potential miRNA-mediated regulation of these genes and SAN fibrosis (Figure 7). Overall, these putative interactions could be utilized to identify candidate miRNAs to determine regulation of predicted fibrosis-associated mRNA targets in the SAN.
Although few studies have sequenced global proteomic profiles in the human heart43, 44, proteomic profiling of human SAN fibroblasts has never been performed. We identified an average of ~15,000 peptide spectral matches across all SAN and RA samples, demonstrating both similar and distinct profiles (Figure 8A). Specifically, expression patterns of many pro-fibrotic and ECM proteins including LTBP2 and POSTN, indicate that common ECM and myofibroblast associated signaling pathways are similarly regulated in both untreated and TGFβ1 treated non-HF and HF SAN and RA (Figure 8B). However, untreated non-HF SAN fibroblasts demonstrated proteomic signatures distinct from that of the non-HF RA, with increased expression in AnnexinA6, Collagen 3A1, Filamin B, STAT2, Talin1 and Vinculin. In contrast, Collagen3A1 was lower in untreated HF SAN cells compared to RA. Higher Collagen3A1 could suggest a more flexible SAN ECM composition in non-HF vs HF. Overall, these results along with those in Figures 2B, C indicate that non-HF SAN fibroblasts may secrete proteins that promote flexibility in the ECM, which may decrease in HF leading to a stiffer, less flexible composition in the HF SAN. Substrate stiffness is known to modulate TGFβ1 signaling by promoting myofibroblast transition45, thereby creating a positive feedback loop in fibrosis progression. Hence a shift towards stiffness in HF SAN ECM could promote myofibroblast differentiation and fibrosis within the SAN, similar to what we found in situ (Figure 4A). Moreover, findings from a porcine SAN ECM study identified both stiffer and elastic components, which could withstand higher tensile forces while reducing mechanical strain on pacemaker myocytes23. These findings suggest that a balance of both stiff and elastic ECM components may be necessary for optimal SAN function, which could be altered in the HF SAN. Furthermore, while increases in mechanical stretch or intra-atrial pressure can increase SAN rhythm, compression or increased arterial blood pressure in SAN artery can slow rhythm46–48. A stiffer composition of the ECM in the HF SAN can impede this physiological response to changes in atrial and/or arterial pressures. Further studies to dissect the ECM protein composition in detail in non-HF and HF human SAN should allow us to confirm these fibroblast-specific findings in determining the role of in situ ECM composition in SAN automaticity and conduction abnormalities.
TGFβ1 induced a remarkably similar pattern of protein expression among most of the selected pro-fibrotic/ECM genes in untreated non-HF SAN and RA fibroblasts, with the specific exception of Collagen3A1, which was upregulated in the non-HF SAN but not RA TGFβ1 cells (Figure 8B, 8C). These data suggest that activated non-HF SAN myofibroblasts may be different from their RA counterparts in secretion of Collagen3A1 associated with less rigid ECM properties. Upregulation of POSTN and LTBP2 in non-HF SAN fibroblasts and Cadherin2 and Integrin-α4 in HF SAN fibroblasts post-TGFβ1 may imply inherently different gene programs in untreated non-HF and HF cells that could in turn promote differentiation into distinct myofibroblasts. These protein signatures are also supported by GO and KEGG analyses of SAN untreated vs TGFβ1 datasets (Figures 8D, E). Overall, our data suggest that relative to untreated fibroblasts, activated non-HF SAN myofibroblasts are prone to increased secretion of pro-fibrotic proteins related to ECM and fibrosis, while HF SAN myofibroblasts suggest pathologically phenotypes consistent with cardiomyopathy.
Limitations
Our study was performed on a relatively small sample of intact adult, primarily male human SANs and HF hearts, which may not be representative of all human hearts. Non-cardiac comorbidities and lifestyle modifications, including alcohol consumption and drug abuse as well as presence of implanted cardiac devices, and SAN dysfunctions in HF group may affect the results. Based on the availability of ex-vivo hearts for SAN tissue and cells collections49 at the OSU, we used donor hearts with complete SAN pacemaker complex without cardiac dysfunctions for non-failing group, and failing hearts from transplant surgeries for the HF group. Importantly, only those samples that met quality control and all inclusion criteria were retained for analyses (Figure IV in the Data Supplement).
Conclusions
In human HF SAN, proliferating fibroblasts, myofibroblasts, ECM with stiffer properties and POSTN-positive fibrotic islands increase, which create intranodal structural barriers that could interrupt normal automaticity and conduction. Furthermore, in non-HF SAN fibroblasts, molecular and protein markers are different from RA fibroblasts, which predisposes to SAN-specific impairments in ECM flexibility and myofibroblast differentiation. This comprehensive atlas of molecular and protein targets establishes a valuable human SAN fibroblast-specific proteo-whole transcriptome database that can be utilized to identify molecular and protein signatures, signaling pathways, and potential therapeutic targets of SAN fibrosis and cardiac arrhythmias in HF patients.
Supplementary Material
Clinical Perspective.
What is new?
The study provides a quantitative atlas of unique transcriptomic and proteomic signatures of non-failing and failing human sinoatrial node (SAN) fibroblasts.
In the failing human SAN, myofibroblasts, cartilage intermediate layer protein 1 and periostin are found within increased interstitial fibrosis.
Our data provide a comprehensive fibroblast-specific molecular framework to determine the mechanisms and role of SAN-specific extracellular matrix composition in SAN function and dysfunctions.
What are the clinical implications?
These comprehensive human fibroblast-specific datasets can be utilized to define the contribution of intranodal fibrotic content to SAN normal pacemaker function and for designing biological pacemakers.
Isolated fibroblasts identify a novel cellular platform to study SAN and atrial fibrosis and arrhythmias in human heart failure.
Proteo-transcriptomic datasets can be used to identify novel human SAN fibroblast-specific molecular and protein targets to develop anti-fibrotic strategies targeting SAN dysfunctions and atrial arrhythmias.
Acknowledgements:
We thank the Lifeline of Ohio Organ Procurement Organization and the Division of Cardiac Surgery, OSU Wexner Medical Center for providing explanted hearts; the Campus Microscopy and Imaging Facility, OSU, (P30 CA016058, National Cancer Institute); Campus Chemical Instrument Center Mass Spectrometry and Proteomics shared resource, OSU, (NIH P30 CA016058), (NIH S10 OD018056); Dr. Patrick Stevens, Biomedical Informatics Shared Resource, OSU, Ms. Lisa Dorn (Dr. Accornero Lab), Dr. Nehal Patel (Dr. Hund Lab), and members of Janssen and Fedorov Labs for assistance during the project.
Sources of Funding:
Grants from National Institutes of Health (HL115580, HL135109, HL142179, HL134824, HL135754), Leducq Foundation (TNE FANTASY 19CVD03) and the Bob and Corrine Frick Center for Heart Failure and Arrhythmia, OSU.
Non-standard Abbreviations and Acronyms
- HF
Heart failure
- NGS
Next Generation Sequencing
- Non-HF
Non-Heart failure
- RA
Right atria
- SAN
Sinoatrial node
- SND
Sinoatrial node dysfunction
Footnotes
Disclosures: None
Supplemental Materials
References
- 1.Csepe TA, Zhao J, Hansen BJ, Li N, Sul LV, Lim P, Wang Y, Simonetti OP, Kilic A, Mohler PJ et al. Human sinoatrial node structure: 3D microanatomy of sinoatrial conduction pathways. Prog Biophys Mol Biol 2016;120:164–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Opthof T, de Jonge B, Jongsma HJ and Bouman LN. Functional morphology of the mammalian sinuatrial node. Eur Heart J 1987;8:1249–59. [DOI] [PubMed] [Google Scholar]
- 3.Keith A and Flack M. The Form and Nature of the Muscular Connections between the Primary Divisions of the Vertebrate Heart. J Anat Physiol 1907;41:172–89. [PMC free article] [PubMed] [Google Scholar]
- 4.Csepe TA, Kalyanasundaram A, Hansen BJ, Zhao J and Fedorov VV. Fibrosis: a structural modulator of sinoatrial node physiology and dysfunction. Front Physiol 2015;6:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Csepe TA, Zhao J, Sul LV, Wang Y, Hansen BJ, Li N, Ignozzi AJ, Bratasz A, Powell KA, Kilic A, et al. Novel application of 3D contrast-enhanced CMR to define fibrotic structure of the human sinoatrial node in vivo. Eur Heart J Cardiovasc Imaging. 2017;18:862–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shiraishi I, Takamatsu T, Minamikawa T, Onouchi Z and Fujita S. Quantitative histological analysis of the human sinoatrial node during growth and aging. Circulation. 1992;85:2176–84. [DOI] [PubMed] [Google Scholar]
- 7.Kalyanasundaram A, Li N, Hansen BJ, Zhao J and Fedorov VV. Canine and human sinoatrial node: differences and similarities in the structure, function, molecular profiles, and arrhythmia. J Vet Cardiol 2019;22:2–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lou Q, Hansen BJ, Fedorenko O, Csepe TA, Kalyanasundaram A, Li N, Hage LT, Glukhov AV, Billman GE, Weiss R et al. Upregulation of adenosine A1 receptors facilitates sinoatrial node dysfunction in chronic canine heart failure by exacerbating nodal conduction abnormalities revealed by novel dual-sided intramural optical mapping. Circulation. 2014;130:315–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanders P, Kistler PM, Morton JB, Spence SJ and Kalman JM. Remodeling of sinus node function in patients with congestive heart failure: reduction in sinus node reserve. Circulation. 2004;110:897–903. [DOI] [PubMed] [Google Scholar]
- 10.Li N, Hansen BJ, Csepe TA, Zhao J, Ignozzi AJ, Sul LV, Zakharkin SO, Kalyanasundaram A, Davis JP, Biesiadecki BJ et al. Redundant and diverse intranodal pacemakers and conduction pathways protect the human sinoatrial node from failure. Sci Transl Med 2017;9(400):eaam5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li N, Kalyanasundaram A, Hansen BJ, Artiga EJ, Sharma R, Abudulwahed SH, Helfrich KM, Rozenberg G, Wu PJ, Zakharkin S et al. Impaired neuronal sodium channels cause intranodal conduction failure and reentrant arrhythmias in human sinoatrial node. Nat Commun 2020;11:512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Humeres C and Frangogiannis NG. Fibroblasts in the Infarcted, Remodeling, and Failing Heart. JACC Basic Transl Sci 2019;4:449–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frangogiannis NG. Cardiac fibrosis: Cell biological mechanisms, molecular pathways and therapeutic opportunities. Mol Aspects Med 2019;65:70–99. [DOI] [PubMed] [Google Scholar]
- 14.Edgar R, Domrachev M and Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobuss L, Bar C and Thum T. Long Non-coding RNAs: At the Heart of Cardiac Dysfunction? Front Physiol 2019;10:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Micheletti R, Plaisance I, Abraham BJ, Sarre A, Ting CC, Alexanian M, Maric D, Maison D, Nemir M, Young RA et al. The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Sci Transl Med 2017; 9(395):eaai9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peters T, Hermans-Beijnsberger S, Beqqali A, Bitsch N, Nakagawa S, Prasanth KV, de Windt LJ, van Oort RJ, Heymans S and Schroen B. Long Non-Coding RNA Malat-1 Is Dispensable during Pressure Overload-Induced Cardiac Remodeling and Failure in Mice. PLoS One. 2016;11:e0150236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tallquist MD and Molkentin JD. Redefining the identity of cardiac fibroblasts. Nat Rev Cardiol 2017;14:484–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Travers JG, Kamal FA, Robbins J, Yutzey KE and Blaxall BC. Cardiac Fibrosis: The Fibroblast Awakens. Circ Res 2016;118:1021–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Nieuwenhoven FA, Munts C, Op’t Veld RC, Gonzalez A, Diez J, Heymans S, Schroen B and van Bilsen M. Cartilage intermediate layer protein 1 (CILP1): A novel mediator of cardiac extracellular matrix remodelling. Sci Rep 2017;7:16042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao S, Wu H, Xia W, Chen X, Zhu S, Zhang S, Shao Y, Ma W, Yang D and Zhang J. Periostin expression is upregulated and associated with myocardial fibrosis in human failing hearts. J Cardiol 2014;63:373–8. [DOI] [PubMed] [Google Scholar]
- 22.Oka T, Xu J, Kaiser RA, Melendez J, Hambleton M, Sargent MA, Lorts A, Brunskill EW, Dorn GW 2nd, Conway SJ et al. Genetic manipulation of periostin expression reveals a role in cardiac hypertrophy and ventricular remodeling. Circ Res 2007;101:313–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gluck JM, Herren AW, Yechikov S, Kao HKJ, Khan A, Phinney BS, Chiamvimonvat N, Chan JW and Lieu DK. Biochemical and biomechanical properties of the pacemaking sinoatrial node extracellular matrix are distinct from contractile left ventricular matrix. PLoS One. 2017;12:e0185125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camelliti P, Green CR, LeGrice I and Kohl P. Fibroblast network in rabbit sinoatrial node: structural and functional identification of homogeneous and heterogeneous cell coupling. Circ Res 2004;94:828–35. [DOI] [PubMed] [Google Scholar]
- 25.Nagaraju CK, Robinson EL, Abdesselem M, Trenson S, Dries E, Gilbert G, Janssens S, Van Cleemput J, Rega F, Meyns B et al. Myofibroblast Phenotype and Reversibility of Fibrosis in Patients With End-Stage Heart Failure. J Am Coll Cardiol 2019;73:2267–2282. [DOI] [PubMed] [Google Scholar]
- 26.Fu X, Khalil H, Kanisicak O, Boyer JG, Vagnozzi RJ, Maliken BD, Sargent MA, Prasad V, Valiente-Alandi I, Blaxall BC et al. Specialized fibroblast differentiated states underlie scar formation in the infarcted mouse heart. J Clin Invest 2018;128:2127–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schafer S, Viswanathan S, Widjaja AA, Lim WW, Moreno-Moral A, DeLaughter DM, Ng B, Patone G, Chow K, Khin E et al. IL-11 is a crucial determinant of cardiovascular fibrosis. Nature. 2017;552:110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frey N, Richardson JA and Olson EN. Calsarcins, a novel family of sarcomeric calcineurin-binding proteins. Proc Natl Acad Sci U S A. 2000;97:14632–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gladka MM, Molenaar B, de Ruiter H, van der Elst S, Tsui H, Versteeg D, Lacraz GPA, Huibers MMH, van Oudenaarden A and van Rooij E. Single-Cell Sequencing of the Healthy and Diseased Heart Reveals Cytoskeleton-Associated Protein 4 as a New Modulator of Fibroblasts Activation. Circulation. 2018;138:166–180. [DOI] [PubMed] [Google Scholar]
- 30.Osio A, Tan L, Chen SN, Lombardi R, Nagueh SF, Shete S, Roberts R, Willerson JT and Marian AJ. Myozenin 2 is a novel gene for human hypertrophic cardiomyopathy. Circ Res 2007;100:766–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexanian M, Przytycki PF, Micheletti R, Padmanabhan A, Ye L, Travers JG, Teran BG, Duan Q, Ranade SS, Felix F et al. A Transcriptional Switch Governing Fibroblast Plasticity Underlies Reversibility of Chronic Heart Disease. bioRxiv Preprint posted online July 22, 2020; 10.1101/2020.07.21.214874. [DOI] [Google Scholar]
- 32.Leight JL, Wozniak MA, Chen S, Lynch ML and Chen CS. Matrix rigidity regulates a switch between TGF-beta1-induced apoptosis and epithelial-mesenchymal transition. Mol Biol Cell. 2012;23:781–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Y, Feng Z, Jian Z and Xiao Y. Long noncoding RNA TUG1 promotes cardiac fibroblast transformation to myofibroblasts via miR29c in chronic hypoxia. Mol Med Rep 2018;18:3451–3460. [DOI] [PubMed] [Google Scholar]
- 34.Piccoli MT, Bar C and Thum T. Non-coding RNAs as modulators of the cardiac fibroblast phenotype. J Mol Cell Cardiol 2016;92:75–81. [DOI] [PubMed] [Google Scholar]
- 35.Thum T. Noncoding RNAs and myocardial fibrosis. Nat Rev Cardiol 2014;11:655–63. [DOI] [PubMed] [Google Scholar]
- 36.McKinsey TA, Vondriska TM and Wang Y. Epigenomic regulation of heart failure: integrating histone marks, long noncoding RNAs, and chromatin architecture. F1000Res 2018;7:F1000 Faculty Rev–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tao H, Zhang JG, Qin RH, Dai C, Shi P, Yang JJ, Deng ZY and Shi KH. LncRNA GAS5 controls cardiac fibroblast activation and fibrosis by targeting miR-21 via PTEN/MMP-2 signaling pathway. Toxicology. 2017;386:11–18. [DOI] [PubMed] [Google Scholar]
- 38.Kenneweg F, Bang C, Xiao K, Boulanger CM, Loyer X, Mazlan S, Schroen B, Hermans-Beijnsberger S, Foinquinos A, Hirt MN et al. Long Noncoding RNA-Enriched Vesicles Secreted by Hypoxic Cardiomyocytes Drive Cardiac Fibrosis. Mol Ther Nucleic Acids. 2019;18:363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan T, Ooi WF, Qamra A, Cheung A, Ma D, Sundaram GM, Xu C, Xing M, Poon L, Wang J et al. HoxC5 and miR-615–3p target newly evolved genomic regions to repress hTERT and inhibit tumorigenesis. Nat Commun 2018;9:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han X, Yan S, Weijie Z, Feng W, Liuxing W, Mengquan L and Qingxia F. Critical role of miR-10b in transforming growth factor-beta1-induced epithelial-mesenchymal transition in breast cancer. Cancer Gene Ther 2014;21:60–7. [DOI] [PubMed] [Google Scholar]
- 41.Hui W, Ma X, Zan Y, Song L, Zhang S and Dong L. MicroRNA-1292–5p inhibits cell growth, migration and invasion of gastric carcinoma by targeting DEK. Am J Cancer Res 2018;8:1228–1238. [PMC free article] [PubMed] [Google Scholar]
- 42.van Boven N, Kardys I, van Vark LC, Akkerhuis KM, de Ronde MWJ, Khan MAF, Merkus D, Liu Z, Voors AA, Asselbergs FW et al. Serially measured circulating microRNAs and adverse clinical outcomes in patients with acute heart failure. Eur J Heart Fail 2018;20:89–96. [DOI] [PubMed] [Google Scholar]
- 43.Sebastiao MJ, Pereira R, Serra M, Gomes-Alves P and Alves PM. Unveiling Human Cardiac Fibroblast Membrane Proteome. Proteomics. 2018;18:e1700446. [DOI] [PubMed] [Google Scholar]
- 44.Doll S, Dressen M, Geyer PE, Itzhak DN, Braun C, Doppler SA, Meier F, Deutsch MA, Lahm H, Lange R et al. Region and cell-type resolved quantitative proteomic map of the human heart. Nat Commun 2017;8:1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herum KM, Lunde IG, McCulloch AD and Christensen G. The Soft- and Hard-Heartedness of Cardiac Fibroblasts: Mechanotransduction Signaling Pathways in Fibrosis of the Heart. J Clin Med 2017;6(5):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hashimoto K, Tanaka S, Hirata M and Chiba S. Responses of Sino-Atrial Node to Change in Pressure in Sinus Node Artery. Circulation Research. 1967; September;21(3):297–304 [DOI] [PubMed] [Google Scholar]
- 47.James TN and Nadeau RA. Direct perfusion of the sinus node: an experimental model for pharmacologic and electrophysiologic studies of the heart. Henry Ford Hosp Med Bull. 1962;10:21–5. [PubMed] [Google Scholar]
- 48.Lange G, Lu HH, Chang A and Brooks CM. Effect of stretch on the isolated cat sinoatrial node. Am J Physiol 1966;211:1192–6. [DOI] [PubMed] [Google Scholar]
- 49.Mikhailov AV, Kalyanasundaram A, Li N, Scott SS, Artiga EJ, Subr MM, Zhao J, Hansen BJ, Hummel JD and Fedorov VV. Comprehensive evaluation of electrophysiological and 3D structural features of human atrial myocardium with insights on atrial fibrillation maintenance mechanisms. J Mol Cell Cardiol 2020;151:56–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


