Abstract
Intraflagellar transport (IFT) is an evolutionarily conserved mechanism essential for the assembly and maintenance of most eukaryotic cilia and flagella, including mammalian sperm tails. Depletion of IFT27, a component of the IFT complex, in male germ cells results in infertility associated with disrupted sperm flagella structure and motility. Leucine zipper transcription factor-like 1 (LZTFL1) is an IFT27 associated protein. LZTFL1, also known as BBS17, is a Bardet-Biedl syndrome (BBS) associated protein. Patients carrying biallelic variants of LZTFL1 gene exhibit the common BBS phenotypes. The global Lztfl1 knockout mice showed abnormal growth rate and retinal degeneration, typical of BBS phenotype. However, it is not clear if Lztfl1 has a role in male fertility. The LZTFL1 protein is highly and predominantly expressed in mouse testis. During the first wave of spermatogenesis, the protein is only expressed during spermiogenesis phase from the round spermatid stage and displays a cytoplasmic localization with a vesicular distribution pattern. At the elongated spermatid stage, LZTFL1 is present in the developing flagella and appears also close to the manchette. Fertility of Lztfl1 knockout mice was significantly reduced and associated with low sperm motility and a high level of abnormal sperm (astheno-teratozoospermia). In vitro assessment of fertility revealed reduced fertilization and embryonic development when using sperm from homozygous mutant mice. In addition, we observed a significant decrease of the testicular IFT27 protein level in Lztfl1 mutant mice contrasting with a stable expression levels of other IFT proteins, including IFT20, IFT81, IFT88 and IFT140. Overall, our results support strongly the important role of LZTFL1 in mouse spermatogenesis and male fertility.
Keywords: Leucine zipper transcription factor-like 1, Intraflagellar transporter protein 27, Spermatogenesis, Male fertility
Graphical Abstract
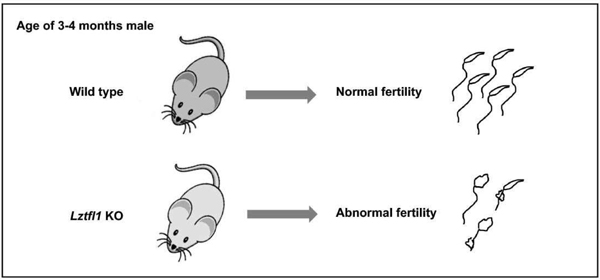
Summary sentence
LZTFL1 is required for mouse spermatogenesis and particularly in maintaining normal sperm function and male fertility.
Introduction
Intraflagellar transport (IFT) is an evolutionarily conserved mechanism for the assembly and maintenance of most eukaryotic cilia and flagella [1]. The IFT complexes are composed of more than 20 unique proteins organized in two subcomplexes called IFT-A and IFT-B. The IFT-A complex mediates the retrograde transport from the cilium tip to the basal body [2, 3]. The IFT-B complex is responsible for the anterograde transport from the basal body to the cilium tip [4]. It has been shown that mutations that affect IFT function result in ciliary defects leading to a range of ciliopathies [5]. Depletion of IFT27 in male germ cells results in male infertility due to disrupted structure and function of sperm flagella. This result suggests that IFT27 plays a crucial role in flagellum formation and establishment of sperm motility [6].
Mutations in the IFT27 gene were discovered in two siblings with Bardet-Biedl syndrome (BBS) [7]. The affected siblings displayed classic BBS phenotypes as described below. Bardet-Biedl syndrome (BBS) is a rare inherited ciliopathy characterized by the association of rod-cone dystrophy, polydactyly, obesity, renal defects, learning difficulties and male infertility [8]. Because ciliary integrity is crucial for numerous pathways of developmental signaling, their dysfunction may impact multiple organs –leading to a syndromic disease like BBS [9]. At present, 25 BBS associated genes have been identified, and deficiency of any of them leads to a ciliopathy [10]. Among these genes, eight (BBS1, 2, 4, 5, 7, 8, 9 and 18) form a complex called the BBSome. The BBSome is described to associate with IFT particles to mediate ciliary trafficking of membrane proteins. It was also shown that IFT27 links the BBSome to the IFT complex for maintenance of the ciliary signaling compartment through its association with LZTFL1 (leucine zipper transcription factor-like 1) [11, 12]. LZTFL1, also called BBS17, is the 17th member of the BBS family [13, 14]. LZTFL1 gene has a genomic location at 3p21.3 [15] which is frequently deleted in numerous types of cancer [16]. LZTFL1 was first identified as a human tumor suppressor [17, 18] and then described to participate in ciliary trafficking and immune synapse formation [19–21]. Mutations in human LZTFL1 gene have been reported in patients displaying typical symptoms of BBS [22, 23].
Recently, Lztfl1 global knockout mouse model was established revealing a phenotype of obesity, retinal degeneration, and abnormal cilia development [24]. However, it was not clear if this gene was also implicated in spermatogenesis, sperm function and male fertility. Given that LZTFL1 interacts with IFT27, a key regulator of sperm flagellum formation and function in sperm, we hypothesized that LZTFL1 would have a similar role during germ cell development in the testis. In this study, we showed that LZTFL1 protein has a similar localization in male germ cells as was found for IFT27. In addition, we showed that Lztfl1 deficient male mice display reduced fertility due to quantitative and qualitative defects of spermatogenesis, including abnormal formation of the flagellum and disruption of the sperm tail function. This study further supports that LZTFL1 is functionally downstream of IFT27, and that other proteins, presumably other BBSome component(s), may partially compensate for loss of LZTFL1 function during male germ cell differentiation, which would lead to a milder reproductive phenotype in Lztfl1 deficient mice.
Materials and methods
Ethics statement
All animal research was executed in compliance with the guidelines of the Wayne State University Institutional Animal Care with the Program Advisory Committee (Protocol number: 18-02-0534).
Yeast two-hybrid experiments
Full-length mouse IFT27 coding sequence was amplified using the following primers: forward: 5’-CATATGGTGAAGCTAGCTGCCAAATG-3’; reverse: 5’-GGATCCCTCACACCAGGGTATGGAAT-3’. After the TA cloning and sequencing, the correct cDNA was subcloned into the NdeI/BamH1 sites of pGBKT7, which was used to screen a Mate & Plate™ Library-Universal Mouse (Normalized) (Clontech, Cat#: 630482) using the stringent protocol according to the manufacturer’s instructions. For direct yeast two-hybrid assay, the coding sequence of the LZTFL1 cDNA was amplified by RT-PCR using the following primers: forward: 5’-GAATTCATGGCAGAGTTGGGCCTAAATGAG-3’ and reverse: 5’-GGATCCCATCTTCAGATTCATATTTTGCCAG-3’. The correct cDNA was cloned into EcoRI/BamH1 sites of pGAD-T7 vector. The yeast was transformed with the indicated plasmids using the Matchmaker™ Yeast Transformation System 2 (Clontech, Cat#: 630439). Two plasmids containing simian virus (SV) 40 large T antigen in pGADT7 and p53 in pGBKT7 were co-transformed into AH109 as a positive control. The AH109 transformants were streaked out in complete drop-out medium (SCM) lacking tryptophan, leucine and histidine to test for histidine prototrophy.
Cell culture and transient transfection
The CHO and COS-1 cells were cultured with DMEM (with 10% fetal bovine serum) at 37°C. The cells were transfected with indicated plasmids using Lipofectamine™ 2000 transfection reagent (Invitrogen). After 48 h transfection, the CHO cells were processed for immunofluorescence and the COS-1 cells were used for co-immunoprecipitation assay.
Localization assay
To make mouse IFT27/pEGFP-N2 plasmid, Ift27 cDNA was amplified using the primer set: forward: 5’-CTCGAGAATGGTGAAGCTAGCTGCCAAATGC-3’; reverse: 5’-GGATCCCCACCAGGGTATGGAATATGTCCAC-3’, and the correct Ift27 cDNA was ligated into pEGFP-N2 vector. Mouse LZTFL1/Myc plasmid was provided by Dr. Natarajan’s laboratory [20]. The IFT27/pEGFP-N2 and LZTFL1/Myc were transfected individually or together into CHO cells, and immunofluorescence was conducted using an anti-MYC antibody.
Co-immunoprecipitation assay
The IFT27/GFP and LZTFL1/Myc plasmids were co-transfected into COS-1 cells by sing Lipofectamine™ 2000 transfection reagent (Invitrogen). After 48 h transfection, the cells were processed for co-immunoprecipitation assay. For co-immunoprecipitation assays, the cells were lysed with IP buffer (Beyotime, Cat No. P0013) for 5 min and centrifuged at 10,000 g for 3~5 min. The supernatant of cell lysates was pre-cleaned with protein A beads at 4°C for 30 min and the pre-cleared lysate was then incubated with anti-MYC antibody at 4°C for 2 h. The mixture was then incubated with protein A beads at 4°C overnight. The beads were washed with IP buffer three times and then were re-suspended in 2x Laemmli sample buffer and heated at 95°C for 5 min. The samples were centrifuged at 3,000 g for 30 s and the supernatant was then subjected to Western blot analysis with MYC and IFT27 antibodies.
Luciferase protein complementation assay
HEK293 cells were grown in 12-well plates and transfected in triplicate with N- and C-luciferase fragments (supplied by Dr. James G Granneman, Wayne State University) fused to LZTFL1 and IFT27 along with various controls as specified in the figure legends. The following primers were used to create the constructs: IFT27/C-Luc forward: 5’-AAGCTTCGATGGTGAAGCTAGCTGCCAAATG-3’, IFT27/C-Luc reverse: 5’-ACCGGTGGCACCAGGGTATGGAATATGTCCAC-3’, N-Luc/LZTFL1 forward: 5’-AAGCTTATGGCAGAGTTGGGCCTAAATGAG-3’, N-Luc/LZTFL1 reverse: 5’-ACCGGTGGATCTTCAGATTCATATTTTGCCAG-3’. After transfection, cells were cultured for 18–24 h. Luciferase activities were measured using the luciferase assay system as described in the work of Dr. James G Granneman [25] and readings were recorded using Veritas microplate luminometer. Experiments were performed three times independently and the results are presented with standard errors.
Real-time PCR
Total tissue RNA was isolated using TRIZOL reagent (QIAGEN), and cDNA was synthesized using a first strand cDNA SensiFASTTM cDNA Synthesis Kit (BIOLINE). To compare Lztfl1 mRNA expression levels in different mouse tissues, real-time fluorescence quantitative PCR (qPCR) was conducted using the following primer pair designed with the GeneScript tool: 5’- TGTCCCAAATACGCCTCTGT-3’; and reverse: 5’- GGATGGCTCTTCTTCCCACT-3’. Mouse Gapdh was used to normalize the expression level.
Immunoblotting
Adult mouse tissue extracts were obtained after lysis with buffer containing 50 mM Tris–HCl pH 8.0, 170 mM NaCl, 1% NP40, 5 mM EDTA, 1 mM DTT and protease inhibitors (Complete mini; Roche diagnostics GmbH)). Protein concentration for lysates was determined using BCA reagent (Sigma-Aldrich), and equal amounts of protein (50 μg/lane) were heated to 95°C for 10 min in sample buffer, loaded onto 10% sodium dodecyl sulfate-polyacrylamide gels, electrophoretically separated, and transferred to polyvinylidene difluoride membranes (Millipore Corporation, Bedford, Mass.). Membranes were blocked (Tris-buffered saline solution containing 5% nonfat dry milk and 0.05% Tween 20 [TBST]) and then incubated with the indicated primary antibodies at 4°C overnight. After being washed in TBST, the blots were incubated with immunoglobulin conjugated to horseradish peroxidase for 1 h at room temperature. After washing, the target proteins were detected with Super Signal chemiluminescent substrate (Pierce). The following primary antibodies were used: Anti-LZTFL1, SIGMA-ALDRICHTM, 1:2,000, Cat No: AV48390-100UL; GAPDH, Santa Cruz Biotechnology, 1:2,000, Cat No: SC-32233. Secondary antibodies include: Anti-Rabbit IgG, Jackson ImmunoResearch, 1:2,000, Cat No: 711166152; Anti-Mouse IgG, Vector Laboratories, 1:2,000, Cat No: DI-2488.
Spermatogenic cells preparation and immunofluorescence
Testes from 3–4-months old mice were dissected and placed in PBS at room temperature to wash off any contaminating material. The tunica albuginea was then removed from each testis and the isolated testicular parenchyma incubated in 5 mL of DMEM containing 0.5 mg/mL collagenase IV and 1.0 μg/mL DNase I (Sigma-Aldrich) with shaking, at 32 °C for 30 min. Then the solution was centrifuged at 1,000 rpm for 5 min under 4°C and the supernatant was discarded. After fixing in 5 mL 4% PFA containing 0.1 M sucrose and shaking gently for 15 min at room temperature, the cell pellet was washed three times with PBS. The cells were carefully resuspended in 2mL PBS then, deposited on positively charged slides at room temperature for 15 minutes. 50 μL of 0.1% Triton X-100 (Sigma-Aldrich) was added to the spermatogenic cells for 5 min at 37 °C for permeabilization and then the cells were blocked with 10% goat serum in PBS. The slides were washed three times with PBS, and the samples were incubated with the primary antibody (Anti-LZTFL1: SIGMA-ALDRICH™, 1:200, Cat No: AV48390-100UL; Anti-α-tubulin: SIGMA, 1:200, Cat No: T9026-2ML) at 4°C overnight. The cells were incubated at room temperature with secondary antibody (Anti-Rabbit IgG, Jackson ImmunoResearch, 1:200, Cat No: 711166152; Anti-Mouse IgG, Vector Laboratories, 1:200, Cat No: DI-2488) at a dilution of 1:200 for 2 hours after washing three times with PBS. Finally, 5 μL DAPI (5 μ/ml Krebs buffer) was added to the fixed cells, covered with a cover slip, and analyzed with the confocal laser-scanning microscopy (Zeiss LSM 700) or a fluorescence microscopy.
Genotyping
Lztfl1 global knockout mice were generated by Dr. Natarajan’s laboratory [24]. Genomic DNA was isolated from the toes of 7-day old mice. Genotype was determined by PCR using the primer sets listed below. KOL 1: 5’-GCTGTCCATTTCTCAGTCGGTG-3’, KOL 2: 5’-CTTCGGAATAGGAACTTCGGTTC-3’, KOR 6: 5’-GCACTTACCACACAAATGTGAGGAC-3’, m-Gapdh 452 Forward: 5’-TAACCTCAGATCAGGGCGGA-3’, m-Gapdh 452 Reverse: 5’-TGTAGGCCAGGTGATGCAAG-3’.
Male fertility test
Sexually mature Lztfl1 KO and control male mice were each mated with a 2–4-months old wild-type female for at least 2 months. The presence of vaginal plugs was checked, and the pregnancy of females was recorded. The number of pups was counted the day after birth. Average litter sizes are presented as the number of total pups born divided by the number of mating cages.
Sperm parameters
After breeding studies, the male mice were euthanized by cervical dislocation following anesthesia. Sperm were collected from the cauda epididymis in 37°C PBS solution and fixed for 15 min at room temperature with 4% formaldehyde. Cells were counted using a hemocytometer chamber under a light microscope, and sperm number was calculated by standard methods. Motility percentages and velocities (average path velocity) were then analyzed using Image J (National Institutes of Health, Bethesda, MD) and the plug-in MTrackJ.
In vitro fertilization (IVF)
Sperm samples were collected from the cauda epididymis, incubated at 37 °C and 5% CO2 for 30–60 minutes, and allowed to capacitate [26]. For in vitro fertilization, sperm samples were collected for the global Lztfl1 KO and wild type males (2 to 6 months of age) as described above and used to fertilize oocytes collected from the oviduct of hormonally stimulated wild type C57BL/6NCr females. The fertilized oocytes were surgically transferred into the oviducts of pseudo pregnant recipient females at the 2-cell stage, approximately 24 hours post-IVF. The medium for sperm collection, capacitation and IVF was purchased from Research Vitro Fertilization Media, K-RVFE-50.
Histological examination of testicular and epididymal tissues
Testes and epididymides of adult mice were collected and fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) at 4°C overnight. The tissues were embedded in paraffin, sectioned at 5μm thickness, deparaffined, and stained with hematoxylin and eosin, using standard procedures. Slides were examined using a BX51 Olympus microscope (Olympus Corp., Melville, NY, Center Valley, PA), and photographs were taken with a ProgRes C14 camera (Jenoptik Laser, Germany).
Transmission electron microscopy
Mouse testes and sperm from adult control and Lztfl1 KO mice were collected after swimming out from the cauda epididymis in 37°C PBS solution followed by centrifugation at 1,000 × g for 10 min under 4°C and re-suspended in fixation buffer (0.2M HEPES, 8mM CaCl2, 8% PFA, 10% Glutaraldehyde, PH7.4). Electron microscopy was performed using the method described previously [6, 27].
Statistical analysis
Graphs were created using Microsoft Excel. Statistical analyses were performed using Student’s t test. P > 0.05 was considered as not significant and by convention *p < 0.05.
Results
LZTFL1, a binding partner of IFT27.
IFT27, a component of the IFT-B complex and essential for spermatogenesis, was used as bait for a yeast two-hybrid screen and LZTFL1 was identified to be a binding partner (Table 1). A direct yeast two-hybrid assay was conducted to confirm the interaction between IFT27 and LZTFL1. Like the positive control, the yeast co-transformed with the two plasmids expressing LZTFL1 and IFT27 grew on the selection medium (Figure 1A), indicating that the two proteins interact in yeast. To further examine interaction of the two proteins, CHO cells were transfected with the plasmids expressing the two proteins. When the CHO cells expressed IFT27/GFP only, the protein was present throughout the cell, including the nucleus (Figure 1B, a). LZTFL1/MYC was present in the cytoplasm (Figure 1B, b). When CHO cells expressed both proteins, IFT27/GFP partially overlapped with LZTFL1/MYC in the cytoplasm (Figure 1B, c). In addition, we transfected COS-1 cells with LZTFL1/Myc and IFT27/GFP expression plasmids and conducted a co-immunoprecipitation assay. The MYC antibody pulled down both Myc-tagged LZTFL1 and GFP-tagged IFT27, suggesting an interaction between these proteins (Figure 1C). To further confirm the LZTFL1/IFT27 interaction, we conducted luciferase complementation assay. Pair of ABHD5 and PLIN1 proteins was used as a positive control [28]. The N-terminal 91 amino acids of G. princeps luciferase were fused to mouse LZTFL1 and the remaining C-terminal fragment to mouse IFT27. As shown in Figure 1D, the individual fusion constructs have virtually no activity when expressed alone in HEK293 cells even though the proteins were expressed (Supplemental Figure 1). Like the positive control pair, co-expression of N-Luc/LZTFL1 and IFT27/C-Luc reconstituted luciferase activity. However, the luciferase activity was not reconstituted when the cells co-expressed either N-Luc/LZTFL1 plus ABHD5/C-Luc or IFT27/C-Luc plus N-Luc/PLIN1 (Figure 1D).
Table 1: List of putative IFT27 binding proteins selected under stringent conditions.
The full-length IFT27 coding sequence was cloned into the Nde/BamH1 sites of pGBKT7, which was used to screen a Mate & Plate™ Library-Universal Mouse (Normalized) (Clontech, Cat#: 630482) according to the manufacturer’s instructions. The yeasts were grown on plates lacking four amino acids (Ade-Leu-His-Trp). LZTFL1 was found to be one of the putative IFT27 binding proteins. The screen also identified IFT25, which has been shown to be an IFT27 binding partner.
| Name | Ncbi number | Frequency |
|---|---|---|
| Mus musculus ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 (Atp8a1), transcript variant 2, mRNA | NM_009727.3 | 27 |
| Mus musculus COP9 (constitutive photomorphogenic) homolog, subunit 5 (Arabidopsis thaliana) (Cops5), transcript variant 1, mRNA | NM_013715.2 | 8 |
| Mus musculus BRO1 domain and CAAX motif containing (Brox), mRNA | NM_027861.2 | 8 |
| Mus musculus signal peptide peptidase like 2A (Sppl2a), mRNA | NM_023220.2 | 7 |
| Mus musculus syndecan binding protein (Sdcbp), transcript variant 2, mRNA | NM_016807.2 | 6 |
| Mus musculus strain mixed chromosome 16, alternate assembly Mm_Celera | AC_000038.1 | 6 |
| Mus musculus solute carrier family 34 (sodium phosphate), member 2 (Slc34a2), transcript variant X1, mRNA | XM_006503807.1 | 5 |
| Mus musculus amyloid beta (A4) precursor protein-binding, family B, member 2 (Apbb2), transcript variant X12, mRNA | XM_006503697.2 | 5 |
| Mus musculus strain mixed chromosome 9, alternate assembly Mm_Celera | AC_000031.1 | 5 |
| Mus musculus nuclear receptor coactivator 7 (Ncoa7), transcript variant X4, mRNA | XM_011243155.1 | 4 |
| Mus musculus lymphocyte cytosolic protein 1 (Lcp1), transcript variant X6, mRNA | XM_006518703.2 | 4 |
| Mus musculus Yip1 domain family, member 4 (Yipf4), mRNA | NM_026417.4 | 4 |
| Mus musculus ATPase, H+ transporting, lysosomal V1 subunit D (Atp6v1d), mRNA | NM_023721.2 | 4 |
| Mus musculus heat shock protein 90, alpha (cytosolic), class A member 1 (Hsp90aa1), mRNA | NM_010480.5 | 4 |
| Mus musculus metal response element binding transcription factor 2 (Mtf2), transcript variant X10, misc_RNA | XR_389262.2 | 4 |
| Mus musculus mitofusin 2 (Mfn2), transcript variant X1, mRNA | XM_006538611.2 | 4 |
| Mus musculus annexin A2 (Anxa2), mRNA | NM_007585.3 | 4 |
| Mus musculus integrin alpha 6 (Itga6), transcript variant X3, mRNA | XM_011239310.1 | 3 |
| Mus musculus synaptotagmin IV (Syt4), mRNA | NM_009308.3 | 3 |
| Mus musculus echinoderm microtubule associated protein like 1 (Eml1), transcript variant 3, mRNA | NM_001286346.1 | 3 |
| Mus musculus homeodomain interacting protein kinase 3 (Hipk3), transcript variant 2, mRNA | NM_001145824.1 | 3 |
| Mus musculus myosin VA (Myo5a), transcript variant X7, mRNA | XM_006510834.2 | 3 |
| Mus musculus predicted gene 5092 (Gm5092), mRNA | XM_011251591.1 | 2 |
| Mus musculus C2 calcium-dependent domain containing 2 (C2cd2), transcript variant X4, mRNA | XM_011246111.1 | 2 |
| Mus musculus zinc finger protein 654 (Zfp654), transcript variant X4, mRNA | XM_011246019.1 | 2 |
| Mus musculus metadherin (Mtdh), transcript variant X6, mRNA | XM_006520153.2 | 2 |
| Mus musculus glycosyltransferase 8 domain containing 1 (Glt8d1), transcript variant X3, mRNA | XM_006519673.2 | 2 |
| Mus musculus death associated protein kinase 1 (Dapk1), transcript variant X2, mRNA | XM_006517371.2 | 2 |
| Mus musculus plastin 1 (I-isoform) (Pls1), transcript variant X1, mRNA | XM_006510746.2 | 2 |
| Mus musculus hook homolog 1 (Drosophila) (Hook1), transcript variant X3, mRNA | XM_006503487.2 | 2 |
| Mus musculus angel homolog 2 (Drosophila) (Angel2), transcript variant X4, mRNA | XM_006497186.2 | 2 |
| Mus musculus transmembrane protein 186 (Tmem186), mRNA | NM_025708.4 | 2 |
| Mus musculus Wolfram syndrome 1 homolog (human) (Wfs1), mRNA | NM_011716.2 | 2 |
| Mus musculus strain mixed chromosome 8, alternate assembly Mm_Celera | AC_000030.1 | 2 |
| Mus musculus strain mixed chromosome 2, alternate assembly Mm_Celera | AC_000024.1 | 2 |
| Mus musculus strain mixed chromosome 1, alternate assembly Mm_Celera | AC_000023.1 | 2 |
| Mus musculus calpain 7 (Capn7), transcript variant X2, misc_RNA | XR_874288.1 | 1 |
| Mus musculus predicted gene, 32444 (Gm32444), ncRNA | XR_867579.1 | 1 |
| Mus musculus TAR DNA binding protein (Tardbp), transcript variant X6, misc_RNA | XR_389859.2 | 1 |
| Mus musculus predicted gene, 35465 (Gm35465), transcript variant X2, ncRNA | XR_387598.2 | 1 |
| Mus musculus O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase) (Ogt), transcript variant X2, mRNA | XM_011247503.1 | 1 |
| Mus musculus IWS1 homolog (S. cerevisiae) (Iws1), transcript variant X3, mRNA | XM_011247009.1 | 1 |
| Mus musculus coiled-coil domain containing 68 (Ccdc68), transcript variant X7, mRNA | XM_011246972.1 | 1 |
| Mus musculus leucine rich repeat containing 9 (Lrrc9), transcript variant X5, mRNA | XM_011244212.1 | 1 |
| Mus musculus signal-induced proliferation-associated 1 like 1 (Sipa1l1), transcript variant X14, mRNA | XM_011244072.1 | 1 |
| Mus musculus androglobin (Adgb), transcript variant X1, mRNA | XM_011243160.1 | 1 |
| Mus musculus SET and MYND domain containing 2 (Smyd2), transcript variant X4, mRNA | XM_011238912.1 | 1 |
| Mus musculus membrane-spanning 4-domains, subfamily A, member 14 (Ms4a14), transcript variant X4, mRNA | XM_006543452.2 | 1 |
| Mus musculus predicted gene 2511 (Gm2511), transcript variant X2, mRNA | XM_006541333.1 | 1 |
| Mus musculus protein tyrosine phosphatase, non-receptor type 3 (Ptpn3), transcript variant X5, mRNA | XM_006538101.2 | 1 |
| Mus musculus tryptophan rich basic protein (Wrb), transcript variant X1, mRNA | XM_006536895.2 | 1 |
| Mus musculus cyclin G associated kinase (Gak), transcript variant X3, mRNA | XM_006534907.2 | 1 |
| Mus musculus interleukin 31 (Il31), transcript variant X1, mRNA | XM_006530498.1 | 1 |
| Mus musculus protein kinase inhibitor, alpha (Pkia), transcript variant X1, mRNA | XM_006530056.1 | 1 |
| Mus musculus Nik related kinase (Nrk), transcript variant X1, mRNA | XM_006528550.2 | 1 |
| Mus musculus GATA binding protein 6 (Gata6), transcript variant X3, mRNA | XM_006525657.2 | 1 |
| Mus musculus dynein, axonemal, heavy chain 8 (Dnah8), transcript variant X4, mRNA | XM_006523592.2 | 1 |
| Mus musculus RIKEN cDNA 2900011O08 gene (2900011O08Rik), transcript variant X1, mRNA | XM_006522482.2 | 1 |
| Mus musculus Fgfr1op N-terminal like (Fopnl), transcript variant X1, mRNA | XM_006522450.2 | 1 |
| Mus musculus zinc finger RNA binding protein (Zfr), transcript variant X3, mRNA | XM_006520059.2 | 1 |
| Mus musculus sarcolemma associated protein (Slmap), transcript variant X15, mRNA | XM_006519738.2 | 1 |
| Mus musculus THO complex 7 homolog (Drosophila) (Thoc7), transcript variant X10, mRNA | XM_006518078.2 | 1 |
| Mus musculus Rho-related BTB domain containing 3 (Rhobtb3), transcript variant X5, mRNA | XM_006517412.2 | 1 |
| Mus musculus jumonji, AT rich interactive domain 2 (Jarid2), transcript variant X5, mRNA | XM_006516860.2 | 1 |
| Mus musculus La ribonucleoprotein domain family, member 4B (Larp4b), transcript variant X5, mRNA | XM_006516478.2 | 1 |
| Mus musculus catsper channel auxiliary subunit beta (Catsperb), transcript variant X2, mRNA | XM_006515930.1 | 1 |
| Mus musculus HCLS1 binding protein 3 (Hs1bp3), transcript variant X1, mRNA | XM_006515161.2 | 1 |
| Mus musculus ubiquitin-conjugating enzyme E2N (Ube2n), transcript variant X2, mRNA | XM_006514344.2 | 1 |
| Mus musculus ankyrin repeat domain 24 (Ankrd24), transcript variant X24, mRNA | XM_006514107.2 | 1 |
| Mus musculus transmembrane and coiled coil domains 3 (Tmcc3), transcript variant X6, mRNA | XM_006513754.2 | 1 |
| Mus musculus leucine zipper transcription factor-like 1 (Lztfl1), transcript variant X2, mRNA | XM_006512403.2 | 1 |
| Mus musculus kinesin family member 9 (Kif9), transcript variant X5, mRNA | XM_006511945.2 | 1 |
| Mus musculus unc-13 homolog C (C. elegans) (Unc13c), transcript variant X7, mRNA | XM_006510934.2 | 1 |
| Mus musculus zinc finger CCCH type containing 12C (Zc3h12c), transcript variant X3, mRNA | XM_006510335.2 | 1 |
| Mus musculus zinc finger protein 426 (Zfp426), transcript variant X14, mRNA | XM_006510182.2 | 1 |
| Mus musculus adenosine monophosphate deaminase 3 (Ampd3), transcript variant X6, mRNA | XM_006507222.2 | 1 |
| Mus musculus FGFR1 oncogene partner 2 (Fgfr1op2), transcript variant X1, mRNA | XM_006507094.1 | 1 |
| Mus musculus acyl-Coenzyme A oxidase 3, pristanoyl (Acox3), transcript variant X4, mRNA | XM_006504198.2 | 1 |
| Mus musculus cytoplasmic polyadenylation element binding protein 2 (Cpeb2), transcript variant X6, mRNA | XM_006503932.2 | 1 |
| Mus musculus heat shock protein family B (small), member 11 (Hspb11, Ift25), transcript variant X2, mRNA | XM_006503425.1 | 1 |
| Mus musculus zinc finger, CCHC domain containing 11 (Zcchc11), transcript variant X1, mRNA | XM_006502978.2 | 1 |
| Mus musculus glutathione S-transferase, mu 1 (Gstm1), transcript variant X1, mRNA | XM_006501017.2 | 1 |
| Mus musculus gelsolin (Gsn), transcript variant X3, mRNA | XM_006497992.2 | 1 |
| Mus musculus abl-interactor 1 (Abi1), transcript variant X3, mRNA | XM_006497619.1 | 1 |
| Mus musculus PHD finger protein 3 (Phf3), transcript variant X7, mRNA | XM_006495840.2 | 1 |
| Mus musculus predicted gene 6644 (Gm6644), non-coding RNA | NR_037965.1 | 1 |
| Mus musculus RIKEN cDNA C130080G10 gene (C130080G10Rik), long non-coding RNA | NR_028422.1 | 1 |
| Mus musculus RIKEN cDNA A230072C01 gene (A230072C01Rik), transcript variant 2, long non-coding RNA | NR_027445.1 | 1 |
| Mus musculus thioredoxin-related transmembrane protein 3 (Tmx3), mRNA | NM_198295.2 | 1 |
| Mus musculus MORN repeat containing 2 (Morn2), mRNA | NM_194269.2 | 1 |
| Mus musculus zinc finger, MIZ-type containing 1 (Zmiz1), mRNA | NM_183208.3 | 1 |
| Mus musculus Son DNA binding protein (Son), transcript variant 1, mRNA | NM_178880.4 | 1 |
| Mus musculus ankyrin repeat and KH domain containing 1 (Ankhd1), mRNA | NM_175375.3 | 1 |
| Mus musculus eukaryotic translation initiation factor 5 (Eif5), transcript variant 1, mRNA | NM_173363.5 | 1 |
| Mus musculus SNAP-associated protein (Snapin), mRNA | NM_133854.3 | 1 |
| Mus musculus hook homolog 1 (Drosophila) (Hook1), mRNA | NM_030014.2 | 1 |
| Mus musculus zinc finger, CCHC domain containing 13 (Zcchc13), mRNA | NM_029158.2 | 1 |
| Mus musculus small nuclear ribonucleoprotein polypeptide G (Snrpg), mRNA | NM_026506.2 | 1 |
| Mus musculus integrin beta 3 binding protein (beta3-endonexin) (Itgb3bp), mRNA | NM_026348.3 | 1 |
| Mus musculus plexin domain containing 2 (Plxdc2), mRNA | NM_026162.6 | 1 |
| Mus musculus cytochrome b5 type B (Cyb5b), mRNA | NM_025558.5 | 1 |
| Mus musculus RAS related protein 1b (Rap1b), mRNA | NM_024457.2 | 1 |
| Mus musculus charged multivesicular body protein 1B (Chmp1b), mRNA | NM_024190.2 | 1 |
| Mus musculus solute carrier family 6 (neurotransmitter transporter), member 14 (Slc6a14), mRNA | NM_020049.4 | 1 |
| Mus musculus phosphatidylethanolamine binding protein 1 (Pebp1), mRNA | NM_018858.2 | 1 |
| Mus musculus sulfotransferase family 1D, member 1 (Sult1d1), mRNA | NM_016771.3 | 1 |
| Mus musculus anterior gradient 2 (Agr2), mRNA | NM_011783.2 | 1 |
| Mus musculus solute carrier family 34 (sodium phosphate), member 2 (Slc34a2), mRNA | NM_011402.3 | 1 |
| Mus musculus 6-pyruvoyl-tetrahydropterin synthase (Pts), mRNA | NM_011220.2 | 1 |
| Mus musculus phosphodiesterase 3B, cGMP-inhibited (Pde3b), mRNA | NM_011055.2 | 1 |
| Mus musculus eukaryotic translation initiation factor 3, subunit A (Eif3a), mRNA | NM_010123.3 | 1 |
| Mus musculus CD24a antigen (Cd24a), mRNA | NM_009846.2 | 1 |
| Mus musculus neural precursor cell expressed, developmentally down-regulated gene 8 (Nedd8), mRNA | NM_008683.3 | 1 |
| Mus musculus inhibitor of growth family, member 1 (Ing1), transcript variant 2, mRNA | NM_001302451.1 | 1 |
| Mus musculus neuron specific gene family member 2 (Nsg2), transcript variant 3, mRNA | NM_001290681.1 | 1 |
| Mus musculus prion protein (Prnp), transcript variant 2, mRNA | NM_001278256.1 | 1 |
| Mus musculus tetraspanin 2 (Tspan2), transcript variant 2, mRNA | NM_001243132.1 | 1 |
| Mus musculus doublecortin-like kinase 1 (Dclk1), transcript variant 7, mRNA | NM_001195540.1 | 1 |
| Mus musculus remodeling and spacing factor 1 (Rsf1), mRNA | NM_001081267.2 | 1 |
| Mus musculus RNA binding protein gene with multiple splicing (Rbpms), transcript variant 2, mRNA | NM_001042674.2 | 1 |
| Mus musculus mitochondrion, complete genome | NC_005089.1 | 1 |
| Mus musculus strain C57BL/6J chromosome 11, GRCm38.p3 C57BL/6J | NC_000077.6 | 1 |
| Mus musculus strain C57BL/6J chromosome 9, GRCm38.p3 C57BL/6J | NC_000075.6 | 1 |
| Mus musculus strain mixed chromosome 17, alternate assembly Mm_Celera | AC_000039.1 | 1 |
| Mus musculus strain mixed chromosome 15, alternate assembly Mm_Celera | AC_000037.1 | 1 |
| Mus musculus strain mixed chromosome 13, alternate assembly Mm_Celera | AC_000035.1 | 1 |
| Mus musculus strain mixed chromosome 7, alternate assembly Mm_Celera | AC_000029.1 | 1 |
| Mus musculus glutamate receptor, ionotropic, AMPA2 (alpha 2) (Gria2), transcript variant X8, misc_RNA | XR_375489.2 | 1 |
| Mus musculus aldehyde dehydrogenase 4 family, member A1 (Aldh4a1), transcript variant X1, mRNA | XM_011250224.1 | 1 |
| Mus musculus metadherin (Mtdh), transcript variant X6, mRNA | XM_006520153.2 | 1 |
| Mus musculus oxysterol binding protein-like 8 (Osbpl8), transcript variant X3, mRNA | XM_006513638.2 | 1 |
| Mus musculus CCR4-NOT transcription complex, subunit 7 (Cnot7), transcript variant X8, mRNA | XM_006509307.2 | 1 |
| Mus musculus interferon induced transmembrane protein 3 (Ifitm3), mRNA | NM_025378.2 | 1 |
| Mus musculus Shc SH2-domain binding protein 1-like (Shcbp1l), mRNA | NM_001033162.2 | 1 |
Figure 1. Identification of LZTFL1 as a binding partner of intraflagellar transport protein 27 (IFT27), a major spermatogenesis regulator.
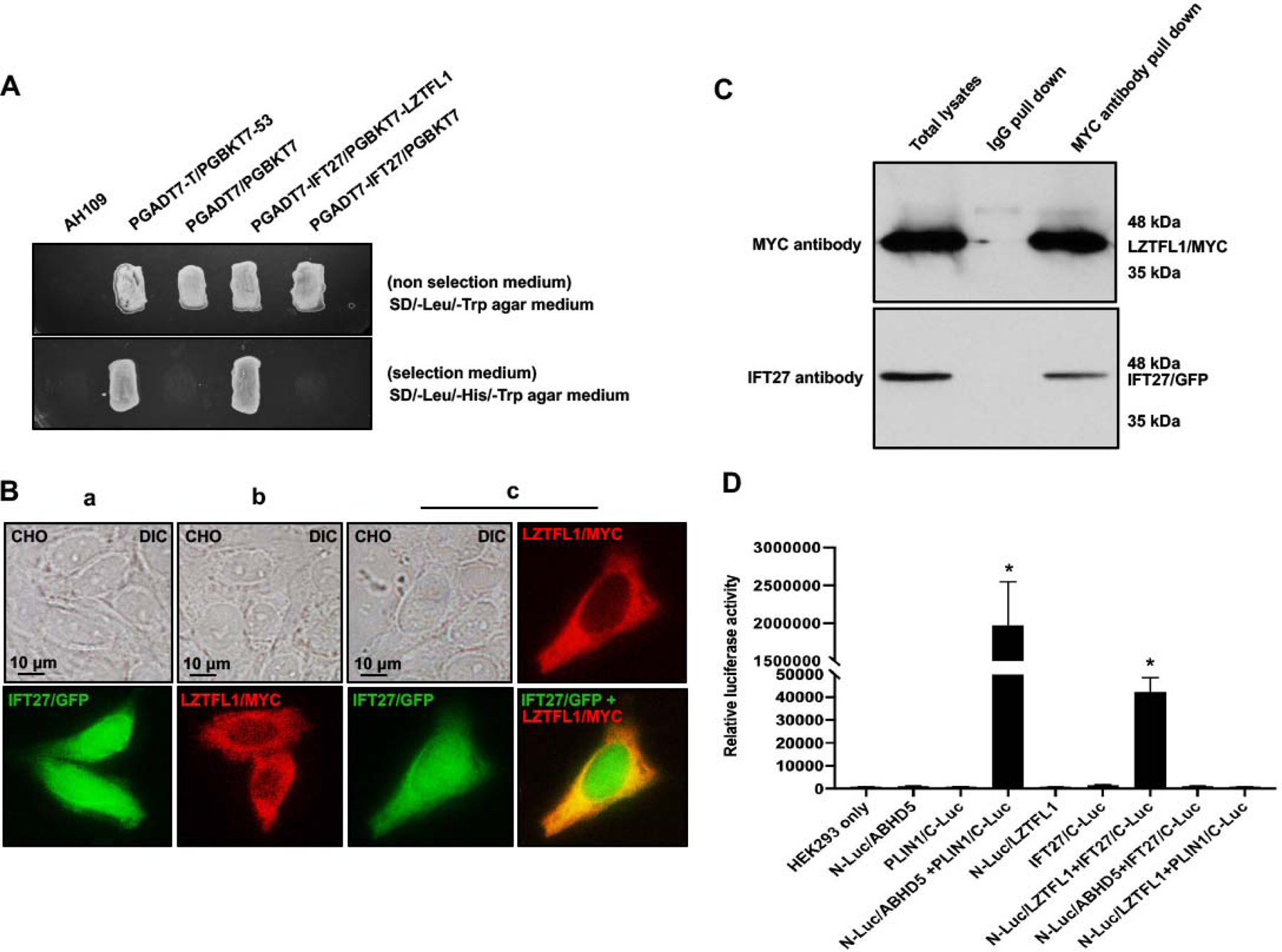
A. Direct yeast two-hybrid assay to examine the interaction between IFT27 and LZTFL1. Pairs of indicated plasmids were co-transformed into AH109 yeast, and the transformed yeast were grown on either selection plates (lacking leucine, histidine and tryptophan) or non-selection plates (lacking leucine and tryptophan). Notice that all the yeast except AH109 grew on the non-selection plate. Yeast expressing IFT27/LZTFL1 and P53/large T antigen pairs grew on selection plate. B. LZTFL1/MYC overlaps with IFT27/GFP in CHO cells. When expressed alone, IFT27/GFP was present in the whole cells (a), and LZTFL1/MYC was present in the cytoplasm (b). When the two proteins were co-expressed, LZTFL1/MYC partially overlapped with IFT27/GFP in the cytoplasm (c). C. Co-immunoprecipitation of LZTFL1/MYC with IFT27/GFP. COS-1 cells were transfected with plasmids to co-express LZTFL1/MYC and IFT27/GFP. The cell lysate was immunoprecipitated with anti-MYC antibody and then analyzed by Western blotting with anti-MYC and anti-IFT27 antibodies. The cell lysate immunoprecipitated with a mouse normal IgG was used as a control. The anti-MYC antibody pulled down both LZTFL1/MYC and IFT27/GFP. D. Interaction of LZTFL1 with IFT27 in HEK293 cells as determined by G. princeps luciferase complementation assay. HEK293 cells were transfected with the indicated plasmids, and luciferase activity was evaluated 24 h after transfection. The cells expressing both N-Luc-ABHD5 and PLIN1-C-Luc was a positive control. Like the positive control, cells expressing both N-Luc/LZTFL1 and IFT27/C-Luc reconstituted activity. * P<0.01 compared to the N-Luc-LZTFL1 or IFT27-C-Luc only.
Mouse LZTFL1 protein and mRNA expression in vivo.
LZTFL1 protein expression in vivo was examined by Western blot analysis. LZTFL1 protein was detected only in the testis when 50ug of total protein was loaded and the protein was examined by less sensitive Pico system with the anti-LZTFL1 antibody concentration 1:2000 (Figure 2A). When 100 μg of total protein was loaded and anti-LZTFL1 antibody was increased to 1:400, LZTFL1 protein was also observed in brain, spleen, lung and liver when examined by the more sensitive Femto system (Supplemental Figure 2). To investigate if LztfL1 mRNA was also expressed in other tissues and to compare Lztfl1 mRNA expression levels, RT-PCR and qPCR were conducted using the cDNAs from eight mouse tissues, including heart, brain, spleen, lung, liver, kidney, muscle, and testis. Lztfl1 mRNA was expressed in all these tissues, but highly abundant in testis (Figure 2B). Dynamic expression of LZTFL1 during the first wave of spermatogenesis was further examined. The protein was expressed as early as postnatal day 16 and expression was dramatically increased at postnatal day 28, corresponding to the differentiation of elongating spermatids in the last phase of spermatogenesis (spermiogenesis) (Figure 2C).
Figure 2. Mouse LZTFL1 protein and mRNA tissue distribution.
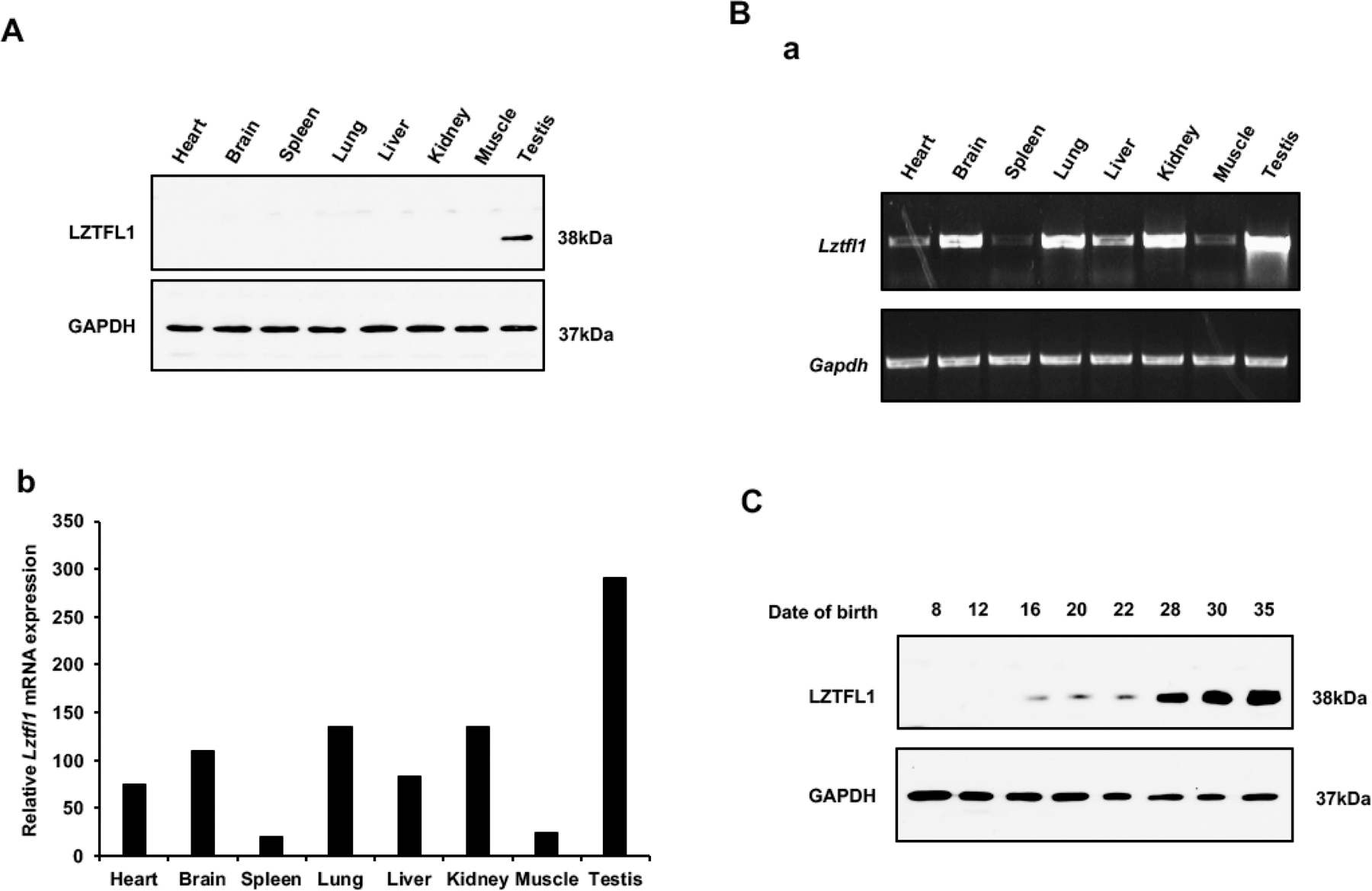
A. Analysis of tissue distribution of mouse LZTFL1 protein by Western blot. 50ug of total protein was loaded and the protein was examined by less sensitive Pico system. The anti-LZTFL1 antibody concentration: 1:2000. LZTFL1 protein is abundant only in the testis.
B. Analysis of relative Lztfl1 mRNA expression in adult mouse tissues by RT-PCR (a) and qPCR (b). Lztfl1 was detected in all tissues, but most abundant in testis. Lztfl1 mRNA expression levels were normalized by Gapdh.
C. LZTFL1 expression levels during the first wave of spermatogenesis. LZTFL1 was detectable from postnatal day 16; its expression was up-regulated at day 28, and further increased on days 30 and 35. Thus, the protein level was elevated during spermiogenesis, the final phase of spermatogenesis.
Localization of LZTFL1 in isolated male germ cells.
LZTFL1 localization in germ cells was examined by immunofluorescence staining using the specific antibody. With confocal microscopic imaging, the protein was observed in the cytoplasm of both spermatocytes and differentiating spermatids. Some signal appeared near the manchette of elongating spermatids, and it was also present in the developing flagellum (Figure 3A, a–c). However, no LZTFL1 protein was present in the developed sperm (Figure 3A, d). Western blot analysis confirmed that LZTFL1 was not present in the epididymal sperm (Figure 3B).
Figure 3. Localization of LZTFL1 in normal male germ cells.
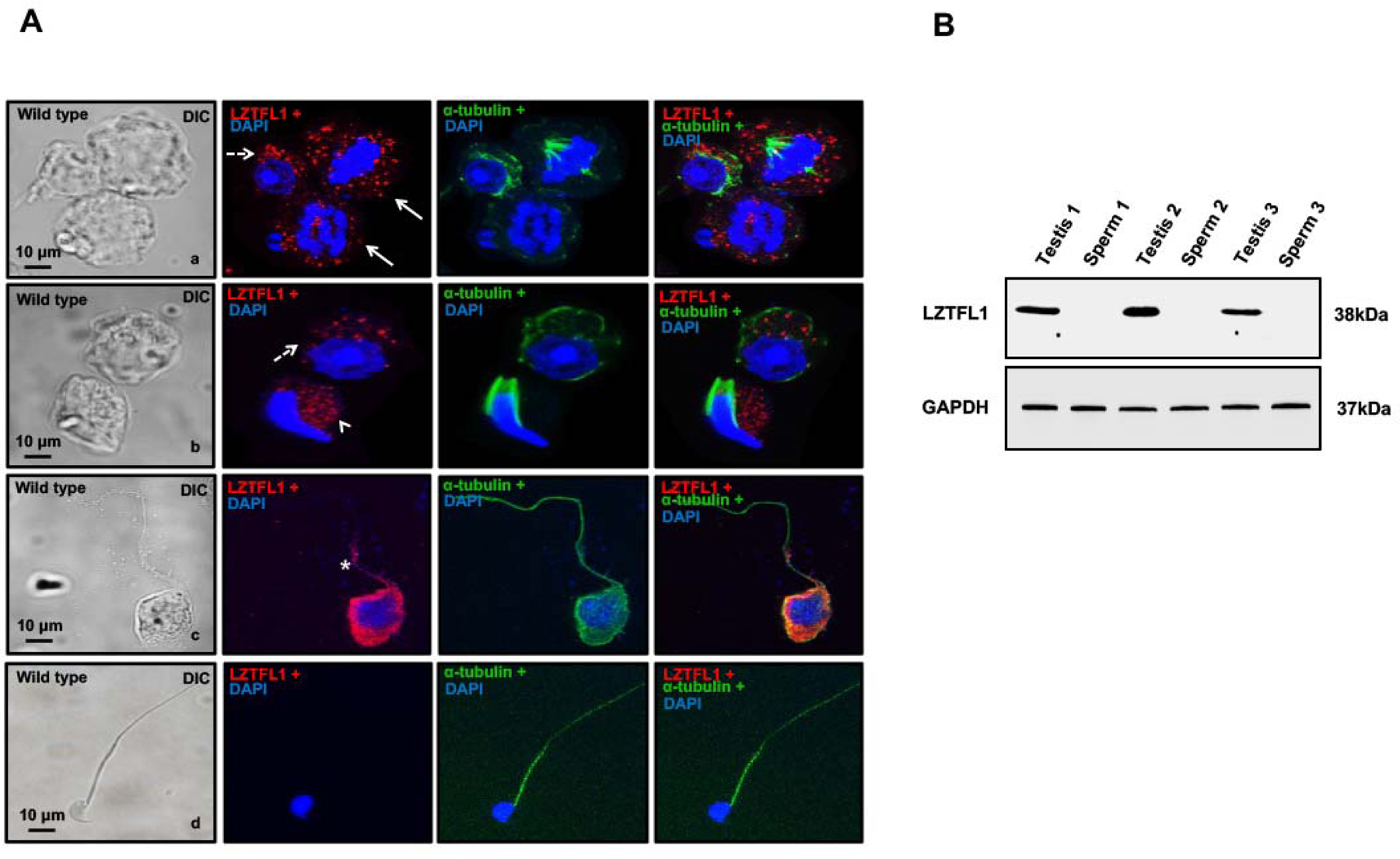
A. Localization of LZTFL1 in isolated germ cells was examined by immunofluorescence staining. LZTFL1 signal was observed in the cytoplasm of spermatocytes (panel a, arrows) and round spermatids (panels a and b, dotted arrows). Some signal appears near the manchette of elongating spermatids (panel b, arrow head), and is also localized in the developing sperm tails (panel c, star). No LZTFL1 signal was observed in the tails of the developed sperm (panel d). Images were taken with a confocal microscope.
B. LZTFL1 is not expressed in the epididymal sperm. Epididymal sperm were collected from cauda epididymis, and Western blot was conducted. LZTFL1 protein was detected in the testis, but not in sperm.
Inactivation of mouse Lztfl1 gene resulted in reduced fertility associated with decreased sperm number and motility and an increase in abnormal sperm.
High expression level of LZTFL1 in the testis, particularly in germ cells, suggests a role for this gene in spermatogenesis. To examine its role, we examined the reproductive phenotype on the Lztfl1 global knockout (Lztfl1 KO) mice. The model was generated by Dr. Natarajan’s laboratory (Supplemental Figure 3A) [24]. The mice were genotyped using specific primer sets (Supplemental Figure 3B). To examine the role of LZTFL1 in male fertility, Lztfl1 KO and control males (≧8-week-old) were bred with wild-type 3–4-month-old fertile females for a two-month breeding test. All tested mice showed normal mating behavior, and vaginal plugs were observed in the females. Lztfl1 KO mice had decreased sperm numbers and showed reduced fertility (Table 2 and Supplemental Figure 4). Examination of total testicular LZTFL1 protein expression by Western blot and immunofluorescence staining revealed the loss of LZTFL1 in the KO mice (Figure 4A, and Supplemental Figure 5). Percentage of motile sperm (Figure 4B) and sperm motility (Figure 4C) were decreased in the KO mice. There changes were accompanied with an increase in abnormal sperm morphology (Figure 4D). To determine the factors that caused the reduced fertility phenotype, spermatozoa morphology was evaluated with bright field microscopy. Spermatozoa from the control mice showed normal morphology, including normal shaped heads and long, smooth tails (Figure 4E, a). However, multiple abnormalities were discovered in the sperm collected from the Lztfl1 KO mice (Figure 4E, b–f), including misshaped heads (Figure 4E, b–d), uneven thickness of the tail (Figure 4E, b, c), and short tails (Figure 4E, d–f). Some sperm had swollen tail tips (Figure 4E, d, f).
Table 2: Fertility, fecundity and sperm count related to control and Lztfl1 KO mice.
To test fertility, adult males and 8 weeks old males were bred with wild-type fertile females for at least two months. Litter size was recorded for each mating and sperm number was counted.
| Genotype | Male fertility | Litter size (n=10) | Sperm count (×106, n=10) |
|---|---|---|---|
| Control | 10/10 | 6.80 ± 2.52 | 7.48 ± 1.33 |
| Lztfl1 KO | 6/10 | 3.130 ± 2.81* | 3.15 ± 2.41* |
Statistically significant differences:
p < 0.05.
Figure 4. Reduced fertility associated with decreased sperm number and abnormally developed sperm in global Lztfl1 KO mice.
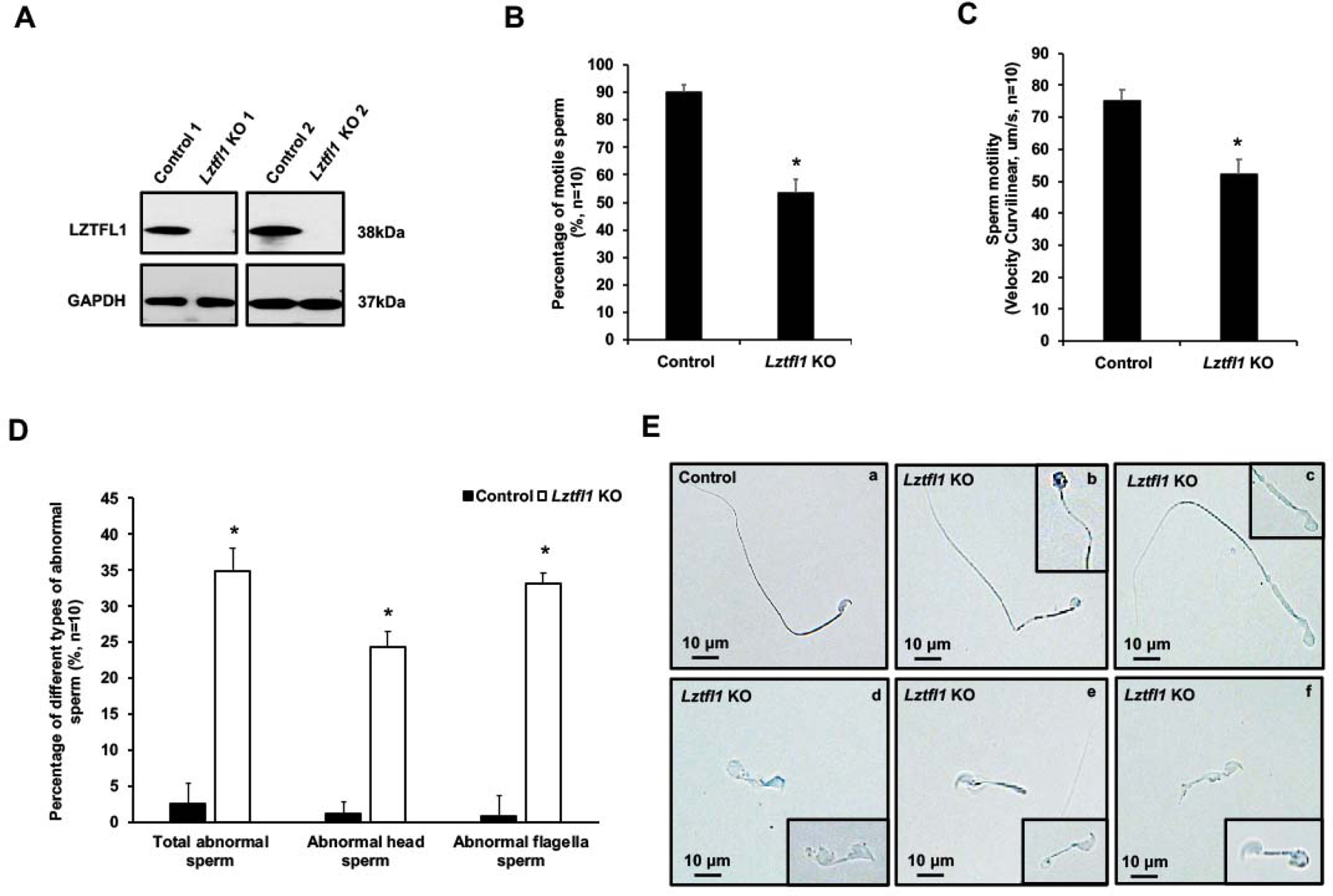
A. Representative Western blot result showing that LZTFL1 protein was missing in the testis of global Lztfl1 KO mice.
B, C. Percentage of motile sperm and sperm motility was reduced in the Lztfl1 KO mice (n=10). Statistically significant differences: *p < 0.05.
D. Quantification of the percentage of different types of abnormal sperm in the control (wild type and heterozygote) and Lztfl1 KO mice (n=10). Statistically significant differences: *p < 0.05.
E. Representative images of epididymal sperm under a light microscopy from the control (heterozygote) (a) and Lztfl1 KO mice (b-f). Sperm from the KO mice showed uneven thickness tail (b, c), misshaped head (b-d), short tail (d-f), and swollen tail tips (d, f).
For in vitro fertilization (IVF), sperm samples were collected from the Lztfl1 KO and wild type males (2 to 6 months of age) and used to fertilize oocytes collected from the oviduct of hormonally stimulated wild type C57BL/6NCr females. Sperm from the KO mice showed a reduction in the rate of IVF compared with sperm from the control mice. However, the live birth rate was not significantly changed (Supplemental Table 1 and Figure 5).
Figure 5. Lztfl1 knockout affects in vitro fertilization (IVF) rate.
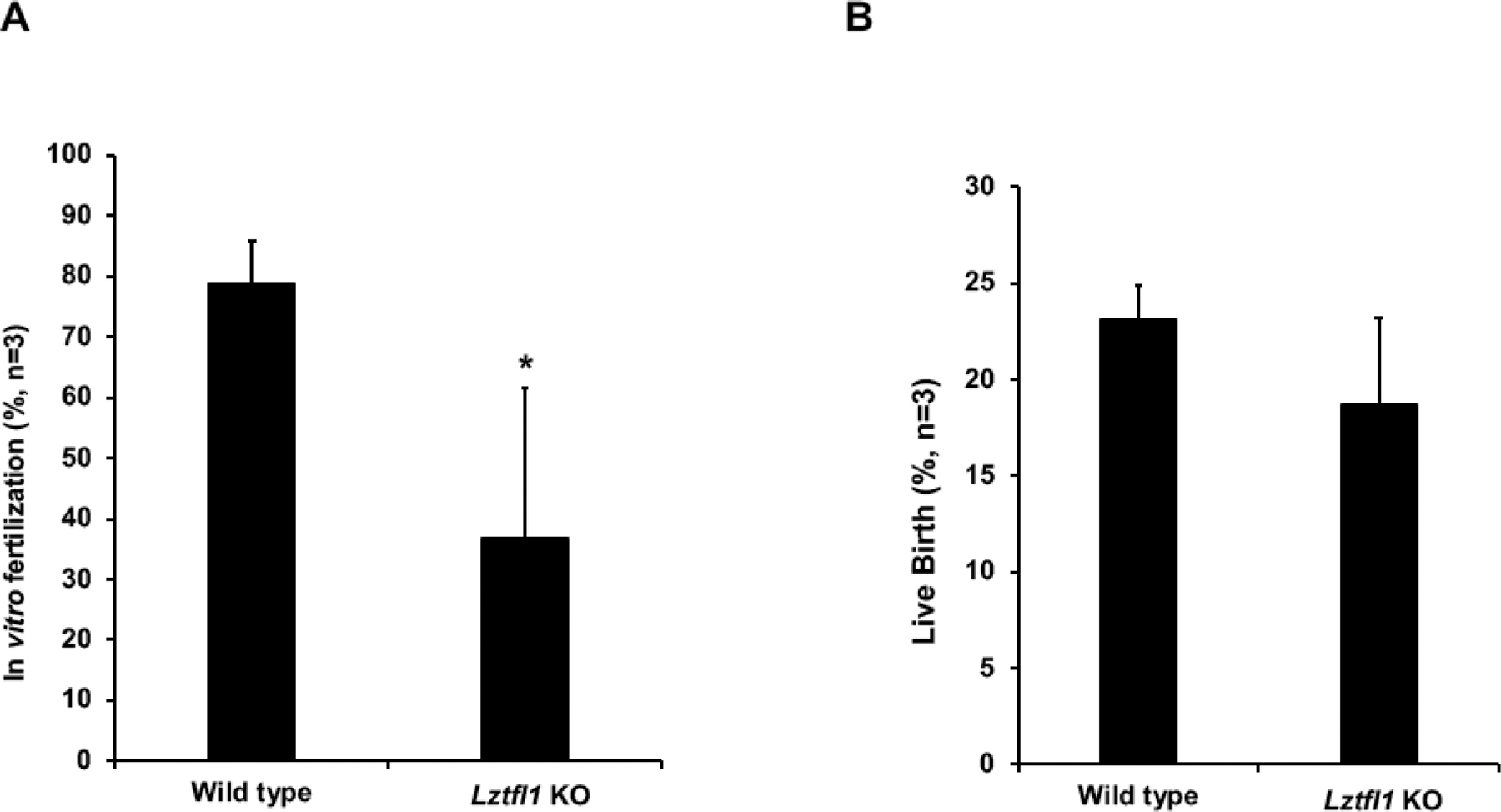
For in vitro fertilization, sperm samples were collected for the Lztfl1 KO and wild type males (2 to 6 months of age, n=3) as described above and used to fertilize oocytes collected from the oviduct of hormonally stimulated wild type C57BL/6NCr females. The fertilized oocytes were surgically transferred into the oviducts of pseudo pregnant recipient females at 2-cell stage, approximately 24 hours post-IVF. The KO mouse sperm showed a reduction in the rate of IVF (A) but not live birth (B), compared with control mice sperm. *p < 0.05.
Spermatogenesis was affected in the Lztfl1 global knockout mice.
Testes of Lztfl1 KO mice exhibited abnormal spermatogenesis, compared to normal seminiferous epithelia observed in the control testes (Figure 6A, a). In the seminiferous epithelium of Lztfl1 KO mice, fewer spermatids reached mature development and elongating spermatids with excess cytoplasm (possibly attached residual bodies) were shed to the lumen (Figure 6A, b, c). Numerous mature sperm were concentrated in the cauda epididymis of control mice (Figure 6B, upper panel), whereas, the Lztfl1 KO mice had fewer sperm and the presence of residual bodies (Rb, arrows) in the epididymal lumen (Figure 6B, lower panel).
Figure 6. Histology and TEM analysis for the adult control (heterozygote) and Lztfl1 KO mice.
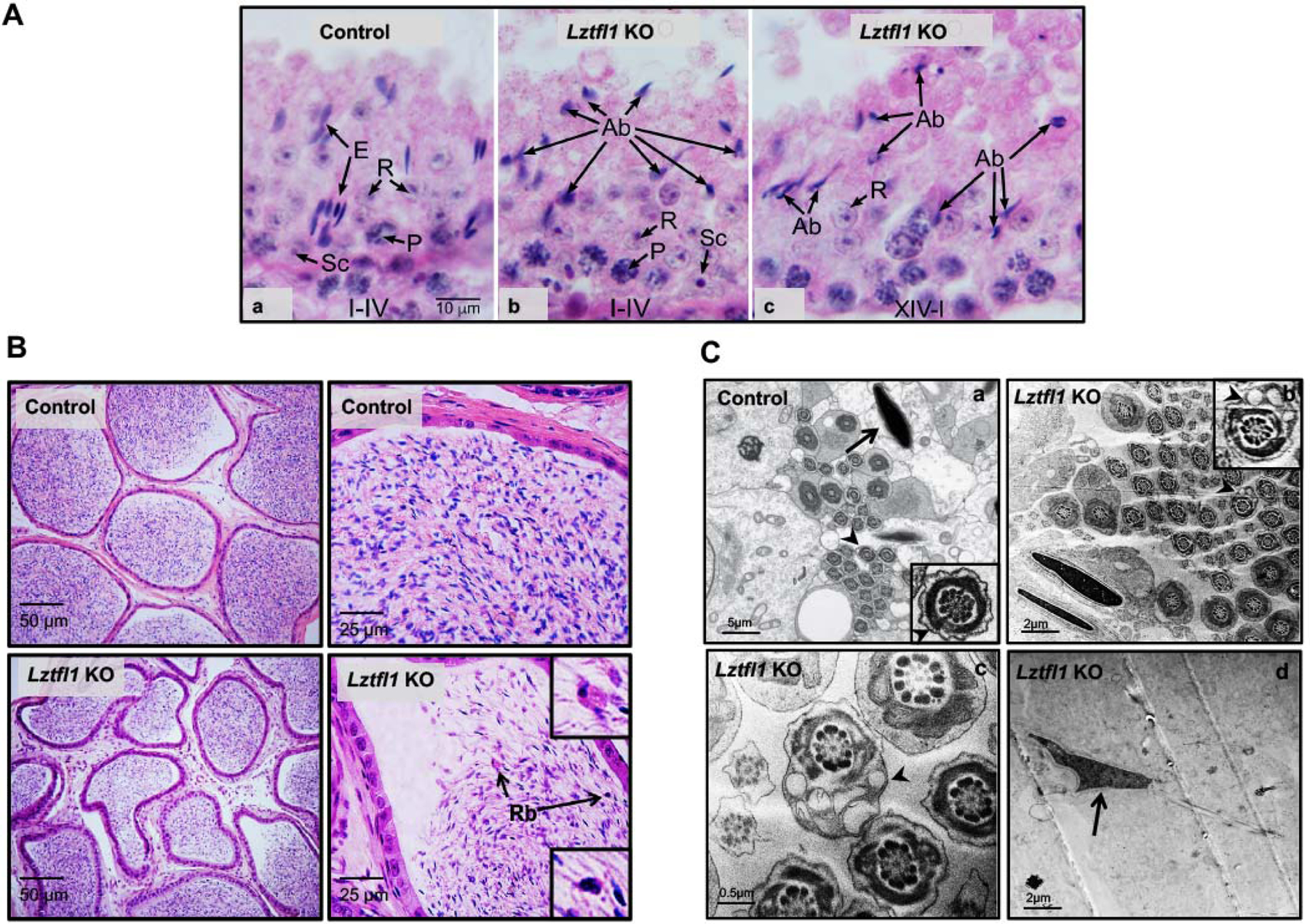
A. Testis from control and Lztfl1 KO mice. (a) In the control testis, stage I-IV tubule shows normal elongated spermatid heads (E) in bundles between the round spermatids (R). Some elongated heads are seen deep within the epithelium, between pachytene spermatocytes (P). Sc, Sertoli cell nucleus. Micron marker for all photos (10 μm). (b-c) Lztfl1 KO testis shows normal round spermatids (R) and pachytene spermatocytes (P), but the seminiferous epithelium contains numerous abnormal elongating spermatids (Ab). With H&E staining of stages XIV-IV, only abnormal elongating spermatid head shapes could be detected.
B. Numerous mature sperm were present in the cauda epididymal lumen of a control mouse (upper panel). But in the Lztfl1 KO mouse, there were fewer sperm in the epididymal lumen and residual bodies (Rb, arrows) appeared to be attached to numerous abnormal sperm (lower panel).
C. TEM images of elongated spermatids from control (a) and Lztfl1 KO (b-d) mice. In the control mice, nuclei have normal elongated shapes with condensed chromatin (arrow) and the flagella contains normal axonemes and fibrous sheath (arrow heads) within the seminiferous tubule. However, in Lztfl1 KO mice, large vacuoles were observed in the sperm flagella (arrow heads in b, c). The arrow in d point to abnormal shapes of nucleus in the Lztfl1 KO mouse.
To investigate the structural basis for abnormal sperm morphology in the Lztfl1 KO, testes from the KO were examined for ultrastructural changes by TEM. Control seminiferous tubules showed elongating spermatids with normal heads forming with condensed chromatin. Control sperm in the lumen had normal axonemal structure in the flagellum, and accessory structures with normal organization (Figure 6C, a). However, Lztfl1 KO testes showed multiple sperm abnormalities, including abnormal heads and tails (Figure 6C, b–d). Abnormal vacuoles (Figure 6C, b, c) and disruption in the fibrous sheath (Supplemental Figure 6A, a) were observed in the Lztfl1 KO sperm flagellum; some sperm had abnormally developed chromatin (Figure 6C, d, Supplemental Figure 6A, b). The core axoneme structure appeared to be normal. The Lztfl1 KO sperm had a normal acrosome and mitochondrial sheath and the chromatin also appeared to have normal condensation (Supplemental Figure 6B).
IFT27 determines LZTFL1 expression level in the testis
LZTFL1 is an IFT27 associated protein. IFT27 is essential for sperm flagella formation, sperm function, and male fertility in mice [6]. The interaction between LZTFL1 and IFT27 propelled us to further examine the functional relationship between these two proteins in vivo. Western blot analysis revealed that testicular IFT27 expression was slightly reduced in Lztfl1 KO mice (Figure 7A). Similarly, testicular LZTFL1 expression levels were significantly reduced in the Ift27 knockout mice (Figure 7B).
Figure 7. IFT27 determines LZTFL1 expression level in the testis.
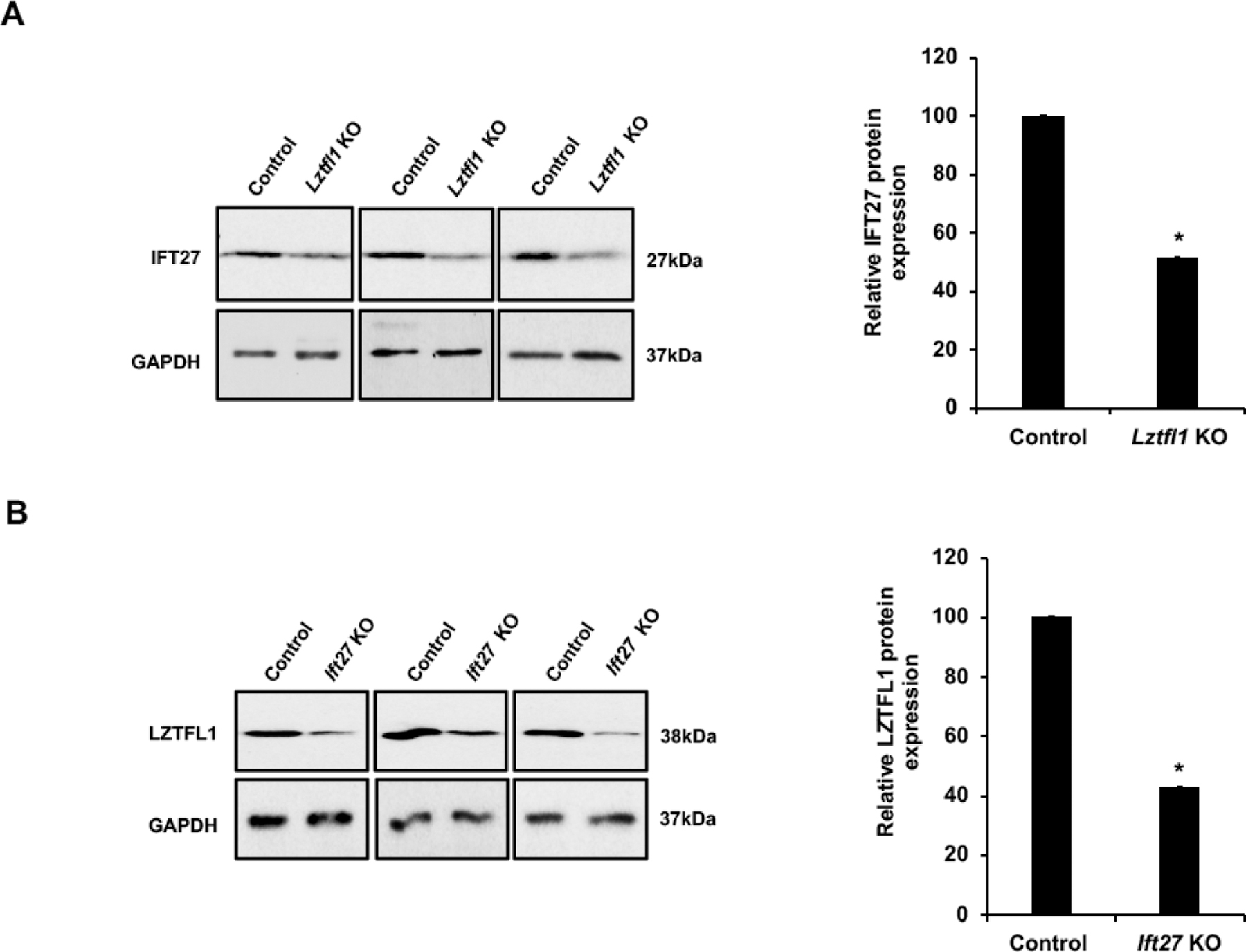
A. Left: Examination the expression level of IFT27 in control (wild type and heterozygote) and Lztfl1 KO mice testes by western blot. Compared with control mice, there was a slight but significant reduction in IFT27 expression level. Right: Quantitative analysis of IFT27 protein expression. Statistically significant differences: *p < 0.05.
B. Left: Examination the expression level of LZTFL1 in control (wild type and heterozygote) and Ift27 conditional knockout mice testes by western blot. Compared with control mice, there was a significant reduction in LZTFL1 expression. Right: Quantitative analysis of LZTFL1 protein expression. Statistically significant differences: *p < 0.05.
Testicular protein levels of other IFT proteins were not changed in the absence of LZTFL1
To examine if the loss of LZTFL1 affected protein levels of other IFT proteins in the testis, we determined the levels of selective IFT proteins by Western blot in the testes of control and Lztfl1 KO mice. Expression levels of IFT-B components, including IFT20, IFT81, IFT88 and IFT140, showed no significant change in the Lztfl1 KO mice (Figure 8).
Figure 8. Expression levels of IFT20, 81, 88 and 140 proteins in the control and Lztfl1KO mice.
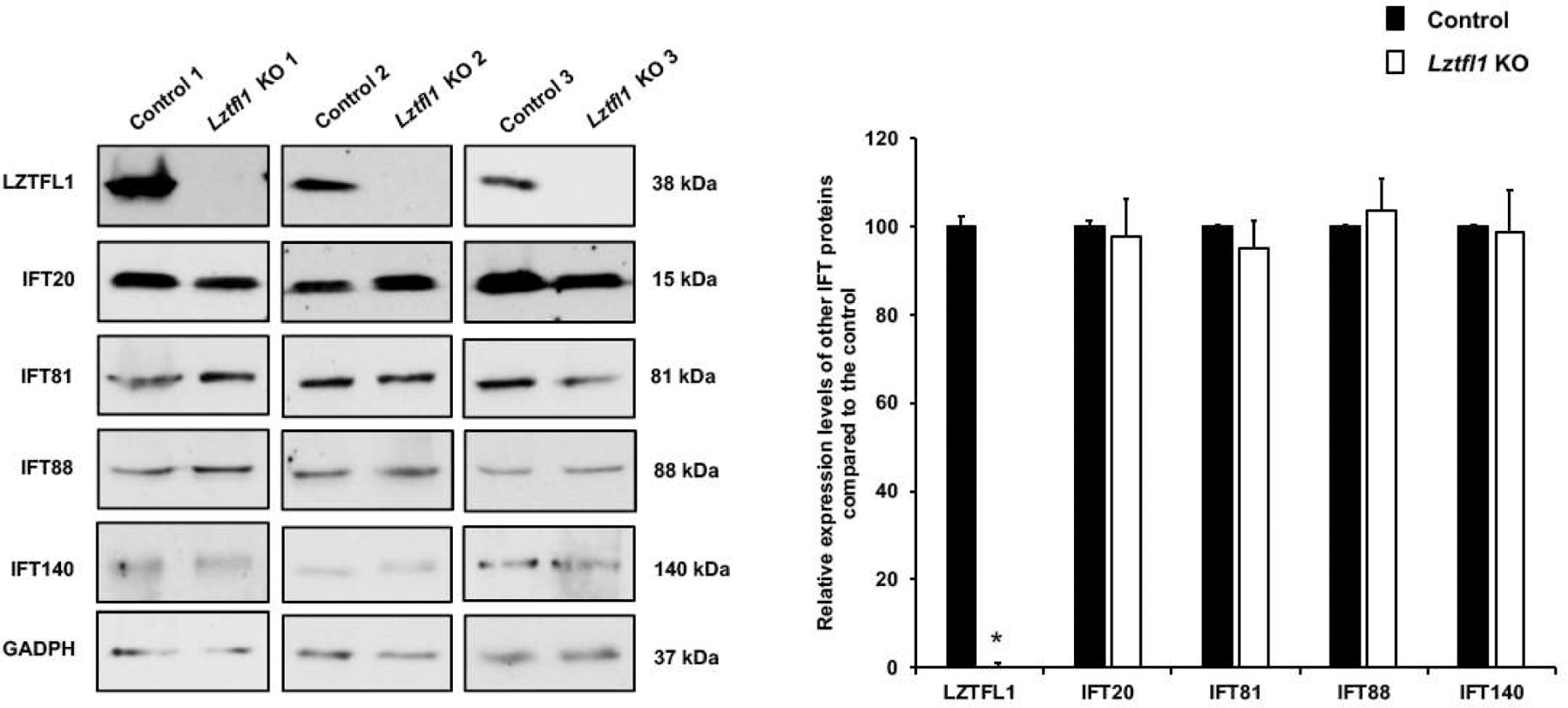
Left: Examination of IFT20, IFT81, IFT88 and IFT140 proteins expression levels in the control (wild type and heterozygote) and global Lztfl1 KO mice by Western blot. Between the control and mutant mice, the expression levels of all selective IFT proteins showed no significant change. Right: Quantitative analysis of selective IFT proteins expression. Statistically significant differences: *p < 0.05.
Discussion
LZTFL1 displayed typical leucine zipper and coil–coil domains, which are conserved among vertebrates. LZTFL1 was initially identified as a tumor suppressor [17] and then found to function as a negative regulator of the Hedgehog pathway [19, 29]. Through the establishment of mice with globally-deleted Lztfl1 or deleted in forebrain tissue, Lztfl1 was also identified as a novel regulator of leptin signaling and obesity in hypothalamic neurons through [24, 30]. Although LZTFL1 has been shown to play important roles in many other somatic systems, its highest expression is in the testis, indicating a potential role in male reproduction. Therefore, we utilized the Lztfl1 KO mouse to investigate the functional relevance of LZTFL1’s in spermatogenesis.
LZTFL1 protein was expressed during spermiogenesis, the last phase of spermatogenesis, corresponding to the differentiation of spermatids. In round spermatids, LZTFL1 signal was localized throughout the cytoplasm; but in elongating spermatids, LZTFL1 was detected near the manchette and in the developing flagellum. Given that IFT27 is also present in the cytoplasm and the developing flagellum and is essential for spermiogenesis, the pattern of LZTFL1 localization strongly suggested that LZTFL1 could function in synergy with IFT27. Mouse IFT components are only present in the developing sperm flagella, but not developed sperm flagella [6, 31]. The same as IFT27, LZTFL1 is only detected in the developing sperm flagella, but not epididymal sperm, suggesting that LZTFL1 coordinates with IFT components for sperm flagella formation.
In this study, we compared the phenotypes observed in Lztfl1 global knockout and Ift27 conditional KO (Ift27 cKO) mice [6]. The male reproductive phenotype of Lztfl1 KO was discovered to be significantly weaker than that observed in Ift27 cKO mice (specific for germ cell deletion). In both animal models, fertility was affected due to some defects in spermiogenesis, although some germ cells completed the spermatogenesis process and formed spermatozoa. Sperm in both Ift27 cKO and Lztfl1 KO mice showed abnormal morphology of the flagellum, particularly in the accessory structures, and some also developed abnormal heads. However, unlike the Ift27 cKO mice, in which only a few sperm were motile but with dramatically reduced motility, most sperm from the Lztfl1 KO mice were motile and showed only slightly reduced motility.
IFT27 is a component of IFT-B [32], and disruption of IFT-B genes usually result in short cilia/flagella [33, 34]. Studies in model organisms, including Chlamydomonas reinhardtii [35] and C. elegans [36] and cultured mammalian cells [34], have shown that the BBSome moves in and out of cilia [19], as well as along the ciliary axoneme together with the IFT particles [37, 38]. Like the conditional Ift27cKO mice, Lztfl1 KO mouse sperm demonstrated defects of the sperm accessory structures, as shown by TEM studies. One possibility is that LZTFL1 and IFT27 together play a role in assembling the sperm accessory structure of the sperm tail, which would differ significantly from their functions in somatic cells, where they do not modulate ciliogenesis. The sperm core axoneme structure was affected in the Ift27 cKO mice [6], but not the Lztfl1 KO mice. Defects of the core axoneme structure can be secondary to the defects of the accessory structures, which were reported in the CABYR mutant mice [39]. The normal looking sperm core axoneme structure in the Lztfl1 KO mice indicates that the defects in the accessory structure were not severe enough to cause defects in the core structure.
In primary cilia, LZTFL1 was found to interact with BBS-9 and to sequester it from its ciliary localization, therefore negatively regulating BBSome ciliary trafficking and hedgehog signaling. LZTFL1 is a BBSome regulator, and failure of BBSome ciliary trafficking is a common cellular feature of BBS, supporting the idea that BBS results from trafficking defects to the cilia membrane. LZTFL1 has been shown to coordinate the interactions between the BBSome and the IFT particle to regulate the removal of Ptch1 and Smo from cilia at the appropriate times during hedgehog signaling [12, 37]. However, it is unknown whether IFT proteins can directly mediate SMO ciliary trafficking or if they are involved indirectly. LZTFL1 is a specific regulator of BBSome ciliary trafficking activity. Other proteins may regulate the activities of the IFT complex or of those of the specific transition zone complex. Precisely how LZTFL1 regulates spermatogenesis is under further investigation.
If LZTFL1 is an IFT downstream molecule [12, 37], it should not be surprising that LZTFL1 expression was reduced in the testis of Ift27 cKO mice. Interestingly, testicular IFT27 level was also slightly reduced in the Lztfl1 KO mice, suggesting that losing either one will affect stability of the IFT27/LZTFL1 complex. Unfortunately, we have not been able to determine if LZTFL1 localization was changed in the absence of IFT27, due to difficulty with immunofluorescence staining on paraffin sections and loss of the of Ift27 cKO mouse line in our laboratory.
In the Ift27 cKO mice, testicular expression of several IFT proteins, including IFT81, were down-regulated [6]. This was not observed in the Lztfl1 KO mice. Besides binding to LZTFL1, IFT27 also coordinates with other IFT proteins to transport cargo proteins for sperm flagellum formation. It is likely that LZTFL1 does not associate with these IFT proteins and it plays less important functions than IFT27 does. Therefore, the Ift27 cKO mice showed stronger phenotype than the Lztfl1 KO mice.
Conclusions
Overall, we studied the role of LZTFL1 in male mice, and compared its role to that of IFT27, its binding partner. Similar to IFT27, LZTFL1 plays an important role in the formation of the sperm flagellum, development of sperm motility and maintenance of male fertility. It appears that formation of the sperm flagellum and assembly of the accessory structures requires the combined effort of IFT27 and LZTFL1. Loss of either protein will result in abnormal spermatid differentiation. Although the male reproductive phenotype is significantly weaker than the conditional Ift27 knockout mouse, our results support the hypothesis that LZTFL1 is a downstream molecule of IFT27, and other BBS components might have redundant functions thus compensating for LZTFL1 loss during male germ cell development.
Supplementary Material
HEK293 cells were transfected plasmids to express IFT27/C-Luc and N-Luc/LZTFL1, and non-transfected HEK293 cells were used as controls. The cell lysates were collected for Western blot analysis by using specific anti-IFT27 and anti-LZTFL1 antibodies. IFT27/C-Luc (left) and N-Luc/LZTFL1 (right) proteins were detected in the transfected HEK293 cells respectively.
100 μg of total protein was loaded and the LZTFL1 protein was examined by a higher sensitive Femto system. The anti-LZTFL1 antibody concentration: 1:400. LZTFL1was highly abundant in the testis, and it was also detected in brain, spleen, lung and liver.
A. Structures of Lztfl1 targeting vector and wild-type and Lztfl1 knockout alleles and locations of primers used in the study. The global Lztfl1 knockout mouse was generated as described by Dr. Natarajan’s laboratory [24]. Briefly, this mouse line was generated via the conventional embryonic stem (ES) cell-based gene-targeting approach, using an ES cell clone in which the Lztfl1 gene was disrupted by inserting a DNA fragment with a LacZ-neo cassette between exons 3 and 4 of Lztfl1. A duplex PCR was set up by using KOL 1 & 2 or KOL 1 & 6 along with Gapdh. KO duplex PCR: KOL1 & 2 and Gapdh. Wild type (WT) duplex PCR: KOL 1& 6 and Gapdh. All samples should have a 452bp Gapdh band whether or not they have a KO or WT band. KO would give a band shorter than 452 bp Gapdh band. Wild type would produce a longer than 452bp Gapdh band. Het would have both KO & WT bands.
B. Representative PCR results showing mice with different genotypes. Upper panel: primer set to analyze Lztfl1 wide type genotyping; lower panel: primer set to detect Lztfl1 knockout genotyping.
Notice that sperm density of the control (heterozygote, Het) mice is higher than those observed in the Lztfl1 KO mice under the same dilution.
Representative images of testicular cells isolated from control (heterozygote) (a-e) and Lztfl1KO mice (f-j) stained with specific anti-LZTFL1 antibody (red), a lectin acrosome marker, peanut agglutinin (PNA) (green) and anti-α-tubulin antibody (green). No specific LZTFL1 signal was detected in the negative control using rabbit IgG in the control (a) and Lztfl1KO mouse (f). LZTFL1, peanut-lectin and α-tubulin staining was observed in spermatids from the germ cells from the control mouse (b-e), but no LZTFL1 staining was observed in germ cells of the Lztfl1 KO mouse (g-j). Spermatocytes were indicated by arrows, round spermatids were indicated by dotted arrows, elongating spermatids were indicated by arrow heads, and developing tails were indicated by stars in all images. Images were taken with a fluorescence microscopy.
A. Disrupted fibrous sheath (arrow heads in a) and abnormally developed chromatin (arrow in b) were observed in the Lztfl1 KO sperm;
B. Normal acrosome and mitochondria sheath in the sperm of Lztfl1 KO mice.
The arrows point to acrosome; the arrow heads point to mitochondria sheath. The dashed arrows point to normal chromatin in the control (a) and Lztfl1 KO (b, c) mice.
Three independent IVF experiments were performed. The number of embryos developed to 2-cell stage and live-born pups after embryo transferred were recorded.
Highlights.
LZTFL1 is required for normal mouse male fertility
Deficiency of LZTFL1 results in abnormal sperm
LZTFL1 is possibly a downstream molecule of IFT27 in mouse testis
Acknowledgments
This research was supported by National Institutes of Health (grant number HD076257), Wayne State University Start-up fund and Wayne State University Research Fund (to ZZ); the National Institute of Allergy and Infectious Disease, in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health (to VN, grant number HHSN261200800001E); National Institutes of Health (DK76229 to JG); INSERM-Bettencourt Foundation; National Science Foundation of China (to LZ, grant number 81671514), Hubei Provincial Colleges and Universities Outstanding Young and Middle-aged Technological Innovation Team Project T (to LZ, grant number 2020003). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest
There is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- 1.KozMINSKI KG, et al. , A motility in the eukaryotic flagellum unrelated to flagellar beating. Proceedings of the National Academy of Sciences, 1993. 90(12): p. 5519–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scholey JM, Intraflagellar transport. Annual review of cell and developmental biology, 2003. 19(1): p. 423–443. [DOI] [PubMed] [Google Scholar]
- 3.Cole DG, The intraflagellar transport machinery of Chlamydomonas reinhardtii. Traffic, 2003. 4(7): p. 435–442. [DOI] [PubMed] [Google Scholar]
- 4.Follit JA, et al. , Characterization of mouse IFT complex B. Cell motility and the cytoskeleton, 2009. 66(8): p. 457–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown JM and Witman GB, Cilia and Diseases. BioScience, 2014. 64(12): p. 1126–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, et al. , Intraflagellar Transporter Protein (IFT27), an IFT25 binding partner, Is Essential For Male Fertility and Spermiogenesis In Mice. Developmental Biology, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aldahmesh MA, et al. , IFT27, encoding a small GTPase component of IFT particles, is mutated in a consanguineous family with Bardet–Biedl syndrome. Human molecular genetics, 2014. 23(12): p. 3307–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forsythe E and Beales PL, Bardet–Biedl syndrome. European journal of human genetics, 2013. 21(1): p. 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tayeh MK, et al. , Genetic interaction between Bardet-Biedl syndrome genes and implications for limb patterning. Human molecular genetics, 2008. 17(13): p. 1956–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rohrschneider K and Bolz HJ, The Bardet-Biedl Syndrome-Diagnosis and Follow-up. Klinische Monatsblatter fur Augenheilkunde, 2020. 237(3): p. 239–247. [DOI] [PubMed] [Google Scholar]
- 11.Nakayama K and Katoh Y, Ciliary protein trafficking mediated by IFT and BBSome complexes with the aid of kinesin-2 and dynein-2 motors. The Journal of Biochemistry, 2018. 163(3): p. 155–164. [DOI] [PubMed] [Google Scholar]
- 12.Eguether T, et al. , IFT27 links the BBSome to IFT for maintenance of the ciliary signaling compartment. Developmental cell, 2014. 31(3): p. 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakurai T, et al. , Involvement of leucine zipper transcription factor-like protein 1 (Lztfl1) in the attenuation of cognitive impairment by exercise training. Biochemical and biophysical research communications, 2011. 416(1–2): p. 125–129. [DOI] [PubMed] [Google Scholar]
- 14.Khan S, et al. , Genetics of human Bardet–Biedl syndrome, an updates. Clinical genetics, 2016. 90(1): p. 3–15. [DOI] [PubMed] [Google Scholar]
- 15.Kiss H, et al. , The LZTFL1 gene is a part of a transcriptional map covering 250 kb within the common eliminated region 1 (C3CER1) in 3p21. 3. Genomics, 2001. 73(1): p. 10–19. [DOI] [PubMed] [Google Scholar]
- 16.Kok K, Naylor SL, and Buys CH, Deletions of the short arm of chromosome 3 in solid tumors and the search for suppressor genes, in Advances in cancer research. 1997, Elsevier. p. 27–92. [DOI] [PubMed] [Google Scholar]
- 17.Wei Q, et al. , Tumor-Suppressive Functions of Leucine Zipper Transcription Factor–Like 1. Cancer research, 2010. 70(7): p. 2942–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, et al. , LZTFL1 suppresses gastric cancer cell migration and invasion through regulating nuclear translocation of β-catenin. Journal of cancer research and clinical oncology, 2014. 140(12): p. 1997–2008. [DOI] [PubMed] [Google Scholar]
- 19.Seo S, et al. , A novel protein LZTFL1 regulates ciliary trafficking of the BBSome and Smoothened. PLoS Genet, 2011. 7(11): p. e1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang H, et al. , LZTFL1 upregulated by all-trans retinoic acid during CD4+ T cell activation enhances IL-5 production. The Journal of Immunology, 2016. 196(3): p. 1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Promchan K and Natarajan V, Leucine zipper transcription factor-like 1 binds adaptor protein complex-1 and 2 and participates in trafficking of transferrin receptor 1. Plos one, 2020. 15(1): p. e0226298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marion V, et al. , Exome sequencing identifies mutations in LZTFL1, a BBSome and smoothened trafficking regulator, in a family with Bardet–Biedl syndrome with situs inversus and insertional polydactyly. Journal of medical genetics, 2012. 49(5): p. 317–321. [DOI] [PubMed] [Google Scholar]
- 23.Schaefer E, et al. , Mesoaxial polydactyly is a major feature in Bardet–Biedl syndrome patients with LZTFL1 (BBS17) mutations. Clinical genetics, 2014. 85(5): p. 476–481. [DOI] [PubMed] [Google Scholar]
- 24.Jiang J, et al. , Depletion of BBS protein LZTFL1 affects growth and causes retinal degeneration in mice. Journal of Genetics and Genomics, 2016. 43(6): p. 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang A, et al. Dynamic interactions of ABHD5 with PNPLA3 regulate triacylglycerol metabolism in brown adipocytes. Nature Metabolism, 2019. 1(5): p. 560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Visconti PE, et al. , Capacitation of mouse spermatozoa. I. Correlation between the capacitation state and protein tyrosine phosphorylation. Development, 1995. 121(4): p. 1129–1137. [DOI] [PubMed] [Google Scholar]
- 27.McDonald K, Osmium ferricyanide fixation improves microfilament preservation and membrane visualization in a variety of animal cell types. Journal of ultrastructure research, 1984. 86(2): p. 107–118. [DOI] [PubMed] [Google Scholar]
- 28.Matthew A, et al. Endogenous and Synthetic ABHD5 Ligands Regulate ABHD5-Perilipin Interactions and Lipolysis in Fat and Muscle. Cell Metabolism, 2015. 22(5): p. 851–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishikawa H and Marshall WF, Ciliogenesis: building the cell’s antenna. Nature reviews Molecular cell biology, 2011. 12(4): p. 222–234. [DOI] [PubMed] [Google Scholar]
- 30.Wei Q, et al. , Lztfl1/BBS17 controls energy homeostasis by regulating the leptin signaling in the hypothalamic neurons. Journal of molecular cell biology, 2018. 10(5): p. 402–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.San Agustin JT, Pazour GJ, and Witman GB, Intraflagellar transport is essential for mammalian spermiogenesis but is absent in mature sperm. Molecular Biology of the Cell, 2015. 26(24):4358–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nachury MV, et al. , A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell, 2007. 129(6): p. 1201–1213. [DOI] [PubMed] [Google Scholar]
- 33.Pedersen LB, Geimer S, and Rosenbaum JL, Dissecting the molecular mechanisms of intraflagellar transport in Chlamydomonas. Current Biology, 2006. 16(5): p. 450–459. [DOI] [PubMed] [Google Scholar]
- 34.Qin H, et al. , Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. The Journal of cell biology, 2004. 164(2): p. 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lechtreck K-F, et al. , The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. Journal of Cell Biology, 2009. 187(7): p. 1117–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei Q, et al. , The BBSome controls IFT assembly and turnaround in cilia. Nature cell biology, 2012. 14(9): p. 950–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ou G, et al. , Functional coordination of intraflagellar transport motors. Nature, 2005. 436(7050): p. 583–587. [DOI] [PubMed] [Google Scholar]
- 38.Wingfield JL, Trafficking of ciliary membrane proteins by the intraflagellar transport/BBSome machinery. Essays in biochemistry, 2018. 62(6): 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Young SA, et al. , CABYR is essential for fibrous sheath integrity and progressive motility in mouse spermatozoa. Journal of cell science, 2016. 129(23): p. 4379–4387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HEK293 cells were transfected plasmids to express IFT27/C-Luc and N-Luc/LZTFL1, and non-transfected HEK293 cells were used as controls. The cell lysates were collected for Western blot analysis by using specific anti-IFT27 and anti-LZTFL1 antibodies. IFT27/C-Luc (left) and N-Luc/LZTFL1 (right) proteins were detected in the transfected HEK293 cells respectively.
100 μg of total protein was loaded and the LZTFL1 protein was examined by a higher sensitive Femto system. The anti-LZTFL1 antibody concentration: 1:400. LZTFL1was highly abundant in the testis, and it was also detected in brain, spleen, lung and liver.
A. Structures of Lztfl1 targeting vector and wild-type and Lztfl1 knockout alleles and locations of primers used in the study. The global Lztfl1 knockout mouse was generated as described by Dr. Natarajan’s laboratory [24]. Briefly, this mouse line was generated via the conventional embryonic stem (ES) cell-based gene-targeting approach, using an ES cell clone in which the Lztfl1 gene was disrupted by inserting a DNA fragment with a LacZ-neo cassette between exons 3 and 4 of Lztfl1. A duplex PCR was set up by using KOL 1 & 2 or KOL 1 & 6 along with Gapdh. KO duplex PCR: KOL1 & 2 and Gapdh. Wild type (WT) duplex PCR: KOL 1& 6 and Gapdh. All samples should have a 452bp Gapdh band whether or not they have a KO or WT band. KO would give a band shorter than 452 bp Gapdh band. Wild type would produce a longer than 452bp Gapdh band. Het would have both KO & WT bands.
B. Representative PCR results showing mice with different genotypes. Upper panel: primer set to analyze Lztfl1 wide type genotyping; lower panel: primer set to detect Lztfl1 knockout genotyping.
Notice that sperm density of the control (heterozygote, Het) mice is higher than those observed in the Lztfl1 KO mice under the same dilution.
Representative images of testicular cells isolated from control (heterozygote) (a-e) and Lztfl1KO mice (f-j) stained with specific anti-LZTFL1 antibody (red), a lectin acrosome marker, peanut agglutinin (PNA) (green) and anti-α-tubulin antibody (green). No specific LZTFL1 signal was detected in the negative control using rabbit IgG in the control (a) and Lztfl1KO mouse (f). LZTFL1, peanut-lectin and α-tubulin staining was observed in spermatids from the germ cells from the control mouse (b-e), but no LZTFL1 staining was observed in germ cells of the Lztfl1 KO mouse (g-j). Spermatocytes were indicated by arrows, round spermatids were indicated by dotted arrows, elongating spermatids were indicated by arrow heads, and developing tails were indicated by stars in all images. Images were taken with a fluorescence microscopy.
A. Disrupted fibrous sheath (arrow heads in a) and abnormally developed chromatin (arrow in b) were observed in the Lztfl1 KO sperm;
B. Normal acrosome and mitochondria sheath in the sperm of Lztfl1 KO mice.
The arrows point to acrosome; the arrow heads point to mitochondria sheath. The dashed arrows point to normal chromatin in the control (a) and Lztfl1 KO (b, c) mice.
Three independent IVF experiments were performed. The number of embryos developed to 2-cell stage and live-born pups after embryo transferred were recorded.


