Abstract
Microbiomes are complex and ubiquitous networks of microorganisms whose seemingly limitless chemical transformations could be harnessed to benefit agriculture, medicine, and biotechnology. The spatial and temporal changes in microbiome composition and function are influenced by a multitude of molecular and ecological factors. This complexity yields both versatility and challenges in designing synthetic microbiomes and perturbing natural microbiomes in controlled, predictable ways. In this review, we describe factors that give rise to emergent spatial and temporal microbiome properties and the meta-omics and computational modeling tools that can be used to understand microbiomes at the cellular and system levels. We also describe strategies for designing and engineering microbiomes to enhance or build novel functions. Throughout the review, we discuss key knowledge and technology gaps for elucidating the networks and deciphering key control points for microbiome engineering and highlight examples where multiple omics and modeling approaches can be integrated to address these gaps.
Keywords: microbiome, synthetic biology, genetic engineering, computational modeling, meta-omics, microbial interaction network
INTRODUCTION
Diverse communities of microorganisms inhabit every known environment, including oceans, soil, the surface and proximity of plants, and the intestines of humans, animals, and insect wing to the myriad functions they perform, from biogeochemical cycling of nutrients to transforming dietary substrates into nutrients for multicellular hosts, microbiomes attract immense attention from industry and academic researchers alike. Efforts to understand and engineer microbiomes frequently require integrated approaches that blur the lines between microbiology, ecology, medicine, computer science, mathematics, and engineering. Natural and synthetic microbiomes that robustly perform target functions could be exploited to address grand challenges in human health, agriculture, bioremediation, and bioprocessing that face society.
Target microbiome engineering goals include the ability to predictably modulate community composition, enhance existing functions, or install novel capabilities. Harnessing the properties of microbiomes remains difficult because we do not yet fully understand the molecular and ecological mechanisms that govern systems-level behaviors, and therefore we lack the capability to predict their multifunctional properties. Microbiomes are immensely complex; they can consist of hundreds to thousands of organisms, exhibit spatial and temporal variability, and establish dynamic feedback loops with the environment. A detailed and quantitative understanding of microbiomes could ultimately inform the design of interventions to predictably modify system properties or guiding principles for how to construct desired community functions from the bottom up. Exploiting and understanding the full functional potential of microbiomes necessitate integration of multiple experimental and computational methodologies that bridge many different disciplines.
Here, we describe tools currently used to understand microbiomes in diverse habitats and outline how meta-omics tools may be used and integrated to characterize microbiome composition and function. We describe representative case studies that showcase the various spatial and temporal scales that influence the composition and collective function of microbiomes, and we describe how interactions between microorganisms can lead to emergent functions that cannot be predicted on the basis of each community member’s behaviors in isolation. We discuss the relative advantages of several microbiome modeling approaches of varying degrees of coarse-graining and highlight recent efforts to integrate multi-omics data and multiscale considerations into a single model. Finally, we present recent successes in microbiome engineering and biocontainment of engineered communities and provide perspectives on how to move the field closer to wide-scale deployment of engineered communities for broad applications in bioenergy, agriculture, and human health. Throughout the review, we highlight opportunities to improve our understanding of the causal links between microbiome composition and function and our ability to engineer them for societal benefit.
Meta-omics: omics analyses (e.g., genomics and transcriptomics) applied to microbial communities to characterize numerous organisms or metabolites at once
Multi-omics: the integration and analysis of multiple omics datasets, often for specified biological samples under a variety of environmental conditions
Biocontainment: measures to prevent escape of genetically engineered organisms or transfer of genes (e.g., antibiotic resistance) to organisms in the environment
MICROBOMES OPERATE ON MANY SPATIAL AND TEMPORAL SCALES
Microbiome functions are driven by myriad abiotic and biotic forces that span multiple length scales and timescales (Figure 1). Abiotic parameters include nutrient availability, oxygen, and pH as well as physical properties of the environment; biotic factors include the complex interactions between different organisms. Microbial interactions are mediated by metabolite transformations, including competition for limited resources and release of compounds that benefit (or inhibit) the growth of other organisms. Microbial ecological strategies are dictated by intracellular networks that sense and respond to environmental parameters and dynamically regulate cellular behaviors. In addition, functional capabilities of microbes can be altered due to changes in their genetic information, which enables evolutionary adaptation. Although microbial communities can be highly sensitive to variations in environmental parameters (e.g., antibiotics or nutrient shifts), the complex web of interactions within microbiomes can enhance resilience to environmental perturbations. In this section we discuss some of the ecological, structural, and functional aspects of microbiomes, spanning from the molecular to the systems level, and the implications for microbiome engineering.
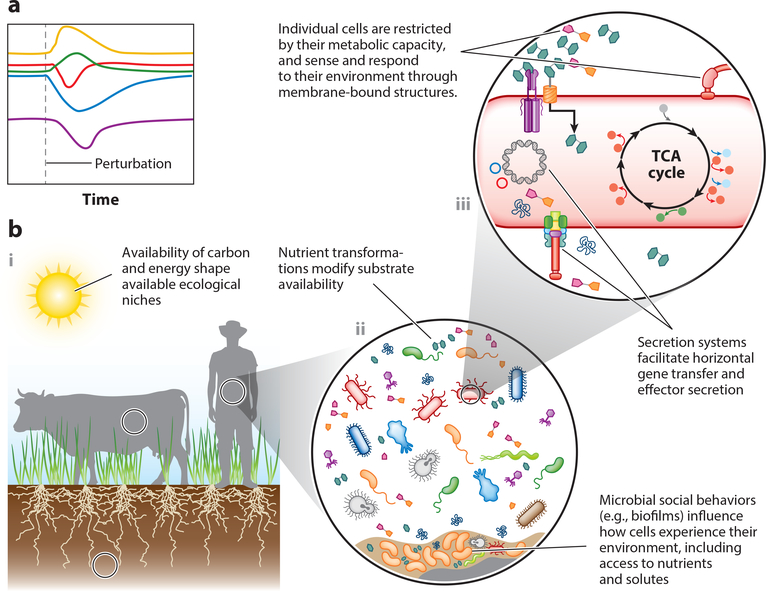
Figure 1.
(a) Microbiome stability is a function of the community’s ability to recover its original functions following a disturbance. Functions could include the production of specific molecules over time. (b) Microbiome composition and function are shaped by multiple spatial and temporal factors. (i) At the broadest scale, availability of carbon and energy defines possible ecological niches within each environment. (ii) Within a community, interspecies interactions, including social behaviors, modulate how cells respond to their environment by modifying substrate availability (i.e., syntrophy or competition), releasing effectors, or occupying available space. Biofilms, in particular, provide resilience to specific environmental perturbations. (iii) Within each cell, function is constrained by individual metabolic capacity, which can be altered through genetic mutations or horizontal gene transfer. Membrane-bound proteins transduce signals and molecules from the environment to the cytosol as well as facilitate secretion of products from the cell to the environment. Figure adapted from images created with BioRender.com.
Resilience to environmental perturbation: the magnitude of a perturbation that a system can tolerate while maintaining specific functional activities
Ecological Niches Govern Structure, Functions, and Resilience of Microbiomes
Microbiome composition in any environment is governed by the availability of ecological niches, and microbes have evolved to exploit nearly all imaginable niches. Niche diversification within microbiomes coupled with functional redundancy can enhance resilience to environmental perturbations. Non-host-associated microbial communities are key drivers of biogeochemical cycles and persist in a broad spectrum of environments on an astounding diversity of substrates (1–3). In host-associated microbiomes such as the mammalian gastrointestinal tract, microbes subsist on dietary substrates consumed by the host or use host-derived nutrients such as mucus (4). Similarly, plant-associated microbes can utilize plant exudates (5, 6). Although microbiomes can be resilient to specific environmental parameters, perturbation in parameters such as nutrient availability can cause dramatic shifts in composition and function, leading to a boom-and-bust cycle of growth of lower-abundance taxa as microbes seize on resource fluctuations, as evidenced by, for example, algal blooms (7). A similar phenomenon is observed when the nutrient landscape changes in the gastrointestinal tract owing to environmental disturbances or host dysfunctions (8).
Ecological niche: the set of parameters that enables growth and survival of individual members of a community
For host-associated microbiomes, millennia of coevolution have established bidirectional interactions far beyond the canonical functions of pathogen resistance and the breakdown and exchange of nutrients. For example, it has become apparent that the gut microbiome is closely associated with neurological disorders such as depression, anxiety, Alzheimer’s disease, and schizophrenia, leading to the recognition of a neurointestinal connection known as the gut-brain axis (9–11), although work remains in parsing the causality of these observed associations (12). Evolutionary theory informs the hypothesis that although many host-associated microbes are symbiotic, they are also under strong selective pressure to compete with other community members even at the cost of their beneficial functions to the host (13, 14).
Microbial Interactions Determine Microbiome Behavior and Stability
Microbial interactions are multidimensional and diverse mechanisms can combine to influence the net effect of one organism on another. At a coarse-grained level, the net effect of an organism on another can be represented as competitive (−/−), mutualist (+/+), predative (+/−), neutral (0/0), amensal (0/−), or commensal (0/+). Positive and negative interactions can be established by diverse molecular mechanisms, including resource competition and release of molecules, that affect growth rates and metabolic activities (15). Microbial interactions change as a function of time due to intracellular networks that sense and respond to environmental shifts. Nevertheless, simplified mathematical models that represent single-organism growth parameters and pairwise microbial interactions have been shown to predict multispecies community behaviors (16–18). How these microbial interaction networks map to community-level properties such as stability, diversity, and function remains largely unresolved. Theoretical work has demonstrated that negative interactions can drive networks toward stability (19), whereas positive interactions have been associated with enhanced diversity and metabolic activities (20, 21). To gain a predictive understanding of how microbial interaction networks are shaped by environmental stimuli and combine to generate community-level properties, we must consider the molecular mechanism of interactions as well as how they change as a function of time and spatial proximity.
Stability: rate of return to a system’s steady state in response to an environmental perturbation
Spatial Organization and Biofilms Are Key Properties of Microbiome Resilience
Microbial populations exhibit heterogenous spatial distributions in natural environments. The spatial organization of a microbiome is a major determinant of systems-level properties, including community metabolism, and response to environmental perturbations, such as antibiotics (22–24). In biofilms, a predominant form of microbial life in nature (25), the occurrence of intracolony channels can mediate the transfer of molecules to cells that are deeply embedded in the matrix (26). Although biofilms can be exploited for specific applications such as lignocellulose valorization (27) and environmental remediation (28), they can be detrimental in the context of human disease (29). Beyond biofilms, microbes can also self-organize to establish complex physical structures in the environment. For example, fungi play an integral role in maintaining the spatial structure of the soil matrix and soil bacteria can use fungal phyla to traverse pores in the soil that would otherwise prohibit dispersal (30–32). Therefore, the spatial organization of a microbiome can be harnessed or targeted to alter system properties.
At a smaller scale, membrane proteins such as receptors and transporters are key drivers of microbiome function and are involved in cellular processes as diverse as sensing and uptake of nutrients, secretion of enzymes and small molecules, virulence, motility, and adhesion. For example, bacterial two-component systems are involved in quorum sensing (33) and chemotaxis (34), and substrate-specific transporter proteins are central to metabolic cross-feeding and flux through biochemical pathways (35). These proteins enable microbial constituents to respond to changes as they enter and exit habitats to which they are well (or ill) adapted, all while maintaining a favorable intracellular environment for metabolism, and they can be engineered to influence substrate utilization, production of target molecules, and formation of biofilms (36–39) (see the sidebar titled Membrane Proteins).
MEMBRANE PROTEINS.
Membrane proteins are at the core of how microorganisms interact with each other and with their environment. All cells navigate their environment by using membrane-embedded receptor proteins to sense and respond to changes in their environment. Typical receptor proteins, such as bacterial two-component systems, have a domain that is exposed to the extracellular environment and that recognizes a ligand with high specificity. Ligand binding induces a conformational shift in the receptor protein that leads to an intracellular response mediated, for example, by enzymatic cascades or by protein–DNA interactions. The extracellular ligands comprise a tremendous variety of ions and molecules (215). Motor proteins allow cells to swim toward nutrients and away from toxins; adhesion proteins facilitate cell–cell connections and adhesion to the environment. Meanwhile, a large variety of transporter proteins enable the efficient and highly selective uptake of nutrients as well as secretion of waste products, enzymes, and effector molecules. About one-quarter of the genes in any typical organism appear to encode integral membrane proteins that are involved in cell development and growth, energy conversions, communication, mobility, defense, and virulence (216, 217).
Investigating and Designing Microbiomes from the Bottom Up and Top Down
Microbiomes can be studied and engineered through two different and complementary approaches: top down and bottom up (15, 40, 41). A top-down approach investigates natural communities by introducing them into highly controlled laboratory environments. Such top-down manipulations can be used to understand microbiome dynamics and functions in response to environmental inputs (e.g., nutrient availability or antibiotic stress) (42). By contrast, isolated species can also be assembled in vitro to form synthetic microbial communities, which have reduced complexity compared with natural microbiomes and greater controllability via manipulation of initial community composition (16, 43). Although molecular and ecological mechanisms of synthetic communities can be more easily dissected, these simplified systems can display reduced temporal stability in composition and/or function, limiting their deployment in real-world environments and for biotechnological applications (41). At the core, if we understand the temporal changes in “which microbe is there” and “which microbe can do what, when, and how,” we become better equipped to tailor microbiomes for, say, medical and agricultural purposes. A promising approach is to combine ecological studies, quantitative measurements, and computational modeling to map the functional potential of microbiomes with increasing resolution (44).
Synthetic microbial community: combination of three or more individual microorganisms (often not isolated from the same habitat) in a single environment
TOOLS FOR UNDERSTANDING MICROBIOMES
To harness the properties of microbiomes, we must develop tools that decipher which microbes have the capability and flexibility to perform specific functions, quantify their functional activities across space and time, and decipher interactions between organisms and between organisms and the environment. High-throughput sequencing has significantly enhanced our ability to investigate microbiome composition and functional activities, as today’s next-generation and emerging so-called third-generation sequencing tools (45) can rapidly process billions of DNA base pairs with continuous read lengths greater than 100 kbp (46) (>2% of the average bacterial genome size). These technologies enable characterization of phylogeny (amplicon sequencing), functional potential (shotgun metagenomics), and gene expression (metatranscriptomics) in thousands of species or synthetic communities simultaneously. Beyond nucleic acid sequencing, metaproteomics and metabolomics can analyze the activities of microbiomes by quantifying the abundance of enzymes that perform key chemical transformations and the metabolites that mediate interspecies interactions (Figure 2). Such meta-omics tools can be applied to quantify the spatial distribution of organisms within microbiomes, characterize low-abundance members and assess cellular heterogeneity, identify the organisms that perform key chemical transformations, and elucidate the web of metabolic interactions that ultimately drives microbiome functions.
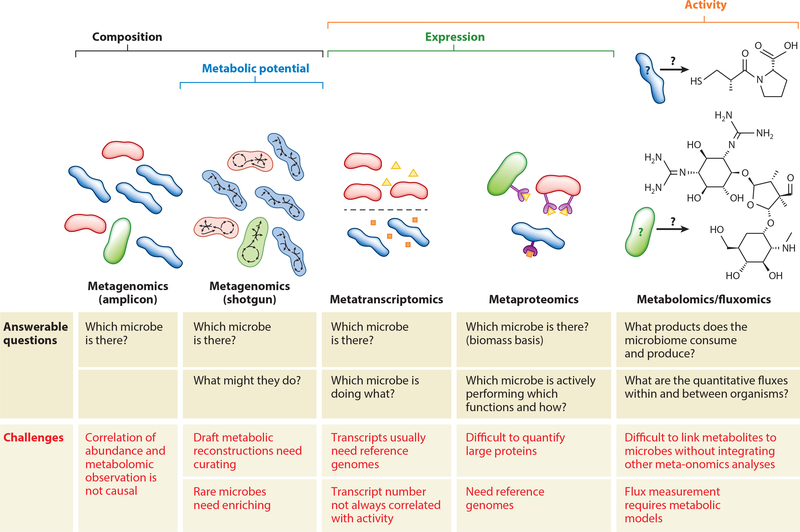
Figure 2.
Different meta-omics tools are suited to answer different questions about microbiome composition and function. Amplicon metagenomics can reveal which organisms are present in a microbiome but not necessarily what each microbe’s role in the community is. Shotgun metagenomics elucidates which microbes are present in the community and what functions they have the capacity to perform. Metatranscriptomics and metaproteomics are necessary to uncover which functions are actually being performed in the community; assigning these transcripts and proteins to the microbes that produced them typically requires high-quality reference genomes or concurrent metagenomics analyses. Metabolomics and fluxomics quantify the chemical composition of the microbiome environment; however, linking metabolites to the microbes that produce or consume them is challenging, even with reference genomes. Linking microbiome composition and function is facilitated by integrating multiple meta-omics techniques, for example, concurrent shotgun metagenomics, metaproteomics, and metabolomics studies to assess which enzymes are producing an observed small molecule of interest and which microbes could produce those enzymes.
Quantifying Microbiome Composition and Functional Potential via Metagenomics
Quantitative measurements of microbial abundance are critical for understanding the spatial and temporal behaviors of microbiomes. Microbiome composition is frequently determined via highly conserved marker genes for ribosomal RNA (rRNA), usually the 16S rRNA gene in prokaryotes and the 18S, 28S, and internal transcribed spacer regions in eukaryotes (47). Amplicon sequencing is particularly useful for characterizing community composition in systems contaminated by host DNA or in samples with low DNA template concentrations. However, these methods provide a genus-level resolution, and sequence-dependent variations in (nominally) universal primer affinity between clades, along with extraction efficiencies of DNA and variation in gene copy numbers (48, 49), can bias abundance results.
In shotgun metagenomics, a library is constructed with all community DNA and the reads can be assembled into genomes of individual species called metagenome-assembled genomes (MAGs). These genome sequences provide simultaneous quantification of the microbiome’s functional potential and its phylogenetic composition. Shotgun metagenomics has several advantages, including finer phylogenetic resolution than amplicon-based sequencing down to the strain level (50–53) and detection of viral DNA, with the trade-off that more reads are needed to confidently quantify more genes. To probe rare microbes with potentially unique functions, researchers have leveraged DNA extraction methodologies targeted for different species to assemble near-complete genomes of bacteria present below 1% relative abundance within a community by employing differential coverage binning (54). In addition, the variations in genome copy number in different regions of the chromosome have been used to infer the bacterial replication rates in natural environments (55), providing key insights into the distribution of metabolic activity states within a community.
Differential coverage binning: extraction of nucleic acid samples by multiple techniques to improve resolution of species and strains
Sequencing-based methods provide relative abundance or compositional data, which presents challenges for statistical analyses and can lead to spurious correlations. Therefore, methods to quantify absolute abundance, including determining correlations between organisms, growth rates, per-cell metabolic activities, or total microbial loads present in a host, are critical for understanding microbiomes. Absolute DNA-based quantification of microbiome composition is a major challenge that has been approached with spike-in (56), quantitative polymerase chain reaction (PCR) (57), flow cytometry (58), and total DNA quantification (59) methods; however, all of these methods have inherent biases and limitations. Recently, amplicon sequencing was coupled with digital PCR in a microfluidic format for absolute DNA-based quantification of microbiome composition with the advantage of evaluating concurrently the limits of both clade detection and clade quantification (60).
Researchers use sequencing methods to study the spatial distribution of clades within a microbiome by sampling from different locations. This approach has been used to map the biogeography of the mammalian gut (61) and to demonstrate how anaerobic digestion communities self-assemble into distinct microbiomes when reactors are connected in series (62). Although the spatial resolution that can be achieved by sampling different locations is limited (~20 μm), micron- and nanometer-level spatial variation can be elucidated by imaging approaches (see the sidebar titled Microbiome Imaging). To uncover how spatial clustering of clades influences community functions and interactions, researchers must elucidate both the spatial distribution of functions and the identity of organisms.
MICROBIOME IMAGING.
Fluorescence in situ hybridization (FISH) and mass spectrometry imaging (MSI) techniques may be used to visualize localization of phylogenetic groups and specific molecules and isotopes, respectively. In FISH, a fluorescent reporter is attached to a probe with a nucleic acid sequence complementary to that of a target sequence in the microbiome, often for ribosomal RNA. The specificity of the FISH probe can vary, targeting an entire kingdom to a single species or genus, and if multiple probes are used for one sample, a different fluorescent reporter can be attached to each probe for simultaneous resolution of numerous microbial clades at the micron scale (218, 219).
In MSI, the spatial organization of both microbes and small molecules within a microbiome can be quantified over nanometer lengths, as reviewed in Reference 220. When integrated with FISH (221) or stable isotope probing (SIP) (222; described below), nanoscale secondary ion mass spectrometry (NanoSIMS) can map the location of both microbes and specific metabolites. In SIP-NanoSIMS, the fate of specific moieties derived from an isotope-labeled substrate can be quantified with nanometer resolution and linked to microbial identity, enabling quantification of localized functional activity within microbiomes.
In vivo imaging of microbiomes in systems such as the human gut promises to improve our understanding of host–microbiota interactions and how these vary in space and time. However, in the gut and other anaerobic systems, such as tumor interiors, microbe labeling with fluorescent proteins is difficult, as most fluorescent proteins require oxygen to develop their chromophores, and anaerobic fluorescent proteins are typically not bright enough to be deployed in vivo. Researchers have overcome these limitations by fluorescently labeling selected gut bacteria and their polysaccharides with bio-orthogonal click chemistry (222a) and by growing and imaging bacterial communities in low oxygen concentrations in which obligate anaerobes survive but chromophores still form (222b). See References 104 and 222c for reviews and perspectives on live-cell anaerobic imaging and related in vivo applications.
By evaluating putative metabolic pathways, as well as which metabolites may be taken up or secreted by certain microbes, researchers can use the information in MAGs to predict the chemical transformations the system is capable of performing. For example, lignocellulose-degrading enzymes in the porcupine gut microbiome were identified via shotgun metagenomics and expressed in Escherichia coli, leading to the discovery of an active endo-1,4-β-xylanase even though the microbe encoding this gene was unknown (63). Accurate predictions of functional potential rely heavily on reference genomes and metagenomics datasets, available in databases such as those outlined in Table 1.
Table 1.
Bioinformatic databases and tools for reference genomes, metabolite databases, genome browsers, and sequence-based membrane protein predictors
| Database/tool | Notes | Reference | |
|---|---|---|---|
| Primary sequence databases | NCBI (GenBank)a | Primary American sequence database NCBI includes more specific databases such as dbSNP for human SNPs, dbVar for larger human variants, and the Sequence Read Archive for raw sequence data, among others. | 201 |
| RefSeq | NCBI-maintained and annotated versions of GenBank assemblies | 202 | |
| ENAa | Primary European sequence database | 203 | |
| DDBJa | Primary Japanese sequence database | 204 | |
| Collections of bioinformatic data with links to primary databases | National Bioscience Database Center | Contains portals to many databases with specific focuses | 205 |
| KEGG | Searchable for links to genomic information in primary databases; includes genomes and metagenomes as well as pathway and reaction information | 206 | |
| MINE | Metabolite reference database searchable by name or structure; shows related biochemical reactions and pathways involving each metabolite | 84 | |
| Annotated genome browsers | IMG | Includes microbiome metagenome browser and is linked to genomic data stored in NCBI | 207 |
| JGI MycoCosm | Genome browser for annotated fungal genomes Contains functional annotation browsing with an emphasis on CAZymes, secondary metabolism clusters, transporters, and transcription factors | 208 | |
| JGI PhycoCosm | Similar to MycoCosm for algal genomes; newer | 209 | |
| JGI Phytozome | Genome browser for annotated plant genomes | 210 | |
| Sequencebased membrane protein predictors | SignalP5.0 | Predicts canonical signal peptides in protein sequences | 211 |
| TMHMM | Predicts transmembrane helices in protein sequences | 212 | |
| TOPCONS | Consensus prediction of transmembrane helices in protein sequences | 213 | |
Part of the International Nucleotide Sequence Database Collaboration (INSDC). BioProject, for collections of biological data related to single efforts like HMP, is shared among the INSDC. Individual projects are accessible through one of the repositories (214).
Abbreviations:DDBJ, DNA Data Bank of Japan; ENA, European Nucleotide Archive; HMP, Human Microbiome Project; IMG, Integrated Microbial Genomes and Microbiomes; JGI, Joint Genome Institute; KEGG, Kyoto Encyclopedia of Genes and Genomes; MINE, Metabolic In Silico Network Expansion; NCBI, National Center for Biotechnology Information; SNP, single nucleotide polymorphism; TMHMM; TOPCONS.
Our ability to collect metagenomics data has outpaced our ability to functionally annotate the data for interpretation of biological context (i.e., which microbe has the capability to do what in the microbiome). In genetically tractable organisms, a powerful approach to functionally annotate genes involves quantification of strain fitness within pooled genome-wide mutant libraries that can be grown in monoculture or coculture (64) across many different environmental conditions (65). However, because most strains lack genetic tools, bioinformatic approaches, including the Integrated Gut Genomes Database and its associated IGGsearch tool (66) and the Distilled and Refined Annotation of Metabolism (DRAM) tool (67), can be used to analyze large sequencing datasets containing potentially thousands of interacting species.
Using Metatranscriptomics to Map Organism Identities to Functional Activities
Metatranscriptomics, by which total community messenger RNA (mRNA) is extracted, reverse transcribed to complementary DNA, and sequenced, provides insight into the potential functions performed by organisms within a community as well as which microbes may be performing them. For example, metagenomics and metatranscriptomics were combined to identify sugar-fermenting and fatty-acid-chain-elongating microbes in an anaerobic bioreactor and to propose routes of metabolite exchange between the clades (42). By quantifying nitrogen fixation transcripts, Gómez-Godínez et al. (68) reported that Azospirillum brasilense was the predominant nitrogen-fixing bacterium in a synthetic consortium of plant growth-promoting bacteria on maize roots. Although metatranscriptomics also generates many unannotated hypothetical genes, the ability to identify genes that change across different conditions greatly facilitates the downstream identification of genes involved in microbiome processes, including the breakdown of lignocellulose or the biogeochemical cycling of elements. Further, investigating the genome-wide transcriptional activity of organisms within a community may guide the development of hypotheses about mechanisms involved in observed microbiome states (e.g., dysbiosis and steady-state recovery after perturbation). For example, a combined metagenomics and metatranscriptomics study of the fecal microbiome of 308 adult men revealed that pathways that were encoded in the genomes of many members of the microbiome were actually transcribed by a small subset of species (69).
Notably, even when reference genomic data are lacking, metatranscriptomics can be used to mine microbiomes for enzymes with a desired function. A transcriptomics survey of anaerobic gut fungi harvested from the intestinal tract of herbivores revealed that these unusual and understudied eukaryotes produce an unrivaled array of biomass-degrading enzymes (70), marking these fungi as attractive targets for sourcing of valuable enzymes (71). Similarly, He et al. (72) identified 125,252 putative CAZymes in a sheep gut microbiome, most of which had less than 75% identity to known proteins in the CAZy database or the National Center for Biotechnology Information (NCBI) database, but 19 of the 30 tested candidates showed cellulase activity when heterologously expressed.
CAZyme: carbohydrate-active enzyme that degrades biomass into smaller, fermentable sugars
In microbes, mRNA transcripts represent less than 10% of the total RNA; therefore, rRNA should typically be removed prior to sequencing. Methods for prokaryotic rRNA depletion vary in efficacy on the basis of microbiome type (biofilm versus planktonic) and composition, and tool development is an active area of research (73). Methods for single-cell prokaryotic rRNA depletion are only beginning to show some success (74, 75) and will be useful for interrogating the unique activities of low-abundance organisms as well as the cell-to-cell heterogeneity of gene expression within a given species that is not observable with bulk methods. The identification of optimal spatial locations and time points to discover ecological driver organisms or novel biochemical pathways mediating microbiome functions remains unresolved. Further, because transcript number does not always correlate with protein abundance or activity, it is difficult to estimate from metatranscriptomics data alone the relative contribution of different metabolic reactions and pathways to the overall function of the microbiome.
Quantifying Microbiome Functional Capabilities via Metaproteomics
Metaproteomics, which studies all proteins recovered from a microbiome sample, can provide critical information about microbiome functional capabilities. Liquid chromatography can be coupled with mass spectrometry (LC-MS) or with tandem mass spectrometry (LC-MS/MS) to detect tens of thousands of peptides in one sample (76, 77). In the future, nanopore-based devices hold promise to revolutionize proteomics and biotechnology by enabling amino acid sequencing of intact proteins, allowing structural characterization of proteins larger than those characterized by LC-MS (78). In addition to differential enzyme expression, metaproteomics may be employed to quantify the abundance of individual organisms in a microbiome on a biomass basis (79), which offers an alternative method for microbiome composition. Indeed, proteinaceous biomass may be a better representation of composition for systems composed of eukaryotic and prokaryotic cells where the size and weight of organisms vary significantly, but may be perhaps more biased in cases where the intracellular protein content varies widely across species.
Beyond prospecting for genes with predicted functions, metaproteomics can be used to identify posttranslational modifications as well as directly quantify the abundance of proteins in a community, which may not necessarily correlate with transcript abundances (80). This approach is greatly improved by integration with metagenomics analysis owing to the difficulties of mapping fragmented peptide sequences to genes. In particular, mining microbiomes for membrane proteins such as transporters is critical for elucidating molecular mechanisms involved in interspecies interactions but remains challenging owing to their low abundance compared with soluble proteins and technically and analytically challenging owing to their hydrophobicity (81).
Metabolomics Reveals the Chemical Repertoire of Microbiomes
Microbes are exquisite chemists and these chemical mediators produced and utilized by constituent community members are a major driving force of microbiome functions. Metabolomics can be used to detect small-molecule metabolites, including intermediates and end products of cellular metabolism. Integration of metabolomics with metagenomics is particularly useful for formulating hypotheses about the role of measured metabolites in interspecies interactions and microbiome functions, as metabolites typically cannot be assigned to specific organisms. Therefore, the integration of other meta-omics tools is necessary to determine which metabolic pathways are active in a community and to hypothesize how different metabolites may be utilized, released, and exchanged to form an integrated community metabolic network.
Many metabolomics approaches employ gas chromatography (GC) instead of LC to precede MS analysis, offering greater chromatographic separation of metabolites. Nuclear magnetic resonance spectroscopy offers an alternative, more quantitative measure of metabolites without the sample preparation and derivatization steps required in MS studies but typically cannot detect metabolites below micromolar concentration (82). Untargeted metabolomics seeks to characterize the structures of as many metabolites present in the sample as possible that can be identified. However, it is impossible to characterize all classes of metabolites with a single solvent and column chemistry, and many metabolites in databases remain unannotated (83). Therefore, strategies must be developed to predict unknown chemical structures and link them to the microbes and biosynthetic pathways. Recently, researchers (84) developed the Pickaxe tool for generating novel metabolites and predicting the enzymes and putative pathways based on Enzyme Commission numbers and the MINE (Metabolic In Silico Network Expansion) database in Table 1.
Enzyme Commission number: a numerical classification of a metabolic enzyme based on the chemical reaction it performs
Investigating Metabolic Flux in Microbial Communities
Metabolic flux analysis (MFA) can quantify the distribution (flux) of carbon in cellular metabolism, directly measuring the activity of metabolic networks. When this method is used, cells are typically exposed to 13C-labeled carbon and the degree of labeling of biomass components like glycogen and proteins is quantified via GC-MS (85). On the basis of these data, researchers use an organism-specific metabolic model to estimate the flux through each pathway by using software such as METRAN. Although MFA has been used to study well-characterized and simplified communities (86), the challenge associated with assigning metabolites to microbes in complex communities has stymied the broader application of MFA. However, analysis of isotope-labeled peptides, which can be mapped to individual microbes with reference genomes by metaproteomics, may unlock MFA for microbiomes (87). Nevertheless, this method may perform best on communities composed of microbes with dissimilar metabolisms or of microbes that can be spatially separated. To quantify interspecies metabolic interactions and community-wide fluxes, researchers must develop metafluxomic protocols and software to translate isotopic labeling and multi-omics data to quantitative descriptions of community metabolic networks (88).
Stable Isotope Probing in Microbiomes
Stable isotope probing (SIP) has emerged as a promising technique to enrich for rare microbes, link microbe identities to functions, and investigate interaction networks within microbiomes. In DNA-SIP or RNA-SIP, isotope-labeled substrates are differentially taken up and incorporated into nucleic acids by community members. After extraction, nucleic acids are fractionated by density to simultaneously enrich for nucleic acids from rare microbes and link the affinity for the labeled substrate to microbe identity (89, 90). For example, DNA-SIP was used to curate a complete genome of a member of the phylum Saccharibacteria with less than 1× coverage in the bulk metagenome and to decipher the metabolite exchange networks within the Saccharibacteria’s surrounding community (91).
A major limitation is that the SIP culturing procedure may not mimic a microbiome’s natural microenvironment. Although most microbes remain uncultivated, meta-omics analyses may elucidate clues for isolating and culturing previously uncharacterized species (66). Nucleic acid–SIP requires that isotopes be incorporated directly into nucleic acids, and the incubation time with the isotope influences the degree of community labeling. For example, short incubation times exclude isotope uptake by slow-growing microbes, and long incubation times may lead to nonspecific cross-feeding of isotopes across the community, skewing which clades initially metabolized the substrate (92, 93).
In principle, SIP can also be used to quantify incorporation of labels into proteins (protein-SIP) (94), metabolites (metabolome-SIP), and phospholipid-derived fatty acids (PLFA-SIP), though linking these to microbe identity requires excellent reference genomes or concurrent metagenomics and/or metatranscriptomics analyses. When integrated with meta-omics analyses, SIP may link microbe identity to function and uncover interspecies metabolite exchange mechanisms and community-wide metabolic networks. For example, protein-SIP coupled with amplicon sequencing and shotgun metagenomics was used to map acetate metabolism to organism identity in anaerobic digester consortia (95). In addition, RNA-SIP, metagenomics, and metatranscriptomics have been integrated to characterize the predominant CO2 fixation pathways and their transcribing microbes in deep-sea hydrothermal vent microbiomes at a range of temperatures (92). Finally, DNA-SIP has been combined with differential coverage binning to enhance resolution of MAGs with specific activity in anaerobic digesters (96). See Reference 97 for a review of SIP applied to in vivo and ex vivo human and animal gut systems.
Microfluidic Devices for Microbiome Fractionation, Analysis, and Cultivation
Microfluidic devices have provided major advances in cell culturing and analysis by enabling micron-level spatial precision, temporal control of environmental inputs, and ultrahigh-throughput analysis of cellular and molecular systems. For example, physical separation of microbes within a microfluidic device can be used to sort clades on the basis of function for single-cell analysis or subsequent cultivation. Using droplet microfluidics, Schaerli & Hollfelder (98) generated picoliter to nanoliter aqueous droplets at kilohertz rates to create millions of compartments to study microbial behaviors. When coupled with DNA barcoding, this approach has enabled processing of more than 50,000 cells per run to sequence and assemble genomes from single cells (99). Microfluidics-enabled sorting of cells with desired genomic traits or metabolic activities is useful for screening strains that display greater enzyme productivity while consuming a millionfold fewer reagents (100).
In addition to single-cell screening, droplet microfluidics has enabled massively parallelized, compartmentalized culturing of human-associated intestinal organisms to enrich for low-fitness community members that would otherwise be outcompeted in a well-mixed culture (101). In addition, droplet microfluidics has been used to decipher pairwise and higher-order interactions in microbial communities across different environmental conditions (102). Coupling droplet microfluidics to a microwell array platform, by way of droplet dye barcoding and fluorescent reporters for strain abundance or metabolic activities, was used to construct and analyze approximately 100,000 subcommunities composed of 19 soil microbes (103). Although these methods enable the characterization of many subcommunities, they have relied largely on fluorescence labeling of specific strains, which is restricted to organisms that can be manipulated with genetic tools. Therefore, label-free methods are needed to more broadly apply these techniques to study diverse communities, including anaerobic systems (104).
Lower-throughput but higher-precision microfluidic devices have been used to investigate the effects of spatial separation on the dynamics of bacterial signaling information transmission and metabolic cross-feeding (105). In addition, microfluidic platforms have also been developed to study host–microbiome interactions (106). The intestine-on-a-chip can recapitulate physiologically relevant microenvironments such as oxygen gradients found within the human intestine as well as mechanical forces reflecting peristalsis (107). Although these techniques can capture critical features to summarize natural microenvironments and enable detailed control of the environment, a major limitation is the complexity of the design of the platforms, which is a barrier to widespread adoption by and deployment to end users.
COMPUTATIONAL MODELS TO PREDICT MICROBIOME DYNAMICS AND FUNCTIONS
Mathematical models, based on ecological, thermodynamic, and biochemical principles, can be used to simulate microbiome population dynamics and metabolic functions on many length scales and timescales. These models range from data-driven differential equation–based models of community composition and interactions to mechanistic genome-scale models of metabolic flux and interspecies metabolite exchange (Figure 3). In the sections below, we discuss relative advantages and limitations of several modeling approaches and how they may be used to enable microbiome engineering.
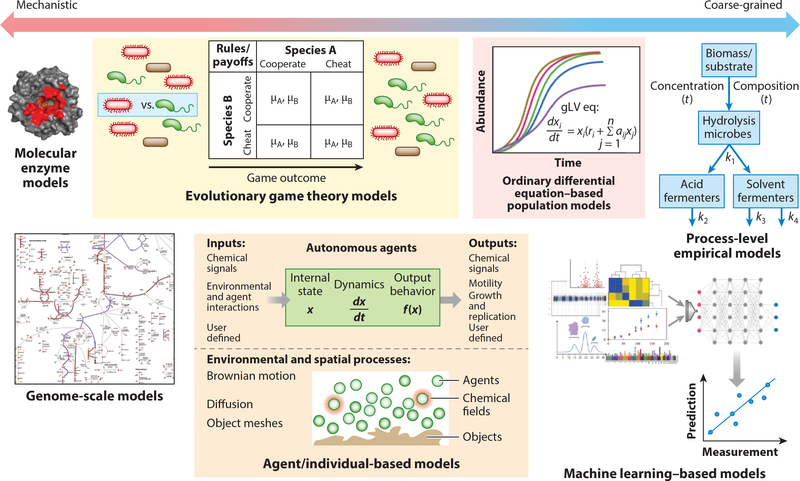
Figure 3.
Microbiomes can be modeled on many scales, and the choice of modeling technique depends on the question at hand. At the most mechanistic level, molecular simulations may be used to model the thermodynamics and kinetics of individual enzymes identified through metaproteomics; however, these are not scalable to encompass the entire microbiome. GEMs enable prediction of the metabolic fluxes and end-product profiles within a microbiome and can offer mechanistic insight into metabolomic observations given high-quality genomic reconstructions and sufficient experimental model validation. Evolutionary game theory models and differential equation–based models are particularly useful when microbiome population dynamics are of the greatest interest, because detailed metabolic reconstructions are not needed for each organism to be modeled. AbMs offer flexibility in that the user may define which inputs and outputs to include in the model and are often the technique of choice when integrating both metabolic and physical interactions between microbes. Data-driven models, including emerging machine learning–based models, offer empirical predictions of microbiome behaviors under specified conditions given appropriate training data. Although less mechanistic than GEMs, machine learning–based models are a pragmatic approach to synthesizing large amounts of different data types into interpretable conclusions, for example, rate constant estimations for process-level models of microbiome function. The structure of the molecular enzyme model is from PDB ID 4QLK. The structure of the AbM is reproduced with permission from Reference 162. The microbes in the evolutionary game theory models panel and the entire machine learning–based models panel were adapted from images created with BioRender.com. Abbreviations: AbM, agent-based model; EGT, evolutionary game theory; GEM, genome-scale model; ODE, ordinary differential equation; PDB, Protein Data Bank.
Ordinary Differential Equation and Evolutionary Game Theory Models of Microbiome Population Dynamics and Interactions
Ordinary differential equation (ODE) models, such as the generalized Lotka–Volterra (gLV) model (16), have been used to model microbiome population dynamics with time series data of absolute organism abundance for parameter estimation and experimental validation (108, 109). In the gLV model, the temporal changes in the abundance of each species are a function of its growth rate and intraspecies and interspecies interactions (110). Modified gLV models have been used to analyze dynamic behaviors, including the response to perturbations such as dilution rate and the response to antibiotics and temperature fluctuations (111, 112). In addition, the inferred parameters of the gLV model can be visualized as an interaction network to examine the distribution of negative and positive interactions and to identify ecological driver species (16). The gLV model could be used to design community cultivation strategies to achieve desired community compositions and stability properties.
Evolutionary game theory (EGT) can also be used to model microbiome population dynamics (as described in 109, 113, 114). The fitness parameters that dictate the outcome of metabolic games in microbiomes can be difficult to estimate, as they are influenced by nonlinear environmental and intracellular conditions. For this purpose, EGT can be integrated with genome-scale models (GEMs) (described in the next section), enabling prediction of interspecies interactions and stable steady-state fluxes and species abundances. The system states at which each microbe locally maximizes its own growth, but not necessarily the global maximum community growth, can be identified as Nash equilibria and evolutionarily steady solutions (a subset of Nash equilibria) (115) or as asymptotically stable solutions to dynamic replicator equations, all of which pose candidates for stable coexistence states that the microbiome could exhibit. Evolutionary stability is a key factor to consider when designing bottom-up communities or altering the composition of a native system. Although gLV and EGT models capture context-dependent pairwise interactions, these models fail to capture higher-order or metabolite-based interactions in the community. Public goods games are, in principle, extendable to multispecies interactions and integrable with other modeling approaches like GEMs; some associated challenges are reviewed in Reference 113.
Public goods game: game in which participants can choose to donate their own resources to improve the fitness of the community
Predicting Microbiome Fluxes and Interactions with Genome-Scale Models and Machine Learning
Mechanistic GEMs can predict microbial behaviors in untested conditions and serve as useful platforms for synthesizing multi-omics data into one comprehensible and interactive format. GEMs mathematically represent an organism’s metabolic pathways (with gene–protein–reaction associations in metadata) as a stoichiometric matrix of reactions and metabolites (116, 117). Through flux balance analysis (FBA) (118) and related techniques, including flux variability analysis (119, 120), GEMs can be used to assess the effects of media and substrate changes and genetic edits on fluxes of target compounds; this approach has been used extensively to guide metabolic engineering (121–123). As metabolic engineers look to cocultures for specialty products (124, 125) and as systems biology is applied to medicine (122, 126), genome-scale modeling of microbiomes is becoming increasingly useful.
Automated Genome-Scale Reconstructions: Scaffolds for Genome-Scale Models Advances in meta-omics tools described above have enabled semiautomated construction of metabolic networks for hundreds of species that compose a microbiome (tools reviewed in 127); however, manual curation is needed to accurately recapitulate metabolism in silico (128). For example, Magnúsdóttir et al. (129) published the AGORA (Assembly of Gut Organisms through Reconstruction and Analysis) database, complete with 773 genome-scale reconstructions of human gut microbes and simulated pairwise microbial interactions when fed different diets. Follow-up studies to this report highlight the distinction between a reconstruction and a context-specific predictive GEM (see the sidebar titled Metabolic Reconstructions and Models) and the importance of identifying and applying appropriate constraints before attempting to simulate metabolism in silico (130, 131).
METABOLIC RECONSTRUCTIONS AND MODELS.
A metabolic reconstruction (sometimes called a genome-scale network reconstruction) is a compact and accessible representation of metabolism linked to the genome and can feasibly be created for an entire microbiome at once given high-quality bioinformatic data, albeit with some experimental and bioinformatic gap-filling (223). A genome-scale model (GEM) based on a reconstruction is used to simulate metabolism. In communities, such simulations are limited to a few well-characterized species at a time, as they require a high-quality GEM for each constituent member; this necessitates that each member be cultivable in isolation in defined media. Importantly, a metabolic simulation will only ever be as accurate as the condition-specific constraints applied to it. These constraints dictate reaction reversibility and impose upper and lower bounds on the flux through each reaction in an organism or community (including transport and interspecies exchange) on the basis of enzyme turnover, nutrient uptake rates, and many other factors (224, 225).
Databases exist for published genome-scale reconstructions such as BiGG (132) and ModelSEED (133). Significant progress has been made in cataloging reconstructions for the human gut microbiome and human body at large (129, 134, 135). These reconstructions serve as templates for context-specific GEMs, such as a known viral infection of a macrophage cell (136) or dysbiosis of the gut (137). In an important step toward universalizing GEMs over different sequence annotation styles, programming languages, and operating systems, Lieven et al. (138) recently published MEMOTE, which scores GEMs for completeness and feasibility. However, owing to the highly variable and environment-dependent nature of enzyme kinetics and transcriptional regulation, the individual constraints that make models predictive cannot yet be generally cataloged this way.
Constraining and Optimizing Community Genome-Scale Models
When a GEM is curated from a genome-scale network reconstruction, integration of multi-omics data and experimental metabolite concentrations (139) facilitates filling in pathway gaps, constraining the solution space (140), and validating and improving simulations of microbiome function. To this end, Pandey et al. (141) published in 2019 REMI (Relative Expression and Metabolomic Integrations) for integrating transcriptomics, thermodynamic, and metabolomics data from differential expression analyses into GEMs, which Hadadi et al. (142) used to simulate the transition of Pseudomonas veronii from exponential to stationary phase, as well as from culture in liquid media to soil, demonstrating advancement in our ability to model microbial adaptation to environmental perturbations.
In addition to flux constraints, choosing and defining the objective function to be optimized are particularly challenging for community models (119). In single-organism FBA systems, the objective function is usually to maximize flux through a biomass-forming reaction based on the organism’s macromolecular composition (i.e., to grow as fast as possible). For multispecies FBA systems, many optimization strategies exist. In so-called supraorganism approaches, the metabolic pathways in all organisms are combined into one stoichiometric matrix, and organisms are partitioned into separate compartments that exchange metabolites (109, 143). Optimization of the weighted community growth rate may require individual species to grow at suboptimal growth rates, which is not always an accurate assumption (especially in competitively interacting systems). Bi-level optimization has been implemented to maximize both individual and community growth rates in algorithms such as OptCom (144) and CASINO (Community and System-Level Interactive Optimization) (145).
Objective function: the function to be optimized to select for particular solutions within a multidimensional solution space
To model microbiome fluxes at steady state, community FBA (cFBA) (146) and SteadyCom (147) impose a fixed community growth rate that is adopted by all organisms; however, this is not always achievable in practice. Alternatively, EGT can be integrated with FBA to predict evolutionarily stable interactions and steady-state flux distributions (148). In other bi-level optimization strategies, a microbe may be predicted to produce a metabolite that benefits the community but does not necessarily maximize its own growth rate. To avoid imposing this forced altruism, Cai et al. (149) developed NECom, which predicts steady-state community fluxes and pairwise interactions by identifying Nash equilibria and removes any influence by the community optimization problem on a microbe’s incentive to secrete a metabolite in community GEMs.
Although highly useful for predicting and describing microbial fluxes and interactions, GEMs require significant time and resources to construct and curate predictive models from automated reconstructions. Because a microbiome’s composition must be fully defined to accurately apply FBA, GEMs are currently limited to bottom-up microbiomes with a few representative species, which often lack long-term stability. Further, experimental measurements of community growth rates and fluxes are critical to GEM validation improvement but remain a challenge to obtain for large (more than three members) communities.
Machine Learning Can Identify Complex Mappings Between Microbiome Inputs and Outputs
Machine learning can be employed to predict and link microbiome composition and functions by using multi-omics data (as reviewed in 150, 151). Machine learning can learn complex and nonlinear relationships between microbiome inputs (e.g., cultivation conditions and initial species compositions) and outputs (e.g., metabolite concentrations, gene expression profiles, and community structure) that may enable design of novel microbial consortia with target functions without the need to rigorously characterize each organism in isolation. A major limitation is that these models may not provide insight into the biological mechanisms that generate the observed microbiome states. To address this challenge, researchers have integrated machine learning with GEMs to extract information from multi-omics experiments and metabolic simulations that is relevant to the engineering objective at hand (152).
Dynamic Models of Microbiome Flux
Dynamic computational models are particularly useful when the time-varying microbiome fluxes or 3D structure is of interest (153). Dynamic FBA (dFBA) has been used extensively to model transient compositions and flux distributions in small (two to three members) microbial communities (see 119 for a review). In these cases, estimation of metabolite uptake and secretion kinetics is particularly important for accurately simulating growth and fluxes. Characterization of transporter membrane protein specificity and influx/efflux kinetics promises to significantly improve both dynamic and steady-state FBA simulations (154, 155) but remains a major challenge (156).
Spatial heterogeneity can be incorporated into community GEMs (153, 157, 158), typically by means of FBA to find each species’ growth rate at each time step in numerical solutions of reaction–diffusion partial differential equations (159). Biofilms, in particular, are increasingly being modeled with spatiotemporal GEMs (21, 158, 160, 161). With the goal of accounting for their moving boundary conditions brought on by film growth and expansion, it is useful to simulate biofilms as collections of individual microbes in so-called agent-based models (AbMs).
Agent-Based Models Simulate Predefined Physical and Metabolic Interactions in Microbiomes
In AbMs, microbes are treated as individuals with specified traits rather than as concentration state variables as in other methods. AbMs can capture gene regulation (162) and metabolic and mechanical interactions between microbes in a community, and they are well suited to model biofilm formation, deformation, and disruption (163). Metabolism can be coarse-grained to allow only for reactions involving exchange with the environment, or AbMs can integrate genome-scale metabolism to compute fluxes with FBA. The latter approach is taken in BacArena, an R package that was demonstrated to model spatiotemporal metabolic interactions among seven human gut microbes (164).
Although they are computationally demanding, AbMs are versatile models that suggest priority experiments for answering specific questions of microbiome function. For example, van Hoek et al. (165) simulated metabolism in the human large intestine with a coupled dFBA–mass transport model of individuals from a supraorganism metabacterium. Although they did not include experimental validation, their model generated hypotheses for the effects of diarrhea (a macroscale system state) on the microscale spatial organization and relative abundance of microbes with various flux profiles, which can be explored experimentally. Similarly, Doloman et al. (166) used a multispecies AbM (without genome-scale metabolism) to predict cultivation conditions that maximize methane productivity in spatially structured anaerobic sludge granules.
ENGINEERING MICROBIOMES
As microbiomes are complex, dynamic, and yet often resistant to change, how can we best modulate and design functional and predictable microbiomes for targeted use? Combining diverse omics tools with computational modeling facilitates the identification of microbial and molecular targets as well as the prediction of system behaviors. The widespread interest in engineering microbiomes for diverse applications is evidenced by the increasing number of biotechnology companies that seek to design microbiome interventions for human health, livestock fitness, and agriculture (Table 2). The approaches through which microbial populations, interactions, and biochemical fluxes may be tuned can be summarized as either modifying existing functional activities or engineering novel functions (Figure 4).
Table 2.
Selected microbiome biotech startups and ongoing projects
| Microbiome | Company | Overview | Project | Stages |
|---|---|---|---|---|
| Plant | Pivot Bio | Nitrogen fixation | Corn supplement | Commercialized |
| AgBiome | Pest control | Fungicide | Commercialized | |
| Boost Biomes | Manage disease of high-value crops | Reduce crop loss in the field and post-harvest | In development | |
| Animal | Native Microbials | Restore highfunctioning lumen | Dairy | Commercialized |
| Other animals | In development | |||
| AnimalBiome | Personalized FMT | Pet dogs and cats | Commercialized | |
| Human | Evelo Biosciences | Monoclonal microbials | COVID-19 | Phase II/III |
| Psoriasis | Phase II | |||
| Triple-negative breast cancer | Preclinical | |||
| Vedanta Biosciences | Defined consortia | Clostridium difficile infection | Phase II | |
| Inflammatory bowel disease | Phase I | |||
| Cancer immunotherapy | Phase I | |||
| Food allergy | Phase I | |||
| Seres Therapeutics | Defined consortia | C. difficile infection | Phase III | |
| Ulcerative colitis | Phase II | |||
| Metastatic melanoma | Phase I | |||
| Synlogic Therapeutics | Smart living biotherapeutics | Phenylketonuria | Phase II | |
| Immune-oncology solid tumors | Phase I |
Abbreviation: COVID-19, coronavirus disease 2019; FMT, fecal microbiota transplantation.
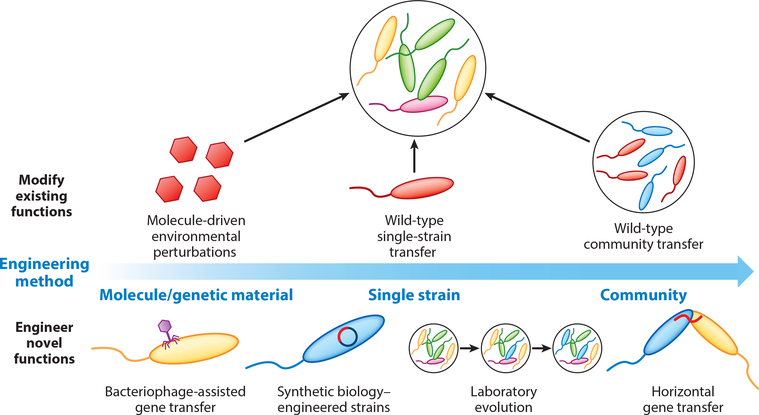
Figure 4.
Microbiome engineering tools can be used to modify existing functions within the community or to introduce novel functions. These tools vary substantially in scale, from modification of the genome of a single organism to introduction of an entirely new engineered community into the microbiome. Highlighted methods include environmental perturbations such as the addition of a molecular antibiotic or a wild-type or engineered strain or community to an existing microbiome. More recently, genetic engineering efforts and adaptive laboratory evolution, including bacteriophage-assisted gene transfer, increasingly sophisticated synthetic biology tools, and targeted horizontal gene transfer, have gained traction.
Microbiome Functions Can Be Modified by External Inputs
Manipulation of environmental factors has been widely used to modify the composition and functions of microbiomes. This strategy has attracted interest from the medical and nutritional fields that aim to tailor microbiomes to enhance host health. In gut models, prebiotics have been used to enrich for specific beneficial microbes. It is well established that diet affects the composition of intestinal microbes (167, 168), and prebiotics are used to increase the abundance of specific organisms associated with beneficial health outcomes, typically by modulating the intake of ingested fermentable dietary fibers (169).
Prebiotics: chemical compounds that promote growth and metabolism of certain microbes to benefit host health
Beyond chemical inputs, microorganisms can be administered to microbiomes with the goal of altering a target function. Several probiotics products, containing selected nonpathogenic and predicted beneficial bacterial strains, mitigate symptoms of gastrointestinal dysbiosis and protect against pathogens (170, 171). In addition to human health, probiotics have been used in animal feed (172), and the addition of rhizobacteria to fertilizer has been shown to promote plant growth (173). However, the efficacy of probiotics remains unclear due to challenges in the stable engraftment into the resident community (174) and to the unpredictable activities of beneficial biochemical pathways within probiotics that are continuously modified by the environment.
Probiotics: live bacteria that promote human gastrointestinal tract health when consumed
The introduction of microbial communities, as opposed to isolated strains, into the intestinal tract of animals or into the roots of crops is an alternative method that has attracted considerable interest from the medical and agricultural communities. For example, fecal microbiota transplantation (FMT), the procedure of transferring a fecal sample from a healthy donor to a patient with dysbiosis to reestablish a functioning microbial community, is highly effective against recurrent Clostridium difficile infections (175). However, FMT remains an investigational procedure and is associated with potential health risks, including transfer of antibiotic-resistant pathogens in the samples, though these are typically screened for in samples from human donors (176). In other words, using black box natural microbiomes may result in the inadvertent transfer of multidrug-resistant organisms and other pathogens (177), spurring interest in exploring carefully designed microbial consortia.
Rationally designed consortia comprise organisms that maintain stability in desired functions over long periods of time. A potentially powerful approach involves using top-down and bottom-up approaches to characterize system properties and responses to environmental inputs and to exploit these insights to guide the design of consortia that perform target functions (15). Both top-down and bottom-up approaches, with the aid of computational and statistical modeling (178), have had some success in introducing desirable functions in animals and plants (179, 180). Several microbiome biotechnology companies, including Vedanta and Seres, have designed microbial consortia in various stages of clinical trials, targeting human-gut-related diseases, from C. difficile infection to ulcerative colitis. (180a) However, the efficacy and effects of prebiotics and probiotics are not yet predictable, as many complex factors, including abiotic factors and interactions between gut bacteria, ultimately determine how an individual microbiome responds to different interventions.
Rewiring Microbial Functions by Genetic Engineering and Laboratory Evolution
Genetic engineering involves modification of the genome of a given organism. These modifications include, for example, deletion of genes to redirect metabolic fluxes and introduction of exogenous genes or entire pathways that allow the organism to make a valuable molecule or utilize a novel substrate. Genetically engineered microorganisms have been deployed into microbial communities associated with both plants and animals (181–183). In addition to metabolic pathway engineering, novel genetic circuits are introduced into engineered bacteria to sense and report disease-associated biomarkers in animal models and the human gut (recently reviewed in 183a). Further, engineered microbes are explored for the prevention and treatment of cancer, for example, through the targeted delivery of enzymes and toxins that inhibit tumor growth (reviewed in 183b). Another method activates the stimulator of interferon genes (STING) pathways for localized immune activation and impact on tumor growth (183c), which is currently in phase I of a clinal trial by Synlogic Therapeutics.
Although many molecular tools exist to modify genomic content of microorganisms, the clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 system has recently gained momentum as a particularly versatile tool (reviewed in 184). An alternative strategy exploits microbial horizontal gene transfer (HGT), which has the potential to influence the functional potential of organisms in situ (185). As revealed by high-resolution genomic data, HGT events occur not only between prokaryotes but also between prokaryotes and eukaryotes (186). HGT can be used to deliver genetic payloads encoding, for example, CRISPR machinery for destruction or incorporation of genes in target cells (187, 188). Lysogenic phages have been similarly explored for the integration of genes into bacterial chromosomes in order to subsequently alter target microbial communities (189, 190). The use of HGT or phage-delivered payloads is particularly attractive for nonmodel microorganisms for which established molecular engineering tools are lacking. Furthermore, the use of established genetic engineering tools that rely on given nucleotide motifs (e.g., CRISPR) may be hampered in microbes that have extreme AT or GC richness (191). Challenges of HGT and phage-based tools for microbiome engineering include mechanisms to enable stable propagation of circuits over time and regulatory elements for the control of gene expression within diverse organisms in response to specific environmental cues.
Horizontal gene transfer (HGT): transfer of genetic material between organisms with different genomes, often aided by membrane proteins and transfer origins
Evolution continuously shapes the properties of microbiomes in unpredictable ways, but efforts aim to harness adaptive laboratory evolution for microbial engineering. Laboratory evolution can be used to select for single strains or communities with desired traits, including improved substrate utilization (192), product titer (193), stress tolerance (194), and controlled shift in host characteristics (e.g., flowering time of host plant) (195). A major challenge in applying directed evolution strategies to modify properties of communities is developing capabilities to predict how microbial communities’ functions evolve toward specific discrete states or a continuum of different states in response to environmental inputs (196). Indeed, there is evidence that community context shapes the evolutionary dynamic trajectories, indicating the critical role of biotic interactions in driving evolutionary processes (197).
Biocontainment Strategies for Microbiome Engineering
All microbiome engineering strategies involving genetically engineered organisms require biocontainment mechanisms that restrict the growth of the organisms within a given environment for a designated period of time. Whereas for some applications long-term colonization may be needed to achieve the desired outcome, in other cases the transient presence of a nonnative organism or community may be sufficient and also provides biocontainment (198). To prevent engineered organisms from escaping into the surrounding ecosystem, researchers can use auxotrophy as a biocontainment strategy (199). One emerging method involves engineering metabolic niches by introducing unique metabolic capabilities and novel molecules that cannot otherwise be metabolized by native members of the microbiome (200). The discovery of unique metabolic niches can be aided by genome sequencing, metabolomics, and detailed characterization of novel metabolic pathways within microbial communities.
CONCLUSION AND PERSPECTIVES
Microbiome engineering should consider the complex interplay and feedback loops between the environment and the resident microbiome, which together drive community dynamics and multifunctional properties. Advancements in our understanding of microbiomes gleaned from ecology and systems biology must be leveraged to develop new strategies to program prescribed functions by using tools from synthetic biology. Top-down design of microbiomes presents challenges in predictably modifying composition and function, and the engineering of stable and highly functional synthetic microbial communities from the bottom up also remains difficult. Development of microbiome engineering that can achieve high-precision, robust, and predictable outcomes hold promise for diverse applications in biotechnology, medicine, agriculture, and the environment. A comprehensive understanding of microbiomes necessitates an integration of multi-omics data as well as quantification of the spatial and ecological factors that contribute to community functions.
SUMMARY POINTS.
Leveraging microbiomes for environmental and societal benefit in agriculture, medicine, and bioproduction demands holistic understanding of the various biochemical, molecular, spatial, and ecological factors that dictate their composition and function.
Continuously improving meta-omics tools allow comprehensive characterization of environmental and cultured samples and elucidate strategies for tailoring the performance of microbial communities.
Computational models of microbiomes can synthesize and explain vast empirical data, as well as predict new strategies to accentuate desired microbiome functions, by varying chemical, biological, and physical parameters such as initial species composition, media composition, and culture parameters.
Experimental validation is irreplaceable for microbiome model validation and improvement; obtaining key metrics such as the stability of target functions over time, growth rates, and fluxes remains a major challenge.
Owing to its complexity, microbiome engineering requires versatile tools that allow the precise introduction of targeted traits into a complex community with predictable outcomes.
ACKNOWLEDGMENTS
M.A.O., S.S., and P.A.L. acknowledge funding support from the National Science Foundation (MCB-1553721), the U.S. Department of Energy (DOE) (DE-SC0020420), the Army Research Office via contract W911NF-19-1-0010, and the California NanoSystems Institute Challenge Grant Program. This work was part of the DOE Joint BioEnergy Institute (http://www.jbei.org) supported by the DOE, Office of Science, Office of Biological and Environmental Research through contract DE-AC02-05CH11231 between the Lawrence Berkeley National Laboratory and the DOE. P.A.L. further acknowledges fellowship funding from the University of California, Santa Barbara, Graduate Research Mentorship Program Fellowship. O.S.V., Y.L., and M.H. were supported by the National Institutes of Health (R35GM124774 and R01EB030340), Army Research Office Young Investigator Award (W911NF-17-1-0296), Multi University Research Initiative (MURI) from the Army Research Office (W911NF-19-1-0269), and DOE (DE-FC02-07ER64494).
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Malik AA, Martiny JBH, Brodie EL, Martiny AC, Treseder KK, Allison SD. 2020. Defining trait-based microbial strategies with consequences for soil carbon cycling under climate change. ISME J. 14:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graham EB, Knelman JE, Schindlbacher A, Siciliano S, Breulmann M, et al. 2016. Microbes as engines of ecosystem function: When does community structure enhance predictions of ecosystem processes? Front. Microbiol 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abatenh E, Gizaw B, Tsegaye Z, Tefera G. 2018. Microbial function on climate change - a review. Environ. Pollut. Clim. Chang 2:147 [Google Scholar]
- 4.Werlang C, Cárcarmo-Oyarce G, Ribbeck K. 2019. Engineering mucus to study and influence the microbiome. Nat. Rev. Mater 4(2):134–45 [Google Scholar]
- 5.Sasse J, Martinoia E, Northen T. 2018. Feed your friends: Do plant exudates shape the root microbiome? Trends Plant Sci. 23(1):25–41 [DOI] [PubMed] [Google Scholar]
- 6.Zhalnina K, Louie KB, Hao Z, Mansoori N, da Rocha UN, et al. 2018. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol 3(4):470–80 [DOI] [PubMed] [Google Scholar]
- 7.Mello FD, Braidy N, Marçal H, Guillemin G, Nabavi SM, Neilan BA. 2018. Mechanisms and effects posed by neurotoxic products of cyanobacteria/microbial eukaryotes/dinoflagellates in algae blooms: a review. Neurotox. Res 33(1):153–67 [DOI] [PubMed] [Google Scholar]
- 8.Francino MP. 2016. Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front. Microbiol 6:1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogt NM, Kerby RL, Dill-McFarland KA, Harding SJ, Merluzzi AP, et al. 2017. Gut microbiome alterations in Alzheimer’s disease. Sci. Rep 7(1):13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt TSB, Raes J, Bork P. 2018. The human gut microbiome: from association to modulation. Cell 172(6):1198–215 [DOI] [PubMed] [Google Scholar]
- 11.Zheng P, Zeng B, Liu M, Chen J, Pan J, et al. 2019. The gut microbiome from patients with schizophrenia modulates the glutamate-glutamine-GABA cycle and schizophrenia-relevant behaviors in mice. Sci. Adv 5(2):eaau8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mukhtar K, Nawaz H, Abid S. 2019. Functional gastrointestinal disorders and gut-brain axis: What does the future hold? World J. Gastroenterol 25(5):552–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster KR, Schluter J, Coyte KZ, Rakoff-Nahoum S. 2017. The evolution of the host microbiome as an ecosystem on a leash. Nature 548(7665):43–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. 2010. Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol 8(1):15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawson CE, Harcombe WR, Hatzenpichler R, Lindemann SR, Löffler FE, et al. 2019. Common principles and best practices for engineering microbiomes. Nat. Rev. Microbiol 17:725–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venturelli OS, Carr AV, Fisher G, Hsu RH, Lau R, et al. 2018. Deciphering microbial interactions in synthetic human gut microbiome communities. Mol. Syst. Biol 14(6):e8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong W, Meldgin DR, Collins JJ, Lu T. 2018. Designing microbial consortia with defined social interactions. Nat. Chem. Biol 14(8):821–29 [DOI] [PubMed] [Google Scholar]
- 18.Abreu CI, Friedman J, Andersen Woltz VL, Gore J. 2019. Mortality causes universal changes in microbial community composition. Nat. Commun 10(1):2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coyte KZ, Schluter J, Foster KR. 2015. The ecology of the microbiome: networks, competition, and stability. Science 350(6261):663–66 [DOI] [PubMed] [Google Scholar]
- 20.Stams AJM, Plugge CM. 2009. Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat. Rev. Microbiol 7(8):568–77 [DOI] [PubMed] [Google Scholar]
- 21.Henson M, Phalak P. 2017. Byproduct cross feeding and community stability in an in silico biofilm model of the gut microbiome. Processes 5(4):13 [Google Scholar]
- 22.Limdi A, Pérez-Escudero A, Li A, Gore J. 2018. Asymmetric migration decreases stability but increases resilience in a heterogeneous metapopulation. Nat. Commun 9(1):2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tropini C, Earle KA, Huang KC, Sonnenburg JL. 2017. The gut microbiome: connecting spatial organization to function. Cell Host Microbe 21(4):433–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan J, Bassler BL. 2019. Surviving as a community: antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe 26(1):15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartmann R, Singh PK, Pearce P, Mok R, Song B, et al. 2019. Emergence of three-dimensional order and structure in growing biofilms. Nat. Phys 15(3):251–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rooney LM, Amos WB, Hoskisson PA, McConnell G. 2020. Intra-colony channels in E. coli function as a nutrient uptake system. ISME J. 14(10):2461–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brethauer S, Shahab RL, Studer MH. 2020. Impacts of biofilms on the conversion of cellulose. Appl. Microbiol. Biotechnol 104:5201–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Catania V, Lopresti F, Cappello S, Scaffaro R, Quatrini P. 2020. Innovative, ecofriendly biosorbent-biodegrading biofilms for bioremediation of oil-contaminated water. New Biotechnol. 58:25–31 [DOI] [PubMed] [Google Scholar]
- 29.Liu Z, Hong C-J, Yang Y, Dai L, Ho CL. 2020. Advances in bacterial biofilm management for maintaining microbiome homeostasis. Biotechnol. J 15(10):e1900320. [DOI] [PubMed] [Google Scholar]
- 30.Ritz K, Young IM. 2004. Interactions between soil structure and fungi. Mycologist 18(2):52–59 [Google Scholar]
- 31.Simon A, Hervé V, Al-Dourobi A, Verrecchia E, Junier P. 2017. An in situ inventory of fungi and their associated migrating bacteria in forest soils using fungal highway columns. FEMS Microbiol. Ecol 93(1):fiw217. [DOI] [PubMed] [Google Scholar]
- 32.Kohlmeier S, Smits THM, Ford RM, Keel C, Harms H, Wick LY. 2005. Taking the fungal highway: mobilization of pollutant-degrading bacteria by fungi. Environ. Sci. Technol 39(12):4640–46 [DOI] [PubMed] [Google Scholar]
- 33.Ng W-L, Bassler BL. 2009. Bacterial quorum-sensing network architectures. Annu. Rev. Genet 43:197–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zschiedrich CP, Keidel V, Szurmant H. 2016. Molecular mechanisms of two-component signal transduction. J. Mol. Biol 428(19):3752–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kell DB, Swainston N, Pir P, Oliver SG. 2015. Membrane transporter engineering in industrial biotechnology and whole cell biocatalysis. Trends Biotechnol. 33(4):237–46 [DOI] [PubMed] [Google Scholar]
- 36.Chen J, Zhu X, Tan Z, Xu H, Tang J, et al. 2014. Activating C4-dicarboxylate transporters DcuB and DcuC for improving succinate production. Appl. Microbiol. Biotechnol 98(5):2197–205 [DOI] [PubMed] [Google Scholar]
- 37.Zhou YJ, Yang W, Wang L, Zhu Z, Zhang S, Zhao ZK. 2013. Engineering NAD+ availability for Escherichia coli whole-cell biocatalysis: a case study for dihydroxyacetone production. Microb. Cell Factor 12(1):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farwick A, Bruder S, Schadeweg V, Oreb M, Boles E. 2014. Engineering of yeast hexose transporters to transport D-xylose without inhibition by D-glucose. PNAS 111(14):5159–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Darbani B, Stovicek V, Van Der Hoek SA, Borodina I. 2019. Engineering energetically efficient transport of dicarboxylic acids in yeast Saccharomyces cerevisiae. PNAS 116(39):19415–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng X, Gilmore SP, O’Malley MA. 2016. Microbial communities for bioprocessing: lessons learned from nature. Curr. Opin. Chem. Eng 14:103–9 [Google Scholar]
- 41.Gilmore SP, Lankiewicz TS, Wilken SE, Brown JL, Sexton JA, et al. 2019. Top-down enrichment guides in formation of synthetic microbial consortia for biomass degradation. ACS Synth. Biol 8(9):2174–85 [DOI] [PubMed] [Google Scholar]
- 42.Scarborough MJ, Lawson CE, Hamilton JJ, Donohue TJ, Noguera DR. 2018. Metatranscriptomic and thermodynamic insights into medium-chain fatty acid production using an anaerobic microbiome. mSystems 3(6):e00221–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gutiérrez N, Garrido D. 2019. Species deletions from microbiome consortia reveal key metabolic interactions between gut microbes. mSystems 4(4):e00185–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson CD, Auchtung JM, Collins J, Britton RA. 2014. Epidemic Clostridium difficile strains demonstrate increased competitive fitness compared to nonepidemic isolates. Infect. Immun 82(7):2815–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Dijk EL, Jaszczyszyn Y, Naquin D, Thermes C. 2018. The third revolution in sequencing technology. Trends Genet. 34(9):666–81 [DOI] [PubMed] [Google Scholar]
- 46.Amarasinghe SL, Su S, Dong X, Zappia L, Ritchie ME, Gouil Q. 2020. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 21(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raja HA, Miller AN, Pearce CJ, Oberlies NH. 2017. Fungal identification using molecular tools: a primer for the natural products research community. J. Nat. Prod 80(3):756–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campanaro S, Treu L, Kougias PG, Zhu X, Angelidaki I. 2018. Taxonomy of anaerobic digestion microbiome reveals biases associated with the applied high throughput sequencing strategies. Sci. Rep 8(1):1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson J-J, Brandon-Mong G-J, Gan H-M, Sing K-W. 2019. High-throughput terrestrial biodiversity assessments: mitochondrial metabarcoding, metagenomics or metatranscriptomics? Mitochondrial DNA Part A 30(1):60–67 [DOI] [PubMed] [Google Scholar]
- 50.Nayfach S, Rodriguez-Mueller B, Garud N, Pollard KS. 2016. An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Res. 26(11):1612–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scholz M, Ward DV, Pasolli E, Tolio T, Zolfo M, et al. 2016. Strain-level microbial epidemiology and population genomics from shotgun metagenomics. Nat. Methods 13(5):435–38 [DOI] [PubMed] [Google Scholar]
- 52.Shi X, Shao C, Luo C, Chu Y, Wang J, et al. 2019. Microfluidics-based enrichment and whole-genome amplification enable strain-level resolution for airway metagenomics. mSystems 4(4):e00198–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Truong DT, Tett A, Pasolli E, Huttenhower C, Segata N. 2017. Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res. 27(4):626–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. 2013. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat. Biotechnol 31(6):533–38 [DOI] [PubMed] [Google Scholar]
- 55.Brown CT, Olm MR, Thomas BC, Banfield JF. 2016. Measurement of bacterial replication rates in microbial communities. Nat. Biotechnol 34(12):1256–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tkacz A, Hortala M, Poole PS. 2018. Absolute quantitation of microbiota abundance in environmental samples. Microbiome 6(1):110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lou J, Yang L, Wang H, Wu L, Xu J. 2018. Assessing soil bacterial community and dynamics by integrated high-throughput absolute abundance quantification. PeerJ 6:e4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vandeputte D, Kathagen G, D’Hoe K, Vieira-Silva S, Valles-Colomer M, et al. 2017. Quantitative microbiome profiling links gut community variation to microbial load. Nature 551(7681):507–11 [DOI] [PubMed] [Google Scholar]
- 59.Contijoch EJ, Britton GJ, Yang C, Mogno I, Li Z, et al. 2019. Gut microbiota density influences host physiology and is shaped by host and microbial factors. eLife 8:e40553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barlow JT, Bogatyrev SR, Ismagilov RF. 2020. A quantitative sequencing framework for absolute abundance measurements of mucosal and lumenal microbial communities. Nat. Commun 11(1):2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sheth RU, Li M, Jiang W, Sims PA, Leong KW, Wang HH. 2019. Spatial metagenomic characterization of microbial biogeography in the gut. Nat. Biotechnol 37(8):877–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fontana A, Campanaro S, Treu L, Kougias PG, Cappa F, et al. 2018. Performance and genome-centric metagenomics of thermophilic single and two-stage anaerobic digesters treating cheese wastes. Water Res. 134:181–91 [DOI] [PubMed] [Google Scholar]
- 63.Thornbury M, Sicheri J, Slaine P, Getz LJ, Finlayson-Trick E, et al. 2019. Characterization of novel lignocellulose-degrading enzymes from the porcupine microbiome using synthetic metagenomics. PLOS ONE 14(1):e0209221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ibberson CB, Stacy A, Fleming D, Dees JL, Rumbaugh K, et al. 2017. Co-infecting microorganisms dramatically alter pathogen gene essentiality during polymicrobial infection. Nat. Microbiol 2:17079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Price MN, Wetmore KM, Waters RJ, Callaghan M, Ray J, et al. 2018. Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 557(7706):503–9 [DOI] [PubMed] [Google Scholar]
- 66.Nayfach S, Shi ZJ, Seshadri R, Pollard KS, Kyrpides NC. 2019. New insights from uncultivated genomes of the global human gut microbiome. Nature 568(7753):505–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shaffer M, Borton MA, McGivern BB, Zayed AA, La Rosa SL, et al. 2020. DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res. 48(16):8883–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gómez-Godínez LJ, Fernandez-Valverde SL, Martinez Romero JC, Martínez-Romero E. 2019. Metatranscriptomics and nitrogen fixation from the rhizoplane of maize plantlets inoculated with a group of PGPRs. Syst. Appl. Microbiol 42(4):517–25 [DOI] [PubMed] [Google Scholar]
- 69.Abu-Ali GS, Mehta RS, Lloyd-Price J, Mallick H, Branck T, et al. 2018. Metatranscriptome of human faecal microbial communities in a cohort of adult men. Nat. Microbiol 3(3):356–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Solomon KV, Haitjema CH, Henske JK, Gilmore SP, Borges-Rivera D, et al. 2016. Early-branching gut fungi possess a large, comprehensive array of biomass-degrading enzymes. Science 351(6278):1192–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seppälä S, Wilken SE, Knop D, Solomon KV, O’Malley MA. 2017. The importance of sourcing enzymes from non-conventional fungi for metabolic engineering and biomass breakdown. Metab. Eng 44:45–59 [DOI] [PubMed] [Google Scholar]
- 72.He B, Jin S, Cao J, Mi L, Wang J. 2019. Metatranscriptomics of the Hu sheep rumen microbiome reveals novel cellulases. Biotechnol. Biofuels 12(1):153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Petrova OE, Garcia-Alcalde F, Zampaloni C, Sauer K. 2017. Comparative evaluation of rRNA depletion procedures for the improved analysis of bacterial biofilm and mixed pathogen culture transcriptomes. Sci. Rep 7(1):41114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fang N, Akinci-Tolun R. 2016. Depletion of ribosomal RNA sequences from single-cell RNA-sequencing library. Curr. Protoc. Mol. Biol 115(1):7.27.1–7.27.20 [DOI] [PubMed] [Google Scholar]
- 75.Wangsanuwat C, Heom KA, Liu E, O’Malley MA, Dey SS. 2020. Efficient and cost-effective bacterial mRNA sequencing from low input samples through ribosomal RNA depletion. BMC Genomics 21(1):717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mueller RS, Pan C. 2013. Sample handling and mass spectrometry for microbial metaproteomic analyses. Methods Enzymol. 531:289–303 [DOI] [PubMed] [Google Scholar]
- 77.Kleiner M 2019. Metaproteomics: much more than measuring gene expression in microbial communities. mSystems 4(3):e00115–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chinappi M, Cecconi F. 2018. Protein sequencing via nanopore based devices: a nanofluidics perspective. J. Phys. Condens. Matter. 30(20):204002. [DOI] [PubMed] [Google Scholar]
- 79.Kleiner M, Thorson E, Sharp CE, Dong X, Liu D, et al. 2017. Assessing species biomass contributions in microbial communities via metaproteomics. Nat. Commun. 8(1):1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Speda J, Jonsson B-H, Carlsson U, Karlsson M. 2017. Metaproteomics-guided selection of targeted enzymes for bioprospecting of mixed microbial communities. Biotechnol. Biofuels 10(1):128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vit O, Petrak J. 2017. Integral membrane proteins in proteomics. How to break open the black box? J. Proteom 153:8–20 [DOI] [PubMed] [Google Scholar]
- 82.Emwas AHM. 2015. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. Methods Mol. Biol 1277:161–93 [DOI] [PubMed] [Google Scholar]
- 83.Zamboni N, Saghatelian A, Patti GJ. 2015. Defining the metabolome: size, flux, and regulation. Mol. Cell 58(4):699–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jeffryes JG, Colastani RL, Elbadawi-Sidhu M, Kind T, Niehaus TD, et al. 2015. MINEs: open access databases of computationally predicted enzyme promiscuity products for untargeted metabolomics. J. Cheminform 7(1):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Long CP, Antoniewicz MR. 2019. High-resolution 13C metabolic flux analysis. Nat. Protoc 14(10):2856–77 [DOI] [PubMed] [Google Scholar]
- 86.Gebreselassie NA, Antoniewicz MR. 2015. 13C-metabolic flux analysis of co-cultures: a novel approach. Metab. Eng 31:132–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ghosh A, Nilmeier J, Weaver D, Adams PD, Keasling JD, et al. 2014. A peptide-based method for 13C metabolic flux analysis in microbial communities. PLOS Comput. Biol 10(9):e1003827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Antoniewicz MR. 2020. A guide to deciphering microbial interactions and metabolic fluxes in microbiome communities. Curr. Opin. Biotechnol 64:230–37 [DOI] [PubMed] [Google Scholar]
- 89.Coyotzi S, Pratscher J, Murrell JC, Neufeld JD. 2016. Targeted metagenomics of active microbial populations with stable-isotope probing. Curr. Opin. Biotechnol 41:1–8 [DOI] [PubMed] [Google Scholar]
- 90.Egert M, Weis S, Schnell S. 2018. RNA-based stable isotope probing (RNA-SIP) to unravel intestinal host-microbe interactions. Methods 149:25–30 [DOI] [PubMed] [Google Scholar]
- 91.Starr EP, Shi S, Blazewicz SJ, Probst AJ, Herman DJ, et al. 2018. Stable isotope informed genome-resolved metagenomics reveals that Saccharibacteria utilize microbially-processed plant-derived carbon. Microbiome 6(1):122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fortunato CS, Huber JA. 2016. Coupled RNA-SIP and metatranscriptomics of active chemolithoautotrophic communities at a deep-sea hydrothermal vent. ISME J. 10(8):1925–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Radajewski S, McDonald IR, Murrell JC. 2003. Stable-isotope probing of nucleic acids: a window to the function of uncultured microorganisms. Curr. Opin. Biotechnol 14(3):296–302 [DOI] [PubMed] [Google Scholar]
- 94.Seifert J, Taubert M, Jehmlich N, Schmidt F, Völker U, et al. 2012. Protein-based stable isotope probing (protein-SIP) in functional metaproteomics. Mass Spectrom. Rev 31(6):683–97 [DOI] [PubMed] [Google Scholar]
- 95.Mosbæk F, Kjeldal H, Mulat DG, Albertsen M, Ward AJ, et al. 2016. Identification of syntrophic acetate-oxidizing bacteria in anaerobic digesters by combined protein-based stable isotope probing and metagenomics. ISME J. 10(10):2405–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ziels RM, Sousa DZ, Stensel HD, Beck DAC. 2018. DNA-SIP based genome-centric metagenomics identifies key long-chain fatty acid-degrading populations in anaerobic digesters with different feeding frequencies. ISME J. 12(1):112–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Berry D, Loy A. 2018. Stable-isotope probing of human and animal microbiome function. Trends Microbiol. 26(12):999–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schaerli Y, Hollfelder F. 2009. The potential of microfluidic water-in-oil droplets in experimental biology. Mol. Biosyst 5(12):1392–404 [DOI] [PubMed] [Google Scholar]
- 99.Lan F, Demaree B, Ahmed N, Abate AR. 2017. Single-cell genome sequencing at ultra-high-throughput with microfluidic droplet barcoding. Nat. Biotechnol 35(7):640–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sjostrom SL, Bai Y, Huang M, Liu Z, Nielsen J, et al. 2014. High-throughput screening for industrial enzyme production hosts by droplet microfluidics. Lab Chip 14(4):806–13 [DOI] [PubMed] [Google Scholar]
- 101.Watterson WJ, Tanyeri M, Watson AR, Cham CM, Shan Y, et al. 2020. Droplet-based high-throughput cultivation for accurate screening of antibiotic resistant gut microbes. eLife 9:e56998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hsu RH, Clark RL, Tan JW, Ahn JC, Gupta S, et al. 2019. Microbial interaction network inference in microfluidic droplets. Cell Syst. 9(3):229–42.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kehe J, Kulesa A, Ortiz A, Ackerman CM, Thakku SG, et al. 2019. Massively parallel screening of synthetic microbial communities. PNAS 116(26):12804–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ozbakir HF, Anderson NT, Fan KC, Mukherjee A. 2020. Beyond the green fluorescent protein: biomolecular reporters for anaerobic and deep-tissue imaging. Bioconjug. Chem 31(2):293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gupta S, Ross TD, Gomez MM, Grant JL, Romero PA, Venturelli OS. 2020. Investigating the dynamics of microbial consortia in spatially structured environments. Nat. Commun 11(1):2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zengler K, Hofmockel K, Baliga NS, Behie SW, Bernstein HC, et al. 2019. EcoFABs: advancing microbiome science through standardized fabricated ecosystems. Nat. Methods 16(7):567–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jalili-Firoozinezhad S, Gazzaniga FS, Calamari EL, Camacho DM, Fadel CW, et al. 2019. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng 3(7):520–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kumar M, Ji B, Zengler K, Nielsen J. 2019. Modelling approaches for studying the microbiome. Nat. Microbiol 4(8):1253–67 [DOI] [PubMed] [Google Scholar]
- 109.Song HS, Cannon WR, Beliaev AS, Konopka A. 2014. Mathematical modeling of microbial community dynamics: a methodological review. Processes 2(4):711–52 [Google Scholar]
- 110.Cao X, Hamilton JJ, Venturelli OS. 2019. Understanding and engineering distributed biochemical pathways in microbial communities. Biochemistry 58(2):94–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stein RR, Bucci V, Toussaint NC, Buffie CG, Rätsch G, et al. 2013. Ecological modeling from time-series inference: insight into dynamics and stability of intestinal microbiota. PLOS Comput. Biol 9(12):e1003388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dam P, Fonseca LL, Konstantinidis KT, Voit EO. 2016. Dynamic models of the complex microbial metapopulation of Lake Mendota. NPJ Syst. Biol. Appl 2(1):16007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pusa T, Wannagat M, Sagot M-F. 2019. Metabolic games. Front. Appl. Math. Stat 5:18 [Google Scholar]
- 114.Frey E 2010. Evolutionary game theory: theoretical concepts and applications to microbial communities. Phys. A 389:4265–98 [Google Scholar]
- 115.Nowak MA. 2006. Evolutionary Dynamics: Exploring the Equations of Life. Cambridge, MA: Harvard Univ. Press [Google Scholar]
- 116.Gu C, Kim GB, Kim WJ, Kim HU, Lee SY. 2019. Current status and applications of genome-scale metabolic models. Genome Biol. 20(1):121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Thiele I, Palsson B. 2010. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc 5(1):93–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Orth JD, Thiele I, Palsson BØ. 2010. What is flux balance analysis? Nat. Biotechnol 28(3):245–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gottstein W, Olivier BG, Bruggeman FJ, Teusink B. 2016. Constraint-based stoichiometric modelling from single organisms to microbial communities. J. R. Soc. Interface 13(124):20160627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gudmundsson S, Thiele I. 2010. Computationally efficient flux variability analysis. BMC Bioinform 11:489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hartmann A, Vila-Santa A, Kallscheuer N, Vogt M, Julien-Laferrière A, et al. 2017. OptPipe - a pipeline for optimizing metabolic engineering targets. BMC Syst. Biol 11(1):143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang C, Hua Q. 2016. Applications of genome-scale metabolic models in biotechnology and systems medicine. Front. Physiol 6:413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kim B, Kim WJ, Kim DI, Lee SY. 2015. Applications of genome-scale metabolic network model in metabolic engineering. J. Ind. Microbiol. Biotechnol 42(3):339–48 [DOI] [PubMed] [Google Scholar]
- 124.Sgobba E, Wendisch VF. 2020. Synthetic microbial consortia for small molecule production. Curr. Opin. Biotechnol 62:72–79 [DOI] [PubMed] [Google Scholar]
- 125.Wang R, Zhao S, Wang Z, Koffas MA. 2019. Recent advances in modular co-culture engineering for synthesis of natural products. Curr. Opin. Biotechnol 2020:65–71 [DOI] [PubMed] [Google Scholar]
- 126.Thiele I, Sahoo S, Heinken A, Hertel J, Heirendt L, et al. 2020. Personalized whole-body models integrate metabolism, physiology, and the gut microbiome. Mol. Syst. Biol 16(5):e8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mendoza SN, Olivier BG, Molenaar D, Teusink B. 2019. A systematic assessment of current genome-scale metabolic reconstruction tools. Genome Biol. 20(1):158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Reimers A-M, Lindhorst H, Waldherr S. 2017. A protocol for generating and exchanging (genome-scale) metabolic resource allocation models. Metabolites 7(3):47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Magnúsdóttir S, Heinken A, Kutt L, Ravcheev DA, Bauer E, et al. 2017. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat. Biotechnol 35(1):81–89 [DOI] [PubMed] [Google Scholar]
- 130.Babaei P, Shoaie S, Ji B, Nielsen J. 2018. Challenges in modeling the human gut microbiome. Nat. Biotechnol 36(8):682–86 [DOI] [PubMed] [Google Scholar]
- 131.Magnúsdóttir S, Heinken A, Fleming RMT, Thiele I. 2018. Reply to “Challenges in modeling the human gut microbiome”. Nat. Biotechnol 36(8):686–91 [DOI] [PubMed] [Google Scholar]
- 132.Norsigian CJ, Pusarla N, McConn JL, Yurkovich JT, Dräger A, et al. 2020. BiGG models 2020: multi-strain genome-scale models and expansion across the phylogenetic tree. Nucleic Acids Res. 48(D1):D402–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Seaver SMD, Liu F, Zhang Q, Jeffryes J, Faria JP, et al. 2020. The ModelSEED Biochemistry Database for the integration of metabolic annotations and the reconstruction, comparison and analysis of metabolic models for plants, fungi and microbes. Nucleic Acids Res. 49(D1):D1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Noronha A, Modamio J, Jarosz Y, Guerard E, Sompairac N, et al. 2019. The Virtual Metabolic Human database: integrating human and gut microbiome metabolism with nutrition and disease. Nucleic Acids Res. 47(D1):D614–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Robinson JL, Kocabaş P, Wang H, Cholley PE, Cook D, et al. 2020. An atlas of human metabolism. Sci. Signal 13(624):eaaz1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Aller S, Scott A, Sarkar-Tyson M, Soyer OS. 2018. Integrated human-virus metabolic stoichiometric modelling predicts host-based antiviral targets against Chikungunya, dengue and Zika viruses. J. R. Soc. Interface 15(146):20180125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kumar M, Ji B, Babaei P, Das P, Lappa D, et al. 2018. Gut microbiota dysbiosis is associated with malnutrition and reduced plasma amino acid levels: lessons from genome-scale metabolic modeling. Metab. Eng 49:128–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lieven C, Beber ME, Olivier BG, Bergmann FT, Ataman M, et al. 2020. MEMOTE for standardized genome-scale metabolic model testing. Nat. Biotechnol 38(3):272–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kuang E, Marney M, Cuevas D, Edwards RA, Forsberg EM. 2020. Towards predicting gut microbial metabolism: integration of flux balance analysis and untargeted metabolomics. Metabolites 10(4):156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tian M, Reed JL. 2018. Integrating proteomic or transcriptomic data into metabolic models using linear bound flux balance analysis. Bioinformatics 34(22):3882–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pandey V, Hadadi N, Hatzimanikatis V. 2019. Enhanced flux prediction by integrating relative expression and relative metabolite abundance into thermodynamically consistent metabolic models. PLOS Comput. Biol 15(5):e1007036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hadadi N, Pandey V, Chiappino-Pepe A, Morales M, Gallart-Ayala H, et al. 2020. Mechanistic insights into bacterial metabolic reprogramming from omics-integrated genome-scale models. NPJ Syst. Biol. Appl 6(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Stolyar S, Van Dien S, Hillesland KL, Pinel N, Lie TJ, et al. 2007. Metabolic modeling of a mutualistic microbial community. Mol. Syst. Biol 3(1):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zomorrodi AR, Maranas CD. 2012. OptCom: a multi-level optimization framework for the metabolic modeling and analysis of microbial communities. PLOS Comput. Biol 8(2):e1002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shoaie S, Ghaffari P, Kovatcheva-Datchary P, Mardinoglu A, Sen P, et al. 2015. Quantifying diet-induced metabolic changes of the human gut microbiome. Cell Metab. 22(2):320–31 [DOI] [PubMed] [Google Scholar]
- 146.Khandelwal RA, Olivier BG, Ling R, Teusink W, Bruggeman BJ. 2013. Community flux balance analysis for microbial consortia at balanced growth. PLOS ONE 8(5):64567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chan SHJ, Simons MN, Maranas CD. 2017. SteadyCom: predicting microbial abundances while ensuring community stability. PLOS Comput. Biol 13(5):e1005539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zomorrodi AR, Segrè D. 2017. Genome-driven evolutionary game theory helps understand the rise of metabolic interdependencies in microbial communities. Nat. Commun 8(1):1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cai J, Tan T, Chan SHJ. 2019. Bridging evolutionary game theory and metabolic models for predicting microbial metabolic interactions. bioRxiv 623173. 10.1101/623173 [DOI] [Google Scholar]
- 150.Zhou Y-H, Gallins P. 2019. A review and tutorial of machine learning methods for microbiome host trait prediction. Front. Genet 10:579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Namkung J 2020. Machine learning methods for microbiome studies. J. Microbiol 58(3):206–16 [DOI] [PubMed] [Google Scholar]
- 152.Zampieri G, Vijayakumar S, Yaneske E, Angione C. 2019. Machine and deep learning meet genome-scale metabolic modeling. PLOS Comput. Biol 15(7):e1007084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Øyås O, Stelling J. 2018. Genome-scale metabolic networks in time and space. Curr. Opin. Syst. Biol 8:51–58 [Google Scholar]
- 154.Zeng H, Yang A. 2020. Bridging substrate intake kinetics and bacterial growth phenotypes with flux balance analysis incorporating proteome allocation. Sci. Rep 10:4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nilsson A, Nielsen J, Palsson BO. 2017. Metabolic models of protein allocation call for the kinetome. Cell Syst. 5(6):538–41 [DOI] [PubMed] [Google Scholar]
- 156.Boyarskiy S, Tullman-Ercek D. 2015. Getting pumped: membrane efflux transporters for enhanced biomolecule production. Curr. Opin. Chem. Biol 28:15–19 [DOI] [PubMed] [Google Scholar]
- 157.Lillington SP, Leggieri PA, Heom KA, O’Malley MA. 2020. Nature’s recyclers: anaerobic microbial communities drive crude biomass deconstruction. Curr. Opin. Biotechnol 62:38–47 [DOI] [PubMed] [Google Scholar]
- 158.Harcombe WR, Riehl WJ, Dukovski I, Granger BR, Betts A, et al. 2014. Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics. Cell Rep. 7(4):1104–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chen J, Gomez JA, Höffner K, Phalak P, Barton PI, Henson MA. 2016. Spatiotemporal modeling of microbial metabolism. BMC Syst. Biol 10(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Patel A, Carlson RP, Henson MA. 2019. In silico metabolic design of two-strain biofilm systems predicts enhanced biomass production and biochemical synthesis. Biotechnol. J 14(7):e1800511. [DOI] [PubMed] [Google Scholar]
- 161.Phalak P, Chen J, Carlson RP, Henson MA. 2016. Metabolic modeling of a chronic wound biofilm consortium predicts spatial partitioning of bacterial species. BMC Syst. Biol 10(1):90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gorochowski TE, Matyjaszkiewicz A, Todd T, Oak N, Kowalska K, et al. 2012. BSim: an agent-based tool for modeling bacterial populations in systems and synthetic biology. PLOS ONE 7(8):e42790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Jayathilake PG, Gupta P, Li B, Madsen C, Oyebamiji O, et al. 2017. A mechanistic individual-based model of microbial communities. PLOS ONE 12(8):e0181965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bauer E, Zimmermann J, Baldini F, Thiele I, Kaleta C. 2017. BacArena: individual-based metabolic modeling of heterogeneous microbes in complex communities. PLOS Comput. Biol 13(5):e1005544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.van Hoek MJA, Merks RMH. 2017. Emergence of microbial diversity due to cross-feeding interactions in a spatial model of gut microbial metabolism. BMC Syst. Biol 11(1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Doloman A, Varghese H, Miller CD, Flann NS. 2017. Modeling de novo granulation of anaerobic sludge. BMC Syst. Biol 11(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, et al. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505(7484):559–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Johnson AJ, Vangay P, Al-Ghalith GA, Hillmann BM, Ward TL, et al. 2019. Daily sampling reveals personalized diet-microbiome associations in humans. Cell Host Microbe 25(6):789–802.e5 [DOI] [PubMed] [Google Scholar]
- 169.Carlson JL, Erickson JM, Lloyd BB, Slavin JL. 2018. Health effects and sources of prebiotic dietary fiber. Curr. Dev. Nutr 2(3):nzy005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Gilijamse PW, Hartstra AV, Levin E, Wortelboer K, Serlie MJ, et al. 2020. Treatment with Anaerobutyricum soehngenii: a pilot study of safety and dose–response effects on glucose metabolism in human subjects with metabolic syndrome. NPJ Biofilms Microbiomes 6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Piewngam P, Zheng Y, Nguyen TH, Dickey SW, Joo HS, et al. 2018. Pathogen elimination by probiotic Bacillus via signalling interference. Nature 562(7728):532–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Puniya AK, Salem AZM, Kumar S, Dagar SS, Griffith GW, et al. 2015. Role of live microbial feed supplements with reference to anaerobic fungi in ruminant productivity: a review. J. Integr. Agric 14(3):550–60 [Google Scholar]
- 173.Bhardwaj D, Ansari M, Sahoo R, Tuteja N. 2014. Biofertilizers function as key player in sustainable agriculture by improving soil fertility, plant tolerance and crop productivity. Microb. Cell Factor 13(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Maldonado-Gómez MX, Martínez I, Bottacini F, O’Callaghan A, Ventura M, et al. 2016. Stable engraftment of Bifidobacterium longum AH1206 in the human gut depends on individualized features of the resident microbiome. Cell Host Microbe 20(4):515–26 [DOI] [PubMed] [Google Scholar]
- 175.Brandt LJ, Aroniadis OC, Mellow M, Kanatzar A, Kelly C, et al. 2012. Long-term follow-up of colonoscopic fecal microbiota transplant for recurrent Clostridium difficile infection. Am. J. Gastroenterol 107(7):1079–87 [DOI] [PubMed] [Google Scholar]
- 176.Gupta S, Allen-Vercoe E, Petrof EO. 2016. Fecal microbiota transplantation: in perspective. Therap. Adv. Gastroenterol 9(2):229–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.DeFilipp Z, Bloom PP, Torres Soto M, Mansour MK, Sater MRA, et al. 2019. Drug-resistant E. coli bacteremia transmitted by fecal microbiota transplant. N. Engl. J. Med 381(21):2043–50 [DOI] [PubMed] [Google Scholar]
- 178.Stein RR, Tanoue T, Szabady RL, Bhattarai SK, Olle B, et al. 2018. Computer-guided design of optimal microbial consortia for immune system modulation. eLife 7:e30916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, et al. 2019. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 565(7741):600–5 [DOI] [PubMed] [Google Scholar]
- 180.Jia Y, Niu C-T, Lu Z-M, Zhang X-J, Chai L-J, et al. 2020. A bottom-up approach to develop a synthetic microbial community model: application for efficient reduced-salt broad bean paste fermentation. Appl. Environ. Microbiol 86(12):e00306–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180a.Jimenez M, Langer R, Traverso G. 2019. Microbial therapeutics: New opportunities for drug delivery. J. Exp. Med 216(5):1005–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hwang IY, Koh E, Wong A, March JC, Bentley WE, et al. 2017. Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nat. Commun 8(1):15028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Thompson JA, Oliveira RA, Djukovic A, Ubeda C, Xavier KB. 2015. Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 10(11):1861–71 [DOI] [PubMed] [Google Scholar]
- 183.Ryu MH, Zhang J, Toth T, Khokhani D, Geddes BA, et al. 2020. Control of nitrogen fixation in bacteria that associate with cereals. Nat. Microbiol 5(2):314–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183a.Tanna T, Ramachanderan R, Platt RJ. 2021. Engineered bacteria to report gut function: technologies and implementation. Curr. Opin. Microbiol 59:24–33 [DOI] [PubMed] [Google Scholar]
- 183b.Sedighi M, Zahedi Bialvaei A, Hamblin MR, Ohadi E, Asadi A, et al. 2019. Therapeutic bacteria to combat cancer; current advances, challenges, and opportunities. Cancer Med. 8(6):3167–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183c.Leventhal DS, Sokolovska A, Li N, Plescia C, Kolodziej SA, et al. 2020. Immunotherapy with engineered bacteria by targeting the STING pathway for anti-tumor immunity. Nat. Commun 11(1):2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Nemudryi AA, Valetdinova KR, Medvedev SP, Zakian SM. 2014. TALEN and CRISPR/Cas genome editing systems: tools of discovery. Acta Nat. 6(22):19–40 [PMC free article] [PubMed] [Google Scholar]
- 185.Sheth RU, Cabral V, Chen SP, Wang HH, Wang HH. 2016. Manipulating bacterial communities by in situ microbiome engineering. Trends Genet. 32(4):189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Haitjema CH, Gilmore SP, Henske JK, Solomon KV, de Groot R, et al. 2017. A parts list for fungal cellulosomes revealed by comparative genomics. Nat. Microbiol 2(8):17087. [DOI] [PubMed] [Google Scholar]
- 187.Hamilton TA, Pellegrino GM, Therrien JA, Ham DT, Bartlett PC, et al. 2019. Efficient inter-species conjugative transfer of a CRISPR nuclease for targeted bacterial killing. Nat. Commun 10(1):4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ronda C, Chen SP, Cabral V, Yaung SJ, Wang HH. 2019. Metagenomic engineering of the mammalian gut microbiome in situ. Nat. Methods 16(2):167–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Howard-Varona C, Hargreaves KR, Abedon ST, Sullivan MB. 2017. Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J. 11(7):1511–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Westwater C, Kasman LM, Schofield DA, Werner PA, Dolan JW, et al. 2003. Use of genetically engineered phage to deliver antimicrobial agents to bacteria: an alternative therapy for treatment of bacterial infections. Antimicrob. Agents Chemother 47(4):1301–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wilken SE, Seppälä S, Lankiewicz TS, Saxena M, Henske JK, et al. 2020. Genomic and proteomic biases inform metabolic engineering strategies for anaerobic fungi. Metab. Eng. Commun 10:e00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wang Y, Fan L, Tuyishime P, Liu J, Zhang K, et al. 2020. Adaptive laboratory evolution enhances methanol tolerance and conversion in engineered Corynebacterium glutamicum. Commun. Biol 3(1):217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Reyes LH, Gomez JM, Kao KC. 2014. Improving carotenoids production in yeast via adaptive laboratory evolution. Metab. Eng 21:26–33 [DOI] [PubMed] [Google Scholar]
- 194.Wang L, Xue C, Wang L, Zhao Q, Wei W, Sun Y. 2016. Strain improvement of Chlorella sp. for phenol biodegradation by adaptive laboratory evolution. Bioresour. Technol 205:264–68 [DOI] [PubMed] [Google Scholar]
- 195.Panke-Buisse K, Poole AC, Goodrich JK, Ley RE, Kao-Kniffin J. 2015. Selection on soil microbiomes reveals reproducible impacts on plant function. ISME J. 9(4):980–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Celiker H, Gore J. 2014. Clustering in community structure across replicate ecosystems following a long-term bacterial evolution experiment. Nat. Commun 5:4643. [DOI] [PubMed] [Google Scholar]
- 197.Scheuerl T, Hopkins M, Nowell RW, Rivett DW, Barraclough TG, Bell T. 2020. Bacterial adaptation is constrained in complex communities. Nat. Commun 11(1):754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lee JW, Chan CTY, Slomovic S, Collins JJ. 2018. Next-generation biocontainment systems for engineered organisms. Nat. Chem. Biol 14(6):530–37 [DOI] [PubMed] [Google Scholar]
- 199.Mandell DJ, Lajoie MJ, Mee MT, Takeuchi R, Kuznetsov G, et al. 2015. Biocontainment of genetically modified organisms by synthetic protein design. Nature 518(7537):55–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Shepherd ES, Deloache WC, Pruss KM, Whitaker WR, Sonnenburg JL. 2018. An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature 557(7705):434–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2016. GenBank. Nucleic Acids Res. 44(D1):D67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44(D1):D733–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Amid C, Alako BTF, Balavenkataraman Kadhirvelu V, Burdett T, Burgin J, et al. 2020. The European Nucleotide Archive in 2019. Nucleic Acids Res. 48(D1):D70–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kodama Y, Mashima J, Kosuge T, Kaminuma E, Ogasawara O, et al. 2018. DNA Data Bank of Japan: 30th anniversary. Nucleic Acids Res. 46(D1):D30–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kawashima S, Katayama T, Hatanaka H, Kushida T, Takagi T. 2018. NBDC RDF portal: a comprehensive repository for semantic data in life sciences. Database 2018:bay123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28(1):27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chen IMA, Chu K, Palaniappan K, Pillay M, Ratner A, et al. 2019. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 47(D1):D666–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Grigoriev IV, Nordberg H, Shabalov I, Aerts A, Cantor M, et al. 2012. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 40(D1):D26–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Nordberg H, Cantor M, Dusheyko S, Hua S, Poliakov A, et al. 2014. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res. 42(D1):D26–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40(D1):D1178–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, et al. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol 37(4):420–23 [DOI] [PubMed] [Google Scholar]
- 212.Krogh A, Larsson B, Von Heijne G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol 305(3):567–80 [DOI] [PubMed] [Google Scholar]
- 213.Bernsel A, Viklund H, Hennerdal A, Elofsson A. 2009. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 37:W465–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Karsch-Mizrachi I, Takagi T, Cochrane G. 2018. The international nucleotide sequence database collaboration. Nucleic Acids Res. 46(D1):D48–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Heldin C-H, Lu B, Evans R, Gutkind JS. 2016. Signals and receptors. Cold Spring Harb. Perspect. Biol 8(4):a005900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Wallin E, Von Heijne G. 2008. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 7(4):1029–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Almén MS, Nordström KJV, Fredriksson R, Schiöth HB. 2009. Mapping the human membrane proteome: a majority of the human membrane proteins can be classified according to function and evolutionary origin. BMC Biol. 7(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.O’Sullivan C, Burrell PC, Pasmore M, Clarke WP, Blackall LL. 2009. Application of flowcell technology for monitoring biofilm development and cellulose degradation in leachate and rumen systems. Bioresour. Technol 100(1):492–96 [DOI] [PubMed] [Google Scholar]
- 219.Welch JLM, Hasegawa Y, McNulty NP, Gordon JI, Borisy GG. 2017. Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. PNAS 114(43):E9105–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Dunham SJB, Ellis JF, Li B, Sweedler JV. 2017. Mass spectrometry imaging of complex microbial communities. Acc. Chem. Res 50(1):96–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Behrens S, Lösekann T, Pett-Ridge J, Weber PK, Ng WO, et al. 2008. Linking microbial phylogeny to metabolic activity at the single-cell level by using enhanced element labelling-catalyzed reporter deposition fluorescence in situ hybridization (EL-FISH) and NanoSIMS. Appl. Environ. Microbiol 74(10):3143–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.McGlynn SE, Chadwick GL, Kempes CP, Orphan VJ. 2015. Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 526(7574):531–35 [DOI] [PubMed] [Google Scholar]
- 222a.Geva-Zatorsky N, Alvarez D, Hudak JE, Reading NC, Erturk-Hasdemir D, et al. 2015. In vivo imaging and tracking of host-microbiota interactions via metabolic labeling of gut anaerobic bacteria. Nat. Med. 21(9):1091–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222b.García-Bayona L, Coyne MJ, Hantman N, Montero-Llopis P, Von SS, et al. 2020. Nanaerobic growth enables direct visualization of dynamic cellular processes in human gut symbionts. PNAS 117(39):24484–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222c.Chia HE, Marsh ENG, Biteen JS. 2019. Extending fluorescence microscopy into anaerobic environments. Curr. Opin. Chem. Biol 51:98–104 [DOI] [PubMed] [Google Scholar]
- 223.Orth JD, Palsson BØ. 2010. Systematizing the generation of missing metabolic knowledge. Biotechnol. Bioeng 107(3):403–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Wiback SJ, Mahadevan R, Palsson BØ. 2004. Using metabolic flux data to further constrain the metabolic solution space and predict internal flux patterns: the Escherichia coli spectrum. Biotechnol. Bioeng 86(3):317–31 [DOI] [PubMed] [Google Scholar]
- 225.Bogaerts P, Gziri KM, Richelle A. 2017. From MFA to FBA: defining linear constraints accounting for overflow metabolism in a macroscopic FBA-based dynamical model of cell cultures in bioreactor. J. Process Control 60:34–47 [Google Scholar]