Abstract
Background
Mandatory Day 2 and Day 8 PCR testing and variant sequencing of international arrivals has been recently introduced by the UK Government to mitigate against cross-border transmission of high-risk SARS-CoV-2 variants.
Methods
SARS-CoV-2 testing and sequencing combines TaqPath CE-IVD COVID-19 RT-PCR with Ion AmpliSeq SARS-CoV-2 Next Generation Sequencing Assay. Retrospective analysis of test trending data was performed from initiation of testing on the 11th March through to the 14th April 2021.
Findings
During this time interval, 203,065 SARS-CoV-2 PCR tests were performed, with 3,855 samples testing positive, giving a prevalence of 1.9%. In total 1,913 SARS-CoV-2 genomes were sequenced from positive cases with Ct values < 30 and 1,635 (85.5%) sequences passed quality metrics for lineage analysis. A high diversity of 49 different SARS-CoV-2 variants were identified, including the VOCs B.1.1.7 (Kent; 80.6%), B.1.351 (South Africa; 4.2%), B.1.617.2 (India; 1.7%), P.1 (Brazil; 0.4%) and B.1.1.7 with E484K (Bristol; 0.2%). Vaccine effectiveness was age-related and dose-dependent, ranging from 5% in > 60 with a single dose to 83% in <60 with both doses of a vaccine. Viral load was variant dependent with the B.1.617.2 showing a 21 fold increase in viral copy number compared to the other variants.
Interpretation
The unexpectedly high prevalence of COVID-19 infection in UK arrivals is associated with a rich diversity of SARS-CoV-2 high risk variants entering the UK including the VOC B.1.617.2. Vaccination does not preclude infection and its effectiveness is significantly age-dependent and impacted by variant type. The rapid high-throughput test and sequence workflow we have adopted is particularly suited to the monitoring of cross border transmission and enables immediate public health interventions.
Funding
Data analysis conducted in this study was limited to secondary use of information previously collected in the course of normal care.
Research in context.
Evidence before this study
In June 2020 Alaska instituted a traveller SARS-CoV-2 cross border testing programme during the re-opening phases of the pandemic and this public health measure is thought to have contributed to Alaska having a lower number of excess deaths when compared with most US states. To mitigate the risk of traveller related import of SARS-CoV-2 immunity-escape variants into the UK, the government has initiated the Day 2 and Day 8 test and sequencing programme for international arrivals.
Added value of this study
Our analysis of SARS-CoV-2 test trending data from the Day 2 and Day 8 programme for international travellers shows that cross border transmission of high risk variants poses a significant risk to the UK population. The unexpectedly high prevalence of COVID-19 infection in travellers (1.9%) was associated with a rich diversity of SARS-CoV-2 high risk variants including the VOC B.1.617.2.
Implications of all the available evidence
International travel exposes countries to a major risk of import of SARS-CoV-2 high risk variants with potential for vaccine immune evasion. Here we demonstrate that genomic surveillance for monitoring of cross border transmission will be a critical public health measure that countries will most likely need to adopt to protect their populations from emerging high risk SARS-CoV-2 variants until the global roll-out of effective vaccines has been achieved.
Alt-text: Unlabelled box
1. Introduction
First identified as a cluster of pneumonia cases in Wuhan, Hubei Province, China, Severe Acute Respiratory Syndrome-related coronavirus 2, SARS-CoV-2 [1], causing the disease known as COVID-19, has since infected over 150 million people, leading to at least 3.3 million deaths worldwide as of 10 May 2021 [2]. SARS-CoV-2 belongs to the SARS-related coronavirus (SARSr-CoV) species of Betacoronavirus, characterised by a large (~30 kb) +ssRNA genome. SARS-CoV-2, in common with all other known human coronaviruses is believed to have a recent zoonotic origin from pangolins [3], bats [4], or an intermediate species [5].
RNA viruses have high mutation rates and these high rates are associated with enhanced virulence and evolvability [6]. Coronaviruses also have a propensity for recombination in co-infection events which further enhances the ability of the virus to undergo evolutionary adaption [7]. These biological characteristics have enabled SARS-CoV-2 to rapidly adapt to human hosts and to potentially undergo vaccine-induced evolution. Indeed, since the outbreak of the pandemic at least 1250 distinct variants have been identified [8]. The first variants found were the ancestral ’S-type’ (later reclassified as A) and ’L-type’ (later reclassified as B), which soon became dominant in Wuhan [9]. The B.1 lineage, which emerged in Northern Italy in February 2020, has been later displaced by the B.1.177 variant during the second wave [10].
B.1.1.7 is a variant of concern (VOC) first identified in Kent, and now dominant in the UK. Characterised by 17 mutations, three spike protein mutations in particular are thought to be the most significant [11]. N501Y is found in the receptor binding domain (RBD) of the spike protein and has been shown to increase its binding affinity to human angiotensin-concerting enzyme 2 (ACE2), the virus’ primary mode of entry into human cells. 69–70del is thought to lead to increased transmissibility in combination with N501Y. P681H is adjacent to the furin cleavage site, potentially affecting membrane fusion [11]. Although the B.1.1.7 variant shows enhanced transmissibility [12], it has not demonstrated more clinical severity [13,14].
A mutation of particular concern is E484K, which reduces antibody binding to the RBD, reducing neutralisation efficacy in some SARS-CoV-2 WT convalescent human sera by > 10-fold [15]. K417 has been found to be 60–100% buried in class 1 antibody paratopes and K417N has also been shown to significantly reduce sera neutralisation [16]. The combination of nearby RBD mutations K417N/T, E484K and N501Y found in B.1.351 (South Africa) and P.1 (Brazil) make these particularly concerning variants. Indeed, evidence suggests both the Moderna and Pfizer vaccines are less effective at neutralising viruses with these mutations and of 17 vaccine-elicited antibodies tested, 9 were at least 10 times less effective against pseudotyped viruses containing E484K, 5 against K417N and 4 against N501Y [17]. Sera from individuals vaccinated twice with the Pfizer vaccine were 14-fold less effective against B.1.351 and 40% of sera from individuals convalescent for > 9 months lacked any activity at all [18]. The E484K mutation has also been identified in B.1.1.7 variants, designated VOC-21FEB-02, as well as B.1.525, a strain first found in the UK, but thought to originate in Nigeria [19]. This mutation has the same effect of reducing vaccine-elicited antibody effectiveness in B.1.1.7 [20].
A further VOC has recently been identified in India, designated B.1.617 and contains 3 clades with different mutation profiles; B.1.617.1 has a spike profile which includes L452R and E484Q; B.1.617.2 has a different profile without E484Q and appears to have undergone recent expansion; B.1.617.3 has L452R and E484Q but is distinct from B.1.617.1 and currently is only detected in a small proportion of sequences. B.1.617.2 is designated VOC-21APR-02 due to recent expansion in the UK, while B.1.617.1 is designated VUI-21APR-01 on the basis of the mutation profile and apparent successful transmission and spread. B.1.617.3 is designated VUI-21APR-03 but remains at low prevalence in the UK [21].
In response to the expanding numbers of high risk variants, mandatory Day 2 and Day 8 PCR testing of international arrivals has been recently introduced by the UK government to mitigate against international importation of high risk variants into the United Kingdom [22]. Stringent minimal performance characteristics have been set by the UK government Department of Health & Social Care, overseen and assessed by the United Kingdom Accreditation Service (UKAS) to ISO:15189 standards for laboratories participating in this programme. These include a SARS-CoV-2 RT-PCR test sensitivity of > 99% with a specificity of > 99% and a limit of detection less than or equal to 1000 SARS-CoV-2 copies per millilitre (≤ 1000 copies/mL). All positive Day 2 samples with cycle threshold (Ct) values of < 30 (defined as equivalent to ~10,000–1000 viral genome copies/mL) must undergo genomic sequencing with minimal sequencing coverage of 50% at ≥ 30x coverage [22]. Travellers to the UK must also undertake pre-departure testing, but this is set at a lower sensitivity of 80%, specificity of 97% and a limit of detection of 100,000 viral copies per mL [23]. This may include different technologies to PCR testing such as Loop-mediated isothermal Amplification (LAMP) or lateral flow tests which can be less sensitive in detecting active infections.
Oncologica was the first UK laboratory to receive approval by UKAS and listing by the Department of Health and Social Care (DHSC) to perform Day 2 and Day 8 testing. The accreditation relates to a novel integrated workflow which combines TaqPath CE-IVD COVID-19 RT-PCR with Ion AmpliSeq SARS-CoV-2 Next Generation Sequencing Assay. This workflow has enabled us to undertake rapid high-throughput testing and sequencing of international arrivals into the UK. Here we have conducted a retrospective analysis of SARS-CoV-2 test trending data for international arrivals to determine the prevalence and variant types entering the UK and correlated SARS-CoV-2 test positive data with vaccination status.
2. Materials and methods
2.1. Sample cohort demographics
Between 11th March and 14th April 2021, 203,065 UK arrivals received mandatory SARS-CoV-2 RT-PCR testing as part of the UK Government guidelines for international travel. Travel declarations including patient age and vaccination status were available for 128,566 samples. The demographics of the study cohort are summarised in Supplementary Table 1. The research conducted in this study was limited to secondary use of information previously collected in the course of normal care (without an intention to use it for research at the time of collection) and therefore does not require REC review. The patients and service users were not identifiable to the research team carrying out trend data analysis. This is also in accordance with guidance issued by the National Research Ethics Service, Health Research Authority, NHS and follows the tenants of the Declaration of Helsinki. Gareth Williams, Keeda-Marie Hardisty, Kaiya Chowdhary and Marco Loddo had access to patient data and Alex Llewelyn and Ruben Brandao had access to anonymised data only.
2.2. Nucleic acid extraction
Nasopharyngeal swab samples were collected in COVIDSafe™ Virus Inactivation Medium (Cat: AL167CST5–1ML, Hi Media; Trafalgar Scientific, UK) and sent to the laboratory at ambient temperature. RNA was extracted using the MagMAX™ Viral/Pathogen II Nucleic Acid Isolation Kit (Cat: A42352; Thermo Fisher Scientific, UK), according to the manufacturer's instructions. Briefly, 400 µL of Viral Transport Medium from each patient swab sample was incubated with binding solution, total nucleic acid magnetic beads and proteinase K. MS2 bacteriophage was added as an internal control for qPCR testing. Beads were washed using kit wash buffer and 80% ethanol and eluted in TE buffer using a Kingfisher Flex instrument (Cat: 5400630; Thermo Fisher Scientific, UK).
2.3. SARS-CoV2 RT-PCR
The TaqPath™ COVID-19 CE-IVD RT-PCR (A48067; Thermofisher Scientific) assay was used for the qualitative detection of SARS-CoV-2 in RNA elutions using QuantStudio™ 5 (Cat: A34322, Thermo Fisher Scientific, UK) instruments according to the manufacturer's instructions. Samples were reported as positive if two or more of the target genes (N gene, S gene and ORF1ab) were amplified with a Ct < 37. Samples were reported as inconclusive if one gene amplifed and negative if all genes were undetectable or had a Ct > 37. Positive samples from day 2 travellers with an N gene Ct < 30, or where N gene amplification is not observed, ORF1ab Ct < 30, were selected for sequencing. Extracts were stored at −80 °C prior to sequencing.
2.4. Ion Torrent semiconductor sequencing
Reverse transcription was carried out using the SuperScript™ VILO™ cDNA Synthesis Kit (Cat: 11754050; Thermo Fisher Scientific, UK) according to the manufacturer's instructions (samples with a Ct < 8 were diluted 1:50 before this step). Library preparation was carried out using the Ion Ampliseq™ Library Kit Plus (Cat: 4488990, Thermo Fisher Scientific, UK) according to the manufacturer's instructions. Briefly, cDNA was amplified using the Ion Ampliseq™ SARS-CoV-2 Research Panel (A47573; Thermofisher, UK) dividing samples into four groups with varying cycle numbers on the basis of their Ct value. Libraries were purified using Ampure XP beads before quantitation by qPCR with the Ion Library TaqMan™ quantitation Kit (Cat: 4468802; Thermo Fisher Scientific, UK). Libraries were normalised to 70pM before pooling and templating onto an Ion 540™ Chip (Cat: A22765, Thermo Fisher Scientific, UK) using an Ion Chef™ (Cat: 4484177, Thermo Fisher Scientific, UK). Sequencing was conducted using an Ion GeneStudio™ S5 Prime System (Cat: A38196, Thermo Fisher Scientific, UK).
2.5. Quality control
Sequencing results were visualised using Torrent Suite version 5.14.0. The overall run metrics were quality controlled using the following parameters according to manufacturer's recommendations: chip loading > 60% with > 45 million reads observed, enrichment > 95%, polyclonal ratio <55%, low quality < 26%, usable reads >30% and aligned bases were ≥ 80%, mean raw accuracy was > 99% and chip test fragment alignment was > 80% (Supplementary Table 2). Sample specific metrics were assessed for quality according to manufacturer's instructions: Mapped reads > 500,000, percentage reads on target > 90%, average base coverage depth > 1200, uniformity of base coverage > 85%, assigned amplicon reads > 90%, amplicons reading end to end > 80%, bases in target regions 30,253, base reads on target > 85%. Specifically, samples with a target base coverage (100x) < 90% were rejected (Supplementary Table 3). Sequences with N count > 1%, average depth <500x or low depth bases% >5% assigned by the SARS_CoV-2_Pangolin (v5.12.1.0) were discarded.
2.6. Data analysis
After quality control, SARS-CoV-2 were aligned to the original Wuhan strain Wuhan/WH04/2020 [24], accession number MT291829, using MAFFT [25]. Sequences were pruned and analysed for Tajima's Neutrality Test using Mega-X [26]. Viral Lineages were assigned with the PANGOLIN package, version 2.4.2 [27]. Phylogenetic trees were produced with IQ-TREE using the automatic ModelFinder with default settings using the Bayesian Information Criterion (BIC) to determine the most appropriate substitution model and a cmax of 10 [28] and visualised with iTOL [29]. Graphs were produced in Microsoft Excel and Graphpad Prism v9.1.0, and to conduct statistical analysis [30], including Šídák's multiple comparisons test [31] for comparing selected means, and Dunnet's multiple comparisons test [32] for comparing a single mean to all other means.
2.7. Role of funding source
Data analysis conducted in this study was limited to secondary use of information previously collected in the course of normal care. No dedicated funding source was allocated for this study.
3. Results
Between 11 March and 14 April 2021, a total of 203,065 international travellers entering the United Kingdom underwent RT-PCR testing for SARS-CoV-2 as part of the mandatory International Travel Testing Programme for UK arrivals. The presence of SARS CoV-2 was detected in 3855 samples (1.9%), and 1913 SARS-CoV-2 genomes were selected from Day 2 travellers for genome sequencing based on Ct < 30 according to Government guidelines. High quality sequencing data and subsequent lineage analysis was obtained for 1635 (85.5%) samples.
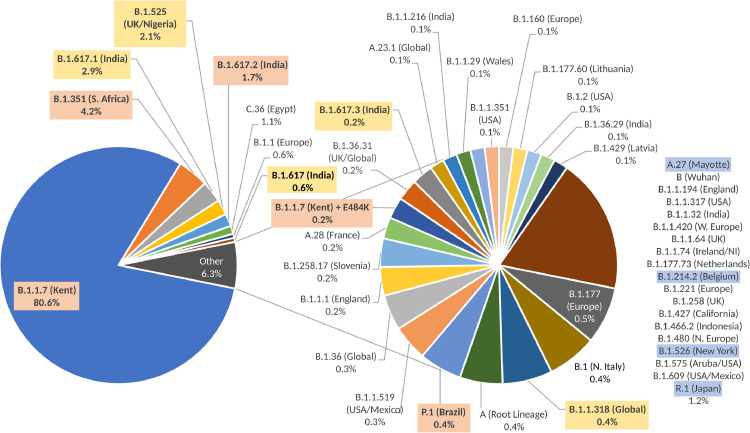
In total, 49 different variants were identified (Fig. 1) with the most prevalent represented by the B.1.1.7 (Kent) variant in more than 80% (n = 1317) of samples sequenced. The second most common was the South African variant B.1.351 (n = 69; 4.2%). In addition, a number of other VOCs including B.1.617.2 (n = 28; 1.7%), P.1 (n = 6; 0.4%), B.1.1.7 with E484K (n = 3; 0.2%) and VUIs B.1.617.1 (n = 49; 2.9%), B.1.617 (n = 9; 0.6%), B.1.617.3 (n = 3; 0.2%), B.1.525 (n = 31; 2.0%) and B.1.1.318 (n = 7; 0.4%) were identified. Single instances of variants under monitoring A.27, B.1.214.2, B.1.526 and R.1 were also identified.
Fig. 1.
Lineage assignments of 1635 SARS-CoV-2 sequences of samples collected from day 2 international arrivals in the UK between 11 March and 14 April 2021. Variants of Concern B.1.1.7 (n = 1317), B.1.1.7 with E484K (n = 3), B.1.351 (n = 69), B.1.617.2 (n = 28), and P.1 (n = 6) are highlighted in orange, Variants under Investigation B.1.617.1 (n = 49), B.1.617 (n = 9), B.1.617.3 (n = 3), B.1.525 (n = 34), and B.1.318 (n = 7) are highlighted in yellow and Variants under Monitoring A.27, B.1.214.2, B.1.526 and R.1 (all n = 1) are highlighted in blue (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
A phylogenetic analysis for all sequences that had been retained in full (n = 1361) was conducted (Supplementary Fig. 1). This showed a clear separate clade containing the majority of variants, which included all B.1.1.7 variants. This analysis confirmed the shared lineage of Indian variants B.1.617.1, B.1.617.2 and B.1.617.3 and showed they form an entirely separate evolutionary branch from the root to all other identified variants. B.1.351 and B.1.525 shared a common ancestor on a separate branch to B.1.1.7.
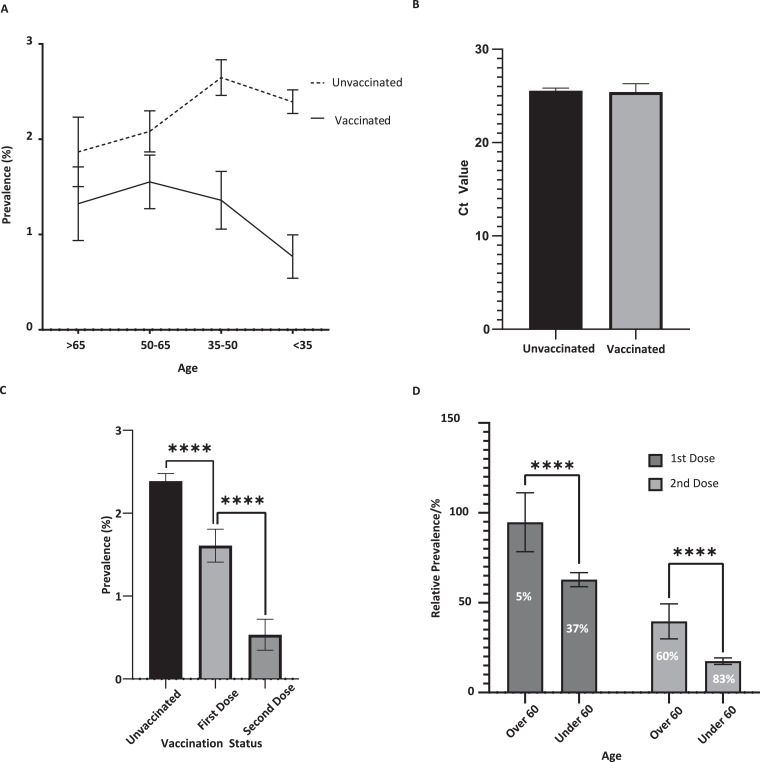
Across all samples tested for which vaccination status and age were known (n = 128,566, 63.3%), the overall prevalence for unvaccinated individuals was 2.4% (n = 2449/105,193), compared to 1.3% (n = 276/21,992) for those that had received at least one vaccine dose, representing a 46% overall vaccine effectiveness (Table 1 and Fig. 2A). Samples from individuals in the lower age groups < 35 and 35–50 exhibited a significant reduction in prevalence within the vaccinated cohort (with a vaccine effectiveness of 67% and 48% respectively; p < 0.00001), while the 50–65 and > 65 age groups exhibited vaccine effectiveness of 25% (p = 0.007) and 27% (p = 0.075) respectively (Table 1 and Fig. 2A).
Table 1.
Prevalence of SARS-CoV-2 infections by age and vaccination status.
| Age Group | Unvaccinated |
Vaccinated |
Vaccine Effectiveness(%) | P Value | ||||
|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Prevalence (%) | Positive | Negative | Prevalence (%) | |||
| <35 | 1284 | 53,710 | 2.3 | 43 | 5592 | 0.8 | 67.3 | < 0.00001 |
| 35–50 | 726 | 27,420 | 2.6 | 75 | 5517 | 1.3 | 48.0 | < 0.00001 |
| 50–65 | 342 | 16,420 | 2.0 | 113 | 7282 | 1.5 | 25.1 | 0.007 |
| 65+ | 97 | 5194 | 1.8 | 45 | 3325 | 1.3 | 27.2 | 0.075 |
| Total | 2449 | 102,744 | 2.4 | 276 | 21,716 | 1.3 | 46.1 | < 0.00001 |
Fig. 2.
(A) Prevalence of SARS-CoV-2 in unvaccinated individuals (n = 105,193) and those who had received at least one dose of a vaccine (n = 21,992) stratified by age with 95% Wald confidence intervals. (B) Ct values of positive cases from vaccinated (n = 274) and unvaccinated (n = 2417) individuals with 95% Wald confidence intervals. (C) Prevalence of SARS-CoV-2 in unvaccinated (n = 106,501) individuals and those who had received one (n = 1 5171) or two (n = 5844) doses of a vaccine with 95% Wald confidence intervals. (D) Relative prevalence of infection in individuals above and below the age of 60 after one (n = 3443 and n = 11,510 respectively) and two (n = 1617 and n = 4112 respectively) doses of a vaccine compared to unvaccinated individuals with Wald 95% confidence intervals. **** indicates p < 0.0001.
Mean Ct values of positive cases from vaccinated (n = 274) and unvaccinated (n = 2417) individuals were 25.42 and 25.55 respectively, indicating no significant difference in the viral loads between these two groups (Fig. 2B).
The effectiveness of a second vaccine dose was significantly greater than the first dose alone. Individuals that had received one dose (n = 15,415) had a prevalence of 1.6% (n = 244), a 32% reduction compared to unvaccinated individuals, while individuals that had received two doses (n = 5844) had a prevalence of 0.5% (n = 31), representing a 78% reduction (Fig. 2C). In individuals who had received a single dose of a vaccine, the effectiveness was 5% in the >60 cohort, compared to 37% in the <60 cohort. While the second dose showed a significant improvement in both the >60 and <60 cohorts, disease prevalence was still higher in twice-vaccinated >60 (13/1617, 0.8%), a decrease of 60%, compared to <60 (17/4112, 0.4%), a decrease of 83% (Fig. 2D).
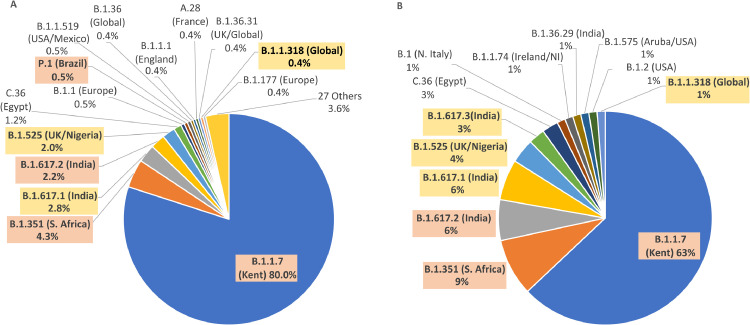
The prevalence of B.1.1.7, B.1.351, B.1.525 and combined B.1.617 variants in vaccinated and unvaccinated individuals was compared using Šídák's multiple comparisons test (Fig. 3). B.1.1.7 had a lower prevalence (p < 0.0001) in vaccinated individuals (n = 51, 63%) compared to unvaccinated individuals (n = 607, 80%), while B.1.617 had a higher prevalence (p = 0.007) in vaccinated individuals (n = 12, 15%) compared to unvaccinated individuals (n = 38, 5%). While B.1.351, B.1.525 both were more prevalent in vaccinated individuals, these did not reach significance.
Fig. 3.
Lineage assignment of SARS-CoV-2 sequences from unvaccinated (A, n = 759) and vaccinated (B, n = 81) day 2 international arrivals in the UK between 11 March and 14 Apr 2021. Variants of Concern B.1.1.7 (A: n = 607, B: n = 51), B.1.351 (A: n = 33, B: n = 7), B.1.617.2 (A: n = 17, B: n = 5) and P.1 (A: n = 4, B: n = 0) are highlighted in orange. Variants under Investigation B.1.617.1 (A: n = 21, B: n = 5 ), B.1.617.3 (A: n = 0, B: n = 3), B.1.525 (A: n = 15, B: n = 3) and B.1.1.318 (A: n = 3, B: n = 1) are highlighted in yellow (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
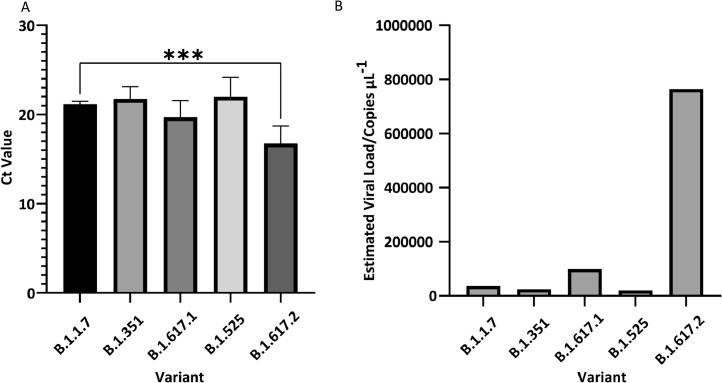
The mean Ct values for the most prevalent variants (n > 20) were compared to B.1.1.7 (Ct = 21.2) using Dunnett's multiple comparisons test (Fig. 4A). B.1.617.2 had a significantly lower Ct value (16.7) compared to B.1.1.7 (p = 0.0004). Notably, this is equivalent to a 21-fold increase in viral load compared to B.1.1.7 (Fig. 4B). The combined B.1.617 variants (B.1.617/.1/.2/.3), which share a common lineage root, also had a lower mean Ct value (18.8), equivalent to a 5.3-fold increase in viral load (p = 0.001) when compared to B.1.1.7.
Fig. 4.
(A) Mean Ct values for 5 most prevalent variants (n > 20) with 95% Wald confidence intervals. *** represents p < 0.001 by Dunnett's multiple comparisons test. (B) Estimated viral loads calculated from mean Ct value.
4. Discussion
The cross-border transmission of SARS-CoV-2 during resumption of international travel is a public health risk to the United Kingdom and could potentially undermine the success of the ongoing vaccination programme. High-risk variants characterised by mutations in the spike protein domain have been shown to reduce neutralisation sensitivity to convalescent sera and monoclonal antibodies and therefore have the potential to bypass the protection afforded by the current vaccines [[15], [16], [17], [18],20]. Here we have shown that the Day 2 and Day 8 UK testing programme is successful in identifying traveller-related introduction of SARS-CoV-2 into the United Kingdom. We have detected a high prevalence (1.9%) of cross-border transmission of SARS-CoV-2 involving a broad range of variants including VOCs, VUIs and monitored variants. The unexpectedly high prevalence of COVID-19 infection may reflect the lower sensitivity requirements for pre-embarkation testing, which includes lateral flow, having lower sensitivity when compared to PCR testing. The B.1.1.7 (Kent) variant was the most commonly detected among UK arrivals (80.6%), mirroring its high prevalence within the UK population [21]. Due to its increased transmissibility, the B.1.1.7 variant has spread from less than 0.1% in November 2020 to more than 95% of new SARS-CoV-2 infections in England in February 2021 [12,33] and has now been detected globally in over 130 countries. The high diversity of 49 different SARS-CoV-2 variants we have identified in international travellers including VOCs and VUIs reflects the rapid biological evolution of SARS-CoV-2 leading to increased replication efficiency and transmissibility and potential for immune evasion [34].
Our data show that vaccination has a major impact in protecting individuals against infection dropping from a prevalence of 2.4% in the unvaccinated population to 1.3% in the vaccinated population. Notably, vaccine effectiveness varies between first and second doses and age demographics. Analysis of vaccinated vs non-vaccinated groups with at least one dose shows overall effectiveness of 46%. However, this was significantly age-related, ranging from 67% in < 35 age group to 27% in the > 65 group. Analysis of the effect of the second dose revealed that vaccine effectiveness was significantly higher following second dose. Overall, one dose had a vaccine effectiveness of 32% compared with 78% in those who received their second dose of the vaccine. In the > 60 age group, the effectiveness for a single dose was 5% compared to 37% in < 60 group. Notably, the effect of the second dose was significantly higher in both age groups with 60% effectiveness in > 60 compared to 83% in < 60.
The high vaccine protection against infection we observed in the younger age groups and effect of vaccine dosage confirms the findings of the SIREN prospective multicentre cohort study in which vaccine effectiveness against infection was 70% 21 days after a single dose and 85% 7 days after two doses [35]. This study relates to healthcare workers immunised with the BNT162b2mRNA (Pfizer-BioNTech) vaccine and involves a relatively young age demographic with a median age of 46.1 years (IQR 36.0–54.1). The reduced vaccine effectiveness we have observed for SARS-CoV-2 with increasing age is in keeping with the development of immunesenescence [36], [37], [38]. This mirrors the reduced prophylactic efficacy of vaccinations against other viral and bacterial pathogens such as influenza [39], hepatitis[40] and pneumococcus [41] in which diminished titre, efficacy and affinity of antibodies is observed as a result of age-related impairment of innate and adaptive immune responses. Optimisation of vaccination protocols for older adults have previously been investigated using a range of approaches including booster doses and improved adjuvants [36]. The self-declaration data did not provide vaccination type used and so it was not possible to determine vaccine effectiveness in relation to individual vaccine types using different formulations. Rigorous assessment of the effectiveness of different vaccine types for preventing infection in > 60 age population will be important going forward in order to select the optimal vaccines to circumvent the constraints imposed by immunesenescence.
The proportion of the B.1.1.7 (Kent) variant in the vaccinated population was significantly reduced compared with the non-vaccinated population, with a corresponding expansion in the relative proportion of high-risk variants including the B.1.351 (South Africa), B.1.525 (Nigeria/UK), and in particular the B.1.617.1, B.1.617.2, B1.617.3 (India) variants. This suggests current vaccines are less effective in preventing infection by high-risk variants when compared to the current dominant B.1.1.7 Kent strain. This is in keeping with studies showing that emerging variants of SARS-CoV-2 can evade neutralising antibodies induced by previous infection or vaccination through mutations in the spike protein, including the receptor-binding domain (RBD) [[15], [16], [17], [18],20]. However, this trend needs to be confirmed in a larger cohort and is the focus of our ongoing analysis of day 2 testing and sequencing data. Notably the viral load in PCR test positive cases was also linked to variant type with B.1.617.2 showing a marked increase in mean viral load (~8.3 × 108 virions/mL) compared to the B.1.1.7 variant (~3.6 × 107 virions/mL). This variant first detected in India is designated as a VOC and we have observed a rapidly increasing prevalence in travellers entering the UK. Day 2 testing and sequencing has revealed an increased prevalence of B.1.617 variants from 5.4% over the period from 11 March-14 April 2021 to 23% over the period 15 April-6 May 2021. Over the same period, the B.1.617.2 variant in particular increased in prevalence from 1.7 to 17% (unpublished data). In relation to the B.1.617 lineage, spike L452R is notable for its ability to induce substantially reduced sensitivity to convalescent sera and for having the greatest increase in binding affinity to ACE2 [42]. B.1.617.1 also features E484Q, a similar mutation to the E484K SNP found in B.1.351, B.1.525 and P.1, which has been demonstrated to mediate immune escape. B.1.617.2 on the other hand contains T478K in place of E484Q. This is a less studied mutation, but a separate in silico analysis has shown it to cause the largest increase in binding affinity to ACE2 of 95 mutations examined [43]. The high risk mutations that characterise the B.1.617 variant may account for the strikingly high viral load we have observed in infected travellers compared with other variants and the increased prevalence in vaccinated individuals. This unusually aggressive biology compared to other VOCs and VUIs is consistent with our phylogenetic analysis showing that variants B.1.617.1, B.1.617.2 and B.1.617.3 form an entirely separate evolutionary branch from the root to all other identified variants.
The integrated workflow combining TaqPath CE-IVD COVID-19 RT-PCR and Ion AmpliSeq SARS-CoV-2 Next Generation Sequencing assay has a number of distinct advantages. The integrated approach allows testing and sequencing to be performed within a 48–72 h turnaround time ensuring that positive cases are detected and sequenced in a time frame that allows effective active contact tracing. This integrated workflow also circumvents the problems associated with referral of PCR test positive cases to a centralised reference laboratory. Referral causes significant delays arising as a consequence of complex logistics and the requirement for the repeat PCR of samples. Interlaboratory transfer of extracted RNA samples impacts on turn-around times and RNA is inherently unstable and so requires temperature controlled transport. The RT-PCR cycle threshold (Ct) values are used to normalise samples for library amplification as part of the next generation sequencing protocols. The reference laboratory will therefore need to repeat RT-PCR on the transferred samples due to potential degradation of the sample and differences in laboratory protocols and instrumentation.
Our positive test and sequencing data is communicated to Public Health England (PHE) as soon as test results are available via the Second Generation Surveillance System (SGSS), a national surveillance system that holds all test results, enabling active follow up of cases and their contacts. The effectiveness of border testing is critically dependent on countries having well developed public health infrastructure enabling test results to be rapidly actioned.
The state instituted traveller SARS-CoV-2 testing program introduced by Alaska in June 2020 during the re-opening phases of the pandemic contributed to Alaska having a lower number of excess deaths when compared with most US states and was the first study to highlight the importance of cross border testing [44].
Here we have shown that test and sequencing for international arrivals is playing a critical role in controlling the import of high-risk variants into the UK. Monitoring cross-border transmission will become an increasing priority for many countries as vaccine programmes reach an advanced stage of roll out. However, the great public health benefits of vaccination programmes are at risk from the increasing numbers of variants of concern emerging as a result of rapidly increasing rates of global infections. The number of new cases each week has “nearly doubled” over the past two months. Continuing globally high transmission rates of SARS-CoV-2 means the potential for the emergence of further variants with greater immunity-evading properties is high. Test and sequencing for international travellers combined with self-quarantine measures will therefore play a critical role in the public health measures required by countries to limit cross-border transmission of SARS-CoV-2 high risk variants and monitor potential future targets for vaccines.
Data sharing statement
The datasets analysed during the current study are provided in the Supplementary File. Further technical details are available from the corresponding author upon reasonable request.
Funding
Data analysis conducted in this study was limited to secondary use of information previously collected in the course of normal care. No dedicated funding source was allocated for this study.
Declaration of Competing Interest
ML and GW are shareholders and company directors of Oncologica UK Ltd. GW, AL, RB, KC, KH and ML are currently employed at Oncologica UK Ltd.
Acknowledgments
The authors thank the technical support staff at Oncologica UK Ltd for their valuable contribution to the study.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.eclinm.2021.101021.
Contributor Information
Gareth H Williams, Email: keeda.hardisty@oncologica.com.
Keeda-Marie Hardisty, Email: keeda.hardisty@oncologica.com.
Appendix. Supplementary materials
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China. N Engl J Med. 2019;382(8):727–733. doi: 10.1056/NEJMoa2001017. http://www.nejm.org/doi/10.1056/NEJMoa2001017 [Internet]2020 Feb 20Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Worldometers.info. COVID-19 Coronavirus pandemic [Internet]. 2021. Available from: https://www.worldometers.info/coronavirus/.
- 3.Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr Biol. 2020;30(7):1346–1351. doi: 10.1016/j.cub.2020.03.022. https://linkinghub.elsevier.com/retrieve/pii/S0960982220303602 [Internet]Apre2. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjee A., Doxey A.C., Mossman K., Irving A.T. Unraveling the zoonotic origin and transmission of SARS-CoV-2. Trends Ecol. Evol. 2021;36(3):180–184. doi: 10.1016/j.tree.2020.12.002. https://linkinghub.elsevier.com/retrieve/pii/S0169534720303487 [Internet]MarAvailable from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dhama K., Patel S.K., Sharun K., Pathak M., Tiwari R., Yatoo M.I. SARS-CoV-2 jumping the species barrier: zoonotic lessons from SARS, MERS and recent advances to combat this pandemic virus. Travel Med. Infect. Dis. 2020;37 doi: 10.1016/j.tmaid.2020.101830. https://linkinghub.elsevier.com/retrieve/pii/S1477893920303264 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elena S.F., Sanjuán R. Adaptive value of high mutation rates of RNA viruses: separating causes from consequences. J. Virol. 2005;79(18):11555–11558. doi: 10.1128/JVI.79.18.11555-11558.2005. https://jvi.asm.org/content/79/18/11555 [Internet]Sep 15Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graham R.L., Baric R.S. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J. Virol. 2010;84(7):3134–3146. doi: 10.1128/JVI.01394-09. https://jvi.asm.org/content/84/7/3134 [Internet]Apr 1Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Á. O'Toole, E. Scher, A. Underwood, B. Jackson, V. Hill, J.T. McCrone, et al. pangolin: lineage assignment in an emerging pandemic as an epidemiological tool [Internet]. 2021. Available from: github.com/cov-lineages/pangolin. [DOI] [PMC free article] [PubMed]
- 9.Tang X., Wu C., Li X., Song Y., Yao X., Wu X. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020;7(6):1012–1023. doi: 10.1093/nsr/nwaa036. https://academic.oup.com/nsr/article/7/6/1012/5775463 [Internet]Jun 1Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.F. Di Giallonardo, I. Puglia, V. Curini, C. Cammà, I. Mangone, P. Calistri, et al. Emergence and spread of SARS-CoV-2 lineages B.1.1.7 and P.1 in Italy. Medrxiv [Preprint] 2021. Available from: https://www.medrxiv.org/content/101101/2021032421254277v1. [DOI] [PMC free article] [PubMed]
- 11.Anderson T. B.1.1.7: What we know about the novel SARS-CoV-2 variant. Am Soc Microbiol. 2021 [Internet] Available from: https://asm.org/Articles/2021/January/B-1-1-7-What-We-Know-About-the-Novel-SARS-CoV-2-Va. [Google Scholar]
- 12.Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021 doi: 10.1126/science.abg3055. https://www.sciencemag.org/lookup/doi/ (80-) [Internet]Mar 3;eabg3055. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Public Health England. Investigation of novel SARS-CoV-2 variant -technical briefing 2 [Internet]. 2020. Available from: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/959361/Technical_Briefing_VOC202012-2_Briefing_2.pdf.
- 14.Frampton D., Rampling T., Cross A., Bailey H., Heaney J., Byott M. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B1.1.7 lineage in London, UK: a whole-genome sequencing and hospital-based cohort study. Lancet Infect Dis. 2021 doi: 10.1016/S1473-3099(21)00170-5. https://linkinghub.elsevier.com/retrieve/pii/S1473309921001705 [Internet]Apr; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29(3):463–476. doi: 10.1016/j.chom.2021.02.003. https://linkinghub.elsevier.com/retrieve/pii/S1931312821000822 [Internet]Mare6. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.C.K. Wibmer, F. Ayres, T. Hermanus, M. Madzivhandila SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med 2021;27: 622-625. Available from: https://www.nature.com/articles/s41591-021-01285-x. [DOI] [PubMed]
- 17.Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591(7851):639–644. doi: 10.1038/s41586-021-03207-w. http://www.nature.com/articles/s41586-021-03207-w [Internet]Mar 25;Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Planas D., Bruel T., Grzelak L., Guivel-Benhassine F., Staropoli I., Porrot F. Sensitivity of infectious SARS-CoV-2 B1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med. 2021 doi: 10.1038/s41591-021-01318-5. http://www.nature.com/articles/s41591-021-01318-5 [Internet]Mar 26; Available from: [DOI] [PubMed] [Google Scholar]
- 19.Public Health England. Investigation of SARS-CoV-2 variants of concern in england-technical briefing 6 [Internet]. 2021. Available from: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/961299/Variants_of_Concern_VOC_Technical_Briefing_6_England-1.pdf.
- 20.Collier D.A., De Marco A., Ferreira I.A.T.M., Meng B., Datir R.P., Walls A.C. Sensitivity of SARS-CoV-2 B1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021 doi: 10.1038/s41586-021-03412-7. http://www.nature.com/articles/s41586-021-03412-7 [Internet]Mar 11; Available from: [DOI] [PubMed] [Google Scholar]
- 21.Public Health England. SARS-CoV-2 variants of concern and variants under investigation in england technical briefing 10 [Internet]. 2021. Available from: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/984274/Variants_of_Concern_VOC_Technical_Briefing_10_England.pdf.
- 22.Department of Health and Social Care. Day 2 and day 8 testing for international arrivals: minimum standards for providers [Internet]. 2021. Available from: https://www.gov.uk/guidance/testing-on-day-2-and-day-8-for-international-arrivals
- 23.Department for Transport. Coronavirus (COVID-19) testing before you travel to England [Internet]. 2021. Available from: https://www.gov.uk/guidance/coronavirus-covid-19-testing-for-people-travelling-to-england.
- 24.Fan H., Qin E., Wu Y., Guo Y., Zhang X., Yong Y. Severe acute respiratory syndrome coronavirus 2 isolate SARS-CoV-2/human/CHN/Wuhan_IME-WH04/2019, complete genome. National Center Biotechnology Information. 2020 https://www.ncbi.nlm.nih.gov/nuccore/MT291829 [Internet] Available from: [Google Scholar]
- 25.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20(4):1160–1166. doi: 10.1093/bib/bbx108. https://academic.oup.com/bib/article/20/4/1160/4106928 [Internet]Jul 19Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stecher G., Tamura K., Kumar S. Molecular Evolutionary genetics analysis (MEGA) for macOS. Russo C, editor. Mol Biol Evol. 2020;37(4):1237–1239. doi: 10.1093/molbev/msz312. https://academic.oup.com/mbe/article/37/4/1237/5697095 [Internet]Apr 1Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rambaut A., Holmes E.C., Á O.’.T., Hill V., McCrone J.T., Ruis C. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5(11):1403–1407. doi: 10.1038/s41564-020-0770-5. http://www.nature.com/articles/s41564-020-0770-5 [Internet]Nov 15Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32(1):268–274. doi: 10.1093/molbev/msu300. https://academic.oup.com/mbe/article-lookup/doi/10.1093/molbev/msu300 [Internet]JanAvailable from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Letunic I., Bork P. Interactive tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021 doi: 10.1093/nar/gkab301/6246398. https://academic.oup.com/nar/advance-article/doi/ [Internet]Apr 22Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graphpad Prism, version 9.1.0 [Software]. San Diego, California, USA: GraphPad software; available from: www.graphpad.com
- 31.Šidák Z. Rectangular confidence regions for the means of multivariate normal distributions. J Am Stat Assoc. 1967;62(318):626–633. doi: 10.1080/01621459.1967.10482935. [Internet]JunAvailable from: http://www.tandfonline.com/doi/abs/ [DOI] [Google Scholar]
- 32.Dunnett C.W. A multiple comparison procedure for comparing several treatments with a control. J Am Stat Assoc. 1955;50(272):1096–1121. doi: 10.1080/01621459.1955.10501294. [Internet]DecAvailable from: http://www.tandfonline.com/doi/abs/ [DOI] [Google Scholar]
- 33.Davies N.G., Jarvis C.I., Edmunds W.J., Jewell N.P., Diaz-Ordaz K., Keogh R.H. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. nature. 2021 doi: 10.1038/s41586-021-03426-1. http://www.nature.com/articles/s41586-021-03426-1 [Internet]Mar 15Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.V'kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19(3):155–170. doi: 10.1038/s41579-020-00468-6. http://www.nature.com/articles/s41579-020-00468-6 [Internet]Mar 28Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hall V.J., Foulkes S., Saei A., Andrews N., Oguti B., Charlett A. COVID-19 vaccine coverage in health-care workers in England and effectiveness of BNT162b2 mRNA vaccine against infection (SIREN): a prospective, multicentre, cohort study. Lancet. 2021;397(10286):1725–1735. doi: 10.1016/S0140-6736(21)00790-X. https://linkinghub.elsevier.com/retrieve/pii/S014067362100790X [Internet]MayAvailable from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lord J.M. The effect of aging of the immune system on vaccination responses. Hum Vaccin Immunother. 2013;9(6):1364–1367. doi: 10.4161/hv.24696. http://www.tandfonline.com/doi/abs/ [Internet].Jun 12Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bajaj V., Gadi N., Spihlman A.P., Wu S.C., Choi C.H., Moulton V.R. Aging, immunity, and COVID-19: how age influences the host immune response to coronavirus infections? Front Physiol. 2021;11 doi: 10.3389/fphys.2020.571416/full. https://www.frontiersin.org/articles/ [Internet]Jan 12Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nikolich-Zugich J., Knox K.S., Rios C.T., Natt B., Bhattacharya D., Fain M.J. SARS-CoV-2 and COVID-19 in older adults: what we may expect regarding pathogenesis, immune responses, and outcomes. GeroScience. 2020;42(2):505–514. doi: 10.1007/s11357-020-00186-0. https://link.springer.com/ [Internet]Apr 10Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.J.C. Seidman, S.A. Richard, C. Viboud, M.A. Miller Quantitative review of antibody response to inactivated seasonal influenza vaccines. influenza other respi viruses 2012 Jan;6 (1):52–62. Available from: http://doi.wiley.com/ 10.1111/j.1750-2659.2011.00268.x. [DOI] [PMC free article] [PubMed]
- 40.Vermeiren A.P.A., Hoebe C.J.P.A., Dukers-Muijrers N.H.T.M. High non-responsiveness of males and the elderly to standard hepatitis B vaccination among a large cohort of healthy employees. J Clin Virol. 2013;58(1):262–264. doi: 10.1016/j.jcv.2013.07.003. https://linkinghub.elsevier.com/retrieve/pii/S1386653213002783 [Internet]SepAvailable from: [DOI] [PubMed] [Google Scholar]
- 41.van Werkhoven C.H., Huijts S.M., Bolkenbaas M., Grobbee D.E., Bonten M.J.M. The impact of Age on the Efficacy of 13-valent pneumococcal conjugate vaccine in elderly. Clin Infect Dis. 2015;61(12):1835–1838. doi: 10.1093/cid/civ686. https://academic.oup.com/cid/article-lookup/doi/ [Internet]Dec 15Available from: [DOI] [PubMed] [Google Scholar]
- 42.Chen J., Wang R., Wang M., Wei G.W. Mutations strengthened SARS-CoV-2 infectivity. J Mol Biol. 2020;432(19):5212–5226. doi: 10.1016/j.jmb.2020.07.009. https://linkinghub.elsevier.com/retrieve/pii/S0022283620304563 [Internet]SepAvailable from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.R. Wang, J. Chen, K. Gao, G.W. Wei Vaccine-escape and fast-growing mutations in the United Kingdom, the United States, Singapore, Spain, South Africa, and other COVID-19-devastated countries. Arxiv [Preprint] 2021; Available from: http://arxiv.org/abs/2103.08023. [DOI] [PMC free article] [PubMed]
- 44.E.C. Ohlson, K.A. Porter, E. Mooring, C. Cutchins, A. Zink, J. McLaughlin Airport traveler testing program for SARS-CoV-2 - Alaska, June-November 2020 [Internet]. Centres for Disease Control and Prevention 2021. Available from: https://www.cdc.gov/mmwr/volumes/70/wr/mm7016a2.htm?s_cid=mm7016a2_w. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.