Abstract
The microbiota plays essential roles in health and disease, in both the intestine and the extra-intestine. Dysbiosis of the gut microbiota causes dysfunction in the intestine, which leads to inflammatory, immune, and infectious diseases. Dysbiosis is also associated with diseases beyond the intestine via microbial translocation or metabolisms. The in situ breast microbiome, which may be sourced from the gut through lactation and sexual contact, could be altered and cause breast diseases. In this review, we summarize the recent progress in understanding the interactions among the gut microbiome, breast microbiome, and breast diseases. We discuss the intestinal microbiota, microbial metabolites, and roles of microbiota in immune system. We emphasize the novel roles and mechanisms of the microbiome (both in situ and gastrointestinal sourced) and bacterial products in the development and progression of breast cancer. The intestinal microbial translocation suggests that the gut microbiome is translocated to the skin and subsequently to the breast tissue. The gut bacterial translocation is also due to the increased intestinal permeability. The breast and intestinal microbiota are important factors in maintaining healthy breasts. Micronutrition queuine (Q) is derived from a de novo synthesized metabolite in bacteria. All human cells use queuine and incorporate it into the wobble anticodon position of specific transfer RNAs. We have demonstrated that Q modification regulates genes critical in tight junctions and migration in human breast cancer cells and a breast tumor model. We further discuss the challenges and future perspectives that can move the field forward for prevention, diagnosis, and treatment of breast diseases.
Keywords: Breast cancer, Breast microbiome, Dysbiosis, Inflammation, Metabolite, Micronutrition, Probiotics, Transfer RNAs
Introduction
The human body harbors far more microbial cells (100 trillion) and viruses (quadrillion) than human cells.1,2 These microbes constitute our microbiota, and the genes that they encode are known as the microbiome. This complex community contains bacteria, eukaryotes, viruses, and archaea, which greatly impact our health and physiology by interacting with one another and the host. The microbiota, as our “forgotten organ”,3 is most heavily colonized in the gastrointestinal tract and plays major roles, such as energy harvesting and storage, the metabolic functions of fermenting and absorbing undigested carbohydrates,4 and interaction with the immune system.5 Far beyond the crucial role in nutrition6 and metabolism in intestine, the gut microbiome is also known to influence the physiological functions of other organs or tissues,7, 8, 9 including the breast.
Breast cancer is currently the major cancer in women in many regions of the world, and its incidence has risen to unprecedented levels in recent decades.10 Breast cancer is broadly categorized into carcinoma in situ (also known as non-invasive carcinoma) and invasive (infiltrating) carcinoma. Breast carcinoma is further sub-classified as either ductal or lobular, as distinguished by the growth patterns and cytological features from the basis. Ductal carcinoma in situ (DCIS) is considerably more common than the lobular carcinoma in situ counterpart and encompasses a heterogeneous group of tumors.11 According to gene expression profiles and immunohistochemistry of hormone receptors [estrogen (ER+) and progesterone (PR+) and HER2 (human epidermal growth factor receptor 2)], breast cancer is categorized into five subtypes: Luminal A, Luminal B, Normal-like, HER2+ without ER or PR (HER2-enriched), and a “basal-like” or triple negative subtype that does not express any of the three receptors.12 Breast cancer is not only the most frequently diagnosed cancer among women worldwide (except for non-melanoma skin cancers) and affects the entire lives of women but is also one of the leading causes of cancer mortality in women.13,14 A complex interaction exists between genes and the environment in breast cancer. However, under perfect conditions of uncommon genetic mutations and environmental exposure, only a fraction of cases develop to cancer.15 Therefore, other undescribed pathways should exist that lead to or promote the development of breast cancer. The bacterial communities within the host, and especially those in the gastrointestinal (GI) tract, could be one of the environmental factors related to breast cancer. It was reported that women with HER2+ breast tumors compared to HER2– ones shown 12–23% lower alpha diversity, Shannon index, lower abundance of Firmicutes and higher abundance of Bacteroidetes in the study performed with 37 breast cancer patients with ER+PR+, ER+PR– and ERPR status.16
This review highlights recent progress in understanding the interactions between breast diseases (primarily breast cancer) and the microbiome in intestine and breast. We summarize the roles and mechanisms of the microbiome (both in situ and GI sourced) in the cause, development, and progression of breast cancer. We also discuss the challenges and future perspectives in the fields of the microbiome and breast diseases.
Basic roles of the gut microbiome in the gastrointestinal (GI) tract and beyond
It is estimated that the human microbiota outnumbers human cells present in our bodies, and this number is increasing with the identification of candidate bacterial species.17,18 The most heavily colonized organ is the GI tract, and the colon alone is estimated to contain over 70% of all of the microbes of the human body.17 Recent studies further indicate that the ratio of bacteria to human cells is about 1:1.19 Once fully developed by microbial acquisition and influenced by host factors, environmental cues, and self-assembly rules exerted by microbes themselves, the microbiome becomes an essential acquired organ that supplies many vital functions to the host. In the absence of gut microbiota or after ablation by treatment with broad-spectrum antibiotics, these functions can be destroyed or significant consequences can occur, such as improper development of the immune system and antibiotic-associated colitis.20
The interactions between the gut microbiome and its host were explored using metagenomic sequencing technologies that enable researchers to study the microbial communities that colonize the body in a culture-independent manner. The rough taxonomic disease associations can be analyzed based on the 16S ribosomal RNA (rRNA) gene amplicon data. Moreover, the robust microbiome disease associations can be assessed with shotgun metagenomics that enable analyses with high taxonomic resolution and analyses of gene functions that were not available in 16S rRNA sequencing.
The microbiota and immune system develop and mature together beginning at birth, or even potentially in the womb, and are probably essential in shaping the immune system response to avoid unwanted immune reactions for intestinal microbial components.21 Dietary changes are known to affect both the composition and function of the gut microbial communities, which in turn can modulate the host's innate and adaptive immune system. Commensals exert profound effects on the development and function of the immune system and therefore likely further influence immune-mediated diseases. Indeed, the prevention, exacerbation or induction of intestinal bacteria in numerous autoimmune, allergic or inflammatory diseases and malignancies have been studied.22 Segmented filamentous bacteria (SFB), an unculturable, spore-forming, and potently immunomodulatory species, was reported to support the develop of autoimmune arthritis and experimental autoimmune encephalomyelitis.22
The primarily general dysbiotic alterations of multiple disease, including colorectal cancer, type 2 diabetes, Parkinson's diseases, ulcerative colitis and Crohn's diseases,23, 24, 25, 26, 27 were studied by comparing the microbiome changes, which highlighted the need to examine the disease specificity of microbiome signatures. Increasing evidence suggests that the reach of the gut microbiome extends beyond the intestine and further affects systemic processes, such as metabolism and organ functions of the brain through the gut–brain axis, cardiovascular system, and others. Furthermore, deficiencies/dysbiosis of the gut microbiota not only lead to intestinal inflammatory and infectious diseases,28 but also are importantly associated with other diseases beyond the GI tract, such as allergic diseases and nervous diseases.
Human gut microbiota contributes to brain function via neural, humoral, and immune pathways and also via the cumulative effects of microbial metabolites. Our studies have been the first to report the associations between dysbiosis and intestinal inflammation in the development of ALS.29,30 Inversely, ALS affects the gastrointestinal tract, as supported by the common complaint of constipation in ALS patients.29,31 Furthermore, we demonstrated that the bacterial product butyrate could slow ALS progression in an ALS animal model.32,33
Overall, changes in the composition and structure of the human microbiota may predispose individuals to many diseases, although the relationships between the microbiota and disease states beyond GI tract are not cause-and-effect relationships. It is clear that the microbiome is an important contributor to disease states, including diseases of the immune system (multiple sclerosis, rheumatoid arthritis, type 1 diabetes, systemic lupus erythematous (SLE)), nervous system (Alzheimer's disease, Parkinson's disease, ALS), respiratory system (asthma, chronic obstructive pulmonary disease, cystic fibrosis), urinary system (chronic kidney disease), and liver (non-alcoholic fatty liver disease, non-alcoholic steatohepatitis, cirrhosis).
Breast microbiome and breast diseases
Breast microbiota in situ
The human breast contains a diverse and unique community of microbiota that is distinct from that found at other body sites, regardless of the sample location within the breast, pregnancy history, presence/absence of breast malignancy, age, country of origin, and sequencing technologies.34 Here, we refer to this community as the “breast microbiome” to differentiate it from the microbiome in other locations of the human body. The bacterial populations in human breast milk and breast tissue play a role in the development of healthy infant gut microbiota and also in the health of women. The diversity of breast bacteria is comparable to that observed in other microbial compartments (e.g., intestine) but is greater than that observed in the vagina.35 Proteobacteria is the most abundant phylum presented in breast tissue from women aged 18 to 90 with (around the tumor) or without cancer, as reported, and another common phylum is the Firmicutes.35 In our recent study, we found Butyrivibrio fibrisolvens-related clones, which belong to the Firmicutes phylum, in breast tumors with a xenograft mouse model.36
The microbiota, one of the bioactive compounds in human milk, have a key role in infant health. Although the origin of the breast milk microbiome is not currently clear, its presence corresponds to a perinatal period that starts during the third trimester of pregnancy and continues through lactation. It has been proposed that the human milk microbiota could derive from the skin of the mother, the oral cavity of the infant during suckling, or the gut of the mother via an entero-mammary pathway.37 Specific microbes are shared among the maternal microbiota/breast milk and infant intestinal microbiota, as well as in other maternal-neonatal niches, including the meconium, placenta and amniotic fluid.37, 38, 39, 40, 41, 42, 43 Butyrate-producing bacteria such as Coprococcus, Faecalibacterium, and Roseburia spp. were found to be shared in maternal feces and human milk.39 The maternal microbial environment impacts the newborn's immune development and consequently the infant's health both early and later life. Dysbiosis of bacteria in the breast could induce mastitis and can further influence infant growth, adequate microbial colonization, immune system maturation, and metabolic development during the breastfeeding period, which in turn, may have an impact on the programming of health later in life.44 Human milk contains short-chain fatty acids (SCFAs), such as butyrate, acetate and formic acid, which play a beneficial role in weight gain and adiposity during infancy.44 Human milk feeding is a new and important link among butyrate production, CAZymes, and atopic diseases in babies.45
Breast microbiota and breast health
The bacterial population in human breast milk and breast tissue plays a role in the development of a healthy infant gastrointestinal microbiota system, and also in the health of the female host. In breast tissue, Proteobacteria is the most abundant phylum, and another common phylum is Firmicutes. The diversity of bacteria in breast tissue is comparable to that observed in other microbial compartments (e.g., the intestine) but greater than that observed in the vagina.35 However, it is currently unclear whether the distribution of bacteria differs at the species level between normal and adjacent tumor tissues.
Using whole genome and transcriptome amplification, a pan-pathogen microarray strategy, and hierarchical cluster analysis of breast cancer samples, Banerjee et al detected unique and common viral, bacterial, fungal and parasitic signatures for each of the breast cancer types found distinct patterns for triple negative and triple positive breast cancer samples, whereas the ER+ and HER2+ samples shared similar microbial signatures.46 These signatures, unique or common to the different breast cancer types, offer a new understanding of the role of microbiome in breast cancer. In a qualitative survey of breast microbiota DNA using breast tumor tissue (ER+ breast cancer) and paired normal adjacent tissue from the same patient, Xuan et al. found that the bacterium Methylobacterium radiotolerans was relatively enriched in tumor tissue and that the bacterium Sphingomonas yanoikuyae was relatively enriched in paired normal tissue. Therefore, the relative abundances of these two bacterial species were inversely correlated in paired normal breast tissue but not in tumor tissue, indicating that dysbiosis is associated with breast cancer.47 Another study described that normal adjacent tissue from women with breast cancer (DCIS, invasive lobular and ductal carcinoma) compared to tissue from healthy controls had higher relative abundances of Bacillus, Enterobacteriaceae and Staphylococcus.48 These findings indicated that the bacteria or their components might influence the local immune microenvironment and that an unrecognized link between dysbiosis and breast cancer might exist. Urbaniak et al. investigated breast tissues collected from sites all around the breast in women age 18 to 90 with (around the tumor) or without cancer, and some subjects had no history of lactation. They detected a diverse population of bacteria, and the principal phylum was Proteocacteria. Although none of the subjects had signs or symptoms of infection, the presence of viable bacteria was confirmed in certain samples via culture.35 Tumors of Asian patients had significantly higher immune score, particular in the luminal B and HER2-enriched subtypes by analyzing gene expression profiles from eight data sets with both Asian (n = 462) and Western (n = 2186) patients who included Luminal A, Luminal B, HER2-enriched, basal and normal-like breast tumor types.49 Because the estrogen and breast local microenvironments are much different in premenopausal and postmenopausal women, the mechanism and related pathways of these microbiomes in breast cancer development and progression should be further studied in the future to offer greater choice in treatment of breast cancer and breast diseases. In breast tissues from patients with mastitis (inflammation of the breast with or without infection), researchers found that opportunistic pathogens, such as the mutually cooperative species Staphylococcus and Corynebacterium, were maintained in balance by normal microbiota.50 Questions still exist as to what role the breast microbiome plays in breast cancer or mastitis. It is likely that the potential source for those bacteria present in breast tissue is translocation from the gastrointestinal tract via the skin/nipple-areolar orifices or blood to reach the breast tissue.51 The potential intestinal bacterial translocation suggests that the gut microbiome is translocated to the skin and subsequently to the breast tissue via nipple-areolar orifices, nipple-oral contact via lactation, and/or sexual contact.52
Dendritic cells might transport bacteria to the breast via gut translocation.53 Bacteria could also translocate from intestine to other tissue or organs, such as the MLNs (mesenteric lymph nodes)54 and liver.55 It is reasonable to envision that a notably amount of bacteria might occasionally breach the gut epithelium and quickly arrive in the breasts through systemic circulation while the majority of intestinal microbiota remain located in the gut lumen and mucus layer. Gut dysbiosis can lead to impaired local, co-regional and systemic immune responses with the breakdown of mucosal barriers and translocation of the gut microbiome to the MLNs and into the peripheral circulation.56
Gut microbiome and breast cancer
Breast cancer development is highly related to the alteration of estrogen. Additionally, immune regulation, endogenous and exogenous substance metabolism, obese status and other factors involved with the gastrointestinal microbiome are all potentially related factors in the cause and development of breast cancer. Peroration of the microbiota in the gut by antibiotics has been shown to have negative impacts on breast cancer outcomes,57 and preexisting dysbiosis initiated by antibiotic treatment was found to be a host-intrinsic regulator of tissue inflammation and tumor cell dissemination in breast cancer.58 Therefore, progression of breast cancer could be affected by estrogen and integration of complicated biological behaviors of the gastrointestinal microbiome.
Gastrointestinal microbiome in estrogen metabolization and breast cancer
Estrogen is one of the most important factors in breast cancer and breast diseases. Estrogen is cycled in the body in complicated pathways involving various tissues and organs. The metabolism of estrogen, including hydroxylation and conjugation, takes place primarily in the liver and involves a pattern of enterohepatic circulation. After excretion in the bile and subsequently into the gastrointestinal tract, conjugated estrogen and its metabolites are deconjugated into a wide array of estrogen metabolites. These metabolites (free estrogens) are reabsorbed in the distal intestine and pass into the portal vein and on to other tissues, including the mammary glands.59 The ratios of estrogen metabolites (such as hydroxylated estrogen estrone and estradiol) to parent estrogens are inversely associated with the risk of breast cancer.60,61 Interestingly, the gastrointestinal microbiome can modulate systemic estrogens, and gut dysbiosis has been linked to postmenopausal breast cancer through interaction with a higher level of circulating estrogen. The deconjugation process of circulatory estrogens is mainly involved in plausible mechanisms for microbiota composition and diversity. It is reported that the ratio of metabolites to parent estrogens was directly associated with whole-tree phylogenetic diversity and the relative abundances of the microbiome.62 In a study including both men and postmenopausal women, researchers found that the richness of the gastrointestinal microbiome was inversely related to fecal estrogen levels.63 A recent study has reported the associations among dietary fiber, fecal microbiota, and estrogen metabolism in postmenopausal women with breast cancer.64 The objective of this cross-sectional study involved 29 newly diagnosed (stage 0-II), post-menopausal breast cancer patients, including both in situ (n = 7) and invasive (n = 22) clinical stage. There is a trend toward and inverse association between soluble fiber and estradiol levels, though no correlations between serum estradiol and estrone levels and species/genera or dietary fiber.64 However, a limited report on the association with systemic estrogen in premenopausal women (for whom the hormone level is highly variable throughout the menstrual cycle) confines the interpretation to the results from non-ovarian estrogen, such as in postmenopausal individuals. Although the similar association was found in premenopausal women, this observation has not been firmly established due to small sample size and greater variation in hormone levels.65 In a pilot study, the researchers found that early menarche (age ≤11) compared with later menarche (age ≥12) was associated with lower OTU, Chao1 index and lower abundance of Firmicutes, in 37 breast cancer patients who were Ⅰ/Ⅱ or Ⅲ stage at diagnosis.16
Microbial and digestive products and breast cancer
The products of digestion and microbial products originating from the intestine play a significant role in the process and development of breast diseases through the system in circulation. It has been reported that numerous bacterial metabolites are either the own metabolites of the microbes (such as short chain fatty acids, lactate, pyruvate) or modified products of the host (such as secondary bile acids, metabolites of aromatic amino acids, redox-modified sex steroids). These metabolites are bioactive and act through various pathways that involve modification of gene expression or modulation of signal transduction in the host. In the body, breast cells cannot directly contact the gut microbiota, but diseases could be influenced by these bacterial metabolites, including secondary bile acids, which are exclusively synthesized by the microbiome. Secondary bile acid could reduce breast cancer cell proliferation and aggressiveness and metastatic potential of primary tumors by inducing the mesenchymal-to-epithelial transition, as shown in both murine and bench experiments.66 Butyrate is important as food for cells lining the mammalian colon (colonocytes) and also serves as a histone deacetylase (HDAC) inhibitor and has attracted considerable interest as a possible regulator of cancer cell death. Sodium butyrate could promote apoptosis and ultrastructural changes67,68 and enhance anti-tumor activity in vitro.69 Sodium butyrate and other HDAC inhibitors have shown promising therapeutic outcomes against triple-negative breast cancer, especially if they are used in combination with other anticancer agents.70
Diet contents and quantity have major roles in shaping the gut microbiota composition and functions. Lifestyle could negatively affect breast cancer by impacting on the gastrointestinal flora and further the microbial and digestive products. Zengul et al. reported that the total dietary fiber is inversely associated with Clostridium hathewayi, while soluble fiber is inversely associated with Clostridium and insoluble fiber is positively associated with Bacteroides uniformis sp. by studying the newly-diagnosed (stage 0-II) and post-menopausal breast cancer patients who were in situ or invasive carcinoma.64 After 2-month of probiotics (Bifidobacterium longum, Lactobacillus rhamnosus) and Mediterranean diet administration, the bacterial species, bacterial diversity and Bacteroidetes-to-Firmicutes ratio were positively adjusted in the intervention group of breast cancer survivors, compared to the controls.71 Dietary supplementation and healthy diet could regulate the gut microbiota structure and function through the interaction with the commensal microbiota and the expression microbial enzymes and metabolites.
Intestinal immune modulation and breast cancer
The intestinal microbiota interact with the immune system of the host in the large surface area of the intestinal lumen. Immune modulation based on the intestinal microbiome should serve as a highly important component in breast cancer development and progression. For example, the systemic counter-interactions between microbes and immune items of IL-6 and neutrophils have been indicated in breast cancer.36,72 It is known that both neutrophils and lymphocytes are modulated by the microbiota and inflammation. The neutrophil-associated immune responses to the GI microbiome significantly impact carcinogenesis progress in non-intestinal tissues, such as mammary glands. Lakritz et al. tested the function of neutrophils in etiopathogenesis in the targeted pathogenic gut microbial infection animal model with a predilection to breast cancer. They depleted FVB-Tg(C3-1-TAg)cJeg/JegJ female mice of neutrophils with anti-Ly-6G antibody, and investigated that the tumor development was even completely blocked in this bacteria-triggered model by gastric gavage with Helicobacter hepaticus.73,74 Inflammation contributes to the breast malignancy. Gut bacteria may withhold the early carcinogenetic processes in the epithelia located distally from the intestine by down-regulating the systemic inflammatory index in the manner of proinflammatory cytokines and inflammatory cells.59,75 Because specific resident gut bacteria initiate mammary gland tumors in patients with GI tract inflammation, microbiota research may reveal novel targets for prevention and therapy of breast cancer.59 Moreover, the regular gastrointestinal microbiome is involved in the maturation of effector CD8+ T-cells (also known as killer T-cells), which are the most potent immune cells that can eradicate HER2/neu+ breast tumor cells.76 The innate immune inflammation associated with intestinal bacterial infections may play an important role in breast cancer. Infection of Rag2-deficient C57BL/6 Apcmin/+ mice with the intestinal bacterial pathogen Helicobacter hepaticus significantly promoted mammary carcinoma in females and enhances intestinal adenoma multiplicity via a tumor necrosis factor α (TNFα)-dependent mechanism.77 As symptoms of aggressiveness and invasiveness in breast cancer, especially hormone receptor-positive (HR+) breast cancer, it is also important to identify the role of intestinal microorganisms in metastatic dissemination of breast cancer.
Toll-like receptors (TRLs) are transmembrane receptors, an essential component of the innate immune system in a pattern recognition receptor family. TLRs recognize pathogen-associated molecular patterns (PAMP), which are molecular products derived from pathogens of bacteria, fungi and viruses. The emerging evidence suggests strong linkages between TLRs and breast cancer. Breast cancer pathologists have systematically investigated the expression of TLRs in breast cancer cell lines and tissues. The expression pattern of expressed TLRs is unique for each cell line, and the levels are different in the same cell line, e.g., expression of TLR2 in MDA-MB-231 is higher than in poorly invasive MCF-7 cells, whereas TLR3 expression was lowest in the MDA-MB-231 cell line.78 TLR2 activation leads to enhanced activity of NF-κB and increased levels of IL-6, TGF-β, VEGF and MMP9, which showed that high invasiveness may be associated with TLR2 expression. Except for the breast cancer cell lines, TLR3, 4, and 9 were also identified as highly expressed at the mRNA level in samples from breast carcinoma patients.78 Invasive ductal carcinoma showed a significant correlation between pathological features and TLR4 expression.78
Dysbiosis of intestinal microbiota and breast cancer
Preexisting dysbiosis in the gut was reported as a host-intrinsic regulator of tissue inflammation and tumor cell dissemination in breast cancer.58 More importantly, the compositional and functional differences in the gut microbial community between postmenopausal breast cancer patients and healthy controls highlighted the importance of the intestinal microbiome in breast cancer.79 Microbial diversity was higher in breast cancer patients than in controls, although relative species abundance in gut microbiota did not differ significantly between premenopausal breast cancer and premenopausal controls.79 A case–control clinical study that examines the association between intestine microbiota dysbiosis and the risk of breast cancer (stages Ⅰ and Ⅱ) is ongoing and is the first to evaluate the contribution to breast cancer of bacteria, archaea, virus and fungi and their alterations.80 Preexisting intestinal dysbiosis induced by antibiotics treatment was reported as a possible host-intrinsic regulator of tissue inflammation and tumor cell dissemination in hormone receptor-positive breast cancer.58
Recently, a study with newly diagnosed breast cancer patients (n = 25) and benign breast disease control patients (n = 25) reported that the abundance of Faecalibacterium was reduced in breast cancer patients, which was negatively correlated with various phosphorylcholines.81 Faecalibacterium prausnitzii, the inhibitor of the secretion of interleukin-6 (IL-6) and the phosphorylation of Janus kinases 2 (JAK2)/signal transducers and activators of transcription 3 (STAT3) in breast cancer cells, was also reported suppression the growth of breast cancer cells through inhibition of IL-6/STAT3 pathway.81 Also, our recent study has shown the potential involvement of the microbiome in breast cancer development. Intestinal bacteria including Lachnospiraceae, Lactobacillus and Alistipes were found to be markedly altered in xenograft nude mice after challenging breast cancer cells.36
Micronutrients are essential elements needed by life in small quantities. They include microminerals and vitamins. Micronutrients from the diet and microbiota are essential to human health. Queuine is derived from a de novo synthesized metabolite in bacteria;82,83 it is considered as micronutrition taken up through the intestine and circulated in the blood. All human cells then use queuine and incorporate it into the wobble anticodon position of specific transfer RNAs (tRNA) by two protein enzymes encoded in our genome.84 We have demonstrated that Q modification regulates genes critical in cell proliferation, tight junctions, and migration in human breast cancer cells in vitro and a breast tumor mouse model in vivo. We found evidence that microbiome was involved in the growth of breast cancer in vivo.36 The correlation of the gut microbiome and breast cancer is expected to aid future diagnostic assays for clinical application. For instance, metabolites (estrogen metabolites) combined with bacteria l biomarkers (Bacteroidetes-to-Firmicutes ratio) might be a new diagnosis method for breast cancer.
Conclusions and future perspectives
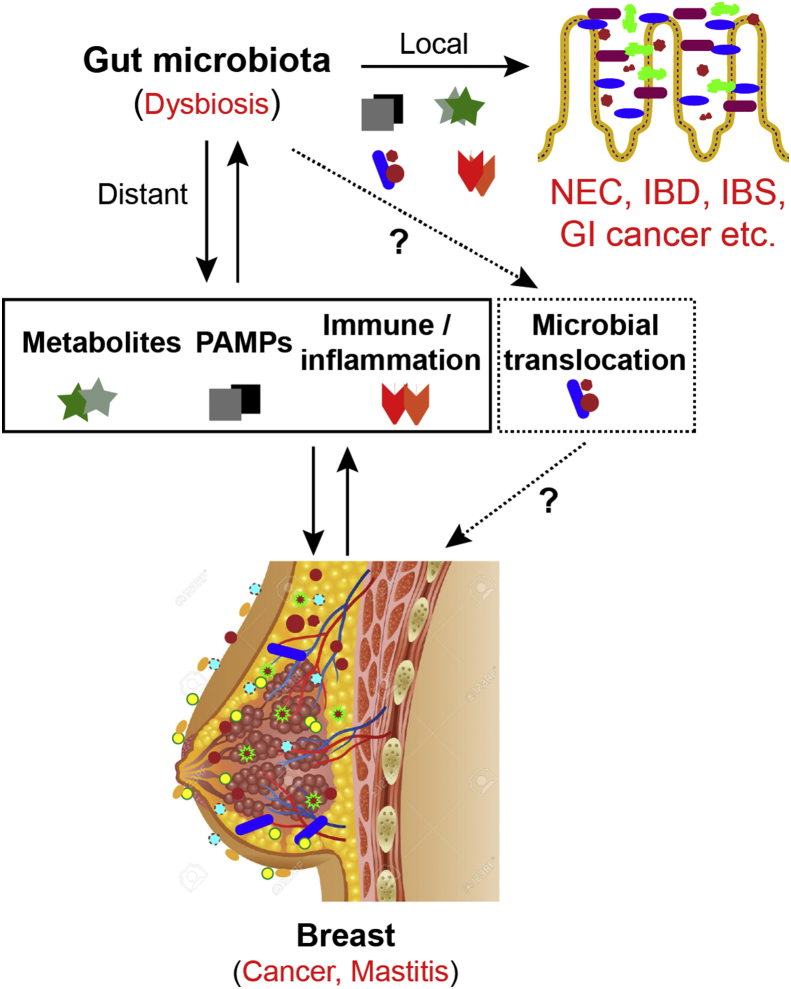
Host-microbiome interactions can promote effects in the gut and in distal organs, such as the breast. These effects are mediated by many microbe-derived effectors, including metabolites (free estrogens, SCFAs, secondary bile acids, and vitamins), immune and inflammatory modulators (TLR, cytokines), and secreted small natural products. Intestinal bacterial translocation may play a marked role in the process and development of breast cancer and other diseases. Dysbiosis of gastrointestinal microbiota not only has consequences for local diseases, but also for the distant breast. Metagenomic sequencing technologies enable us to yield glimpses into the complex and far more completely understood interactions between the gut microbiome and breast cancer. Better understanding of gut microbiome and breast microbiome will provide novel methods of treatment and diagnosis of breast carcinoma by recovering the steady-state of the microbiome (Fig. 1).
Figure 1.
Microbiota-host interactions in local intestine and distant breast. Gut microbiota influence amino acid bioavailability, metabolites (such as secondary bile acids, SCFAs), and PAMPs. Maternal milk plays an important role in establishing the gut bacterial system of the infant. Intestinal dysbiosis is associated with dysfunctions of estrogen enterogastric circulation, altered immunity, and inflammation of the host (TLRs, cytokines). However, the role of intestinal microbiota translocation between the gut and breast in the development and progression of breast disease is still unknown. All of these factors could potentially influence pathogenesis and progression of breast cancer and other breast diseases.
In the future, we need characterize the variability of the microbiota both in situ and in the GI tract. A better understanding of how such variability can result in similar or different functional profiles, and additional integrative studies should consider the interactions among the microbiota, host, and environment. It could enhance many aspects of our daily lives, such as optimizing the composition of infant formulas and offering new tools in our fight against breast cancer and obesity. Finally, understanding the cellular and organismal mechanisms of this microbiome-dependent micronutrient will advance the prevention of breast diseases and improve the life quality of patients. We could potentially manipulate microbiome through prebiotics and probiotics. Overall, we should consider the breast and intestinal microbiota as two important factors in maintaining health and monitoring its changes in breast diseases. Explore and understand the mechanisms of gut microbiome and breast microbiome will provide insights into effective approaches that manipulate microbiome in prevention and treatment of breast diseases.
Authors contribution
JZ: acquisition, analysis and interpretation, drafting of the manuscript;
YX: analysis and interpretation, writing the manuscript for important intellectual content;
JS: study concept and design; analysis and interpretation of data; writing the manuscript for important intellectual content, obtained funding, and supervision.
Conflict of Interests
The authors declare that they have no conflict of interest.
Acknowledgements
We would like to acknowledge the United States National Institute of Diabetes and Digestive and Kidney Diseases [grant numbers R01DK105118-01 and R01DK114126 to Jun Sun]; United States Department of Defense Congressionally Directed Medical Research Programs [grant number BC160450 to Jun Sun]; and the UIC Cancer Center funds to Jun Sun. The study sponsors play no role in the study design, data collection, analysis, and interpretation of data.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Whitman W.B., Coleman D.C., Wiebe W.J. Prokaryotes: the unseen majority. Proc Natl Acad Sci. U S A. 1998;95(12):6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clemente J.C., Ursell L.K., Parfrey L.W., Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148(6):1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Hara A.M., Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7(7):688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gill S.R., Pop M., Deboy R.T. Metagenomic analysis of the human distal gut microbiome. Science (New York, N.Y.). 2006;312(5778):1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chow J., Lee S.M., Shen Y., Khosravi A., Mazmanian S.K. Host-bacterial symbiosis in health and disease. Adv Immunol. 2010;107:243–274. doi: 10.1016/B978-0-12-381300-8.00008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song M., Chan A.T., Sun J. Influence of the gut microbiome, diet, and environment on risk of colorectal cancer. Gastroenterology. 2020;158(2):322–340. doi: 10.1053/j.gastro.2019.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brestoff J.R., Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14(7):676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruff W.E., Kriegel M.A. Autoimmune host-microbiota interactions at barrier sites and beyond. Trends Mol Med. 2015;21(4):233–244. doi: 10.1016/j.molmed.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sommer F., Backhed F. The gut microbiota—masters of host development and physiology. Nat Rev Microbiol. 2013;11(4):227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 10.Darbre P.D., Fernandez M.F. Environmental oestrogens and breast cancer: long-term low-dose effects of mixtures of various chemical combinations. J Epidemiol Commun Health. 2013;67(3):203–205. doi: 10.1136/jech-2012-201362. [DOI] [PubMed] [Google Scholar]
- 11.Malhotra G.K., Zhao X., Band H., Band V. Histological, molecular and functional subtypes of breast cancers. Canc Biol Ther. 2010;10(10):955–960. doi: 10.4161/cbt.10.10.13879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vuong D., Simpson P.T., Green B., Cummings M.C., Lakhani S.R. Molecular classification of breast cancer. Virchows Arch Int J Pathol. 2014;465(1):1–14. doi: 10.1007/s00428-014-1593-7. [DOI] [PubMed] [Google Scholar]
- 13.Ghoncheh M., Pournamdar Z., Salehiniya H. Incidence and mortality and epidemiology of breast cancer in the world. Asian Pac J Cancer Prev APJCP. 2016;17(S3):43–46. doi: 10.7314/apjcp.2016.17.s3.43. [DOI] [PubMed] [Google Scholar]
- 14.Global Cancer Observatory W. 2018. Cancer Population Fact Sheets.http://gco.iarc.fr/today/fact-sheets-populations [Google Scholar]
- 15.Rudolph A., Chang-Claude J., Schmidt M.K. Gene-environment interaction and risk of breast cancer. Br J Cancer. 2016;114(2):125–133. doi: 10.1038/bjc.2015.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu A.H., Tseng C., Vigen C. Gut microbiome associations with breast cancer risk factors and tumor characteristics: a pilot study. Breast Canc Res Treat. 2020;182(2):451–463. doi: 10.1007/s10549-020-05702-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun J., Dudeja P.K. Springer; New York: 2018. Mechanisms Underlying Host–Microbiome Interactions in Pathophysiology of Human Diseases. [Google Scholar]
- 18.Almeida A., Mitchell A.L., Boland M. A new genomic blueprint of the human gut microbiota. Nature. 2019;568(7753):499–504. doi: 10.1038/s41586-019-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sender R., Fuchs S., Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell. 2016;164(3):337–340. doi: 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 20.Sun J., Chang E.B. Exploring gut microbes in human health and disease: pushing the envelope. Genes Dis. 2014;1(2):132–139. doi: 10.1016/j.gendis.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomkovich S., Jobin C. Microbiota and host immune responses: a love-hate relationship. Immunology. 2016;147(1):1–10. doi: 10.1111/imm.12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vieira S.M., Pagovich O.E., Kriegel M.A. Diet, microbiota and autoimmune diseases. Lupus. 2014;23(6):518–526. doi: 10.1177/0961203313501401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bedarf J.R., Hildebrand F., Coelho L.P. Functional implications of microbial and viral gut metagenome changes in early stage L-DOPA-naive Parkinson's disease patients. Genome Med. 2017;9(1) doi: 10.1186/s13073-017-0428-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlsson F.H., Tremaroli V., Nookaew I. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498(7452):99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 25.Qin J., Li R., Raes J. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schirmer M., Franzosa E.A., Lloyd-Price J. Dynamics of metatranscription in the inflammatory bowel disease gut microbiome. Nat Microbiol. 2018;3(3):337–346. doi: 10.1038/s41564-017-0089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wirbel J., Pyl P.T., Kartal E. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med. 2019;25(4):679–689. doi: 10.1038/s41591-019-0406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bakke D., Sun J. Ancient nuclear receptor VDR with new functions: microbiome and inflammation. Inflamm Bowel Dis. 2018;24(6):1149–1154. doi: 10.1093/ibd/izy092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu S., Yi J., Zhang Y.G., Zhou J., Sun J. Leaky intestine and impaired microbiome in an amyotrophic lateral sclerosis mouse model. Physiol Rep. 2015;3(4) doi: 10.14814/phy2.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rowin J., Xia Y., Jung B., Sun J. Gut inflammation and dysbiosis in human motor neuron disease. Physiol Rep. 2017;5(18) doi: 10.14814/phy2.13443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herdewyn S., Cirillo C., Van Den Bosch L., Robberecht W., Vanden Berghe P., Van Damme P. Prevention of intestinal obstruction reveals progressive neurodegeneration in mutant TDP-43 (A315T) mice. Mol Neurodegener. 2014;9 doi: 10.1186/1750-1326-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y-g., Wu S., Yi J. Target intestinal microbiota to alleviate disease progression in amyotrophic lateral sclerosis. Clin Therapeut. 2017;39(2):322–336. doi: 10.1016/j.clinthera.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou J., Sun J. Does the gut drive amyotrophic lateral sclerosis progress? Future Med. 2015;5(5):375–378. doi: 10.2217/nmt.15.38. [DOI] [PubMed] [Google Scholar]
- 34.Younes J.A., Lievens E., Hummelen R., van der Westen R., Reid G., Petrova M.I. Women and their microbes: the unexpected friendship. Trends Microbiol. 2018;26(1):16–32. doi: 10.1016/j.tim.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 35.Urbaniak C., Cummins J., Brackstone M. Microbiota of human breast tissue. Appl Environ Microbiol. 2014;80(10):3007–3014. doi: 10.1128/AEM.00242-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J., Lu R., Zhang Y. tRNA queuosine modification enzyme modulates the growth and microbiome recruitment to breast tumors. Cancers. 2020;12(3) doi: 10.3390/cancers12030628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gomez-Gallego C., Garcia-Mantrana I., Salminen S., Collado M.C. The human milk microbiome and factors influencing its composition and activity. Semin Fetal Neonatal Med. 2016;21(6):400–405. doi: 10.1016/j.siny.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 38.Benito D., Lozano C., Jiménez E. Characterization of Staphylococcus aureus strains isolated from faeces of healthy neonates and potential mother-to-infant microbial transmission through breastfeeding. FEMS Microbiol Ecol. 2015;91(3) doi: 10.1093/femsec/fiv007. [DOI] [PubMed] [Google Scholar]
- 39.Collado M.C., Rautava S., Aakko J., Isolauri E., Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep. 2016;6 doi: 10.1038/srep23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jost T., Lacroix C., Braegger C.P., Rochat F., Chassard C. Vertical mother–neonate transfer of maternal gut bacteria via breastfeeding. Environ Microbiol. 2014;16(9):2891–2904. doi: 10.1111/1462-2920.12238. [DOI] [PubMed] [Google Scholar]
- 41.Makino H., Kushiro A., Ishikawa E. Mother-to-infant transmission of intestinal bifidobacterial strains has an impact on the early development of vaginally delivered infant's microbiota. PLoS One. 2013;8(11) doi: 10.1371/journal.pone.0078331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makino H., Martin R., Ishikawa E. Multilocus sequence typing of bifidobacterial strains from infant's faeces and human milk: are bifidobacteria being sustainably shared during breastfeeding? Benef Microbes. 2015;6(4):563–572. doi: 10.3920/BM2014.0082. [DOI] [PubMed] [Google Scholar]
- 43.Martín V., Maldonado-Barragán A., Moles L. Sharing of bacterial strains between breast milk and infant feces. J Hum Lactation. 2012;28(1):36–44. doi: 10.1177/0890334411424729. [DOI] [PubMed] [Google Scholar]
- 44.Prentice P.M., Schoemaker M.H., Vervoort J. Human Milk Short-Chain Fatty Acid Composition Is Associated with Adiposity Outcomes in Infants. J Nutr. 2019;149(5):716–722. doi: 10.1093/jn/nxy320. [DOI] [PubMed] [Google Scholar]
- 45.Macia L., Mackay C.R. Dysfunctional microbiota with reduced capacity to produce butyrate as a basis for allergic diseases. J Allergy Clin Immunol. 2019;144(6):1513–1515. doi: 10.1016/j.jaci.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 46.Banerjee S., Tian T., Wei Z. Distinct microbial signatures associated with different breast cancer types. Front Microbiol. 2018;9 doi: 10.3389/fmicb.2018.00951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xuan C., Shamonki J.M., Chung A. Microbial dysbiosis is associated with human breast cancer. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0083744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Urbaniak C., Gloor G.B., Brackstone M., Scott L., Tangney M., Reid G. The microbiota of breast tissue and its association with breast cancer. Appl Environ Microbiol. 2016;82(16):5039–5048. doi: 10.1128/AEM.01235-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen C.H., Lu Y.S., Cheng A.L. Disparity in tumor immune microenvironment of breast cancer and prognostic impact: Asian versus Western populations. Oncol. 2020;25(1) doi: 10.1634/theoncologist.2019-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sam Ma Z., Guan Q., Ye C., Zhang C., Foster J.A., Forney L.J. Network analysis suggests a potentially 'evil' alliance of opportunistic pathogens inhibited by a cooperative network in human milk bacterial communities. Sci Rep. 2015;5 doi: 10.1038/srep08275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fernandez M.F., Reina-Perez I., Astorga J.M., Rodriguez-Carrillo A., Plaza-Diaz J., Fontana L. Breast cancer and its relationship with the microbiota. Int J Environ Res Publ Health. 2018;15(8) doi: 10.3390/ijerph15081747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hieken T.J., Chen J., Hoskin T.L. The microbiome of aseptically collected human breast tissue in benign and malignant disease. Sci Rep. 2016;6 doi: 10.1038/srep30751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rescigno M., Urbano M., Valzasina B. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001;2(4):361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- 54.Adiliaghdam F., Almpani M., Gharedaghi M.H., Najibi M., Hodin R.A., Rahme L.G. Targeting bacterial quorum sensing shows promise in improving intestinal barrier function following burnsite infection. Mol Med Rep. 2019;19(5):4057–4066. doi: 10.3892/mmr.2019.10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma H.D., Zhao Z.B., Ma W.T. Gut microbiota translocation promotes autoimmune cholangitis. J Autoimmun. 2018;95:47–57. doi: 10.1016/j.jaut.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gopalakrishnan V., Helmink B.A., Spencer C.N., Reuben A., Wargo J.A. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Canc Cell. 2018;33(4):570–580. doi: 10.1016/j.ccell.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kirkup B., McKee A., Makin K. Perturbation of the gut microbiota by antibiotics results in accelerated breast tumour growth and metabolic dysregulation. bioRxiv. 2019 Jan 1:553602. [Google Scholar]
- 58.Rosean C.B., Bostic R.R., Ferey J.C. Pre-existing commensal dysbiosis is a host-intrinsic regulator of tissue inflammation and tumor cell dissemination in hormone receptor-positive breast cancer. Cancer Res. 2019;79(14):3662–3675. doi: 10.1158/0008-5472.CAN-18-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang J., Tan Q., Fu Q. Gastrointestinal microbiome and breast cancer: correlations, mechanisms and potential clinical implications. Breast Cancer (Tokyo, Japan) 2017;24(2):220–228. doi: 10.1007/s12282-016-0734-z. [DOI] [PubMed] [Google Scholar]
- 60.Falk R.T., Brinton L.A., Dorgan J.F. Relationship of serum estrogens and estrogen metabolites to postmenopausal breast cancer risk: a nested case-control study. Breast Cancer Res. 2013;15(2) doi: 10.1186/bcr3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fuhrman B.J., Schairer C., Gail M.H. Estrogen metabolism and risk of breast cancer in postmenopausal women. J Natl Cancer Inst. 2012;104(4):326–339. doi: 10.1093/jnci/djr531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fuhrman B.J., Feigelson H.S., Flores R. Associations of the fecal microbiome with urinary estrogens and estrogen metabolites in postmenopausal women. J Clin Endocrinol Metab. 2014;99(12):4632–4640. doi: 10.1210/jc.2014-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Flores R., Shi J., Fuhrman B. Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: a cross-sectional study. J Transl Med. 2012;10 doi: 10.1186/1479-5876-10-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zengul A.G., Demark-Wahnefried W., Barnes S. Associations between dietary fiber, the fecal microbiota and estrogen metabolism in postmenopausal women with breast cancer. Nutr Canc. 2020:1–10. doi: 10.1080/01635581.2020.1784444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Samavat H., Kurzer M.S. Estrogen metabolism and breast cancer. Canc Lett. 2015;356(2 Pt A):231–243. doi: 10.1016/j.canlet.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miko E., Vida A., Kovacs T. Lithocholic acid, a bacterial metabolite reduces breast cancer cell proliferation and aggressiveness. Biochim Biophys Acta Bioenerg. 2018;1859(9):958–974. doi: 10.1016/j.bbabio.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 67.Salimi V., Shahsavari Z., Safizadeh B., Hosseini A., Khademian N., Tavakoli-Yaraki M. Sodium butyrate promotes apoptosis in breast cancer cells through reactive oxygen species (ROS) formation and mitochondrial impairment. Lipids Health Dis. 2017;16(1) doi: 10.1186/s12944-017-0593-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Y., Hu P.C., Ma Y.B. Sodium butyrate-induced apoptosis and ultrastructural changes in MCF-7 breast cancer cells. Ultrastruct Pathol. 2016;40(4):200–204. doi: 10.3109/01913123.2016.1170083. [DOI] [PubMed] [Google Scholar]
- 69.Lee K.M., Lee M., Lee J. Enhanced anti-tumor activity and cytotoxic effect on cancer stem cell population of metformin-butyrate compared with metformin HCl in breast cancer. Oncotarget. 2016;7(25):38500–38512. doi: 10.18632/oncotarget.9522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garmpis N., Damaskos C., Garmpi A. Histone deacetylases as new therapeutic targets in triple-negative breast cancer: progress and promises. Cancer Genomics Proteomics. 2017;14(5):299–313. doi: 10.21873/cgp.20041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pellegrini M., Ippolito M., Monge T. Gut microbiota composition after diet and probiotics in overweight breast cancer survivors: a randomized open-label pilot intervention trial. Nutrition. 2020 doi: 10.1016/j.nut.2020.110749. [DOI] [PubMed] [Google Scholar]
- 72.Rutkowski M.R., Stephen T.L., Svoronos N. Microbially driven TLR5-dependent signaling governs distal malignant progression through tumor-promoting inflammation. Cancer Cell. 2015;27(1):27–40. doi: 10.1016/j.ccell.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lakritz J.R., Poutahidis T., Mirabal S. Gut bacteria require neutrophils to promote mammary tumorigenesis. Oncotarget. 2015;6(11) doi: 10.18632/oncotarget.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Erdman S.E., Poutahidis T., Tomczak M. CD4+ CD25+ regulatory T lymphocytes inhibit microbially induced colon cancer in Rag2-deficient mice. Am J Pathol. 2003;162(2):691–702. doi: 10.1016/S0002-9440(10)63863-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Erdman S.E., Poutahidis T. Gut bacteria and cancer. Biochim Biophys Acta Rev Cancer. 2015;1856(1):86–90. doi: 10.1016/j.bbcan.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gritzapis A.D., Voutsas I.F., Lekka E. Identification of a novel immunogenic HLA-A∗0201-binding epitope of HER-2/neu with potent antitumor properties. J Immunol. 2008;181(1):146–154. doi: 10.4049/jimmunol.181.1.146. [DOI] [PubMed] [Google Scholar]
- 77.Rao V.P., Poutahidis T., Ge Z. Innate immune inflammatory response against enteric bacteria Helicobacter hepaticus induces mammary adenocarcinoma in mice. Canc Res. 2006;66(15):7395–7400. doi: 10.1158/0008-5472.CAN-06-0558. [DOI] [PubMed] [Google Scholar]
- 78.Bhatelia K., Singh K., Singh R. TLRs: linking inflammation and breast cancer. Cell Signal. 2014;26(11):2350–2357. doi: 10.1016/j.cellsig.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 79.Zhu J., Liao M., Yao Z. Breast cancer in postmenopausal women is associated with an altered gut metagenome. Microbiome. 2018;6(1) doi: 10.1186/s40168-018-0515-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Plaza-Diaz J., Alvarez-Mercado A.I., Ruiz-Marin C.M. Association of breast and gut microbiota dysbiosis and the risk of breast cancer: a case-control clinical study. BMC Cancer. 2019;19(1):495. doi: 10.1186/s12885-019-5660-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ma J., Sun L., Liu Y. Alter between gut bacteria and blood metabolites and the anti-tumor effects of Faecalibacterium prausnitzii in breast cancer. BMC Microbiol. 2020;20:1–19. doi: 10.1186/s12866-020-01739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fergus C., Barnes D., Alqasem M.A., Kelly V.P. The queuine micronutrient: charting a course from microbe to man. Nutrients. 2015;7(4):2897–2929. doi: 10.3390/nu7042897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zaborske J.M., DuMont V.L., Wallace E.W., Pan T., Aquadro C.F., Drummond D.A. A nutrient-driven tRNA modification alters translational fidelity and genome-wide protein coding across an animal genus. PLoS Biol. 2014;12(12) doi: 10.1371/journal.pbio.1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.White R.J. RNA polymerase III transcription and cancer. Oncogene. 2004;23(18):3208–3216. doi: 10.1038/sj.onc.1207547. [DOI] [PubMed] [Google Scholar]