Abstract
Objective
Integrated, real-time data are crucial to evaluate translational efforts to accelerate innovation into care. Too often, however, needed data are fragmented in disparate systems. The South Carolina Clinical & Translational Research Institute at the Medical University of South Carolina (MUSC) developed and implemented a universal study identifier—the Research Master Identifier (RMID)—for tracking research studies across disparate systems and a data warehouse-inspired model—the Research Integrated Network of Systems (RINS)—for integrating data from those systems.
Materials and Methods
In 2017, MUSC began requiring the use of RMIDs in informatics systems that support human subject studies. We developed a web-based tool to create RMIDs and application programming interfaces to synchronize research records and visualize linkages to protocols across systems. Selected data from these disparate systems were extracted and merged nightly into an enterprise data mart, and performance dashboards were created to monitor key translational processes.
Results
Within 4 years, 5513 RMIDs were created. Among these were 726 (13%) bridged systems needed to evaluate research study performance, and 982 (18%) linked to the electronic health records, enabling patient-level reporting.
Discussion
Barriers posed by data fragmentation to assessment of program impact have largely been eliminated at MUSC through the requirement for an RMID, its distribution via RINS to disparate systems, and mapping of system-level data to a single integrated data mart.
Conclusion
By applying data warehousing principles to federate data at the “study” level, the RINS project reduced data fragmentation and promoted research systems integration.
Keywords: learning health system, clinical data warehouse, health information interoperability, application programming interfaces
INTRODUCTION
Many academic medical centers have seen the promise of the learning health system, which leverages integrated, real-time data to improve patient care and inform other quality improvement and cost-saving efforts.1–11 However, learning health system implementation has been hampered by the fragmentation of data across siloed systems.12,13 To address the issue, institutions have often turned to tools for enterprise application integration14–16 that link their electronic health records (EHRs) to other proprietary and homegrown systems. Data from these systems are then exported into an enterprise clinical data warehouse (CDW) that provides rich and comprehensive information for performance improvement initiatives.
Although perhaps better known for its application to patient care17–20 and hospital business performance,21 the concept of a learning system is also highly relevant to clinical and translational research22–24 and has been embraced by the Clinical and Translational Science Awards (CTSA) program.25,26 The CTSA program has recognized that comprehensive data on research performance and robust evaluation mechanisms will be necessary to document how efforts of its hubs have sped up and otherwise enhanced the translation of discovery into clinical care. A serious obstacle to such a learning system for research is the fragmentation of relevant data across disparate research systems. For instance, such fragmentation can make it very difficult to track study activation timelines, participant recruitment rates, and financial performance. This, in turn, can result in inefficiencies and redundant efforts in clinical trials, which can negatively impact an institution’s bottom line, dampening its support for research.4,9,27 Although bioinformatics tools have been created to address this fragmentation, most are intended to streamline the conduct of clinical research and not to assess the success of translational interventions.
To address this gap, the South Carolina Clinical & Translational Research (SCTR) Institute, the CTSA hub with an academic home at the Medical University of South Carolina (MUSC), has applied data warehousing principles to create a much-needed “meta” evaluation tool, the Research Integrated Network of Systems (RINS), for assessing the success of translational research initiatives, in particular those aimed at improving the efficiency of clinical trials. RINS adopts a federated rather than centralized model to integration, linking disparate research systems while enabling each area of research administration to continue to use existing “best of breed” systems that are most suited to their operations. RINS uses a unique study identifier—the Research Master Identifier (RMID)—to track each clinical trial or study across research systems integrated by RINS and extracts granular, study-specific data from those systems into an integrated research data mart. User-friendly dashboards and reports were developed using a business intelligence tool to provide visualizations of data to university and SCTR leadership and staff to guide their performance improvement initiatives or to assess the success of past interventions to improve efficiency.
MATERIALS AND METHODS
Inception and implementation of the Research Master Identifier
In October 2016, SCTR invited a broad cross-section of RMID stakeholders, including research subject matter experts, biomedical informaticians, and systems engineers to join the RINS working group that would spearhead the initiative. Initial goals of the group were to integrate and harmonize fragmented research data, interface systems, decrease duplicative data entry, and ensure data integrity.
In January 2017, the first version of the RMID application, built using a Ruby on Rails framework,28 was launched. The application enabled research teams to register their studies with a system-generated unique identifier that facilitated tracking across existing research systems. Required data fields for the RMID record were identified (primary investigator [PI], department, long title, short title, funding source, and study type) and a source of truth designated for each.
As a homegrown system, RMID could be tailored to the needs of our institution and users and integrated in phases. In the initial phase, RMID was implemented in 3 research systems required for new human-subject research studies. Gradually, RMID was integrated into more systems and finally was required for all studies.
The RINS working group met every 2 weeks to troubleshoot problems as they arose and provide guidance as the tool evolved and its usage grew. During implementation, it provided a forum for discussing how best to handle roadblocks, such as grants with multiple research protocols, duplicative data entry within each research system, and duplicative RMIDs for the same protocol. These issues were promptly addressed using a team science approach with the subject matter experts and systems engineers sitting in the same session. Since implementation, the group has continued to meet to receive feedback on how the tool can be optimized for its users. Stakeholders from colleges, departments, and specific groups present “case studies” illustrating a bug or need for improved functionality in RINS, or they request reports and dashboards to support metric tracking and reporting.
Data model for the Research Integrated Network of Systems
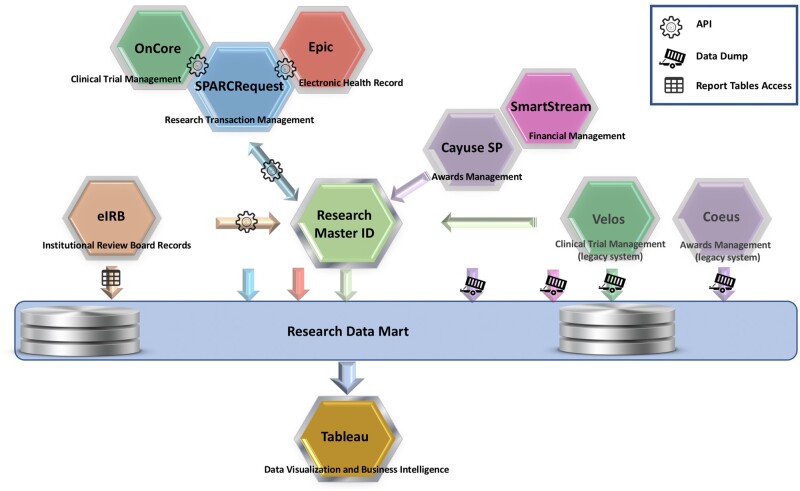
As of October 2020, RINS links SPARCRequest,28,29 an MUSC-created open source research transaction management system that has been adopted by 12 CTSA and Clinical and Translational Research (CTR) hubs, with the institution’s EHR (Epic Systems, Verona, WI),30 electronic Institutional Review Board (eIRB; Click),31 and systems for grants award management (Coeus, retired; Cayuse SP, implemented early 2020)32,33 and expenditure tracking (SmartStream).34 In addition, links were developed to a clinical trial management system (CTMS) used for cancer trials (Velos)35 and its replacement, an enterprise-wide CTMS (OnCore, implemented late 2020).36 The overall integration of systems is shown in Figure 1. RINS has been sufficiently flexible to allow integration of the new CTMS and grants award system without losing the historical data from the legacy systems.
Figure 1.
The Research Master ID is the centerpiece that enables the Research Integrated Network of Systems to bridge research systems at the Medical University of South Carolina via RESTFul application programming interfaces (APIs) and then to export select information from those systems into a research data mart.
Abbreviations: API, application programming interface; eIRB, electronic institutional review board.
Currently, RMIDs are required for all human-subject protocols in SPARCRequest and Cayuse and for submission of all protocols to the eIRB. For preclinical studies lacking RMIDs, we use alternative linking methods. For instance, 1 of the unique RINS identifiers, such as the SPARC ID, can be used to create indirect linkages between the various study numbers, providing an alternative pathway for bridging study data.
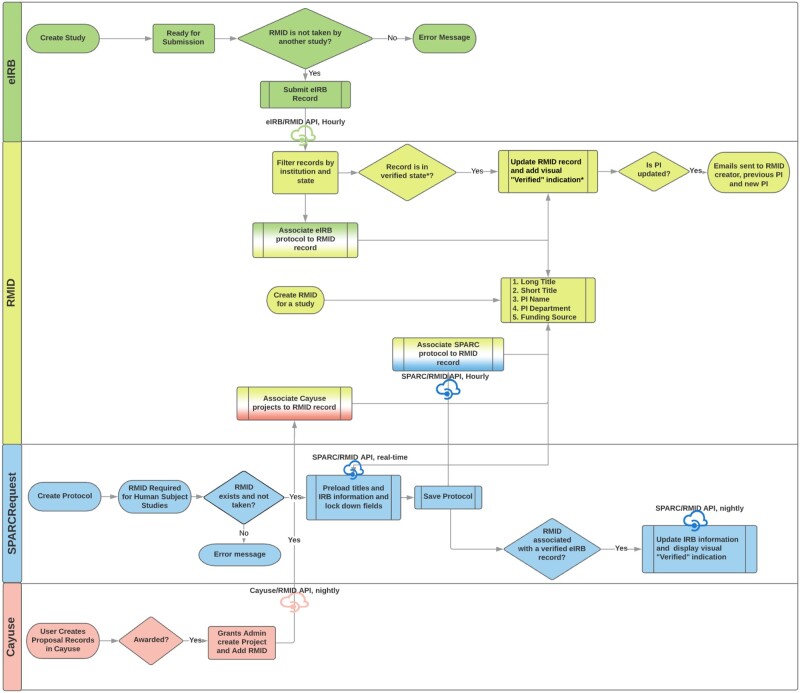
We then built middleware to integrate the RMID with institution-owned research systems and added validations onto commercial systems (Figure 2). For example, with the institution-owned SPARCRequest, the RMID/SPARC application programming interface (API) was developed to pull into SPARC any updates to the eIRB-approved protocol, such as titles and key study dates, via the RMID entered into the eIRB system. Although we could not achieve a real-time data integration via API with the Click eIRB system, we were able to build validations into the rules to prevent users from submitting a protocol with a duplicative RMID for review.
Figure 2.
Flow charts illustrating the Research Master ID (RMID) and its application programming interfaces (APIs) during study record maintenance in the Research Integrated Network of Systems.
Abbreviations: eIRB, electronic institutional review board; PI, principal investigator; SPARCRequest, services, pricing, & application for research centers.
The APIs were made bidirectional, enabling information from these research systems to feed into the RMID system and be distributed to other linked systems when needed. A “record of truth” was designated for each data field in the RMID system, preventing creation and propagation of errors in data entry across systems. For instance, Click was designated as the record of truth for protocol-level information (ie, short title, long title, and PI) and Coeus/Cayuse for financial data.
Web-based front-end user interface and Open Source license
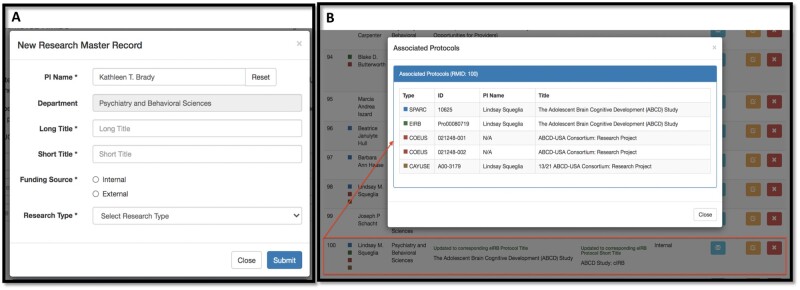
Users create a new RMID or retrieve an existing one using a web-based interface. Figure 3A shows the pop-up window for creating a new research master record, with the 6 required data fields. For data accuracy, if the PI entered for an RMID record exists in the institutional faculty database, the department is pulled into the window automatically, with a gray background indicating that it is automatically populated and noneditable. In this way, the number of required fields is reduced to 5 for most records, with departmental data provided directly from the record-of-truth source.
Figure 3.
Front-end user interface of the Research Master ID application. Shown here are the window for creating a new Research Master Record (A) and another window displaying the inventory of all systems associated with the selected RMID record (B).
After an RMID record has been created (and utilized) in 1 of the research systems, an inventory of its associations with all systems is created automatically and displayed within an hour to the front-end user. Figure 3B shows an example of an RMID record that has been associated with 5 records in 4 research systems (one for SPARCRequest, 1 for eIRB, 2 for Coeus, and 1 for Cayuse). Color legends are used to represent different systems to facilitate interpretation. This front-end display gives study team users an easy way to track their research project through its life cycle as it proceeds through development, compliance, funding, and fruition.
With the mindset of sharable coding and system integration,37 we made the RMID application open source on GitHub38 with a 3-Clause BSD license in November 2020. The 3-Clause BSD license allows others to download, use, or modify the code repository for private use or distribution with a clause that prohibits others from using the name of the project or its contributors to promote derived products without written consent.
Extract, transform, load (ETL) into the research data mart for metrics reporting
The overall architecture used RINS as an information bus. A subset of source data from research and clinical systems connected to RINS is extracted nightly and stored in a relational database. Tables are created to store data from each system individually. Data sources are added to or deleted from the data mart as current research support systems begin using RMID in the workflow, a new research support system is brought to campus, or legacy systems are discontinued. Thus, leadership maintains the ability to use legacy data from discontinued systems, as well as to link and integrate data in a variety of ways, depending on the desired outcome of a particular project or report. Tables are refreshed nightly to maintain synchronization with data in ground truth systems.
The SCTR/BMIC teams also created reports and performance dashboards using the data visualization software Tableau39 for university, college, departmental, and research leaders. Institutional metrics drove the original dashboards and included study activation timelines, financial performance, and recruitment tracking. SCTR/BMIC teams are now also developing dashboards for operational staff, such as SCTR program managers, study team members, source system experts, and research support staff. When issues arise with functionality or data quality, an expert is consulted who is well versed in the source data of the system in question. Once the report is finalized, it continues to be monitored by all parties for accuracy and uptake.
The Tableau reports provide users with various filters, parameters, and e-mail alert functionalities that they can use to customize the report to their individual needs without requiring additional data analyst time. This functionality also ensures that a single dashboard can be used for multiple reporting purposes. For instance, university and SCTR leaders can use the metrics provided by the Tableau dashboards to assess the performance of the research enterprise as a whole and look for opportunities for improvement, while SCTR managers can assess the performance of their programs, and their staff can drill down to look for underperforming studies in need of SCTR service support.
RESULTS
Growth of RMID utilization
Since its implementation in January 2017, RMID use has spread rapidly across MUSC’s research community. This was achieved via a phased-in approach that required RMID for all new human-subject studies for eIRB submissions and SPARCRequest research protocols in the first two years. It was then required for all eIRB submissions and human-subject award renewals in Coeus/Cayuse in 2019, with more stakeholders and user groups coming on board. As shown in Figure 4, as of October 2020, 5513 RMIDs have been created, linking SPARCRequest, eIRB, Coeus, Cayuse, and Epic; 4801 RMIDs have been associated with at least 1 of the 4 types of research systems.
Figure 4.
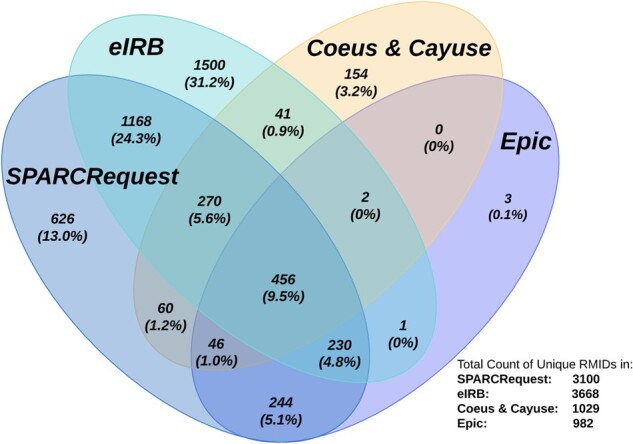
Venn diagram showing the number (percentage) of Research Master IDs shared among different research systems at the Medical University of South Carolina as of October 13, 2020. The denominator used to determine all percentages was the total number of RMIDs associated with at least 1 research system (4801). Calculations were performed using an online tool by BioinfoGP group.40
Among all the existing RMIDs, 726 (13%) can be linked across SPARCRequest, eIRB, and financial awards systems (Coeus and Cayuse) to determine the startup time of a study and collect financial award, revenue, and expenditure data; and 982 (18%) link to Epic, MUSC’s EHR, enabling patient-level reporting for research study performance evaluations. In-depth, multidimensional metrics reporting is possible for 456 (8%) RMIDs that hit all 4 types of research systems.
As of October 2020, 4418 unique users have logged into the system (see Table 1 for detailed information). As the RMID system has matured, the number of research staff user accounts has increased, with a corresponding decrease in customer service needs.
Table 1.
Research Master ID website user accounts analysisa
| Role/Position | Total Unique Users, No. (%) | Total Login Count, No. (%) |
|---|---|---|
| Principal investigator | 968 (22) | 2286 (21) |
| Research staff | 3435 (78) | 7621 (70) |
| RMID administrator | 15 (0.3) | 1046 (10) |
| Total | 4418 | 10 953 |
User account analytics are for the period between January 29, 2017 and October 13, 2020. Percentage values may add up to more than 100% due to rounding.
Reduced data duplication and improved data integrity
RINS includes a robust RMID search function to help prevent the creation of duplicative RMIDs. Should a duplicative RMID be assigned, safeguards exist to help identify and remove it. RMID uses a combination of the PI’s name and long title of the research project to identify a unique study. The source of truth for both is the eIRB, and these fields in the linked systems (ie, RMID and SPARC) update automatically to match those in the eIRB. When a PI name is updated in accordance with the approved eIRB record, an e-mail is sent to notify the previous PI. A Tableau report dashboard was developed to identify duplicative RMIDs using the PI/long title combination and other “red flags.” These duplicative RMIDs are then reviewed and resolved.
As shown in Table 2, among the 5513 RMID records created between January 29, 2017 and October 13, 2020, 174 RMIDs were removed from the system by the creator, PI of the study, or an RMID administrative user. Due to the observed phenomenon of records deletion, we implemented a “Removed RMID” web page and tracking mechanism in July 2019 to document user-removed RMIDs and the reason for deletion. Of the 57 RMIDs deleted with reasons recorded since then, 50 (88%) were deleted due to duplicative entry, and 7 (12%) were deleted due to study termination.
Table 2.
Duplicative records identified via RMIDa
| Potential Duplicate Source Category | Time Frame | No. (%) |
|---|---|---|
| Total removed RMIDs | 1/29/2017–10/13/2020 | 174 (3) |
| Removed RMIDs with recorded reasons | 7/2/2019–10/13/2020 | 57 (1) |
| Unassociated RMID records | As of 10/13/2020 | 651 (12) |
| Merged duplicative SPARCRequest protocolsb | 7/5/2019–10/13/2020 | 90 |
Percentages are calculated using 5 513, the total number of Research Master Identifiers (RMIDs), as the denominator. Percentage values may add up to more than 100% due to rounding.
The merged duplicative SPARCRequest protocols do not all result in removed RMIDs.
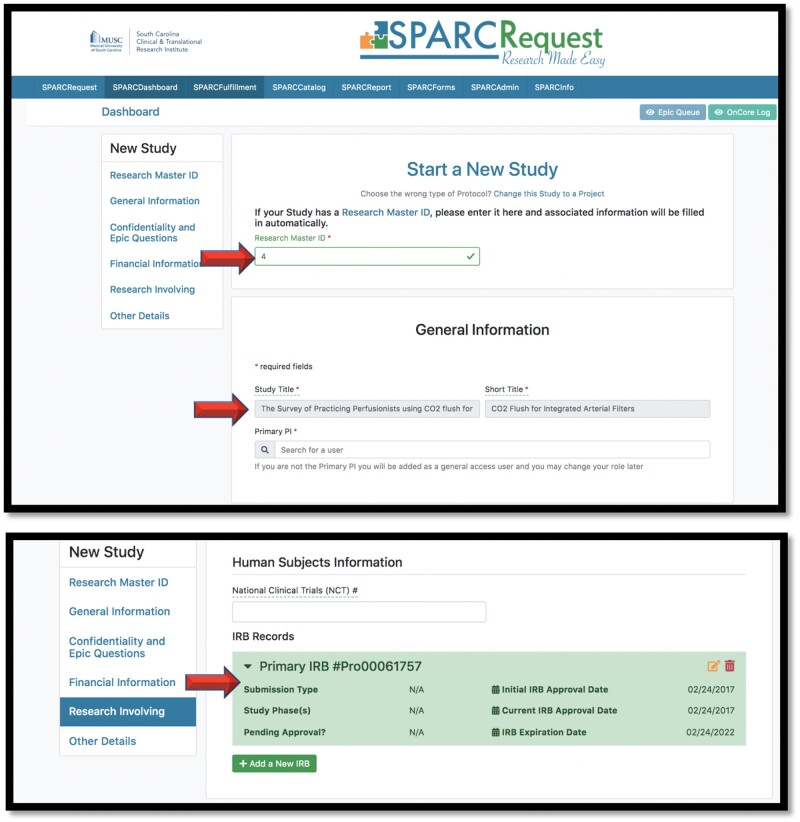
In addition to validations and APIs built within the RMID application, the RINS team also initiated and built APIs into SPARCRequest to fill out fields automatically and in real time when an RMID is entered on a study. Figure 5 shows screenshots of the user interface for creating a protocol in SPARCRequest. If there are other records associated with the protocol’s RMID, the SPARC/RMID API autofills data fields such as study title, short title, eIRB record number, approval dates, and expiration date.
Figure 5.
Screenshots from SPARCRequest showing fields that are automatically filled via the SPARC/RMID API.
Abbreviations: API, application programming interface; RMID, Research Master ID; SPARC (SPARCRequest): services, pricing, & application for research centers.
Metrics dashboards and informed performance improvement
Of the many Tableau performance dashboards and reports that have been built since December 2017 using RINS data, Table 3 lists the most frequently used. As of October 2020, these Tableau reports have been viewed a total of 2964 times by RINS working group members and institutional and CTSA leaders for their routine metrics reporting. As the RINS project evolves, demand for more department-specific metrics has been increasing, and a prioritized workflow has been established to accommodate that demand. Using the Tableau dashboards, university and CTSA leadership can track the overall financial performance of the research enterprise, investigate metrics on a particular type of clinical trial or translational intervention, or drill down on a single underperforming study.
Table 3.
Usage of Tableau reports created for RINS
| Report Name | Description | Number of Dashboards | Number of Views |
|---|---|---|---|
| RMID records summary | Usage summary and validation of RMIDs in 4 systems | 7 | 455 |
| Potential duplicative RMIDs | Identification of duplicative RMIDs for review and processing by system administrators | 1 | 209 |
| Patient accrual dashboard | Study enrollment timeline and patient recruitment ratio | 1 | 135 |
| Industry IRB studies | Turnaround time from eIRB approval to first revenue received | 3 | 1044 |
| Invoicing phase report | Facilitation of Office of Clinical Research invoice management—includes data from SPARCRequest and financial systems | 5 | 1121 |
Abbreviations: eIRB, electronic institutional review board; RMID, Research Master Identifier; RINS, Research Integrated Network of Systems.
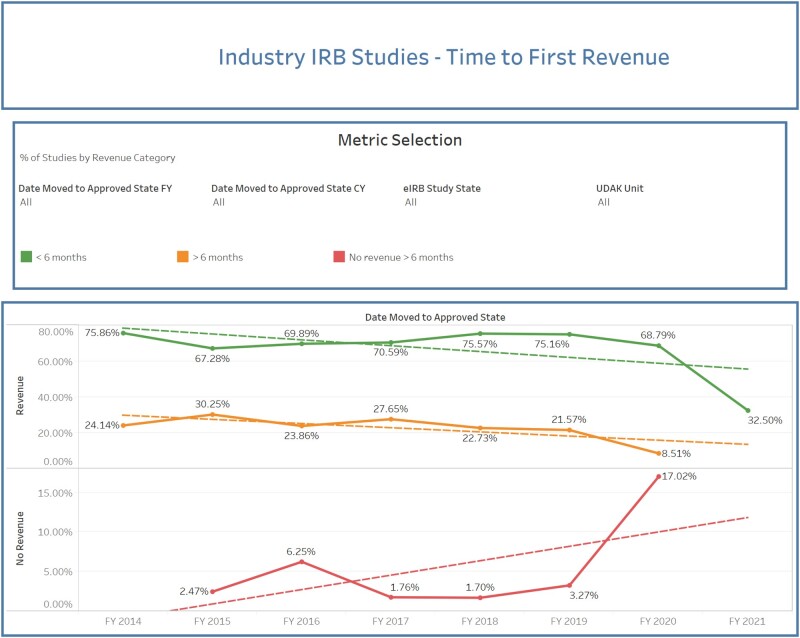
For instance, leaders frequently use a dashboard that relies on data from the eIRB (study activation date) and a financial system to track the time to first revenue for corporate trials (Figure 6). Typically, startup costs should be recouped from the sponsor within the first 6 months of a study. Leadership depends on this summary view to identify early on which studies may be at risk for not bringing in revenue within that timeline and to assist those study teams in working with corporate sponsors to achieve success. Trials that have never received startup funds from the sponsor can also be identified, enabling SCTR staff to provide additional training or support to those study teams and recover revenue from the sponsor even after the initial startup. This report can also be used to compare historic and current metrics to assess the effects of enterprise changes to systems and policies.
Figure 6.
Performance dashboard showing time to first revenue for industry studies. Studies with no revenue are indicated in red, those with revenue after 6 months in orange, and those with revenue in less than 6 months in green. Solid lines in the graph connect the data points, and dotted lines show the trends using linear fitting.
Abbreviations: CY, current year; eIRB, electronic institutional review board; FY, fiscal year; UDAK = financial account number.
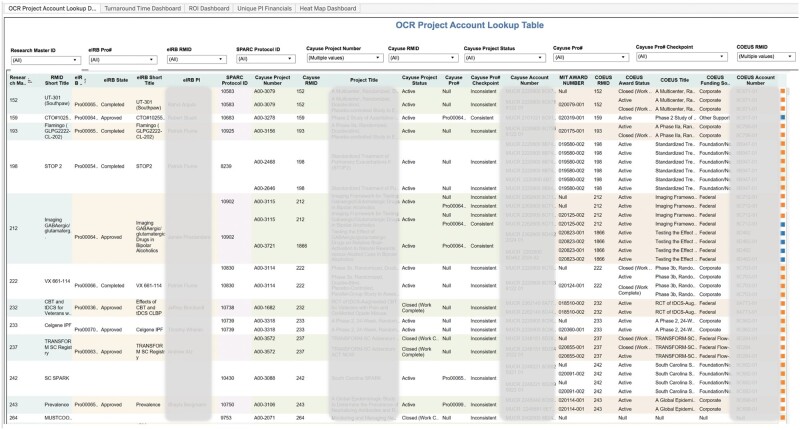
As another example, the project account lookup dashboard provides comprehensive information on a study’s regulatory compliance and financial status by linking data across the eIRB, grants award, and SCTR service-tracking systems (Figure 7). It is used to identify the industry sponsor for any given study, simplifying billing and thereby aiding research service providers to accelerate their invoicing process. It also enables leaders to monitor the award and financial account status of all studies involving human subjects at a glance.
Figure 7.
The project account lookup dashboard shows the linkage among research systems using Research Master ID records, with different colors of columns indicating different research systems.
Abbreviation: OCR, Office of Clinical Research.
DISCUSSION
For any academic medical institution or CTSA that can require adoption of a universal study identifier, RINS offers an elegant, minimally disruptive and dynamic solution for tracking and evaluating clinical research metrics for the continuous performance and process improvement envisioned for a learning system. RINS relies on a unique study identifier (RMID), state-of-the art RESTful APIs, and data warehousing principles to integrate translational research systems and their data in support of a translational research learning system. It enables each research area to continue to use “best of breed” systems while also offering robust data integration. RINS provides CTSA the granular, integrated, study-level data they need for pinpointing studies in need of support and for metrics reporting about the success of their translational interventions to the National Center for Advancing Translational Sciences.
Applying data warehousing principles to translational research
Clinical data warehouses have for decades been the cornerstone of efforts by academic medical centers to create a learning health system, providing the integrated data on patient care needed to track key quality and cost indicators and target and evaluate the success of performance improvement initiatives.41–44 Bioinformatics programs have been created45 to mine the data in these CDWs to better investigate a wide variety of diseases and to improve overall patient care.46,47
More recently, the value of CDWs for translational research has been recognized.48 In an attempt to leverage integrated data to enhance the conduct of translational research, institutions have employed CTSA-developed bioinformatics platforms ,49,50 adapted proprietary clinical enterprise business intelligence tools,51 or created their own integration solutions.52–56 A popular application of these tools has been the identification of clinical trial cohorts using CDW data.53,57
Two of these systems bear particular mention in relation to RINS. Like RINS, the Stanford Translational Research Integrated Database Environment (STRIDE)53 adopts a single-identifier approach but uses a patient instead of a study identifier to facilitate integration of data across systems. The Clinical Research Administration (CLARA)56 at the University of Arkansas for Medical Sciences is a centralized platform that integrates functionalities and data from a legacy eIRB and clinical research information management system to enable tracking of a research study from IRB submission to approval and postapproval regulatory monitoring. Like RINS, it also provides integrated data from a number of these systems to target and assess performance improvement, leading to streamlining of the study approval process. However, RINS provides access to data extracted from a larger number of research systems than CLARA, including an awards system, enabling a trial to be traced to its underlying grant award and thereby facilitating reporting to funding agencies.
RINS differs most from STRIDE and CLARA in its federated approach.58–61 By linking the disparate systems via the RMID and RESTful APIs, RINS creates what Haas61 has called a “virtual data warehouse” without having to incur the costs or face the logistical challenges of restructuring and moving data from these systems. Relevant data can simply be extracted nightly from the linked systems, without the need for restructuring, into a clinical research data mart using ETL jobs. Because the deployment of RINS involved the linking of existing systems, it caused minimal disruptions to workflow and necessitated little staff retraining. RINS’ federated model has also made it possible to integrate new systems or sunset old ones while preserving legacy data, as it did when we transitioned CTMS systems from Velos to OnCore.
Linking the systems also helps identify and resolve any discrepancies in the data and, through automatic population of linked records with information from the RMID record, to prevent inaccurate data entry. Tableau reports provide a feedback loop for quality control, enabling systems administrators to review and address outdated, inaccurate, or duplicative data. In short, RINS provides opportunities for data cleansing at both the front end (construction of the APIs) and back end (report feedback loop).
RINS is a powerful tool for not only linking research systems but also extracting and integrating their data so that they can be used to evaluate the effectiveness of translational initiatives. As such, it promotes a translational research learning system, valuing continuous process and performance improvement. The RMID code repository is open source, making it easily sharable with any potential adopters.
Challenges and lessons learned
The integration of RMID and its APIs with research systems is bidirectional: its adoption requires not only the support of a centralized research office, but also that of subject matter experts and stakeholders who must add the unique RMID into their systems and workflows. We have learned that this process requires intensive communication and goal alignment, and the RINS working group has been invaluable in this regard.
In addition, the adoption of RMID is not simply adding a new number to an existing system; it involves the infrastructural alignment between systems and identification of the correct user groups to weigh in on those changes. Reluctance to change and workflow confusion are potential barriers to this integration. The rapid uptake of RINS at our institution could have been due in large part to the already widespread usage of an eIRB and the MUSC-created SPARCRequest. The importance of these 2 electronic resources to the rapid growth of RINS at MUSC suggests that uptake might be more difficult at institutions that had not previously digitized most of their clinical research services or did not have access to informatics expertise.
Future directions
MUSC has recently implemented an enterprise-wide CTMS (OnCore) and is in the process of integrating it into RINS. Once the process is complete, RINS will integrate OnCore data with information in existing clinical research administration data systems, enhancing SCTR’s capacity to report on CTSA common metrics. This implementation marks an important milestone in RINS development and is the subject of a manuscript in preparation.
Currently, Tableau dashboards are used primarily by university and SCTR leaders. The RINS team is working to create access authorizations and dashboards that will enable colleges, departments, and ultimately study teams to monitor the progress of their own studies so that they can adapt as necessary to improve performance.
We have presented a summary of the RINS integration model to the SPARCRequest Open-Source Consortium, which consists of 12 CTSA and CTR hubs comprising 27 institutions. These open-source partners expressed interest in the RINS model, with some particularly interested in the APIs that have already been built between SPARC and a number of proprietary clinical research systems (eg, Epic, Click, REDCap,62 and OnCore). We will continue to use the SPARCRequest Open-Source Consortium as a forum for promoting the RINS integration model as an evaluation environment to more institutions.
CONCLUSION
RINS offers an elegant solution to the problem of fragmented research data by linking research systems via a unique identifier (RMID) and real-time or near–real-time APIs, extracting data nightly to a research data mart, and employing a business intelligence tool to create user-friendly dashboards that facilitate translational research performance improvement initiatives. RINS is a flexible, federated solution that provides the integrated, granular, study-level data necessary to assess translational interventions and tools while enabling research teams to continue to use best-of-breed systems for their operations. It is highly adaptable, easily enabling new systems to be integrated or old ones to be replaced to meet changing needs. This flexibility ensures that it can continue to provide university and CTSA leadership with the real-time, integrated, high-quality data they need to realize the potential of a translational research learning system to optimize CTSA interventions and tools.
FUNDING
This publication was supported, in part, by a grant (UL1 TR001450) from the National Center for Advancing Translational Sciences to the South Carolina Clinical and Translational Research Institute at the Medical University of South Carolina. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
AUTHOR CONTRIBUTIONS
LAL conceived the idea of the Research Master ID and Research Integrated Network of Systems, and RRS, WH, KGK, and AMC have all been integral to RMID, APIs, and Data Mart ETL design and implementation. WH and KGK are also Tableau dashboard developers for this project. LAL, RRS, and JSO are leadership representatives at the RINS working group and review Tableau dashboard requirements. The first draft of the manuscript was written by WH, KGK, and KKM and revised critically for important intellectual content by all authors. All coauthors approve of the final version to be published and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
ACKNOWLEDGMENTS
The authors of this manuscript would like to express their appreciation to Leila Forney, Kyle Hutson, the MUSC SUCCESS Center and the Office of Clinical Research, and all members of the MUSC BMIC Ruby development teams for their contribution to this project.
DATA AVAILABILITY STATEMENT
The data underlying this article are available in the article. The RMID code repository is open source and available on GitHub at https://github.com/sparc-request/research-master-id.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1. Rodrigues RJ. Information systems: the key to evidence-based health practice. Bull World Health Organ 2000; 78 (11): 1344–51. 2001/01/06]. [PMC free article] [PubMed] [Google Scholar]
- 2. Kruse CS, Goswamy R, Raval Y, Marawi S.. Challenges and opportunities of big data in health care: a systematic review. JMIR Med Inform 2016; 4 (4): e38.2016/11/23]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Luo J, Wu M, Gopukumar D, Zhao Y.. Big data application in biomedical research and health care: a literature review. Biomed Inform Insights 2016; 8: BII.S31559.1-10 2016/02/05]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Institute of Medicine (US). Forum on Drug Discovery, Development, and Translation. Transforming Clinical Research in the United States: Challenges and Opportunities: Workshop Summary. Washington, DC: National Academies Press; 2010. [PubMed] [Google Scholar]
- 5. Embi PJ, Payne PR.. Clinical research informatics: challenges, opportunities, and definition for an emerging domain. J Am Med Inform Assoc 2009; 16 (3): 316–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morain SR, Kass NE, Grossmann C.. What allows a health care system to become a learning health care system: results from interviews with health system leaders. Learn Health Sys 2017; 1 (1): e10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sung NS, Crowley WF Jr, Genel M, et al. Central challenges facing the national clinical research enterprise. JAMA 2003; 289 (10): 1278–87. [DOI] [PubMed] [Google Scholar]
- 8. Olsen LA, Aisner D, McGinnis JM, eds. The Learning Healthcare System: Workshop Summary. Washington, DC: The National Academies Press; 2007. [PubMed] [Google Scholar]
- 9.McGinnis JM, Olsen LA, Goolsby WA, Grossman C. Institute of Medicine and National Academy of Engineering. Engineering a Learning Healthcare System: A Look at the Future: Workshop Summary. Washington, DC: The National Academies Press; 2011. [Google Scholar]
- 10. Friedman CP, Wong AK, Blumenthal D.. Achieving a nationwide learning health system. Sci Transl Med 2010; 2 (57): 57cm29. [DOI] [PubMed] [Google Scholar]
- 11. Payne PR, Embi PJ, Sen CK.. Translational informatics: enabling high-throughput research paradigms. Physiol Genomics 2009; 39 (3): 131–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. El Fadly A, Rance B, Lucas N, et al. Integrating clinical research with the healthcare enterprise: from the re-use project to the EHR4CR platform. J Biomed Inform 2011; 44: S94–102. [DOI] [PubMed] [Google Scholar]
- 13. Payne PR, Johnson SB, Starren JB, Tilson HH, Dowdy D.. Breaking the translational barriers: the value of integrating biomedical informatics and translational research. J Investig Med 2005; 53 (4): 192–200. [DOI] [PubMed] [Google Scholar]
- 14. Khoumbati K, Themistocleous M, Irani Z.. Investigating enterprise application integration benefits and barriers in healthcare organisations: an exploratory case study. IJEH 2006; 2 (1): 66–78. [DOI] [PubMed] [Google Scholar]
- 15. Khoumbati K, Themistocleous M, Irani Z . Integration Technology Adoption in Healthcare Organisations: A Case for Enterprise Application Integration. In: proceedings of the 38th Annual Hawaii International Conference on System Sciences; January 3–6, 2005; Big Island, HI, USA. ieeexplore.ieee.org.
- 16. Khoumbati K, Themistocleous M, Irani Z.. Evaluating the adoption of enterprise application integration in health-care organizations. J Manage Inform Syst 2006; 22 (4): 69–108. [Google Scholar]
- 17. Krumholz HM. Big data and new knowledge in medicine: the thinking, training, and tools needed for a learning health system. Health Aff (Millwood) 2014; 33 (7): 1163–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weiner MG, Embi PJ.. Toward reuse of clinical data for research and quality improvement: the end of the beginning? Ann Intern Med 2009; 151 (5): 359–60. [DOI] [PubMed] [Google Scholar]
- 19. Safran C, Bloomrosen M, Hammond WE, et al. Toward a national framework for the secondary use of health data: an American Medical Informatics Association white paper. J Am Med Inform Assoc 2007; 14 (1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jensen PB, Jensen LJ, Brunak S.. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet 2012; 13 (6): 395–405. [DOI] [PubMed] [Google Scholar]
- 21. Ferguson TB Jr. The Institute of Medicine committee report “best care at lower cost: the path to continuously learning health care.” Circ Cardiovasc Qual Outcomes 2012; 5 (6): e93–4. [DOI] [PubMed] [Google Scholar]
- 22. Katzan IL, Rudick RA.. Time to integrate clinical and research informatics. Sci Transl Med 2012; 4 (162): 162fs41. [DOI] [PubMed] [Google Scholar]
- 23. Riley WT, Glasgow RE, Etheredge L, Abernethy AP.. Rapid, responsive, relevant (r3) research: a call for a rapid learning health research enterprise. Clin Transl Med 2013; 2 (1): 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gelijns AC, Gabriel SE.. Looking beyond translation–integrating clinical research with medical practice. N Engl J Med 2012; 366 (18): 1659–61. [DOI] [PubMed] [Google Scholar]
- 25. Dilts DM, Rosenblum D, Trochim WM.. A virtual national laboratory for reengineering clinical translational science. Sci Transl Med 2012; 4 (118): 118cm2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leshner AI, Terry SF, Schultz AM, et al. , eds. The CTSA Program at NIH: Opportunities for Advancing Clinical and Translational Research. Washington, DC: National Academies Press; 2013. [PubMed] [Google Scholar]
- 27. Duley L, Gillman A, Duggan M, et al. What are the main inefficiencies in trial conduct: a survey of UKCRC registered clinical trials units in the UK. Trials 2018; 19 (1): 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. He W, Sampson R, Obeid J, et al. Dissemination and continuous improvement of a CTSA-based software platform, SPARCRequest, using an open-source governance model. J Clin Trans Sci 2019; 3 (5): 227–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sampson RR, Glenn JL, Cates AM, Scott MD, Obeid JS.. SPARC: a multi-institutional integrated web-based research management system. AMIA Jt Summits Transl Sci Proc 2013; 2013: 230. [PubMed] [Google Scholar]
- 30.Epic. https://www.epic.com/Accessed October 22, 2020
- 31.Health sciences south carolina. https://eirb.healthsciencessc.org/. Accessed October 22, 2020
- 32.Mit kuali coeus. https://kc.mit.edu Accessed October 22, 2020
- 33.Era software. https://cayuse.com Accessed October 22, 2020
- 34.Smartstream technologies. https://www.smartstream-stp.com Accessed October 22, 2020
- 35.Velos eresearch. https://www.wcgclinical.com/services/velos-eresearch Accessed October 22, 2020
- 36.Oncore ctms enterprise research system, clinical research software. https://www.advarra.com/oncore-enterprise-research-ctms/ Accessed January 17, 2021
- 37. Obeid JS, Tarczy-Hornoch P, Harris PA, et al. Sustainability considerations for clinical and translational research informatics infrastructure. J Clin Trans Sci 2018; 2 (5): 267–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Research master id code repository. https://github.com/sparc-request/research-master-id Accessed January 18, 2020
- 39.Tableau software. https://www.tableau.com/about/mission Accessed May14, 2020
- 40. Collazos J. Venny 2.1.0. An interactive tool for comparing lists with venn’s diagrams. https://bioinfogp.cnb.csic.es/tools/venny Accessed October 22, 2020
- 41. Turley CB, Obeid J, Larsen R, et al. Leveraging a statewide clinical data warehouse to expand boundaries of the learning health system. eGEMs 2016; 4 (1): 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Grant A, Moshyk A, Diab H, et al. Integrating feedback from a clinical data warehouse into practice organisation. Int J Med Inform 2006; 75 (3-4): 232–9. [DOI] [PubMed] [Google Scholar]
- 43. Jannot AS, Zapletal E, Avillach P, Mamzer MF, Burgun A, Degoulet P.. The Georges Pompidou University Hospital clinical data warehouse: a 8-years follow-up experience. Int J Med Inform 2017; 102: 21–8. [DOI] [PubMed] [Google Scholar]
- 44. Wisniewski MF, Kieszkowski P, Zagorski BM, Trick WE, Sommers M, Weinstein RA.. Development of a clinical data warehouse for hospital infection control. J Am Med Inform Assoc 2003; 10 (5): 454–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fahy BG, Balke CW, Umberger GH, et al. Crossing the chasm: information technology to biomedical informatics. J Investig Med 2011; 59 (5): 768–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Karami M, Rahimi A, Shahmirzadi AH.. Clinical data warehouse: an effective tool to create intelligence in disease management. Health Care Manag 2017; 36 (4): 380–4. [DOI] [PubMed] [Google Scholar]
- 47. Prather JC, Lobach DF, Goodwin LK, Hales JW, Hage ML, Hammond WE.. Medical data mining: knowledge discovery in a clinical data warehouse. Proc AMIA Annu Fall Symp 1997; 4: 101–5. [PMC free article] [PubMed] [Google Scholar]
- 48. Campion TR, Craven CK, Dorr DA, Knosp BM.. Understanding enterprise data warehouses to support clinical and translational research. J Am Med Inform Assoc 2020; 27 (9): 1352–8. 2020/07/18]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kohane IS, Churchill SE, Murphy SN.. A translational engine at the national scale: informatics for integrating biology and the bedside. J Am Med Inform Assoc 2012; 19 (2): 181–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc 2010; 17 (2): 124–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Roth JA, Goebel N, Sakoparnig T; the PATREC Study Group, et al. Secondary use of routine data in hospitals: description of a scalable analytical platform based on a business intelligence system. JAMIA Open 2018; 1 (2): 172–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stead WW, Miller RA, Musen MA, Hersh WR.. Integration and beyond: linking information from disparate sources and into workflow. J Am Med Inform Assoc 2000; 7 (2): 135–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lowe HJ, Ferris TA, Hernandez PM, Weber SC.. STRIDE–an integrated standards-based translational research informatics platform. AMIA Annu Symp Proc 2009; 2009: 391–5. [PMC free article] [PubMed] [Google Scholar]
- 54. Wade TD, Hum RC, Murphy JR.. A dimensional bus model for integrating clinical and research data. J Am Med Inform Assoc 2011; 18 (Supplement 1): i96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zapletal E, Rodon N, Grabar N, Degoulet P.. Methodology of integration of a clinical data warehouse with a clinical information system: the HEGP case. Stud Health Technol Inform 2010; 160 (Pt 1): 193–7. [PubMed] [Google Scholar]
- 56. Bian J, Xie M, Hogan W, et al. CLARA: an integrated clinical research administration system. J Am Med Inform Assoc 2014; 21 (e2): e369–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Horvath MM, Rusincovitch SA, Brinson S, Shang HC, Evans S, Ferranti JM.. Modular design, application architecture, and usage of a self-service model for enterprise data delivery: the Duke enterprise data unified content explorer (DEDUCE). J Biomed Inform 2014; 52: 231–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ganslandt T, Kunzmann U, Diesch K, Palffy P, Prokosch HU.. Semantic challenges in database federation: lessons learned. Stud Health Technol Inform 2005; 116: 551–6. [PubMed] [Google Scholar]
- 59. Marenco L, Wang TY, Shepherd G, Miller PL, Nadkarni P.. Qis: A framework for biomedical database federation. J Am Med Inform Assoc 2004; 11 (6): 523–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kemp GJ, Angelopoulos N, Gray PM.. Architecture of a mediator for a bioinformatics database federation. IEEE Trans Inform Technol Biomed 2002; 6 (2): 116–22. [DOI] [PubMed] [Google Scholar]
- 61. Haas LM, Lin ET, Roth MA.. Data integration through database federation. IBM Syst J 2002; 41 (4): 578–96. [Google Scholar]
- 62. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG.. Research electronic data capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42 (2): 377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article are available in the article. The RMID code repository is open source and available on GitHub at https://github.com/sparc-request/research-master-id.