Abstract
Fusion peptides (FP) are prominent hydrophobic segments of viral fusion proteins that play critical roles in viral entry. FPs interact with and insert into the host lipid membranes, triggering conformational changes in the viral protein that leads to the viral-cell fusion. Multiple membrane-active domains from the severe acute respiratory syndrome (SARS) coronavirus (CoV) spike protein have been reported to act as the functional fusion peptide such as the peptide sequence located between the S1/S2 and S2’ cleavage sites (FP1), the S2’-adjacent fusion peptide domain (FP2), and the internal FP sequence (cIFP). Using a combined biophysical approach, we demonstrated that the α-helical coiled-coil-forming internal cIFP displayed the highest membrane fusion and permeabilizing activities along with membrane ordering effect in phosphatidylcholine (PC)/phosphatidylglycerol (PG) unilamellar vesicles compared to the other two N-proximal fusion peptide counterparts. While the FP1 sequence displayed intermediate membranotropic activities, the well-conserved FP2 peptide was substantially less effective in promoting fusion, leakage, and membrane ordering in PC/PG model membranes. Furthermore, Ca2+ did not enhance the FP2-induced lipid mixing activity in PC/phosphatidylserine/cholesterol lipid membranes, despite its strong erythrocyte membrane perturbation. Nonetheless, we found that the three putative SARS-CoV membrane-active fusion peptide sequences here studied altered the physical properties of model and erythrocyte membranes to different extents. The importance of the distinct membranotropic and biological activities of all SARS-CoV fusion peptide domains and the pronounced effect of the internal fusion peptide sequence to the whole spike-mediated membrane fusion process are discussed.
Keywords: Fusion peptide, SARS-CoV, SARS-CoV-2, Lipid-protein interaction, COVID, Viral fusion, Membrane protein
Graphical abstract
1. Introduction
Human coronaviruses (HCoV) are known since the 1960s [1], [2]. To date, seven HCoV strains capable of infecting humans were identified: HCoV-229E, HCoV-NL63, HCoV-OC43, HCoV-HKU1, severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and SARS-CoV-2. The first four are endemic and usually cause mild to moderate symptoms in humans, while infection by the latter three HCoV can result in serious outcomes. SARS-CoV was responsible for the SARS outbreak in China in 2002-2003, provoking 774 deaths worldwide (10% case fatality rate) [3]. MERS-CoV, a SARS-CoV-related coronavirus, is the causative agent of the respiratory syndrome called MERS that broke out in Saudi Arabia in 2012, causing 886 deaths (35% case fatality rate) [4]. The third severe HCoV is SARS-CoV-2, which was identified in Wuhan, China, in 2019, and is the causative agent of the 2019 coronavirus disease (COVID-19), that resulted in millions of deaths worldwide [5]. The high transmission of the SARS-CoV-2 associated with the difficulty of early diagnosis, and the possible infection by asymptomatic individuals, contributed to the COVID-19 outbreak worldwide, driving the current pandemic.
Coronavirus spike (S) glycoproteins are responsible for catalyzing the fusion between viral and cell membranes [3], [5], [6]. Like other class I viral fusion proteins, such as the human immunodeficiency virus gp41 and the influenza virus hemagglutinin, different functional segments of the S protein play essential roles in the membrane fusion process [7]. Upon receptor binding through the N-terminal S1 subunit of the S protein (Fig. 1 ), both SARS-CoV and SARS-CoV-2 enter cells via two routes depending upon protease availability: an “early” plasma membrane entry route, which is triggered by exogeneous or membrane-bound proteases, and a “late” endosome entry pathway, in which the spike can be activated by endosomal cathepsins [6], [8]. Either entry pathway, receptor binding and proteolytic priming drive multiple conformational changes in the C-terminal S2 subunit that expose functional membrane-active domains for interaction with target membranes (Fig. 1) [9], [10], [11], [12], [13], [14]. Among them, the so-called fusion peptides (FP) are pivotal in viral entry. FPs interact with the host cell membranes and significantly change their ordering, fluidity, curvature, and hydration properties, driving the refolding of the S trimer coiled-coil core, ultimately leading to membrane fusion [13], [14], [15], [16], [17], [18], [19], [20].
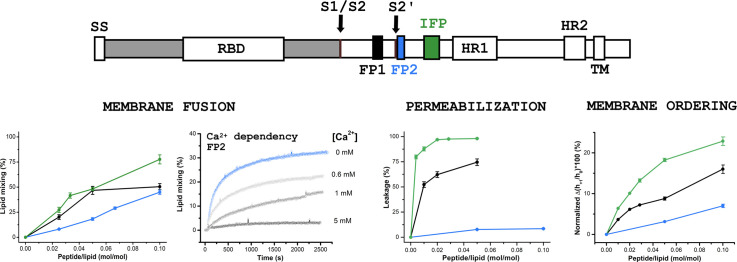
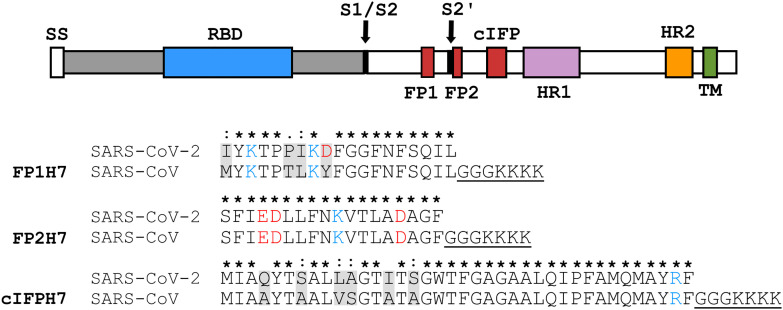
Fig. 1.
Schematic representation of the SARS-CoV spike glycoprotein containing the fusion peptide sequences. Top. The primary structure of the spike protein contains functionally relevant domains both in the N-terminal S1 subunit (gray), such as the receptor binding domain (RBD), and in the C-terminal S2 fusion subunit (white), such as the fusion peptide sequences (FP1, FP2, and cIFP), the heptad repeats 1 (HR1) and 2 (HR2), and the transmembrane domain (TM). The locations of the cleavage sites S1/S2 and S2’ are shown. Bottom. Sequence of the designed host-guest SARS-CoV fusion peptides used in the present study and sequence alignment with the SARS-CoV-2 fusion peptide domains (Accession numbers: AAP13441.1 for SARS-CoV-Urbani, and QHD43416.1 for SARS-CoV-2-Wuhan-Hu-1). Asterisks represent fully conserved residues, whereas colons and periods correspond to residues exhibiting strongly and weakly similar properties, respectively. Negatively (positively) charged residues are shown in red (blue). The host H7 sequence is underlined. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The FPs of many class I viral fusion proteins reside at the N-terminus of the S2 subunit, near or immediately adjacent to the protein proteolytic site. On the other hand, FPs from the Ebola virus glycoprotein or class II and class III viral glycoproteins are internally located within the polypeptide chain [21], [22]. Interestingly, three FP candidates have been identified in the S2 subunit of the SARS-CoV S protein: a highly conserved sequence across the Coronaviridae, corresponding to residues 798–815 (FP2) [23], and two less conserved sequences across the family but well conserved across lineage B betacoronaviruses, corresponding to residues 770–788 (FP1) [9] and 873–888 (IFP) [10]. FP1 and FP2 are located proximal or adjacent to the proteolytic sites R667 and R797 [24], respectively, whereas IFP is located between FP2 and the coiled-coil-forming heptad-repeat HR1 domain [24], [25] (Fig. 1). It has also been shown that a sequence C-terminally adjacent to FP2, comprising residues 816–835, also exhibits FP properties and acts, together with FP2, as an extended bipartite fusion “platform” [17]. The possible existence of different membrane-active domains displaying fusogenic properties in the spike protein of coronaviruses is indicative of a complex coronavirus membrane fusion mechanism.
The exact location of the putative internal membrane fusion peptide sequence has been a matter of debate. Using a 16/18-mer peptide library derived from the SARS-CoV S glycoprotein, Guillén and coworkers have shown that the 873–888 segment displays a significant membranotropic effect [10], later confirmed by a series of biophysical studies [14], [26]. On the other hand, based on the Wimley and White interfacial hydrophobicity scale, Sainz and colleagues identified the 864–886 segment as an internal membrane-interacting sequence of the SARS-CoV S protein [9]. Biophysical studies have demonstrated the ability of this peptide to partition into membranes causing permeabilization and membrane fusion [9]. In addition, a mutagenesis analysis performed on the predicted SARS-CoV fusion peptide sequence [27] highlighted the importance of the 852–881 segment for the S-mediated cell-cell fusion since mutations within this particular sequence severely interfered in the protein ability to induce cell-cell fusion [25]. A fourth peptide segment of the SARS-CoV S protein, corresponding to the 855–880 residues, was proposed by Ou et al. as the possible FP [28]. The authors identified the putative FP-containing regions of the spike proteins of representative betacoronaviruses using a predictor of protein transmembrane domains. They performed a systematic mutagenesis analysis of the predicted FPs. Mutations within the 855–880 sequence severely affected cell-cell fusion, highlighting the importance of this internal hydrophobic sequence in the S-mediated viral-cell fusion [28]. Therefore, due to these conflicting results with overlapping sequences, we decided to investigate the membranotropic properties of a combined segment containing the aforementioned internal FP sequences. We thus designated the 851–888 sequence as the complete internal fusion peptide, or cIFP (Fig. 1). The corresponding cIFP and N-terminal FP sequences from the SARS-CoV-2 S protein are also shown in Fig. 1 to illustrate the degree of sequence conservation. The primary structure of the cIFP displays a high content of alanine and glycine residues (~ 40%), intermediate hydrophobicity, and a fully conserved charged residue, which are characteristic of class I fusion peptides [29]. It also exhibits features of internal fusion peptides, such as the internal location and the presence of a fully conserved proline residue within the Coronaviridae family [28], which may confer the peptide a higher degree of conformational flexibility [30], [31].
Although some earlier data attributed to FP1 the highest fusogenic activity among the FP1, FP2, and IFP sequences [9], [13], [23], liposomes of different lipid compositions were used in those studies, thus limiting a more definitive assessment of the peptide fusogenicity. Moreover, it has been recently shown that the 16-amino acid residue long IFP16, corresponding to the 873–888 segment, is more fusogenic than FP1 in cholesterol-containing membranes [32]. Therefore, it is imperative to investigate the activity of the SARS-CoV fusion peptide sequences in the same lipid and environmental conditions.
Here, we characterize the interaction of three membrane fusion peptide sequences from the SARS-CoV S glycoprotein (FP1, FP2, and cIFP) with model and biological cell membranes. To do so, we made use of a combined approach employing fluorescence spectroscopy assays to probe the permeabilization and membrane fusion activity of the peptides, circular dichroism (CD) to monitor the peptide secondary structure in membrane mimetics, and electron spin resonance (ESR) to investigate changes in the membrane packing and fluidity. In particular, ESR has been extensively used to report changes in the structural dynamics of the lipids upon interaction with membrane-active agents [33], [34], [35], [36], [37], [38]. It has also been particularly useful in the viral fusion peptide research since the FP-induced ordering of lipid bilayers, as reflected by the ESR-derived order parameter, is a crucial step in the viral membrane fusion process [15], [16], [17], [18], [39], [40], [41]. We have previously shown that both SARS-CoV FP1 and IFP16 promote ordering and induce negative curvature of lipid bilayers composed of anionic lipids and generate curvature stresses on phosphatidylethanolamine membranes [15]. By measuring the intramembrane water content using the pulsed ESR technique called electron spin echo envelope modulation (ESEEM), we have also reported that the FP1- and IFP16-induced membrane ordering is correlated to membrane dehydration, giving new insights into the membrane fusion mechanism mediated by the SARS-CoV S fusion peptides [15].
On the other hand, FP2 exerts a membrane ordering effect dependent on Ca2+ binding to the peptide negatively charged residues [17], [40], [42]. However, the fusogenic activity of FP2 in lipid model membranes has not been so far evaluated in the presence of calcium. Therefore, we also investigated the calcium requirement for FP2 fusogenicity. Moreover, we evaluated the capacity of the SARS-CoV fusion peptides to permeabilize and aggregate erythrocytes and perturb the thermal stability of the erythrocyte ghost membrane proteins, used as models of biological membranes. The significance of the different membranotropic and biological activities of the SARS-CoV membrane fusion peptides, the reported higher fusogenicity of the complete internal FP, and the lack of enhancement of the FP2-mediated membrane fusion activity in the presence of Ca2+ are discussed. Due to the high degree of sequence conservation with the SARS-CoV-2 fusion peptides (Fig. 1), our data can also shed light on the possible action mechanism of the corresponding SARS-CoV-2 membrane fusion peptides.
2. Materials and methods
2.1. Materials
The peptides were synthesized by GenScript (Piscataway, USA) and GenOne Biotechnologies (Rio de Janeiro, Brazil). All peptides shown in Fig. 1 were acetylated at the N-terminus and amidated at the C-terminus and contained a guest -GGGKKKK hydrophilic sequence (H7) added to their C-terminus to increase solubility. It has been shown that the addition of the H7 tag does not perturb the structure and function of the fusion peptides [17], [43], [44]. The lysophospholipids 1-palmitoyl-2-hydroxy-sn-glycero-3-phosphocholine (16:0 lyso-PC or LPC) and 1-palmitoyl-2-hydroxy-sn-glycero-3-phospho-(1'-rac-glycerol) (16:0 lyso-PG or LPG), the phospholipids 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (POPG), 1,2-dimyristoyl-sn-glycero-phosphatidylcholine (DMPC), 1,2-dimyristoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (DMPG), the fluorophores N-lissamine rhodamine B 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (RhB-PE) and N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (NBD-PE), and the spin labels 1,2-dipalmitoyl-sn-glycero-3-phospho(tempo)choline (DPPTC) and 1-palmitoyl-2-stearoyl(n-doxyl)-sn-glycero-3-phosphocholine (n-PCSL, where n = 5 and 14) were purchased from Avanti Polar Lipids, Inc. (Alabaster, USA). Triton X-100, HEPES, and the fluorescent probe calcein were purchased from Sigma-Aldrich (São Paulo, Brazil). The peptide concentration was determined spectrophotometrically by using their theoretical molar extinction coefficients. The solubilization of cIFPH7 required 20% acetonitrile (ACN). Control experiments were performed with the same amount of ACN used in the cIFPH7 measurements and subtracted from them when needed. All reagents were used without further purification.
2.2. Circular dichroism studies
CD experiments were performed at 20 °C on a Jasco J-815 spectropolarimeter using a 1-mm path-length quartz cell. The bandwidth and response time were set, respectively, to 1 nm and 2 s. Spectra of the peptides (10 μM) in buffer solution (20 mM sodium phosphate, pH 5.0 or 7.4) and in the presence of 10 mM of LPC or LPG micelles were recorded at 50 nm/min over the wavelength range 190–270 nm. The resultant spectra were averaged over 8 to 16 consecutive scans. The spectra of the control samples (buffer or micelles), acquired under the same conditions, were subtracted from the averaged spectra from the peptides and converted to mean residue molar ellipticity. The fractional helical content, f H, was estimated by the mean residue ellipticity at 222 nm (θ 222), calculated using the limiting θ 222 values for all-helical (θ H) and all-disordered (θ C) conformations and the following equations: f H = (θ 222 – θ C)/(θ H – θ C), θC = 2220–53 T, and θ H = (250 T – 44,000)(1–3/n), where T is the temperature in Celsius, and n is the number of amino acid residues of the peptide [45].
2.3. Liposome preparation
Multilamellar vesicles (MLV) were prepared by mixing appropriate amounts of phospholipids dissolved in chloroform or chloroform/methanol 1:1 (v/v) stock solutions in a glass tube, drying under a N2 stream, and keeping under vacuum overnight to remove traces of organic solvent. The lipid film was hydrated with the appropriate buffer at a temperature above the melting transition of the lipids and subjected to five freeze-thaw cycles.
For the leakage experiments, the lipid films were hydrated with 10 mM HEPES buffer, pH 7.4, containing 35 mM of the fluorescent probe calcein. After freezing and thawing, the resulting MLVs were extruded using polycarbonate filters with a pore size of 100 nm (Nuclepore Corporation, Pleasanton, USA) to obtain large unilamellar vesicles (LUV). Non-encapsulated calcein was separated from the calcein-loaded liposome suspension through a Sephadex G-75 filtration column (Sigma-Aldrich, São Paulo, Brazil). Vesicles were eluted in 10 mM HEPES, pH 7.4, 100 mM NaCl. Lipid concentration was determined by measuring the lipid phosphorus content as described elsewhere [46]. Size distribution was confirmed by dynamic light scattering (DLS) experiments using a Zetasizer μV (Malvern Instruments Limited, Worcestershire, UK).
For the lipid mixing and the ESR assays, small unilamellar vesicles (SUV) were prepared as follows: either 1 mol% of the lipid fluorophores NBD-PE and RhB-PE in chloroform/methanol 1/1 (v/v) for fluorescence or 0.5 mol% of spin-labeled lipids for ESR were mixed with phospholipids in organic solvent and MLVs were prepared as described above. The lipid film was hydrated either with 20 mM sodium phosphate buffer at pH 5.0 and 7.4 or with 10 mM HEPES/MES, pH 5.0, 150 mM NaCl. The freeze-thawed MLVs were then sonicated in ice using an Ultrasonic Dismembrator Model 500 (ThermoFisher Scientific, Waltham, USA) for 20 min at a 10% duty cycle. The titanium particles released from the tip were removed by centrifugation at 15,700g for 10 min. DLS experiments confirmed the size distribution of the resultant SUVs. Unlabeled SUVs for the lipid mixing and vesicle aggregation assays were prepared using the same procedure as the labeled ones.
2.4. Liposome aggregation
Peptide-induced liposome aggregation was monitored by changes in the optical density at 405 nm. The absorbance was recorded on a SpectraMax i3 multimode plate reader (Molecular Devices, San Jose, USA) using a 96-well microplate containing 100 μL of a 100-μM SUV suspension with and without the peptides at different lipid-to-peptide molar ratios. The experiments were carried out at 25 °C in triplicates.
2.5. Fluorescence measurements
Leakage and phospholipid mixing assays were performed in an ISS K2 spectrofluorimeter (Analytical and Biomedical Instruments, Champaign, USA) using a fluorescence quartz cuvette of 1-cm path length. Peptides were added into a 1-ml vesicle solution under constant stirring.
2.6. Leakage assays
Membrane permeabilization was investigated by adding appropriate amounts of peptides into 50 μM calcein-loaded LUVs. The release of the liposome internal content due to peptide-induced membrane rupture was monitored at 25 °C by changes in the probe fluorescence intensity. The excitation and emission wavelengths were set to 490 nm and 520 nm, respectively. The percentage of leakage of the intraliposomal calcein was calculated with the following equation:
where F 0 and F 100 correspond to, respectively, the fluorescence intensities of the calcein-encapsulated liposomes before peptide addition and after addition of 1% (v/v) of Triton X-100 to obtain 100% leakage.
2.7. Phospholipid mixing
Membrane fusion was monitored by the decrease of Förster resonance energy transfer (FRET) between two lipid fluorophores and was based on the probe dilution assay [47]. In this assay, an unlabeled population of LUVs is mixed with a labeled population containing 1 mol% of the fluorophores NBD-PE and RhB-PE at a 4:1 unlabeled:labeled molar ratio with 150 μM total lipid. The two lipid compositions used were: POPC/POPG 3/2 (mol/mol) and POPC/POPS/Chol 3/1/1 (mol/mol/mol). Fusion of the probe-containing vesicles with the unlabeled ones allows the new vesicle formed to exchange their lipids, resulting in a decrease in the FRET. Thus, the kinetics and extent of membrane fusion can be measured by the decrease of the emission intensity of the acceptor, RhB-PE, or by the increase of the emission intensity of the donor, NBD-PE. We chose the latter with the excitation and emission wavelengths set to 467 nm and 530 nm, respectively. The percentage of lipid mixing was calculated with the following equation:
where F 0 and F 100 correspond to the fluorescence intensities of the probe-labeled liposomes before peptide addition and after addition of 1% (v/v) of Triton X-100.
2.8. Electron spin resonance (ESR) and spectral fitting
X-band (9.5 GHz) ESR experiments were performed at 30 °C on a Varian E-109 spectrometer. Lipid and lipid/peptide samples were transferred to glass capillaries (1.5 mm I.D.) and spun at 15,700 g for 15 min. The capillaries were immersed into a mineral oil bath in a quartz tube for the measurements. A homemade temperature control unit coupled to the spectrometer (accuracy of ~0.2 °C) controlled the temperature. The spectra were recorded with a modulation amplitude of either 0.5 or 1.0 G and a microwave power of 5 mW. Nonlinear least-squares (NLLS) fitting of selected ESR spectra was carried out using the Multicomponent LabView (National Instruments, Austin, USA) [48], [49] as described elsewhere [38], [50].
2.9. Hemolytic and hemagglutination activities of the SARS fusion peptides
Freshly collected blood from a healthy human donor was washed three times with phosphate buffered saline (PBS) pH 7.4. A suspension of erythrocytes at 1% (v/v) was prepared in PBS and used in the hemolytic and hemagglutination assays. The 1% suspension of erythrocytes was incubated with the peptides FP1H7, FP2H7, and cIFPH7 at different concentrations (1–50 μM) for 1 h at 37 °C. The release of hemoglobin assessed the hemolytic activity. After the incubation time, the samples were centrifuged at 3000g for 5 min, and the absorbance of hemoglobin was measured at 405 nm on a SpectraMax i3 multimode plate reader (Molecular Devices, San Jose, USA). The percentage of hemolysis was calculated considering 100% hemolysis of the samples incubated with 1% Triton X-100 after subtracting the absorbance of the control samples incubated with PBS or acetonitrile. The hemagglutinating activity of the peptides was observed in 96-well microtiter U-plates after 1 h incubation time. Images of the hemagglutination process were acquired in an optical microscope Olympus BX53. In this experiment, the peptides were added to the erythrocyte suspension at desired concentrations, and 10 μL aliquots were placed on glass slides, covered with coverslips, and immediately imaged.
2.10. Differential scanning calorimetry (DSC) experiments
Erythrocyte ghost samples were prepared as described in Felizatti et al. [35] . Peptide/ghost samples were obtained by adding the desired peptide concentration into 100 mg/mL erythrocyte ghost in PBS solution. The samples were incubated for 20 min at 37 °C. The DSC thermograms were recorded in a VP-DSC MicroCal MicroCalorimeter (Microcal, Northampton, USA) using a heating rate of 60 °C/h. Analysis of the thermograms was performed with the MicroCal Origin software.
3. Results
3.1. SARS-CoV membrane fusion peptides promote extensive membrane disruption
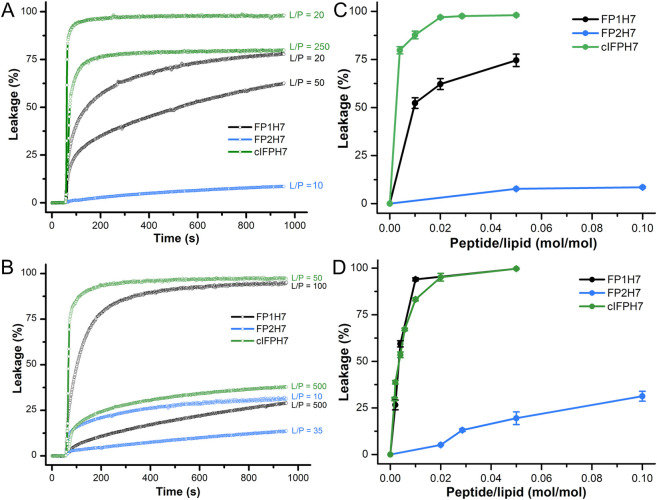
It has been suggested that the fusogenic properties of viral fusion peptides are strongly correlated with their capacity to destabilize lipid membranes [51]. Thus, the ability of the SARS-CoV fusion peptides to elicit membrane permeabilization was measured by the release of the fluorescent probe calcein encapsulated at a self-quenching concentration in the internal aqueous phase of the liposomes. Fig. 2A and B show representative data illustrating the kinetics of intraliposomal calcein release upon peptide addition to the zwitterionic POPC and the anionic POPC/POPG 3/2 (mol/mol) LUVs, respectively. The extent of membrane rupture of the probe-loaded liposomes is shown in Fig. 2C and D. Both FP1H7 and cIFPH7 promoted extensive leakage of the liposome internal contents in a concentration-dependent manner. The ability of cIFPH7 to destabilize the zwitterionic LUVs and to cause membrane rupture was higher than that of FP1H7 (Fig. 2C, Table 1 ). On the other hand, the permeabilizing effects of both cIFPH7 and FP1H7 in anionic LUVs were virtually similar (Fig. 2D, Table 1). At a lipid-to-peptide molar ratio (L/P) of 20, both cIFPH7 and FP1H7 induced ~100% leakage of calcein from POPC/POPG liposomes. On the other hand, in POPC membranes, cIFPH7 promoted leakage of ~98% of the total internal calcein content, whereas FP1H7 caused 75% leakage (Table 1). FP2H7 displayed the smallest membrane destabilization effect, inducing only ~8% leakage of POPC and ~ 19% of POPC/POPG liposomes at 5 mol% (Table 1).
Fig. 2.
Membrane permeabilization induced by the SARS-CoV fusion peptides. Representative data on the kinetics of the leakage of calcein encapsulated in 50 μM of (A) POPC and (B) POPC/POPG 3/2 (mol/mol) at pH 7.4 and at different lipid-to-peptide molar ratios (L/P). (C, D) The extent of membrane permeabilization as a function of peptide-to-lipid molar ratio for POPC (panel C) and for POPC/POPG (panel D) lipid vesicles. The peptides were added to the vesicle solution at ~50 s. For each measurement, Triton X-100 (1%) was added to the sample to achieve 100% of calcein release.
Table 1.
Percentage of membrane permeabilization (leakage) and membrane hemifusion (lipid mixing) induced by 5 mol% (L/P = 20) of the putative SARS-CoV fusion peptides. Total lipid concentrations: 50 μM for the permeabilization assays and 150 μM for the lipid mixing assays.
| Sample | POPC pH 7.4 |
POPC/POPG pH 7.4 |
POPC/POPG pH 5.0 |
|
|---|---|---|---|---|
| Leakage (%) | Leakage (%) | Fusion (%) | Fusion (%) | |
| FP1H7 | 75 (3) | 99.7 (0.3) | 59 (3) | 47 (4) |
| FP2H7 | 7.7 (0.3) | 19 (3) | 15 (2) | 18 (1) |
| cIFPH7 | 98.0 (0.5) | 99.7 (0.3) | 99 (1) | 48 (2) |
Standard deviations from duplicate or triplicate measurements are shown in parenthesis.
3.2. The complete internal fusion peptide elicits membrane fusion at greater extents
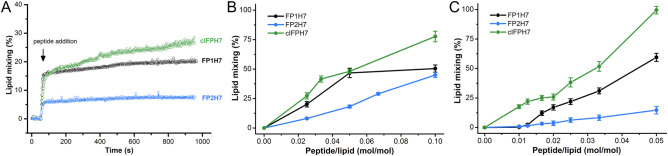
Since the peptides promoted differential membrane disruption activity, we tested their capacity to promote the fusion of lipid model membranes. Thus, the fusogenicity of the SARS peptides was probed by investigating their ability to induce lipid mixing of labeled and unlabeled liposomes. To do so, we monitored the decrease of the resonance energy transfer between the fluorescent lipids NBD-PE and RhoB-PE, both incorporated in the labeled vesicles, due to the dilution of the probes upon lipid mixing with the unlabeled liposomes [47]. This type of assay probes membrane hemifusion, which is characterized by mixing lipids from the outer monolayers of the interacting vesicles. A complete fusion would be achieved with pore formation, allowing for mixing the vesicle's internal contents. Since the hemifusion state precedes fusion pore formation, phospholipid mixing assays are used to probe the fusogenic activity of fusion peptides [13], [23], [52].
We chose the POPC/POPG 3/2 (mol/mol) lipid composition for these membrane fusion assays because the peptides promoted the largest permeabilizing activity in anionic membranes (Fig. 2). Additionally, since SARS-CoV entry may occur through an endosomal pathway [6], [8], we also probed the peptides fusogenicity at an environmental pH mimicking the acidic conditions of the endosomes. The kinetics of phospholipid mixing of POPC/POPG SUVs induced by the SARS peptides was recorded by monitoring the increase of the NBD-PE emission intensity, and representative data are illustrated in Fig. 3A. At L/P = 40, both cIFPH7 (~ 27.3%) and FP1H7 (~ 20%) were more fusogenic than FP2H7 (~ 8%) at pH 5.0. The extent of the peptide-mediated membrane fusion of POPC/POPG SUVs at acidic and neutral pH is shown in Fig. 3B and C, respectively. All peptides induced lipid mixing in a concentration- and pH-dependent manner. Irrespective of the pH, cIFPH7 exhibited the largest lipid mixing effect, while FP2H7 displayed the smallest membrane fusion activity among all peptides. Greater extents of lipid mixing were observed for FP1H7 and cIFPH7 at pH 7.4 compared to the acidic environment (Table 1). On the other hand, the fusogenic activity of FP2H7 was slightly increased in an acidic environment (Table 1).
Fig. 3.
Membrane fusion elicited by the SARS-CoV fusion peptides. (A) Representative kinetics data of phospholipid mixing induced by addition of 2.5 mol% (L/P = 40) peptides measured by the dilution of the probes NBD-PE and Rho-PE from labeled vesicles (30 μM) into unlabeled POPC/POPG 3/2 (mol/mol) LUVs (120 μM) at pH 5.0. For each measurement, the peptides were added to the vesicle solution at the time indicated by the arrow. Triton X-100 (1%) was added to the sample to achieve 100% probe dilution. (B, C) Extent of lipid mixing as a function of peptide-to-lipid molar ratio for POPC/POPG 3/2 (mol/mol) lipid vesicles at pH 5.0 (panel B) and 7.4 (panel C).
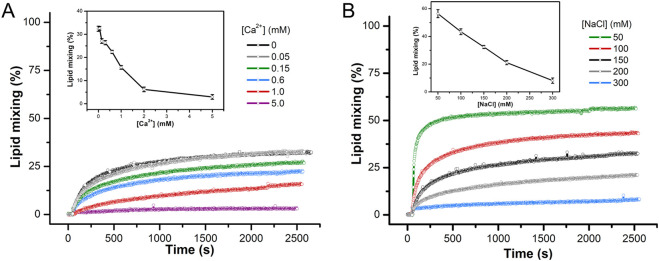
3.3. The fusogenicity of the FP2H7 peptide is not enhanced by Ca2+ binding
The FP2 peptide was the least effective in promoting membrane fusion and leakage in the conditions tested. However, it has been shown that the binding of Ca2+ ions triggers FP2 membrane insertion and membrane order to the FP2 Asp and Glu residues [17], [40], [53], and this binding is highly specific [42]. Therefore, it is tempting to investigate whether the low membrane fusion activity displayed by FP2H7 would be enhanced by Ca2+ binding. We only tested FP2H7 since the primary structures of both FP1H7 and cIFPH7 lack negatively charged residues (Fig. 1), thus not exhibiting putative metal-binding sites. Furthermore, no enhancement of the peptide-mediated lipid mixing was previously observed in the presence of Ca2+ for FP1 and an IFP construct [9].
For the FP2H7-membrane fusion assays, we used LUVs comprised of 60% POPC, 20% of the anionic POPS, and 20% cholesterol (molar ratio), a lipid model membrane used in other fusion peptide studies [23], [40], [53]. The Ca2+-dependent SARS-CoV FP2H7 membrane insertion and membrane-ordering effect are virtually identical in POPS- and POPG-containing membranes [17], [42]. The experiments were performed at pH 5.0 due to the higher FP2H7 fusogenic activity in cholesterol-containing membranes in an acidic environment, as previously reported [23].
In the absence of calcium, FP2H7 promoted about 32% of lipid mixing in our POPC/POPS/Chol 3/1/1 LUVs, similar to previously obtained with a POPC/POPS/Chol 1/3/1 model membrane under the same conditions [23]. The addition of increasing Ca2+ concentrations to the buffer containing the vesicles decreased the rate of fusion and the ability of FP2H7 to mediate lipid mixing (Fig. 4A). Pre-incubation of calcium with FP2H7 yielded similar results. Our assays showed that the binding of Ca2+ ions to FP2H7 did not enhance the peptide fusion activity. On the contrary, the presence of Ca2+ in the medium decreased FP2H7 fusogenicity putatively by charge screening [54]. We then tested whether this effect was electrostatic by nature by changing the buffer ionic strength. The addition of NaCl did decrease the extent of lipid mixing, whereas reduction of salt concentration yielded greater extents of membrane fusion (Fig. 4B). Our results suggest that FP2H7 fusogenicity is not only pH-dependent (previous section) but also ionic strength-dependent, and Ca2+ binding to the peptide sites does not play a role in the peptide fusion activity.
Fig. 4.
FP2H7 mediated lipid mixing is not enhanced in the presence of Ca2+. (A) Representative kinetics data of phospholipid mixing induced by addition of 5 mol% FP2H7 (L/P = 20) measured by the dilution of the probes NBD-PE and Rho-PE from labeled vesicles (30 μM) into unlabeled POPC/POPS/Chol 3/1/1 (molar ratio) LUVs (120 μM) at pH 5.0 in the absence and presence of Ca2+. Box. Extent of lipid mixing as a function of Ca2+ concentration. Buffer used: 10 mM MES/HEPES, 150 mM NaCl, pH 5.0. (B) Kinetics of lipid mixing induced by addition of 5 mol% FP2H7 (L/P = 20) into a mixture of labeled and unlabeled POPC/POPS/Chol 3/1/1 (molar ratio) LUVs at different NaCl concentrations. Inset. Extent of lipid mixing as a function of NaCl concentration.
3.4. The complete internal FP adopts a coiled-coil helical structure, whereas the N-terminal FPs acquire monomeric helical conformations in lipid micelles
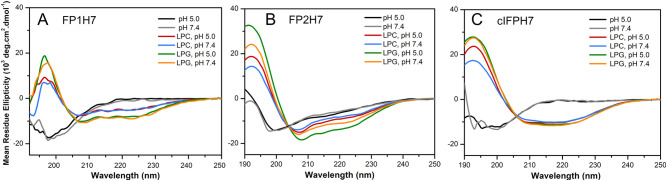
The propensity of the fusion peptides to acquire ordered conformations in the presence of lipid micelles was assessed by CD spectroscopy. The advantages of using micelles in structural studies of fusion peptides are three-fold: (i) they provide an excellent membrane mimetic environment for peptide folding [55]; (ii) the high curvature of detergent micelles mimics the strongly curved lipid bilayer states involved in the whole membrane fusion process [56]; and (iii) it avoids vesicle aggregation elicited by the peptides (Fig. S1).
Fig. 5 illustrates the CD spectra of the SARS-CoV fusion peptides in solution and micelles comprised of the zwitterionic lysolipid 16:0 phosphatidylcholine (LPC) or the anionic 16:0 phosphatidylglycerol (LPG) at pH 5.0 and 7.4 [57]. The negative band around 198–200 nm indicates a disordered conformation of the peptides in solution at both pH values. In the presence of the membrane mimetics, the negative bands at ~208 nm and 222 nm in the spectra of the peptides are characteristic of α-helical conformations (Fig. 5). These results indicate that all peptides adopted ordered α-helical secondary structures in LPC and LPG micelles at both pH values. The helicity of the peptides was higher in LPG, implying that peptide secondary structure formation might be enhanced by the electrostatic interaction between the peptides and the negatively charged LPG micelles (Table 2 ).
Fig. 5.
Peptide conformation in solution and bound to membrane mimetics. CD spectra of the peptides (A) FP1H7, (B) FP2H7, and (C) cIFPH7 in solution and incubated with 10 mM LPC or LPG micelles at pH 5.0 or 7.4. Peptide concentration was 10 μM. Spectra were recorded at 20 °C.
Table 2.
Percentage of helical content and average number of helical residues (Nα) of the SARS-CoV membrane fusion peptides (10 μM) bound to the lipid micelles (10 mM) obtained from the CD spectra. For the cIFPH7, the ratio of the molar ellipticities at 222 nm and 208 nm is also shown. The percentage of helical content was calculated as shown in Methods.
| Sample | FP1H7 |
FP2H7 |
cIFPH7 |
||||
|---|---|---|---|---|---|---|---|
| % | Nα | % | Nα | % | Nα | [θ]222/[θ]208 | |
| LPC, pH 5.0 | 18 ± 1 | 4.7 | 28 ± 1 | 6.9 | 29 ± 1 | 13.0 | 1.00 |
| LPC, pH 7.4 | 18 ± 1 | 4.7 | 25 ± 1 | 6.2 | 29 ± 1 | 13.0 | 1.11 |
| LPG, pH 5.0 | 26 ± 2 | 6.7 | 40 ± 1 | 10.1 | 32 ± 1 | 14.5 | 1.08 |
| LPG, pH 7.4 | 28 ± 2 | 7.3 | 33 ± 1 | 8.3 | 32 ± 1 | 14.5 | 1.10 |
Interestingly, the ratio of the molar ellipticities at 222 nm and 208 nm, i.e. [θ]222nm/[θ]208nm, exceeded 1 for the cIFPH7 (varied from 1.00 to 1.11, Table 2), suggesting a coiled-coil formation [58]. However, [θ]222/[θ]208 < 1 in trifluoroethanol/water solutions (Fig. S2), indicating disruption of the coiled-coil helices [59]. This result suggests that the cIFPH7 quaternary structure formation is triggered by membrane binding. Additionally, a pH-triggered secondary structure formation was only observed for FP2H7 (Fig. 5B). In an acidic environment, the α-helical content slightly increased from 25 to 28% in LPC and from 33 to 40% in LPG micelles (Table 2). The structural change may be caused by protonation of the FP2H7 negatively charged Asp and Glu residues. The other peptides exhibited virtually no pH-dependent conformational changes (Table 2, Fig. 5). The more remarkable ability of the peptides to acquire higher content of ordered conformations in anionic membranes compared to zwitterionic ones and the pH-dependency of the FP2H7 conformation might be functionally relevant to the viral membrane fusion process [13], [14], [15], [23].
3.5. The complete internal fusion peptide promotes more extensive membrane surface ordering
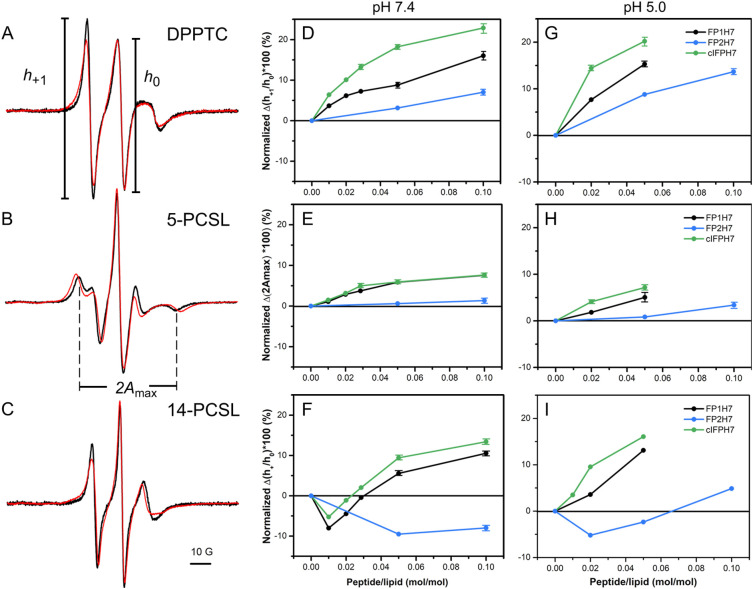
Our membrane permeabilization and lipid mixing assays indicate that cIFPH7 disturbs lipid bilayers more effectively than the other peptides. We then used electron spin resonance (ESR) to investigate the peptide-mediated changes in the local ordering and mobility of the lipids from the polar headgroup region down to the bilayer center [60]. To do so, we used nitroxide-labeled lipids that report on alterations at the lipid/water interface (DPPTC) and different depths along the hydrophobic core (5-PCSL and 14-PCSL) of SUVs composed of a mixture of the zwitterionic phospholipid DMPC and the anionic phospholipid DMPG. Saturated PC and PG phospholipids have been used in previous viral fusion peptide studies [15], [18], [61].
Fig. 6A–C illustrates representative ESR spectra of the spin-labeled lipids in the absence and presence of cIFPH7 in the physiologically relevant liquid crystalline phase of anionic DMPC/DMPG 3/2 (mol/mol) lipid vesicles. The peptide was added into the solution containing preformed SUVs to mimic the peptide interaction with the outer leaflet of the lipid bilayer, as previously reported [15]. Qualitatively, peptide binding to the membrane caused extensive line broadening, yielding a reduction in the amplitude of low field resonance lines (h +1) of DPPTC and 14-PCSL spectra (Fig. 6A and C) and an increase in the outer hyperfine splitting of the 5-PCSL spectra (2A max, Fig. 6B). Only cIFPH7 and FP1H7 significantly changed the ESR lineshape of all spectra in a concentration-dependent manner (Fig. S3). The higher the peptide concentration (lower L/P ratio), the less intense is the h +1 of the DPPTC and 14-PCSL spectra, and the more significant is the 2A max of the 5-PCSL spectra. These results indicate that both FP1H7 and cIFPH7 increased membrane ordering and reduced the lipid chain rotational mobility [15]. On the other hand, FP2H7 caused only marginal spectral changes.
Fig. 6.
SARS-CoV membrane peptides promote ordering on anionic lipid bilayers. Representative ESR spectra of DPPTC (A), 5-PCSL (B), and 14-PCSL (C) embedded in 5 mM DMPC/DMPG 3/2 (mol/mol) SUVs in the fluid phase (30 °C) and at neutral pH in the absence (black) and presence (red) of 5 mol% of cIFPH7. The parameters defined on the spectra were used for spectral analysis shown in the panels on the right. The spectra were normalized by the height of the central line (h0). (D—I) Percentage of spectral change as a function of peptide-to-lipid molar ratio obtained from the analysis of the ESR spectra of DPPTC (D, G), 5-PCSL (E, H), and 14-PCSL (F, I) at pH 7.4 (D, E, F) and 5.0 (G, H, I). The plots are shown as the percentage of h+1/h0 reduction of the indicated line amplitudes of both DPPTC (panel A) and 14-PCSL (panel C) spectra and the percentage of 2Amax increase of the 5-PCSL spectra (panel B) induced by the peptides. Positive values indicate membrane ordering, whereas negative values imply lipid disordering. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To quantitatively characterize the peptide-induced perturbation in the dynamic structure of the DMPC/DMPG SUVs, we used parameters defined on the ESR spectra that can provide valuable information about the spin probe's mobility in phospholipid bilayers [60], [62]. The ratio of the amplitudes of the low (h +1) and central (h 0) field lines of the ESR spectra of DPPTC and 14-PCSL and the outer hyperfine splitting, 2A max, of the 5-PCSL ESR spectrum (Fig. 6A-C) are sensitive to the nitroxide dynamics and membrane packing. The higher the h +1/h 0 and the lower the 2A max values, the higher the spin-label mobility and the less ordered is the lipid chain [60], [62]. To describe the spectral differences quantitatively, we calculated the percentage of h +1/h 0 reduction and the percentage of 2A max increase so that positive values denote an increase of the membrane ordering (Fig. 6D–I).
At the headgroup level and neutral pH, cIFPH7 promoted the most prominent ordering effect. Indeed, 5 mol% of cIFPH7 altered the h +1/h 0 parameter by ca. 18%, whereas 10 mol% of FP2H7 caused only a slight 7% alteration in the h +1/h 0 (Fig. 6D). Nonlinear least-squares spectral simulations indicate that the order parameter S 0 increased from 0.38 in the pure membrane to 0.43 for FP2H7, 0.45 for FP1H7, and 0.49 for cIFPH7 (Fig. S4 and Table S1). In the nonpolar core of the membrane, both FP1H7 and cIFPH7 also affected the bilayer packing and membrane fluidity more significantly than FP2H7. However, the perturbations were lower than those caused at the lipid/water interface (Fig. 6E and F).
Interestingly, the h +1/h 0 parameter obtained from the 14-PCSL ESR spectra in the presence of FP1H7 and cIFPH7 initially increased at low peptide concentrations and then decreased with further peptide binding to the lipid vesicles (Fig. 6F). This result suggests that the interaction of those peptides with the lipid bilayers yields opposing effects in the acyl chain mobility in the lipid bilayer center: the mobility (i) increases at low peptide concentration, and (ii) decreases upon further peptide binding. This ordering effect in the bilayer center at high peptide concentrations is consistent with our previous work using FP1 and IFP16 (without the H7 tag) [15]. On the other hand, the binding of FP2H7 promoted a disordering effect in the bilayer center for all concentrations used. Taken together, these results indicate a differential and concentration-dependent membrane insertion depth of the peptides. At low concentration, all peptides may be located in a shallow position at the membrane surface, acting as lipid spacer, thus promoting a disordering effect in the bilayer center. At higher concentrations, both FP1H7 and cIFPH7 penetrated deeper into the membrane, causing an ordering effect in the bilayer center.
All peptides induced slight changes in the acidic environment, mainly in the DPPTC and 14-PCSL ESR spectra (Fig. 6G and I). In particular, the pH-dependent peptide-induced ordering effect in the membrane surface was higher for FP2H7 than for the other peptides (Fig. 6G). Interestingly, at a very high peptide-to-lipid molar ratio (10 mol%), FP2H7 enhanced the membrane packing of the bilayer midplane, a strikingly different effect than that induced at neutral pH (Fig. 6F). On the other hand, an increase of membrane packing in the bilayer center was observed for all concentrations of both cIFPH7 and FP1H7 (Fig. 6I), different from the result obtained at pH 7.4 (Fig. 6F). These results suggest a putative pH-dependent transition of the peptide topology or a pH-triggered insertion depth in DMPC/DMPG membranes.
We also tested pure zwitterionic DMPC to investigate whether the peptide-induced ordering of the anionic DMPC/DMPG was due to the negative surface charge provided by DMPG. Our ESR results showed that only cIFPH7 could significantly modify the ordering of the lipid head group and acyl chain of DMPC (Supplementary Fig. S5), despite the extensive permeabilization of PC membranes caused by both cIFPH7 and FP1H7 (Fig. 2B). The cIFPH7-induced ordering effect on DMPC was much less pronounced than those observed in negatively charged vesicles. This result indicates that lipid charge does play a role in modulating the effect of the SARS fusion peptides on the ordering and fluidity of lipid model membranes.
3.6. SARS membrane fusion peptides promote opposing hemolytic and hemagglutinating activities
Previous studies with other class I fusion peptides, such as those of influenza hemagglutinin and HIV-1 gp41, have demonstrated the correlation between hemolytic and hemagglutinating activities of synthetic peptides their ability to promote membrane fusion [63], [64], [65]. Since SARS peptides can promote hemifusion, leakage, and lipid aggregation (Fig. S1) of model membranes to different extents, it is tempting to evaluate their potential to elicit permeabilization and agglutination of natural biological membranes, such as those of erythrocytes.
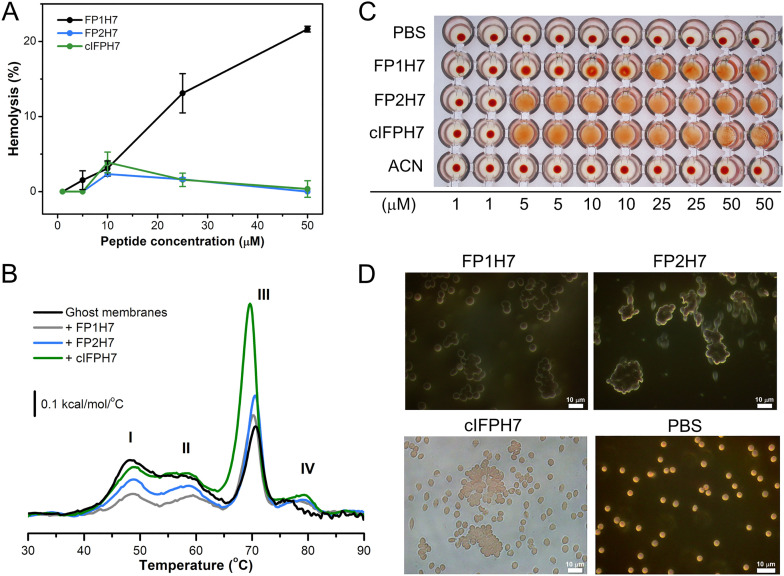
Fig. 7A shows that only FP1H7 exhibited significant hemolytic activity (13% at 25 μM and 21% at 50 μM) compared to the other SARS peptides, but smaller than those found for the fusion peptides from HIV-1 gp41 (70% at 40 μM) [64] and influenza hemagglutinin (~ 40% at 35 μM) [44]. FP2H7 and cIFPH7 induced mildly to none hemolysis up to 50 μM. Despite their low permeabilization activity on erythrocytes, the SARS-CoV fusion peptides interacted with erythrocyte ghosts and perturbed the thermal stability of the ghost membrane proteins (Fig. 7B), as previously shown for other peptides [35]. Our DSC data show that cIFPH7 altered both the enthalpy change and the denaturation temperature of the anion-transporting domain of the band 3 protein (peak III) more significantly than the other peptides [66]. On the other hand, FP1H7 and FP2H7 were more effective (FP1H7 > FP2H7) in reducing the transition enthalpies of a set of different ghost membrane proteins ascribed to peaks I and II (see Fig. 7B caption).
Fig. 7.
Biological activities of the SARS-CoV membrane fusion peptides. (A) Percentage of hemolysis as a function of peptide concentration. 100% hemolysis was achieved by adding 1% Triton X-100. (B) Representative DSC thermograms of erythrocyte ghosts in the absence and presence of 20 μM of FP1H7, FP2H7, and cIFPH7. The thermograms show four endothermic transitions corresponding to the melting of different erythrocyte ghost proteins. Besides peak IV, the other three transitions have been assigned to the denaturation of the following proteins [66]: (I) bands 1 and 2 of the spectrin complex (~ 48 °C); (II) bands 2.1, 4.1, and 4.2 along with some glycophorins (~ 58 °C); and (III) the anion-transporting domain of the band 3 protein (~ 70 °C). (C) Hemagglutination assays with different concentrations of SARS peptides (1 to 50 μM). The red dots represent the deposition of the red blood cells onto the bottom of the wells, indicating a negative hemagglutination reaction. Absence of the red dot means positive reaction. PBS and ACN (acetonitrile) were used as negative controls. (D) Optical microscopy imaging of the SARS peptides hemagglutinating activity. Erythrocyte suspensions were incubated with FP1H7 (50 μM), FP2H7 (25 μM), and cIFPH7 (25 μM) and readily imaged. The data shown are representative of two independent experiments at the selected concentration. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To gain further insights into the effects of the SARS fusion peptides on the properties of a cell membrane model, we also evaluated the ability of the peptides to promote the agglutination of erythrocytes. Both FP2H7 and cIFPH7 promoted hemagglutination at low concentrations (Fig. 7C). On the other hand, FP1H7 exhibited hemagglutinating activity only at high concentrations. The high hemagglutinating activity of cIFPH7 cannot be attributed to the acetonitrile (ACN) amount used for peptide solubilization (Fig. 7C). The relative aggregation potency of the peptides followed the order cIFPH7 ~ FP2H7 > FP1H7. Indeed, when analyzed under a light microscope, it is evidenced the enhanced ability of FP2H7 and cIFPH7 to promote erythrocyte aggregation as compared to FP1H7 (Fig. 7D).
Despite being only qualitative, our results confer to cIFPH7 and FP2H7 the highest hemagglutinating activities since the minimal peptide concentration required to aggregate the erythrocytes was 5 μM. Furthermore, a roughly inverse relation between hemolytic and hemagglutinating activities was noticed for the SARS-CoV membrane peptides. Those that exhibited the highest erythrocyte aggregating potency (cIFPH7 and FP2H7) exhibited substantially less hemolytic activity. At the highest concentration used, though, FP1H7 promoted both hemolysis and hemagglutination.
4. Discussion
Membrane fusion is a critical step in the infectivity of enveloped viruses. The surface-attached fusion glycoproteins possess multiple functional domains that play essential roles in the viral-host membrane fusion process (Fig. 1, top). Among them, fusion peptides constitute key components of the viral fusion machinery. Upon receptor binding and S priming, FPs are triggered to interact with cell membranes, initiating the contact between the host and viral lipid bilayers. FPs insert into the target cell and establish an anchor that bridges both membranes via the S protein. FP binding to the host membrane is thought to drive refolding of the viral fusion protein into a trimeric hairpin conformation, leading to the merge of the viral and host lipid bilayers and fusion pore formation [7], [56]. Accumulating evidence supports the view that interaction studies of synthetic peptides corresponding to the isolated viral FP domains with lipid model membranes can provide valuable information regarding the membrane fusion mechanism induced by the entire viral fusion glycoprotein [52], [67], [68]. Here, we explored the membrane activity of three peptides whose sequences are intended to reproduce the proposed fusion peptide sequences found in the spike protein of the SARS-CoV (Fig. 1, top). The high level of sequence identity between these peptides and the corresponding sequences found in the spike protein of the SARS-CoV-2 suggest that our results can be fairly extended to the latter case.
We demonstrated that all membrane-active fusion peptide sequences (FP1, FP2, and cIFP) studied here adopted α-helical conformations in the membrane (Fig. 5) and had the ability to induce membrane permeabilization (Fig. 2), lipid mixing (Fig. 3, Fig. 4), vesicle aggregation (Fig. S1), and membrane ordering (Fig. 6) to different extents. They also interacted with ghost and erythrocyte membranes, promoting hemolysis, hemagglutination, and perturbation of the thermal stability of the erythrocyte membrane proteins (Fig. 7). We found that the 38-residue long internal fusion peptide, cIFP, exhibited much more potent fusogenic and membranotropic activities on lipid model membranes than the other N-terminal FP1 and FP2 fusion peptides at both neutral and acidic pH. This finding highlights the importance of cIFP sequence to the SARS-CoV S-mediated fusion between the viral envelope and the plasma or the endosomal membranes since coronavirus entry might occur via a neutral-pH plasma membrane route or through the low-pH endosomal pathway depending upon the target cell type [6], [8].
We have previously reported the functional plasticity of the SARS fusion peptides FP1 and IFP16, a short, 16-amino acid residue internal FP corresponding to the C-terminal half of cIFP [15]. We showed that both peptides perturbed lipid model membranes by promoting lipid packing, head group ordering, membrane dehydration, and membrane curvature and that FP1 disturbed the bilayers more effectively than IFP16 [15]. On the other hand, biochemical, biophysical, and mutagenesis analysis revealed that mutations in the region corresponding to the N-terminal half of the complete cIFP sequence used here strongly affected cell-cell fusion and viral entry [25], [28]. Our biophysical studies also pointed out that the cIFP N-terminal half played a critical role in mediating higher membranotropic and biological activities than the other peptides. Our results showed that the H7-tagged IFP16, or IFP16H7, promoted lower leakage, lipid mixing, and hemagglutinating activities than the complete cIFPH7 under the same conditions (Fig. S6). Therefore, the use of the complete sequence of the cIFP allowed us to pinpoint the contributions of the N-terminal part of the extended internal fusion peptide to its functional membrane-interacting properties.
On the other hand, the well-conserved fusion peptide sequence immediately downstream from the S2’ cleavage site, FP2, was comparatively less potent than FP1 and cIFP but exhibited higher helical content and membrane activities in acidic compared to neutral environments, which might be physiologically important in the context of the endosomal pathway for viral entry.
Using ESR of nitroxide-labeled lipids incorporated in fluid bilayer membranes, we also showed an FP-induced ordering effect in the order cIFPH7 > FP1H7 > FP2H7, observed both at acidic and neutral pH values (Fig. 6). The membrane order was determined by changes in parameters defined on the ESR spectra, which was further confirmed by calculating the order parameter S 0 via spectral simulations of selected spectra. The lipid order parameter is an excellent indicator of the functional membrane activity of viral fusion peptides [15], [16], [17], [18], [39], [40], [41], [42]. The increase of the lipid head groups’ order parameter indicates membrane dehydration [15], [69]. Therefore, the ordering effect caused by different viral FPs in the lipid head group and the acyl chain region suggests the removal of water molecules from the membrane surface, which may reduce the hydration repulsion between two approaching lipid bilayers, thus triggering the fusion process. Our ESR data clearly showed that the complete internal fusion peptide sequence promoted the most significant ordering effect under the experimental conditions tested and may significantly contribute to the coronavirus membrane fusion process.
The influence of ions, especially calcium, in regulating fusion processes is of particular interest [39], [70]. In the case of fusion peptides derived from the S protein of SARS-CoV, only FP2 exhibited a Ca2+ dependency of the peptide conformation and the FP-induced membrane activity [9], [17]. It has been previously shown, also by ESR of spin-labeled lipids, that the membrane ordering promoted by FP2 was strongly dependent on and highly specific to Ca2+ ions [17], [42]. This Ca2+-mediated bilayer ordering effect has also been reported for the corresponding FP2 sequences of the MERS-CoV and SARS-CoV-2 S proteins [40], [42]. By isothermal titration calorimetry, the same authors showed that FP2 had one binding site for Ca2+, and the adjoining C-terminal sequence, comprising residues 816 to 835, displayed a second binding site [17]. Surprisingly, we observed no enhancement in the lipid mixing activity of FP2H7 in the presence of Ca2+ ions, despite the primary structure of the FP2H7 here used contained a single Ca2+ binding site and the reported Ca2+ dependency of peptide-induced membrane ordering. This result contrasts with that obtained with the Ebola fusion peptide, for which a Ca2+-mediated enhancement of both lipid mixing and membrane ordering was observed [39], [54]. Our lipid mixing assays showed an ionic strength dependency of the FP2H7 fusogenicity since increasing Ca2+ or NaCl concentrations in the medium downregulated the peptide membrane fusion activity. Reconciling the apparent lack of correlation between membrane ordering and lipid mixing activity for the fully conserved FP2 sequence from both SARS-CoV and SARS-CoV-2 S proteins (Fig. 1) will require further investigation. Nonetheless, the FP2-induced membrane ordering effect triggered by Ca2+ binding to the peptide may also contribute along with the other peptides to the fusion reaction since it can promote membrane dehydration, helping to lower the hydration repulsion between the approaching viral envelope and the host cell lipid bilayer.
We also showed that the FP1H7, FP2H7, and cIFPH7 were disordered in solution but underwent a transition to α-helical structures in lipid micelles. This result is in agreement with the literature for FP2 and for different IFP sequences, in diverse nonpolar environments [14], [17], [23], [28], [71], [72]. A nuclear magnetic resonance structural characterization of FP1 in detergent micelles has been conducted and reported a V-shaped helical conformation for FP1 [72]. On the other hand, most structural studies have shown that FP1 (without the H7 tag) adopts β-strand conformations both in aqueous and TFE solutions and bound to lipid bilayer membranes [9], [13], [73], [74]. Our CD data indicated that adding the poly-lysine tag at the FP1 C-terminus provided a solubilizing platform for the peptide, reducing the FP1 tendency to form β-like aggregates. This result may be important in the context of the whole SARS-CoV S protein. The FP1 conformation in the spike prefusion state is completely disordered. In contrast, it becomes α-helical in lipid micelles [55], [72], [75], whose curved shapes may mimic the highly curved bilayer conformation of the fusion pore state.
Interestingly, the sequence of the most membrane-active and fusogenic cIFP peptide displays several consecutive GxxxG-like motifs (G can be substituted by A or S; X, any residue), which are also observed in the IFP sequence of the SARS-CoV-2 S protein (Fig. 1) and the corresponding IFP sequences from spike proteins within the Coronaviridae family [28]. Moreover, the presence of GxxxG-like motifs seems to play critical roles in the structure and function of class I fusion peptides such as those from influenza and HIV-I [75], [76]. Mutations of the glycine, alanine or serine residues from GxxxG-like motifs of the IFP segment of the MERS-CoV S protein completely abrogated cell-cell fusion, whereas Gly-to-Ala substitutions only mildly affected cell-cell fusion [28]. The importance of such motif lies in the fact that it facilitates interhelical interactions in membranes [77], [78], such as those taking place between fusion peptides, promoting FP dimerization, or between FPs and transmembrane domains (TMD, Fig. 1) or FPs and membrane-proximal external regions (MPER), promoting FP-TMD or FP-MPER associations [19], [79], [80]. Our CD data obtained with the complete internal FP indicated a membrane-triggered cIFPH7 homoassociation in the form of α-helical coiled coils. It has also been shown that a 23-residue long IFP (IFP23, 866–888 sequence range) encompassing GxxxG-like motifs heteromerizes with the SARS-CoV spike MPER, forming a quaternary structure that might facilitate the stabilization of the hemifusion state and the formation of the fusion pore [26], [73]. Therefore, since the IFP23 primary structure overlaps with the cIFP C-terminal half, this study further highlights the importance of the complete internal fusion peptide C-terminus. Taken together, our results, along with data from the literature, evidence the functional relevance of both C- and N-termini halves of the complete internal fusion peptide both in the early and late stages of the SARS-CoV membrane fusion process.
Our studies with ghost and erythrocyte membrane models showed that the interaction of the membrane fusion peptides with biological membranes was much more complex than in lipid model membranes since the erythrocyte structural and membrane proteins were also perturbed, as shown by our DSC assays (Fig. 7B). Therefore, different from what has been found for HIV-1 and influenza FPs [63], [64], [65], a direct correlation between the membranotropic activity of the SARS-CoV fusion peptides and their biological activities was not possible, indicating a more complex behavior. For instance, FP2H7 promoted as many multicellular aggregates as cIFPH7 but exhibited substantially less fusion and vesicle aggregation. On the other hand, FP1H7 induced the highest hemolytic activity while displayed intermediate permeabilizing capacity. Interestingly, though, all peptides seemed to play complementary biological activities since those that promoted extensive hemagglutination disturbed erythrocyte integrity.
Similarly, the peptide which perturbed most the thermal stability of the anion-transporting domain of the band 3 protein (corresponding to the peak III in the DSC thermogram, Fig. 7B) caused only a marginal disturbance in the denaturation of the ghost membrane proteins comprising peaks I and II. Whether the peptides interacted directly with the ghost proteins or indirectly perturbed them via interaction with the ghost membrane bilayer was not in the scope of the present work and will require further investigation. Regardless, it is remarkable that synthetic membrane peptides corresponding to fusion domains of viral fusion proteins can promote high levels of aggregation of biological membranes and perturb the thermal stability of the membrane proteins of such cell models [63], [64], [65].
To summarize, here we showed that a 38-residue long internal fusion peptide sequence from the SARS-CoV spike glycoprotein displayed higher membranotropic and biological activities than the N-proximal FP sequences under the experimental conditions tested. We hypothesize that all membrane-active FP sequences from SARS-CoV S protein, and likely those from SARS-CoV-2 given the degree of sequence conservation (Fig. 1), play complementary roles in the fusion process. Depending on physiological conditions such as pH, salt, Ca2+ presence, proteolytic priming, and lipid composition [13], [14], [32], [68], [74], [81], the SARS-CoV membrane fusion peptides may act independently, but likely synergistically [26], in order to promote the necessary membrane remodeling and restructuring critical for the SARS-CoV spike-mediated viral-cell membrane fusion.
5. Conclusion
Using a combined biophysical approach, we investigated the interaction of three fusion peptide sequences from the SARS-CoV spike glycoprotein with model and biological membranes. Our findings revealed that the internal fusion peptide, cIFP, was more efficient in promoting membrane activity than the other N-proximal fusion peptides at both neutral and acidic pH. This result suggests that cIFP may disturb both cellular and endosome membranes, highlighting its importance to both early and late viral entry pathways. The cIFP formed a coiled-coil α-helical structure in model membranes, putatively due to the presence of several GxxxG-like motifs in its primary structure. The presence of such motifs may also be important to a possible heteroassociation of cIFP with the pretransmembrane domain of the S protein, highlighting the relevance of the internal fusion peptide also to the late stages of viral entry.
Our data also revealed that the fusogenicity of the well-conserved FP2 sequence was ionic strength dependent and that Ca2+ downregulated the FP2 fusion activity. However, FP2 induced extensive aggregation of biological membranes. Our hypothesis is that different hydrophobic segments from the spike protein with characteristics of fusion peptides may act independently from each other and likely play complementary roles in the viral membrane fusion. The possible existence of both N-terminal and internal fusion peptide domains may be advantageous since it may facilitate the contact of the SARS-CoV fusion S2 subunit with the host membrane, thus triggering more efficiently the necessary fusion-relevant membrane changes of distinct cell types under different physiological conditions such as pH, salt, Ca2+, and lipid composition.
CRediT authorship contribution statement
L.G.M.B., A.E.Z, and A.J.C.F. conceived and designed experimental work. L.G.M.B., A.E.Z., and A.P.F. performed the experiments. L.G.M.B. and A.J.C.F. wrote the paper. All authors reviewed the manuscript.
Declaration of competing interest
ANTONIO J C FILHO reports financial support and equipment, drugs, or supplies were provided by State of Sao Paulo Research Foundation.
Acknowledgments
The authors acknowledge the Brazilian agencies FAPESP (Grant no. 2008/57910-0, 2010/17668-2, 2015/18390-5, 2015/50366-7), CNPq (Grant no. 573607/2008-7, 306682/2018-4), and CAPES for financially supporting this work. L.G.M.B. and A.E.Z. acknowledge FAPESP for post-doctoral fellowships (Grants No. 2014/00206-0 and 2013/207154). A.P.F. acknowledges CAPES for the scholarship (Grant code: 001). The authors would also like to thank the “Sérgio Mascarenhas” Biophysics and Structural Biology Group at Sao Carlos Physics Institute of the University of Sao Paulo for allowing access to the biophysical facility.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbamem.2021.183697.
Appendix A. Supplementary data
Supplementary material
Data availability
Data will be made available on request.
References
- 1.Hamre D., Procknow J.J. A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- 2.McIntosh K., Dees J.H., Becker W.B., Kapikian A.Z., Chanock R.M. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc. Natl. Acad. Sci. U. S. A. 1967;57:933–940. doi: 10.1073/pnas.57.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Bankamp B., Maher K., Chen M., Tong S., Tamin A., Lowe L., Frace M., Derisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C.T., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., Mccaustland K., Olsen-rasmussen M., Fouchier R., Osterhaus A.D.M.E., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science (80- ) 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 4.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/nejmoa1211721. [DOI] [PubMed] [Google Scholar]
- 5.C.N.C.I. and R. Team. Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang T., Bidon M., Jaimes J.A., Whittaker G.R., Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antivir. Res. 2020;178 doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrison S.C. Viral membrane fusion. Nat. Struct. Mol. Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sainz B., Rausch J.M., Gallaher W.R., Garry R.F., Wimley W.C. Identification and characterization of the putative fusion peptide of the severe acute respiratory syndrome-associated coronavirus spike protein. J. Virol. 2005;79:7195–7206. doi: 10.1128/JVI.79.11.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guillén J., Pérez-Berná A.J., Moreno M.R., Villalaín J. Identification of the membrane-active regions of the severe acute respiratory syndrome coronavirus spike membrane glycoprotein using a 16 / 18-mer peptide scan: implications for the viral fusion mechanism. J. Virol. 2005;79:1743–1752. doi: 10.1128/JVI.79.3.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guille J., Moreno M.R., Pe A.J., Bernabeu A. Interaction of a peptide from the pre-transmembrane domain of the severe acute respiratory syndrome coronavirus spike protein with phospholipid membranes. J. Phys. Chem. B. 2007;111:13714–13725. doi: 10.1021/jp073675y. [DOI] [PubMed] [Google Scholar]
- 12.Guillén J., De Almeida R.F.M., Prieto M., Villalaín J. Interaction of a peptide corresponding to the loop domain of the S2 SARS-CoV virus protein with model membranes. Mol. Membr. Biol. 2009;26:236–248. doi: 10.1080/09687680902926203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guillén J., De Almeida R.F.M., Prieto M., Villalaín J. Structural and dynamic characterization of the interaction of the putative fusion peptide of the S2 SARS-CoV virus protein with lipid membranes. J. Phys. Chem. B. 2008;112:6997–7007. doi: 10.1021/jp7118229. [DOI] [PubMed] [Google Scholar]
- 14.Guillén J., Pérez-Berná A.J., Moreno M.R., Villalaín J. A second SARS-CoV S2 glycoprotein internal membrane-active peptide. Biophysical characterization and membrane interaction. Biochemistry. 2008;47:8214–8224. doi: 10.1021/bi800814q. [DOI] [PubMed] [Google Scholar]
- 15.Basso L.G.M., Vicente E.F., Crusca E., Cilli E.M., Costa-Filho A.J. SARS-CoV fusion peptides induce membrane surface ordering and curvature. Sci. Rep. 2016;6:37131. doi: 10.1038/srep37131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai A.L., Freed J.H. HIV gp41 fusion peptide increases membrane ordering in a cholesterol-dependent fashion. Biophys. J. 2014;106:172–181. doi: 10.1016/j.bpj.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai A.L., Millet J.K., Daniel S., Freed J.H., Whittaker G.R. The SARS-CoV fusion peptide forms an extended bipartite fusion platform that perturbs membrane order in a calcium- dependent manner. J. Mol. Biol. 2017;429:3875–3892. doi: 10.1016/j.jmb.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ge M., Freed J.H. Fusion peptide from influenza hemagglutinin increases membrane surface order: an electron-spin resonance study. Biophys. J. 2009;96:4925–4934. doi: 10.1016/j.bpj.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donald J.E., Zhang Y., Fiorin G., Carnevale V., Slochower D.R., Gai F., Klein M.L., DeGrado W.F. Transmembrane orientation and possible role of the fusogenic peptide from parainfluenza virus 5 (PIV5) in promoting fusion. Proc. Natl. Acad. Sci. U. S. A. 2011;108:3958–3963. doi: 10.1073/pnas.1019668108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao H., Hong M. Conformation and lipid interaction of the fusion peptide of the paramyxovirus PIV5 in anionic and negative-curvature membranes from solid-state NMR. J. Am. Chem. Soc. 2014;136:2611–2624. doi: 10.1021/ja4121956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roche S., Bressanelli S., Rey F.A., Gaudin Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science (80-. ) 2006;313:187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- 22.Heldwein E.E., Lou H., Bender F.C., Cohen G.H., Eisenberg R.J., Harrison S.C. Crystal structure of glycoprotein B from herpes simplex virus 1. Science (80-.) 2006;313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- 23.Madu I.G., Roth S.L., Belouzard S., Whittaker G.R. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J. Virol. 2009;83:7411–7421. doi: 10.1128/JVI.00079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petit C.M., Melancon J.M., Chouljenko V.N., Colgrove R., Farzan M., Knipe D.M., Kousoulas K.G. Genetic analysis of the SARS-coronavirus spike glycoprotein functional domains involved in cell-surface expression and cell-to-cell fusion. Virology. 2005;341:215–230. doi: 10.1016/j.virol.2005.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guillén J., Kinnunen P.K.J., Villalaín J. Membrane insertion of the three main membranotropic sequences from SARS-CoV S2 glycoprotein. Biochim. Biophys. Acta Biomembr. 2008;1778:2765–2774. doi: 10.1016/j.bbamem.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bosch B.J., Martina B.E.E., Van Der Zee R., Lepault J., Haijema B.J., Versluis C., Heck A.J.R., De Groot R., Osterhaus A.D.M.E., Rottier P.J.M. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. U. S. A. 2004;101:8455–8460. doi: 10.1073/pnas.0400576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ou X., Zheng W., Shan Y., Mu Z., Dominguez S.R., Holmes K.V., Qian Z. Identification of the fusion peptide-containing region in betacoronavirus spike glycoproteins. J. Virol. 2016;90:5586–5600. doi: 10.1128/JVI.00015-16.Editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin I., Ruysschaert J. Common properties of fusion peptides from diverse systems. Biosci. Rep. 2000;20:483–500. doi: 10.1023/A:1010454803579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delos S.E., Gilbert J.M., White J.M. The central proline of an internal viral fusion peptide serves two important roles. J. Virol. 2000;74:1686–1693. doi: 10.1128/JVI.74.4.1686-1693.2000.Updated. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gregory S.M., Harada E., Liang B., Delos S.E., White J.M., Tamm L.K. Structure and function of the complete internal fusion loop from ebolavirus glycoprotein 2. Proc. Natl. Acad. Sci. U. S. A. 2011;108:11211–11216. doi: 10.1073/pnas.1104760108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pattnaik G.P., Bhattacharjya S., Chakraborty H. Enhanced cholesterol-dependent hemifusion by internal fusion peptide 1 of SARS Coronavirus-2 compared to its N-terminal counterpart. Biochemistry. 2021;60:559–562. doi: 10.1021/acs.biochem.1c00046. [DOI] [PubMed] [Google Scholar]
- 33.Barroso R.P., Basso L.G.M., Costa-Filho A.J. Interactions of the antimalarial amodiaquine with lipid model membranes. Chem. Phys. Lipids. 2015;186:68–78. doi: 10.1016/j.chemphyslip.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 34.Sastre D.E., Basso L.G.M., Trastoy B., Cifuente J.O., Contreras X., Gueiros-Filho F., de Mendoza D., Navarro M.V.A.S., Guerin M.E. Membrane fluidity adjusts the insertion of the transacylase PlsX to regulate phospholipid biosynthesis in gram-positive bacteria. J. Biol. Chem. 2020;295:2136–2147. doi: 10.1074/jbc.RA119.011122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Felizatti A.P., Zeraik A.E., Basso L.G.M., Kumagai P.S., Lopes J.L.S., Wallace B.A., Araujo A.P.U., DeMarco R. Interactions of amphipathic a-helical MEG proteins from Schistosoma mansoni with membranes. Biochim. Biophys. Acta Biomembr. 2020;1862 doi: 10.1016/j.bbamem.2019.183173. [DOI] [PubMed] [Google Scholar]
- 36.Dyszy F., Pinto A.P.A., Araújo A.P.U., Costa-Filho A.J. Probing the interaction of brain fatty acid binding protein (B-FABP) with model membranes. PLoS One. 2013;8 doi: 10.1371/journal.pone.0060198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Couto S.G., Cristina Nonato M., Costa-Filho A.J. Defects in vesicle core induced by escherichia coli dihydroorotate dehydrogenase. Biophys. J. 2008;94:1746–1753. doi: 10.1529/biophysj.107.120055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vieira E.D., Basso L.G.M., Costa-filho A.J. Non-linear van’t hoff behavior in pulmonary surfactant model membranes. Biochim. Biophys. Acta Biomembr. 1859;2017:1133–1143. doi: 10.1016/j.bbamem.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 39.Nathan L., Lai A.L., Millet J.K., Straus M.R., Freed J.H., Whittaker G.R., Daniel S. Calcium ions directly interact with the ebola virus fusion peptide to promote structure-function changes that enhance infection. ACS Infect. Dis. 2020;6:250–260. doi: 10.1021/acsinfecdis.9b00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Straus M.R., Tang T., Lai A.L., Flegel A., Bidon M., Freed J.H., Daniel S., Whittaker G.R. Ca2 ions promote fusion of Middle East respiratory syndrome coronavirus with host cells and increase infectivity. J. Virol. 2020;94 doi: 10.1128/JVI.00426-20. (e00426-20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pinello J.F., Lai A.L., Millet J.K., Cassidy-Hanley D., Freed J.H., Clark T.G. Structure-function studies link class II viral fusogens with the ancestral gamete fusion protein HAP2. Curr. Biol. 2017;27:651–660. doi: 10.1016/j.cub.2017.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lai A.L., Freed J.H. SARS-CoV-2 fusion peptide has a greater membrane perturbating effect than SARS-CoV with highly specific dependence on Ca2. J. Mol. Biol. 2021;433 doi: 10.1016/j.jmb.2021.166946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lai A.L., Park H., White J.M., Tamm L.K. Fusion peptide of influenza hemagglutinin requires a fixed angle boomerang structure for activity. J. Biol. Chem. 2006;281:5760–5770. doi: 10.1074/jbc.M512280200. [DOI] [PubMed] [Google Scholar]
- 44.Han X., Tamm L.K. A host-guest system to study structure-function relationships of membrane fusion peptides. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13097–13102. doi: 10.1073/pnas.230212097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crusca E., Basso L.G.M., Altei W.F., Marchetto R. Biophysical characterization and antitumor activity of synthetic pantinin peptides from scorpion's venom. Biochim. Biophys. Acta Biomembr. 1860;2018:2155–2165. doi: 10.1016/j.bbamem.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 46.McClare C.W.F. An accurate and convenient organic phosphorus assay. Anal. Biochem. 1971;39:527–530. doi: 10.1016/0003-2697(71)90443-X. [DOI] [PubMed] [Google Scholar]
- 47.Struck D.K., Hoekstra D., Pagano R.E. Use of resonance energy transfer to monitor membrane fusion. Biochemistry. 1981;20:4093–4099. doi: 10.1021/bi00517a023. [DOI] [PubMed] [Google Scholar]
- 48.Budil D.E., Lee S., Saxena S., Freed J.H. Nonlinear-least-squares analysis of slow-motion EPR spectra in one and two dimensions using a modified Levenberg–Marquardt algorithm. J. Magn. Reson. Ser. A. 1996;120:155–189. [Google Scholar]
- 49.C. Altenbach , LabVIEW programs for the analysis of EPR data, (n.d.). http://www.biochemistry.ucla.edu/biochem/Faculty/Hubbell/.
- 50.Basso L.G.M., Rodrigues R.Z., Naal R.M.Z.G., Costa-filho A.J. Effects of the antimalarial drug primaquine on the dynamic structure of lipid model membranes. Biochim. Biophys. Acta Biomembr. 1808;2011:55–64. doi: 10.1016/j.bbamem.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 51.Lau W.L., Ege D.S., Lear J.D., Hammer D.A., Degrado W.F. Oligomerization of fusogenic peptides promotes membrane fusion by enhancing membrane destabilization. Biophys. J. 2004;86:272–284. doi: 10.1016/S0006-3495(04)74103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nieva L., Agirre A. Are fusion peptides a good model to study viral cell fusion? Biochim. Biophys. Acta. 2003;1614:104–115. doi: 10.1016/S0005-2736(03)00168-8. [DOI] [PubMed] [Google Scholar]
- 53.Khelashvili G., Plante A., Doktorova M., Weinstein H. Ca2 -dependent mechanism of membrane insertion and destabilization by the SARS-CoV-2 fusion peptide. Biophys. J. 2021;120:1105–1119. doi: 10.1016/j.bpj.2021.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suárez T., Gómara M.J., Goñi F.M., Mingarro I., Muga A., Pérez-Payá E., Nieva J.L. Calcium-dependent conformational changes of membrane-bound ebola fusion peptide drive vesicle fusion. FEBS Lett. 2003;535:23–28. doi: 10.1016/S0014-5793(02)03847-4. [DOI] [PubMed] [Google Scholar]
- 55.Han X., Bushweller J.H., Cafiso D.S., Tamm L.K. Membrane structure and fusion-triggering conformational change of the fusion domain from influenza hemagglutinin. Nat. Struct. Biol. 2001;8:715–720. doi: 10.1038/90434. [DOI] [PubMed] [Google Scholar]
- 56.Kielian M. Mechanisms of virus membrane fusion proteins. Annu. Rev. Virol. 2014;1:171–189. doi: 10.1146/annurev-virology-031413-085521. [DOI] [PubMed] [Google Scholar]
- 57.Oliver R.C., Lipfert J., Fox D.A., Lo R.H., Kim J.J., Doniach S., Columbus L. Tuning micelle dimensions and properties with binary surfactant mixtures. Langmuir. 2014;30:13353–13361. doi: 10.1021/la503458n. [DOI] [PubMed] [Google Scholar]
- 58.Lau S.Y.M., Taneja A.K., Hodges R.S. Synthesis of a model protein of defined secondary and quaternary structure. effect of chain length on the stabilization and formation of two-stranded alpha-helical coiled-coils. J. Biol. Chem. 1984;259:13253–13261. doi: 10.1016/S0021-9258(18)90686-1. [DOI] [PubMed] [Google Scholar]
- 59.Choy N., Raussens V., Narayanaswami V. Inter-molecular coiled-coil formation in human apolipoprotein E C-terminal domain. J. Mol. Biol. 2003;334:527–539. doi: 10.1016/j.jmb.2003.09.059. [DOI] [PubMed] [Google Scholar]
- 60.Basso L.G.M., Mendes L.F.S., Costa-Filho A.J. The two sides of a lipid-protein story. Biophys. Rev. 2016;8:171–191. doi: 10.1007/s12551-016-0199-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lai A.L., Freed J.H. The interaction between influenza HA fusion peptide and transmembrane domain affects membrane structure. Biophys. J. 2015;109:2523–2536. doi: 10.1016/j.bpj.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rozenfeld J.H.K., Duarte E.L., Oliveira T.R., Lamy M.T. Structural insights on biologically relevant cationic membranes by ESR spectroscopy. Biophys. Rev. 2017;9:633–647. doi: 10.1007/s12551-017-0304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mobley P.W., Lee H.F., Curtain C.C., Kirkpatrick A., Waring A.J., Gordon L.M. The amino-terminal peptide of HIV-1 glycoprotein 41 fuses human erythrocytes. Biochim. Biophys. Acta Mol. basis Dis. 1995;1271:304–314. doi: 10.1016/0925-4439(95)00048-9. [DOI] [PubMed] [Google Scholar]
- 64.Mobley P.W., Waring A.J., Sherman M.A., Gordon L.M. Membrane interactions of the synthetic N-terminal peptide of HIV-1 gp41 and its structural analogs. Biochim. Biophys. Acta. 1999;1418:1–18. doi: 10.1016/s0005-2736(99)00014-0. [DOI] [PubMed] [Google Scholar]
- 65.Wharton S.A., Martin S.R., Ruigrok R.W., Skehel J.J., Wiley D.C. Membrane fusion by peptide analogues of influenza virus haemagglutinin. J. Gen. Virol. 1988;69:1847–1857. doi: 10.1099/0022-1317-69-8-1847. [DOI] [PubMed] [Google Scholar]
- 66.Hernández-Hernández A., Rodríguez M.C., López-Revuelta A., Sánchez-Gallego J.I., Shnyrov V., Llanillo M., Sánchez-Yagüe J. Alterations in erythrocyte membrane protein composition in advanced non-small cell lung cancer. Blood Cells Mol. Dis. 2006;36:355–363. doi: 10.1016/j.bcmd.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 67.Apellániz B., Huarte N., Largo E., Nieva J.L. The three lives of viral fusion peptides. Chem. Phys. Lipids. 2014;181:40–55. doi: 10.1016/j.chemphyslip.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang S.T., Kiessling V., Simmons J.A., White J.M., Tamm L.K. HIV gp41-mediated membrane fusion occurs at edges of cholesterol-rich lipid domains. Nat. Chem. Biol. 2015;11:424–431. doi: 10.1038/nchembio.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ge M., Freed J.H. Hydration, structure, and molecular interactions in the headgroup region of dioleoylphosphatidylcholine bilayers: an electron spin resonance study. Biophys. J. 2003;85:4023–4040. doi: 10.1016/S0006-3495(03)74816-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dubé M., Etienne L., Fels M., Kielian M. Calcium-dependent rubella virus fusion occurs in early endosomes. J. Virol. 2016;90:6303–6313. doi: 10.1128/JVI.00634-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Basso L.G.M., Park S.H., Costa-Filho A.J., Opella S.J. Structures, dynamics, and functions of viral membrane proteins by NMR. Biophys. J. 2020;114:237a. doi: 10.1016/j.bpj.2017.11.1319. [DOI] [Google Scholar]
- 72.Mahajan M., Bhattacharjya S. NMR structures and localization of the potential fusion peptides and the pre-transmembrane region of SARS-CoV: implications in membrane fusion. Biochim. Biophys. Acta Biomembr. 1848;2015:721–730. doi: 10.1016/j.bbamem.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liao Y., Zhang S.M., Neo T.L., Tam J.P. Tryptophan-dependent membrane interaction and heteromerization with the internal fusion peptide by the membrane proximal external region of SARS-CoV spike protein. Biochemistry. 2015;54:1819–1830. doi: 10.1021/bi501352u. [DOI] [PubMed] [Google Scholar]
- 74.Meher G., Bhattacharjya S., Chakraborty H. Membrane cholesterol modulates oligomeric status and peptide-membrane interaction of severe acute respiratory syndrome coronavirus fusion peptide. J. Phys. Chem. B. 2019;123:10654–10662. doi: 10.1021/acs.jpcb.9b08455. [DOI] [PubMed] [Google Scholar]
- 75.Lorieau J.L., Louis J.M., Bax A. The complete influenza hemagglutinin fusion domain adopts a tight helical hairpin arrangement at the lipid: water interface. Proc. Natl. Acad. Sci. U. S. A. 2010;107:11341–11346. doi: 10.1073/pnas.1006142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Faingold O., Cohen T., Shai Y. A GxxxG-like motif within HIV-1 fusion peptide is critical to its immunosuppressant activity, structure, and interaction with the transmembrane domain of the T-cell receptor. J. Biol. Chem. 2012;287:33503–33511. doi: 10.1074/jbc.M112.370817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Russ W.P., Engelman D.M. The GxxxG motif: a framework for transmembrane helix-helix association. J. Mol. Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 78.Kleiger G., Grothe R., Mallick P., Eisenberg D. GXXXG and AXXXA : common R -helical interaction motifs in proteins, particularly in extremophiles †. Biochemistry. 2002;41:5990–5997. doi: 10.1021/bi0200763. [DOI] [PubMed] [Google Scholar]
- 79.Reuven E.M., Dadon Y., Viard M., Manukovsky N., Blumenthal R., Shai Y. HIV-1 gp41 transmembrane domain interacts with the fusion peptide: implication in lipid mixing and inhibition of Virus-Cell fusion. Biochemistry. 2012;51:2867–2878. doi: 10.1021/bi201721r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee J., Nyenhuis D.A., Nelson E.A., Cafiso D.S., White J.M., Tamm L.K. Structure of the ebola virus envelope protein MPER/TM domain and its interaction with the fusion loop explains their fusion activity. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E7987–E7996. doi: 10.1073/pnas.1708052114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meher G., Chakraborty H. Membrane composition modulates fusion by altering membrane properties and fusion peptide structure. J. Membr. Biol. 2019;252:261–272. doi: 10.1007/s00232-019-00064-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available on request.