Abstract
The fields of application of functional proteomics are not limited to the study of protein–protein interactions; they also extend to those involving protein complexes that bind DNA or RNA. These interactions affect fundamental processes such as replication, transcription, and repair in the case of DNA, as well as transport, translation, splicing, and silencing in the case of RNA. Analytical or preparative experimental approaches, both in vivo and in vitro, have been developed to isolate and identify DNA/RNA binding proteins by exploiting the advantage of the affinity shown by these proteins toward a specific oligonucleotide sequence. The present review proposes an overview of the approaches most commonly employed in proteomics applications for the identification of nucleic acid-binding proteins, such as affinity purification (AP) protocols, EMSA, chromatin purification methods, and CRISPR-based chromatin affinity purification, which are generally associated with mass spectrometry methodologies for the unbiased protein identification.
Keywords: proteomics, mass spectrometry, DNA−protein interactions, RNA−protein interactions, pull-down, EMSA, ChIP, CRISPR-Cas9
1. Introduction
“Life is a relationship between molecules, not a property of any one molecule. So is, therefore, disease, which endangers life,” wrote Zuckerkandl and Pauling1,2 in their chapter on “Molecular disease, evolution and genic heterogeneity”. Several years have passed, and the molecular mechanisms underlying many diseases and the interactions between molecules in healthy and diseased organisms are still poorly understood and unclear. Therefore, we are still very far from understanding and elucidating the complex network of interactions taking place in living organisms.3 In the cell, biological functions are not exerted by single individual proteins, but by transient complexes they form, interacting each other or with other molecules such as nucleic acids4−7 and metabolites.8,9 Thus, the study of proteins and their interactions is essential to understand their roles within the cell and to elucidate the organization of functional networks.
A complete description of cellular processes is then strictly dependent on a clear definition of the complexes that take part in the molecular mechanisms and the individual protein components involved in these functional entities. The association of an unknown protein with partners belonging to a specific complex involved in a particular mechanism might strongly suggest its biological function.10 Furthermore, a detailed description of cell signaling pathways could greatly benefit from the elucidation of interactions in vivo.11
Modern functional proteomic studies are not solely addressed to the study of protein–protein interactions but also to the investigation of the interactions between multiprotein complexes and nucleic acids, thus to define both the biological functions of specific proteins and their influence on nucleic acids dependent events.12 It is well-known that cell processes involving DNA (i.e., replication, transcription, processing, repair, specific package, DNA rearrangement, etc.) as well as RNAs (splicing, transport, translation, silencing, etc.) are the result of the constant interaction between functional nucleic acids and specific proteins.13
The evolution selected two different ways of protein–DNA binding: a nonspecific manner, as occurring in histones–DNA interaction, and a very selective and specific mode, in which the protein recognition site is strictly nucleotide sequence-dependent.14 The same modalities belong also to RNA–protein interactions, as amply illustrated by Guenther et al. in their work, where the meaning of “specific” and “non-specific” of RNA–protein interactions and their complexity is well illustrated. The authors highlight the presence of specific regions of RNA (RNA Binding Domain) that exclusively bind classes of “specific” proteins as well as proteins that perform as many important functions although in absence of specific binding capabilities.15,16 Both binding modes at the level of DNA affect gene expression: the former through epigenetic modifications, the latter through the recognition and binding of protein factors on specific nucleotide stretch (often consisting of palindromic sequences of at least 12 nucleotides).17−20
The impairment of either process, the modulation of histones modifications, or the recruitment time- and space-specific of protein complexes on their nucleotide-binding site, is often involved in the onset of particularly serious pathologies.21−23 Already in 1940, the biologist Conrad Hal Waddington, introducing the term epigenetics, emphasized the importance of the interactions of genes with the surrounding environment for the understanding of a specific phenotype. Hence the need to elucidate the mechanisms underlying these complex phenomena and the development of techniques to be applied for their investigation.24,25 Therefore, in these past years, we have assisted in the implementation of a large number of analytical methods for studying both the profiling of epigenetics modification in different conditions (i.e., physiological vs pathological)26−29 and the isolation and identifications of specific DNA/RNA binding protein complexes.30−33
The highest number of experimental approaches, either analytical or preparative, both in vivo and in vitro, were developed to isolate and identify DNA/RNA binding protein complexes by tacking advantage from their affinity toward a specific oligonucleotide sequence. In proteomics experimental workflows, these affinity-based isolation methods are usually coupled to advanced mass spectrometry (MS) methodologies, for protein identification.34,35
Nowadays, many strategies have been borrowed from molecular biology protocols and then improved for high-throughput -omics approaches. Many of them rely on in vitro investigations, such as affinity purification and electrophoresis mobility shift assay (EMSA). Conversely, chromatin immunoprecipitation (ChIP) based protocols aim for the isolation and identification of both protein partners and nucleic acid targets of a specific protein “bait” in ex vivo approaches. Finally, newborn techniques based on CRISPR-Cas9 technology have been set up for the isolation and identification of proteins interacting with a specific genomic locus in vivo. For each mentioned strategy, several variants have been finely tuned to respond to specific biological problems and to be successfully applied for RNA binding protein investigation fields, too. This review provides an excursus on the main strategies developed in the field of protein–nucleic acid interactions: starting from classical biochemical and/or molecular biological approaches, we focused mainly on those, which coupled with mass spectrometry methodologies, have found the largest applications in the field of -omics sciences. For each approach, the points of strength and drawbacks will be critically treated.
2. Probing DNA/RNA–Protein Interactions by in Vitro Affinity Procedures
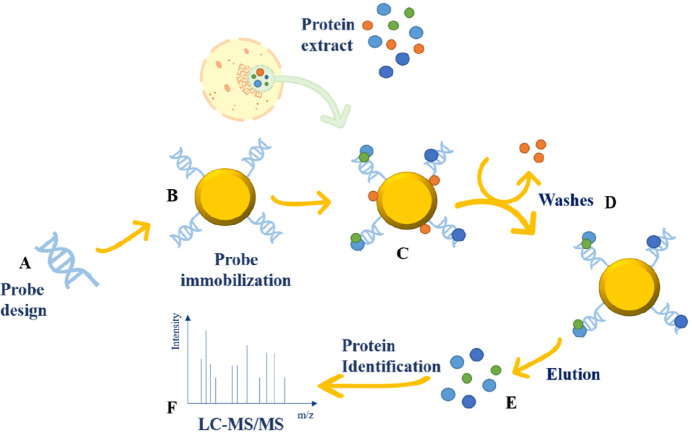
The unbiased procedure defined as affinity purification–mass spectrometry (AP-MS) is based on the combination of affinity isolation strategies with MS procedures and constitutes an important branch of the functional proteomics approach.36−38 The AP-MS procedure exploits the intrinsic affinity of DNA/RNA binding proteins for a specific oligonucleotide sequence which is used as bait to fish the protein partners out from the cellular extract. In a generic protocol, the protein extract is incubated with the oligonucleotide bait immobilized onto insoluble support. Following several washes proteins, specifically retained by the bait, are eluted and identified by mass spectrometry (Figure 1).
Figure 1.
Schematic representation of DNA-pull-down assay workflow. (A) Probe design and synthesis. (B) Chemical or affinity immobilization of the probe on an insoluble solid support. (C) Incubation of nuclear extract with specific oligonucleotide for partners isolation. (D) Washings to remove unspecific proteins. (E) Elution of oligonucleotide interacting proteins. (F) Protein identification by mass spectrometry-based approaches.
The crucial point in the entire procedure relies on the design of the nucleotide fragment. The length of the probe is chosen according to the number of nucleotides composing the sequence of interest. An optimal bait length is difficult to fix. It would strictly depend on the aim of the study in terms of how narrow the fishing has to be carried out. Generally, longer probes that also include sequences outside the binding region might increase the retention of unspecific proteins lacking the affinity for the consensus sequence. Nevertheless, a longer probe including different regulatory regions can contribute to a cooperative and more stable interaction between DNA/RNA and large multiprotein complexes.38−40 On the contrary, too-short probes might affect the stability of protein binding. In order to improve the interaction of the bait with transcriptional factors, which generally are under-expressed and only transiently bind their nucleic acid targets, the oligonucleotide consensus sequence can be multimerized to provide multiple copies of the specific sequence thus increasing the number of potential interacting regions onto the same bait.41 In addition, for both DNA and more frequently for RNA probes, the establishment of secondary stable structures have to be considered. Finally, in the design of the oligonucleotide target, the presence of a spacer sequence to properly outdistance the solid support and the binding site of interest, reducing the steric hindrance, has also to be taken into account.42 Several procedures have been explored to bind the oligonucleotide bait to the polymeric support, with two methods being the most successful. The nucleotide sequence can be covalently bound by chemical cross-links on a suitably derivatized resin;43 alternatively, the oligonucleotide bait can be labeled with biotin and immobilized on streptavidin-coated beads.44−46 Binding buffer composition also exerts a critical role in favoring specific DNA–protein interactions. Physicochemical characteristics including pH value, the concentration of mono- and bivalent ions, the addition of specific DNA competitors to reduce nonspecific binding (poly dI, AD, etc.), and finally the amount of glycerol have to be strictly controlled.47
The occurrence of nonspecific interactions constitutes a not negligible drawback, which must be faced using the following precautions: (i) stringent washes using at high salt concentrations, (ii) oligonucleotide competitors, like salmon sperm, lacking the target sequence and able to capture background proteins,48,49 and (iii) a precleaning phase. The latter relies on a preincubation of protein extract with a not specific oligonucleotide showing the same nucleotide composition of the bait, but modified in specific sites or simply with a random sequence.38 However, the discrimination between the true and false interactors is a longstanding central theme in all applications of functional proteomics. More recently, quantitative mass spectrometry-based approaches relied on label-free,35 stable isotope labeling by amino acids in cell culture (SILAC),50 or tandem mass tag (TMT)51 quantification methodologies have been employed to distinguish random interactions from effective ones. In particular, the false positives are expected to be equally abundant in both conditions, while the true binders will be quantitatively prevalent in the presence of the bait.52 Finally, software such as CRAPome can provide information on proteins more frequently identified as background in each specific affinity purification experimental system as well as in different cell lines, allowing the recognition and, therefore, the elimination of proteins considered false positives.38,53
The “fishing” strategy has found great application in studies addressed to the isolation and identification of nucleic acid-binding proteins involved in fundamental biological processes such as transcription, translation, and/or splicing regulation.54−57 In one of the first application, indeed, Medugno and collaborators identified the interaction between the negative cis-element (AldA-NRE) and ZNF224, a Kruppel-like zinc finger transcription repressor factor as a key step in modulating transcription of the human and mouse aldolase A (AldA) gene during the cell cycle and differentiation.58 Since then, affinity purification strategies have been further developed and largely applied to the investigation of several biological processes involving key DNA–protein interactions leading to the discovery of previously unknown crucial DNA binding proteins. The nature of the regulatory complex bound to the proximal promoter region to regulate EPHX1 expression was explored using a biotinylated oligonucleotide encompassing this region in conjunction with mass spectrometric analysis. A 4-component regulatory complex including the inhibitory factor PSF, CAR, RXR and HNF-4α was identified.59
Nasrullah et al. identified tripartite motif-containing protein 25 (TRIM25) as a Caspase-2 mRNA-binding protein in colon carcinoma cells. TRIM25, known to be an E3 ubiquitin ligase, seems to exert important tumorigenic functions, controlling metastatic gene signatures both at the transcriptional and post-transcriptional levels. In particular, although TRIM25 lacks any typical RNA binding domains (RBDs), it can bind and affect the processing and stability of the specific mRNA, acting as a negative regulator of caspase-2 translation. By unveiling this TRIM25 unexpected function, a novel mechanism of drug resistance in human colorectal carcinoma cells associated with the activity of TRIM25 as an inhibitor of chemotherapeutic drug-induced apoptosis was described.60
Analogously to DNA, RNA molecules, interact with proteins to perform a variety of functions in living cells. Pisa et al. employed AP-MS to isolate novel cap-binding factors that might be involved in translational control of specific mRNAs within the growing Drosophila oocyte, by using m7GTP- derivatized Sepharose beads as bait. About 30 putative interactors were identified, including Hsp90 which was able to bind the translational repressor Cup in vitro, suggesting for Cup novel multiple functions during egg chamber development during Drosophila oogenesis.61
RNA pull-down experiments in which the RNA bait was covalently immobilized using adipic acid dehydrazide derivatized beads were applied to the identification of specific ENPP1-3′ UTR binding proteins revealing that N-acetylgalactosaminyltransferase 2 gene (GALNT2) was a novel factor involved in the modulation of ENPP1 expression as well as insulin signaling and action in human liver HepG2 cells.62 The same adipic acid dihydrazide-agarose beads were used to bind in vitro transcribed RNA probes containing either the wild type or mutated sequence to explore the molecular mechanisms underlying splicing defects in the DMD gene. Incubation with a cellular extract followed by mass spectrometry analyses identified proteins that display differential binding affinities for the wild type and mutant RNA probes.63
Slightly different variants of the AP-MS approach were designed to address specific biological problems involving functional RNAs. Ray and collaborators introduced AptA–MS (aptamer affinity–mass spectrometry), a robust strategy involving a specific, high-affinity RNA aptamer against green fluorescent protein (GFP) to identify the interactors of a GFP-tagged protein. This approach led to the identification of molecular chaperones and translation elongation factors that interact with human heat shock factor 1 (HSF1). This technique provides a significant enrichment in terms of sensitivity, in evolutionarily different organisms, and allows identification of PTMs without the need for specific enrichments.64
The identification of the protein partners of bacterial small noncoding RNAs was performed by an optimized affinity chromatography protocol that enables purification of in vivo formed sRNA–protein complexes. The desired sRNAs were tagged with the MS2 aptamer which is affinity-captured by the MS2-MBP protein conjugated to an amylose resin leading to the recovery of the RNA chaperone Hfq associated with the strictly Hfq-dependent AbcR2 trans-sRNA.65
An RNA/DNA hybrid formed by the In-1 transcript and a 5′-biotinylated DNA oligonucleotide complementary to the upstream exon sequence was used to probe HeLa nuclear extracts for spliceosome investigation. The hybrid probe bound with the nuclear proteins was coupled to streptavidin magnetic beads and the retained proteins were identified by mass spectrometry highlighting the occurrence of canonical spliceosome core components belonging to the spliceosomal B-complex.66 Analogously, a sequence-specific biotinylated peptide nucleic acid (PNA)-neamine hybrid targeted to HCV RNA was developed for the in situ capture of cellular and viral factors associated with HCV leading to the identification of both cellular factors including transcriptional regulators, RNA helicases, DEAD-box proteins, and translational regulators and three viral proteins (NS5B, NS5A, and NS3–4a protease-helicase) associated with the viral genome.67
Recently, G-quadruplex has also been used in pull-down experiments. Guanine quadruple helices, or G-quadruplex, are guanine tetramers stacked together forming a helical structure within the DNA/RNA molecule. They spontaneously form in guanine-rich regions of DNA/RNA giving rise to a variety of conformationally different quadruple helices depending on various factors. G-quadruplex motifs are known to be involved in several biological processes including the mechanisms of initiation of DNA replication and the maintenance of genomic stability by interacting with proteins like chaperones and DNA helicases. Santi Mestre-Fos et al. used G-quadruplex bait in pull-down experiments to identify proteins that specifically recognize these structures in rRNA and applied the SILAC approach for protein quantification. They identified several G-quadruplex linked proteins including helicases (DDX3, CNBP, DDX21, DDX17) and heterogeneous nuclear ribonucleoproteins.68
Tatsuo Serikawa and collaborators performing pull-down and SILAC experiments identify and quantify proteins that have an affinity for four different G-quadruplex motifs located in mRNAs of the cancer-related genes Bcl-2, NRAS, MMP16, and ARPC2. Some of the proteins identified by mass spectrometry appear to be involved in processes of the fine-tuning of translation but are also relevant to the regulation of mRNA maturation and may play an important role in tumor biology.33
Although the identification and quantification of DNA/RNA interacting proteins are necessary to understand the biological role of these associations, also the strength of the interactions plays a central role in protein complex characterization. Dissociation constants (Kd) of in vitro one-by-one interactions are usually measured by classic biochemical analyses (i.e., isothermal titration calorimetry, surface plasmon resonance, fluorescence polarization, fluorescence resonance energy transfer). Makowski et al. proposed an innovative assay in which DNA affinity purification was coupled with tandem mass tag (TMT) labeling to measure the apparent KdApp values for the identified interactors in pull-down experiments carried out at different bait concentration.51 They calculated the protein DNA-bounded fraction by comparing the ion signal of each protein in a single pulldown with that recorded in saturation condition of oligonucleotide bait (micromolar concentration). The KdApp values were calculated by plotting DNA concentration and the bound fractions in a Hill-like curve. The curve also allowed to filter out nonspecific interactions, since background proteins would display randomly distributed signal ratios near 1:1 for all titration points in comparison to control pulldown.
3. Electrophoresis Mobility Shift Assay Mass Spectrometry (EMSA-MS)
Since 1981, the electrophoresis mobility shift assay (EMSA) on either polyacrylamide or agarose gel has constituted the most largely employed biochemical procedure to detect in vitro DNA–protein interactions, for the simplicity of the procedure, its low cost, and the speed of execution.
The binding of a specific protein to a stretch of DNA (probe) can easily be verified by monitoring the delay (shift) of the oligonucleotide probe following its binding to proteins in comparison with the free probe on a native electrophoretic gel. Moreover, these experiments can also monitor the formation of multiple component complexes simply by observing the supershift originated in the gel electrophoresis by the addition of protein components one at a time.69 In the beginning, the electrophoresis shift was usually detected either by using P32 radiolabeled oligonucleotide probes70−72 or staining the gel with ethidium bromide.73,74 However, nowadays, these methods have completely been replaced by fluorophores (Cy3, Cy5)75 or biotin tagged nucleotides76 or by the employment of fluorescent dyes.77
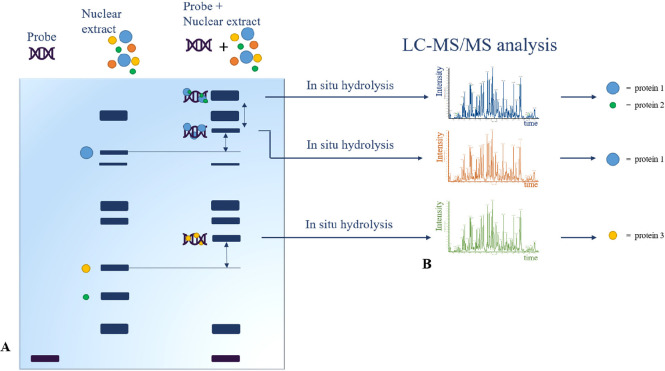
In the EMSA classical approach, a recombinant and purified form of the putative DNA- binding protein is incubated with the probe and the interaction is confirmed by the occurrence of a shift in electrophoretic mobility. Alternatively, the protein extract can be challenged with the oligonucleotide probe, and, in the presence of a mobility shift, a specific antibody is further added to confirm the identity of the DNA binding protein. In both cases, knowledge of the protein under investigation is a crucial prerequisite.78 More recently, an unbiased approach was developed by coupling the classic EMSA assay with advanced mass spectrometry methodology for the identification of DNA-binding proteins (EMSA/MS). The EMSA/MS procedure combined the simplicity and effectiveness of the EMSA experiments with the ability of high sensitivity, high-resolution mass spectrometry to identify all the proteins interacting with the probe in an unbiased operative mode. Following the EMSA experiment, the shifted band containing the probe-protein complex is excised from the gel and the protein components are identified by tryptic digest and nanoLC-MS/MS analysis of the resulting peptide mixture (Figure 2). Although only a tiny amount of material is usually employed in the EMSA assay, identification of probe binding proteins is quite straightforward due to the high sensitivity of modern mass spectrometers.79,80
Figure 2.
Schematic representation of EMSA-MS experiment. (A) Nuclear proteins are incubated with an oligonucleotide probe and bands showing an electrophoretic mobility shift in comparison to the control are in situ hydrolyzed by trypsin and (B) proteins identified by LC-MS/MS approach. Here the control is the nuclear extract alone (second lane).
The focal point of the entire procedure is the choice of the correct negative control to discriminate effective probe binding proteins from false positives, i.e., proteins that display the same electrophoretic mobility of the probe-protein complex. This is especially true when an entire cellular extract is incubated with the probe. A convincing negative control can be obtained by loading the same amount of protein extract on a separate gel lane in the absence of the probe or using an oligonucleotide of the same length and a randomized sequence. For each band excised from the sample lane, an analogous band with the same electrophoretic mobility is picked up from the control lane. Proteins identified both in the sample and the control are discarded.81
Möller and co-workers used the electrophoretic mobility shift assay in combination with mass spectrometry to elucidate the molecular genetic bases of the last unresolved blood group system by identifying proteins able to bind the enhancer region rs311103G of the Xga human allele. Mobility shifts were noted after the addition of a nuclear extract to the oligonucleotide and the individual components in the probe/protein complexes were identified by tandem LC-MS/MS leading to the identification of GATA1 protein.82
Analogously, EMSA/MS was performed to identify proteins binding to the ZNF423 single nucleotide polymorphisms (SNP), a potential biomarker for response to selective estrogen receptor modulators (SERMs) therapy for breast cancer prevention. After EMSA, the shifted gel bands indicating specific DNA–protein interactions were isolated and submitted to mass spectrometry analysis identification. Calmodulin-like protein 3 (CALML3) was identified as a key sensor of this SNP and a coregulator of ERα, which contributes to differential gene transcription regulation in an estrogen and SERM-dependent fashion.83
A slightly different approach was used by Fusco et al. in their investigation on proteins associated with F55, a transcription repressor belonging to S. solfataricus (S.so) spindle-shaped virus 1 (SSV1) when the transcription factor is bound to its specific DNA promoter sequence. When the probe was incubated with the S.so protein extract containing F55, two different delayed bands were clearly detected by fluorescence. Western Blot assay revealed the presence of F55 in only one of the two bands that were in situ digested with trypsin and the proteins identified by a nanoLC-MS/MS-based strategy. Among the putative F55 interactors, RadA, a homologue of E. coliRecA, was identified, suggesting that the archaeal molecular components F55 and RadA are functional homologues of bacteriophage λ (factor CI) and Escherichia coli (RecA) system.84
4. Chromatin Purification Methods Coupled with Mass Spectrometry
Despite a considerable amount of biochemical data obtained with in vitro experimental systems, detailed information about the interactions between transcription factors and their targets in vivo has been obtained following the introduction of chromatin immunoprecipitation (ChIP) technique (Figure 3).85 According to ChIP protocol, the in vivo interactions between transcriptional factors or other DNA binding proteins are stabilized by chemical cross-links. The protein–DNA complexes are then immunoprecipitated using a specific antibody for the protein of interest following sonication. In a classical approach, proteins are completely digested and the immunoprecipitated DNA regions are identified by PCR amplification and/or DNA sequencing (ChIP-seq) (Figure 3G).86 The ChIP procedure combined with the knowledge of the human genome opened incredible new scenarios allowing the researchers to deeply understand how transcriptional factors affect crucial cellular processes through the identification of their target genes.87,47
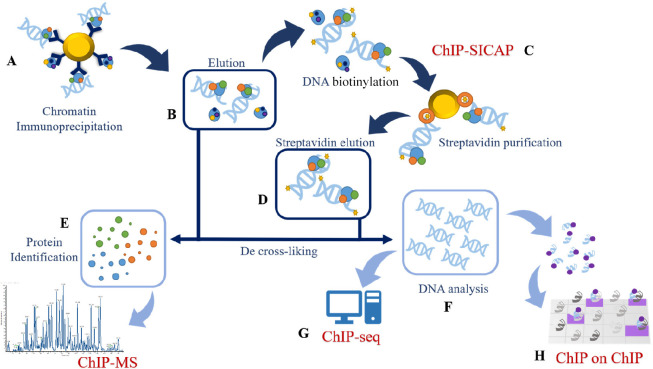
Figure 3.
A generic representation of ChIP experiments. (A) After chromatin immunoprecipitation, (B) DNA–protein complexes are eluted. (C) For only DNA–protein complex isolation, ChIP-SICAP experiment is possible by DNA biotinylation and streptavidin purification, and then (D) elution and de-cross-linking. Finally, protein identification (E) was carried out by mass spectrometry approach (ChIP-MS) and DNA analysis (F), by (G) sequencing (ChIP-seq) or (H) by hybridization with a pull of fluorescence probe (ChIP on ChIP).
As well as for AP-MS, even for a ChIP experiment, a control experiment must be designed. It could consist in the use of the solid support only, called “beads-only” control, which does not involve antibodies and provides solely the nonspecific adsorption due to the beads used for the experiment.88 A more stringent control includes a nonspecific antibody, to be used in a parallel ChIP experiment, as that employed by Collas and collaborators to demonstrate the binding specificity of Oct4 on the NANOG promoter in pluripotent carcinoma cells.88
Since the introduction of the ChIP technique, new upgrades have been developed like the Re-ChIP method, in which two antibodies directed against two different antigens are used, one after the other, allowing the identification of DNA fragments where two protein factors are simultaneously bound;89−92 or the ChIP-on-ChIP variant, in which immunoprecipitated DNA is used as a probe to hybridize a slide containing fragments of genomic DNA (chips) leading to the identification of new targets (Figure 3H). Initially developed in yeast, ChIP-on-ChIP is today successfully applied to human systems.93−96 As mentioned above, the ChIP procedure constituted a fantastic improvement in the investigation of DNA–protein interactions both in vitro and in vivo. However, although this approach was born to be addressed to the identification of DNA target regions where a specific protein factor is bound to, in the last years, the attention was moved also toward the investigation of the additional proteins contemporarily present on the same oligonucleotide sequences. Nowadays, it is well-known that DNA binding proteins are embedded within multiprotein complexes to fulfill their biological role. No information can be provided by the classical ChIP procedures on the other individual components of these functional complexes. Only recently, with the advent of functional proteomics, a fundamental modification of the ChIP procedure, defined ChIP-MS97,98 or rapid immunoprecipitation mass spectrometry of endogenous protein (RIME) has been proposed, with the aim to provide simultaneous identification of the specific DNA target sequences and the protein components of the functional complex bound to those regions.99−101
According to these aims, the ChIP protocol was adapted to allow protein identification. The DNA binding protein complex is allowed to bind its DNA target sequence in vivo and then covalently cross-linked to the DNA stretch. The stabilized complex is immunoprecipitated following the classical ChIP procedure and then either enzymatically digested with trypsin directly on the beads (RIME) or eluted, de-cross-linked, and digested with trypsin (ChIP-MS) (Figure 3E). In both cases, the DNA target is amplified for sequencing, whereas the peptide mixture is directly analyzed by nanoLC-MS/MS to provide protein identification.102
Hwang and co-workers used the chromatin immunoprecipitation procedure coupled to mass spectrometry (ChIP-MS) to identify β-catenin-interacting proteins within a multifunctional protein that might be involved in transcriptional regulation in rat inner medullary collecting duct (IMCD). Several β-catenin-binding proteins were identified, including several known β-catenin-binding partners as well as novel interacting proteins among which Taf1, Jup, Tdrd3, Cdh1, Cenpj, and several histones were involved in transcriptional regulation.103
A large systematic investigation of proteins that bind transcriptional enhancers and promoters in embryonic stem cells was carried out by exploiting chromatin immunoprecipitations (ChIP) by using antibodies for characteristic histone modifications and identification of associated proteins using mass spectrometry.104−107 The ChIP-MS method provided a detailed read-out of the transcriptional landscape representative of the investigated cell type, leading to the identification of several protein factors, most of which drive reprogramming to pluripotent stem cells100 such as Oct4, Esrrb, Klf5, Mycn, and Dppa2.
A functional proteomic experiment based on ChIP-MS was designed to understand the molecular mechanisms that underlie the involvement of CBX7 in cancer progression. CBX7, a component of the polycomb repressive complex (PRC1), can positively or negatively regulate the expression of genes involved in cell proliferation and cancer progression, including E-cadherin. Using the ChIP-MS approach, Federico et al. demonstrated that CBX7 effectively interacts with histone deacetylase 2 (HDAC2)92 and with protein arginine methyltransferase 1 (PRMT1)91 on the E-cadherin promoter. These findings demonstrated that CBX7 activity on E-cadherin promoter is strictly related to several enzymes involved in epigenetic modifications and belonging to both so-called writers (i.e., PMRT1) and erasers (i.e., HDAC2) categories.
The main drawbacks in the ChIP-MS procedure lay in a large number of contaminants due to the possible capture and identification of nonchromatin-associated complexes like those involving proteins or transcription factors that form different complexes on and off chromatin in response to different stimuli. Rafiee et al. proposed a modified version of the ChIP-MS protocol to specifically identify proteins in their DNA-bound state. The method combines ChIP with selective isolation of chromatin-associated proteins (SICAP) (Figure 3C) followed by mass spectrometry to identify chromatin-bound partners of a protein of interest. DNA protein complexes are cross-linked by formaldehyde, and fixed chromatin is immunoprecipitated with a suitable antibody and fragmented by sonication. DNA fragments are then biotinylated and chromatin is retrieved along with interacting proteins on streptavidin beads. Following extensive washing, the cross-link is reversed and proteins are trypsin digested and identified by mass spectrometry.108
The effectiveness of ChIP-SICAP was demonstrated by characterizing the chromatin-bound network around Oct4, Sox2, and Nanog in mouse ESCs, the so-called OSN system, leading to the discovery of Trim24 as a component of the pluripotency network. ChIP-SICAP uniquely benefits from the double purification of protein–DNA complexes, accomplished by subsequent ChIP of the protein of interest and pull-down of biotinylated DNA allowing the exclusive capture of protein complexes bound to DNA.109
In general, ChIP based methods consist of protein-centric approaches since the purification strategies described are tailored according to the identity of the protein of interest that interacts with DNA/RNA. Conversely, alternative DNA/RNA-centric approaches have been developed in order to isolate protein complexes by fishing them from the specific genomic locus. In the proteomics of isolated chromatin segments (PICh)20 strategy, the formaldehyde-cross-linked protein/chromatin complexes are isolated through nucleic acid hybridization and identified by mass spectrometry methodologies. A specific capture probe called locked nucleic acid (LNA), consisting of an RNA nucleotide sequence in which the ribose moiety is modified with an extra bridge connecting the 2′ oxygen and 4′ carbon with increased 3′-exonucleolytic stability and improved hybridization affinity,110 is employed for soluble chromatin hybridization and then for the isolation of the specific genomic locus of interest. The probe is also tagged with a desthiobiotin that allows its affinity purification.
Déjardin et al. developed and used the PICh strategy to purify and analyze protein complexes bound to two distinct types of the telomere, as a proof-of-principle, in three human cell lines: HeLa S3, HeLa 1.2.11, and WI38-VA13 ALT. By designing a specific probe for the PICh experiment and a scrambled probe as control, they were able to isolate the telomeric regions of interest and identified 85% of the proteins known to be associated with them.20
Additional analogous strategies for comprehensive identification of RNA-binding proteins (ChIRP), in capture hybridization analysis (CHART-MS)111,112 or RNA antisense purification (RAP-MS)113 were set up to isolate RNA or ncRNA binding proteins, by employing DNA (for CHART, ChIRP) or transcribing DNA into RNA (for RAP)114 hybridization capture-based approach. The main difference among these techniques consists in probe design: in ChIRP and RAP techniques, a pool of oligonucleotides that cover the full length of the RNA target is used, while in CHART-MS only a few shorter probes are required.114,115In vivo, RNA–protein interactions are chemically cross-linked and purified using biotinylated oligonucleotides complementary to the RNA stretch of interest. Coprecipitated proteins are eluted and identified by mass spectrometry. This approach resulted effective for both abundant housekeeping and relatively low expressed RNAs.116,117 ChIRP-MS analysis allowed the identification of multiple splicing factors in nuclear stress bodies (nSBs) containing long noncoding RNAs, including serine and arginine-rich pre-mRNA splicing factors (SRSFs) which affect splicing patterns.118 The same strategy was employed to explore carcinogenic mechanisms involving long noncoding RNA SNHG6 in colorectal carcinoma onset, leading to the identification of several proteins involved in spliceosomes and mRNA processing.119
CHART-MS strategy was used by West et al. to elucidate genomic binding partners of two human lncRNAs, NEAT1 (nuclear enriched abundant transcript 1) and MALAT1 (metastasis-associated lung adenocarcinoma transcript 1). In this study, they showed that NEAT1 and MALAT1 bind multiple active genes. The two lncRNA, when colocalized on the same DNA region, displayed different protein partners indicating different but synergistic functional roles.111
RAP (RNA antisense purification) is a biochemical method described in 2015 by Engreitz and co-workers120 that enables mapping of RNA interactions with chromatin. As for other several examples reported in this review, in its original version, this approach aimed for the identification of DNA loci interacting with the target RNA by using high-throughput DNA sequencing. Three years later, the same research group proposed an upgrade of the method called RAP-MS, in which the RNA antisense purification was coupled with mass spectrometry for the identification of proteins directly interacting with a specific RNA molecule.113
RAP-MS strategy uses ultraviolet light to cross-link and stabilize only direct protein interactions and is coupled with SILAC protein quantification.121 Wanowska et al. investigate the effect of ENST00000501665.2, OIP5-AS1 (OIP5 Antisense RNA 1) splicing variant, on Opa interacting protein 5 (OIP5) expression with RAP-MS strategy. In HEK293 cells, they demonstrate that ENST00000501665.2 is a positive regulator of OIP5 expression by binding SMARCA4, a component of the SWI/SNF complex and facilitating the interaction between SWI/SNF chromatin remodeling complex and OIP5 promoter.122
5. CRISPR-Based Chromatin Affinity Purification–Mass Spectrometry (CRISPR-ChAP-MS)
Recently, another locus-specific strategy exploits the CRISPR (regularly clustered interspaced palindromic repeats)/dCas9 system has been developed. A specific locus is targeted using nuclease-deficient Cas9 (dCas9) (often labeled with a terminal protein or peptide tag, e.g., FLAG, myc, etc.) in combination with a specific guide RNA (gRNA). Normally, the gRNA is designed to bring the dCas9 upstream to the locus of interest. Then, an experiment resembling chromatin immunoprecipitation is carried out: following the cross-linking and sonication procedure, dCas9 is immunoprecipitate bound to chromatin through the gRNA anchor. De-cross-linked proteins are then identified by mass spectrometry methodologies in a combined approach known as CRISPR-ChAP-MS.123 In the latter, the optimal experimental control is provided by the same cell line transfected with dCas9 in the absence of gRNA.
Waldrip et al. introduced the CRISPR-ChAP-MS approach to isolate the protein complex specifically linked to the GAL1 promoter in yeast under transcriptionally active conditions. Cells were treated with formaldehyde to stabilize protein–DNA interactions, chromatin was sheared to fragments, and the target chromatin region harboring the Gal1 promoter was specifically located using the CRISPR-Cas9 system. The complex was affinity purified by a Protein A tagged version of Cas9 together with the proteins gathered at the promoter that was then eluted and identified by mass spectrometry123 (Figure 4).
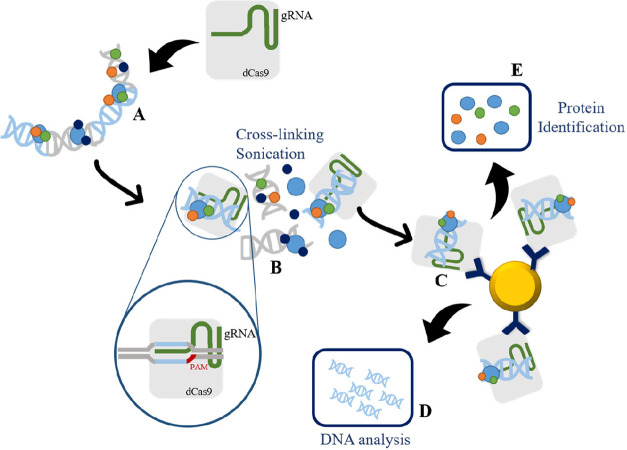
Figure 4.
Schematic workflow of CRISPR-ChAP-MS experiment. (A) Overexpression of dCas9 and a specific RNA guide (gRNA), (B) binding to the specific promoter, cross-linking and sonication. (C) Immunoprecipitation of Cas9 for the isolation of DNA–protein complexes of the promoter of interest. After de-cross-linking, DNA (D) and protein (E) elution are carried out.
The affinity enrichment strategy CRISPR-ChAP-MS was used to define how transcription from the arsenic response locus is regulated in an arsenic-dependent manner in budding yeast.
This locus constitutes a conserved pathway ranging from prokaryotes to higher eukaryotes. CRISPR-ChAP-MS was applied to the promoter regions of the activated arsenic response locus and the proteomic characterization of the targeted protein complex uncovered 40 nuclear-annotated proteins. Among these, the histone acetyltransferase SAGA and the chromatin remodeling complex SWI/SNF required for the locus activation were identified, providing key insight into the mechanisms of transcriptional activation required for the detoxification of arsenic from the cell.124
CRISPR-ChAP-MS was also successfully applied for application in mammalian cell models.
MACC1 (colon cancer-associated metastasis 1) is a protein that induces metastasis in colon cancer. The mechanisms by which its expression is transcriptionally regulated is still not fully known. Huang and collaborators exploited the CRISPR-ChAP-MS technique to identify proteins interacting with MACC1 promoter. They use the catalytically inactive 3xFLAG tagged dCas9 along with a gRNA to target and isolate the promoter region of the MACC1 gene.125 The c-JUN transcription factor was found physically bound to the MACC1 promoter and able to upregulate its expression.
In addition to the basic approach, many following versions have been enriched with further experimental steps and then have been proposed. For instance, a new hybrid approach (CasID)126 was introduced by Schmidtmann et al. and was based on BirA* and dCas9 to label by biotinylation protein components present at specific DNA sequence within a 10 nm range, thus to map also transient interactions. By using the construct BirA*-dCas9-eGFP, Schmidtmann and collaborators identified, for example, TERF2, TINF2, ACD that are known to directly bind telomeric DNA but also new chromatin factors, demonstrating that CasID is a robust method to investigate native protein environment at specific genomic loci. This approach provides a more detailed view of complexes that involve specific chromatin loci under dynamic and functional aspects if compared to ChIP. On the basis of the integration of CRISP technology and the proximity labeling enzyme APEX2 (dCas9-APEX2), Gao and collaborators proposed the new method C-BERST127 as an alternative to CasID.
Another valid strategy inspired by CRISPR-dead Cas9 was introduced by Yi and collaborators and was called CARPID128 (CRISPR-assisted RNA–protein interaction detection). It has been proposed to overcome the limitations of many other techniques used to elucidate the interactions between lncRNAs (noncoding RNA consisting of more than 200 nucleotides in length) and proteins in living cells. They designed a gRNA array composed of two gRNA sequences spaced by a 30-nucleotide direct repeat to target 2 adjacent loci on the same lncRNA transcript which could offer both greater specificity of targeting and a reduction in background noise. Yi et al. used an X-inactive specific transcript (XIST), a well-studied mammalian lncRNA, to validate the efficiency of the method. Besides already known protein partners, several novel XIST-interacting proteins have been identified; in particular, TAF15 (transcription activators) and SNF2L (repressive factors) that confirmed previous models of XIST-mediated X chromosome inactivation (XCI). This technique allowed them to confirm that it is an excellent method for identifying RNA–protein interactions but still has limitations and should be considered complementary and associated with other types of investigations.128
More recently, another Cas enzyme, i.e., Cas13, has been introduced to purify RNA-specific targets and identify proteins associated with an endogenous RNA within the RNA proximity proteomic methods (CBRPP),129 a new RNA-centric method.
6. Conclusions
Over the years, the advancement of technologies and the epochal changes in the way of looking at the biological world typical of -omic approaches have allowed the comprehension of many cell processes as well as the definition of new scenarios hitherto unknown. The combination of functional proteomics experiments coupled with mass spectrometry for the isolation and identification of protein complexes on specific DNA/RNA regions or investigation of epigenetic modifications both as profiling and at the level of gene promoters have had the largest impact in the elucidation of processes concerning gene regulation, splicing, translation, and so on. The findings deriving from these high throughput studies have allowed the understanding of the complex series of events, most of them also involved in the onset of several diseases.
From a methodological point of view, affinity purification (AP), ChIP, EMSA, CRISPR, and their variations provide complementary information.130−132 Among these methods, those in vitro, such as AP and EMSA, are more approachable and cheaper, although they suffer of all limitations associated with in vitro approaches. Others, i.e., DNA/RNA-centered ChIP and CRISPR-based, provide more detailed clues relative to a single genomic locus analysis in vivo but require more elaborate cell systems to be developed. Despite that the latter are time-consuming and expensive procedures to be set up, they are the only that allow the investigation of different DNA/RNA interactomes, modulated in vivo by different stimuli.133 All the described procedures might be enriched by the integration with structural data, such as the analyses of contact interfaces at the amino acid level,134 or the identification of post-translational modifications (PTMs) and how they tune the binding to DNA/RNA,135 and finally with innovative strategies for the global analysis of all putative DNA/RNA binding proteins.136 Overall, mass spectrometry is and could further be a master technology for all the present and future applications in the investigation of the nucleic acid interactome.
Glossary
Abbreviation
- AP-MS
affinity purification–mass spectrometry
- AP
affinity purification
- AptA-MS
aptamer affinity–mass spectrometry
- CALML3
Calmodulin-like protein 3
- CHART-MS
capture hybridization analysis of RNA targets–mass spectrometry
- ChIP
chromatin immunoprecipitation mass spectrometry
- ChIP-MS
chromatin immunoprecipitation–mass spectrometry
- CRISPR
clustered regularly interspaced short palindromic repeats
- ChIRP-MS
comprehensive identification of RNA-binding proteins–mass spectrometry
- CARPID
CRISPR-assisted RNA–protein interaction detection
- CRISPR-ChAP-MS
CRISPR-based chromatin affinity purification–mass spectrometry
- CRISPR-ChAP
CRISPR-based chromatin affinity purification
- Kd
dissociation constants
- EMSA
electrophoresis mobility shift assay
- EMSA-MS
electrophoresis mobility shift assay–mass spectrometry
- GFP
green fluorescent protein
- HSF1
heat shock factor 1
- IMCD
inner medullary collecting duct
- lncRNAs
noncoding RNA
- LNA
locked nucleic acid
- MACC1
colon cancer-associated metastasis 1
- MALAT1
metastasis-associated lung adenocarcinoma transcript 1
- GALNT2
N-acetylgalactosaminyltransferase 2 gene
- NEAT1
nuclear enriched abundant transcript 1
- nSBs
nuclear stress bodies
- PNA
peptide nucleic acid
- PRC
polycomb repressive complex
- PCR
polymerase chain reaction
- PRMT1
protein arginine methyltransferase 1
- PICh
proteomics of isolated chromatin segments
- RIME
rapid immunoprecipitation mass spectrometry of endogenous protein
- RAP-MS
RNA antisense purification–mass spectrometry
- RBDs
RNA binding domains
- S.so
S. solfataricus
- SERMs
selective estrogen receptor modulators
- SICAP
selective isolation of chromatin-associated proteins
- SRSFs
serine and arginine-rich pre-mRNA splicing factors
- SNP
single nucleotide polymorphisms
- SSV1
spindle-shaped virus 1
- SILAC
stable isotope labeling by amino acids in cell culture
- TMT
tandem mass tag
- CBX7
chromobox protein homologue 7
- TRIM25
tripartite motif-containing protein 25
- ssDNA
single-stranded DNA
- SSB
DNA binding proteins
- XIST
X-inactive specific transcript.
The authors declare no competing financial interest.
References
- Zuckerkandl E.; Pauling L. B.. Molecular disease, evolution, and genetic heterogeneity. In Horizons in Biochemistry; Kasha M., Pullman B., Eds.; Academic Press: New York, 1962; pp 189–225. [Google Scholar]
- Zuckerkandl E.; Pauling L. Molecules as Documents of Evolutionary History. J. Theor. Biol. 1965, 8 (2), 357–366. 10.1016/0022-5193(65)90083-4. [DOI] [PubMed] [Google Scholar]
- Kann M. G. Protein Interactions and Disease: Computational Approaches to Uncover the Etiology of Diseases. Briefings Bioinf. 2007, 8 (5), 333–346. 10.1093/bib/bbm031. [DOI] [PubMed] [Google Scholar]
- Preeti P.; Sabeeha H.; Shandar A., Protein-DNA Interactions. In Encyclopedia of Bioinformatics and Computational Biology; Academic Press, 2019; pp 142–154. [Google Scholar]
- Chowdhury N.; Bagchi A. An Overview of DNA-Protein Interactions. Curr. Chem. Biol. 2016, 9, 73. 10.2174/2212796809666151022202255. [DOI] [Google Scholar]
- Balcerak A.; Trebinska-Stryjewska A.; Konopinski R.; Wakula M.; Grzybowska E. A. RNA-Protein Interactions: Disorder, Moonlighting and Junk Contribute to Eukaryotic Complexity. Open Biol. 2019, 9 (6), 190096. 10.1098/rsob.190096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Re A.; Joshi T.; Kulberkyte E.; Morris Q.; Workman C. T. RNA-Protein Interactions: An Overview. Methods Mol. Biol. 2014, 1097, 491–521. 10.1007/978-1-62703-709-9_23. [DOI] [PubMed] [Google Scholar]
- Matsuda R.; Bi C.; Anguizola J.; Sobansky M.; Rodriguez E.; Vargas Badilla J.; Zheng X.; Hage B.; Hage D. S. Studies of Metabolite-Protein Interactions: A Review. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2014, 966, 48–58. 10.1016/j.jchromb.2013.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian-Azodi M; Abdolreza Mortazavi-Tabatabaei S.; Mansouri V.; Vafaee R. Metabolite- protein interaction (MPI) network analysis of obsessive compulsive disorder (OCD) from reported metabolites. Arvand J. Health Med. Sci. 2016, 1 (2), 112–120. 10.22631/ajhms.2016.43223. [DOI] [Google Scholar]
- Budayeva H. G.; Cristea I. M. A Mass Spectrometry View of Stable and Transient Protein Interactions. Adv. Exp. Med. Biol. 2014, 806, 263–282. 10.1007/978-3-319-06068-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobucci I.; Monaco V.; Cozzolino F.; Monti M. From Classical to New Generation Approaches: An Excursus of -Omics Methods for Investigation of Protein-Protein Interaction Networks. J. Proteomics 2021, 230, 103990. 10.1016/j.jprot.2020.103990. [DOI] [PubMed] [Google Scholar]
- Hudson W. H.; Ortlund E. A. The Structure, Function and Evolution of Proteins That Bind DNA and RNA. Nat. Rev. Mol. Cell Biol. 2014, 15 (11), 749–760. 10.1038/nrm3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Brown S. J. BindN: A Web-Based Tool for Efficient Prediction of DNA and RNA Binding Sites in Amino Acid Sequences. Nucleic Acids Res. 2006, 34 (Web Server issue), W243–W248. 10.1093/nar/gkl298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M.; Guo J.-T. New Insights into Protein-DNA Binding Specificity from Hydrogen Bond Based Comparative Study. Nucleic Acids Res. 2019, 47 (21), 11103–11113. 10.1093/nar/gkz963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E.; Harris M. E. Specificity and Nonspecificity in RNA-Protein Interactions. Nat. Rev. Mol. Cell Biol. 2015, 16 (9), 533–544. 10.1038/nrm4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther U.-P.; Yandek L. E.; Niland C. N.; Campbell F. E.; Anderson D.; Anderson V. E.; Harris M. E.; Jankowsky E. Hidden Specificity in an Apparently Nonspecific RNA-Binding Protein. Nature 2013, 502 (7471), 385–388. 10.1038/nature12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval-Pérez A.; Berger R. M. L.; Garaizar A.; Farr S. E.; Brehm M. A.; König G.; Schneider S. W.; Collepardo-Guevara R.; Huck V.; Rädler J. O.; Aponte-Santamaría C. DNA Binds to a Specific Site of the Adhesive Blood-Protein von Willebrand Factor Guided by Electrostatic Interactions. Nucleic Acids Res. 2020, 48 (13), 7333–7344. 10.1093/nar/gkaa466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Fan J.-S.; Li S.; Yang Y.; Sun P.; Zhu Q.; Wang J.; Jiang B.; Yang D.; Liu M. Structural Basis of DNA Binding to Human YB-1 Cold Shock Domain Regulated by Phosphorylation. Nucleic Acids Res. 2020, 48 (16), 9361–9371. 10.1093/nar/gkaa619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iftode C.; Daniely Y.; Borowiec J. A. Replication Protein A (RPA): The Eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 1999, 34 (3), 141–180. 10.1080/10409239991209255. [DOI] [PubMed] [Google Scholar]
- Déjardin J.; Kingston R. E. Purification of Proteins Associated with Specific Genomic Loci. Cell 2009, 136 (1), 175–186. 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes M. T.; Almeida-Lousada H.; Castelo-Branco P. Histone modifications in diseases. Histone Modifications in Therapy 2020, 20, 1–15. 10.1016/B978-0-12-816422-8.00001-5. [DOI] [Google Scholar]
- Reichert N.; Choukrallah M.-A.; Matthias P. Multiple Roles of Class I HDACs in Proliferation, Differentiation, and Development. Cell. Mol. Life Sci. 2012, 69 (13), 2173–2187. 10.1007/s00018-012-0921-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonelli R.; Jiménez C.; Riley M.; Servidei T.; Riccardi R.; Soriano A.; Roma J.; Martínez-Saez E.; Martini M.; Ruggiero A.; Moreno L.; Sánchez de Toledo J.; Gallego S.; Bové J.; Hooker J. M.; Segura M. F. CN133, a Novel Brain-Penetrating Histone Deacetylase Inhibitor, Hampers Tumor Growth in Patient-Derived Pediatric Posterior Fossa Ependymoma Models. Cancers 2020, 12 (7), 1922. 10.3390/cancers12071922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräff J.; Kim D.; Dobbin M. M.; Tsai L.-H. Epigenetic Regulation of Gene Expression in Physiological and Pathological Brain Processes. Physiol. Rev. 2011, 91 (2), 603–649. 10.1152/physrev.00012.2010. [DOI] [PubMed] [Google Scholar]
- Navis A. R.Organisers and Genes (1940), by Conrad Hal Waddington. Embryo Project Encyclopedia (2007–10–30). http://embryo.asu.edu/handle/10776/1685.
- Cozzolino F.; Iacobucci I.; Monaco V.; Angrisano T.; Monti M. Lysines Acetylome and Methylome Profiling of H3 and H4 Histones in Trichostatin A-Treated Stem Cells. Int. J. Mol. Sci. 2021, 22 (4), 2063. 10.3390/ijms22042063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murarka P.; Srivastava P. An Improved Method for the Isolation and Identification of Unknown Proteins That Bind to Known DNA Sequences by Affinity Capture and Mass Spectrometry. PLoS One 2018, 13 (8), e0202602 10.1371/journal.pone.0202602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choux C.; Petazzi P.; Sanchez-Delgado M.; Hernandez Mora J. R.; Monteagudo A.; Sagot P.; Monk D.; Fauque P. The Hypomethylation of Imprinted Genes in IVF/ICSI Placenta Samples Is Associated with Concomitant Changes in Histone Modifications. Epigenetics 2020, 15 (12), 1386–1395. 10.1080/15592294.2020.1783168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.; Garcia B. A. Combining Genomic and Proteomic Approaches for Epigenetics Research. Epigenomics 2013, 5 (4), 439–452. 10.2217/epi.13.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y.-H.; Huang H. Advances in the Study of Protein-DNA Interaction. Amino Acids 2012, 43 (3), 1141–1146. 10.1007/s00726-012-1377-9. [DOI] [PubMed] [Google Scholar]
- Ascano M.; Gerstberger S.; Tuschl T. Multi-Disciplinary Methods to Define RNA-Protein Interactions and Regulatory Networks. Curr. Opin. Genet. Dev. 2013, 23 (1), 20–28. 10.1016/j.gde.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro D. G.; Clarke T.; Davies C. C.; Bailey D. Identifying Novel Protein Interactions: Proteomic Methods, Optimisation Approaches and Data Analysis Pipelines. Methods 2016, 95, 46–54. 10.1016/j.ymeth.2015.08.022. [DOI] [PubMed] [Google Scholar]
- Serikawa T.; Spanos C.; von Hacht A.; Budisa N.; Rappsilber J.; Kurreck J. Comprehensive Identification of Proteins Binding to RNA G-Quadruplex Motifs in the 5′ UTR of Tumor-Associated MRNAs. Biochimie 2018, 144, 169–184. 10.1016/j.biochi.2017.11.003. [DOI] [PubMed] [Google Scholar]
- Gingras A.-C.; Aebersold R.; Raught B. Advances in Protein Complex Analysis Using Mass Spectrometry. J. Physiol. 2005, 563, 11–21. 10.1113/jphysiol.2004.080440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriel N. L.; Heunis T.; Sampson S. L.; Gey van Pittius N. C.; Williams M. J.; Warren R. M. Identifying Nucleic Acid-Associated Proteins in Mycobacterium Smegmatis by Mass Spectrometry-Based Proteomics. BMC Mol. Cell Biol. 2020, 21 (1), 19. 10.1186/s12860-020-00261-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenarcic E. M.; Ziehr B. J.; Moorman N. J. An Unbiased Proteomics Approach to Identify Human Cytomegalovirus RNA-Associated Proteins. Virology 2015, 481, 13–23. 10.1016/j.virol.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butter F.; Scheibe M.; Mörl M.; Mann M. Unbiased RNA-Protein Interaction Screen by Quantitative Proteomics. Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (26), 10626–10631. 10.1073/pnas.0812099106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacheny A.; Dieu M.; Arnould T.; Renard P. Mass Spectrometry-Based Identification of Proteins Interacting with Nucleic Acids. J. Proteomics 2013, 94, 89–109. 10.1016/j.jprot.2013.09.011. [DOI] [PubMed] [Google Scholar]
- Babayeva N. D.; Baranovskaya O. I.; Tahirov T. H. Structural Basis of Ets1 Cooperative Binding to Widely Separated Sites on Promoter DNA. PLoS One 2012, 7 (3), e33698 10.1371/journal.pone.0033698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler G.; Butter F.; Mann M. A SILAC-Based DNA Protein Interaction Screen That Identifies Candidate Binding Proteins to Functional DNA Elements. Genome Res. 2008, 19 (2), 284–293. 10.1101/gr.081711.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. A.; Pastrana C. L.; Butterer A.; Pernstich C.; Gwynn E. J.; Sobott F.; Moreno-Herrero F.; Dillingham M. S. Specific and Non-Specific Interactions of ParB with DNA: Implications for Chromosome Segregation. Nucleic Acids Res. 2015, 43 (2), 719–731. 10.1093/nar/gku1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hégarat N.; Cardoso G. M.; Rusconi F.; Francois J.-C.; Praseuth D. Analytical Biochemistry of DNA--Protein Assemblies from Crude Cell Extracts. Nucleic Acids Res. 2007, 35 (13), e92 10.1093/nar/gkm490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y.; Jarrett H. W. Method for Trapping Affinity Chromatography of Transcription Factors Using Aldehyde-Hydrazide Coupling to Agarose. Anal. Biochem. 2015, 482, 1–6. 10.1016/j.ab.2015.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan T.; Smith S. G. J.; Gaughan K.; Oglesby I. K.; O’Neill S.; McElvaney N. G.; Greene C. M. Isolation and Identification of Cell-Specific MicroRNAs Targeting a Messenger RNA Using a Biotinylated Anti-Sense Oligonucleotide Capture Affinity Technique. Nucleic Acids Res. 2013, 41 (6), e71 10.1093/nar/gks1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B.; Nath S. K. Identification of Proteins Interacting with Single Nucleotide Polymorphisms (SNPs) by DNA Pull-Down Assay. Methods Mol. Biol. 2019, 1855, 355–362. 10.1007/978-1-4939-8793-1_30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Pogar T.; Liang S.; Hague L. K.; Lutz C. S. Specific Trans-Acting Proteins Interact with Auxiliary RNA Polyadenylation Elements in the COX-2 3′-UTR. RNA 2007, 13 (7), 1103–1115. 10.1261/rna.577707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey B.; Thukral S.; Krishnan S.; Chakrobarty M.; Gupta S.; Manghani C.; Rani V. DNA-Protein Interactions: Methods for Detection and Analysis. Mol. Cell. Biochem. 2012, 365 (1–2), 279–299. 10.1007/s11010-012-1269-z. [DOI] [PubMed] [Google Scholar]
- LaSarre B.; Federle M. J. EMSA Analysis of DNA Binding By Rgg Proteins. Bio-Protoc. 2013, 10.21769/BioProtoc.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadgil H.; Jarrett H. W. Oligonucleotide Trapping Method for Purification of Transcription Factors. J. Chromatogr. A 2002, 966 (1–2), 99–110. 10.1016/S0021-9673(02)00738-0. [DOI] [PubMed] [Google Scholar]
- Rees J. S.; Lilley K. S.; Jackson A. P. SILAC-IPAC: A Quantitative Method for Distinguishing Genuine from Non-Specific Components of Protein Complexes by Parallel Affinity Capture. J. Proteomics 2015, 115, 143–156. 10.1016/j.jprot.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowski M. M.; Gräwe C.; Foster B. M.; Nguyen N. V.; Bartke T.; Vermeulen M. Global Profiling of Protein-DNA and Protein-Nucleosome Binding Affinities Using Quantitative Mass Spectrometry. Nat. Commun. 2018, 9 (1), 1653. 10.1038/s41467-018-04084-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcilla M.; Albar J. P. Quantitative Proteomics: A Strategic Ally to Map Protein Interaction Networks. IUBMB Life 2013, 65 (1), 9–16. 10.1002/iub.1081. [DOI] [PubMed] [Google Scholar]
- Mellacheruvu D.; Wright Z.; Couzens A. L.; Lambert J.-P.; St-Denis N. A.; Li T.; Miteva Y. V.; Hauri S.; Sardiu M. E.; Low T. Y.; Halim V. A.; Bagshaw R. D.; Hubner N. C.; Al-Hakim A.; Bouchard A.; Faubert D.; Fermin D.; Dunham W. H.; Goudreault M.; Lin Z.-Y.; Badillo B. G.; Pawson T.; Durocher D.; Coulombe B.; Aebersold R.; Superti-Furga G.; Colinge J.; Heck A. J. R.; Choi H.; Gstaiger M.; Mohammed S.; Cristea I. M.; Bennett K. L.; Washburn M. P.; Raught B.; Ewing R. M.; Gingras A.-C.; Nesvizhskii A. I. The CRAPome: A Contaminant Repository for Affinity Purification-Mass Spectrometry Data. Nat. Methods 2013, 10 (8), 730–736. 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling G.; Hauer C. R.; Gronostajski R. M.; Pentecost B. T.; Ding X. Transcriptional Regulation of Rat CYP2A3 by Nuclear Factor 1: Identification of a Novel NFI-A Isoform, and Evidence for Tissue-Selective Interaction of NFI with the CYP2A3 Promoter in Vivo. J. Biol. Chem. 2004, 279 (27), 27888–27895. 10.1074/jbc.M403705200. [DOI] [PubMed] [Google Scholar]
- Dhawan L.; Liu B.; Pytlak A.; Kulshrestha S.; Blaxall B. C.; Taubman M. B. Y-Box Binding Protein 1 and RNase UK114 Mediate Monocyte Chemoattractant Protein 1 MRNA Stability in Vascular Smooth Muscle Cells. Mol. Cell. Biol. 2012, 32 (18), 3768–3775. 10.1128/MCB.00846-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNair L.; Xiao S.; Miletic D.; Ghani M.; Julien J.-P.; Keith J.; Zinman L.; Rogaeva E.; Robertson J. MTHFSD and DDX58 Are Novel RNA-Binding Proteins Abnormally Regulated in Amyotrophic Lateral Sclerosis. Brain 2016, 139, 86–100. 10.1093/brain/awv308. [DOI] [PubMed] [Google Scholar]
- Dhar S. K.; Zhang J.; Gal J.; Xu Y.; Miao L.; Lynn B. C.; Zhu H.; Kasarskis E. J.; St Clair D. K. FUsed in Sarcoma Is a Novel Regulator of Manganese Superoxide Dismutase Gene Transcription. Antioxid. Redox Signaling 2014, 20 (10), 1550–1566. 10.1089/ars.2012.4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medugno L.; Costanzo P.; Lupo A.; Monti M.; Florio F.; Pucci P.; Izzo P. A Novel Zinc Finger Transcriptional Repressor, ZNF224, Interacts with the Negative Regulatory Element (AldA-NRE) and Inhibits Gene Expression. FEBS Lett. 2003, 534 (1–3), 93–100. 10.1016/S0014-5793(02)03783-3. [DOI] [PubMed] [Google Scholar]
- Peng H.; Zhu Q.-S.; Zhong S.; Levy D. Transcription of the Human Microsomal Epoxide Hydrolase Gene (EPHX1) Is Regulated by an HNF-4α/CAR/RXR/PSF Complex. Biochim. Biophys. Acta, Gene Regul. Mech. 2013, 1829 (10), 1000–1009. 10.1016/j.bbagrm.2013.05.003. [DOI] [PubMed] [Google Scholar]
- Nasrullah U.; Haeussler K.; Biyanee A.; Wittig I.; Pfeilschifter J.; Eberhardt W. Identification of TRIM25 as a Negative Regulator of Caspase-2 Expression Reveals a Novel Target for Sensitizing Colon Carcinoma Cells to Intrinsic Apoptosis. Cells 2019, 8 (12), 1622. 10.3390/cells8121622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisa V.; Cozzolino M.; Gargiulo S.; Ottone C.; Piccioni F.; Monti M.; Gigliotti S.; Talamo F.; Graziani F.; Pucci P.; Verrotti A. C. The Molecular Chaperone Hsp90 Is a Component of the Cap-Binding Complex and Interacts with the Translational Repressor Cup during Drosophila Oogenesis. Gene 2009, 432 (1–2), 67–74. 10.1016/j.gene.2008.11.025. [DOI] [PubMed] [Google Scholar]
- Marucci A.; Cozzolino F.; Dimatteo C.; Monti M.; Pucci P.; Trischitta V.; Di Paola R. Role of GALNT2 in the Modulation of ENPP1 Expression, and Insulin Signaling and Action: GALNT2: A Novel Modulator of Insulin Signaling. Biochim. Biophys. Acta, Mol. Cell Res. 2013, 1833 (6), 1388–1395. 10.1016/j.bbamcr.2013.02.032. [DOI] [PubMed] [Google Scholar]
- Miro J.; Bourgeois C. F.; Claustres M.; Koenig M.; Tuffery-Giraud S. Identification of Splicing Factors Involved in DMD Exon Skipping Events Using an In Vitro RNA Binding Assay. Methods Mol. Biol. 2018, 1687, 157–169. 10.1007/978-1-4939-7374-3_11. [DOI] [PubMed] [Google Scholar]
- Ray J.; Kruse A.; Ozer A.; Kajitani T.; Johnson R.; MacCoss M.; Heck M.; Lis J. T. RNA Aptamer Capture of Macromolecular Complexes for Mass Spectrometry Analysis. Nucleic Acids Res. 2020, 48 (15), e90 10.1093/nar/gkaa542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robledo M.; Matia-González A. M.; García-Tomsig N. I.; Jiménez-Zurdo J. I. Identification of Small RNA-Protein Partners in Plant Symbiotic Bacteria. Methods Mol. Biol. 2018, 1737, 351–370. 10.1007/978-1-4939-7634-8_20. [DOI] [PubMed] [Google Scholar]
- Martínez-Salazar M.; López-Urrutia E.; Arechaga-Ocampo E.; Bonilla-Moreno R.; Martínez-Castillo M.; Díaz-Hernández J.; Del Moral-Hernández O.; Cedillo-Barrón L.; Martines-Juarez V.; De Nova-Ocampo M.; Valdes J.; Berumen J.; Villegas-Sepúlveda N. Biochemical and Proteomic Analysis of Spliceosome Factors Interacting with Intron-1 of Human Papillomavirus Type-16. J. Proteomics 2014, 111, 184–197. 10.1016/j.jprot.2014.07.029. [DOI] [PubMed] [Google Scholar]
- Upadhyay A.; Dixit U.; Manvar D.; Chaturvedi N.; Pandey V. N. Affinity Capture and Identification of Host Cell Factors Associated with Hepatitis C Virus (+) Strand Subgenomic RNA. Mol. Cell. Proteomics MCP 2013, 12 (6), 1539–1552. 10.1074/mcp.M112.017020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestre-Fos S.; Penev P. I.; Suttapitugsakul S.; Hu M.; Ito C.; Petrov A. S.; Wartell R. M.; Wu R.; Williams L. D. G-Quadruplexes in Human Ribosomal RNA. J. Mol. Biol. 2019, 431 (10), 1940–1955. 10.1016/j.jmb.2019.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ream J. A.; Lewis L. K.; Lewis K. A. Rapid Agarose Gel Electrophoretic Mobility Shift Assay for Quantitating Protein: RNA Interactions. Anal. Biochem. 2016, 511, 36–41. 10.1016/j.ab.2016.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin-Laprade D.; Burrus V. Electrophoretic Mobility Shift Assay Using Radiolabeled DNA Probes. Methods Mol. Biol. 2015, 1334, 1–15. 10.1007/978-1-4939-2877-4_1. [DOI] [PubMed] [Google Scholar]
- Butturini E.; Cozzolino F.; Boriero D.; Carcereri de Prati A.; Monti M.; Rossin M.; Canetti D.; Cellini B.; Pucci P.; Mariotto S. S-Glutathionylation Exerts Opposing Roles in the Regulation of STAT1 and STAT3 Signaling in Reactive Microglia. Free Radical Biol. Med. 2018, 117, 191–201. 10.1016/j.freeradbiomed.2018.02.005. [DOI] [PubMed] [Google Scholar]
- Butturini E.; Gotte G.; Dell’Orco D.; Chiavegato G.; Marino V.; Canetti D.; Cozzolino F.; Monti M.; Pucci P.; Mariotto S. Intermolecular Disulfide Bond Influences Unphosphorylated STAT3 Dimerization and Function. Biochem. J. 2016, 473 (19), 3205–3219. 10.1042/BCJ20160294. [DOI] [PubMed] [Google Scholar]
- Boon M.; De Zitter E.; De Smet J.; Wagemans J.; Voet M.; Pennemann F. L.; Schalck T.; Kuznedelov K.; Severinov K.; Van Meervelt L.; De Maeyer M.; Lavigne R. Drc”, a Structurally Novel SsDNA-Binding Transcription Regulator of N4-Related Bacterial Viruses. Nucleic Acids Res. 2019, 48 (1), 445–459. 10.1093/nar/gkz1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman L. M.; Fried M. G. Electrophoretic Mobility Shift Assay (EMSA) for Detecting Protein-Nucleic Acid Interactions. Nat. Protoc. 2007, 2 (8), 1849–1861. 10.1038/nprot.2007.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien N.; Herman J.-P. LUEGO: A Cost and Time Saving Gel Shift Procedure. BioTechniques 2011, 51 (4), 267–269. 10.2144/000113751. [DOI] [PubMed] [Google Scholar]
- Yu L.; He W.; Xie J.; Guo R.; Ni J.; Zhang X.; Xu Q.; Wang C.; Yue Q.; Li F.; Luo M.; Sun B.; Ye L.; Zheng K. In Vitro Biochemical Assays Using Biotin Labels to Study Protein-Nucleic Acid Interactions. J. Visualized Exp. 2019, 149. 10.3791/59830. [DOI] [PubMed] [Google Scholar]
- Hsieh Y.-W.; Alqadah A.; Chuang C.-F. An Optimized Protocol for Electrophoretic Mobility Shift Assay Using Infrared Fluorescent Dye-Labeled Oligonucleotides. J. Visualized Exp. 2016, 117. 10.3791/54863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parés-Matos E. I. Electrophoretic Mobility-Shift and Super-Shift Assays for Studies and Characterization of Protein-DNA Complexes. Methods Mol. Biol. 2013, 977, 159–167. 10.1007/978-1-62703-284-1_12. [DOI] [PubMed] [Google Scholar]
- Woo A. J.; Dods J. S.; Susanto E.; Ulgiati D.; Abraham L. J. A Proteomics Approach for the Identification of DNA Binding Activities Observed in the Electrophoretic Mobility Shift Assay. Mol. Cell. Proteomics MCP 2002, 1 (6), 472–478. 10.1074/mcp.T200003-MCP200. [DOI] [PubMed] [Google Scholar]
- Stenger D.; Gruissem W.; Baginsky S. Mass Spectrometric Identification of RNA Binding Proteins from Dried EMSA Gels. J. Proteome Res. 2004, 3 (3), 662–664. 10.1021/pr049966q. [DOI] [PubMed] [Google Scholar]
- Zhang Z.; Huang Q.; Wang Z.; Zou J.; Yu Z.; Strauss J. F. Iii; Zhang Z. Elongin B Is a Binding Partner of the Male Germ Cell Nuclear Speckle Protein Sperm-Associated Antigen 16S (SPAG16S) and Is Regulated Post-Transcriptionally in the Testis. Reprod., Fertil. Dev. 2019, 31 (5), 962–971. 10.1071/RD18303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller M.; Lee Y. Q.; Vidovic K.; Kjellström S.; Björkman L.; Storry J. R.; Olsson M. L. Disruption of a GATA1-Binding Motif Upstream of XG/PBDX Abolishes Xga Expression and Resolves the Xg Blood Group System. Blood 2018, 132 (3), 334–338. 10.1182/blood-2018-03-842542. [DOI] [PubMed] [Google Scholar]
- Qin S.; Ingle J. N.; Liu M.; Yu J.; Wickerham D. L.; Kubo M.; Weinshilboum R. M.; Wang L. Calmodulin-like Protein 3 Is an Estrogen Receptor Alpha Coregulator for Gene Expression and Drug Response in a SNP, Estrogen, and SERM-Dependent Fashion. Breast Cancer Res. 2017, 19 (1), 95. 10.1186/s13058-017-0890-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco S.; Aulitto M.; Iacobucci I.; Crocamo G.; Pucci P.; Bartolucci S.; Monti M.; Contursi P. The Interaction between the F55 Virus-Encoded Transcription Regulator and the RadA Host Recombinase Reveals a Common Strategy in Archaea and Bacteria to Sense the UV-Induced Damage to the Host DNA. Biochim. Biophys. Acta, Gene Regul. Mech. 2020, 1863 (5), 194493. 10.1016/j.bbagrm.2020.194493. [DOI] [PubMed] [Google Scholar]
- Weinmann A. S.; Farnham P. J. Identification of Unknown Target Genes of Human Transcription Factors Using Chromatin Immunoprecipitation. Methods 2002, 26 (1), 37–47. 10.1016/S1046-2023(02)00006-3. [DOI] [PubMed] [Google Scholar]
- Nelson J. D.; Denisenko O.; Bomsztyk K. Protocol for the Fast Chromatin Immunoprecipitation (ChIP) Method. Nat. Protoc. 2006, 1 (1), 179–185. 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- Schuch R.; Agelopoulos K.; Neumann A.; Brandt B.; Bürger H.; Korsching E. Site-Specific Chromatin Immunoprecipitation: A Selective Method to Individually Analyze Neighboring Transcription Factor Binding Sites in Vivo. BMC Res. Notes 2012, 5, 109. 10.1186/1756-0500-5-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collas P. The Current State of Chromatin Immunoprecipitation. Mol. Biotechnol. 2010, 45 (1), 87–100. 10.1007/s12033-009-9239-8. [DOI] [PubMed] [Google Scholar]
- Beischlag T. V.; Prefontaine G. G.; Hankinson O. ChIP-Re-ChIP: Co-Occupancy Analysis by Sequential Chromatin Immunoprecipitation. Methods Mol. Biol. 2018, 1689, 103–112. 10.1007/978-1-4939-7380-4_9. [DOI] [PubMed] [Google Scholar]
- Truax A. D.; Greer S. F. ChIP and Re-ChIP Assays: Investigating Interactions between Regulatory Proteins, Histone Modifications, and the DNA Sequences to Which They Bind. Methods Mol. Biol. 2012, 809, 175–188. 10.1007/978-1-61779-376-9_12. [DOI] [PubMed] [Google Scholar]
- Federico A.; Sepe R.; Cozzolino F.; Piccolo C.; Iannone C.; Iacobucci I.; Pucci P.; Monti M.; Fusco A. The Complex CBX7-PRMT1 Has a Critical Role in Regulating E-Cadherin Gene Expression and Cell Migration. Biochim. Biophys. Acta, Gene Regul. Mech. 2019, 1862 (4), 509–521. 10.1016/j.bbagrm.2019.02.006. [DOI] [PubMed] [Google Scholar]
- Federico A.; Pallante P.; Bianco M.; Ferraro A.; Esposito F.; Monti M.; Cozzolino M.; Keller S.; Fedele M.; Leone V.; Troncone G.; Chiariotti L.; Pucci P.; Fusco A. Chromobox Protein Homologue 7 Protein, with Decreased Expression in Human Carcinomas, Positively Regulates E-Cadherin Expression by Interacting with the Histone Deacetylase 2 Protein. Cancer Res. 2009, 69 (17), 7079–7087. 10.1158/0008-5472.CAN-09-1542. [DOI] [PubMed] [Google Scholar]
- Soares M. A. F.; Castro D. S. Chromatin Immunoprecipitation from Mouse Embryonic Tissue or Adherent Cells in Culture, Followed by Next-Generation Sequencing. Methods Mol. Biol. 2018, 1689, 53–63. 10.1007/978-1-4939-7380-4_5. [DOI] [PubMed] [Google Scholar]
- Buck M. J.; Lieb J. D. ChIP-Chip: Considerations for the Design, Analysis, and Application of Genome-Wide Chromatin Immunoprecipitation Experiments. Genomics 2004, 83 (3), 349–360. 10.1016/j.ygeno.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Pillai S.; Chellappan S. P. ChIP on Chip Assays: Genome-Wide Analysis of Transcription Factor Binding and Histone Modifications. Methods Mol. Biol. 2009, 523, 341–366. 10.1007/978-1-59745-190-1_23. [DOI] [PubMed] [Google Scholar]
- Pillai S.; Chellappan S. P. ChIP on Chip and ChIP-Seq Assays: Genome-Wide Analysis of Transcription Factor Binding and Histone Modifications. Methods Mol. Biol. 2015, 1288, 447–472. 10.1007/978-1-4939-2474-5_26. [DOI] [PubMed] [Google Scholar]
- Engelen E.; Brandsma J. H.; Moen M. J.; Signorile L.; Dekkers D. H. W.; Demmers J.; Kockx C. E. M.; Ozgür Z.; van IJcken W. F. J.; van den Berg D. L. C.; Poot R. A. Proteins That Bind Regulatory Regions Identified by Histone Modification Chromatin Immunoprecipitations and Mass Spectrometry. Nat. Commun. 2015, 6, 7155. 10.1038/ncomms8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guezennec X.; Brinkman A. B.; Vermeulen M.; Denissov S. G.; Gazziola C.; Lohrum M. E.; Stunnenberg H. G. Targeted Discovery Tools: Proteomics and Chromatin Immunoprecipitation-on-Chip. BJU Int. 2005, 96, 16–22. 10.1111/j.1464-410X.2005.05942.x. [DOI] [PubMed] [Google Scholar]
- Rafiee M.-R.; Girardot C.; Sigismondo G.; Krijgsveld J. Expanding the Circuitry of Pluripotency by Selective Isolation of Chromatin-Associated Proteins. Mol. Cell 2016, 64 (3), 624–635. 10.1016/j.molcel.2016.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierer M.; Mann M. Proteomics to Study DNA-Bound and Chromatin-Associated Gene Regulatory Complexes. Hum. Mol. Genet. 2016, 25 (R2), R106–R114. 10.1093/hmg/ddw208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed H.; Taylor C.; Brown G. D.; Papachristou E. K.; Carroll J. S.; D’Santos C. S. Rapid Immunoprecipitation Mass Spectrometry of Endogenous Proteins (RIME) for Analysis of Chromatin Complexes. Nat. Protoc. 2016, 11 (2), 316–326. 10.1038/nprot.2016.020. [DOI] [PubMed] [Google Scholar]
- Paltoglou S.; Das R.; Townley S. L.; Hickey T. E.; Tarulli G. A.; Coutinho I.; Fernandes R.; Hanson A. R.; Denis I.; Carroll J. S.; Dehm S. M.; Raj G. V.; Plymate S. R.; Tilley W. D.; Selth L. A. Novel Androgen Receptor Coregulator GRHL2 Exerts Both Oncogenic and Antimetastatic Functions in Prostate Cancer. Cancer Res. 2017, 77 (13), 3417–3430. 10.1158/0008-5472.CAN-16-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J. R.; Chou C.-L.; Medvar B.; Knepper M. A.; Jung H. J. Identification of β-Catenin-Interacting Proteins in Nuclear Fractions of Native Rat Collecting Duct Cells. Am. J. Physiol. Renal Physiol. 2017, 313 (1), F30–F46. 10.1152/ajprenal.00054.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldi M.; Mari T.; Nicosia L.; Musiani D.; Sigismondo G.; Cuomo A.; Pavesi G.; Bonaldi T. Chromatin Proteomics Reveals Novel Combinatorial Histone Modification Signatures That Mark Distinct Subpopulations of Macrophage Enhancers. Nucleic Acids Res. 2017, 45 (21), 12195–12213. 10.1093/nar/gkx821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldi M.; Bonaldi T. The Proteomic Investigation of Chromatin Functional Domains Reveals Novel Synergisms among Distinct Heterochromatin Components. Mol. Cell. Proteomics MCP 2013, 12 (3), 764–780. 10.1074/mcp.M112.024307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X.; Dadon D. B.; Abraham B. J.; Lee T. I.; Jaenisch R.; Bradner J. E.; Young R. A. Chromatin Proteomic Profiling Reveals Novel Proteins Associated with Histone-Marked Genomic Regions. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (12), 3841–3846. 10.1073/pnas.1502971112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser A.; Krüger T.; Eiselt G.; Bechler J.; Kniemeyer O.; Huber O.; Schmidt M. Identification of PARP-1, Histone H1 and SIRT-1 as New Regulators of Breast Cancer-Related Aromatase Promoter I.3/II. Cells 2020, 9 (2), 427. 10.3390/cells9020427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaddek D.; Lamond A. I. Unlocking the Chromatin Code by Deciphering Protein-DNA Interactions. Mol. Syst. Biol. 2016, 12 (11), 887. 10.15252/msb.20167332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiee M.-R.; Sigismondo G.; Kalxdorf M.; Förster L.; Brügger B.; Béthune J.; Krijgsveld J. Protease-Resistant Streptavidin for Interaction Proteomics. Mol. Syst. Biol. 2020, 16 (5), e9370 10.15252/msb.20199370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S. K.; Koshkin A. A.; Wengel J.; Nielsen P. LNA (Locked Nucleic Acids): Synthesis and High-Affinity Nucleic Acid Recognition. Chem. Commun. 1998, (4), 455–456. 10.1039/a708608c. [DOI] [Google Scholar]
- West J. A.; Davis C. P.; Sunwoo H.; Simon M. D.; Sadreyev R. I.; Wang P. I.; Tolstorukov M. Y.; Kingston R. E. The Long Noncoding RNAs NEAT1 and MALAT1 Bind Active Chromatin Sites. Mol. Cell 2014, 55 (5), 791–802. 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M. D.; Wang C. I.; Kharchenko P. V.; West J. A.; Chapman B. A.; Alekseyenko A. A.; Borowsky M. L.; Kuroda M. I.; Kingston R. E. The Genomic Binding Sites of a Noncoding RNA. Proc. Natl. Acad. Sci. U. S. A. 2011, 108 (51), 20497–20502. 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh C. A.; Guttman M.. RAP-MS: A Method to Identify Proteins That Interact Directly with a Specific RNA Molecule in Cells. In RNA Detection; Gaspar I., Ed.; Methods Mol. Biol.; Springer: New York, NY, 2018; Vol. 1649, pp 473–488. 10.1007/978-1-4939-7213-5_31. [DOI] [PubMed] [Google Scholar]
- Machyna M.; Simon M. D. Catching RNAs on Chromatin Using Hybridization Capture Methods. Briefings Funct. Genomics 2018, 17 (2), 96–103. 10.1093/bfgp/elx038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Wen L.; Zhu H. Unveiling the Hidden Function of Long Non-Coding RNA by Identifying Its Major Partner-Protein. Cell Biosci. 2015, 5, 59. 10.1186/s13578-015-0050-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C.; Chang H. Y. ChIRP-MS: RNA-Directed Proteomic Discovery. Methods Mol. Biol. 2018, 1861, 37–45. 10.1007/978-1-4939-8766-5_3. [DOI] [PubMed] [Google Scholar]
- Chu C.; Zhang Q. C.; da Rocha S. T.; Flynn R. A.; Bharadwaj M.; Calabrese J. M.; Magnuson T.; Heard E.; Chang H. Y. Systematic Discovery of Xist RNA Binding Proteins. Cell 2015, 161 (2), 404–416. 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya K.; Adachi S.; Natsume T.; Iwakiri J.; Terai G.; Asai K.; Hirose T. LncRNA-Dependent Nuclear Stress Bodies Promote Intron Retention through SR Protein Phosphorylation. EMBO J. 2020, 39 (3), e102729 10.15252/embj.2019102729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X.; Lan Z.; Lai Q.; Li A.; Liu S.; Wang X. LncRNA SNHG6 Plays an Oncogenic Role in Colorectal Cancer and Can Be Used as a Prognostic Biomarker for Solid Tumors. J. Cell. Physiol. 2020, 235 (10), 7620–7634. 10.1002/jcp.29672. [DOI] [PubMed] [Google Scholar]
- Engreitz J.; Lander E. S.; Guttman M.. RNA Antisense Purification (RAP) for Mapping RNA Interactions with Chromatin. In Nuclear Bodies and Noncoding RNAs; Nakagawa S., Hirose T., Eds.; Methods Mol. Biol.; Springer: New York, NY, 2015; Vol. 1262, pp 183–197. 10.1007/978-1-4939-2253-6_11. [DOI] [PubMed] [Google Scholar]
- McHugh C. A.; Guttman M. RAP-MS: A Method to Identify Proteins That Interact Directly with a Specific RNA Molecule in Cells. Methods Mol. Biol. 2018, 1649, 473–488. 10.1007/978-1-4939-7213-5_31. [DOI] [PubMed] [Google Scholar]
- Wanowska E.; Kubiak M.; Makałowska I.; Szcześniak M. W. A Chromatin-Associated Splicing Isoform of OIP5-AS1 Acts in Cis to Regulate the OIP5 Oncogene. RNA Biol. 2021, 1–12. 10.1080/15476286.2021.1871816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldrip Z. J.; Byrum S. D.; Storey A. J.; Gao J.; Byrd A. K.; Mackintosh S. G.; Wahls W. P.; Taverna S. D.; Raney K. D.; Tackett A. J. A CRISPR-Based Approach for Proteomic Analysis of a Single Genomic Locus. Epigenetics 2014, 9 (9), 1207–1211. 10.4161/epi.29919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West K. L.; Byrum S. D.; Mackintosh S. G.; Edmondson R. D.; Taverna S. D.; Tackett A. J. Proteomic Characterization of the Arsenic Response Locus in S. Cerevisiae. Epigenetics 2019, 14 (2), 130–145. 10.1080/15592294.2019.1580110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.; Xiang Y.; Xie Z.; Cai Y.; Yang Q.; Huang H.; Chen Z.; Xiao Z.; He Q. Mass Spectrometry-Based Proteomic Capture of Proteins Bound to the MACC1 Promoter in Colon Cancer. Clin. Exp. Metastasis 2020, 37 (4), 477–487. 10.1007/s10585-020-10045-z. [DOI] [PubMed] [Google Scholar]
- Schmidtmann E.; Anton T.; Rombaut P.; Herzog F.; Leonhardt H. Determination of Local Chromatin Composition by CasID. Nucl. Austin Tex 2016, 7 (5), 476–484. 10.1080/19491034.2016.1239000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X. D.; Tu L.-C.; Mir A.; Rodriguez T.; Ding Y.; Leszyk J.; Dekker J.; Shaffer S. A.; Zhu L. J.; Wolfe S. A.; Sontheimer E. J. C-BERST: Defining Subnuclear Proteomic Landscapes at Genomic Elements with DCas9-APEX2. Nat. Methods 2018, 15 (6), 433–436. 10.1038/s41592-018-0006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W.; Li J.; Zhu X.; Wang X.; Fan L.; Sun W.; Liao L.; Zhang J.; Li X.; Ye J.; Chen F.; Taipale J.; Chan K. M.; Zhang L.; Yan J. CRISPR-Assisted Detection of RNA-Protein Interactions in Living Cells. Nat. Methods 2020, 17 (7), 685–688. 10.1038/s41592-020-0866-0. [DOI] [PubMed] [Google Scholar]
- Li Y.; Liu S.; Cao L.; Luo Y.; Du H.; Li S.; Zhang Z.; Guo X.; Tian W.; Wong C. C.; You F. CBRPP: A New RNA-Centric Method to Study RNA-Protein Interactions. RNA Biol. 2021, 1–14. 10.1080/15476286.2021.1873620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer M.; Levin M.; Butter F.; Scheibe M. Quantitative Proteomics to Identify Nuclear RNA-Binding Proteins of Malat1. Int. J. Mol. Sci. 2020, 21 (3), 1166. 10.3390/ijms21031166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen S.; Wei Y.; Zen C.; Xiong W.; Niu Y.; Zhao Y. Long Non-Coding RNA NEAT1 Promotes Bone Metastasis of Prostate Cancer through N6-Methyladenosine. Mol. Cancer 2020, 19 (1), 171. 10.1186/s12943-020-01293-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q.; Zhang S.-Y.; Zhao J.-F.; Han X.-G.; Wang H.-B.; Sun M.-L. HIF-1α or HOTTIP/CTCF Promotes Head and Neck Squamous Cell Carcinoma Progression and Drug Resistance by Targeting HOXA9. Mol. Ther.--Nucleic Acids 2020, 20, 164–175. 10.1016/j.omtn.2019.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M.; Déjardin J. Publisher Correction: Locus-Specific Chromatin Isolation. Nat. Rev. Mol. Cell Biol. 2020, 21 (5), 300. 10.1038/s41580-020-0222-3. [DOI] [PubMed] [Google Scholar]
- Stützer A.; Welp L. M.; Raabe M.; Sachsenberg T.; Kappert C.; Wulf A.; Lau A. M.; David S.-S.; Chernev A.; Kramer K.; Politis A.; Kohlbacher O.; Fischle W.; Urlaub H. Analysis of Protein-DNA Interactions in Chromatin by UV Induced Cross-Linking and Mass Spectrometry. Nat. Commun. 2020, 11 (1), 5250. 10.1038/s41467-020-19047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z.; Li J.; Liu T.; Zhang Z.; Qin W.; Qian X. A New Tandem Enrichment Strategy for the Simultaneous Profiling of O-GlcNAcylation and Phosphorylation in RNA-Binding Proteome. Analyst 2021, 146 (4), 1188–1197. 10.1039/D0AN02305A. [DOI] [PubMed] [Google Scholar]
- Zhang Z.; Liu T.; Dong H.; Li J.; Sun H.; Qian X.; Qin W. An RNA Tagging Approach for System-Wide RNA-Binding Proteome Profiling and Dynamics Investigation upon Transcription Inhibition. Nucleic Acids Res. 2021, 10.1093/nar/gkab156. [DOI] [PMC free article] [PubMed] [Google Scholar]