ABSTRACT
Eicosanoids are lipid-based signaling molecules that play a unique role in innate immune responses. The multiple types of eicosanoids, such as prostaglandins (PGs) and leukotrienes (LTs), allow the innate immune cells to respond rapidly to bacterial invaders. Bacterial pathogens alter cyclooxygenase (COX)-derived prostaglandins (PGs) in macrophages, such as PGE2 15d-PGJ2, and lipoxygenase (LOX)-derived leukotriene LTB4, which has chemotactic functions. The PG synthesis and secretion are regulated by substrate availability of arachidonic acid and by the COX-2 enzyme, and the expression of this protein is regulated at multiple levels, both transcriptionally and posttranscriptionally. Bacterial pathogens use virulence strategies such as type three secretion systems (T3SSs) to deliver virulence factors altering the expression of eicosanoid-specific biosynthetic enzymes, thereby modulating the host response to bacterial lipopolysaccharides (LPS). Recent advances have identified a novel role of eicosanoids in inflammasome activation during intracellular infection with bacterial pathogens. Specifically, PGE2 was found to enhance inflammasome activation, driving the formation of pore-induced intracellular traps (PITs), thus trapping bacteria from escaping the dying cell. Finally, eicosanoids and IL-1β released from macrophages are implicated in the efferocytosis of neighboring neutrophils. Neutrophils play an essential role in phagocytosing and degrading PITs and associated bacteria to restore homeostasis. This review focuses on the novel functions of host-derived eicosanoids in the host-pathogen interactions.
KEYWORDS: bacterial infection, inflammasome, inflammation, lipid metabolism, prostaglandins
INTRODUCTION
Nonsteroidal anti-inflammatory drugs (NSAIDs) are some of the most taken over-the-counter pain relievers. While these medications are taken to alleviate the symptoms of viral and bacterial infections (1, 2), NSAIDs can have severe side effects, including gastrointestinal and cardiovascular complications (3–6). Many NSAIDs act by inhibiting cyclooxygenase (COX)-1/-2 enzymes, which biosynthesize prostaglandins (PGs) and other bioactive lipids that downstream trigger pain or elevated temperature (1, 2, 5). PGs belong to the class of eicosanoids, which are immunomodulatory lipids generated via the oxidation of arachidonic acid (AA) or other polyunsaturated fatty acids (PUFAs) (7). Eicosanoids vary in function and structure and act on discrete cellular receptors designated EP receptors (8, 9). As a result, a wide variety of physiological responses are possible depending on the generation of specific eicosanoids and the expression of individual eicosanoid receptors (10–14). Eicosanoids have a complex function in the innate immune response: in some cases, eicosanoids support inflammation, such as enhancing pro-interleukin-1β (pro-IL-1β) biosynthesis, and in others, eicosanoids block inflammatory processes, for instance, by destabilizing tumor necrosis factor alpha (TNF-α) transcripts (15–19). Despite what is known, eicosanoid functions in bacterial infections remain elusive, and it appears that depending on the pathogen, unique mechanisms lead to the perturbations of the eicosanoid metabolism in the host cell. This review focuses on the function of distinct classes of eicosanoids such as PGE2, 15d-PGJ2, and LTA4/LTB4 in the conserved host response to bacterial LPS.
REGULATION OF EICOSANOID BIOSYNTHESIS
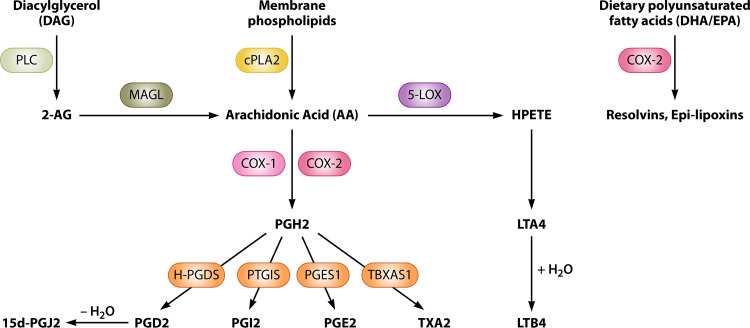
Arachidonic acid, AA, is a 20-carbon fatty acid, and it serves as a primary eicosanoid precursor in the cells. AA is not freely available in the cell, and it is instead associated with cellular membranes. However, under activation of Toll-like receptor 4 (TLR4) by bacterial LPS and other stimuli, membrane phospholipids are cleaved by cPLA2 to produce free AA (20–22) (Fig. 1). Host mitogen-activated protein kinases (MAPK) are rapidly phosphorylated in response to lipid-A of bacterial LPS binding to host TLR4, leading to activation of transcription factors such as nuclear factor-κB (NF-κβ) that drives pro-inflammatory cytokine biosynthesis (22, 23). The cPLA2 enzyme is activated via phosphorylation by host MAPKs at serine 505 residue, leading to activation of cPLA2 and consequent AA liberation (20, 24, 25). Cytosolic AA can then be further metabolized to oxygenated derivatives, eicosanoids, which include prostanoids (prostaglandins, PGs, and thromboxanes), leukotrienes (LTs), lipoxins (LXs), and epoxyeicosatrienoic acids (EETs) (26). In addition to AA, other PUFAs can also be used as eicosanoid precursors, including ω6-derived PUFAs from di-homo-γ-linolenic acid (DGLA) or ω3-derived PUFAs from α-linolenic acid (ALA), for instance, eicosapentaenoic acid (EPA). PGs and LTs are generated via the oxidation of AA by cyclooxygenase (COX) or lipo-oxygenase (LOX) enzymes, respectively.
FIG 1.
Biosynthesis of various prostaglandins and leukotrienes. Membrane phospholipids are converted to AA by the PLA2 enzyme. Alternatively, AA can be synthesized from diacylglycerol (DAG), where DAG is converted by PLC to 2-AG intermediate, followed by MAGL degradation of 2-AG to AA. AA serves as a substrate of COX-1 and COX-2, which convert AA to PGH2. Alternatively, AA is used by the 5-LOX enzyme to produce leukotrienes. PGH2 is converted to PGE2 by prostaglandin E synthase 1 (PGES-1) or PGD2 by hematopoietic prostaglandin D synthase (H-PGDS). PGD2 undergoes a dehydration reaction to form 15d-PGJ2. LTA4 is derived from 5-LOX generated HPETE. LTA4 can be processed further to LTB4 via a hydrolysis reaction. Various other eicosanoids, such as resolvins and other lipoxins, are generated from the oxidation of polyunsaturated fatty acids other than AA, such as DHA and EPA.
Two different cyclooxygenase enzymes are responsible for converting AA into the various prostaglandins, COX-1 and COX-2. Human COX-1 and COX-2 are homodimers and share 60% sequence similarity (27). These enzymes contain two different active sites, a peroxidase and a cyclooxygenase active site, flanking a central heme group (27, 28). The peroxidase center catalyzes the removal of two electrons from Tyr-385 residue in the cyclooxygenase active site to produce a tyrosyl radical that can catalyze the cyclooxygenase reaction. Due to their functions in regulating inflammation, COX enzymes are targets of nonsteroidal anti-inflammatory drugs (NSAIDs) such as aspirin or ibuprofen, which function by inhibiting pro-inflammatory PG synthesis and thus result in decreased pain, inflammation, and fever (29). Different NSAIDs have distinct selectivity for COX isoenzymes. For example, aspirin covalently modifies Ser-530 residue in the active site of both COX isoenzymes, preventing PG synthesis, while other COX inhibitors such as ibuprofen are reversible inhibitors of both COX enzyme functions (reviewed in reference 30). In addition, while aspirin and ibuprofen are nonselective COX inhibitors, celecoxib and etoricoxib have high selectivity toward COX-2 and therefore have reduced adverse effects related to gastrointestinal tract (31, 32). Under biological conditions, AA is first reduced to the hydroperoxy arachidonate metabolite PGG2 (33) and then further reduced to PGH2 by the peroxidase activity of COX-1/2 (34). PGH2 is converted rapidly to biologically active prostanoids by specific synthases and acts on discrete receptors designated EP receptors, which exist at various levels in particular tissues (35–38). These prostanoids include prostaglandin E2 (PGE2) synthesized by prostaglandin E synthase (PGES), prostacyclin (PGI2) synthesized by prostaglandin I synthase (PGIS), prostaglandin D2 (PGD2) synthesized by prostaglandin D synthase (PGDS), prostaglandin F2α (PGF2α) synthesized by prostaglandin F synthase (PGFS), and thromboxane A2 (TXA2) generated by thromboxane A synthase (TXAS) (7). Further, an anti-inflammatory prostaglandin 15d-PGJ2 is synthesized from PGD2 via its dehydration to PGJ2, which is also converted to 15d-PGJ2 or Δ-12-PGJ2 (7).
Leukotrienes are generated from 5-lipoxygenase (arachidonate-5-lipoxygenase; 5-LOX)-mediated conversion of AA to 5-hydroperoxyeicosatetraenoic acid (5-HPETE) intermediate (39), which is then rapidly reduced to 5-hydroxyeicosatetraenoic acid (5-HETE) (Fig. 1). However, 5-HPETE can be alternatively converted by 5-LOX to an unstable structure called leukotriene A4 (LTA4), which can be further modified to generate leukotriene B4 (LTB4) by LTA4 hydrolase, or to leukotriene C4 (LTC4) by LTC4 synthase, respectively. LTB4 and LTC4 molecules are mainly produced by immune cells. Following the export of these LTs, LTC4 is metabolized to LTD4 by gamma-glutamyl transpeptidase, while LTD4 can be modified to LTE4 by cysteinyl glycinase.
The availability of AA and the cell’s physiological state dictate the LT and PG levels at the same time since COX and LOX enzymes both utilize AA as a substrate for PG and LT biosynthesis, respectively. Hence, a delicate balance between PG and LT synthesis is maintained upon various stimuli, resulting in specific physiological responses. Bacterial LPS and subsequent inflammation associated with these infections serve as such stimuli contributing to COX and LOX activation. During bacterial infection, pathogen-associated molecular patterns (PAMPs) are recognized by membrane-bound Toll-like receptors (TLRs) or other pattern recognition receptors (PRRs), as well as by intracellular cytosolic nucleotide oligomerization domain (NOD)-like receptors (NLRs) (40). Pro-inflammatory signaling is not always desirable for a pathogen. Certain bacteria aim to evade the immune system by inducing changes to the bacterial lipopolysaccharide (LPS), a component of Gram-negative bacteria’s outer membrane, impairing TLR signaling (41–44). LPS is a potent inducer of PGE2 biosynthesis and is involved in a wide variety of bacterial infections (Table 1). Gram-negative bacterial LPS comprises multiple elements, including lipid-A, inner core, outer core, and O antigen components. Lipid-A is a potent endotoxin that binds specifically to TLR4 and, along with the help of MyD88, activates transcriptional factors such as NF-κB to upregulate pro-inflammatory cytokine production as well as COX-2 expression, the major biosynthetic enzyme of PGE2 (21–23). The binding of lipid-A to TLR4 activates MyD88, which results in phosphorylation of host mitogen-activated protein kinases (MAPKs), resulting in the subsequent phosphorylation of IκB kinase (IKK), which results in derepression of NF-κB, allowing its translocation to the nucleus to upregulate gene transcription (23). It is unclear if PGE2 biosynthesis in response to LPS-TLR4 is due to lipid-A or other parts of bacterial LPS since rough or smooth mutants of Salmonella enterica serovar Typhimurium lacking O antigen or characterized by an impaired lipid-A biosynthesis all induce PGE2 biosynthesis (19). Chemical inhibitors of the MAPK cascade during LPS treatment alone or several Gram-negative bacterial infection models have been reported to abrogate PGE2 biosynthesis, potentially indicating a conserved host response mechanism of PGE2 biosynthesis in response to Gram-negative bacterial infection (45). In contrast, Gram-positive bacteria lack LPS, but they contain teichoic and lipoteichoic acids as well as a thick peptidoglycan layer, and these pathogens typically do not induce large levels of PGE2 biosynthesis except for Staphylococcus aureus infection (46, 47).
TABLE 1.
The effect of bacterial infections on eicosanoid production and function of eicosanoids in bacterial infections
| Organism | The effect of infection on eicosanoids | The effect of eicosanoids on infection | Reference(s) |
|---|---|---|---|
| Burkholderia pseudomallei | Increase in PGE2 | PGE2 has detrimental effects on the survival of infected mice | 155 |
| Campylobacter jejuni | Increase in PGE2 | 156 | |
| Escherichia coli | Increase in PGE2 | PGE2 increase by colibactin-producing E. coli functions in colon cancer tumorigenesis; inhibition of COX-2 enhances infection with extraintestinal E. coli | 51, 134, 137 |
| Francisella tularensis | Increase in PGE2 | PGE2 has detrimental effects on the infected host via downregulation of Th1 immunity | 157, 158 |
| Helicobacter pylori | Increase in PGE2 | PGE2 contributes to chronic inflammation but also cancer cell growth and proliferation | 130, 131, 133 |
| Klebsiella pneumoniae | LTB4 promotes phagocytosis, enhances ROS-dependent NADPH oxidase activation | 111 | |
| Legionella pneumophila | Increase in PGE2 | 159 | |
| Mycobacterium tuberculosis | Degradation of LTB4 to its inactive form 12-oxo-LTB4; decrease in PGE2 | LTA4H deficiency downregulates TNF-α; COX-2 inhibition leading to reduced PGE2 enhances phagocytosis of M. tuberculosis | 109, 124, 125 |
| Pseudomonas aeruginosa | Increase in PGE2 | COX-2-deficient mice have increased survival rates during infection | 160–162 |
| Salmonella enterica | Increase in PGE2 by SPI-2 T3SS factors | PGE2 affects macrophage polarization | 19, 57, 123 |
| Shigella flexneri | PGE2 levels are elevated in the stool of infected patients and in rabbit ileal loops | 52, 148, 149 | |
| Staphylococcus aureus | Increase in PGE2 | PGE2 promotes the growth and biofilm formation of S. aureus and enhances the attachment of bacterium to the human fibronectin | 47 |
| Yersinia enterocolitica | Increase in PGE2 but downregulation of PGE2 by T3SS virulence factors | PGE2 affects macrophage polarization | 19 |
The immunomodulatory effects of PGs and LTs depend on the expression of cognate receptors on recipient cells, which varies depending on the cell population and the cells’ physiological state. For example, macrophages generate a wide variety of PGs and LTs in response to TLR stimuli, but the biosynthesized levels of specific eicosanoids change across macrophages from different sources and under distinct physiological states (12). While COX-1 is constitutively expressed in almost all tissue types, COX-2 is inducible by inflammatory cytokines and TLR activation (48). The 5′-UTR (untranslated region) of the COX-2 gene includes binding sites for several transcription factors such as NF-κB, AP-1, CRE, and others (49). COX-2 is also regulated by posttranscriptional processes due to instability sequences in the 3′-UTR. RNA-binding proteins that recognize AU-rich elements (AREs) are hypothesized to control COX-2 mRNA stability, such as CUGBP2 or HuR, which binds to COX-2 mRNA and prevents translation (50). Mitogenic inhibitors can also influence COX-2 expression in a protein kinase C-p38 MAPK (mitogen-activated protein kinase)-dependent manner (41–44). TLR4 signaling is in particular essential for COX-2 upregulation. COX-2 and PGE2 biosynthesis is upregulated in response to a wide variety of Gram-negative pathogens, including Salmonella enterica serovar Typhimurium, Shigella flexneri, Helicobacter pylori, Yersinia enterocolitica, and Escherichia coli, which will be covered in following sections (14, 19, 50–53).
The leukotriene biosynthesis is also enzymatically controlled by several proteins. Specific LOX enzymes are present in different cells to metabolize AA, including 5-LOX in leukocytes, 12-LOX in platelets, or 15-LOX in endothelial cells. LTB4 is a chemoattractant for polymorphonuclear leukocytes (PMNs), such as neutrophils and eosinophils (54, 55). Accordingly, 5-LOX and LTB4 are heavily regulated in immune cells. For instance, pro-inflammatory or M1 macrophages secrete high levels of LTB4, whereas anti-inflammatory M2 macrophages express lipoxins, which aid in immune suppression. 5-LOX is inactive in resting cells and becomes activated by increasing intracellular calcium or phosphorylation (56, 57).
THE FUNCTION OF EICOSANOIDS IN THE REGULATION OF INFLAMMATORY PROCESSES IN RESPONSE TO BACTERIAL INFECTIONS
Eicosanoids regulate pro-inflammatory processes, often associated with bacterial infections. Bacterial products (PAMPs) and danger-associated molecular patterns (DAMPs) are recognized by both intracellular and extracellular receptors expressed by the host cell, such as macrophages. PAMPs include LPS, flagellin, and the needle and rod protein of the bacterial T3SS, while DAMPs include elevated ATP levels or K+ influx (58–60). These components are recognized by typically extracellular membrane-bound TLRs or cytosolic NOD-like receptors (NLR). The initial stimulus triggers the oligomerization of NLRs with an adapter protein, ASC, to form the inflammasome (61). Five well-studied receptors capable of creating inflammasomes in response to bacterial infection and noncanonical inflammasomes that detect bacterial LPS and activate caspase-11 are NLRP1, NLRP3, NLRC4, AIM2, and pyrin (61). For example, during S. Typhimurium infection, NLRP3 and NLRC4 inflammasomes are upregulated and activated to produce a large quantity of IL-1β (62).
In contrast, other pathogens such as Mycobacterium tuberculosis and Yersinia pestis actively repress inflammasome formation by employing virulence factors and impairing IL-1β signaling during infection (63, 64). Inflammasomes are a hallmark of pyroptosis, a form of pro-inflammatory programmed cell death that leads to the rapid clearance of bacteria by removing intracellular replication niches and enhancing chemotaxis of neutrophils to the site of infection (51). Recent studies have investigated PGs’ role in modulating the activity of inflammasomes and will be discussed below. Relatively few categories of eicosanoids have been extensively studied in the context of infections, whereas PGs and LTs are some of the best-characterized eicosanoids due to their roles in inflammatory conditions, such as arthritis and bacterial sepsis (65). Below, we will discuss specific functions of eicosanoids in inflammasome activation, phagocytosis, and efferocytosis, giving specific examples of bacteria that modulate the biosynthesis of eicosanoids. Finally, we will review the outcomes of eicosanoid biosynthesis changes in these infections, focusing on gastrointestinal pathogens.
The role of PGE2 and 15d-PGJ2 during inflammasome activation and inflammation.
PGE2 is one of the most significant eicosanoids released during infection with Gram-negative pathogens (Table 1). PGE2 stimulates the production of pro-inflammatory cytokines such as IL-1β, IL-23/17, or IL-8 while limiting inflammation by controlling TNF-α levels and inducing IL-10 secretion (19, 66). These unique functions of PGE2 in the stimulation of specific cytokine profiles emphasize the importance of this molecule in maintaining homeostasis during infection. Bacterial components such as LPS or virulence factors can induce transcriptional changes that enhance COX-2 expression, resulting in the biosynthesis and secretion of PGE2 (19, 21). PGE2 acts on neighboring immune cells by binding to discrete G-protein coupled receptors, designated EP receptors 1 to 4, each with different downstream consequences in response to activation (as extensively reviewed in reference 8). Once bound, the G-linked receptors control levels of secondary messengers in the cell, such as cAMP, IP3, and Ca2+, which induce transcriptional changes via NF-κB or C-JUN (67–70). Gene regulation by EP receptor signaling has a wide range of effects on host cells, including shifts in macrophage polarization or downregulation of bactericidal capabilities in neutrophils by downregulating NADPH oxidase and reducing the generation of reactive oxygen species (ROS) (19, 71–73).
PGE2 has also been reported as having a pro-inflammatory role in inflammasome activation in pyroptosis that is often present during bacterial infections. For instance, PGE2 increases inflammation by leading to an elevated IL-1β expression, which is subsequently cleft by caspase-1 and secreted and in this way can reach neighboring cells (15, 74). The binding of IL-1β to IL-1R also increases the expression of the COX-2 enzyme, creating a positive feedback loop by stimulating host MAPK-extracellular signal-regulated kinase (ERK) signaling-mediated transcriptional changes (15). Additionally, increased PGE2 biosynthesis is associated with decreased phagocytosis (67). The killing ability of macrophages is also reduced by PGE2, for instance, by diminished nitric oxide synthesis in macrophages, leading to larger bacterial loads during some infections, such as Mycobacterium tuberculosis (75). As such, PGE2 has become increasingly acknowledged as an essential factor in determining the fate of the host during bacterial infection and has made PGE2 an appealing option for antimicrobial treatments targeting COX-2 or EP receptor inhibition (76–79). PGE2 signaling via the EP3 receptor increases activation of inflammasome in some infections. Specifically, PGE2 signaling through the EP3 receptor leads to activation of the NLRC4 inflammasome during infection with Anaplasma phagocytophilum, a bacterium that causes anaplasmosis (80).
Although EP2 and EP4 receptors also have been shown to be involved in inflammasome activation, reports of PGE2 function in mediating inflammasome activation through EP2 and EP4 have been conflicting. For example, PGE2 inhibits NLRP3 inflammasome by signaling through the EP4 receptor (69). The EP4 receptor activates adenylate cyclase, increasing intracellular cAMP levels via phosphoinositide 3-kinase (PI3K), leading to the downregulation of NLRP3 (16). Interestingly, EP2 receptor agonists failed to produce a similar result in dampening NLRP3 activation, even though PGE2 has a much stronger affinity for the EP2 receptor. The EP2 receptor is more strongly associated with the cAMP production by adenylate cyclase than is the EP4 receptor, thus leading to more considerable changes in cAMP intracellular concentrations (81). In zebrafish, macrophage uptake of apoptotic neutrophils was found to be required for the resolution of inflammation. In this model, PGE2 acts via EP4 receptors to drive inflammation resolution by reverse migration (82). Other reports have suggested that PGE2 and IL-1β form a positive feedback loop, which may be downstream of EP2 or EP4 (79). A receptor-specific response to PGE2 has been reported multiple times, emphasizing that the functions of EP2 and EP4 receptors have to be studied more extensively (70, 79, 83, 84).
Apart from PGE2, there are other PGs that affect inflammasome activity and inflammation, such as 15d-PGJ2. 15d-PGJ2 is a cyclopentane prostaglandin formed via the dehydration of PGD2 and has been shown to activate PPARγ promoting adipocyte and monocyte differentiation (85–87). 15d-PGJ2 also inhibits inflammatory gene expressions such as inducible NO synthase and tumor necrosis factor α (TNF-α) (88–90). Additionally, 15d-PGJ2 inhibits multiple steps in the NF-κB signaling pathway by directly modifying critical cysteine residues in IKK-β kinase and the DNA-binding domains of NF-κB subunits (91). 15d-PGJ2 also inhibits inflammasome activation by inhibiting caspase-1 activation by the NLRP1 and NLRP3 inflammasomes in response to anthrax lethal toxin in a murine infection model (92). The negative effect of 15d-PGJ2 on inflammasome activation is independent of canonical signaling pathways activated by 15d-PGJ2. Instead, 15d-PGJ2 induces a cell state that does not process caspase-1 to its active form, indicating a unique form of inflammasome inhibition (92). Additionally, exogenous administration of 15d-PGJ2 appears to be anti-inflammatory in additional mouse models by inhibiting NLRP3-dependent peritonitis (93).
The function of 15d-PGJ2 during bacterial infection models has become a new interest among researchers considering its ability to interact with PPARγ. In a brain abscess inflammation model, microglial activation by Staphylococcus aureus (S. aureus), one of the main etiologic agents of brain abscess in humans, was selectively attenuated by 15d-PGJ2 (94). 15d-PGJ2 inhibited increases in pro-inflammatory cytokines IL-1β and TNF-α, immunological markers such as TLR2, CD14, MHC class II, CD40 expression, and a variety of chemokines in microglial infected with S. aureus (94). Another study found similar anti-inflammatory effects for 15d-PGJ2 in an astrocyte infection model of S. aureus but interestingly was able to function independently of PPARγ since inhibition of inflammation was still observed in PPARγ-deficient astrocytes (95). 115d-PGJ2 also has been reported to play an anti-inflammatory role in several sepsis models, including Salmonella enterica serovar Typhimurium (S. Typhimurium) and Escherichia coli infections (96–98). In vivo infection with S. Typhimurium led to increased 15d-PGJ2 biosynthesis in the spleen and livers of infected mice (99), and follow-up studies using RAW macrophages, bone marrow-derived macrophages, and J774 cells indicate that 15d-PGJ2 is essential for reducing bacterial colonization (100). 15d-PGJ2 suppresses RANTES expression by inhibited NADPH oxidase activation in Helicobacter pylori-infected gastric epithelial cells (101). Ultimately, these studies suggest an important anti-inflammatory role for 15d-PGJ2 during inflammasome activation and inflammation, which indicates a potential use of 15d-PGJ2 in a variety of infectious diseases.
Functions of LTs in phagocytosis and efferocytosis.
Among the LTs, LTB4 is most involved in the early phases of inflammation during infection. For instance, these eicosanoids act as factors that recruit neutrophils to phagocytize and degrade invading pathogens (102). LTB4 is a potent activator and chemoattractant of neutrophils and is biosynthesized and secreted primarily by polymorphonuclear leukocytes in macrophages and monocytes in response to bacterial infection (103, 104). LTB4 works with other chemokine gradients such as C5a to attract neutrophils to the site of infection (105). LTB4 signaling, through its cognate receptor BLT1, is critical for neutrophil migration to the foci of damage (106). For example, in Wistar rats, neutrophil migration induced by IL-1β is lost when animals are treated with MK866, a LOX-5 inhibitor (107). In a methicillin-resistant S. aureus (MRSA) skin infection model, LTB4 promotes cutaneous abscess formation absent in LTB4R1-deficient mice (108). Interestingly, an ointment containing LTB4 acted synergistically with the antibiotic mupirocin in lowering MRSA bacterial numbers in cutaneous abscesses, thereby indicating LTB4 as a novel potential therapeutic option (108). In the zebrafish model of Mycobacterium tuberculosis infection, LTB4 appears to be detrimental to host defense (109). LTA4H (which converts LTA4 to LTB4) overexpression results in increased TNF-α secretion and increased M. tuberculosis bacterial numbers (110). LTB4 also plays an important role in promoting phagocytosis (14, 104). Exogenous LTB4 restores the phagocytic ability of 5-LOX-deficient neutrophils for serum opsonized Klebsiella pneumoniae (111). Once phagocytized, neutrophils utilize reactive oxygen species (ROS) to kill invading pathogens. LTB4 enhances ROS-dependent NADPH oxidase activation via phosphorylation of the cytosolic subunit p47phox (112). LTB4 also enhances MyD88 expression, which is required for NF-κB-mediated activation of pro-inflammatory cytokines (23). Upon PAMP or DAMP recognition, TLRs recruit Toll/IL-1R domain (TIR) adaptor molecules such as MyD88 or TRIF to initiate downstream signaling events, dictating the type of host response. LPS recognition and binding to TLR4 induce MyD88- and TRIF-dependent signaling pathways involving multiple host-activated mitogen kinases (MAPKs) such as MEK, JNK, and p38 kinases, which function by altering translocation of transcription factors to the nucleus to bind to and alter gene transcription during MyD88-dependent responses. For example, activation of host MyD88-dependent MAPKs results in derepression of NF-κB signaling by inhibiting IKK repression of NF-κB by direct phosphorylation of IKK, allowing NF-κB to migrate to the nucleus and initiate transcription of pro-inflammatory cytokines such as IL-1β and TNF-α (NF-κB has been extensively reviewed, e.g., see reference 113). These cytokines are important for the host immune response to invasive pathogens.
LTs are critical for efferocytosis of neutrophils to resolve inflammation, but paradoxically high levels of LTB4 are associated with chronic inflammatory disorders such as type-1 and type-2 diabetes or arthritis (114–116). Paradoxically, LTB4 plays a critical role in bacterial clearance but also contributes to long-term inflammatory disorders. Aberrant leukotriene production is hypothesized to alter immune cell function, which results in the hyperproductivity of inflammatory cytokines such as TNF-α, IL-8, and Il-6 during chronic inflammation (117–119). A genetic screen discovered that mutants of the Ita4h locus in zebrafish resulted in altered LT4AH expression, which caused increased susceptibility to Mycobacterium marinum infection (120). LTA4H deficiency leads to anti-inflammatory activity in the host, which happens due to aberrant TNF-α production, damaging the host since a moderate level of inflammation is typically required to resolve infection (120). Accordingly, bacterial numbers were lower in wild-type zebrafish than in LTA4H-deficient zebrafish (120). In an additional study, Streptococcus iniae infection of zebrafish with deficiencies in LTB4 production due to loss of LTB4H expression resulted in a loss of macrophage aggregation and increased susceptibility to disease (121). The resulting phenotype was reversible when exogenous LTB4 was restored, indicating the importance of functional LTB4 during bacterial infection (120). LTB4 signaling also contributes to bacterial sepsis during endotoxic shock (122). For instance, BLT1/2-dependent signaling pathways mediated the expression of IL-17, IL-6, and IL-1β, key cytokines for the development of this endotoxic shock, via NF-κB activation in the LPS-induced endotoxic shock mouse model (122).
Modulation of host eicosanoids by bacterial agents causing gastrointestinal diseases.
PGE2 is rapidly generated in response to infections with Gram-negative bacteria such as Salmonella enterica, Escherichia coli, Chlamydia trachomatis, Legionella pneumophila (19, 46, 51, 57, 123), acid-fast bacterium Mycobacterium tuberculosis (109, 124), and the Gram-positive bacteria Staphylococcus aureus (46, 47). Some pathogens,0 such as Yersinia enterocolitica and Mycobacterium tuberculosis, have mechanisms to counteract PGE2 biosyntheses, (19, 125). While mounting evidence suggests that bacteria can use virulence factors to increase or decrease PGE2 production, specific mechanisms that regulate eicosanoid levels are mostly unknown. Although several pathogens have been reported to stimulate eicosanoid biosynthesis during infection (Table 1), in this review, we focus on pathogens that primarily cause gastrointestinal diseases (124).
Helicobacter pylori. Helicobacter pylori is among the most prevalent infectious disease-causing pathogens, being present in approximately half of the world population, although this microorganism often persists in the host without inducing disease (126). H. pylori uses a variety of virulence factors, including outer membrane proteins (OMPs) such as OipA, toxins like CagA, and a type 4 secretion system (T4SS), and thus induces severe inflammation and damage in the host (127). However, H. pylori is also proposed to have favorable immunomodulatory properties in the case of asthma and skin allergies by reducing inflammation (128). H. pylori has recently been postulated to activate the NLRP3 inflammasome in murine bone marrow-derived dendritic cells (129). H. pylori LPS signals through the TLR4-MyD88 axis to drive IL-1β transcription, which in turn results in increases IL-1β secretion upon inflammasome activation in macrophages. In dendritic cells, H. pylori urease B subunit (UreB) mutant was unable to trigger inflammasome activation (129). What triggers the oligomerization of the NLRs to form the inflammasome in H. pylori infection is still unclear. H. pylori infection also induces COX-2 expression in gastric epithelial cells, where it first comes in contact with the stomach by activating the epidermal growth factor receptor (130) in a T4SS-dependent fashion, leading to robust production of PGE2, which contributes to chronic inflammation. PGE2 supports cancer cell growth and proliferation, which are associated with H. pylori infection. Treatment with COX-2/EGFR inhibitors leads to a decrease in gastric tumorigenesis (131). Moreover, 15d-PGJ2 is reportedly downregulated upon infection with H. pylori and even more so in gastric tumors associated with H. pylori infection (132). These observations suggest that control of homeostatic imbalance between the PGE2 and 15d-PGJ2 levels could counteract gastric cancer tumorigenesis. Other studies in monocytes and macrophages have indicated an important role of host microRNA-155 in increasing COX-2, TNF-α, and IL-23 during H. pylori infection (133). The mechanism by which H. pylori induces miRNA-155 is unknown.
Escherichia coli. E. coli is one of the most abundant species of bacteria found in the gut of mammals. Depending on the species, E. coli has various pathogenicity levels, from the highly pathogenic pedestal-forming E. coli (EPEC) or uropathogenic E. coli (UPEC) to commensal E. coli found in the gut of most mammals. EPEC, UPEC, and commensal E. coli induce PGE2 secretion, but more invasive strains contribute a more robust PGE2 response (134). Using a T3SS, EPEC induces COX-2 expression via the virulence protein EspT independent of LPS or passive recognition by the host in an ERK1/2-dependent manner (51). Some E. coli strains belonging to the B2 phylogroup and producing colibactin are capable of inducing COX-2 and are hypothesized to have a role in colon cancer tumorigenesis (135). Macrophages are heavily involved in tumor infiltration and removal (136). Macrophages affected by colibactin-producing E. coli enhance the tumorigenesis of colon cancers and are more resistant to killing by THP-1 macrophages (135).
Extraintestinal pathogenic Escherichia coli (ExPEC) can colonize sites outside the gastrointestinal tract, and as a comparison to EPEC, ExPEC infection also upregulates COX-2, which happens in the TLR4-dependent manner (137). However, in ExPEC infection, COX-2 inhibition enhances the infection, leading to an increased bacterial burden in blood, liver, lung, spleen, and brain (137). The mechanism by which COX-2 downregulation has a deleterious effect on the host depends on autophagy, which is a process that the host can direct against bacterial pathogens, such as Salmonella (138). In ExPEC infection, COX-2 inhibition leads to a decrease in macrophage autophagy, and inhibition of the autophagy process leads to increased bacterial survival in the cells. In contrast, enhanced autophagy leads to decreased bacterial survival in macrophages. Hence, COX-2 upregulation can prime the activation of autophagy in macrophages, thereby facilitating clearance of this bacterium (137).
Uropathogenic E. coli also increases COX-2 transcription and secretion of PGE2 during urinary tract infections (UTIs) (139). The type 1 fimbriae of uropathogenic Escherichia coli (UPEC) have been described as important for establishing bladder infections and urinary tract infections (UTI) (139). Induction of COX-2 in infected human bladder 5637 cells by UPEC is mediated via host TLR4-dependent activation of MAPKs such as JNK, p38, and ERK, and inhibition of transcription factors AP-1 and NF-κB resulted in abrogation of COX-2 promoter activity and expression (139).
Salmonella enterica. S. enterica Typhimurium is a foodborne pathogen which causes severe inflammation upon infection. Salmonella’s virulence originates in at least two different Salmonella pathogenicity islands (SPI) designated SPI-1 and SPI-2, which control the infection process by encoded protein effectors. The T3SS of S. Typhimurium injects host macrophages with virulence factors that induce COX-2 expression. For example, individual deletions of genes carried by the SPI-2 revealed SpiC as a significant contributor to COX-2-based prostaglandin biosynthesis via activation of the ERK1/2 pathway (57). Interestingly, COX-2 inhibition does not impair Salmonella’s ability to invade and replicate in host cells, but it enhances host survival during later infection stages. Our group identified an essential role for PGE2 production in inflammasome activation with S. Typhimurium (19). Consistent with other studies, infection with wild-type S. Typhimurium leads to an increase in PGE2 biosynthesis and upregulation of COX-2 compared to infection with ssaV-deficient mutant, which has an impaired SPI-2. PGE2 priming of macrophages results in a robust inflammasome upregulation in macrophages infected with S. Typhimurium, signified by an enhanced IL-1β secretion. This PGE2-mediated effect on inflammasome in infected cells is most likely a result of PGE2 signaling via EP4 receptor, which effectively leads to increased transcription of IL-1β in human THP-1 macrophages. Following the inflammasome oligomerization, a greater pool of immature IL-1β is converted to mature IL-1β, which drives the inflammation and may play a beneficial role for the pathogen during later stages of infection (19). As another outcome, PGE2 also alters macrophage polarization following the infection with Salmonella, suggesting that other functions of macrophages are also changed by PGE2 (19).
Another PG that is enhanced by Salmonella Typhimurium infection is 15-deoxy-PGJ2 (15d-PGJ2). This lipid is released from macrophages during infection with Salmonella Typhimurium and exerts anti-inflammatory effects. 15d-PGJ2 acts on two different PDG2 receptors, DP1 and DP2, with a preference for the DP2 receptor (140), which is found primarily on Th2 cells, T cytotoxic cells, eosinophils, and basophils and plays a role in chemotactic migration (141). Interestingly, 15d-PGJ2 is known to be active in the cytosol, binding to the intranuclear receptor PPARγ leading to reduced inflammatory cytokine gene expression (142). Many of the effects attributed to PPARγ activation contribute to the production of anti-inflammatory cytokines in macrophages. For example, 15d-PGJ2 inhibits TNF-α, IL-1β, and inducible nitric oxide synthase in vitro (88).
Yersinia enterocolitica. Y. enterocolitica is a foodborne pathogen that resides almost exclusively in the lymphoid tissue. This bacterium resists the host immune system’s activation by virulence factors encoded within its 70-kb virulence plasmid designated pYV. Yersinia’s T3SS encodes several adhesins, including YadA and virulence proteins designated Yops (143). Elevation of the temperature to 37°C increases the expression of genes involved in pathogenesis such as the T3SS, loss of motility by flagellar downregulation, and upregulation of Yops (144).
The pYV virulence plasmid plays a crucial role in modulating eicosanoid responses to infection (19). While COX-2 is slightly increased upon infection with Y. enterocolitica, the Y. enterocolitica devoid of pYV virulence plasmid leads to an extensive induction of COX-2 mRNA in infected macrophages, suggesting that some yet unidentified effectors encoded by Yersinia play a role in COX-2 downregulation (19). The function of this downregulation of COX-2 signaling by Y. enterocolitica is unclear. The baseline activation of COX-2 by wild-type Y. enterocolitica may be due to LPS-TLR-4 signaling, which is reduced in the case of Y. pestis infection due to changes in LPS composition but appears active in Y. enterocolitica infection (145). PGE2 is also secreted from macrophages in response to Y. enterocolitica, which depends on the presence or absence of the pYV virulence plasmid (19). Triple-quadruple mass spectrometry analysis of media from macrophages infected with Y. enterocolitica revealed large quantities of PGE2 in response to a pYV mutant compared to those in response to the wild type. Exogenous addition of PGE2 before infection with Y. enterocolitica resulted in a strong inflammasome response via EP4 stimulation and decreased bacterial load 2, 24, and 48 h postinfection (19). In summary, Y. enterocolitica downregulated PGE2 biosynthesis, which might be a possible pathogen strategy to skew the macrophage function.
Shigella flexneri. Another pathogen proposed to alter eicosanoid biosynthesis upon infection is Shigella flexneri. S. flexneri is a highly inflammatory intracellular pathogen that tends to cause bloody stool and diarrhea during shigellosis. S. flexneri expresses virulence genes from a conserved virulence plasmid and encodes multiple secretion systems, including the T3SS (146). Both pathogens activate the inflammasome and are known to release IL-1β and IL-18. However, S. flexneri is also known to inhibit the release of pro-inflammatory cytokines, such as IL-8, by inhibiting the NF-κB pathway (147). Due to the conserved nature of the T3SS and inflammasome activation, it is unsurprising that PGE2 levels are elevated in the stools of patients suffering from shigellosis T3SSs (146). Both pathogens activate the inflammasome and are known to release IL-1β and IL-18. However, S. flexneri is also known to inhibit the release of pro-inflammatory cytokines, such as IL-8, by inhibiting the NF-κB pathway (147). Due to the conserved nature of the T3SS and inflammasome activation, it is unsurprising that PGE2 levels are elevated in the stool of patients suffering from shigellosis (148). Another study performed by Sansonetti et al. also indicated that COX-2 transcripts are strongly induced in the rabbit ileal loops infected with S. flexneri (149). It has also been shown that S. flexneri leads to the upregulation of COX-2 transcription in Peyer’s patches of the infected rabbit ligated intestinal loops and that this upregulation of COX-2 mRNA is abrogated by secretory IgA (SIgA) (52). HeLa cells infected with ΔospF S. flexneri exhibit increased COX-2 mRNA compared to that of cells infected with wild-type strains (150). OspF/OspB are involved in the activation and sustained phosphorylation of host MAPKs, including p38, JNK, and ERK signaling pathways, in response to LPS-TLR4 interactions during infection, which lead to cPLA2 activation. cPLA2 activation is the first step in PGE2 biosynthesis and is necessary for AA synthesis prior to conversion to PGE2 by COX-2 (151).
NOVEL ROLES FOR PGE2 AND LTB4 DURING BACTERIA-DRIVEN PYROPTOSIS: PORE-INDUCED INTRACELLULAR TRAPS
During pyroptosis, the inflammasome activates caspase-1, which activates other enzymes via proteolytic cleavage. One such target, gasdermin D (GSDMD), is cleaved to produce an N-terminal product that triggers inflammatory cell death. The N-terminal product oligomerizes into the cell membrane to form pores visible by microscopy (152). High affinity for negatively charged lipid heads allows the pore to be formed via binding of GSDMD and subsequent insertion. The pore formation disrupts the sodium-potassium gradient across the plasma membrane, resulting in a massive influx of sodium into the cell, causing intracellular changes and eventual cell death.
One of the most recent discoveries pertaining to pyroptosis is that this form of cellular death is associated with the formation of cell corpse consisting of insoluble components, which trap bacteria inside the macrophage. The formation of these pore-induced intracellular traps (PITs) is hypothesized to destroy the replicative niche the pathogen is trying to use and immobilize the invading bacteria. Neutrophils are then recruited via PGs and LTs to phagocytize and degrade the dead cell, including the trapped bacteria (153). It appears that pyroptosis damages the bacteria but is not sufficient for the complete clearance of the pathogens. The efferocytosis of neighboring neutrophils by PGs and LTs is critical in controlling bacterial escape from PITs.
While undergoing pyroptosis, large amounts of eicosanoids are released from cells, such as PGE2 (7) and the chemotactic leukotriene LTB4, hypothesized to play a role in neutrophil migration toward newly formed PITs. Studies done by Jorgenson et al. have demonstrated that neutrophil migration is influenced by inflammasome activation products like IL-1β and IL-18 (154). By using an engineered strain of S. Typhimurium expressing flagellin during intracellular infection, it was determined that IL-1β and IL-18 work cooperatively to clear the bacterium in vivo. Interestingly, inhibition of individual COX enzymes or leukotriene generating lipooxygenases (LOXs) did not affect bacterial clearance, while simultaneous inhibition of both enzymes resulted in impaired clearance. It is possible that optimal neutrophil migration is not specific to essential prostaglandins or leukotrienes and that the loss of one of these lipids is complemented by the other. Future studies likely will further investigate the mechanism behind PIT-mediated clearance by neutrophils and roles for eicosanoids in this process.
CONCLUSION AND FUTURE DIRECTIONS
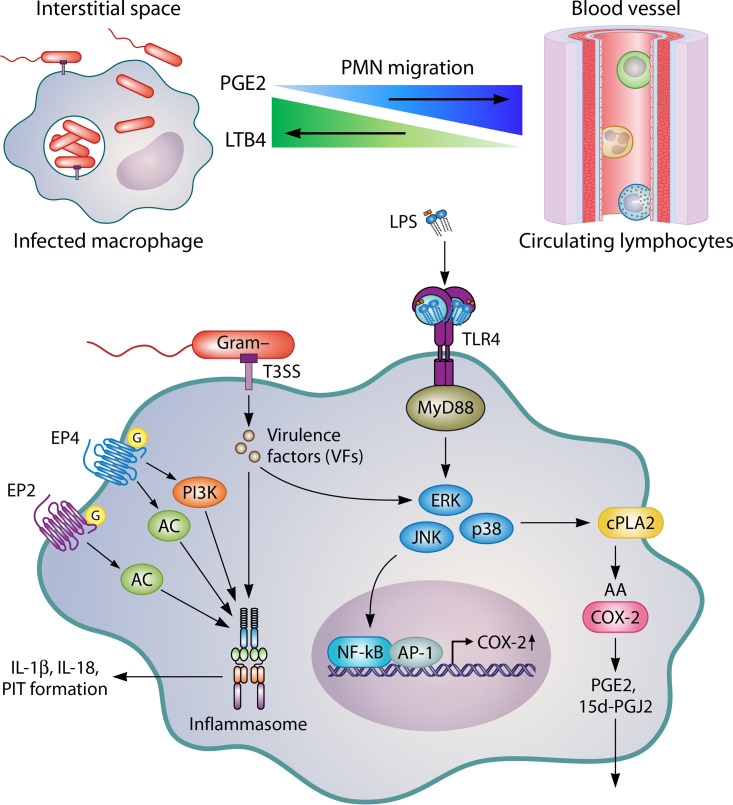
Bacteria-modulated changes in eicosanoid biosynthesis play a vital role during pyroptosis or inflammatory cell death (Fig. 2). Evidence is mounting to support a conserved mechanism used by Gram-negative pathogens. S. Typhimurium, EPEC, and S. flexneri use T3SSs to inject virulence factors into host cells that alter COX-2 transcription. By increasing COX-2 levels, PGE2, one of the most predominant COX-2-derived PGs, is released in large quantities during infection. PGE2 enhances inflammasome activation, secretion of IL-1β and IL-18, and formation of PITs, which are associated with pyroptosis. Upon cell death, live bacteria are trapped in the cell matrix (called PITs), preventing escape from the dying cell. For complete clearance, neutrophils must migrate to PITs and subject them to efferocytosis to degrade the dying cell and immobilized bacteria. COX-2 induction by Gram-negative pathogens may seem counterintuitive considering how PGE2 enhances inflammasome activation, but the secretion of IL-1β, IL-18, and PIT formation alone is not sufficient for the clearance of Salmonella infection in vivo. PGE2 may inhibit normal leukocyte function by lowering the killing abilities of neutrophils and macrophages, such as inhibiting phagocytosis and reactive oxygen species (ROS) generation. Studies with COX-2-deficient mice or animals treated with COX-2 inhibitors display enhanced survival rates during later stages of S. Typhimurium infection. Future studies should focus on the role eicosanoids play in modulating the phagocytic and killing ability of neutrophils toward PITs formed during infection with Gram-negative pathogens and discovering new pathogens that alter eicosanoid production.
FIG 2.
Gram-negative pathogens such as S. Typhimurium and EPEC use T3SS to inject virulence factors such as EspT and SpiC that activate NF-κB to induce COX-2 expression. This activation of COX-2, in turn, leads to the production of prostaglandins, such as PGE2, which act via discrete EP receptors EP2 and EP4 to enhance inflammasome activation during pyroptosis. Inflammasome activation further drives the formation of PITs, which trap live bacteria. Neutrophils are recruited to the site of infection via eicosanoids and phagocytize PITs to clear the invaders.
Biographies

Austin E. F. Sheppe received a B.S. degree in biomedical sciences from the University of Central Florida. He continued his studies as a doctoral student at the University of Florida under the mentorship of Dr. Mariola Edelmann. His dissertation research focused on elucidating novel roles of host-derived bioactive lipids such as inflammatory prostaglandins and anti-inflammatory endocannabinoids and their role in preventing/exacerbating disease. He graduated with a Ph.D. in 2021 and has since taken a position as a postdoctoral research associate in Dr. Aria Eshraghi's lab where his work involves characterizing novel host-pathogen interactions of intracellular bacteria. His career goals include understanding the roles of bioactive lipids and developing novel host-directed therapeutics for patients with chronic inflammatory diseases and enteric infections.

Mariola J. Edelmann obtained a master’s degree in biology in 2005 from the Jagiellonian University in Krakow, Poland. After a brief postgraduate appointment at the University of Oxford, she obtained a Ph.D. in clinical medicine from the Nuffield Department of Clinical Medicine at the University of Oxford, completing her studies in 2010 under the mentorship of Dr. Benedikt Kessler. Next, she obtained a position at Mississippi State University, where she began work on extracellular signaling in enteric infections involving bioactive lipids such as prostaglandins and nano-scale extracellular vesicles. Since 2015 she has joined the Department of Microbiology and Cell Science at the University of Florida, where she has a federally funded program focused on developing novel host-directed therapies against Gram-negative infections and the functions of exosomes in enteric infections.
Contributor Information
Mariola J. Edelmann, Email: medelmann@ufl.edu.
Karen M. Ottemann, University of California, Santa Cruz
REFERENCES
- 1.Mikaeloff Y, Kezouh A, Suissa S. 2008. Nonsteroidal anti-inflammatory drug use and the risk of severe skin and soft tissue complications in patients with varicella or zoster disease. Br J Clin Pharmacol 65:203–209. 10.1111/j.1365-2125.2007.02997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Janabi AA. 2010. In vitro antibacterial activity of Ibuprofen and acetaminophen. J Glob Infect Dis 2:105–108. 10.4103/0974-777X.62880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fries JF, Miller SR, Spitz PW, Williams CA, Hubert HB, Bloch DA. 1990. Identification of patients at risk for gastropathy associated with NSAID use. J Rheumatol Suppl 20:12–19. [PubMed] [Google Scholar]
- 4.Trelle S, Reichenbach S, Wandel S, Hildebrand P, Tschannen B, Villiger PM, Egger M, Jüni P. 2011. Cardiovascular safety of non-steroidal anti-inflammatory drugs: network meta-analysis. BMJ 342:c7086. 10.1136/bmj.c7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rainsford KD. 2009. Ibuprofen: pharmacology, efficacy and safety. Inflammopharmacology 17:275–342. 10.1007/s10787-009-0016-x. [DOI] [PubMed] [Google Scholar]
- 6.Scheiman JM. 2013. The use of proton pump inhibitors in treating and preventing NSAID-induced mucosal damage. Arthritis Res Ther 15 Suppl 3:S5. 10.1186/ar4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.von Moltke J, Trinidad NJ, Moayeri M, Kintzer AF, Wang SB, van Rooijen N, Brown CR, Krantz BA, Leppla SH, Gronert K, Vance RE. 2012. Rapid induction of inflammatory lipid mediators by the inflammasome in vivo. Nature 490:107–111. 10.1038/nature11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawahara K, Hohjoh H, Inazumi T, Tsuchiya S, Sugimoto Y. 2015. Prostaglandin E2-induced inflammation: relevance of prostaglandin E receptors. Biochim Biophys Acta 1851:414–421. 10.1016/j.bbalip.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 9.Regan JW. 2003. EP2 and EP4 prostanoid receptor signaling. Life Sci 74:143–153. 10.1016/j.lfs.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 10.Arosh JA, Banu SK, Chapdelaine P, Fortier MA. 2004. Temporal and tissue-specific expression of prostaglandin receptors EP2, EP3, EP4, FP, and cyclooxygenases 1 and 2 in uterus and fetal membranes during bovine pregnancy. Endocrinology 145:407–417. 10.1210/en.2003-1007. [DOI] [PubMed] [Google Scholar]
- 11.Qian H, Luo N, Chi Y. 2012. Aging-shifted prostaglandin profile in endothelium as a factor in cardiovascular disorders. J Aging Res 2012:121390. 10.1155/2012/121390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Norris PC, Reichart D, Dumlao DS, Glass CK, Dennis EA. 2011. Specificity of eicosanoid production depends on the TLR-4-stimulated macrophage phenotype. J Leukoc Biol 90:563–574. 10.1189/jlb.0311153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Norris PC, Gosselin D, Reichart D, Glass CK, Dennis EA. 2014. Phospholipase A2 regulates eicosanoid class switching during inflammasome activation. Proc Natl Acad Sci U S A 111:12746–12751. 10.1073/pnas.1404372111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Werz O, Gerstmeier J, Libreros S, De la Rosa X, Werner M, Norris PC, Chiang N, Serhan CN. 2018. Human macrophages differentially produce specific resolvin or leukotriene signals that depend on bacterial pathogenicity. Nat Commun 9:59. 10.1038/s41467-017-02538-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zasłona Z, Pålsson-McDermott EM, Menon D, Haneklaus M, Flis E, Prendeville H, Corcoran SE, Peters-Golden M, O'Neill LAJ. 2017. The induction of Pro-IL-1β by lipopolysaccharide requires endogenous prostaglandin E2 production. J Immunol 198:3558–3564. 10.4049/jimmunol.1602072. [DOI] [PubMed] [Google Scholar]
- 16.Kunkel SL, Spengler M, May MA, Spengler R, Larrick J, Remick D. 1988. Prostaglandin E2 regulates macrophage-derived tumor necrosis factor gene expression. J Biol Chem 263:5380–5384. 10.1016/S0021-9258(18)60727-6. [DOI] [PubMed] [Google Scholar]
- 17.Renz H, Gong JH, Schmidt A, Nain M, Gemsa D. 1988. Release of tumor necrosis factor-alpha from macrophages. Enhancement and suppression are dose-dependently regulated by prostaglandin E2 and cyclic nucleotides. J Immunol 141:2388–2393. [PubMed] [Google Scholar]
- 18.Stafford JB, Marnett LJ. 2008. Prostaglandin E2 inhibits tumor necrosis factor-alpha RNA through PKA type I. Biochem Biophys Res Commun 366:104–109. 10.1016/j.bbrc.2007.11.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheppe AEF, Kummari E, Walker A, Richards A, Hui WW, Lee JH, Mangum L, Borazjani A, Ross MK, Edelmann MJ. 2018. PGE2 Augments inflammasome activation and M1 polarization in macrophages infected with Salmonella Typhimurium and Yersinia enterocolitica. Front Microbiol 9:2447. 10.3389/fmicb.2018.02447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ambs P, Baccarini M, Fitzke E, Dieter P. 1995. Role of cytosolic phospholipase A2 in arachidonic acid release of rat-liver macrophages: regulation by Ca2+ and phosphorylation. Biochem J 311:189–195. 10.1042/bj3110189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salvi V, Vaira X, Gianello V, Vermi W, Bugatti M, Sozzani S, Bosisio D. 2016. TLR signalling pathways diverge in their ability to induce PGE2. Mediators Inflamm 2016:5678046. 10.1155/2016/5678046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi HY, Shelhamer JH. 2005. Toll-like receptor 4 signaling regulates cytosolic phospholipase A2 activation and lipid generation in lipopolysaccharide-stimulated macrophages. J Biol Chem 280:38969–38975. 10.1074/jbc.M509352200. [DOI] [PubMed] [Google Scholar]
- 23.Sakai J, Cammarota E, Wright JA, Cicuta P, Gottschalk RA, Li N, Fraser IDC, Bryant CE. 2017. Lipopolysaccharide-induced NF-κB nuclear translocation is primarily dependent on MyD88, but TNFα expression requires TRIF and MyD88. Sci Rep 7:1428. 10.1038/s41598-017-01600-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu J, Weng YI, Simonyi A, Krugh BW, Liao Z, Weisman GA, Sun GY, Simoni A. 2002. Role of PKC and MAPK in cytosolic PLA2 phosphorylation and arachadonic acid release in primary murine astrocytes. J Neurochem 83:259–270. 10.1046/j.1471-4159.2002.01145.x. [DOI] [PubMed] [Google Scholar]
- 25.Lin LL, Wartmann M, Lin AY, Knopf JL, Seth A, Davis RJ. 1993. cPLA2 is phosphorylated and activated by MAP kinase. Cell 72:269–278. 10.1016/0092-8674(93)90666-E. [DOI] [PubMed] [Google Scholar]
- 26.Harizi H, Corcuff JB, Gualde N. 2008. Arachidonic-acid-derived eicosanoids: roles in biology and immunopathology. Trends Mol Med 14:461–469. 10.1016/j.molmed.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Fitzpatrick FA. 2004. Cyclooxygenase enzymes: regulation and function. CPD 10:577–588. 10.2174/1381612043453144. [DOI] [PubMed] [Google Scholar]
- 28.Luong C, Miller A, Barnett J, Chow J, Ramesha C, Browner MF. 1996. Flexibility of the NSAID binding site in the structure of human cyclooxygenase-2. Nat Struct Biol 3:927–933. 10.1038/nsb1196-927. [DOI] [PubMed] [Google Scholar]
- 29.Vane JR. 1971. Inhibition of prostaglandin synthesis as a mechanism of action for aspirin-like drugs. Nat New Biol 231:232–235. 10.1038/newbio231232a0. [DOI] [PubMed] [Google Scholar]
- 30.Prusakiewicz JJ, Duggan KC, Rouzer CA, Marnett LJ. 2009. Differential sensitivity and mechanism of inhibition of COX-2 oxygenation of arachidonic acid and 2-arachidonoylglycerol by ibuprofen and mefenamic acid. Biochemistry 48:7353–7355. 10.1021/bi900999z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tacconelli S, Capone ML, Sciulli MG, Ricciotti E, Patrignani P. 2002. The biochemical selectivity of novel COX-2 inhibitors in whole blood assays of COX-isozyme activity. Curr Med Res Opin 18:503–511. 10.1185/030079902125001335. [DOI] [PubMed] [Google Scholar]
- 32.Warner TD, Giuliano F, Vojnovic I, Bukasa A, Mitchell JA, Vane JR. 1999. Nonsteroid drug selectivities for cyclo-oxygenase-1 rather than cyclo-oxygenase-2 are associated with human gastrointestinal toxicity: a full in vitro analysis. Proc Natl Acad Sci U S A 96:7563–7568. 10.1073/pnas.96.13.7563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pagels WR, Sachs RJ, Marnett LJ, Dewitt DL, Day JS, Smith WL. 1983. Immunochemical evidence for the involvement of prostaglandin H synthase in hydroperoxide-dependent oxidations by ram seminal vesicle microsomes. J Biol Chem 258:6517–6523. 10.1016/S0021-9258(18)32442-6. [DOI] [PubMed] [Google Scholar]
- 34.Hamberg M, Samuelsson B. 1973. Detection and isolation of an endoperoxide intermediate in prostaglandin biosynthesis. Proc Natl Acad Sci U S A 70:899–903. 10.1073/pnas.70.3.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sugimoto Y, Narumiya S. 2007. Prostaglandin E receptors. J Biol Chem 282:11613–11617. 10.1074/jbc.R600038200. [DOI] [PubMed] [Google Scholar]
- 36.Sugimoto Y, Namba T, Honda A, Hayashi Y, Negishi M, Ichikawa A, Narumiya S. 1992. Cloning and expression of a cDNA for mouse prostaglandin E receptor EP3 subtype. J Biol Chem 267:6463–6466. 10.1016/S0021-9258(19)50448-3. [DOI] [PubMed] [Google Scholar]
- 37.Watabe A, Sugimoto Y, Honda A, Irie A, Namba T, Negishi M, Ito S, Narumiya S, Ichikawa A. 1993. Cloning and expression of cDNA for a mouse EP1 subtype of prostaglandin E receptor. J Biol Chem 268:20175–20178. 10.1016/S0021-9258(20)80710-8. [DOI] [PubMed] [Google Scholar]
- 38.Honda A, Sugimoto Y, Namba T, Watabe A, Irie A, Negishi M, Narumiya S, Ichikawa A. 1993. Cloning and expression of a cDNA for mouse prostaglandin E receptor EP2 subtype. J Biol Chem 268:7759–7762. 10.1016/S0021-9258(18)53022-2. [DOI] [PubMed] [Google Scholar]
- 39.Rådmark O, Werz O, Steinhilber D, Samuelsson B. 2015. 5-Lipoxygenase, a key enzyme for leukotriene biosynthesis in health and disease. Biochim Biophys Acta 1851:331–339. 10.1016/j.bbalip.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 40.Zhou H, Coveney AP, Wu M, Huang J, Blankson S, Zhao H, O'Leary DP, Bai Z, Li Y, Redmond HP, Wang JH, Wang J. 2018. Activation of both TLR and NOD signaling confers host innate immunity-mediated protection against microbial infection. Front Immunol 9:3082. 10.3389/fimmu.2018.03082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsuura M, Takahashi H, Watanabe H, Saito S, Kawahara K. 2010. Immunomodulatory effects of Yersinia pestis lipopolysaccharides on human macrophages. Clin Vaccine Immunol 17:49–55. 10.1128/CVI.00336-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, Wang Z, Chen J, Ernst RK, Wang X. 2013. Influence of lipid A acylation pattern on membrane permeability and innate immune stimulation. Mar Drugs 11:3197–3208. 10.3390/md11093197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moffatt JH, Harper M, Harrison P, Hale JD, Vinogradov E, Seemann T, Henry R, Crane B, St Michael F, Cox AD, Adler B, Nation RL, Li J, Boyce JD. 2010. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother 54:4971–4977. 10.1128/AAC.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simpson BW, Trent MS. 2019. Pushing the envelope: LPS modifications and their consequences. Nat Rev Microbiol 17:403–416. 10.1038/s41579-019-0201-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scherle PA, Jones EA, Favata MF, Daulerio AJ, Covington MB, Nurnberg SA, Magolda RL, Trzaskos JM. 1998. Inhibition of MAP kinase kinase prevents cytokine and prostaglandin E2 production in lipopolysaccharide-stimulated monocytes. J Immunol 161:5681–5686. [PubMed] [Google Scholar]
- 46.Hessle CC, Andersson B, Wold AE. 2003. Gram-negative, but not Gram-positive, bacteria elicit strong PGE2 production in human monocytes. Inflammation 27:329–332. 10.1023/B:IFLA.0000006700.41614.21. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Ren B, Zhou X, Liu S, Zhou Y, Li B, Jiang Y, Li M, Feng M, Cheng L. 2017. Growth and adherence of Staphylococcus aureus were enhanced through the PGE2 produced by the activated COX-2/PGE2 pathway of infected oral epithelial cells. PLoS One 12:e0177166. 10.1371/journal.pone.0177166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vane JR, Bakhle YS, Botting RM. 1998. Cyclooxygenases 1 and 2. Annu Rev Pharmacol Toxicol 38:97–120. 10.1146/annurev.pharmtox.38.1.97. [DOI] [PubMed] [Google Scholar]
- 49.Kang YJ, Wingerd BA, Arakawa T, Smith WL. 2006. Cyclooxygenase-2 gene transcription in a macrophage model of inflammation. J Immunol 177:8111–8122. 10.4049/jimmunol.177.11.8111. [DOI] [PubMed] [Google Scholar]
- 50.Fukata M, Chen A, Klepper A, Krishnareddy S, Vamadevan AS, Thomas LS, Xu R, Inoue H, Arditi M, Dannenberg AJ, Abreu MT. 2006. Cox-2 is regulated by Toll-like receptor-4 (TLR4) signaling: role in proliferation and apoptosis in the intestine. Gastroenterology 131:862–877. 10.1053/j.gastro.2006.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raymond B, Crepin VF, Collins JW, Frankel G. 2011. The WxxxE effector EspT triggers expression of immune mediators in an Erk/JNK and NF-κB-dependent manner. Cell Microbiol 13:1881–1893. 10.1111/j.1462-5822.2011.01666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boullier S, Tanguy M, Kadaoui KA, Caubet C, Sansonetti P, Corthésy B, Phalipon A. 2009. Secretory IgA-mediated neutralization of Shigella flexneri prevents intestinal tissue destruction by down-regulating inflammatory circuits. J Immunol 183:5879–5885. 10.4049/jimmunol.0901838. [DOI] [PubMed] [Google Scholar]
- 53.Cho SO, Lim JW, Kim KH, Kim H. 2010. Involvement of Ras and AP-1 in Helicobacter pylori-induced expression of COX-2 and iNOS in gastric epithelial AGS cells. Dig Dis Sci 55:988–996. 10.1007/s10620-009-0828-y. [DOI] [PubMed] [Google Scholar]
- 54.Pal K, Feng X, Steinke JW, Burdick MD, Shim YM, Sung SS, Teague WG, Borish L. 2019. Leukotriene A4 hydrolase activation and leukotriene B4 production by eosinophils in severe asthma. Am J Respir Cell Mol Biol 60:413–419. 10.1165/rcmb.2018-0175OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Partsch G, Schwarzer C, Neumüller J, Dunky A, Petera P, Bröll H, Ittner G, Jantsch S. 1989. Modulation of the migration and chemotaxis of PMN cells by hyaluronic acid. Z Rheumatol 48:123–128. [PubMed] [Google Scholar]
- 56.Resta-Lenert S, Barrett KE. 2002. Enteroinvasive bacteria alter barrier and transport properties of human intestinal epithelium: role of iNOS and COX-2. Gastroenterology 122:1070–1087. 10.1053/gast.2002.32372. [DOI] [PubMed] [Google Scholar]
- 57.Uchiya K, Nikai T. 2004. Salmonella enterica serovar Typhimurium infection induces cyclooxygenase 2 expression in macrophages: involvement of Salmonella pathogenicity island 2. Infect Immun 72:6860–6869. 10.1128/IAI.72.12.6860-6869.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Medici NP, Rashid M, Bliska JB. 2019. Characterization of pyrin dephosphorylation and inflammasome activation in macrophages as triggered by the Yersinia effectors YopE and YopT. Infect Immun 87. 10.1128/IAI.00822-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reyes Ruiz VM, Ramirez J, Naseer N, Palacio NM, Siddarthan IJ, Yan BM, Boyer MA, Pensinger DA, Sauer JD, Shin S. 2017. Broad detection of bacterial type III secretion system and flagellin proteins by the human NAIP/NLRC4 inflammasome. Proc Natl Acad Sci U S A 114:13242–13247. 10.1073/pnas.1710433114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zwack EE, Snyder AG, Wynosky-Dolfi MA, Ruthel G, Philip NH, Marketon MM, Francis MS, Bliska JB, Brodsky IE. 2015. Inflammasome activation in response to the Yersinia type III secretion system requires hyperinjection of translocon proteins YopB and YopD. mBio 6:e02095-14. 10.1128/mBio.02095-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lamkanfi M, Dixit VM. 2014. Mechanisms and functions of inflammasomes. Cell 157:1013–1022. 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 62.Qu Y, Misaghi S, Newton K, Maltzman A, Izrael-Tomasevic A, Arnott D, Dixit VM. 2016. NLRP3 recruitment by NLRC4 during Salmonella infection. J Exp Med 213:877–885. 10.1084/jem.20132234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Atkinson S, Williams P. 2016. Yersinia virulence factors - a sophisticated arsenal for combating host defences. F1000Res 5:1370. 10.12688/f1000research.8466.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jayaraman P, Sada-Ovalle I, Nishimura T, Anderson AC, Kuchroo VK, Remold HG, Behar SM. 2013. IL-1β promotes antimicrobial immunity in macrophages by regulating TNFR signaling and caspase-3 activation. J Immunol 190:4196–4204. 10.4049/jimmunol.1202688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ozer EK, Goktas MT, Kilinc I, Bariskaner H, Ugurluoglu C, Iskit AB. 2017. Celecoxib administration reduced mortality, mesenteric hypoperfusion, aortic dysfunction and multiple organ injury in septic rats. Biomed Pharmacother 86:583–589. 10.1016/j.biopha.2016.11.102. [DOI] [PubMed] [Google Scholar]
- 66.Cheon H, Rho YH, Choi SJ, Lee YH, Song GG, Sohn J, Won NH, Ji JD. 2006. Prostaglandin E2 augments IL-10 signaling and function. J Immunol 177:1092–1100. 10.4049/jimmunol.177.2.1092. [DOI] [PubMed] [Google Scholar]
- 67.Aronoff DM, Canetti C, Peters-Golden M. 2004. Prostaglandin E2 inhibits alveolar macrophage phagocytosis through an E-prostanoid 2 receptor-mediated increase in intracellular cyclic AMP. J Immunol 173:559–565. 10.4049/jimmunol.173.1.559. [DOI] [PubMed] [Google Scholar]
- 68.Chen Y, Hughes-Fulford M. 2000. Prostaglandin E2 and the protein kinase A pathway mediate arachidonic acid induction of c-fos in human prostate cancer cells. Br J Cancer 82:2000–2006. 10.1054/bjoc.2000.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sokolowska M, Chen LY, Liu Y, Martinez-Anton A, Qi HY, Logun C, Alsaaty S, Park YH, Kastner DL, Chae JJ, Shelhamer JH. 2015. Prostaglandin E2 inhibits NLRP3 inflammasome activation through EP4 receptor and intracellular cyclic AMP in human macrophages. J Immunol 194:5472–5487. 10.4049/jimmunol.1401343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fujino H, Salvi S, Regan JW. 2005. Differential regulation of phosphorylation of the cAMP response element-binding protein after activation of EP2 and EP4 prostanoid receptors by prostaglandin E2. Mol Pharmacol 68:251–259. 10.1124/mol.105.011833. [DOI] [PubMed] [Google Scholar]
- 71.Luan B, Yoon YS, Le Lay J, Kaestner KH, Hedrick S, Montminy M. 2015. CREB pathway links PGE2 signaling with macrophage polarization. Proc Natl Acad Sci U S A 112:15642–15647. 10.1073/pnas.1519644112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Serezani CH, Chung J, Ballinger MN, Moore BB, Aronoff DM, Peters-Golden M. 2007. Prostaglandin E2 suppresses bacterial killing in alveolar macrophages by inhibiting NADPH oxidase. Am J Respir Cell Mol Biol 37:562–570. 10.1165/rcmb.2007-0153OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruffell D, Mourkioti F, Gambardella A, Kirstetter P, Lopez RG, Rosenthal N, Nerlov C. 2009. A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc Natl Acad Sci U S A 106:17475–17480. 10.1073/pnas.0908641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Toda Y, Tsukada J, Misago M, Kominato Y, Auron PE, Tanaka Y. 2002. Autocrine induction of the human pro-IL-1beta gene promoter by IL-1beta in monocytes. J Immunol 168:1984–1991. 10.4049/jimmunol.168.4.1984. [DOI] [PubMed] [Google Scholar]
- 75.Marotta P, Sautebin L, Di Rosa M. 1992. Modulation of the induction of nitric oxide synthase by eicosanoids in the murine macrophage cell line J774. Br J Pharmacol 107:640–641. 10.1111/j.1476-5381.1992.tb14499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dejani NN, Orlando AB, Niño VE, Penteado LA, Verdan FF, Bazzano JMR, Codo AC, Salina ACG, Saraiva AC, Avelar MR, Spolidorio LC, Serezani CH, Medeiros AI. 2018. Intestinal host defense outcome is dictated by PGE. Proc Natl Acad Sci U S A 115:E8469–E8478. 10.1073/pnas.1722016115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ikeh MAC, Fidel PL, Noverr MC. 2017. Prostaglandin E. Antimicrob Agents Chemother 62. 10.1128/AAC.01920-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Salina AC, Souza TP, Serezani CH, Medeiros AI. 2017. Efferocytosis-induced prostaglandin E2 production impairs alveolar macrophage effector functions during Streptococcus pneumoniae infection. Innate Immun 23:219–227. 10.1177/1753425916684934. [DOI] [PubMed] [Google Scholar]
- 79.Nishimura T, Zhao X, Gan H, Koyasu S, Remold HG. 2013. The prostaglandin E2 receptor EP4 is integral to a positive feedback loop for prostaglandin E2 production in human macrophages infected with Mycobacterium tuberculosis. FASEB J 27:3827–3836. 10.1096/fj.13-228858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang X, Shaw DK, Hammond HL, Sutterwala FS, Rayamajhi M, Shirey KA, Perkins DJ, Bonventre JV, Velayutham TS, Evans SM, Rodino KG, VieBrock L, Scanlon KM, Carbonetti NH, Carlyon JA, Miao EA, McBride JW, Kotsyfakis M, Pedra JH. 2016. The prostaglandin E2-EP3 receptor axis regulates anaplasma phagocytophilum-mediated NLRC4 inflammasome activation. PLoS Pathog 12:e1005803. 10.1371/journal.ppat.1005803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dey I, Lejeune M, Chadee K. 2006. Prostaglandin E2 receptor distribution and function in the gastrointestinal tract. Br J Pharmacol 149:611–623. 10.1038/sj.bjp.0706923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Loynes CA, Lee JA, Robertson AL, Steel MJ, Ellett F, Feng Y, Levy BD, Whyte MKB, Renshaw SA. 2018. PGE2 production at sites of tissue injury promotes an anti-inflammatory neutrophil phenotype and determines the outcome of inflammation resolution in vivo. Sci Adv 4:eaar8320. 10.1126/sciadv.aar8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gomi K, Zhu FG, Marshall JS. 2000. Prostaglandin E2 selectively enhances the IgE-mediated production of IL-6 and granulocyte-macrophage colony-stimulating factor by mast cells through an EP1/EP3-dependent mechanism. J Immunol 165:6545–6552. 10.4049/jimmunol.165.11.6545. [DOI] [PubMed] [Google Scholar]
- 84.Nishigaki N, Negishi M, Ichikawa A. 1996. Two Gs-coupled prostaglandin E receptor subtypes, EP2 and EP4, differ in desensitization and sensitivity to the metabolic inactivation of the agonist. Mol Pharmacol 50:1031–1037. [PubMed] [Google Scholar]
- 85.Forman BM, Tontonoz P, Chen J, Brun RP, Spiegelman BM, Evans RM. 1995. 15-Deoxy-delta 12, 14-prostaglandin J2 is a ligand for the adipocyte determination factor PPAR gamma. Cell 83:803–812. 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 86.Nagy L, Tontonoz P, Alvarez JG, Chen H, Evans RM. 1998. Oxidized LDL regulates macrophage gene expression through ligand activation of PPARgamma. Cell 93:229–240. 10.1016/s0092-8674(00)81574-3. [DOI] [PubMed] [Google Scholar]
- 87.Tontonoz P, Nagy L, Alvarez JG, Thomazy VA, Evans RM. 1998. PPARgamma promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell 93:241–252. 10.1016/s0092-8674(00)81575-5. [DOI] [PubMed] [Google Scholar]
- 88.Ricote M, Li AC, Willson TM, Kelly CJ, Glass CK. 1998. The peroxisome proliferator-activated receptor-gamma is a negative regulator of macrophage activation. Nature 391:79–82. 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- 89.Jiang C, Ting AT, Seed B. 1998. PPAR-gamma agonists inhibit production of monocyte inflammatory cytokines. Nature 391:82–86. 10.1038/34184. [DOI] [PubMed] [Google Scholar]
- 90.Marx N, Schönbeck U, Lazar MA, Libby P, Plutzky J. 1998. Peroxisome proliferator-activated receptor gamma activators inhibit gene expression and migration in human vascular smooth muscle cells. Circ Res 83:1097–1103. 10.1161/01.res.83.11.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Straus DS, Pascual G, Li M, Welch JS, Ricote M, Hsiang CH, Sengchanthalangsy LL, Ghosh G, Glass CK. 2000. 15-deoxy-delta 12,14-prostaglandin J2 inhibits multiple steps in the NF-kappa B signaling pathway. Proc Natl Acad Sci U S A 97:4844–4849. 10.1073/pnas.97.9.4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Maier NK, Leppla SH, Moayeri M. 2015. The cyclopentenone prostaglandin 15d-PGJ2 inhibits the NLRP1 and NLRP3 inflammasomes. J Immunol 194:2776–2785. 10.4049/jimmunol.1401611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Surh YJ, Na HK, Park JM, Lee HN, Kim W, Yoon IS, Kim DD. 2011. 15-Deoxy-Δ12,14-prostaglandin J2, an electrophilic lipid mediator of anti-inflammatory and pro-resolving signaling. Biochem Pharmacol 82:1335–1351. 10.1016/j.bcp.2011.07.100. [DOI] [PubMed] [Google Scholar]
- 94.Kielian T, McMahon M, Bearden ED, Baldwin AC, Drew PD, Esen N. 2004. S. aureus-dependent microglial activation is selectively attenuated by the cyclopentenone prostaglandin 15-deoxy-Delta12,14- prostaglandin J2 (15d-PGJ2). J Neurochem 90:1163–1172. 10.1111/j.1471-4159.2004.02579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Phulwani NK, Feinstein DL, Gavrilyuk V, Akar C, Kielian T. 2006. 15-deoxy-Delta12,14-prostaglandin J2 (15d-PGJ2) and ciglitazone modulate Staphylococcus aureus-dependent astrocyte activation primarily through a PPAR-gamma-independent pathway. J Neurochem 99:1389–1402. 10.1111/j.1471-4159.2006.04183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zingarelli B, Sheehan M, Hake PW, O'Connor M, Denenberg A, Cook JA. 2003. Peroxisome proliferator activator receptor-gamma ligands, 15-deoxy-Delta(12,14)-prostaglandin J2 and ciglitazone, reduce systemic inflammation in polymicrobial sepsis by modulation of signal transduction pathways. J Immunol 171:6827–6837. 10.4049/jimmunol.171.12.6827. [DOI] [PubMed] [Google Scholar]
- 97.Dugo L, Collin M, Cuzzocrea S, Thiemermann C. 2004. 15d-prostaglandin J2 reduces multiple organ failure caused by wall-fragment of Gram-positive and Gram-negative bacteria. Eur J Pharmacol 498:295–301. 10.1016/j.ejphar.2004.07.074. [DOI] [PubMed] [Google Scholar]
- 98.Guyton K, Zingarelli B, Ashton S, Teti G, Tempel G, Reilly C, Gilkeson G, Halushka P, Cook J. 2003. Peroxisome proliferator-activated receptor-gamma agonists modulate macrophage activation by gram-negative and gram-positive bacterial stimuli. Shock 20:56–62. 10.1097/01.shk.0000070903.21762.f8. [DOI] [PubMed] [Google Scholar]
- 99.Antunes LC, Arena ET, Menendez A, Han J, Ferreira RB, Buckner MM, Lolic P, Madilao LL, Bohlmann J, Borchers CH, Finlay BB. 2011. Impact of salmonella infection on host hormone metabolism revealed by metabolomics. Infect Immun 79:1759–1769. 10.1128/IAI.01373-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Buckner MM, Antunes LC, Gill N, Russell SL, Shames SR, Finlay BB. 2013. 15-Deoxy-Δ12,14-prostaglandin J2 inhibits macrophage colonization by Salmonella enterica serovar Typhimurium. PLoS One 8:e69759. 10.1371/journal.pone.0069759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cha B, Lim JW, Kim KH, Kim H. 2011. 15-deoxy-D12,14-prostaglandin J2 suppresses RANTES expression by inhibiting NADPH oxidase activation in Helicobacter pylori-infected gastric epithelial cells. J Physiol Pharmacol 62:167–174. [PubMed] [Google Scholar]
- 102.Serhan CN, Chiang N, Dalli J, Levy BD. 2015. Lipid mediators in the resolution of inflammation. Cold Spring Harb Perspect Biol 7:a016311. 10.1101/cshperspect.a016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sala A, Bolla M, Zarini S, Müller-Peddinghaus R, Folco G. 1996. Release of leukotriene A4 versus leukotriene B4 from human polymorphonuclear leukocytes. J Biol Chem 271:17944–17948. 10.1074/jbc.271.30.17944. [DOI] [PubMed] [Google Scholar]
- 104.Zhang Y, Olson RM, Brown CR. 2017. Macrophage LTB4 drives efficient phagocytosis of Borrelia burgdorferi via BLT1 or BLT2. J Lipid Res 58:494–503. 10.1194/jlr.M068882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sadik CD, Miyabe Y, Sezin T, Luster AD. 2018. The critical role of C5a as an initiator of neutrophil-mediated autoimmune inflammation of the joint and skin. Semin Immunol 37:21–29. 10.1016/j.smim.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 106.Subramanian BC, Moissoglu K, Parent CA. 2018. The LTB-BLT1 axis regulates the polarized trafficking of chemoattractant GPCRs during neutrophil chemotaxis. J Cell Sci 131:jcs217422. 10.1242/jcs.217422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Oliveira SH, Canetti C, Ribeiro RA, Cunha FQ. 2008. Neutrophil migration induced by IL-1beta depends upon LTB4 released by macrophages and upon TNF-alpha and IL-1beta released by mast cells. Inflammation 31:36–46. 10.1007/s10753-007-9047-x. [DOI] [PubMed] [Google Scholar]
- 108.Brandt SL, Klopfenstein N, Wang S, Winfree S, McCarthy BP, Territo PR, Miller L, Serezani CH. 2018. Macrophage-derived LTB4 promotes abscess formation and clearance of Staphylococcus aureus skin infection in mice. PLoS Pathog 14:e1007244. 10.1371/journal.ppat.1007244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sorgi CA, Soares EM, Rosada RS, Bitencourt CS, Zoccal KF, Pereira PAT, Fontanari C, Brandão I, Masson AP, Ramos SG, Silva CL, Frantz FG, Faccioli LH. 2020. Eicosanoid pathway on host resistance and inflammation during Mycobacterium tuberculosis infection is comprised by LTB. Biochim Biophys Acta Mol Basis Dis 1866:165574. 10.1016/j.bbadis.2019.165574. [DOI] [PubMed] [Google Scholar]
- 110.Tobin DM, Roca FJ, Oh SF, McFarland R, Vickery TW, Ray JP, Ko DC, Zou Y, Bang ND, Chau TT, Vary JC, Hawn TR, Dunstan SJ, Farrar JJ, Thwaites GE, King MC, Serhan CN, Ramakrishnan L. 2012. Host genotype-specific therapies can optimize the inflammatory response to mycobacterial infections. Cell 148:434–446. 10.1016/j.cell.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mancuso P, Standiford TJ, Marshall T, Peters-Golden M. 1998. 5-Lipoxygenase reaction products modulate alveolar macrophage phagocytosis of Klebsiella pneumoniae. Infect Immun 66:5140–5146. 10.1128/IAI.66.11.5140-5146.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Serezani CH, Aronoff DM, Jancar S, Peters-Golden M. 2005. Leukotriene B4 mediates p47phox phosphorylation and membrane translocation in polyunsaturated fatty acid-stimulated neutrophils. J Leukoc Biol 78:976–984. 10.1189/jlb.1004587. [DOI] [PubMed] [Google Scholar]
- 113.Zhang Q, Lenardo MJ, Baltimore D. 2017. 30 years of NF-κB: a blossoming of relevance to human pathobiology. Cell 168:37–57. 10.1016/j.cell.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Griffiths RJ, Pettipher ER, Koch K, Farrell CA, Breslow R, Conklyn MJ, Smith MA, Hackman BC, Wimberly DJ, Milici AJ. 1995. Leukotriene B4 plays a critical role in the progression of collagen-induced arthritis. Proc Natl Acad Sci U S A 92:517–521. 10.1073/pnas.92.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ramalho T, Filgueiras L, Silva-Jr IA, Pessoa AFM, Jancar S. 2018. Impaired wound healing in type 1 diabetes is dependent on 5-lipoxygenase products. Sci Rep 8:14164. 10.1038/s41598-018-32589-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Freire MO, Dalli J, Serhan CN, Van Dyke TE. 2017. Neutrophil resolvin E1 receptor expression and function in type 2 diabetes. J Immunol 198:718–728. 10.4049/jimmunol.1601543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Goldman G, Welbourn R, Kobzik L, Valeri CR, Shepro D, Hechtman HB. 1993. Lavage with leukotriene B4 induces lung generation of tumor necrosis factor-alpha that in turn mediates neutrophil diapedesis. Surgery 113:297–303. [PubMed] [Google Scholar]
- 118.Kuhns DB, Nelson EL, Alvord WG, Gallin JI. 2001. Fibrinogen induces IL-8 synthesis in human neutrophils stimulated with formyl-methionyl-leucyl-phenylalanine or leukotriene B(4). J Immunol 167:2869–2878. 10.4049/jimmunol.167.5.2869. [DOI] [PubMed] [Google Scholar]
- 119.Brach MA, de Vos S, Arnold C, Gruss HJ, Mertelsmann R, Herrmann F. 1992. Leukotriene B4 transcriptionally activates interleukin-6 expression involving NK-chi B and NF-IL6. Eur J Immunol 22:2705–2711. 10.1002/eji.1830221034. [DOI] [PubMed] [Google Scholar]
- 120.Tobin DM, Vary JC, Ray JP, Walsh GS, Dunstan SJ, Bang ND, Hagge DA, Khadge S, King MC, Hawn TR, Moens CB, Ramakrishnan L. 2010. The lta4h locus modulates susceptibility to mycobacterial infection in zebrafish and humans. Cell 140:717–730. 10.1016/j.cell.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vincent WJB, Harvie EA, Sauer JD, Huttenlocher A. 2017. Neutrophil derived LTB4 induces macrophage aggregation in response to encapsulated Streptococcus iniae infection. PLoS One 12:e0179574. 10.1371/journal.pone.0179574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kwon SY, Ro M, Kim JH. 2019. Mediatory roles of leukotriene B. Sci Rep 9:5936. 10.1038/s41598-019-42410-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bowman CC, Bost KL. 2004. Cyclooxygenase-2-mediated prostaglandin E2 production in mesenteric lymph nodes and in cultured macrophages and dendritic cells after infection with Salmonella. J Immunol 172:2469–2475. 10.4049/jimmunol.172.4.2469. [DOI] [PubMed] [Google Scholar]
- 124.Knight M, Braverman J, Asfaha K, Gronert K, Stanley S. 2018. Lipid droplet formation in Mycobacterium tuberculosis infected macrophages requires IFN-γ/HIF-1α signaling and supports host defense. PLoS Pathog 14:e1006874. 10.1371/journal.ppat.1006874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen M, Divangahi M, Gan H, Shin DS, Hong S, Lee DM, Serhan CN, Behar SM, Remold HG. 2008. Lipid mediators in innate immunity against tuberculosis: opposing roles of PGE2 and LXA4 in the induction of macrophage death. J Exp Med 205:2791–2801. 10.1084/jem.20080767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Carter FP, Frankson T, Pintard J, Edgecombe B. 2011. Seroprevalence of Helicobacter pylori infection in adults in the Bahamas. West Indian Med J 60:662–665. [PubMed] [Google Scholar]
- 127.Kao CY, Sheu BS, Wu JJ. 2016. Helicobacter pylori infection: an overview of bacterial virulence factors and pathogenesis. Biomed J 39:14–23. 10.1016/j.bj.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen Y, Blaser MJ. 2007. Inverse associations of Helicobacter pylori with asthma and allergy. Arch Intern Med 167:821–827. 10.1001/archinte.167.8.821. [DOI] [PubMed] [Google Scholar]
- 129.Koch KN, Müller A. 2015. Helicobacter pylori activates the TLR2/NLRP3/caspase-1/IL-18 axis to induce regulatory T-cells, establish persistent infection and promote tolerance to allergens. Gut Microbes 6:382–387. 10.1080/19490976.2015.1105427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sierra JC, Hobbs S, Chaturvedi R, Yan F, Wilson KT, Peek RM, Polk DB. 2013. Induction of COX-2 expression by Helicobacter pylori is mediated by activation of epidermal growth factor receptor in gastric epithelial cells. Am J Physiol Gastrointest Liver Physiol 305:G196–203. 10.1152/ajpgi.00495.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lan C, Yang L, Fan L, Zhang Y, Wang J, Guo GJ, Wan S, Yang S, Wang R, Fang D. 2013. Celecoxib inhibits Helicobacter pylori-induced invasion of gastric cancer cells through an adenine nucleotide translocator-dependent mechanism. Anticancer Agents Med Chem 13:1267–1272. 10.2174/18715206113139990324. [DOI] [PubMed] [Google Scholar]
- 132.Zhao J, Wen S, Wang X, Zhang Z. 2017. Helicobacter pylori modulates cyclooxygenase-2 and 15-hydroxy prostaglandin dehydrogenase in gastric cancer. Oncol Lett 14:5519–5525. 10.3892/ol.2017.6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yao Y, Li G, Wu J, Zhang X, Wang J. 2015. Inflammatory response of macrophages cultured with Helicobacter pylori strains was regulated by miR-155. Int J Clin Exp Pathol 8:4545–4554. [PMC free article] [PubMed] [Google Scholar]
- 134.Eckmann L, Stenson WF, Savidge TC, Lowe DC, Barrett KE, Fierer J, Smith JR, Kagnoff MF. 1997. Role of intestinal epithelial cells in the host secretory response to infection by invasive bacteria. Bacterial entry induces epithelial prostaglandin h synthase-2 expression and prostaglandin E2 and F2alpha production. J Clin Invest 100:296–309. 10.1172/JCI119535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Raisch J, Rolhion N, Dubois A, Darfeuille-Michaud A, Bringer MA. 2015. Intracellular colon cancer-associated Escherichia coli promote protumoral activities of human macrophages by inducing sustained COX-2 expression. Lab Invest 95:296–307. 10.1038/labinvest.2014.161. [DOI] [PubMed] [Google Scholar]
- 136.Bonnotte B, Larmonier N, Favre N, Fromentin A, Moutet M, Martin M, Gurbuxani S, Solary E, Chauffert B, Martin F. 2001. Identification of tumor-infiltrating macrophages as the killers of tumor cells after immunization in a rat model system. J Immunol 167:5077–5083. 10.4049/jimmunol.167.9.5077. [DOI] [PubMed] [Google Scholar]
- 137.Ren H, Chen X, Jiang F, Li G. 2020. Cyclooxygenase-2 inhibition reduces autophagy of macrophages enhancing extraintestinal pathogenic. Front Microbiol 11:708. 10.3389/fmicb.2020.00708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Noad J, von der Malsburg A, Pathe C, Michel MA, Komander D, Randow F. 2017. LUBAC-synthesized linear ubiquitin chains restrict cytosol-invading bacteria by activating autophagy and NF-κB. Nat Microbiol 2:17063. 10.1038/nmicrobiol.2017.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chen TC, Tsai JP, Huang HJ, Teng CC, Chien SJ, Kuo HC, Huang WS, Chen CN. 2011. Regulation of cyclooxygenase-2 expression in human bladder epithelial cells infected with type I fimbriated uropathogenic E. coli. Cell Microbiol 13:1703–1713. 10.1111/j.1462-5822.2011.01650.x. [DOI] [PubMed] [Google Scholar]
- 140.Wright DH, Metters KM, Abramovitz M, Ford-Hutchinson AW. 1998. Characterization of the recombinant human prostanoid DP receptor and identification of L-644,698, a novel selective DP agonist. Br J Pharmacol 123:1317–1324. 10.1038/sj.bjp.0701708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tanaka K, Hirai H, Takano S, Nakamura M, Nagata K. 2004. Effects of prostaglandin D2 on helper T cell functions. Biochem Biophys Res Commun 316:1009–1014. 10.1016/j.bbrc.2004.02.151. [DOI] [PubMed] [Google Scholar]
- 142.Ide T, Egan K, Bell-Parikh LC, FitzGerald GA. 2003. Activation of nuclear receptors by prostaglandins. Thromb Res 110:311–315. 10.1016/S0049-3848(03)00418-3. [DOI] [PubMed] [Google Scholar]
- 143.Bliska JB, Wang X, Viboud GI, Brodsky IE. 2013. Modulation of innate immune responses by Yersinia type III secretion system translocators and effectors. Cell Microbiol 15:1622–1631. 10.1111/cmi.12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dhar MS, Virdi JS. 2014. Strategies used by Yersinia enterocolitica to evade killing by the host: thinking beyond Yops. Microbes Infect 16:87–95. 10.1016/j.micinf.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 145.Montminy SW, Khan N, McGrath S, Walkowicz MJ, Sharp F, Conlon JE, Fukase K, Kusumoto S, Sweet C, Miyake K, Akira S, Cotter RJ, Goguen JD, Lien E. 2006. Virulence factors of Yersinia pestis are overcome by a strong lipopolysaccharide response. Nat Immunol 7:1066–1073. 10.1038/ni1386. [DOI] [PubMed] [Google Scholar]
- 146.Mattock E, Blocker AJ. 2017. How do the virulence factors of Shigella work together to cause disease? Front Cell Infect Microbiol 7:64. 10.3389/fcimb.2017.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ashida H, Ogawa M, Mimuro H, Kobayashi T, Sanada T, Sasakawa C. 2011. Shigella are versatile mucosal pathogens that circumvent the host innate immune system. Curr Opin Immunol 23:448–455. 10.1016/j.coi.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 148.Raqib R, Mia SM, Qadri F, Alam TI, Alam NH, Chowdhury AK, Mathan MM, Andersson J. 2000. Innate immune responses in children and adults with Shigellosis. Infect Immun 68:3620–3629. 10.1128/iai.68.6.3620-3629.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Schnupf P, Sansonetti PJ. 2012. Quantitative RT-PCR profiling of the rabbit immune response: assessment of acute Shigella flexneri infection. PLoS One 7:e36446. 10.1371/journal.pone.0036446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lippmann J, Gwinner F, Rey C, Tamir U, Law HK, Schwikowski B, Enninga J. 2015. Bacterial internalization, localization, and effectors shape the epithelial immune response during Shigella flexneri infection. Infect Immun 83:3624–3637. 10.1128/IAI.00574-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ambrosi C, Pompili M, Scribano D, Limongi D, Petrucca A, Cannavacciuolo S, Schippa S, Zagaglia C, Grossi M, Nicoletti M. 2015. The Shigella flexneri OspB effector: an early immunomodulator. Int J Med Microbiol 305:75–84. 10.1016/j.ijmm.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 152.Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, Lieberman J. 2016. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535:153–158. 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jorgensen I, Zhang Y, Krantz BA, Miao EA. 2016. Pyroptosis triggers pore-induced intracellular traps (PITs) that capture bacteria and lead to their clearance by efferocytosis. J Exp Med 213:2113–2128. 10.1084/jem.20151613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jorgensen I, Lopez JP, Laufer SA, Miao EA. 2016. IL-1β, IL-18, and eicosanoids promote neutrophil recruitment to pore-induced intracellular traps following pyroptosis. Eur J Immunol 46:2761–2766. 10.1002/eji.201646647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wilson WJ, Afzali MF, Cummings JE, Legare ME, Tjalkens RB, Allen CP, Slayden RA, Hanneman WH. 2016. Immune modulation as an effective adjunct post-exposure therapeutic for B. pseudomallei. PLoS Negl Trop Dis 10:e0005065. 10.1371/journal.pntd.0005065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Everest PH, Cole AT, Hawkey CJ, Knutton S, Goossens H, Butzler JP, Ketley JM, Williams PH. 1993. Roles of leukotriene B4, prostaglandin E2, and cyclic AMP in Campylobacter jejuni-induced intestinal fluid secretion. Infect Immun 61:4885–4887. 10.1128/IAI.61.11.4885-4887.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Navratil AR, Brummett AM, Bryan JD, Woolard MD. 2014. Francisella tularensis LVS induction of prostaglandin biosynthesis by infected macrophages requires specific host phospholipases and lipid phosphatases. Infect Immun 82:3299–3311. 10.1128/IAI.02060-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Woolard MD, Hensley LL, Kawula TH, Frelinger JA. 2008. Respiratory Francisella tularensis live vaccine strain infection induces Th17 cells and prostaglandin E2, which inhibits generation of gamma interferon-positive T cells. Infect Immun 76:2651–2659. 10.1128/IAI.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.N'Guessan PD, Etouem MO, Schmeck B, Hocke AC, Scharf S, Vardarova K, Opitz B, Flieger A, Suttorp N, Hippenstiel S. 2007. Legionella pneumophila-induced PKCalpha-, MAPK-, and NF-kappaB-dependent COX-2 expression in human lung epithelium. Am J Physiol Lung Cell Mol Physiol 292:L267–L77. 10.1152/ajplung.00100.2006. [DOI] [PubMed] [Google Scholar]
- 160.Hurley BP, Pirzai W, Mumy KL, Gronert K, McCormick BA. 2011. Selective eicosanoid-generating capacity of cytoplasmic phospholipase A2 in Pseudomonas aeruginosa-infected epithelial cells. Am J Physiol Lung Cell Mol Physiol 300:L286–294. 10.1152/ajplung.00147.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sadikot RT, Zeng H, Azim AC, Joo M, Dey SK, Breyer RM, Peebles RS, Blackwell TS, Christman JW. 2007. Bacterial clearance of Pseudomonas aeruginosa is enhanced by the inhibition of COX-2. Eur J Immunol 37:1001–1009. 10.1002/eji.200636636. [DOI] [PubMed] [Google Scholar]
- 162.Saliba AM, Nascimento DO, Silva MC, Assis MC, Gayer CR, Raymond B, Coelho MG, Marques EA, Touqui L, Albano RM, Lopes UG, Paiva DD, Bozza PT, Plotkowski MC. 2005. Eicosanoid-mediated proinflammatory activity of Pseudomonas aeruginosa ExoU. Cell Microbiol 7:1811–1822. 10.1111/j.1462-5822.2005.00635.x. [DOI] [PubMed] [Google Scholar]