Abstract
Brain extracellular matrix (ECM) structure mediates many aspects of neural development and function. Probing structural changes in brain ECM could thus provide insights into mechanisms of neurodevelopment, the loss of neural function in response to injury, and the detrimental effects of pathological aging and neurological disease. We demonstrate the ability to probe changes in brain ECM microstructure using multiple particle tracking (MPT). We performed MPT of colloidally stable polystyrene nanoparticles in organotypic rat brain slices collected from rats aged 14–70 days old. Our analysis revealed an inverse relationship between nanoparticle diffusive ability in the brain extracellular space and age. Additionally, the distribution of effective ECM pore sizes in the cortex shifted to smaller pores throughout development. We used the raw data and features extracted from nanoparticle trajectories to train a boosted decision tree capable of predicting chronological age with high accuracy. Collectively, this work demonstrates the utility of combining MPT with machine learning for measuring changes in brain ECM structure and predicting associated complex features such as chronological age. This will enable further understanding of the roles brain ECM play in development and aging and the specific mechanisms through which injuries cause aberrant neuronal function. Additionally, this approach has the potential to develop machine learning models capable of detecting the presence of injury or indicating the extent of injury based on changes in the brain microenvironment microstructure.
Keywords: particle tracking, diffusion, extracellular matrix, machine learning, microstructure, boosted decision tree
Graphical Abstract
The extracellular spaces (ECS) of brain tissue are home to the brain extracellular matrix (ECM), a heterogeneous collection of proteoglycans, tenascins, and a hyaluronic acid backbone that can either be free floating, tethered to cellular surfaces, or condensed to form specific structures.1,2 The ability to organize into specific structures allows brain ECM to perform special roles in both development and normal brain function. For example, the formation of highly condensed perineuronal nets (PNNs), which envelop the soma and proximal dendrites of certain populations of neurons, helps regulate plasticity and protects neurons from harmful processes like excitotoxicity and oxidative stress.3,4 Similarly, the ECM structure that wraps around brain endothelial cells, known as the basement membrane, is a critical functional and structural component of the blood–brain barrier (BBB) and neurovascular unit.5,6 Brain ECM is also highly dynamic, and the ability to assemble, disband, and reorganize is required for the development of proper neuronal circuitry and helps facilitate repair in response to injury.7–12 The structural integrity of PNNs is thought to be impacted by many neurological diseases, including epilepsy, schizophrenia, and stroke.12–15 Likewise, many brain injuries are associated with changes in basement membrane protein expression, including stroke16,17 and traumatic brain injury.18,19 Probing real-time changes in the ECM microstructure, particularly changes that occur locally at the cellular level in living tissue, remains an ongoing challenge. The limited ability to assess ECM structural changes prevents a complete understanding of the role ECM plays in neurodevelopment and the mechanisms through which brain injuries impair neuronal function.
To address these knowledge gaps, we need to characterize both spatial and temporal changes in ECM structure, which requires a technique that can probe extracellular dynamics in real-time at the microscale. Electron microscopy achieves the spatial resolution necessary but often requires tissue fixation, and different fixation techniques can lead to significantly different estimates of ECS-related parameters.20 Fluorescent-based staining can be used to label specific components of brain ECM, but features commonly quantified from fluorescent images provide no direct insight into physical and geometric properties of the local environment, like viscosity and ECM pore size.21,22 Atomic force microscopy has also been used to quantify mechanical properties of brain ECM23 but only provides a surface-level analysis, preventing insight into the microrheological properties in the bulk of the tissue. Diffusion-based techniques, such as real-time iontophoresis24–27 and integrative optical imaging,28–30 have also been used to quantify ECS parameters, specifically ECS volume fraction and tortuosity. These methods report an average value for a given volume of brain tissue, however, and are limited in providing the resolution necessary to study ECS heterogeneity at the submicron level, where variation is known to exist.31 Multiple and single particle tracking techniques have thus gained traction across the field of biophysics, as they provide ways to study ECS properties and ECM dynamics with submicron resolution.31–35
Multiple particle tracking (MPT) is a technique that leverages fluorescent microscopy to capture the motion of nanoparticles in real-time. A strength of MPT is that the microscopic behavior of hundreds to thousands of individual particles is tracked simultaneously, while retaining single particle resolution. The motions exhibited by particles provide information about the environment in which they reside, and the ability to track the movement of individual particles provides high spatial resolution. MPT has already been leveraged to characterize structural features of many biological domains, including the vitreous of the eye,36 various mucosal membranes,37–40 and intracellular environments.41–43 MPT and single nanoparticle tracking have been used to better estimate the average spacing of brain ECS31,33 and evaluate the diffusive ability of many nanoparticle-based drug delivery platforms in the brain.33,44,45
An additional advantage of MPT is the sheer amount of data it generates, with experiments typically producing anywhere between 102 and 105 total trajectories. Because of this, machine learning methods have already started to be incorporated into the MPT workflow to explore otherwise hidden trends in data and make predictions. Wagner et al. demonstrated the ability to predict motion type (confined, directed, anomalous, normal) using random forest classifiers trained on trajectory feature data sets,46 and others have employed artificial neural networks to predict agarose gel stiffness and in vitro cell uptake of nanoparticles.47 The initial success of this approach is promising and raises the question as to whether complex biological variables like the presence or extent of injury can be predicted.
The main findings we present herein are two-fold. We first demonstrate the ability of MPT to characterize changes in brain ECM structure at varying stages of neurodevelopment and provide potential explanations as to why these changes are observed. We then implement extreme gradient boosting (XGBoost) to generate boosted decision tree classifiers capable of predicting age from nanoparticle trajectory feature data. Collectively, these results support the potential of MPT in probing ECM-related mechanisms of disease progression that cause aberrant neuronal function. Additionally, MPT can be applied to enhance understanding of the specific roles ECM plays during development and aging. This work also further validates the utility of combining machine learning practices with MPT. The ability to develop models capable of predicting a complex biological variable like age based solely on nanoparticle diffusion information is beneficial, as natural extensions of this work include exploring whether tracking data can predict mechanisms of disease pathology or brain injury severity, initially in model organisms but eventually in human specimens.
RESULTS AND DISCUSSION
Nanoparticle Diffusive Ability and Effective Brain ECM Mesh Size Decrease with Age in the Developing Rat Brain.
The critical period of synaptogenesis, neural circuit formation, and refinement in rodent cortex can vary depending on region but typically begins in the first 1–2 weeks postnatal and extends another 3–4 weeks before transitioning to a more stable adult state.48–50 Brain ECM is thought to be involved in nearly all stages of neural development, including the period of elevated synapse formation and plasticity.11,51,52 In addition to distinct ECM protein expression during the critical period, neural stem cell behavior in vitro differs depending on the composition of the ECM.53 We wanted to leverage MPT to probe ECM structure changes during this time. For this, we collected organotypic hemispheric brain slices from rats at varying ages, from postnatal (P) day 14, 21, 28, 35, and 70 (P14, P21, P28, P35, and P70) Sprague–Dawley (SD) rats. We performed MPT in the cortex of slices from each group using near-neutral, poly(ethylene glycol) (PEG)-coated 40 nm polystyrene nanoparticles (PS–PEG) (Table S1). At 20–24 h postslice preparation, the nanoparticles were added and videos of particle Brownian motions were collected with high-resolution MPT (Figure 1A,B). Nanoparticle trajectories were quantified using a custom Python package54 that leverages TrackMate, an open source Fiji plugin for the tracking of single particles (Figure 1C).55 Forty nanometer PS–PEG nanoparticles were chosen for two reasons: First, their 51 nm hydrodynamic diameter falls below the most recently reported mean—150 ± 40 nm (mean ± standard deviation)31—and median—270 [130–560 nm] (median [interquartile range])56—values of brain ECS width. Second, PEG-coated PS nanoparticles have demonstrated an ability to evade adhesive interactions with various cellular and ECM-associated components33 and remain stable in physiologically relevant conditions such as artificial cerebrospinal fluid.57 By minimizing electrostatic and hydrophobic interactions while remaining colloidally stable, the motion these particles exhibit is predominantly impacted by local fluid properties of the brain ECS and structural properties of the local ECM.
Figure 1.
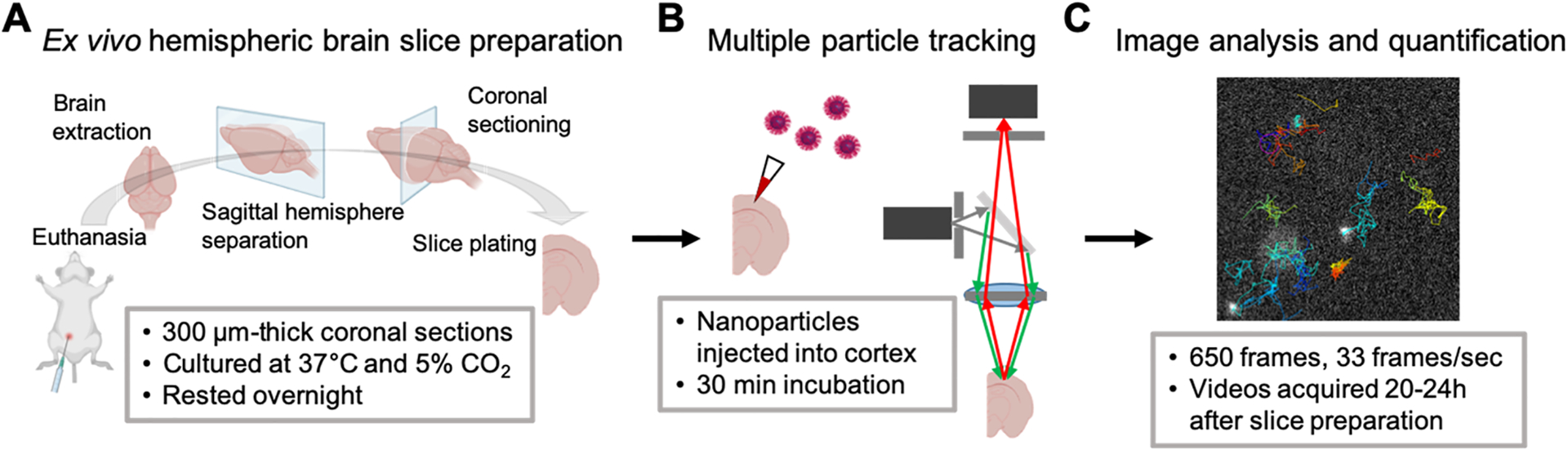
Multiple particle tracking in ex vivo hemispheric brain slices. (A) Following euthanasia, brains were extracted, split into hemispheres sagittally, and sectioned coronally before being plated on cell culture inserts and cultured at 37 °C and 5% CO2. Slices were allowed to rest overnight. (B) Forty nanometer PS–PEG nanoparticles were injected into the cortex and allowed to incubate for 30 min prior to video acquisition via high-speed fluorescence microscopy. Videos were acquired 20–24 h postslice preparation. (C) Videos were analyzed in ImageJ to track nanoparticle Brownian motions and quantify mean squared displacements and effective diffusion coefficients. Created with BioRender.com.
Tracking revealed an inverse relationship between nanoparticle diffusive ability in the cortex and brain age. Raw MPT data come in the form of nanoparticle trajectories or x–y positions over time. By calculating nanoparticle mean squared displacements at various lag times, the Einstein–Smoluchowski relation can be used to obtain effective diffusion coefficients in the brain (Db,eff) for each nanoparticle in the population. Distributions of Db,eff values shift to lower values as the cortex develops (Figure 2A). Significant differences in Db,eff values exist between all groups (p = 0.05), with median (95% CI) Db,eff values of 2.71 (2.64–2.81), 1.42 (1.39–1.45), 1.05 (1.00–1.09), 0.705 (0.692–0.721), and 0.376 (0.369–0.383) μm2/s for P14, P21, P28, P35, and P70, respectively (Figure 2B). Compared to theoretical diffusion coefficients in a representative free medium of artificial cerebral spinal fluid (DACSF), diffusivity in cortical ECM (Db,eff) decreased 5-, 9-, 12-, 18, and 34-fold in P14, P21, P28, P35, and P70 aged brains, respectively (Figure 2B). To estimate the distribution of effective ECM pore sizes, we fit the Amsden obstruction-scaling model for entangled and cross-linked hydrogels58,59 to the Db,eff data in Figure 2A. As brains aged, the distribution of ECM pore sizes in the cortex shifted to smaller pores (Figure 2C–G). The average effective pore size (ζ) decreased from 76.8 nm in P14 brains to 36.0 nm in P70 brains (Figure 2B).
Figure 2.
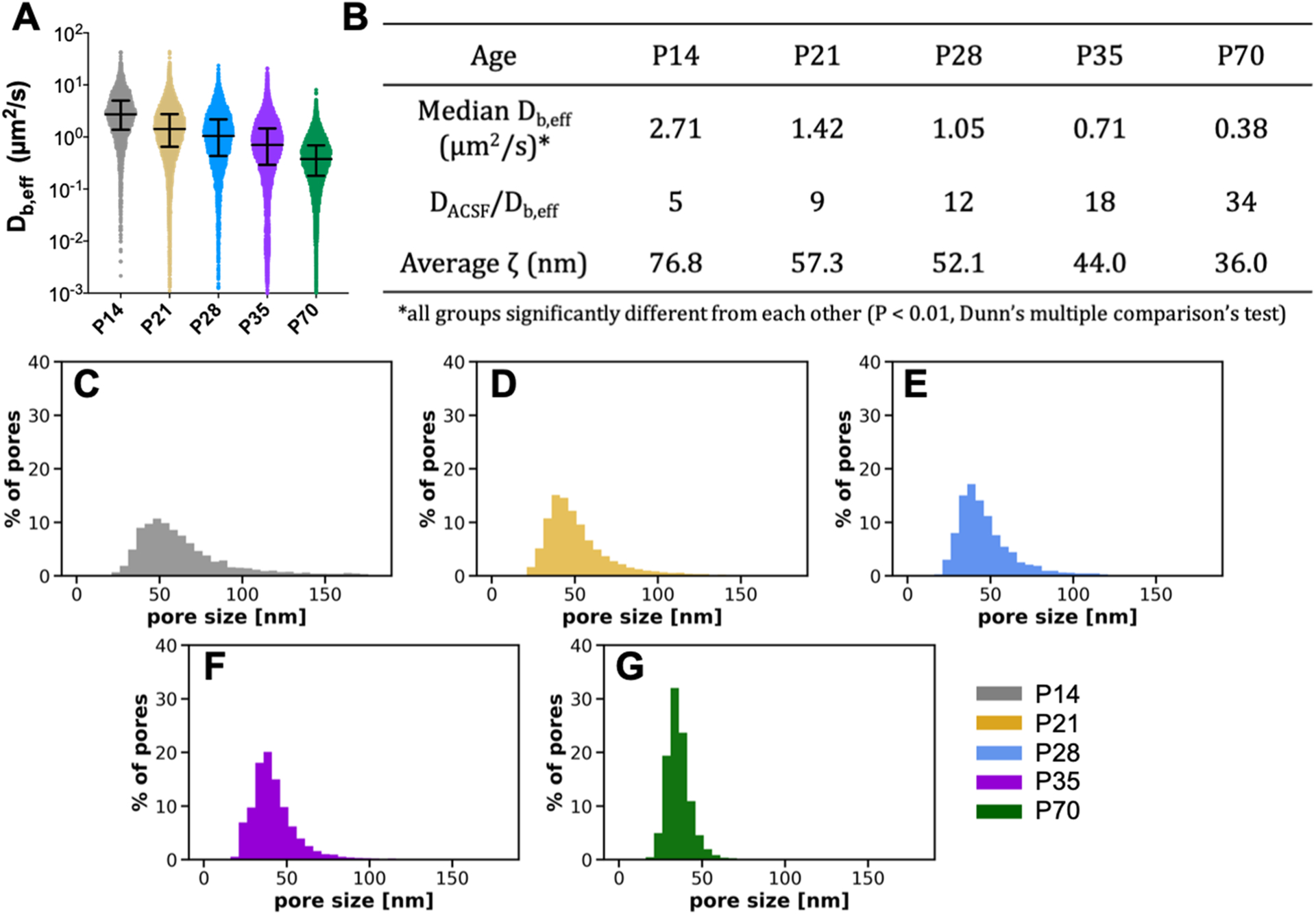
Nanoparticle diffusivity and effective ECM mesh size in the cortex decrease with age in the developing rat brain. (A) Distribution of Db,eff values at a 0.33 s lag time for each postnatal age. Error bars provide the median Db,eff value and interquartile range. (B) Median Db,eff (μm2/s), ratio of diffusivity in artificial cerebral spinal fluid to diffusivity in cortical ECM (DACSF/Db,eff), and average effective ECM pore size (ζ) for each age. Asterisk (*) denotes significant differences, using a Dunn’s test, between groups after adjusting for multiple comparisons (p < 0.01). (C–G) Distributions of effective ECM pore sizes (ζ) for each age.
Inducing Changes in Brain ECM in Rat Brain Tissue Ex Vivo.
One likely contributor to the decreased diffusivity observed with increasing age is an increase in the number of physical obstructions present in the ECS. PNNs, a densely cross-linked and entangled form of brain ECM, begin forming early in life and increase in abundance as the critical period of neurodevelopment ends.11,12,60 We confirm an increased presence of PNNs from P14–P70 (Figure S1). An increase in the presence of these dense aggregates of insoluble protein and glycans likely hinders extracellular diffusion by increasing the number of physical obstructions present and reducing the effective size of brain ECM pores. To test this hypothesis, we designed experiments to determine if exogenously induced breakdown of brain-specific ECM structures would lead to changes in nanoparticle diffusion. With brain slices taken from P35 rats, an age at which PNNs are present in the cortex,11 we first demonstrate the ability to induce ECM breakdown ex vivo. P35 brain slices were treated with either chondroitinase ABC (ChABC, 0.4 U/mL) or hyaluronidase (HYase, 35 U/mL), two enzymes known to degrade components of brain ECM.8,61,62 Brain slices treated with enzyme-free slice culture media (SCM) served as the negative control (nontreated, NT). We monitored the presence of PNNs following treatment by staining with a fluorescently labeled wisteria floribunda agglutinin (WFA) lectin at 15, 30, 45, and 120 min post-treatment (Figure 3A). The PNN structure was completely lost in the cortex within 120 min of enzyme treatment (Figure 3B). PNN structures in nontreated brain slices were unaffected over the experimental window (Figure 3B).
Figure 3.
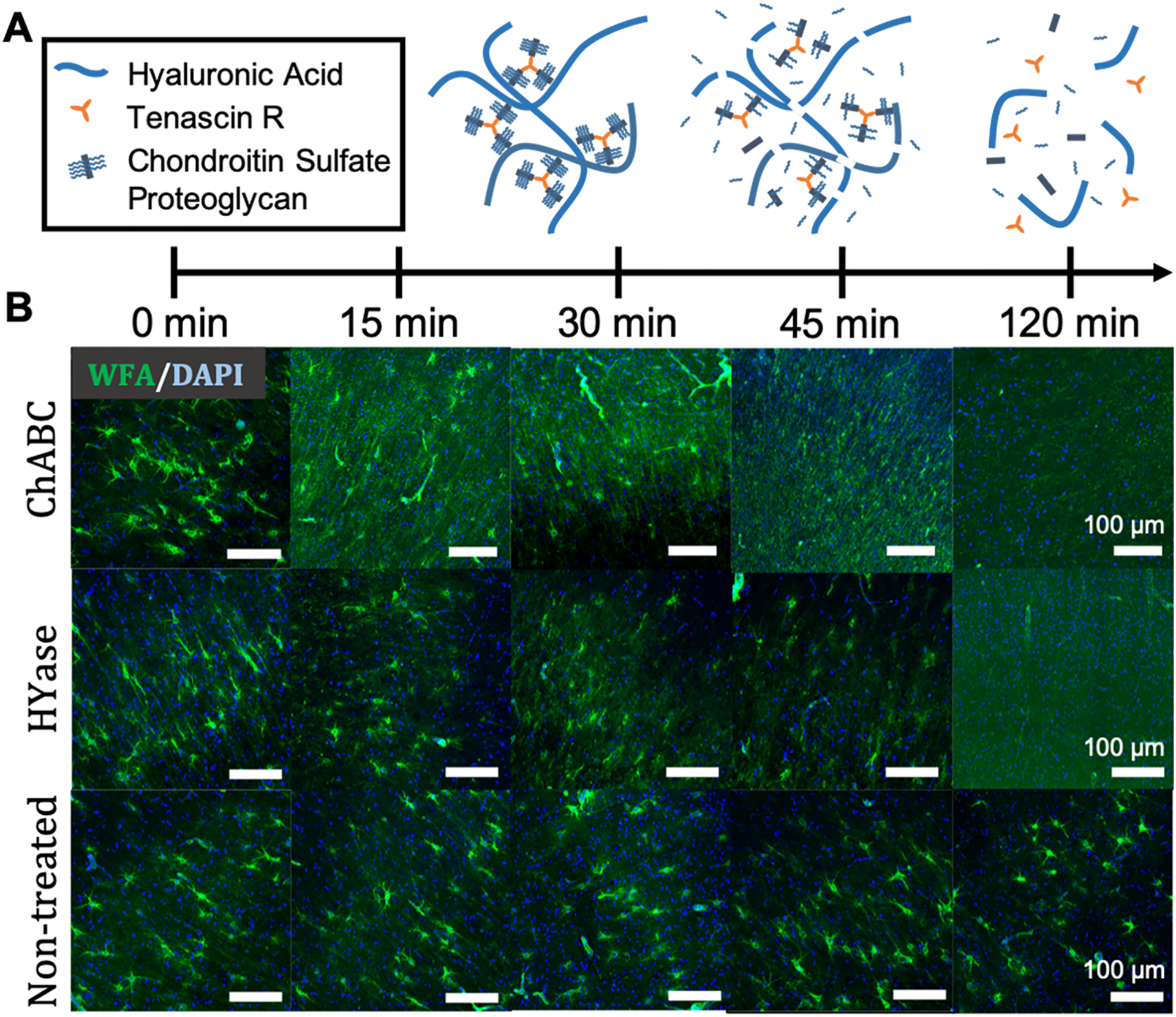
PNN structures degrade within 2 h following treatment with either ChABC or HYase. (A) Schematic representation of PNN breakdown following treatment with HYase or ChABC. (B) Representative 20× magnification images taken from the cortex of P35 rat brain slices receiving one of three treatments (HYase, ChABC, or SCM). PNNs were stained with WFA (green) and cell nuclei stained with DAPI (blue). Rows represent treatment group. Columns represent treatment time. Scale bars: 100 μm.
MPT in Enzyme-Treated Rat Brain Slices Ex Vivo.
Having identified 2 h as sufficient for complete loss of PNN structure in brain slices treated with either ChABC (0.4 U/mL) or HYase (35 U/mL), we next investigated whether changes in the diffusion of 40 nm PS–PEG nanoparticles—the same nanoparticles used in the age-dependent study (Table S1)—would result from these induced ECM structural changes. PS–PEG nanoparticles are not expected to be affected by newly free components of brain ECM or the applied enzymes due to their high PEG surface density (>9 PEG chains per 100 nm2)63 which prevents protein adsorption.64 Ex vivo MPT in the cortex revealed that nanoparticle populations explore a greater area, move faster, and have increased diffusivities when navigating enzyme-treated brain tissue. Representative trajectory maps generated from a single video taken in a nontreated, ChABC-treated, and HYase-treated brain slice are provided in Figure 4A. Despite each map containing around the same number of total trajectories (1478, 1593, and 1732 for nontreated, ChABC-treated, and HYase-treated, respectively), nanoparticles surveyed a greater area when diffusing in slices treated with ECM-degrading enzymes. The ranges of Db,eff values were similar for all treatment groups, but the median Db,eff value was greater in magnitude for both enzyme-treated groups compared to the nontreated control (Figure 4B). The median Db,eff at a 0.33 s lag time was 1.34 and 1.20 μm2/s for ChABC- and HYase-treated slices, respectively, compared to 0.67 μm2/s for nontreated slices. This represented a 19-, 10-, and 11-fold decrease compared to diffusivity in a representative free medium of artificial cerebral spinal fluid (DACSF) (Figure 4C). The Db,eff data were also used to estimate how the distribution of effective ECM pore sizes changes in response to ECM degradation via ChABC or HYase. The distribution of ECM pore sizes in the cortex shifted to larger pores in both instances (Figure S2). The average ζ increased from 43.4 nm in nontreated P35 brains to 56.1 and 54.4 nm in ChABC- and HYase-treated P35 brains, respectively (Figure 4C).
Figure 4.
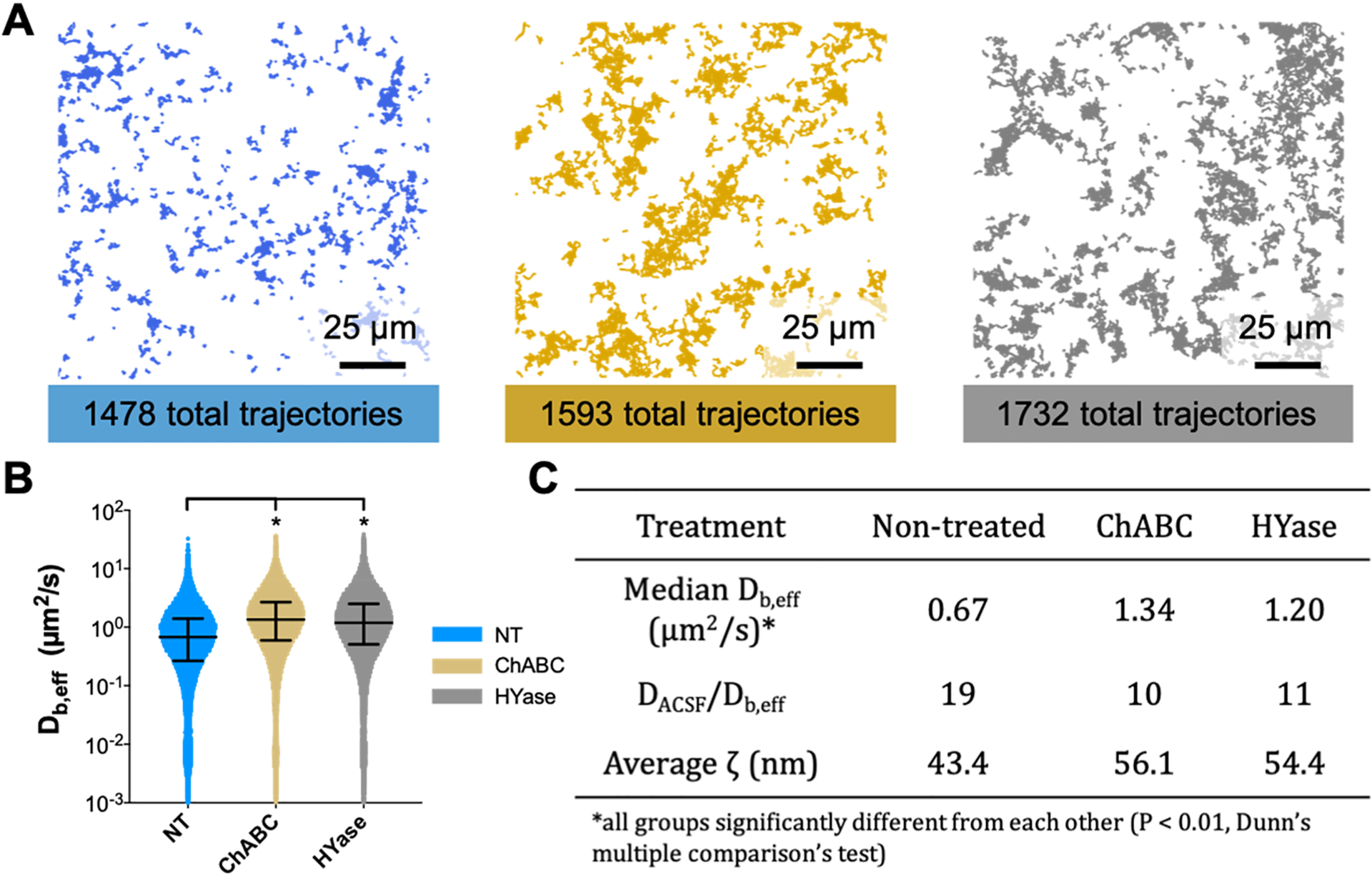
Nanoparticles survey a greater area of the brain microenvironment and diffuse faster following treatment with ECM-degrading enzymes. (A) Representative trajectory maps generated from MPT experiments carried out in nontreated (blue), ChABC-treated (gold), and HYase-treated (gray) P35 brain slices ex vivo. The total number of trajectories in each group is provided. Scale bar: 25 μm. (B) Distribution of Db,eff values at a 0.33 s lag time for each treatment group. Error bars provide the median Db,eff value and interquartile range. (C) Median Db,eff (μm2/s), ratio of diffusivity in artificial cerebral spinal fluid to diffusivity in cortical ECM (DACSF/Db,eff), and average effective ECM pore size (ζ) for each group. Asterisk (*) denotes significant differences (Dunn’s test) between groups after adjusting for multiple comparisons (p < 0.01).
To account for biological variability slice to slice and animal to animal, MPT experiments were performed using four separate rats, all within the same developmental range (P35–P38). Significant differences in nanoparticle diffusive ability existed across treatment groups, independent of both slice and animal. If the trajectories from all videos taken in all slices and all animals are compiled into one data set, nanoparticle populations diffusing in ChABC-treated and HYase-treated slices have median Db,eff values significantly larger than those diffusing in nontreated slices (Figure 4B). If data are instead split by animal, significant differences in Db,eff remain between treated and nontreated groups (Figure S2).
The extracellular environment of cortical brain tissue changes throughout neurodevelopment, and our findings demonstrate MPT is capable of detecting changes in extracellular nanoparticle diffusion with age (Figure 2A). There are many factors that potentially contribute to the decreased diffusivity observed with age. First, there are changes in the organization and density of ECM structures, like PNNs, that occur throughout development. An enzyme-induced breakdown of PNNs leads to increased nanoparticle diffusivity, along with an increase in the distribution of effective ECM pore sizes (Figure 4). The evidence thus suggests that changes in the number of steric obstructions present in the ECS plays a role in decreasing extracellular diffusivity with age, but it remains difficult to label it as the sole contributor. This hypothesis is also supported by previous work, albeit in different models of brain tissue, that demonstrates enhanced diffusion of various probes following chondroitin sulfate or hyaluronic acid breakdown.31,34,65 The incorporation of high-temporal resolution confocal or two-photon microscopy into the MPT technique for simultaneous fluorescent microscopy would allow future work targeted at tracking the behavior of nanoparticles away from, in close proximity too, and interacting directly with PNNs and help confirm the conclusions drawn herein.
In addition to steric interactions with ECM constituents and cellular surfaces, nanoparticles are also subject to hydrodynamic interactions brought about by the narrow channels of brain ECS and pores present in ECM structures.66,67 Diffusion theory tells us that increased drag is experienced when a diffusing particle’s size approaches the width of the pore it is navigating.68 Existing work by Lehmenkühler et al. found that ECS volume fraction decreases from ~40% at P2–P4 to about 27% at P10–P11 and then steadily declines to an adult value of ~22% by P21;69 however, it is still unknown if volume fraction relates directly to ECS channel width, especially given the interconnectedness of ECS channels. We did see a shift in effective ECM pore size distributions to smaller pores when our data were fit to the Amsden obstruction-scaling model (Figure 2C–G), but our analysis only included one nanoparticle size (40 nm). The analysis becomes more robust when diffusion data are pooled across nanoparticles of varying size.33 Similar to the effect of increased physical obstructions with age, it is likely the decrease in nanoparticle diffusivity observed with age is partly due to the narrowing of ECS channels, but it is difficult to know the exact extent at which it contributes. Changes in the magnitude of certain geometric features associated with the raw trajectory data, such as trajectory aspect ratio, elongation, or straightness, could also be indicative of a reduction in ECS width. To know definitively, MPT would have to be carried out in controlled environments where brain ECS pore size can be tuned precisely, and the impact of pore size on each specific trajectory feature could be investigated in greater detail.
Interestingly, a notable portion of ECM pores were predicted to be smaller than 40 nm, the particle diameter, especially in the P70 age group (Figure 2C–G). There are two main reasons for this. First, only a single probe size was included in this study. It is known that a significant portion of ECS channels in the brain are larger than 100 nm in diameter,33,35,56 and that pores greater than 1 μm in diameter do exist.56 There is potential that a 40 nm probe will not fully explore the entirety of the space it resides within the tracking window. Additionally, a 40 nm particle within a large ECM-filled pore might not diffuse as readily as it would in a free medium, due to nonsterically imposed hindrances to diffusion. This provides a second explanation for the apparent underestimation of the distribution of pore sizes. The Amsden obstruction-scaling model assumes the probe is completely inert to its environment and only impacted by steric obstructions.59 While the PEGylation reaction used to synthesize the probe particles achieves a dense PEG surface coating63 and results in near-neutral zeta-potential (Table S1), there is still potential that electrostatic and hydrophobic forces are affecting their movement in tissue. By neglecting these nonsteric interactions, the model likely underestimates pore sizes. Future work would benefit from quantifying the motions of multiple sizes of nanoparticle probes, as has been done previously.32,33 This would also provide insight into whether the observed relationship between particle diffusivity and chronological age is consistent regardless of probe size. Similar particle tracking methodologies have been used by others to demonstrate elevated diffusivity and an increase in ECS dimensions following treatment with hyaluronidase, and these experiments used single-walled carbon nanotubes (SWCNTs) ranging from ~490 to 780 nm in one dimension (length; SWCNTs do have a diameter in the range of a nanometer).31,70 The alignment of results, between our studies using 40 nm PS–PEG nanoparticles in hyaluronidase-treated brain slices (Figure 4) and previous experiments using 490–780 nm in length SWCNTs,31 suggests that alterations in ECM structure are detectable by a range of probe sizes. We thus expect the trend of decreased extracellular diffusivity with developmental age to hold regardless of probe size and probe material, so long as the applied probe can penetrate brain tissue effectively.31,33–35,44,71–73
Finally, changes in the viscosity of the extracellular fluid could also influence nanoparticle diffusion, although viscosity is thought to be less influential than ECS geometry as particle size approaches the width of the ECS.74 Extracellular fluid viscosity can vary depending on fluctuations in the concentration of extracellular components, such as proteins, neuromodulators, extracellular vesicles, glycans, and metabolic waste. PNN-specific chondroitin sulfate proteoglycan (CSPG) mRNA expression profiles in the rat cortex have been shown to vary during the first 6 weeks of life,75 and the total amount of hyaluronic acid, a ligand for CSPGs, also fluctuates during development before eventually stabilizing in adulthood.76 Both hyaluronic acid and CSPGs spend time in matrix-immobilized states, however, so their impact on extracellular fluid viscosity likely varies depending on current ECM dynamics. Any non-cell-adhered, free floating aggregates of ECM would certainly influence viscosity. Collectively, both ECM-associated and non-ECM-associated components can be in flux during development. It is therefore plausible that the composition and consequently viscosity of the extracellular fluid varies, as well. Whether these changes are significant enough to alter nanoparticle diffusion in a notable way is difficult to determine—it would require a technique capable of probing the viscosity of the extracellular fluid in vivo at the microscale, and diffusion-based strategies that leverage nanosized probes are confounded by other factors, such as steric hindrances and hydrodynamic factors.
The decreased diffusivity of nanoparticles in brain ECS as brains age is likely due to a combination of all three factors: narrowing of ECS channels and shifts to smaller ECM pores, an increase in the number of physical obstructions present in the ECS, and changes in extracellular fluid viscosity. The ability to delineate the significance of each will remain a challenge until methods can be developed to assay each individually. There is also potential that cellular changes contribute to changes in the extracellular diffusion of nanosized probes. In the mouse cortex specifically, axonal and dendritic arborization is actively occurring during the first 3 weeks postnatal77 and potentially continues into the later stages of development. Glial cells are also active participants in many neurodevelopment processes, including synapse formation, pruning, and plasticity, and thus their current morphology, activation state, and density could influence local ECS properties. Brain ECM helps facilitate communication between neurons and glial cells, so any neuronal- or glial-specific change in cellular dynamics, morphology, and activity would likely result in a change in brain ECM that impacts local extracellular diffusion.
Similar to existing, well-established methods of fluorescence single particle tracking,78 our ex vivo MPT technique allows the tracking of individual particles that are present over the duration of a high frame-rate video. The scale of the age-dependent study carried out herein, which included 15 videos per age group with each video containing hundreds to thousands of individual trajectories, provided an abundant amount of data in the form of nanoparticle x–y positions over time. These trajectory data sets represent an obvious target for the incorporation of machine learning algorithms; we were interested to see if models could actually predict a complex biological variable such as age based solely off features associated with the trajectory of an individual nanoparticle.
Boosted Decision Trees Generated from MPT Feature Data Far Outperform the Accuracy of a Random Guess for Predicting Chronological Age.
Using the XGBoost (eXtreme Gradient Boosting) software library,79 we trained boosted decision tree classifiers on the age-dependent MPT data to determine if the incorporation of machine learning could result in predictive power. Prior to model training, the amount of features the algorithm could access was enhanced by calculating additional trajectory features to complement the Deff values already available. A total of 39 features were computed, some based on trajectory geometry (aspect ratio and straightness, for example) and some based on traditional diffusion theory (anomalous diffusion exponent and MSD ratio, for example). Trajectory features were adopted from previous literature47 but scaled by introducing additional, local-averaged features; Table S2 contains a full list of features and feature calculations. Using a subset of the original age-dependent feature data set, we trained a boosted decision tree targeted to classify between P14, P21, P28, P35, and P70 age groups. The resulting model achieves a total predictive accuracy of 60.09% when tested on a separate subset of data, triple the accuracy of a random guess (20.00%) (Figure 5A). The weighted average precision and recall are also notable, at 58.28 and 60.09%, respectively. The model most accurately predicted P14 and P70 ages (recall of 74.47 and 84.40%, respectively) and performed worse in the intermediate age groups (Figure 5B). Recall for the P21, P28, and P35 ages was 48.08, 28.44, and 53.02%, respectively. However, even the lowest recall of 28.44% is still greater than a one in five random guess. Of the entire population of P70 trajectories included in the test data set (348 in total), 26 trajectories were incorrectly classified (17 predicted as P35 and 9 predicted as P28) (Figure 5B), and 79 trajectories of the entire P14 population (out of 324) were mislabeled.
Figure 5.
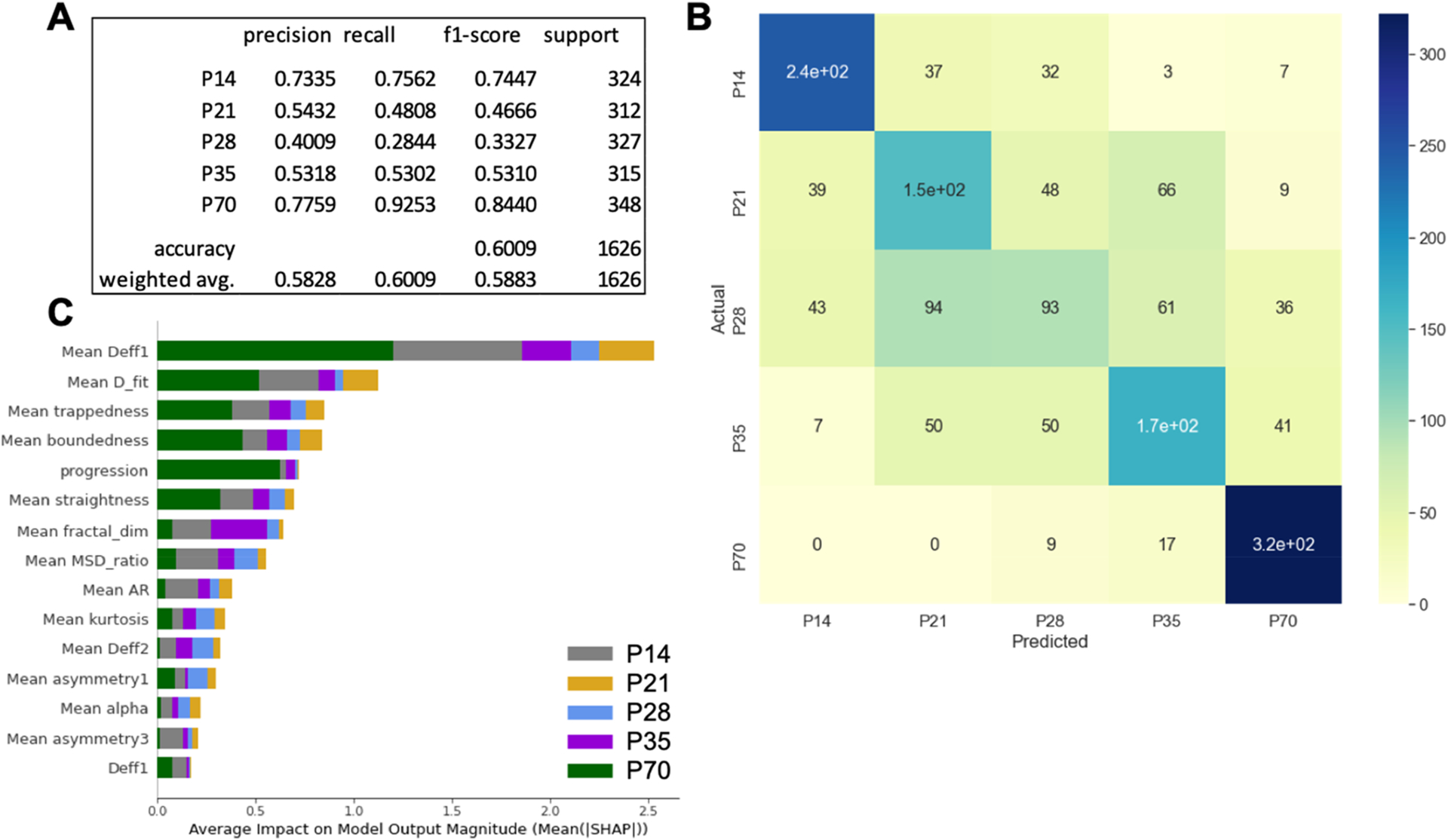
Boosted decision tree evaluation metrics, confusion matrix, and SHAP summary plot. (A) Precision, recall, f1-score, and support for each of the five classes along with overall model accuracy and weighted average precision, recall, and f1-score. (B) Confusion matrix for test data set comparing predicted age versus actual age. The colorbar reflects the number of trajectories in each bin. (C) SHAP summary plot for the 5-age classifier. Included are the top 15 most influential features. Contributions from each age are provided by the bar color: P14 = gray, P21 = gold, P28 = blue, P35 = purple, P70 = green.
A distinct advantage of decision-tree-based classifiers is the ability to quantify the importance of each feature to the classification being made. To identify features that impact the model most significantly, a summary plot of Shapley Additive exPlanation (SHAP) values80,81 was created for the 5-age predictor (Figure 5C). Interestingly, four of the top five and 13 of the top 15 features are local-averaged features. The top five feature dependencies are mean Deff at 0.33 s (Mean Deff1), mean fitted diffusion coefficient (D_fit), mean trappedness, mean boundedness, and progression (Figure 5C). Both Mean Deff1 and D_fit are calculated using diffusion theory and are based on particle mean square displacements at varying lag times, while mean trappedness, mean boundedness, and progression relate to the geometry of the trajectory.
To understand how feature impact varies depending on feature magnitude, we generated SHAP dependence plots for the four most influential features in the 5-age boosted decision tree classifier (Figure 6A–D). SHAP dependence plots show the SHAP value associated with each trajectory fed into the model (grouped by age) and how that SHAP value varies with feature magnitude. Positive SHAP values increase the likelihood of prediction, whereas negative SHAP values decrease the likelihood of prediction. For example, when trajectories from the P70 data set possess small values of mean Deff, mean Deff has a positive SHAP value and thus increases the likelihood the model classifies the trajectory as P70. As mean Deff increases, SHAP values decrease toward zero and eventually go negative, meaning mean Deff becomes progressively less influential in the prediction (Figure 6A). The opposite trend is observed for the P14 age group (Figure 6A). SHAP dependence plots for mean Dfit, mean trappedness, and mean boundedness are presented in Figure 3B–D. In general, when trajectories from the P70 age group were slower moving, defined by a small mean Deff and Dfit, and more confined, defined by a small mean trappedness and boundedness, the model leveraged those features to make the prediction. The opposite is observed for the P14 age group, with the intermediate ages behaving between the extrema.
Figure 6.
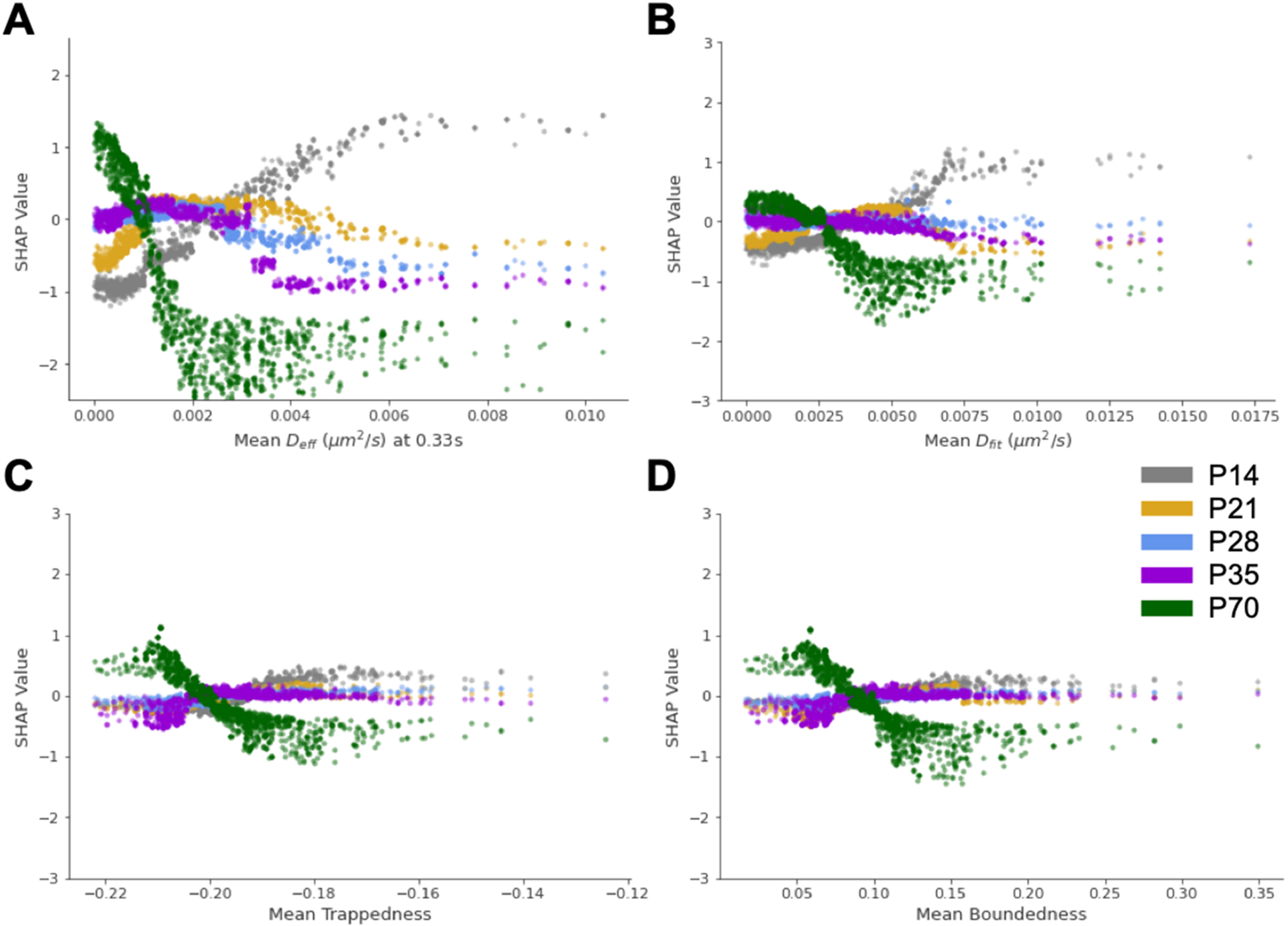
SHAP dependence plots for the four most influential features in the 5-age boosted decision tree classifier. SHAP dependence plots for (A) mean Deff, (B) mean Dfit, (C) mean trappedness, and (D) mean boundedness. Color scheme: P14 = gray, P21 = gold, P28 = blue, P35 = purple, P70 = green.
SHAP summary plots, like the one presented in Figure 5C, can also be split and used to rank the most influential features for predicting each of the discrete classes (Figure 7A–E). These summary plots combine feature importance with feature effects. Each point on the summary plot is a SHAP value for a specific feature and instance. The position on the y-axis is determined by the feature and the x-axis by SHAP value. The color represents the magnitude of the feature from low (blue) to high (red). Local-averaged features make up the entirety of the top five features for four of the five age groups. The only age to include a standard not locally averaged feature was P70, where progression is the second most influential feature. These SHAP summary plots provide insight into why the extreme ages were predicted with the highest accuracy. In the P14 and P70 age groups, SHAP values were typically clustered at two separate locations along the axis of SHAP magnitude. The clusters for a given feature tended to either be on separate sides of the neutral value of 0.0 or have one cluster much closer to 0.0 (Figure 7A,E). While this clustering behavior was also observed for certain features within the intermediate age groups, the magnitude of the SHAP value where clustering occurred was greater for the P14 and P70 groups. Additionally, SHAP values were, in general, closer to the neutral value of 0.0 for the intermediate ages. SHAP values ranged from −1.1 to 1.6 and −2.6 to 1.4 for the P14 and P70 data sets, respectively, while the widest range for the intermediate ages, the P35 group, only spanned from −1.0 to 0.5 (Figure 7A–E).
Figure 7.
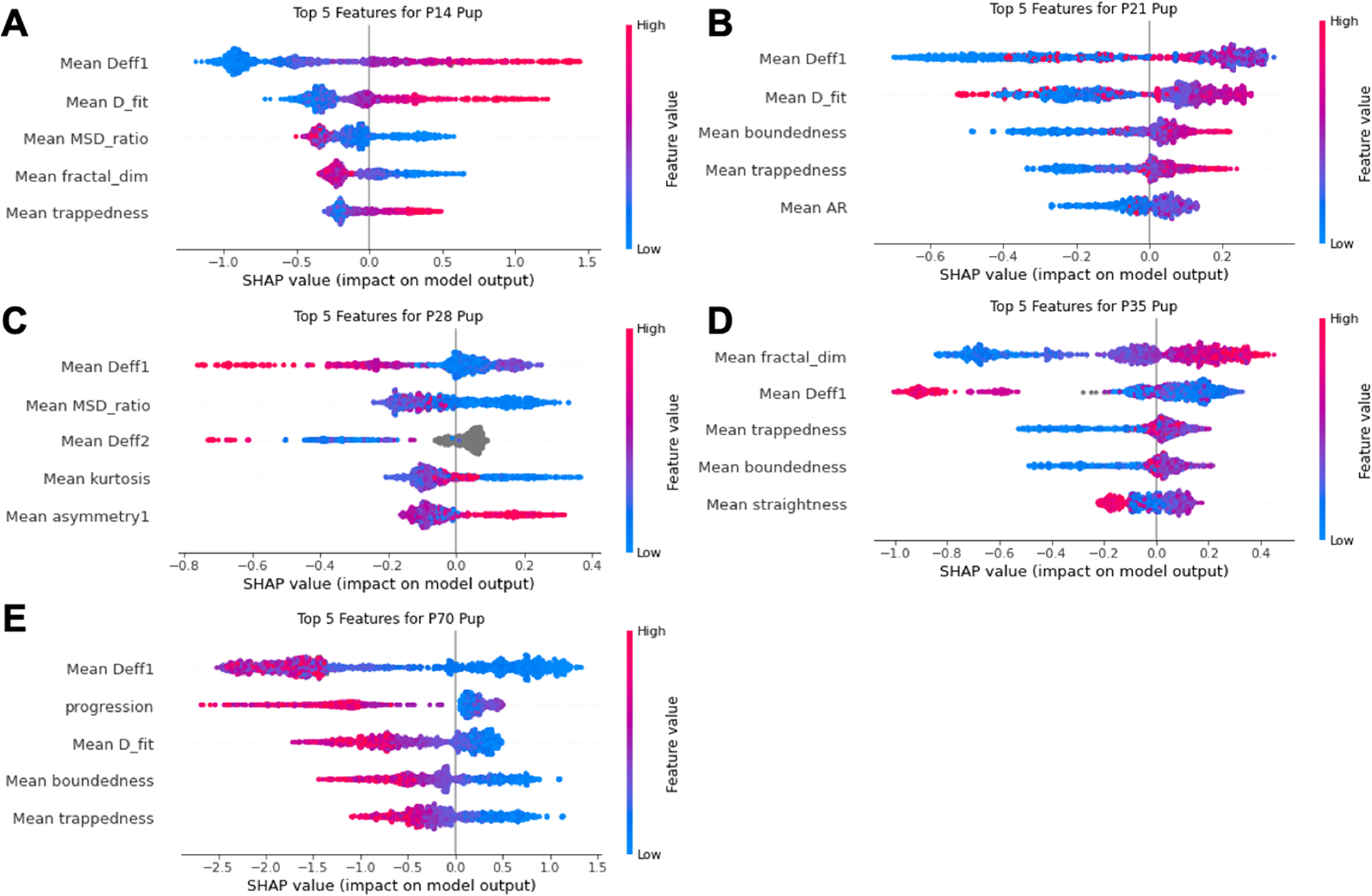
SHAP summary plots for each age of the 5-age boosted decision tree classifier. SHAP summary plots of the five most influential features for predicting the (A) P14, (B) P21, (C) P28, (D) P35, and (E) P70 age groups. Colorbar provides feature magnitude from low (blue) to high (red).
Reducing Resolution to Three Ages (P14, P35, and P70) Increases Classifier Accuracy to >86%.
Our 5-age boosted decision tree classifier struggled most at predicting the intermediate age groups (P21, P28, and P35) (Figure 5). To determine how accurate a model could be if resolution was reduced, we removed the P21 and P28 data sets from our analysis and trained a boosted decision tree classifier using data from the remaining groups (P14, P35, and P70). With these wider age gaps, we expected the differences in trajectory features from group to group to be larger and more readily leveraged by the machine learning algorithm. The new model far outperformed the 5-age predictor. The new 3-age model achieves an 86.64% accuracy overall, with individual recalls of 90.86, 76.72, and 92.92% for the P14, P35, and P70 groups, respectively (Figure 8A). This represents a significant increase in overall accuracy compared to the 5-age classifier (60.09%, Figure 5A). Similar to the 5-age classifier, the model was better at predicting the extreme ages (P14 and P70) than the intermediate age (P35). Of the 1609 P14 trajectories used to test the model, 147 were incorrectly classified (Figure 8B) and 101 of the 1498 P70 trajectories were mislabeled (Figure 8B). The P35 group had the lowest precision, recall, and f1-score, and of the 1632 trajectories tested, 380 were incorrectly classified (with 200 trajectories classified as P14 and 180 trajectories classified as P70).
Figure 8.
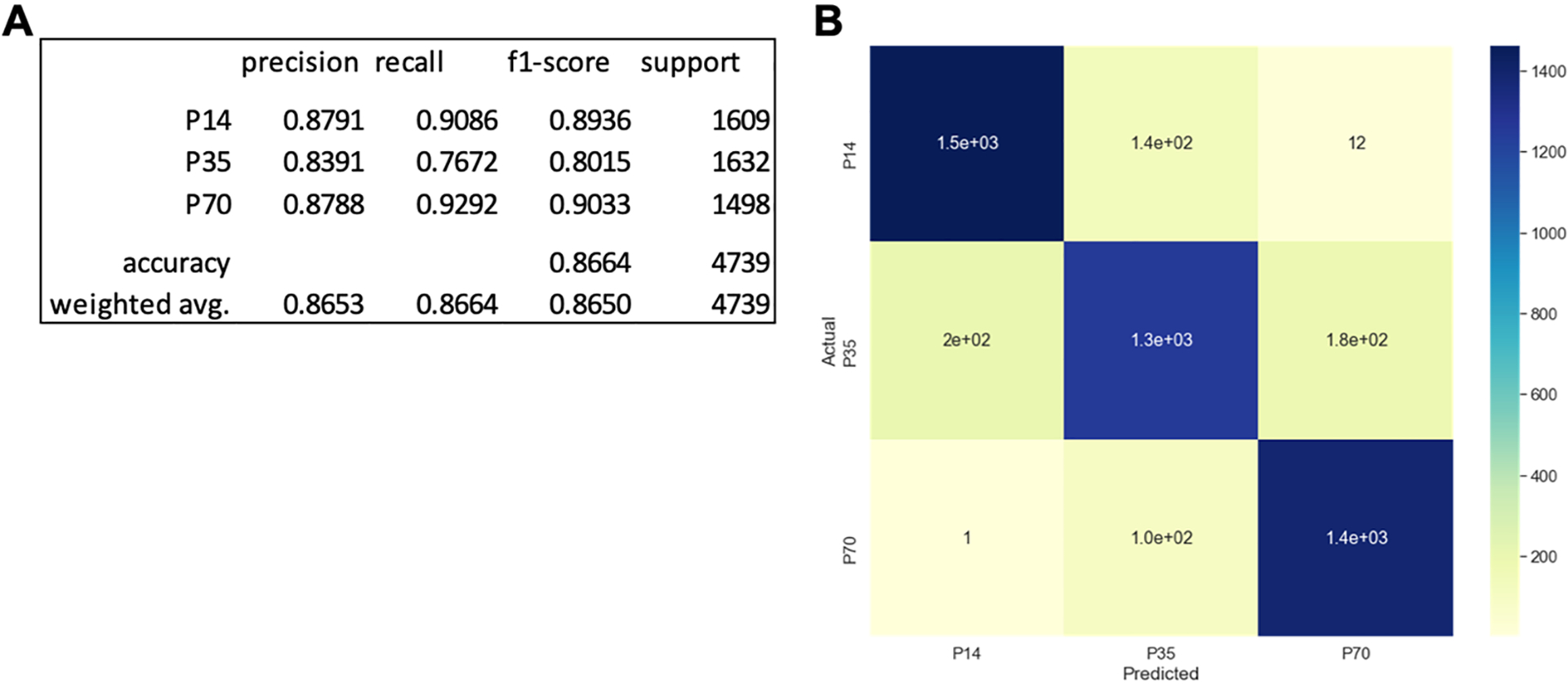
Reducing resolution to a 3-class prediction between P14, P35, and P70 increases model accuracy. (A) Precision, recall, f1-score, and support for each of the three classes along with overall model accuracy and weighted average precision, recall, and f1-score. (B) Confusion matrix for test data set comparing predicted age versus actual age. The colorbar reflects the number of trajectories in each bin.
SHAP feature importance plots, both for the overall model (Figure S3) and specific age groups (Figure S4), were generated along with SHAP dependence plots for the four most influential features in the 3-age classifier (Figure S3). Similar to the 5-age classifier, local-averaged features dominated the feature importance plots. Local-averaged features accounted for four of the top five and 13 of the top 15 most influential features overall (Figure S3). When focusing instead on the most influential features for predicting specific ages, local-averaged features remained dominant. The five most influential features for P14 and P35 prediction were all local-averaged features, and only one of the top five for the P70 group was not local-averaged (Figure S4).
In addition to showing changes in nanoparticle diffusivity with brain age, we also demonstrated the effectiveness of combining machine learning algorithms with biological MPT data sets. Through XGBoost, we generated boosted decision tree classifiers capable of predicting age with high accuracy. The model trained to classify trajectories between all five age groups (P14, P21, P28, P35, P70) predicted age with a 60.09% average accuracy (Figure 5), and the 3-age classifier was 86.64% accurate (Figure 8). To date, machine learning in combination with trajectory data has predominantly been used to predict motion type; the majority of this existing work has relied on artificial or simulated trajectories to train the model rather than experimentally collected trajectory data from particle tracking methodologies.46,82,83 MPT trajectory data have been used to train classifiers capable of predicting nanoparticle probe properties, such as size and surface charge,46,84 with high accuracy, but the focus herein was to allow prediction of a biological variable such as age. Based on existing MPT data that show differences in nanoparticle diffusivity and motion type depending on the duration of oxygen–glucose deprivation in rat brain slices,85 this approach can be expanded to a wide range of biological factors, such as the presence or extent of brain injury.
A distinct advantage of decision-tree-based classifiers like XGBoost is the ability to quantify individual feature importance. Through the use of SHAP summary plots, the five most important trajectory features for predicting chronological age between all five age groups were determined to be mean Deff at 0.33 s (Mean Deff1), mean fitted diffusion coefficient (D_fit), mean trappedness, mean boundedness, and progression. There is potential that certain features are indicative of specific interactions the nanoparticle experiences in the biological environment. For example, a shift in trajectory trappedness can represent nanoparticle uptake into a more confined environment like cytoplasm or lysosome, or an increase in efficiency can be indicative of particles being actively transported across a membrane via directed motion.46 One thing that remained consistent across models and age groups was the significance of local-averaged features to model prediction. These made up four of the top five most influential features for the 5-age classifier. Brain ECS is known to contain randomly distributed void spaces, commonly referred to as “lakes”, connected by highly tortuous channels of variable widths.30,67,86 If a nanoparticle is present in a “lake”, neighboring trajectories are likely experiencing a similar environment and exhibiting comparable behavior. Alternatively, behavior is expected to be more heterogeneous across neighboring trajectories if a nanoparticle is diffusing in an ECS channel whose width varies spatially. The distribution and density of these “lakes”, along with the fraction of the extracellular volume of which they consist, likely varies throughout neurodevelopment. It is thus unclear whether these differences are linked to the differences observed in local-averaged features as a function of age, but we do know that the significance of features varies depending on the age being predicted (Figure 7) and following the elimination of P21 and P28 groups (Figure S3). Regardless, local-averaged features were the most influential in predicting brain age from boosted decision tree classifiers and promote the importance of including statistical features in future applications of machine learning to MPT data sets.
While the 5-age and 3-age classifiers were able to achieve an average accuracy of 60.09 and 86.64%, respectively, which both far exceed the accuracy of a random guess, there is still room for improvement, especially when predicting the intermediate age groups. The 5-age classifier only accurately recalled 48.08, 28.44, and 53.02% of the P21, P28, and P35 age groups, respectively. In instances where brain microstructure undergoes changes from week-to-week, and potentially day-to-day, like elevated periods of plasticity during neurodevelopment, being able to predict age with single day resolution would be ideal. The incorporation of MPT data from additional nanoparticle sizes and types, the generation of extra features, and the implementation of alternative machine learning algorithms are all potential ways to improve model performance in future iterations. However, it is encouraging that even the worst-performing age group (P28) still outperformed a random guess (20.00% for 5-class prediction).
The MPT studies carried out herein were performed in the motor and somatosensory cortex. One limitation to this study is a lack of control over the cortical layer in which MPT was performed and that experiments are carried out ex vivo. In humans and rodents, the cortex consists of six layers, each having different cell populations87 and timelines of increased excitatory and inhibitory activity.88 ECM protein expression can also vary from layer to layer.89 To determine if MPT is sensitive enough to detect intercortical differences, studies will need to be run in each of the six cortical layers in the rat or in more advanced species, such as the ferret, where cortical macrostructure is more aligned with the human brain.90 There is potential that the processing steps required to generate ex vivo brain slices alter the innate structure of brain ECM. However, Nance et al. has previously demonstrated good agreement between the behavior of 40, 100, and 200 nm PS–PEG nanoparticles in mouse brain in vivo and rat brain tissue ex vivo.33 Additionally, organotypic brain slices have been used extensively to study the diffusion of various substances,33,84,85,91 neural electrophysiology,92 and cell migration.93,94 Sources of error stemming from slice preparation, if present at all, would likely only exist in the older age groups, which have less documented cases of their use and more limited culturing viability. Regardless, this study demonstrates the utility of combining MPT with machine learning to predict the chronological age of rat brain slices using features generated solely from nanoparticle trajectories.
The success of this approach lends itself to numerous avenues of future study. Of particular interest would be studies aimed at better understanding the mechanisms of neurodegeneration, whether it is induced by a traumatic event such as stroke or traumatic brain injury, or disorders associated with aging, like Alzheimer’s disease and Parkinson’s disease. Neurodegeneration is typically associated with the activation of neuroinflammatory pathways, which triggers the release of ECM-degrading matrix metalloproteinases (MMPs).95,96 In addition to the onset of neuroinflammation, astrocytes take part in a process known as glial scarring, where they migrate to areas of neuronal death, proliferate, and deposit CSPGs in an attempt to wall off damaged areas and prevent the propagation of injury.97,98 The upregulation of ECM-degrading MMPs will inevitably cause changes to the extracellular environment but so should the processes associated with glial scarring. It is thus difficult to predict whether neurodegeneration would enhance or reduce extracellular diffusion, and whether that trend would be consistent across various injuries and disorders. It likely depends on both proximity to the core of an injury, such as in stroke or traumatic brain injury and, in the case of neurodegeneration, timing following the onset of pathological processes. Initial work suggests an increase in diffusive ability, however. In an oxygen–glucose deprivation model of ischemic injury, Joseph and Liao et al. found that extracellular nanoparticle diffusion in both the cortex and striatum was enhanced following injury.85 Similarly, in an α-synuclein-induced model of neurodegeneration, Soria et al. observed an increase in extracellular diffusivity when compared to that of control animals.34 However, these represent the results of only two studies; more work is needed before any definitive conclusions can be drawn on the impact of neurodegeneration on diffusion and MPT analysis of brain ECM structure.
An added bonus of the particle tracking technique is that multiple types of probes and particle dynamics can be studied. This allows researchers to identify key design parameters for specific injuries or disorders, such as the max hydrodynamic diameter of the therapeutic vehicle, for example. There is also potential to leverage an enhanced understanding of the barriers to extracellular diffusion to engineer ways to overcome diffusional barriers. One such option is to include or co-administer an ECM-altering substance along with a therapeutic payload.99–101 The ECM-altering substance can specifically alter the diseased brain microenvironment in a way that enhances tissue penetration and achieves greater distribution of the therapeutic throughout the injured region, resulting in more efficacious outcomes.
CONCLUSION
MPT is capable of detecting changes in brain ECM that occur during neurodevelopment and in response to enzymatic digestion of the ECM. When paired with machine learning algorithms such as XGBoost, we can generate models capable of predicting associated biological factors such as age with high accuracy, and these predications are made based solely on features associated with a given trajectory. By using decision-tree-based classifiers, the influence of individual features can be quantified and ranked. Local-averaged features were the most influential when predicting brain age, but further work is necessary to understand what the magnitude of certain features elucidates about the underlying biology. In addition to neurodevelopment, the ECM plays many critical roles in maintaining homeostasis and is thought to be involved in the pathophysiology of many neurological diseases and brain injuries. The application of MPT in studying ECM structure could explicitly define mechanisms involved in neurological disease progression and reveal additional targets for therapeutic intervention. Additionally, MPT can enhance our baseline understanding of the structure–function relationships of the brain under normal physiological conditions and has the potential to become used as one marker of neurological disease severity.
MATERIALS AND METHODS
Organotypic Hemispheric Brain Slice Preparation.
Brain slices were prepared from male SD rat pups at varying ages, depending on the specific study. This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (NIH). All of the animals were handled according to approved institutional animal care and use committee (IACUC) protocols (#4383-02) of the University of Washington. The University of Washington has an approved Animal Welfare Assurance (#A3464-01) on file with the NIH Office of Laboratory Animal Welfare (OLAW), is registered with the United States Department of Agriculture (USDA, certificate #91-R-0001), and is accredited by AAALAC International. Every effort was made to minimize suffering. Following euthanasia, brains were extracted, immersed in room temperature (22 °C) dissection media, and cut into hemispheres with a razor blade. Coronal slices (300 μm thick) were prepared from each hemisphere using a Mcllwain tissue chopper (Ted Pella, Redding, CA).47 Individual slices were plated on 30 mm cell culture inserts (MilliporeSigma, Burlington, MA) in nontreated 6-well plates (USA Scientific, Ocala, FL). Prior to plating, 6-well plates were filled with 1 mL of SCM. Slices were incubated in sterile conditions at 37 °C and 5% CO2. For a more detailed, step-by-step procedure of slice preparation, and buffer recipes, refer to SI Appendix, SI Experimental Procedures.
Nanoparticle Preparation and Characterization.
Fluorescent carboxylate (COOH)-modified polystyrene latex (PS) nanoparticles (PS-COOH) (40 nm) (Fisher Scientific, Hampton, NH) were covalently modified with methoxy (MeO)-poly(ethylene glycol) (PEG)-amine (NH2) (5 kDa MW, Creative PEG Works, Winston-Salem, NC) by carboxyl amine reaction.63 The hydrodynamic diameter and polydispersity index of the resulting PEG-conjugated fluorescent nanoparticles were measured via dynamic light scattering and the ζ-potential by laser Doppler anemometry. Refer to SI Appendix, SI Experimental Procedures for more detailed reaction conditions and characterization.
Multiple Particle Tracking in Organotypic Brain Slices Ex Vivo.
All MPT studies were performed between 20 and 24 h postslice preparation. Slices were imaged in a temperature-controlled incubation chamber maintained at 37 °C, 5% CO2, and 80% humidity. Thirty minutes prior to video acquisition, injections of 40 nm PS–PEG nanoparticles diluted in 1× phosphate-buffered saline (PBS) were carried out in each slice using a 10 μL glass syringe (model 701, cemented needle, 26-gauge, Hamilton Company, Reno, NV). A total of five 0.5 μL injections were made in the cortex of each slice. For the study involving the degradation of PNNs ex vivo, particle injections were made 90 min after treatment was applied, and videos were collected following a 30 min incubation. In total, MPT was performed 120 min after treatment with either HYase, ChABC, or SCM.
A total of five videos were collected from the motor and somatosensory cortex of each slice. Videos were collected at 33 fps with a 100× magnification objective (oil immersion, 1.45 numerical aperture, Nikon Instruments, Melville, NY) for 651 frames via fluorescent microscopy using a cMOS camera (Hamamatsu Photonics, Bridgewater, NJ) mounted on a confocal microscope. Nanoparticle trajectories, trajectory MSDs, and Db,eff were calculated via diff_classifier (https://github.com/ccurtis7/diff_classifier), a Python package developed within our group.54
For enzyme-induced PNN breakdown experiments, three brain slices for each treatment group (ChABC, HYase, and NT) were taken from each of the four animals used. Collecting five videos from each slice resulted in a total of 60 videos and >60,000 total trajectories per treatment group. For age-dependent MPT, a total of 15 videos were taken from three slices at each age. This resulted in >4900 total trajectories per group.
XGBoost Predictive Model for Age-Related Data Classification.
XGBoost is a type of boosted decision tree in which the algorithm sequentially builds itself using multiple weak prediction models. Every tree produced in the series is fit to a modified weighted version of the original data set. This sequential method continues until a set number of learners has been created or until the model converges within the exponential loss function. Prediction is then made by calculating the weighted average of all produced learners.102 XGBoost specifically incorporates regularization into its algorithm to control overfitting the data during training. It incorporates a distinct objective function that encourages simple models and decreases variance.103
A total of 64,452 samples were rebalanced using under-sampling into five even sets of 6194 data points for each age classification. Training and testing data sets were randomly sampled from the age data with a training/testing split percentage of 90 to 10%. Features were chosen and calculated based on the geometry of the trajectory using feature calculation algorithms on diff_classifier. This includes asymmetry, anomalous exponent, aspect ratio, elongation, boundedness, fractal dimension, efficiency, straightness, kurtosis, and MSD ratio. Extra features were created based on the immediate surrounding data. Mean values of each calculated feature were calculated and used in prediction. In total, 39 different features were used. A comprehensive list and description of every feature used can be found in the Supporting Information (Table S2). The XGBoost model was trained using a max depth of 6, an eta of 0.005, a gamma of 5, a subsample of 0.6, and a colsample_bytree of 0.6.
To better understand the individual contribution to overall prediction, shapely additive explanations were calculated for every feature. SHAP is based on the theoretically optimal use of shapley values, which are a feature’s contribution to the prediction, :
in which E(βjXj) is the mean effect estimate for feature j. The contribution is the difference in the feature effect and the average effect. SHAP values were used to create summary and dependency plots of the top features in prediction of each age category. The summary plot shows the average impact of each feature on prediction output calculated by the mean absolute SHAP values:
This importance value differs from other importance calculations due to its basis on magnitude of feature attributions. Analysis for age-related data can be found on the ECM-MPT-Predictive_Age_Data repository (https://github.com/dash2927/ECM-MPT-Predictive_Age_Data).
Characterizing the Timeline of Enzyme-Induced Perineuronal Net Breakdown in Organotypic Rat Brain Slices Ex Vivo.
All experiments were initiated between 20 and 24 h postslice preparation and used litter-matched male rats to reduce biological variability. Slices were treated with either ChABC (0.4 U/mL), HYase (35 U/mL), or SCM (NT). At the initial time point, 200 μL of a given treatment was applied to each brain slice and returned to the incubator. One brain slice from each treatment group was removed and fixed at 15, 30, 45, and 120 min post-treatment, resulting in a total of four slices per treatment condition. Slices were stained with 500 μL of 1× PBS containing 10 μg/mL fluorescein-labeled WFA lectin for 12 h at 4 °C. Cell nuclei were stained with 1 μg/mL DAPI for 30 min. All imaging was performed using a confocal microscope (Nikon Instruments). Three representative images were taken at 20× magnification from the cortex of each brain slice at each time point. For specific details on ChABC, HYase, and NT working solution preparation and staining buffers, see SI Appendix, SI Experimental Procedures.
Immunohistochemistry and Lectin Staining on Fixed Rat Brain Slices.
Following euthanasia, SD rats were perfused with sterile 1× PBS. Brains were immediately extracted and placed in 10% formalin phosphate buffer for 24 h at 4 °C. Brains went through a 30% sucrose gradient to be frozen and sectioned into 30 μm thick coronal sections using a Leica CM1950 cryostat (Leica Biosystems, Buffalo Grove, IL). Sections were first incubated with rabbit anti-parvalbumin (anti-PVA, Abcam ab11427, Cambridge, UK) at a 1:100 dilution in 1× PBS containing 1% Triton X-100 (MilliporeSigma), 3% donkey serum (MilliporeSigma), and 10 μg/mL WFA for 6 h at room temperature (22 °C). Following a wash step, a 1:500 dilution of Alexa Fluor 568-labeled donkey anti-rabbit IgG (ThermoFisher) in 1× PBS containing 1% Triton X-100 and 10 μg/mL WFA was applied to sections for 2 h. Cellular nuclei were stained with a 1 μg/mL solution of DAPI in 1× PBS for 15 min. Following a final wash, microscope slides were mounted with a glass coverslip using Wako antifade media (Vector Laboratories) and stored at −20 °C until imaged. Sections were imaged using a confocal microscope.
Statistical Analysis.
All statistical analyses were carried out in GraphPad Prism (GraphPad Software Inc., version 8.2.0). For all tests run, differences were defined as statistically significant at p < 0.05. The D’Agostino–Pearson omnibus K2 test was used to test for normality. If we were unable to reject the null hypothesis that data were sampled from a population that follows a Gaussian distribution, we ran Brown–Forsythe and Welch ANOVA tests. We used Dunnett T3 to correct for multiple comparisons. If we were able to reject the null hypothesis that the data were taken from a normally distributed population, we used the Kruskal–Wallis test for significance. In these instances, we applied Dunn’s method to correct for multiple comparisons.
Data and Code Availability.
All data presented herein can be provided upon request. All code is available on github, with links included in the methods.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Dr. Chad Curtis, who established the Python-based analysis pipeline that was used to track and quantify nanoparticle trajectories, and Dr. David Beck for his guidance on machine algorithm development, training, testing, and validation. This work was supported by the National Institute of General Medical Sciences (Grant No. R35 GM124677) and the Data Sciences Initiative in the Department of Chemical Engineering at the University of Washington. This project received partial support from the Data Intensive Research Enabling Clean Technology (DIRECT) NSF National Research Traineeship (DGE-1633216).
Footnotes
The authors declare no competing financial interest.
An earlier version of this work appeared as a preprint: McKenna, M.; Shackelford, D.; Ferreira Pontes, H.; Ball, B. K.; Gao, T.; Nance, E. Multiple particle tracking detects changes in brain extracellular matrix structure and predicts neurodevelopmental age; https://www.biorxiv.org/content/10.1101/2020.04.20.050112v1 (accessed April 30, 2021).
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.1c00394.
Supplemental experimental procedures, figures, and tables (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acsnano.1c00394
REFERENCES
- (1).Zimmermann DR; Dours-Zimmermann MT Extracellular Matrix of the Central Nervous System: From Neglect to Challenge. Histochem. Cell Biol 2008, 130, 635–53. [DOI] [PubMed] [Google Scholar]
- (2).Krishnaswamy VR; Benbenishty A; Blinder P; Sagi I Demystifying the Extracellular Matrix and Its Proteolytic Remodeling in the Brain: Structural and Functional Insights. Cell. Mol. Life Sci 2019, 76, 3229–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Okamoto M; Mori S; Endo H A Protective Action of Chondroitin Sulfate Proteoglycans against Neuronal Cell Death Induced by Glutamate. Brain Res. 1994, 637, 57–67. [DOI] [PubMed] [Google Scholar]
- (4).Cabungcal JH; Steullet P; Morishita H; Kraftsik R; Cuenod M; Hensch TK; Do KQ Perineuronal Nets Protect Fast-Spiking Interneurons against Oxidative Stress. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 9130–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Thomsen MS; Routhe LJ; Moos T The Vascular Basement Membrane in the Healthy and Pathological Brain. J. Cereb. Blood Flow Metab 2017, 37, 3300–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Xu L; Nirwane A; Yao Y Basement Membrane and Blood-Brain Barrier. Stroke Vasc Neurol 2019, 4, 78–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Barritt AW; Davies M; Marchand F; Hartley R; Grist J; Yip P; McMahon SB; Bradbury EJ Chondroitinase Abc Promotes Sprouting of Intact and Injured Spinal Systems after Spinal Cord Injury. J. Neurosci 2006, 26, 10856–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Carstens KE; Phillips ML; Pozzo-Miller L; Weinberg RJ; Dudek SM Perineuronal Nets Suppress Plasticity of Excitatory Synapses on Ca2 Pyramidal Neurons. J. Neurosci 2016, 36, 6312–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Carulli D; Pizzorusso T; Kwok JC; Putignano E; Poli A; Forostyak S; Andrews MR; Deepa SS; Glant TT; Fawcett JW Animals Lacking Link Protein Have Attenuated Perineuronal Nets and Persistent Plasticity. Brain 2010, 133, 2331–47. [DOI] [PubMed] [Google Scholar]
- (10).Massey JM; Hubscher CH; Wagoner MR; Decker JA; Amps J; Silver J; Onifer SM Chondroitinase Abc Digestion of the Perineuronal Net Promotes Functional Collateral Sprouting in the Cuneate Nucleus after Cervical Spinal Cord Injury. J. Neurosci 2006, 26, 4406–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Pizzorusso T; Medini P; Berardi N; Chierzi S; Fawcett JW; Maffei L Reactivation of Ocular Dominance Plasticity in the Adult Visual Cortex. Science 2002, 298, 1248–51. [DOI] [PubMed] [Google Scholar]
- (12).Sorg BA; Berretta S; Blacktop JM; Fawcett JW; Kitagawa H; Kwok JC; Miquel M Casting a Wide Net: Role of Perineuronal Nets in Neural Plasticity. J. Neurosci 2016, 36, 11459–11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Wen TH; Binder DK; Ethell IM; Razak KA The Perineuronal ‘Safety’ Net? Perineuronal Net Abnormalities in Neurological Disorders. Front. Mol. Neurosci 2018, 11, 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Dzyubenko E; Manrique-Castano D; Kleinschnitz C; Faissner A; Hermann DM Topological Remodeling of Cortical Perineuronal Nets in Focal Cerebral Ischemia and Mild Hypoperfusion. Matrix Biol. 2018, 74, 121–132. [DOI] [PubMed] [Google Scholar]
- (15).Pantazopoulos H; Berretta S In Sickness and in Health: Perineuronal Nets and Synaptic Plasticity in Psychiatric Disorders. Neural Plast 2016, 2016, 9847696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Fukuda S; Fini CA; Mabuchi T; Koziol JA; Eggleston LL Jr.; del Zoppo GJ Focal Cerebral Ischemia Induces Active Proteases That Degrade Microvascular Matrix. Stroke 2004, 35, 998–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Hamann GF; Okada Y; Fitridge R; del Zoppo GJ Microvascular Basal Lamina Antigens Disappear During Cerebral Ischemia and Reperfusion. Stroke 1995, 26, 2120–6. [DOI] [PubMed] [Google Scholar]
- (18).Jullienne A; Roberts JM; Pop V; Murphy MP; Head E; Bix GJ; Badaut J Juvenile Traumatic Brain Injury Induces Long-Term Perivascular Matrix Changes Alongside Amyloid-Beta Accumulation. J. Cereb. Blood Flow Metab 2014, 34, 1637–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Muellner A; Benz M; Kloss CU; Mautes A; Burggraf D; Hamann GF Microvascular Basal Lamina Antigen Loss after Traumatic Brain Injury in the Rat. J. Neurotrauma 2003, 20, 745–54. [DOI] [PubMed] [Google Scholar]
- (20).Korogod N; Petersen CC; Knott GW Ultrastructural Analysis of Adult Mouse Neocortex Comparing Aldehyde Perfusion with Cryo Fixation. eLife 2015, 4, No. e05793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Lipachev N; Arnst N; Melnikova A; Jaalinoja H; Kochneva A; Zhigalov A; Kulesskaya N; Aganov AV; Mavlikeev M; Rauvala H; Kiyasov AP; Paveliev M Quantitative Changes in Perineuronal Nets in Development and Posttraumatic Condition. J. Mol. Histol 2019, 50, 203–216. [DOI] [PubMed] [Google Scholar]
- (22).Rowlands D; Lensjo KK; Dinh T; Yang S; Andrews MR; Hafting T; Fyhn M; Fawcett JW; Dick G Aggrecan Directs Extracellular Matrix-Mediated Neuronal Plasticity. J. Neurosci 2018, 38, 10102–10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Moeendarbary E; Weber IP; Sheridan GK; Koser DE; Soleman S; Haenzi B; Bradbury EJ; Fawcett J; Franze K The Soft Mechanical Signature of Glial Scars in the Central Nervous System. Nat. Commun 2017, 8, 14787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Nicholson C Ion-Selective Microelectrodes and Diffusion Measurements as Tools to Explore the Brain Cell Microenvironment. J. Neurosci. Methods 1993, 48, 199–213. [DOI] [PubMed] [Google Scholar]
- (25).Nicholson C Diffusion and Related Transport Mechanisms in Brain Tissue. Rep. Prog. Phys 2001, 64, 815–884. [Google Scholar]
- (26).Nicholson C; Phillips JM Ion Diffusion Modified by Tortuosity and Volume Fraction in the Extracellular Microenvironment of the Rat Cerebellum. J. Physiol 1981, 321, 225–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Nicholson C; Phillips JM; Gardner-Medwin AR Diffusion from an Iontophoretic Point Source in the Brain: Role of Tortuosity and Volume Fraction. Brain Res. 1979, 169, 580–4. [DOI] [PubMed] [Google Scholar]
- (28).Hrabetova S; Nicholson C Biophysical Properties of Brain Extracellular Space Explored with Ion-Selective Microelectrodes, Integrative Optical Imaging and Related Techniques. In Electrochemical Methods for Neuroscience; Michael AC, Borland LM, Eds.; CRC Press: Boca Raton, FL, 2007. [PubMed] [Google Scholar]
- (29).Nicholson C; Tao L Hindered Diffusion of High Molecular Weight Compounds in Brain Extracellular Microenvironment Measured with Integrative Optical Imaging. Biophys. J 1993, 65, 2277–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Thorne RG; Nicholson C In Vivo Diffusion Analysis with Quantum Dots and Dextrans Predicts the Width of Brain Extracellular Space. Proc. Natl. Acad. Sci. U. S. A 2006, 103, 5567–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Godin AG; Varela JA; Gao Z; Danne N; Dupuis JP; Lounis B; Groc L; Cognet L Single-Nanotube Tracking Reveals the Nanoscale Organization of the Extracellular Space in the Live Brain. Nat. Nanotechnol 2017, 12, 238–243. [DOI] [PubMed] [Google Scholar]
- (32).Nance E; Timbie K; Miller GW; Song J; Louttit C; Klibanov AL; Shih TY; Swaminathan G; Tamargo RJ; Woodworth GF; Hanes J; Price RJ Non-Invasive Delivery of Stealth, Brain-Penetrating Nanoparticles across the Blood-Brain Barrier Using Mri-Guided Focused Ultrasound. J. Controlled Release 2014, 189, 123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Nance EA; Woodworth GF; Sailor KA; Shih TY; Xu Q; Swaminathan G; Xiang D; Eberhart C; Hanes J A Dense Poly(Ethylene Glycol) Coating Improves Penetration of Large Polymeric Nanoparticles within Brain Tissue. Sci. Transl. Med 2012, 4, 149ra119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Soria FN; Paviolo C; Doudnikoff E; Arotcarena ML; Lee A; Danne N; Mandal AK; Gosset P; Dehay B; Groc L; Cognet L; Bezard E Synucleinopathy Alters Nanoscale Organization and Diffusion in the Brain Extracellular Space through Hyaluronan Remodeling. Nat. Commun 2020, 11, 3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Paviolo C; Soria FN; Ferreira JS; Lee A; Groc L; Bezard E; Cognet L Nanoscale Exploration of the Extracellular Space in the Live Brain by Combining Single Carbon Nanotube Tracking and Super-Resolution Imaging Analysis. Methods 2020, 174, 91–99. [DOI] [PubMed] [Google Scholar]
- (36).Xu Q; Boylan NJ; Suk JS; Wang YY; Nance EA; Yang JC; McDonnell PJ; Cone RA; Duh EJ; Hanes J Nanoparticle Diffusion in, and Microrheology of, the Bovine Vitreous ex Vivo. J. Controlled Release 2013, 167, 76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Suk JS; Lai SK; Wang YY; Ensign LM; Zeitlin PL; Boyle MP; Hanes J The Penetration of Fresh Undiluted Sputum Expectorated by Cystic Fibrosis Patients by Non-Adhesive Polymer Nanoparticles. Biomaterials 2009, 30, 2591–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Macierzanka A; Mackie AR; Bajka BH; Rigby NM; Nau F; Dupont D Transport of Particles in Intestinal Mucus under Simulated Infant and Adult Physiological Conditions: Impact of Mucus Structure and Extracellular DNA. PLoS One 2014, 9, No. e95274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Wang YY; Lai SK; Suk JS; Pace A; Cone R; Hanes J Addressing the Peg Mucoadhesivity Paradox to Engineer Nanoparticles That “Slip” through the Human Mucus Barrier. Angew. Chem., Int. Ed 2008, 47, 9726–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Lai SK; O’Hanlon DE; Harrold S; Man ST; Wang YY; Cone R; Hanes J Rapid Transport of Large Polymeric Nanoparticles in Fresh Undiluted Human Mucus. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 1482–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Suk JS; Suh J; Lai SK; Hanes J Quantifying the Intracellular Transport of Viral and Nonviral Gene Vectors in Primary Neurons. Exp Biol. Med. (Maywood) 2007, 232, 461–469. [PubMed] [Google Scholar]
- (42).Xiao PJ; Samulski RJ Cytoplasmic Trafficking, Endosomal Escape, and Perinuclear Accumulation of Adeno-Associated Virus Type 2 Particles Are Facilitated by Microtubule Network. J. Virol 2012, 86, 10462–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Suh J; Wirtz D; Hanes J Efficient Active Transport of Gene Nanocarriers to the Cell Nucleus. Proc. Natl. Acad. Sci. U. S. A 2003, 100, 3878–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Nance E; Zhang C; Shih TY; Xu Q; Schuster BS; Hanes J Brain-Penetrating Nanoparticles Improve Paclitaxel Efficacy in Malignant Glioma Following Local Administration. ACS Nano 2014, 8, 10655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Joseph A; Wood T; Chen CC; Corry K; Snyder JM; Juul SE; Parikh P; Nance E Curcumin-Loaded Polymeric Nanoparticles for Neuro-Protection in Neonatal Rats with Hypoxic-Ischemic Encephalopathy. Nano Res. 2018, 11, 5670–5688. [Google Scholar]
- (46).Wagner T; Kroll A; Haramagatti CR; Lipinski HG; Wiemann M Classification and Segmentation of Nanoparticle Diffusion Trajectories in Cellular Micro Environments. PLoS One 2017, 12, No. e0170165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Curtis C; McKenna M; Pontes H; Toghani D; Choe A; Nance E Predicting in Situ Nanoparticle Behavior Using Multiple Particle Tracking and Artificial Neural Networks. Nanoscale 2019, 11, 22515–22530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Benoit J; Ayoub AE; Rakic P Transcriptomics of Critical Period of Visual Cortical Plasticity in Mice. Proc. Natl. Acad. Sci. U. S. A 2015, 112, 8094–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Espinosa JS; Stryker MP Development and Plasticity of the Primary Visual Cortex. Neuron 2012, 75, 230–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Gordon JA; Stryker MP Experience-Dependent Plasticity of Binocular Responses in the Primary Visual Cortex of the Mouse. J. Neurosci 1996, 16, 3274–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Dityatev A; Rusakov DA Molecular Signals of Plasticity at the Tetrapartite Synapse. Curr. Opin. Neurobiol 2011, 21, 353–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Long KR; Huttner WB How the Extracellular Matrix Shapes Neural Development. Open Biol. 2019, 9, 180216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Sood D; Cairns DM; Dabbi JM; Ramakrishnan C; Deisseroth K; Black LD 3rd; Santaniello S; Kaplan DL Functional Maturation of Human Neural Stem Cells in a 3d Bioengineered Brain Model Enriched with Fetal Brain-Derived Matrix. Sci. Rep 2019, 9, 17874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Curtis C; Rokem A; Nance E Diff_Classifier: Parallelization of Multi-Particle Tracking Video Analyses. J. Open Source Softw 2019, 4, 989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Tinevez JY; Perry N; Schindelin J; Hoopes GM; Reynolds GD; Laplantine E; Bednarek SY; Shorte SL; Eliceiri KW Trackmate: An Open and Extensible Platform for Single-Particle Tracking. Methods 2017, 115, 80–90. [DOI] [PubMed] [Google Scholar]
- (56).Tonnesen J; Inavalli V; Nagerl UV Super-Resolution Imaging of the Extracellular Space in Living Brain Tissue. Cell 2018, 172, 1108–1121. [DOI] [PubMed] [Google Scholar]
- (57).Curtis C; Toghani D; Wong B; Nance E Colloidal Stability as a Determinant of Nanoparticle Behavior in the Brain. Colloids Surf., B 2018, 170, 673–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Amsden B Solute Diffusion within Hydrogels. Mechanisms and Models. Macromolecules 1998, 31, 8382–8395. [Google Scholar]
- (59).Amsden B An Obstruction-Scaling Model for Diffusion in Homogeneous Hydrogels. Macromolecules 1999, 32, 874–879. [Google Scholar]
- (60).Lensjø KK; Christensen AC; Tennøe S; Fyhn M; Hafting T Differential Expression and Cell-Type Specificity of Perineuronal Nets in Hippocampus, Medial Entorhinal Cortex, and Visual Cortex Examined in the Rat and Mouse. eNeuro 2017, 4, e0379–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Chu P; Abraham R; Budhu K; Khan U; De Marco Garcia N; Brumberg JC The Impact of Perineuronal Net Digestion Using Chondroitinase Abc on the Intrinsic Physiology of Cortical Neurons. Neuroscience 2018, 388, 23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Vedunova M; Sakharnova T; Mitroshina E; Perminova M; Pimashkin A; Zakharov Y; Dityatev A; Mukhina I Seizure-Like Activity in Hyaluronidase-Treated Dissociated Hippocampal Cultures. Front. Cell. Neurosci 2013, 7, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Nance E Brain-Penetrating Nanoparticles for Analysis of the Brain Microenvironment. Methods Mol. Biol 2017, 1570, 91–104. [DOI] [PubMed] [Google Scholar]
- (64).Perry JL; Reuter KG; Kai MP; Herlihy KP; Jones SW; Luft JC; Napier M; Bear JE; DeSimone JM Pegylated Print Nanoparticles: The Impact of Peg Density on Protein Binding, Macrophage Association, Biodistribution, and Pharmacokinetics. Nano Lett. 2012, 12, 5304–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Hrabetova S; Masri D; Tao L; Xiao F; Nicholson C Calcium Diffusion Enhanced after Cleavage of Negatively Charged Components of Brain Extracellular Matrix by Chondroitinase Abc. J. Physiol 2009, 587, 4029–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Nicholson C; Sykova E Extracellular Space Structure Revealed by Diffusion Analysis. Trends Neurosci. 1998, 21, 207–15. [DOI] [PubMed] [Google Scholar]
- (67).Sykova E; Nicholson C Diffusion in Brain Extracellular Space. Physiol. Rev 2008, 88, 1277–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Deen W Hindered Transport of Large Molecules in Liquid-Filled Pores. AIChE J. 1987, 33, 1409–1425. [Google Scholar]
- (69).Lehmenkuhler A; Sykova E; Svoboda J; Zilles K; Nicholson C Extracellular Space Parameters in the Rat Neocortex and Subcortical White Matter During Postnatal Development Determined by Diffusion Analysis. Neuroscience 1993, 55, 339–51. [DOI] [PubMed] [Google Scholar]
- (70).Saifuddin N; Raziah AZ; Junizah AR Carbon Nanotubes: A Review on Structure and Their Interaction with Proteins. J. Chem 2013, 2013, 676815. [Google Scholar]
- (71).Beyene AG; Delevich K; Del Bonis-O’Donnell JT; Piekarski DJ; Lin WC; Thomas AW; Yang SJ; Kosillo P; Yang D; Prounis GS; Wilbrecht L; Landry MP Imaging Striatal Dopamine Release Using a Nongenetically Encoded near Infrared Fluorescent Catecholamine Nanosensor. Sci. Adv 2019, 5, No. eaaw3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Zhang M; Bishop BP; Thompson NL; Hildahl K; Dang B; Mironchuk O; Chen N; Aoki R; Holmberg VC; Nance E Quantum Dot Cellular Uptake and Toxicity in the Developing Brain: Implications for Use as Imaging Probes. Nanoscale Adv. 2019, 1, 3424–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Nance E; Zhang F; Mishra MK; Zhang Z; Kambhampati SP; Kannan RM; Kannan S Nanoscale Effects in Dendrimer-Mediated Targeting of Neuroinflammation. Biomaterials 2016, 101, 96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Rusakov DA; Kullmann DM Geometric and Viscous Components of the Tortuosity of the Extracellular Space in the Brain. Proc. Natl. Acad. Sci. U. S. A 1998, 95, 8975–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Gao R; Wang M; Lin J; Hu L; Li Z; Chen C; Yuan L Spatiotemporal Expression Patterns of Chondroitin Sulfate Proteoglycan Mrnas in the Developing Rat Brain. NeuroReport 2018, 29, 517–523. [DOI] [PubMed] [Google Scholar]
- (76).Margolis RU; Margolis RK; Chang LB; Preti C Glycosaminoglycans of Brain During Development. Biochemistry 1975, 14, 85–8. [DOI] [PubMed] [Google Scholar]
- (77).Larsen DD; Callaway EM Development of Layer-Specific Axonal Arborizations in Mouse Primary Somatosensory Cortex. J. Comp. Neurol 2006, 494, 398–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Braeckmans K; Buyens K; Bouquet W; Vervaet C; Joye P; De Vos F; Plawinski L; Doeuvre L; Angles-Cano E; Sanders NN; Demeester J; De Smedt SC Sizing Nanomatter in Biological Fluids by Fluorescence Single Particle Tracking. Nano Lett. 2010, 10, 4435–42. [DOI] [PubMed] [Google Scholar]
- (79).Chen T; Guestrin C Xgboost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, August 13–17, 2016; Vol. 1, pp 785–794. [Google Scholar]
- (80).Lundberg SM; Erion GG; Lee S Consistent Individualized Feature Attribution for Tree Ensembles. 2019, https://github.com/slundberg/shap. https://arxiv.org/abs/1802.03888 (accessed November 15, 2020).
- (81).Lundberg SM; Lee S A Unified Approach to Interpreting Model Predictions. 2017, https://arxiv.org/abs/1705.07874 (accessed November 15, 2020).
- (82).Granik N; Weiss LE; Nehme E; Levin M; Chein M; Perlson E; Roichman Y; Shechtman Y Single-Particle Diffusion Characterization by Deep Learning. Biophys. J 2019, 117, 185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Muñoz-Gil G; Garcia-March AG; Manzo C; Martín-Guerrero JD; Lewenstein M Single Trajectory Characterization via Machine Learning. New J. Phys 2020, 22, No. 013010. [Google Scholar]
- (84).Curtis C; McKenna M; Pontes H; Toghani D; Choe A; Nance E Predicting in Situ Nanoparticle Behavior Using Multiple Particle Tracking and Artificial Neural Networks. Nanoscale 2019, 11, 22515–22530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Joseph A; Liao R; Zhang M; Helmbrecht H; McKenna M; Filteau JR; Nance E Nanoparticle-Microglial Interaction in the Ischemic Brain Is Modulated by Injury Duration and Treatment. Bioengineering & Translational Medicine 2020, 5, No. e10175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Vanharreveld A; Crowell J; Malhotra SK A Study of Extracellular Space in Central Nervous Tissue by Freeze-Substitution. J. Cell Biol 1965, 25, 117–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Watakabe A Comparative Molecular Neuroanatomy of Mammalian Neocortex: What Can Gene Expression Tell Us About Areas and Layers? Dev Growth Differ 2009, 51, 343–54. [DOI] [PubMed] [Google Scholar]
- (88).Kroon T; van Hugte E; van Linge L; Mansvelder HD; Meredith RM Early Postnatal Development of Pyramidal Neurons across Layers of the Mouse Medial Prefrontal Cortex. Sci. Rep 2019, 9, 5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Dauth S; Grevesse T; Pantazopoulos H; Campbell PH; Maoz BM; Berretta S; Parker KK Extracellular Matrix Protein Expression Is Brain Region Dependent. J. Comp. Neurol 2016, 524, 1309–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Barnette AR; Neil JJ; Kroenke CD; Griffith JL; Epstein AA; Bayly PV; Knutsen AK; Inder TE Characterization of Brain Development in the Ferret via Mri. Pediatr. Res 2009, 66, 80–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Joseph AI; Edwards RL; Luis PB; Presley SH; Porter NA; Schneider C Stability and Anti-Inflammatory Activity of the Reduction-Resistant Curcumin Analog, 2,6-Dimethyl-Curcumin. Org. Biomol. Chem 2018, 16, 3273–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Ibata Y; Piccoli F; Pappas GD; Lajtha A An Electron Microscopic and Biochemical Study on the Effect of Cyanide and Low Na+ on Rat Brain Slices. Brain Res. 1971, 30, 137–58. [DOI] [PubMed] [Google Scholar]
- (93).Chaichana KL; Capilla-Gonzalez V; Gonzalez-Perez O; Pradilla G; Han J; Olivi A; Brem H; Garcia-Verdugo JM; Quinones-Hinojosa A Preservation of Glial Cytoarchitecture from ex Vivo Human Tumor and Non-Tumor Cerebral Cortical Explants: A Human Model to Study Neurological Diseases. J. Neurosci. Methods 2007, 164, 261–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).de Bouard S; Christov C; Guillamo JS; Kassar-Duchossoy L; Palfi S; Leguerinel C; Masset M; Cohen-Hagenauer O; Peschanski M; Lefrancois T Invasion of Human Glioma Biopsy Specimens in Cultures of Rodent Brain Slices: A Quantitative Analysis. J. Neurosurg 2002, 97, 169–76. [DOI] [PubMed] [Google Scholar]
- (95).Amor S; Puentes F; Baker D; van der Valk P Inflammation in Neurodegenerative Diseases. Immunology 2010, 129, 154–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Rosenberg GA Matrix Metalloproteinases and Their Multiple Roles in Neurodegenerative Diseases. Lancet Neurol. 2009, 8, 205–16. [DOI] [PubMed] [Google Scholar]
- (97).Adams KL; Gallo V The Diversity and Disparity of the Glial Scar. Nat. Neurosci 2018, 21, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).D’Ambrosi N; Apolloni S Fibrotic Scar in Neurodegenerative Diseases. Front. Immunol 2020, 11, 1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Bookbinder LH; Hofer A; Haller MF; Zepeda ML; Keller GA; Lim JE; Edgington TS; Shepard HM; Patton JS; Frost GI A Recombinant Human Enzyme for Enhanced Interstitial Transport of Therapeutics. J. Controlled Release 2006, 114, 230–41. [DOI] [PubMed] [Google Scholar]
- (100).Danielyan L; Schafer R; von Ameln-Mayerhofer A; Buadze M; Geisler J; Klopfer T; Burkhardt U; Proksch B; Verleysdonk S; Ayturan M; Buniatian GH; Gleiter CH; Frey WH 2nd Intranasal Delivery of Cells to the Brain. Eur. J. Cell Biol 2009, 88, 315–24. [DOI] [PubMed] [Google Scholar]
- (101).Wanisch K; Kovac S; Schorge S Tackling Obstacles for Gene Therapy Targeting Neurons: Disrupting Perineural Nets with Hyaluronidase Improves Transduction. PLoS One 2013, 8, No. e53269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Hastie T; Tibshirani R; Friedman J The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer: New York, 2009; Vol. 2. [Google Scholar]
- (103).Orzechowski P; La Cava W; Moore JH Where Are We Now? A Large Benchmark Study of Recent Symbolic Regression Methods. GECCO ‘18. In Genetic and Evolutionary Computation Conference, Kyoto, Japan, July 15–19, 2018; Vol. 1, pp 1–12. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data presented herein can be provided upon request. All code is available on github, with links included in the methods.


