Summary
Astrocytes are recognized to be a heterogeneous population of cells that differ morphologically, functionally, and molecularly. Whether this heterogeneity results from generation of distinct astrocyte cell lineages, each functionally specialized to perform specific tasks, remains an open question. In this study, we used RNA sequencing analysis to determine the global transcriptome profile of the Olig2-expressing astrocyte subtype (Olig2-AS), a specific spinal astrocyte subtype that segregates early during development from Olig2 progenitors and differs from other spinal astrocytes by the expression of Olig2. We identified 245 differentially expressed genes. Among them, 135 exhibit higher levels of expression when compared with other populations of spinal astrocytes, indicating that these genes can serve as a “unique” functional signature of Olig2-AS. Among them, we identify two genes, inka2 and kcnip3, as specific molecular markers of the Olig2-AS in the P7 spinal cord. Our work thus reveals that Olig2 progenitors produce a unique spinal astrocyte subtype.
Subject areas: Cellular neuroscience, Omics, Transcriptomics
Graphical abstract
Highlights
-
•
Efficient method to isolate Olig2-AS from other spinal glial cells
-
•
Provide astrocyte subtype transcriptome from the post-natal spinal cord
-
•
Identification of two specific markers of the Olig2-AS
-
•
Bioinformatics identifies functional specificity of Olig2-AS in synapse regulation
Cellular neuroscience; Omics; Transcriptomics
Introduction
Astrocytes are the largest class of glial cells in the mammalian central nervous system (CNS) and are recognized as essential for a wide variety of complex functions. Astrocytes supply energy metabolites to neurons, regulate the blood flow, and the blood-brain barrier and control the levels of extracellular ions, neurotransmitters, and fluids (Allen and Lyons, 2018). The expression of functional receptors on their plasma membrane also allows astrocytes to sense neurotransmitters from nearby synaptic sites and to respond to neurons by the release of various gliotransmitters, which can influence neuronal and synaptic functions (Allen and Eroglu, 2017). A major still unsolved issue regarding astrocytes is whether the diversity of functions they assume is shared by all astrocytes or is achieved by specialized astrocyte subtypes dedicated to perform specific functions in the CNS. Based on their morphology and location, astrocytes have long been divided into two distinct classes, protoplasmic astrocytes found in the gray matter and fibrous astrocytes found in the white matter. However, interest for these cells over the last decades has provided an increasing amount of evidence for a far greater heterogeneity of astrocytes (Zhang and Barres, 2010). Based on differences in morphology and marker protein expression, different populations of astrocytes, displaying both inter- and intra-regional differences, have been identified in the gray and white matters of the rodent brain and spinal cord (Chaboub and Deneen 2013; Khakh and Sofroniew, 2015; Ben Haim and Rowitch, 2017; Pestana et al., 2020). Recently, genome-wide transcriptome profiles of astrocytes provided additional insights into astrocyte heterogeneity, resulting in an unprecedented amount of data that offer opportunities to study astrocyte phenotypes and functions in health and disease (Pestana et al., 2020). These studies revealed molecular diversity of astrocytes, across brain regions (Doyle et al., 2008; Morel et al., 2017; Boisvert et al., 2018; Clarke et al., 2018; Zeisel et al., 2018; Batiuk et al., 2020; Lozzi et al., 2020) and also within regions (Zeisel et al., 2015; Gokce et al., 2016; Chai et al., 2017; John Lin et al., 2017; Saunders et al., 2018; Morel et al., 2019; Bayraktar et al., 2020; Batiuk et al., 2020). Gene expression profiling also revealed regionally distinct molecular differences in astrocytes populating the dorsal and ventral spinal cord (Molofsky et al., 2014). However, there still remains major gap in our understanding about the identification and overall characterization of different astroglial cell populations.
Developmental studies, by revealing that positional identity during embryogenesis is an organizing feature of astrocyte diversity, have also proved to be very insightful for our understanding of origin and possible function of astrocyte heterogeneity. Pioneer studies performed in the embryonic spinal cord identified that distinct astrocyte subtypes are generated from separate classes of progenitor cells specified early during patterning of the neural tube (Muroyama et al., 2005; Hochstim et al., 2008). During this process, morphogen factors regulate expression of sets of transcription factors throughout the dorsoventral axis of the neural tube, thus subdividing it into distinct neural progenitor domains, each dedicated to generate specific neuronal subtypes (Dessaud et al., 2008). Gliogenesis is initiated later and, at these later phases, domain organization of the spinal cord still plays an instructive role for the generation of glial cell subtypes (Ben Haim and Rowitch, 2017). This principle was first established from the observation that oligodendrocyte precursor cells (OPC) first emerge from a discrete region in the ventral neural tube, named the pMN domain, which, earlier on, gives rise to motor neurons (MN). Progenitor cells of the pMN domain are characterized by the expression of the transcription factor Olig2, which is required for establishment of the domain and is also recognized as a cell-intrinsic determinant essential for the specification of both MNs and OPCs (Rowitch and Kriegstein, 2010). Astrocytes were first recognized to arise from pMN-adjacent but non-overlapping progenitor domains, and three subtypes of white matter spinal astrocytes, termed VA1, VA2, and VA3, originating from separate domains of the ventral progenitor zone, have been identified (Bayraktar et al., 2015). Importantly, lineage tracing studies have shown that astrocyte precursors (APs) migrate in a restricted segmental fashion and are regionally allocated into the spinal cord according to the dorsoventral position of their domains of origin (Tsai et al., 2012). A similar regional specification mechanism linked to the ultimate astrocyte spatial position in the adult CNS has been shown to occur in the forebrain (Bayraktar et al., 2015). However, the question remains as to whether patterning mechanisms might establish a template for generation of the functional diversity of mature astrocytes. In favor of this assumption, region-restricted astrocyte subsets have been reported to assume specialized functions in the ventral spinal cord (Molofsky et al., 2014; Kelley et al., 2018).
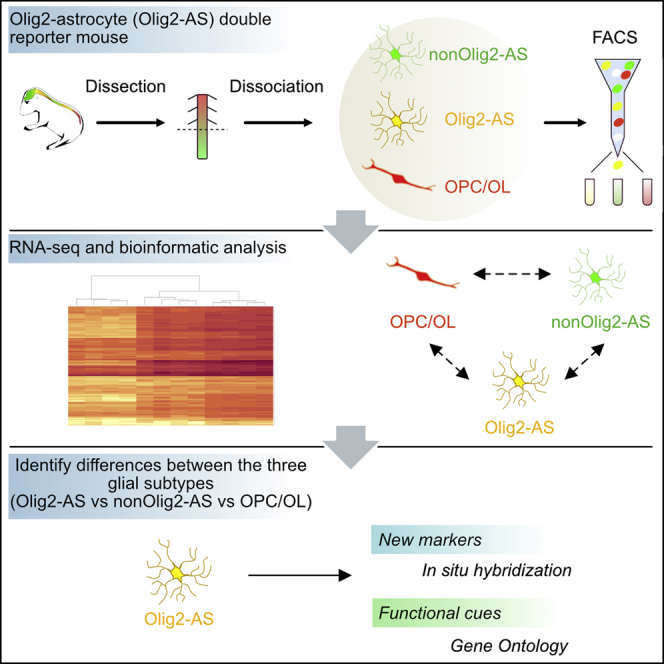
In a previous work, we identified a regionalized astrocyte sub-type populating the gray matter of the spinal cord that we named the Olig2-AS, for Olig2-astrocytes because they originate from Olig2 progenitor cells of the pMN domain, as do MNs and OPCs, and also because they retain expression of Olig2 as they differentiate into mature astrocytes (Ohayon et al., 2019). An important aspect of the identification of Olig2-AS is that these astrocytes can be distinguished from other spinal astrocytes at post-natal stages by the expression of Olig2. Thus, their identification offered a unique opportunity to study molecular characteristics of one specific subtype of spinal cord astrocytes. In this study, we developed a fluorescence-activated cell sorting (FACS)-based approach to purify Olig2-AS from other astrocyte populations as well as from oligodendroglial cells. We then used RNA sequencing (RNA-seq) to generate high-resolution transcriptome databases and identify the molecular signature of the Olig2-AS. Overall, our study reveals molecular insight into the nature of astrocyte diversity in the spinal cord.
Results
Selective labeling of Olig2-AS by the expression of fluorescent reporter proteins in double transgenic mice
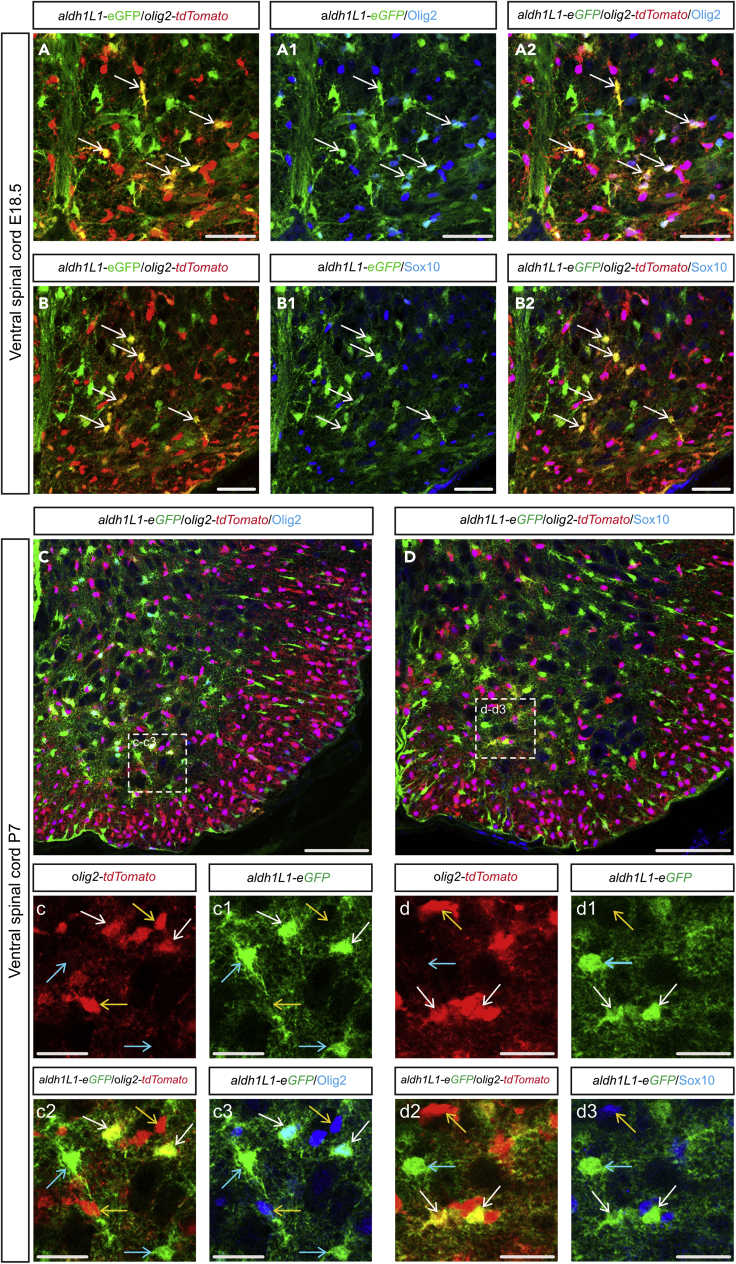
The prerequisite for characterizing further Olig2-AS cells was the need to isolate them from the two other main glial cell populations, namely, the astrocytes that do not express Olig2 (nonOlig2-AS) and cells of the oligodendroglial lineage, i.e., OPC and differentiated oligodendrocytes (OL). For this purpose, we developed a double transgenic mouse model in which all the three cell types can be distinguished by specific expression of fluorescent reporter proteins. To specifically color code these cells, we took advantage of the combined expression of Aldh1L1 and Olig2 in the Olig2-AS (Ohayon et al., 2019). We thus turned to the transgenic aldh1L1-eGFP mouse line (Heintz, 2004) in which aldh1L1 promoter drives eGFP-targeted expression in all astrocytes (Cahoy et al., 2008) and to an olig2-tdTomato mouse line that expresses the red fluorescent protein tdTomato (tdT) under the control of olig2 regulatory sequences (GENSAT). Because the latter mouse line had not yet been characterized, we first compared expression of tdT with that of the endogenous Olig2 protein performing immunodetection of Olig2 on E13.5 embryonic and P7 post-natal spinal cord sections. In these experiments, we also used Sox10, an early and reliable marker of OPC and OL in mouse (Kuhlbrodt et al., 1998; Zhou et al., 2000; Claus Stolt et al., 2002). At E13.5, Olig2 expression was detected in pMN progenitor cells as well as in two distinct cell populations that have emigrated in the mantle zone, comprising Sox10+ OPC and Olig2-AS that do not express Sox10 (Figure S1A). We found that most, if not all, mantle zone tdT+ cells were positive for the Olig2 immunostaining (Figure S1A, a-a5). Similarly, at P7, Olig2 immunostaining colocalized with tdT, and cells co-expressing tdT and Olig2 but not Sox10 were detected in the gray matter (Figure S1B, b-b5). The olig2-tdTomato mouse thus faithfully reports endogenous expression of Olig2, validating the use of this mouse line to label all spinal cord Olig2-expressing cells, including the Olig2-AS. We then crossed these mice with aldh1L1-eGFP individuals and examined the expression patterns of the reporter proteins. In these mice, Olig2-AS were expected to co-express tdT and eGFP, whereas nonOlig2-AS and OPC/OL were expected to only express eGFP and tdT, respectively. At E18.5, a stage in which Olig2-AS have already been produced in the ventral spinal cord (Ohayon et al., 2019), we were able to distinguish tdT+, eGFP+, and tdT+/eGFP+ cells in the ventral gray matter (Figures 1A and 1B). The same three cell populations were detected at P7 (Figures 1C-c2 and D-d2). Confirming that tdT+/eGFP+ cells indeed correspond to the Olig2-AS, we found that these cells were immunostained for Olig2 but not for Sox10 at either E18.5 (Figure 1A-A2, B-B2) or P7 (Figure 1C-c3, D-d3). Cell counting further indicated that tdT+/eGFP+ Olig2-AS represent 49% of the eGFP+ astrocyte population and 19% of the tdT+ cell population (Figure S2A).
Figure 1.
Detection of Olig2-AS in aldh1L1-GFP/olig2-tdTomato double transgenic mice
(A–D) Here and in all subsequent panels, images show transverse sections of hemi-ventral spinal cord. (A–d3) Combined detection of eGFP (green), td-Tomato (red), and Olig2 (blue, A–A2; C–c3) or Sox10 (blue, B–B2, D–d3) on aldh1L1-eGFP/olig2-tdTomato transgenic mice at E18.5 (A–B2) and P7 (C–d3). Horizontal sets in A–B2 present successively GFP and tdTomato staining, Olig2 (A1) or Sox10 (B1) and GFP staining, and the merged image. Images (c–c3) and (d–d3) show higher magnification of the areas framed in (C and D), respectively, and show successively tdTomato (C and D) and eGFP (c1 and d1) staining, double tdTomato and eGFP staining (c2 and d2), and Olig2 (c3) or Sox10 (d3) and eGFP staining. Colored arrows point to OPC/OL (yellow), Olig2-AS (white), and nonOlig2-AS (blue). Scale bars, 100 μm in (C and D); 50 μm in (A–A2) and (B–B2); and 25 μm in (c–c3) and (d–d3).
See also Figures S1 and S2.
Overall, these data validate the olig2-tdTomato/aldh1L1-eGFP double transgenic mouse line as a suitable model to specifically color code the Olig2-AS and thus to distinguish these cells from nonOlig2-AS and OPC/OL in the developing and post-natal spinal cords.
Efficient RNA-seq-based segregation analysis of Olig2-AS, nonOlig2-AS, and OPC/OL
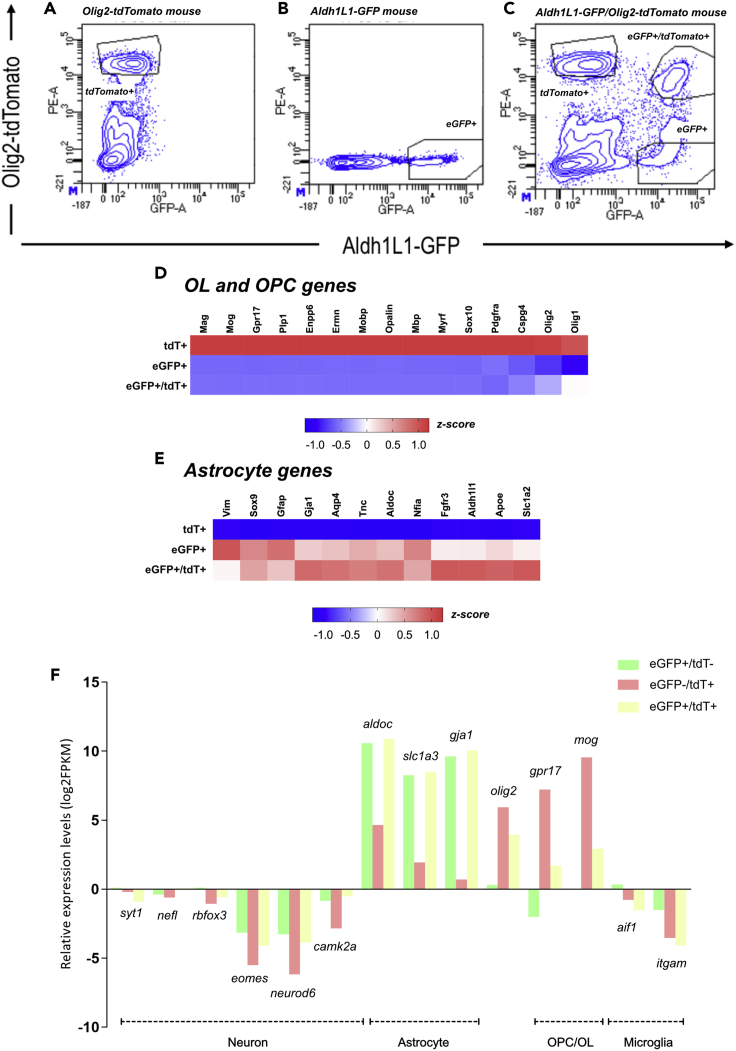
A first step toward the identification of differentially expressed genes between Olig2-AS and nonOlig2-AS or OPC/OL was to purify the three differentially labeled cell populations. For this purpose, spinal cord cells were dissociated at P7 (Figure S3A), a stage that has been proved to be suitable for the purification of astrocytes with minimal activation and when, even if astrocytes are not fully differentiated, their gene expression profiles closely resemble that of mature astrocytes (Cahoy et al., 2008). As a first approach, spinal cords from aldh1L1-eGFP and olig2-tdTomato simple transgenic mice were processed into single-cell suspensions and further sorted by FACS relying on either eGFP or tdT expression (Figures 2A and 2B). To get high purity of the desired population, we next performed stringent gating on these samples, and these gates were further applied to all experiments (Figures 2A and 2B). We next sorted cells dissociated from the spinal cords of aldh1L1-eGFP/olig2-tdTomato double transgenic mice by FACS (Figure 2C). This procedure was reproduced four times, each time with different mouse litters, to ultimately get four replicates of tdT+, eGFP+, and eGFP+/tdT+ cell type populations, each cell replicate being obtained from two distinct individuals. Cell counting of the dissociated cell populations showed similar representation of the three cell types as that found by counting cells in vivo (Figure S2). We then extracted RNA from the purified cell populations and performed RNA-seq. The transcriptional profiles of each cell type replicate were then compared. To assess the reproducibility of our data and conservation across biological replicates, we calculated correlations across all RNA-seq samples. We indeed found high correlations among cell type replicates (correlation coefficient R > 0.92, Figure S3B). Principal-component analysis and hierarchical clustering data further showed good discrimination between the different cell type replicates according to mRNA expression profiles (Figures S3C and S3D). Our results, available online as a resource through GEO accession number GSE158517 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE158517), indicated that gene expression was similar in each group, thus providing a dataset that we can use to find out the molecular specificity of the three glial cell populations.
Figure 2.
FACS purification and transcriptome analysis of eGFP+/tdT-, tdT+/eGFP-, and eGFP+/tdT + cell populations
(A–F) (A and B) Representative FACS plots showing the gating strategy. Gates for tdT+ (A) and eGFP+ (B) cells were defined from cell populations purified from olig2-tdTomato and aldh1l1-eGFP single transgenic mice, respectively. (C) Representative FACS plot of the purification of tdT+/eGFP-, eGFP+/tdT-, and tdT+/eGFP + cell populations from P7 aldh1l1-eGFP/olig2-tdTomato double transgenic mice. (D and E) Heatmap profiles presented as Z score of established oligodendrocyte- (D) and astrocyte- (E) specific genes in tdT+/eGFP-, eGFP+/tdT-, and eGFP+/tdT+ FACS-sorted cell populations. (F) Comparison of neuronal, astroglial, oligodendroglial, and microglial gene expression levels (by log2FPKM values) in the three cell populations. Average FPKM values of four biological replicates are shown.
See also Figure S3.
To validate the purity of the isolated cell types, we next probed the transcriptome data for the expression of eGFP and tdT for each cell type replicate. In agreement with the expected expression profiles for the two reporter proteins in each cell type replicate, we found high expression levels of eGFP in cells sorted on eGFP or eGFP and tdT expression and of tdT in cells sorted on tdT or tdT and eGFP expression (Figure S3E). Analysis of RNA-seq data for cell-specific mRNA demonstrates enrichment for oligodendroglial markers in tdT+ cells and for astroglial markers both in eGFP+ and eGFP+/tdT+ cells (Figures 2D and 2E). We then probed the transcriptome data for expression levels of well-known cell type-specific genes for astrocytes, oligodendroglial cells, neurons, and microglia (Cahoy et al., 2008; Zhang et al., 2014). Fragments per kilobase of transcript per million mapped reads (FPKM) values of astrocyte genes, such as aldoC, gja1, and slc1a3, were generally high in the eGFP+ and in the eGFP+/tdT+ cell populations, whereas oligodendroglial genes, such as mog and gpr17, displayed a high FPKM value in the tdT+ cell population (log2FPKM>7, Table S1, Figure 2F) but were undetectable or had extremely low expression levels in the remaining cell populations (log2 FPKM<2, Table S1, Figure 2F). Expectedly, expression of olig2 was high in tdT+ and eGFP+/tdT+ cells but quite undetectable in eGFP+ cells. FPKM values of the neuronal genes syt1, nefl, neurod6, rbfox3, eomes, and camk2a were found negative (log2 FPKM<0, Table S1) in all the three cell populations (Figure 2F). The specific microglial genes aif1 and itgam genes were also found negative (log2 FPKM<0, Table S1) in all the three cell populations (Figure 2F).
Together, these data confirmed the purity of the various isolated cell types and established the feasibility of constructing a high-quality transcriptome database representative of Olig2-AS (eGFP+/tdT+), nonOlig2-AS (eGFP+/tdT-), and OPC/OL (eGFP-/tdT+).
A specific Olig2-AS molecular signature revealed by RNA-seq analysis
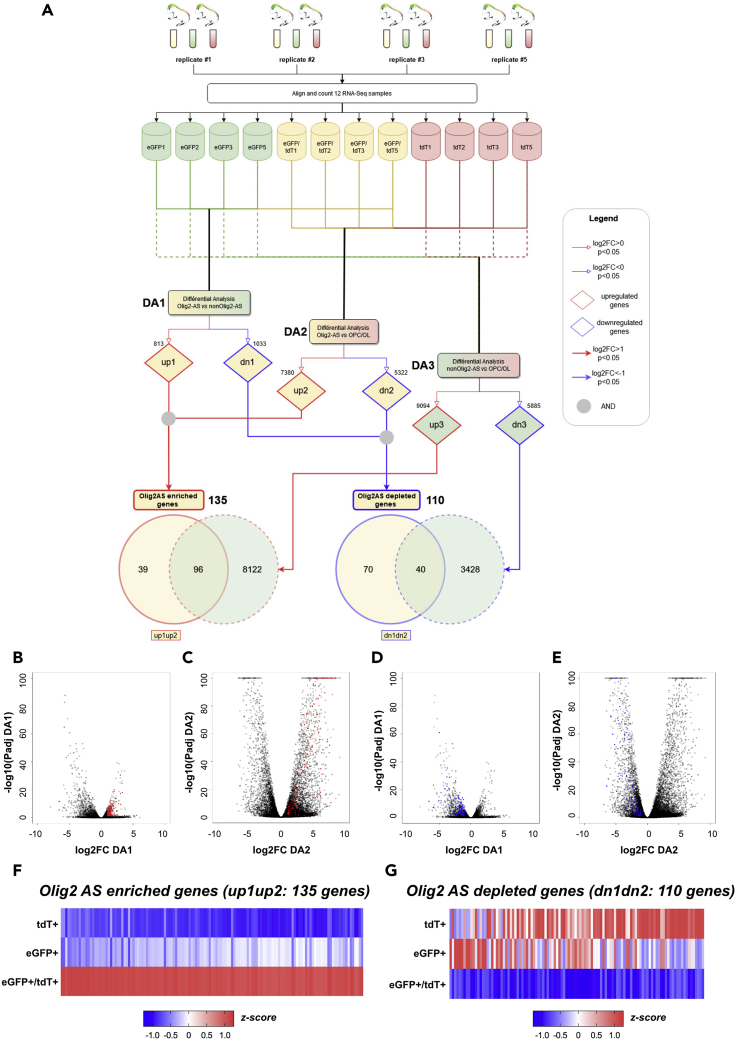
To identify an expression profile that distinguishes Olig2-AS from the two other glial cell types, we carried out a two-step comparative analysis (Figure 3A). In the first step, each cell population was pairwise compared to identify genes up- or down-regulated in Olig2AS compared with nonOlig2AS and OPC/OL and in nonOligAS compared with OPC/OL. Then, to identify genes that distinguishes the Olig2-AS from both of the two other glial cell types, genes found up- or down-regulated in the pairwise comparisons were in turn compared. Differential analyses of RNA-seq were performed using DESeq2 with normalized counts (Love et al., 2014). In pairwise analyses, genes with a log2foldchange (log2FC) > 0 and p < 0.05 or log2FC < 0 and p < 0.05 were considered to be significantly up- or down-regulated, respectively. We first got three pairwise differential analysis (DA): between Olig2-AS and nonOlig2-AS (DA1), between Olig2-AS and OPC/OL (DA2), and between nonOlig2-AS and OPC/OL (DA3) (Figure 3A). DA1 comparison showed a relatively low number of genes displaying differential values as only 813 and 1,033 genes were found up- and down-regulated, respectively (Figures 3A and S4A). In contrast, a large number of genes showed significant differential expression values in DA2 (7,380 up-regulated and 5,322 down-regulated genes) and DA3 (9,094 up-regulated and 5,885 down-regulated genes) in which Olig2-AS and nonOlig2-AS were compared with OPC/OL, respectively (Figures 3A, S4B, and S4C). Thus, and as expected, these data indicated that Olig2-AS and nonOlig2-AS share many similarities but largely differ from OPC/OL. We then performed the second selection process aimed at identifying genes specifically enriched in Olig2AS, i.e., genes up-regulated in both DA1 (Olig2-AS versus nonOlig2-AS) and DA2 (Olig2-AS versus OPC/OL). In this process, the threshold was adjusted to log2FC > 1 and padj<0.05. Analysis of the relative expression values then revealed a set of 135 highly expressed genes (up1up2, Figures 3A–3C and 3F and Table S2). We considered these genes as “signature” genes as they together provide a unique profile of the Olig2-AS. Notably, among these genes, 39 were found up-regulated in the Olig2-AS but not in the nonOlig2-AS compared with OPC/OL (compare up1up2 and up3 in Figures 3A and S5), indicating that these genes are good candidates to represent specific molecular markers of the Olig2-AS. Within this short list, only few genes, such as ptn, grm5, or lrtm2, are recognized as astroglial genes (González-Castillo et al., 2015; Sun et al., 2013; Chaboub et al., 2016), whereas the majority had not previously been identified as cell type specific.
Figure 3.
Identification of Olig2-AS molecular signature by differential analysis
(A–G) (A) Flowchart representing the treatment and two-step comparative analyses performed on the RNA-seq data. The top half of the chart shows the 4 cell replicates of Olig2AS (yellow), nonOlig2AS (green), and OPC/OL (red), each obtained from the dissociation of two spinal cords isolated at P7. The bottom half represents the two consecutive comparisons performed between the three glial cell subtypes, and the numbers of genes found up-regulated (up, red) or down-regulated (dn, blue) in each pairwise analysis are indicated for each comparison. Numbers of genes found up-regulated (up1up2) or down-regulated (dn1dn2) in both DA1 and DA2 are indicated in solid line red and blue circles, respectively. Dashed line circles represent genes found up-regulated (red) or down-regulated (blue) in the DA3 comparison. Note that 39 genes were found up-regulated and 70 were found down-regulated specifically in Olig2AS compared with the two other glial cell populations. (B–E) Volcano plots of differentially expressed genes representing the log2FC and –log10padj from the differential analyses DA1 (B, C) and DA2 (D, E). Red dots in (B and C) represent enriched genes (up1up2; padj<0.05 and a log2FC > 1). Blue dots in (D and E) represent down-regulated genes (dn1dn2; padj<0.05 and a log2FC < 1). (F and G) Heatmap analysis representing the 135 genes up-regulated (F) and the 110 genes down-regulated (G) in Olig2-AS. All heatmaps are presented as Z score.
See also Figures S4, S5, and S6.
A similar analysis was performed for the genes down-regulated specifically in the Olig2-AS, adjusting the threshold to log2FC < −1 and padj<0.05. This yielded a list of 110 genes down-regulated both in DA1 and DA2 (Figures 3A, 3D, 3E, and 3G and Table S3). As noted earlier, among these genes, 70 were found down-regulated in the Olig2-AS but not in the nonOlig2-AS compared with the OPC/OL (Figures 3A and S6). This analysis thus revealed a set of genes whose expression is repressed specifically in the Olig2-AS subtype, thus reinforcing the view that the molecular identity of Olig2-AS differs from that of nonOlig2-AS.
Validation of RNA-seq results by in situ hybridization and identification of Olig2-AS-specific markers
The bioinformatics analysis has generated a list of 135 genes enriched in the Olig2-AS, including 39 potential molecular markers of Olig2-AS. To bring support on the reliability of the results, we again sorted by FACS P7 spinal cord cells of aldh1L1-GFP/olig2-tdTomato mice and performed qRT-PCR using RNA samples prepared from each glial cell population. We first analyzed the expression of the three markers aldh1L1, mbp, and olig2. Expectedly, data showed expression of aldh1L1 in nonOlig2-AS (eGFP+/tdT-) and Olig2-AS (eGFP+/tdT+) but not in OPC/OL (eGFP-/tdT+), of mbp only in OPC/OL, and of olig2 in Olig2-AS and in OPC/OL (Figure S7A). We next detected the expression levels of three genes, inka2, slc7a10, and grm3, which we found enriched in Olig2-AS based on RNA-seq data. In agreement with RNA-seq data, Olig2-AS cells showed increased levels of all the three genes compared with nonOlig2-AS and OPC/OL, and fold change values obtained comparing Olig2-AS and nonOlig2-AS were very similar to the ones obtained by RNA-seq (inka2: 2.12 versus 2.1; slc7a10: 1.79 versus 1.39; grm3: 1.73 versus 1.7; Figures S7B–S7D).
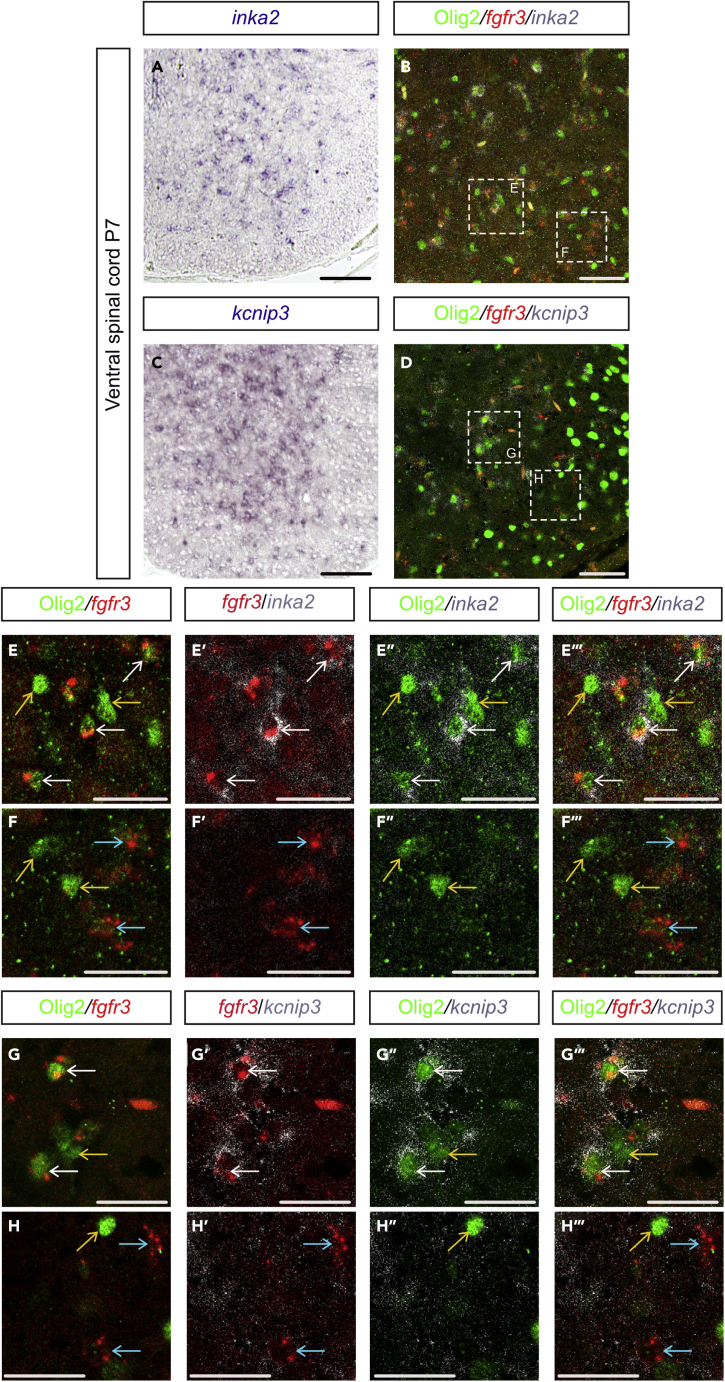
For further validation, we selected, from the short list of 39 genes, two genes, inka2 and kcnip3, encoding for the Inka Box Actin Regulator 2 protein and the voltage-dependent K+ channel interacting protein 3 (KCNIP3), respectively, and analyzed their expression pattern by in situ hybridization on P7 spinal cord sections. Motivating the choice of inka2 and kcnip3, expression of these genes in astrocytes had not been previously reported, making them possible new markers. A second reason for the choice of inka2 is that this gene had been reported to be an oligodendroglial-specific gene (Iwasaki et al., 2015), which seemed at first in contradiction with our transcriptomic data. Our data showed that both inka2 and kcnip3 mRNA were detected in cells scattered in the ventral spinal gray matter, whereas only few positive cells were found in the white matter, thus revealing cells displaying a distribution very reminiscent to that of Olig2-AS (Figures 4A and 4C). To confirm that these cells indeed are Olig2-AS, we next combined detection of inka2 or kcnip3 mRNAs with immunodetection of Olig2 and in situ localization of the fgfr3 mRNA, an early and specific hallmark of astroglial cells (Pringle et al., 2003; Ohayon et al., 2019). Our data showed that cells expressing inka2 and kcnip3 were positive for both Olig2 and fgfr3 staining (Figures 4B and 4D–4H‴), validating expression of these two genes in Olig2-AS. By contrast, expression of these genes was not detected in either OPC/OL cells expressing Olig2 but not fgfr3 or in nonOlig2-AS identified by the expression of fgfr3 (Figures 4E, 4F‴, 4G, and 4H‴). Thus, inka2 and kcnip3 specifically mark Olig2-AS in the P7 spinal cord. Because inka2 has previously been reported to be an OPC marker in the developing spinal cord (Iwasaki et al., 2015), we examined its expression, together with that of Olig2 and fgfr3, in the E14.5 embryonic spinal cord. As observed at P7, we found that inka2-positive cells all express the fgfr3 mRNA, and comparison with the Olig2 staining showed inka2+/fgfr3+ cells positive for the Olig2 staining intermingled with inka2+/fgfr3+ cells that do not express Olig2 (Figure S8). In these experiments, we never found cells co-expressing inka2 and Olig2 but not fgfr3. These data thus indicate that, during development, expression of inka2 marks AP cells but not OPC and that, unlike at post-natal stage P7, inka2 expression is not only restricted to the Olig2-AS sub-type but also marks at least a subset of nonOlig2-AS precursor cells. We thus conclude that the preferential expression of inka2 in Olig2-AS observed at P7 results from the down-regulation of this gene during the maturation period of nonOlig2-AS.
Figure 4.
The up-regulated genes inka2 and kcnip3 are specifically expressed in Olig2-AS
(A–H) (A and C) Expression profiles of inka2 (A) and kcnip3 (C) mRNAs at P7. (B and D) Immunodetection of Olig2 (green) combined with detection of fgfr3 (red) and inka2 (gray, B) or kcnip3 (gray, D) mRNAs in the ventral spinal cord at P7. (E–H‴) (E–F‴) and (G–H‴) show higher magnification of the areas framed in (B and D), respectively, and show successively combined Olig2 immunostaining and detection of fgfr3 mRNA (E, F, G, H), combined detection of fgfr3 and inka2 (E′, F′) or kcnip3 (G′, H′) mRNA, combined Olig2 immunostaining and detection of inka2 (E″, F″) or kcnip3 mRNA (G″, H″), and the merged images (E‴, F‴, G‴, H‴). Colored arrows point to OPC/OL (yellow), Olig2-AS (white), and nonOlig2-AS (blue). Scale bars, 100 μm in (A and C), 50 μm in (B and D), and 25 μm in (E–H‴).
See also Figures S8 and S9.
Still focused on validating the RNA-seq, we selected one gene among the 70 genes found down-regulated in Olig2-AS (Figure S6A). The selected gene was efnb3, a member of the ephrin gene family, previously reported as being expressed in post-natal myelinating OLs of the mouse spinal cord (Benson et al., 2005). Accordingly, we preferentially detected the efnb3 mRNA in cells located in the white matter of the P7 spinal cord (Figure S9A). Detection of efnb3 mRNA was next combined with immunodetection of Olig2 and in situ localization of the fgfr3 mRNA. We found no fgfr3+/Olig2+ cell positive for the efnb3 mRNA staining in the gray matter (Figure S9B, c-c2), but, as expected, we detected numerous efnb3+/Olig2+ cells negative for the fgfr3 staining in the white matter (Figure S9B, d-d2). These data thus reinforce the conclusion that the RNA-seq approach reliably identifies genes down-regulated in Olig2-AS.
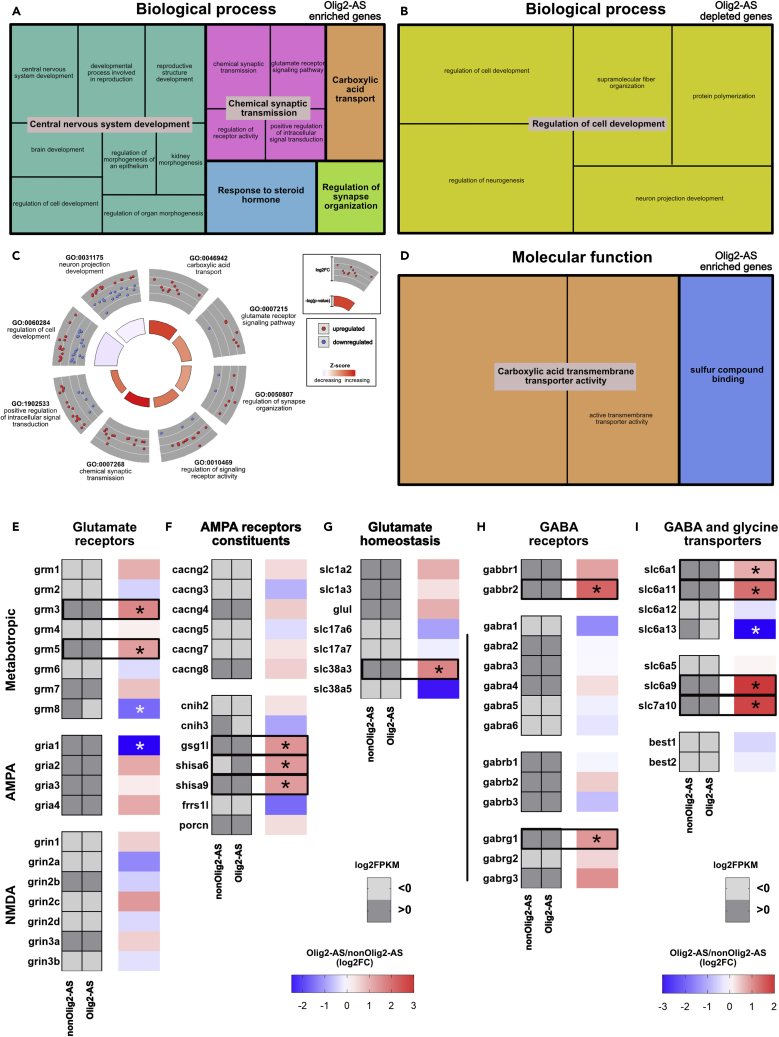
Functional analysis of genes differentially expressed in Olig2-AS
To get insights on the biological meaning behind the list of the 135 and 110 genes enriched and decreased in Olig2-AS, respectively, we performed Gene Ontology (GO) term-enrichment analysis (GO summaries). In terms of biological processes, Olig2-AS-enriched genes were classified into 5 functional groups: “central nervous system development,” “chemical synaptic transmission,” “carboxylic acid transport,” “regulation of synapse organization,” and “response to steroid hormone” (Figure 5A, Table 1, and S4). Among these 5 GO terms, 3, including “chemical synaptic transmission,” “carboxylic acid transport,” and “regulation of synapse organization,” can be related to synaptic activity (Figure 5A). On the other hand, GO analysis performed on down-regulated genes displays a strong association with “regulation of cell development,” including “neuron projection development” (Figure 5B, Tables 1 and S4). To get more details, circular visualization of the GO results was performed selecting synaptic-related and cell development-related terms (Figure 5C). Data indicated that, among the 135 genes enriched in Olig2-AS, one-third are associated with GO synapse-related terms (45 genes), whereas only 9 among the 110 down-regulated genes are associated with these terms (Figure 5C, Table 1). Instead, down-regulated genes appear mostly associated to cell development terms (Figure 5C, Table 1). GO annotation results thus pointed to a fair number of genes that are dominant in Olig2-AS and could be related to synapse organization and to modulation of synaptic activity. Finally, GO analysis of genes up-regulated in Olig2-AS performed in terms of molecular function (MF) again pointed to synaptic related functions such as “carboxylic acid transmembrane transporter activity,” including “active transmembrane transporter activity” and “sulfur compound binding” (Figure 5D, Table 1). Among these genes, we identified genes coding for the Glutamate Metabotropic Receptors 3 and 5 (grm3 and grm5), the GABA receptor subunits (gabbr1 and gabbr2), the glutamine transporter SNAT3 (slc38a3), and the glycine and D-serine transporter SLC7A10 (slc7a10). To go further and have a broader view of the molecular identity of Olig2-AS, we next completed this analysis by comparing expression levels in Olig2-AS and in nonOlig2-AS of additional genes encoding for neurotransmitter receptors and transporters. Results of this comparison, including some genes identified through the GO analysis, are shown in Figures 5E–5I. We found that 3 (grm3/5/7) of the 8 genes coding for glutamate metabotropic receptors are expressed in both Olig2-AS and nonOlig2-AS with significant enrichment for grm3 and grm5 in Olig2-AS (Figure 5E). Data indicated that Olig2-AS and nonOlig2-AS also differ by grm8 expression, which appears significantly enriched in the nonOlig2-AS (Figure 5E). Similar analysis performed for genes coding for ionotropic glutamate receptors indicated that only 2 genes encoding for NMDA subunits (grin2b, grin3a) are expressed in spinal astrocytes with no difference in their expression levels (Figure 5E). All of the 4 genes coding for AMPA receptors (gria1-4) were found expressed in spinal astrocytes with no significant difference in their expression levels, except for gria1, which is specifically enriched in the nonOlig2-AS (Figure 5E). We also analyzed genes coding for AMPA receptor interacting proteins, which are known to modulate localization and functional properties of the receptors (Jacobi and von Engelhardt, 2018; Mölders et al., 2018), and found enrichment for 3 of them (gsg1l, shisa6, and shisa9) in Olig2-AS compared with nonOlig2-AS (Figure 5F). We further analyzed a set of genes whose products are known to control glutamate homeostasis. We found expression in spinal astrocytes of the glutamate transporters GLT1 (slc1a2) and GLAST-1 (slc1a3), the glutamine synthetase GLUL (glul), and the sodium-dependent glutamine transporters SN2 (slc38a5) and SN1 (slc38a3), the latter being the only gene found enriched in Olig2-AS compared with nonOlig2-AS (Figure 5G). By contrast, and in agreement with recent reports that question the expression of vesicular glutamate transporters in brain astrocytes (Cahoy et al., 2008; Li et al., 2013), our data provide no evidence that VGlut1 (slc17a7) and VGlut2 (slc17a6) are expressed in spinal astrocytes (Figure 5G). We also found expression of a majority of genes known to encode for different subunits of GABA receptors both in Olig2-AS and nonOlig2-AS (Figure 5H). Noticeably, 2 genes, encoding for the metabotropic (gabbr2) and ionotropic (gabrg1) GABA receptors, were found enriched in Olig2-AS (Figure 5H). Three GABA transporters GAT1/3/4 (slc6a1, slc6a13, slc6a11) as well as 2 glycine transporters (slc6a9 and slc7a10) were also found expressed in the two astrocyte subsets with significant enrichment in Olig2-AS of slc6a1 and slc6a11, coding for GABA transporters, and of slc6a9 and slc7a10, coding for glycine transporters (Figure 5I). Together, these analyses, pointing to differential expression in Olig2-AS and nonOlig2-AS of genes involved in synaptic transmission with a set of 12 genes (grm3, grm5, gsg1l, shisa6, shisa9, slc38a3, gabbr2, gabrg1, slc6a1, slc6a11, slc6a9, and slc7a10) enriched in Olig2-AS and 3 genes (grm8, gria1, and slc6a13) conversely enriched in nonOlig2-AS, provide clues as to how the Olig2AS might differ in their ability to dynamically modulate synapse function.
Figure 5.
Functional enrichment analysis of Olig2-AS differentially expressed genes
(A–D) Treemap representations of Gene Ontology (GO) (A, B, and D) and circular visualization of gene-annotation enrichment (GOcircle, C) analyses performed on Olig2-AS differentially expressed genes. GO analyses were performed from genes specifically up-regulated (A, D) or down-regulated (B) in Olig2-AS and are presented in terms of Biological Process (A, B) and Molecular Function (D). Significant GO terms (smaller squares) were grouped according to their parental ontology to highlight highly represented functions. GOcircle in (C) represents scaled scatterplots for genes found up-regulated (red dots) and down-regulated (blue dots) in Olig2AS and associated with the most-enriched biological processes indicated by GO terms. The outer to inner layers of the gray circle indicate the relative fold-change of gene expression (log2FC, from higher to lower values). The color code in inner circle indicates the Z score that defines the likelihood of a process being decreased (blue) or increased (red) in Olig2-AS, and its height is representative of the p value for each GO term.
(E–I) Heatmaps depicting abundance of mRNA encoding for glutamate receptors (E), AMPA receptors constituents (F), glutamate transporters and enzyme (G), GABA receptors (H), and GABA and glycine transporters (I). Heatmaps in gray represent mRNA expression levels (log2FPKM) in nonOlig2-AS and Olig2-AS. Light and dark grays indicate values lower and above zero, respectively. Adjacent heatmaps illustrate fold-change in abundance of mRNA in Olig2-AS versus nonOlig2-AS (log2FC). Black and white asterisks indicate significant up-regulation and down-regulation in Olig2-AS, respectively.
Table 1.
Gene ontology of all 245 differentially expressed genes in Olig2-AS
| Category | ID | Term | adj_pval | Up-regulated genes in Olig2-AS | Down-regulated genes in Olig2-AS |
|---|---|---|---|---|---|
| BP | GO:0046942 | Carboxylic acid transport | 0.000458 | slc38a3,slc13a3,acsl6,slc13a5,slc6a9,slco1c1,slc6a11,slc6a1,slc7a10,pla2g3 | |
| BP | GO:0007268 | Chemical synaptic transmission | 0.00518 | gabrg1,grm3,etv5,egfr,shc3,shisa9,car2,slc6a9,ptn,slc6a1,rasgrf1,lzts1,grm5,shisa6 | |
| BP | GO:0007215 | Glutamate receptor signaling pathway | 0.00614 | grm3,shisa9,rasgrf1,gsg1l,grm5,trpm3 | arc |
| BP | GO:0050807 | Regulation of synapse organization | 0.0367 | ephb3,etv5,slit1,ptn,fzd9,shisa6,lrtm2 | arc, c1ql3 |
| BP | GO:0010469 | Regulation of signaling receptor activity | 0.0391 | bmp7,cxcl14,wnt7b,shisa9,bmp15,hbegf,il33,ptn,rasgrf1,heyl,fgf1,gsg1l | arc,bmp2,ifnk |
| BP | GO:1902533 | Positive regulation of intracellular signal transduction | 0.041 | egfr,wnt7b,hbegf,pde8a,slc30a10,myoc,cth,adgrg1,rasgrf1,prr5,fgf1,myorg,hes5,grm5, fzd10 | bmp2,stox1,cdon, lpar1, rit2,tnfrsf19 |
| BP | GO:0060284 | Regulation of cell development | 2.62 × 10−6 | ephb3,bmp7,etv5,acsl6,ptprg,wnt7b,slit1,myoc,ptn,adgrg1,heyl, lzts1,hes5,grm5,ttpa | efnb3,hap1,cobl, enpp2,arc,grb14,frzb,bmp2,pak3,slit2,dixdc1,prtg, cdon,lpar1,map1b,cntn1,rit2 |
| BP | GO:0031175 | Neuron projection development | 0.00013 | ephb,bmp7,etv4,egfr, acsl6,ptprg,wnt7b,boc,slit1,myoc,ptn,rasgrf1,lzts1,hes5 | efnb3,hap1,cobl, apod,arc,uchl1,pak3,slit2,mmp2,dixdc1,lpar1,crtac1,map1b,cntn1,rit2 |
| MF | GO:0046943 | Carboxylic acid transmembrane transporter activity | 0.000104 | slc38a3,slc13a3,slc13a5,slc6a9,slco1c1,slc6a11,slc6a1,slc7a10 | |
| MF | GO:0022804 | Active transmembrane transporter activity Sulfur compound binding |
0.0029 | slc38a3,slc13a3,slc13a5,tap2,slc6a9,abcb9,slco1c1,slc6a11,slc6a1,slc7a10 | |
| MF | GO:1901681 | Sulfur compound binding | 0.00368 | bmp7,ogdhl,cbs,hbegf, slit1,ptn,adgrg1,fgf1,gstm1 |
Adj_pval, adjusted p value; BP, biological process; MF, molecular function.
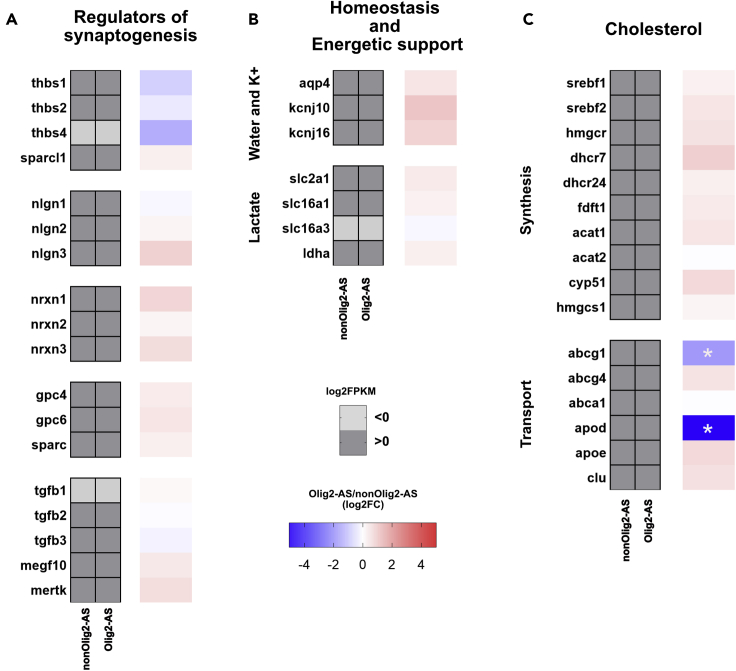
Additional functional (dis)similarities between Olig2-AS and nonOlig2-AS
Beyond their function in regulating synaptic activity, astrocytes are also known to carry out important functions in synapse formation and elimination (Chung et al., 2015) and in regulating homeostasis and energetic support (Sofroniew and Vinters, 2010). They are also known to be a primary source of lipid synthesis in the brain and, in particular, to provide neurons with cholesterol, which is essential for presynaptic vesicle formation (Kıray et al., 2016). None of these functions were found explicitly over- or under-represented in the above-mentioned GO term enrichment analysis. However, and in particular because this analysis was performed from a relatively limited list of genes, we examined the possibility that Olig2-AS and nonOlig2-AS might differ by expression levels of genes involved in these functions. We first analyzed the expression levels of genes coding for key players of synapse formation, i.e., Thrombospondins (thbs 1, 2, and 4), SPARC-like protein 1/Hevin (sparcl1), SPARC (sparc) (Christopherson et al., 2005; Kucukdereli et al., 2011), Neuroligins (nlgn1-3), Neurexins (nrxn1-3) (Stogsdill et al., 2017; Uchigashima et al., 2019), and Glypicans (gpc4, gpc6) (Allen et al., 2012), or synapse elimination, i.e., members of the Transforming growth factor β family (tgfb1-3), the tyrosine kinase receptor MERTK (mertk), and the protein MEGF10 (megf10) (Chung et al., 2013). All these genes, except thbs4 and tgfb1, were found expressed in Olig2-AS and nonOlig2-AS with no significant difference in their expression levels, if not a trend toward down-regulation of the 3 thbs genes in the Olig2-AS (Figure 6A). Similar analyses were then performed for a set of genes whose products are known to control brain homeostasis and energetic support, i.e., the Aquaporin-4 water channel (aqp4), the inwardly rectifying potassium channels Kir4.1 (kcnj10) and Kir5.1 (kcnj16) (Simard and Nedergaard, 2004), the glucose transporter GLUT1 (slc2a1), and components of the lactate shuttle (slc6a1, slc16a3, ldha) (Tekkök et al., 2005). Again, all these genes, except slc16a3, were found expressed both in Olig2-AS and nonOlig2-AS with no significant difference in their expression levels (Figure 6B). Finally, we examined the expression levels of a set of genes involved in cholesterol synthesis, i.e., different enzymes, HMGCR (hmgcr), DHCR7 (dhcr7), DHCR24 (dhcr24), FDFT1 (fdft1), ACAT1 (acat1), ACAT2 (acat2), and CYP51 (cyp51) (Pfrieger and Ungerer, 2011), and the two transcription factors regulating lipid synthesis, Srebf1 (srebf1) and Srebf2 (srebf2). All these genes were expressed in Olig2-AS and nonOlig2-AS, and no significant difference was found in their expression levels (Figure 6C). However, some differences can be observed when looking at the expression of the cholesterol-binding molecules Apolipoprotein D (apod), Apolipoprotein E (apoe), and Apolipoprotein J (clu) and of specific transporters, i.e., ATP-binding cassette (ABC) transporters abcg1, abcg4, and abca1 (Pfrieger and Ungerer, 2011). Among genes found expressed by both Olig2-AS and nonOlig2-AS, 2, abcg1 and apod appeared specifically down-regulated in Olig2-AS (Figure 6C). Taken together, these analyses did not reveal major differences between Olig2-AS and nonOlig2-AS in relation to these important physiological functions of astrocytes.
Figure 6.
Functional similarities between Olig2-AS and nonOlig2-AS
(A–C) Heatmaps depicting abundance of mRNA encoding for genes involved in synaptogenesis (A), homeostasis and energetic support (B), and cholesterol metabolism (C). Heatmaps in gray represent mRNA expression levels (log2FPKM) in nonOlig2-AS and Olig2-AS. Light and dark grays indicate values less than and greater than zero, respectively. Adjacent heatmaps illustrate fold-change in abundance of mRNA in Olig2-AS versus nonOlig2-AS (log2FC). White asterisks indicate significant down-regulation in Olig2-AS.
Discussion
In recent years, transcriptome profiling has been widely used to unravel astrocyte heterogeneity, and a large amount of data supporting molecular astrocyte diversity has been provided. However, how this diversity is defined, i.e., does it result from specific developmental transcriptional programs and/or from later refinement by local environmental cues remains an open question. In this context, the main novelty of our work is that it allows performing relationship between molecular identity of a particular astrocyte subset and its developmental origin. The method we set up made it possible to isolate the Olig2-AS a sub-population of spinal cord astrocytes that originate from Olig2/pMN progenitors (Ohayon et al., 2019); from the other spinal glial cell populations, i.e., OPC/OL, also generated by Olig2/pMN progenitors; and other astrocyte subsets that do not express Olig2 (nonOlig2-AS) and are generated by more dorsal progenitors, which express patterning factors distinct from Olig2 (Bayraktar et al., 2015). Comparison of RNA-seq transcriptome datasets obtained for the three cell types revealed a unique Olig2-AS molecular profile, thus providing a specific gene expression signature of an astrocyte subtype whose identity is tied to its embryonic origin.
A major limitation in understanding astrocyte diversity is the lack of specific markers to prospectively identify and isolate the distinct astrocyte subtypes. A first important finding of our work is thus the identification of two highly specific expression markers of Olig2-AS in the post-natal spinal cord, kcnip3 and inka2, both entering the list of genes composing the Olig2-AS molecular signature. KCNIP3 is a multifunctional protein known to interact with the Kv4 channels and modulate A-type potassium currents in a Ca2+-dependent manner and also to function as a calcium-regulated transcriptional repressor (Frank An et al., 2000; Ledo et al., 2000; Anderson et al., 2010a, 2010b). Inka Box Actin Regulator 2 is known to inhibit the serine/threonine-protein kinase PAK4 (Iwasaki et al., 2015). Expression of inka2 in Olig2-positive cells of the developing spinal cord has previously been reported, and this observation led, at this moment, to the conclusion that expression of this gene marks cells of the oligodendroglial lineage (Iwasaki et al., 2015). Our present data show that co-expression of inka2 and Olig2 instead marks the Olig2-AS, which also expresses Olig2 from the earliest stage of their generation in the ventral spinal cord (Ohayon et al., 2019).
Additional exploration into the molecular signature of the Olig2-AS points to a list of enriched genes that encode for secreted factors, transcriptional regulators, and factors associated with synapse function. First on this list is ptn encoding for Pleiotrophin (PTN), a secreted growth factor with multiple functions during neural development, also known to be up-regulated in astrocytes in response to injury and to act as a neurotrophic factor for spinal MN (Mi et al., 2007; González-Castillo et al., 2015). Another important result of our work is that we identify a number of transcriptional regulators (ERRα, Zfp219, Kcnip3, Irx2, Etv5, HES5, Etv4, Heyl, Epas-1), all likely contributing to the control of the unique expression profile observed in Olig2-AS. To our knowledge, most of them have not previously been linked with astrocyte identity or function. It may be noted that HES5 is known to enhance astrogliogenesis (Bansod et al., 2017) and that Etv5 (ERM) has recently been reported to be differentially expressed in cortical astrocytes (Batiuk et al., 2020). Finally, the most noteworthy aspect is enrichment of a set of genes encoding for neurotransmitter receptors and transporters, mainly related to the glutamatergic and GABAergic signaling, opening the possibility that Olig2-AS might be endowed with particular functions in regulating synaptic activity in the ventral gray matter of the spinal cord. The proposal that local ventral horn astrocytes might ensure specialized function is not a new idea. Previous work from D. Rowitch and collaborators has indeed highlighted the presence, in this region, of a specialized astrocyte subset that plays a critical role to selectively maintain the physiological properties and function of MN subtypes (Molofsky et al., 2014; Kelley et al., 2018). Of note, these astrocytes were found enriched in transcripts for kcnj10, which encode for the K+ channel Kir4.1 (Kelley et al., 2018). Although kcnj10 was not listed in the Olig2-AS molecular signature, this gene was found enriched with a fold change value of 0.98 compared with nonOlig2-AS, i.e., a value just below the arbitrary fold change cutoffs of >1 that we have chosen to apply. This observation, together with the close association of Olig2-AS and MNs (Ohayon et al., 2019), leaves open the possibility that the Olig2-AS corresponds to those astrocytes identified for their discrete molecular function in supporting alpha MN physiology. Otherwise, it is also interesting to note the recent identification of a regionally and functionally distinct astrocyte subset, located in cortical layer V of the cerebral cortex (Miller et al., 2019), which presents notable similarities with Olig2-AS in the sense that it was found enriched in olig2 and lgr6 transcripts, the latter being one of the 135 genes enriched in Olig2-AS. These cortical astrocytes were also found enriched in kcnj10 and ndp (Norrin) transcripts, that, as previously mentioned for kcnj10, were also found enriched in Olig2-AS but not included in the Olig2-AS molecular signature because of a fold change value below the cutoffs of >1. Although still very speculative, and because layer V of the cerebral cortex contains corticospinal neurons that regulate voluntary motor control (Anderson et al., 2010a, 2010b), it is tempting to hypothesize that Olig2 expression in astrocytes, either in the spinal cord or in the cerebral cortex, might confer them specific functional properties to control motor circuits. Olig2-expressing progenitors have been reported to also generate astrocyte during brain development (Marshall et al., 2005; Cai et al., 2007; Griemsmann et al., 2015; Tatsumi et al., 2018; Clavreul et al., 2019) opening the possibility that the embryonic origin of astrocytes might also influence their diversity in the brain.
Astrocyte interaction with neurons has become recognized as a driver of astrocyte diversity (Khakh and Deneen, 2019). The possibility that neurons might exert an influence on the differential gene expression patterns we observed between Olig2AS and nonOlig2AS cannot be excluded. However, there are several arguments against these differences resulting only from neuronal local environment. The three genes, slc1a2 (GLT1) and slc1a3 (GLAST, Figure 5G) as well as Gja1 (Figure 2), whose expression levels have been reported to depend on neurons (Swanson et al., 1997, Hasel et al., 2017), were not found differentially expressed in Olig2AS and nonOlig2AS. In the same line, most of the Olig2AS “signature” genes (Figure S5), including the two Olig2AS markers inka2 and kcnip3, and all but three of the differentially expressed genes encoding for neurotransmitter receptors and transporters (Figures 5E–5I) were not found to depend on neuronal influence in the recent transcriptomic study of Hasel and collaborators (2017). Furthermore, the secreted factor Sonic Hedgehog (Shh), which has previously been identified as a regulator of astrocyte identity and function in the cerebellum (Farmer et al., 2016), is unlikely to be involved in generating differences between Olig2AS and nonOlig2AS because our study shows no evidence for differential expression of genes related to this signaling pathway.
In conclusion, our work provides the first transcriptomic study of an astrocyte subtype whose molecular identity can be linked to its developmental origin. We make available transcriptome dataset of this particular astrocyte subset (Olig2-AS) as well as that of other spinal cord astrocytes and oligodendroglial cells while they are maturating in the post-natal spinal cord. Analysis of the 135 genes that compose the molecular signature of Olig2-AS reveals specific markers of this astrocyte subtype and provides a wealth of information to further unravel to what extent the developmental origin of an astrocyte subtype might influence its functional specialization.
Limitations of study
This study has not dissected in detail the functional specificity of the Olig2-AS; this was mainly based on the interpretation of the RNA-seq data and the GO analysis. Gene knockout models of some candidate genes involved in synaptic regulation that are up-regulated in Olig2-AS represent a necessary model to understand the functional relevance of this subtype of astrocytes in synaptic regulation.
In terms of new candidates' genes differentially expressed in Olig2-AS, even if we had brought validation by conventional in situ hybridization and qPCR, we have also been unfortunate in adapting the RNAscope technique, a quantitative in situ hybridization procedure. This approach is not compatible either with reporter protein detection or with Olig2 immunostaining.
A last point to raise is the intra-heterogeneity of the Olig2-AS; the bulk RNA-seq we perform on this subpopulation could hide a more in-depth heterogeneity, and the use of scRNA-seq on purified Olig2-AS could answer this question.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Olig2 antibody | Millipore | Cat#AB9610; RRID: AB_570666 |
| Goat anti-Sox10 antibody | Santa Cruz Biotechnology | Cat#sc-17342; RRID: AB_2195374 |
| AP conjugated anti-DIG antibody | Sigma-Aldrich | Cat#11093274910; RRID: AB_2734716 |
| POD conjugated anti-Fluo antibody | Sigma-Aldrich | Cat#11426346910; RRID: AB_840257 |
| Donkey anti-rabbit IgG AlexaFluor-488 | ThermoFisher Scientific | Cat#A-21206; RRID: AB_2535792 |
| Donkey anti-goat IgG AlexaFluor-488 | ThermoFisher Scientific | Cat#A-11055; RRID: AB_2534102 |
| Donkey anti-rabbit IgG AlexaFluor-647 | ThermoFisher Scientific | Cat#A-31573; RRID: AB_2536183 |
| Donkey anti-goat IgG AlexaFluor-647 | ThermoFisher Scientific | Cat#A-21447; RRID: AB_141844 |
| AlexaFluor 488 Tyramide kit | ThermoFisher Scientific | Molecular Probes Cat#B-40912 |
| AlexaFluor 555 Tyramide kit | ThermoFisher Scientific | Molecular Probes Cat#B-40913 |
| Chemicals, peptides, and recombinant proteins | ||
| Paraformaldehyde | Sigma-Aldrich | Cat#158127 |
| Phosphate-buffer saline (PBS) | Sigma-Aldrich | Cat#D1408 |
| Sucrose | Sigma-Aldrich | Cat#S0389 |
| OCT media | Sakura | Cat#4583 |
| Triton X-100 | Sigma-Aldrich | Cat#T8787 |
| Tween-20 | Euromedex | Cat#2001-A |
| Sheep serum | Sigma-Aldrich | Cat#S2263 |
| FBS | ThermoFisher Scientific | Gibco Cat#A31604-01 |
| Superfrost Plus microscope slides | Thermoscientific | Cat#J1800AMNT |
| 40μm cell strainer | Falcon | Cat#352340 |
| NBT | Sigma-Aldrich | Cat#10760994001 |
| BCIP | Sigma-Aldrich | Cat# N6639 |
| Trizol | Thermofisher Scientific | Cat#15596026 |
| RNase/DNase-free water | Bio-RAD | Cat#W4502 |
| RNasin | Roche | Cat#N2511 |
| DTT | Promega | Cat#P1171 |
| Evagreen | Bio-RAD | Cat# 1725204 |
| Critical commercial assays | ||
| Papain dissociation system | Worthington Biochemical | Cat#LK003150 |
| Deposited data | ||
| Raw and analyzed data | This paper | GEO: GSE158517 |
| Experimental models: Organisms/strains | ||
| Mouse: Tg(Aldh1l1-eGFP/Rpl10a) JD130Htz | GENSAT, Kindly provided by Dr. N. Rouach, Paris, FRANCE | RRID: IMSR_JAX:030247 |
| Mouse: Olig2-tdtomato | GENSAT, Tg(Olig2-tdTomato)TH39Gsat | MGI: 5311714 http://www.informatics.jax.org/allele/MGI:5311714#imsr |
| Oligonucleotides | ||
| Primer for genotyping GFP-Fw |
This paper | 5’-CGCACCATCTTCTTCAAGGACGAC-3’ |
| Primer for genotyping GFP-Rev |
This paper | 5’-AACTCCAGCAGGACCATGTGATCG-3’ |
| Primer for genotyping Tomato-Fw | This paper | 5’-CTGTTCCTGTACGGCATGG-3’ |
| Primer for genotyping Tomato-Rev | This paper | 5’-GGCATTAAAGCAGCGTATCC-3’ |
| Primers for qPCR | See Table S5 | N/A |
| Software and algorithms | ||
| STAR aligner | Dobin et al., 2012 | https://github.com/alexdobin/STAR |
| FastQC | Babraham Bioinfomatics | RRID:SCR_014583 https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| HTSeq count | Anders et al., 2014 | https://htseq.readthedocs.io/en/master/ |
| DeSeq2 | R-Bioconductor (Love et al., 2014) |
RRID:SCR_015687 https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Bioconductor (3.10) | R-Bioconductor | www.biocondutor.org |
| GOsummaries | R-Bioconductor (Kolde and Vilo, 2015) |
https://www.bioconductor.org/packages/release/bioc/html/GOsummaries.html |
| Revigo | (Supek et al., 2011) | RRID:SCR_005825 http://revigo.irb.hr/ |
| ggplot2 | CRAN (Wickham, 2016) |
RRID:SCR_014601 https://ggplot2.tidyverse.org |
| GOplot | CRAN (Walter et al., 2015) |
https://wencke.github.io/ |
| Cufflinks (v2.2.1) | Trapnell et al., 2010 | RRID:SCR_014597 http://cole-trapnell-lab.github.io/cufflinks |
| Heatmap.2 | R-Bioconductor | |
| ImageJ | NIH | https://imagej.nih.gov |
| Leica LAS X | Leica systems | RRID:SCR_013673 https://www.leica-microsystems.com |
| Prism8 | Graphpad Software | RRID:SCR_002798 https://www.graphpad.com/ |
| Affinity Photo | Serif | RRID:SCR_016951 https://affinity.serif.com |
| Affinity Designer | Serif | https://affinity.serif.com |
| R (3.6.0) | The R foundation | https://www.r-project.org/ |
| Other | ||
| Cryostat CM1950 | Leica | RRID:SCR_018061 |
| Tissue-chopper | Mc Ilwain | RRID:SCR_015798 |
| Nikon Eclipse 80i microscope | Nikon | |
| Leica SP5 confocal | Leica | |
| Leica SP8 confocal | Leica | |
| CFX Maestro BioRad | Bio-RAD | Cat#185-5096 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed and will be fulfilled by the Lead Contact, David Ohayon (david-robert.ohayon@univ-tlse3.fr).
Materials availability
This study did not generate new unique materials.
Data and code availability
RNA-seq data and processed files generated during this study are available through Gene Expression Omnibus (GEO) database. Accession number is GSE158517.
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE158517
No unpublished custom code, software or algorithm was used in this study. Freely available software and algorithms used for analysis are listed in the key resources table. All custom scripts and data contained in this manuscript are available upon request from the Lead Contact.
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.
Experimental model and subject details
Mouse strains
All procedures were performed in agreement with the European Community guiding principles on the care and use of animals (Scientific Procedures) Act, 1987 and approved by the national Animal Care and Ethics Committee (APAFIS#20396) following Directive 2010/63/EU. Olig2-tdTomato mice were generated by the GENSAT project [Tg(olig2-tdTomato) TH39Gsat] and were genotyped using the following primers (Tomato-Fw: 5’-CTGTTCCTGTACGGCATGG-3’ and Tomato-Rev: 5’-GGCATTAAAGCAGCGTATCC-3’). Aldh1L1-GFP (GENSAT) transgenic mice were genotyped as previously reported (Gong et al., 2003; Heintz, 2004; Cahoy et al., 2008). All mice were maintained on a 12h light/dark cycle with food and water ad libitum. Both males and females were used for all experiments. All mice were maintained on a C57BL/6 background.
Method details
Dissociation procedure
Spinal cords from aldh1L1-GFP/olig2-tdTomato mice were harvested at postnatal day 7 (P7), only the brachial and thoracic levels were isolated. DRGs and meninges were then removed and spinal cord explants were sliced into 100 μm sections using a tissue-chopper (Mc Ilwain). The tissue was enzymatically dissociated to make a suspension of single cells as described previously (Cahoy et al., 2008). Briefly, the tissues were dissociated with papain (20U/mL, Worthington Biochemical, Cat#LK003150) for 60 minutes at 37°C in bicarbonate-buffered Earle’s balanced salt solution and 0,005% of DNaseI (Worthington Biochemical, Cat#LK003150). The papain solution was equilibrated with 5% CO2 and 95% O2 before and during papain treatment. Then, the tissue was mechanically dissociated by gentle trituration with a 10mL pipette. The cloudy cell suspension was collected and centrifuged at 300 x g for 5 minutes. The cell pellet was resuspended in a low concentration inhibitor solution with DNase, BSA 1mg/mL and ovomucoid 1mg/mL (Worthington Biochemical, Cat#LK003150). Then, the cell suspension was layered on top of 5mL of a high concentration inhibitor solution with BSA 10mg/mL and ovomucoid 10mg/mL and centrifuged at 70 x g for 6 minutes. The cell pellet was then resuspended in 300μL of PBS and 10% FBS (Gibco, Cat# A31604-01) and filtered through a 40μm cell strainer (Falcon, Cat#352340) to remove any remaining clumps of tissue.
FACS and RNA preparation
Dissociated cells from two mouse pups coming from the same litter were pooled. Cells were sorted using a FACS Aria Fusion BSL1 at the IPBS FACS-platform. Dead cells and debris were gated by their low forward scatter area (FSC-A) and high side scatter area (SSC-A). Following that, two successive gating approaches were done to exclude doublets, first a forward scatter height (FSC-H) vs forward scatter width (FSC-W) and then a side scatter height (SSC-H) vs side scatter width (SSC-W). Then gating for fluorescence was performed, the three awaited cells population were then identified based on their fluorescence using a PE-A vs GFP-A: high eGFP fluorescence for the nonOlig2-AS; high tdTomato fluorescence for the OPC/OL and high eGFP and tdTomato fluorescence for the Olig2-AS. Purified cells were harvested by centrifugation at 2000xg for 5 minutes and cell pellets were processed for RNA extraction. The whole procedure for cell suspension preparation and FACS sorting was completed in 3-4 hours. After sorting, RNA was isolated from the cell pellet using standard Trizol reagent (Thermofisher, Cat.#15596026) with glycogen added as carrier according to manufacturer instructions and eluted into 35μL RNase-free water. Eluted RNAs were then stored at -80°C. RNA quality and integrity (RIN) were verified on an Agilent 2100 Bioanalyzer. The whole procedure going from the dissociation to RNA preparation has been performed four times, each time on a different mouse litter.
RNA-seq library generation and sequencing
For each cell population (tdT-/eGFP+; tdT+/eGFP-; tdT+/eGFP+) that was subjected to RNA-seq analysis, FACS isolated cells were used from pooled samples; each cell population was sequenced in quadriplicate. Library preparation and RNA sequencing on the twelve samples were performed by Beijing Genomics Institute (BGI, Hong Kong, China) using a BGISEQ-500 RNA-seq platform (paired-end sequencing, 100 bp reads, 30M reads/sample). The quality of each raw sequencing file (fastq) was verified with FastQC. All files were aligned to the reference mouse genome (Ensembl mus musculus GRCm38.91; gtf(mm10)) using STAR aligner (Version Galaxy Tool: V2.1; Dobin et al., 2012). Read count per sample was computed using HT-seq count (Version Galaxy Tool: V1.0; Anders et al., 2014). Then raw count table was cleaned, we decided to keep gene having an averaged raw read count per sample higher or equal to 1.
Differential analysis
This analysis was applied per pairs of cell populations using DESeq2 (V1.22.2; Love et al., 2014), available as an R package in bioconductor (www.biocondutor.org), to normalize raw read count using RLE methods generating the log2 fold change (log2FC) values. This procedure has led to three different differential analysis (DA), between Olig2-AS and nonOlig2-AS cells (DA1), between Olig2-AS and OPC/OL (DA2) and between nonOlig2-AS and OPC/OL (DA3). Mouse ENSEMBL IDs from each sample were then converted to mouse official gene names using Biomart (Durinck et al., 2005), available also as an R package in bioconductor. We used principal component analysis (PCA) and hierarchical clustering to cluster samples based on their expression levels. Data representation of the PCA was generated using the ggplot2 package (https://ggplot2.tidyverse.org). Euclidean distances between the cell types were determined for the top 20% most variable mRNA and plotted using the heatmap.2 package in R software.
Sample correlation
Following RNA-seq data preprocessing, we computed the pairwise Pearson correlations between each sample for the expression levels of all detected mRNAs to determine the reproducibility between our samples. We then used the heatmap.2 package for graphical representation of correlations, without sample clustering.
Gene ontology
Gene ontology was performed using the GO summaries package (V3.11; Kolde and Vilo, 2015) on genes up-regulated (Table S2) and down-regulated in the Olig2-AS (Table S3). Those results were visualized using the REVIGO graphical tool (http://revigo.irb.hr/) and GOplot (V1.0.2; Walter et al., 2015).
Quantitative PCR
RNA was extracted from cell pellets using Trizol reagent (Thermofisher, Cat.#15596026) according to the manufacturer’s instructions. 1 μg RNA was used for reverse transcription. Genomic DNA was degraded with 1 μl DNase (Cat#M6101, Promega) for 20 min at 37°C in 20 μl RNase/DNase-free water (Cat#W4502, Bio-RAD), and the reaction was stopped by adding 1 μl stop solution under heat inactivation at 65°C for 10 min. 2 μl dNTPs (10 mM; Promega) and 2 μl oligodTs (100 mM, idtDNA) were added for 5 min at 65°C, then 8 μl of 5 × buffer, 2 μl RNasin (Cat#N2511, Roche), and 4 μl of 100 mM DTT (Cat#P1171, Promega) were added for 2 min at 42°C. The mix was divided into equal volumes in a reverse-transcriptase–negative control tube with addition of 1 μl water and in a reverse-transcriptase–positive tube with 1 μl superscript enzyme and placed at 42°C for 1 hr. The reaction was stopped at 70°C for 15 min, and cDNAs were diluted (10-, 100-, and 1,000-fold) and processed for quantitative PCR in triplicate for each dilution. 10 μl diluted cDNA was mixed with 10 μl premix Evagreen (Cat# 1725204, Bio-RAD) containing 1μM of each primer, and the PCR program was run for 40 cycles on a CFX Maestro BioRad Realtime system (Cat#185-5096, Bio-RAD). mRNA relative expression levels were calculated using the 2-ΔΔCts method (Livak and Schmittgen, 2001). Primers are listed in Table S5.
Tissue collection and processing
Spinal cords at the brachial level were isolated from E13.5 mouse embryos to P7 mouse pups and fixed in 4% paraformaldehyde (PFA, Sigma, Cat#158127) in phosphate-buffered saline (PBS) overnight at 4°C. Tissues were then sectioned at 15 μm to 25 μm using a cryostat (Leica CM1950) after cryoprotection in 20% sucrose (Sigma, Cat# S0389) in PBS and freezing in OCT media (Sakura, CatN: 4583) on the surface of dry ice.
In situ RNA hybridization (ISH) and immunofluorescent (IF) staining
Double in situ hybridizations were performed on 25 μm frozen sections. Digoxigenin- or Fluorescein-labeled antisens RNA probes for inka2, efnb3, fgfr3 and kcnip3 were synthesized using DIG- or Fluorescein-labelling kit (Roche) according to the manufacturer’s instructions. One DIG- and one Fluo probe were mixed in hybridization buffer and applied to frozen sections. After hybridization at 65°C overnight, sections were washed twice in 50% Formamide, 1× SSC, 0.1% Tween-20 for 30 min at 65°C, twice in MABT buffer for 30 min before blocking in blocking buffer (MABT, 2% blocking reagent from Roche, 20% inactivated sheep serum) for 2 h at room temperature. Then, the sections were incubated to a 1:500 dilution of anti-Fluo-POD antibody (Sigma, Cat#11426346910) in blocking buffer overnight at 4°C. After washing three times in PBS with 0,1% Tween 20 (PBST), the bound Fluo-probe was revealed by using fluorescent tyramide AlexaFluor 555 (Thermo Fisher Scientific, Cat#B-40913) used at 1:100 dilution into the amplification diluent during 30 minutes at room temperature. Slides were then washed three times in PBST and incubated in blocking buffer for one hour at room temperature. Sections were then incubated overnight with alkaline phosphatase (AP) conjugated anti-DIG antibody (Sigma, Cat#11093274910) diluted 1:2000 in blocking buffer. After three washes with MABT for 10 minutes, sections were washed four times in a pre-developing buffer (100mM Tris pH 9,5; 100mM NaCl; 50mM MgCl2) during 5 minutes. The bound DIG-probe was revealed using NBT/BCIP (Sigma, Cat#10760994001 and Cat# N6639) diluted in pre-developing buffer. For triple staining, in which immunofluorescence was followed by double in situ hybridization, at the end of NBP/BCIP incubation, sections were washed three times with PBST and then incubated for one hour at room temperature with IF blocking buffer (4% goat serum in PBST). Then, sections were incubated to a 1:500 dilution of rabbit anti-Olig2 in IF blocking buffer overnight at 4°C. After washing three times in PBST, the sections are incubated with a fluorophore-conjugated secondary antibody at 1:500 dilution for one hour at room temperature. Sections were then washed in PBST and mounted with moviol and coverslip. Multiple immunofluorescent stainings were performed as previously reported (Ohayon et al., 2019). Antibodies used in this study were as follows: rabbit anti-Olig2 (Millipore, Cat#AB9610) and goat anti-Sox10 (Santa Cruz Biotechnology, Cat#sc-17342), Alexa Fluor-488 or Alexa Fluor-647-conjugated secondary antibodies (Thermo Fisher Scientific).
Imaging
Confocal images were acquired from tissue sections using Leica SP5 or SP8 confocal microscopes and were always represented as single optical plane sections. All confocal images showing hemi-ventral spinal cord were acquired using a x20 objective, while higher magnification images were obtained using a x40 objective. Images of simple ISHs were collected with Nikon digital camera DXM1200C on a Nikon Eclipse 80i microscope. Double ISH stainings and double ISH-immunofluorescence stainings were imaged on Leica SP5 and SP8 confocal microscope according to the method of Trinh et al. (Trinh et al., 2007) which allows acquiring in the same optical plane a high resolution confocal image of NBT/BCIP stain in the near infrared range together with the immunofluorescence or TSA signals. Images were processed (size adjustment, luminosity and contrast, and merging or separating layers) using Affinity Photo software (Serif) and ImageJ.
Quantification and statistical analysis
Figures and representations
Figure 2F: Barplot of the relative expression levels (log2FPKM) of CNS cell type specific genes. Graph built using Prism7 (Graphpad).
FPKM values were generated using the Cufflinks (v2.2.1; Trapnell et al., 2010) software (http://cole-trapnell-lab.github.io/cufflinks)
Figures S2B and S2C: Histogram representing the proportion of the different glial cell populations based on their specific fluorescent markers both in vivo and after FACS sorting. Graph built using Prism7 (Graphpad)
Figure S3E: Barplot of the relative expression levels (log2FPKM) of eGFP and tdTomato transcripts in each replicate. Graph built using Prism7 (Graphpad)
Figure 3A: Flowchart representing the treatment and two-step comparative analysis performed on the RNA-seq data. Flowchart built by using the web application draw.io (www.draw.io)
Figures 3B–3E and S4A–S4C: Volcano plots were built using the ggplot2 package on the differentially expressed genes representing the log2FC and –log10Padj from the differential analysis DA1, DA2 and DA3.
Figures 2D, 2E, 3F, 3G, S5A, S5B, S6A, and S6B: Heatmap representation data (comparisons between the three glial cell populations) were reduced and scaled to generate z-score
Figures 5A-5B-5D: Treemaps were created using the REVIGO graphical tool (http://revigo.irb.hr/)
Figure 5C: Circular visualization of gene-annotation enrichment analysis (GOcircle) with the 245 specifically differentially regulated genes in Olig2-AS (135 genes up-regulated + 110 genes down-regulated).
Figures 5E–5I and 6A–6C: Expression level measurements in Olig2-AS and non-Olig2AS, heatmap are based upon log2FPKM values.
Figures 5E–5I and 6A–6C: Heatmap representation data (comparison between Olig2-AS and nonOlig2-AS) are based upon log2FC.
All the heatmaps were made with Prism 7 (Graphpad).
Table description and cutoffs
Gene lists in supplementary tables were generated as follows:
Table S2: genes enriched in Olig2-AS (up1up2), with a log2FC>1 and padj<0.05 both in DA1 and in DA2.
Table S3: genes down-regulated in Olig2-AS (dn1dn2), with log2FC<-1 and padj<0.05 both in DA1 and in DA2.
Table S4: gene ontology analysis (BP) on genes enriched in Olig2-AS (log2FC>1 and padj<0.05 both in DA1 and in DA2) and in gene depleted in Olig2-AS (log2FC<-1 and padj<0.05 both in DA1 and in DA2).
Quantification
Cell counts performed on spinal cord sections were done using ImageJ software and obtained from 4 animals and at least five spinal cord sections of brachial spinal cord (anterior limb level) have been counted for each individual. (eGFP+/tdT+, eGFP+/tdT- and eGFP-/tdT+ cells were counted on the whole spinal cord (both grey and white matter). Quantifications are expressed as the mean ratio ± SEM of each cell type on either the total population of eGFP+ cells or the total population of tdT+ cells counted per optical spinal cord section. Representativeness of FACS-sorted cells was calculated using FACSAria Fusion BSL1 before applying stringent fluorescence gating strategy. This has been performed on ten separated experiments, each one corresponding to one dissociated spinal cord harvested from a P7 aldh1L1-GFP/olig2-tdTomato pup (n=10).
qPCR statistic
All statistical analyses were performed using GraphPad Prism9. All values were plotted as the mean ± SEM. For multiple groups (> 2), one-way ANOVA was used to analyze the variance, followed by a Tukey post hoc test to compare multiple groups. For two-group comparison, Student's t-test was used. Statistical significance was tested at a 95% (p < .05) confidence level and was depicted with an asterisk (∗p < .05; ∗∗p < .01; ∗∗∗p < .001).
Acknowledgments
We especially thank N. Rouach and P. Durbec for sharing of Aldh1L1-GFP and Olig2-tdTomato mice, respectively, and E. Nasser and P. Viana for the technical support at the IPBS-FACS platform. We acknowledge I. Néant, A. Davy, and S. Sakakiba for the gift of the kcnip3, efnb3, and inka2 probes, respectively, and A. Davy, W. Richardson, and JP Hugnot for critical reading of the manuscript. We thank the ABC facility and ANEXPLO for housing mice and the Toulouse Regional Imaging Platform (TRI) for technical assistance in confocal microscopy. Work in C.S.’ laboratory was supported by grants from ANR (ANR-15-CE16-0014-02), ARC (PJA20161204698), ARSEP, CNRS, and University of Toulouse.
Author contributions
Conceptualization, D.O. and C.S.; methodology, D.O.; software, D.O. and M.A.; investigation, D.O., N.E., and T.J.; writing – Original draft, D.O. and C.S.; writing – review and editing, D.O. and C.S.; funding acquisition, D.O. and C.S.; supervision, D.O. and C.S.
Declaration of interests
The authors declare that they have no competing interests.
Published: July 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102806.
Supplemental information
Expression values for all RNA-seq data generated in the study, presented as fragments per kilobase of transcript per million mapped reads (FPKM), for all the glial subpopulations in each replicate.
List of genes enriched in Olig2-AS that present a log2FC > 1 and padj<0.05 in both DA1 and DA2.
List of genes depleted in Olig2-AS that present a log2FC < −1 and padj<0.05 in both DA1 and DA2.
Gene Ontology analysis (BP) from GO summaries on genes enriched in Olig2-AS (log2FC > 1 and padj<0.05 both in DA1 and in DA2) and in genes depleted in Olig2-AS (log2FC < −1 and padj<0.05 both in DA1 and in DA2). The table indicates the list of GO terms enriched, the list of genes involved in a given GO term, and the p value. Only terms with p < 0.05 are shown.
References
- Allen N.J., Bennett M.L., Foo L.C., Wang G.X., Chakraborty C., Smith S.J., Barres B.A. Astrocyte glypicans 4 and 6 promote formation of excitatory synapses via GluA1 AMPA receptors. Nature. 2012 doi: 10.1038/nature11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen N.J., Eroglu C. Cell Biology of astrocyte-synapse interactions. Neuron. 2017 doi: 10.1016/j.neuron.2017.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen N.J., Lyons D.A. Glia as architects of central nervous system formation and function. Science. 2018 doi: 10.1126/science.aat0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. HTSeq - a Python framework to work with high-throughput sequencing data. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C.T., Sheets P.L., Kiritani T., Shepherd G.M.G. Sublayer-specific microcircuits of corticospinal and corticostriatal neurons in motor cortex. Nat. Neurosci. 2010 doi: 10.1038/nn.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D., Mehaffey W.H., Iftinca M., Rehak R., Engbers J.D.T., Hameed S., Zamponi G.W., Turner R.W. Regulation of neuronal activity by Cav3-Kv4 channel signaling complexes. Nat. Neurosci. 2010 doi: 10.1038/nn.2493. [DOI] [PubMed] [Google Scholar]
- Bansod S., Kageyama R., Ohtsuka T. Development; 2017. Hes5 Regulates the Transition Timing of Neurogenesis and Gliogenesis in Mammalian Neocortical Development. [DOI] [PubMed] [Google Scholar]
- Batiuk M.Y., Martirosyan A., Wahis J., de Vin F., Marneffe C., Kusserow C., Koeppen J., Viana J.F., Oliveira J.F., Voet T. Identification of region-specific astrocyte subtypes at single cell resolution. Nat. Commun. 2020 doi: 10.1038/s41467-019-14198-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayraktar O.A., Bartels T., Holmqvist S., Kleshchevnikov V., Martirosyan A., Polioudakis D., Ben Haim L., Young A.M.H., Batiuk M.Y., Prakash K. Astrocyte layers in the mammalian cerebral cortex revealed by a single-cell in situ transcriptomic map. Nat. Neurosci. 2020 doi: 10.1038/s41593-020-0602-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayraktar O.A., Fuentealba L.C., Alvarez-Buylla A., Rowitch D.H. Astrocyte development and heterogeneity. Cold Spring Harbor Perspect. Biol. 2015;7 doi: 10.1101/cshperspect.a020362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Haim L., Rowitch D.H. Functional diversity of astrocytes in neural circuit regulation. Nat. Rev. Neurosci. 2016 doi: 10.1038/nrn.2016.159. [DOI] [PubMed] [Google Scholar]
- Benson, M.D., Romero, M.I., Lush, M.E., Lu, Q.R., Henkemeyer, M. and Parada, L.F. (2005). Ephrin-B3 is a myelin-based inhibitor of neurite outgrowth. Proceedings of the National Academy of Sciences of the United States of America. 10.1073/pnas.0504021102. [DOI] [PMC free article] [PubMed]
- Boisvert M.M., Erikson G.A., Shokhirev M.N., Allen N.J. The aging astrocyte transcriptome from multiple regions of the mouse brain. Cell Rep. 2018 doi: 10.1016/j.celrep.2017.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy J.D., Emery B., Kaushal A., Foo L.C., Zamanian J.L., Christopherson K.S., Xing Y., Lubischer J.L., Krieg P.A., Krupenko S.A. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 2008 doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, J., Chen, Y., Cai, W.H., Hurlock, E.C., Wu, H., Kernie, S.G., Parada, L.F., Lu, Q.R. 10.1242/dev.02847. [DOI]
- Chaboub L.S., Deneen B. Developmental origins of astrocyte heterogeneity: the final frontier of CNS development. Developmental Neurosci. 2013 doi: 10.1159/000343723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaboub L.S., Manalo J.M., Lee H.K., Glasgow S.M., Chen F., Kawasaki Y., Akiyama T., Kuo C.T., Creighton C.J., Mohila C.A., Deneen B. Temporal profiling of astrocyte precursors reveals parallel roles for Asef during development and after injury. J. Neurosci. 2016;36:11904–11917. doi: 10.1523/JNEUROSCI.1658-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai H., Diaz-Castro B., Shigetomi E., Monte E., Octeau J.C., Yu X., Cohn W., Rajendran P.S., Vondriska T.M., Whitelegge J.P. Neural circuit-specialized astrocytes: transcriptomic, proteomic, morphological, and functional evidence. Neuron. 2017 doi: 10.1016/j.neuron.2017.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopherson K.S., Ullian E.M., Stokes C.C.A., Mullowney C.E., Hell J.W., Agah A., Lawler J., Mosher D.F., Bornstein P., Barres B.A. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005 doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Chung W.S., Allen N.J., Eroglu C. Astrocytes control synapse formation, function, and elimination. Cold Spring Harbor Perspect. Biol. 2015 doi: 10.1101/cshperspect.a020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W.S., Clarke L.E., Wang G.X., Stafford B.K., Sher A., Chakraborty C., Joung J., Foo L.C., Thompson A., Chen C. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature. 2013 doi: 10.1038/nature12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L.E., Liddelow S.A., Chakraborty C., Münch A.E., Heiman M., Barres B.A. Normal aging induces A1-like astrocyte reactivity. Proc. Natl. Acad. Sci. United States America. 2018 doi: 10.1073/pnas.1800165115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus Stolt C., Rehberg S., Ader M., Lommes P., Riethmacher D., Schachner M., Bartsch U., Wegner M. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Development. 2002 doi: 10.1101/gad.215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavreul S., Abdeladim L., Hernández-Garzón E., Niculescu D., Durand J., Ieng S.H., Barry R., Bonvento G., Beaurepaire E., Livet J. Cortical astrocytes develop in a plastic manner at both clonal and cellular levels. Nat. Commun. 2019 doi: 10.1038/s41467-019-12791-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessaud E., McMahon A.P., Briscoe J. Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development. 2008 doi: 10.1242/dev.009324. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.a, Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. RNA-STAR: ultrafast universal spliced sequences aligner: supplementary materials. Bioinformatics. 2012 doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J.P., Dougherty J.D., Heiman M., Schmidt E.F., Stevens T.R., Ma G., Bupp S., Shrestha P., Shah R.D., Doughty M.L. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008 doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S., Moreau Y., Kasprzyk A., Davis S., De Moor B., Brazma A., Huber W. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005 doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- Farmer W.T., Abrahamsson T., Chierzi S., Lui C., Zaelzer C., Jones E.V. Neurons diversify astrocytes in the adult brain through sonic hedgehog signaling. Science. 2016 doi: 10.1126/science.aab3103. [DOI] [PubMed] [Google Scholar]
- Frank An W., Bowlby M.R., Betty M., Cao J., Ling H.P., Mendoza G., Hinson J.W., Mattsson K.I., Strassle B.W., Trimme J.S., Rhodes K.J. Modulation of A-type potassium channels by a family of calcium sensors. Nature. 2000 doi: 10.1038/35000592. [DOI] [PubMed] [Google Scholar]
- Gokce O., Stanley G.M., Treutlein B., Neff N.F., Camp J.G., Malenka R.C., Rothwell P.E., Fuccillo M.V., Südhof T.C., Quake S.R. Cellular taxonomy of the mouse striatum as revealed by single-cell RNA-seq. Cell Rep. 2016 doi: 10.1016/j.celrep.2016.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S., Zheng C., Doughty M.L., Losos K., Didkovsky N., Schambra U.B., Nowak N.J., Joyner A., Leblanc G., Hatten M.E., Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003 doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- González-Castillo C., Ortuño-Sahagún D., Guzmán-Brambila C., Pallàs M., Rojas-Mayorquín A.E. Pleiotrophin as a central nervous system neuromodulator, evidences from the hippocampus. Front. Cell Neurosci. 2015 doi: 10.3389/fncel.2014.00443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griemsmann S., Höft S.P., Bedner P., Zhang J., Von Staden E., Beinhauer A., Degen J., Dublin P., Cope D.W., Richter N. Characterization of panglial gap junction networks in the thalamus, neocortex, and hippocampus reveals a unique population of glial cells. Cereb. Cortex. 2015 doi: 10.1093/cercor/bhu157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasel P., Dando O., Jiwaji Z., Baxter P., Todd A.C., Heron S. Neurons and neuronal activity control gene expression in astrocytes to regulate their development and metabolism. Nat. Commun. 2017 doi: 10.1038/ncomms15132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N. Gene expression nervous system atlas (GENSAT) Nat. Neurosci. 2004 doi: 10.1038/nn0504-483. [DOI] [PubMed] [Google Scholar]
- Hochstim C., Deneen B., Lukaszewicz A., Zhou Q., Anderson D.J. Identification of positionally distinct astrocyte subtypes whose identities are specified by a homeodomain code. Cell. 2008;133:510–522. doi: 10.1016/j.cell.2008.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki Y., Yumoto T., Sakakibara S.I. Expression profiles of inka2 in the murine nervous system. Gene Expr. Patterns. 2015;19:83–97. doi: 10.1016/j.gep.2015.08.002. [DOI] [PubMed] [Google Scholar]
- Jacobi E., von Engelhardt J. AMPA receptor complex constituents: control of receptor assembly, membrane trafficking and subcellular localization. Mol. Cell Neurosci. 2018 doi: 10.1016/j.mcn.2018.05.008. [DOI] [PubMed] [Google Scholar]
- John Lin C.C., Yu K., Hatcher A., Huang T.W., Lee H.K., Carlson J., Weston M.C., Chen F., Zhang Y., Zhu W. Identification of diverse astrocyte populations and their malignant analogs. Nat. Neurosci. 2017 doi: 10.1038/nn.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley K.W., Ben Haim L., Schirmer L., Tyzack G.E., Tolman M., Miller J.G., Tsai H.H., Chang S.M., Molofsky A.V., Yang Y. Kir4.1-dependent astrocyte-fast motor neuron interactions are required for peak strength. Neuron. 2018;98:306–319.e7. doi: 10.1016/j.neuron.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh B.S., Deneen B. The emerging nature of astrocyte diversity. Annu. Rev. Neurosci. 2019 doi: 10.1146/annurev-neuro-070918-050443. [DOI] [PubMed] [Google Scholar]
- Khakh B.S., Sofroniew M.V. Diversity of astrocyte functions and phenotypes in neural circuits. Nat. Neurosci. 2015 doi: 10.1038/nn.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kıray H., Lindsay S.L., Hosseinzadeh S., Barnett S.C. The multifaceted role of astrocytes in regulating myelination. Exp. Neurol. 2016;283:541–549. doi: 10.1016/j.expneurol.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R., Vilo J. GOsummaries: an R package for visual functional annotation of experimental data. F1000Research. 2015 doi: 10.12688/f1000research.6925.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukdereli H., Allen N.J., Lee A.T., Feng A., Ozlu M.I., Conatser L.M., Chakraborty C., Workman G., Weaver M., Sage E.H. Control of excitatory CNS synaptogenesis by astrocyte-secreted proteins hevin and SPARC. Proc. Natl. Acad. Sci. United States America. 2011 doi: 10.1073/pnas.1104977108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlbrodt K., Herbarth B., Sock E., Hermans-Borgmeyer I., Wegner M. Sox10, a novel transcriptional modulator in glial cells. J. Neurosci. 1998 doi: 10.1523/jneurosci.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledo F., Link W.A., Carrión A.M., Echeverria V., Mellström B., Naranjo J.R. The DREAM-DRE interaction: key nucleotides and dominant negative mutants. Biochim. Biophys. Acta - Mol. Cell Res. 2000 doi: 10.1016/S0167-4889(00)00092-6. [DOI] [PubMed] [Google Scholar]
- Li D., Hérault K., Silm K., Evrard A., Wojcik S., Oheim M. Lack of evidence for vesicular glutamate transporter expression in mouse astrocytes. J. Neurosci. 2013 doi: 10.1523/JNEUROSCI.3667-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001 doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Love M.I., Anders S., Huber W. Differential analysis of count data - the DESeq2 package. Genome Biol. 2014 doi: 10.1186/s13059-014-0550-8. [DOI] [Google Scholar]
- Lozzi B., Huang T.W., Sardar D., Huang A.Y.S., Deneen B. Regionally distinct astrocytes display unique transcription factor profiles in the adult brain. Front. Neurosci. 2020 doi: 10.3389/fnins.2020.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall C.A.G., Novitch B.G., Goldman J.E. Olig2 directs astrocyte and oligodendrocyte formation in postnatal subventricular zone cells. J. Neurosci. 2005 doi: 10.1523/JNEUROSCI.1924-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi R., Chen W., Höke A. Pleiotrophin is a neurotrophic factor for spinal motor neurons. Proc. Natl. Acad. Sci. United States America. 2007 doi: 10.1073/pnas.0603243104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S.J., Philips T., Kim N., Dastgheyb R., Chen Z., Hsieh Y.C., Daigle J.G., Datta M., Chew J., Vidensky S. Molecularly defined cortical astroglia subpopulation modulates neurons via secretion of Norrin. Nat. Neurosci. 2019;22:741–752. doi: 10.1038/s41593-019-0366-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mölders A., Koch A., Menke R., Klöcker N. Heterogeneity of the astrocytic AMPA-receptor transcriptome. GLIA. 2018 doi: 10.1002/glia.23514. [DOI] [PubMed] [Google Scholar]
- Molofsky A.V., Kelley K.W., Tsai H.H., Redmond S.A., Chang S.M., Madireddy L., Chan J.R., Baranzini S.E., Ullian E.M., Rowitch D.H. Astrocyte-encoded positional cues maintain sensorimotor circuit integrity. Nature. 2014;509:189–194. doi: 10.1038/nature13161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel L., Chiang M.S.R., Higashimori H., Shoneye T., Iyer L.K., Yelick J., Tai A., Yang Y. Molecular and functional properties of regional astrocytes in the adult brain. J. Neurosci. 2017 doi: 10.1523/JNEUROSCI.3956-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel L., Men Y., Chiang M.S.R., Tian Y., Jin S., Yelick J., Higashimori H., Yang Y. Intracortical astrocyte subpopulations defined by astrocyte reporter Mice in the adult brain. Glia. 2019;67:171–181. doi: 10.1002/glia.23545. [DOI] [PubMed] [Google Scholar]
- Muroyama Y., Fujiwara Y., Orkin S.H., Rowitch D.H. Specification of astrocytes by bHLH protein SCL in a restricted region of the neural tube. Nature. 2005 doi: 10.1038/nature04139. [DOI] [PubMed] [Google Scholar]
- Ohayon D., Escalas N., Cochard P., Glise B., Danesin C., Soula C. Sulfatase 2 promotes generation of a spinal cord astrocyte subtype that stands out through the expression of Olig2. GLIA. 2019;67 doi: 10.1002/glia.23621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestana F., Edwards-Faret G., Belgard T.G., Martirosyan A., Holt M.G. No longer underappreciated: the emerging concept of astrocyte heterogeneity in neuroscience. Brain Sci. 2020 doi: 10.3390/brainsci10030168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfrieger F.W., Ungerer N. Cholesterol metabolism in neurons and astrocytes. Prog. Lipid Res. 2011;50:357–371. doi: 10.1016/j.plipres.2011.06.002. [DOI] [PubMed] [Google Scholar]
- Pringle N.P., Yu W.P., Howell M., Colvin J.S., Ornitz D.M., Richardson W.D. Fgfr3 expression by astrocytes and their precursors: evidence that astrocytes and oligodendrocytes originate in distinct neuroepithelial domains. Development. 2003 doi: 10.1242/dev.00184. [DOI] [PubMed] [Google Scholar]
- Rowitch D.H., Kriegstein A.R. Developmental genetics of vertebrate glial-cell specification. Nature. 2010;468:214–222. doi: 10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- Saunders A., Macosko E.Z., Wysoker A., Goldman M., Krienen F.M., de Rivera H., Bien E., Baum M., Bortolin L., Wang S. Molecular diversity and specializations among the cells of the adult mouse brain. Cell. 2018 doi: 10.1016/j.cell.2018.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard M., Nedergaard M. The neurobiology of glia in the context of water and ion homeostasis. Neuroscience. 2004 doi: 10.1016/j.neuroscience.2004.09.053. [DOI] [PubMed] [Google Scholar]
- Sofroniew M.V., Vinters H.V. Astrocytes: Biology and pathology. Acta Neuropathologica. 2010 doi: 10.1007/s00401-009-0619-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stogsdill J.A., Ramirez J., Liu D., Kim Y.H., Baldwin K.T., Enustun E., Ejikeme T., Ji R.R., Eroglu C. Astrocytic neuroligins control astrocyte morphogenesis and synaptogenesis. Nature. 2017;551:192–197. doi: 10.1038/nature24638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., McConnell E., Pare J.F., Xu Q., Chen M., Peng W., Lovatt D., Han X., Smith Y., Nedergaard M. Glutamate-dependent neuroglial calcium signaling differs between young and adult brain. Science. 2013 doi: 10.1126/science.1226740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F., Bošnjak M., Škunca N., Šmuc T. Revigo summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011 doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R.A., Liu J., Miller J.W., Rothstein J.D., Farrell K., Stein B.A., Longuemare M.C. Neuronal regulation of glutamate transporter subtype expression in astrocytes. J. Neurosci. 1997 doi: 10.1523/jneurosci.17-03-00932.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi K., Isonishi A., Yamasaki M., Kawabe Y., Morita-Takemura S., Nakahara K., Terada Y., Shinjo T., Okuda H., Tanaka T. Olig2-Lineage astrocytes: A distinct subtype of astrocytes that differs from GFAP astrocytes. Front. Neuroanat. 2018 doi: 10.3389/fnana.2018.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekkök S.B., Brown A.M., Westenbroek R., Pellerin L., Ransom B.R. Transfer of glycogen-derived lactate from astrocytes to axons via specific monocarboxylate transporters supports mouse optic nerve activity. J. Neurosci. Res. 2005 doi: 10.1002/jnr.20573. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., Van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010 doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh L.A., McCutchen M.D., Bonner-Fraser M., Fraser S.E., Bumm L.A., McCauley D.W. Fluorescent in situ hybridization employing the conventional NBT/BCIP chromogenic stain. BioTechniques. 2007 doi: 10.2144/000112476. [DOI] [PubMed] [Google Scholar]
- Tsai H.H., Li H., Fuentealba L.C., Molofsky A.V., Taveira-Marques R., Zhuang H., Tenney A., Murnen A.T., Fancy S.P.J., Merkle F. Regional astrocyte allocation regulates CNS synaptogenesis and repair. Science. 2012;337:358–362. doi: 10.1126/science.1222381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchigashima M., Cheung A., Suh J., Watanabe M., Futai K. Differential expression of neurexin genes in the mouse brain. J. Comp. Neurol. 2019;527:1940–1965. doi: 10.1002/cne.24664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter W., Sánchez-Cabo F., Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 2015 doi: 10.1093/bioinformatics/btv300. [DOI] [PubMed] [Google Scholar]
- Wickham H. Media. Second Edition. Springer; 2016. ggplot2: Elegant Graphics for Data Analysis. [DOI] [Google Scholar]
- Zeisel A., Hochgerner H., Lönnerberg P., Johnsson A., Memic F., van der Zwan J., Häring M., Braun E., Borm L.E., La Manno G. Molecular architecture of the mouse nervous system. Cell. 2018 doi: 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A., M͡oz-Manchado A.B., Codeluppi S., Lönnerberg P., Manno G. La, Juréus A., Marques S., Munguba H., He L., Betsholtz C. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015 doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Barres B.A. Astrocyte heterogeneity: an underappreciated topic in neurobiology. Curr. Opin. Neurobiol. 2010 doi: 10.1016/j.conb.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Chen K., Sloan S.A., Bennett M.L., Scholze A.R., O’Keeffe S., Phatnani H.P., Guarnieri P., Caneda C., Ruderisch N. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014 doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Wang S., Anderson D.J. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron. 2000 doi: 10.1016/S0896-6273(00)80898-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression values for all RNA-seq data generated in the study, presented as fragments per kilobase of transcript per million mapped reads (FPKM), for all the glial subpopulations in each replicate.
List of genes enriched in Olig2-AS that present a log2FC > 1 and padj<0.05 in both DA1 and DA2.
List of genes depleted in Olig2-AS that present a log2FC < −1 and padj<0.05 in both DA1 and DA2.
Gene Ontology analysis (BP) from GO summaries on genes enriched in Olig2-AS (log2FC > 1 and padj<0.05 both in DA1 and in DA2) and in genes depleted in Olig2-AS (log2FC < −1 and padj<0.05 both in DA1 and in DA2). The table indicates the list of GO terms enriched, the list of genes involved in a given GO term, and the p value. Only terms with p < 0.05 are shown.
Data Availability Statement
RNA-seq data and processed files generated during this study are available through Gene Expression Omnibus (GEO) database. Accession number is GSE158517.
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE158517
No unpublished custom code, software or algorithm was used in this study. Freely available software and algorithms used for analysis are listed in the key resources table. All custom scripts and data contained in this manuscript are available upon request from the Lead Contact.
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.