Abstract
Strategies to counteract or prevent emerging drug resistance are crucial for the design of next-generation antimalarials. In the past, resistant parasites were generally identified following treatment failures in patients and compounds would have to be abandoned late in development. An early understanding of how candidate therapeutics lose efficacy as parasites evolve resistance is important to facilitate drug design and improve resistance detection and monitoring up to the post-registration phase. We describe a new strategy to assess resistance to antimalarial compounds as early as possible in preclinical development by leveraging tools to define the Plasmodium falciparum resistome, predict potential resistance risks of clinical failure for candidate therapeutics, and inform decisions to guide antimalarial drug development.
Keywords: Malaria, Plasmodium falciparum, resistance, drug discovery
The Urgent Need to Predict and Manage Risks of Parasite Resistance to Antimalarials
Like many other antimicrobials, great successes with antimalarials have often been their downfall: their extensive and at times suboptimal use has favored the evolution and dissemination of drug-resistant P. falciparum parasites. Once again, multidrug resistance threatens malaria control and elimination [1]. Medicines for Malaria Venture (MMV) i was established in 1999 specifically to redress the diminishing number of reliable treatments caused by the emergence and global spread of parasites resistant to first-line therapies. By 2003, several artemisinin (ART)-based combination therapies (ACTs, see Glossary) were introduced [2]. ACTs combine a short-acting artemisinin derivative that rapidly reduces parasite numbers in patients with a longer-acting partner drug that eliminates remaining parasites via a different mode of action. The widespread adoption of ACTs has been an important contributor to the nearly 60% reduction in malaria mortality and morbidity rates over the subsequent 12 years [1]. However, ART resistance has emerged, manifesting as slower rates of parasite clearance in treated patients. Mutations that reduce the efficacy of partner drugs have also now emerged and have severely impacted the clinical efficacy of ACTs against P. falciparum parasite populations in Southeast Asia [3]. Some of these mutations have recently been reported in East Africa [4,5], Papua New Guinea [6] and French Guiana [7] yet there is limited evidence of slow parasite clearance after ACT treatment in this region [8–13]. Were ACT resistance to spread in Africa, the impact could be devastating [14,15]. Fortunately, ACTs remain currently effective at curing patients in this continent.
New antimalarials that kill multidrug-resistant (MDR) P. falciparum are a high priority [16] and their discovery and development form a core mission for MMV ii. The expectation is that compounds with novel modes of action, and high barriers against the emergence of resistance, must constitute the next generation of antimalarial combinations. Many of these compounds have overcome important drug development hurdles and are aligned with established target candidate and product profiles [17]. However, for most of their targets, resistance has been identified as a concern based on in vitro selections with cultured asexual blood stage (ABS) parasites and their characterization [18] (Table 1). In vitro and ex vivo studies focusing on P. falciparum ABS parasites have helped assess the risk of resistance to new antimalarials, although, until recently the information was often obtained only late in development.
Table 1.
Historical and current MMV portfolio of clinical antimalarial projects and their in vitro resistance-associated parameters.
| Molecular target | Project/Compound | Stage in portfolio | Targeted parasite stage | Resistance associated characteristics | ||||
|---|---|---|---|---|---|---|---|---|
| Resistance gene(s) | EC50 fold Pressure | MIR | EC50 fold shift | Ref. | ||||
| PfATP4 | SJ733 | Patient exploratory - halted (Eisai/St Jude/Kentucky) | ABS, transmission | pfatp4 (Pf3D7_1211900) | 5 | 9 | 3 – 7 | [70] |
| Cipargamin (KAE609) | Patient exploratory | ABS, transmission | pfatp4 (Pf3D7_1211900) | 3 | 7 | 7 – 10 | [71] | |
| PfDHODH | DSM265 | Patient exploratory - halted | ABS, prophylaxis | pfdhodh (Pf3D7_0603300) | 3 | 5.5 | 3.5 | [26] |
| PfDHFR | P218 | Human volunteers | ABS, transmission, prophylaxis | pfdhfr (PF3D7_041720) | 3 | 8 | 3 | N/A |
| PfPi4K | MMV390048 | Patient exploratory - halted | ABS, transmission, prophylaxis | pfpi4k (Pf3D7_0509800) | 3 | 6 | 3 – 7 | [57] |
| PfEF2 | M5717 (DDD107498) | Human volunteers | ABS, transmission, prophylaxis | pfeef2 (Pf3D7_1451100) | 5 | 6 | 7 –5500 | [41] |
| Pf Cyt-bc1 | Atovaquone | Approved | Prophylaxis | Pfcytb (mal_mito_3) | 3 | 7 | 10 – 60 | N/A |
| PfACoAS | Pantothenate series | Preclinical development | ABS, transmission | pfAcCoaS (Pf3D7_0627800) | 3 | 9 | 15 | [72] |
| Unknown | Ganaplacide (KAF156) | Patient exploratory | ABS, transmission, prophylaxis | pfcarl (Pf3D7_0321900) pfugt (Pf3D7_1113300) pfact (Pf3D7_1036800) | 3 | 8 | 10 | [56] |
MIR: in vitro logarithmic value of the minimum inoculum for resistance obtained in Pf Dd2 with a drug pressure corresponding to a designated multiple of the EC50 (or more recently EC90); DHODH, dihydroorotate dehydrogenase; DHFR, dihydrofolate reductase; PI4K, phosphatidylinositol-4-kinase; EF2, elongation factor 2; AcCoAS, acetyl-CoA synthetase; N/A: non-available. The current portfolio is listed in Resources ii, on 20.12.2020. Some compounds displaying log10MIR3xEC50 > 9 in the MMV portfolio are not described here because the data are unpublished to date.
Here we propose a strategy to expedite the early prediction of resistance for compounds designed to treat blood-stage malaria. The plan is based on research insights, lessons learned over the past decade, and consultation with experts. Current experimental approaches are discussed, together with future directions and key knowledge gaps that need to be addressed to identify compounds with potential risks of rapidly selecting for clinical resistance.
Complexity of antimalarial resistance
The emergence of resistance to antimalarials is the result of a complex interplay between the drug administered to infected patients and the exposure of the parasite as it transitions through its life cycle in the human host and the mosquito vector (Figure 1A). Parasites are far more numerous in the blood of the patient than during transmission to or from mosquitoes (Figure 1B). Single point mutations, some able to confer resistance, occur randomly all the time, at a frequency of ~1 to 5 per 109 ABS parasites [19,20]. Amplification of drug and solute efflux transporter genes such as pfmdr1, which can confer resistance across a variety of antimalarial chemotypes, can occur at a frequency close to 1 per 107 parasites. However, such a resistance mechanism only leads to a few-fold decrease in drug potency. During a human blood stage infection, the parasite count can reach 1012, providing opportunities for a few resistant parasites to survive drug exposure and continue their life cycle, if transmitted. In contrast, transmission from an infected human to mosquitoes requires a very small number of gametocytes. These gametocytes are ~100-fold fewer in number than ABS parasites and thus face lower resistance selection pressure [18]. Defining the genetic barrier to resistance, i.e. the potential for a compound to select for resistance [21] at the earliest possible stage in drug development is a key step in evaluating the risk that resistant parasites may be selected rapidly in clinical use.
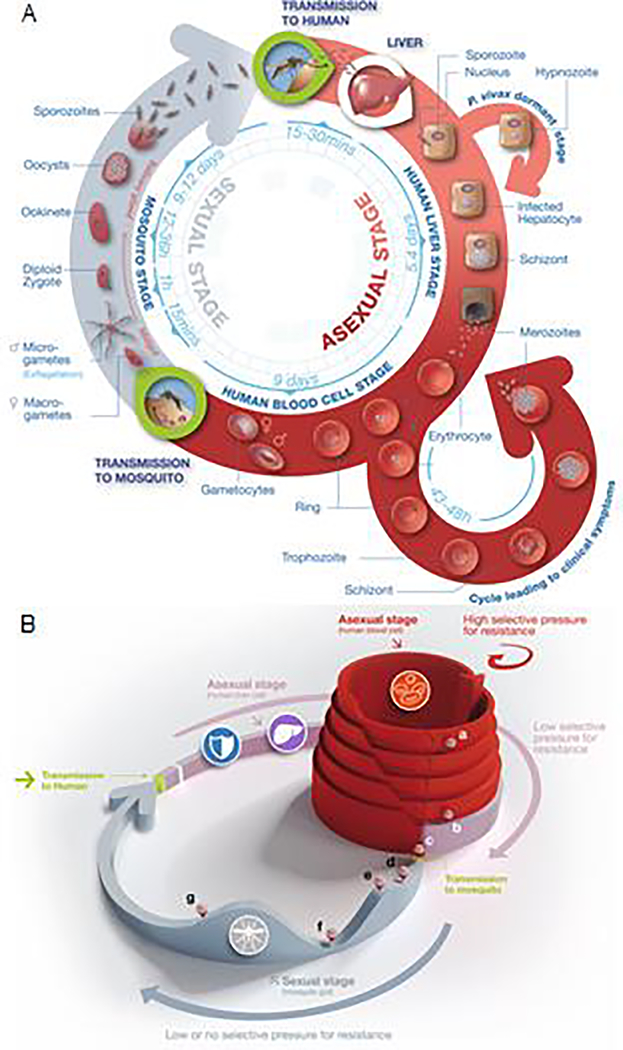
Figure 1. Life cycle of malaria parasites.
(A) Approximatively 20 minutes after human infection by Plasmodium sporozoites (top right), these forms reach the liver, infect hepatocytes and develop into liver schizonts over a ~5–7 day period, before bursting and liberating thousands of merozoites into the blood. Merozoites infect erythrocytes and develop successively into rings, trophozoites and schizonts that liberate merozoites (bottom right). At each asexual blood stage cycle, ~1% of trophozoites commit to producing sexual stage male and female gametocytes. Gametocytes differentiate in five successive stages, the first four being sequestered and the final one being liberated into the bloodstream. This stage V gametocyte can then be ingested by a mosquito during a blood meal. Once in the mosquito midgut, male and female gametocytes transform into gametes before mating and forming a diploid zygote. This zygote transforms into a motile ookinete that escapes the midgut contents and forms an oocyst under the basal lamina, wherein meiosis occurs leading the production of haploid sporozoites. Rupture of the mature oocyst liberates thousands of sporozoites (top left) that migrate to the mosquito salivary glands, ready to be injected into a human host during the next mosquito blood meal. Modified from Delves et al. [73]. (B) The three-dimensional aspect of this figure highlights differences in the parasite population size throughout the stages of the life cycle. At the liver stage fewer than 100 sporozoites reach the liver and only a few will successfully produce liver schizonts inside hepatocytes (b). At this stage the pressure of selection for mutated parasites leading to drug resistance is very low. Once the erythrocytic stage reaches its maximum after several proliferative cycles (a), the population of these asexual blood stage parasites can reach 1012 in a patient. At this stage, the patient can develop severe malaria. This high number of parasites creates a high selective pressure for the emergence of resistance. Following differentiation into the non-proliferating gametocytes (c), the subsequent less abundant forms, i.e. micro- and macro-gametes (d), ookinetes (e), oocysts (f) and sporozoites (g), do not enable a high risk for the emergence of resistance. Modified from Leroy et al. [43].
Assessing antimalarial resistance
There is no validated method to precisely predict the frequency, genotype and phenotype of clinical resistance to a particular compound during preclinical development, i.e. before the compound has been tested in humans. Currently, this prediction largely relies on the ability of a drug candidate to kill MDR P. falciparum parasites in the laboratory in parallel with in vitro resistance selection studies to identify causal de novo mutations and/or gene amplifications [17,22]. Standardized in vitro studies using a range of parasite inocula (105 to 109) under drug pressure allow the determination of the minimum inoculum for resistance (MIR). This value can be used as a surrogate to estimate the potential for resistance to develop against a given candidate compound. The choice of drug concentration used in these selections directly impacts both the MIR and the genotype and phenotype of resistant parasites. Therefore, it is important to perform several of these in vitro studies in parallel using a range of compound concentrations.
Recently, safe protocols for clinical trials in healthy human volunteers have been pioneered (volunteer infection studies, VIS). Healthy volunteers are infected with a known number of parasites (typically Good Manufacturing Process-produced P. falciparum 3D7) and are monitored in a hospital setting to document compound safety and efficacy [23,24]. For these early-stage phase Ia and IIa trials, blood samples are collected and, in cases the agent does not completely clear all parasites, the patients are cured with a different drug. Blood samples collected before initiating this rescue treatment contain the surviving parasites, whose genotypes and phenotypes are examined to identify the reasons for incomplete parasite clearance. These VIS provide a catalyst for drug development and complement more traditional human clinical trials. In instances where these parasites carry a mutation in a particular parasite gene and exhibit lower susceptibility in vitro to the test agent, this information can be compared with data from earlier in vitro resistance selection studies.
Two examples illustrate the utility of this two-pronged strategy. First, in the Phase IIa clinical study of the DHODH inhibitor DSM265, persistent parasites were observed in two Peruvian patients ~25 days after the initial treatment. Resistance to DSM265 was confirmed in both cases, based on their genotype: pfdhodh G181S and C276Y substitutions were detected in parasites from both patients [25]; and their phenotype: a >10-fold decrease in drug susceptibility was observed when mutant parasites were assayed in vitro. Earlier in vitro studies had suggested that there might be an elevated risk of resistance with this drug, as the MIR at a 3×EC50 selection concentration averaged 2×106 for DSM265 across several parasite strains [26]. Second, at the opposite extreme, the ongoing VIS trials of MMV253/ZY yielded no recrudescent parasites displaying altered genotypes or phenotypes, consistent with the inability to select for resistance to MMV253/ZY in vitro in MIR studies. These congruent conclusions of in vitro and human volunteer studies suggest that parasites resistant to DSM265 would likely be selected quickly in the field, but that parasites resistant to MMV253/ZY would be a rare occurrence. These two approaches can therefore be useful early indicators of the risk of resistance to new antimalarials.
The current consensus is that if drug-resistant parasites are observed in vitro, they will inexorably appear in the clinic: sooner if compound is used as monotherapy, or later if administered in combination – with the longevity dependent on the choice of partner(s). Given this consensus and historical and recent clinical findings, it is unwise to ignore MIR and related preclinical data. On the other hand, it is counterproductive to impose criteria so stringent that only candidates with neither observable cross resistance nor a resistance marker (so-called ‘irresistibles’) are progressed [27]. Instead, it is both important to down-prioritize those with the greatest intrinsic risk, and to accept those other candidates along with a well-characterized resistance risk assessment that facilitates further combination decision making. In summary, compounds generating resistance in vitro can still play a critical role in treatment as part of a well-chosen combination regimen [28]. One scenario would be to combine a compound with a defined resistance risk with an ‘irresistible’ to mitigate the risk.
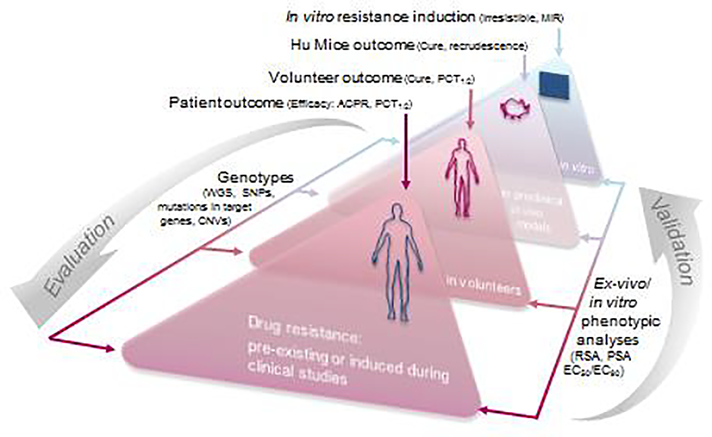
For a new antimalarial a “resistance triangle” can be constructed that connects clinical efficacy outcome, in vitro phenotype of a recrudescent parasite population (i.e. EC50), and genotype (Figure 2). Indeed, it is crucial that for each new clinical study, both genotypic and phenotypic (ex vivo / in vitro) analyses are conducted systematically on the baseline parasite population prior to drug administration as well as on recrudescent parasites post drug administration. This integrated approach is important to assess key factors that may influence clinical efficacy, such as recrudescence and cure rates linked to Adequate Clinical and Parasitological Response (ACPR), parasite clearance half-life, baseline parasitemia, and drug concentration. This resistance triangle approach can be applied at all stages of the drug discovery pipeline, including preclinical in vivo and in vitro resistance selection studies (in the latter case, the MIR would represent a proxy for “clinical outcome”, as defined above). This allows an iterative refinement of the methodology to assess the risk of drug resistance during early development and how this should be managed. Decision making on drug candidates and combination choices can continuously improve, as new clinical data become available.
Figure 2. Resistance risk validation and evaluation, an iterative process between the bedside and the bench.
Learning from the clinical observations collected over the past two decades, our proposed new strategy for the evaluation of resistance risks relies on the “resistance triangle”. First, clinical outcomes, phenotypic studies and genotypic analyses are compiled from P. falciparum-infected patients and volunteers, through in vivo models, down to in vitro experiments. Resistance is declared when parasites from clinical recrudescences harbor validated resistant genetic markers and exhibit decreased in vitro sensitivity to the drug/compound. ACPR: adequate clinical and parasitological response; PCT: parasite clearance time; MIR: minimum inoculum for resistance; WGS: whole-genome sequencing; SNP: single nucleotide polymorphism; CNV: copy number variation; RSA: ring-survival assay, PSA: piperaquine survival assay.
Assessing drug resistance in discovery
A key pillar of MMV’s resistance strategy relies on assessing the ability of new compounds to select for resistance in ABS parasites. This has successfully led to the identification of drug resistance mechanisms related to specific parasite genes and mutations [29,30] and the analysis of the frequency of resistance selection by compounds in the discovery pipeline. Technological advances make it possible to study resistance determinants using whole-genome sequencing (WGS), metabolomics [30–32], proteomics [33] or other biophysical approaches such as cellular thermal shift assays (CETSA) [34] or thermal proteome profiling (TPP) [27]. Through collaboration with the Malaria Drug Accelerator (MalDA; funded by the Bill & Melinda Gates Foundation) these multiple technologies are applied in a concerted effort to identify novel mechanisms of resistance and characterize new antimalarial drug targets [35].
This resistance strategy has been recently revised to 1) establish clear selection criteria to down-prioritize early chemical series with high risks of resistance, and certainly by the Late-Lead stage; and 2) assess the resistance risk for preclinical candidates and thus the extent to which mitigation strategies are required. This two-step process requires three main workstreams to be deployed in parallel. First, the potential of portfolio compounds to generate resistant parasites is assessed in vitro. Second, early potential for cross-resistance between new compounds and known antimalarials as well as related common resistance mechanisms is evaluated using a standardized panel of MDR strains. Finally, the activity of portfolio compounds against Plasmodium parasites in circulation is verified by testing lead compounds against contemporary field isolates from Africa, Southeast Asia and South America.
In vitro selection of resistant parasites
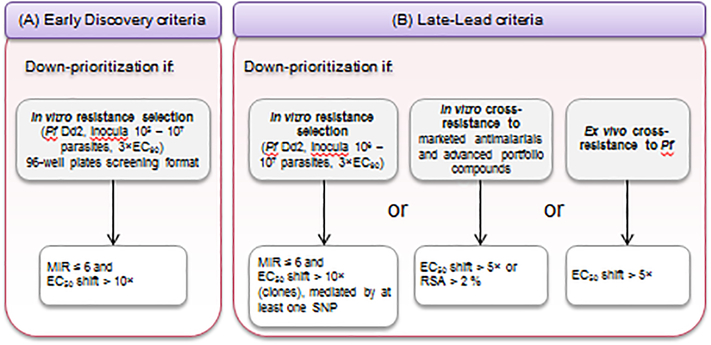
Until recently, our standard in vitro resistance selection method consisted of applying a constant drug pressure (typically 3 to 5×EC50) for up to 60 days [36]. Now, a selection pressure of 3×EC90 is applied to 106 parasites of the Asian MDR P. falciparum Dd2 strain (B2 clone). Ongoing studies show that this condition can generate higher MIRs and select for higher levels of resistance compared to the earlier EC50 conditions (Figure 3A and 3B). The choice of 3×EC90 also helps standardize conditions between compounds that either act rapidly or show a delayed onset of parasiticidal action. Dd2-B2 is preferred as this is a rapidly proliferating and extensively characterized parasite line used regularly for selection studies [18]. Early lead compounds with logMIRDd2, 3×EC90 ≤ 6 (i.e. ≤106 parasites) and EC50 shifts >10-fold will be down-prioritized (Figure 3A). These values are generally observed when resistance is mediated by point mutations in the target gene rather than gene amplification. Then, the inoculum range is refined, and in vitro cross resistance studies are performed with currently marketed antimalarials via EC50 determination and recrudescence analyses in the Ring Stage Assay (RSA) when relevant. Late leads will not be progressed further if one of the three following conditions is met: 1) a MIR ≤ 6 and an EC50 shift > 10-fold; 2) cross resistance with existing antimalarials is observed with either an EC50 shift > 5-fold compared with the sensitive parental line or parasite survival > 2% in the RSA; or 3) a median EC50 increase > 5-fold is observed in ex vivo P. falciparum field isolates compared to laboratory strains, when tested on the same platform in the same assay (Figure 3B). Resistant clones resulting from the MIR selections are also submitted for WGS or selected target gene sequencing. Gene editing with CRISPR/Cas9 technology or transgene expression can then be used to demonstrate that the mutated candidate gene is causal for resistance [37–41].
Figure 3. Multicriteria assessment of the risk of resistance and mitigation strategy.
Resistance is evaluated according to the stage of advancement in the pipeline of the compound/series. Early series (A) and Late-Lead compounds (B) are down-prioritized in the event of a MIR value ≤ 6 (i.e. resistance obtained in ≤ 106 parasites), and an EC50 fold shift > 10, generated in vitro in P. falciparum Dd2 parasites subjected to a selection pressure of 3×EC90. Additionally, Late-Leads showing a cross-resistance greater than a 5-fold EC50 shift compared with marketed antimalarials or advanced MMV portfolio compounds, or a median EC50 with P. falciparum isolates 5-fold higher compared with values obtained with sensitive laboratory strains, will be down-prioritized.
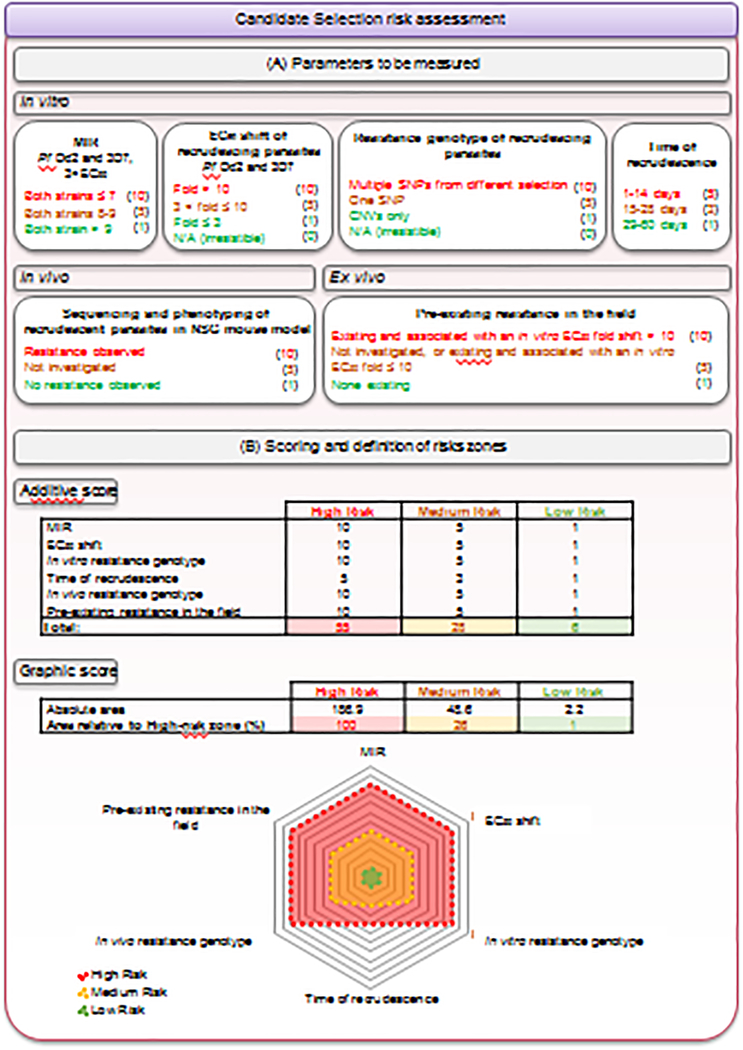
At the candidate selection stage (Figure 4A), in vitro selections are performed on Dd2 parasites with low (3×EC50) and higher (3×EC90) selection concentrations, at parasite inocula up to 109. These studies are useful to gauge the concentration-MIR correlation. In vitro selections are also performed on the 3D7 strain with 3×EC90 to compared results with Dd2 selections. These results also help translation to the in vivo NSG mouse model [42] and Phase Ib VIS studies, which both use adapted 3D7 parasites. Resistant clones are phenotyped, and given a statistically significant EC50 or EC90 shift, also genotyped. Pre-existing resistance in the field is evaluated and the time of recrudescence of each in vitro selection recorded as it could inform on how rapidly resistance may develop in patients [22]. Finally, the DNA sequence and phenotype of recrudescent parasites from the highest dose used in in vivo studies in NSG mice infected with P. falciparum parasites is analyzed.
Figure 4. Quantitative assessment of the risk of resistance.
(A) The risk of resistance is assessed for each compound by integrating various in vitro (log10MIR, EC50 fold shift, resistance genotype and time of recrudescence in resistance selection experiments on at least two P. falciparum strains), in vivo (resistance-conferring mutation(s) in resistant recrudescing parasites from humanized mice efficacy studies) and ex-vivo (pre-existing resistance in the field) parameters. Each parameter is assigned a score corresponding to a high (red), medium (yellow) or low (green) risk, as depicted in parentheses. The weighting of the parameters varies according to their relevance. For instance, the day of recrudescence during in vitro selections was attributed a lower weight, to acknowledge the fact that whether this event occurs or not is more important than the timing. In contrast with the in vitro resistance genotype, where a clear distinction between SNPs and CNVs is made, both alterations are considered equally important in the in vivo studies. Indeed, while CNVs are more frequent in vitro, usually leading to small EC50 shifts (i.e., < 5-fold), they can remain clinically relevant as evidenced with pfmdr1 copy number and mefloquine. Some data, particularly from in vivo studies, and the pre-existence of resistance conferring-mutations in the field, are not always available. To acknowledge the risk of such a knowledge gap and encourage teams to obtain these data, a medium score of 5 is attributed to the “not investigated” category. Publicly available information on genome sequence diversity is constantly increasing. While a thorough investigation at a given time point may reveal no concerns, updated information should be regularly considered for a given compound even after candidate declaration and during clinical development. Of note, some criteria, such as the EC50 fold shift, the time of recrudescence, or the Log10MIR value, may spread over two categories. In this case, we select the median score between the two categories. (B) Two complementary approaches are then used to classify compounds into a global risk zone: a numerical approach based on an additive score and a graphic assessment based on the relative area of a spider chart overlapping with the total high-risk area. A compound with a cumulative score (method A) up to 6 is classified in the low-risk zone, between 7 and 28 is in the medium-risk zone, and 29 or above is in the high-risk zone. A compound with an area (method B) covering up to 1% of the high-risk area is classified in the low-risk zone, between 1 and 26% is in the medium-risk zone, and above 26% is in the high-risk zone. As shown in Supplementary Figure 1, both approaches lead to the same outcome.
Thus, rather than applying fixed resistance criteria, at the candidate selection step a detailed resistance risk assessment profile is delivered for each new preclinical candidate (Figure 4B and Supplementary Figure S1) and is used to guide further development and combination strategies.
Cross-resistance studies
Novel molecules intended for development must not be cross-resistant with existing therapies (EC50 shift < 5-fold). This is evaluated by measuring the potency of compounds against a panel of sensitive and MDR P. falciparum strains harboring common alleles conferring resistance to licensed antimalarials [43]. Mechanisms of resistance to established antimalarials have been extensively reviewed [18,44,45]. In brief summary, these can be due to: 1) mutations in target genes (e.g. pfcytb or pfdhfr/pfdhps) [46,47]; 2) mutations in or amplification of drug transporter genes (including pfcrt and pfmdr1) [48]; 3) mutations that reduce proteotoxic stress (Pfkelch13) [49]; or 4) other recently revealed polygenic mechanisms such as resistance to piperaquine that is associated with specific mutations in pfcrt accompanied by duplication of the tandem pfpm 2 and pfpm3 genes [50–53].
Hits are first declared after demonstrating unaltered potency against a panel of MDR P. falciparum strains that represent the most commonly mutated alleles of pfdhfr, pfdhps, pfcrt and pfmdr1, as well as amplified pfpm2 that is detected in malaria-endemic areas [54]. This panel contains the strains 7G8 from South America, and K1, Dd2, TM90C2B [55] and PH1263-C [52] from Southeast Asia that collectively represent resistance to antimalarials including chloroquine, pyrimethamine, atovaquone, ART, and piperaquine (Supplementary Table S1). To evaluate potential cross-resistance of compounds in the hit-to-lead phase with compounds in development, we employ a set of engineered P. falciparum mutated lines resistant to KAF156 [56], MMV048 [57], M5717 (also known as DDD107498) [41], DSM265 [25], or ELQ300 [58] (Supplementary Table S2). Of note, pfcarl is likely to be a mediator of pleiotropic drug resistance [40,59]. Cross-resistance studies with ART and more broadly with any endoperoxide is performed using the RSA [60].
Ex vivo drug susceptibility testing of late leads on Plasmodium field isolates
Beyond P. falciparum laboratory strains, it is critical to demonstrate the efficacy of lead compounds against contemporary clinical isolates of diverse geographic origins (EC50 shift < 5-fold). Ideally these isolates should be genotyped against a panel of known drug targets and drug resistance markers. In this context, MMV has recently emphasized testing compounds more widely on field parasites including P. falciparum in Uganda (and soon also in Burkina Faso), Brazil and Cambodia; P. ovale in Mali, P. malariae in Ghana, and P. vivax in Cambodia and Brazil, as well as isolates resistant to both ART and piperaquine in Southeast Asia. Among these, only P. falciparum is amenable to long-term ABS in vitro culture and therefore demonstration of drug susceptibility relies exclusively on the ex vivo testing of clinical isolates [17,61,62].
Analysis of genetic determinants of resistance in sequenced parasite genomes
Genomic databases such as those curated at the Wellcome Sanger Institute, the Broad Institute and Oxford University’s Pf3K iii can be used to search for pre-existing resistance mutations or in some instances gene amplifications identified from in vitro selection studies or present in clinical isolates of geographically diverse origins [63,64]. This analysis is performed as soon as a set of resistance mutations/gene amplifications is associated to a new compound. Additionally, the nucleotide diversity of genes should be examined to expand our understanding of the mutational space for a given compound [63]. The prevalence of target gene variants should ideally be associated with ex vivo EC50 values, although, unfortunately, corresponding parasites are rarely available for phenotypic analyses. Alternatively, the impact of gene target polymorphisms on compound EC50 values should be established with isogenic parasites engineered using gene editing approaches. While it is difficult to define a threshold of pre-existing resistance above which a drug candidate should be disqualified for further development, an understanding of resistant allele frequencies across geographical regions can benefit drug resistance mitigation and monitoring strategies.
Concluding remarks
The evolution, transmission and fixation of antimalarial resistance is driven by the intricate interplay of several factors, including the rate of mutation or gene amplification, the strength of the selective pressure, the level of resistance, and the fitness cost imposed by point mutations or amplification of a resistance determinant. Mutations in other genes can compensate for initial fitness defects while having a low or neutral effect on resistance levels [65]. Clonal interference [66] and the environmental context are additional factors that complicate the prediction of resistance development (see Outstanding Questions and Box 1).
Outstanding questions.
How to develop the next generation of antimalarial drugs with a high barrier against resistance?
What additional factors should be considered to refine the prediction of resistance development in the field?
How to strengthen the predictive aspect of the preclinical workflow to better assess the risk of resistance in patients?
How could the malaria humanized mouse model be used to increase the translational quality of the assessment of resistance risks?
Would the selection criteria described here apply to the discovery of new compounds for chemoprotection?
Is resistance a feature of the molecule or the target or both? Should a deprioritized compound condemn its target?
Box 1. Current gaps and limitations.
Transmission potential of resistant mutants. The transmissibility of resistant parasite clones can be evaluated by assays supporting the TCP5 strategy [17] (i.e. gametocyte assays, DGFA, and SMFA) that measure the viability and transmissibility of mutant versus wild-type parasites in the absence or presence of antimalarial drugs or candidates. The major limitation remains the availability of mutants derived from a parasite strain that is proficient for transmission. Indeed, most resistant lines derived from the Dd2 or 3D7 strains cannot produce gametocytes and infect mosquitoes and therefore are of little use for studying the impact of resistance-conferring mutations on transmission. These mutations can be engineered in a transmissible strain like NF54, with the limitation that this strain can readily lose its ability to produce mature infectious gametocytes in vitro [68]. Nonetheless, the value of this approach was demonstrated [69] in atovaquone-resistant parasites that failed to transmit. If resistant stage V gametocytes cannot transmit, then the resistance risk is dramatically reduced.
Evaluation of fitness-compensatory mutations. Fitness experiments with resistant versus sensitive parasites require multiple growth cycles in media, during which compensatory changes may be selected. WGS of competing strains should be performed to distinguish between compensatory mutations associated with a fitness loss resulting from a resistance-conferring mutation and losses that may arise during in vitro culture. This analysis requires a robust genome analytics pipeline and expertise. This approach is limited by the impossibility to reconstitute in vitro the parasite’s genomic diversity observed in the field, and therefore the in vitro assessment of fitness adaptation can only be interpreted as a surrogate for the field situation. Currently, fitness is not a considered criterion in the MMV progression pipeline.
Translation of the in vitro MIR into the field. What does a specific MIR portend for the future probability of resistance development in the clinic? Ideally, a MIRDd2, 3×EC90 >12 should be sought because ABS P. falciparum parasites typically reach 1010−1012 in a symptomatic individual. However, technical restrictions limit the number of ABS parasites that can be routinely cultured in a flask to 1–2×109, thus giving only an incomplete view of the resistance frequency. Today, based on the observations made on multiple preclinical antimalarials, a MIR equal to or greater than 9 seems to assure a sufficiently high barrier to resistance. Antimalarials with a MIR of 7 to 9 despite having higher resistance risks could play important roles in combination treatments but the resistance risk will need to be managed.
New insights about mechanisms of resistance have rapidly emerged over the past decade and can now be used to guide antimalarial discovery and development. A more integrated approach to the assessment of evolution of resistance at all phases of drug development should enable improved clinical predictions, help design studies, and inform decision-making throughout the drug development process. In particular, understanding the propensity for resistance selection in vitro is increasingly important to assess risks in progression and development of new hits and leads [18]. Interestingly, examples of different compounds sharing the same target yet showing different resistance profiles have started to accumulate and support the idea of not systematically de-emphasizing a particular target or pathway when a related compound has been deprioritized. Investigating whether resistance is implicated in parasite recrudescence in human clinical trials is key for future development efforts. Such an approach focuses on 1) whether there are resistance-associated mutations in the genomes of parasites isolated from patients prior to or after drug dosing; 2) the ex vivo potency (EC50) shift of the drugs (ideally on baseline and recrudescent parasites) used to treat patients; and 3) clinical outcomes. At the translational research level, it is expected that generating resistance by applying sub-optimal drug pressure to P. falciparum parasites infecting humanized NSG mice will help to explore the entire range of possible resistance mechanisms that could ultimately affect drug efficacy in humans. This approach is currently under investigation, both regarding resistance to PfDHODH and ART inhibitors [67]. Moreover, adapting field parasites to infect humanized mice could allow preclinical candidates to be tested under conditions that better reflect parasites currently in circulation.
Over time, it is expected that an analysis of clinical outcomes and associated data will contribute to an improved prediction of the observed resistance risk in patients compared to the risk defined on the basis of in vitro resistance selection experiments. Given the current need to protect endemic populations against post treatment reinfections, a similar reflection will be needed on resistance selection criteria to apply to candidate chemoprotective compounds currently in the discovery phase. Although the parasite burden in the liver of infected people remains several orders of magnitude lower than the symptomatic one in blood, potential post-treatment prophylaxis approach targeting blood stage parasites could well necessitate criteria not so different to those described above. At the clinical level, we continue to scrutinize for evidence of resistance emerging to current antimalarial therapies (artemether-lumefantrine, dihydroartemisinin-piperaquine and pyronaridine-artesunate), especially in Africa. The need to optimize the antimalarial pipeline through the early identification of resistance liabilities is reinforced by the plateau in morbidity and mortality rates observed since 2015 globally, and the increasing prevalence of parasites that carry known markers of resistance. Developing the next generation of antimalarial drug combinations that have longevity to withstand selective pressures is one of the key priorities in global infectious diseases, and one that the malaria research and clinical community has eagerly embraced.
Supplementary Material
Highlights.
Artemisinin-based combination therapies are first-line treatments for uncomplicated Plasmodium falciparum malaria. However, during the last decade partial resistance to artemisinins, mediated by PfK13 mutations, has spread across Southeast Asia and emerged in South America, the Western Pacific and Africa. The emergence of resistance to partner drugs has led to frequent treatment failures in Southeast Asia.
The development of new antimalarials with a high barrier to resistance is an urgent and critical need and should be guided by a standardized and streamlined assessment of potential resistance liabilities.
We propose a novel strategy to assess drug resistance risks early in the development pipeline and to help prioritize novel antimalarials with a low potential for resistance. We also review approaches to gain a greater understanding of resistance risks and guide future combination decisions.
Acknowledgments
We thank Drs. Dyann F. Wirth, Philip J. Rosenthal, Daniel E. Goldberg, Rick Fairhurst, and Stephen Ward for expert discussions and Dr. Carol Sibley for expert discussions and editing. We are grateful to our colleagues Drs Stephan Duparc, Fiona Macintyre and Farouk Chughlay for their feedback. We thank Drs. Sergio Wittlin and Christian Scheurer for reference cross-resistance data. We recognize the graceful contribution of Pierre Chassany (Comstone) for the design of Figure 1. D.A.F. gratefully acknowledges funding support from MMV, the Department of Defense (W81XWH-15-2-0033 and W81XWH-19-1-0086), the Bill & Melinda Gates Foundation (OPP1201387) and the NIH (R01 AI109023, R37 AI50234, R01 124678).
Glossary
- Artemisinin-based combination therapy (ACT)
recommended first line of treatment for P. falciparum uncomplicated malaria. A fast acting, short lived, artemisinin derivative, such as dihydroartemisinin, artesunate or artemether, is combined with a longer lasting drug from a different chemical/mechanistic class. Partner drugs include lumefantrine, mefloquine, amodiaquine, sulfadoxine/pyrimethamine, piperaquine or chlorproguanil/dapsone. Such a combination promotes high efficacy, fast action and a reduced likelihood of resistance development.
- Clonal interference
competition for survival between different asexually reproducing clones. As multiples clones interfere with each other in an asexually replicating population, a beneficial mutation, likely to be fixed if occurring alone, may fail to do so, or even be lost if other beneficial mutations arise in clones of the same population.
- Copy number variation (CNV)
genomic structural variation resulting in a difference in the number of copies of a specific gene between parasites.
- Dual gamete formation assay (DGFA)
in vitro assay that identifies transmission-blocking compounds that affect the functional viability of the P. falciparum mature male and female stage V gametocytes and inhibit the process of gametogenesis.
- EC50
Effective concentration of a compound that inhibits the growth of P. falciparum ABS parasites by 50%. This value, often documented as IC50, is obtained from concentration-response growth assays conducted with parasites cultured in vitro and exposed to the compound for 72 hr or 48 hr.
- Late-Lead
a chemical compound with biological activity and pharmacological properties likely to be suitable for therapeutic applications but that can still require optimization.
- Minimum inoculum of resistance (MIR)
The minimum number of cultured P. falciparum ABS parasites required to yield resistance following exposure to a defined drug concentration (currently 3×EC90). The MIR is preferably obtained early in the antimalarial drug discovery process. The MIR is a function of the selection pressure and is reported as a log10 of the minimum parasite inoculum at which resistance can be selected (e.g. MIR=6 implies 106 parasites are required at a minimum to obtain resistance to the selection concentration).
- Multidrug-resistant (MDR) Plasmodium parasite
a parasite strain showing resistance to several categories of antimalarial compounds, i.e., with distinct modes of actions.
- NOD SCID gamma (NSG) mouse
a highly immunodeficient laboratory mouse strain, lacking mature T cells, B cells and Natural Killer cells. This model allows engraftment with human erythrocytes to study P. falciparum infection and drug efficacy in vivo.
- Single nucleotide polymorphism (SNP)
a DNA sequence variation occurring when a single nucleotide in the genome differs between members of a species or paired chromosomes in an individual. Drug resistance in P. falciparum is often a result of one or more non-synonymous SNPs that cause amino acid substitutions.
- Standard membrane feeding assay (SMFA)
the current gold standard test for transmission-blocking compounds. Classically, stage V gametocytes are fed to female Anopheles mosquitoes in the absence or presence of variable concentrations of an experimental compound. The capacity to block transmission is assessed by counting the number of oocysts present in the mosquito midgut 7 to 10 days later.
- Target candidate profile (TCP)
a description of the ideal properties of molecules as candidate antimalarial medicines, defined by MMV to guide early antimalarial drug discovery.
Footnotes
Declaration of interests
The authors Drs Maëlle Duffey, Jeremy N. Burrows, Timothy N.C. Wells and Didier Leroy are employees of Medicines for Malaria Venture (Geneva).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.World Health Organization (2016) Global technical strategy for malaria 2016–2030, World Health Organization. [Google Scholar]
- 2.Bennett A et al. (2017) Population coverage of artemisinin-based combination treatment in children younger than 5 years with fever and Plasmodium falciparum infection in Africa, 2003–2015: a modelling study using data from national surveys. The Lancet Global Health 5, e418–e427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marapana D and Cowman AF (2020) Uncovering the ART of antimalarial resistance. Science 367, 22–23 [DOI] [PubMed] [Google Scholar]
- 4.Uwimana A et al. (2020) Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nature Medicine 26, 1602–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asua V et al. (2021) Changing prevalence of potential mediators of aminoquinoline, antifolate, and artemisinin resistance across Uganda. The Journal of Infectious Diseases 223, 985–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miotto O et al. (2020) Emergence of artemisinin-resistant Plasmodium falciparum with kelch13 C580Y mutations on the island of New Guinea. PLoS Pathogens 16, e1009133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathieu LC et al. (2020) Local emergence in amazonia of Plasmodium falciparum K13 C580Y mutants associated with in vitro artemisinin resistance. eLife 9, 1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dokomajilar C et al. (2006) Selection of Plasmodium falciparum pfmdr1 alleles following therapy with artemether-lumefantrine in an area of Uganda where malaria is highly endemic. Antimicrobial agents and chemotherapy 50, 1893–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Humphreys GS et al. (2007) Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrobial agents and chemotherapy 51, 991–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Happi CT et al. (2009) Selection of Plasmodium falciparum multidrug resistance gene 1 alleles in asexual stages and gametocytes by artemether-lumefantrine in Nigerian children with uncomplicated falciparum malaria. Antimicrobial agents and chemotherapy 53, 888–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeka A et al. (2016) Artesunate/amodiaquine versus artemether/lumefantrine for the treatment of uncomplicated malaria in Uganda: a randomized trial. The Journal of infectious diseases 213, 1134–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plucinski MM et al. (2017) Efficacy of artemether–lumefantrine, artesunate–amodiaquine, and dihydroartemisinin–piperaquine for treatment of uncomplicated Plasmodium falciparum malaria in Angola, 2015. Malaria Journal 16, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matrevi SA et al. (2019) Plasmodium falciparum kelch propeller polymorphisms in clinical isolates from Ghana from 2007 to 2016. Antimicrobial Agents and Chemotherapy 63, e00802–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lubell Y et al. (2014) Artemisinin resistance – modelling the potential human and economic costs. Malaria Journal 13, 452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conrad MD and Rosenthal PJ (2019) Antimalarial drug resistance in Africa: the calm before the storm? The Lancet Infectious Diseases 19, e338–e351 [DOI] [PubMed] [Google Scholar]
- 16.Access to Medicine Foundation (2018) Antimicrobial resistance benchmark 2018. First independent assessment of pharmaceutical company action on AMR
- 17.Burrows JN et al. (2017) New developments in anti-malarial target candidate and product profiles. Malaria Journal 16, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blasco B et al. (2017) Antimalarial drug resistance: linking Plasmodium falciparum parasite biology to the clinic. Nature medicine 23, 917–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bopp SER et al. (2013) Mitotic evolution of Plasmodium falciparum shows a stable core genome but recombination in antigen families. PLoS genetics 9, e1003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee AH and Fidock DA (2016) Evidence of a mild mutator phenotype in Cambodian Plasmodium falciparum malaria parasites. PloS one 11, e0154166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leroy D (2017) How to tackle antimalarial resistance? EMBO molecular medicine 9, 133–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding XC et al. (2012) A framework for assessing the risk of resistance for antimalarials in development. Malaria journal 11, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCarthy JS et al. (2011) A pilot randomised trial of induced blood-stage Plasmodium falciparum infections in healthy volunteers for testing efficacy of new antimalarial drugs. PLoS ONE 6, e21914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watts RE et al. (2020) Safety and parasite clearance of artemisinin-resistant Plasmodium falciparum infection: A pilot and a randomised volunteer infection study in Australia. PLoS Medicine 17, e1003203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Llanos-Cuentas A et al. (2018) Antimalarial activity of single-dose DSM265, a novel Plasmodium dihydroorotate dehydrogenase inhibitor, in patients with uncomplicated Plasmodium falciparum or Plasmodium vivax malaria infection: a proof-of-concept, open-label, phase. The Lancet. Infectious diseases 18, 874–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phillips MA et al. (2015) A long-duration dihydroorotate dehydrogenase inhibitor (DSM265) for prevention and treatment of malaria. Science translational medicine 7, 296ra111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carolino K and Winzeler EA (2020) The antimalarial resistome – finding new drug targets and their modes of action. Current Opinion in Microbiology 57, 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Rycker M et al. (2020) Setting Our Sights on Infectious Diseases. ACS Infectious Diseases 6, 3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carlton JM (2018) Malaria parasite evolution in a test tube. Science 359, 159–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cowell AN et al. (2018) Mapping the malaria parasite druggable genome by using in vitro evolution and chemogenomics. Science 359, 191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allman EL et al. (2016) Metabolomic profiling of the Malaria Box reveals antimalarial target pathways. Antimicrobial agents and chemotherapy 60, 6635–6649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Creek DJ et al. (2016) Metabolomics-based screening of the Malaria Box reveals both novel and established mechanisms of action. Antimicrobial Agents and Chemotherapy 60, 6650–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cooper RA and Carucci DJ (2004) Proteomic Approaches to Studying Drug Targets and Resistance in Plasmodium. Current drug targets. Infectious disorders 4, 41–51 [DOI] [PubMed] [Google Scholar]
- 34.Dziekan JM et al. (2020) Cellular thermal shift assay for the identification of drug–target interactions in the Plasmodium falciparum proteome. Nature Protocols 15, 1881–1921 [DOI] [PubMed] [Google Scholar]
- 35.Yang T et al. (2021) MalDA, accelerating malaria drug discovery. Trends in Parasitology DOI: 10.1016/j.pt.2021.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng CL and Fidock DA (2019) Plasmodium falciparum in vitro drug resistance selections and gene editing. Methods in molecular biology (Clifton, N.J.) 2013, 123–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghorbal M et al. (2014) Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nature biotechnology 32, 819–21 [DOI] [PubMed] [Google Scholar]
- 38.Ng CL et al. (2016) CRISPR-Cas9-modified pfmdr1 protects Plasmodium falciparum asexual blood stages and gametocytes against a class of piperazine-containing compounds but potentiates artemisinin-based combination therapy partner drugs. Molecular Microbiology 101, 381–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vanaerschot M et al. (2017) Hexahydroquinolines are antimalarial candidates with potent blood-stage and transmission-blocking activity. Nature microbiology 2, 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.LaMonte G et al. (2016) Mutations in the Plasmodium falciparum cyclic amine resistance locus (PfCARL) confer multidrug resistance. mBio 7, e00696–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baragaña B et al. (2015) A novel multiple-stage antimalarial agent that inhibits protein synthesis. Nature 522, 315–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiménez-Díaz MB et al. (2009) Improved murine model of malaria using Plasmodium falciparum competent strains and non-myelodepleted NOD-scid IL2Rγnull mice engrafted with human erythrocytes. Antimicrobial Agents and Chemotherapy 53, 4533–4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leroy D et al. (2014) Defining the biology component of the drug discovery strategy for malaria eradication. Trends in Parasitology 30, 478–490 [DOI] [PubMed] [Google Scholar]
- 44.Miller LH et al. (2013) Malaria biology and disease pathogenesis: insights for new treatments. Nature Medicine 19, 156–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haldar K et al. (2018) Drug resistance in Plasmodium. Nature Reviews Microbiology 16, 156–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gregson A and Plowe CV (2005) Mechanisms of resistance of malaria parasites to antifolates. Pharmacological reviews 57, 117–45 [DOI] [PubMed] [Google Scholar]
- 47.Fisher N et al. (2012) Cytochrome b mutation Y268S conferring atovaquone resistance phenotype in malaria parasite results in reduced parasite bc1 catalytic turnover and protein expression. The Journal of biological chemistry 287, 9731–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valderramos SG and Fidock DA (2006) Transporters involved in resistance to antimalarial drugs. Trends in Pharmacological Sciences 27, 594–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ariey F et al. (2014) A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505, 50–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amato R et al. (2017) Genetic markers associated with dihydroartemisinin–piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype–phenotype association study. The Lancet Infectious Diseases 17, 164–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Witkowski B et al. (2017) A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype–genotype association study. The Lancet Infectious Diseases 17, 174–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ross LS et al. (2018) Emerging Southeast Asian PfCRT mutations confer Plasmodium falciparum resistance to the first-line antimalarial piperaquine. Nature Communications 9, 3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim J et al. (2019) Structure and drug resistance of the Plasmodium falciparum transporter PfCRT. Nature 576, 315–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chugh M et al. (2015) Identification and deconvolution of cross-resistance signals from antimalarial compounds using multidrug-resistant Plasmodium falciparum strains. Antimicrobial agents and chemotherapy 59, 1110–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Canfield CJJ et al. (1995) Interactions of atovaquone with other antimalarial drugs against Plasmodium falciparum in vitro. Experimental Parasitology 80, 373–381 [DOI] [PubMed] [Google Scholar]
- 56.Kuhen KL et al. (2014) KAF156 is an antimalarial clinical candidate with potential for use in prophylaxis, treatment, and prevention of disease transmission. Antimicrobial Agents and Chemotherapy 58, 5060–5067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paquet T et al. (2017) Antimalarial efficacy of MMV390048, an inhibitor of Plasmodium phosphatidylinositol 4-kinase. Science translational medicine 9, eaad9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stickles AM et al. (2015) Subtle changes in endochin-like quinolone structure alter the site of inhibition within the cytochrome bc1 complex of Plasmodium falciparum. Antimicrobial agents and chemotherapy 59, 1977–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Magistrado PA et al. (2016) Plasmodium falciparum Cyclic cmine resistance locus (PfCARL), a resistance mechanism for two distinct compound classes. ACS infectious diseases 2, 816–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Witkowski B et al. (2013) Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. The Lancet Infectious Diseases 13, 1043–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Schalkwyk DA et al. (2017) Comparison of the susceptibility of Plasmodium knowlesi and Plasmodium falciparum to antimalarial agents. The Journal of antimicrobial chemotherapy 72, 3051–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rangel GW et al. (2018) Enhanced ex vivo Plasmodium vivax intraerythrocytic enrichment and maturation for rapid and sensitive parasite growth assays. Antimicrobial Agents and Chemotherapy 62, e02519–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gomes AR et al. (2017) Genetic diversity of next generation antimalarial targets: A baseline for drug resistance surveillance programmes. International Journal for Parasitology: Drugs and Drug Resistance 7, 174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pearson RD et al. (2019) An open dataset of Plasmodium falciparum genome variation in 7,000 worldwide samples. DOI: 10.1101/824730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petersen I et al. (2015) Balancing drug resistance and growth rates via compensatory mutations in the Plasmodium falciparum chloroquine resistance transporter. Molecular microbiology 97, 381–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bushman M et al. (2016) Within-host competition and drug resistance in the human malaria parasite Plasmodium falciparum. Proceedings. Biological sciences 283, 20153038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mandt REK et al. (2019) In vitro selection predicts malaria parasite resistance to dihydroorotate dehydrogenase inhibitors in a mouse infection model. Science translational medicine 11, eaav1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Delves MJ et al. (2016) Routine in vitro culture of P. falciparum gametocytes to evaluate novel transmission-blocking interventions. Nature protocols 11, 1668–80 [DOI] [PubMed] [Google Scholar]
- 69.Goodman CD et al. (2016) Parasites resistant to the antimalarial atovaquone fail to transmit by mosquitoes. Science 352, 349–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jiménez-Díaz MB et al. (2014) (+)-SJ733, a clinical candidate for malaria that acts through ATP4 to induce rapid host-mediated clearance of Plasmodium. Proceedings of the National Academy of Sciences of the United States of America 111, E5455–E5462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vaidya AB et al. (2014) Pyrazoleamide compounds are potent antimalarials that target Na(+) homeostasis in intraerythrocytic Plasmodium falciparum. Nature communications 5, 5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schalkwijk J et al. (2019) Antimalarial pantothenamide metabolites target acetyl-coenzyme A biosynthesis in Plasmodium falciparum. Science Translational Medicine 11, eaas9917. [DOI] [PubMed] [Google Scholar]
- 73.Delves M et al. (2012) The activities of current antimalarial drugs on the life cycle stages of Plasmodium: a comparative study with human and rodent parasites. PLoS medicine 9, e1001169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.