Bomba-Warczak et al. use stable isotope labeling of mice and tandem mass spectrometry to discover a pool of mitochondrial proteins persisting for months in tissues containing postmitotic cells. Long-lived proteins are enriched at cristae. Crosslinking analysis revealed limited exchange of subunits throughout their lifetime.
Abstract
Long-lived proteins (LLPs) have recently emerged as vital components of intracellular structures whose function is coupled to long-term stability. Mitochondria are multifaceted organelles, and their function hinges on efficient proteome renewal and replacement. Here, using metabolic stable isotope labeling of mice combined with mass spectrometry (MS)–based proteomic analysis, we demonstrate remarkable longevity for a subset of the mitochondrial proteome. We discovered that mitochondrial LLPs (mt-LLPs) can persist for months in tissues harboring long-lived cells, such as brain and heart. Our analysis revealed enrichment of mt-LLPs within the inner mitochondrial membrane, specifically in the cristae subcompartment, and demonstrates that the mitochondrial proteome is not turned over in bulk. Pioneering cross-linking experiments revealed that mt-LLPs are spatially restricted and copreserved within protein OXPHOS complexes, with limited subunit exchange throughout their lifetimes. This study provides an explanation for the exceptional mitochondrial protein lifetimes and supports the concept that LLPs provide key structural stability to multiple large and dynamic intracellular structures.
Graphical Abstract
Introduction
Interest in long-lived proteins (LLPs) with lifetimes of months or more has been growing in recent years due to the development of proteomic strategies enabling large-scale measurements of in vivo protein turnover. LLPs exist in many organisms, are present in multiple tissue types, and can be as old as the organism itself (Savas et al., 2012; Truscott et al., 2016). Most known LLPs form discrete structures that provide long-term architectural stability within cells and at extracellular sites. Recently, we discovered that nucleoporins (NUPs), which form the core of the nuclear pore complex, are extremely long lived in rodent brains and persist in neurons for the entire lifetime of animals (Savas et al., 2012; Toyama et al., 2013). This long-term persistence of long-lived NUPs can be both detrimental and vital to the survival of the cell. On the one hand, because of their longevity, LLPs are likely to accumulate damage over their extended lifetimes and thus create a layer of vulnerability. On the other hand, frequent or impaired exchange of the nuclear pore complex core subunits, for example, could transiently compromise the nuclear envelope, resulting in loss of nuclear/cytoplasmic compartmentalization (D’Angelo et al., 2009).
Intriguingly, in our previous discovery-based proteomic analysis (Toyama et al., 2013), we also identified a discrete number of mitochondrial proteins that last for >6 mo. This is substantially longer than in any previous report on the longevity of the mitochondrial proteome, since the average half-life of mitochondrial proteins in the brain has been estimated to be <3 wk (Fornasiero et al., 2018). Efficient protein turnover is an essential homeostatic process ensuring a functional mitochondrial proteome throughout the cell’s lifetime (Cambridge et al., 2011; Goldberg, 2003). Consequently, cells have an intricate multilevel quality control system aimed at maintaining a pristine mitochondrial proteome. These processes include selective mitophagy, constitutive autophagy, mitochondrial-derived vesicles, and proteasomal degradation (Glynn, 2017; Misgeld and Schwarz, 2017; Pickles et al., 2018; Soubannier et al., 2012; Youle and Narendra, 2011). Therefore, the discovery of mitochondrial proteins that avoid turnover amid such elaborate quality control and persist for months in healthy tissues is unexpected and of potential importance.
In the present study, we report a multiscale characterization of the long-lived mitochondrial proteome in several mouse tissues using dynamic metabolic stable isotope labeling and mass spectrometry (MS)–based proteomic analysis. Our findings uncover striking tissue differences, with a select panel of mitochondrial proteins persisting for >4 mo in tissues rich in long-lived cells. Closer examination of these long-lived mitochondrial proteins, termed mt-LLPs, revealed significant enrichment of proteins associated with the inner mitochondria membrane (IMM), specifically in complexes and pathways associated with cristae membranes. Interestingly, similar to other architecturally stable and long-lived structures, cristae are also recognized for their highly defined and elaborate ultrastructure (Cogliati et al., 2016). We discovered that old subunits of oxidative phosphorylation (OXPHOS) complexes integrate into both individual and higher-order complexes. At the same time, protein subunits within OXPHOS complexes show remarkable persistence, suggesting that, once assembled, OXPHOS complexes are preserved with little to no subunit exchange. Altogether, we report that inner membrane mitochondrial proteins in the heart and brain have remarkable stability and support mitochondrial ultrastructure.
Results and discussion
Mitochondrial proteins are exceptionally long lived in tissues containing enduring postmitotic cells
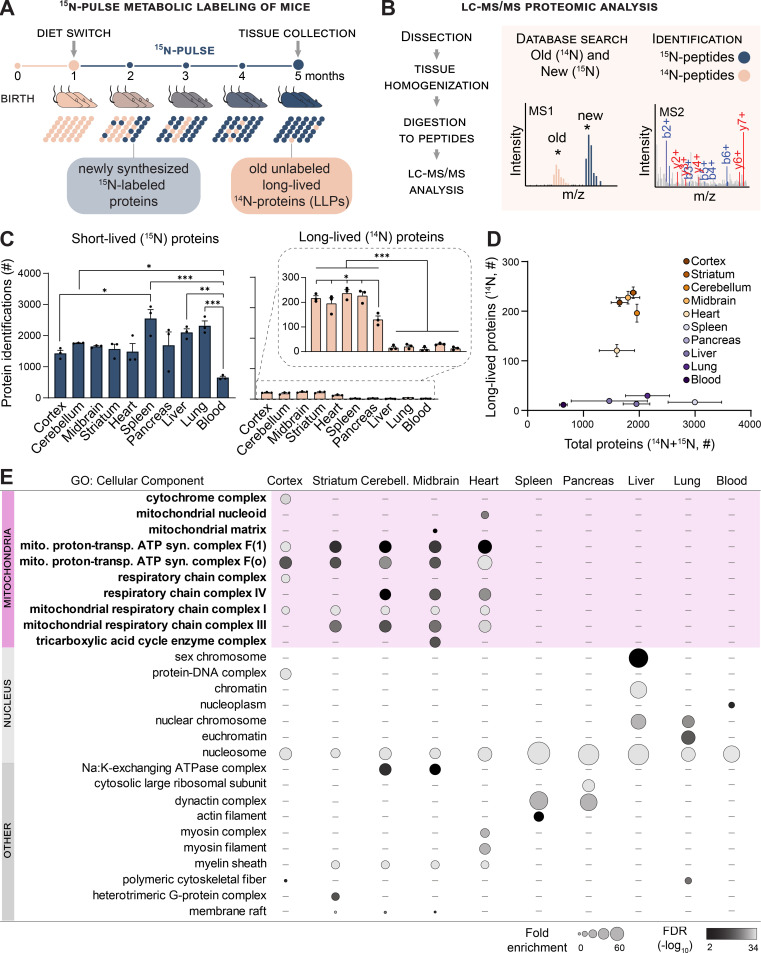
We previously discovered a small pool of intracellular proteins with exceptionally long life spans in mammalian brains (Savas et al., 2012; Toyama et al., 2013). While the most prominent LLPs identified in the brain localize to the nuclei of postmitotic neurons, our analysis revealed that select mitochondrial proteins, albeit to a lesser degree, also exhibit exceptional persistence (Toyama et al., 2013). In the present study, we set out to investigate these proteins with a focused in-depth proteomic analysis of tissues from mice metabolically pulse-labeled with the stable isotope of nitrogen-15 (15N; Savas et al., 2016). To accommodate the apparent shorter lifetimes of mitochondrial proteins as compared with the nuclear LLPs, we modified our previously published protocol by shortening the 15N isotope–labeling period to 4 mo (Fig. 1 A). Guided by our previous findings, we analyzed a panel of tissues enriched in terminally differentiated, long-lived cells (cortex, cerebellum, midbrain, striatum, and heart) and a set of proliferative tissues (spleen, pancreas, liver, lung, and blood). We then used liquid chromatography (LC)-tandem MS–based proteomics to identify and quantify 15N- and 14N-containing peptides belonging to the newly synthesized and old protein pools, respectively (Fig. 1 B).
Figure 1.
Mitochondrial proteins are exceptionally longlived in tissues containing enduring postmitotic cells. (A) Whole-animal metabolic pulse 15N labeling scheme to identify and measure LLPs (≥4 mo) in mice. (B) Analytical workflow illustrating how the dissected tissues were homogenized and digested to peptides, and 15N and 14N peptides were identified and quantitated with LC-MS/MS–based proteomics. (C) Summary of protein identifications. Left plot indicates the number of 15N proteins identified (i.e., new, ≤4 mo old), right plot indicates 14N proteins identified (i.e., ≥4 mo old). (D) Tissues enriched with long-lived cells selectively harbor LLPs despite a similar number of total proteins identified in most tissues. (E) GO analysis of the LLPs in tissues that contain enduring postmitotic cells revealed that terms related to mitochondria (mito.) are significantly enriched. syn., synthesis; trans., transport. Data in C and D are mean ± SEM; in C–E, n = three mice for each tissue analyzed. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by Kruskal–Wallis ANOVA with Tukey’s multiple comparisons test. See also Fig. S1 and Table S1.
In agreement with the average protein half-life in mouse tissues being on the scale of days (Fornasiero et al., 2018), after 4 mo of 15N labeling, most identified proteins consisted entirely of 15N (Fig. 1 C, blue graph). However, in all tissue extracts examined, we also identified a small pool of old peptides composed entirely of 14N, representing a population of proteins that persisted without turnover (i.e., absence of 15N incorporation) for ≥4 mo (Fig. 1 C, tan graph, and Fig. S1 A). Globally, we found that tissues harboring long-lived cells have a significantly higher ratio of long-lived/short-lived proteins compared with proliferating tissues(11.4 ± 2.6% vs. 1.1 ± 0.7%). To visualize the relationship between the total number of proteins identified and our ability to detect old peptides by MS/MS, we plotted these values in each tissue examined. Notably, while there was no correlation between the overall protein abundance and the identification of proteins as long lived, we found a strong trend between the presence of LLPs and tissues enriched in terminally differentiated long-lived cells (Fig. 1 D).
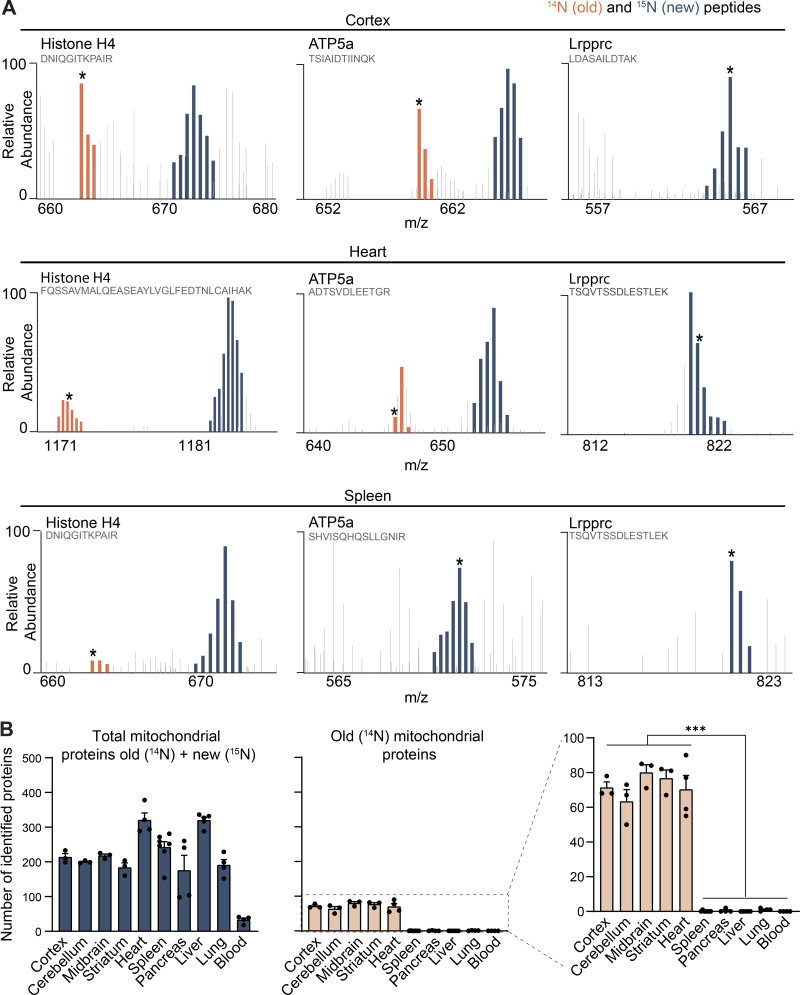
Figure S1.
Mitochondrial proteins are exceptionally longlived in tissues containing postmitotic cells. Related to Fig. 1. (A) Annotated representative raw MS1 scans across the indicated m/z ranges after 4 mo of 15N-dynamic pulse. Distinct peptides for indicated proteins; dark blue, 15N peptide peaks; orange, 14N; gray, other peptides. Starred peaks (*) were identified by MS/MS. (B) Number of identified mitochondrial proteins (blue) along with mt-LLPs (orange), based on MitoCarta3.0; mt-LLPs are only present in the brain and heart tissues. Data are mean ± SEM; three to five biological replicates per tissue. ***, P < 0.001 by Kruskal–Wallis ANOVA with Tukey’s multiple comparisons test.
To investigate the possibility that the LLPs in each tissue are associated with common cellular structures, we performed gene ontology (GO) cellular component enrichment analysis (Fig. 1 E and Table S1). By comparing the pool of old proteins to the total collection of proteins identified within each tissue, we found significant enrichment for terms related to mitochondrion, exclusively in brain (cortex, cerebellum, midbrain, and striatum) and heart tissues. The absence of mt-LLPs in the other tissues examined was not due to disproportional identification of mitochondrial proteins. Excluding blood, which contains mitochondria-depleted red blood cells, we identified on average 235 ± 63 mitochondrial proteins across all of the tissues analyzed (Fig. S1 B). Hence, a select panel of mitochondrial proteins with exceptional life spans are enriched exclusively in the brain and heart.
LLPs are preferentially associated with the IMM
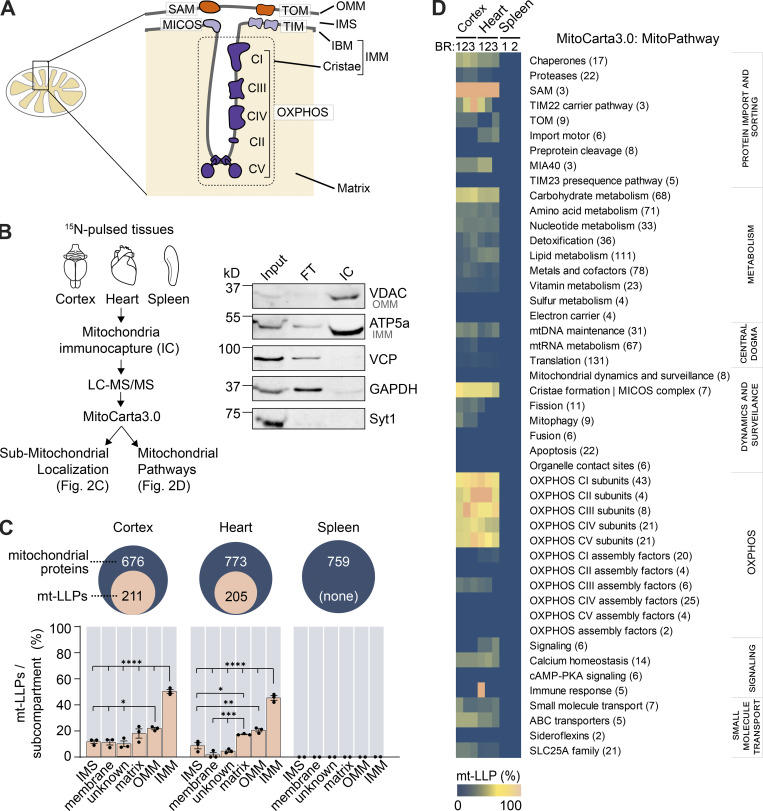
Mitochondria are multifaceted organelles that play vital roles in energy production, metabolism, calcium homeostasis, and cell death pathways (Clapham, 2007; Contreras et al., 2010; Halliwell, 2001; Nicholls, 2002). Their functional versatility is intimately linked to their elaborate morphology, with two noncontiguous membranes (outer [OMM] and inner [IMM]), cristae invaginations, matrix, and intermembrane space (IMS; Fig. 2 A). Considering this high degree of structural and functional complexity, we sought to investigate whether mt-LLPs localize to any specific mitochondrial domain or functional pathway.
Figure 2.
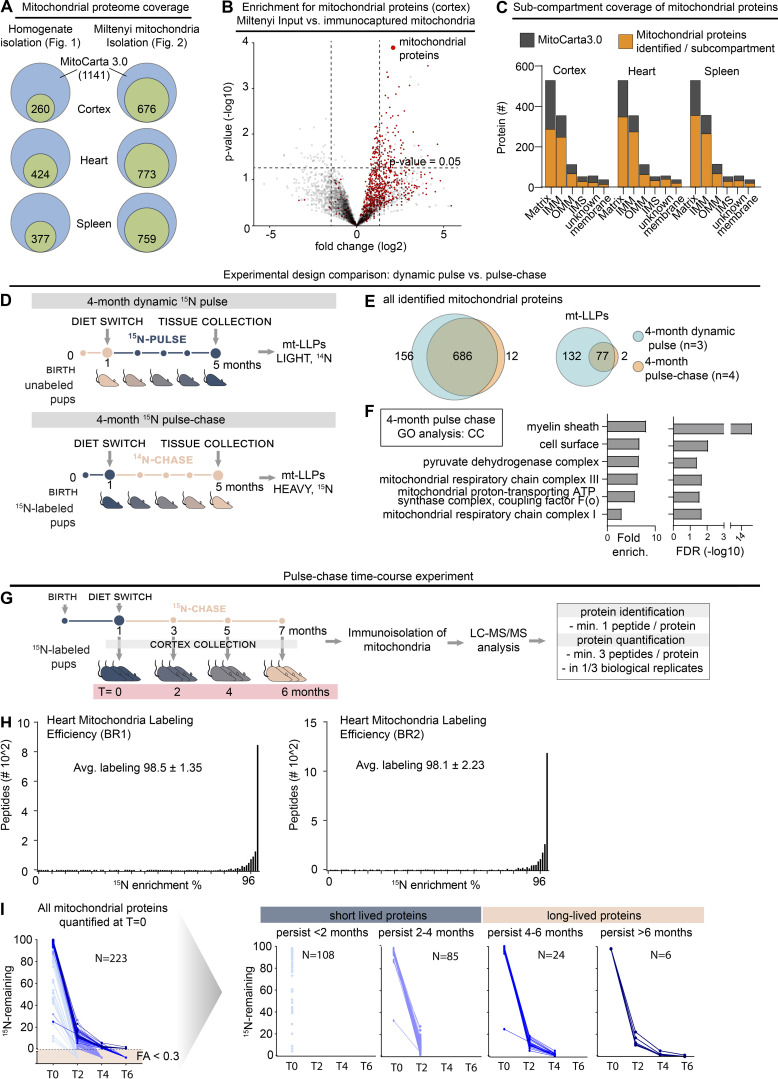
LLPs are preferentially associated with the IMM. (A) Schematic illustrating mitochondrial subcompartmentalization and relevant protein complexes. (B) Workflow to immune-capture mitochondria. Cortex, heart, and spleen from 15N-pulsed mice were homogenized using a gentleMACS system, and dissociated mitochondria were purified by immuno-capture using anti-Tom20 antibodies (Miltenyi Biotec). Purified mitochondria were analyzed using LC-MS/MS, mitochondrial proteins were filtered using MitoCarta 3.0, and mt-LLPs were grouped and examined based on their submitochondrial localization and Mitochondrial Pathway assignments. FT, flow through; IC, immunocapture; IBM, inner boundary membrane. (C) The total number of mitochondrial proteins identified based on 15N and 14N MS/MS spectral matches per tissue (blue) along with the number of identified 14N mt-LLPs (tan). mt-LLPs are enriched at IMM in both heart and cortical extracts, but not in the other subcompartments. None of the mitochondrial proteins in spleen were identified as mt-LLPs. (D) MitoPathway analysis of mt-LLPs. mt-LLPs are enriched in several protein complexes including OXPHOS, Tim22, TOM, SAM, and the cristae formation (i.e., MICOS complex). BR, biological replicate. Data in C are mean ± SEM; for C and D, n = 3 mice for cortex and heart, 2 for spleen. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by Kruskal–Wallis ANOVA with Tukey’s multiple comparisons test. See also Fig. S2 and Table S2.
We focused our analysis on two tissues enriched in mt-LLPs (cortex and heart) and one control tissue (spleen) that, while rich in mitochondria, lacks mt-LLPs. To begin our investigation into the distribution of mt-LLPs we shifted our analysis from tissue homogenates to immuno-captured mitochondria (Fecher et al., 2019), which enabled deeper analytical coverage of the mitochondrial proteome (Fig. 2 B and Fig. S2, A and B). Using LC-MS/MS–based proteomics, we identified 676 mitochondrial proteins in extracts from the cortex, 773 from the heart, and 759 from the spleen based on MitoCarta3.0 (Rath et al., 2021; Fig. 2 C and Table S2). Of these proteins, 211 in the cortex and 205 in the heart were identified based on both heavy (15N, new) and light (14N, old) protein database searches. Even though the total number of mitochondrial proteins identified in the spleen was higher than in the cortex and heart, consistent with our initial findings, our proteomic analysis did not identify a single 14N spectra mapping to a mitochondrial protein in spleen. We note that although previously published estimations of protein turnover in mammals found mitochondrial proteins to have longer half-lives than the global average (i.e., 20 versus 10.4 d; Fornasiero et al., 2018), these estimated protein lifetimes are far shorter than the observed lifetimes reported here. Moreover, while we identified >200 proteins that persist for >4 mo in the cortex, Fornasiero et al. (2018) reported only 35 proteins as LLPs (Table S2). This discrepancy is likely due to the difference in labeling methods (15N versus 13C6-lysine), the reliance on mathematical modeling of lysine pools, and vastly different labeling periods (i.e., 4 mo versus 3–6 wk), all of which explain the previous underestimated lifetime specifically for proteins with exceptional lifetimes.
Figure S2.
LLPs are preferentially associated with the IMM. Related to Fig. 2. (A) Coverage of mitochondrial proteome across immuno-isolated mitochondria. Cumulative number of proteins identified shown. n = 2–4 mice. (B) Immuno-isolated mitochondria are substantially enriched in mitochondrial proteins. n = 4 mice. (C) Submitochondrial distribution of identified mitochondrial proteins from cortical, heart, and spleen tissue extracts. Identified proteins are shown in orange; MitoCarta3.0 proteins are shown in gray. n = 3 mice. (D) Schematic illustrating two-generation whole-animal metabolic 15N-dynamic pulse compared with 14N pulse-chase experiment design. (E) Venn diagram comparing proteins identified in the 15N-dynamic pulse and 14N pulse-chase experiments. enrich., enrichment. (F) GO analysis of the mt-LLPs identified after 4 mo of 14N pulse-chase showing an enrichment for terms related to mitochondria. See also Table S1. (G) Time course pulse-chase experiment of 15N-labeled mice (born to 15N-labeled females) that were chased with 14N diet for 0, 2, 4, and 6 mo. Note that the 4-mo 14N pulse chase time point shown here represents the same dataset as in panels D and E. (H) Metabolic labeling efficiency of second-generation pups analyzed at t = 0 (at weaning) in the 14N pulse-chase experiments. Average peptide labeling percentage is indicated ± SD. (I) Summary of protein degradation trends across chase periods. Only proteins identified at t = 0 which were then tracked through the time course are shown. n = 3–4 mice.
To examine the distribution of mt-LLPs within mitochondria, we began by extracting each identified protein’s submitochondrial localization assignment from MitoCarta 3.0 (Table S2). First, we established that there are no significant differences in coverage of mitochondrial proteome across the subcompartments of cortical, heart, and spleen mitochondria (Fig. S2 C). Next, we determined how many of the proteins within each subcompartment were identified in both 14N and 15N searches. We found that 50.3% and 45.3% of IMM-localized proteins in the cortex and heart, respectively, can persist for ≥4 mo (Fig. 2 C). This enrichment of mt-LLPs in IMM was significantly higher than the fraction of mt-LLPs localized to matrix, OMM, or IMS in both heart and cortex. Since our proteomic analysis shows that mitochondrial proteins in the spleen do not persist past 4 mo, the distribution analysis could not be performed for this tissue.
Further, to gain additional insight into mt-LLPs, we used the new feature of MitoCarta3.0 termed MitoPathways, which provides mitochondria-centric pathway annotations (Rath et al., 2021). We categorized all of the identified proteins according to their MitoPathway assignments and calculated mt-LLP enrichment in each major pathway (Fig. 2 D). Consistent with our initial GO analysis (Fig. 1 E), we found an enrichment of mt-LLPs in “OXPHOS subunits,” but not “OXPHOS assembly factors” in both cortex and heart. Moreover, we found that mt-LLPs are selectively associated with the “cristae formation pathway/MICOS (mitochondrial contact site and cristae organizing system) complex,” “SAM (sorting and assembly machinery),” “TIM22 carrier pathway,” and “chaperones,” including cristae-localized Prohibitins and IMS-localized “small Tims,” and “carbohydrate metabolism,” specifically the cristae-associated TCA cycle.
To confirm our findings, we performed a label-swap experiment using brain cortices from an independent cohort of 15N-labeled mice (born to 15N-labeled females) that we chased with 14N-chow diet for 4 mo (Fig. S2 D). In this pulse-chase paradigm, 15N-labeled peptides represent the old protein pool, while 14N peptides represent the newly synthesized pool of proteins. First, we compared the 4-mo chase with 4-mo dynamic-pulse data for cortex, and we found an overlap of 80.3% in the identified mitochondrial proteins (Fig. S2 E). Notably, 97.5% of mt-LLPs identified at this time point in the pulse-chase experiment were also identified as mt-LLPs in the dynamic-pulse experiment. However, in the label-swap experiment, in general, we identified fewer mt-LLPs, which is consistent with our previously published findings (Hark et al., 2021; Table S3). GO analysis of mt-LLPs identified in the label-swap experiment revealed enrichment of a set of terms similar to the 15N-dynamic pulse experimental design, further demonstrating consistency between the two paradigms (Fig. S2 F and Table S1). To extend our analysis, we tracked the longevity of proteins after 0, 2, 4, and 6 mo of chase (Fig. S2 G). First, we confirmed efficient labeling of the proteome. In the pups born to 15N-labeled dams, at the time of weaning (t = 0), we achieved an average labeling efficiency of ≥98.3% (Fig. S2 H). We found that 47.7% of mitochondrial proteins were completely turned over within 2 mo, and another 18.7% in 2–4 mo, leaving just 10.9% of mitochondrial proteins persisting for 4–6 mo (Fig. S2 I). Taken together, these results provide independent confirmation of the mt-LLPs identified in the dynamic 15N-pulse analysis paradigm.
Exceptional longevity of mitochondrial cristae-associated proteins
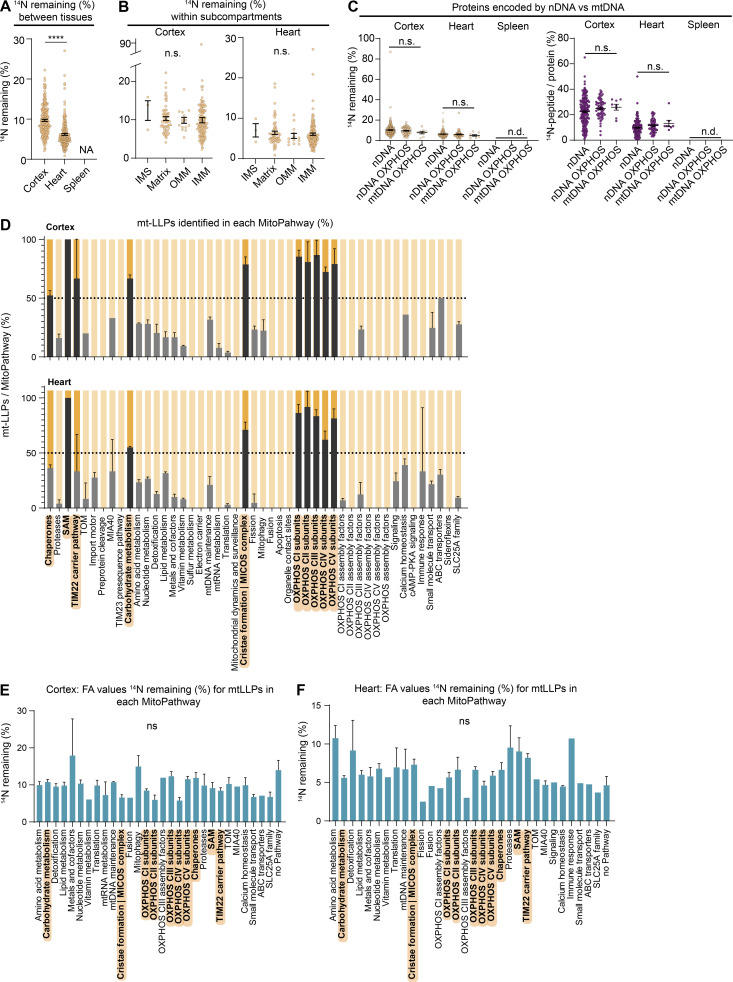
Our proteomic characterization of mt-LLPs in cortex and heart tissues incorporates measures of both old and new peptides, and the results indicate that only a fraction of each protein pool persists for ≥4 mo. Therefore, we set out to determine the quantity of each mt-LLP remaining after 4 mo of 15N-pulse. To accomplish this, we calculated the fractional abundance (FA; 14N remaining, 14N/[14N + 15N]) for each mt-LLP in cortex and heart using reconstructed MS1 chromatograms from LC-MS/MS analysis (Park et al., 2008). Overall, we found that, on average, 10.2 ± 6.6% and 6.2 ± 3.3% mt-LLPs persisted in cortex and heart for ≥4 mo, indicating significantly longer lifetimes of mt-LLPs in the cortex as compared with heart (Fig. S3 A). We next plotted the 14N-remaining values for mt-LLPs across subcompartments and found no significant differences in FA values between mitochondrial compartments within heart and cortical tissue extracts (Fig. S3 B). Moreover, we found no significant difference between the quantities of mt-LLPs encoded by nuclear versus mitochondrial DNA, suggesting that protein longevity is not dictated by genome localization (Fig. S3 C).
Figure S3.
Exceptional longevity of mitochondrial cristae-associated proteins. Related to Fig. 3. (A) Overall FA values of cortical, heart, and spleen mt-LLPs. (B) Mt-LLP FA values do not vary significantly within mitochondrial subcompartments. (C) Comparison of FA values and spectral counts for mt-LLPs encoded by nDNA versus mtDNA. (D) Average number of mt-LLPs identified per MitoPathway in cortex and heart, represented as percentage ± SEM. MitoPathways with enrichment of ≥50% (dotted line) of mt-LLPs are highlighted in hard gray/orange. (E and F) Average FA values for mt-LLPs identified in MitoPathway. Only MitoPathways for which mt-LLPs were identified are shown. Data points in A–F, n = 4 mice; mean ± SEM. ****, P < 0.0001 by Kruskal–Wallis ANOVA with Tukey’s multiple comparisons test.
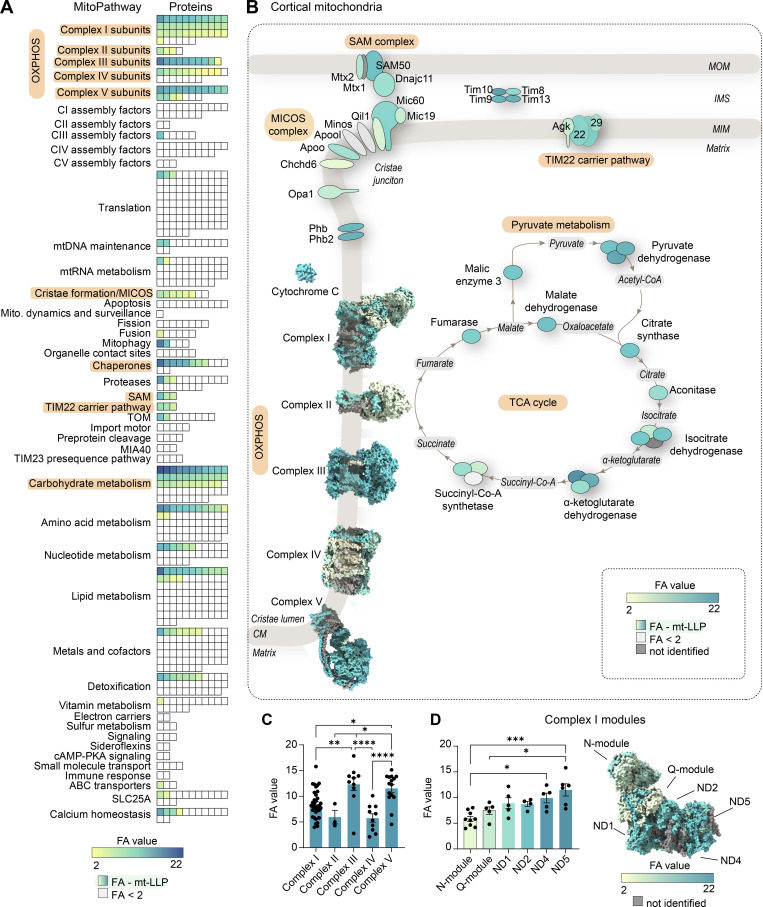
To obtain a more high-resolution view of the long-lived mitochondrial proteome landscape, we extracted all the mitochondrial proteins identified in immuno-isolations from cortex according to their MitoPathway assignments. First, to illustrate the enrichment of mt-LLPs in IMM-associated pathways, we graphed the average number of mt-LLPs identified in each MitoPathway (Fig. S3 D). Next, we represented all of the mitochondrial proteins identified according to both the MitoPathway and their FA values (Fig. 3 A). The mt-LLP FA is represented according to its 14N-remaining value; the lowest calculated value is 2.11 for mt-Co3 in complex III, and the highest is 20.2 for Bdh1 within the “carbohydrate metabolism” pathway. Short-lived 15N proteins (life spans <4 mo) that were identified in our proteomic analysis, but not quantified and lacking FA values, are represented in white. Finally, we illustrated the localization patterns of mt-LLPs from MitoPathways for which ≥50% of proteins were identified as long lived, along with their FA values, reinforcing the finding that mt-LLPs are enriched at IMM (Fig. 3 B). On a global scale, we found no significant differences in the average FA values between the various MitoPathways (Fig. S3, E and F). However, when we compared OXPHOS complexes, we found complex III and V to have significantly more longer-lived proteins compared with complexes I, II, and IV (Fig. 3 C). We also analyzed individual complex I modules and found that, consistent with previously published data (Szczepanowska et al., 2020), N and Q modules have fewer LLPs and are in general shorter-lived compared with the other modules composing the complex (Fig. 3 D).
Figure 3.
Exceptional longevity of mitochondrial cristae-associated proteins. (A) All mitochondrial (mito.) proteins identified in cortical extracts sorted according to their assigned MitoPathway. For proteins that were identified as mt-LLPs in the 14N search, we calculated the average FA values, shown in yellow-to-blue scale. For proteins that were not identified in light search, FA values could not be computed, and these proteins are displayed with white boxes. The lowest calculated FA value was 2.11; non–long-lived proteins are indicated with FA <2. (B) Illustration of major pathways, which showed enrichment of ≥50% in mt-LLPs (orange) with coloring based on FA, in cortex. (C) The pools of long-lived complex III and V are significantly larger than those of complexes I, II, and IV. Each dot represent a protein: complex I, 34; complex II, 3; complex III, 10; complex IV, 10; complex V, 15 subunits. (D) N- and Q-modules of complex I are turned over more rapidly than the rest of the complex. Each dot represent a protein: N-module, 9; Q-module, 5; ND1, 5; ND2, 4; ND4, 5; ND5, 6 subunits. Data in C and D are mean ± SEM; n = 3 mice. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by Kruskal–Wallis ANOVA with Tukey’s multiple comparisons test. See also Table S2 and Fig. S3.
The identification of mito-LLPs in MitoPathways associated with IMM and cristae membranes raises several key questions regarding their long-term maintenance and the cellular mechanisms that shield them from degradation. MICOS complex proteins, Opa1, SAM–mitochondrial IMS-bridging complex, and prohibitin-1 and -2 are known to play crucial roles in establishment, maintenance, and structural stability of cristae invaginations (Campello and Scorrano, 2010; Cogliati et al., 2016; Ding et al., 2015; Frezza et al., 2006; Ott et al., 2012; Scorrano et al., 2002). Additionally, a number of proteins whose function is unrelated to cristae maintenance, but rather are physically associated with cristae membranes, were also found to be long lived: namely, cytochrome C, which is a soluble electron carrier protein that is present in the enclosed lumen of the cristae invagination compartment, as well as enzymes of the TCA cycle, one of which also functions as OXPHOS complex II. Furthermore, Tim22 has been shown to interact with both the TOM complex and the MICOS complex, which physically positions potentially long-lived TOM-Tim22 pathway at cristae junctions (Callegari et al., 2019). This enrichment of mt-LLPs at cristae membranes and pathways associated with cristae suggests that, similar to other LLPs, long-lived mitochondrial proteins could play a crucial structural role in the stabilization of mitochondrial ultrastructure. Frequent exchange of mt-LLPs could compromise cristae integrity, and consequently destabilize mitochondria, which would be detrimental to cell survival. Indeed, cristae membrane instability has been linked to mitochondrial dysfunction, and inhibition of cristae remodeling protects mitochondria in a variety of pathological conditions (Cogliati et al., 2013; Quintana-Cabrera et al., 2018; Varanita et al., 2015). This hypothesis is further supported by recent work showing that individual cristae within the same mitochondrion exist as electrically isolated entities, implying that the neighboring cristae are functionally independent (Wolf et al., 2019). It is plausible that during mitochondrial fusion-fission, which segregates damaged parts of organelles for degradation, whole cristae along with their immediate surroundings can be preserved or spared and thus persist for months in long-lived cells. Together, our data suggest that the structural stability of cristae might rely not only on an elaborate interplay between the cristae-shaping proteins, but also on the long-term persistence of mt-LLPs, and consequently the cristae compartment as a whole.
Long-lived subunits of OXPHOS complexes integrate into individual and higher-order complexes
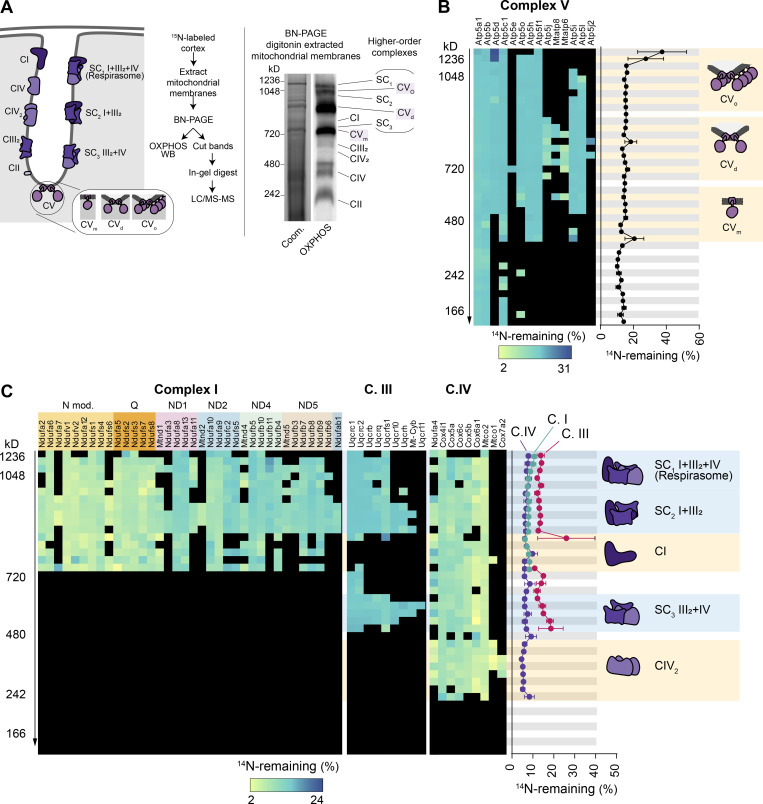
Mitochondrial complexes I, III, and IV exist as both individual complexes and higher-order supercomplexes (Cogliati et al., 2016; Schägger and Pfeiffer, 2000) of defined composition and stoichiometry (Fig. S4 A). Similarly, complex V forms dimers (CVd) and oligomers (CVo) that line the apex of cristae membranes and are key structural determinants of the highly curved cristae morphology (Paumard et al., 2002; Strauss et al., 2008). Our findings thus far show that a fraction of OXPHOS proteins persist for ≥4 mo in mouse cortex and heart tissues. This prompts the question of how the older versions of OXPHOS proteins are integrated and retained within the complexes and across their corresponding supercomplexes. To explore this idea, we performed complexome profiling analysis (Jha et al., 2016; Szczepanowska et al., 2020) on extracted mitochondria from cortices of 15N-pulsed mice (Fig. S4 A). We separated native OXPHOS complexes with BN-PAGE, sliced the gel lane based on molecular weight, and measured the FA values for each identified protein subunit of complexes I, II, III, IV, and V with LC-MS/MS (Fig. S4, B and C). The overall lifetimes of OXPHOS subunits obtained via BN-PAGE separation were consistent with FA values calculated with our previously presented methods of mitochondrial isolations. Furthermore, we found a remarkably uniform distribution of old proteins within individual complexes as well as the various arrangements of complexes. This suggests that while the overall longevity of different OXPHOS complexes is heterogeneous, the LLPs are uniformly distributed within both individual complexes and their higher-order structures.
Figure S4.
Long-lived subunits of OXPHOS complexes integrate into individual and higher-order complexes. (A) Solubilized cortical mitochondria extracts isolated from 15N-pulsed mice separated by BN-PAGE to resolve mitochondrial complexes and supercomplexes. Representative Coomassie (Coom.)-stained BN-PAGE gel and Western blot probed with an OXPHOS antibody cocktail. Mitochondrial complexes and supercomplexes are annotated. (B and C) 40 gel slices excised from each gel lane were subjected to in-gel trypsin digestion and LC-MS/MS analysis. FA values were calculated for each protein in a given complex and plotted. C.I, complex I; C.II, complex II; C.III, complex III; C.IV, complex IV; CVm, complex V monomer; CVd, complex V dimer; CVo, complex V oligomer; SC, supercomplex. Data points in B and C, n = 4 mice; mean ± SEM.
Limited mixing and exchange of mt-LLPs
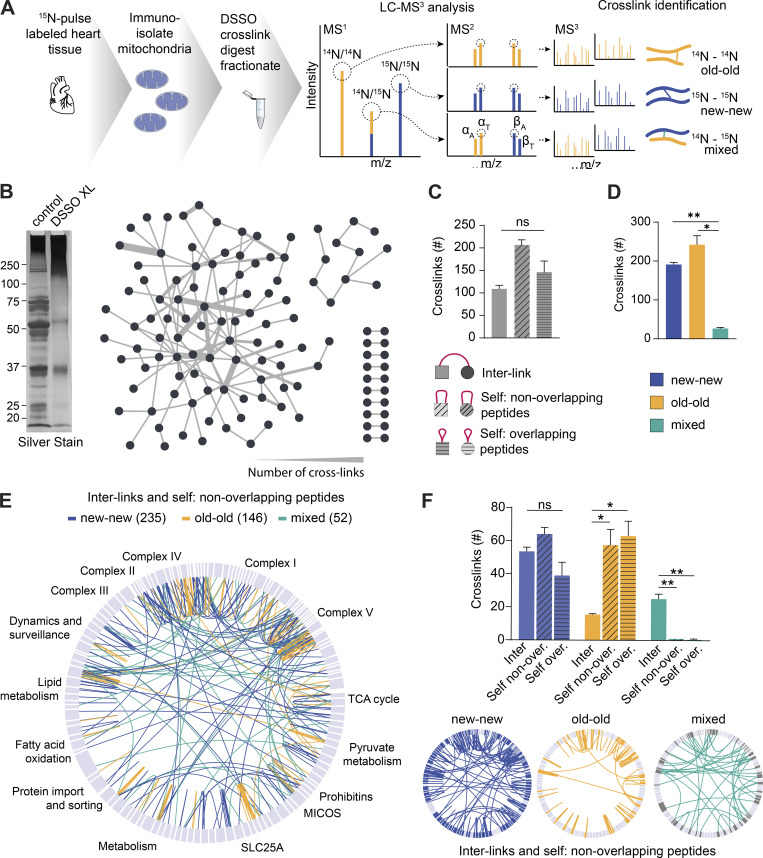
The exceptional stability of mt-LLPs prompts the question of how they distribute within individual mitochondria. Mitochondrial fusion-fission events inevitably lead to content dispersal and mixing over a period of months. Consequently, mt-LLPs could be scattered in random patterns or maintained in the same complexes and nano-environments. Thus far, our experiments have been unable to determine whether old and new proteins are in close physical proximity or spatially separated. To investigate this question, we performed pioneering cross-linking experiments on intact immuno-isolated mitochondria from heart extracts of dynamically 15N-pulse–labeled mice (Liu et al., 2018). We used the MS-cleavable cross-linker disuccinimidyl sulfoxide (DSSO; Kao et al., 2011) with Orbitrap Tribrid MS2-MS3 based analysis. The IP2 proteomic bioinformatic toolbox allowed us to probe interactions between new (15N) and old (14N) mitochondrial proteins (Fig. 4 A). We hypothesized that if mt-LLPs are randomly dispersed, we would predominantly identify mixed cross-links, i.e., between 15N and 14N peptides. Conversely, if mt-LLPs are co-maintained in the same complexes throughout their lifetimes, we would expect to identify more homo-isotopic, i.e., 15N-15N (new-new) and 14N-14N (old-old), cross-links.
Figure 4.
Limited mixing and exchange of mt-LLPs. (A) Schematic illustrating DSSO cross-linking of mitochondria isolated from 15N-pulse–labeled heart tissue extracts, sample processing, and MS1/MS2/MS3 analysis. (B) Silver-stained gel of control and cross-linked heart mitochondrial isolations. DSSO was added at a concentration of 1 mM; control was treated with DMSO only. Overview of cross-links between different mitochondrial proteins identified in the heart. (C) Number of cross-links identified in heart mitochondria according to the cross-link type. (D) Number of cross-links identified in heart mitochondria according to the peptide isotope build. (E) Map of the mitochondrial protein cross-links. 15N-15N (new-new) peptide cross-links are shown in blue; 14N-14N (old-old) peptide cross-links are shown in yellow; mixed 15N-14N (new-old) peptide cross-links are shown in green. (F) Summary of identified cross-linked peptides broken down by cross-link type and nitrogen isotype. Maps illustrating cross-linked peptides according to nitrogen isotype. In B, E, and F, maps were generated in XiView proteins categorized based on MitoCarta3.0. Data in C, D, and F are mean ± SEM; n = 2 mice. *, P < 0.05; **, P < 0.01, by Kruskal–Wallis ANOVA with Tukey’s multiple comparisons test. See also Fig. S5.
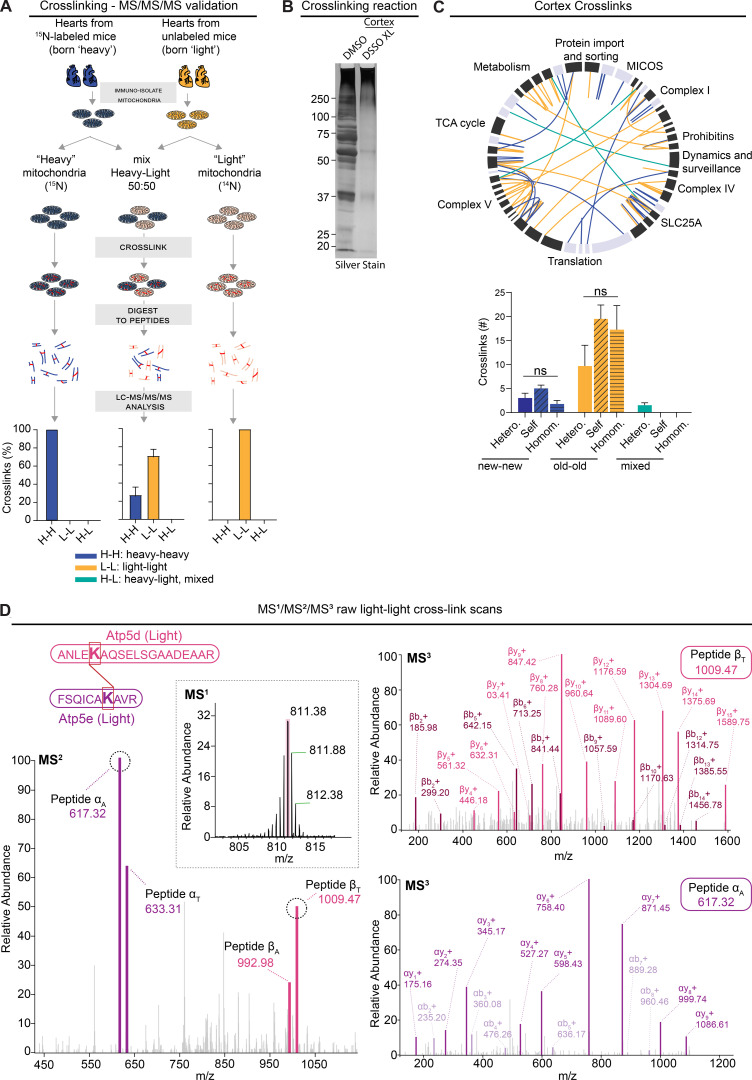
To assess the reliability of our novel approach, we devised a series of control experiments where we cross-linked mitochondria isolated from hearts from either fully 15N-labeled mice (i.e., mice born to 15N-labeled moms and kept on 15N feed throughout life) or unlabeled mice (14N, unlabeled; Fig. S5 A). We searched these experiments against both heavy and light protein databases. Subsequently, as expected, all of the identified cross-links in both conditions were homo-isotopic, that is between either two 15N-peptides or two 14N-peptides. Next, we mixed heavy and light mitochondria 1:1 and repeated the cross-linking. Again, we found that all identified cross-links were homo-isotopic. Overall, these results demonstrate that we are able to not only identify cross-links in both heavy and light database searches, but also provide evidence that heavy-light cross-links are not technical artifacts.
Figure S5.
Limited mixing and exchange of mt-LLPs. Related to Figs. 4 and 5. (A) Schematic illustrating control experiments used to assess the rigor of our new method of cross-link analysis of 15N dynamic-labeling experiments. Plot indicates percentage of heavy-heavy, light-light, or heavy-light. (B) Silver-stained gel of cross-linked cortical and heart mitochondrial isolations showing a shift toward higher molecular weights after cross-linking with 1 mM DSSO. (C) XiView map of cortical cross-links. Bar graph indicates identified cross-linked peptides broken down by class and nitrogen isotype. Hetero., interlink; self, self-link (nonoverlapping peptide); homom., self-link (overlapping peptides). (D) Representative example of high-resolution MS2 and MS3 spectra used to identify 14N-14N cross-linked ≥4-mo-old Atp5d and Atp5e peptides. Mean ± SEM; A, n = 2 mice; C, n = 4 mice; data not significant (n.s.) by Kruskal–Wallis ANOVA with Tukey’s multiple comparisons test.
Next, we examined cross-linked peptides identified in mitochondria isolated from 15N-pulsed hearts and cortices. In the heart mitochondria, we identified a total of 919 cross-linked peptides, of which 218 were interlinks, i.e., between two different proteins (Fig. 4, B and C, and Table S4). The remainder of the identified cross-links were self-links: specifically, 410 were between two nonoverlapping, and 291, overlapping peptides of the same protein. When we analyzed the cross-links based on their 15N- or 14N-peptide identification, we uncovered that 41.6% of cross-links formed between two new (15N-15N) peptides and another 52.7% formed between two old (14N-14N) peptides (Fig. 4, D and E). Consequently, only 5.7% of cross-links identified were formed between one old and one new peptide. While the number of cross-links between the different cross-link types did not significantly vary for new-new cross-links, for old-old cross-links, we detected significantly fewer interlinks as compared with both types of self-links (Fig. 4 F). This is likely due to the significantly lower number of old proteins, which as we already showed, accounts for only 6.2 ± 3.3% of the heart’s protein population (Fig. S3 A). Similar patterns of cross-links were observed in mitochondria isolated from cortical extracts, with 99.1% of cross-links identified formed between same isotype, and just 0.9% between old and new peptides (Fig. S5, B and C). Altogether, these data strongly suggest that mt-LLPs are not randomly dispersed throughout individual mitochondria, but are rather spatially restricted and co-preserved for months in brains and hearts of mice.
Assembled OXPHOS complexes are copreserved with limited subunit exchange for months
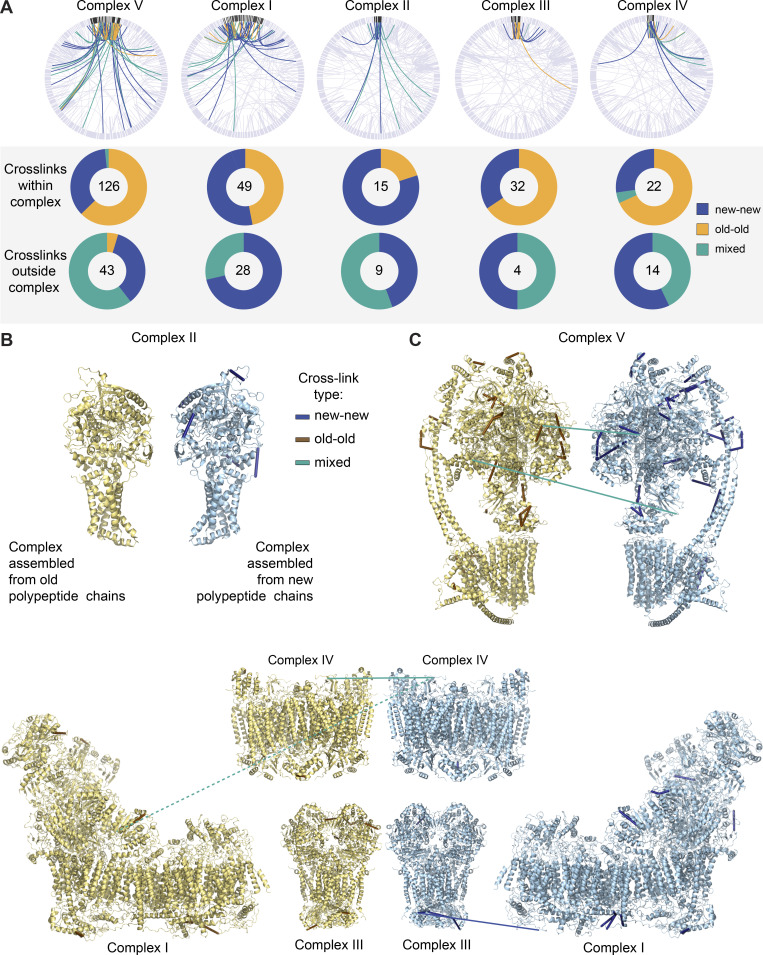
OXPHOS complexes, the most prominent of mt-LLPs, are multiprotein ensembles with complex biogenesis, assembly, and degradation dynamics (Szczepanowska and Trifunovic, 2021). While the assembly of these complexes is known to involve sequential incorporation of preformed modules, the exact mechanisms governing these processes remain to be elucidated. Moreover, it is unknown whether preformed modules or individual subunits are recycled and thus intermixed with newly synthesized proteins. Therefore, we performed in-depth structural analysis to examine the interactions between old and new OXPHOS proteins. Overall, we found that within individual complexes, the cross-links are nearly exclusively between either old-old or new-new peptides, with just two mixed cross-links in complex V (i.e., 1.5%) and one in complex IV (i.e., 4.5%; Fig. 5 A). Interestingly, both of these complexes are known to dimerize; therefore, the mixed cross-links could indicate dimers between one old and one new complex. Cross-links between individual complexes and other mitochondrial proteins, however, formed predominantly between either new-old peptides or two new peptides. The exception was two cross-links formed between old Atp5l and old Hadha and between old Atp5k and old Dld. These data suggest that while old OXPHOS complexes interact with the newly synthesized pool of mitochondrial proteins, the subunits within individual complexes are copreserved with limited turnover or exchange.
Figure 5.
OXPHOS complexes are copreserved with limited subunit exchange for months. (A) Total cross-links within individual OXPHOS complexes are predominantly between two peptides of the same isotope build (new-new or old-old). Cross-links between OXPHOS complexes and other mitochondrial proteins are mostly between new-new or new-old peptides but are rarely formed between two old peptides. (B) Inter- and self- (nonoverlapping peptides) cross-links identified in new and old complex II mapped to high-resolution protein structures (Protein Data Bank [PDB] accession no. 1ZOY). No cross-links were identified between old-old or old-new complex II proteins. Yellow structure illustrates complex build from old, 14N, peptides; blue structure illustrates complex build from new, 15N peptides. (C) Inter- and self- (nonoverlapping peptides) cross-links identified in new and old complex V mapped to high-resolution protein structures (PDB accession no. 6J5I). Only two cross-links of mixed isotypes were identified (teal). (D) PDB structure of complexes I (accession no. 6G2J), III (accession no. 1BGY), and IV (accession no. 3X2Q), which can assemble into respirasome supercomplexes. Inter- and self- (nonoverlapping peptides) cross-links between new or old proteins, as well as intercomplex cross-links, are shown. Blue indicates cross-links between two new peptides, and teal represents cross-links between new and old peptides. Dotted line represents a cross-link between complex I and IV, which is outside of cross-linker constraints. Maps were generated in XIView. n = 2 mice. See also Fig. S6.
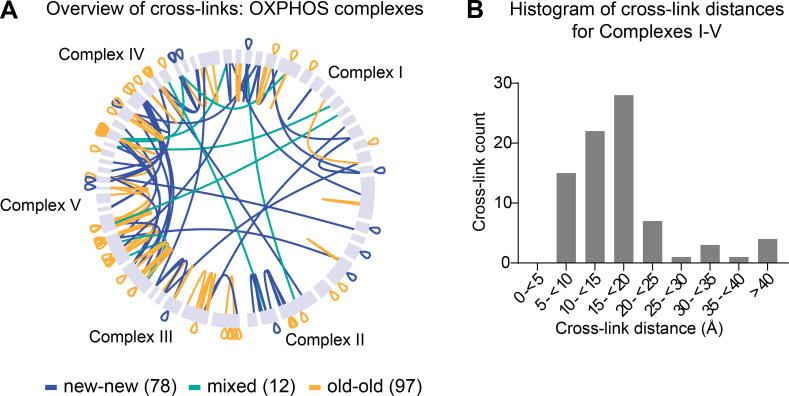
Next, we mapped the identified new-new, old-old, and mixed cross-links onto available high-resolution structures of complex I (Agip et al., 2018), II (Sun et al., 2005), III (Iwata et al., 1998), IV (Yano et al., 2015), and V (Gu et al., 2019; Fig. 5, B–D). First, we found that of the 81 cross-links that mapped within these structures, 77 had a corresponding distance of <40 Å (Merkley et al., 2014), indicating that these cross-links can be regarded as structurally confirmed (Fig. S6 B). The four long-range cross-links may either be false positives or indicate alternative interactions between subunits or complexes. Also, none of the cross-links identified within complexes transverse the IMM, indicating that cross-linked mitochondria were intact. Cross-links mapped to the high-resolution structures further illustrate the limited subunit exchange within complexes, since identified cross-links formed nearly exclusively within peptides of the same nitrogen build. We also identified three intercomplex links between complexes I, III, and IV, which, as mentioned above, form the respirasome supercomplex. Two of the links were mixed, i.e., formed between one old and one new peptide, and one was formed between two new peptides (Fig. 5 D). Altogether, these data suggest that OXPHOS complexes, once assembled, can remain intact throughout their lifetimes, at least in the heart. In contrast, cross-links formed between complexes and between other proteins within mitochondria were predominantly new-new or mixed, with very few cross-links identified between old OXPHOS and other proteins. Therefore, the spatial preservation of mt-LLPs, as shown by our cross-linking experiments, suggests that mt-LLPs are not scattered through the mitochondrion protein network, but rather exist in close proximity for months.
Figure S6.
Assembled OXPHOS complexes are copreserved with limited subunit exchange for months. Related to Fig. 5. (A) Overview of inter- and self (overlapping and nonoverlapping) cross-links between subunits of OXPHOS complexes. (B) Histogram of cross-link distances shown in angstroms extracted from cross-links mapped to high-resolution structures of complexes I–V. A and B, n = 2 mice.
Taken together, our discovery-based investigation uncovered that a subset of the mitochondrial proteome is long lived in tissues enriched in terminally differentiated, long-lived cells. Unexpectedly, we found that mitochondrial LLPs are concentrated at the IMM and persist for months with limited exchange, potentially due to compartmentalization and restricted spatial dissociation. While lifelong stability of NUPs and chromatin-associated proteins in postmitotic cells has been previously reported, this is the first study examining an analogous cytosolic phenomenon. The exchange and renewal of nuclear envelope–associated proteins occurs concurrently with envelope membrane dispersal during cell division (Horbay and Bilyy, 2016; Mishra and Chan, 2014). However, mitochondria are highly dynamic organelles that are remodeled through fission, fusion, biogenesis, and mitophagy in a mitosis-dependent and -independent manner. Our findings provide a new example of a nonnuclear compartment in which, in spite of continual remodeling, a subset of protein constituents persist for months. We propose that like long-lived NUPs that provide structural stability to the core of nuclear pore complexes, the exceptional longevity of mt-LLPs plays an essential role in the long-term stabilization of the mitochondrial cristae.
In summary, our study identifies long-lived mitochondrial proteins as previously underappreciated pillars of mitochondrial architecture, standing guard to long-lasting cristae integrity. Future research into the mechanisms responsible for maintaining mt-LLPs will be crucial to untangle their contribution to the mitochondrial network homeostasis and survival of long-lived cells.
Materials and methods
Ethics statement
All animal care and experiment protocols in this study were conducted under the guidelines set by the National Institutes of Health Guide for the Care and Use of Laboratory Animals handbook. The protocols were reviewed and approved by the Animal Care and Use Committee of Northwestern University (protocols IS00004793 and IS00010858).
Materials and reagents
Antibodies used were total OXPHOS Blue Native (BN) WB antibody cocktail (mouse monoclonal, 1:1,000; ab110412; Abcam; RRID:AB_2847807), anti-VDAC1 (rabbit polyclonal, 1:500, ab34726; Abcam; RRID:AB_778788), anti-VCP (rabbit polyclonal, 1:1,000; 111740; Abcam; RRID:AB_10861709), goat anti-rabbit IgG HRP (1:10,000; 32260; Thermo Fisher Scientific; RRID:AB_1965959), and goat anti-mouse IgG HRP (1:10,000; 32230; Thermo Fisher Scientific; RRID:AB_1965958). BN-PAGE reagents were prepared using an Invitrogen NativePAGE Sample Prep Kit (BN2008), run on 3–12% Bis-Tris NativePAGE gels (BN1001; Invitrogen); running buffers were prepared using NativePAGE running buffer kit (BN2007; Invitrogen); and gels were transferred using NuPAGE transfer buffer (NP00061; Invitrogen) onto polyvinylidene difluoride membranes (1620264; Bio-Rad). Other reagents were Halt Phosphatase Inhibitor cocktail (1:100; Thermo Fisher Scientific; 78420), DSSO (A33545; Thermo Fisher Scientific), DMSO (D2650; Sigma-Aldrich), sequencing-grade modified trypsin (V5280; Promega), ProteaseMAX (V2072; Promega), Tris(2-carboxyethyl)phosphine (TCEP; C4706; Sigma-Aldrich), iodoacetamide (IAA; I1149; Sigma-Aldrich), Lys-C Protease (PI90307; Thermo Fisher Scientific), and DTT (DSD11000; DOT Scientific).
Stable 15N isotope labeling of mammals
15N isotope labeling was accomplished essentially as previously described (Savas et al., 2016; Savas et al., 2012). Briefly, FVB mice of both sexes were purchased immediately after weaning. After a week-long acclimation, mice were placed on ad libitum 15N diet (MF-SpirulinaN-IR; Cambridge Isotopes). For the dynamic labeling experiments (single-generation), mice stayed on the 15N diet for 4 mo and were sacrificed. For pulse-chase 15N labeling (two-generation), an unlabeled male was placed with a labeled female and removed once the female was pregnant. The pups born to the labeled female remained on 15N milk until weaning (28 d) and then were chased with 14N-feed diet for the indicated periods of time.
Tissue harvesting and mitochondria isolation
Mice were anesthetized with 3% isoflurane followed by acute decapitation. Indicated tissues were harvested, flash frozen in a dry ice/ethanol bath, and stored at −80°C. For data presented in Fig. 1, tissues were thawed on ice, finely chopped, and homogenized in Precellys24 in HEENK solution (10 mm HEPES, 1 mm EDTA, 1 mm EGTA, 10 mm NaCl, 150 mm KCl, pH 7.1, ∼300 mOsm/l with Halt protease inhibitor cocktail, and 1 mM PMSF; Liu et al., 2018). For experiments requiring purified mitochondria, indicated tissues were thawed on ice, and mitochondria were extracted according to the Miltenyi Biotec Mitochondria Extraction–Tissue Isolation Protocol (130-097-340). Mitochondria were further immune-isolated as described by Fecher et al. (2019), with minor modifications. In brief, extracted mitochondria were diluted in Separation Buffer (Miltenyi Biotec) and incubated with 50 µl microbeads coated with mouse IgG1 antibodies against Tom22 (130-096-946; Miltenyi Biotec) for 1 h at 4°C, with rotation. Mitochondria-microbeads were separated using an LC column (130-042-401; Miltenyi Biotec) in a magnetic QuadroMACS Separator (130-090-976; Miltenyi Biotec) according to the manufacturer’s instructions. Immuno-isolated mitochondria were pelleted by centrifugation for 3 min at 12,000 g, washed twice with HEENK buffer, and used in subsequent experiments.
BN-PAGE
BN-PAGE was performed according to a previously published protocol (Jha et al., 2016). Crude membrane fractions were thawed on ice and spun fast (9,000 g for 5 min at 4°C). Pellets were resuspended in native PAGE sample buffer cocktail with 8 g/g digitonin concentration and incubated on ice for 20 min. Solubilized mitochondria were spun (20,000 g for 10 min at 4°C), and supernatant was transferred into a new tube. Coomassie G-250 was added, such that the final G-250 concentration was one fourth of the detergent concentration. Samples were loaded into the NB-PAGE gels, the inner chamber was filled with dark cathode buffer, and clear anode buffer was used in the outside chamber. Gels were run at 150 V for 30 min, at which time the buffer in the inner chamber was switched to clear running buffer, and the gel was run at 250 V for an additional 60 min. In all cases, electrophoresis was performed on ice.
Immunoblotting
BN-PAGE gels were transferred onto polyvinylidene fluoride membrane (70 min at 120 V), using a Bio-Rad TransBlot module and NuPAGE transfer buffer. Membranes were washed in 8% acetic acid solution (5 min with gentle shaking), washed with water (2 × 5 min), and air dried. Dried membranes were incubated in 100% methanol (3 × 5 min) to remove the Coomassie blue and washed with water (3 × 5 min). SDS-PAGE membranes and BN-PAGE membranes were blocked (10% milk for 60 min) and incubated with primary and HRP-coupled secondary antibodies (1% milk for 60 min each). Membranes were washed in TBS, 1% Tween in between and after antibody incubation steps (3 × 5 min). Immunoblots were imaged on a Bio-Rad Chemidoc XRS system.
Cross-linking
Cross-linking was performed according to a previously published protocol, with some modifications (Liu et al., 2018). Immunoisolated mitochondria were resuspended in HEENK plus 1 mM DSSO (resuspended freshly in anhydrous DMSO to 50 mM) and incubated at RT for 1 h with gentle rotation. Control samples were incubated with an equal volume of DMSO. The reaction was stopped with Tris (final concentration 100 mM, pH 8.0), and spun fast (9,000 g for 5 min at 4°C). Pellet was resuspended in 1 ml HEENK with 100 mM Tris, pH 8.0, and incubated for 10 min at RT. Cross-linked mitochondria were split into two aliquots: 100 µl dedicated to silver stain and 900 µl for MS analysis. Samples were spun fast (9,000 g for 5 min at 4°C), and pellets were stored at −80°C.
MS sample preparation: Total homogenate fractions
Total homogenate samples were precipitated using the chloroform/methanol method, denatured with 8 M urea (in a 50-mM ammonium bicarbonate [ABC] vortex 1 h at RT), and processed with ProteaseMAX according to the manufacturer’s protocol. Samples were reduced with 5 mM Tris(2-carboxyethyl)phosphine (TCEP; vortexed for 1 h at RT), alkylated in the dark with 10 mM IAA (20 min), diluted with 50 mM ABC, and quenched with 25 mM TCEP. Samples were digested with sequencing-grade modified trypsin overnight at 37°C with shaking, spun down (15,000 g for 15 min at RT), placed in a new tube, and acidified with trifluoroacetic acid (TFA) to a final concentration of 0.1%. 25 µg of each sample was desalted with Pierce C18 spin columns (89873; Thermo Fisher Scientific) per the manufacturer’s instructions and dried down with vacuum centrifugation for future MS analysis.
MS sample preparation: Immuno-isolated mitochondria
Immuno-isolated mitochondria were resuspended in 20 mM Tris-HCl, pH 8.0, incubated with 4 mM DTT at 56°C for 30 min, and alkylated with 10 mM IAA for 20 min in the dark. Samples were digested with sequencing-grade modified trypsin overnight at 37°C with shaking, spun down (15,000 g for 15 min at RT), placed in a new tube, and acidified with TFA to a final concentration of 0.1%. 100 µg of digested and acidified sample was fractionated using a High pH Reversed-Phase Peptide Fractionation Kit (84868; Pierce). Fractions were step eluted in 300 µl buffer of increasing acetonitrile (ACN) concentrations with decreasing concentrations of triethylamine (0.1%) per the manufacturer. Samples were dried down with vacuum centrifugation for future MS analysis.
MS sample preparation: GeLC/MS on BN-PAGE gels
BN-PAGE gels were fixed in 40% methanol/10% acetic acid solution (10 min), destained in 8% acetic acid (30 min), and rinsed with ultrapure water (3 × 5 min). Gels were scanned using a Bio-Rad Chemidoc XRS system, and appropriate areas of the gel were cut out, chopped into 1 × 1–mm cubes, and processed for in-gel digestion. Gel pieces were incubated in 10 mM TCEP (in 50 mM ABC for 1 h at 37°C). Liquid was replaced by 50 mM IAA (in 50 mM ABC for 45 min at RT in the dark), followed by 50 mM TCEP (in 50 mM ABC for 30 min at RT). Gel pieces were washed with 50 mM ABC (3×) and digested with sequencing-grade modified trypsin (1 µg in 50 mM ABC, overnight at 37°C, with shaking). The following day, supernatant was collected into a new tube, and the gel pieces were subjected to three rounds of incubations with 50% ACN and 5% FA solution (30 min at RT, with shaking). Supernatant was collected after each incubation, combined, and dried down with vacuum centrifugation. Samples were resuspended in 0.5% TFA, desalted with Pierce C18 spin columns (89873; Thermo Fisher Scientific) per the manufacturer’s instructions, and dried down with vacuum centrifugation for future MS analysis.
MS sample preparation: cross-linking
Cross-linked mitochondria isolations were solubilized in 20 mM Tris-HCl, pH 8.0, reduced with 4 mM DTT (30 min at 56°C), alkylated with 8 mM IAA (30 min at RT in the dark), and sequentially proteolysed with Lys-C (4 h at 37°C; enzyme-to-protein ratio of 1:75 [wt/wt]) and trypsin (overnight at 37°C; enzyme-to-protein ratio of 1:100 [wt/wt]). Protein digests were spun down, acidified, and fractionated using a High pH Reversed-Phase Peptide Fractionation Kit. Fractions were step-eluted into eight fractions in 300 µl buffer of increasing ACN concentrations, with decreasing concentrations of triethylamine (0.1%) per the manufacturer. Samples were dried down with vacuum centrifugation for future MS analysis.
MS analysis
Dried samples were resuspended in 20 µl buffer A (94.875% H2O with 5% ACN and 0.125% FA) and 3 µg, as determined by microBCA assay (23235; Thermo Fisher Scientific), of each fraction or sample was loaded via autosampler with either Thermo EASY nLC 100 UPLC or UltiMate 3000 HPLC pump, onto a vented Pepmap 100, 75 µm × 2 cm, nanoViper trap column coupled to a nanoViper analytical column (Thermo Fisher Scientific) with a stainless steel emitter tip assembled on the Nanospray Flex Ion Source with a spray voltage of 2,000 V. A coupled Orbitrap Fusion was used to generate MS data. Buffer A contained 94.785% H2O with 5% ACN and 0.125% FA, and buffer B contained 99.875% ACN with 0.125% FA.
For total homogenate, mitochondria isolations, and GeLC/MS experiments, the MS parameters were as follows: ion transfer tube temp, 300°C; Easy-IC internal mass calibration; default charge state, 2; cycle time, 3 s; detector type set to Orbitrap; 60K resolution; wide quad isolation; mass range, normal; scan range, 300–1,500 m/z; maximum injection time, 50 ms; AGC (automatic gain control) target, 200,000; microscans, 1; S-lens RF level, 60; without source fragmentation; and datatype, positive and centroid; MIPS set as on; included charge states, 2–6 (reject unassigned); dynamic exclusion enabled, with n = 1 for 30- and 45-s exclusion duration at 10 ppm for high and low; precursor selection decision, most intense, top 20; isolation window, 1.6; scan range, auto normal; first mass, 110; and collision energy, 30%. For collision-induced dissociation (CID): detector type, ion trap; OT resolution, 30K; IT scan rate, rapid; maximum injection time, 75 ms; AGC target, 10,000; Q, 0.25; and inject ions for all available parallelizable time.
For cross-linking experiments, the CID-MS2-MS3 acquisition strategy was used, and the above MS parameters were modified as follows: for MS scans, the scan range was set to 300–1800 m/z, with AGC target set to 4.0 × 105. For the MS/MS scans, the resolution was set to 30,000, AGC target was set to 5.0 × 104, and maximum injection time was set to 100 ms. We limited selection for MS/MS to peptides with charge states 2–8. MS1 and MS2 scans were acquired in the Orbitrap, whereas MS3 scans were acquired in the ion trap. Mass difference–dependent CID-MS3 acquisitions were triggered if a mass difference of 31.9721 D was observed in the CID-MS2 spectrum; scans were performed on the more intense ion in the pair, allowing for different ion charge states. Precursor ion exclusion was set to 5 ppm high and low. The normalized collision energy was set to 25% for CID-MS2 scans and 35% for CID-MS3 scans.
For the total homogenate experiments, the chromatographic run was 4.5 h in total, per fraction, with the following profile of buffer B: 2% for 7 min, 2–7% for 1 min, 7–10% for 5 min, 10–25% for 160 min, 25–33% for 40 min, 33–50% for 7 min, 50–95% for 5 min, 95% for 15 min, and back to 2% for the remaining 30 min. For the GeLC/MS and isolated mitochondria experiments, the chromatographic run was 2.5 h in total with the following profile of buffer B: 2–8% for 6 min, 8–24% for 64 min, 24–36% for 20 min, 36–55% for 10 min, 55–95% for 10 min, 95% for 10 min, and back to 2% for the remaining 30 min. For the cross-linking experiments, the chromatographic run was 2.5 h per fraction, with the following profile of buffer B: 2–8% for 6 min, 8–30% for 39 min, 30–36% for 25 min, 36–50% for 30 min, 50–95% for 10 min, 95% for 10 min, and back to 2% for the remaining 30 min.
MS data analysis and quantification
Protein identification/quantification and analysis were performed with the Integrated Proteomics Pipeline–IP2 (Integrated Proteomics Applications) using ProLuCID (Eng et al., 1994; Xu et al., 2015), DTASelect2 (Cociorva et al., 2007; Tabb et al., 2002), Census, and QuantCompare. Spectral raw files were extracted into MS1 and MS2 files (and MS3, when appropriate) using RawConverter v1.0.0.0. The tandem mass spectra were searched against databases for their relevant species (UniProt mouse protein database downloaded on March 25, 2014, and MitoCarta3.0 database for cross-linking experiments; Rath et al., 2021). Searched spectra were matched to sequences using the ProLuCID/SEQUEST algorithm (ProLuCID v3.1) with 50 ppm peptide mass tolerance for precursor ions and 600 ppm for fragment ions. ProLuCID searches included all fully and half-tryptic peptide candidates that fell within the mass tolerance window and had unlimited miscleavages. Carbamidomethylation (+57.02146 D) of cysteine was considered as a static modification. For cross-linking experiments, the following differential modifications were added: +86.98263 D and +54.01056 D, both exclusively on lysine residues.
Peptide/spectrum matches were assessed in DTASelect2 using the cross-correlation score (XCorr) and normalized difference in cross-correlation scores (DeltaCN). For total homogenate, isolated mitochondria, and cortex GeLC/MS experiments, each protein identified was required to have a minimum of three peptides (−p 3) of minimal length of six amino acid residues. For cross-linking and heart GeLC/MS experiments, each protein identified was required to have a minimum of one peptide (−p 1) of minimal length of six amino acid residues. The false discovery rate (FDR) was set to 1% at the protein level for all experiments. Peptide probabilities and FDR were calculated based on a target/decoy database containing the reversed sequences of all the proteins appended to the target database (Peng et al., 2003). Each dataset was searched twice, once against light (14N) and then against heavy (15N) protein databases, as described previously (Savas et al., 2012). In the light searches, all the amino acid residues were considered to contain only 14N nitrogen, while in the heavy searches, all the amino acid residues were considered to contain only 15N nitrogen. After the results from ProLuCID were filtered using DTASelect2, the assembled search result file was used to obtain quantitative ratios between 14N and 15N using Census software (MacCoss et al., 2003; Park et al., 2008).
LLPs were identified as previously described, with modifications (Savas et al., 2012). Briefly, for a protein to be considered long lived, it had to be identified in our heavy/light search by at least one long-lived peptide (14N-peptides for dynamic labeling experiments or 15N-peptides for two-generation labeling experiments). Peptide ratio measurements were filtered in Census based on a correlation threshold, and only peptides with correlation coefficients >0.5 were used for further analysis. For singleton analysis, we required the 14N/15N ratio to be >5.0 and the threshold score to be >0.5. Identified peptides were further filtered based on their average peptide enrichment, which was set to 0.9, and peptide profile score, which was set to 0.8. Proteins were identified as long lived only if they had more than three long-lived peptides that passed the above filtering (except for cross-linking experiments, in which one peptide was required). FA values were calculated according to the following formula: for 15N values, FA = 100 × [1/(1 + AR)]; for 14N values, FA = 100 – {100 × [1/(1 + AR)]}, where AR is area ratio.
For the identification of cross-linked peptides, raw MS1/MS2/MS3 files were used as input to identify cross-links in IP2. All MS2 and MS3 spectra were searched twice, once against the light (14N) and then against the heavy (15N) protein database, as described above (MitoCarta3.0; Rath et al., 2021). Only peptides with differential modifications were considered (+86.982635 D and +54.01056 D, both on lysine residues). Peptide lists were extracted and sorted according to the scan number. To be considered cross-linked, at least one MS2 fragment ion (per peptide in the putative pair) had to be selected and identified (i.e., α-S or α-L and β-S or β-L), in a consecutive MS3 scan. MS3, MS2 and MS1 scans were extracted from raw files using RawQuant (Kovalchik et al., 2018), a Python package, and cross-referenced with search files to confirm that all MS3 scans within putative cross-links mapped to the same MS1 scan. Cross-links were reported at a protein 1% FDR in each fraction based on a target–decoy calculation strategy. The NCBI standalone Blast program (Camacho et al., 2009; v2.11) was used to align all identified cross-linked peptides against a protein database created from Uniport database. The Blast alignment output for each peptide and the position at modified amino acids in the peptides was used to find the exact amino acid position where the cross-link occurred between proteins. Cross-link network maps were generated using xiView (Combe et al., 2015).
PDB structures
The following PDB structures were used: complex I, 6G2J; complex II, 1ZOY; complex III, 3X2Q; complex IV, 1BGY; and complex V, 6J5I. Molecular drawings were made using Pymol (Schrodinger).
GO analysis
GO analysis was performed using Pantherdb (Mi et al., 2019). The query was defined as proteins identified as long lived in the analyzed tissue (based on 14N-peptide identification), and the reference was defined as all proteins identified in the same tissue analyzed (14N- and 15N-peptide identification).
Statistical analysis
Statistical analyses were conducted using GraphPad Prism, v8. Multiple group comparisons were made using Kruskal–Wallis ANOVA with Tukey’s multiple comparisons test between each group. Data were considered significant when P < 0.05 (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P <0.0001). Normality of data distribution was determined by Shapiro–Wilk test.
Online supplemental material
Fig. S1 provides additional MS data showing that mitochondrial proteins are exceptionally long lived in tissues containing postmitotic cells. Fig. S2 displays additional evidence of deep analytical coverage of the mitochondrial proteome and shows that LLPs are preferentially associated with the mitochondrial inner membrane in an independent 15N pulse-chase experiment. Fig. S3 provides additional evidence that long-lived mitochondrial proteins are associated with the cristae. Fig. S4 shows that long-lived subunits of OXPHOS complexes integrate into individual and higher-order complexes based on BN-PAGE with MS analysis. Fig. S5 provides additional evidence from cross-linking analysis that mt-LLPs have limited mixing and exchange over months. Table S1 shows the significantly overrepresented cellular component terms from GO analysis using a query of specific LLPs versus all protein identifications. Table S2 provides a summary of mitochondrial proteins identified in 15N-pulse–labeled mouse cortex, heart, and spleen extracts. Table S3provides a summary of mitochondrial proteins identified in 15N-pulse–chase time courses. Table S4shows the results from cross-linking experiments of immuno-captured mitochondria from 15N-pulse–labeled cortical and heart tissues.
Supplementary Material
lists significantly overrepresented cellular component terms from GO analysis using a query of specific LLPs versus all protein identifications (reference list) in a given tissue.
is a summary of mitochondrial proteins identified in 15N-pulse–labeled mouse cortex, heart, and spleen extracts.
is a summary of mitochondrial proteins identified in 15N-pulse–chase time course.
is a summary of results from cross-linking experiments of immunocaptured mitochondria from 15N-pulse–labeled cortical and heart tissues.
Acknowledgments
We would like to thank the members of the Savas laboratory, Yvette Wong, Ge Gao, Jason Vevea, Erin Seifert, and members of the Francesca Duncan laboratory for discussion and comments related to this manuscript.
E. Bomba-Warczak was supported by the National Institutes of Health (F32 NS106812, National Institute of Neurological Disorders and Stroke); this research was also supported by R21HD098498 (National Institute of Child Health and Human Development), R21NS107761 (National Institute of Neurological Disorders and Stroke), and R01AG061787 (National Institute of Aging) to J.N. Savas. This work used resources of the Northwestern University Structural Biology Facility, which is generously supported by a National Cancer Institute CCSG P30 CA060553 grant awarded to the Robert H. Lurie Comprehensive Cancer Center.
The authors declare no competing financial interests.
Author contributions: E. Bomba-Warczak and J.N. Savas conceptualized the experiments and cowrote the manuscript. E. Bomba-Warczak performed data curation, analysis, validation, visualization, and methodology. T.J. Hark and S.L. Edassery performed data curation, analysis, and validation. E. Bomba-Warczak and J.N. Savas acquired funding for the research.
Data availability
All data generated or analyzed during this study are included in the manuscript and supporting files. Raw MS data has also been deposited at MassIVE under accession number MSV000087712 and Proteome Exchange under accession number PXD027060.
References
- Agip, A.A., Blaza J.N., Bridges H.R., Viscomi C., Rawson S., Muench S.P., and Hirst J.. 2018. Cryo-EM structures of complex I from mouse heart mitochondria in two biochemically defined states. Nat. Struct. Mol. Biol. 25:548–556. 10.1038/s41594-018-0073-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callegari, S., Müller T., Schulz C., Lenz C., Jans D.C., Wissel M., Opazo F., Rizzoli S.O., Jakobs S., Urlaub H., et al. 2019. A MICOS-TIM22 Association Promotes Carrier Import into Human Mitochondria. J. Mol. Biol. 431:2835–2851. 10.1016/j.jmb.2019.05.015 [DOI] [PubMed] [Google Scholar]
- Camacho, C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., and Madden T.L.. 2009. BLAST+: architecture and applications. BMC Bioinformatics. 10:421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambridge, S.B., Gnad F., Nguyen C., Bermejo J.L., Krüger M., and Mann M.. 2011. Systems-wide proteomic analysis in mammalian cells reveals conserved, functional protein turnover. J. Proteome Res. 10:5275–5284. 10.1021/pr101183k [DOI] [PubMed] [Google Scholar]
- Campello, S., and Scorrano L.. 2010. Mitochondrial shape changes: orchestrating cell pathophysiology. EMBO Rep. 11:678–684. 10.1038/embor.2010.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham, D.E.2007. Calcium signaling. Cell. 131:1047–1058. 10.1016/j.cell.2007.11.028 [DOI] [PubMed] [Google Scholar]
- Cociorva, D., Tabb D.L., and Yates J.R.. 2007. Validation of tandem mass spectrometry database search results using DTASelect. Curr. Protoc. Bioinformatics. Chapter 13:4. [DOI] [PubMed] [Google Scholar]
- Cogliati, S., Frezza C., Soriano M.E., Varanita T., Quintana-Cabrera R., Corrado M., Cipolat S., Costa V., Casarin A., Gomes L.C., et al. 2013. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell. 155:160–171. 10.1016/j.cell.2013.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliati, S., Enriquez J.A., and Scorrano L.. 2016. Mitochondrial Cristae: Where Beauty Meets Functionality. Trends Biochem. Sci. 41:261–273. 10.1016/j.tibs.2016.01.001 [DOI] [PubMed] [Google Scholar]
- Combe, C.W., Fischer L., and Rappsilber J.. 2015. xiNET: cross-link network maps with residue resolution. Mol. Cell. Proteomics. 14:1137–1147. 10.1074/mcp.O114.042259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras, L., Drago I., Zampese E., and Pozzan T.. 2010. Mitochondria: the calcium connection. Biochim. Biophys. Acta. 1797:607–618. 10.1016/j.bbabio.2010.05.005 [DOI] [PubMed] [Google Scholar]
- D’Angelo, M.A., Raices M., Panowski S.H., and Hetzer M.W.. 2009. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 136:284–295. 10.1016/j.cell.2008.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, C., Wu Z., Huang L., Wang Y., Xue J., Chen S., Deng Z., Wang L., Song Z., and Chen S.. 2015. Mitofilin and CHCHD6 physically interact with Sam50 to sustain cristae structure. Sci. Rep. 5:16064. 10.1038/srep16064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng, J.K., McCormack A.L., and Yates J.R.. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976–989. 10.1016/1044-0305(94)80016-2 [DOI] [PubMed] [Google Scholar]
- Fecher, C., Trovò L., Müller S.A., Snaidero N., Wettmarshausen J., Heink S., Ortiz O., Wagner I., Kühn R., Hartmann J., et al. 2019. Cell-type-specific profiling of brain mitochondria reveals functional and molecular diversity. Nat. Neurosci. 22:1731–1742. 10.1038/s41593-019-0479-z [DOI] [PubMed] [Google Scholar]
- Fornasiero, E.F., Mandad S., Wildhagen H., Alevra M., Rammner B., Keihani S., Opazo F., Urban I., Ischebeck T., Sakib M.S., et al. 2018. Precisely measured protein lifetimes in the mouse brain reveal differences across tissues and subcellular fractions. Nat. Commun. 9:4230. 10.1038/s41467-018-06519-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezza, C., Cipolat S., Martins de Brito O., Micaroni M., Beznoussenko G.V., Rudka T., Bartoli D., Polishuck R.S., Danial N.N., De Strooper B., and Scorrano L.. 2006. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 126:177–189. 10.1016/j.cell.2006.06.025 [DOI] [PubMed] [Google Scholar]
- Glynn, S.E.2017. Multifunctional Mitochondrial AAA Proteases. Front. Mol. Biosci. 4:34. 10.3389/fmolb.2017.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, A.L.2003. Protein degradation and protection against misfolded or damaged proteins. Nature. 426:895–899. 10.1038/nature02263 [DOI] [PubMed] [Google Scholar]
- Gu, J., Zhang L., Zong S., Guo R., Liu T., Yi J., Wang P., Zhuo W., and Yang M.. 2019. Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1. Science. 364:1068–1075. 10.1126/science.aaw4852 [DOI] [PubMed] [Google Scholar]
- Halliwell, B.2001. Role of free radicals in the neurodegenerative diseases: therapeutic implications for antioxidant treatment. Drugs Aging. 18:685–716. 10.2165/00002512-200118090-00004 [DOI] [PubMed] [Google Scholar]
- Hark, T.J., Rao N.R., Castillon C., Basta T., Smukowski S., Bao H., Upadhyay A., Bomba-Warczak E., Nomura T., O’Toole E.T., et al. 2021. Pulse-Chase Proteomics of the App Knockin Mouse Models of Alzheimer’s Disease Reveals that Synaptic Dysfunction Originates in Presynaptic Terminals. Cell Syst. 12:141–158.e9. 10.1016/j.cels.2020.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horbay, R., and Bilyy R.. 2016. Mitochondrial dynamics during cell cycling. Apoptosis. 21:1327–1335. 10.1007/s10495-016-1295-5 [DOI] [PubMed] [Google Scholar]
- Iwata, S., Lee J.W., Okada K., Lee J.K., Iwata M., Rasmussen B., Link T.A., Ramaswamy S., and Jap B.K.. 1998. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science. 281:64–71. 10.1126/science.281.5373.64 [DOI] [PubMed] [Google Scholar]
- Jha, P., Wang X., and Auwerx J.. 2016. Analysis of Mitochondrial Respiratory Chain Supercomplexes Using Blue Native Polyacrylamide Gel Electrophoresis (BN-PAGE). Curr. Protoc. Mouse Biol. 6:1–14. 10.1002/9780470942390.mo150182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao, A., Chiu C.L., Vellucci D., Yang Y., Patel V.R., Guan S., Randall A., Baldi P., Rychnovsky S.D., and Huang L.. 2011. Development of a novel cross-linking strategy for fast and accurate identification of cross-linked peptides of protein complexes. Mol. Cell. Proteomics. 10:M110.002170. 10.1074/mcp.M110.002212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchik, K.A., Moggridge S., Chen D.D.Y., Morin G.B., and Hughes C.S.. 2018. Parsing and Quantification of Raw Orbitrap Mass Spectrometer Data Using RawQuant. J. Proteome Res. 17:2237–2247. 10.1021/acs.jproteome.8b00072 [DOI] [PubMed] [Google Scholar]
- Liu, F., Lössl P., Rabbitts B.M., Balaban R.S., and Heck A.J.R.. 2018. The interactome of intact mitochondria by cross-linking mass spectrometry provides evidence for coexisting respiratory supercomplexes. Mol. Cell. Proteomics. 17:216–232. 10.1074/mcp.RA117.000470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCoss, M.J., Wu C.C., Liu H., Sadygov R., and Yates J.R. III. 2003. A correlation algorithm for the automated quantitative analysis of shotgun proteomics data. Anal. Chem. 75:6912–6921. 10.1021/ac034790h [DOI] [PubMed] [Google Scholar]
- Merkley, E.D., Rysavy S., Kahraman A., Hafen R.P., Daggett V., and Adkins J.N.. 2014. Distance restraints from crosslinking mass spectrometry: mining a molecular dynamics simulation database to evaluate lysine-lysine distances. Protein Sci. 23:747–759. 10.1002/pro.2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi, H., Muruganujan A., Ebert D., Huang X., and Thomas P.D.. 2019. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47(D1):D419–D426. 10.1093/nar/gky1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misgeld, T., and Schwarz T.L.. 2017. Mitostasis in Neurons: Maintaining Mitochondria in an Extended Cellular Architecture. Neuron. 96:651–666. 10.1016/j.neuron.2017.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, P., and Chan D.C.. 2014. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol. 15:634–646. 10.1038/nrm3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls, D.G.2002. Mitochondrial function and dysfunction in the cell: its relevance to aging and aging-related disease. Int. J. Biochem. Cell Biol. 34:1372–1381. 10.1016/S1357-2725(02)00077-8 [DOI] [PubMed] [Google Scholar]
- Ott, C., Ross K., Straub S., Thiede B., Götz M., Goosmann C., Krischke M., Mueller M.J., Krohne G., Rudel T., and Kozjak-Pavlovic V.. 2012. Sam50 functions in mitochondrial intermembrane space bridging and biogenesis of respiratory complexes. Mol. Cell. Biol. 32:1173–1188. 10.1128/MCB.06388-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.K., Venable J.D., Xu T., and Yates J.R. III. 2008. A quantitative analysis software tool for mass spectrometry-based proteomics. Nat. Methods. 5:319–322. 10.1038/nmeth.1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paumard, P., Vaillier J., Coulary B., Schaeffer J., Soubannier V., Mueller D.M., Brèthes D., di Rago J.P., and Velours J.. 2002. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 21:221–230. 10.1093/emboj/21.3.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., Elias J.E., Thoreen C.C., Licklider L.J., and Gygi S.P.. 2003. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2:43–50. 10.1021/pr025556v [DOI] [PubMed] [Google Scholar]
- Pickles, S., Vigié P., and Youle R.J.. 2018. Mitophagy and Quality Control Mechanisms in Mitochondrial Maintenance. Curr. Biol. 28:R170–R185. 10.1016/j.cub.2018.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Cabrera, R., Quirin C., Glytsou C., Corrado M., Urbani A., Pellattiero A., Calvo E., Vázquez J., Enríquez J.A., Gerle C., et al. 2018. The cristae modulator Optic atrophy 1 requires mitochondrial ATP synthase oligomers to safeguard mitochondrial function. Nat. Commun. 9:3399. 10.1038/s41467-018-05655-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath, S., Sharma R., Gupta R., Ast T., Chan C., Durham T.J., Goodman R.P., Grabarek Z., Haas M.E., Hung W.H.W., et al. 2021. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 49(D1):D1541–D1547. 10.1093/nar/gkaa1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas, J.N., Toyama B.H., Xu T., Yates J.R. III, and Hetzer M.W.. 2012. Extremely long-lived nuclear pore proteins in the rat brain. Science. 335:942. 10.1126/science.1217421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas, J.N., Park S.K., and Yates J.R. III. 2016. Proteomic Analysis of Protein Turnover by Metabolic Whole Rodent Pulse-Chase Isotopic Labeling and Shotgun Mass Spectrometry Analysis. Methods Mol. Biol. 1410:293–304. 10.1007/978-1-4939-3524-6_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger, H., and Pfeiffer K.. 2000. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19:1777–1783. 10.1093/emboj/19.8.1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorrano, L., Ashiya M., Buttle K., Weiler S., Oakes S.A., Mannella C.A., and Korsmeyer S.J.. 2002. A distinct pathway remodels mitochondrial cristae and mobilizes cytochrome c during apoptosis. Dev. Cell. 2:55–67. 10.1016/S1534-5807(01)00116-2 [DOI] [PubMed] [Google Scholar]
- Soubannier, V., McLelland G.L., Zunino R., Braschi E., Rippstein P., Fon E.A., and McBride H.M.. 2012. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr. Biol. 22:135–141. 10.1016/j.cub.2011.11.057 [DOI] [PubMed] [Google Scholar]
- Strauss, M., Hofhaus G., Schröder R.R., and Kühlbrandt W.. 2008. Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 27:1154–1160. 10.1038/emboj.2008.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, F., Huo X., Zhai Y., Wang A., Xu J., Su D., Bartlam M., and Rao Z.. 2005. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell. 121:1043–1057. 10.1016/j.cell.2005.05.025 [DOI] [PubMed] [Google Scholar]
- Szczepanowska, K., and Trifunovic A.. 2021. Tune instead of destroy: How proteolysis keeps OXPHOS in shape. Biochim. Biophys. Acta Bioenerg. 1862:148365. 10.1016/j.bbabio.2020.148365 [DOI] [PubMed] [Google Scholar]
- Szczepanowska, K., Senft K., Heidler J., Herholz M., Kukat A., Höhne M.N., Hofsetz E., Becker C., Kaspar S., Giese H., et al. 2020. A salvage pathway maintains highly functional respiratory complex I. Nat. Commun. 11:1643. 10.1038/s41467-020-15467-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb, D.L., McDonald W.H., and Yates J.R. III. 2002. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1:21–26. 10.1021/pr015504q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama, B.H., Savas J.N., Park S.K., Harris M.S., Ingolia N.T., Yates J.R. III, and Hetzer M.W.. 2013. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell. 154:971–982. 10.1016/j.cell.2013.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott, R.J.W., Schey K.L., and Friedrich M.G.. 2016. Old Proteins in Man: A Field in its Infancy. Trends Biochem. Sci. 41:654–664. 10.1016/j.tibs.2016.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varanita, T., Soriano M.E., Romanello V., Zaglia T., Quintana-Cabrera R., Semenzato M., Menabò R., Costa V., Civiletto G., Pesce P., et al. 2015. The OPA1-dependent mitochondrial cristae remodeling pathway controls atrophic, apoptotic, and ischemic tissue damage. Cell Metab. 21:834–844. 10.1016/j.cmet.2015.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, D.M., Segawa M., Kondadi A.K., Anand R., Bailey S.T., Reichert A.S., van der Bliek A.M., Shackelford D.B., Liesa M., and Shirihai O.S.. 2019. Individual cristae within the same mitochondrion display different membrane potentials and are functionally independent. EMBO J. 38:e101056. 10.15252/embj.2018101056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, T., Park S.K., Venable J.D., Wohlschlegel J.A., Diedrich J.K., Cociorva D., Lu B., Liao L., Hewel J., Han X., et al. 2015. ProLuCID: An improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J. Proteomics. 129:16–24. 10.1016/j.jprot.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, N., Muramoto K., Mochizuki M., Shinzawa-Itoh K., Yamashita E., Yoshikawa S., and Tsukihara T.. 2015. X-ray structure of cyanide-bound bovine heart cytochrome c oxidase in the fully oxidized state at 2.0 Å resolution. Acta Crystallogr. F Struct. Biol. Commun. 71:726–730. 10.1107/S2053230X15007025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle, R.J., and Narendra D.P.. 2011. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12:9–14. 10.1038/nrm3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists significantly overrepresented cellular component terms from GO analysis using a query of specific LLPs versus all protein identifications (reference list) in a given tissue.
is a summary of mitochondrial proteins identified in 15N-pulse–labeled mouse cortex, heart, and spleen extracts.
is a summary of mitochondrial proteins identified in 15N-pulse–chase time course.
is a summary of results from cross-linking experiments of immunocaptured mitochondria from 15N-pulse–labeled cortical and heart tissues.
Data Availability Statement
All data generated or analyzed during this study are included in the manuscript and supporting files. Raw MS data has also been deposited at MassIVE under accession number MSV000087712 and Proteome Exchange under accession number PXD027060.