Abstract
Stochastic gains and losses of DNA methylation at CG dinucleotides are a frequent occurrence in plants. These spontaneous “epimutations” occur at a rate that is 100,000 times higher than the genetic mutation rate, are effectively neutral at the genome-wide scale and are stably inherited across mitotic and meiotic cell divisions. Mathematical models have been extraordinarily successful at describing how epimutations accumulate in plant genomes over time, making this process one of the most predictable epigenetic phenomena to date. Here, we propose that their high rate and effective neutrality make epimutations a powerful new molecular clock for timing evolutionary events of the recent past, as well as for age-dating of long-lived perennials such as trees.
Keywords: Molecular clock, epimutations, DNA methylation, phylogenetics, aging, evolution
Spontaneous epimutations: from nuisance to utility
DNA cytosine methylation (mC) is a conserved base modification in eukaryotes. It contributes mainly to the silencing of transposable elements (TEs) and repeat sequences, but is also found in some constitutively expressed genes. Plant methylomes (see Glossary) are remarkably stable across development [1], environmental conditions [2–7] and generations [8–13]. The faithful maintenance of mC is carried out by a number of designated pathways that are well characterized at the molecular level [14]. However, mC fidelity is not perfect. Analogous to DNA mutations, mistakes in the maintenance of mC can arise stochastically [12,13], both at the level of individual cytosines [12,13,15,16] as well as at the level of larger regions (i.e. clusters of cytosines) [10,12,17,18]. This phenomenon has been termed “spontaneous epimutation” [19]. Once acquired, these stochastic changes are often stably inherited across mitotic cell divisions, and even pass through the germline to subsequent generations. Estimates in a number of plant species indicate that the rate of spontaneous epimutations is orders of magnitude higher than the genetic mutation rate per unit time [15–17]. Hence, epigenetic variation in plants arises much more rapidly than genetic variation.
A handful of experimental studies have been able to link spontaneous epimutations with heritable morphological and developmental traits [20–27]. These observations continue to generate much excitement, because they point to an alternative molecular substrate for adaptive evolution [28] and possibly also for selective breeding [29]. But reported associations between heritable epimutations and phenotypes remain scarce. This shortage of evidence suggests that the vast majority of these events are functionally inconsequential, and thus irrelevant from an evolutionary and agricultural perspective [30]. Multigenerational surveys of mC corroborate this concern: Estimates show that the accumulation of spontaneous epimutations is effectively neutral at the genome-wide scale [15–18]. In light of this, it seems tempting to dismiss these stochastic events as a nuisance and to treat them as mere “molecular noise” in plant genomic studies.
Here we offer an alternative perspective. We propose that it is precisely their effective neutrality and high stochastic rate that make spontaneous epimutations a powerful new tool for plant biology: they define a fast-ticking molecular clock, which can be used to address a number of important questions in phylogenetics, population genetics and developmental biology. This clock’s fast tick rate offers a unique window into the recent past, a segment of time that has been largely inaccessible to classical DNA mutation clocks. Applications include the dating of recent lineage divergence, inference of demographic changes, and the age-estimation of long-lived perennials such as trees. In this opinion paper, we summarize the molecular basis and clock-like properties of spontaneous epimutations and discuss how one can exploit these properties in a modeling framework.
The molecular basis and somatic origin of epimutations
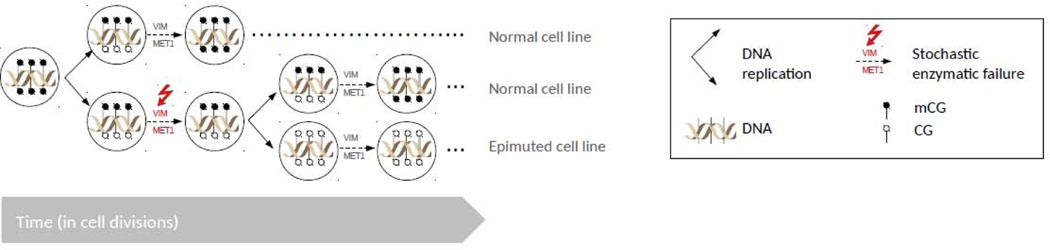
Although plants methylate cytosines extensively in three different sequence contexts (CG, CHG and CHH, where H is nucleotide A, T or C), the inheritance of stochastic methylation changes is mainly restricted to CG dinucleotides [15,16,31]. This follows directly from how CG methylation is maintained. During DNA replication, hemimethylated CG sites are recognized by the VARIANT IN METHYLATION family of proteins and recruit METHYLTRANSFERASE 1 to catalyze CG methylation on the newly synthesized strand by way of “template-copying” [14]. Enzymatic failure or offtarget methyltransferase activity can thus lead to permanent methylation losses or gains in daughter cells and their decedent cell lineages (Fig. I). By contrast, non-CG sites (i.e. CHG and CHH) are preferentially targeted by de novo methylation pathways, so that any methylation losses can be ‘corrected’ by re-targeting these sites at some later point in time in a replication independent manner.
Fig. I: Model of the molecular basis of spontaneous somatic CG epimutations.
CG epimutations can arise spontaneously as a result of stochastic enzymatic failure of the MET1 methyltransferase. Shown are two rounds of cell division. During DNA replication, hemimethylated CG sites are recognized by the VARIANT IN METHYLATION family of proteins and recruit METHYLTRANSFERASE 1 to catalyze CG méthylation on the newly synthesized strand by way of “template-copying”. Enzymatic failure or off-target methyltransferase activity can thus lead to permanent méthylation losses or gains in daughter cells and their decedent cell lineages (Le. epimutated cell line). For simplicity, only CG méthylation loss is shown.
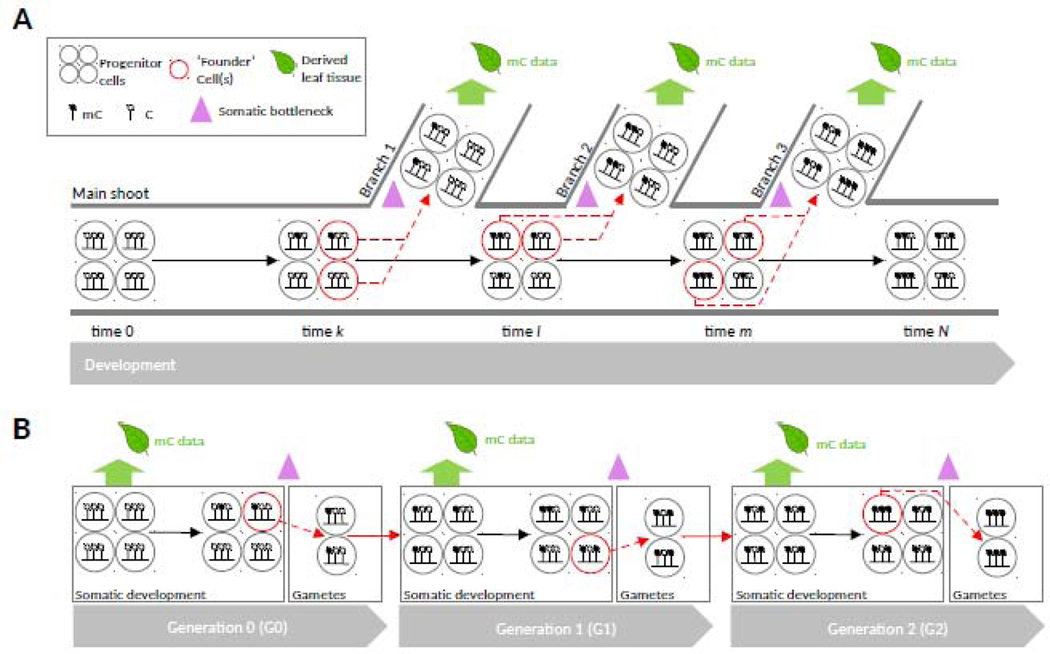
The somatic consequences of CG maintenance errors should be especially visible when they originate in shoot apical meristems (SAM), a small population of stem cells that give rise to all above-ground plant structures. Since only a small number of meristematic cells act as “founders” to new lateral branches [32], leaves and flowers, the finite sampling of these cells leads to fixation of CG epimutations in newly formed vegetative lineages. This process can be described as “somatic epigenetic drift” (Fig. IIA) and can lead to extensive chimerism [33]. Evidence for this is seen in perennials, where methylation changes seem to arise sequentially in nested sectors of the plant [18,34]. Similar observations have been made at the level of rare somatic DNA nucleotide mutations [18,35–39], indicating that both genetic and epigenetic stochastic changes have a common meristematic origin. Plants lack a dedicated germline unlike animals. Instead, cells that eventually give rise to the gametes are also derived from SAM precursors quite late in development [40]. This means that somatically acquired CG epimutations are frequently inherited to the next generation (Fig. IIB). Stable passage through the generation barrier requires that the CG methylome is not extensively reprogrammed in the sex cells and early zygote, which is indeed the case in plants [41].
Fig. II: Somatic accumulation and transgenerational inheritance of CG epimutations in plants.
(A) The failure to maintain CG methylaiion during the mitotic divisions of SAM-derived progenitor ceils leads to spontaneous somatic epimutations that are inherited to lateral branches. Shown here are only spontaneous gains of methylation on a haploid genome, for simplicity Since only a small number of progenitor cells act as “founders” to new lateral branches, the finite sampling of these cells forces CG epimutations through a strong bottleneck at each branch point. As a result, they eventually become “fixed” in newly formed vegetative lineages through “somatic drift” This process leads to increased mC divergence between leaves originating from different terminal branches (e.g. leaf methylomes from Branch 1 and 2 are more similar than those from Branch 1 and 3). (B) Cells that give raise to the gametes are aJso derived from SAM precursors quite late in development This means that somatically acquired CG epimutations can be inherited to the next generation Stable passage through the generation barrier is possible in plants, as the CG methylome is not extensively reprogrammed in the sex cells and early zygote. At each generation, only two gametes from a single germline (males or female) are shown for simplicity
Epimutations have clock-like properties: the evidence
There is mounting evidence that CG epimutations accumulate in plant genomes in a clock-like fashion (see below). This is true for developmental as well as evolutionary time-scales. Mathematical models have been extraordinarily successful at describing these accumulation dynamics, making this stochastic process one of the most predictable epigenetic phenomena to date. To ease the discussion we will henceforth refer to “CG epimutations” as simply “epimutations”; but it should be kept in mind that the phenomena we are describing do not readily extend to non-CG contexts.
Developmental time-scales
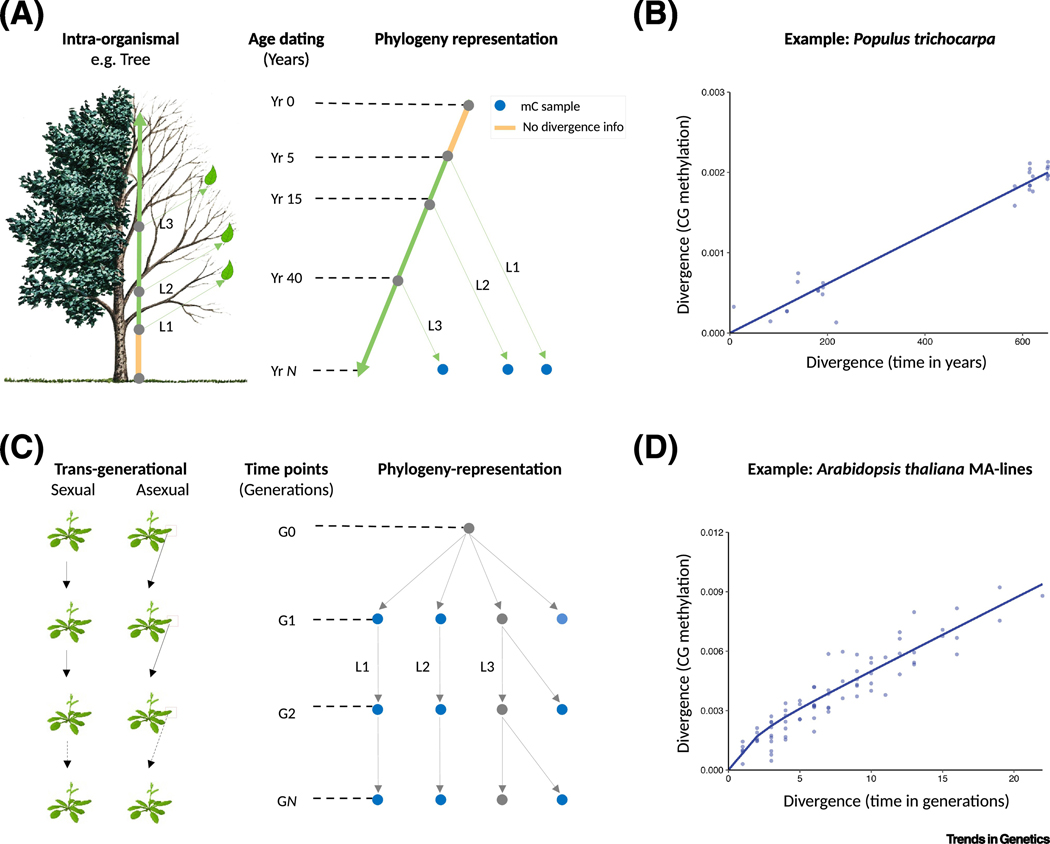
Recent developmental insights into the clock-like properties of epimutations have come from the analysis of trees. Due to their exceptional longevity, trees have emerged as model systems for studying somatic (epi)mutational processes in plants [18,35–38]. A key advance in this regard has been the realization that a tree’s branching structure can be treated as an intra-organismal phylogeny, with the leaves representing the endpoints of somatically evolving vegetative lineages [42] (Fig. IIIA). Genomic or epigenomic measurements collected on the leaves, or adjacent axillary meristems, are therefore amenable to phylogenetic inference methods [16,38]. In poplar (Populus trichocarpa), for instance, it was shown that methylomes of leaves sampled from different branches of a single tree diverge approximately linearly with developmental distance (in years) [16,18] (Fig. IIIB). This distance could be calculated by tracing back the ages of the branches to their most recent common branch point (Fig. IIIA). The branch ages, in this case, were known from coring data (i.e. tree ring counts). Phylogenetic analyses revealed that the rate of methylation divergence over developmental time is consistent with a neutral epimutation process, which seems to depend only on the stochastic methylation gain and loss rates at individual CG sites [16,18]. Global estimates of these per site rates were 1.8 × 10−6 and 5.8 × 10−6 per year per haploid genome, respectively. These estimates somewhat mask the considerable variation in epimutation rates across genomic features, with gene bodies showing the highest and TEs the lowest rates [16,18]. Nonetheless, it is safe to say that, on average, the somatic epimutation rate is about four orders of magnitude higher than the somatic nucleotide mutation rate (1.3 × 10−10). These large rate differences explain why methylome diversity arises much more rapidly than genome diversity within the tree crown.
Fig. III: Epimutations have clock-like properties.
(A) A tree can interpreted as an intra-organismal phylogeny. The topology (i.e branching structure) is typical known and the branch points and branch lengths can be dated by coring. Only three branches are highlighted (L1, to L3) for simplicity. Leaf mC measurements can be obtained and used as the basis to calculate CG methylation divergence. Similarly, divergence times (in years) for pairs of leaves can be calculated by tracing back the ages of the branches to their most recent common branch point. This can only be done down to the tree’s earliest branch point (in this case, Yr = 5) but not to earlier time points (orange segment). Note: tree image was modified from www.photos.com (B) Data from Poplar tricocarpa is shown (Hotmeister 2020, Shahryary 2020). CH methylation gain and loss rates at individual CG sites. (C) Construction of multi-generational (GO to GN) mutation accumulation (MA) lines through sexual (selfing or sibling mating) or asexual (clonal) propagation. The different lineages (L1 to L3) can be represented as a phylogeny. The branch point times and the branch lengths are typically known, a priori, from the experimental design. mC sampling can be performed at selected generations, either from plant material of direct progenitors or from siblings of those progenitors. (D) Data from an A. thaliana MA-line (MA3) is shown (Shahryary 2020). CG methylation between individuals/lineages diverge approximately linearly with divergence time (in generations). Divergence increases according to a neutral epimutation process, and depends only on the stochastic methylation gain and loss rates at individual CG sites as well as the Mendelian segregation and fixation of de novo epimutations
Transgenerational and evolutionary time-scales
Similar clock-like properties have emerged from transgenerational studies. The most comprehensive demonstration of this comes from analyses of selfing-derived Arabidopsis (A. thaliana) mutation accumulation (MA) lines. MA lines descent from a single isogenic founder and are independently propagated for many generations [43] (Fig. IIIC). Similar to the branching structure of a tree, as described above, the kinship among the different lines can be represented as a phylogeny (Fig. IIIC), whose topology is known, a priori, as the branchpoint times and the branch lengths are deliberately chosen as part of the experimental design. Coupled with multigenerational methylome measurements, MA-lines thus permit ‘real-time’, rather than retrospective, observations of epimutational processes in a well-defined phylogenetic context. In agreement with the developmental findings in trees, global CG methylation divergence among the lines increases steadily as a function of divergence time (in generations) (Fig. IIID). This transgenerational divergence can be predicted with remarkable accuracy on the basis of a neutral epimutation model, accounting for up to 90% of the variance [15,16]. Again, the underlying prediction model is relatively simple. It depends only on the methylation gain and loss rates at individual CG sites in each line, as well as the Mendelian segregation and fixation of de novo epimutations. Global estimates of these gain and loss rates are 2.5 × 10−4 and 6.3 × 10−4, per generation per haploid genome [15]; although, as in poplar, there is considerable variation in rates across genomic features [15–17]. For comparison, the per generation DNA nucleotide mutation rate in A. thaliana MA lines is about 10−9; that is, five orders of magnitude higher [44,45]. Comparable insights have been obtained with different experimental designs, species (Arabidopsis, rice, Dandelion), and types of reproduction (sexual and asexual), indicating that these clock-like properties are relatively robust [9,12,13,15,16,46–49].
Beyond the experimental time-scales probed by MA studies, the clock-like accumulation of epimutations is also visible over evolutionary time where it seems to have a major role in shaping plant CG methylation diversity [9,50,51]. Formal support for this comes from methylation site frequency spectrum (mSFS) studies of natural populations of A. thaliana [50,52] and maize [53]. Analysis of the genic (CG) mSFS revealed that methylome diversity is most consistent with a neutral or near neutral model. Furthermore, in A. thaliana, the estimated ratio of population epimutation gain and loss rates was found to be approximately similar to that in experimental MA lines [50]. The latter observation suggests that epimutation rates are stable over long time periods, at least within the same species. This is remarkable, considering that extensive genetic variations, too, arise over these time-scales [54], which could potentially perturb CG methylation patterns through cis or trans-acting SNPs or structural variants [51,55–57].
Exploiting the clock-like properties of epimutations
The above discussion highlights several important properties about epimutations that are worth reiterating here: 1. epimutations accumulate neutrally over developmental and evolutionary time, at least at the genome-wide scale, 2. they occur at a rate that is orders of magnitude higher than the genetic mutation rate, and 3. they cause rapid methylome divergence among plant or cell lineages. An emerging question is how to best exploit these properties as a genomic tool in plant biology. This question is not just about possible applications, but also about how to integrate epimutation data into existing analytical frameworks. Here we will highlight two applications, namely, the use of epimutations as a molecular clock to time evolutionary events in the recent past and to estimate the age of long-lived plants.
Epimutations as an aging clock
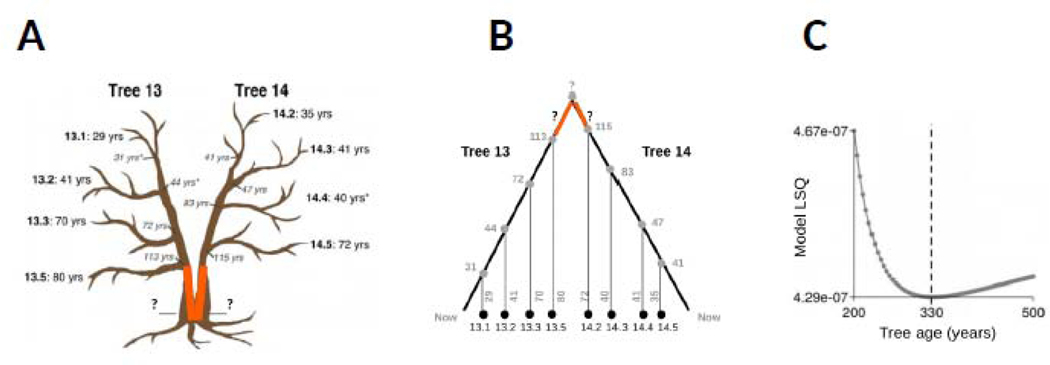
The first direct indication that epimutations can be employed as an accurate time keeper has come from the abovementioned proof-of-principle study in poplar [16,18] (see Fig. IVA). A fortunate limitation of that study was that the tree topology could only be partly dated. That is, the age of the bottom sector of the tree was unknown because it could not be determined by coring. This initially prevented the calculation of divergence times between leaves across the two main stems (Fig. IVB). To bypass this problem, the unknown age of the bottom sector was included as an additional parameter in the model fitting. This yielded an unbiased age estimate of the complete tree of 330 years (Fig. IVC), which agreed well with circumference-based estimates [16]. Thus, the accumulation of somatic epimutations seems to encode sufficient temporal information to serve as an aging-clock in trees. Clearly, the generality of this conclusion needs to be solidified in future studies by examining different tree species as well as other long-lived perennials. Such efforts are underway.
Fig. IV: Proof-of-principle evidence that CG epimutations act as an aging clock in trees.
Results from the study of Shahryary et al. (2020) are shown. Figures were slightly modified from that manuscript. (A) A single poplar (P. trichocarpa) tree was analyzed. Tree 13 and 14 are two main stems that have diverged early in development. Four branches from each tree were chosen and aged by coring. Shown are the coring sites along with the coring-based branch ages. Age coring proved technically challenging at the bottom of the tree and led to unintelligible ring counts (orange segment). An educated guess, based on diameter measurements, placed the age of the tree between 250 and 350 years. (B) The tree can be presented as an intra-organismal phylogeny. Leaf methylomes were collected from each of the selected branches. (C) A phylogenetic discrete time Markov model fitted to the global CG methylation divergence data of the complete tree data treating tree age (i.e. the age of the orange segment) as an unknown parameter. Model residual (LSQ) was minimized at an age of 330 years. This unbiased estimate fit well within the circumference-based estimated age range.
Looking ahead, a logical extension of the above approach is to calibrate epimutation rates beforehand, and use them to estimate branch and stem ages freely. These calibrated rates need to be obtained only once by analysing a species-specific “reference tree” for which extensive DNA methylation and coring data has been collected. Application of these rates to any new tree would merely require that the topology of that tree is known. This latter requirement is trivial since the (surviving) branching structure of a tree can be easily reconstructed from observation. An interesting exception, however, is when the observed topology does not reflect the developmental history of the tree. This can happen when there is extensive dormancy in the initiation of lateral branches. We propose that it may be possible to detect dormancy, retrospectively and without coring data, on the basis of epimutation divergence data alone. Dormant branches would show up as incongruencies between the observed tree topology and the one reconstructed on the basis of DNA methylation measurements. It should be clear that the development of similar approaches using de novo somatic mutations, instead of epimutations, would be challenging. The relatively small number of somatic SNPs that are detectable in even very old trees [18,36–38,58] would probably not yield enough temporal resolution for accurate statistical inferences.
A key limitation with the above epimutation-based age-estimator in trees is that it requires DNA methylation divergence data as input. This implies that the age of a tree can only be determined down to its oldest (viable) branch point. For trees that have formed early lateral branches or two dominant stems (e.g. Fig. IVA), the age of the oldest branch point is approximately equivalent to the total age of the tree. In other situations where this is not the case, it may be possible to invoke species-specific growth models [59–61] and infer the overall age of the tree by interpolating the length of the missing bottom segment (i.e. from root to first branch point).
DNA methylation (DNAme) aging clocks are already extensively used in the mammalian field [62]. However, these types of clocks differ fundamentally from the CG epimutation-based clock observed in trees (Table I). A central difference is that DNAme clocks in mammals are designed to track ‘systematic’, age-related changes of CG methylation levels at a few hundert pre-selected loci [62]. These changes are directional (i.e. proggressive transition from hypo- to hypermethylation or vice versa), highly reproducible, and are best viewed as a molecular read-out of cellular aging. By contrast, CG epimutation-based clocks quantify ‘random’ CG methylation changes, which can only be reliably captured by tracking the methylation status of millions of CG sites throughout the genome. Although these latter changes are reproducible globally, in terms of aggregate measures like epimutation rates or mCG divergence, at the level of individual CG sites they are not. Table I highlights these differences in more detail.
Tabel I: DNAme clocks vs. Epimutation-based aging clocks.
The CG epimutation-based aging clock described for trees differ fundamentally from the DNA methylation-based (DNAme) clocks used in the mammalian field. This table summarizes theri differences along several key dimensions. Current data on CG epimutation-based aging clocks in trees is still limited, and an assessment of their properties is therefore somewhat hypothetical. To indicate this uncertainty, we use parentheses, “( )”, in places. The properties of DNAme clocks have been reviewed by Horvath and Raj [62]. Their summary has informed the construction of the “DNAme clocks in mammals” column. When necessary, additional references were used as indicated.
| Clock properties and data requirements | DNAme clocks, in mammals | Epimuialion clocks in Uses |
|---|---|---|
| Minimum Nb. of input CGs for clock | few (1 to 513 target CGs) |
Many (> 1C°, but depends on epimutation rates) |
| Quantification | Weighted mean CG methylation levels + model | CG methyiatior divergence f model |
| Nature of age-rela;ed mCG change | Systematic (reproducible, directional change from hypo- to typermetiiylaton. or vice versa, as function of age) |
Random (stochastic CG gains and losses throughout agng) |
| Dependence on Nb. oi mHotic divisions | No | Yes |
| Clock at equilibrium | No (follows from the Uirectonalry of die changes) |
(Yes) |
| Clock reset at every generation | Yes | (No) |
| Accurate across genotypes | Yes (by construction) | unknown |
| Acturate across species | Yes (by construction) Refs: [90. 91] |
unlikely |
| Minimum # of samples required | 1 sample (individual) | >> 1 sample (individual) (precise number is still undear) |
| Tissue requirement | Wide range of tissues possible | Leaves, stems, buds |
Epimutations as an evolutionary clock
The basic idea behind using epimutations as an age-estimator in trees can be readily transferred into the evolutionary arena. Instead of age-dating stems and branches of an actual tree, the task would be to estimate the timing of evolutionary events along a phylogenetic or coalescent tree. A priori, there is no reason why this should not work, given the evidence discussed above. Molecular clocks already play an instrumental role in phylogenetics. Classically, these clocks are built around DNA sequence data. They work by counting DNA nucleotide substitutions between species to derive a measure of genetic distances [63]. If substitution rates are constant over time (and lineages) these distances are directly proportional to divergence times. Absolute dates, in terms of years or generations, can be obtained by calibrating substitution rates against the fossil record or else by using experimentally estimated mutation rates as a proxy [64].
However, molecular clocks are typically applied to species trees that involve divergence times in the order of millions of years [65]. Flowering plants (angiosperms), for instance, have diverged as far back as 200 million years ago [66]. Over these long phylogenetic time-scales, epimutations are unlikely to provide any useful information (Fig. VB). The reason for this is that high epimutation rates result in frequent ‘reversions’ [67,68] and homoplasy, which lead to rapid saturation in divergence estimates [50]. That is, branch points that lie far back in time cannot be confidently separated temporally. Moreover, major genome rearrangements, such as polyploidization or whole genome duplication events, can lead to extensive methylome remodeling [69–75], and thus to a breakdown of the clock assumption.
Fig. V: Epimutations as an evolutionary clock.
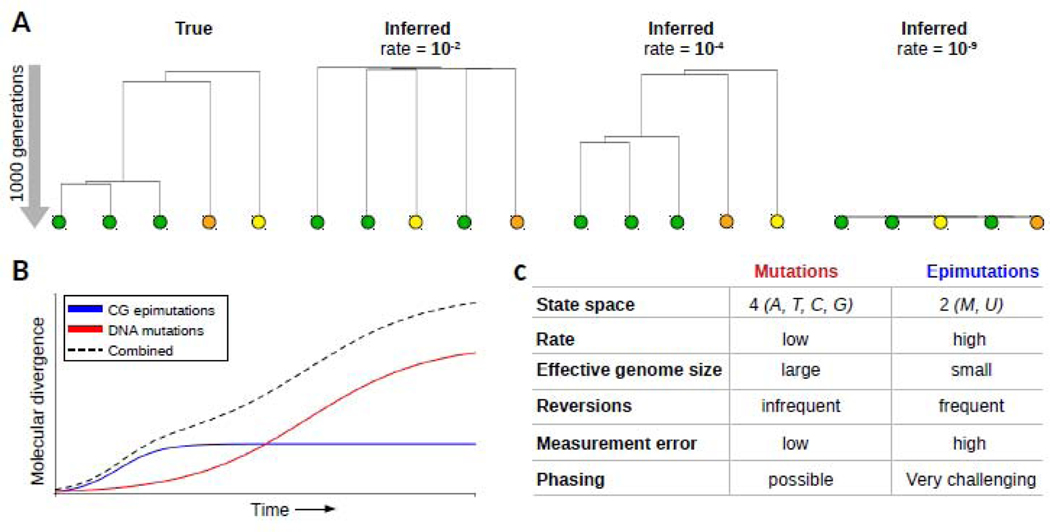
(A) The inclusion of epimutation data can improve the temporal resolution of phylogenetic analyses of the recent past To illustrate this we simulated an ultmetric tree (true, left) with five lineages and a depth of 1000 generations. Conditional on the tree topology, we simulated a donally reproducing species forward in time, and introduced epimutations at a rate of 10J, 10- and 10−5, per site, per haploid genome per generation. mCG divergence observed between the tips were used as input to construct a neighbor-joining tree for each scenario. On this time-scale, a high rate (10−2) or a low rate (10−9) lead to incorrect inferences of topology and relative branch lengths With a rate of 10−4, which is the approximate rate estimated in plants, the epimutation data contained sufficient information to reconstruct the true tree With calibrated epimutation rates, it will be possible to estimate branch ages and branch point times in recent phylogemes The color of the circles emphasize the original cluster membership of samples in the true tree (B) Schematic showing that CG epimutations diverge relatively quickly over time and saturate early By contrast, DNA mutations accumulate slowly and saturate late Both molecular markers capture different segments of evolutionary time. Future methods should integrate CG epimutations and DNA mutations to cover the full time-scales. (C) The incorperation of epimutation data into existing phylogenetic and population genetic models will be challenging as epimutations differ in key aspects from DNA mutations. These differences should inform the development of new models.
In contrast to phylogenetic (macroevolutionary) time-scales, the clock-like properties of epimutations could be highly informative for analyzing microevolutionary (population genetic) time scales, ranging from decades to a few thousand years (Fig. VA). This is a segment of time, where DNA sequenced-based information is limited due to the slow rate at which new mutations (i.e. genetic variations) arise (Fig. VA–B). Many important evolutionary events can occur in this time-frame, particularly in the presence of rapidly changing environmental conditions [76]. Such events could include shifts in demography due to bottlenecks, extinction, range expansion, population differentiation, as well as transitions in mating systems (e.g. the emergence of selfing-compatibility). The integration of epimutation data into coalescent models would be one way forward to to detect such events at the molecular level and to place them on an evolutionary timeline.
Furthermore, modified versions of classical phylogenetic models could also be employed to estimate divergence times of clonal lineages in species that reproduce, continuously, by fragmentation or budding. Analytically, this would require a similar inference approach as the one used for age-dating (actual) trees, except that the underlying phylogeny (i.e. topology) needs to be inferred along with divergence times (Box 1, SI text). Again, known epimutation rates are needed here, since there is no fossil record to calibrate against over these short time-scales. The reconstruction of clonal phylogenies on the basis of DNA methylation data is already used in the cancer field. However, the main focus there is on inferring the topology of tumor cell lineages rather than on estimating their branching times [77–82]. Still, selected elements of these methods could be transferred to the plant world.
Box 1: Incorporating epimutation data into evolutionary inference models
Incorporation into phylogenetic models
The substitution model is at the core of phylogeny/genealogy inference and connects molecular data and divergence times [83]. It is usually applied to macroevolutionary data and assumes that mutations have been fixed in each species. However, these models can also be used to reconstruct clonal lineage evolution, which could occur over “ultra-micro evolutionary” timescales [84,85]. The incorporation of epimutation data into these models could provide increased temporal resolution (Fig. VA). A discrete time Markov model has been proposed to model epigenetic divergence among diploid clonal lineages in situations where underlying topology is known [16]. In this case, the transitions probabilities were defined for epigenotypes UU (homozygous unmethylated), UM (epi-heterozygous) and MM (homozygous methylated). However, simultaneous inference of the underlying topology is a much harder problem. One solution is to reformulate the problem as a continuous Markov process with a transition rate matrix (substitution matrix) for epigenotype. In Table S1 we proposed a set of substitution matrices, which lean on classical phylogenetic models. These could be incorporated into existing maximum likelihood or Bayesian inference softwares. Estimates for divergence times in such models can be obtained by supplying experimentally calibrated transition rates, which are - themselves - functions of the stochastic methylation gain and loss rates. We illustrate a working example of this in SI text. If phased data were available, the above diploid epigenotype model could be further simplified (Table S1). However, phased methylation data is difficult to obtain from WGBS data due to measurement uncertainties.
Incorporation into population genetics models
Apart from difficulties in obtaining phased methylation data, additional theoretical challenges arise from the high epimutation rate and relatively small number of effective epimutable sites (a subset of all CG sites). These properties violate the infinite-sites model (ISM) (i.e. “no site can be hit twice”), which is at the heart of many population genetic theories and methods, such as estimating fixation rates, site frequency spectra (SFS), natural selection and demographic histories. Violations to the ISM have also been recognized for mtDNA, microsatellite markers and in hyper-diversity populations [86][87][67]. To address this, the finite-sites model (FSM) has been proposed [87][88][89][67]. Recently, the FSM has been incorporated into site frequency spectrum (SFS) models, and successfully applied to plant methylation diversity data [50,52,53]. Similar extensions to coalescent methods, such as the Sequential Markov Coalescent (SMC) model are currently emerging.
A central challenge ahead will be the methodological integration of epimutation data into existing phylogenetic and coalescent models (Box 1). This is not a trivial task. There are key properties that set DNA mutations apart from epimutations, including their ‘mutation’ spectrum, measurement uncertainty, the availability of phased haplotype data, etc. (Fig. VC). Ultimately, models that integrate both DNA sequence and DNA methylation data into a single inference framework may be most successful at accurately dating evolutionary events across time-scales (Fig. VB).
Concluding remarks and future perspective
As a result of their high stochastic rate and functional neutrality, spontaneous epimutations have received relatively little attention in the plant community. Yet, from a statistical standpoint, these stochastic events are one of the most predictable epigenetic phenomena to date. Here, we hope to have shown that these unique properties make spontaneous epimutations a powerful new molecular clock with a wide range of applications in plant biology. In contrast to classical DNA mutation-based clocks, this new clock offers novel insights into the recent evolutionary past of species/populations and can also be used to estimate the age of long-lived perennials such as trees. Furthermore, a formal understanding of the neutral process by which epigenetic diversity arises in plant populations provides a framework for detecting signatures of selection on rare, but potentially adaptive, epigenetic mutations. From an evolutionary perspective, the potential relevance of these insights cannot be overstated. In an era of climate change, plant biodiversity is transforming rapidly [76]. The ability to monitor effective population sizes, demographic changes, selection, bottlenecks etc. at the molecular level over short time-scales will contribute to our basic understanding and management of this diversity going into the future. Further research into the molecular properties of epimutation-based clocks, their integration into existing evolutionary models, and their applications define a novel research program for years to come (see Outstanding Questions).
Outstanding Questions Box
How universal is the clock-like accumulation of CG epimutations across plant species?
How robust are CG epimutation rates across environmental conditions, genetic backgrounds, mating systems and time?
When using CG epimutation as a molecular clock, what level of accuracy can be achieved in age-dating long-lived perennials or in timing evolutionary events?
Are some genomic regions more clock-like than others (i.e. less susceptible to measurement noise, or external perturbations)? If yes, what are the sequence features and functional properties of such genomic regions?
What is the optimal temporal window (in the past) where epimutation-based clocks work best?
Can epimutation data be incorporated into existing phylogenetic and population genetic software packages?
How can the clock-like properties of CG epimutations be reconciled with current research programs into the adaptive basis of DNA methylation in plants?
Supplementary Material
Highlights
In plants, stochastic gains and losses of CG methylation can be inherited across mitotic and meiotic cell divisions.
These stochastic events are called spontaneous (CG) “epimutations”. They occur at a rate that is about four to five orders of magnitude higher than the DNA mutation rate per unit time.
Methylome surveys in multigenerational experiments as well as in long-lived trees show that epimutations are neutral at the genome-wide scale, and that they accumulate in plant genomes in a “clock-like” fashion.
Emerging evidence indicates that these “clock-like” properties can be exploited for reconstructing/timing recent evolutionary events, as well as for age-dating long-lived perennials.
Acknowledgements
We thank Hanno Schäfer, Aurélien Tellier, Thibaut Sellinger, Thorsten Reusch, Liang Liu and Hans Pretzsch for their feedback, Lutz Johannes and Rashmi Hazarika for help with the figures. FJ and RJS acknowledge support from the Technical University of Munich-Institute for Advanced Study funded by the German Excellent Initiative and the European Seventh Framework Programme under grant agreement no. 291763. This work was supported by the National Institutes of Health (R01GM134682) to RJS.
Glossary
- Methylome
The set of nucleic acid methylation modifications in an organism’s genome or in a particular cell
- Molecular clock
A tool that is traditionally used in evolutionary biology to measure evolutionary change over time at the molecular level. It is based on the theory that specific DNA sequences or the proteins they encode spontaneously mutate at constant rates. This information can be used to estimate how long ago two related organisms diverged from a common ancestor
- Chimerism
A chimera is an individual, organ, or part consisting of tissues of diverse (epi)genetic constitution
- Epigenome
The complete set of chemical compounds that modify the expression and function of an organisms’ genome or in a particular cell
- Homoplasy
Correspondence or similarity in form or function between parts of different species or lineages that are not attributable to common ancestry. In the text, it refers more specifically to the similarity in DNA methylation states at a locus that originates from independent epimutations, rather than from shared ancestry
- SFS
In population genetics, the allele frequency spectrum, sometimes called the site frequency spectrum (SFS), is the distribution of the allele frequencies of a given set of loci (often SNPs) in a population or sample. The methylation site frequency spectrum (mSFS) is its epigenetic counterpart, where ‘epiallele’ frequencies are used instead
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kawakatsu T. et al. (2016) Unique cell-type-specific patterns of DNA methylation in the root meristem. Nature Plants 2, 16058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wibowo A. et al. (2016) Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity. Elife 5, e13546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ganguly DR et al. (2018) Maintenance of pre-existing DNA methylation states through recurring excess-light stress. Plant Cell Environ. 41, 1657–1672 [DOI] [PubMed] [Google Scholar]
- 4.Stelpflug SC et al. (2014) Consistent and heritable alterations of DNA methylation are induced by tissue culture in maize. Genetics 198, 209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stroud H. et al. (2013) Plants regenerated from tissue culture contain stable epigenome changes in rice. Elife 2, e00354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji L. et al. (2019) Genome-Wide Reinforcement of DNA Methylation Occurs during Somatic Embryogenesis in Soybean. Plant Cell 31, 2315–2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borges F. et al. (2021) Loss of Small-RNA-Directed DNA Methylation in the Plant Cell Cycle Promotes Germline Reprogramming and Somaclonal Variation. Curr. Biol 31, 591–600.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eichten SR et al. (2013) Epigenetic and Genetic Influences on DNA Methylation Variation in Maize Populations. Plant Cell 25, 2783–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagmann J. et al. (2015) Century-scale Methylome Stability in a Recently Diverged Arabidopsis thaliana Lineage. PLoS Genet. 11, e1004920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofmeister BT et al. (2017) Stable inheritance of DNA methylation allows creation of epigenotype maps and the study of epiallele inheritance patterns in the absence of genetic variation. Genome Biol. 18, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colomé-Tatché M. et al. (2012) Features of the Arabidopsis recombination landscape resulting from the combined loss of sequence variation and DNA methylation. Proc. Natl. Acad. Sci. U. S. A 109, 16240–16245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Becker C. et al. (2011) Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 480, 245–249 [DOI] [PubMed] [Google Scholar]
- 13.Schmitz RJ et al. (2011) Transgenerational epigenetic instability is a source of novel methylation variants. Science 334, 369–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Law JA and Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet 11, 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Graaf A. et al. (2015) Rate, spectrum, and evolutionary dynamics of spontaneous epimutations. Proc. Natl. Acad. Sci. U. S. A 112, 6676–6681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shahryary Y. et al. (2020) AlphaBeta: computational inference of epimutation rates and spectra from high-throughput DNA methylation data in plants. Genome Biol. 21, 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denkena J. et al. Region-level Epimutation Rates in Arabidopsis thaliana. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmeister BT et al. (2020) A genome assembly and the somatic genetic and epigenetic mutation rate in a wild long-lived perennial Populus trichocarpa. Genome Biol. 21, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johannes F. and Schmitz RJ (2019) Spontaneous epimutations in plants. New Phytol. 221, 1253–1259 [DOI] [PubMed] [Google Scholar]
- 20.Cubas P. et al. (1999) An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401, 157–161 [DOI] [PubMed] [Google Scholar]
- 21.Manning K. et al. (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet 38, 948–952 [DOI] [PubMed] [Google Scholar]
- 22.Ong-Abdullah M. et al. (2015) Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm. Nature 525, 533–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gouil Q. and Baulcombe DC (2018) Paramutation-like features of multiple natural epialleles in tomato. BMC Genomics 19, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miura K. et al. (2009) A metastable DWARF1 epigenetic mutant affecting plant stature in rice. Proc. Natl. Acad. Sci. U. S. A 106, 11218–11223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stam M. et al. (2002) Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev. 16, 1906–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soppe WJ et al. (2000) The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol. Cell 6, 791–802 [DOI] [PubMed] [Google Scholar]
- 27.Jacobsen SE and Meyerowitz EM (1997) Hypermethylated SUPERMAN epigenetic alleles in arabidopsis. Science 277, 1100–1103 [DOI] [PubMed] [Google Scholar]
- 28.Richards CL et al. (2017) Ecological plant epigenetics: Evidence from model and non-model species, and the way forward. Ecol. Lett 20, 1576–1590 [DOI] [PubMed] [Google Scholar]
- 29.Springer NM and Schmitz RJ Exploiting induced and natural epigenetic variation for crop improvement., Nature Reviews Genetics, 18. (2017), 563–575 [DOI] [PubMed] [Google Scholar]
- 30.Charlesworth D. et al. (2017) The sources of adaptive variation. Proc. Biol. Sci 284, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Picard CL and Gehring M. (2017) Proximal methylation features associated with nonrandom changes in gene body methylation. Genome Biol. 18, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burian A. et al. (2016) Patterns of Stem Cell Divisions Contribute to Plant Longevity. Curr. Biol 26, 1385–1394 [DOI] [PubMed] [Google Scholar]
- 33.Szymkowiak EJ and Sussex IM (1996) WHAT CHIMERAS CAN TELL US ABOUT PLANT DEVELOPMENT. Annu. Rev. Plant Physiol. Plant Mol. Biol 47, 351–376 [DOI] [PubMed] [Google Scholar]
- 34.Herrera CM et al. (2021) Lifetime genealogical divergence within plants leads to epigenetic mosaicism in the shrub Lavandula latifolia (Lamiaceae). New Phytol. DOI: 10.1111/nph.17257 [DOI] [PubMed] [Google Scholar]
- 35.Wang L. et al. (2019) The architecture of intra-organism mutation rate variation in plants. PLoS Biol. 17, e3000191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanlon VCT et al. (2019) Somatic mutations substantially increase the per-generation mutation rate in the conifer. Evol Lett 3, 348–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmid-Siegert E. et al. (2017) Low number of fixed somatic mutations in a long-lived oak tree. Nature Plants 3, 926. [DOI] [PubMed] [Google Scholar]
- 38.Orr AJ et al. (2020) A phylogenomic approach reveals a low somatic mutation rate in a long-lived plant. Proceedings of the Royal Society B: Biological Sciences 287, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu L. et al. (2020) Somatic genetic drift and multilevel selection in a clonal seagrass. Nat Ecol Evol 4, 952–962 [DOI] [PubMed] [Google Scholar]
- 40.McDaniel CN and Poethig RS (1988) Cell-lineage patterns in the shoot apical meristem of the germinating maize embryo. Planta 175, 13–22 [DOI] [PubMed] [Google Scholar]
- 41.Kawashima T. and Berger F. (2014) Epigenetic reprogramming in plant sexual reproduction. Nat. Rev. Genet 15, 613–624 [DOI] [PubMed] [Google Scholar]
- 42.Pineda-Krch M. and Fagerstrom T. (1999) On the potential for evolutionary change in meristematic cell lineages through intraorganismal selection. J. Evol. Biol 12, 681–688 [Google Scholar]
- 43.Shaw FH et al. (2002) A comprehensive model of mutations affecting fitness and inferences for Arabidopsis thaliana. Evolution 56, 453–463 [DOI] [PubMed] [Google Scholar]
- 44.Ossowski S. et al. (2010) The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327, 92–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weng M-L et al. (2019) Fine-Grained Analysis of Spontaneous Mutation Spectrum and Frequency in Arabidopsis thaliana. Genetics 211, 703–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang C. et al. (2014) Environmentally responsive genome-wide accumulation of de novo Arabidopsis thaliana mutations and epimutations. Genome Res. 24, 1821–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ganguly DR et al. (2017) The Arabidopsis DNA Methylome Is Stable under Transgenerational Drought Stress. Plant Physiol. 175, 1893–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng X. et al. (2017) Transgenerational epimutations induced by multi-generation drought imposition mediate rice plant’s adaptation to drought condition. Sci. Rep 7, 39843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yadav NS et al. 02-Dec-(2020), Multigenerational exposure to heat stress induces phenotypic resilience, and genetic and epigenetic variations in Arabidopsis thaliana offspring., Cold Spring Harbor Laboratory, 2020.11.30.405365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vidalis A. et al. (2016) Methylome evolution in plants. Genome Biol. 17, 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawakatsu T. et al. (2016) Epigenomic Diversity in a Global Collection of Arabidopsis thaliana Accessions. Cell 166, 492–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muyle A. et al. (2020) Investigation Gene body methylation is under selection in Arabidopsis thaliana. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu G. et al. (2020) Evolutionary and functional genomics of DNA methylation in maize domestication and improvement. Nat. Commun 11, 5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.1001 Genomes Consortium. Electronic address: magnus.nordborg@gmi.oeaw.ac.at and 1001 Genomes Consortium (2016) 1,135 Genomes Reveal the Global Pattern of Polymorphism in Arabidopsis thaliana. Cell 166, 481–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmitz RJ et al. (2013) Patterns of population epigenomic diversity. Nature 495, 193–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dubin MJ et al. (2015) DNA methylation in Arabidopsis has a genetic basis and shows evidence of local adaptation. Elife 4, e05255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y. et al. (2020) Natural variation in DNA methylation homeostasis and the emergence of epialleles. Proceedings of the National Academy of Sciences DOI: 10.1073/pnas.1918172117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Plomion C. et al. (2018) Oak genome reveals facets of long lifespan. Nat Plants 4, 440–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pretzsch H. et al. (2002) The single tree-based stand simulator SILVA: construction, application and evaluation. For. Ecol. Manage 162, 3–21 [Google Scholar]
- 60.Pretzsch H. et al. (2015) Representation of species mixing in forest growth models. A review and perspective. Ecol. Modell 313, 276–292 [Google Scholar]
- 61.Pretzsch H. et al. (2008) Models for forest ecosystem management: a European perspective. Ann. Bot 101, 1065–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Horvath S. and Raj K. (2018) DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet 19, 371. [DOI] [PubMed] [Google Scholar]
- 63.Bromham L. and Penny D. (2003) The modern molecular clock. Nat. Rev. Genet 4, 216. [DOI] [PubMed] [Google Scholar]
- 64.Tiley GP et al. (2020) Molecular Clocks without Rocks: New Solutions for Old Problems. Trends Genet. 36, 845–856 [DOI] [PubMed] [Google Scholar]
- 65.Morris JL et al. (2018) The timescale of early land plant evolution. Proc. Natl. Acad. Sci. U. S. A 115, E2274–E2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H-T et al. (2019) Origin of angiosperms and the puzzle of the Jurassic gap. Nat Plants 5, 461–470 [DOI] [PubMed] [Google Scholar]
- 67.Charlesworth B. and Jain K. (2014) Purifying selection, drift, and reversible mutation with arbitrarily high mutation rates. Genetics 198, 1587–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang J. and Fan C. (2014) A neutrality test for detecting selection on DNA methylation using single methylation polymorphism frequency spectrum. Genome Biol. Evol 7, 154–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seymour DK et al. (2014) Evolution of DNA methylation patterns in the Brassicaceae is driven by differences in genome organization. PLoS Genet. 10, e1004785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim KD et al. (2015) A Comparative Epigenomic Analysis of Polyploidy-Derived Genes in Soybean and Common Bean. Plant Physiol. 168, 1433–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang J. et al. (2015) Autotetraploid rice methylome analysis reveals methylation variation of transposable elements and their effects on gene expression. Proc. Natl. Acad. Sci. U. S. A 112, E7022–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shi T. et al. (2020) Distinct Expression and Methylation Patterns for Genes with Different Fates following a Single Whole-Genome Duplication in Flowering Plants. Mol. Biol. Evol 37, 2394–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Edger PP et al. (2017) Subgenome Dominance in an Interspecific Hybrid, Synthetic Allopolyploid, and a 140-Year-Old Naturally Established Neo-Allopolyploid Monkeyflower. Plant Cell 29, 2150–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li N. et al. (2019) DNA methylation repatterning accompanying hybridization, whole genome doubling and homoeolog exchange in nascent segmental rice allotetraploids. New Phytol. 223, 979–992 [DOI] [PubMed] [Google Scholar]
- 75.Taudt A. et al. (2016) Genetic sources of population epigenomic variation. Nat. Rev. Genet 17, 319–332 [DOI] [PubMed] [Google Scholar]
- 76.Parmesan C. and Hanley ME (2015) Plants and climate change: complexities and surprises. Ann. Bot 116, 849–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mazor T. et al. (2015) DNA Methylation and Somatic Mutations Converge on the Cell Cycle and Define Similar Evolutionary Histories in Brain Tumors. Cancer Cell 28, 307–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brocks D. et al. (2014) Intratumor DNA methylation heterogeneity reflects clonal evolution in aggressive prostate cancer. Cell Rep. 8, 798–806 [DOI] [PubMed] [Google Scholar]
- 79.Hua X. et al. (2020) Genetic and epigenetic intratumor heterogeneity impacts prognosis of lung adenocarcinoma. Nat. Commun 11, 2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gaiti F. et al. (2019) Epigenetic evolution and lineage histories of chronic lymphocytic leukaemia. Nature [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Capra JA and Kostka D. (2014) Modeling DNA methylation dynamics with approaches from phylogenetics. Bioinformatics 30, i408–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schwartz R. and Schäffer AA (2017) The evolution of tumour phylogenetics: principles and practice. Nat. Rev. Genet 18, 213–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang Z. (2014) Molecular Evolution: A Statistical Approach, Oxford University Press. [Google Scholar]
- 84.Wu C-I et al. (2016) The Ecology and Evolution of Cancer: The Ultra-Microevolutionary Process. Annu. Rev. Genet 50, 347–369 [DOI] [PubMed] [Google Scholar]
- 85.Ling S. et al. (2015) Extremely high genetic diversity in a single tumor points to prevalence of non-Darwinian cell evolution. Proc. Natl. Acad. Sci. U. S. A 112, E6496–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Misawa K. and Tajima F. (1997) Estimation of the amount of DNA polymorphism when the neutral mutation rate varies among sites. Genetics 147, 1959–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang Z. (1996) Statistical Properties of a DNA Sample Under the Finite-Sites Model. Genetics 144, 1941–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Deng H-W and Fu Y-X The Effects of Variable Mutation Rates Across Sites on the Phylogenetic Estimation of Effective Population Size or Mutation Rate of DNA Sequences., Genetics, 144. (1996), 1271–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jenkins PA and Song YS (2011) The effect of recurrent mutation on the frequency spectrum of a segregating site and the age of an allele. Theor. Popul. Biol 80, 158–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wilkinson GS et al. (2021) DNA methylation predicts age and provides insight into exceptional longevity of bats. Nat. Commun 12, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.MAMMALIAN METHYLATION CONSORTIUM et al. 19-Jan-(2021), Universal DNA methylation age across mammalian tissues., Cold Spring Harbor Laboratory, 2021.01.18.426733 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.