Abstract
Fungal biotechnology is set to play a keystone role in the emerging bioeconomy, notably to address pollution issues arising from human activities. Because they preserve biological diversity, Biological Resource Centres are considered as critical infrastructures to support the development of biotechnological solutions. Here, we report the first large-scale phenotyping of more than 1,000 fungal strains with evaluation of their growth and degradation potential towards five industrial, human-designed and recalcitrant compounds, including two synthetic dyes, two lignocellulose-derived compounds and a synthetic plastic polymer. We draw a functional map over the phylogenetic diversity of Basidiomycota and Ascomycota, to guide the selection of fungal taxa to be tested for dedicated biotechnological applications. We evidence a functional diversity at all taxonomic ranks, including between strains of a same species. Beyond demonstrating the tremendous potential of filamentous fungi, our results pave the avenue for further functional exploration to solve the ever-growing issue of ecosystems pollution.
Subject terms: Environmental biotechnology, Fungal biology
Navarro et al. present a culture-based approach to the degradation of industrial products and by-products by assessing >1,000 fungal strains. Using growth-assay-screening and a large sample of fungal phenotypes, they explore the functional differences across strains and species to demonstrate the potential of filamentous fungi in breaking down dyes and industrial by-products with lignocellulose and plastic bases.
Introduction
Over the past 200 years, humankind has engineered processes and commodities that have greatly contributed to the life standards we enjoy today. However, this progress has come with a cost we are just beginning to assess. Plastics, dyes, and additives are widely used in the food, agricultural, medical, transport, textile, cosmetic, and electronic industries. These compounds have been designed to be resistant to degradation and are thus difficult to recycle, which poses today major challenges. The emerging bioeconomy1 and in particular white biotechnologies are set to play a cornerstone role in future societies, notably through the discovery and engineering of efficient biocatalysts (enzymes and microorganisms)2. The fungal kingdom contains an estimated diversity ranging between 2.2 and 3.8 million species3 and is considered as a remarkable reservoir of enzymes and biotechnological solutions4. The diversity of fungal lifestyles (saprobic, symbiotic, or parasitic), together with the diverse properties of their hosts (e.g., plants, insects, animals), are key drivers of the evolution of their extraordinary biocatalytic potential. Notably, plant-decaying filamentous fungi display the unique capacity to degrade efficiently lignocellulose, a notoriously recalcitrant biopolymer. The underlying biochemical strategies have inspired many industrial processes for plant biomass valorization into fermentable sugars and other biomolecules for second-generation biofuels, bioproducts, and biomaterials5–7. Noting the analogy of challenges intrinsic to both lignocellulosic and human-designed polymers (e.g., crystallinity, insolubility, heterogeneity, toxicity), fungi, and enzymes thereof represent a yet untapped reservoir of biological tools to be discovered and harnessed to face new challenges (e.g., plastics bioconversion).
As defined in 2001 by the Organization for Economic Cooperation and Development, the creation and maintenance of Biological Resource Centres (BRC) is of great importance to the scientific community to tackle global challenges of modern societies8. In France, the CIRM-CF (Centre International de Ressources Microbiennes – Champignons Filamenteux, https://www.cirm-fungi.fr) is a French BRC created in 2006 and gathering representatives of the fungal diversity and associated molecular and physiological information. The CIRM-CF preserves and studies not only >2800 fungal original strains of biotechnological interest, mainly saprobic species growing on plant materials, but also strains from polluted agro-industrial sites.
Here we present the first large-scale phenotyping study to assess the potential of this fungal collection toward five non-natural compounds used at industrial scales, including dyes from the textile industry, lignosulfonates (LGSs) from the pulp and paper industry, soluble polyurethane from the plastic industry, and microcrystalline cellulose used in the food, pharmaceutical, and cosmetic industries. We describe the functional phenotyping (growth and degradation assays) of >1000 strains belonging to 26 orders, 78 families, and 231 different genera, according to the Mycobank Database9–11. The functional diversity was analyzed in regard to the ecology of the species and the taxonomy at the family, species, or intraspecies levels. Our work unveils the high biotechnological potential of fungal diversity for the development of sustainable solutions to preserve our planet.
Results
Building of the CIRM-CF fungal collection
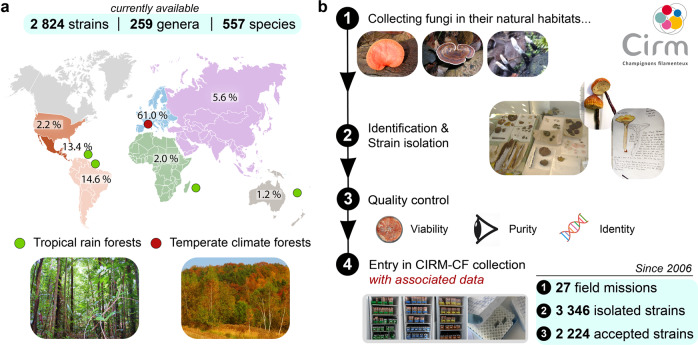
As of May 2021, the CIRM-CF collection contains 2824 fungal strains, from 259 genera and 557 species (Fig. 1a). The collection results from our efforts deployed since 2006 to acquire and preserve the fungal diversity living in French territories (Fig. 1b), through field collection (in natural habitats) and strain deposits by mycologists. As shown in Fig. 1b, the CIRM-CF took part in 27 field collecting expeditions in tropical rainforests (mainly French Guiana, Martinique, Guadeloupe) as well as in temperate forests. As a result, in collaboration with a consortium of expert mycologists (from Universities and learned societies), we have carried out the macro- and micro-morphological identification and strain isolation of 3346 candidate strains. In order to ensure that high-quality BRC standards are met, we checked the purity and the viability of candidate strains by three successive sub-cultures before entering them in the collection. Moreover, we systematically checked strain identity (initially deduced from morphological identification keys) using molecular authentication such as the genotyping of one barcoding gene (usually ITS1-5.8S-ITS2). After applying these selection filters, we rejected about one-third of isolated strains due to (i) impurity, (ii) poor viability, or (iii) when the molecular information was not in agreement with specimen morphological identification. This elevated exclusion rate highlights the technical hurdles and challenges pertaining to proper cultivation and/or isolation of pure fungal strains from natural habitats. After authentication, the fungal strains are maintained with 3 different storage modes (cryopreservation under liquid nitrogen, water, and oil-submerged cultures at 4 °C) in different locations and the viability of the cryopreserved cultures are checked after 6 months.
Fig. 1. Diversity and enrichment strategy of the CIRM-CF fungal collection.
a Overview of the taxonomic and geographic diversity of the 2824 (as of May, 2021) available fungal strains preserved in the CIRM-CF. b Workflow of strains enrichment in the CIRM-CF collection from 2006 to 2020, with fungi coming from mainly France and its overseas territories. The first step consists in collecting fungi in their natural habitat. Second, each collected sample is registered and identified by expert mycologists. The strain isolations are directly performed in a field laboratory, by the CIRM-CF or by associated mycologists from Universities and French learned societies. Finally, the viability, the purity, and the identity of each strain are checked. When the strains are viable, pure, and when the molecular information is in accordance with the morphological identification, the strains can enter the CIRM-CF collection.
From the complete collection of the CIRM-CF, a total of 1031 fungal strains were selected for this functional study. These 1031 strains were selected to proportionally represent the taxonomic diversity of the strains available in the CIRM-CF collection. Precisely, this set encompasses 400 species, 99% from the Ascomycota and Basidiomycota phyla, and spanning 26 orders, 78 families (43 Basidiomycota, 31 Ascomycota, and 4 Mucoromycota families), and 231 genera (135 Basidiomycota, 91 Ascomycota, 5 Mucoromycota genera; Supplementary Data 1 and Supplementary Table 1). In all, 68% of these strains were collected in French territories, 40% of which coming from overseas territories (mainly French Guiana and Martinique). Of note, fungal orders for which at least 10 strains were tested (shown in bold characters in Supplementary Table 1) encompass 365 different species. Furthermore, although the strain sampling of some orders could appear as downsized (e.g., 20 vs 392 strains in the Gloeophyllales and Polyporales orders, respectively), the diversity sampling is still ensured since these seemingly downsized orders usually contain less families (e.g., 1 vs 18 families, respectively)12,13. We underscore that the CIRM-CF collection is dedicated to filamentous fungi able to decay plant biomass transformation, the represented fungi are thus almost exclusively saprobic (growing on wood particularly).
Set-up of the large-scale multi-phenotyping
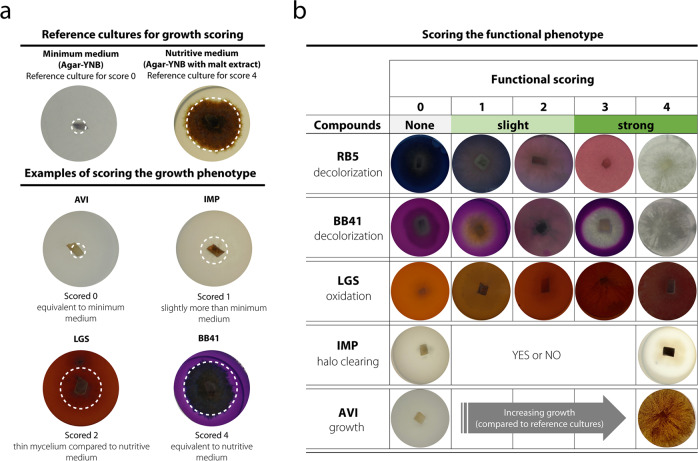
To assess the degrading potential of the fungal strains toward molecules engineered by mankind (Fig. 2), we selected five compounds known to be highly recalcitrant to degradation14–17: two different synthetic azo dyes, Reactive Black 5 (RB5) and Basic Blue 41 (BB41), which are among the most commonly used dyes in the textile industry, and thus major environmental toxic pollutants; soluble LGS as a pollutant coproduct from the pulp and paper industry; Impranil® DLN-SD (IMP) as a soluble polyurethane used in plastic and textile industries; and microcrystalline cellulose (Avicel®PH-101 (AVI)), a recalcitrant refined wood pulp, notably widely used in the food industry as texturizer (Supplementary Fig. 1). For each strain, a six-well plate was used to assess simultaneously the different growth conditions (Fig. 2a), including (i) a positive control culture on malt medium and (ii) each of the five tested compounds suspended in agar-yeast nitrogen base (YNB) minimum medium (except for RB5 and BB41, vide infra). Of note, a “negative” control culture on agar-YNB minimum medium (devoid of additional carbon source) was systematically assessed in parallel. After an incubation period of 13 days at 25 °C, different phenotypes were observed: the decolorization of RB5 and BB41, phenol oxidation of LGS, clearing halo formation of IMP, and growth on AVI. The biosorption (i.e., absorption on the mycelium) of the dyes was not considered as decolorization. To obtain semi-quantitative information on the efficiency of degradation, we attributed fungal growth scores (from 0 to 4; Fig. 2a) and degradation scores (from 0 to 4; Fig. 2b), henceforth called “functional phenotype scores,” as follows: for fungal growth (mycelium diameter and density) scoring, a score of 0 was attributed when no growth difference was observed between a given tested condition and the agar plate control (Fig. 2a). When growth was similar to cultures on malt agar plate (positive control), a score of 4 was attributed. Intermediate growth was attributed intermediate score (from 1 to 3). Regarding functional phenotype scores (Fig. 2b), scores for azo dye (RB5 and BB41) decolorization and LGS phenol oxidation were 0 (no decolorization/oxidation), 1 (light), 2 (medium), 3 (strong), or 4 (maximum), whereas scores for clearing halo formation on IMP were 0 (negative) or 4 (positive). For Avicel, the functional phenotype score was the growth score (raw data are available in Supplementary Data 2).
Fig. 2. Functional screening methodology.
a The growth phenotype was scored by comparing the mycelium growth in a given condition (i.e., agar-YNB + tested compound) vs the two following controls setting the boundaries of the growth scale: agar-YNB plate (score 0) and malt agar plate (score 4). Note that for RB5 and BB41 the agar plates contained 0.5% malt (vs 1.5% malt in positive control plate) to sustain growth. The white dotted circle indicates the fungal growth diameter (which is not always visible on pictures). b Functional phenotype scores (ranging from 0 to 4) reflect the extent of decolorization (on the industrial dyes Reactive Black 5 (RB5) and Basic Blue 41 (BB41)), browning upon phenol oxidation (on lignosulfonate (LGS)), clearing halo formation (on the soluble polyurethane Impranil® DLN (IMP)) or growth (on Avicel (AVI)). A score of 0 means no decolorization/degradation/oxidation. The pictures show examples for the different scores.
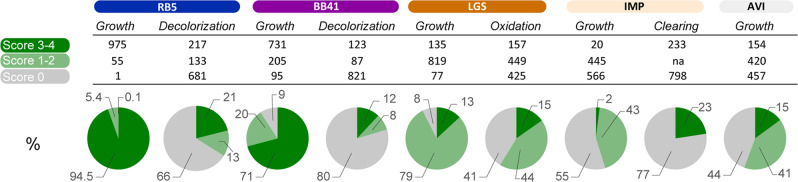
For RB5 and BB41, preliminary tests indicated that the minimum medium needed to be supplemented with malt extract, as otherwise no fungal growth could be observed. Therefore, for these two substrates fungal growth cannot be associated with RB5 and BB41 being used as carbon source. Most fungal strains were able to grow (scores 1 to 4) on malt agar plates containing RB5 (99.9%) and BB41 (91%) (Fig. 3). Yet, the relatively inferior growth (scores ≤2) for a significant number of strains (5.5% for RB5 and 29% for BB41), compared to the positive control and scored 4 strains, points at inhibitory growth properties of these azo-dyes, particularly for BB41. Furthermore, growth was clearly not correlated to decolorization potential as only 34% (RB5) and 20% (BB41) of the strains significantly achieved decolorization (Fig. 3). Note that biosorption hits were excluded from the decolorizing strain count. Regarding LGS, most strains (92%) were able to grow, albeit the growth extent remained limited as only 13% reached the scores 3 and 4, most probably due to the difficulty to assimilate LGS that was used as sole carbon source in the culture medium. Yet, 59% of the strains managed to oxidize phenols from LGS (observed by browning). As to the crystalline cellulose, a substrate that could be expected to be readily degraded by saprotrophic fungi, only 15% of strains succeeded to significantly grow. Regarding IMP, our large-scale phenotyping experiment allowed to evidence a few set of strains (2%) showing a significant growth (score ≥3) (and 43% of the strains showed a limited growth), indicating that IMP was the most recalcitrant compound tested in the present study. Nonetheless, 23% of the strains succeeded to form a clearing halo, underlying that limited growth could be sufficient to produce degrading enzymes.
Fig. 3. Global phenotype scoring of the 1031 filamentous strains.
The figure shows for each substrate the number of strains displaying growth and either decolorization (for RB5 and BB41), or oxidation (for LGS) or clearing (for IMP), clustered by score intensity (0, 1–2, and 3–4), see Fig. 2 legend for details on score definition. The pie charts show the relative distribution of scores categories (100% = 1031 strains). AVI Avicel, BB5 Basic Blue 41, IMP Impranil, LGS lignosulfonate, na not applicable, RB5 Reactive Black 5.
Functional phenotypes are not correlated with growth capacities
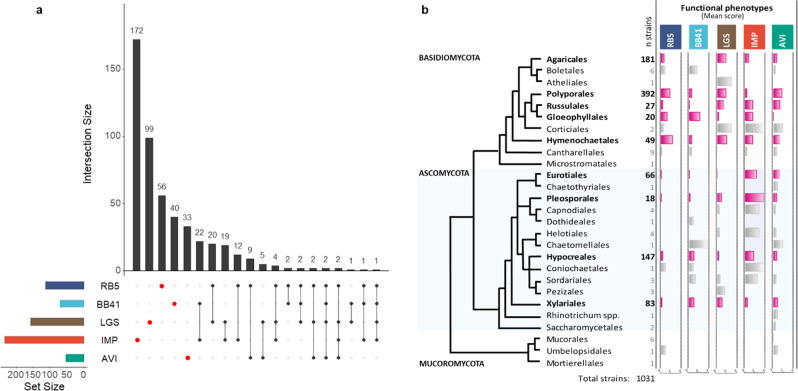
As mentioned above, we did not observe any correlation between growth capacities and (i) decolorization of azo dyes, (ii) oxidation of LGS, or (iii) clearing of IMP (Supplementary Fig. 2a). This result suggests that extensive fungal biomass development is not required for degrading the targeted compounds. In an attempt to identify potential correlations between the phenotypes, we focused our attention on the best-performing strains (with the highest score of 4) for each phenotype (Supplementary Data 2) and generated an UpSet plot (Fig. 4a). Remarkably, most of the best-performing strains were shown to have a single, preferred target compound, with little overlap observed between functional phenotypes (Fig. 4a). Strikingly, this observation was also true for RB5 and BB41, two azo dyes characterized by different chemical structures and number of azo bonds (Supplementary Fig. 1). This lack of correlation between phenotypes suggests that the best-performing strains make use of specialized enzymatic activities not universally shared. Regarding the phylogenetic distribution of the observed phenotypes (Fig. 4b), for orders in which >10 strains could be characterized (shown in bold characters/pink histograms in Fig. 4b), the following general trend stood out: Basidiomycota appear to degrade or modify a broader range of compounds than Ascomycota. RB5 was predominantly decolorized by Basidiomycota, whereas BB41 was decolorized by both Basidiomycota and Ascomycota (mainly Gloeophyllales, Hypocreales, and Xylariales orders). The oxidation of LGS was mainly observed for Basidiomycota, although some Ascomycota showed relatively high mean scores (Pleosporales, Hypocreales, Xylariales). The clearing activity on IMP was observed in a large array of fungal orders with Ascomycota fungi (Pleosporales, Eurotiales, Hypocreales) displaying the highest scores. Finally, the Polyporales and Russulales orders, belonging to Basidiomycota phylum, showed the best mean scores for growth on microcrystalline cellulose.
Fig. 4. Substrate-wise and phylogeny-wise distribution of the phenotypic scores of the 1031 tested fungal strains.
a UpSet plots illustrating quantitative intersection of the sets of strains showing the highest score (score 4) for each functional phenotype (the colored bars on the left-hand side indicate the total number of strains that scored a 4 for each compound). For instance, looking at IMP, out of 233 “score 4” strains, 172 scored a 4 exclusively on IMP (and not on any other compound), while 22 scored a 4 on both IMP and BB41. The figure shows that high scoring strains with a single phenotype represent the majority. b Mean scores of multi-phenotyping of 1031 natural strains belonging to 26 fungal orders mapped onto the phylogenetic tree. The numbers of analyzed strains (n samples) are indicated in the first column. Horizontal histograms show the mean scores for each fungal order for each phenotype, ranging from 0 to 4. Note that these scores reflect a functional phenotype (i.e., decolorization (RB5, BB41), oxidation (LGS), degradation (IMP), or growth (AVI)). Growth phenotypes for all strains/compounds couples are provided in Supplementary Data 2. Histograms colored in pink indicate orders for which more than ten strains were analyzed (otherwise shown in gray). The topology of the tree was built according to recent phylogenies of Fungi57–59.
Fungi display a great functional diversity at every taxonomic rank
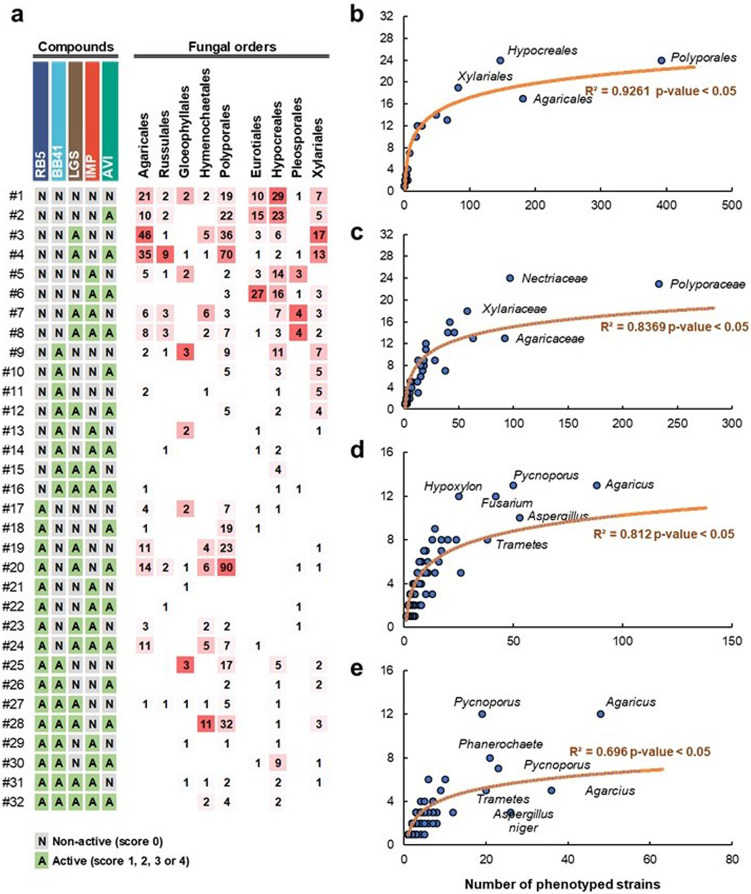
The global analysis presented herein above highlighted that most of the best-performing strains display a main phenotype, although minor “activity” toward other targets could be detected. To probe the order/species-dependent functional diversity, we categorized each target compound/strain couple as either non-active (“N,” i.e., when score was 0) or as active (“A,” i.e., for scores from 1 to 4). Given that 5 compounds were tested, this 2-level categorization yields in theory 25 possible phenotype combinations (Fig. 5a). Interestingly, the 32 theoretically possible functional profiles were all observed at least once, albeit profiles with single and dual phenotypes were preponderant. We observed that some profiles were more frequent than others such as the functional profile #3 (LGS oxidation) and #4 (LGS oxidation and growth on microcrystalline cellulose). Taking the Russulales order as an example, while profile #4 gathers one-third of the tested strains, the remaining two-thirds are distributed across 11 different profiles, indicating the presence of a significant functional diversity within a single order.
Fig. 5. Overview of the functional diversity at different taxonomic levels.
a Distribution of functional profiles at the order level. The two-level categorization of non-active (N) and active (A) substrate–strain couples evaluated on 5 different substrates yields a theoretical maximum of 32 profiles. For each fungal order, the numbers provided in the matrix represent the number of strains with a given profile (for each fungal order, a color gradient from white (only one strain) to red (maximum number of strains) has been applied to show the distribution of functional profiles in the order). As an example, 46 strains from the Agaricales order displayed the profile #3 (LGS oxidation only). b–e Logarithmic regression between the number of functional profiles observed (x axis) within a taxonomic rank (indicated in the figure) and the number of analyzed strains (y axis) in this rank, at the order (b), family (c), genus (d), and species (e) levels.
To delineate the “strain effect” on the observed functional diversity, we then analyzed to which extent the number of tested strains impacted the number of detected functional profiles (Fig. 5b–e). Unsurprisingly, at the order level, the higher the number of tested strains, the more the functional profiles were observed (Fig. 5b). We also observed a similar correlation at the family (Fig. 5c) and genus (Fig. 5d) levels. Astonishingly, a high functional diversity was also noticed between strains belonging to a same species (Fig. 5e). Depending on the species, 3–12 distinct profiles were observed when >10 strains of the same species were screened. For instance, 12 distinct profiles were observed with the phenotyping of 19 strains of Pycnoporus cinnabarinus or the phenotyping of 49 strains of Agaricus bisporus. Using the non-linear regressions shown in Fig. 5b–e, and provided that good enough taxonomic coverage/species diversity is ensured, we estimated the minimal set of strains to be screened to cover a functional diversity as large as possible to be 150, 75, 40, and 20 strains at the order, family, genus, and species level, respectively.
A roadmap for the selection of fungal families for dedicated applications
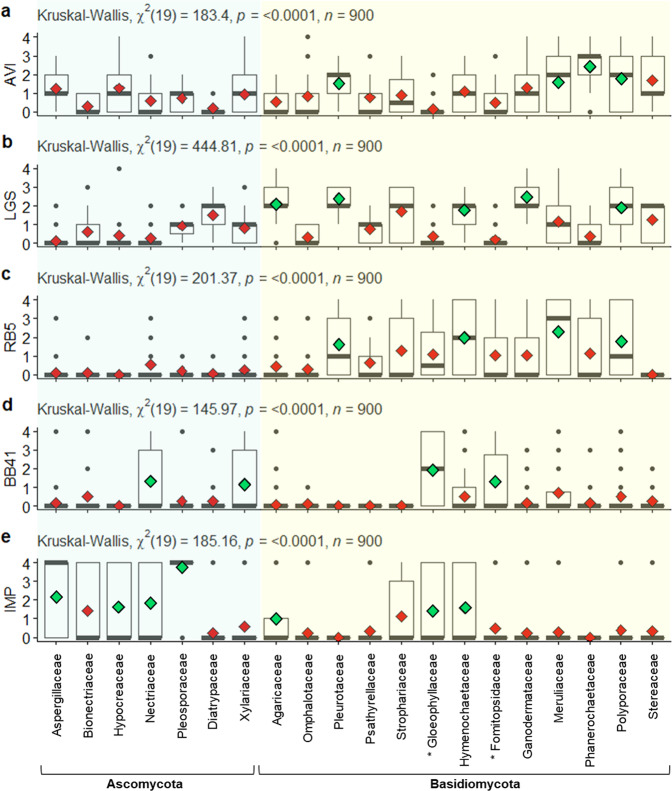
Among the wide diversity offered by the fungal kingdom, identifying the most suitable fungal family(ies) for a dedicated application is a true challenge. Here we set off to provide knowledge-based selection guidelines regarding the degradation of the five selected industrial compounds and applications thereof. To this end, we analyzed the statistical distribution of the functional phenotype scores within fungal families with satisfying diversity coverage (i.e., >10 tested strains), representing a total of 900 fungal strains (Fig. 6 and Supplementary Data 3). Horizontal reading of Fig. 6 shows that some compounds are preferentially targeted by specific fungal families (e.g., RB5 by Basidiomycota families) while vertical analysis informs that some fungal families display a marked preference for specific industrial compounds (e.g., Phanerochaetaceae with AVI). Strikingly, almost all families revealed the presence of high-score outliers, highlighting that even fungal families with seemingly overall poor activity on a particular compound can contain a few efficient strains.
Fig. 6. Box plots showing the fungal family-dependent distribution of phenotypic scores.
For each fungal family, and each target compound (a–e), the median score (thick gray bars) and mean score (red or green diamonds) are shown. For a given target compound, green diamonds show families with a mean score significantly higher than other families. Outliers are represented by dots. Families of brown-rot fungi are marked by an asterisk. For each target compound, non-parametric Kruskal–Wallis test indicates (p value and χ2) significative differences between fungal families. Wilcoxon signed-rank test were computed to compare paired data (i.e., family to family; p values are shown in Supplementary Data 3).
In detail, regarding microcrystalline cellulose, out of the 154 strains that showed growth, 142 belong to the Basidiomycota phylum (Fig. 6a), with a significantly higher prevalence of strains from the Phanerochaetaceae, Polyporaceae, and Meruliaceae families. We underscore that strains from the phlebioid clade, within the order Polyporales (Phanerochaetaceae, Irpicaceae, Meruliaceae)12,18, show the highest mean scores of growth on microcrystalline cellulose. For Ascomycota, the Hypocreaceae, Aspergillaceae, and Xylariaceae families stood out with numerous strains able to slightly grow on microcrystalline cellulose (median score ≤2).
Concerning LGS oxidation, families from lignicolous Basidiomycota showed relatively good mean scores, with highest prevalence being observed for strains from the Ganodermataceae, Pleurotaceae, Agaricaceae, Polyporaceae, and Hymenochaetaceae families (Fig. 6b). In contrast, the mean scores obtained by brown-rot families (Gloeophyllaceae, Fomitopsidaceae) were very low. This observation is consistent with the acknowledged limited abilities of brown-rot fungi to degrade or modify lignin in nature (see “Discussion”).
Regarding the decolorization of RB5 (Fig. 6c), stark differences between the fungal phyla were observed, Basidiomycota being in general much more efficient than Ascomycota. In particular, the Basidiomycota families Pleurotaceae (69% of tested Pleurotaceae), Meruliaceae (66%), Hymenochaetaceae (63%), and Polyporaceae (56%) showed the strongest prevalence for decolorizing RB5. The fact that families of wood decaying Basidiomycota were overall the most efficient for RB5 decolorizing is in line with previous observations made on synthetic dyes19. For Ascomycota, a few strains were outstanding, in particular among the Fusarium species (Nectriaceae). Strikingly, the taxonomic distribution of phenotypes observed on the second tested dye, BB41, was totally different from that of RB5 (Fig. 6d), Gloeophyllaceae, Nectriaceae, Xylariaceae, and Fomitopsidaceae families displaying the best mean scores.
Finally, concerning the commercial polyester polyurethane IMP, we were surprised to observe that all families except two had at least one strain able to efficiently clear IMP (Fig. 6e). Indeed, Fig. 6e shows outliers in all families, except the Pleurotaceae and Phanerochaetaceae families. In general, Ascomycota were more efficient (higher mean and median scores) than Basidiomycota at degrading such artificial polymer. In particular, the Pleosporaceae family showed the best mean score, with 14 out of the 15 tested strains clearing IMP with success, followed by the Aspergillaceae family that displayed >50% of positive strains. It is noteworthy that genera containing pathogenic species (Botrytis, Sclerotinia, Alternaria, Phoma, Colletotrichum, Mycosphaerella, Verticillium, Trichothecium, Fusarium…) showed a propensity to be good IMP degraders. Yet, some Basidiomycota families (in particular Hymenochaetaceae, Gloeophyllaceae, Strophariaceae, Agaricaceae) also emerged (because of few strains from Phellinus (sensu stricto), Tropicoporus, Gloeophyllum, Heterobasidion, Amylosporus, Trichaptum, Laetisaria, Agrocybe, and Agaricus genera) as potential good candidates to uncover novel polyesterases (see “Discussion”).
Discussion
Fungi are set to play a cornerstone role in the success of the emerging bioeconomy, in particular to propose innovative biotechnological solutions to pollution issues originating from human-designed compounds. Intensive fungal genome sequencing programs carried out in the past decade teach us that a tremendous diversity in terms of fungal enzyme activities and mechanisms is to be expected20. However, functional exploration of such diversity is still very limited and selecting the most appropriate fungal strains for a dedicated application remains a true challenge. Here, by carrying out a large-scale phenotyping of 1031 fungal strains, on 5 different industrial, non-natural compounds, we (i) map the biotechnological potential of filamentous fungi, (ii) demonstrate that functional diversity can be observed down to the intra-species level, and (iii) provide guidelines for the selection of fungal families/species for selected applications. Such endeavor would not have been possible without the existence of fungal BRCs, which, due to a time-consuming and painstaking work, collect, acquire, authenticate, preserve, and finely characterize their biological materials. Compared to other larger or taxonomically more diverse BRCs, such as the CBS-KNAW (Westerdijk Institute, Netherlands), the IBT Fungal collection (DTU, Denmark), the CABI collection (United Kingdom BRC network), or the BCCM/MUCL (Catholic University of Leuven, Belgium), the “CIRM-CF” has the particularity to be a more focused collection, rich in wood-decaying filamentous fungi. While we believe that this collection is well suited to search for activities on recalcitrant (industrial) compounds, it is also likely that the evaluation of other collections, following the methodology outlined here, could reveal alternative interesting taxonomic ranks, especially if different types of compounds are to be targeted.
Our comparative study lays the foundations for future studies insofar as the observed growth and degrading phenotypes allows highlighting strains with promising, yet-to-be explored enzymatic potential. Among our target substrates, we chose two plant biomass-derived compounds, LGS and AVI, serving as proxy of plant components (lignin and cellulose) that are at the heart of the biorefinery. On the one hand, lignin is still seen as a very recalcitrant polymer for which fungal biotechnology can bring solutions for its efficient conversion. On the other hand, microcrystalline cellulose AVI is already used in various fields (e.g., food, pharmaceutical and cosmetic industries) and the search for new strains efficient at its degradation will prove useful in the future circular bioeconomy context. Here we show that white-rot fungi (e.g., Ganodermataceae, Polyporaceae, Hymenochaetaceae) displayed the best scores for LGS oxidation, which is consistent with their abilities to degrade and mineralize lignin in nature21. In contrast, brown-rot fungi showed a poor ability to oxidize LGS. This observation can be rationalized by the fact that, unlike white-rot fungi that can mineralize lignin22, brown-rot fungi can only modify it, notably via non-enzymatic reactions (e.g., Fenton chemistry)23,24. During evolution, brown-rot fungi have lost their lignin-degrading peroxidase genes25 and possess only one or two genes coding for non-ligninolytic, low-redox potential general peroxidases26,27. Brown-rot fungi also possess on average three laccases25,28, which are, however, usually inefficient on syringyl unit, the main constituent of LGS29,30. Furthermore, brown-rot fungi (e.g., the Gloeophyllaceae and Fomitopsidaceae families) hardly thrive on AVI as carbon source, most likely because of their poor enzymatic arsenal related to crystalline cellulose degradation31,32.
Synthetic dyes, such as BB41 and RB5, are known to be a serious source of water pollution, as various industries from developing countries (e.g., textile industry) have been reported to often dump their waste streams in lakes, rivers, or in the sea. Fungi represent good candidates for bioremediation strategies insofar as they are considered as microorganisms highly robust and tolerant to high concentrations of pollutants. Here, we showed that despite their poor efficiency on LGS and AVI, brown-rot fungi proved to be able to decolorize synthetic dyes, notably BB41. It is known that fungal laccases (with redox mediators) and peroxidases can bleach RB533–35, whereas only peroxidases have hitherto been shown to discolor BB4136,37. Brown-rot fungi being devoid of ligninolytic peroxidases25 probably act on BB41 via other systems, involving either Fenton or other types of heme-peroxidases38. For instance, chloroperoxidases, which are produced by brown-rot fungi39,40, are able to efficiently oxidize synthetic dyes41 and lignin42,43. More globally, the observed drastically different taxonomic distribution of BB41 vs RB5 decolorizing strains suggests the involvement of (enzymatic) mechanisms not universally shared by the different fungal families.
The afterlife fate of plastics is a major concern of modern societies. Plastics represent a large class of synthetic polymers, including two main categories, namely, thermoplastics (e.g., polyethylene, polystyrene) and thermoset plastics (e.g., polyurethanes). Here we chose IMP as a proxy of the latter category. IMP is a polyester–polyurethane polymer from Bayer Corporation used for textile, leather, and aircraft fabric coatings44,45. For this first large-scale study, we chose IMP because its main chain is composed of heteroatoms, making it more susceptible to be degraded by wild-type fungal strains. IMP is an opaque milky suspension that becomes transparent upon degradation by urethanases and/or polyesterases, such as cutinases46–48. In our study, Ascomycota, and in particular phytopathogenic species, appeared to be more amenable to degrade IMP, which may be related to the capacity of phytopathogenic fungi to penetrate living-plant surfaces via the recruitment of cutinases49,50. This observation is consistent with previous studies showing several Ascomycota strains able to form a clearing halo from the IMP46,47,51. However, the fact that non-pathogenic Basidiomycota strains, notably from Hymenochaetaceae and Gloeophyllaceae families, were able to degrade the IMP is more surprising as their genomes contain very few or even no genes encoding cutinases (Pfam domain: PF01083; Supplementary Table 2). These observations augur the presence of new cutinases or polyurethanases yet to be uncovered. Furthermore, building on this work, we will tackle harder to degrade plastics in future studies.
Beyond providing functional leads and revealing fungal strains of interest, our study highlighted various hurdles underlying the maintenance and functional characterization of such a large collection. Notably, considerable research efforts are needed to address the difficulties of capture, isolation, and cultivation of fungal diversity from natural environments. Notably, we underscored the crucial role of strain authentication, in particular for freshly isolated strains as well as strains originating from ancient collections.
Our study proposes a methodological pipeline, exemplified by the heat-mapped numerical scoring system based on active vs non-active strain categorization (Fig. 5), which allows to evaluate the functional diversity at different taxonomic ranks. Nevertheless, we draw the attention toward the fact that the observed functional diversity is dependent on the inherent biological diversity of the tested taxonomic group. The seemingly broader functional diversity of Polyporales observed here (Fig. 5) should thus be interpreted with caution given the prevalence of this order in our fungal collection. Also, determining the appropriate number of strains to be screened to get a representative overview of the functional diversity at a given taxonomic rank is far from being obvious. For instance, our results demonstrated here that, to capture the largest functional diversity, one may need to assess up to 20 strains within a single species. In the same vein, the “strain effect” should not be overlooked, as illustrated by the numerous outliers frequently observed within each fungal taxonomic group, which may represent promising strains.
In a nutshell, searching for novel fungal biocatalysts adapted to new biotech applications requires an a priori-free screening covering a large diversity, both at different taxonomic ranks and at the intraspecies level. We foresee that fungal BRCs, by sampling in accordance to the Nagoya protocol (access to resources and benefit-sharing)52,53, and preserving large taxonomic and intraspecies diversity, along with evolving screening methods, can play a major role in the discovery and engineering of bio-inspired solutions to address the various challenges of this century.
Methods
Chemicals and reagents
Reactive Black (RB5; dye content 50%), lignosulfonic acid sodium salt (LGS; purity 90%), and Avicel PH-101 (AVI) were purchased from Sigma-Aldrich (Saint-Quentin-Fallavier, France). Basic Blue 41 (BB41, dye content 40%) was purchased from Setaş company (Turkey) and Impranil DLN-SD (IMP, purity 40%) from Covestro (Leverkusen, Germany). Malt extract was purchased from Duchefa Biochemie (Haarlem, Netherlands), agar from Becton Dickinson (Grenoble, France), and YNB from Fischer Scientific (Illkirch, France). Most other chemicals were purchased from Sigma-Aldrich unless stated otherwise.
Collection of fungi and deposit of strains in the CIRM-CF
The CIRM-CF collection was established in 2006, resulting from the fusion of the collection of filamentous fungi of agro-industrial interest already held by our laboratory (BBF) and existing collections from different organizations: the French National Research Institute for Agriculture, Food and Environment (INRAE), the National Museum of Natural History (MNHN), the French Agricultural research and international cooperation organization (CIRAD), and the French Universities of Pharmacy (Paris Descartes, UGA Université Grenoble-Alpes). Since 2007, the CIRM-CF collection is regularly enriched and diversified via field-collecting in natural habitats, carried out in mainland France, and also in overseas territories, i.e., Guadeloupe, Martinique, French New Caledonia, and French Guiana. The initial identification of field-collected fungi, based on macro- and micro-morphological features, is carried out by our consortium of expert mycologists from Universities and learned societies.
Strain isolation, authentication, and preservation
Following initial identification (vide supra), isolation were performed by transferring tissues from collected sporocarps onto agar plates containing either malt agar 2% (MA2; malt extract, Duchefa Biochemie, Haarlem, Netherlands) for basidiomycetes or potato dextrose agar (PDA; Sigma-Aldrich, Saint-Quentin-Fallavier, France) for ascomycetes, and both supplemented with antibiotics (0.025% (w/v) Chloramphenicol and 0.04% Gentamicin). Then, at least three successive subcultures were performed using the same media without antibiotics to obtain visibly pure cultures.
To check the identity of candidate strains, genomic DNA was extracted from approximately 100 mg of fresh mycelia using the NucleoSpin PlantII Kit (Macherey–Nagel, France). Then, for each strain, at least one barcoding gene (mainly rDNA-ITS) or species-specific genes were amplified by PCR. To this end, the following primers were used: ITS1/ITS4 and ITS5/ITS4 for rDNA-ITS amplification54, TEF1α-983-F-CF2/TEF1α-2218-R-CR2 for TEF-1α region amplification55. Following a protocol adapted from Lomascolo et al.56, the PCR reaction (50 µL total volume) contained 50 ng genomic DNA and 1.5 U Expand™ High Fidelity PCR master mix and primers (0.3 µM) (Roche, France). DNA amplification was then performed in a Mastercycler Nexus GSX1 (Eppendorf, Montesson, France) using the following sequence: 1 cycle at 94 °C for 2 min; 10 cycles of 94 °C for 30 s/55 °C for 90 s/72 °C for 1 min; 20 cycles of 94 °C for 90 s/Tannealing°C for 90 s/72 °C for 1 min; then 1 cycle at 72 °C for 7 min. Annealing temperatures (Tannealing) were 51 °C (for ITS1/ITS4 amplification), 53 °C (for ITS5/ITS4 amplification), and 51 °C (for TEF1α-983-F-CF2/TEF1α-2218-R-CR2 amplification). The PCR products were sequenced by Genewiz (Leipzig, Germany). For the identification of A. bisporus and Fusarium strains, we used species-specific primers (all primers are provided in Supplementary Table 3) to probe the presence of species-specific genes by qPCR (CFX96 Touch Real-Time PCR, Bio-Rad, Marnes-la-Coquette, France). The qPCR reaction (20 µL total volume) contained 50 ng genomic DNA and 10 µL SsoAdvanced™ Universal SYBR® Green Supermix (Biorad, Marnes-la-coquette, France). The thermocycle was conducted according to the manufacturer’s recommendation.
Strains were entered in the CIRM-CF collection only upon agreement of both the molecular authentication and identification based on macro- and micro-morphological features. Subsequently, the authenticated strains were maintained using 3 storage modes, including cryo-conservation (under vapor phase of liquid nitrogen at −196 °C), water, and oil-submerged agar slant (at 4 °C). The most appropriate storage mode being species-dependent, the viability of stored strains was systematically controlled after 6 months post-entry in the collection. This know-how is available to the scientific community, through the CIRM-CF website (https://www.cirm-fungi.fr).
Multiphenotyping assay
The 1031 assessed strains were selected in the CIRM-CF collection with the objective to not only cover the taxonomic diversity but also fairly represent the proportion of strains available within the different fungal families. For precultures, all fungal strains were cultivated in petri dishes containing either PDA or MA2 for ascomycetes or basidiomycetes, respectively. The preculture plates were incubated between 7 and 15 days, at 25 °C or 30 °C (see CIRM-CF website for details). For each strain, the preculture was used to inoculate a 6-well plate (Greiner bio-one, Dutscher, Brumath, France), in which each well contained 6 mL of minimum medium (1.5 % (w/v) of agar supplemented by YNB (6.7 g/L final)) and one of the following mixtures: 1.5% (w/v) of malt extract (well #1, positive growth control); 0.5% (w/v) of malt extract supplemented with 0.02% (w/v final concentration) of either RB5 (well #2) or BB41 (well #3); 0.5% (v/v) IMP (well #4); 1.5% (w/v) LGS (well #5); 1.5% (w/v) AVI (well #6). In parallel, for each strain we prepared a “negative” control culture on minimum medium agar-YNB. RB5 and BB41 solutions were filter-sterilized before addition to autoclaved agar-malt medium. Note that we added malt in RB5 and BB41 wells as otherwise no fungal growth could be observed. All media had a similar pH (ca. pH 5). Inoculation of each well was done by placing a mycelia-overgrown agar plug (of standardized size, 0.3 × 0.3 cm) into the center of a well. After inoculation, the plates were incubated during 13 days at 25 °C. As shown in Fig. 2, a scoring rule (from 0 to 4) was defined to estimate growth and functional phenotypes after 13 days of culture. Growth was estimated by visual comparison of growth diameter and density of mycelia with culture on agar-YNB plate (“negative” control, score = 0) and with culture on malt agar plate (positive control, score = 4). Examples of extreme scores (0 and 4) are shown in Fig. 2a. Note that dyes biosorption was not considered as decolorization. As the scoring method can be subjective, all the measures were collected by only two inter-trained persons.
Statistics and reproducibility
To investigate the significance of differences between fungal families on each target compound, non-parametric Kruskal–Wallis test and Wilcoxon signed-rank test were performed using the R package. First, for each fungal family/target compound couples, the statistical data (mean, median, standard deviation, quartiles) were computed. Kruskal–Wallis test was performed to obtain a p value indicating the significant differences between all families. As the samplings were different between fungal families, the size effects are always large to moderate. To circumvent this effect, family pairwise comparison was further carried out with Wilcoxon signed-rank tests. Statistical information is given in Supplementary Data 3.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank members of the Fungal Biodiversity and Biotechnological Labs (UMR 1163-BBF) who assisted with the project, especially Craig B. Faulds for his advises concerning the fungal decolorization of dyes, and Hayat Hage for her constructive discussion concerning R programing. We acknowledge industrial companies for furnishing Impranil (Covestro, Germany) and Basic Blue 41 (Seitas, Turkey). We thank Anaïs Bonzino for her help of the figure design. We acknowledge funding from the INRAE Institute (CNOC), from the Sud-PACA Region, and from the French National Research Agency (ANR, program E-TRICEL ANR-07-BIOE-006). We acknowledge support from the CIRM Network and the National Research Infrastructure AgroBRC RARe (Biological Agronomic Resources: https://www.agrobrc-rare.org/agrobrc-rare_eng/). We acknowledge all the French institutions who deposited fungal strains in the CIRM-CF collection (INRAe, Grenoble-Alpes University, Lille University, MNHN, Paris University, CIRAD…) as well as depositor mycologists and collaborators: Christophe Billette, Marc Ducousso, Noémie Jacques, Guo Lin, Eliane Louisanna, Magalie Moinard, Heidy Schimann, Alba Zaremsky, Mingzhe Zhang, and the French Mycological Society (SMF). Financial help from various organisms (French Forestry Office – ONF; Martinique Territorial Administration; Regional Nature Park of Martinique – PNRM; Regional Environment Direction of Guadeloupe and Martinique - DEAL) to support field trips in the French West Indies is also acknowledged.
Author contributions
D.N., D.C., and A.F. conceived the idea and designed the phenotyping method; A.F. established the growing strategy of the CIRM-CF; D.C., S.T., and D.N. conducted the experiments and collected the data; P.C., R.C., C.D., F.R.-F., J.F., J.D., J.G., C.L., P.-A.M., B.R., L.S., S.W., D.N., S.G., and A.F. collected, identified, and deposited fungal specimens or strains in the CIRM-CF collection; D.C., S.T, M.H., L.L.-M., D.N., and A.F. authenticated strains by molecular typing before entering in the CIRM-CF collection; D.N. and D.C. analyzed the data; D.N and A.F. directed the CIRM-CF; D.N. and B.B. wrote the initial draft and revised versions of the manuscript, with contributions from J.-G.B., M.-N.R., and A.F. All authors read and revised the final version of the manuscript.
Data availability
All data are available within the article and its Supporting Information File and from the corresponding author upon reasonable request. A reporting summary for this Article is available as a Supplementary Information file. Supplementary Data files are available online on the FigShare platform (10.6084/m9.figshare.14742810). The CIRM-CF (“Centre International de Ressources Microbiennes – Champignons Filamenteux”) repository can be freely accessed at https://www.cirm-fungi.fr.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary handling editor: Luke R. Grinham.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
David Navarro, Email: david.navarro@inrae.fr.
Bastien Bissaro, Email: bastien.bissaro@inrae.fr.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-021-02401-w.
References
- 1.van Renssen S. A bioeconomy to fight climate change. Nat. Clim. Change. 2014;4:951–953. doi: 10.1038/nclimate2419. [DOI] [Google Scholar]
- 2.Heux S, Meynial-Salles I, O’Donohue MJ, Dumon C. White biotechnology: state of the art strategies for the development of biocatalysts for biorefining. Biotechnol. Adv. 2015;33:1653–1670. doi: 10.1016/j.biotechadv.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Hawksworth, D. L. & Lücking, R. Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol. Spectr. 10.1128/microbiolspec.FUNK-0052-2016 (2017). [DOI] [PubMed]
- 4.Meyer V, et al. Growing a circular economy with fungal biotechnology: a white paper. Fungal Biol. Biotechnol. 2020;7:5. doi: 10.1186/s40694-020-00095-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hülsey MJ, Yang H, Yan N. Sustainable routes for the synthesis of renewable heteroatom-containing chemicals. ACS Sustain. Chem. Eng. 2018;6:5694–5707. doi: 10.1021/acssuschemeng.8b00612. [DOI] [Google Scholar]
- 6.Lynd LR, et al. How biotech can transform biofuels. Nat. Biotechnol. 2008;26:169–172. doi: 10.1038/nbt0208-169. [DOI] [PubMed] [Google Scholar]
- 7.Graham-Rowe D. Agriculture: beyond food versus fuel. Nature. 2011;474:S6–S8. doi: 10.1038/474S06a. [DOI] [PubMed] [Google Scholar]
- 8.OECD. Biological Resource Centres: Underpinning the Future of Life Sciences and Biotechnology (OECD, 2021).
- 9.Robert V, et al. MycoBank gearing up for new horizons. IMA Fungus. 2013;4:371–379. doi: 10.5598/imafungus.2013.04.02.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crous, P., Stalpers, J. & Robert, V. MycoBank: an online initiative to launch mycology into the 21st century. Stud. Mycol. 50, 19–22 (2004).
- 11.Robert, V., Stegehuis, G. & Stalpers, J. The MycoBank engine and related databases. www.mycobank.org (2005).
- 12.Justo A, et al. A revised family-level classification of the Polyporales (Basidiomycota) Fungal Biol. 2017;121:798–824. doi: 10.1016/j.funbio.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Sandoval R, Wang Z, Binder M, Hibbett DS. Molecular phylogenetics of the Gloeophyllales and relative ages of clades of Agaricomycotina producing a brown rot. Mycologia. 2011;103:510–524. doi: 10.3852/10-209. [DOI] [PubMed] [Google Scholar]
- 14.Forgacs E, Cserháti T, Oros G. Removal of synthetic dyes from wastewaters: a review. Environ. Int. 2004;30:953–971. doi: 10.1016/j.envint.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Ruiz‐Dueñas FJ, Martínez ÁT. Microbial degradation of lignin: how a bulky recalcitrant polymer is efficiently recycled in nature and how we can take advantage of this. Microb. Biotechnol. 2009;2:164–177. doi: 10.1111/j.1751-7915.2008.00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Himmel ME, et al. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science. 2007;315:804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 17.Mahajan N, Gupta P. New insights into the microbial degradation of polyurethanes. RSC Adv. 2015;5:41839–41854. doi: 10.1039/C5RA04589D. [DOI] [Google Scholar]
- 18.Binder M, et al. Phylogenetic and phylogenomic overview of the Polyporales. Mycologia. 2013;105:1350–1373. doi: 10.3852/13-003. [DOI] [PubMed] [Google Scholar]
- 19.Rodríguez Couto S. Dye removal by immobilised fungi. Biotechnol. Adv. 2009;27:227–235. doi: 10.1016/j.biotechadv.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 20.Grigoriev IV, et al. Fueling the future with fungal genomics. Mycology. 2011;2:192–209. [Google Scholar]
- 21.Lundell TK, Mäkelä MR, Hildén K. Lignin-modifying enzymes in filamentous basidiomycetes - ecological, functional and phylogenetic review. J. Basic Microbiol. 2010;50:5–20. doi: 10.1002/jobm.200900338. [DOI] [PubMed] [Google Scholar]
- 22.Del Cerro C. et al. Intracellular pathways for lignin catabolism in white-rot fungi. Proc. Natl Acad. Sci. USA118, e2017381118 (2021). [DOI] [PMC free article] [PubMed]
- 23.Eastwood DC, et al. The plant cell wall–decomposing machinery underlies the functional diversity of forest fungi. Science. 2011;333:762–765. doi: 10.1126/science.1205411. [DOI] [PubMed] [Google Scholar]
- 24.Arantes, V. & Goodell, B. in Deterioration and Protection of Sustainable Biomaterials, Vol. 1158 (eds Schultz, T. P., Goodell, B. & Nicholas, D. D.) 3–21 (American Chemical Society, 2014).
- 25.Riley R, et al. Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc. Natl Acad. Sci. USA. 2014;111:9923–9928. doi: 10.1073/pnas.1400592111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruiz-Dueñas FJ, et al. Lignin-degrading peroxidases in Polyporales: an evolutionary survey based on 10 sequenced genomes. Mycologia. 2013;105:1428–1444. doi: 10.3852/13-059. [DOI] [PubMed] [Google Scholar]
- 27.Hage, H. et al. Gene family expansions and transcriptome signatures uncover fungal adaptations to wood decay. Environ. Microbiol. 10.1111/1462-2920.15423 (2021). [DOI] [PMC free article] [PubMed]
- 28.Bissaro B, Várnai A, Røhr ÅK, Eijsink VGH. Oxidoreductases and reactive oxygen species in conversion of lignocellulosic biomass. Microbiol. Mol. Biol. Rev. 2018;82:e00029-18. doi: 10.1128/MMBR.00029-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D’Souza TM, Boominathan K, Reddy CA. Isolation of laccase gene-specific sequences from white rot and brown rot fungi by PCR. Appl. Environ. Microbiol. 1996;62:3739–3744. doi: 10.1128/aem.62.10.3739-3744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.An H, Xiao T, Fan H, Wei D. Molecular characterization of a novel thermostable laccase PPLCC2 from the brown rot fungus Postia placenta MAD-698-R. Electron. J. Biotechnol. 2015;18:451–458. doi: 10.1016/j.ejbt.2015.09.004. [DOI] [Google Scholar]
- 31.Floudas D, et al. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science. 2012;336:1715–1719. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- 32.Floudas D, et al. Uncovering the hidden diversity of litter-decomposition mechanisms in mushroom-forming fungi. ISME J. 2020;14:2046–2059. doi: 10.1038/s41396-020-0667-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ben Ali W, et al. Screening of five marine-derived fungal strains for their potential to produce oxidases with laccase activities suitable for biotechnological applications. BMC Biotechnol. 2020;20:27. doi: 10.1186/s12896-020-00617-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wikee S, et al. Characterization and dye decolorization potential of two laccases from the marine-derived fungus Pestalotiopsis sp. Int. J. Mol. Sci. 2019;20:1864. doi: 10.3390/ijms20081864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uzan E, et al. High redox potential laccases from the ligninolytic fungi Pycnoporus coccineus and Pycnoporus sanguineus suitable for white biotechnology: from gene cloning to enzyme characterization and applications. J. Appl. Microbiol. 2010;108:2199–2213. doi: 10.1111/j.1365-2672.2009.04623.x. [DOI] [PubMed] [Google Scholar]
- 36.Celebia M, Kayac MA, Altikatogluc M, Yildirima H. Removal of cationic dye from textile industry wastewater with using enzyme, fungus and polymer. Online J. Sci. Technol. 2013;3:2. [Google Scholar]
- 37.Urra J, Sepúlveda L, Contreras E, Palma C. Screening of static culture and comparison of batch and continuous culture for the textile dye biological decolorization by Phanerochaete chrysosporium. Braz. J. Chem. Eng. 2006;23:281–290. doi: 10.1590/S0104-66322006000300002. [DOI] [Google Scholar]
- 38.Janusz G, et al. Lignin degradation: microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol. Rev. 2017;41:941–962. doi: 10.1093/femsre/fux049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sista Kameshwar AK, Qin W. Systematic metadata analysis of brown rot fungi gene expression data reveals the genes involved in Fenton’s reaction and wood decay process. Mycology. 2019;11:22–37. doi: 10.1080/21501203.2019.1703052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hofrichter M, Ullrich R, Pecyna MJ, Liers C, Lundell T. New and classic families of secreted fungal heme peroxidases. Appl. Microbiol. Biotechnol. 2010;87:871–897. doi: 10.1007/s00253-010-2633-0. [DOI] [PubMed] [Google Scholar]
- 41.Jin X, Li S, Long N, Zhang R. Improved biodegradation of synthetic azo dye by anionic cross-linking of chloroperoxidase on ZnO/SiO2 nanocomposite support. Appl. Biochem. Biotechnol. 2018;184:1009–1023. doi: 10.1007/s12010-017-2607-0. [DOI] [PubMed] [Google Scholar]
- 42.Ortiz-Bermúdez P, Srebotnik E, Hammel KE. Chlorination and cleavage of lignin structures by fungal chloroperoxidases. Appl. Environ. Microbiol. 2003;69:5015–5018. doi: 10.1128/AEM.69.8.5015-5018.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ortiz-Bermúdez P, Hirth KC, Srebotnik E, Hammel KE. Chlorination of lignin by ubiquitous fungi has a likely role in global organochlorine production. Proc. Natl Acad. Sci. USA. 2007;104:3895–3900. doi: 10.1073/pnas.0610074104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Windemuth E, Gensel H, Kramer M. Impranil, a new raw material for textile finishing. Melliand Textilber. 1956;37:843–846. [Google Scholar]
- 45.Traeubel H. Polyurethane dispersions. J. Am. Leather Chem. Assoc. 1988;83:317–326. [Google Scholar]
- 46.Russell JR, et al. Biodegradation of polyester polyurethane by endophytic fungi. Appl. Environ. Microbiol. 2011;77:6076–6084. doi: 10.1128/AEM.00521-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Biffinger JC, et al. The applicability of Impranil®DLN for gauging the biodegradation of polyurethanes. Polym. Degrad. Stab. 2015;120:178–185. doi: 10.1016/j.polymdegradstab.2015.06.020. [DOI] [Google Scholar]
- 48.Molitor R, et al. Agar plate-based screening methods for the identification of polyester hydrolysis by Pseudomonas species. Microb. Biotechnol. 2020;13:274–284. doi: 10.1111/1751-7915.13418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, Chen J, Li D-W, Zheng L, Huang J. CglCUT1 gene required for cutinase activity and pathogenicity of Colletotrichum gloeosporioides causing anthracnose of Camellia oleifera. Eur. J. Plant Pathol. 2017;147:103–114. doi: 10.1007/s10658-016-0983-x. [DOI] [Google Scholar]
- 50.Davies KA, et al. Evidence for a role of cutinase in pathogenicity of Pyrenopeziza brassicae on brassicas. Physiol. Mol. Plant Pathol. 2000;57:63–75. doi: 10.1006/pmpp.2000.0282. [DOI] [Google Scholar]
- 51.Brunner I, Fischer M, Rüthi J, Stierli B, Frey B. Ability of fungi isolated from plastic debris floating in the shoreline of a lake to degrade plastics. PLoS ONE. 2018;13:e0202047. doi: 10.1371/journal.pone.0202047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Overmann J. Significance and future role of microbial resource centers. Syst. Appl. Microbiol. 2015;38:258–265. doi: 10.1016/j.syapm.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 53.Smith D, da Silva M, Jackson J, Lyal C. Explanation of the Nagoya Protocol on Access and Benefit Sharing and its implication for microbiology. Microbiology. 2017;163:289–296. doi: 10.1099/mic.0.000425. [DOI] [PubMed] [Google Scholar]
- 54.White, T. J., Bruns, T., Lee, S. & Taylor, J. in PCR Protocols 315–322 (Elsevier, 1990).
- 55.Stielow JB, et al. One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia Mol. Phylogeny Evol. Fungi. 2015;35:242–263. doi: 10.3767/003158515X689135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lomascolo A, et al. Molecular clustering of Pycnoporus strains from various geographic origins and isolation of monokaryotic strains for laccase hyperproduction. Mycol. Res. 2002;106:1193–1203. doi: 10.1017/S0953756202006494. [DOI] [Google Scholar]
- 57.Varga T, et al. Megaphylogeny resolves global patterns of mushroom evolution. Nat. Ecol. Evol. 2019;3:668–678. doi: 10.1038/s41559-019-0834-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hibbett DS, et al. A higher-level phylogenetic classification of the Fungi. Mycol. Res. 2007;111:509–547. doi: 10.1016/j.mycres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 59.Spatafora JW, et al. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia. 2016;108:1028–1046. doi: 10.3852/16-042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available within the article and its Supporting Information File and from the corresponding author upon reasonable request. A reporting summary for this Article is available as a Supplementary Information file. Supplementary Data files are available online on the FigShare platform (10.6084/m9.figshare.14742810). The CIRM-CF (“Centre International de Ressources Microbiennes – Champignons Filamenteux”) repository can be freely accessed at https://www.cirm-fungi.fr.