Abstract
Lactic acid bacteria (LAB) are a representative probiotic. As the dominant flora in the human intestinal tract, LAB can regulate the balance of human intestinal flora and improve host health. The purpose of this study was to isolate and screen LAB that are well suited to the intestinal characteristics of the Chinese population, with excellent probiotics and high antibacterial activity. After 16S ribosomal RNA (rRNA) homology and phylogenetic tree analysis, potential probiotics were tested for their antibacterial activity, resistance to artificial gastrointestinal fluid and drugs, surface hydrophobicity, and safety. Three strains of LAB with acid resistance, bile salt resistance, epithelial cell adhesion, and no multidrug resistance were selected: Lactobacillus salivarius, Leuconostoc lactis, and Lactobacillus paracasei. Analysis of the antibacterial active substances in the three strains and their fermentation broths revealed that the main antibacterial substances of L. lactis were organic acids, whereas those of L. salivarius and L. paracasei were organic acids and bacteriocins with broad-spectrum antibacterial activity. These three strains of probiotic LAB with high antibacterial activity were identified as bacterial resources that could potentially be used to develop probiotic preparations for the prevention and treatment of intestinal diseases caused by intestinal pathogens.
Keywords: Intestinal lactic acid bacteria, Screening, Probiotic, Bacteriostatic substance
1 Introduction
With changes in people's living environments and diet, many new diseases have begun to appear and seriously threaten human health (Khanna and Tosh, 2014). The microbial flora in the intestine are diverse and complex, and participate in various activities of the host, such as nutrition metabolism, immune regulation, and defense against pathogens (Jubair et al., 2018). Therefore, the intestinal microecosystem is closely related to human health and diseases. In recent years, regulating the intestinal flora has gradually become a new method for the prevention and treatment of some diseases. Probiotics are widely used for disease prevention, treatment, and repair due to their safety, reliability, and excellent performance (Krzych-Fałta et al., 2018). The probiotic effect mainly involves inhibiting the activity of pathogenic bacteria, preventing bacterial diarrhea, relieving constipation, and maintaining intestinal microecological balance (Yerlikaya, 2019). Lactic acid bacteria (LAB) and bifidobacteria have been widely used in medicine as the main constituents of probiotic dietary supplements and probiotic nonprescription drugs. In the context of the concept of "big health" and its industrialization, it is particularly important to develop new types of intestinal probiotics with excellent performance to avoid the various problems caused by the use of antibiotics.
The criteria for screening probiotics are very strict; probiotics must have a tolerance to the digestive juice secreted by the human digestive system such that a sufficient amount of live bacteria can reach the intestines after passing through the digestive system (Giraffa et al., 2010). Candidate probiotic strains must resist acidic environments and bile acids, adhere to and colonize the intestine, and be safe in animals (Tuomola et al., 2001). Probiotic strains need to exhibit a certain degree of process resistance during preparation, resistance to heat, acid, salt, cold, and other environmental stress changes, and maintain a certain level of biological activity. In addition, it is necessary to evaluate the safety of selected probiotic strains (Lähteinen et al., 2010). Although the risk of probiotic infection is very low, individuals with weak immune systems can be infected. Probiotics may produce harmful substances during their metabolism. The antibiotic resistance of probiotics also needs to be considered, as some resistance genes can spread between probiotic strains and even between probiotics and pathogenic bacteria, posing a serious threat to human health (Alshammari et al., 2019).
Most LAB can inhibit the activity of pathogenic and spoilage microorganisms mainly because they secrete and produce antibacterial substances, such as organic acids, bacteriocins, and hydrogen peroxide (Zheng and Slavik, 1999). Organic acid is an important antibacterial compound produced by LAB. Hydrophobic organic acid that is not dissociated is highly soluble in fat, facilitating penetration of the cell membranes of pathogenic microorganisms for entry into the cytoplasm where these molecules destroy the dissociated bacterial cells and release their contents (Otero and Nader-Macías, 2006). Gram-positive bacteriocins primarily affect the stability of the target cell membrane by inhibiting the formation of the cell wall of pathogenic bacteria or creating pores in the membrane, thereby causing cell death (Pei et al., 2018). Some Gram-positive bacterial bacteriocins can also induce autolysis of target cells, thereby exerting antibacterial effects (Kordel et al., 1989). Nisin was the first bacteriocin approved for use in food and industrial production, but it has a narrow antibacterial spectrum and inhibits only Gram-positive bacteria, thus limiting its application (Kim et al., 2019). After LAB colonize a host's intestines, they will generate many superoxide anion free radicals, which can be converted into less toxic hydrogen peroxide by respiration. LAB do not possess catalase, which produces hydrogen peroxide in the environment (Shahandashti et al., 2016). Continuous accumulation of hydrogen peroxide inhibits the growth of other bacteria. Some LAB that undergo heterogeneous fermentation produce CO2 by metabolizing hexose, reducing the O2 concentration in the environment, and generating an anaerobic environment, which inhibits the growth of harmful aerobic bacteria (Aymes et al., 1999). LAB can also metabolize citric acid to produce diacetyl through glycolysis, which can inhibit a variety of harmful microorganisms (Marko, 2009).
Few probiotics have been researched and developed specifically for the characteristics of the Chinese intestinal tract. In this study, we isolated LAB with high antibacterial activity from the intestines of healthy Chinese people. We used molecular biology methods to determine the taxonomic status of potential probiotic LAB strains, and then conducted a preliminary study on the antibacterial substances secreted by bacteriostatic LAB. The aim of this study was to screen and obtain probiotic strains that are well suited to Chinese intestinal characteristics, and which will inhibit intestinal pathogenic bacteria. Such strains may have application in future trials of safety and production performance. In addition, we aimed to provide ideas and experience for the research and development of intestinal probiotic strains in China.
2 Materials and methods
2.1. Sample collection
Fecal samples were collected from ten healthy adults, five males and five females, aged (25±3) years, in Harbin, Heilongjiang Province, China. Sample collection was conducted in compliance with local rules and with the approval of national authorities, on the basis of informed consent of subjects. One fecal sample was collected from each subject. The volunteers who provided samples had no history of gastrointestinal disease, did not take any antibiotics within one month before sample collection, and did not take probiotic products within one week before sample collection. Each sample was collected with a sterilized stool sampler. The sample was placed in a freezer at –20 °C, and the experimental isolation of LAB was performed as soon as possible.
2.2. Isolation of intestinal LAB
The fecal samples were diluted by the conventional plate dilution method. One gram of sample was homogenized with 9 mL of 9 g/L sterile physiological saline to make an initial dilution (1×10-1). Serial dilutions were made for each sample and 100 μL of the appropriate dilutions (1×10-5, 1×10-6, and 1×10-7) were spread on plates in triplicate on MRS (de Man Rogosa Sharpe) medium (Hopebio Company, Qingdao, China) containing 0.02 g/mL CaCO3, and incubated in an anaerobic atmosphere at 37 °C for 36–48 h (Liu et al., 2012). Individual colonies that produced a calcareous zone were selected. Single colonies were selected, inoculated into MRS, and then cultured at 37 °C for 24–48 h (Fečkaninová et al., 2019). Bacteria with differences in cell morphology and arrangement were inoculated in solid MRS and observed under a microscope (DM500, Leica Microsystems, Wetzlar, Germany) for the presence of more than one bacterial strain. If more than one strain was present, the sample was inoculated on solid MRS. Then, a single colony was selected after culturing, and the process was repeated until a mixture of bacteria no longer appeared in microscopic examination. Strains with the same cell morphology and arrangement were subjected to a catalase test, and catalase-negative strains were subcultured with 25% glycerol for preservation and stored at -20 °C.
2.3. Identification of intestinal LAB
Bacterial genomic DNA was extracted according to the instructions of TIANamp Bacteria DNA kit (Tiangen Biochemical Technology Company, Beijing, China). Extracted genomic DNA was used as a template. The following universal bacterial 16S ribosomal RNA (rRNA) gene amplification primers were used to amplify the 16S rRNA gene sequence of each target strain: forward 27F (5'-AGAGTTTGATCCTGGCTCAG-3') and reverse 1492R (5'-GGTTACCTTGTTACGACTT-3'). The amplified products were sequenced after electrophoresis on a 1.0% (10 g/L) agarose gel (Cocolin et al., 2001). Basic local alignment search tool (BLAST; Delphi, the National Center for Biotechnology Information, USA) was used to analyze the homology between the determined sequence and the 16S rRNA gene sequence in the GenBank database, and MEGA 6.5 software (Embarcadero Company, USA) was used to construct a phylogenetic tree (Chavagnat et al., 2002).
2.4. Screening of antibacterial LAB
The Oxford cup double-layer agar diffusion method was used to screen for LAB strains with antibacterial effects (Nuobariene et al., 2015). Escherichia coli ATCC 25922 and Staphylococcus aureus ATCC 25923 were selected as indicator bacteria. Briefly, 2% (20 g/L) agar medium was poured on as the first layer of the culture dish, and the Oxford cup was gently placed on the agar after solidification. Then, 0.7% (7 g/L) Mueller-Hinton broth (MHB) with indicator bacteria was poured on as the second layer. The Oxford cup was removed after solidification. After standing for 30 min, the prepared fermentation supernatant of the tested strain was added to each Oxford cup hole and incubated at 37 °C under an anaerobic condition for 20–24 h. The diameter of the inhibition zone was determined.
2.5. Evaluation of potential probiotics of intestinal LAB
2.5.1. Tolerance to artificial gastric juice and intestinal juice
Phosphate-buffered saline (PBS; Sigma Company, USA) was used to prepare pepsin (1:10 000 (volume ratio); Sigma Company, USA) as a solution at a concentration of 3 mg/mL. The pH was adjusted to 3.0 to prepare artificial gastric juice, and the juice was filtered and sterilized using a 0.22-μm microporous membrane. A trypsin (1:250 (volume ratio); Sigma Company, USA) solution was prepared with a concentration of 1 mg/mL using PBS. Then, 0.3% (3 g/L) bovine bile salt (Aladdin Bio-Chem Technology Company, Shanghai, China) was added and the pH value was adjusted to 8.0 to prepare artificial intestinal juice, which was filtered and sterilized through a 0.22-μm microporous membrane before use. Then, the selected bacteriostatic LAB strains were activated and inoculated into MRS, and cultured at 37 °C under an anaerobic condition for 18 h. The bacteria were collected by centrifugation (8000 r/min, 10 min) and washed twice with PBS. The bacteria were suspended in artificial gastric juice at pH 3.0 and cultured for 0 or 2 h, and the number of viable bacteria was determined using the spreading plate counting method (Jalilpour et al., 2017). The bacteria were also suspended in artificial intestinal juice at pH 8.0, and the number of viable bacteria was determined by the spreading plate counting method after 0, 2, or 4 h of culture. Survival rate=(number of viable bacteria after treatment)/(number of untreated viable bacteria)×100%.
2.5.2. Surface hydrophobicity test
Activated LAB strains were inoculated into MRS and cultured at 37 °C under an anaerobic condition for 18 h. The bacteria were collected by centrifugation (8000 r/min, 10 min) and washed twice with PBS. A suitable amount of PBS was added to create a bacterial suspension, and 2 mL of the suspension was mixed with 0.4 mL of xylene by vortexing for 120 s. The water phase was removed, and the absorbance at a wavelength of 600 nm was measured (Reuben et al., 2020). The decrease in absorbance of the water phase was used as a measure of the surface hydrophobicity (H, %). The following formula was used: H=(1-A/A 0)×100%, where A 0 and A are the absorbance before and after extraction with xylene, respectively.
2.6. Evaluation of the antibiotic resistance of intestinal LAB
After the LAB strains were activated, the MRS agar medium was poured into a plate, and then poured into soft agar (0.7% (7 g/L) agar) containing 10% (0.1 g/mL) bacterial solution at an appropriate concentration after solidification. After coagulation, a drug sensitivity test paper (Wenzhou Kangtai Biological Technology Co., Ltd., Wenzhou, China) was gently placed on the culture medium under aseptic conditions and incubated at 37 °C for 24 h. The diameter of the inhibition zone was observed and measured. Using S. aureus ATCC 25923 as the quality control bacteria and referring to the drug sensitivity test paper standards published by the American Association for Clinical and Laboratory Standardization (Table 1), the resistance of the tested strains to six antibiotics was determined (Kılıç and Karahan, 2010).
Table 1.
Antimicrobial resistance criteria for inhibitory results against six antibiotics
| Drug sensitivity test paper |
Drug content of paper (μg/tablet) |
Judgment standard of inhibition zone diameter (mm) | ||
|---|---|---|---|---|
| R | I | S | ||
| Cephalexin | 30 | ≤14 | 15‒17 | ≥18 |
| Tetracycline | 30 | ≤14 | 15‒18 | ≥19 |
| Erythromycin | 15 | ≤13 | 14‒22 | ≥23 |
| Vancomycin | 30 | ≤11 | 12‒14 | ≥15 |
| Streptomycin | 10 | ≤11 | 12‒14 | ≥15 |
| Gentamicin | 10 | ≤12 | 13‒14 | ≥15 |
R: resistant to antibiotics; I: moderately sensitive to antibiotics; S: sensitive to antibiotics.
2.7. Analyses of antibacterial substances and properties of probiotic LAB
2.7.1. Effect of enzyme treatment on the antibacterial activity of LAB fermentation broth
The pH of the concentrated fermentation broth was adjusted to the optimum pH for catalase (7.0), pepsin (3.0), trypsin (8.2), papain (7.0), or proteinase K (8.0) (Sigma Company, USA). Final concentrations of 5 mg/mL catalase and 1 mg/mL protease were added, and the pH was adjusted back to the initial value after 2 h in a 37 °C water bath. The Oxford cup double-layer agar diffusion method was used to determine the inhibitory effect of the treated fermentation broth on E. coli ATCC 25922. The original fermentation broth without enzyme treatment was used as a blank control to determine the effect of enzyme treatment on the antibacterial active substances in the fermentation broth (Svetoch et al., 2011).
2.7.2. Influence of pH on the antibacterial activity of LAB fermentation broth
The original pH of the fermentation broth was recorded, and the pH of the concentrated fermentation broth was adjusted to 2.0, 3.0, 4.0, 5.0, 6.0, or 7.0. The Oxford cup double-layer agar diffusion method was again used to determine the inhibitory effect of the treated fermentation broth on E. coli ATCC 25922. The fermentation broth without pH adjustment was used as a control to determine the effect of pH on the antibacterial active substances in the fermentation broth.
2.7.3. Effect of heat treatment on the antibacterial activity of LAB fermentation broth
The concentrated fermentation broth was heated at 60, 80, or 100 °C in a water bath for 1 h and then the temperature was increased to 121 °C for 30 min. The Oxford cup double-layer agar diffusion method was used to determine the inhibitory effect of the treated fermentation broth on E. coli ATCC 25922. The fermentation broth without heat treatment served as a blank control to determine the effect of heat treatment on the antibacterial active substances in the fermentation broth.
2.8. Purification of antibacterial substances
Preliminary extraction of protein-like active bacteriostatic substances that may exist in the test strains was achieved using the two-phase method (Lappe et al., 2012). Using E. coli ATCC 25922 as the indicator bacterium, the Oxford cup double-layer agar diffusion method was used to verify the extraction effect (Sant'Anna et al., 2016). The total mass of the fixed two-phase extraction system was 50.0 g, and 20% polyethylene glycol 4000 (PEG4000; Qingdao Haibo Biotechnology Co., Ltd., Qingdao, China), 0.2 g/mL (NH4)2SO4 (Tianjin Kemiou Chemical Reagent Co., Ltd., Tianjin, China), and 1% surfactant Triton X-100 (Beijing Aoboxing Biology Technology Co., Ltd., Beijing, China) were added to the original fermentation broth without centrifugation. The pH of the system was adjusted, and the upper and lower phases were separated after centrifugation (4000 r/min, 5 min). After concentration, the optimal extraction pH was determined by measuring the protein partition coefficient and antibacterial activity of the two phases at different pH values. The following formula was used to calculate the protein partition coefficient (K): K=(total protein concentration in the extract phase)/(total protein concentration in the non-extract phase).
2.9. Prediction of molecular weight of bacteriostatic substances and determination of antibacterial spectrum
Bacteriocins have a low molecular weight. Tricine-sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE; Beijing Aoboxing Biology Technology Co., Ltd., Beijing, China) is suitable for separating proteins with molecular weights in the range of 1–100 kDa, and the separation effect is better for proteins with molecular weights less than 30 kDa (Milioni et al., 2015). Each concentrated sample to be tested was mixed with the loading buffer at a volume ratio of 1:1, boiled for 10 min, and cooled before loading. After tricine-SDS-PAGE electrophoresis, the electrophoresis gel was placed in the fixing solution (water:isopropanol:glacial acetic acid=7:2:1, volume ratio). Then, the samples were stained with Coomassie Brilliant Blue (Shanghai Yuanye Biological Technology Co., Ltd., Shanghai, China) and repeatedly decolorized until the background was clear. Finally, the results were photographed using a gel imager (Biorad ChemiDoc XRS, Bio-Rad Laboratories, Inc., USA).
Using pathogenic bacteria stored in the laboratory as indicator bacteria, a bacteriostatic test was performed according to the test method in Section 2.4 to determine the antibacterial spectrum of the bacteriostatic substance in the fermentation broth of the test strain.
2.10. Statistical analysis
All experiments in this study were carried out three times, and the results are expressed as the mean±standard deviation (SD). SPSS 20.0 software was used for data analysis, and Origin 8.5 software to generate charts. Independent-sample t-tests were used to determine statistical significance. Values of P<0.05 were considered to be statistically significant.
3 Results
3.1. Isolation and preliminary identification of intestinal LAB
A total of 114 strains that produced calcareous circles on MRS agar medium containing 20 g/L CaCO3 were isolated from the 10 stool samples. Sixteen strains showed negative reactions by Gram staining, and 18 strains showed positive reactions in the catalase test. The remaining 80 strains with positive Gram staining results and negative reactions for catalase were provisionally determined to be LAB.
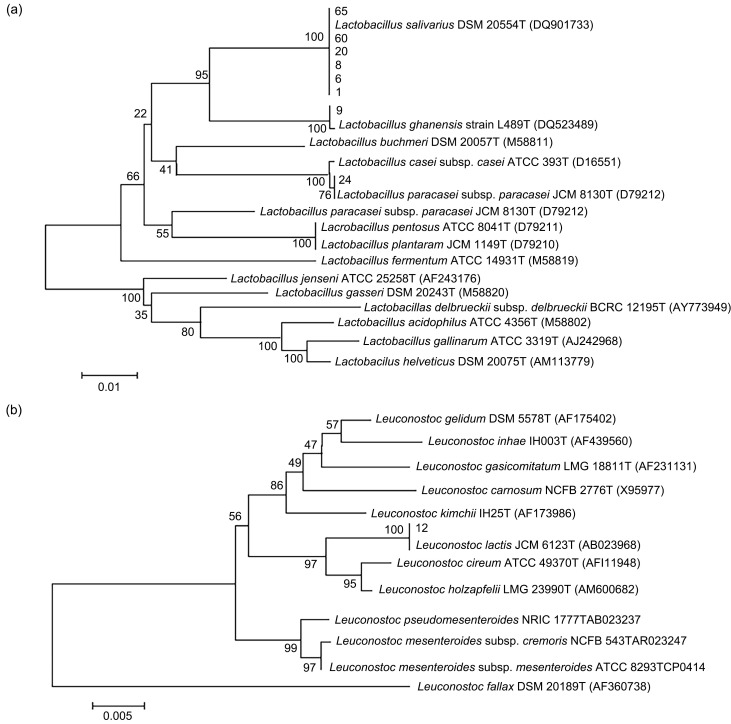
The results of the identification of LAB by their 16S rRNA sequence are shown in Table S1. The phylogenetic tree of LAB constructed based on their 16S rRNA gene sequences is shown in Fig. 1. The strains included Lactobacillus (Fig. 1a) and Leuconostoc (Fig. 1b). The establishment of a phylogenetic tree confirmed that the 80 strains isolated were all LAB and belonged to a total of 12 species from five genera, including Lactobacillus, Lactococcus, Leuconostoc, Enterococcus, and Streptococcus. According to evolutionary affinity, one or several representative strains were selected from each species for subsequent tests. The selected representative strains were: strains 2, 11 (Lactococcus garvieae), 17 (Enterococcus thailandicus), 28, 32, 29 (Streptococcus salivarius), 30, 78 (Enterococcus faecium), 33 (Enterococcus hirae), 59 (Enterococcus durans), 63 (Enterococcus faecalis); strains 1, 6, 8, 20, 60, 65, 9, 24 (Lactobacillus) and strain 12 (Leuconostoc). A total of 20 strains were selected to test bacteriostatic properties.
Fig. 1. Phylogenetic trees of lactic acid bacteria constructedbased on 16S rRNA gene sequences of Lactobacillus (a) and Leuconostoc (b).
3.2. Screening of antibacterial LAB
E. coli ATCC 25922 and S. aureus ATCC 29213 were selected as indicator bacteria, and the bacteriostatic properties of the 20 LAB strains were tested using the Oxford cup double-layer agar diffusion method. The results are shown in Table S2. According to the selection criteria, the 20 strains of LAB showed different degrees of inhibition on the two indicator bacteria. Strains 1 (Lactobacillus salivarius), 9 (Lactobacillus ghanensis), 12 (Leuconostoc lactis), 24 (Lactobacillus paracasei), and 63 (Enterococcus faecalis) exhibited strong antibacterial activity.
3.3. Analysis of potential probiotics of antibacterial LAB
3.3.1. Analysis of tolerance to gastric juice
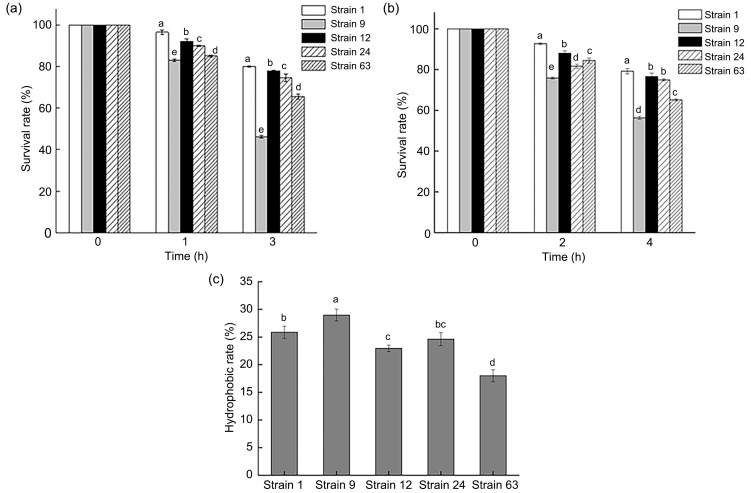
The tolerance of five strains of LAB with good antibacterial ability to artificial gastric juice at pH 3.0 is shown in Fig. 2a. After 1 h of incubation in the juice, the five strains all exhibited good tolerance, and the survival rate was greater than 80%. Strain 1 had the best tolerance, with a survival rate of 96.63%, which was significantly higher than those of the other four strains (P<0.05). After 3 h of incubation, the five strains exhibited different tolerances to artificial gastric juice, but the differences were similar to those noted after 1 h of incubation. Strain 1 again showed the best tolerance, with a survival rate of 80.16%. The survival rate of strain 9 was 46.72%, much lower than those of the other four LAB strains (P<0.05). Combining the 1 and 3 h culture results, strains 1, 12, and 24 showed improved tolerance to artificial gastric juice at pH 3.0.
Fig. 2. Potential probiotics of five lactic acid bacteria. (a) Tolerance to artificial gastric fluid; (b) Tolerance to 3 g/L bile salts; (c) Surface hydrophobicity. Data are expressed as mean±standard deviation (n=3). Different letters indicate significant difference (P<0.05) among strains.
3.3.2. Analysis of tolerance to intestinal juice
The tolerance of the five LAB strains to artificial intestinal juice containing 3 g/L bile salt is shown in Fig. 2b. After 2 h of incubation in the juice, the five strains exhibited different tolerances. Strain 1 had the best tolerance, with a survival rate of 92.67%, which was significantly higher than those of the other four LAB strains (P<0.05). After 4 h of incubation, the survival rate of strain 1 was again the highest at 79.21%, and the survival rate of strain 9 was the lowest at 56.36%. Based on the 2 and 4 h culture results, strains 1, 12, and 24 exhibited improved tolerance to artificial intestinal juice.
3.3.3. Surface hydrophobicity analysis
Surface hydrophobicity can indirectly indicate cell adhesion. The surface hydrophobicity of the five LAB strains varied significantly (P<0.05; Fig. 2c). The surface hydrophobicity of strain 9 was significantly higher than that of the other strains. Strains 1, 12, 24, and 63 exhibited relatively low hydrophobicity.
3.4. Analysis of antimicrobial resistance of LAB
The diameter of the bacteriostatic circle produced by the quality control strains used in the test was within the quality control range, demonstrating that the medium and drugs used in this test were valid. The drug resistance results of the five LAB strains to different antibiotics are shown in Table 2. All five strains exhibited resistance to at least one antibiotic. Strains 1 and 24 exhibited resistance to two, strain 12 to three, and strains 9 and 24 exhibited multiple drug resistance. The strains exhibited resistance rates of 100% to gentamicin, 80% to streptomycin, 60% to vancomycin, 40% to tetracycline, 20% to erythromycin, and 0% to cefalexin.
Table 2.
Resistance to different antibiotics of five lactic acid bacteria
| Drug sensitivity test paper | Inhibition zone diameter of lactic acid bacteria (mm) | ||||
|---|---|---|---|---|---|
| Strain 1 | Strain 9 | Strain 12 | Strain 24 | Strain 63 | |
| Cephalexin |
15.89±0.19 I |
15.34±0.21 I |
16.98±0.09 I |
15.56±0.05 I |
15.12±0.03 I |
| Tetracycline |
26.53±0.02 S |
11.15±0.19 R |
21.46±0.31 S |
38.41±0.07 S |
11.28±0.17 R |
| Erythromycin |
21.83±0.24 I |
21.68±0.11 I |
21.37±0.05 I |
36.08±0.04 S |
10.37±0.15 R |
| Vancomycin |
17.86±0.21 S |
- R |
- R |
- R |
18.24±0.07 S |
| Streptomycin | -R |
- R |
- R |
11.21±0.02 I |
- R |
| Gentamicin |
10.11±0.14 R |
- R |
11.34±0.14 R |
10.33±0.02 R |
- R |
"-" represents the non-inhibitory circle. S: sensitive to antibiotics; I: moderately sensitive to antibiotics; R: resistant to antibiotics. Data are expressed as mean±standard deviation (n=3).
3.5. Analyses of active ingredients and properties of antibacterial substances
Comprehensive evaluation of the survival rate in artificial gastric juice and intestinal juice, surface hydrophobicity and antibiotic resistance of the five strains resulted in the selection of strain 1 (L. salivarius), strain 12 (L. lactis), and strain 24 (L. paracasei) for further testing. For these strains with good probiotics and no multidrug resistance, the antibacterial substances in the fermentation broth were studied.
3.5.1. Effect of enzyme treatment on the antibacterial activity of fermentation broth
The effect of different enzyme treatments on the antibacterial activity of three strains of LAB fermentation broths is shown in Table 3. Compared with the control group, after catalase treatment, there was no significant difference in the diameter of the inhibition zone of the three LAB strains (P>0.05), indicating that hydrogen peroxide was not the main antibacterial active substance. After protease treatment, the antibacterial activity of the strains against E. coli ATCC 25922 changed to variable degrees. No significant difference in antibacterial activity was noted between strain 12 and the control group (P>0.05). We hypothesized that no protein antibacterial substance was present in the fermentation broth of strain 12, or that protein antibacterial substances were not the main effective substances for antibacterial activity. The antibacterial activity of strains 1 and 24 was significantly reduced compared with that of the control group (P<0.05). We speculated that there were protein antibacterial substances in the fermentation broths of these LAB strains that exhibited antibacterial activity against the test pathogenic bacteria.
Table 3.
Effect of different enzyme treatments on antibacterial activity
| Enzyme type | Inhibition zone diameter of lactic acid bacteria (mm) | ||
|---|---|---|---|
| Strain 1 | Strain 12 | Strain 24 | |
| Catalase | 18.45±0.18a | 17.88±0.25a | 23.57±0.79a |
| Pepsin | 12.66±0.29c | 18.08±0.23a | 15.25±0.71bc |
| Trypsin | 11.82±0.27c | 17.57±0.29a | 13.66±0.39cd |
| Papain | 14.00±0.18b | 17.99±0.46a | 16.32±0.41b |
| Proteinase K | 12.04±0.63c | 17.87±0.28a | 13.35±0.09d |
| Control | 20.59±0.30a | 18.56±0.23a | 25.14±0.46a |
The diameter of the Oxford cup was 8 mm. Data are expressed as mean±standard deviation (n=3). Different letters in column indicate significant difference (P<0.05) among enzyme types.
3.5.2. Influence of pH on the antibacterial activity of fermentation broth
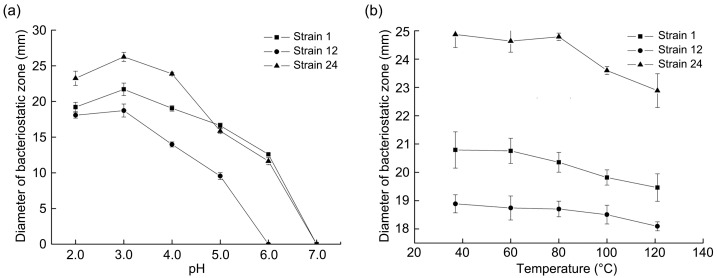
The effect of different pH values on the antibacterial activity of LAB fermentation broth is shown in Fig. 3a. The initial pH value and antibacterial activity of LAB fermentation broths are shown in Table S3. Table S3 shows that the three LAB strains had strong acid production ability. As the pH increased, the antibacterial activity of the three LAB strains exhibited a downward trend (Fig. 3a). After adjusting the pH of strain 12 to be greater than the pH value of the original fermentation broth, the antibacterial activity began to decrease. When the pH was greater than 5.0, the antibacterial activity gradually disappeared, indicating that the antibacterial activity of the fermentation broth of strain 12 was greatly affected by organic acids. For strains 1 and 24, when the pH was 3.0, the antibacterial activity was highest. Their activity levels were greater than that of the original fermentation broth, indicating that the fermentation broths of strains 1 and 24 contained active antibacterial organic acids. Other substances in the fermentation broths of strains 1 and 24 also participated in the antibacterial effect, and the antibacterial substance exhibited good activity in an acidic environment.
Fig. 3. Effects of different pH values (a) and heat treatment (b) on the antibacterial activity of fermentation broths. Data are expressed as the mean±standard deviation (n=3).
3.5.3. Effect of heat treatment on the antibacterial activity of fermentation broth
The effect of heat treatment on the antibacterial activity of the fermentation broth is shown in Fig. 3b. High temperature treatment had minimal effect on the antibacterial activity of the fermentation broth of the three LAB strains, and no obvious change in antibacterial activity was noted at 80 °C. The antibacterial active substances in the fermentation broths of the three LAB exhibited thermal stability. The antibacterial substances of strain 1 (L. salivarius) and strain 24 (L. paracasei) were sensitive to alkaline environments and proteases. The thermal stability results obtained in this test were consistent with the properties of proteins. These results revealed that protein bacteriostatic substances were found in the fermentation broths of the two bacteria, and these bacteriostatic protein substances were provisionally classed as bacteriocins.
3.6. Analysis of the results of crude extraction of antibacterial substances
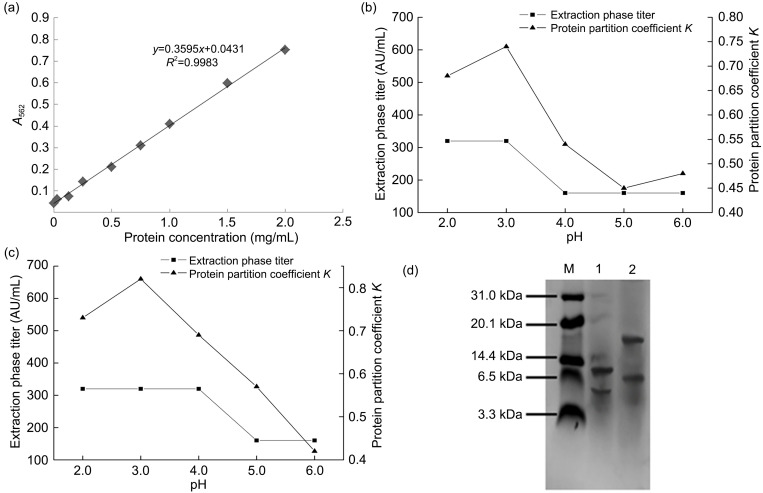
After diluting the upper and lower phases with double distilled H2O to an appropriate value, a protein standard curve (Fig. 4a) was used to determine the protein content, and the titers of the extracted phase in AU/mL were calculated. The results are shown in Figs. 4b and 4c. Both strains 1 and 24 exhibited maximum protein partition coefficient K values at pH 3.0. As the pH increased, the K of strain 1 first increased, then decreased and subsequently increased. The K value was lowest at pH 5.0. The K value of strain 24 first increased, then decreased, and was lowest at pH 6.0. In an acidic environment, the K values of the two strains were both less than 1. We hypothesized that the isoelectric points of the bacteriocins secreted by the two strains were in the alkaline region. According to the extraction situation and titer determination, the optimal extraction of strains 1 and 24 bacteriocins was achieved at pH 3.0, and the titer was 320 AU/mL.
Fig. 4. Analysis and identification of crude extraction results of antibacterial substances. (a) Standard curve of protein concentrations; (b) Effect of pH on the biaqueous extraction of bacteriocins produced by strain 1; (c) Effect of pH on extraction of bacteriostatic protein from strain 24; (d) Tricine-sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE) of antibacterial proteins extracted using the two-phase method. A 562: absorbance at 562 nm; M: marker; 1: strain 1; 2: strain 24.
3.7. Determination of the molecular weight of bacteriostatic substances and analysis of bacteriostasis
Results of tricine-SDS-PAGE electrophoresis of the bacteriocins crudely extracted from the fermentation broths of strains 1 and 24 by aqueous two-phase extraction are shown in Fig. 4d. Both strains produced clearer bands after electrophoretic staining and decolorization, demonstrating that their fermentation broths contained protein antibacterial substances. The two lanes in Fig. 4d clearly show the presence of bands of less than 10 kDa, indicating that the bacteriostatic substance in the fermentation broths of strains 1 and 24 was a small molecule antimicrobial peptide, namely, a bacteriocin.
Common Gram-positive and Gram-negative bacteria stored in the laboratory were selected, and the bacteriostatic spectra of bacteriocins produced by strains 1 and 24 were analyzed. The results are shown in Table 4. The crude bacteriocins of both strains exhibited inhibitory effects on most Gram-positive bacteria (S. aureus, Staphylococcus epidermidis, Listeria monocytogenes) and Gram-negative bacteria (E. coli, Salmonella typhimurium, Salmonella pullorum, Salmonella enteric, Enterobacter sakazakii), indicating that the bacteriocins produced by the two strains exhibit broad-spectrum bacteriostasis.
Table 4.
Bacteriostatic spectrum of crude bacteriocins
| Strain name | Inhibition zone diameter of crude bacteriocin (mm) | |
|---|---|---|
| Strain 1 | Strain 24 | |
| Gram-negative bacteria | ||
| Escherichia coli ATCC 25922 | ++ | ++ |
| E. coli UB1005 | ++ | ++ |
| Salmonella typhimurium C77-31 | ++ | ++ |
| S. typhimurium ATCC 14028 | ++ | ++ |
| Salmonella pullorum C 79-13 | +++ | ++ |
| S. typhimurium CMCC 50071 | ++ | ++ |
| Salmonella enteric CMCC 47020 | ++ | ++ |
| Enterobacter sakazakii ATCC 25944 | ++ | ++ |
| Gram-positive bacteria | ||
| Staphylococcus aureus ATCC 25923 | ++ | ++ |
| S. aureus ATCC 29213 | ++ | ++ |
| S. aureus CMCC 26074 | ++ | ++ |
| Staphylococcus epidermidis ATCC 12228 | ++ | ++ |
| Listeria monocytogenes CMCC 54004 | +++ | ++ |
| Bacillus cereus CMCC 6303 | - | - |
The outside diameter of Oxford cup was 8 mm. "-" means that non-inhibitory circle or inhibitory circle diameter is ≤8 mm; "+" means that diameter is 8‒10 mm; "++" means that diameter is 10‒15 mm; and "+++" means that diameter is >15 mm.
4 Discussion
The intestines of human adults are homes to about 400 to 500 types of bacteria. LAB are the easiest type of potential probiotic bacteria to isolate. The commonly used MRS medium separation method for isolating LAB was selected. Various studies have demonstrated that this method is relatively simple and feasible for separating LAB (le Barz et al., 2019). In this study, a total of 80 strains of LAB were isolated, of which 7 were bacilli and 73 were cocci. According to the analysis and identification of their 16S rRNA gene sequences and phylogenetic tree, the obtained LAB strains could be assigned to 5 genera and 12 species. In this study, three different species of Lactobacillus were finally isolated, including two probiotic species approved for food use in China, L. salivarius and L. paracasei. This study used feces as a source for the separation and purification of probiotics (Zhai et al., 2020). The method is simple, effective, and feasible, and can be further promoted and applied under limited conditions.
A variety of antibacterial active substances are produced during the fermentation process of LAB, including organic acids, hydrogen peroxide, and bacteriocins. These substances can inhibit the growth of spoilage and pathogenic microorganisms (Hammami et al., 2009). Current research mainly involves screening LAB with high antibacterial activity using an in vitro antagonism test. In this study, the common Gram-negative pathogenic bacterium E. coli ATCC 25922 and the Gram-positive pathogenic bacterium S. aureus ATCC 25923 were selected as indicator bacteria, and LAB with high antibacterial activity were screened using the Oxford cup double-layer agar diffusion method. The test identified a total of five strains of LAB with good inhibitory effects on the two indicator bacteria. The bacteriostatic active substances in the fermentation broths of these five strains exhibited broad-spectrum bacteriostasis and have the potential to be applied to the food and medical fields. Therefore, these five LAB strains were selected for follow-up research.
To colonize the intestinal tract, LAB must pass through the gastric juice environment. The pH of gastric juice is about 1.3–2.0 when fasting, and about 3.0–5.0 after ingesting food. A probiotic must survive for 1.5–2 h at pH 3.0. The concentrations of bile salts in the small intestine of normal humans are between 0.3 and 3.0 g/L (Amorim et al., 2018). LAB must colonize the small intestine to produce a probiotic effect and need to tolerate bile salts. High concentrations of bile salts are antagonistic to the growth of probiotics because they alter the permeability of the cell membrane, decompose membrane proteins, and cause cell content to flow out, thereby inducing cell rupture and death (Begley et al., 2005). The artificial gastric juice test showed that the five tested LAB exhibited good tolerance after being cultured for 1 h, and the survival rate of the LAB exceeded 80%. After 3 h of culture, the five tested LAB maintained good tolerance. These effects may be related to the proton pump dynamic mechanism, acid tolerance reaction mechanism, macromolecule protection and repair mechanism, or acid generation resistance mechanism (Cotter and Hill, 2003). Differences in cell membranes and cell density may also affect the acid resistance of LAB. Moreover, the LAB isolated and screened in this experiment showed less acid resistance than those isolated from fermented foods in other studies (Shafei et al., 2000; Liong and Shah, 2005). Thus, LAB selected from the neutral environment of the intestinal tract exhibited weaker acid resistance compared with those isolated from the acidic environment, indicating that LAB acid resistance involves different mechanisms (Usman and Hosono, 1999; Zhang et al., 2008). In this study, three strains of LAB that survived in the artificial gastrointestinal fluid environment were identified: strain 1 (L. salivarius), strain 12 (L. lactis), and strain 24 (L. paracasei). Therefore, these three strains were selected for further research on antibacterial active substances.
The long-term abuse of antibiotics has caused antibiotic pollution to increase daily. Antibiotics and resistant bacteria remaining in food, the environment, and livestock may eventually be transferred to the human body through the food chain (Bansal et al., 2019). Therefore, it is necessary to evaluate the drug resistance of LAB isolated from the human intestine. Numerous studies on the antibiotic resistance of LAB have been performed at home and abroad. Results have shown that the resistance of LAB to vancomycin shows large differences between genera. Specifically, Lactococcus and Streptococcus exhibit no resistance to vancomycin, whereas Bacillus exhibits high resistance (Gad et al., 2014). Our results are consistent with the above research results. Among the five strains of LAB in this study, three were resistant to vancomycin, and neither E. faecalis nor L. lactis was resistant. The results showed that none of multidrug-resistant strains that emerged from the five LAB strains was safe for future applications in food, medicine, or animal husbandry industries.
The fermentation process of LAB can produce organic acids, hydrogen peroxide, bacteriocins, and other antibacterial active substances, which can inhibit the growth of spoilage and pathogenic microorganisms ( Hols et al., 2019). Analysis of the antibacterial substances produced by strains typically involves treatment of fermentation broth using multiple conditions to determine the influence of the broth on antibacterial activity (Özogul and Hamed, 2018). The results of this study showed that after treatment with different conditions, none of the bacteriostatic substances in the fermentation supernatants of the three strains of LAB was hydrogen peroxide. The antibacterial activity of the fermentation broth of strain 12 was minimally affected by proteases, but was more sensitive to changes in pH, indicating that its main antibacterial active substances were organic acids. The antibacterial active substances in the fermentation broths of strains 1 and 24 were more sensitive to changes in protease and pH values, and exhibited thermal stability. The main antibacterial substances of strains 1 and 24 were organic acids and proteins, and the protein substances exhibited good antibacterial activity under acidic conditions. We hypothesized that the protein antibacterial substances were bacteriocins (Simha et al., 2012).
Bacteriocins are a class of extracellular secretions of proteins with antibacterial effects that contain both simple polypeptides and complex macromolecular proteins (Garsa et al., 2014). The aqueous two-phase method is a newly proposed rapid and effective method for crude bacteriocin extraction (Sant'Anna et al., 2016). Protein bacteriostatic substances are hydrophobic, exhibit different charging properties at different pH values, and adsorb to and dissociate bacterial cells in the environment at different pH values. This makes the selection of the extraction pH of the aqueous two-phase system very important. This study found that the volatility of the titer was not completely proportional to the K value. This may be because the AU/mL titer calculation method, as a discontinuous semiquantitative method, had low calculation accuracy, leading to variability in the results.
In this study, two clear bands were obtained through electrophoresis experiments, indicating that the aqueous two-phase extraction method had a good crude extraction effect and that the fermentation broths of strains 1 and 24 contained protein antibacterial substances. This finding is consistent with the findings of Sant'Anna et al. (2016). The crude samples of the two strains inhibited the selected Gram-positive and Gram-negative bacteria, indicating that the bacteriocin produced by the two strains exhibited broad-spectrum antibacterial activity. However, the antibacterial activity of the two strains against Bacillus cereus was not strong. This may have been due to the large amount of protein, carbohydrates, and lipids wrapped in the outer layer of Bacillus, hindering the effect of bacteriocins (Hols et al., 2019). The results of this study showed that the bacteriostatic substance in the fermentation broths of strains 1 and 24 was a small molecule, antimicrobial peptide with a molecular weight of less than 10 kDa, which produced a significant bacteriostatic effect on L. monocytogenes. This protein was presumed to be a Class II bacteriocin (Rolhion et al., 2019). This finding was consistent with the conclusion that bacteriocins of L. salivarius and Lactobacillus casei belong to Class II (Hu et al., 2018; Cárdenas et al., 2019).
5 Conclusions
In this study, three strains of LAB with good potential beneficial effects and safety were isolated and screened from the feces of healthy Chinese adults. These LAB exhibited good antibacterial activity against E. coli ATCC 25922 and S. aureus ATCC 25923. The main antibacterial substances of L. lactis were organic acids, and those of L. salivarius and L. paracasei were organic acids and bacteriocins with broad-spectrum antibacterial activity. This discovery provides bacterial resources and application potential for the development of agents for the prevention and treatment of diseases caused by intestinal pathogenic bacteria, and is beneficial for promoting the development of new functional foods.
Supplementary information
Acknowledgments
This work was supported by the National Key Research and Development Program of China (No. 2016YFD0400605).
Author contributions
Zhijing LIU performed the experimental research and data analysis, and wrote and edited the manuscript. Cong XU, Ran TIAN, and Wan WANG contributed to the study design and data analysis of the manuscript. Jiage MA, Liya GU, and Fei LIU contributed to writing and editing of the manuscript. Zhanmei JIANG and Juncai HOU contributed to the study design, writing and editing of the manuscript. All authors have read and approved the final manuscript and, therefore, have full access to all the data in the study and take responsibility for the integrity and security of the data.
Compliance with ethics guidelines
Zhijing LIU, Cong XU, Ran TIAN, Wan WANG, Jiage MA, Liya GU, Fei LIU, Zhanmei JIANG, and Juncai HOU declare that they have no conflict of interest.
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008 (5). Informed consent was obtained from all persons for being included in the study.
References
- Alshammari E, Patel M, Sachidanandan M, et al. , 2019. Potential evaluation and health fostering intrinsic traits of novel probiotic strain Enterococcus durans F3 isolated from the gut of fresh water fish Catla catla . Food Sci Anim Resour, 39(5): 844-861. 10.5851/kosfa.2019.e57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorim JC, Piccoli RH, Duarte WF, 2018. Probiotic potential of yeasts isolated from pineapple and their use in the elaboration of potentially functional fermented beverages. Food Res Int, 107: 518-527. 10.1016/j.foodres.2018.02.054 [DOI] [PubMed] [Google Scholar]
- Aymes F, Monnet C, Corrieu G, 1999. Effect of α-acetolactate decarboxylase inactivation on α-acetolactate and diacetyl production by Lactococcus lactis subsp. lactis biovar diacetylactis. J Biosci Bioeng, 87(1): 87-92. 10.1016/s1389-1723(99)80013-9 [DOI] [PubMed] [Google Scholar]
- Bansal R, Jain A, Goyal M, et al. , 2019. Antibiotic abuse during endodontic treatment: a contributing factor to antibiotic resistance. J Family Med Prim Care, 8(11): 3518-3524. 10.4103/jfmpc.jfmpc_768_19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley M, Gahan CGM, Hill C, 2005. The interaction between bacteria and bile. FEMS Microbiol Rev, 29(4): 625-651. 10.1016/j.femsre.2004.09.003 [DOI] [PubMed] [Google Scholar]
- Cárdenas N, Martín V, Arroyo R, et al. , 2019. Prevention of recurrent acute otitis media in children through the use of Lactobacillus salivarius PS7, a target-specific probiotic strain. Nutrients, 11(2): 376. 10.3390/nu11020376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavagnat F, Haueter M, Jimeno J, et al. , 2002. Comparison of partial tuf gene sequences for the identification of lactobacilli. FEMS Microbiol Lett, 217(2): 177-183. 10.1111/j.1574-6968.2002.tb11472.x [DOI] [PubMed] [Google Scholar]
- Cocolin L, Manzano M, Cantoni C, et al. , 2001. Denaturing gradient gel electrophoresis analysis of the 16S rRNA gene V1 region to monitor dynamic changes in the bacterial population during fermentation of Italian sausages. Appl Environ Microbiol, 67(11): 5113-5121. 10.1128/aem.67.11.5113-5121.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter PD, Hill C, 2003. Surviving the acid test: responses of Gram-positive bacteria to low pH. Microbiol Mol Biol Rev, 67(3): 429-453. 10.1128/mmbr.67.3.429-453.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fečkaninová A, Koščová J, Mudroňová D, et al. , 2019. Characterization of two novel lactic acid bacteria isolated from the intestine of rainbow trout (Oncorhynchus mykiss, Walbaum) in Slovakia. Aquaculture, 506: 294-301. 10.1016/j.aquaculture.2019.03.026 [DOI] [Google Scholar]
- Gad GFM, Abdel-Hamid AM, Farag ZSH, 2014. Antibiotic resistance in lactic acid bacteria isolated from some pharmaceutical and dairy products. Braz J Microbiol, 45(1): 25-33. 10.1590/s1517-83822014000100005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsa AK, Kumariya R, Sood SK, et al. , 2014. Bacteriocin production and different strategies for their recovery and purification. Probiotics Antimicrob Proteins, 6(1): 47-58. 10.1007/s12602-013-9153-z [DOI] [PubMed] [Google Scholar]
- Giraffa G, Chanishvili N, Widyastuti Y, 2010. Importance of lactobacilli in food and feed biotechnology. Res Microbiol, 161(6): 480-487. 10.1016/j.resmic.2010.03.001 [DOI] [PubMed] [Google Scholar]
- Hammami I, Rhouma A, Jaouadi B, et al. , 2009. Optimization and biochemical characterization of a bacteriocin from a newly isolated Bacillus subtilis strain 14B for biocontrol of Agrobacterium spp. strains. Lett Appl Microbiol, 48(2): 253-260. 10.1111/j.1472-765X.2008.02524.x [DOI] [PubMed] [Google Scholar]
- Hols P, Ledesma-García L, Gabant P, et al. , 2019. Mobilization of microbiota commensals and their bacteriocins for therapeutics. Trends Microbiol, 27(8): 690-702. 10.1016/j.tim.2019.03.007 [DOI] [PubMed] [Google Scholar]
- Hu J, Ma LB, Nie YF, et al. , 2018. A microbiota-derived bacteriocin targets the host to confer diarrhea resistance in early-weaned piglets. Cell Host Microbe, 24(6): 817-832.e8. 10.1016/j.chom.2018.11.006 [DOI] [PubMed] [Google Scholar]
- Jalilpour Y, Abdollahzade B, ParviziFard G, et al. , 2017. A simple route for preparation of pH-sensitive hydrogels by using egg white proteins in alginate scaffold for the encapsulation of probiotics. Ars Pharm, 58(3): 127-136. 10.4321/s2340-98942017000300006 [DOI] [Google Scholar]
- Jubair WK, Hendrickson JD, Severs EL, et al. , 2018. Modulation of inflammatory arthritis in mice by gut microbiota through mucosal inflammation and autoantibody generation. Arthritis Rheumatol, 70(8): 1220-1233. 10.1002/art.40490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna S, Tosh PK, 2014. A clinician’s primer on the role of the microbiome in human health and disease. Mayo Clin Proc, 89(1): 107-114. 10.1016/j.mayocp.2013.10.011 [DOI] [PubMed] [Google Scholar]
- Kılıç GB, Karahan AG, 2010. Identification of lactic acid bacteria isolated from the fecal samples of healthy humans and patients with dyspepsia, and determination of their pH, bile, and antibiotic tolerance properties. J Mol Microbiol Biotechnol, 18(4): 220-229. 10.1159/000319597 [DOI] [PubMed] [Google Scholar]
- Kim SG, Becattini S, Moody TU, et al. , 2019. Microbiota-derived lantibiotic restores resistance against vancomycin-resistant Enterococcus . Nature, 572(7771): 665-669. 10.1038/s41586-019-1501-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordel M, Schüller F, Sahl HG, 1989. Interaction of the pore forming-peptide antibiotics Pep 5, nisin and subtilin with non-energized liposomes. FEBS Lett, 244(1): 99-102. 10.1016/0014-5793(89)81171-8 [DOI] [PubMed] [Google Scholar]
- Krzych-Fałta E, Furmańczyk K, Tomaszewska A, et al. , 2018. Probiotics: myths or facts about their role in allergy prevention. Adv Clin Exp Med, 27(1): 119-124. 10.17219/acem/65476 [DOI] [PubMed] [Google Scholar]
- Lähteinen T, Malinen E, Koort JMK, et al. , 2010. Probiotic properties of Lactobacillus isolates originating from porcine intestine and feces. Anaerobe, 16(3): 293-300. 10.1016/j.anaerobe.2009.08.002 [DOI] [PubMed] [Google Scholar]
- Lappe R, Sant'Anna V, Brandelli A, 2012. Extraction of the antimicrobial peptide cerein 8A by aqueous two-phase systems and aqueous two-phase micellar systems. Nat Prod Res, 26(23): 2259-2265. 10.1080/14786419.2011.652958 [DOI] [PubMed] [Google Scholar]
- le Barz M, Daniel N, Varin TV, et al. , 2019. In vivoscreening of multiple bacterial strains identifies Lactobacillus rhamnosus Lb102 and Bifidobacterium animalis ssp. lactis Bf141 as probiotics that improve metabolic disorders in a mouse model of obesity. FASEB J, 33(4): 4921-4935. 10.1096/fj.201801672R [DOI] [PubMed] [Google Scholar]
- Liong MT, Shah NP, 2005. Acid and bile tolerance and cholesterol removal ability of lactobacilli strains. J Dairy Sci, 88(1): 55-66. 10.3168/jds.S0022-0302(05)72662-X [DOI] [PubMed] [Google Scholar]
- Liu WJ, Bao QH, Jirimutu, et al. , 2012. Isolation and identification of lactic acid bacteria from Tarag in Eastern Inner Mongolia of China by 16S rRNA sequences and DGGE analysis. Microbiol Res, 167(2): 110-115. 10.1016/j.micres.2011.05.001 [DOI] [PubMed] [Google Scholar]
- Marko D, 2009. Food-borne mycotoxins. Mol Nutr Food Res, 53(4): 421-421. 10.1002/mnfr.200990009 [DOI] [PubMed] [Google Scholar]
- Milioni C, Martínez B, Degl'Innocenti S, et al. , 2015. A novel bacteriocin produced by Lactobacillus plantarum LpU4 as a valuable candidate for biopreservation in artisanal raw milk cheese. Dairy Sci Technol, 95(4): 479-494. 10.1007/s13594-015-0230-9 [DOI] [Google Scholar]
- Nuobariene L, Cizeikiene D, Gradzeviciute E, et al. , 2015. Phytase-active lactic acid bacteria from sourdoughs: isolation and identification. LWT-Food Sci Technol, 63(1): 766-772. 10.1016/j.lwt.2015.03.018 [DOI] [Google Scholar]
- Otero MC, Nader-Macías ME, 2006. Inhibition of Staphylococcus aureus by H2O2-producing Lactobacillus gasseri isolated from the vaginal tract of cattle. Anim Reprod Sci, 96(2): 35-46. 10.1016/j.anireprosci.2005.11.004 [DOI] [PubMed] [Google Scholar]
- Özogul F, Hamed I, 2018. The importance of lactic acid bacteria for the prevention of bacterial growth and their biogenic amines formation: a review. Crit Rev Food Sci Nutr, 58(10): 1660-1670. 10.1080/10408398.2016.1277972 [DOI] [PubMed] [Google Scholar]
- Pei JJ, Li XS, Han H, et al. , 2018. Purification and characterization of plantaricin SLG1, a novel bacteriocin produced by Lb. plantarum isolated from yak cheese. Food Control, 84: 111-117. 10.1016/j.foodcont.2017.07.034 [DOI] [Google Scholar]
- Reuben RC, Roy PC, Sarkar SL, et al. , 2020. Characterization and evaluation of lactic acid bacteria from indigenous raw milk for potential probiotic properties. J Dairy Sci, 103(2): 1223-1237. 10.3168/jds.2019-17092 [DOI] [PubMed] [Google Scholar]
- Rolhion N, Chassaing B, Nahori MA, et al. , 2019. A Listeria monocytogenes bacteriocin can target the commensal prevotella copri and modulate intestinal infection. Cell Host Microbe, 26(5): 691-701. 10.1016/j.chom.2019.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sant'Anna V, Folmer Correa AP, de Souza da Motta A, et al. , 2016. Liquid-liquid extraction of antimicrobial peptide P34 by aqueous two-phase and micellar systems. Prep Biochem Biotechnol, 46(8): 838-843. 10.1080/10826068.2016.1141301 [DOI] [PubMed] [Google Scholar]
- Shafei HA, Sabour HA, Ibrahim N, et al. , 2000. Isolation, screening and characterization of bacteriocin-producing lactic acid bacteria isolated from traditional fermented food. Microbiol Res, 154(4): 321-331. 10.1016/S0944-5013(00)80006-3 [DOI] [PubMed] [Google Scholar]
- Shahandashti RV, Kermanshahi RK, Ghadam P, 2016. The inhibitory effect of bacteriocin produced by Lactobacillus acidophilus ATCC 4356 and Lactobacillus plantarum ATCC 8014 on planktonic cells and biofilms of Serratia marcescens . Turk J Med Sci, 46(4): 1188-1196. 10.3906/sag-1505-51 [DOI] [PubMed] [Google Scholar]
- Simha BV, Sood SK, Kumariya R, et al. , 2012. Simple and rapid purification of pediocin PA-1 from Pediococcus pentosaceous NCDC 273 suitable for industrial application. Microbiol Res, 167(9): 544-549. 10.1016/j.micres.2012.01.001 [DOI] [PubMed] [Google Scholar]
- Svetoch EA, Eruslanov BV, Levchuk VP, et al. , 2011. Isolation of Lactobacillus salivarius 1077 (NRRL B-50053) and characterization of its bacteriocin, including the antimicrobial activity spectrum. Appl Environ Microbiol, 77(8): 2749-2754. 10.1128/aem.02481-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuomola E, Crittenden R, Playne M, et al. , 2001. Quality assurance criteria for probiotic bacteria. Am J Clin Nutr, 73(2): 393s-398s. 10.1093/ajcn/73.2.393s [DOI] [PubMed] [Google Scholar]
- Usman, Hosono A, 1999. Bile tolerance, taurocholate deconjugation, and binding of cholesterol by Lactobacillus gasseri strains. J Dairy Sci, 82(2): 243-248. 10.3168/jds.S0022-0302(99)75229-X [DOI] [PubMed] [Google Scholar]
- Yerlikaya O, 2019. Probiotic potential and biochemical and technological properties of Lactococcus lactis ssp. lactis strains isolated from raw milk and kefir grains. J Dairy Sci, 102(1): 124-134. 10.3168/jds.2018-14983 [DOI] [PubMed] [Google Scholar]
- Zhai QX, Shen XD, Cen S, et al. , 2020. Screening of Lactobacillus salivarius strains from the feces of Chinese populations and the evaluation of their effects against intestinal inflammation in mice. Food Funct, 11(1): 221-235. 10.1039/c9fo02116g [DOI] [PubMed] [Google Scholar]
- Zhang M, Hang XM, Fan XB, et al. , 2008. Characterization and selection of Lactobacillus strains for their effect on bile tolerance, taurocholate deconjugation and cholesterol removal. World J Microbiol Biotechnol, 24(1): 7-14. 10.1007/s11274-007-9431-6 [DOI] [Google Scholar]
- Zheng G, Slavik MF, 1999. Isolation, partial purification and characterization of a bacteriocin produced by a newly isolated Bacillus subtilis strain. Lett Appl Microbiol, 28(5): 363-367. 10.1046/j.1365-2672.1999.00545.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.