Significance
NK cells have been observed to be present within most solid tumors, but the antitumor activity of the intratumoral NK cells has been unclear. In this study, we examined the entire spectrum of innate lymphoid cells within primary human tumors and demonstrate that peripheral NK cells in the tumor microenvironment differentiate into heterogeneous cell states, resulting in either a hyporesponsive NK cell subset or a highly active NK cell phenotype that closely resembles intraepithelial ILC1s and that has potent antitumor properties. Importantly, this differentiation into ieILC1-like cells occurs when NK cells are cocultured with epithelial cells, providing important information for NK cell immunotherapy approaches.
Keywords: ILC, natural killer cells, intratumoral, ieILC1, HNSCC
Abstract
Natural killer (NK) cells comprise one subset of the innate lymphoid cell (ILC) family. Despite reported antitumor functions of NK cells, their tangible contribution to tumor control in humans remains controversial. This is due to incomplete understanding of the NK cell states within the tumor microenvironment (TME). Here, we demonstrate that peripheral circulating NK cells differentiate down two divergent pathways within the TME, resulting in different end states. One resembles intraepithelial ILC1s (ieILC1) and possesses potent in vivo antitumor activity. The other expresses genes associated with immune hyporesponsiveness and has poor antitumor functional capacity. Interleukin-15 (IL-15) and direct contact between the tumor cells and NK cells are required for the differentiation into CD49a+CD103+ cells, resembling ieILC1s. These data explain the similarity between ieILC1s and tissue-resident NK cells, provide insight into the origin of ieILC1s, and identify the ieILC1-like cell state within the TME to be the NK cell phenotype with the greatest antitumor activity. Because the proportions of the different ILC states vary between tumors, these findings provide a resource for the clinical study of innate immune responses against tumors and the design of novel therapy.
The role of NK cells in host protection against human tumor formation and growth has been unclear. Intratumoral NK cells, found in most solid tumors, are often dysfunctional and, under certain conditions, can promote tumor growth (1, 2). Conversely, the detection of activated NK cells within tumors, particularly in head and neck squamous cell carcinoma (HNSCC), has also been associated with significantly better clinical outcomes (3–5). Previous studies examining NK cells within human tumors have focused on narrow definitions based on limited surface marker profiles. The emerging recognition of plasticity between NK cells and other ILC family members has suggested that a broader examination of ILCs within solid tumors may provide insight into the different intratumoral NK cell subsets and their relationships.
The ILC family of lymphocytes plays diverse roles in tissue homeostasis and host defense (6). Although ILCs do not express recombined antigen receptors found on T cells and do not undergo clonal expansion or selection when stimulated, they can be grouped into subsets that are akin to subsets of T cells. Based on the cytokines they produce and the transcription factors they express, ILC subsets are categorized into group 1 ILC, group 2 ILC, and group 3 ILC, which are considered analogous to the CD4+ T helper cell Th1, Th2, and Th17 cell subsets, respectively. Specifically, group I ILC includes ILC1s, intraepithelial ILC1s (ieILC1s) and NK cells, which produce IFNγ and TNFα and are dependent on the transcription factor T-BET, similar to Th1 cells (7–10). ILC2s produce IL-4, IL-5, and IL-13 and express GATA-3, similar to Th2 cells (11–14). ILC3s produce IL-17 and IL-22, similar to Th17 cells, and are dependent on the transcription factor RORγt (15–17).
NK cells are considered to be a member of the group 1 ILC (18), but, unlike ILC1s, NK cells express granzymes and perforin and have cytolytic activity, making them analogous to cytotoxic CD8+ T cells. The expression of the transcription factor EOMES by NK cells, in addition to T-BET, also distinguishes NK cells from ILC1s. Finally, unlike ILC1s which are primarily tissue resident, NK cells both circulate in the peripheral blood and can reside within tissue compartments, including solid tumors. Within humans, the conventional NK (cNK) cells that circulate in the blood are primarily CD56dimCD3–CD19– cells. These peripheral NK cells can be distinguished from tissue-resident NK (trNK) cells, which resemble CD56bright cells (19) and express a combination of tissue residency markers, including the chemokine receptor CXCR6, integrin α1 (ITGA1, CD49a), and integrin αE (ITGAE, CD103), depending on particular tissue type and location (18, 20). Another member of the group 1 ILC is ieILC1 cells, which are characterized by the expression of surface markers CD49a, CD103, and NKp44 (9). Because ieILC1s express both EOMES and T-BET, some have proposed that these cells are more closely related to NK cells than classic helper ILC1s (18, 21). The ieILC1s, which reside in mucosal and tonsillar tissue, also express CXCR6, similar to trNK cells described in other tissues such as the bone marrow (BM), spleen, and liver (18, 22).
There is evidence for plasticity between ILC subsets, which can be induced by changes in the tissue microenvironment (23–29). Within murine group 1 ILC, there is evidence for plasticity between NK cells and ILC1-like cells (30–33), and TGF-β signaling is involved in the conversion of NK cells into ILC1-like cells. However, the significance of this plasticity in disease contexts is not well understood. For instance, in murine transplantable tumor models using B16 melanoma and MCA-induced sarcomas, the TGF-β−mediated conversion of NK cells to an intermediate ILC1 (intILC1) state resulted in poor control of tumor growth (32). In contrast, in the murine MMTV-PyMT mammary tumor model, a CD49ahiCD103hi ILC1-like population was demonstrated to have a potent immunosurveillance function (31). These findings indicate that a better understanding of the different ILC states within human tumors will identify key immune cell subsets that have important influence on tumor growth and behavior.
It is only recently that the role of ILCs in human cancer has begun to be studied, and there is still very little that is understood (34). The evidence, in mice, that peripheral NK cells can convert into ILC1-like cells within the tumor microenvironment (32, 33) has not been reported in humans. In this study, we investigated the landscape of the ILC subsets in human HNSCC by single-cell RNA sequencing (scRNA-seq) of sorted ILCs from primary tumors. We describe a spectrum of ILC states that are present within the tumors and demonstrate that peripheral circulating NK cells differentiate into two divergent terminal states in the presence of tumor cells and IL-15. One state is an NK cell subset that resembles CD49a+CD103+ ieILC1s with potent antitumor functional capacity, and the other is an NR4A-expressing CD49a– NK cell subset with poor antitumor functional capacity. The understanding of these two NK cell states within human tumors may have important implications for prognostication and therapy, and the results from this study provide an important resource for the future investigation of NK cells in human cancer.
Results
Human HNSCC Contains Heterogeneous ILC Subsets and States.
Using scRNA-seq, we took an unbiased approach to evaluate the heterogeneity of ILC states within the tumor microenvironment of human HNSCC. We obtained histologically confirmed HNSCC tumor samples from patients undergoing tumor resection, as well as lymph node tissue and peripheral blood (Table 1). These HNSCC tumors are all of epithelial origin, and the tumor sites included the mucosa of the oral cavity or oropharynx. Some were associated with the human papilloma virus (HPV), as determined by either HPV in situ hybridization or by p16 staining, which is a surrogate marker of HPV infection in these tumors. There was no correlation of CD49a or CD103 expression with tumor site or HPV status. We also obtained peripheral blood from healthy volunteers. The tissue and blood were processed into single-cell suspensions (Fig. 1A) to sort comprehensively all ILCs, including NK cells, for scRNA-seq library preparation and sequencing. Human peripheral NK cells (also referred as conventional NK cells) are phenotypically defined as CD56+ lymphocytes lacking expression of CD3 and CD19. Although ILC1s, ILC2s, and ILC3s are characterized by expression of CD127 (IL-7Rγ), some subsets express CD56 (18). Likewise, some NK cells, particularly CD56bright NK cells, express CD127 (18). Thus, to include all intratumoral ILCs and NK cells, we sorted lineage-negative lymphocytes expressing CD56 and/or CD127 for our analysis (SI Appendix, Fig. 1A).
Table 1.
Patient and tumor characteristics of the samples used for scRNA-seq
| Sample ID | Tumor site | Pathological staging of cancers | P16 Immunostain | HPV in situ hybridization | Ethnicity | Sex | Diagnosis | Age, y (in 2019) | |
| T (primary tumor) | N (metastatic node) | ||||||||
| S-SCC-7202 | Oral cavity | pT3 | pN2c | Positive | N.A. | Asian | F | New diagnosis | 49 |
| S-SCC-7204 | Oropharynx | pT2 | pN2 | Positive | N.A. | White | M | New diagnosis | 43 |
| S-SCC-7205 | Oral cavity | pT2 | pN2b | Negative | Positive for high-risk HPV | White | F | New diagnosis | 72 |
| S-SCC-7207 | Oropharynx | pT1 | pN0 | Positive | N.A. | White | M | New diagnosis | 83 |
| S-SCC-7211 | Oral cavity | pT4a | pN3b | Negative | N.A. | Asian | M | New diagnosis | 73 |
| S-SCC-7257 | Oral cavity | pT3 | pN2b | N.A. | N.A. | White | M | New diagnosis | 62 |
| S-SCC-7267 | Oral cavity | pT4a | pN3 | Negative | N.A. | Other (non-Hispanic or Latino) | M | New diagnosis | 57 |
| S-SCC-7329 | Oral cavity | pT4a | pN1 | Negative | N.A. | Hispanic or Latino | M | New diagnosis | 51 |
N.A., not available.
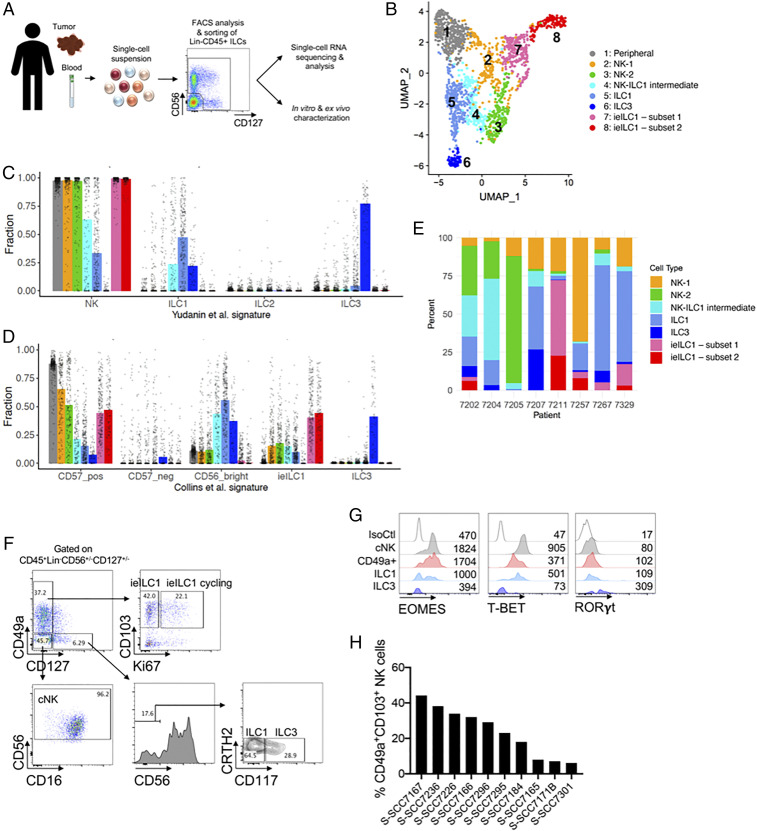
Fig. 1.
Heterogeneity of ILCs in human HNSCC. (A) Workflow of the ILC characterization from blood, primary tumors, or metastatic lymph nodes. Dissociated cell suspensions were sorted as Lineage- CD56+CD127+, CD56+CD127−, and CD56−CD127+ single cells, in order to capture the full diversity of ILCs, and used for scRNA-seq or function analysis. Cartoon image adaptations credit: Servier Medical Art, licensed under CC BY 3.0. (B) UMAP of scRNA-seq profiles of human intratumoral and peripheral ILCs shows eight distinct clusters. (C and D) Deconvoluted compositions of single cells within each ILC cluster based on gene expression signatures derived from bulk-sorted cells. CIBERSORTx fractions were determined for each single-cell gene expression library in this study, based on gene expression signatures derived from (C) Yudanin et al. (36), comprising bulk RNA-seq libraries of human NK, ILC1, ILC2, and ILC3 cells from lymphoid and mucosal tissue sites, or (D) Collins et al. (37), comprising bulk RNA-seq libraries of human NK cells (CD56bright, CD56dimCD57pos, CD56dimCD57neg) from peripheral blood, as well as ieILC1 and ILC3 cells isolated from tonsil tissue. The median deconvoluted fraction of each signature for single cells in each ILC cluster is represented by vertical bars. (E) Distribution of ILC clusters across tumor tissue samples obtained from eight different patients. (F–H) Cell suspensions of primary human HNSCC tumor specimens were stained for surface and intracellular markers and analyzed by flow cytometry. (F) Analysis of CD45+Lin−CD56+/−CD127+/− cells demonstrates the presence of ieILC1, ieILC1 cycling, ILC1, ILC3, and cNK cell populations. (G) Expression levels of transcription factors EOMES, T-BET, and RORγt in cNK cells, CD49a+ cells, ILC1s, and ILC3s. Dot plots and histograms show representative examples of the phenotype observed on ILCs from at least three donors. (H) Heterogeneity in the percentage of intratumoral CD49a+CD103+ cells among different HNSCC patients.
Dimension reduction of the scRNA-seq data illustrated eight distinct clusters of ILCs (Fig. 1B). Cell indexing of the fluorescence-activated cell sorter (FACS)-sorted cells revealed that the CD127+ cells were primarily within clusters 5 and 6, whereas the CD56+CD127– cells were distributed among all the clusters (SI Appendix, Fig. 1B and Fig. 1B). Further, the cells sorted from the peripheral blood clustered separately (cluster 1, peripheral NK cluster) from cells sorted from the tumor and normal lymph node tissue (SI Appendix, Fig. 1C). In order to understand the different intratumoral clusters, we estimated the fractional compositions of the scRNA-seq profiles by CIBERSORTx (35), using previously published gene signature matrices derived from bulk RNA-seq profiles (36, 37) of sorted human ILC subsets (Fig. 1 C and D). Using this approach, we were able to objectively assign identities to the seven different intratumoral clusters in an unbiased manner. We identified two NK cell clusters (clusters 2 and 3, designated NK-1 and NK-2), one ILC1 cluster (cluster 5), one ILC3 cluster (cluster 6), two ieILC1 clusters (clusters 7 and 8), and one cluster that appeared to represent a transition state between NK cell and ILC1 (cluster 4, designated NK-ILC1 intermediate) (Fig. 1 B–D). While cells from all the clusters were present in all the patient samples, there was a significant heterogeneity in the relative proportion of each population between patient samples (Fig. 1E and SI Appendix, Fig. 1D).
The presence of ILC subsets within primary HNSCC tumors was further confirmed by flow cytometry (Fig. 1F and SI Appendix, Fig. 1 E and F). Within the CD45+Lin–CD127+CD56– compartment, we found CRTH2–CD117– cells consistent with ILC1s, and CRTH2–CD117+ cells consistent with ILC3s. The latter were EOMES–T-BET–RORγt+ (Fig. 1G). Similar to our scRNA-seq data, there was no evidence of the presence of CD127+CRTH2+ ILC2s in the tumors. Within the NK cell gate (CD45+Lin–CD56+), we observed cells expressing CD49a and CD103 that were EOMEShi T-BETint RORγt–, consistent with ieILC1s (Fig. 1 F and G and SI Appendix, Fig. 1 E and F). Some of these cells expressed the proliferation marker Ki-67 (Fig. 1 F and SI Appendix, Fig. 1G), suggesting that a subset of ieILC1s was actively cycling. Also, intratumoral CD56+CD49a+ ieILC1 cells expressed high levels of CD69, suggesting an activated and/or tissue-resident state (SI Appendix, Fig. 1H). Similar to the scRNA-sequencing analysis (Fig. 1E and SI Appendix, Fig. 1D), we observed significant heterogeneity in the proportion of ieILC1 cells between different patient samples by flow cytometry (Fig. 1H and SI Appendix, Fig. 1 I and J).
Intratumoral ILC Populations Display Distinctive Functional and Effector States.
In order to study the function and possible relationship between populations, we first analyzed the gene expression profiles among the different populations (Dataset S1). Peripheral NK cells were characterized by expression of FGFBP2, CXC3R1, and FCGR3A (encoding CD16) (Fig. 2A). The NK-1 population had a signature of type I interferon (IFN) activation, whereas the NK-2 population had a signature notable for expression of the NR4A2 orphan nuclear receptor transcription factor gene (also known as NURR1) (Fig. 2 A and B), which was recently shown to negatively regulate T cell responsiveness (38–40). Both the ILC1 and ILC3 clusters expressed IL7R (CD127) and SELL (CD62L) (Fig. 2 A and B). The ILC3 cluster expressed the canonical genes RORC, KIT, IL23R, and AHR (Fig. 2 A and B and SI Appendix, Fig. 2A). We observed an intermediate NK-ILC1 population with genes that were also expressed in the NK-2 and ILC1 clusters (Fig. 2A), including AREG (SI Appendix, Fig. 2A), which has been observed to be up-regulated in in CD56bright NK cells (41). CIBERSORTx fractional compositions showed that the gene expression profiles of cells in the NK-1, ILC1, and NK-ILC1 intermediate clusters were more similar to CD56bright NK cells when compared to CD56dim NK cells (SI Appendix, Fig. 2B).
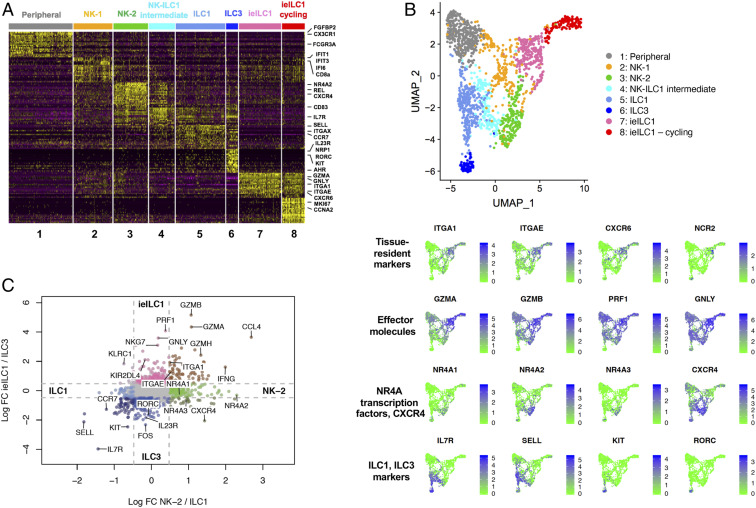
Fig. 2.
Intratumoral ILCs have distinct effector states. (A) Heat map of the top 20 differentially expressed genes (up-regulated) of each cluster, ranked by adjusted P value. (B) Feature plots showing the expression of tissue-resident markers (ITGA1, ITGAE, CXCR6, NCR2), cytotoxic effector molecules (GZMA, GZMB, PRF1, GNLY), markers for the NK-2 group (NR4A transcription factors, CXCR4), markers for ILC1 and ILC3 cells (IL7R, SELL) and for ILC3 cells (KIT, RORC), among the ILC clusters in the UMAP shown in Fig. 1B. (C) Log fold change plot showing gene expression differences between the intratumoral ILC populations. Colored dots (as opposed to gray) represent transcripts expressed at least threefold higher in one subset versus another.
Both ieILC1 clusters expressed genes previously used to define the ieILC1 population (9, 37), that is, ITGAE (CD103), ITGA1 (CD49a), CXCR6, and NCR2 (NKp44) (Fig. 2 A and B). One ieILC1 cluster, which we termed “ieILC1-cycling,” highly expressed genes associated with active proliferation such as CCNA2 (cyclin A2) and MKI67 (Ki-67) (Fig. 2A). This was consistent with our flow cytometry observations of a subset of intratumoral ieILC1s that expressed Ki-67 (Fig. 1F and SI Appendix, Fig. 1G).
Consistent with the previously proposed hypothesis that ieILC1s are related to NK cells (18), we found that all ieILC1, NK-1, NK-2, and peripheral NK cell clusters expressed NK-related genes GZMA (granzyme A), GZMB (granzyme B), GZMH (granzyme H), PRF1 (perforin), and GNLY (granulysin) (Fig. 2). However, the log fold change plots and the feature plots showed that the ieILC1s had the highest levels of GZMA, GZMB, PRF1, and GNLY (Fig. 2 B and C and SI Appendix, Fig. 2C), suggesting that these cells may have high cytolytic capacity. In contrast, the NK-2 cluster had relatively lower expression of effector molecule genes (GZMA, GZMB, and PRF1) and was notable for high expression of NR4A1 and NR4A2 (Fig. 2). Thus, among the intratumoral NK cell-related subsets, the ieILC1 subset appeared to have the greatest cytolytic capacity, whereas the NK-2 subset expressed genes suggesting a dysfunctional phenotype.
NK Cells Have Two Distinct Intratumoral Differentiation Trajectories, Leading to Either an ieILC1 Phenotype or an NK-2 Phenotype.
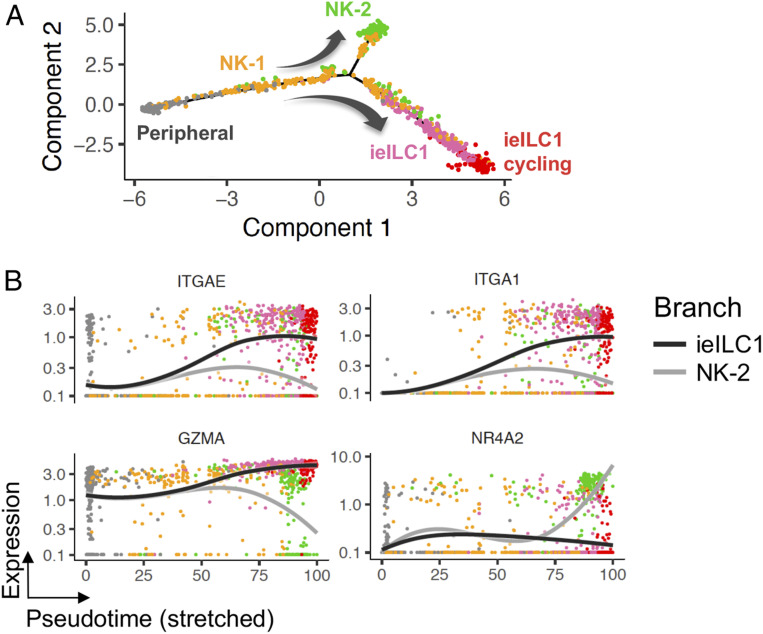
The tumor microenvironment is known to affect the gene expression of immune cells, including their effector function phenotypes (42). In order to understand the relationship between the peripheral cNK cells and the intratumoral NK cell subsets, we used pseudotime ordering (43) of cells that had primarily NK cell signatures identified by CIBERSORTx deconvolution of the subset clusters (Fig. 1 C and D). This enabled the inference of potential trajectories of NK cell differentiation among the different cell states seen in our scRNA-seq data. Relative to the cNK cells, two divergent trajectories, passing through an intermediate NK-1 population, were observed: one resulting in the ieILC1 state and another resulting in the NR4A2-expressing NK-2 state (Fig. 3 and SI Appendix, Fig. 3).
Fig. 3.
Intratumoral ILCs have distinct differentiation trajectories. (A) Pseudotime trajectories of cells that had primarily NK cell signatures, determined by CIBERSORTx deconvolution of the subset clusters, demonstrated two possible differentiation trajectories from peripheral NK cells: an end state of either the NK-2 phenotype or the ieILC1 phenotype. (B) Expression of selected differentially expressed genes along the NK-2 and ieILC1 pseudotime trajectories. Curves represent Loess-smoothed regression of the gene expression along each trajectory.
Peripheral NK Cells Can Differentiate into CD49a+CD103+ Cells and NR4A-Expressing NK Cells In Vitro.
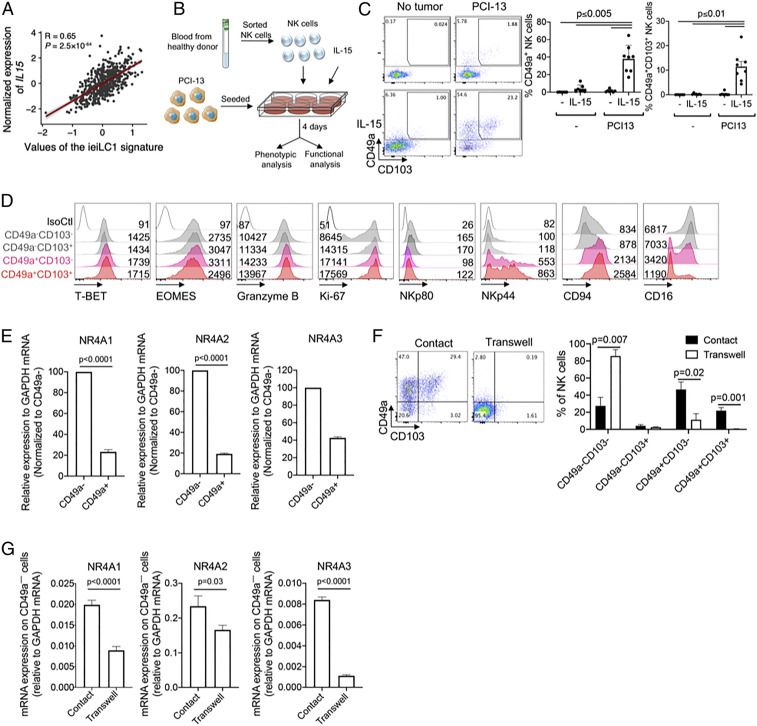
The scRNA-seq data prompted us to investigate the plasticity of cNK cells to differentiate into ieILC1-like cells or NK-2−like cells in vitro. Our analysis of The Cancer Genome Atlas (TCGA) database showed that the expression of IL-15 in HNSCC tumors was highly correlated with top genes expressed by the ieILC1 cluster (Fig. 4A) but not with the NK-2 gene signature (SI Appendix, Fig. 4A). Given that IL-15 is an important cytokine for NK cells and that it is involved in the generation of CD49ahiCD103hi ILC1-like cells in the murine MMTV-PyMT mammary tumor model (31), we hypothesized that IL-15 may be involved in the differentiation of NK cells into ieILC1-like cells in human tumors. In vitro coculture of peripheral cNK cells, which lack expression of CD49a and CD103 (SI Appendix, Fig. 4B), with HNSCC cells resulted in the up-regulation of CD49a and CD103 on a subset of NK cells, and this up-regulation was highly dependent on the presence of IL-15 (Fig. 4 B and C). In addition to HNSCC cells, other tumor cell lines also induced an ieILC1-like phenotype with variable efficiencies (SI Appendix, Fig. 4C). Similar to our observation of primary intratumoral CD49a+ ieILC1 cells (SI Appendix, Fig. 1 E, F, and J), CD103 tended to be coexpressed with CD49a following in vitro differentiation (Fig. 4C and SI Appendix, Fig. 4C). Phenotypically, in vitro differentiated CD56+CD49a+CD103+ cells were NKp44+NKp80–CD94+CD16–GzmB+ and expressed T-BET and EOMES (Fig. 4D). The phenotype of CD49a+CD103+ cells was similar to that of CD49a+CD103– cells, suggesting that, overall, in vitro differentiated CD49a+ cells were phenotypically similar to primary intratumoral ieILC1s (SI Appendix, Figs. 1 F and G and 2 A and B). In contrast, the phenotype of CD49a–CD103+ cells was similar to that of CD49a–CD103– cells (Fig. 4D), and CD49a– cells were characterized by high expression of NR4A genes, particularly NR4A2 (Fig. 4E), similar to the NK-2 cells we identified in primary tumors (Fig. 2 A and B). Importantly, the differentiation of NK cells into CD49a+CD103+ cells required contact with the tumor cells (Fig. 4 F and G), suggesting the involvement of unknown receptor−ligand interactions.
Fig. 4.
Peripheral NK cells differentiate into two states in the presence of IL-15 and tumor cells, in vitro. (A) Correlation between IL15 gene expression in human HNSCC tumors from the TGCA database and an ieILC1 gene expression signature obtained from the scRNA-seq data. (B) Schematic for in vitro differentiation of peripheral NK cells into ieILC1-like cells. PCI-13 cells were seeded into tissue culture plates; then peripheral NK cells were sorted and added to the culture (NK cell:tumor cell = 10:1) with or without IL-15 (10 ng/mL). After 4 d, NK cells were harvested for phenotypic and functional analysis. Cartoon image adaptations credit: Servier Medical Art, licensed under CC BY 3.0. (C) Expression of CD49a and CD103 on gated CD45+Lin−CD56+ NK cells, following coculture of NK cells with PCI-13 and IL-15 (10 ng/mL) or control conditions. (Left) Dot plots show a representative sample. (Right) Cumulative results from eight independent experiments, each using NK cells from different donors. (D) After coculture, NK cells were assessed for their expression of T-BET, EOMES, Granzyme B, Ki-67, NKp80, NKp44, CD94, and CD16. Gating is on CD49a–CD103–, CD49a–CD103+, CD49a+CD103–, and CD49a+CD103+ cells. Histograms show representative examples of the phenotype observed on cells from at least four donors. (E) Sorted CD49a+ and CD49a− NK cells were assessed for expression of NR4A1, NR4A2, and NR4A3 by qRT-PCR. Data are representative of four independent experiments, each using NK cells from a different donor. (F and G) Peripheral NK cells were cultured with PCI-13 cells and IL-15 either in contact or in a transwell system. (F) (Left) Representative sample, and (Right) aggregate results showing the expression of CD49a and CD103 on gated CD45+Lin–CD56+ NK cells following the coculture with IL-15, from four independent experiments, each using NK cells from different donors. (G) Sorted CD49a– NK cells from contact or transwell cultures were assessed for expression of NR4A1, NR4A2, and NR4A3 by qRT-PCR. Graphs show cumulative results from three independent experiments, each using NK cells from a different donor. Graphs (C, E–G) are expressed as mean ± SEM; paired t test analysis was done, and P values are shown.
Immature NK Cells Preferentially Differentiate into CD49a+CD103+ Cells.
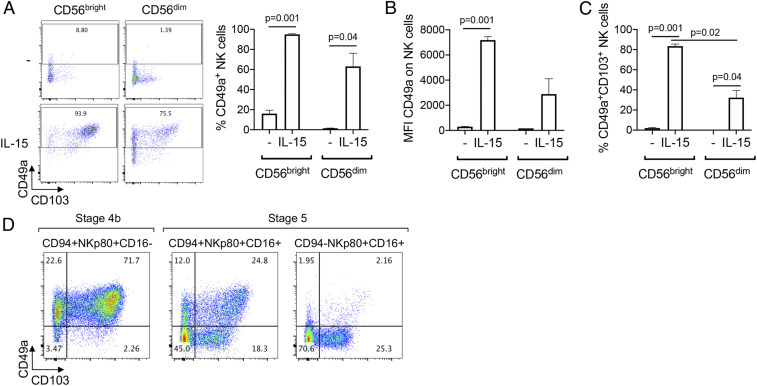
Peripheral NK cells are divided into two main subsets: CD56dim and CD56bright NK cells, which differ in their function and maturation status. CD56bright NK cells have an immature phenotype compared to CD56dim NK cells, and represent a minor subset in peripheral blood but are highly enriched in tissues, including malignant tissues (44). We assessed whether there was a difference in the plasticity of peripheral CD56bright and CD56dim NK cells to differentiate into the ieILC1-like cells. We observed that CD56bright NK cells up-regulated CD49a to a greater extent than CD56dim NK cells when cultured with HNSCC cells and IL-15 (Fig. 5 A and B), and the percentage of CD49a+CD103+ cells was higher following culture with CD56bright NK cells compared to CD56dim NK cells (Fig. 5C).
Fig. 5.
Immature NK cells preferentially differentiate into ieILC1-like cells. (A–C) CD56bright or CD56dim NK cells were sorted from peripheral blood of healthy donors and cocultured with PCI-13 with or without IL-15 (10 ng/mL), as described in Fig. 4B. (A) (Left) Dot plots show a representative example of the expression of CD49a and CD103 on NK cells after the 4-d coculture. Graphs show cumulative results of (Right) the percentage and (B) Mean fluorescence intensity (MFI) of CD49a, and (C) the percentage of CD49a+CD103+ cells, after the 4-d coculture, from three independent experiments, each using NK cells from a different donor. Graphs were expressed as mean ± SEM, and P values are shown. (D) Stage 4b (CD94+NKp80+CD16−) and two populations of stage 5 NK cells (CD94+NKp80+CD16+ and CD94−NKp80+CD16+) were sorted from peripheral blood of healthy donors and cocultured with PCI-13 and IL-15 (10 ng/mL), as described in Fig. 4B. Dot plots show a representative example, from three independent experiments, of the expression of CD49a and CD103 on NK cells after the 4-d coculture.
NK cells develop through a multistage process in the BM, secondary lymphoid tissues, and peripheral blood (45). In the peripheral blood, there are three distinct latter stages of NK cell development that are observed: stage 4b (CD94+NKp80+CD16−CD57−), stage 5 (CD94+/−NKp80+CD16+CD57−), and stage 6 (CD94−NKp80+CD16+CD57+), which represents fully differentiated cells. While most CD56bright NK cells are stage 4b, some are stage 5. Therefore, in order to study more accurately the degree of plasticity of peripheral NK cells and their ability to take on characteristics of ieILC1s, we assessed differences between stage 4b and stage 5 NK cells. We sorted stage 4b cells and two distinct subsets of stage 5 cells, with the latter being distinguished by the expression of CD94 (CD94+ stage 5 cells being less mature than CD94– stage 5 cells) and cultured them with HNSCC and IL-15 (Fig. 5D). We found that the capacity of stage 4b cells to differentiate into CD49a+CD103+ was much greater than that of stage 5 CD94+, which, in turn, was much greater than the capacity of stage 5 CD94− cells. Overall, this indicates that circulating immature NK cells preferentially differentiate into ieILC1-like cells, compared to mature NK cells.
Since CD103 has been shown to be a TGF-β−imprinted gene (46) and because TGF-β is involved in NK-ILC1 differentiation (32, 33, 47), we assessed whether TGF-β played a role in the induction of the ieILC1 phenotype in vitro. In support of this, we found that TGFβ could be detected in cultured supernatants of HNSCC tumor cells and that inhibition of TGF-β signaling reduced the proportion of ieILC1-like cells that were seen after NK cells are cocultured with the tumor cells (SI Appendix, Fig. 5). Although culture of NK cells with IL-15 and soluble TGF-β (in the absence of tumor cells) was able to induce the expression of CD49a and CD103 (SI Appendix, Fig. 5C), in accordance with previous observations (48), the appearance of ieILC1-like cells, when NK cells are cocultured with HNSCC cells, required contact between the cells (Fig. 4F). Membrane-bound TGF-β could not be detected on the PCI-13 cells; thus, another unknown mechanism related to TGF-β signaling appears to be involved.
CD49a+ ieILC1-Like Cells Display Greater Effector Responses than CD49a– NK Cells after Stimulation.
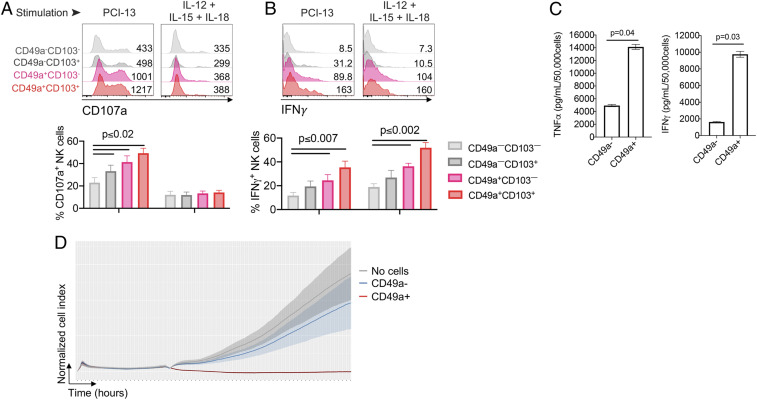
Next, we analyzed the functional response of the in vitro differentiated ieILC1-like cells (CD49a+) and NK-2−like cells (CD49a–). Consistent with the ieILC1 clusters having the greatest expression of GZMA, GZMB, PRF1, and IFNG among the intratumoral ILC clusters (Fig. 2), the in vitro-differentiated ieILC1-like cells (particularly CD49a+CD103+ cells) degranulated more, as evidenced by higher CD107a expression (Fig. 6A), and produced greater amounts of IFNγ and TNFα (Fig. 6 B and C), compared to NK-2−like cells (particularly CD49a–CD103– cells) in response to tumor cells (PCI-13), cytokines (IL-12, IL-15, and IL-18), and phorbol 12-myristate 13-acetate (PMA) and ionomycin. In line with a higher degranulation (Fig. 6A), CD49a+ ieILC1-like cells killed HNSCC tumor cells more effectively than did the CD49a− NK-2−like cells derived from the same coculture (Fig. 6D). Overall, this indicates that ieILC1-like cells and NK-2−like cells derived from peripheral cNK cells have distinct functional capacities.
Fig. 6.
CD49a+ ieILC1-like NK cells have higher functional activity than CD49a– NK cells. (A and B) Analysis of the functional response of NK cells following coculture with PCI-13 and IL15, according to the expression of CD49a and CD103. Following coculture, NK cells were stimulated with either PCI-13 tumor cells or cytokines (IL12 + IL15 + IL18), and the (A) degranulation (CD107a surface expression) and (B) intracellular IFNγ production were determined by flow cytometry. Results are gated on CD49a–CD103–, CD49a–CD103+, CD49a+CD103–, and CD49a+CD103+ populations. (Top) Representative samples, and (Bottom) cumulative results from three independent experiments, each using NK cells from different donors. (C) Sorted CD49a+ and CD49a– NK cells were stimulated with PMA and ionomycin for 12 h, and supernatants were assessed for TNFα and IFNγ by Luminex Immuno-Assay. Data are representative of four independent experiments, each using NK cells from a different donor. Graphs (A–C) are expressed as mean ± SEM; paired t test analysis was done, and P values are shown. (D) Peripheral NK cells were cultured with PCI-13 and IL-15 (10 ng/mL), described in Fig. 4B. After 4 d in culture, CD49a– or CD49a+ ieILC1-like cells were sorted, and their cytolytic activity was tested using xCELLigence assay (E:T = 1:1). Graph shows the normalized cell index over time, representative of three independent experiments, each using NK cells from different donors.
Conventional NK Cells Differentiate into CD49a+CD103+ Cells in the Tumor Microenvironment and Have Potent Antitumor Activity.
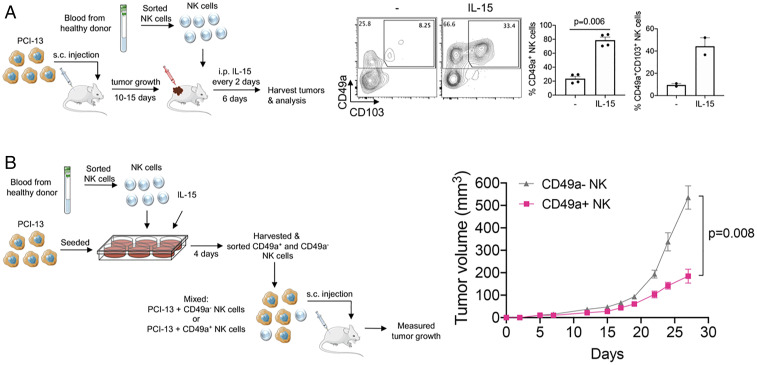
Considering the in vitro differentiation of CD49a+CD103+ cells (Fig. 4 B and C), we hypothesized that IL-15 would promote the differentiation of cNK cells into ieILC1-like cells in vivo. Human HNSCC tumor cells were used to establish tumors in the subcutaneous compartment of immunodeficient NOD-scid IL2Rgammanull (NSG) mice, which lack T cells, B cells, and NK cells. Peripheral CD56+ NK cells (SI Appendix, Fig. 6A) or CD56bright NK cells (Fig. 7A), sorted from healthy volunteers, were directly injected into the tumors, and the mice were treated with or without IL-15. After 6 d, tumors were harvested, and we assessed the intratumoral NK cells by flow cytometry. We observed that a subset of NK cells had up-regulated the ieILC1 markers CD49a and CD103 and that this was dependent on exogenous administration of IL-15 to the mice (Fig. 7A and SI Appendix, Fig. 6A). Similar to our in vitro findings (Fig. 6 A–C), we observed that, in the presence of IL-15, the in vivo-differentiated CD49a+ cells produced higher amounts of IFNγ compared to the CD49a– cells following stimulation (SI Appendix, Fig. 6B).
Fig. 7.
CD49a+ ieILC1-like cells have potent antitumor activity. (A) (Left) Schematic of the experimental design for studying the differentiation of human peripheral NK cells in vivo. PCI-13 cells were subcutaneously injected into the flanks of NSG mice. After 10 d to 15 d, when the tumors were palpable, FACS-sorted peripheral Lin–CD56+CD94+CD16–CD49a– NK cells from healthy human donors were injected into the growing tumors. These mice were then split into two groups, and one group was administered 5 μg of human IL-15 intraperitoneally (i.p.) per mouse, every 2 d. After 6 d, tumors were harvested and dissociated, and the intratumoral NK cells were characterized by flow cytometry. (Right) Expression of CD49a and CD103 on intratumoral NK cells of a representative example is shown. Bar graphs show cumulative results of the percentages of the CD49a+ and CD49a+CD103+ cells (gated on CD45+Lin–CD56+ cells). Graphs are expressed as mean ± SEM; paired t test analysis was done, and P value is shown. (B) (Left) Schematic of approach used to assess the antitumor activity of in vitro-differentiated ieILC1-like cells. NK cells were cocultured with PCI-13 cells and IL-15 (10 ng/mL) for 4 d to induce differentiation. Then, CD49a+ and CD49a– NK cell populations were sorted, and each sorted NK cell population was mixed with PCI-13 (NK cells:PCI-13 cells = 1:2.5). The cell mixtures were injected subcutaneously (s.c.) into the flanks of NSG mice, and tumor growth (n = 8 per condition) was measured. (Right) Tumor growth curves are representative of three independent experiments. Cartoon image adaptations credit: Servier Medical Art, licensed under CC BY 3.0.
Because CD49a+ ieILC1-like cells produce higher levels of IFNγ, TNFα, and granzymes than CD49a– cells and are able to degranulate more readily upon stimulation (Fig. 6 A–C), we hypothesized that CD49a+ cells would be able to control tumor growth in vivo better than CD49a– NK cells. To assess this, in vitro-differentiated CD49a+ or CD49a– NK cells were FACS sorted and mixed separately with HNSCC tumor cells. Equal numbers of total cells of these separate mixes were injected into the subcutaneous compartment of NSG mice (Fig. 7B). We observed that CD49a+ ieILC1-like cells were able to control tumor growth to a significantly greater degree than the CD49a– NK cells derived from the same in vitro culture. Together, our data indicate that the tumor microenvironment can shape the plasticity of NK cells by polarizing them either into an ieILC1-like phenotype or an NK-2−like phenotype, which may have opposing effects on the tumor and clinical behavior (see SI Appendix, Fig. 7).
Discussion
In this study, we comprehensively assessed the panorama of intratumoral ILCs, including NK cells, in human HNSCC using an unbiased scRNA-seq approach and found seven cell states, distinct from peripheral blood NK cells (Fig. 1B). Previous tumor scRNA-seq studies included NK cells among many different cell types (other immune cells, tumor cells, and stromal cells); however, because the NK cell/ILC compartment in tumors is relatively small (∼1 to 5% of the immune cells), insights into intratumoral NK cell/ILC biology have been limited. A recent scRNA-seq study in melanoma included only CD56+ NK cells, limiting the spectrum of ILC subsets, and relied heavily on integration of gene expression profiles between samples in their analysis (49). Here, despite having a larger number of source patients and a greater diversity of samples, we did not require any batch correction or rely on any integrative approaches in our analysis of the entire ILC population.
Our study revealed that, within the tumor microenvironment, peripheral cNK cells transition through an intermediate NK-1 state and can differentiate into two distinct and divergent terminal states. One is a CD49a+CD103+ ieILC1-like subset with high granzyme and perforin expression and high capacity to control tumor growth, and the other is an NR4A2-expressing CD49a– subset with low capacity to control tumor growth.
Mechanistically, we found that the differentiation of cNK cells into ieILC1-like cells required IL-15 and was dependent on contact between NK cells and HNSCC cells. Also, we found that immature NK cells (i.e., CD56bright stage 4b cells) have the greatest potential to differentiate into ieILC1-like cells among the circulating NK cells. In accordance with previous studies showing that TGF-β is involved in the imprinting of CD103, and the differentiation of cNK into ILC1 cells (32, 33, 48), we found that TGF-β signaling in the NK cells plays a role in the acquisition of the ieILC1-like phenotype. This observation is somewhat surprising, given that the ieILC1-like cells obtained under our conditions display much high effector functions and that TGF-β has been shown to have suppressive effects on NK cell function (50). It is possible that the combination of TGF-β with IL-15 has quite different effects on NK cells than TGF-β alone, similar to the role of TGF-β in the differentiation of both Treg cells and Th17 cells (51).
An ieILC1-like NK cell subset has been observed by mass cytometry studies of human lung and colorectal tumors previously; however, in that study, the ieILC1-like cells had lower expression of granzyme B and perforin compared to a CD103– NK cell population (52). It is possible that the CD103– cells in the lung are predominantly peripheral blood NK cells, given the highly vascularized nature of the lung tissue. It is also possible that different tumor types (e.g., squamous cell carcinoma vs. adenocarcinoma) induce different ieILC1 functional states. Interestingly, the CD49a+CD103+ ieILC1-like population that we observed in HNSCC resembled the murine CD49ahiCD103hiGzmBhi ILC1-like tissue-resident lymphocyte population observed in the tumor microenvironment of MMTV-PyMT tumors (31). Similar to the human ieILC1-like cells that we identified here, the murine ILC1-like population also exhibited potent cytotoxicity and capacity to control tumor growth. Among the two ieILC1 clusters that we observed, one had a distinctive cycling phenotype, which may be reflective of greater IL-15 signaling or other stimuli present within the tumor microenvironment.
NR4A2 was recently demonstrated to confer a hyporesponsive phenotype in cytotoxic T cells and to promote Treg differentiation in CD4+ T cells (38–40, 53, 54). Our finding that NR4A2+CD49a– NK cells show poor tumor control in vivo is consistent with the inhibitory function of NR4A2 on T cells. While the underlying mechanism remains to be determined, it is interesting to note that NR4A2+CD49a– NK cells have significantly lower IFNγ and TNFα expression. In future studies, it will be important to understand the role of NR4A2 in NK cell function and the tumor-associated factors governing the differentiation of peripheral NK cells into divergent NK cell states.
Materials and Methods
Study Design.
We used a total of eight primary tumor samples and four paired nodal metastases from HNSCC patients, as well as peripheral blood mononuclear cells (PBMC) from four HNSCC patients and two healthy volunteers for scRNA-seq analysis. In vitro experiments used a sample size of at least three different donors per experiment. In vitro and in vivo experiments were replicated at least three times. No data were excluded from analysis.
The goal of the study was to examine the spectrum of ILCs present within the tumor microenvironment of human HNSCC. For this, primary tissue samples were obtained from patients consented at Stanford University. Our prespecified hypothesis was that all known ILCs would be observed; however, multiple altered states of NK cells, in addition to conventionally defined ILC1s and ILC3s, were found. These data led to the hypothesis that NK cells differentiated in the tumor microenvironment, which we assessed in vitro and in vivo using commercially available cell lines and blood from healthy donors.
Human Blood and Tumor Tissue Sample Processing.
Deidentified blood and fresh tumor tissue specimens, with clinical annotation, were obtained through the Stanford Tissue Bank from patients undergoing surgical resection of HNSCC at the Stanford Hospital, Stanford, CA, after informed consent, in accordance with an Institutional Review Board (IRB)-approved protocol. Human tumor tissue specimens were minced into small fragments and mechanically dissociated using a gentleMACS Dissociator (Miltenyi Biotec). The digested tissue was filtered through a 70-μm cell strainer (Falcon; Cat# 352350) to obtain a single-cell suspension. Human blood was layered on Ficoll-Paque Premium (GE Healthcare; Cat# 17544202), then centrifuged at 400 × g (no breaks) for 20 min at room temperature (RT). The layer containing PBMC was harvested carefully and washed twice with phosphate-buffered saline (PBS) (Mediatech; Cat# 21-040-CV) to obtain a single-cell suspension. For in vitro and in vivo NK cell differentiation, blood from healthy donors was obtained from the Stanford Blood Center, in accordance with a protocol approved by the IRB at Stanford University, and NK cells were enriched using RosetteSep NK Cell Enrichment Mixture (Stem Cell Technologies; Cat# 15065) according to manufacturer’s instructions.
Harvested xenograft tumors growing in NSG mice were minced into small fragments, which were mechanically dissociated using a gentleMACS Dissociator (Miltenyi Biotec) followed by enzymatic digestion using Collagenase/Hyaluronidase (Stemcell Technologies; Cat# 07912) for 30 min at RT with constant rotation. Then, the mixture was filtered with a 70-μm cell strainer (Falcon; Cat# 352350) to obtain single-cell suspension.
Cell Lines and In Vitro NK Cell Differentiation.
The human HNSCC cell lines, PCI-13 (Research Resource Identifier [RRID]: CVCL_C182) and UM-SCC-6 (RRID: CVCL_7773), were obtained from the University of Pittsburgh and the University of Michigan, respectively. The human HNSCC cell line, SCC4 (RRID: CVCL_1684), and the human melanoma cell line, SK-MEL-028 (RRID: CVCL_0526), were obtained from American Type Culture Collection. Cells were cultured in Dulbecco’s modified Eagle’s medium/F-12 50/50 (Corning; Cat# 10-092-CV) supplemented with 10% heat-inactivated fetal bovine serum (Omega Scientific; Cat# FB-21) and 1% Pen Strep (Gibco; Cat# 15140-122) and were passaged without reaching confluence. Before the coculture with NK cells, the tumor cells were harvested using TrypLE Express Enzyme (Gibco; Cat# 12-604-039).
For the study of in vitro differentiation of NK cells, tumor cells were first seeded into six-well tissue culture plates (E&K Scientific; Cat# EK-27160) at 5 × 105 cells per well. Then, sorted NK cells were added to the culture at an effector-to-target ratio of 10:1. Cells were cultured in RPMI 1640 (Corning; Cat# 10-040-CV) supplemented with 10% heat-inactivated fetal bovine serum (Omega Scientific; Cat# FB-21), 1% Pen Strep (Gibco; Cat# 15140-122), 55 µM 2-Mercaptoethanol (Gibco; Cat# 21985-023), 1× MEM Non-Essential Amino acids (Gibco; Cat# 11140-050), 1 mM Sodium Pyruvate (Gibco; Cat# 11360-070), and 10 mM Hepes (Gibco; Cat# 25-060-Cl). Where mentioned, the culture media was supplemented with 10 ng/mL of human recombinant IL-15 (BioLegend; Cat# 570306 and the BRB Preclinical Biologics Repository at National Cancer Institute), 10 ng/mL of human recombinant TGF-β1 (BioLegend; Cat# 580702), 10 µM of SB 431542 (Tocris; Cat# 1614), or 10 µM of ITD 1 (Tocris; Cat# 5068).
Mice and In Vivo NK Cell Differentiation.
NSG mice were housed and bred in an animal facility under standard temperature, humidity, and timed lighting conditions and were provided with mouse chow and water ad libitum. All animal experimentation protocols were approved by the Stanford University Animal Care and Use Committee. Mice of ages 7 wk to 10 wk were used in our experiments.
One million PCI-13 cells were injected subcutaneously in the flank of the mouse, two injections per mouse. After 10 d to 15 d, when the tumors reached a group average of 200 mm3 to 300 mm3, mice were sublethally irradiated 300cG using a Faxitron irradiator (Faxitron). Then, 2 × 106 to 5 × 106 sorted NK cells were injected intratumorally, per tumor. Where indicated, human recombinant IL-15 (BioLegend; Cat# 570306) was injected intraperitoneally on the day of NK cell intratumoral injection and every 2 d thereafter. After 6 d, tumors were harvested and processed (as previously described), and the function and phenotype of the intratumor NK cells was studied.
Functional Characterization of NK Cells.
NK cells were stimulated in vitro for 4 h with either PCI-13 (E:T = 1:1); a mixture of cytokines containing IL-12 (10 ng/mL), IL15 (10 ng/mL), and IL-18 (50 ng/mL); PMA (at 50 ng/mL) and ionomycin (500 ng/mL); or control condition. At 20 min into the incubation, protein transport inhibitors (BD; Cat# 555029 and #554724) and anti-CD107 antibody were added to the culture. After the stimulation, NK cells were harvested, surface stained, and fixed, and intracellular content on IFNγ was determined by intracellular staining.
In vivo antitumor potential of NK cells was determined by mixing PCI-13 with sorted NK cells (NK cells:PCI-13 cells ratio of 1:2.5), and the mixture was injected subcutaneously in the flanks of NSG mice; tumor growth was measured over time.
To compare the killing capacity of NK cells, xCELLigence Real-time Cellular Analysis (Agilent Technologies) was performed. On day 0, the xCELLigence microtiter plate was loaded with 10,000 target cells; on day 1, 10,000 sorted effector cells (at E:T ratio of 1:1) were loaded into wells in the presence of IL-15 (10 ng/mL). The cytolytic activity was measured based on the electric impedance (cell index) between the biosensors, and the obtained cell index values were normalized.
Cytokine Secretion.
Sorted NK cells were stimulated with PMA (50 ng/mL) and ionomycin (500 ng/mL) for 12 h, in 96-well round-bottom plates, at 50,000 NK cells per well in duplicate. Then, supernatants were harvested, frozen at −80 °C, and analyzed by 62-plex Luminex Immuno-Assay by the Human Immune Monitoring Center facility at Stanford University.
PCI-13 cells were cultured for 3 d, in 96-well flat-bottom plates, at 104 cells per well, in duplicate. Then, culture supernatants were harvested, frozen at −80 °C, and analyzed by 62-plex Luminex Immuno-Assay by the Human Immune Monitoring Center facility at Stanford University.
Antibody Staining for Phenotypic Analysis and Cell Sorting.
The list of antibodies used to sort and phenotype NK cells and ILCs is available in SI Appendix, Table 1. Starting from single-cell suspensions, either after dissociation, gradient centrifugation, or in vitro culture, cells were first washed with FACS buffer (PBS [Mediatech; Cat# 21–040-CV] containing 2% heat-inactivated fetal bovine serum [Omega Scientific; Cat# FB-21], 1 mM (ethylenedinitrilo)tetraacetic acid [Invitrogen; Cat# 15575-038], and 1% Pen Strep [Gibco; Cat# 15140-122]). Then, the cells were stained with Fixable Viability Dye eFluor 780 (eBioscience; Cat# 65-0865-18) for 20 min at 4 °C in the dark. Cells were then washed with FACS buffer, stained with the desired mixture of antibodies for surface markers for 30 min at 4 °C in the dark, and washed with FACS buffer. Then, the cells were resuspended in either FACS buffer for single-cell sorting for scRNA-seq or NK cell culture media for bulk sorting for in vivo and in vitro experiments, or were fixed and permeabilized using the Foxp3/Transcription Factor Staining Buffer Set (eBioscience; Cat# 00-5523-00) followed by intracellular staining according to manufacturer’s protocol for FACS analysis.
For the scRNA-seq study, live Lineage(CD3/CD19/CD20/CD14/CD34/CD68)− CD56+/− CD127+/− (SI Appendix, Fig. 1) lymphocytes were single-cell sorted into 96-well skirted PCR plates (Bio-Rad; Cat# HSP9631) containing 4 μL of lysis buffer per well, using a FACSAria Fusion instrument (BD). For some tumors, CD45 was included to help specify the lymphocyte population. Lysis buffer (55) consisted of 0.1 μL of oligo dT30VN at 100 μM (IDT), 0.4 μL of deoxynucleoside triphosphate mix at 25 mM (Invitrogen; Cat# 10297-018), 0.1 μL of “spike-ins” from the External RNA Controls Consortium at 1:600,000 dilution (Thermo Fisher Scientific; Cat# 4456739), 0.038 μL of 10% Triton X-100 (Thermo Fisher Scientific; Cat# 85111), 0.1 μL of RNase inhibitor (Clontech; Cat# 2313B), and 3.262 μL of nuclease free water (IDT; Cat# 11-04-02-01).
For the in vitro and in vivo differentiation study, live peripheral NK cells, defined as Lineage(CD3/CD19/CD20/CD14/CD34/CD68)−CD56+, were sorted (SI Appendix, Fig. 8), using either a FACSAria Fusion or a FACSAria II instrument (BD). For phenotypic analysis, sample acquisition was done using either an LSR Fortessa (BD) or a FACSAria Fusion instrument (BD). Flow cytometric analysis of acquired data was done using FlowJo 10.6.1 software (BD).
qRT-PCR.
Sorted cells were frozen on TriZol Reagent (Life Technologies; Cat# 15596018). RNA was extracted with the RNeasy mini kit (QIAGEN; Cat# 74104), and complementary DNA (cDNA) was made using the Maxima First Strand cDNA Synthesis kit (Thermo Fisher Scientific; Cat# K1642). Quantitative gene expression was performed using the Taqman Gene Expression Assay with the recommended best coverage primers for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), NR4A1, NR4A2, and NR4A3 (Thermo Fisher Scientific). Each gene expression assessment was measured in triplicate. Gene expression was determined relative to GAPDH messenger RNA (mRNA) expression using the formula 2(−(NR4A-GAPDH)). In some cases, NR4A gene expression was normalized to that of CD49a– NK cells (which was set to one), to allow easy comparison with the mRNA expression on other NK cell population.
Preparation of Single-Cell Libraries and Sequencing.
Sorted single cells were processed for scRNA-seq library preparation using the previously described Smart-seq2 protocol (55). Briefly, the RNA from each cell was reverse transcribed and PCR amplified (25 cycles). Amplified cDNA product was purified using 0.55× Agencourt AMPure XP Beads (Beckman Coulter; Cat# A63880) on Biomek FX Automated Workstation (Beckman Coulter). A 2-μL aliquot of purified cDNA sample from each well was analyzed for quality on a 5300 Fragment Analyzer with 96 capillary array (Agilent), using High Sensitivity NGS Fragment kit-DNF-474 (Agilent). Plates with a minimum DNA concentration of 0.2 ng/μL per well (average range 0.5 ng/μL to 1.5 ng/μL) were used for the final library preparation. First, the cDNA concentration was normalized with nuclease free water (IDT; Cat# 11-04-02-01) to a concentration less than or equal to 1 ng per well and transferred to a 384-well low volume serial dilution plate (TTP Labtech; Cat# 4150-05828) using the Mosquito ×1 (TTP Labtech). Then, library preparation steps as per Smart-seq2 protocol were performed using Nextera XT DNA Library Prep Kit (Illumina; FC-131-1096) in a 384-Well Rigi-Plate PCR Microplate (TTP Labtech; Cat# PCR-384-RGD-C) using Mosquito HTS (TTP Labtech). One-microliter volumes of indexed libraries from each well of the 384-well plate were pooled together and purified using 0.8× Agencourt AMPure XP Beads (Beckman Coulter; Cat# A63880). Concentration of the purified pooled library was determined using the Bioanalyzer (Agilent). The pooled library was run on the HiSeq 2500 high-output v4 sequencer (Illumina), with 2 × 125 bp read length. One 384-cell pooled library was run on one lane of the HiSeq 2500, with an average output of 1 × 106 reads per cell.
Alignment and Generation of Gene Matrices.
As ERCC RNA spike-in mixes were used during library preparation, the human hg19 genome was concatenated with ERCC92 sequences (https://www.thermofisher.com/order/catalog/product/4456739#/4456739) to create a combined reference genome. Similarly, annotations for the human hg19 genome were concatenated with annotations for the ERCC92 sequences. Raw reads were mapped to this combined reference genome by STAR aligner (v2.7.0f) (56). Mapping parameters were as per ENCODE RNA-Seq parameters (https://github.com/ENCODE-DCC/long-rna-seq-pipeline/blob/master/DAC/STAR_RSEM.sh), with the exception of “—outFilterMultimapNmax 1,” to avoid any multimapping. PCR duplicates were marked with Picard (http://broadinstitute.github.io/picard), using the MarkDuplicates function at its default settings. Gene expression features were then quantified using the featureCounts function from Subread (v1.6.4) (57).
Downstream analysis was performed in R (v3.4.4). The fraction of transcriptome-assigned reads assigned to mitochondria was determined by 13 mitochondrial genes as previously described (https://scrnaseq-course.cog.sanger.ac.uk/website/index.html) (58). Gene expression libraries were filtered using all of the following criteria: percent uniquely mapped > 40%, library size > 100,000 counts, ERCC fraction < 0.8, mitochondrial fraction < 0.1, and number of represented Ensembl features > 1,200 per library (SI Appendix, Fig. 9).
Library Size Normalization, Dimensional Reduction, and Visualization.
Further downstream processing was performed using the R package Seurat (59), starting from the gene expression matrix comprising raw counts. Specific parameters included the following: 1) LogNormalization was performed using a scale factor of 10,000. 2) Variable features were identified using the “vst” selection method. 3) The statistical significance of principal components was determined by the JackStraw function. 4) Both JackStraw and Elbow plots were used to determine the significant principal components for further evaluation.
An initial round of clustering was performed using the top 12 principal components at a resolution of 0.9. Visualization was performed using the Uniform Manifold Approximation and Projection (UMAP) dimensional reduction technique. Ten clusters were identified, including three clusters comprising CD3+/CD4+ T cells and CD86+ antigen-presenting cells (SI Appendix, Fig. 10). Cells from these three contaminating clusters were removed. The remaining cells, comprising NK cells and ILCs, were reanalyzed starting from the gene expression matrix comprising raw counts. Clustering was performed using the top 12 principal components at a resolution of 0.8, resulting in eight distinct UMAP clusters. Each cell was annotated by its tissue of origin, patient source, and CD56 and CD127 protein expression based on flow cytometry data.
Gene expression markers (differentially expressed genes) representative of each cluster were identified by the Seurat “FindAllMarkers” function using the Wilcoxon Rank Sum test and fulfilling the following thresholds: 1) minimum detection fraction of 0.25 and 2) log fold change of at least 0.25. The top 20 differentially expressed genes for each cluster were represented in a heat map. For fold change comparisons between two clusters, the “FindMarkers” function was used. For the log fold change plots, genes with a fold change of three or greater and an adjusted P value of < 0.05 were considered as significant.
Deconvolution of Gene Expression Profiles.
In order to assign phenotypic identities to each single-cell cluster, CIBERSORTx, a gene signature deconvolution approach, was used (35, 60).
RNA-seq gene expression profiles from Yudanin et al. (36) comprising raw counts of bulk-sorted human NK, ILC1, ILC2, and ILC3 cells from the ileum, jejunum, and spleen were downloaded from the Gene Expression Omnibus (GSE126107). Reads per kilobase million (RPKM) values were obtained using the “rpkm” function of the edgeR package (61).
The log2-normalized RPKM gene expression profiles of bulk-sorted peripheral human NK cells (CD56 bright, CD56dim/CD57pos, CD56dim/CD57neg), as well as ieILCs and ILC3s from human tonsil (GSE112813), were obtained from the supplemental information accompanying Collins et al. (37) The log2-normalized RPKM gene expression matrix was converted to RPKM for consistency.
A reference signature matrix was created for each of the two datasets using the CIBERSORTx web interface (https://cibersortx.stanford.edu), using the default parameters (kappa = 999, q value = 0.01, number of genes 300 to 500). The fractional composition of each single cell was imputed using this signature matrix, with “B-mode” batch correction enabled to account for the different gene expression profiling techniques, and quantile normalization disabled as recommended by the authors.
Pseudotime Analysis.
Pseudotime analysis was performed using the R package Monocle version 3.0 (43). The log-normalized gene expression matrix for the top 50 differentially expressed genes of each cluster (total of 400 genes) was used as input. Dimension reduction was performed using the reduceDimension function and the DDRtree reduction method with a maximum of two components. Pseudotime was determined by the orderCells function selecting the peripheral cluster as the root state.
The expression of marker genes along the ieILC1 and NK-2 trajectories was plotted using the plot_genes_branched_pseudotime function, with the curves representing Loess-smoothed expression of marker genes along each trajectory.
Correlation of IL-15 Gene Expression and ieILC1 Signature.
The expression values of IL15 were correlated with the values of an ieILC1 signature, across head and neck (HNSC) primary tumor samples from TCGA (n = 522). The transcripts-per-million (TPM) counts of IL15 and each of the top 20 ieILC1 marker genes used for generating the ieILC1 signature were log-transformed using formula , and scaled to mean zero and unit variance across samples. The values of the ieILC1 signature were obtained by averaging the log-transformed and scaled values of the top 20 differentially expressed genes in the ieILC1 cluster.
Statistical Analysis for In Vitro and In Vivo Experiments.
Results are expressed as mean ± SE. For statistical analysis, matched-pairs t test, two-tailed with a 95% confidence level, was performed using Prism software version 8.3.1 (GraphPad Software, LLC). P values are shown when statistically difference was reached.
Supplementary Material
Acknowledgments
Our studies were supported by the Stanford University Human Immune Monitoring Core for Luminex Immuno-Assays, the Lokey Stem Cell Research Building Flow Cytometry core facility for NK cell sorting and flow cytometric analysis, and the Stanford Cancer Institute Tissue Bank for procurement of tumor samples and blood. Cartoons were made with free stock illustrations provided by Servier Medical Art. This work was supported by funding from the NIH (Grants R01CA158516, R35DE030054, and U54CA209971) to J.B.S. and the National Medical Research Council, Singapore (Grants CIRG18nov-0045, Healthcare Research Scholarship: 0001/2014, to J.K.T.).
Footnotes
Competing interest statement: A patent application for differentiating innate lymphoid cells for immunotherapy was filed by Stanford University. J.B.S. is the scientific cofounder and member of the scientific advisory board of Indapta Therapeutics; however, the science presented here is not related to the focus of the company. U.Y.M.-N. is the founder of Conference Fund; however, the science presented here is not related to the focus of the company.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2101169118/-/DCSupplemental.
Data Availability
The mapped BAM files from the blood and tumor specimens of the HNSCC patients have been uploaded to the database of Genotypes and Phenotypes (dbGaP Study Accession: phs002002.v1.p1, “Landscape of Intratumoral NK Cell and ILC in Head and Neck Squamous Cell Carcinoma”, Study ID: 38031).
Code Availability
The source code, reference files and processing pipeline to reproduce our analyses can be accessed at https://github.com/josh-tay/HNSCC_NK_scRNA.
References
- 1.Bruno A., Ferlazzo G., Albini A., Noonan D. M., A think tank of TINK/TANKs: Tumor-infiltrating/tumor-associated natural killer cells in tumor progression and angiogenesis. J. Natl. Cancer Inst. 106, dju200 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rocca Y. S., et al., Altered phenotype in peripheral blood and tumor-associated NK cells from colorectal cancer patients. Innate Immun. 19, 76–85 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Villegas F. R., et al., Prognostic significance of tumor infiltrating natural killer cells subset CD57 in patients with squamous cell lung cancer. Lung Cancer 35, 23–28 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Mandal R., et al., The head and neck cancer immune landscape and its immunotherapeutic implications. JCI Insight 1, e89829 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gentles A. J., et al., The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 21, 938–945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diefenbach A., Gnafakis S., Shomrat O., Innate lymphoid cell-epithelial cell modules sustain intestinal homeostasis. Immunity 52, 452–463 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Bernink J. H., et al., Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat. Immunol. 14, 221–229 (2013). [DOI] [PubMed] [Google Scholar]
- 8.McKenzie A. N. J., Spits H., Eberl G., Innate lymphoid cells in inflammation and immunity. Immunity 41, 366–374 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Fuchs A., et al., Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells. Immunity 38, 769–781 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Serafini N., Vosshenrich C. A., Di Santo J. P., Transcriptional regulation of innate lymphoid cell fate. Nat. Rev. Immunol. 15, 415–428 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Serafini N., et al., Gata3 drives development of RORγt+ group 3 innate lymphoid cells. J. Exp. Med. 211, 199–208 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moro K., et al., Innate production of TH2 cytokines by adipose tissue-associated c-Kit+Sca-1+ lymphoid cells. Nature 463, 540–544 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Neill D. R., et al., Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature 464, 1367–1370 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price A. E., et al., Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proc. Natl. Acad. Sci. U.S.A. 107, 11489–11494 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Satoh-Takayama N., et al., Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity 29, 958–970 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Sanos S. L., et al., RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat. Immunol. 10, 83–91 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luci C., et al., Influence of the transcription factor RORgammat on the development of NKp46+ cell populations in gut and skin. Nat. Immunol. 10, 75–82 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Riggan L., Freud A. G., O’Sullivan T. E., True detective: Unraveling Group 1 innate lymphocyte heterogeneity. Trends Immunol. 40, 909–921 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melsen J. E., Lugthart G., Lankester A. C., Schilham M. W., Human circulating and tissue-resident CD56bright natural killer cell populations. Front. Immunol. 7, 262 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dogra P., et al., Tissue determinants of human NK cell development, function, and residence. Cell 180, 749–763.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simoni Y., Newell E. W., Dissecting human ILC heterogeneity: More than just three subsets. Immunology 153, 297–303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hudspeth K., et al., Human liver-resident CD56bright/CD16neg NK cells are retained within hepatic sinusoids via the engagement of CCR5 and CXCR6 pathways. J. Autoimmun. 66, 40–50 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernink J. H., et al., Interleukin-12 and -23 control plasticity of CD127+ Group 1 and Group 3 innate lymphoid cells in the intestinal lamina propria. Immunity 43, 146–160 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Raykova A., et al., Interleukins 12 and 15 induce cytotoxicity and early NK-cell differentiation in type 3 innate lymphoid cells. Blood Adv. 1, 2679–2691 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koh J., et al., IL23-producing human lung cancer cells promote tumor growth via conversion of Innate Lymphoid Cell 1 (ILC1) into ILC3. Clin. Cancer Res. 25, 4026–4037 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Cella M., et al., Subsets of ILC3-ILC1-like cells generate a diversity spectrum of innate lymphoid cells in human mucosal tissues. Nat. Immunol. 20, 980–991 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bal S. M., et al., IL-1β, IL-4 and IL-12 control the fate of group 2 innate lymphoid cells in human airway inflammation in the lungs. Nat. Immunol. 17, 636–645 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Ohne Y., et al., IL-1 is a critical regulator of group 2 innate lymphoid cell function and plasticity. Nat. Immunol. 17, 646–655 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Silver J. S., et al., Inflammatory triggers associated with exacerbations of COPD orchestrate plasticity of group 2 innate lymphoid cells in the lungs. Nat. Immunol. 17, 626–635 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cuff A. O., et al., The obese liver environment mediates conversion of NK cells to a less cytotoxic ILC1-like phenotype. Front. Immunol. 10, 2180 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dadi S., et al., Cancer immunosurveillance by tissue-resident innate lymphoid cells and innate-like T cells. Cell 164, 365–377 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao Y., et al., Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat. Immunol. 18, 1004–1015 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Cortez V. S., et al., SMAD4 impedes the conversion of NK cells into ILC1-like cells by curtailing non-canonical TGF-β signaling. Nat. Immunol. 18, 995–1003 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vacca P., et al., Human natural killer cells and other innate lymphoid cells in cancer: Friends or foes? Immunol. Lett. 201, 14–19 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Newman A. M., et al., Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 37, 773–782 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yudanin N. A., et al., Spatial and temporal mapping of human innate lymphoid cells reveals elements of tissue specificity. Immunity 50, 505–519.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collins P. L., et al., Gene regulatory programs conferring phenotypic identities to human NK cells. Cell 176, 348–360.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J., et al., NR4A transcription factors limit CAR T cell function in solid tumours. Nature 567, 530–534 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seo H., et al., TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc. Natl. Acad. Sci. U.S.A. 116, 12410–12415 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu X., et al., Genome-wide analysis identifies NR4A1 as a key mediator of T cell dysfunction. Nature 567, 525–529 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanna J., et al., Novel insights on human NK cells’ immunological modalities revealed by gene expression profiling. J. Immunol. 173, 6547–6563 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Scharping N. E., et al., The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T cell metabolic insufficiency and dysfunction. Immunity 45, 374–388 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trapnell C., et al., The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Michel T., et al., Human CD56bright NK cells: An update. J. Immunol. 196, 2923–2931 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Freud A. G., et al., NKp80 defines a critical step during human natural killer cell development. Cell Rep. 16, 379–391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hadley G. A., et al., Regulation of the epithelial cell-specific integrin, CD103, by human CD8+ cytolytic T lymphocytes. Transplantation 67, 1418–1425 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Cerdeira A. S., et al., Conversion of peripheral blood NK cells to a decidual NK-like phenotype by a cocktail of defined factors. J. Immunol. 190, 3939–3948 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hawke L. G., Mitchell B. Z., Ormiston M. L., TGF-β and IL-15 synergize through MAPK pathways to drive the conversion of human NK cells to an innate lymphoid cell 1-like phenotype. J. Immunol. 204, 3171–3181 (2020). [DOI] [PubMed] [Google Scholar]
- 49.de Andrade L. F., et al., Discovery of specialized NK cell populations infiltrating human melanoma metastases. JCI Insight 4, e133103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viel S., et al., TGF-β inhibits the activation and functions of NK cells by repressing the mTOR pathway. Sci. Signal. 9, ra19 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Sanjabi S., Oh S. A., Li M. O., Regulation of the immune response by TGF-β: From conception to autoimmunity and infection. Cold Spring Harb. Perspect. Biol. 9, a022236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simoni Y., et al., Human innate lymphoid cell subsets possess tissue-type based heterogeneity in phenotype and frequency. Immunity 48, 1060 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Hibino S., et al., Inhibition of Nr4a receptors enhances antitumor immunity by breaking Treg-mediated immune tolerance. Cancer Res. 78, 3027–3040 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Sekiya T., et al., The nuclear orphan receptor Nr4a2 induces Foxp3 and regulates differentiation of CD4+ T cells. Nat. Commun. 2, 269 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Picelli S., et al., Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Dobin A., et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liao Y., Smyth G. K., Shi W., The subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tung P. Y., et al., Batch effects and the effective design of single-cell gene expression studies. Sci. Rep. 7, 39921 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Newman A. M., et al., Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCarthy D. J., Chen Y., Smyth G. K., Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mapped BAM files from the blood and tumor specimens of the HNSCC patients have been uploaded to the database of Genotypes and Phenotypes (dbGaP Study Accession: phs002002.v1.p1, “Landscape of Intratumoral NK Cell and ILC in Head and Neck Squamous Cell Carcinoma”, Study ID: 38031).
The source code, reference files and processing pipeline to reproduce our analyses can be accessed at https://github.com/josh-tay/HNSCC_NK_scRNA.