Significance
Voltage-gated sodium channels initiate electric signals in cell communications. The S4–S5 linker between the voltage-sensing and pore modules transmits depolarization signals to trigger channel activation. The mechanisms of this action, however, remain elusive. By combining biophysical and computational approaches, we identify a critical residue, T140, in the S4–S5 linker of the bacterial sodium channel NaChBac, which plays a pivotal role in channel activation and drug modulation of slow inactivation. Specifically, we discovered conformation-dependent drug binding at this site and unveiled a toggling mode of action by T140, which switches interaction partners with different S6 residues to regulate channel activation and slow inactivation. These observations suggest the possibility of conformation-specific drugs targeting the gating machinery of voltage-gated ion channels.
Keywords: general anesthesia, propofol, NaChBac, NMR, saturation transfer difference
Abstract
Voltage-gated sodium (NaV) channels control excitable cell functions. While structural investigations have revealed conformation details of different functional states, the mechanisms of both activation and slow inactivation remain unclear. Here, we identify residue T140 in the S4–S5 linker of the bacterial voltage-gated sodium channel NaChBac as critical for channel activation and drug effects on inactivation. Mutations at T140 either attenuate activation or render the channel nonfunctional. Propofol, a clinical anesthetic known to inhibit NaChBac by promoting slow inactivation, binds to a pocket between the S4–S5 linker and S6 helix in a conformation-dependent manner. Using 19F-NMR to quantify site-specific binding by saturation transfer differences (STDs), we found strong STDs in inactivated, but not activated, NaChBac. Molecular dynamics simulations show a highly dynamic pocket in the activated conformation, limiting STD buildup. In contrast, drug binding to this pocket promotes and stabilizes the inactivated states. Our results provide direct experimental evidence showing distinctly different associations between the S4–S5 linker and S6 helix in activated and inactivated states. Specifically, an exchange occurs between interaction partners T140 and N234 of the same subunit in activation, and T140 and N225 of the domain-swapped subunit in slow inactivation. The drug action on slow inactivation of prokaryotic NaV channels seems to have a mechanism similar to the recently proposed “door-wedge” action of the isoleucine-phenylalanine-methionine (IFM) motif on the fast inactivation of eukaryotic NaV channels. Elucidating this gating mechanism points to a possible direction for conformation-dependent drug development.
Voltage-gated sodium (NaV) channels control the initiation and propagation of the action potentials in excitable cells, including most neurons, myocytes, and endocrine cells (1). They convert chemical to electrical signaling by responding to cell membrane depolarization and are responsible for maintaining the normal physiological functions of vital organs such as the heart and brain. Naturally occurring mutations in NaV channels are often associated with pathophysiological conditions (for a recent review, see ref. 2). Dysfunction of NaV channels is related to various diseases, including epilepsy (3), cardiac arrhythmia (4), myotonias (5), and chronic pain syndromes (6). Understanding how NaV channels function will help uncover their physiological and pathophysiological roles and at the same time facilitate the development of pharmaceuticals to target these channel proteins.
The functional cycle of NaV channels involves transitions from resting to activated states by opening a conducting passage for Na+ ions to flow into the cells upon membrane depolarization. During prolonged depolarizations, NaV channels undergo fast and slow conformational changes known as inactivation to close the activation gate, leading to functional states that cannot be reactivated or conduct ions (7, 8). Upon membrane repolarization, the channels transition back to the fully closed resting state in a process known as deactivation (9). In eukaryotic cells, the gating process is achieved by a pore-forming α-subunit, which is a single polypeptide chain folded into four homologous but nonidentical repeats in a pseudosymmetric arrangement. Each repeat is composed of four transmembrane helices (S1 to S4) as the voltage-sensing module (VSM) and two additional transmembrane helices (S5 and S6) as the pore module (PM). The PMs from the four repeats aggregate around a vestibule having a funnel-shaped ion selectivity filter (SF) near the extracellular entrance, a large water-filled cavity in the middle of the membrane, and an activation gate near the intracellular exit. Prokaryotic NaV channels, as exemplified by the ancestral bacterial voltage-gated sodium channel NaChBac from Bacillus halodurans (10), are composed of four smaller and often identical subunits, each contributing a VSM and PM along with the SF to a tetrameric channel (Fig. 1). In both prokaryotic and eukaryotic NaV channels, each of the VSMs and PMs is connected by a helical segment called the S4–S5 linker (7).
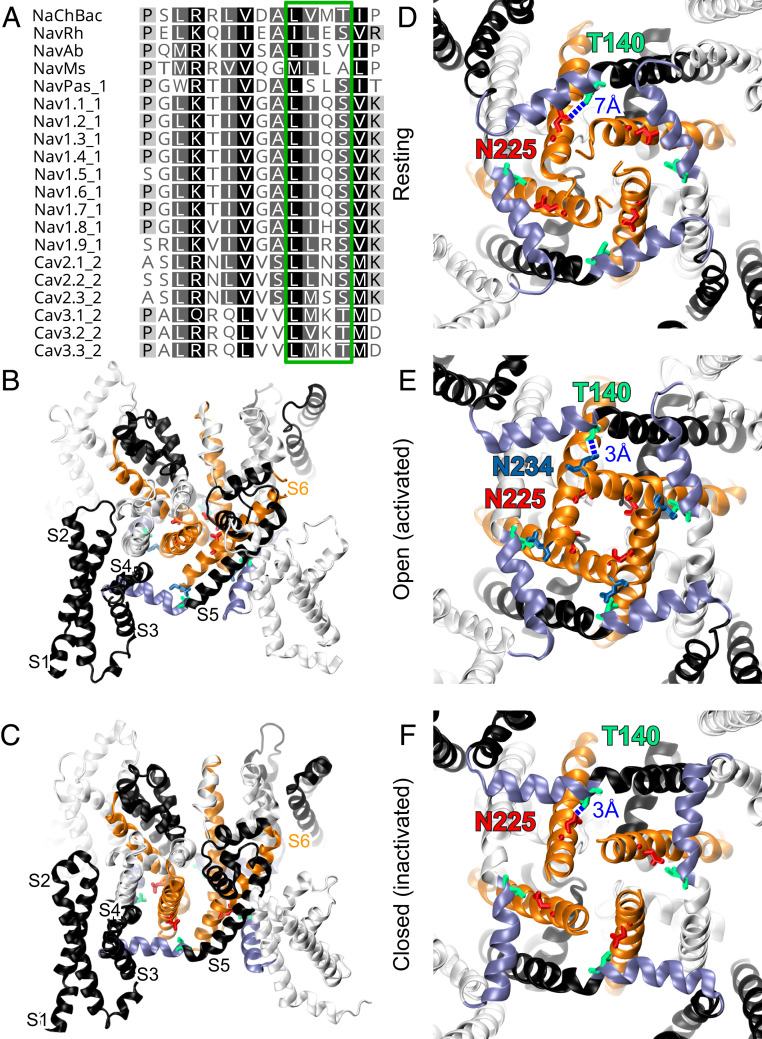
Fig. 1.
Structural models of NaChBac showing pivotal position of T140 in the S4–S5 linker relative to S6 and activation gate. (A) Sequence alignment of S4–S5 linker among NaV and CaV channels. T140 of NaChBac belongs to the LXXS/T motif (green box) at the C-terminal end of the linker, where possible interactions with the S6 helix occur. Overview of aligned (B) open (activated) and (C) closed (inactivated) structural models showing different orientations of S6 helices (orange) between S4–S5 linkers (purple) from the same subunits and the S6 helix of the adjacent subunits. Cytosolic views of NaChBac in (D) resting, (E) open, and (F) closed conformations showing the orientations of the S4–S5 linkers (purple) and S6 helices (orange) in the tetrameric channel. Critical residues T140 (green), N225 (red), and N234 (blue) and their approximate distances are highlighted in blue. Note the hydrogen bonding between the side chains of T140 and N225 in the inactivated state.
Significant advances have been made in recent years in the structural characterization of prokaryotic and eukaryotic NaV channels (11–21). The first successful crystal structure of a NaV channel at atomic resolution was that from the bacteria Arcobacter butzleri (NavAb) (11), which was captured with the VSM in the activated conformation and the PM in a nonconducting and presumably pre-open or inactivated conformation. Subsequently, high-resolution structures of NavAb (22) and of NavRh from Rickettsiales sp. HIMB114 (12), which is an ortholog of NaChBac, were solved in putatively inactivated states. Major breakthroughs in cryo-electron microscopy (cryo-EM) have led to several high-resolution NaV channel structures, including NavPaS from American cockroaches (Periplaneta americana) in a nonconducting conformation (13), as well as NaChBac (21), Nav1.2 (19), and Nav1.7 (20) structures in presumably inactivated states. Because no crystallization or cryo-EM experiments to date have been attempted at nonzero membrane potentials and the experimental time is often longer than the timescale of inactivation, capturing NaV channels in either the true resting or open-channel conformations is technically challenging. A truncated NavAb structure (23) and a full-length NavMs (from Magnetococcus marinus) structure (15) were reported to be in possible open conformations. Two cryo-EM structures of Nav1.4 from human and electric eel (17, 18) were captured with the activation gate held open by an amphiphile-like molecule, albeit the conformation states of these structures are yet to be determined. By using mutations that shift the activation voltage to positive values (hyper-depolarization shift) along with an engineered disulfide bond to lock the S4 helix of the VSMs in the “down” position, a cryo-EM structure of NavAb in a resting state was recently reported (16). These high-resolution experimental structures corresponding to different functional states provide an array of robust structural templates for homology modeling of other NaV channels.
Except for its lack of a fast inactivation state, NaChBac shares many gating features with eukaryotic channels. It is ideally suited for studying NaV channel function because of its easy availability in large quantity and high purity from heterologous expressions in bacteria and mammalian cells (24–29). It also has favorable gating kinetics for accurate electrorheology investigation into the transitions among different functional states (24). Since NaV channels control the axonal propagation of the electric signals and the presynaptic release of neurotransmitters, they are essential for cell–cell communications in peripheral and central nervous systems. It has been proposed recently (30) that sensory information flow in the nervous systems is governed by a nondeterministic process where thresholding of signal propagation, like that regulated by NaV currents, controls the emergence and loss of consciousness. Indeed, a growing body of evidence has pointed to the NaV channels as one of the important molecular targets for general anesthetics (8, 27, 28, 31–34). We showed previously that propofol, a clinically used intravenous general anesthetic, binds to three different sites in NaChBac (31). Among these sites, we hypothesized that a binding cleft in the VSM contributes to the acceleration of activation, the nonspecific binding within the central pore cavity facilitates inactivation, and the binding in a pocket between the S4–S5 linker and S6 has a dual role of promoting activation-coupled inactivation, leading to the net effect of channel inhibition (31).
In this study, we investigate the role of the S4–S5 linker and its relationship to the movement of the S6 helix in channel activation and drug effects on slow inactivation. We show that T140 in the S4–S5 linker of NaChBac is a pivotal residue in controlling NaChBac gating. By placing a fluorine probe at this site and using 19F-NMR to measure site-specific binding, we show that propofol binds to this pocket in a conformation-dependent manner. Computational simulations show that propofol fits into the amphiphilic pocket between T140 and the N225 from the domain-swapped subunit in the inactivation conformation, and propofol binding to T140 accelerates and stabilizes the inactivation transition. The experimental and computational results reveal a mechanism of T140 switching interaction partners from the activated state to the inactivated state. Drugs that can selectively stabilize one of the interacting partners have the potential to control the gating machinery of the NaV channels.
Results
T140 in the S4–S5 Linker Is Crucial for Activation.
The S4–S5 linker transmits the voltage-sensing S4 movement in the VSM to channel opening through conformational changes in the PM. This linker typically starts with a Pro residue to create an elbow at the end of the S4 helix (16) and extends into the adjacent subunit in a domain-swapping tetrameric arrangement. The helical repeats of the linker residues facing the pore axis have a high amino acid similarity in the NaV and CaV channels (Fig. 1A), particularly with a conserved amphipathic LXXS/T motif near the C-terminal end of the linker. To use the available structural information to gain insights into how the S4–S5 linker influences state transitions, we made homology models of NaChBac structures using templates from experimental NaV channel structures in putatively resting, open, and inactivated states. In these structural models, the difference in the spatial relationship between the S4–S5 linker and the S6 helices in the tetrameric arrangement revealed possible interactions between the LXXS/T motif and the S6 helix that are state dependent (Fig. 1 B–F): T140, the Thr residue in the LXXT motif of NaChBac, engages in fewer residue–residue interactions with S6 helices in the resting state, but likely forms side-chain hydrogen bonds with S6 in both the open activated (Fig. 1 B and E) and closed inactivated conformations Fig. 1 C and F. A highly conserved and functionally essential residue in the S6 helix, corresponding to N225 in NaChBac, is adjacent to T140 in the inactivated state (Fig. 1F). In contrast, the open structure model shows a π-bulge in the S6 helix after residue T220, resulting in the C-terminal half of the S6 helix swinging into the space between the S4–S5 linker and the domain-swapped S6 helix (Fig. 1B). With S6 intervening between the S4–S5 linker and domain-swapped S6, T140 becomes distanced from the domain-swapped N225 but adjacent to N234 from the same subunit (Fig. 1E).
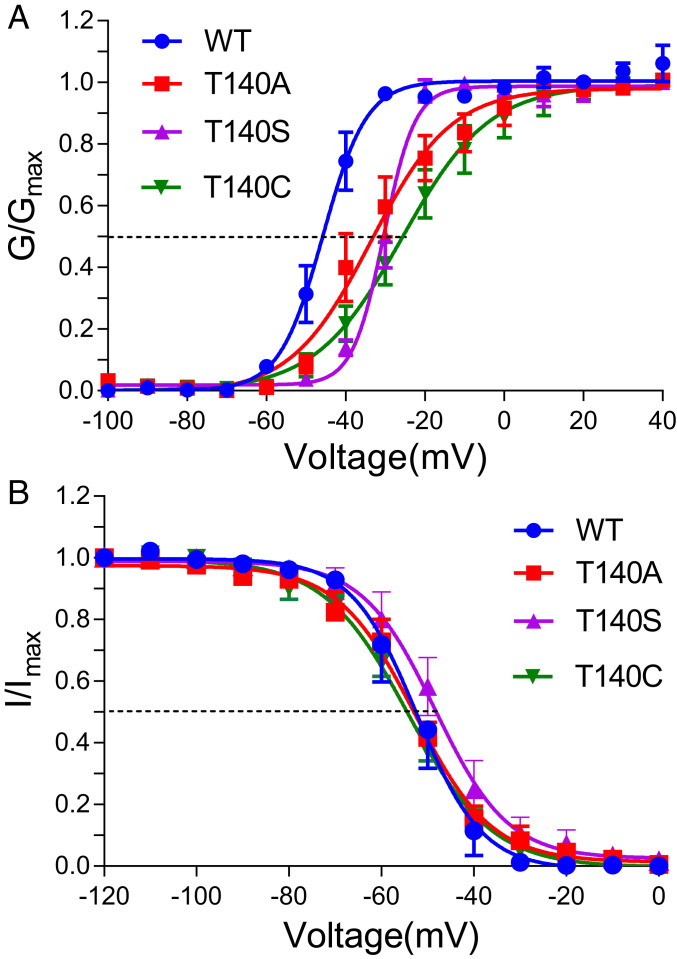
To validate these structure-based predictions, we first tried to establish the dependence of channel gating on possible interactions between the S4–S5 linker and the S6 helix. We mutated T140 to G, A, S, C, I, F, and Y to vary the side-chain volume and hydrophobicity. Among these mutants, ranging from the smallest possible side chain to bulky hydrophobic or hydrophilic side chains, T140G, T140I, T140F, and T140Y render the channel completely nonfunctional, with no measurable activation from –100 to +290 mV. The other three mutants, T140A, T140S, T140C, along with the wild-type (WT) channel, show current responses to varying voltages (SI Appendix, Fig. S1). Compared with the WT, the functional mutants show a sizable right shift in G–V activation curves (Fig. 2A and SI Appendix, Table S1), indicating a decreased opening propensity of the mutants. Moreover, T140A and T140C showed a more gradual activation voltage dependence, suggesting changes in the opening kinetics. While T140S shows a significant right shift in the G–V dependence, the slope is comparable to that of the WT. These results indicate that hydrogen bonding with the side-chain hydroxyl group in the WT T140 or mutant T140S in the LXXS/T motif occupying the pocket between the S4–S5 linker and the S6 helix is critical for the channel to transition from resting to activated conformation. More specifically, this hydrogen bonding appears to play an important role in maintaining the steep voltage dependence of channel activation.
Fig. 2.
Steady-state activation and slow inactivation in functional NaChBac T140 mutants. (A) The steady-state activation G–V curves of the T140S, T140A, and T140C mutants show right shifts relative to that of the WT. Note a significant decrease in the slopes of the T140A and T140C curves, indicating a gradual voltage dependence of activation in these mutants. (B) Steady-state inactivation I–V curves of the same T140 mutants compared with the WT. Only T140S shows a slight right shift in V1/2in, but this change is not statistically significant. The side-chain conformation of T140 is critical for the transition from the resting to activation state but not for the initiation of slow inactivation. Data are shown as mean ± SEM from n = 8–10 independent measurements. The solid lines are best fit to the data using SI Appendix, Eq. S2 or Eq. S3, with the best fitting parameters listed in SI Appendix, Table S1.
Slow Inactivation Is Not Altered by Mutations at T140.
Unlike activation, the three functional mutations at T140 have negligible effects on the steady-state slow inactivation when compared with the WT (Fig. 2B and SI Appendix, Table S1). There is no significant difference in V1/2in and the slope factor of the steady-state slow inactivation between the WT and the functional mutants. By comparing the structural models for the putatively open and inactivated states, especially the different orientations of N225 side chain (Fig. 1), it seems that slow inactivation involves the straightening of the S6 helix by eliminating the π-bulge after residue T220, resulting in a rotation of the S6 helix to move the N225 side chain from partially facing the pore to facing away from the pore. The absence of mutation effects at T140 on inactivation suggests that in the open state, interactions between T140 and their surrounding residues do not contribute substantially to the initiation of the slow inactivation process.
Mutations at T140 Affect Propofol Modulation of NaChBac Functions.
The different activation and inactivation sensitivities to mutations at T140 suggest that NaChBac gating depends on the state conformation within the pocket between the S4–S5 linker and the S6 helix. We previously showed that this pocket is also a part of a critical propofol binding site (31). To probe into the nature of propofol action at this site and to further elucidate how the interactions within the pocket alter channel gating, we investigated propofol modulation of activation and slow inactivation in the three functional T140 mutants and compared them with the WT channel. Fig. 3 A–D shows that propofol, at a clinically relevant concentration of 1 µM, promotes channel activation in WT and T140S, with a significant left shift of the G–V curves (ΔV1/2 = –9.3 ± 2.9 and –10.0 ± 2.3 mV, respectively; SI Appendix, Table S2). In contrast, the same concentration of propofol produces negligible or small changes in the activation of T140A (–0.74 ± 1.6 mV) and T140C (–2.4 ± 0.8 mV). Both Thr and Ser have side chain hydroxyl groups, which appears to be necessary in mediating propofol effects on activation. It should be noted, however, that having a side chain hydroxyl group is not a sufficient condition for the observed propofol effects: The T140Y mutation, with a bulky hydroxyl-containing hydrophilic side chain, renders the channel nonfunctional, irrespective of the presence or absence of propofol.
Fig. 3.
Propofol modulation of steady-state activation and slow inactivation in functional NaChBac T140 mutants. Steady-state activation G–V curves (A–D) and slow inactivation I–V curves (E–H) of the indicated WT NaChBac and functional T140 mutants in the presence (red) and absence (blue) of 1 µM propofol. V1/2a and V1/2in values and propofol-induced changes are given in SI Appendix, Tables S1 and S2, respectively. Compared with the WT NaChBac, T140 mutations minimized propofol effects, with the exception of the activation of T140S and inactivation of T140C. Data are shown as mean ± SEM from n > 6 independent measurements.
Propofol effects on steady-state inactivation show a different pattern: 1 µM propofol promotes steady-state inactivation in WT and T140C but has negligible effects on inactivation of T140A and T140S (Fig. 3 E–H and SI Appendix, Table S2). The removal of propofol effects on both activation and slow inactivation by the T140A mutation, which has a smaller side-chain volume than residues in the other three functional channels, suggest that 1) steric fitting of propofol in the amphipathic pocket, which would be compromised by this mutation, dominates the observed propofol actions on NaChBac; and 2) propofol binding at two other previously identified sites [i.e., a cleft in VSM and the central pore (31)], which should be minimally affected by the T140A mutation, plays only an auxiliary role in modulating NaChBac function. The contrasting propofol effects on T140S and T140C, whose side chains are sterically similar, suggest that the propofol’s modes of action on activation and inactivation are different: Propofol promotes the resting-to-open transition in T140S but not in T140C, whereas it accelerates the open-to-inactivation transition in T140C but not in T140S.
Propofol Binding to the S4–S5 Linker Is Conformation Dependent.
To understand the observed differences in the mutation- and propofol-induced changes in activation and slow inactivation, we further investigated the conformation-dependent propofol binding in the pocket between the S4–S5 linker and S6 helix by using an additional T220A mutation in both the WT and T140C channels. The T220A mutation has been shown to abolish slow inactivation in NaChBac (26), which we also confirmed for the double mutant T140C/T220A by electrophysiology (SI Appendix, Fig. S2). It is believed that replacing the pore-facing hydrophilic T220 with hydrophobic Ala residue in the S6 helix stabilizes the π-bulge at F221, prepositioning the S6 helix to assume an activated conformation. This is evidenced by the large left shifts of the activation V1/2 of T220A relative to the WT (–64.7 vs. –45.7 mV; SI Appendix, Table S1) and of T140C/T220A relative to T140C (–34.6 vs. –25.8 mV).
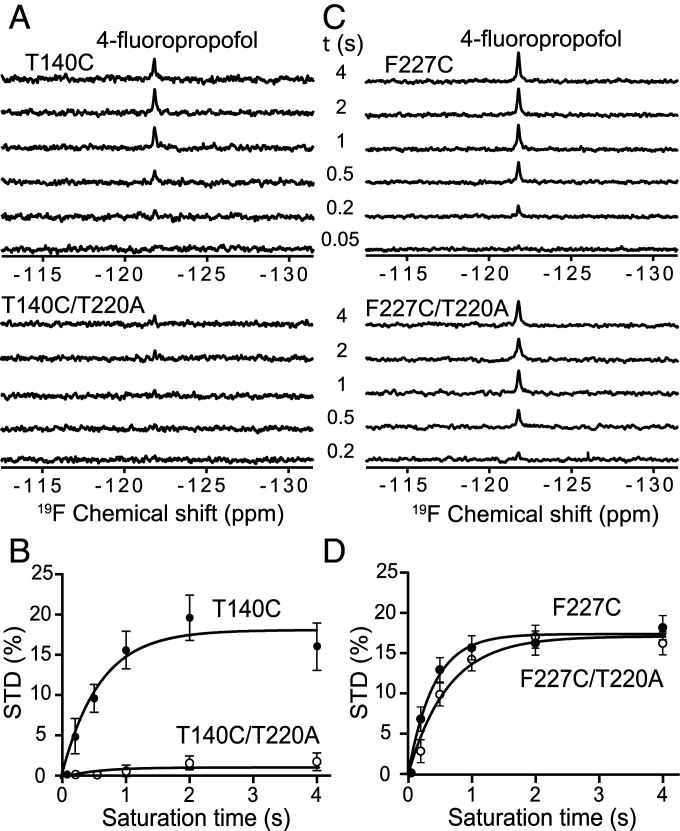
To measure the site-specific propofol binding, we covalently attached a small, fluorinated probe, 3-bromo-1,1,1-trifluoroacetone (BTFA), to the side chain of T140C in both the T140C and T140C/T220A mutants and measured the intermolecular 19F-NMR saturation transfer difference (STD) spectra of 4-fluoro-propofol (4fp), a fluorinated propofol. Intermolecular STD quantifies the cross-relaxation between two nuclei that are in a close and stable contact [<7 Å (35)], hence providing a direct measure of site-specific binding. We previously showed that 4fp and propofol have nearly identical anesthetizing potencies in tadpoles and have similar effects on the activation and slow inactivation of the WT NaChBac (31). In this study, we also confirmed that effects of 4fp and propofol on the T140C mutant were not statistically different (SI Appendix, Fig. S3), validating the use of 4fp as an excellent surrogate for propofol. Since the NMR experiments were conducted in the absence of a cross-membrane potential (i.e., depolarized), we expect that the T140C mutant was in an inactivated state under NMR experimental conditions, whereas the double mutant T140C/T220A, due to mutation-induced abolition of inactivation, was in a pre-open or an activated state. In our experiments, the 19F-labeling efficiency was 10–50%, which is sufficient for STD measurements as unlabeled protein contributes no NMR signal. The measured cross-relaxation by STD between 4fp and the BTFA label was distinctly different in T140C and T140C/T220A (Fig. 4A). A strong saturation transfer was observed from BTFA to 4fp in T140C, with an STDmax of 18.1% and a relatively large 19F intermolecular cross relaxation rate (σ) of ∼0.31 s–1 (Fig. 4B and SI Appendix, Table S3). In the T140C/T220A double mutant, which is devoid of slow inactivation, the saturation transfer between BTFA and 4fp was greatly diminished, with STDmax at merely ∼2% and the cross-relaxation rate unmeasurable. To confirm that this conformation dependence is specific to T140, we labeled the nonspecific propofol binding site in the central cavity at F227C and used the F227C/T220A double mutant as the corresponding control. The cross-relaxations between BTFA and 4fp in F227C and F227C/T220A are essentially identical (Figs. 4 C and D and SI Appendix, Table S3). We conclude that the specific propofol binding pocket between the S4–S5 linker and S6 helix near T140 is conformation dependent: Stable propofol binding, which ensures intermolecular STD buildup, occurs in only the inactivated state, whereas in a channel devoid of slow inactivation, the binding pocket is either removed due to conformation changes or the pocket is too dynamic to allow significant intermolecular STD buildup.
Fig. 4.
Conformation-dependent 4-fluoropropofol binding to T140. (A) Stack plots of 19F-NMR saturation transfer difference (STD) spectra of 4fp at the indicated saturation times from a 19F probe labeled at T140C in an inactivated state and in the T140C/T220A double mutant devoid of slow inactivation. (B) Plot of normalized STD from A as a function of saturation time. Sizable cross-relaxation between 4fp and T140 is quantifiable only in the inactivated state and negligible in the double-mutant channel that is unable to inactivate. (C and D) Corresponding control experiments with a 19F probe labeled at the nonspecific 4fp binding site near F227C and the inactivation-disabled double mutant F227C/T220A. In B and D, solid lines are best fit to the data using SI Appendix, Eq. S5. Error bars are derived from the signal-to-noise ratios of the STD spectra. Fitting parameters are listed in SI Appendix, Table S3.
Interactions within the Pocket between the S4–S5 Linker and the S6 Helix Regulate Channel Gating.
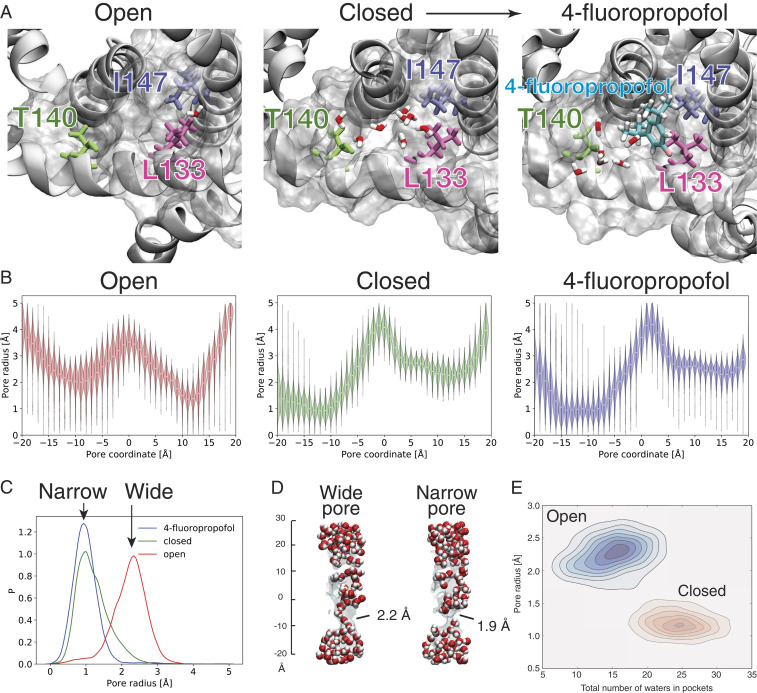
To further explore the molecular mechanisms underlying how the interactions within the S4–S5 linker pocket regulate channel gating, we conducted molecular dynamics (MD) simulations using the open (activated) and closed (inactivated) structure models of NaChBac; the closed structure was also simulated in the presence of 4fp. We defined the two ends of the amphiphilic pocket by a triad of residues L133 and T140 from the S4–S5 linker and I147 from the adjacent subunit (Fig. 5A). The hydrophobic moiety of the pocket is marked by L133 and I147 (SI Appendix, Fig. S4A), and the hydrophilic moiety is marked by T140 and, in the inactivated configuration, N225. We first tested the structural stability of 4fp in the pocket by measuring the minimum among all atom-to-atom distances between a single 4fp molecule and residues L133 and I147 for 160 ns of MD simulations in the inactivated conformation (SI Appendix, Fig. S4B). For the entire simulation period, 4fp remains steadily close (∼2.5 Å) to these residues.
Fig. 5.
Mechanistic association between the drug-binding pocket at T140 and channel activation state. (A) Representative snapshots from MD simulations of NaChBac channels in open (activated, Left), closed (inactivated, Middle), and 4fp-bound (Right) states. The amphipathic drug-binding pocket is surrounded by L133 and T140 in the S4–S5 linker and L147 from the adjacent subunit. The pocket is water-filled in the closed and 4fp-bound states but has a greatly reduced volume in the activated state. (B) The pore radius profiles, averaged from all MD simulation frames, along the channel axis in the open (Left), closed (Middle), and 4fp-bound (Right) structures. The activation gate is located from z = –15 Å to z = –6 Å. The lower bound corresponds to the pore entrance and is lined by the four symmetry-related residues I231. At lower values of z, the pore lining helix S6 is solvent exposed and, in the open structure, partially unfolded and highly fluctuating. (C) The radius distribution functions are calculated from each instantaneous channel configuration in the MD simulations and averaged from z = –15 Å to z = –6 Å for open (red), closed (green), and 4fp-bound (blue) structures. Note that 4fp binding locks the channel in a tightly closed configuration with a narrower radius distribution width than the apo activated and inactivated states. (D) Pore water profiles reveal that a wider pore (Left) allows passage of water, and the narrow pore (Right) does not. (E) Two-dimensional probability distribution of the number of water molecules in the T140 pocket and the pore radius at the activation gate. The darker colors correspond to a higher probability. The red and blue tones refer to the closed and open state simulations, respectively.
To establish a connection between 4fp binding and the open (activated) or closed (inactivated) state, we calculated the pore radius profile along the channel axis (Fig. 5B). In all cases, the first local radius minimum (near pore coordinate –10 Å) corresponds to the activation gate, the second to the SF (near pore coordinate +10 Å), and the local maximum to the internal pore cavity (near pore coordinate 0 Å). Importantly, the average pore radius at the activation gate is larger in the open conformation (Fig. 5 B, Left) compared with the closed (Fig. 5 B, Middle) and the 4fp-bound (Fig. 5 B, Right) conformations.
We then analyzed the radius distribution at the activation gate over the course of the MD simulations. For each molecular configuration, we defined the radius of the activation gate as the spatial average over the pore coordinates between –15 and –6 Å (Fig. 5B). As expected, the activated and inactivated configurations show a relatively large (∼2.5-Å) and small (∼1-Å) gate radius, respectively (Fig. 5C). Importantly, wider pores are hydrated and allow permeation of water molecules, while the narrow ones do not (Fig. 5D). Interestingly, the 4fp-bound conformation shows the same average value of the gate as the closed channel but with significantly smaller fluctuations (a narrower distribution width). Hence, the probability of observing an instantaneous configuration with a wide pore is greatly decreased in the 4fp-bound configurations. These results indicate that the binding of 4fp to the pocket between the S4–S5 linker and the S6 helix “locks” the channel in the closed (inactivated) conformation by reducing fluctuations of the activation gate.
We further set out to investigate the mechanistic link between drug binding and pore radius. We noticed that, in the apo configurations, the 4fp binding site is often occupied by water molecules (Fig. 5A). The number of occupying water molecules varies as the binding pocket undergoes structural fluctuations. Since the four binding pockets in the tetrameric NaChBac form a collar surrounding the pore near the activation gate, we hypothesized that fluctuations that increase the binding pocket volume would tighten the collar and decrease the pore radius at the activation gate, or vice versa. To test this, we measured the number of water molecules within 6 Å from the reference residues L133 and I147, which mark the distal end of the pocket from T140, for each frame in the simulations. Fig. 5E shows the two-dimensional probability distributions as a function of the pore radius and the number of water molecules in the pocket. In the open conformation (Fig. 5 E, Upper), a peak distribution of wider pore radii (∼2.5 Å) is associated with fewer numbers of water molecules (∼15) in the pocket, whereas in the closed conformation (Fig. 5E, Lower), the probability distribution is associated with a narrower gate radius of ∼ 1 Å and ∼25 water molecules in the pocket. We interpret this anticorrelation between the radius and the number of water molecules in the pocket as supporting evidence that as the channel fluctuates toward the closed (inactivated) conformation, the size of the pocket increases. Indeed, for the closed conformation (Fig. 5 E, Lower), only one population is present in the probability distribution, with a gate radius of ∼1 Å and ∼25 water molecules in the pocket.
To experimentally verify that modifying the size of the binding pocket can impact inactivation, we covalently conjugated 8-(chloromercuri)-2-dibenzofuransulfonic acid (CBFS) to the cysteine thiol group to partially occupy the hydrophilic end of the pocket at T140C (Fig. 5 A, Right). In the Cys-free WT control, in which no conjugation occurred, CBFS produced negligible effects on both activation and inactivation (SI Appendix, Fig. S5 A and B), and propofol again shifted the activation and inactivation curves in the hyperpolarization direction. In T140C, in which CBFS occupied the hydrophilic moiety of the pocket, the activation curve exhibits a slight left shift relative to the already right-shifted T140C activation, similar to the minimal propofol effects observed in the unconjugated T140C mutant. Like propofol binding, the inactivation curve also left-shifted significantly due to covalent CBFS conjugation. Interestingly, because the pocket geometry is modified by CBFS, a small modulation by propofol is now measurable in the activation of the CBFS-conjugated T140C mutant (SI Appendix, Fig. S5C). More importantly, the addition of propofol in the presence of CBFS can further stabilize the pocket, leading to an additional left shift of the steady-state inactivation (SI Appendix, Fig. S5D). The effects of propofol on V1/2a and V1/2in with and without CBFS conjugation are summarized in SI Appendix, Table S4.
Discussion
In this study, we demonstrated the dependence of NaChBac gating on residue T140 in the S4–S5 linker. Mutating this residue profoundly changed NaChBac’s steady-state activation, and several amino acids with side chains too large or too small compared with Thr, irrespective of hydrophobicity, rendered the channel nonfunctional. In mutations that retained channel function, the voltage dependence of activation was shifted to the right relative to the WT channel while slow inactivation was unchanged. By analyzing the cryo-EM structure of NaChBac in an inactivated state (21) and homology models of NaChBac in the resting and putatively open states, it is clear that T140 is situated at a pivotal location where its side chains in the tetrameric channel pinch an amphipathic collar surrounding the crossing of the S6 helices at the level of the activation gate. The collar consists of the inward-facing residues of the S4–S5 linker within four buried pockets of the tetramer, and these pockets create spaces between the S4–S5 linkers and the adjacent S6 helices for iris-like S6 movement upon gating (23). The outward movement of S4 helices in response to membrane depolarization has been proposed to cause conformational change in the S4–S5 linker to loosen the collar (16), allowing S6 movement. The MD simulations in this study show that the pockets are partially water-filled, and the number of trapped water molecules is inversely correlated with the averaged radius of the pore at the activation gate (Fig. 5E). While our simulations do not sample full transitions between the open and closed pore conformations and we could not directly observe an association between opening/closing of the pore gate and wetting/dewetting of the pockets, it is conceivable that tightening or loosening of the collar is coupled to the compression of the pockets. The hydrophilic side chain of T140, with its hydrogen-bonding propensity for the trapped water molecules in the amphipathic pockets, plays a critical role both in facilitating the loosening of the collar to allow the iris-like movement of S6 into the spaces and in establishing specific interactions with S6 in a conformation-dependent manner. Specifically, the T140 side chain toggles between two possible interaction partners in the S6 helices upon gating: In the activated state, T140 of each S4–S5 linker interacts with N234 from the S6 helix of the same subunit, whereas in the inactivated states, T140 releases N234 from the same subunit and establishes a hydrogen bond with N225 from the domain-swapped S6 helix. The contrasting mutation effects on activation and slow inactivation underscore the conformational distinctions of the pockets in different functional states.
Unlike the mutations at T140, a complementary mutation N225D in the S6 helix was found not to affect channel activation but profoundly shift the steady-state inactivation to the left (29). This, combined with the results in the current study, supports the notion that the S4–S5 linker initiates the conformation transition from the resting to the activation state by creating spaces for S6 kinking movement, whereas slow inactivation involves subsequent S6 helix rotation to orient N225 to face the S4–S5 linker to stabilize activation-coupled inactivation.
The amphipathic pockets at T140 are also sites for propofol binding (31). Examination of snapshots from the MD simulation trajectories in this study suggests that propofol binding drives the trapped water molecules to the hydrophilic moiety of the pocket (Fig. 5 A, Right), which is at the C-terminal end of the LXXS/T motif. Amino acids Thr and Ser, with side-chain hydroxyl groups, allow favorable interaction with the water molecules, promoting conformational dynamics of the region. This may explain the propofol-induced left shifts of the steady-state activation in the WT and the T140S mutant. In contrast, the absence of the side-chain hydroxyl group in the T140A and T140C mutations likely limits the interactions with the trapped water and possibly also reduces propofol binding, hence resulting in smaller propofol effects on activation (SI Appendix, Table S2). It should be noted that propofol promotion of activation and the absence of 4fp NMR STD in the activated state are not in contradiction with each other. Propofol promotion of activation requires propofol binding to the pocket in the resting state (not necessarily in the activated state) to lower the energy barrier for activation. Since acceleration occurs with residues having side-chain hydroxyl group, it is likely that in the resting state, propofol increases the dynamics of the pocket along with the trapped water to allow easy establishment of hydrogen bonding between T140 or S140 with N234 to facilitate channel activation, resulting in a conformation as captured in the open-state structure (Fig. 1E). The importance of hydroxyl group and hydrogen bonding propensity in the molecular target recognition for specific anesthetic–protein interactions has been examined at the atomic resolution in several propofol-bound proteins (36).
Propofol binding also serves as a molecular probe to examine the conformational transition of S6 helices relative to S4–S5 linkers between activated and inactivated states. Although this transition is implicated in various structural models under different crystallization and cryo-EM conditions, our NMR STD experiments in a physiologically relevant environment (particularly with respect to temperature) provided direct experimental evidence showing highly specific interactions between bound propofol and T140C in the inactivated state and the removal of such interactions upon channel activation. Notably, the absence of STD buildup in the activated state can be interpreted as propofol being pushed out of the binding pockets by S6 moving into the pockets or the binding sites becoming too dynamic in the activated state to sustain magnetization transfer. Both scenarios are possible based on the structural models and MD simulations, and the current STD experiments cannot distinguish these two possibilities. In either case, however, propofol binding in the pockets between the S4–S5 linkers and S6 helices is less frequent and less stable in the open, activated channel.
Conformation-dependent propofol binding is also indirectly reflected in the different propofol effects in different mutants. Propofol promotes channel activation in T140S but not in T140C, and it accelerates activation-coupled inactivation in T140C but not in T140S (Fig. 3). The side chains of Ser and Cys are sterically similar except for the difference in size and the hydrogen-bonding propensity between the side-chain hydroxyl and thiol groups. While both mutations render the S4–S5 linker less effective in activating the channel than the WT Thr, as reflected in the sizable right shifts in activation curves (Fig. 2A), the contrasting propofol effects in these two mutants suggest the possibility that propofol binding depends on the side-chain hydrogen bonding propensity and on the conformation difference between the resting and inactivated states at this location. Both hydrogen-bonding propensity and strength are expected to be higher with T140S than with T140C. It is also well documented that the torsion angle χ1 distribution of Thr is strongly skewed to the g+ conformation (300°) due to the steric hinderance of the side-chain methyl group in helical and buried regions (37), which is the case for the inward-facing surface of the S4–S5 linker. The side-chain torsion angle is considerably more flexible in Ser but highly restricted in Cys. These changes in the opposite directions in the two mutants seem to suggest a delicate balance between conformation dynamics in the pockets and the steric fitting of the drug molecule to stabilize certain specific interactions, including hydrogen bonding, for the functional effects (16). Conformation-dependent propofol binding and differential propofol effects on channel potentiation and inhibition have been recently demonstrated in a bacterial pentameric ligand-gated ion channel (38).
The electrophysiology measurements and MD simulations in this study also suggest that propofol occupation in the T140 pockets between the S4–S5 linkers and S6 helices promotes transition to inactivated states. A high degree of intermolecular STD buildup between 4fp and a fluorine probe fixed on T140C in the inactivated state but not in the activated state is a strong indication that propofol binding stabilizes the inactivated conformation, allowing intermolecular cross-relaxation (Fig. 4). It is generally believed that prokaryotic NaV channels do not display fast inactivation because of the lack of the isoleucine-phenylalanine-methionine (IFM) motif, which was originally thought to act as a “plug” to quickly block the ion passage upon channel activation. However, the new high-resolution Nav1.4 structures (17, 18) revealed an alternative “door-wedge” mechanism for fast inactivation in eukaryotic NaV channels, in which the IFM motif acts as a “wedge” to insert allosterically into the space between the S4–S5III linker and S6IV helix, exactly at the corner between L1153 and N1591, corresponding to the space between T140 and N225 in NaChBac. In NaV1.4 structures, the IFM wedge forces S6IV to move into other S6 helices to close the gate (18). The same mechanism seems to play a role in the propofol action to inhibit NaChBac by accelerating channel inactivation. The insertion of propofol expands the water-filled collar to tighten the activation gate (Fig. 5 B and C). This shared mechanism between drug action on slow inactivation of NaChBac and IFM wedging in fast inactivation of eukaryotic NaV channels points to a direction of strategies for drug discovery, namely by directly targeting the gating machinery of the voltage-gated sodium channels for state-dependent modulations.
The door-wedge mechanism of drug effects on slow inactivation is also indirectly supported by the different effects between large side-chain mutations and postexpression modifications via chemical labeling or conjugation. Although BTFA is a small addition to the T140C side chain, CBFS addition is rather bulky. Mutations to amino acids with bulky side chains render the channel nonfunctional or unable to be activated, yet conjugating a large chemical group like CBFS did not completely abolish the channel function, as shown in SI Appendix, Fig. S5 C and D. One possible explanation of this difference is that when all four side chains at T140 in a tetramer are mutated to a large amino acid, the channel either cannot fold correctly or the collar becomes so tight that there is no room for activation. In postexpression chemical modifications, however, it is likely that only some of the subunits in the tetramer are chemically modified, mimicking the situation in the eukaryotic channel where only one subunit being wedged is sufficient to cause fast inactivation. This explanation is consistent with the 10–50% labeling efficiency as determined in our experiments.
In conclusion, residue T140 near the C-terminal end of the S4–S5 linkers in the ancestral voltage-gated sodium channel NaChBac regulates how the VSM communicates with the PM to activate the Na+ channel upon membrane depolarization. The intravenous general anesthetic propofol binds to a collar of amphipathic pockets at this residue in a state-dependent manner. Stable and site-specific interactions between propofol and T140 can be observed and quantified in the inactivated state but not in the activated state. Acceleration and stabilization of NaChBac slow inactivation by propofol share the same site as the recently proposed door-wedge mechanism observed in the fast inactivation of eukaryotic NaV channels. Although fast inactivation is absent in bacterial NaV channels, the “modulability” of the space between the S4–S5 linkers and S6 helices seems to be evolutionarily conserved, allowing for rational designs of modulators as conformation-specific drugs to regulate the function of NaV channels.
Materials and Methods
Experimental details are given in SI Appendix. All chemicals and biological reagents were obtained from suppliers with accreditation to meet ISO/IEC 17025, ISO Guide 34, or ISO 17034 quality standard. NaChBac was transiently expressed in HEK-293 cells, and whole-cell patch-clamping experiments were performed using an IonFlux Mercury HT automated electrophysiology machine. Electrophysiology data and statistics were processed and analyzed using Clampfit and GraphPad. 19F-NMR experiments were performed using a Bruker Biospin Avance 600 spectrometer, equipped with a 19F cryoprobe. 19F-NMR STD spectra were acquired by collecting alternating on- and off-resonance 19F spectra with saturation irradiations at −83.8 and −45.00 ppm, respectively. Intermolecular STD buildup from protein to ligand was determined using time-dependent saturation of 0.05, 0.2, 0.5, 1.0, 2.0, and 4.0 s in a randomized order. Structure models of NaChBac in different functional states were developed using experimental structures of prokaryotic and eukaryotic NaV channels as the homology-model templates. These models were individually embedded in a fully hydrated 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine lipid bilayer in either the presence or absence of 4fp. 4fp was included exclusively in the simulation of the closed conformation as the pockets in the open state were not wide enough to fit a 4fp molecule, and thus the docking algorithm did not return any binding pose. MD simulations were carried out using NAMD 2.12 with the CHARMM36 all-atom potential energy functions for protein and phospholipids, and the TIP3P potential for water molecules. The structural parameters and partial atomic charges for 4fp were modeled using the CHARMM general force field (CGenFF) web server. Molecular systems were equilibrated for ∼2 ns with decreasing harmonic restraints on the protein atoms, the pore ions, and the water molecules localized in the SF. MD simulations were run for 160 ns with each system using 2-fs time steps under constant pressure (1 atm) and temperature (300 K) conditions.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the NIH (P01GM055876 and R01GM049202) and by the Department of Anesthesiology and Perioperative Medicine at the University of Pittsburgh. J.X. acknowledges the Shanghai Jiaotong University and Ruijin Hospital for supporting her in studying abroad as part of the international exchange program.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102285118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Yu F. H., Catterall W. A., Overview of the voltage-gated sodium channel family. Genome Biol. 4, 207 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang W., Liu M., Yan S. F., Yan N., Structure-based assessment of disease-related mutations in human voltage-gated sodium channels. Protein Cell 8, 401–438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richards K. L., et al., Selective NaV1.1 activation rescues Dravet syndrome mice from seizures and premature death. Proc. Natl. Acad. Sci. U.S.A. 115, E8077–E8085 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pei Z., Xiao Y., Meng J., Hudmon A., Cummins T. R., Cardiac sodium channel palmitoylation regulates channel availability and myocyte excitability with implications for arrhythmia generation. Nat. Commun. 7, 12035 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fusco C., et al., New phenotype and neonatal onset of sodium channel myotonia in a child with a novel mutation of SCN4A gene. Brain Dev. 37, 891–893 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Meents J. E., et al., The role of Nav1.7 in human nociceptors: Insights from human induced pluripotent stem cell-derived sensory neurons of erythromelalgia patients. Pain 160, 1327–1341 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Catterall W. A., Forty years of sodium channels: Structure, function, pharmacology, and epilepsy. Neurochem. Res. 42, 2495–2504 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang E., Granata D., Eckenhoff R. G., Carnevale V., Covarrubias M., Propofol inhibits prokaryotic voltage-gated Na+ channels by promoting activation-coupled inactivation. J. Gen. Physiol. 150, 1299–1316 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ulbricht W., Sodium channel inactivation: Molecular determinants and modulation. Physiol. Rev. 85, 1271–1301 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Ren D., et al., A prokaryotic voltage-gated sodium channel. Science 294, 2372–2375 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Payandeh J., Scheuer T., Zheng N., Catterall W. A., The crystal structure of a voltage-gated sodium channel. Nature 475, 353–358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X., et al., Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature 486, 130–134 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen H., et al., Structure of a eukaryotic voltage-gated sodium channel at near-atomic resolution. Science 355, eaal4326 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Shen H., et al., Structural basis for the modulation of voltage-gated sodium channels by animal toxins. Science 362, eaau2596 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Sula A., et al., The complete structure of an activated open sodium channel. Nat. Commun. 8, 14205 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wisedchaisri G., et al., Resting-state structure and gating mechanism of a voltage-gated sodium channel. Cell 178, 993–1003.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan X., et al., Structure of the human voltage-gated sodium channel Nav1.4 in complex with β1. Science 362, eaau2486 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Yan Z., et al., Structure of the Nav1.4-β1 complex from electric eel. Cell 170, 470–482.e11 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Pan X., et al., Molecular basis for pore blockade of human Na+ channel Nav1.2 by the μ-conotoxin KIIIA. Science 363, 1309–1313 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Shen H., Liu D., Wu K., Lei J., Yan N., Structures of human Nav1.7 channel in complex with auxiliary subunits and animal toxins. Science 363, 1303–1308 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Gao S., et al., Employing NaChBac for cryo-EM analysis of toxin action on voltage-gated Na+ channels in nanodisc. Proc. Natl. Acad. Sci. U.S.A. 117, 14187–14193 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Payandeh J., Gamal El-Din T. M., Scheuer T., Zheng N., Catterall W. A., Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature 486, 135–139 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lenaeus M. J., et al., Structures of closed and open states of a voltage-gated sodium channel. Proc. Natl. Acad. Sci. U.S.A. 114, E3051–E3060 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ouyang W., Jih T. Y., Zhang T. T., Correa A. M., Hemmings H. C. Jr, Isoflurane inhibits NaChBac, a prokaryotic voltage-gated sodium channel. J. Pharmacol. Exp. Ther. 322, 1076–1083 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Lee S., Goodchild S. J., Ahern C. A., Local anesthetic inhibition of a bacterial sodium channel. J. Gen. Physiol. 139, 507–516 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S., Goodchild S. J., Ahern C. A., Molecular and functional determinants of local anesthetic inhibition of NaChBac. Channels (Austin) 6, 403–406 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barber A. F., Carnevale V., Klein M. L., Eckenhoff R. G., Covarrubias M., Modulation of a voltage-gated Na+ channel by sevoflurane involves multiple sites and distinct mechanisms. Proc. Natl. Acad. Sci. U.S.A. 111, 6726–6731 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinde M. N., et al., Fluorine-19 NMR and computational quantification of isoflurane binding to the voltage-gated sodium channel NaChBac. Proc. Natl. Acad. Sci. U.S.A. 113, 13762–13767 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Reilly A. O., et al., Mutagenesis of the NaChBac sodium channel discloses a functional role for a conserved S6 asparagine. Eur. Biophys. J. 46, 665–674 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou D. W., Mowrey D. D., Tang P., Xu Y., Percolation model of sensory transmission and loss of consciousness under general anesthesia. Phys. Rev. Lett. 115, 108103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y., et al., Propofol inhibits the voltage-gated sodium channel NaChBac at multiple sites. J. Gen. Physiol. 150, 1317–1331 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Covarrubias M., Barber A. F., Carnevale V., Treptow W., Eckenhoff R. G., Mechanistic insights into the modulation of voltage-gated ion channels by inhalational anesthetics. Biophys. J. 109, 2003–2011 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sand R. M., Gingrich K. J., Macharadze T., Herold K. F., Hemmings H. C. Jr, Isoflurane modulates activation and inactivation gating of the prokaryotic Na+ channel NaChBac. J. Gen. Physiol. 149, 623–638 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rehberg B., Duch D. S., Suppression of central nervous system sodium channels by propofol. Anesthesiology 91, 512–520 (1999). [DOI] [PubMed] [Google Scholar]
- 35.Jayalakshmi V., Rama Krishna N., CORCEMA refinement of the bound ligand conformation within the protein binding pocket in reversibly forming weak complexes using STD-NMR intensities. J. Magn. Reson. 168, 36–45 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Qiu L., et al., The role of the hydroxyl group in propofol-protein target recognition: Insights from ONIOM studies. J. Phys. Chem. B 121, 5883–5896 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Gray T. M., Matthews B. W., Intrahelical hydrogen bonding of serine, threonine and cysteine residues within alpha-helices and its relevance to membrane-bound proteins. J. Mol. Biol. 175, 75–81 (1984). [DOI] [PubMed] [Google Scholar]
- 38.Heusser S. A., et al., Allosteric potentiation of a ligand-gated ion channel is mediated by access to a deep membrane-facing cavity. Proc. Natl. Acad. Sci. U.S.A. 115, 10672–10677 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.