Abstract
Phase separation is emerging as a paradigm that explains the self-assembly and organization of membrane-less bodies in the cell. Recent advances show that this principle also extends to nucleoprotein complexes, including DNA-based structures. We discuss here recent observations on the role of phase separation in genome organization across the evolutionary spectrum from bacteria to mammals. These findings suggest that molecular interactions amongst many DNA-binding proteins evolved to form a variety of biomolecular condensates with distinct material properties that contribute to genome organization and function. We suggest that phase separation contributes to genome organization across evolution and that the resulting phase behavior of genomes may underlie regulatory mechanisms and disease.
Keywords: Biomolecular condensates, phase separation, genome organization, evolution, transcription
Phase separation organizes proteins and nucleic acids
The interior of the cell is highly spatially organized. The cell relies on lipid membranes and cytoskeletal fibers to arrange and stabilize its contents. However, the cell also contains many membrane-less organelles which lack these organizational features, yet persist as cohesive, dynamic structures. Many of these bodies assemble and maintain their organization via the process of phase separation, which is reminiscent of the immiscible behavior between oil and water [1, 2] (Figure 1A). Such cellular bodies appear as droplets within the cell, each representing a distinct phase, and collectively, are referred to as biomolecular condensates [3]. Well-studied examples of phase separated sub-cellular structures include P-granules [4] and stress granules [5] in the cytoplasm, and the nucleolus [6], histone locus bodies [7], and nuclear speckles [8] in the nucleus [9]. While these examples are comprised of protein and RNA, similar phase behavior also occurs with protein and DNA complexes.
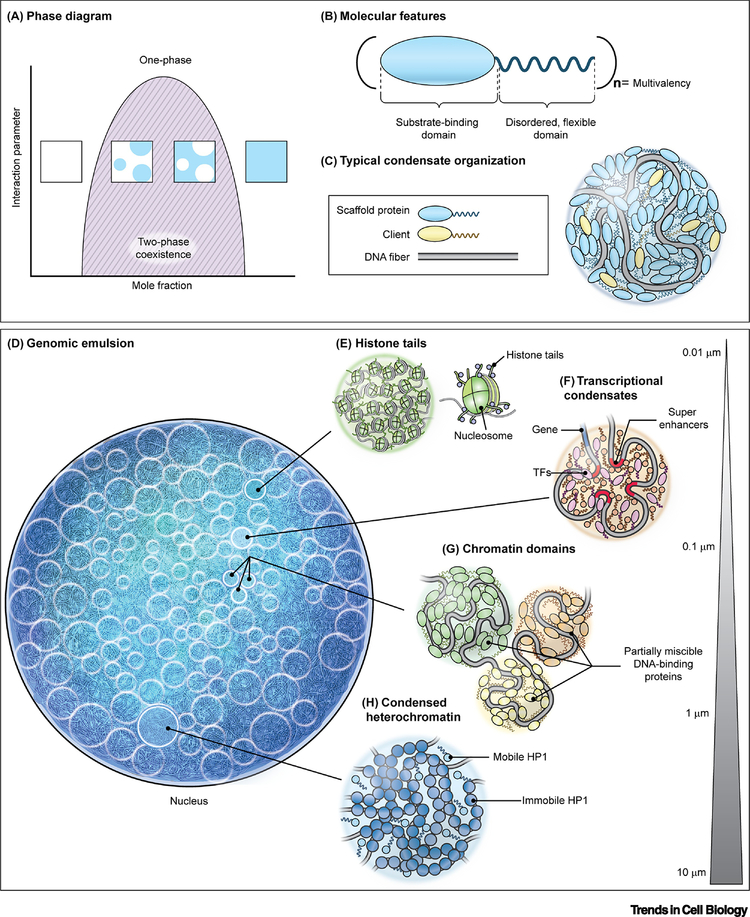
Figure 1. Molecular driving forces of phase separation of the mammalian nuclear genome.
(A) Phase diagram for a simple, two-component system where an interaction parameter, such as temperature, salt concentration, pH, etc., is plotted as a function of concentration of one of the components. The dark solid line represents the boundary between a single, homogenous phase and the two-phase coexistence region. Insets show representative examples of the solution in different regions of the phase diagram. (B) Schematic of a typical phase separating protein. Many phase separating proteins contain substrate-binding (i.e. DNA-binding, protein-interaction) domains and unstructured, disordered domains that can both be arranged in (n) multiplicities, giving rise to multivalent interactions. (C) Representative arrangement of DNA-binding phase separating proteins to form condensates in association with DNA. (D) The nucleus as an emulsion of DNA condensates. (E-H) Levels of phase separation in the nucleus include interactions between histone tails as part of nucleosomal arrays (E), transcriptional condensates involving the interaction of enhancer elements and transcription factors (F), partial miscibility or immiscibility between neighboring chromatin domains (G), and large gene-silenced regions compacted in heterochromatin (H). Bar represents typical length scale of interactions.
Biomolecular phase separation generally occurs via multiple, but weak, interactions between proteins and/or (ribo)nucleic acids to favor de-mixing from a single, homogenous phase into multiple phases (Figure 1A). Typically, proteins that drive phase separation are highly modular in that they contain multiple structured, interacting domains that include self-interacting domains, such as dimerization domains [10], nucleic acid binding domains [11], and/or protein interaction domains [12, 13] (Figure 1B). Additionally, phase separating proteins are typically interspersed with unstructured sequences, including intrinsically disordered regions (IDRs), which add flexibility to the protein chain (Figure 1B) [13]. Together, these domains allow for networks of homo- and hetero-typic interactions among multiple biomolecular species, and in this way, form condensates [14]. Moreover, the long DNA fiber also contributes to multivalency, particularly through its repetitive DNA sequences [15, 16]. Commonly, one or a few biomolecular species are the main drivers of phase separation of a condensate, thereby acting as scaffolds, which can further recruit other biomolecules, or clients, to partition into the droplets, but which cannot condense on their own (Figure 1C) [17]. The summation of these weak interactions among biopolymers promotes phase separation, yet, importantly, allowing for dynamic rearrangements that frequently confer dynamic, liquid-like behavior [3].
Typically, de-mixing molecules in solution undergo liquid-liquid phase separation (LLPS) [1, 18]. However, with increasingly complex systems, as in cells, phase behavior can deviate from classic LLPS [19] and can exhibit more nuanced properties such as multi-phase behavior [6], liquid-solid phase transitions [20] and gel-like states [21], which we collectively refer to as phase separation.
Here, we survey the emerging role of phase separation among proteins and DNA, and how phase separation may contribute to the organization of genetic material in diverse organisms, including bacterial, mitochondrial and nuclear genomes.
The mammalian genome as an emulsion of condensates
Genomes are organized and function via their interactions with DNA-binding proteins and associating factors and also via chromatin-chromatin interactions [22]. Phase separation can influence the higher-order structure of the nuclear genome across several hierarchical levels of organization (Figure 1D–H).
Histone tails and nucleosomal arrays
The DNA double helix locally wraps around histone octamers to form nucleosomes, which in turn assemble into a chromatin fiber (Figure 1E) [22]. Histones contain disordered tails that protrude from the nucleosome core and are post-translationally modified to mark epigenetic states. Akin to the IDRs of biomolecular condensates, histone tails are sufficient to drive phase separation as seen with the C-terminal tail of histone H1 (CH1), which forms a complex coacervate with DNA in vitro [23], and with larger assemblies of small nucleosome arrays containing linker histone H1, whose tails are required for droplet formation (Figure 1E) [24]. Moreover, post-translational modifications (PTMs) of the tails significantly influence their phase behavior: CH1 phosphorylation reduces propensity for phase separation with DNA [23] and acetylation leads to droplet dissolution [24]. Furthermore, bromodomain-containing proteins bind to acetylated nucleosomes, leading to de-mixing of acetylated droplets from unmodified nucleosomal droplets [24]. PTMs may not only serve as mediators of epigenetic control, but may also allow multiple nucleosomal condensates to stably coexist in the nucleus (Figure 1D) [25].
Transcriptional condensates
Transcription requires the coordinated local action of a large number of proteins, many with DNA-binding properties (Figure 1F). Nuclear transcription sites represent many defining features of condensates, including dynamic recovery after photobleaching, fusion events, and reversibility (Figure 1F) [26]. Many proteins associated with the transcription machinery, including RNA Polymerase II (Pol II) itself, contain multiple modular domains as well as IDRs that can phase separate into liquid-like droplets in vitro and in vivo [11, 27, 28]. However, the biomolecular features of their IDRs vary; for example, TAF15 is enriched in charged residues [29], whereas the carboxy-terminal domain (CTD) of Pol II contains heptapeptide repeats that can be phosphorylated [30]. CTD hyper-phosphorylation influences the phase behavior to coincide with the transition between transcription initiation and splicing [31]. Moreover, the Pol II CTD lengths and sequence motifs are selectively conserved along evolutionary lineages [32], and engineered CTDs that contain varying copies of the consensus heptapeptide differ in levels of partitioning and dynamics in live cells [33].
Gene activity is often controlled by upstream regulatory regions referred to as enhancers or super-enhancers, if present in multiple copies [34]. An emerging model has been proposed in which these sequences may exert their function by acting as nucleation sites for the formation of large, phase-separated transcriptional condensates, which contain high concentrations of transcriptional machinery components and transcriptional regulators (Figure 1F) [11, 34]. Consistently, transcriptional co-activators, such as BRD4 and MED1, assemble into dynamic puncta at sites of super-enhancers that are enriched in Pol II in live cells [26, 28]. The unique physicochemical environment provided by the condensate may enhance transcription [26, 28, 29]. Such properties have been proposed to contribute to the characteristic bursting behavior of many genes by which they undergo rapid cycles of high activity and inactivity [35]. In fact, the concentrated production of RNA transcripts associated with these transcriptional condensates may serve a regulatory role in their stability, since high RNA levels lead to droplet dissolution in vitro and in vivo [36].
Chromatin domains
The interactions of histone tails and other chromatin proteins have been suggested to facilitate the formation and maintenance of larger chromatin domains, such as topologically associating domains (TADs), by promoting short-range polymer-polymer interactions (Figure 1G) [22, 37]. Indeed, super-resolution microscopy reveals that nucleosomes pack to form domains of ~200–300 nm and move as units, primarily stabilized by cohesin and nucleosomal interactions [38]. These domains tend to contain cohesin and active histone modifications that envelope a core enriched in repressive modifications [39]. Consistently, purified cohesin and DNA complexes phase separate into droplet-like structures in vitro [40]. However, in vivo nucleosomal domains can persist after cohesin depletion, suggesting the additional contribution of cohesin-independent mechanisms to maintain their condensed structure [39]. These sub-compartments or chromatin domains could effectively represent distinct phases within the nucleus [41], and the immiscibility between neighboring chromatin domains could explain how they stably maintain their identity and composition (Figure 1G).
Higher-order chromatin structure
Genomic regions are typically classified as either euchromatin which represents regions that are actively transcribed or heterochromatin containing regions that are permanently repressed. One key protein known to coat and organize DNA in heterochromatin is heterochromatin protein 1α (HP1α). HP1α phase separates in vitro and forms condensates in live cells [42, 43]. HP1α has two structural domains, which allow for a variety of interactions that collectively contribute to its phase behavior. The chromoshadow domain undergoes homodimerization and is separated by a hinge region from the chromodomain that interacts specifically with methylated histone 3, lysin 9 (H3K9me); the two domains are flanked by disordered N- and C-tails. Modulated by phosphorylation, HP1 engages with the H3K9me2 and 3 writer SUV39H1 and with the TRIM28 scaffolding protein in live cells and forms multi-component droplets in vitro that closely resemble properties of heterochromatin [12, 42]. Indeed, the collective condensation of HP1α around DNA provides mechanical protection, preventing DNA from rearranging in response to tensile force [44]. Another example of heterochromatin phase separation includes the Polycomb protein chromobox 2 (CBX2), which forms liquid-like droplets via its IDR [45]. Together, these results suggest phase separation driven by multiple, weak interactions, including between histone tails and heterochromatin-associated proteins, underlie heterochromatin formation and cohesion [12].
Phase separation may also contribute to the segregation of heterochromatin and euchromatin on a larger scale due to preferential homotypic interactions between eu- and heterochromatin phases in the nucleus [22] (Figure 1H). The pattern of segregation is also be affected by the interaction of eu- and heterochromatin with spatial constraints such as the nucleolus and the nuclear periphery as suggested by the observation that nuclear lamina components determine the location of heterochromatin relative to the nuclear periphery [22, 46].
Other nucleoprotein condensates
Other nucleoprotein complexes likely also arise from phase separation. During the cell cycle, genomes undergo significant condensation into highly compact mitotic chromosomes followed by dissolution into a relatively more diffuse state of chromatin in the intact nucleus. Many of the reversible changes to condensation/dissolution state of many membrane-less organelles during mitosis are mediated by the kinase DYRK3, which serves as a dissolvase that solubilizes the mitotic cytoplasm [47]. During mitosis, the proliferation marker protein Ki-67 acts as a polymeric surfactant that envelopes the condensed mitotic chromosomes [48]. Specifically, Ki-67 forms a brush-like coating along the chromosome that stabilizes its structure both sterically and electrostatically [48] and reduces partitioning of cytoplasmic components [49]. Indeed, other features of mitotic chromosomes further maintain this condensed state, such as the centromere, which contains a borealin subunit as part of the chromosomal passenger complex (CPC) that phase separates in vitro [50]. Inactivation of the second X chromosome in female cells occurs via the non-coding Xist RNA and associated proteins that assemble into droplets, reminiscent of the behavior of heterochromatin [51, 52]. Furthermore, assemblies of DNA repair factors spontaneously assemble upon DNA damage into DNA-damage-response foci [53]. Replication compartments may also represent distinct phases as the replication machinery, including Origin Recognition Complex components, Cdc6 and Cdt1, contains evolutionarily conserved IDRs and forms liquid-like foci on DNA origins [54].
Limits of phase separation mechanisms in genome organization
Important to note is that phase separation of chromatin into liquid-like bodies may not be a universal nor the exclusive physical mechanism of genome organization [55, 56]. For example, mouse heterochromatin does not exhibit canonical features of phase separation in vivo, as the heterochromatin compaction state does not appear to be influenced by HP1 levels as would be expected in a phase separation model [57]. Moreover, nucleosomal arrays can be experimentally varied to exhibit a range of material properties [55]. They can condense into solid-like structures under MgCl2-buffer conditions in vitro, and fluidity can be restored for example by reducing agents [55]. Similarly, limited recovery after photobleaching of fluorescently labelled chromatin in live cells suggests little rearrangement of the DNA fiber itself, while associated proteins are able to exchange [55]. These dynamics point to solid-like features of chromatin on longer-length scales [55], but the dynamics may also be limited by the long-diffusion times associated with the considerable length of the DNA fiber compared to that of proteins. These collective interactions propagate to give rise to chromatin scaffold that contributes mechanical integrity to the nucleus, yet can still support local assembly of condensates [55]. In addition, other competing mechanisms are likely at play, giving rise to the multiple time and length scales involved in eu-and heterochromatin organization [56].
Organization of the mitochondrial genome by phase separation
Mammalian cells contain a second, much smaller, genome: the mitochondrial genome (mt-genome). The mt-genome consists of a 16 kb circular DNA (mtDNA) and exists as hundreds of copies per cell [58]. Unlike the assembly of nuclear DNA via histones, mtDNA is organized by a set of mitochondrial proteins, forming dynamic, membrane-less nucleoprotein complexes. These mt-nucleoids are normally maintained to be ∼100 nm in size (Figure 2A), but can assemble and coarsen into larger structures [58].
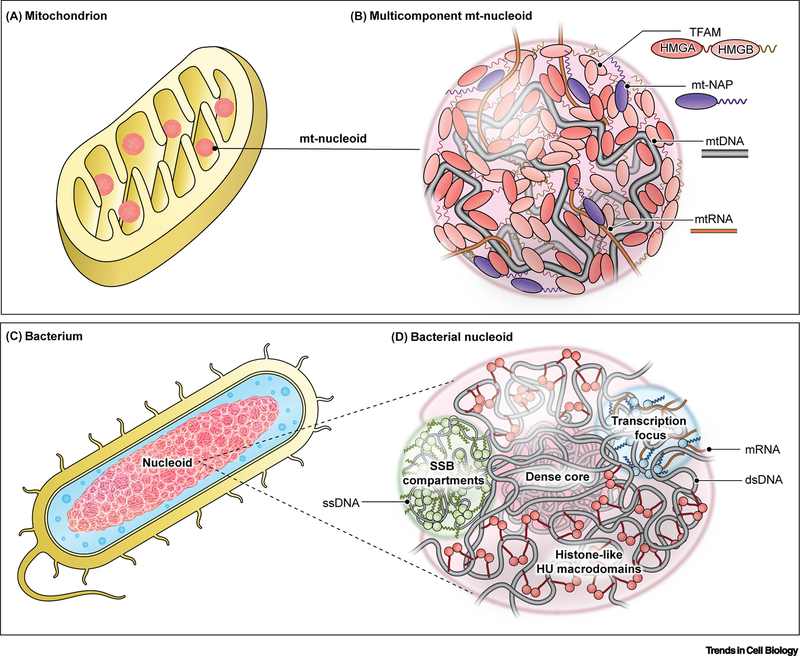
Figure 2. Conservation of phase separation in organization of nucleoids.
(A,B) Phase separation interactions drive the organization of the mitochondrial genome in a mitochondrion (A) into nucleoprotein complexes called mitochondrial nucleoids (B). (B) Mitochondrial Transcription Factor A (TFAM): primary nucleoid architectural protein containing two HMG domains and two IDRs (linker and C-tail); mt-nucleoid associated proteins (mt-NAPs): core proteins associated with mtDNA involved in transcription (TFB2M, POLRMT, mTERF) and replication (mtSSB, POLG1/2, TOP1MT), many of which have DNA-binding domains and intrinsically disordered regions; mtDNA: mitochondrial genome consisting of ∼16 kB circular DNA existing as ∼1–2 copies per nucleoid; mtRNA: mitochondrial RNA transcribed directly from the mitochondrial genome inside the condensate but later associates into separate RNP condensates. (C,D) The larger bacterial genome is analogously organized as a phase-separated nucleoid (D) in a bacterium (C). HU: one of the major architectural, histone-like proteins that exists as a dimer with flexible β-sheet arms that package DNA into a dense core surrounded by less dense phase of DNA and associated proteins; Transcriptional foci: dynamic condensates comprised of RNA polymerase (RNAP) and other transcription factors, many of which contain IDRs; SSB compartments: single-stranded DNA binding protein (SSB) forms a tetramer around single-stranded DNA with protruding intrinsically disordered linkers that are capped by conserved C-terminal peptide motif.
The major mt-nucleoid architectural protein is the mitochondrial transcription factor A (TFAM), which binds and bends DNA via its two high mobility group (HMG) domains that are separated by a disordered linker and flanked by a disordered C-tail (Figure 2B) [59], rendering TFAM highly flexible [60]. These molecular features allow TFAM to spontaneously self-assemble into viscoelastic droplets in vitro [61]. mtDNA partitions into TFAM droplets with heterogenous localization consistent with the organization seen in live cells [61]. The heterogeneity may be due to intrinsic differences in the two HMG domains, which are not physicochemically identical as they bind DNA with varying affinity [62] and arrange DNA in inverse localizations within the droplet [61]. Interestingly, HMG domains are abundant and highly conserved in many nuclear proteins, suggesting that similar phase behavior and organization may occur in nuclear genomes [63].
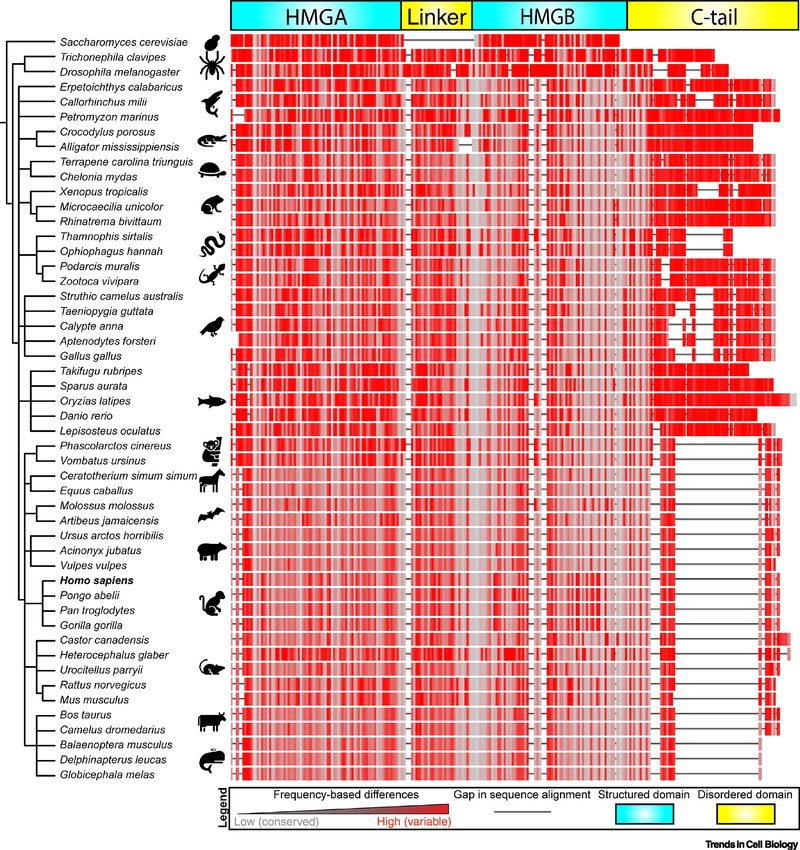
Phase separation may not be unique to human mt-nucleoids. Yeast S. cerevisiae package their mt-genome with Abf2, a TFAM homologue that lacks a prominent disordered C-tail (Figure 3) [64, 65]. mtDNA of other organisms diverge from that of a single, small plasmid, and one notable example is from unicellular flagellates that contain thousands of copies of catenated circular DNA (kinetoplasts, ∼10 μm in length) [66]. Interestingly, kinetoplast proteins have highly basic tails, which are reminiscent of those in nuclear histone proteins, allowing for a conserved role of phase separation in kinetoplast organization [67]. Furthermore, evolutionary analysis of the mt-nucleoid protein TFAM indicates that the structured domains (HMGA, HMGB) tend be more conserved than disordered regions (linker, C-tail), in line with reduced evolutionary pressure of disordered regions to maintain a specific sequence (Figure 3). Moreover, observed systematic changes to disordered regions, and not to structured domains, along the phylogenetic tree, such as the significant truncation of the C-tail in mammals, raise the intriguing possibility that changes to TFAM evolved to adjust the phase behavior of mt-nucleoids to maximize evolutionary fitness of organisms under various environmental conditions.
Figure 3. Phylogenetic tree comparing the evolution of TFAM’s modular domains.
FASTA sequences were obtained for fifty representative organisms for TFAM and/or the homologue Abf2 (S. cerevisiae). Organisms were grouped using NCBI Taxonomy Common Tree algorithm, and the generated tree was visualized using the EMBL Interactive Tree of Life (iTOL) tool. Sequences were aligned using the NCBI Cobalt algorithm using default settings, and color coding was assigned based on frequency-based differences, where red indicates highly variable regions with high frequency of mutations and grey indicates highly conserved regions with low frequency of mutations. Sequence gaps are indicated by solid black lines.
Phase behavior of genomes in the context of evolution
Mitochondria are thought to have arisen via endosymbiosis of prokaryotic progenitors, suggesting mitochondrial and bacterial genomes are evolutionarily related. The involvement of phase separation in the organization of not only nuclear and mitochondrial genomes in mammals, but also of genomes from other phylogenetic kingdoms, thus, raises the possibility that phase separation is an evolutionarily conserved mechanism of genome organization.
The bacterial genome
Prokaryotes lack membrane-bound organelles that are common in eukaryotes. Bacterial genomes consist of a single, circular plasmid that is organized by abundant, diverse histone-like proteins, but unlike in mammalian nuclear genomes, bacterial genomes do not assemble into orderly-spaced nucleosomes [68]. Instead, the bacterial genome forms a nucleoid, analogous to the mitochondrial nucleoid (Figure 2C) [69], and bacterial nucleoids frequently undergo large conformational changes in response to growth and environmental conditions [70]. Early observations [71] and more recent work suggest that phase separation may play a role in organization of bacterial nucleoids [72–76] (Figure 2).
Bacterial nucleoids are organized by nucleoid associated proteins (NAPs). In stationary phase, E. coli nucleoids are enriched in the DNA-binding protein from starved cells (Dps), which promotes the compaction of the nucleoid, while loss of Dps leads to a more expanded nucleoid that permeates the bacterial volume in a manner consistent with condensate formation [73]. One of the most prominent histone-like proteins is HU, which binds non-specifically to DNA to widely cover the bacterial chromosome (Figure 2D) [68]. HU has two subunits, which associate as homo- or heterodimers, providing multivalency and containing flexible β-sheet arms that interact with DNA [77]. The nucleoid is most diffuse during lag phase, but transitions into a highly compact state consisting of a dense core surrounded by flexible chromatin during the exponential phase, and is followed by an intermediary state during stationary phase [77]. While the effects of Dps and HU on bacterial genomes are consistent with a role of phase separation in nucleoid organization, neither Dps nor HU have been fully demonstrated to phase separate. In addition, NAPs tend to be more structured than their eukaryotic counterparts [78], suggesting other molecular interactions besides disordered regions may determine their behavior.
Several bacterial proteins involved in active processes such as transcription and replication demonstrate signatures of phase behavior [72, 79, 80]. Although the bacterial RNAP lacks a disordered CTD characteristic of eukaryotic Pol II [81], RNAP localizes to discrete foci, which are typically found along the periphery of the nucleoid and exhibit phase behavior [72, 79]. Other bacterial transcription factors, including NusA and Fis, undergo phase separation in vitro, and several additional proteins contain disordered domains, such as the highly expressed H-NS and DksA [72]. Phase separation may also be relevant for bacterial genome replication, since the tetrameric single-stranded DNA-binding protein (SSB) recruits single stranded DNA (ssDNA) and other SSB-interacting partners, which assemble into dynamic condensates in vitro (Figure 2F) [80].
Although these studies point to a general role of phase separation in some aspects of mitochondrial and bacterial nucleoid organization, their respective NAPs are not conserved [82], suggesting that the ability of DNA-binding proteins to phase separate has evolved multiple times.
Potential roles of phase separation in other genomes
Many other forms of genome organization exist across phylogenetic kingdoms that may involve phase separation. The plant nuclear genome is dramatically larger (∼100-fold) than the mammalian genome. As similar organizational principles are shared between the two, including chromatin loops, TADs, chromosome territories, and heterochromatin [83], phase separation has been implicated in plant genomes. Indeed, many plant DNA binding proteins are modular and are enriched in disordered domains [84], and the plant protein ADCP1 (triple tandem Agenet protein) reads H3K9me, associates into gene-silenced phases in cells, and forms droplets with nucleosomal arrays in vitro similarly to HP1 [85]. These observations point to phase separation playing a putative role in organizing plant genomes and to convergent evolution of this property [83, 86].
Phase separation in modulation of genome organization and function
Several models have been proposed for how RNA-condensates can affect biological function, such as sequestration of mRNA in stress granules or outward flux of processed rRNA in the multiphase nucleolus [87]. However, more nuanced implications for genome organization and function emerge for DNA-condensates, whose physics tend to be governed by the long, polymeric nature of DNA (Figure 4).
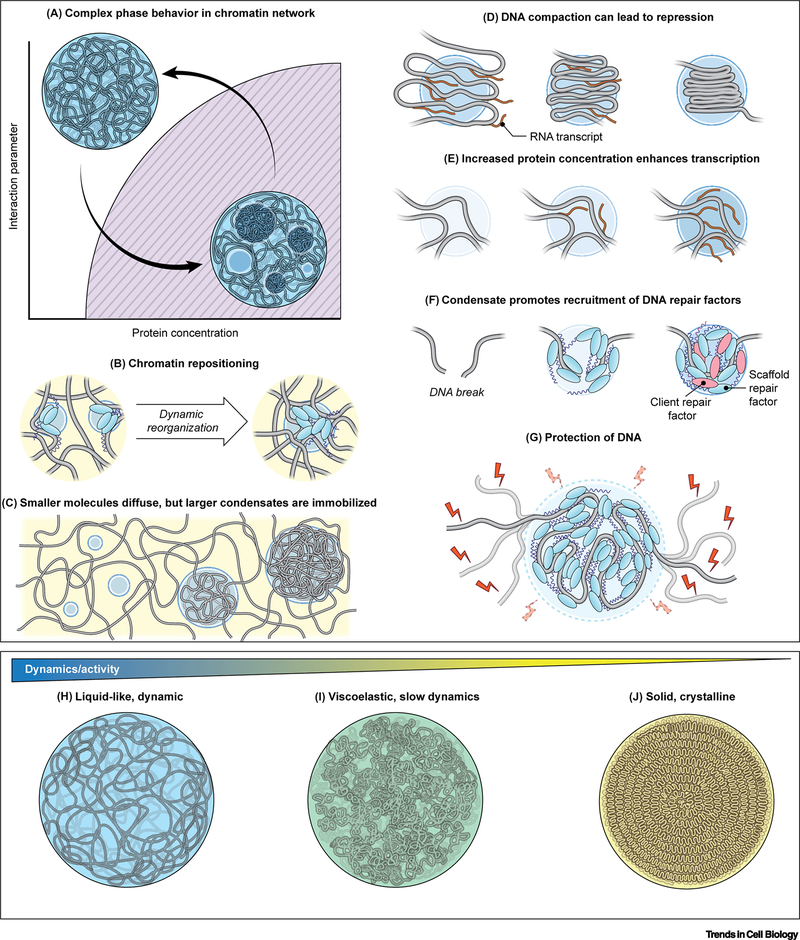
Figure 4. Regulation of phase separation in genome organization and function.
(A) Phase diagram describing phase separation of a DNA condensate within the cell nucleus. There is an interplay between organization of the chromatin and droplet coarsening. (B) Droplets nucleate around specific genomic loci, which are brought together as droplets grow and coarsen, while other genomic loci are excluded from the droplet. (C) Individual proteins and small condensates can diffuse and rearrange in the nucleus, while larger condensates are immobilized within the chromatin network. (D) Droplets concentrate DNA which may lead to transcriptional repression. (E) Droplets concentrate proteins, such as transcription factors, which can alter reaction kinetics, leading to changes in gene activity. (F) In DNA repair, droplets nucleate scaffold proteins such as 53BP1 that later recruit client proteins which partition into the droplet to augments DNA repair. (G) The viscous physicochemical environment within the droplet stabilizes stabilize the chromatin fiber from thermal fluctuations and excludes immiscible molecules from entering, such as potentially damaging reactive oxygen species (ROS). (H-J) DNA condensates exhibit a spectrum of material properties: highly dynamic, liquid-like behavior (H), intermediary, viscoelastic behavior (I), and solid-like, crystalline behavior (J). These material properties correlate with the level of biological activity, such as transcriptional bursting, controlled processing, or genome preservation, respectively.
Phase separation in chromatin positioning
Condensate formation in the nucleus can be a reversible process, involving cycles of assembly and disassembly in response to changes in the cellular state (Figure 4A). Typically, DNA condensates nucleate around genomic loci and coarsen with time (Figure 4A) [88], which may also lead to rearrangements of the chromatin fiber (Figure 4A). In support, upon optogenetically-induced condensation, protein-bound segments of DNA come together to form a condensate (Figure 4B) [88], and chromatin rearrangements can persist after polycomb complex PRC1 condensate dissolution, suggesting the chromatin does not elastically return to its original conformation, but is stabilized by the repressive marks accrued while in the condensate [89]. Similar entrapment may be the foundation for the formation of super-enhancer clusters by coactivator proteins into transcriptional condensates [28]. The dynamic assembly and disassembly of nuclear condensates in a population of cells may explain, in part, the known spatial variability of genomic loci [90, 91] and their characteristically confined motion [92].
Ultimately, thermodynamic equilibrium would favor a fully de-mixed nucleus containing a single, cohesive phase of each DNA condensate. However, the nucleus may not reach such a de-mixed state as many non-thermal, ATP-dependent processes occur in the interior [93, 94], while the nucleus may also be kinetically or sterically trapped, preventing the sampling of all configurations necessary to reach equilibrium (Figure 4C). Thus, the nucleus may effectively represent an emulsion of condensates formed by various chromatin domains (Figure 1D). While individual proteins or small nucleoprotein complexes can diffuse rapidly across the nucleus [95], condensates exhibit highly sub-diffusive motion within the nucleus [96], since they are part of the larger continuous chromatin fiber that is further confined by larger architectural elements of the nucleus, such as nuclear bodies and the nuclear lamina [22].
Phase separation as a means to localize and coordinate function
One characteristic feature of condensates is their ability to concentrate protein and DNA, thereby controlling their local composition (Figure 4D–G). Increasing the content of DNA relative to protein impedes the relaxation of condensates into a round droplets in vitro [44, 61] and can translate to inaccessible DNA states, which are consistent with compact, gene-silenced heterochromatic regions (Figure 4D) [10, 42]. Similarly, viral RNA genomes may be compacted into droplets as observed for the SARS-CoV-2 genome by its nucleoplasmid (N) protein [97], suggesting the interactions that drive phase separation may facilitate tight packaging of various genomes.
High protein concentrations inside of droplets may also enhance reaction rates (Figure 4E) [34]. For example, hyperosmotic stress triggers condensation of the transcriptional coactivator Yes-associated protein (YAP) in the nucleus, recruiting other transcription factors to increase transcription of essential proliferative genes [98]. Optogenetic approaches also show increased RNA transcription upon light-induced clustering of transcriptional components [29]. By concentrating transcriptional machinery into condensates, the cell may be able to react more robustly in response to stimuli to carry out specific transcriptional programs.
The self-organization of select factors into DNA-based condensates also allows for precise spatiotemporal control of nuclear functions. DNA repair factors rapidly assemble into phase separated foci at double strand breaks (DSBs). The condensates seem to be initially seeded by short-lived PARylation of regions around DSBs [99] and DSBs are subsequently recognized by the DNA repair factor 53BP1, which acts as a scaffold protein for recruitment of additional clients, such as p53, for signal activation (Figure 4F) [53]. Another potential role of condensates may be the exclusion of immiscible factors from entering DNA repair foci [99] as well as damaging agents, including ROS, which may invoke further DNA damage (Figure 4G).
Phase separation in genome activity
DNA condensates exhibit a range of physicochemical properties: while the long DNA fiber remains immobilized in condensates, proteins associate with varying affinities and dynamics [55]. It is tempting to speculate that the continuum of material properties may influence the degree of biological activity along the DNA fiber (Figure 4H–J). More liquid, dynamic characteristics as found in transcriptional condensates allow higher accessibility and transcriptional activity along the chromatin fiber (Figure 4H) [26, 28]. In contrast, the DNA condensates associated with gene-silenced regions, such as heterochromatin, have been associated with a large immobile fraction of protein within the DNA condensate (Figure 4I) [43], indicative of reduced dynamics or of viscoelastic behavior. At the extreme end of the spectrum, the ability of DNA to exist in solid-like or crystalline states may be advantageous in maintaining the sequence-integrity of the genome (Figure 4J). DNA can take on a multitude of crystalline morphologies under various in vitro conditions [100–102] and DNA forms highly condensed, liquid-crystals in some bacteria and in sperm nuclei [103–106]. Furthermore, viral genomes, such as that of herpesvirus, actively thread DNA using an ATP-dependent motor into highly crystalline organization within their capsids, by which DNA may be perpetually preserved [107, 108]. These observations underscore how the emergent material properties within the condensate affect genome function.
Anomalies in DNA-condensate phase behavior in disease
Dysregulation of phase behavior of biomolecular condensates is thought to underlie disease [87]. A prominent example is how mutations in the FUS protein drive an aberrant liquid-to-solid transition implicated in neurodegenerative diseases [20]. By extension, perturbation of the phase behavior of DNA-condensates could be pathogenic [87, 109, 110].
Cancer is associated with increased DNA damage and altered transcriptional activity, particularly of oncogenes, leading to changes in genome organization. Phase separation may contribute to these functionally by influencing (dis)assembly and properties of condensates in localizing and affecting function, in particular transcription [76]. An example is the EWS-FLI1 fusion protein in Ewing’s sarcoma in which the prion-like domain (PLD) of the RNA-binding protein EWS1R is joined to the transcription factor FL1. The fusion of the PLD to FL1 triggers de novo formation of condensates, which can recruit and concentrate factors to tumor-specific enhancers [111]. Similar LSD-translocation and condensation of heterogenous nuclear ribonucleoproteins (hnRNPs) from the FET family (FUS, EWSR1, and TAF15) may be at play in other cancers [112]. Oncoproteins that drive leukemia progression analogously involve the fusion of NUP98’s intrinsically disordered N-terminus preferentially to C-termini of other phase-separating proteins, resulting in nuclear puncta formation and altered gene expression [113]. Additionally, mutations to the tumor suppressor POZ protein (SPOP) cause cancer in part by leading to the dissolution of enzymatically functional SPOP condensates [114]. Furthermore, cancer cells can lengthen their telomeres independent of telomerase via alternative lengthening of telomeres (ALT) in ALT-associated promyelocytic leukemia nuclear bodies, whose formation involves phase separation around telomeres and enrichment of DNA repair factors [115]. Understanding the molecular interactions that govern phase behavior in the genome may also be relevant for clinical applications as some cancer therapeutics are enriched in DNA-based condensates and thus affect drug pharmacodynamics [116].
Viruses are known to significantly alter genome organization upon infection. For example, herpesvirus pushes the host nuclear genome to the periphery to allow for coalescence of viral replication compartments (RCs) in the nuclear interior [117]. The resulting de-mixed RCs are able to accumulate host Pol II and other DNA-binding proteins, but their physicochemical properties do not follow classic LLPS as there is no diffusion barrier between phases as would be expected [118]. Epstein-Barr virus has two transcription factors EBNA2 and EBNALP that assemble into condensates, and inside of nuclei, these condensates are concentrated in super-enhancers to aid in transcription of viral genes [119]. Infection with influenza virus is associated with high levels of transcription elongation that in turn favors the transition of heterochromatin into euchromatin [120]. Comparing the nature of the physical mechanisms, be they phase separation or other forms, of viral-induced genomic compaction could shed light on the particular advantages for each class of virus [19].
Concluding Remarks
Biological phase separation may represent a fundamental behavior of biopolymers (DNA, RNA, protein) [121]. Indeed, the long, polymeric nature of DNA combined with the modular proteins that bind and package DNA endow genomes with molecular features that favor phase separation. The ubiquity of phase separation, especially in distantly related organisms, suggests phase separation is an evolutionarily conserved mechanism in genome organization with implications for preservation of DNA sequence integrity and gene function. Future studies will explore how these interactions drive phase separation of the genome, and how they relate to other modes of DNA organization, to give rise to emergent material properties that affect genomic activity. A further major direction in this field will be to determine how cells regulate the phase behavior of their genomes in response to vicissitudes of their environment and long-term over the course of evolution. An evolutionary framework for phase separation of genomes should serve as a springboard for future hypothesis-driven studies to uncover the fundamental mechanisms and functional consequences of genome organization (see Outstanding Questions box).
Outstanding questions:
How have phase separation mechanisms, and architectural genome proteins, evolved to accommodate size and complexity of various genomes (bacterial, mitochondrial, chloroplast, nuclear, germline, viral)?
How do specific molecular interactions lead to promoting evolutionary fitness versus contributing to disease processes?
How do non-thermal activity and complexity of the crowded cellular environment influence the phase behavior and consequently function of DNA-condensates?
How does the role of phase separation compare to the other modes of DNA compaction and organization across the phylogenetic tree of life?
Has phase separation been actively selected for or is it an innate feature of proteins and DNA confined to cellular milieu?
How can comparing differences in protein-mediated phase separation across evolution give insight into fundamental principles for engineering de novo condensates and developing biomimetic therapeutics?
Highlights.
Protein-mediated phase separation contributes to the self-assembly and organization of genome features, such as domains and compartments.
Evidence for phase separation in overall genome organization exists across the tree of life.
DNA-based biomolecular condensates have a wide range of emergent material properties that may have functional consequences, including for transcription and maintenance of DNA integrity.
Dysregulation of genomic phase behavior may reduce organismal fitness and contribute to disease.
Acknowledgements
The Misteli lab is supported by funding from the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute, and Center for Cancer Research (1- ZIA-BC010309) and 4D Nucleome Common Fund (U54 DK107980; UM1 HG011593); MF is supported by a Postdoctoral Research Associate Training (PRAT) fellowship from the National Institute of General Medical Sciences (NIGMS, 1Fi2GM128585–01). We would like to thank Stephanie Weber for discussion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- 1.Hyman AA et al. (2014) Liquid-liquid phase separation in biology. Annu. Rev. Cell. Dev. Bi 30, 39–58. [DOI] [PubMed] [Google Scholar]
- 2.Brangwynne CP et al. (2015) Polymer physics of intracellular phase transitions. Nat. Phys 11 (11), 899–904. [Google Scholar]
- 3.Banani SF et al. (2017) Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell. Bio 18 (5), 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brangwynne CP et al. (2009) Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324 (5935), 1729–1732. [DOI] [PubMed] [Google Scholar]
- 5.Molliex A et al. (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163 (1), 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feric M et al. (2016) Coexisting liquid phases underlie nucleolar subcompartments. Cell 165 (7), 1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hur W et al. (2020) CDK-regulated phase separation seeded by histone genes ensures precise growth and function of Histone Locus Bodies. Dev. Cell 54 (3), 379–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu H et al. (2018) Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558 (7709), 318–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabari BR et al. (2020) Biomolecular condensates in the nucleus. Trends Biochem. Sci 45 (11), 961–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanulli S et al. (2019) HP1 reshapes nucleosome core to promote heterochromatin phase separation. Nature 575 (7782), 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boija A et al. (2018) Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175 (7), 1842–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L et al. (2019) Histone modifications regulate chromatin compartmentalization by contributing to a phase separation mechanism. Mol. Cell 76 (4), 646–659. [DOI] [PubMed] [Google Scholar]
- 13.Dignon GL et al. (2020) Biomolecular Phase Separation: From Molecular Driving Forces to Macroscopic Properties. Annu. Rev. Phys. Chem 71, 53–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peran I and Mittag T (2020) Molecular structure in biomolecular condensates. Curr. Opin. Struc. Biol 60, 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang S-J (2017) Potential role of phase separation of repetitive DNA in chromosomal organization. Genes-Basel 8 (10), 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall AC et al. (2019) Phase separation as a melting pot for DNA repeats. Trends Genet 35 (8), 589–600. [DOI] [PubMed] [Google Scholar]
- 17.Banani SF et al. (2016) Compositional control of phase-separated cellular bodies. Cell 166 (3), 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi J-M et al. (2020) Physical principles underlying the complex biology of intracellular phase transitions. Annu. Rev. Biophys 49, 107–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber SC (2019) Evidence for and against liquid-liquid phase separation in the nucleus. Non-coding RNA 5 (4), 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel A et al. (2015) A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162 (5), 1066–1077. [DOI] [PubMed] [Google Scholar]
- 21.Shin Y et al. (2017) Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets. Cell 168 (1–2), 159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Misteli T (2020) The self-organizing genome: Principles of genome architecture and function. Cell 183 (1), 28–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turner AL et al. (2018) Highly disordered histone H1— DNA model complexes and their condensates. P. Natl. Acad. Sci. USA 115 (47), 11964–11969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibson BA et al. (2019) Organization of chromatin by intrinsic and regulated phase separation. Cell 179 (2), 470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allis CD and Jenuwein T (2016) The molecular hallmarks of epigenetic control. Nat. Rev. Genet 17 (8), 487. [DOI] [PubMed] [Google Scholar]
- 26.Cho W-K et al. (2018) Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361 (6400), 412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chong S et al. (2018) Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361 (6400), eaar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sabari BR et al. (2018) Coactivator condensation at super-enhancers links phase separation and gene control. Science 361 (6400), eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei M-T et al. (2020) Nucleated transcriptional condensates amplify gene expression. Nat. Cell Biol 22 (10), 1187–1196. [DOI] [PubMed] [Google Scholar]
- 30.Boehning M et al. (2018) RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol 25 (9), 833–840. [DOI] [PubMed] [Google Scholar]
- 31.Guo YE et al. (2019) Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572 (7770), 543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang C and Stiller JW (2014) Evolutionary diversity and taxon-specific modifications of the RNA polymerase II C-terminal domain. P. Natl. Acad. Sci. USA 111 (16), 5920–5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu F et al. (2019) The C-terminal domain of RNA polymerase II is a multivalent targeting sequence that supports drosophila development with only consensus heptads. Mol. Cell 73 (6), 1232–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hnisz D et al. (2017) A phase separation model for transcriptional control. Cell 169 (1), 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tunnacliffe E and Chubb JR (2020) What is a transcriptional burst? Trends Genet 36 (4), 288–297. [DOI] [PubMed] [Google Scholar]
- 36.Henninger JE et al. (2021) RNA-mediated feedback control of transcriptional condensates. Cell 184 (1), 207–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nuebler J et al. (2018) Chromatin organization by an interplay of loop extrusion and compartmental segregation. P. Natl. Acad. Sci. USA 115 (29), E6697–E6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nozaki T et al. (2017) Dynamic organization of chromatin domains revealed by super-resolution live-cell imaging. Mol. Cell 67 (2), 282–293. [DOI] [PubMed] [Google Scholar]
- 39.Miron E et al. (2020) Chromatin arranges in chains of mesoscale domains with nanoscale functional topography independent of cohesin. Sci. Adv 6 (39), eaba8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryu J-K et al. (2020) Phase separation induced by cohesin SMC protein complexes. bioRxiv, 10.1101/2020.06.13.149716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Erdel F and Rippe K (2018) Formation of chromatin subcompartments by phase separation. Biophys. J 114 (10), 2262–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larson AG et al. (2017) Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547 (7662), 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strom AR et al. (2017) Phase separation drives heterochromatin domain formation. Nature 547 (7662), 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keenen MM et al. (2020) HP1 proteins compact DNA into mechanically and positionally stable phase separated domains. bioRxiv, 10.1101/2020.10.30.362772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tatavosian R et al. (2019) Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation. J. Biol. Chem 294 (5), 1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Falk M et al. (2019) Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature 570 (7761), 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rai AK et al. (2018) Kinase-controlled phase transition of membraneless organelles in mitosis. Nature 559 (7713), 211–216. [DOI] [PubMed] [Google Scholar]
- 48.Cuylen S et al. (2016) Ki-67 acts as a biological surfactant to disperse mitotic chromosomes. Nature 535 (7611), 308–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cuylen-Haering S et al. (2020) Chromosome clustering by Ki-67 excludes cytoplasm during nuclear assembly. Nature 587 (7833), 285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trivedi P et al. (2019) The inner centromere is a biomolecular condensate scaffolded by the chromosomal passenger complex. Nat. Cell Biol 21 (9), 1127–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cerase A et al. (2019) Phase separation drives X-chromosome inactivation: a hypothesis. Nat. Struct. Mol. Biol 26 (5), 331–334. [DOI] [PubMed] [Google Scholar]
- 52.Pandya-Jones A et al. (2020) A protein assembly mediates Xist localization and gene silencing. Nature 587 (7832), 145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kilic S et al. (2019) Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J 38 (16), e101379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parker MW et al. (2019) A new class of disordered elements controls DNA replication through initiator self-assembly. Elife 8, e48562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strickfaden H et al. (2020) Condensed chromatin behaves like a solid on the mesoscale in vitro and in living cells. Cell 183 (7), 1772–1784. [DOI] [PubMed] [Google Scholar]
- 56.Sanulli S and Narlikar GJ (2020) Liquid-like interactions in heterochromatin: Implications for mechanism and regulation. Curr. Opin. Cell Biol 64, 90–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Erdel F et al. (2020) Mouse heterochromatin adopts digital compaction states without showing hallmarks of HP1-driven liquid-liquid phase separation. Mol. Cell 78 (2), 236–249. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen XJ and Butow RA (2005) The organization and inheritance of the mitochondrial genome. Nat. Rev. Genet 6 (11), 815–825. [DOI] [PubMed] [Google Scholar]
- 59.Ngo HB et al. (2011) The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nat. Struct. Mol. Biol 18 (11), 1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rubio-Cosials A et al. (2011) Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nat. Struct. Mol. Biol 18 (11), 1281. [DOI] [PubMed] [Google Scholar]
- 61.Feric M et al. (2019) Self-assembly of multi-component mitochondrial nucleoids via phase separation. bioRxiv, 10.1101/822858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wong TS et al. (2009) Biophysical characterizations of human mitochondrial transcription factor A and its binding to tumor suppressor p53. Nucleic Acids Res 37 (20), 6765–6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Štros M et al. (2007) The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell. Mol. Life Sci 64 (19–20), 2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brewer LR et al. (2003) Packaging of single DNA molecules by the yeast mitochondrial protein Abf2p. Biophys. J 85 (4), 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miyakawa I et al. (1987) Isolation of morphologically intact mitochondrial nucleoids from the yeast, Saccharomyces cerevisiae. J. Cell. Sci 88 (4), 431–439. [DOI] [PubMed] [Google Scholar]
- 66.Lukeš J et al. (2002) Kinetoplast DNA network: evolution of an improbable structure. Eukaryot. Cell 1 (4), 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu CW et al. (1996) Nucleus-encoded histone H1-like proteins are associated with kinetoplast DNA in the trypanosomatid Crithidia fasciculata. Mol. Cell. Biol 16 (2), 564–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dillon SC and Dorman CJ (2010) Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol 8 (3), 185–195. [DOI] [PubMed] [Google Scholar]
- 69.Wang X et al. (2013) Organization and segregation of bacterial chromosomes. Nat. Rev. Genet 14 (3), 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dame RT et al. (2019) Chromosome organization in bacteria: mechanistic insights into genome structure and function. Nat. Rev. Genet 21 (4), 227–242. [DOI] [PubMed] [Google Scholar]
- 71.Odijk T (1998) Osmotic compaction of supercoiled DNA into a bacterial nucleoid. Biophys. Chem 73 (1–2), 23–29. [DOI] [PubMed] [Google Scholar]
- 72.Ladouceur A-M et al. (2020) Clusters of bacterial RNA polymerase are biomolecular condensates that assemble through liquid-liquid phase separation. P. Natl. Acad. Sci. USA 117 (31), 18540–18549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Janissen R et al. (2018) Global DNA compaction in stationary-phase bacteria does not affect transcription. Cell 174 (5), 1188–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Joyeux M (2018) A segregative phase separation scenario of the formation of the bacterial nucleoid. Soft Matter 14 (36), 7368–7381. [DOI] [PubMed] [Google Scholar]
- 75.Cohan MC and Pappu RV (2020) Making the Case for Disordered Proteins and Biomolecular Condensates in Bacteria. Trends Biochem. Sci 45 (8), 668–680. [DOI] [PubMed] [Google Scholar]
- 76.Azaldegui CA et al. (2020) The emergence of phase separation as an organizing principle in bacteria. Biophys. J S0006–3495 (20), 30734–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Remesh SG et al. (2020) Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling. Nat. Commun 11 (1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hirata A et al. (2008) The X-ray crystal structure of RNA polymerase from Archaea. Nature 451 (7180), 851–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stracy M et al. (2015) Live-cell superresolution microscopy reveals the organization of RNA polymerase in the bacterial nucleoid. P. Natl. Acad. Sci. USA 112 (32), E4390–E4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Harami GM et al. (2020) Phase separation by ssDNA binding protein controlled via protein- protein and protein- DNA interactions. P. Natl. Acad. Sci. USA 117 (42), 26206–26217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bernecky C et al. (2016) Structure of transcribing mammalian RNA polymerase II. Nature 529 (7587), 551–554. [DOI] [PubMed] [Google Scholar]
- 82.Kucej M and Butow RA (2007) Evolutionary tinkering with mitochondrial nucleoids. Trends Cell. Biol 17 (12), 586–592. [DOI] [PubMed] [Google Scholar]
- 83.Stam M et al. (2019) 3D genome organization: a role for phase separation and loop extrusion? Curr. Opin. Plant Biol 48, 36–46. [DOI] [PubMed] [Google Scholar]
- 84.Pontvianne F and Liu C (2020) Chromatin domains in space and their functional implications. Curr. Opin. Plant Biol 54, 1–10. [DOI] [PubMed] [Google Scholar]
- 85.Zhao S et al. (2019) Plant HP1 protein ADCP1 links multivalent H3K9 methylation readout to heterochromatin formation. Cell Res 29 (1), 54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang N and Liu C (2019) Implications of liquid—liquid phase separation in plant chromatin organization and transcriptional control. Curr. Opin. Genet. Dev 55, 59–65. [DOI] [PubMed] [Google Scholar]
- 87.Shin Y and Brangwynne CP (2017) Liquid phase condensation in cell physiology and disease. Science 357 (6357), eaaf4382. [DOI] [PubMed] [Google Scholar]
- 88.Shin Y et al. (2018) Liquid nuclear condensates mechanically sense and restructure the genome. Cell 175 (6), 1481–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Eeftens JM et al. (2020) Epigenetic memory as a time integral over prior history of Polycomb phase separation. bioRxiv, 10.1101/2020.08.19.254706. [DOI] [Google Scholar]
- 90.Finn EH and Misteli T (2019) Molecular basis and biological function of variability in spatial genome organization. Science 365 (6457), eaaw9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Finn EH et al. (2019) Extensive heterogeneity and intrinsic variation in spatial genome organization. Cell 176 (6), 1502–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen B et al. (2013) Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155 (7), 1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou CY et al. (2016) Mechanisms of ATP-dependent chromatin remodeling motors. Annu. Rev. Biophys 45, 153–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bowman GD (2010) Mechanisms of ATP-dependent nucleosome sliding. Curr. Opin. Struc. Biol 20 (1), 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Misteli T (2001) Protein dynamics: implications for nuclear architecture and gene expression. Science 291 (5505), 843–847. [DOI] [PubMed] [Google Scholar]
- 96.Lee DSW et al. (2021) Chromatin mechanics dictates subdiffusion and coarsening dynamics of embedded condensates. Nat. Phys, 10.1038/s41567-020-01125-8. [DOI] [Google Scholar]
- 97.Cubuk J et al. (2020) The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. bioRxiv, 2020.06.17.158121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cai D et al. (2019) Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. Nat. Cell. Biol 21 (12), 1578–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Altmeyer M et al. (2015) Liquid demixing of intrinsically disordered proteins is seeded by poly (ADP-ribose). Nat. Commun 6 (1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Siavashpouri M et al. (2017) Molecular engineering of chiral colloidal liquid crystals using DNA origami. Nat. Mater 16 (8), 849–856. [DOI] [PubMed] [Google Scholar]
- 101.Laramy CR et al. (2019) Crystal engineering with DNA. Nat. Rev. Mater 4 (3), 201–224. [Google Scholar]
- 102.Winfree E et al. (1998) Design and self-assembly of two-dimensional DNA crystals. Nature 394 (6693), 539–544. [DOI] [PubMed] [Google Scholar]
- 103.Brach K et al. (2018) Photochemical analysis of structural transitions in DNA liquid crystals reveals differences in spatial structure of DNA molecules organized in liquid crystalline form. Sci. Rep-UK 8 (1), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Livolant F et al. (1989) The highly concentrated liquid-crystalline phase of DNA is columnar hexagonal. Nature 339 (6227), 724–726. [DOI] [PubMed] [Google Scholar]
- 105.Cha YJ et al. (2019) Microstructure arrays of DNA using topographic control. Nat. Commun 10 (1), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tarafder AK et al. (2020) Phage liquid crystalline droplets form occlusive sheaths that encapsulate and protect infectious rod-shaped bacteria. P. Natl. Acad. Sci. USA 117 (9), 4724–4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu Y-T et al. (2019) Cryo-EM structures of herpes simplex virus type 1 portal vertex and packaged genome. Nature 570 (7760), 257–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yang Y et al. (2020) Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation. Protein Cell 11 (5), 339–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Misteli T (2010) Higher-order genome organization in human disease. CSH. Perspect. Biol 2 (8), a000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Alberti S and Dormann D (2019) Liquid—liquid phase separation in disease. Annu. Rev. Genet 53, 171–194. [DOI] [PubMed] [Google Scholar]
- 111.Boulay G et al. (2017) Cancer-specific retargeting of BAF complexes by a prion-like domain. Cell 171 (1), 163–178. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jiang S et al. (2020) Protein phase separation and its role in tumorigenesis. Elife 9, e60264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Terlecki-Zaniewicz S et al. (2021) Biomolecular condensation of NUP98 fusion proteins drives leukemogenic gene expression. Nat. Rev. Mol. Cell. Biol, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bouchard JJ et al. (2018) Cancer mutations of the tumor suppressor SPOP disrupt the formation of active, phase-separated compartments. Mol. Cell 72 (1), 19–36. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang H et al. (2020) Nuclear body phase separation drives telomere clustering in ALT cancer cells. Mol. Biol. Cell 31 (18), 2048–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Klein IA et al. (2020) Partitioning of cancer therapeutics in nuclear condensates. Science 368 (6497), 1386–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bosse JB et al. (2015) Remodeling nuclear architecture allows efficient transport of herpesvirus capsids by diffusion. P. Natl. Acad. Sci. USA 112 (42), E5725–E5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.McSwiggen DT et al. (2019) Evidence for DNA-mediated nuclear compartmentalization distinct from phase separation. Elife 8, e47098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Peng Q et al. (2020) Phase separation of Epstein-Barr Virus EBNA2 and Its Coactivator EBNALP controls gene expression. J. Virol 94 (7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Heinz S et al. (2018) Transcription elongation can affect genome 3D structure. Cell 174 (6), 1522–1536. e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Keating CD (2012) Aqueous phase separation as a possible route to compartmentalization of biological molecules. Accounts Chem. Res 45 (12), 2114–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]