Summary
Objectives: Cachexia is a systemic metabolic disorder characterized by loss of fat and muscle mass, which disproportionately impacts patients with gastrointestinal malignancies such as pancreatic cancer. While the immunologic shifts contributing to the development of other adipose tissue (AT) pathologies such as obesity have been well described, the immune microenvironment has not been studied in the context of cachexia.
Methods: We performed bulk RNA-sequencing, cytokine arrays, and flow cytometry to characterize the immune landscape of visceral AT (VAT) in the setting of pancreatic and colorectal cancers.
Results: The cachexia inducing factor IL-6 is strongly elevated in the wasting VAT of cancer bearing mice, but the regulatory type 2 immune landscape which characterizes healthy VAT is maintained. Pathologic skewing toward Th1 and Th17 inflammation is absent. Similarly, the VAT of patients with colorectal cancer is characterized by a Th2 signature with abundant IL-33 and eotaxin-2, albeit also with high levels of IL-6.
Conclusions: Wasting AT during the development of cachexia may not undergo drastic changes in immune composition like those seen in obese AT. Our approach provides a framework for future immunologic analyses of cancer associated cachexia.
Keywords: adipose tissue, cachexia, pancreatic cancer
Graphical Abstract
Graphical Abstract.
Introduction
Cachexia is a metabolic syndrome characterized by adipose wasting and loss of skeletal muscle mass observed in the setting of cancer and other chronic diseases such as chronic obstructive pulmonary disease and heart failure [1]. The prevalence of cancer-associated cachexia (CAC) is variable by cancer type, with particularly high rates observed in patients with pancreatic ductal adenocarcinoma (PDAC) [1–3]. Cachexia is a significant morbidity in PDAC, both reducing the quality of life and limiting the ability of patients to receive optimal doses of standard of care chemotherapies [4]. Current CAC treatments aim to improve symptoms via lifestyle changes, antiemetic drugs, and appetite stimulants [5]. There are currently no therapies approved for the prevention or reversal of cachexia, emphasizing the need to better understand the mechanisms underlying this disease.
CAC is by definition a systemic disease, affecting muscles and adipose tissue (AT) throughout the body in addition to other tissues such as the liver and brain. Accordingly, previous work to understand the mechanisms underlying CAC has identified circulating molecules that are elevated in the context of cancer, some of which are released directly by tumor cells. This has led to the identification of inflammatory proteins such as TNFα (originally named ‘cachectin’) [6] and IL-6 as potential drivers of CAC in both humans and animal models. IL-6 blockade has shown promise in reversing CAC phenotypes in murine models and in patient case reports [7–9], but this therapeutic benefit is inconsistent and has not been observed in prospective clinical trials. Further, cachexia has been observed in murine PDAC models prior to elevations in circulating IL-6, and IL-6 levels are not associated with the degree of weight loss in human patients with PDAC [10]. Taken together, these findings emphasize that the mechanisms driving CAC in PDAC likely extend beyond IL-6.
Metabolic alterations in the blood and target tissues of CAC have been extensively studied. For example, plasma metabolomic studies have identified elevations in plasma amino acids due to muscle breakdown early in the course of human and murine PDAC development [11]. Various circulating factors can promote excessive autophagy and proteasomal activation in skeletal muscle along with contractile dysfunction, which together contribute to progressive muscle atrophy [12–14]. Pathologic elevations in lipolysis drive fat loss [1, 15, 16], and the beiging of distant white AT has been observed in several murine models of cancer as well as multiple human cancer types [8]. This adipose wasting may actually be a prerequisite for muscle atrophy, although the exact mechanisms of this dependence are not yet understood [17, 18].
More recently, there has been interest in exploring changes in systemic immunity in the context of CAC. For example, an elevation of the neutrophil to lymphocyte ratio in peripheral blood was associated with higher rates of cachexia in a cohort of 50 stage III/IV colon, lung, and prostate cancer patients [19]. Interestingly, in a murine model of PDAC, neutrophils are recruited into the central nervous system via the CCR2/CCL2 axis early in the course of disease, and blocking this neutrophil infiltration can prevent the subsequent development of CAC [20]. However, the AT immune microenvironment has not been well characterized in CAC, with only limited data exploring how cellular infiltrates and cytokine profiles within AT change in the setting of CAC [21, 22].
On the contrary, the immune microenvironment of AT has been well studied in the healthy state and in other pathologic conditions such as obesity and diabetes [23]. Healthy AT is characterized by a regulatory type 2 immune signature which suppresses inflammation and maintains adipocyte function [23]. At the center of this homeostatic network are anti-inflammatory AT macrophages (ATMs), which are supported by cytokines such as IL-4 and IL-13 produced by other resident cells including group 2 innate lymphoid cells (ILC2s) and eosinophils [24–26]. Regulatory T cells (Tregs) in AT and muscle express ST2, the receptor for IL-33, and require this IL-33/ST2 axis for their development and maintenance over time [27, 28]. In genetic and diet-induced models of obesity, as well as in human subjects, the adipose landscape shifts in favor of inflammation-promoting cells such as neutrophils, inflammatory ATMs, Th17 cells, and IFNγ-producing Th1 and CD8+ T cells [23, 29], with a concomitant reduction in the regulatory cell types including eosinophils [30], iNKT cells [31], and Tregs [27]. Interestingly, while immunotherapies are now mainstays in the treatment of malignancies, autoimmune diseases, and infectious diseases [32, 33], targeted modulation of this adipose inflammation is not yet applied clinically to treat metabolic diseases such as obesity, diabetes, or cachexia.
Here we aim to assess the immune phenotype of wasting AT during the development of CAC. We compare the immune landscape in visceral AT (VAT) of healthy mice to that of mice bearing orthotopic pancreatic tumors which are known to induce AT wasting and ultimately a CAC-like phenotype. Our results suggest that the type 2 immune signature of healthy AT is maintained in wasting AT and in the VAT of patients with colorectal cancer. Additionally, we observed high levels of IL-6 in the AT of cancer bearing mice and humans. We propose that similar analyses should be performed to more fully identify and characterize immunologic shifts that occur in AT and other target organs during and after the development of CAC in preclinical models and in patients experiencing CAC.
Material and methods
Animal care
Animals were housed at the Dana-Farber Cancer Institute and were maintained according to protocols approved by the DFCI Institute Committee on Animal Care and Use. C57BL/6J and Balb/c mice were purchased from Jackson Labs.
Cell lines
KPC.1 cells were a kind gift of Dr. Anirban Maitra and were maintained in RPMI 1640 (Gibco #11875-093) supplemented with 10% fetal bovine serum (FBS), 1× MEM Non-Essential Amino Acids (Gibco #11140-050), 1× GlutaMAX (Gibco #35050–061), 1000 Units/ml Penicillin (Gibco #15140-122), 1 mg/ml Streptomycin (Gibco #15140-122), 1 mM Sodium Pyruvate (Gibco #11360-070), and 0.0008% β-mercaptoethanol (Sigma #M3148). CT26 cells were purchased from the American Type Culture Collection (ATCC) and were maintained in complete DMEM.
Protein isolation from whole adipose tissue and skeletal muscle
Gonadal white adipose tissue (WAT), inguinal WAT, interscapular brown adipose tissue (BAT), and quadriceps muscles were isolated from mice and flash frozen in liquid nitrogen. Tissues were powderized using a mortar and pestle, and powder was suspended in RIPA buffer supplemented with the Halt Protease and Phosphatase Inhibitor cocktail (ThermoFisher #78442). Samples were shaken at 4°C for 30 minutes with vortexing after 15 and 30 minutes, then centrifuged for 15 minutes at 4°C at 13,300 g, and the supernatant collected. Protein concentration was measured using the Micro BCA Assay (ThermoFisher #23235).
Protein isolation from stromovascular fraction for immunoblot
BAT and WAT were isolated, and each tissue was transferred into a well of a 6-well tissue culture plate. Tissue was finely minced with scissors, and 2 ml of 2 mg/ml type II collagenase from Clostridium histolyticum (Sigma C6885) was added to each well. The tissue suspension was transferred into a 15-ml falcon tube and then digested for 30 minutes at 37°C with shaking at 250 rpm and vortexing every 10 minutes. After digestion, samples were diluted with 4 ml of RPMI containing 10% FBS and then filtered through a 70 µm strainer. The filtered mixture was centrifuged at 350 g for 5 minutes at 4°C, which leads to the separation of mature adipocytes (floating fraction) from the stromovascular fraction (SVF) (pellet). The supernatant was aspirated to leave behind the SVF pellet, which was then suspended in RIPA buffer supplemented with the Halt Protease and Phosphatase Inhibitor cocktail (ThermoFisher #78442). Protein isolation and quantification were performed as described above.
Western blots
Twenty micrograms of tissue lysate were loaded per lane on a 12% Tris-glycine SDS PAGE gel, and transferred onto polyvinylidene difluoride membranes. Prior to blotting, membranes were blocked with 5% bovine serum albumin (BSA) (powder from Cell Signaling Technologies #9998S) in Tris-buffered saline + 0.1% Tween 20 (TBST) unless specifically noted otherwise. Membranes were blotted with primary antibodies (diluted in blocking buffer) at 4°C overnight, washed three times for 5 minutes with TBST, blotted with horseradish peroxidase (HRP)-conjugated secondary antibodies (diluted in blocking buffer) at room temperature for 1 hour, and then washed three times for 5 minutes with TBST. Samples were then incubated with 1 ml of Western Lightning Plus-ECL detection reagent (PerkinElmer #NEL103E001EA) and imaged using a Bio-Rad ChemiDoc Imaging system. The following antibodies were used for western blotting: goat polyclonal anti-PDL1 (R&D Systems #AF1019), rabbit monoclonal anti-UCP1 (clone D9D6X; CST #14670), HRP-conjugated rabbit anti-beta actin (clone 13E5; CST #5125), HRP-conjugated goat anti-rabbit IgG (CST #7074), and HRP-conjugated rabbit anti-goat IgG (Invitrogen #61–1620).
Analysis of adipose SVF by flow cytometry
SVF was isolated from adipose tissue as described above. Briefly, tissues were minced, digested, filtered, and centrifuged to separate adipocytes from the SVF. After removing the adipocyte-containing supernatant, the SVF was washed once with 1 ml cold phosphate-buffered saline (PBS). Samples were ACK lysed with 1 ml ACK Lysis Buffer for 2 minutes, diluted in 1 ml PBS, and centrifuged at 350 g for 5 minutes at 4°C. Samples were washed once more with 1 ml cold PBS and then stained for flow cytometry for 20 minutes at 4°C. After staining, samples were diluted with 1 ml cold PBS and centrifuged at 350 g for 5 minutes at 4°C, then washed twice with 1 ml cold PBS. After the final wash, samples were fixed in 1% formalin (in PBS) and analyzed on a Sony SP6800 spectral flow cytometer. Analysis and quantification of flow cytometry data was performed in FlowJo 10. The following antibodies against mouse antigens were purchased from BioLegend for flow cytometry: anti-CD45 BV711 (clone 30-F11), anti-CD11b Pacific Blue (clone M1/70), anti-CD11b APC-Cy7 (clone M1/70), anti-CD11c FITC (clone N418), anti-F4/80 APC (clone BM8), anti-Gr1 PE-Cy7 (clone RB6-8C5), anti-Ly6G BV570 (clone 1A8), anti-B220 BV605 (clone RA3-6B2), anti-CD3 PE (clone 17A2), anti-CD3 BV605 (clone 17A2), anti-TCRβ AlexaFluor 488 (clone H57-597), anti-TCRγδ APC (clone GL3), anti-CD4 BV421 (clone GK1.5), anti-CD8a Pacific Blue (clone 53–6.7). Anti-SiglecF BV421 (clone E50-2440) was purchased from BD Biosciences. The following antibodies against human antigens were purchased from BioLegend for flow cytometry: anti-CD45 Pacific Blue (clone HI30), anti-CD45 FITC (clone 2D1), anti-CD15 Pacific Blue (clone MMA), anti-CD15 PE (clone MMA), anti-SIGLEC8 PE-Cy7 (clone 7C9). Anti-CCR3 PE (clone 5E8) was purchased from BD Biosciences.
Cloning and expression of VHH nanobodies
The B3-, and VHH control 1B7-IFNg coding sequences were sub-cloned into the mammalian expression vector pVRC and transiently transfected using polyethyleneimine into HEK293F cells cultured in FreeStyle media (ThermoFisher #12338018). Media containing secreted protein was harvested 6 days following transfection by centrifugation at 8000 g for 20 minutes at 4°C, then loaded onto a HiTrap NiNTA column (GE Healthcare) and washed with 50 mM Tris, pH 8, 150 mM NaCl and 10 mM imidazole. Protein was eluted in 50 mM Tris, pH 8, 150 mM NaCl, 500 mM imidazole and 10% glycerol, then loaded onto a Superdex 200 16/600 column (GE Healthcare) in 50 mM Tris, pH 8, 150 mM NaCl, 10% glycerol. Recombinant VHH purity was assessed by SDS-PAGE, and peak fractions were recovered and concentrated with an Amicon 10,000 KDa MWCO filtration unit (Millipore), and stored at −80°C.
In vivo delivery of nanobody-cytokine fusions
Mice were injected intraperitoneally with 200 µl of PBS, a nanobody control (1B7), or a nanobody-cytokine fusion (1B7-IFNγ, B3-IFNγ) once daily for five days. 40 µg of B3, or the molar equivalents of 1B7-IFNγ (88 µg) or B3-IFNγ (85.7 µg) were injected daily. Mice were euthanized and tissues isolated 24 hours after the last injection for analysis by qPCR and immunoblot.
Orthotopic pancreatic tumors
Orthotopic surgeries were performed as described previously [34]. Briefly, female C57BL/6J mice were anesthetized with a ketamine/xylazine cocktail, shaved on the left flank, and the surgical site cleaned with ethanol and betadine. An incision was made in the skin and peritoneum, and the pancreas externalized with forceps. KPC cells were resuspended in PBS and mixed 1:1 by volume with Matrigel (Corning) for a total of 50,000 cells per 15 µl. Sham mice received PBS mixed with Matrigel instead of tumor cells. The cell suspension was kept on ice and drawn into a chilled insulin syringe. Cells were then injected into the tail of the pancreas, and a bubble was observed. Mice that showed signs of leakage were removed from the experiment. The pancreas was left external to the body cavity for 1 minute with the mice on a warming pad to solidify the Matrigel. The pancreas was then reinserted, peritoneum sutured with one stitch of absorbable suture, and the skin stapled with a sterile wound clip. Mice were given analgesia (Buprenex) and monitored post-surgery according to protocols approved by the Dana-Farber IACUC. Mice were sacrificed 28 days post-surgery unless otherwise indicated. Tumors weights were obtained at the time of sacrifice and subtracted from total body weight to obtain a net body weight.
Cytokine concentration measurements
Protein was isolated from adipose tissue as described above. Murine samples were sent to Eve Technologies for analysis by the Mouse Cytokine Array/Chemokine Array 31-Plex Panel and the Mouse Cytokine Array Th17 Discovery 12-Plex Panel. Human samples were sent to Eve Technologies for analysis by the Human Cytokine Array/Chemokine Array 71-Plex Panel. Cytokine concentrations (in pg/ml) were normalized to total protein concentration for comparison between samples.
Assessment of cachexia models
Tumors were implanted as described above. Mouse weights were collected prior to surgery and recorded as the initial body mass. After 28 days, mouse weights were collected and recorded as the final total body mass. Mice were then euthanized and tumors were isolated along with the right-sided VAT (gonadal fat pad) and quadriceps muscle. For each mouse, the weights of the tumor, VAT, and quadriceps were recorded. The tumor mass was subtracted from the final total body mass to obtain the final net body mass. The percent change in body mass was then calculated as % Change = (Final Net Body Mass − Initial Body Mass)/Initial Body Mass. The VAT mass and quadriceps mass were normalized to the initial body mass for display and comparisons between groups.
RNA-sequencing
Adipose tissue was isolated from mice and immediately flash frozen in liquid nitrogen. Samples were then stored at −80°C. RNA was subsequently isolated using the QIAGEN RNeasy Plus Mini Kit (Qiagen #74134). Briefly, we added 350 µl of RLT Buffer supplemented with 3.5 µl β-mercaptoethanol (Sigma #M3148), and then crushed the tissue in a 1.5-ml tube using a plastic pestle. Samples were sequentially transferred to QIA Shredder columns, genomic DNA eliminator columns, and RNA isolation columns. RNA was eluted using 50 µl of nuclease-free water.
RNA was sent to GeneWiz for library preparation and RNA-sequencing. Paired-end 150 bp single-index sequencing was performed using an Illumina HiSeq system. Transcript quantification was performed with Salmon [35], using quasi-mapping mode with selective alignment (validateMappings). We set the rangeFactorizationBins to 4 and passed both the seqBias and gcBias flags to correct for sequence-specific bias and fragment-level GC bias, respectively. Transcript quantifications were collapsed to gene-level quantifications using the tximport package [36] and the Ensembl-based annotation package EnsDb.Mmusculus.v79. Differential expression analysis was then performed using DESeq2 [37] with fold change shrinkage by ashr [38]. For downstream analysis and visualization, only genes with a mean TPM (transcripts per million) > 2 in VAT from both sham-operated and tumor-bearing mice were considered. Plots were made in R using the ggplot2 and heatmap.2 packages.
Gene Set Enrichment Analysis
Gene Set Enrichment Analysis (GSEA) was performed using the fgsea package in R, with genes ranked according to their fold change between cancer bearing and sham-operated mice. Ribosomal genes were excluded from this analysis. KEGG and Reactome gene sets containing human gene names were obtained from MSigDB [39–41], and human gene symbols were converted to their corresponding mouse gene symbols using biomaRt. Plots were made in R using the plotEnrichment function (from the fgsea package) and the ggplot2 package.
Enzyme-linked immunosorbent assays
Clear 96-well flat bottom polystyrene high bind microplates (Corning #9018) were incubated overnight with 100 µl of goat anti-mouse IgG (H+L), Human ads-UNLB (Southern Biotech #1031-01) diluted 1:500 in coating buffer (1L H2O + 3.03g Na2CO3 + 6g NaHCO3, pH adjusted to 9.6), washed three times with PBST (PBS + 0.5% Tween20), blocked with 200 µl of blocking buffer (PBS + 10% FBS) for 1 hour at room temperature, and washed three times with PBST. Samples were then added to the plates (in technical triplicates) at a 1:1000 dilution in blocking buffer and incubated for 2 hours at room temperature. Plates were washed three times with PBST, and then incubated with 100 µl of HRP-conjugated detection antibodies diluted 1:5000 in blocking buffer. Secondary antibodies were goat anti-mouse IgG1-HRP (Southern Biotech #1071-05) and goat anti-mouse IgG2b-HRP (Southern Biotech #1091-05). After a one-hour incubation in the dark, plates were washed five times with PBST. Plates were then developed with 100 µl of 3,3’,5,5’-Tetramethylbenzidine (Sigma #T8665) for up to 30 minutes until a modest color change was detected, at which time 50 µl of 1M HCl was added to quench the reaction. Plates were analyzed on a PerkinElmer Envision Multimode Plate Reader for absorbance at 450 nm. Protein concentrations were calculated based on a standard curve generated on the same plate.
RT-qPCR
RNA was isolated as described above and quantified using a DeNovix DS-11+ Spectrophotometer. Five hundred grams of RNA was used for reverse transcription using the iScript Reverse Transcription Supermix (Bio-Rad #1708840). The resultant cDNA was then diluted 1:10 in nuclease-free water. For qPCR, we added 7.5 µl of SsoAdvanced Universal SYBR Green Supermix (Bio-Rad #1725270), 4 µl of diluted cDNA, 1.5 µl of a primer mix containing forward and reverse primers at 3 µM each, and 2 µl of nuclease free water per well in a 96-well plate. Cycling conditions were set as follows on a BioRad CFX Connect Real-Time System (Bio-Rad #1855200): (1) 98°C for 30 seconds, (2) 98°C for 10 seconds, (3) 60°C for 30 seconds, (4) measure fluorescence in each well, (5) repeat steps 2–4 for 40 cycles. Samples were analyzed as technical duplicates. Primer sequences were as follows: 5′-TGCTGCATAATCAGCTACGG-3′ (Pdl1 forward) and 5′-CCACGGAAATTCTCTGGTTG-3′ (Pdl1 reverse), 5′-CAGAGGAAAGAGAGAAAGTCC-3′ (Irf1 forward) and 5′-CACACGGTGACAGTGCTGG-3′ (Irf1 reverse), 5′-TGTCCACCTTCCAGCAGATGT-3′ (beta-Actin forward) and 5′-AGCTCAGTAACAGTCCGCCTAG-3′ (beta-Actin reverse). Primers were synthesized as DNA oligos by Integrated DNA Technologies.
Cytospin
For mouse cytospin, eosinophils were sorted into 1.5 ml tubes by fluorescence-activated cell sorting from adipose tissue as CD45+SiglecF+Gr1- cells. The sorted cells were centrifuged at 300 g for 5 minutes at 4°C, washed once with 1 ml of cold PBS, centrifuged at 300 g for 5 minutes at 4°C, and resuspended in 100 ml of cold PBS. This 100 ml cell suspension was loaded into a cytofunnel and spun down onto a microscope slide using a Thermo Scientific Cytospin 4 Cytocentrifuge (Thermo #A78300003) at 800 rpm on medium acceleration for 3 minutes at room temperature. Slides were immediately fixed in 100% methanol for 5 minutes and then stored at 4°C until their subsequent staining.
For human cytospin, 100 µl of the whole stromovascular fraction (SVF) suspension was loaded into a cytofunnel. Cytospin and cell fixation was performed as described above.
Hematoxylin and eosin staining of cytospin samples
Harris hematoxylin was filtered through filter paper and added to a staining jar. Slides were incubated in the hematoxylin solution for 5 minutes, removed, washed with distilled water for 30 seconds, and then transferred to a jar containing a 2.5% Eosin Y solution for 3 minutes. After removing the eosin solution, the slides were dehydrated through three rounds of 95% ethanol (20 seconds each) and three rounds of xylene (20 seconds each). After drying, slides were mounted with a coverslip and imaged under a light microscope.
Statistical analyses
Comparisons of body mass, protein concentration (enzyme-linked immunosorbent assay [ELISA], cytokine array), gene expression (RT-qPCR), and cell proportions (flow cytometry) between sham-operated and tumor-bearing mice were performed using the nonparametric Mann–Whitney U test. Correction for multiple hypothesis testing was performed using the two-stage step-up method by Benjamini, Krieger, and Yekutieli. Comparisons of gene expression (RT-qPCR) between mice injected with PBS, control nanobody, or nanobody fusions were performed using two-way analysis of variances (ANOVAs), with correction for multiple hypothesis testing obtained using the two-stage step-up method by Benjamini, Krieger, and Yekutieli. Parametric tests were applied here due to the very small sample size of the control group (n = 2). These statistical analyses were performed using Prism 9.
RNA-seq analysis was performed in R. The statistical models used for differential expression analysis in DESeq2 and for GSEA have been described previously [37, 39]. Briefly, DESeq2 fits a generalized linear model (GLM) for each gene in a counts matrix modeled using the negative binomial distribution and then tests whether each gene’s GLM coefficient differs significantly from zero. GSEA calculates an enrichment score for any gene set based on a running sum calculated while walking down a ranked list of genes (here, ranked by fold change), and then tests for the statistical significance of this score using an empirical permutation approach followed by correction for multiple hypothesis testing.
Consent forms and ethical inclusion of patients
Written and informed consent was obtained from all patients who participated in this study under Mass General Brigham Institutional Review Board. All patients complied with and met ethical regulations included in our protocol.
Results
Orthotopic pancreatic tumors induce reductions in visceral adipose mass
In order to study the immune alterations in CAC target tissues, we first identified preclinical models that resulted in the onset of AT wasting and other CAC-like phenotypes. The genetically engineered KPC (LSL-KrasG12D/+;LSL-Trp53R172H/+;Pdx-1-Cre) model of pancreatic cancer was previously generated to express mutant KRAS under the control of the pancreas-specific Pdx1 promoter on a p53-mutant background [42]. These mice faithfully develop pancreatic tumors that closely resemble the human disease by pathology, and they develop cachexia phenotypes including loss of muscle and adipose mass [8, 43]. Further, orthotopic implantation of KPC tumors can induce CAC phenotypes including a particularly profound reduction in adiposity compared to baseline [44].
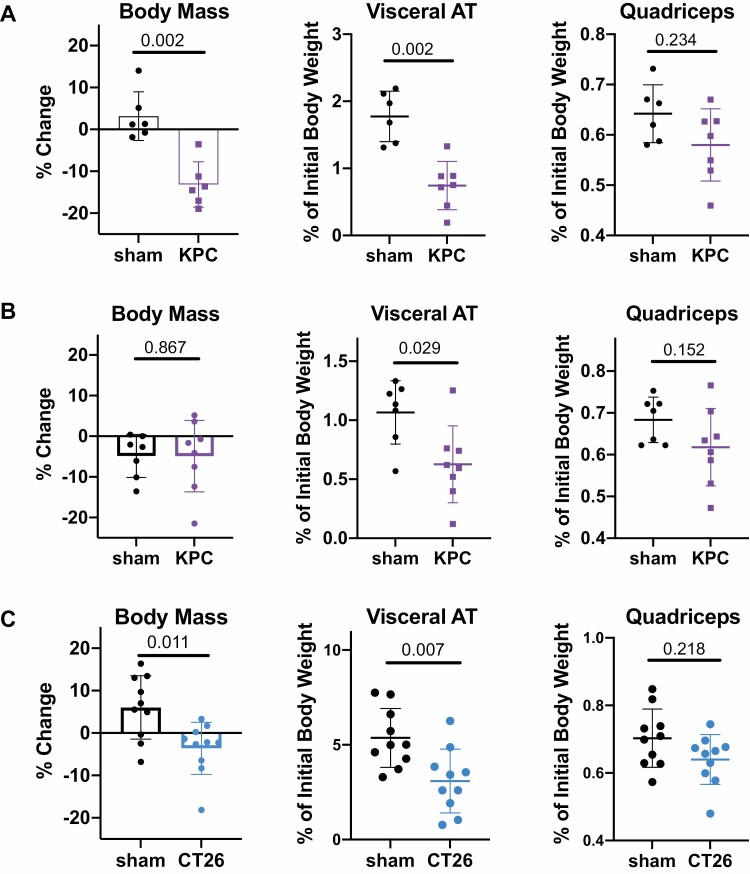
We first tested whether orthotopic KPC tumors similarly induce cachexia in our own hands, with a particular interest in the effect on VAT mass. We found that in 20 week old female mice with starting weights of at least 25 g, tumor-bearing mice showed significant reductions in body mass and VAT mass along with a trending decrease in muscle mass compared to sham-operated mice after four weeks (Fig. 1A). Female mice implanted with tumors at 7 weeks of age exhibited a significant reduction in VAT mass compared to sham operated controls after four weeks, although total body mass and skeletal muscle mass were unchanged (Fig. 1B). We also tested the CT26 model of colorectal cancer, another established model of CAC [45], and found that subcutaneous CT26 tumors induced reductions in total body and VAT mass along with a trending decrease in muscle mass (Fig. 1C). In subsequent experiments, we focused primarily on mice implanted with orthotopic KPC tumors at 7 weeks in order to study the immune environment in wasting AT of cancer bearing mice prior to their development of outright cachexia (i.e. decreases in muscle mass and total body mass in addition to AT mass).
Figure 1.
Models of cancer associated cachexia and adipose tissue wasting. Summary of changes in body mass, visceral adipose tissue (AT) mass, and quadriceps muscle mass in three tested models of CAC. (A) 50,000 KPC cells were mixed with Matrigel and surgically implanted into the pancreas of 20-week-old female C57BL/6J mice versus sham surgery. Body weight was monitored over time, and mice were harvested 28 days post-surgery. (B) Orthotopic KPC tumors were implanted as in panel (A) into 7-week old female C57BL/6J recipient mice versus sham surgery. Net body weight, adipose and muscle weights were determined at 28 days post-surgery. (C) PBS (‘sham’) or 250,000 CT26 cells (‘CT26’) were implanted subcutaneously in 7-week-old female Balb/c mice. Net body weight, adipose and muscle weights were determined at 21 days post-implantation. Statistical analyses were performed using the Mann–Whitney test; mean ± SD is shown.
RNA-sequencing shows upregulation of immune-related genes in AT of tumor-bearing mice
To understand whether immunologic changes occur in the VAT during the development of CAC, we subjected 7-week-old mice to sham surgery or implanted orthotopic KPC tumors into the pancreas and harvested the VAT after 4 weeks to perform RNA-sequencing (RNA-seq), cytokine array, antibody quantification, and flow cytometry (Fig. 2A, Supplementary Figs S1 and S2). We also assessed the immune infiltrates in AT from mice inoculated with subcutaneous CT26 tumors compared to mice injected with PBS to determine whether any cellular changes observed are consistent across cancer types (Supplementary Fig. S3).
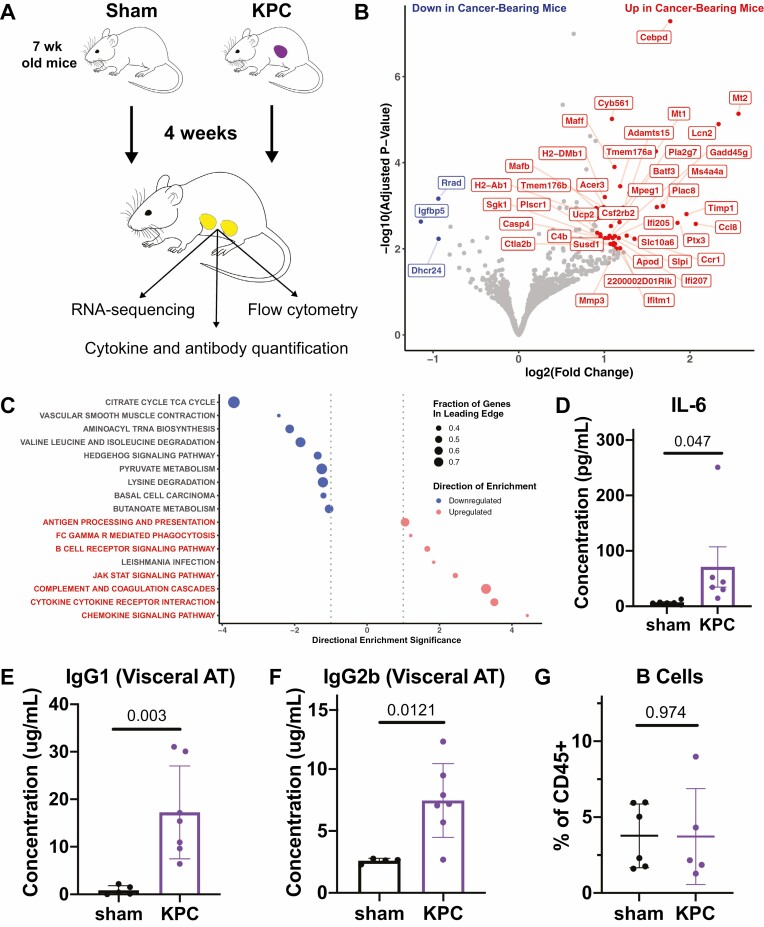
Figure 2.
Immune-related transcripts and proteins are upregulated in adipose tissue in the setting of cancer. (A) Schematic describing the approach to assessing immunologic changes in VAT in cancer bearing mice. Note that RNA-sequencing and cytokine quantification were performed using VAT samples from the same mice, while flow cytometry and antibody quantification were performed using VAT samples from two separate experiments. Mice were 7 weeks old at the time of tumor inoculation. (B) Volcano plot highlighting genes which are most strongly upregulated (subset shown in red) and downregulated (subset shown in blue) in VAT of cancer bearing mice compared to that of sham-operated mice. (C) Bubble plot showing KEGG pathways enriched based on RNA-sequencing data depicted in (B). Immune-related pathways that are upregulated are highlighted in red. (D) IL-6 quantification by cytokine array shows elevation in VAT of cancer bearing mice. (E–F) Quantification of total IgG1 (E) and IgG2b (F) by ELISA in VAT of cancer bearing and sham-operated mice. (G) Quantification of B cells as a fraction of total CD45+ cells in VAT by flow cytometry. Statistical analyses were performed using the Mann–Whitney test; mean ± SD is shown.
From RNA-seq, we identified 161 and 27 genes which were significantly upregulated and downregulated (adjusted P-value <= 0.05) in the AT of KPC tumor-bearing mice compared to sham operated mice, respectively (Fig. 2B). The upregulated genes included MHC class II (e.g. H2-Ab1, H2-DMb1), interferon-inducible genes (e.g. Ifi207, Ifitm1, Ifi205), complement-related genes (e.g. C4b), and canonical markers of myeloid cells (e.g. Batf3, Mafb). The upregulation of Mt1 and Mt2 in the VAT of cancer bearing mice was also notable, as upregulation of these genes in WAT was previously reported in the context of fasting [46] and β3 adrenergic receptor agonism [47].
We performed GSEA using the KEGG and Reactome gene sets to identify differential expression patterns at a pathway level. This analysis highlighted a number of immune-related gene sets among the most strongly upregulated including pathways related to chemokine and cytokine signaling, JAK-STAT signaling, B cell receptor signaling, and antigen presentation (Fig. 2C, Supplementary Fig. S4). These pathway enrichments could be due to upregulation of the respective transcripts in one or more constituent cell types (e.g. adipocytes, stromal cells, or immune cells) or from an increase in the proportion of immune cells contributing to the total RNA content.
Among the cytokines analyzed, we found that IL-6 was most strongly elevated in the VAT of cancer bearing mice (Fig. 2D; Supplementary Table S1), which is consistent with observations from serum in other PDAC and CAC models as well as in human PDAC patients [8, 48]. IgG antibodies (both IgG1 and IgG2a) were also elevated in the AT of cancer bearing mice (Fig. 2E and F), which to our knowledge has not been previously reported. This increased antibody titer was not associated with an increase in the B cell fraction of AT-infiltrating CD45+ cells (Fig. 2G), and both IgG1 and IgG2a were increased in other tissues of cancer bearing mice, including subcutaneous AT, skeletal muscle, and brown AT (Supplementary Fig. S5A–C). Further, albumin levels were similar in VAT and muscle protein lysates from sham and cancer bearing mice (Supplementary Fig. S6). These data suggest that increased tissue IgG is likely not a consequence of local antibody production, increased vascular permeability, or excessive tissue contamination with blood. Other possibilities include selective uptake or retention of antibodies in these peripheral tissues by cells expressing Fc receptors, or increased antibody titers in circulation.
Together, this dataset identifies differentially regulated gene sets and provides a framework for future studies investigating potentially relevant pathways in AT wasting during the development of CAC.
The Th1 shift observed in obese AT is not present in the setting of cachexia
The finding that both type I and II interferon signaling pathways were upregulated in the VAT of cancer bearing mice (Fig. 3A and B, Supplementary Fig. S4) prompted us to investigate whether this reflected a shift toward IFNγ-high Th1 immunity as is observed in other AT pathologies such as obesity [23], versus an increase in hematopoietic cells expressing IFNγ response genes (Fig. 3C). At the protein level, we found that IFNγ and other Th1 cytokines and chemokines (TNFα, IL-2, CXCL9, CXCL10) were not significantly elevated in the AT of cancer bearing mice, suggesting that this transcriptional signature likely did not represent a bonafide shift toward Th1 inflammation (Fig. 3D–H; Supplementary Table S1). Consistent with these findings, there were no significant changes in the frequency of CD8+ T cells or dendritic cells in VAT of cancer bearing mice (Fig. 3I and J). There was also no difference in the total T cell fraction of VAT immune infiltrates in cachectic mice with CT26 tumors (Supplementary Fig. S7A).
Figure 3.
Th1 inflammation is not evident in the adipose tissue of cancer bearing mice. (A–B) Gene set enrichment plots depicting the upregulation of both IFNγ (A) and IFNα/β (B) signaling pathways in VAT of cancer bearing mice. (C) Heatmap depicting sample-level expression of genes in the IFNγ signaling pathway which are most strongly upregulated in the VAT of cancer bearing mice. (D–H) Cytokine array quantifications of Th1-related cytokines including IFNγ (D), TNFα (E), IL-2 (F), CXCL9 (G), and CXCL10 (H). (I-J) Quantification of cell types by flow cytometry, including CD8+ T cells (I) and CD11c+ dendritic cells (J). Statistical analyses were performed using the Mann–Whitney test; mean ± SD is shown.
To further explore whether cancer-associated AT wasting was associated with a Th1 response, we assessed PD-L1 expression in the AT of sham and tumor-bearing mice. PD-L1 is an IFNγ-inducible gene in multiple cell types including tumor cells and immune cells. We previously used a PD-L1-specific alpaca antibody fragment (VHH) to demonstrate PD-L1 expression in BAT in mice [49]. These VHHs, which are smaller than conventional antibodies and therefore have better tissue penetration, can also be linked to therapeutic cytokines such as IFNγ for local delivery [34]. Here, we confirmed by immunoblotting that PD-L1 protein is expressed predominantly in adipocytes from both BAT and VAT, consistent with our previous in depth analyses of PD-L1 expression on brown adipocytes [49] (Fig. 4A). To determine whether PD-L1 is induced by IFNγ signaling in AT, we treated non-tumor-bearing mice with intraperitoneal injections of PBS, a control VHH (1B7), a PDL1-targeted VHH-IFNγ fusion (B3-IFNγ), and a control VHH-IFNγ fusion (1B7-IFNγ) [34]. We confirmed that PD-L1 is induced at the protein level in both BAT and VAT upon in vivo treatment with B3-IFNγ and 1B7-IFNγ (Fig. 4B and C), and further confirmed the induction of IFNγ signaling by RT-qPCR for both Pdl1 and Irf1 (Fig. 4D and E). However, Pdl1 was not transcriptionally upregulated in VAT of cancer bearing mice as assessed by RNA-seq (Fig. 4F), nor was PD-L1 induced at the protein level in BAT or VAT of cancer bearing mice in a separate experiment (Fig. 4G and H). This concordant lack of PD-L1 induction provides further evidence for the lack of true Th1 skewing in wasting AT.
Figure 4.
PD-L1 expression in adipose tissue is regulated by IFNγ but is not increased in the setting of cancer. (A) Immunoblot depicting the expression of brown fat marker uncoupling protein 1 (UCP1), PD-L1, and beta actin in the whole tissue lysate (‘whole’) and stromovascular fraction (‘SVF’) of brown adipose tissue (BAT) and visceral adipose tissue (VAT). Samples were taken from Pdl1+/- (‘WT’) and Pdl1-/- (‘KO’) mice. The band corresponding to PD-L1 is indicated with arrows. (B-C) Western blot depicting the expression of PD-L1 and beta actin in VAT (B) and BAT (C) isolated 24 hours after the systemic administration of PBS, 1B7, B3-IFNγ, or 1B7-IFNγ. (D-E) RT-qPCR analysis depicting the expression of Irf1 and Pdl1 in the VAT (D) and BAT (E) isolated 24 hours after the systemic administration of PBS, 1B7, B3-IFNγ, or 1B7-IFNγ. The normalized expression ratio (NER) relative to the housekeeping gene beta-Actin is shown. (F) Comparison of Pdl1 expression by RNA-seq in VAT of sham-operated and KPC tumor-bearing mice. Values plotted are DESeq2-derived normalized expression values. The difference in Pdl1 expression between these groups was not statistically significant (P = 0.71). (G–H) Western blot depicting the expression of PD-L1 and beta actin in VAT (G) and BAT (H) of sham-operated and KPC tumor-bearing mice. Statistical analyses in (D) and (E) were performed using an ordinary two-way ANOVA with Dunnett’s multiple comparisons test; mean ± SD is shown. Immunoblot in (A) was blocked with 3% BSA in PBST, while subsequent PD-L1 blots were blocked with 5% BSA in PBST.
Th17 immunity is not increased in the VAT of cancer bearing mice
Th17 cells are rare in the AT of healthy mice but increase in the setting of obesity and contribute to pathologic obesity-associated inflammation [50]. To evaluate whether the Th17 milieu in AT changes with cancer, we measured the concentration of relevant cytokines including IL-17A, IL-17E (IL-25), IL-17F, IL-21, IL-22, IL-22, IL-23, IL-27, IL-31, CCL20 (MIP-3a), and lymphotoxin alpha (LT-a) (Supplementary Table S1). IL-23 and LT-a were significantly lower in VAT of cancer bearing mice compared to healthy VAT, and several other cytokines trended toward a reduction including IL-31 and IL-17E (Fig. 5A).
Figure 5.
Th17 immunity is not increased in the adipose tissue of cancer bearing mice. (A) Cytokine array quantifications of Th17 cytokines in VAT of sham-operated and cancer bearing mice. Separate plots are shown for those with high, medium, and low concentrations to improve visualization. (B) Quantification of γδ T cells in VAT as a fraction of all CD45+ cells (left) and as a fraction of T cells (right) defined by CD3 expression. (C) Cytokine array quantification of CXCL1 (KC), the murine homologue of the human neutrophil chemoattractant IL-8. (D) Quantification of neutrophils in VAT as a fraction of all CD45+ cells. Statistical analyses were performed using the Mann–Whitney test; mean ± SD is shown.
We also analyzed the γδ T cell population in VAT, as γδ T cells can drive Th17 inflammation in response to IL-23 [50] and are known to regulate AT homeostasis via production of IL-17A [29]. Consistent with the observed cytokine reductions, γδ T cells tended to be less abundant in the VAT of cancer bearing mice as a fraction of total CD45+ cells or T cells, although these reductions did not reach statistical significance (Fig. 5B). Since Th17 inflammation also promotes neutrophil recruitment, we investigated whether neutrophils or their chemoattractants were altered in cancer bearing mice. The amount of CXCL1 and the fraction of neutrophils were similar in the VAT of sham-operated and cancer bearing mice (Fig. 5C and D). Similarly, there was no change in the neutrophil fraction of CD45+ cells in cachectic mice with CT26 tumors compared to sham injected mice (Supplementary Fig. S7B). We thus conclude that there is not a shift toward Th17 inflammation in wasting VAT of mice which are developing or have already developed CAC.
Th2 immunity is maintained in wasting AT of cancer bearing mice
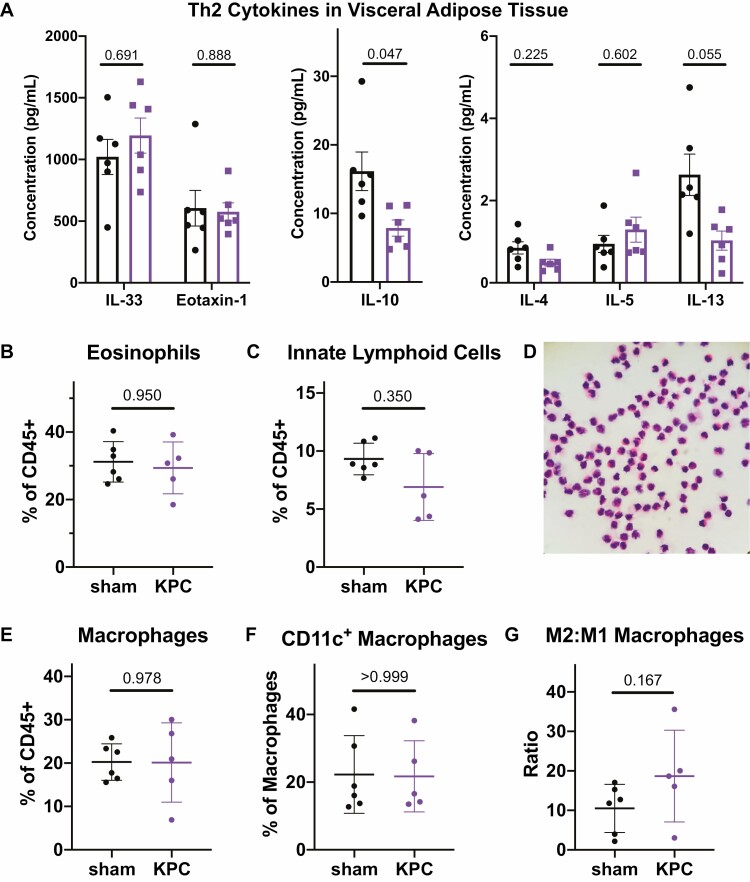
To assess whether the regulatory milieu that characterizes AT in the healthy state is altered in the setting of cancer, we measured the concentrations of Th2 cytokines including IL-4, IL-5, IL-10, IL-13, IL-33, and eotaxin-1 (Fig. 6A; Supplementary Table S1). IL-4 and IL-5 were detected at low levels with no discernable difference between the groups, while IL-10 and IL-13 showed slight reductions in the VAT of cancer bearing mice. Both IL-33 and eotaxin-1 were present at high levels in healthy VAT and were not altered in cancer bearing mice.
Figure 6.
Th2 immunity is maintained in adipose tissue of cancer bearing mice. (A) Cytokine array quantifications of Th2 cytokines in VAT of sham-operated and cancer bearing mice. Separate plots are shown for those with high, medium, and low concentrations to improve visualization. (B–C) Quantification of eosinophils (B) and innate lymphoid cells (C) as a fraction of total CD45+ cells in VAT by flow cytometry. (D) Hematoxylin and eosin staining of cytospin with sorted CD45+CD11b+SiglecF+ cells from VAT, confirming the eosinophil identity of the cell population defined by this gate in flow cytometry. (E) Quantification of macrophages (CD11b+F4/80hi) as a fraction of total CD45+ cells in VAT by flow cytometry. (F) Quantification of inflammatory CD11c+ macrophages in VAT as a fraction of total macrophages. (G) Ratio of M2 to M1 macrophages in VAT, where M1 phenotype is defined by CD11c expression. Statistical analyses were performed using the Mann–Whitney test; mean ± SD is shown.
Consistent with the high concentration of eotaxin-1 and IL-33, we found robust populations of VAT eosinophils and ILCs which were present at similar frequencies in both groups (Fig. 6B and C). We confirmed the identity of the rather large population of eosinophils (~30% of all CD45+ cells) by hematoxylin and eosin (H&E) staining on a cytospin of sorted CD45+CD11b+SiglecF+ cells from VAT (Fig. 6D). VAT macrophages comprised a similar fraction of total CD45+ cells in sham and tumor-bearing mice, and macrophages in both groups displayed a predominantly anti-inflammatory phenotype as determined by the lack of CD11c expression (Fig. 6E–G) [51]. In cachectic mice with CT26 tumors, there was a slight reduction in the eosinophil fraction of VAT CD45+ cells and no change in the macrophage fraction (Supplementary Fig. S7C,D). This contrasting reduction in eosinophils observed here versus the maintained eosinophil population in KPC tumor-bearing mice could reflect a difference in the tumor models used (i.e. KPC vs. CT26), a generalizable difference in the cancer types studied (i.e. pancreatic cancer versus colorectal cancer), or a difference in the degree of systemic disease (i.e. isolated AT wasting versus outright cachexia).
AT of human cancer patients is characterized by a Th2 immune environment
To determine whether our findings in murine models of cancer associated AT wasting and cachexia are applicable in humans, we analyzed VAT from colorectal cancer patients by cytokine profiling and flow cytometry. Consistent with our preclinical data, the most abundant cytokines included IL-6, IL-33, and eotaxin-2 (Fig. 7A). Eotaxin-1 and eotaxin-3 were notably absent in human VAT (Fig. 7A and Supplementary Table S2), a pattern departing from our observations in the murine VAT where eotaxin-1 is detected at high levels (Fig. 6A). Consistent with the abundance of eotaxin-2 and IL-33, we identified eosinophils across several human VAT samples by flow cytometry for SIGLEC8 (Fig. 7B) and for CCR3 (Fig. 7C), and we confirmed their presence by cytospin and H&E staining of whole stromovascular fraction preparations (Fig. 7D). Th1 cytokines such as IFNγ, TNFα, and IL-2 were either undetectable or present at very low levels in these human VAT samples (Fig. 7A), indicating a lack of Th1 skewing in the AT of cancer patients. In summary, these results are consistent with our mouse models and suggest that the Th2 environment in human AT is maintained in the setting of cancer.
Figure 7.
Th2 immunity is present in the adipose tissue of human cancer patients. (A) Cytokine array shows high levels of IL-6, IL-33, and eotaxin-2 in VAT from humans with colorectal cancer (n = 5), with low levels of eotaxin-1, IFNγ, and TNFα detected. Mean ± SD is shown. (B–C) Flow cytometry of human VAT stromovascular fraction (SVF) captures eosinophil population defined by coexpression of CD45, CD15, and SIGLEC8 (B) or CD45, CD15, and CCR3 (C). Each panel corresponds to a unique patient. (D) Hematoxylin and eosin staining of whole SVF from human VAT identifies eosinophils among other tissue-resident immune cells.
Discussion
CAC is a significant morbidity among patients with pancreatic cancer and other tumor types, but our understanding of the mechanisms driving this disease is incomplete [1]. Many prior mechanistic studies have focused on identifying proteins that are elevated in serum in the context of CAC-inducing tumors and studying the effects of those proteins on parenchymal cells in cachexia target organs. In this way, we have learned how factors such as IL-6, TNFα, activin, and others can promote autophagy and proteasomal degradation leading to myocyte atrophy, activate lipolysis and beiging processes in adipocytes, or reduce ketogenesis in hepatocytes [6, 8, 12, 48, 52]. However, recent studies have highlighted cellular immune changes in the periphery [19] and in the central nervous system [20] that may contribute to the development of CAC. Interestingly, while the AT immune landscape in health and disease has been actively investigated [23, 53], whether it changes during or contributes to the development of CAC is still unknown.
Our results indicate that the regulatory Th2 signature is maintained in AT that is wasting in the context of tumor progression, in contrast to the strong inflammatory skewing observed in other AT pathologies such as obesity and diabetes [23]. Our flow cytometry analyses did not identify changes in proportions of the surveyed cell types in cancer bearing mice, but whether the functional profile of resident immune cells such as eosinophils or macrophages change in the setting of CAC still needs to be determined. It is notable that our RNA-seq analysis identified the complement system among the most strongly upregulated gene sets in VAT of cancer bearing mice (Fig. 2C). Previous single-cell RNA-seq studies have identified subsets of complement-high macrophages [54, 55], and we may be detecting a shift toward this macrophage state rather than a shift along the classically described M1/M2 axis.
In light of the fact that obese AT is skewed toward Th1 and Th17 inflammation and away from regulatory Th2 immunity [23, 29], the absence of these shifts in cachexia may not seem surprising. Indeed, in some ways obesity and cachexia can be viewed as opposite ends on a spectrum of adipose homeostasis. While obesity is characterized by a positive energy balance with excess deposition of AT often in the context of increased food intake [56], cachexia is a disease of negative energy balance with loss of AT often in the setting of reduced food intake [57]. Defective thermogenesis has been suggested as an underlying mechanism of obesity, while activation of thermogenesis has been reported in cachectic AT and suggested as a therapeutic modality for obesity [8, 58]. As type 2 immune circuits involving eosinophils, innate lymphoid cells, and regulatory macrophages are known to maintain thermogenic capacity in adipocytes [24–26], we may not necessarily expect these cell types to decline in cachexia as they do in obese AT.
Obesity and cachexia also have important similarities that warrant consideration. For example, a hallmark of obesity is the resulting insulin resistance that critically drives the onset of type 2 diabetes [59]. Under healthy circumstances insulin stimulates lipogenesis and inhibits lipolysis in AT, and accordingly increased basal lipolysis due to insulin resistance is observed in obesity [60]. Insulin resistance has also been suggested to contribute to the pathophysiology of CAC [61], and this may be especially important in the context of PDAC patients, in whom cachexia and hyperglycemia can occur even months or years prior to the diagnosis of cancer, respectively. And while hyperglycemia in PDAC patients may be partially a consequence of pancreatic beta cell failure or loss, the fact that new-onset hyperglycemia occurs years prior to diagnosis in patients with small tumors which are not yet detectable by radiological imaging suggests that systemic changes in insulin sensitivity also occur in PDAC patients [2, 62–64]. As many of the immunologic changes described in obese AT are thought to drive insulin resistance, similar changes could occur in cachectic AT as well. While we did not observe notable shifts in the AT immune composition in our model, this does not rule out the possibility of such changes in other CAC models or cachexia patients.
Increased IL-6 in the VAT of cancer bearing mice was among the strongest changes observed in our experiments, and it was highly abundant in the VAT of colorectal cancer patients. IL-6 is elevated in the serum of cancer patients, and its local abundance in VAT suggests that it could mediate cachexia via direct interactions with cells in this tissue. Our findings suggest that even a robust local elevation in IL-6 does not orchestrate major immunologic shifts that contribute to AT wasting, but it is possible that IL-6 affects adipocytes directly to drive this process. Unlike IL-6, the cachectin TNFα was only detected at very low levels in VAT of cancer bearing mice and cancer patients, suggesting that the cachexia-inducing effects of TNFα are likely not mediated locally in AT. It is worth noting that various other secreted proteins have also been implicated in the pathogenesis of cancer associated AT wasting and cachexia. For example, PTHrP signaling in AT has been shown to drive browning and loss of AT along with muscle wasting in models of cachexia secondary to cancer and renal failure. While we did not quantify the amount of other known cachexia inducing factors such as PTHrP at the protein level, such factors were not transcriptionally elevated in AT of cancer bearing mice.
We also observed a strong increase in IgG levels in the VAT of cancer bearing mice. The consistent fraction of B cells in VAT and the finding of elevated IgG at other sites suggests that this is not due to increased local antibody production. Further, the lack of a corresponding increase in tissue albumin levels suggests that the IgG increase is not simply a consequence of increased vascular permeability, excessive vascularity, or blood contamination of our tissue samples. Other potential mechanisms include enhanced selective transport of IgG into peripheral tissues or IgG retention in tissues by Fc-receptor expressing cells. The consequences of antibody deposition in CAC target tissues are unknown. Macrophages and dendritic cells express high levels of Fcγ receptors, and interactions between these receptors and IgG (alone or in immune complexes) can affect the activation state of these cells [65]. Whether increased tissue IgG levels shape the functional immune landscape of VAT or contribute to the pathogenesis of CAC should be investigated.
This study has implications for the field of immunotherapy in both oncology and metabolic disease. First, there is an established association in cancer patients between adiposity and responsiveness to immune checkpoint blockade [66–68]. The mechanisms underlying this association are not clear, but it could reflect an effect of these treatments on AT-resident immune cells. Indeed, pre-clinical data supports a role for AT as a reservoir for memory T cells which can drive recall responses to protect against viral infection [69]. Whether a similar axis exists to coordinate anti-tumor immune responses is not established. Second, anti-PDL1 therapy is approved for several cancer types including lung, breast, and bladder cancers [70]. T cell activation in response to immune checkpoint blockade can increase IFNγ production and signaling both intratumorally and systemically. The baseline expression of PD-L1 in AT and its significant capacity for induction by IFNγ treatment which we demonstrated here may be pharmacodynamically relevant, as AT could serve as a sink for anti-PDL1 monoclonal antibodies. Finally, while modulation of the AT microenvironment has been studied pre-clinically to reverse metabolic phenotypes such as obesity and diabetes, its potential utility in the treatment of cachexia has been relatively understudied to date.
There are several limitations of this study. First, only female mice were evaluated, which we selected for their more consistent body weights and ease of randomization. However, as PDAC and CAC are more common in males [71, 72], further studies are warranted to characterize whether sex impacts the AT immune environment in the setting of cancer. Second, our experiments did not address whether obesity, which is a common comorbidity in PDAC patients, impacts the development of cachexia or associated immunologic shifts in AT. Third, our analyses of human AT were limited to patients with colorectal cancer and relied on published literature for a description of healthy AT. Finally, while we did not observe alterations in the frequencies of the most prominent immune cell types, there could be differences in more granular cell subsets which would be revealed by techniques such as single cell transcriptional profiling.
Our study represents a comprehensive characterization of the visceral adipose immune landscape in healthy and cancer bearing mice, and it informs us of the human adipose cytokine profile in the setting of cancer. We find that IL-6 and IgG are strongly elevated in the wasting AT of mice with PDAC, but the Th2 immune environment is generally maintained at the transcript, protein, and cellular levels. We also demonstrate that PD-L1 can be induced by IFNγ in both white and brown adipose tissue, although it is not induced by the presence of a tumor. Whether adipocyte PD-L1 expression increases in cancer patients receiving immunotherapy remains an important question. Our integrated analysis of RNA-sequencing, cytokine array profiling, and flow cytometry can serve as a framework for subsequent evaluations of tissues affected by CAC in preclinical models and cancer patients.
Supplementary material
Supplementary data are available at Immunotherapy Advances online.
Figure S1. Flow cytometry gating strategy for myeloid panel in murine visceral adipose tissue. Cells quantified include eosinophils, neutrophils, macrophages (M1 and M2), dendritic cells, B cells, T cells, and innate lymphoid cells.
Figure S2. Flow cytometry gating strategy for lymphoid panel in murine visceral adipose tissue. Cells quantified include B cells and T cell subsets, including γδ T cells, CD4+ αβ T cells, and CD8+ αβ T cells.
Figure S3. Flow cytometry gating strategy for visceral adipose tissue in CT26 tumor bearing mice. Cells quantified include total CD45+ cells, eosinophils, neutrophils (Ly6G+), macrophages, and T cells.
Figure S4. Gene set enrichment analysis of RNA-sequencing data using Reactome gene sets. Several immune-related pathways are upregulated, including type I and II interferon signaling and the complement cascade (highlighted in red).
Figure S5. IgG1 is elevated in sites other than visceral adipose tissue in mice bearing orthotopic KPC pancreatic tumors. (A) Quantification of IgG1 levels by ELISA in whole tissue protein lysates from subcutaneous adipose tissue (A), quadriceps muscle (B), and brown adipose tissue (C). Statistical analyses were performed using the Mann-Whitney test; mean ± SD is shown.
Figure S6. Albumin levels are similar in AT and skeletal muscle from sham operated and KPC tumor bearing mice. Albumin was assessed by immunoblot in whole tissue lysates of visceral adipose tissue (VAT) and quadriceps muscle (Quad). Arrow indicates the band corresponding to albumin. 20 mg of protein were loaded per sample.
Figure S7. Quantification of cell types in VAT of sham operated and CT26 tumor bearing mice by flow cytometry. (A) T cells were defined as CD45+SigF-Ly6G-CD11b-F4/80-CD3+. (B) Neutrophils were defined as CD45+Ly6G+SigF-. (C) Eosinophils were defined as CD45+SigF+Ly6G-. (D) Macrophages were defined as CD45+Ly6G-SigF-CD11b+F4/80+.
Supplemental Table 1. Summary of cytokine abundance in VAT from sham and KPC tumor bearing mice. For sham and KPC groups (n = 6 each), the average and standard deviation of the normalized concentrations are given. Cytokine concentrations were obtained in units of pg/mL. For each sample, these concentrations were normalized to the total protein concentration from the whole tissue lysate such that the reported quantities reflect the relative amount of each cytokine in the tissue.
Supplemental Table 2. Summary of cytokine abundance in VAT from colorectal cancer patients. The mean, median, and standard deviation of the normalized concentrations are given for each cytokine (n = 5). Cytokine concentrations were obtained in units of pg/mL. For each sample, these concentrations were normalized to the total protein concentration from the whole tissue lysate such that the reported quantities reflect the relative amount of each cytokine in the tissue.
Acknowledgements
The Editor-in-Chief, Tim Elliott, and handling editor, Adriana Bonomo, would like to thank the following reviewer, Kristyn Gumpper, and an anonymous reviewer, for their contribution to the publication of this article.
Glossary
Abbreviations
- AT
adipose tissue
- ATM
adipose tissue macrophage
- BAT
brown adipose tissue
- CAC
cancer-associated cachexia
- ELISA
enzyme-linked immunosorbent assay
- GLM
generalized linear model
- GSEA
Gene Set Enrichment Analysis
- ILC
innate lymphoid cell
- PDAC
pancreatic ductal adenocarcinoma
- SVF
stromovascular fraction
- Th1
T helper 1
- Th2
T helper 2
- Th17
T helper 17
- VAT
visceral adipose tissue
- WAT
white adipose tissue
Author contributions
P.L.: Conceptualization, Data curation, Investigation, Formal analysis, Methodology, Visualization, Funding acquisition, Writing – original draft. A.C.: Investigation, Writing – review and editing. A.M.U.: Investigation, Writing – review and editing. S.J.C.: Investigation, Writing – review and editing. T.S.: Project administration, Writing – review and editing. G.B.: Supervision, Resources, Writing – review and editing. M.D.: Conceptualization, Supervision, Writing – review and editing. S.K.D.: Conceptualization, Supervision, Funding acquisition, Writing – review and editing. M.H.: Conceptualization, Data curation, Investigation, Formal analysis, Methodology, Funding acquisition, Visualization, Supervision, Writing – review and editing.
Funding
P.L. was funded by the PL was funded by the National Institute of General Medical Sciences (T32GM007753) and the National Cancer Institute (T32CA207021). A.C. was funded by the Giovanni Armenise-Harvard Foundation. A.M.U. was funded by NIH T32DK007191. S.J.C. was funded by the Claudia Adams Barr Foundation. S.K.D. was funded by the Emerson Foundation, the Hale Center for Pancreatic Cancer Research, and by NIH U01 CA224146-01. M.H. was funded by the German Research Foundation (DFG; project number: 398222819).
Conflict of interest
The authors have no competing interests to declare.
Data availability
Raw RNA-sequencing data is deposited in the NCBI Gene Expression Omnibus under accession number GEO: GSE171402 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE171402). Data from mouse and human cytokine arrays are available with group level mean, standard deviation, and sample count in Table S1 and Table S2, respectively.
References
- 1. Baracos VE, Martin L, Korc Met al. Cancer-associated cachexia. Nature Reviews Disease Primers 2018;4:17105. 10.1038/nrdp.2017.105 [DOI] [PubMed] [Google Scholar]
- 2. Hendifar AE, Chang JI, Huang BZet al. Cachexia, and not obesity, prior to pancreatic cancer diagnosis worsens survival and is negated by chemotherapy. J Gastrointest Oncol 2018;99(1):17–23. 10.21037/jgo.2017.11.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bachmann J, Büchler MW, Friess Het al. Cachexia in patients with chronic pancreatitis and pancreatic cancer: impact on survival and outcome. Nutr Cancer 2013;65(6):827–33. 10.1080/01635581.2013.804580 [DOI] [PubMed] [Google Scholar]
- 4. Nemer L, Krishna SG, Shah ZKet al. Predictors of pancreatic cancer-associated weight loss and nutritional interventions. Pancreas 2017;46(9):1152–7. 10.1097/MPA.0000000000000898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Argils JM, López-Soriano FJ, Stemmler Bet al. Therapeutic strategies against cancer cachexia. Eur J Transl Myol 2019;29(1):7960. 10.4081/ejtm.2019.7960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Torti FM, Dieckmann B, Beutler Bet al. A macrophage factor inhibits adipocyte gene expression: an in vitro model of cachexia. Science 1985;229(4716):867–9. 10.1126/science.3839597 [DOI] [PubMed] [Google Scholar]
- 7. Miller A, McLeod L, Alhayyani S. et al. Blockade of the IL-6 trans-signalling/STAT3 axis suppresses cachexia in Kras-induced lung adenocarcinoma. Oncogene 2017;36(21):3059–66. 10.1038/onc.2016.437 [DOI] [PubMed] [Google Scholar]
- 8. Petruzzelli M, Schweiger M, Schreiber Ret al. A switch from white to brown fat increases energy expenditure in cancer-associated cachexia. Cell Metab 2014;20(3):433–47. 10.1016/j.cmet.2014.06.011 [DOI] [PubMed] [Google Scholar]
- 9. Hirata H, Tetsumoto S, Kijima T. et al. Favorable responses to tocilizumab in two patients with cancer-related cachexia. J Pain Symptom Manage 2013;46(2):e9–e13. 10.1016/j.jpainsymman.2013.01.009 [DOI] [PubMed] [Google Scholar]
- 10. Ramsey ML, Talbert E, Ahn Det al. Circulating interleukin-6 is associated with disease progression, but not cachexia in pancreatic cancer. Pancreatology 2019;19(1):80–7. 10.1016/j.pan.2018.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mayers JR, Wu C, Clish CBet al. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat Med 2014;20(10):1193–8. 10.1038/nm.3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Acharyya S, Ladner KJ, Nelsen LLet al. Cancer cachexia is regulated by selective targeting of skeletal muscle gene products. J Clin Invest 2004;114(3):370–8. 10.1172/JCI20174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aversa Z, Pin F, Lucia Set al. Autophagy is induced in the skeletal muscle of cachectic cancer patients. Sci Rep 2016;6(1):30340. 10.1038/srep30340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pettersen K, Andersen S, Degen Set al. Cancer cachexia associates with a systemic autophagy-inducing activity mimicked by cancer cell-derived IL-6 trans-signaling. Sci Rep 2017;7:2046. 10.1038/s41598-017-02088-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bing C, Trayhurn P. Regulation of adipose tissue metabolism in cancer cachexia. Curr Opin Clin Nutr Metab Care 2008;11(3):201–7. 10.1097/MCO.0b013e3282f948e2 [DOI] [PubMed] [Google Scholar]
- 16. Zuijdgeest-van Leeuwen SD, van den Berg JW, Wattimena JLet al. Lipolysis and lipid oxidation in weight-losing cancer patients and healthy subjects. Metabolism 2000;49(7):931–6. 10.1053/meta.2000.6740 [DOI] [PubMed] [Google Scholar]
- 17. Das SK, Eder S, Schauer Set al. Adipose triglyceride lipase contributes to cancer-associated cachexia. Science 2011;333(6039):233–8. 10.1126/science.1198973 [DOI] [PubMed] [Google Scholar]
- 18. Kir S, White JP, Kleiner Set al. Tumour-derived PTH-related protein triggers adipose tissue browning and cancer cachexia. Nature 2014;513(7516):100–4. 10.1038/nature13528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barker T, Fulde G, Moulton Bet al. An elevated neutrophil-to-lymphocyte ratio associates with weight loss and cachexia in cancer. Sci Rep 2020;10(1):7535. 10.1038/s41598-020-64282-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Burfeind KG, Zhu X, Norgard MAet al. Circulating myeloid cells invade the central nervous system to mediate cachexia during pancreatic cancer. 2020; 9. 10.7554/eLife.54095 Available from: https://elifesciences.org/articles/54095 (3 December 2020, date last accessed). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Erdem M, Möckel D, Jumpertz Set al. Macrophages protect against loss of adipose tissue during cancer cachexia. J Cachexia Sarcopenia Muscle 2019;10(5):1128–42. 10.1002/jcsm.12450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. de Matos-Neto EM, Lima JD, de Pereira WOet al. Systemic inflammation in cachexia - is tumor cytokine expression profile the culprit? Front Immunol 2015;6:629. 10.3389/fimmu.2015.00629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kane H, Lynch L. Innate immune control of adipose tissue homeostasis. Trends Immunol 2019;40(9):857–72. 10.1016/j.it.2019.07.006 [DOI] [PubMed] [Google Scholar]
- 24. Qiu Y, Nguyen KD, Odegaard JIet al. Eosinophils and type 2 cytokine signaling in macrophages orchestrate development of functional beige fat. Cell 2014;157(6):1292–308. 10.1016/j.cell.2014.03.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee MW, Odegaard JI, Mukundan Let al. Activated type 2 innate lymphoid cells regulate beige fat biogenesis. Cell 2015;160(0):74–87. 10.1016/j.cell.2014.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Knights AJ, Vohralik EJ, Houweling PJet al. Eosinophil function in adipose tissue is regulated by Krüppel-like factor 3 (KLF3). Nat Commun 2020;11(1):2922. 10.1038/s41467-020-16758-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vasanthakumar A, Moro K, Xin Aet al. The transcriptional regulators IRF4, BATF and IL-33 orchestrate development and maintenance of adipose tissue-resident regulatory T cells. Nat Immunol 2015;16(3):276–85. 10.1038/ni.3085 [DOI] [PubMed] [Google Scholar]
- 28. Burzyn D, Kuswanto W, Kolodin D. et al. A special population of regulatory T cells potentiates muscle repair. Cell 2013;155(6):1282–95. 10.1016/j.cell.2013.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kohlgruber AC, Gal-Oz ST, LaMarche NMet al. γδ T cells producing interleukin-17A regulate adipose regulatory T cell homeostasis and thermogenesis. Nat Immunol 2018;19(5):464–74. 10.1038/s41590-018-0094-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu D, Molofsky AB, Liang HEet al. Eosinophils sustain adipose alternatively activated macrophages associated with glucose homeostasis. Science 2011;332(6026):243–7. 10.1126/science.1201475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lynch L, Michelet X, Zhang Set al. Regulatory iNKT cells lack expression of the transcription factor PLZF and control the homeostasis of T(reg) cells and macrophages in adipose tissue. Nat Immunol 2015;16(1):85–95. 10.1038/ni.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Melief CJM. The future of immunotherapy. Immunother Advanc. 2021;1(1). 10.1093/immadv/ltaa005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ramamurthy D, Nundalall T, Cingo Set al. Recent advances in immunotherapies against infectious diseases. Immunother Advanc. 2021;1(1). 10.1093/immadv/ltaa007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dougan M, Ingram JR, Jeong HJet al. Targeting cytokine therapy to the pancreatic tumor microenvironment using PD-L1-specific VHHs. Cancer Immunol Res 2018;6(4):389–401. 10.1158/2326-6066.CIR-17-0495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Patro R, Duggal G, Love MIet al. Salmon: fast and bias-aware quantification of transcript expression using dual-phase inference. Nat Methods 2017;14(4):417. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 2015;4:1521. 10.12688/f1000research.7563.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15(12):550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stephens M. False discovery rates: a new deal. Biostatistics 2017;18(2):275–94. 10.1093/biostatistics/kxw041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Subramanian A, Tamayo P, Mootha VKet al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102(43):15545–50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liberzon A, Subramanian A, Pinchback Ret al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011; 27(12):1739–40. 10.1093/bioinformatics/btr260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jassal B, Matthews L, Viteri Get al. The reactome pathway knowledgebase. Nucleic Acids Res 2020;48(D1):D498–503. 10.1093/nar/gkz1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hingorani SR, Wang L, Multani ASet al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005;7(5):469–83. 10.1016/j.ccr.2005.04.023 [DOI] [PubMed] [Google Scholar]
- 43. Roberts EW, Deonarine A, Jones JOet al. Depletion of stromal cells expressing fibroblast activation protein-α from skeletal muscle and bone marrow results in cachexia and anemia. J Exp Med 2013;210(6):1137–51. 10.1084/jem.20122344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Michaelis KA, Zhu X, Burfeind KGet al. Establishment and characterization of a novel murine model of pancreatic cancer cachexia. J Cachexia Sarcopenia Muscle 2017;8(5):824–38. 10.1002/jcsm.12225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hatakeyama S, Summermatter S, Jourdain Met al. ActRII blockade protects mice from cancer cachexia and prolongs survival in the presence of anti-cancer treatments. Skelet Muscle 2016;6:26. 10.1186/s13395-016-0098-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Szrok S, Stelmanska E, Turyn Jet al. Metallothioneins 1 and 2, but not 3, are regulated by nutritional status in rat white adipose tissue. Genes Nutr 2016;11(1):18. 10.1186/s12263-016-0533-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Trayhurn P, Duncan JS, Wood AMet al. Metallothionein gene expression and secretion in white adipose tissue. Am J Physiol Regul Integr Comp Physiol 2000;279(6):R2329–35. 10.1152/ajpregu.2000.279.6.R2329 [DOI] [PubMed] [Google Scholar]
- 48. Flint TR, Janowitz T, Connell CMet al. Tumor-induced IL-6 reprograms host metabolism to suppress anti-tumor immunity. Cell Metab 2016;24(5):672–84. 10.1016/j.cmet.2016.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ingram JR, Dougan M, Rashidian Met al. PD-L1 is an activation-independent marker of brown adipocytes. Nat Commun 2017;8(1):647. 10.1038/s41467-017-00799-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McGeachy MJ, Cua DJ, Gaffen SL. The IL-17 family of cytokines in health and disease. Immunity 2019;50(4):892–906. 10.1016/j.immuni.2019.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ono Y, Nagai M, Yoshino Oet al. CD11c+ M1-like macrophages (MΦs) but not CD206+ M2-like MΦ are involved in folliculogenesis in mice ovary. Sci Rep. 2018;8(8171). 10.1038/s41598-018-25837-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhong X, Pons M, Poirier Cet al. The systemic activin response to pancreatic cancer: implications for effective cancer cachexia therapy. J Cachexia Sarcopenia Muscle 2019;10(5):1083–101. 10.1002/jcsm.12461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Weisberg SP, McCann D, Desai Met al. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest 2003;112(12):1796–808. 10.1172/JCI19246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Qian J, Olbrecht S, Boeckx Bet al. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res 2020;30(9):745–62. 10.1038/s41422-020-0355-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. MacParland SA, Liu JC, Ma XZet al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun 2018;9(1):4383. 10.1038/s41467-018-06318-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Heymsfield SB, Wadden TA. Mechanisms, pathophysiology, and management of obesity. N Engl J Med 2017;376(3):1492. 10.1056/NEJMra1514009 [DOI] [PubMed] [Google Scholar]
- 57. Ezeoke CC, Morley JE. Pathophysiology of anorexia in the cancer cachexia syndrome. J Cachexia Sarcopenia Muscle 2015;6(4):287. 10.1002/jcsm.12059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Betz MJ, Enerbäck S. Targeting thermogenesis in brown fat and muscle to treat obesity and metabolic disease. Nat Rev Endocrinol 2018;14(2):77–87. 10.1038/nrendo.2017.132 [DOI] [PubMed] [Google Scholar]
- 59. Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 2006;444(7121):840–6. 10.1038/nature05482 [DOI] [PubMed] [Google Scholar]
- 60. Duncan RE, Ahmadian M, Jaworski Ket al. Regulation of lipolysis in adipocytes. Annu Rev Nutr 2007;27:79–101. 10.1146/annurev.nutr.27.061406.093734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dev R, Bruera E, Dalal S. Insulin resistance and body composition in cancer patients. Ann Oncol. 2018;29(suppl_2): ii18 – ii26. 10.1093/annonc/mdx815 [DOI] [PubMed] [Google Scholar]
- 62. Sharma A, Smyrk TC, Levy MJet al. Fasting blood glucose levels provide estimate of duration and progression of pancreatic cancer before diagnosis. Gastroenterology 2018;155(2):490–500.e2. 10.1053/j.gastro.2018.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chari ST, Leibson CL, Rabe KGet al. Pancreatic cancer-associated diabetes mellitus: prevalence and temporal association with diagnosis of cancer. Gastroenterology 2008;134(1):95–101. 10.1053/j.gastro.2007.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pelaez-Luna M, Takahashi N, Fletcher JGet al. Resectability of presymptomatic pancreatic cancer and its relationship to onset of diabetes: a retrospective review of CT scans and fasting glucose values prior to diagnosis. Am J Gastroenterol 2007;102(10):2157–63. 10.1111/j.1572-0241.2007.01480.x [DOI] [PubMed] [Google Scholar]
- 65. Guilliams M, Bruhns P, Saeys Yet al. The function of Fcγ receptors in dendritic cells and macrophages. Nat Rev Immunol 2014;14(2):94–108. 10.1038/nri3582 [DOI] [PubMed] [Google Scholar]
- 66. McQuade JL, Daniel CR, Hess KRet al. Association of body-mass index and outcomes in patients with metastatic melanoma treated with targeted therapy, immunotherapy, or chemotherapy: a retrospective, multicohort analysis. Lancet Oncol 2018;19(3):310–22. 10.1016/S1470-2045(18)30078-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang Z, Aguilar EG, Luna JIet al. Paradoxical effects of obesity on T cell function during tumor progression and PD-1 checkpoint blockade. Nat Med 2019;25(1):141–51. 10.1038/s41591-018-0221-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cortellini A, Bersanelli M, Buti Set al. A multicenter study of body mass index in cancer patients treated with anti-PD-1/PD-L1 immune checkpoint inhibitors: when overweight becomes favorable. J Immunother Cancer 2019;7(1):57. 10.1186/s40425-019-0527-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Han SJ, Glatman Zaretsky A, Andrade-Oliveira Vet al. The white adipose tissue is a reservoir for memory T cells and promotes protective memory responses to infection. Immunity 2017;47(6):1154–1168.e6. 10.1016/j.immuni.2017.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Akinleye A, Rasool Z. Immune checkpoint inhibitors of PD-L1 as cancer therapeutics. J Hematol Oncol 2019;12(1):92. 10.1186/s13045-019-0779-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Becker AE, Hernandez YG, Frucht Het al. Pancreatic ductal adenocarcinoma: risk factors, screening, and early detection. World J Gastroenterol 2014;20(32):11182–98. 10.3748/wjg.v20.i32.11182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhong X, Zimmers TA. Sex differences in cancer cachexia. Curr Osteoporos Rep 2020;18(6):646–54. 10.1007/s11914-020-00628-w [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw RNA-sequencing data is deposited in the NCBI Gene Expression Omnibus under accession number GEO: GSE171402 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE171402). Data from mouse and human cytokine arrays are available with group level mean, standard deviation, and sample count in Table S1 and Table S2, respectively.