Abstract
Genetically identical individuals in bacterial populations can display significant phenotypic variability. This variability can be functional, for example by allowing a fraction of stress prepared cells to survive an otherwise lethal stress. The optimal fraction of stress prepared cells depends on environmental conditions. However, how bacterial populations modulate their level of phenotypic variability remains unclear. Here we show that the alternative sigma factor σV circuit in Bacillus subtilis generates functional phenotypic variability that can be tuned by stress level, environmental history and genetic perturbations. Using single‐cell time‐lapse microscopy and microfluidics, we find the fraction of cells that immediately activate σV under lysozyme stress depends on stress level and on a transcriptional memory of previous stress. Iteration between model and experiment reveals that this tunability can be explained by the autoregulatory feedback structure of the sigV operon. As predicted by the model, genetic perturbations to the operon also modulate the response variability. The conserved sigma‐anti‐sigma autoregulation motif is thus a simple mechanism for bacterial populations to modulate their heterogeneity based on their environment.
Keywords: Bacillus subtilis, microbial systems biology, single‐cell time‐lapse microscopy, stochastic gene expression, stress priming
Subject Categories: Microbiology, Virology & Host Pathogen Interaction
A combination of single‐cell imaging and mathematical modelling reveals a simple mechanism for bacterial populations to modulate their heterogeneity based on their environment and environmental history.

Introduction
Cells live in a changeable environment and experience a wide range of environmental stresses. Bacterial populations have evolved strategies to survive these stresses. One strategy is for all cells to immediately respond to stress with the activation of the relevant stress response pathway (Hilker et al, 2016). Alternatively, a bacterial population can generate a broad range of cellular states, which allows it to hedge its bets against the changeable environment (Veening et al, 2008b). Noise in gene expression has been proposed as a mechanism for generating phenotypic variability in genetically identical cells (Raj & van Oudenaarden, 2008; Martins & Locke, 2015). This phenotypic variability has also been shown to be affected by changes in the cellular environment, such as a shift in stress level or growth conditions (Megerle et al, 2008; de Jong et al, 2012; Mitosch et al, 2019), as well as by previous ‘priming’ stresses (Mitosch et al, 2017). However, how the bacterial population regulates individual cell decisions to modulate the fraction of cells that enter an alternative transcriptional state remains unclear (Fig 1A).
Figure 1. σV is activated heterogeneously in response to lysozyme stress.
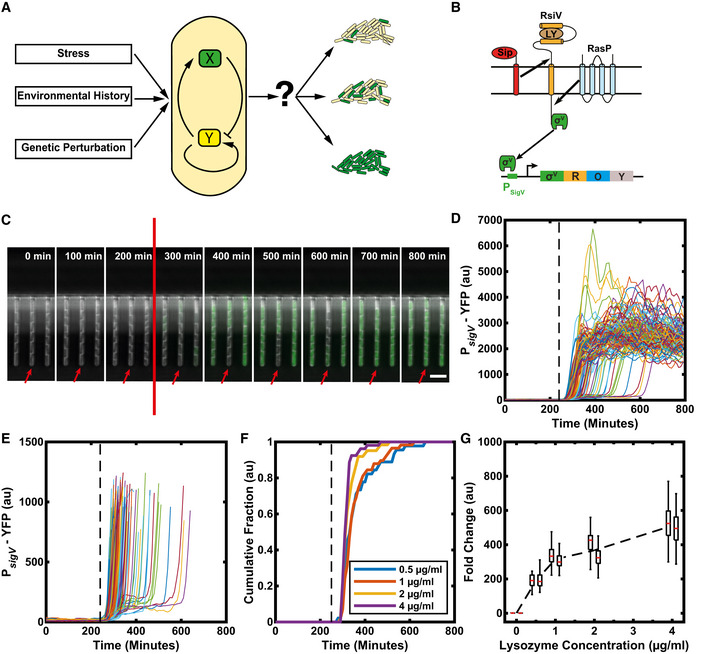
-
AIt is unclear how genetic circuits (centre) tune phenotypic variability in a bacterial population in response to genetic perturbation, environmental stress and history (left) to modulate the fraction of cells that activate a given pathway (right).
-
BSchematic of the σV circuit. In its inactive form, σV is bound to its anti‐sigma factor RsiV. If there is lysozyme present in the environment, RsiV binds to lysozyme and undergoes a conformational change. Only then the proteases SipS/ SipT and RasP can cleave RsiV to release σV. Once σV is released from RsiV, it can bind to RNA polymerase to redirect transcription to the σV regulon. σV can initiate transcription of its own operon which includes sigV (σV), rsiV (R), oatA (O, one of the main lysozyme resistance genes) and yrhK (Y, unknown function).
-
CTime‐lapse microscopy of cells containing a P sigV ‐YFP promoter reporter reveals heterogeneous activation of σV in response to lysozyme stress. The stress (red line) was added between the 200 and 300 min time points (at 240 min). The red arrow highlights a cell with a delayed activation of σV. Scale bar: 5 μm.
-
DTime traces of mean YFP fluorescence per cell. Each trace represents a single‐cell lineage's response to 1 µg/ml lysozyme. (109 traces from two independent experiments). Lysozyme added at the black dashed line.
-
EFigure D replotted to allow examination of variable activation times.
-
FThe observed heterogeneity is reduced with increasing stress levels. Each line represents the cumulative fraction of cells (N = ˜ 50) with P sigV ‐YFP values that are higher than the half maximum of their final values (representing cells that have activated).
-
GThe fold change in mean YFP fluorescence increases with increasing stress levels, shown for two biological repeats (adjacent boxplots). Each day's fold change distribution consisted of > 40 manually corrected single‐cell traces. On each box, the central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The lower and higher whiskers of boxplot are extended to the first quartile minus 1.5 * interquartile range and the third quartile plus 1.5 * interquartile range, respectively. The black dashed line is the mean fold change.
Data information: For more information on the number of repeats, please see Appendix Table S3.
The σV mediated lysozyme stress response pathway in Bacillus subtilis is an ideal model system to examine how bacterial populations can tune their phenotypic variability. σV is an extracytoplasmic function (ECF) alternative sigma factor. Alternative sigma factors replace the ‘housekeeping’ sigma factor, σA, in the RNA polymerase holoenzyme and redirect it to regulons that control distinct regulatory programmes. They have already been shown to display a high level of gene expression variability in B. subtilis (Locke et al, 2011; Young et al, 2013; Cabeen et al, 2017; Park et al, 2018), and the σV activation pathway is both well characterized and specific to one stress condition, which greatly simplifies analysis of its activation.
σV is the only pathway activated in response to C‐type lysozyme (Guariglia‐Oropeza & Helmann, 2011; Ho et al, 2011; Ho & Ellermeier, 2012). Lysozyme is produced by animals as part of their innate immune system and kills bacteria by cleaving the peptidoglycan of the cell wall between the N‐acetylmuramic acid residue and the β‐(1,4)‐linked N‐acetylglucosamine (Lal & Caplan, 2011). In its inactive form, σV is bound to its anti‐sigma factor RsiV, a transmembrane protein (Fig 1B). If lysozyme is present, RsiV binds to lysozyme (Hastie et al, 2014; Hastie et al, 2016) and activates a signal transduction cascade to release σV. First RsiV undergoes a conformational change that allows signalling peptidases to cleave RsiV at site‐1 (Hastie et al, 2014; Castro et al, 2018; Lewerke et al, 2018) (Fig 1B). Bacillus subtilis has five type 1 signal peptidases, of which the two major peptidases are SipS and SipT (Tjalsma et al, 1998). Either SipS or SipT is sufficient for site‐1 cleavage (Castro et al, 2018; Ho & Ellermeier, 2019). The truncated RsiV can then be cleaved by RasP (a site‐2 protease), which results in the release of σV (Hastie et al, 2013; Hastie et al, 2014) (Fig 1B).
Once σV is released it can bind to RNA polymerase and redirect transcription to the σV regulon. The regulon contains genes that allow adaptation to lysozyme stress, such as the penicillin‐binding protein PbpX and the d‐alanyl‐d‐alanine carrier protein ligase DltA. Both these genes are also part of other sigma factor pathways (Guariglia‐Oropeza & Helmann, 2011; Ho et al, 2011). Like many alternative sigma factors, one of σV's targets is its own operon. The sigV operon includes sigV (the gene that codes for σV) and its anti‐sigma factor rsiV, and so its activation results in ‘mixed’ positive and negative feedback loops. In addition, the operon contains oatA, one of the main lysozyme resistance genes and yrhK, which is of unknown function (Hastie & Ellermeier, 2016). OatA contributes to lysozyme adaptation by transferring an acetyl group to the C‐6‐OH position of N‐acetylmuramic acid in the peptidoglycan, thus blocking cell wall cleavage by lysozyme (Bernard et al, 2011; Ho et al, 2011).
Although the molecular mechanisms underlying σV activation by lysozyme have been elucidated, previous work was carried out using bulk experiments that average out single‐cell dynamics (Guariglia‐Oropeza & Helmann, 2011; Ho et al, 2011) and mask cell‐to‐cell heterogeneity. This heterogeneity can be crucial in identifying and distinguishing between regulatory strategies (Munsky et al, 2012). In this study, we used single‐cell time‐lapse microscopy of fluorescent reporters for σV activity to characterize σV induction dynamics in individual cells in response to lysozyme stress. We found that upon induction by sub‐lethal levels of lysozyme, σV is activated heterogeneously, with some cells activating σV rapidly, whereas some cell lineages do not activate σV for multiple generations. This heterogeneity is functional, as cells that respond to an initial sub‐lethal stress are more likely to survive a subsequent lethal stress of lysozyme. Through experiment and modelling, we found that these dynamics can be explained solely by the autoregulatory feedback of σV on its own operon. Our model predicted that the observed heterogeneity could be tuned by environmental history and by genetic perturbations, which we confirmed experimentally. The conserved sigma‐anti‐sigma autoregulation motif is thus a simple mechanism for bacterial populations to tune their levels of phenotypic variability.
Results
Individual cells activate σV heterogeneously under lysozyme stress
To characterize σV activation dynamics, we first constructed a B. subtilis reporter strain containing a chromosomally integrated fluorescent reporter for σV activity, P sigV ‐YFP. This strain also contained a reporter for the housekeeping sigma factor σA (P trpE ‐RFP). P trpE ‐RFP expression was used as a constitutive control and to aid image analysis (Materials and Methods). We then used time‐lapse microscopy to examine σV activity in individual cells grown in the mother machine microfluidic device (Wang et al, 2010). This device allows long‐term tracking of hundreds of individual cells trapped at the ends of channels. It also allows for fast switching between media conditions during imaging (Materials and Methods). We first measured σV expression dynamics in response to a step change to a sub‐lethal concentration of 1 µg/ml lysozyme (Fig 1C and D, Appendix Fig S1 and Fig EV1 and Movie EV1). The first cells begin to respond to the lysozyme stress with raised P sigV ‐YFP levels within two frames (20 min), suggesting rapid activation given the ˜ 15 min maturation time of the YFP reporter protein (Appendix Fig S2). However, our time‐lapse movies revealed that P sigV ‐YFP was heterogeneously activated at the single‐cell level (Fig 1C). While 20% of cells reached the half maximum of σV activity within 70 min of being exposed to lysozyme, it took 200 min (approximately four generations) for 90% of all cells to activate σV (Fig 1D and Appendix Fig S3B).
Figure EV1. With increasing stress levels, the heterogeneity in σV activation times is reduced.
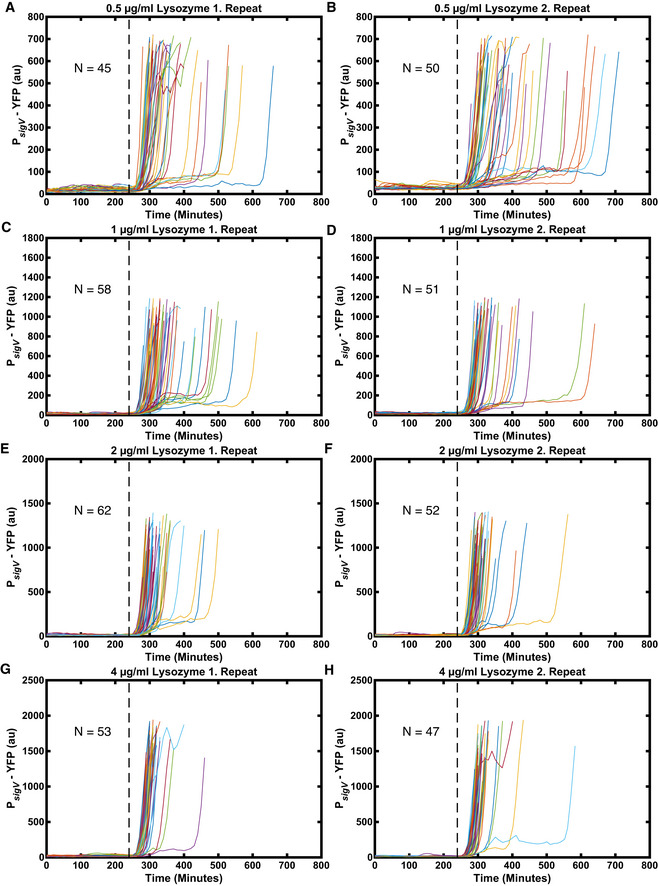
-
A, BSingle‐cell traces in response to 0.5 µg/ml lysozyme.
-
C, DSingle‐cell traces in response to 1 µg/ml lysozyme.
-
E, FSingle‐cell traces in response to 2 µg/ml lysozyme.
-
G, HSingle‐cell traces in response to 4 µg/ml lysozyme.
Data information: The data are the same as in n For more information on the number of repeats, please see Appendix Table S4.
To test whether the observed heterogeneous activation of σV in response to lysozyme was due to the growth conditions in the mother machine, we investigated σV activation dynamics in liquid culture (Appendix Fig S4) and using an alternative microfluidic device (Appendix Fig S5). Both conditions showed similar heterogeneous σV activation to that observed in the mother machine. Therefore, the observed heterogeneity in P sigV ‐YFP is independent of the experimental setup. Previous work has shown that heterogeneous gene expression could be due to intrinsic noise in the expression of the P sigV ‐YFP reporter (Elowitz et al, 2002), rather than due to heterogeneous σV activity. To test whether the observed heterogeneity was due to the intrinsic variability of the P sigV ‐YFP reporter, we constructed a strain containing chromosomally integrated P sigV ‐YFP and P sigV ‐mTurq reporters and exposed it to 1 µg/ml lysozyme in liquid culture. In these snapshot experiments, the expression of P sigV ‐YFP and P sigV ‐mTurq was highly correlated (R 2 = 0.85) (Appendix Fig S6). Thus, the observed heterogeneity in P sigV ‐YFP reflects changes in σv activity and not intrinsic variability of the P sigV ‐YFP promoter.
Next, we asked whether the observed heterogeneity in P sigV ‐YFP is modulated by the level of lysozyme applied. We examined P sigV ‐YFP expression after the application of 0.5, 1, 2 and 4 µg/ml lysozyme. These values were all below the previously reported minimal growth inhibitory concentration of 6.25 µg/ml (Ho et al, 2011) and led to a transient decrease in growth rate (Appendix Fig S7). To measure the distribution of σV activation times, for each time point we calculated the fraction of cells that had crossed the half‐maximum of their respective final σV value (meaning the cell had activated σV). We found that when increasing the lysozyme concentration from 0.5 to 4 µg/ml the heterogeneity in σV activity was reduced (Figs 1F and EV1 and Appendix Fig S8). The time for 90% of cells to activate their σV pathway decreased from 300 min (approximately six generations) for 0.5 µg/ml to 100 min (approximately two generations) for 4 µg/ml lysozyme (Appendix Fig S3). At the same time, the fold change in induction between the unstressed σV activity and the steady‐state σV activity under lysozyme stress increased from ˜ 190 for 0.5 μg/ml to ˜ 520 for 4 μg/ml (Fig 1G and Appendix Fig S1). We also observed that under a 4 µg/ml concentration of lysozyme some cells (8 and 21% in two different repeat experiments) appeared sick and were wider than usual cells. These cells also overshot their new σV activity steady‐state before relaxing to it. We removed these cells from our analysis (Appendix Fig S9), although including them did not affect our results (Appendix Fig S10). We also observed that the fraction of cells activating σV increased with increasing lysozyme in the alternative microfluidic device, although in this device movies were stopped before all cells activated due to crowding of the cells in the chip (Appendix Fig S5F). Taken together, our results reveal that the level of phenotypic variability in σV activation times is tuned by stress levels, with the heterogeneity in σV activation reduced as lysozyme levels increase.
We examined whether the differences in individual cell states before applying a lysozyme stress could predict how a cell responds to the stress. There was no correlation between either growth rate or cell size before lysozyme application and response time after lysozyme application (Appendix Figs S11 and S12). Alternative sigma factors in B. subtilis display gene expression variability even in the absence of stress (Locke et al, 2011; Park et al, 2018). We found that, in the absence of stress, the coefficient of variation of P sigV ‐YFP was over four times higher than that of the constitutive control (0.65 ± 0.29 vs 0.14 ± 0.05) (Appendix Fig S13). Cells which had elevated levels of P sigV ‐YFP before the addition of stress were more likely to activate σV instantaneously (Fig EV2 and Appendix Fig S14). However, some cells with low P sigV ‐YFP levels also activate σV instantaneously (Fig EV2 and Appendix Fig S14). Therefore, while noise in P sigV ‐YFP expression before stress contributes to the observed heterogeneity in σV activation after the addition of lysozyme, it is not the only cause.
Figure EV2. Cells with high P sigV‐YFP before stress activate σV rapidly.
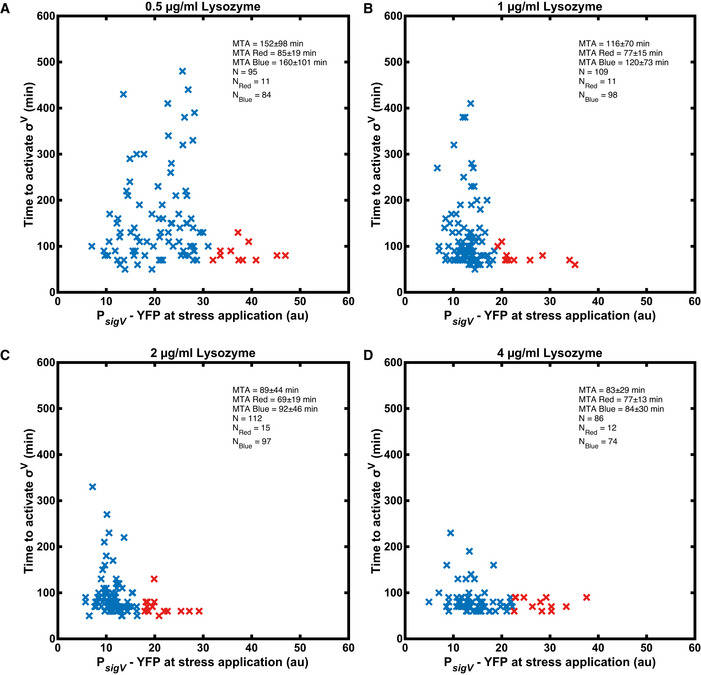
-
AActivation time for 0.5 µg/ml lysozyme.
-
BActivation time for 1 µg/ml lysozyme.
-
CActivation time for 2 µg/ml lysozyme.
-
DActivation time for 4 µg/ml lysozyme.
Data information: All shown data in the plots are from two biological repeats. For more information on the number of repeats, please see Appendix Table S4.
Given the heterogeneity in σV activation times, we examined whether activating σV early had any effect on the survival against future lethal concentrations of lysozyme. Cells that were exposed to 20 μg/ml of lysozyme for 20 min all died within 1 h (Fig 2 and Appendix Fig S15). However, if the cells were first exposed to a priming stress of 1 μg/ml of lysozyme for 30 min, which heterogeneously induced σV, and then subsequently to 20 μg/ml of lysozyme for 20 min, some cells survived the high lysozyme stress (Fig 2A). We chose this priming stress level and duration as previous experiments had shown (Appendix Fig S4) that it ensured heterogeneous activation of σV, with a large fraction of cells not having turned on before the second higher stress. We found that the short exposure to sub‐lethal concentrations of lysozyme (during the priming stress) improved survival to subsequent lethal stress levels from 0 to 12.5 ± 2.7% (See Materials and Methods, Fig 2B and Appendix Fig S15). Cells that survived until the end of the movie, 280 min after the 20 μg/ml of lysozyme, had on average 1.57 ± 0.33 fold higher P sigV ‐YFP levels than perishing cells immediately before the application of the lethal concentration of lysozyme (Fig 2C).
Figure 2. Rapid activation of σV after a first stress application increases survival after a second higher stress.
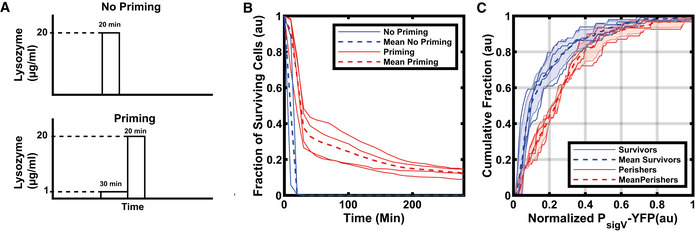
-
ASchematic of lysozyme application. Cells are either exposed directly to a high concentration of lysozyme (20 µg/ml) for 20 min (top) or exposed first to a short (30 min) lysozyme priming stress (1 µg/ml) before exposure to the higher concentration (bottom).
-
BA priming (30 min) stress of 1 µg/ml lysozyme followed by the high lysozyme stress (20 µg/ml) improves survival. The solid blue lines are the biological repeats for the no priming experiment (n = 2) whereas the solid red lines are the biological repeats for the priming experiment (n = 4). (Total number of cells shown for No Priming, N = 2,013 and Priming, N = 4,937).
-
CSurviving cells have higher P sigV ‐YFP levels after the initial priming stress (1 µg/ml) than perishing cells. The cumulative distributions were normalized by their maximum σV activity and baseline subtracted. Each dashed line is the mean of experiments from n = 4 biological repeats. The shaded areas correspond to the mean ± s.d. For more information on the number of repeats and cell numbers, please see the supplementary text.
Data information: For more information on the number of repeats, please see Appendix Table S3.
The heterogeneous activation of P sigV ‐YFP is solely due to σV and its anti‐sigma factor RsiV
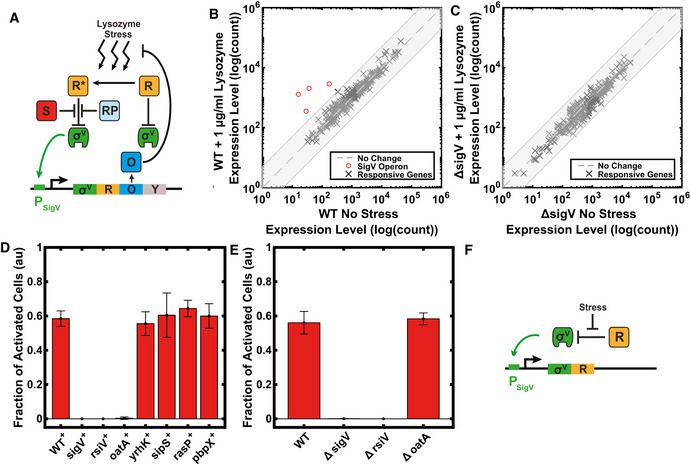
We next attempted to understand how the single‐cell activation dynamics of the σV pathway are generated. First, to test whether the heterogeneity that we observed in σV activation times was due to lysozyme stress activating different stress response pathways, we analysed the genome‐wide transcriptional response of cells to 1 μg/ml lysozyme. We carried out RNA‐seq 30 min after the addition of stress in the wild type and in the ΔsigV knockout. We chose this stress level and duration as we had seen in previous experiments that it resulted in heterogeneous σV activation (Appendix Fig S4). As previously reported, only the sigV operon was strongly (> 5 fold induction) induced by lysozyme (Guariglia‐Oropeza & Helmann, 2011) in the wild type (Fig 3B). The lysozyme resistance gene pbpX, which is part of the sigV regulon, was also upregulated in the WT (4.9 fold induction), but was not upregulated in the ΔsigV background (0.88 fold induction), consistent with the known role of sigV in its activation (Guariglia‐Oropeza & Helmann, 2011). The ΔsigV strain did not show any genes with strong (> 5 fold induction) induction (Fig 3C). DltA, which is known to contribute to lysozyme resistance and is part of the sigV regulon (Guariglia‐Oropeza & Helmann, 2011), was not upregulated in the WT or the ΔsigV strain. Our results suggest that no other pathway is strongly induced by 1 µg/ml lysozyme. We therefore hypothesized that the observed P sigV ‐YFP heterogeneity is due to the σV circuit itself (Ho et al, 2011; Hastie et al, 2013; Hastie et al, 2014).
Figure 3. The observed σV heterogeneity can be explained by a simplified σV circuit.
-
ASchematic of the σV circuit. In the figure, R (orange) is the anti‐sigma factor RsiV, R* (orange) is RsiV bound to lysozyme, S (red) is signalling peptidase, RP (light blue) is RasP the site‐2 protease, O (blue) is OatA, and Y (grey) is YrhK. For more information on the activation mechanism, see Fig 1B.
-
B, CRNA‐seq experiment on WT (JLB130) and ΔsigV (JLB154) strains, showing quantification of the absolute expression of individual genes in the presence and absence of lysozyme stress. The shaded grey box represents a ± 5 fold change. DEseq was used to identify genes which were differentially expressed with a 5% P‐value cut‐off between the WT and ΔsigV mutant in response to lysozyme treatment (default DEseq test was used). (B) Only the sigV operon is strongly activated (> 5 fold change) in response to lysozyme stress in WT (JLB130), as previously reported (Guariglia‐Oropeza & Helmann, 2011). (C) No genes were strongly upregulated in ΔsigV (JLB154) by lysozyme stress.
-
DEffect of the overexpression of individual components of the σV pathway (Biological repeats: WT: n = 8, sigV +: n = 4, rsiV +: n = 3 oatA +: n = 4, yrhK +: n = 3, sipS +: n = 3 and rasP +: n = 3, pbpX +: n = 6) on the fraction of activated cells. Only overexpression of sigV, rsiV or oatA changed the observed dynamics compared to WT. The histograms of the shown data are shown in Appendix Fig S16.
-
EDeleting oatA did not alter the σV activation dynamics. However, deleting sigV or rsiV resulted in no further activation of σV in response to 1 ug/ml lysozyme. n ≥ 3 biological repeats for all data shown. The histograms of the shown data are shown in Appendix Fig S17.
-
FSchematic of simplified σV circuit with only σV and RsiV, where σV (green) activates its own expression and that of its anti‐sigma RsiV (orange, R).
Data information: Bars correspond to the mean ± s.d. For more information on the number of repeats, please see Appendix Table S3.
To test which components of the σV circuit play a role in the heterogeneous activation of σV, we constructed IPTG‐inducible overexpression constructs of all genes in the sigV operon (sigV, rsiV, oatA, yrhK), as well as genes in the σV circuit (sipS, rasP) and the sigV regulon (pbpX) in a P sigV ‐YFP reporter background (Fig 3A). We then investigated σV activation dynamics under lysozyme stress after full induction of each circuit component. To do this, we took single‐cell snapshots of P sigV ‐YFP expression of cells grown in liquid culture. The cells were either unstressed or were stressed for 30 min with 1 μg/ml lysozyme. As a metric for the variability in activation time of σV in response to lysozyme, we calculated the fraction of cells with P sigV ‐YFP levels larger than a threshold above the unstressed level of each strain (See Materials and Methods). Single‐cell snapshots of P sigV ‐YFP expression revealed that overexpressing either yrhK, sipS, rasP or pbpX only had a minor effect on the fraction of activated σV cells (Fig 3D) or on the level of P sigV ‐YFP expression (Appendix Fig S16). For cells overexpressing sigV itself, no further activation of σV occurred on the addition of lysozyme (Fig 3D). This was because P sigV ‐YFP expression was already at a high level prior to stress (Appendix Fig S16). Conversely, overexpression of rsiV caused cells not to express P sigV ‐YFP at all, preventing activation of all cells on addition of lysozyme (Fig 3D and Appendix Fig S16). Finally, overexpressing oatA, which blocks lysozyme cleavage of the peptidoglycan, shuts off the activation of σV (Fig 3D and Appendix Fig S16). However, when we increased the lysozyme concentration to 20 μg/ml in the presence of oatA overexpression, the heterogeneous expression of P sigV ‐YFP reappeared (Fig EV3), suggesting oatA is not responsible for the heterogeneous activation dynamics. We repeated the experiment for rsiV overexpression, but increased RsiV did not increase protect against lysozyme as a concentration of 20 μg/ml lysozyme killed all cells.
Figure EV3. Overexpressing oatA shuts off σV activation.
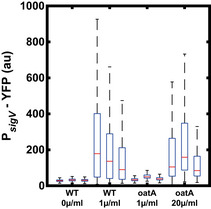
Increasing stress levels from 1 to 20 µg/ml recaptures the WT σV activation dynamics when oatA is overexpressed. n = 3 biological repeats for all data shown. On each box, the central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The lower and higher whiskers of boxplot are extended to the first quartile minus 1.5 * interquartile range and the third quartile plus 1.5 * interquartile range, respectively.
Data information: For more information on the number of repeats, please see Appendix Table S4.
To further validate the importance of sigV and rsiV as compared to oatA for the observed heterogeneity in σV activation, we constructed deletion mutants of sigV, rsiV and oatA. Only the deletion of oatA left the activation σV dynamics unchanged (Fig 3E). In the sigV mutant, P sigV ‐YFP levels did not increase in response to lysozyme stress (Fig 3C and Appendix Fig S17). Deleting rsiV caused all cells to have high P sigV ‐YFP expression even before the addition of lysozyme and the addition of lysozyme did not activate the system any further (Appendix Fig S17). These findings suggest that the heterogeneity in σV activation only depends on σV and its anti‐sigma factor RsiV (Fig 3F). We found that the leakiness of the inducible sigV construct (P hyperspank ‐sigV, without addition of IPTG) increased the fraction of activated cells on the addition of lysozyme, as well as causing an increase in the steady‐state levels of P sigV ‐YFP before and after stress (Appendix Figs S18 and S19). The leakiness of the RsiV construct (P hyperspank ‐rsiV) caused the opposite effect. These effects were still apparent for inducible constructs with lower leakiness (P spank‐sigV, P spank ‐rsiV (Appendix Figs S18 and S19)), confirming that sigV activation dynamics are sensitive to these two system components.
Mathematical modelling reveals that P sigV ‐YFP expression dynamics can be explained by the ‘mixed’ σV autoregulatory feedback loop
Based on our transcriptome and overexpression analysis, we constructed a model of a simplified sigV operon consisting of sigV and its anti‐sigma factor rsiV, with positive autoregulation of the operon by active σV (Fig 3F). We did not include oatA, as it is not required for the heterogeneous activation of σV (Fig EV3). RsiV binds σV to form an inactive complex σV‐RsiV. On stress application, RsiV is degraded and σV is free to activate the operon. The model was simulated using Gillespie‐type stochastic simulations, tracking the changes in copy number of each species of the system by simulating the individual reaction events (Gillespie, 1977). With the exception of the production of σV and RsiV (which were implemented through a Hill function of σV activity with a Hill coefficient of 2), all reactions were modelled using mass action kinetics (See Materials and Methods). A Hill coefficient > 1 was required to generate heterogeneous activation dynamics as a degree of ultrasensitivity is needed in the system to amplify the response to molecule fluctuations. We note that other networks, including a single positive feedback loop, can generate heterogeneous activation dynamics (See Materials and Methods and Appendix Fig S20), but that this is the simplest model that can simulate the role of both σV and RsiV in the regulation of sigV activation.
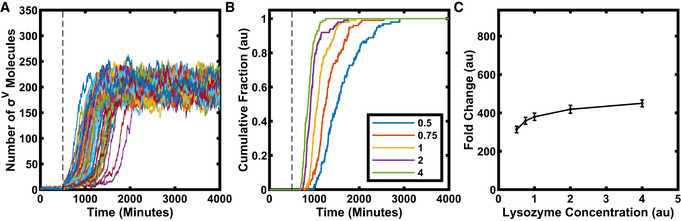
We searched a range of biologically feasible parameters and were able to find parameters that capture the heterogeneous σV activation in response to lysozyme stress (Figs 4A and EV4). The sigV operon components were assumed to be stable, so the dilution rate was set to approximately match the division rate observed in experiments. Simulations yielded plausible copy numbers for the number of sigma factor molecules (Jishage & Ishihama, 1995; Jishage et al, 1996). In addition, we verified that the heterogeneous activation behaviour is robust to perturbations to the system parameters (Appendix Fig S21). Using our model, we were able to recreate several aspects of the experimental data, including the dependency of induction time (Fig 4B) and steady‐state levels of sigV expression on levels of lysozyme stress (Fig 4C). Finally, the heterogeneous activation dynamics were also modulated by small increases in the baseline production rate of either σV or RsiV (Appendix Fig S22), qualitatively matching the effects of the leakiness of the inducible σV or RsiV construct observed in experiment (Appendix Fig S18).
Figure 4. A model of the simplified σV circuit captures experimental results.
-
ASimulations display heterogeneous σV activation after lysozyme stress (N = 100). Dashed vertical line marks point of stress addition (parameter L changed from 0 to 1) in simulation.
-
BIn simulations, cells activate σV faster as stress levels (the value of the L parameter) are increased, as observed experimentally (N = 100 simulations for each stress level: 0.5, 0.75, 1, 2, 4).
-
CWith increasing stress levels, the steady‐state level of σV activity increases, as observed experimentally. Bars correspond to the mean ± s.d. (N = 100 simulations for each lysozyme stress level).
Data information: For more information on the number of repeats, please see Appendix Table S3.
Figure EV4. With increasing stress levels, the heterogeneity in activation is reduced in the mathematical model.
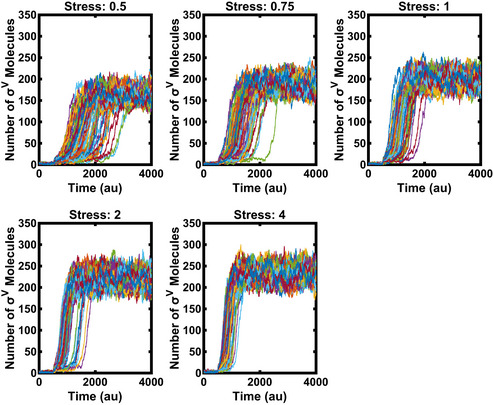
Each line corresponds to one simulated cell. The stress was added (parameter L changed from 0 to 1) at time point 500 (N = 100).
Data information: For more information on the number of repeats, please see Appendix Table S4.
Our model consists of a mixed positive and negative feedback loop. We tested the requirements of this feedback for the dynamics by modelling a feedback‐broken system, with constitutive expression of sigV and rsiV. For a range of constitutive expression, the dynamic range of P sigV ‐YFP expression for the feedback‐broken system on addition of lysozyme was less than that of the WT system (Fig EV5A and Appendix Fig S23). This reflected the role of the feedback loop in amplifying the system dynamics. To test this prediction experimentally, we constructed a strain with no autoregulation of the sigV operon by knocking out the sigV operon and replacing it with a sigV operon driven by an inducible promoter. This allowed us to study the system at different steady‐state expression levels (by varying IPTG induction level) to the WT system. We found that the fold change induction of the WT on addition of lysozyme was at least 4.5 times higher than that observed in the inducible operon strain, regardless of the IPTG induction level (Fig EV5B and Appendix Fig S24), matching the behaviour observed in our model (Fig EV5A).
Figure EV5. The sigV feedback loop increases the dynamic range of the circuit.
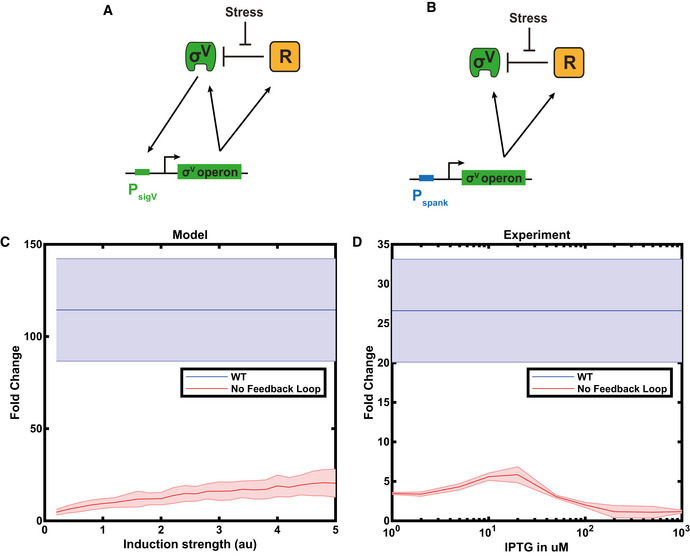
-
ASketch of WT sigV circuit. Where R stands of RsiV.
-
BSketch of rewired circuit with deleted positive feedback loop. Where R stands for RsiV and Pspank for the IPTG inducible promoter spank.
-
CThe fold change in sigV expression pre/post 1 μg/ml lysozyme stress was calculated from model simulations of the WT system (blue line) and a feedback‐broken system where the levels of the sigV operon are inducible (red line). The shaded areas represent one standard deviation of mean. N = 100 simulation.
-
DThe experimental fold change for the WT, and the feedback‐broken strain for IPTG values between 0 to 1,000 μM. The blue line is the WT fold change, and the red line is the fold change of ΔsigVrsiVoatAyrhK P spank ‐sigVrsiVoatAyrhK. The shaded areas represent one standard deviation of mean. n = 3 biological repeats for all data shown.
Data information: For more information on the number of repeats, please see Appendix Table S4.
Phenotypic variability is tuned by doubling copy numbers of sigV operon genes
To further test the assumptions of our model, we predicted the effect on σV activation dynamics of introducing a second copy of each component of the sigV operon (sigV, rsiV or both sigV and rsiV), with each component driven by the sigV promoter (Fig 5A). Our model predicted that these perturbations would modulate the heterogeneity of the system's response to lysozyme stress. A second copy of rsiV meant that large fluctuations in sigV expression were required to kick the system into the high σV expression state (Fig 5B and Appendix Fig S25). This increased the cell‐to‐cell variability in response times to lysozyme stress, as compared to the WT in the simulations (Appendix Fig S25). Conversely, with a second copy of sigV, or a second copy of both sigV and rsiV, smaller fluctuations in sigV expression are required to kick the system into the high σV expression state. Cell‐to‐cell variability in response times to lysozyme therefore decreases in these simulations (Fig 5 and Appendix Fig S25). In fact, a second copy of sigV caused an increase in the levels of σV expression even before the addition of lysozyme in the simulations (Appendix Fig S25).
Figure 5. Phenotypic variability is tuned by additional copies of components of the σV circuit.
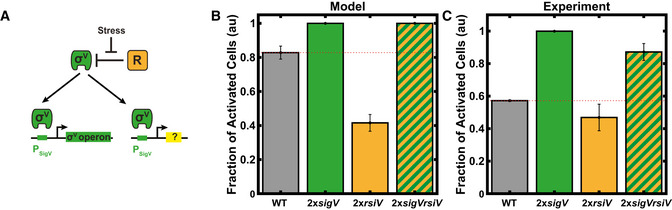
-
ASchematic of genetic perturbations to σV circuit. Second copies of σV components were added under the control of the P sigV promoter. ? = P sigV‐sigV (2xsigV), P sigV ‐sigVrsiV (2xsigV‐rsiV) or P sigV‐rsiV (2xrsiV).
-
BModel simulations predict that the 2xsigV or 2xsigV‐rsiV strains have a homogenous σV response to lysozyme, while the 2xrsiV strain has increased heterogeneity. Bars correspond to mean (N = 999 simulations) ± s.d. (1,000 bootstraps).
-
CAs predicted, the 2xsigV or 2xsigV‐rsiV strains have a homogenous σV response to 1 µg/ml lysozyme, while the 2xrsiV strain has increased heterogeneity. Each bar plot is the average of three biological repeats. The bars correspond to the mean ± s.d.
Data information: For both (B) and (C), bars represent the fraction of cells that have activated σV 30 min after treatment with lysozyme. σV activated cells were defined as those cells that had passed a threshold of the WT mean before lysozyme application plus six standard deviations. For more information on the number of repeats, please see Appendix Table S3.
To test these predictions, we constructed strains containing a second copy of either sigV, rsiV or sigVrsiV driven by the sigV promoter. As predicted by our model, snapshots of sigV expression after induction by 1 μg/ml of lysozyme revealed that a second copy of sigV or sigVrsiV reduced the observed heterogeneity in σV activation, while a second copy of rsiV caused an increase in the heterogeneity in σV activation (Fig 5C and Appendix Fig S26). Finally, adding a second copy of sigV alone caused the activation of σV even in the absence of stress, as predicted. These results validate our model assumptions and also show that the heterogeneity in sigV activation is easily tuned by simple genetic perturbations.
The heterogeneous activation dynamics of σ V are dependent on environmental history
Given that the σV activation dynamics appear sensitive to the baseline levels of σV, RsiV and the σV‐RsiV complex, any perturbations to these levels should affect σV activation times. We hypothesized that cells should have elevated levels of sigV operon components after a lysozyme stress is removed due to the recent high expression of the operon components. The elevated levels of σV stored in stabilized σV‐RsiV complexes should cause all cells to respond immediately to a subsequent reapplication of lysozyme. We investigated this hypothesis in our model. We simulated the application of lysozyme stress, which was then removed for the equivalent duration of several cell cycles. The same level of stress was then reapplied and the heterogeneity in σV activation disappeared (Fig 6A and Appendix Fig S27). However, as the delay before the second application of lysozyme was increased (allowing system components to relax to pre‐stress levels), the heterogeneity gradually returned (Fig 6B and C and Appendix Fig S27). Thus, our simulations predict the existence of a temporary molecular memory of the environmental history.
Figure 6. The σV circuit has a memory of previous stress.
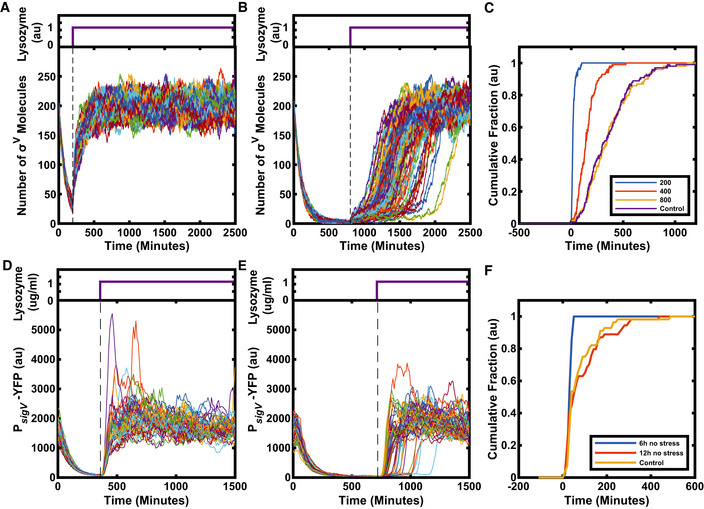
-
ASimulations predict that after a break in stress, on reapplication of stress all cells turn on σV immediately and homogeneously (N = 99).
-
B, CWith increasing intervals between stresses, response heterogeneity reappears in the model, indicating a loss of memory. (C) Each line represents the cumulative fraction of cells (N = 99) with σV concentration values that are higher than the half maximum of their final values (representing cells that have activated). If the interval between stress is increased long enough, (800 au), the heterogeneity is the same as if the cells had not experienced a prior lysozyme stress (control).
-
DExperiments confirm memory in the σV circuit. For short intervals between stress applications, cells respond immediately and homogeneously. Each line corresponds to one of 48 single‐cell traces.
-
E, FFor very long intervals between stress (12 h), the heterogeneity is the same as if the cells had not experienced a prior lysozyme stress (control). N > 50 single‐cell traces.
Data information: For more information on the number of repeats, please see Appendix Table S3.
To test whether the σV circuit exhibits such a memory, we grew cells in the mother machine device under 1 μg/ml lysozyme stress before removing the stress for 6 h (approximately seven cell cycles). All cells stopped activating σV as soon as lysozyme was removed, as indicated by the decay in YFP fluorescence (Fig 6D and Movie EV2). The decay in fluorescence was tightly synchronized across the population and YFP decayed with a half‐life time of 51 ± 1 min, which was similar to the cell cycle time of 51 ± 13 min. When reapplying a 1 µg/ml lysozyme stress after 6 h (approximately seven generations), all cells responded instantaneously. As predicted by the model, the heterogeneity in σV activation was lost (Fig 6D, Appendix Fig S28A and Movie EV2). Also as predicted by the model, increasing the duration of the break in stress to 12 h (approximately 14 generations) resulted in heterogeneous activation dynamics similar to those observed in response to the first application of lysozyme stress (Fig 6E and F, Appendix Fig S28B and C and Movie EV3). This return to a heterogeneous response reflects the system's loss of transcriptional memory through reduction in the sigV operon component concentrations to pre‐stress exposure levels. These results held for two different versions of the mother machine microfluidic device (Appendix Fig S29). Taken together, our results show how the autoregulatory σV circuit can generate heterogeneous activation dynamics that can be tuned by stress level, genetic perturbations and the environmental history of the cell.
Discussion
Here, we report a general mechanism for a bacterial population to tune its phenotypic variability based on stress levels, genetic architecture and environmental history. Using quantitative single‐cell microscopy and microfluidics, we found that the activation of the alternative sigma factor σV in response to lysozyme stress was heterogeneous. While some cells activated their σV pathway immediately, others could take up to six generations to activate their σV pathway. The observed phenotypic variability plays a functional role, as cells that respond to a sub‐lethal stress were more likely to survive a subsequent higher stress application (Fig 2). Through experiments and modelling, we found that this heterogeneity could be understood by the ‘mixed’ positive and negative feedback of σV activating both itself and its anti‐sigma factor RsiV. Alternative sigma factors are a common regulatory system in prokaryotes, often controlling stress response and virulence pathways. The ‘mixed’ feedback loop is also a common motif, suggesting that this motif can be a general mechanism for bacterial populations to tune their phenotypic variability.
Our modelling and experiments found that recent exposure to lysozyme stress modulates the heterogeneity observed on a second stress application, even though the system turns off after removal of the first lysozyme stress. The key to this behaviour appears to be the ‘mixed’ feedback loop, as it allows amplified levels of inactive σV‐RsiV complexes in each cell after stress. These complexes can be cleaved by a subsequent addition of lysozyme, releasing σV to activate its operon. Similar transcriptional memories of previous stress have been observed in bacterial systems, although typically not to modulate phenotypic heterogeneity. For example, other pathways such as the heat stress response in B. subtilis (Runde et al, 2014) or the oxidative stress response in yeast (Kelley & Ideker, 2009) have been shown to have a transcriptional memory of past conditions. Often this transcriptional memory is facilitated through an autoregulatory positive feedback loop that can lock the cell into an ON state that is heritable through cell divisions (Novick & Weiner, 1957; Biggar & Crabtree, 2001; Xiong & Ferrell, 2003; Acar et al, 2005; Hashimoto et al, 2013; Lambert & Kussell, 2014). However, it is difficult for a single positive feedback loop to allow the system to be OFF but primed for future stress, as we find to be the case for the ‘mixed’ feedback loop in the σV pathway. Dilution during growth causes heterogeneous activation of sigV to eventually return when levels of σV ‐RsiV drop to pre‐stress levels, so the memory is qualitatively different from that generated by a positive feedback loop locking a system ON. However, we find that the sigV transcriptional memory remains for several generations. In the future, it will be important to investigate whether the ‘mixed’ feedback loop also tunes the levels of phenotypic diversity by environmental history in other systems. Interestingly, computational studies have proposed that a ‘mixed’ feedback loop structure in the MAR operon in Escherichia coli allows the tuning of the fraction of cells prepared to survive antibiotic exposure (Garcia‐Bernardo & Dunlop, 2013). Additionally, the mixed feedback loop mechanism could be compared to other mechanisms proposed to allow the modulation of phenotypic variability, such as multi‐site phosphorylation (Libby et al, 2019) or threshold‐based mechanisms in toxin–antitoxin modules (Rotem et al, 2010).
While simple, our model allows qualitative matches to data. In future, it will be important to increase the complexity of the model to make more precise predictions of the behaviour of the sigV system. One aspect of the system that can be modelled in more detail is how noise in gene expression is generated in the circuit. In our model, we do not model transcription and translation separately and assume uncorrelated noise for each reaction channel. A more detailed model could involve characterizing the noise in terms of bursts of transcription and translation (Friedman et al, 2006). In turn, this would require experiments to characterize the noise in transcription, such as single‐molecule FISH (Raj & Oudenaarden, 2008). Our assumption of uncorrelated noise is also a simplification as, for example, we have modelled the degradation events as uncorrelated, which may not hold as these are primarily caused by dilution. Additionally, the system requires ultrasensitivity (in the form of a Hill coefficient n > 1 in the operon production term) in order to amplify molecule fluctuations. While other sigma factor response systems have been shown to utilize ultrasensitivity (Narula et al, 2012; Narula et al, 2016), there is no known source of it in the σV circuit (neither the binding of σV to RsiV, nor its operon, is cooperative). Further research should examine possible sources of ultrasensitivity in the circuit, one possibility being sigma factor competition for RNA polymerase (Park et al, 2018).
Cells that activate sigV quickly after a priming stress have a higher chance of surviving a subsequent high stress (Fig 2B). This points to a potential benefit of early activation of sigV. Future work should examine the costs of early sigV activation to see if the heterogeneous activation dynamics we observe represent a bet‐hedging strategy (Veening et al, 2008a; Veening et al, 2008b), where early responding cells suffer a fitness penalty in return for protection against future stress. We do not observe a growth rate difference in cells that activate sigV earlier compared to later, suggesting that early responders are not suffering a growth rate penalty. However, it is possible that we are missing small growth rate effects, as the time resolution of our mother machine experiments only allows approximately 5 time points to be measured per cell cycle. Our experiments do indicate that high constitutive expression of sigV or oatA reduces the growth rate in bulk culture (Appendix Fig S30). It could also be that the fitness penalty is due to early sigV activation blocking cells from responding to stress with other alternative sigma factors, as it appears alternative sigma factors compete for RNA polymerase (Nyström, 2004; Park et al, 2018). Evolution experiments under changeable stressful environments could reveal whether the heterogeneity in activation and transcriptional memory that we observe evolve to optimally match the external environment.
We have found that a combination of noise in gene expression and a mixed feedback loop can generate tunable phenotypic diversity in an alternative sigma factor circuit. Our work will be of utility for synthetic biology, where alternative sigma factors are a promising system for engineering orthogonal gene circuits (Rhodius et al, 2013; Bervoets et al, 2018; Pinto et al, 2018). Going forward, it will be important to observe whether alternative sigma factors more generally are used as a mechanism for bacterial populations to tune phenotypic diversity. This is particularly the case given that alternative sigma factors often control pathways critical to pathogenicity and resistance to antibiotics. Noise in sigma factor activity has already been observed in multiple alternative sigma factor circuits in B. subtilis (Locke et al, 2011; Narula et al, 2016; Park et al, 2018), as well as for the general stress response sigma factor in E. coli (Patange et al, 2018). Additionally, an alternative sigma factor plays a role in generating phenotypic diversity in Mycobacteria (Sureka et al, 2008; Ghosh et al, 2011). Two other obvious candidates for further study are the pathogens Enterococcus faecalis and Clostridioides difficile (Ho & Ellermeier, 2019), which also have a σV pathway that is responsive to lysozyme.
Materials and Methods
Strains and media
All strains are derivatives of the PY79 background strain (Appendix Table S1). Deletions were generated by replacing genes of interest with an antibiotic resistance cassette by recombination of a linear DNA fragment homologous to the region of interest. All strains had a ΔytvA::neo deletion insertion. YtvA is a blue light sensor, and it was deleted to avoid any activation of the general stress response pathway σB by the microscope illumination (Gaidenko et al, 2006; Locke et al, 2011). For strains used in mother machine experiments, cells were made immotile by inserting a ΔhaG:erm deletion cassette in order to improve the loading into the microfluidic device. Additionally, all strains contained a house keeping σA promoter‐driven mCherry for segmentation and as a constitutive control (Locke et al, 2011).
Cells were routinely grown in Spizizen's Minimal Media (SMM) (Spizizen, 1958). It contained 50 µg/ml tryptophan as an amino acid source and 0.5% glucose as a carbon source. Cultures were started from frozen stock in SMM and grown overnight at 30°C to an OD between 0.3 and 0.8. The overnight cultures were resuspended to an OD of 0.01 and regrown to an OD of 0.1 at 37°C.
Plasmids
Escherichia coli strain DH5α was used to clone all plasmids. The cloning was done with a combination of non‐ligase‐dependent cloning and standard molecular cloning techniques using Clontech In‐Fusion Advantage PCR Cloning kits. Plasmids were chromosomally integrated into the PY79 background via double crossover using standard techniques. The list below provides a description of the used plasmids, with details on selection marker and integration position/cassette given at the beginning. Note that all plasmids below replicate in E. coli but not in B. subtilis.
-
ppsB::P trpE ‐mCherry PhleoR
This plasmid was used to provide uniform expression of mCherry from a σA‐dependent promoter, enabling automatic image segmentation (cell identification) in time‐lapse movie analysis. A minimal σA promoter from the trpE gene was cloned into a vector with ppsB homology regions (Locke et al, 2011). The original integration vector was a gift from A. Eldar (Eldar et al, 2009).
-
sacA::P sigV ‐YFP CmR
The target promoter of sigV was cloned into the EcoRI/BamHI sites of AEC12 (Eldar et al, 2009) (gift from M. Elowitz, CalTech).
-
amyE::P sigV ‐mTurq SpectR
The target promoter of sigV and the mTurq gene from GL‐FP‐31 (gift from E. Gardner, Harvard) were cloned into the sites EcoRI/BAmHI and BamHI/HindIII of pdL30 (Locke et al, 2011).
-
amyE::P hyperspan k ‐X SpectR
Where X is sigV, rsiV, yrhK, oatA, rasP, sipS, pbpX or YFP. The coding region of the genes along with a 5′ transcriptional terminator was cloned downstream of the hyperspank IPTG‐inducible promoter in plasmid pDR‐111 (gift from D. Rudner, Harvard).
-
amyE::P spank ‐X SpectR
Where X is sigV, rsiV, yrhK, oatA, rasP, sipS, pbpX or YFP. The coding region of the genes along with a 5′ transcriptional terminator was cloned downstream of the spank IPTG‐inducible promoter in plasmid pDR‐110 (gift from D. Rudner, Harvard).
-
amyE::P sigV ‐X SpectR
The coding region of (where X is sigV, rsiV or sigVrsiV) along with a 5′ transcriptional terminator was cloned downstream of the sigV promoter.
Microscopy
A Nikon inverted Ti‐E microscope (Nikon, Amsterdam, Netherlands) with a Nikon Perfect Focus System (PFS) hardware auto‐focus was used. All images were acquired using either a Photometrics CoolSnap HQ2 CCD camera (Photometrics, Tucson, AZ, USA) or a Photometrics Prime sCMOS camera (Photometrics, Tucson, AZ, USA). The microscope stage was enclosed in an incubator (Solent Scientific, Segensworth, UK) which was set to 37°C for all experiments. Illumination was provided by a LED lamp (CoolLED, Andover, UK). Epifluorescence was provided by a Lumencore Solar II light engine (Lumencore, Beaverton, OR, USA). Chroma filters (Chroma, Bellows Falls, USA) #41027 for the RFP channel, #49003 for the YFP channel and #49001 for the CFP channel were used. All experiments were done with a phase 100x Plan Apo (NA 1.4) objective (Nikon, Amsterdam, Netherlands). Metamorph (Molecular Device, Sunnyvale, CA, USA) controlled the camera, the motorized stage (Nikon, Amsterdam, Netherlands) and the microscope.
Snapshots
Agarose pads were made by pipetting 1 ml of 1.5% (wt/vol) low melt agarose (Merck, Darmstadt, Germany) dissolved in PBS onto a 22 mm2 cover glass slide. Immediately after pipetting, another cover glass slide was placed on top of the agarose creating a “sandwich” with the agarose in the middle. Once the agarose had solidified, the top cover glass was removed and the agarose was cut into squares of approximately 5 mm × 5 mm with a scalpel.
Once the cultures had reached an OD of 0.1, they were split into aliquots of 1 ml to which different concentrations of lysozyme from hen eggs white (Sigma Aldrich, St. Louis, MO, USA) were added. The aliquots were then incubated at 37°C for 30 min. For experiments with IPTG‐inducible strains, 1 mM of IPTG was added to the aliquots for 60 min before addition of lysozyme, in order to allow for full induction of the promoter before stress. Different concentrations of lysozyme were then applied for 30 min before snapshot measurements. For snapshots, 2 µl of cell culture was pipetted onto the pads, which were left to dry and then laid face down onto a cover glass‐bottom dish (#HBSt‐5040, WillCO‐dish, Amsterdam, Netherlands). The glass dish was sealed with parafilm and put under the microscope to image. Single‐cell data were extracted using custom MATLAB (Mathworks, Natick, USA) scripts based on the Schnitzcells package (Young et al, 2011).
CellAsic experiments
Overnight cultures were grown and then resuspended to an OD of 0.01 as described above. Once the cultures had reached an OD of 0.1, they were resuspended to an OD of 0.001 for loading into CellAsic B04 microfluidic chips (Merck, Darmstadt, Germany). Cells were loaded into the microfluidic chips with a pressure of 4–6.5 psi for 2–4 s. The following lysozyme concentrations were investigated with the CellAsic setup: 0, 0.5, 1 and 4 µg/ml. Fresh media was perfused into the chip with a pressure of 1 psi. After 60 min of growth in standard SMM, the media was switched to media containing lysozyme. Cells were imaged at regular intervals (every 10 min), and the acquired movies were analysed with the standard Schnitzcells package for MATLAB (Young et al, 2011).
Microfluidics
Wafer fabrication
We used two different microfluidic designs of the mother machine:
All the mother machine data in this paper were acquired with a microfluidic design based on the original mother machine (Wang et al, 2010) unless stated otherwise. As a substrate for the PDMS chip fabrication, we used an epoxy master which was a kind gift from the Jun laboratory at the University of California, San Diego, USA (Taheri‐Araghi et al, 2015).
For data shown in Appendix Fig S29, a different mother machine design was used. It was based on a mother machine design first described by Norman et al (2013). The main difference to the Jun laboratory design is that the channels in which cells grew were longer and that they have a shallow side channel for better perfusion of media to the cells. As a substrate for the PDMS chip fabrication, we used an epoxy master which was a kind gift from the Elowitz laboratory at the California Institute of Technology, Pasadena, USA (Park et al, 2018).
Soft lithography
Sylgard 184 polydimethylsiloxane (PDMS) from Dow Corning (Midland, MI, USA) was used to replica‐mould the microfluidic devices. A 10:1 (base:curing agent) PDMS mixture was cast onto the epoxy master and cured overnight at 65°C. In the next morning, the chips were cut out and the inlets were punched using a 0.75 mm biopsy puncher (#504529, WPI, Sarasota, FL, USA). To remove any uncured PDMS from the chips, these were chemically treated in a pentane (Sigma Aldrich, St. Louis, Mo, USA) bath for 90 min and then washed twice for 90 min in acetone (Sigma Aldrich, St. Louis, Mo, USA). The chips were then left to dry overnight in the fume hood and were ready to use the next day (Gruenberger et al, 2013).
Experimental preparation
The chips were plasma bonded to a glass‐bottom dish (HBSt‐5040, Willco Wells, Amsterdam Netherlands) by treating the chip and the glass dish surfaces with a plasma (Femto Plasma System, Diener, Ebhausen, Germany) for 12 s at a power of 30 W and a pressure of 0.35 mbar. The chips were then put into the oven for 10 min at 65°C, in order to strengthen the bonding. In the meantime, a bovine serum albumin (BSA) passivation buffer was prepared at a concentration of 20 mg/ml in SMM. The chips were then passivated with this buffer for 1 h at 37°C.
Cells were grown overnight as for the other movie experiments. 10 ml of the regrown cultures with an OD of 0.1 was spun down for 10 min at 3,000 g (4,000 rpm) and resuspended in 0.1 ml to a final OD of 10. This cell suspension was loaded into the growth channels by spinning the chip for 5 min at 3,000 rpm with a spin coater (Polos SPIN150i, SPS, Harlem, the Netherland).
Setup of microfluidic mother machine experiments
Perfusion was provided by a Fluigent pressure pump setup. Briefly, reservoirs were pressurized using a computer controllable pressure controller (MFCS‐EZ, 1 bar, Villejuif, Fluigent, France). A rotary selection valve (M‐Switch, Fluigent, Villejuif, France) allowed for switching between different reservoirs. The flow rate was set to 1 ml/h and by using a flow sensor (M‐Flow Sensor, Fluigent, Villejuif, France) which feeds back onto the pressure controller.
For the experiments on the memory in the σV response, syringe pumps (Legato 200, KD Scientific, Holliston, USA) were used. Switching was provided by an electronic switching valve (MXP9960, Rheodyne, Lake Forest, USA). The flow rate remained at 1 ml/h.
The analysis of all movies was done using a modified version of Schnitzcells (Young et al, 2011; Park et al, 2018) optimized for mother machine experiments.
Memory experiments
For all shown memory experiments, the JLB221 strain (ΔespH) was used instead of JLB130, unless stated otherwise. This was done in order to prevent clogging of the microfluidic chip during these long‐term experiments due to the production of extracellular matrix. The two strains had the same P sigV ‐YFP dynamics in response to lysozyme (Appendix Figs S31 and S32).
Cells were inoculated in SMM and grown overnight to an OD of 0.3–0.8. In the morning, the cells were resuspended to an OD of 0.01 in SMM + 1 μg/ml lysozyme and grown to an OD of 0.1. Cells were then loaded as described in the “Experimental preparation” section.
In the mother machine, the cells were grown for 4 h with 1 μg/ml lysozyme before removing the stress. The stress was then reapplied after 6 or 12 h.
Growth curve determination
Cultures were started from frozen stock in SMM and grown overnight at 30°C. The overnight cultures were resuspended to an OD of 0.02 and grown for 2 h at 37°C before adding 1 mM of IPTG. The cells were then grown for another hour before adjusting the OD to 0.1 and aliquoting the culture into 2 ml samples (with one 2 ml sample used for each time point), and then, the samples were returned to grow at 37°C. A sample was taken every hour, and the OD600 was determined.
Data analysis
Cumulative activation time
The activation time in mother machine experiments was calculated as the time between switching from SMM to lysozyme and the time point when then P sigV ‐YFP passed its half maximum (Appendix Fig S1). The cumulative fraction was then calculated as the cumulative fraction of cells with P sigV ‐YFP values higher than the half maximum of their final values (representing cells that have activated) (Appendix Figs S1 and S8).
Removal of overshooting cells
For 4 μg/ml lysozyme, the P sigV ‐YFP activity overshot its steady‐state activity. Overshooting cells were sick and also wider. Thus, overshooting cells could be removed based on their width. First, the single‐cell width traces were smoothed with a Gaussian filter, and their maximum value was determined. All single‐cell traces with a maximum width larger than the mean width + 6 sigma of cells before the stress were removed (Appendix Figs S9 and S10).
Growth rate
The instantaneous growth rate as shown in Appendix Fig S11 was calculated as in (Martins et al, 2018):
| (1) |
where len i is the length of the cell at frame i, and Δt is the time between subsequent frames taken from the image metadata. Growth rates were only calculated within one cell cycle and not over a division.
Calculation of surviving cells in priming experiment
The lysis curve (Fig 2B) was calculated as the fraction of surviving cells 280 min after the addition of 20 µg/ml lysozyme. We only examined the survival rate of the top three cells in each channel at the time of the addition of 20 µg/ml lysozyme to avoid problems with cells being washed out of the chip during the 280 min after the stress. Only one surviving lineage (the longest) was counted from each of these top three cells, to avoid cell division artificially increasing the number of survivors. The survival rate at the end of the movie using this method was 12.5 ± 2.7% (Fig 2B). To verify this method, we also used an alternative approach. In this second method, the end of each channel was approximated using the average cell position before a cell left the channel in the ˜ 100 frames before the stress was added. Cells that at their last recorded position after the addition of 20 µg/ml lysozyme stress were within six standard deviations of the end of the channel were removed from the analysis as they were assumed to have left the channel rather than died. This second approach gave qualitatively similar results, with a survival rate of 13.6 ± 3.1%.
Statistical analysis of the effects of higher P sigV ‐YFP levels before lysozyme stress application on response time
As the distributions in Appendix Fig S14 were not normal, we use the non‐parametric Kolmogorov–Smirnov test (ks test) to verify the null hypothesis that cells with higher P sigV ‐YFP values before lysozyme stress application would have the same activation time distribution as cells with lower P sigV ‐YFP expression. Cells with high P sigV ‐YFP were defined as cells with a YFP fluorescence higher than the mean YFP fluorescence before the addition of lysozyme plus one standard deviation. All other cells were defined as low P sigV ‐YFP. A P‐value below 0.05 was used to reject the null hypothesis. The ks test was performed using the built in MATLAB function kstest2.
We found that for 0.5, 1 and 2 μg/ml lysozyme the null hypothesis was rejected. Thus, cells with higher P sigV ‐YFP values before lysozyme stress application had a shorter activation time. For 4 μg/ml lysozyme, the null hypothesis was accepted. Therefore, cells with higher P sigV ‐YFP values before lysozyme stress application had the same activation time as cells with lower P sigV ‐YFP values.
Calculation of fraction of activated cells from snapshots
Cells which, after the application of lysozyme stress, had a higher mean P sigV ‐YFP expression than the mean P sigV ‐YFP expression plus six standard deviations before stress were defined as having activated. The fraction of activated cells was then calculated as the number of cells which had activated normalized by the total number of cells (Figs 3 and 5).
RNA‐seq experiment
Lysozyme treatment before RNA extraction
Overnight cultures were prepared as for snapshot or movie experiments. Briefly, cultures were started from frozen stock in SMM and grown overnight at 30°C to an OD between 0.3 and 0.8. The overnight cultures were resuspended to an OD of 0.01 and regrown to an OD of 0.1 at 37°C. Once the cultures had reached an OD of 0.1, they were split into aliquots of 10 ml to which either 0 or 1 µg/ml lysozyme from hen eggs white (Sigma Aldrich, St. Louis, MO, USA) were added. The aliquots were then incubated at 37°C for 30 min.
RNA extraction and library preparation
10 ml of B. subtilis at OD 0.1–0.2 was centrifuged in 15‐ml falcon tubes for 10 min at 3,000 g (4,000 rpm). Cell pellets were resuspended in 1 ml of Qiagen RNAprotect Bacteria Reagent and flash frozen in liquid nitrogen.
Defrosted cells were pelleted by a quick centrifugation. After removing the supernatant, cells were resuspended in 1 ml buffer RLT from Qiagen RNeasy kit. Resuspended cells were transferred in a Fastprep Lysing Matrix B tube (MP Bio) and processed in Fastprep apparatus 45 s at speed 6.5 M/s. 700 µl of the supernatant, containing lysed cells, was transferred to a new microcentrifuge tube, to which 500 µl of absolute ethanol was added. After vortexing, the lysate was transferred to a RNeasy spin column and centrifuged 15 s at > 9,400 g (10,000 rpm). RNA purification was then carried out following the instructions of the Qiagen RNeasy kit. RNA quality and integrity were assessed on the Agilent 2200 TapeStation, and RNA concentration was assessed using Qubit RNA HS assay kit. Library preparation was performed using ScriptSeq™ Complete Kit (Illumina, BB1224), for 2 µg of high integrity total RNA (RIN > 8). The libraries were sequenced on a NextSeq500 using paired‐end sequencing of 75 bp in length.
RNA‐seq analysis
The raw reads were analysed using a combination of publicly available software and in‐house scripts. We first assessed the quality of reads using FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were aligned to the B. subtilis PY79 transcriptome (NCIB no.CP006881) using Salmon (Patro et al, 2017). Read counts for each gene were imported using the tximport R package (Soneson et al, 2015). Genes differentially expressed (P‐value < 5%) between the WT and ΔsigV mutant, or in response to lysozyme treatment were identified using the DESeq2 R package (Love et al, 2014). The RNA‐seq data of our study can be found here (Gene Expression Omnibus: GSE171761, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE171761).
Mathematical model
We constructed a mathematical model of the σV circuit that was based on the core components that we found experimentally to modulate the heterogeneity in σV activation (Fig 3). The model consists of σV, RsiV and the σV‐RsiV complex. Free σV that is not bound in a complex activates the production of both σV and RsiV. Lysozyme stress was modelled as activating the cleavage of the σV‐RsiV complex, releasing σV while degrading RsiV. We note that although heterogeneous activation can be simulated through just a positive feedback loop, (Tiwari et al, 2011; Frigola et al, 2012) (Appendix Fig S20), our model is the simplest model that could be compared to the mutations including both sigV and rsiV (Fig 5).
Model reactions
We used the following set of biochemical reactions to model the dynamics of σV, RsiV and σV‐RsiV.
Production:
∅ → σV + RsiV
The production rate of σV and RsiV was assumed to follow a Hill function, where the production rate of both σV and RsiV were identical and activated by σV. The production rate is as follows: v 0 + v*(σV)ⁿ/((σV)ⁿ + Kⁿ), where v 0 (= 0.1 molecules/min, is the operon leakage), v (= 2.5 molecules/min, is the maximal operon activity), K (= 60 molecules, is the apparent dissociation constant) and n (= 2, is the Hill coefficient) are parameters.
Dilution/Degradation:
σV → ∅.
RsiV → ∅
σV‐RsiV → ∅
We assumed a constant and identical dilution rate for all three components of the system. The dilution rate is set by the parameter kdeg (= 0.01 min−1).
Binding/Dissociation:
σV + RsiV → σV‐RsiV
σV‐RsiV → σV + RsiV
We assumed that σV and RsiV would bind to each other to form a complex and that this complex could dissociate. We also assumed the binding rate to be higher than the dissociation rate. The binding rate is set by the parameter kB (= 10 molecules−1 min−1) and the dissociation rate by the parameter kD (=5 min−1).
Cleavage:
σV‐RsiV → σV
We assumed that the σV/RsiV complex would be cleaved by lysozyme, with σV getting released and RsiV degraded. The cleavage rate is set by the product of the two parameters L (which attains variable values, is the amount of lysozyme stress experienced by the system) and kC (= 0.05 min−1, is the base cleavage rate of the complex). Splitting the rate into two parameters allows setting more natural lysozyme input values (such as 1 and 2).
Model implementation
The model was implemented in the Julia programming language using the Catalyst.jl modelling package. Simulations were made using the DifferentialEquations.jl package (Rackauckas & Nie, 2017b). To account for the stochastic nature of the system, we use a Gillespie‐type model (Gillespie, 1977). Here, we tracked the copy numbers of the three components of the system (σV, RsiV and σV‐RsiV), and their change due to the individual reaction events. We used Gillespie's direct stochastic simulation algorithm to determine the time to, and which, the next reaction event in the simulation should be. We used DifferentialEquations.jl's SSAStepper method to simulate the model. For more details, please see the implementations in the files provided. The model parameters were hand‐picked by carrying out a coarse‐grained search of feasible values. The degradation rate was set to be similar to what would be produced by the bacteria division rate in the experiments. This yields plausible copy numbers for the number of sigma factor molecules (Jishage & Ishihama, 1995; Jishage et al, 1996). See Appendix Table S2 for parameter values. The heterogeneous activation behaviour is robust with respect to perturbations to the selected parameters (Appendix Fig S21). Finally, we also note that a continuous SDE model, based on the chemical Langevin equations, could reproduce the features of the system, (Appendix Fig S33), suggesting that that behaviour does not depend on choice of model approach (Gillespie, 2000; Rackauckas & Nie, 2017a).
Author contributions
CPS, TEL and JCWL conceived and designed the study. CPS, TEL, BMCM and JCWL analysed and interpreted the data and wrote the article. CPS, VK and TL performed the experiments. SC did the RNA‐seq experiments. CV and TS constructed strains. TEL and BMCM developed the mathematical model.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Movie EV1
Movie EV2
Movie EV3
Review Process File
Acknowledgements
We thank John Sauls and Professor Suckjoon Jun for the kind gift of a mother machine epoxy mould. We thank Jin Park for help with the mother machine analysis code, Michael Elowitz for sharing plasmids, Craig Ellermeier for sharing strains, critical reading of the manuscript and helpful discussions, Hugo Tavares for help with RNA‐seq analysis, Katie Abley for critical reading of the manuscript and Niklas Korsbo for helpful discussions. This research was made possible by the award of a European Research Council under the European Union's Seventh Framework Programme (FP/2007‐2013)/ERC Grant Agreement 338060. The work in the Locke laboratory is further supported by a fellowship from the Gatsby Foundation (GAT3272/GLC). Torkel Loman has received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska‐Curie grant agreement No. 721456.
Mol Syst Biol. (2021) 17: e9832
Data availability
The RNA‐seq data produced in this study can be found here:
Gene Expression Omnibus GSE171761: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE171761.
Model simulation, data analysis and figure plotting code, as well as source data for main and extended view figures can be found here: https://gitlab.com/slcu/teamJL/schwall_etal_msb_2021.
References
- Acar M, Becskei A, van Oudenaarden A (2005) Enhancement of cellular memory by reducing stochastic transitions. Nature 435: 228–232 [DOI] [PubMed] [Google Scholar]
- Bernard E, Rolain T, Courtin P, Guillot A, Langella P, Hols P, Chapot‐Chartier M‐P (2011) Characterization of O‐acetylation of N‐acetylglucosamine: a novel structural variation of bacterial peptidoglycan. J Biol Chem 286: 23950–23958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bervoets I, Van Brempt M, Van Nerom K, Van Hove B, Maertens J, De Mey M, Charlier D (2018) A sigma factor toolbox for orthogonal gene expression in Escherichia coli . Nucleic Acids Res 46: 2133–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggar SR, Crabtree GR (2001) Cell signaling can direct either binary or graded transcriptional responses. EMBO J 20: 3167–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabeen MT, Russell JR, Paulsson J, Losick R (2017) Use of a microfluidic platform to uncover basic features of energy and environmental stress responses in individual cells of Bacillus subtilis . PLoS Genet 13: e1006901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro AN, Lewerke LT, Hastie JL, Ellermeier CD (2018) Signal peptidase is necessary and sufficient for site 1 cleavage of RsiV in Bacillus subtilis in response to lysozyme. J Bacteriol 200: e00663‐17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldar A, Chary VK, Xenopoulos P, Fontes ME, Losón OC, Dworkin J, Piggot PJ, Elowitz MB (2009) Partial penetrance facilitates developmental evolution in bacteria. Nature 460: 510–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS (2002) Stochastic gene expression in a single cell. Science 297: 1183–1186 [DOI] [PubMed] [Google Scholar]
- Friedman N, Cai L, Xie XS (2006) Linking stochastic dynamics to population distribution: an analytical framework of gene expression. Phys Rev Lett 97: 168302 [DOI] [PubMed] [Google Scholar]
- Frigola D, Casanellas L, Sancho JM, Ibañes M (2012) Asymmetric stochastic switching driven by intrinsic molecular noise. PLoS One 7: e31407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidenko TA, Kim T‐J, Weigel AL, Brody MS, Price CW (2006) The blue‐light receptor YtvA acts in the environmental stress signaling pathway of Bacillus subtilis . J Bacteriol 188: 6387–6395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Bernardo J, Dunlop MJ (2013) Tunable stochastic pulsing in the Escherichia coli multiple antibiotic resistance network from interlinked positive and negative feedback loops. PLoS Comput Biol 9: e1003229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Sureka K, Ghosh B, Bose I, Basu J, Kundu M (2011) Phenotypic heterogeneity in mycobacterial stringent response. BMC Syst Biol 5: 18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie DT (1977) Exact stochastic simulation of coupled chemical reactions. J Phys Chem 81: 2340–2361 [Google Scholar]
- Gillespie DT (2000) The chemical Langevin equation. J Chem Phys 113: 297 [Google Scholar]
- Gruenberger A, Probst C, Heyer A, Wiechert W, Frunzke J, Kohlheyer D (2013) Microfluidic picoliter bioreactor for microbial single‐cell analysis: fabrication, system setup, and operation. J Vis Exp 50560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guariglia‐Oropeza V, Helmann JD (2011) Bacillus subtilis σV confers lysozyme resistance by activation of two cell wall modification pathways, peptidoglycan O‐acetylation and D‐alanylation of teichoic acids. J Bacteriol 193: 6223–6232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Seki T, Matsuoka S, Hara H, Asai K, Sadaie Y, Matsumoto K (2013) Induction of extracytoplasmic function sigma factors in Bacillus subtilis cells with defects in lipoteichoic acid synthesis. Microbiology (Reading, Engl) 159: 23–35 [DOI] [PubMed] [Google Scholar]
- Hastie JL, Williams KB, Ellermeier CD (2013) The activity of σV, an extracytoplasmic function σ factor of Bacillus subtilis, is controlled by regulated proteolysis of the anti‐σ factor RsiV. J Bacteriol 195: 3135–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie JL, Williams KB, Sepúlveda C, Houtman JC, Forest KT, Ellermeier CD (2014) Evidence of a bacterial receptor for lysozyme: binding of lysozyme to the anti‐σ factor RsiV controls activation of the ecf σ factor σV. PLoS Genet 10: e1004643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie JL, Ellermeier CD (2016) Proteolytic activation of extra cytoplasmic function (ECF) σ factors, In Stress and Environmental Regulation of Gene Expression and Adaptation in bacteria de, Bruijn FJ (ed.), pp 344–351. Hoboken, NJ: John Wiley, Sons Inc. [Google Scholar]
- Hastie JL, Williams KB, Bohr LL, Houtman JC, Gakhar L, Ellermeier CD (2016) The Anti‐sigma factor RsiV Is a bacterial receptor for lysozyme: co‐crystal structure determination and demonstration that binding of lysozyme to RsiV is required for σV activation. PLoS Genet 12: e1006287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilker M, Schwachtje J, Baier M, Balazadeh S, Bäurle I, Geiselhardt S, Hincha DK, Kunze R, Mueller‐Roeber B, Rillig MC et al (2016) Priming and memory of stress responses in organisms lacking a nervous system. Biol Rev Camb Philos Soc 91: 1118–1133 [DOI] [PubMed] [Google Scholar]
- Ho TD, Hastie JL, Intile PJ, Ellermeier CD (2011) The Bacillus subtilis extracytoplasmic function σ factor σV is induced by lysozyme and provides resistance to lysozyme. J Bacteriol 193: 6215–6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TD, Ellermeier CD (2012) Extra cytoplasmic function σ factor activation. Curr Opin Microbiol 15: 182–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TD, Ellermeier CD (2019) Activation of the extracytoplasmic function σ factor σV by lysozyme. Mol Microbiol 112: 410–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jishage M, Ishihama A (1995) Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of sigma 70 and sigma 38. J Bacteriol 177: 6832–6835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jishage M, Iwata A, Ueda S, Ishihama A (1996) Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J Bacteriol 178: 5447–5451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong IG , Veening J‐W, Kuipers OP (2012) Single cell analysis of gene expression patterns during carbon starvation in Bacillus subtilis reveals large phenotypic variation. Environ Microbiol 14: 3110–3121 [DOI] [PubMed] [Google Scholar]
- Kelley R, Ideker T (2009) Genome‐wide fitness and expression profiling implicate Mga2 in adaptation to hydrogen peroxide. PLoS Genet 5: e1000488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal M, Caplan M (2011) Regulated intramembrane proteolysis: signaling pathways and biological functions. Physiology (Bethesda) 26: 34–44 [DOI] [PubMed] [Google Scholar]
- Lambert G, Kussell E (2014) Memory and fitness optimization of bacteria under fluctuating environments. PLoS Genet 10: e1004556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewerke LT, Kies PJ, Müh U, Ellermeier CD (2018) Bacterial sensing: a putative amphipathic helix in RsiV is the switch for activating σV in response to lysozyme. PLoS Genet 14: e1007527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby EA, Reuveni S, Dworkin J (2019) Multisite phosphorylation drives phenotypic variation in (p)ppGpp synthetase‐dependent antibiotic tolerance. Nat Commun 10: 5133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JCW, Young JW, Fontes M, Hernández Jiménez MJ, Elowitz MB (2011) Stochastic pulse regulation in bacterial stress response. Science 334: 366–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol 15: 550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins BMC, Locke JCW (2015) Microbial individuality: how single‐cell heterogeneity enables population level strategies. Curr Opin Microbiol 24: 104–112 [DOI] [PubMed] [Google Scholar]
- Martins BMC, Tooke AK, Thomas P, Locke JCW (2018) Cell size control driven by the circadian clock and environment in cyanobacteria. Proc Natl Acad Sci USA 115: E11415–E11424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megerle JA, Fritz G, Gerland U, Jung K, Rädler JO (2008) Timing and dynamics of single cell gene expression in the arabinose utilization system. Biophys J 95: 2103–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitosch K, Rieckh G, Bollenbach T (2017) Noisy response to antibiotic stress predicts subsequent single‐cell survival in an acidic environment. Cell Syst. 4: 393–403.e5 [DOI] [PubMed] [Google Scholar]
- Mitosch K, Rieckh G, Bollenbach T (2019) Temporal order and precision of complex stress responses in individual bacteria. Mol Syst Biol 15: e8470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munsky B, Neuert G, van Oudenaarden A (2012) Using gene expression noise to understand gene regulation. Science 336: 183–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narula J, Tiwari A, Igoshin OA (2016) Role of autoregulation and relative synthesis of operon partners in alternative sigma factor networks. PLoS Comput Biol 12: e1005267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman TM, Lord ND, Paulsson J, Losick R (2013) Memory and modularity in cell‐fate decision making. Nature 503: 481–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick A, Weiner M (1957) Enzyme induction as an all‐or‐none phenomenon. Proc Natl Acad Sci USA 43: 553–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyström T (2004) Growth versus maintenance: a trade‐off dictated by RNA polymerase availability and sigma factor competition? Mol Microbiol 54: 855–862 [DOI] [PubMed] [Google Scholar]
- Park J, Dies M, Lin Y, Hormoz S, Smith‐Unna SE, Quinodoz S, Hernández‐Jiménez MJ, Garcia‐Ojalvo J, Locke JCW, Elowitz MB (2018) Molecular time sharing through dynamic pulsing in single cells. Cell Syst 6: 216–229.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patange O, Schwall C, Jones M, Villava C, Griffith DA, Phillips A, Locke JCW (2018) Escherichia coli can survive stress by noisy growth modulation. Nat Commun 9: 5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C (2017) Salmon provides fast and bias‐aware quantification of transcript expression. Nat Methods 14: 417–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Vecchione S, Wu H, Mauri M, Mascher T, Fritz G (2018) Engineering orthogonal synthetic timer circuits based on extracytoplasmic function σ factors. Nucleic Acids Res 46: 7450–7464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackauckas C, Nie Q (2017a) Adaptive methods for stochastic differential equations via natural embeddings and rejection sampling with memory. Discrete Continuous Dyn Syst Ser B 22: 2731–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackauckas C, Nie Q (2017b) DifferentialEquations.jl – a performant and feature‐rich ecosystem for solving differential equations in Julia. J Open Res Softw 5: 15 [Google Scholar]
- Raj A, van Oudenaarden A (2008) Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135: 216–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodius VA, Segall‐Shapiro TH, Sharon BD, Ghodasara A, Orlova E, Tabakh H, Burkhardt DH, Clancy K, Peterson TC, Gross CA et al (2013) Design of orthogonal genetic switches based on a crosstalk map of σs, anti‐σs, and promoters. Mol Syst Biol 9: 702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem E, Loinger A, Ronin I, Levin‐Reisman I, Gabay C, Shoresh N, Biham O, Balaban NQ (2010) Regulation of phenotypic variability by a threshold‐based mechanism underlies bacterial persistence. Proc Natl Acad Sci USA 107: 12541–12546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runde S, Molière N, Heinz A, Maisonneuve E, Janczikowski A, Elsholz AKW, Gerth U, Hecker M, Turgay K (2014) The role of thiol oxidative stress response in heat‐induced protein aggregate formation during thermotolerance in Bacillus subtilis . Mol Microbiol 91: 1036–1052 [DOI] [PubMed] [Google Scholar]
- Soneson C, Love MI, Robinson MD (2015) Differential analyses for RNA‐seq: transcript‐level estimates improve gene‐level inferences. F1000Research 4: 152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spizizen J (1958) Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc Natl Acad Sci USA 44: 1072–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureka K, Ghosh B, Dasgupta A, Basu J, Kundu M, Bose I (2008) Positive feedback and noise activate the stringent response regulator rel in mycobacteria. PLoS One 3: e1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri‐Araghi S, Bradde S, Sauls JT, Hill NS, Levin PA, Paulsson J, Vergassola M, Jun S (2015) Cell‐size control and homeostasis in bacteria. Curr Biol 25: 385–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari A, Ray JCJ, Narula J, Igoshin OA (2011) Bistable responses in bacterial genetic networks: designs and dynamical consequences. Math Biosci 231: 76–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjalsma H, Bolhuis A, van Roosmalen ML , Wiegert T, Schumann W, Broekhuizen CP, Quax WJ, Venema G, Bron S, van Dijl JM (1998) Functional analysis of the secretory precursor processing machinery of Bacillus subtilis: identification of a eubacterial homolog of archaeal and eukaryotic signal peptidases. Genes Dev 12: 2318–2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening J‐W, Smits WK, Kuipers OP (2008a) Bistability, epigenetics, and bet‐hedging in bacteria. Annu Rev Microbiol 62: 193–210 [DOI] [PubMed] [Google Scholar]
- Veening J‐W, Stewart EJ, Berngruber TW, Taddei F, Kuipers OP, Hamoen LW (2008b) Bet‐hedging and epigenetic inheritance in bacterial cell development. Proc Natl Acad Sci USA 105: 4393–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Robert L, Pelletier J, Dang WL, Taddei F, Wright A, Jun S (2010) Robust growth of Escherichia coli . Curr Biol 20: 1099–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W, Ferrell JE (2003) A positive‐feedback‐based bistable “memory module” that governs a cell fate decision. Nature 426: 460–465 [DOI] [PubMed] [Google Scholar]
- Young JW, Locke JCW, Altinok A, Rosenfeld N, Bacarian T, Swain PS, Mjolsness E, Elowitz MB (2011) Measuring single‐cell gene expression dynamics in bacteria using fluorescence time‐lapse microscopy. Nat Protoc 7: 80–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JW, Locke JCW, Elowitz MB (2013) Rate of environmental change determines stress response specificity. Proc Natl Acad Sci USA 110: 4140–4145 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Movie EV1
Movie EV2
Movie EV3
Review Process File
Data Availability Statement
The RNA‐seq data produced in this study can be found here:
Gene Expression Omnibus GSE171761: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE171761.
Model simulation, data analysis and figure plotting code, as well as source data for main and extended view figures can be found here: https://gitlab.com/slcu/teamJL/schwall_etal_msb_2021.


